- Key Laboratory of Rubber Biology, Ministry of Agriculture, Rubber Research Institute, Chinese Academy of Tropical Agricultural Science, Danzhou, China
Rubber trees are economically important tropical tree species and the major source of natural rubber, which is an essential industrial material. This tropical perennial tree is susceptible to cold stress and other abiotic stresses, especially in the marginal northern tropics. Recent years, the genome sequencing and RNA-seq projects produced huge amount of sequence data, which greatly facilitated the functional genomics study. However, the characterization of individual functional gene is in urgent demands, especially for those involved in stress resistance. Here we identified and characterized the rubber tree gene ErbB-3 binding protein 1, which undergoes changes in expression in response to cold, drought stress and ABA treatment. HbEBP1 overexpression (OE) in Arabidopsis increased organ size, facilitated root growth and increased adult leaf number by delaying the vegetative-to-reproductive transition. In addition, HbEBP1 OE enhanced the resistance of the Arabidopsis plants to freezing and drought stress, demonstrating that this gene participates in the regulation of abiotic stress resistance. RD29a, RD22 and CYCD3;1 expression was also greatly enhanced by HbEBP1 OE, which explains its regulatory roles in organ size and stress resistance. The regulation of drought stress resistance is a novel function identified in plant EBP1 genes, which expands our understanding of the roles of EBP1 gene in response to the environment. Our results provide information that may lead to the use of HbEBP1 in genetically engineered crops to increase both biomass and abiotic stress resistance.
Introduction
ErbB-3 Binding Protein 1 is a member of the proliferation-associated 2G4 protein (PA2G4) family (Lamartine et al., 1997; Lessor et al., 2000; Xia et al., 2001; Squatrito et al., 2004). This recently identified transcription factor is involved in multiple pathways such as the cell cycle, protein translation, rRNA synthesis and cell proliferation (Squatrito et al., 2006; Sun et al., 2012; Figeac et al., 2014; Liu et al., 2014), although detailed mechanisms for each of these activities have yet to be determined. In humans, HsEBP1 binds dsRNA to form part of the ribonucleoproein (RNP) complexes via association with different rRNA species (Squatrito et al., 2004). EBP1 also associates with mature ribosomes and suppresses the phosphorylation of the eukaryotic initiation factor 2 alpha under stress condition, and thus is possibly involved in the control of protein translation (Squatrito et al., 2006). In T cells, EBP1 is required for the regulation of ribosomal RNA synthesis (Nguyen et al., 2015). EBP1 also functions as a nuclear cell survival factor that interacts with Akt and PKC to inhibit apoptosis (Ahn et al., 2006).
EBP1 encodes two isoforms, p48 and p42; p48 enhances cell growth, whereas p42 stimulates cell proliferation (Liu et al., 2006). Ectopic expression of EBP1, however, inhibits proliferation and induces differentiation in the breast carcinoma cell lines AU565 (Lessor et al., 2000). EBP1 is also up-regulated in the sciatic nerve after crushing of the nerve, where it is believed to be involved in the differentiation and migration of Schwann cells (Liu et al., 2014). Recently, EBP1 was reported to bind Anxa2 and negatively regulate the proliferation and invasion of breast cancer cells (Zhang et al., 2015). Knockdown of EBP1 inhibits both the proliferation and differentiation of resident stem cells of skeletal muscle, which cannot be rescued by ErbB3 over-expression, suggesting that EBP1 controls the proliferation and differentiation of muscle stem cells (Figeac et al., 2014).
EBP1 is also functionally conserved in plants. The expression of plant EBP1 is tightly regulated and is remarkably correlated with organ growth in a dose-dependent manner (Horváth et al., 2006). Plant EBP1 is also required for the expression of CyclinD3;1, ribonucleotide reductase 2 and the cyclin-dependent kinase B1;1 genes and thus is involved in cell cycle regulation, as shown in Solanum tuberosum and Arabidopsis thaliana (Horváth et al., 2006). EBP1 from Ammopiptanthus mongolicus is up-regulated by cold, and it’s OE in Escherichia coli and Arabidopsis confers cold and freezing tolerance by accelerating ribosome biogenesis and the translation of transcriptional factors and downstream functional proteins that are induced under cold stress (Cao et al., 2008). In maize, ZmEBP1 gene has an overdominant expression pattern in the immature ears of hybrids that exhibit heterosis (Wang et al., 2016). The OE of ZmEBP1 in Arabidopsis increases organ size by accelerating cell proliferation. Thus the EBP1 genes have conserved functions across eukaryotes.
Rubber trees (Hevea brasiliensis) are tropical perennial trees that are susceptible to cold stress (Cheng et al., 2015). In China and other regions in the northern edge of the tropics, rubber tree plantations suffer from cold stress in the winter. Although some cold-resistant clones have been bred in China, there is an urgent demand for additional clones with stronger cold resistance. The development of transgenic technique in rubber trees may help to meet this demand more quickly (Montoro et al., 2003). Although the high-quality rubber tree genome was recently released (Tang et al., 2016), the functional genes that related to stress resistance are still to be identified individually (Cheng et al., 2015). Previously, we constructed a cold- induced full-length cDNA library (Cheng et al., 2008), in which HbEBP1 was highly abundant. Further characterization revealed that HbEBP1 responded to cold and drought stress and ABA treatment. Here we characterized HbEBP1 and overexpressed it in Arabidopsis, which showed that this gene regulates organ size and resistance to drought and cold stresses in rubber trees.
Materials and Methods
Plant Material, Preparation of Transgenic Plants
The 1-year-old rubber tree clone 93–114 was maintained as described (Cheng et al., 2015). Arabidopsis thaliana ecotype Columbia-0 (Col-0) was used as wild-type in this study. Seeds were surface sterilized and sowed in pots (10 cm × 10 cm × 10 cm) containing a mixture of soil and vermiculite (3:1 v/v). After 4 days imbibitions at 4°C, the plates and pots were transferred to a growth chamber at a constant temperature of 20°C under 16 h light (125 μmol m-2 s-1)/8 h dark cycles.
To generate the Arabidopsis plants that overexpress HbEBP1 (HbEBP1 OE), the coding region of HbEBP1 was amplified using primers 5′-TCAAAGCTGTAAGCTTATGTCGG-3′ and 5′-CATAAGAATTCCATACAAGGT-3′. The coding sequence fragment was then subcloned between the HindIII and EcoRI sites in pXCS-HAStrep plasmid. Arabidopsis ecotype Col-0 plants were transformed according to the floral dip method (Clough and Bent, 1998) using Agrobacterium tumefaciens strain GV3101(pMP90RK).
Manipulation of DNA, RNA Isolation, and Expression Analysis
Isolation of rubber tree DNA and RNA was carried out as described (Zewei An, 2012). For northern blotting, a HbEBP1 probe was prepared from full-length cDNA with DIG Northern Starter Kit (Roche, USA). Twenty micrograms of total RNA was run on a denature 1.5% agarose gel and the blot was then transferred onto an Amersham HybondTM-N+ nylon membrane (GE Healthcare, USA). Hybridization and detection were carried out according to the manufacturer’s instructions.
Total RNA was extracted from Arabidopsis seedlings using Qiagen RNeasy Plant Mini Kit (Qiagen, USA). For quantitative RT-PCR (Q-PCR) analysis, the RNA samples were treated with DNase I to remove possible DNA contamination. Two micrograms of total RNA was used for each detection. The first-strand cDNA was synthesized with SuperScript III according to the manufacturer’s instructions (Invitrogen, USA). Q-PCR was carried out using IQ SYBR Green Supermix (Bio-Rad, USA) with gene-specific primers as listed in Supplemental Table S1. PCR was performed on a Bio-Rad Iq5 RT PCR instrument using the following program: 95°C for 3 min, followed by 40 cycles of 95°C for 15 s, 60°C for 30 s and 72°C for 30 s.
Bioinformatic and Phylogenetic Analysis
Multiple sequence alignments were performed using ClustalW2 at the EBI ClustalW server1 using the default parameters (Larkin et al., 2007). The coding sequence was predicted using Bioedit software and confirmed by using BLASTP program at NCBI BLAST server2. The protein secondary domains were predicted with Interproscan program (Jones et al., 2014) at EBI server3. MEGA software (version 6.06) was used for phylogenetic analysis (Tamura et al., 2013). A phylogenetic tree was constructed using the neighbor-joining method with bootstrap test. The default parameters were used for the construction.
Southern Blotting
Southern blotting analysis was carried out as described (Cheng et al., 2015). Briefly, 20 μg Arabidopsis genomic DNA was digested with EcoRI. The DNA was resolved on a 0.8% agarose gel and transferred onto an Amersham HybondTM-N+ nylon membrane (GE Healthcare, USA); the blot was cross-linked under 0.12 J/cm2 UV irradiation. Then the blot was hybridized at 42°C with Digoxigenin-labeled probe for 12 h. After two stringent washes at 68°C, the signal was detected using DIG Nucleic Acid Detection Kit (Roche, USA).
Cold and Drought Stress and ABA Treatment
Cold and drought stress and ABA treatment in rubber trees were carried out as described (Cheng et al., 2015). To measure cold resistance in Arabidopsis, an electrolyte leakage analysis was performed as described (Cheng et al., 2013). Briefly, electrolyte leakage was measured using leaves from 2–week-old seedlings that had been frozen to -7°C at a cooling rate of 1°C/h from -1°C with an A28F Thermo Fisher temperature-controlled water bath (Thermo Scientific, USA).
To calculate dehydration rate, leaves were detached from 2-week-old seedlings, weighed and placed on dry filter paper in a drying chamber. The samples were weighed at 30-min intervals and the dehydration rate was calculated as the ratio of the remaining weight to the initial weight at each time point.
The drought tolerance test was carried out by sowing seeds in pots containing vermiculite. Irrigation was performed daily by adding half Hoagland nutrient solution in the tray until all the liquid was absorbed. When the seedlings were 2 weeks old, irrigation was withheld for 1 week. At least three pots of seedlings were observed for each genotype in this test.
For cold treatment in rubber tree, the seedlings were transferred to a 4°C cold culture room. Drought was carried out by detaching the leaf from the seedlings and keeping on the filter paper at room temperature. For ABA treatment, 100 μmol/L ABA solution was sprayed onto the seedlings until the liquid dropped from the leaf. Then the seedlings were covered with a plastic bag. The leaf sample was collected at indicated time points and stored in liquid nitrogen immediately.
Developmental Phenotypic Analyses
The leaf development phases were determined by observing abaxial trichome production as described (Willmann and Poethig, 2011). The emergence of rosette leaves was recorded every day from day 7 after planting based on the criteria that they could be recognized with the naked eyes. Flowering time was recorded as the day on which flower primordia were visible without the aid of a microscope. Leaf length and blade length and width were measured as described (Willmann and Poethig, 2011). For leaf area measurements, the leaves were carefully detached from the seedlings and pasted on a sheet of A4 paper and then were scanned. The images were then analyzed with Image J software (version 1.45s) to calculate leaf area (Schneider et al., 2012).
For root length measurement, seeds were surface sterilized and sown onto the surface of a square plate containing half MS agar medium, and were then cultured vertically. Photos were taken daily, and root length on each day was measured with a ruler. At least 20 individual seedlings were analyzed from each line in these studies.
For measurement of the leaf epidermal cells, ten Arabidopsis plants were selected randomly. The area of the fully expanded seventh leaf was measured at about 25 days after germination in each transgenic line and Col-0 plants. The detail measurement was performed according to the method reported (Wang et al., 2016). Total cell number per leaf was calculated by dividing the leaf area by the average cell size.
Statistical Analysis
All results are presented as the mean ± standard error of the mean (SEM) from five biological replicates for the stress experiments or from the number of replicates indicated. Statistical analysis was performed by using the Student’s t-test, and p < 0.05 was recognized as significant.
Results
Characterization of HbEBP1
We previously identified a cDNA clone that putatively encodes a proliferation-associated 2G4 family member from the cold induced full-length cDNA library (Cheng et al., 2008). This cDNA fragment is 1462 bp in length, and contains an 1188 bp coding region (Figure 1A). The predicted peptide is highly similar to EBP1 proteins in other plant species and was therefore designated as HbEBP1. The deduced HbEBP1 amino acid sequence contains a proliferation-associated PA2G4-like domain, an aminopeptidase APP_MetAP domain and a methionine aminopeptidase (MAP) domain (Supplementary Figure S1). The HbEBP1 gene sequence was then deposited into the GenBank (Accession No: KX661028). A phenogram generated by MEGA analysis demonstrated that EBP1 proteins are highly conserved in eukaryotic species and that EBP1s from plant and animal species are clustered respectively. Rubber tree EBP1 is highly similar to EBP1 from Ricinus communis (accession no. XP_002530504.1) and Jatropha curcas (accession no. XP_012089820.1), which also belong to the Euphorbiaceae family (Figure 1B).
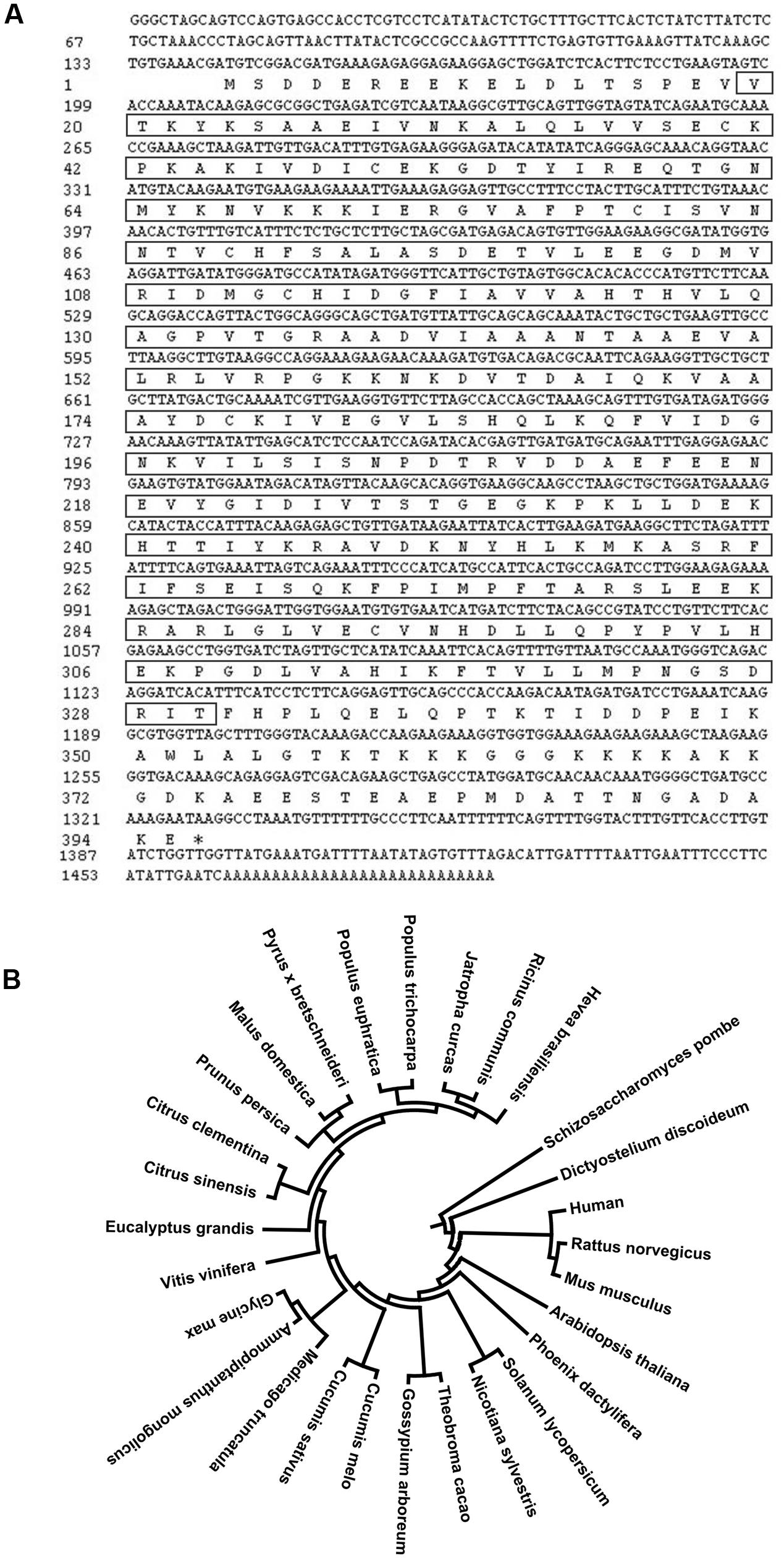
FIGURE 1. (A) Full-length cDNA sequence of rubber tree ErbB-3 binding protein 1 (HbEBP1) and its deduced encoding peptide. The sequence in the square frame corresponds to the deduced MAP domain. (B) Phylogenetic tree showing similarity of the deduced H. brasiliensis EBP1 protein sequence with EBP1s from other plant species. EBP1 peptide sequences of each species were downloaded from GenBank based on the following accession numbers: Ricinus communis, XP_002530504.1; Jatropha curcas, XP_012089820.1; Populus trichocarpa, XP_002308228.2; Populus euphratica, XP_011046861.1; Pyrus × bretschneideri, XP_009376423.1; Malus domestica, XP_008362268.1; Prunus persica, XP_007205303.1; Citrus clementina, XP_006429434.1; Citrus sinensis, KDO56711.1; Eucalyptus grandis, XP_010037691.1; Vitis vinifera, XP_002262986.2; Glycine max, XP_003529167.1; Ammopiptanthus mongolicus, ABF66654.1; Medicago truncatula, XP_003623305.1; Cucumis melo, XP_008442393.1; Cucumis sativus, XP_004137704.1; Gossypium arboreum, KHG25370.1; Theobroma cacao, XP_007026646.1; Nicotiana sylvestris, XP_009789408.1; Solanum lycopersicum, XP_004242516.1; Phoenix dactylifera, XP_008805004.1; Arabidopsis thaliana, Q96327.1; Mus musculus, P50580.3; Rattus norvegicus, Q6AYD3.1; Homo sapiens, Q9UQ80.3; Dictyostelium discoideum, Q1ZXG4.1; Schizosaccharomyces pombe, Q09184.1. The phylogenetic tree was constructed using a MEGA 6.06 software.
HbEBP1 Expression Is Affected by Cold and Drought Stress and ABA Treatment
The gene expression profiles under abiotic stress and ABA treatment were analyzed using northern blotting. HbEBP1 expression was moderately expressed under ambient cultural conditions and this expression increased when rubber tree plants were subjected to cold, drought or ABA exposure. HbEBP1 transcripts increased notably after 4 h of treatment with cold stress and reached their highest level at 8 h, with some decrease after 24 h of treatment (Figure 2). A similar expression pattern was found when the seedlings were subjected to drought stress, i.e., induction occurred after 4 h of treatment and reached its highest level after 8 h (Figure 2). During ABA treatment, HbEBP1 expression showed some decrease during the first 4 h, followed by an increase after 8 h (Figure 2). EBP1 genes accumulated in response to auxin in Arabidopsis and tomato, and their expression correlated with genes involved in ribosome biogenesis and function (Zhang et al., 2005; Horváth et al., 2006). The responses toward abiotic stress and ABA suggest that HbEBP1 may be involved in the regulation of abiotic stress resistance in rubber trees.
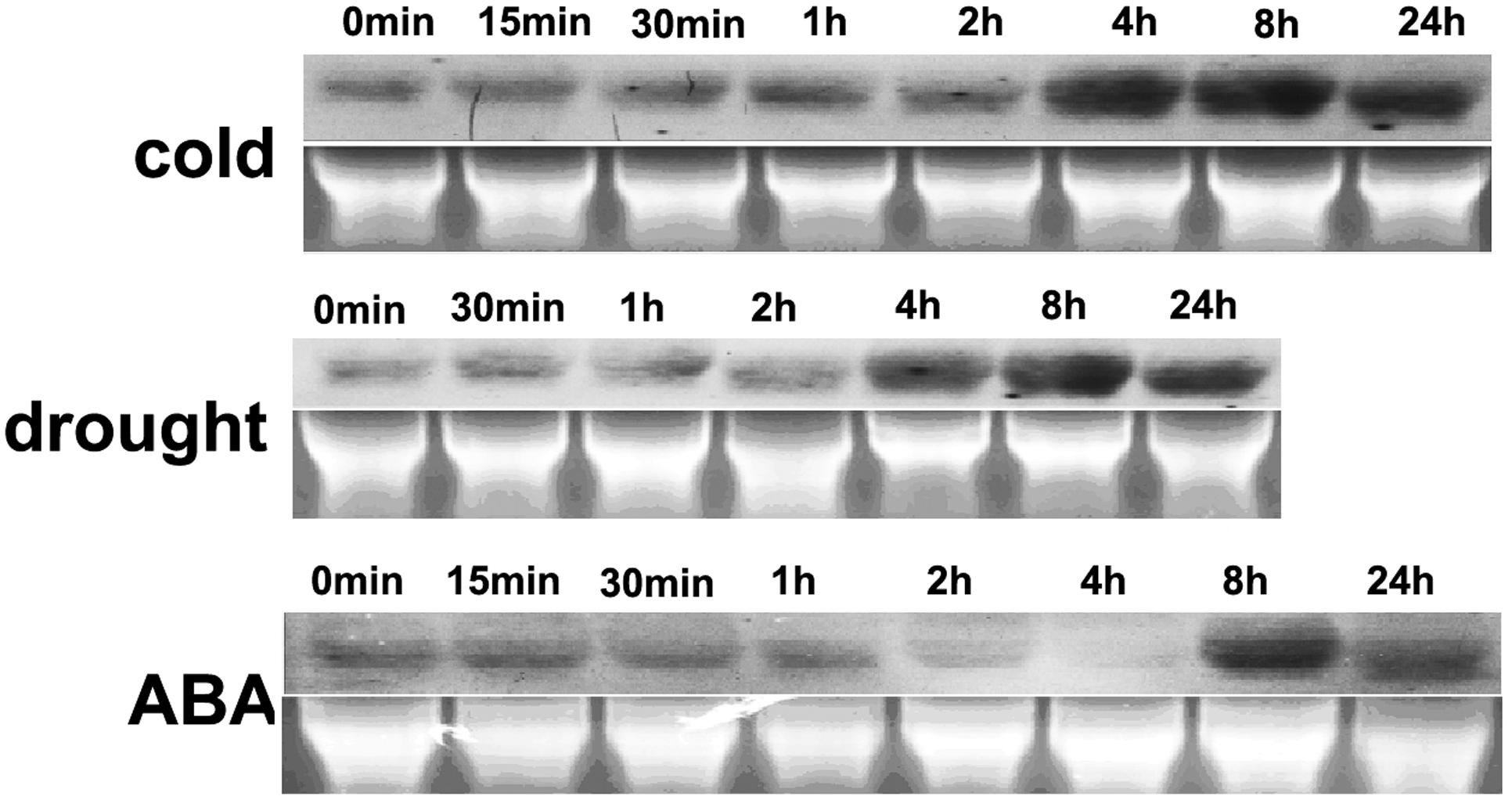
FIGURE 2. Changes in HbEBP1 expression in response to cold and drought stress and ABA treatment. Total RNA from the leaves of rubber tree seedlings treated with cold, drought or ABA for the indicated time was used for northern blotting With probes prepared from full-length HbEBP1 cDNA. Ethidium bromide–stained rRNA was used as an internal standard to monitor equal loading of total RNA.
Overexpression of HbEBP1 in Arabidopsis Leads to Enlarged Organ Size
The EBP1 genes regulate organ size by stimulating both cell proliferation and expansion via the regulation of RBR1 levels (Horváth et al., 2006). To validate these functions, HbEBP1 was overexpressed in Arabidopsis using the floral dip method. PCR amplification was used to confirm gene transfer. Among tens of HbEBP1 OE lines, we selected OE5, OE18 and OE28, each of which contained a single-copy T-DNA insertion and showed high and stable expression, for further study (Figure 3A). The T3 generation seedlings that harbored homozygous T-DNA insertions were used for further analysis. Northern blotting was used to detect HbEBP1 expression in the transgenic lines. As shown in Figure 3B, the OE5, OE18 and OE28 lines displayed comparable high expression. As expected, OE of HbEBP1 led to markedly enlarged organ size in Arabidopsis seedlings. Leaves from all three OE lines were notably larger as compared with leaves from the Col-0 seedlings (Figure 3C).
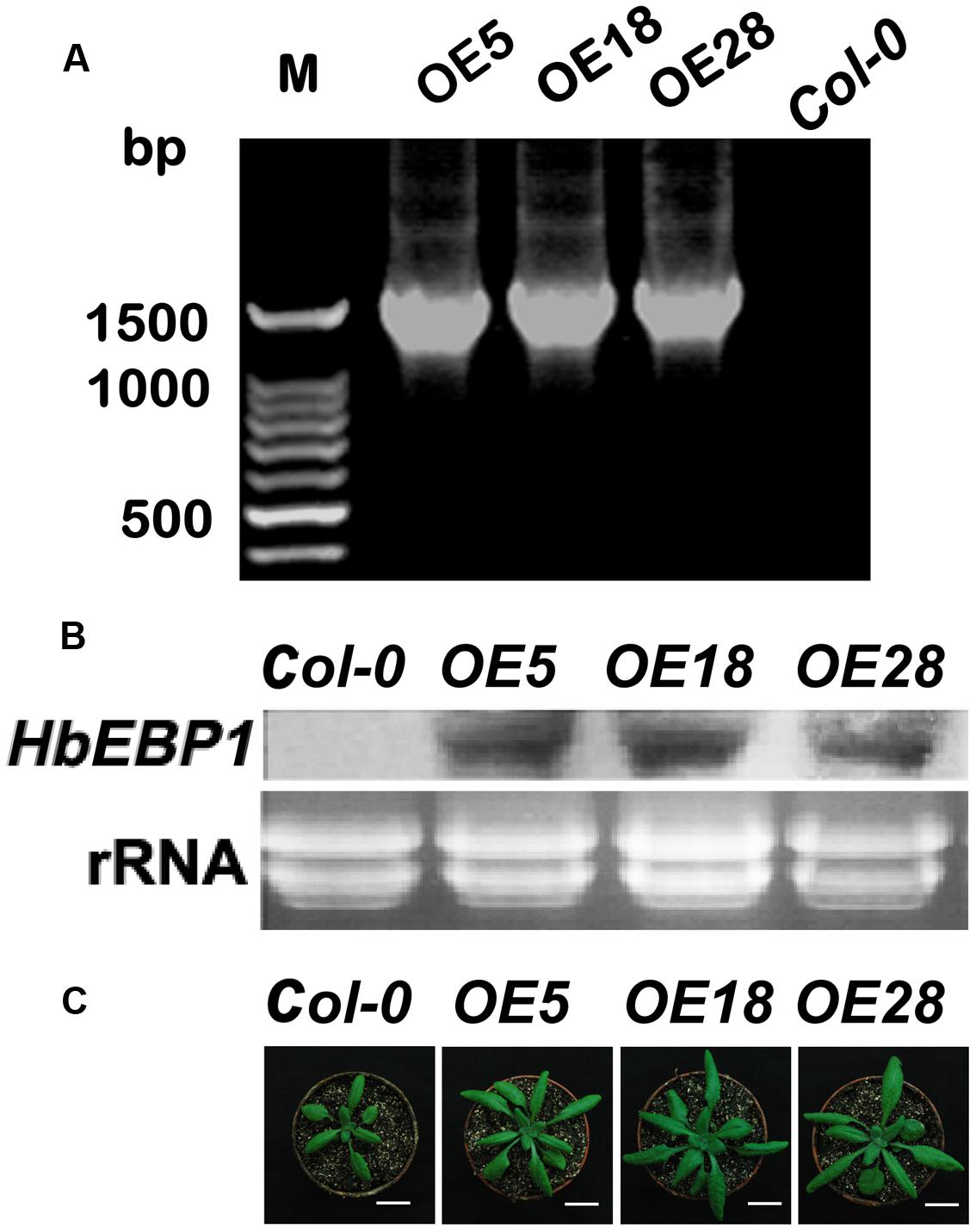
FIGURE 3. HbEBP1 overexpression in Arabidopsis. (A) PCR of HbEBP1 in transgenic plants. (B) HbEBP1 expression in the OE5, OE18 and OE28 lines. HbEBP1 expression was detected by northern blotting analysis using HbEBP1 probe. (C) Twenty-day-old seedlings from the Col-0, OE5, OE18 and OE28 lines showed enlarged rosette leaves. Bar = 2 cm.
The area of the first adult rosette leaf (normally the seventh leaf) was measured. As demonstrated in Figures 4A,B, the area of the seventh leaf was significantly larger in OE5, OE18 and OE28 lines, as compared with that in the Col-0 plants (p < 0.01). The wild-type plants had an average leaf area of 1.43 ± 0.25 cm2, whereas the average leaf area from OE5, OE18 and OE28 plant was 2.72 ± 0.36, 4.04 ± 0.44 and 3.12 ± 0.49 cm2 respectively. In addition, the HbEBP1 OE lines developed more rosette leaves than did the wild type Col-0 seedlings (Figure 4B). We further measured the cell size and calculated total cell numbers. The results showed that the OE leaves have more cell numbers than the Col-0, while the cell size is comparable (Supplementary Figure S4). These results demonstrated HbEBP1 OE promotes cell proliferation in transgenic lines.
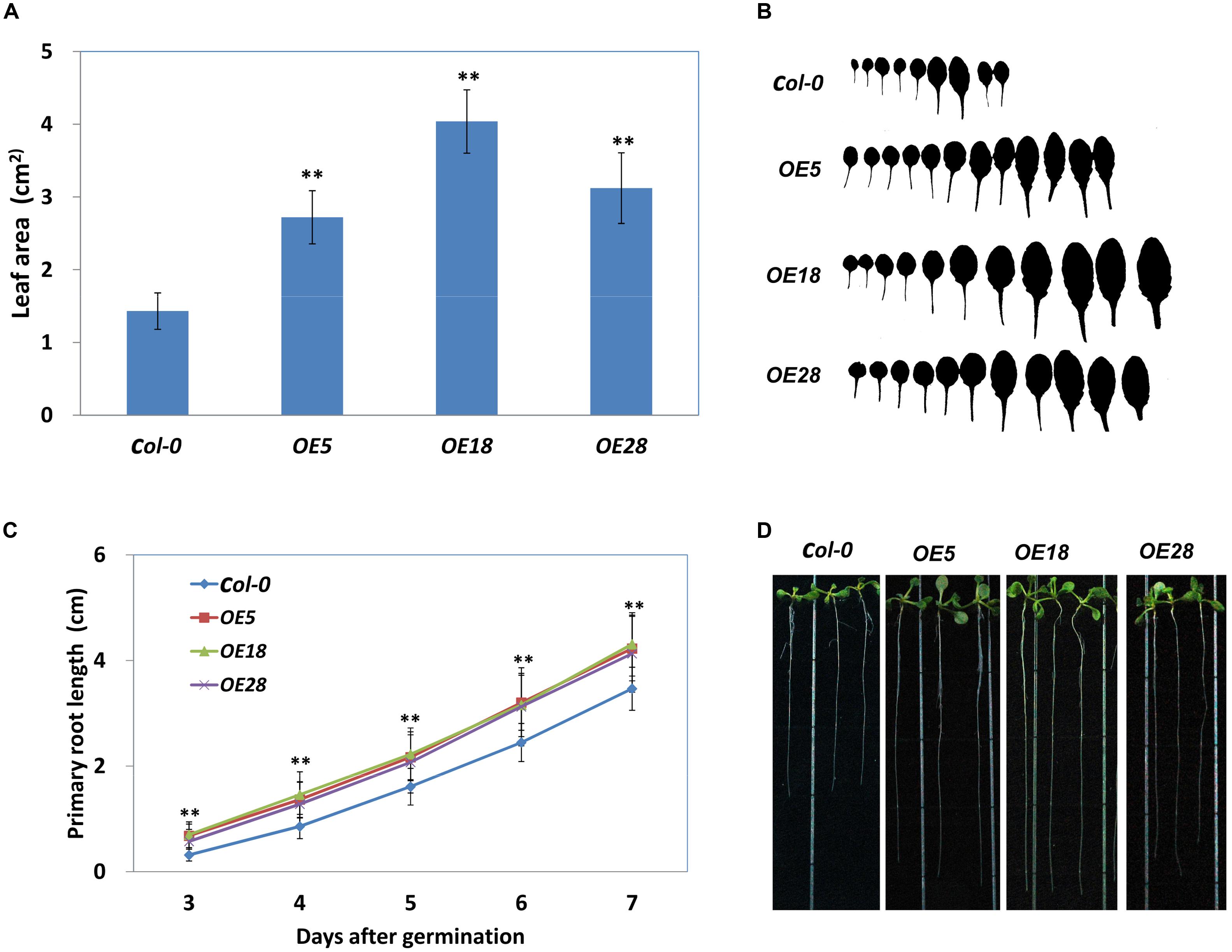
FIGURE 4. Enlarged organ size in Arabidopsis plants that overexpress HbEBP1. (A) Leaf area of the first adult-stage leaf (the seventh leaf) in the Col-0, OE5, OE18 and OE28 lines. Leaves from at least 20 individual seedlings were measured for each line. (B) The morphology of rosette leaves from the Col-0, OE5, OE18 and OE28 seedlings arranged from left to right in the order of initiation. (C) Primary root elongation was faster in the OE5, OE18 and OE28 lines. The seedlings were cultured vertically, and the root length was measured daily. Roots from at least 20 individual seedlings were measured for each line. (D) Representative seedlings show the primary root length at 8 days after germination. Data in (A,C) were presented as mean ± standard error of mean of at least 20 individual seedlings. Student’s t-test was carried out for statistical analysis. Significant difference was defined by ∗∗p < 0.01.
Similar results were observed in the roots. Primary root length was measured daily, and roots were found to grow faster in HbEBP1 transgenic seedlings. The OE5, OE18 and OE28 lines all displayed significantly faster root growth than did the Col-0 seedlings from 3 days after germination (p < 0.01) (Figure 4C). By 8 days after germination, the HbEBP1 OE lines had developed obviously longer primary roots than did the wild-type Col-0 seedlings (Figure 4D). At 2 weeks of age, the root systems were more complex in the HbEBP1 OE lines as compared with those in the Col-0 plants (Supplementary Figure S2). Thus HbEBP1 OE facilitated root development in Arabidopsis. The obviously larger leaves and longer primary roots in HbEBP1 OE plants suggested that HbEBP1 has a conserved function related to the regulation of organ size in H. brasiliensis.
Overexpression of HbEBP1 in Arabidopsis Leads to a Delayed Vegetative-to-Reproductive Transition and an Increased Adult Leaves
The HbEBP1 OE lines displayed a late flowering phenotype. Under long-day conditions, the flower meristem was visible at 22.2 ± 0.9 days after planting for the Col-0 plants. In contrast, for OE5, OE18 and OE28 lines, the flower meristem was visible at 25.8 ± 1.0, 24.8 ± 0.7 and 23.6 ± 1.1 days respectively. The flower meristems in the HbEBP1 OE lines were induced significantly later when compared with those from the wild-type plants (p < 0.05) (Figure 5A).
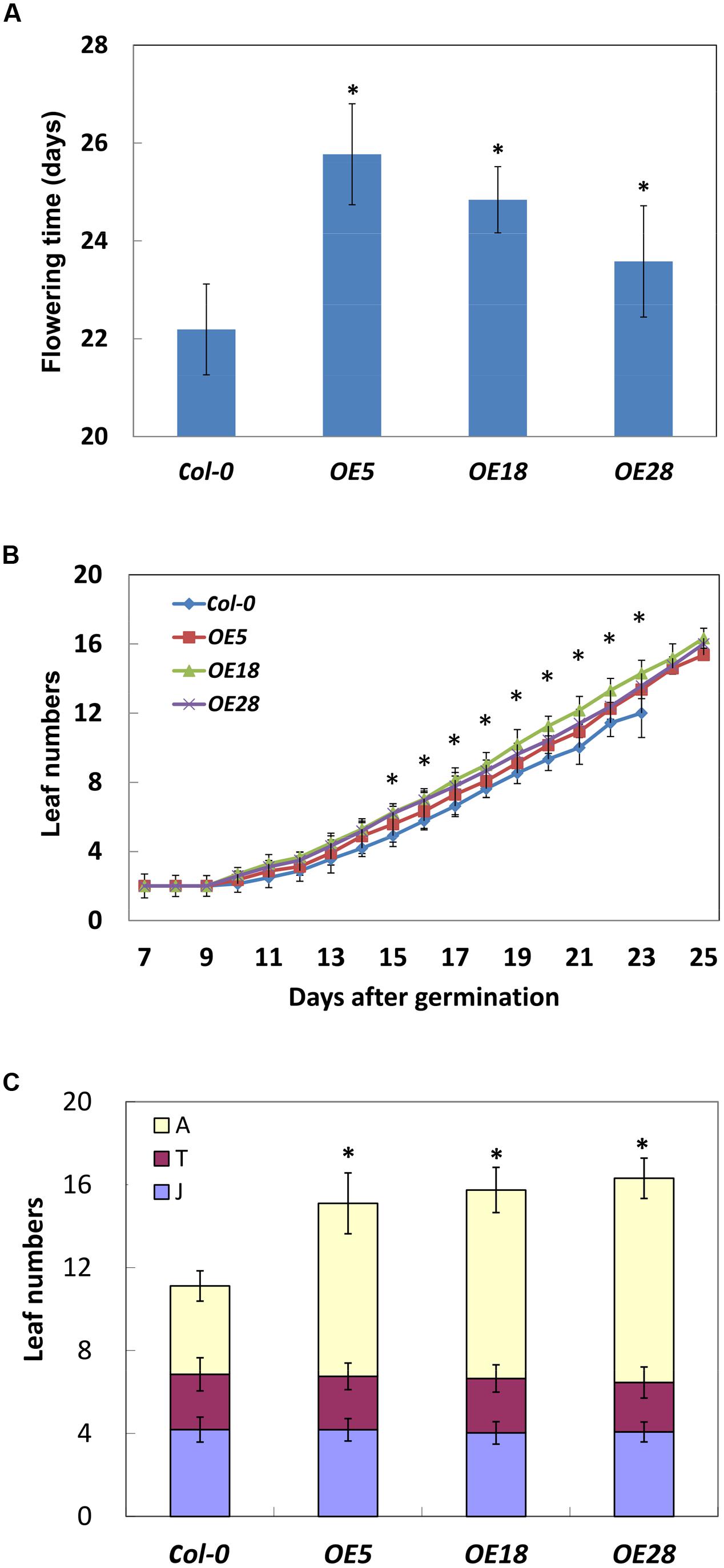
FIGURE 5. HbEBP1 overexpression delayed the vegetative-to-reproductive transition in Arabidopsis. (A) Days needed for the Col-0, OE5, OE18 and OE28 plants to produce visible flower meristems. (B) The leaf emergence rate is faster in the OE5, OE18 and OE28 lines than in Col-0 plants. Leaf numbers were counted daily with naked eyes. (C) The number of rosette leaves in the phases of juvenile (J), juvenile-to-adult transition (T) and adult (A) leaves in the Col-0, OE5, OE18 and OE28 lines. Data were presented as mean ± SEM from at least twenty individual seedlings. Student’s t-test was carried out for statistical analysis. Significant difference was defined by ∗p < 0.05.
Late flowering prolonged the vegetative growth period in HbEBP1 OE seedlings, which allowed the plants to grow more rosette leaves. An average of 15 or 16 leaves developed before floral induction for HbEBP1 OE lines (25 days), whereas the number for the Col-0 plants was 12 (22 days) (Figure 5B). We carefully counted rosette leaf number daily, and found that HbEBP1 OE plants developed rosette leaves faster than did the Col-0 plants. As shown in Figure 5B, all the HbEBP1 OE lines grew significantly more leaves than Col-0 from 15 days after germination (p < 0.01). Thus HbEBP1 OE promoted leaf growth in Arabidopsis.
Late flowering is a result of prolonged vegetative growth, in which the juvenile-to-adult and vegetative-to-reproductive phase transitions are delayed (Martínez-Zapater et al., 1995; Poethig, 2003). To examine if the phase transitions were delayed in HbEBP1 OE lines, leaf morphology was investigated at different phases. The leaf developmental stages were determined by observing the onset and the distribution of abaxial trichomes, leaf length and leaf shape (blade length/width ratio) (Telfer et al., 1997). The OE5, OE18 and OE28 lines had a comparable number of juvenile and juvenile-to-adult transition leaves relative to the Col-0 plants (Figure 5C). Both the HbEBP1 OE and wild-type plants developed about four juvenile and two juvenile-to-adult transition leaves. However, the number of leaves that developed during the adult phase was significantly higher for OE5, OE18 and OE28 plants (8.35 ± 1.47, 9.09 ± 1.09, and 9.85 ± 0.97, respectively; p < 0.01), whereas adult leaf number for the Col-0 plants was 4.26 ± 0.73 (Figure 5C). Presumably the increase in the number of adult leaves and their enlarged size (Figures 4A,B) prolonged vegetative growth in the HbEBP1 OE lines.
Overexpression of HbEBP1 in Arabidopsis Enhances Resistance to Abiotic Stress
HbEBP1 transcription was affected by cold and drought stress and ABA treatment, suggesting that this gene may be involved in abiotic resistance in plants. To assess its involvement in drought resistance, the dehydration rates were calculated for detached leaves. The OE18 and OE28 leaves dehydrated significantly slower than did the wild-type leaves from 2 to 8 h. For the OE5 leaves, the dehydration rate was significantly slower from 6 to 8 h (Figure 6A). Thus HbEBP1 OE plants might lose water more slowly than Col-0 plants under drought conditions.
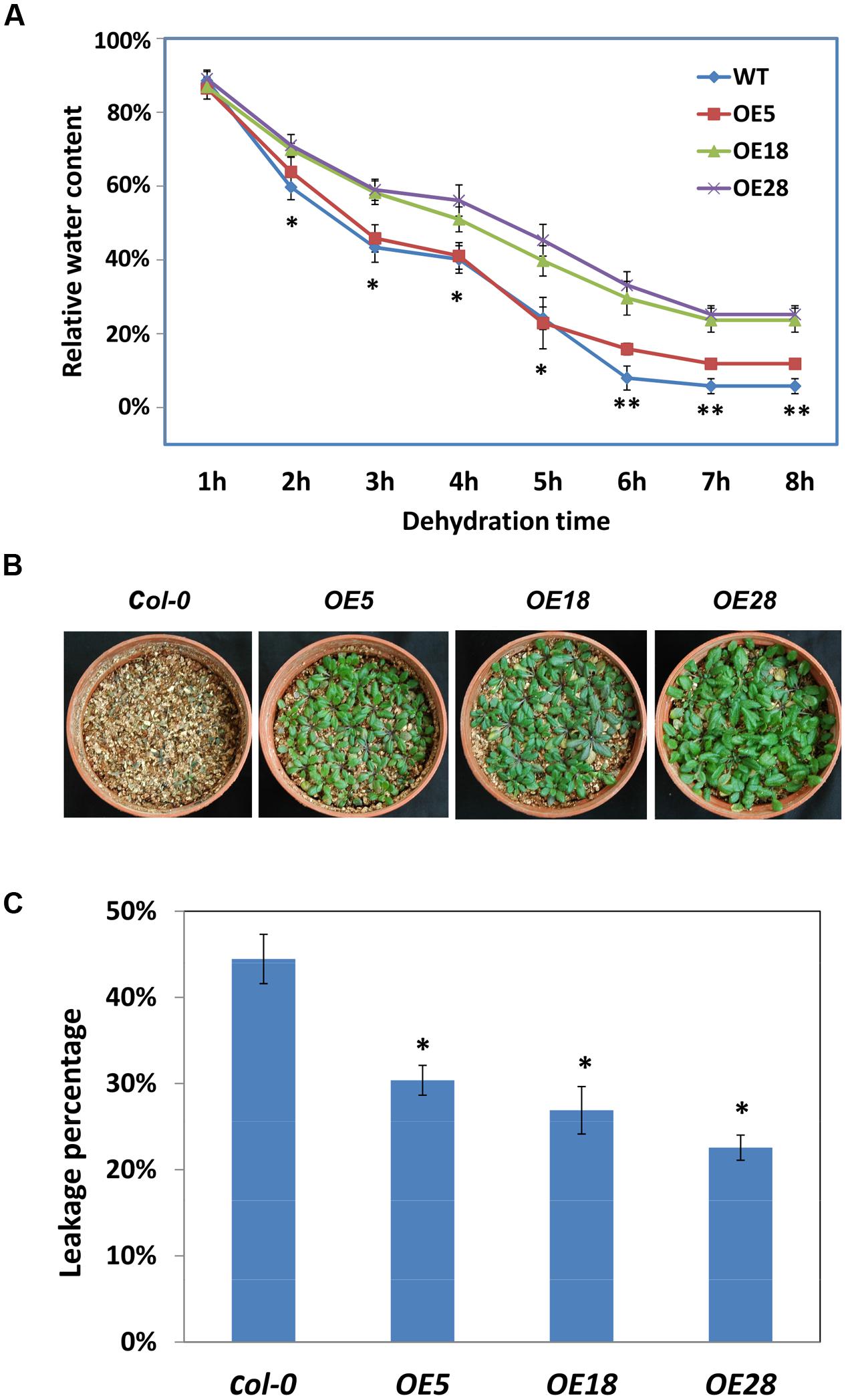
FIGURE 6. HbEBP1 overexpression conferred drought and cold resistance in Arabidopsis. (A) The dehydration rates of leaves from the Col-0, OE5, OE18 and OE28 seedlings. An asterisk (∗)indicates a significant difference between Col-0 and the individual OE18 and OE28 lines, whereas a double asterisk (∗∗) indicates a significant difference between Col-0 and each individual HbEBP1 OE line. (B) Representative seedlings of the Col-0, OE5, OE18 and OE28 lines after irrigation were withheld for 7 days. The seedlings were grown in a pot containing vermiculite, and the drought stress treatment was conducted with 2-week-old seedlings by withdrawing water for 1 week. The photos were taken 3 days after re-irrigation. (C) Electrolyte leakage of leaves from the Col-0, OE5, OE18 and OE28 lines. Electrolyte leakage was measured using leaves from 2-week-old seedlings frozen to -7°C at a cooling rate of 1°C/h from -1°C. An asterisk (∗) indicates a significant difference between each individual HbEBP1 OE line and the wild type Col-0 seedlings. Data in (A,C) are presented as the mean ± SEM from five biological replicates. Significance was determined by the Student’s t-test at the probability levels of p < 0.05. Comparisons were made between Col-0 and each individual OE line.
To evaluate drought tolerance in vivo, 2-week-old HbEBP1 OE and Col-0 seedlings were subjected to drought treatment by withdrawing water for 1 week. The HbEBP1 OE5, OE18 and OE28 lines showed a more resistant phenotype as compared with the wild-type plants (Figure 6B). After 7 days of withheld irrigation, all the Col-0 plants were wilted and the rosette leaves had become chlorotic, whereas the HbEBP1 OE lines had leaf blades that green and turgid, although their petioles were purple and displayed some aspects of a drought stressed phenotype (Figure 6B).
Similar results were obtained when HbEBP1 OE plants were subjected to cold stress. Electrolyte leakage analysis was used to evaluate cold resistance. Significantly less leakage was detected in the HbEBP1 OE plants when compared with the Col-0 plants (p < 0.05). In this analysis, the control plants had a leakage rate of 44.5%, whereas the rates were 30.4, 26.9, and 22.6% for OE5, OE18 and OE28 lines, respectively. HbEBP1 OE enhanced resistance to drought and cold stress in Arabidopsis, suggesting that this gene may function as a positive regulator of abiotic stress resistance in rubber trees.
HbEBP1 Overexpression Increased CYCD3, RD22 and RD29a Transcripts in Arabidopsis
EBP1 regulates organ size by promoting cell cycle transitions during leaf development (Horváth et al., 2006). The expression of the critical cell cycle regulator of the G1 to S and G2 to M transitions, CYCD3;1, was examined by Q-PCR (Dewitte et al., 2007; Blomme et al., 2014; Collins et al., 2015). Two-week-old seedlings were used for the analysis, and gene expression was quantified by comparing the fold changes in expression between the genes of interest and the reference ACTIN7. In the HbEBP1 OE lines, CYCD3;1 expression were significantly increased by 1.7- to 2.1- fold as compared with that in the Col-0 plants (Figure 7A). This effect may explain why HbEBP1 OE led to enlarged organ size in Arabidopsis.
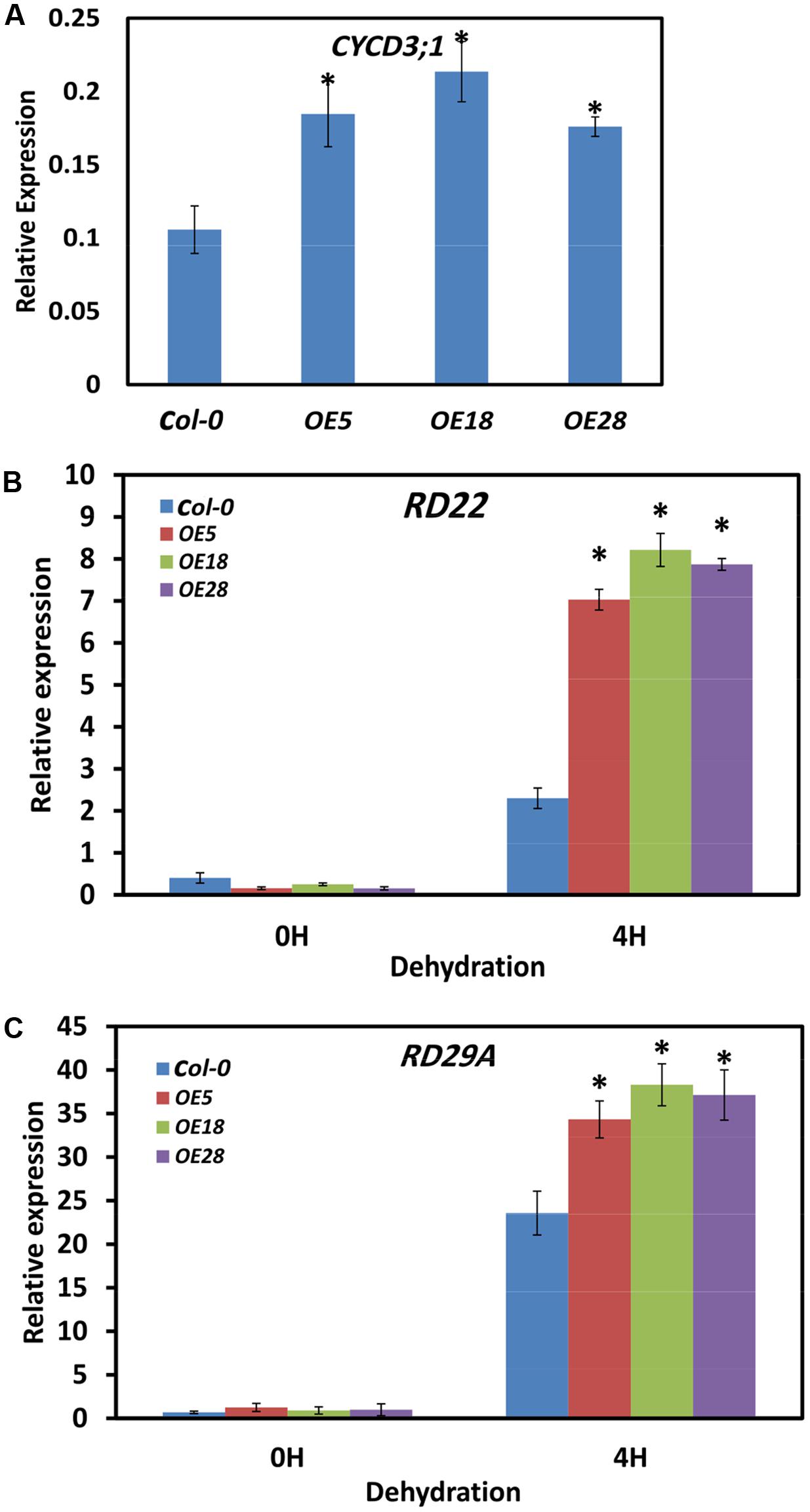
FIGURE 7. HbEBP1 overexpression promoted the expression of CYCD3;1, RD22 and RD29a in Arabidopsis. (A–C) Quantification of expression of (A) AtCYCD3;1, (B) AtRD22 and (C) AtRD29a by Q-PCR in the Col-0, OE5, OE18 and OE28 plants. For the dehydration treatment, the detached leaves were subjected to dehydration under the controlled condition for 4 h. Total RNA was extracted from 2-week-old seedling samples and Q-PCR was performed with gene-specific primers as listed in Supplementary Table S1. The transcript levels were normalized to that of ACTIN7. Two micrograms of total RNA was used for each sample analysis, and the data are presented as mean ± SEM from three biological replicates. An asterisk (∗) indicates a significant difference (p < 0.05) between the HbEBP1 OE lines and Col-0 in (A), or between the HbEBP1 OE lines and Col-0 after 4 h of dehydration treatment in (B,C). Significance was determined by Student’s t-test at the probability levels of p < 0.05.
To unravel the mechanism by which HbEBP1 OE enhances drought resistance in Arabidopsis, expression of the drought-responsive genes RD22 and RD29a was examined (Yamaguchi-Shinozaki and Shinozaki, 1993b, 1994; Msanne et al., 2011; Harshavardhan et al., 2014). Leaves were detached from 2-week-old seedlings and subjected to dehydration for 4 h. Then the expression of dehydration-responsive genes was detected. In the seedlings without dehydration, RD22 and RD29a expression was very low in both the Col-0 and HbEBP1 OE lines and showed no significant differences (p > 0.05; Figures 7B,C). After 4 h of dehydration, RD22 and RD29a expression was highly induced in both the wild-type Col-0 and HbEBP1 OE lines, but the HbEBP1 OE lines accumulated higher levels of transcripts than did the wild type plants. For RD22, the expression levels were 3.0-, 3.5- and 3.4- fold for OE5, OE18 and OE28 lines, respectively, when compared with that in the Col-0 seedlings (Figure 7B). For RD29a, expression was increased by 45, 62, and 57% in OE5, OE28 and OE28 lines, respectively, relative to wild type (Figure 7C). The genes CBF1, CBF2 and CBF3, which regulate RD29a (Yamaguchi-Shinozaki and Shinozaki, 1994; Stockinger et al., 1997; Jaglo-Ottosen et al., 1998; Liu et al., 1998; Medina et al., 1999), were also tested. Their expression did not differ between the wild-type and OE line seedlings (Supplementary Figure S3). These findings are similar to the reported effects of AmEBP1 (Cao et al., 2008), demonstrating that the HbEBP1 gene regulates drought resistance by other but not CBF pathway.
Discussion
In plants, EBP1 regulates cell growth in a dose-dependent manner and requires the involvement of auxin (Horváth et al., 2006). The OE of EBP1 often results in the enlargement of organs (Horváth et al., 2006; Wang et al., 2016). The OE of maize EBP1 increases organ size by promoting cell proliferation in Arabidopsis (Wang et al., 2016). In this study, the HbEBP1 OE lines develop leaves faster than the wild type seedlings (Figure 5B), and they have more cell number per leaf (Supplementary Figure S4), indicating that cell proliferation is accelerated in the HbEBP1 OE plants. HbEBP1 OE increased the expression of CYCD3;1 in Arabidopsis, suggesting that HbEBP1 has a conserved function in regulating the cell cycle. However, we also noticed that the HbEBP1 OE lines exhibited a late flowering phenotype, which means that these plants had a longer vegetative growth phase. In plants, developmental progress includes distinct phases and a number of developmental transitions during their life cycle. The transitions between phases are controlled by several intrinsic and extrinsic cues (Amasino, 2010; Huijser and Schmid, 2011). When a plant passes through the juvenile-to-adult transition, it gains reproductive competency and gets ready for flowering (Poethig, 2003; Huijser and Schmid, 2011; Srikanth and Schmid, 2011). HbEBP1 OE lines, with their late flowering phenotype, had more time to grow before passing through this transition. This is consistent with our observations concerning rosette leaf development in HbEBP1 OE plants: the juvenile leaves did not differ from those in the wild-type seedlings, whereas the adult leaves had an accelerated rate of emergence, were present in higher numbers and had larger leaf blades (Figures 4 and 5). As a result, the accelerated cell proliferation and longer vegetative growth period increased both the leaf number and size.
Ammopiptanthus mongolicus EBP1 is induced by cold, and confers enhanced cold tolerance when expressed in E. coli, as well as notably increased freezing survival when expressed in Arabidopsis (Cao et al., 2008). In this study, the rubber tree EBP1 was also induced by cold, drought and ABA treatment (Figure 2), OE of HbEBP1 conferred increased drought and freezing tolerance in Arabidopsis (Figure 6). Phenotypic characterization indicated that the HbEBP1 OE lines had relatively slower dehydration rates as compared with wild-type seedlings. In addition, the OE lines had better-developed root systems (Figure 4D, Supplementary Figure S2). These developmental features may also contribute to the robust drought resistance in the HbEBP1 OE lines.
Gene expression analysis indicated that RD29a and RD22 expression was enhanced in the HbEBP1 OE lines. RD29a and RD22 are involved in drought stress in plants (Yamaguchi-Shinozaki and Shinozaki, 1993a,b; Thomashow, 1999). The up-regulation of RD29a and RD22 thus suggested that HbEBP1 gene is involved in the regulation of drought resistance. We also analyzed the expression of CBF1–3, regulators of RD29a (Yamaguchi-Shinozaki and Shinozaki, 1994; Stockinger et al., 1997; Jaglo-Ottosen et al., 1998; Liu et al., 1998; Medina et al., 1999), The absence of an effect on their expression in the HbEBP1 OE lines (Supplementary Figure S3) is consistent with the effects of AmEBP1 (Cao et al., 2008), demonstrating that HbEBP1 regulates drought resistance by a mechanism that does not involve the CBF pathway.
Although the detailed regulatory pathway is still unknown, there has been speculation that EBP1 functions as a MAP and forms complexes with small molecules to accelerate protein processing after translation, which is essential for the plant to respond to abiotic stress (Cao et al., 2008). However, HbEBP1 when expressed in E. coli did not exhibit MAP activity in vitro (Cheng et al., 2016). This is consistent with the EBP1 crystal structure, which has the conserved pita bread fold of MAPs, although the protein lacks the characteristic enzymatic activity (Kowalinski et al., 2007; Monie et al., 2007). Therefore, even though EBP1 binds with dsRNA to form part of RNP complexes via association with different rRNA species in human cells (Squatrito et al., 2004), HbEBP1 may not possess the MAP activity needed to accelerate protein processing, which involves cutting off the first methionine from the peptide after translation in eukaryotic organisms (Datta, 2000). As EBP1 interacts with a number of proteins and RNAs that are involved in either transcription regulation or translation control (Squatrito et al., 2004, 2006; Ahn et al., 2006; Zhang et al., 2015), the MAP domain in HbEBP1 is more likely to function as a protein- or RNA-interacting motif (Monie et al., 2007).
In summary, the expression of an EBP1 from tropical woody plants was shown to be induced by drought and cold stress treatment. HbEBP1 was also able to promote drought and cold resistance, in addition to growth and organ size, in transgenic Arabidopsis plants. HbEBP1 OE also increased the expression of drought resistance–related RD22 and RD29a and of the cell cycle CYCD3;1. The regulation of drought resistance is a novel function identified in plant EBP1 genes. Although the detailed regulatory mechanism has yet to be determined, HbEBP1 may be useful in genetically engineered crops to increase both organ size (biomass) and abiotic stress resistance at the same time.
Conclusion
The study reported here, for the first time, describes the identification and characterization of an ErbB-3 binding protein 1 gene from H. brasiliensis. HbEBP1 is conserved at the level of its amino acid sequence and protein domains with other reported EBP1s. When subjected to abiotic stress or ABA treatment, HbEBP1 expression was induced, suggesting its roles in cold and drought resistance. Further transgenic experiment indicated that HbEBP1 has conserved functions in enlarging organ size. In addition, HbEBP1 OE delayed flowering time and the vegetative-to-reproductive transition and increased the adult leaf number. More importantly, the HbEBP1 OE lines showed enhanced resistance to freezing and drought stress, which is a novel function identified in plant EBP1 genes. Further analysis revealed that CYCD3;1, RD29a and RD22 expression was promoted in the HbEBP1 OE lines, which helps to explain the regulatory roles of HbEBP1. Taking together, HbEBP1 may be useful in genetically engineered crops to increase both organ size (biomass) and abiotic stresses resistance at the same time.
Author Contributions
HC and HH designed the experiments; JZ and XC conducted the experiments; HC wrote the manuscript draft; HH discussed the results and finalized the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
This work was financially supported by the National Natural Science Foundation of China (grant No.: 31301072)
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01703/full#supplementary-material
Abbreviations
ABA, abscisic acid; EBP1, ErbB-3 binding protein 1; OE, overexpression.
Footnotes
- ^ http://www.ebi.ac.uk/Tools/msa/clustalw2/
- ^ http://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^ http://www.ebi.ac.uk/Tools/pfa/iprscan5/
References
Ahn, J.-Y., Liu, X., Liu, Z., Pereira, L., Cheng, D., Peng, J., et al. (2006). Nuclear Akt associates with PKC-phosphorylated Ebp1, preventing DNA fragmentation by inhibition of caspase-activated DNase. EMBO J. 25, 2083–2095. doi: 10.1038/sj.emboj.7601111
Amasino, R. (2010). Seasonal and developmental timing of flowering. Plant J. 61, 1001–1013. doi: 10.1111/j.1365-313X.2010.04148.x
Blomme, J., Inzé, D., and Gonzalez, N. (2014). The cell-cycle interactome: a source of growth regulators? J. Exp. Bot. 65, 2715–2730. doi: 10.1093/jxb/ert388
Cao, P., Song, J., Zhou, C., Weng, M., Liu, J., Wang, F., et al. (2008). Characterization of multiple cold induced genes from Ammopiptanthus mongolicus and functional analyses of gene AmEBP1. Plant Mol. Biol. 69, 529–539. doi: 10.1007/s11103-008-9434-1
Cheng, H., Cai, H., An, Z., and Huang, H. (2013). Comparative proteomics analysis revealed increased expression of photosynthetic proteins in transgenic tobacco by overexpression of AtCBF1 gene. Plant Omics 6, 240–245.
Cheng, H., Cai, H., Fu, H., An, Z., Fang, J., Hu, Y., et al. (2015). Functional characterization of Hevea brasiliensis CRT/DRE binding factor 1 gene revealed regulation potential in the CBF pathway of tropical perennial tree. PLoS ONE 10:e0137634. doi: 10.1371/journal.pone.0137634
Cheng, H., Cai, H., and Huang, H. (2008). Construction of full-length cDNA library in rubber tree under cold stress. Chin. J. Trop. Crops 41, 410–414.
Cheng, H., Zhu, J., An, Z., Fang, J., Hu, Y., and Huang, H. (2016). The characterization, prokaryotic expression and MetAP activity analysis of Hevea brasiliensis HbEBP1. Chin. J. Trop. Crops 37, 317–324.
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Collins, C., Maruthi, N. M., and Jahn, C. E. (2015). CYCD3 D-type cyclins regulate cambial cell proliferation and secondary growth in Arabidopsis. J. Exp. Bot. 66, 4595–4606. doi: 10.1093/jxb/erv218
Datta, B. (2000). MAPs and POEP of the roads from prokaryotic to eukaryotic kingdoms. Biochimie 82, 95–107. doi: 10.1016/S0300-9084(00)00383-7
Dewitte, W., Scofield, S., Alcasabas, A. A., Maughan, S. C., Menges, M., Braun, N., et al. (2007). Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc. Natl. Acad. Sci. U.S.A. 104, 14537–14542. doi: 10.1073/pnas.0704166104
Figeac, N., Serralbo, O., Marcelle, C., and Zammit, P. S. (2014). ErbB3 binding protein-1 (Ebp1) controls proliferation and myogenic differentiation of muscle stem cells. Dev. Biol. 386, 135–151. doi: 10.1016/j.ydbio.2013.11.017
Harshavardhan, V. T., Van Son, L., Seiler, C., Junker, A., Weigelt-Fischer, K., Klukas, C., et al. (2014). AtRD22 and AtUSPL1, members of the plant-specific BURP domain family involved in Arabidopsis thaliana drought tolerance. PLoS ONE 9:e110065. doi: 10.1371/journal.pone.0110065
Horváth, B. M., Magyar, Z., Zhang, Y., Hamburger, A. W., Bakó, L., Visser, R. G. F., et al. (2006). EBP1 regulates organ size through cell growth and proliferation in plants. EMBO J. 25, 4909–4920. doi: 10.1038/sj.emboj.7601362
Huijser, P., and Schmid, M. (2011). The control of developmental phase transitions in plants. Development 138, 4117–4129. doi: 10.1242/dev.063511
Jaglo-Ottosen, K. R., Gilmour, S. J., Zarka, D. G., Schabenberger, O., and Thomashow, M. F. (1998). Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280, 104–106. doi: 10.1126/science.280.5360.104
Jones, P., Binns, D., Chang, H.-Y., Fraser, M., Li, W., McAnulla, C., et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240. doi: 10.1093/bioinformatics/btu031
Kowalinski, E., Bange, G., Bradatsch, B., Hurt, E., Wild, K., and Sinning, I. (2007). The crystal structure of Ebp1 reveals a methionine aminopeptidase fold as binding platform for multiple interactions. FEBS Lett. 581, 4450–4454. doi: 10.1016/j.febslet.2007.08.024
Lamartine, J., Seri, M., Cinti, R., Heitzmann, F., Creaven, M., Radomski, N., et al. (1997). Molecular cloning and mapping of a human cDNA (PA2G4) that encodes a protein highly homologous to the mouse cell cycle protein p38-2G4. Cytogenet. Cell Genet. 78, 31–35. doi: 10.1159/000134621
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Lessor, T. J., Yoo, J.-Y., Xia, X., Woodford, N., and Hamburger, A. W. (2000). Ectopic expression of the ErbB-3 binding protein Ebp1 inhibits growth and induces differentiation of human breast cancer cell lines. J. Cell. Physiol. 183, 321–329. doi: 10.1002/(SICI)1097-4652(200006)183:3<321::AID-JCP4>3.0.CO;2-O
Liu, Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Yamaguchi-Shinozaki, K., et al. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10, 1391–1406. doi: 10.1105/tpc.10.8.1391
Liu, Y., Liu, Y., Cao, J., Zhu, X., Nie, X., Yao, L., et al. (2014). Upregulated expression of Ebp1 contributes to schwann cell differentiation and migration after sciatic nerve crush. J. Mol. Neurosci. 54, 602–613. doi: 10.1007/s12031-014-0331-6
Liu, Z., Ahn, J.-Y., Liu, X., and Ye, K. (2006). Ebp1 isoforms distinctively regulate cell survival and differentiation. Proc. Natl. Acad. Sci. U.S.A. 103, 10917–10922. doi: 10.1073/pnas.0602923103
Martínez-Zapater, J. M., Jarillo, J. A., Cruz-Alvarez, M., Roldán, M., and Salinas, J. (1995). Arabidopsis late-flowering fve mutants are affected in both vegetative and reproductive development. Plant J. 7, 543–551. doi: 10.1046/j.1365-313X.1995.7040543.x
Medina, J., Bargues, M., Terol, J., Perez-Alonso, M., and Salinas, J. (1999). The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiol. 119, 463–470. doi: 10.1104/pp.119.2.463
Monie, T. P., Perrin, A. J., Birtley, J. R., Sweeney, T. R., Karakasiliotis, I., Chaudhry, Y., et al. (2007). Structural insights into the transcriptional and translational roles of Ebp1. EMBO J. 26, 3936–3944. doi: 10.1038/sj.emboj.7601817
Montoro, P., Rattana, W., Pujade-Renaud, V., Michaux-Ferrière, N., Monkolsook, Y., Kanthapura, R., et al. (2003). Production of Hevea brasiliensis transgenic embryogenic callus lines by Agrobacterium tumefaciens: roles of calcium. Plant Cell Rep. 21, 1095–1102. doi: 10.1007/s00299-003-0632-7
Msanne, J., Lin, J., Stone, J. M., and Awada, T. (2011). Characterization of abiotic stress-responsive Arabidopsis thaliana RD29A and RD29B genes and evaluation of transgenes. Planta 234, 97–107. doi: 10.1007/s00425-011-1387-y
Nguyen, L. X. T., Lee, Y., Urbani, L., Utz, P. J., Hamburger, A. W., Sunwoo, J. B., et al. (2015). Regulation of ribosomal RNA synthesis in T cells: requirement for GTP and Ebp1. Blood 125, 2519–2529. doi: 10.1182/blood-2014-12-616433
Poethig, R. S. (2003). Phase change and the regulation of developmental timing in plants. Science 301, 334–336. doi: 10.1126/science.1085328
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. doi: 10.1038/nmeth.2089
Squatrito, M., Mancino, M., Donzelli, M., Areces, L. B., and Draetta, G. F. (2004). EBP1 is a nucleolar growth-regulating protein that is part of pre-ribosomal ribonucleoprotein complexes. Oncogene 23, 4454–4465. doi: 10.1038/sj.onc.1207579
Squatrito, M., Mancino, M., Sala, L., and Draetta, G. F. (2006). Ebp1 is a dsRNA-binding protein associated with ribosomes that modulates eIF2α phosphorylation. Biochem. Biophys. Res. Commun. 344, 859–868. doi: 10.1016/j.bbrc.2006.03.205
Srikanth, A., and Schmid, M. (2011). Regulation of flowering time: all roads lead to Rome. Cell. Mol. Life Sci. 68, 2013–2037. doi: 10.1007/s00018-011-0673-y
Stockinger, E. J., Gilmour, S. J., and Thomashow, M. F. (1997). Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. U. S. A. 94, 1035–1040. doi: 10.1073/pnas.94.3.1035
Sun, J., Luo, Y., Tian, Z., Gu, L., Xia, S. C., and Yu, Y. (2012). Expression of ERBB3 binding protein 1 (EBP1) in salivary adenoid cystic carcinoma and its clinicopathological relevance. BMC Cancer 12:499. doi: 10.1186/1471-2407-12-499
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tang, C., Yang, M., Fang, Y., Luo, Y., Gao, S., Xiao, X., et al. (2016). The rubber tree genome reveals new insights into rubber production and species adaptation. Nat. Plants 2, 16073. doi: 10.1038/nplants.2016.73
Telfer, A., Bollman, K. M., and Poethig, R. S. (1997). Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development 124, 645–654.
Thomashow, M. F. (1999). PLANT COLD ACCLIMATION: freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 571–599. doi: 10.1146/annurev.arplant.50.1.571
Wang, T., Sui, Z., Liu, X., Li, Y., Li, H., Xing, J., et al. (2016). Ectopic expression of a maize hybrid up-regulated gene, ErbB-3 binding Protein 1 (ZmEBP1), increases organ size by promoting cell proliferation in Arabidopsis. Plant Sci. 243, 23–34. doi: 10.1016/j.plantsci.2015.11.002
Willmann, M. R., and Poethig, R. S. (2011). The effect of the floral repressor FLC on the timing and progression of vegetative phase change in Arabidopsis. Development 138, 677–685. doi: 10.1242/dev.057448
Xia, X., Lessor, T. J., Zhang, Y., Woodford, N., and Hamburger, A. W. (2001). Analysis of the expression pattern of Ebp1, an ErbB-3-binding protein. Biochem. Biophys. Res. Commun. 289, 240–244. doi: 10.1006/bbrc.2001.5942
Yamaguchi-Shinozaki, K., and Shinozaki, K. (1993a). Arabidopsis DNA encoding two desiccation-responsive rd29 genes. Plant Physiol. 101, 1119–1120. doi: 10.1104/pp.101.3.1119
Yamaguchi-Shinozaki, K., and Shinozaki, K. (1993b). The plant hormone abscisic acid mediates the drought-induced expression but not the seed-specific expression of rd22, a gene responsive to dehydration stress in Arabidopsis thaliana. Mol. Gen. Genet. 238, 17–25.
Yamaguchi-Shinozaki, K., and Shinozaki, K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6, 251–264. doi: 10.1105/tpc.6.2.251
Zewei An (2012). Co-extraction of high-quality RNA and DNA from rubber tree (Hevea brasiliensis). Afr. J. Biotechnol. 11, 9308–9314. doi: 10.5897/AJB12.136
Zhang, F., Liu, Y., Wang, Z., Sun, X., Yuan, J., Wang, T., et al. (2015). A novel Anxa2-interacting protein Ebp1 inhibits cancer proliferation and invasion by suppressing Anxa2 protein level. Mol. Cell. Endocrinol. 411, 75–85. doi: 10.1016/j.mce.2015.04.013
Keywords: drought stress, Hevea brasiliensis, ErbB-3 Binding Protein 1, Arabidopsis, organ size, cell cycle
Citation: Cheng H, Chen X, Zhu J and Huang H (2016) Overexpression of a Hevea brasiliensis ErbB-3 Binding protein 1 Gene Increases Drought Tolerance and Organ Size in Arabidopsis. Front. Plant Sci. 7:1703. doi: 10.3389/fpls.2016.01703
Received: 10 August 2016; Accepted: 28 October 2016;
Published: 14 November 2016.
Edited by:
Keqiang Wu, National Taiwan University, TaiwanReviewed by:
Vidhu Aniruddha Sane, National Botanical Research Institute (CSIR), IndiaKyoung Hee Nam, Sookmyung Women’s University, South Korea
Copyright © 2016 Cheng, Chen, Zhu and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Han Cheng, Zm9yY2hlbmdAZ21haWwuY29t Huasun Huang, aHVhbmdodWFzdW5AZ21haWwuY29t
 Han Cheng
Han Cheng Xiang Chen
Xiang Chen