- 1National Center for Soybean Improvement, Key Laboratory of Biology and Genetic Improvement of Soybean Ministry of Agriculture P.R. China, State Key Laboratory of Crop Genetics and Germplasm Enhancement, Nanjing Agricultural University, Nanjing, China
- 2College of Agro-grassland Science, Nanjing Agricultural University, Nanjing, China
MADS-domain proteins are important transcription factors involved in many aspects of plant reproductive development. In this study, a MADS-box gene, Glycine max AGAMOUS-LIKE1 (GmAGL1), was isolated from soybean flower. The transcript of GmAGL1 was expressed in flowers and pods of different stages in soybean and was highly expressed in carpels. GmAGL1 is a nucleus-localized transcription factor and can interact directly with SEP-like proteins in soybean flowers. Ectopic overexpression of GmAGL1 resulted in the absence of petals in Arabidopsis. Moreover, morphological changes in the valves were observed in 35S:GmAGL1 Arabidopsis fruits that dehisced before the seeds reached full maturity. GmAGL1 was found to be sufficient to activate the expression of Arabidopsis ALC, IND, STK, SEP1, and SEP3. Therefore, our data suggest that GmAGL1 may play important roles in both floral organ identity and fruit dehiscence.
Introduction
Angiosperms, or flowering plants, are the most diverse and numerous groups of plants. Despite their diversity, the most remarkable features distinguishing them from gymnosperms are their flowers and the production of fruits that contain seeds. Most flowers are composed of four types of floral organs with external sterile organs (sepals and petals) surrounding the reproductive structures (stamens and carpels) located in the center (Robles and Pelaz, 2005). The fertilized carpels give rise to the fruits, which protect the developing seeds and ultimately disperse the mature seeds into the environment. The vast majority of crops in the world are angiosperms, therefore, the regulation of flower and fruit development directly affects the economic benefits in agricultural production. Understanding the genetic factors regulating flower and fruit patterning may help to improve crop breeding.
Flower development has been the subject of intensive study for over the past 20 years. These studies led to the establishment of the well-known ABCDE model that explained the genetic regulation of floral organ identity determination. This model proposes that five classes of genes (A, B, C, D, and E) act in a combinatorial way to specify the distinct floral organs (Theissen and Saedler, 2001). The A+E protein complex determines sepals, the A+B+E complex specifies petals, the B+C+E complex specifies stamens, the C+E complex specifies carpels and the D+E complex determines the ovules (Colombo et al., 1995; Theissen and Saedler, 2001). In Arabidopsis, the class A genes are APETALA1/2 (AP1/2) (Mandel et al., 1992; Jofuku et al., 1994); the class B genes are AP3 (Jack et al., 1992) and PISTILLATA (PI) (Goto and Meyerowitz, 1994); the class C gene is AGAMOUS (AG) (Yanofsky et al., 1990); the class D genes are SEEDSTICK (STK) and SHATTERPROOF1/2 (SHP1/2) (Favaro et al., 2003); and the class E genes are SEPALLATA1/2/3/4 (SEP1/2/3/4) (Pelaz et al., 2000). Interestingly, all of these floral organ identity genes except AP2 belong to the MADS-box gene family. Moreover, orthologs of these genes have been found in many other species, such as other eudicots, monocots, and even in gymnosperms (Robles and Pelaz, 2005; Arora et al., 2007; Katahata et al., 2014).
The genetic networks controlling fruit patterning are well-characterized in many plants (White, 2002; Xue et al., 2012; Karlova et al., 2014). In the model plant Arabidopsis, the fruit is a dry pod derived from two fused carpels called a silique. During dry fruit development, one main process is the differentiation of tissues required for dehiscence (Robles and Pelaz, 2005). Many genes related to fruit dehiscence have been identified. The MADS-box transcription factors SHP1 and SHP2 (previously named AGL1 and AGL5) are the primary regulators of dry fruit dehiscence (Liljegren et al., 2000). Although SHP genes have been shown to specify carpel identity in a transcriptional complex (Favaro et al., 2003), they are best known for their functions in the differentiation of the dehiscence zone and the lignification of adjacent cells (Liljegren et al., 2000). Mutations in the SHP genes lead to indehiscent fruits, thus, inhibiting the seed dispersal process. SHP genes act at the top of the genetic regulatory hierarchy in valve margin formation and positively regulate INDEHISCENT (IND) and ALCATRAZ (ALC), which are two bHLH transcription factors also required for correct valve margin development (Rajani and Sundaresan, 2001; Liljegren et al., 2004). The expression of SHP1, SHP2, IND, and ALC in the valve margins is negatively controlled by FRUITFUL (FUL). As a MADS-box transcription factor, FUL is responsible for both ovary growth and valve cell differentiation (Ferrandiz et al., 2000; Liljegren et al., 2004).
Soybean (Glycine max [L.] Merr.) is an economically important global crop and it is now the main source of vegetable oil and protein. The development of flowers and pods directly affects seed yield and quality. Additionally, soybean is a self-pollinated crop, the anther and stigma of which are enclosed by the wing flap and keel. This floral morphology causes difficulties in cross-pollination and prevents crossing within or between individuals, which is unfavorable for soybean hybrid breeding (Huang et al., 2014). Therefore, genetic modification of the perianth is a priority in soybean hybrid breeding. The studies on MADS-box genes related to floral morphology will facilitate the production of valuable plant materials that have potential applications in soybean hybrid breeding. Based on a previous study on gene expression analysis by Affymetrix Gene Chip (Huang et al., 2009), a Glycine max AGAMOUS-LIKE1 gene (Probe ID: Gma.11881.1.A1_at) was found to predominantly accumulate in soybean flowers and pods, indicating its role in floral organs and fruit development (Ma et al., 1991; Flanagan et al., 1996). We isolated GmAGL1 from soybean flower and characterized its spatial and temporal expression patterns. As a MADS domain protein, GmAGL1 is a nucleus-localized transcription factor and functions in a multimeric complex with SEP-like proteins. GmAGL1 was sufficient to activate the expression of ALC, IND, STK, SEP1, and SEP3 in Arabidopsis. The ectopic overexpression of GmAGL1 resulted in an abnormal floral organ identity in Arabidopsis, in which the petals were completely absent. Moreover, morphological changes in the valves were observed in 35S:GmAGL1 fruits that dehisced before the seeds reached full maturity.
Materials and Methods
Plant Materials and Growth
Soybean plants (cv. Jackson) were grown under field conditions in Nanjing Agricultural University, China. Young leaves, roots, stems and shoot apical meristems (SAMs) were collected at the third euphylis expanding stage. Flowers were harvested at different stages from tiny buds to mature flowers. Four types of floral organs were obtained from mature flowers. The developing pods were harvested at 5, 10, 20, 30, and 40 DAF (day after flowering). Seeds at 20 DAF were dissected to collect the seed coat, embryo and cotyledon under a dissection microscope with surgical blades and tweezers. All of these samples were lyophilized and stored at -80°C until used.
Arabidopsis thaliana, ecotype Columbia-0, was used for GmAGL1 ectopic expression experiments. All plants were grown in a growth room at 22°C with 16 h light/8 h dark.
Isolation of GmAGL1 Gene by RACE
Based on the sequence information from NCBI1, the fragment (GenBank accession No: AW433203) was found to have an incomplete ORF. The rapid amplification of cDNA using the end (RACE) technique was employed to identify both the 3′- and 5′-ends of GmAGL1 cDNA with a SMARTTM RACE cDNA Amplification Kit (Clontech, USA). The 3′-end was amplified from flower cDNA by two nested PCR reactions, using the gene specific primer 3′-GSP (5′-GAGAAAGCACAACAACGGCAACAG-3′) for the first round PCR and another gene specific primer 3′-NGSP (5′-GCGAGTCAACCATACCTCCA-3′) for nested PCR. For 5′-end amplification, the two primers used were 5′-GSP (5′-TGGAGGTATGGTTGACTCGCACA-3′) and 5′-NGSP (5′-CCGTTGTTGTGCTTTCTCGTGTTC-3′). All the PCR products were gel purified, cloned into pGEM®-T easy vectors (Promega, USA) and sequenced (Invitrogen, Shanghai). According to the RACE results, the primer pair (sense: 5′-GCATAACACCAAAGAACTAC-3′ and anti-sense: 5′-TCACGAAACATAGGACGATT-3′) was used to isolate the intact ORF of GmAGL1 by RT-PCR.
Sequence and Phylogenetic Analysis
The ORF of GmAGL1 and its deduced protein sequence were analyzed by BioXM (ver 2.6). Conserved domains were analyzed by SMART2. The sequences of another published MADS-box genes were obtained from NCBI using BLASTP3. Multiple alignments were conducted with ClustalX 2.0 and viewed with GeneDOC. Sequences selected for the phylogenetic analysis were GmAGL1, the ABCDE classes of MADS-box proteins from Arabidopsis and previously reported AG homologs from several species. The accession numbers of the protein sequences are listed in Supplementary S2. The ML (Maximum Likelihood) phylogenetic tree was constructed using MEGA 6.0 with the following parameters: bootstrap method with 1,000 replications, “Jones-Taylor-Thornton (JTT)” as the substitution model, “complete deletion” for gaps/missing data treatment and other parameters with a default value.
Gene Expression Analysis
Total RNA was isolated using an RNA Plant Extraction Kit (TIANGEN, China) and reverse transcribed by AMV reverse transcriptase (Takara, Japan) as described in the manufacturer’s instructions. GmAGL1 specific primers were synthesized as follows: sense: 5′-GCTGAACACGAGAAAGCACA-3′; anti-sense: 5′-GGCACTCTCCTTCACGAAAC-3′. Semi-quantitative RT-PCR assay was performed as previously described (Huang et al., 2006). Real-time PCR was carried out with a Bio-RAD iQ5 real-time PCR system (Bio-Rad, USA) using SYBR® Green Real-time PCR Master Mix QPK-201 (Toyobo Japan). The Tubulin gene (GenBank accession No: AY907703) was quantified to normalize the amount of total transcript. The relative expression of GmAGL1 was calculated according to the method of 2-ΔΔCt (Livak and Schmittgen, 2001).
For the expression analysis of MADS-box genes in transgenic Arabidopsis, total RNA was prepared from the flowers of the wild-type and 35S:GmAGL1 plants. The Arabidopsis TIP4 gene was used as an internal control. The primers for each MADS-box gene are shown in Supplementary Table S2.
Subcellular Localization of GmAGL1
The coding region of GmAGL1 was amplified with a sense primer (5′-CTAGTCTAGAATGGAATTTCCCAACGAAGC-3′) containing an Xba I site and an anti-sense primer (5′-CGCGGATCCGACAAGTTGAAGAGCAGTCTGGTC-3′) containing a BamH I site. The PCR product was correctly inserted into the Xba I and BamH I sites of vector pAN580, generating a GmAGL1:GFP in-frame fusion protein. This construct was then introduced into Arabidopsis leaf protoplasts via PEG-mediated transformation. The protoplasts were incubated under weak light for 12 h to 16 h and observed with a LSM 700 exciter confocal laser scanning microscope (Carl Zeiss). The excitation wavelength used for GFP was 488 nm.
Ectopic Expression in Arabidopsis
The plant overexpression vector pMDC32 was used for Arabidopsis transformation and contained a double 35S CaMV promoter. The GmAGL1 ORF from the start to stop codons was cloned into pMDC32 by GatewayTM Technology (Invitrogen Shanghai). Col-0 plants were transformed with this construct by Agrobacterium tumefaciens using the floral dip method. Transgenic seeds were germinated on solid MS medium containing 20 μg/mL Hygromycin B. Resistant seedlings were transferred to soil and further analyzed by PCR and qRT-PCR.
Scanning Electron Microscopy
Fruits from wild-type and 35S:GmAGL1 Arabidopsis plants were vacuum infiltrated with 4% glutaraldehyde in 25 mM phosphate buffer (pH 7.0) for 10 min and fixed with fresh solution for 16 h at 4°C, washed subsequently in 25 mM phosphate buffer (pH 7.0) and incubated for 4 h in 1% osmic acid. Samples were dehydrated gradually in an ethanol series of 30, 50, 70, 80, 95, and 100% and then critical point dried in liquid CO2. Dried samples were covered with gold in a Nanotech sputter coater and examined with a scanning electron microscope (Philips SEM-505).
Yeast Two-Hybrid Assay
To screen the GmAGL1 interaction proteins in vivo, the yeast two-hybrid (Y2H) screen of the soybean cDNA library was performed by the ProQuestTM Two-Hybrid System (Invitrogen). The coding region of GmAGL1 was recombined in a pDEST32 vector that carried a GAL4 DNA-binding domain to generate the bait construct. The Y2H cDNA library prepared from soybean flowers was used as prey and co-transformed with the bait into MaV203 yeast competent cells according to the manufacturer’s protocol. Transformants were then cultured on SC-Leu-Trp-His-master plates supplemented with 40 mM of 3-aminotriazole (3-AT). The positive clones were verified by retransformation.
Bimolecular Fluorescence Complementation (BiFC)
The coding sequences for GmAGL1, GmSEP1 (GenBank accession No: DQ159905) (Huang et al., 2009) and GmSEP3 (GenBank accession No: AJ878424) (Huang et al., 2014), were cloned into vector pUC-SPYCE (C-YFP, 156–239 amino acid) to generate the C-terminal in-frame fusions with C-YFP, whereas GmAGL1 coding sequences were introduced into pUC-SPYNE (N-YFP, 1–155 amino acid) to form N-terminal in-frame fusions with N-YFP. The plasmids were co-transformed into onion epidermal cells to verify the interaction between proteins by gold particle bombardment, with a concentration ratio of 1: 1. Two days after bombardment, imaging of co-expressed YFP fragment signals was examined with a confocal fluorescence microscope. The primers used for bimolecular fluorescence complementation (BiFC) are shown in Supplementary Table S3.
Results
Isolation and Sequence Analysis of AGAMOUS-LIKE1 from Soybean
The complete GmAGL1 including 729 bp of ORF (GenBank accession No: KY321171) was isolated from soybean flower cDNA by RT-PCR and sequenced (Supplementary S1). GmAGL1 encodes a predicted protein of 242 amino acids with a calculated molecular mass of 27.90 kDa and a pI of 9.55. GmAGL1 contained the conserved domains that characterize MADS-box proteins: MADS domain, I domain and K domain (Figure 1). Although the C-terminal region was highly divergent, there were two short conserved motifs: the AG motifs I and II (Figure 1). Alignment analysis of amino acid sequences showed that GmAGL1 shared 86% identity with M8 (Pisum sativum, AAX69070) and 84% identity with LjAGL1 (Lotus japonicus, AAX13305). Compared with other well-known MADS-box proteins, GmAGL1 was 72% identical to SHP1/2 of Arabidopsis (Liljegren et al., 2000) and 78% identical to PPERSHP of Peach (Tani et al., 2007).
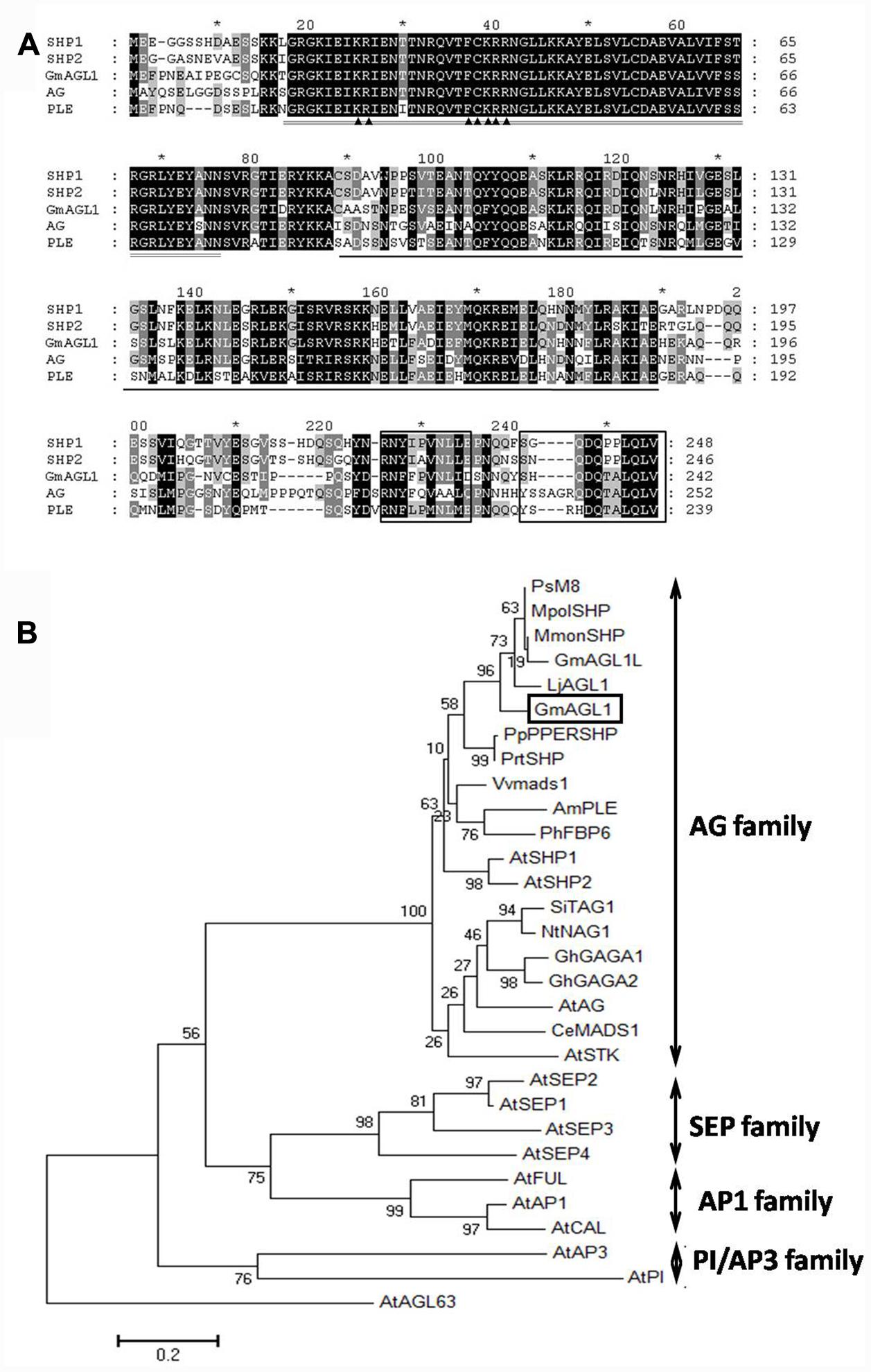
FIGURE 1. Sequence alignment and phylogenetic analysis of GmAGL1. (A) Clustal X alignment of the deduced amino acid sequence of GmAGL1 and other AG proteins: AG (AEE84111), SHP1 (AEE79829), SHP2 (AAU82070) and PLE (AAB25101). The highly conserved region, MADS domain, is double-underlined. The K domain is underlined. The AG motifs I and II are boxed at the C terminus. ▲ marks the bipartite NLS. (B) Phylogenetic relationships of ABCDE class proteins in Arabidopsis and AG proteins in another species. The phylogenetic tree was generated from full-length amino acid sequences by MEGA 6 using the Maximum Likelihood method. Am (Antirrhinum majus), At (Arabidopsis thaliana), Ce (Cymbidium ensifolium), Gh (Gerbera hybrid), Gm (Glycine max), Lj (Lotus japonicus), Mmon (Medicago monspeliaca), Mpol (Medicago polyceratia), Nt (Nicotiana tabacum), Ph (Petunia hybrida), Pp (Prunus persica), Prt (Prunus triloba), Ps (Pisum sativum), Si (Solanum lycopersicum), and Vv (Vitis vinifera). AGL63 was used as an outgroup. The accession numbers of the protein sequences are listed in Supplementary S2.
To determine the evolutionary relationship of GmAGL1 with other known ABCDE class proteins from Arabidopsis, we used the overall amino acid sequences were used for phylogenetic analysis. GmAGL1 is located in the AGAMOUS (AG) subfamily that comprises the conserved euAG and PLENA (PLE) lineages in core eudicots (Figure 1) (Kramer et al., 2004). GmAGL1 was highly homologous to PLE proteins in legumes and Arabidopsis, suggesting that it belonged to PLE lineage and may function as a class D gene in the ABCDE model.
Expression Patterns of the GmAGL1 during Reproductive Growth
GmAGL1 is specifically expressed in flowers and pods of different stages (Figures 2C–F), but not in roots, stems, and leaves (Figure 2A). Its expression was detected at early stages and increased gradually with the process of flower and pod development (Figures 2C–F). In floral organs, GmAGL1 transcripts showed strong expression in carpels and less expression in sepals (Figure 2B). In the developing seed, GmAGL1 was exclusively expressed in the seed coat, which was derived from ovule integument after fertilization (Supplementary Figure S1). The gene expression profile indicates that GmAGL1 may play important roles in carpel and pod development.
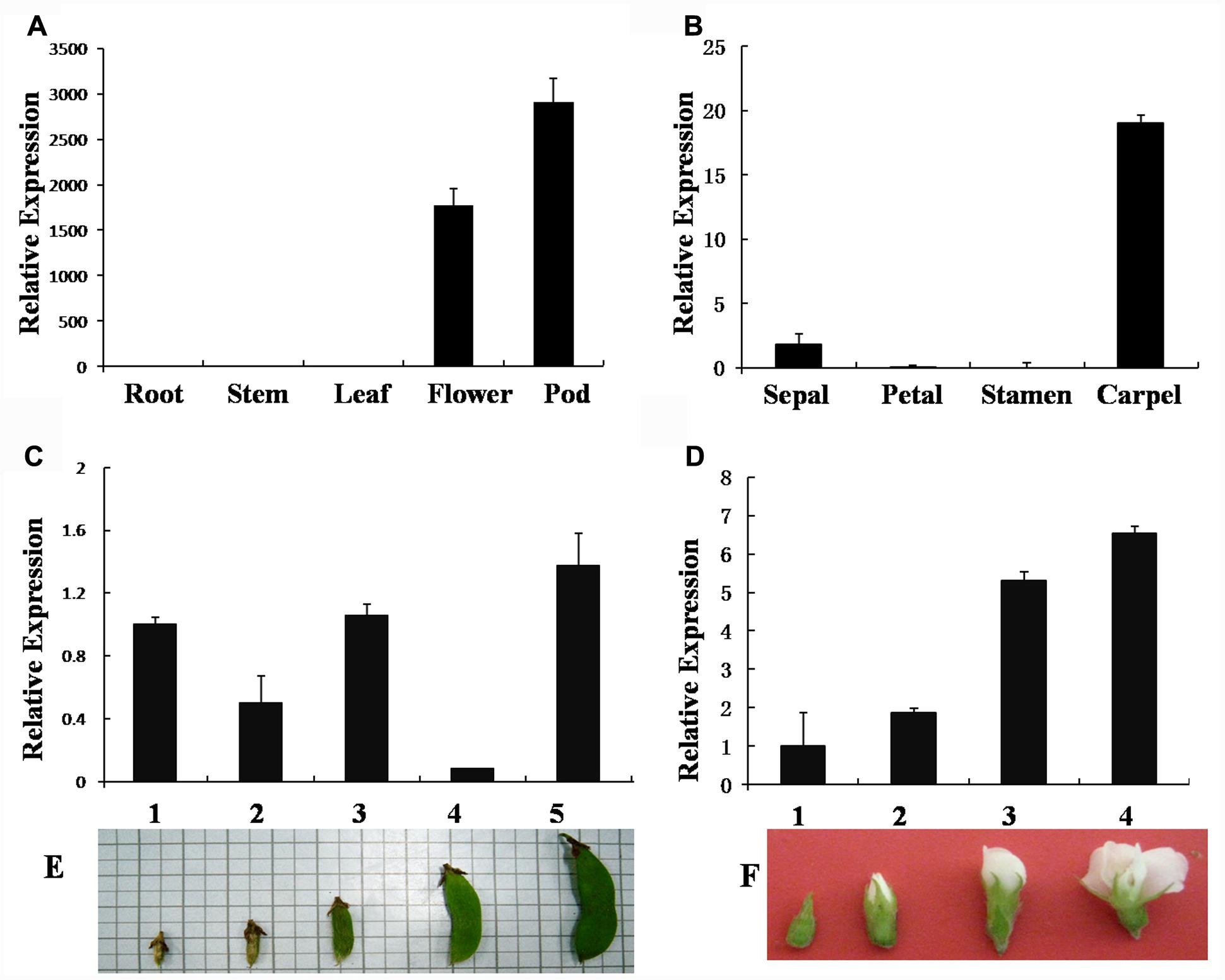
FIGURE 2. Real-time PCR analysis of GmAGL1 expression in different organs and at different developmental stages. (A) Gene expression profile of GmAGL1 in vegetative organs, flowers and pods. (B) Expression profile of GmAGL1 in floral organs. (C) Expression profile of GmAGL1 in developing pods at the five developmental stages (1–5) illustrated in (E). (D) Expression profile of GmAGL1 in developing flowers at the four developmental stages (1–4) illustrated in (F). The error bars represent SD based on three replicates.
Nuclear Localization of GmAGL1
Because the highly conserved MADS domain contains a putative nuclear localization signal (NLS) (Immink et al., 2002), GmAGL1 was predicted to be localized in the nucleus (Figure 1). To confirm the localization in plant cells, GmAGL1 was fused with GFP, under the control of a 35S promoter, and transiently expressed in Arabidopsis protoplasts. Confocal green fluorescence imaging revealed that GmAGL1–GFP fusion protein was located in the nucleus, whereas free GFPs, 35S:GFP, were distributed throughout the whole cell (Figure 3).
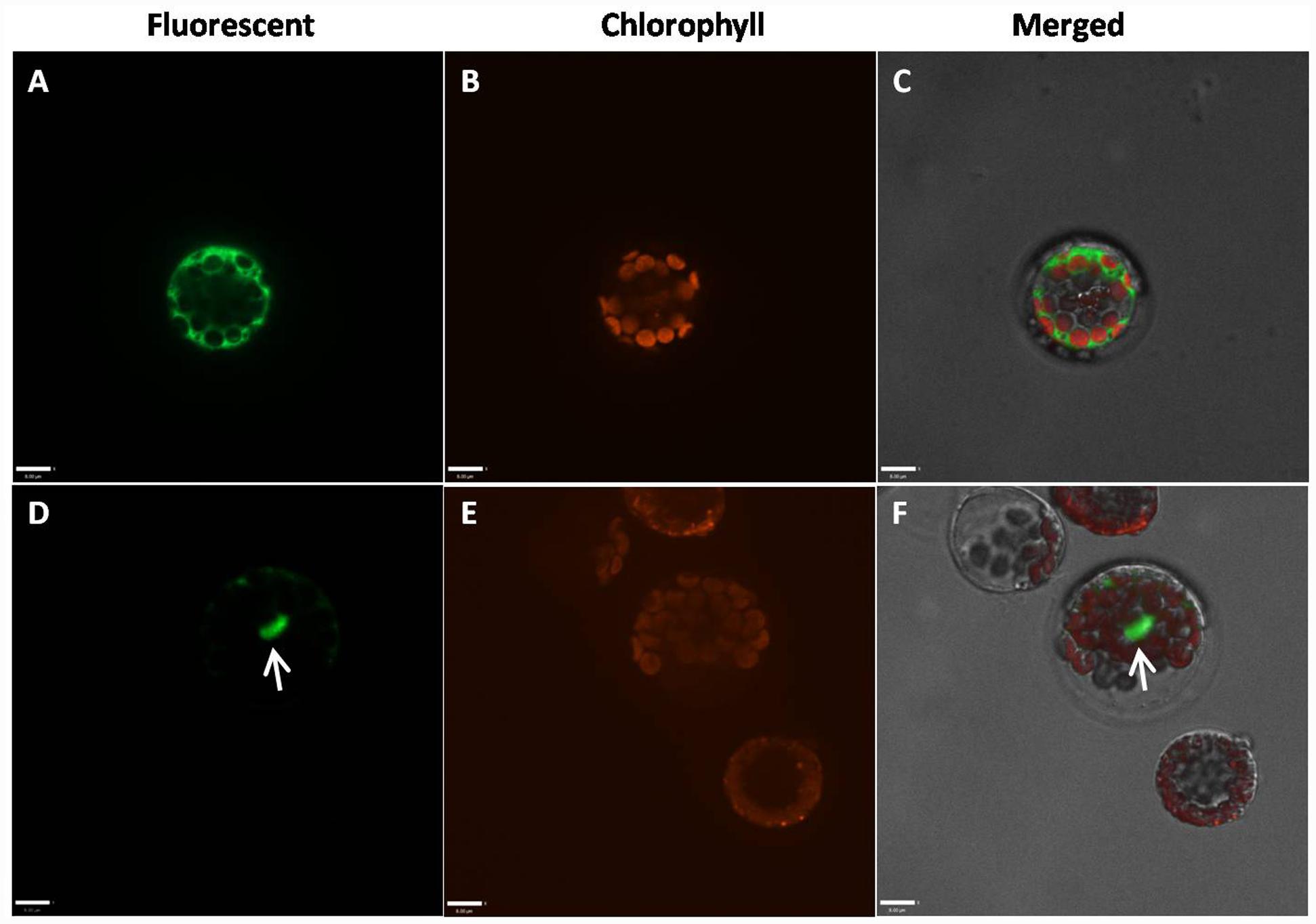
FIGURE 3. Subcellular localization of GmAGL1. GmAGL1-GFP fusion proteins were driven by CaMV 35S promoter and transiently expressed in Arabidopsis leaf protoplasts. Photographs were obtained with a confocal microscope. (A–C) are images of 35S:GFP as controls; (D–F) are images of GmAGL1-GFP. The arrow shows the location of GmAGL1. Scale bars = 8 μm.
Phenotypic Changes in GmAGL1 Ectopic Expression Lines
To characterize the function of GmAGL1 further, we examined transgenic Arabidopsis plants that were constitutively expressed GmAGL1 under the control of the cauliflower mosaic virus 35S promoter (35S:GmAGL1). The overexpression of GmAGL1 significantly affected the development of the transgenic Arabidopsis, which flowered substantially earlier than the flowers of wild-type plants (Figure 4A). The number of leaves produced in the transgenic lines at bolting was significantly different from that in the wild-type plants. The transgenic lines produced on average 9 leaves before flowering, whereas wild-type plants produced approximately 17 leaves (Figure 4B). Flowers from plants highly expressing GmAGL1 showed an abnormal morphology, in which all the petals were absent (Figures 5A–D). After fertilization, the senescence and abscission of sepals were significantly delayed in transgenic plants, and green unabscised sepals were observed even in the matured siliques (Figure 5E).
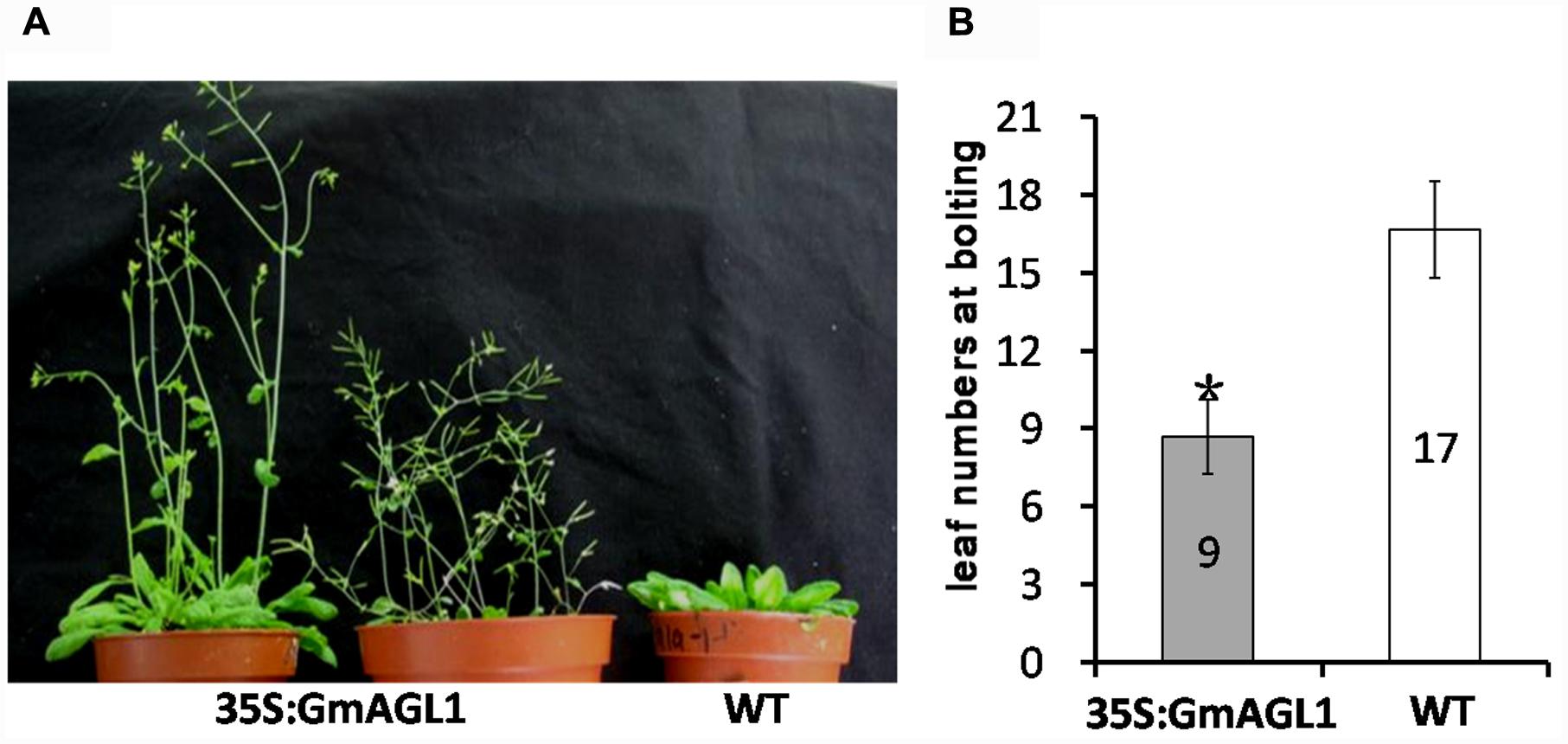
FIGURE 4. Early flowering in 35S:GmAGL1 transgenic Arabidopsis plants. (A) Image of wild-type plants (WT) and transgenic plants (35S:GmAGL1) overexpressing GmAGL1 at 6 weeks after germination. (B) Comparison of leaf numbers at bolting between 35S: GmAGL1 and WT plants. Values correspond to the average leaf numbers at bolting. Error bars represent the standard deviation. An asterisk indicates a significant difference (t-test, P < 0.01) between transgenic and wild-type plants.

FIGURE 5. Abnormal morphology of flowers and siliques in 35S:GmAGL1 transgenic Arabidopsis plants. (A,B) The wild-type flowers at anthesis showing four petals surrounding the stamen and stigma. (C,D) 35S:GmAGL1 flowers. Note the complete absence of petals. (E) The siliques from three 35S:GmAGL1 lines (1–3) are short and yellowish-green with unabscised sepals, compared to those from wild-type (WT). (F) The closer view of the siliques. (G) The silique of 35S:GmAGL1 dehisced before the valves turned yellow and the seeds reach full maturity, which was much earlier than in wild-type (WT).
Moreover, phenotypic differences between the developing fruits of 35S:GmAGL1 and wild-type plants were also observed. 35S:GmAGL1 fruits were shorter in length and appeared yellowish-green compared to the long and dark green color of wild-type fruits (Figure 5E). The valve margins of 35S:GmAGL1 fruits were more remarkable than those of wild-type fruits (Figure 5F). To gain a clear observation, scanning electron microscopy was used to examine the morphological changes in the fruits. The outer epidermis of 35S:GmAGL1 fruit valves differed from that of wild-type fruit valves. The valve margins were significantly wider and thinner compared to wild-type fruits (Figures 6A–D). The microscopic cross section of the transgenic siliques cracking are showed a less condensed structure and reduced thickness (Figures 6E–H). In the premature fruits, dehiscence occurred in the valve regions while the seeds were still fresh and green, resulting in early seed dispersal (Figure 5G). These gain-of-function analyses suggest that GmAGL1 has a direct role in promoting valve development and regulating fruit elongation and dehiscence.
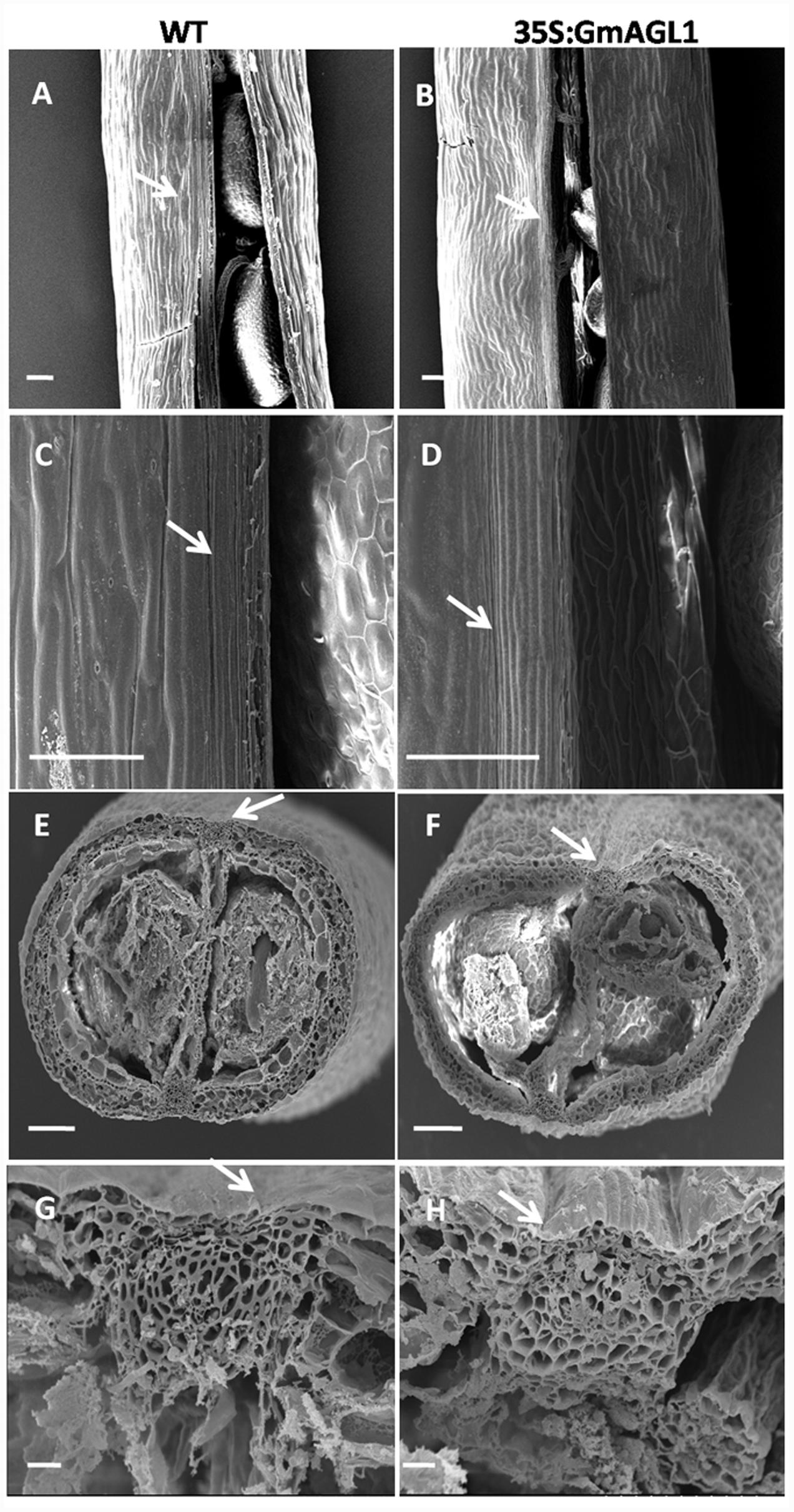
FIGURE 6. Scanning electron micrograph of 35S:GmAGL1 transgenic Arabidopsis silique. (A–D) The epidermal cells morphology in silique of wild-type and 35S:GmAGL1. (E–H) The cross sections cellular structures in siliques of wild-type and 35S:GmAGL1 plants. Arrows mark the dehiscence zones. Scale bar = 100 μm.
Expression Analysis of Genes in Transgenic Arabidopsis
GmAGL1 was localized to the nuclear compartments of plant cells, indicating that it might regulate gene expression as a transcription factor. Based on the function of GmAGL1, as revealed by its constitutive expression, we studied the expression of five genes involved in flowering and fruit development in the 35S:GmAGL1 flowers. It was observed that the Arabidopsis homologs of the fruit dehiscence related genes, ALC, IND and STK, showed more accumulation in 35S:GmAGL1 compared to wild-type plants (Figure 7). As will be described later in the Y2H screen, GmAGL1 interacted with SEP homologs. The expression levels of both SEP1 and SEP3 were increased in the 35S:GmAGL1 Arabidopsis (Figure 7). These results suggested that GmAGL1 might directly regulate the expression of these genes to control flower development and fruit dehiscence.
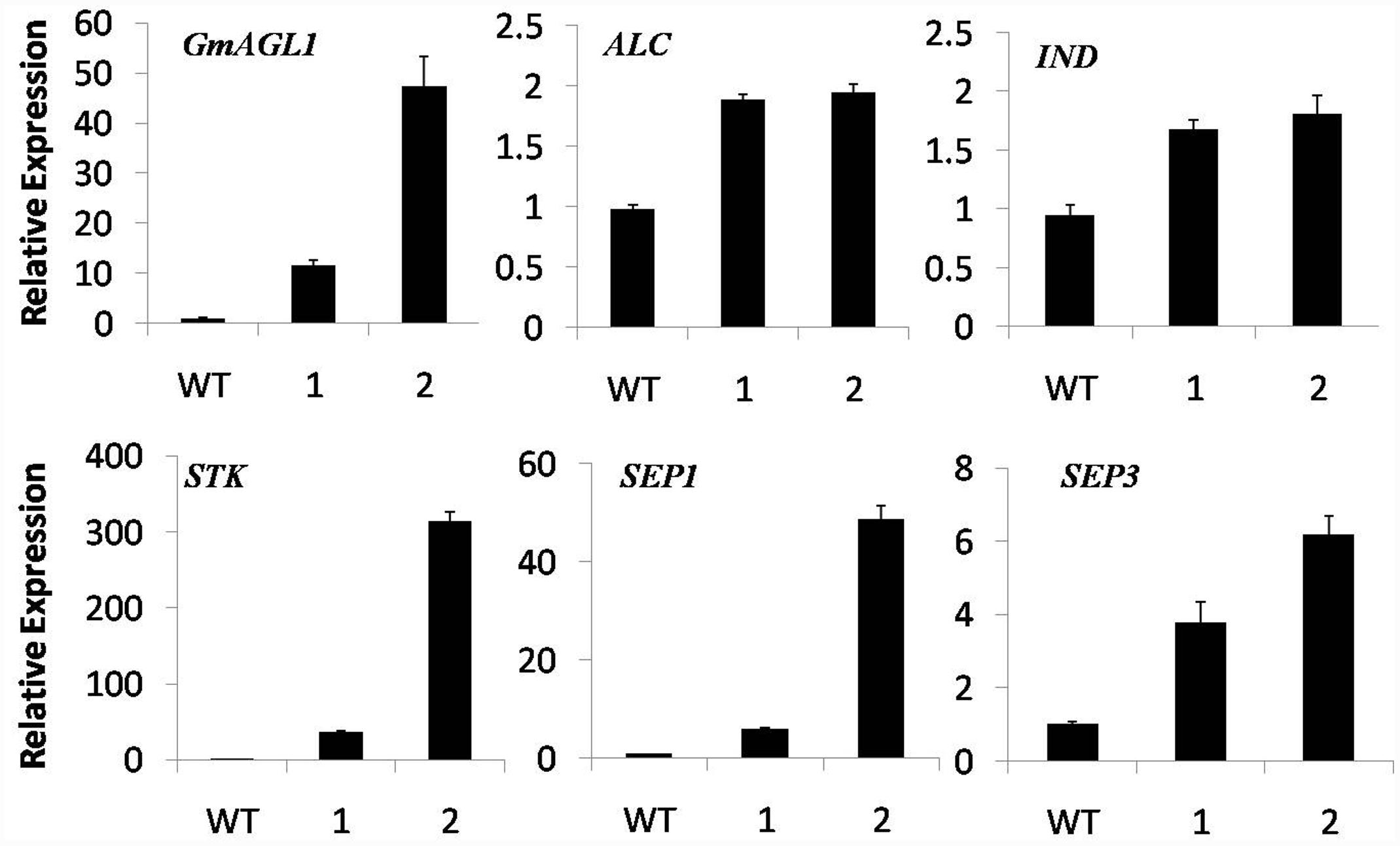
FIGURE 7. Real-time PCR analysis of flowering and fruit dehiscence related gene expression in 35S:GmAGL1 transgenic Arabidopsis flowers. The TIP4 gene was used as an internal control. The primers for quantitative RT-PCR are shown in Supplementary Table S2. The error bars represent SD based on three replicates.
Screening of GmAGL1 Interaction Proteins in Flowers
We applied a GAL4-based experiment to screen for the proteins that interact directly with GmAGL1. As shown in Supplementary Figure S2, the yeast cells containing BD-GmAGL1 could not grow on the SC-Leu-Try-His+40 mM 3AT medium, nor could the negative self-activation control. This suggests that GmAGL1 exhibits no transactivation activity and can be used as bait to perform a Y2H screen. As a result, we identified four proteins as interacting proteins of GmAGL1, which were named Interaction Protein 1 to Interaction Protein 4 (IP1∼4) (Supplementary Table S1). Two of these were SEP1-like proteins (IP1/3), one was a SEP3-like protein (IP2), and one was a putative CHUP1 protein (IP4). IP1 (Glyma18g50900), IP2 (Glyma08g11120), and IP3 (Glyma08g27670) were highly similar to homologs of SEP in the MADS-box gene family. This result suggests that GmAGL1 and SEP-like proteins in soybean can interact in specific manners and form macromolecular complexes to regulate flower and fruit development.
GmAGL1 Interacts with Soybean SEP Homologs in the Nucleus
The interactions between GmAGL1 and SEP homologs were further confirmed in vivo by BiFC assay. Two SEP functional homologs studied in soybean (Huang et al., 2009, 2014) were inserted into pUC-SPYCE (GmSEP1/3-C-YFP). Using particle bombardment, GmAGL1-N-YFP and GmSEP1/3-C-YFP were co-transformed into onion epidermal cells. Fluorescence confocal microscopy showed that BiFC signals were present in the nuclear compartments of transformed cells (Figure 8). GmAGL1-N-YFP and pUC-SPYCE, pUC-SPYNE and GmSEP1/3-C-YFP, pUC-SPYNE and pUC-SPYCE – as negative controls – were separately bombarded into onion epidermal cells, after which no fluorescence was detected (Supplementary Figure S3). These experiments demonstrated that GmAGL1 interacted with soybean SEP homologs in the nucleus.
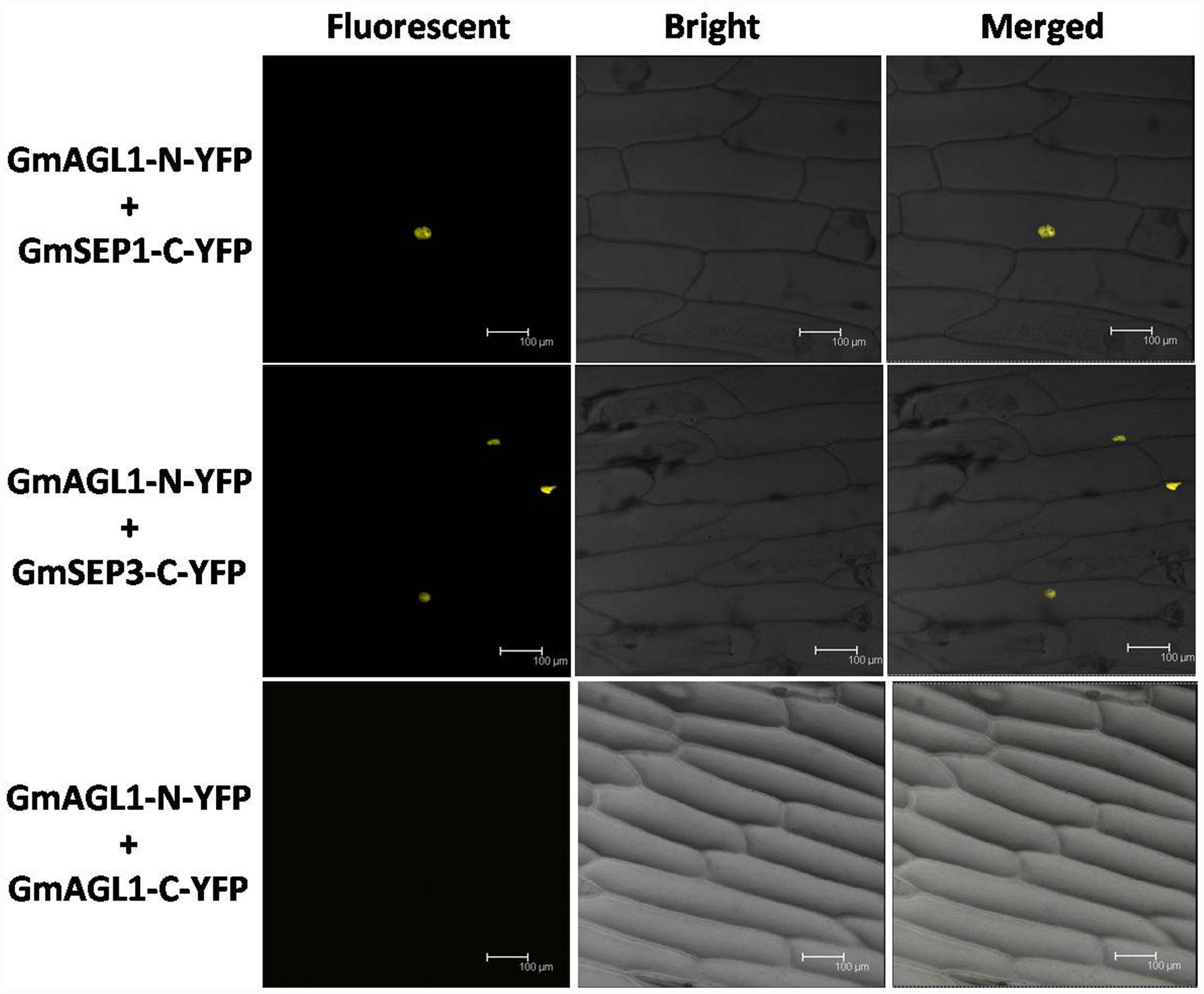
FIGURE 8. Bimolecular fluorescence complementation (BiFC) analysis of GmAGL1 interactions in planta. Fluorescence was observed in the nuclear compartment of onion epidermal cells and results from complementation of the N-terminal part of the YFP fused with GmAGL1 (GmAGL1-N-YFP) with the C-terminal part of the YFP fused with soybean SEP homologs (GmSEP1/3-C-YFP). No fluorescence was observed when GmAGL1-N-YFP was co-expressed with GmAGL1-C-YFP. Bright-field images, YFP epifluorescence images and overlay images of the same cells are shown. Scale bar = 100 μm.
GmAGL1 Cannot Form Homodimers
The BIFC system was also used to detect whether GmAGL1 can form homodimers. Full-length GmAGL1 sequences were inserted into pUC-SPYNE and pUC-SPYCE to construct the recombinant plasmids GmAGL1-N-YFP and GmAGL1-C-YFP, respectively, where GmAGL1 was in-frame fused with the N-terminus and C-terminus of YFP. GmAGL1-N-YFP and GmAGL1-C-YFP were co-expressed in onion epidermal cells through particle bombardment, but no YFP signals were found (Figure 8). These results indicated that GmAGL1 did not interact with itself in plant cells, i.e., GmAGL1 cannot form homodimers.
Discussion
The MADS-box transcription factor family is a large family that controls flower and fruit patterning in plants. To date, many members of this family have been identified and extensively studied in different plant species. However, much less is known about their functions in the developmental processes of soybean (Huang et al., 2014). In a previous study, genome wide expression profiles of soybean genes were investigated by Affymetrix Gene Chip in roots, leaves, flowers and pods (Huang et al., 2009). Some MADS-box genes were found to be primarily expressed in both flowers and pods. The roles of these genes are still not clear and require further functional analysis. In this study, we functionally characterized a soybean MADS-box gene, GmAGL1.
GmAGL1 is a MIKCc type MADS-box protein encoded by genomic DNA, as it possesses a modular structure where the MADS (M) domain is followed by an intervening (I), a keratin (K) and a C-terminal (C) domain (Theissen et al., 1996; Kaufmann et al., 2005). Two short regions of high conservation, the AG motifs I and II, were identified in the variable C domain. These two motifs are conserved in the AG subfamily (Kramer et al., 2004). Ectopic expression of an AG protein lacking the C-terminal produced ag-like flowers in transgenic Arabidopsis, indicating that these AG motifs are required for the correct function of AG proteins in plant development (Mizukami et al., 1996). Phylogenetic analyses also showed that GmAGL1 was clustered into the AG subfamily and closely allied with the PLE lineage. Members of the AG subfamily have been characterized from angiosperms as master regulators of stamen, carpel and ovule identities. They play important roles after fertilization in the developing fruits and seeds. These studies also demonstrated that AG-like genes retain functional conservation within flowering plants (Dreni and Kater, 2014). The functional conservation of AG homologs suggests that GmAGL1 might have central roles in regulating soybean floral organ identity and pod development.
Previous results showed that GmAGL1 was exclusively expressed in flowers and pods. We found that GmAGL1 was specifically expressed in carpels, pod walls and seed coats, but only weak expression was seen in sepals. The transcript was detected at all stages of flower and pod development. The expression pattern of GmAGL1 was similar to that observed for other AG-like genes. For instance, the transcript of PPERSHP was detected primarily in carpels and accumulated throughout fruit development from full anthesis until fruit harvest (Tani et al., 2007). AGL1 was preferentially expressed in particular regions of the gynoecium and ovule, only during and after floral development (Flanagan et al., 1996). The spatial and temporal expression profiles of AG-like genes are closely aligned with their conserved functions in carpel and pod development.
Consistent with a role as a transcription factor, GmAGL1 was localized in the nucleus and contained putative DNA-binding domain in the N-terminus. Nevertheless, none of the putative motifs related to transcriptional activation were found in the conserved domain analysis of GmAGL1. The full length of GmAGL1 also exhibited inactivity in the self-activation test in the Y2H system. Thus far, the ability of transactivation has not been described in AGL1 proteins. Interestingly, the proteins that directly interact with GmAGL1 were obtained from a soybean flower cDNA library by a yeast two-hybrid screen. It was also reported that AP1 and SEP3 added transcriptional activator activity to PI–AP3 and AG by the formation of complexes (Honma and Goto, 2001). Therefore, we predict that GmAGL1 might allow transcriptional regulation of target genes as an activator in a combinatorial way by interacting with other factors.
It is well-known that MADS domain proteins do not exert their functions as monomers, but rather they form multimeric protein complexes with other proteins (de Folter et al., 2005; Immink et al., 2010). In this study, a screen of a cDNA expression library with the yeast two-hybrid GAL4 system was performed to unravel the protein–protein interaction network for GmAGL1. As a result, three putative MADS-box transcription factors were identified as GmAGL1 interaction partners from soybean flowers. Interestingly, all these proteins belonged to the SEP subfamily, indicating that the function of GmAGL1 in promoting carpel identity may be based on a biochemical interaction with SEP proteins. In Arabidopsis, three SEP factors, SEP1, SEP2 and SEP3, which are closely related and functionally redundant, were necessary to determine the identities of petals, stamens, and carpels (Pelaz et al., 2000). In addition, protein-protein interaction studies have revealed that MADS-box proteins are dependent for their function of floral organ identity on the interaction with SEP proteins (Honma and Goto, 2001). These findings contributed to the proposal of the genetic ABCDE model for flower development, which demonstrated that the members of SEP proteins (E-class) act as bridges allowing the formation of higher order complexes (Theissen and Saedler, 2001). The study also showed that the complexes composed of AGL1 and SEP homologous proteins are probably able to promote carpel identity in the absence of AG and AP2 (Favaro et al., 2003).
To investigate the biological function in planta, we overexpressed GmAGL1 in transgenic Arabidopsis. We observed flowers with abnormal morphology wherein the petals were absent. The senescence and abscission of sepals were delayed in the developing transgenic siliques. These observations suggested that GmAGL1 may have a similar activity to the C function gene AG, whose overexpression caused homeotic conversion of perianth organs by suppressing A function genes (Mizukami and Ma, 1995). In Arabidopsis, SHP1/2 have retained the ability to substitute for AG activity, as the flowers constitutively expressing SHP1 or SHP2 showed a phenotype similar to those constitutively expressing AG (Liljegren et al., 2000). The introduction of 35S:SHP2 into ag mutants was sufficient to rescue stamen and carpel development (Pinyopich et al., 2003). Many studies have shown that the overexpression of AG-like genes would alter floral organ identities in the two perianth whorls (Liljegren et al., 2000; Boss et al., 2001; Vrebalov et al., 2009; Gimenez et al., 2010). In addition, GmAGL1 interacted with soybean SEP-like proteins and promoted SEP expression in Arabidopsis. SEP-like proteins are necessary to determine the identities of petals, stamens and carpels (Pelaz et al., 2000; Huang et al., 2014). GmAGL1 might interfere with the activity of petal specification genes through competition for interacting partners.
In general, fruits are derived directly from the carpels. Therefore, any mutation that affects carpel development has an effect on fruit development. GmAGL1 is also important for fruit development. The overexpression of GmAGL1 in Arabidopsis, which has a dry dehiscent fruit similar to the soybean pod, resulted in striking phenotypic effects in the 35S:GmAGL1 lines fruits, which were short, yellowish-green and early dehiscent. Microscopy revealed that valve margins were more visibly constricted in the developing transgenic fruits, which led to the dehiscence of valves before the seeds reach full maturity. In Arabidopsis, there has been significant progress in the molecular mechanisms of fruit dehiscence. ALC and IND encode two bHLH transcription factors related to Arabidopsis pod shatter, which is regulated by SHP in the genetic regulatory hierarchy (Rajani and Sundaresan, 2001; Liljegren et al., 2004). STK is a D-type MADS-box gene that is required for normal development of the funiculus, an umbilical-cord-like structure that connects the developing seed to the fruit, and for dispersal of the seeds when the fruit matures (Pinyopich et al., 2003). As expected, the overexpression of GmAGL1 activated the expression of native ALC, IND and STK, which might be the molecular mechanism for the phenotypic effects of transgenic Arabidopsis. The functional studies of other AG-like genes gave very similar results, not only in Arabidopsis (Liljegren et al., 2000) but also in tobacco (Fourquin and Ferrandiz, 2012) and tomato (Gimenez et al., 2010), suggesting that AG-like genes may play a prominent role late in fruit development and dehiscence that is generally conserved in other eudicots. Pod dehiscence is a major cause of yield loss in mechanical harvesting of soybeans, especially in several countries in tropical and sub-tropical regions (Tiwari and Bhatia, 1995). Our results indicate that further analysis of the molecular network underlying fruit dehiscence may contribute to the potential genetic manipulation of pod shattering in soybean plants.
Author Contributions
FH and DY conceived this project. YC and TW designed and performed the sequence characterization, expression profile and protein interaction of GmAGL1. GX, HY, and XZ conducted the plant transformation and microscopy analysis. YC and FH wrote the article. YS and DY contributed to scientific discussions and critical revision of manuscript. All authors reviewed the final manuscript.
Funding
This work is supported in part by the National Basic Research Program of China project from the Ministry of Agriculture of China for Transgenic Research (2015ZX08004-003), the National Basic Research Program of China (973 Program) (2010CB125906), the National Natural Science Foundation of China (31371644, 31571688, 31601324), and the Jiangsu Collaborative Innovation Center for Modern Crop Production (JCIC-MCP).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00175/full#supplementary-material
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
BD, binding domain; GFP, green fluorescent protein; ORF, open reading frame; Y2H, yeast two hybrid.
Footnotes
- ^ http://www.ncbi.nlm.nih.gov
- ^ http://smart.embl-heidelberg.de/
- ^ http://www.ncbi.nlm.nih.gov/blast
References
Arora, R., Agarwal, P., Ray, S., Singh, A. K., Singh, V. P., Tyagi, A. K., et al. (2007). MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 8:242. doi: 10.1186/1471-2164-8-242
Boss, P. K., Vivier, M., Matsumoto, S., Dry, I. B., and Thomas, M. R. (2001). A cDNA from grapevine (Vitis vinifera L.), which shows homology to AGAMOUS and SHATTERPROOF, is not only expressed in flowers but also throughout berry development. Plant Mol. Biol. 45, 541–553. doi: 10.1023/A:1010634132156
Colombo, L., Franken, J., Koetje, E., van Went, J., Dons, H. J., Angenent, G. C., et al. (1995). The Petunia mads box gene Fbp11 determines ovule identity. Plant Cell 7, 1859–1868. doi: 10.2307/3870193
de Folter, S., Immink, R. G., Kieffer, M., Parenicová, L., Henz, S. R., Weigel, D., et al. (2005). Comprehensive interaction map of the Arabidopsis MADS Box transcription factors. Plant Cell 17, 1424–1433. doi: 10.1105/tpc.105.031831
Dreni, L., and Kater, M. M. (2014). MADS reloaded: evolution of the AGAMOUS subfamily genes. New Phytol. 201, 717–732. doi: 10.1111/nph.12555
Favaro, R., Pinyopich, A., Battaglia, R., Kooiker, M., Borghi, L., Ditta, G., et al. (2003). MADS-box protein complexes control carpel and ovule development in Arabidopsis. Plant Cell 15, 2603–2611. doi: 10.1105/tpc.015123
Ferrandiz, C., Liljegren, S. J., and Yanofsky, M. F. (2000). Negative regulation of the SHATTERPROOF genes by FRUITFULL during Arabidopsis fruit development. Science 289, 436–438. doi: 10.1126/science.289.5478.436
Flanagan, C. A., Hu, Y., and Ma, H. (1996). Specific expression of the AGL1 MADS-box gene suggests regulatory functions in Arabidopsis gynoecium and ovule development. Plant J. 10, 343–353. doi: 10.1046/j.1365-313X.1996.10020343.x
Fourquin, C., and Ferrandiz, C. (2012). Functional analyses of AGAMOUS family members in Nicotiana benthamiana clarify the evolution of early and late roles of C-function genes in eudicots. Plant J. 71, 990–1001. doi: 10.1111/j.1365-313X.2012.05046.x
Gimenez, E., Pineda, B., Capel, J., Antón, M. T., Atarés, A., Pérez-Martín, F., et al. (2010). Functional analysis of the arlequin mutant corroborates the essential role of the ARLEQUIN/TAGL1 gene during reproductive development of tomato. PLoS ONE 5:e14427. doi: 10.1371/journal.pone.0014427
Goto, K., and Meyerowitz, E. M. (1994). Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 8, 1548–1560. doi: 10.1101/gad.8.13.1548
Honma, T., and Goto, K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409, 525–529. doi: 10.1038/35054083
Huang, F., Chi, Y., Gai, J., and Yu, D. (2009). Identification of transcription factors predominantly expressed in soybean flowers and characterization of GmSEP1 encoding a SEPALLATA1-like protein. Gene 438, 40–48. doi: 10.1016/j.gene.2009.03.002
Huang, F., Chi, Y., Meng, Q., Gai, J., and Yu, D. (2006). GmZFP1 encoding a single zinc finger protein is expressed with enhancement in reproductive organs and late seed development in soybean (Glycine max). Mol. Biol. Rep. 33, 279–285. doi: 10.1007/s11033-006-9012-z
Huang, F., Xu, G., Chi, Y., Liu, H., Xue, Q., and Zhao, T. (2014). A soybean MADS-box protein modulates floral organ numbers, petal identity and sterility. BMC Plant Biol. 14:89. doi: 10.1186/1471-2229-14-89
Immink, R. G., Gadella, T. W. Jr., Ferrario, S., Busscher, M., and Angenent, G. C. (2002). Analysis of MADS box protein-protein interactions in living plant cells. Proc. Natl. Acad. Sci. U.S.A. 99, 2416–2421. doi: 10.1073/pnas.042677699
Immink, R. G., Kaufmann, K., and Angenent, G. C. (2010). The ‘ABC’ of MADS domain protein behaviour and interactions. Semin. Cell Dev. Biol . 21, 87–93. doi: 10.1016/j.semcdb.2009.10.004
Jack, T., Brockman, L. L., and Meyerowitz, E. M. (1992). The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68, 683–697. doi: 10.1016/0092-8674(92)90144-2
Jofuku, K. D., den Boer, B. G., Van Montagu, M., and Okamuro, J. K. (1994). Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6, 1211–1225. doi: 10.1105/tpc.6.9.1211
Karlova, R., Chapman, N., David, K., Angenent, G. C., Seymour, G. B., and de Maagd, R. A. (2014). Transcriptional control of fleshy fruit development and ripening. J. Exp. Bot. 65, 4527–4541. doi: 10.1093/jxb/eru316
Katahata, S. I., Futamura, N., Igasaki, T., and Shinohara, K. (2014). Functional analysis of SOC1-like and AGL6-like MADS-box genes of the gymnosperm Cryptomeria japonica. Tree Genet. Genomes 10, 317–327. doi: 10.1007/s11295-013-0686-9
Kaufmann, K., Melzer, R., and Theissen, G. (2005). MIKC-type MADS-domain proteins: structural modularity, protein interactions and network evolution in land plants. Gene 347, 183–198. doi: 10.1016/j.gene.2004.12.014
Kramer, E. M., Jaramillo, M. A., and Di Stilio, V. S. (2004). Patterns of gene duplication and functional evolution during the diversification of the AGAMOUS subfamily of MADS box genes in angiosperms. Genetics 166, 1011–1023. doi: 10.1534/genetics.166.2.1011
Liljegren, S. J., Ditta, G. S., Eshed, Y., Savidge, B., Bowman, J. L., and Yanofsky, M. F. (2000). SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404, 766–770. doi: 10.1038/35008089
Liljegren, S. J., Roeder, A. H., Kempin, S. A., Gremski, K., Østergaard, L., Guimil, S., et al. (2004). Control of fruit patterning in Arabidopsis by INDEHISCENT. Cell 116, 843–853. doi: 10.1016/S0092-8674(04)00217-X
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, H., Yanofsky, M. F., and Meyerowitz, E. M. (1991). AGL1-AGL6, an Arabidopsis gene family with similarity to floral homeotic and transcription factor genes. Genes Dev. 5, 484–495. doi: 10.1101/gad.5.3.484
Mandel, M. A., Gustafson-Brown, C., Savidge, B., and Yanofsky, M. F. (1992). Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360, 273–277. doi: 10.1038/360273a0
Mizukami, Y., Huang, H., Tudor, M., Hu, Y., and Ma, H. (1996). Functional domains of the floral regulator AGAMOUS: characterization of the DNA binding domain and analysis of dominant negative mutations. Plant Cell 8, 831–845. doi: 10.1105/tpc.8.5.831
Mizukami, Y., and Ma, H. (1995). Separation of Ag function in floral meristem determinacy from that in reproductive organ identity by expressing antisense Ag Rna. Plant Mol. Biol. 28, 767–784. doi: 10.1007/BF00042064
Pelaz, S., Ditta, G. S., Baumann, E., Wisman, E., and Yanofsky, M. F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405, 200–203. doi: 10.1038/35012103
Pinyopich, A., Ditta, G. S., Savidge, B., Liljegren, S. J., Baumann, E., Wisman, E., et al. (2003). Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424, 85–88. doi: 10.1038/nature01741
Rajani, S., and Sundaresan, V. (2001). The Arabidopsis myc/bHLH gene ALCATRAZ enables cell separation in fruit dehiscence. Curr. Biol. 11, 1914–1922. doi: 10.1016/S0960-9822(01)00593-0
Robles, P., and Pelaz, S. (2005). “Flower and fruit development in Arabidopsis thaliana.” Int. J. Dev. Biol. 49, 633–643. doi: 10.1387/ijdb.052020pr
Tani, E., Polidoros, A. N., and Tsaftaris, A. S. (2007). Characterization and expression analysis of FRUITFULL- and SHATTERPROOF-like genes from peach (Prunus persica) and their role in split-pit formation. Tree Physiol. 27, 649–659. doi: 10.1093/treephys/27.5.649
Theissen, G., Kim, J. T., and Saedler, H. (1996). Classification and phylogeny of the MADS-box multigene family suggest defined roles of MADS-box gene subfamilies in the morphological evolution of eukaryotes. J. Mol. Evol. 43, 484–516. doi: 10.1007/BF02337521
Theissen, G., and Saedler, H. (2001). Plant biology. Floral quartets. Nature 409, 469–471. doi: 10.1038/35054172
Tiwari, S. P., and Bhatia, V. S. (1995). Characters of pod anatomy associated with resistance to pod-shattering in soybean. Ann. Bot. 76, 483–485. doi: 10.1006/anbo.1995.1123
Vrebalov, J., Pan, I. L., Arroyo, A. J. M., McQuinn, R., Chung, M., Poole, M., et al. (2009). Fleshy fruit expansion and ripening are regulated by the tomato SHATTERPROOF gene TAGL1. Plant Cell 21, 3041–3062. doi: 10.1105/tpc.109.066936
White, P. J. (2002). Recent advances in fruit development and ripening: an overview. J. Exp. Bot. 53, 1995–2000. doi: 10.1093/jxb/erf105
Xue, L. J., Zhang, J. J., and Xue, H. W. (2012). Genome-wide analysis of the complex transcriptional networks of rice developing seeds. PLoS ONE 7:e31081. doi: 10.1371/journal.pone.0031081
Keywords: AGAMOUS-LIKE1, soybean, fruit dehiscence, floral organ identity genes, protein interactions
Citation: Chi Y, Wang T, Xu G, Yang H, Zeng X, Shen Y, Yu D and Huang F (2017) GmAGL1, a MADS-Box Gene from Soybean, Is Involved in Floral Organ Identity and Fruit Dehiscence. Front. Plant Sci. 8:175. doi: 10.3389/fpls.2017.00175
Received: 01 July 2016; Accepted: 27 January 2017;
Published: 09 February 2017.
Edited by:
Susana Araújo, Instituto de Tecnologia Química e Biológica António Xavier (ITQB-NOVA), PortugalReviewed by:
Ana Maria Rocha De Almeida, University of California, Berkeley, USAJiayin Pang, University of Western Australia, Australia
Autar Krishen Mattoo, United States Department of Agriculture, USA
Copyright © 2017 Chi, Wang, Xu, Yang, Zeng, Shen, Yu and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fang Huang, Zmh1YW5nQG5qYXUuZWR1LmNu
 Yingjun Chi
Yingjun Chi Tingting Wang1
Tingting Wang1 Hui Yang
Hui Yang Xuanrui Zeng
Xuanrui Zeng Fang Huang
Fang Huang