- College of Life Sciences, Nanjing Normal University, Nanjing, China
Orchidaceae (orchids) is the largest family in the monocots, including about 25,000 species in 880 genera and five subfamilies. Many orchids are highly valued for their beautiful and long-lasting flowers. However, the phylogenetic relationships among the five orchid subfamilies remain unresolved. The major dispute centers on whether the three one-stamened subfamilies, Epidendroideae, Orchidoideae, and Vanilloideae, are monophyletic or paraphyletic. Moreover, structural changes in the plastid genome (plastome) and the effective genetic loci at the species-level phylogenetics of orchids have rarely been documented. In this study, we compared 53 orchid plastomes, including four newly sequenced ones, that represent four remote genera: Dendrobium, Goodyera, Paphiopedilum, and Vanilla. These differ from one another not only in their lengths of inverted repeats and small single copy regions but also in their retention of ndh genes. Comparative analyses of the plastomes revealed that the expansion of inverted repeats in Paphiopedilum and Vanilla is associated with a loss of ndh genes. In orchid plastomes, mutational hotspots are genus specific. After having carefully examined the data, we propose that the three loci 5′trnK-rps16, trnS-trnG, and rps16-trnQ might be powerful markers for genera within Epidendroideae, and clpP-psbB and rps16-trnQ might be markers for genera within Cypripedioideae. After analyses of a partitioned dataset, we found that our plastid phylogenomic trees were congruent in a topology where two one-stamened subfamilies (i.e., Epidendroideae and Orchidoideae) were sisters to a multi-stamened subfamily (i.e., Cypripedioideae) rather than to the other one-stamened subfamily (Vanilloideae), suggesting that the living one-stamened orchids are paraphyletic.
Introduction
The Orchidaceae (orchids), the largest family in the monocots (Chase et al., 2003), includes approximately 25,000 species in 880 genera and five subfamilies. They diverged from other monocots around 112 million years ago (Givnish et al., 2015). Orchids are distributed mainly in tropical and subtropical forests. Their flowers (see Figure 1) are characterized by enlarged petals (called labella) and a cylindrical structure of fused gynoecium and anther (together called the column). The latter facilitates pollination by insects (Ramírez et al., 2007) and is distinctive and attractive to humans.
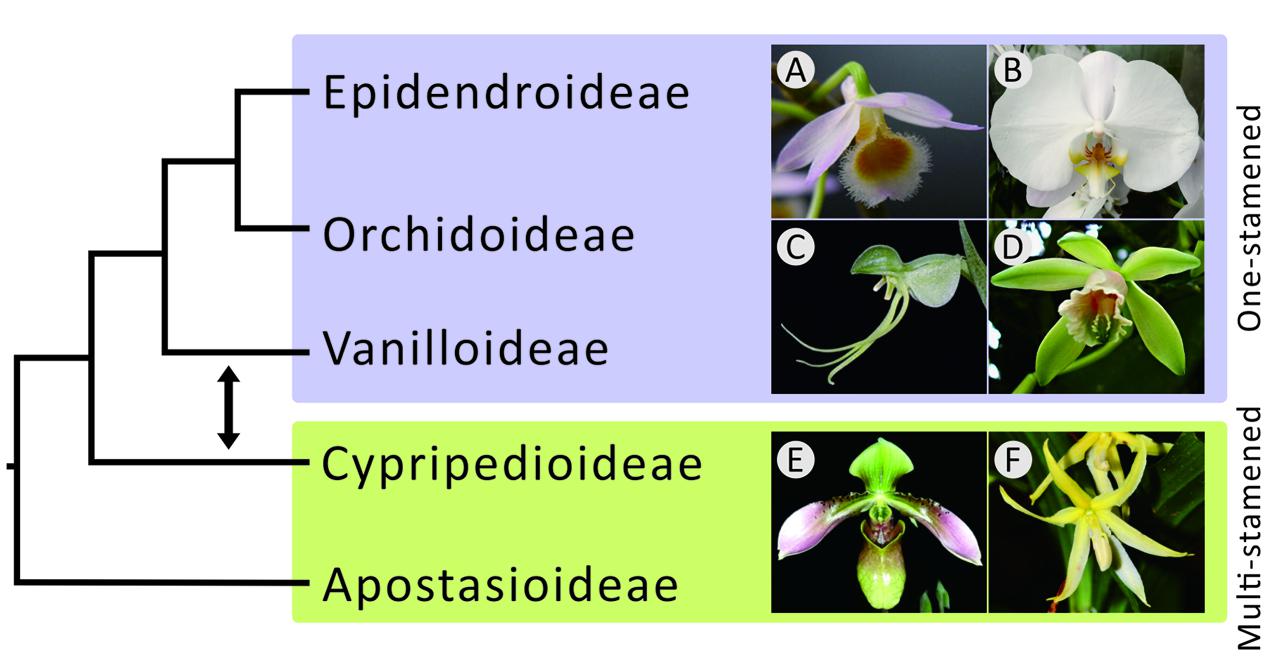
FIGURE 1. Phylogenetic relationships among the five subfamilies of orchids. The topology on the left was suggested by morphological studies that claimed a monophyletic relationship among one-stamen orchids. The double-headed arrow between Vanilloideae and Cypripedioideae shows that the positions of these two subfamilies were exchanged in molecular studies, highlighting the controversy between morphological and molecular evidence. The photos on the left are (A) Dendrobium and (B) Phalaenopsis of Epidendroideae, (C) Habenaria of Orchidoideae, (D) Vanilla of Vanilloideae, (E) Paphiopedilum of Cypripedioideae, and (F) Apostasia of Apostasioideae.
Studies on the phylogenetic relationships among the five orchid subfamilies, Apostasioideae, Cypripedioideae, Epidendroideae, Orchidoideae, and Vanilloideae (sesu Cameron), however, have long been controversial. A major debate centers on the evolution of the number of stamens. Previously, one-stamened flowers had been considered morphologically more advanced than two- and three-stamened ones (Vermeulen, 1966; Rasmussen, 1985; Szlachetko, 1995). Therefore, the three one-stamened subfamilies, Epidendroideae, Orchidoideae, and Vanilloideae, were thought to have once consisted of a super-subfamily separate from the other two subfamilies (Apostasioideae and Cypripedioideae) in early morphological studies (Rasmussen, 1985; Szlachetko, 1995). However, recent molecular studies (e.g., Górniak et al., 2010; Givnish et al., 2015; Kim H.T. et al., 2015; Lin et al., 2015) have concluded that Epidendroideae and Orchidoideae are more closely related to Cypripedioideae than to Vanilloideae (Figure 1), implying that one-stamened orchids are paraphyletic and that the analogous one-stamened floral structures are the consequence of convergent evolution.
Complete plastid genome (plastome) sequences of seed plants are useful and cost-effective for phylogenetic and evolutionary studies, because of their mostly uniparental inheritance, dense gene content, and slower evolutionary rate of change compared to nuclear genomes (Wolfe et al., 1987; Drouin et al., 2008; Smith, 2015). These features make it possible to obtain plastome sequences from the total genomic DNA using next-generation sequencing technologies (Nock et al., 2011; Kim K. et al., 2015). Plastid phylogenomics, which comparatively analyzes combined plastid genes, has been widely employed in the reassessment of several open phylogenetic questions. For instance, major groups of angiosperms (e.g., Jansen et al., 2007; Moore et al., 2010; Goremykin et al., 2013) and gymnosperms (e.g., Lin et al., 2010; Zhong et al., 2011; Wu et al., 2013) have been reassessed using this method.
The plastomes of seed plants typically contain four parts: two large inverted repeats (IRs) that separate the remaining regions into a large single copy and a small single copy (SSC) region. The boundaries of IRs are dynamic and are considered mutational hotspots as well as genetic markers useful for studying phylogenetic questions (Wu et al., 2007; Wang et al., 2008; Downie and Jansen, 2015). For example, comparative analyses of a few orchid plastomes have revealed diverse patterns of junctions between IR and SSC regions (Chang et al., 2006; Jheng et al., 2012; Kim H.T. et al., 2015). Mycoheterotrophic orchids, which do not photosynthesize, lack functional ndh genes (Delannoy et al., 2011; Logacheva et al., 2011; Barrett and Davis, 2012). However, comparative studies of plastomes have also found variable loss/retention of ndh genes loss among different photosynthetic orchid species; for example, only ndhB in Oncidium (Wu et al., 2010) and ndhE, J, and C in Cymbidium have been predicted to encode functional ndh proteins (Yang et al., 2013). Nevertheless, the mechanisms that underlie the relationship between shifts in IR boundaries in orchids and the variable loss/retention of ndh genes are not clear.
Plastomes are ideal resources for selecting the mutational hotspots of various lineages. For example, the locus matK has been employed to identify orchid species (Lahaye et al., 2008). Neubig et al. (2009) considered the locus ycf1 as a more appropriate one for low-level phylogenetic studies of orchids, because the sequence variability of this locus is greater than the universal loci rbcL and matK. Recently, the comparative plastomic method has become available for mutational hotspot selection, which uses at least two complete plastomes within the study genus to screen for the most informative regions (e.g., Ahmed et al., 2013; Downie and Jansen, 2015). This has been surveyed in many groups of flowering plants. For instance, Ahmed et al. (2013) assessed mutational hotspots in Colocasia plastomes. Downie and Jansen (2015) proposed that the combination trnD-trnY+trnE-trnT might present the greatest number of informative characters among Apiaceae species. However, plastome-wide investigation of mutational hotspots has not been conducted for all of the five orchid subfamilies.
The diverse patterns of junctions between IR and SSC regions and the independent loss of ndh genes have attracted the intense attention of researchers, leading to the publication of numerous orchid plastomes (e.g., Chang et al., 2006; Jheng et al., 2012; Yang et al., 2013). However, most plastomes that have been investigated belong to the Epidendroideae subfamily. To evaluate the evolution of the orchid subfamilies and their plastomes on the basis of a more comprehensive sampling, we examined the plastomes of three key orchid species from the Orchidoideae, Cypripedioideae, and Vanilloideae subfamilies and 67 plastid coding genes from Neuwiedia singapureana (Apostasioideae) and Cypripedium japonicum (Cypripedioideae). In addition, for Dendrobium, one of the largest and most economically important genera in Orchidaceae, only a few plastomes have been sequenced (e.g., Luo et al., 2014). Therefore, the plastome of Dendrobium moniliforme was also sequenced, which might be helpful for the exploration of mutational hotspots. In this study, we surveyed the plastomes of 41 orchid species. Including our four newly sequenced plastomes, we sampled 14 diverse genera from all of the five orchid subfamilies. We compared their structural changes and mutational hotspots to identify the level of orchid species. Furthermore, previously proposed relationships among the five orchid subfamilies were re-examined on the basis of plastid phylogenomics. An evolutionary scenario for orchid stamens was created.
Materials and Methods
Plant Materials and DNA Extraction
For each species, 2 g of fresh leaves was harvested from individuals of Cypripedium japonicum Thunb, Dendrobium huoshanense, D. loddigesii Rolfe, D. moniliforme (L.) Sw., Goodyera schlechtendaliana Rchb. f., Neuwiedia zollingeri var. singapureana (Wall. ex Baker), Paphiopedilum armeniaclum, and Vanilla aphylla Blume that had been cultivated in the greenhouse of Nanjing Normal University. Total DNA was extracted using a Qiagen DNeasy plant mini Kit (Qiagen, Germany). The quality of obtained DNA was measured on a NanoDrop 8000 Spectrophotometer (Thermo Scientific, Wilmington, DE, USA). DNA samples that passed the threshold (DNA concentrations greater than 300 ng/μL, A260/A280 = 1.8-2.0, and A260/A230 larger than 1.7) were collected for next-generation sequencing.
Plastome Sequencing, Assembly, Finishing, and Annotation
With the development of next-generation sequencing technologies, many high-throughput methods can now be used to obtain complete plastome sequences, using the Illumina platform whole-genome sequence (e.g., Kim K. et al., 2015). In this study, plastome assembly was done using the CLC Genomics Workbench 6.0.1 (CLC Bio, Aarhus, Denmark). For each species, approximately 3.7 Gb 73 bp pair-end reads were sequenced on an Illumina Genome Analyzer 2000 at Beijing Genomics Institute. After the adaptors were removed, reads were trimmed with an error probability < 0.05 and de novo assembled on the CLC Genomics Workbench 6.0.1. Contigs that had >30× sequencing depths were searched using blastn against the plastomic sequences of G. fumata (NC_026773). Matched contigs with E values < 10-10 were designated plastomic contigs. For D. moniliforme, G. schlechtendaliana, P. armeniaclum, and V. aphylla, gaps between plastomic contigs were closed using sequences of amplicons obtained via PCR with specific primers. The boundaries of IRs were confirmed by PCR assays. Genes were predicted using DOGMA (Wyman et al., 2004) and tRNAscan-SE 1.21 (Schattner et al., 2005). The exact boundaries of predicted genes were confirmed by aligning them with their orthologs from other orchid species.
Identification of Syntenic Loci and Counts of SSR Elements
The comparative plastomic method, which uses at least two complete plastomes within the study genus, is now available for mutational hotspot selection (e.g., Ahmed et al., 2013; Shaw et al., 2014). However, only 10 orchid genera contain two or more plastomes (2 in Bletilla, 5 in Corallorhiza, 11 in Cymbidium, 2 in Dendrobium, 2 in Masdevallia, 3 in Phalaenopsis, 4 in Goodyera, 2 in Cypripedium, 2 in Paphiopedilum, and 2 in Vanilla) (Supplementary Table S1). Therefore, sequences of intergenic and intronic loci (hereafter, “non-coding loci”) were retrieved from those 32 plastomes. Loci smaller than 150 bp were excluded. We identified syntenic loci on the basis of their neighboring genes; i.e., loci that are flanked by the same genes/exons in the 32 sampled plastomes were identified as syntenic. Simple sequence repeat (SSR) elements located in the syntenic loci were detected using GMATo with the criteria that the “min length” for mono-nucleotide and multi-nucleotide SSRs was set to 8 and 5 units, respectively (Wang et al., 2013). Then we counted all of the SSR elements of syntenic loci for each genus, and the polymorphic SSR loci were counted one time.
Estimation of Genetic Variability
To assess the sequence variability (SV) between congeneric species, the syntenic loci we identified were used for estimation and comparison. In total, we compared 84 pairs of congeneric species, including 1, 15, 55, 3, 1, 3, 6, 1, 1, and 1 pairs within Bletilla, Corallorhiza, Cymbidium, Dendrobium, Masdevallia, Phalaenopsis, Goodyera, Cypripedium, Paphiopedilum, and Vanilla, respectively. The sequences compared between each pair were aligned using MUSCLE 3.8.31 (Edgar, 2004) with the “refining” option. Pairwise nucleotide mutations and indel events were counted using DnaSP v5 (Librado and Rozas, 2009), with the exclusion of indels at the 5′- and 3′-ends of alignments. We adopted the methods of Shaw et al. (2005) and Downie and Jansen (2015) for calculating the SV of the examined loci. The formula was as follows: SV = (number of nucleotide mutations + the number of indel events)/(number of conserved sites + the number of nucleotide mutations + the number of indel events) × 100%.
Construction of Phylogenetic Trees
In orchids, the loss of plastid ndh genes has independently occurred among different genera (Lin et al., 2015). The truncation or complete loss of ndh genes can lead to difficulties in alignment, caused by the high rates of nucleotide substitution or the presence of many deletions and insertions. Therefore, we only extracted sequences of 67 plastid protein-coding genes common to 53 orchids and eight other monocots (Supplementary Table S1) (without ndh genes). Sequence alignments were performed using MUSCLE. After removing all gaps and ambiguous sites, the alignments of the 67 genes were concatenated using SequenceMatrix 1.8 (Vaidya et al., 2011). The partitions of the 67 concatenated genes were analyzed using PartitionFinder v1.1.1 (Lanfear et al., 2012) and the result was incorporated into construction of maximum likelihood (ML) and Bayesian inference (BI) trees using RAxML 8.0.2 (Stamatakis, 2014) and MrBayes 3.2 (Ronquist et al., 2012), respectively. Fritillaria taipaiensis and Lilium longiflorum were designated as outgroups. The robustness of the ML tree was estimated using 1,000 bootstrap replicates. We performed two independent MCMC runs in analyses of the BI tree. Each run yielded one million generations. We collected one tree per 1,000 generations. The initial 25% of the collected trees were discarded as burn-in, and the remaining ones were used to estimate posterior probability.
Results
Sequencing and Plastome Assembly
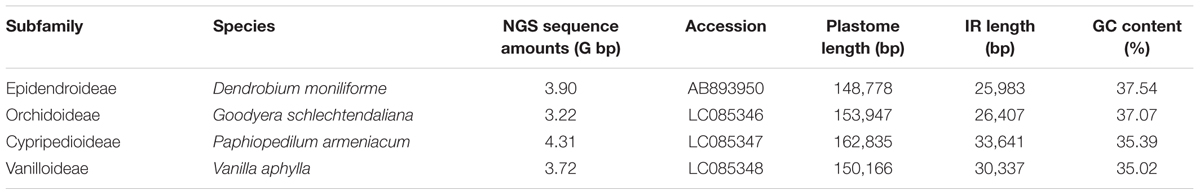
The Illumina paired-end sequencing produced approximately 3.7 Gb of 73 bp pair-end reads for each species. The de novo assembly included 39,055 contigs for D. moniliforme, 44,793 contigs for G. schlechtendaliana, 10,613 contigs for P. armeniacum, and 15,676 contigs for V. aphylla. After comparisons were conducted using the plastomic sequences of G. fumata (NC_026773), 38 contigs were obtained with E values < 10-10 and mean coverage depth > 30× for D. moniliforme, 83 contigs for G. schlechtendaliana, 87 contigs for P. armeniacum, and 47 contigs for V. aphylla. Two contigs (length = 86,931 bp and 35,926 bp) resulted in a nearly complete draft genome for D. moniliforme. Four contigs longer than 20 kb were used for G. schlechtendaliana assembly, and five contigs (longer than 20 kb) and three contigs (longer than 40 kb) were used to assemble the plastomes of P. armeniacum and V. aphylla, respectively. After assembly and gap closure, four complete plastomes were obtained.
Plastome Features of Four Newly Sequenced Orchids
The newly sequenced plastomes of D. moniliforme (Epidendroideae), G. schlechtendaliana (Orchidoideae), P. armeniacum (Cypripedioideae), and V. aphylla (Vanilloideae) are circular with IRs (i.e., IRA and IRB) separated by large single-copy and SSC regions (Figure 2). They range in size from 148,778 to 162,835 bp, with a GC content between 35.02 and 37.54% (Table 1). These four orchid plastomes differ in IR lengths, from 25,983 to 33,641 bp. The genes located in the IRs also vary greatly. The IRs of both D. moniliforme and G. schlechtendaliana contain a small fraction of the gene ycf1. By contrast, the IR of P. armeniacum encompasses three genes, ycf1, rps15, and psaC. That of V. aphylla includes ycf1, rps15, trnL, and a partial segment of ccsA, although the ycf1 has mutated with numerous stop codons (Figure 2).
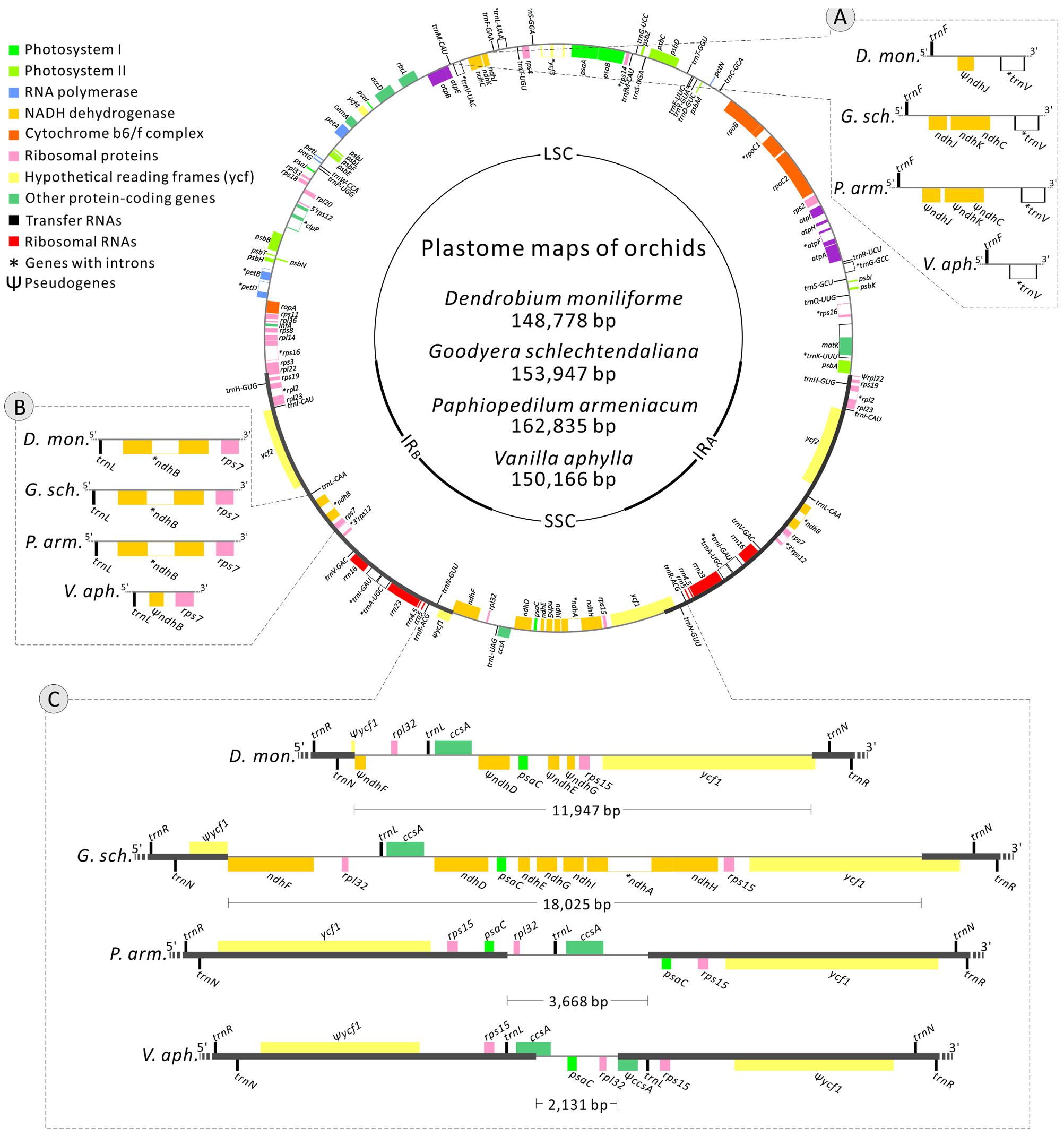
FIGURE 2. Plastomes of Dendrobium moniliforme, Goodyera schlechtendaliana, Paphiopedilum armeniacum, and Vanilla aphylla. Only the plastome of G. schlechtendaliana is illustrated and shown as a circular map, in which genes outside and inside the circle are transcribed clockwise and counterclockwise, respectively. Three major distinct regions among the four plastomes are compared and depicted in the blocks (A–C). Note that in block (C), differences in the expansion of IRs and loss of ndh genes have resulted in highly variable lengths of SSC regions among the four orchid plastomes sampled.
Among the four orchid plastomes sequenced, only G. schlechtendaliana contains a full set of 11 plastid ndh genes. By contrast, D. moniliforme and P. armeniacum have truncated or lost 10 of them (only the ndhB gene is functional; Figure 2). No functional ndh gene was detected in the plastome of V. aphylla (Figure 2). Because the four sequenced orchids belong to four different subfamilies, their distinct loss/retention of ndh genes reflects multiple events of plastid gene loss during their evolution. Furthermore, the loss of plastid ndh genes and the expansion of IRs have caused drastic reductions in SSC regions. For example, the length of the SSC region in V. aphylla is only about one-eighth of that in G. schlechtendaliana (Figure 2).
Compared to the plastomes, which have full sets of ndh genes (Supplementary Table S2), the IR/SSC boundaries of Vanilla and Paphiopedilum have expanded to a highly anomalous position. We determined the degree of IR expansion/contraction based on the length of a region from the 5′ end of the ycf1 gene to the IR and SSC junction. A two-sided Mann–Whitney test showed that the IR lengths in Vanilla and Paphiopedilum plastomes were significantly different from plastomes with 11 functional ndh genes (P < 0.05). These results indicate that ndh genes play an important role in IR/SSC junction stability.
Diverse Mutational Hotspots among Orchid Genera
We identified 68 syntenic non-coding loci. SSRs within these loci were also counted (Supplementary Table S3). In these plastomes, the ratios of SSR-containing loci to SSR-lacking loci are significantly (two-sided Fisher’s exact test, all P < 0.05) higher in the SC than in the IR regions, indicating that the distribution of SSRs is dependent on their locations in plastomes. The mean values of pairwise interspecific SV were compared and the results are shown in Supplementary Table S3. The 68 syntenic intergenic spacers and introns were sorted into SC and IR loci, depending on their location. Our analysis, using a two-sided Mann–Whitney test, indicated that in all of the 10 orchid genera examined, the SC loci have significantly greater SV than the IR loci (all P < 0.05) (Supplementary Table S3). Moreover, mean SV was inversely correlated with mean GC content in all of the 10 orchid genera (Spearman’s r = -0.496 to -0.80, all P < 0.01), suggesting that highly mutated loci have also evolved toward the enrichment of AT nucleotides. To determine whether the evolution of SV is also conserved among the orchid genera, we conducted a correlation test of the 68 syntenic loci between and within different subfamilies. Except for Cymbidium vs. Bletilla (Spearman’s r = 0.767, P < 0.01), the SV values of the 68 syntenic loci within Epidendroideae were statistically uncorrelated (Corallorhiza vs. Phalaenopsis, r = 0.238, P > 0.05), slightly correlated (r = 0.238 to 0.488, all P < 0.05), or intermediately correlated (r = 0.513 to 0.675, all P < 0.05). Moreover, no strongly correlated relationship between different subfamilies was detected (Corallorhiza vs. Vanilla, r = 0.086, P < 0.05, others r = 0.243 to 0.675, all P < 0.05). These results suggest that in orchid plastomes, the evolution of SV is genus specific.
Figure 3 shows the top 10 loci that have the greatest SV for each genus. These loci are exclusively mutational hotspots. Obviously, most of them contain SSRs. However, none of the hotspots is common to all 10 genera, and only four of them (5′trnK-rps16, trnS-trnG, clpP-psbB, and rps16-trnQ) are present in more than four examined genera of Epidendroideae. In addition, among these four hotspots, only two, clpP-psbB and rps16-trnQ, are present in the two genera of Cypripedioideae. Therefore, we conclude that plastomic mutational hotspots are diverse among orchid genera.
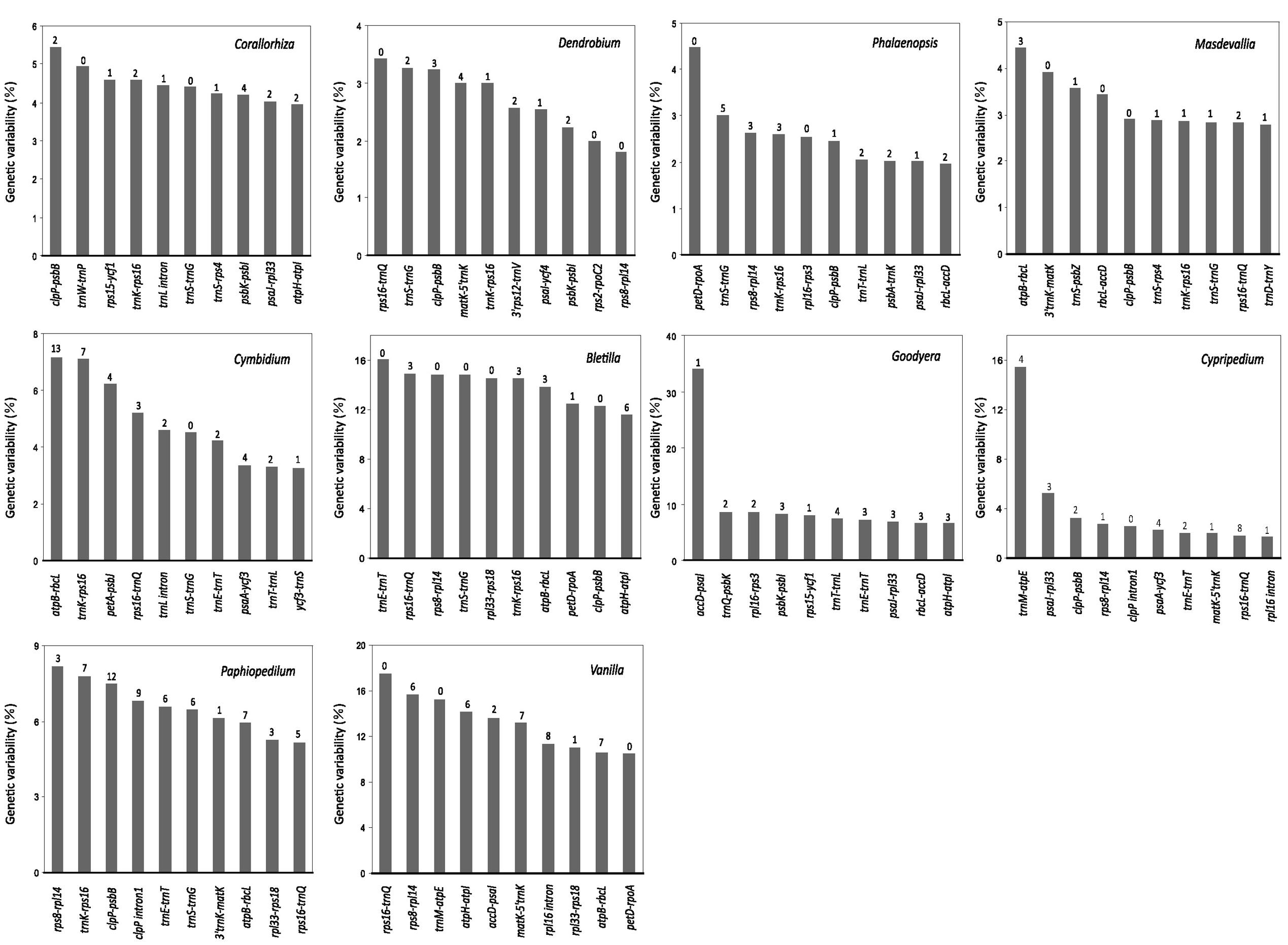
FIGURE 3. The 10 syntenic intergenic and intronic loci with the highest genetic variability in the plastome from each of the 10 examined orchid genera. The Arabic numeral shown on the top of each gray bar denotes the number of SSR elements within the corresponding locus. The SD values of Corallorhiza, Cymbidium, Dendrobium, Phalaenopsis, and Goodyera are shown in Supplementary Table S3.
Phylogenetic Relationships among the Five Orchid Subfamilies
We performed phylogenetic analyses of 67 concatenated plastid genes with or without partitions of sequences. Both ML and BI trees recovered a monophyly of the five orchid subfamilies, irrespective of whether or not the partitions of sequences were incorporated (Figure 4 and Supplementary Table S3). However, without partitioning, the ML and BI trees were inconsistent in the inferred positions of Neottia and Epipogium. As shown in Supplementary Figure S1, the ML tree placed both Neottia and Epipogium within Epidendroideae, whereas these two genera are placed as sister to Orchidoideae rather than to other species of Epidendroideae in the BI tree. However, with partitioning (Supplementary Table S4), both trees congruently placed Neottia and Epipogium within Epidendroideae (Figure 4 and Supplementary Table S4) and the trees no longer contradicted each other in their topologies (Figure 4 and Supplementary Table S1).
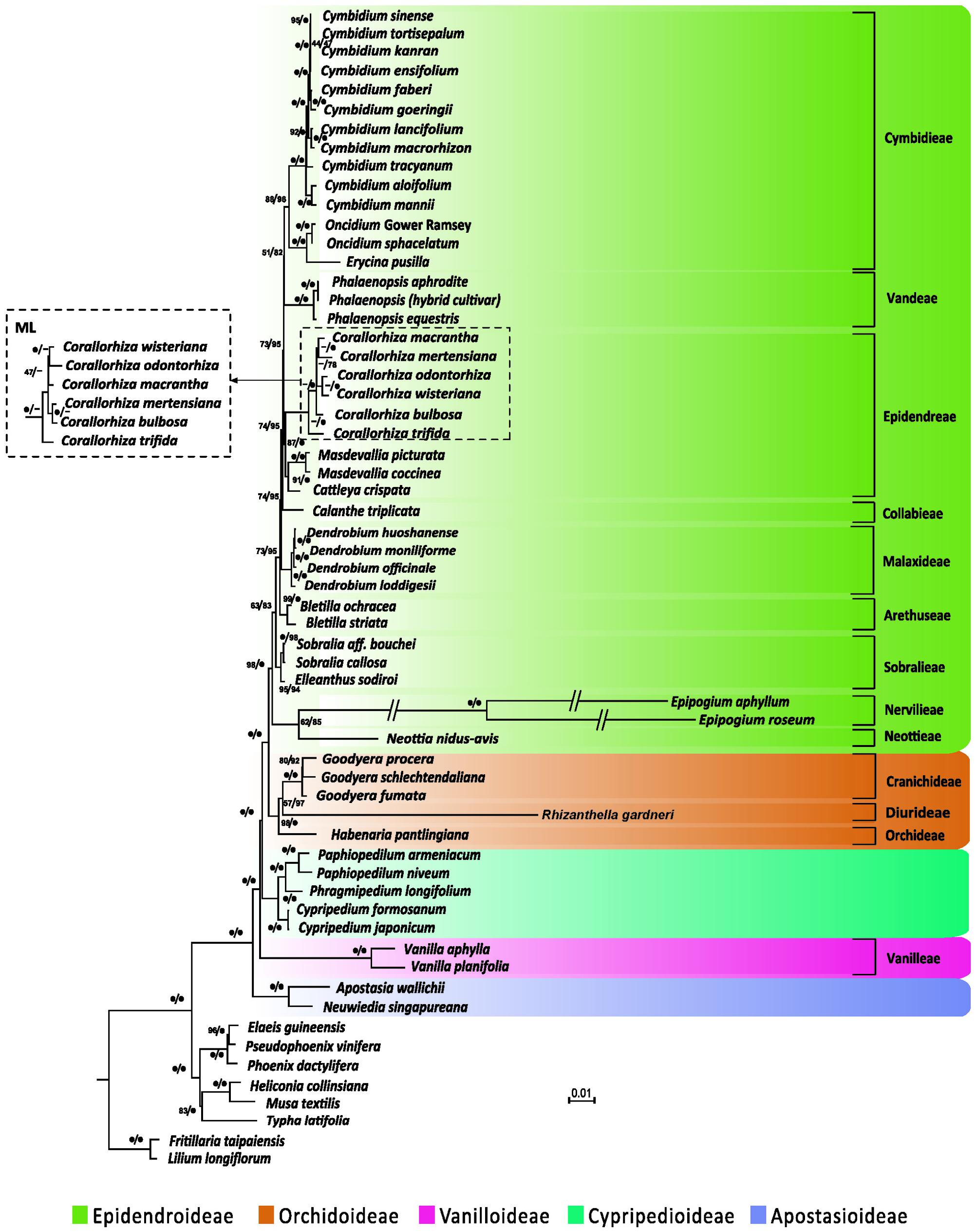
FIGURE 4. Plastid phylogenomics of orchid subfamilies based on partitioned analyses. Only the BI tree is shown, because its topology is nearly identical to that of the obtained ML tree. The only differences are the relationships among Corallorhiza species, which are shown in the dashed box. Clade support values in the tree are bootstrap supports/posterior probabilities in percent from RAxML/Mayas. 100% support; -conflict between the ML tree and the BI tree. Branches marked with // are shortened by 50%.
The Apostasioideae, whose members have multiple stamens, is the earliest diverging subfamily of orchids. The Vanilloideae, although consisting of one-stamened members, is closely related to a multi-stamened subfamily (i.e., Cypripedioideae) rather than to the other two one-stamened subfamilies (i.e., Epidendroideae and Orchidoideae). Therefore, on the basis of our phylogenetic analyses, the one-stamened orchids are paraphyletic. This suggests that the number of stamens evolved independently at least twice during the evolution of orchids.
Discussion
Expansion/Contraction of IRs is Associated with the Loss of ndh Genes
In orchids, the lost plastid ndh genes were previously hypothesized to be functionally replaced by their putative nuclear homologs of plastid origin (Chang et al., 2006). However, analyses of nuclear transcriptome have failed to detect the complete set of the 11 ndh genes in some orchid species whose plastids lack ndh genes (Johnson et al., 2012; Su et al., 2013; Tsai et al., 2013; Lin et al., 2015). Considering the notable variation in the loss/retention of ndh genes among the four newly elucidated plastomes (Figure 2), we suggest that multiple ndh loss events have occurred during the evolution of orchids. This is in good agreement with the proposition that loss of ndh genes has occurred independently among orchid genera and subfamilies (Kim H.T. et al., 2015; Lin et al., 2015).
Plastid ndh proteins form a complex that joins cyclic electron transport at photosystem I (Martín and Sabater, 2010). Based on the existence of the PGR5-dependent cyclic electron transport pathway and the fact that no deleterious effects have been observed in ndh-deficient mutants growing under favorable conditions, Ruhlman et al. (2015) deduced that plastid ndh genes might be dispensable in contemporary plants. Nevertheless, the loss of ndh genes has led to structural changes in the plastomes of orchids, as shown by Kim H.T. et al. (2015) and us here.
The structure and gene content of the mycoheterotrophy orchid plastome are highly variable in photosynthetic orchids (Delannoy et al., 2011; Logacheva et al., 2011; Barrett and Davis, 2012). Our comparative analyses found that the Vanilla and Paphiopedilum plastomes have also expanded their IRs to a highly anomalous position. However, the mechanisms that underlie shifts of their IR boundaries are not clear. Kim H.T. et al. (2015) proposed that the instability of the IR/SSC junctions in orchids is strongly correlated with the deletion of the ndhF gene. Our two-sided Mann–Whitney test also indicated that ndh genes play an important role in IR/SSC junction stability. The loss of the full set of 11 plastid ndh genes has been reported in diverse photosynthetic seed plants, such as gnetophytes (Braukmann et al., 2009; Wu et al., 2009, 2011), orchids (Chang et al., 2006; Wu et al., 2010; Pan et al., 2012; Lin et al., 2015), pines (Braukmann et al., 2009; Wu et al., 2013), slender naiads (Peredo et al., 2013), and saguaros (Sanderson et al., 2015). With the exception of pines and saguaros, which have only one IR copy, all lineages mentioned above have expanded their IRs to include some genes (i.e., ycf1, rpl32, trnL, rps15, psaC, or ccsA) that are usually situated in the SSC regions of seed plant plastomes. Given that plastid ndh genes were helpful for the instability of IR/SSC junctions and that the expansion of IRs is common to ndh-deleted plastomes, we infer that shifts of IR boundaries might be associated with the variable loss of ndh genes in the Vanilla and Paphiopedilum plastomes. As the expansion of IRs directly causes gene duplication (Figure 2), whether it can benefit adaptation is worthy of further investigation.
Diverse Mutational Hotspots among Orchid Genera
It has been shown that plastome-wide comparisons facilitate the screening of mutational hotspots used for intraspecies discrimination (Ahmed et al., 2013; Hsu et al., 2014) and phylogenetic studies at the species level (Shaw et al., 2014; Downie and Jansen, 2015). In orchids, numerous non-coding plastid loci have been shown to be useful in species-level phylogenetic studies. For instance, the non-coding loci rpl32-trnL, trnE-trnT, trnH-psbA, trnK-rps16, and trnT-trnL may be markers for identifying species of Cymbidium (Yang et al., 2013), and trnS-trnG, psaC-ndhE, clpP-psbB, rpl16 intron, rpoB-trnC, trnT-psbD, rbcL-accD, rpl32-trnL, ccsA-ndhD, and ndhC-trnV may be useful for Phalaenopsis (Shaw et al., 2014). However, none of these was detected in IRs of the orchid plastomes, nor were the loci we report in Figure 3. These data reinforce the view that plastid substitution rates are considerably lower in IRs than in SC regions (Wolfe et al., 1987; Smith, 2015; Wu and Chaw, 2015).
Different mutational hotspots have been used for phylogenetic and identification analyses in orchid species, however, these have not been proposed to be diverse mutational hotspots shared among orchid genera (e.g., Neubig et al., 2009; Yang et al., 2013; Luo et al., 2014). In this study, our correlation analyses revealed that SV between and within different orchid subfamilies are uncorrelated or not strongly correlated, which suggests that in orchid plastomes, the evolution of SV is genus specific (Supplementary Table S3). Furthermore, the top 10 loci we screened as most likely containing the highest degrees of genetic variability in orchids are quite diversified (Figure 3). These results indicate that a weakly evolving locus in one orchid genus might be a quite variable locus in another genus.
Although highly variable mutational hotspots are diverse among orchid genera, we propose that the three loci 5′trnK-rps16, trnS-trnG, and rps16-trnQ might be powerful markers for genera within Epidendroideae, and clpP-psbB and rps16-trnQ might be useful for Cypripedioideae, for two reasons. The three hotspots 5′trnK-rps16, trnS-trnG, and rps16-trnQ are present in more than four genera of Epidendroideae, and the hotspots clpP-psbB and rps16-trnQ are present in the two examined genera of Cypripedioideae (Figure 3). Additionally, among these hotspots, 5′trnK-rps16, trnS-trnG, and rps16-trnQ are located in one of the three most variable plastome regions, matK to 3′trnG, as determined by Shaw et al. (2014) based on comparisons of non-coding loci among different plant genera. Our results are in good agreement with Shaw et al. (2014) and Downie and Jansen (2015). These authors identified the same highly variable loci in several disparate plant lineages.
Our findings will be helpful in identifying mutational hotspots representing the orchid subfamily level. However, there is still an indispensable need for the careful discovery and characterization of loci specific to all genera in orchids.
Evolution of Orchid Subfamilies
However, molecular phylogenetic studies of orchid to date have failed to agree on the placement of Cypripedioideae and Vanilloideae (Cameron et al., 1999; Freudenstein and Chase, 2001; Cameron, 2004, 2007; Górniak et al., 2010; Givnish et al., 2015; Kim H.T. et al., 2015). Recently, Givnish et al. (2015) and Kim H.T. et al. (2015) reconstructed ML trees of 39 and 14 orchid genera, respectively, using the concatenated nucleotide sequences of plastid genes. The results of our tree-based partitioned analyses (Figure 4) are congruent with theirs in the relationships among the five orchid subfamilies and in the monophyly of the clade Epidendroideae–Orchidoideae–Cypripedioideae. The monophyly of Epidendroideae, Orchidoideae, and Cypripedioideae, which is strongly supported in our plastid phylogenomics analyses, is also consistent with a number of previous studies that used increased taxon sampling or different molecular markers (e.g., Cozzolino and Widmer, 2005; Górniak et al., 2010; Lin et al., 2015). Neither their nor our results support the view suggested by morphological studies (Vermeulen, 1966; Rasmussen, 1985; Szlachetko, 1995) that the one-stamened subfamilies Epidendroideae, Orchidoideae, and Vanilloideae are monophyletic. Moreover, morphological studies have also shown that, in development, the single fertile anthers of the Vanilloideae are not homologous to those of the Orchidoideae and Epidendroideae (Freudenstein et al., 2002). Therefore, we conclude that one-stamened orchids are paraphyletic.
The generic relationships found in our partitioned analyses within Epidendroideae are largely congruent with those of recent studies (e.g., Górniak et al., 2010; Freudenstein and Chase, 2015; Givnish et al., 2015; Kim H.T. et al., 2015), with only some genera being weakly supported. This may be attributable to the absence of numerous photosynthesis-related genes from Corallorhiza, Epipogium, and Neottia, resulting in large amounts of missing data for these taxa (Kim H.T. et al., 2015). Moreover, incomplete taxon sampling has likely also resulted in the inconsistent placement of the tribe of Epidendreae and Sobralieae found in previous studies (Górniak et al., 2010; Freudenstein and Chase, 2015; Givnish et al., 2015) and ours. We anticipate that increases in the breadth of taxon sampling will improve the resolution of orchid phylogenies.
Conclusion
This study reports the complete plastome sequences of four orchid species: D. moniliforme, G. schlechtendaliana, P. armeniaclum, and V. aphylla. These four plastomes differ in their shifts of IR boundaries and the variable loss/retention of ndh genes. A two-sided Mann–Whitney test indicated that ndh genes play an important role in IR/SSC junction stability. Our comparative analyses suggested that shifts of IR boundaries might be associated with the variable loss of ndh genes in Vanilla and Paphiopedilum plastomes. Moreover, our analyses revealed that the mutational hotspots among orchid genera were highly variable. After careful examination of the data, we propose that the three loci 5′trnK-rps16, trnS-trnG, and rps16-trnQ could be powerful markers for genera within Epidendroideae, and clpP-psbB and rps16-trnQ could be useful for Cypripedioideae. We used different orchid species and plastid genes than those used in previous studies, however, both our and previous results indicate that the one-stamened orchids are paraphyletic. The data presented here will be helpful for settling phylogenetic relationships among the five orchid subfamilies. They will provide an informative and valuable genetic resource of orchid species for studies of changes in plastome structure, selection of mutational hotspots, and reconstruction of phylogenetic trees.
Author Contributions
XD: designed the study. SZ, JS, and WL: performed the experiments with regular advice from XD and ZN. ZN and QX: analyzed the data. ZN: wrote the manuscript. All authors approved the final version of the manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (Grant No. 31170300 and 31670330) and the Priority Academic Program Development of Jiangsu Higher Education Institutions to XD.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Mr. Huang Mingzhong (Chinese Academy of Tropical Agricultural Sciences) for his assistance collecting orchid samples and photos. We also thank Professor Chaw Shu-Miaw and Dr. Wu Chung-Shien from Academia Sinica for providing suggestions and comments.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00715/full#supplementary-material
FIGURE S1 | Plastid phylogenomics of orchid subfamilies based on partitioned analyses. Only bootstrap supports and posterior probabilities in percentages less than 100 are shown on the trees.
References
Ahmed, I., Matthews, P. J., Biggs, P. J., Naeem, M., Mclenachan, P. A., and Lockhart, P. J. (2013). Identification of chloroplast genome loci suitable for high-resolution phylogeographic studies of Colocasia esculenta (L.) Schott (Araceae) and closely related taxa. Mol. Ecol. Resour. 13, 929–937. doi: 10.1111/1755-0998.12128
Barrett, C. F., and Davis, J. I. (2012). The plastid genome of the mycoheterotrophic Corallorhiza striata (Orchidaceae) is in the relatively early stages of degradation. Am. J. Bot. 99, 1513–1523. doi: 10.3732/ajb.1200256
Braukmann, T. W., Kuzmina, M., and Stefanović, S. (2009). Loss of all plastid ndh genes in Gnetales and conifers: extent and evolutionary significance for the seed plant phylogeny. Curr. Genet. 55, 323–337. doi: 10.1007/s00294-009-0249-7
Cameron, K. M. (2004). Utility of plastid psaB gene sequences for investigating intrafamilial relationships within Orchidaceae. Mol. Phylogenet. Evol. 31, 1157–1180. doi: 10.1016/j.ympev.2003.10.010
Cameron, K. M. (2007). “Molecular phylogenetics of Orchidaceae: the first decade of DNA sequencing,” in Orchid Biology Reviews and Perspectives, Vol. IX, eds J. Arditti, T. Kull, and K. M. Cameron (New York, NY: The New York Botanical Garden Press), 163–200.
Cameron, K. M., Chase, M. W., Whitten, W. M., Kores, P. J., Jarrell, D. C., Albert, V. A., et al. (1999). A phylogenetic analysis of the Orchidaceae: evidence from rbcL nucleotide sequences. Am. J. Bot. 86, 208–224. doi: 10.2307/2656938
Chang, C. C., Lin, H. C., Lin, I. P., Chow, T. Y., Chen, H. H., Chen, W. H., et al. (2006). The chloroplast genome of Phalaenopsis aphrodite (Orchidaceae): comparative analysis of evolutionary rate with that of grasses and its phylogenetic implications. Mol. Biol. Evol. 23, 279–291. doi: 10.1093/molbev/msj029
Chase, M. W., Cameron, K. M., Barrett, R. L., and Freudenstein, J. V. (2003). “DNA data and Orchidaceae systematics: a new phylogenetic classification,” in Orchid Conservation, eds K. W. Dixon, S. P. Kell, R. L. Barrett, and P. J. Cribb (Kota Kinabalu: Natural History Press), 69–89.
Cozzolino, S., and Widmer, A. (2005). Orchid diversity: an evolutionary consequence of deception? Trends Ecol. Evol. 20, 487–494.
Delannoy, E., Fujii, S., Colas des Francs-Small, C., Brundrett, M., and Small, I. (2011). Rampant gene loss in the underground orchid Rhizanthella gardneri highlights evolutionary constraints on plastid genomes. Mol. Biol. Evol. 28, 2077–2086. doi: 10.1093/molbev/msr028
Downie, S. R., and Jansen, R. K. (2015). A comparative analysis of whole plastid genomes from the Apiales: expansion and contraction of the inverted repeat, mitochondrial to plastid transfer of DNA, and identification of highly divergent noncoding regions. Syst. Bot. 40, 336–351. doi: 10.1600/036364415X686620
Drouin, G., Daoud, H., and Xia, J. (2008). Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. Mol. Phylogenet. Evol. 49, 827–831. doi: 10.1016/j.ympev.2008.09.009
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Freudenstein, J. V., and Chase, M. W. (2001). Analysis of mitochondrial nad1b-c intron sequences in Orchidaceae: utility and coding of length-change characters. Syst. Bot. 26, 643–657.
Freudenstein, J. V., and Chase, M. W. (2015). Phylogenetic relationships in Epidendroideae (Orchidaceae), one of the great flowering plant radiations: progressive specialization and diversification. Ann. Bot. 115, 665–681. doi: 10.1093/aob/mcu253
Freudenstein, J. V., Harris, E. M., and Rasmussen, F. N. (2002). The evolution of anther morphology in orchids: incumbent anthers, superposed pollinia, and the vandoid complex. Am. J. Bot. 89, 1747–1755. doi: 10.3732/ajb.89.11.1747
Givnish, T. J., Spalink, D., Ames, M., Lyon, S. P., Hunter, S. J., Zuluaga, A., et al. (2015). Orchid phylogenomics and multiple drivers of their extraordinary diversification. Proc. Biol. Sci. B 282, 2108–2111. doi: 10.1098/rspb.2015.1553
Goremykin, V. V., Nikiforova, S. V., Biggs, P. J., Zhong, B., Delange, P., Martin, W., et al. (2013). The evolutionary root of flowering plants. Syst. Biol. 62, 50–61. doi: 10.1093/sysbio/sys070
Górniak, M., Paun, O., and Chase, M. W. (2010). Phylogenetic relationships with Orchidaceae based on a low-copy nuclear-coding gene, Xdh: congruence with organellar and nuclear ribosomal DNA results. Mol. Phylogenet. Evol. 56, 784–795. doi: 10.1016/j.ympev.2010.03.003
Hsu, C. Y., Wu, C. S., and Chaw, S. M. (2014). Ancient nuclear plastid DNA in the yew family (taxaceae). Genome Biol. Evol. 6, 2111–2121. doi: 10.1093/gbe/evu165
Jansen, R. K., Cai, Z., Raubeson, L. A., Daniell, H., Depamphilis, C. W., Leebens-Mack, J., et al. (2007). Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. U.S.A. 104, 19369–19374. doi: 10.1073/pnas.0709121104
Jheng, C. F., Chen, T. C., Lin, J. Y., Chen, T. C., Wu, W. L., and Chang, C. C. (2012). The comparative chloroplast genomic analysis of photosynthetic orchids and developing DNA markers to distinguish Phalaenopsis, orchids. Plant Sci. 190, 62–73. doi: 10.1016/j.plantsci.2012.04.001
Johnson, M. T., Carpenter, E. J., Tian, Z., Bruskiewich, R., Burris, J. N., Carrigan, C. T., et al. (2012). Evaluating methods for isolating total RNA and predicting the success of sequencing phylogenetically diverse plant transcriptomes. PLoS ONE 7:e50226. doi: 10.1371/journal.pone.0050226
Kim, H. T., Kim, J. S., Moore, M. J., Neubig, K. M., Williams, N. H., Whitten, W. M., et al. (2015). Seven new complete plastome sequences reveal rampant independent loss of the ndh gene family across orchids and associated instability of the inverted repeat/small single-copy region boundaries. PLoS ONE 10:e0142215. doi: 10.1371/journal.pone.0142215
Kim, K., Lee, S. C., Lee, J., Yu, Y., Yang, T. J., Choi, B. S., et al. (2015). Complete chloroplast and ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species. Sci. Rep. 5:15655. doi: 10.1038/srep15655
Lahaye, R., van der Bank, M., Bogarin, D., Warner, J., Pupulin, F., Gigot, G., et al. (2008). DNA barcoding the floras of biodiversity hotspots. Proc. Natl. Acad. Sci. U.S.A. 105, 2923–2928. doi: 10.1073/pnas.0709936105
Lanfear, R., Calcott, B., Ho, S. Y., and Guindon, S. (2012). Partitionfinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29, 1695–1701. doi: 10.1093/molbev/mss020
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Lin, C. P., Huang, J. P., Wu, C. S., Hsu, C. Y., and Chaw, S. M. (2010). Comparative chloroplast genomics reveals the evolution of Pinaceae genera and subfamilies. Genome Biol. Evol. 2, 504–517. doi: 10.1093/gbe/evq036
Lin, C. S., Chen, J. J., Huang, Y. T., Chan, M. T., Daniell, H., Chang, W. J., et al. (2015). The location and translocation of ndh genes of chloroplast origin in the Orchidaceae family. Sci. Rep. 5:9040. doi: 10.1038/srep09040
Logacheva, M. D., Schelkunov, M. I., and Penin, A. A. (2011). Sequencing and analysis of plastid genome in mycoheterotrophic orchid Neottia nidus-avis. Genome Biol. Evol. 3, 1296–1303. doi: 10.1093/gbe/evr102
Luo, J., Hou, B. W., Niu, Z. T., Liu, W., Xue, Q. Y., and Ding, X. Y. (2014). Comparative chloroplast genomes of photosynthetic orchids: insights into evolution of the orchidaceae and development of molecular markers for phylogenetic applications. PLoS ONE 9:e99016. doi: 10.1371/journal.pone.0099016
Martín, M., and Sabater, B. (2010). Plastid ndh genes in plant evolution. Plant Physiol. Biochem. 48, 636–645. doi: 10.1016/j.plaphy.2010.04.009
Moore, M. J., Soltis, P. S., Bell, C. D., Burleigh, J. G., and Soltis, D. E. (2010). Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc. Natl. Acad. Sci. U.S.A. 107, 4623–4628. doi: 10.1073/pnas.0907801107
Neubig, K. M., Whitten, W. M., Carlsward, B. S., Blanco, M. A., Endara, L., Williams, N. H., et al. (2009). Phylogenetic utility of ycf1 in orchids: a plastid gene more variable than matK. Plant Syst. Evol. 277, 75–84. doi: 10.1007/s00606-008-0105-0
Nock, C. J., Waters, D. L. E., Edwards, M. A., Bowen, S. G., Rice, N., Cordeiro, G. M., et al. (2011). Chloroplast genome sequences from total DNA for plant identification. Plant Biotechnol. J. 9, 328–333. doi: 10.1111/j.1467-7652.2010.00558.x
Pan, I. C., Liao, D. C., Wu, F. H., Daniell, H., Singh, N. D., Chang, C., et al. (2012). Complete chloroplast genome sequence of an orchid model plant candidate: Erycina pusilla apply in tropical Oncidium breeding. PLoS ONE 7:e34738. doi: 10.1371/journal.pone.0034738
Peredo, E. L., King, U. M., and Les, D. H. (2013). The plastid genome of Najas flexilis: adaptation to submersed environments is accompanied by the complete loss of the NDH complex in an aquatic angiosperm. PLoS ONE 8:e68591. doi: 10.1371/journal.pone.0068591
Ramírez, S. R., Gravendeel, B., Singer, R. B., Marshall, C. R., and Pierce, N. E. (2007). Dating the origin of the Orchidaceae from a fossil orchid with its pollinator. Nature 448, 1042–1045. doi: 10.1038/nature06039
Rasmussen, F. N. (1985). “The families of the monocotyledones - structure, evolution and taxonomy,” in Orchids, eds R. Dahlgren, H. T. Cliford, and P. F. Yeo (Berlin: Springer Verlag Press), 249–274.
Ronquist, F., Teslenko, M., Mark, P. V. D., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Ruhlman, T. A., Chang, W. J., Chen, J. J., Huang, Y. T., Chan, M. T., Jin, Z., et al. (2015). NDH expression marks major transitions in plant evolution and reveals coordinate intracellular gene loss. BMC Plant Biol. 15:100. doi: 10.1186/s12870-015-0484-7
Sanderson, M. J., Copetti, D., Búrquez, A., Bustamante, E., Charboneau, J. L., Eguiarte, L. E., et al. (2015). Exceptional reduction of the plastid genome of saguaro cactus (Carnegiea gigantea): loss of the ndh gene suite and inverted repeat. Am. J. Bot. 102, 1115–1127. doi: 10.3732/ajb.1500184
Schattner, P., Brooks, A. N., and Lowe, T. M. (2005). The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33, W686–W689. doi: 10.1093/nar/gki366
Shaw, J., Lickey, E. B., Beck, J. T., Farmer, S. B., Liu, W., Miller, J., et al. (2005). The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am. J. Bot. 92, 142–166. doi: 10.3732/ajb.92.1.142
Shaw, J., Shafer, H. L., Leonard, O. R., Kovach, M. J., Schorr, M., and Morris, A. B. (2014). Chloroplast DNA sequence utility for the lowest phylogenetic and phylogeographic inferences in angiosperms: the tortoise and the hare IV. Am. J. Bot. 101, 1987–2004. doi: 10.3732/ajb.1400398
Smith, D. R. (2015). Mutation rates in plastid genomes: they are lower than you might think. Genome Biol. Evol. 7, 1227–1234. doi: 10.1093/gbe/evv069
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Su, C., Chao, Y. T., Yen, S. H., Chen, C. Y., Chen, W. C., Chang, Y. C. A., et al. (2013). Orchidstra: an integrated orchid functional genomics database. Plant Cell Physiol. 54, e11. doi: 10.1093/pcp/pct004
Tsai, W. C., Fu, C. H., Hsiao, Y. Y., Huang, Y. M., Chen, L. J., Wang, M., et al. (2013). OrchidBase 2.0: comprehensive collection of Orchidaceae floral transcriptomes. Plant Cell Physiol. 54, e7. doi: 10.1093/pcp/pcs187
Vaidya, G., Lohman, D. J., and Meier, R. (2011). SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27, 171–180. doi: 10.1111/j.1096-0031.2010.00329.x
Wang, R. J., Cheng, C. L., Chang, C. C., Wu, C. L., Su, T. M., and Chaw, S. M. (2008). Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots. BMC Evol. Biol. 8:36. doi: 10.1186/1471-2148-8-36
Wang, X., Lu, P., and Luo, Z. (2013). GMATo: a novel tool for the identification and analysis of microsatellites in large genomes. Bioinformation 9, 541–544. doi: 10.6026/97320630009541
Wolfe, K. H., Li, W. H., and Sharp, P. M. (1987). Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl. Acad. Sci. U.S.A. 84, 9054–9058. doi: 10.1073/pnas.84.24.9054
Wu, C. S., and Chaw, S. M. (2015). Evolutionary stasis in cycad plastomes and the first case of plastome GC-biased gene conversion. Genome Biol. Evol. 7, 2000–2009. doi: 10.1093/gbe/evv125
Wu, C. S., Chaw, S. M., and Huang, Y. Y. (2013). Chloroplast phylogenomics indicates that Ginkgo biloba is sister to cycads. Genome Biol. Evol. 5, 243–254. doi: 10.1093/gbe/evt001
Wu, C. S., Lai, Y. T., Lin, C. P., Wang, Y. N., and Chaw, S. M. (2009). Evolution of reduced and compact chloroplast genomes (cpDNAs) in gnetophytes: selection toward a lower-cost strategy. Mol. Phylogenet. Evol. 52, 115–124. doi: 10.1016/j.ympev.2008.12.026
Wu, C. S., Wang, Y. N., Hsu, C. Y., Lin, C. P., and Chaw, S. M. (2011). Loss of different inverted repeat copies from the chloroplast genomes of Pinaceae and Cupressophytes and influence of heterotachy on the evaluation of gymnosperm phylogeny. Genome Biol. Evol. 3, 1284–1295. doi: 10.1093/gbe/evr095
Wu, C. S., Wang, Y. N., Liu, S. M., and Chaw, S. M. (2007). Chloroplast genome (cpDNA) of Cycas taitungensis and 56 cp protein-coding genes of gnetum parvifolium: insights into cpdna evolution and phylogeny of extant seed plants. Mol. Biol. Evol. 24, 1366–1379. doi: 10.1093/molbev/msm059
Wu, F. H., Chan, M. T., Liao, D. C., Hsu, C. T., Lee, Y. W., Daniell, H., et al. (2010). Complete chloroplast genome of Oncidium Gower Ramsey and evaluation of molecular markers for identification and breeding in Oncidiinae. BMC Plant Biol. 10:68. doi: 10.1186/1471-2229-10-68
Wyman, S. K., Jansen, R. K., and Boore, J. L. (2004). Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20, 3252–3255. doi: 10.1093/bioinformatics/bth352
Yang, J. B., Tang, M., Li, H. T., Zhang, Z. R., and Li, D. Z. (2013). Complete chloroplast genome of the genus Cymbidium: lights into the species identification, phylogenetic implications and population genetic analyses. BMC Evol. Biol. 13:84. doi: 10.1186/1471-2148-13-84
Keywords: orchids, plastome, ndh gene, mutational hotspots, plastid phylogenomic
Citation: Niu Z, Xue Q, Zhu S, Sun J, Liu W and Ding X (2017) The Complete Plastome Sequences of Four Orchid Species: Insights into the Evolution of the Orchidaceae and the Utility of Plastomic Mutational Hotspots. Front. Plant Sci. 8:715. doi: 10.3389/fpls.2017.00715
Received: 10 October 2016; Accepted: 18 April 2017;
Published: 03 May 2017.
Edited by:
Xiaowu Wang, Biotechnology Research Institute (CAAS), ChinaReviewed by:
Shuhua Yang, Institute of Vegetables and Flowers (CAAS), ChinaTae-Jin Yang, Seoul National University, South Korea
Copyright © 2017 Niu, Xue, Zhu, Sun, Liu and Ding. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoyu Ding, ZGluZ3h5bmpAMjYzLm5ldA==
 Zhitao Niu
Zhitao Niu Xiaoyu Ding
Xiaoyu Ding