- 1Key Laboratory of Tropical Plant Resources and Sustainable Use, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Kunming, China
- 2College of Life Sciences, University of Chinese Academy of Sciences, Beijing, China
SNF 1-RELATED PROTEIN KINASE 2 (SnRK2) is a family of plant-specific protein kinases which is the key regulator of hyper-osmotic stress signaling and abscisic acid (ABA)-dependent development in various plants. Among the rice subclass-I and -II SnRK2s, osmotic stress/ABA–activated protein kinase 2 (SAPK2) may be the primary mediator of ABA signaling. However, SAPK2 has not been comprehensively characterized. In this study, we elucidated the functional properties of SAPK2 using loss-of-function mutants produced with the CRISPR/Cas9 system. The SAPK2 expression level was strongly upregulated by drought, high-salinity, and polyethylene glycol (PEG) treatments. The sapk2 mutants exhibited an ABA-insensitive phenotype during the germination and post-germination stages, suggesting that SAPK2 had a pivotal role related to ABA-mediated seed dormancy. The sapk2 mutants were more sensitive to drought stress and reactive oxygen species (ROS) than the wild-type plants, indicating that SAPK2 was important for responses to drought conditions in rice. An additional investigation revealed that SAPK2 increased drought tolerance in the following two ways: (i) by reducing water loss via the accumulation of compatible solutes, promoting stomatal closure, and upregulating the expression levels of stress-response genes such as OsRab16b, OsRab21, OsbZIP23, OsLEA3, OsOREB1 and slow anion channel (SLAC)-associated genes such as OsSLAC1 and OsSLAC7; (ii) by inducing the expression of antioxidant enzyme genes to promote ROS-scavenging abilities that will ultimately decrease ROS damages. Moreover, we also observed that SAPK2 significantly increased the tolerance of rice plants to salt and PEG stresses. These findings imply that SAPK2 is a potential candidate gene for future crop improvement studies.
Introduction
Rice (Oryza sativa) is a cereal that is a staple food crop for many people worldwide. The primary challenge for increasing rice production is overcoming a global water shortage, which can severely limit rice yields (Zhang, 2007). Drought stress induces various morphological and physiological changes in plants. Under drought or salinity stress conditions, H2O2 and abscisic acid (ABA) are often generated in many biological systems.
In plants, drought and salinity stresses usually cause oxidative damage via the production of reactive oxygen species (ROS), such as H2O2 and superoxide (Zhu, 2001; Mittler, 2002; Xiong and Zhu, 2002). High ROS-scavenging activities decrease the overaccumulation of ROS in plants, thereby inhibiting the onset of programmed cell death (Mittler, 2002; Farooq et al., 2009; Hou et al., 2009). Plant responses to the overaccumulation of ROS involve two mechanisms. The main ROS-scavenging mechanism depends on the activation of antioxidant enzymes, including superoxide dismutase (SOD), ascorbate peroxidase (APX), and catalase (CAT). The other mechanism relies on non-enzymatic molecular compounds such as ascorbic acid (AsA) (Jabs et al., 1996; Mittler, 2002; Xiong and Zhu, 2002; Farooq et al., 2009; Hou et al., 2009; Das and Roychoudhury, 2014).
Abscisic acid has a central role in seed dormancy, germination, and the acquisition of desiccation tolerance (Wasilewska et al., 2008). Since ABA was discovered in the 1960s, many details regarding the physiological importance of this hormone have been documented (Weiner et al., 2010). The ABA signaling pathway has been characterized as a signaling complex consisting of PYR/PYL/RCAR-type ABA receptors, the type A protein phosphatase 2Cs (PP2Cs), and sucrose non-fermenting 1-related protein kinases 2 (SnRK2s) (Fujii et al., 2009; Ma et al., 2009; Melcher et al., 2009). In the absence of ABA, the ABA signaling pathway is repressed via dephosphorylation and inactivation of SnRK2s by PP2Cs. As ABA accumulates and is perceived, RCAR/PYR receptors bind to PP2Cs and SnRK2s are released. The SnRK2s then phosphorylate downstream substrates, and activate ABA-responsive gene expression or other responses (Fujii et al., 2009; Ma et al., 2009; Park et al., 2009; Santiago et al., 2009; Umezawa et al., 2009).
Previous studies have confirmed that protein phosphorylation and dephosphorylation play crucial roles in abiotic stress responses (Sopory and Munshi, 1998; Umezawa et al., 2013). SNF 1-related protein kinases (SnRKs), which constitute a plant-specific kinase group, take their name from their fungal homolog SNF1 (AMP-activated protein kinase). In plants, the SnRK family comprises 38 members which can be subdivided into three sub-families: SnRK1, SnRK2, and SnRK3. Compelling evidence suggests that SnRK2s are involved in ABA and/or stress signaling pathways. Ten plant-specific SnRK2s have been identified in Arabidopsis thaliana (SnRK2.1-2.10) and rice (SAPK1-10) (Yoshida et al., 2002; Hrabak et al., 2003; Kobayashi et al., 2004; Halford and Hey, 2009). The ten members of SnRK2s are divided into three subclasses based on their domain structures. In A. thaliana, subclasses I contains SnRK2.1, SnRK2.4, SnRK2.5, SnRK2.9 and SnRK2.10; subclasses II contains SnRK2.7 and SnRK2.8; subclasses III contains SnRK2.2, SnRK2.3, and SnRK2.6. In A. thaliana, all SnRK2 except SnRK2.9 were activated by hyperosmotic stress, and five members are activated by ABA (Boudsocq et al., 2004). Three members of subclasses III, SRK2E/OST1/SnRK2.6, SRK2D/SnRK2.2 and SRK2I/SnRK2.3, have been sufficiently studied. The snrk2.2/2.3 double mutant, but not snrk2.2 or snrk2.3 single mutants, showed insensitive to ABA in seed germination and root growth inhibition (Fujii et al., 2007). The snrk2.6 (ost1) mutant was insensitive to ABA and did not exhibit ABA induced stomatal closure (Mustilli et al., 2002). In the snrk2.2/3/6 triple mutant all examined ABA responses are blocked (Fujii et al., 2009; Fujita et al., 2009). These results demonstrate that SnRK2.2, 2.3, and 2.6 are functional redundancy in ABA regulation of guard cells, seed germination and seedling growth. All members of SnRK2 subclass I, SnRK2.1/SRK2G, SnRK2.4/SRK2.4A, SnRK2.5/SRK2H and SnRK2.10/SRK2B are activated by osmotic stress, but not by ABA (Boudsocq et al., 2004). SnRK2.4 and SnRK2.10 were primarily identified in a screen for PA-binding proteins (Testerink et al., 2004). McLoughlin et al. (2012) proved that SnRK2.4 and SnRK2.10 were rapidly activated in response to salt and involved in the maintenance of root system architecture under saline conditions. Protein kinase SnRK2.10/SRK2B was involved in drought tolerance by phosphorylating a conserved motif found in the stress-related dehydrin (LEA) proteins (Vlad et al., 2008). SnRK2.4 was shown to play a role for membrane association in the response to salt stress (McLoughlin et al., 2012). Members of SnRK2s subclass II, SRK2F/SnRK2.7 and SRK2C/SnRK2.8, were strongly activated by hyperosmotic stress but weakly activated by ABA in A. thaliana and the activation of SRK2C/SnRK2.8 and SRK2F/SnRK2.7 by ABA was considerably lower than that of SRK2D/E/I (SnRK2.2/2.3/2.6) (Boudsocq et al., 2004, 2007). SRK2C/SnRK2.8 positively regulated drought tolerance in A. thaliana roots, and overexpression of SRK2.8-GFP increased drought tolerance due to up-regulation of stress-responsive gene in A. thaliana (Umezawa et al., 2004). SRK2C/SnRK2.8 was also shown to function in metabolic processes in plant growth and overexpression of SRK2C/SnRK2.8 enhanced plant growth in A. thaliana (Shin et al., 2007). Under drought stress, the wild type, snrk2.7, snrk2.8, and the snrk2.7/2.8 double mutant, did not show any clear differences in survival or visible damage, but microarray analysis suggested that subclass II SnRK2s regulate some drought-responsive genes involving ABA responsive element binding transcription factors (AREB/ABF) and their targets (Mizoguchi et al., 2010). These studies suggest subclass II SnRK2s play important roles in drought stress signaling in A. thaliana.
All 10 osmotic stress/ABA–activated protein kinases (SAPKs), which encode SnRK2 family protein kinases in the rice genome, are activated by hyperosmotic stress, while SAPKs 8–10 are also strongly activated by ABA (Kobayashi et al., 2004). In rice, subclasses I, II, and III of SnRK2s, contain SAPK3 through SAPK7, SAPK1 and SAPK2, and SAPK8 through SAPK10, respectively (Kobayashi et al., 2004). Many studies have demonstrated that subclasses I and III of SnRK2 family members are important for stress responses and development in rice. For example, Diédhiou et al. (2008) reported that the overexpression of SAPK4 increased salt tolerance by changing the expression level of genes related to ion homeostasis and oxidative stress responses. Chae et al. (2007) revealed that ectopic expression of OSRK1 (SAPK6) in transgenic tobacco plants resulted in reduced sensitivity to ABA during seed germination and root elongation. Nam et al. (2012) proved that the overexpression of OSRK1 (SAPK6) enhanced phosphorylation activities and salt-sensitive root growth in rice. Lin et al. (2015) observed that SAPK8, 9 and 10 phosphorylated the Tiller Enhancer protein and inhibited the interaction between this protein and OsPYL10. Furthermore, SAPK9 was reported to positively regulate ABA-mediated stress signaling pathways in rice (Dey et al., 2016). Kim et al. (2012) proved that the constitutive expression of OsPYL/RCAR5 upstream of SAPK2 leaded to an ABA-hypersensitive phenotype during seed germination and early seedling growth in rice. Recently, studies indicated that OsbZIP23 and OsbZIP46 were phosphorylated by SAPKs to increase drought stress tolerance in rice (Tang et al., 2012; Zong et al., 2016). These data indicate that SAPK2 is probably crucial for ABA-mediated abiotic stress responses and developmental controls in rice, although SAPK2 has not been fully characterized. Therefore, clarifying the specific function of SAPK2 in rice is necessary for developing abiotic stress-tolerant transgenic rice.
In this study, we used the CRISPR/Cas9 system to generate sapk2 mutants. Expression analyses revealed that SAPK2 expression was strongly induced by drought, NaCl, and polyethylene glycol (PEG) treatments, but not by ABA. Germination assays proved that the sapk2 mutants exhibited an ABA-insensitive phenotype during the germination and post-germination stages. Drought tolerance assays indicated that sapk2 mutant lines were sensitive to drought conditions, with lower survival rates than the wide-type plants. The mutants also exhibited greater water loss, lower proline and soluble sugar contents, higher proportions of fully open stomata, higher ROS levels, and lower antioxidant enzyme activities. Quantitative reverse transcription polymerase chain reaction (qRT-PCR) analyses indicated that SAPK2 improved drought tolerance by enhancing the expression of stress-responsive and antioxidant enzyme genes in rice. We also observed that SAPK2 increased the tolerance of rice to salt and PEG stresses. Overall, these results confirmed that SAPK2 was an important positive regulator of ABA-dependent abiotic stress responses and developmental controls in rice.
Materials and Methods
Mutant Generation Methods
We employed the CRISPR/Cas9 system to generate sapk2 mutants. The CRISPR/Cas9 plasmid was designed according to the protocol described previously (Liang et al., 2016). Concisely, the third coding exon of SAPK2 was selected for guide RNA design. Double-strand DNA generated by annealing the oligo pairs, and then was cloned into the pYLCRISPR/Cas9Pubi-H vector. Rice (O. sativa L. japonica.) was used for transformation. Then transgenetic seedlings were kept in growth chamber at 28°C under long-day conditions (14 h light/10 h dark cycles). For mutation detection, genomic DNA extracted from mutant seedlings (all plant) were used for PCR. Then PCR products (sequence is in Supplementary Table S3) were identified by comparing the 19-bp gRNA target sequences (tagttatggaatatgctgc) to the rice reference genome (sequence is in Supplementary Table S3) (Liu et al., 2015). In the T0 generation, we collected 20 hygromycin-resistant plants. Based on mutation detection results, we identified two independent homozygous mutant lines in the T1 generation, which we named sapk2-1 and sapk2-7.
The primers used for CRISPR/Cas9 (U3-SAPK2-F and U3-SAPK2-R) and mutation detection (SAPK2-u3-F and SAPK2-u3-R) were listed in the Supplementary Table S1.
Plant Growth Conditions and Stress Treatments
Two sapk2 mutant lines (sapk2-1 and sapk2-7) and wide type rice (O. sativa L. japonica.) were selected for the experimental works. All the experiments were performed using the seeds of same harvest and storage conditions. The sterilized seeds from different genotypes were germinated on half-strength Murashige and Skoog (MS) medium simultaneously and kept in growth chamber at 28°C under long-day conditions (14 h light/10 h dark cycles). After 1 week germination, seedlings were then transplanted into the same pot with same amount of soil for drought stress or transferred into half-strength liquid medium for PEG and NaCl treatments.
To detect the transcript level of target genes under different stresses, wide type plants were grown in the same condition for 4 weeks. Then seedlings were treated with ABA or other different abiotic stresses. We performed ABA treatment by spraying leaves with 0.1 mM ABA. For drought stress, seedlings were exposed to the air without water supply. For PEG and NaCl treatments, Seedlings were transferred into half-strength liquid medium supplemented with 25% PEG6000 (m/v) or 175mM NaCl. With these treatments, leaves were sampled at 0, 3, 6, 12, and 24 h.
RNA Extraction and qRT-PCR Analyses
For the qRT-PCR analysis, we used the same method as described (Jiang et al., 2016). Total RNA was isolated from rice seedlings using the TriZol reagent (Invitrogen). The cDNAs were obtained by using Superscript II in accordance with manufacturer’s instructions (Invitrogen). The qRT-PCR analysis was performed using SYBR Premix Ex Taq kit (Takara).
At least three independent biological experiments were conducted (three independent samples were conducted for each experiment and three technological replications in every independent experiment). One representative result was displayed here. Gene-specific primers used in qRT-PCR analysis were listed in Supplementary Table S1.
GUS Staining of SAPK2
For GUS reporter analysis, the putative promoters of SAPK2 were amplified from genomic DNA using primers Promoter-SAPK2-F and Promoter-SAPK2-R (Supplementary Table S1). The fused ProSAPK2-GUS was cloned into the P1300 vector. Samples from sapk2 mutant lines and wide type plants were harvested before and after drought stress, GUS staining was detected as described (He et al., 2014).
Germination and Early Seedling Growth Assays
To test seed germination, seeds from sapk2 mutant lines and wide type plants were surface-sterilized (75% ethanol for 5min, 40% NaClO for 30 min) and subsequently were washed at least five times with sterilized water. Then sterilized seeds were planted on half-strength MS agar medium containing 0, 2, or 5 μM ABA. Germination was recognized as complete when the coleoptiles were 5 mm long according the reported protocol (Lin et al., 2015). The germination rates were assessed at designated time (i.e., 0, 36, 48, 60, 72, 84, 96, 108, and 120 h). To test the growth performance at post-germination stage, the seeds of sapk2 mutant lines and wide type plants were germinated on half-strength MS agar medium simultaneously. After 3 days, the seedlings were transferred to half-strength MS agar medium supplemented with 0, 2, or 5 μM ABA for further growth. Twenty seedlings were used to measure the shoot and root length. All these experiments were repeated three times, with 30 seeds per sample. Three independent experimental replications were conducted.
Drought Tolerance Assays
After germinated simultaneously on half-strength MS agar medium for 7 days, 4-week-old seedlings of sapk2 mutant lines and wide type plants were transplanted into the same pot with the same amount of soil. The drought phenotype was identified after withdrawing water for 7 days and re-watering for 7 days. Three independent experiments repeated at the same time and a representative result was displayed. Three independent experimental replications were conducted.
Analyses for Water Loss Rates and Electrolyte Leakage, Content of Proline (Pro), Soluble Sugar and Malondialdehyde (MDA), Staining of Trypan Blue, 3, 3′-Diaminobenzidine (DAB), and Nitroblue Tetrazolium (NBT)
For the measurement of water loss rates (WLRs), detached flag leaves of sapk2 mutants and wide type plants were placed in weighing dishes and left on the laboratory bench with light at room temperature. Fresh weight was weighed at indicated times (0–200 min). WLR was expressed as a percentage of initial fresh weight.
The relative ion leakage was checked following the method of Clarke et al. (2004) with slight modifications. Approximately 1g detached flag leaves from sapk2 mutants and wide type plants were cut into 5 mm length and placed in test tubes containing 10 ml deionized water. The tubes were covered with plastic caps and placed in a water bath maintained at the constant temperature of 22°C for 2 h. The conductance of H2O was measured by conductivity meter (HORIBA TWIN COND B-173).
The content of free proline in leaves was estimated as described (Bates et al., 1973) with slight modifications. Approximately 0.5 g leaf segments from mutants and wide type plants are ground into powder with liquid nitrogen, and then homogenized with 10 ml of 3% sulphosalicylic acid in tube. Then centrifuged at 3,000 × g for 20 min and collect the supernatant. 2 ml of supernatant was reacted with 2 ml acid ninhydrin and 2 ml glacial acetic acid in a test tube at 100°C for 1 h. Following cooled on ice. And then the absorbance at 520 was measured by spectrophotometer. L-Pro was used as a standard to calculate the proline concentration.
Total soluble sugar content in leaves was determined using anthrone reagent (Zhang and Huang, 2013). Approximately 0.5 g dried leaf segments from mutants and wide type plants were ground into powder with liquid nitrogen, and then homogenized with 2 ml 80% ethanol in shaker at 200 rpm in 50 ml tube for 1 h. Following centrifuge at 6,000 × g for 10 min, and then transfer as much supernatant as possible into a new 5 ml tube. Add equal volume of chloroform, completely mix, and then centrifuge at 12,000 × g for 10 min. The aqueous part is transferred to a new tube, 50 μl of each is mixed with 4.95 ml anthrone reagent and then boiled for 15 min. Measure the optical density of glucose standards at 620 nm by spectrophotometer.
The content of MDA in leaves was detected as described (Heath and Packer, 1968) with slight modifications. Approximately 1g leaf segments from mutants and wide type plants were homogenized in 10 ml of 10% trichloroacetic (v/v) and centrifuged at 5,000 × g for 10 min. Following 2 ml of supernatant was reacted with 2 ml thiobarbituric acid in a test tube at 100°C for 15 min, quickly cooled on ice, and the absorbance at 532 was measured by spectrophotometer. The MDA content was expressed as nmol g-1 FW.
Trypan Blue, DAB and NBT Staining were performed as described (Jiang et al., 2016).
For the above assays, consistent results were obtained from at least three experiments, and the result from one experimental was exhibited here. Average of three replicates was calculated to represent each data point.
Analyses of Rice Stomata
Leaf samples from sapk2 mutant lines and wide type plants (harvested before and after drought stress) were fixed by white nail polish blotting. The stomata conductance was measured following the described method (Dey et al., 2016) with modifications. The stomatal pictures were obtained using a laser confocal microscope (Nikon A1, Japan), Percentages of three levels of stomata (completely open, partially open and completely closed stomata) in sapk2 mutants and wild type plants were calculated before and after drought stress. (n = 98 stomata for S2-1, n = 95 stomata for S2-7, n = 98 stomata for wild type plants).
Three independent experiments repeated at the same time and a representative result was displayed. Three independent experimental replications were conducted.
Oxidative Enzyme Activity Measurement
The activities SOD, POD and CAT were performed as described (Jiang et al., 2016) with slight modifications. The 4-week-old seedlings of mutants and wide type plants were withheld water for 7 days, and then leaf segments (0.5 g) from control and dehydration lines were homogenized using a chilled mortar and pestle in 5 mL of 50 mM sodium phosphate buffer (pH7.8) supplemented with 1% polyvinylpyrrolidone and 10mM β-mercaptoethanol in an ice-cold mortar. After centrifugation (13,000 × g) for 15min at 4°C, transfer as much supernatant as possible into a new 5 ml tube. The supernatant was used to identify SOD, POD and CAT activity levels. Enzymes activities of the SOD, peroxidases (POD) and CAT were shown by U/mg protein.
Total SOD activity in the supernatant was assayed by measuring the oxidation of epinephrine at 475 nm by observing increase in absorbance monitored using spectrophotometer.
The supernatant was used for POD assay by measuring the oxidation of guaiacol to tetra guaiacol at 470 nm by observing increase in absorbance monitored. A 0.01 absorbance increase per min at 470 nm was defined as one unit of POD activity.
The supernatant was used for CAT assay by measuring the decomposition of H2O2 at 240 nm by observing decrease in absorbance using spectrophotometer.
The content of AsA was detected using commercial assay kits purchased from Nanjing Jiancheng Bioengineering Institute (Nanjing, China). The 4-week-old seedlings of mutants and wide type plants withheld water for 7 days. According to the manual, leaf segments (0.5 g) from control and dehydration lines were homogenized using a chilled mortar and pestle in 5 mL of 5% (w/v) m-phosphoric acid. Following centrifugation at 10,000 × g for 15 min at 4°C, the supernatant was used for the determination of AsA. Following 2 ml of supernatant was reacted with specified dosage of R1, R2, R3, and R4 (According to the manual) in a test tube at 22°C for 30 min, quickly cooled on ice, and the absorbance at 536 was measured.
The data points which presented activities of enzymes were the average of three replicates. At least three experiments were performed, and the results were consistent. The result from one set of experiments was presented here.
Salt and Osmotic Tolerance Assays
The sterilized seeds of sapk2 mutants and wide type plants germinated on half-strength MS medium simultaneously for 3 days. Seedlings were then transplanted in half-strength liquid MS medium for 4 weeks. For NaCl and PEG tolerance assays, Seedlings were then transferred into half-strength MS liquid medium supplemented with 175mM NaCl or 25% PEG6000 (m/v) for 3 days. After 3 days treatment, the seedlings were transferred into fresh half-strength MS liquid medium to recover for 7 days.
For NaCl and PEG treatments, the sapk2 mutant plants and wide type plants were transplanted in the same pot. Three independent experiments repeated at the same time and a representative result displayed here. Three independent experimental replications were conducted.
Statistical Analyses
The experiments were arranged in a completely randomized design with at least three replicates for each treatment. Excel 2010 was used for making charts. Significant difference among different treatments was identified by an analysis of variance by using SigmaPlot10.0. The data represent mean ± standard error (SE) of three independent experiments.
Results
SAPK2 Expression Profile and Protein Localization
SAPK2 is a member of the rice SnRK2 II subfamily. The SAPK2 open reading frame consists of 1,020 bp and encodes a 339-amino acid polypeptide (annotation identified in Introduction to the Rice Genome Annotation Project. database: LOC_Os07g429401). We analyzed the SAPK2 expression profiles under different stress conditions to clarify the physiological functions of SAPK2. The qRT-PCR analyses used to determine the SAPK2 transcriptional responses to various abiotic stresses revealed that SAPK2 expression was not induced by ABA (Figure 1A). In contrast, SAPK2 expression was considerably upregulated by exposures to drought, NaCl, and PEG (Figure 1A).
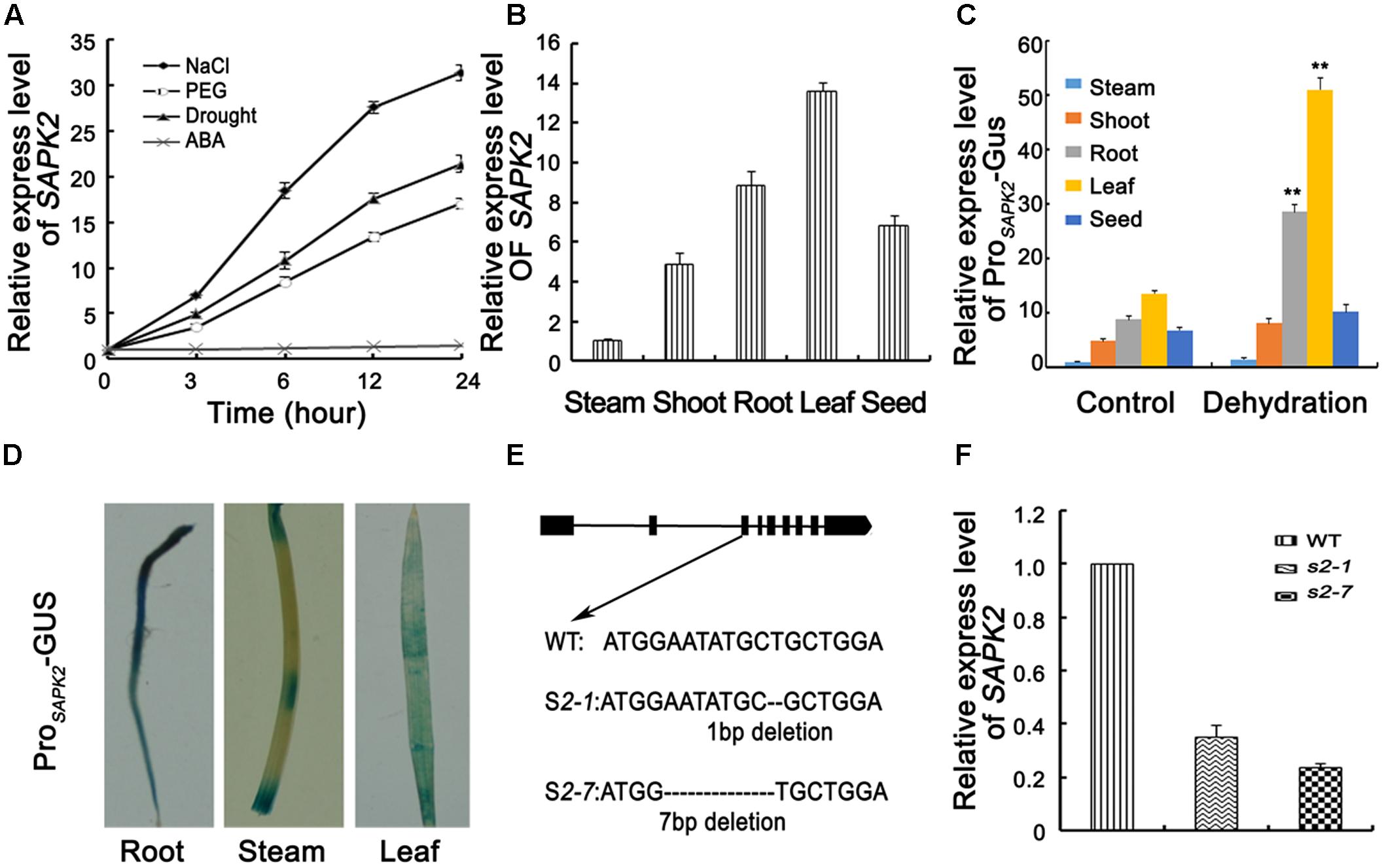
FIGURE 1. Expression analyses of SAPK2 and Mutation Detection. (A) Analyses of SAPK2 expression level under drought, NaCl, ABA, and PEG treatments. The expression level was assessed by qRT-PCR. (B) Expression level of SAPK2 in different tissues by qPCR analyses. (C) Expression level of ProSAPK2-GUS in response to drought in different tissues by qPCR analyses. (D) GUS staining of ProSAPK2-GUS in different tissues expressing the GUS reporter gene under the control of SAPK2 promoter. (E) Diagram showing that sgRNAs targets of SAPK2 were used in the CRISPR/Cas9 system to generate sapk2 mutants. Two independent sapk2 mutant lines with different deletions were obtained. (F) Accumulation of SAPK2 transcripts in sapk2 mutants and wide type plants. Transcript accumulation was assessed by qRT-PCR. Error bars indicate the SD of three biological replicates. ∗∗indicate statistically significant differences between mutant lines and wild type plants (P < 0.01).
We investigated the tissue specificity of SAPK2 expression via qRT-PCR analyses using total RNA isolated from various tissues (i.e., stem, shoot, root, leave, and seed). The SAPK2 expression level was highest in leaves, followed by the roots (Figure 1B). We also investigated ProSAPK2-Gus expression in response to drought in different tissue via qRT-PCR analyses using total RNA isolated from various tissues (i.e., stem, shoot, root, leave, and seed). The ProSAPK2-Gus expression level was highest in leaves, followed by roots without drought stress (Figure 1C). Under drought stress, the significant increase was in leaves (4.7 folds) and roots (3.2 folds) (Figure 1C). Tissue-specific SAPK2 expression in response to drought was also assessed by expressing the β-glucuronidase reporter gene. Consistent with the results of the qRT-PCR analyses, β-glucuronidase signals were abundant in leaves and roots without drought stress (Figure 1D and Supplementary Figure S2). Furthermore, β-glucuronidase signals increased significantly in leaves and roots under drought stress (Supplementary Figure S2).
To confirm the specific SAPK2 function, we employed the CRISPR/Cas9 system to generate sapk2 mutants, which we named sapk2-1 and sapk2-7. The sapk2-1 plants contained a 1-bp deletion in the third exon of SAPK2, while sapk2-7 plants carried a 7-bp deletion in this exon (Figure 1E). The SAPK2 expression level was much lower in sapk2 mutants than in wild-type plants (Figure 1F). Furthermore, we also identified that the nucleotide sequences of SAPK2 CDS from sapk2-1 and sapk2-7 mutant had frame shift mutation. We found that amino acid sequences of SAPK2 predicted based on these nucleotide sequences are only 106 and 104 in sapk2-1 and sapk2-7, respectively (Supplementary Table S2) caused by premature termination. A previous study revealed that Ser-158, Thr-159, and Thr-162 were essential for SAPK1 activity or activation (Kobayashi et al., 2004). We considered Ser-158, Thr-159, and Thr-162 are also essential for activity or activation of SAPK2, because the kinase domains of SAPK1 and SAPK2 are highly conserved (92% amino acid identity). These results indicate that the sapk2-1 and sapk2-7 mutant lines lacked SAPK2.
SAPK2 Affects Seed Germination and Early Seeding Growth in an ABA-Dependent Manner
Abscisic acid is crucial for preventing germination and inhibiting early seedling growth (Finkelstein et al., 2002). Because SAPK2 influences the ABA signal transduction pathway in rice (Kim et al., 2012), we tested the ABA sensitivity of sapk2 mutant lines during the germination and post-germination stages.
For the germination assay, seeds from two independent mutant lines (i.e., sapk2-1 and sapk2-7) and wide-type plants were simultaneously sown on half-strength MS agar medium containing 0, 2, or 5 μM ABA. The germination rate was calculated at specific time points after seeds were imbibed. In the absence of ABA, there was no significant difference between sapk2 mutants and wide-type plants (Figures 2A,B). However, the germination rates for the samples treated with 2 μM ABA were about 90% for the wild-type seeds and nearly 100% for the sapk2 mutant lines (i.e., the sapk2-1 and sapk2-7 seed germination rates were about 98 and 99%, respectively). At 5 μM ABA, the germination rates of sapk2 mutant lines were significantly higher than that of wide-type seeds (i.e., about 81 and 87% of sapk2-1 and sapk2-7 seeds germinated, while only about 52% of wild-type seeds germinated) (Figures 2A,B). These results indicated that the seeds of sapk2 mutant lines were mostly insensitive to ABA during the germination stage.
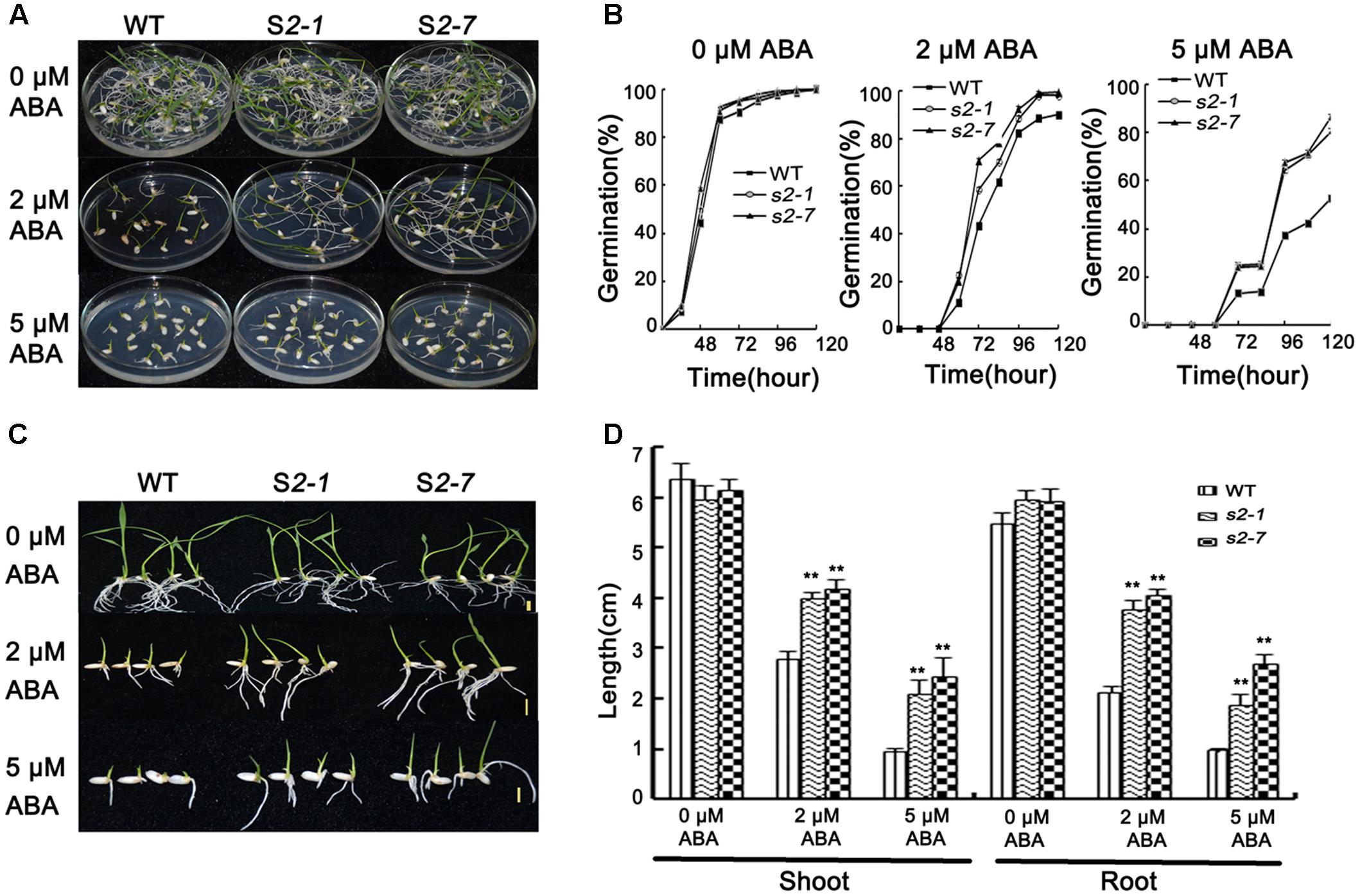
FIGURE 2. Abscisic acid sensitivity assays. (A) Germination phenotype of seeds from sapk2 mutants and wide type plants grown on MS agar medium containing 0, 2, or 5 μM ABA at 5 day after imbibition. (B) Germination rates corresponding to (A). (C) Growth performance of seedlings from sapk2 mutants and wide type plants grown on MS agar medium containing 0, 2, or 5 μM ABA during the post-germination stage. (D) Shoot and root length corresponding to (C). Error bars indicate the SD of three biological replicates. ∗∗indicate statistically significant differences between mutant lines and wild type plants (P < 0.01).
The insensitivity of sapk2 mutant lines to ABA was also confirmed during the post-germination stage. Wild-type and sapk2 mutant seeds were simultaneously germinated on half-strength MS agar medium for 3 days, and then seedlings were transferred to the same medium supplemented with 0, 2, or 5 μM ABA. Shoot and root lengths were measured 7 days after seedlings were transferred. There was no significant difference between wild-type and sapk2 mutant plants when the medium lacked ABA. In contrast, in media with 2 or 5 μM ABA, the sapk2 mutants had significantly longer roots and shoots than wild-type plants (P < 0.01) (Figures 2C,D). These findings suggest that sapk2 mutant lines are relatively insensitive to ABA after the germination stage.
These results imply that SAPK2 is a positive regulator of ABA-dependent germination and post-germination development, and is required for optimal seed germination.
SAPK2 Is Important for Dehydration Tolerance
The qRT-PCR data revealed that SAPK2 expression was strongly induced by drought stress (Figure 1A). Thus, we examined the drought stress responses of SAPK2. Four-week-old plants were exposed to drought conditions by withholding water for 7 days. The survival rate (%) was determined after a 7-day rewatering period. After the 1-week drought treatment, the dehydration symptoms were slighter in wild-type plants than in sapk2 mutants (Figure 3A and Supplementary Figure S1). When plants were rewatered, a significantly higher survival rate was observed for the wide-type plants (45.5%) than the sapk2 mutants (i.e., the sapk2-1 and sapk2-7 survival rates were only 6.5 and 9.1%, respectively) (P < 0.01) (Figure 3B).
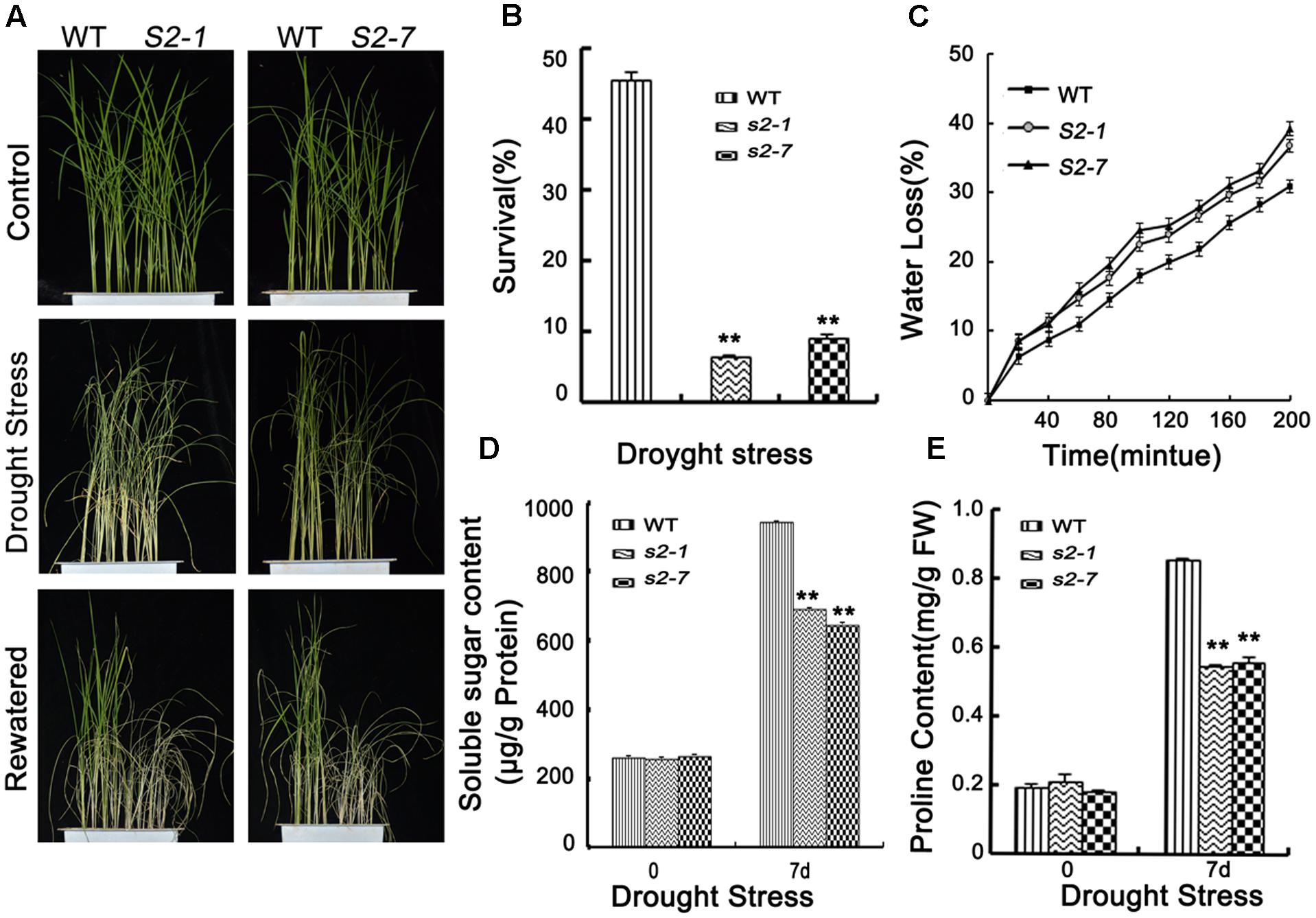
FIGURE 3. Drought stress tolerance assays. (A) Phenotype of sapk2 mutants and wild-type plants before drought stress, after drought stress and re-watered for 7 days. Drought stress analysis was repeated three times. In each repeated experiment, at least 40 plants were used for each individual line. One representative picture was shown. (B) Survival rates corresponding to (A). (C) Water loss rates in the leaves segregated from sapk2 mutants and wild type plants under normal conditions. (D) The contents of soluble sugar and (E) proline in leaf tissues sampled from sapk2 mutants and wild-type plants before and after drought stress. Error bars indicate the SD of three independent experiments. ∗∗indicate statistically significant differences between mutant lines and wild type plants (P < 0.01). FW, fresh weight.
Transpiration water loss is an important factor related to drought tolerance. Thus, we determined the leaf WLR as described by Jiang et al. (2016). We observed the leaves of both sapk2 mutant lines lost water faster than the wild-type leaves (Figure 3C). Given that WLRs were higher in two mutant lines than in wide type plants, we further investigated whether the WLR was affected by the stomatal density or aperture. We then assessed the stomatal density, and determined that there were no significant difference between sapk2 mutants and wild-type plants (Figure 4B). For stomatal aperture, there were also no significant difference between sapk2 mutants and wild-type plants under normal condition (Figure 4A). However, under drought condition, only about 36% of stomata were completely closed in sapk2 mutant lines (i.e., 36.7% for sapk2-1 and 38.8% for sapk2-7); while nearly 50% of stomata were completely closed in wild-type plants (Figure 4A). Additionally, the proportions of completely open stomata for both sapk2 mutant lines were higher than that of wild-type plants (i.e., 25.1% for sapk2-1, 24.5% for sapk2-7, and 19.6% for wild-type plants). Moreover, the percentage of partially open stomata was also higher for the sapk2 mutant lines (i.e., 38.2% for sapk2-1, 36.7% for sapk2-7, and 30.9% for wild-type plants) (Figure 4A). These results suggest that SAPK2 promotes the closure of stomata result in the lower WLR, which may be critical to adapt to drought stress in rice.
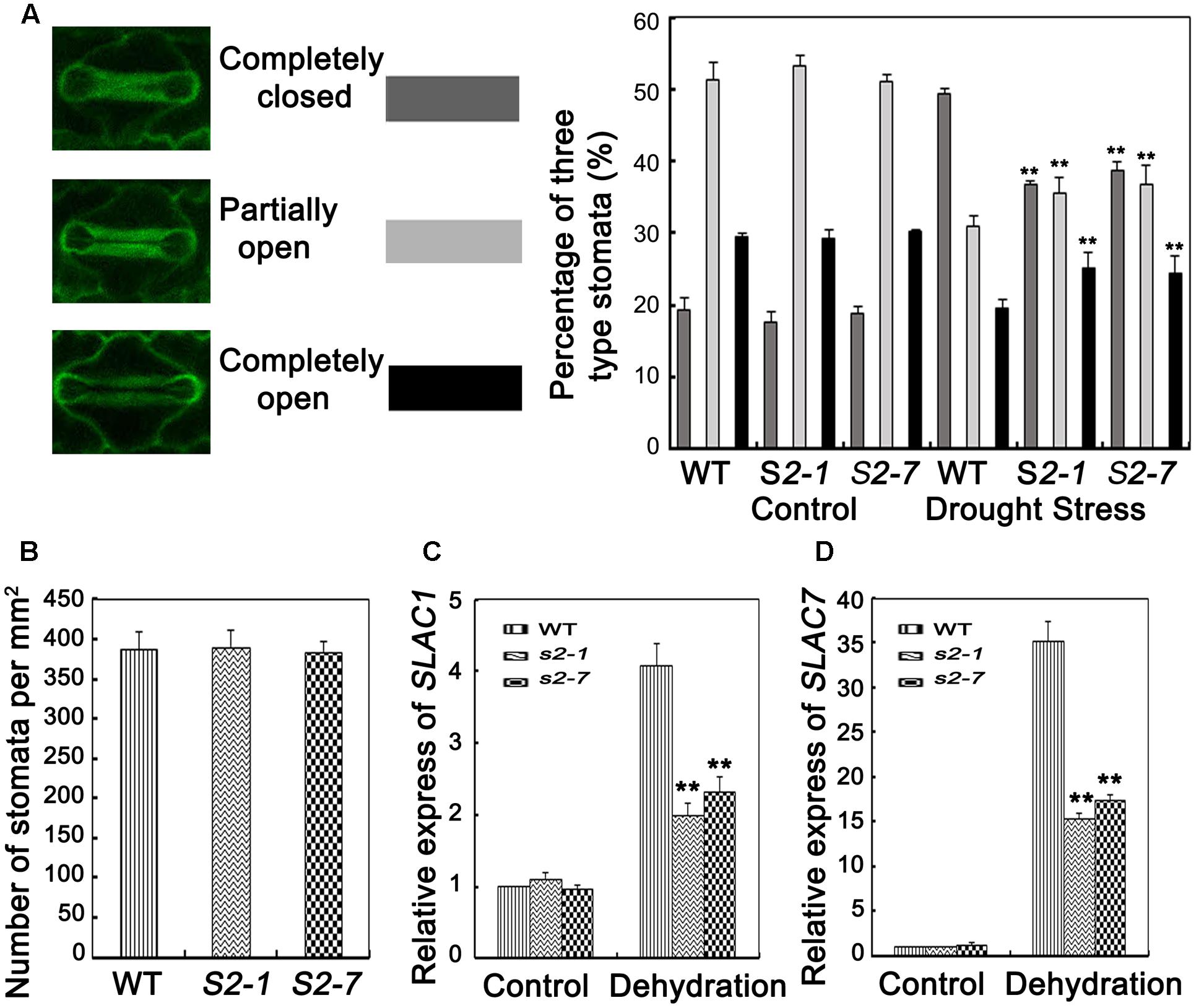
FIGURE 4. Analyses of stomata closure and relative expression level of slow anion channel (SLAC)-associated genes. (A) Percentages of three levels of stomata (completely open, partially open, and completely closed stomata) in sapk2 mutants and wild type plants were calculated before and after drought stress (n = 98 stomata for S2–1, n = 95 stomata for S2–7, n = 98 stomata for wild type plants). (B) Stomata numbers per mm2 in 10 leaves from sapk2 mutants and wild type plants, respectively, were counted to indicate stoma densities. (C) Transcript level of stomata genes OsSLAC1 and (D) OsSLAC7 in sapk2 mutants and wild type plants before and after drought stress. Error bars indicate the SD of three independent experiments. ∗∗indicate statistically significant differences between mutant lines and wild type plants (P < 0.01).
To confirm our results, we also verified that OsSLAC1 and OsSLAC7 expression levels were significantly higher in wild-type plants than in sapk2 mutants (Figures 4C,D). These results suggest that SAPK2 induces stomatal closure by upregulating the expression of slow anion channel (SLAC)-associated genes.
To clarify the mechanisms by which SAPK2 induces dehydration tolerance, we further compared the proline and soluble sugar contents of wild-type and sapk2 mutant plants. We observed no significant difference in proline and soluble sugar contents between the wild-type and sapk2 mutant plants under normal conditions. However, under drought stress, proline and soluble sugar contents were lower in sapk2 mutants than in wild-type plants (Figures 3D,E). These results strongly suggest that SAPK2 corresponds to the accumulation of compatible solutes, such as proline and soluble sugars, in response to drought conditions in rice.
Overall, our results indicate that SAPK2 was involved in rice drought tolerance response by decreasing water loss through osmotic adjustments, stomatal closure, and upregulation of SLAC gene expression levels.
Expression of Stress-Responsive Genes in sapk2 Mutants
To comprehensively characterize SAPK2 functions in drought-stressed plants, we investigated the expression levels of several drought-related genes in sapk2 mutants, including a pyrroline-5-carboxylate synthetase gene (OsP5CS), as well as OsbZIP23, OsLEA3, OsRab16b, OsRab21, OsOREB1 and OsDREB1A, which all function downstream of SAPKs in the ABA-signaling pathway.
Proline biosynthesis is catalyzed by OsP5CS. According to our qRT-PCR data, the OsP5CS expression level was much lower in sapk2 mutants than in wild-type plants under drought conditions (Figure 5D). We also examined the expression of another five well-characterized drought resistance-related genes (i.e., OsRab21, OsRab16b, OsbZIP23, OsOREB1, and OsLEA3) and one control gene OsDREB1A. All their expression levels except OsDREB1A were significantly lower in sapk2 mutants than in wild-type plants after the drought treatment (Figures 5A–C,E,F and Supplementary Figure S3). These results suggest that SAPK2 upregulate the expression of some stress-responsive genes under drought stress.
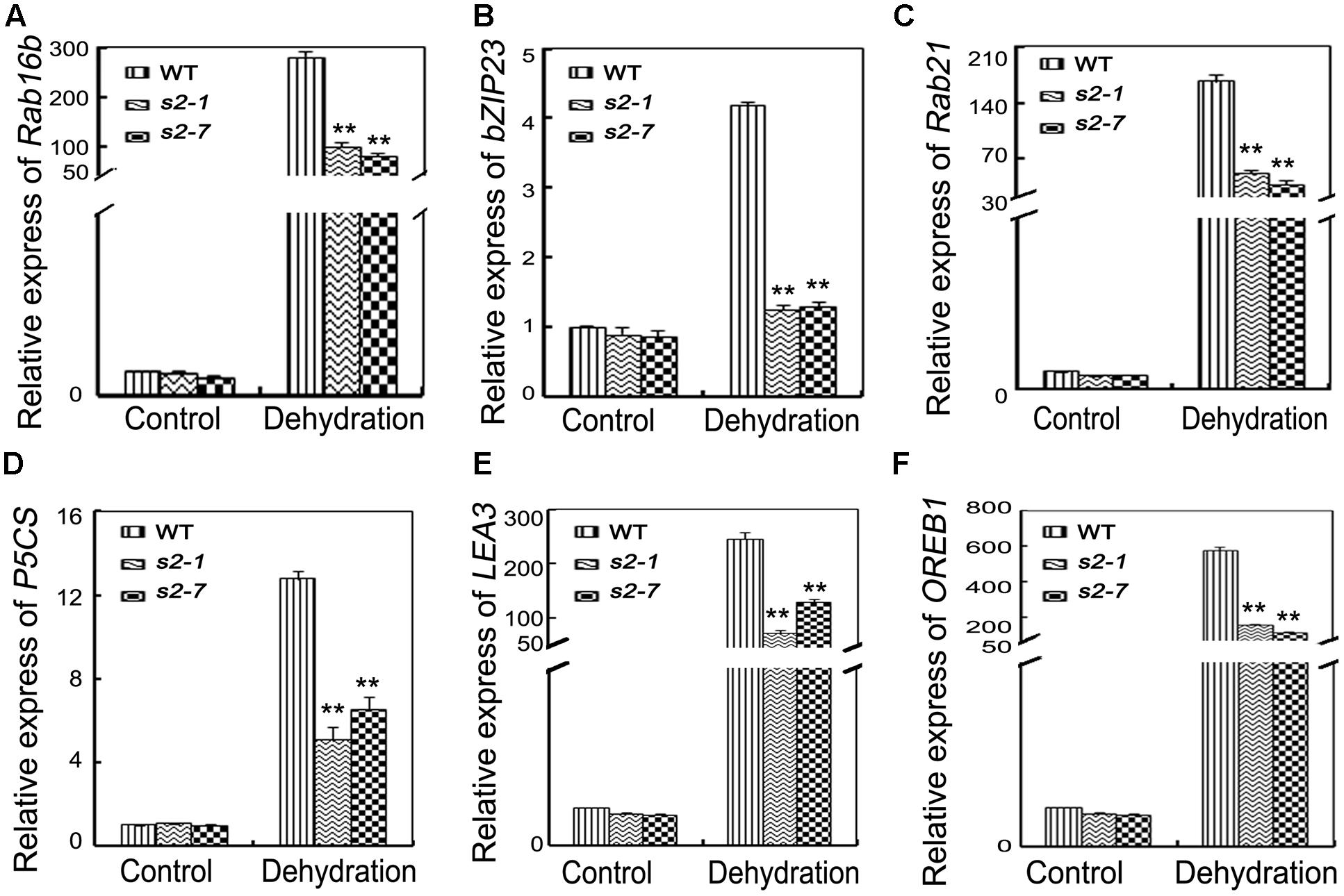
FIGURE 5. Expression of stress-response genes. (A–F) Relative expression levels of OsRab16b, OsbZIP23, OsRab21, OsP5CS, OsLEA3, and OsOREB1 in the leaves of 4-week-old mutants and wide type plants before and after drought stress. Error bars indicate SD from three independent RNA extracts. ∗∗indicate statistically significant differences between mutant lines and wild type plants (P < 0.01).
Reactive Oxygen Species-Induced Damage in sapk2 Mutant Plants
Because sapk2 mutants exhibited reduced drought tolerance, we assessed whether SAPK2 affects drought tolerance via ROS detoxification. The leaves of wild-type and sapk2 mutant plants began to produce brownish lesions after the 7-day drought treatment (Figure 6A). However, the lesions were more severe on the sapk2 mutant leaves. This result was consistent with that of the leaf cell death analysis based on trypan blue staining (Figure 6B).
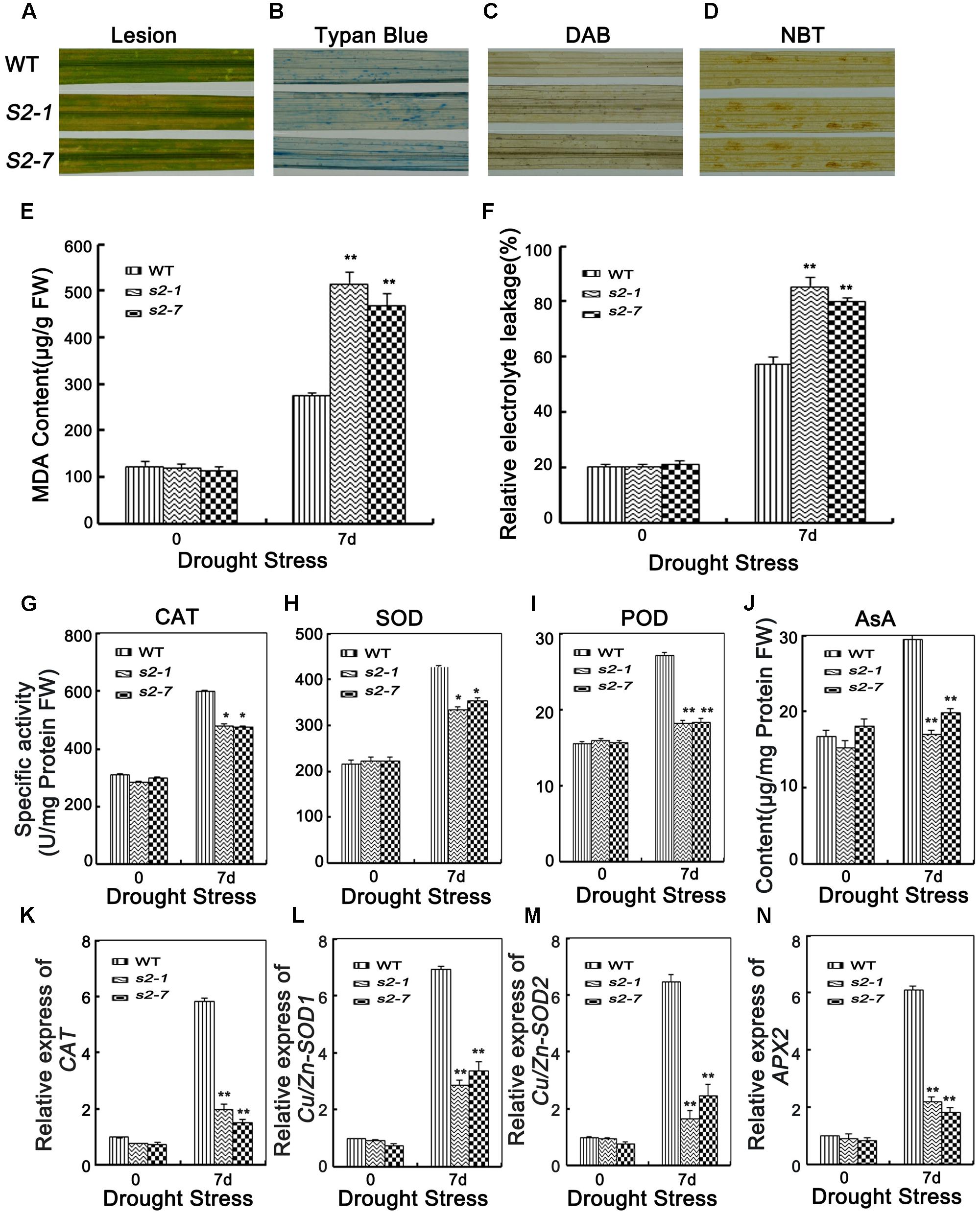
FIGURE 6. Analyses of ROS damage, ROS-scavenging ability and the expression level of oxidative enzymes genes. (A) Phenotype of brownish lesions after 7 days drought stress. (B) Phenotype of Typan blue staining after 7 days drought stress. (C) Phenotype of DAB staining after 7 days drought stress. (D) Phenotype of NBT staining after 7 days drought stress. (E) MDA content in the leaves of 4-week-old mutants and wide type plants before and after drought stress. FW, fresh weight. (F) Relative electrolyte leakage in the leaves of 4-week-old mutants and wide type plants before and after drought stress. (G–I) POX, SOD, and CAT activities in the leaves of 4-week-old mutants and wide type plants. (J) The content of AsA in the leaves of 4-week-old mutants and wide type plants. (K) Relative expression of oxidative enzymes genes CAT, (L) Cu/Zn-SOD1, (M) Cu/Zn-SOD2, and (N) APX2 in the leaves of 4-week-old mutants and wide type plants. Error bars indicate SD from three independent RNA extracts. ∗indicate statistically significant differences between mutant lines and wild type plants (P < 0.05).
To detect H2O2 and superoxide damage, leaves of wild-type and sapk2 mutant plants were stained with DAB and NBT, respectively, after 7 days of drought stress. Considerably more brown spots caused by H2O2 and superoxide were observed in the sapk2 mutants than in the wild-type plants (Figures 6C,D). These results suggest that the sapk2 mutants are unable to efficiently remove the H2O2 and superoxide produced under drought conditions.
Malondialdehyde (MDA) is the product of lipid peroxidation resulting from ROS activities and a stress-specific molecular marker indicating the extent of cellular damage (Mittler, 2002). To confirm that SAPK2 improves drought tolerance through ROS detoxification, we examined MDA contents. There were no significant differences in the MDA contents of sapk2 mutants and wild-type plants prior to the drought treatment. Among the drought-stressed plants, the sapk2 mutant lines accumulated more MDA than the wild-type plants (Figure 6E). Moreover, electrolyte leakage is widely used as an indicator of membrane damage in studies of stress-induced injuries and stress tolerance in plants (Lee and Zhu, 2010). Thus, we also measured electrolyte leakage due to drought stress, and determined that the leakage was greater in sapk2 mutants (i.e., > 80%) than in wild-type plants (i.e., about 60%) (Figure 6F). These results imply that the decreased drought tolerance of the sapk2 mutants is likely a consequence of increased cell membrane damage caused by ROS.
Decreased ROS-Scavenging Ability and Expression of Antioxidant Enzyme Genes in sapk2 Mutants
Given the enhanced ROS-induced damage of the sapk2 mutants, we compared the SOD, peroxidase (POD), and CAT activities, as well as the AsA contents between sapk2 mutants and wild-type plants. Under normal growth conditions, there were no significant differences between sapk2 mutants and wild-type plants regarding POD, SOD, and CAT activities, and AsA contents. However, after a 7-day drought treatment, the antioxidant enzyme activities and AsA contents were significantly lower in sapk2 mutant lines than in wild-type plants (Figures 6G–J). These results indicate that ROS-scavenging is inhibited in the sapk2 mutant lines, resulting in an accumulation of ROS damage.
To confirm that in the sapk2 mutant lines ROS-scavenging was suppressed, we assessed the expression levels of several antioxidant genes. Under drought conditions, the OsCAT, OsCu/Zn-SOD1, OsCu/Zn-SOD2, and OsAPX2 transcript levels were up-regulated in wild-type and sapk2 mutant plants, but the expression levels were significantly higher in the wild-type plants (Figures 6K–N). These results were consistent with the observed higher antioxidant enzyme activities in wild-type plants than in sapk2 mutants, and further confirmed that SAPK2 limits ROS-induced damage by promoting the expression of antioxidant enzyme genes.
Decreased Tolerance of sapk2 Mutants to NaCl and Polyethylene Glycol
Considering SAPK2 expression was strongly induced by NaCl and PEG (Figure 1A), we explored SAPK2 functions in plants treated with NaCl or PEG. For the PEG tolerance assays, we observed the phenotypes of sapk2 mutants and wild-type plants grown in half-strength liquid MS medium supplemented with 25% PEG-6000 for 3 days. Survival rates were calculated after plants were allowed to recover in fresh half-strength liquid MS medium for 7 days. The sapk2 mutant plants exhibited a more severe PEG stress phenotype, with a much lower survival rates than wild-type plants (i.e., 96.3% for wild-type plants, 28.1% for sapk2-1, and 39.6% for sapk2-7) (Figures 7A,B). For the NaCl stress assays, we observed the phenotypes of sapk2 mutants and wild-type plants grown in half-strength liquid MS medium containing 175 mM NaCl for 3 days. The survival rates were assessed after plants recovered in fresh half-strength liquid MS medium for 7 days. The sapk2 mutants were more badly damaged by salt stress, and had significantly lower survival rates than the wild-type plants (i.e., survival rates: 70.4% for wild-type plants, 32.6% for sapk2-1, and 17% for sapk2-7) (Figures 7C,D). These results suggested that the loss of function of SAPK2 decreased osmotic stress tolerance such as NaCl, PEG and drought stress in rice.
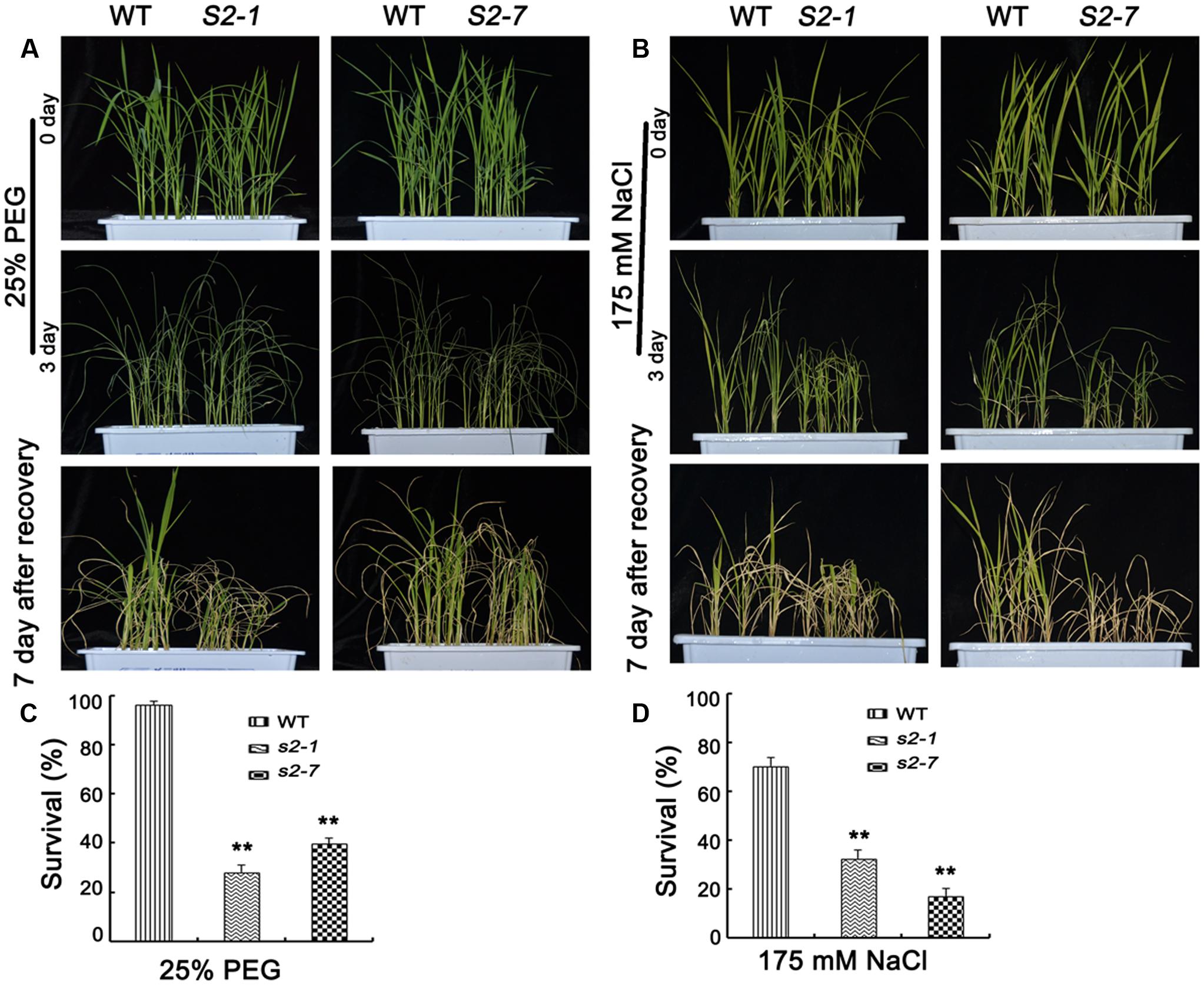
FIGURE 7. Reduced NaCl and PEG tolerance in sapk2 mutant plants. (A) Phenotype before 25% PEG stress, after 25% PEG stress and recovery for 7 days of sapk2 mutants and wild-type plants. (B) Survival rates corresponding to (A). (C) Phenotype before 175 mM NaCl stress, after 175 mM NaCl stress and recovery for 7 days of sapk2 mutants and wild-type plants. (D) Survival rates corresponding to (C). Analysis for PEG and NaCl stress was repeated three times. In each repeated experiment, at least 40 plants were used for each individual line. One representative picture was shown here. Error bars indicate SD from three independent RNA extracts. ∗∗indicate statistically significant differences between mutant lines and wild type plants (P < 0.01).
Discussion
Known as crucial positive regulators of ABA and osmotic stress responses in A. thaliana and rice, SnRK2s play important roles in various processes. SAPK2 is a member of SnRK2 subclass II in rice. However, little information is available regarding the functional properties of rice SAPK2. In this study, we characterized the functional properties of SAPK2 by developing loss-of-function mutants using the CRISPR/Cas9 system.
A previous study revealed that ABA does not upregulate the expression of SAPK2 or activate the encoded protein (Kobayashi et al., 2004). We confirmed that SAPK2 expression was not induced by an ABA treatment. Researchers have determined that OsPYL/RCAR5, functioning upstream of SAPK2 with mechanisms similar to A. thaliana PYL/RCARs, modulates germination and early seedling growth in rice (Kim et al., 2012). This suggests that SAPK2 is crucial for ABA-dependent developmental controls in rice. Therefore, we characterized the ABA sensitivity of sapk2 mutant lines regarding developmental controls. The sapk2 mutant plants exhibited significantly decreased sensitivity to ABA during the germination and post-germination stages (Figure 2). These results imply that SAPK2 has a significant role in the ABA signaling pathway during the initial development stages in rice, which is very different from the function of subclass II SnRK2 in A. thaliana. Because subclass II SnRK2 is not so important for ABA signaling compared to subclass III in A. thaliana.
SRK2C/SnRK2.8, which is homologous to rice SAPK2, was expressed abundantly in roots and weakly in leaves (Umezawa et al., 2004). This tissue-specific localization is consistent with the results that SRK2C/SnRK2.8 is a positive regulator of drought tolerance in A. thaliana roots. However, our data indicate that the expression level of SAPK2 is highest in leaves, followed by the roots (Figures 1B–D), which is consistent with the expression patterns observed for other drought- and salt-tolerance genes as well as other SnRK2 family members (Xiang et al., 2007, 2008; Mao et al., 2010; Tian et al., 2013; Dey et al., 2016). This tissue-specific expression pattern further suggests that SAPK2 may function similar to SAPK8 or SAPK9 in rice when plants exposed to osmotic stress, possibly by regulating the stomatal closure (Dey et al., 2016; Sun et al., 2016).
Our results revealed that SAPK2 expression was strongly induced by drought, NaCl, and PEG (Figure 1A) in rice, implying that SAPK2 may have critical roles related to osmotic stress tolerance. Mizoguchi et al. (2010) observed that snrk2.8 mutant did not show any clear differences in survival or visible damage compare with WT under drought stress. However, we observed that sapk2 mutant plants were sensitive to drought stress, with relatively low survival rates and high WLRs (Figures 3A–C). The increased WLR is not favorable for drought tolerance in the sapk2 mutant lines. In response to drought stress, stomata often close to limit water loss by transpiration. Previous studies confirmed that A. thaliana OST1/SnRK2.6 and rice SAPK8 and SAPK9 affect ABA-regulated stomatal movements (Mustilli et al., 2002; Dey et al., 2016; Sun et al., 2016). Additionally, OsSLAC1 and OsSLAC7 have recently been identified as stomatal proteins involved in anion transport (Fan et al., 2015; Sun et al., 2016). We observed that, under drought stress, sapk2 mutants had larger proportions of completely and partially open stomata than wild-type plants (Figure 4A). Furthermore, the OsSLAC1 and OsSLAC7 expression levels were significantly higher in wild-type plants than in sapk2 mutants after drought stress treatment (Figures 4C,D). Thus, SAPK2 may be responsible for activating FABA-induced stomatal closure, similar to how SAPK8 functions as an essential activator of OsSLAC1 via phosphorylations (Sun et al., 2016). This possibility should be investigated in a future study. By contrast, SRK2C/SnRK2.8 functioning very different from SAPK2. The water-loss levels of srk2c mutants were similar to that of WT, and overexpression of SRK2C/SnRK2.8 did not affect stomatal regulation (Umezawa et al., 2004). Such limited phenotype of SRK2C/SnRK2.8 may be due to tissue-specific localization of SRK2C in root tips. These results suggest that SRK2C/SnRK2.8 may not be involved in stomatal response and mediated signaling should be quite different from SAPK2.
Plants evolved two central physiological mechanisms to decrease water loss under drought conditions: osmotic adjustment and stomatal closure. Zhang et al. (2011) confirmed that accumulated proline and soluble sugar could stabilize subcellular structures and facilitate cell recovery from damages due to abiotic stress. Our results of proline and soluble sugar content analyses (Figures 3D,E) suggest that the sensitivity to drought and the low survival rates of sapk2 mutants may be because of several physiological traits, including excessive water loss and low proline and soluble sugar contents. Additionally, the significantly decreased OsP5CS transcript levels in sapk2 mutants were consistent with the low proline content of these plants under drought conditions (Figure 5D). These results indicate that SAPK2 positive regulate rice drought tolerance by decreasing water loss through osmotic adjustments.
OREB1 is activated by SAPK2 in rice (Kim et al., 2012). OsbZIP23 is a member of bZIP transcription factor family, and confers ABA-dependent drought and salinity tolerance in rice. OsLEA3 is a late embryogenesis abundant protein that protects cell membrane structures and improves drought tolerance (Xiao et al., 2007). The transcription of OsRab21 and OsRab16b, which encode dehydrin proteins, is responsive to abiotic stress treatments (Yamaguchi-Shinozaki et al., 1990). The expression of OsDREB1A was induced by cold and used as one control gene in our study (Dubouzet et al., 2003). In our study, except OsDREB1A, the transcript levels of OsRab16b, OsRab21, OsbZIP23, OsLEA3, and OsOREB1 were significantly lower in sapk2 mutants than in wild-type plants under drought conditions. These results imply that SAPK2 has an important role related to ABA-dependent dehydration tolerance, and may serve as a transcriptional activator of these genes in the ABA signaling pathway of rice. Moreover, there is accumulating evidence that most of the transcriptional activators are localized in the nucleus, which is where SAPK2 localizes (Kim et al., 2012).
Reactive oxygen species causes lipid peroxidation, which results in the production of MDA. High MDA levels are toxic to plant cells and cause programmed cell death (Hou et al., 2009). Major ROS-scavenging mechanisms of plants include SOD, APX, and CAT (Das and Roychoudhury, 2014). The balance between SOD and APX or CAT activities in cells is crucial for determining the steady-state level of superoxide radicals and H2O2 (Mittler, 2002). Among drought-treated plants, the sapk2 mutants exhibited high levels of superoxide, H2O2, and cell death according to NBT, DAB, and trypan blue staining, respectively (Figures 6A–D). The mutants also exhibited higher MDA contents and relative electrolyte leakage (Figures 6E,F). Our results indicate that drought-stressed sapk2 mutants develop lesions because of their inhibited ability to detoxify ROS, which should due to decreased SOD, POD, and CAT activities as well as low AsA contents (Figures 6G–J) through decreasing transcription of antioxidant enzyme genes (i.e., CAT, SOD, and APX2) (Figures 6K–N). These results confirm that SAPK2 protects plants from oxidative damages by enhancing ROS detoxification.
Our results indicate that SAPK2 also increases rice tolerance to NaCl and PEG stresses (Figure 7). Recent studies confirmed that the overexpression of SAPK9 may significantly enhance drought tolerance, while also increasing grain yields in drought-stressed rice (Dey et al., 2016). Shin et al. (2007) reported that overexpression of SRK2C/SnRK2.8 enhanced plant growth in A. thaliana. We observed that under normal conditions, the wild-type and sapk2 mutant plants developed similarly, suggesting that SAPK2 may be useful for improving crop yields under drought conditions. However, we did not analyze the effects of SAPK2 on productivity. Future studies should focus on the grain yields of mutant plants under drought conditions. The findings presented herein expand our knowledge regarding the role of SAPK2 as a key kinase in the ABA signaling pathway of rice. This increased characterization may be relevant for developing new rice cultivars with enhanced tolerance to drought and salt stress.
Author Contributions
DL and DY designed the experiments. DL performed the experiments and wrote the manuscript. DL, HW, and GL analyzed the data and edited the article. All authors read and approved the final article.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (U1202264), the Strategic Priority Research Program of the Chinese Academy of Sciences (Class A) (XDA08020203), and the program for Innovative Research Team of Yunnan Province (2014HC017).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00993/full#supplementary-material
FIGURE S1 | Replicates of drought stress experiments. (A) First replicate. Phenotype of sapk2 mutants and wild-type plants before drought stress, after drought stress and re-watered for 7 days. (B) Second replicate. Drought stress analysis was repeated three times. In each repeated experiment, at least 40 plants were used for each individual line.
FIGURE S2 | GUS Staining of SAPK2. (A) GUS staining of ProSAPK2-GUS in different tissues with or without drought stress in root, (B) steam, (C) leaf.
FIGURE S3 | Expression of stress-response genes OsDREB1A. Relative expression levels of OsDREB1A in the leaves of 4-week-old mutants and wide type plants before and after drought stress. Error bars indicate SD from three independent RNA extracts.
Footnotes
References
Bates, L., Waldren, R., and Teare, I. (1973). Rapid determination of free proline for water-stress studies. Plant Soil 39, 205–207. doi: 10.1016/j.dental.2010.07.006
Boudsocq, M., Barbier-Brygoo, H., and Laurière, C. (2004). Identification of nine sucrose nonfermenting1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J. Biol. Chem. 279, 41758–41766. doi: 10.1074/jbc.M405259200
Boudsocq, M., Droillard, M. J., Barbier-Brygoo, H., and Laurière, C. (2007). Different phosphorylation mechanisms are involved in the activation of sucrose non-fermenting 1 related protein kinases 2 by osmotic stresses and abscisic acid. Plant Mol. Biol. 63, 491–503. doi: 10.1007/s11103-006-9103-1
Chae, M. J., Lee, J. S., Nam, M. H., Cho, K., Hong, J. Y., Yi, S. A., et al. (2007). A rice dehydration-inducible SNF1-related protein kinase 2 phosphorylates an abscisic acid responsive element-binding factor and associates with ABA signaling. Plant Mol. Biol. 63, 151–169. doi: 10.1007/s11103-006-9079-x
Clarke, S. M., Mur, L. A. J., Wood, J. E., and Scott, I. M. (2004). Salicylic acid dependent signaling promotes basal thermotolerance but is not essential for acquired thermotolerance in Arabidopsis thaliana. Plant J. 33, 432–447. doi: 10.1111/j.1365-313X.2004.02054.x
Das, K., and Roychoudhury, A. (2014). Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 3:53. doi: 10.3389/fenvs.2014.00053
Dey, A., Samanta, M. K., Gayen, S., and Maiti, M. K. (2016). The sucrose non-fermenting 1-related kinase 2 gene SAPK9 improves drought tolerance and grain yield in rice by modulating cellular osmotic potential, stomatal closure and stress-responsive gene expression. BMC Plant Biol. 16:158. doi: 10.1186/s12870-016-0845-x
Diédhiou, C. J., Popova, O. V., Dietz, K. J., and Golldack, D. (2008). The SNF1-type serine-threonine protein kinase SAPK4 regulates stress-responsive gene expression in rice. BMC Plant Biol. 8:49. doi: 10.1186/1471-2229-8-49
Dubouzet, J. G., Sakuma, Y., Ito, Y., Kasuga, M., Dubouzet, E. G., and Miura, S. (2003). OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 33, 751–763. doi: 10.1046/j.1365-313X.2003.01661.x
Fan, X., Wu, J., Chen, T., Tie, W., Chen, H., Zhou, F., et al. (2015). Loss-of-function mutation of rice SLAC7 decreases chloroplast stability and induces a photoprotection mechanism in rice. J. Integr. Plant Biol. 57, 1063–1077. doi: 10.1111/jipb.12350
Farooq, M., Wahid, A., Lee, D. J., Ito, O., and Siddique, K. H. M. (2009). Advances in drought resistance of rice. Crit. Rev. Plant Sci. 28, 199–217. doi: 10.1080/07352680902952173
Finkelstein, R. R., Gampala, S. S., and Rock, C. D. (2002). Abscisic acid signaling in seeds and seedlings. Plant Cell 14(Suppl.), S15–S45.
Fujii, H., Chinnusamy, V., Rodrigues, A., Rubio, S., Antoni, R., Park, S. Y., et al. (2009). In vitro reconstitution of an abscisic acid signaling pathway. Nature 462, 660–664. doi: 10.1038/nature08599
Fujii, H., Verslues, P. E., and Zhu, J. K. (2007). Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19, 485–494. doi: 10.1105/tpc.106.048538
Fujita, Y., Nakashima, K., Yoshida, T., Katagiri, T., Kidokoro, S., Kanamori, N., et al. (2009). Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 50, 2123–2132. doi: 10.1093/pcp/pcp147
Halford, N. G., and Hey, S. J. (2009). Snf1-related protein kinases (SnRKs) act within an intricate network that links metabolic and stress signaling in plants. Biochem. J. 419, 247–259. doi: 10.1042/BJ20082408
He, H., Liang, G., Li, Y., Wang, F., and Yu, D. Q. (2014). Two young miRNAs originating from target duplication mediate nitrogen starvation adaptation via regulation of glucosinolate synthesis in Arabidopsis thaliana. Plant Physiol. 164, 853–865. doi: 10.1104/pp.113.228635
Heath, R. L., and Packer, L. (1968). Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 125, 189–198. doi: 10.1016/0003-9861(68)90654-1
Hou, X., Xie, K., Yao, J., Qi, Z., and Xiong, L. (2009). A homolog of human ski-interacting protein in rice positively regulates cell viability and stress tolerance. Proc. Natl. Acad. Sci. U.S.A. 106, 6410–6415. doi: 10.1073/pnas.0901940106
Hrabak, E. M., Chan, C. W. M., Gribskov, M., Harper, J. F., Choi, J. H., Halford, N., et al. (2003). The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 132, 666–680. doi: 10.1104/pp.102.011999
Jabs, T., Dietrich, R. A., and Dangl, J. L. (1996). Initiation of runaway cell death in an Arabidopsis mutant by extracellular superoxide. Science 273, 1853–1856. doi: 10.1126/science.273.5283.1853
Jiang, Y. J., Qiu, Y. P., Hu, Y. R., and Yu, D. Q. (2016). Heterologous expression of AtWRKY57 confers drought tolerance in Oryza sativa. Front. Plant Sci. 7:145. doi: 10.3389/fpls.2016.00145
Kim, H., Hwang, H., Hong, J. W., Lee, Y. N., Ahn, I. P., Yoon, I. S., et al. (2012). A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth. J. Exp. Bot. 63, 1013–1024. doi: 10.1093/jxb/err338
Kobayashi, Y., Yamamoto, S., Minami, H., Kagaya, Y., and Hattori, T. (2004). Differential activation of the rice sucrose nonfermenting1–related protein kinase2 family by hyperosmotic stress and abscisic acid. Plant Cell 16, 1163–1177. doi: 10.1105/tpc.019943
Lee, B. H., and Zhu, J. K. (2010). Phenotypic analysis of Arabidopsis mutants: electrolyte leakage after freezing stress. Cold Spring Harb. Protoc. 2010:db.rot4970. doi: 10.1101/pdb.prot4970
Liang, G., Zhang, H. M., Lou, D. J., and Yu, D. Q. (2016). Selection of highly efficient sgRNAs for CRISPR/Cas9-based plant genome editing. Sci. Rep. 6:21451. doi: 10.1038/srep21451
Lin, Q., Wu, F., Sheng, P., Zhang, Z., Zhang, X., Guo, X., et al. (2015). The SnRK2-APC/CTE regulatory module mediates the antagonistic action of gibberellic acid and abscisic acid pathways. Nat. Commun. 6:7981. doi: 10.1038/ncomms8981
Liu, W., Xie, X., Ma, X., Li, J., Chen, J., and Liu, Y. G. (2015). DSDecode: a web-based tool for decoding of sequencing chromatograms for genotyping of targeted mutations. Mol. Plant 8, 1431–1433. doi: 10.1016/j.molp.2015.05.009
Ma, Y., Szostkiewicz, I., Korte, A., Moes, D., Yang, Y., Christmann, A., et al. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068. doi: 10.1126/science.1172408
Mao, X., Zhang, H., Tian, S., Chang, X., and Jing, R. (2010). TaSnRK2.4, an SNF1- type serine/threonine protein kinase of wheat (Triticum aestivum L.), confers enhanced multistress tolerance in Arabidopsis. J. Exp. Bot. 61, 683–696. doi: 10.1093/jxb/erp331
McLoughlin, F., Galvan-Ampudia, C. S., Julkowska, M. M., Caarls, L., van der Does, D., and Lauriere, C. (2012). The Snf1-related protein kinases SnRK2.4 and SnRK2.10 are involved in maintenance of root system architecture during salt stress. Plant J. 72, 436–449. doi: 10.1111/j.1365-313X.2012.05089.x
Melcher, K., Ng, L. M., Zhou, X. E., Soon, F. F., Xu, Y., Suino-Powell, K. M., et al. (2009). A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature 462, 602–608. doi: 10.1038/nature08613
Mittler, R. (2002). Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 7, 405–410. doi: 10.1016/S1360-1385(02)02312-9
Mizoguchi, M., Umezawa, T., Nakashima, K., Kidokoro, S., Takasaki, H., Fujita, Y., et al. (2010). Two closely related subclass II SnRK2 protein kinases cooperatively regulate drought-inducible gene expression. Plant Cell Physiol. 51, 842–847. doi: 10.1093/pcp/pcq041
Mustilli, A. C., Merlot, S., Vavasseur, A., Fenzi, F., and Giraudat, J. (2002). Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14, 3089–3099. doi: 10.1105/tpc.007906
Nam, M. H., Huh, S. M., Kim, K. M., Park, W. J., Seo, J. B., Cho, K., et al. (2012). Comparative proteomic analysis of early salt stress-responsive proteins in roots of SnRK2 transgenic rice. Proteome Sci. 10:25. doi: 10.1186/1477-5956-10-25
Park, S., Fung, P., Nishimura, N., Jensen, D. R., Fujii, H., Zhao, Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071. doi: 10.1126/science.1173041
Santiago, J., Dupeux, F., Round, A., Antoni, R., Park, S. Y., Jamin, M., et al. (2009). The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 462, 665–668. doi: 10.1038/nature08591
Shin, R., Alvarez, S., Burch, A. Y., Jez, J. M., and Schachtman, D. P. (2007). Phosphoproteomic identification of targets of the Arabidopsis sucrose nonfermenting-like kinase SnRK2.8 reveals a connection to metabolic processes. Proc. Natl. Acad. Sci. U.S.A. 104, 6460–6465. doi: 10.1073/pnas.0610208104
Sopory, S. K., and Munshi, M. (1998). Protein kinases and phosphatases and their role in cellular signaling in plants. Crit. Rev. Plant Sci. 17, 245–318. doi: 10.1080/07352689891304230
Sun, S. J., Qi, G. N., Gao, Q. F., Wang, H. Q., Yao, F. Y., Hussain, J., et al. (2016). Protein kinase OsSAPK8 functions as an essential activator of S-type anion channel OsSLAC1, which is nitrate-selective in rice. Planta 243, 489–500. doi: 10.1007/s00425-015-2418-x
Tang, N., Zhang, H., Li, X., Xiao, J., and Xiong, L. (2012). Constitutive activation of transcription factor OsbZIP46 improves drought tolerance in rice. Plant Physiol. 158, 1755–1768. doi: 10.1104/pp.111.190389
Testerink, C., Dekker, H. L., Lim, Z. Y., Johns, M. K., Holmes, A. B., and Koster, C. G. (2004). Isolation and identification of phosphatidic acid targets from plants. Plant J. 39, 527–536. doi: 10.1111/j.1365-313X.2004.02152.x
Tian, S., Mao, X., Zhang, H., Chen, S., Zhai, C., Yang, S., et al. (2013). Cloning and characterization of TaSnRK2.3, a novel SnRK2 gene in common wheat. J. Exp. Bot. 64, 2063–2080. doi: 10.1093/jxb/ert072
Umezawa, T., Sugiyama, N., Mizoguchi, M., Hayashi, S., Myouga, F., Yamaguchi-Shinozaki, K., et al. (2009). Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 17588–17593. doi: 10.1073/pnas.0907095106
Umezawa, T., Sugiyama, N., Takahashi, F., Anderson, J. C., Ishihama, Y., Peck, S. C., et al. (2013). Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci. Signal. 6, rs8. doi: 10.1126/scisignal.2003509
Umezawa, T., Yoshida, R., Maruyama, K., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2004). SRK2C, a SNF1 -related protein kinase 2, improves drought tolerance by controlling stress -responsive gene expression in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 101, 17306–17311. doi: 10.1073/pnas.0407758101
Vlad, F., Turk, B. E., Peynot, P., Leung, J., and Merlot, S. (2008). A versatile strategy to define the phosphorylation preferences of plant protein kinases and screen for putative substrates. Plant J. 55, 104–117. doi: 10.1111/j.1365-313X.2008.03488.x
Wasilewska, A., Vlad, F., Sirichandra, C., Redko, Y., Jammes, F., Valon, C., et al. (2008). An update on abscisic acid signaling in plants and more. Mol. Plant 1, 198–217. doi: 10.1093/mp/ssm022
Weiner, J. J., Peterson, F. C., Volkman, B. F., and Cutler, S. R. (2010). Structural and functional insights into core ABA signaling. Curr. Opin. Plant Biol. 13, 495–502. doi: 10.1016/j.pbi.2010.09.007
Xiang, Y., Huang, Y., and Xiong, L. (2007). Characterization of stress -responsive CIPK genes in rice for stress tolerance improvement. Plant Physiol. 144, 1416–1428. doi: 10.1104/pp.107.101295
Xiang, Y., Tang, N., Du, H., Ye, H., and Xiong, L. (2008). Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol. 148, 1938–1952. doi: 10.1104/pp.108.128199
Xiao, B., Huang, Y., Tang, N., and Xiong, L. (2007). Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theor. Appl. Genet. 115, 35–46. doi: 10.1007/s00122-007-0538-9
Xiong, L., and Zhu, J. K. (2002). Molecular and genetic aspects of plant responses to osmotic stress. Plant Cell Environ. 25, 131–139. doi: 10.1046/j.1365-3040.2002.00782.x
Yamaguchi-Shinozaki, K., Mundy, J., and Chua, N. H. (1990). Four tightly linked rab genes are differentially expressed in rice. Plant Mol. Biol. 14, 29–39. doi: 10.1007/BF00015652
Yoshida, R., Hobo, T., Ichimura, K., Mizoguchi, T., Takahashi, F., Aronso, J., et al. (2002). ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol. 43, 1473–1483. doi: 10.1093/pcp/pcf188
Zhang, L., Xiao, S., Li, W., Feng, W., Li, J., Wu, Z., et al. (2011). Overexpression of a Harpin-encoding gene hrf1 in rice enhances drought tolerance. J. Exp. Bot. 62, 4229–4238. doi: 10.1093/jxb/err131
Zhang, Q. (2007). Strategies for developing Green Super Rice. Proc. Natl. Acad. Sci. U.S.A. 104, 16402–16409. doi: 10.1073/pnas.0708013104
Zhang, Z., and Huang, R. (2013). Analysis of Malondialdehyde, Chlorophyll, Proline, Soluble sugar, Glutathione content in seedling. Bioprotocol 3:e817. doi: 10.21769/BioProtoc.817
Zhu, J. K. (2001). Plant salt tolerance. Trends Plant Sci. 6, 66–72. doi: 10.1016/S1360-1385(00)01838-0
Keywords: SAPK2, abscisic acid (ABA), rice, CRISPR/Cas9 system, germination, drought tolerance, reactive oxygen species (ROS)
Citation: Lou D, Wang H, Liang G and Yu D (2017) OsSAPK2 Confers Abscisic Acid Sensitivity and Tolerance to Drought Stress in Rice. Front. Plant Sci. 8:993. doi: 10.3389/fpls.2017.00993
Received: 25 January 2017; Accepted: 26 May 2017;
Published: 13 June 2017.
Edited by:
Paul E. Verslues, Academia Sinica, TaiwanReviewed by:
Omar Borsani, University of the Republic, UruguayTaishi Umezawa, Tokyo University of Agriculture and Technology, Japan
Copyright © 2017 Lou, Wang, Liang and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Diqiu Yu, eWRxQHh0YmcuYWMuY24=
 Dengji Lou1,2
Dengji Lou1,2 Gang Liang
Gang Liang Diqiu Yu
Diqiu Yu