- School of Biological Sciences, University of Essex, Colchester, United Kingdom
Systems studies of drought stress in resurrection plants and other xerophytes are rapidly identifying a large number of genes, proteins and metabolites that respond to severe drought stress or desiccation. This has provided insight into drought resistance mechanisms, which allow xerophytes to persist under such extreme environmental conditions. Some of the mechanisms that ensure cellular protection during severe dehydration appear to be unique to desert species, while many other stress signaling pathways are in common with well-studied model and crop species. However, despite the identification of many desiccation inducible genes, there are few “gene-to-field” examples that have led to improved drought tolerance and yield stability derived from resurrection plants, and only few examples have emerged from model species. This has led to many critical reviews on the merit of the experimental approaches and the type of plants used to study drought resistance mechanisms. This article discusses the long-standing arguments between the ecophysiology and molecular biology communities, on how to “drought-proof” future crop varieties. It concludes that a more positive and inclusive dialogue between the different disciplines is needed, to allow us to move forward in a much more constructive way.
Introduction
Four hundred and fifty million years of land plant evolution has generated biological complexity, which has allowed plants to adapt to terrestrial environments, ranging from extreme cold environments in the Arctic and Antarctica, high salinity environments to extreme temperature changes and drought conditions in desert environments (von Willert et al., 1990; Alberdi et al., 2002; Amtmann et al., 2005). Plants that inhabit these environments are collectively called extremophiles, which harbor a range of different mechanisms that allow them to withstand these extreme environments.
During the green revolution of late twentieth century, breeding dramatically modified plant architecture, and yield improvements were made by selecting for characteristics such as rapid growth and a reduction of vegetative biomass in favor of fruit and seed production (Pingali, 2012). However, artificial selection for yield inadvertently reduced diversity, resulting in the loss of abiotic stress tolerance, with crop species likely to be more sensitive to abiotic stress compared to their wild ancestors (Mayrose et al., 2011; Koziol et al., 2012). Abiotic stresses dramatically reduce crop yields posing a threat to food security (Boyer, 1982; Cramer et al., 2011), and with the worldwide population growth expected to reach 9.7 billion people by 2050 (United Nations Department of Economic Social Affairs, 2017), the demand for global crop production is expected to double by 2050 (Tilman et al., 2011). This problem is likely to be exacerbated in the future by climate change (Mittler and Blumwald, 2010; Lesk et al., 2016). Furthermore future expansion of agricultural area is likely to occur in drylands and deserts (Millennium Ecosystem Assessment, 2005; Millenium Ecosystem Assessment, 2010), and new solutions to meet the world's future food security by improving crop yields is vital to not only prevent losses where crops are currently grown but also to cultivate them on more marginal land (Foley et al., 2011).
Interestingly, just as the plant stress community is beginning to tackle the molecular mechanisms of combined stress tolerances in models, crops and extremophiles, crop ecophysiologists are reassessing how misconceptions of stress resistance mechanisms may be avoided, advocating the need for clear physiological frameworks to meaningfully integrate the wealth of genetic response data. This article will focus on the efforts being made to understand dehydration resistance mechanisms in extremophile and model plants, and discuss the prospects of unlocking the genetic codes and mechanisms of extremophiles in the battle for stress tolerant crops.
Why are Resurrection Plants so Superb at Surviving Drought Stress?
Until recently, our knowledge of the molecular mechanisms of stress tolerance in extremophiles was relatively limited, but with the onset of next generation sequencing technologies, the number genome and transcriptome datasets of extremophiles has steadily increased (Oh et al., 2012; Dinakar and Bartels, 2013). In this context, halophytes have traditionally attracted more attention at the molecular level, partly because they include highly salt tolerant close relatives of Arabidopsis thaliana, such as Thellungiella parvula and Eutrema salsugineum (Dassanayake et al., 2011; Wu et al., 2012), allowing for direct comparisons of stress tolerance mechanisms such as salt, cold, heat, drought, and freezing tolerances already widely studied in Arabidopsis (Lee et al., 2012; Koch and German, 2013). E. salsugineum also harbors tolerances to low soil nitrogen (Kant et al., 2008), high boron levels (Lamdan et al., 2012), low phosphate levels (Velasco et al., 2016) and heat stress (Higashi et al., 2013), which is an exciting prospect for translating multiple stress tolerance traits to other plant species, including agronomically relevant Brassicaceae.
The interest in vegetative desiccation tolerance is illustrated by the large the number of publications relating to their ecology, physiology and molecular mechanism over the last 20 years, with 700 publications containing a reference to desiccation tolerance in plants, of which 257 were linked to resurrection plants (https://www.ncbi.nlm.nih.gov/pubmed/, search terms: desiccation tolerance, resurrection plants, last searched 3/2/18). With more than 130 known varieties, resurrection plants are mostly found in arid and semi-arid environments, (Gaff, 1971, 1977), and are probably the best studied of the xerophytes. Common to all resurrection plants is a vegetative desiccation tolerance (Oliver et al., 2000), and a small number of different species have been extensively studied with the aim to identify the underlying molecular mechanisms (Farrant, 2000; Bartels and Salamini, 2001; Cooper, 2002; Bartels, 2005; Farrant et al., 2015). Resurrection plants rapidly respond to water deficiency by switching into a “stress mode” that leads to a complete inhibition of photosynthesis and an overproduction of reactive oxygen species (ROS) in the chloroplasts due to excess light energy. ROS subsequently oxidize proteins and lipids, damage DNA and RNA, and ultimately lead to programmed cell death (Farrant et al., 2003).
Transcriptome analysis of Craterostigma plantagineum and Haberlea rhodopensis (Rodriguez et al., 2010; Gechev et al., 2013; Giarola et al., 2017) under early dehydration, desiccation and subsequent rehydration revealed common genetic pathways among desiccation-tolerant species. Resurrection plants undergo different stages during desiccation, which are accompanied by distinct physiological, metabolic, and molecular changes extensively reviewed by Farrant et al. (2015). Essentially, during the early dehydration stage, photosynthesis is shut down, ABA dependent responses to water stress become prominent, there is increased activity of antioxidants and a redirection of metabolism to the increased formation of sucrose and oligosaccharides as osmo-protectants (Rodriguez et al., 2010; Farrant et al., 2015). The late stages of drying are indicated by increased expression of proteins involved in signal transduction, altering sugar metabolism and genes encoding classical stress-associated proteins such as early light-inducible (ELIPs), Heat Shock Proteins (HSPs) and Late Embryogenesis Abundant (LEAs) proteins (Gechev et al., 2013; Farrant et al., 2015; Costa et al., 2017). This reprogramming of metabolism is driven by the induction of known stress responsive transcription factors, such as NACs, NF-Ys, HSFs, and WRKYs (Gechev et al., 2013; Farrant et al., 2015; Costa et al., 2017). It appears that resurrection plants are generally in a constantly “primed state,” with high basal levels of protective sugars, antioxidants and defense proteins, allowing a rapid and strong response during desiccation (Rodriguez et al., 2010; Gechev et al., 2013; Costa et al., 2016).
What are the Lessons Learned From Resurrection Plants?
Many of the genes identified in the above transcriptome studies have also been identified in drought responses of Arabidopsis and other plant models (Tripathi et al., 2014; Bechtold et al., 2016), suggesting that common signaling pathways are in operation. The advantages of Arabidopsis as a model for plant molecular biology and the role it played and still plays in investigating abiotic stress response pathways is undisputed (Provart et al., 2016). Many of the stress signaling pathways identified are now known to be general responses that appear to be conserved in many higher plant species (Boscaiu et al., 2012; Provart et al., 2016).
However, there are counterarguments which suggest that too much emphasis is being placed on investigating unsuitable experimental models, such as Arabidopsis (Boscaiu et al., 2012). For example, Arabidopsis and many crop plants die at leaf water potentials of around −3 MPa (van der Weele et al., 2000; Fitter and Hay, 2002). Consequently, Arabidopsis may not be appropriate to identify dehydration tolerance pathways. Yet most of the biochemical and molecular studies on plant responses to abiotic stress have been carried out using Arabidopsis (Bressan et al., 2009), which has resulted in the identification and isolation of stress-tolerance genes, some of which have been used in modulating crop stress tolerance with varying success (reviewed by Mittler and Blumwald, 2010; Varshney et al., 2011). Especially stress responsive transcription factors (TFs) such as the AP2/EREBP family DREBs, MYB, WRKY, NAC, bHLH, and bZIPs have attracted attention due to their important roles in plant stress responses and improved stress tolerance phenotypes when overexpressed in Arabidopsis and crop plants alike (reviewed by Wang et al., 2016).
From many of these transgenic studies it is evident that TFs have conserved functions across species boundaries including resurrection plants (see above), and many of the Arabidopsis TFs have been shown to confer stress tolerances in unrelated crop species and vice versa (Jiang et al., 2011; Wang et al., 2016). However, the bottom line is that currently few abiotic stress tolerant, high yielding crops are grown in our fields utilizing the genes identified from the many biochemical and molecular analyses carried out on Arabidopsis and/or model crops reviewed by (Mickelbart et al., 2015; Ricroch and Hénard-Damave, 2016).
This has raised several questions with regard to the experimental conditions applied in gene discovery studies, the appropriateness of the stress phenotypes being assessed (Blum and Tuberosa, 2018), or whether common stress signaling pathways are indeed the best targets (Boscaiu et al., 2012). While vegetative desiccation tolerance in resurrection plants is clearly at the extreme end of the stress survival spectrum, it is argued that extremophile species may act as a source of novel genes for the genetic improvement of stress tolerance in crops (Boscaiu et al., 2012). Yet many of the TF families identified in Arabidopsis have also been identified in resurrection plants in response to drought stress (see above; Gechev et al., 2013; Farrant et al., 2015; Costa et al., 2017). It is now generally accepted that many of these common signaling pathways are functional in xerophytes adapted to arid or semi-arid conditions (Farrant et al., 2015; Costa et al., 2016, 2017). Therefore, the argument regarding the suitability of stress sensitive model species vs. xerophytes/resurrection plants in the pursuit to study drought responses is unresolved, and the question remains how studies on resurrection plants are going to lead us to novel genes and stress signaling pathways, when so far, many transcriptome studies have mostly delivered on general stress pathways?
Interestingly, transcriptome studies of resurrection plants not only found a high proportion of unknown transcripts (33% C. plantagineum and ~40% H. rhodopensis; Rodriguez et al., 2010; Gechev et al., 2013), but in the case of the C. plantagineum transcriptome, also identified many taxonomically restricted genes (TRGs) and non-protein coding RNAs (ncRNAs) (Giarola et al., 2014; Giarola and Bartels, 2015). TRGs are known to code for new traits required for the adaptation of organisms to particular environmental conditions (Johnson and Tsutsui, 2011), and it has been suggested that these may harbor the potential for novel gene discovery linked to desiccation tolerance.
Early attempts to utilize TRGs such as desiccation induced proteins from C. plantagineum to improve drought tolerance in tobacco failed (Iturriaga et al., 1992), while recent examples successfully used tonoplast cation/H+ antiporter and H+ pyrophophatases genes from the xerophyte Zygophyllum xanthoxylum to enhance stress tolerance in alfalfa and Lotus corniculatus (Bao et al., 2014, 2016). These studies suggested that improved “systems wide approaches” of large datasets, together with comparative genomics that aim to identify whole network-based homologies between species could be more successful in discovering novel genes/pathways that underlie differences and similarities across species (Farrant et al., 2015).
The Continuous Argument—Are We Studying the Right System?
There also appears to be a lack of synergy between different disciplines with physiology, ecophysiology on the one hand, and genetics and molecular biology on the other. Perhaps the problem is not only the type of plant we study, but also how we study stress tolerance mechanisms in general? Crop physiologists rightly argue that molecular mechanisms of plant survival traits are often studied in isolation to physiological responses, whether this is carried out in model species such as Arabidopsis (Blum, 2005), or in extremophiles such as the resurrection plants (Blum and Tuberosa, 2018). This may be confounded by experimental setups that are not fit for purpose or comparable to plant responses under field conditions, for example pot grown vs. field grown plants (Passioura J. B., 2006; Poorter et al., 2012).
Desiccation tolerance in resurrection plants is a survival trait, and survival traits after any given stress have generally been a popular phenotype for gene discovery and gene function in model species (Woo et al., 2008; Skirycz et al., 2011), and drought resistance is often assessed under quite severe conditions in which plant survival is scored after a prolonged period of soil drying. Yet, even with what superficially appears to be simple phenotype (i.e., survival); the mechanisms ensuring drought survival are often no being fully assessed (Blum and Tuberosa, 2018). Plant survival after a period of drought stress, can either be due to dehydration avoidance or dehydration tolerance (Levitt, 1980). Avoidance strategies are observed in plants that maintain high plant water status due to osmotic adjustment during dehydration, while tolerance usually result from a delayed mortality in response to low plant water status (Levitt, 1980). In resurrection plants, the desiccation survival trait is due to delayed mortality and suppression of drought-related senescence pathways (Griffiths et al., 2014). Therefore, when phenotyping for dehydration survival there is a need to distinguish between dehydration avoidance and dehydration tolerance, and how this may affect survival and recovery. Without this distinction, it has been argued that molecular studies performed to identify the genes that underlie this trait might be biased. For example, in gene-expression studies, RNA is sampled after a set period of dehydration, or at a given relative soil water content, and often the assumption is made that all genotypes are therefore subjected to the same level of stress (Des Marais et al., 2012). Without additional information regarding the physiological- or plant water status this could result in artifacts, where differences in plant water status on the day of sampling are not adequately taken into account. Consequently, an in depth understanding of the physiological basis of the phenotype is essential to avoid misinterpretations (Zhang et al., 2014; Bechtold et al., 2016). While there have clearly been some deficiencies with molecular biology approaches in many molecular/genomic centric studies used to study drought stress, recent critics of the topic fail to mention the progress made by molecular biologists to address some of these early short-comings in the area. In recent years, much more effort has been placed on linking plant genomic events to the metabolic status and the rate of plant dehydration especially in model species, such as Arabidopsis, Medicago truncatula and rice (Harb et al., 2010; Des Marais et al., 2012; Lasky et al., 2014; Zhang et al., 2014; Bechtold et al., 2016; Wilkins et al., 2016), resurrection plants (Farrant et al., 2015) and other extremophiles (Brinker et al., 2010). Often these changes are recorded along a gradient of relative water content, plant water potential, plant physiological or metabolic changes (Brinker et al., 2010; Farrant and Moore, 2011; Zhang et al., 2014; Farrant et al., 2015; Bechtold et al., 2016). For example, a significant relationship between water potential and the number of differentially expressed genes was observed in a progressive drought time-series experiments, allowing the clear distinction between early and late dehydration responses at the transcriptional as well as at the physiological level (Bechtold et al., 2016). More importantly, the switch between early and late dehydration responses coincided with a breakpoint in the soil dehydration profile, and this breakpoint clearly differs between natural accessions of Arabidopsis (Ferguson et al., 2018), suggesting difference in dehydration strategies and potentially drought resistance strategies.
In crop plants drought survival through delayed mortality is unlikely to be a suitable option due to expected growth and yield penalties, and therefore questions regarding the usefulness of survival traits in achieving meaningful improvements still remain (Passioura J., 2006; Skirycz and Inzé, 2010; Blum, 2011; Skirycz et al., 2011). Resurrection plants, although excellent models to investigate drought tolerance strategies associated with delayed mortality, may therefore not be appropriate to investigate avoidance strategies associated with the maintenance of photosynthesis and growth. Consequently, plant species adapted to arid and semi-arid environments that avoid desiccation may be more useful models to study the phenomena of stress tolerance under extreme environmental conditions. One such species is the C3 desert plant Rhazya stricta, which is common in arid zones at elevations of 100–700 m above sea level, and overcomes water restriction through long tap roots to access water in underground river beds (Batanouny and Baeshin, 1983).
The Need to Study Xerophytes in Their Native Environment
Extensive physiological phenotyping under native growth conditions is essential due to the extreme conditions experienced in their native environments. Furthermore, growth conditions in control environments often do not fully reflect the conditions experienced in desert environments, especially with regards to the prevailing light conditions (Table 1), even though light is one of the main contributing factors of chloroplast damage during desiccation (Farrant et al., 2003). Relatively few studies have attempted transcriptome analysis from plants grown in their native environments (Table 1), and even fewer have underpinned these studies with extensive physiological characterization of these plants in their native habitats (Yates et al., 2014).
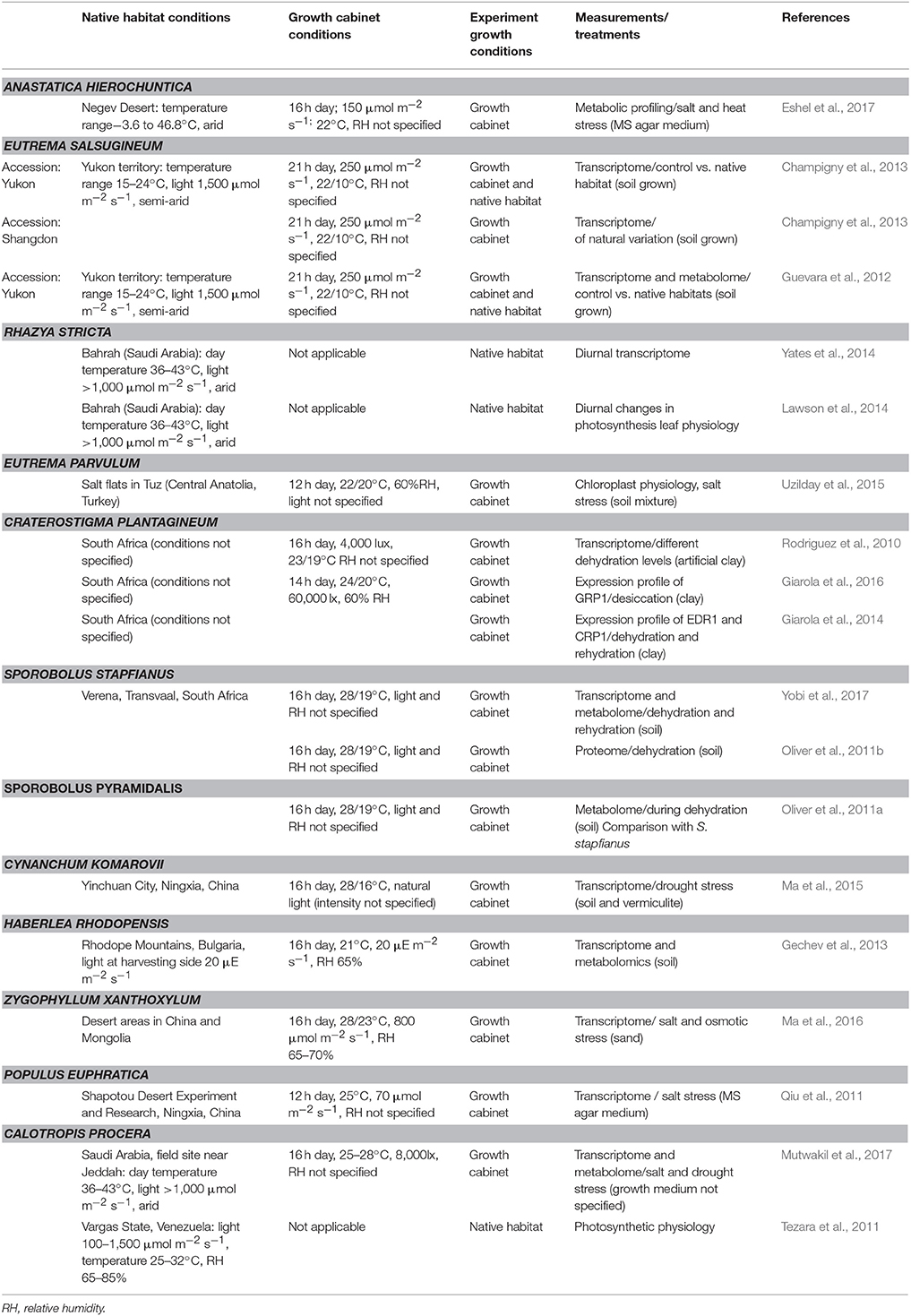
Table 1. Comparison of growth conditions in transcriptome, proteome, metabolome, and physiological studies of extremophiles.
For example, physiological and transcriptome analysis of R. stricta in its native desert environment revealed that R. stricta maintains growth and high photosynthetic rates at leaf temperatures as high as 43°C, light intensities >1,000 μmol m−1 s−1 and high vapor pressure deficits (VPDs; Lawson et al., 2014), and is one of the few desert C3 shrubs that achieve such high photosynthetic yields at leaf temperatures above 40°C (Mooney et al., 1978; Tezara et al., 2011; Rivas et al., 2017). Gene expression and gene sequence analysis identified two RUBISCO ACTIVASE isoforms, that are likely responsible for the maintenance of high Rubisco activities and photosynthetic rates under these extreme conditions (Lawson et al., 2014). Interestingly, in this very hot and arid environment, R. strica is able to maintain an adequate water supply in order to maintain a high and constant photosynthetic activity (Lawson et al., 2014). While strictly speaking R. stricta may not suffer from drought stress in those circumstances, it nevertheless gives us an insight into extreme thermo- and high light tolerance in arid environments, which are often part of drought stress conditions experienced in the field. Importantly, by combining detailed physiological analysis with genetic investigations, it was possible to identify some potentially different physiological adaptation mechanisms that go beyond the usual TFs and chaperones mentioned above. Interestingly, a detailed study of the diurnal transcriptome analysis of R. stricta, identified considerable overlap with C. plantagineum and P. euphratica such as cysteine proteases and raffinose synthesis genes, highlighting more general protective mechanisms against high temperature (Yates et al., 2014). Furthermore, gene families specific to R. stricta, such as photosynthesis and respiration associated genes, were differentially expressed at midday during a diurnal period responding to VPD, temperature and light levels (Yates et al., 2014). These changes coincided with changes in photosynthetic physiology (Lawson et al., 2014; Yates et al., 2014). Importantly, a number of these unique protein families were found to have diverged from their homologs in other species (Yates et al., 2014). Therefore, C3 species from arid/semi-arid environments that do not undergo a dormant state, could provide novel gene targets responsible for maintaining photosynthesis more commonly associated with dehydration avoidance.
Conclusions
With unprecedented access to increasing genome information and transcriptome datasets from a variety of plants, coupled with tools to analyse and compare these datasets, we are beginning to identify gene families and gene regulatory networks behind complex traits such as dehydration tolerance. It is only by developing a more realistic framework in which to study drought resistance mechanism that we will make progress in understanding how plant productivity can be maximized under water limiting conditions. For this to happen, a more positive and inclusive dialogue between the different disciplines, all primarily concerned with “drought-proofing” future crop varieties, is essential in order to move forward in a much more constructive way.
Author Contributions
The author confirms being the sole contributor of this work and approved it for publication.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Alberdi, M., Bravo, L. A., Gutiérrez, A., Gidekel, M., and Corcuera, L. J. (2002). Ecophysiology of Antarctic vascular plants. Physiol. Plant 115, 479–486. doi: 10.1034/j.1399-3054.2002.1150401.x
Amtmann, A., Bohnert, H. J., and Bressan, R. A (2005). Abiotic Stress and plant genome evolution search for new models. Plant Physiol. 138, 127–130. doi: 10.1104/pp.105.059972
Bao, A. K., Du, B. Q., Touil, L., Kang, P., Wang, Q. L., and Wang, S. M. (2016). Co-expression of tonoplast Cation/H+antiporter and H+-pyrophosphatase from xerophyte Zygophyllum xanthoxylum improves alfalfa plant growth under salinity, drought and field conditions. Plant Biotechnol. J. 14, 964–975. doi: 10.1111/pbi.12451
Bao, A. K., Wang, Y. W., Xi, J. J., Liu, C., Zhang, J. L., and Wang, S. M. (2014). Co-expression of xerophyte Zygophyllum xanthoxylum ZxNHX and ZxVP1-1 enhances salt and drought tolerance in transgenic Lotus corniculatus by increasing cations accumulation. Funct. Plant Biol. 41, 203–214. doi: 10.1071/FP13106
Bartels, D. (2005). Desiccation tolerance studied in the resurrection plant Craterostigma plantagineum. Integr. Comp. Biol. 45, 696–701. doi: 10.1093/icb/45.5.696
Bartels, D., and Salamini, F. (2001). Desiccation tolerance in the resurrection plant Craterostigma plantagineum. a contribution to the study of drought tolerance at the molecular level. Plant Physiol. 127, 1346–1353. doi: 10.1104/pp.010765
Batanouny, K. H., and Baeshin, N. A. (1983). Plant communities along the Medina-Badr road across the Hejaz mountains, Saudi Arabia. Vegetatio 53, 33–43. doi: 10.1007/BF00039769
Bechtold, U., Penfold, C. A., Jenkins, D. J., Legaie, R., Moore, J. D., Lawson, T., et al. (2016). Time-series transcriptomics reveals that AGAMOUS-LIKE22 affects primary metabolism and developmental processes in drought-stressed arabidopsis. Plant Cell 28, 345–366. doi: 10.1105/tpc.15.00910
Blum, A. (2005). Drought resistance, water-use efficiency, and yield potential—are they compatible, dissonant, or mutually exclusive? Aust. J. Agric. Res. 56, 1159–1168. doi: 10.1071/AR05069
Blum, A., and Tuberosa, R. (2018). Dehydration survival of crop plants and its measurement. J. Exp. Bot. 69:erx445. doi: 10.1093/jxb/erx445
Boscaiu, M., Donat, P., Llinares, J., and Vicente, O. (2012). Stress-tolerant wild plants: a source of knowledge and biotechnological tools for the genetic improvement of stress tolerance in crop plants. Not. Bot. Horti Agrobot. Cluj-Napoca 40, 323–327. doi: 10.15835/nbha4028199
Boyer, J. S. (1982). Plant productivity and environment. Science 218, 443–448. doi: 10.1126/science.218.4571.443
Bressan, R., Bohnert, H., and Zhu, J. K. (2009). Abiotic stress tolerance: from gene discovery in model organisms to crop improvement. Mol. Plant 2, 1–2. doi: 10.1093/mp/ssn097
Brinker, M., Brosché, M., Vinocur, B., Abo-Ogiala, A., Fayyaz, P., Janz, D., et al. (2010). Linking the salt transcriptome with physiological responses of a salt-resistant populus species as a strategy to identify genes important for stress acclimation. Plant Physiol. 154, 1697–1709. doi: 10.1104/pp.110.164152
Champigny, M. J., Sung, W. W., Catana, V., Salwan, R., Summers, P. S., Dudley, S. A., et al. (2013). RNA-Seq effectively monitors gene expression in Eutrema salsugineum plants growing in an extreme natural habitat and in controlled growth cabinet conditions. BMC Genomics 14:578. doi: 10.1186/1471-2164-14-578
Cooper, K. (2002). Recovery of the resurrection plant Craterostigma wilmsii from desiccation: protection versus repair. J. Exp. Bot. 53, 1805–1813. doi: 10.1093/jxb/erf028
Costa, M. C. D., Farrant, J. M., Oliver, M. J., Ligterink, W., Buitink, J., and Hilhorst, H. M. W. (2016). Key genes involved in desiccation tolerance and dormancy across life forms. Plant Sci. 251, 162–168. doi: 10.1016/j.plantsci.2016.02.001
Costa, M. D., Artur, M. A. S., Maia, J., Jonkheer, E., Derks, M. L., Nijveen, H., et al. (2017). A footprint of desiccation tolerance in the genome of Xerophyta viscosa. Nat. Plants 3, 17038. doi: 10.1038/nplants.2017.38
Cramer, G. R., Urano, K., Delrot, S., Pezzotti, M., and Shinozaki, K. (2011). Effects of abiotic stress on plants: a systems biology perspective. BMC Plant Biol. 11:163. doi: 10.1186/1471-2229-11-163
Dassanayake, M., Oh, D. H., Haas, J. S., Hernandez, A., Hong, H., Ali, S., et al. (2011). The genome of the extremophile crucifer Thellungiella parvula. Nat. Genet. 43, 913–918. doi: 10.1038/ng.889
Des Marais, D. L., McKay, J. K., Richards, J. H., Sen, S., Wayne, T., and Juenger, T. E. (2012). Physiological genomics of response to soil drying in diverse Arabidopsis accessions. Plant Cell 24, 893–914. doi: 10.1105/tpc.112.096180
Dinakar, C., and Bartels, D. (2013). Desiccation tolerance in resurrection plants: new insights from transcriptome, proteome and metabolome analysis. Front. Plant Sci. 4:482. doi: 10.3389/fpls.2013.00482
Eshel, G., Shaked, R., Kazachkova, Y., Khan, A., Eppel, A., Cisneros, A., et al. (2017). Anastatica hierochuntica, an Arabidopsis Desert relative, is tolerant to multiple abiotic stresses and exhibits species-specific and common stress tolerance strategies with its halophytic relative, Eutrema (Thellungiella) salsugineum. Front. Plant Sci. 7:992. doi: 10.3389/fpls.2016.01992
Farrant, J. M. (2000). A comparison of mechanisms of desiccation tolerance among three angiosperm resurrection plant species. Plant Ecol. 151, 29–39. doi: 10.1023/A:1026534305831
Farrant, J. M., and Moore, J. P. (2011). Programming desiccation-tolerance: from plants to seeds to resurrection plants. Curr. Opin. Plant Biol. 14, 340–345. doi: 10.1016/j.pbi.2011.03.018
Farrant, J. M., Cooper, K., Hilgart, A., Abdalla, K. O., Bentley, J., Thomson, J. A., et al. (2015). A molecular physiological review of vegetative desiccation tolerance in the resurrection plant Xerophyta viscosa (Baker). Planta 242, 407–426. doi: 10.1007/s00425-015-2320-6
Farrant, J. M., Vander Willigen, C., Loffell, D. A., Bartsch, S., and Whittaker, A. (2003). An investigation into the role of light during desiccation of three angiosperm resurrection plants. Plant Cell Environ. 26, 1275–1286. doi: 10.1046/j.0016-8025.2003.01052.x
Ferguson, J. N., Humphry, M., Lawson, T., Oliver, B., and Bechtold, U. (2018). Natural variation of life-history traits, water use, and drought responses in Arabidopsis. Plant Direct. 2:e00035. doi: 10.1002/pld3.35
Fitter, A., and Hay, R. (2002). 4 - Water BT - Environmental Physiology of Plants, 3rd Edn. London: Academic Press.
Foley, J. A., Ramankutty, N., Brauman, K. A., Cassidy, E. S., Gerber, J. S., Johnston, M., et al. (2011). Solutions for a cultivated planet. Nature 478, 337–342. doi: 10.1038/nature10452
Gaff, D. F. (1971). Desiccation-tolerant flowering plants in southern Africa. Science 174, 1033–1034. doi: 10.1126/science.174.4013.1033
Gaff, D. F. (1977). Desiccation tolerant vascular plants of southern Africa. Oecologia 31, 95–109. doi: 10.1007/BF00348713
Gechev, T. S., Benina, M., Obata, T., Tohge, T., Sujeeth, N., Minkov, I., et al. (2013). Molecular mechanisms of desiccation tolerance in the resurrection glacial relic Haberlea rhodopensis. Cell. Mol. Life Sci. 70, 689–709. doi: 10.1007/s00018-012-1155-6
Giarola, V., and Bartels, D. (2015). What can we learn from the transcriptome of the resurrection plant Craterostigma plantagineum? Planta 242, 427–434. doi: 10.1007/s00425-015-2327-z
Giarola, V., Hou, Q., and Bartels, D. (2017). Angiosperm plant Desiccation tolerance: hints from transcriptomics and genome sequencing. Trends Plant Sci. 22, 705–717. doi: 10.1016/j.tplants.2017.05.007
Giarola, V., Krey, S., Frerichs, A., and Bartels, D. (2014). Taxonomically restricted genes of Craterostigma plantagineum are modulated in their expression during dehydration and rehydration. Planta 241, 193–208. doi: 10.1007/s00425-014-2175-2
Giarola, V., Krey, S., von den Driesch, B., and Bartels, D. (2016). The Craterostigma plantagineum glycine-rich protein CpGRP1 interacts with a cell wall-associated protein kinase 1 (CpWAK1) and accumulates in leaf cell walls during dehydration. New Phytol. 210, 535–550. doi: 10.1111/nph.13766
Griffiths, C. A., Gaff, D. F., and Neale, A. D. (2014). Drying without senescence in resurrection plants. Front. Plant Sci. 5:36. doi: 10.3389/fpls.2014.00036
Guevara, D. R., Champigny, M. J., Tattersall, A., Dedrick, J., Wong, C. E., Li, Y., et al. (2012). Transcriptomic and metabolomic analysis of Yukon Thellungiella plants grown in cabinets and their natural habitat show phenotypic plasticity. BMC Plant Biol. 12:175. doi: 10.1186/1471-2229-12-175
Harb, A., Krishnan, A., Ambavaram, M. M., and Pereira, A. (2010). Molecular and physiological analysis of drought stress in Arabidopsis reveals early responses leading to acclimation in plant growth. Plant Physiol. 154, 1254–1271. doi: 10.1104/pp.110.161752
Higashi, Y., Ohama, N., Ishikawa, T., Katori, T., Shimura, A., Kusakabe, K., et al. (2013). HsfA1d, a protein identified via FOX hunting using thellungiella salsuginea cDNAs improves heat tolerance by regulating heat-stress-responsive gene expression. Mol. Plant 6, 411–422. doi: 10.1093/mp/sst024
Iturriaga, G., Schneider, K., Salamini, F., and Bartels, D. (1992). Expression of desiccation-related proteins from the resurrection plant Craterostigma plantagineum in transgenic tobacco. Plant Mol. Biol. 20, 555–558. doi: 10.1007/BF00040614
Jiang, F., Wang, F., Wu, Z., Li, Y., Shi, G., Hu, J., et al. (2011). Components of the Arabidopsis CBF cold-response pathway are conserved in non-heading Chinese cabbage. Plant Mol. Biol. Rep. 29, 525–532. doi: 10.1007/s11105-010-0256-3
Johnson, B. R., and Tsutsui, N. D. (2011). Taxonomically restricted genes are associated with the evolution of sociality in the honey bee. BMC Genomics 12:164. doi: 10.1186/1471-2164-12-164
Kant, S., Bi, Y. M., Weretilnyk, E., Barak, S., and Rothstein, S. J. (2008). The Arabidopsis halophytic relative Thellungiella halophila Tolerates nitrogen-limiting conditions by maintaining growth, nitrogen uptake, and assimilation. Plant Physiol. 147, 1168–1180. doi: 10.1104/pp.108.118125
Koch, M. A., and German, D. A. (2013). Taxonomy and systematics are key to biological information: Arabidopsis, Eutrema (Thellungiella), Noccaea and Schrenkiella (Brassicaceae) as examples. Front. Plant Sci. 4:267. doi: 10.3389/fpls.2013.00267
Koziol, L., Rieseberg, L. H., Kane, N., and Bever, J. D. (2012). Reduced drought tolerance during domestication and the evolution of weediness results from tolerance-growth trade-offs. Evolution 66, 3803–3814. doi: 10.1111/j.1558-5646.2012.01718.x
Lamdan, N. L., Attia, Z., Moran, N., and Moshelion, M. (2012). The Arabidopsis-related halophyte Thellungiella halophila: boron tolerance via boron complexation with metabolites? Plant Cell Environ. 35, 735–746. doi: 10.1111/j.1365-3040.2011.02447.x
Lasky, J. R., Des Marais, D. L., Lowry, D. B., Povolotskaya, I., McKay, J. K., Richards, J. H., et al. (2014). Natural variation in abiotic stress responsive gene expression and local adaptation to climate in Arabidopsis thaliana. Mol. Biol. Evol. 31, 2283–2296. doi: 10.1093/molbev/msu170
Lawson, T., Davey, P. A., Yates, S. A., Bechtold, U., Baeshen, M., Baeshen, N., et al. (2014). C3 photosynthesis in the desert plant Rhazya stricta is fully functional at high temperatures and light intensities. New Phytol. 201, 862–873. doi: 10.1111/nph.12559
Lee, Y. P., Babakov, A., de Boer, B., Zuther, E., and Hincha, D. K. (2012). Comparison of freezing tolerance, compatible solutes and polyamines in geographically diverse collections of Thellungiella sp. and Arabidopsis thaliana accessions. BMC Plant Biol. 12:131. doi: 10.1186/1471-2229-12-131
Lesk, C., Rowhani, P., and Ramankutty, N. (2016). Influence of extreme weather disasters on global crop production. Nature 529, 84–87. doi: 10.1038/nature16467
Levitt, J. (1980). “Responses of plants to environmental stresses,” in Water, Radiation, Salt, and Other Stresses, Vol 2, edn 2 (New York, NY: Academic Press), 607.
Ma, Q., Bao, A. K., Chai, W. W., Wang, W. Y., Zhang, J. L., Li, Y. X., et al. (2016). Transcriptomic analysis of the succulent xerophyte Zygophyllum xanthoxylum in response to salt treatment and osmotic stress. Plant Soil 402, 1–19. doi: 10.1007/s11104-016-2809-1
Ma, X., Wang, P., Zhou, S., Sun, Y., Liu, N., Li, X., et al. (2015). De novo transcriptome sequencing and comprehensive analysis of the drought-responsive genes in the desert plant Cynanchum komarovii. BMC Genomics 16, 753. doi: 10.1186/s12864-015-1873-x
Mayrose, M., Kane, N. C., Mayrose, I., Dlugosch, K. M., and Rieseberg, L. H. (2011). Increased growth in sunflower correlates with reduced defences and altered gene expression in response to biotic and abiotic stress. Mol. Ecol. 20, 4683–4694. doi: 10.1111/j.1365-294X.2011.05301.x
Mickelbart, M. V., Hasegawa, P. M., and Bailey-Serres, J. (2015). Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat. Rev. Genet. 16, 237–251. doi: 10.1038/nrg3901
Mittler, R., and Blumwald, E. (2010). Genetic engineering for modern agriculture: challenges and perspectives. Annu. Rev. Plant Biol. 61, 443–462. doi: 10.1146/annurev-arplant-042809-112116
Mooney, H., Björkman, O., and Collatz, G. (1978). Photosynthetic acclimation to temperature in the desert shrub, Larrea divaricata. I. carbon dioxide exchange characteristics of intact leaves. Plant Physiol. 61, 406–410. doi: 10.1104/pp.61.3.406
Mutwakil, M. Z., Hajrah, N. H., Atef, A., Edris, S., Sabir, M. J., Al-Ghamdi, A. K., et al. (2017). Transcriptomic and metabolic responses of Calotropis procera to salt and drought stress. BMC Plant Biol. 17, 231. doi: 10.1186/s12870-017-1155-7
Oh, D. H., Dassanayake, M., Bohnert, H. J., and Cheeseman, J. M. (2012). Life at the extreme: Lessons from the genome. Genome Biol. 13:241. doi: 10.1186/gb-2012-13-3-241
Oliver, M. J., Guo, L., Alexander, D. C., Ryals, J. A., Wone, B. W., and Cushman, J. C. (2011a). A sister group contrast using untargeted global metabolomic analysis delineates the biochemical regulation underlying desiccation tolerance in Sporobolus stapfianus. Plant Cell 23, 1231–1248. doi: 10.1105/tpc.110.082800
Oliver, M. J., Jain, R., Balbuena, T. S., Agrawal, G., Gasulla, F., and Thelen, J. J. (2011b). Proteome analysis of leaves of the desiccation-tolerant grass, Sporobolus stapfianus, in response to dehydration. Phytochemistry 72, 1273–1284. doi: 10.1016/j.phytochem.2010.10.020
Oliver, M. J., Tuba, Z., and Mishler, B. D. (2000). The evolution of vegetative desiccation tolerance in land plants. Plant Ecol. 151, 85–100. doi: 10.1023/A:1026550808557
Passioura, J. (2006). Increasing crop productivity when water is scarce - from breeding to field management. Agric. Water Manag. 176–196. doi: 10.1016/j.agwat.2005.07.012
Passioura, J. B. (2006). Viewpoint: the perils of pot experiments. Funct. Plant Biol. 33, 1075–1079. doi: 10.1071/FP06223
Pingali, P. (2012). Green revolution:impacts, limits, and the path ahead. Proc. Natl. Acad. Sci. U.S.A. 109, 12302–12308. doi: 10.1073/pnas.0912953109
Poorter, H., Bühler, J., Van Dusschoten, D., Climent, J., and Postma, J. A. (2012). Pot size matters: a meta-analysis of the effects of rooting volume on plant growth. Funct. Plant Biol. 39, 839–850. doi: 10.1071/FP12049
Provart, N. J., Alonso, J., Assmann, S. M., Bergmann, D., Brady, S. M., Brkljacic, J., et al. (2016). 50 years of Arabidopsis research: highlights and future directions. New Phytol. 209, 921–944. doi: 10.1111/nph.13687
Qiu, Q., Ma, T., Hu, Q., Liu, B., Wu, Y., Zhou, H., et al. (2011). Genome-scale transcriptome analysis of the desert poplar, Populus euphratica. Tree Physiol. 31, 452–461. doi: 10.1093/treephys/tpr015
Ricroch, A. E., and Hénard-Damave, M. C. (2016). Next biotech plants: new traits, crops, developers and technologies for addressing global challenges. Crit. Rev. Biotechnol. 36, 675–690. doi: 10.3109/07388551.2015.1004521
Rivas, R., Frosi, G., Ramos, D. G., Pereira, S., Benko-Iseppon, A. M., and Santos, M. G. (2017). Photosynthetic limitation and mechanisms of photoprotection under drought and recovery of Calotropis procera, an evergreen C3from arid regions. Plant Physiol. Biochem. 118, 589–599. doi: 10.1016/j.plaphy.2017.07.026
Rodriguez, M. C., Edsgärd, D., Hussain, S. S., Alquezar, D., Rasmussen, M., Gilbert, T., et al. (2010). Transcriptomes of the desiccation-tolerant resurrection plant Craterostigma plantagineum. Plant J. 63, 212–228. doi: 10.1111/j.1365-313X.2010.04243.x
Skirycz, A., and Inzé, D. (2010). More from less: plant growth under limited water. Curr. Opin. Biotechnol. 21, 197–203. doi: 10.1016/j.copbio.2010.03.002
Skirycz, A., Vandenbroucke, K., Clauw, P., Maleux, K., De Meyer, B., Dhondt, S., et al. (2011). Survival and growth of Arabidopsis plants given limited water are not equal. Nat. Biotechnol. 29, 212–214. doi: 10.1038/nbt.1800
Tezara, W., Colombo, R., Coronel, I., and Marín, O. (2011). Water relations and photosynthetic capacity of two species of Calotropis in a tropical semi-arid ecosystem. Ann. Bot. 107, 397–405. doi: 10.1093/aob/mcq245
Tilman, D., Balzer, C., Hill, J., and Befort, B. L. (2011). Global food demand and the sustainable intensification of agriculture. Proc. Natl. Acad. Sci. U.S.A. 108, 20260–20264. doi: 10.1073/pnas.1116437108
Tripathi, P., Rabara, R. C., and Rushton, P. J. (2014). A systems biology perspective on the role of WRKY transcription factors in drought responses in plants. Planta 239, 255–266. doi: 10.1007/s00425-013-1985-y
United Nations Department of Economic and Social Affairs, P. D. (2017). World population prospects the 2017 revision key findings and advance tables. World Popul. Prospect. 2017, 1–46. doi: 10.1017/CBO9781107415324.004
Uzilday, B., Ozgur, R., Sekmen, A. H., Yildiztugay, E., and Turkan, I. (2015). Changes in the alternative electron sinks and antioxidant defence in chloroplasts of the extreme halophyte Eutrema parvulum (Thellungiella parvula) under salinity. Ann. Bot. 115, 449–463. doi: 10.1093/aob/mcu184
van der Weele, C. M., Spollen, W. G., Sharp, R. E., and Baskin, T. I. (2000). Growth of Arabidopsis thaliana seedlings under water deficit studied by control of water potential in nutrient-agar media. J. Exp. Bot. 51, 1555–1562. doi: 10.1093/jexbot/51.350.1555
Varshney, R. K., Bansal, K. C., Aggarwal, P. K., Datta, S. K., and Craufurd, P. Q. (2011). Agricultural biotechnology for crop improvement in a variable climate: hope or hype? Trends Plant Sci. 16, 363–371. doi: 10.1016/j.tplants.2011.03.004
Velasco, V. M., Mansbridge, J., Bremner, S., Carruthers, K., Summers, P. S., Sung, W. W., et al. (2016). Acclimation of the crucifer Eutrema salsugineum to phosphate limitation is associated with constitutively high expression of phosphate-starvation genes. Plant Cell Environ. 39, 1818–1834. doi: 10.1111/pce.12750
von Willert, D. J., Eller, B. M., Werger, M. J. A., and Brinckmann, E. (1990). Desert succulents and their life strategies. Vegetatio 90, 133–143. doi: 10.1007/BF00033023
Wang, H., Wang, H., Shao, H., and Tang, X. (2016). Recent advances in utilizing transcription factors to improve plant abiotic stress tolerance by transgenic technology. Front. Plant Sci. 7:67. doi: 10.3389/fpls.2016.00067
Wilkins, O., Hafemeister, C., Plessis, A., Holloway-Phillips, M. M., Pham, G. M., Nicotra, A. B., et al. (2016). EGRINs (Environmental Gene Regulatory Influence Networks) in rice that function in the response to water deficit, high temperature, and agricultural environments. Plant Cell. 28, 365–2384. doi: 10.1105/tpc.16.00158
Woo, N. S., Badger, M. R., and Pogson, B. J. (2008). A rapid, non-invasive procedure for quantitative assessment of drought survival using chlorophyll fluorescence. Plant Methods 4:27. doi: 10.1186/1746-4811-4-27
Wu, H. J., Zhang, Z., Wang, J. Y., Oh, D. H., Dassanayake, M., Liu, B., et al. (2012). Insights into salt tolerance from the genome of Thellungiella salsuginea. Proc. Natl. Acad. Sci. U.S.A. 109, 12219–12224. doi: 10.1073/pnas.1209954109
Yates, S. A., Chernukhin, I., Alvarez-Fernandez, R., Bechtold, U., Baeshen, M., Baeshen, N., et al. (2014). The temporal foliar transcriptome of the perennial C3 desert plant Rhazya stricta in its natural environment. BMC Plant Biol. 14:2. doi: 10.1186/1471-2229-14-2
Yobi, A., Schlauch, K. A., Tillett, R. L., Yim, W. C., Espinoza, C., Wone, B. W., et al. (2017). Sporobolus stapfianus: insights into desiccation tolerance in the resurrection grasses from linking transcriptomics to metabolomics. BMC Plant Biol. 17:67. doi: 10.1186/s12870-017-1013-7
Keywords: extremophiles, Arabidopsis, drought survival, drought tolerance, drought avoidance
Citation: Bechtold U (2018) Plant Life in Extreme Environments: How Do You Improve Drought Tolerance? Front. Plant Sci. 9:543. doi: 10.3389/fpls.2018.00543
Received: 20 February 2018; Accepted: 09 April 2018;
Published: 15 May 2018.
Edited by:
Nicholas Provart, University of Toronto, CanadaReviewed by:
Mauro Guida Santos, Universidade Federal de Pernambuco, BrazilFulai Liu, University of Copenhagen, Denmark
Copyright © 2018 Bechtold. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ulrike Bechtold, dWJlY2hAZXNzZXguYWMudWs=
 Ulrike Bechtold
Ulrike Bechtold