- 1Key Laboratory of Cotton and Rapeseed, Ministry of Agriculture, Institute of Industrial Crops, Jiangsu Academy of Agricultural Sciences, Nanjing, China
- 2Provincial Key Laboratory of Agrobiology, Jiangsu Academy of Agricultural Sciences, Nanjing, China
Verticillium wilt (VW) is a soil-borne fungal disease that is caused by Verticillium dahliae Kleb and seriously damages cotton production annually in China. To date, many efforts have been made to improve the resistance of upland cotton against VW, but little progress has been achieved because of a lack of resistant upland cotton to VW. G. barbadense is known to carry high resistance to VW; however, it is difficult to transfer the resistance trait from G. barbadense to upland cotton because of linkage drag and distortion in the interspecific hybrid. In this study, a chromosomal segment introgression line (CSIL), SuVR043, containing a single and homozygous chromosome segment of G. barbadense cv. H7124 D04 (Chr 22), was created and used to construct an F2 population for mapping of VW resistance quantitative trait loci (QTLs) in the greenhouse. Two major resistance QTLs against nondefoliating V. dahliae isolate Bp2, called qVW-Bp2-1 and qVW-Bp2-2, which were flanked by the markers cgr6409-ZHX37 and ZHX57-ZHX70 and explained an average of 16.38 and 22.36% of the observed phenotypic variation, respectively, were detected in three independent replicate experiments. The genetic distances from cgr6409 to ZHX37 and from ZHX57 to ZHX70 were 2.4 and 0.8 cM, respectively. By analyzing the genome sequence of the qVW-Bp2-1 and qVW-Bp2-2 regions, we determined that the accurate physical distances from cgr6409 to ZHX37 and from ZHX57 to ZHX70 in the G. barbadense genome are 254 and 140 kb, and that those spans 36 and 20 putative genes, respectively. The results of the expression analysis showed significant differences in the expression profiles of GbCYP450, GbTMEM214, and GbRLK among G. barbadense cv. H7124, CSIL SuVR043 and G. hirsutum acc. Sumian 8 at different times after inoculation with V. dahliae isolate Bp2. Virus-induced gene silencing (VIGS) analysis showed that silencing of GbCYP450 and GbTMEM214 decreased H7124 and CSIL SuVR043 resistance to VW. These results form a solid foundation for fine mapping and cloning of resistance genes in the substituted segment and will provide valuable assistance in future efforts to breed for VW resistance.
Introduction
Cotton (Gossypium spp.) is widely cultivated for the important economic value of its fiber. Verticillium wilt (VW), caused by Verticillium dahliae (V. dahliae), is the major limiting factor of cotton producers and leads to yield loss every year. V. dahliae is a soil-borne pathogen of vascular plants and has a broad host range (over 200 dicotyledonous plant species) (Fradin and Thomma, 2006), different strains vary in virulence against different hosts. A great deal of research in comparative genomics has revealed that the V. dahliae can degrade plant cell walls by the expansion of some CAZyme families, and that flexible genomic islands containing TE and LS genes contribute to the development of virulence and host adaptation by the De et al. (2013). In China, V. dahliae strains that have cotton as their main host are divided into defoliating and nondefoliating strains. The main differences between them are that the defoliating strains develop into epidemics earlier and more rapidly and cause significantly greater yield losses than the nondefoliating strains at equal inoculum densities (BejaranoAlcazar et al., 1995). In addition, the defoliating strains can cause to be completely defoliated, whereas the nondefoliating strains cause only chlorosis and mild wilting of leaves, not defoliation (Schnathorst and Mathre, 1966). The recent nation-wide distribution of large quantities of cotton seed without implementation of proper quarantine procedures before distribution may have contributed to the widespread dissemination of the defoliating type in major cotton cultivating areas (Xia et al., 1998). VW is particularly difficult to prevent in cotton, and no efficient method has been developed to control this disease. Therefore, breeding and planting resistant varieties to VW is the most effective and economical method to control the epidemic. Along with a lack of immune or highly resistant germplasm, the genetic and molecular mechanisms of cotton resistance to V. dahliae infection are poorly understood, and conventional breeding for VW resistance has not been successful (Cai et al., 2009). Creating VW-resistant cotton germplasm through genetic engineering will be the main research objective in the future. The fine mapping and cloning of resistant genes will be the key to achieve this goal.
The resistance mechanism of cotton to VW is very complicated and involves a number of signaling pathways and biochemical substances (Fradin and Thomma, 2006). Previous studies have revealed that a series of basal defense responses and accumulated phytoalexin, including terpenoids and phenylpropanoid, play an important role in the response of cotton to VW infection. In addition, lignin and lignin-like phenolic polymers also serve important functions (Mace et al., 1990; Gayoso et al., 2010; Xu et al., 2011). Signaling transduction pathways are known to play central roles in the perception of attacks and activation of plant defense responses. To date, some results have indicated that reactive oxygen species, salicylic acid (SA), jasmonic acid (JA), ethylene (ET), brassinosteroids (BRs), spermine, and camalexin take part in the resistance of cotton to VW (Johansson et al., 2006; Gao W. et al., 2013; Mo et al., 2015; Guo et al., 2016). To reveal the resistance mechanism of cotton against VW, researchers have studied a number of related gene, Ve1 in tomato is the most famous such gene and endows some tomato varieties with high VW resistance (Kawchuk et al., 2001; Fradin et al., 2009, 2011); its resistance mechanism has been investigated by many researchers (Fradin et al., 2009; Gayoso et al., 2010; Jonge et al., 2012; Zhang Z. et al., 2013). However, this gene does not improve the resistance of cotton to V. dahliae (Liu et al., 2014; Song et al., 2017). A series of homologous Ve genes in cotton have also been identified, but no reports have shown they were successfully used in cotton breeding (Zhang et al., 2011; Zhang J. F. et al., 2012).
In addition to the Ve gene, several other genes have been reported to improve resistance to VW, including GbTLP1 (Munis et al., 2010), GbCAD1, GbSSI2 (Gao W. et al., 2013), GbSTK (Zhang Y. et al., 2013), GbWRKY1 (Li et al., 2014), GbRLK (Zhao et al., 2015), GhPAO (Mo et al., 2015), Gbvdr5 (Yang Y. W. et al., 2015), GbERF1-like (Guo et al., 2016), GhSAMDC (Mo et al., 2016), GbNRX1 (Li et al., 2016), GhLMM (Chai et al., 2017), and more. Additionally, some exogenous genes can also improve resistance to VW in transgenic cotton, including GAFPs (Wang et al., 2016), Hpa1Xoo (Miao et al., 2010), NaD1 (Gaspar et al., 2014), p35 (Tian et al., 2010), and Hcm1 (Zhang et al., 2016). In contrast to the traditional hybridization-based approaches, high-throughput sequencing technologies, referred to as RNA-Seq and proteomics, provide a large amount of information regarding the resistance mechanisms of cotton against VW (Xu et al., 2011; Gao W. et al., 2013). The Agrobacterium-mediated VIGS assay is another method for analyzing genes that confer resistance to VW. To data, several genes have been shown to participate in VW resistance, including GhNDR1, GhMKK2, and GhBAK1 (Gao et al., 2011; Gao X. et al., 2013).
With respect to the inheritance of resistance and improving genetic resistance to VW, researchers have carried out many studies on the loci encoding disease resistance traits by using different materials, different genetic groups or different identifying environments (Yang et al., 2008; Ning et al., 2013; Wang et al., 2014; Zhang J. F. et al., 2015; Shi et al., 2016; Palanga et al., 2017). To date, many VW resistance quantitative trait loci (QTLs) have been reported, and most of them have been detected in G. barbadense × G. hirsutum populations (Zhang J. F. et al., 2015). G. hirsutum is the most widely planted cotton species in China and has been used as the subject of most genetic studies and breeding efforts. It accounts for more than 95% of annual cotton production worldwide (National Cotton Council, http://www.cotton.org/, 2006), but most commercial cultivars of the species are susceptible or only tolerant to VW (Cai et al., 2009). G. barbadense is characterized by extra-long-staple cotton compared to G. hirsutum. Many G. barbadense genotypes are known to possess a high level of resistance to VW (Wilhelm et al., 1974; Zhang J. F. et al., 2012, 2015; Zhou et al., 2014). Hence, cotton breeders have made great efforts to introgress the resistance genetic fragment(s) or gene(s) from G. barbadense to G. hirsutum, with the expectation of breeding new commercial cotton cultivars with both high yield and high VW resistance. However, most of these resistance fragment(s) or gene(s) have not been successfully transferred into commercial upland cotton due to the linkage drag between resistance and undesired agronomic traits, as well as distortion in segregation of the interspecific hybrids (Cai et al., 2009; Zhang J. F. et al., 2015).
To order to break down the linkage drag and distortion in interspecific hybrids, a set of chromosomal segment introgression lines (CSILs) have been developed by crossing G. hirsutum and G. barbadense (Wang et al., 2008, 2014; Zhai et al., 2016). Because CSILs carry introgressed chromosomal segments on the same genetic background, these lines provide the ideal material for studying the genetic functions of chromosomal segments, particularly for QTL mapping. To data, fine mapping of QTLs using CSILs has been reported in different plants, including tomato, rape, rice, and cotton (Zamir, 2001). In this study, G. hirsutum acc. Sumian 8 was used as the recipient parent and G. barbadense cv. H7124 as the donor. By marker-assisted selection (MAS), we created the highly VW-resistant stain CSIL SuVR043, featuring the introgression of G. barbadense cv. H7124 chromosome 22 (D04). For mapping of the VW resistance QTLs in this region, we developed a population of 1,100 individuals and constructed a linkage group with an average genetic distance of 0.35 cM between loci in this study; two major resistance QTLs for V. dahliae nondefoliating isolate Bp2, qVW-Bp2-1, and qVW-Bp2-2, were detected on the introgressed chromosome segment in three independently replicated experiments. The candidate genes in the QTL regions were selected according to analysis of gene sequence divergences, screening of gene expression patterns and VIGS analysis. The results of this study establish a foundation for fine mapping and cloning of the resistance gene in the future.
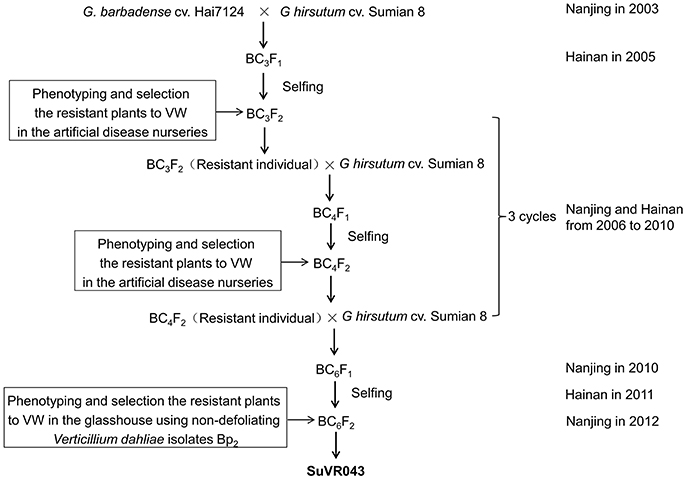
Materials and Methods
Plant Materials and Development of Cotton CSIL SuVR043
G. hirsutum cv. Sumian 8 was bred by the cotton foundation seed farm in Taicang, China, and released by the Jiangsu Crops Variety Approval Committee in 1988; this cultivar is susceptible to VW. G. barbadense cv. H7124, grown extensively and serving as a widely used VW-resistant cultivar for genetic study and breeding in China, is the offspring of a selected individual in studies of the inheritance of resistance to V. dahliae. The creation scheme for CSIL SuVR043 is described in Figure 1. The BC3F1 was generated by 3 cycles of crossing using Sumian 8 as the recipient and H7124 as the donor parent, conducted from 2003 to 2005 at the Cotton Breeding Station of Jiangsu Academy of Agricultural Sciences; BC3F1 plants were grown at Hainan Plant Experiment Station in the winter of 2005 and were self-pollinated to produce the BC3F2 generation. The BC3F2 generation was planted in the artificial disease nurseries in Nanjing, and then resistant plants were selected and backcrossed using Sumian 8 as the male parent. In the winter of the same year, the hybrids were grown at Hainan Plant Experiment Station and self-pollinated to produce the generation for disease resistance identification in the artificial disease nurseries of Nanjing in the following year. Continuous identification, backcrossing and selfing were conducted for 3 cycles to acquire the BC6F1 generation from 2006 to 2010. In the winter of 2012, the BC6F2 generation grown in a greenhouse was inoculated with the nondefoliating V. dahliae isolate Bp2 and 7 resistant plants were selected. Meanwhile, the foreground and background of these 7 resistant plants were selected using 470 simple sequence repeat (SSR) markers distributed on 26 cotton chromosomes based on G. hirsutum×G. barbadense maps (Zhao et al., 2012), and one plant was identified to contain a single and homozygous G. barbadense introgressed chromosome segment; this plant was named SuVR043 (Supplementary Figure 1), and its resistance was higher significantly than that of Sumian 8 in the three subsequent VW resistance evaluation experiments. In 2014, SuVR043 and Sumian 8 were crossed in Nanjing, and 58 F1 seeds were planted and self-pollinated in Hainan to produce F2 progeny. In 2015, the F2 population, containing 1,100 individuals, was planted in Nanjing and self-pollinated to produce F2:3 family seeds.
VW Resistance Evaluation Under Greenhouse Conditions
Resistance evaluation under greenhouse conditions was also conducted in three independently repeated experiments from October 2015 to April 2016 using the Chinese national standard method. Three independently repeated experiments were performed in three different greenhouses at the same time. In every experiment, the parents and F2:3 family lines were planted in a completely randomized design with three replications. For each replicate, the disease grades of at least 15 plants were counted for every F2:3 family line. Sulfuric-acid-delinted seeds of each F2:3 line and the parents lines were germinated in paper cups (250 mL) containing a soil mixture of vermiculite and peat (2:1 volume ratio) in three greenhouses. The temperature of the greenhouses was controlled to approximately 26°C in the daytime and 22°C at night. When cotton seedlings developed two true leaves stage, the bottom of the paper cup was torn out, and the cup was placed into a new paper cup with 20 ml conidial suspensions (2 × 107 conidia/ml), after which the seedlings were kept at a similar temperature and a high humidity environment. Thirty-five days after inoculation, disease symptoms were scored as described in a previous report (Ning et al., 2013). The disease grades of 0, 1, 2, 3, and 4 for leaf disease symptoms were defined as follows: 0 indicates a healthy plant with no disease symptoms; 1 indicates ≤25.0% of leaves exhibited disease symptoms; 2 indicates 25.1–50.0% of leaves exhibiting disease symptoms; 3 indicates 50.1–75.0% of leaves exhibiting disease symptoms; 4 indicates >75.0% of leaf surfaces exhibiting disease symptoms, with plants completely defoliated or dead. The VW disease index was calculated using the following formula:
The DI is the VW disease index of each F2:3 family, X is the disease grade (from 0 to 4), Y is the number of plants with the corresponding disease grade, and N is the total number of investigated plants from each F2:3 family.
In this study, the nondefoliating V. dahliae isolate Bp2 was used as a representative of medium virulence varieties in the Yangtze River cotton-growing region. The V. dahliae isolate was grown on solid potato sucrose agar culture medium at room temperature (25°C) for 7 d, then inoculated into liquid potato sucrose culture medium and oscillated for 7 d, filtered using 2-layers of gauze and diluted to a final concentration of 2 × 107 conidia/ml.
V. dahliae Biomass Quantification in Plants
CSIL SuVR043, Sumian 8 and H7124 were inoculated with V. dahliae isolate Bp2 when they had developed two true leaves, as described above. At 21 d after inoculation, the lower 2 cm of the stems of CSIL SuVR043, Sumian 8, and H7124 were harvested from at least 20 plants and flash frozen in liquid nitrogen. The samples were mixed and ground to powder, approximately 100 mg of which was used for DNA isolation. Quantitative reverse-transcriptase polymerase chain reaction (qRT-PCR) was conducted with an ABI PRISM 7500 PCR machine (Applied Biosystems, USA) with the SYBR Premix Ex Taq II kit (TaKaRa, Japan, Code No. RR820A). For V. dahliae biomass measurement, the internal transcribed spacer region of the ribosomal DNA was targeted to generate a 200 bp amplicon using the fungus-specific primer ITS1-F (5-AAAGTTTTAATGGTTCGCTAAGA-3) (Gardes and Bruns, 1993) and the V. dahliae-specific reverse primer ST-VE1-R (5-CTTGGTCATTTAGAGGAAGTAA-3) (Lievens et al., 2006). EF-1α (Acc.No.AJ223969) from cotton was used as an internal control for normalization of the different DNA samples.
DNA Extraction, Development of SSR Primer Pairs and Molecular Marker Analysis
DNA was extracted from leaf samples of 1100 F2 plants using the CTAB extraction procedure as described by Paterson et al. (1993). Some of the microsatellite primer pairs used in this experiment are available in the Cotton Marker Database (CMD) (https://www.cottongen.org). Based on the selected result of the foreground and background of CSIL SuVR043, we extracted the sequence of the introgression segment and designed SSR primer pairs using the recently released cotton D genome sequence as a reference. The SSR loci were identified and characterized using the program SSR locator (da Maia et al., 2008). Forward and reverse flanking primer pairs were designed using Primer 3 (Rozen and Skaletsky, 2000). The details of the SSR primers development strategy were as described by Lu et al. (2010). PCR amplifications were conducted on an Eastwin PCR machine (ETC811) and performed according to the method described in Zhang et al. (2000); the PCR products were electrophoresed on a 10 % non-denaturing polyacrylamide gel. DNA fragments were visualized by staining with silver nitrate, following the procedure of Zhang et al. (2000).
QTL Analysis
The genetic linkage map and orientation of the SSR markers were determined using the software JoinMap version 3.0 with a logarithm of the odds (LOD) value of 3.0 (Ooijen and Voorrips, 2001). The conversion of recombination frequencies into centimorgans (cM) was calculated with the Kosambi map function (Kosambi, 2016). The graphic representation of the linkage groups was created using MapChart 2.2 (Voorrips, 2002). QTLs were identified by composite interval mapping (Zeng, 1994) using Windows QTL Cartographer 2.5 (Wang et al., 2012), and an LOD threshold of 2.5 was used in the F2 population.
Gene Prediction and Annotation in the Target Region
The linkage markers with QTLs were anchored to the cotton reference genome sequence of the tetraploid G. hirsutum and G. barbadense with alignments of E-value < le-10 using BLASTN (Altschul et al., 1990). The sequences were first extracted from the tetraploid G. hirsutum and G. barbadense and were then uploaded into a local Blast2Go system (Conesa et al., 2005) to perform functional annotation.
RNA Extraction and Candidate Gene Expression Analysis qRT-PCR
Seedlings of H7124, Sumian 8, and CSIL SuVR043 displaying two true leaves stage were inoculated with V. dahliae spore suspension, and mock-inoculated plants dipped in sterile water were used as the control. The roots of samples were harvested at time intervals of 0, 24, 48, 96, and 144 h from inoculation and were immediately frozen in liquid nitrogen for subsequent determination of expression responses. Total RNA was extracted from cotton root samples using an E.Z.N.ATM Plant RNA Kit (Omega, Norcross, GA, USA; cat: R6827-01). First-strand cDNA was synthesized in a final reaction volume of 20 μl containing 2 μg of RNA according to the instructions of the PrimeScript™ RT Reagent Kit with gDNA Eraser Kit (TaKaRa, Japan, Code No. RR047A). The synthesized cDNAs were utilized as templates in the following qRT-PCR reactions. All qRT-PCR reactions were performed on an ABI PRISM 7500 Real-Time PCR System using the SYBR Premix Ex Taq II kit (TaKaRa, Japan, Code No. RR820A) according to the manufacturer's instructions. Three biological replicates were conducted for each qRT-PCR reaction. The cotton Histone 3 gene (accession number: AF024716) was used as an internal reference. The relative expression level was calculated as 2−ΔΔΔCT; ΔΔΔCT = [(Ctgene−CtHis)xh–(Ctgene−CtHis)0h]V.D−[(Ctgene−CtHis)xh−(Ctgene−CtHis)0h]CK. qRT-PCR specific primers are listed in Supplementary Table 1 and were designed using Beacon Designer 7.0 from Premier Biosoft International, Palo Alto, CA.
VIGS in Cotton Followed by V. dahliae Inoculation
The fragments of Cytochrome p450 78a5-like (GbCYP450), Transmembrane protein 214a (GbTMEM214) and Receptor-like protein kinase (GbRLK) genes were amplified by PCR using primer pairs with the restriction sites for tobacco rattle virus (TRV) vectors shown in Supplementary Table 2 from the cDNA of G. barbadense cv. H7124 and were digested with the corresponding restriction enzyme and then ligated into the TRV:00 plasmid to generate pTRV2:GbCYP450, pTRV2:GbTMEM214, and pTRV2:GbRLK. pTRV2:GbCLA1 (Cloroplastos alterados 1), causing an albino plant phenotype was used as a positive control. These constructs and pTRV1 were transformed into Agrobacterium tumefaciens strain GV3101 by electroporation. Agrobacterium cultures containing pTRV2:Target gene and pTRV1 were grown overnight at 28°C in liquid LB medium containing 50 mg/L kanamycin and 25 mg/L rifampicin to an OD of approximately 0.8. The cells were pelleted by centrifugation at 1,180 g at room temperature for 5 min and resuspended in an infiltration culture containing 10 mM MgCl2, 10 mM MES and 200 μM acetosyringone. Cell suspensions containing pTRV2:Target gene and pTRV1 were incubated at room temperature for at least 3 h and then mixed at a 1:1 ratio and infiltrated into the two fully expanded cotyledons of G. barbadense cv. Hai 7124 and CSIL SuVR043 seedlings. The treated seedlings were then grown in a controlled-environment chamber at 25°C with a 16/8 h light/dark photoperiod cycle and a relative humidity of 80%. When the albefaction phenotype of the leaves occurred 2 weeks after infiltration with pTRV2:GbCLA1, the expression of target genes in all plantlets treated by VIGS was checked by qRT-PCR with specific primers. Then, the positive plantlets with silent target genes were inoculated with V. dahliae as previously described in this paper. The disease index was calculated from 20 plants per treatment in each of three repetitions. The standard of VW resistance evaluation for each treatment was evaluated as described previously in this paper.
Results
Genome-Wide Analysis of G. hirsutum cv. CSIL SuVR043 With SSR Markers
In a previous study, 470 SSR markers that were approximately evenly spaced across the cotton genome based on G. hirsutum × G. barbadense maps were used to analyze the foreground and background of CSIL SuVR043 (Zhao et al., 2012), and this study detected one SSR marker (NAU3392) that was polymorphic between G. hirsutum cv. Sumian 8 and CSIL SuVR043. We constructed the F2 population containing 176 individuals of CSIL SuVR043 × Sumian 8. Based on the F2:3 phenotypes of the resistance to V. dahliae strain Bp2, the result of the single-marker analysis showed the resistance locus was linked to marker NAU3392 with a p value of 2.3 × 10−12. Subsequently, we used 3100 SSR markers to further select the background of CSIL SuVR043, of which 1,750 markers were mapped and distributed on 26 cotton chromosomes based on the published genetic map of G. hirsutum × G. barbadense (Zhao et al., 2012), and identified four makers (NAU3392, NAU3791, NAU5294, and JESPR220) to be polymorphic between CSIL SuVR043 and Sumian 8. Based on the G. hirsutum × G. barbadense maps, these four makers (NAU3392, NAU3791, NAU5294, and JESPR220) were mapped and were found to be distributed on Chr 22 (D04) of the cotton genome. In 2013, we selected all 86 SSR markers of Chr 22 group based on G. hirsutum × G. barbadense maps (Zhao et al., 2012) to analyze the background of CSIL SuVR043 and identified that eight makers (NAU3392, NAU6992, NAU6993, NAU3791, cgr6409, NAU5294, JESPR220, and NAU7290) showed identical banding patterns between G. barbadense L. cv. H 7124 and CSIL SuVR043, in contrast to the polymorphic patterns between CSIL SuVR043 and G. hirsutum L. cv. Sumian 8, and 78 other SSR markers like NAU476 and NAU2291 showed identical banding patterns between G. hirsutum L. cv. Sumian 8 and CSIL SuVR043 (Supplementary Figure 1, Supplementary Table 3). Then, through the continuous two self-generations and the marker selection, we ensured the introgressed segment to be homozygous. The above results showed that CSIL SuVR043 contained a single and homozygous chromosome introgressed segment of G. barbadense cv. H7124 Chr 22 (D04).
Comparison of Agronomic Characters and Vw Resistance Performance Between Sumian8 and CSIL SuVR043
To learn more about the characteristics of CSIL SuVR043, we compared the main agronomic traits, fiber quality and disease resistance of CSIL SuVR043, Sumian 8 and H7124. The results showed that there were no significant differences in main agronomic traits and fiber quality except for VW resistance between CSIL SuVR043 and Sumian 8 (Figure 2A, Table 1). This indicated that the chromosome introgressed segment from G. barbadense cv. H7124 Chr 22 (D04) significantly increased CSIL SuVR043 resistance to VW. Meanwhile, we detected the biomass of V. dahliae in CSIL SuVR043, Sumian 8, and H7124 after 35 days inoculation using qRT-PCR. The results showed that the proliferation of V. dahliae in Sumian 8 was significantly higher than that in CSIL SuVR043 and H7124. This suggested that CSIL SuVR043 and H7124 could inhibit the spread of V. dahliae in vivo compared to Sumian 8 (Figure 2B).
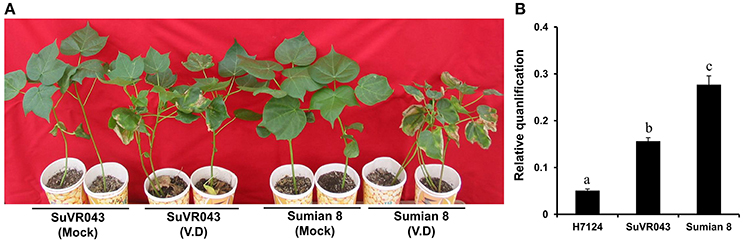
Figure 2. Resistance phenotypes and detection of V. dahliae biomass of two parents, CSILSuVR043 and Sumian 8, after artificial VW inoculation in 2014 in Nanjing. (A) The resistance phenotype of CSILSuVR043 and Sumian 8. The picture was taken 4 weeks after inoculation. (B) Detection of V. dahliae biomass in CSILSuVR043, Sumian 8 and H7124 using qRT-PCR. DNA was extracted from the lower 2 cm of stems 21 days after inoculation with V. dahliae. The relative average fungal biomass is shown with standard errors. The letter indicates significant differences according to Duncan's multiple range tests (p < 0.05).
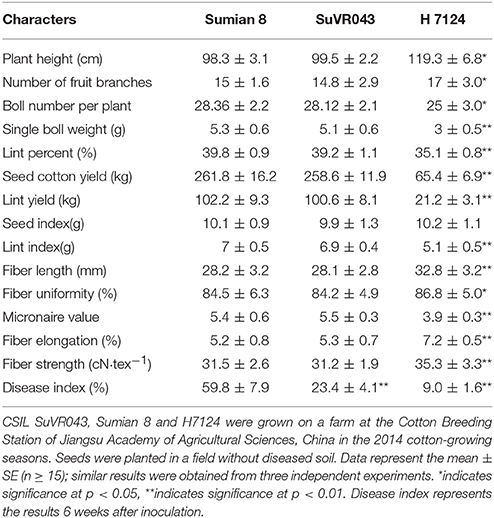
Table 1. CSILSuVR043, Sumian 8 and H7124 mean values for agronomic and fiber traits and resistance to VW.
Development of SSR Primer Pairs and Construction of a Genetic Map
Based on G. hirsutum × G. barbadense maps (Zhao et al., 2012), the introgressed chromosome segment was flanked by NAU476 and NAU2291. To further fine map the VW resistance QTL in the introgressed chromosome segment from G. barbadense cv. H7124, we searched the sequence of 10 markers (NAU476, NAU3392, NAU6992, NAU6993, NAU3791, cgr6409, NAU5294 JESPR220, NAU7290, and NAU2291) against the complete genome sequence of G. raimondii. Among them, the markers NAU476 and NAU2291 were adjacent to NAU3392 and NAU7290, respectively, and were not polymorphic between CSIL SuVR043 and Sumian 8. The search result showed that the forward and reverse sequences of seven markers (NAU6992, NAU6993, NAU3791, cgr6409, JESPR220, NAU7290, and NAU2291) were located in the genome, but the reverse sequence of marker NAU3392, the forward sequence of marker NAU476 and the reverse sequence for marker NAU5294 were not (Supplementary Table 4). We extracted the sequence from the reverse sequence of marker NAU2291 to the reverse sequence of the marker NAU476 in G. raimondii genome (32304099–35343399) and developed 189 SSR primer pairs (Supplementary Table 5). Among them, 42 SSR markers showed polymorphisms between CSIL SuVR043 and Sumian 8. Using 49 SSRs showing polymorphisms, we genotyped 1,100 individuals and constructed a linkage group using MapChart 2.2. The group included 49 markers and 32 loci and spanned 11.1 cM. There were 8 loci containing at least 2 co-segregating markers.
Phenotypic Evaluations of the Parents and F2 Population Resistance to VW in Greenhouse Investigations
VW resistance evaluation experiments under greenhouse conditions were performed from October 2015 to April 2016 in three different greenhouses. The plant disease response in both parental lines and F2:3 family lines were evaluated in terms of leaf symptoms under greenhouse conditions, and the distribution of different resistance levels frequencies of the parents and F2:3 family lines are shown in Figure 3, Table 2. The evaluations of VW resistance for the F2:3 family lines were performed three times in the greenhouse using V. dahliae isolate Bp2. The DI values of the F2:3 family lines varied from 0.00 to 89.71% in the first experiment, from 2.6 to 100% in the second experiment, and from 0.00 to 100% in the third experiment. The broad-sense heritabilities were 0.69, 0.64, and 0.72 in the three independent repeated experiments, respectively. The mean DI value of the F2:3 family lines was 25.32% in the first experiment, which was close to that of the resistant parent SuVR043 (20.24%), and in the second and third experiments, the mean DI values were 43.03 and 50.86%, respectively, which were close to that of the midparent (40.53 and 54.29%).
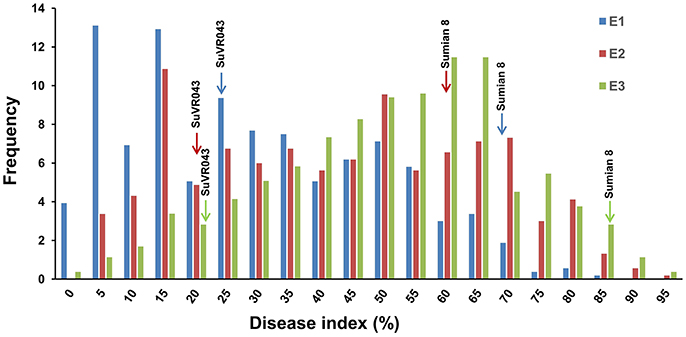
Figure 3. Frequency distributions of disease index for three independently repeated VW resistance evaluation experiments in the greenhouse. E1, E2, and E3 denote three VW resistance evaluation experiments that were performed from October 2015 to April 2016 in three different greenhouses.
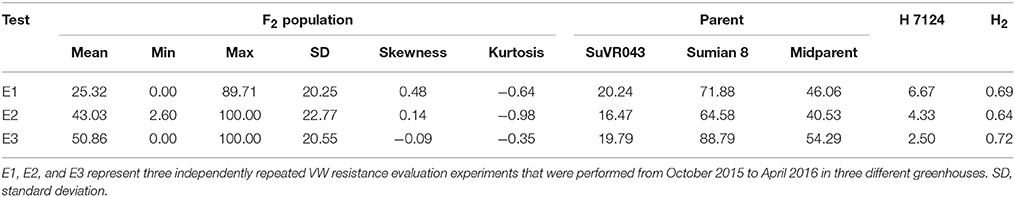
Table 2. Descriptive statistics and broad-sense heritability (H2) of disease index in greenhouse tests.
QTL Detection of VW Resistance by Composite Interval Mapping
Three VW resistance QTLs were detected on the introgressed chromosome segment using V. dahliae nondefoliating isolate Bp2 as shown in Table 3. The QTLs qVW-Bp2-1 and qVW-Bp2-2 flanked by the markers cgr6409-ZHX37 and ZHX57-ZHX70 were detected in three independent repeated experiments, with average explained phenotypic variances of 16.38 and 22.36% and LOD scores ranging from 3.2 to 8.8 and 4.9 to 9.6, respectively. The QTL qVW-Bp2-3 flanked by the markers ZHX102 and NAU5294 was detected only in one experiment with an average explained phenotypic variance of 5.1% and an LOD value of 4.7. Taken together, the results showed that there are two major resistance QTLs for V. dahliae nondefoliating isolate Bp2 on the introgressed chromosome segment.
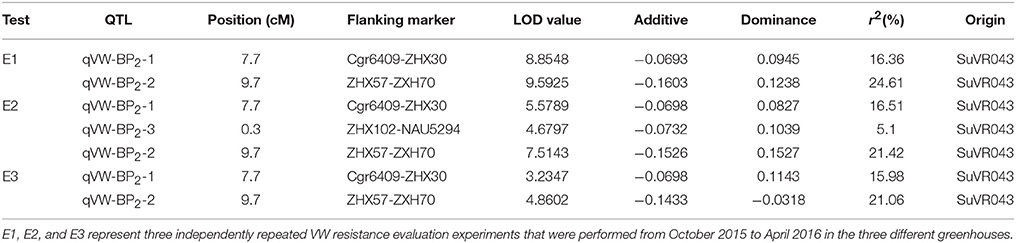
Table 3. QTL analyses for VW resistance detected by composite interval mapping in the F2 generations under greenhouse conditions.
Prediction of Candidate Genes Relevant to the VW Resistance QTL
Based on the available G. hirsutum (Zhang T. et al., 2015) and G. barbadense (Liu et al., 2015) genome sequences, 49 marker sequences were used for alignment analysis. First, we searched the sequence of 49 markers on the linkage group against the complete genome sequence of G. hirsutum cv. TM-1 and G. barbadense cv. Xinhai 21. The results showed the forward and reverse sequence of 34 markers were located in the G. barbadense cv Xinhai 21 genome, excluding markers JESPR220, ZHX-124, ZHX-27, ZHX-23, NAU6693, ZHX-25, ZHX-29, ZHX-30, ZHX-31, NAU3392, ZHX-59, ZHX-62, ZHX-68, ZHX-78, and ZHX-88 (Figure 4). The genetic orders of the 34 loci were consistent with their physical orders in the G. barbadense genome except for markers ZHX-6, ZHX-106, and ZHX-108. For G. hirsutum cv TM-1, the forward and reverse sequences of 49 markers were all located in the genome, and the genetic orders of these loci were consistent with their physical orders in the G. hirsutum cv TM-1 genome except for ZHX-37 and ZHX-39, which were located on scaffold3990_D04. The key markers cgr6409 and ZHX37 closely linked to the qVW-Bp2-1 locus, and ZHX57 and ZHX70 closely linked to the qVW-Bp2-2 locus in the local map, showed genetic and physical locations consistent with the G. barbadense genome. The precise physical distances between cgr6409-ZHX37 and ZHX57-ZHX70 in the G. barbadense genome were 254 and 140 kb, respectively. There were 36 and 20 putative genes in the qVW-Bp2-1 and qVW-Bp2-2 regions of the G. barbadense genome, respectively (Table 4), and there were 49 and 19 putative genes in the qVW-Bp2-1 and qVW-Bp2-2 regions of the G. hirsutum cv. TM-1 genome, respectively (Supplementary Table 6). Through comparing gene information of the qVW-Bp2-1 and qVW-Bp2-2 regions in the G. barbadense and G. hirsutum genomes, the results showed that six unique genes belonged to G. barbadense cv. Xinhai 21 in the Cgr6409-ZHX37 region. These are ORF17 (Chaperone protein dnaj-related), ORF18 (REF SRPP-like protein at1g67360), ORF19 (Cytochrome P450 78a5-like), ORF29 (Sodium transporter HTK1-like), ORF30 (Sodium transporter HTK1-like), and ORF36 (Histone H1). The open reading frame (ORF) lengths of the six genes were different, including ORF13 (Pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16), ORF14 (Ran bp2 NZF zinc finger-like superfamily protein isoform partial), ORF16 (alpha-ketoglutarate-dependent dioxygenase alkB), ORF21 (Duf493 family protein), ORF25 (B-ZIP transcription isoform 2), and ORF34 (Transmembrane protein 214a). In the qVW-Bp2-2 region, the four unique genes in G. barbadense cv. Xinhai 21 included ORF3 (ZZ-type zinc finger-containing protein isoform 2), ORF7 (probable receptor-like protein kinase at1g67000-like), ORF10 (6-phosphogluconate decarboxylating 3), and ORF18 (Malectin receptor protein kinase family).
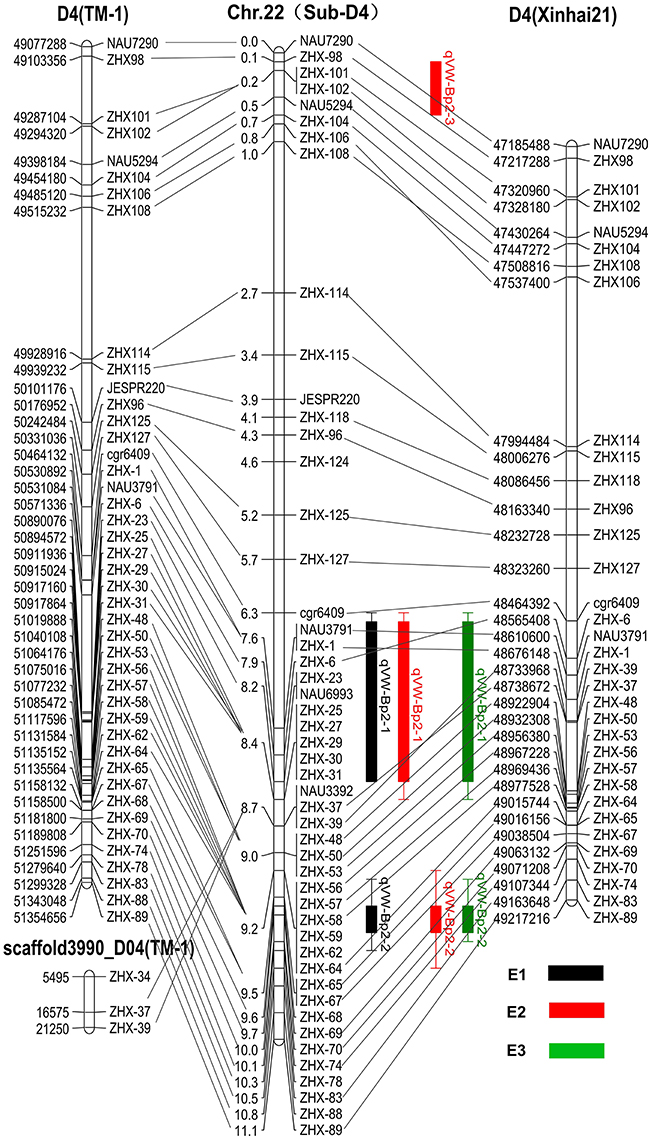
Figure 4. Comparison of genetic and physical maps of CSILSuVR043; mapping the major QTL for VW resistance. The middle map is the linkage group. The right and left maps are the physical maps based on the position of the sequence of markers on the linkage group against the complete genome sequence of G. hirsutum cv. TM-1 and G. barbadense cv. Xinhai 21. The lines connect the same marker locus on different maps. E1, E2, and E3 represent three VW resistance evaluation experiments in three different greenhouses.
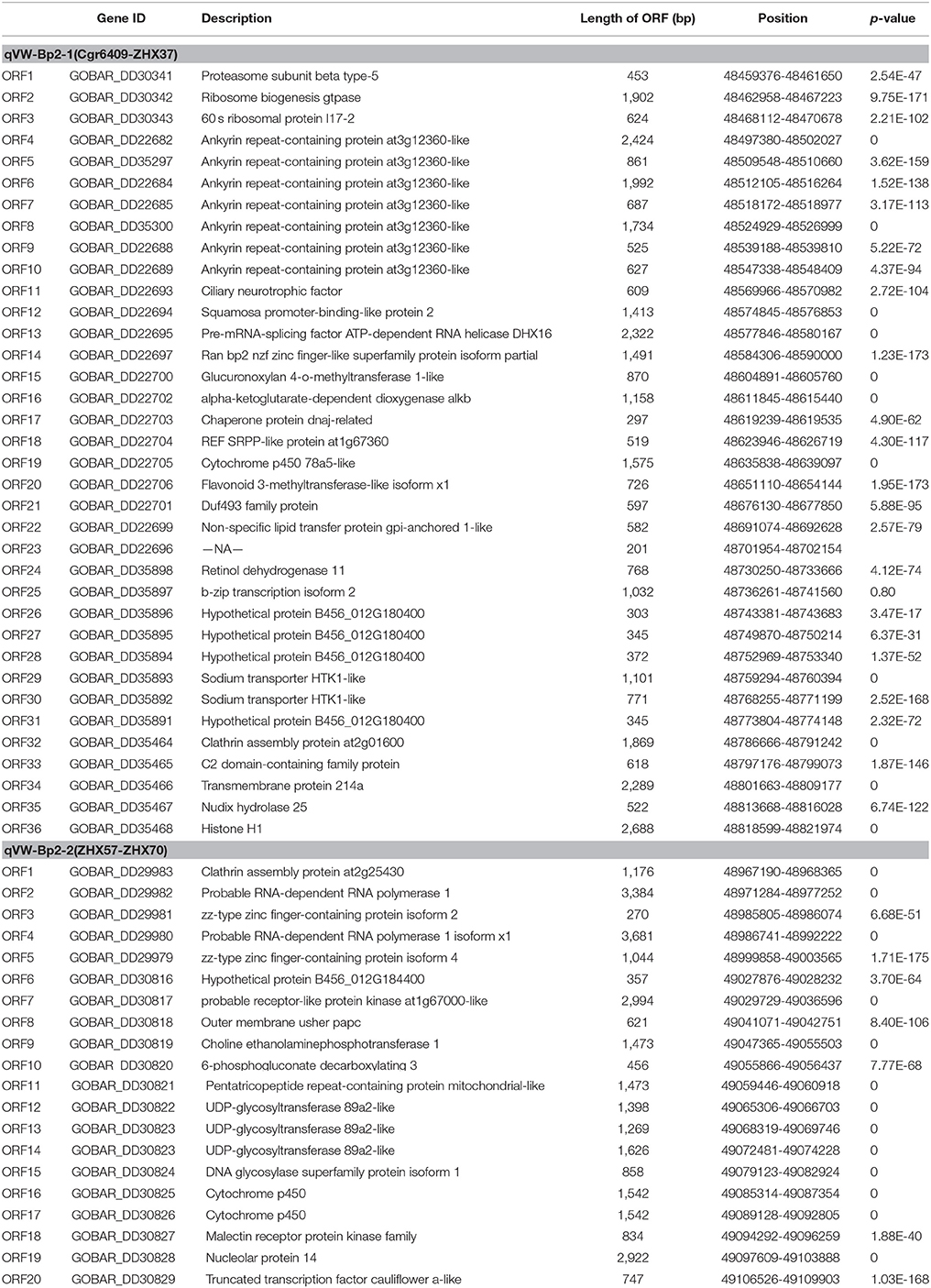
Table 4. Information on genes included in the qVW-Bp2-1 region flanked by the markers Cgr6409 and ZHX37 and the qVW-Bp2-2 region flanked by the markers ZHX57 and ZHX70 in G. barbadense L.cv.Xinhai 21.
Expression Analysis of Candidate Genes
For analysis of the candidate VW resistance genes in the qVW-Bp2-1 and qVW-Bp2-2 regions, 11 genes were selected based on previous reports, sequence difference and gene ontology analysis, and analyzed their expression profiles by qRT-PCR using root tissue with V. dahliae treatment at different stages of CSIL SuVR043, G. barbadense cv. H7124 and G. hirsutum cv. Sumian 8. In the qVW-Bp2-1 region, the expression pattern of Cytochrome P450 78a5-like was similar, and the expression levels increased 3.6, 2.6, and 7.7-fold at 96 h after inoculation and then decreased rapidly in CSIL SuVR043, H7124 and Sumian 8, respectively. Transmembrane protein 214a expression began to increase by 4.4 and 13.1-fold and reached the maximum value at 24 h and decreased rapidly at 48 h after inoculation in CSIL SuVR043 and H7124, however, only increase just by 2.3-fold at 144 h after inoculation in Sumian 8. The expression levels of two genes, B-Zip transcription 2 and Ran bp2 NZF zinc isoform finger-like superfamily protein isoform 1 were changed in response to V. dahliae only in CSIL SuVR043 and Sumian 8, but were not changed in H7124. The expression levels of Histone H1-like were changed only in Sumian 8, and the expression levels of Ref srpp-like protein at1g67360 were not changed in response to V. dahliae among the three cotton varieties (Figure 5A). In the qVW-Bp2-2 region, Cytochrome P450 expression began to rise at 24 h after inoculation, decreased at 48 h, and then reached a maximum at 96 h in CSIL SuVR043 and H7124. In Sumian 8, the expression of Cytochrome P450 began to increase at 48 h, and the up- regulation time was shorter than that of CSIL SuVR043 and H7124. The expression level of Probable receptor-like protein kinase was increased by 1.4- and 2.3-fold at 24 h after inoculation in CSIL SuVR043 and H7124, but was not changed in Sumian 8. The expression levels of 6-phosphogluconate decarboxylating 3 were changed in response to V. dahliae only in CSIL SuVR043 and Sumian 8 and were not changed in H7124. The expression levels of ZZ-type zinc finger–containing protein were not changed in response to V. dahliae among the three cotton varieties (Figure 5B). These results showed that Cytochrome P450, Transmembrane protein 214a and probable receptor-like protein kinase may play important roles in CSIL SuVR043 resistance to VW.
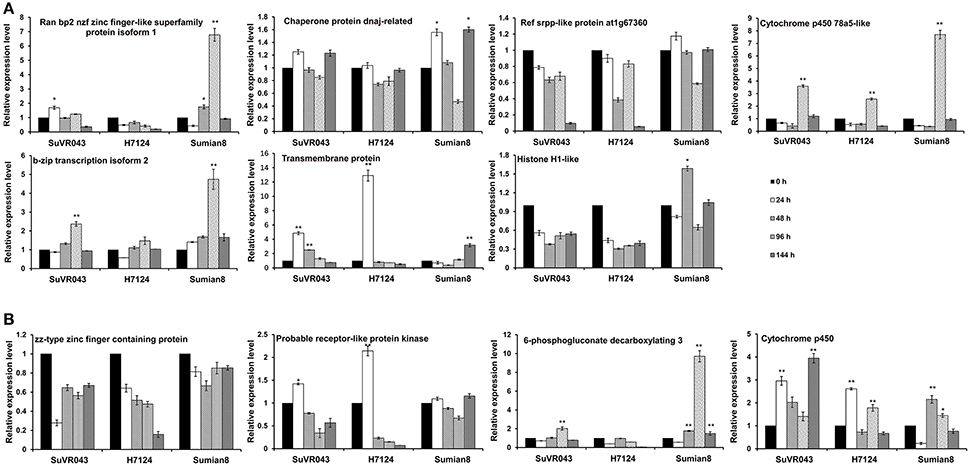
Figure 5. Expression analysis of candidate genes in the qVW-Bp2-1 (A) and qVW-Bp2-2 (B) regions. *Indicates significance at P < 0.05, **Indicates significance at P < 0.01.
Functional Analysis of Candidate Genes by VIGS
To further reduce the range of candidate genes in the introgressed G. barbadense chromosome segment, we applied VIGS to analyze the roles of Cytochrome P450 (GbCYP450), Transmembrane protein 214a (GbTMEM214) and probable receptor-like protein kinase (GbRLK) in resistance to VW based on the results of expression analysis. V. dahliae isolate Bp2 was inoculated into GbCYP450-deficient, GbTMEM214-deficient, GbRLK-deficient cotton lines. The results of the RT-PCR analysis showed that the expression levels of GbCYP450, GbTMEM214 and GbRLK decreased significantly in seedlings 2 weeks after VIGS infiltration in H7124 and CSIL SuVR043. The disease indices were calculated after 4 weeks of inoculation. For the gene GbCYP450, the disease index of H7124 and CSIL SuVR043 with GbCYP450 deficiency reached 41.7 and 53.8% after VIGS infiltration compared to 10.3% (H7124, control plant), 11.7% (H7124, pTRV2:00), 20.3% (CSIL SuVR043, control plant), and 21.7% (CSIL SuVR043, pTRV2:00). For GbTMEM214, the disease indices of H7124 and CSIL SuVR043 with GbTMEM214 deficiency reached 38.9 and 75.9% after VIGS infiltration. For GbRLK, the disease indices of H7124 and CSIL SuVR043 with GbRLK-deficient were 13.3 and 27.1% after VIGS infiltration, and no significant differences were noted compared to the control (data not shown). The results showed that H7124 and CSIL SuVR043 with silenced GbCYP450 and GbTMEM214 had decreased resistance to VW. This indicates that GbTMEM214 and GbCYP450 in this study played important roles in CSIL SuVR043 resistance to VW and are candidate genes in the qVW-Bp2-1 and qVW-Bp2-2 regions, respectively (Figure 6).
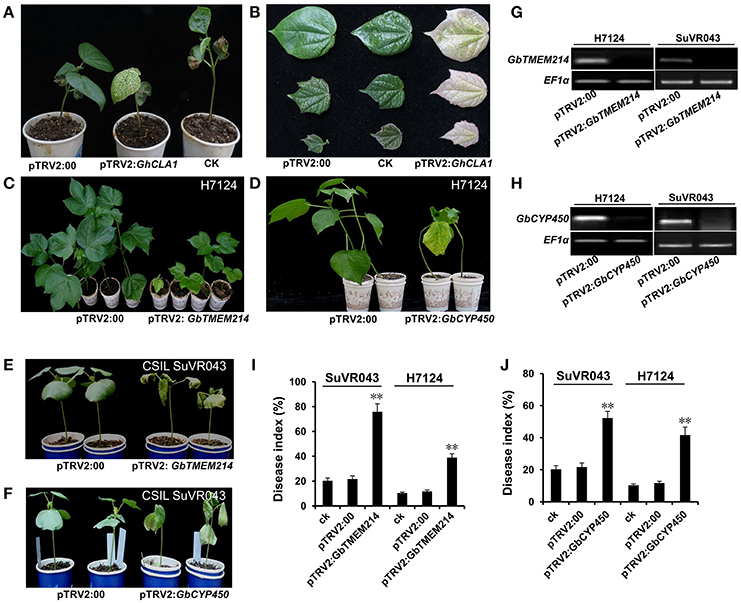
Figure 6. Resistance function analysis of the GbCYP450 and GbTMEM214 genes by VIGS. (A) The phenotype of the cotton plants 14 days after VIGS. The untreated cotton was used as the CK. The empty vector pTRV2:00 was used as a negative control. The cotton gene GhCLA1 was used as a positive control; VIGS of GhCLA1 results in a phenotype of white leaves. (B) The phenotypes of the cotton leaves 14, 21, and 28 d after VIGS with the empty vector pTRV2:00, no treatment, and VIGS with the vector pTRV2-GhCLA1. (C,D) The phenotypes of H7124 under infection by V. dahliae isolate BP2 after VIGS with Agrobacterium carrying pTRV2:GbTMEM214, pTRV2:GbCYP450 and pTRV2:00. The photos were taken at 42 days after V. dahliae inoculation. (E,F) The phenotypes of CSIL SuVR043 under infection by V. dahliae isolate BP2 after VIGS with Agrobacterium carrying pTRV2:GbTMEM214, pTRV2:GbCYP450, and pTRV2:00. Photos were taken at 42 d after V. dahliae inoculation. (G,H) RT-PCR analysis of the expression levels of the genes GbTMEM214 and GbCYP450 in the silenced lines. (I,J) The disease index of plants with silenced GbTMEM214 and GbCYP450 genes. The results were evaluated at 28 d after V. dahliae inoculation, with three replications containing at least 20 plants each. Asterisks indicate statistically significant differences, as determined by Student's t-test (**p < 0.01).
Discussion
Many G. barbadense genotypes have been known to carry high levels of resistance to VW (Wilhelm et al., 1974; Zhang J. F. et al., 2012; Zhou et al., 2014), In particular, G. barbadense cv. H7124 is grown extensively and is a widely used VW-resistant cultivar in China for analysis of the inheritance of resistance, mapping of resistance QTL, cloning of resistance genes and breeding. To date, many VW resistance QTLs have been detected on G. barbadense cv. H7124 chromosomes (Yang et al., 2008; Wang et al., 2014), but its resistance has not been successfully transferred into commercial upland cotton due to the linkage drag. In this study, we developed a G. barbadense cv. H7124 CSIL SuVR043 with higher resistance to VW than the recurrent parents through the multiple cycles of backcross and disease resistance identification, and there were no significant differences between CSIL SuVR043 and the recurrent parents in agronomic characters including fiber quality and yield. This indicated that the chromosome fragments of CSIL SuVR043 from G. barbadense cv. H7124 could significantly improve the resistance of upland cotton to VW and that CSIL SuVR043 could be as an ideal material for breeding the the resistance benefits of introgression from G. barbadense into upland cotton.
The identification of VW resistance QTLs is difficult. To date, a total of 306 disease resistance QTLs have been reported, and 193 QTLs are associated with VW resistance (Zhang J. F. et al., 2015). Most VW resistance QTLs could not be detected repeatedly in different environmental conditions or repeated experiments. There are a number of factors that contribute to this phenomenon. First, most VW resistance QTLs have low contributions to VW resistance (Zhang J. F. et al., 2015; Palanga et al., 2017). To increase the marker density of the genetic map when mapping QTLs, the varieties with the greater genetic background difference were selected to be the parents. This minimized the contribution values due to the background genetic effects. Furthermore, an artificial grading system for resistance evaluation makes it very difficult to quantitatively and accurately evaluate the phenotypic features of plants responses to VW; any experimental errors in VW resistance screening may lead to failure to detect the QTLs for VW resistance (Zhang J. F. et al., 2015). Second, VW infections are highly sensitive to environmental factors. Most VW resistance QTLs have specialization to the different V. dahliae isolates, and the distinct gene(s) control the resistance to the different V. dahliae isolates. Many mapping of VW resistance QTL used the different V. dahliae isolates or evaluated the phenotype of resistance in different environment (Yang et al., 2008; Wang et al., 2014). The pathogenicity and sporulation ability of the same V. dahliae isolate were different in the different culture conditions, especially for V. dahliae isolates present in the field, V. dahliae is a mixture of many strains and the fungal community structure is constantly changed in response to the changes in the environment (Xia et al., 1998). It is possible to obtain different results for disease resistance QTLs identification in different environments or using different strains. Thirdly, the low genome coverage of molecular markers in many mapping studies has limited the detection of QTLs genome-wide at a high resolution (Zhang J. F. et al., 2015; Palanga et al., 2017).
In this study, qVW-Bp2-1 and qVW-Bp2-2 were both detected stably in three independently repeated experiments, and the average explained phenotypic variances and position of qVW-Bp2-1 and qVW-Bp2-2 (15.98–16.51% for qVW-Bp2-1 and 21.06–24.61% for qVW-Bp2-2) were relatively fixed. This means that these two QTLs for VW resistance were accurate and stable. The development of CSIL SuVR043 was the key for the QTL identification in this study. Due to CSIL SuVR043 contained one substituted segments from the donor G. barbadense cv. H7124, all the genetic background variation between CSIL SuVR043 and G. hirsutum cv. Sumian 8 is associated with the substituted segment. This circumstance minimizes background genetic effects and allows more reliable QTL detection and evaluation of high contribution values (Zamir, 2001; Wang et al., 2008, 2014). Meanwhile, Precautions were needed to increase the accuracy of our data and thus to reduce experimental errors. We performed the three independently repeated experiments in three different greenhouses at the same time and inoculated the plants using the same batch of V. dahliae. In every experiment the parents and F2:3 family lines were planted in a completely randomized design with three replications. For each replicate, the disease grades of at least 15 plants were counted for every F2:3 family line in the three independent repeated experiments. The high heritability (0.64–0.72) in the resistance investigation (Figure 3) suggested that the phenotypic variability of three independently repeated experiments was not subject to environmental effects. For increasing the density of the genetic map, we developed 189 SSR primer pairs based on the G. raimondii genome sequence (32304099–35343399), and used an F2 population (1,100 individuals) to construct a linkage group which containing 49 SSRs and 32 loci, spanning 11.1 cM and covering the whole substituted segment. The average genetic distance between loci was just 0.35 cM. The construction of the high-density genetic map circumvented the problem of low genome coverage of the genetic map, and was another key factor for detection of two loci in the substituted segment,
In the past, some researchers have also detected VW resistance QTLs in Chr 22 (D04). To date, at least nine resistance QTLs on Chr 22 (D04) have been reported; Additionally, one resistance QTL cluster linked with markers NAU2235, BNL3873, CIR218, CIR122, and NAU2291 were identified by meta-analysis of VW (Zhang J. F. et al., 2015). Further comparing the sequences of markers NAU2235, BNL3873, CIR218, CIR122, and NAU2291 against the complete genome sequence of G. raimondii, we found that the markers BNL3873, CIR218, CIR122, and NAU2291 were located on the introgressed chromosome segment or were neighboring it (Supplementary Table 7). Palanga et al. also identified 3 resistance QTL clusters explaining 3.7–8.6% on Chromosome 22 (D04) (Palanga et al., 2017), indicating that there was a resistance-related QTL in the target region of this study. Yang et al. (2008) used G. barbadense cv. H7124 and G. hirsutum cv. Junmian 1 as material to construct an F2 population and mapped a VW resistance QTL, which explained 20.3% of the phenotypic variation, to the region NAU3437-NAU3392 of Chr. 22 for the non-defoliating V. dahliae isolate Bp2,. Wang et al. (2014) used the interspecific CSILs to map a VW resistance QTL linked to the marker JESPR220 on Chromosome 22 (D04) under greenhouse conditions; the QTL explained 2.2% of the phenotypic variation. In this study, we used CSIL SuVR043 with its published resistance QTL region to construct F2 population plants and detected two major resistance QTLs (qVW-Bp2-1 and qVW-Bp2-2) for the nondefoliating V. dahliae isolate Bp2 on the introgressed chromosome segment. The key marker NAU3392 was located on the qVW-Bp2-1 region between the markers cgr6409 and ZHX37 in our genetic map. The region qVW-Bp2-2 was adjacent to qVW-Bp2-1, and the distance between them was just 1.5 cM. Therefore, we speculated that qVW-Bp2-1 and qVW-Bp2-2 may be the same resistance locus detected by Yang et al. (2008). Due to the large population and increased number of linked markers in this study, the resistance QTL locus detected by Yang et al. (2008) was separated into two loci in our results. In this study, the qVW-Bp2-1 and qVW-Bp2-2 regions contained 14 and 11 markers, respectively; most of these markers were co-separated and restricted positioning was reduced. Therefore, we expected to further narrow the field of resistance loci by expanding the population.
V. dahliae of cotton is divided mainly into defoliating and nondefoliating isolates, and each isolate also includes many races. Significant differences in pathogenicity exist among different races (Short et al., 2015); therefore, breeding broad-spectrum disease resistant cotton varieties will be the main solution to the cotton production problems caused by V. dahliae. To date, some scholars have detected some broad-spectrum disease resistance loci (Ning et al., 2013; Wang et al., 2014), but most resistant QTLs are specific to certain races. Therefore, it is probably an appropriate choice to breed broad-spectrum resistant cotton by polymerization of the different pathogen resistance QTLs through marker-assisted selection. In this study, resistance loci were detected for the nondefoliating isolate Bp2, but it was not determined whether these resistance loci are effective against other strains.
In recent decades, many genes related to cotton VW resistance have been cloned, but these were obtained by analyzing differences in transcript profiles or by homologous cloning (Xu et al., 2011, 2014; Zhang B. L. et al., 2012). In this study, we identified three candidate genes by analyzing the sequence and expression of genes in two QTL regions following QTL mapping. Those genes encoded GbTMEM214, GbCYP450, and GbRLK, and their expression levels in CSIL SuVR043 and G. barbadense cv. H7124 were significantly different from those in G. hirsutum cv. Sumian 8. The results of functional analysis of the three candidate genes by VIGS showed that resistance to VW of CSIL SuVR043 and G. barbadense cv. H7124 was significantly reduced after silencing GbTMEM214 and GbCYP450, but not GbRLK. This result suggests that GbRLK might not be a key gene in the resistance pathways or may be complemented by other genes.
The transmembrane protein (TMEM) family has been known to be involved in many physiological processes, such as intercellular and intracellular signal transduction, related disease immunity, tumor occurrence and development, ion channels, mediating cell chemotaxis, adhesion, apoptosis, autophagy, and more (Katoh and Katoh, 2004; Miller, 2006; Gregersen et al., 2014; Lees et al., 2017). However, there have been few reports on the function of TMEM family members in plants, indicating that their study in plants is still in its infancy. Previous studies have shown that the TMEM214 gene was involved in the process of apoptosis in human cells and in signal transduction of cancer cells (Li C. et al., 2013). In this study, the ORF length of GbTMEM214 gene was different in the released genome sequences of G. hirsutum cv. TM-1 and G. barbadense cv. Xinhai21 (Supplementary Table 8). The results of location, expression and VIGS analysis suggested that the GbTMEM214 gene might play a key role in the VW resistance process in cotton. Then, it is possible to provide some perspective for the future research on the function of the TMEM gene family in plants and a new candidate gene for VW resistance research in cotton. CYP450, widely present in microbes, plants and animals is a large protein family and is divided into many subfamilies. According to existing reports, the CYP450 protein is involved in the synthesis of a variety of substances, including not only lignin, suberin, keratin and other macromolecules but also hormones, signal molecules and other small molecules (Werckreichhart et al., 2001). Some CYP450 genes also play important roles in plant disease resistance (Smigocki and Wilson, 2004; Wang et al., 2004; Li W. Q. et al., 2013; Sun et al., 2014; Xu et al., 2014; Yang L. et al., 2015). In this study, GbCYP450 belongs to the CYP72A subfamily by analysis of a phylogenetic analysis of CYP450 proteins in Arabidopsis using the software MEGA5.0, and the copy number of GbCYP450 gene was different based on the released genome sequences of G. hirsutum cv. TM-1 and G. barbadense cv. Xinhai21 (Supplementary Table 8). Meanwhile, the expression and VIGS analysis also showed that GbCYP450 also plays an important role in VW resistance.
Conclusion
In this study, CSIL SuVR043 containing a single and homozygous chromosome introgressed segment of G. barbadense cv. H7124 Chr 22 (D04) was used to construct an F2 population for mapping of VW resistance QTL. We constructed a linkage group with 32 loci and an average of 0.35 cM genetic distance between loci using an F2 population containing 1,100 individuals. Two major resistance QTLs for V. dahliae isolate Bp2, qVW-Bp2-1, and qVW-Bp2-2, were detected in three independently repeated experiments, which explained an average of 16.38 and 22.36%, respectively, of the observed phenotypic variation. The sequence, gene expression and VIGS analysis revealed that two candidate genes, GbCYP450 and GbTMEM214, might play the key roles in the VW resistance for SuVR043. Hence, these results provide solid foundation for fine mapping and cloning of resistance genes on the substituted segment in the future.
Author Contributions
JZ and SX: designed the experiments and wrote the manuscript; JZ: collected the genotype and phenotype data; JL and QW: collected the phenotype data; JX: collected the genotype data; LZ: analyzed the data.
Funding
The work was supported by the National Key Research and Development Program of China (2016YFD0100203), the National Natural Science Foundation of China (31401727), the Priority Research and Development Project of Jiangsu Province (BE2015353) and the Natural Science Foundation of Jiangsu Province (BK20170597).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Dr. Ling Lin and Xin Zhang (Institute of Plant Protection, Jiangsu Academy of Agricultural Sciences) for providing the V. dahliae isolate.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00682/full#supplementary-material
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
BejaranoAlcazar, J., Melerovara, J. M., Blancolopez, M. A., and Jimenezdiaz, R. M. (1995). Influence of inoculum density of defoliating and nondefoliating pathotypes of Verticillium dahliae on epidemics of Verticillium wilt of cotton in southern Spain. Phytopathology 85, 1474–1481. doi: 10.1094/Phyto-85-1474
Cai, Y. F., He, X. H., Mo, J. C., Sun, Q., Yang, J. P., and Liu, J. G. (2009). Molecular research and genetic engineering of resistance to Verticillium wilt in cotton: a review. Afr. J. Biotechnol. 8, 7363–7372. doi: 10.5897/AJB2009.000-9571
Chai, Q. C., Shang, X. G., Wu, S., Zhu, G. Z., Cheng, C. Z., Cai, C. P., et al. (2017). 5-Aminolevulinic acid dehydratase gene dosage affects programmed cell death and immunity. Plant Physiol. 175, 511–528. doi: 10.1104/pp.17.00816
Conesa, A., Gotz, S., Garcia-Gomez, J. M., Terol, J., Talon, M., and Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
da Maia, L. C., Palmieri, D. A., De Souza, V. Q., Kopp, M. M., De Carvalho, F. I., and Costa De Oliveira, A. (2008). SSR locator: tool for simple sequence repeat discovery integrated with primer design and PCR simulation. Int. J. Plant Genomics 2008:412696. doi: 10.1155/2008/412696
De, J. R., Bolton, M. D., Kombrink, A., Gc, V. D. B., Yadeta, K. A., and Thomma, B. P. (2013). Extensive chromosomal reshuffling drives evolution of virulence in an asexual pathogen. Genome Res. 23, 1271–1282. doi: 10.1101/gr.152660.112
Fradin, E. F., Abd-El-Haliem, A., Masini, L., Van Den Berg, G. C. M., Joosten, M. H., and Thomma, B. P. (2011). Interfamily transfer of tomato Ve1 mediates Verticillium resistance in Arabidopsis. Plant Physiol. 156, 2255–2265. doi: 10.1104/pp.111.180067
Fradin, E. F., and Thomma, B. P. (2006). Physiology and molecular aspects of Verticillium wilt diseases caused by V-dahliae and V-albo-atrum. Mol. Plant Pathol. 7, 71–86. doi: 10.1111/j.1364-3703.2006.00323.x
Fradin, E. F., Zhang, Z., Ayala, J. C. J., Castroverde, C. D. M., Nazar, R. N., Robb, J., et al. (2009). Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1. Plant Physiol. 150, 320–332. doi: 10.1104/pp.109.136762
Gao, W., Long, L., Zhu, L. F., Xu, L., Gao, W. H., Sun, L. Q., et al. (2013). Proteomic and virus-induced gene silencing (VIGS) analyses reveal that gossypol, brassinosteroids, and jasmonic acid contribute to the resistance of cotton to Verticillium dahliae. Mol. Cell. Proteom. 12, 3690–3703. doi: 10.1074/mcp.M113.031013
Gao, X., Li, F., Li, M., Kianinejad, A. S., Dever, J. K., Wheeler, T. A., et al. (2013). Cotton GhBAK1 mediates Verticillium wilt resistance and cell death. J. Integr. Plant Biol. 55, 586–596. doi: 10.1111/jipb.12064
Gao, X., Wheeler, T., Li, Z., Kenerley, C. M., He, P., and Shan, L. (2011). Silencing GhNDR1 and GhMKK2 compromises cotton resistance to Verticillium wilt. Plant J. 66, 293–305. doi: 10.1111/j.1365-313X.2011.04491.x
Gardes, M., and Bruns, T. D. (1993). ITS primers with enhanced specificity for basidiomycetes–application to the identification of mycorrhizae and rusts. Mol. Ecol. 2, 113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x
Gaspar, Y. M., Mckenna, J. A., Mcginness, B. S., Hinch, J., Poon, S., Connelly, A. A., et al. (2014). Field resistance to Fusarium oxysporum and Verticillium dahliae in transgenic cotton expressing the plant defensin NaD1. J. Exp. Bot. 65, 1541–1550. doi: 10.1093/jxb/eru021
Gayoso, C., Pomar, F., Novo-Uzal, E., Merino, F., and De Ilarduya, O. M. (2010). The Ve-mediated resistance response of the tomato to Verticillium dahliae involves H2O2, peroxidase and lignins and drives PAL gene expression. BMC Plant Biol. 10:232. doi: 10.1186/1471-2229-10-232
Gregersen, N. O., Buttenschon, H. N., Hedemand, A., Dahl, H. A., Kristensen, A. S., Clementsen, B., et al. (2014). Are TMEM genes potential candidate genes for panic disorder? Psychiatr. Genet. 24, 37–41. doi: 10.1097/YPG.0000000000000022
Guo, W. F., Jin, L., Miao, Y. H., He, X., Hu, Q., Guo, K., et al. (2016). An ethylene response-related factor, GbERF1-like, from Gossypium barbadense improves resistance to Verticillium dahliae via activating lignin synthesis. Plant Mol. Biol. 91, 305–318. doi: 10.1007/s11103-016-0467-6
Johansson, A., Staal, J., and Dixelius, C. (2006). Early responses in the Arabidopsis-verticillium longisporum pathosystem are dependent on NDR1, JA- and ET-associated signals via cytosolic NPR1 and RFO1. Mol. Plant Microbe Interact. 19, 958–969. doi: 10.1094/MPMI-19-0958
Jonge, R. D., Esse, H. P. V., Maruthachalam, K., Bolton, M. D., Santhanam, P., Saber, M. K., et al. (2012). Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc. Natl. Acad. Sci. U.S.A. 109, 5110–5115. doi: 10.1073/pnas.1119623109
Katoh, M., and Katoh, M. (2004). Characterization of human TMEM16G gene in silico. Int. J. Mol. Med. 14, 759–764. doi: 10.3892/ijmm.14.4.759
Kawchuk, L. M., Hachey, J., Lynch, D. R., Kulcsar, F., Van Rooijen, G., Waterer, D. R., et al. (2001). Tomato Ve disease resistance genes encode cell surface-like receptors. Proc. Natl. Acad. Sci. U.S.A. 98, 6511–6515. doi: 10.1073/pnas.091114198
Kosambi, D. D. (2016). The Estimation of Map Distances from Recombination Values. New Delhi: Springer India.
Lees, J. A., Messa, M., Sun, E. W., Wheeler, H., Torta, F., Wenk, M. R., et al. (2017). Lipid transport by TMEM24 at ER-plasma membrane contacts regulates pulsatile insulin secretion. Science 355:eaah6171. doi: 10.1126/science.aah6171
Li, C., He, X., Luo, X. Y., Xu, L., Liu, L. L., Min, L., et al. (2014). Cotton WRKY1 mediates the plant defense-to-development transition during infection of cotton by Verticillium dahliae by Activating JASMONATE ZIM-DOMAIN1 expression. Plant Physiol. 166, 2179–2194. doi: 10.1104/pp.114.246694
Li, C., Wei, J., Li, Y., He, X., Zhou, Q., Yan, J., et al. (2013). Transmembrane protein 214 (TMEM214) mediates endoplasmic reticulum stress-induced caspase 4 enzyme activation and apoptosis. J. Biol. Chem. 288, 17908–17917. doi: 10.1074/jbc.M113.458836
Li, W. Q., Shao, M., Yang, J., Zhong, W. G., Okada, K., Yamane, H., et al. (2013). Oscyp71Z2 involves diterpenoid phytoalexin biosynthesis that contributes to bacterial blight resistance in rice. Plant Sci. 207, 98–107. doi: 10.1016/j.plantsci.2013.02.005
Li, Y. B., Han, L. B., Wang, H. Y., Zhang, J., Sun, S. T., Feng, D. Q., et al. (2016). The thioredoxin GbNRX1 plays a crucial role in homeostasis of apoplastic reactive oxygen species in response to Verticillium dahliae infection in cotton. Plant Physiol. 170, 2392–2406. doi: 10.1104/pp.15.01930
Lievens, B., Brouwer, M., Vanachter, A. C. R. C., Cammue, B. P. A., and Thomma, B. P. H. J. (2006). Real-time PCR for detection and quantification of fungal and oomycete tomato pathogens in plant and soil samples. Plant Sci. 171, 155–165. doi: 10.1016/j.plantsci.2006.03.009
Liu, L. L., Zhang, W. W., Zhou, Y., Miao, Y. H., Lian, X. U., Liu, M., et al. (2014). Resistance of cotton and tomato to Verticillium dahliae from cotton is independent on Ve1. Sci. Sin. 44, 803–814. doi: 10.1360/052014-90
Liu, X., Zhao, B., Zheng, H. J., Hu, Y., Lu, G., Yang, C. Q., et al. (2015). Gossypium barbadense genome sequence provides insight into the evolution of extra-long staple fiber and specialized metabolites. Sci. Rep. 5:14139. doi: 10.1038/srep14139
Lu, Y. D., Cai, C. P., Wang, L., Lin, S. Y., Zhao, L. A., Tian, L. L., et al. (2010). Mining, characterization, and exploitation of EST-derived microsatellites in Gossypium barbadense. Chin. Sci. Bull. 55, 1889–1893. doi: 10.1007/s11434-010-3230-4
Mace, M. E., Stipanovic, R. D., and Bell, A. A. (1990). Relation between sensitivity to terpenoid phytoalexins and virulence to cotton of Verticillium-Dahliae strains. Pestic. Biochem. Physiol. 36, 79–82. doi: 10.1016/0048-3575(90)90024-V
Miao, W. G., Wang, X. B., Song, C. F., Wang, Y., Ren, Y. H., and Wang, J. S. (2010). Transcriptome analysis of Hpa1(Xoo) transformed cotton revealed constitutive expression of genes in multiple signalling pathways related to disease resistance. J. Exp. Bot. 61, 4263–4275. doi: 10.1093/jxb/erq227
Miller, C. (2006). ClC chloride channels viewed through a transporter lens. Nature 440, 484–489. doi: 10.1038/nature04713
Mo, H. J., Sun, Y. X., Zhu, X. L., Wang, X. F., Zhang, Y., Yang, J., et al. (2016). Cotton S-adenosylmethionine decarboxylase-mediated spermine biosynthesis is required for salicylic acid- and leucine-correlated signaling in the defense response to Verticillium dahliae. Planta 243, 1023–1039. doi: 10.1007/s00425-015-2463-5
Mo, H. J., Wang, X. F., Zhang, Y., Zhang, G. Y., Zhang, J. F., and Ma, Z. Y. (2015). Cotton polyamine oxidase is required for spermine and camalexin signalling in the defence response to Verticillium dahliae. Plant J. 83, 962–975. doi: 10.1111/tpj.12941
Munis, M. F. H., Tu, L. L., Deng, F. L., Tan, J. F., Xu, L., Xu, S. C., et al. (2010). A thaumatin-like protein gene involved in cotton fiber secondary cell wall development enhances resistance against Verticillium dahliae and other stresses in transgenic tobacco. Biochem. Biophys. Res. Commun. 393, 38–44. doi: 10.1016/j.bbrc.2010.01.069
Ning, Z. Y., Zhao, R., Chen, H., Ai, N. J., Zhang, X., Zhao, J., et al. (2013). Molecular tagging of a major quantitative trait locus for broad-spectrum resistance to Verticillium wilt in upland cotton cultivar prema. Crop Sci. 53, 2304–2312. doi: 10.2135/cropsci2012.12.0694
Ooijen, J. W. V., and Voorrips, R. E. (2001). JoinMap 3.0 Software for the Calculation of Genetic Linkage Maps. Wageningen: Plant Research International.
Palanga, K. K., Jamshed, M., Rashid, M. H. O., Gong, J., Li, J., Iqbal, M. S., et al. (2017). Quantitative trait locus mapping for Verticillium wilt resistance in an upland cotton recombinant inbred line using SNP-based high density genetic map. Front. Plant Sci. 8:382. doi: 10.3389/fpls.2017.00382
Paterson, A. H., Brubaker, C. L., and Wendel, J. F. (1993). A rapid method for extraction of cotton (Gossypium spp.) genomic DNA suitable for RFLP or PCR analysis. Plant Mol. Biol. Rep. 11, 122–127. doi: 10.1007/BF02670470
Rozen, S., and Skaletsky, H. (2000). Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132, 365–386. doi: 10.1385/1-59259-192-2:365
Schnathorst, W. C., and Mathre, D. E. (1966). Host range and differentiation of a severe form of Verticillium albo-atrum in Cotton. Phytopathology 56, 1155–1161.
Shi, Y. Z., Zhang, B. C., Liu, A. Y., Li, W. T., Li, J. W., Lu, Q. W., et al. (2016). Quantitative trait loci analysis of Verticillium wilt resistance in interspecific backcross populations of Gossypium hirsutum x Gossypium barbadense. BMC Genomics 17:877. doi: 10.1186/s12864-016-3128-x
Short, D. P. G., Gurung, S., Gladieux, P., Inderbitzin, P., Atallah, Z. K., Nigro, F., et al. (2015). Globally invading populations of the fungal plant pathogen Verticillium dahliae are dominated by multiple divergent lineages. Environ. Microbiol. 17, 2824–2840. doi: 10.1111/1462-2920.12789
Smigocki, A. C., and Wilson, D. (2004). Pest and disease resistance enhanced by heterologous suppression of a Nicotiana plumbaginifolia cytochrome P450 gene CYP72A2. Biotechnol. Lett. 26, 1809–1814. doi: 10.1007/s10529-004-4615-8
Song, Y., Liu, L., Wang, Y., Valkenburg, D. J., Zhang, X., Zhu, L., et al. (2017). Transfer of tomato immune receptor Ve1 confers Ave1-dependent Verticillium resistance in tobacco and cotton. Plant Biotechnol. J. 16, 638–648. doi: 10.1111/pbi.12804
Sun, L. Q., Zhu, L. F., Xu, L., Yuan, D. J., Min, L., and Zhang, X. L. (2014). Cotton cytochrome P450 CYP82D regulates systemic cell death by modulating the octadecanoid pathway. Nat. Commun. 5:5372. doi: 10.1038/ncomms6372
Tian, J. A., Zhang, X. Y., Liang, B. G., Li, S. W., Wu, Z. X., Wang, Q. H., et al. (2010). Expression of baculovirus anti-apoptotic genes p35 and op-iap in cotton (Gossypium hirsutum L.) enhances tolerance to Verticillium wilt. PLoS ONE 5:e14218. doi: 10.1371/journal.pone.0014218
Voorrips, R. E. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78. doi: 10.1093/jhered/93.1.77
Wang, P., Ding, Y. Z., Lu, Q. X., Guo, W. Z., and Zhang, T. Z. (2008). Development of Gossypium barbadense chromosome segment substitution lines in the genetic standard line TM-1 of Gossypium hirsutum. Chin. Sci. Bull. 53, 1512–1517. doi: 10.1007/s11434-008-0220-x
Wang, P., Ning, Z., Lin, L., Chen, H., Mei, H., Zhao, J., et al. (2014). Genetic dissection of tetraploid cotton resistant to Verticillium wilt using interspecific chromosome segment introgression lines. Crop J. 2, 278–288. doi: 10.1016/j.cj.2014.06.007
Wang, S., Basten, C., and Zeng, Z. (2012). Windows QTL Cartographer 2.5. Raleigh, NC: Department of Statistics, North Carolina State University. Available online at http://statgen.ncsu.edu/qtlcart/WQTLCart.htm
Wang, Y., Li, Q., and He, Z. (2004). Blast fungus-induction and developmental and tissue-specific expression of a rice P450 CYP72A gene cluster. Chin. Sci. Bull. 49, 131–135. doi: 10.1360/03wc0183
Wang, Y. Q., Liang, C. Z., Wu, S. J., Zhang, X. Y., Tang, J. Y., Jian, G. L., et al. (2016). Significant improvement of cotton Verticillium wilt resistance by manipulating the expression of Gastrodia antifungal proteins. Mol. Plant 9, 1436–1439. doi: 10.1016/j.molp.2016.06.013
Werckreichhart, D., Bak, S., and Paquette, S. (2001). Cytochromes P450. Arabidopsis Book 1:e0028. doi: 10.1199/tab.0028
Wilhelm, S., Sagen, J. E., and Tietz, H. (1974). Resistance to Verticillium wilt in cotton - sources, techniques of identification, inheritance trends, and resistance potential of multiline cultivars. Phytopathology 64, 924–931. doi: 10.1094/Phyto-64-924
Xia, Z. J., Achar, P. N., and Gu, B. K. (1998). Vegetative compatibility groupings of Verticillium dahliae from cotton in mainland China. Eur. J. Plant Pathol. 104, 871–876. doi: 10.1023/A:1008628209867
Xu, L., Zhang, W. W., He, X., Liu, M., Zhang, K., Shaban, M., et al. (2014). Functional characterization of cotton genes responsive to Verticillium dahliae through bioinformatics and reverse genetics strategies. J. Exp. Bot. 65, 6679–6692. doi: 10.1093/jxb/eru393
Xu, L., Zhu, L. F., Tu, L. L., Liu, L. L., Yuan, D. J., Jin, L., et al. (2011). Lignin metabolism has a central role in the resistance of cotton to the wilt fungus Verticillium dahliae as revealed by RNA-Seq-dependent transcriptional analysis and histochemistry. J. Exp. Bot. 62, 5607–5621. doi: 10.1093/jxb/err245
Yang, C., Guo, W. Z., Li, G. Y., Gao, F., Lin, S. S., and Zhang, T. Z. (2008). QTLs mapping for Verticillium wilt resistance at seedling and maturity stages in Gossypium barbadense L. Plant Sci. 174, 290–298. doi: 10.1016/j.plantsci.2007.11.016
Yang, L., Shi, C., Mu, X. Y., Liu, C., Shi, K., Zhu, W. J., et al. (2015). Cloning and expression of a wild eggplant cytochrome P450 gene, StoCYP77A2, involved in plant resistance to Verticillium dahliae. Plant Biotechnol. Rep. 9, 167–177. doi: 10.1007/s11816-015-0355-6
Yang, Y. W., Ling, X. T., Chen, T. Z., Cai, L. W., Liu, T. L., Wang, J. Y., et al. (2015). A Cotton Gbvdr5 Gene encoding a leucine-rich-repeat receptor-like protein confers resistance to Verticillium dahliae in transgenic arabidopsis and upland cotton. Plant Mol. Biol. Rep. 33, 987–1001. doi: 10.1007/s11105-014-0810-5
Zamir, D. (2001). Improving plant breeding with exotic genetic libraries. Nat. Rev. Genet. 2, 983–989. doi: 10.1038/35103590
Zhai, H. C., Gong, W. K., Tan, Y. N., Liu, A. Y., Song, W. W., Li, J. W., et al. (2016). Identification of chromosome segment substitution lines of Gossypium barbadense Introgressed in G. hirsutum and quantitative trait locus mapping for fiber quality and yield traits. PLoS ONE 11:e0159101. doi: 10.1371/journal.pone.0159101
Zhang, B. L., Yang, Y. W., Chen, T. Z., Yu, W. G., Liu, T. L., Li, H. J., et al. (2012). Island cotton Gbve1 gene encoding a receptor-like protein confers resistance to both defoliating and non-defoliating isolates of Verticillium dahliae. PLoS ONE 7:e51091. doi: 10.1371/journal.pone.0051091
Zhang, J. F., Sanogo, S., Flynn, R., Baral, J. B., Bajaj, S., Hughs, S. E., et al. (2012). Germplasm evaluation and transfer of Verticillium wilt resistance from Pima (Gossypium barbadense) to upland cotton (G. hirsutum). Euphytica 187, 147–160. doi: 10.1007/s10681-011-0549-0
Zhang, J. F., Yu, J. W., Pei, W. F., Li, X. L., Said, J., Song, M. Z., et al. (2015). Genetic analysis of Verticillium wilt resistance in a backcross inbred line population and a meta-analysis of quantitative trait loci for disease resistance in cotton. BMC Genomics 16:557. doi: 10.1186/s12864-015-1682-2
Zhang, J., Wu, Y. T., Guo, W. Z., and Zhang, T. Z. (2000). Fast screening of miscrosatellite markers in cotton with PAGE/silver staining. Cotton Sci. Sin. 12, 267–269. doi: 10.3969/j.issn.1002-7807.2000.05.012
Zhang, T., Hu, Y., Jiang, W., Fang, L., Guan, X., Chen, J., et al. (2015). Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 33, 531–537. doi: 10.1038/nbt.3207
Zhang, Y., Wang, X. F., Li, Y. Y., Wu, L. Z., Zhou, H. M., Zhang, G. Y., et al. (2013). Ectopic expression of a novel Ser/Thr protein kinase from cotton (Gossypium barbadense), enhances resistance to Verticillium dahliae infection and oxidative stress in Arabidopsis. Plant Cell Rep. 32, 1703–1713. doi: 10.1007/s00299-013-1481-7
Zhang, Y., Wang, X. F., Yang, S., Chi, J., Zhang, G. Y., and Ma, Z. Y. (2011). Cloning and characterization of a Verticillium wilt resistance gene from Gossypium barbadense and functional analysis in Arabidopsis thaliana. Plant Cell Rep. 30, 2085–2096. doi: 10.1007/s00299-011-1115-x
Zhang, Z., Van Esse, H. P., Van, D. M., Fradin, E. F., Liu, C. M., and Thomma, B. P. (2013). Ve1-mediated resistance against Verticillium does not involve a hypersensitive response in Arabidopsis. Mol. Plant Pathol. 14, 719–727. doi: 10.1111/mpp.12042
Zhang, Z. Y., Zhao, J., Ding, L. Y., Zou, L. F., Li, Y. R., Chen, G. Y., et al. (2016). Constitutive expression of a novel antimicrobial protein, Hcm1, confers resistance to both Verticillium and Fusarium wilts in cotton. Sci. Rep. 6:20773. doi: 10.1038/srep20773
Zhao, J., Zhang, Z. Y., Gao, Y. L., Zhou, L., Fang, L., Chen, X. D., et al. (2015). Overexpression of GbRLK, a putative receptor-like kinase gene, improved cotton tolerance to Verticillium wilt. Sci. Rep. 5:15048. doi: 10.1038/srep15048
Zhao, L., Lv, Y. D., Cai, C. P., Tong, X. C., Chen, X. D., Zhang, W., et al. (2012). Toward allotetraploid cotton genome assembly: integration of a high-density molecular genetic linkage map with DNA sequence information. BMC Genomics 13:539. doi: 10.1186/1471-2164-13-539
Keywords: cotton, Verticillium wilt, CSIL, quantitative trait loci (QTLs), resistance gene
Citation: Zhao J, Liu J, Xu J, Zhao L, Wu Q and Xiao S (2018) Quantitative Trait Locus Mapping and Candidate Gene Analysis for Verticillium Wilt Resistance Using Gossypium barbadense Chromosomal Segment Introgressed Line. Front. Plant Sci. 9:682. doi: 10.3389/fpls.2018.00682
Received: 23 January 2018; Accepted: 03 May 2018;
Published: 30 May 2018.
Edited by:
Petr Smýkal, Palacký University, Olomouc, CzechiaReviewed by:
Umesh K. Reddy, West Virginia State University, United StatesHossein Borhan, Agriculture and Agri-Food Canada (AAFC), Canada
Copyright © 2018 Zhao, Liu, Xu, Zhao, Wu and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Songhua Xiao, bmp4c2hAc2luYS5jb20=
 Jun Zhao
Jun Zhao Jianguang Liu1
Jianguang Liu1 Jianwen Xu
Jianwen Xu Songhua Xiao
Songhua Xiao