- State Key Laboratory of Crop Biology, College of Life Sciences, Shandong Agricultural University, Shandong, China
Protein disulfide isomerase (PDI) catalyzes the conversion of thiol-disulfide and plays an important role in various physiological events in animals. A PDI (OaPDI) from a tropical plant was detailed studied and it was found to be involved in response of biotic stress (Gruber et al., 2007). However, the activities of PDI related to physiological functions in plants are poorly understood. In the present study, a homolog of human PDI in Arabidopsis (AtPDI1), encoded by the gene (At3g54960), was characterized. The recombinant AtPDI1 protein had disulfide isomerase activity in vitro and two pairs of conservative cysteines in catalytic domains play a crucial role in the PDI activities. Expression of AtPDI1 in Escherichia coli significantly enhanced stress tolerance of cells and the mutations of critical cysteines almost lose this function. In plants, AtPDI1 was strongly induced by abiotic stresses and exogenous abscisic acid. An Arabidopsis AtPDI1 knockdown mutant (pdi1) and overexpression lines of transgenic plants obtained by this investigation were used to further examine the function of AtPDI1. The mutant line was more sensitive to stresses than the wild-type, while overexpressing AtPDI1 increased tolerance of seedlings to abiotic stresses, with a higher germination ratio and longer length of roots than the wild-type. Our results suggested AtPDI1 played roles in anti-stresses in Arabidopsis, which relate to the activities of PDI.
Introduction
Protein disulfide isomerase (PDI, EC 5.3.4.1) is a thiol-disulfide oxidoreductase, catalyzing oxidation, reduction, and isomerization of target proteins in protein folding and unfolding (Wilkinson and Gilbert, 2005; Ali and Mutus, 2014; Meng et al., 2015). Some PDIs also display chaperone activities, helping protein correct folding (Narindrasorasak et al., 2003; Wilkinson and Gilbert, 2005). However, some play opposite functions, for example, human PDI (hPDI) catalyzing extensive intermolecular disulfide cross-linking of lysozyme into large, inactive aggregates (Wilkinson and Gilbert, 2005; Khan et al., 2016). As a classical PDI, hPDI (Gene ID: 5034) is composed of four modular domains (a, b, b′, a′) and endoplasmic reticulum (ER) retention signal (KDEL sequence) at carboxyl terminus (Alanen et al., 2003). The domain a and a′ are catalytic domains sharing homology to thioredoxin domain with a di-cysteine motif (CXXC motif), respectively, while domain b and b′ play roles in keeping the tertiary structures of the whole molecule in U-shape (Kemmink et al., 1997; Byrne et al., 2009). Generally, more than a dozen members of PDIs exist in animal cells and some of them are involved in unfolded protein response (UPR) of persistent stress in ER, which is closely related to the development and progression of diseases like diabetes and neurodegenerative diseases (Schröder and Kaufman, 2005; Honjo et al., 2014). Elevated expressions of some PDIs have been found in patients suffering from lung cancer or melanoma (Xu et al., 2012).
Family members of plant PDIs are also identified in the genome in different plants, for example, 12 members for Oryza sativa, 9 for Triticum aestivum L., 32 in Brassica rapa ssp. pekinensis, and 22 for Zea mays (Houston et al., 2005; D’Aloisio et al., 2010; Onda and Kobori, 2016; Kayum et al., 2017). Similar to how animal PDIs catalyze the exchange of disulfide, plant PDIs are found to be involved in the unfolded and refolded protein response (Takemoto et al., 2002; Lu and Christopher, 2008; Kimura et al., 2015; Onda and Kobori, 2016; Peng et al., 2017), maturation of the embryo sac (Wang et al., 2008), the programmed cell death of endothelial cells in developing seeds, and response to biotic stress (Gruber et al., 2007; Ondzighi and Staehelin, 2008). The multiplicity and structural difference of plants PDIs suggest that they may serve both specialized and overlapping functions.
Though plants do not suffer animal-specific diseases, plants are more susceptible to stresses in the process of growth and development, due to their immobility. Individual plant PDIs are found to be up-regulated under abiotic stresses. PDIs from Brachypodium distachyon L. and Brassica rapa ssp. pekinensis were up-regulated under abiotic stresses, suggesting their involvement in multiple stress responses (Zhu et al., 2014; Kayum et al., 2017). A PDI from Jatropha curcas could notably enhance cold resistance of yeast cells (Wang et al., 2015). PDI from Oldenlandia affinis (OaPDI), with activity of PDI, is involved in the oxidative folding of a cystine knot defense protein (kalata B1), suggesting OaPDI takes part in antibiotic stress (Gruber et al., 2007).
As a model plant, Arabidopsis thaliana has 12 or more PDI-related members (Houston et al., 2005; Lu and Christopher, 2008). Arabidopsis PDIs are classified into three groups based on polypeptide length, presence of signal peptide and ER retention signal, and composition of thioredoxin domains (Lu and Christopher, 2008). There are three members group III (AtPDI9–AtPDI11) and two members in group I (AtPDI7, AtPDI12), all of them lacking b and b′ domains. Group II is the largest group with six members, all of them containing classical four domains. AtPDI8 contains only three domains (a, b, b′), not belonging to any of the above groups.
As a member in group II, AtPDI1, which contains classical four domains (a, b, b′, a′) with C-terminal ER-anchoring signal. Different from hPDI, AtPDI1 has an additional sequence at N-terminus which is predicted as possible signal peptide targeting to chloroplast. GFP-fused experimental evidences showed that AtPDI1 is located in ER (Yuen et al., 2013), while AtPDI1 is also identified in chloroplast by 2D-proteome (Armbruster et al., 2009; Kieselbach, 2013). Up-regulation of AtPDI1 induced by stress suggested it may play roles in stress responses (Lu and Christopher, 2008). Whether the potential anti-stress function of AtPDI1 is related to its activity of PDI is unknown. In this study, the purified recombinant AtPDI1 and cysteine-mutations were used to examine enzymatic activity. To further investigate the function in plants, overexpression lines of AtPDI1 were obtained by transgenic technique. Different responses to stresses were observed among wild-type, overexpression lines, and a knockdown mutant (pdi1). This study will provide a clue about the function of the AtPDI1 in anti-stresses.
Materials and Methods
Bioinformatics Analyses of AtPDI1
The sequences of AtPDI1, plant PDIs, and hPDI were obtained from the NCBI database1. Multiple sequence alignment was performed with CLC sequence viewer. The three-dimensional structure of AtPDI1 was obtained by homology modeling using the website SWISS-MODEL2. The crystal structure of the hPDI was downloaded from the PDB database3. All structure images were output by PyMOL software.
Cloning of AtPDI1 cDNA and Construction of Expression Plasmids
The cDNA sequence of AtPDI1 was amplified by the RT-PCR method, using Arabidopsis leaves treated by NaCl. The sequence was inserted into pET-30a expression plasmid which contained His-tagged. Six mutations of conserved cysteines were amplified using the recombinant vector as the template, which change cysteine to alanine at positions 128, 131, 467, and 470 of AtPDI1 amino acid sequence. All the primers used in this investigation were listed in Supplementary Table S1. All recombinant plasmid were confirmed by Shanghai Sunny Biotechnology Co. (Shanghai, China), and the recombinant vectors were transformed into Escherichia coli BL21 cell.
Expression and Purification of Recombinant Proteins
Transformed E. coli of BL21 cell were grown until OD600 of 0.4–0.6, induced by 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG) for 2–4 h at 37°C, as done in similar studies (Alergand et al., 2006; Gruber et al., 2007; Wittenberg et al., 2014). The soluble fraction of cells by ultrasonication was purified by affinity chromatography (Ni2+-Sepharose) using 150 mM imidazole as elution buffer (Fu et al., 2008). Purity and molecular mass of the AtPDI1 proteins was confirmed using SDS–PAGE (Supplementary Figure S1). The protein concentrations were determined using a NanoDrop Spectrometer (ND-1000 Spectrophotometer, Peqlab).
Assay Protein Disulfide Isomerase of Recombinant Proteins
The disulfide bond isomerase activity of recombinant proteins was measured as described previously (Walker et al., 1996). The denatured RNase (reduced RNase) was prepared as described previously (Shin and Scheraga, 2000; Fu and Zhu, 2009). Scrambled RNase A (20 μg/mL) was incubated with or without recombinant proteins into 1 mL solution containing 1 mM GSH, 0.2 mM GSSG, 0.5 mM cCMP, 2 mM EDTA, and 100 mM Tris–HCl performed at 25°C. The formation of active RNase was measured spectrophotometrically by continuous monitoring hydrolysis of the RNase substrate, cCMP, at 296 nm (Walker et al., 1996).
Cell Growth in Stress Medium
Survival test on LB solid medium was carried out to ascertain the function of AtPDI1 in E. coli. Transformed E. coli of BL21 cell were grown till OD600 of 0.6, induced by 0.5 mM IPTG for 2 h at 37°C (Du et al., 2014). Cultures were diluted to OD600 of 0.6, and then diluted to 10-2, 10-3, and 10-4. For stress treatments, different dilution cultures were spotted on LB medium supplemented with 0.5 M NaCl, 0.3 mM H2O2, and 0.6 M mannitol, and then incubated at 37°C for 16 h.
Growth analysis in LB solution culture was used to further determine cell growth under abiotic stress. Transformed E. coli of BL21 cell were grown till OD600 of 0.6, induced by 0.5 mM IPTG for 2 h at 37°C and then 100 μl cultures were inoculated in 20 mL LB solution medium containing mannitol, H2O2 or NaCl, and incubated at 37°C on a rotary shaker (200 rpm). The bacterial suspension was harvested at every 2 h till 12 h and OD600 was measured. Each experiment was carried out in three biological replicates.
Stress Treatments to Plants, RNA Extraction, and Quantitative Real-Time PCR (qRT-PCR) Analysis
Normally grown 3-week-old Arabidopsis seedlings were treated with solution containing 10 mM H2O2, 20% PEG, 200 mM NaCl, or 100 μM exogenous ABA, respectively, for different times, with water at room temperature as a control. The leaves were collected at different treatment times and the total RNA was extracted from the material. Qualified RNA was used to synthesize cDNA with a cDNA synthesis kit. The GAPDH gene from Arabidopsis was used as an internal control for expression normalization in the quantitative real-time PCR (qRT-PCR) as similar investigations did and the relative expression levels were calculated by the method of 2-ΔΔCt. The specific primers for amplifying GAPDH and AtPDI1 genes were used for qRT-PCR and are listed in Supplementary Table S1. The relative expression levels of AtPDI1 in stressed samples (12, 24, and 36 h) were compared to the controls (0 h) with Student’s t-test at significance levels of ∗P < 0.05 and ∗∗P < 0.01. Each reaction was carried out in three biological replicates.
Plasmids for Plant Transformation
The full-length sequence of AtPDI1 was fused to plant transformation plasmid pROKII-GFP. The promoter sequence of AtPDI1 (2 kb upstream the gene) was cloned and β-glucuronidase (GUS) gene was fused to plant transformation plasmid pBI121. Plant transformation plasmid was transformed into Agrobacterium GV3101, which was used for floral dip transformation. Transgenic plants were selected for kanamycin resistance and verified by genomic PCR with specific primers (Supplementary Table S1). The homozygous lines were obtained. For the GUS assays, 2-weeks-old transgenic Arabidopsis T3 seedlings were exposed to 100 μM ABA, 20% PEG, 200 mM NaCl or 10 mM H2O2 for 9 h, as done in similar studies (Jia H. et al., 2015).
Plant Materials and Growth Conditions
Arabidopsis thaliana Columbia ecotype (Col-0) seeds were sown on Murashige and Skoog (MS) agar medium in greenhouse conditions at 22°C with a 16 h light/8 h dark cycle (light intensity of 200 μmol/m2/s; relative humidity of 60–75%). Two lines of overexpressing lines and a knockdown AtPDI1 mutant (pdi1-1) by T-DNA insertion line purchased from the Arabidopsis biological resource center4 were used for analysis of plants under stresses. For growth on 1/2 MS medium, the seeds of wild-type, T-DNA insertion mutant and overexpression lines were surface-sterilized with 70% ethanol and 2.6% bleach for 5 and 10 min, respectively. Then, seeds were washed for more than 10 mins with sterilized water containing detergent Tween-20. The washed seeds were allowed to germinate on 1/2 MS agar medium, or added 0.6 μM ABA, 200 mM NaCl, 300 mM mannitol, and 10 mM H2O2, respectively. The concentrations of ABA, NaCl, mannitol, and H2O2 were determined based on the similar investigations (Jia F. et al., 2015; Gallino et al., 2018). The concentrations used in the present study were stronger than general studies (Brini et al., 2007; Wang et al., 2017). In the root length experiment, the washed seeds were laid in 1/2 MS as above described. All the seeds were photographed after 10 days of growth and the seeds germination rate. The seedling root lengths were counted after 10 days of growth. Each experiment was carried out in three biological replicates.
Results
Bioinformatics Analyses on Subcellular Location of AtPDI1
The cDNA sequence of AtPDI1 (also named as AtPDIL1–3) was downloaded from the Arabidopsis information resource (TAIR) (accession number At3g54960), with a complete open reading frame (ORF) of 1740 bp. The ORF encoded a 579-amino acid protein with molecular mass of 64.21 kD and a predicted isoelectric point of 4.74. AtPDI1 contained the classical four domains (a, b, b′, a′) and C-terminal ER-anchoring signal (-KEDL- sequence). Multiple sequence alignment of AtPDI1 with plant PDIs and hPDI showed considerable conserved amino acid sequences, with 50 and 70% similarity of hPDI and OaPDI, respectively (Figure 1A). However, it was a different case with OaPDI, as AtPDI1 contains additional sequence in N-terminus, which is predicted as signal peptide. Three-dimensional modeling structure of the AtPDI1 suggested that four domains (a, b, b′, a′) of AtPDI1 were arranged in the U-shape, similar with other animal and plant classical PDIs. The a and a′ domains of AtPDI1 were at the open end of the U-shape with several α helix and β sheets, while b and b′ were located at the bottom, maintaining the U-shape. The catlytic sites of C-X-X-C motif of AtPDI1were located in a and a′ domains, respectively (Figure 1B). This modeling structure of AtPDI1 was reasonable based on checking dihedral angle, since 94% of amino acid residues were located in the energy stable regions, using Ramachandran plot (Figure 1C).
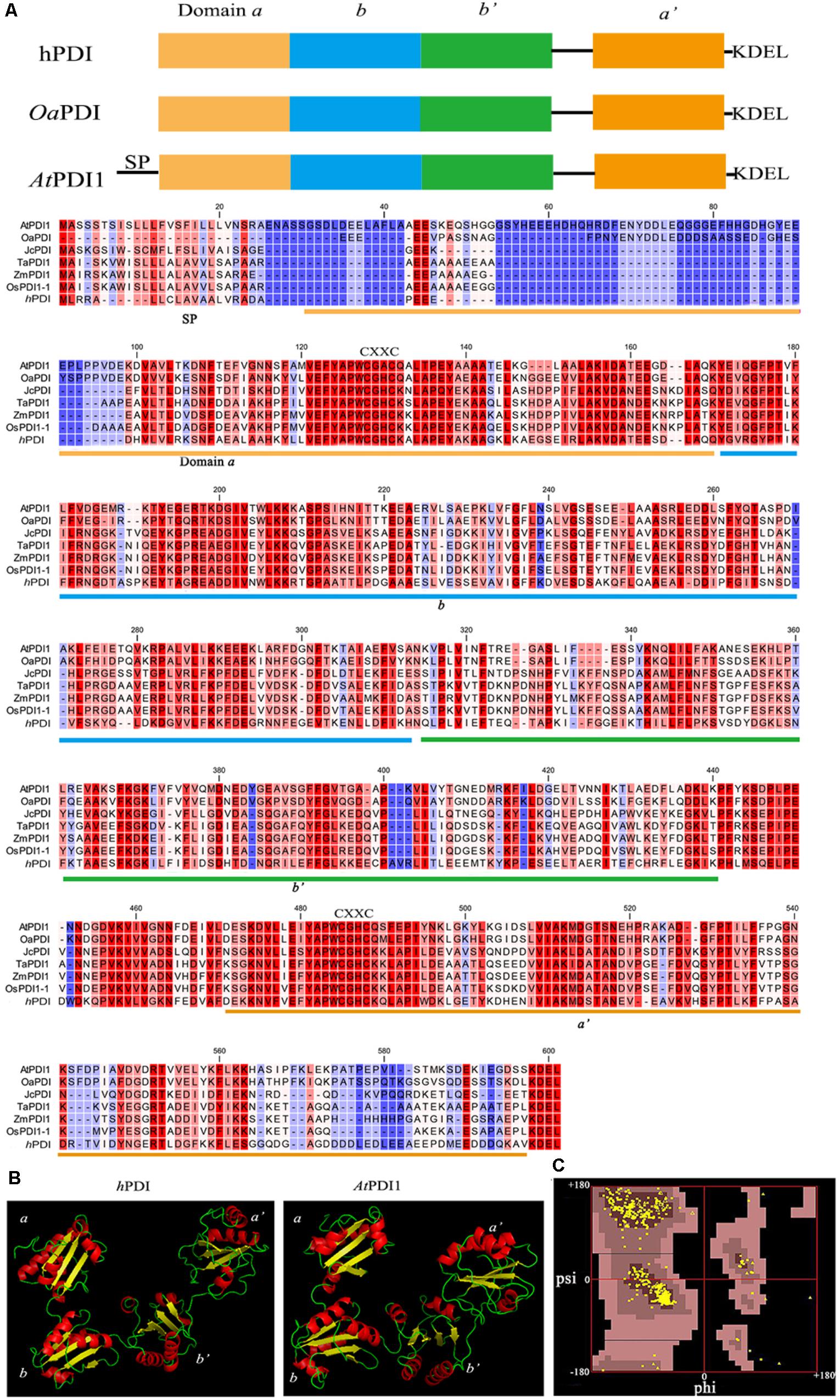
FIGURE 1. Domain structure and alignment of PDI proteins. (A) The domain structures and multiple sequence alignment of AtPDI1, plant PDIs, and hPDI. SP, signal peptide. (B) 3D structure of AtPDI1 constructed by homology-based modeling. (C) Ramachandran map of the AtPDI1 modeling structure, Phi, and Psi represent the dihedral angle of amino acid residues.
As to the subcellular localization of AtPDI1, we expressed the recombinant plasmids 35S:AtPDI1-GFP in epidermal cells of tobacco leaf transiently. The GFP signals were almost co-localized with ER-specific label (mCherry) (Supplementary Figure S2), indicating that AtPDI1 protein was predominantly retained within the ER in epidermal cells of tobacco leaves.
Recombinant AtPDI1 Had Activities of Protein Disulfide Isomerase
The cDNA of AtPDI1 was successfully cloned by the RT-PCR method, using RNA extracted from leaves treated by NaCl, instead of that from normal growth leaves, which suggested the expression of AtPDI1 was up-regulated under stresses. The qRT-PCR experiment showed the expression of AtPDI1 was up-regulated sixfold under NaCl treatment (Figure 5A). The purified recombinant AtPDI1 expressed in E. coli was used to examine enzymatic activity by using scramble RNase A. The initial rate of the reaction was promoted as the concentration of AtPDI1 increased, which was AtPDI1 concentration dependent (Figure 2A). When Cys in thioredoxin motif (Cys-X-X-Cys) are mutated to alanines, the isomerase activities of AtPDI1 decreased 30–70%, which suggested that four Cys play an important role in the isomerase activities of AtPDI1 (Figure 2B).
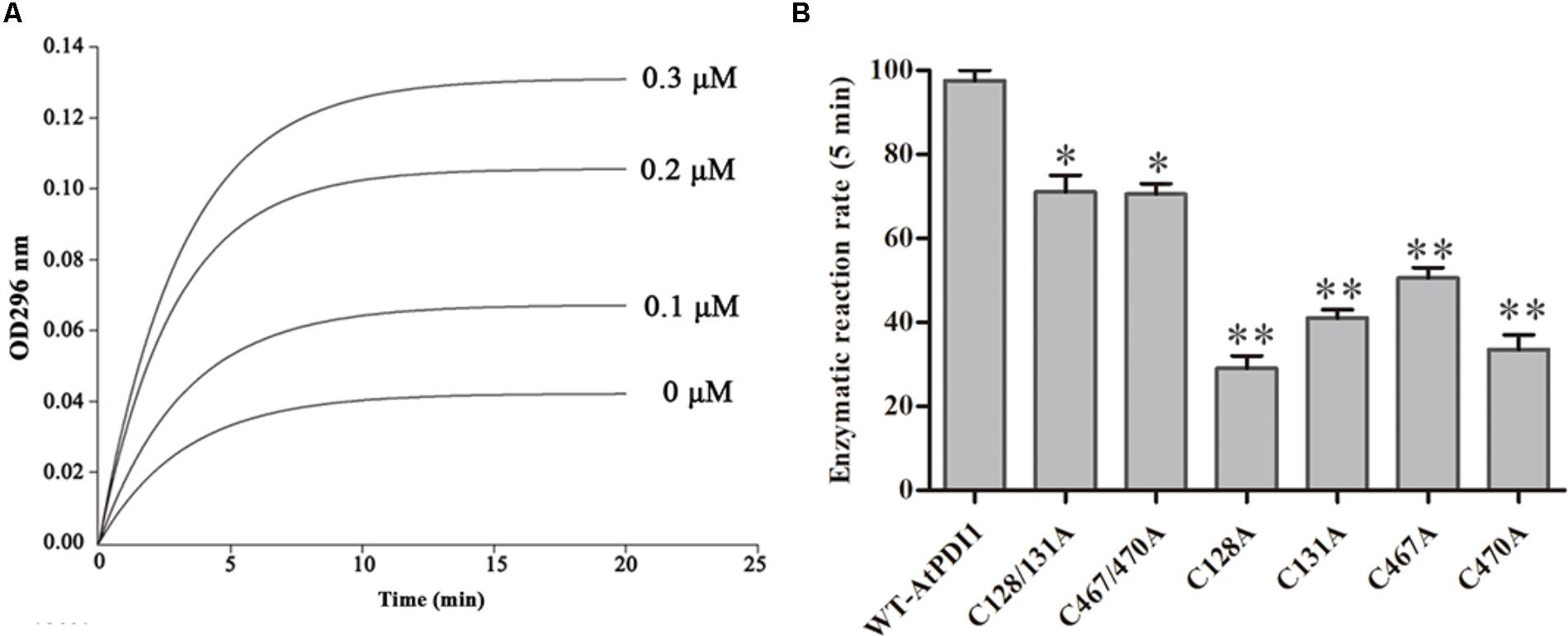
FIGURE 2. The protein isomerase activity of AtPDI1. (A) Continuous assay of the activities with different concentration of recombinant AtPDI1. (B) Relative isomerase activities of WT–AtPDI1 and mutations. Results are represented as mean values of enzymatic reaction rate of three biological replicates. Error bars indicate standard deviation and asterisks indicate statistical significance levels (Student’s t-test; ∗P < 0.05; ∗∗P < 0.01).
Heterogeneous Expression of AtPDI1 in E. coli Enhanced Cell’s Resistance to Abiotic Stresses
The stress medium containing different concentrations of osmolytes or H2O2 was used to check the resistance of E. coli cells transformed the plasmid containing AtPDI1 gene, the empty vector as the controls. No significant differences were observed regarding the growth of the cells on non-stress medium among AtPDI1 and the empty vector. On the medium with NaCl, mannitol or H2O2, the colony number showed significant differences, much more colonies with AtPDI1 than that with empty vector, when same concentration of cultures were inoculated on stress medium plates, respectively (Figure 3A). In liquid fermentation, the growth rate of the E. coli with AtPDI1 also remarkably outperformed the control, the time to reach the logarithmic growth phase was much shorter than that with vector (Figure 3B). The results suggested that AtPDI1 conferred strong stress-tolerance to cells against abiotic stresses. Defective mutations of cysteines of AtPDI1 almost lose the anti-stress activities (Figure 4), which suggested that AtPDI1 isomerase activity and anti-stress function were closely related.
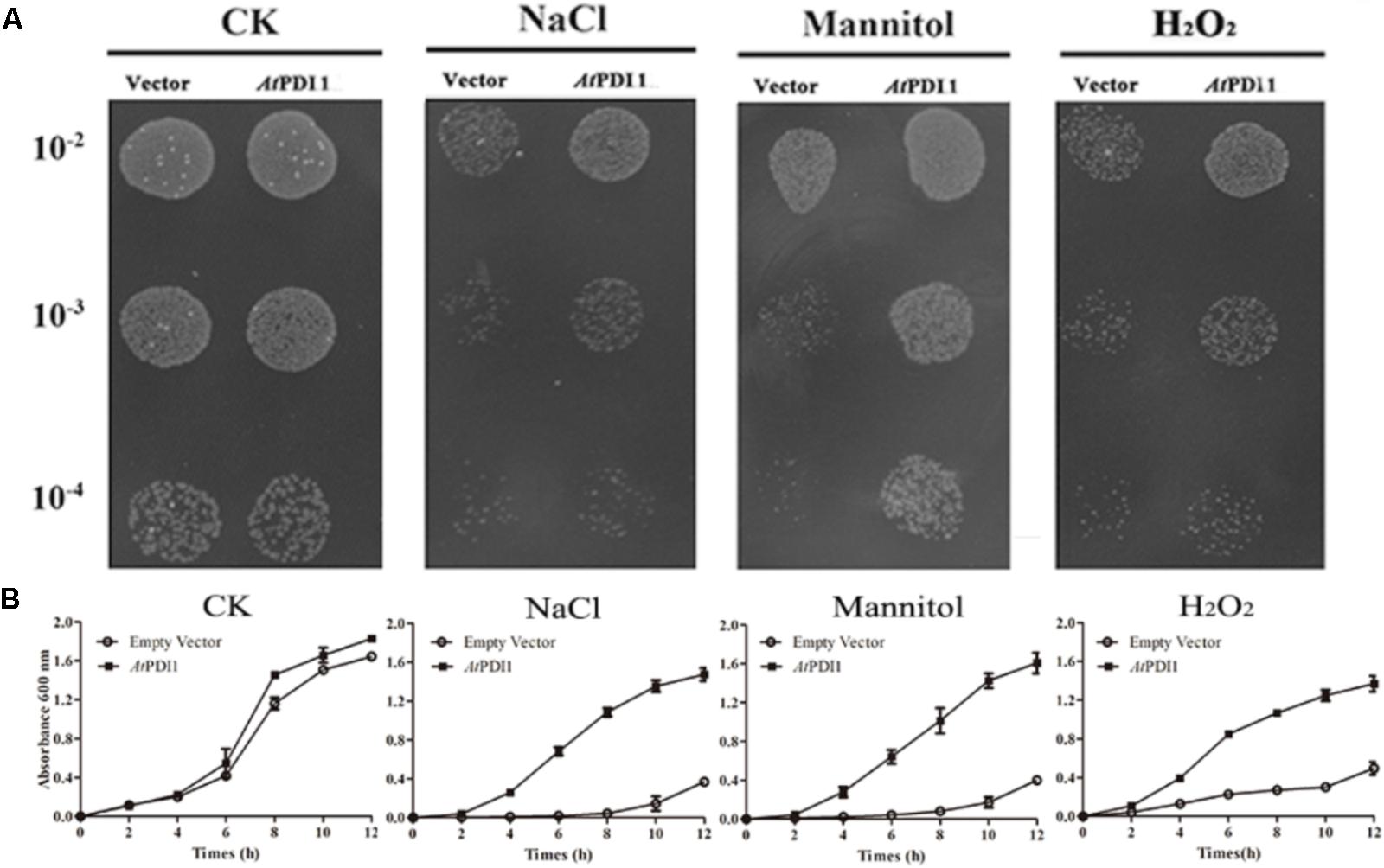
FIGURE 3. Survival test of E. coli cell carrying AtPDI1 or empty vector on various stress conditions. (A) 10 μl cultures from 10-2 to 10-4 dilutions were spotted on plates treated with various concentrations of NaCl, mannitol or H2O2. (B) E. coli cell was cultivated in LB solution medium supplemented with NaCl, mannitol or H2O2. OD600 was recorded at 2 h interval up to 12 h. Results are represented as mean values of three biological replicates and error bars indicate standard deviation.
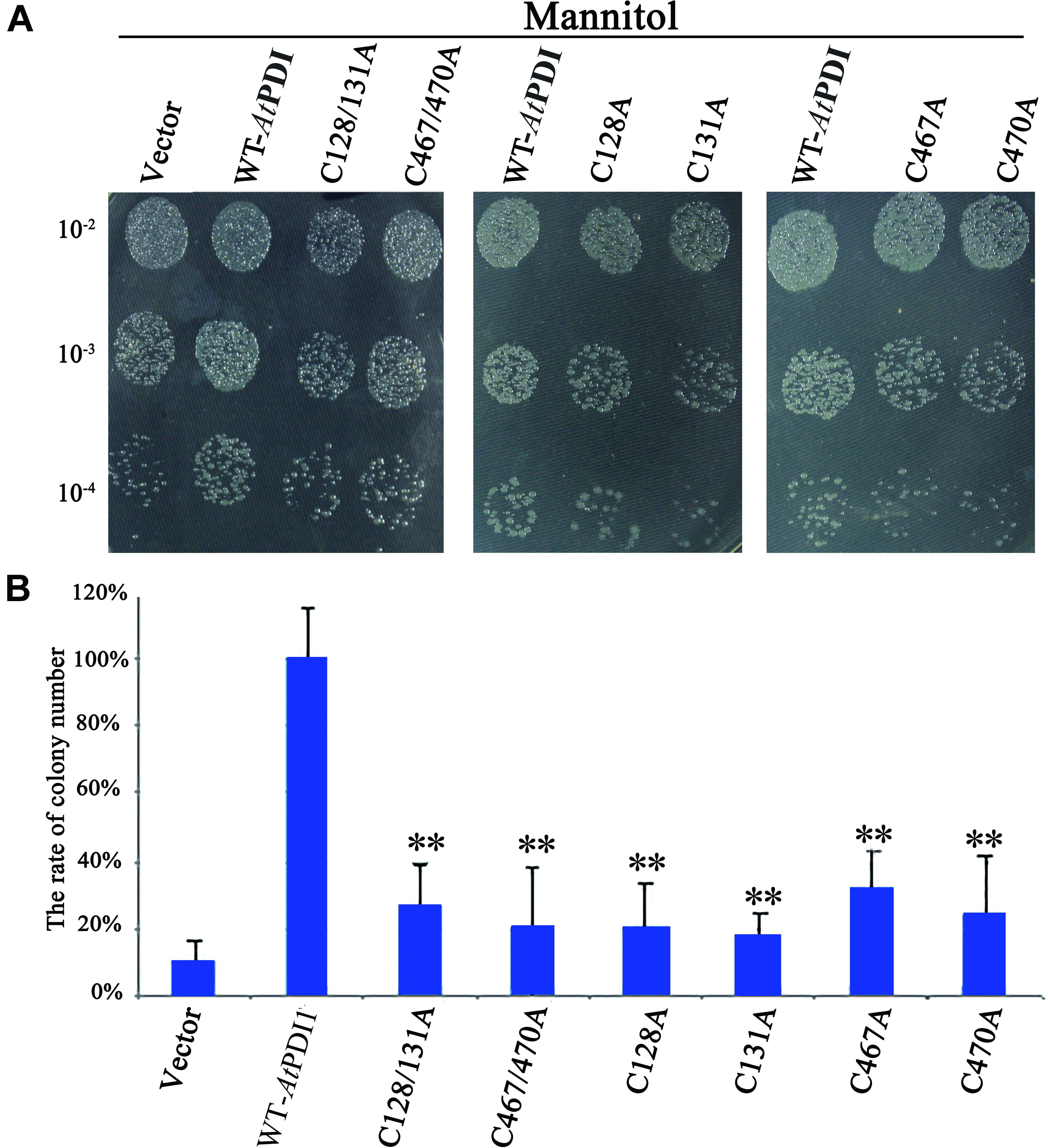
FIGURE 4. Survival test of E. coli cell carrying AtPDI1, mutant AtPDI1 or empty vector on stress conditions. (A) 10 μ l cultures from 10-2 to 10-4 dilutions were spotted on plates treated with mannitol. (B) The colony numbers appearing on stress medium were counted in 10-4 concentration. Each experiment was carried out in three biological replicates. Results are represented as mean values of three independent replicates and error bars indicate standard deviation. Asterisks show statistical significance levels (Student’s t-test; ∗P < 0.05; ∗∗P < 0.01).
The Expression of AtPDI1 Was Up-Regulated by Stresses in Plants
To determine the response of AtPDI1 in plants, 3-week-old Arabidopsis seedlings were treated with H2O2, PEG, NaCl or exogenous ABA, respectively. The expression of AtPDI1, checked by qRT-PCR, was up-regulated under different stresses, approximately four- to ninefold compared with control condition in leaves (Figure 5A). The expression peak of AtPDI1 was reached at 12–24 h under different treatments.
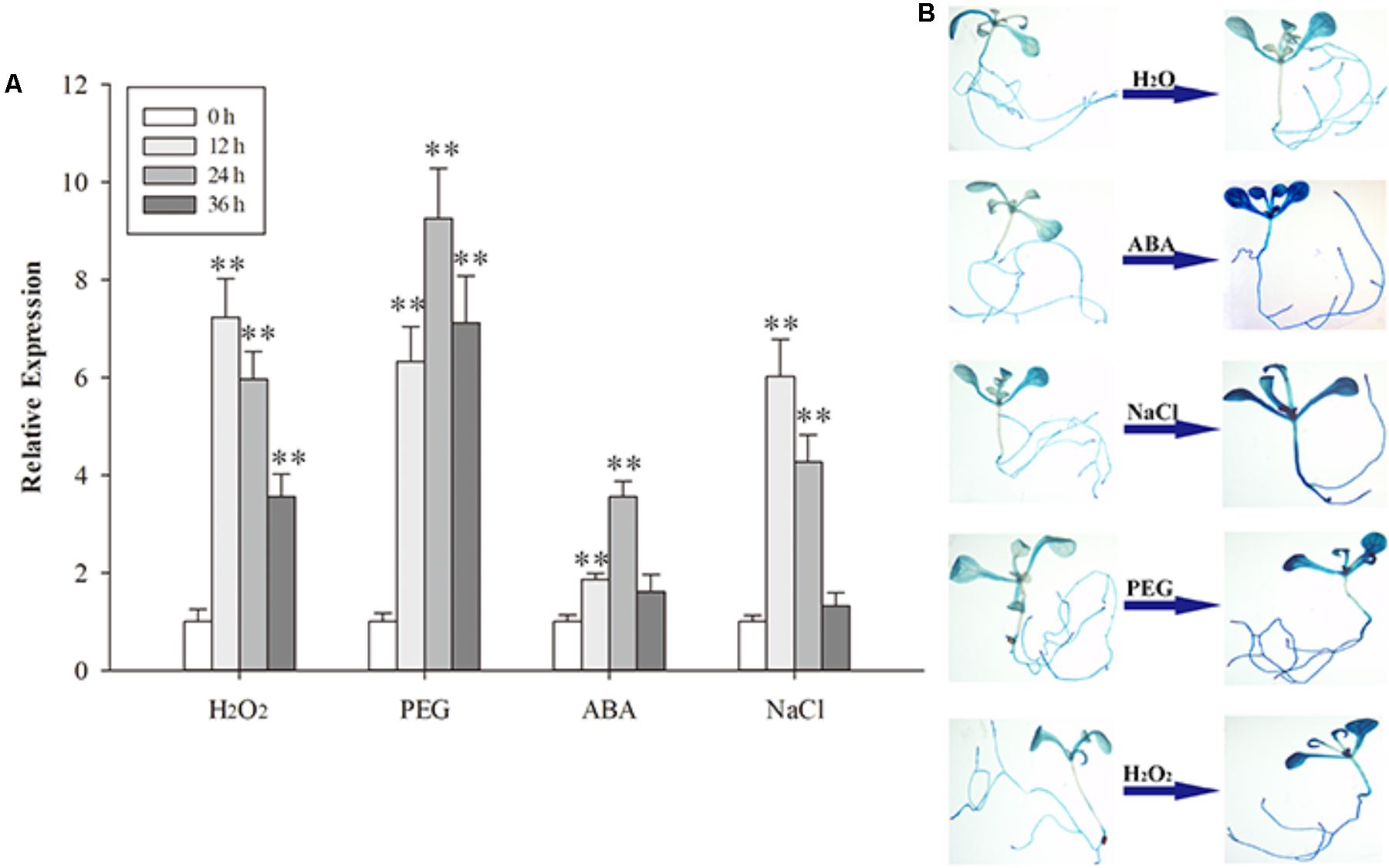
FIGURE 5. The expression of AtPDI1 under different stresses (A) and the AtPDI1 promoter-glucuronidase (GUS) expression pattern in transgenic Arabidopsis plants under various stresses (B). For (A), results are represented as mean values of three independent replicates and error bars indicate standard deviation. The statistical significance level is shown using asterisks (Student’s t-test; ∗P < 0.05; ∗∗P < 0.01).
To further test the response of the AtPDI1 to stresses, the promoter sequence of AtPDI1 (2 kb upstream the gene) was cloned and linked with GUS gene and transformed into Arabidopsis. Four independent transgenic Arabidopsis T3 lines were used for GUS histochemical staining assays. The GUS signal of seedlings were significantly enhanced after NaCl, PEG or H2O2 treatments, respectively, compared with H2O treatment (Figure 5B). Both the above results suggest that AtPDI1 was a stress-inducible gene and up-regulated expression may participate in defensive responses to stresses.
Up- and Down-Expression of AtPDI1 Affected Abilities of Anti-resistance of Seedlings
Independent T3 transgenic plants of overexpressed AtPDI1 driven by the CaMV 35S promoter were generated. A knockdown AtPDI1 mutant (pdi1-1) by T-DNA insertion line was purchased from the Arabidopsis biological resource center5. Under normal conditions, no significant differences were observed in the growth among the wild-type, transgenic, and pdi1-1 plants (Figure 6). However, under stress treatments (0.6 μM ABA, 200 mM NaCl, 300 mM mannitol, and 10 mM H2O2), the seeds of the overexpression lines (OE1 and OE2) germinated much earlier and showed significantly higher germination rates than that of wild type (WT), while the pdi1-1 mutant was later and lower than that of WT under same stress treatments (Figure 6 and Supplementary Figure S3). Similar tendency of root length of young seedlings were observed among OE lines, WT, and pdi1-1 mutant (Figure 7 and Supplementary Figure S4). However, no obvious differences of phenotype were observed among OE lines, WT, and pdi1-1 mutant at mature growth stages under stresses (data was not shown). The results showed that AtPDI1 play important roles in stress-tolerance during seed germination and the roots growth of young seedlings.
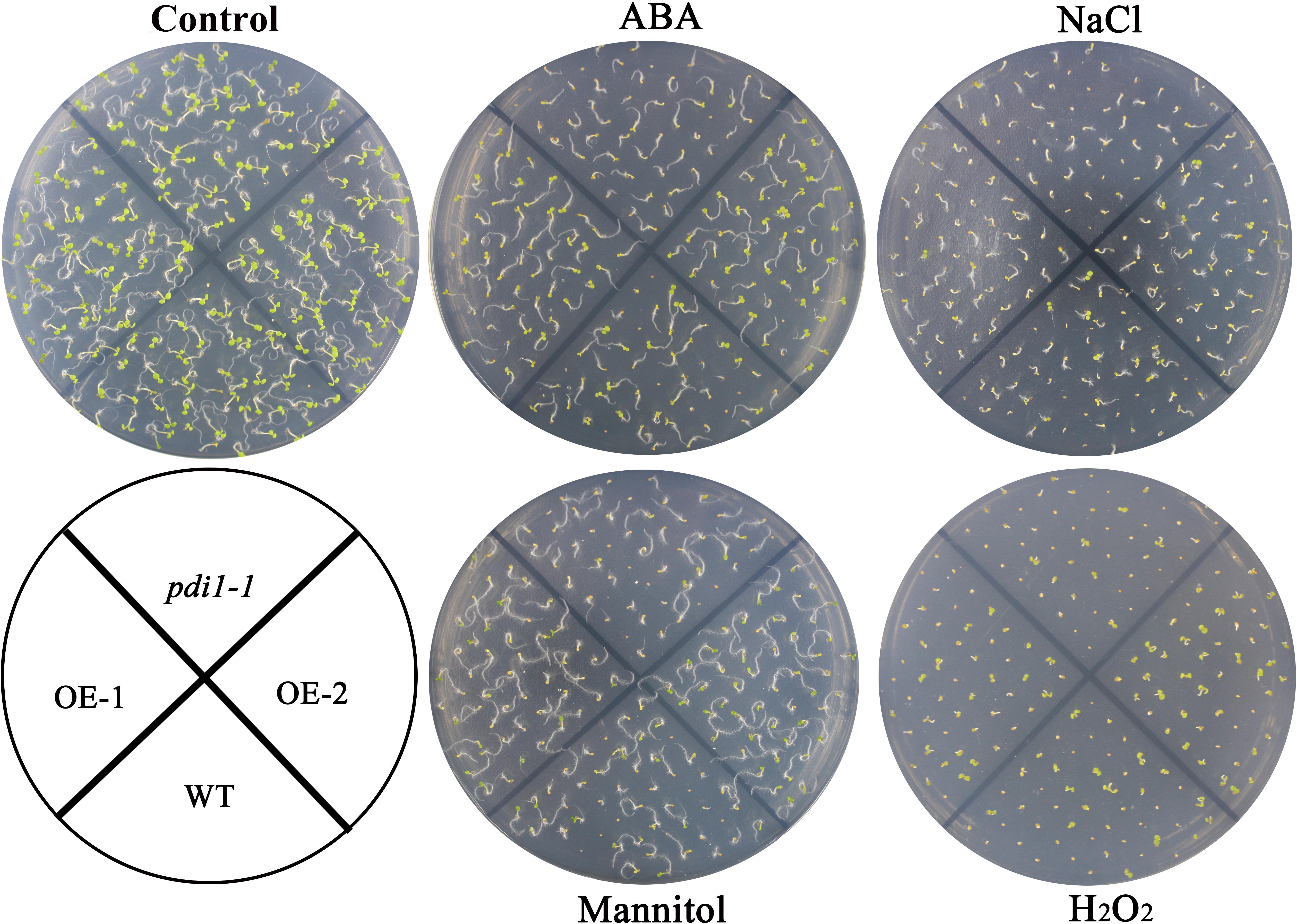
FIGURE 6. Germination phenotype of the wild type (WT), pdi, and AtPDI1-overexpressing lines under 1/2 MS medium containing ABA (0.6 μM), H2O2 (10 mM), NaCl (200 mM), or mannitol (300 mM), respectively. Three independent experiments were conducted and each phenotype included 50 seeds.
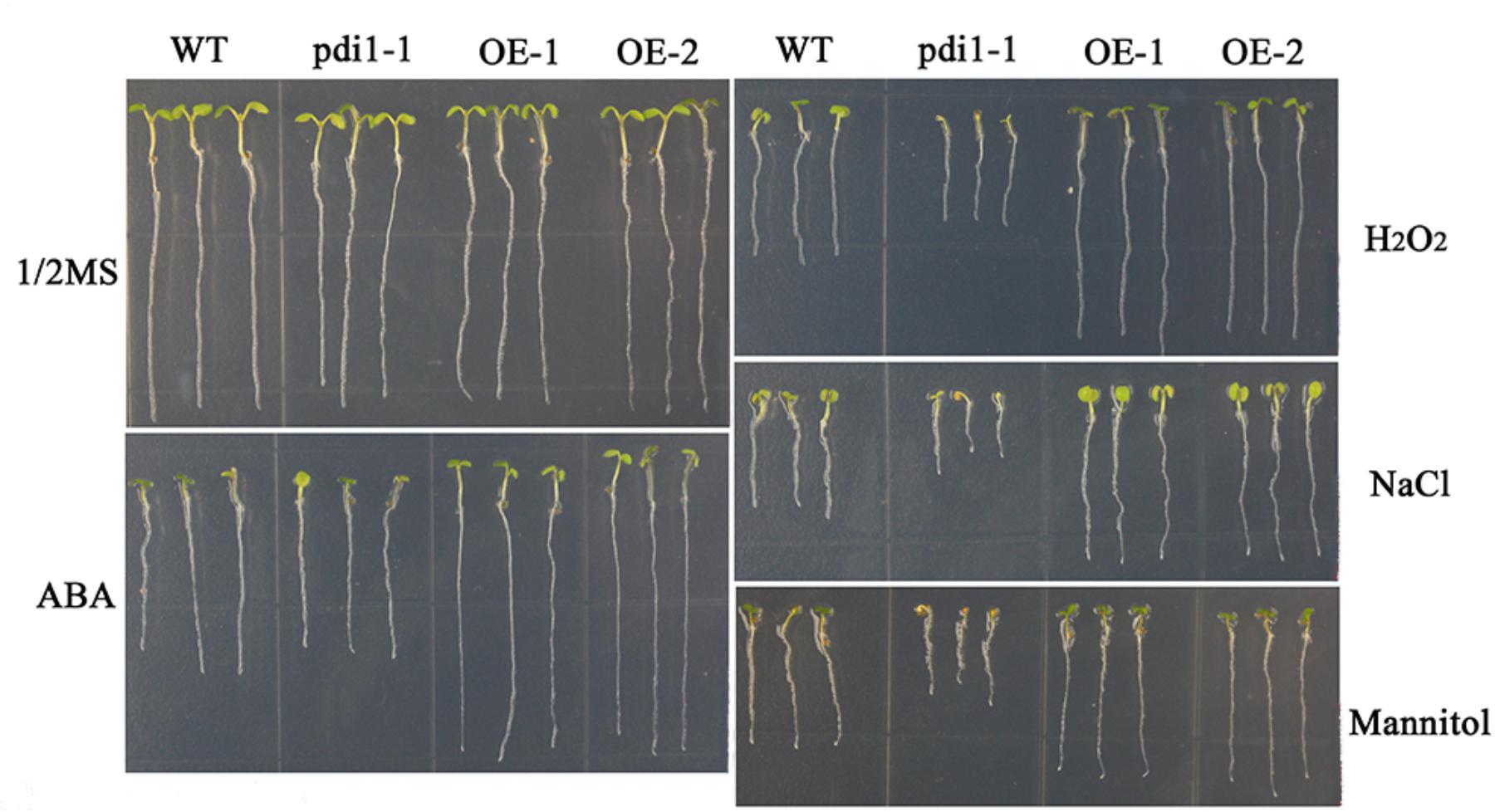
FIGURE 7. The roots lengths of WT, pdi, and AtPDI1-transgenic lines in stress-free (Control) or stress-containing MS medium. The root lengths of wild-type, pdi, and AtPDI1 transgenic seedlings were determined after 14 days on basal medium supplemented with ABA, H2O2, NaCl, or mannitol. Each experiment was carried out in three biological replicates.
Discussion
When animals and plants are exposed to various stresses, misfolded or unfolded proteins are accumulated, while a series of signal transduction pathways are activated, such as UPR to response the stress (Schröder and Kaufman, 2005). In animals, PDIs existing in ER are key proteins during UPR (Schröder and Kaufman, 2005; Ali and Mutus, 2014). In plants, the relations between ER stress and PDIs is poorly studied. Lu and Christopher (2008) treated seedlings with chemical inducers of ER stress and found that six of twelve Arabidopsis PDIs, including AtPDI1, were up-regulated. Three of them (AtPDI1, AtPDI5, and AtPDI6) have classical constitution of domains (a, b, b′, a′) (Supplementary Figure S5). No homologous b and b′ fold domain could be found in the other three members (AtPDI9, AtPDI10, and AtPDI11). All six members have signal peptide sequence at N-terminus and predominantly retained within the ER (Supplementary Figure S5), while they belong to different groups (Lu and Christopher, 2008). Our results showed that AtPDI1 predominantly existed in ER and can catalyze disulfide bond formation, which suggested that it played roles in UPR. The T-DNA knockout Arabidopsis mutant (pdi1-1) was sensitive to stresses, but overexpression AtPDI1 in WT could significantly enhance the stress resistance of plants. Discovering which proteins interact with AtPDI1 and then alleviate the protein damages of stress-elicited oxidative stress needs to be elucidated.
Proteomic analyses of chloroplasts showed that AtPDI1 was also localized in the chloroplast (Wittenberg and Danon, 2008). In chloroplast, there are at least one third proteins whose activities are regulated by thiol-disulfide conversions (Kieselbach, 2013). Compared with intensive studies about reduction of disulfide bonds in chloroplasts, only one thylakoid protein vitamin K epoxide reductase (AtVKOR/LTO1) has been demonstrated to catalyze the formation of the disulfide bonds in luminal proteins (Feng et al., 2011; Lu et al., 2011; Kieselbach, 2013). However, in chloroplast stroma, which protein catalyzes the formation of the disulfide bond has been unclear until now. AtPDI1 is regarded as the most likely candidate (Kieselbach, 2013). In this study, AtPDI1 was confirmed to have activities of PDI and two pairs of cysteins in domain a and a′ were critical for catalyzing function, just like other classical animal and plant PDIs. AtPDI1 may have alternative form without ER retention signal by different edit processing according to the National Center for Biotechnology Information (NCBI). However, our results showed that this form of AtPDI1 was still predominately targeted to ER (Supplementary Figure S2). Whether AtPDI1 play important roles in chloroplast needs to be elucidated.
In summary, AtPDI1 was involved in the abiotic stress processes, which was related to its activity of disulfide isomerase. The mechanisms of AtPDI1 used to regulate anti-stress ability and related physiological functions have yet to be investigated.
Author Contributions
XW, ZZ, and XL designed the experiments. ZZ, XL, RL, and LY performed the experiments. XL, RL, and YD analyzed the data. XW, XL, and ZZ wrote the paper. All authors read and approved the manuscript.
Funding
This work was supported by Special Research Fund of Public Welfare of China Agricultural Ministry (201303093).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00913/full#supplementary-material
Footnotes
- ^ https://www.ncbi.nlm.nih.gov/gene/
- ^ https://www.swissmodel.expasy.org/
- ^ http://www.rcsb.org/pdb/home/home.do
- ^ https://www.arabidopsis.org/abrc/index.jsp
- ^ https://www.arabidopsis.org/abrc/index.jsp
References
Alanen, H. I., Salo, K. E., Pekkala, M., Siekkinen, H. M., Pirneskoski, A., and Ruddock, L. W. (2003). Defining the domain boundaries of the human protein disulfide isomerases. Antioxid. Redox Signal. 5, 367–374. doi: 10.1089/152308603768295096
Alergand, T., Peledzehavi, H., Katz, Y., and Danon, A. (2006). The chloroplast protein disulfide isomerase RB60 reacts with a regulatory disulfide of the RNA-binding protein RB47. Plant Cell Physiol. 47, 540–548. doi: 10.1093/pcp/pcj023
Ali, H. K., and Mutus, B. (2014). Protein disulfide isomerase a multifunctional protein with multiple physiological roles. Front. Chem. 2:70. doi: 10.3389/fchem.2014.00070
Armbruster, U., Hertle, A., Makarenko, E., Zühlke, J., Pribil, M., Dietzmann, A., et al. (2009). Chloroplast proteins without cleavable transit peptides: rare exceptions or a major constituent of the chloroplast proteome? Mol. Plant 2, 1325–1335. doi: 10.1093/mp/ssp082
Brini, F., Hanin, M., Lumbreras, V., Amara, I., Khoudi, H., Hassairi, A., et al. (2007). Overexpression of wheat dehydrin DHN-5 enhances tolerance to salt and osmotic stress in Arabidopsis thaliana. Plant Cell Rep. 26, 2017–2026. doi: 10.1007/s00299-007-0412-x
Byrne, L. J., Sidhu, A., Wallis, A. K., Ruddock, L. W., Freedman, R. B., Howard, M. J., et al. (2009). Mapping of the ligand-binding site on the b′ domain of human PDI: interaction with peptide ligands and the x-linker region. Biochem. J. 423, 209–217. doi: 10.1042/BJ20090565
D’Aloisio, E., Paolacci, A. R., Dhanapal, A. P., Tanzarella, O. A., Porceddu, E., and Ciaffi, M. (2010). The protein disulfide isomerase gene family in bread wheat (T. aestivum L.). BMC Plant Biol. 10:101. doi: 10.1186/1471-2229-10-101
Du, F., Xu, J. N., Zhan, C. Y., Yu, Z. B., and Wang, X. Y. (2014). An obesity-like gene MdTLP7 from apple (Malus × domestica) enhances abiotic stress tolerance. Biochem. Biophys. Res. Commun. 445, 394–397. doi: 10.1016/j.bbrc.2014.02.005
Feng, W. K., Wang, L., Lu, Y., and Wang, X. Y. (2011). A protein oxidase catalysing disulfide bond formation is localized to the chloroplast thylakoids. FEBS J. 278, 3419–3430. doi: 10.1111/j.1742-4658.2011.08265.x
Fu, X., and Zhu, B. T. (2009). Human pancreas-specific protein disulfide isomerase homolog (PDIp) is redox-regulated through formation of an inter-subunit disulfide bond. Arch. Biochem. Biophys. 485, 1–9. doi: 10.1016/j.abb.2008.12.021
Fu, X., Wang, P., and Zhu, B. T. (2008). Protein disulfide isomerase is a multifunctional regulator of estrogenic status in target cells. J. Steroid Biochem. Mol. Biol. 112, 127–137. doi: 10.1016/j.jsbmb.2008.09.005
Gallino, J. P., Ruibal, C., Casaretto, E., Fleitas, A. L., Bonnecarrère, V., Borsani, O., et al. (2018). A dehydration-induced eukaryotic translation initiation factor iso4G identified in a slow wilting soybean cultivar enhances abiotic stress tolerance in Arabidopsis. Front. Plant Sci. 9:262. doi: 10.3389/fpls.2018.00262
Gruber, C. W., Cemazar, M., Clark, R. J., Horibe, T., Renda, R. F., Anderson, M. A., et al. (2007). A novel plant protein-disulfide isomerase involved in the oxidative folding of cystine knot defense proteins. J. Biol. Chem. 282, 20435–20446. doi: 10.1074/jbc.M700018200
Honjo, Y., Horibe, T., Torisawa, A., Ito, H., Nakanishi, A., Mori, H., et al. (2014). Protein disulfide isomerase P5-immunopositive inclusions in patients with alzheimer’s disease. J. Alzheimers Dis. 38, 601–609. doi: 10.3233/JAD-130632
Houston, N. L., Fan, C., Xiang, J. Q., Schulze, J. M., Jung, R., and Boston, R. S. (2005). Phylogenetic analyses identify 10 classes of the protein disulfide isomerase family in plants, including single-domain protein disulfide isomerase-related proteins. Plant Physiol. 137, 762–778. doi: 10.1104/pp.104.056507
Jia, F., Wang, C., Huang, J., Yang, G., Wu, C., and Zheng, C. (2015). SCF E3 ligase PP2-B11 plays a positive role in response to salt stress in Arabidopsis. J. Exp. Bot. 66, 4683–4697. doi: 10.1093/jxb/erv245
Jia, H., Wang, C., Wang, F., Liu, S., Li, G., and Guo, X. (2015). GhWRKY68 reduces resistance to salt and drought in transgenic Nicotiana benthamiana. PLoS One 10:e0120646. doi: 10.1371/journal.pone.0120646
Kayum, M. A., Park, J. I., Nath, U. K., Saha, G., Biswas, M. K., Kim, H. T., et al. (2017). Genome-wide characterization and expression profiling of PDI family gene reveals function as abiotic and biotic stress tolerance in Chinese cabbage (Brassica rapa ssp. pekinensis). BMC Genomics 18:885. doi: 10.1186/s12864-017-4277-2
Kemmink, J., Darby, N. J., Dijkstra, K., Nilges, M., and Creighton, T. E. (1997). The folding catalyst protein disulfide isomerase is constructed of active and inactive thioredoxin modules. Curr. Biol. 7, 239–245. doi: 10.1016/S0960-9822(06)00119-9
Khan, R. H., Siddiqi, M. K., and Salahuddin, P. (2016). Protein disulfide isomerase: structure, mechanism of oxidative protein folding and multiple functional roles. J. Biochem. Mol. Biol. 2, 173–179.
Kieselbach, T. (2013). Oxidative folding in chloroplasts. Antioxid. Redox Signal. 19, 72–82. doi: 10.1089/ars.2012.4582
Kimura, S., Higashino, Y., Kitao, Y., Masuda, T., and Urade, R. (2015). Expression and characterization of protein disulfide isomerase family proteins in bread wheat. BMC Plant Biol. 15:73. doi: 10.1186/s12870-015-0460-2
Lu, D. P., and Christopher, D. A. (2008). Endoplasmic reticulum stress activates the expression of a sub-group of protein disulfide isomerase genes and AtbZIP60 modulates the response in Arabidopsis thaliana. Mol. Genet. Genomics 280, 199–210. doi: 10.1007/s00438-008-0356-z
Lu, Y., Hall, D., and Last, R. (2011). A small zinc finger thylakoid protein plays a role in maintenance of photosystem II in Arabidopsis thaliana. Plant Cell 23, 1861–1875. doi: 10.1105/tpc.111.085456
Meng, Y., Zhang, Q., Zhang, M., Gu, B., Huang, G., Wang, Q., et al. (2015). The protein disulfide isomerase 1 of Phytophthora parasitica (PpPDI1) is associated with the haustoria-like structures and contributes to plant infection. Front. Plant Sci. 6:632. doi: 10.3389/fpls.2015.00632
Narindrasorasak, S., Ping, Y., and Sarkar, B. (2003). Protein disulfide isomerase, a multifunctional protein chaperone, shows copper-binding activity. Biochem. Biophys. Res. Commun. 311, 405–414. doi: 10.1016/j.bbrc.2003.09.226
Onda, Y., and Kobori, Y. (2016). Differential activity of rice protein disulfide isomerase (PDI) family members for disulfide bond formation and reduction. FEBS Open Bio. 4, 730–734. doi: 10.1016/j.fob.2014.07.007
Ondzighi, C. A., and Staehelin, L. A. (2008). Arabidopsis protein disulfide isomerase-5 inhibits cysteine proteases during trafficking to vacuoles before programmed cell death of the endothelium in developing seeds W. Plant Cell 20, 2205–2220. doi: 10.1105/tpc.108.058339
Peng, R. H., Qiu, J., Tian, Y. S., Gao, J. J., Han, H. J., Fu, X. Y., et al. (2017). Disulfide isomerase-like protein AtPDIL1–2 is a good candidate for trichlorophenol phytodetoxification. Sci. Rep. 7:40130. doi: 10.1038/srep40130
Schröder, M., and Kaufman, R. J. (2005). ER stress and the unfolded protein response. Mutat. Res. 569, 29–63. doi: 10.1016/j.mrfmmm.2004.06.056
Shin, H. C., and Scheraga, H. A. (2000). Catalysis of the oxidative folding of bovine pancreatic ribonuclease a by protein disulfide isomerase. J. Mol. Biol. 300, 995–1003. doi: 10.1006/jmbi.2000.3928
Takemoto, Y., Coughlan, S. J., Okita, T. W., Satoh, H., Ogawa, M., and Kumamaru, T. (2002). The rice mutant esp2 greatly accumulates the glutelin precursor and deletes the protein disulfide isomerase. Plant Physiol. 128, 1212–1222. doi: 10.1104/pp.010624
Walker, K. W., And, M. M. L., and Gilbert, H. F. (1996). Catalysis of oxidative protein folding by mutants of protein disulfide isomerase with a single active-site cysteine. Biochemistry 35, 1972–1980. doi: 10.1021/bi952157n
Wang, H., Boavida, L. C., Ron, M., and Mccormick, S. (2008). Truncation of a protein disulfide isomerase, PDIL2-1, delays embryo sac maturation and disrupts pollen tube guidance in Arabidopsis thaliana. Plant Cell 20, 3300–3311. doi: 10.1105/tpc.108.062919
Wang, H. B., Zou, Z. R., and Gong, M. (2015). Molecular cloning, expression analysis, and preliminarily functional characterization of the gene encoding protein disulfide isomerase from Jatropha curcas. Appl. Biochem. Biotechnol. 176, 428–439. doi: 10.1007/s12010-015-1585-3
Wang, L., Li, Z., Lu, M., and Wang, Y. (2017). ThNAC13, a NAC transcription factor from Tamarix hispida, confers salt and osmotic stress tolerance to transgenic Tamarix and Arabidopsis. Front. Plant Sci. 8:635. doi: 10.3389/fpls.2017.00635
Wittenberg, G., and Danon, A. (2008). Disulfide bond formation in chloroplasts: formation of disulfide bonds in signaling chloroplast proteins. Plant Sci. 175, 459–466. doi: 10.1016/j.plantsci.2008.05.011
Wittenberg, G., Levitan, A., Klein, T., Dangoor, I., Keren, N., and Danon, A. (2014). Knockdown of the Arabidopsis thaliana chloroplast protein disulfide isomerase 6 results in reduced levels of photoinhibition and increased D1 synthesis in high light. Plant J. 78, 1003–1013. doi: 10.1111/tpj.12525
Xu, S., Butkevich, A. N., Yamada, R., Zhou, Y., Debnath, B., Duncan, R., et al. (2012). Discovery of an orally active small-molecule irreversible inhibitor of protein disulfide isomerase for ovarian cancer treatment. Proc. Natl. Acad. Sci. U.S.A. 109, 16348–16353. doi: 10.1073/pnas.1205226109
Yuen, C. Y., Matsumoto, K. O., and Christopher, D. A. (2013). Variation in the subcellular localization and protein folding activity among Arabidopsis thaliana homologs of protein disulfide isomerase. Biomolecules 3, 848–869. doi: 10.3390/biom3040848
Keywords: PDI, isomerase activities, stress resistance, critical amino acids, endoplasmic reticulum
Citation: Zhang Z, Liu X, Li R, Yuan L, Dai Y and Wang X (2018) Identification and Functional Analysis of a Protein Disulfide Isomerase (AtPDI1) in Arabidopsis thaliana. Front. Plant Sci. 9:913. doi: 10.3389/fpls.2018.00913
Received: 09 January 2018; Accepted: 08 June 2018;
Published: 19 July 2018.
Edited by:
Ruth Grene, Virginia Polytechnic Institute and State University, United StatesReviewed by:
Ping Lan, Institute of Soil Science (CAS), ChinaMiyako Kusano, University of Tsukuba, Japan
Copyright © 2018 Zhang, Liu, Li, Yuan, Dai and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoyun Wang, eHl1bndhbmdAc2RhdS5lZHUuY24=
†These authors have contributed equally to this work.
 Zhengrong Zhang
Zhengrong Zhang Xin Liu
Xin Liu Rong Li
Rong Li Li Yuan
Li Yuan Yaqing Dai
Yaqing Dai Xiaoyun Wang
Xiaoyun Wang