- 1State Key Laboratory for Conservation and Utilization of Subtropical Agro-Bioresources – Key Laboratory of Plant Functional Genomics and Biotechnology of Guangdong Provincial Higher Education Institutions, College of Life Sciences, South China Agricultural University, Guangzhou, China
- 2Laboratory Center of Basic Biology and Biotechnology, Education Department of Guangdong Province, Zhongkai University of Agriculture and Engineering, Guangzhou, China
- 3Key Laboratory of Crop Stress Biology for Arid Areas, College of Agronomy, Northwest A&F University, Yangling, China
- 4State Key Laboratory of Biocontrol and Guangdong Provincial Key Laboratory of Plant Resources, School of Life Sciences, Sun Yat-sen University, Guangzhou, China
Cuticle waxes, which are primarily comprised of very-long-chain (VLC) alkanes, play an important role in plant reproductive development. ECERIFERUM1 (CER1) is recognized as the core element for VLC alkane biosynthesis in Arabidopsis (Arabidopsis thaliana). However, genes involved in the VLC alkane biosynthesis in rice remain unclear, and the alkane-form pathway in rice has still to be further explored. Here, we show that OsCER1, a homology of CER1, functions in VLC alkanes biosynthesis, which also could regulate anther development and plastids differentiation in rice. OsCER1 was highly expressed in the tapetum (stage 10) and bicellular pollen cells (stage 11). The decreased content of VLC alkanes (C25 and C27) in the OsCER1 knocked down plants as well as the increased content of C27 alkanes in the OsCER1 overexpression plants indicates that OsCER1 participates in VLC alkane biosynthesis. Downregulation of OsCER1 in rice led to sterility, and fewer amyloplasts within the mature pollen grains. In addition, the downregulation of OsCER1 in rice caused delayed tapetal programmed cell death and abnormal development of plastids in the tapetal cells. Furthermore, significantly altered levels of expression of genes involved in the pollen development were exhibited in the OsCER1 knocked down plants. These results indicate that OsCER1 is critical for VLC alkanes biosynthesis, plastids differentiation, and pollen development. This work provides insights into the VLC alkanes biosynthesis in anther development in rice.
Introduction
In flowering plants, successful male reproductive development within the anther includes critical developmental events such as meristem specification, sporogenous cell differentiation, meiosis, and mitosis (Scott et al., 2004; Blackmore et al., 2007; Wilson and Zhang, 2009). Rice (Oryza sativa L.) is one of the most important food crops in the world. Abnormal reproductive development has significantly influenced rice production. Rice anthers have four lobes in which the primordial cells differentiate into reproductive pollen mother cells. Four somatic cell layers surround these cells: the tapetum, the middle layer, the endothecium, and the epidermis, which are arranged from the innermost to the outermost layer (Goldberger et al., 1995; Zhang and Yang, 2014).
Covering the epidermal cells, the rice anther has a reticulate cuticle, which plays a protective role in pollen development (Li and Zhang, 2010). The anther cuticle is a thin, continuous hydrophobic layer that coats the outermost surface of anthers and is mainly composed of epicuticular waxes and a cutin matrix (Kunst and Samuels, 2003; Jung et al., 2006; Nawrath, 2006; Pollard et al., 2008). Cuticular waxes are mostly composed of very-long-chain fatty acids (VLCFAs) derivatives, including primary and secondary alcohols, aldehydes, alkanes, ketones, and esters (Kunst and Samuels, 2003; Buschhaus and Jetter, 2011). Abnormal cuticular waxes of anthers frequently accompany aborted pollen development (Jung et al., 2006; Jessen et al., 2011; Leide et al., 2011; Smirnova et al., 2013; Haslam et al., 2015).
The biosynthesis of VLCFAs occurs in two stages. First, de novo fatty acids (FAs), such as C16 and C18 FAs, are synthesized in the plastid stroma and are catalyzed by the FA synthase complex using acetyl-CoA as the starting substrate. The majority of long-chain fatty acids (LCFAs) serve as central intermediates in catalytic steps, such as elongation and acyl editing (Haslam and Kunst, 2013), and some LCFAs remain in plastids integrated into polar lipids, such as glycolipids and phospholipids (Li-Beisson et al., 2013).
The second stage of VLCFA biosynthesis is the elongation of C16 and C18 FAs into VLCFAs consisting of C20 to C34 chains. The synthesis of VLCFAs is catalyzed by fatty acyl-CoA elongase in the endoplasmic reticulum (ER) (Hooker et al., 2002; Efremova et al., 2004; Bach et al., 2008; Joubes et al., 2008; Beaudoin et al., 2009). Subsequently, extended VLCFAs are modified into diverse VLCFAs derivatives, the main components of wax, via two pathways, namely, the alcohol-forming (acyl reduction) pathway and the alkane-forming (decarbonylation) pathway. The alcohol-forming pathway is involved in the production of very-long-chain (VLC) primary alcohols catalyzed by a fatty acyl-CoA reductase. The alkane-forming pathway yields VLC alkanes, aldehydes, secondary alcohols, and ketones (Jenks et al., 1995; Rashotte et al., 2001). Among these metabolites, VLC alkanes predominate in the wax structure, and account for 50–70% of the total wax content in numerous different plants species (Kunst and Samuels, 2003; Kosma et al., 2009; Kunst and Samuels, 2009). The VLC alkanes play critical roles in various plant physiological processes, notably in plant fertility and resistance to drought stress (Aarts et al., 1995; Kosma et al., 2009; Bourdenx et al., 2011).
Recently, several genes function in the metabolism of VLCFAs and wax were discovered (Kunst and Samuels, 2003; Buschhaus and Jetter, 2011), and several eceriferum (cer) mutations in Arabidopsis affect wax composition, suggesting that CER proteins play important roles in wax biosynthesis (Jenks et al., 1995; Lai et al., 2007). For instance, CER1 and CER3, which are localized in the ER, may form a complex that catalyzes the synthesis of VLC alkanes from VLC acyl-CoAs via a two-step reaction (Bernard et al., 2012). The GLOSSY1 (GL1) gene in maize (Zea mays), a homolog of CER3, affects the biosynthesis of epicuticular waxes on the surfaces of seedling leaves (Sturaro et al., 2005). CsCER1 and CsWAX2 in Cucumis are involved in the biosynthesis of VLC alkanes and influence pollen fertility (Wang et al., 2015a,b). It is important to understand the VLC biosynthesis pathway in rice at the molecular level because rice is one of the most important food crops worldwide. Recent studies have reported that Wax Crystal-Sparse Leaf 2 (WSL2), OsGL1-2, OsGL1-3, Wax-deficient Anther 1 (WDA1), and OsGL1-6 are homologs of CER involved in the wax biosynthesis in rice (Jung et al., 2006; Qin et al., 2011; Mao et al., 2012; Zhou et al., 2013, 2015). However, the alkane-form pathway in rice still needs to be explored, and whether other unreported CER homologs in rice are involved in VLC alkanes biosynthesis remains unclear.
Plastids where de novo fatty acids synthesis occurred play crucial roles in successful anther development. During anther development, proplastids in tapetal cells undergo division during the early stages of microsporogenesis and subsequently develop into elaioplasts that are associated with biosynthesis of the lipids that tapetum cells secrete into the anther locule to contribute to pollen wall formation (Clément and Pacini, 2001). For instance, No Exine Formation 1 (NEF1) is a plastid integral membrane protein that plays an important role in tapetum degeneration, plastid development, and lipid accumulation, and effects fertility in Arabidopsis (Ariizumi et al., 2004). However, whether there are lipid signals from plastids that regulate anther development and how these pathways accomplish this regulatory role remain unclear.
In this article, we isolated a homologous protein of CER1, OsCER1, and characterized its function in rice. OsCER1 is preferentially expressed in tapetum cells at stage 10 and in bicellular pollens at stage 11 of anther development. We obtained several independent OsCER1 downregulated lines, and found that these plants displayed low fertility, including abnormal anther development with smaller and fewer amyloplasts within the mature pollens. We also observed abnormal plastids differentiation in the tapetum. Furthermore, we provide evidence for the pivotal role of OsCER1 in the biosynthesis of VLC alkanes and in the tapetum degeneration during anther development in rice.
Materials and Methods
Plant Materials and Growth Conditions
The rice variety Zhonghua 11 (Oryza sativa L. ssp. japonica cv. Zhonghua 11) was used for all experiments in this study. The wild-type (WT) and transgenic plants were grown in a paddy field at South China Agricultural University. The developmental stages of the spikelets were determined according to spikelet length and anther morphology as described by Zhang et al. (2011).
Phylogenetic Analysis
Sequence alignment of OsCER1 and its related proteins from rice and other species was performed using Clustal Omega from EMBL-EBI1 to construct a phylogenetic tree, using default parameters. The phylogenetic tree was constructed using the neighbor-joining method as implemented in the MEGA program version 3.0 with 1,000 bootstrap replications2. CER1 and OsCER1 were aligned with ClustalW3, followed by manual alignment in conserved active His-rich motifs.
Generating OsCER1 Antisense-RNA and Overexpression Transgenic Plants
The OsCER1 antisense transgene driven by the OsCER1 promoter was constructed as described by Li J. et al. (2011). A 560-bp fragment of the OsCER1 cDNA was amplified using reverse transcription-polymerase chain reaction (RT-PCR) from Zhonghua 11 with gene-specific OsCER1-A F/R primers (Supplementary Table 4). The PCR product was inserted in reverse orientation into the multiple cloning site of pCAMBIA1380. A 2,036-bp genomic fragment located upstream of the annotated ATG start codon of OsCER1 was isolated from Zhonghua 11 genomic DNA using OsCER1-P F and OsCER1-P R primers (Supplementary Table 4) and then inserted upstream of the antisense fragment. A 1,860-bp ORF of OsCER1 cDNA was amplified using RT-PCR from Zhonghua 11 with gene-specific OsCER1-OE F/R primers (Supplementary Table 4) and inserted into the multiple cloning site of pYLpox driven by Ubi promoter. Then constructs were transformed into Agrobacterium tumefaciens strain EHA105. Transgenic plants were obtained as described by Li J. et al. (2011). Two homozygous OsCER1 antisense transgenic (OsCER1A) lines, OsCER1A 3-6 and OsCER1A 9-1, and two homozygous OsCER1 overexpression transgenic lines, OsCER1 overexpression 3 (OV-3) and OsCER1 overexpression 6 (OV-6) were obtained and used for relevant analysis.
Southern Bolt and Quantitative Reverse Transcription-PCR (qRT-PCR) Analysis
Genomic DNA was extracted with the plant genomic DNA miniprep Kit (Sigma, United States) from the 3–5 g mature leaves of WT and the nine OsCER1 antisense-RNA transgenic lines, respectively. To generate a Hpt gene-specific probe for the Southern bolt, a 651-bp Hpt fragment was amplified using the Hpt F and Hpt R primers (Supplementary Table 4) and labeled with a PCR DIG labeling mix (Roche, Diagnostics). The Southern bolt procedures were performed as previously described by Ariizumi et al. (2004). Total RNA was isolated from rice tissues using TRIzol reagent (Invitrogen, United States), including shoots, roots, and spikelets with anthers at different stages. First-strand cDNA synthesis was performed using a Takara PrimeScript first-strand cDNA synthesis kit (Takara Bio Inc.), and the cDNA was then used for qRT-PCR, which was performed in triplicate using SsoFast EvaGreen Supermix (Bio-Rad, United States). All primers for qRT-PCR are listed in Supplementary Table 4. OsActin1 was used as reference for normalization of mRNA expression.
RNA in situ Hybridization
Rice anthers from stages 6 to 11 were collected. To generate gene-specific and control probes, a 613-bp OsCER1 fragment was amplified using the OsCER1 HIS F and OsCER1 HIS R primers (Supplementary Table 4) and inserted into a pEASY-Blunt vector (Transgen, Beijing, China). The probes were transcribed in vitro from the T7 promoter using RNA polymerase as provided in the DIG RNA labeling kit (Roche, Indianapolis, IN, United States). The following RNA in situ hybridization procedures were performed as previously described by Kouchi and Hata (1993).
Terminal Deoxynucleotidyl Transferase-Mediated dUTP Nick-End Labeling (TUNEL) Assay
For the TUNEL assay, 6-μm sections of anthers were prepared and the following procedures were performed using the TUNEL apoptosis detection kit (DeadEnd Fluorometric TUNEL System, Promega). Slides were stained with propidium iodide (PI). Green GFP signals (excitation wavelength: 488 nm) and the red PI signals (excitation wavelength: 543 nm) were assessed using a confocal laser-scanning microscope (LSM510 Mera; Carl Zeiss).
Microscopy Analysis
Anthers from mature flowers were dissected, and pollen grains were stained with iodine/potassium iodide (I2/KI) staining (0.2% iodine and 2% potassium iodide) and observed under a brightfield microscope (Zeiss, Axio Observer D1, Germany). To investigate pollen germination in vitro, mature pollen grains were germinated on a glass slide at 33°C for 30 min in a pollen germination medium composed of 2.0% (m/v) agar, 30% (m/v) sucrose, 50 mM KNO3, 400 mM H3BO3, and 400 mM MgSO4⋅7H2O. The pollen grains were then observed under a brightfield microscope (Zeiss, Axio Observer D1).
Anthers of the WT and OsCER1A plants at different stages were fixed in 3.7% formaldehyde/acetic acid (v/v) and resin-embedded semi-thin sections (epoxy resin PolyBed 812, Polysciences) were made and observed as described by Zhu et al. (2011). For transmission electron microscopy (TEM), anthers were fixed in sodium phosphate buffer (pH 7.2) containing 4% (w/v) paraformaldehyde and 2.5% (v/v) glutaraldehyde and sectioned as described by Schulz et al. (1993). The sections were observed using an electron microscope (TECNAI G2 12, FEI) at 100–120 kV. For scanning electron microscopy (SEM), anthers were fixed in sodium phosphate buffer (pH 7.2) containing 4% (w/v) paraformaldehyde and 2.5% (v/v) glutaraldehyde. The following SEM procedures were performed as previously described by Zhou et al. (2015).
Subcellular Localization
The CaMV35S promoter, eGFP fragments, and NOS terminator were successively cloned into pUC18 to construct the eGFP transient expression (GFP) vectors. The signal peptide fragment of OsCER1 was amplified using the primer pair OsCER1-eGFP F, OsCER1-eGFP R (Supplementary Table 4) and then subcloned into the GFP vector between the CaMV35S promoter and the eGFP gene, in-frame with the N-terminus of eGFP being driven by the CaMV35S promoter. Protoplasts from the leaf sheaths of 14-day-old rice seedlings were prepared, and transient expression was conducted following the protocol described by Zhou et al. (2015).
WAX Profiling
For the anthers wax composition studies, waxes were extracted from 10 to 20 mg of freeze-dried anthers and analyzed as described by Zhou et al. (2015). For the leaves wax composition analysis, 20–50 mg of freeze-dried flag leaves were harvested in a Teflon-lined screw cap glass tube, extracted, and analyzed following the method of Zhou et al. (2015).
Accession Numbers
The sequences used in this study were downloaded from GenBank with the following accession numbers: CER1 (D64155.1), CER3 (X95962), BnCER1 (KF724897.1), CsCER1 (KJ461885), CYP704B2 (Os03g07250), Defective Pollen Wall (DPW) (EU971135), GL1 (U37428.1), Rice ATP binding cassette subfamily G 15/ Post-meiotic deficient anther 1 (OsABCG15/PDA1) (Os06g40550), OsC6 (Os11g37280), OsNEF1 (Os11g32470), OsGL1-3 (AK070469), WSL2 (AK060786), OsGL1-2 (AK066569), OsCER1 (AK066386), WDA1 (AK100751), OsGL1-6 (AK068166), OsRAFTIN1 (Os08g38810), Tapetum Degeneration Retardation (TDR) (Os02g02820), and TDR Interacting Protein 2 (TIP2) (Os01g18870).
Results
OsCER1 Is Homologous to CER1
Phylogenetic reconstruction indicated that CER1 and other 10 homologous proteins of CER1 could be classified into two clades; CER1 and CER3 belonging to clades I and II, respectively (Figure 1A and Supplementary Data Set 1). The gene Os02g0621300 located on rice chromosome 2 is a homology of CER1, thus we named this gene OsCER1. The full-length 2,481-bp cDNA clone of OsCER1 contains an open reading frame of 1,860 bp in length, which encodes a 619-amino acid protein (XP_015627618.1) with a molecular mass of 71 kD. Putative conserved domains include a fatty acid hydroxylase superfamily domain (amino acids 139–253) and a WAX2 C-terminal domain (amino acids 453–615) (Islam et al., 2009). OsCER1 was grouped into clade I, and shared 57% sequence identity with CER1. Alignment of CER1 and OsCER1 showed that their predicted amino acid sequences contained the same three conserved histidine-rich (His-rich) motifs (Figure 1B) that are likely critical for alkane synthesis function (Bernard et al., 2012).
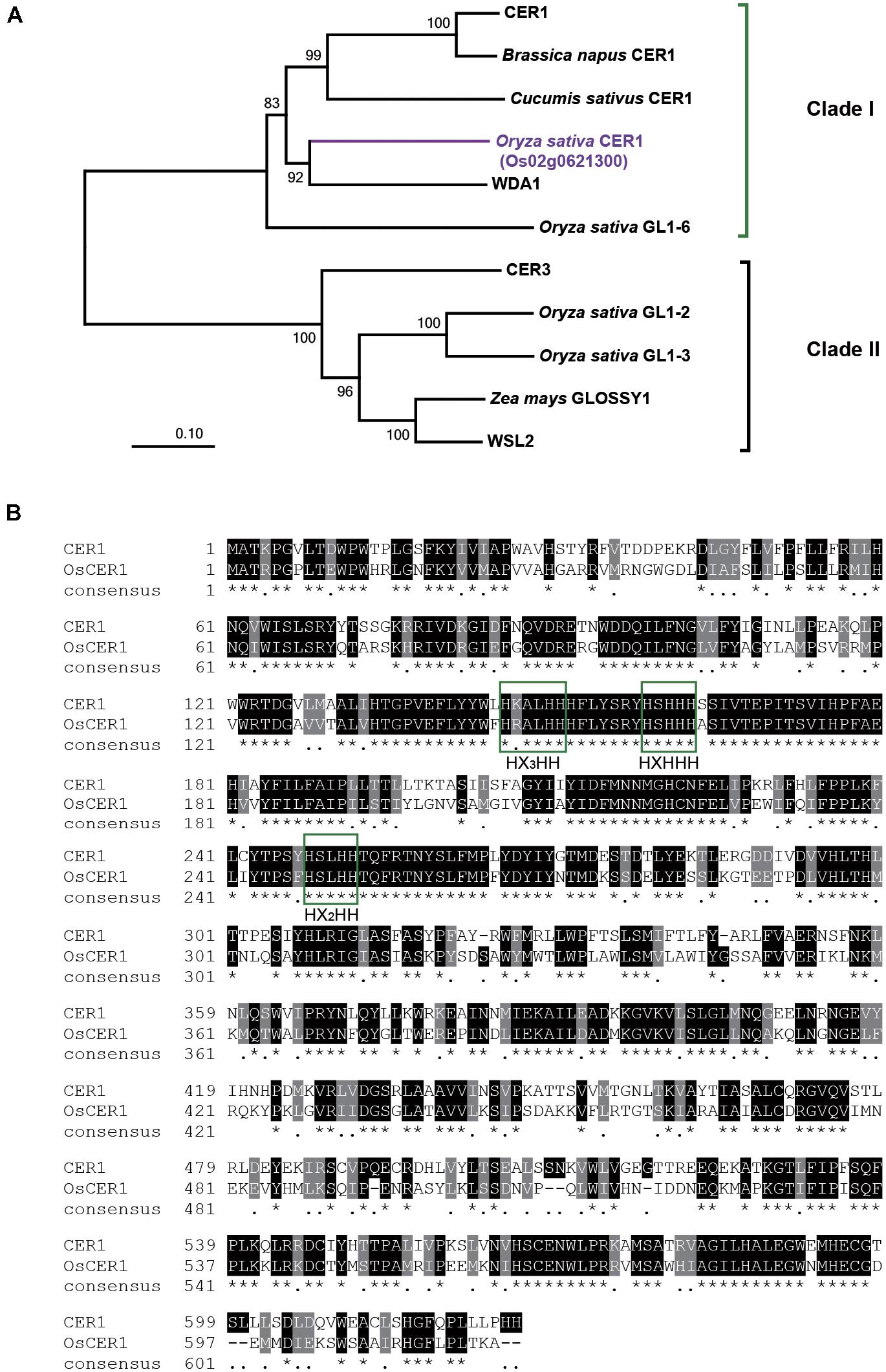
FIGURE 1. Phylogenic analysis and sequence alignment of OsCER1. (A) Phylogenetic analysis of the OsCER1 protein and 10 OsCER1 related proteins. Bootstrap values are shown on the branches. The scale bar represents the number of amino acid substitutions per site. The value of 0.01 on the scale bar indicates 1 amino acid substitution per 100 residues. The numbers near the branches refer to the bootstrap value of the neighbor-joining phylogenetic tree. The length of the branches is proportional to the amino acid variation rates. (B) Sequence alignment between CER1 and OsCER1. Identical and similar residues are shown on black and gray backgrounds, respectively. Green boxes indicate the three conserved active His-rich motifs; asterisks (∗) indicate the identical residues in all sequences, black dots (.) indicate the semi-conserved substitutions, and dashes (–) indicate gaps in the alignment.
OsCER1 Is Mainly Expressed in Tapetum and Microspores
To further characterize the expression pattern of OsCER1, we measured its transcript levels in different tissues by qRT-PCR. In rice, anther development can be divided into 14 stages based on morphological characteristics (Zhang et al., 2011). OsCER1 was highly expressed in the panicles, particularly in the spikelets within anthers at stages 10 and 11, whereas its expression level was lower in the vegetative organs (Figure 2A). This finding indicates that OsCER1 might be involved in panicle development in rice. To explore the spatiotemporal expression of OsCER1 during anther development, RNA in situ hybridization of the WT anthers was performed. Rare OsCER1 signals were detectable in anthers from stages 6 to 9 (Supplementary Figure 1), which refer to the stages from microspore mother cells generation to microspores generation. However, strong RNA in situ hybridization signals were detected in the tapetum cells with swollen structure at stage 10 (Figure 2B). Also, a high OsCER1 expression level was detected in bicellular pollens undergoing the first mitosis at stage 11 (Figure 2D). In the negative control, hybridization with the OsCER1 sense probe did not generate any detectable signals (Figures 2C,E). These results demonstrate that OsCER1 is highly expressed in tapetum cells and bicellular pollens of anthers at stages 10 and 11, respectively.
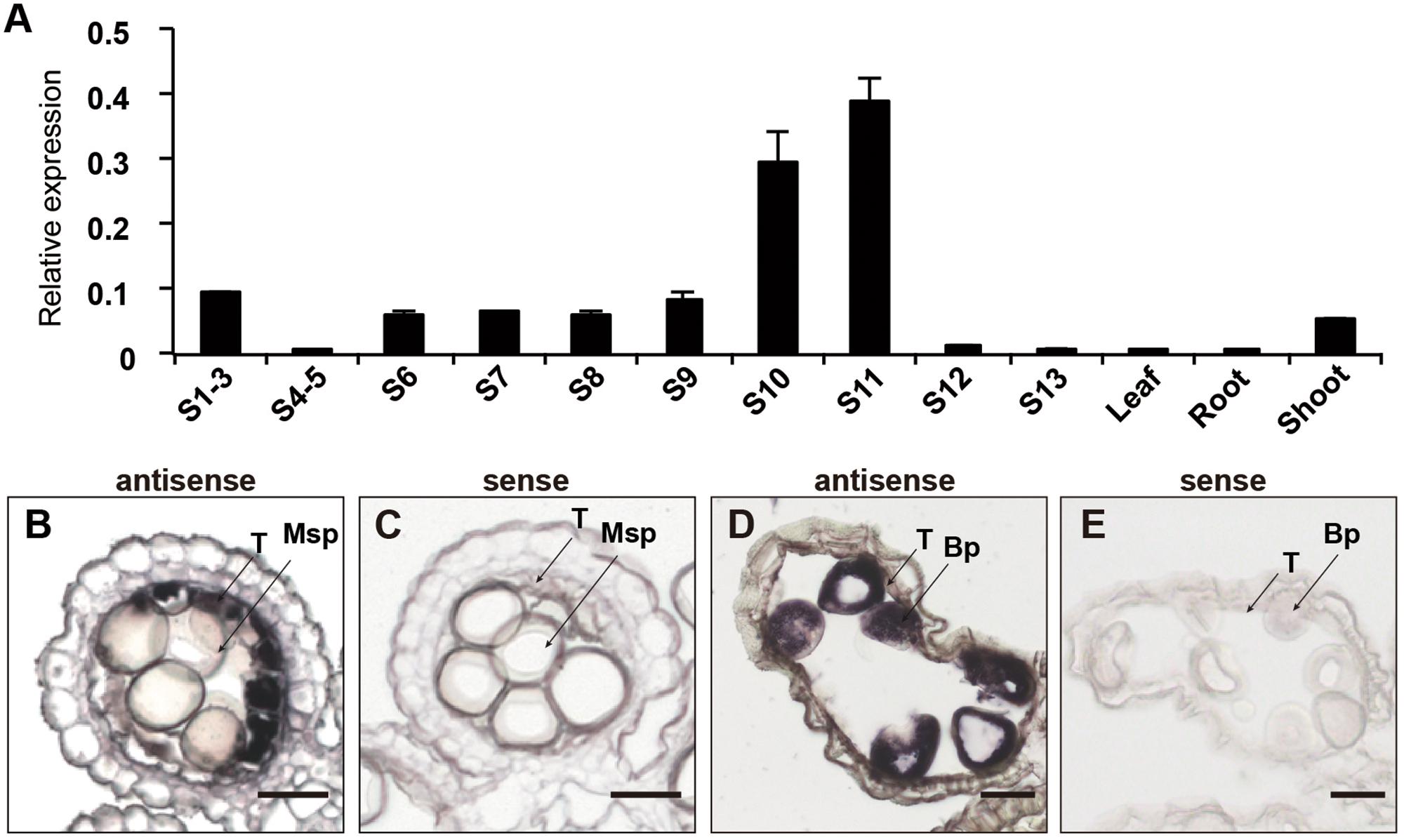
FIGURE 2. The expression patterns of OsCER1. (A) Spatial and temporal expression analysis of OsCER1 in spikelets at various anther developmental stages by qRT-PCR. S1-3, before stage 3; S4-5, from stages 4 to 5; S6, stage 6; S7, stage 7; S8, stage 8; S9, stage 9; S10, stage 10; S11, stage 11; S12, stage12; S13 stage13. OsActin1 was used as an internal control. Each reaction had three biological repeats and error bars indicate the standard deviations (SD). (B–E) In situ hybridization of OsCER1 in WT anthers. The anthers at stage 10 (B) and stage 11 (D) hybridized with an OsCER1 antisense probe. The anthers at stage 10 (C) and stage 11 (E) hybridized with an OsCER1 sense probe. Bp, bicellular pollen; Msp, microspore; T, tapetum. Bars = 50 μm in (B–E).
Defects in Anther Development in Plants Expressing an OsCER1 Antisense Construct
To analyze the biological function of OsCER1, we constructed an OsCER1 antisense vector containing a gene-specific OsCER1 fragment driven by the native promoter and transformed this construct into Zhonghua 11 calli (Hiei et al., 1994; Li J. et al., 2011). A total of nine independent OsCER1A lines were obtained and identified by Southern blotting (Supplementary Figure 2). Furthermore, we obtained two transgenic homozygous lines through genetic screening, lines OsCER1A 3-6 and OsCER1A 9-1, which exhibited lower OsCER1 transcript levels as determined by semi-quantitative RT-PCR (Supplementary Figure 3). These two lines were then used in the subsequent experiments, and the OsCER1A 3-6 plants showed the lowest OsCER1 expression.

FIGURE 3. Comparison of the WT plants and OsCER1A lines. (A–C) Comparison of seed setting rate between the WT (A), OsCER1A 3-6 (B), and OsCER1A 9-1 (C) plants showing low seed setting rate in the OsCER1A plants. (D–F) A WT spikelet (D), an OsCER1A 3-6 spikelet (E), and an OsCER1A 9-1 spikelet (F) after removing the lemma and palea showing no obvious difference in stamens with each other. (G) Mature pollen grains stained with 1% I2/KI solution in the WT plants. (H,I) Abnormal pollen grains stained with 1% I2/KI solution in the OsCER1A 3-6 (H) and OsCER1A 9-1 (I) plants showing lightly stained pollen grains. (J–L) In vitro pollen germination of the WT (J), OsCER1A 3-6 (K) and OsCER1A 9-1 (L) plants. St, stamen. Bars = 10 cm in (A–C); 100 μm in (D–F); 50 μm in (G–L).
The plants of OsCER1A 3-6 and OsCER1A 9-1 at maturity stage were slightly smaller than the WT plants (Supplementary Figure 4). The most remarkable feature of the OsCER1A plants was the lower seed setting. The seed setting of the OsCER1A 3-6 plants only 50.14%, which is lower than that of the OsCER1A 9-1 plants (54.79%) and significantly reduced compared to that of the WT plants (94.20%) (Figures 3A–C and Supplementary Table 1). The anthers of the OsCER1A 3-6 and OsCER1A 9-1 plants also showed a normal stamen morphology (Figures 3D–F) and SEM showed slightly higher concentration of wax crystal in the surface of OsCER1A anthers and no obvious alteration in OsCER1A pollens morphology (Supplementary Figure 5). To explore the reason of sterility, we examined pollen viability and pollen tube growth by I2/KI staining and in vitro germination analysis, respectively. The pollen grains of the OsCER1A 3-6 and OsCER1A 9-1 showed lighter staining than that of the WT (Figures 3G–I), suggesting lower pollen viability in the OsCER1A plants. Additionally, statistical analysis of the in vitro germination demonstrated that none of the OsCER1A 3-6 and OsCER1A 9-1 pollens were able to germinate normal pollen-tubes compared to the 42.33% normal pollen-tube rate of the WT pollens germination (Figures 3J–L). These results strongly indicate that a decrease in OsCER1 expression affects pollen fertility and pollen germination.
To examine defects in anther development in the OsCER1A plants, we evaluated transverse semi-thin sections of the WT and OsCER1A anthers by light microscopy. At stage 8, the dyad cells underwent normal meiosis and produced tetrads, and tapetum displayed shrunk and vacuolated cytoplasm of the WT anthers (Figure 4A). However, the tetrads became abnormally vacuolated in the OsCER1A 3-6 and OsCER1A 9-1 anthers (Figures 4F,K), and tapetal cells exhibit non-vacuoles of the OsCER1A 3-6 anthers (Figure 4F) and small vacuoles of the OsCER1A 9-1 anthers (Figure 4K). At stage 9, new microspores were generated and released from the tetrads, and the vacuoles in the tapetal cells were reabsorbed and the cytoplasm appeared highly condensed with intense staining of the WT anthers (Figure 4B). In contrast, the thicker tapetum was observed in the OsCER1A 3-6 and OsCER1A 9-1 anthers at the same stage (Figures 4G,L). At stage 10, the microspores exhibited a round-shape within the enlarging vacuoles and the tapetum continued to degenerate, forming swollen and lighter stained tapetal cells in the WT anthers (Figure 4C), whereas darkly stained tapetal cells appeared in the OsCER1A 3-6 and OsCER1A 9-1 anthers at this stage (Figures 4H,M). At stage 11, the microspores underwent the first mitosis, and the tapetal layer continued to degenerate and formed large, clear vacuoles in the WT anthers (Figure 4D); the small vacuoles could be observed in the OsCER1A 9-1 tapetal cells (Figure 4N), whereas the tapetal cells still showed intense staining cytoplasm in the OsCER1A 3-6 anthers (Figure 4I). Subsequently, the mature pollen grains of the OsCER1A 3-6 and OsCER1A 9-1 plants were abnormally deeply stained compared with that of the WT plants (Figures 4E,J,O). These results suggest that the OsCER1A plants underwent abnormal tapetum and pollen development.
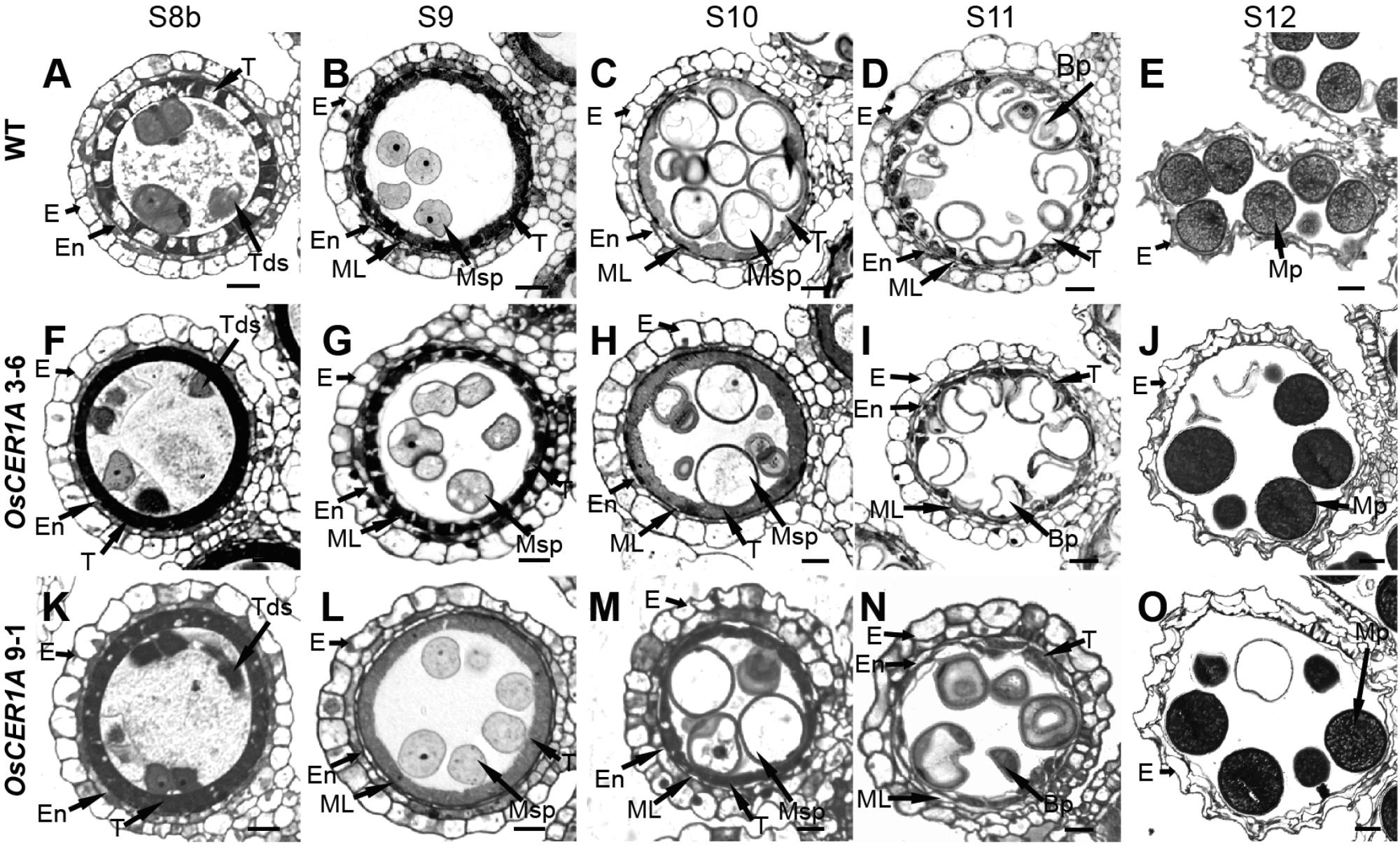
FIGURE 4. Comparison of transverse semi-thin sections of anthers from the WT plants and OsCER1A lines. (A–E) Transverse semi-thin sections of a single locule of the WT anthers at stage 8b (S8b) (A), stage 9 (S9) (B), stage 10 (S10) (C), stage 11 (S11) (D), and stage 12 (S12) (E). (F–J) Transverse semi-thin sections of a single locule of the OsCER1A 3-6 anthers at S8b (F), S9 (G), S10 (H), S11 (I), and S12 (J). (K–O) Transverse semi-thin sections of a single locule of the OsCER1A 9-1 anthers at S8b (K), S9 (L), S10 (M), S11 (N), and S12 (O). Bp, bicellular pollen; E, epidermis; En, endothecium; ML, middle layer; MP, mature pollen; Msp, microspore; T, tapetum; Tds, tetrads. Bars = 20 μm.
To further characterize the details of the defect in the tapetal cells of the OsCER1A plants, the ultra-structural features of the tapetal cells were assessed by TEM. From stages 8b to 11, the tapetum of the WT plants showed signs of degeneration (Figures 5A–D). In contrast, the tapetal cells of the OsCER1A 3-6 and OsCER1A 9-1 plants showed the state of condensed cytoplasm had been maintained from stages 8b to 11 (Figures 5E–L). In particular, at stage 11, obviously large vacuoles and fragmented cytoplasm were observed in the tapetal cells of the WT anthers (Figure 5D). A densely stained cytoplasm full of the whole tapetal cells and a dense cytoplasm with small vacuoles within tapetal cells were observed in the OsCER1A 3-6 and OsCER1A 9-1 plants, respectively (Figures 5H,L). These findings suggest that the tapetal cells of the OsCER1A plants underwent abnormal degeneration compared to that in the WT plants.
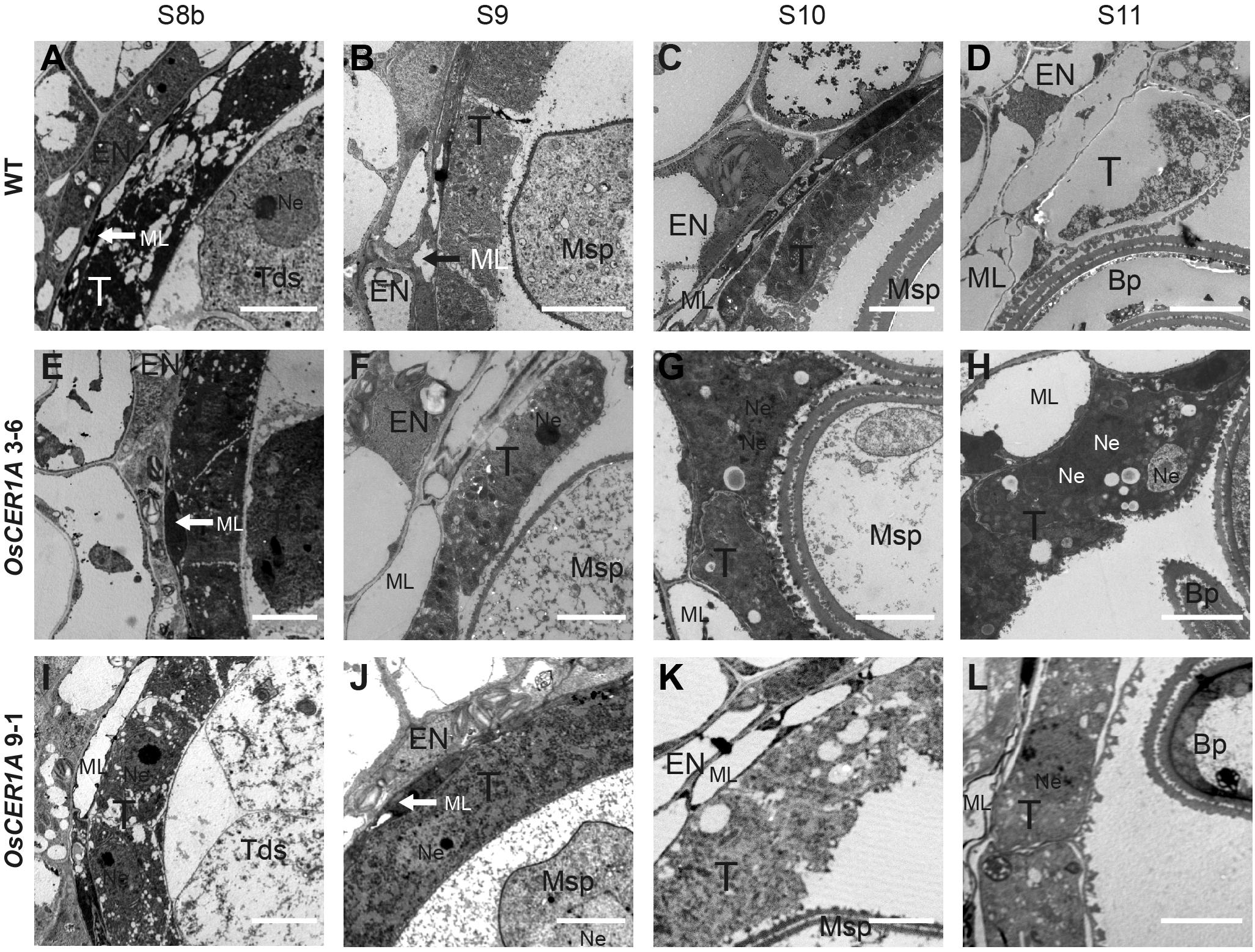
FIGURE 5. Comparison of transmission electron microscopy (TEM) of anthers from the WT plants and OsCER1A lines. (A–D) TEM analysis of the WT anthers at stage 8b (S8b) (A), stage 9 (S9) (B), stage 10 (S10) (C), and stage 11 (S11) (D). (E–H) TEM analysis of the OsCER1A 3-6 anthers at S8b (E), S9 (F), S10 (G), and S11 (H). (I–L) TEM analysis of the OsCER1A 9-1 anthers at S8b (I), S9 (J), S10 (K), and S11 (L). Bp, bicellular pollen; En, endothecium; ML, middle layer; Msp, microspore; Ne, nucleus; T, tapetum; Tds, tetrads. Bars = 10 μm.
Delayed Programmed Cell Death (PCD) in Tapetum of OsCER1A Plants
In rice, PCD is necessary for anther development, and abnormal PCD in tapetal cells may cause male sterility (Parish and Li, 2010; Qin et al., 2011; Shi et al., 2011; Tan et al., 2012). The hallmarks of PCD include DNA fragmentation, mitochondrial, and cytoskeletal disintegration, cellular condensation, and nuclear condensation (Parish and Li, 2010). The above TEM observations in the tapetum indicate that the tapetum degeneration in the OsCER1A plants was affected during anther development. To determine the effect of OsCER1 downregulation on tapetum degeneration and PCD during anther development, we examined DNA fragmentation of the WT, OsCER1A 3-6 and OsCER1A 9-1 tapetum using the TUNEL assay. In the WT tapetal cells, TUNEL-positive signals were first detected at stage 8a (Figures 6A,B), and the signals were strongest at stage 8b (Figure 6C). Subsequently, the signals in the tapetal cells gradually weakened from stage 9 to stage 10 in the WT tapetal cells (Figures 6D,E). However, in the OsCER1A plants, no TUNEL signals were observed in the OsCER1A 3-6 and OsCER1A 9-1 tapetal cells at stages 7 (Figures 6F,K) and 8a (Figures 6G,L). At stage 8b, no apoptotic characteristics were detected in the OsCER1A 3-6 tapetal cells (Figure 6H), and weakly TUNEL signals were detected in the OsCER1A 9-1 tapetal cells (Figure 6M). Stronger but fewer TUNEL-positive signals and several intense TUNEL-positive signals were observed in the OsCER1A 3-6 and OsCER1A 9-1 tapetal cells at stage 9, respectively (Figures 6I,N). Until stage 10, signals in the OsCER1A 3-6 and OsCER1A 9-1 tapetal cells remained distinct (Figures 6J,O). These results suggest the initiation of tapetal PCD in the OsCER1A 3-6 and OsCER1A 9-1 tapetum was delayed from stage 8a to stage 9 and stage 8b, respectively. These results suggest that the abnormal non-vacuolization in the OsCER1A tapetum during stage 8b was coupled with delayed tapetal PCD may result in male sterility.
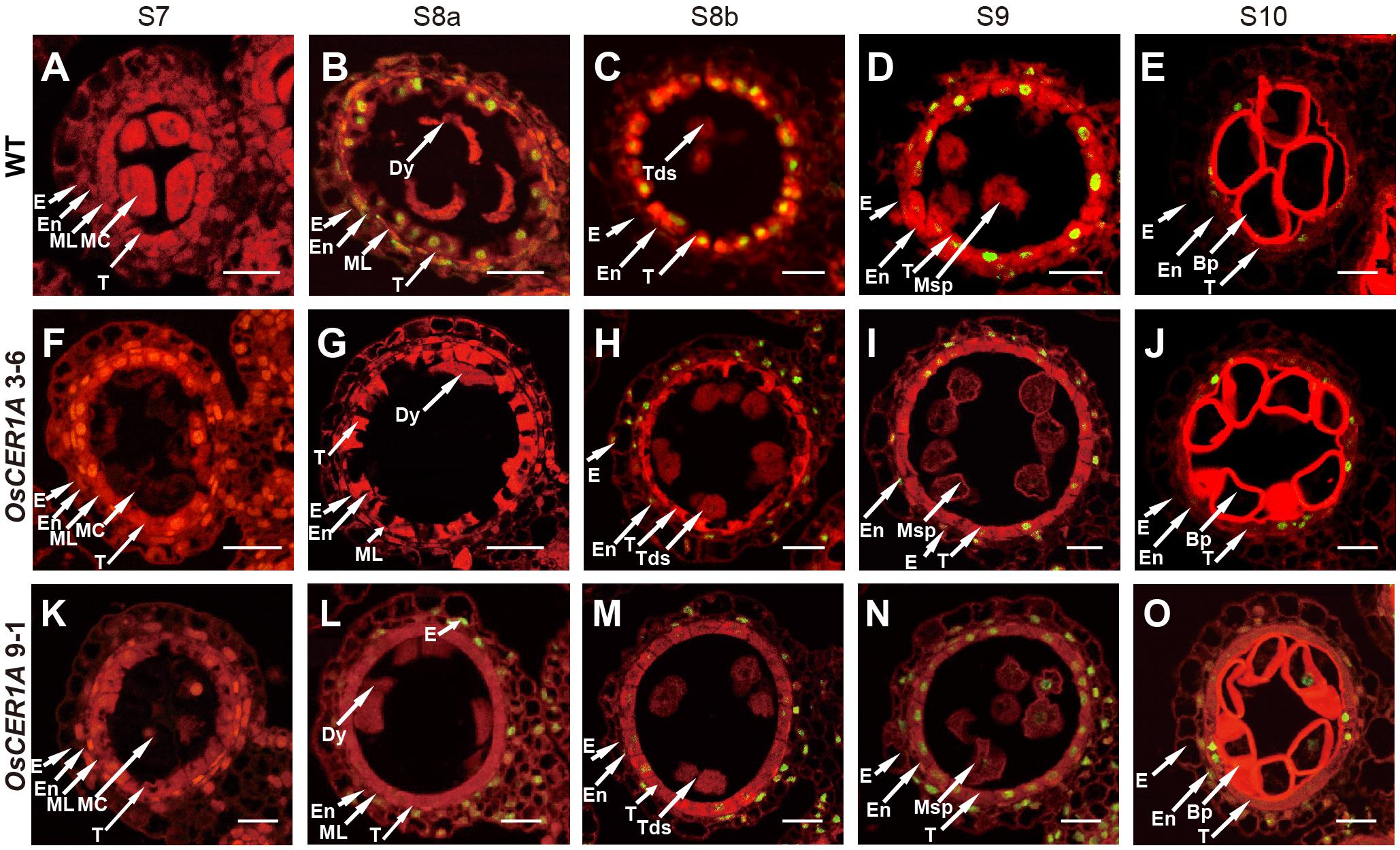
FIGURE 6. Programmed cell death (PCD) defects in the tapetal cells of the OsCER1A lines. (A–E) TUNEL assay of the WT anthers at stage 7 (S7) (A), stage 8a (S8a) (B), stage 8b (S8b) (C), stage 9 (S9) (D), and stage 10 (S10) (E). (F–J) TUNEL assay of the OsCER1A 3-6 anthers at S7 (F), S8a (G), S8b (H), S9 (I), and S10 (J). (K–O) TUNEL assay of the OsCER1A 9-1 anthers at S7 (K), S8a (L), S8b (M), S9 (N), and S10 (O). The red fluorescence showed tissues stained with propidium iodide and the yellow fluorescence showed TUNEL-positive nuclei. Bp, bicellular pollen; Dy, dyad cell; E, epidermis; En, endothecium; MC, meiotic cell; ML, middle layer; Msp, microspore; Tds, tetrads; T, tapetum. Bars = 50 μm.
Defects in Plastids During Anther Development in the OsCER1A Plants
Abnormal tapetal elaioplasts were also detected in the OsCER1A plants at stage 11 (Figures 5H,L). TEM was used to observe tapetal cells and pollens at higher magnification, which exhibited significant differences in plastid structure in the OsCER1A anthers relative to that in the WT anthers (Figure 7). The proplastids in the WT tapetal cells at stage 10 were beginning to develop into larger elaioplasts (Figure 7A). In contrast, in the OsCER1A 3-6 and OsCER1A 9-1 tapetal cells the plastids were observed without elaioplast-like shape at this stage (Figures 7B,C). At stage 11 in the WT anthers, the tapetal cells had generated into cellular debris and contained the large completely differentiated elaioplasts (Figure 7D). However, at this stage the OsCER1A 3-6 tapetal cells remained filled with cytoplasm including plastids that lacked a distinct boundary and a typical elaioplast structure, and in the OsCER1A 9-1 plants, tapetal cytoplasm was still dense including elaioplasts with abnormal structure (Figures 7E,F). In the WT plants, mature pollens were filled with amyloplasts that differentiated from proplastids (Figure 7G). In contrast, in the OsCER1A 3-6 and OsCER1A 9-1 plants, fewer amyloplasts were observed within the mature pollen grains and most of them exhibited abnormal morphology (Figures 7H,I). Taken together, these results suggest that the downregulation of OsCER1 disrupts the development of elaioplasts and amyloplasts within tapetal cells and in microspores, respectively.
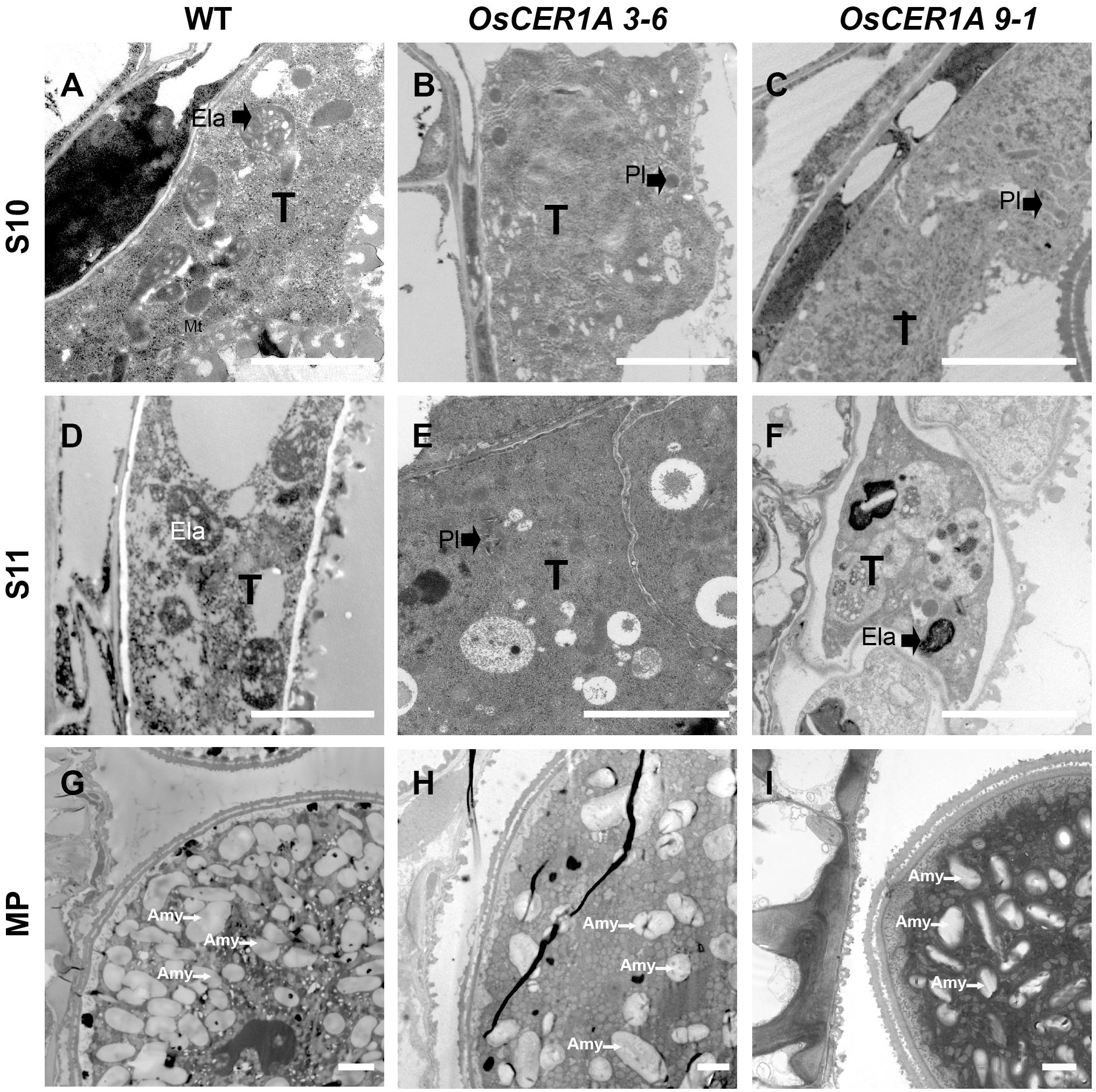
FIGURE 7. TEM of tapetum cells at stage 10 (S10), at stage 11 (S11) and mature pollen (MP) in the WT plants and OsCER1A lines. (A–C) Tapetal regions at S10 of the WT (A), OsCER1A 3-6 (B), and OsCER1A 9-1 (C) plants. (D–F) Tapetum regions at S11 of the WT (D), OsCER1A 3-6 (E), and OsCER1A 9-1 (F) plants. (G–I) Pollen cross sections of the WT (G), OsCER1A 3-6 (H), and OsCER1A 9-1 (I) plants. Amy, amyloplast; Ela, elaioplast; Pl, plastid; T, tapetum. Bars = 2 μm.
OsCER1 Is Involved in VLC Alkanes Biosynthesis
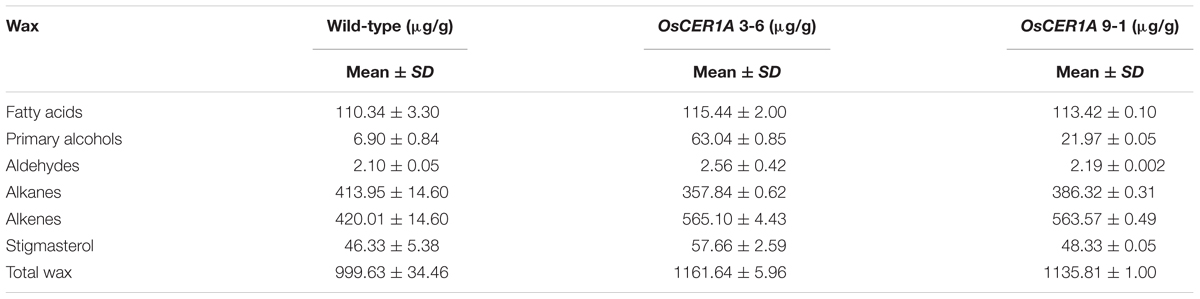
In Arabidopsis, CER1 is involved in VLC alkanes biosynthesis (Bourdenx et al., 2011; Bernard et al., 2012). Our bioinformatics analysis showed that OsCER1 possesses three conserved His-rich motifs and shares high sequence identity with CER1. The subcellular localization of the signal peptide of OsCER1 showed that the ER localization of the GFP fusion protein was detected by the co-localization of GFP fluorescence and KDEL-MCherry (ER marker) fluorescence (Supplementary Figure 6) (Zhou et al., 2015), which indicating that OsCER1 was localized in the ER. These results suggest that OsCER1 may also be involved in the biosynthesis of VLC alkanes in rice. To investigate the alteration of wax profiles in the OsCER1A anthers, we measured the composition of chloroform-extractable cuticular wax in the mature anthers using gas chromatography-mass spectrometry (GC-MS). In the WT anthers, total wax content of dry, mature anthers was 999.63 μg/g, whereas in the OsCER1A lines, total wax increased by 16.2% to 1,161.64 μg/g in the OsCER1A 3-6 anthers and by 13.6% to 1135.81 μg/g in the OsCER1A 9-1 anthers (Table 1). Compared to the WT anthers, the alkene content in the OsCER1A 3-6 and OsCER1A 9-1 anthers was increased by 34.54% and by 43.18%, respectively (Table 1). The primary alcohol content was increased by 813.62% and by 218.41% in the OsCER1A 3-6 and OsCER1A 9-1 anthers, respectively (Table 1). However, alkane content decreased by 13.55% and 6.67% in the OsCER1A 3-6 and OsCER1A 9-1 anthers, respectively (Table 1). A significant reduction of alkanes C25 (45.26% and 21.27%) and C27 (35.46% and 12.38%) and a dramatic increase in corresponding primary alcohols C26 (573.26% and 146.1%) and C28 (2,494.40% and 477.79%) were detected in the OsCER1A 3-6 and OsCER1A 9-1 anthers, respectively (Figure 8A and Supplementary Table 2). These results indicate that OsCER1 may function in alkane biosynthesis.
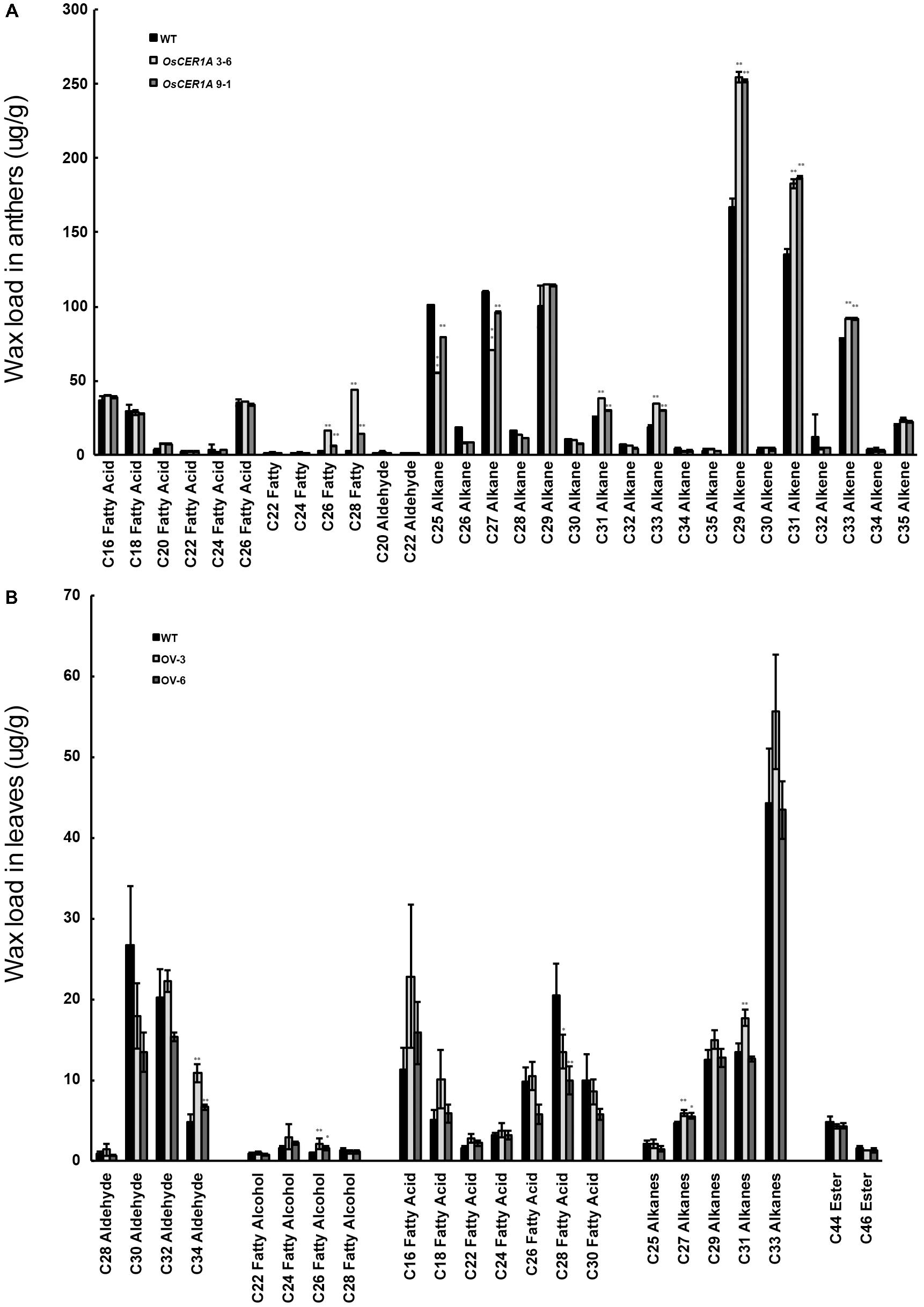
FIGURE 8. Analysis of wax constituents. (A) Wax constituents in whole anthers at stage 12 in the WT (Black bars), OsCER1A 3-6 (slightly gray bars) and OsCER1A 9-1 (darkly gray bars) plants. (B) Wax constituents in leaves in the WT (black bars), OsCER1 overexpression 3 (OV-3) (slight gray bars) and OsCER1 overexpression 6 (OV-6) (darkly gray bars) plants. Error bars indicate SD (n = 5). Statistical significance of differences between WT and transgenic plants means is indicated by ∗p < 0.05 and ∗∗p < 0.01.
To confirm whether OsCER1 is involved in VLC alkanes biosynthesis, we obtained 2 homozygous OsCER1 overexpression lines, OV-3 and OV-6, which exhibited higher OsCER1 transcript levels, as determined by qRT-PCR (Supplementary Figure 7). These two lines were then used in the subsequent wax composition analysis, and the OsCER1 expression of OV-3 exhibited higher than that of OV-6. To avoid the effect of native OsCER1, we selected leaves that showed minimal native OsCER1 expression for wax composition analysis. Compared to the WT leaves, the C27, C29, C31, C33 VLC alkanes increased in the OV-3 leaves, and C27 alkanes were increased in the OV-6 leaves (Figure 8B). The C27 alkanes were showed a significant increase (24.71% and 16.77%) in the OV-3 and OV-6 leaves, respectively (Figure 8B and Supplementary Table 3). We also detected a significant reduction of the corresponding fatty acid C28 (34.17% and 51.54%) in the OV-3 and OV-6 leaves, respectively (Figure 8B and Supplementary Table 3). The reduction in the content of VLC alkanes of the OsCER1A plants and the increase in the content of VLC alkanes of OsCER1 overexpression plants suggest that OsCER1 functions in VLC alkanes biosynthesis.
Altered Gene Expression Affects Lipid Metabolism and Tapetum Development in OsCER1A Plants
The reduced fertility and altered lipid profiles of the OsCER1A plants implied that some genes that were critical for anther development and lipid metabolism might be affected. Therefore, we used qRT-PCR to examine five genes involved in lipid metabolism and anther development, namely, WDA1 (Jung et al., 2006; Islam et al., 2009), OsABCG15/PDA1 (Qin et al., 2013; Zhu et al., 2013), OsC6 (Zhang et al., 2010), OsRAFTIN1 (Wang et al., 2003), and CYP704B2 (Li et al., 2010) in the WT, OsCER1A 3-6, and OsCER1A 9-1 spikelets with anthers from stages 6 to 10. Among these genes, OsCER1 showed 63% identity with WDA1, which also belonged to the clade I (Figure 1A) in rice. The expression level of WDA1, which is essential for anther wall and pollen development, significantly increased in OsCER1A spikelets with anthers at stage 7 (Figure 9A). An increase in the expression of OsABCG15/PDA1, which encodes a lipid-soluble precursor transporter, was observed at stage 9 (Figure 9B), whereas an upregulation of OsC6, which is required for transport of lipophilic materials, was detected at stage 10 in the OsCER1A plants (Figure 9C). CYP7042B2 and OsRAFTIN1 are important for exine formation during pollen development. The expression pattern of CYP704B2 was similar to that of WT plants from stages 6 to 8, but at stages 9 and 10, the expression levels of CYP704B2 significantly decreased (Figure 9D). OsRAFTIN1 expression increased at stages 9 and 10 in the OsCER1A plants (Figure 9E).
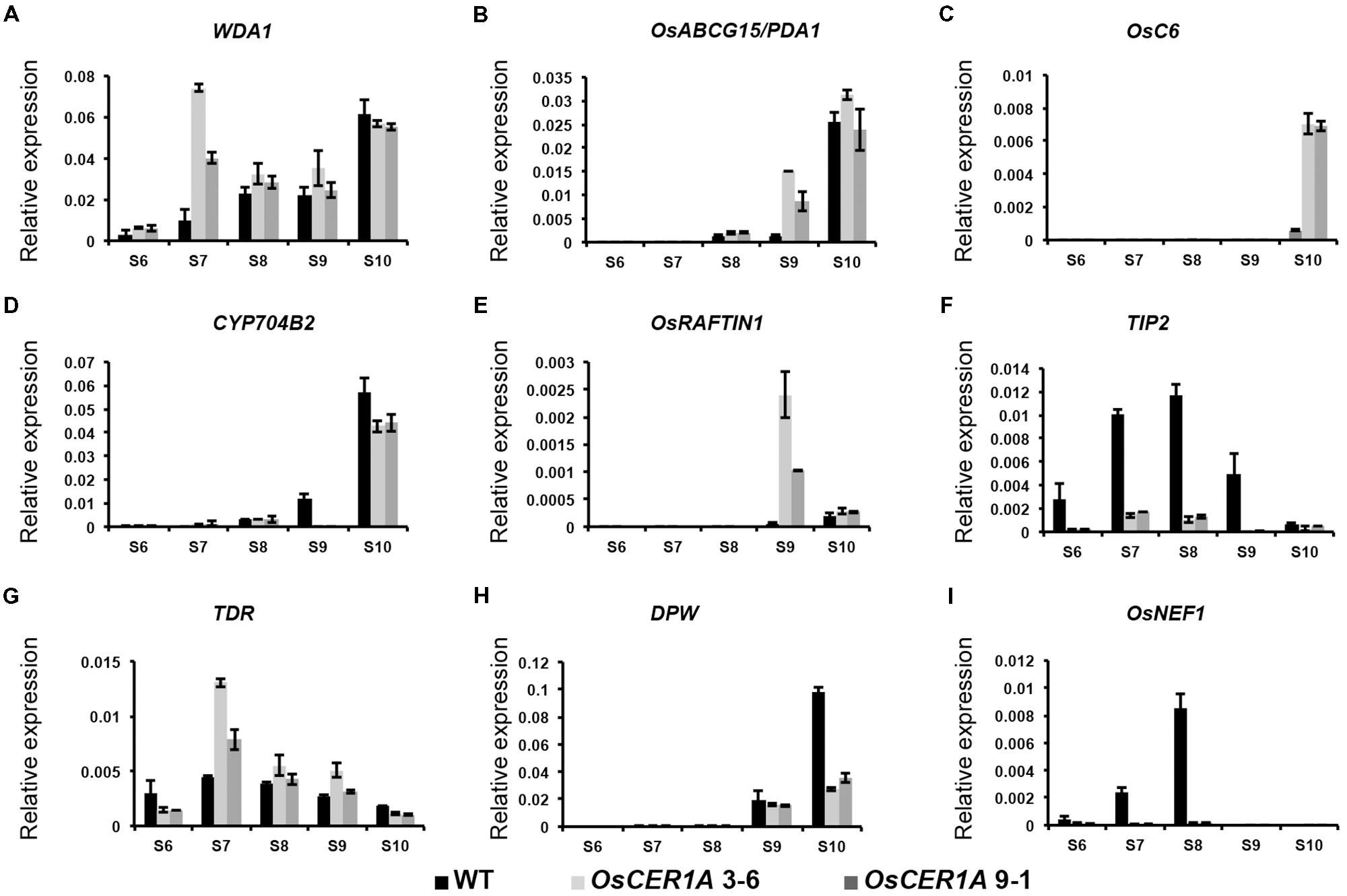
FIGURE 9. Expression analysis of genes related to lipid metabolism and pollen wall biosynthesis in the WT and OsCER1A lines by qRT-PCR. (A–I) Expression analysis of WDA1 (A), OsABCG15/PDA1 (B), OsC6 (C), CYP704B2 (D), OsRAFTIN1 (E), TIP2 (F), TDR (G), DPW (H), and OsNEF1 (I) in the WT, OsCER1A 3-6 and OsCER1A 9-1 plants with anthers from stages 6 to 10. OsActin1 was used as a control. Black bars represent the WT plants. Slightly gray bars represent the OsCER1A 3-6 plants. Darkly gray bars represent the OsCER1A 9-1. Each reaction had three biological repeats. Error bars indicate SD (n = 4). Gene names are abbreviated as follows: DPW, Defective Pollen Wall; OsABCG15/PDA1, Rice ATP binding cassette subfamily G 15/ Post-meiotic deficient anther 1; OsNEF1, Rice No Exine Formation 1; TDR, Tapetum Degeneration Retardation; TIP2, TDR Interacting Protein 2; WDA1, Wax-deficient Anther 1.
The abnormal differentiation of tapetal cells in the OsCER1A plants implied that some genes critical for tapetum degeneration could be affected. We analyzed two basic helix-loop-helix (bHLH) transcription factor genes, TIP2 and TDR (Zhang et al., 2008; Yi et al., 2012, 2016; Fu et al., 2014). TIP2 functions in tapetum differentiation during early anther development, and TIP2 transcript levels significantly decreased from stage 6 to stage 10 in the OsCER1A plants (Figure 9F). TDR functions downstream of TIP2, and unexpectedly, TDR transcript levels significantly increased from stages 7 to 8 (Figure 9G). These results suggest that OsCER1 is involved in the regulation of tapetum development probably not via the TIP2-TDR pathway.
Because plastids development in the OsCER1A plants was blocked, we analyzed two plastidial genes, DPW and OsNEF1. DPW is a plastid-localized fatty acid reductase that is involved in primary fatty alcohol synthesis and pollen wall formation (Shi et al., 2011). The expression of DPW started to decrease at stage 9 in the OsCER1A plants, suggesting that DPW is affected by the downregulation of OsCER1 (Figure 9H). OsNEF1 shares high identity (up to 60%) with Arabidopsis NEF1, which encodes a regulator of primexine formation that is localized in plastids (Ariizumi et al., 2004; Shi et al., 2011). The expression level of OsNEF1 significantly decreased at stages 6, 7, and 8 (Figure 9I), indicating that the downregulation of OsCER1 also results in a decrease in OsNEF1 expression.
Discussion
OsCER1 Is Associated With VLC Alkanes Biosynthesis
Cuticular wax coats the surface of anthers, and the disruption of anther wax synthesis is frequently accompanied by abnormal pollen development (Jung et al., 2006; Jessen et al., 2011; Leide et al., 2011; Smirnova et al., 2013; Haslam et al., 2015). VLC alkanes and other VLCFA derivatives are produced via the alkane-formation pathway (Greer et al., 2007; Bernard et al., 2012; Wang et al., 2015b). There was strong evidence for the involvement of Arabidopsis CER1 in VLC alkanes biosynthesis (Aarts et al., 1995; Bourdenx et al., 2011). Arabidopsis CER1 contains three His-rich motifs (HX3H, HX2HH, and HX2HH, in which X stands for any amino acid) that are critical for its function in alkane synthesis (Aarts et al., 1995; Shanklin and Cahoon, 1998; Bourdenx et al., 2011). CsCER1, a CER1 homolog in cucumber sharing 61% identity with CER1, also contains three His-rich motifs (Wang et al., 2015b). Overexpression of CsCER1 resulted in significant changes in VLC alkanes production, indicating that CsCER1 also plays a role in VLC alkanes biosynthesis (Wang et al., 2015b). However, genes involved in VLC alkane forming have not been reported in rice, and the VLC alkanes biosynthesis pathway has not been elucidated. The present study determined that full-length OsCER1 shares 57% sequence identity with CER1; OsCER1 also contains the three conserved His-rich motifs. Furthermore, the proportion of C25 and C27 alkanes decreased in the OsCER1A plants. Accordingly, the concentration of C27 alkanes increased in the OsCER1 overexpression plants. These results suggest that the synthesis of some species of VLC alkanes decreased when OsCER1 was knocked down, and increased when OsCER1 was overexpressed. Based on these findings, we suggest that OsCER1 is involved in VLC alkanes biosynthesis and may be mainly involved in C27 alkanes in rice.
OsCER1 Is Involved in a Distinctive Wax Synthesis Pathway
In the present study OsCER1 deficiency was also associated with an increase in the concentration of total waxes, alkenes, and primary alcohols, and with a decrease in the concentration of alkanes. Mutants of genes homologous to OsCER1 in rice, including WSL2, OsGL1-2, OsGL1-3, WDA1, and OsGL1-6, exhibited decreased total waxes, alkenes, primary alcohols, and alkanes (Jung et al., 2006; Qin et al., 2011; Mao et al., 2012; Zhou et al., 2013, 2015). Among these genes, WDA1 is sharing the highest identity (to 63%) with OsCER1. WDA1 is involved in wax synthesis and pollen development in rice, and loss-of-function mutant wda1 exhibits significant reduction in the contents of all wax components (fatty acids, alcohols, alkanes, and alkene) (Jung et al., 2006). However, in our OsCER1A plants, only the contents of C25 and C27 alkane significantly decreased, whereas contents of C31 and C33 alkanes markedly increased. There is a possibility that the antisense construct intended for non-specifically targets OsCER1 and WDA1. If this were the case, then the decrease in VLC alkane content in OsCER1A plants could be an indirect effect of double knockdown of OsCER1 and WDA1. To exclude such a possibility, qRT-PCR was performed using the spikelets with anthers from stages 6 to 10 of the WT and OsCER1A plants. A significant increase in the expression of WDA1 was observed at stage 7, whereas no distinct changes were observed at other anther development stages (Figure 9A). These findings suggest that OsCER1 may participate in a different wax synthesis pathway other than WDA1 and other reported homologies in rice.
OsCER1 Is Required for Plastid Development in Anthers
Plastids play crucial roles for successful anther development in angiosperms with different morphologies at different developmental stages. In rice tapetum plastids begin as proplastids and differentiate into elaioplasts, which are involved in the biosynthesis of tapetal lipids; in turn, these contribute to lipidic pollen wall formation (Lévesque-Lemay et al., 2016). Amyloplasts are terminally differentiated plastids that serve as sites for starch synthesis and storage in mature pollen grains (Matsushima et al., 2014). However, the mechanisms of plastid development during anther development are not well understood. In the OsCER1A plants, we observed abnormal plastids differentiation and alterations in the structure of the plastid in tapetal cells. These results reveal that decreased OsCER1 could affect plastid development. Furthermore, a significant decrease in the number of amyloplasts was observed in mature pollen grains of the OsCER1A plants. A recent study showed that VLCFAs play an essential role in plastid division, and reduced levels of VLCFAs lead to a decrease in the number of plastids (Nobusawa and Umeda, 2012). Wax analysis showed that OsCER1 knockdown plants disrupt the homeostasis of VLCFAs and their derivatives, which in turn could cause alterations in plastid division and development, thereby affecting the quantity of amyloplasts in mature pollens, which in turn influences pollen viability.
Plastid Functions in Anther Development
De novo FA synthesis occurs in plastids, and lipid metabolism also affects plastid development. Furthermore, plastid-related lipid metabolism in the tapetum can influence anther development through aliphatic compound synthesis and secretion. In Arabidopsis, fatty acid export 1 (FAX1) has been localized to the inner envelope of chloroplasts and is involved in the transport of plastid fatty acids; in addition, fax1 mutants produced shorter siliques containing almost no seeds (Li et al., 2015). In Brassica napus, Tic40 is a membrane-anchored co-chaperone homolog localized in the inner membrane of chloroplast that is mainly expressed in tapetum cells and microsporocytes. The tic40 mutant exhibits delayed tapetum PCD and abnormal tapetal cell expansion and division (Dun et al., 2011). Plastid genes could also affect anther development through sugar signaling, such as glyceraldehyde-3-phosphate dehydrogenase (GAPCp) was localized in plastid, and gapcp1gapcp2 double mutant displayed the sterility in Arabidopsis (Muñoz-Bertomeu et al., 2010). However, the role of plastids in anther development remains elusive.
Tapetum PCD is a precise, genetically controlled cellular process that is regulated by several transcription factors. For instance, the altered expression of some bHLH transcription factors strongly influences tapetum PCD in rice; these bHLH transcription factors include Undeveloped Tapetum 1 (UDT1/bHLH164), TDR1, TIP2, and Eternal Tapetum 1 (EAT1/DTD1/bHLH141) (Li et al., 2006; Ji et al., 2013; Niu et al., 2013; Fu et al., 2014; Ko et al., 2014). In rice, several other genes independent of the ROS and bHLH pathways also regulate tapetum PCD, including Persistent Tapetal Cell 1 (PTC1), Apoptosis inhibitor 5 (API5), and Microspore and Tapetum Regulator1 (MTR1) (Li H. et al., 2011, Li X. et al., 2011; Tan et al., 2012; Cao et al., 2015). In addition to the PCD-related genes, various other genes are involved in tapetum development and degeneration in rice such as Histone Monoubiquitination (OsHUB1/OsHUB2), Defective Tapetum and Meiocytes 1 (DTM1), and Anther Development F-box (ADF) (Aarts et al., 1995; Jung et al., 2005; Yi et al., 2012; Li et al., 2015; Cao et al., 2015). These genes form an extensive network that controls tapetum differentiation and PCD. The present studies had shown a delayed tapetum degeneration with a downregulation of TIP2 but an upregulation of TDR in the OsCER1A plants. However, our results showed the opposite alterations in the primary alcohols and alkenes in OsCER1A plants compared to tdr mutants (Li et al., 2006). These findings suggest that delayed tapetum degeneration in OsCER1A plants may not via the TIP2-TDR pathway. However, whether plastids are involved in the delayed tapetum degeneration in OsCER1A plants and the potential roles of plastids in the regulation of tapetum PCD remain elusive.
In the present study, the reduced fertility OsCER1A plants exhibited abnormal plastid development, and delayed tapetum PCD during anther development. Alterations in the expression levels of genes involved in lipid metabolism, and plastidial genes were observed in the OsCER1A plants. This suggests that OsCER1, a protein involved in VLC alkanes biosynthesis, possibly generates some lipidic signals that impact genes involved in plastids development and tapetum PCD then affects anther development in rice.
Author Contributions
CZ and HZ designed the project and wrote the manuscript. EN, LZ, JL, DJ, ZW, SZ, HQ, YZ, CW, SX, and ZL performed the experiments and analyzed the data.
Funding
This work was supported by grants from the National Basic Research Program of China (2016YFD0100405, 2013CB126905, and 2013CBA01401), Genetically Modified Breeding Major Projects (2016ZX08001-004-001-003), the National Natural Science Foundation of China (31371225 and 31571255), and the Scientific Research Project of Guangzhou City (201504010004).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01217/full#supplementary-material
Footnotes
- ^http://www.ebi.ac.uk/Tools/msa/clustalo/
- ^http://www.megasoftware.net/index.html
- ^http://www.genome.jp/tools/clustalw/
References
Aarts, M. G., Keijzer, C. J., Stiekema, W. J., and Pereira, A. (1995). Molecular characterization of the CER1 gene of Arabidopsis involved in epicuticular wax biosynthesis and pollen fertility. Plant Cell 7, 2115–2127. doi: 10.1105/tpc.7.12.2115
Ariizumi, T., Hatakeyama, K., Hinata, K., Inatsugi, R., Nishida, I., Sato, S., et al. (2004). Disruption of the novel plant protein NEF1 affects lipid accumulation in the plastids of the tapetum and exine formation of pollen resulting in male sterility in Arabidopsis thaliana. Plant J. 39, 170–181. doi: 10.1111/j.1365-313X.2004.02118.x
Bach, L., Michaelson, L. V., Haslam, R., Bellec, Y., Gissot, L., Marion, J., et al. (2008). The very-long-chain hydroxy fatty acyl-CoA dehydratase PASTICCINO2 is essential and limiting for plant development. Proc. Natl. Acad. Sci. U.S.A. 105, 14727–14731. doi: 10.1073/pnas.0805089105
Beaudoin, F., Wu, X., Li, F., Haslam, R. P., Markham, J. E., Zheng, H., et al. (2009). Functional characterization of the Arabidopsis β-ketoacyl-coenzyme a reductase candidates of the fatty acid elongase. Plant Physiol. 150, 1174–1191. doi: 10.1104/pp.109.137497
Bernard, A., Domergue, F., Pascal, S., Jetter, R., Renne, C., Faure, J. D., et al. (2012). Reconstitution of plant alkane biosynthesis in yeast demonstrates that Arabidopsis ECERIFERUM1 and ECERIFERUM3 are core components of a very-long-chain alkane synthesis complex. Plant Cell 24, 3106–3118. doi: 10.1105/tpc.112.099796
Blackmore, S., Wortley, A. H., Skvarla, J. J., and Rowley, J. R. (2007). Pollen wall development in flowering plants. New Phytol. 174, 483–498. doi: 10.1111/j.1469-8137.2007.02060.x
Bourdenx, B., Bernard, A., Domergue, F., Pascal, S., Léger, A., Roby, D., et al. (2011). Overexpression of Arabidopsis ECERIFERUM1 promotes wax very-long-chain alkane biosynthesis and influences plant response to biotic and abiotic stresses. Plant Physiol. 156, 1532–2548. doi: 10.1104/pp.111.172320
Buschhaus, C., and Jetter, R. (2011). Composition differences between epicuticular and intracuticular wax substructures: how do plants seal their epidermal surfaces? J. Exp. Bot. 62, 841–853. doi: 10.1093/jxb/erq366
Cao, H., Li, X., Wang, Z., Ding, M., Sun, Y., Dong, F., et al. (2015). Histone H2B Monoubiquitination Mediated by HISTONE MONOUBIQUITINATION1 and HISTONE MONOUBIQUITINATION2 is involved in anther development by regulating tapetum degradation-related genes in rice. Plant Physiol. 168, 1389–1405. doi: 10.1104/pp.114.256578
Clément, C., and Pacini, E. (2001). Anther plastids in angiosperms. Bot. Rev. 67, 54–73. doi: 10.1007/BF02857849
Dun, X., Zhou, Z., Xia, S., Wen, J., Yi, B., Shen, J., et al. (2011). BnaC.Tic40 a plastid inner membrane translocon originating from Brassica oleracea is essential for tapetal function and microspore development in Brassica napus. Plant J. 68, 532–545. doi: 10.1111/j.1365-313X.2011.04708.x
Efremova, N., Schreiber, L., Bar, S., Heidmann, I., Huijser, P., Wellesen, K., et al. (2004). Functional conservation and maintenance of expression pattern of FIDDLEHEAD-like genes in Arabidopsis and Antirrhinum. Plant Mol. Biol. 56, 821–837. doi: 10.1007/s11103-004-5576-y
Fu, Z., Yu, J., Cheng, X., Zong, X., Xu, J., Chen, M., et al. (2014). The rice basic helix-loop-helix transcription factor TDR INTERACTING PROTEIN2 is a central switch in early anther development. Plant Cell 26, 1512–1524. doi: 10.1105/tpc.114.123745
Goldberger, J. J., Smith, J. R., Kim, Y. H., Damle, R. S., and Kadish, A. H. (1995). Effect of increased drive-train stimulus intensity on dispersion of ventricular refractoriness. Circulation 92, 875–880. doi: 10.1161/01.CIR.92.4.875
Greer, S., Wen, M., Bird, D., Wu, X., Samuels, L., Kunst, L., et al. (2007). The cytochrome P450 enzyme CYP96A15 is the midchain alkane hydroxylase responsible for formation of secondary alcohols and ketones in stem cuticular wax of Arabidopsis. Plant Physiol. 145, 653–667. doi: 10.1104/pp.107.107300
Haslam, T. M., Haslam, R., Thoraval, D., Pascal, S., Delude, C., Domergue, F., et al. (2015). ECERIFERUM2-LIKE proteins have unique biochemical and physiological functions in very-long-chain fatty acid elongation. Plant Physiol. 167, 682–692. doi: 10.1104/pp.114.253195
Haslam, T. M., and Kunst, L. (2013). Extending the story of very-long-chain fatty acid elongation. Plant Sci. 210, 93–107. doi: 10.1016/j.plantsci.2013.05.008
Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282. doi: 10.1046/j.1365-313X
Hooker, T. S., Millar, A. A., and Kunst, L. (2002). Significance of the expression of the CER6 condensing enzyme for cuticular wax production in Arabidopsis. Plant Physiol. 129, 1568–1580. doi: 10.1104/pp.003707
Islam, M. A., Du, H., Ning, J., Ye, H., and Xiong, L. (2009). Characterization of glossy1-homologous genes in rice involved in leaf wax accumulation and drought resistance. Plant Mol. Biol. 70, 443–456. doi: 10.1007/s11103-009-9483-0
Jenks, M. A., Tuttle, H. A., Eigenbrode, S. D., and Feldmann, K. A. (1995). Leaf epicuticular waxes of the eceriferum mutants in Arabidopsis. Plant Physiol. 108, 369–377. doi: 10.1104/pp.108.1.369
Jessen, D., Olbrich, A., Knufer, J., Krüger, A., Hoppert, M., Polle, A., et al. (2011). Combined activity of LACS1 and LACS4 is required for proper pollen coat formation in Arabidopsis. Plant J. 68, 715–726. doi: 10.1111/j.1365-313X
Ji, C., Li, H., Chen, L., Xie, M., Wang, F., Chen, Y., et al. (2013). A novel rice bHLH transcription factor, DTD, acts coordinately with TDR in controlling tapetum function and pollen development. Mol. Plant 6, 1715–1718. doi: 10.1093/mp/sst046
Joubes, J., Raffaele, S., Bourdenx, B., Garcia, C., Laroche-Traineau, J., Moreau, P., et al. (2008). The VLCFA elongase gene family in Arabidopsis thaliana: phylogenetic analysis 3D modelling and expression profiling. Plant Mol. Biol. 67, 547–566. doi: 10.1007/s11103-008-9339-z
Jung, K. H., Han, M. J., Lee, D. Y., Lee, Y. S., Schreiber, L., Franke, R., et al. (2006). Wax-deficient anther1 is involved in cuticle and wax production in rice anther walls and is required for pollen development. Plant Cell 18, 3015–3032. doi: 10.1105/tpc.106.042044
Jung, K. H., Han, M. J., Lee, Y. S., Kim, Y. W., Hwang, I., Kim, M. J., et al. (2005). Rice undeveloped tapetum1 is a major regulator of early tapetum development. Plant Cell 17, 2705–2722. doi: 10.1105/tpc.105.034090
Ko, S. S., Li, M. J., Sun-Ben, Ku, M., Ho, Y. C., Lin, Y. J., et al. (2014). The bHLH142 transcription factor coordinates with TDR1 to modulate the expression of EAT1 and regulate pollen development in rice. Plant Cell 26, 2486–2504. doi: 10.1105/tpc.114.126292
Kosma, D. K., Bourdenx, B., Bernard, A., Parsons, E. P., Lü, S., Joubès, J., et al. (2009). The impact of water deficiency on leaf cuticle lipids of Arabidopsis. Plant Physiol. 151, 1918–1929. doi: 10.1104/pp.109.141911a
Kouchi, H., and Hata, S. (1993). Isolation and characterization of novel nodulin cDNAs representing genes expressed at early stages of soybean nodule development. Mol. Gen. Genet. 238, 106–119. doi: 10.1007/BF00279537
Kunst, L., and Samuels, A. L. (2003). Biosynthesis and secretion of plant cuticular wax. Prog. Lipid Res. 42, 1–80.
Kunst, L., and Samuels, L. (2009). Plant cuticles shine: advances in wax biosynthesis and export. Curr. Opin. Plant Biol. 12, 721–727. doi: 10.1016/j.pbi.2009.09.009
Lai, C., Kunst, L., and Jetter, R. (2007). Composition of alkyl esters in the cuticular wax on inflorescence stems of Arabidopsis thaliana cer mutants. Plant J. 50, 189–196. doi: 10.1111/j.1365-313X
Leide, J., Hildebrandt, U., Vogg, G., and Riederer, M. (2011). The positional sterile (ps) mutation affects cuticular transpiration and wax biosynthesis of tomato fruits. J. Plant Physiol. 168, 871–877. doi: 10.1016/j.jplph.2010.11.014
Lévesque-Lemay, M., Chabot, D., Hubbard, K., Chan, J. K., Miller, S., and Robert, L. S. (2016). Tapetal oleosins play an essential role in tapetosome formation and protein relocation to the pollen coat. New Phytol. 209, 691–704. doi: 10.1111/nph.13611
Li, H., Pinot, F., Sauveplane, V., Werck-Reichhart, D., Diehl, P., Schreiber, L., et al. (2010). Cytochrome P450 family member CYP704B2 catalyzes the ω-hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. Plant Cell 22, 173–190. doi: 10.1105/tpc.109.070326
Li, H., Yuan, Z., Vizcay-Barrena, G., Yang, C., Liang, W., Zong, J., et al. (2011). PERSISTENT TAPETAL CELL1 ecodes a PHD-Finger protein that is required for tapetal cell death and pollen development in rice. Plant Physiol. 156, 615–630. doi: 10.1104/pp.111.175760
Li, J., Jiang, D., Zhou, H., Li, F., Yang, J., Hong, L., et al. (2011). Expression of RNA-interference/antisense transgenes by the cognate promoters of target genes is a better gene-silencing strategy to study gene functions in rice. PLoS One 6:e17444. doi: 10.1371/journal.pone.0017444
Li, X., Gao, X., Wei, Y., Deng, L., Ouyang, Y., Chen, G., et al. (2011). Rice APOPTOSIS INHIBITOR5 coupled with two DEAD-Box adenosine 5′-triphosphate-dependent RNA helicases regulates tapetum degeneration. Plant Cell 23, 1416–1434. doi: 10.1105/tpc.110.082636
Li, H., and Zhang, D. (2010). Biosynthesis of anther cuticle and pollen exine in rice. Plant Signal. Behav. 5, 1121–1123. doi: 10.4161/psb.5.9.12562
Li, N., Gügel, I. L., Giavalisco, P., Zeisler, V., Schreiber, L., Soll, J., et al. (2015). FAX1, a novel membrane protein mediating plastid fatty acid export. PLoS Biol. 13:e1002053. doi: 10.1371/journal.pbio.1002053
Li, N., Zhang, D. S., Liu, H. S., Yin, C. S., Li, X. X., Liang, W. Q., et al. (2006). The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18, 2999–3014. doi: 10.1105/tpc.106.044107
Li-Beisson, Y., Shorrosh, B., Beisson, F., Andersson, M. X., Arondel, V., Bates, P. D., et al. (2013). Acyl-lipid metabolism. Arabidopsis Book. 11:e0161. doi: 10.1199/tab.0161
Mao, B., Cheng, Z., Lei, C., Xu, F., Gao, S., Ren, Y., et al. (2012). Wax crystal-sparse leaf2 a rice homologue of WAX2/GL1 is involved in synthesis of leaf cuticular wax. Planta 235, 39–52. doi: 10.1007/s00425-011-1481-1
Matsushima, R., Maekawa, M., Kusano, M., Kondo, H., Fujita, N., Kawagoe, Y., et al. (2014). amyloplast-localized SUBSTANDARD STARCH GRAIN4 protein influences the size of starch grains in rice endosperm. Plant Physiol. 164, 623–636. doi: 10.1104/pp.113.229591
Muñoz-Bertomeu, J., Cascales-Miñana, B., Irles-Segura, A., Mateu, I., Nunes-Nesi, A., Fernie, A. R., et al. (2010). The plastidial glyceraldehyde-3-phosphate dehydrogenase is critical for viable pollen development in Arabidopsis. Plant Physiol. 152, 1830–1841. doi: 10.1104/pp.109.150458
Nawrath, C. (2006). Unraveling the complex network of cuticular structure and function. Curr. Opin. Plant Biol. 9, 281–287. doi: 10.1016/j.pbi.2006.03.001
Niu, N., Liang, W., Yang, X., Jin, W., Wilson, Z. A., Hu, J., et al. (2013). EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice. Nat. Commun. 4:1445. doi: 10.1038/ncomms2396
Nobusawa, T., and Umeda, M. (2012). Very-long-chain fatty acids have an essential role in plastid division by controlling Z-ring formation in Arabidopsis thaliana. Genes Cells 17, 709–719. doi: 10.1111/j.1365-2443.2012.01619.x
Parish, R. W., and Li, S. F. (2010). Death of a tapetum: a programme of developmental altruism. Plant Sci. 178, 73–89. doi: 10.1016/j.plantsci.2009.11.001
Pollard, M., Beisson, F., Li, Y., and Ohlrogge, J. B. (2008). Building lipid barriers: biosynthesis of cutin and suberin. Trends Plant Sci. 13, 236–246. doi: 10.1016/j.tplants.2008.03.003
Qin, B. X., Tang, D., Huang, J., Li, M., Wu, X. R., Lu, L. L., et al. (2011). Rice OsGL1-1 is involved in leaf cuticular wax and cuticle membrane. Mol. Plant. 4, 985–995.
Qin, P., Tu, B., Wang, Y., Luchang, D., Quilichini, T., Li, T., et al. (2013). ABCG15 encodes an ABC transporter protein and is essential for post-meiotic anther and pollen exine development in rice. Plant Cell Physiol. 54, 138–154. doi: 10.1093/pcp/pcs162
Rashotte, A. M., Jenks, M. A., and Feldmann, K. A. (2001). Cuticular waxes on eceriferum mutants of Arabidopsis thaliana. Phytochemistry 57, 115–123. doi: 10.1016/S0031-9422(00)00513-6
Schulz, P. J., Cross, J. W., and Almeida, E. (1993). Chemical agents that inhibit pollen development: effects of the phenyl-cinnoline carboxylates SC-1058 and SC-1271 on the ultrastructure of developing wheat anthers (Triticum aestivum L. var Yecora rojò). Sex Plant Reprod. 6, 108–212. doi: 10.1007/BF00227655
Scott, R. J., Spielman, M., and Dickinson, H. G. (2004). Stamen structure and function. Plant Cell 16, S46–S60. doi: 10.1105/tpc.017012
Shanklin, J., and Cahoon, E. B. (1998). Desaturation and related modifications of fatty acids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 611–641. doi: 10.1146/annurev.arplant.49.1.611
Shi, J., Tan, H., Yu, X. H., Liu, Y., Liang, W., Ranathunge, K., et al. (2011). Defective pollen wall is required for anther and microspore development in rice and encodes a fatty acyl carrier protein reductase. Plant Cell 23, 2225–2246. doi: 10.1105/tpc.111.087528
Smirnova, A., Leide, J., and Riederer, M. (2013). Deficiency in a very-long-chain fatty acid β-Ketoacyl-Coenzyme a synthase of tomato impairs microgametogenesis and causes floral organ fusion. Plant Physiol. 161, 196–209. doi: 10.1104/pp.112.206656
Sturaro, M., Hartings, H., Schmelzer, E., Velasco, R., Salamini, F., and Motto, M. (2005). Cloning and characterization of GLOSSY1 a maize gene involved in cuticle membrane and wax production. Plant Physiol. 138, 478–489. doi: 10.1104/pp.104.058164
Tan, H., Liang, W., Hu, J., and Zhang, D. (2012). MTR1 encodes a secretory fasciclin glycoprotein required for male reproductive development in rice. Dev. Cell 22, 1127–1137. doi: 10.1016/j.devcel.2012.04.011
Wang, A., Xia, Q., Xie, W., Datla, R., and Selvaraj, J. (2003). The classical Ubisch bodies carry a sporophytically produced structural protein (RAFTIN) that is essential for pollen development. Proc. Natl. Acad. Sci. U.S.A. 100, 14487–14492. doi: 10.1073/pnas.2231254100
Wang, W., Liu, X., Gai, X., Ren, J., Liu, X., Cai, Y., et al. (2015a). Cucumis sativus L. WAX2 plays a pivotal role in wax biosynthesis influencing pollen fertility and plant biotic and abiotic stress responses. Plant Cell Physiol. 56, 1339–1354. doi: 10.1093/pcp/pcv052
Wang, W., Zhang, Y., Xu, C., Ren, J., Liu, X., Black, K., et al. (2015b). Cucumber ECERIFERUM1 (CsCER1) which influences the cuticle properties and drought tolerance of cucumber plays a key role in VLC alkanes biosynthesis. Plant Mol. Biol. 87, 219–233. doi: 10.1007/s11103-014-0271-0
Wilson, Z. A., and Zhang, D. B. (2009). From Arabidopsis to rice: pathways in pollen development. J. Exp. Bot. 60, 1479–1492. doi: 10.1093/jxb/erp095
Yi, J., Kim, S. R., Lee, D. Y., Moon, S., Lee, Y. S., Jung, K. H., et al. (2012). The rice gene DEFECTIVE TAPETUM AND MEIOCYTES 1 (DTM1) is required for early tapetum development and meiosis. Plant J. 70, 256–270. doi: 10.1111/j.1365-313X.2011.04864.x
Yi, J., Moon, S., Lee, Y. S., Zhu, L., Liang, W., Zhang, D., et al. (2016). Defective tapetum cell death 1 (DTC1) regulates ROS levels by binding to metallothionein during tapetum degeneration. Plant Physiol. 170, 1611–1623. doi: 10.1104/pp.15.01561
Zhang, D., Liang, W., Yin, C., Zong, J., Gu, F., and Zhang, D. (2010). OsC6 encoding a lipid transfer protein is required for postmeiotic anther development in rice. Plant Physiol. 154, 149–162. doi: 10.1104/pp.110.158865
Zhang, D., Luo, X., and Zhu, L. (2011). Cytological analysis and genetic control of rice anther development. J. Genet. Genom. 38, 379–390. doi: 10.1016/j.jgg.2011.08.001
Zhang, D., and Yang, L. (2014). Specification of tapetum and microsporocyte cells within the anther. Curr. Opin. Plant Biol. 17, 49–55. doi: 10.1016/j.pbi.2013.11.001
Zhang, D.-S., Liang, W.-Q., Yuan, Z., Li, N., Shi, J., Wang, J., et al. (2008). Tapetum degeneration retardation is critical for aliphatic metabolism and gene regulation during rice pollen development. Mol. Plant 1, 599–610. doi: 10.1093/mp/ssn028
Zhou, L., Ni, E., Yang, J., Zhou, H., Liang, H., Li, J., et al. (2013). Rice OsGL1-6 is involved in leaf cuticular wax accumulation and drought resistance. PLoS One 8:e65139. doi: 10.1371/journal.pone.0065139
Zhou, X., Li, L., Xiang, J., Gao, G., Xu, F., Liu, A., et al. (2015). OsGL1-3 is involved in cuticular wax biosynthesis and tolerance to water deficit in rice. PLoS One 10:e116676. doi: 10.1371/journal.pone.0116676
Zhu, L., Shi, J., Zhao, G., Zhang, D., and Liang, W. (2013). Post-meiotic deficient anther1 (PDA1) encodes an ABC transporter required for the development of anther cuticle and pollen exine in rice. J. Plant Biol. 56, 59–68. doi: 10.1007/s12374-013-0902-z
Keywords: Oryza sativa L., male sterility, plastid development, tapetum degeneration, anther cuticle wax
Citation: Ni E, Zhou L, Li J, Jiang D, Wang Z, Zheng S, Qi H, Zhou Y, Wang C, Xiao S, Liu Z, Zhou H and Zhuang C (2018) OsCER1 Plays a Pivotal Role in Very-Long-Chain Alkane Biosynthesis and Affects Plastid Development and Programmed Cell Death of Tapetum in Rice (Oryza sativa L.). Front. Plant Sci. 9:1217. doi: 10.3389/fpls.2018.01217
Received: 23 April 2018; Accepted: 30 July 2018;
Published: 06 September 2018.
Edited by:
Zhong-Nan Yang, Shanghai Normal University, ChinaCopyright © 2018 Ni, Zhou, Li, Jiang, Wang, Zheng, Qi, Zhou, Wang, Xiao, Liu, Zhou and Zhuang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hai Zhou, aGFpX3pob3UxMjdAMTI2LmNvbQ== Chuxiong Zhuang, emh1YW5nY3hAc2NhdS5lZHUuY24=
†These authors have contributed equally to this work
 Erdong Ni
Erdong Ni Lingyan Zhou
Lingyan Zhou Jing Li
Jing Li Dagang Jiang
Dagang Jiang Zhonghua Wang
Zhonghua Wang Shaoyan Zheng
Shaoyan Zheng Hua Qi
Hua Qi Ying Zhou
Ying Zhou Cimei Wang
Cimei Wang Shi Xiao
Shi Xiao Zhenlan Liu
Zhenlan Liu Hai Zhou
Hai Zhou Chuxiong Zhuang
Chuxiong Zhuang