- 1Independent Researcher, Hyderabad, India
- 2The UWA Institute of Agriculture, The University of Western Australia, Perth, WA, Australia
- 3Department of Crop Sciences, College of Agricultural and Marine Sciences, Sultan Qaboos University, Al Khoud, Oman
- 4University of Agriculture, Faisalabad, Pakistan
- 5CGIAR Research Program on Climate Change, Agriculture and Food Security (CCAFS), International Livestock Research Institute (ILRI), Nairobi, Kenya
- 6Department of Plant Breeding, Swedish University of Agricultural Sciences, Alnarp, Sweden
Drought and heat in dryland agriculture challenge the enhancement of crop productivity and threaten global food security. This review is centered on harnessing genetic variation through biotechnology-led approaches to select for increased productivity and stress tolerance that will enhance crop adaptation in dryland environments. Peer-reviewed literature, mostly from the last decade and involving experiments with at least two seasons’ data, form the basis of this review. It begins by highlighting the adverse impact of the increasing intensity and duration of drought and heat stress due to global warming on crop productivity and its impact on food and nutritional security in dryland environments. This is followed by (1) an overview of the physiological and molecular basis of plant adaptation to elevated CO2 (eCO2), drought, and heat stress; (2) the critical role of high-throughput phenotyping platforms to study phenomes and genomes to increase breeding efficiency; (3) opportunities to enhance stress tolerance and productivity in food crops (cereals and grain legumes) by deploying biotechnology-led approaches [pyramiding quantitative trait loci (QTL), genomic selection, marker-assisted recurrent selection, epigenetic variation, genome editing, and transgene) and inducing flowering independent of environmental clues to match the length of growing season; (4) opportunities to increase productivity in C3 crops by harnessing novel variations (genes and network) in crops’ (C3, C4) germplasm pools associated with increased photosynthesis; and (5) the adoption, impact, risk assessment, and enabling policy environments to scale up the adoption of seed-technology to enhance food and nutritional security. This synthesis of technological innovations and insights in seed-based technology offers crop genetic enhancers further opportunities to increase crop productivity in dryland environments.
Introduction
Global warming overall is predicted to have negative effects on agricultural output and productivity. The current world population of 7.2 billion people is projected to increase to between 9.6 and 12.3 billion by 2100, mostly occurring in Africa and South Asia (Gerland et al., 2014). Current yield growth trends are insufficient to match the growing demand for food (Ray et al., 2013). Hence, meeting the dietary and nutritional requirements of an increasing population remains an enormous challenge.
Changes in weather pattern, erratic and uncertain rainfall, and rises in temperature have led to more frequent and longer dry spells and heat waves in many places. In addition to impacting crop growth and yields, such changes may affect crop performance by increasing infestations of insects and other pests, disease epidemics, and weed prevalence, and decreasing pollinating insects (Myers et al., 2017). Declining food quality and increased risk of food and feed contaminated with mycotoxin-producing fungi have the potential to adversely impact human and animal health (Dwivedi et al., 2013).
Agriculture accounts for 70% of freshwater use by humans, which is expected to increase by 17% by 2025 (Molden, 2007). Underground aquifers are rapidly being depleted due to excessive water use (Morison et al., 2008). The acreage of dryland agriculture will expand due to global warming (Challinor et al., 2014). Thus, drought and heat stress present critical challenges for enhancing crop productivity to ensure global food security (Lesk et al., 2016).
There are more than 160 million hectares of dryland cereal and legume crops worldwide. The regions where these crops are grown are prone to drought and heat stress, have limiting soil constraints, contain half of the global population, and account for 60% of the planet’s poor and malnourished people (Hyman et al., 2016). A recent study involving six major crop commodity groups in 131 countries reported losses from 1961–2014 of 1.6% in cereal production, 0.5% in oil crops, 0.6% in pulses, 0.2% in fruits, and 0.09% in vegetables due to drought and heat disasters. Estimated global production losses were US$ 237 billion (using 2004–2006 as a baseline), with the United States, the former Soviet Union, India, China, and Australia being the most adversely affected nations (Mehrabi and Ramankutty, 2017).
Three crop groups (cereals, legumes, and roots/tubers) are the major source of human food worldwide (1271.4 million tons), with root/tuber crops contributing to 65% (825 million t), cereals 26% (332 million t), and legumes 9% (114 million t) (http://www.fao.org/faostat/en/#data/QC, accessed on June 12, 2018). The major cereal crops are narley (Hordeum vulgare L.), maize (Zea mays L.), pearl millet (Pennisetum glaucum (L.) R.Br), rice (Oryza sativa L.), sorghum (Sorghum bicolor (L.) Moench) and wheat (Triticum aestivum L.). The major legume crops are chickpea (Cicer arietinum L.), common bean (Phaseolus vulgaris L.), cowpea (Vigna unguiculata (L.) Walp.), faba bean (Vicia faba L.), groundnut (Arachis hypogaea L.), lentil (Lens culinaris Medikus), peas (Pisum sativum L.), pigeonpea (Cajanus cajan (L.) Millsp.), and soybean (Glycine max (L.) Merr.). Groundnut and soybean are also the main sources of edible oil. Both cereals and legumes are covered in this review because they not only represent worldwide the most important crops, but both are also very similar in their responses to drought and heat stress (Nuñez Barrios et al., 2005; Prasad et al., 2008; Lopes et al., 2011; Daryanto et al., 2017).
In comparison to recent reviews with focus on understanding physiological and genetic basis of drought and heat stress tolerance (Kaushal et al., 2016; Dwivedi et al., 2017; Sita et al., 2017), assessing impact of drought stress on productivity of food crops at global level (Daryanto et al., 2017) and on germplasm mining using SNPs-based arrays and GWAS to identify SNPs/candidate genes associated with plant phenology, architecture, productivity, and stress tolerance (Dwivedi et al., 2017), this review has the unique focus on the use of genetic and genomic resources to enhancing stress [drought, heat, elevated CO2 (eCO2)] tolerance and productivity using biotech-led approaches in addition to the uptake and adoption of seed-based technology to enhancing food and nutritional security in dryland environments.
Synthesis
Physiological Basis of Adaptation to Stress Tolerance
Drought and Heat
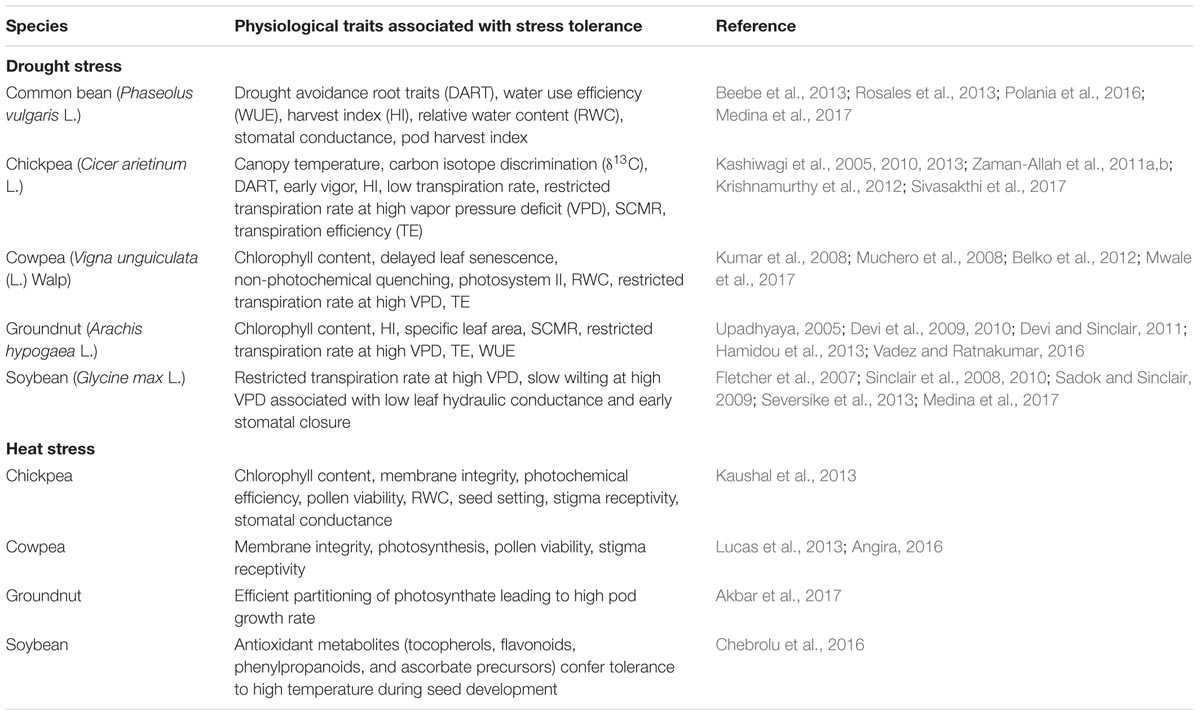
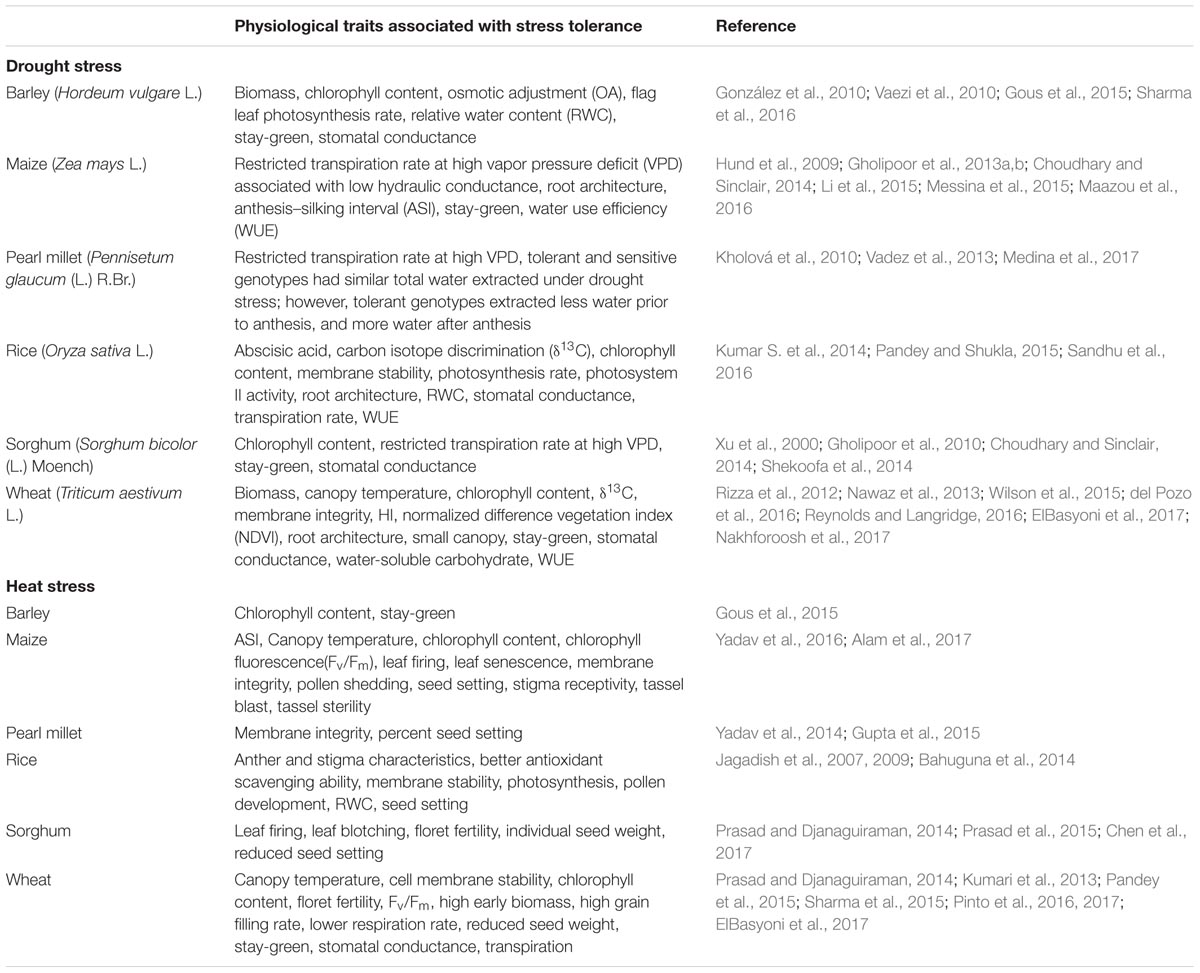
Natural variation in response to drought or heat stress among and within crop species has been reported elsewhere (Dwivedi et al., 2010, 2013; Kaushal et al., 2016; Sita et al., 2017). The major physiological traits associated with drought or heat stress tolerance, both in cereals and legumes, include abscisic acid, canopy temperature, chlorophyll content, chlorophyll fluorescence (Fv/Fm), early vigor, harvest index (HI), membrane integrity, normalized difference vegetation index (NDVI), osmotic adjustment, photochemical efficiency, relative water content, restricted transpiration at high vapor pressure deficit (RT-HVPD) controlled by variation in hydraulic conductance and stomatal conductance, root architecture, stomatal conductance, stay-green (Stg), total transpiration (Tr), transpiration efficiency (TE), and water use efficiency (WUE; Tables 1, 2). Differences in the root profile, Stg, Tr, TE, HI, WUE, and RT-HVPD, among others, were associated with enhanced adaptation to drought and heat stress. Furthermore, the surrogate traits such as carbon isotope discrimination (δ13C), specific leaf area, and SPAD chlorophyll meter reading (SCMR) that determine WUE have been deployed in many breeding programs to enhance drought tolerance in both cereals and legumes (Rebetzke et al., 2002; Condon et al., 2004; Tausz-Posch et al., 2012; Cossani and Reynolds, 2015; Reynolds and Langridge, 2016).
Drought and heat stress often occur together. Is tolerance to both genetically distinct from tolerance to either? Prasad et al. (2011) reported that heat reduced photosynthesis more than drought in wheat, while the lowest leaf photosynthesis observed was due to combined heat and drought stress. Each stress had similar effects on spikelet fertility, grain number, and yield (between 48 and 56%), while their combined effect was higher than their additive effects for chlorophyll content, grain number, and HI (Prasad et al., 2011). In maize, tolerance to combined drought and heat stress is due to distinct genes (Cairns et al., 2013). In chickpea, drought and heat stress individually damaged membranes and decreased cellular oxidizing ability, stomatal conductance, PSII function, and leaf chlorophyll content, with greater damage noted with the combined stress (Awasthi et al., 2014). Furthermore, a variable response to Rubisco (ribulose-1,5-biphosphate/oxygenase) activity was noted, which increased with heat, declined with drought, and declined significantly with both. Sucrose and starch concentrations declined significantly with combined stress. Drought stress had a greater effect on grain yield than heat stress, and the accessions showed partial cross-tolerance (Awasthi et al., 2014). In lentil, heat stress reduced seed yield more than drought stress, while the combined stress severely reduced seed size and seed weight, largely due to a reduction in sucrose and starch-synthesizing enzymes. The combined stress, however, had less effect on drought- and heat-tolerant lines, probably due to a partial cross-tolerance to the two stresses (Sehgal et al., 2017). Thus, the interactions between drought and heat stress should be considered when addressing these stresses in breeding programs.
Elevated CO2 (eCO2)
Atmospheric carbon dioxide (CO2) may increase from the current level (∼400 μmol mol-1) to 600 μmol mol-1 by 2050 (Ciais et al., 2013). eCO2 is associated with increased global warming (de Conto et al., 2012). Natural variation in grain yield in response to eCO2 has been noted in trials with cereals and grain legumes in controlled greenhouses or free-air CO2 enrichment (FACE) systems (Table 3), indicating the feasibility of exploiting genetic variation to develop cultivars responsive to eCO2. Reduced subsets (Frankel, 1984; Upadhyaya and Ortiz, 2001), representing the diversity of the entire collection of a given species in a genebank, are available for most cereal and grain legume crops (Dwivedi et al., 2006), and are excellent resources for identifying CO2-responsive germplasm.
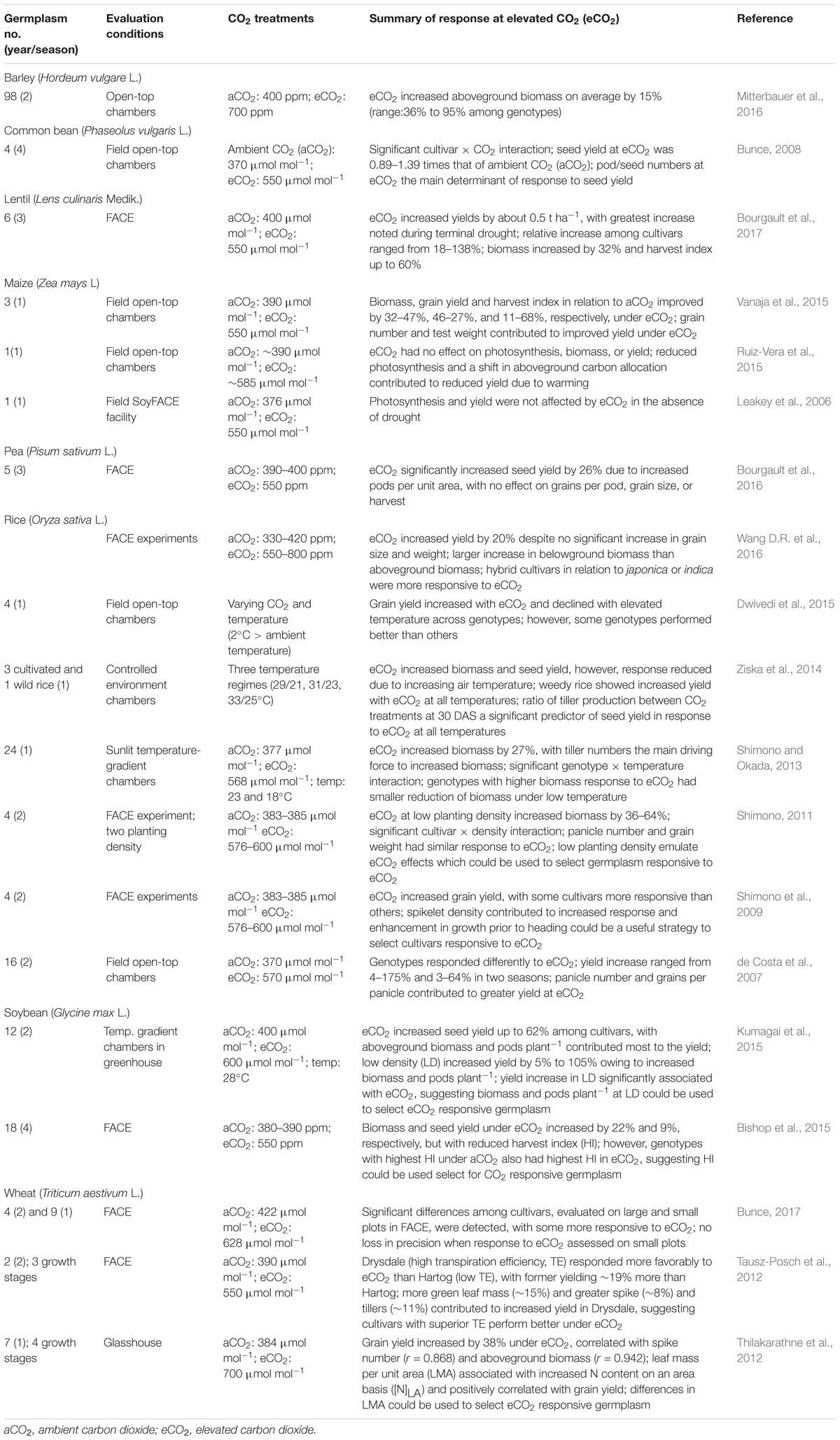
TABLE 3. Cultivars response to elevated carbon dioxide (eCO2) relative to ambient carbon dioxide (aCO2) in barley, common bean, lentil, maize, pea, rice, soybean, and wheat from 2007–2017.
Establishing and operating CO2-fumigation facilities are costly. An alternative method is needed where large numbers of germplasm can be accommodated to assess genotype responses under eCO2. In multi-year and multi-location trials involving 127 diverse rice accessions, two planting densities (normal and wider spacing), Shimono et al. (2014) identified two japonica types most responsive to wider spacing as candidates to support the pre-screening CO2-responsiveness in a FACE system and temperature-gradient chambers under eCO2. The results from FACE confirmed that responsiveness to wider spacing density, as measured by changes in panicles plant-1, is positively associated with responsiveness to eCO2 across both temperate and tropical surroundings, suggesting that wider spacing density in rice would be a useful prescreen for testing diverse germplasm at low cost for responsiveness to eCO2 (Shimono et al., 2014). Finlay–Wilkinson’s regression coefficient (RC) approach has also been suggested as a preliminary evaluation to select accessions responsive to eCO2. In rice and soybean RCs and the responsiveness of yield to eCO2 were positively associated (Kumagai et al., 2016), suggesting that RCs may be initially used to discard accessions not responsive to eCO2. The reason for these relationships may be due to the high CO2-responsive accessions exhibiting high plasticity in resource-rich environments (Shimono, 2011; Shimono et al., 2014; Kumagai et al., 2015). Additional research is required to assess the validity of these methods in diverse crops.
Biomass, HI, heads per unit area, spikelet density, grains per panicle, and grain weight under eCO2 environments contributed to increased yield in cereals (Shimono et al., 2009; Shimono, 2011; Tausz-Posch et al., 2012; Thilakarathne et al., 2012; Shimono and Okada, 2013; Bunce, 2017), while increased biomass, HI, and pods and grains per plant enhanced yields in legumes (Bunce, 2008; Bishop et al., 2015; Kumagai et al., 2015; Bourgault et al., 2016, 2017). A detailed meta-analysis involving 79 species and eight reproductive traits showed that CO2 enrichment (500–800 μl l-1) across all species produced a greater proportion of flowers (19%), fruits (18%), and grains (16%), increased individual grain weight (4%) and test weight (25%), and lowered grain N (–14%). Crops and wild relatives showed a similar response for biomass; however, crops under eCO2 had higher HI and yielded more fruits (28% vs. 4%) and grains (21% vs. 4%) than the wild relatives. Thus, crops were more responsive to eCO2 than wild relatives (Jablonski et al., 2002).
Elevated CO2 is associated with increased atmospheric temperature. Both eCO2 and heat stress strongly affect crop production. There is a need to understand the genotypic response to such an interaction to identify genotypes responsive to eCO2 and heat stress. Rice and wheat grown under two levels of CO2 (ambient and enriched up to 500 μml mol-1) and temperature (ambient and increased 1.5–2.0°C) in FACE systems increased grain yield, while an increase in temperature reduced yield (Cai et al., 2016). These authors noted a differential response in the treatment combining eCO2 and temperature: greater reductions (17–35%) in wheat than rice (10–12%), and the number of filled grains per unit area accounted for most of the effect of eCO2 and temperature. Similar antagonistic effects of eCO2 and high temperature were noted among diverse rice accessions (Wang D.R. et al., 2016). Rising CO2 may compensate for losses due to drought in some situations. However, a recent study – conducted over eight years under varying precipitation and with year-to-year variation in weather at a unique open-air field facility – revealed that stimulation of soybean yield by eCO2 diminished to zero as drought intensified and eCO2 interacted with drought to modify stomatal function and canopy energy balance (Gray et al., 2016). eCO2 did not stimulate photosynthesis, biomass or grain yield, whereas the treatment combining eCO2 and high temperature reduced grain yield in maize (Ruiz-Vera et al., 2015). The above examples show the negative impact of rising CO2 on crop productivity due to changes in the intensity of drought and heat stress. Careful selection of crop cultivars responsive to eCO2 under drought and heat stress is needed, therefore, to minimize the negative impacts from interacting climatic factors in key crop production areas.
Molecular Basis of Adaptation to Stress Tolerance
Drought
Legumes
A “quantitative trait loci (QTL) hotspot” region bearing 12 QTL for drought tolerance explained up to 58% of the variation in chickpea (Varshney et al., 2013). Further research involving this region unlocked two subregions enriched with 23 genes, of which four were functionally validated for drought tolerance in chickpea (Kale et al., 2015). Eighteen single nucleotide polymorphisms (SNPs) from five genes (ERECTA, ASR, DREB, CAP2, and AMDH) significantly contributed to enhanced drought and heat tolerance in chickpea (Thudi et al., 2014b). Roorkiwal et al. (2014) generated 1.3 Mbp of sequence data on 300 chickpea reference set accessions (Upadhyaya et al., 2008) using 10 genes known to confer drought adaptation, and noted 79 SNPs and 41 indels in nine genes, with the maximum number of SNPs in ASR gene, grouped into two distinct haplotypes.
In common bean, 19 WRKY genes, 11 downregulated and eight upregulated, responded to drought stress (Wu et al., 2017) and eight significant marker-trait associations under drought stress were noted on chromosome 9 and 11 (Hoyos-Villegas et al., 2017).
The stay-green QTL Dro-1, Dro-3, and Dro-7 were noted in recombinant inbred lines and diverse germplasm, with a map resolution equal to or below ≤3.2 cM, suggesting valuable targets for marker-assisted genetic enhancement in cowpea (Muchero et al., 2013). Furthermore, the co-location of a stay-green trait with biomass and grain yield QTL (Muchero et al., 2009) opens up the opportunity to select for stay-green at the seedling stage as a rapid screening tool for selection of terminal drought tolerance in cowpea.
In pigeonpea (C. cajan (L.) Millsp.), U-Box protein-coding genes, uspA domain genes, CHX gene, and an uncharacterized protein gene (C. cajan08737) contribute to drought adaptation (Sinha et al., 2016).
Anderson et al. (2016) reported SNPs-based genetic variation associated with high temperature, low precipitation, or differing soil pH among wild soybeans. Accessions possessing related adaptation alleles could be deployed for soybean breeding. Environmental association analyses involving wild barley accessions unraveled QTL on chromosome 2H and 5H, which were related to temperature and precipitation, respectively (Fang et al., 2014). Multiple SNPs associated with drought and heat stress assessed by variations in total chlorophyll content, photochemical reflectance index, and δ13C among germplasm were found in soybean (Dwivedi et al., 2017). The NAC gene family enhances stress tolerance in plants. Eight of the 28 GmNAC genes are reportedly upregulated in drought-tolerant soybeans, which could be used in breeding to enhance abiotic stress adaptation (Hussain et al., 2017). GmWRKY27 interacts with GmMYB174 to enhance adaptation to drought stress in soybean (Wang F. et al., 2015). Wild soybean accessions contributed SNPs related to variability in reduced precipitation, heat stress, and soil pH (Anderson et al., 2016).
Cereals
In barley, 17 constitutively expressed (exclusively in tolerant accessions) and 18 drought-responsive (differential expression under drought among tolerant and intolerant accessions) genes have been reported (Guo et al., 2009). This suggests that both types contribute adaptation to drought stress in barley. A gene expression study involving barley accessions with varying levels of drought response unraveled 34 genes at the reproductive stage that were exclusively expressed in drought-tolerant accessions (Guo et al., 2009). P5CS1 is the key gene involved in the biosynthesis of proline and is significantly induced under water-deficit conditions. Xia et al. (2017) detected 41 polymorphisms (16 SNPs and 25 indels) at the HvP5CS1 locus among barley accessions, with 13 distinct haplotypes; of these, five polymorphisms in HvP5CS1 were significantly (P < 0.001) associated with drought tolerance. Wild barley provided new genes and differentially expressed alleles for adaptation to drought. These genes appear to be more conserved than non-associated genes, while those tolerance genes evolved more rapidly than other drought-associated genes (Hübner et al., 2015).
Anthesis–silking interval (ASI), ears per plant, plant height, and stay-green traits were correlated with drought tolerance in maize (Messmer et al., 2009, 2011). Chromosome 3 bears a QTL hotspot region (∼8 Mb) harboring QTL for traits related to drought tolerance (Almeida et al., 2014). Many drought-tolerant candidate genes (271) and non-synonymous protein-coding SNPs (nsSNPs) (524), with most nsSNPs in bin 1.07 region harboring known drought-tolerant QTL, are reportedly involved in physiological and metabolic pathways in response to water deficit in maize (Xu et al., 2014). Li et al. (2016) detected numerous SNPs, located in 354 candidate genes, associated with drought-related traits. More recently, 77 SNPs associated with ten drought-responsive transcription factors involved in stomatal closure, root development, hormonal signaling, and photosynthesis were found in maize (Shikha et al., 2017). In a multi-environment study, Millet et al. (2016) noted a pattern of QTL effects expressed as functions of environmental variables, of which eight were associated with tolerance to heat and 12 with drought in maize.
A SNP related to a putative acetyl CoA carboxylase gene and an indel associated with a putative chlorophyll a/b binding protein gene, near a major drought-tolerant QTL, are linked with stay-green, grain yield, and HI in pearl millet under drought (Dwivedi et al., 2017).
In rice, several large-effect QTL have been reported in multiple genetic backgrounds and production environments (Kumar A. et al., 2014). In a study involving N22 (drought tolerant) and IR64 (drought intolerant) rice cultivars, Shankar et al. (2016) detected 801 transcripts that were exclusively expressed in N22 under stress conditions, with a larger number encoding NAC and DBP transcription factors. Transcripts encoding for thioredoxin and involved in phenylpropanoid metabolism were upregulated in N22. This suggests that cultivar-specific stress-responsive transcripts may serve as a useful resource to explore novel candidate genes for abiotic stress adaptation in rice. Plant phenotypic plasticity refers to producing a distinct phenotype in response to changing environments (Nicotra et al., 2010). Seventy-six SNPs and 233 priori candidate genes associated with root plasticity traits regulate root growth and development, which upon validation can be deployed in breeding to enhance rice adaptation to drought (Kadam et al., 2017). Mining allelic variability in the OsDREB1F gene in wild rice (Oryza nivara S. D. Sharma and Shastry) unlocked three SNPs conferring drought tolerance, which may be deployed to enhance drought stress adaptation in rice (Singh et al., 2015). Using image-based traits (i-traits) – non-destructive phenotyping as a measure of response to drought – Guo et al. (2018) noted high heritability and large variation and discovered previously unknown genes associated with drought resistance. For example, OsPP15 is associated with a hub of i-traits, whose role was further confirmed by genetic transformation.
The reduction in canopy size associated with Stg QTL in sorghum reduces water demand prior to anthesis, thereby saving more water for plants during post-anthesis. Over expression of P5CS2, mapped within the Stg1 QTL (Subudhi et al., 2000), in the stay-green line relative to the senescent line was correlated with higher proline (Johnson et al., 2015). Polymorphisms at cis-elements in the putative promoter region of P5CS2 probably caused a difference in the expression of this gene, which may facilitate sorghum improvement by marker-assisted selection (MAS). Fracasso et al. (2017) assessed the gene expression dynamics of five drought-related genes in sorghum and found increased expression levels of all genes under drought in tolerant compared with sensitive genotypes, and identified three candidate genes (SbCA, SbERECTA, and SbDHN) that may be deployed to assess drought adaptation in sorghum. Five to nine SNPs near the 14 candidate genes conferred heat stress tolerance in sorghum (Chen et al., 2017).
Variation in water-soluble carbohydrates in stems confers stress tolerance in wheat (Blum, 1998). Polymorphisms of trait-associated SNPs unraveled eight candidate genes grouped into two distinct classes – defense response proteins and proteins triggered by environmental stress – which provide opportunities for selecting for stress tolerance in wheat (Dong et al., 2016). Meta-analysis unraveled 43 co-localized QTL for both drought and heat stress, and 20 drought and two heat stress QTL (Acuña-Galindo et al., 2015). Furthermore, integration of 137 SNP markers for drought- and heat-responsive candidate genes identified 50 candidate SNPs within MQTL confidence intervals, including genes involved in sugar metabolism, reactive oxygen species scavenging, and abscisic-acid-induced stomatal closure (Acuña-Galindo et al., 2015). More recently, three major QTL for drought-responsive traits, one each for days to anthesis (QDa.ccsu-5A.1), plant height (QHt.ccsu-5A), and 100-grain weight (QTgw.ccsu-7A), were reported under rainfed conditions among double haploid lines in wheat (Gahlaut et al., 2017). The maximum quantum efficiency of photosystem II, measured as Fv/Fm, is the most widely used parameter for the rapid non-destructive measurement of stress response in plants. Major QTL on chromosome 3B (two) and chromosome 1D (one), explaining 13–35% of the variation for Fv/Fm, involved in heat stress response were noted in wheat (Sharma et al., 2017).
Emmer wheat (Triticum dicoccum Schrank ex Schübl) – the durum wheat (T. durum Desf) ancestor – harbors a rich source of allelic diversity for abiotic stress adaptation (Peng et al., 2012). Near-isogenic lines (NILs) of durum and bread (T. aestivum L.) wheats containing wild emmer QTL alleles enhanced productivity and yield stability across environments, thereby enriching their gene pools with diversity for improving drought adaptation (Merchuk-Ovnat et al., 2016). Wheatgrass (Agropyron elongatum (Host) P. Beauv.) and wild barley (Hordeum spontaneum (K. Koch) Thell.) also provided genes for drought adaptation: KNAT3 and SERK1 in wheat (Placido et al., 2013) and Hsdr4 in barley (H. vulgare L.) (Suprunova et al., 2007).
Heat
Legumes
Tolerance to heat stress is associated with browning (Hbs) of seed coats in cowpea. Five QTL associated with heat stress tolerance accounting for 11–18% phenotypic variation were tagged with 48 SNP in cowpea (Lucas et al., 2013). Hbs-1 contributed 28–73% phenotypic variation associated with heat stress tolerance and segregated with SNPs, 1_0032 and 1_1128 (Pottorff et al., 2014).
Cereals
Genomic regions on chromosome 2H and 7H at elevated temperature and 17 root/shoot trait QTL have been found in barley (Dwivedi et al., 2017).
Six heat stress tolerant and 112 heat-responsive candidate genes were co-located within two QTL hotspot regions (Frey et al., 2015) – reported to contain three heat tolerance and 23 heat-responsive genes – for grain yield under heat stress in maize (Frey et al., 2016). Of interest is the heat tolerance gene GRMZM2G324886, present in both QTL hotspot regions, which encodes a calcicyclin-binding protein involved in calcium signaling in response to external stress (Frey et al., 2016).
Using NILs differing in heat sensitivity and cDNA-AFLP analysis, Liao et al. (2012) reported 54 transcript-derived fragments (TDFs), 45 of which were mapped to rice chromosomes, mostly in heat-tolerant lines. Twenty-eight of these homologous sequences encoding proteins were associated with signal transduction, oxidation, transcriptional regulation, transport, and metabolism; thus, they are good candidates for studying the molecular basis underlying adaptation to heat stress in rice. Five to 14 loci including qHTSF4.1, qSTIPSS9.1, and qSTIY5.1/SSIY5.2, which are major QTL associated with heat tolerance at the reproductive stage, are known in rice (Lafarge et al., 2017; Shanmugavadivel et al., 2017). A novel QTL on chromosome 9 for percent spikelet fertility at the reproductive stage and one known QTL on chromosome 5 for heat tolerance mapped to 331 kbp genomic regions, comprised 65 and 54 genes, respectively (Shanmugavadivel et al., 2017).
Elevated CO2 (eCO2)
The eCO2 in the atmosphere, which enhances CO2 fixation, plant growth, and production (Xu et al., 2013), is the major driving force behind global warming. The global surface temperature may rise between 2.6 and 4.8°C by the end of this century, depending on greenhouse gas emission pathway (IPCC, 2013). Stomata on the surface of plant leaves balance the uptake of CO2 with evaporation. How plants regulate their stomata in response to environmental stimuli may have important implications for future crop production. Stomata responds to eCO2 by partial closing, and large variation in response to eCO2 was noted among crop species [see section “Elevated Carbon Dioxide (eCO2)”]. The decrease in stomatal conductance, the rate at which CO2 enters the leaves, under eCO2 reduces water consumption and increases WUE. Stomatal function is regulated by genetic variation such as stomatal development, density, and trade-off, and molecular mechanisms regulating guard cell movement under eCO2, and its interactive effects with environmental factors such as vapor pressure deficit, temperature, CO2 concentrations, and light either alone or in combination (Xu et al., 2016). The genetic basis for inter- and intraspecific variation in the response of stomata under eCO2 and the mechanisms for sensing and responding to eCO2 are poorly understood in plants. To date, 16 genes (HIC, GTL1, EPF2, STOMAGEN, OST1, CA1, CA4, SCAP1, HT1, APRC2, SLAC1, PATROL1, AtALMTA12/QAC1, QAC1, AtABCB14, and ROP2), two (EP3 and SLAC1) in rice, and one (ROP2) in faba bean, controlling stomatal development and movement response to eCO2 in Arabidopsis, have been discovered (Xu et al., 2016). Johansson et al. (2015) noted two genetic loci and candidate genes involved in the stomatal response to eCO2 in Arabidopsis. Furthermore, a MATE-type transporter associated with eCO2 concentration in the repression of HT1, which negatively regulates CO2-induced stomatal closing, was found in Arabidopsis (Tian et al., 2015). It was also shown that ABA in guard cells or their precursors mediate the CO2-induced stomatal density response in Arabidopsis (Chater et al., 2015).
High-Throughput Phenotyping to Study Phenomes and Genomes and Their Use in Plant Breeding
High-throughput precision phenotyping (HPP) facilitates measurements within a wide spectrum of light reflectance wavelengths that should correlate significantly with a trait of interest to assess phenotypes (White et al., 2012; Yang et al., 2013). HPP and other phenomic methods and tools are becoming available to predict edible yield, nitrogen use efficiency, host plant resistance in early development stages, as well as for large-scale multi-environment trials (Furbank and Tester, 2011; Araus and Cairns, 2014). Phenomic platforms were first validated for their use in biotrons, growth chambers, and screenhouses, or for field testing using cameras attached to tractors and harvesters. Nonetheless, HPP remains limited in large-scale field trials because of the related costs and logistics involved in its implementation. The availability of relatively cheap unmanned aerial vehicles provides an opportunity for developing and adopting HPP to screen many genotypes evaluated in field trials. Adopting new technologies with efficient phenotyping may lead to larger genetic gains and reduce the time needed to breed climate-resilient cultivars. Phenomic tools such as digital imaging allow the screening of physiological traits that are difficult to measure in the field. Likewise, HPP platforms are facilitating functional genetics as noted in rice (Yang et al., 2013). Today selections are made that consider both the genotype and the resulting phenotype, which increases the speed and efficiency of selection.
Sir Ronald A. Fisher, Sewall G. Wright, and J. B. S. “Jack” Haldane defined quantitative genetic models that included various genes with large or small effects as well as non-genetic factors influencing the phenotype of complex traits, using variance components, resemblance between relatives, and the mathematical theory of selection, respectively (Ortiz, 2015). Large datasets of precise phenotypic measurements in early generation testing and large multi-environment trials are becoming available after using HPP methods to measure canopy temperature, or to obtain spectral reflectance indices and associated information, which can be used to predict grain yield and its components, as well as abiotic stress tolerance and host plant resistance to pathogens and pests.
Biometrics explains complex quantitative traits with continuous variation to understand the action of multi-genes and the genotype by environment interaction controlling phenotypic variation measured across environments. Furthermore, data from genome sequencing on genetic resources and breeding lines or populations along with HPP are increasing the knowledge base regarding specific genomic regions, loci, and alleles therein, haplotypes, linkage disequilibrium blocks, and gene networks contributing to specific phenotypes (Xu et al., 2012). This approach has changed plant breeding by enlarging it to a three-dimensional concept that includes genotype, phenotype and environment. Moreover, prediction models using high-density DNA markers offer promising results in terms of prediction accuracy (Abera Desta and Ortiz, 2014). Genomic prediction (GP) based on genotyping with genome-wide SNPs along with pedigree and phenotypic data is indeed powerful for capturing small genetic effects across the genome, thus allowing the prediction of breeding values or the genetic merit of an individual. Breeding values result from the parents’ average component and the Mendelian sampling of the parents’ genes. Genetic gains due to the response to selection depend on the accuracy and time taken to estimate the Mendelian sampling. The factors affecting the accuracy of GP are model performance, sample size and relatedness, DNA marker density, gene effects, trait heritability, and genetic architecture. Bayesian statistics can effectively estimate breeding values and may be coupled with geo-statistics for modeling multi-environment trials.
Biotechnology-Led Methods to Enhance Crop Adaptation to eCO2, Drought, and Heat Stress
Limited progress has been made on enhancing abiotic stress adaptation in crops through crossbreeding largely because of low heritability, genetic (epistasis) and genotype by environment interactions, and multigenic effects (Dwivedi et al., 2010). Figure 1 shows how biotechnology may assist in developing cereal and legume-bred germplasm. Considerable progress has been made in developing genetic resources (i.e., diversity panels to identify new sources of variations and mapping populations to unravel the molecular basis of trait expressions) and genomic resources (i.e., high-density SNPs arrays, genetic maps, and reference genomes to unravel QTL and candidate genes associated with agronomically beneficial traits) for crop breeding. Transgenic research has moved from discovering protocols, promoters (constitutive or tissue-specific), and transgenes to developing appropriate technology to generate stress-tolerant GM crops without yield penalty under optimum agronomic conditions. Both genomic-assisted breeding and transgenic approaches can enhance the adaptation of cereal and legume crops to drought and heat stress, as detailed here.
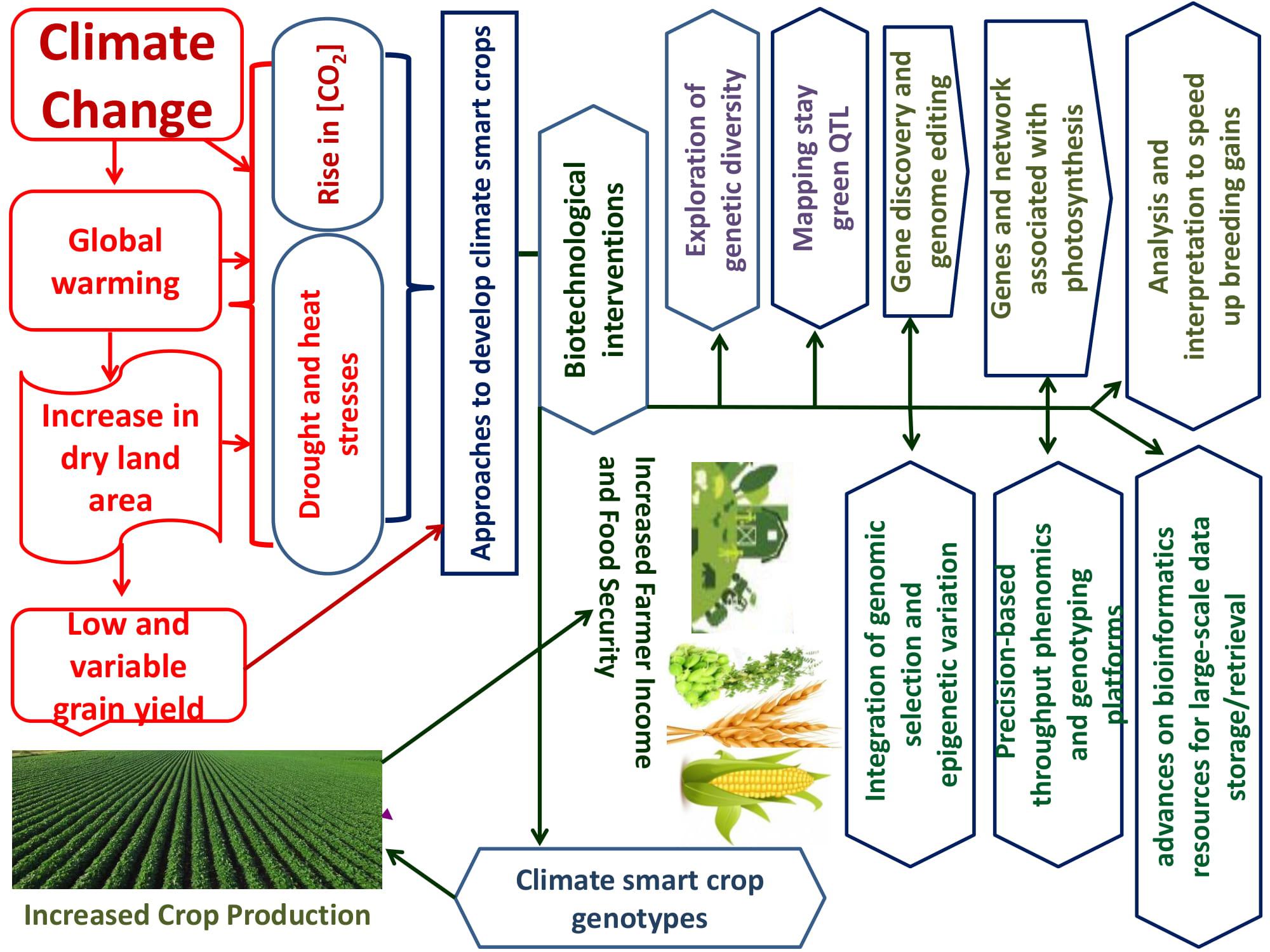
FIGURE 1. Biotechnological innovations to improve cereal and legume production in dryland environment under climate change.
Pyramiding Quantitative Trait Loci
Drought Stress in Grain Legumes
A few chickpea introgression lines (ILs) harboring drought-tolerant QTL for root traits produced deeper roots, better root length density, and higher root dry weight than their recurrent parent J 11 or drought-tolerant donor parent ICC 4958 (Varshney et al., 2014), with some lines in national trials prior to release in India (Thudi et al., 2014a). Three backcross-derived cowpea (V. unguiculata L. Walp.) lines containing drought-tolerant QTL under water-deficit conditions produced on average 48% more seed yield (990–1185 kg ha-1) than the drought-tolerant cultivar Gorom (728 kg ha-1) in Burkina Faso (Batieno et al., 2016).
Drought Stress in Cereals
Six of 14 large-effect QTL associated with drought adaptation in rice were effective across genetic backgrounds and environments (Kumar S. et al., 2014). Pyramiding large-effect QTL enhanced drought adaptation of rice cultivars in Asia (Uga et al., 2013; Shamsudin et al., 2016; Dixit et al., 2017). Lines with two-qDTY combinations were more drought tolerant and productive than lines with three-qDTY combinations, suggesting a differential synergistic relationship among QTL (Shamsudin et al., 2016).
NILs containing Stg QTL in sorghum increased water uptake during grain filling relative to RT × 7000, and produced higher biomass, grain numbers, and grain yield without yield penalty across Australian environments (Borrell et al., 2014). In India, ILs containing Stg QTL in R16 (highly senescent but adapted to the post-rainy season) and ICSV 111 (dual purpose, grain and sweet sorghum types, open-pollinated cultivar) genetic backgrounds improved stover and grain yields as well as stover quality, with no tradeoff (Dryland Cereals and Archival Report, 2012–2013). A large-scale MABC program involving promising ILs and SSRs flanking Stg QTL was initiated to transfer stay-green trait into other sorghum cultivars adapted to the post-rainy season in India. Likewise, 1-2 Stg QTL was successfully transferred via marker-assisted breeding into a farmer-preferred sorghum cultivar E36-1 in Kenya (Ngugi et al., 2013).
The IL S42IL-121 containing major QTL from wild barley (H. vulgare ssp. spontaneum) produced 36% more biomass under drought-stressed conditions than recurrent parent Scarlet (Honsdorf et al., 2014). ILs containing major drought-tolerant QTL in pearl millet (Pennisetum glaucum (L.) R. Br) in the genetic background of a drought-sensitive parent (H 77/833-2) tolerated drought better and significantly out-yielded testcross hybrids made with the original recurrent parent under varying water deficits (Serraj et al., 2005).
Heat Stress in Cereals
Oryza officinalis contributed the major QTL qEMF3 (on chromosome 3) for flower opening time in rice. NILs containing qEMF3 in IR64 genetic background shifted flower opening time 1.5–2.0 h earlier in the tropics, thereby escaping heat stress (≥35°C causes spikelet sterility) at flowering. Thus, qEMF3 has the potential to advance flower opening time in current cultivars to mitigate the effect of heat stress at flowering (Hirabayashi et al., 2015). O. glaberrima contributed a major QTL for heat tolerance Thermotolerance1 (TT1), that protects cells from heat stress. NILs containing TT1 allele had higher thermotolerance than the recurrent parent (WYJ) at flowering and grain filling (Li X. M. et al., 2015). Thus, pyramiding qEMF3 and TT1 is expected to enhance adaptation to heat stress in rice.
Drought Tolerance in Marker-Aided Breeding in Maize
Ziyomo and Bernardo (2013) noted that indirect selection based on secondary traits or grain yield in maize under irrigation was not more efficient than selecting directly for grain yield under drought, while genomic selection (GS) was more efficient, with predicted relative efficiency of 1.24, for grain yield under water deficit. ASI, ear girth, ear length, 100-kernel weight, and grain yield in maize, on the other hand, had the highest prediction accuracies (0.95–0.97) across drought-stressed environments (Shikha et al., 2017). Furthermore, 77 SNPs distributed across the genomes were associated with ten drought-responsive transcription factors related to stomatal closure, root development, hormonal signaling, and photosynthesis (Shikha et al., 2017). A more recent study in maize revealed higher prediction accuracy of GS than phenotypic variance explained by the sum of QTL for individual traits following MAS. This suggests that using QTL-MAS in forward breeding will enrich allelic frequency for a few desired traits with strong additive QTL in early selection cycles while GS-MAS will additionally capture alleles with smaller additive effects (Cerrudo et al., 2018).
Marker-assisted recurrent selection (MARS) has been successfully employed to enhance genetic gains in maize, with greater genetic gains (105 kg ha-1 year-1) in well-watered (WW) than water-stressed (51 kg ha-1 year-1) plants in sub-Saharan Africa (Beyene et al., 2015). Likewise, test crosses derived from the second selection cycle (C2) had higher grain yield under drought stress than those coming from the original population, with a relative genetic gain of 7% per cycle, while test crosses of inbred S1 lines from C2 had an average genetic gain of 1% per cycle under WW and 3% per cycle under rainfed (Bankole et al., 2017). Hence, both GS and MARS were more effective at increasing genetic gain in maize under both irrigated and drought-stressed environments.
Epigenetic Variation and Abiotic Stress Adaptation
Epigenetics is “the study of mitotically and/or meiotically heritable changes in gene function that cannot be explained by changes in DNA sequences” (Russo et al., 1996). Epigenetic changes contribute to gene expression under environmental stress and are regulated by processes such as DNA methylation, histone modification, and RNA interference (RNAi; Meyer, 2015). Increasing evidence shows that changes arising from DNA methylation or histone acetylation are heritable (Zheng et al., 2013; Wang W. et al., 2016) and may be exploited in plant breeding. Cytosine methylation polymorphism, as indicated by hypermethylation or hypomethylation under stress (Gayacharan and Joel, 2013), plays a key role in the expression of genes in plants (Shen et al., 2012). Hypermethylation and hypomethylation of DNA under drought are indicators of stress susceptibility and tolerance, respectively. The most commonly used assays to assess cytosine DNA methylation are methylation-sensitive amplified polymorphism (Xiong et al., 1999), bisulfite sequencing (Li et al., 2010), and methylated DNA immunoprecipitation sequencing (Taiwo et al., 2012).
DNA Methylation
DNA methylation is chemical modification arising from addition of a methyl group to the nitrogenous base in the DNA strand in a sequence-specific manner (Law and Jacobsen, 2010). Cytosine DNA methylation is a major epigenetic modification affecting gene regulation in response to environmental stress. Drought-induced genome-wide epigenetic modification accounted for ∼12% of the methylation differences in the rice genome, which were genotype-, tissue-, and developmental stage-specific (Wang et al., 2011), while Xia et al. (2016) noted greater DNA methylation (52.9–54.3%) under drought stress among diverse rice ecotypes adapted to upland (rainfed) and lowland (irrigated) sites. Moreover, adaptation-specific highly divergent epiloci confer adaptation to drought in both rice ecotypes (Xia et al., 2016). Gayacharan and Joel (2013) noted a positive correlation between spikelet sterility and methylation (%), whereas panicle length and weight, seeds per panicle, 100-seed weight, and grain yield were negatively correlated, suggesting the role of epigenetic regulation in grain-yield-attributing traits in response to drought in rice.
The molecular basis of DNA methylation at the genome level may unravel the regulatory mechanisms associated with abiotic stress adaptation. Wang W. et al. (2016) detected 1190 differentially methylated regions (DMRs) between drought-tolerant (IL_DK151) and intolerant (IR 64) lines under irrigation. The IL_DK151 plants under drought had more stable methylome (only 92 drought-induced DMRs) than IR64 plants (506 DMRs). Epigenetic regulation of drought responses was associated with changes in DNA methylation of genotype-specific genes. A set of 12 and 23 DMR-associated genes were differentially expressed in IL_DK151 and IR 64, respectively, under drought (Wang W. et al., 2016). Previous research found numerous DMRs among rice cultivars with distinct responses to drought and salinity, many of which were associated with the differential expression of genes associated with stress tolerance (Garg et al., 2015). Five of the six drought-stressed DMRs reported in faba bean (V. faba L.) had higher expression in drought tolerant (Bachar) than sensitive (F177) cultivars, suggesting their role in faba bean drought tolerance (Abid et al., 2017). DNA methylation polymorphism can, therefore, be effectively deployed to develop crops with enhanced drought tolerance.
Histone Modification
Histones are proteins associated with DNA in the nucleus that condense it into chromatin. Histone modifications may regulate gene expression in plants under abiotic stress. Post-translational modification of histones includes acetylation, methylation, phosphorylation, and ubiquitination (Chinnusamy and Zhu, 2009). Transcriptionally active chromatin is always associated with histone hyperacetylation, while inactive chromatin region is associated with deacetylated histone (Kim et al., 2008).
A change in the promoter region of cell cycle genes showing specific patterns of histone acetylation increased the total acetylation level under stress in maize roots. Differences in acetylation states (either hyper- or hypo-acetylation) of specific lysine sites on H3 and H4 tails of the promoter regions in cell cycle genes affect their expression under stress. Overexpression of ZmDREB2A enhances tolerance to osmotic stress in Arabidopsis (Qin et al., 2007). Furthermore, Zhao et al. (2014) noted that osmotic stress activates the transcription of the ZmDREB2A gene by increasing the levels of acetylated histones H3K9 and H4K5 associated with the ZmDREB2A promoter region.
Histone acetyltransferases (HATs) and histone deacetylases (HDACs) are involved in histone acetylation homeostasis. Eight HATs, grouped into four families, are known in rice. When investigating the response of four HATs, one from each family in rice, Fang et al. (2014) noted that drought significantly increased the expression of OsHAC703, OsHAG703, OsHAF701, and OsHAM701. They further showed that the acetylation level on certain lysine sites of H3 and H4 increased with OsHATs expression, suggesting that OsHATs is involved in the response of rice to drought. Previous research also identified that the modification in H3K4me3 was positively correlated with a subset of genes showing changes both in modification and expression under drought (Zong et al., 2013).
Programmed cell death (PCD) plays a prominent role in plants during development and in response to stress (Danon et al., 2000; Jones, 2001). Wang P. et al. (2015) showed significantly increased levels of total acetylation of histones H3K9, H4K5, and H3, whereas the di-methylation level of histone H3K4 remained unchanged and H3K9 decreased in maize seedlings under heat stress. Furthermore, maize seedlings treated with “trichostatin A” resulted in histone hyperacetylation caused by increased levels of the superoxide anion (O2-) within the cell, leading to PCD in association with histone modification changes under heat stress in maize leaves.
RNA Interference (RNAi)
MicroRNAs (miRNAs) are single-stranded RNA molecules (20–24 nt) that affect the expression of genes at the post-transcriptional level and are involved in plant development and a wide array of stress responses (Zhang et al., 2006). Many of the drought and heat stress-responsive miRNAs and their target genes associated with plant development, metabolism, and stress tolerance has been found in cereals and grain legumes. Drought-associated miRNAs include 11 in cowpea (Barrera-Figueroa et al., 2011), five in maize (Sheng et al., 2015), 18 each in rice (Barrera-Figueroa et al., 2012) and sorghum (Katiyar et al., 2015), and 71 in durum wheat (Liu et al., 2015). In contrast, the heat stress responsive miRNAs include eight in common bean (Jyothi et al., 2015), 11 and 26 in rice (Liu et al., 2017; Mangrauthia et al., 2017), and 6 and 12 in wheat (Xin et al., 2010; Kumar et al., 2015). Changes in miRNAs under stress correlate well with increased expression of stress-related genes in tolerant genotypes. For example, Liu et al. (2017) reported eight target genes that correspond with 26 miRNAs within the four QTL regions associated with heat stress in heat-tolerant rice. This relationship of miRNAs with their target genes, however, may not be one-to-one (i.e., negative or positive) and may vary between miRNAs of the same gene family or even for the same target miRNAs at various development stages (Lopez-Gomollon et al., 2012; Zhang et al., 2012; Kumar et al., 2015). Identification of differentially abundant miRNAs in stress-tolerant germplasm may thus provide molecular evidence that miRNAs contribute to stress tolerance and so are good candidates for enhancing stress tolerance in crops by transgenic breeding.
Genome Editing to Harness Novel Allelic Variations in Stress Adaptation
Genome editing brings changes in DNA that are smaller than those from genetic engineering (Ainsworth, 2015). Rice and wheat showing host plant resistance to pathogens (Wang et al., 2014; Wang F. et al., 2016) or potato with improved cold storage and processing (Clasen et al., 2016) became available using genome editing.
Targeted modification of specific genes using CRISPR and CRISPR-associated proteinP9 (Cas9) leads to useful genetic variations for improving plants (Feng et al., 2013). Indeed, plant breeding enterprises are beginning to exploit genome editing with CRISP/Cas9 to develop genetically enhanced seed-embedded technology showing new traits. Avoiding off-target mutations during genome editing may be achieved using protocols to eliminate off-target effects and improve the specificity of the CRISPR/Cas-9 system (Pattanayak et al., 2013).
The OPEN STOMATA2 (OST2) gene encodes the major plasma membrane H+-ATPase (AHA1) in Arabidopsis (Palmgren, 2001). Using CRISPR/Cas9 and the tissue-specific promoter EF1 (germline- and meristematic cell-specific), Osakabe et al. (2016) generated a new mutant allele for OST2/AHA1 (ost2_crispr-1) without off-target effects in Arabidopsis. Such plants did not show any significant differences in plant growth compared to wild-type (WT) plants. Furthermore, ost2_crispr-1 plants exhibited higher leaf temperature and lower water loss than WTs, suggesting enhanced stomatal closure upon abiotic stress.
Maize plants overexpressing ARGOS8 produced high grain yields under water deficit (Shi et al., 2015). Endogenous expression of ARGOS8 mRNA in maize is low and spatially non-uniform. Using CRISPR/Cas9 technology and the GOS2 promoter, Shi et al. (2017) obtained heritable novel variants with enhanced expression of ARGOS8 transcripts relative to the native allele in all maize tissues tested. Furthermore, such variants produced higher grain yield than their WT under water deficit at flowering, without any grain yield penalty under irrigation.
Rodríguez-Leal et al. (2017) recently proposed the engineering of beneficial quantitative variation for plant breeding through genome editing of promoters that leads to the generation of diverse cis regulatory alleles. They successfully generated tomato mutants from genome editing with variation in fruit size, inflorescence branching, and plant architecture. Their findings provide the underpinning for dissecting intricate interactions between gene regulatory changes and quantitative phenotypic variation, including adaptation to stress-prone environments.
Inducing Flowering Independent of Environmental Clues to Match Growing Season
West Africa and South Asia are among the regions worst affected due to climate change and variability effects. Farmers in West Africa have experienced a shortening of the rainy season. Climate models have projected the late onset, early cessation of rainfall, and reduction in length of growing season, which will negatively impact agriculture in West Africa (Sarr, 2012) although there is high uncertainty about shifting rainfall patterns and amounts. In South Asia, by the end of the 21st century, models project a delay in the start of rainy season by 5–15 days and an overall weakening of the summer monsoon precipitation (Ashfaq et al., 2009). A reduction in the length of the growing season has direct impacts on the adaptation of current cultivars. There is thus a continued need to breed cultivars adapted to a shortened growing season. An alternative to crossbreeding is to engineer flowering time along with the external application of flower-inducing compounds (Ionescue et al., 2017).
Enhanced expression of the Ghd7 gene under extended photoperiod delays flowering and increases plant height and panicle size in rice (Xue et al., 2008). Such orthologous floral repressor systems also exist in maize (ZmCCT) and sorghum (SbPRR37 and SbGHD7) (Hung et al., 2012; Yang et al., 2014). Okada et al. (2017) produced non-flowering rice after overexpressing the Ghd7 gene that inhibits environmentally induced spontaneous flowering. Thereafter, they isolated a promoter responsive to agrochemical spraying. Next, Hd3a was introduced under the control of an agrochemical-responsive promoter, which was already co-transformed with Ghd7. Such plants reacted to chemical spraying by beginning their floral transition, thus flowering earlier than the non-transgenic control, with no adverse impact on plant growth and development. This result shows that flowering time can be manipulated independently from the environment in which the plant grows, leading to the possibility of engineering crops with suitable growth in different climates (Okada et al., 2017).
Pyramiding QTL has been successful for combining tolerance to drought and heat stress into bred-germplasm for both cereals and legumes. Both GS and MARS, in contrast to QTL pyramiding by MAS, have been more effective at enhancing gains in maize productivity, both in drought-stressed and irrigated environments. The proof of concept also indicated the usefulness of deploying epigenetic variation or genomic editing to create new variation for agronomic traits and drought tolerance in some crops. As noted above, use of agrochemicals to induce flowering independent of environmental signals has also been effective, as evidenced in rice, thereby suggesting the utility of this approach in other crops.
Stress-Tolerant Genetically Modified Crops
Plant adaptation to abiotic stress is achieved by genes associated with stress tolerance. The coordinated involvement of these genes and their interaction with the timing and intensity of the stress make it difficult for plant breeders to develop stress-tolerant crops by crossbreeding. Advances in genomics are allowing researchers to identify tissue-specific expression of genes under abiotic stress. The transfer and expression of such candidate genes into crops through genetic engineering offer considerable promise.
The overexpression of transgenes enhances abiotic stress adaptation, both in model plants and cereals (Dwivedi et al., 2010). Grain yield penalty is often associated with the introduction of transgenes that improve plant responses to abiotic stress adaptation (Campos et al., 2004). Overexpression of HvEPF1 in barley, TPP in maize, OsERF48, OsKAT2, AtGolS2, OsERF71, OsNAC9, and OsNAC10 in rice, AtGoLS2 in soybean, and SeCspA and TaNAC69-1 in wheat enhances drought adaptation and increases grain yield under field drought, without yield penalty under WW conditions (Table 4). Overexpression of OsMYB55 enhances adaptation to drought and heat stress in maize and rice (El-Kereamy et al., 2012; Casaretto et al., 2016). Furthermore, OsMYB55 under individual or combined (drought and heat) stress during or after recovery of stress produced more plant biomass with less leaf damage in maize (Casaretto et al., 2016). It appears that using a tissue-specific promoter (i.e., root-specific or seed-specific), rather than whole body (plant)-specific promoters, containing a transgene had no or relatively fewer adverse effects on plant growth and development, leading to no yield penalty.
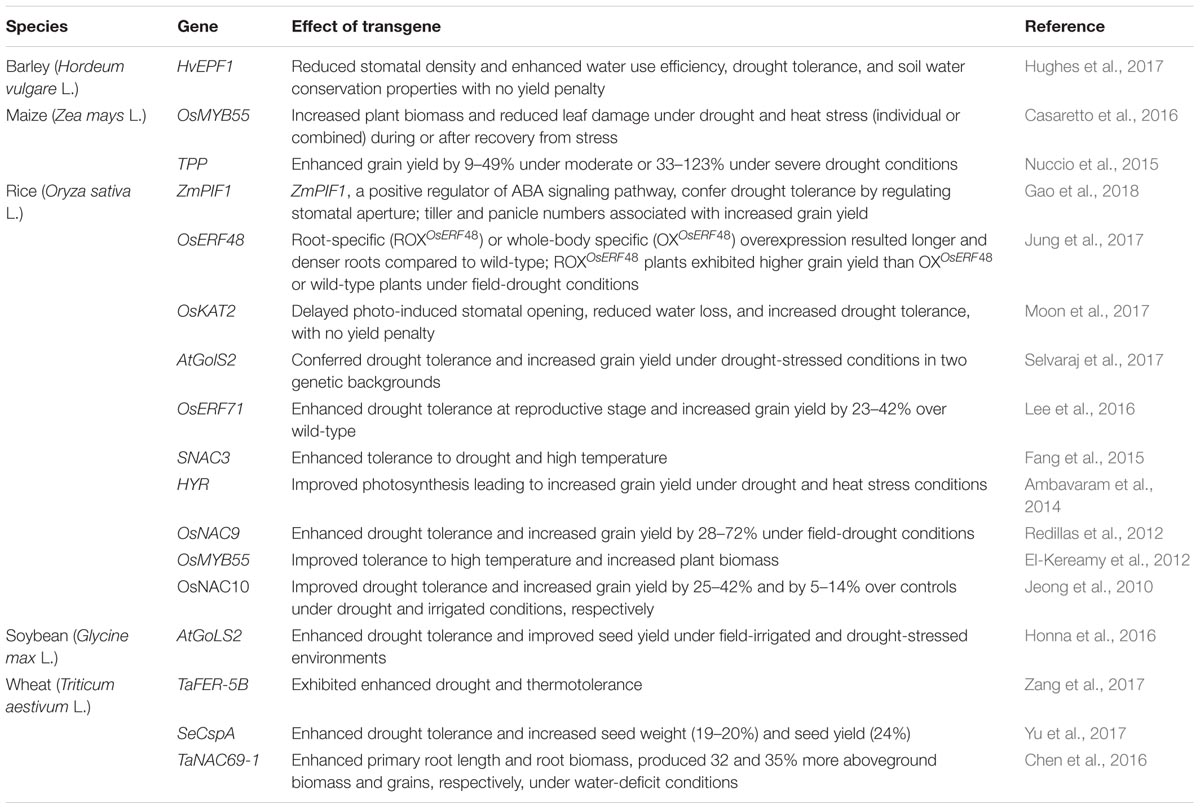
TABLE 4. Overexpression of transgenes enhances drought adaptation and increases grain yields under field-drought environments in barley, maize, rice, soybean, and wheat from 2010–2018.
Selection criteria in breeding programs will benefit from insights on the molecular and physiological basis for adaptation to increased grain yield under stress. For example, HvEPF1 significantly reduced stomatal density but enhanced WUE and soil water conservation properties, thus enhancing drought adaptation and productivity in barley (Hughes et al., 2017). Likewise, improving sucrose supply or altering sucrose metabolism in developing ear spikelets improved kernel set in maize under drought (Zinselmeier et al., 1995; Boyer and Westgate, 2004). Furthermore, EPP increased sucrose content in spikelet and kernel set in maize, leading to increased HI and grain yield under drought, without yield penalty under WW conditions (Nuccio et al., 2015). Similarly, in rice, OKAT2 delayed photo-induced stomatal opening and enhanced drought adaptation without grain yield penalty (Moon et al., 2017), while tolerance to heat stress seems to be related to enhanced amino acid metabolism by OsMYB55 (El-Kereamy et al., 2012).
Environmental stress reduces photosynthetic carbon metabolism (PCM) and limits grain yield in cereals. For example, increased expression of HYR, which is associated with PCM, enhanced photosynthesis, leading to high grain yield in rice under stress (Ambavaram et al., 2014). Overexpression of OsERF48, OsERF71, OsNAC9, and OsNAC10 enhanced grain yield under water scarcity by altering root architecture (Jeong et al., 2010; Redillas et al., 2012; Lee et al., 2016; Jung et al., 2017). In contrast, increased grain yield in transgenic rice overexpressing AtGolS2 under drought was related to higher panicle numbers, grain fertility, and biomass (Selvaraj et al., 2017). Enhanced thermotolerance due to overexpression of TaFER-5B in wheat was due to ROS scavenging (Zang et al., 2017). Overexpression of TaNAC69-1 under drought promoted root elongation in drying soils, producing more aboveground biomass and grain in wheat (Chen et al., 2016), while overexpression of SeCspA under drought increased test weight and grain yield (Yu et al., 2017).
Monsanto’s “DroughtGuard” maize (MON87460) expressing Bacillus subtilis cold shock protein B (cspB) (Castiglioni et al., 2008) which produced 6% higher grain yield across years than the control (Monsanto hybrid NH6212) under water-limited conditions (Nemali et al., 2015) was cultivated on ≥50,000 ha in the United States during the first year after release (Marshall, 2014). New genes conferring drought and heat stress adaptation are being discovered and the experimental area sown to drought or heat stress tolerant crops containing transgene(s) is expected to grow as more countries become involved in the development and field testing of such crops. However, the major concern with biotech crops is food safety for humans and animals. A rigorous assessment based on robust, science-based criteria should be in place to assess biosafety issues associated with the use of such crops.
Unlike in the past when the induction of transgenes was often associated with adverse impacts on plant growth and productivity, the proof of concept has been demonstrated for the development of stress tolerant genetically modified crops without yield penalty under agronomically optimal environments, as evidenced both in cereals and legumes.
Genetic Variation and Candidate Genes Associated With Photosynthetic Traits
Photosynthesis is a complex process whereby green plants use sunlight to synthesize nutrients (carbohydrates) from carbon dioxide (CO2) and water, in the process, generating oxygen as a by-product. The process of converting light energy into chemical energy to produce biomass is highly efficient. Over the years, there has been increased understanding of the processes that limit photosynthetic efficiency (PE), paving the way to manipulate light energy into carbon gain in plants. Future increases in biomass production may, therefore, depend largely on increases in PE, with a focus on harnessing cross species (C3 vs. C4 crops) as well within-species differences, as detailed below.
Harnessing Novel Variation for Photosynthesis Traits
Plant genetic resources are a valuable foundation of natural genetic variation for photosynthetic traits. Rubisco is a target for improving photosynthesis. Prins et al. (2016) measured Rubisco carboxylation velocity (Vc), Michaelis–Menten constants for CO2 (Kc) and O2 (Ko), and specificity factor (Sc/o) for Rubisco diversity in vitro in 25 accessions of the Triticeae tribe at 25 and 35°C and noted a positive association between Vc and Kc and negative association between Vc and Sc/o. Rubisco from barley increased photosynthesis at 25 and 35°C while Rubisco from jointed goatgrass (Aegilops cylindrica Host) increased photosynthesis at 25°C. Thus, naturally occurring Rubisco with superior properties among the Triticeae tribe may be exploited to develop wheats with enhanced photosynthesis and productivity. The CO2 fixation rate (Kccat) for Rubisco from the C4 grasses (Paniceae tribe) with NAD, NADP-ME, and PCK photosynthetic pathways was twofold greater than the kccat of Rubisco from NAD-ME species across all temperatures, while the declining response of CO2/O2 specificity with increasing temperature was less pronounced for PCK and NADP-ME Rubisco, which would be advantageous in warm climates relative to the NAD-ME grasses (Sharwood et al., 2016). Rubisco from certain C4 species could thus enhance carbon gain and resource use efficiency in C3 crops.
Driever et al. (2014) reported significant variation in photosynthetic capacity, biomass, and grain yield among 64 wheat accessions, although they noted an inconsistent correlation between the flag leaf photosynthetic capacity and grain yield across accessions, thus suggesting scope for further improvement in photosynthetic capacity. Moreover, Carmo-Silva et al. (2017) compared flag leaf photosynthetic traits, crop development, and grain yield in the same cultivars used by Driever et al. (2014) and noted that pre-anthesis and post-anthesis photosynthetic traits in the field were positively associated with grain yield and HI. Moderate to high heritability for photosynthetic traits suggests that phenotypic variation can be deployed to enhance photosynthesis in wheat.
Gu et al. (2012a) noted significant genetic variation for stomatal conductance (Gs), mesophyll conductance (Gm), electron transport capacity (Jmax), and Rubisco carboxylation capacity (Vcmax) in rice, although drought and leaf age contributed for a greater proportion of phenotypic variation. The differences in light-saturated photosynthesis and TE were mainly associated with variation in Gs and Gm. Further research revealed that a known photosynthesis-QTL is associated with differences in Gs, Gm, Jmax, and Vcmax at flowering. One to three QTL associated with eight photosynthetic traits contributed 7–30% of the phenotypic variation, with some QTL near the SSR marker RM410 consistent across development stages and moisture regimes (Gu et al., 2012b). Gu et al. (2014) noted a 25% increase in genetic variation in leaf photosynthetic rate (Pr) corresponding to a 22–29% increase in crop biomass across locations and years. More recently, Qu et al. (2017) detected large variation in photosynthetic parameters among diverse rice germplasm across environments, and noted that Pr under low light intensity is correlated with biomass accumulation, and the narrow-sense heritability is high, suggesting its potential in breeding for improving biomass and yield potential in rice.
Natural variation in the CO2 assimilation rate is another promising strategy to identify genes contributing to higher photosynthesis. Adachi et al. (2017) fine mapped a QTL Carbon Assimilation Rate8 (CAR8), identical to rice flowering QTL DTH/Ghd8/LHD1, that controls flag leaf nitrogen content, Gs and Pr in rice. QTL GPS (GREEN FOR PHOTOSYNTHESIS), identical to NAL1 (a gene that controls lateral leaf growth in rice), controls photosynthesis rate by regulating carboxylation efficiency. The high-photosynthesis allele of GPS is a partial loss-of-function allele of NAL1, which increases mesophyll cells between vascular bundles, leading to thickened leaves, and pleiotropically enhances Pr with no adverse impact on plant productivity (Takai et al., 2013). Further research on molecular functions of GPS and CAR8 may contribute to enhanced photosynthesis in rice and possibly other crops.
Understanding the genetic basis of PE may contribute to enhanced seed yield. Basu et al. (2018) reported 16 SNPs linked to major QTLs underlying 16 candidate genes associated with PE and seed yield in chickpea. Furthermore, they delineated superior haplotypes from a chlorophyll A-B binding protein-coding gene, Timing of a CAB Expression1, as the most potential candidates for enhancing PE and seed yield.
Enhancing Gm may increase WUE and photosynthetic water use efficiency (WUEp). Gm refers to the rate of CO2 diffusion from substomatal cavities to the site of carboxylation. To improve WUE, Gm should increase without concomitantly increasing Gs. Across species, a 24-fold variation was noted in Gm (Tomás et al., 2013). Within-species differences in Gm were also noticed, e.g., 60% in rice (Gu et al., 2012a), two to threefold in wheat (Jahan et al., 2014; Barbour et al., 2016), and twofold in soybean (Tomeo and Rosenthal, 2017). A QTL for Gm on chromosome 3A contributes to 9% variation in wheat (Barbour et al., 2016). Tomeo and Rosenthal (2017) found a positive correlation between Gm and Pr in soybean; however, it impedes positive scaling between Gm and WUE. Both Pr and Gm increased with increasing leaf mass, suggesting the potential to increase photosynthesis and Gm by selecting for greater leaf mass in soybean (Tomeo and Rosenthal, 2017).
WUEp refers to carbon fixed in photosynthesis per unit of water transpired (Lawson and Blatt, 2014). ILs with a high density of hairs on leaves containing a hairy leaf gene BLANKET LEAF from O. nivara in IR24 had a warmer leaf surface, and lower net photosynthesis rate (Pn), transpiration rate (Tr), and Gs, but higher WUEp, suggesting that BLANKET LEAF increases WUEp when evaluated under moderate to high light intensities. Hence, it could be deployed to improve WUEP in rice (Hamaoka et al., 2017). ILs containing a genomic region from O. rufigopon in KMR3 showed significant variation in Pn, Tr, TE (Pn/Tr), and carboxylation efficiency (Pn/Ci). Hamaoka et al. (2017) further noted positive correlations involving Pn with Tr, Gs, Pn/Ci, and total canopy dry matter, and several ILs had higher pn. Thus, ILs with greater Pn are potential sources for breeding rice cultivars with enhanced biomass and yield (Haritha et al., 2017).
Transforming C3 Plants With C4 Photosynthesis
C4 plants depending on the mechanisms of decarboxylation of C4 acids in bundle sheath cells are grouped as NADP-malic enzyme (NADP-ME), NAD-malic enzyme (NAD-ME), and PEP carboxykinase (PCK) biochemical types (Hatch, 1987). Genus Flaveria consists of species that perform C3, C4, and C3–C4 (intermediate) photosynthesis. All Flaveria species are closely related and therefore may unravel key mutations in the evolution from C3 to C4 photosynthesis (Monson et al., 1986; Brown et al., 2005). Fixation of CO2 in C3-plants is catalyzed by the enzyme Rubisco. However, Rubisco is prone to energy loss due to photorespiration under high temperatures. In contrast, C4 plants have developed an additional CO2 concentration mechanism to minimize this energy loss, which enables them to adapt to abiotic stresses (Peterhänsel and Maurino, 2011; Sage et al., 2012). The key enzyme in the C4 photosynthesis pathway is phosphoenolpyruvate carboxylase that, during the evolution of C4 photosynthesis, increased its kinetic efficiency and reduced its sensitivity to feedback inhibitors (malate, aspartate). A single point mutation from “arginine” at the inhibitor site (884) to “glycine” in the active site (774) reduces inhibitor affinity and enables phosphoenolpyruvate carboxylases to participate in the C4 photosynthesis pathway (Paulus et al., 2013).
Amaranth (Amaranthus palmeri S.Wats.) is the first dicotyledonous NAD-ME-type C4 species possessing “Kranz anatomy” (i.e., tightly arranged, thick-walled, vascular bundle sheath cells, and surrounding mesophyll cells with airspaces) with Pn similar to C4 species. It is highly resource-use efficient and tolerant to drought, heat, and salinity (El-Sharkawy, 2016). Amaranthus species showed large genetic variation in structural, biochemical, and physiological traits associated with photosynthesis and resource use efficiency; specific leaf weights (SLW) ranged from 20–34.2 g m-2, chlorophyll (Chl) 0.28–0.51 g m-2, nitrogen (N) 53.2–114.1 mmol m-2, Pn 19.7–40.5 μmol m-2 s-1, Gs 165.7–245.6 mmol m-2 s-1, photosynthetic nitrogen use efficiency (PNUE) 260–458 μmol m-1 Ns-1, and WUEp 5.6–10.4 mmol mol-1 (El-Sharkawy, 2016). Pn was positively correlated with Gs, N and Chl of leaves, weakly correlated with SLW, and no correlation with leaf structural traits (Tsutsumi et al., 2017). Thus, Amaranthus species have characteristic physiological, anatomical, and biochemical traits, including high Pn efficiency, which are similar to C4 crops. Knowledge gained from Amaranthus may assist crop genetic enhancers to rapidly develop food, feed, and bioenergy crops with improved productivity in drylands.
To date, a set of 128 C4 specific genes, which are expressed in bundle sheath or mesophyll cells, are known (Ding et al., 2015). About 63% of these genes showed differential expression patterns between C4 [maize, sorghum, green foxtail millet (Setaria italica (L.) P. Beauvois)] and C3 (rice) species, while a smaller proportion had either novel (18%) or elevated (20%) expression patterns in C4 plants, which may have been involved in the evolutionary transition from C3 to C4 photosynthesis. Their differential gene expression is a novel genomic resource to establish C4 pathways in C3 crops.
Wheat is a C3 crop. Rangan et al. (2016a,b) discovered a C4 pathway restricted to wheat grain and identified six genes (PPc, aat, mdh, me2, gpt, ppd), specific for C4 photosynthesis and only expressed in the photosynthetic pericarp of the caryopsis of wheat exhibiting dimorphism related to the mesophyll- and bundle sheat cells of the chloroplasts in the leaf of C4 plants. Hence, it is feasible to genetically engineer C4 photosynthesis in wheat due to the available expression of the whole pathway in its grain, though some researchers are skeptical of this approach.
Maize is a classical C4 plant. The foliar leaf blade is a Kranz (C4) type, while the husk leaf sheath is a non-kranz (C3) type (Wang et al., 2013). Sixty-four of the 124 C4 specific genes showed differential expression between the foliar leaf blade and husk leaf sheath, of which 57 were highly expressed in the foliar leaf blade but not the leaf primordia, suggesting their involvement mainly in C4 photosynthesis in leaves (Ding et al., 2015). Further research on gene expression in husk leaf sheath tissues may unlock candidate genes associated with C3-type photosynthesis and the subsequent comparison and cross-talk between candidate genes (C4 and C3 photosynthetic pathways in maize) may unravel a cascade of genes and networks to guide further improvements in photosynthesis and productivity.
The introduction of Krantz anatomy into C3 leaves is a major challenge. The genes that control Krantz anatomy are unknown. Screening large-scale mutagenized populations identified variants with altered leaf anatomical structure, e.g., maize mutants with bundle sheath and mesophyll sheath specific pathways, sorghum mutants with variation in vein spacing, or rice mutants with closure vein spacing (Hall et al., 1998; Covshoff et al., 2008; Kajala et al., 2011; Rizal et al., 2015). More importantly, Scarecrow and Shr1 which regulate structural differentiation of the root endodermis also contribute to the development of Kranz anatomy in maize (Slewinski et al., 2012, 2014). Thus, the discovery of such structural variation in leaf and Scarecrow and Shr1 genes may provide an opportunity to introduce Krantz anatomy into C3 leaves.
Research to transform rice with C4 photosynthesis began in the last decade (Covshoff and Hibberd, 2012; von Caemmerer et al., 2012; Babege and Magule, 2017). While researchers are getting closer to unraveling the complex C4 photosynthesis, the pathways for such transformation will not be easy and may take another decade before this innovation can be used to introduce C4 photosynthesis into C3 crops.
Sourcing diverse germplasm pools has unraveled natural genetic variation for photosynthetic traits, both in cereals and legumes. A few QTLs and candidate genes associated with enhanced PE are known in chickpea, rice, and wheat. Significant knowledge on Krantz anatomy and key genes (and their differential expression between C3 and C4 plants) and pathways involved in the evolutionary transition from C3 to C4 photosynthesis have been unlocked. The discovery of a C4 pathway in wheat grain (or C3 pathway in maize husk) may further unravel a cascade of genes and networks to improve photosynthesis and productivity. The introduction of Krantz anatomy into C3 leaves is a major challenge. However, the discovery of genetic variants with altered leaf anatomical structure or genes (Scarecrow and Shr1) regulating structural differences in the root epidermis may provide opportunities for introducing Krantz anatomy into C3 leaves.
Adoption, Impact, Risk Assessment, and Enabling Policy Environments
Genetically enhanced seed-embedded technology is an important innovation leading to gains in crop productivity. Both crossbreeding and biotechnology have been used to enhance tolerance to drought and heat stress in food crops. Bred-cultivars may substantially reduce projected yield losses under climate change in the tropics and subtropics (Challinor et al., 2014) while improving food self-sufficiency of smallholders and increasing their incomes without the need to cultivate extra land. Changing crop cultivars will continue to be among the first lines of defense for improving productivity and resilience in farming systems (Thornton et al., 2017).
Drought tolerant maize (DTM) cultivars developed by crossbreeding have been released in sub-Saharan Africa, while in South Asia, efforts are underway to scale up cultivation of heat stress tolerant maize hybrids. Likewise, a genetically modified drought-tolerant maize-derived hybrid, DroughtGuard (MON 87460), has been approved for cultivation in the United States (Marshall, 2014). DTM hybrids may bring benefits ranging from US$362 to 590 million for consumers and producers and could reduce poverty by 5% within 13 countries in sub-Saharan Africa (Kostandini et al., 2013). Wossen et al. (2017) showed that the adoption of DTM cultivars in rural Nigeria increased grain yields by 13.3%, and reduced grain yield variance and risk by 53 and 81% among adopters, respectively. Furthermore, productivity gains and risk reduction due to adoption reduced the incidence of poverty by 12.9% and increased the probability of food security among adopters by 83.3%. In rural Zimbabwe, adoption of DTM hybrids significantly enhanced maize productivity, allowing farmers to set aside enough produce for personal household consumption and excess for sale (Makate et al., 2017).
Adoption rates of improved seeds, regardless of their origin, in areas where those seeds are available can be as high as 85% among smallholder farmers, if awareness is high (Kyazze and Kristjanson, 2011). Many situations exist, however, where few farmers have access to improved seeds in the developing world. In sub-Saharan Africa, for example, 68–97% of seed grown by smallholder farmers comes from informal sources or local markets (McGuire and Sperling, 2015). Seed availability thus constitutes a primary barrier to the adoption of bred cultivars (Fisher et al., 2015; Lunduka et al., 2017).
DTM hybrid cultivars with big cob size, good tip cover, resistance to lodging, and good grain quality are the most preferred types in Eastern and Southern Africa (Setimala et al., 2017). Female farmers have much lower adoption of DTM, mainly due to differences in resource access, notably land, agricultural information, and credit. Male farmer age or economic status (poor or rich) has no significant influence on adoption of DTM. Young but poor female household heads are least likely to adopt DTM, while wives strongly influence the adoption of DTM on plots controlled by their husbands (Fisher and Carr, 2015). Are the farmers willing to invest more to purchase DTM seeds? Kassie et al. (2017) noted that farmers in rural Zimbabwe are willing to pay three to seven times higher premiums for drought-tolerant hybrids giving an additional metric ton of yield per acre and possessing bigger cob size, covered cob tip, and larger grains.
The environmental risks associated with the cultivation of MON 87460 are no different from the risks associated with crossbred maize (Sammons et al., 2014). Are GM crops safe for human consumption? No major differences on compositional profile were noted between GM and their counterpart, non-GM crops (Venkatesh et al., 2015; Herman et al., 2017); minor differences, however, typically reflect changes associated with crossbreeding practices (Venkatesh et al., 2015; Herman et al., 2017). Thus, GM crops are as safe and nutritious as currently consumed food crops. Robust and science-based criteria should, however, be in place to assess any biosafety issues associated with the use of such transgenic crops.
Improved understanding of how policy environments can support the widespread adoption of bred cultivars is greatly needed. There is only limited quantitative information available to guide national investment and policy choices (Vermeulen et al., 2016). Relatively small investments in cultivar development and seed multiplication now can save substantial costs later (Cacho et al., 2016), while the cost may be enormous if investment is delayed in maize breeding and seed systems (Challinor et al., 2016). It seems considerable work remains to be done with respect to the kinds of enabling policies and informational environment needed in different contexts, if adoption of bred cultivars by smallholder farmers at the scales required is to occur in the coming decades.
Conclusion and Recommendations
Today’s agriculture is faced with increasing intensity of drought and heat stress worldwide. Greenhouse gases are the major source of global warming. The positive impact of eCO2 on crop productivity may be nullified or even overwhelmed by increased drought and heat stress. Developing crops that are tolerant to stress and responsive to eCO2 is a considerable challenge. While research on drought has long been the focus, genetic and physiological research on plant responses to eCO2 and heat stress has gained momentum. Tolerance to drought and heat stress is under distinct genetic control. Accessions and traits responsive to eCO2 (increased photosynthesis) or stress tolerance have been found in germplasm collections. The knowledge gained about the physiological and molecular bases of stress tolerance has facilitated greater access and intake of stress-tolerant germplasm by cross breeding. Advances in high-throughput genotyping and precision-based phenotyping platforms as well as the parallel development of bioinformatic resources and tools to handle large datasets are enabling crop genetic enhancers to accelerate introgression of candidate genes associated with stress tolerance into improved genetic backgrounds. Many breeding programs are routinely using these resources, which increase the speed and efficiency of selection. A few drought or heat stress-tolerant cultivars of cereals and legumes, developed by crossbreeding, are becoming popular among farmers in stress-prone environments. How are stress-tolerant crops making an impact on food and nutritional security across the globe? DTM cultivars out-yielded commercial controls on farmers’ fields, with significant gains under stress, showing the huge potential for uptake of DTM seed in Africa. However, barriers to adoption exist, such as lack of seed availability, inadequate information, resource crunch, high seed cost, and consumer preference, that must be overcome for greater uptake of seed-embedded technology to enhance food and nutritional security in the developing world.
Today, agricultural production worldwide is affected by climate change. Landraces and crop wild relatives are unique genetic resources to meet current and new challenges for farming in stressful environments. However, plant breeders are reluctant to use these unique genetic resources in breeding largely due to fear of loss of co-adapted gene complexes, linkage drag, and the protracted time needed to develop cultivars. Considerable knowledge gap also exists about crop response to eCO2, for example, the variance in results between controlled environment and FACE experiments, discovering plant traits most responsive to eCO2, or selection criterion to be adapted in breeding for the development of crop cultivars responsive to eCO2. Future research should focus on (i) capturing and harnessing variation in landraces and crop wild relatives, (ii) cost-effective precision-based high-throughput phenotyping platforms amenable to small-scale breeding programs, (iii) use of image-based traits (i-traits) to measure response to stress, (iv) traits and methodology to access response to selection under eCO2 conditions, (v) the molecular basis of plant response to selection under eCO2 conditions, (vi) traits, genetics, and methods to select for enhanced photosynthesis, (vii) genes and networks to unravel crosstalk involving drought × heat, genotype × eCO2, drought × heat × eCO2, and genotype × drought × heat × eCO2, (viii) haplotype-specific associations and protein–protein interactions to delineate natural alleles and superior haplotypes, and (ix) phenotyping and sequencing wild and weedy relative genomes of crop species to identify and relate differences with sequence variation. A focus on these issues can be expected to increase breeding efficiency to develop crops responsive to eCO2, stress tolerance and productivity.
Author Contributions
All co-authors participated in planning, outlining, and writing sections of the manuscript under the lead of SD, while RO took care of the overall editing of various drafts of the manuscript after interacting with all co-authors.
Funding
PT acknowledges funding to CCAFS from CGIAR Fund Donors and through bilateral funding agreements. RO acknowledges funding from the Swedish Research Council (Vetenskapsrådet, VR, Sweden) U-forsk project “Genomisk förutsägelse att leverera värme tolerant vete till Senegal avrinningsområde.”
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer SW and the handling Editor declared their shared affiliation.
Acknowledgments
SD is grateful to Ramesh Kotnana of the Knowledge Sharing and Innovation Program of the International Crops Research Institute for the Semi-Arid Tropics (ICRISAT, Patancheru, India) for arranging reprints on relevant subjects as a valuable resource for shaping this article. We are grateful to the three reviewers for their helpful suggestions on improving the manuscript.
References
Abera Desta, Z., and Ortiz, R. (2014). Genomic selection: genome-wide breeding value prediction in plant improvement. Trends Plant Sci. 19, 592–601. doi: 10.1016/j.tplants.2014.05.006
Abid, G., Mingeot, D., Muhovski, Y., Mergeai, G., Aouida, M., Abdelkarim, S., et al. (2017). Analysis of DNA methylation patterns associated with stress response in faba bean (Vicia faba L.) using methylation-sensitive amplification polymorphism (MSAP). Environ. Exp. Bot. 142, 34–44. doi: 10.1016/j.envexpbot.2017.08.004
Acuña-Galindo, M. A., Mason, R. E., Subramanian, N. K., and Hays, D. B. (2015). Meta-analysis of wheat QTL regions associated with adaptation to drought and heat stress. Crop Sci. 55, 477–492. doi: 10.2135/cropsci2013.11.0793
Adachi, S., Yoshikawa, K., Yamanouchi, U., Tanabata, T., Sun, J., Okkawa, T., et al. (2017). Fine mapping of carbon assimilation rate 8, a quantitative trait locus for flag leaf nitrogen content, stomatal conductance and photosynthesis in rice. Front. Plant Sci. 8:60. doi: 10.3389/fpls.2017.00060
Akbar, A., Manohar, S. S., Variath, M. T., Kurapati, S., and Pasupuleti, J. (2017). Efficient partitioning of assimilates in stress-tolerant groundnut genotypes under high-temperature stress. Agronomy 7:30. doi: 10.3390/agronomy7010030
Alam, M. A., Seetharam, K., Zaidi, P. H., Dinesh, A., Vinayan, M. T., and Nath, U. K. (2017). Dissecting heat stress tolerance in tropical maize (Zea mays L.). Field Crops Res. 204, 110–119. doi: 10.1016/j.fcr.2017.01.006
Almeida, G. D., Nair, S., Borém, A., Cairns, J., Trachsel, S., Ribaut, J.-M., et al. (2014). Molecular mapping across three populations reveals a QTL hotspot region on chromosome 3 for secondary traits associated with drought tolerance in tropical maize. Mol. Breed. 34, 701–715. doi: 10.1007/s11032-014-0068-5
Ambavaram, M. M. R., Basu, S., Krishnan, A., Ramegowda, V., Batlang, U., Rahman, L., et al. (2014). Coordinated regulation of photosynthesis in rice increases yield and tolerance to environmental stress. Nat. Commun. 5:5302. doi: 10.1038/ncomms6302
Anderson, J. E., Kono, T. J. Y., Stupar, R. M., Kantar, M. B., and Morrell, P. L. (2016). Environmental association analyses identify candidates for abiotic stress tolerance in Glycine soja, the wild progenitor of cultivated soybean. G3 6, 835–843. doi: 10.1534/g3.116.026914
Angira, B. (2016). Genetic and Physiological Studies of Heat Tolerance in Cowpea. Ph.D. dissertation, College Station: TX, Texas A & M University.
Araus, J. L., and Cairns, J. E. (2014). Field high-throughput phenotyping: the new plant breeding frontier. Trends Plant Sci. 19, 52–61. doi: 10.1016/j.tplants.2013.09.008
Ashfaq, M., Shi, Y., Tung, W.-W., Trapp, R. J., Gao, X., and Pal, J. S. (2009). Suppression of south Asian summer monsoon precipitation in the 21st century. Geophys. Res. Lett. 36:L01704. doi: 10.1029/2008GL036500
Awasthi, R., Kaushal, N., Vadez, V., Turner, N. C., Berger, J., Siddique, K. H. M., et al. (2014). Individual and combined effects of transient drought and heat stress on carbon assimilation and seed filling in chickpea. Funct. Plant Biol. 41, 1148–1167. doi: 10.1071/FP13340
Babege, T., and Magule, T. (2017). Molecular and genomic tools for improving photosynthetic efficiency of C3 species in the changing climates. J. Plant Biol. Soil Health 4:4.
Bahuguna, R. N., Jha, J., Pal, M., Shah, D., Lawas, L. M. F., Khetarpal, S., et al. (2014). Physiological and biochemical characterization of NERICA-L-44: a novel source of heat tolerance at the vegetative and reproductive stages in rice. Physiol. Plant. 154, 543–559. doi: 10.1111/ppl.12299
Bankole, F., Menkir, A., Olaoye, G., Crossa, J., Hearne, S., Unachukwu, N., et al. (2017). Genetic gains in yield related traits under drought stress and favorable environments in a maize population improved using marker-assisted recurrent selection. Front. Plant Sci. 8:808. doi: 10.3389/fpls.2017.00808
Barbour, M. M., Bachmann, S., Bansal, U., Bariana, H., and Sharp, P. (2016). Genetic control of mesophyll conductance in common wheat. New Phytol. 209, 461–465. doi: 10.1111/nph.13628
Barrera-Figueroa, B. E., Gao, L., Diop, N. N., Wu, Z., Ehlers, J. D., Roberts, P. A., et al. (2011). Identification and comparative analysis of drought-associated microRNAs in two cowpea genotypes. BMC Plant Biol. 11:127. doi: 10.1186/1471-2229-11-127
Barrera-Figueroa, B. E., Gao, L., Wu, Z., Zhou, X., Zhu, J., Jin, H., et al. (2012). High throughput sequencing reveals novel and abiotic stress-regulated microRNAs in the inflorescences of rice. BMC Plant Biol. 12:132. doi: 10.1186/1471-2229-12-132
Basu, U., Bajaj, D., Sharma, A., Malik, N., Daware, A., Narnoliya, L., et al. (2018). Genetic dissection of photosynthetic efficiency traits for enhancing seed yield in chickpea. Plant Cell Environ. [Epub ahead of print]. doi: 10.1111/pce.13319
Batieno, B. J., Danquah, E., Tignrgro, J.-B., Hyunh, B.-L., Drabo, I., Close, T. J., et al. (2016). Application of marker-assisted backcrossing to improve cowpea (Vigna unguiculata L. Walp) for drought tolerance. J. Plant Breed. Crop Sci. 8, 273–286. doi: 10.5897/JPBCS2016.0607
Beebe, S. E., Rao, I. M., Blair, M. W., and Acosta-Gallegos, J. A. (2013). Phenotyping common beans for adaptation to drought. Front. Physiol. 4:35. doi: 10.3389/fphys.2013.00035
Belko, N., Zaman-Allah, M., Cisse, N., Diop, N. D., Zombre, G., Ehler, J. D., et al. (2012). Low soil moisture threshold for transpiration decline under water deficit correlates with lower canopy conductance and higher transpiration efficiency in drought tolerant cowpea. Funct. Plant Biol. 39, 306–322. doi: 10.1071/FP11282
Beyene, Y., Semagn, K., Crossa, J., Mugo, S., Atlin, G. N., Tarekegne, A., et al. (2015). Improving maize grain yield under drought stress and non-stress environments in Sub-Saharan Africa using marker-assisted recurrent selection. Crop Sci. 56, 344–353. doi: 10.2135/cropsci2015.02.0135
Bishop, K. A., Beltzelberger, A. M., Long, S. P., and Ainsworth, E. A. (2015). Is there potential to adapt soybean (Glycine max Merr.) to future [CO2]? An analysis of yield response of 18 genotypes in free-air CO2 enrichment. Plant Cell Environ. 38, 1765–1774. doi: 10.1111/pce.12443
Blum, A. (1998). Improving wheat grain filling under stress by stem reserve mobilization. Euphytica 100, 77–83. doi: 10.1023/A:1018303922482
Borrell, A. K., van Oosterom, E. J., Mullet, J. E., George-Jaeggli, B., Jordan, D. R., Klein, P. E., et al. (2014). Stay-green alleles individually enhance grain yield in sorghum under drought by modifying canopy development and water uptake patterns. New Phytol. 203, 817–830. doi: 10.1111/nph.12869
Bourgault, M., Brand, J., Tausz, M., and Fitzgerald, G. J. (2016). Yield, growth and grain nitrogen response to elevated CO2 of five field pea (Pisum sativum L.) cultivars in a low rainfall environment. Field Crops Res. 196, 1–9. doi: 10.1016/j.fcr.2016.04.011
Bourgault, M., Brand, J., Tausz-Posch, S., Armstrong, R. D., O’Leary, G. L., Fitzgerald, G. J., et al. (2017). Yield, growth and grain nitrogen response to elevated CO2 in six lentil (Lens culinaris) cultivars grown under Free Air CO2 Enrichment (FACE) in a semi-arid environment. Eur. J. Agron. 87, 50–58. doi: 10.1016/j.eja.2017.05.003
Boyer, J. S., and Westgate, M. E. (2004). Grain yields with limited water. J. Exp. Bot. 55, 2385–2394. doi: 10.1093/jxb/erh219
Brown, N. J., Parsley, K., and Hibberd, J. M. (2005). The future of C4 research - maize, Flaveria or Cleome? Trends Plant Sci. 10, 215–221.
Bunce, J. A. (2008). Contrasting responses of seed yield to elevated carbon dioxide under field conditions within Phaseolus vulgaris. Agric. Ecosyst. Environ. 128, 219–224. doi: 10.1016/j.agee.2008.06.003
Bunce, J. A. (2017). Using FACE systems to screen wheat cultivars for yield increases at elevated CO2. Agronomy 7:20. doi: 10.3390/agronomy7010020
Cacho, O., Moss, J., Thornton, P. K., Herrero, M., Henderson, B., and Bodirsky, B. (2016). Adaptation Pathways for Vulnerable Areas. Background Report for The State of Food and Agriculture 2016 (SOFA): Climate Change, Agriculture and Food Security. Rome: FAO.
Cai, C., Yin, X., He, S., Jiang, W., Si, C., Struik, P. C., et al. (2016). Responses of wheat and rice to factorial combinations of ambient and elevated CO2 and temperature in FACE experiments. Glob. Chang. Biol. 22, 856–874. doi: 10.1111/gcb.13065
Cairns, J. E., Cross, J., Zaidi, P. H., Grudlyoma, P., Sanchez, C., Araus, J. L., et al. (2013). Identification of drought, heat, and combined drought and heat tolerant donors in maize. Crop Sci. 53, 1335–1346. doi: 10.2135/cropsci2012.09.0545
Campos, H., Cooper, M., Habben, J. E., Edmeades, G. O., and Schussler, J. R. (2004). Improving drought tolerance in maize: a view from industry. Field Crops Res. 90, 19–34. doi: 10.1016/j.fcr.2004.07.003
Carmo-Silva, E., Andralojc, P. J., Scales, J. C., Driever, S. M., Mead, A., Lawson, T., et al. (2017). Phenotyping of field-grown wheat in the UK highlights contribution of light response of photosynthesis and flag leaf longevity to grain yield. J. Exp. Bot. 68, 3473–3486. doi: 10.1093/jxb/erx169
Casaretto, J. A., El-kereamy, A., Zeng, B., Stiegelmeyer, S. M., Chen, X., Bi, Y.-M., et al. (2016). Expression of OsMYB55 in maize activates stress-responsive genes and enhances heat and drought tolerance. BMC Genomics 17:312. doi: 10.1186/s12864-016-2659-5
Castiglioni, P., Warner, D., Bensen, R. J., Anstrom, D. C., Harrison, J., Stoecker, M., et al. (2008). Bacterial RNA chaperones confer abiotic stress tolerance in plants and improved grain yield in maize under water-limited conditions. Plant Physiol. 147, 446–455. doi: 10.1104/pp.108.118828
Cerrudo, D., Cao, S., Yuan, Y., Martinez, C., Suarez, E. A., Babu, R., et al. (2018). Genomic selection outperforms marker-assisted selection for grain yield and physiological traits in a maize doubled haploid population across water treatments. Front. Plant Sci. 9:366. doi: 10.3389/fpls.2018.00366
Challinor, A., Watson, J., Lobell, D., Howden, S., Smith, D., and Chhetri, N. (2014). A meta-analysis of crop yield under climate change and adaptation. Nat. Clim. Chang. 4, 287–291. doi: 10.1038/nature13809
Challinor, A. J., Koehler, A. K., Ramirez-Villegas, J., Whitfield, S., and Das, B. (2016). Current warming will reduce yields unless maize breeding and seed systems adapt immediately. Nat. Clim. Chang. 6, 954–958. doi: 10.1038/nclimate3061
Chater, C., Peng, K., Movahedi, M., Dunn, J. A., Walker, H. J., Liang, Y. K., et al. (2015). Elevated CO2-induced response in stomata require ABA and ABA signaling. Curr. Biol. 25, 2709–2716. doi: 10.1016/j.cub.2015.09.013
Chebrolu, K. K., Fritschi, F. B., Ye, S., Gillman, J., Krishnan, H., James, R. H., et al. (2016). Impact of heat stress during seed development on soybean seed metabolome. Metabolomics 12:28. doi: 10.1007/s11306-015-0941-1
Chen, D., Richardson, T., McIntyre, C. L., Rae, A. L., and Xue, G.-P. (2016). Drought upregulated TaNAC69-1 is a transcriptional repressor of TaSHY2 and TalAA7, and enhances root length and biomass in wheat. Plant Cell Physiol. 57, 2076–2090. doi: 10.1093/pcp/pcw126
Chen, J., Chopra, R., Hayes, C., Morris, G., Marla, S., Burke, J., et al. (2017). Genome-wide association study of developing leaves heat tolerance during vegetative growth stages in a sorghum association mapping panel. Plant Genome 10, 1–5. doi: 10.3835/plantgenome2016.09.0091
Chinnusamy, V., and Zhu, J.-K. (2009). Epigenetic regulation of stress responses in plants. Curr. Opin. Plant Biol. 12, 133–139. doi: 10.1016/j.pbi.2008.12.006
Choudhary, S., and Sinclair, T. R. (2014). Hydraulic conductance differences among sorghum genotypes to explain variation in restricted transpiration rates. Funct. Plant Biol. 41, 270–275. doi: 10.1071/FP13246
Ciais, P., Sabine, C., Bala, G., Bopp, L., Brovkin, V., Canadell, J., et al. (2013). “Carbon and other biogeochemical cycles,” in Climate Change 2013: The Physical Science Basis, eds T. F. Stocker, D. Qin, G. K. Plattner, M. Tignor, S. K. Allen, J. Boschung, et al. (Cambridge: Cambridge University Press).
Clasen, B., Stoddard, T. J., Luo, S., Demorest, Z. L., Li, J., Cedrone, F., et al. (2016). Improving cold storage and processing traits in potato through targeted gene knockout. Plant Biotechnol. J. 14, 169–176. doi: 10.1111/pbi.12370
Condon, A. G., Richards, R. A., Rebetzke, G. J., and Farquhar, G. D. (2004). Breeding for high water use efficiency. J. Exp. Bot. 55, 2447–2460. doi: 10.1093/jxb/erh277
Cossani, C. M., and Reynolds, M. P. (2015). Heat stress adaptation in elite lines derived from synthetic hexaploid wheat. Crop Sci. 55, 2719–2735. doi: 10.2135/cropsci2015.02.0092
Covshoff, S., and Hibberd, J. M. (2012). Integrating C4 photosynthesis into C3 crops to increase yield potential. Curr. Opin. Biotechnol. 23, 209–214. doi: 10.1016/j.copbio.2011.12.011
Covshoff, S., Majeran, W., Liu, P., Kolkman, J. M., van Wijk, K. J., and Brutnell, T. P. (2008). Deregulation of maize C4 photosynthetic development in a mesophyll cell-defective mutant. Plant Physiol. 146, 1469–1481. doi: 10.1104/pp.107.113423
Danon, A., Delorme, V., Mailhac, N., and Gallois, P. (2000). Plant programmed cell death: a common way to die. Plant Physiol. Biochem. 38, 647–655. doi: 10.1016/S0981-9428(00)01178-5
Daryanto, S., Wang, L., and Jacinthe, P.-A. (2017). Global synthesis of drought effects on cereal, legume, tuber and root crop production: a review. Agric. Water Manage. 179, 18–33. doi: 10.1016/j.agwat.2016.04.022
de Conto, R. M., Galeotti, S., Pagani, M., Tracy, D., Schaefer, K., and Zhang, T. (2012). Past extreme warming events linked to massive carbon release from thawing permafrost. Nature 484, 87–91. doi: 10.1038/nature10929
de Costa, W. J. A. M., Weerakoon, W. M. W., Chinthaka, K. G. R., Herath, H. M. L. K., and Abeywardena, R. M. I. (2007). Genotypic variation in the response of rice (Oryza sativa L.) to increased atmospheric carbon dioxide and its physiological basis. J. Agron. Crop Sci. 193, 117–130. doi: 10.1111/j.1439-037X.2007.00255.x
del Pozo, A., Yáñz, A., Matus, I. A., Tapia, G., Castillo, D., Sanchez-Jardón, L., et al. (2016). Physiological traits associated with wheat yield potential and performance under water-stress in a Mediterranean environment. Front. Plant Sci. 7:987. doi: 10.3389/fpls.2016.00987
Devi, M. J., and Sinclair, T. R. (2011). Diversity in drought traits among commercial southeastern US peanut cultivars. Int. J. Agron. 2011:754658. doi: 10.1155/2011/754658
Devi, M. J., Sinclair, T. R., and Vadez, V. (2010). Genotypic variation in peanut for transpiration response to vapor deficit. Crop Sci. 50, 191–196. doi: 10.2135/cropsci2009.04.0220
Devi, M. J., Sinclair, T. R., Vadez, V., and Krishnamurthy, L. (2009). Peanut genotypic variation in transpiration efficiency and decreased transpiration during progressive soil drying. Field Crops Res. 114, 280–285. doi: 10.1016/j.fcr.2009.08.012
Ding, Z., Weissmann, S., Wang, M., Du, B., Huang, L., Wang, L., et al. (2015). Identification of photosynthesis-associated C4 candidate genes through comparative leaf gradient transcriptome in multiple lineages of C3 and C4 species. PLoS One 10:e0140629. doi: 10.1371/journal.pone.0140629
Dixit, S., Yadaw, R. B., Mishra, K. K., and Kumar, A. (2017). Marker-assisted breeding to develop the drought-tolerant version of Sabitri, a popular variety from Nepal. Euphytica 213:184. doi: 10.1007/s10681-017-1976-3
Dong, Y., Liu, J., Zhang, Y., Geng, H., Rasheed, A., Xiao, Y., et al. (2016). Genome wide association of stem water soluble carbohydrates in bread wheat. PLoS One 11:e0164293. doi: 10.1371/journal.pone.0164293
Driever, S. M., Lawson, T., Andralojc, P. J., Raines, C. A., and Parry, M. A. J. (2014). Natural variation in photosynthetic capacity, growth, and yield in 64 field-grown wheat genotypes. J. Exp. Bot. 65, 4959–4973. doi: 10.1093/jxb/eru253
Dryland Cereals and Archival Report (2012–2013). International Crops Research Institute for the Semi-Arid Tropics. Patancheru: ICRISAT, 328.
Dwivedi, S. K., Kumar, S., Prakash, V., Mondal, S., and Mishra, J. S. (2015). Influence of rising atmospheric CO2 concentrations and temperature on morpho-physiological traits and yield of rice genotypes in sub humid climate of eastern India. Am. J. Plant Sci. 6, 2339–2349. doi: 10.4236/ajps.2015.614237
Dwivedi, S. L., Sahrawat, K., Upadhyaya, H., and Ortiz, R. (2013). Food, nutrition and agrobiodiversity under global climate change. Adv. Agron. 120, 1–128. doi: 10.3389/fpls.2016.00850
Dwivedi, S. L., Scheben, A., Edwards, D., Spillane, C., and Ortiz, R. (2017). Assessing and exploiting functional diversity in germplasm pools to enhance abiotic stress adaptation and yield in cereals and food legumes. Front. Plant Sci. 8:1461. doi: 10.3389/fpls.2017.01461
Dwivedi, S. L., Upadhyaya, H. D., Balaji, J., Buhariwalla, H. K., Blair, M. W., Ortiz, R., et al. (2006). Using genomics to exploit grain legume biodiversity in crop improvement. Plant Breed. Rev. 26, 171–357.
Dwivedi, S. L., Upadhyaya, H. D., Subudhi, P., Gehring, C., Bajic, V., and Ortiz, R. (2010). Enhancing abiotic stress tolerance in cereals through breeding and transgenic interventions. Plant Breed. Rev. 33, 31–114. doi: 10.1002/9780470535486.ch2
ElBasyoni, I., Saadalla, M., Baenziger, S., Bockelman, H., and Morsy, S. (2017). Cell membrane stability and association mapping for drought and heat tolerance in a worldwide wheat collection. Sustainability 9:1606. doi: 10.3390/su9091606
El-Kereamy, A., Bi, Y.-M., Ranathunge, K., Beatty, P. H., Good, A. G., and Rothstein, S. J. (2012). The rice R2R3-MYB transcription factor OsMYB55 is involved in the tolerance to high temperature and modulates amino acid metabolism. PLoS One 7:e52030. doi: 10.1371/journal.pone.0052030
El-Sharkawy, M. A. (2016). Prospects of photosynthetic research for increasing agricultural productivity, with emphasis on the tropical C4 Amaranthus and the cassava C3-C4 crops. Photosynthetica 54, 161–184. doi: 10.1007/s11099-016-0204-z
Fang, H., Liu, X., Thorn, G., Duan, J., and Tian, L. (2014). Expression analysis of histone acetyltransferases in rice under drought stress. Biochem. Biophys. Res. Commun. 443, 400–405. doi: 10.1016/j.bbrc.2013.11.102
Fang, Y., Liao, K., Du, H., Xu, Y., Song, H., Li, X., et al. (2015). A stress-responsive NAC transcription factor SNAC3 confers heat and drought tolerance through modulation of reactive oxygen species in rice. J. Exp. Bot. 66, 6803–6817. doi: 10.1093/jxb/erv386
Feng, Z., Zhang, B., Ding, W., Liu, X., Yang, D.-L., Wei, P., et al. (2013). Efficient genome editing in plants using a CRISPR/Cas-9 system. Cell Res. 23, 1229–1232. doi: 10.1007/s11103-017-0599-3
Fisher, M., Abate, T., Lunduka, R. W., Asnake, W., Alemayehu, Y., and Madulu, R. B. (2015). Drought tolerant maize for farmer adaptation to drought in sub-Saharan Africa: determinants of adoption in eastern and southern Africa. Clim. Chan. 133, 283–299. doi: 10.1007/s10584-015-1459-2
Fisher, M., and Carr, E. R. (2015). The influence of gendered roles and responsibilities on the adoption of technologies that mitigate drought risk: the case of drought-tolerant maize seed in Eastern Uganda. Glob. Environ. Chang. 35, 82–92. doi: 10.1016/j.gloenvcha.2015.08.009
Fletcher, A. L., Sinclair, T. R., and Allen, L. H. Jr. (2007). Transpiration responses to vapor pressure deficit in well-watered “slow-wilting” and commercial soybean. Environ. Exp. Bot. 61, 145–151. doi: 10.1016/j.envexpbot.2007.05.004
Fracasso, A., Magnanini, E., Marocco, A., and Amaducci, S. (2017). Real time determination of photosynthesis, transpiration, water-use efficiency and gene expression of two Sorghum bicolor (Moench) genotypes subjected to dry-down. Front. Plant Sci. 8:932. doi: 10.3389/fpls.2017.00932
Frankel, O. H. (1984). “Genetic manipulations: impact on man and society,” in Genetic Perspective of Germplasm Conservation, eds W. K. Arber, K. Llimensee, W. J. Peacock, and P. Starlinger (Cambridge: Cambridge University Press), 161–470.
Frey, F. P., Presterl, T., Lecoq, P., Orlik, A., and Stich, B. (2016). First step to understand heat tolerance of temperate maize at adult stage: identification of QTL across multiple environments with connected segregating populations. Theor. Appl. Genet. 129, 945–961. doi: 10.1007/s00122-016-2674-6
Frey, F. P., Urbany, C., Hüttel, B., Reinhardt, R., and Stich, B. (2015). Genome-wide expression profiling and phenotypic evaluation of European maize inbreds at seedling stage in response to heat stress. BMC Genomics 16:123. doi: 10.1186/s12864-015-1282-1
Furbank, R. T., and Tester, M. (2011). Phenomics technologies to relieve the phenotyping bottleneck. Trends Plant Sci. 16, 635–644. doi: 10.1016/j.tplants.2011.09.005
Gahlaut, V., Jaiswal, V., Tyagi, B. S., Singh, G., Sareen, S., and Balyan, H. S. (2017). QTL mapping for nine drought-responsive agronomic traits in bread wheat under irrigated and rainfed environments. PLoS One 12:e0182857. doi: 10.1371/journal.pone.0182857
Gao, Y., Wu, M., Zhang, M., Jiang, W., Ren, X., Liang, E., et al. (2018). A maize phytochrome-interacting factors protein ZmPIF1 enhances drought tolerance by inducing stomatal closure and improves grain yield in Oryza sativa. Plant Biotechnol. J. 16, 1375–1387. doi: 10.1111/pbi.12878
Garg, R., Chevala, V. V. S. N., Shankar, R., and Jain, M. (2015). Divergent DNA methylation patterns associated with gene expression in rice cultivars with contrasting drought and salinity response. Sci. Rep. 5:14922. doi: 10.1038/srep14922
Gayacharan, and Joel, A. J. (2013). Epigenetic responses to drought stress in rice (Oryza sativa L.). Physiol. Mol. Biol. Plants 19, 379–387. doi: 10.1007/s12298-013-0176-4
Gerland, P., Raftery, A. E., Ševèíková, H., Li, N., Gu, D., Spoorenberg, T., et al. (2014). World population stabilization unlikely this century. Science 346, 234–237. doi: 10.1126/science.1257469
Gholipoor, M., Choudhary, S., Sinclair, T. R., Messina, C. D., and Cooper, M. (2013a). Transpiration response of maize hybrids to atmospheric vapor pressure deficit. J. Agron. Crop Sci. 199, 155–160. doi: 10.1111/jac.12010
Gholipoor, M., Sinclair, T. R., Raza, M. A. S., Löffler, C., Cooper, M., and Messina, C. D. (2013b). Maize hybrid variability for transpiration decrease with progressive soil drying. J. Agron. Crop Sci. 199, 23–29. doi: 10.1111/j.1439-037X.2012.00530.x
Gholipoor, M., Prasad, P. V. V., Mutava, R. N., and Sinclair, T. R. (2010). Genetic variability of transpiration response to vapor pressure deficit among sorghum genotypes. Field Crops Res. 119, 85–90. doi: 10.1016/j.fcr.2010.06.018
González, A., Bermejo, V., and Gimeno, B. S. (2010). Effect of different physiological traits on grain yield in barley grown under irrigated and terminal water deficit conditions. J. Agric. Sci. 148, 319–328. doi: 10.1017/S0021859610000031
Gous, P. W., Meder, R., and Fox, G. P. (2015). Near infrared spectral assessment of stay-green barley genotypes under heat stress. J. Near Infrared Spectrosc. 23, 145–153. doi: 10.1255/jnirs.1163
Gray, S. B., Dermody, O., Klein, S. P., Locke, A. M., McGreath, J. M., Paul, R. E., et al. (2016). Intensifying drought eliminates the expected benefits of elevated carbon dioxide for soybean. Nat. Plants 2:16132. doi: 10.1038/NPLANTS.2016.132
Gu, J., Yin, X., Stomph, T.-J., and Struik, P. C. (2014). Can exploiting natural genetic variation in leaf photosynthesis contribute to increasing rice productivity? A simulation study. Plant Cell Environ. 37, 22–34. doi: 10.111/pce.12173
Gu, J., Yin, X., Stomph, T.-J., Wabg, H., and Struik, P. C. (2012a). Physiological basis of genetic variation in leaf photosynthesis among rice (Oryza sativa L.) introgression lines under drought and well-watered conditions. J. Exp. Bot. 63, 5173–5153. doi: 10.1093/jxb/ers170
Gu, J., Yin, X., Struik, P. C., Stomph, T. J., and Wang, H. (2012b). Using chromosome introgression lines to map quantitative trait loci for photosynthesis parameters in rice (Oryza sativa L.) leaves under drought and well-watered field conditions. J. Exp. Bot. 63, 455–469. doi: 10.1093/jxb/err292
Guo, P., Baum, M., Grando, S., Ceccarelli, S., Bai, G., Li, R., et al. (2009). Differentially expressed genes between drought-tolerant and drought-sensitive barley genotypes in response to drought stress during the reproductive stage. J. Exp. Bot. 60, 3531–3544. doi: 10.1093/jxb/erp194
Guo, Z., Yang, W., Chang, Y., Ma, X., Tu, H., Xiong, F., et al. (2018). Genome-wide association studies of image traits reveal the genetic architecture of drought resistance in rice. Mol. Plant 11, 789–805. doi: 10.1016/j.molp.2018.03.018
Gupta, S. K., Rai, K. N., Singh, P., and Verma, Y. S. (2015). Seed set variability under high temperatures during flowering period in pearl millet (Pennisetum glaucum L. (R.) Br.). Field Crops Res. 171, 41–53. doi: 10.1016/j.fcr.2014.11.005
Hall, L. N., Roth, R., Brutnell, T. P., and Langdale, J. A. (1998). Cellular differentiation in maize leaf is disrupted by bundle sheath defective mutations in Zea mays. Symp. Soc. Exp. Biol. 51, 27–31.
Hamaoka, N., Yasui, H., Yamagata, Y., Inoue, Y., Furuya, N., Araki, T., et al. (2017). A hairy-leaf gene, BLANKET LEAF, of wild Oryza nivara increases photosynthetic water use efficiency in rice. Rice 10:20. doi: 10.1186/s12284-017-0158-1
Hamidou, F., Halilou, O., and Vadez, V. (2013). Assessment of groundnut under combined heat and drought stress. J. Agron. Crop Sci. 199, 1–11. doi: 10.1111/j.1439-037X.2012.00518.x
Haritha, G., Vishnukiran, T., Yugandhar, P., Sarla, N., and Subrahmanyam, D. (2017). Introgression from Oryza rufipogon increase photosynthetic efficiency of KMR3 rice lines. Rice Sci. 24, 85–96. doi: 10.1016/j.rsci.2016.07.006
Hatch, M. D. (1987). C4 photosynthesis: a unique blend of modified biochemistry, anatomy and ultrastructure. Biochim. Biophys. Acta 895, 81–106. doi: 10.1016/S0304-4173(87)80009-5
Herman, R. A., Fast, B. J., Scherer, P. N., Brune, A. M., de Cerqueira, D. T., and Schafer, B. W. (2017). Stacking transgenic event DAS-Ø15Ø7-1 alters maize composition less than traditional breeding. Plant Biotechnol. 15, 1264–1272. doi: 10.1111/pbi.12713
Hirabayashi, H., Sasaki, K., Kambe, T., Gannaban, R. B., Miras, M. A., Mendioro, M. S., et al. (2015). qEMF3, a novel QTL for the early-morning flowering trait from wild rice, Oryza officinalis, to mitigate heat stress damage at flowering in rice, O. sativa. J. Exp. Bot. 66, 1227–1236. doi: 10.1093/jxb/eru474
Honna, P. T., Fuganti-Pagliarini, R., Ferreira, L. C., Molinari, M. D. C., Marin, S. R. R., de Oliveira, M. C. N., et al. (2016). Molecular, physiological, and agronomical characterization, in greenhouse and field conditions, of soybean plants genetically modified with AtGolS2 gene for drought tolerance. Mol. Breed. 36:157. doi: 10.1007/s11032-016-0570-z
Honsdorf, N., March, T. J., Berger, B., Tester, M., and Pillen, K. (2014). High throughput phenotyping to detect drought tolerance QTL in wild barley introgression lines. PLoS One 9:e97047. doi: 10.1371/journal.pone.0097047
Hoyos-Villegas, V., Song, Q., and Kelly, J. D. (2017). Genome-wide association analysis for drought tolerance and associated traits in common bean. Plant Genome 10:1 doi: 10.3835/plantgenome2015.12.0122
Hübner, S., Korol, A. B., and Schmid, K. J. (2015). RNA-seq analysis identified genes associated with differential reproductive success under drought-stress in accessions of wild barley Hordeum spontaneum. BMC Plant Biol. 15:134. doi: 10.1186/s12870-015-0528-z
Hughes, J., Hepworth, C., Dutton, C., Dunn, J. A., Hunt, L., Stephens, J., et al. (2017). Reducing stomatal density in barley improves drought tolerance without impacting on yield. Plant Physiol. 174, 776–787. doi: 10.1104/pp.16.01844
Hund, A., Ruta, N., and Liedgens, M. (2009). Rooting depth and water use efficiency of tropical maize inbred lines, differing in drought tolerance. Plant Soil 318, 311–325. doi: 10.1007/s11104-008-9843-6
Hung, H. Y., Shannon, L. M., Tian, F., Bradbury, P. J., Chen, C., Flint-Garcia, S. A., et al. (2012). ZmCCT and the genetic basis of day-length adaptation underlying the post-domestication spread of maize. Proc. Natl. Acad. Sci. U.S.A. 109, E1913–E1921. doi: 10.1073/pnas.1203189109
Hussain, R. M., Ali, M., Feng, X., and Li, X. (2017). The essence of NAC gene family to the cultivation of drought-resistant soybean (Glycine max) cultivars. BMC Plant Biol. 17:55. doi: 10.1186/s12870-017-1001-y
Hyman, G., Barona, E., Biradar, C., Guevara, E., Dixon, J., Beebe, S., et al. (2016). Priority regions for research on dryland cereals and legumes. F1000Res. 5:885. doi: 10.12688/f1000research.8657.1
Ionescue, I. A., Møller, B. L., and Sánchez-Pérez, R. (2017). Chemical control of flowering. J. Exp. Bot. 68, 369–382. doi: 10.1093/jxb/erw427
IPCC. (2013). “Summary for policy makers. In climate change 2013: the physical science basis,” in Proceedings of the Contribution of Working Group 1 to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change, eds D. Qin, T. F. Stocker, G. K. Plattner, M. Tignor, S. K. Allen, et al. (Cambridge: Cambrideg University Press).
Jablonski, L. M., Wang, X., and Curtis, P. S. (2002). Plant reproduction under elevated CO2 conditions: a meta-analysis of reports on 79 crop and wild species. New Phytol. 156, 9–26. doi: 10.1046/j.1469-8137.2002.00494.x
Jagadish, S. V. K., Craufurd, P. Q., and Wheeler, T. R. (2007). Phenotyping parents of mapping populations of rice for heat tolerance during anthesis. Crop Sci. 48, 1140–1146. doi: 10.2135/cropsci2007.10.0559
Jagadish, S. V. K., Muthurajan, R., Oane, R., Wheeler, T. R., Heuer, S., Bennett, J., et al. (2009). Physiological and proteomic approaches to address heat tolerance during anthesis in rice (Oryza sativa L.). J. Exp. Bot. 61, 143–156. doi: 10.1093/jxb/erp289
Jahan, E., Amthor, J. S., Farquhar, G. D., Trethowan, R., and Barbour, M. M. (2014). Variation in mesophyll conductance among Australian wheat genotypes. Funct. Plant Biol. 41, 568–580. doi: 10.1071/FP13254
Jeong, J. S., Kim, Y. S., Baek, K. H., Jung, H., Ha, S.-W., Choi, Y. D., et al. (2010). Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 153, 185–197. doi: 10.1104/pp.110.154773
Johansson, K. S. L., El-Soda, M., Nilsson, A. K., Andersson, M. X., and Fredin, J. U. (2015). Molecular basis of variation in stomatal responsiveness to elevated CO2. Procedia Environ. Sci. 29:210. doi: 10.1016/j.proenv.2015.07.266
Johnson, S. M., Cummins, I., Lim, F. L., Slabas, A. R., and Knight, M. R. (2015). Transcriptomic analysis comparing stay-green and senescent Sorghum bicolor lines identifies a role for proline biosynthesis in the stay-green trait. J. Exp. Bot. 66, 7061–7073. doi: 10.1093/jxb/erv405
Jones, A. M. (2001). Programmed cell death in development and defense. Plant Physiol. 125, 94–97. doi: 10.1104/pp.125.1.94
Jung, H., Chung, P. J., Park, S.-H., Reillas, M. C. F. R., Kim, Y. S., Suh, J.-W., et al. (2017). Overexpression of OsERF48 causes regulation of OsCML16, a calmodulin-like protein gene that enhances root growth and drought tolerance. Plant Biotechnol. J. 15, 1295–1308. doi: 10.1111/pbi.12716
Jyothi, M. N., Rai, D. V., and Babu, R. N. (2015). Identification and characterization of high temperature stress responsive novel miRNAs in French bean (Phaseolus vulgaris). Appl. Biochem. Biotechnol. 176, 835–849. doi: 10.1007/s12010-015-1614-2
Kadam, N. N., Tamilselvan, A., Lawas, L. M. F., Quinones, C., Bahuguna, R. N., Thomson, M. J., et al. (2017). Genetic control of plasticity in root morphology and anatomy of rice in response to water deficit. Plant Physiol. 174, 2302–2315. doi: 10.1104/pp.17.00500
Kajala, K., Covshoff, S., Karki, S., Woodfield, H., Tolley, B. J., Dionora, M. J. A., et al. (2011). Strategies for engineering a two-celled C4 photosynthetic pathway into rice. J. Exp. Bot. 62, 3001–3010. doi: 10.1093/jxb/err022
Kale, S. M., Jaganathan, D., Ruperao, P., Chen, C., Punna, R., Kudapa, H., et al. (2015). Prioritization of candidate genes in “QTL-hotspot” region for drought tolerance in chickpea (Cicer arietinum). Sci. Rep. 5:15296. doi: 10.1038/srep15296
Kashiwagi, J., Krishnamurthy, L., Gaur, P. M., Upadhyaya, H. D., Varshney, R. K., and Tobita, S. (2013). Traits of relevance to improve yield under terminal drought stress in chickpea (C. arietinum L.). Field Crops Res. 145, 88–95. doi: 10.1016/j.fcr.2013.02.011
Kashiwagi, J., Krishnamurthy, L., Upadhyaya, H. D., Krishna, H., Chandra, S., Vadez, V., et al. (2005). Genetic variability of drought-avoidance root traits in the mini-core germplasm collection of chickpea (Cicer arietinum L.). Euphytica 146, 213–222. doi: 10.1007/s10681-005-9007-1
Kashiwagi, J., Upadhyaya, H. D., and Krishnamurthy, L. (2010). Significance and genetic diversity of SPAD chlorophyll meter reading in chickpea germplasm in the semi- arid environments. J. Food Legumes 23, 99–105.
Kassie, G. T., Abdulai, A., Greene, W. H., Shiferaw, B., Abate, T., Tarekegne, A., et al. (2017). Model preference and willingness to pay for drought tolerance (DT) in maize in rural Zimbabwe. World Dev. 94, 465–477. doi: 10.1016/j.worlddev.2017.02.008
Katiyar, A., Smita, S., Muthusamy, S. K., Chinnusamy, V., Pandey, D. M., and Bansal, K. C. (2015). Identification of novel drought-responsive microRNAs and trans-acting siRNAs from Sorghum bicolor (L.) Moench by high-throughput sequencing analysis. Front. Plant Sci. 6:506. doi: 10.3389/fpls.2015.00506
Kaushal, N., Awasthi, A., Gupta, K., Gaur, P., Siddique, K. H. M., and Nayar, H. (2013). Heat-stress-induced reproductive failures in chickpea (Cicer arietinum) are associated with impaired sucrose metabolism in leaves and anthers. Funct. Plant Biol. 40, 1334–1349. doi: 10.1071/FP13082
Kaushal, N., Bhandari, K., Siddique, K. H. M., and Nayyar, H. (2016). Food crops face rising temperatures: an overview of responses, adaptive mechanisms, and approaches to improve heat tolerance. Cogent Food Agric. 2:1134380. doi: 10.1080/23311932.2015.1134380
Kholová, J., Hash, C. T., Kakkera, A., Koěová, M., and Vadez, V. (2010). Constitutive water conservation mechanisms are correlated with terminal drought tolerance of pearl millet (Pennisetum glaucum). J. Exp. Bot. 61, 369–377. doi: 10.1093/jxb/erp314
Kim, J. M., To, T. K., Ishida, J., Morosawa, T., and Kawashima, M. (2008). Alterations of lysine modifications on the histone H3 N-tail under drought stress conditions in Arabidopsis thaliana. Plant Cell Physiol. 49, 1580–1588. doi: 10.1093/pcp/pcn133
Kostandini, G., La Rovere, R., and Abdoulaye, T. (2013). Potential impacts of increasing average yields and reducing maize yield variability in Africa. Food Policy 43, 213–226. doi: 10.1016/j.foodpol.2013.09.007
Krishnamurthy, L., Kashiwagi, J., Tobita, S., Ito, O., Upadhyaya, H. D., Gowda, C. L. L., et al. (2012). Variation in carbon isotope discrimination and its relationship with harvest index in the reference collection of chickpea germplasm. Funct. Plant Biol. 40, 1350–1361. doi: 10.1071/FP13088
Kumagai, E., Aoki, N., Masuya, Y., and Shimono, H. (2015). Phenotypic plasticity conditions the response of soybean seed yield to elevated atmospheric CO2 concentration. Plant Physiol. 169, 2021–2029. doi: 10.1104/pp.15.00980
Kumagai, E., Homma, K., Kuroda, E., and Shimono, H. (2016). Finlay-Wilkinson’s regression coefficient as a prescreening criterion for yield responsiveness to elevated atmospheric CO2 concentration in crops. Physiol. Plant. 158, 312–317. doi: 10.1111/ppl.12468
Kumar, A., Dixit, S., Ram, T., Yadav, R. B., Mishra, K. K., and Mandal, N. P. (2014). Breeding high-yielding drought-tolerant rice: genetic variations and conventional and molecular approaches. J. Exp. Bot. 65, 6265–6278. doi: 10.1093/jxb/eru363
Kumar, S., Dwivedi, S. K., Singh, S. S., Bhatt, B. P., Mehta, P., Elanchezhian, R., et al. (2014). Morpho-physiological traits associated with reproductive stage drought tolerance of rice (Oryza sativa L.) genotypes under rainfed condition of eastern Indo-Gangetic Plain. Indian J. Plant Physiol. 19, 87–93. doi: 10.1007/s40502-014-0075-x
Kumar, A., Sharma, K. D., and Kumar, D. (2008). Traits for screening and selection of cowpea genotypes for drought tolerance at early stages of breeding. J. Agric. Rural Dev. Trop. Subtrop. 109, 191–199.
Kumar, R. R., Pathak, H., Sharma, S. K., Kala, Y. K., Nirjal, M. K., Singh, G. P., et al. (2015). Novel and conserved heat-responsive microRNAs in wheat (Triticum aestivum L.). Funct. Integr. Genomics 15, 323–348. doi: 10.1007/s10142-014-0421-0
Kumari, M., Pudake, R. N., Singh, V. P., and Joshi, A. K. (2013). Association of stay-green trait with canopy temperature depression and yield traits under terminal heat stress in wheat (Triticum aestivum L.). Euphytica 190, 87–97. doi: 10.1007/s10681-012-0780-3
Kyazze, F. B., and Kristjanson, P. (2011). Summary of Baseline Household Survey Results: Rakai District, South Central Uganda. Copenhagen: Agriculture and Food Security.
Lafarge, T., Bueno, C., Frouin, J., Jacquin, L., Courtois, B., and Ahmadi, N. (2017). Genome-wide association analysis for heat tolerance at flowering detected a large set of genes involved in adaptation to thermal and other stresses. PLoS One 12:e0171254. doi: 10.1371/journal.pone.0171254
Law, J. A., and Jacobsen, S. E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204–220. doi: 10.1038/nrg2719
Lawson, T., and Blatt, M. (2014). Stomatal size, speed and responsiveness impact on photosynthesis and water use efficiency. Plant Physiol. 164, 1556–1570. doi: 10.1104/pp.114.237107
Leakey, A. D. B., Uribelarrea, M., Ainsworth, E. A., Naidu, S. L., Rogers, A., Ort, D. R., et al. (2006). Photosynthesis, productivity, and yield of maize are not affected by open-air elevation of CO2 concentration in the absence of drought. Plant Physiol. 140, 779–790. doi: 10.1104/pp.105.073957
Lee, D.-K., Jung, H., Jang, G., Jeong, J. S., Kim, Y. S., Ha, H.-S., et al. (2016). Overexpression of OsERF71 transcription factor alters rice root structure and drought resistance. Plant Physiol. 172, 575–588. doi: 10.1104/pp.16.00379
Lesk, C., Rowhani, P., and Ramankutty, N. (2016). Influence of extreme weather disasters on global crop production. Nature 529, 84–87. doi: 10.1038/nature16467
Li, C., Sun, B., Li, Y., Liu, C., Wu, X., Zhang, D., et al. (2016). Numerous genetic loci identified for drought tolerance in the maize nested association mapping populations. BMC Genomics 17:894. doi: 10.1186/s12864-016-3170-8
Li, N., Ye, M., Li, Y., Yan, Z., Butcher, L. M., Sun, J., et al. (2010). Whole genome DNA methylation analysis based on high throughput sequencing technology. Methods 52, 203–212. doi: 10.1016/j.ymeth.2010.04.009
Li, R., Zeng, Y., Xu, J., Wang, Q., Wu, F., Cao, M., et al. (2015). Genetic variation for maize root architecture in response to drought stress at the seedling stage. Breed. Sci. 65, 298–307. doi: 10.1270/jsbbs.65.298
Li, X. M., Chao, D.-Y., Wu, Y., Huang, X., Chen, K., Cui, L.-G., et al. (2015). Natural allele of a proteasome α2 subunit gene contribute to thermotolerance and adaptation of African rice. Nat. Genet. 47, 827–833. doi: 10.1038/ng.3305
Liao, J.-L., Zhang, H.-Y., Liu, J.-B., Zhong, P.-A., and Huang, Y.-J. (2012). Identification of candidate genes related to rice grain weight under high-temperature stress. Plant Sci. 196, 32–43. doi: 10.1016/j.plantsci.2012.07.013
Liu, H., Searle, L. R., Watson-Haigh, N. S., Baumann, U., Mather, D. E., Able, A. J., et al. (2015). Genome-wide identification of microRNAs in leaves and the developing head of four durum genotypes during water deficit stress. PLoS One 10:e142799. doi: 10.1371/journal.pone.0142799
Liu, Q., Yang, T., Yu, T., Zhang, S., Mao, X., Zhao, J., et al. (2017). Integrating small RNA sequencing with QTL mapping for identification of miRNAs and their target genes associated with heat tolerance at the flowering stage in rice. Front. Plant Sci. 8:43. doi: 10.3389/fpls.2017.00043
Lopes, M. S., Araus, J. L., Van Heereden, P. D., and Foyer, C. H. (2011). Enhancing drought tolerance in C4 crops. J. Exp. Bot. 62, 3135–3153. doi: 10.1093/jxb/err105
Lopez-Gomollon, S., Mohorianu, I., Szittya, G., Moulton, V., and Dalmay, T. (2012). Diverse correlation patterns between mRNAs and their targets during tomato fruit development indicates different modes of mRNAs actions. Planta 236, 1875–1887. doi: 10.1007/s00425-012-1734-7
Lucas, M. R., Ehler, J. D., Huynh, B.-L., Diop, N.-N., Roberts, P. A., and Close, T. J. (2013). Markers for breeding heat-tolerant cowpea. Mol. Breed. 31, 529–536. doi: 10.1007/s11032-012-9810-z
Lunduka, R. M., Mateva, K. I., Magorokosho, C., and Manjeru, P. (2017). Impact of adoption of drought-tolerant maize varieties on total maize production in south eastern Zimbabwe. Clim. Dev. doi: 10.1080/17565529.2017.1372269
Maazou, A. R. S., Tu, J., Qiu, J., and Liu, Z. (2016). Breeding for drought tolerance in maize (Zea mays L.). Am. J. Plant Sci. 7, 1858–1870. doi: 10.4236/ajps.2016.714172
Makate, C., Wang, R., Makate, M., and Mango, N. (2017). Impact of drought tolerant maize adoption on maize productivity, sales and consumption in rural Zimbabwe. Agrekon 56, 67–81. doi: 10.1080/03031853.2017.1283241
Mangrauthia, S. K., Bhogireddy, S., Agarwal, S., Prasanth, V. V., Voleti, S. R., Neelamraju, S., et al. (2017). Genome wide changes in microRNA expression during short and prolonged heat stress and recovery in contrasting rice cultivars. J. Exp. Bot. 68, 2399–2412. doi: 10.1093/jxb/erx111
Marshall, A. (2014). Drought-tolerant varieties begin global march. Nat. Biotechnol. 32:308. doi: 10.1038/nbt.2875
McGuire, S., and Sperling, L. (2015). Seed systems smallholder farmers use. Food Secur. 8, 179–195. doi: 10.1007/s12571-015-0528-8
Medina, S., Gupta, S. K., and Vadez, V. (2017). Transpiration response and growth in pearl millet parental lines and hybrids bred for contrasting rainfall environments. Front. Plant Sci. 8:1846. doi: 10.3389/fpls.2017.01846
Mehrabi, Z., and Ramankutty, N. (2017). The cost of heat waves and droughts for global crop production. bioRxiv [Preprint]. doi: 10.1101/188151
Merchuk-Ovnat, L., Barak, V., Fahima, T., Ordon, F., Lidzbarsky, G. A., Krugman, T., et al. (2016). Ancestral QTL alleles from wild emmer wheat improve drought resistance and productivity in modern wheat cultivars. Front. Plant Sci. 7:452. doi: 10.3389/fpls.2016.00452
Messina, C. D., Sinclair, T. R., Hammer, G. L., Curan, D., Thompson, J., Oler, Z., et al. (2015). Limited-transpiration trait may increase maize drought tolerance in the US corn belt. Agron. J. 107, 1978–1986. doi: 10.2134/agronj15.0016
Messmer, R., Fracheboud, Y., Bänziger, M., Stamp, P., and Ribaut, J. M. (2011). Drought stress and tropical maize: QTL for leaf greenness, plant senescence, and root capacitance. Field Crops Res. 124, 93–103. doi: 10.1016/j.fcr.2011.06.010
Messmer, R., Fracheboud, Y., Bänziger, M., Vargas, M., Stamp, P., and Ribaut, J. M. (2009). Drought stress and tropical maize: QTL-by-environment interaction and stability of QTL across environments for yield components and secondary traits. Theor. Appl. Genet. 119, 913–930. doi: 10.1007/s00122-009-1099-x
Meyer, P. (2015). Epigenetic variation and environmental change. J. Exp. Bot. 66, 3541–3548. doi: 10.1093/jxb/eru502
Millet, E. J., Welcker, C., Kruijer, W., Negro, S., Coupel-Ledru, A., Nicolar, S. D., et al. (2016). Genome-wide analysis of yield in Europe: allelic effects vary with drought and heat scenarios. Plant Physiol. 172, 749–764. doi: 10.1104/pp.16.00621
Mitterbauer, E., Enders, M., Bender, J., Erbs, M., Habekuß, A., Kilian, B., et al. (2016). Growth response of 98 barley (Hordeum vulgare L.) genotypes to elevated CO2 and identification of related quantitative trait loci using genome-wide association studies. Plant Breed. 136, 483–497. doi: 10.1111/pbr.12501
Molden, D. (ed.). (2007). Water for Food, Water for Life: A Comprehensive Assessment of Water Management in Agriculture. London: Earthscan.
Monson, R. K., Moore, B. D., Ku, M. S. B., and Edwards, G. E. (1986). Cofunction of C3-photosynthetic and C4 photosynthetic pathways in C3, C4 and C3-C4 intermediate Flaveria species. Planta 168, 493–502. doi: 10.1007/BF00392268
Moon, S.-J., Kim, H. Y., Hwang, H., Kim, J.-A., Lee, Y., Min, M. K., et al. (2017). A dominant negative OsKAT2 mutant delays light-induced stomatal opening and improves drought tolerance without yield penalty in rice. Front. Plant Sci. 8:772. doi: 10.3389/fpls.2017.00772
Morison, J. I. L., Baker, N. R., Mullineaux, P. M., and Davies, W. J. (2008). Improving water use in crop production. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363, 639–658. doi: 10.1098/rstb.2007.2175
Muchero, W., Ehlers, J., Close, T., and Roberts, P. A. (2009). Mapping QTL for drought stress-induced premature senescence and maturity in cowpea [Vigna unguiculata(L) Walp.]. Theor. Appl. Genet. 118, 849–863. doi: 10.1007/s00122-008-0944-7
Muchero, W., Ehlers, J. D., and Roberts, P. A. (2008). Seedling stage drought-induced phenotypes and drought-responsive genes in diverse cowpea genotypes. Crop Sci. 48, 541–552. doi: 10.2135/cropsci2007.07.0397
Muchero, W., Roberts, P. A., Diop, N. N., Drabo, I., Close, T. J., Muranaka, S., et al. (2013). Genetic architecture of delayed senescence, biomass, and grain yield under drought stress in cowpea. PLoS One 8:e70041. doi: 10.1371/journal.pone.0070041
Mwale, S. E., Ochwo-Ssemakula, M., Sadik, K., Achola, E., Okul, V., Gibson, P., et al. (2017). Response of cowpea genotypes to drought stress in Uganda. Am. J. Plant Sci. 8, 720–733. doi: 10.4236/ajps.2017.84050
Myers, S. S., Smith, M. R., Guth, S., Golden, C. D., Vaitla, B., Mueller, N. D., et al. (2017). Climate change and global food systems: potential impacts on food security and undernutrition. Annu. Rev. Public Health 38, 259–277. doi: 10.1146/annurev-publhealth-031816-044356
Nakhforoosh, A., Bodewein, T., Fiorani, F., and Bodner, G. (2017). Identification of water use strategy at early growth stages in durum wheat from shoot phenotyping and physiological measurements. Front. Plant Sci. 7:1155. doi: 10.3389/fpls.2016.01155
Nawaz, A., Farooq, M., Cheema, S. A., Yasmeen, A., and Wahid, A. (2013). Stay green character at grain filling ensures resistance against terminal drought in wheat. Int. J. Agric. Biol. 15, 1272–1276.
Nemali, K. S., Bonin, C., Dohleman, F. G., Stephens, M., Reeves, W. R., Nelson, D. E., et al. (2015). Physiological response related to increased grain yield under drought in the first biotechnology-derived drought tolerant maize. Plant Cell Environ. 38, 1866–1880. doi: 10.1111/pce.12446
Ngugi, K., Kimani, W., Kiambi, D., and Mutitu, E. W. (2013). Improving drought tolerance in Sorghum bicolor L. Moench: marker-assisted transfer of the stay-green quantitative trait loci (QTL) from a characterized donor source into a local farmer variety. Int. J. Sci. Res. Knowl. 1, 154–162. doi: 10.12983/ijsrk-2013-p154-162
Nicotra, A. B., Atkin, O. K., Bonser, S. P., Davidson, A. M., Finnegan, E. J., Mathesius, U., et al. (2010). Plant phenotypic plasticity in a changing climate. Trends Plant Sci. 15, 684–692. doi: 10.1016/j.tplants.2010.09.008
Nuccio, M. L., Wu, J., Mowers, R., Zhou, H.-P., Meghji, M., Primavesi, L. F., et al. (2015). Expression of trehalose-6-phosphate-phosphatase in maize ears improves yield in well-watered and drought conditions. Nat. Biotechnol. 33, 862–869. doi: 10.1038/nbt.3277
Nuñez Barrios, A., Hoogenboom, G., and Nesmith, D. S. (2005). Drought stress and the distribution of vegetative and reproductive traits of a bean cultivar. Sci. Agric. 62, 18–22. doi: 10.1590/S0103-90162005000100004
Okada, R., Nemoto, Y., Endo-Higashi, N., and Izawa, T. (2017). Synthetic control of flowering in rice independent of the cultivation environment. Nat. Plants 3:17039. doi: 10.1038/nplants.2017.39
Ortiz, R. (2015). Plant Breeding in the Omics Era. New York, NY: Springer. doi: 10.1007/978-3-319-20532-8
Osakabe, Y., Watanabe, T., Sugano, S. S., Ueta, R., Ishihara, R., Shinozaki, K., et al. (2016). Optimization of CRISPR/Cas9 genome editing to modify abiotic stress responses in plants. Sci. Rep. 6:26685. doi: 10.1038/srep26685
Palmgren, M. G. (2001). Plant plasma membrane H+-ATPases: powerhouses for nutrient uptake. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 817–845. doi: 10.1146/annurev.arplant.52.1.817
Pandey, G. C., Mamrutha, H. M., Tiwari, R., Sareen, S., Bhatia, S., Siwach, P., et al. (2015). Physiological traits associated with heat tolerance in bread wheat (Triticum aestivum L.). Physiol. Mol. Biol. Plants 21, 93–99. doi: 10.1007/s12298-014-0267-x
Pandey, V., and Shukla, A. (2015). Acclimation and tolerance strategies of rice under drought stress. Rice Sci. 22, 147–161. doi: 10.1016/j.plaphy.2017.03.018
Pattanayak, V., Lin, S., Guilinger, J. P., Ma, E., Doudna, J. A., and Liu, D. R. (2013). High throughput profiling of off-target DNA cleavage reveals RNA programmed Cas-9 nuclease activity. Nat. Biotechnol. 31, 839–843. doi: 10.1038/nbt.2673
Paulus, J. K., Schlieper, D., and Groth, G. (2013). Greater efficiency of photosynthetic carbon fixation due to single amino acid substitution. Nat. Commun. 4:1518. doi: 10.1038/ncomms2504
Peng, J., Sun, D., Peng, Y., and Nevo, E. (2012). Gene discovery in Triticum dicoccoides, the direct progenitor of cultivated wheat. Cereal Res. Commun. 41, 1–22. doi: 10.1556/CRC.2012.0030
Peterhänsel, C., and Maurino, V. G. (2011). Photorespiration redesigned. Plant Physiol. 155, 49–55. doi: 10.1104/pp.110.165019
Pinto, R. S., Lopes, M. S., Collins, N. C., and Reynolds, M. P. (2016). Modelling and genetic dissection of staygreen under heat stress. Theor. Appl. Genet. 129, 2055–2074. doi: 10.1007/s00122-016-2757-4
Pinto, R. S., Morero, G., and Reynolds, M. (2017). Identification of heat-tolerant wheat lines showing genetic variation in leaf respiration and other physiological traits. Euphytica 213:76. doi: 10.1007/s10681-017-1858-8
Placido, D. F., Campbell, M. T., Folsom, J. J., Cui, X. P., Kruger, G. R., Baenziger, P. S., et al. (2013). Introgression of novel traits from a wild wheat relative improves drought adaptation in wheat. Plant Physiol. 161, 1806–1819. doi: 10.1104/pp.113.214262
Polania, J., Rao, I. M., Cajiao, C., Rivera, M., Raatz, B., and Beebe, S. (2016). Physiological traits associated with drought resistance in Andean and Mesoamerican genotypes of common bean (Phaseolus vulgaris L.). Euphytica 210, 17–29. doi: 10.1007/s10681-016-1691-5
Pottorff, M., Roberts, P. A., Close, T. J., Lonardi, S., Wanamaker, S., and Ehlers, J. D. (2014). Identification of candidate genes and molecular markers for heat-induced brown discoloration of seed coats in cowpea (Vigna unguiculata). BMC Genomics 15:328. doi: 10.1186/1471-2164-15-328
Prasad, P. V. V., and Djanaguiraman, M. (2014). Response of floret fertility and individual grain weight of wheat to high temperature stress: sensitive stages and thresholds for temperature and duration. J. Exp. Bot. 41, 1261–1269. doi: 10.1071/FP14061
Prasad, P., Staggenborg, S., and Ristic, Z. (2008). “Impacts of drought and/or heat stress on physiological, developmental, growth, and yield process of crop plants,” in Response of Crops to Limited Water: Understanding and Modelling Water Stress Effects on Plant Growth Processes, (Madison, WI: American Society Agronomy, Inc), 301–355.
Prasad, P. V. V., Djanaguiraman, M., Perumal, R., and Ciampitti, I. A. (2015). Impact of high temperature stress on floret fertility and individual grain weight of grain sorghum: sensitive stages and thresholds for temperature and duration. Front. Plant Sci. 6:820. doi: 10.3389/fpls.2015.00820
Prasad, P. V. V., Pisipati, S. R., Momèiloviæ, I., and Ristic, Z. (2011). Independent and combined effects of high temperature and drought stress during grain filling on plant yield and chloroplast EF-Tu expression in spring wheat. J. Agron. Crop Sci. 197, 430–441. doi: 10.1111/j.1439-037X.2011.00477.x
Prins, A., Orr, D. J., Andralojc, P. J., Reynolds, M. P., Carmo-Silva, E., and Parry, M. A. J. (2016). Rubisco catalytic properties of wild and domesticated relatives provide scope for improving wheat photosynthesis. J. Exp. Bot. 67, 1827–1838. doi: 10.1093/jxb/erv574
Qin, F., Kakimoto, M., Sakuma, Y., Maruyama, K., Osakabe, Y., Tran, L. S., et al. (2007). Regulation and functional analysis of ZmDREB2A in response to drought and heat stresses in Zea mays L. Plant J. 50, 54–69. doi: 10.1111/j.1365-313X.2007.03034.x
Qu, M., Zheng, G., Hamdani, S., Essmine, J., Song, Q., Wang, H., et al. (2017). Leaf photosynthetic parameters related to biomass accumulation in a global rice diversity survey. Plant Physiol. 175, 248–258. doi: 10.1104/pp.17.00332
Rangan, P., Furtado, A., and Henry, R. J. (2016a). Commentary: new evidence for grain specific C4 photosynthesis in wheat. Front. Plant Sci. 7:1537. doi: 10.3389/fpls.2016.01537
Rangan, P., Furtado, A., and Henry, R. J. (2016b). New evidence for grain specific C4 photosynthesis in wheat. Sci. Rep. 6:31721. doi: 10.1038/srep31721
Ray, D. K., Mueller, N. D., West, P. C., and Foley, J. A. (2013). Yield trends are insufficient to double global crop production by 2050. PLoS One 8:e66428. doi: 10.1371/journal.pone.0066428
Rebetzke, G. J., Condon, A. G., Richards, R. A., and Farquhar, G. D. (2002). Selection for reduced carbon isotope discrimination increases aerial biomass and grain yield of rainfed bread wheat. Crop Sci. 42, 739–745. doi: 10.2135/cropsci2002.0739
Redillas, M. C. F. R., Jeong, J. S., Kim, Y. S., Jung, H., Bang, S. W., Choi, Y. D., et al. (2012). The overexpression of OcNAC9 alters the root architecture of rice plants enhancing drought resistance and grain yield under field conditions. Plant Biotechnol. J. 10, 792–805. doi: 10.1111/j.1467-7652.2012.00697.x
Reynolds, M., and Langridge, P. (2016). Physiological breeding. Curr. Opin. Plant Biol. 31, 162–171. doi: 10.1016/j.pbi.2016.04.005
Rizal, G., Thakur, V., Dionora, J., Karki, S., Wanchana, S., Acebron, K., et al. (2015). Two forward genetic screens for vein density mutants in sorghum converge on a cytochrome P450 gene in the brassinosteroid pathway. Plant J. 84, 257–266. doi: 10.1111/tpj.13007
Rizza, F., Ghashghaie, J., Meyer, S., Mattu, L., Mastrangelo, A. M., and Badeck, F.-W. (2012). Constitutive differences in water use efficiency between two durum wheat cultivars. Field Crops Res. 125, 49–60. doi: 10.1016/j.fcr.2011.09.001
Rodríguez-Leal, D., Lemmon, Z. L., Man, J., Bartlett, M. E., and Lippman, Z. B. (2017). Engineering quantitative trait variation for crop improvement by genome editing. Cell 171, 470–480. doi: 10.1016/j.cell.2017.08.030
Roorkiwal, M., Nayak, S. N., Thudi, M., Upadhyaya, H. D., Brunel, D., Mournet, P., et al. (2014). Allele diversity for abiotic stress responsive candidate genes in chickpea reference set using gene based SNP markers. Front. Plant Sci. 5:248. doi: 10.3389/fpls.2014.00248
Rosales, M. A., Cueller-Ortiz, S. M., de la Paz Arrieta-Montiel, M., Acosta-Gallegos, J., and Covarrubias, A. A. (2013). Physiological traits related to terminal drought resistance in common bean (Phaseolus vulgaris L.). J. Sci. Food Agric. 93, 324–331. doi: 10.1002/jsfa.5761
Ruiz-Vera, U. M., Siebers, M. H., Drag, D. W., Ort, D. R., and Bernacchi, C. J. (2015). Canopy warming caused photosynthetic acclimation and reduced seed yield in maize grown at ambient and elevated [CO2]. Glob. Chang. Biol. 21, 4237–4249. doi: 10.1111/gcb.13013
Russo, V. E., Martienssen, R. A., and Riggs, A. D. (1996). Epigenetic Mechanism of Gene Regulation. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Sadok, W., and Sinclair, T. R. (2009). Genetic variability of transpiration response to vapor pressure deficit among soybean cultivars. Crop Sci. 49, 955–956. doi: 10.2135/cropsci2008.09.0560
Sage, R. F., Sage, T. L., and Kocacinar, F. (2012). Photorespiration and the evolution of C4 photosynthesis. Annu. Rev. Plant Physiol. 63, 17.1–17.29.
Sammons, B., Whitsel, J., Stork, L. G., Reeves, W., and Horak, M. (2014). Characterization of drought tolerant maize MON 87460 for use in environmental risk assessment. Crop Sci. 54, 719–729. doi: 10.2135/cropsci2013.07.0452
Sandhu, N., Raman, K. A., Torres, R. O., Audebert, A., Dardou, A., and Kumar, A. (2016). Rice root architectural plasticity traits and genetic regions for adaptability to variable cultivation and stress conditions. Plant Physiol. 171, 2562–2576. doi: 10.1104/pp.16.00705
Sarr, B. (2012). Present and future climate change in the semi-arid region of West Africa: a crucial input for practical adaptation in agriculture. Atmos. Sci. Lett. 13, 108–112. doi: 10.1002/asl.368
Sehgal, A., Sita, K., Kumar, J., Kumar, S., Singh, S., Siddique, K. H. M., et al. (2017). Effects of drought, heat and their interaction on the growth, yield and photosynthetic function of lentil (Lens culinaris Medikus) genotypes varying in heat and drought sensitivity. Front. Plant Sci. 8:1776. doi: 10.3389/fpls.2017.01776
Selvaraj, M. G., Ishizaki, T., Valencia, M., Ogawa, S., Dedicova, B., Ogata, T., et al. (2017). Overexpression of an Arabidopsis thaliana galactinol synthase gene improves drought tolerance in transgenic rice and increased grain yield in the field. Plant Biotechnol. J. 15, 1465–1477. doi: 10.1111/pbi.12731
Serraj, R., Hash, C. T., Rizvi, S. M. H., Sharma, A., Yadav, R. S., and Bidinger, F. R. (2005). Recent advances in marker-assisted selection for drought tolerance in pearl millet. Plant Prod. Sci. 8, 334–337. doi: 10.1626/pps.8.334
Setimala, P. S., Magorokosho, C., Lunduka, R., Gasura, E., Makumbi, D., Tarekegne, A., et al. (2017). On-farm yield gains with stress-tolerant maize in Eastern and Southern Africa. Agron. J. 109, 406–417. doi: 10.2134/agronj2015.0540
Seversike, T. M., Sermons, S. M., Sinclair, T. R., Carter, T. E., and Rufry, T. W. (2013). Temperature interactions with transpiration response to vapor pressure deficit among cultivated and wild soybean genotypes. Physiol. Plant. 148, 62–73. doi: 10.1111/j.1399-3054.2012.01693.x
Shamsudin, N. A. A., Mallikarjuna Swamy, B. P., Ratnam, W., Cruz, M. T. S., Sandhu, N., Raman, A. K., et al. (2016). Pyramiding of drought yield QTLs into a high-quality Malaysian rice cultivar MRQ74 improves yield under reproductive stage drought. Rice 9:21. doi: 10.1186/s12284-016-0093-6
Shankar, R., Bhattacharjee, A., and Jain, M. (2016). Transcriptome analysis in different rice cultivars provides novel insights into desiccation and salinity stress responses. Sci. Rep. 5:23719. doi: 10.1038/srep23719
Shanmugavadivel, P. S., Mithra, S. V., Prakash, C., Ramkumar, M. K., Tiwari, R., and Mohapatra, T. (2017). High resolution mapping of QTLs for heat tolerance in rice using a 5K SNP array. Rice 10:28. doi: 10.1186/s12284-017-0167-0
Sharma, D. K., Andersen, S. B., Ottosen, C.-T., and Rosenqvist, E. (2015). Wheat cultivars selected for high Fv/Fm under heat stress maintain high photosynthesis, total chlorophyll, stomatal conductance, transpiration and dry matter. Physiol. Plant. 153, 284–298. doi: 10.1111/ppl.12245
Sharma, D. K., Torp, A. M., Rosenqvist, E., Ottosen, C.-T., and Andersen, S. B. (2017). QTLs and potential candidate genes for heat stress tolerance identified from the mapping populations specifically segregating for Fv/Fm in wheat. Front. Plant Sci. 8:1668. doi: 10.3389/fpls.2017.01668
Sharma, K. D., Kumar, A., and Verma, S. R. (2016). Variations in physiological traits as screening tool for drought tolerance in barley (Hordeum vulgare L.). Indian J. Plant Physiol. 21, 93–100. doi: 10.1007/s40502-016-0207-6
Sharwood, R. E., Ghannoum, O., Kapralov, M. V., Gunn, L. H., and Whitney, S. M. (2016). Temperature responses of Rubisco from Paniceae grasses provide opportunities for improving C3 photosynthesis. Nat. Plant 2:16186. doi: 10.1038/nplants.2016.186
Shekoofa, A., Balota, M., and Sinclair, T. R. (2014). Limited-transpiration trait evaluated in growth chamber and field for sorghum genotypes. Environ. Exp. Bot. 99, 175–179. doi: 10.1016/j.envexpbot.2013.11.018
Shen, H., Li, J., Chen, W., Wang, X., Guo, L., Peng, Z., et al. (2012). Genome-wide analysis of DNA methylation and gene expression changes in two Arabidopsis ecotypes and their reciprocal hybrids. Plant Cell 24, 875–892. doi: 10.1105/tpc.111.094870
Sheng, L., Chai, W., Gong, X., Zhou, L., Cai, R., Li, X., et al. (2015). Identification and characterization of novel mRNAs involved in different genetic backgrounds. Int. J. Biol. Sci. 11, 781–793. doi: 10.7150/ijbs.11619
Shi, J., Gao, H., Wang, H., Lafitte, H. R., Archibald, R. L., Yang, M., et al. (2017). ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 15, 207–216. doi: 10.1111/pbi.12603
Shi, J., Habben, J. E., Archibald, R. L., Drumond, B. J., Chamberlin, M. A., Williams, R. W., et al. (2015). Overexpression of ARGOS genes modifies plant sensitivity to ethylene, leading to improved drought tolerance in both Arabidopsis and maize. Plant Physiol. 169, 266–282. doi: 10.1104/pp.15.00780
Shikha, M., Kanika, A., Rao, A. R., Mallikarjuna, M. G., Gupta, H. S., and Nepolean, T. (2017). Genomic selection for drought tolerance using genome-wide SNPs in maize. Front. Plant Sci. 8:550. doi: 10.3385/fpls.2017.00550
Shimono, H. (2011). Rice genotypes that respond strongly to elevated CO2 also respond strongly to low planting density. Agric. Ecosyst. Environ. 141, 240–243. doi: 10.1016/j.agee.2011.02.028
Shimono, H., and Okada, M. (2013). Plasticity of rice tiller production is related to genotypic variation in the biomass response to elevated atmospheric CO2 concentration and low temperatures during vegetative growth. Environ. Exp. Bot. 87, 227–234. doi: 10.1016/j.envexpbot.2012.11.008
Shimono, H., Okada, M., Yamakawa, Y., Nakamura, H., Kobayashi, K., and Hasegawa, T. (2009). Genotypic variation in rice yield enhancement by elevated CO2 relates to growth before heading, and not to maturity groups. J. Exp. Bot. 60, 523–532. doi: 10.1093/jxb/ern288
Shimono, H., Ozaki, Y., Jagadish, K. S. V., Sakai, H., Usui, Y., Hasegawa, T., et al. (2014). Planting geometry as a pre-screening technique for identifying CO2-responsive rice genotypes: a case study of panicle number. Physiol. Plant. 152, 520–528. doi: 10.1111/ppl.12202
Sinclair, T. R., Messina, C. D., Beatty, A., and Samples, M. (2010). Assessment across United States of the benefits of altered soybean drought traits. Agron. J. 102, 475–482. doi: 10.2134/agronj2009.0195
Sinclair, T. R., Zwieniecki, M. A., and Holbrook, N. M. (2008). Low leaf hydraulic conductance associated with drought tolerance in soybean. Physiol. Plant. 132, 446–451. doi: 10.1111/j.1399-3054.2007.01028.x
Singh, B. P., Jayaswal, P. K., Singh, B., Singh, P. K., Kumar, V., Mishra, F., et al. (2015). Natural allelic diversity in OsDREB1F gene in the Indian wild rice germplasm led to ascertain its association with drought tolerance. Plant Cell Rep. 34, 993–1004. doi: 10.1007/s00299-015-1760-6
Sinha, P., Pazhamala, L. T., Singh, V. K., Saxena, R. K., Krishnamurthy, L., Azam, S., et al. (2016). Identification and validation of selected universal stress protein domain containing drought-responsive genes in pigeonpea (Cajanus cajan). Front. Plant Sci. 6:1065. doi: 10.3380/fpls.2015.01065
Sita, K., Sehgal, A., HanumanthaRao, B., Nair, R. M., Prasad, P. V. V., Kumar, S., et al. (2017). Food legumes and rising temperatures: effects, adaptive functional mechanisms specific to reproductive growth stage and strategies to improve heat tolerance. Front. Plant Sci. 8:1658. doi: 10.3389/flps.2017.01658
Sivasakthi, K., Tharanya, M., Kholová, J., Muriuki, R. W., Thirunalasundari, T., and Vadez, V. (2017). Chickpea genotypes contrasting for vigor and canopy conductance also differ in their dependence on different water transport pathways. Front. Plant Sci. 8:1663. doi: 10.3389/fpls.2017.01663
Slewinski, T. L., Anderson, A. A., Price, S., Withee, J. R., Gallagher, K., and Turgeon, R. (2014). Short-Root1 plays a role in the development of vascular tissue and Kranz anatomy in maize leaves. Mol. Plants 7, 1388–1392. doi: 10.1093/mp/ssu036
Slewinski, T. L., Anderson, A. A., Zhang, C., and Turgeon, R. (2012). Scarecrow plays a role in establishing Kranz anatomy in maize leaves. Plant Cell Physiol. 53, 2030–2037. doi: 10.1093/pcp/pcs147
Subudhi, P. K., Rosenow, D. T., and Nguyen, H. T. (2000). Quantitative trait loci for the stay green trait in sorghum (Sorghum bicolor L. Moench): consistency across genetic backgrounds and environments. Theor. Appl. Genet. 101, 733–741. doi: 10.1007/s001220051538
Suprunova, T., Krugman, T., Distelfeld, A., Fahima, T., Nevo, E., and Korol, A. (2007). Identification of a novel gene (Hsdr4) involved in water stress tolerance in wild barley. Plant Mol. Biol. 64, 17–34. doi: 10.1007/s11103-006-9131-x
Taiwo, O., Wilson, G. A., Morris, T., Seisenberger, S., Reik, W., Pearce, D., et al. (2012). Methylome analysis using MeDIP-seq with low DNA concentrations. Nat. Protoc. 7, 617–636. doi: 10.1038/nprot.2012.012
Takai, T., Adachi, S., Taguchi-Shiobara, F., Sanoh-Arai, Y., Iwasawa, N., Yoshinaga, S., et al. (2013). A natural variant of NAL1, selected in high yield rice breeding program, pleiotropically increases photosynthesis rate. Rice 3:2149. doi: 10.1038/srep02149
Tausz-Posch, S., Seneweera, S., Norton, R. M., Fitzgerald, G. J., and Tausz, M. (2012). Can a wheat cultivar with high transpiration efficiency maintain its yield advantage over a near-isogenic cultivar under elevated CO2? Field Crops Res. 133, 160–166. doi: 10.1016/j.fcr.2012.04.007
Thilakarathne, C. L., Tausz-Posch, S., Cane, K., Norton, R. M., Tausz, M., and Seneweera, S. (2012). Intraspecific variation in growth and yield response to elevated CO2 in wheat depends on the differences of leaf mass per unit area. Funct. Plant Biol. 40, 185–194. doi: 10.1071/FP12057
Thornton, P. K., Rosenstock, T., Förch, W., Lamanna, C., Bell, P., Henderson, B., et al. (2017). “A qualitative evaluation of CSA options in mixed crop-livestock systems in developing countries,” in Climate Smart Agriculture: Building Resilience to Climate Change, eds L. Lipper, N. McCarthy, D. Zilberman, S. Asfaw, and G. Branca (Berlin: Springer), 385–423.
Thudi, M., Gaur, P. M., Krishnamurthy, L., Mir, R. R., Kudapa, H., Fikre, A., et al. (2014a). Genomics-assisted breeding for drought tolerance in chickpea. Funct. Plant Biol. 41, 1178–1190. doi: 10.1071/FP13318
Thudi, M., Upadhyaya, H. D., Rathore, A., Gaur, P. M., Krishnamurthy, L., Roorkiwal, M., et al. (2014b). Genetic dissection of drought and heat tolerance in chickpea through genome-wide and candidate gene-based association mapping approaches. PLoS One 9:e96758. doi: 10.1371/journal.pone.0096758
Tian, W., Hou, C., Ren, Z., Pan, Y., Jia, J., Zhang, H., et al. (2015). A molecular pathway for CO2 response in Arabidopsis guard cells. Nat. Commun. 6:6057. doi: 10.1038/ncomms7057
Tomás, M., Flexas, J., Copolovici, L., Galmés, J., Hallik, L., Medrano, H., et al. (2013). Importance of leaf anatomy in determining mesophyll diffusion conductance to CO2 across species: quantitative limitations and scaling up by models. J. Exp. Bot. 64, 2269–2281. doi: 10.1093/jxb/ert086
Tomeo, N. J., and Rosenthal, D. M. (2017). Variable mesophyll conductance among soybean cultivars sets a tradeoff between photosynthesis and water use efficiency. Plant Physiol. 174, 241–257. doi: 10.1104/pp.16.01940
Tsutsumi, N., Tohya, M., Nakashima, T., and Ueno, O. (2017). Variations in structural, biochemical, and physiological traits of photosynthesis and resource use efficiency in Amaranthus species (NAD-ME-type C4). Plant Prod. Sci. 20, 300–312. doi: 10.1080/1343943X.2017.1320948
Uga, Y., Sugimoto, K., Ogawa, S., Rane, J., Ishitani, M., Hara, N., et al. (2013). Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 45, 1097–1102. doi: 10.1038/ng.2725
Upadhyaya, H. D. (2005). Variability for drought resistance related traits in the mini core collection of peanut. Crop Sci. 45, 1432–1440. doi: 10.2135/cropsci2004.0389
Upadhyaya, H. D., Dwivedi, S. L., Baum, M., Varshney, R. K., Sripada, M. U., Gowda, C. L. L., et al. (2008). Genetic structure, diversity, and allelic richness in composite collection and reference set in chickpea (Cicer arietinum L.). BMC Plant Biol. 8:106. doi: 10.1186/1471-2229-8-106
Upadhyaya, H. D., and Ortiz, R. (2001). A mini core subset for capturing diversity and promoting utilization of chickpea genetic resources in crop improvement. Theor. Appl. Genet. 102, 1292–1298. doi: 10.1007/s00122-001-0556-y
Vadez, V., Kholová, J., Yadav, R. S., and Hash, C. T. (2013). Small temporal differences in water uptake among varieties of pearl millet (Pennisetum glaucum) are critical for grain yield under terminal drought. Plant Soil 371, 447–462. doi: 10.1007/s11104-013-1706-0
Vadez, V., and Ratnakumar, P. (2016). High transpiration efficiency increases pod yield under intermittent drought in dry and hot atmospheric conditions but less so under wetter and cooler conditions in groundnut (Arachis hypogaea L.). Field Crops Res. 193, 16–23. doi: 10.1016/j.fcr.2016.03.001
Vaezi, B., Bavei, V., and Shiran, B. (2010). Screening of barley genotypes for drought tolerance by agro-physiological traits in field condition. Afr. J. Agric. Res. 5, 881–892.
Vanaja, M., Maheshwari, M., Jyoti Laxmi, N., Satish, P., Yadav, S. K., Salini, K., et al. (2015). Variability in growth and yield response of maize genotypes at elevated CO2 concentration. Adv. Plant Agric. Res. 2:e00042.
Varshney, R. K., Gaur, P. M., Chamarthi, S. K., Krishnamurthy, L., Tripathi, S., Kashiwagi, J., et al. (2013). Fast-track introgression of ‘QTL-hotspot’ for root traits and other drought tolerance traits in J11, an elite and leading variety of chickpea. Plant Genome 6, 1–9. doi: 10.3835/plantgenome2013.07.0022
Varshney, R. K., Thudi, M., Nayak, S. N., Gaur, P. M., Kashiwagi, J., Krishnamurthy, L., et al. (2014). Genetic dissection of drought tolerance in chickpea (Cicer arietinum). Theor. Appl. Genet. 127, 445–462. doi: 10.1007/s00122-013-2230-6
Venkatesh, T. V., Cook, K., Liu, B., Perez, T., Willse, A., Tichich, R., et al. (2015). Compositional differences between near-isogenic GM and conventional maize hybrids are associated with backcrossing practices in conventional breeding. Plant Biotechnol. J. 13, 200–210. doi: 10.1111/pbi.12248
Vermeulen, S., Richards, M., De Pinto, A., Ferrarese, D., Läderach, P., Lan, L., et al. (2016). The Economic Advantage: Assessing the Value of Climate-Change Actions in Agriculture. Rome: IFAD.
von Caemmerer, S., Quick, W. P., and Furbank, R. T. (2012). The development of C4 rice: current progress and future challenges. Science 336, 1671–1672. doi: 10.1126/science.1220177
Wang, D. R., Bunce, J. A., Tomecek, M. B., Gealy, D., Mcclung, A., McCouch, S. R., et al. (2016). Evidence for divergence of response in indica, japonica, and wild rice to high CO2 × temperature interaction. Glob. Chang. Biol. 22, 2620–2632. doi: 10.1111/gcb.13279
Wang, W.-S., Pan, Y.-J., Zhao, X.-Q., Dwivedi, D., Zhu, L.-H., Ali, J., et al. (2011). Drought-induced site-specific DNA methylation and its association with drought tolerance in rice (Oryza sativa L.). J. Exp. Bot. 62, 1951–1960. doi: 10.1093/jxb/erq391
Wang, W., Qin, Q., Sun, F., Wang, Y., Xu, D., Li, Z., et al. (2016). Genome-wide differences in DNA methylation changes in two contrasting rice genotypes in response to drought conditions. Front. Plant Sci. 7:1675. doi: 10.3389/fpls.2016.01675
Wang, F., Wang, C. L., Liu, P. Q., Lei, C. L., Hao, W., Gao, Y., et al. (2016). Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the EFR transcription factor gene OsERF922. PLoS One 11:e0154027. doi: 10.1371/journal.pone.0154027
Wang, F., Chen, H.-W., Li, Q.-T., Wei, W., Li, W., Zhang, W.-K., et al. (2015). GmWRKY27 interacts with GmMYB174 to reduce expression of GmNAC29 for stress tolerance in soybean plants. Plant J. 83, 224–236. doi: 10.1111/tpj.12879
Wang, P., Zhao, L., Hou, H., Zhang, H., Huang, Y., Wang, Y., et al. (2015). Epigenetic changes are associated with programmed cell death induced by heat stress in seedling leaves of Zea mays. Plant Cell Physiol. 56, 965–976. doi: 10.1093/pcp/pcv023
Wang, L., Czedik-Eysenberg, A., Mertz, R. A., Tohge, T., Nunes-Nesi, A., Arrivault, S., et al. (2013). Comparative analysis of C4 and C3 photosynthesis in developing leaves of maize and rice. Nat. Biotechnol. 32, 1158–1164. doi: 10.1038/nbt.3019
Wang, Y., Cheng, X., Shan, Q., Zhang, Y., Liu, J., and Gao, C. (2014). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951. doi: 10.1038/nbt.2969
White, J. W., Andrade-Sanchez, P., Gore, M. A., Bronson, K. F., Coffelt, T. A., and Conley, M. M. (2012). Field-based phenomics for plant genetics research. Field Crops Res. 133, 101–112. doi: 10.1016/j.fcr.2012.04.003
Wilson, P. B., Rebetzke, G. J., and Condon, A. G. (2015). Of growing importance: combining greater early vigour and transpiration efficiency for wheat in variable rainfed environments. Funct. Plant Biol. 42, 1107–1115. doi: 10.1071/FP15228
Wossen, T., Abdoulaye, T., Alene, A., Feleke, S., Menkir, A., and Manyong, V. (2017). Measuring the impacts of adaptation strategies to drought stress: the case of drought tolerant maize varieties. J. Environ. Manage. 203, 106–113. doi: 10.1016/j.jenvman.2017.06.058
Wu, J., Chen, J., Wang, L., and Wang, S. (2017). Genome-wide investigation of WRKY transcription factors involved in terminal drought stress response in common bean. Front. Plant Sci. 8:380. doi: 10.3389/fpls.2017.00380
Xia, H., Huang, W., Xiong, J., Tao, T., Zheng, X., Wei, H., et al. (2016). Adaptive epigenetic differentiation between upland and lowland rice ecotypes revealed by methylation-sensitive amplified polymorphism. PLoS One 11:e0157810. doi: 10.1371/journal.pone.0157810
Xia, Y., Li, R., Bai, G., Siddique, K. H. M., Varshney, R. K., and Baum, M. (2017). Genetic variations of HvP5CS1 and their association with drought tolerance related traits in barley (Hordeum vulgare L.). Sci. Rep. 7:7870. doi: 10.1038/s41598-017-08393-0
Xin, M., Wang, Y., Yao, Y., Xie, C., Peng, H., Ni, Z., et al. (2010). Diverse set of microRNAs are responsive to powdery mildew infection and heat stress in wheat (Triticum aestivum L.). BMC Plant Biol. 10:123. doi: 10.1186/1471-2229-10-123
Xiong, I. Z., Xu, C. G., Maroof, M. A. S., and Zhang, Q. F. (1999). Patterns of cytosine methylation in elite rice hybrid and its parental lines, detected by a methylation sensitive amplification polymorphism technique. Mol. Gen. Genet. 261, 439–446. doi: 10.1007/s004380050986
Xu, J., Yuan, Y., Xu, Y., Zhang, G., Guo, X., Wu, F., et al. (2014). Identification of candidate genes for drought tolerance by whole-genome resequencing in maize. BMC Plant Biol. 14:83. doi: 10.1186/1471-2229-14-83
Xu, W., Rosenow, D. T., and Nguyen, H. T. (2000). Stay green trait in grain sorghum: relationship between visual rating and leaf chlorophyll concentration. Plant Breed. 119, 365–367. doi: 10.1046/j.1439-0523.2000.00506.x
Xu, Y., Lu, Y., Xie, C., Gao, S., Wan, J., and Prasanna, B. M. (2012). Whole-genome strategies for marker-assisted plant breeding. Mol. Breed. 29, 833–854. doi: 10.1007/s11032-012-9699-6
Xu, Z., Jiang, Y., Jia, B., and Zhou, G. (2016). Elevated-CO2 response of stomata and its dependence on environmental factors. Front. Plant Sci. 7:657. doi: 10.3389/fpls.20116.00657
Xu, Z. Z., Shimizu, H., Yagasaki, Y., Ito, S., Zheng, Y. R., and Zhou, G. S. (2013). Interactive effects of elevated CO2, drought and warming on plants. J. Plant Growth Regul. 32, 692–707. doi: 10.1007/s00344-013-9337-5
Xue, W., Xing, Y., Weng, X., Zhao, Y., Tang, W., Wang, L., et al. (2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40, 761–767. doi: 10.1038/ng.143
Yadav, A. K., Arya, R. K., and Narwal, M. S. (2014). Screening of pearl millet F1 hybrids for heat tolerance at early seedling stage. Adv. Agric. 2014:231301. doi: 10.1155/2014/231301
Yadav, S. K., Tiwari, Y. K., Singh, V., Patil, A. A., Shanker, A. K., and Lakshmi, N. J. (2016). Physiological and biochemical basis of extended and sudden heat stress tolerance in maize. Proc. Nat. Acad. Sci. India B Biol. Sci. 88, 249–263. doi: 10.1007/s40011-016-0752-9
Yang, S., Murphy, R. L., Morishige, D. T., Klein, P. E., Rooney, W. L., and Mullet, J. E. (2014). Sorghum phytochrome B inhibits flowering in long days by activating expression of SbPRR37 and Sb GHD7, repressors of SbEHD1, SbCN8 and SbCN12. PLoS One 9:e105352. doi: 10.1371/journal.pone.0105352
Yang, W., Duan, L., Che, G., Xiong, L., and Liu, Q. (2013). Plant phenomics and high-throughput phenotyping: accelerating rice functional genomics using multidisciplinary technologies. Curr. Opin. Plant Biol. 16, 180–187. doi: 10.1016/j.pbi.2013.03.005
Yu, T.-F., Xu, Z.-S., Guo, J.-K., Wang, Y.-X., Abernathy, B., Fu, J.-D., et al. (2017). Improved drought tolerance in wheat plants overexpressing a synthetic bacterial cold shock protein gene SeCspA. Sci. Rep. 7:44050. doi: 10.1038/srep44050
Zaman-Allah, M., Jenkinson, D. M., and Vadez, V. (2011a). A conservative pattern of water use, rather than deep or profuse rooting, is critical for the terminal drought tolerance of chickpeas. J. Exp. Bot. 62, 4239–4252. doi: 10.1093/jxb/err139
Zaman-Allah, M., Jenkinson, D. M., and Vadez, V. (2011b). Chickpea genotypes contrasting for seed yield under terminal drought stress in the field differ for traits related to control of water use. Funct. Plant Biol. 38, 270–281. doi: 10.1071/FP10244
Zang, X., Geng, X., Wang, F., Liu, Z., Zhang, L., Zhao, Y., et al. (2017). Overexpression of wheat ferritin gene TaFER-5B enhances tolerance to heat stress and other abiotic stresses associated with ROS Scavenging. BMC Plant Biol. 17:14. doi: 10.1186/s12870-016-0958-2
Zhang, B., Pan, X., Cobb, G. P., and Anderson, T. A. (2006). Plant microRNA: a small regulatory molecule with big impact. Dev. Biol. 289, 3–16. doi: 10.1016/j.ydbio.2005.10.036
Zhang, J. Z., Ai, X. Y., Guo, W. W., Peng, S. A., Deng, X. X., and Hu, C. G. (2012). Identification of mRNAs and their target genes using deep sequencing and degradome analysis in trifoliate orange (Poncirus trifoliata L. Ref). Mol. Biotechnol. 51, 44–57. doi: 10.1007/s12033-011-9439-x
Zhao, L., Wang, P., Yan, S., Gao, F., Li, H., Hou, H., et al. (2014). Promoter-associated histone acetylation is involved in the osmotic stress-induced transcriptional regulation of the maize ZmDREB2A gene. Physiol. Plant. 151, 459–467. doi: 10.1111/ppl.12136
Zheng, X., Chen, L., Li, M., Xia, H., Wang, P., Li, T., et al. (2013). Transgenerational variations in DNA methylation induced by drought stress in two rice varieties with distinguished differences in drought resistance. PLoS One 8:e080253. doi: 10.1371/journal.pone.0080253
Zinselmeier, C., Lauer, M. J., and Boyer, J. S. (1995). Reversing drought-induced losses in grain yield: sucrose maintains embryo growth in maize. Crop Sci. 35, 1390–1400. doi: 10.2135/cropsci1995.0011183X003500050022x
Ziska, L. H., Tomecek, M. B., and Gealy, D. R. (2014). Assessment of cultivated and wild, weedy rice lines to concurrent changes in CO2 concentration and air temperature: determining traits for enhanced seed yield with increasing atmospheric CO2. Funct. Plant Biol. 41, 236–243. doi: 10.1071/FP13155
Ziyomo, C., and Bernardo, R. (2013). Drought tolerance in maize: indirect selection through secondary traits versus genome-wide selection. Crop Sci. 52, 1269–1275. doi: 10.2135/cropsci2012.11.0651
Keywords: adoption and impact, applied genomics, gene editing, global warming, phenomics, photosynthesis, stress-tolerant cultivar, transgene
Citation: Dwivedi SL, Siddique KHM, Farooq M, Thornton PK and Ortiz R (2018) Using Biotechnology-Led Approaches to Uplift Cereal and Food Legume Yields in Dryland Environments. Front. Plant Sci. 9:1249. doi: 10.3389/fpls.2018.01249
Received: 03 May 2018; Accepted: 06 August 2018;
Published: 27 August 2018.
Edited by:
Hussein Shimelis, University of KwaZulu-Natal, South AfricaReviewed by:
Karl Kunert, University of Pretoria, South AfricaNemera Shargie, Agricultural Research Council of South Africa (ARC-SA), South Africa
Sileshi Gudeta Weldesemayat, University of KwaZulu-Natal, South Africa
Copyright © 2018 Dwivedi, Siddique, Farooq, Thornton and Ortiz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rodomiro Ortiz, cm9kb21pcm8ub3J0aXpAc2x1LnNl
 Sangam L. Dwivedi
Sangam L. Dwivedi Kadambot H. M. Siddique
Kadambot H. M. Siddique Muhammad Farooq
Muhammad Farooq Philip K. Thornton
Philip K. Thornton Rodomiro Ortiz
Rodomiro Ortiz