- College of Life Science, Engineering Research Center of the Chinese Ministry of Education for Bioreactor and Pharmaceutical Development, Jilin Agricultural University, Changchun, China
Carthamus tinctorius L., commonly known as safflower, is an important oilseed crop containing oil bodies. Oil bodies are intracellular organelles in plant cells for storing triacylglycerols (TAGs) and sterol esters. Oleosins are the most important surface proteins of the oil bodies. We predicted and retrieved the sequences of eight putative C. tinctorius oleosin (Ctoleosin) genes from the genome database of safflower. The bioinformatics analyses revealed the size of their open reading frames ranging from 414 to 675 bp, encoding 137 to 224 aa polypeptides with predicted molecular weights of 14.812 to 22.155 kDa, all containing the typical “proline knot” motif. Reverse transcription quantitative polymerase chain reaction (RT-qPCR) determined the spatiotemporal expression pattern of Ctoleosin genes, which gradually increased and peaked during flowering and seed ripening, and decreased thereafter. To validate their role in plant development, we transformed and overexpressed these eight putative Ctoleosin genes in Arabidopsis. Overexpressing Ctoleosins did not affect leaf size, although silique length was altered. Arabidopsis transformed with Ctoleosin3, 4, and 5 grew longer siliques than did the wild-type plants, without altering seed quantity. The 100-grain weight of the transgenic Arabidopsis seeds was slightly more than that of the wild-type seeds. The seed germination rates of the plants overexpressing Ctoleosin4 and 6 were slightly lower as compared with that of the wild-type Arabidopsis, whereas that in the other transgenic lines were higher than that in the wild-type plants. The overexpression of Ctoleosin genes elevated the oil content in the seeds of transgenic Arabidopsis. Our findings not only provide an approach for increasing the oil content, but also for elucidating the intricate mechanisms of oil body synthesis.
Introduction
Oil crop seeds store lipids, primarily triacylglycerols (TAGs), which provide energy for seed germination and seedling growth. The TAGs are contained in specialized organelles called oil bodies (Huang, 1996; Napier et al., 1996; Murphy, 2001), which are present in the seed, pollen, and tapetum of higher plants (Tzen et al., 1993; Huang, 1996). These oil bodies, which have a diameter of 0.2–0.5 μm (Murphy, 2012; Pasaribu et al., 2014), are enclosed in a single layer of phospholipids, unlike most other organelles, which are enclosed in a bilayer membrane (Tzen et al., 1992, 1993; Wu et al., 2010). This single membrane is stabilized by a collection of embedded proteins, three classes of which have been identified, namely oleosin, caleosin, and steroleosin (Lin and Tzen, 2004; Hanano et al., 2006; Tzen, 2012). Among oil body-associated proteins, oleosins, which are the major class, are basic proteins with a molecular weight of about 10–24 kDa (Qu and Huang, 1990). The amino acid sequences of all the oleosins so far studied can be divided into three distinct structural domains: N-terminal amphipathic, central hydrophobic, and C-terminal amphipathic. The hydrophobic domain consists of approximately 70 amino acids and it is highly conserved between species. It is therefore likely to be essential for oleosin for integration into the hydrophobic core of the oil body (Lacey et al., 1998). Both the N- and C-terminal hydrophilic ends of oleosin are much less conserved in their amino acid sequence and located on the surface of the oil body, such that their steric hindrance and electronegative repulsion provide stability to the single phospholipid layer (Huang, 1992; Tzen et al., 1992, 1993; Peng et al., 2003; Purkrtova et al., 2008; Hyun et al., 2013). Oleosins can also modulate oil body size and stability (Huang, 1992; Frandsen et al., 2001; Siloto et al., 2006; Jolivet et al., 2009, 2013; David et al., 2013; Hyun et al., 2013).
To date, a number of cDNA and genomic clones encoding seed oleosins have been isolated from Zea mays, Glycine max, Arabidopsis thaliana, Sesamum indicum, Brassica napus, Helianthus annuus, and other plant species (Keddie et al., 1992; Beaudoin and Napier, 2000; Alexander et al., 2002; Roux et al., 2004; Xu et al., 2004; Chapman et al., 2012). The genes encoding oleosins have been sequenced in many important oilseed crops but they have not yet been assayed in C. tinctorius L. Safflower is an oilseed crop of semi-arid regions and occupies a unique position among the oil seed crops due to the high linoleic content of its seed oil (Nikam and Shitole, 1999). Identification of the Ctoleosin genes and their functions would be of great significance in breeding. Previously, our team accomplished the de novo transcriptome assembly of safflower from which we predicted putative genes for oleosins (Li et al., 2012). Subsequently, we also sequenced the safflower genome, but the data has not yet been released. In this study, eight putative Ctoleosin genes retrieved from the safflower genome database were characterized. The expression levels of these Ctoleosin genes in different tissues and developmental stages of seeds and flowering were analyzed by RT-qPCR. In addition, the overexpression vector for each gene was constructed and transformed into Arabidopsis. The regulatory effect of Ctoleosin genes on oil body size and oil content was determined in transgenic Arabidopsis.
Materials and Methods
Plant Materials
The C. tinctorius L. (JiHong No. 1) seeds were purchased from Xinjiang Province of China. They were cultivated in the experimental field of Jilin Agricultural University, Changchun, China. The different tissues of safflower were collected, which included cotyledons, hypocotyls, leaves, stems, flowers, roots, and seeds. The safflower seeds were collected after flowering on day 4 (DAF4), 8 (DAF8), 12 (DAF12), 16 (DAF16), 20 (DAF20), 24 (DAF24), 28 (DAF28), and 32 (DAF32). These samples were immediately frozen in liquid nitrogen and stored at −80°C.
Sequence Analysis and Prediction of Ctoleosin Genes
We predicted and analyzed the sequences of Ctoleosin genes from the whole-genome sequence (WGS) database of safflower, which we have accomplished earlier. The physical and chemical properties of the predicted Ctoleosin proteins were analyzed by ProtParam online tools1 and the transmembrane domain was predicted by TMHMM2.02. The predicted Ctoleosin sequences were checked for the presence of the conserved oleosin domain (PF01277) using Pfam3 and SMART4. Arabidopsis oleosin sequences were collected from The Arabidopsis Information Resource (TAIR) 10.0, whereas the other oleosin sequences were obtained from NCBI5. The oleosin protein sequences were utilized to identify homologous peptides through BLASTP searches (e-value cut-off of 1.0). A phylogenetic tree was constructed by MEGA5.1 using the default parameters (Sarmiento et al., 1997). Analyses of the conserved motifs of Ctoleosin sequences were carried out using MEME6 with default parameters (Tamura et al., 2011).
RNA Extraction and cDNA Synthesis
The safflower seeds (0.2 mg) were ground in liquid nitrogen and total RNA was extracted from the various collected tissues and developing seeds using TRIzol (Invitrogen, Carlsbad, CA, United States), according to the manufacturer’s protocols. The RNA quality was determined based on OD260/280 values by NanoDrop 2000 (Thermo Fisher Scientific, Beijing, China) and its integrity was detected by 1.2% agarose gel electrophoresis. The total RNA (1 μg) was reverse transcribed into cDNA by the PrimeScript RT Reagent Kit with gDNA eraser (Takara, Japan), following the manufacturer’s protocols, and the cDNA was stored at −20°C.
Reverse Transcription Quantitative Polymerase Chain Reaction
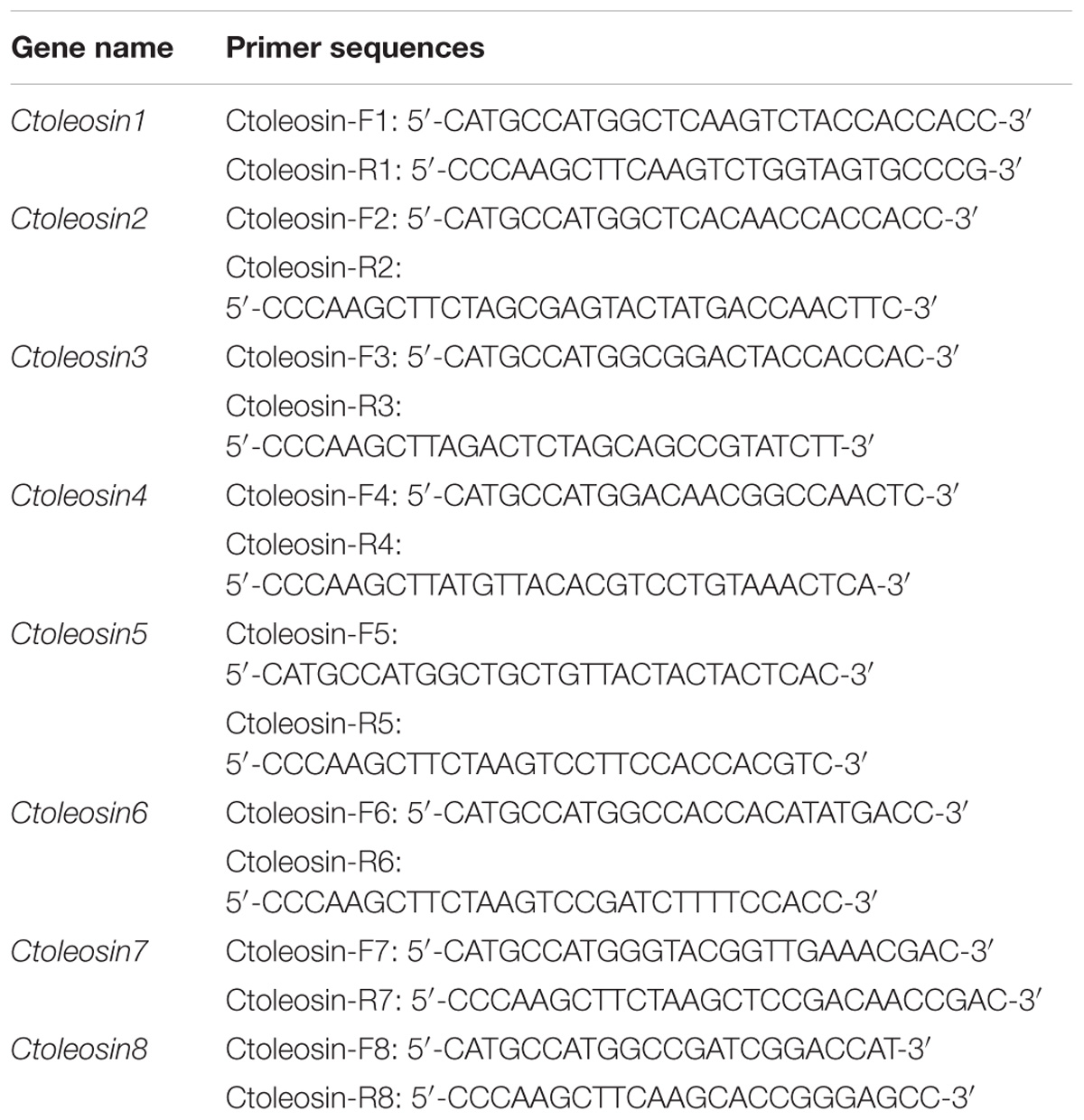
Reverse transcription quantitative polymerase chain reaction (RT-qPCR) was carried out using the target gene-specific primers (Table 1) and SYBR Premix Ex TaqTM kit (Takara, Japan) on Stratagene Mx3000P thermocycler. The six housekeeping reference genes ACT, EF1a, GAPDH, UBI, TUA, and TUB were selected as references for expression analysis in different tissues, and the stability of their expression was evaluated by geNorm and NormFinder software. Ctoleosin genes were cloned using a template of cDNA through gene-specific primers. Each reaction was performed in 15 μL reaction mixtures, containing 7.5 μL SYBR Premix Ex Taq, 0.3 μL ROX Reference Dye, 0.3 μL of each gene-specific Primer, 1.5 μL of cDNA, and 5.1 μL ddH2O. The PCR profile was set as follows: pre-denaturation at 95°C for 5 min; followed by 40 cycles of 95°C for 20 s and annealing at 62°C for 30 s. The fold-change in relative expression level was calculated using the 2−ΔΔCT method.
Construction of Over Expression Vector
Ctoleosin cDNA was amplified by PCR from safflower using gene-specific primers (Table 2) with NcoI and HindIII restriction sites, and the PCR product was cloned into the respective site of vector pOTB, which was supplied by the Jilin Agricultural University, China. The pOTB-Ctoleosins recombinant plasmids were created, which included the phaseolin promoter, Ctoleosin genes, the phaseolin terminator, CaMV35S promoter, bar gene as the selection marker gene, and NOS terminator. The binary vectors pOTB-Ctoleosins were further verified by PCR and NcoI/HindIII restriction analysis, and then transformed into Agrobacterium tumefaciens EHA105 competent cells (Hofgen and Willmitzer, 1988; Bailey et al., 2009). The recombinant Agrobacterium lines were used to transform Arabidopsis plants.
Generation of Transgenic Arabidopsis
Arabidopsis seeds were sown in soil, grown for 2 days in the dark and then kept in 16 h photoperiod at 23°C after germination. After 40 days growth, the plants were transformed through the floral dip method and then were harvested for T1 seeds, which was done using 1% basta. The T2 seeds were obtained and bred sequentially until T3 transgenic seeds were harvested.
Analysis of Morphological Phenotypes
The morphological phenotypes of transgenic Arabidopsis were photographed by a digital camera (Nikon, Tokyo, Japan) and an inverted microscope (Olympus IX51, Japan). The phenotypes of leaves, siliques, and seeds of Arabidopsis were analyzed from the captured images. The silique lengths, 100-grain weights, and germination rates were measured and statistically analyzed. The above experiments were performed for three biological replicates. Statistical analysis was assessed using the one-way Analysis of Variance (ANOVA), significance level p < 0.05∗, p < 0.01∗∗ and p < 0.001∗∗∗.
Purification of Arabidopsis Oil Bodies
Arabidopsis seeds (20 mg) were initially soaked in deionized water (1:5) for 24 h, transferred to 200 μL phosphate-buffered saline (PBS, pH 7.5) and finely ground with a mortar and pestle (Yang et al., 2017). The mixture was filtered, re-dispersed in PBS at pH 7.5, followed by centrifugation at 12,000 × g for 20 min to remove the debris. The residual oil bodies were collected, dispersed in 200 μL PBS solution, and centrifuged at 12,000 × g and 4°C for 20 min to collect the oil bodies free of extraneous impurities. The recovered pure oil bodies were stored at 4°C.
Fluorescence Microscopy
The pure oil bodies were diluted using PBS and mixed prior to measurements to ensure their homogeneity. A stock solution of Nile Red (5 mg/L, Sigma, United States) was prepared in absolute ethyl alcohol. The oil body suspensions were stained with an aqueous solution of Nile Red (0.5 mg/L) to visualize neutral lipids and placed in dark for 30 min at 24°C. The stained oil bodies were observed at the magnification of 40× under the fluorescence microscope.
Measurement of the Oil Body Diameter
The oil bodies were diluted with deionized water to an oil body content of 0.01 weight percentage. The diameter of the oil body was measured by a laser light scattering instrument (PSS NiComp 380ZLS, United States) which ranged from 0.001 to 5 μm. The SD values for oil body suspensions were calculated from every independent experiment. Statistical analysis of the measurements was performed using one-way ANOVA at ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Lipid Content Detection
For the measurement of lipid content, we added 2 mL of methanol and 4 mL of chloroform to 30 mg of the ground dry seed powder. We mixed the solution for 2 min and treated with ultrasonication for 30 min. The supernatant liquid was transferred to a 20 mL test tube, and 4 mL of trichloromethane solution was added to it. The mixture was centrifuged for 5 min at 12000 × g and the supernatant was collected. Chloroform methanol solution was added to the supernatant and set for 30 min. Finally, the oil layer was collected into a glass vial. The weights of oil and glass vial were determined separately. The formula for measuring oil content was as follows:
where, R is the oil content (%), m1 is the total weight of oil and glass vial, m2 is the weight of glass vial, and m is the weight of dry seed powders. Statistical analysis was assessed using the one-way ANOVA at ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Statistical Analysis
These experiments were performed on three biological replicates and the results were visualized using the GraphPad Prism 6.01 software (Inc., La Jolla, CA, United States). Statistical analysis of the measurements was performed using the one-way ANOVA at ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Results
In silico Analysis of the Identified Ctoleosin Gene Family
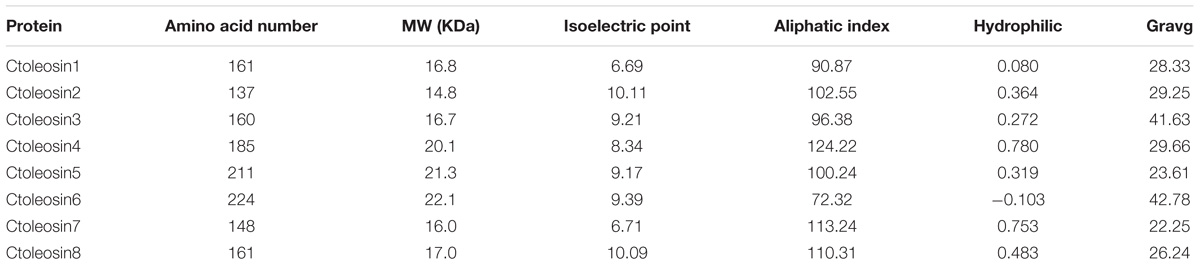
Ctoleosin genes were searched in safflower genome database. They were named as Ctoleosin1 to 8 according to their homology. Their predicted molecular weights were 16.8, 14.8, 16.7, 20.1, 21.3, 22.1, 16.0, and 17.0 kDa. Their theoretical isoelectric points were 6.69, 10.11, 9.21, 8.34, 9.17, 9.39, 6.71, and 10.09. The stability index ranged from 23.61 to 42.78 and the aliphatic index from 90.87 to 113.24 (Table 3). Most of the Ctoleosins were concentrated in the endoplasmic reticulum, whereas a few Ctoleosin proteins were found in the cytoplasm, Golgi apparatus, and plasma membrane. There were two transmembrane regions in Ctoleosin5 and 6, whereas the rest had three transmembrane regions in Figure 2B.
Phylogenetic and Structural Domain Analysis of Ctoleosin
Using MEGA5.1, we generated a phylogenetic tree from the aligned Ctoleosin sequences of Arabidopsis, Helianthus annuus, Zea mays L., Brassica oleracea L., and Oryza sativa to reveal their evolutionary relationship (Figure 1). Ctoleosin1 showed higher homology with the 14.9 kDa oleosin of Brassica oleracea. Ctoleosin2 and 3 were similar to 18.5 kDa oleosin of Brassica oleracea. Ctoleosin4 was similar to oleosin1-like of Brassica oleracea. Ctoleosin5 showed more similarity with 16.4 kDa oleosin of Helianthus annuus. Ctoleosin7 and 8 were similar to 16 kDa oleosin of Helianthus annuus. Ctoleosin proteins had the same structure as that of the other species; for instance, a hydrophilic N-terminal variable region, a characteristic conserved hydrophobic region, contained a typical “proline knot” motif (PX5SPX3P), and an amphoteric C-terminal variable region (Figures 2A,C).
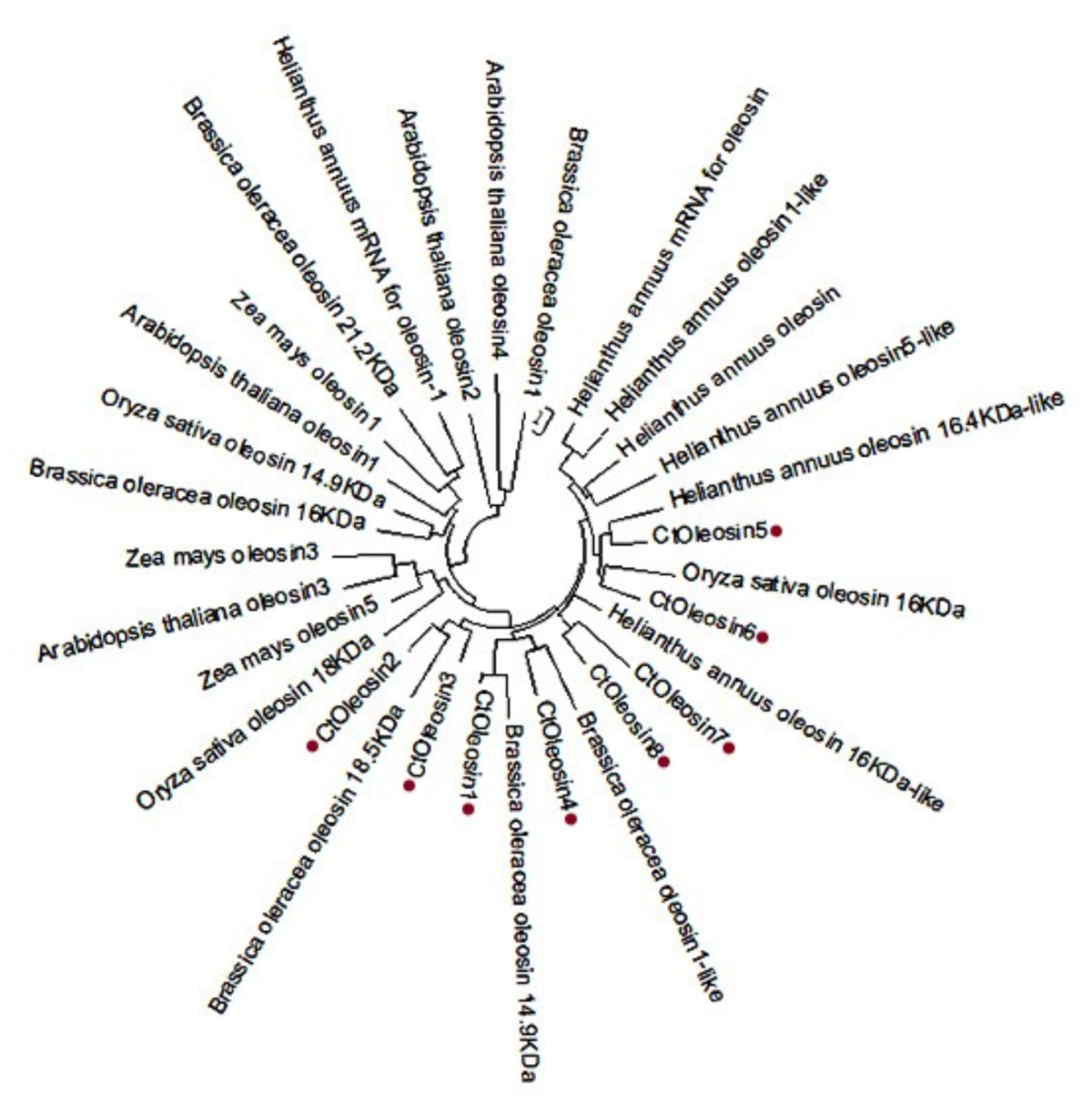
FIGURE 1. Phylogenetic tree of oleosin genes includes the Ctoleosin genes and oleosin genes from Arabidopsis, Helianthus annuus, Zea mays L., Brassica oleracea L., and Oryza sativa.
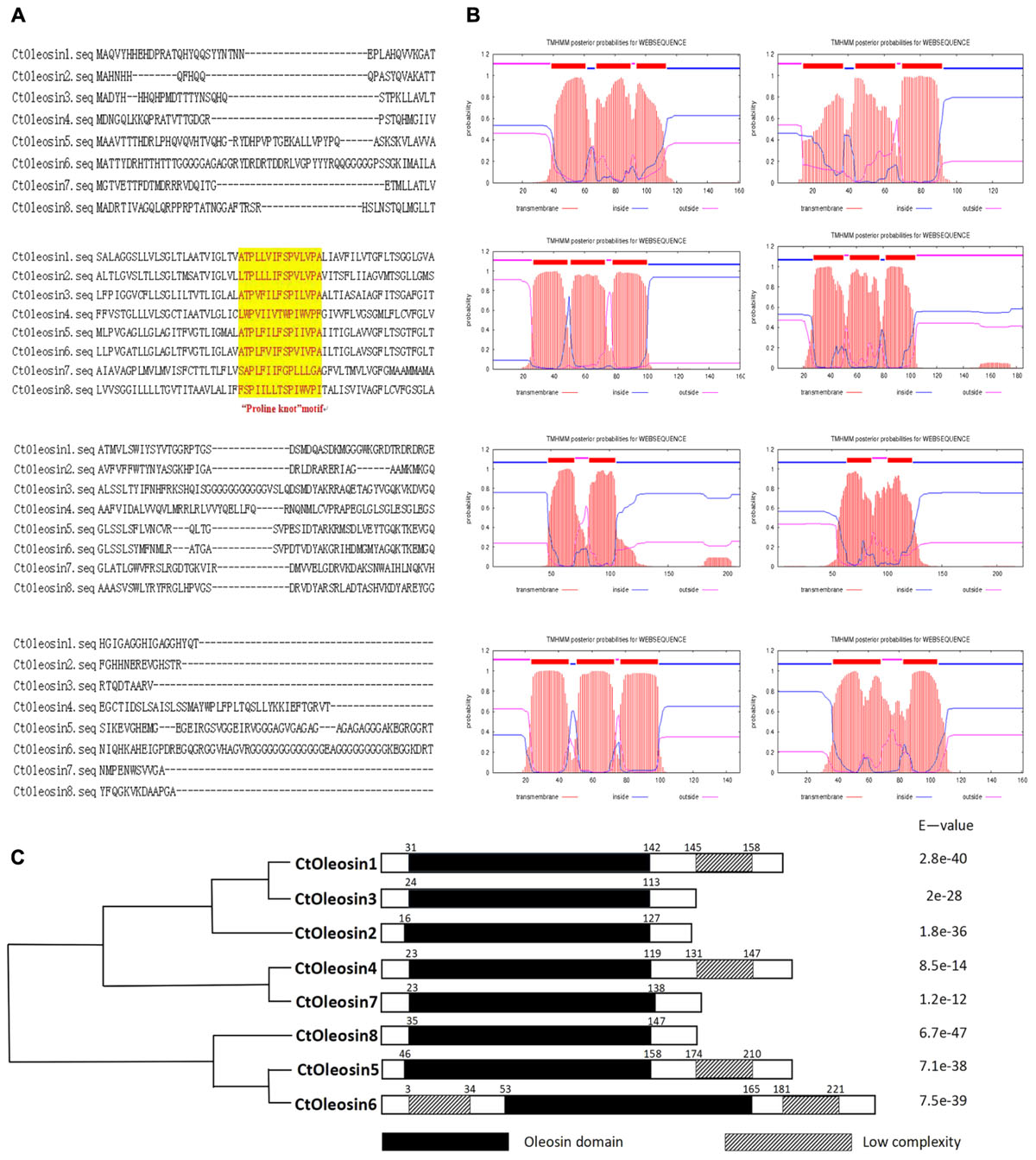
FIGURE 2. Domain analysis of Ctoleosins. (A) Structural domain analysis of the 8 Ctoleosin proteins revealed that they all possess the typical proline knot motif (shaded in yellow). (B) Transmembrane domain analysis predicted two transmembrane regions in Ctoleosin5 and 6 while the rest Ctoleosins had three regions. (C) Conserved oleosin domain (PF01277) analysis of 8 Ctoleosins unveiled that all of them are consistent with the oleosin domain.
RT-qPCR Deciphered the Spatiotemporal Expression Pattern of Ctoleosins in Safflower Tissues
RT-qPCR was performed to determine the transcript levels of Ctoleosin genes in various tissues of safflower. The most stable reference gene screened by geNorm software was EF1α. Ctoleosin genes were barely expressed in roots, stems, cotyledons, hypocotyls, and leaves, but their expression level was higher in flowers and seeds (Figure 3). The expression level of Ctoleosin1 in flowers was 1.29-, 18.7-, 5.95-, 2.45-, 4.68-, 1.95-, and 3.74-fold higher than that of Ctoleosin2, 3, 4, 5, 6, 7, and 8, respectively. The expression of Ctoleosin1, 5, 6, and 7 was higher than that of Ctoleosin2, 3, 4, and 8 on DAF 32.
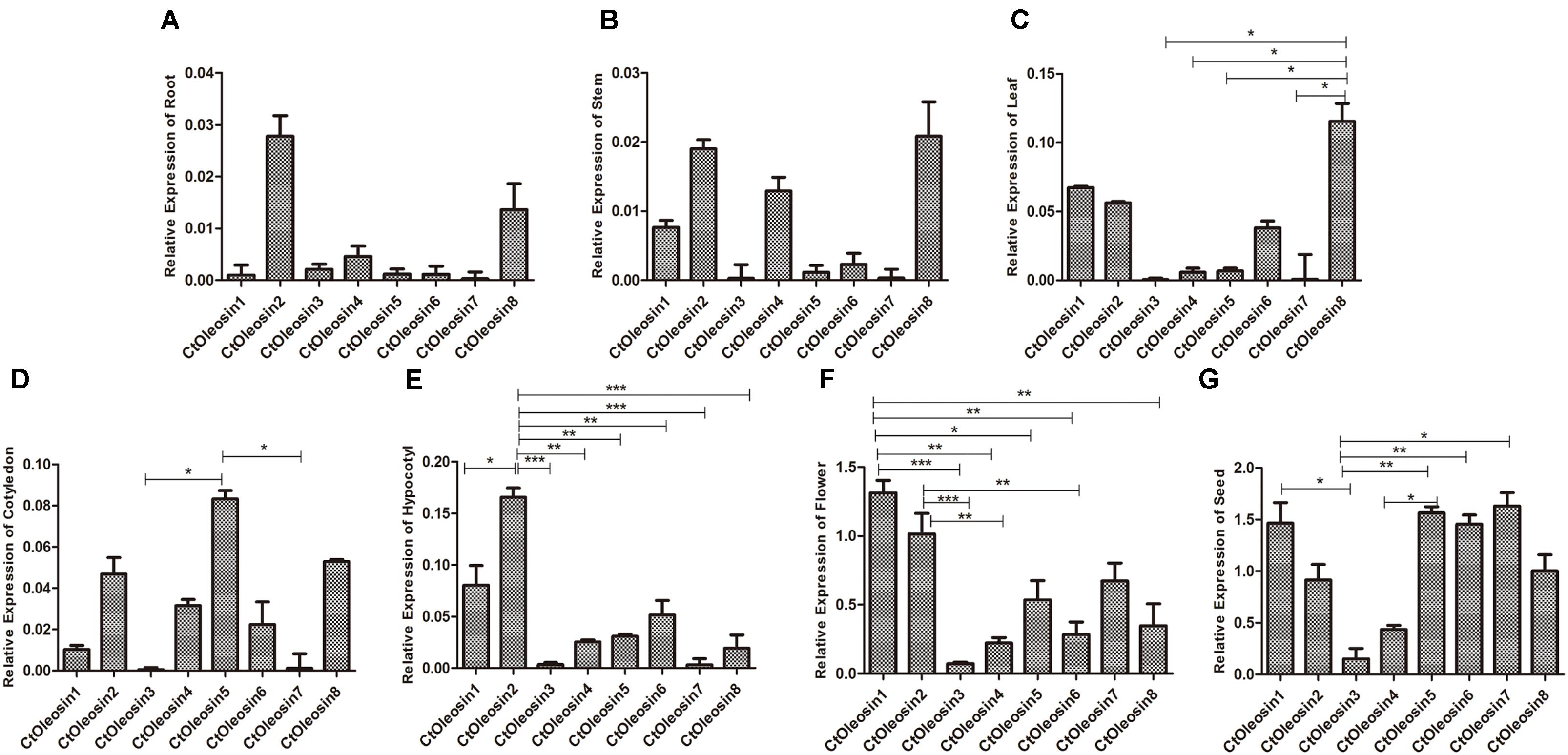
FIGURE 3. Expression of Ctoleosin genes in various tissues. The relative expression levels of Ctoleosin genes in root (A), stem (B), leaf (C), cotyledon (D), hypocotyl (E), flower (F) and seed (G), which were shown in Y-axis, were compared with various tissues. In all graphs, values are average of three biological replicates ± SD, different asterisks indicate significant different applying ANOVA (∗p < 0.05; ∗∗p < 0.01; and ∗∗∗p < 0.001).
Ctoleosin Genes Expressed in Different Stages After Flowering of Safflower
We analyzed the expression pattern of Ctoleosin genes in different stages after flowering on days 4, 8, 12, 16, 20, 24, 28, and 32. Ctoleosin genes were expressed in all the eight developing stages studied, but their transcript levels were lower at the initial stage of endosperm formation. As the seeds matured, the transcript levels of Ctoleosin genes increased progressively and peaked on DAF 28; thereafter, they declined slightly on DAF 32 (Figure 4). The expression level of Ctoleosin5 was reached 1.08-, 1.13-, 1.17-, 1.21-, 1.22-, 1.14-, and 1.23-fold of that of Ctoleosin1, 2, 3, 4, 6, 7, and 8, respectively, on DAF 28.
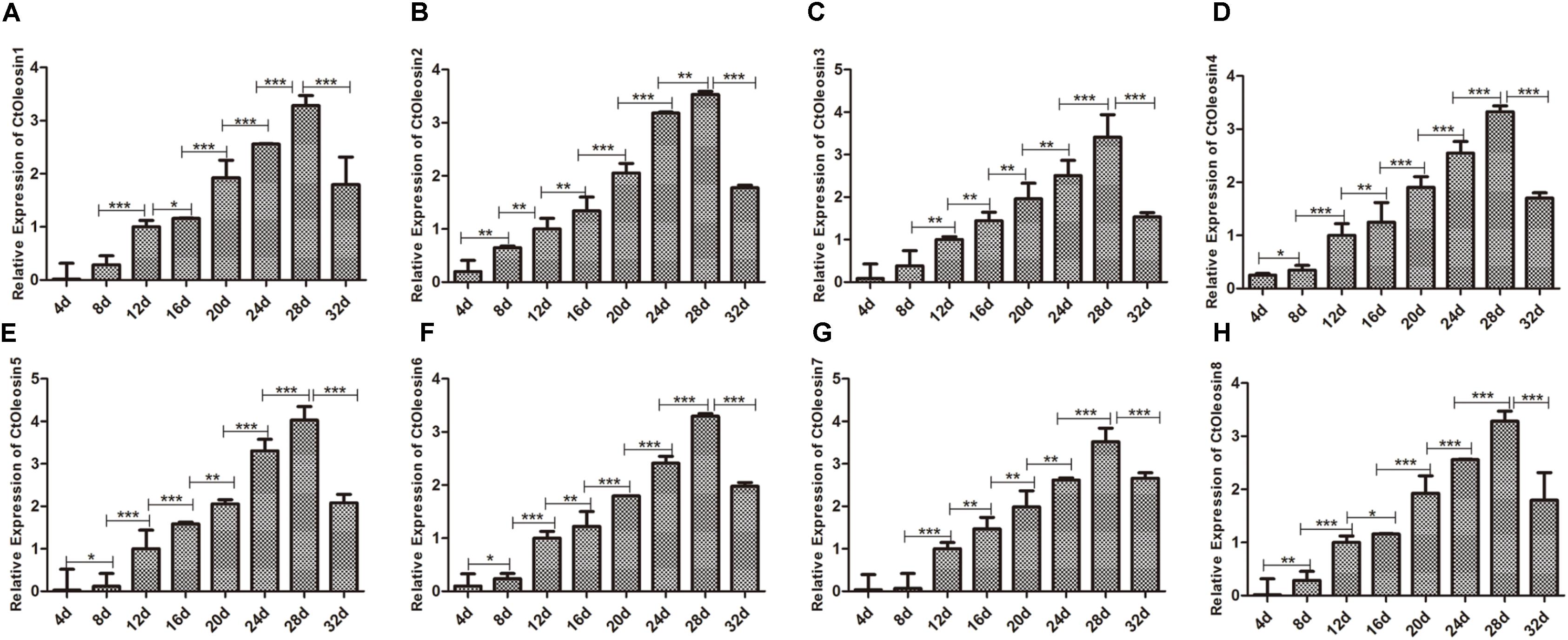
FIGURE 4. Expression of Ctoleosin genes in safflower at different stages after flowering. The relative expression levels of Ctoleosin1 (A), 2 (B), 3 (C), 4 (D), 5 (E), 6 (F), 7 (G), and 8 (H) during different stages after flowering. Values are average of three replicates ± SD. Asterisks indicate significant difference applying ANOVA (∗p < 0.05; ∗∗p < 0.01; and ∗∗∗p < 0.001).
Ctoleosin Transgene Overexpression in Transgenic Arabidopsis Seeds
Higher transcript levels of Ctoleosin were found in transgenic seeds than that in the seeds of wild-type Arabidopsis (Figure 5). Transcript level of Ctoleosin2 was the highest, followed by that of Ctoleosin7, 3, 4, and 5 in transgenic Arabidopsis seeds. Expression levels of Ctoleosin1 and 6 were the lowest in transgenic Arabidopsis seeds. Ctoleosin genes were not expressed in wild-type Arabidopsis seeds.
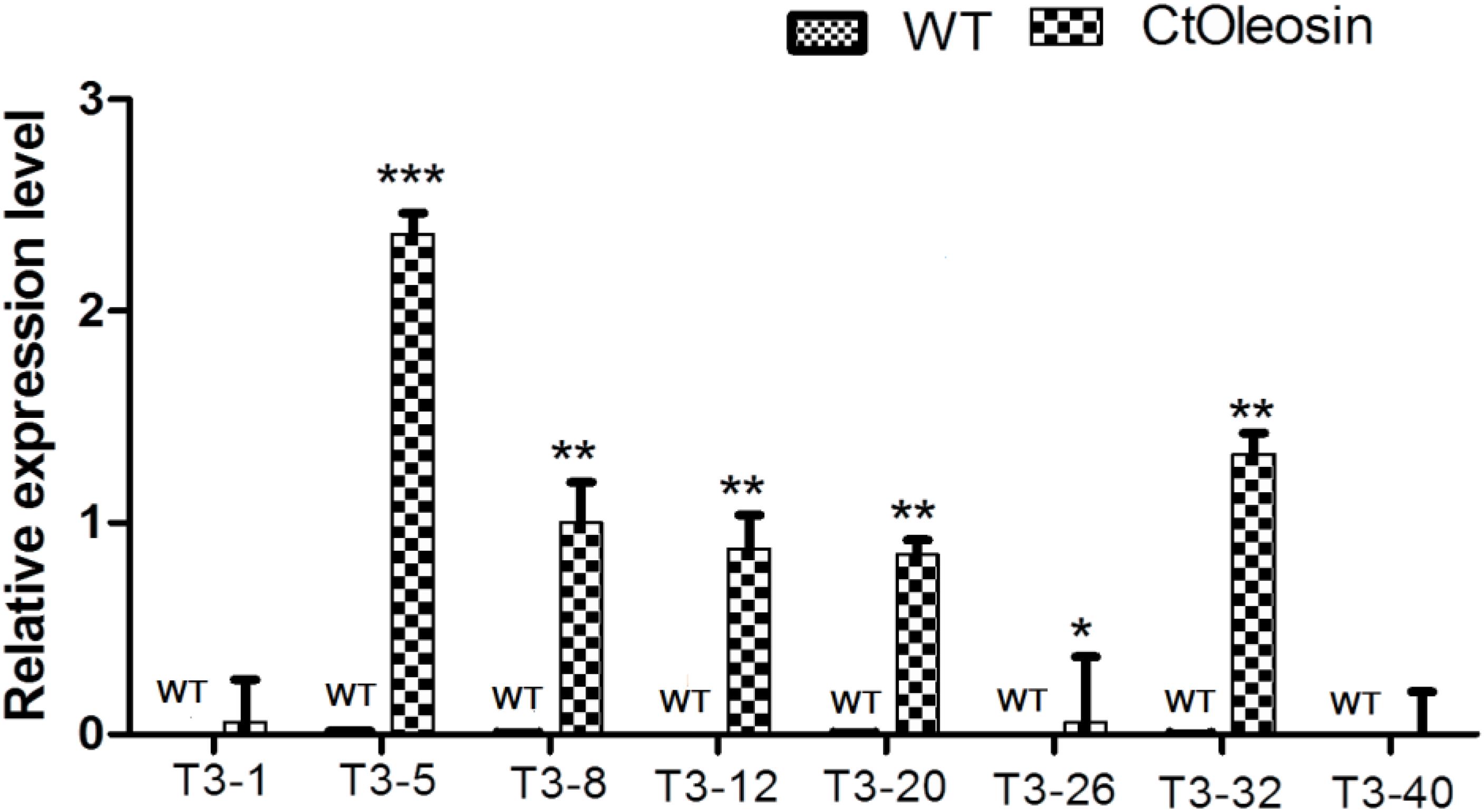
FIGURE 5. The expression analysis of Ctoleosin genes in wild-type Arabidopsis seeds and transgenic Arabidopsis seeds (Ctoleosin1, 2, 3, 4, 5, 6, 7, and 8). Values are average of three biological replicates ± SD. T3-1: T3 transgenic line for Ctoleosin1; T3-5: T3 transgenic line for Ctoleosin2; T3-8: T3 transgenic line for Ctoleosin3; T3-12: T3 transgenic line for Ctoleosin4; T3-20: T3 transgenic line for Ctoleosin5; T3-26: T3 transgenic line for Ctoleosin6; T3-32: T3 transgenic line for Ctoleosin7; and T3-40: T3 transgenic line for Ctoleosin8. Asterisks indicate significant difference applying ANOVA (∗P < 0.05; ∗∗P < 0.01; and ∗∗∗P < 0.001).
Ctoleosin Affected Seed Germination in Arabidopsis
The eight Ctoleosin genes were overexpressed in Arabidopsis under the seed-specific promoter of Phaseolus vulgaris. The basta-resistant Arabidopsis plants were further validated by PCR with genomic DNA as a template. Fifteen transgenic plants were selected and eight transgenic homozygous lines, whose Ctoleosin transcript levels were higher than other seven transgenic plants, were used for morphological observations. As compared with the wild type Arabidopsis, leaf size was not significantly altered in the transgenic Arabidopsis lines (Figure 6A), and silique length increased only in the lines expressing Ctoleosin3, 4, and 5 (Figures 6B,D). In addition, seed quantity was not significantly altered (Figure 6C). The 100-grain weight of transgenic seeds was slightly more than that of the wild-type Arabidopsis (Figure 6E). The seed germination rates of Ctoleosin4- and 6-overexpressing lines were slightly lower than those of the wild-type Arabidopsis. In contrast, the germination rates of the lines overexpressing Ctoleosin1–3, 5, and 7 were higher than of the wild-type Arabidopsis, whereas the germination rate of Ctoleosin8 did not differ from that of the wild-type (Figure 6F).
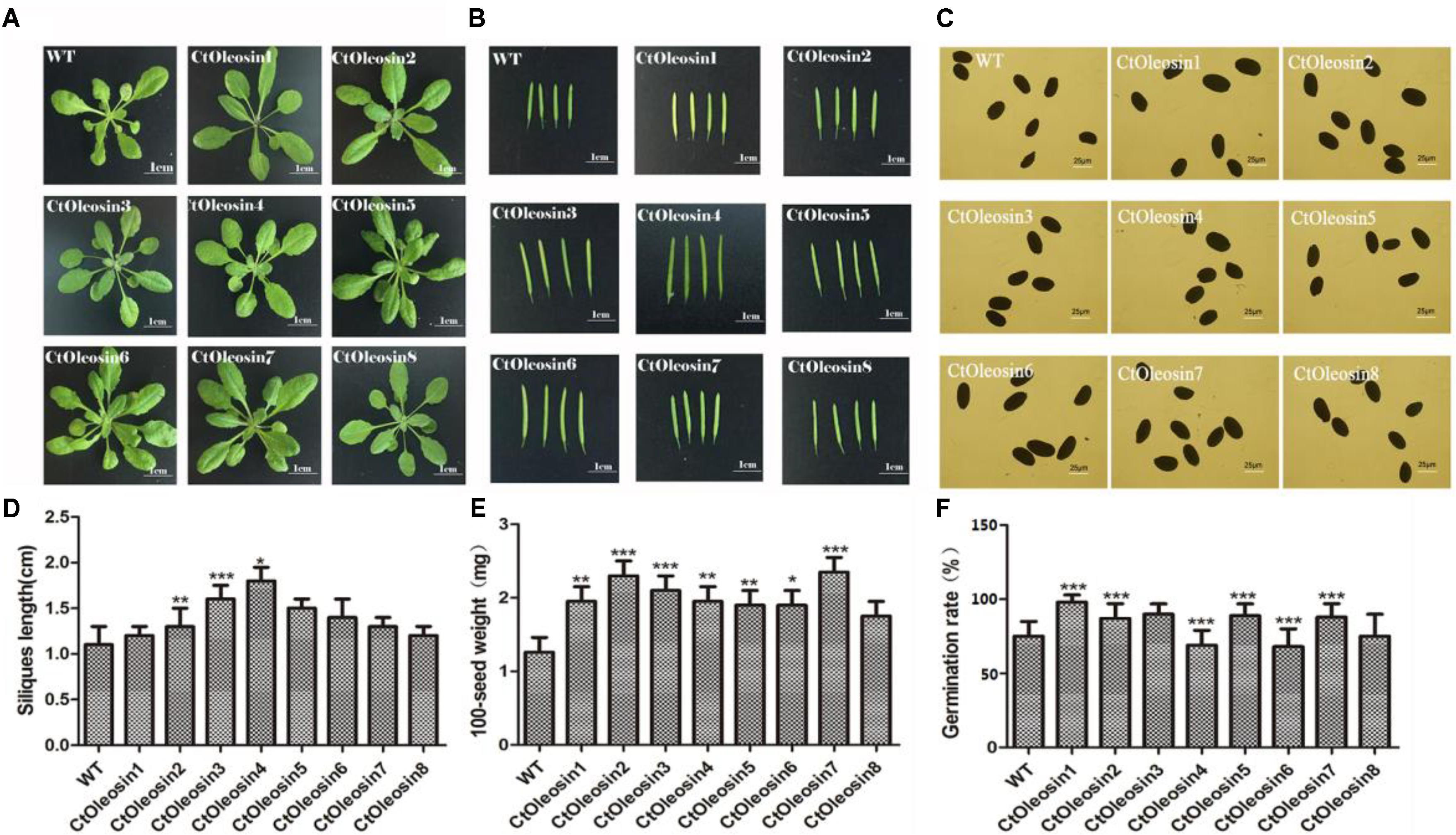
FIGURE 6. Overexpressing Ctoleosin genes in Arabidopsis affected tissue development of transgenic plants. (A) Leave; (B) siliques; (C) seeds; (D) the siliques length of the transgenic Arabidopsis; (E) The 100-grain weight of the transgenic seeds; and (F) The germination rate of transgenic seeds. Values are average of three biological replicates ± SD. Asterisks indicate significant difference applying ANOVA (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001).
Ctoleosin Genes Altered Oil Body Size and Oil Content in Arabidopsis Seeds
In order to demonstrate that oleosin can regulate the size of the oil body in seeds, we determined the diameter of oil bodies. The oil body diameter was 5.6 μm in wild-type Arabidopsis, whereas they were 2.1, 1.9, 4.5, 3.7, 2.5, 3.5, 2.4, and 3.7 μm, respectively, in lines expressing Ctoleosin1 through Ctoleosin8 (Figure 7B). The diameter of the oil bodies in transgenic Arabidopsis seeds was less than that of the wild-type plants. The oil bodies were spherical and dispersed uniformly in transgenic seeds (Figure 7A). The oil content of transgenic seeds was higher than that of the wild-type seeds. The oil content of lines expressing Ctoleosin2 and 3 was higher than that of the other transformants (Figure 7C). Accumulation of Ctoleosin not only determined the size of the oil bodies in seeds, but also regulated their oil content.
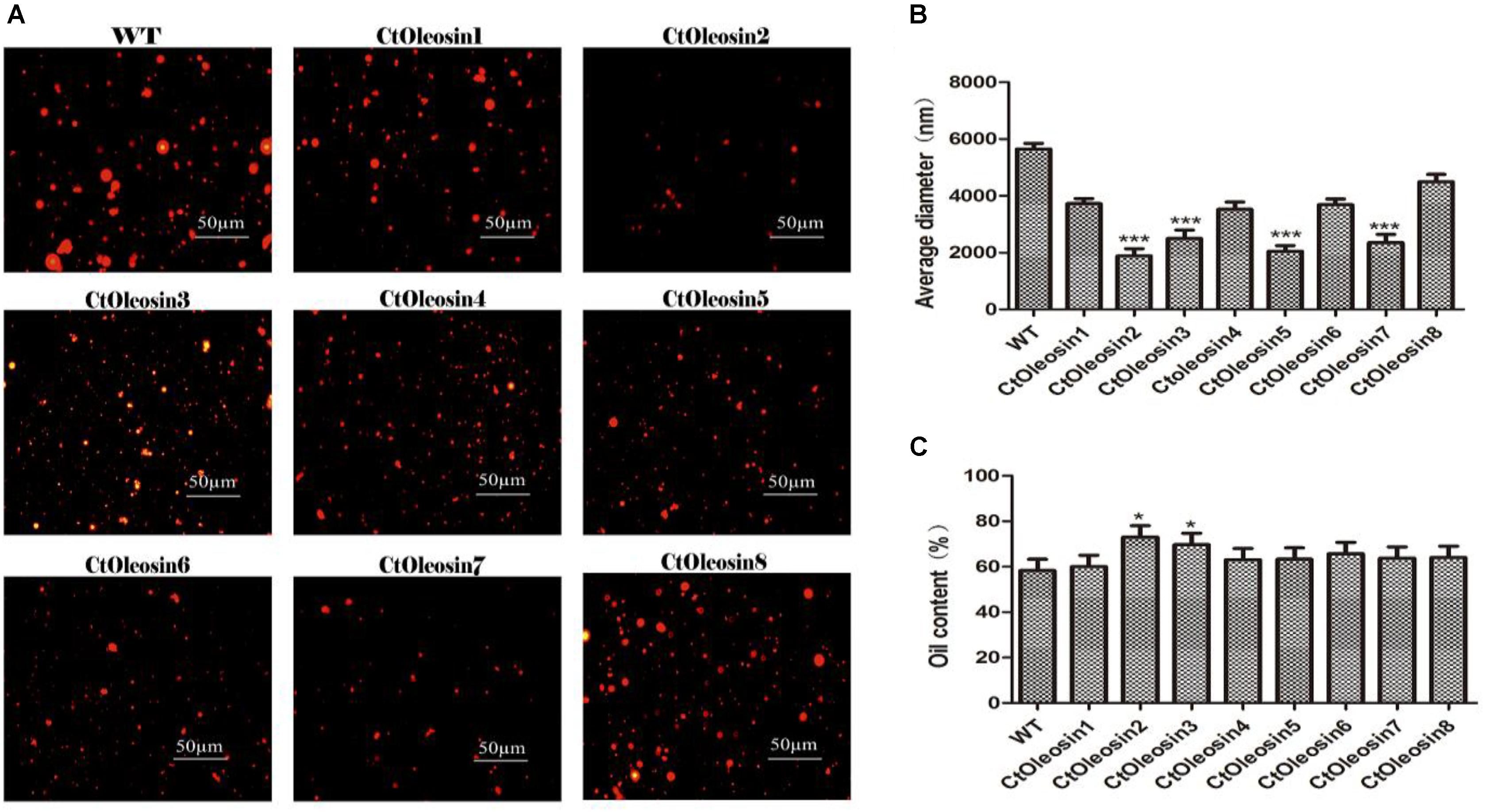
FIGURE 7. Ctoleosin genes altered the oil body size in transgenic Arabidopsis seeds. (A) The oil body structure in transgenic seeds under the microscope; (B) The particle sizes of oil bodies in transgenic seeds. Hydrodynamic diameter expressed by average diameter, size distribution. The average of triplicates was obtained by software. (C) The oil content of transgenic seeds. Values are average of three biological replicates ± SD. Asterisks indicate significant difference applying ANOVA (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001).
Discussion
Oilseeds store lipids in oil bodies, which are relatively simple organelles, consisting of a matrix of TAG coated with a phospholipid monolayer embedded with oleosins (Siloto et al., 2006). Oil bodies are formed via an endoplasmic reticulum (ER)-budding process during seed development (Sarmiento et al., 1997; Hsieh and Huang, 2004; Wu et al., 2010) and they are detected as early as the heart stage of embryo development (Siloto et al., 2006; Gallardo et al., 2016). Besides seeds, oil bodies are found in many different tissues and organs (Siloto et al., 2006; Song et al., 2017). Oleosins appear to play an important role in oil seeds, which are the major proteins associated with oil bodies, usually present as two or more isoforms. They have similar structural properties that include a long hydrophobic core organized around a proline knot (Abell et al., 1997). A high degree of similarity is present in but not restricted to the hydrophobic domain and proline knot motif, both of which are essential for the correct targeting of the oil body (van Rooijen and Moloney, 1995; Abell et al., 1997). Ctoleosins have a hydrophobic domain and proline knot motif, and this structure can stabilize the oil body. It is presumed that oleosins accumulate throughout seed development (Siloto et al., 2006; Song et al., 2017), and that they stabilize the oil body by steric hindrance and electronegative repulsion (Tzen et al., 1993; Wu et al., 2010). Moreover, they prevent oil body coalescence during the process of seed maturation and affect the final size of oil bodies (Cummins et al., 1993; Leprince and Hoekstra, 1998; Schmidt and Herman, 2008). Siloto et al. have clearly demonstrated that oleosin accumulation regulates the size of oil body by studying the effect of reduced oleosin accumulation on seed germination and TAG accumulation (Siloto et al., 2006). Oleosin silencing in Arabidopsis resulted in the formation of enlarged oil bodies as compared with that of the wild-type plants (He and Wu, 2009). It has been suggested that the size of the oil bodies is controlled by the relative contents of the oleosin (Ting et al., 1996; Shimada et al., 2008; Wu et al., 2010).
In previous studies, when the major oleosin was suppressed in Arabidopsis seeds, oil bodies were found to be larger and TAG accumulation levels were reduced (Siloto et al., 2006; Shimada et al., 2008). Several lines of evidence support this statement, including the variability of oleosins and oil body size in maize lines with different oil content (He and Wu, 2009; Wu et al., 2010). In the present study, we verified that Ctoleosin genes decrease oil body sizes and alter oil content by overexpressing the genes of the Ctoleosin family in transgenic Arabidopsis seeds. These results indicate that Ctoleosin genes play an important role in altering oil body size and oil content. Simultaneously, the introduction of exogenous oleosin in the transgenic lines indicates that it can replace the modified oleosin on the surface of oil body. Replacing natural oleosins by recombinant modified oleosin can provide novel insights into the targeting mechanisms, TAG sequestration in oil bodies, and in vivo features of the oil body surface (Siloto et al., 2006). This is certainly useful in many studies on oleosin function and molecular interactions. The use of modified oleosin as oil body platforms for the production of recombinant exogenous proteins has wide prospects of application.
Author Contributions
JY and HaiL conceived and designed the experiments. YL and MC conducted most of the experiments. KJ, YY, MN, HaoL, WQ, XL, and LD participated in experiments and data collection. All authors read and approved the final manuscript.
Funding
This research was financially supported by National Natural Science Foundation of China (No. 31801396).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
- ^ http://www.expasy.org/
- ^ http://www.cbs.dtu.dk/Services/TMHMM
- ^ http://pfam.xfam.org/
- ^ http://smart.embl-Heidelberg.de/
- ^ https://www.ncbi.nlm.nih.gov/
- ^ http://meme-suite.org/tools/meme
References
Abell, B. M., Holbrook, L. A., Abenes, M., Murphy, D. J., Hills, M. J., and Moloney, M. M. (1997). Role of the proline knot motif in oleosin endoplasmic reticulum topology and oil body targeting. Plant Cell. 9, 1481–1493. doi: 10.1105/tpc.9.8.1481
Alexander, L. G., Sessions, R. B., Clarke, A. R., Tatham, A. S., Shewry, P. R., and Napier, J. A. (2002). Characterization and modelling of the hydrophobic domain of a sunflower oleosin. Planta 214, 546–551. doi: 10.1007/s004250100655
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, 202–208. doi: 10.1093/nar/gkp335
Beaudoin, F., and Napier, J. A. (2000). The targeting and accumulation of ectopically expressed oleosin in non-seed tissues of Arabidopsis thaliana. Planta 210, 439–445. doi: 10.1007/PL00008152
Chapman, K. D., Dyer, J. M., and Mullen, R. T. (2012). Biogenesis and functions of lipid droplets in plants: thematic review series: lipid droplet synthesis and metabolism: from yeast to man. J. Lipid Res. 53, 215–226. doi: 10.1194/jlr.R021436
Cummins, I., Hills, M. J., Ross, J. H., Hobbs, D. H., Watson, M. D., and Murphy, D. J. (1993). Differential, temporal and spatial expression of genes involved in storage oil and oleosin accumulation in developing rapeseed embryos: implications for the role of oleosins and the mechanisms of oil-body formation. Plant Mol. Biol. 23, 1015–1027. doi: 10.1007/BF00021816
David, A., Yadav, S., and Bhatla, S. C. (2013). “Plant oil bodies and oleosins: structure, function and biotechnological applications,” in Bionanotechnology: Biological Self-Assembly and its Applications, ed. B. H. A. Rehm (Poole: Caister Academic Press).
Frandsen, G. L., Mundy, J., and Tzen, J. T. C. (2001). Oil bodies and their associated proteins, oleosin and caleosin. Physiol. Plant. 112, 301–307. doi: 10.1034/j.1399-3054.2001.1120301.x
Gallardo, K., Jolivet, P., Vernoud, V., Canonge, M., Larre, C., and Chardot, T. (2016). “Storage cells-oil and protein bodies,” in Molecular Cell Biology of the Growth and Differentiation of Plant Cells, ed. R. J. Rose (London: CRC Press), 362–381.
Hanano, A., Burcklen, M., Flenet, M., Ivancich, A., Louwagie, M., Garin, J., et al. (2006). Plant seed peroxygenase is an original heme-oxygenase with an EF-hand calcium binding motif. J. Biol. Chem. 281, 33140–33151. doi: 10.1074/jbc.M605395200
He, Y. Q., and Wu, Y. (2009). Oil body biogenesis during Brassica napus embryogenesis. J. Integr. Plant Biol. 51, 792–799. doi: 10.1111/j.1744-7909.2009.00851
Hofgen, R., and Willmitzer, L. (1988). Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res. 16:9877. doi: 10.1093/nar/16.20.9877
Hsieh, K., and Huang, A. H. (2004). Endoplasmic reticulum, oleosins and oils in seeds and taperum cells. Plant Physiol. 136, 3427–3434. doi: 10.1104/pp.104.051060
Huang, A. H. (1992). Oil bodies and oleosins in seeds. Annu. Rev. Plant Biol. 43, 177–200. doi: 10.1146/annurev.pp.43.060192.001141
Huang, A. H. C. (1996). Oleosin and oil bodies in seeds and other organs. Plant Physiol. 110, 1055–1061. doi: 10.1104/pp.110.4.1055
Hyun, T. K., Kumar, D., Cho, Y. Y., and Hyun, H. N. (2013). Computational identification and phylogenetic analysis of the oil-body structural proteins, oleosin and caleosin in castor bean and flax. Gene 515, 454–460. doi: 10.1016/j.gene.2012.11.065
Jolivet, P., Acevedo, F., Boulard, C., d’Andréa, S., and Faure, J. D. (2013). Crop seed oil bodies: from challenges in protein identification to an emerging picture of the oil body proteome. Proteomics 13, 1836–1849. doi: 10.1002/pmic.201200431
Jolivet, P., Boulard, C., Bellamy, A., Larré, C., Barre, M., Rogniaux, H., et al. (2009). Protein composition of oil bodies from mature Brassica napus seed. Proteomics 9, 3268–3284. doi: 10.1002/pmic.200800449
Keddie, J. S., Hubner, G., Slocomber, S. P., Jarvis, R. P., Cummins, L., Edwards, E. W., et al. (1992). Cloning and characterization of an oleosin gene from Brassica napus. Plant Mol. Biol. 19, 443–453. doi: 10.1007/BF00023392
Lacey, D. J., Wellner, N., Beaudoin, F., Napier, J. A., and Shewry, P. R. (1998). Secondary structure of oleosins in oilbodies isolated from seeds of safflower (Carthamus tinctorius, L.) and sunflower (Helianthus annuus, L.). J. Biol. Chem. 334, 469–477.
Leprince, O., and Hoekstra, F. A. (1998). The responses of cytochrome redox state and energy metabolism to dehydration support a role for cytoplasmic viscosity in desiccation tolerance. Plant Physiol. 118, 1253–1264. doi: 10.1104/pp.118.4.1253
Li, H., Dong, Y., Yang, J., Liu, X., Wang, Y., Yao, N., et al. (2012). De novo transcriptome of safflower and the identification of putative genes for oleosin and the biosynthesis of flavonoids. PLoS One 7:e30987. doi: 10.1371/journal.pone.0030987
Lin, L. J., and Tzen, J. (2004). Two distinct steroleosins are present in seed oil bodies. Plant Physiol. Biochem. 42, 601–608. doi: 10.1016/j.plaphy.2004.06.006
Murphy, D. J. (2001). The biogenesis and functions of lipid bodies in animals, plant and mircroorganisms. Prog. Lipid Res. 40, 325–438. doi: 10.1016/S0163-7827(01)00013-3
Murphy, D. J. (2012). The dynamic roles of intracellular lipid droplets: from archaea to mammals. Protoplasma 249, 541–585. doi: 10.1007/s00709-011-0329-7
Napier, J. A., Stobart, A. K., and Shewry, P. R. (1996). The structure and biogenesis of plant oil bodies: the role of the ER membrane and the oleosin class of proteins. Plant Mol. Biol. 31, 945–956. doi: 10.1007/BF00040714
Nikam, T. D., and Shitole, M. G. (1999). In vitro culture of safflower, L.cv. Bhima: initiation, growth optimization and organogenesis. Plant Cell Tissue Organ Cult. 55, 15–22. doi: 10.1023/A:1026493616991
Pasaribu, B., Chung, T. Y., Chen, C. S., Wang, S. L., Jiang, P. L., and Tzen, J. T. C. (2014). Identification of caleosin and two oleosin isoforms in oil bodies of pine megagametophytes. Plant Physiol. Biochem. 82, 142–150. doi: 10.1016/j.plaphy.2014.05.015
Peng, C. C., Lin, I. P., Lin, C. K., and Tzen, J. T. C. (2003). Size and stability of reconstituted sesame oil bodies. Biotechnol. Prog. 19, 1623–1626. doi: 10.1021/bp034129z
Purkrtova, Z., Jolivet, P., Miquel, M., and Chardot, T. (2008). Structure and function of seed lipid body associated proteins. C. R. Biol. 331, 746–754. doi: 10.1016/j.crvi.2008.07.016
Qu, R. D., and Huang, A. H. (1990). Oleosin kD 18 on the surface of oil bodies in maize. Genomic and cDNA sequences and the deduced protein structure. J. Biol. Chem. 265, 2238–2243.
Roux, E., Baumberger, S., Axelos, M. A., and Chardot, T. (2004). Oleosins of Arabidopsis thaliana: expression in Escherichia coli, purification, and functional properties. J. Agric. Food Chem. 52, 5245–5249. doi: 10.1021/jf0355046
Sarmiento, C., Ross, J. H. E., Herman, E., and MurPhy, D. J. (1997). Expression and subcellular targeting of a soybean oleosin in transgenie rapeseed. Implications for the mehanism of oil-body formation in seeds. Plant J. 11, 783–796. doi: 10.1046/j.1365-313X.1997.11040783.x
Schmidt, M. A., and Herman, E. M. (2008). Suppression of soybean oleosin produces micro-oil bodies that aggregate into oil body/ER complexes. Mol. Plant 6, 910–924. doi: 10.1093/mp/ssn049
Shimada, T. L., Shimada, T., Takahashi, H., Fukao, Y., and Nishimura, I. H. (2008). A novel role for oleosins in freezing tolerance of oilseeds in Arabidopsis thaliana. Plant J. 55, 798–809. doi: 10.1111/j.1365-313X.2008.03553.x
Siloto, R. M. P., Findlay, K., Lopez, V. A., Yeung, E. C., Nykifork, C. L., and Moloney, M. M. (2006). The accumulation of oleosins determines the size of seed oil bodies in Arabidopsis. Plant Cell 18, 1961–1974. doi: 10.1105/tpc.106.041269
Song, Y. H., Wang, X. D., and Rose, R. J. (2017). Oil body biogenesis and biotechnology in legume seeds. Plant Cell Rep. 36, 1519–1532. doi: 10.1007/s00299-017-2201-5
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 10, 2731–2739. doi: 10.1093/molbev/msr121
Ting, J. T. L., Lee, K., Ratnayake, C., Balsamo, K. A., and Huang, A. H. (1996). Oleosin genes in maize kernels having diverse oil contents are constitutively expressed independent of oil content. Size and shape of intracellular oil bodies are determined by the oleosins/oils ratio. Planta 199, 158–165. doi: 10.1007/BF00196892
Tzen, J. T. (2012). Integral proteins in plant oil bodies. Int. Sch. Res. Notices 3, 263270–263286. doi: 10.5402/2012/173954
Tzen, J. T., Lie, G. C., and Huang, A. H. (1992). Characterization of the charged components and their topology on the surface of plant seed oil seed oil bodies. J. Biol. Chem. 267, 15626–15634.
Tzen, J. T. C., Cao, Y. Z., Laurent, P., Ratnayake, C., and Huang, A. H. C. (1993). Lipids, proteins and structure of seed oil bodies from diverse species. Plant Physiol. 101, 267–276. doi: 10.1104/pp.101.1.267
van Rooijen, G. J., and Moloney, M. M. (1995). Structural requirements of oleosin domains for subcellular targeting to the oil body. Plant Physiol. 109, 1353–1361. doi: 10.1104/pp.109.4.1353
Wu, Y. Y., Chou, Y. R., Wang, C. S., Tseng, T. H., Chen, L. J., and Tzen, J. T. C. (2010). Different effects on triacyglycerol packaging to oil bodies in transgenic rice seeds by specifically eliminating one of their two oleosin isoforms. Plant Physiol. Biochem. 48, 81–89. doi: 10.1016/j.plaphy.2009.12.004
Xu, M. Y., Liu, D. H., and Li, G. Q. (2004). Cloning of soybean 24kD oleosin gene and its transient expression as a carrier for foreign protein. Agr. Sci. China 3, 321–329.
Keywords: oil body, Ctoleosin, expression, morphological phenotypes, oil content
Citation: Lu Y, Chi M, Li L, Li H, Noman M, Yang Y, Ji K, Lan X, Qiang W, Du L, Li H and Yang J (2018) Genome-Wide Identification, Expression Profiling, and Functional Validation of Oleosin Gene Family in Carthamus tinctorius L. Front. Plant Sci. 9:1393. doi: 10.3389/fpls.2018.01393
Received: 16 April 2018; Accepted: 03 September 2018;
Published: 18 October 2018.
Edited by:
Agnieszka Ludwików, Adam Mickiewicz University in Poznań, PolandReviewed by:
Rosemary White, Commonwealth Scientific and Industrial Research Organisation (CSIRO), AustraliaAbdulsamie Hanano, Atomic Energy Commission of Syria, Syria
Copyright © 2018 Lu, Chi, Li, Li, Noman, Yang, Ji, Lan, Qiang, Du, Li and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing Yang, eWFuZ2ppbmc1MTIyMDEwQDE2My5jb20= Haiyan Li, aHlsaTk5QDE2My5jb20=
†These authors have contributed equally to this work
 Yubin Lu†
Yubin Lu† Muhammad Noman
Muhammad Noman Jing Yang
Jing Yang