- 1Cereal Crops Research Unit, Red River Valley Agricultural Research Center, United States Department of Agriculture–Agricultural Research Service, Fargo, ND, United States
- 2Department of Plant Sciences, North Dakota State University, Fargo, ND, United States
- 3Plant Science Research Unit, United States Department of Agriculture–Agricultural Research Service, Raleigh, NC, United States
- 4Department of Plant Pathology, North Dakota State University, Fargo, ND, United States
- 5Wheat Health, Genetics, and Quality Research Unit, United States Department of Agriculture–Agricultural Research Service, Pullman, WA, United States
- 6Department of Plant Pathology, Kansas State University, Manhattan, KS, United States
Aegilops markgrafii (Greuter) Hammer is an important source of genes for resistance to abiotic stresses and diseases in wheat (Triticum aestivum L.). A series of six wheat ‘Alcedo’-Ae. markgrafii chromosome disomic addition lines, designated as AI(B), AII(C), AIII(D), AV(E), AIV(F), and AVIII(G) carrying the Ae. markgrafii chromosomes B, C, D, E, F, and G, respectively, were tested with SSR markers to establish homoeologous relationships to wheat and identify markers useful in chromosome engineering. The addition lines were evaluated for resistance to rust and powdery mildew diseases. The parents Alcedo and Ae. markgrafii accession ‘S740-69’ were tested with 1500 SSR primer pairs and 935 polymorphic markers were identified. After selecting for robust markers and confirming the polymorphisms on the addition lines, 132 markers were considered useful for engineering and establishing homoeologous relationships. Based on the marker analysis, we concluded that the chromosomes B, C, D, E, F, and G belong to wheat homoeologous groups 2, 5, 6, 7, 3, and 4, respectively. Also, we observed chromosomal rearrangements in several addition lines. When tested with 20 isolates of powdery mildew pathogen (Blumeria graminis f. sp. tritici) from five geographic regions of the United States, four addition lines [AIII(D), AV(E), AIV(F), and AVIII(G)] showed resistance to some isolates, with addition line AV(E) being resistant to 19 of 20 isolates. The addition lines were tested with two races (TDBJ and TNBJ) of the leaf rust pathogen (Puccinia triticina), and only addition line AI(B) exhibited resistance at a level comparable to the Ae. markgrafii parent. Addition lines AII(C) and AIII(D) had been previously identified as resistant to the Ug99 race group of the stem rust pathogen (Puccinia graminis f. sp. tritici). The addition lines were also tested for resistance to six United States races (PSTv-4, PSTv-14, PSTv-37, PSTv-40, PSTv-51, and PSTv-198) of the stripe rust pathogen (Puccinia striiformis f. sp. tritici); we found no resistance either in Alcedo or any of the addition lines. The homoeologous relationships of the chromosomes in the addition lines, molecular markers located on each chromosome, and disease resistance associated with each chromosome will allow for chromosome engineering of the resistance genes.
Introduction
Aegilops markgrafii (Greuter) Hammer (synonym Ae. caudata L., 2n = 2x = 14, genome CC), is one of the most important diploid wild relatives of wheat (Triticum aestivum L., 2n = 6x = 42, AABBDD genomes) because it carries resistance to powdery mildew [caused by Blumeria graminis f. sp. tritici (DC.) Speer], leaf rust (Puccinia triticina Erikss.), stem rust (Puccinia graminis Pers.: Pers. f. sp. tritici Eriks. and E. Henn.) and stripe rust (Puccinia striiformis Westend. f. sp. tritici Eriks.) (Valkoun et al., 1985; Dyck et al., 1990; Schubert and Blüthner, 1995; Xu et al., 2009; Weidner et al., 2012). A set of chromosome disomic addition lines carrying Ae. markgrafii accession ‘S 740-69’ chromosomes B, C, D, E, F, and G in wheat variety ‘Alcedo’ were developed by Schubert and Blüthner (1992, 1995). This set of disomic addition lines can serve as an alternate and direct genetic source for wheat germplasm enhancement. An addition line for chromosome A is absent from this set, and Niu et al. (2011) found that none of the six addition lines carried high-molecular-weight glutenins from Ae. markgrafii, suggesting that chromosome A may be homoeologous to group 1. Friebe et al. (1992) noted results from unpublished studies which support the conclusion that chromosome A belongs to group 1. Danilova et al. (2014, 2017) determined that chromosome A of Ae. markgrafii is homoeologous to the group 1 chromosomes of wheat by using fluorescence in situ hybridization (FISH) with cDNA probes.
Xu et al. (2009) identified two Alcedo-Ae. markgrafii S740-69 addition lines, AII(C) and AIII(D), that conditioned resistance to the Ug99 race group of the stem rust pathogen, the most virulent races appearing in Africa. To transfer these alien genes from the addition lines to wheat in a short period of time, detailed information concerning the homoeology between wheat and the added Ae. markgrafii chromosomes will be very useful. There are several ways to establish the homoeologous relationships between wheat and its wild relatives, including C-banding (Friebe et al., 1992), isozyme analysis (Schmidt et al., 1993), molecular marker analysis (Peil et al., 1998), sequential fluorescence in situ hybridization (FISH), and genomic in situ hybridization (GISH) (Xu et al., 2016). In addition, marker assisted selection has become a useful tool for the gene introgression process (Niu et al., 2011). Friebe et al. (1992) and Schmidt et al. (1993) used isozymes and the C-banding technique, respectively, to determine homoeologous relationships of the six addition lines, and determined that the chromosome in lines AII(C), AIII(D), and AIV(F) belonged to group 5, 6, and 3, respectively, but homoeologous relationships of chromosomes in lines AV(E), AI(B), and AVIII(G) were not clearly established. Peil et al. (1998) tested 88 SSR markers and identified only 20 that were useful to distinguish the Ae. markgrafii chromosomes; and because the marker number was less than 4 for each chromosome, the results did not indicate homoeology. In addition to homoeologous relationships of each added chromosome, knowledge of the Alcedo genetic background is needed. For example, Alcedo is a major donor of stripe rust resistance (Jagger et al., 2011), and in attempting to transfer stripe rust resistance from Ae. markgrafii, detailed information about Alcedo is important to ensure that the stripe rust resistance is from Ae. markgrafii and not Alcedo.
Simple sequence repeats (SSRs) have become very useful and desirable molecular markers because they are often codominant, highly reproducible, frequent in most eukaryotes, and have high allelic diversity (Mohan et al., 1997). With the development of sequencing technology, more and more SSRs (over 3000) are available for marker analysis in wheat, and many genetic maps featuring SSR markers are available for reference. Screening for polymorphisms between the parents using additional SSRs will help to determine the homoeologous relationships and the polymorphic markers can subsequently be used for marker-assisted gene introgression. Sequential FISH and GISH will produce additional chromosome constitution information for the addition lines. Our objectives in this study were to use additional SSR markers and sequential FISH and GISH to characterize Alcedo and its six Ae. markgrafii addition lines, determine the homoeologous relationships of the chromosomes, and develop useful SSR markers for marker assisted selection.
Materials and Methods
Plant Material
Wheat cultivar ‘Alcedo,’ Ae. markgrafii (Greuter) Hammer (accession S740-69), the Alcedo-Ae. markgrafii amphiploid (W0492), and six Alcedo-Ae. markgrafii S740-69 disomic addition lines AI, AII, AIII, AV, AIV, and AVIII carrying the Ae. markgrafii chromosomes B, C, D, E, F, and G, respectively (Schmidt et al., 1993) were used for this study. A line carrying chromosome A was not available for this study. The original seed stocks of these lines were kindly provided by Dr. Richard R.-C. Wang, USDA-ARS Forage and Range Research Laboratory, Logan, UT, United States.
Fluorescence in situ Hybridization
Root-tips of plants were prepared for FISH following the procedure described by Xu et al. (2016). This included pretreatment of root tips in ice water for 20–24 h, fixation in ethanol-acetic acid (3:1 ratio), pretreatment in 1% acetocarmine, and squashing on a slide using 45% acetic acid. Slides were examined to select samples with good preparations, and cover glasses were removed. Prepared slides were incubated in 100 μg/mL RNase in 2× saline sodium citrate (SSC) at 37°C for 1 h, then denatured in 70% formamide in 2× SSC at 72°C for 2 min followed by dehydration in a chilled graded ethanol series (70%, 95%, and 100%) at -20°C each for 5 min.
Multi-color FISH was carried out with two probes, pAS1 carrying about 1 kb of repeat sequence from Ae. tauschii Cosson (Rayburn and Gill, 1986) and labeled with digoxigenin-11-dUTP (Roche Diagnostics, Mannheim, Germany), and pSC119.2 carrying a highly repeated sequence from rye (Secale cereale L.) and labeled with biotin-16-dUTP (Enzo Life Sciences, Inc., Farmingdale, NY, United States). The two probes were equally mixed before hybridization and then added to the hybridization mix (15 μL formamide, 6 μL dextran sulfate, 3 μL 20× SSC, and 3 μL single stranded DNA). Fifteen microliters of hybridization mix were added to each slide and slides were covered with cover slips for incubation overnight. The slides were then washed as described by Xu et al. (2016). The fluorescent signals were detected with anti-digoxigenin-rhodamine (Roche Diagnostics) and fluorescein isothiocyanate-conjugated avidin (FITC-avidin) (Vector Laboratories, Inc., Burlingame, CA, United States) for both probes. The slides were mounted with VECTORSHIELD Antifade Mounting Medium (Vector Laboratories) containing 4′,6-diamidino-2-phenylindole (DAPI) (Sigma, St. Louis, MO, United States). The slides were examined under a Zeiss Axioplan 2 Imaging Research Microscope (Carl Zeiss Light Microscopy, Germany). The GISH images were captured using an Axiocam HRm CCD (charge-coupled device) camera (Carl Zeiss Light Microscopy) and analyzed using imaging software AxioVision Release 4.5 (Carl Zeiss Light Microscopy).
Genomic in situ Hybridization
After FISH, the slides were washed in 0.1× SSC with 0.5% formamide three times each for 10 min at 42°C, then 2× SSC twice each for 10 min, then in 4× SSC overnight at room temperature. The slides were sequentially dehydrated in 70%, 95%, and 100% ethanol each for 5 min. Total genomic DNA from Ae. markgrafii was used as probe and labeled with biotin-16-dUTP by nick translation (Enzo Life Sciences, Inc.). Sheared genomic DNA from Chinese Spring was used for blocking. Detailed procedures of the chromosome preparation and hybridization were previously described by Xu et al. (2016). GISH signals were detected with FITC-avidin (Vector Laboratories). The slides were mounted with VECTORSHIELD Antifade Mounting Medium (Vector Laboratories) containing propidium iodide (PI) (Vector Laboratories) and were observed under the Zeiss Axioplan 2 Imaging Research Microscope for GISH analysis as described above. Photographs were captured using the Axiocam HRm CCD camera and analyzed using the imaging software AxioVision Release 4.5 for GISH analysis as described above.
Karyotype Analysis of Ae. markgrafii Chromosomes
The chromosome spreads from GISH and FISH analyses were used for karyotypic analysis of each of the Ae. markgrafii chromosomes in the six addition lines. Each of the Ae. markgrafii chromosomes was measured for lengths of short and long arms from at least 20 cells using the “Measure Length” tool in the imaging software AxioVision Release 4.5 (Carl Zeiss Light Microscopy). Total length of each Ae. markgrafii chromosome was calculated by adding the averages of long and short arms. The arm ratio (long arm/short arm) of each Ae. markgrafii chromosome was calculated from the lengths of short and long arms.
SSR Marker Analysis
DNA extraction from fresh leaves and SSR marker genotyping were carried out according to the procedure outlined by Niu et al. (2011). Markers studied included SSRs from the BARC (Song et al., 2005), GWM (Röder et al., 1998), WMC (Somers et al., 2004), CFA (Sourdille et al., 2003), GDM (Pestsova et al., 2000), CFD (Guyomarc’h et al., 2002), DuPw (Eujayl et al., 2002), KSM (Yu et al., 2004), CNL (Yu et al., 2004), and AC (Barkley et al., 2006) groups. Markers were assigned to chromosomes and chromosome groups based on locations reported in the citations or based on a search for the markers in the GrainGenes database1. DNA fragments were amplified by polymerase chain reaction (PCR) at an annealing temperature of 50°C and labeled with four different fluorescent dyes (6-FAM, VIC, NED, and PET). Amplified PCR products were separated by capillary electrophoresis using the ABI 3130xl Genetic Analyzer (Applied Biosystems, Forster City, CA, United States) according to the procedures of Chao et al. (2007). The genotype calls were analyzed using GeneMapper software v3.7 (Applied Biosystems).
Evaluation of Ae. markgrafii Addition Lines for Resistance to Leaf Rust, Stripe Rust, and Powdery Mildew
Wheat cultivar Alcedo, Ae. markgrafii accession S740-69, Alcedo-Ae. markgrafii amphiploid W0492, six disomic addition lines, and Chinese Spring were included in tests for resistance to leaf rust, stripe rust, and powdery mildew.
Leaf rust resistance was evaluated at North Dakota State University (Fargo, ND, United States) followed the procedures of Kertho et al. (2015). Two P. triticina races, TDBJ+Lr21&Lr28 and TNBJ, were used to evaluate the genotypes. Race TDBJ+Lr21&Lr28 produces a high infection type on Lr21 and Lr28, while TNBJ has a high infection type on Lr9. The experiment was conducted using a randomized complete block design with two replicates, with the entire experiment being repeated for each race as described by Kertho et al. (2015). Approximately five seeds per genotype were planted in a greenhouse set at 22°C/18°C (day/night) with 16-h photoperiod. Ten-day-old seedlings were inoculated by spraying fresh urediniospores suspended in a light mineral oil (Soltrol-170, Phillips Petroleum, Bartlesville, OK, United States). Following inoculation, plants were placed into a darkened dew chamber maintained at 20°C for 16–24 h. Following the incubation period, the plants were removed to a greenhouse maintained at 20°C with a normal 16/8 h day/night photoperiod. Genotypes were scored for infection types (ITs) at 12–14 days post inoculation using the 0–4 scale (McIntosh et al., 1995). Infection types of 2 or lower were considered resistant, and ITs 3 or higher were considered susceptible.
Resistance to stripe rust was evaluated at USDA-ARS, Wheat Health, Genetics, and Quality Research Unit, Pullman, WA, United States, using six P. striiformis f. sp. tritici races, PSTv-4, PSTv-14, PSTv-37, PSTv-40, PSTv-51, and PSTV-198 (Wan and Chen, 2014; Wan et al., 2016). For each race test, 5–10 seeds per line were planted and seedlings at the two-leaf stage were uniformly inoculated with a mixture of urediniospores with talc at a 1:20 ratio and kept in a dew chamber for 24 h at 10°C and 100% relative humidity without light. The inoculated plants were them moved to a growth chamber at a diurnal temperature cycle gradually changing from 4°C at 2:00 am to 20°C at 2:00 pm and a diurnal cycle of 8 h dark/16 light corresponding to the low/high temperature cycle (Wan and Chen, 2014). Plants were scored 20 days post inoculation using the 0–9 scale of Line and Qayoum (1992).
Powdery mildew tests were conducted at the USDA-ARS, Plant Science Research Unit, Raleigh, NC, using the detached-leaf method as described by Worthington et al. (2014). Twenty isolates representing differing United States regional virulence profiles were used for tests. These isolates originated from nine US states of the Southeast, Mid-Atlantic, Great Lakes, and Great Plains wheat growing regions. To simplify presentation, two Montana isolates are included as “Great Plains” isolates despite originating west of the Great Plains because they exhibit a similar virulence profile to isolates from the Great Plains (Cowger et al., 2018). Inoculations were performed following Worthington et al. (2014). In brief, two 1.5-cm detached leaf segments from each genotype were floated on 0.5% water agar containing 50 mg L-1 benzimidazole in a Petri plate. Each plate also contained four replicate leaf segments of a susceptible wheat cultivar as a positive control. Four replicate Petri plates per host genotype were inoculated with each isolate of B. graminis f. sp. tritici. The plates were then placed in a growth chamber set to 18°C with an 11-h photoperiod. Disease reactions were scored 10 days post-inoculation using the 0–9 scale of Leath and Heun (1990). Reactions were then classified as resistant (R), intermediate (I), or susceptible (S) based on whether the predominant reaction among the replicate plates was <4, 4 to 6, or >6, respectively.
Results
GISH and FISH Analysis of Six Alcedo-Ae. markgrafii Disomic Addition Lines
The six Alcedo-Ae. markgrafii disomic addition lines, AI(B) through AVIII(G), were examined for differences in spike morphologies (Figure 1). We observed that AI(B) is unique in its non-free threshing spikes, AII(C) has large glumes, AIII(D) and AV(E) have awns, and AIV(F) has club spikes with brittle rachis. Lines AV(E) and AVIII(G) have sterile spikelets on the top and in the upper half portion of the spikes, respectively (Table 1).

FIGURE 1. The morphology of the spikes of Alcedo, the amphiploid between Alcedo and Aegilops markgrafii, and six wheat addition lines carrying the alien chromosomes from Ae. markgrafii. 1, amphiploid, 2, AI(B); 3, AII(C); 4, AIII(D); 5, AV(E); 6, AIV(F); 7, AVIII(G); 8, Alcedo.
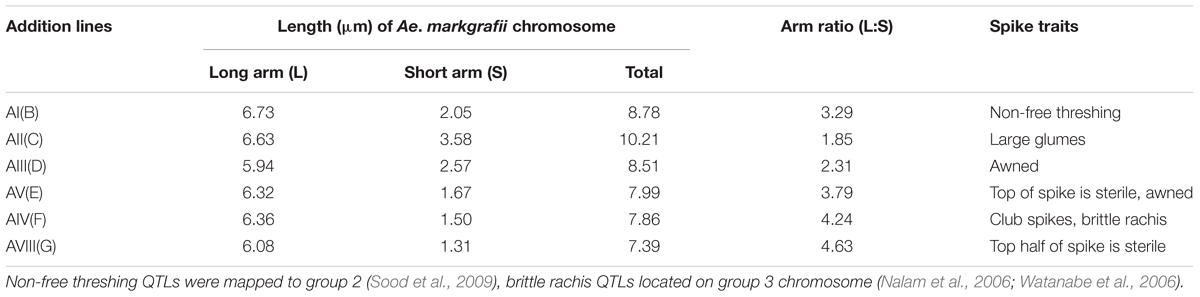
TABLE 1. Karyotypic characteristics of Aegilops markgrafii chromosomes and the spike agronomic traits of the six Alcedo-Ae. markgrafii S740-69 disomic addition lines.
The GISH analysis (Figure 2) showed that all six lines had a mitotic chromosome number of 2n = 44, and in each case, only one pair of chromosomes showed a distinct green coloration (arrows) compared to the red coloration of the remaining 42 chromosomes. No structural abnormalities were observed on any of the chromosomes. These results indicated that each addition line carried only one intact chromosome pair from Ae. markgrafii. Karyotypic characteristics of six Ae. markgrafii chromosomes are listed in Table 1. The long arm to short arm ratios of the Ae. markgrafii chromosomes B, C, D, E, F, and G were 3.29, 1.85, 2.31, 3.79, 4.24, and 4.63, respectively (Table 1). The FISH results showed that the Ae. markgrafii chromosomes (arrows) in all the additions had the pSC119.2 hybridization signals in the telomeric regions in either one or both arms (Figure 2). The general morphologies of the six Ae. markgrafii chromosomes from GISH/FISH analysis are consistent with those from the N- and C-banded karyotypes reported by Schubert et al. (1987) and Friebe et al. (1992), respectively. By comparing to the reference karyotype developed based on Ae. markgrafii accession MvGB428 (Molnár et al., 2015), we found that only chromosomes C and F had the identical pSC119.2 band patterns as chromosomes 5C and 3C, respectively.
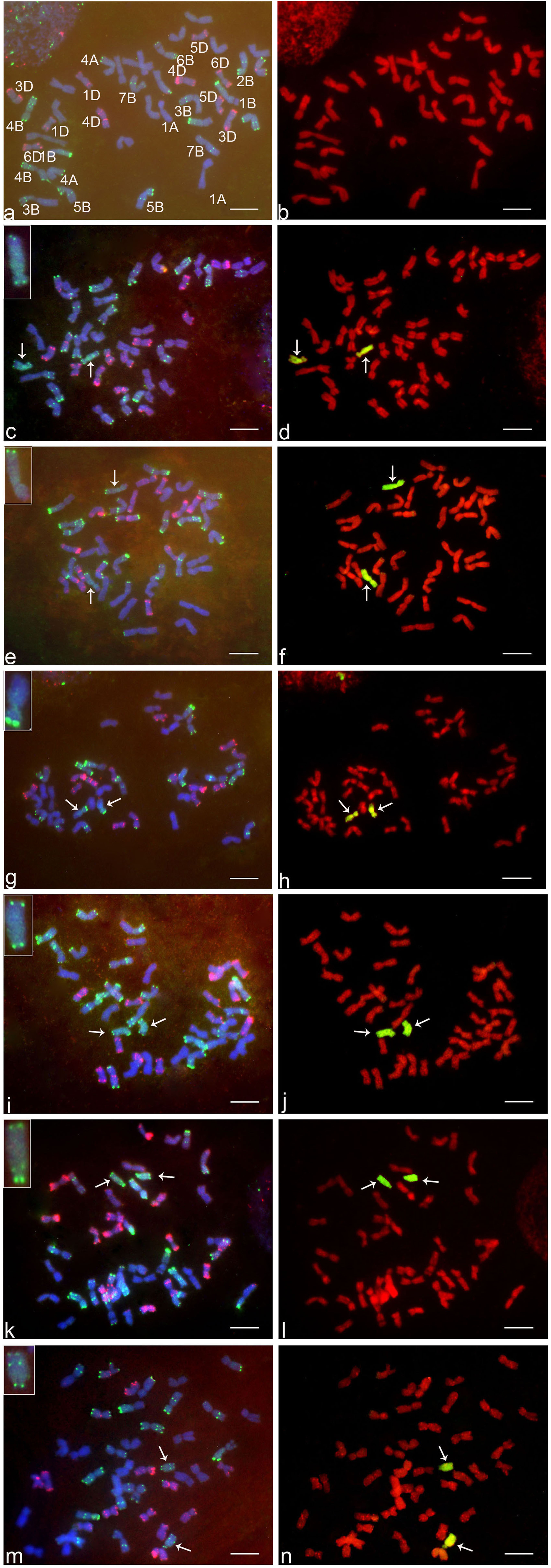
FIGURE 2. FISH and GISH on the somatic metaphase chromosomes of six addition lines and their wheat parent ‘Alcedo.’ The left side of figure (a,c,e,g,i,k,m) are FISH results, where red indicates pAs1 hybridization sites detected by rhodamine fluorescence and green indicates pSc119.2 hybridization sites detected by FITC fluorescence. The right sides of figure (b,d,f,h,j,l,n) are GISH results, where green indicates Aegilops markgrafii chromatin detected by FITC fluorescence. a and b, Alcedo; c and d, AI(B); e and f, AII(C); g and h, AIII(D); i and j, AV(E); k and l, AIV(F); m and n, AVIII(G). Arrows indicate the alien chromosomes. Bar represents 10 μm.
Identification of SSR Markers Associated to Ae. markgrafii Chromosomes
In this study, 1,500 SSR primer pairs were used to detect polymorphism between the parents, Alcedo and Ae. markgrafii accession S740-69. Nine hundred and thirty-five pairs of SSRs (62.3%) amplified polymorphic bands. From those polymorphic primer pairs, SSRs located on group 1 chromosomes, the majority of the SSRs that produced dominant bands and SSRs that produced weak bands were eliminated from further analyses. As a result, only 234 robust SSRs were selected for analysis of the Alcedo-Ae. markgrafii addition lines. These SSRs were comprised of 27 BARCs, 58 GWMs, 72 WMCs, 14 CFAs, 16 GDMs, 34 CFDs, 5 DuPws, 4 KSMs, 3 CNL, and 1 AC. These SSRs belonged to six homoeologous groups, 52 to group 2, 45 to group 3, 35 to group 4, 47 to group 5, 24 group to 6, and 31 to group 7. Analysis of the Ae. markgrafii addition lines resulted in the elimination of additional SSRs. As a result, only 132 SSRs were polymorphic between the addition lines and Alcedo (Supplementary Table 1). However, many of these SSRs mapped to multiple groups (Supplementary Table 1), and therefore, in the summarized distribution of the SSRs to chromosome groups, it appears that there are more than 132 polymorphic SSRs (Table 2).
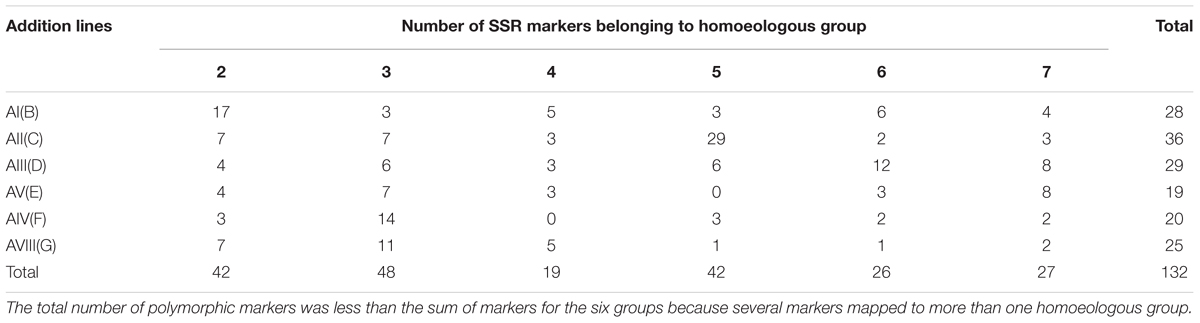
TABLE 2. The distribution of the polymorphic SSR marker belonging to different homoeologous groups in six Alcedo-Aegilops markgrafii disomic addition lines.
The assignment of Ae. markgrafii chromosomes to homoeologous groups was determined based on the distribution of the polymorphic SSR markers among the addition lines. Of the 28 polymorphic markers identified for addition line AI(B) (Table 2), 17 (61%) mapped to group 2 chromosomes, suggesting that the alien chromosome in the AI(B) addition line belongs to group 2. Similar comparisons for the other five addition lines clearly indicate that the Ae. markgrafii chromosomes in the lines AII(C), AIII(D), and AV(F) belongs to groups 5, 6, and 3, respectively. The chromosome in line AV(E) might belong to group 7 or group 3, and the G addition chromosome might belong to group 2, 3, or 4 (Table 3).
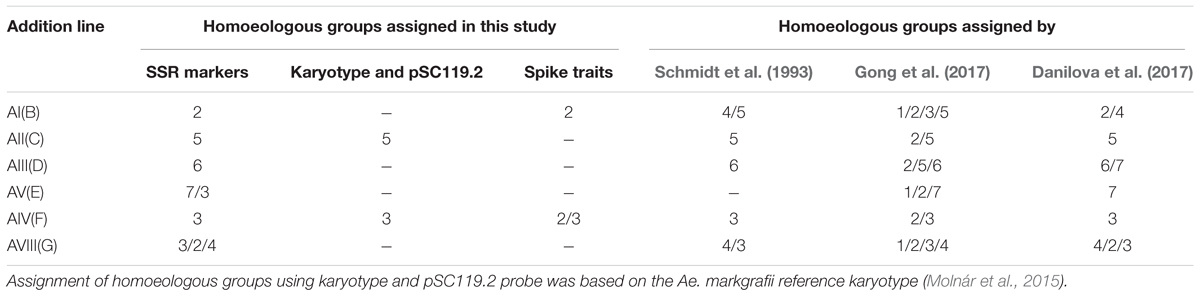
TABLE 3. Assignment of homoeologous groups of six Aegilops markgrafii chromosomes derived from six Alcedo-Ae. markgrafii disomic addition lines.
Reactions of Alcedo-Ae. markgrafii Addition Lines to Leaf Rust, Stripe Rust, and Powdery Mildew
The addition lines were tested with two races (TDBJ and TNBJ) of the leaf rust pathogen (Table 4). As expected, Chinese Spring was susceptible to both races. Alcedo had an intermediate (2+3) or resistant (2) IT to TDBJ and TNBJ, respectively; indicating that it carries at least one leaf rust resistance gene. The Ae. markgrafii parent (S740-69) was highly resistant to both races, with an immune response. For the six disomic addition lines, only AI(B) exhibited resistance, with a level of resistance similar to S740-69. The B chromosome appears to be a good source of leaf rust resistance.
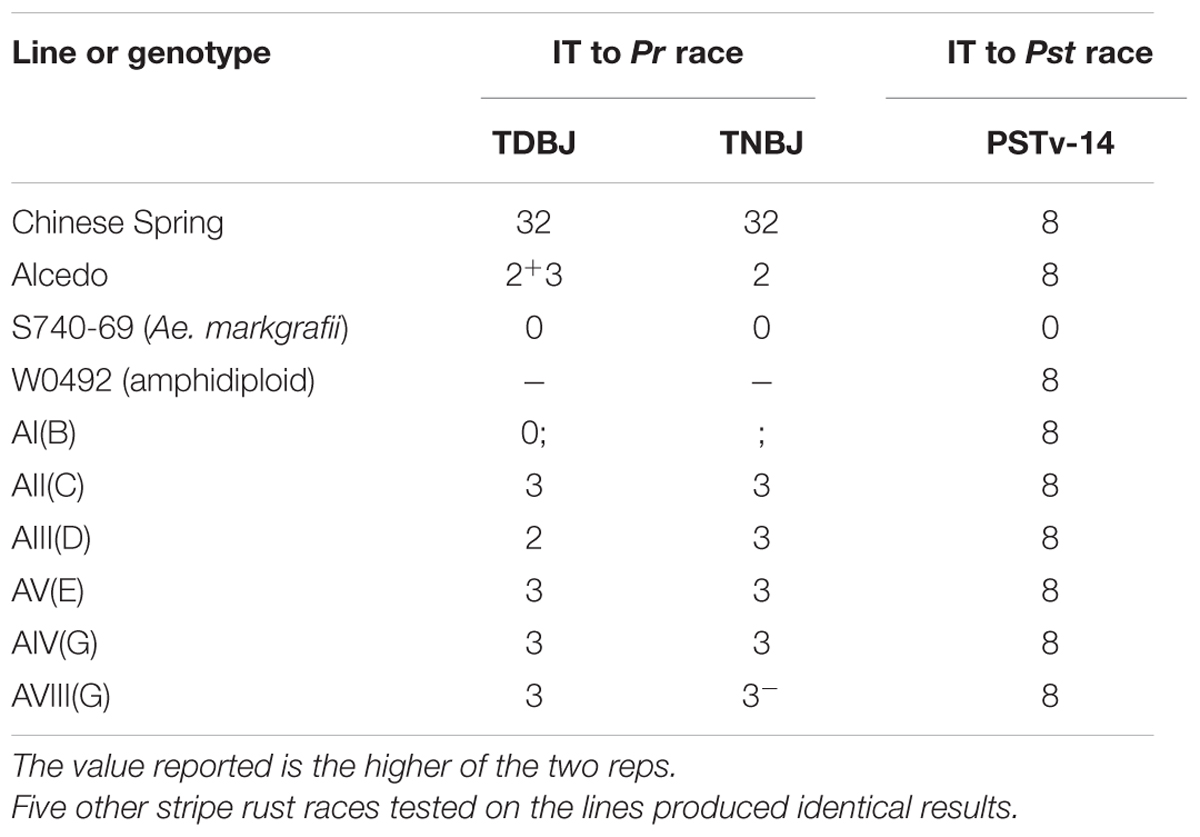
TABLE 4. Infection types (IT) observed on Aegilops markgrafii addition lines when tested with two races of the leaf rust pathogen (Puccinia tritcina, Pr) and one race of stripe rust pathogen (Puccinia striiformis f. sp. tritici, Pst).
The addition lines and parents were tested with six races of the stripe rust pathogen (Table 4). Although Alcedo had been reported to carry two genes for stripe rust resistance, Alcedo was observed to be highly susceptible (IT 8) to all six US races. For the remaining parents and addition lines, the Ae. markgrafii parent (S740-69) was immune, but all other lines were highly susceptible. The amphidiploid W0492 was included in the stripe rust tests, and it expressed an IT similar to Alcedo and all addition lines; and this indicated that the resistance in S740-69 could not be confirmed to any of the seven Ae. markgrafii chromosomes.
When tested with powdery mildew (Table 5), Chinese Spring was susceptible to all 20 isolates, while Alcedo was susceptible to 19 isolates and had an intermediate reaction to isolate MTG1-1a. In contrast, S740-69 was resistant to all 20 isolates, indicating Ae. markgrafii was an excellent source of powdery mildew resistance. Among the addition lines, AI(B) and AII(C) were susceptible to almost all isolates, indicating that resistance was not contributed by the B and C chromosomes. The remaining four addition lines had varying levels of resistance. Line AV(E) had resistance to 19 of 20 isolates and an intermediate reaction to isolate MSG-D-1-5. Line AIII(D) was resistant to all eight Great Plains isolates, but had a mixture of R, I, and S reactions to isolates from the other geographical regions. Line AIV(F) also had resistance to all Great Plains isolates, but AIV(F) had a different mixture of R, I, and S reactions to other isolates as compared to AIII(D). AVIII(G) had a mixture of reactions without regard to the region of origin and had more intermediate reactions than the other addition lines. In summary, the E chromosome conditioned resistance to nearly all isolates, the D and F chromosomes conditioned resistance to the Great Plains isolates and some isolates from other regions, and the G chromosome conditioned resistance to some isolates without regard to the region of origin.
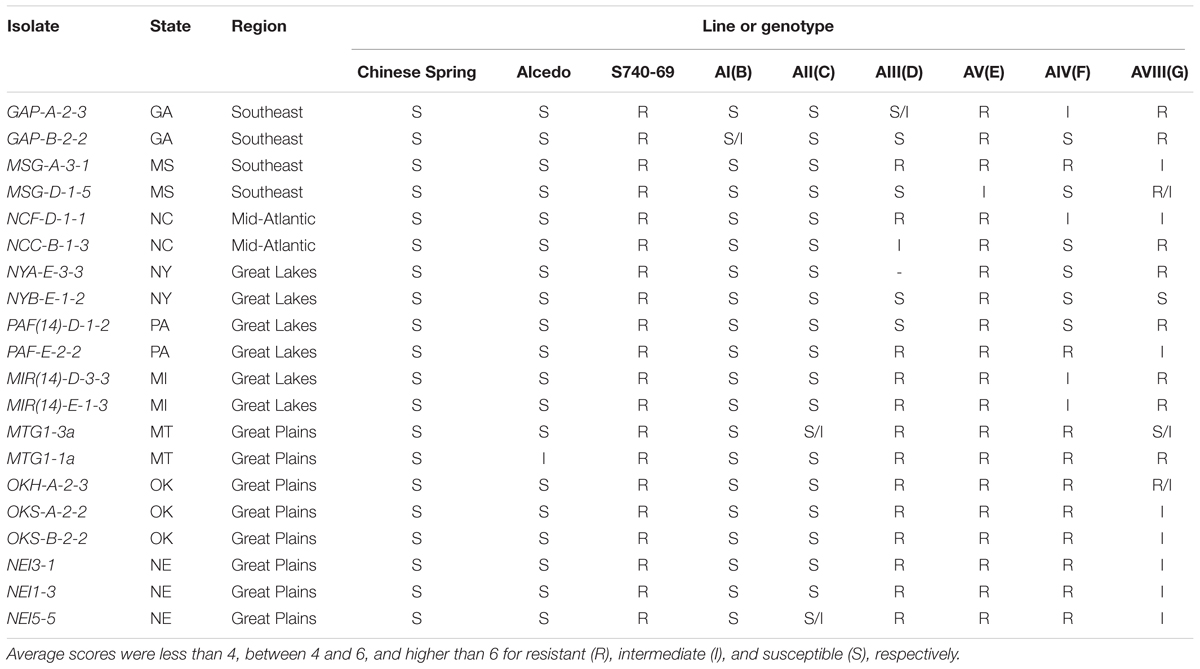
TABLE 5. Reactions of six Alcedo-Aegilops markgrafii addition lines, their parents, and the Chinese Spring check when tested with 20 isolates of powdery mildew pathogen collected from nine states and four regions of the United States.
Discussion
In assigning SSRs to specific Ae. markgrafii chromosomes, the addition lines must exhibit a high level of homogeneity relative to Alcedo to exclude detection of polymorphisms on the wheat chromosomes. Friebe et al. (1992) concluded from C-banding results that the Alcedo-Ae. markgrafii addition lines were not in a pure Alcedo background. This is supported by the results of Niu et al. (2011), who studied HMW glutenin subunits in the six addition lines. They found addition line AIII(D) differed from Alcedo at all three Glu-1 loci, which indicated an additional wheat genotype in the parentage of AIII(D) rather than the presence of biotypes in Alcedo. Variability in the wheat background of the addition lines complicates determination of the origin of the observed polymorphisms. For example, in AIII(D) the evidence suggests that the Ae. markgrafii chromosome is homoeologous to group 6, but additional markers also mapped to all the other chromosome groups.
The observed variability in the assignment of molecular markers to chromosomes indicates the presence of chromosomal rearrangements. Studies by Danilova et al. (2017) and Gong et al. (2017) found that the Alcedo-Ae. markgrafii additions lines carried several inversions and translocations. While both studies found a high level of rearrangement, the two studies did not agree on the rearrangements carried by each chromosome (Table 3). For example, Danilova et al. (2017) concluded that chromosome D was mainly a group 6 chromosome with the long arm telomere composed of a translocated 7CL telomeric region. In contrast, Gong et al. (2017) concluded that the rearrangements in chromosome D involved chromosomes 2C, 5C, and 6C.
For each addition line, we observed markers that were not associated with the homoeologous group identified for that line. For example, of the 28 polymorphic markers associated with addition line AI(B), 17 were group 2 markers, and 11 markers were therefore not associated with group 2. There is more than one possible explanation for the markers that do not fit with the alien chromosome. Some of these markers may represent polymorphisms present in the addition lines that were not eliminated during backcrossing to Alcedo, and therefore these markers would not be associated with the alien chromosome. Some may actually be associated with the alien chromosome but have simply not been previously identified to that homoeologous group. Finally, some may be associated with the alien chromosome, but the rearrangements present results in markers being identified with multiple homoeologous groups. For example, Danilova et al. (2017) concluded that the Ae. markgrafii chromosome D carried a group 6/7 rearrangement, and 19 of the 29 markers we observed for AIII(D) would fit with this rearrangement. Similarly, Danilova et al. (2017) concluded that Ae. markgrafii chromosome G carried a 4/2/3 rearrangement, and 21 of the 25 markers we observed fit this rearrangement. Therefore, our results seem to fit well with the conclusions of Danilova et al. (2017). However, considering the high levels of rearrangements in the Ae. markgrafii genome, it is possible that the differences in the present study from Danilova et al. (2017) and Gong et al. (2017) may represent observational differences, with additional rearrangements yet to be discovered. The GISH and FISH analysis showed that the Ae. markgrafii chromosomes in AII(C) and AIV(F) (Figure 2) are morphologically most like chromosomes 5C and 3C of the reference karyotype (Molnár et al., 2015), respectively. Taken together, the spike traits (Table 1), molecular marker data (Table 2), and FISH and GISH analyses (Figure 2) indicated that Ae. markgrafii chromosomes in AI(B), AII(C), AIII(D), AV(E), AIV(F), and AVIII(G) belong to groups 2, 5, 6, 7, 3, and 4, respectively.
Spike traits were recorded for each Ae. markgrafii addition line, and in two instances, the observed trait (Table 1) corresponded with the molecular marker data. In AI(B), spikes were non-free threshing. Genes for tenacious glume (Tg) have been identified on group 2 chromosomes (Simonetti et al., 1999; Jantasuriyarat et al., 2004; Sood et al., 2009; Faris et al., 2014; Katkout et al., 2014). Genes for brittle rachis have been located to group 3 chromosomes (Nalam et al., 2006; Watanabe et al., 2006). These observations agree with the conclusion that Ae. markgrafii chromosomes B and F are homoeologous to group 2 and 3, respectively. Other spike traits did not yield useful homoeology information. For example, large glumes and club spikes were associated with chromosomes C (group 5) and F (group 3), respectively. The large glume trait of T. polonicum has been mapped to group 7 chromosomes (Watanabe, 1999), while the club spike trait is a group 2 characteristic (Johnson et al., 2008). The failure to observe corresponding results between the molecular data and the morphological traits may represent either incomplete knowledge of the trait, impurity of the Alcedo background, or may indicate that the chromosomes in question carry chromosomal rearrangements.
We observed resistance to leaf rust conditioned by chromosome B of Ae. markgrafii. In contrast, Gong et al. (2017) found that chromosome D conditioned resistance to leaf rust, while chromosome B provided no leaf rust resistance. It is possible that the differences in these two studies may represent differential response to races. However, Iqbal et al. (2007) transferred leaf rust resistance from Ae. markgrafii to wheat chromosome arm 2AS, and they noted that chromosome B was the likely source of this gene.
Alcedo has been reported to carry two major and two minor genes conferring adult-plant resistance to stripe rust (Jagger et al., 2011). When we tested the addition lines for resistance to six US races of P. striiformis f. sp. tritici in the seedling stage, we observed a susceptible reaction on Alcedo, all addition lines, and the amphidiploid. Our results were consistent with Jagger et al. (2011) that Alcedo was susceptible in the seedling stages to the United Kingdom isolates used in the field tests. However, the seedling tests could not detect the adult-plant resistance in Alcedo. Nevertheless, the seedling data showed that the addition lines did not get any genes from Ae. markgrafii for all-stage resistance against the current predominant and most virulent races in the United States. Further tests of the lines with the races at the adult-plant stage or in the field are needed to determine if the addition lines inherited any adult-plant resistance genes from Alcedo and/or from Ae. markgrafii.
For resistance to powdery mildew, Gong et al. (2017) tested the six addition lines and parents using mixed races of the pathogen in China. They identified only line AV(E) as carrying resistance from Ae. markgrafii. However, we found that four addition lines, AIII(D), AV(E), AIV(F), and AVIII(G), carried powdery mildew resistance and Ae. markgrafii accession S740-69 was resistant to all 20 isolates tested in our study. The resistance conferred by chromosomes D, F, and G was generally confined to isolates originating from a geographical region and thus restricts their adaptability. The E chromosome conferred resistance to 19 of the 20 powdery mildew isolates in the test, making it particularly attractive for alien gene introgression.
We report here tests for powdery mildew, stripe rust, and leaf rust resistance. The previous study of Xu et al. (2009) identified chromosomes C and D as carrying resistance to the Ug99 race group of the stem rust pathogen. Therefore, each of the six addition lines carries resistance to at least one fungal disease, making this a rich resource for gene introgression. Alien gene introgression is very valuable for introduction of new traits into wheat. Historically these introgressions were the product of homoeologous recombination or radiation induced chromosomal breakage which usually required standard cytogenetic techniques. With the incorporation of molecular markers as a tool to select recombinants, induced homoeologous recombination is much more effective than techniques that relied on cytogenetic observation. This study therefore identifies not only which lines carry disease resistance genes, but also identifies markers that can be used to detect recombination. By using the SSR markers associated with Ae. markgrafii chromosome D, we recently introgressed a new gene for Ug99 resistance from AIII(D) into common wheat (Xu et al., 2017).
Author Contributions
SX conceived and planned this study. ZN, DK, and SC conducted marker analysis. ZN, XWC, BF, BG, and SX conducted molecular and cytogenetic analysis on alien chromosomes. RW and CC performed assay for powdery mildew resistance. MB and JR conducted leaf rust test. XMC conducted stripe rust test. ZN, DK, and SX wrote the manuscript. All authors reviewed and edited the manuscript.
Funding
This research was supported in part by funds to SX provided through a grant from the Bill & Melinda Gates Foundation and United Kingdom Department for International Development to Cornell University for the Borlaug Global Rust Initiative (BGRI) Durable Rust Resistance in Wheat (DRRW) Project and the USDA-ARS CRIS Project No. 5442-22000-080-037-00D.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Authors thank Mary Osenga for technical support in SSR marker analysis. All opinions expressed in this paper are the author’s and do not necessarily reflect the policies and views of USDA. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the United States Department of Agriculture. USDA is an equal opportunity provider and employer.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01616/full#supplementary-material
Footnotes
References
Barkley, N. A., Roose, M. L., Krueger, R. R., and Federici, C. T. (2006). Assessing genetic diversity and population structure in a citrus germplasm collection utilizing simple sequence repeat markers (SSRs). Theor. Appl. Genet. 112, 1519–1531. doi: 10.1007/s00122-006-0255-9
Chao, S., Zhang, W., Dubcovsky, J., and Sorrells, M. (2007). Evaluation of genetic diversity and genome-wide linkage disequilibrium among U.S. wheat (Triticum aestivum L.) germplasm representing different market classes. Crop Sci. 47, 1018–1030. doi: 10.2135/cropsci2006.06.0434
Cowger, C., Mehra, L., Arellano, C., Meyers, E., and Murphy, J. P. (2018). Virulence differences in Blumeria graminis f. sp. tritici from the Central and Eastern United States. Phytopathology 108, 402–411. doi: 10.1094/PHYTO-06-17-0211-R
Danilova, T. V., Akhunova, A. R., Akhunov, E. D., Friebe, B., and Gill, B. S. (2017). Major structural genomic alterations can be associated with hybrid speciation in Aegilops markgrafii (Triticeae). Plant J. 92, 317–330. doi: 10.1111/tpj.13657
Danilova, T. V., Friebe, B., and Gill, B. S. (2014). Development of a wheat single gene FISH map for analyzing homoeologous relationship and chromosomal rearrangements within the Triticeae. Theor. Appl. Genet. 127, 715–730. doi: 10.1007/s00122-013-2253-z
Dyck, P. L., Kerber, E. R., and Martens, J. W. (1990). Transfer of a gene for stem rust resistance from Aegilops caudata to common wheat. Can. J. Plant Sci. 70, 931–934. doi: 10.4141/cjps90-114
Eujayl, I., Sorrells, M. E., Baum, M., Wolters, P., and Powell, W. (2002). Isolation of EST-derived microsatellite markers for genotyping the A and B genomes of wheat. Theor. Appl. Genet. 104, 399–407. doi: 10.1007/s001220100738
Faris, J. D., Zhang, Z., and Chao, S. (2014). Map-based analysis of the tenacious glume gene Tg-B1 of wild emmer and its role in wheat domestication. Gene 542, 198–208. doi: 10.1016/j.gene.2014.03.034
Friebe, B., Schubert, V., Blüthner, W. D., and Hammer, K. (1992). C-banding pattern and polymorphism of Aegilops caudata and chromosomal constitutions of the amphiploid T. aestivum-Ae. caudata and six derived chromosome addition lines. Theor. Appl. Genet. 83, 589–596. doi: 10.1007/BF00226902
Gong, W., Han, R., Li, H., Song, J., Yan, H., Li, G., et al. (2017). Agronomic traits and molecular marker identification of wheat-Aegilops caudata addition lines. Front. Plant Sci. 8:1743. doi: 10.3389/fpls.2017.01743
Guyomarc’h, H., Sourdille, P., Charmet, G., Edwards, K. J., and Bernard, M. (2002). Characterisation of polymorphic microsatellite markers from Aegilops tauschii and transferability to the D-genome of bread wheat. Theor. Appl. Genet. 104, 1164–1172. doi: 10.1007/s00122-001-0827-7
Iqbal, N., Eticha, F., Khlestkina, E. K., Weidner, A., Röder, M. S., and Börner, A. (2007). The use of simple sequence repeat (SSR) markers to identify and map alien segments carrying genes for effective resistance to leaf rust in bread wheat. Plant Gen. Res. 5, 100–103. doi: 10.1017/S1479262107672311
Jagger, L. J., Newell, C., Berry, S. T., MacCormack, R., and Boyd, L. A. (2011). The genetic characterization of stripe rust resistance in the German wheat cultivar Alcedo. Theor. Appl. Genet. 122, 723–733. doi: 10.1007/s00122-010-1481-8
Jantasuriyarat, C., Vales, M. I., Watson, C. J. W., and Riera-Lizarazu, O. (2004). Identification and mapping of genetic loci affecting the free-threshing habit and spike compactness in wheat (Triticum aestivum L.). Theor. Appl. Genet. 108, 261–273. doi: 10.1007/s00122-003-1432-8
Johnson, E. B., Nalam, V. J., Zemetra, R. S., and Riera-Lizarazu, O. (2008). Mapping the compactum locus in wheat (Triticum aestivum L.) and its relationship to other spike morphology genes of the Triticeae. Euphytica 163, 193–201. doi: 10.1007/s10681-007-9628-7
Katkout, M., Kishii, M., Kawaura, K., Mishina, K., Sakuma, S., Umeda, K., et al. (2014). QTL analysis of genetic loci affecting domestication-related spike characters in common wheat. Genes Genet. Sys. 89, 121–131. doi: 10.1266/ggs.89.121
Kertho, A., Mamidi, S., Bonman, J. M., McClean, P. E., and Acevedo, M. (2015). Genome-wide association mapping for resistance to leaf and stripe rust in winter-habit hexaploid wheat landraces. PLoS One 10:e0129580. doi: 10.1371/journal.pone.0129580
Leath, S., and Heun, M. (1990). Identification of powdery mildew resistance genes in cultivars of soft red winter wheat. Plant Dis. 74, 747–752. doi: 10.1094/PD-74-0747
Line, R. F., and Qayoum, A. (1992). Virulence, Aggressiveness, Evolution, and Distribution of Races of Puccinia striiformis (the Cause of Stripe Rust of Wheat) in North America, 1968–87. Washington DC: US Department of Agriculture, Agricultural Research Service.
McIntosh, R. A., Wellings, C. R., and Park, R. F. (1995). Wheat Rusts: an Atlas of Resistance Genes. East Melbourne, VIC: CSIRO Publications, 205. doi: 10.1007/978-94-011-0083-0
Mohan, M., Nair, S., Bhagwat, A., Krishna, T. G., Yano, M., Bhatio, C. R., et al. (1997). Genome mapping, molecular markers and marker-assisted selection in crop plants. Mol. Breed. 3, 87–103. doi: 10.1023/A:1009651919792
Molnár, I., Vrána, J., Farkas, A., Kubaláková, M., Cseh, A., Molnár-Láng, M., et al. (2015). Flow sorting of C-genome chromosomes from wild relatives of wheat Aegilops markgrafii, Ae. triuncialis and Ae. cylindrica, and their molecular organization. Ann. Bot. 116, 189–200. doi: 10.1093/aob/mcv073
Nalam, V. J., Vales, M. I, Watson, C. J. W., Kianian, S. F., and Riera-Lizarazu, O. (2006). Map-based analysis of genes affecting the brittle rachis character in tetraploid wheat (Triticum turgidum L.). Theor. Appl. Genet. 112, 3736–3781. doi: 10.1007/s00122-005-0140-y
Niu, Z., Klindworth, D. L., Friesen, T. L., Chao, S., Jin, Y., Cai, X., et al. (2011). Targeted introgression of a wheat stem rust resistance gene by DNA marker-assisted chromosome engineering. Genetics 187, 1011–1021. doi: 10.1534/genetics.110.123588
Niu, Z. X., Klindworth, D. L., Wang, R. R.-C., Jauhar, P. P., Larkin, P. J., and Xu, S. S. (2011). Characterization of HMW glutenin subunits in Thinopyrum intermedium, Th bessarabicum, Lophopyrum elongatum, Aegilops markgrafii and their addition lines in wheat. Crop Sci. 51, 667–677. doi: 10.2135/cropsci2010.04.0235
Peil, A., Korzun, V., Schubert, V., Schumann, E., Weber, W. E., and Röder, M. S. (1998). The application of wheat microsatellites to identify disomic Triticum aestivum-Aegilops markgrafii addition lines. Theor. Appl. Genet. 96, 138–146. doi: 10.1007/s001220050720
Pestsova, E., Ganal, M. W., and Röder, M. S. (2000). Isolation and mapping of microsatellite markers specific for the D genome of bread wheat. Genome 43, 689–697. doi: 10.1139/g00-042
Rayburn, A. S., and Gill, B. S. (1986). Molecular identification of the D-genome chromosomes of wheat. J. Hered. 77, 253–255. doi: 10.1093/oxfordjournals.jhered.a110231
Röder, M. S., Korzun, V., Wendehake, K., Plaschke, J., Tixier, M.-H., Leroy, P., et al. (1998). A microsatellite map of wheat. Genetics 149, 2007–2023.
Schmidt, J.-C., Schubert, V., and Blüthner, W. D. (1993). Use of isozymes to characterize Triticum aestivum – Aegilops markgrafii addition lines. Biochem. Physiol. Pflanzen 188, 385–392. doi: 10.1016/S0015-3796(11)80141-1
Schubert, V., and Blüthner, W. D. (1992). Zerlegung des Genoms von Aegilops markgrafii mit Hilfe von chromosomalen Additions-linien. Kühn Archiv. 86, 38–46.
Schubert, V., and Blüthner, W. D. (1995). “Triticum aestivum-Aegilops markgrafii addition lines: production and morphology,” in Proceedings of the 8th International. Wheat Genetic Symposium, eds Z. S. Li and Z. Y. Xin (Beijing: China Agricultural Scientech Press), 421–425.
Schubert, V., Blüthner, W. D., and Schlegel, R. (1987). N-banding and Feulgen karyogram of Aegilops markgrafii/(Greuter) Hammer var. markgrafii. Cer. Res. Commun. 15, 317–320.
Simonetti, M. C., Bellomo, M. P., Laghetti, G., Perrino, P., Simeone, R., and Blanco, A. (1999). Quantitative trait loci affecting free-threshing habit in tetraploid wheats. Genet. Res. Crop Evol. 46, 267–271. doi: 10.1023/A:1008602009133
Somers, D. J., Isaac, P., and Edwards, K. (2004). A high density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 109, 1105–1114. doi: 10.1007/s00122-004-1740-7
Song, Q. J., Shi, J. R., Singh, S., Fickus, E. W., Costa, J. M., Lewis, J., et al. (2005). Development and mapping of microsatellite (SSR) markers in wheat. Theor. Appl. Genet. 110, 550–560. doi: 10.1007/s00122-004-1871-x
Sood, S., Kuraparthy, V., Bai, G., and Gill, B. S. (2009). The major threshability genes soft glume (sog) and tenacious glume (Tg), of diploid and polyploid wheat, trace their origin to independent mutations at non-orthologous loci. Theor. Appl. Genet. 119, 341–351. doi: 10.1007/s00122-009-1043-0
Sourdille, P., Cadalen, T., Guyomarc’h, H., Snape, J. W., Perretant, M. R., Charmet, G., et al. (2003). An update of the Courtot-Chinese Spring intervarietal molecular marker linkage map for the QTL detection of agronomic traits in wheat. Theor. Appl. Genet. 106, 530–538. doi: 10.1007/s00122-002-1044-8
Valkoun, J., Hammer, K., Kuèerová, D., and Bartoš, P. (1985). Disease resistance in the genus Aegilops L.- stem rust, leaf rust, stripe rust, and powdery mildew. Kulturpflanze 33, 133–153. doi: 10.1007/BF01997267
Wan, A. M., and Chen, X. M. (2014). Virulence characterization of Puccinia striiformis f. sp. tritici using a new set of Yr single-gene line differentials in the United States in 2010. Plant Dis. 98, 1534–1542. doi: 10.1094/PDIS-01-14-0071-RE
Wan, A. M., Chen, X. M., and Yuen, J. (2016). Races of Puccinia striiformis f. sp. tritici in the United States in 2011 and 2012 and comparison with races in 2010. Plant Dis. 100, 966–975. doi: 10.1094/PDIS-10-15-1122-RE
Watanabe, N. (1999). Genetic control of the long glume phenotype in tetraploid wheat by homoeologous chromosomes. Euphytica 106, 39–43. doi: 10.1023/A:1003589117853
Watanabe, N., Fujii, Y., Kato, N., Ban, T., and Martinek, P. (2006). Microsatellite mapping of the genes for brittle rachis on homoeologous group 3 chromosomes in tetraploid and hexaploid wheats. J. Appl. Genet. 47, 93–98. doi: 10.1007/BF03194606
Weidner, A., Röder, M. S., and Börner, A. (2012). Mapping wheat powdery mildew resistance derived from Aegilops markgrafii. Plant Genet. Res. 10, 137–140. doi: 10.1017/S1479262112000123
Worthington, M., Lyerly, J., Petersen, S., Brown-Guedira, G., Marshall, D., Cowger, C., et al. (2014). MlUM15: an Aegilops neglecta-derived powdery mildew resistance gene in common wheat. Crop Sci. 54, 1397–1406. doi: 10.2135/cropsci2013.09.0634
Xu, S., Klindworth, D., Niu, Z., Zhang, Q., Chao, S., Friesen, T., et al. (2017). “Introgression of a new stem rust resistance gene from Aegilops markgrafii into wheat,” in Proceedings of 13th International Wheat Genetics Symposium, eds H. Buertmayr, C. Lang-Mladek, B. Steiner, S. Michel, M. Buertmayr, M. Lemmens, et al. (Vienna: University of Natural Resources and Life Sciences), 38–39.
Xu, S. S., Jin, Y., Klindworth, D. L., Wang, R. R.-C., and Cai, X. (2009). Evaluation and characterization of seedling resistances to stem rust Ug99 races in wheat-alien species derivatives. Crop Sci. 49, 2167–2175. doi: 10.2135/cropsci2009.02.0074
Xu, S. S., Liu, Z., Zhang, Q., Niu, Z., Jan, C.-C., and Cai, X. (2016). “Chromosome painting by GISH and multi-color FISH,” in Plant Cytogenetics. Methods in Molecular Biology, Vol. 1429, eds S. F. Kianian and P. M. A. Kianian (New York, NY: Springer Science), 7–21. doi: 10.1007/978-1-4939-3622-9_2
Keywords: wheat, homoeology, chromosome engineering, molecular markers, alien introgression, stripe rust, leaf rust, powdery mildew
Citation: Niu Z, Chao S, Cai X, Whetten RB, Breiland M, Cowger C, Chen X, Friebe B, Gill BS, Rasmussen JB, Klindworth DL and Xu SS (2018) Molecular and Cytogenetic Characterization of Six Wheat-Aegilops markgrafii Disomic Addition Lines and Their Resistance to Rusts and Powdery Mildew. Front. Plant Sci. 9:1616. doi: 10.3389/fpls.2018.01616
Received: 23 August 2018; Accepted: 18 October 2018;
Published: 08 November 2018.
Edited by:
István Molnár, Centre for Agricultural Research (MTA), HungaryReviewed by:
Michał Tomasz Kwiatek, Poznań University of Life Sciences, PolandGeorge Fedak, Agriculture and Agri-Food Canada (AAFC), Canada
Copyright © 2018 Niu, Chao, Cai, Whetten, Breiland, Cowger, Chen, Friebe, Gill, Rasmussen, Klindworth and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daryl L. Klindworth, ZGFyeWwua2xpbmR3b3J0aEBhcnMudXNkYS5nb3Y= Steven S. Xu, c3RldmVuLnh1QGFycy51c2RhLmdvdg==
 Zhixia Niu1
Zhixia Niu1 Rebecca B. Whetten
Rebecca B. Whetten Xianming Chen
Xianming Chen Bernd Friebe
Bernd Friebe Bikram S. Gill
Bikram S. Gill Daryl L. Klindworth
Daryl L. Klindworth Steven S. Xu
Steven S. Xu