- 1Key Laboratory of Bioactive Substances and Resources Utilization of Chinese Herbal Medicine, Ministry of Education, Institute of Medicinal Plant Development, Chinese Academy of Medicinal Science and Peking Union Medicinal College, Beijing, China
- 2NHP Research Alliance, Biodiversity Institute of Ontario (BIO), University of Guelph, Guelph, ON, Canada
- 3Key Laboratory of Beijing for Identification and Safety Evaluation of Chinese Medicine, Institute of Chinese Materia Medica, China Academy of Chinese Medical Sciences, Beijing, China
Cistanches Herba is a medicinal plant that has tonification properties and is commonly used in Asia. Owing to the imbalance between supply and demand, adulterants are frequently added for profit. However, there is no regulatory oversight because quality control tools are not sufficient for identifying heavily processed products. Thus, a novel molecular tool based on nucleotide signatures and species-specific primers was developed. The ITS2 regions from 251 Cistanches Herba and adulterant samples were sequenced. On the basis of SNP sites, four nucleotide signatures within 30~37 bp and six species-specific primers were developed, and they were validated by artificial experimental mixtures consisting of six different species and different ratios. This method was also applied to detect 66 Cistanches Herba products on the market, including extracts and Chinese patent medicines. The results demonstrated the utility of nucleotide signatures in identifying adulterants in mixtures. The market study revealed 36.4% adulteration: 19.7% involved adulteration with Cynomorium songaricum or Cistanche sinensis, and 16.7% involved substitution with Cy. songaricum, Ci. sinensis, or Boschniakia rossica. The results also revealed that Cy. songaricum was the most common adulterant in the market. Thus, we recommend the use of species-specific nucleotide signatures for regulating adulteration and verifying the quality assurance of medicinal product supply chains, especially for processed products whose DNA is degraded.
Introduction
Cistanches Herba (Rou Cong Rong) is a well-known Pharmacopoeia-recorded medicine in Asia (Chinese Pharmacopoeia Commission, 2015; Japan Pharmacopeial Convention, 2016); this medicine is derived from the dried succulent stems of Cistanche deserticola Y. C. Ma or Cistanche tubulosa (Schenk) Wight. Cistanches Herba has been used for more than 3,000 years as a superior tonic, as it is not toxic and can be taken for long periods of time (Li et al., 2016). Furthermore, Cistanches Herba was bestowed with the honor of being named “Desert Ginseng” because of its great medicinal value, especially in strengthening male sexual function (Zhang and Su, 2014; Gu et al., 2016). There are more than 100 Chinese patent medicines recorded in the Chinese Pharmacopoeia Commission (2015) and in other local official promulgated standards (Wang et al., 2012). As the population of elderly individuals increases, there is considerable demand for Cistanches Herba and its medicinal products. However, raw material resources are becoming increasingly scarce. In fact, the two original species of Cistanches Herba have been added to the China Plant Red Data Book as state-protected wild plants (category II) (Fu, 1991). The medicinal materials on the market are mainly cultivated in northwestern China.
Owing to the considerable imbalance between the supply and demand of Cistanches Herba, many adulterants have entered the market; these adulterants are inconsistent with standards and can threaten drug security. The known adulterants include the dried succulent stems of Cynomorium songaricum Rupr. (Cynomorii Herba, Suo Yang in Chinese), Cistanche sinensis Beck, Orobanche coerulescens Stephan, and Boschniakia rossica (Cham. et Schlecht.) Fedtsch. et Flerov (Sun et al., 2012). These adulterants have morphological characteristics similar to those of Cistanches Herba, making traditional taxonomic identification difficult, particularly after the material is processed into medicinal products. Microscopic identification is not available because Cistanches Herba has no definitive or unique microscopic characteristics. Current analytical chemistry tools are not sufficient for detecting adulteration of Cistanches Herba because similar compounds also exist within the known adulterants Ci. salsa (Lei et al., 2001; Chen et al., 2007) and Ci. sinensis (Liu et al., 2013). Therefore, the development of a rapid molecular method for the authentication of Cistanches Herba and its products is urgently needed for proper quality control systems in Chinese patent medicine and other medicinal product industries.
Chen et al. first suggested internal transcribed spacer 2 (ITS2) as a universal barcode for medicinal plants (Chen et al., 2010). Sun et al. verified the ITS2 region as a preferable DNA barcode for identifying Cistanches Herba and its adulterants (Sun et al., 2012). However, ITS2 cannot be used to distinguish Chinese patent medicine with degraded DNA. Recently, an increasing number of studies have shown that the “mini barcode” is a useful method for amplifying degraded DNA (Hajibabaei et al., 2006; Meusnier et al., 2008; Dubey et al., 2011; Lo et al., 2015). However, nucleotide signatures are more appropriate than mini barcodes, as the formers refer to one or more nucleotides that are unique to one species and can be effectively utilized by many molecular techniques, such as DNA probes, microfluidics and loop-mediated isothermal amplification (de Boer et al., 2015). Han et al. developed nucleotide signatures for Panax ginseng, Angelica sinensis and Lonicera japonica and successfully identified the associated Chinese patent medicines (Liu et al., 2016; Wang et al., 2016a; Gao et al., 2017).
The goal of our research focused on the development of nucleotide signatures for the identification of adulterants in functional products containing Cistanches Herba. Specifically, we (1) developed four nucleotide signatures and six specific primer pairs for differentiating authentic Cistanches Herba and known adulterants, (2) validated the four nucleotide signatures in experiments including mixtures of known taxonomic vouchers used to prepare Cistanches Herba products that contain both authentic and adulterated ingredients, and (3) performed a market survey of 66 Cistanches Herba products via their nucleotide signatures.
Materials and Methods
Sample Collection and Preparation
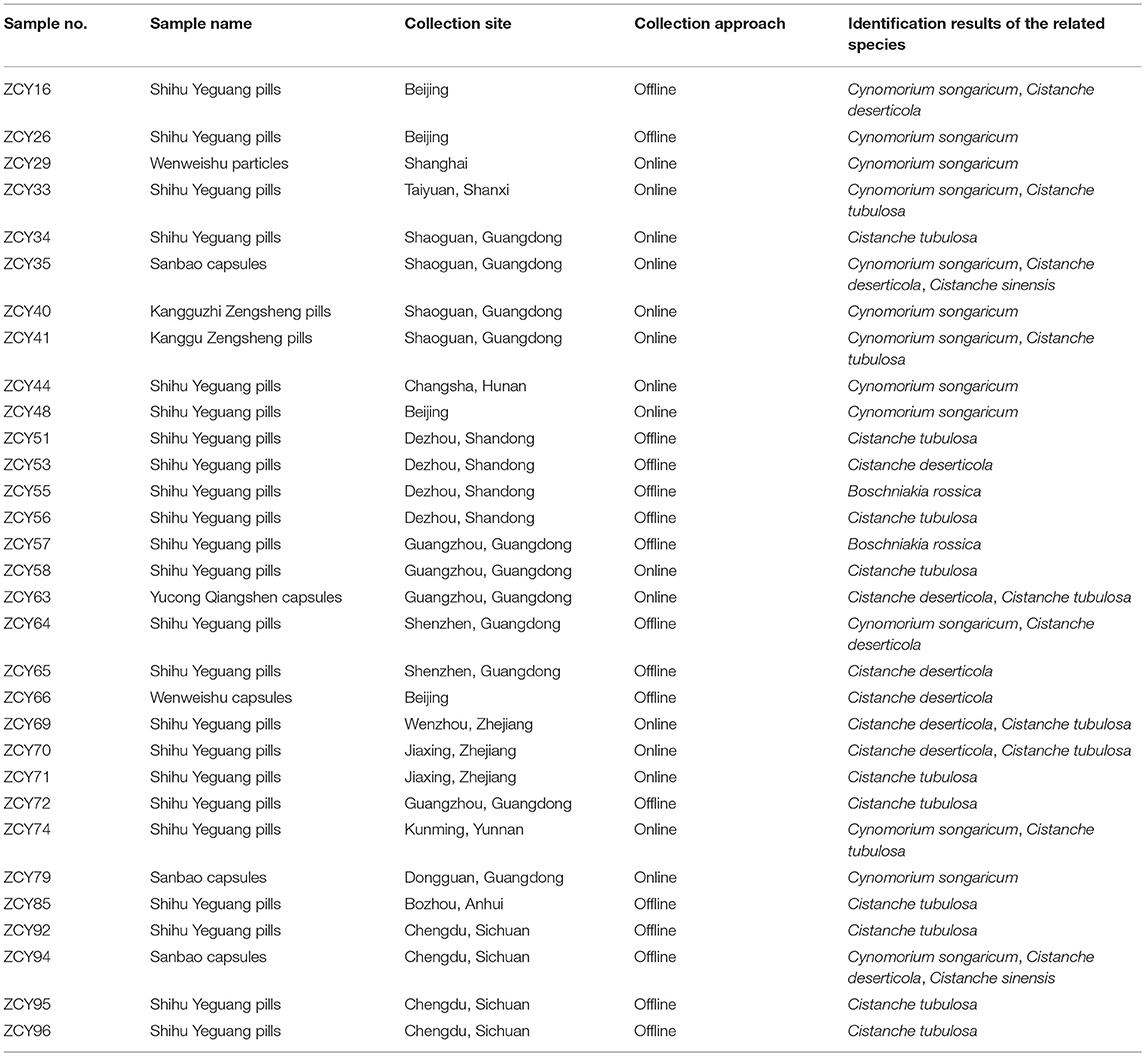
In total, 251 samples were collected from Inner Mongolia, Xinjiang, and Ningxia, among other areas; these samples included 214 Cistanches Herba and 37 adulterants and are detailed in Supplementary Table S1. Corresponding voucher samples were validated by taxonomists and deposited in the Herbarium of the Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences, Beijing, China. A total of 35 batches of powders, slices and extracts of Cistanches Herba were purchased from online stores and brick-and-mortar drugstores in Beijing and Chengdu (Table 1). In total, 31 batches of Chinese patent medicine containing Cistanches Herba were purchased from different drugstores (Table 2), and the declared compositions of different Chinese patent medicines are shown in Supplementary Table S3. The different morphological characteristics of different dose forms are shown in Figure 1.
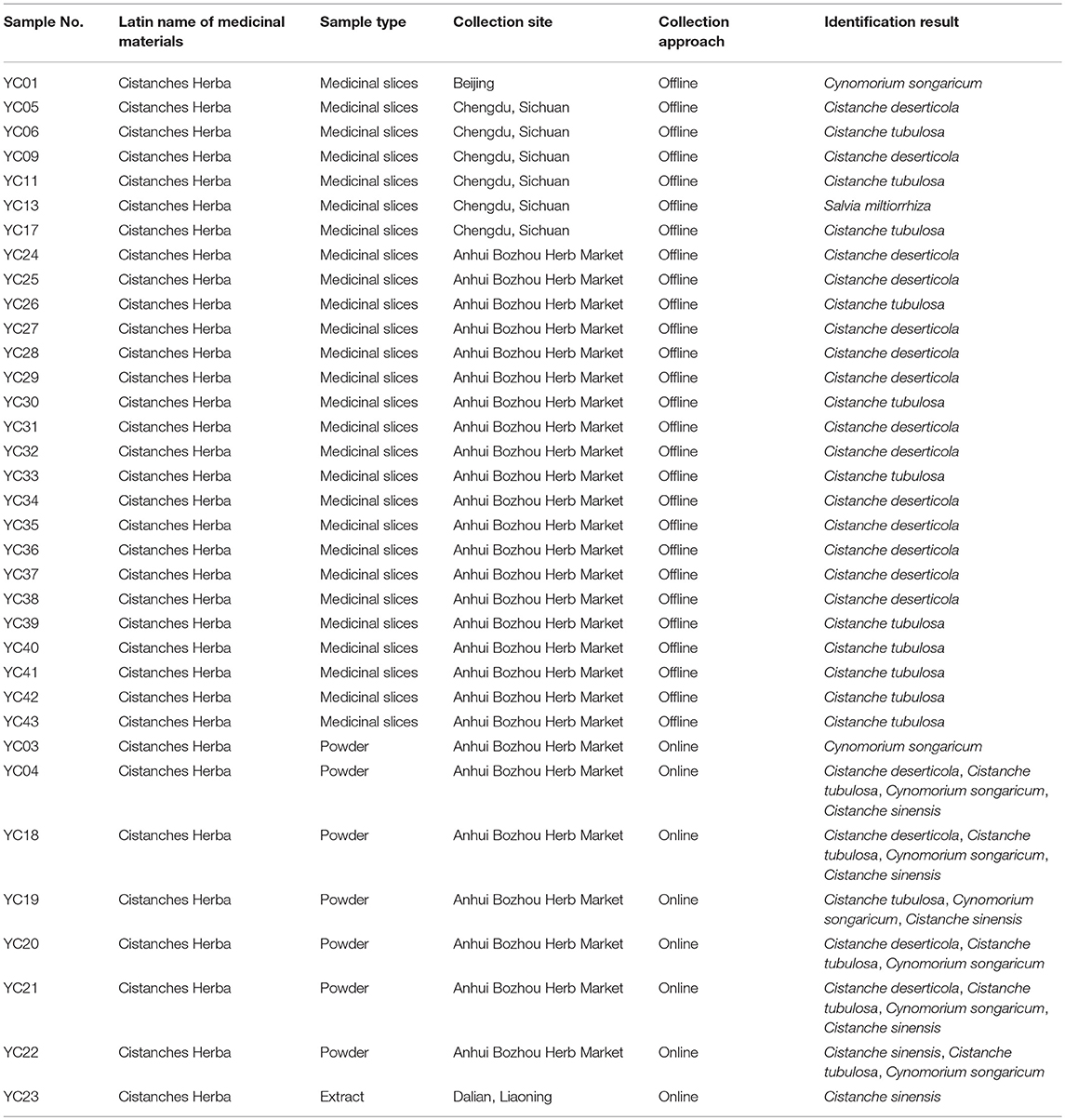
Table 1. Sample information and identification results of the 35 powders, medicinal slices, and extract.

Figure 1. Different morphological characteristics of different dose forms of Cistanches Herba products. (A) Particles (ZCY29, Wenweishu particles); (B) Capsules (ZCY35, Sanbao capsules); (C) Water honeyed pills (ZCY70, Shihu Yeguang pills); (D) Large honeyed pills (ZCY48, Shihu Yeguang pills); (E) Extracts (YC23).
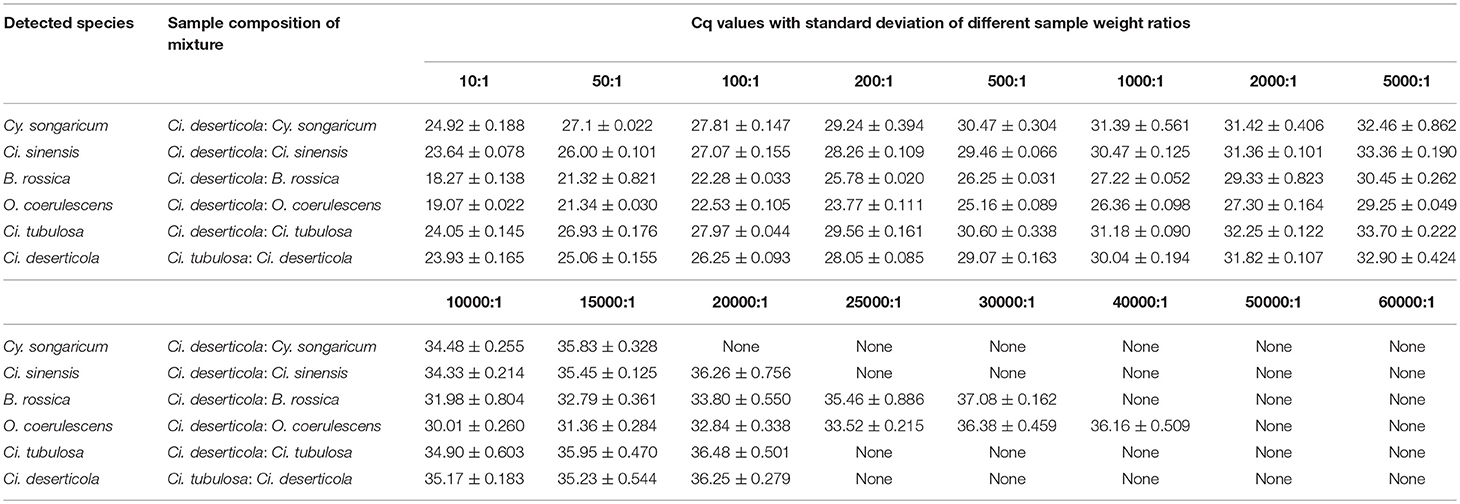
Mixed samples: Powders of Ci. deserticola, Ci. tubulosa, Cy. songaricum, Ci. sinensis, B. rossica, and O. coerulescens were artificially mixed in different combinations (at a ratio of 1:1) before extraction. The details of these mixed samples are shown in the legend of Figure 3. In addition, the powders of four adulterants were mixed with the genuine Ci. deserticola at different weight ratios: 10:1, 50:1, 100:1, 200:1, 500:1, 1000:1, 2000:1, 5000:1, 10000:1, 15000:1, 20000:1, 25000:1, 30000:1, 40000:1, 50000:1 and 60000:1 (Table 3). And the two genuine products are mixed in the same proportions.
Decoction: Slices of Ci. deserticola and Cy. songaricum were used to prepare the decoction. The slices (10 g) were boiled in 300 mL of double-distilled water for 30, 60, 90, 120, 150, 180, 210, and 240 min and then used for DNA extraction.
DNA Extraction, Polymerase Chain Reaction (PCR) Amplification and Sequencing
Specimens, decoction, and mixed samples: The samples (40–50 mg) were ground into fine powders via a Retsch MM400 laboratory mixer mill (Retsch Co., Germany) at a frequency of 30 Hz. The genomic DNA was subsequently extracted with a Plant Universal Genomic DNA Kit (Tiangen Biotech Beijing Co., China) according to the manufacturer's instructions. ITS2 was amplified by the universal primers 2F/3R (Chen et al., 2010).
Extract and Chinese patent medicines: Samples (40–50 mg) were collected into a tube and then ground via a Retsch MM400 laboratory mixer mill (Retsch Co.). Ten samples were collected in parallel per batch. The Chinese patent medicine powder was washed with 700 μL of prewash buffer [100 mM Tris-HCl, pH 8.0; 20 mM ethylenediaminetetraacetic acid (EDTA), pH 8.0; 700 mM NaCl; 2% polyvinylpyrrolidone (PVP)-40; and 0.4% β-mercaptoethanol] several times until the supernatant was clear and colorless, after which the mixture was centrifuged at 7500 × g for 5 min at room temperature. The precipitate was subsequently used to extract the genomic DNA via the Plant Universal Genomic DNA Kit (Tiangen Biotech Beijing Co.) according to the manufacturer's instructions. In the end, DNA from each batch was concentrated into one tube.
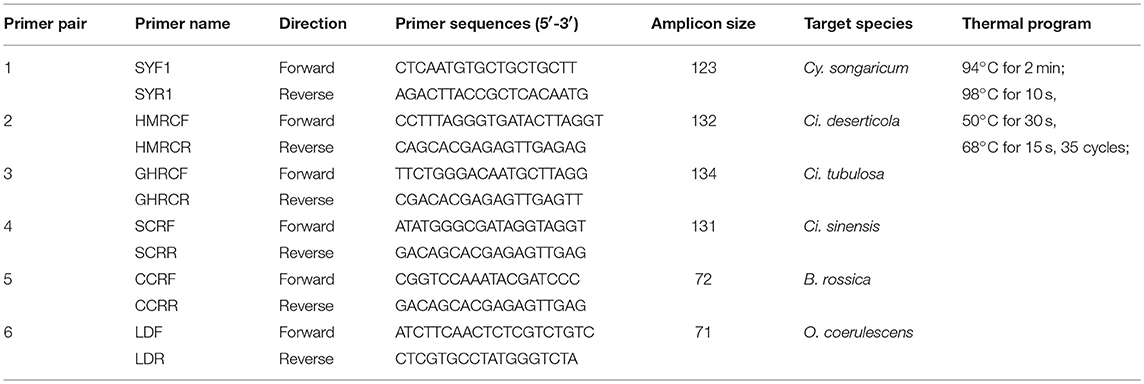
Six species-specific primer pairs—SYF1/SYR1, HMRCF/HMRCR, GHRCF/GHRCR, CCRF/CCRR, SCRF/SCRR, and LDF/LDR—were designed via Primer Premier 6.0 software (Premier Co., Canada) to amplify Cistanches Herba and its adulterants (the details are shown in Table 4). PCR was performed in a 50 μL-volume reaction containing 1 μL of KOD FX (Toyobo Co., Japan), 25 μL of 2 × PCR buffer, 10 μL of dNTPs (2 mM), 1.5 μL of each primer (10 μM), and 4 μL (~50 ng) of DNA template; the remaining volume consisted of double-distilled water. The reactions were performed by a thermal cycler (Veriti™ 96-Well Thermal Cycler, Applied Biosystems Co., USA); the thermal programs are listed in Table 4. The PCR products were examined via 3% agarose gel electrophoresis and purified for bidirectional sequencing with an ABI 3730XL sequencer (Applied Biosystems Co.) in accordance with the Sanger sequencing method. Then, the sensitivity of the six primers was tested via the quantitative real-time PCR (qRT-PCR) assay with a CFX96 Real-time System (Bio-rad Lab., USA). The cycle thresholds were automatically calculated by the system via the “PCR Baseline Subtracted Curve Fit” model. qRT-PCR was performed in a 15 μL-volume reaction containing 7.5 μL of SYBR® Premix Ex Taq™ (Tli RNaseH Plus) (Takara Bio Co., Japan), 1.0 μL of each primer (10 μM), and 1.0 μL of DNA template with the remaining volume consisting of double-distilled water. The qRT-PCR was performed with three technical replicates based on which standard error was calculated.
Sequence analysis: The sequences were edited and manually assembled via CodonCode Aligner 5.1.4 (CodonCode Co., USA). ITS sequences from GenBank were annotated via the Hidden Markov model (HMM) to obtain the ITS2 sequences (Keller et al., 2009). The sequences were then aligned by MEGA 5.0 software via the “Muscle” alignment method (Edgar, 2004; Tamura et al., 2011).
Results
Development of Nucleotide Signatures and Species-Specific Primer Pairs for Cistanches Herba and Adulterants
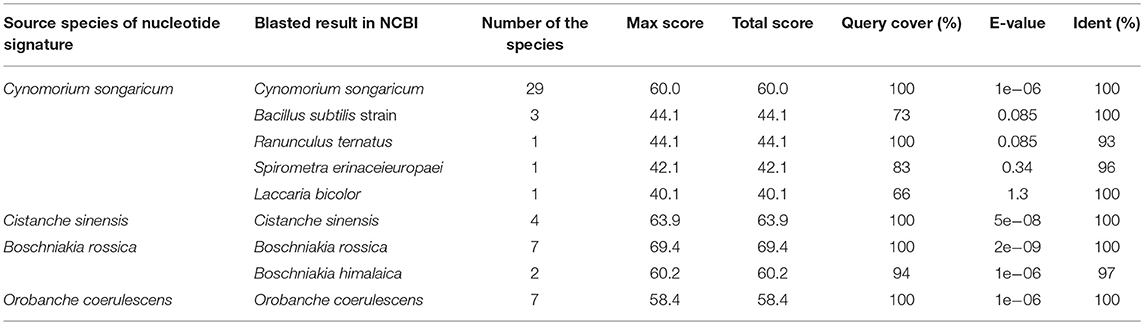
The PCR amplification and sequencing success rates of the 251 samples were 100% when the primer pair 2F/3R was used. The aligned length of the Cy. songaricum ITS2 sequences was 229 bp (Supplementary Figure S1). Analysis of the sequences from the herbarium species and those of closely related species retrieved from GenBank (Supplementary Table S2) revealed two single nucleotide polymorphism (SNP) sites for Cy. songaricum. On the basis of the SNPs, one Cy. songaricum-specific 30-bp nucleotide signature (5′-caattatttg aggtgcattg taagaagcgt-3') was developed (Figure 2A). Basic Local Alignment Search Tool (BLAST) results in NCBI demonstrated that this nucleotide signature was unique to Cy. songaricum (Table 5). With the similar analysis of the sequences from closely related species, one to two SNPs were discovered from Ci. sinensis, B. rossica and O. coerulescens. On the basis of the SNPs, the nucleotide signatures for the other three adulterants were also developed similarly, including a 34 bp signature (5′-cgatggtctc ccgtgcgcga ggatgcacgg ccgg-3′) for Ci. sinensis, a 37 bp signature (5′-acactggcct cccgtgcgca acgacgtgcg gccggtc-3') for B. rossica, and a 31 bp signature (5′-gtctgtcgtg tcggatggtg ttgcttgttg g-3′) for O. coerulescens (Figures 2B–D). BLAST analysis in NCBI also revealed that these nucleotide signatures were specific and not present in any other species (Table 5).
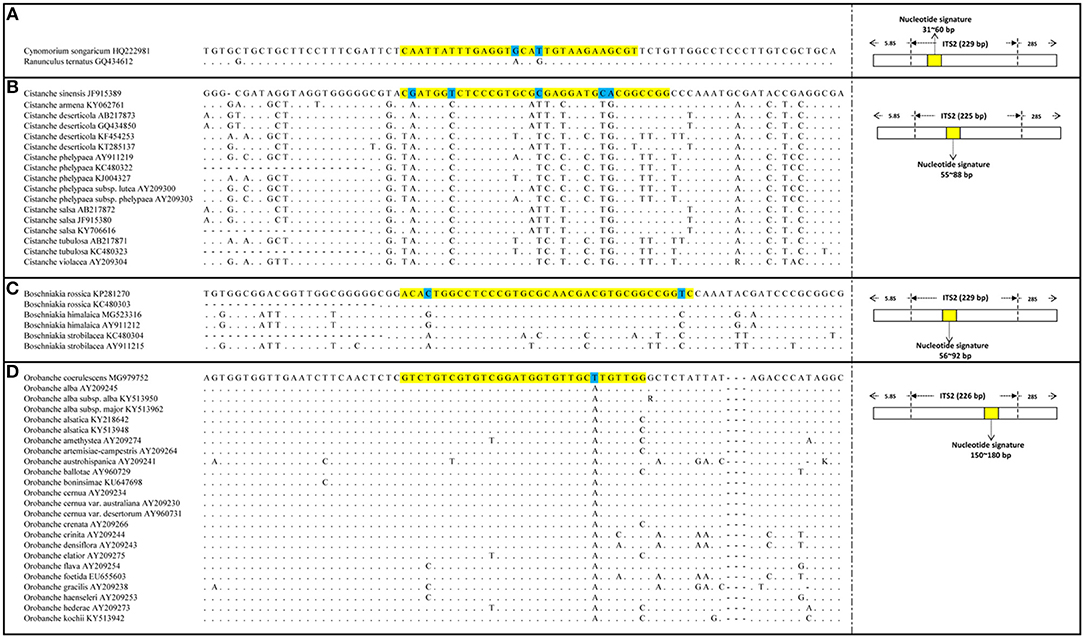
Figure 2. DNA sequence alignment results and SNP sites of the four nucleotide signatures. (A) Alignment of Cynomorium songaricum nucleotide signature with its region location in ITS2; (B) Alignment of Cistanche sinensis nucleotide signatures with its region location in ITS2; (C) Alignment of Boschniakia rossica nucleotide signature with its region location in ITS2; (D) Alignment of Orobanche coerulescens nucleotide signature with its region location in ITS2. The highlighted regions represent nucleotide signature regions and the marked bases represent the SNP sites of each nucleotide signature, the dots represent identical nucleotides.
Cistanches Herba and its adulterants could be amplified simultaneously from mixtures via the 2F/3R universal primer pair. Thus, we designed species-specific primers for nucleotide signature amplification by aligning the ITS2 sequences. Four specific primer pairs—SYF1/SYR1, CCRF/CCRR, SCRF/SCRR, and LDF/LDR—were designed to amplify the nucleotide signatures of Cy. songaricum, B. rossica, Ci. sinensis, and O. coerulescens, respectively (Table 4). The lengths of the amplicons were 123, 72, 131, and 71 bp, respectively.
In addition, a total of 214 ITS2 sequences from experimental Cistanches Herba materials were analyzed. Two short specific primers—HMRCF/HMRCR and GHRCF/GHRCR for Ci. deserticola and Ci. tubulosa, respectively—were designed to amplify the short regions of the degraded samples(Table 4). The lengths of the amplicons were 132 and 134 bp, respectively.
Validation of the Nucleotide Signature and Species-Specific Primer Method Based on Artificial Mixtures and Decoction
The amplification efficiencies of the new primer pairs were validated from the mixture. PCR products were obtained via each primer pair for each targeted species, as shown in Figure 3. For instance, the fifth mixture sample is a mixture of Cistanches Herba and its four adulterants, and each species could be amplified with the primers SYF1/SYR1, SCRF/SCRR, CCRF/CCRR, LDF/LDR, HMRCF/HMRCR, and GHRCF/GHRCR. Moreover, the amplification regions were sequenced, and the nucleotide signatures were observed within the target sequences. Furthermore, to measure the sensitivity, mixtures of two of the six samples were created at different weight ratios and the sample with lower proportion was amplified by specific primer pairs via qRT-PCR. As shown in Table 3, cq values of B. rossica or O. coerulescens could be obtained even the proportion of samples was 30000:1 or 40000:1, and the amplification results of Ci. sinensis, Ci. tubulosa, and Ci deserticola could be detected when the ratio was 20000:1. But no detectable results could be obtained for Cy. songaricum when it constituted the proportion of 20000:1.
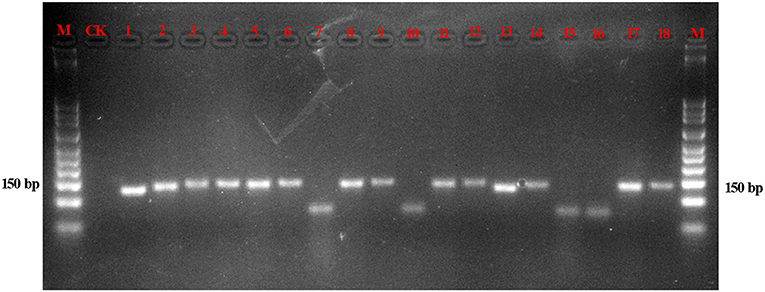
Figure 3. Gel image of the products of PCR amplification of the mixed powders. With the exception of the lanes marked M (marker) and CK (negative control), the other 13 lanes contain the PCR products of the mixtures. Lanes 1–3 are a mixture of Cy. songaricum, Ci. deserticola and Ci. tubulosa amplified with the primers SYF1/SYR1, HMRCF/HMRCR, and GHRCF/GHRCR, respectively; lanes 4–6 are a mixture of Ci. sinensis, Ci. deserticola, and Ci. tubulosa amplified with the primers SCRF/SCRR, HMRCF/HMRCR, and GHRCF/GHRCR, respectively; lanes 7–9 are a mixture of B. rossica, Ci. deserticola, and Ci. tubulosa amplified with the primers CCRF/CCRR, HMRCF/HMRCR, and GHRCF/GHRCR, respectively; lanes 10–12 are a mixture of O. coerulescens, Ci. deserticola, and Ci. tubulosa amplified with the primers LDF/LDR, HMRCF/HMRCR, and GHCRF/GHRCR, respectively; and lanes 13–18 are a mixture of Cy. songaricum, Ci. sinensis, B. rossica, O. coerulescens, Ci. deserticola, and Ci. tubulosa with the primers SYF1/SYR1, SCRF/SCRR, CCRF/CCRR, LDF/LDR, HMRCF/HMRCR, and GHCRF/GHRCR, respectively.
To verify whether the nucleotide signature method functions with processed materials, decoctions of Ci. deserticola and Cy. songaricum were prepared. The results showed that the short barcode from Ci. deserticola could be amplified, even after the samples were boiled for 210 min. In addition, the nucleotide signature of Cy. songaricum could be amplified after the samples were boiled for 150 min, while no PCR products were detected after the samples were boiled for 210 or 240 min (Figure 4). The sequencing results demonstrated that the short nucleotide signature was successfully obtained from the decoction.
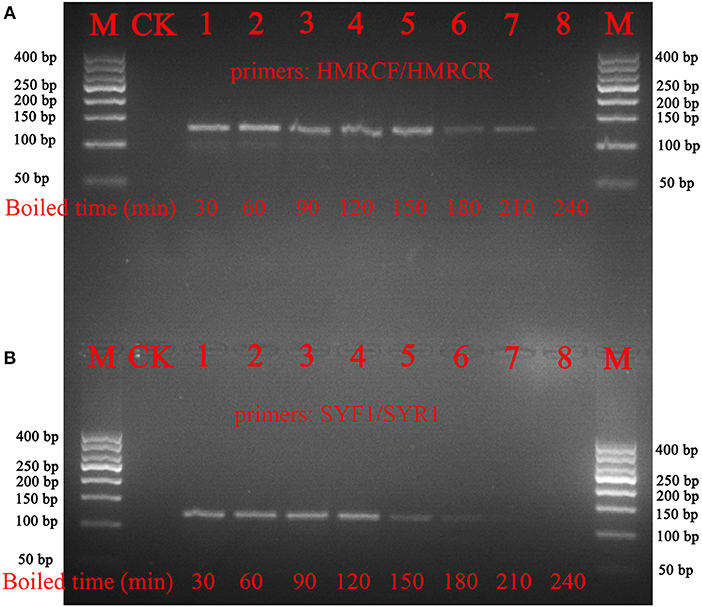
Figure 4. Gel images of the PCR amplifications of the boiled samples of Ci. deserticola and Cy. Songaricum. (A) Ci. deserticola; (B) Cy. Songaricum. With the exception of the lanes marked M (marker) and CK (negative control), the remaining 8 lanes contain the PCR products from the samples boiled for 30, 60, 90, 120, 150, 180, 210, and 240 min, respectively.
Market Survey Of Adulteration Via Nucleotide Signature and Specific Primers
The above method was applied for the detection of Cistanches Herba products on the market. Thirty five batches of Cistanches Herba slices, powders and extracts were amplified and sequenced by using six designed specific primers (the agarose gel electrophoresis results are shown in Supplementary Figure S2 and the sequences are listed in Supplementary Data Sheet 1). Analysis of the sequences via their nucleotide signatures revealed that five batches of slices were authentic. One slice batch and one powder batch were substituted with Cy. songaricum, and one extract was substituted with Ci. sinensis. The other six batches of powders were mixtures: five were adulterated with Cy. songaricum and Ci. sinensis, and one was adulterated with Cy. songaricum (Table 1). In addition, one slice batch was substituted with Salvia miltiorrhiza.
The Cistanches Herba in Chinese patent medicines is subjected to various processes that make authentication difficult. The ability of the six primer pairs to amplify the species-specific nucleotide signature regions from the Chinese patent medicines was tested (the agarose gel electrophoresis results are shown in Supplementary Figure S2). Most Chinese patent medicines contain components of different types of species. For example, Shihu Yeguang pills (ZCY34) contain 25 ingredients, including Cistanches Herba (Rou Cong Rong), Dendrobii Caulis (Shi Hu), and Ginseng Radix et Rhizoma (Ren Shen). From these ingredients, Cistanches Herba could be amplified specifically via the primer pair GHRCF/GHRCR. Direct sequencing of the PCR products revealed very clean traces. However, a visible band could not be obtained with the primer pair HMRCF/HMRCR.
Another example is Kangguzhi Zengsheng pills (ZCY44). There are eight ingredients in addition to Cistanches Herba in this Chinese patent medicine, but there were no visible bands obtained by either GHRCF/GHRCR or HMRCF/HMRCR, which meant that no Cistanches Herba was present. However, the adulterant region of Cy. songaricum was successfully amplified by the specific primer pair SYF1/SYR1. The sequencing results demonstrated the short nucleotide signature of Cy. songaricum was successfully obtained.
By seeking the nucleotide signatures developed in this study, we found that 15 of 31 Chinese patent medicines labeled as containing Cistanches Herba instead contained adulterants, including eight counterfeit ingredients and seven adulterants (Table 2). For example, six batches were replaced with Cy. songaricum, including three batches of Shihu Yeguang pills, one batch of Kangguzhi Zengsheng pills, one batch of Sanbao capsules, and one batch of Wenweishu particles.
Moreover, different batches from the same manufacturer produced somewhat different results. For example, among three batches from one manufacturer (ZCY16, ZCY69, and ZCY71), one batch comprised a mixture of Ci. deserticola and Cy. songaricum, one batch comprised a mixture of Ci. deserticola and Ci. tubulosa, and one batch contained only Ci. tubulosa. Two batches from another manufacturer (ZCY44 and ZCY70) also differed: one batch contained Cy. songaricum, whereas the other batch contained Ci. deserticola and Ci. tubulosa.
Cistanches Herba was detected in 23 of the 31 Chinese patent medicines tested (Table 2), and only 16 samples were authentic, e.g., without adulterants or counterfeit ingredients. Ci. tubulosa was detected in 16 batches of Chinese patent medicines, and Ci. deserticola was detected in 10 batches. O. coerulescens was not detected in any of the products (Table 2).
Discussion
Necessity of Developing a New Method for Monitoring Commercially Available Medicinal Products Containing Cistanches Herba
Cistanches Herba is a tonic that is widely used in restorative Chinese patent medicines and other medicinal products. However, the quality control of Chinese patent medicines presents great challenge due to the diversity and complexity of the ingredients. Due to the lack of regulatory oversight, there is considerable opportunity for product adulteration or counterfeiting. In addition, all products should be processed in accordance with the Pharmacopoeia or other standards; adulterating or counterfeiting is not permitted during processing. Thus, varieties of quality control methods have been established, such as multi-heart-cutting two-dimensional liquid chromatography (Yao et al., 2015), near-infrared reflectance spectroscopy (Zhang and Su, 2014; Zhang et al., 2015) and liquid chromatography-mass spectrometry (Wang et al., 2016b). However, the analytical chemistry methods currently in the Chinese Pharmacopoeia Commission (2015) cannot be used to authenticate all of the ingredients in Chinese patent medicines or to detect the presence of adulterant ingredients. Moreover, studies have shown that targeted metabolites in plants are altered during product processing, resulting in considerable variability in test results or complete failure of test methods (Ananingsih et al., 2013). Thus, molecular tools such as species-specific nucleotide signatures are poised to reinforce quality control systems against the risk of fraudulent product substitution and adulteration and inclusion of unlabeled ingredients.
Although ITS/ITS2 is considered a high-efficiency tool for the identification of herbal medicines, these sequences cannot be amplified from highly processed samples (Newmaster et al., 2013; de Boer et al., 2015). Wang et al. reported that ITS2 could not be amplified from Angelicae Sinensis Radix extract or decoctions boiled for more than 120 min (Wang et al., 2016a). According to traditional technologies and the Chinese Pharmacopoeia Commission (2015), Cistanches Herba is always highly processed to increase its medicinal efficacy; these processes, include oven drying, salting and steaming with wine (Zou et al., 2017), which lead to DNA degradation. In addition, various excipients are added during processing, such as honey, starch, and dextrin. If these excipients are not removed completely, the purity of DNA will be affected. For example, the following manufacturing process is used to generate Cistanches Herba-containing Sanbao capsules: “Boil the medicinal slices for 1.5 h twice, combine the decoctions and filter the mixture. Concentrate the filtrate to a relative density of 1.20~1.25 (at 80°C). Add other ground powders and combine them to obtain a homogeneous mixture. Next, dry the mixture at 60°C, and then grind it into a fine powder.” However, after the production process described above, there could be difficulties during the DNA extraction of Chinese patent medicines, and long fragments might not be amplified from the degraded DNA, which would prevent the identification of adulterants. Thus, to ensure the quality and purity of DNA, we added additional steps before the genomic DNA extraction, including washing with prewash buffer and eluting ten parallel tubes into one tube for each batch.
Although all the Chinese patent medicines used in the present study contained 6–25 ingredients, the primer pairs developed could specifically amplify the sequences of the adulterants in these Chinese patent medicines. Direct sequencing of the PCR products showed clean trace files. Thus, this nucleotide signature method is capable of identifying both authentic species ingredients and adulterants and should broaden the application of DNA-based molecular diagnostic tools for market supervision.
Nucleotide Signatures for the Effective Identification of Cistanches Herba Products
Molecular tools that utilize PCR technology are very promising for medicinal product authentication within quality control systems. The successful application of the primers for identifying DNA-degraded adulterants from Cistanches Herba suggests that a PCR-based detection method could be used widely. In the Chinese herbal medicine market, the price of authentic Cistanches Herba species ingredients is more than five times higher than the prices of its adulterants. Our results showed that Cy. songaricum is the most common adulterant of Cistanches Herba on the market, followed by Ci. sinensis. Cy. songaricum was added because these medicines share similar morphological characteristics. In addition, the chemical composition of Ci. sinensis is similar to that of Cistanches Herba. As quality control markers for Cistanches Herba extracts, echinacoside and acteoside can be inexpensively extracted from Ci. sinensis. Thus, some pharmaceutical factories use Ci. sinensis as a substitute in the production of Cistanches Herba extracts. Taken together, the results of this study indicate that there is considerable fraud in the market for medicinal products.
Adulteration in Chinese patent medicine is similar to that found in other countries. Similar levels of adulteration have been recorded in North America (Newmaster et al., 2013), Europe (Raclariu et al., 2017), and Asia (Cheng et al., 2014; Shanmughanandhan et al., 2016; Gao et al., 2017). In this study, the adulterated rate of Chinese patent medicines was approximately 48.4%, with only 16 of the 31 samples being authentic Cistanches Herba. In addition, we speculated that the different results produced in products from the same manufacturer could be attributed to differences in the qualities of the different batches of Chinese medicine materials. Therefore, to control the quality of Chinese patent medicines, the raw materials should be authenticated before being processed into products.
Adulteration of Cistanches Herba has traditionally been associated with issues of supply and demand of raw materials. Ci. deserticola and Ci. tubulosa are the two original plants currently used to formulate Cistanches Herba. However, Ci. deserticola is the only original species in traditional authentic Cistanches Herba listed in the Chinese Pharmacopoeia Commission (2000), in which Ci. tubulosa is identified as an adulterant. Owing to the shortage of Ci. deserticola resources, Ci. tubulosa has been listed as a supplement in the Chinese Pharmacopoeia since 2005 (Jiang and Tu, 2009). Until recently, the prices of these herbs have markedly differed; Ci. tubulosa has been much less expensive than Ci. deserticola because there is a much larger supply of the former. Here, our results showed that Ci. tubulosa is more widely used in commercially available Cistanches Herba products.
In conclusion, the nucleotide signatures and PCR-based methods developed in this study may serve as useful tools for the medicinal product industry to authenticate ingredients and detect adulterants in Cistanches Herba products. In accordance of the sensitivity result, even if the proportion of adulterant was one in ten thousand, it can be detected via qRT-PCR. It means that once a nucleotide signature is detected in Cistanches Herba-containing functional products, it could be identified as an adulterant or counterfeit ingredient. A novel solution for detecting counterfeit ingredients or adulterated Cistanches Herba was provided that was not previously available via chemical detection methods in the Chinese Pharmacopoeia. In addition, this method could be used to validate increasing types of medicine and to broaden the applications of DNA-based molecular diagnostic tools for market supervision.
Author Contributions
JH conceived the study and participated in its design. XW, RX, and JC contributed samples and performed the experiments. XW analyzed the data. XW, JH, ZZ, S-GN, JS, and SC drafted the manuscript. All authors have read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China [grant number 81673552], the CAMS Innovation Fund for Medical Sciences [grant number 2016-I2 M-3-016], and the United Fund Key Project of the National Natural Science Foundation of China [grant number U1403224].
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank our colleagues who helped with the sample collection, identification, laboratory work and manuscript preparation, including Chaokui Sun, Dianyun Hou, and Piao Zhang.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01643/full#supplementary-material
Supplementary Figure S1. The alignment result of ITS2 sequence from six species.
Supplementary Figure S2. Agarose gel electrophoresis of all samples.
Supplementary Table S1. Sampling information of Cistanches Herba and its adulterants.
Supplementary Table S2. Sequence information of related species downloaded from GenBank.
Supplementary Table S3. The declared compositions of different Chinese patent medicine samples.
Supplementary Data Sheet S1. All the sequences obtained in this study.
References
Ananingsih, V. K., Sharma, A., and Zhou, W. (2013). Green tea catechins during food processing and storage: a review on stability and detection. Food Res. Int. 50, 469–479. doi: 10.1016/j.foodres.2011.03.004
Chen, H., Jing, F. C., Li, C. L., Tu, P. F., Zheng, Q. S., and Wang, Z. H. (2007). Echinacoside prevents the striatal extracellular levels of monoamine neurotransmitters from diminution in 6-hydroxydopamine lesion rats. J. Ethnopharmacol. 114, 285–289. doi: 10.1016/j.jep.2007.07.035
Chen, S., Yao, H., Han, J., Liu, C., Song, J., Shi, L., et al. (2010). Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE 5:e8613. doi: 10.1371/journal.pone.0008613
Cheng, X., Su, X., Chen, X., Zhao, H., Bo, C., Xu, J., et al. (2014). Biological ingredient analysis of traditional Chinese medicine preparation based on highthroughput sequencing: the story for Liuwei Dihuang Wan. Sci. Rep. 4:5147. doi: 10.1038/srep05147
Chinese Pharmacopoeia Commission. (2000). Pharmacopoeia of the People's Republic of China. Part I. Beijing: China Medical Science Press.
Chinese Pharmacopoeia Commission. (2015). Pharmacopoeia of the People's Republic of China. Part I. Beijing: China Medical Science Press.
de Boer, H. J., Ichim, M. C., and Newmaster, S. G. (2015). DNA barcoding and pharmacovigilance of herbal medicines. Drug Saf. 38, 611–620. doi: 10.1007/s40264-015-0306-8
Dubey, B., Meganathan, P. R., and Haque, I. (2011). DNA mini-barcoding: an approach for forensic identification of some endangered Indian snake species. Forensic. Sci. Int. Genet. 5, 181–184. doi: 10.1016/j.fsigen.2010.03.001
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Gao, Z., Liu, Y., Wang, X., Song, J., Chen, S., Ragupathy, S., et al. (2017). Derivative technology of DNA barcoding (nucleotide signature and SNP double peak methods) detects adulterants and substitution in Chinese patent medicines. Sci. Rep. 7:5858. doi: 10.1038/s41598-017-05892-y
Gu, C. M., Yang, X. Y., and Huang, L. F. (2016). Cistanches Herba: a neuropharmacology review. Front. Pharmacol. 7:289. doi: 10.3389/fphar.2016.00289
Hajibabaei, M., Smith, M. A., Janzen, D. H., Rodriguez, J. J., Whitfield, J. B., and Hebert, P. D. N. (2006). A minimalist barcode can identify a specimen whose DNA is degraded. Mol. Ecol. Notes 6, 959–964. doi: 10.1111/j.1471-8286.2006.01470.x
Japan Pharmacopeial Convention. (2016). The Japanese Pharmacopoeia, 17th Edn. Tokyo: The Ministry of Health, Labour and Welfare of Japan, 1831–1832.
Jiang, Y., and Tu, P. F. (2009). Analysis of chemical constituents in Cistanche species. J. Chromatogr. A 1216, 1970–1979. doi: 10.1016/j.chroma.2008.07.031
Keller, A., Schleicher, T., Schultz, J., Müller, T., Dandekar, T., and Wolf, M. (2009). 5.8S-28S rRNA interaction and HMM-based ITS2 annotation. Gene 430, 50–57. doi: 10.1016/j.gene.2008.10.012
Lei, L., Yang, F., Zhang, T., Tu, P., Wu, L., and Ito, Y. (2001). Preparative isolation and purification of acteoside and 2'-acetyl acteoside from Cistanches salsa (C.A. Mey.) G. Beck by high-speed counter-current chromatography. J. Chromatogr. A 912, 181–185. doi: 10.1016/S0021-9673(01)00583-0
Li, Z., Lin, H., Gu, L., Gao, J., and Tzeng, C. M. (2016). Herba Cistanche (Rou Cong-Rong): one of the best pharmaceutical gifts of traditional Chinese medicine. Front. Pharmacol. 7:41. doi: 10.3389/fphar.2016.00041
Liu, X. M., Li, J., Jiang, Y., Zhao, M. B., and Tu, P. F. (2013). Chemical constituents from Cistanche sinensis (Orobanchaceae). Biochem. Syst. Ecol. 47, 21–24. doi: 10.1016/j.bse.2012.09.003
Liu, Y., Wang, X., Wang, L., Chen, X., Pang, X., and Han, J. (2016). A nucleotide signature for the identification of American ginseng and its products. Front. Plant Sci. 7:319. doi: 10.3389/fpls.2016.00319
Lo, Y. T., Li, M., and Shaw, P. C. (2015). Identification of constituent herbs in ginseng decoctions by DNA markers. Chin. Med. 10:1. doi: 10.1186/s13020-015-0029-x
Meusnier, I., Singer, G. A., Landry, J. F., Hickey, D. A., Hebert, P. D., and Hajibabaei, M. (2008). A universal DNA mini-barcode for biodiversity analysis. BMC Genomics 9:214. doi: 10.1186/1471-2164-9-214
Newmaster, S. G., Grguric, M., Shanmughanandhan, D., Ramalingam, S., and Ragupathy, S. (2013). DNA barcoding detects contamination and substitution in North American herbal products. BMC. Med. 11:222. doi: 10.1186/1741-7015-11-222
Raclariu, A. C., Mocan, A., Popa, M. O., Vlase, L., Ichim, M. C., Crisan, G., et al. (2017). Veronica officinalis product authentication using DNA metabarcoding and HPLC-MS reveals widespread adulteration with Veronica chamaedrys. Front. Pharmacol. 8:378. doi: 10.3389/fphar.2017.00378
Shanmughanandhan, D., Ragupathy, S., Newmaster, S. G., Mohanasundaram, S., and Sathishkumar, R. (2016). Estimating herbal product authentication and adulteration in India using a vouchered, DNA-based biological reference material library. Drug Saf. 39, 1211–1227. doi: 10.1007/s40264-016-0459-0
Sun, Z. Y., Song, J. Y., Yao, H., and Han, J. P. (2012). Molecular identification of Cistanches Herba and its adulterants based on nrITS2 sequence. J. Med. Plants Res. 6, 1041–1045. doi: 10.5897/JMPR11.1115
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Wang, Q., Song, W., Qiao, X., Ji, S., Kuang, Y., Zhang, Z. X., et al. (2016b). Simultaneous quantification of 50 bioactive compounds of the traditional Chinese medicine formula Gegen-Qinlian decoction using ultra-high performance liquid chromatography coupled with tandem mass spectrometry. J. Chromatogr. A 1454, 15–25. doi: 10.1016/j.chroma.2016.05.056
Wang, T., Zhang, X. Y., and Xie, W. Y. (2012). Cistanche deserticola YC Ma, “Desert ginseng”: a review. Am. J. Chin. Med. 40, 1123–1141. doi: 10.1142/S0192415X12500838
Wang, X., Liu, Y., Wang, L., Han, J., and Chen, S. (2016a). A nucleotide signature for the identification of Angelicae Sinensis Radix (Danggui) and its products. Sci. Rep. 6:34940. doi: 10.1038/srep34940
Yao, C. L., Yang, W. Z., Wu, W. Y., Da, J., Hou, J. J., Zhang, J. X., et al. (2015). Simultaneous quantitation of five Panax notoginseng saponins by multi heart-cutting two-dimensional liquid chromatography: method development and application to the quality control of eight Notoginseng containing Chinese patent medicines. J. Chromatogr. A 1402, 71–81. doi: 10.1016/j.chroma.2015.05.015
Zhang, C., and Su, J. (2014). Application of near infrared spectroscopy to the analysis and fast quality assessment of traditional Chinese medicinal products. Acta. Pharm. Sin. B 4, 182–192. doi: 10.1016/j.apsb.2014.04.001
Zhang, W., Qu, Z., Wang, Y., Yao, C., Bai, X., Bian, S., et al. (2015). Near-infrared reflectance spectroscopy (NIRS) for rapid determination of ginsenoside Rg1 and Re in Chinese patent medicine Naosaitong pill. Spectrochim. Acta. A Mol. Biomol. Spectrosc. 139, 184–188. doi: 10.1016/j.saa.2014.11.111
Keywords: Cistanches Herba, Chinese patent medicine, nucleotide signature, degraded DNA, medicine quality control
Citation: Wang X, Xu R, Chen J, Song J, Newmaster S-G, Han J, Zhang Z and Chen S (2018) Detection of Cistanches Herba (Rou Cong Rong) Medicinal Products Using Species-Specific Nucleotide Signatures. Front. Plant Sci. 9:1643. doi: 10.3389/fpls.2018.01643
Received: 27 February 2018; Accepted: 23 October 2018;
Published: 13 November 2018.
Edited by:
Caroline Howard, Medicines and Healthcare Products Regulatory Agency, United KingdomReviewed by:
Catherine Anne Kidner, University of Edinburgh, United KingdomAnna Paola Casazza, Istituto di Biologia e Biotecnologia Agraria (IBBA), Italy
Copyright © 2018 Wang, Xu, Chen, Song, Newmaster, Han, Zhang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian-ping Han, anBoYW5AaW1wbGFkLmFjLmNu
Zheng Zhang, emhhbmd6aGVuZzMyMUAxMjYuY29t
 Xiao-yue Wang
Xiao-yue Wang Rong Xu
Rong Xu Jun Chen
Jun Chen Jing-yuan Song1
Jing-yuan Song1 Steven-G Newmaster
Steven-G Newmaster Jian-ping Han
Jian-ping Han Zheng Zhang
Zheng Zhang Shi-lin Chen
Shi-lin Chen