- 1Chongqing Engineering Research Center for Rapeseed, College of Agronomy and Biotechnology, Southwest University, Chongqing, China
- 2Academy of Agricultural Sciences, Southwest University, Chongqing, China
Brassica napus L. is a widely cultivated oil crop and provides important resources of edible vegetable oil, and its quality is determined by fatty acid composition and content. To explain the genetic basis and identify more minor loci for fatty acid content, the multi-locus random-SNP-effect mixed linear model (mrMLM) was used to identify genomic regions associated with fatty acid content in a genetically diverse population of 435 rapeseed accessions, including 77 winter-type, 55 spring-type, and 303 semi-winter-type accessions grown in different environments. A total of 149 quantitative trait nucleotides (QTNs) were found to be associated with fatty acid content and composition, including 34 QTNs that overlapped with the previously reported loci, and 115 novel QTNs. Of these, 35 novel QTNs, located on chromosome A01, A02, A03, A05, A06, A09, A10, and C02, respectively, were repeatedly detected across different environments. Subsequently, we annotated 95 putative candidate genes by BlastP analysis using sequences from Arabidopsis thaliana homologs of the identified regions. The candidate genes included 34 environmentally-insensitive genes (e.g., CER4, DGK2, KCS17, KCS18, MYB4, and TT16) and 61 environment-sensitive genes (e.g., FAB1, FAD6, FAD7, KCR1, KCS9, KCS12, and TT1) as well as genes invloved in the fatty acid biosynthesis. Among these, BnaA08g08280D and BnaC03g60080D differed in genomic sequence between the high- and low-oleic acid lines, and might thus be the novel alleles regulating oleic acid content. Furthermore, RT-qPCR analysis of these genes showed differential expression levels during seed development. Our results highlight the practical and scientific value of mrMLM or QTN detection and the accuracy of linking specific QTNs to fatty acid content, and suggest a useful strategy to improve the fatty acid content of B. napus seeds by molecular marker-assisted breeding.
Introduction
Rapeseed (Brassica napus L.) is one of the most important oil crops in the world, providing not only edible vegetable oil but also its potential use in lubricants and biofuels (Saeidnia and Gohari, 2012). However, the physical, chemical, and nutritional qualities of rapeseed oil depend mainly on its fatty acid composition, which consists approximately of 60% oleic acid (C18:1), 4% palmitic acid (16:0), and 2% stearic acid (18:0) (Bauer et al., 2015; Wen et al., 2015). Rapeseed oil is considered by many nutritionists to be ideal for human nutrition and superior to many other plant oils (Zhao et al., 2008; Qu et al., 2017), as it can be heated to high temperatures without smoking (Miller et al., 1987), and reduces levels of undesirable low-density lipoprotein cholesterol in the blood plasma, reducing the risk of arteriosclerosis (Chang and Huang, 1998). Optimizing the fatty acid composition is an important breeding objective for rapeseed cultivar development.
In B. napus, fatty acid metabolism is influenced by both genotype and environmental factors. Efforts to improve the oil quality have yielded many high oleic acid Brassica lines, including B. rapa (Tanhuanpää et al., 1996), B. carinata (Velasco et al., 1997), and B. napus (Pleines and Friedt, 1989; Fei et al., 2012). Further, oleic acid concentrations >70% have already been achieved in rapeseed through hybrid breeding methods (Zhang et al., 2009). Fatty acid content is a typical quantitative trait controlled by multiple genes that regulate its desaturation (Wang et al., 2015; Chen et al., 2018), and numerous quantitative trait loci (QTLs) for fatty acids have been mapped to all 19 chromosomes of B. napus, with most being found on chromosomes A01, A02, A03, A08, A10, C03, A04, A07, A09, C01, C06, and C08 (Burns et al., 2003; Zhao et al., 2008; Liu and Li, 2014; Bauer et al., 2015; Lee et al., 2015; Teh, 2015; Wen et al., 2015; Javed et al., 2016). With the increasing availability of whole-genome-sequences and SNP array development, association mapping represents a powerful approach for dissecting the genetic basis of complex quantitative traits at high resolution, which could significantly increase the precision of estimating QTL locations (Meuwissen and Goddard, 2000). Recently, genome-wide association studies (GWAS) have been performed to detect the genetic variation associated with important agronomic traits in rapeseed using the Illumina Infinium Brassica 60K SNP array (Delourme et al., 2013; Li et al., 2014; Lu et al., 2014; Hatzig et al., 2015; Luo et al., 2015), including seed weight and quality (Li et al., 2014), seed oil content in a panel of 521 rapeseed accessions (Liu et al., 2016), and the composition of seven fatty acids (Qu et al., 2017). Although these studies have revealed loci for associated with fatty acid traits, no beneficial alleles have been detected within the B. napus accessions.
Numerous studies showed that FATTY ACID DESATURASE 2 (FAD2) is the major gene responsible for the desaturation of oleic acid to linolenic acid (Hu et al., 2006; Peng et al., 2010; Yang et al., 2012), and four paralogs of FAD2 were previously identified in B. napus (Scheffler et al., 1997; Yang et al., 2012). These paralogs were mainly expressed in the developing seeds, suggesting possible roles in controlling oleic acid content in B. napus (Xiao et al., 2008). In addition, KCS18, is known to play a crucial role in regulating erucic acid biosynthesis in B. napus (Wang et al., 2008; Wu et al., 2008; Li et al., 2014). However, the identified QTL were not cloned and undertaken for contributing to the minor fatty acids. Furthermore, the genetic basis of fatty acid synthesis is still unclear.
The multi-locus random-SNP-effect mixed linear model (mrMLM) is emerged as a powerful tool for quantitative trait nucleotide (QTN) detection and QTN effect estimation for complex traits (Wang et al., 2016; Li et al., 2017; Chang et al., 2018; Peng et al., 2018). For example, Li et al. (2017) detected 38 significantly-associated loci and identified numerous highly-promising candidate genes (e.g., TAC1, SGR1, SGR3, and SGR5), for branch angle across 472 rapeseed accessions. Zhang et al. (2018) identified 127 significant QTNs for stalk lodging resistance-related traits using mrMLM in a population of 257 maize inbred lines. As reported by Ma et al. (2018), 127 significant QTNs with maize embryonic callus regenerative capacity were identified in a population of 144 maize inbred lines, and many candidate genes were reported to relate with auxin transport, cell fate, seed germination, or embryo development, respectively. In the present study, we analyzed the fatty acid composition in 77 winter varieties, 55 spring varieties, and 303 semi-winter varieties of rapeseed grown in three environments, and genotyped all of the accessions using the high-through Brassica 60K SNP array (Clarke et al., 2016). Then, 32,543 SNPs from the 60K SNP array were used for genome-wide association analysis usingmrMLM. In total, 149 QTNs were identified using mrMLM, suggesting that this is an effective model for identifying candidate genes underlying complex traits. Subsequently, 95 candidate genes were annotated using BlastP against A. thaliana homologs, providing insight into the genetic control of fatty acid content in B. napus. Furthermore, novel fatty acid content-associated SNPs identified here may be useful for marker-based breeding programs aimed at improving the fatty acid content of B. napus seeds.
Materials and Methods
Plant Materials
A diversity panel consisting of 55 spring, 77 winter, and 303 semi-winter rapeseed accessions (B. napus; Supplementary Table S1) was used for the association analysis. These accessions were grown in three growing seasons (2015–2016, 2016–2017, and 2017–2018) in Beibei (106.38°E, 29.84°N), Chongqing, China. Three rows of 10–12 plants per accession were established in the experimental fields with a randomized complete block design and three replications. Self-pollinated seeds were harvested from plants at complete physiological maturity and used for the fatty acid analysis.
Fatty Acid Measurement and Statistical Analysis
Seeds (200 mg) were homogenized with a pestle and extracted in 2 mL petroleum ether:ether (1:1) for 40 min, and methylated with 1 mL KOH/methanol (0.4 mol L−1). The supernatants separated by adding distilled water were identified by gas-liquid chromatography on a Model GC-2010 (Shimadzu, Kyoto, Japan). Chromatographic analysis was carried out using a fused silica capillary column DB-WAX (30 m × 0.246 mm × 0.25 um) with default parameters (Qu et al., 2017). Fatty acid profiles were calculated as a percentage of total fatty acids in the seeds, and optimized with an R script of the best linear unbiased prediction (BLUP) (Merk et al., 2012). The resulting values for each accession were used in the association analysis. All experiments were performed in triplicate, and the mean, standard deviation (SD), coefficient of variation (CV), and minimum (Min) and maximum (Max) values of the oleic acid content were calculated using SPSS 15.0 (IBM Corp, Armonk, NJ, USA).
SNP Genotyping Data Acquisition and Analysis
The methods used for SNP genotyping and mapping were described in previous reports of Li et al. (2014) and Liu et al. (2016). Using the Brassica 60K Illumina® Infinium SNP array (Clarke et al., 2016), the genotype of each accession was generated at the National Subcenter of Rapeseed Improvement in Wuhan (Huazhong Agricultural University, Wuhan, China). The low quality SNPs (call frequency < 0.9 and a minor allele frequency ≤ 0.05) were filtered in all accessions. In addition, SNPs not accurately mapped to the B. napus genome were excluded. The probe sequences of 52,157 SNPs were used to perform a local BlastN search against the B. napus “Darmor-bzh” reference genome (version 4.1, http://www.genoscope.cns.fr/brassicanapus/data/; Chalhoub et al., 2014) using our previously published method (Wei et al., 2015). In total, 32,543 SNPs were analyzed further.
The Q matrix of population structure was calculated by a Bayesian model-based analysis in STRUCTURE 2.1 (Pritchard et al., 2000) with published parameters of Falush et al. (2003) and Qu et al. (2017). The optimal number of K values (K = 2; Supplementary Figure S1) was determined using the Evanno method (Evanno et al., 2005). The Q matrix was selected as the fixed covariate in the subsequent association analysis (Gajardo et al., 2015). To visualize genetic relatedness among all genotypes, the principal component analysis (PCA) was constructed using the GCTA tool (Yang et al., 2011). The relative kinship matrix for each association panel was calculated using SPAGeDi (Hardy and Vekemans, 2002), and the negative values were defined as zero between two individuals, following the method of Yu et al. (2006).
Genome-Wide Association Analysis
The mrMLM significantly improved the power and precision of the GWAS, which was previously used in B. napus (Li et al., 2017). Therefore, the multi-locus GWAS method (mrMLM, https://cran.r-project.org/web/packages/mrMLM.GUI/index.html) was performed to evaluate the trait-SNP association analysis in this study (Wang et al., 2016). Moreover, the phenotypic and genotypic datasets, kinship (K), and population structure (Q) were imported into the R package mrMLM, and significantly associated SNPs were identified by mrMLM with the critical log of odds (LOD) score of 3.0 (p = 0.0002) (Wang et al., 2016). The QTNs were named using the nomenclature described by McCouch et al. (1997). For example, q-C16:0-A03-1 indicated the first locus located on chromosome A03 associated with palmitic acid.
Candidate Gene Prediction
Candidate genes were identified using significant SNP markers, which were detected using mrMLM (Wang et al., 2016). The association regions and 100-kb region upstream or downstream of peak SNPs associated with fatty acid content were identified based on the physical distance of chromosomes of significant associated-trait SNPs in the B. napus “Darmor-bzh” genome (version 4.1; Chalhoub et al., 2014). Subsequently, putative candidate genes were predicted according to the annotation of the SNP-tagged genome regions and confirmed by BlastP searches against the Arabidopsis genome with an E-value ≤ 1E-10. The function of these candidate genes was further annotated using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (https://www.genome.jp/kegg/pathway.html). Highly-orthologous genes involved in fatty acid biosynthesis were analyzed further, which were defined as the environment-insensitive and -sensitive genes according to the frequency detected between the different ecological genotypes and environments. To identify the directed functional genes for fatty acid, sequences of these candidate genes, isolated from plants with higher- and lower fatty acid levels, were aligned using ClustalW (Thompson et al., 1994) implemented in Geneious 4.8.5 software (Biomatters, Auckland, New Zealand).
Analysis of the Expression Profiles of Candidate Fatty Acid-Associated Genes
Total RNA was extracted from the seeds of B. napus cultivar Zhongshuang No. 11 (ZS11) at 15 developmental stages (3–49 days after pollination) using the RNAprep Pure Plant Kit (Tiangen Biotech, Beijing, China), following the manufacturer's instructions. The cDNA library construction and RNA sequencing were performed using Novogene Bioinformatics Technology (Beijing, China). Transcriptome sequencing datasets were deposited in the BioProject database (BioProject ID PRJNA358784). The data were analyzed as previously described (Qu et al., 2015), and the expression profiles of the candidate genes were quantified in terms of fragments per kilobase of exon per million mapped fragments (FPKM), using Cufflinks with default parameters (Trapnell et al., 2012). These transcriptome datasets were previously deemed suitable for selecting candidate genes (Zhou et al., 2017). Candidate genes were validated using RT-qPCR analysis. Three biological replicates and three technical replicates were performed on a CFX96 Real-Time PCR system (Bio-Rad, Laboratories, Hercules, CA, USA), and the expression levels of candidate genes were calculated using the 2−ΔΔCt method (Zhou et al., 2017). Hence, the expression values of the 106 candidate genes were normalized by Log2 (expression values). Heatmaps of the candidate genes were drafted using HemI 1.0 (http://hemi.biocuckoo.org/). The specific primer sequences used in this study were obtained from the qPCR Primer Database (Lu et al., 2017) and are listed in Supplementary Table S3.
Results
Phenotypic Variation and Correlation Among Different Rapeseed Genotypes
Extensive variation in fatty acid content was observed between rapeseed plants of different genotypes grown in over 3 years (Table 1). The content of palmitic acid, stearic acid and linolenic acid varied slightly among the different ecotypic rapeseed accession at different years. For example, the mean palmitic acid content varied from 2.53 to 5.49% in spring rapeseed, 2.66 to 5.78% in winter rapeseed, and 2.69 to 6.10% in semi-winter rapeseed. The stearic acid content varied from 0.24 to 2.94% in spring rapeseed, 0.05 to 3.98% in winter rapeseed, and 0.09 to 2.89% in semi-winter rapeseed. The linolenic acid content varied from 4.90 to 14.61% in spring rapeseed, 2.15 to 11.41% in winter rapeseed, and 6.54 to 12.31% in semi-winter rapeseed. However, considerable quantitative variation was found for the content of oleic acid, linoleic acid, eicosenoic acid, and erucic acid. For instance, the mean oleic acid content ranged from 14.49 to 72.21% in spring rapeseed, 10.65 to 76.19% in winter rapeseed, and 7.91 to 83.00% in semi-winter rapeseed; the linoleic acid content ranged from 10.98 to 28.32%, 10.07 to 27.61%, and 10.41 to 28.07% in spring, winter, and semi-winter rapeseed, respectively, the eicosenoic acid content were ranged from 1.92 to 17.16%, 1.12 to 18.05%, and 0.22 to 22.34% in spring, winter and semi-winter rapeseed, respectively, and the erucic acid content ranged from 0 to 53.41 μmol g−1, 0 to 52.83 μmol g−1, and 0 to 48.58 μmol g−1 in spring, winter and semi-winter rapeseed, respectively (Table 1). Moreover, the largest CV (coefficient of variation) was found among the oleic acid, eicosenoic acid, and erucic acid content in different ecotypic rapeseed at different environments, ranging from 25.33 to 40.62, 50.00 to 183.00%, and 54.28 to 199.83%, respectively (Table 1), indicating that extensive variation was widely detected in the panel of accessions. In addition, the phenotypic values of fatty acid content were displayed among the ecotypic rapeseed accessions at different years (Figures 1A–U, Table 1). Of these, the palmitic acid, stearic acid, linoleic acid, and linolenic acid content were normally distributed, but the eicosenoic, oleic, and erucic acids content were skewed for three genotypic populations in different years (Figures 1A–U). Plants with higher oleic acid content were more common than those with lower content for each ecological type of rapeseed (Figures 1G–I). Analysis of variance (ANOVA) was performed among the spring, winter, and semi-winter rapeseed ecological types in different years, and showed that genotype and environment have significant effects on the fatty acid content of rapeseed (Table 1).
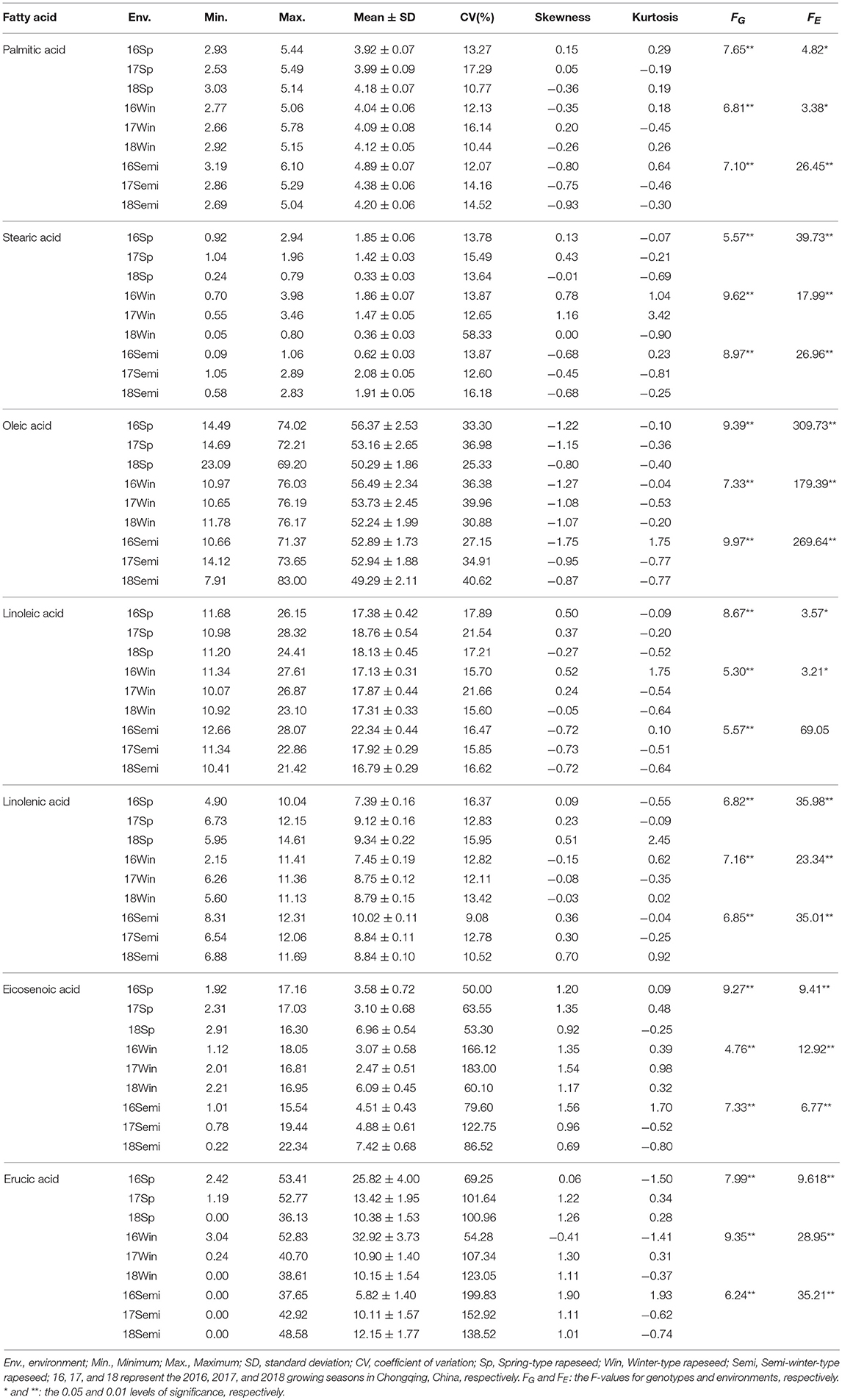
Table 1. Statistical analysis of fatty acid content in different ecological types of rapeseed grown in different environments.
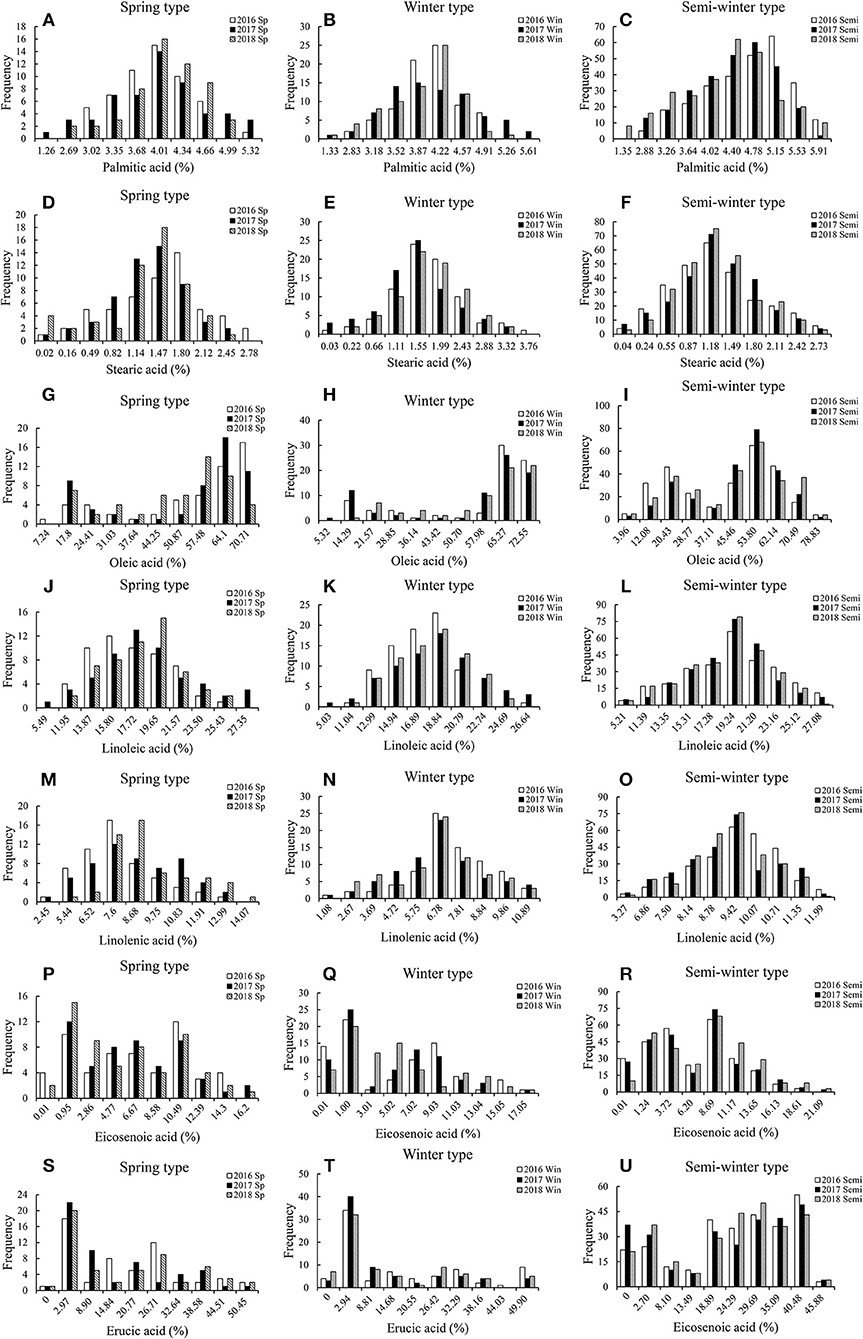
Figure 1. The frequency distribution of fatty acid contents in the different rapeseed accessions grown in different environments. The percentage indicates the proportion of the total dry weight of the seed represented by fatty acid composition. Sp, Spring-type rapeseed; Win, Winter-type rapeseed; Semi, Semi-winter-type rapeseed; 16, 17, and 18 represent the 2016, 2017, and 2018 growing seasons in Chongqing, China. (A–C) The frequency distribution of Palmitic acid contents in Spring-type, Winter-type, and Semi-winter-type rapeseed; (D–F) The frequency distribution of Stearic acid contents in Spring-type, Winter-type, and Semi-winter-type rapeseed; (G–I) The frequency distribution of Oleic acid contents in Spring-type, Winter-type, and Semi-winter-type rapeseed; (J–L) The frequency distribution of Linoleic acid contents in Spring-type, Winter-type, and Semi-winter-type rapeseed; (M–O) The frequency distribution of Linolenic acid contents in Spring-type, Winter-type, and Semi-winter-type rapeseed; (P–R) The frequency distribution of Eicosenoic acid contents in Spring-type, Winter-type, and Semi-winter-type rapeseed; (S–U) The frequency distribution of Erucic acid contents in Spring-type, Winter-type, and Semi-winter-type rapeseed.
Genome-Wide Association Analysis of Fatty Acid via mrMLM
For palmitic acid (C16:0) content, 11 QTNs were detected on chromosomes A01, A03, A04, A06, A07, A08, C01, and C03, respectively (Table 2). Of these, three consensus QTNs (q-C16:0-A06, q-C16:0-A08-2, and q-C16:0-C03-2) were commonly detected for palmitic acid among different ecotypic rapeseed and ecotypic rapeseed cultivated in different years, providing useful information for searching for candidate genes associated with palmitic acid biosynthesis. However, q-C16:0-A01, q-C16:0-A04, q-C16:0-A06, and q-C16:0-A07 were mainly found in the spring-type and all 3 years, and other QTNs were detected among different ecotypic rapeseed and years (Table 2).
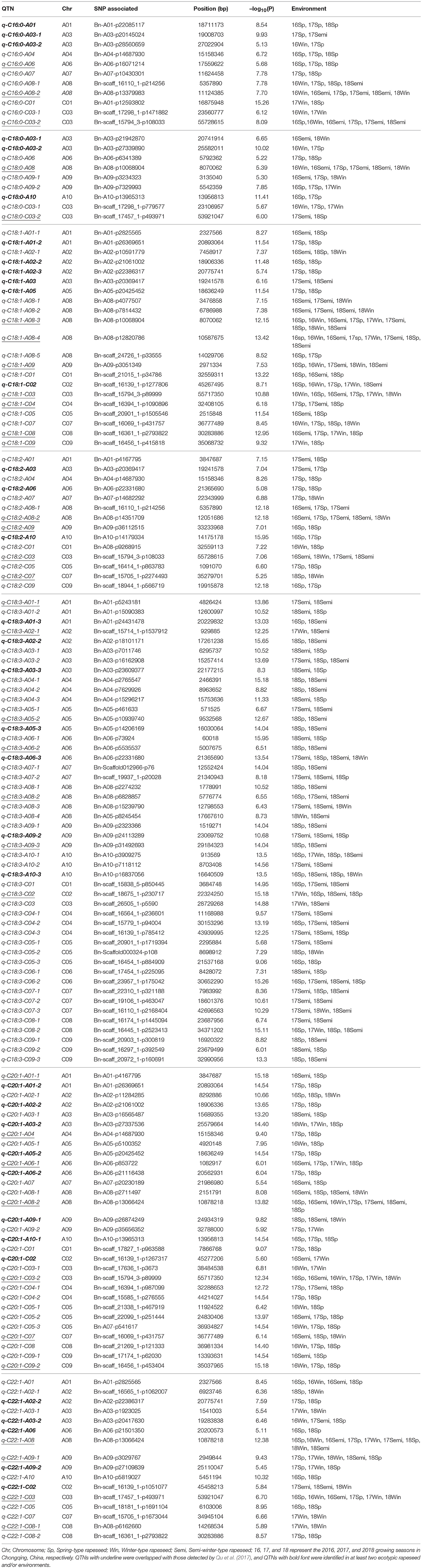
Table 2. Quantitative trait nucleotides (QTNs) associated with fatty acid content in B. napus accessions grown in different environments.
For stearic acid (C18:0) content, a total of 9 QTNs were resolved and distributed on A03, A06, A08, A09, A10, and C03, respectively (Table 3). Among them, two QTNs, q-C18:0-A08, and q-C18:0-C03-2, were detected in different ecotypic rapeseed and environments, and others were detected in different ecotypic rapeseed grown in at least two different environments.
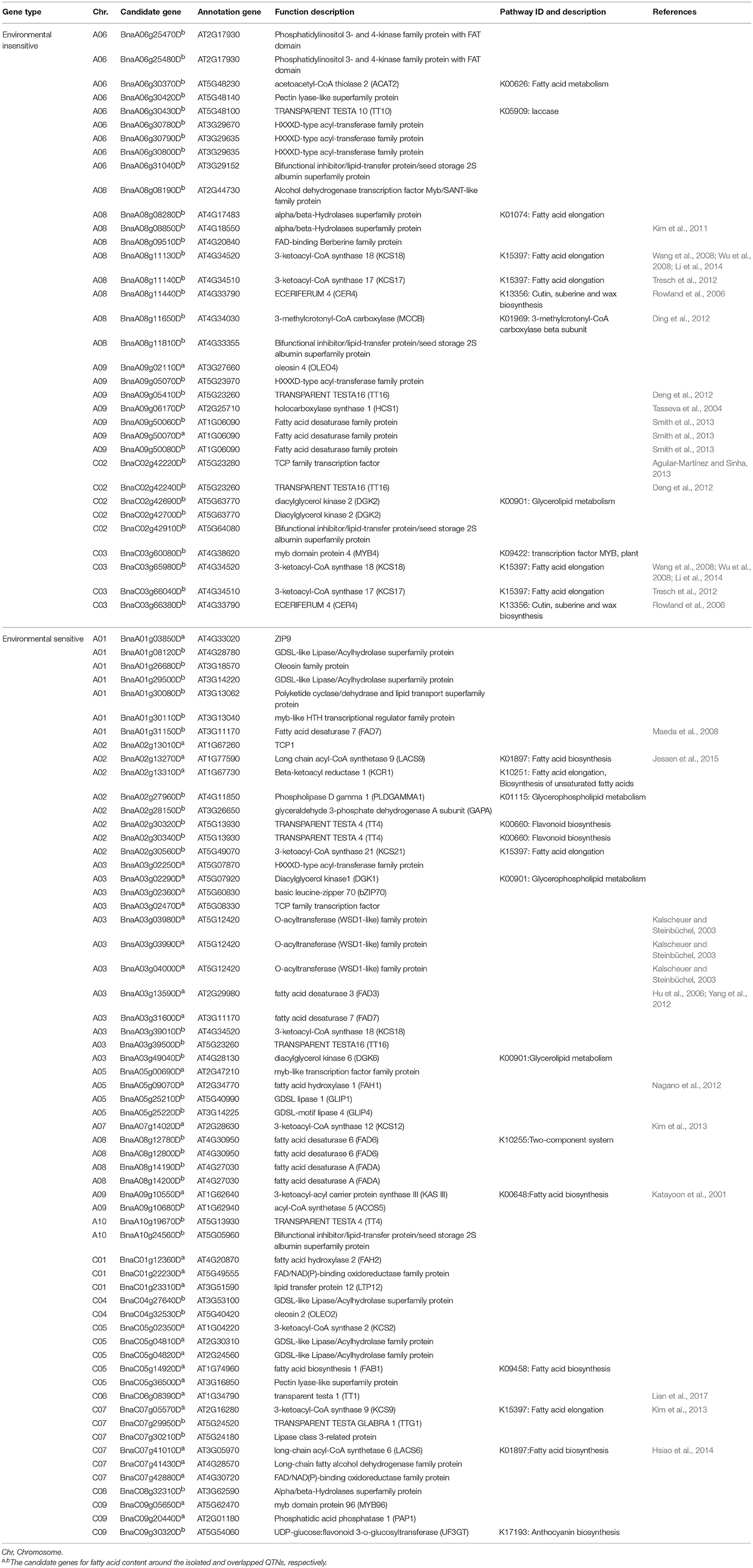
Table 3. Summary of candidate genes associated with fatty acid biosynthesis in significant association regions.
For oleic acid (C18:1) content, 21 QTNs were detected and distributed throughout of the B. napus genome, including chromosomes A01, A02, A03, A05, A08, A09, C01, C02, C03, C04, C05, C07, C08, and C09, respectively (Table 2). Of these, seven QTNs (q-C18:1-A08-3, q-C18:1-A08-4, q-C18:1-A09, q-C18:1-C03, q-C18:1-C04, q-C18:1-C08, and q-C18:1-C09) were co-localized in the same genomic regions of A08, A09, C03, C04, C08, and C09 using mrMLM and the PCA+K model (Qu et al., 2017). These seven QTNs were considered the major candidate regions for oleic acid content.
For linoleic acid (C18:2) content, fourteen QTNs were detected and mapped on chromosomes A01, A03, A04, A06, A07, A08, A09, A10, C01, C03, C05, C07, and C09, respectively (Table 2). Of these, q-C18:2-A08-2, q-C18:2-A09, q-C18:2-C03, and q-C18:2-C07 were identified in our previous research (Qu et al., 2017).
For linolenic acid (C18:3) content, a total 48 QTNs were found and covered almost the whole B. napus genome (Table 2). Among them, seven highly identical QTNs distributed on chromosome A01, A02, A05, A06, A08, A09, and C02, along with other minor loci could be identified in different ecotypic rapeseed accessions in at least 1 year of growth (Table 2).
For eicosenoic acid (C20:1) content, 30 QTNs were obtained, including seven QTNs that overlapped with previously published QTNs (Qu et al., 2017), and were distributed on chromosome A01, A04, A06, A08, C03, C07, and C09, respectively (Table 2). The novel loci for eicosenoic acid content also displayed marked variation among the different ecotypic rapeseeds and environments.
For erucic acid (C22:1) content, 16 QTNs were detected and mapped on chromosome A01, A02, A03, A06, A08, A09, A10, C02, C03, C05, C07, and C08, respectively (Table 2). Of these, two QTNs (q-C22:1-A08 and q-C22:1-C03) had been widely considered as the major genetic regions of A08 and C03, consistent with the findings of published works (Li et al., 2014; Lee et al., 2015; Xu et al., 2015; Qu et al., 2017). In addition, two QTNs (q-C22:1-A09-1 and q-C22:1-C08-1) associated with erucic acid content were also detected in different ecotypic rapeseed and environments (Table 2), indicating that mrMLM is a powerful and accurate tool to detect QTNs and estimate the effect of QTNs on complex traits.
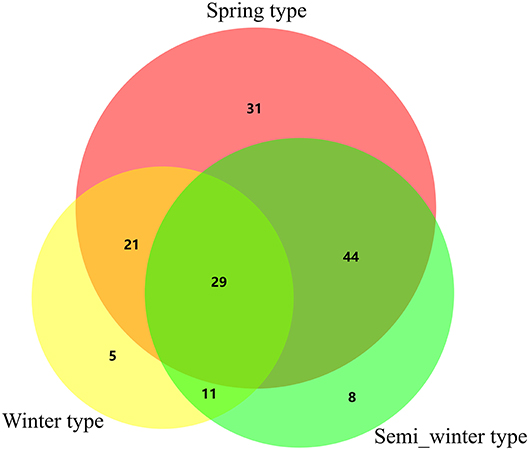
In all, 149 QTNs associated with fatty acid content were detected using mrMLM (Table 2, Supplementary Table S2), while only 62 associated regions were detected using the PCA + K model in TASSEL 5.2.1 (Qu et al., 2017). Among these, 34 QTNs were overlapped, including the association regions on A08 and C03, which had been widely reported previously (Wang et al., 2015; Liu et al., 2016; Qu et al., 2017), indicating that these results were credible and reproducible. In addition, 115 novel loci were identified for fatty acids via mrMLM compared with MLM (PCA+K; Table 2), indicating that a multi-locus random effect MLM method was better able to detect QTNs of complex quantitative traits. Furthermore, of these, 30.43% novel QTNs (35/115) were repeatedly detected for fatty acid content among different ecotypic rapeseed accessions and environments, located on chromosome A01, A02, A03, A05, A06, A09, A10, and C02, respectively (bold in Table 2), and other novel QTNs (80/115) were found for fatty acid content in a single environment. Among these QTNs, 29 were simultaneously detected in three ecotypic rapeseed, with greater QTN variation in spring and semi-winter rapeseed (Figure 2, Supplementary Table S2). Our results provideinsight into the mechanism underlying fatty acid composition, and lay the foundation for marker assisted selection in breeding projects for improved rapeseed genotypes with high oil quality and an ideal fatty acid composition.
Identification of Candidate Genes
To predict candidate genes for loci significantly associated with fatty acid content, the reported and repeadly detected novel QTNs were used to confirm the genomic regions in the B. napus “Darmor v4.1” reference genome (Chalhoub et al., 2014). We identified five environment-insensitive and fifteen environment-sensitive association regions and screened for candidate genes within these regions. Subsequently, we extracted gene sequences within the GWAS peaks in candidate association regions, and identified 95 putative genes that possibly influence fatty acid content (Table 3). Of these, 63.16% candidate genes (60/95) were screened in the overlapping and repeatedly detected association regions, while the remaining genes (35/95) were detected on the single QTN regions (Table 3). Using peak SNPs on A08 and C03 (Bn-A08-p13066424 and Bn-scaff_15794_3-p89999, respectively), 9 and 4 candidate genes were selected in the association regions on each chromosome, respectively (Table 3). BnaA08g11130D and BnaC03g65980D are putative paralogs of 3-ketoacyl-CoA synthase 18 (KCS18), while BnaA08g11140D and BnaC03g66040D are putative paralogs of KCS17 (Table 3), based on comparisons of the physical positions of genes associated with erucic acid traits in a GWAS (Wu et al., 2008; Li et al., 2014; Lee et al., 2015; Xu et al., 2015), indicating that there is a strong correlation between GWAS peak regions and candidate genes. In addition, the putative candidate genes were characterized and annotated, such as 3-methylcrotonyl-CoA carboxylase (MCCB, BnaA08g11650D), TRANSPARENT TESTA16 (TT16, BnaA09g05410D, and BnaC02g42240D), and MYB4 (BnaC03g60080D), which might be environment-insensitive genes located on chromosome A06, A09, and C02 respectively (Table 3). Of these, 17 genes had been identified that might be involved in the fatty acid pathway, and 12 members were annotated for fatty acid metabolism in KEGG analysis (Table 3).
The expression of candidate genes in low-frequency association regions identified in this study were influenced by the genotype and environments, and 61 environment-sensitive genes were obtained by comparing these regions with the B. napus reference genome (Chalhoub et al., 2014), including GDSL (GDSL-like Lipase), GAPA (Glyceraldehyde 3-phosphate dehydrogenase A), KCS21, FAD3, FAD7, FAD6 fatty acid biosynthesis 1 (FAB1), acyl-activating enzyme 17 (AAE17), long chain acyl-CoA synthetase 9 (LACS9), oleosin 2 (OLEO2), beta-ketoacyl reductase 1 (KCR1), and trigalactosyldiacylglycerol2 (TGD2) (Table 3). Of these, some genes (e.g., TT16 and TT1) were predicted to be associated with oleic acid content in B. napus (Lian et al., 2017; Qu et al., 2017); however, novel loci were also identified, including MYB67, OLEO2, KCS21, FAD3, KCR1, TT1, and TGD2 (Table 3). Among these genes, 12 putative gene members were identified in previous research, and 14 gene members were enriched in fatty acid pathways in the KEGG database (Table 3).
Oleic acid is a monounsaturated fat beneficial for human health that contributes to the nutritional and economic value of rapeseed oil. To provide insight into the genetic control of oleic acid content, we therefore aligned 95 gene sequences from the different rapeseed accessions, and identified nucleotides in the intronic regions of BnaA08g08280D and BnaC03g60080D that show significant differences between the high- and low-oleic acid lines (Figure 3).
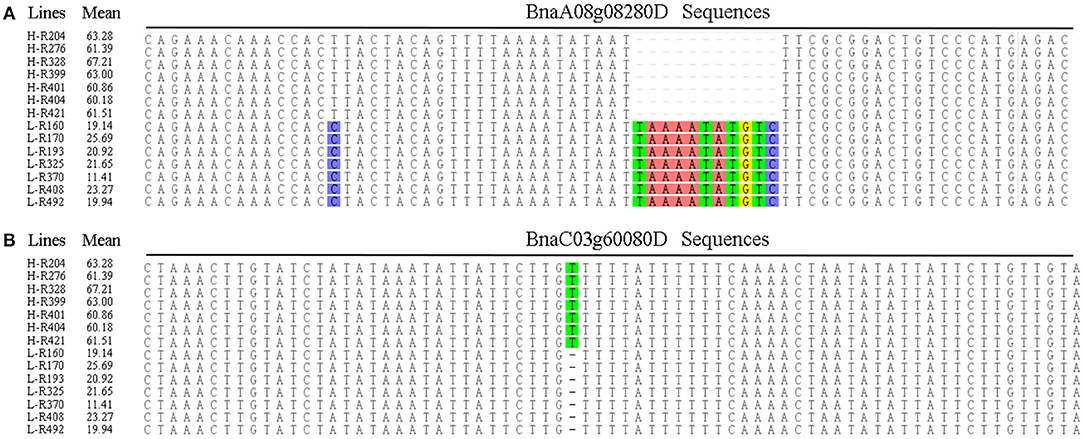
Figure 3. Multiple alignments of candidate gene sequences between the high- and low-oleic acid B. napus accessions. (A) BnaA08g08280D; (B) BnaC03g60080D. H-number and L-number indicates the high- and low-oleic acid B. napus accessions, respectively. The oleic aicd contents represents by the mean values.
Expression Patterns of Candidate Genes
We assessed the relative expression levels of the candidate genes during seed development of B. napus variety ZS11 (Figures 4, 5), which had a high oleic acid content and low erucic acid. The expression levels of the environment-insensitive genes showed no obvious variation during seed development, but KCS18 (BnaA08g11130D and BnaC03g65980D), and BnaC02g42910D showed higher expression levels during the middle stages of seed development (Figure 4), indicating that they might contribute to the accumulation of oleic acid during the middle stages of seed development. In addition, TT16 (BnaA09g05410D and BnaC02g42240D) and BnaC02g42240D were expressed at high levels in the early stages of seed development, while other genes showed low expression levels throughout seed development (Figure 4).
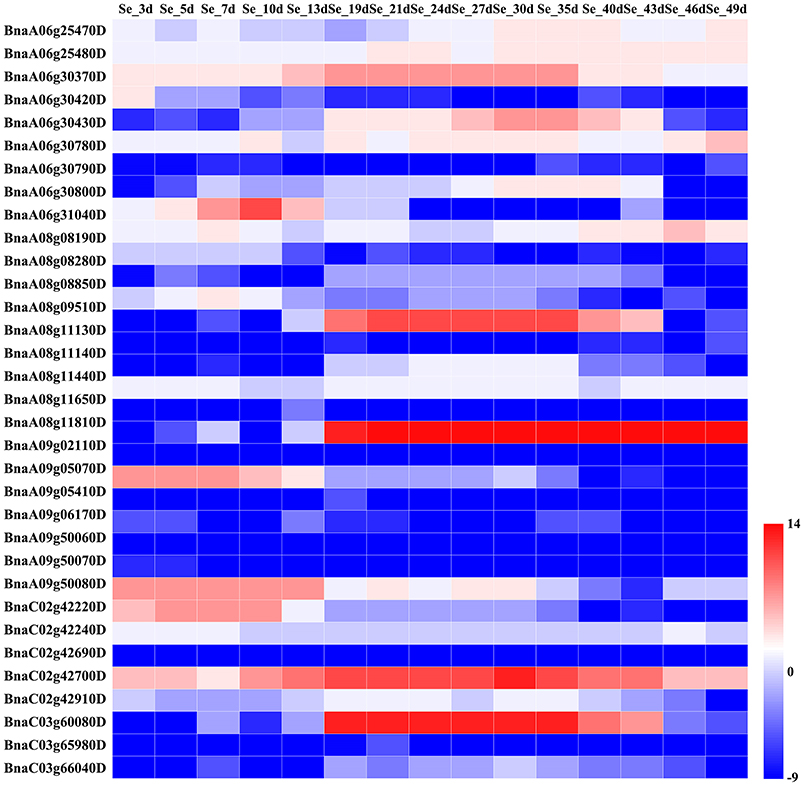
Figure 4. Comparative expression analysis of environment-insensitive genes associated with fatty acid content during seed development. The abbreviations above the heatmap indicate the different developmental stages of the seeds from B. napus ZS11 (defined in Supplementary Table S4). The expression values of the candidate genes were calculated using three biological replicates with three technical replicates and normalized by Log2 (mean expression values). The “scale” function in R was used to normalize the relative expression levels (R = log2/mean expression values). The heatmap was generated using Heatmap Illustrator 1.0 (HemI 1.0).
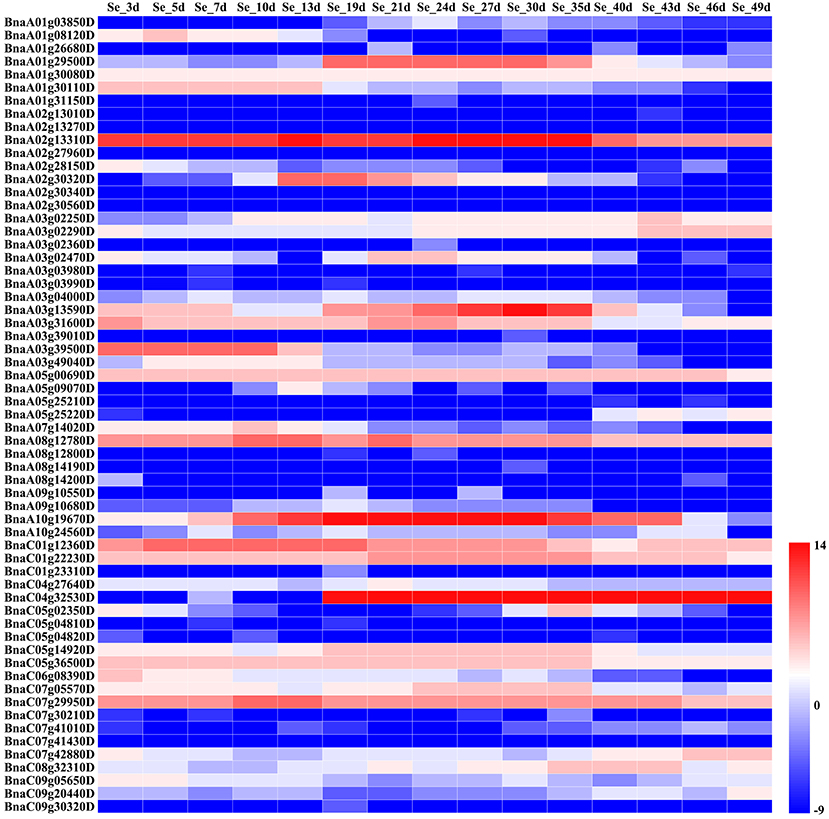
Figure 5. Comparative expression analysis of environment-sensitive genes associated with the fatty acid content during seed development. The abbreviations above the heatmap indicate the different developmental stages of the seeds from B. napus ZS11 (defined in Supplementary Table S5). The expression values of the candidate genes were calculated using three biological replicates with three technical replicates and normalized by Log2 (mean expression values). The “scale” function in R was used to normalize the relative expression levels (R = log2/mean expression values). The heatmap was generated using Heatmap Illustrator 1.0 (HemI 1.0).
However, we found that the expression levels of the environment-sensitive genes varied throughout seed development (Figure 5). For example, OLEO2 (BnaC04g32530D) was highly expressed in the middle and late stages of seed development, while the expression of KCS9 (BnaC07g05570D) peaked in the early and middle stages (Figure 5). In addition, BnaA01g29500D and TT4 (BnaA02g30320D) were mainly expressed in the middle stages of seed development, but KCR1 (BnaA02g13310D) and TTG1 (BnaC07g29950D) showed high expression levels throughout seed development (Figure 5). Furthermore, other genes displayed different patterns of expression throughout seed development.
Discussion
In B. napus, seeds fatty acids are mainly composed of palmitic, stearic, oleic, linoleic, linolenic eicosenoic, and erucic acids, which determine the rapeseed oil quality. Enhancing the oleic acid content and quality of rapeseed through modifying its fatty acid composition has become an important breeding goal. However, previous studies identified the effect of putative fatty acid genes and the interaction of genotype and environment on the fatty acid content of rapeseed (Zhao, 2002; Zhao et al., 2005, 2008; Wen et al., 2015). Here, we report that fatty acid content also varies significantly among different rapeseed ecological types (spring, winter, and semi-winter rapeseed varieties) and environments (Figure 1, Table 1), indicating the complexity of the biosynthetic processes underlying fatty acid content in rapeseed. Interestingly, accessions with high oleic acid content were common amongst the different rapeseed ecological types (Figures 1G–I), possibly because these accessions are artificially selected in breeding projects aimed at producing rapeseed with high oleic acid content. Therefore, identifying the relationship between favorable alleles and environments will be beneficial for improving the fatty acid content of rapeseed.
With the development of genome sequencing and computational technologies, the Illumina Infinium Brassica 60K SNP array has been developed and widely used for the genome-wide association analysis of B. napus as well as the analysis of some trait-associated genomic regions and candidate genes (Li et al., 2014; Qian et al., 2014; Gajardo et al., 2015; Hatzig et al., 2015; Lee et al., 2015; Wei et al., 2015; Gacek et al., 2017; Qu et al., 2017). Furthermore, the MLM (Q+K and PCA+K) was found to be a powerful model for GWAS in these previous studies (Yu et al., 2006; Zhao et al., 2011; Xu et al., 2015; Li et al., 2016; Qu et al., 2017). In the present study, the mrMLM was employed for a GWAS, which is confirmed as a precise and powerful tool for analyzing phenotypic and genotypic information derived from numerous accessions and SNPs (Wang et al., 2016; Li et al., 2017). In the present study, 149 QTNs significantly associated with fatty acid content were identified using the mrMLM (Table 2, and Supplementary Table S3). Of these, 34 associated SNPs overlapped with those obtained using MLM (Qu et al., 2017), indicating that the association analysis was reliable; however, eight novel association regions containing 35 QTNs were simultaneously detected among different ecotypic rapeseed grown in different environments, strongly suggesting that mrMLM is more powerful to detect SNPs associated with complex traits than MLM in GWAS. In addition, 29 QTNs for fatty acids were simultaneously detected in spring, winter and semi-winter rapeseed (Figure 2), indicating that the orthologous genes for fatty acid might be better identified using these singinficant QTNs. Furthermore, more QTNs associated with fatty acids were identified from the sping and semi-winter type than in winter rapeseed (Figure 2), indicating that the fatty acids were associated with their genotype. These results might be helpful for elucidating the mechanism that determines fatty acid composition in B. napus.
In B. napus, fatty acid variation is controlled by multiple genes (Zhao et al., 2008; Wen et al., 2015). In this study, we categorized the candidate genes as either environment-insensitive or -sensitive genes, according to the published results and their detection frequency between the rapeseed genotypes grown in the different environments. A total of 95 candidate genes were identified with known functions in the fatty acid biosynthesis pathway. Of these, 34 were environment-insensitive genes (Table 3), including KCS18, which is known to play a crucial role in regulating erucic acid biosynthesis in B. napus (Wang et al., 2008; Wu et al., 2008; Li et al., 2014), and the putative KCS18 paralogs BnaA08g11130D (Chromosome A08) and BnaC03g65980D (Chromosome C03), which are most highly expressed during the middle to late stages of seed development (Figure 4), suggesting that they are key genes regulating the accumulation of fatty acids in rapeseed oil. In addition, CER4 encodes an alcohol-forming fatty acyl-coenzyme A reductase (Rowland et al., 2006), and KCS17 is known to be involved in the biosynthesis of saturated fatty acids (Tresch et al., 2012) and would therefore be expected to be more highly expressed during seed development. We found that the putative orthologs of KCS17 (BnaA08g11140D and BnaC03g66040D) and CER4 (BnaC03g66380D) were associated with fatty acid content in B. napus, but the expression of BnaA08g11140D and BnaC03g66040D, putative KCS17 paralogs, was downregulated during seed development (Figure 4), suggesting that functional segregation exists among the different paralogs of the ancestral KCS17 gene, which has been reported previously in the B. napus genome (Chalhoub et al., 2014). In addition, the environment-insensitive genes, including DGK2 (BnaC02g42690D and BnaC02g42700D), HCS1 (BnaA09g06170D), MYB4 (BnaC03g60080D), and TT16 (BnaA09g05410D and BnaC02g42240D), might also been involved in fatty acid biosynthesis (Tasseva et al., 2004; Deng et al., 2012; Yang et al., 2012; Chen et al., 2013). Most of the environment-insensitive genes were steadily expressed throughout seed development (Figure 4), and these might be the major factors regulating oleic acid accumulation in B. napus. Furthermore, 12 environment-insensitive genes were enriched in the fatty acid biosynthesis pathway according to KEGG pathway analysis (Table 3). Importantly, the nucleotide sequences of BnaA08g08280D and BnaC03g60080D differed between the high- and low-oleic acid lines (Figure 3), but these differences were all located in the intronic regions. Furthermore, 61 environment-sensitive genes showed wide variation in expression during seed development (Table 3, Figure 5), and these could be divided into early, middle, and late expression genes, respectively. For example, BnaA06g31040D showed high expression levels in the early stages of seed development, KCS9 (BnaC07g05570D) peaked at the early and middle stages, OLEO2 (BnaC04g32530D) had high expression during the middle and late stages, and BnaA01g29500D, and TT4 (BnaA02g30320D) expression markedly increased in the middle stages (Table 3, Figure 5). Several other candidate genes, including putative homologs of FAD (3, 6, and 7), KCS (9, 12, and 21), KCR1, and LACS9, were found to be associated with fatty acid biosynthesis (Peng et al., 2010; Tresch et al., 2012; Yang et al., 2012; Lai et al., 2017; Shi et al., 2017), but their contribution remains to be confirmed by further studies (Table 3). In addition, 14 gene members were involved in the fatty acid metabolism confirmed by KEGG database analysis (Table 3). However, there is no clear evidence indicating that these genes control the fatty acid content in B. napus.
In summary, 149 QTNs for fatty acid content (including 34 reported and 115 novel loci) were detected, strongly demonstrating that mrMLM is a powerful and suitable tool for detecting QTNs for fatty acid content in rapeseed. Among these putative candidate genes, 63.16% (60/95) and 36.84% (35/95) were distributed on overlapping and isolated QTNs, respectively. Based on the pervious reports, 29 genes are involved in the fatty acid biosynthesis, and 26 gene members were enriched in the fatty acid pathway by the KEGG pathway database, indicating that mrMLM is an accurate tool to estimate the effect of QTNs on complex traits. Whether these genes exert significant regulatory effects on the fatty acid content of the seeds remains to be investigated. Hence, further studies are needed. Our findings provide useful candidate genes for the marker-assisted selection and breeding of rapeseed lines with increased oleic acid content in the seed.
Author Contributions
CMQ and JL conceived and designed the experiments; MG and XH conducted the experiments; ZX, XX, LJ, and KL collected and analyzed the data; SW, and LJ made sequence alignment; YL, LW, and RW carried out the field experiments; MG, MZ, and CLQ wrote the manuscript; JL and CMQ reviewed the manuscript.
Funding
This work was supported by the National Key Research and Development Plan (2016YFD0101007 and 2016YFD0100202), the Department of Agriculture projects of modern agricultural technology system (CARS-12), National Science Foundation of China (31401412, 31571701), the 111 Project (B12006), Chongqing Basic Scientific and Advanced Technology Research (cstc2016shms-ztzx80010, cstc2015jcyjBX0001 and cstc2017jcyjAX0321), and Fundamental Research Funds for the Central Universities (XDJK2016B030).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would also like to thank Kathy Farquharson for critical reading of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01872/full#supplementary-material
Supplementary Figure S1. Subpopulations in the spring, winter, and semi-winter B. napus accessions, determined using a principal coordinate analysis (PCA).
Supplementary Table S1. List of rapeseed accessions used.
Supplementary Table S2. Sum of SNPs in candidate regions for fatty acid content in B. napus accessions grown in different environments.
Supplementary Table S3. Primers used to amplify candidate genes and reference genes via RT-qPCR analysis.
Supplementary Table S4. Heatmap of the expression levels of the B. napus environment-insensitive genes at different stages of seed development.
Supplementary Table S5. Heatmap of the expression levels of the B. napus environment-sensitive genes at different stages of seed development.
References
Aguilar-Martínez, J. A., and Sinha, N. (2013). Analysis of the role of Arabidopsis class I TCP genes AtTCP7, AtTCP8, AtTCP22, and AtTCP23 in leaf development. Front. Plant Sci. 4:406. doi: 10.3389/fpls.2013.00406
Bauer, B., Kostik, V., and Gjorgjeska, B. (2015). Fatty acid composition of seed oil obtained from different canola varieties. Farm. Glas. 71, 1–7.
Burns, M., Barnes, S., Bowman, J., Clarke, M., Werner, C., and Kearsey, M. (2003). QTL analysis of an intervarietal set of substitution lines in Brassica napus: (i) Seed oil content and fatty acid composition. Heredity 90, 39–48. doi: 10.1038/sj.hdy.6800176
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A., Tang, H., Wang, X., et al. (2014). Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953. doi: 10.1126/science.1253435
Chang, F., Guo, C., Sun, F., Zhang, J., Wang, Z., Kong, J., et al. (2018). Genome-wide association studies for dynamic plant height and number of nodes on the main stem in summer sowing soybeans. Front. Plant Sci. 9:e1184. doi: 10.3389/fpls.2018.01184
Chang, N. W., and Huang, P. C. (1998). Effects of the ratio of polyunsaturated and monounsaturated fatty acid to saturated fatty acid on rat plasma and liver lipid concentrations. Lipids 33, 481–487. doi: 10.1007/s11745-998-0231-9
Chen, F., Zhang, W., Yu, K., Sun, L., Gao, J., Zhou, X., et al. (2018). Unconditional and conditional QTL analyses of seed fatty acid composition in Brassica napus L. BMC Plant Biol. 18:49. doi: 10.1186/s12870-018-1268-7
Chen, X., Chou, H.-H., and Wurtele, E. S. (2013). Holocarboxylase synthetase 1 physically interacts with histone H3 in Arabidopsis. Scientifica 2013:983501. doi: 10.1155/2013/983501
Clarke, W. E., Higgins, E. E., Plieske, J., Wieseke, R., Sidebottom, C., Khedikar, Y., et al. (2016). A high-density SNP genotyping array for Brassica napus and its ancestral diploid species based on optimised selection of single-locus markers in the allotetraploid genome. Theor. Appl. Genet. 129, 1887–1899. doi: 10.1007/s00122-016-2746-7
Delourme, R., Falentin, C., Fomeju, B. F., Boillot, M., Lassalle, G., André, I., et al. (2013). High-density SNP-based genetic map development and linkage disequilibrium assessment in Brassica napus L. BMC Genomics 14:120. doi: 10.1186/1471-2164-14-120
Deng, W., Chen, G., Peng, F., Truksa, M., Snyder, C. L., and Weselake, R. J. (2012). Transparent testa 16 plays multiple roles in plant development and is involved in lipid synthesis and embryo development in canola. Plant Physiol. 160, 978–989. doi: 10.1104/pp.112.198713
Ding, G., Che, P., Ilarslan, H., Wurtele, E. S., and Nikolau, B. J. (2012). Genetic dissection of methylcrotonyl CoA carboxylase indicates a complex role for mitochondrial leucine catabolism during seed development and germination. Plant J. 70, 562–577. doi: 10.1111/j.1365-313X.2011.04893.x
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software structure: a simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Falush, D., Stephens, M., and Pritchard, J. K. (2003). Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164, 1567–1587. doi: 10.3410/f.1015548.197423
Fei, W., Wu, X., Li, Q., Chen, F., Hou, S., Fan, Z., et al. (2012). Genetic analysis of high oleic acid mutation materials in Brassica napus. Chin. Agric. Sci. Bull. 28, 176–180. doi: 10.3969/j.issn.1000-6850.2012.01.034
Gacek, K., Bayer, P. E., Bartkowiakbroda, I., Szala, L., Bocianowski, J., Edwards, D., et al. (2017). Genome-wide association study of genetic control of seed fatty acid biosynthesis in Brassica napus. Front. Plan. Sci. 7:e2062. doi: 10.3389/fpls.2016.02062
Gajardo, H. A., Wittkop, B., Soto-Cerda, B., Higgins, E. E., Parkin, I. A., Snowdon, R. J., et al. (2015). Association mapping of seed quality traits in Brassica napus L. using GWAS and candidate QTL approaches. Mol. Breeding 35, 1–19. doi: 10.1007/s11032-015-0340-3
Hardy, O. J., and Vekemans, X. (2002). SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol. Ecol. Notes 2, 618–620. doi: 10.1046/j.1471-8286.2002.00305.x
Hatzig, S. V., Frisch, M., Breuer, F., Nesi, N., Ducournau, S., Wagner, M. H., et al. (2015). Genome-wide association mapping unravels the genetic control of seed germination and vigor in Brassica napus. Front. Plant Sci. 6, 221. doi: 10.3389/fpls.2015.00221
Hsiao, A. S., Haslam, R. P., Michaelson, L. V., Liao, P., Napier, J. A., and Chye, M. L. (2014). Gene expression in plant lipid metabolism in Arabidopsis seedlings. PLoS ONE 9:e107372. doi: 10.1371/journal.pone.0107372
Hu, X., Sullivan-Gilbert, M., Gupta, M., and Thompson, S. A. (2006). Mapping of the loci controlling oleic and linolenic acid contents and development of fad2 and fad3 allele-specific markers in canola (Brassica napus L.). Theor. Appl. Genet. 113, 497–507. doi: 10.1007/s00122-006-0315-1
Javed, N., Geng, J., Tahir, M., Mcvetty, P. B. E., Li, G., and Duncan, R. W. (2016). Identification of QTL influencing seed oil content, fatty acid profile and days to flowering in Brassica napus L. Euphytica 207, 191–211. doi: 10.1007/s10681-015-1565-2
Jessen, D., Roth, C., Wiermer, M., and Fulda, M. (2015). Two activities of Long-Chain Acyl-Coenzyme A Synthetase are involved in lipid trafficking between the endoplasmic reticulum and the plastid in Arabidopsis. Plant Physiol. 167, 351–366. doi: 10.1104/pp.114.250365
Kalscheuer, R., and Steinbüchel, A. (2003). A novel bifunctional wax ester synthase/acyl-CoA:diacylglycerol acyltransferase mediates wax ester and triacylglycerol biosynthesis in Acinetobacter calcoaceticus ADP1. J. Biol. Chem. 278, 8075–8082. doi: 10.1074/jbc.M210533200
Katayoon, D., Patricia, E., and James, B. (2001). Overexpression of 3-ketoacyl-acyl-carrier protein synthase IIIs in plants reduces the rate of lipid synthesis 1. Plant Physiol. 125, 1103–1114. doi: 10.1104/pp.125.2.1103
Kim, E. Y., Seo, Y. S., and Kim, W. T. (2011). AtDSEL, an Arabidopsis cytosolic DAD1-like acylhydrolase, is involved in negative regulation of storage oil mobilization during seedling establishment. J. Plant Physiol. 168, 1705–1709. doi: 10.1016/j.jplph.2011.03.004
Kim, J., Jung, J. H., Lee, S. B., Go, Y. S., Kim, H. J., Cahoon, R., et al. (2013). Arabidopsis 3-ketoacyl-coenzyme a synthase 9 is involved in the synthesis of tetracosanoic acids as precursors of cuticular waxes, suberins, sphingolipids, and phospholipids. Plant Physiol. 162, 567–580. doi: 10.1104/pp.112.210450
Lai, C. P., Huang, L. M., Chen, L. F. O., Chan, M. T., and Shaw, J. F. (2017). Genome-wide analysis of GDSL-type esterases/lipases in Arabidopsis. Plant Mol. Biol. 95, 181–197. doi: 10.1007/s11103-017-0648-y
Lee, S., Jang, M. S., Jeon, E. J., Yun, K. Y., and Kim, S. (2015). “QTL Analysis for erucic acid and oleic acid content in Brassica napus using F2 population,” in: Plant and Animal Genome XXIII Conference (San Diego, CA), 823.
Li, F., Chen, B., Xu, K., Gao, G., Yan, G., Qiao, J., et al. (2016). A genome-wide association study of plant height and primary branch number in Rapeseed (Brassica napus). Plant Sci. 242, 169–177. doi: 10.1016/j.plantsci.2015.05.012
Li, F., Chen, B., Xu, K., Wu, J., Song, W., Bancroft, I., et al. (2014). Genome-wide association study dissects the genetic architecture of seed weight and seed quality in rapeseed (Brassica napus L.). DNA Res. 21, 355–367. doi: 10.1093/dnares/dsu002
Li, H., Zhang, L., Hu, J., Zhang, F., Chen, B., Xu, K., et al. (2017). Genome-wide association mapping reveals the genetic control underlying branch angle in rapeseed (Brassica napus L.). Front. Plant Sci. 8:1054. doi: 10.3389/fpls.2017.01054
Lian, J., Lu, X., Yin, N., Ma, L., Jing, L., Xue, L., et al. (2017). Silencing of BnTT1 family genes affects seed flavonoid biosynthesis and alters seed fatty acid composition in Brassica napus. Plant Sci. 254, 32–47. doi: 10.1016/j.plantsci.2016.10.012
Liu, L. Z., and Li, J. N. (2014). QTL mapping of oleic acid, linolenic acid and erucic acid content in Brassica napus by using the high density snp genetic map. Sci. Agric. Sin. 47, 24–32. doi: 10.3864/j.issn.0578-1752.2014.01.003
Liu, S., Fan, C., Li, J., Cai, G., Yang, Q., Wu, J., et al. (2016). A genome-wide association study reveals novel elite allelic variations in seed oil content of Brassica napus. Theor. Appl. Genet. 129, 1203–1215. doi: 10.1007/s00122-016-2697-z
Lu, G., Harper, A. L., Trick, M., Morgan, C., Fraser, F., O'neill, C., et al. (2014). Associative transcriptomics study dissects the genetic architecture of seed glucosinolate content in Brassica napus. DNA Res. 21, 613–625. doi: 10.1093/dnares/dsu024
Lu, K., Li, T., He, J., Chang, W., Zhang, R., Liu, M., et al. (2017). qPrimerDB: a thermodynamics-based gene-specific qPCR primer database for 147 organisms. Nucleic Acids Res. 46, D1229–D1236. doi: 10.1093/nar/gkx725
Luo, X., Ma, C., Yue, Y., Hu, K., Li, Y., Duan, Z., et al. (2015). Unravelling the complex trait of harvest index in rapeseed (Brassica napus L.) with association mapping. BMC Genomics 16:379. doi: 10.1186/s12864-015-1607-0
Ma, L., Liu, M., Yan, Y., Qing, C., Zhang, X., Zhang, Y., et al. (2018). Genetic dissection of maize embryonic callus regenerative capacity using multi-locus genome-wide association studies. Front. Plant Sci. 9:561. doi: 10.3389/fpls.2018.00561
Maeda, H., Sage, T. L., Isaac, G., Welti, R., and Dellapenna, D. (2008). Tocopherols modulate extraplastidic polyunsaturated fatty acid metabolism in Arabidopsis at low temperature. Plant Cell 20, 452–470. doi: 10.1105/tpc.107.054718
McCouch, S. R., Cho, Y. G., Yano, M., Paul, E., Blinstrub, M., Morishima, H., et al. (1997). Report on QTL nomenclature. Rice Genet. Newsl. 14, 11–13. doi: 10.1007/s10142-013-0328-1
Merk, H. L., Yarnes, S. C., Deynze, V., Tong, N., Menda, N., Mueller, L. A., et al. (2012). Trait diversity and potential for selection indices based on variation among regionally adapted processing tomato germplasm. J. Am. Soc. Hort. Sci. 137, 427–437.
Meuwissen, T., and Goddard, M. (2000). Fine mapping of quantitative trait loci using linkage disequilibria with closely linked marker loci. Genetics 155, 421–430.
Miller, J. F., Zimmerman, D. C., and Vick, B. A. (1987). Genetic control of high oleic acid content in sunflower oil. Crop Sci. 27, 923–926. doi: 10.2135/cropsci1987.0011183X002700050019x
Nagano, M., Takahara, K., Fujimoto, M., Tsutsumi, N., Uchimiya, H., and Kawai-Yamada, M. (2012). Arabidopsis sphingolipid fatty acid 2-hydroxylases (AtFAH1 and AtFAH2) are functionally differentiated in fatty acid 2-hydroxylation and stress responses. Plant Physiol. 159, 1138–1148. doi: 10.1104/pp.112.199547
Peng, Q., Hu, Y., Wei, R., Zhang, Y., Guan, C., Ruan, Y., et al. (2010). Simultaneous silencing of FAD2 and FAE1 genes affects both oleic acid and erucic acid contents in Brassica napus seeds. Plant Cell Rep. 29, 317–325. doi: 10.1007/s00299-010-0823-y
Peng, Y., Liu, H., Chen, J., Shi, T., Zhang, C., Sun, D., et al. (2018). Genome-wide association studies of free amino acid levels by six multi-locus models in bread wheat. Front. Plant Sci. 9:1196. doi: 10.3389/fpls.2018.01196
Pleines, S., and Friedt, W. (1989). Genetic control of linolenic acid concentration in seed oil of rapeseed (Brassica napus L.). Theor. Appl. Genet. 78, 793–797. doi: 10.1007/BF00266660
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959. doi: 10.1111/j.1471-8286.2007.01758.x
Qian, L., Qian, W., and Snowdon, R. J. (2014). Sub-genomic selection patterns as a signature of breeding in the allopolyploid Brassica napus genome. BMC Genomics 15:1. doi: 10.1186/1471-2164-15-1170
Qu, C., Fu, F., Liu, M., Zhao, H., Liu, C., Li, J., et al. (2015). Comparative transcriptome analysis of recessive male sterility (RGMS) in sterile and fertile Brassica napus Lines. PLoS ONE 10:e0144118. doi: 10.1371/journal.pone.0144118
Qu, C., Li, J., Fu, F., Zhao, H., Lu, K., Wei, L., et al. (2017). Genome-wide association mapping and Identification of candidate genes for fatty acid composition in Brassica napus L. using SNP markers. BMC Genomics 18:232. doi: 10.1186/s12864-017-3607-8
Rowland, O., Zheng, H., Hepworth, S. R., Lam, P., Jetter, R., and Kunst, L. (2006). CER4 encodes an alcohol-forming fatty acyl-coenzyme a reductase involved in cuticular wax production in Arabidopsis. Plant Physiol. 142, 866–877. doi: 10.1104/pp.106.086785
Saeidnia, S., and Gohari, A. R. (2012). Importance of Brassica napus as a medicinal food plant. J. Med. Plants Res. 6, 2700–2703. doi: 10.5897/JMPR11.1103
Scheffler, J. A., Sharpe, A. G., Schmidt, H., Sperling, P., Iap, P., Luhs, W., et al. (1997). Desaturase multigene families of Brassica napus arose through genome duplication. Theor. Appl. Genet. 94, 583–591. doi: 10.1007/s001220050454
Shi, J., Lang, C., Wang, F., Wu, X., Liu, R., Zheng, T., et al. (2017). Depressed expression of FAE1 and FAD2 genes modifies fatty acid profiles and storage compounds accumulation in Brassica napus seeds. Plant Sci. 263, 177–182. doi: 10.1016/j.plantsci.2017.07.014
Smith, M. A., Dauk, M., Ramadan, H., Yang, H., Seamons, L. E., Haslam, R. P., et al. (2013). Involvement of Arabidopsis ACYL-COENZYME A DESATURASE-LIKE2 (At2g31360) in the biosynthesis of the very-long-chain monounsaturated fatty acid components of membrane lipids. Plant Physiol. 161, 81–96. doi: 10.1104/pp.112.202325
Tanhuanpää, P. K., Vilkki, J. P., and Vilkki, H. J. (1996). Mapping of a QTL for oleic acid concentration in spring turnip rape (Brassica rapa ssp. oleifera). Theor. Appl. Genet. 92, 952–956. doi: 10.1007/BF00224034
Tasseva, G., De Virville, J. D., Cantrel, C., Moreau, F., and Zachowski, A. (2004). Changes in the endoplasmic reticulum lipid properties in response to low temperature in Brassica napus. Plant Physiol. Biochem. 42, 811–822. doi: 10.1016/j.plaphy.2004.10.001
Teh, L. S. (2015). Genetic Variation and Inheritance of Phytosterol and Oil Content in Winter Oilseed Rape (Brassica napus L.). Available online at http://hdl.handle.net/11858/00-1735-0000-0022-5D9B-E
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Trapnell, C., Roberts, A., Goff, L., Pertea, G., Kim, D., Kelley, D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578. doi: 10.1038/nprot.2012.016
Tresch, S., Heilmann, M., Christiansen, N., Looser, R., and Grossmann, K. (2012). Inhibition of saturated very-long-chain fatty acid biosynthesis by mefluidide and perfluidone, selective inhibitors of 3-ketoacyl-CoA synthases. Phytochemistry 76, 162–171. doi: 10.1016/j.phytochem.2011.12.023
Velasco, L., Fernandezmartinez, J. M., and Ade, H. (1997). Induced variability for C18 unsaturated fatty acids in Ethiopian mustard. Can. J. Plant Sci. 77, 91–95. doi: 10.4141/P96-025
Wang, N., Wang, Y., Tian, F., King, G. J., Zhang, C., Long, Y., et al. (2008). A functional genomics resource for Brassica napus: development of an EMS mutagenized population and discovery of FAE1 point mutations by TILLING. New Phytol. 180, 751–765. doi: 10.1111/j.1469-8137.2008.02619.x
Wang, S. B., Feng, J. Y., Ren, W. L., Huang, B., Zhou, L., Wen, Y. J., et al. (2016). Improving power and accuracy of genome-wide association studies via a multi-locus mixed linear model methodology. Sci. Rep. 6:19444. doi: 10.1038/srep19444
Wang, X., Long, Y., Yin, Y., Zhang, C., Gan, L., Liu, L., et al. (2015). New insights into the genetic networks affecting seed fatty acid concentrations in Brassica napus. BMC Plant Biol. 15:91. doi: 10.1186/s12870-015-0475-8
Wei, L., Jian, H., Lu, K., Filardo, F., Yin, N., Liu, L., et al. (2015). Genome-wide association analysis and differential expression analysis of resistance to Sclerotinia stem rot in Brassica napus. Plant Biotechnol. J. 14, 1368–1380. doi: 10.1111/pbi.12501
Wen, J., Xu, J., Long, Y., Xu, H., Wu, J., Meng, J., et al. (2015). Mapping QTLs controlling beneficial fatty acids based on the embryo and maternal plant genomes in Brassica napus L. J. Am. Oil Chem. Soc. 92, 541–552. doi: 10.1007/s11746-015-2618-3
Wu, G., Wu, Y., Xiao, L., Li, X., and Lu, C. (2008). Zero erucic acid trait of rapeseed (Brassica napus L.) results from a deletion of four base pairs in the fatty acid elongase 1 gene. Theor. Appl. Genet. 116, 491–499. doi: 10.1007/s00122-007-0685-z
Xiao, G., Zhang, H., Peng, Q., and Guan, C. (2008). Screening and analysis of multiple copy of oleate desaturase gene (fad2) in Brassica napus. Acta Agronomica Sinica 34, 1563–1568. doi: 10.3321/j.issn:0496-3490.2008.09.011
Xu, J., Long, Y., Wu, J., Xu, H., Zhao, Z., Wen, J., et al. (2015). QTL identification on two genetic systems for rapeseed glucosinolate and erucic acid contents over two seasons. Euphytica 205, 1–11. doi: 10.1007/s10681-015-1379-2
Yang, J., Lee, S. H., Goddard, M. E., and Visscher, P. M. (2011). GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82. doi: 10.1016/j.ajhg.2010.11.011
Yang, Q., Fan, C., Guo, Z., Qin, J., Wu, J., Li, Q., et al. (2012). Identification of FAD2 and FAD3 genes in Brassica napus genome and development of allele-specific markers for high oleic and low linolenic acid contents. Theor. Appl. Genet. 125, 715–729. doi: 10.1007/s00122-012-1863-1
Yu, J., Pressoir, G., Briggs, W. H., Vroh Bi, I., Yamasaki, M., Doebley, J. F., et al. (2006). A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38, 203–208. doi: 10.1038/ng1702
Zhang, Y., Liu, P., Zhang, X., Zheng, Q., Chen, M., Ge, F., et al. (2018). Multi-Locus genome-wide association study reveals the genetic architecture of stalk lodging resistance-related traits in maize. Front. Plant Sci. 9:611. doi: 10.3389/fpls.2018.00611
Zhang, Z., Xiao, G., Tan, T., and Guan, C. (2009). Advances on high oleic acid oilseed rape breeding and prospects. Crops 24, 1–6. doi: 10.16035/j.issn.1001-7238.2009.05.05
Zhao, J. (2002). QTLs for Oil Content and Their Relationships to Other Agronomic Traits in an European × Chinese Oilseed Rape Population. Germany: Diss. Grorg-Agust University of Goettingen.
Zhao, J., Becker, H. C., Zhang, D., Zhang, Y., and Ecke, W. (2005). Oil content in a European × Chinese rapeseed population: QTL with additive and epistatic effects and their genotype-environment interactions. Crop Sci. 45, 51–59. doi: 10.2135/cropsci2005.0051
Zhao, J., Dimov, Z., Becker, H. C., Ecke, W., and Möllers, C. (2008). Mapping QTL controlling fatty acid composition in a doubled haploid rapeseed population segregating for oil content. Mol. Breed. 21, 115–125. doi: 10.1007/s11032-007-9113-y
Zhao, K., Tung, C. W., Eizenga, G. C., Wright, M. H., Ali, M. L., Price, A. H., et al. (2011). Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat. Commun. 2:467. doi: 10.1038/ncomms1467
Keywords: Brassica napus L., candidate genes, GWAS, mrMLM, fatty acid content
Citation: Guan M, Huang X, Xiao Z, Jia L, Wang S, Zhu M, Qiao C, Wei L, Xu X, Liang Y, Wang R, Lu K, Li J and Qu C (2019) Association Mapping Analysis of Fatty Acid Content in Different Ecotypic Rapeseed Using mrMLM. Front. Plant Sci. 9:1872. doi: 10.3389/fpls.2018.01872
Received: 14 June 2018; Accepted: 04 December 2018;
Published: 04 January 2019.
Edited by:
Yuan-Ming Zhang, Huazhong Agricultural University, ChinaReviewed by:
Guo-Bo Chen, Zhejiang Provincial People's Hospital, ChinaChunyu Zhang, Huazhong Agricultural University, China
Jinjin Jiang, Yangzhou University, China
Copyright © 2019 Guan, Huang, Xiao, Jia, Wang, Zhu, Qiao, Wei, Xu, Liang, Wang, Lu, Li and Qu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiana Li, bGpuMTk1MEBzd3UuZWR1LmNu
Cunmin Qu, ZHJxdWN1bm1pbkBzd3UuZWR1LmNu
†These authors have contributed equally to this work
 Mingwei Guan
Mingwei Guan Xiaohu Huang1,2†
Xiaohu Huang1,2† Ledong Jia
Ledong Jia Meichen Zhu
Meichen Zhu Cailin Qiao
Cailin Qiao Lijuan Wei
Lijuan Wei Xinfu Xu
Xinfu Xu Ying Liang
Ying Liang Kun Lu
Kun Lu Jiana Li
Jiana Li Cunmin Qu
Cunmin Qu