- 1State Key Laboratory of Rice Biology, Key Laboratory of Rice Biology and Genetic Breeding of Ministry of Agriculture, China National Rice Research Institute, Hangzhou, China
- 2The Collaborative Innovation Center of Southern Grain and Oil Crops, Agricultural College of Hunan Agricultural University, Changsha, China
Rice yield is closely related to plant leaf shape and chlorophyll content. In this study, we isolated and identified a narrow and rolled leaf mutant, temporarily named nrl3 with darker green leaves. Histological analysis showed that nrl3 has a reduced number of vascular bundles and undergoes abnormal abaxial sclerenchymatous cell differentiation. The NRL3 mutant phenotype was controlled by a single recessive gene, fine-mapped to a 221 kb interval between Indel3 and RM2322 on Chr3. There are 42 ORF in this interval. Sequencing identified an SNP mutant leading to a premature stop in ORF 18, the candidate gene. Bioinformation analysis indicated that NRL3 encodes a novel protein with unknown function. NRL3 is localized in cytoplasm, membrane and nucleus. Expression analysis of nrl3 showed that genes involved in chlorophyll synthesis were significantly up-regulated while those involved in chlorophyll degradation and programmed cell death (PCD) were significantly down-regulated. The expression levels of photosynthesis genes were also affected. Y2H and BIFC assays indicated that NRL3 interacts directly with NAL9/VYL to regulate leaf morphology in rice. Thus, NRL3 plays an important role in leaf morphogenesis and chlorophyll accumulation, and can be used as a new gene resource for constructing improved rice.
Introduction
Rice is an important crop that provides nutrition and energy to the global population. In recent years, the global demand for rice has increased quickly, but production has risen by less than 1%. Therefore, increasing rice yield is the ultimate goal of rice breeding. Rice leaf is the main photosynthesis organ. Leaf shape affects the efficiency of light capture and energy conversion. Appropriate leaf shape can improve plant photosynthesis efficiency and increase plant yield (Tsukaya, 2006; Micol, 2009).
Many studies have characterized genes which control the rolling and width of rice leaves. The process of leaf development in rice has been divided into four parts: leaf primordia formation, polarity establishment, tissue differentiation and leaf extension (Fan and Liang, 2014). Establishment of polarity is an important process that affects leaf morphology by regulating the adaxial (inward) and abaxial (outward) rolling of leaves (Fan and Liang, 2014). Research in Arabidopsis thaliana and maize showed that transcription factors and small RNAs are involved in regulation during establishment and maintenance of leaf adaxial–abaxial axis polarities (Moon and Hake, 2011). HD-ZIPIII family genes, such as PHB, PHV (McConnell et al., 2001), and REV (Otsuga et al., 2001), negatively regulated by microRNA miR165 and miR166, have been proved to regulate organ polarity, vascular development, and meristem function (Mallory et al., 2004; Kidner and Martienssen, 2004; Prigge et al., 2005). Further studies of PHABULOSA (PHB), PHAVOLUTA (PHV), and REVOLUTA (REV) in A. thaliana have shown that these genes exhibit a polar distribution expression pattern in the lateral organs which as an important regulatory factor for the establishment and maintenance of the adaxial features of these organs (Prigge et al., 2005). In addition, the YAB2 and YAB3 genes (Eshed et al., 2004) belonging to the YABBY family in rice, and MILKWEED POD1 (MWP1) (Candela et al., 2008) belonging to the KANADI family in maize, play important roles in the development of abaxial cells.
There is substantial evidence that defects in the establishment of polarity have a major impact on leaf rolling. Recently, a number of genes involved in the establishment of leaf polarity in rice have been identified. For example, SHALLOT-LIKE1 (SLL1), part of the KANADI family and having homology with KANADI family genes of A. thaliana, encodes a SHAQKYF class MYB family transcription factor regulating programmed cell death (PCD) of mesophyll cells, and affecting abaxial features of leaves (Zhang et al., 2009). OsAGO7 encodes an Argonaute (AGO) family protein, over-expression of which in rice causes upward rolling leaves (Shi et al., 2007). ADL1 gives rise to a plant-specific calpain-like cysteine proteinase which participates in leaf adaxial–abaxial axis maintenance, which the mutant showed abaxially rolled leaves (Hibara et al., 2009).
In addition to the establishment of polarity, the shape of bulliform cells also has an important role in rolling of leaves (Alvarez et al., 2008). Rolling-leaf 14 (RL14) encodes a 2OG-Fe (II) oxygenase family protein which regulates curling of leaves by affecting the formation of secondary walls. Mutants with an altered secondary wall composition exhibit changes in the shape of the bulliform cells, which affects water transport in the leaf (Fang et al., 2012). SRL1 encodes a Putative Glycosylphosphatidylinositol-Anchored Protein that inhibits the formation of bulliform cells by negatively regulating the expression of genes encoding vacuolar H+-ATPase subunit and H+-pyrophosphatase, thereby modifying leaf curl (Xiang et al., 2012).
Cell division and expansion are essential phases in the conversion of leaf primordia to mature leaves (Gonzalez et al., 2012), determining leaf size (Fujino et al., 2008). Many mutants showing abnormal leaf size have been characterized, caused by defects in cell division and cell expansion. For instance, Slender Leaf 1 (SLE1) encodes cellulose synthase-like D4, regulating cell proliferation during M phase and participated in cell wall formation, the mutant of which showed narrow and rolling leaves (Li M. et al., 2009; Wu et al., 2010; Yoshikawa et al., 2013). NAL2 and NAL3 are paralogs which encode the same OsWOX3A transcriptional activator. Their double mutant nal2/3 showed narrow and rolling leaves, and fewer veins (Cho et al., 2013, 2016). NAL1 encodes a trypsin-like serine/cysteine protease, affecting polar auxin transport and vascular patterns of rice, and participates in cell division (Qi et al., 2008; Jiang et al., 2015).
In the present study, we isolated and identified a narrow and rolled leaf mutant, nrl3. Compared with the wild-type, nrl3 showed darker green leaves at the tillering stage and with shorter panicles and longer, narrower seeds. Histological analysis showed that nrl3 had a decreased number of vascular bundles and defects of the abaxial sclerenchymatous cells which, respectively, caused leaf narrowed and rolling in rice. Map-based cloning showed that NRL3 encodes a novel protein with unknown function that can interact with NAL9/VYL directly to regulate leaf morphology in rice. In depth analysis of NRL3 can enhance our understanding of leaf morphogenesis and provides new genetic resources for improving rice.
Materials and Methods
Plant Materials and Growth Conditions
The nrl3 line was from a library of Oryza sativa subsp. indica cv. Zhongjiazao 17 (YK17) mutants obtained by EMS treatment. The F2 population for gene mapping was generated by crossing between nrl3 and tropical japonica rice variety D50. Plants were grown under natural conditions in the experimental fields of China National Rice Research Institute in Fuyang, Hangzhou, China.
Measurement of LRI and Net Photosynthetic Rate
Leaf rolling index (LRI) was determined for the upper leaf by measuring Lw (the maximum width of the expanded leaf blade) and Ln (the natural distance of the leaf blade margins at the same position). Then LRI(%) = (Lw-Ln)/Lw∗100 (Shi et al., 2007). Photosynthetic rate and transpiration rate were measured at the heading stage from 12: 00 to 13: 00 with a Li-6400 portable photosynthesis apparatus. Measurements of LRI, net photosynthetic rate and transpiration rates of wild-type and the nrl3 mutant were repeated at least three times, each replication used five independent plants.
Map-Based Cloning of the NRL3 Gene
To map the NRL3 locus, plants with narrow and rolling leaves were selected from the F2 population. First, we used both parents and 20 F2 individuals with the mutant phenotype for linkage analysis of NRL3. More than 116 polymorphic SSR markers evenly distributed on the whole rice genome were employed. Then a further 936 recessive individuals with the mutant phenotype were selected from the F2 population to fine map the NRL3 locus. SSR and Indel markers were developed based on nucleotide polymorphisms between YK17 and D50 in the corresponding regions (Supplementary Table S1). PCR products of candidate genes were amplified from both nrl3 and YK17 genomic DNA for sequencing. The sequencing results were analyzed using DNAMAN.
Scanning Electron Microscopy
The leaves of the wild-type and the nrl3 mutant at the heading stage were collected, fixed in 2.5% glutaraldehyde solution at 4°C overnight, and rinsed with 0.1 M phosphate buffer (pH 7.4). Then, the samples were dehydrated in a graded ethanol series, 20 min for each step, followed by substitution with isoamyl acetate. The samples were critical-point dried and sputter-coated with gold. The samples were observed and photographed using a scanning electron microscopy (XL30, Philips, United States).
Paraffin Sections
The fresh tissues were first fixed in formalin-glacial acetic acid-alcohol solution (FAA: 70% ethanol, acetic acid, formaldehyde; 16:1:1) for 24–48 h. The fixed tissues were dehydrated by passage through different concentrations of alcohol (75%, 85%, 95%, and absolute ethanol) and finally embedded in paraffin. Cross-sections (5–12 μm) were cut and stained with safranin and then observed under a microscope (3D HISTECH).
RNA Extraction and qRT-PCR Analysis
Total RNA was extracted from different plant tissues (root, stem, leaf, leaf sheath, panicle, and seeds) of wild-type and nrl3 using Trizol reagent (Life Technologies). The first strand cDNA was reverse-transcribed by using First Stand cDNA Synthesis Kit (TOYOBO). PCR program was: 95°C for 30 s, 95°C for 5 s, 58°C for 30 s, 72°C for 15 s, totally 40 cycles. Quantitative real-time PCR determination was based on the SYBR Green Real-time PCR Master Mix (Toyobo). Primers for quantitative real-time PCR (qRT-PCR) analysis are listed in Supplementary Table S1. The rice actin gene (Os03g0718150) was used as internal control. All the materials used for RNA extraction were from mixing three independent plants.
Plasmid Construction and Rice Transformation
For over-expression of the nrl3 mutant, the wild-type NRL3 cDNA sequence from YK17 was cloned using infusion primers: 5′-TTACTTCTGCACTAGGTACCATGGGTTTCATGTCAGCGA-3′ and 5′-TGGCTAGCGTTAACACTAGTCTGGGCACGATATGCAGCC-3′, and the PCR product was constructed into binary vector pCAMBIA1390 driven by the UBIQUTIN1 promoter (Clontech1). The CRISPR target of NRL3 (CAGCTCTGCGCCCAAGCTCGCGG) was cloned into Cas9/gRNA Vector (VK005-01, Beijing Viewsolid Biotech Co., Ltd.2). These plasmids were introduced into Agrobacterium tumefaciens strain EHA105. The overexpression vector was transformed into the nrl3 mutant, and the CRISPR/Cas9 vector was transformed into wild-type plants. Genotypes of the independent transgenic lines were determined by PCR amplification of the specific transgenic fragment (all primers used for plasmid construction are listed in Supplementary Table S1).
Subcellular Localization of NRL3
The full-length coding sequence of wild-type NRL3 from YK17, without a termination codon, was amplified by PCR and subcloned into pAN580 vector to obtain the NRL3-GFP construct. The fusion plasmid was transiently co-expressed in rice protoplasts using polyethylene glycol according to the protocols described previously (Chen et al., 2006). After incubating for 36 h at 28°C in the dark, GFP fluorescence in the protoplast cells was detected using a Zeiss LSM510 laser scanning confocal microscopy (Karl Zeiss, Jena, Carl Zeiss AG, Germany). The free pAN580 vector was used as control.
Determination of Chlorophyll Content
Fresh leaves of wild-type and nrl3 rice plants at heading stage were used to determine chlorophyll content according to the previous described method (Wu et al., 2007). Aliquots of 0.2 g fresh leaves from wild-type and nrl3 were cut into 1–2 mm pieces, immersed in 25 ml ethanol, placed in the dark for 48 h, and then centrifuged at 8000 g for 10 min. A DU800 spectrophotometer (Beckman Coulter, United States) was used to measure absorption of the supernatant at 665 and 649 nm.
Yeast Two-Hybrid Analysis
The full-length coding region of NRL3 without a termination codon was amplified by PCR and cloned into pGBKT7 vector. Two-Hybrid Library Screening Using Yeast Mating was used to find protein interactions with NRL3, applying the protocol described by the manufacturer with little adjustments (Matchmaker® Gold Yeast Two-Hybrid System User Manual, PT4084-1 (092413), Clontech). After sequencing for identification of the interacting protein, full-length cDNA of VYL without a termination codon (also named NAL9, LOC_Os03g29810) was amplified using infusion primers for construction into PGADT7 vector. Yeast transformation and selection procedures were carried out according to the manufacturer’s instructions [YeastmakerTM Yeast Transformation system 2 User Manual, PT1172-1 (PR8Y2629), Clontech].
BIFC Assay
The full-length cDNA fragments of NRL3 and VYL without stop codons were, respectively, cloned into binary vectors pSPYNE and pSPYCE to obtain pNRL3-YFPNE and pVYL-YFPCE. These plasmids were transiently expressed in tobacco leaves using A. tumefaciens strain EHA105 following the method described by Xiang et al. (2012). A confocal laser scanning microscope (Zeiss LSM710) was used to detect YFP fluorescent signals after 84 h post-transfection. Primers used in this assay are listed in Supplementary Table S1.
Results
Characterization of nrl3 Mutants
The mutant nrl3 was identified from a library produced using ethyl methanesulfonate (EMS) on an indica rice variety Zhongjiazao 17 (YK17). The nrl3 plants exhibited narrow and rolled leaves over the whole duration of growth. They had darker green leaves at the tillering stage which became more obvious with plant growth (Figure 1A and Supplementary Figures S5A,B). Statistical analysis showed that the LRI of nrl3 (50%) was over three times that of wild-type (15%) (Figure 1E), and the flag leaf width of nrl3 was reduced by approximately 47% (Figure 1F). Investigation of agronomic traits revealed no obvious differences in the plant height and effective tillers number between wild-type and nrl3 (Supplementary Figures S1A,C), but panicle length was significantly decreased in nrl3 (Figure 1B and Supplementary Figure S1B). Finally, the grain width and 1000-grain weight of nrl3 were significantly decreased (Figure 1G and Supplementary Figure S1D), while the grain length of nrl3 was significantly increased (Figures 1C,H) compared with the wild-type. Therefore, mutation in NRL3 leads to multiple changes in plant agronomic traits.
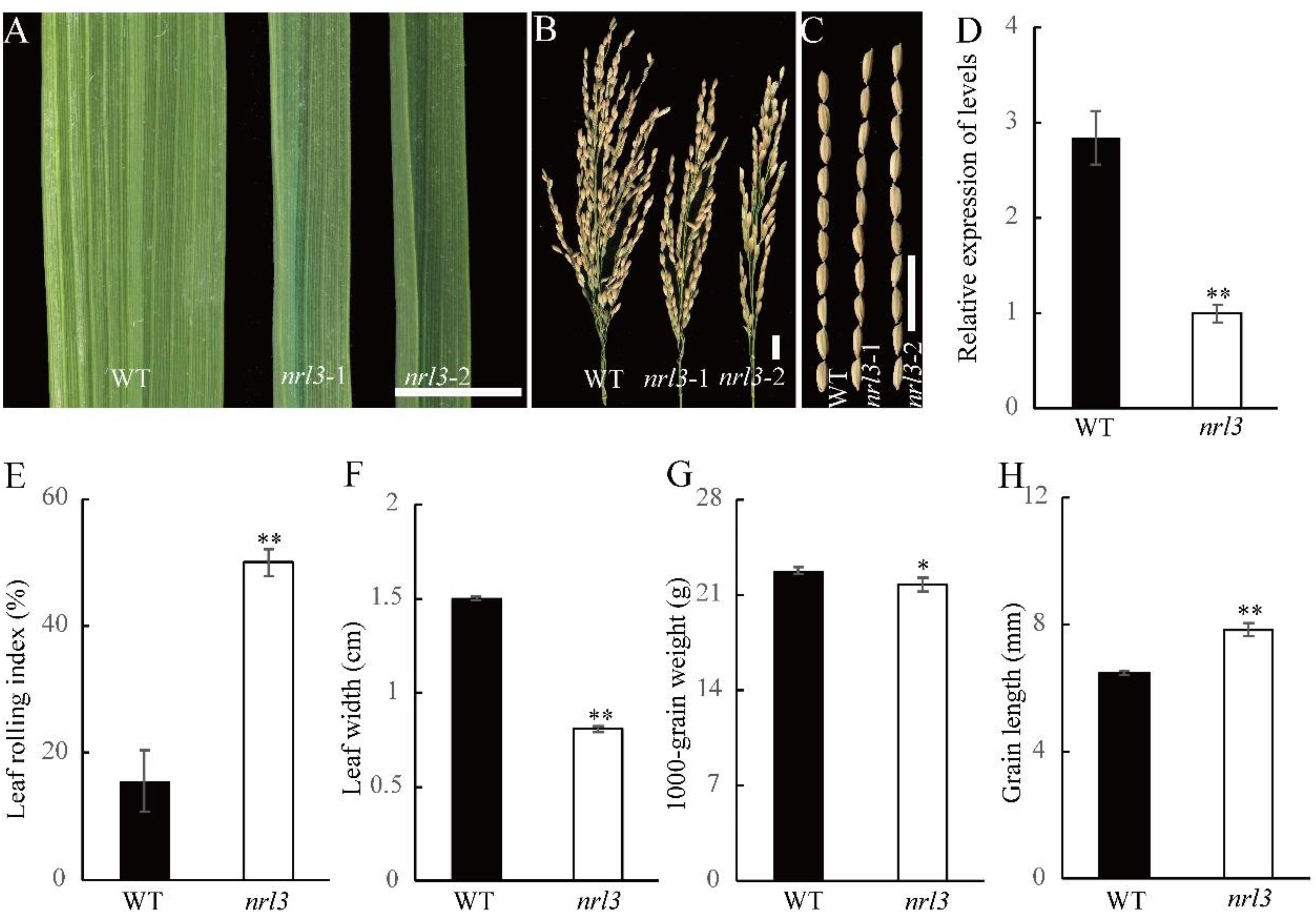
Figure 1. Phenotype the nrl3 mutant. (A) Upper leaf of WT, nrl3-1, and nrl3-2. (B) Panicle length of WT, nrl3-1, and the nrl3-2 mutant. (C) Grain length of WT, nrl3-1, and nrl3-2. Bar: 1 cm (A–C). (D) Expression analysis of NRL3 in WT and nrl3 mutant plants. RNA was isolated from leaves at tillering stage of WT and nrl3 of three independent plants. Data are shown as means ± SD for three biological replicates. (E) Leaf rolling index (LRI) of the flag leaf of WT and nrl3 of three independent plants. (F) Leaf width of the flag leaf of WT and the nrl3 mutant plant of three independent plants. Data are shown as means ± SD for three biological replicates. (G) Thousand-grain weight of WT and nrl3. (H) Grain length of WT and nrl3. Data are shown as mean ± SD from three biological replicates, each replication was not less than 200 seeds. Asterisks indicate statistical significance as determined by Student’s t-test (∗P < 0.05, ∗∗P < 0.01).
Defects in Sclerenchymatous Cells and Reduction in the Number of Vascular Bundles Cause the Rolled and Narrow Leaf in nrl3
To explore the reasons for the narrow and rolled leaves in nrl3 mutant, we carried out an analysis of cross-sections in the flag leaf. Compared with the wild-type, the nrl3 had fewer large and small veins. There were on average only six large veins in the nrl3 mutant, half the number in the wild-type (Figure 2A). Furthermore, in nrl3 mutant there were on average three small veins between each two neighboring large veins, compared to six in the wild-type (Figure 2B). Observed the cross-sections, we found that the development of abaxial sclerenchymatous cells was defective in the region of some small veins in nrl3 mutant, although the adaxial sides were normal (Figure 2C). In addition, we found that the air cavity and parenchymal cell of the nrl3 leaves differed from the wild-type. The mutant displayed the number of parenchyma cells on the large veins was decreased (Figure 2D). To confirm our results, we observed the abaxial side and adaxial side of the leaves by scanning electron microscopy (SEM). This showed defects in the sclerenchymatous cells on the abaxial side of the nrl3 leaves, while the adaxial side was similar to the wild-type (Figures 2E,F).
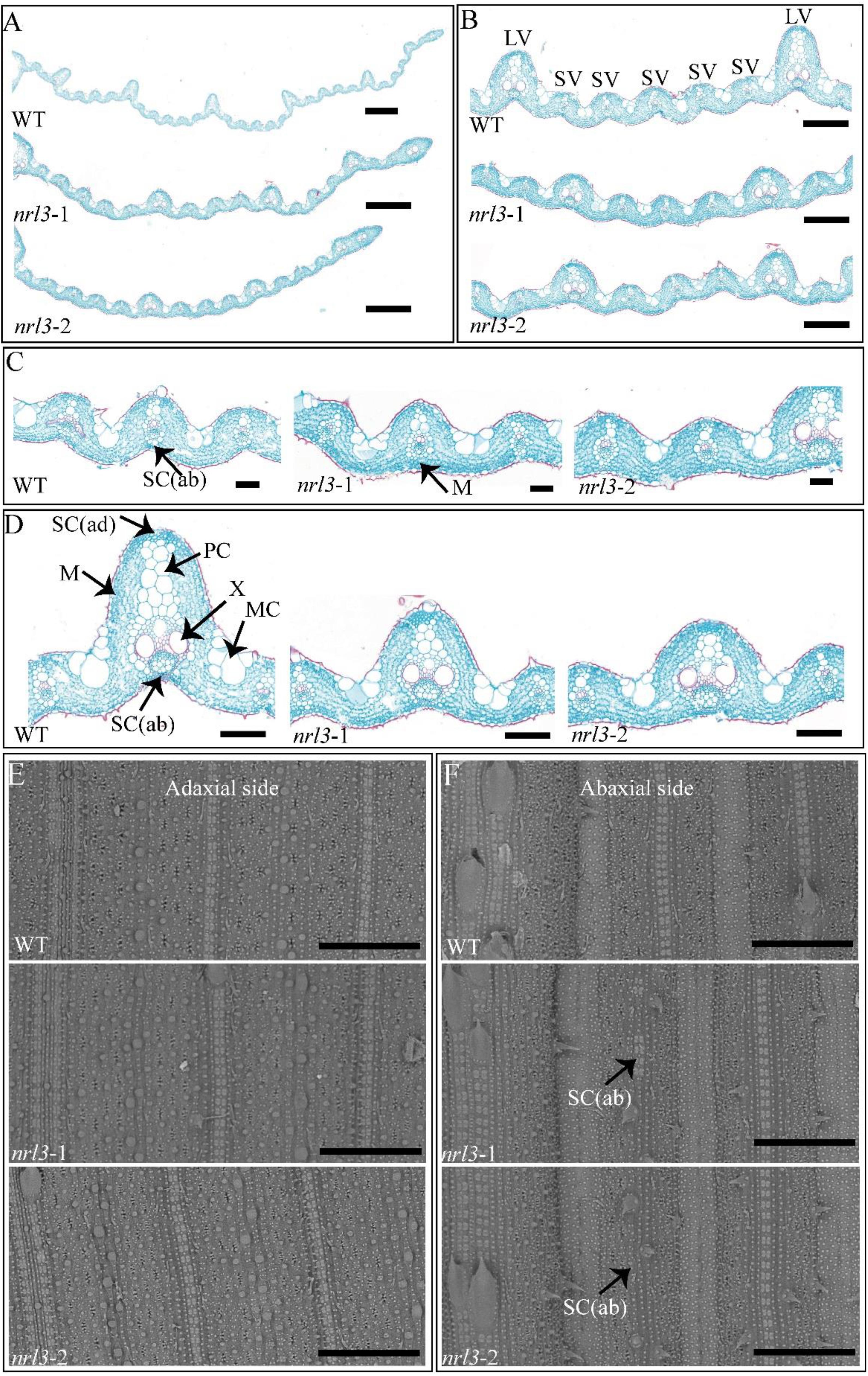
Figure 2. Cytological analysis of WT and nrl3. (A–D) Cross sections of leaves at the heading stage from WT, nrl3-1, and nrl3-2. (E,F) Scanning electron microscopy of the adaxial and abaxial sides of WT, nrl3-1, and nrl3-2 leaves. LV, large veins; SV, small veins; SC(ab), abaxial sclerenchyma cell; SC(ad), adaxial sclerenchyma cell; MC, motor cell; M, mesophyll cell; X, xylem; PC, parenchymal cell; Bar: 500 μm (A), 200 μm (B), 50 μm (C), 100 μm (D), 300 μm (E,F).
Map-Based Cloning of NRL3
In order to identify the inheritance of NRL3, map-based cloning was conducted using a F2 population produced by crossing the nrl3 with D50 (tropical japonica variety). Genetic analysis showed that the plant number ratio for wild-type and nrl3 individuals in the F2 population was 3:1 (185 wild-type plants and 68 mutant plants; x2 = 0.4756 < x20.05,1 = 3.84), indicated that the phenotype of nrl3 is controlled by a single recessive gene. The locus of NRL3 was first mapped on the short arm of chromosome 3 between the simple sequence repeat (SSR) markers RM7197 and RM14898 (Figure 3A). To fine-map NRL3, 936 F2 individuals with the nrl3 phenotype were chosen for genotyping, which further narrowed the mapping interval down to a 221-kb region between Indel marker Indel 3 and SSR marker RM2322 (Figure 3A). According to the Rice Genome Annotation Project database3, there are 42 ORFs in this interval, which were selected for sequencing analysis. We found a single substitution of a guanine (G) with an adenine (A) on the 8th exon of LOC_Os03g19520 (Figure 3A), but no mutation in any of the other predicted ORFs. This G→A substitution leads to the premature termination of NRL3 (Supplementary Figure S2).
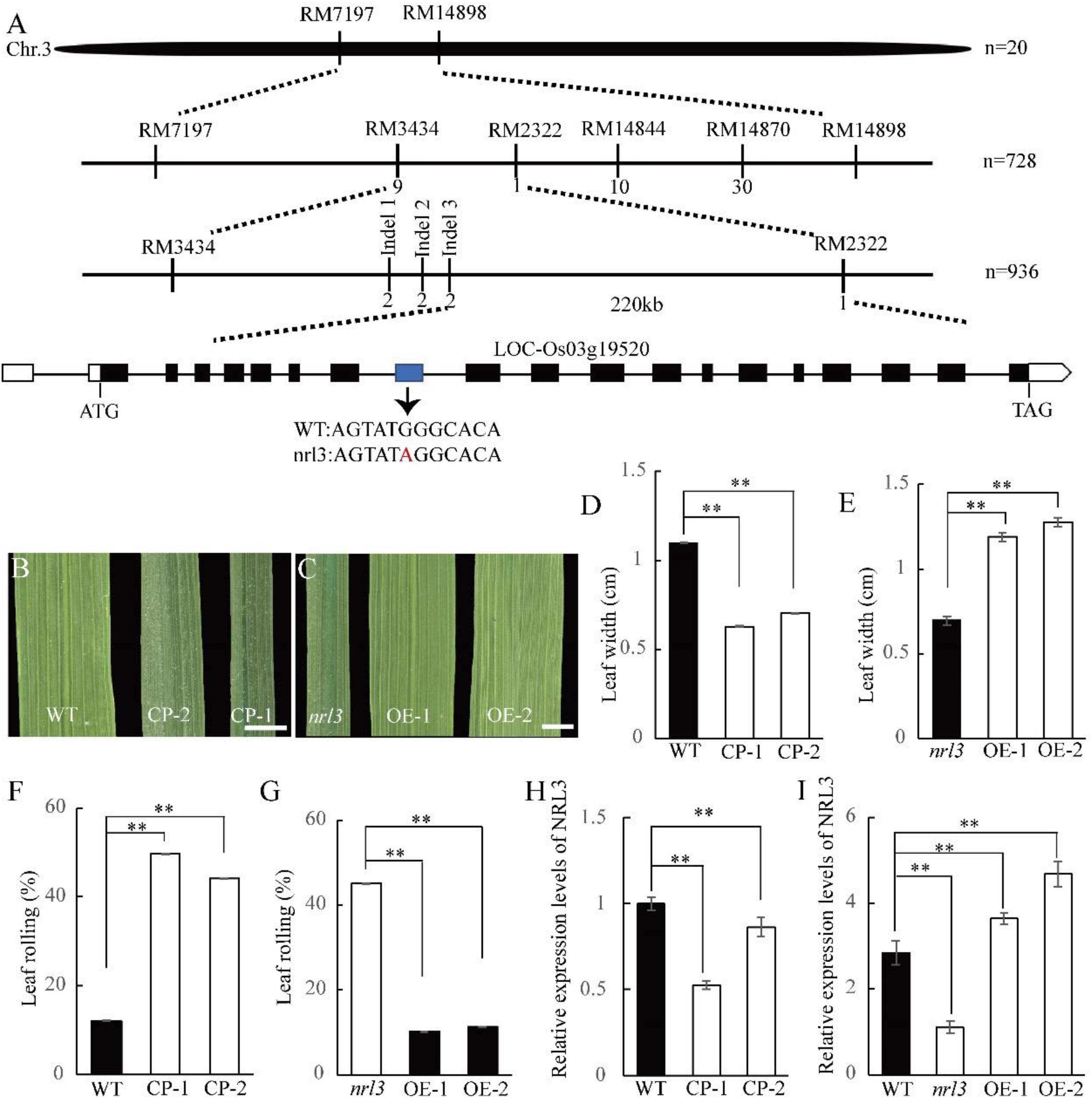
Figure 3. Map-based cloning of NRL3. (A) Map of NRL3 locus in a 221 kb region between markers Indel 3 and RM2322 on chromosome 3 (Chr3). (B) The leaf phenotype of knock-out transgenic plants (CP). Bar: 1 cm. (C) The phenotype of overexpression transgenic plants (OE). Bar: 1 cm. (D,E) Upper leaf width of knock-out transgenic plants (D) and overexpression transgenic plants (E). (F,G) Upper leaf rolling in knock-out transgenic plants (F) and overexpression transgenic plants (G). (H) Expression levels of NRL3 in knock-out transgenic plants. (I) Expression level of NRL3 in overexpression transgenic plants. RNA was isolated from leaves at tillering stage of wild-type, nrl3 and transgenic plants. Data are shown as means ± SD from three replicates. The asterisks indicate statistical significance between the WT and the nrl3 mutant, as determined by Student’s t-test (∗∗P < 0.01).
To confirm LOC_Os03g19520 is the target gene of NRL3, vectors carrying the full-length coding region of NRL3 driven by the UBIQUTIN1 promoter, or knock-out vector using the Crispr/Cas9 system, were constructed and introduced into nrl3 and wild-type, respectively. We found all of the over-expression plants were able to rescue the narrow and rolled leaf phenotype of nrl3, with increased leaf width, decreased LRI, and a lighter green color (Figures 3C,E,G,I). Furthermore, after knock-out of LOC_Os03g19520 in wild-type, the resulting phenotype is similar to nrl3 (Figures 1D, 3B,D,F,H). Thus LOC_Os03g19520 was confirmed as the target gene of NRL3. Protein sequence analysis showed NRL3 encoded a novel protein with unknown function. Phylogenetic analysis revealed that NRL3 protein may have a conserved function in both monocotyledons and dicotyledons (Supplementary Figure S4).
Expression Pattern and Subcellular Localization of NRL3
Quantitative real-time PCR analysis suggested that NRL3 was constitutively expressed in all tested organs, with the highest levels in leaf sheaths (Figure 4A). NRL3-GFP fusion protein was ubiquitous in protoplasts, suggesting that NRL3 may be found in the nucleus, membrane and cytoplasm. The green fluorescence of NRL3-GFP was coincident with the autofluorescence of chlorophyll (Figure 4B).
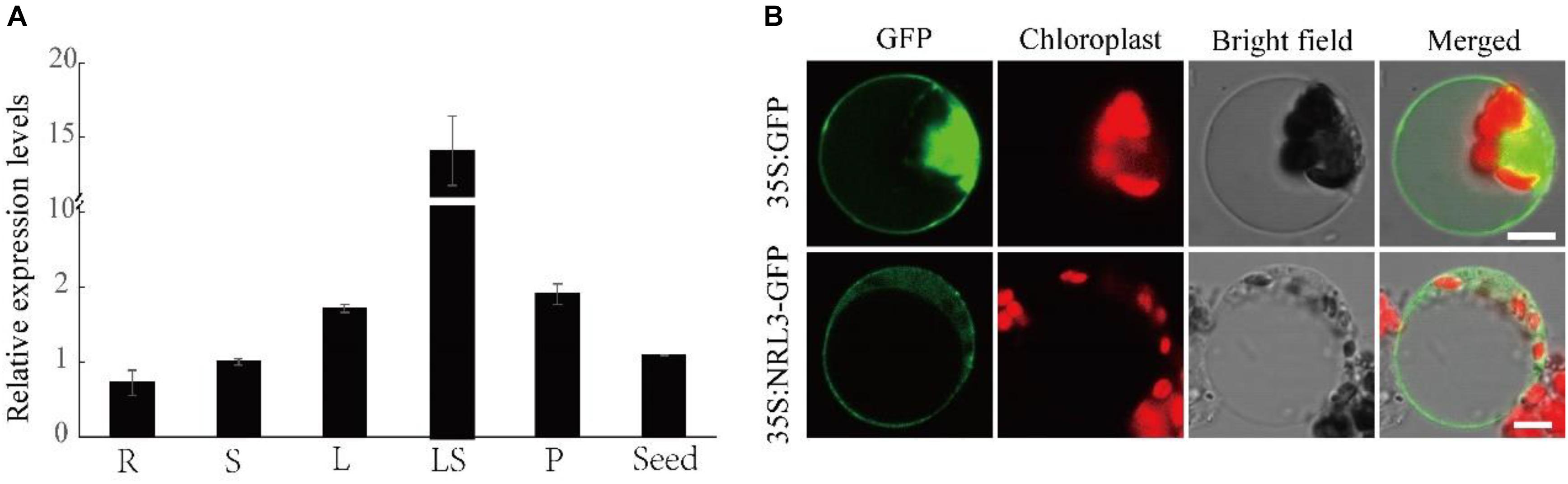
Figure 4. Expression pattern and subcellular localization of NRL3. (A) qRT-PCR analysis of NRL3 expression level in various tissues. R, root; S, stem; L, leaf; LS, leaf sheath; P, panicle. (B) Subcellular localization of NRL3 in rice protoplast cells. Free 35S: GFP, which localized to the cytoplasm, membrane and nucleus, was used as a control; Full-length NRL3 fusion protein (35 S: NRL3-GFP) similarly localized to the cytoplasm, membrane and nucleus. At 36 h after transformation, rice protoplast cells were observed using a confocal laser scanning microscope. GFP signals (Green), chlorophyll autofluorescence (Red), bright-field images, and the merged images of GFP signal and chlorophyll signals are shown in each panel. Bars: 5 μm.
NRL3 Negatively Regulates Chlorophyll Synthesis and Affects Chlorophyll Degradation
Leaves of nrl3 were darker green, and the contents of chlorophyll a (chla) and chlorophyll b (chlb) were significantly increased in its flag leaves (Figures 1B, 5A). Furthermore, chlorophyll content was decreased in the NRL3 over-expression lines, but increased in the knock-out lines (Figures 5B,C). In addition, nrl3 exhibited a significantly lower photosynthetic rate and higher transpiration rate than the wild-type, and the expression levels of photosynthesis-related genes was significantly down-regulated (Supplementary Figures S3A–C). Therefore, the expression of chlorophyll biosynthesis-related genes was analyzed via qRT-PCR. Compared with wild-type the expression in nrl3 of most chlorophyll biosynthetic genes was significantly up-regulated, including: HEMC (Porphobilinogen deaminase, LOC_Os02g07230), HEME (Uroporphyrinogen decarboxylase 1, LOC_Os01g43390), URO (Uroporphyrinogen decarboxylase 2, LOC_Os03g21900), HEMF (Coproporphyrinogen III oxidase, LOC_Os04g52130), CHLD (Mg chelatase D subunit, LOC_Os03g59640), CHLH (Mg chelatase H subunit, LOC_Os03g20700), CHLI (Mg chelatase I subunit, LOC_Os03g36540), PORA (Protochlorophyllide reductase A, LOC_Os04g58200), and CHLG (Chlorophyll synthase, LOC_Os05g28200). The other genes we examined were unchanged in nrl3: HEMA (Glutamyl-t RNA reductase, LOC_Os10g35840), HEML (Glutamate-1-semialdehyde 2,1-aminomutase, LOC_Os08g41990), HEMB (Delta-aminolevulinic acid dehydratase, LOC_Os06g49110), CHLM (Mg-protoporphyrin IX methyltransferase, LOC_Os06g04150), and DVR (Divinyl chlorophyllide a 8-vinyl-reductase, LOC_Os03g22780) (Figure 5E). In addition we examined the expression of chlorophyll degradation-related genes, and found the following groups significant decreased in nrl3 and significant up-regulated in OE-1 and OE-2 (Figure 5D): NYC1 (Chlorophyll b reductase gene, LOC_Os01g12710), NYC3 (α/β hydrolase-fold family protein, LOC_Os06g24730), NOL (Chlorophyll b reductase gene, LOC_Os03g45194), PAO (pheophorbide a oxygenase, LOC_Os03g05310), SGR (stay green gene, LOC_Os09g36200), and CLH (chlorophyllase-2, LOC_Os10g28370).
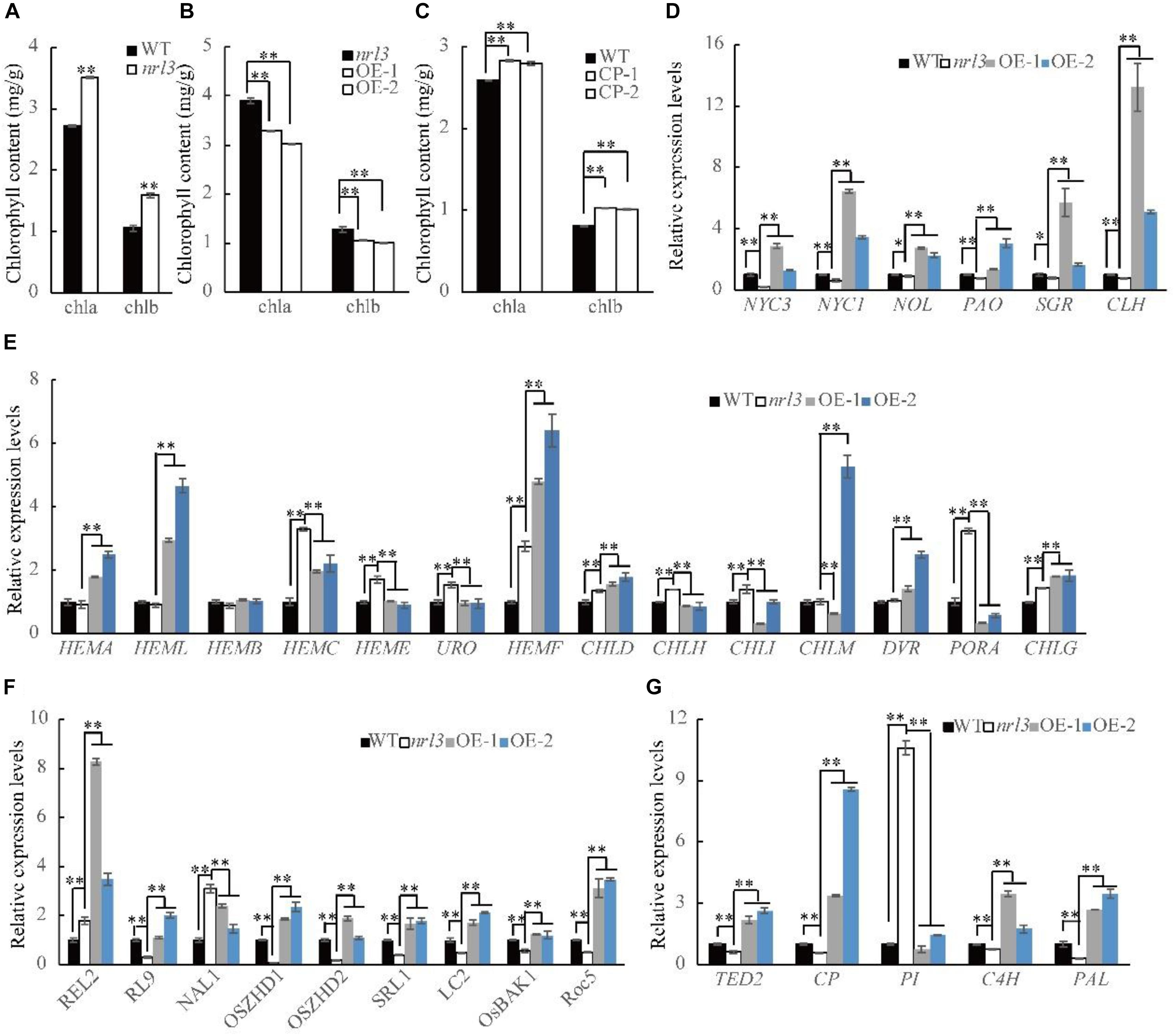
Figure 5. Chlorophyll contents and the transcriptional changes of genes in WT, nrl3 and transgenic plants. (A) Chlorophyll content of leaves in WT and nrl3 mutant at heading stage. Chla, Chlorophyll a; Chlb, Chlorophyll b. (B) Chlorophyll content of leaves in nrl3 and OE mutant at heading stage. (C) Chlorophyll content of leaves in nrl3 and CP mutant at tillering stage. (D) Relative expression of genes involved in chlorophyll degradation in WT, nrl3, OE-1, and OE-2. (E) Relative expression of genes involved in chlorophyll biosynthetic genes in WT, nrl3, OE-1, and OE-2. (F) Expression levels of genes associated with leaf morphology development in WT, nrl3, OE-1, and OE-2. (G) Relative expression levels of PCD genes. RNA was isolated from leaves at heading stage of WT, the nrl3 mutant, OE-1, and OE-2. Data are shown as means ± SD for three biological replicates. The asterisk indicates statistical significance determined by Student’s t-test (∗P < 0.05; ∗∗P < 0.01).
Genes Associated With Leaf Morphology Development Were Affected in nrl3
Quantitative real-time PCR analyses were performed to investigate the relationship between NRL3 and other leaf morphology regulatory genes. The results showed that REL2 (rolled and erect leaf 2, Yang et al., 2016) and NAL1 (trypsin-like serine/cysteine protease, Qi et al., 2008; Jiang et al., 2015) was significantly up-regulated in the nrl3. In contrast, RL9 (a GARP protein gene, Yan et al., 2008), OsZHD1 and OsZHD2 (Zn-finger transcription factor, Figueiredo et al., 2012; Xu et al., 2014), SRL1 (glycosylphosphatidylinositol-anchored protein, Xiang et al., 2012), LC2 (vernalization-insensitive-3-like protein, Wang et al., 2013), ROC5 (homeodomain leucine zipper class IV gene, Zou et al., 2011), and OsBAK1 (SERK-family receptor-like protein kinase, Li D. et al., 2009) were significantly down-regulated in nrl3 (Figure 5F). In addition, compared to nrl3, the expression of most genes in OE-1 and OE-2 was up-regulated (Figure 5F).
nrl3 Causes Leaf Rolling by Affecting PCD of Abaxial Mesophyll Cells
Previous studies have shown that PCD is involved in the formation of sclerenchymatous cells. Thus, we examined in wild-type and nrl3 the expression levels of rice genes which share highly identity with TE-PCD marker genes from Zinnia or other species. Results showed that TED2 (LOC_Os10g41170), CP (LOC_Os08g44270), C4H (LOC_Os05g25640), and PAL (LOC_Os05g35290) were significantly down-regulated, while PI (LOC_Os05g06780) was significantly up-regulated (Figure 5G). In addition, compared to nrl3 the expression of most genes in OE-1 and OE-2 was significant up-regulated (Figure 5G). According to previous studies we know that TED2 is the marker gene of stage two of PCD, with CP, C4H, PI, and PAL marking stage three. Together, these results indicate that the effect of NRL3 on development of sclerenchymatous cells may be via regulation of PCD in abaxial mesophyll cells.
NRL3 and VYL Proteins Form a Complex
NRL3 gene was predicted to encode a protein of unknown function. To explore its mode of action we screened the yeast two-hybrid (Y2H) library of rice seedlings. Using NRL3 as bait, we screened and sequenced many independent fragments interacting with NRL3. The sequencing results showed that the NAL9/VYL gene matched with partial clones. In rice, VYL encodes ATP-dependent Clp protease proteolytic subunit protein, which plays an important role in rice chloroplast biosynthesis and leaf development. The vyl mutant displayed narrow foliage, pale green leaf phenotypes throughout the growth period (Dong et al., 2013; Li et al., 2013). The yeast two-hybrid assay (Y2H) showed that NRL3 directly interacted with VYL (Figure 6A). In addition, bimolecular fluorescence complementation (BIFC) showed that NRL3 and VYL interact on the membrane (Figure 6B). In summary, there is a strong interaction between NRL3 and VYL. VYL forms a complex with ClpP4, ClpP5, ClpP, and ClpP3, and plays important roles in the biosynthesis of chloroplasts and in leaf development, while ClpP4, ClpP5, ClpT, ClpP3, and VYL have similar expression patterns (Dong et al., 2013). Consequently we examined the expression levels of ClpP4, ClpP5, ClpT, ClpP3, and VYL in WT, nrl3, OE-1, and OE-2, and we found that expression of ClpP4, ClpT, ClpP3, and VYL was significantly down-regulated in nrl3 and significantly up-regulated in OE-1 and OE-2 (Figure 6C). In addition, the Y2H results showed that NRL3 did not interact with ClpP3, ClpP4, ClpP5, or ClpPT (Figure 7A).
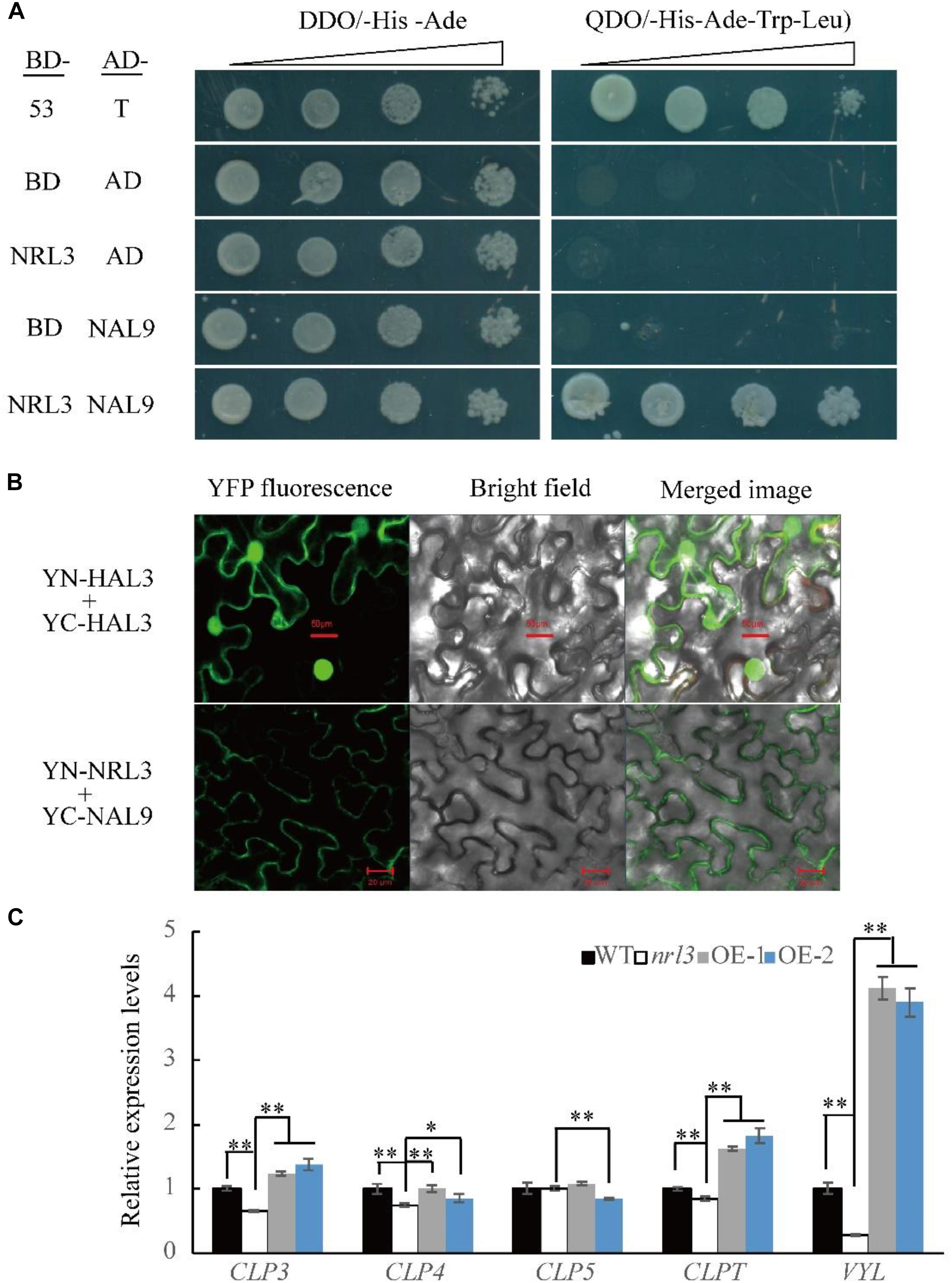
Figure 6. NRL3 interaction with NAL9/VYL. (A) Yeast two-hybrid assays showed interaction between NRL3 and VYL. The interactions between PGBKT7-53 and PGADT7-T, PGBKT7, and PGADT7 were used as the positive control and negative control, respectively. (B) BIFC assay confirmed the interaction of NRL3 and VYL (Bar = 20 μm). The interaction of YN-HAL3 and YC-HAL3 was used as positive control (Bar = 50 μm). (C) Expression of CLP3, CLP4, CLP5, CLPT, and VYL/NAL9 in WT, nrl3, OE-1, and OE-2. Data are means ± SD for three biological replicates. The asterisk indicates the difference determined by Student’s t-test (∗P < 0.05; ∗∗P < 0.01).
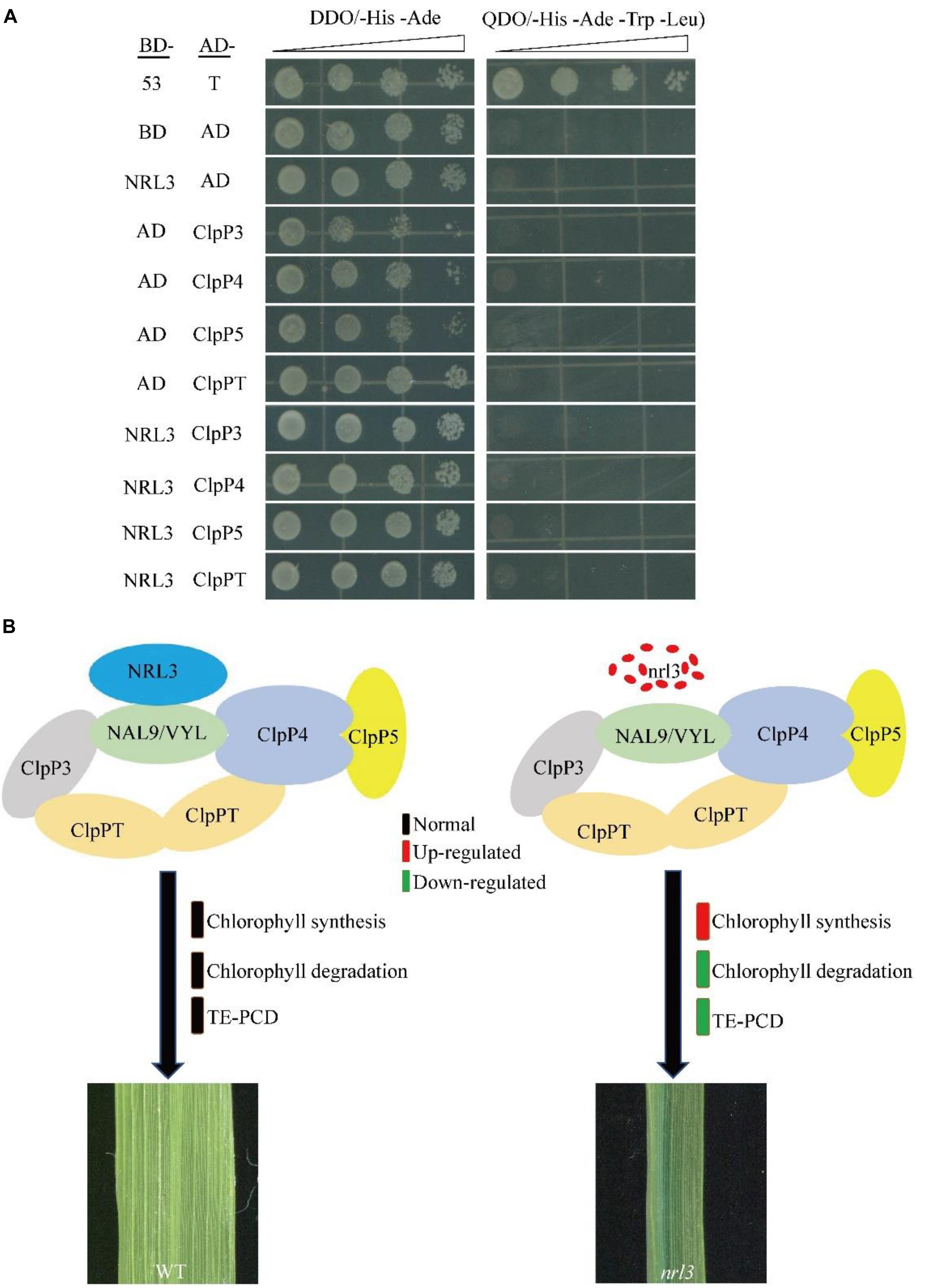
Figure 7. Y2H analysis of NRL3, ClpP3, ClpP4, ClpP5, and ClpPT, and our model showing the role NRL3 plays in regulating leaf development and chlorophyll degradation. (A) Yeast two-hybrid assays showing the absence of interaction between NRL3 and ClpP3, ClpP4, ClpP5, and ClpPT. The interactions between PGBKT7-53 and PGADT7-T, PGBKT7, and PGADT7 were used as the positive control and negative control, respectively. (B) Model showing the important role NRL3 plays in regulating leaf development and chlorophyll degradation.
Discussion
Leaf morphology is an important agronomic trait. Changes in leaf shape impact on photosynthesis efficiency, with consequences for rice yield. Mutations of leaf morphology have been divided into two main categories, narrow leaves and rolled leaves. Factors affecting leaf shape in rice have been divided into eight categories related to changes in cell structure, with differences in the number, size and pattern of bulliform cells, mesophyll cells, sclerenchymatous cells, and vascular bundles (Zou et al., 2014). In our study, we isolated and identified a narrow and rolled leaf mutant, nrl3. It displays defects in sclerenchymatous cells and a reduced number of vascular bundles.
Sclerenchymatous cells play a crucial role in maintaining normal leaf morphology, and defects lead to loss of support for leaves, resulted in leaf rolling. SLL1/RL9 regulates abaxial sclerenchymatous cell formation to modulate leaf rolling, down-regulate expression of which showed defect in abaxial-side sclerenchymatous cells (Zhang et al., 2009). The semi-rolled leaf2 (srl2) and narrow and rolled leaf (nrl2) mutant, which is allelic to nrl3, showed defective development of the sclerenchymatous cells and rolled leaves (Liu et al., 2016; Zhao et al., 2016). In our study, the nrl3 mutant showed inward rolling leaves, and statistical analysis showed that the LRI increased about 33% compared to wild-type (Figure 1E). Histological analysis indicated that the abaxial-side sclerenchymatous cells were defective (Figures 2C,F). The SLL1/RL9 was significantly down-regulated in nrl3 (Figure 5F). Sclerenchymatous cells originate from the differentiation of mesophyll cells via PCD, and are lignified dead cells with thickened secondary cell walls (Zhang et al., 2009). In the process of differentiation, abnormalities in PCD of mesophyll cells can lead to sclerenchymatous cell dysgenesis (Zhang et al., 2009). We found that the nrl3 mutant had defective sclerenchymatous cells with TE-PCD-related genes TED2, CP, C4H, and PAL significantly down-regulated (Figure 5G). This indicates nrl3 undergoes abnormal PCD during the process of mesophyll cell differentiation to sclerenchymatous cells, which would finally cause the observed defects in the development of sclerenchymatous cells, and produce rolled leaves.
Leaf size is controlled by cell division and expansion, and the obvious feature of narrow leaf mutants in rice is reduction in the number of vascular bundles (Cho et al., 2014). The narrow leaf (nal9) mutant showed narrow leaves and reduction in the number of large and small vascular bundles, caused by defective cell proliferation (Li et al., 2013). Mutation of ABNORMAL VASCULAR BUNDLES (AVB), allelic to nrl3, gave rise to narrow leaves by affecting the cell division pattern during lateral primordia development (Ma et al., 2017). The (NARROW LEAF 1) NAL1 encodes trypsin-like serine/cysteine protease, down-regulate expression of which in wild-type affects polar auxin transport and narrow leaf (Qi et al., 2008; Jiang et al., 2015). Auxin acts as a signal for cell division, cell expansion and cell differentiation, which play a crucial part in regulating leaf primordia and cell growth in development of leaf (Fujino et al., 2008). In our study, nrl3 showed narrow leaves and the number of vascular bundles was reduced to only half that of the wild-type, and the transcription expression of NAL1 was signification alter in nrl3 (Figure 5F). Thus, the NRL3 gene may regulate leaf narrowing and the number of vascular bundles by influencing the regulation of auxin, and finally have effection on cell division. Moreover, OsZHD1, OsZHD2, Roc5, and REL2 that involved in leaf development, the mutants of which caused the changes of the number and the abnormal arrangement of bulliform cells in leaf were resulted in rolling leaves. SRL1 regulates leaf morphology by negatively regulating the expression of genes encoding vacuolar H+-ATPase subunit and H+-pyrophosphatase. The transcription expression of these genes showed signification change in nrl3 (Figure 5F). These results suggested that NRL3 may synergistically with these genes to regulate leaf morphological development.
Chlorophyll plays an important role in the photosynthesis of plants. In general, chlorophyll content is closely related to photosynthesis competence. However, the “non-functional stay green” mutants, caused by defective degradation of chlorophyll, have much higher chlorophyll content but also lower photosynthetic capacity (Thomas and Howarth, 2000). Stay Green Rice (SGR) encodes senescence-inducible chloroplast stay-green protein, with a defect in chlorophyll degradation and the mutant showed darker green leaves with loss of photosynthetic competence (Jiang et al., 2007). Both NON-YELLOW COLORING1 (NYC1) and NYC1-LIKE (NOL) genes encode mutated chlorophyll b reductase, with defective chlorophyll degradation pathways causing leaves to stay green but with decreased photosynthetic competence (Kusaba et al., 2007; Sato et al., 2009). Non-yellow coloring 3 (nyc3) plants exhibited a similar phenotype to the above mutants, also with defective chlorophyll degradation (Morita et al., 2009). In our study, the nrl3 mutant showed darker green leaves at the tiller stage, with much higher chlorophyll content than wild-type, but the photosynthetic rate was significantly decreased (Figures 1B, 5A and Supplementary Figure S3A). Expression analysis showed the photosynthesis-related genes NADH, RbcS, PsaA, Cab1R, and Cab2R and chlorophyll degradation related genes NYC3, NYC1, NOL, PAO, SGR, and CLH were all significantly down-regulated in nrl3, but the chlorophyll biosynthesis genes were significantly up-regulated, such as HEME, URO, CHLH, CHLI, and PORA (Supplementary Figure S3C and Figures 5D,E). We hypothesize that the chlorophyll degradation pathway defect leads to the increasing accumulation of non-functional chlorophyll and lower photosynthesis competence, but feedback within the plant accelerates chlorophyll synthesis to compensate for the reduction in functional chlorophyll.
Interestingly, we found that NRL3 protein interacts with VYL/NAL9 protein, an ATP-dependent Clp protease proteolytic subunit affecting leaf size and chloroplast biosynthesis. In higher plants, Clp acts as a protein degradation apparatus to regulating the quantity and quality of protein, which consists of ClpP protein, ClpR protein, ClpT protein, and ClpC protein (Adam et al., 2006; Sakamoto, 2006). Clp protease is a highly conserved protein, containing ClpP1, ClpP4 and other components, regulating plant development and chloroplast biogenesis (Schirmer et al., 1996). Down-regulation of ClpP1 gave rise to slender leaf shape and chloroplast dysplasia (Shikanai et al., 2001), while knock-down ClpP4 showed pale-green leaves and abnormal development of mesophyll cell (Zheng et al., 2006). In our study, NRL3 can interacted with VYL/NAL9 directly, but not with ClpP3, ClpP4, ClpP5, and ClpPT, which the five components can form a complex. Previous reports showed that the complex containing VYL, ClpP3, ClpP4, ClpP5, and ClpPT can be functioned as a protein degradation apparatus to regulate chloroplast biogenesis and leaf development (Dong et al., 2013; Li et al., 2013). Based on these data, we speculated that NRL3 may be a component of the complex, and regulating the leaf development and chlorophyll degradation by participate the process of protein degradation, and finally, we propose a complex model by which NRL3 affects leaf development and chlorophyll degradation (Figure 7B), but the accurate regulatory mechanism of this complex needs further investigation.
Author Contributions
All authors contributed to the creation of the new mutant materials, overall design, and planning of the research.
Funding
This research was financially supported by China National Key Research and Development Program (2016YFD0101801 and 2017 YFD0100302), National Natural Science Foundation of China (No. 31501285), The Major Scientific and Technological Projects of Agricultural new varieties of Zhejiang province, China (2016C02050-2), and the National S&T Major Project (2016ZX08001006).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00175/full#supplementary-material
Footnotes
- ^http://www.clontech.com
- ^http://www.v-solid.com/
- ^http://rice.plantbiology.msu.edu/cgi-bin/gbrowse/rice/
References
Adam, Z., Rudella, A., and van Wijk, K. J. (2006). Recent advances in the study of Clp, FtsH and other proteases located in chloroplasts. Curr. Opin. Plant Biol. 9, 234–240. doi: 10.1016/j.pbi.2006.03.010
Alvarez, J. M., Rocha, J. F., and Machado, S. R. (2008). Bulliform cells in Loudetiopsis chrysothrix (Nees) Conert and Tristachya leiostachya Nees (Poaceae): structure in relation to function. Braz. Arch. Biol. Technol. 55, 113–119. doi: 10.1590/S1516-89132008000100014
Candela, H., Johnston, R., Gerhold, A., Foster, T., and Hake, S. (2008). The milkweed pod1 gene encodes a KANADI protein that is required for abaxial / adaxial patterning in maize leaves. Plant Cell 20, 2073–2087. doi: 10.1105/tpc.108.059709
Chen, S., Tao, L., Zeng, L., Vega-Sanchez, M. E., Umemura, K., and Wang, G. L. (2006). A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol. Plant Pathol. 7, 417–427. doi: 10.1111/j.1364-3703.2006.00346.x
Cho, S. H., Kang, K., Lee, S. H., Lee, I. J., and Paek, N. C. (2016). OsWOX3A is involved in negative feedback regulation of the gibberellic acid biosynthetic pathway in rice (Oryza sativa). J. Exp. Bot. 67, 1677–1687. doi: 10.1093/jxb/erv559
Cho, S. H., Yoo, S. C., Zhang, H., Lim, J. H., and Paek, N. C. (2014). Rice NARROW LEAF1 regulates leaf and adventitious root development. Plant Mol. Biol. Rep. 32, 270–281. doi: 10.1007/s11105-013-0675-z
Cho, S. H., Yoo, S. C., Zhang, H., Pandeya, D., Koh, H. J., Hwang, J. Y., et al. (2013). The rice NARROW LEAF2 and NARROW LEAF3 loci encode WUSCHEL-related homeobox 3A (OsWOX3A) and function in leaf, spikelet, tiller and lateral root development. New Phytol. 198, 1071–1084. doi: 10.1111/nph.12231
Dong, H., Fei, G. L., Wu, C. Y., Wu, F. Q., Sun, Y. Y., Chen, M. J., et al. (2013). A rice virescent-yellow leaf mutant reveals new insights into the role and assembly of plastid caseinolytic protease in higher plants. Plant Physiol. 162, 1867–1880. doi: 10.1104/pp.113.217604
Eshed, Y., Izhaki, A., Baum, S. F., Floyd, S. K., and Bowman, J. L. (2004). Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development 131, 2997–3006. doi: 10.1242/dev.01186
Fan, Y. B., and Liang, W. Q. (2014). Research progress on the mechanism of leaf polarity establishment in rice. J. Shanghai Jiao Tong Univ. 32, 16–22.
Fang, L., Zhao, F., Cong, Y., Sang, X., Du, Q., Wang, D., et al. (2012). Rolling-leaf14 is a 2OG-Fe (II) oxygenase family protein that modulates rice leaf rolling by affecting secondary cell wall formation in leaves. Plant Biotechnol. J. 10, 524–532. doi: 10.1111/j.1467-7652.2012.00679.x
Figueiredo, D. D., Barros, P. M., Cordeiro, A. M., Serra, T. S., Lourenço, T., Chander, S., et al. (2012). Seven zinc-finger transcription factors are novel regulators of the stress responsive gene OsDREB1B. J. Exp. Bot. 63, 3643–3656. doi: 10.1093/jxb/ers035
Fujino, K., Matsuda, Y., Ozawa, K., Nishimura, T., Koshiba, T., Fraaije, M. W., et al. (2008). NARROW LEAF 7 controls leaf shape mediated by auxin in rice. Mol. Genet. Genom. 279, 499–507. doi: 10.1007/s00438-008-0328-3
Gonzalez, N., Vanhaeren, H., and Inze, D. (2012). Leaf size control: complex coordination of cell division and expansion. Trends Plant Sci. 17, 332–340. doi: 10.1016/j.tplants.2012.02.003
Hibara, K., Obara, M., Hayashida, E., Abe, M., Ishimaru, T., Satoh, H., et al. (2009). The ADAXIALIZED LEAF1 gene functions in leaf and embryonic pattern formation in rice. Dev. Biol. 334, 345–354. doi: 10.1016/j.ydbio.2009.07.042
Jiang, D., Fang, J., Lou, L., Zhao, J., Yuan, S., Yin, L., et al. (2015). Characterization of a null allelic mutant of the rice NAL1 gene reveals its role in regulating cell division. PLoS One 10:e0118169. doi: 10.1371/journal.pone.0118169
Jiang, H., Li, M., Liang, N., Yan, N., Wei, Y., Xu, X., et al. (2007). Molecular cloning and function analysis of the stay green gene in rice. Plant J. 52, 197–209. doi: 10.1111/j.1365-313X.2007.03221.x
Kidner, C. A., and Martienssen, R. A. (2004). Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 428, 81–84. doi: 10.1038/nature02366
Kusaba, M., Ito, H., Morita, R., Iida, S., Sato, Y., Fujimoto, M., et al. (2007). Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell 19, 1362–1375. doi: 10.1105/tpc.106.042911
Li, D., Wang, L., Wang, M., Xu, Y. Y., Luo, W., Liu, Y. J., et al. (2009). Engineering OsBAK1 gene as a molecular tool to improve rice architecture for high yield. Plant Biotechnol. J. 7, 791–806. doi: 10.1111/j.1467-7652.2009.00444.x
Li, M., Xiong, G., Li, R., Cui, J., Tang, D., Zhang, B., et al. (2009). Rice cellulose synthase-like D4 is essential for normal cell-wall biosynthesis and plant growth. Plant J. 60, 1055–1069. doi: 10.1111/j.1365-313X.2009.04022.x
Li, W., Wu, C., Hu, G., Xing, L., Qian, W., Si, H., et al. (2013). Characterization and fine mapping of a novel rice narrow leaf mutant nal9. J. Integr. Plant Biol. 55, 1016–1025. doi: 10.1111/jipb.12098
Liu, X., Li, M., Liu, K., Tang, D., Sun, M., Li, Y., et al. (2016). Semi-Rolled Leaf2 modulates rice leaf rolling by regulating abaxial side cell differentiation. J. Exp. Bot. 67, 2139–2150. doi: 10.1093/jxb/erw029
Ma, L., Sang, X., Zhang, T., Yu, Z., Li, Y., Zhao, F., et al. (2017). Abnormal vascular bundles regulates cell proliferation and procambium cell establishment during aerial organ development in rice. New Phytol. 213, 275–286. doi: 10.1111/nph.14142
Mallory, A. C., Reinhart, B. J., Jones-Rhoades, M. W., Tang, G., Zamore, P. D., Barton, M. K., et al. (2004). MicroRNA control of PHABULOSA in leaf development: importance of pairing to the mircroRNA 5′region. EMBO J. 23, 3356–3364. doi: 10.1038/sj.emboj.7600340
McConnell, J. R., Emery, J., Eshed, Y., Bao, N., Bowman, J., and Barton, M. K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411, 709–713. doi: 10.1038/35079635
Micol, J. L. (2009). Leaf development: time to turn over a new leaf? Curr. Opin. Plant Biol. 12, 9–16. doi: 10.1016/j.pbi.2008.11.001
Moon, J., and Hake, S. (2011). How a leaf gets its shape. Curr. Opin. Plant Biol 14, 1–7. doi: 10.1016/j.pbi.2010.08.012
Morita, R., Sato, Y., Masuda, Y., Nishimura, M., and Kusaba, M. (2009). Defect in non-yellow coloring 3, an α/β hydrolase-fold family protein, causes a stay-green phenotype during leaf senescence in rice. Plant J. 59, 940–952. doi: 10.1111/j.1365-313X.2009.03919.x
Otsuga, D., DeGuzman, B., Prigge, M. J., Drews, G. N., and Clark, S. E. (2001). REVOLUTA regulates meristem initiation at lateral positions. Plant J. 25, 223–236. doi: 10.1046/j.1365-313x.2001.00959.x
Prigge, M. J., Otsuga, D., Alonso, J. M., Ecker, J. R., Drews, G. N., and Clark, S. E. (2005). Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17, 61–76. doi: 10.1105/tpc.104.026161
Qi, J., Qian, Q., Bu, Q., Li, S., Chen, Q., Sun, J., et al. (2008). Mutation of the rice narrow leaf1 gene, which encodes a novel protein, affects vein patterning and polar auxin transport. Plant Physiol. 147, 1947–1959. doi: 10.1104/pp.108.118778
Sakamoto, W. (2006). Protein degradation machineries in plastids. Annu. Rev. Plant Biol. 57, 599–621. doi: 10.1146/annurev.arplant.57.032905.105401
Sato, Y., Morita, R., Katsuma, S., Nishimura, M., Tanaka, A., and Kusaba, M. (2009). Two short-chain dehydrogenase/reductases, NON-YELLOW COLORING 1 and NYC1-LIKE, are required for chlorophyll b and light-harvesting complex II degradation during senescence in rice. Plant J. 57, 120–131. doi: 10.1111/j.1365-313X.2008.03670.x
Schirmer, E. C., Glover, J. R., Singer, M. A., and Lindquist, S. (1996). HSP100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem. Sci. 21, 289–296. doi: 10.1016/S0968-0004(96)10038-4
Shi, Z., Wang, J., Wan, X., Shen, G., Wang, X., and Zhang, J. (2007). Over-expression of rice OsAGO7 gene induces upward curling of the leaf blade that enhanced erect-leaf habit. Planta 226, 99–108. doi: 10.1007/s00425-006-0472-0
Shikanai, T., Shimizu, K., Ueda, K., Nishimura, Y., Kuroiwa, T., and Hashimoto, T. (2001). The chloroplast clpP gene, encoding a proteolytic subunit of ATP-dependent protease, is indispensable for chloroplast development in tobacco. Plant Cell Physiol. 42, 264–273. doi: 10.1093/pcp/pce031
Thomas, H., and Howarth, C. J. (2000). Five ways to stay green. J. Exp. Bot. 51, 329–337. doi: 10.1093/jexbot/51.suppl_1.329
Tsukaya, H. (2006). Mechanism of leaf-shape determination. Annu. Rev. Plant Biol. 57, 477–496. doi: 10.1146/annurev.arplant.57.032905.105320
Wang, J., Hu, J., Qian, Q., and Xue, H. W. (2013). LC2 and OsVIL2 promote rice flowering by photoperoid-induced epigenetic silencing of OsLF. Mol. Plant 6, 514–527. doi: 10.1093/mp/sss096
Wu, C., Fu, Y., Hu, G., Si, H., Cheng, S., and Liu, W. (2010). Isolation and characterization of a rice mutant with narrow and rolled leaves. Planta 232, 313–324. doi: 10.1007/s00425-010-1180-3
Wu, Z., Zhang, X., He, B., Diao, L., Sheng, S., Wang, J., et al. (2007). A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis. Plant Physiol. 145, 29–40. doi: 10.1104/pp.107.100321
Xiang, J.-J., Zhang, G.-H., Qian, Q., and Xue, H.-W. (2012). SEMI-ROLLED LEAF1 encodes a putative glycosylphosphatidylinositol-anchored protein and modulates rice leaf rolling by regulating the formation of bulliform cells. Plant Physiol. 159, 1488–1500. doi: 10.1104/pp.112.199968
Xu, Y., Wang, Y., Long, Q., Huang, J., Wang, Y., Zhou, K., et al. (2014). Overexpression of OsZHD1, a zinc finger homeodomain class homeobox transcription factor, induces abaxially curled and drooping leaf in rice. Planta 239, 803–816. doi: 10.1007/s00425-013-2009-7
Yan, S., Yan, C. J., Zeng, X. H., Yang, Y. C., Fang, Y. W., Tian, C. Y., et al. (2008). ROLLED LEAF 9, encoding a GARP protein, regulates the leaf abaxial cell fate in rice. Plant Mol. Biol. 68, 239–250. doi: 10.1007/s11103-008-9365-x
Yang, S. Q., Li, W. Q., Miao, H., Gan, P. F., Qiao, L., Chang, Y. L., et al. (2016). REL2, A gene encoding an unknown function protein which contains DUF630 and DUF632 domains controls leaf rolling in rice. Rice 9:37. doi: 10.1186/s12284-016-0105-6
Yoshikawa, T., Eiguchi, M., Hibara, K., Ito, J., and Nagato, Y. (2013). Rice SLENDER LEAF 1 gene encodes cellulose synthase-like D4 and is specifically expressed in M-phase cells to regulate cell proliferation. J. Exp. Bot. 64, 2049–2061. doi: 10.1093/jxb/ert060
Zhang, G. H., Xu, Q., Zhu, X. D., Qian, Q., and Xue, H. W. (2009). SHALLOT-LIKE1 is a KANADI transcription factor that modulates rice leaf rolling by regulating leaf abaxial cell development. Plant Cell 21, 719–735. doi: 10.1105/tpc.108.061457
Zhao, S., Zhao, L., Liu, F., Wu, Y., Zhu, Z., Sun, C., et al. (2016). NARROW AND ROLLED LEAF 2 regulates leaf shape, male fertility, and seed size in rice. J. Integr. Plant Biol. 58, 983–996. doi: 10.1111/jipb.12503
Zheng, B., MacDonald, T. M., Sutinen, S., Hurry, V., and Clarke, A. K. (2006). A nuclear-encoded ClpP subunit of the chloroplast ATP-dependent Clp protease is essential for early development in Arabidopsis thaliana. Planta 224, 1103–1115. doi: 10.1007/s00425-006-0292-2
Zou, L. P., Sun, X. H., Zhang, Z. G., Liu, P., Wu, J. X., Tian, C. J., et al. (2011). Leaf rolling controlled by the homeodomain leucine zipper class IV gene Roc5 in rice. Plant Physiol. 156, 1589–1602. doi: 10.1104/pp.111.176016
Keywords: nrl3, narrow and rolled leaf, leaf morphogenesis, chlorophyll accumulation, rice (Oryza sativa L.), improvement
Citation: Chen W, Sheng Z, Cai Y, Li Q, Wei X, Xie L, Jiao G, Shao G, Tang S, Wang J and Hu P (2019) Rice Morphogenesis and Chlorophyll Accumulation Is Regulated by the Protein Encoded by NRL3 and Its Interaction With NAL9. Front. Plant Sci. 10:175. doi: 10.3389/fpls.2019.00175
Received: 25 February 2018; Accepted: 04 February 2019;
Published: 19 February 2019.
Edited by:
Antonio Ferrante, University of Milan, ItalyReviewed by:
Xuan Tran Dang, Hiroshima University, JapanZhiguo Zhang, Biotechnology Research Institute (CAAS), China
Copyright © 2019 Chen, Sheng, Cai, Li, Wei, Xie, Jiao, Shao, Tang, Wang and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianlong Wang, d2psOTY3OEAxMjYuY29t Peisong Hu, cGVpc29uZ2h1QDEyNi5jb20=
†These authors have contributed equally to this work
 Wei Chen1,2†
Wei Chen1,2† Peisong Hu
Peisong Hu