- 1CAS Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China
- 2School of Pharmaceutical Sciences and Yunnan Key Laboratory of Pharmacology for Natural Products, Kunming Medical University, Kunming, China
Ligustici Rhizoma et Radix (LReR), an important Chinese medicine known as “Gao-ben,” refers to Ligusticum sinense Oliv. or Ligusticum jeholense Nakai et Kitag. However, a number of other species are commonly sold as “Gao-ben” in the herbal medicine market, which may result in a series of quality control problems and inconsistent therapeutic effects. The “Gao-ben” is commonly sold sliced and dried, making traditional identification methods difficult. Here, the mini barcode ITS2 region was examined on 68 samples representing LReR and 7 potential adulterant or substitute species. The results showed 100% success rates of PCR and sequencing and the existence of a barcoding gap. The neighbor-joining (NJ) tree indicated that all the tested samples could be exactly identified. The ITS2 secondary structure revealed a clear difference between true “Gao-ben” and three adulterant species. We therefore recommend the use of ITS2 as a mini barcode for distinguishing between closely or distantly related plant species that may be used in Chinese medicine.
Introduction
Apiaceae, the 16th-largest flowering plant family, comprises more than 3,540 species in 446 genera (Mabberley, 1997). It is a well-known and economically important plant family in medicine, spices, vegetables and ornamental gardening. The roots and rhizomes of Ligusticum sinense Oliv. or Ligusticum jeholense Nakai et Kitag form a widely used traditional Chinese medicine, known as “Ligustici Rhizoma et Radix” (LReR), or “Gao-ben” in Chinese (Commission, 2015). It is commonly used to treat colds, trapped wind, headaches and rheumatic arthralgia (Commission, 2015). It has been reported to exhibit many beneficial properties such as analgesic, antipyretic, anti-inflammatory and anticonvulsive activities, plus antimicrobial and antioxidant effects (Wang et al., 2011). As a result, this herb has attracted more and more attention in the medical field, and have been widely used in clinical therapies.
However, LReR is easily confused with other herbs, causing potential mistakes in treatment. Certain species that are closely related or morphologically similar to LReR are frequently used as local remedies in various regions due to geographical and historical factors. For instance, Meeboldia yunnanensis (H. Wolff) Constance & F. T. Pu, Ligusticum delavayi Franchet and Ligusticum pteridophyllum Franchet are often used in folk medicine in southwestern China, but their function and efficacy are not quite the same (Li et al., 2001). Additionally, Conioselinum vaginatum (Spreng.) Thell., Ligusticum tenuissimum (Nakai) Kitagawa, Sium suave Walter and Ligusticum acuminatum Franch. are also sold as “Gao-ben” in medicinal markets. Using morphology-based or chemical identification methods to identify Rhizoma et Radix herbs is difficult, especially when the plant is sliced and dried, as they often are for selling (Hon et al., 2003; Joshi et al., 2004; Xue et al., 2006; Mishra et al., 2016). Therefore, a simple, inexpensive and effective method for distinguishing between the above species is urgently needed.
DNA barcoding is a new taxonomic method that uses one or a few short, standard genomic DNA region(s) for rapid, reliable and effective species identification (Floyd et al., 2002; Hebert et al., 2003). For identifying traditional herbal medicines, however, the commonly used three-barcode system has not always been effective, because matK and rbcL are difficult to amplify, especially from powdered products where DNA degradation is very common. The internal transcribed spacer 2 (ITS2) region of nuclear ribosomal DNA might be a promising standardized region to barcode medicinal plants, due to its relatively short length, consistent performance in distinguishing closely related species, and ease of amplification with a single set of universal primers (Chen et al., 2010). In cells, ITS2 has conserved nucleotide motifs which play a role in forming a three dimensional structure, and hence transformation into a functional complex before conversion to a mature rRNA (Hall, 1999; Lalev and Nazar, 1999). Compared with previous sequence alignments based only on nucleotide similarity, the ITS2 conserved nucleotide motifs permit multiple sequence alignments, from which a more homologous overall alignment can be generated (Kjer, 1995; Coleman and Mai, 1997). Additionally, the secondary structure is maintained by the mutual between base-pairs that are canonical (GC, AU), non-canonical stable (GU) and unstable (AC) (Elgavish et al., 2001; Leontis and Westhof, 2001). Theses paired and unpaired ITS2 structures provide extra molecular morphological features that can greatly improve taxonomic classification (Telford et al., 2005; Keller et al., 2010).
In this study, we use ITS2 sequence and secondary structure information to determine whether genuine “Gao-ben” can be distinguished from the most commonly used adulterants and substitutes. We also discuss the possibility that ITS2 might play a regulatory role in the herbal medicine market in the future.
Materials and Methods
Plant Materials
A total of 43 samples belonging to eight species, covering the two original species of LReR (L. sinense and L. jeholense) and seven potential substitutes (L. tenuissimum, L. pteridophyllum, L. acuminatum, S. suave, M. yunnanensis, and C. vaginatum), were collected from fields. Voucher specimens were deposited in the Kunming Institute of Botany, Chinese Academy of Sciences (KUN), and Chengdu Institute of Biology, Chinese Academy of Sciences (CDBI) (Supplementary Table S1). To determine possible infraspecific molecular variation, each species was represented by at least two individuals. Furthermore, 25 commercial “Gao-ben” products were purchased from different medicinal markets in China (Supplementary Table S1).
Laboratory Protocols
Before DNA extraction, the surface of the medicinal materials was wiped with 75% ethanol, then ground into powder with a grinder. Genomic DNA was extracted using the modified CTAB procedure of Doyle and Doyle (1987). The primers BEL-2 (5′-GATGCGGAGATTGGCCCCCCGTGC -3′) and BEL-3 (5′-GACGCTTCTCCAGACTACAAT -3′) were used to amplify the complete ITS2 region (Chiou et al., 2007). The PCR parameters were as follows: Initial denaturation for 3 min at 94°C, followed by 36 cycles of denaturation (94°, 45 s), annealing (55°C, 1 min) and extension (72°C, 2 min), and a final extension for 7 min at 72°C. Purified PCR products were sequenced in both directions with the primers used for PCR amplification on an ABI 3730 automated sequencer (Applied Biosystems, Foster City, CA, United States) in Sangon Biotech Corporation (Shanghai, China).
Data Analysis
Newly generated sequences were initially edited and assembled using SeqMan of the DNASTAR 5.01 software package (DNASTAR, Inc., Madison, United States). The ITS2 region was annotated using the Hidden Markov Model (HMM) (Keller et al., 2009) to delete the conserved 5.8 and 28S sections (Koetschan et al., 2012). ITS2 secondary structures of all investigated taxa were predicted by homology modeling in the ITS2 Database (identity matrix and 75% threshold for the helix transfer1) (Koetschan et al., 2012). This method usually results in several alternative folding patterns for the same ITS2 sequence. The true folding pattern corresponds to the secondary structure model of Mai and Coleman, and was well supported by compensatory base changes (CBCs) and hemi compensatory base changes (hCBCs) revealed by comparisons among related taxa (Mai and Coleman, 1997). Sequences with homologous structure were automatically and synchronously aligned using 4SALE 1.7 (Seibel et al., 2006, 2008). Genetic distances were calculated according to the kimura-2-parameter (K2P) model using MEGA 7.0 software (Kumar et al., 2016). A neighbor-joining (NJ) tree was constructed and bootstrap tests were performed using 1000 replicates to separate the sampled species via MEGA 7.0 (Kumar et al., 2016). CBCs are substitutions in two positions that retain pairing, i.e., G = C ↔ C = G, A = U ↔ U = A. The proposed ITS2 secondary structure was examined for CBCs with the CBCAnalyzer option (Wolf et al., 2005) implemented in 4SALE, whereas hCBCs (pair ↔ non-pair, i.e., G = C ↔ G = U) were observed manually.
Results
Amplification, Sequencing, and Sequences Characteristics
The success rate of the ITS2 PCR amplification and sequencing was 100% (Table 1). All high-quality sequences were submitted to GenBank (Table 1). The ITS2 sequence showed minor length variation across all samples, ranging from 220 bp (M. yunnanensis) to 226 (S. suave). The length of 15 L. sinense and L. jeholense individuals was 222 bp, and the average GC content was 54.9%. The ITS2 sequence lengths of L. acuminatum, L. delavayi, L. pteridophyllum, L. tenuissimum, C. vaginatum, M. yunnanensis, and S. suave were 223, 223, 223, 223, 222, 220, and 226 bp, respectively. The corresponding average GC content of these adulterants varied from 53.2 to 57.9%. The aligned length of 227 bp exhibited 84 variable sites, a rate of 37.0% (Table 1). Therefore, the ITS2 sequences for the sampled species were relatively variable.
Intra/Interspecific Distance, Barcoding Gap, and NJ Tree
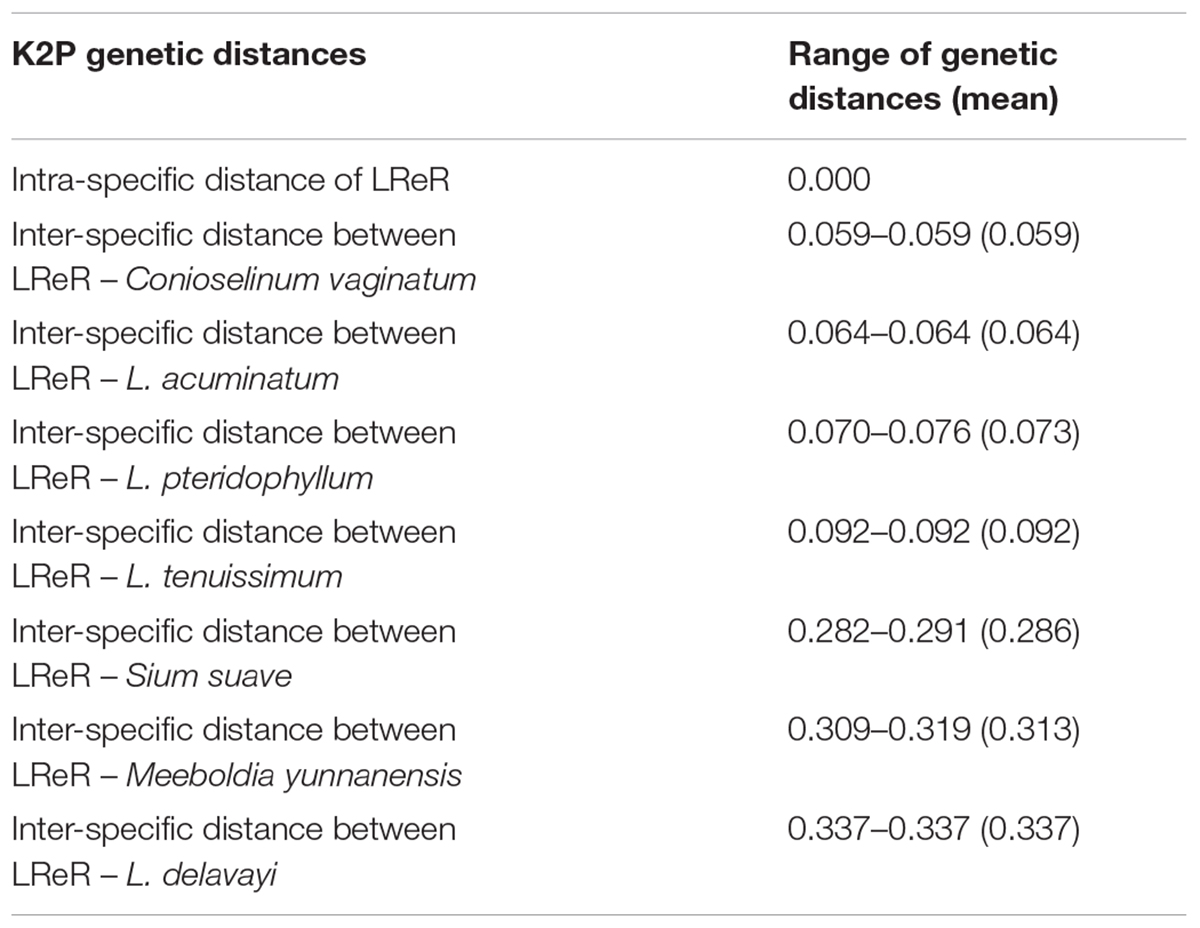
All individuals of L. sinense and L. jeholense were identical for ITS2, sharing a single haplotype. According to the K2P model, the average interspecific distance between these and the adulterant species was 0.175, with the maximum interspecific distance being 0.337 from L. delavayi, and the minimum being 0.059 from C. vaginatum (Table 2 and Figure 1A). The NJ tree showed that 15 individual samples were determined to be L. jeholense and L. sinense clustered together in a highly supported clade (Bootstrap value = 99) that was separated from the commonly used substitutes and adulterants such as L. pteridophyllum, L. acuminatum, L. tenuissimum, C. vaginatum, S. suave, M. yunnanensis, and L. delavayi (Figure 2A). There was therefore a high interspecific variation and an obvious barcoding gap was noted (Figure 1B).
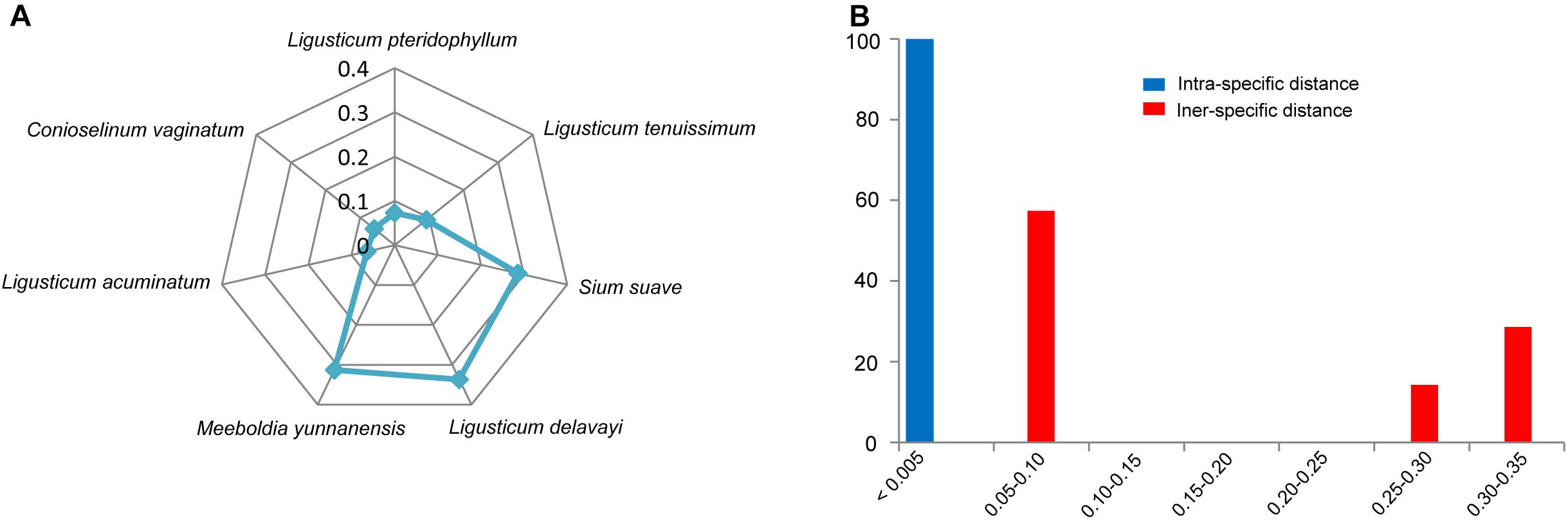
Figure 1. (A) Genetic distances from LReR to its adulterants and substitutes. (B) Relative distribution of interspecific divergence between LReR and its adulterants and substitutes and intraspecific variation in the ITS2 region using K2P genetic distance.
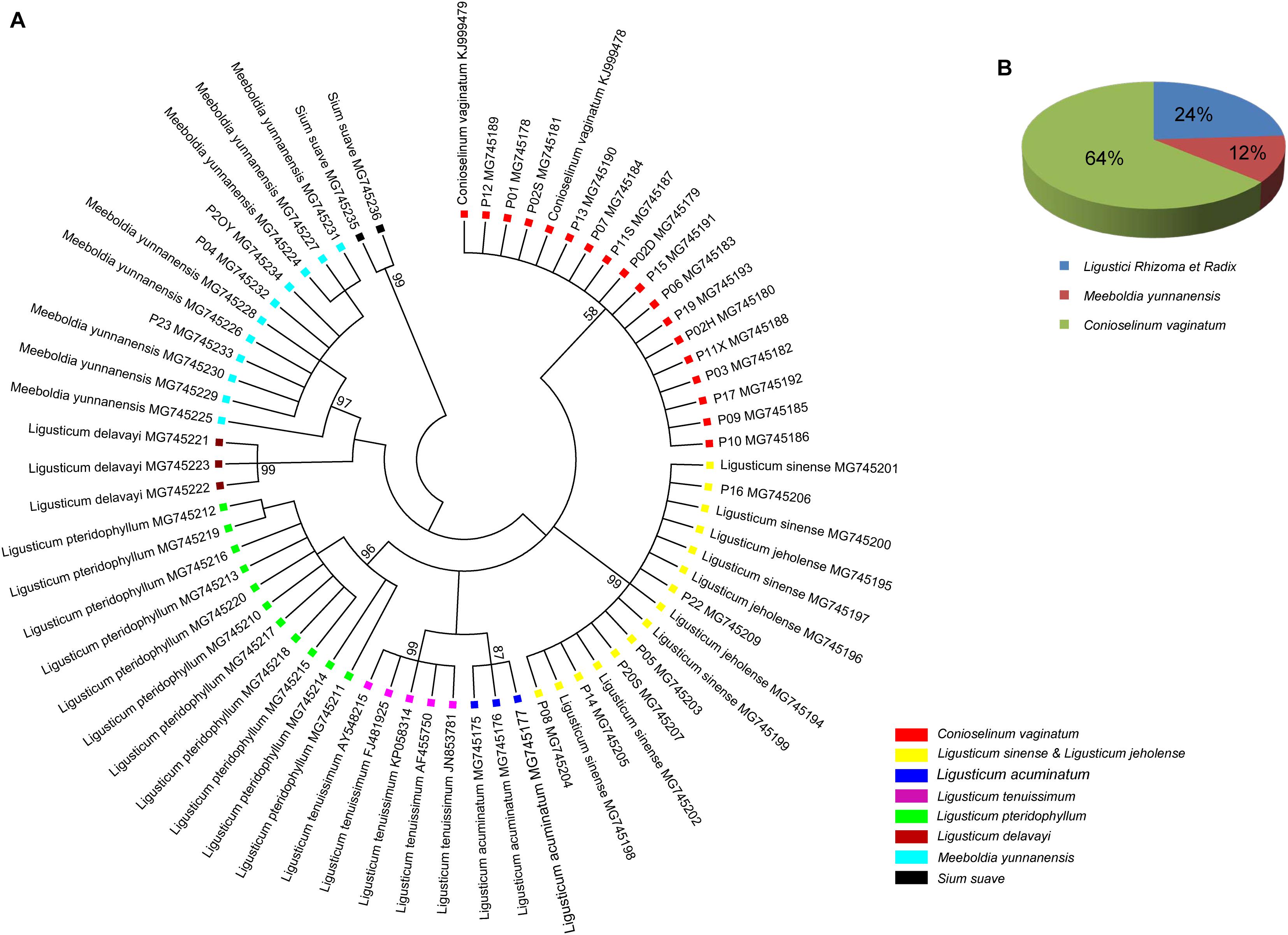
Figure 2. (A) NJ tree of LReR and its adulterants and substitutes. Bootstrap scores (1000) are shown (≥50%) for each branch. (B) The percentage of the commercial “Gao-ben” products identified by barcode ITS2 in this study.
Of the 25 commercial “Gao-ben” products, six samples were identified as LReR, accounting for 24%, while the remainder were identified as M. yunnanensis (12%), and C. vaginatum (64%) (Figure 2B).
Analysis of the Secondary Structure
ITS2 secondary structures obtained for all species examined fold into the common core structure known for eukaryotes, made up of four helices, the third being the longest (Supplementary Figures S1A–H; Mai and Coleman, 1997; Coleman, 2003, 2007, 2015; Schultz et al., 2005). Figure 3 visualizes a 51% consensus structure. Sequence motifs include a U-U mismatch in helix II, an A-rich conserved spacer between helices II and III, and a UGGU motif 5′ side to the apex of helix III (Figure 3). In comparable portions of the secondary structure, most CBCs, and hCBCs observed between LReR and its adulterants are in helices III, I, and II, with a few in helix IV (Table 3). L. sinense, L. jeholense, L. tenuissimum, L. acuminatum, C. vaginatum, and L. pteridophyllum form a group without any CBC in conserved ITS2 regions (i.e., in helices II and III), from which the group of L. delavayi, M. yunnanensis, and S. Suave may be distinguished by the presence of at least one CBC in these regions.
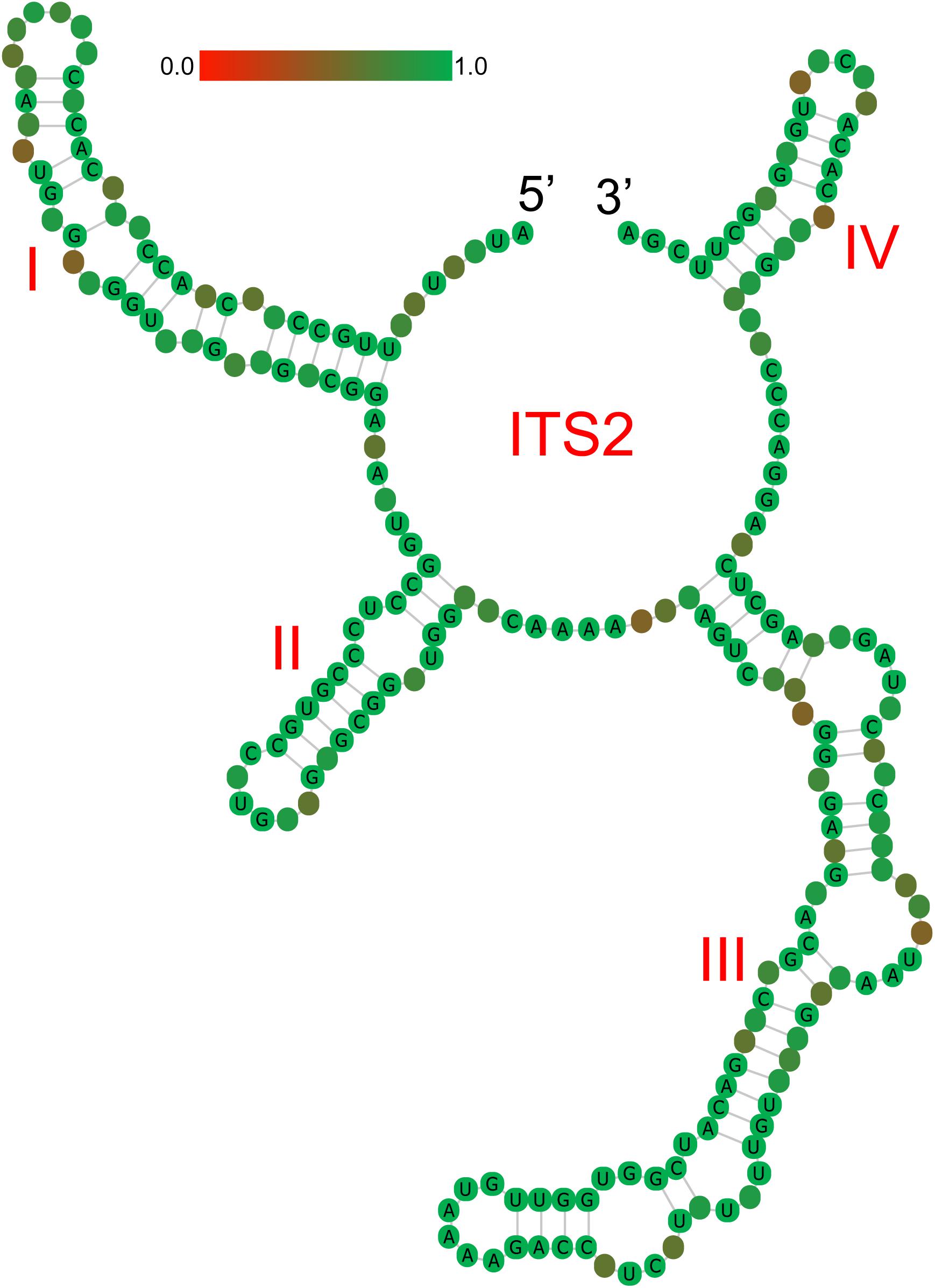
Figure 3. Consensus secondary structure (51%) for all samples without gaps. Helices are numbered I–IV. Sequence conservation is indicated from red/brownish (not conserved) to green (conserved).
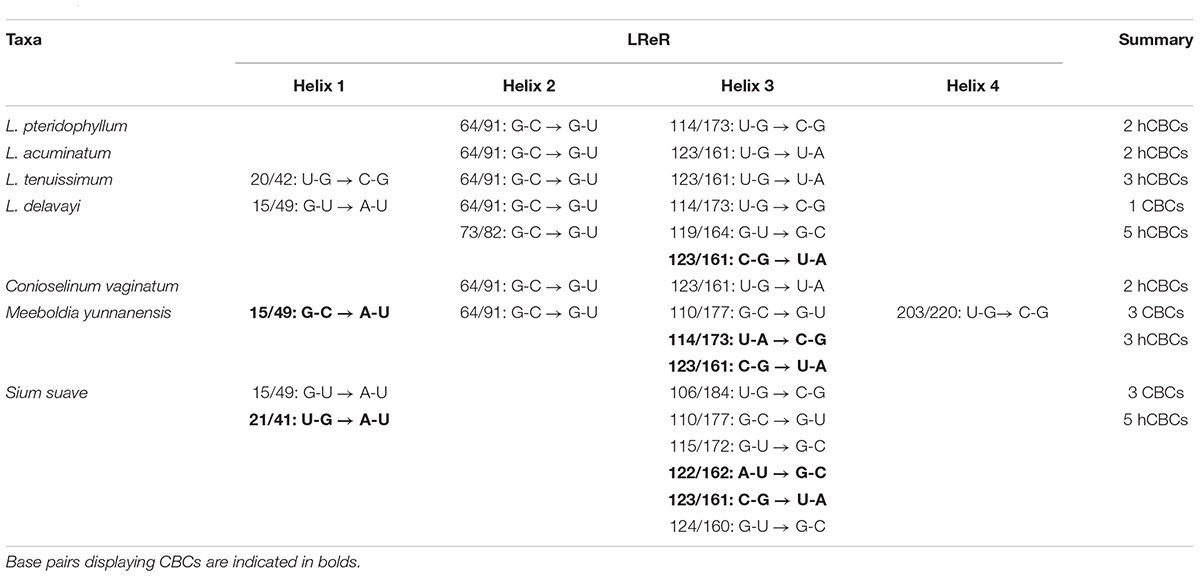
Table 3. Occurrence/frequency of CBCs and hCBCs between LReR (Ligusticum jeholense/L. sinense) and its adulterants and substitutes.
Discussion
Identification Capability of ITS2 for LReR
In Chinese Pharmacopeia, only L. sinense and L. jeholense are listed as LReR for their very similar chemical compositions and the near identical use and efficacy (Commission, 2015). Morphologically, both species share many similar characters, e.g., ternate-2- or 3-pinnate blade, ultimate segment margins irregularly serrate, and long and reflexed styles. Geographically, L. jeholense is in northern China, while L. sinense occurs more widely, but does not overlap with L. jeholense (Sheh and Watson, 2005). Both our present ITS2 (Table 2) and unpublished data including psbA-trnH, matK, and rbcL show no interspecific variation between these two species. Therefore, we consider that L. sinense and L. jeholense are close relatives, or the latter represents a vicariant geographical element.
According to the criterion proposed by Coleman (2003) and coworkers, presence/absence of even a single CBC in the conserved areas of helices II and III of ITS2 is associated with incompatibility/inability to hybridize, thus establishing the boundaries between biological species and populations. In contrast, hCBCs in the conserved parts as well as changes in the less conserved regions (e.g., in helices I and IV) do not correlate with interbreeding ability. LReR can be distinguished from L. delavayi, M. yunnanensis, and S. suave by at least one CBC in the conserved ITS2 regions (i.e., in helices II and III) (Table 3 and Supplementary Figures S1F–H). M. yunnanensis and L. delavayi are widely used as “Huang Gao-ben” in Yunnan Province, whereas S. suave has also been sold as “Gao-ben” in medicinal markets. It is difficult to distinguish them from LReR when they are dried, sliced, and shredded, but our ITS2 analysis indicated that they are phylogenetically distant species (Figures 1A, 2A and Table 2). Moreover, these three species can be distinguished from the true LReR by the presence of at least one CBC in the conserved ITS2 regions (i.e., in helices II and III) (Table 3). Results from epidermal analysis (Zhou and Liu, 2018), together with that from cytological evidence (Zhou et al., 2008) and molecular phylogenetics (Zhou et al., 2009), indicated that these are not close relatives of L. sinense. We therefore recommend that L. delavayi, M. yunnanensis, and S. Suave should be marketed under their original herbal medicinal name.
Other genetically close relatives of the LReR plant group include L. pteridophyllum, L. tenuissimum, L. acuminatum, and C. vaginatum (Figures 1A, 2A and Table 2). L. pteridophyllum is another herbal medicine widely used in Yunnan in the name of “Hei Gao-ben” and has been regarded as an adulterant of Peucedanum praeruptorum Dunn (Rao et al., 1995). In our analysis, all accessions of L. pteridophyllum comprised a strongly supported clade, having a sequence divergence value of 0.073 with LReR (Figures 1A, 2A and Table 2). According to Ye et al. (2004) the chemical composition of L. pteridophyllum is similar to LReR, so, considering it as a regional substitute seems to be reasonable. L. acuminatum had a sequence divergence value of 0.064 from LReR, for which it is used as a regional substitute in western Sichuan (Sheh and Watson, 2005), so further research is needed to determine whether it can be regarded as an effective “Gao-ben” substitute.
Conioselinum vaginatum, known as “Xinjiang Gao-ben,” is found mainly in the Tian and Altai mountains of Xinjiang and western Junggar mountains in central Asia and western Siberia (Sheh and Watson, 2005), and is widely cultivated. Chemical analysis showed that C. vaginatum contains 16 compounds, including ligustilide, ferulic acid, and myristic ether, which are the same as in L. jeholense (Li, 2013); however, its pharmacological action remains controversial (Dai, 1988; Li et al., 2013). Given that the annual demand for LReR exceeds 3500 tons, which exceeds the natural production capacity (Ding, 2010), research on the pharmacological efficacy of C. vaginatum is urgently needed, to determine whether C. vaginatum could be an effective substitute.
Potential Application of ITS2 in the Authentication of Medicinal Materials
The trade in crude drugs has surged globally, generating annual revenues over US $60 billion (Newmaster et al., 2013). There are strong financial incentives for dishonest merchants to use adulterants and substitutes intentionally, leading to treatments not working as advertised, and posing serious risks to the health of consumers (Newmaster et al., 2013). Accurate and rapid species authentication is the best way to combat this. Traditional methods usually require taxonomists, who are few in number, and moreover a fairly complete specimen, meaning they will not work on plant fragments sold as. The ITS2 barcode presented here solves this problem. The ITS2 is region is easy to amplify and sequence, has a short length, and reveals high interspecific variation (Chiou et al., 2007; China Plant Bol Group et al., 2011). While the ITS2 nucleotide sequences evolve quickly, their secondary structures are maintained by certain conserved motifs (Hershkovitz and Zimmer, 1996), which are very useful for sequence alignment (Mai and Coleman, 1997), especially when possible species are spread across many families, as is the case for some Traditional Chinese Medicines (Zhang et al., 2015). Meanwhile, the secondary structures of ITS2 can provide additional molecular morphological characteristics for better species discrimination (Grajales et al., 2007; Gu et al., 2013). Much research has been carried out using ITS2 to regulate the herbal medicine market (Zhang et al., 2015, 2016; Zhao et al., 2015; Zhu et al., 2017), and the current paper shows its effectiveness for LReR or “Gao-ben.” Twenty-five commercial “Gao-ben” samples fell into three clades, corresponding to L. sinense + L. jeholense, C. vaginatum, and M. yunnanensis (Figure 2A). Surprisingly, more than half of the samples were derived from C. vaginatum. As mentioned above, whether C. vaginatum can be substituted for genuine “Gao-ben” is still controversial. Although M. yunnanensis is mainly cultivated and used in Yunnan province in the name of “Huang Gao-ben,” its adulteration or substitution can cause confusion in identification and in therapeutic efficacy.
ITS2 has been proved to vary in sequence and secondary structure in a way that highly correlates with species taxonomy. Müller et al. (2007) compared the ITS2 secondary structure of 1373 species to their nearest relatives, and observed that in 93% of cases, if two taxa are different somewhere in their ITS2 by one CBC, they would be classified as different species. This criterion has been less commonly used for herbal medicine authentication. However, in our study, presence of a CBC distinguishes genuine “Gao-ben” from three other species, i.e., L. delavayi, M. yunnanensis, and S. suave, at least the last of which is sometimes sold as “Gao-ben.” So, as a rapid, inexpensive, and informative DNA barcode, ITS2 could be widely used to regulate the herbal medicine market.
Conclusion
Traditional Chinese medicine is vulnerable to the replacement of the correct and most effective species with others that may be closely or distantly related. ITS2 could be an ideal candidate marker for authentication from both divergences of primary sequences and variations in secondary structures. This method is suitable for the identification of raw medicinal materials, but it is unsuitable for the authentication of heavily processed materials in which DNA degradation frequently occurs. A promising direction suitable for authentication of degraded material would be to combine the next generation sequencing (NGS)-based and species-specific PCR based methods (such as nucleotide signatures). During the process, knowing the secondary structure of ITS2 can help to locate the positions of the short motifs, that is well conserved within the species and develop nucleotide signatures.
Author Contributions
Z-wL, Y-zG, and JZ collected the samples and carried out the experiments. Z-wL and JZ analyzed the data, conceived and designed the study, and wrote the manuscript. All authors have read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 31460052) and the United Research Foundation of Yunnan Science and Technology Department-Kunming Medical University (No. 2015FB014).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Richard I. Milne from The University of Edinburgh and David E. Boufford from Harvard University Herbaria for language polishing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00429/full#supplementary-material
FIGURE S1 | Secondary structure of ITS2 in Ligustici Rhizoma et Radix (LReR) and its adulterants and substitutes. (A) LReR, (B) L. acuminatum, (C) L. pteridophyllum, (D) L. tenuissimum, (E) Conioselinum vaginatum, (F) L. delavayi, (G) Sium suave, and (H) Meeboldia yunnanensis. Red and blue arrows show the site of the compensatory base changes (CBCs) and hemi compensatory base changes (hCBCs) between LReR and its adulterants and substitutes, respectively.
TABLE S1 | Detailed information of samples used in this study.
Footnotes
References
Chen, S., Yao, H., Han, J., Liu, C., Song, J., Shi, L., et al. (2010). Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS One 5:e8613. doi: 10.1371/journal.pone.0008613
China Plant Bol Group, Li, D. Z., Gao, L. M., Li, H. T., Wang, H., Ge, X. J., et al. (2011). Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc. Natl. Acad. Sci. U.S.A. 108, 19641–19646. doi: 10.1073/pnas.1104551108
Chiou, S. J., Yen, J. H., Fang, C. L., Chen, H. L., and Lin, T. Y. (2007). Authentication of medicinal herbs using PCR-amplified ITS2 with specific primers. Planta Med. 73, 1421–1426. doi: 10.1055/s-2007-990227
Coleman, A. W. (2003). ITS2 is a double-edged tool for eukaryote evolutionary comparisons. Trends Genet. 19, 370–375. doi: 10.1016/S0168-9525(03)00118-5
Coleman, A. W. (2007). Pan-eukaryote ITS2 homologies revealed by RNA secondary structure. Nucleic Acids Res. 35, 3322–3329. doi: 10.1093/nar/gkm233
Coleman, A. W. (2015). Nuclear rRNA transcript processing versus internal transcribed spacer secondary structure. Trends Genet. 31, 157–163. doi: 10.1016/j.tig.2015.01.002
Coleman, A. W., and Mai, J. C. (1997). Ribosomal DNA ITS-1 and ITS-2 sequence comparisons as a tool for predicting genetic relatedness. J. Mol. Evol. 45, 168–177. doi: 10.1007/PL00006217
Commission, S. P. (2015). Pharmacopoeia of the People’s Republic of China Part I. Beijing: China Medical Science and Technology Press.
Dai, B. (1988). Comparison of chemical constituents of essential oil from four species of Gaoben (Ligusticum) by GC-MS analysis. Acta Pharm. Sin. 23, 361–369.
Ding, L. W. (2010). The depletion price of Ligusticum resources has gone up all the way. Modern Chin. Med. 12, 46–48.
Doyle, J. J., and Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf issue. Phytochem. Bull. 19, 11–15.
Elgavish, T., Cannone, J. J., Lee, J. C., Harvey, S. C., and Gutell, R. R. (2001). AA.AG@Helix. ends: A:A and A:G base-pairs at the ends of 16S and 23S rRNA helices. J. Mol. Biol. 310, 735–753. doi: 10.1006/jmbi.2001.4807
Floyd, R., Abebe, E., Papert, A., and Blaxter, M. (2002). Molecular barcodes for soil nematode identification. Mol. Ecol. 11, 839–850. doi: 10.1046/j.1365-294X.2002.01485.x
Grajales, A., Aguilar, C., and Sanchez, J. A. (2007). ). Phylogenetic reconstruction using secondary structures of internal transcribed spacer 2 (ITS2, rDNA): finding the molecular and morphological gap in Caribbean gorgonian corals. BMC Evol. Biol. 7:90. doi: 10.1186/1471-2148-7-90
Gu, W., Song, J., Cao, Y., Sun, Q., Yao, H., Wu, Q., et al. (2013). Application of the ITS2 region for barcoding medicinal plants of selaginellaceae in pteridophyta. PLoS One 8:e67818. doi: 10.1371/journal.pone.0067818
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Sym. Ser. 41, 95–98.
Hebert, P. D. N., Cywinska, A., Ball, S. L., and Dewaard, J. R. (2003). Biological identifications through DNA barcodes. Proc. Biol. Sci. 270, 313–321. doi: 10.1098/rspb.2002.2218
Hershkovitz, M. A., and Zimmer, E. A. (1996). Conservation patterns in angiosperm rDNA ITS2 sequences. Nucleic Acids Res. 24, 2857–2867. doi: 10.1093/nar/24.15.2857
Hon, C. C., Chow, Y. C., Zeng, F. Y., and Leung, F. C. C. (2003). Genetic authentication of ginseng and other traditional Chinese medicine. Acta Pharm. Sin. 24, 841–846.
Joshi, K., Chavan, P., Warude, D., and Patwardhan, B. (2004). Molecular markers in herbal drug technology. Curr. Sci. 87, 159–165.
Keller, A., Förster, F., Müller, T., Dandekar, T., Schultz, J., and Wolf, M. (2010). Including RNA secondary structures improves accuracy and robustness in reconstruction of phylogenetic trees. Biol. Direct 5:4. doi: 10.1186/1745-6150-5-4
Keller, A., Schleicher, T., Schultz, J., Müller, T., Dandekar, T., and Wolf, M. (2009). 5.8S-28S rRNA interaction and HMM-based ITS2 annotation. Gene 430, 50–57. doi: 10.1016/j.gene.2008.10.012
Kjer, K. M. (1995). Use of rRNA secondary structure in phylogenetic studies to identify homologous positions: an example of alignment and data presentation from the frogs. Mol. Phylogenet. Evol. 4, 314–330. doi: 10.1006/mpev.1995.1028
Koetschan, C., Hackl, T., Müller, T., Wolf, M., Förster, F., and Schultz, J. (2012). ITS2 database IV: interactive taxon sampling for internal transcribed spacer 2 based phylogenies. Mol. Phylogenet. Evol. 63, 585–588. doi: 10.1016/j.ympev.2012.01.026
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lalev, A. I., and Nazar, R. N. (1999). Structural equivalence in the transcribed spacers of pre-rRNA transcripts in Schizosaccharomyces pombe. Nucleic Acids Res. 27, 3071–3078. doi: 10.1093/nar/27.15.3071
Leontis, N. B., and Westhof, E. (2001). Geometric nomenclature and classification of RNA base pairs. RNA 7, 499–512. doi: 10.1017/S1355838201002515
Li, X. B. (2013). Study on the Chemical Constituents of Conioselinui Rhizoma et Radix and Quality Evaluation of “Gaoben” Herbs. [Master/ dissertation].Peking Union Medical College: [Beijing]
Li, X. B., Qi, Y. D., Liu, H. T., Zhang, J., Zhang, Z., Zhang, B. G., et al. (2013). Chemical constituents of Conioselinum vaginatum. Chin. J. Chin. Mater. Med. 38, 1543–1547.
Li, Y. S., Ye, X. W., Liao, X. R., Zhang, M., and Wang, Z. T. (2001). Textual studies of TCM gaoben (Ligusticum L.) habitually prescribing in Yunnan province. Chin. Tradit. Herbal Drugs 32, 257–259.
Mabberley, D. J. (1997). THE PLANT-BOOK: A Portable Dictionary Of The Vascular Plants (2nf Edn). Cambridge: Cambridge University Press.
Mai, J. C., and Coleman, A. W. (1997). The internal transcribed spacer 2 exhibits a common secondary structure in green algae and flowering plants. J. Mol. Evol. 44, 258–271. doi: 10.1007/PL00006143
Mishra, P., Kumar, A., Nagireddy, A., Mani, D. N., Shukla, A. K., Tiwari, R., et al. (2016). DNA barcoding: an efficient tool to overcome authentication challenges in the herbal market. Plant Biotechnol. J. 14, 8–21. doi: 10.1111/pbi.12419
Müller, T., Philippi, N., Dandekar, T., Schultz, J., and Wolf, M. (2007). Distinguishing species. RNA 13, 1469–1472. doi: 10.1261/rna.617107
Newmaster, S. G., Grguric, M., Shanmughanandhan, D., Ramalingam, S., and Ragupathy, S. (2013). DNA barcoding detects contamination and substitution in North American herbal products. BMC Med. 11:222. doi: 10.1186/1741-7015-11-222
Rao, G. X., Liu, Q. X., Dai, Z. J., Yang, Q., and Dai, W. S. (1995). Textual research for the traditional Chinese medicine radix peucedani and discussion of its modern varieties. J. Yunnan Univ. Tradit. Chin. Med. 18, 1–6.
Schultz, J., Maisel, S., Gerlach, D., Müller, T., and Wolf, M. (2005). A common core of secondary structure of the internal transcribed spacer 2 (ITS2) throughout the Eukaryota. RNA 11, 361–364. doi: 10.1261/rna.7204505
Seibel, P. N., Müller, T., and Dandekar, T. (2008). Synchronous visual analysis and editing of RNA sequence and secondary structure alignments using 4SALE. BMC Res. Notes 1:91. doi: 10.1186/1756-0500-1-91
Seibel, P. N., Muller, T., Dandekar, T., Schultz, J., and Wolf, M. (2006). 4SALE - A tool for synchronous RNA sequence and secondary structure alignment and editing. BMC Bioinform. 7:498. doi: 10.1186/1471-2105-7-498
Sheh, M. L., and Watson, M. F. (2005). “Apiaceae lindley,” in Flora of China, eds Z. Y. Wu and P. H. Raven (Beijing: Science Press), 1–205.
Telford, M. J., Wise, M. J., and Gowri-Shankar, V. (2005). Consideration of RNA secondary structure significantly improves likelihood-based estimates of phylogeny: examples from the bilateria. Mol. Biol. Evol. 22, 1129–1136. doi: 10.1093/molbev/msi099
Wang, J. H., Xu, L., Yang, L., Liu, Z. L., and Zhou, L. G. (2011). Composition, antibacterial and antioxidant activities of essential oils from Ligusticum sinense and L. jeholense (Umbelliferae) from China. Rec. Nat. Prod. 5, 314–318.
Wolf, M., Friedrich, J., Dandekar, T., and Müller, T. (2005). CBCAnalyzer: inferring phylogenies based on compensatory base changes in RNA secondary structures. In Silico Biol. 5, 291–294.
Xue, C. Y., Li, D. Z., Lu, J. M., Yang, J. B., and Liu, J. Q. (2006). Molecular authentication of the traditional Tibetan medicinal plant Swertia mussotii. Planta Med. 72, 1223–1226. doi: 10.1055/s-2006-951695
Ye, X. W., Zhao, Q., Li, Y. S., Liao, X. R., and Xue, Y. M. (2004). Analysis on chemical constituents of volatile oil from Heigaoben- Ligusticum pteridophyllum Franch. J. Yunnan Univ. Tradit. Chin. Med. 27, 40–41.
Zhang, W., Yang, S., Zhao, H., and Huang, L. Q. (2016). Using the ITS2 sequence-structure as a DNA mini-barcode: a case study in authenticating the traditional medicine “Fang Feng”. Biochem. Syst. Ecol. 69, 188–194. doi: 10.1038/s41598-017-09769-y
Zhang, W., Yuan, Y., Yang, S., Huang, J. J., and Huang, L. Q. (2015). ITS2 secondary structure improves discrimination between medicinal “Mu Tong” species when using DNA barcoding. PLoS One 10:e0131185. doi: 10.1371/journal.pone.0131185
Zhao, S., Chen, X., Song, J., Pang, X., and Chen, S. (2015). Internal transcribed spacer 2 barcode: a good tool for identifying Acanthopanacis cortex. Front. Plant Sci. 6:840. doi: 10.3389/fpls.2015.00840
Zhou, J., Gong, X., Downie, S. R., and Peng, H. (2009). Towards a more robust molecular phylogeny of Chinese Apiaceae subfamily Apioideae: additional evidence from nrDNA ITS and cpDNA intron (rpl16 and rps16) sequences. Mol. Phylogenet. Evol. 53, 56–68. doi: 10.1016/j.ympev.2009.05.029
Zhou, J., and Liu, Z. W. (2018). Comparative morphology of the leaf epidermis in Ligusticum (Apiaceae) from china. Am. J. Plant Sci. 9, 1105–1123. doi: 10.4236/ajps.2018.96084
Zhou, J., Pu, F. D., Peng, H. J., Pan, Y. Z., and Gong, X. (2008). Karyological studies of ten Ligusticum species (Apiaceae) from the hengduan mountains region of china. Caryologia 61, 333–341. doi: 10.1080/00087114.2008.10589644
Keywords: apiaceae, DNA barcoding, Ligustici Rhizoma et Radix, ITS2, secondary structures
Citation: Liu Z-w, Gao Y-z and Zhou J (2019) Molecular Authentication of the Medicinal Species of Ligusticum (Ligustici Rhizoma et Radix, “Gao-ben”) by Integrating Non-coding Internal Transcribed Spacer 2 (ITS2) and Its Secondary Structure. Front. Plant Sci. 10:429. doi: 10.3389/fpls.2019.00429
Received: 04 August 2018; Accepted: 21 March 2019;
Published: 09 April 2019.
Edited by:
Caroline Howard, Medicines and Healthcare Products Regulatory Agency, United KingdomReviewed by:
Adrian Slater, De Montfort University, United KingdomNatalia V. Ivanova, University of Guelph, Canada
Copyright © 2019 Liu, Gao and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing Zhou, emhvdWppbmdfYXBpYWNlYWVAMTYzLmNvbQ==
 Zhen-wen Liu
Zhen-wen Liu Yu-zhen Gao2
Yu-zhen Gao2