- 1College of Agronomy, Henan Agricultural University, Zhengzhou, China
- 2Department of Botany, National Museum of Natural History, Smithsonian Institution, Washington, DC, United States
The familial placement of Wightia has been controversial in the Lamiales, and the genus is currently placed in Paulowniaceae in APG IV. Phylogenetic analyses of Wightia and its close relatives in Lamiales are conducted using sequences of the complete chloroplast genomes as well as sequence data from nine chloroplast DNA regions (atpB, matK, ndhF, psbBTNH, rbcL, rps4, rps16 intron, trnL-F, and trnV-atpE) and one mitochondrial gene rps3. The maximum likelihood and Bayesian analyses do not support a close relationship between Wightia and Paulownia of Paulowniaceae; instead the enigmatic Wightia is sister to Phrymaceae with strong support in all analyses. Hence Wightia should not be placed in Paulowniaceae. Because morphological data show Wightia’s affinity to both Phrymaceae and Paulowniaceae and prior nrITS data suggest its sister relationship to Paulownia of Paulowniaceae, it is likely that Wightia may have had a hybrid origin between early lineages of Phrymaceae and Paulowniaceae. It is therefore the best to exclude Wightia from Paulowniaceae and place the genus as unassigned until further nuclear data to test the hybrid hypothesis. The seven species of Paulownia constitute a monophyletic group, and Paulowniaceae is supported to be a monogeneric family, consistent with a series of morphological and floral development characters. The genus Brandisia, which was sometimes regarded as a close relative of Wightia, is supported to be nested within Orobanchaceae, as sister to Pterygiella. This sister relationship can be corroborated by fruit, seed and pollen morphological characters.
Introduction
The familial placement of Wightia Wall. has been controversial. Wightia includes two species distributed mainly in Burma, India, Malaysia, Nepal, Vietnam, and Yunnan of China (van Steenis, 1949; Maheshwari, 1961; Hong et al., 1998). It was initially placed in Bignoniaceae based on the characters such as seeds that are winged but without endosperm and extra-floral nectarines under the leaf surface (Wallich, 1830; Hallier, 1903; Campbell, 1930; Li, 1947; Lawrence, 1951; Maheshwari, 1961). It was also included in Scrophulariaceae sensu lato on the basis of its two locular ovaries, and fruit dehiscing characters (Bentham and Hooker, 1876; Pennell, 1920; van Steenis, 1949; Willis, 1955; Hu, 1959; Hong et al., 1998). Recently Fischer (2004) mentioned that Wightia, together with Brandisia Hook. f. & Thomson, constituted the tribe Wightieae of Paulowniaceae of Nakai (1949). Stevens (2001) onward and APG IV (2016) placed Wightia and Paulownia Siebold & Zucc. as members of Paulowniaceae.
Paulownia consists of seven species native to eastern Asia and it is widely distributed in China and cultivated worldwide (Hong et al., 1998). The genus is well-known for its economic importance as the sources for rapidly growing timbers (Hu, 1959). The woody genus Paulownia was usually placed in Scrophulariaceae s.l. (Bentham and Hooker, 1876; Wettstein, 1891; Takhtajan, 1997; Hong et al., 1998), but it was sometimes positioned between Scrophulariaceae s.l. and Bignoniaceae (Campbell, 1930; Armstrong, 1985) or treated as a member of Bignoniaceae (Hallier, 1903; Takhtajan, 1980; Cronquist, 1981), although Nakai (1949) treated it as its own family, Paulowniaceae.
Brandisia comprises eleven species which are distributed in subtropical eastern Asia (Hong et al., 1998). The genus was placed in the tribe Cheloneae (Bentham and Hooker, 1876) or together with Wightia and Paulownia in Paulownieae (Tsoong, 1979) in Scrophulariaceae s.l., but some workers speculated that it may belong to other families, such as Loganiaceae, Solanaceae, Bignoniaceae, Pedaliaceae, Myoporaceae, and Verbenaceae (Campbell, 1930; Li, 1944, 1947).
Molecular phylogenetic analyses have revealed that the traditionally circumscribed Scrophulariaceae s.l. is polyphyletic (Olmstead and Reeves, 1995; Olmstead et al., 2001; Oxelman et al., 2005; Tank et al., 2006). These studies have resulted in circumscriptions and new descriptions of families to encompass the monophyletic lineages that were recovered in Lamiales. Paulownia was tentatively elevated to the monogeneric family Paulowniaceae (Beardsley and Olmstead, 2002), but its monophyly has not been tested because previous phylogenetic studies often sampled only P. tomentosa. Based on molecular data [rps16 intron, trnL-F region and nuclear ribosomal internal transcribed spacer (nrITS)] and chemical evidence, Zhou et al. (2014) argued that Wightia is closely allied to Paulowniaceae. However, there was phylogenetic conflict between nrITS and plastid loci (rps16 intron and trnL-F region) concerning the position of Wightia. Wightia was sister to Paulownia in the ITS tree, but it formed a clade with Phrymaceae in the chloroplast (combined trnL-F and rps16) tree. Zhou et al. (2014) also found that Brandisia (sampling only one species Brandisia swinglei) is not closely related to Wightia, instead the latter is nested within Orobanchaceae. Other molecular systematic studies (Oxelman et al., 2005; Bennett and Mathews, 2006; McNeal et al., 2013) also showed Brandisia (sampling only one species B. hancei) as a member of Orobanchaceae. However, the accurate systematic position of Brandisia within Orobanchaceae is uncertain. Brandisia was sister to the clade [Rhinantheae (Pedicularideae, Buchnereae)] in the nuclear PHYA gene topology with moderate support (BS = 72; 75, ML support) (Bennett and Mathews, 2006; McNeal et al., 2013) and in the nrITS tree (PP = 0.82 only) with weak support (Yu et al., 2018). However, it was sister to Pterygiella in nrITS and plastid (matK and rps2) trees both with low support (McNeal et al., 2013), or in the combined plastid (matk, rbcL, rps2, rps16, trnK-matK, and trnH-psbA) tree with moderate support (BS = 69, ML support; PP = 0.96) (Yu et al., 2018). The topological conflict between McNeal et al. (2013) and Yu et al. (2018) results with nrITS may be attributed to the difference in taxon sampling in these studies. With the majority of Brandisia species not included in the previous molecular phylogenetic studies, McNeal et al. (2013) suggested that further work on the genus is desirable because it occupies a pivotal place in the phylogeny, as the likely sister of the clade [Rhinantheae (Pedicularideae, Buchnereae)], which contains most species diversity in Orobanchaceae.
It is necessary to increase the sampling of key putative relatives of Wightia and include additional DNA characters in order to accurately determine the familial placement of Wightia in Lamiales. In recent years, the chloroplast genomes have been widely used to resolve difficult phylogenetic relationships in plants (e.g., Jansen et al., 2007; Zhang et al., 2015; Xu et al., 2017; Wen et al., 2018). This study is conducted with a comprehensive sampling of putative relatives of Wightia in Lamiales s.l. and using nine chloroplast DNA regions (atpB, matK, ndhF, psbBTNH, rbcL, rps4, rps16 intron, trnL-F, and trnV-atpE) and one mitochondrial gene rps3 that have been shown to be particularly informative in the Lamiales s.l. (Refulio-Rodriguez and Olmstead, 2014). We further conduct a second phylogenetic analysis using a selected sampling of the putative relatives of Wightia in Lamiales based on chloroplast genome sequences. The complete chloroplast (cp) genome sequences of Wightia, Brandisia, Mazus, and Phryma are herein reported for the first time. The goals of this study are to (1) test the familial placement of Wightia, and (2) determine the taxonomic composition of Paulowniaceae by broadly sampling species of Paulownia, Brandisia, as well as Phrymaceae, Mazaceae, and Orobanchaceae in Lamiales.
Materials and Methods
Taxon Sampling
The taxon sampling consisted of 110 samples representing all the families of Lamiales. One species of Wightia, all seven species of Paulownia and six species of Brandisia were sampled for the first time in this study. In addition, data of the remaining samples in this study are mostly from McNeal et al. (2013), Refulio-Rodriguez and Olmstead (2014), and Yu et al. (2018). We also selected 22 representative genera of nine major clades in Orobanchaceae (McNeal et al., 2013; Yu et al., 2018) to test the placement of Brandisia. Two species: Solanum lycopersicum L. (Solanaceae) and Gelsemium sempervirens (L.) J.St.-Hil. (Gelseminaceae) were selected as outgroups based on Refulio-Rodriguez and Olmstead (2014). The chloroplast genome data of four species (including Wightia speciosissima, B. swinglei, Mazus pumilus, and Phryma leptostachya subsp. asiatica) were reported for the first time in this study. We selected one species of Wightia, two species of Paulownia (Paulowniaceae), two genera of Gesneriaceae, four genera of Plantaginaceae, one genus of Scrophulariaceae sensu stricto, two genera of Phrymaceae, two genera of Mazaceae, 16 representative genera of eight major clades (McNeal et al., 2013; Yu et al., 2018) in Orobanchaceae, two genera of Acanthaceae, three genera of Bignoniaceae, five genera of Lamiaceae, and one genus of Pedaliaceae, one genus of Verbenaceae, and two genera of Lentibulariaceae. Solanum bulbocastanum Dunal and Hyoscyamus niger L. of Solanaceae were selected as outgroups. Voucher specimens are deposited in the Herbarium of Henan Agricultural University (HEAC). Voucher information and GenBank accession numbers of the sequences used in this study are provided in Supplementary Tables S1, S2.
DNA Extraction, PCR Amplification, and Sequencing
Total genomic DNA was extracted from leaf tissue samples preserved in silica gel or leaves removed from herbarium specimens using the modified 2 × CTAB method (Doyle and Doyle, 1987) and the Plant Genomic DNA Kit (DP305) from Tiangen Biotech (Beijing) Co., Ltd., China. We sequenced nine plastid regions, including six coding regions (atpB, matK, ndhF, psbBTNH, rbcL, and rps4), three noncoding regions (rps16 intron, trnL-F intron and spacer, and trnV-atpE spacer), and one mitochondrial coding region (rps3). The primer information of PCR amplification and amplification reactions for all chloroplast and the mitochondrial genes were as in Refulio-Rodriguez and Olmstead (2014). We included available DNA sequences of the above mentioned gene regions from GenBank.
Amplification reactions for all ten genes were run according to the following steps: (1) a denaturing step at 94°C for 45 s, (2) 35 cycles with a denaturing step at 94°C for 45 s, and an annealing step at 52°C for 45 s, an extension step at 72°C for 90 s, and (3) a final extension at 72°C for 10 min. Missing sequences are a consequence of amplification failure or lack of DNA availability. PCR products were purified with a PCR purification kit (UNIQ-10, Sangon, Shanghai, China). Sequencing primers were the same as amplification primers. Sequencing was performed on an ABI 3730xl DNA sequencer (Applied Biosystems) by Sunbiotech Co., Ltd., Beijing.
Chloroplast Genome Sequencing, Assembly, and Annotation
DNA samples were randomly fragmented into 400–600 bp fragments using an ultrasonicator. An Illumina paired-end DNA library with 500-bp insert size was constructed using a NEBNext® UltraTM DNA Library Prep Kit following the manufacturer’s instructions. Paired-end sequencing (2 × 150 bp) was conducted on an Illumina HiSeq X platform.
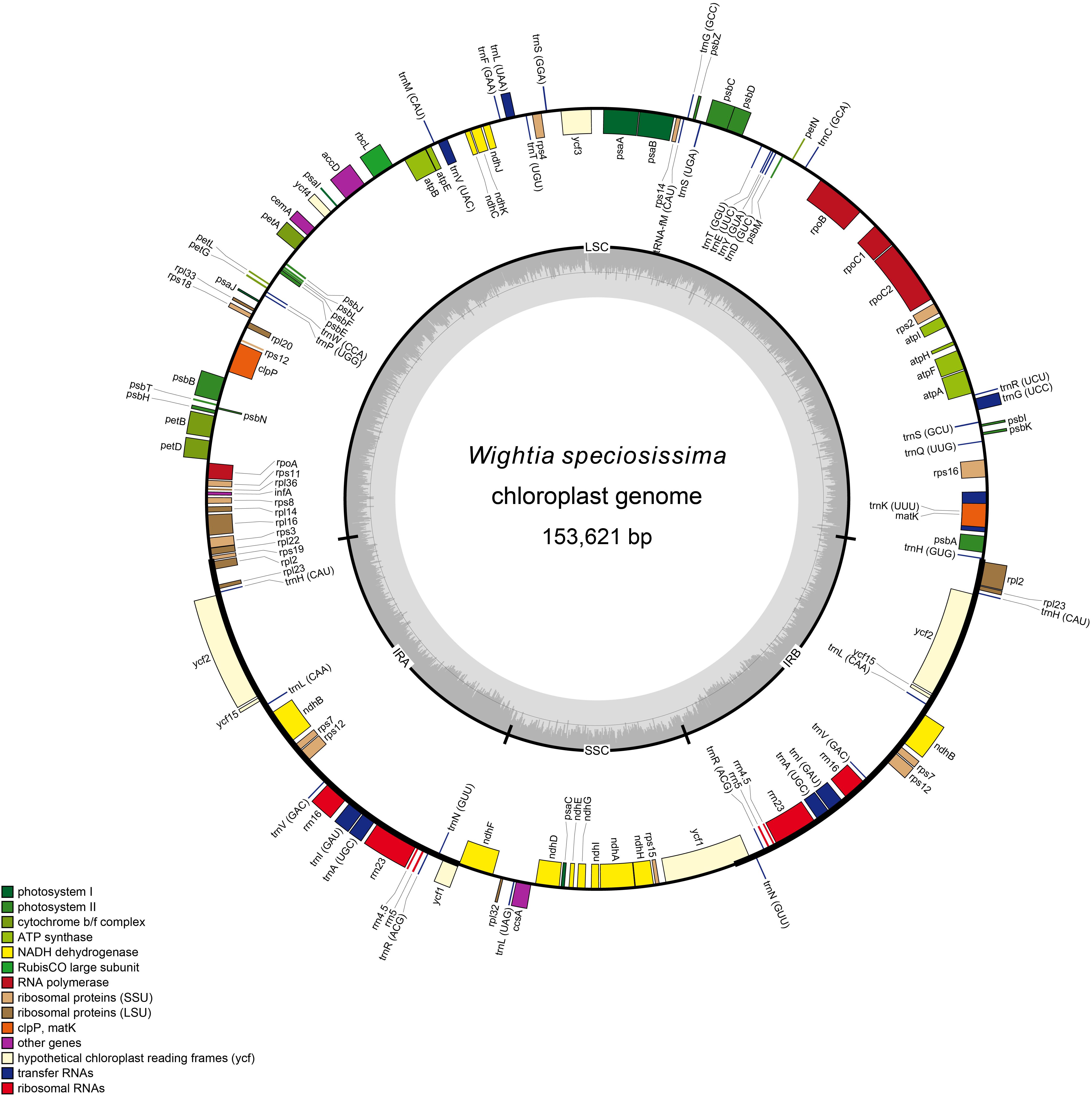
The paired-end reads were qualitatively assessed and initially assembled with SPAdes 3.6.1 (Bankevich et al., 2012), using k-mer ranging from 57 to 99. Contigs of low sequencing depths were discarded. The remaining contigs may contain the information not only from the chloroplast genome but also from the nuclear genome and the mitochondrial genome. Next, chloroplast genome sequence contigs were selected from SPAdes software by performing a BLAST search using the Triaenophora shennongjiaensis X. D. Li, Y. Y. Zan & J. Q. Li chloroplast genome sequence as a reference (GenBank accession number: MH071405) (Xia and Wen, 2018). The selected contigs were further assembled with Sequencher 5.4.5 (Gene Codes, Ann Arbor, MI, United States). Small gaps in the assemblies were bridged with specific primers designed for PCR based on their flanking sequences and then by Sanger sequencing. Based on the reference chloroplast genome, the four junctions between the inverted repeats (IRs) and single copy regions were checked by amplification with specific primers followed by Sanger sequencing (Dong W. et al., 2013). Chloroplast genome annotation was performed with Plann (Huang and Cronk, 2015) using the T. shennongjiaensis reference sequence from GenBank. The annotated GenBank files were used to construct the circular plastid genome maps with the online program Organellar Genome DRAW (OGDRAW) (Lohse et al., 2013) and then the annotated cp genome sequences were submitted to GenBank with the accession number MK381318 (W. speciosissima), MK381315 (B. swinglei), MK381316 (M. pumilus), and MK381317 (P. leptostachya subsp. asiatica).
Sequence Alignment and Phylogenetic Analysis
Initial automated alignments of the individual genes were made using the MAFFT (Katoh and Standley, 2013) with the E-INS-I algorithm in Geneious. The data from nine chloroplast regions, and the mitochondrial gene rps3 were analyzed separately. The chloroplast genome sequences were performed based on the all common protein coding genes (PCGs) (Ycf1 gene was excluded because of high diversity in Lamiales). Gaps were treated as missing data. The data matrix combining all 10 genes, and combining PCGs of chloroplast genome were performed by using both maximum likelihood (ML) and Bayesian inference (BI) methods.
The ML analyses were conducted using RAxML (version 8.2; Stamatakis, 2014). These analyses used the GTR substitution model with gamma-distributed rate heterogeneity among sites and the proportion of invariable sites estimated from the data. The concatenated plastid dataset was partitioned by gene. Support values for the node and clade were estimated from 1000 bootstrap replicates. BI analyses were performed using MrBayes vers. 3.2.6 (Ronquist et al., 2012). The Markovchain Monte Carlo (MCMC) analysis was run in MrBayes for 10, 000, 000 generations for each dataset. We checked for stationarity in Tracer version1.4 (Drummond and Rambaut, 2007) by confirming an ESS of greater than 200 for all parameters and by visually inspecting the distributions of the sampled states. The first 25% of samples were discarded as burn-in (Huelsenbeck and Ronquist, 2001) and the remaining trees were used to generate a majority-rule consensus tree. The ML tree and BI tree were visualized using FigTree version 1.4.2.
Results
Chloroplast Genome Features
Using an Illumina HiSeq X System, samples of the four taxa: W. speciosissima, B. swinglei, M. pumilus and P. leptostachya subsp. asiatica, were sequenced via the genome skimming approach (Zhang et al., 2015; Zimmer and Wen, 2015), producing 26, 779, 390; 29, 029, 530; 39, 331, 112; and 7, 460, 482 paired end raw reads (150 bp average read lengths).
The chloroplast genomes of W. speciosissima, B. swinglei, M. pumilus and P. leptostachya subsp. asiatica had a total sequence length of 153, 621 bp, 155, 344 bp, 153, 034 bp, and 153, 167 bp, respectively. The chloroplast genomes showed a typical quadripartite structure, consisting of a pair of IRs (25, 797 bp, 26, 498 bp, 25, 872 bp, and 25,456 bp, respectively) separated by the LSC (84, 393 bp, 84, 650 bp, 83, 839 bp, and 84,877 bp, respectively) and SSC (17, 634 bp, 17, 698 bp, 17, 451 bp, and 17, 378 bp, respectively) regions. For the four chloroplast genomes (W. speciosissima, B. swinglei, M. pumilus and P. leptostachya subsp. asiatica), the average GC content was 37.7, 38.1, 37.8, and 37.7%, respectively (Table 1). The annotated chloroplast genomes of four species were represented in four circular maps (Figure 1 and Supplementary Figures S1–S3). The chloroplast genomes of W. speciosissima and B. swinglei harbored 112 different genes, including 78 protein-coding genes, 30 tRNA genes and 4 rRNA genes. The chloroplast genome of M. pumilus harbored 114 different genes, including 81 protein-coding genes, 29 tRNA genes, and 4 rRNA genes. The chloroplast genome of P. leptostachya subsp. asiatica had 112 different genes, including 79 protein-coding genes, 29 tRNA genes, and 4 rRNA genes.
Phylogenetic Analyses Using Nine Chloroplast Regions and One Mitochondrial Gene
The 9-marker chloroplast regions combined data matrix consists of 14,789 bp in aligned length. The length of alignment, number of parsimony informative characters and PIC percentage (%) of each gene were shown in Supplementary Table S3. The chloroplast sequence data provided good resolution in the phylogenetic analyses overall. The topologies based on ML and Bayesian methods were both highly supported (Figure 2). Wightia is sister to Phrymaceae [Bootstrap (BS) = 84; posterior probability (PP) = 1.00]. In Orobanchaceae, nine clades (including Brandisia group and Pterygiella group) are well resolved (PP = 1.00). Orobanchaceae is sister to Paulowniaceae (BS = 98; PP = 1.00), and the Orobanchaceae – Paulowniaceae clade is sister to Phrymaceae (BS = 86; PP = 1.00), with the Orobanchaceae – Paulowniaceae – Phrymaceae clade then sister to Mazaceae (BS = 78; PP = 1.00). All seven species in Paulownia constitute monophyletic group with the maximum support (BS = 100; PP = 1.00). The six sampled species of Brandisia form one clade (BS = 100; PP = 1.00) that is nested within Orobanchaceae. Brandisia is sister to Pterygiella (BS = 61; PP = 0.91), and the Brandisia – Pterygiella clade is sister to Rhinantheae (BS = 56; PP = 0.80). Within Paulownia, P. fortune is sister to Paulownia × taiwaniana with BS = 78 and PP = 1.00, and P. elongata is sister to P. catalpifolia with BS = 89 and PP = 1.00.
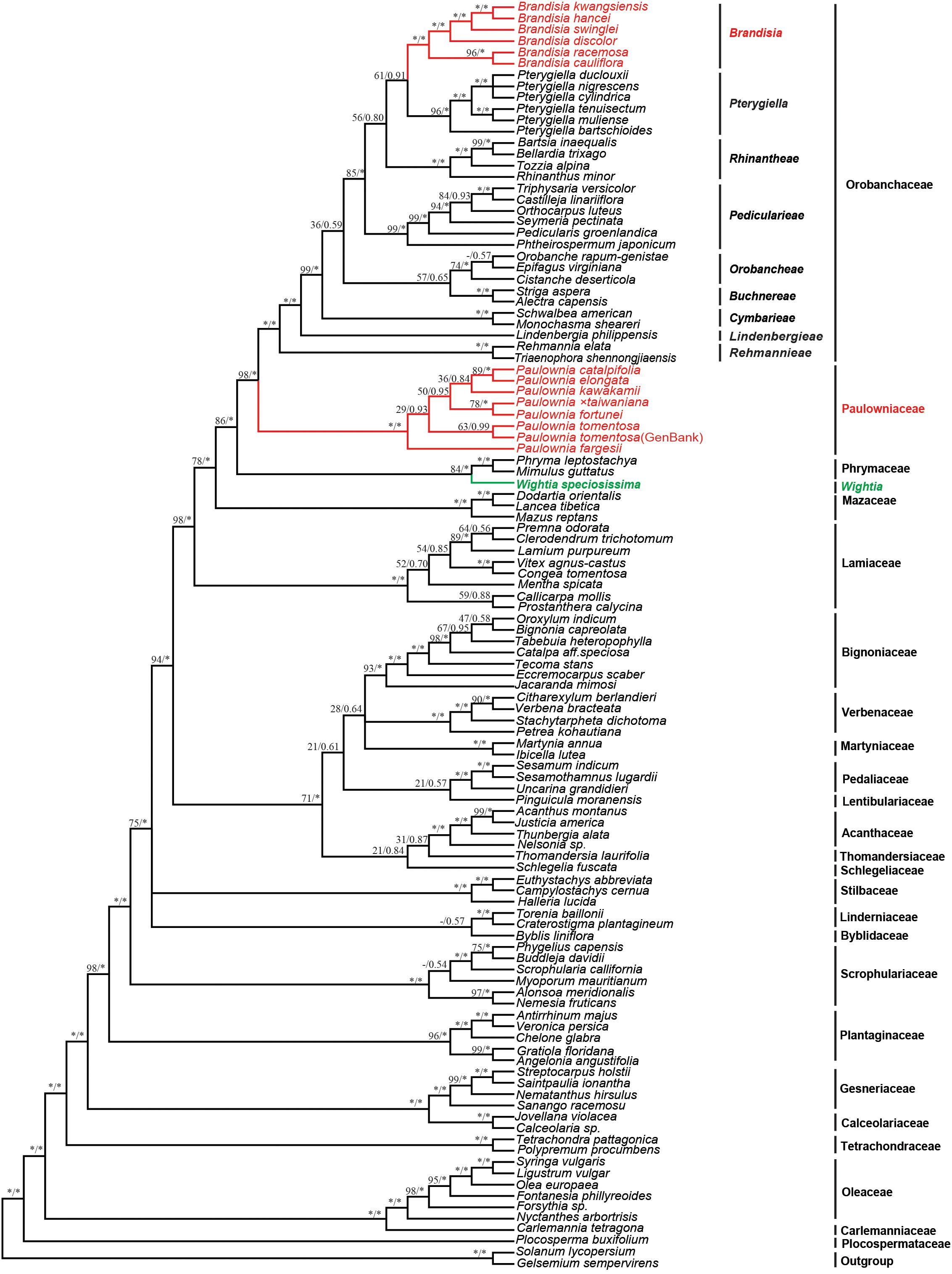
Figure 2. Bayesian tree based on the combined nine plastid markers (atpB, matK, ndhF, psbBTNH, rbcL, rps4, rps16 intron, trnL-F, and trnV-atpE). Number above branches are ML bootstrap values/Bayesian posterior. An asterisk indicates bootstrap value of 100 or posterior probability of 1.00; a hyphen (-) indicates the branch was not obtained in the ML bootstrap consensus.
The mitochondria gene rps3 data matrix consists of 1561 bp in aligned length. The length of alignment, number of parsimony informative characters and PIC percentage (%) of mitochondria gene rps3 were showed in Supplementary Table S3. The topologies based on ML and Bayesian methods showed in Supplementary Figure S4. Wightia is sister to one clade including Phrymaceae and Mazaceae (BS = 65; PP = 0.95). Paulowniaceae is sister to Wightia – Phrymaceae-Mazaceae clade (PP = 0.89), and then sister to Orobanchaceae (BS = 92; PP = 1.00). All seven species in Paulownia constitute monophyletic group with the maximum support (BS = 99; PP = 1.00). The six sampled species of Brandisia form one clade (BS = 62; PP = 0.97).
Phylogenetic Analyses Using Chloroplast Genome Sequences
The 79 protein-coding plastid genes sequence (PCGs) from the chloroplast genome of Wightia and its related taxa in the Lamiales were shown in the Supplementary Table S4. The data set from PCGs of the whole chloroplast genome provided the best resolution in the phylogenetic analyses with high bootstrap support values. The topologies based on the ML and BI methods were highly supported and congruent. Wightia is sister to Phrymaceae (BS = 100; PP = 1.00). The two sampled species of Paulownia form a clade with the maximum support (BS = 100; PP = 1.00). Brandisia is included in Orobanchaceae, which is sister to Rhinantheae (BS = 94; PP = 1.00). Paulownia is sister to Orobanchaceae with BS = 99 and PP = 1.00. Orobanchaceae and Paulowniaceae (Paulownia only) are then sister to Phrymaceae with high support (BS = 99; PP = 1.00). Mazaceae is sister to Lamiaceae with moderate support (BS = 65; PP = 0.95).
Discussion
On the Familial Placement of Wightia
The analyses based on DNA sequences from the 10 combined chloroplast and mitochondrial regions and the complete chloroplast genome data do not support a clade of Wightia and Paulownia. Even though Wightia was placed variously in Scrophulariaceae s.l., Bignoniaceae or Paulowniaceae in Lamiales (Wallich, 1830; Bentham and Hooker, 1876; Hallier, 1903; Pennell, 1920; Campbell, 1930; Li, 1947; van Steenis, 1949; Lawrence, 1951; Willis, 1955; Hu, 1959; Maheshwari, 1961; Hong et al., 1998; Fischer, 2004), our results support Wightia as sister to Phrymaceae (BS = 84,100; PP = 1.00,1.00; Figures 2, 3) or sister to the Phrymaceae - Mazaceae clade (BS = 65; PP = 0.95; Supplementary Figure S4). The Wightia-Phrymaceae clade is then sister to the large clade that includes Orobanchaceae and Paulowniaceae with high support (Figures 2, 3).
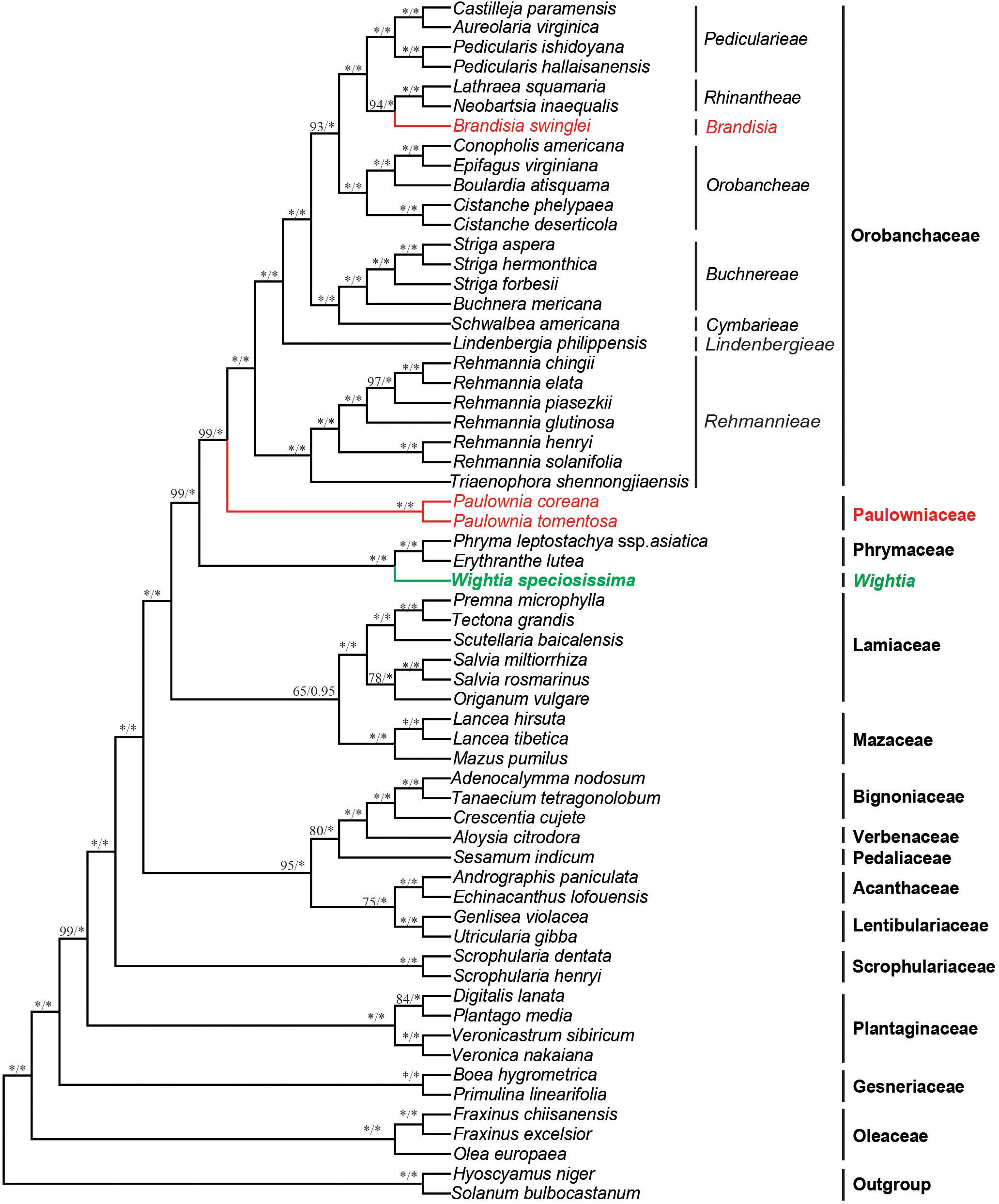
Figure 3. Bayesian tree inferred from the sequence of the protein coding genes (PCGs) of the chloroplast genome data. Numbers above branches are ML bootstrap values/Bayesian posterior probabilities. An asterisk indicates bootstrap value of 100 or posterior probability of 1.00.
The sister relationship between Wightia and Phrymaceae was initially reported by Zhou et al. (2014) using chloroplast trnL-F region and rps16 intron. Because their nrITS tree showed Wightia as sister to Paulownia with moderate support, the authors placed Wightia in Paulowniaceae (Zhou et al., 2014). The placement of Wightia in Paulowniaceae was followed by Stevens (2001 onward) and APG IV (2016). Our results clearly show that Wightia should not be simply placed in Paulowniaceae. Wightia species are hemiepiphytic and evergreen lianas which are distinctively different from taxa of other families in Lamiales (Fischer, 2004). Wightia and Phrymaceae share a series of morphological characters, such as two lateral bracteoles at the base of the pedicel just above the subtending bract, and capsules oblong-ovoid or narrowly ellipsoid (Hong et al., 1998; Fischer, 2004). In addition, the pollen of Wightia also shares the type IIb character of pollen in Mimulus of Phrymaceae in the following respects: trocolporate, exine microreticulate, and mean polar axis 25–35 μm (Argue, 1980; Wei, 1989). These morphological characters thus support a close relationship between Wightia and Phrymaceae. However, several other morphological characters and chemical constituents also suggest a possible close relationship between Wightia and Paulownia. They both bear large, entire, opposite leaves, lateral or terminal thyrses, thick and smooth calyx tubes, and two-valved capsules with many winged seeds (Maheshwari, 1961; Zhou et al., 2014). In light of the morphological evidence that shows that Wightia shares important characters with both Phrymaceae and Paulowniaceae, we propose that the incongruent position of Wightia based on nrITS and chloroplast sequences (c.f., Zhou et al., 2014; this study) might be caused by a likely hybrid origin of Wightia involving early lineages associated with Phrymaceae and Paulowniaceae. Further studies by sampling both species of Wightia and utilizing more nuclear sequences are needed as the next step to better understand the evolutionary history and the taxonomic position of Wightia, especially testing its hybrid origin hypothesis. If the hybrid origin is confirmed, it is the best to recognize Wightia as a distinct family Wightiaceae. At present, we suggest treating Wightia conservatively as unassigned at the familial level until further evidence from the nuclear genome. It is likely that Wightia may need to be placed in its own family.
Our results support recognizing Paulowniaceae as the monogeneric family (Nakai, 1949; Beardsley and Olmstead, 2002; Erbar and Gülden, 2011). The monogeneric Paulowniaceae possesses a series of diagnostic features which distinguish it from other families in Lamiales, i.e., tree habit, woody fruit, the unidirectional initiation of calyx, and corolla lobes from the adaxial to the abaxial side, late sympetaly petal but ledges connecting the petal arise relatively early, tubular stigma with papillae inside a dilated chamber (Erbar and Gülden, 2011) and the plain surface of the placenta with distinct long and slightly angular structures (Rebernig and Weber, 2007). Erbar and Gülden (2011) noted that Paulownia tomentosa shows the unidirectional initiation of calyx and corolla lobes from the adaxial to the abaxial side and plain surface of the placenta with distinct long and slightly angular structures, which are unusual in Lamiales. Hence they argued for the isolated position of the monogeneric family Paulowniaceae.
The position of Mazaceae (Reveal, 2011) still needs to be further explored. Mazaceae is sister to the clade containing Orobanchaceae, Paulowniaceae, and Phrymaceae in the tree based on nine chloroplast markers (Figure 2), consistent with Xia et al. (2009), Schäferhoff et al. (2010), and Refulio-Rodriguez and Olmstead (2014). But in the tree based on complete chloroplast genome, Mazaceae is sister to Lamiaceae with bootstrap support BS = 65 and PP = 0.95. This inconsistence may be caused by the limited sampling of Mazaceae and Phrymaceae in the chloroplast genome data set. Nevertheless, the Mazaceae position as sister to the clade of Orobachaceae, Paulowniaceae, and Phrymaceae had only 50% ML bootstrap support and 0.62 PP in Schäferhoff et al. (2010), and 34% MP bootstrap support in Refulio-Rodriguez and Olmstead (2014). The systematic position of Mazaceae deserves further test with greater taxon sampling in Mazaceae and its putative relatives (Phrymaceae) and by using chloroplast genomes and more nuclear markers.
Species Relationships Within Paulowniaceae
Within the monogeneric Paulowniaceae, our results shed some insights into the evolution of the genus Paulownia, as we sampled all seven species of the genus. Paulownia fortunei is shown to be sister to P × taiwaniana with high support (Figure 2). Paulownia × taiwaniana was reported as a natural hybrid species between P. fortunei and P. kawakamii with the latter as the maternal parent based on RAPD markers, chloroplast RFLP data (using one restriction enzyme only) and morphology (Lin and Wang, 1991; Wang et al., 1994). However, P. kawakamii as the maternal parent of P × taiwaniana is not supported by our results. Our results suggest P. fortunei as the likely maternal parent of P × taiwaniana, because the two species are sisters in the tree based on the maternally inherited chloroplast data (Figure 2). Furthermore, Paulownia elongata is sister to P. catalpifolia with high support. The close relationship between P. elongata and P. catalpifolia is also supported morphologically by their shared pubescent capsules, and calyx lobes shorter than tube (Hu, 1959; Hong et al., 1998). Our present study is the first to confirm the monophyly of Paulownia with all species sampled. But our analysis is limited to chloroplast and mitochondrial data. Fast-evolving, biparently inherited nuclear markers are needed to disentangle the species relationships of Paulownia and test potential hybrid speciation in the genus (Wang et al., 1994), using dense sampling of populations in the genus.
On the Position of Brandisia Within Orobanchaceae
Our study sampled six species of Brandisia, and shows that the genus is nested within Orobanchaceae. Inclusion of Brandisia within Orobanchaceae is consistent with the result of recent phylogenetic studies (Oxelman et al., 2005; Bennett and Mathews, 2006; McNeal et al., 2013; Yu et al., 2018). Brandisia as part of Orobanchaceae is supported by its capsules having half or partly exserted from the persistent calyx tubes and its hemiparasitic habits (Chin, 1979; Zhang, 1990; Hong et al., 1998; Xia et al., 2009). Yet the phylogenetic position of Brandisia within Orobanchaceae has been controversial in previous studies (Oxelman et al., 2005; Bennett and Mathews, 2006; McNeal et al., 2013; Yu et al., 2018). By increasing the sampling of Brandisia, the phylogenetic tree (Figure 2) using nine chloroplast gene regions showed that Brandisia is sister to Pterygiella in Orobanchaceae (BS = 61; PP = 0.91) which is consistent with the result of Yu et al. (2018), and Rhinantheae is sister to the clade containing Brandisia and Pterygiella in Orobanchaceae (BS = 56; PP = 0.80). Without sampling Pterygiella, our analyses using complete chloroplast genome data placed Brandisia as sister to Rhinantheae in Orobanchaceae with BS = 94 and PP = 1.00 (Figure 3). Fruit and seed characters also showed close relationships between Brandisia and Pterygiella, as both have eglandular hairs on the surface of capsules and reticulate seeds (Dong L.-N. et al., 2013; Dong et al., 2015). Furthermore, the pollen grains of Brandisia, Pterygiella and most genera in Rhinantheae are commonly tricolpate, medium-sized, circular or subcircular, and having long and acute-ended colpi with a granulate membrane, supported the close relationships among them (Wei, 1989; Lu et al., 2007).
The six species we sampled in Brandisia formed a monophyletic group with maximum support in the combined chloroplast and mitochondrial gene tree (Figure 2). Brandisia has a series of morphological characters which are distinctive from other genera in Orobanchaceae, such as infundibular corollas with strongly reflexed corolla lobes, and anthers with dense hairs (Ren et al., 2018). Ren et al. (2018) reported that the anther hairs function as a secondary pollen presentation mechanism and play a key role in restricting pollen loss after anther dehiscence, hence facilitating reproductive fitness in delayed selfing in Brandisia hancei. The secondary pollen presentation on anthers hairs is unique and may turn out to be a synapomorphy of Brandisia.
In conclusion, our results argue that Wightia should be removed from Paulowniaceae. As Wightia may be of hybrid origin between early lineages of Phrymaceae and Paulowniaceae, we suggest treating Wightia conservatively as unassigned at the familial level. If its hybrid origin is confirmed with further nuclear data, Wightia may need to be recognized as its own family. The systematic position of Mazaceae deserves further studies. Brandisia is sister to Pterygiella in Orobanchaceae, whichis corroborated by fruit and seed characters, pollen morphology and molecular data.
Author Contributions
ZX and JW conceived the study and interpreted the results. ZX and ZG collected and analyzed the data.
Funding
This study was supported by grants from the National Natural Science Foundation of China (Grant Nos. U1404302 and 31770370), the Scientific Research Projects of Henan Provincial Education Department (Grant No. 18A360006), and the China Scholarship Council (Grant No. 20175087, supporting ZX’s research visit to the Smithsonian Institution, Washington, DC, United States).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Yan Liu, Wen-Bing Yu, Qiang Zhang, Yang Dong, Peng-Wei Li, and Jia-Mei Li for their kind help in the field and, or providing samples for this study; and to Chen Ren, Yuan Xu, Bing-Bing Liu, and Ze-Long Nie for their assistance in data analyses in the Laboratory of Analytical Biology, the National Museum of Natural History of the Smithsonian Institution.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00528/full#supplementary-material
FIGURE S1 | Gene map of the chloroplast genome of Brandisia swinglei.
FIGURE S2 | Gene map of the chloroplast genome of Mazus pumilus.
FIGURE S3 | Gene map of the chloroplast genome of Phryma leptostachya subsp. asiatica.
FIGURE S4 | Bayesian tree inferred from the sequence of one mitochondrial gene rps3 data. ML bootstrap values (>50%)/Bayesian posterior probabilities (>0.50) are shown above branches. An asterisk indicates bootstrap value of 100 or posterior probability of 1.00; a hyphen (-) indicates the branch was not obtained in the ML bootstrap consensus.
TABLE S1 | Species included in this study, with assignment to order (where appropriate), family, origin and voucher information (herbarium acronym), and GenBank accession numbers (chloroplast atpB, matK, ndhF, psbBTNH, rbcL, rps4, rps16 intron, trnL-F, and trnV-atpE, and mitochondrial rps3). “na” indicates no sequence available for this region for the accession.
TABLE S2 | Voucher information and GenBank Accessions of chloroplast genomes used in this study.
TABLE S3 | The nine chloroplast DNA regions and one mitochondrial gene used in the phylogenetic analyses of Lamiales.
TABLE S4 | The 79 protein-coding plastid genes used in the phylogenetic analyses of Lamiales.
References
Apg, I. V. (2016). An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 181, 1–20.
Argue, C. L. (1980). Pollen morphology in the genus Mimulus (Scrophulariaceae) and its taxonomic significance. Am. J. Bot. 67, 68–87.
Armstrong, J. E. (1985). The delimitation of Bignoniaceae and Scrophulariaceae based on floral anatomy and the placement of problem genera. Am. J. Bot. 72, 755–766.
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Beardsley, P. M., and Olmstead, R. G. (2002). Redefining Phrymaceae: the placement of Mimulus, tribe Mimuleae, and Phryma. Am. J. Bot. 89, 1093–1102. doi: 10.3732/ajb.89.7.1093
Bennett, J., and Mathews, S. (2006). Phylogeny of the parasitic plant family Orobanchaceae inferred from Phytochrome A. Am. J. Bot. 93, 1039–1051. doi: 10.3732/ajb.93.7.1039
Bentham, G., and Hooker, J. D. (1876). “Scrophulariaceae,” in Genera Plantarum, Vol. 2, eds G. Bentham and J. D. Hooker (London: Williams and Norgaate), 913–980.
Chin, T.-L. (1979). “Rehmannia and Triaenophora,” in Flora Reipublicae Popularis Sinicae, Part 2, Scrophulariaceae, Vol. 67, eds P. C. Tsoong and H. P. Yang (Beijing: Science Press), 212–222.
Cronquist, A. (1981). An Integrated System of Classification of Flowering Plants. New York, NY: Columbia University Press.
Dong, L.-N., Wang, H., Wortley, A. H., Li, D.-Z., and Lu, L. (2015). Fruit and seed morphology in some representative genera of tribe Rhinantheae sensu lato (Orobanchaceae) and related taxa. Plant Syst. Evol. 301, 479–500.
Dong, L.-N., Wang, H., Wortley, A. H., Lu, L., and Li, D.-Z. (2013). Phylogenetic relationships in the Pterygiella complex (Orobanchaceae) inferred from molecular and morphological evidence. Bot. J. Linn. Soc. 171, 491–507.
Dong, W., Xu, C., Cheng, T., Lin, K., and Zhou, S. L. (2013). Sequencing angiosperm plastid genomes made easy: a complete set of universal primers and a case study on the phylogeny of Saxifragales. Genome Biol. Evol. 5, 989–997. doi: 10.1093/gbe/evt063
Doyle, J. J., and Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15.
Drummond, A. J., and Rambaut, A. (2007). BEAST: bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. doi: 10.1186/1471-2148-7-214
Erbar, C., and Gülden, C. (2011). Ontogeny of the flowers in Paulownia tomentosa-A contribution to the recognition of the resurrected monogeneric family Paulowniaceae. Flora 206, 205–218.
Fischer, E. (2004). “Scrophulariaceae,” in The Families and Genera of Vascular Plants. Vol. 7. Flowering Plants, Dicotyledons: Lamiales (Except Acanthaceae Including Avicenniaceae), ed. J. W. Kaderei (Berlin: Springer).
Hallier, H. (1903). Ueber die abgrenzung und verwandtschaft der einzelnen sippen bei den scrophularineen. Bull. Herb. Boiss. 3, 181–207.
Hong, D.-Y., Yang, H.-B., Jin, C.-L., and Holmgren, N. H. (1998). “Scrophulariaceae,” in Flora of China Editorial Committee, Vol. 18, eds Z. Y. Wu and P. H. Raven (Beijing: Missouri Botanical Garden Press).
Huang, D. I., and Cronk, Q. C. B. (2015). Plann: a command-line application for annotating plastome sequences. Appl. Plant Sci. 3, as.1500026. doi: 10.3732/apps.1500026
Huelsenbeck, J. P., and Ronquist, F. (2001). MRBAYES: bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755.
Jansen, R. K., Cai, Z., Raubeson, L. A., Daniell, H., Leebens-Mack, J., Müller, K. F., et al. (2007). Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. U.S.A. 104, 19369–19374.
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Lin, T.-P., and Wang, Y.-S. (1991). Paulownia taiwaniana, a hybrid between P. fortunei and P. kawakamii (Scrophulariaceae). Plant Syst. Evol. 178, 259–269.
Lohse, M., Drechsel, O., Kahlau, S., and Bock, R. (2013). Organellar Genome DRAW- a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41, W575–W581.
Lu, L., Wang, H., Blackmore, S., Li, D.-Z., and Dong, L.-N. (2007). Pollen morphology of the tribe Rhinantheae (Orobanchaceae) and its systematic significances. Plant Syst. Evol. 268, 177–198.
Maheshwari, J. K. (1961). The genus Wightia Wall. in India with a discussion on its systematic position. Bull. Bot. Surv. India 3, 31–35.
McNeal, J. R., Bennett, J. R., Wolfe, A. D., and Mathews, S. (2013). Phylogeny and origins of holoparasitism in Orobanchaceae. Am. J. Bot. 100, 971–983. doi: 10.3732/ajb.1200448
Nakai, T. (1949). Classes, ordinae, familiae, subfamilieae, tribus, genera nova quae attinentad plantas Koreanas. J. Jap. Bot. 24, 8–14.
Olmstead, R. G., dePamphilis, C. W., Wolfe, A. D., Young, N. D., Elisons, W. J., and Reeves, P. A. (2001). Disintegration of the Scrophulariaceae. Am. J. Bot. 88, 348–361.
Olmstead, R. G., and Reeves, P. A. (1995). Evidence for the polyphyly of the Scrophulariaceae based on chloroplast rbcL and ndhF sequences. Ann. Mo. Bot. Gard. 82, 176–193.
Oxelman, B., Kornhall, P., Olmstead, R. G., and Bremer, B. (2005). Further disintegration of Scrophulariaceae. Taxon 54, 411–425.
Pennell, F. W. (1920). Scrophulariaceae of the south-eastern United States. Proc. Acad. Nat. Sci. Philad. 71, 224–291. doi: 10.3109/17482960903273056
Rebernig, C. A., and Weber, A. (2007). Diversity, development and systematic significance of seed pedestals in Scrophulariaceae (s.l.). Bot. Jahrb. Syst. 127, 133–150.
Refulio-Rodriguez, N. F., and Olmstead, R. G. (2014). Phylogeny of lamiidae. Am. J. Bot. 101, 287–299.
Ren, Y.-Q., Deng, G.-Y., Li, X.-P., Marczewski, T., and Ma, Y.-P. (2018). Secondary pollen presentation on anther hairs enhances reproductive fitness in Brandisia hancei, a protogynous perennial with autonomous selfing. Plant Ecol. Divers. 11, 373–381.
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. ( (2012). MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Schäferhoff, B., Fleischmann, A., Fischer, E., Albach, D. C., Borsch, T., Heubl, G., et al. (2010). Towards resolving Lamiales relationships: insights from rapidly evolving chloroplast sequences. BMC Evol. Biol. 10:352. doi: 10.1186/1471-2148-10-352
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stevens, P. F. (2001). Angiosperm Phylogeny Website. Version. 14, July 2017.Google Scholar
Takhtajan, A. L. (1980). Outline of the classification of flowering plants (Magnoliophyta). Bot. Rev. 46, 225–359.
Takhtajan, A. L. (1997). Diversity and Classification of Flowering Plants. New York, NY: Columbia University Press.
Tank, D. C., Beardsley, P. M., Kelchner, S. A., and Olmstead, R. G. (2006). Review of the systematics of Scrophulariaceae s.l. and their current disposition. Aust. Syst. Bot. 19, 289–307.
Tsoong, P.-C. (1979). “Paulownia,” in Flora Reipublicae Popularis Sinicae, Vol. 67, Part 2, Scrophulariaceae, eds P. C. Tsoong and H. P. Yang (Beijing: Science Press), 17–46.
van Steenis, C. G. G. J. (1949). Notes on the genus Wightia (Scrophulariaceae). Bull. Bot. Gard. Buitenzorg. 18, 213–227. doi: 10.11646/zootaxa.3734.1.1
Wang, W.-Y., Pai, R.-C., Lai, C.-C., and Lin, T.-P. (1994). Molecular evidence for the hybrid origin of Paulownia taiwaniana based on RAPD markers and RFLP of chloroplast DNA. Theor. Appl. Genet. 89, 271–275. doi: 10.1007/BF00225153
Wei, Z.-X. (1989). Pollen morphology of Wightia and its taxonomic significance. Acta Bot. Yunnan. 1, 65–70.
Wen, J., Harris, A. J., Kalburgi, Y., Zhang, N., Xu, Y., Zheng, W., et al. (2018). Chloroplast phylogenomics of the New World grape species (Vitis, Vitaceae). J. Syst. Evol. 56, 297–308. doi: 10.1016/j.ympev.2011.11.015
Wettstein, R. (1891). “Scrophulariaceae,” in Die Natürlichen Pflanzenfamilien IV/3B, eds A. Engler and K. Prantl (Leipzig: Engelmann), 39–107.
Willis, J. C. (1955). A Dictionary of the Flowering Plants and Ferns. Cambridge: Cambridge University Press.
Xia, Z., Wang, Y.-Z., and Smith, J. F. (2009). Familial placement and relations of Rehmannia and Triaenophora (Scrophulariaceae s. l.) inferred from five gene regions. Am. J. Bot. 96, 519–530. doi: 10.3732/ajb.0800195
Xia, Z., and Wen, J. (2018). The complete chloroplast genome of the endangered species Triaenophora shennongjiaensis (Orobanchaceae s.l.). Mitochondr. DNA B 3, 506–507.
Xu, C., Dong, W.-P., Li, W.-Q., Lu, Y.-Z., Xie, X.-M., Jin, X.-B., et al. (2017). Comparative analysis of six Lagerstroemia complete chloroplast genomes. Front. Plant Sci. 8:15. doi: 10.3389/fpls.2017.00015
Yu, W.-B., Randle, C.-P., Lu, L., Wang, H., Yang, J.-B., dePamphilis, C. W., et al. (2018). The hemiparasitic plant Phtheirospermum (Orobanchaceae) is polyphyletic and contains cryptic species in the Hengduan Mountains of Southwest China. Front. Plant Sci. 9:142. doi: 10.3389/fpls.2018.00142
Zhang, N., Wen, J., and Zimmer, E. A. (2015). Congruent deep relationships in the grape family (Vitaceae) based on sequences of chloroplast genomes and mitochondrial genes via genome skimming. PLoS One 10:e0144701. doi: 10.1371/journal.pone.0144701
Zhang, Z.-Y. (1990). “Orobanchaceae,” in Flora Reipublicae Popularis Sinicae, Vol. 69, ed. W. T. Wang (Beijing: Science Press), 69–124.
Zhou, Q.-M., Jensen, S.-R., Liu, G.-L., Wang, S., and Li, H.-Q. (2014). Familial placement of Wightia (Lamiales). Plant Syst. Evol. 300, 2009–2017.
Keywords: Wightia, Paulownia, Brandisia, familial placement, phylogenetic relationship, Scrophulariaceae
Citation: Xia Z, Wen J and Gao Z (2019) Does the Enigmatic Wightia Belong to Paulowniaceae (Lamiales)? Front. Plant Sci. 10:528. doi: 10.3389/fpls.2019.00528
Received: 03 January 2019; Accepted: 05 April 2019;
Published: 30 April 2019.
Edited by:
Stefan Wanke, Dresden University of Technology, GermanyReviewed by:
Luiz Henrique Martins Fonseca, University of São Paulo, BrazilIsabel Larridon, Royal Botanic Gardens, Kew, United Kingdom
Copyright © 2019 Xia, Wen and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhi Xia, eGlhemhpZW1haWxAMTI2LmNvbQ== Jun Wen, d2VuakBzaS5lZHU= Zhiming Gao, R2FvemhpbWluZzY3MkBzaG91LmNvbQ==
 Zhi Xia
Zhi Xia Jun Wen
Jun Wen Zhiming Gao1*
Zhiming Gao1*