Abstract
Glutathione transferases (GSTs) belong to a ubiquitous multigenic family of enzymes involved in diverse biological processes including xenobiotic detoxification and secondary metabolism. A canonical GST is formed by two domains, the N-terminal one adopting a thioredoxin (TRX) fold and the C-terminal one an all-helical structure. The most recent genomic and phylogenetic analysis based on this domain organization allowed the classification of the GST family into 14 classes in terrestrial plants. These GSTs are further distinguished based on the presence of the ancestral cysteine (Cys-GSTs) present in TRX family proteins or on its substitution by a serine (Ser-GSTs). Cys-GSTs catalyze the reduction of dehydroascorbate and deglutathionylation reactions whereas Ser-GSTs catalyze glutathione conjugation reactions and eventually have peroxidase activity, both activities being important for stress tolerance or herbicide detoxification. Through non-catalytic, so-called ligandin properties, numerous plant GSTs also participate in the binding and transport of small heterocyclic ligands such as flavonoids including anthocyanins, and polyphenols. So far, this function has likely been underestimated compared to the other documented roles of GSTs. In this review, we compiled data concerning the known enzymatic and structural properties as well as the biochemical and physiological functions associated to plant GSTs having a conserved serine in their active site.
Introduction
Glutathione transferases (GSTs), formerly glutathione S-transferases, constitute a ubiquitous multigenic superfamily of enzymes that conjugate the tripeptide glutathione (γ-Glu-Cys-Gly) on a broad range of molecules. They catalyze the nucleophilic attack of reduced glutathione (GSH) on the electrophilic centers of these molecules. The omnipresence of these enzymes in all types of organisms highlights an ancient origin as well as fundamental functions preserved during evolution. GSTs were discovered in the early 1960s through their GSH-conjugating activity in cellular extracts from rat liver incubated with sulfobromophthalein, chloronitrobenzenes or halogenated aromatic molecules (Booth et al., 1961; Combes and Stakelum, 1961). Later on, this conjugating activity was identified from plant extracts (sorghum and corn) using herbicides like atrazine or triazine derivatives (Frear and Swanson, 1970; Lamoureux et al., 1970). The interest for these GSH-conjugation reactions in plants was high in the 1980s, particularly concerning the enzymatic properties of cereal GSTs in connection with the detoxification of herbicides (Shah et al., 1986; Wiegand et al., 1986). Accordingly, GSTs are generally strongly induced in response to biotic and abiotic stresses, which coincides with cellular roles in primary and secondary metabolisms, in stress tolerance or cell signaling, and in xenobiotic detoxification by acting as phase II enzymes (Jakoby, 1978; Wiegand et al., 1986; Gonzalez et al., 2018). During the detoxification process, GSTs, which represent the most important classes of conjugating enzymes, conjugate phase I-activated molecules (or toxic molecules that are already activated) with GSH. Conjugation reactions are only performed by specific GST members, i.e., those having usually a conserved serine or a tyrosine (in mammals) in their active site. Those having notably a cysteine residue lack this property. The conjugation step has several benefits in the detoxification process, including a decrease of the reactivity and toxicity of the molecules, as well as an increase of their solubility. Likewise, the addition of large anionic groups such as GSH detoxifies reactive electrophiles and produces polar molecules that cannot diffuse across membranes. These molecules are then recognized and actively transported by ATP-binding cassette transporters (ABC-transporters), also known as phase III proteins (Keppler, 1999). ABC transporters carry out the ATP-dependent transport of a large variety of hydrophobic molecules and thus participate in exocytosis (animals) or sequestration in the vacuole and/or in the cell wall (plants and fungi) of phase II products (Coleman et al., 1997).
Even though most of the studies published over the past years focused on GSTs catalyzing the addition of glutathione, other catalytic activities have been described. For instance, numerous GSTs act as GSH-dependent peroxidases by reducing organic hydroperoxides (Tang and Tu, 1994; Marrs, 1996; Hurst et al., 1998) whereas others perform isomerisation reactions. The zeta GSTs catalyze the cis-trans isomerisation of maleylacetoacetate into fumarylacetoacetate, as part of the tyrosine degradation pathway (Thom et al., 2001; Fernandez-Canon et al., 2002). In addition to their involvement in GSH-conjugation, several GSTs also catalyze the opposite reaction, i.e., the removal of glutathione from small molecules. This reaction will be referred to as deglutathionylation. Note that this term is also used for the reduction of glutathione adducts on protein cysteine residues, as catalyzed by another family of GSH-dependent proteins called glutaredoxins (GRXs) (Rouhier et al., 2008). Although human GSTO1-1 was shown to catalyze the deglutathionylation of peptides/proteins such as glutathionylated β-actin (Menon and Board, 2013), this has been rarely observed among GSTs. The capacity of catalyzing deglutathionylation reactions is linked to the existence of a catalytic cysteinyl residue instead of the active site serine or tyrosine residues. This residue is notably present in the bacterial-specific Beta GSTs (GSTBs); in Omega GSTs (GSTOs) found in mammals, insects, and fungi (Board et al., 2000; Kim et al., 2006; Yamamoto et al., 2009; Meux et al., 2013); in the plant-specific Lambda GSTs (GSTLs); and in glutathionyl-hydroquinone reductases (GHRs), also known as Xi GSTs (GSTX), which are found in bacteria, fungi, archaea, and plants (Xun et al., 2010; Meux et al., 2011; Lallement et al., 2015; Schwartz et al., 2016). However, with a few exceptions, the physiological role of these enzymes is poorly documented. In addition to being involved in deglutathionylation, as mentioned above, human GSTOs may be involved in arsenic biotransformation, reducing methyl and dimethyl arsenate forms (Zakharyan et al., 2001; Burmeister et al., 2008). Plant GSTLs may be involved in the metabolism or trafficking of flavonoids (Dixon and Edwards, 2010b). GHRs are involved in the catabolism of chlorinated quinones and in lignin degradation through the deglutathionylation of metabolic intermediates (Reddy and Gold, 2001; Masai et al., 2003; Huang et al., 2008; Meux et al., 2011).
In addition to these catalytic properties, some GSTs possess the property to bind ligands also referred to as ligandin properties. It consists of the binding of small hydrophobic molecules either at the catalytic site or in a specific ligandin site (L-site) for their transport or storage. This non-catalytic property has been documented in plants for the transport/binding of hydrophobic xenobiotic molecules, of endogenous compounds such as oxylipins and flavonoids (anthocyanins, proanthocyanidins) as well as of phytohormones such as auxin and cytokinin (Zettl et al., 1994; Gonneau et al., 1998), suggesting a possible role of GSTs in cell signaling and/or in plant growth and development (Smith et al., 2003; Kitamura et al., 2004; Gong et al., 2005; Moons, 2005; Ahmad et al., 2017).
Overall, although the biochemical (catalytic and ligandin) properties of representative members from almost all GST classes have been studied, sometimes extensively, the physiological role of most of them remains to be identified, essentially because the existence of several close isoforms in given classes may have hampered their characterization by reverse-genetic approaches. Therefore, having focused recently on the biochemical and structural properties of Cys-GSTs (Lallement et al., 2014), the objective of this review is to inventory the known properties and functions of Ser-GSTs in photosynthetic organisms.
History and Classification of the GSTs
Over the past years, the GST classification has constantly evolved concomitantly to the increase of the genomic resources available, and to the identification and characterization of new isoforms and classes. First discovered in rat, GSTs were characterized initially in mammals and subsequently in insects, plants, fungi, and bacteria. In mammals, GSTs were originally classified as cytosolic, mitochondrial and membrane-associated GSTs, the latter being subdivided into microsomal GSTs and leukotriene C4 synthetases (Kraus, 1980; Hayes and Pulford, 1995). The same three subfamilies were renamed later as soluble GSTs, kappa GSTs and membrane-associated proteins in eicosanoids and glutathione metabolism (MAPEG), respectively (Jakobsson et al., 1999). However, on the basis of their immunological cross-reactivity and sequence relatedness, mammalian GSTs were also classified into the alpha, mu, pi, sigma, theta, and zeta classes (Mannervik et al., 1985; Dixon et al., 1998; Hayes and McLellan, 1999). At the time, most non-mammalian GSTs were placed in the heterogeneous theta class (Buetler and Eaton, 1992). For plant GSTs, the first classification introduced was based on sequence analogy and on the intron-exon structure of the genes. Subsequently, three and then four distinct types of plant GSTs were recognized including type I (GSTs with herbicide-detoxifying activity), type II (GSTs close to the mammalian zeta GSTs), type III (consisted mainly of auxin-induced GSTs), and type IV (GSTs similar to classical mammalian theta enzymes) isoforms (Droog et al., 1995; Droog, 1997). In fact, with the accumulation of biochemically characterized plant GSTs in the late 1990s, it appeared that some plant GSTs clearly grouped with specific mammalian GSTs, whereas others seemed plant-specific. Together with the release of the genome of Arabidopsis thaliana, this contributed to the establishment of a refined phylogenetic classification in plants using the principle of Greek-letter designations, which was widely used for non-plant GSTs (Dixon et al., 1998). GSTs are designated by using a 2 letter-code corresponding to the species (At for A. thaliana) followed by the 3 letters “GST,” a Greek or Latin letter designating the class, and a number distinguishing members of the same class. Thus, in A. thaliana, isoform 1 of the Phi (F) class is designated by “AtGSTF1”. This classification introduced in plants the Phi (replacing former Type I), Zeta (replacing former Type II), Tau (replacing former Type III), and Theta (replacing former Type IV) classes as well as two groups more distantly related to other known plant GSTs forming the Lambda (L) and dehydroascorbate reductase (DHAR) classes (Dixon et al., 2002). The last phylogenetic study performed a few years ago using well-annotated genomes of terrestrial plants (A. thaliana, Hordeum vulgare, Oryza sativa, Physcomitrella patens, Pinus tabulaeformis, Populus trichocarpa, and Solanum lycopersicum) and selecting only proteins possessing the two regular N- and C-terminal domains (see below), led to the identification of 14 GST classes: phi (F), tau (U), theta (T), zeta (Z), lambda (L), hemerythrin (H), iota (I), ure2p, glutathionyl-hydroquinone reductase (GHR), elongation factor 1B gamma (EF1Bγ), DHAR, tetrachlorohydroquinone dehalogenase (TCHQD), metaxin, and microsomal prostaglandin E synthase type-2 (mPGES-2) (Lallement et al., 2014). Some of these classes are found among different kingdoms, such as Zeta or Theta classes whereas Lambda, Tau and DHAR classes are specific to plants. The Phi class is sometimes presented in the literature as specific to the plant kingdom but similar sequences have been identified in some fungi, bacteria, and protists (Morel et al., 2013; Munyampundu et al., 2016). Although it has some limitations, the primary sequence remains to date the most convenient criterion for classifying these proteins.
The evolutionary history of GSTs seems relatively complex and several scenarios have been proposed. Because Theta class GSTs were present in bacteria, the first model of evolution, dating from the early 1990s, proposed that canonical (soluble) GSTs of plants, animals, and fungi have evolved from this ancestral gene as a result of duplications followed by divergent evolution (Pemble and Taylor, 1992). In subsequent years, this model was discarded by taking into account the biochemical properties including the nature of the catalytic residue, but also the oligomeric state of the proteins, and their tridimensional structure when solved (Frova, 2006; Mashiyama et al., 2014). The structural data notably showed that the N-terminal domain of soluble GSTs adopted a TRX fold, suggesting that the evolutionary history of soluble GSTs is linked to one of the TRX superfamily members. In this model, soluble GSTs were proposed to have evolved from a TRX/GRX ancestor to which a C-terminal helical domain has been added. Subsequent major transitions are the result of the dimerization of some GSTs, the replacement of the ancestral catalytic cysteine by a serine, and finally the change of this residue by a tyrosine in many mammalian GST classes. Although these major steps likely remain true, the current model is still incomplete, as it does not include the most recently identified classes such as mPGES2, GHR, Metaxin, Hemerythrin, Iota, and Ure2p, just to cite classes present in plants.
Gene Organization and Distribution of Ser-GSTs in Eukaryote Photosynthetic Organisms
Among the 14 classes previously identified in terrestrial plants (Lallement et al., 2014), only five classes (Tau, Phi, Zeta, Theta, and TCHQD) clearly contain members possessing a conserved serine in their active site, even though this serine is absent in some isoforms. The DHAR, Hemerythrin, Iota, Lambda, GHR, mPGES2, and metaxin classes belong to the Cys-GSTs, as they primarily contain members possessing a conserved cysteine in their active site. For the EF1Bγ and Ure2p classes, the nature of the residue promoting GSH activation remains uncertain. Although this classification is based on the primary sequences, the recent release of several plant genomes allowed for its correlation with the intron-exon structure of GST-encoding genes as analyzed in P. trichocarpa (Lan et al., 2009), P. patens (Liu et al., 2013), Capsella rubella (He et al., 2016), S. lycopersicum (Islam et al., 2017), Ipomoea batatas (Ding et al., 2017), and Brassica rapa (Khan et al., 2018). Indeed, the number of exons is generally conserved for genes belonging to the same class, e.g., 1 or 2 for genes encoding GSTUs, 3 for GSTFs, 9 or 10 for GSTZs, 7 for GSTTs, and 2 for TCHQDs.
Hence, combining the gene structure analysis with protein motifs specific to GST classes, sequence alignments and phylogenetic trees provide a robust view of the Ser-GST gene copy number present in a given organism. A comparative genomic analysis was carried out using 39 sequenced photosynthetic organisms available in Phytozome database [version 12 (Goodstein et al., 2012)] including 3 chlorophytes, 1 bryophyte, 1 lycophyte, and 34 angiosperms; clearly extending previous genomic surveys (Table 1; Ding et al., 2017; Monticolo et al., 2017; Plomion et al., 2018). 1859 sequences were retrieved by BLASTp (Basic Local Alignment Search Tool) using A. thaliana GST sequences as queries and standard parameters. It is worth noting that chlorophytes but not terrestrial plants contain Tyr-GST isoforms (respectively, 6, 4, and 7 in Chlamydomonas reinhardtii, Micromonas pusilla, and Volvox carteri) also shared by animals. The presence of such isoforms likely compensates the absence or low number of Ser-GSTs in these organisms (1 isoform for both C. reinhardtii and V. carteri, 0 for M. pusilla). In the following paragraphs, we emphasize the major features of the different Ser-GST classes, i.e., gene content and protein sequence characteristics.
Table 1
| GSTU | GSTF | GSTT | GSTZ | TCHQD | |
|---|---|---|---|---|---|
| Chlorophyte | |||||
| Chlamydomonas reinhardtii v5.5 | 0 | 0 | 1 | 0 | 0 |
| Volvox carteri v2.1 | 0 | 0 | 1 | 0 | 0 |
| Micromonas pusilla CCMP1545 v3.0 | 0 | 0 | 0 (∗) | 0 | 0 |
| Embryophyte | |||||
| Physcomitrella patens v3.3 | 0 (∗) | 9 | 2 | 1 | 5 (∗) |
| Tracheophyte | |||||
| Selaginella moellendorffii v1.0 | 38 | 1 | 3 | 2 | 1 |
| Angiosperm | |||||
| Amborella trichopoda v1.0 | 22 | 4 | 1 | 2 | 1 |
| Grass | |||||
| Brachypodium distachyon v3.1 | 40 | 21 | 1 | 3 | 1 |
| Oryza sativa v7_JGI | 45 | 16 | 1 | 4 | 1 |
| Panicoideae | |||||
| Setaria italica v2.2 | 48 | 16 | 1 | 5 | 1 |
| Sorghum bicolor v3.1.1 | 53 | 17 | 2 | 4 | 1 |
| Zea mays Ensembl-18 | 34 | 10 | 2 | 2 | 1 |
| Eudicot | |||||
| Aquilegia coerulea v3.1 | 24 | 29 | 3 | 2 | 1 |
| Pentapetalae | |||||
| Asterid | |||||
| Mimulus guttatus v2.0 | 17 | 5 | 2 | 0 (∗) | 1 |
| Solanum lycopersicum iTAG2.4 | 45 | 4 | 3 | 2 | 1 |
| Solanum tuberosum v4.03 | 50 | 4 | 1 | 2 | 1 |
| Rosid | |||||
| Eucalyptus grandis v2.0 | 62 | 19 | 1 | 2 | 7 (∗) |
| Vitis vinifera Genoscope.12X | 36 | 8 | 1 | 3 | 1 |
| Quercus robur | 62 | 12 | 1 | 2 | 1 |
| Poplar-Malvidae | |||||
| Malpighiales | |||||
| Linum usitatissimum v1.0 | 30 | 11 | 4 | 3 | 2 |
| Manihot esculenta v6.1 | 44 | 8 | 4 | 2 | 1 |
| Populus trichocarpa v3.0 | 54 | 8 | 2 | 2 | 1 |
| Ricinus communis v0.1 | 31 | 4 | 3 | 2 | 1 |
| SBM | |||||
| Citrus | |||||
| Citrus sinensis v1.1 | 25 | 6 | 1 | 2 | 1 |
| Citrus clementina v1.0 | 42 | 8 | 2 | 3 | 1 |
| Brassicales-Malvales | |||||
| Theobroma cacao v1.1 | 36 | 9 | 1 | 2 | 1 |
| Brassicaceae | |||||
| Arabidopsis lyrata v2.1 | 29 | 13 | 1 | 2 | 1 |
| Arabidopsis thaliana TAIR10 | 28 | 13 | 3 | 2 | 1 |
| Boechera stricta v1.2 | 27 | 12 | 1 | 1 | 1 |
| Brassica rapa FPsc v1.3 | 38 | 20 | 2 | 2 | 1 |
| Capsella grandiflora v1.1 | 21 | 10 | 1 | 2 | 1 |
| Capsella rubella v1.0 | 26 | 12 | 1 | 1 | 1 |
| Eutrema salsugineum v1.0 | 24 | 11 | 1 | 2 | 1 |
| Fabidae | |||||
| Nitrogen-fixing | |||||
| Cucumis sativus v1.0 | 24 | 3 | 1 | 2 | 1 |
| Fragaria vesca v1.1 | 28 | 5 | 1 | 2 | 1 |
| Glycine max Wm82.a2.v1 | 50 | 10 | 3 | 3 | 2 |
| Malus domestica v1.0 | 34 | 10 | 1 | 4 | 1 |
| Medicago truncatula Mt4.0v1 | 47 | 10 | 2 | 2 | 1 |
| Phaseolus vulgaris v2.1 | 24 | 12 | 2 | 2 | 2 |
| Prunus persica v2.1 | 47 | 9 | 1 | 2 | 1 |
Ser-GST gene content in sequenced chlorophytes and embryophytes.
Sequences have been retrieved from Phytozome v12.1, a Joint Genome Institute database. (∗) Unusually high/divergent gene copy number or absence of a given gene for a few specific organisms must be regarded with cautious as this may originate from bad genome assemblies or annotation problems or from remaining pseudogenes or gene alleles.
GSTs Theta (GSTTs)
In addition to plants, GSTTs are also found in animals, insects, fungi, and bacteria; suggesting that this class appeared early during evolution (Coggan et al., 2002; Bryant et al., 2006; Skopelitou et al., 2012; Han et al., 2016; Shao et al., 2017). In photosynthetic organisms, the number of GSTT genes ranges from 1 to 4 (Table 1). According to its early appearance, this is the only Ser-GST class present in the chlorophytes (green algae) analyzed. Its absence in M. pusilla might be due to a gene loss event during evolution, unless there are annotation problems. In organisms having 2 or more GSTT genes, the genes are often organized in cluster such as in A. thaliana (Dixon et al., 2002), Linum usitatissimum, Manihot esculenta or Ricinus communis suggesting that tandem duplication(s) occurred during evolution from an ancestral gene. Whether the resulting proteins have diverged in function remains to be explored. The GSTT proteins are generally about 250 amino acids long. The conserved serine is found around position 10 in a conserved SQPS active site signature, which (with a few exceptions) is conserved among mammals (SQPC) and insects (S[Q/A]PC). At the subcellular level, these proteins have a peroxisomal localization, which is consistent with the presence of C-terminal SK[I/M] targeting motif (Dixon et al., 2009). Peroxisomes are multifunctional organelles involved notably in the β-oxidation of fatty acids in plants, a catabolic pathway contributing in particular to the production of acetyl-CoA, NADH, and FADH2, but also many lipid peroxides from polyunsaturated fatty acids that are the likely physiological substrates of GSTTs. In vitro, these enzymes exhibit a weak GSH-conjugation activity against conventional model substrates but they have a high GSH-dependent peroxidase activity toward linoleic acid peroxides (Dixon and Edwards, 2009).
GSTs Zeta (GSTZs)
In addition to plants, GSTZs are also present in bacteria, fungi, and animals, even though this is often as a reduced number of isoforms. A possible reason is their specific involvement in a general process, the tyrosine catabolism (Edwards et al., 2011). In photosynthetic organisms used for the present analysis (Table 1), the number of GSTZ genes ranges from 1 to 5 but we could not find them in chlorophytes. They are often found as tandem duplicates in genomes such as in A. thaliana, C. rubella, or O. sativa.
At the protein level, GSTZs are about 225 amino acids long. The serine is located around position 20 and is included in a conserved SSC(S/A) active site signature, the first serine being the residue necessary for the GSH-conjugation reaction. The catalytic mechanism of GSTZs differs from other GSTs in that GSH is initially conjugated to the cis double bond of maleylacetoacetate allowing the isomerisation reaction, before being eliminated in a second step allowing the formation of fumarylacetoacetate (Thom et al., 2001). Thus, it is assumed that the conserved cysteine performs the deglutathionylation of the intermediate product at the manner of Cys-GSTs or as proposed also for TCHQDs (see below). However, bacterial GSTZs lacking this cysteine catalyze the same reaction.
Tetrachlorohydroquinone Dehalogenases (TCHQDs)
TCHQDs have been identified in animals, fungi and plants. Plant genomes usually contain a single gene but 2, 5, and 7 TCHQD-encoding genes were identified in Glycine max, P. patens, and Eucalyptus grandis, respectively (Table 1). At the protein level, TCHQDs are on average 265 amino acids long. These proteins were first discovered in the soil bacterium Sphingobium chlorophenolicum, that is able to use pentachlorophenol, a fungicide used in wood preservation, as a carbon source. During the enzymatic degradation of pentachlorophenol, this bacterial TCHQD (PcpC) catalyzes the reductive dehalogenation of tetrachlorohydroquinone to trichlorohydroquinone and then to dichlorohydroquinone (Xun et al., 1992). PcpC possesses a peculiar SCIS signature containing both a serine and a cysteine. Accordingly, it reduces chloroquinones in two steps. The first step requires the serine in the GSH-conjugation of the quinone causing the departure of a chloride ion. The second step is the removal of the glutathione moiety from the quinone, a reaction performed by a nucleophilic attack of the cysteine (Willett and Copley, 1996; Kiefer and Copley, 2002). In plant proteins, there is no cysteine in the signature (often SLDS) (Lallement et al., 2014). They should therefore not be able to carry out deglutathionylation steps and may have different substrates or reaction mechanisms. Another possibility is that other GST isoforms such as GHRs substitute to TCHQDs as they are able to catalyze quinone deglutathionylation (Lallement et al., 2015).
GSTs Phi (GSTFs)
The GSTF genes are found in all terrestrial non-vascular or vascular plants that have been analyzed, but are absent in green algae/chlorophytes, suggesting important functions for terrestrial life. The gene content is very variable between species as it ranges from 1 in Selaginella moellendorffii to 29 in Aquilegia coerulea (Table 1). In almost all genomes, a large part of these genes is organized as clusters indicating repetitive, species-specific duplications.
At the protein level, GSTFs are about 215 amino acids long and the serine is located around position 12. Given the higher number of isoforms compared to the above-described classes, the four residue signature (reminiscent of the TRX/GRX family) is more variable. Most isoforms have the conserved serine but a few, exemplified by PtGSTF8 (AVCP), AtGSTF11 (AANP), or AtGSTF12/TT19 (AACP) are lacking it. In fact, the presence of the serine is not mandatory for the GSH-conjugation reaction as shown in vitro using poplar GSTFs (Pégeot et al., 2017). Several subgroups have been distinguished previously, according notably to this signature but also depending on the presence of N- or C-terminal extensions (Pégeot et al., 2014). Also, it was observed that some isoforms containing a cysteine exhibit a more diversified activity profile, as they possess deglutathionylation activity in addition to the peroxidase and GSH-conjugation activities (Pégeot et al., 2017).
GSTs Tau (GSTUs)
In light of current genomic resources, GSTUs form a plant specific class as is also the case for DHARs and GSTLs; two Cys-GST classes. Except in rare instances, such as in Triticum aestivum and A. coerulea, which contains respectively 38 and 29 GSTFs vs 26 and 24 GSTUs (Gallé et al., 2009), the GSTU class represents the largest GST class. From the absence of GSTU in green algae and in the mosses P. patens and Sphagnum fallax, the presence of only 2 GSTUs in another moss: Marchantia polymorpha, but the presence of 38 GSTUs in the bryophyte S. moellendorffii, we conclude that these genes have rapidly and dramatically expanded between bryophytes and lycophytes. They became predominant in vascular plants, being supposedly required for novel functions associated to the lifestyle of these plants. In angiosperms, the GSTU gene content is variable and range from 21 (C. grandiflora) to 62 (Quercus robur and E. grandis) (Table 1). The phylogenetic analysis of this family indicates that large clades are formed by proteins from the same species pointing to the fact that species-specific expansions occurred (Plomion et al., 2018). They normally correspond to genomic clusters produced by several successive tandem duplication events as exemplified in poplar, A. thaliana or O. sativa (Wagner et al., 2002; Soranzo et al., 2004; Lan et al., 2009). Accordingly, it is difficult to define strict orthologs for a given isoform among the different species and to determine what is the set of GSTU ancestors shared by angiosperms.
Overall, the GSTU and GSTF classes represent around 75% of all GST genes as in Q. robur (62 out of 88 genes) (Plomion et al., 2018) or A. thaliana (41 out of 55 genes) (Dixon and Edwards, 2010a). For both classes, the expansion, specific genomic organization and high sequence similarity among duplicated members have important implications. One consequence may be the existence of functional redundancy between isoforms making it difficult to study the biological functions of a particular gene using reverse-genetic approaches. However, another consequence may be that upon duplication, some of the new gene copies, which have been less subject to evolutionary pressure and have accumulated mutations, have likely acquired structural and functional diversity. The truth is certainly in between and this remains to be experimentally addressed in a more exhaustive manner.
Biochemical Properties and Catalytic Activities of Ser-GSTs
GSTs are versatile enzymes, accommodating diverse substrates/ligands in the active site or L-sites (Table 2), and catalyzing diverse enzymatic reactions as a function of the active site signature (Chronopoulou et al., 2017a). Besides the so-called ligandin function, Ser-GSTs catalyze GSH-conjugation reactions on numerous types of substrates, the reduction of organic hydroperoxides or substrate isomerisation whereas Cys-GSTs rather catalyze opposite reactions including the reduction of glutathione conjugates. For the latter aspect, we invite the reader to refer to the recent review describing Cys-GST properties (Lallement et al., 2014). For all the catalytic activities, the high reactivity of a cysteine residue (either from GSH or from the polypeptide) plays a central role in the biochemical properties carried out by GSTs. Noteworthy, some Ser-GSTs belonging to the GSTZ, TCHQD, or GSTF classes possess a cysteine residue in the catalytic center, which confers them dual activity profile.
Table 2
| Isoform | Organism | Ligands | References |
|---|---|---|---|
| Bronze-2 or Bz2 (GSTU) | Zea mays | Cyanidin-3-glucoside | Marrs et al., 1995 |
| ZmGSTI-I, ZmGSTI-II, ZmGST II-II, ZmGST III-III (GSTU et GSTF) | Zea mays | Protoporphyrin IX, mesoporphyrin, coproporphyrin, uroporphyrin, Mg-protoporphyrin | Lederer and Böger, 2003 |
| ZmGSTU1 | Zea mays | Uroporphyrin, pentacarboxyl porphyrin, harderoporphyrin-SG, coproporphyrin, heme B | Dixon et al., 2008 |
| ZmGSTF1 | Zea mays | Gibberellic acid, indole-3-butyric acid, 2-naphtoxyacetic acid, 2,4-dichlorophenoxyacetic acid, kinetin, quercetin, luteolin | Axarli et al., 2004 |
| AtGSTU7 | Arabidopsis thaliana | Protoporphyrin-SG, myristoyl-glutathione | Dixon and Edwards, 2009, 2018 |
| AtGSTU19 | Arabidopsis thaliana | Harderoporphyrin-SG, chlorogenic acid, 10-S-glutathionyl-12-oxo-phytodienoic acid, oxylipin-SG, 3-methylindolyl glutathionyl disulfide, 12-oxo-phytodienoic acid (OPDA) | Dixon and Edwards, 2009, 2018 |
| Transparent testa 19 or TT19 (GSTF) | Arabidopsis thaliana | Anthocyanin | Kitamura et al., 2004 |
| AtGSTF2, AtGSTF3 | Arabidopsis thaliana | Norharmane, harmane, lumichrome, indole-3-aldehyde, quercetin-3-O-rhamnoside | Dixon et al., 2011a |
| AtGSTF2 | Arabidopsis thaliana | Indole-3-aldehyde, camalexin, quercetrin, quercetin | Ahmad et al., 2017 |
| AtGSTF2 | Arabidopsis thaliana | Grossamide K-SG, cannabisin, 10-S-glutathionyl-12-oxo-phytodienoic acid, kaempferol-3,7,4′-trimethylether, quercetin-3,7,3′,4′-tetramethylether | Dixon and Edwards, 2018 |
| Anthocyanin9 or An9 (GSTF) | Petunia hybrida | Anthocyanin | Alfenito, 1998 |
| Anthocyanin9 or An9 (GSTF) | Petunia hybrida | Isoquercitrin, quercetin, cyanidin, luteolin | Mueller et al., 2000 |
| GST | Hyoscyamus muticus | Indole-3-acetic acid | Bilang et al., 1993 |
| CkmGST3 (GSTF) | Cyclamen persicum ×Cyclamen purpurascens | Anthocyanin | Kitamura et al., 2012 |
| VvGST1 (GSTU), VvGST4 (GSTF) | Vitis vinifera | Anthocyanin | Conn et al., 2008 |
| VvGSTU2 | Vitis vinifera | Trans-resveratrol | Martínez-Márquez et al., 2017 |
| Flavonoid3 or Fl3 | Dianthus caryophyllus | Anthocyanin | Larsen et al., 2003 |
| PfGST1 (GSTF) | Perilla frutescens | Anthocyanin | Yamazaki et al., 2008 |
| AtGSTU9, AtGSTU10 | Arabidopsis thaliana | fatty acyl (C14,C16,C17, C18) | Dixon and Edwards, 2009 |
| AtGSTU25, AtGSTU28 | Arabidopsis thaliana | fatty acyl (C6,C8,C10,C12,C14) | Dixon and Edwards, 2009 |
| AtGSTF6 | Arabidopsis thaliana | Indole-3-acetonitrile | Su et al., 2011 |
| AtGSTU13 | Arabidopsis thaliana | Indole-3-ylmethyl-ITC (indole glucosinolate) | Piślewska-Bednarek et al., 2018 |
Ligands of plant Ser-GSTs identified.
GSH-Conjugating Activity
Most Ser-GSTs catalyze the conjugation of GSH onto electrophilic compounds including aromatic, aliphatic or heterocyclic compounds (Deponte, 2013). This conjugation requires the binding of a GSH molecule in the glutathione binding site (G-site). In most cases, the presence of a specific residue, most often a cysteine, serine, or tyrosine at the vicinity of the cysteine of the bound GSH, induces a decrease in the thiol pKa (Board and Menon, 2013; Deponte, 2013). This pKa, usually around 9, is lowered to approximately 6.5 or even less, promoting the formation of a nucleophilic (reactive) thiolate group in vivo, which is able to perform a nucleophilic attack on a nearby electrophilic substrate. In other words, the conserved serine in the active site of Ser-GSTs does not play the role of the catalytic residue; rather, this is carried out by the thiolate group of GSH. The nucleophilic attack initiated by GSH occurs either by a substitution (e.g., on a chlorine atom) or by the reduction of an electron acceptor via a Michael’s addition (Deponte, 2013). Several model substrates are used to measure GSH-conjugation activity, the most common being the 1-chloro-2,4-dinitrobenzene (CDNB). The detoxification of herbicides (e.g., atrazine and fluorodifen) and pesticides (e.g., alachlor and metolachlor) through GSH-conjugation has been extensively studied over the years, notably using GSTs from crops (Gronwald and Plaisance, 1998; Cummins et al., 2003; Cho et al., 2007).
Peroxidase Activity
In the cells, hydroperoxides are reduced by several families of enzymes, in particular thiol peroxidases including the peroxiredoxin (PRX) and glutathione peroxidase-like protein (GPXL) families (Rouhier and Jacquot, 2005). Some GSTs also exhibit peroxidase activity, as they are able to reduce (hydro)peroxides into alcohols concomitant to the oxidation of GSH into GSSG. In this reaction, deprotonated GSH bound to the G-site of the enzyme induces a nucleophilic substitution of the bond between the two oxygen atoms of the hydroperoxyl group (R-OOH), leading to the release of GSOR and an OH group that is protonated into H2O. GSOR is then cleaved into GSSG and ROH by a second nucleophilic substitution by the GSH molecule according to a mechanism that is not yet fully elucidated (Deponte, 2013). The GSTs performing this reaction have catalytic efficiencies (102 to 104 M-1⋅s-1) measured in steady-state conditions in the range of those of TRX- and GRX-dependent thiol peroxidases (Pégeot et al., 2017). A major difference is, however, the rate of the first step (i.e., peroxide reduction) that occurs at turnover numbers up to 107 s-1 for some thiol peroxidases. The contribution of GSTs in the reduction of cellular hydroperoxides remains poorly described but it might be important because these are almost the only GSH-dependent peroxidases, most thiol peroxidases being dependent on GRXs or TRXs, including GPXLs (Rouhier and Jacquot, 2005; Navrot et al., 2006). Also, their contribution appears to be different depending on the organisms and the subcellular compartments considered. Among Ser-GSTs, only those belonging to the Phi, Tau, and Theta classes are able to catalyze such reactions, peroxisomal GSTTs being likely specialized in the reduction of fatty acid peroxides (Dixon et al., 2009).
Isomerase Activity
Some GSTs, in particular from the Zeta class, catalyze the GSH-dependent isomerisation of specific metabolites, such as the cis-trans isomerisation of maleylacetoacetate into fumarylacetoacetate occurring during the penultimate step of tyrosine catabolism in eukaryotes (Fernández-Cañón and Peñalva, 1998; Thom et al., 2001; Fernandez-Canon et al., 2002; Edwards et al., 2011). In some bacteria, GSTZs function as maleylpyruvate isomerases (Marsh et al., 2008) and catalyze the isomerisation of maleylpyruvate into fumarylpyruvate through the third committed step in the degradation of salicylate to the metabolites pyruvate and fumarate via gentisate. Contrary to other activities described above, GSH is not considered as a substrate but as a cofactor because it is not consumed during the reaction (Litwack et al., 1971; Marsh et al., 2008).
Non-enzymatic Binding and Intracellular Transport
In addition to their catalytic function, GSTs also serve as non-enzymatic carrier proteins or ligandins (Habig et al., 1974; Mannervik and Danielson, 1988). The term “ligandin” was historically associated to proteins characterized in animals, which were able to bind a wide range of hydrophobic ligands, such as steroids, heme and its degradation product bilirubin, carcinogens, and bile salts. These proteins turned out to be GSTs from the Alpha and Mu classes (Levi and Arias, 1969; Ketley et al., 1975). Since then, plant GSTUs and GSTFs were found to bind several tetrapyrroles, e.g., protoporphyrin IX (Proto IX), Mg-protoporphyrin but also bacterial porphyrin derivatives identified upon expression of recombinant maize GSTUs (Lederer and Böger, 2005; Dixon et al., 2008). However, it is not clear whether this is strictly a ligandin function as Zea mays GSTU1 is able to catalyze the conjugation of GSH on protoporphyrinogen IX and harderoporphyrinogen (Dixon et al., 2008). Another example, likely the best described, concerns anthocyanins, which are phenolic antioxidant compounds conferring the colors we commonly associate with fruits and flowers. These molecules are transiently bound/transported by GSTs before their release to ABC-type transporters for vacuolar sequestration. This is documented for both Tau and Phi class members, i.e., Bz2 (GSTU) from maize, AN9, TT19, and VvGST4 (GSTFs) from Petunia hybrida, A. thaliana and Vitis vinifera, respectively (Marrs, 1996; Alfenito, 1998; Mueller et al., 2000; Kitamura et al., 2004; Conn et al., 2008; Gomez et al., 2011; Momose et al., 2013; Zhao, 2015). Other molecules were isolated using ligand fishing approaches (Table 2). GSTU2 from V. vinifera binds trans-resveratrol, a polyphenol transported from the cells into extracellular medium and conferring antimicrobial properties (Martínez-Márquez et al., 2017). GSTF2 and GSTF3 from A. thaliana were described in vitro to bind various ligands such as lumichrome, harmane, norharmane, indole-3-aldehyde, camalexin, and quercetin-3-O-rhamnoside (Dixon et al., 2011a) but the physiological significance remains unknown. In addition to a carrier function, it may be that the non-enzymatic binding of molecules prevents their oxidative damage, thus ensuring a protective effect (Mueller et al., 2000).
Structural Characteristics of Plant Ser-GSTs
Considering the high number of Ser-GSTs in plants, only a few structures have been solved so far: 27 for GSTUs, 15 for GSTFs, 1 for GSTZ, and none for TCHQD and GSTT (Table 3). All these plant Ser-GSTs are homodimeric enzymes in which each protomer of approximately 23–30 kDa contains two domains with a catalytic center at the interface. The N-terminal domain adopts the typical TRX-fold (with β1α1β2α2β3β4α3 topology) and the C-terminal domain is a bundle of at least five helices (α4 to α8) (Figure 1). GSTUs have an additional α9 helix that is oriented toward the active site without occluding it (Thom et al., 2002). In GSTFs, the α6-α7 connection systematically includes a small helix (α6′) (Reinemer et al., 1996; Pégeot et al., 2014, 2017). The atomic model of the only GSTZ crystal structure, that of A. thaliana GSTZ1, is incomplete between helices α4 and α5, which hinders the accurate determination of the active site (Thom et al., 2001). Within a class, the variable regions are often close to the active site and involved in the binding of the electrophilic substrate. In GSTUs, these regions include helix α9 and the segment from roughly the C-terminal end of α4 to the N-terminal end of α5 (Valenzuela-Chavira et al., 2017). In GSTFs, they include this segment and the connection β2-β3, which, in maize GSTF3, was supposed to move upon binding of the substrate in the active site (Neuefeind et al., 1997a). This connection is also involved in the dimer stabilization (see below).
Table 3
| Class | Organism | Name | Ligand | PDB Entry | References |
|---|---|---|---|---|---|
| Arabidopsis thaliana | AtGSTF2 | GTXa (1GNW)b, FOEa (1BX9)b, 7WBa (5A5K)b, QUEa (5A4V)b, I3Aa (5A4U)b, QCTa (5A4W)b | 1GNW, 1BX9, 5A5K, 5A4V, 5A4U, 5A4W | Reinemer et al., 1996; Prade et al., 1998; Ahmad et al., 2017 | |
| Phi | Populus trichocarpa | PtGSTF1 | GSHa (4RI6)b, GSHa (4RI7)b | 4RI6, 4RI7c | Pégeot et al., 2014 |
| PtGSTF2 | 5EY6 | Pégeot et al., 2017 | |||
| PtGSTF5 | GSHa | 5F05 | |||
| PtGSTF7 | GSHa | 5F06 | |||
| PtGSTF8 | GSHa | 5F07 | |||
| Zea mays | ZmGSTF1 | CYWa (1AXD)b, ATAa (1BYE)b | 1AXD, 1BYE | Neuefeind et al., 1997a; Prade et al., 1998 | |
| ZmGSTF3 | 1AW9 | Neuefeind et al., 1997b | |||
| Triticum aestivum | TaGSTU4-4 | GTXa | 1GWC | Thom et al., 2002 | |
| Arabidopsis thaliana | AtGSTU20/ FIP1 | GSHa | 5ECS, 5ECR, 5ECQ, 5ECP, 5ECO, 5ECN, 5ECM, 5ECL, 5ECK, 5ECI, 5ECH | Chen et al., 2017 | |
| AtGSTU23 | GSHa (6EP7)b | 6EP6, 6EP7, 5O84 | Tossounian et al., 2018d | ||
| AtGSTU25 | GSSG | 5G5A | d | ||
| Tau | Glycine max | GmGSTU4 | GTBa (2VO4, 5AGY)b, GSHa (4TOP)b | 2VO4, 4TOP, 5AGYe | Axarli et al., 2009a,b; Burmeister et al., 2008 |
| GmGSTU10-10 | GS8a | 4CHS | Skopelitou et al., 2015 | ||
| Mangifera indica | MiGSTU | GSHa (5G5F), GTXa (5KEJ) | 5G5E, 5G5F, 5KEJ | Valenzuela-Chavira et al., 2017 | |
| Oryza sativa subsp. japonica | OsGSTU1 | GSHa | 1OYJ | Dixon et al., 2003 | |
| Populus trichocarpa | PtGSTU30 | GSHa | 5J4U, 5J5Nf | Yang et al., 2019d | |
| Ricinus communis | EFI-501866 | 4J2F | d | ||
| Zeta | Arabidopsis thaliana | GSTZ1 | 1E6B | Thom et al., 2001 | |
Crystal structures of Ser-GSTs from plants.
aPDB ligand codes: GSH, Glutathione; GTX, S-hexylglutathione; FOE, FOE-4053-glutathione conjugate; 7WB, Camalexin; QUE, Quercetin; I3A, Indole-3-aldehyde; QCT, Quercetrin; CYW, Lactoylglutathione; ATA, Atrazine glutathione conjugate; GTB, S-(P-Nitrobenzyl)glutathione; GS8, S-Hydroxy-glutathione. bPDB, entry where the ligand is present. c4RI7: crystal structure of an isoform of PtGSTF1, which contains mutation of S13 to C. dcrystal structures only available in the PDB. e5AGY: crystal structure of an isoform of GmGSTU4, which contains mutations of I183 to V, Q46 to K, R38 to Q, and W114 to C. f5J5N: crystal structure of an isoform of PtGSTU30, which contains mutation of R39 to W.
FIGURE 1
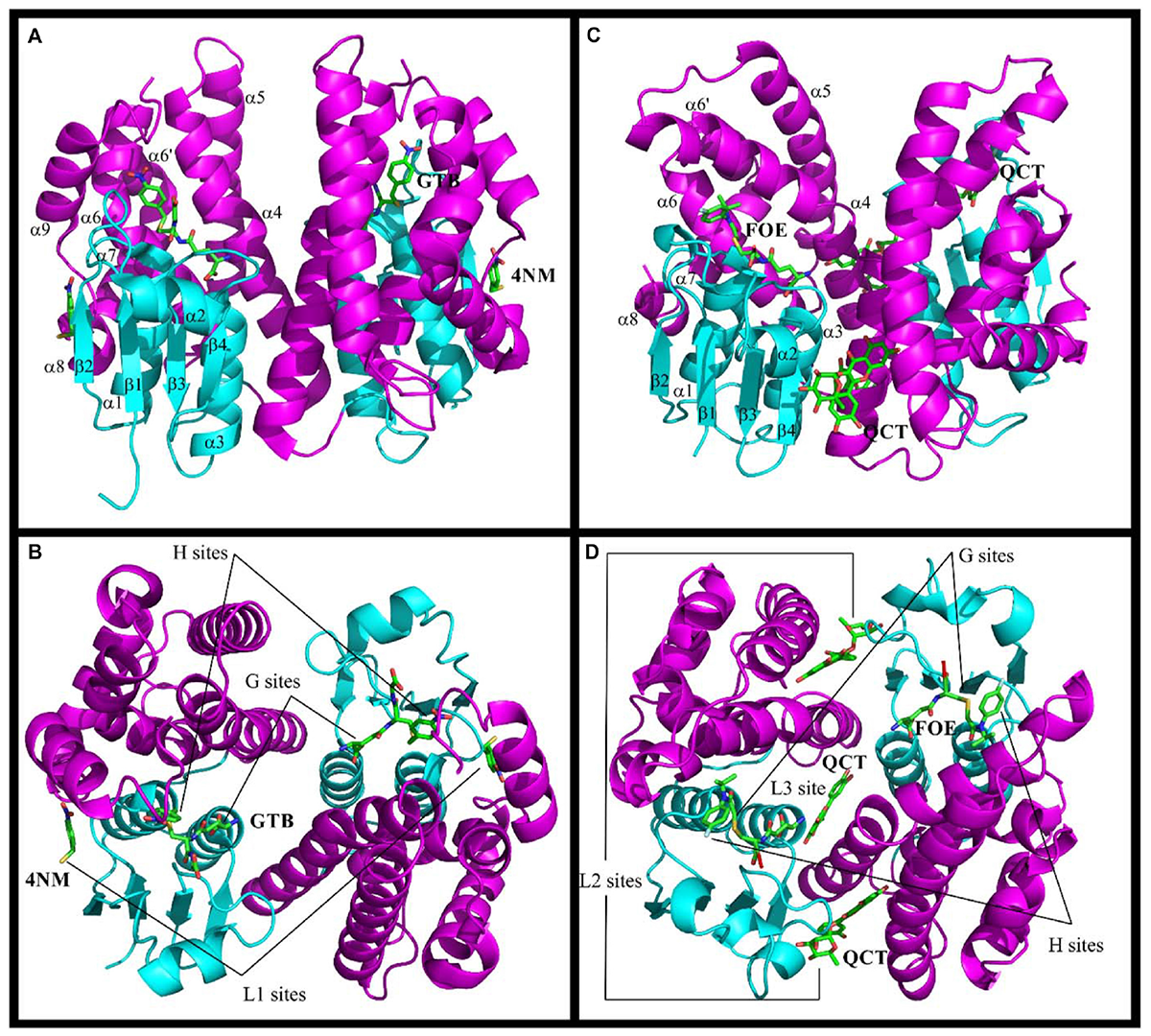
Structures of Ser-GSTs from plants highlighting the location of ligand-binding sites. (A–D) schematic structure of the GmGSTU4 and AtGSTF2 dimers, respectively. (C,D) illustrate the complexes formed between AtGSTF2 and FOE (1BX9) or QCT (5A4W). The secondary structures and the location of the ligand-binding sites are labeled. The TRX domain is in cyan and the C-terminal domain is in magenta. The labeled ligands are: GTB, S-(P-Nitrobenzyl)glutathione; 4NM, 4-Nitrophenyl methanethiol; FOE, FOE-4053-glutathione conjugate; QCT, Quercetrin.
Concerning dimerization, the GSTU dimer has an open V-shaped configuration with 2200 Å2 of the accessible surface that is buried at the interface, comparable with that of GSTOs (2000 Å2), but smaller than that of GSTFs (2700 Å2) and most other classes of GSTs (2800–3400 Å2) (Axarli et al., 2010). The monomers are related by a two-fold symmetry where the N-terminal domain of one subunit cross-interacts with the C-terminal domain of the second one, and vice versa. The contact regions are the loop α2-β2, the strand β3 and the helix α3 of one monomer and the helices α4 and α5 of the other. The dimerization interface involves hydrophobic surface patches and a particular lock-and-key motif in which the side-chain of an aliphatic or aromatic residue extends across the dimer interface (Val52 in TaGSTU4 and Phe53 in PtGSTF1, Table 3). In GSTUs, conserved salt bridges close to the dyad axis bind both subunits. In GSTFs, the number and the nature of the polar interactions vary significantly from one isoform to another. Indeed, a single hydrogen bond connects the two subunits of AtGSTF2 (Reinemer et al., 1996) whereas nine are found in PtGSTF1 (Pégeot et al., 2014).
The GST catalytic center is usually divided in two distinct functional regions, a hydrophilic G-site for binding glutathione, and an adjacent hydrophobic H-site for accommodating electrophilic substrates. The anchoring residues of the G-site are well conserved among all GSTs probably because of their high specificity for glutathione. These residues are highlighted in the structural alignments (Figure 2). In Ser-GSTs, the GSH thiol group is normally hydrogen bonded to the hydroxyl group of the catalytic serine (Ser13 in PtGSTF1) (Pégeot et al., 2014). However, this serine is important but not mandatory for GSH-conjugating reactions as concluded from mutagenesis studies or its absence in some GSTFs (Pégeot et al., 2014). In poplar GSTFs, nearby hydroxylated residues present in the active site signature (often STxT) could be involved in GSH activation (Pégeot et al., 2017). Generally speaking, the H-site is built from elements from both the N- and C-terminal domains. The observed variations reflect the broad electrophilic-substrate specificities of the different GST isoforms/classes. Only AtGSTF2 and ZmGSTF1 crystal structures were obtained in the presence of herbicidal-glutathione conjugates (Figure 1; Prade et al., 1998). In other cases, the H-sites have been defined from the presence of inhibitors such as S-hexylglutathione or molecules from the crystallization medium. In the large majority of cases, the putative H-site residues are hydrophobic in nature. In GSTFs, the H-site involves residues located around the catalytic serine (N-terminal end of helix α1), in the loop β2-α2 and in the C-terminal end of helix α4. In GSTUs, residues from two additional regions are concerned, namely the helix α6 and the additional helix α9. A conserved tryptophan is present in the helix α6 of GSTUs (Trp171 in TaGSTU4) (Thom et al., 2002). In GSTFs, aromatic residues identified by mutagenesis studies have been clearly demonstrated as participating in the affinity toward electrophilic substrates (Axarli et al., 2004; Dixon et al., 2011a).
FIGURE 2

Structure-based sequence alignments of Tau class (A) and Phi class (B) GSTs from plants. The sequence alignment was generated with Chimera (Pettersen et al., 2004) and manually adjusted. Crystal structures and sequences are available at the Protein Data Bank (http://www.rcsb.org): 1GWC for TaGSTU4, 5ECS for AtGSTU20, 6E6P for AtGSTU23, 5G5A for AtGSTU25, 2VO4 for GmGSTU4, 4CHS for GmGSTU10, 5G5E for MiGSTU1, 1OYJ for OsGSTU1, 5J4U for PtGSTU30, 4J2F for RcGSTU1, 1GNW for AtGSTF2, 4RI6 for PtGSTF1, 5EY6 for PtGSTF2, 5F05 for PtGSTF5, 5F06 for PtGSTF7, 5F07 for PtGSTF8, 1AXD for ZmGSTF1, and 1AW9 for ZmGSTF3. Secondary structures are labeled and shown using arrows (β-strands) and squiggles (helices). The active site serine, the invariant proline and the quasi-invariant aspartic acid are in bold type, colored white, highlighted black, and marked with  . Residues that participate in dimer stabilization via strong polar interactions are in bold and marked with ∗. Residues involved in binding glutathione (G-site) are in bold type, highlighted yellow, and marked with
. Residues that participate in dimer stabilization via strong polar interactions are in bold and marked with ∗. Residues involved in binding glutathione (G-site) are in bold type, highlighted yellow, and marked with  . Residues of the characterized H-sites are in bold type, highlighted green, and marked with
. Residues of the characterized H-sites are in bold type, highlighted green, and marked with  . Residues of the L1-site (GmGSTU4, 2VO4) are in bold type, highlighted red, and marked with
. Residues of the L1-site (GmGSTU4, 2VO4) are in bold type, highlighted red, and marked with  . Residues of the L2-site (AtGSTF2, 5A4U, 5A4V, and 5A4W) are in bold type, highlighted blue, and marked with
. Residues of the L2-site (AtGSTF2, 5A4U, 5A4V, and 5A4W) are in bold type, highlighted blue, and marked with  . Residues of the L3-site (AtGSTF2, 5A4K, 5A4U, and 5A4W) are in bold type, highlighted pink, and marked with
. Residues of the L3-site (AtGSTF2, 5A4K, 5A4U, and 5A4W) are in bold type, highlighted pink, and marked with  .
.
The structures of GSTs include other important regions that are associated with non-catalytic functions. AtGSTU20, also named FIP1 (FIN219-interacting protein 1) because it interacts with the jasmonate-amido synthetase FIN219, participates in the jasmonate signaling response under far-red light conditions (Chen et al., 2007, 2017). In a crystallographic study, it was shown that the formation of the FIP1-FIN219 complex results in the reorientation of the FIN219 C-terminal domain, which appears crucial for improving jasmonoyl-isoleucine biosynthesis. However, the overall conformation of AtGSTU20 is not altered and the FIN219-binding region includes the C-terminal α6 to α8 helices. Interestingly, the authors noted that some of the contact residues are well conserved in GSTUs and GSTs of other species (Figure 2; Chen et al., 2017). For their ligandin function, GSTs bind a wide range of compounds in a non-catalytic manner at so-called L-sites, which are often distinct from the active site. Three different L-sites were described in GSTs from plants (Figure 1). The structural analysis of GmGSTU4-4 revealed the presence of one molecule of (4-nitrophenyl)methanethiol in each subunit in a hydrophobic surface pocket (L1-site) (Axarli et al., 2016). The bottom and walls of the L1-site are lined with residues from α1, β2, and α8. The main binding residues are conserved in GSTUs. The crystal structures of AtGSTF2 in complex with two indole derivatives and two flavonoids revealed two other ligand-binding sites (L2 and L3) (Ahmad et al., 2017) extending the observation of tight protein-ligand interactions (Kd < 1 μM) by isothermal titration calorimetry (Dixon et al., 2011a). The L2 site is situated between helices α4 and α7 in each monomer whereas the L3 site is located at the base of the dimer interface involving helices α3 of one subunit and α4 of its neighbor (Figure 1). All ligands are stabilized mainly through hydrophobic interactions (Ahmad et al., 2017). Coupled to biochemical evidence, the presence of these non-catalytic L-sites in GSTUs and GSTFs suggest that at least some of them should function in the transport of endogenous metabolites (Dixon et al., 2011a). However, the residues forming these L-sites are difficult to identify because they are not well-conserved among plant GSTs (Ahmad et al., 2017).
Gene Expression of Ser-GSTs in A. thaliana
Analyzing the transcript abundance of these GSTs could be helpful to understand the possible redundancy between close/duplicated isoforms as well as to give clues about their functions in the absence of molecular and genetic information. In fact, it is quite well documented in many species that the expression of GSTF and GSTU genes is often induced in response to environmental constraints. This includes heavy-metal exposure (Moons, 2003; Ahsan et al., 2008; Reid et al., 2013; Tripathi et al., 2014), salinity, heat, cold, drought (Jha et al., 2011; Tiwari et al., 2016; Yang et al., 2016; Xu et al., 2018; Srivastava et al., 2019), or biotic interactions such as pathogenic interaction (Rinaldi et al., 2007; Skopelitou et al., 2015; Kao et al., 2016). The expression of several GSTF and GSTU genes is also enhanced in response to phytohormones including abscisic acid, auxin, ethylene, methyl jasmonate and salicylic acid, to herbicides and to herbicide safeners, and more generally to treatments leading to an oxidative stress (DeRidder, 2002; Wagner et al., 2002; Lieberherr et al., 2003; Smith et al., 2003, 2004; Sappl et al., 2004, 2009; Chen et al., 2012; Chronopoulou et al., 2017b). Thus, using A. thaliana as a representative organism, the transcript abundance of 44 out of the 47 Ser-GST genes was retrieved from the AtGenExpress datasets. The expression profiles of GSTF2 (At4g02520), GSTF7 (At1g02920), and TCHQD (At1g77290) were not available and are therefore not present in this gene expression analysis.
First, we examined the expression profiles of Ser-GST genes in the context of a developmental time-course in Arabidopsis (Figure 3), using the AtGenExpress Developmental Set (Schmid et al., 2005). In this case, the transcript abundance of each gene was standardized using z-score transformation (a form of normalization that is particularly useful when comparing samples from diverse treatments/tissue backgrounds), and arranged by classes. One can clearly see that most GSTFs and GSTUs, as well as the two GSTZs, have their highest expression in roots. However, for the three GSTTs, the highest expression is found in samples spanning seed development (siliques/seeds), particularly in those containing isolated maturing seeds. In addition, these GSTTs also exhibited increased transcript abundance during developmental leaf senescence (DLS), however, this cannot be relied upon too strongly as only one time point is included in this dataset. Interestingly, within the GSTF family, GSTF5 is only highly expressed at the two first stages of siliques/seeds (siliques bearing developing seeds), a specificity that is also found for GSTF12, albeit the transcript abundance also peaks during DLS. The GSTF3, 6, 8, 9, 10, and 11 genes are also expressed in leaves and in whole plant stages, but to a much lower level than in roots. As already said, most of the GSTUs display their highest expression levels in roots, however, GSTU4, 9, 15, and 16 are exclusively and strongly expressed at the siliques/seeds stages. An exception is GSTU23, which exhibits its highest transcript abundance in both roots and siliques/seeds stages. Finally, many Ser-GSTs appear to have transient expression during whole plant, leaf, flower, and stem development, which could suggest very specific functions in response to developmental cues.
FIGURE 3
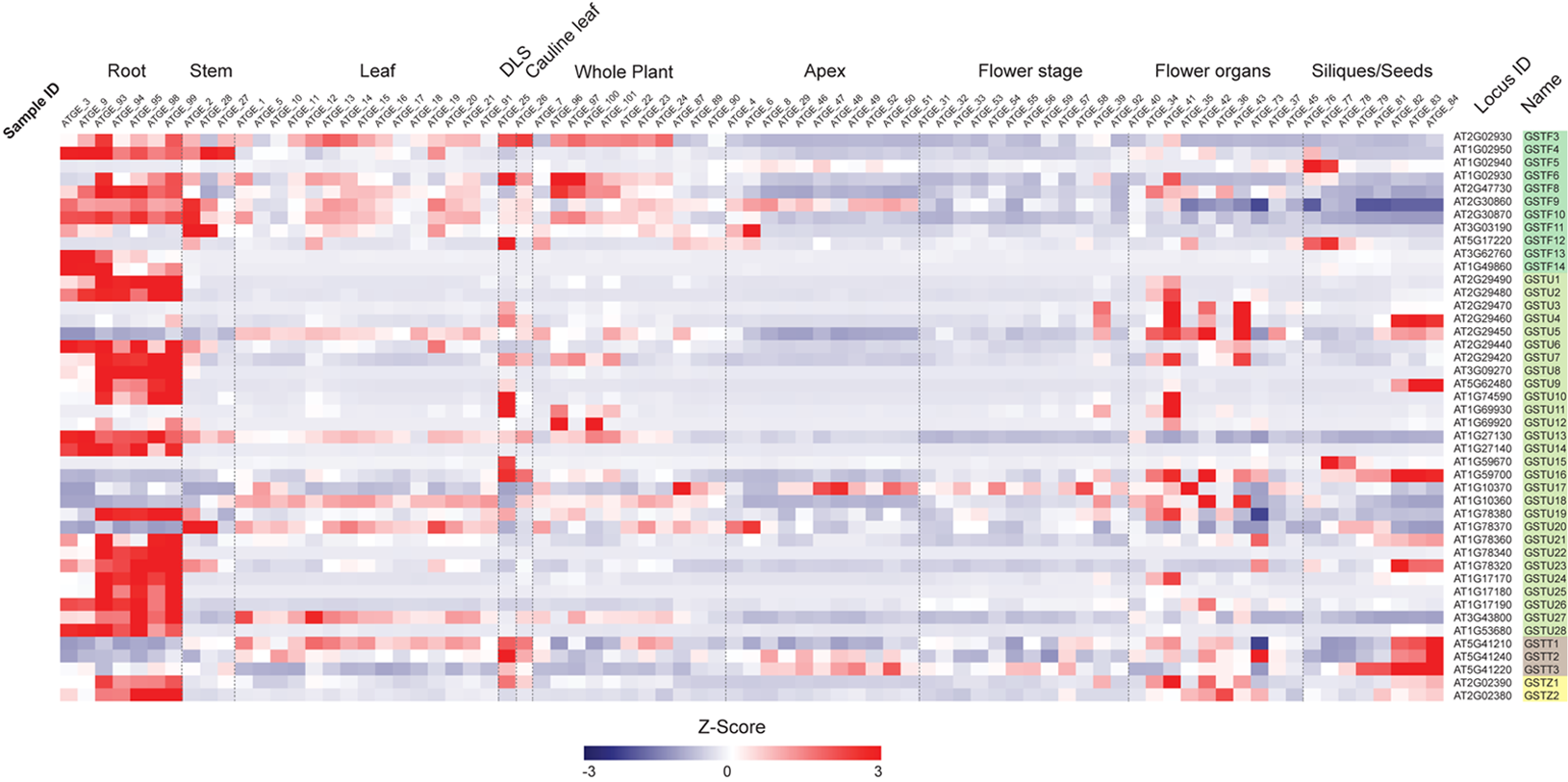
Transcript abundance of 44 Ser-GST genes during Arabidopsis development. Microarray experimental data (generated using Affymetrix ATH1 GeneChip arrays) of Arabidopsis Development (AtGenExpress Developmental Expression Atlas) described by Schmid et al. (2005) were obtained from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/). Intensity values of replicates were averaged and z-score transformed across the following ten developmental conditions: roots, stems, rosette leaves, developmental leaf senescence (DLS), cauline leaf, whole plant, apex, flowering stages, flower organs, and siliques/seeds. The data were then imported into The Institute for Genomic Research Multiple Experiment Viewer (MVE) and hierarchically clustered using average linkage based on Euclidean distance. Gene families of each gene are indicated using the following color key: tau, light green; phi, medium green; theta, gray; and zeta, yellow.
In a second stage of analysis, we examined the expression profile of these same 44 Ser-GSTs in response to a number of abiotic stresses [AtGenExpress Stress Set; (Kilian et al., 2007)]. Data are presented as a log2 fold-change of the stress treatment (at a given time point) versus its respective control sample, and the entire dataset is hierarchically clustered using Euclidean distance (Figure 4). This analysis clearly demonstrates that most Ser-GSTs strongly respond to stresses applied to the roots and thus, substantiates the high expression profiles observed in roots from the developmental stage analysis (Figure 3). Interestingly, GSTs present in clusters 3, 4, 5, 6, and 7 exhibit high transcript abundance in response to temperature changes in roots (cold and heat). That said, the five GSTs comprising clusters 3, 4, and 5 additionally exhibit high transcript abundance in response to almost all stresses, in both aerial and subterranean tissues. Also of note here, whereas the 2 GSTs of cluster 1, (GSTF11 and GSTU20) have an overall low fold-change of their transcript abundance in response to all stresses as compared to most of the other GSTs, the 6 GSTs included in cluster 2 seem to consistently respond to osmotic changes, salt and drought stresses in both root and aerial parts. Altogether, this clearly indicates that Ser-GSTs are involved in the molecular responses to several environmental cues, both biotic and abiotic, a fact that is discussed further in the next chapter.
FIGURE 4
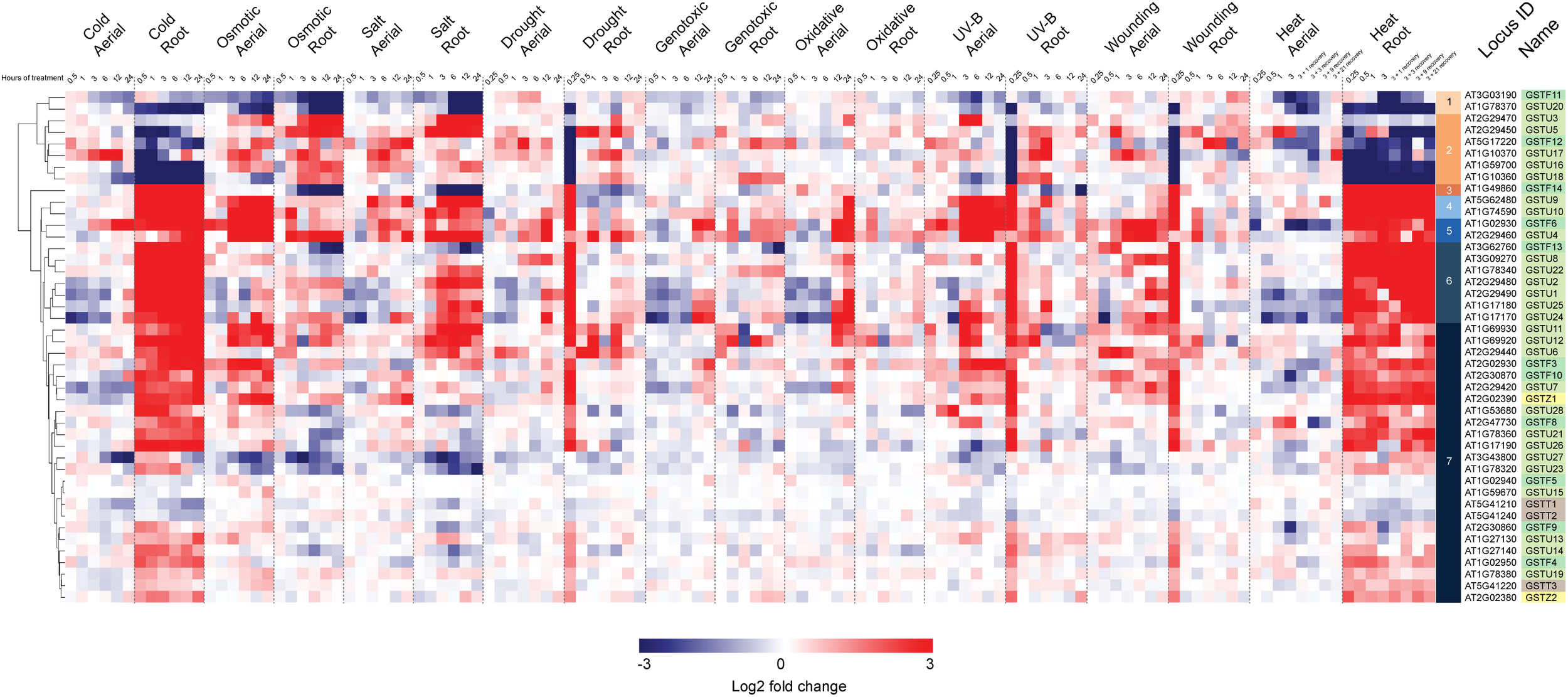
Hierarchical clustering of log2 fold changes of 44 Ser-GST genes in response to abiotic stresses. Microarray experimental data (generated using Affymetrix ATH1 GeneChip arrays), described by Kilian et al. (2007), were obtained from the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/). Intensity values of replicates were averaged and z-score transformed across nine stresses (cold, osmotic, salt, drought, genotoxic, UV, wounding, and heat) grouped as either aerial or root tissue, and further defined according to a time course of exposure in hours. Hierarchical clustering was carried out by average linkage based on Euclidean distance using in the Multiple Experiment Viewer (MEV) analysis package, resulting in the defining of 7 clusters. Gene families of each gene was indicated using the following color key: tau, light green; phi, medium green; theta, gray; and zeta, yellow.
Physiological Roles of Ser-GSTs in A. thaliana
As highlighted above, Ser-GSTs constitute the largest group of GSTs in plants. Although several molecular and biochemical studies have shed light on their tridimensional structures, biochemical properties, and enzymatic activities, very little is known about the actual roles that these proteins play in planta. This lack of knowledge might reflect the functional redundancy that most probably exists between these GSTs (Sappl et al., 2009; Rahantaniaina et al., 2017). Nevertheless, the use of the model plant A. thaliana has aided in deciphering the role played by some of the 47 Ser-GSTs (13 GSTFs, 28 GSTUs, 1 TCHQD, 2 GSTZs, and 3 GSTTs) (Table 4; Wagner et al., 2002; Dixon and Edwards, 2010a).
Table 4
| GST | Gene ID | Acronym | Known role | Mutant characterized | References |
|---|---|---|---|---|---|
| GSTU1 | At2g29490 | GST19 | None | ||
| GSTU2 | At2g29480 | GST20 | None | ||
| GSTU3 | At2g29470 | GST21 | None | ||
| GSTU4 | At2g29460 | GST22 | None | ||
| GSTU5 | At2g29450 | GSTU1, AT103-1A | UV radiation acclimation | GSTU5-OE | Liu and Li, 2002 |
| Excess light acclimation response | None | Lv et al., 2015 | |||
| GSTU6 | At2g29440 | GST24 | None | ||
| GSTU7 | At2g29420 | GST25 | Part of the lipid stress response | None | Stotz et al., 2013 |
| GSTU8 | At3g09270 | None | |||
| GSTU9 | At5g62480 | GST14, GST14B | Salt and drought stress response | None | Ambrosone et al., 2015 |
| GSTU10 | At1g74590 | None | |||
| GSTU11 | At1g69930 | None | |||
| GSTU12 | At1g69920 | None | |||
| GSTU13 | At1g27130 | GST12 | Indole glucosinolate biosynthesis / Response to fungal pathogens (E. pisi, C. gloeosporioides and P. cucumerin) | gstu13 | Piślewska-Bednarek et al., 2018 |
| GSTU14 | At1g27140 | GST13 | Part of the excess light acclimation response | None | Lv et al., 2015 |
| GSTU15 | At1g59670 | None | |||
| GSTU16 | At1g59700 | Part of the plant response to fungal pathogens (V. dahliae) | None | Pantelides et al., 2010 | |
| GSTU17 | At1g10370 | ERD9, GST30, GST30B | Drought and salt stress response | GSTU17-OE and gstu17 | Chen et al., 2012 |
| Light response / Seedling development / Root elongation | GSTU17-OE and gstu17 | Jiang et al., 2010 | |||
| GSTU18 | At1g10360 | GST29 | Part of the lipid stress response | None | Mueller et al., 2008 |
| GSTU19 | At1g78380 | GST8 | Drought, salt and methyl viologen stress response | GSTU19-OE | Xu et al., 2016 |
| GSTU20 | At1g78370 | FIP1 | Light response / Seedling development / Flowering time | GSTU20-OE and gstu20 | Chen et al., 2007 |
| GSTU21 | At1g78360 | None | |||
| GSTU22 | At1g78340 | None | |||
| GSTU23 | At1g78320 | None | |||
| GSTU24 | At1g17170 | GST | Lipid stress response | None | Mueller et al., 2008 |
| Xenobiotic detoxification (TNT, 2,4,6-trinitrotoluene) | GSTU24-OE | Gunning et al., 2014 | |||
| GSTU25 | At1g17180 | Xenobiotic detoxification (TNT, 2,4,6-trinitrotoluene) | GSTU25-OE | Gunning et al., 2014 | |
| GSTU26 | At1g17190 | Xenobiotic detoxification (herbicides) | None | Nutricati et al., 2006 | |
| GSTU27 | At3g43800 | None | |||
| GSTU28 | At1g53680 | None | |||
| GSTF2 | At4g02520 | ATPM24, GST2 | Response to bacterial inoculation (P. syringae) | None | Lieberherr et al., 2003 |
| Response to bacterial volatiles (B. subtilis) | None | Kwon et al., 2010 | |||
| GSTF3 | At2g02930 | GST16 | None | ||
| GSTF4 | At1g02950 | GST31 | None | ||
| GSTF5 | At1g02940 | None | |||
| GSTF6 | At1g02930 | ERD11, GST1, GSTF3 | Response to bacterial inoculation (P. syringae) | None | Lieberherr et al., 2003 |
| Modulation of plant metabolism in response to oxidative stress | gstf6 gstf7 gstf9 gstf10 RNAi lines | Sappl et al., 2009 | |||
| Camalexin biosynthesis (conjugation of GSH with IAN) | GSTF6-OE and gstf6 | Su et al., 2011 | |||
| GSTF7 | At1g02920 | GST11, GSTF8 | Modulation of plant metabolism in response to oxidative stress | gstf6 gstf7 gstf9 gstf10 RNAi lines | Sappl et al., 2009 |
| GSTF8 | At2g47730 | GST6, GSTF5, GSTF6 | Part of the lipid stress response | None | Mueller et al., 2008 |
| Response to fungal (R. solani) and bacterial (P. syringae) pathogens | None | Gleason et al., 2011 | |||
| Response to fungal pathogens (F. oxysporum) | None | Thatcher et al., 2015 | |||
| GSTF9 | At2g30860 | GLUTTR, GSTF7 | Xenobiotic detoxification (CDNB, 1-chloro-2,4-dinitrobenzene) | None | Nutricati et al., 2006 |
| Indole glucosinolate biosynthesis | None | Wentzell et al., 2007 | |||
| Modulation of plant metabolism in response to oxidative stress | gstf6 gstf7 gstf9 gstf10 RNAi lines | Sappl et al., 2009 | |||
| Salt stress response | gstf9 | Horváth et al., 2015 | |||
| GSTF10 | At2g30870 | Indole glucosinolate biosynthesis | None | Wentzell et al., 2007 | |
| Drought and salt stress response | GSTF10-OE and gstf10 RNA lines | Ryu et al., 2009 | |||
| Modulation of plant metabolism in response to oxidative stress | gstf6 gstf7 gstf9 gstf10 RNAi lines | Sappl et al., 2009 | |||
| GSTF11 | At3g03190 | GSTF6 | Aliphatic glucosinolate biosynthesis | None | Wentzell et al., 2007 |
| GSTF12 | At5g17220 | TT19 | Flavonoid storage (anthocyanins and proanthocyanidins) | gstfl12/tt19 | Kitamura et al., 2004; Sun et al., 2012 |
| Response to fungal pathogens (V. dahliae) | None | Pantelides et al., 2010 | |||
| GSTF13 | At3g62760 | None | |||
| GSTF14 | At1g49860 | Response to virus infection (BSCTV, beet severe curly top virus) | None | Yang et al., 2013 | |
| GSTT1 | At5g41210 | GST10 | None | ||
| GSTT2 | At5g41240 | GST10B | Systemic acquired resistance | gstt2 | Banday and Nandi, 2018 |
| GSTT3 | At5g41220 | GST10C | None | ||
| GSTZ1 | At2g02390 | GST18, GSTZ1, MAAI | Tyrosine catabolism | None | Edwards and Dixon, 2000 |
| GSTZ2 | At2g02380 | None | |||
| TCHQD | At1g77290 | None | |||
Diversity of Ser-GST functions.
GSTUs were first associated with plant xenobiotic detoxification, in particular the detoxification of herbicides. It was proposed that high GSTU activity, due to a high endogenous level of expression, was at the center of the observed differential sensitivity to herbicides between cereals (e.g., maize, wheat, and rice) and weeds (Cummins et al., 2011). In A. thaliana, it was proposed that AtGSTU26, whose expression is induced in response to chloroacetanilide herbicide treatments, could participate in the detoxification of these harmful chemicals by catalyzing their glutathionylation (Nutricati et al., 2006). The xenobiotic detoxification activity of GSTU extends to other chemicals. For instance, a recent study focusing on a major worldwide military pollutant, namely the 2,4,6-trinitrotoluene (TNT), highlighted the role played by GSTUs in detoxifying this harmful and highly persistent pollutant by catalyzing its GSH-conjugation (Gunning et al., 2014). In this study, the authors showed that overexpressing AtGSTU24 and AtGSTU25, two genes whose expression is induced by TNT, was sufficient to enhance the ability of A. thaliana plants to withstand and detoxify TNT.
In addition to their role in detoxifying xenobiotic compounds, A. thaliana GSTUs were associated with the response to environmental cues. One of the best examples is the response to light signals. This was first demonstrated with GSTU20, a gene whose expression is induced under far-red irradiation and inhibited by phytochrome A (phyA) mutation (Chen et al., 2007). The characterization of gain- and loss-of-function mutant lines suggested the key role that GSTU20 plays in regulating cell elongation and flowering time in response to light (Chen et al., 2007). Similarly, GSTU17, whose expression is regulated by different photoreceptors (especially phyA), participates in the modulation of several aspects of seedling development (e.g., hypocotyl elongation, root development, anthocyanin accumulation) (Jiang et al., 2010). Two other GSTUs were also associated with the A. thaliana response to light stress, namely GSTU5 and GSTU14 (Liu and Li, 2002; Lv et al., 2015).
GSTU17 also plays a role in the response to drought and salt stresses (Chen et al., 2012). The gstu17 mutation confers a higher tolerance to drought and salt stresses when compared to wild-type plants that could be attributed to an increased accumulation of GSH and ABA within the plant tissues. In this process, GSTU17 acts as a negative component of the stress-mediated signal transduction pathways. Conversely, the overexpression of GSTU19 in A. thaliana plants confers tolerance to drought and salt stresses (Xu et al., 2016). The fact that the overexpression of GSTU19 also confers an increased tolerance to methyl viologen (a pro-oxidant compound) together with increased activity of antioxidant enzymes indicates that GSTU19 may be involved in counteracting the oxidative damages associated with drought or salt stresses. Additionally, the A. thaliana response to drought and salt stresses, mediated by the AtRGGA RNA-binding protein, involves GSTU9 (Ambrosone et al., 2015).
GSTUs are also involved in plant response to biotic stresses. A recent example is the observation that gstu13 mutants display an enhanced susceptibility, when compared to wild-type plants, toward the fungal pathogens Erysiphe pisi, Colletotrichum gloeosporioides, and Plectosphaerella cucumerina (Piślewska-Bednarek et al., 2018). It was demonstrated that this phenotype is essentially due to a decrease in the biosynthesis of indole glucosinolates (IG; defense-related compounds found in Brassicaceae), where GSTU13 plays a catalytic role in conjugating GSH to IG precursors, affecting the innate immune system of A. thaliana plants. GSTU16 is another member that was proposed to be part of the plant response to fungal pathogens (e.g., Verticillium dahliae) (Pantelides et al., 2010). However, the mechanism by which GSTU16 participates in this response still needs to be addressed.
Both biotic and abiotic stresses lead to the formation of non-enzymatically formed oxylipins, such as phytoprostanes, resulting from the oxidation of several types of lipids (most probably in response to the accumulation of stress-mediated free radicals and reactive oxygen species). These compounds serve as signaling molecules to adapt the plant response to environmental constraints, but may also enhance oxidative stress damages. Thus, their homeostasis should be tightly regulated. Interestingly, it has been shown that GSH-conjugation participates in this process, in particular in response to a Pseudomonas syringae infection (Mueller et al., 2008). In addition, the expression of several GSTU genes was shown to be responsive to phytoprostanes (i.e., GSTU7, GSTU18, and GSTU24) indicating that they may regulate phytoprostane homeostasis or be involved in the general detoxification pathways (Mueller et al., 2008; Stotz et al., 2013).
GSTFs constitute the second largest class of Ser-GSTs in plants, with 13 members in A. thaliana (Wagner et al., 2002). The GSTF1 sequence that was initially described (Bartling et al., 1993) is no longer present in the final reference genome of this plant.
As GSTUs, GSTFs are associated with the plant response to various abiotic and biotic stresses. For instance, A. thaliana gstf9 and gstf10 mutants are more sensitive to a salt stress than wild-type plants (Ryu et al., 2009; Horváth et al., 2015). In contrast, overexpression of GSTF10 confers higher tolerance to salt. Together these data indicate that the A. thaliana GSTF9 and GSTF10 play a positive role in the plant response to a salt stress. However, several reports indicate that the role of GSTF9 and GSTF10 extends beyond. GSTF10 was proposed to play a role in modulating developmental processes, such as the brassinosteroid-independent spontaneous cell death, a mechanism that is mediated by the production of reactive oxygen species (Ryu et al., 2009). GSTF9 and GSTF10, as well as GSTF11, are also involved in the biosynthesis of glucosinolates (Wentzell et al., 2007). Whether GSTF9, GSTF10 and GSTF11 act in concert with GSTU13 in this process (Piślewska-Bednarek et al., 2018) remains to be investigated. GSTF9 and GSTF10, together with GSTF6 and GSTF7, also play a role in limiting the metabolic changes that arise during oxidative stress (Sappl et al., 2009).
Additionally, GSTF6 activity is required for the biosynthesis of camalexin, the main phytoalexin (i.e., secondary metabolite with antimicrobial activity) present in A. thaliana (Su et al., 2011). The proposed role of GSTF6 in this process is to catalyze the conjugation of GSH on indole-3-acetonitrile (IAN), leading to the formation of GSH-IAN, the main precursor of camalexin biosynthesis. The above-mentioned functional roles of GSTF6 are in agreement with its first proposed biological function, which was to participate in defense mechanisms against pathogens (e.g., P. syringae), together with GSTF2 (Lieberherr et al., 2003; Kwon et al., 2010). GSTF8, GSTF12, and GSTF14 activities are associated with the A. thaliana response to various pathogens, including viruses (Pantelides et al., 2010; Gleason et al., 2011; Yang et al., 2013; Thatcher et al., 2015). GSTF8 and GSTF9 have been identified as potential actors of the lipid stress response and xenobiotic detoxification machinery, respectively (Nutricati et al., 2006; Mueller et al., 2008).
GSTF12 is another member of the phi class whose function has been extensively studies. GSTF12 is also known as TRANSPARENT TESTA 19 (TT19) and plays a key role in the control of anthocyanin and proanthocyanidin vacuolar accumulation in A. thaliana vegetative tissues and seed testa, respectively (Kitamura et al., 2004). In this process, TT19 acts as a carrier to convey these cytosolic flavonoids to the tonoplasts (Sun et al., 2012).
No more physiological information is available for other Ser-GST classes, namely GSTT, GSTZ, and TCHQD (3, 2, and 1 members in A. thaliana, respectively). A. thaliana GSTT2 seems involved in the modulation of systemic acquired resistance by altering the expression of key genes involved in this process through epigenetic modifications (Banday and Nandi, 2018). According to its capacity to catalyze the isomerisation of maleylacetoacetate into fumarylacetoacetate in vitro, AtGSTZ1 is likely involved in tyrosine catabolism, as demonstrated in animals (Edwards and Dixon, 2000). However, there is no mutant described so far and there is a second gene in A. thaliana, the biochemical properties of the protein having not yet been explored. Moreover, AtGSTZ1 was also able to catalyze the GSH-dependent dehalogenation of dichloroacetic acid to glyoxylic acid, suggesting other possible functions in planta. Finally, there is to date no clear function attributed to the sole member of the TCHQD class, either in A. thaliana or in another photosynthetic organisms.
Conclusion
The GST gene family was subject to a huge genetic expansion in terrestrial plants, with an average number of GSTs around 58 but also in some particular fungi (up to 45 isoforms) (Morel et al., 2013) compared to the 6–17 genes found in bacteria, yeast or mammals. This is linked to expansion within one or several classes as GSTU in plants. Genomic organization and phylogenetic analyses indicate that most duplication events responsible for the expansion of the GSTF and GSTU classes are species-specific. Among Ser-GSTs in plants, the GSTFs and GSTUs are highly represented, and have more diversified primary sequences and catalytic signatures compared to GSTs from the theta, zeta and TCHQD classes. This is likely at the origin of their broader range of activities and set of accommodated substrates/ligands.
According to their stress-inducible expression, it is documented that many GSTFs and GSTUs have functions connected to secondary metabolism, as exemplified by their implication in the vacuolar sequestration of anthocyanin, in the biosynthesis of camalexin, and/or in the binding of biosynthesis intermediates (porphyrin derivatives) or cellular by-products (oxylipins). Their implication in xenobiotic detoxification, such as herbicides, has likely little to do with their ancestral functions, that still remain to be delineated in many cases. However, this has led to the development of biotechnological applications in agriculture and environmental sciences [see the review in this research topic by Perperopoulou et al. (2018)]. The GSTs being targeted for the development of transgenic plants are linked to conferring tolerances against biotic and abiotic stresses or expressing engineered xenobiotic metabolizing enzymes for the bioremediation and detoxification of agrochemicals and pollutants.
To reveal the functions of Ser-GSTs, future studies will have to take into account the redundancy that most probably exists within and between the different classes of Ser-GSTs, which has limited the otherwise powerful reverse-genetic strategy. In order to inventory molecules able to bind GSTs, in vivo and in vitro ligand-fishing approaches have been used successfully in several cases (Dixon et al., 2011b; Dixon and Edwards, 2018). However, determining the nature of the identified molecules and whether, when and how they are conjugated with GSH or just bound to GSTs is a technically challenging and time-consuming task. For instance, out of the 43 structures of Ser-GSTs available to date in the Protein Data Bank, only 6 are solved with bound ligands. Hence, the validation of the physiological relevance of the detected interactions remains a major challenge.
Statements
Author contributions
ES-G, AH, and NR carried out the in silico genome analyses. SL and OK performed the transcriptomic analyses. ClD and MS performed structural data analyses. KR and ChD compiled data presented in Table 4. All authors participated to the writing of the manuscript.
Funding
The UMR1136 is supported by a grant overseen by the French National Research Agency (ANR) as part of the “Investissements d’Avenir” program (ANR-11-LABX-0002-01, Lab of Excellence ARBRE). OK is supported by the Swedish Research Council “VetenskapsRådet” (grant: 621-2014-4688) and the Kempe Foundations (Gunnar Öquist Fellowship). AH, ChD, NR, and KR are supported by the French National Research Agency (ANR-17-CE20-0008-01).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1
AhmadL.RylottE. L.BruceN. C.EdwardsR.GroganG. (2017). Structural evidence for Arabidopsis glutathione transferase At GSTF2 functioning as a transporter of small organic ligands.FEBS Open Bio7122–132. 10.1002/2211-5463.12168
2
AhsanN.LeeD.-G.AlamI.KimP. J.LeeJ. J.AhnY.-O.et al (2008). Comparative proteomic study of arsenic-induced differentially expressed proteins in rice roots reveals glutathione plays a central role during As stress.Proteomics83561–3576. 10.1002/pmic.200701189
3
AlfenitoM. R. (1998). Functional complementation of anthocyanin sequestration in the vacuole by widely divergent glutathione S-transferases.Plant Cell Online101135–1150. 10.1105/tpc.10.7.1135
4
AmbrosoneA.BatelliG.NurcatoR.AuriliaV.PunzoP.BangarusamyD. K.et al (2015). The Arabidopsis RNA-binding protein AtRGGA regulates tolerance to salt and drought stress.Plant Physiol.168292–306. 10.1104/pp.114.255802
5
AxarliI.DhavalaP.PapageorgiouA. C.LabrouN. E. (2009a). Crystallographic and functional characterization of the fluorodifen-inducible glutathione transferase from Glycine max reveals an active site topography suited for diphenylether herbicides and a novel L-site.J. Mol. Biol.385984–1002. 10.1016/j.jmb.2008.10.084
6
AxarliI.DhavalaP.PapageorgiouA. C.LabrouN. E. (2009b). Crystal structure of Glycine max glutathione transferase in complex with glutathione: investigation of the mechanism operating by the Tau class glutathione transferases.Biochem. J.422247–256. 10.1042/BJ20090224
7
AxarliI.GeorgiadouC.DhavalaP.PapageorgiouA. C.LabrouN. E. (2010). Investigation of the role of conserved residues Ser13, Asn48 and Pro49 in the catalytic mechanism of the tau class glutathione transferase from Glycine max.Biochim. Biophys. Acta (BBA) – Proteins Proteom.1804662–667. 10.1016/j.bbapap.2009.10.016
8
AxarliI.MuletaA. W.VlachakisD.KossidaS.KotziaG.MaltezosA.et al (2016). Directed evolution of Tau class glutathione transferases reveals a site that regulates catalytic efficiency and masks co-operativity.Biochem. J.473559–570. 10.1042/BJ20150930
9
AxarliI. A.RigdenD. J.LabrouN. E. (2004). Characterization of the ligandin site of maize glutathione S-transferase I.Biochem. J.382885–893. 10.1042/BJ20040298
10
BandayZ. Z.NandiA. K. (2018). Arabidopsis thaliana GLUTATHIONE- S -TRANSFERASE THETA 2 interacts with RSI1/FLD to activate systemic acquired resistance: GSTT2 interacts with FLD and regulates SAR.Mol. Plant Pathol.19464–475. 10.1111/mpp.12538
11
BartlingD.RadzioR.SteinerU.WeilerE. W. (1993). A glutathione S-transferase with glutathione-peroxidase activity from Arabidopsis thaliana. Molecular cloning and functional characterization.Eur. J. Biochem.216579–586. 10.1111/j.1432-1033.1993.tb18177.x
12
BilangJ.MacdonaldH.KingP. J.SturmA. (1993). A soluble auxin-binding protein from Hyoscyamus muticus is a glutathione S-transferase.Plant Physiol.10229–34. 10.1104/pp.102.1.29
13
BoardP. G.CogganM.ChelvanayagamG.EastealS.JermiinL. S.SchulteG. K.et al (2000). Identification, characterization, and crystal structure of the omega class glutathione transferases.J. Biol. Chem.27524798–24806. 10.1074/jbc.M001706200
14
BoardP. G.MenonD. (2013). Glutathione transferases, regulators of cellular metabolism and physiology.Biochim. Biophys. Acta (BBA) – Gen. Subjects18303267–3288. 10.1016/j.bbagen.2012.11.019
15
BoothJ.BoylandE.SimsP. (1961). An enzyme from rat liver catalysing conjugations with glutathione.Biochem. J.79516–524. 10.1042/bj0790516
16
BryantD.CumminsI.DixonD. P.EdwardsR. (2006). Cloning and characterization of a theta class glutathione transferase from the potato pathogen Phytophthora infestans.Phytochemistry671427–1434. 10.1016/j.phytochem.2006.05.012
17
BuetlerT. M.EatonD. L. (1992). Complementary DNA cloning, messenger RNA expression, and induction of alpha-class glutathione S-transferases in mouse tissues.Cancer Res.52314–318.
18
BurmeisterC.LüersenK.HeinickA.HusseinA.DomagalskiM.WalterR. D.et al (2008). Oxidative stress in Caenorhabditis elegans?: protective effects of the Omega class glutathione transferase (GSTO-1).FASEB J.22343–354. 10.1096/fj.06-7426com
19
ChenC.-Y.HoS.-S.KuoT.-Y.HsiehH.-L.ChengY.-S. (2017). Structural basis of jasmonate-amido synthetase FIN219 in complex with glutathione S-transferase FIP1 during the JA signal regulation.Proc. Natl. Acad. Sci. U.S.A.114E1815–E1824. 10.1073/pnas.1609980114
20
ChenI.-C.HuangI.-C.LiuM.-J.WangZ.-G.ChungS.-S.HsiehH.-L. (2007). Glutathione S-transferase interacting with far-red insensitive 219 is involved in phytochrome A-mediated signaling in Arabidopsis.Plant Physiol.1431189–1202. 10.1104/pp.106.094185
21
ChenJ.-H.JiangH.-W.HsiehE.-J.ChenH.-Y.ChienC.-T.HsiehH.-L.et al (2012). Drought and salt stress tolerance of an arabidopsis glutathione S-transferase U17 knockout mutant are attributed to the combined effect of glutathione and abscisic acid.Plant Physiol.158340–351. 10.1104/pp.111.181875
22
ChoH.-Y.LeeH. J.KongK.-H. (2007). A phi class glutathione S-transferase from Oryza sativa (OsGSTF5): molecular cloning, expression and biochemical characteristics.J. Biochem. Mol. Biol.40511–516. 10.5483/bmbrep.2007.40.4.511
23
ChronopoulouE.AtayaF. S.PouliouF.PerperopoulouF.GeorgakisN.Nianiou-ObeidatI.et al (2017a). “Structure, evolution and functional roles of plant glutathione transferases,” in Glutathione in Plant Growth, Development, and Stress Tolerance, edsHossainM. A.MostofaM. G.Diaz-VivancosP.BurrittD. J.FujitaM.TranL.-S. P. (Cham: Springer International Publishing), 195–213. 10.1007/978-3-319-66682-2_9
24
ChronopoulouE.GeorgakisN.Nianiou-ObeidatI.MadesisP.PerperopoulouF.PouliouF.et al (2017b). “Plant glutathione transferases in abiotic stress response and herbicide resistance,” in Glutathione in Plant Growth, Development, and Stress Tolerance, edsHossainM. A.MostofaM. G.Diaz-VivancosP.BurrittD. J.FujitaM.TranL.-S. P. (Cham: Springer International Publishing), 215–233. 10.1007/978-3-319-66682-2_10
25
CogganM.FlanaganJ. U.ParkerM. W.VichaiV.PearsonW. R.BoardP. G. (2002). Identification and characterization of GSTT3, a third murine theta class glutathione transferase.Biochem. J.366323–332. 10.1042/bj20011878
26
ColemanJ.Blake-KalffM.DaviesE. (1997). Detoxification of xenobiotics by plants: chemical modification and vacuolar compartmentation.Trends Plant Sci.2144–151. 10.1016/S1360-1385(97)01019-4
27
CombesB.StakelumG. S. (1961). A liver enzyme that conjugates sulfobromophtalein sodium with glutathione.J. Clin. Investig.40981–988. 10.1172/JCI104337
28
ConnS.CurtinC.BézierA.FrancoC.ZhangW. (2008). Purification, molecular cloning, and characterization of glutathione S-transferases (GSTs) from pigmented Vitis vinifera L. cell suspension cultures as putative anthocyanin transport proteins.J. Exp. Bot.593621–3634. 10.1093/jxb/ern217
29
CumminsI.DixonD. P.Freitag-PohlS.SkipseyM.EdwardsR. (2011). Multiple roles for plant glutathione transferases in xenobiotic detoxification.Drug Metabol. Rev.43266–280. 10.3109/03602532.2011.552910
30
CumminsI.O’HaganD.JablonkaiI.ColeD. J.HehnA.Werck-ReichhartD.et al (2003). Cloning, characterization and regulation of a family of phi class glutathione transferases from wheat.Plant Mol. Biol.52591–603.
31
DeponteM. (2013). Glutathione catalysis and the reaction mechanisms of glutathione-dependent enzymes.Biochim. Biophys. Acta (BBA) – Gen. Subjects18303217–3266. 10.1016/j.bbagen.2012.09.018
32
DeRidderB. P. (2002). Induction of glutathione S-transferases in Arabidopsis by herbicide safeners.Plant Physiol.1301497–1505. 10.1104/pp.010066
33
DingN.WangA.ZhangX.WuY.WangR.CuiH.et al (2017). Identification and analysis of glutathione S-transferase gene family in sweet potato reveal divergent GST-mediated networks in aboveground and underground tissues in response to abiotic stresses.BMC Plant Biol.17:225. 10.1186/s12870-017-1179-z
34
DixonD. P.CumminsI.ColeD. J.EdwardsR. (1998). Glutathione-mediated detoxification systems in plants.Curr. Opin. Plant Biol.1258–266. 10.1016/S1369-5266(98)80114-3
35
DixonD. P.EdwardsR. (2009). Selective binding of glutathione conjugates of fatty acid derivatives by plant glutathione transferases.J. Biol. Chem.28421249–21256. 10.1074/jbc.M109.020107
36
DixonD. P.EdwardsR. (2010a). Glutathione transferases.Arabidopsis Book8:e0131. 10.1199/tab.0131
37
DixonD. P.EdwardsR. (2010b). Roles for stress-inducible lambda glutathione transferases in flavonoid metabolism in plants as identified by ligand fishing.J. Biol. Chem.28536322–36329. 10.1074/jbc.M110.164806
38
DixonD. P.EdwardsR. (2018). Protein-ligand fishing in planta for biologically active natural products using glutathione transferases.Front. Plant Sci.9:1659. 10.3389/fpls.2018.01659
39
DixonD. P.HawkinsT.HusseyP. J.EdwardsR. (2009). Enzyme activities and subcellular localization of members of the Arabidopsis glutathione transferase superfamily.J. Exp. Bot.601207–1218. 10.1093/jxb/ern365
40
DixonD. P.LapthornA.EdwardsR. (2002). Plant glutathione transferases.Genome Biol.3:REVIEWS3004.
41
DixonD. P.LapthornA.MadesisP.MuddE. A.DayA.EdwardsR. (2008). Binding and glutathione conjugation of porphyrinogens by plant glutathione transferases.J. Biol. Chem.28320268–20276. 10.1074/jbc.M802026200
42
DixonD. P.McEwenA. G.LapthornA. J.EdwardsR. (2003). Forced evolution of a herbicide detoxifying glutathione transferase.J. Biol. Chem.27823930–23935. 10.1074/jbc.M303620200
43
DixonD. P.SellarsJ. D.EdwardsR. (2011a). The Arabidopsis phi class glutathione transferase At GSTF2: binding and regulation by biologically active heterocyclic ligands.Biochem. J.43863–70. 10.1042/BJ20101884
44
DixonD. P.SteelP. G.EdwardsR. (2011b). Roles for glutathione transferases in antioxidant recycling.Plant Signal. Behav.61223–1227. 10.4161/psb.6.8.16253
45
DroogF. (1997). Plant glutathione S-transferases, a tale of theta and tau.J. Plant Growth Regul.1695–107. 10.1007/PL00006984
46
DroogF. N. J.HooykaasP. J. J.Van Der ZaalB. J. (1995). 2,4-Dichlorophenoxyacetic acid and related chlorinated compounds inhibit two auxin-regulated type-III tobacco glutathione S-transferases.Plant Physiol.1071139–1146. 10.1104/pp.107.4.1139
47
EdwardsR.DixonD. P. (2000). The role of glutathione transferases in herbicide metabolism.Environ. Fate Saf. Manage. Agrochem.19216–222.
48
EdwardsT. E.BryanC. M.LeiblyD. J.DieterichS. H.AbendrothJ.SankaranB.et al (2011). Structures of a putative ζ-class glutathione-S-transferase from the pathogenic fungus Coccidioides immitis.Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun.671038–1043. 10.1107/S1744309111009493
49
Fernandez-CanonJ. M.BaetscherM. W.FinegoldM.BurlingameT.GibsonK. M.GrompeM. (2002). Maleylacetoacetate isomerase (MAAI/GSTZ)-deficient mice reveal a glutathione-dependent nonenzymatic bypass in tyrosine catabolism.Mol. Cell. Biol.224943–4951. 10.1128/MCB.22.13.4943-4951.2002
50
Fernández-CañónJ. M.PeñalvaM. A. (1998). Characterization of a fungal maleylacetoacetate isomerase gene and identification of its human homologue.J. Biol. Chem.273329–337. 10.1074/jbc.273.1.329
51
FrearD. S.SwansonH. R. (1970). Biosynthesis of S-(4-ethylamino-6-isopropylamino- 2-s-triazino) glutathione: partial purification and properties of a glutathione S-transferase from corn.Phytochemistry92123–2132. 10.1016/S0031-9422(00)85377-7
52
FrovaC. (2006). Glutathione transferases in the genomics era: new insights and perspectives.Biomol. Eng.23149–169. 10.1016/j.bioeng.2006.05.020
53
GalléÁCsiszárJ.SecenjiM.GuóthA.CseuzL.TariI.et al (2009). Glutathione transferase activity and expression patterns during grain filling in flag leaves of wheat genotypes differing in drought tolerance: response to water deficit.J. Plant Physiol.1661878–1891. 10.1016/j.jplph.2009.05.016
54
GleasonC.HuangS.ThatcherL. F.FoleyR. C.AndersonC. R.CarrollA. J.et al (2011). Mitochondrial complex II has a key role in mitochondrial-derived reactive oxygen species influence on plant stress gene regulation and defense.Proc. Natl. Acad. Sci. U.S.A.10810768–10773. 10.1073/pnas.1016060108
55
GomezC.ConejeroG.TorregrosaL.CheynierV.TerrierN.AgeorgesA. (2011). In vivo grapevine anthocyanin transport involves vesicle-mediated trafficking and the contribution of anthoMATE transporters and GST: anthocyanin trafficking in grapevine.Plant J.67960–970. 10.1111/j.1365-313X.2011.04648.x
56
GongH.JiaoY.HuW.PuaE. (2005). Expression of glutathione-S-transferase and its role in plant growth and development in vivo and shoot morphogenesis in vitro.Plant Mol. Biol.5753–66. 10.1007/s11103-004-4516-1
57
GonneauM.MornetR.LaloueM. (1998). A Nicotiana plumbaginifolia protein labeled with an azido cytokinin agonist is a glutathione S-transferase.Physiol. Plant.103114–124. 10.1034/j.1399-3054.1998.1030114.x
58
GonzalezD.FraichardS.GrasseinP.DelarueP.SenetP.NicolaïA.et al (2018). Characterization of a Drosophila glutathione transferase involved in isothiocyanate detoxification.Insect Biochem. Mol. Biol.9533–43. 10.1016/j.ibmb.2018.03.004
59
GoodsteinD. M.ShuS.HowsonR.NeupaneR.HayesR. D.FazoJ.et al (2012). Phytozome: a comparative platform for green plant genomics.Nucleic Acids Res.40D1178–D1186. 10.1093/nar/gkr944
60
GronwaldJ. W.PlaisanceK. L. (1998). Isolation and characterization of glutathione S-transferase isozymes from sorghum.Plant Physiol.117877–892. 10.1104/pp.117.3.877
61
GunningV.TzafestasK.SparrowH.JohnstonE. J.BrentnallA. S.PottsJ. R.et al (2014). Arabidopsis glutathione transferases U24 and U25 exhibit a range of detoxification activities with the environmental pollutant and explosive, 2,4,6-trinitrotoluene.Plant Physiol.165854–865. 10.1104/pp.114.237180
62
HabigW. H.PabstM. J.FleischnerG.GatmaitanZ.AriasI. M.JakobyW. B. (1974). The identity of glutathione S-transferase B with ligandin, a major binding protein of liver.Proc. Natl. Acad. Sci. U.S.A.713879–3882. 10.1073/pnas.71.10.3879
63
HanJ.-B.LiG.-Q.WanP.-J.ZhuT.-T.MengQ.-W. (2016). Identification of glutathione S-transferase genes in Leptinotarsa decemlineata and their expression patterns under stress of three insecticides.Pesticide Biochem. Physiol.13326–34. 10.1016/j.pestbp.2016.03.008
64
HayesJ. D.McLellanL. I. (1999). Glutathione and glutathione-dependent enzymes represent a co-ordinately regulated defence against oxidative stress.Free Radic. Res.31273–300. 10.1080/10715769900300851
65
HayesJ. D.PulfordD. J. (1995). The glut athione S-transferase supergene family: regulation of GST and the contribution of the lsoenzymes to cancer chemoprotection and drug resistance part I.Crit. Rev. Biochem. Mol. Biol.30445–520. 10.3109/10409239509083491
66
HeG.GuanC.-N.ChenQ.-X.GouX.-J.LiuW.ZengQ.-Y.et al (2016). Genome-wide analysis of the glutathione S-transferase gene family in capsella rubella: identification, expression, and biochemical functions.Front. Plant Sci.7:1325. 10.3389/fpls.2016.01325
67
HorváthE.BelaK.PapdiC.GalléÁSzabadosL.TariI.et al (2015). The role of Arabidopsis glutathione transferase F9 gene under oxidative stress in seedlings.Acta Biol. Hungarica66406–418. 10.1556/018.66.2015.4.5
68
HuangY.XunR.ChenG.XunL. (2008). Maintenance role of a glutathionyl-hydroquinone lyase (PcpF) in pentachlorophenol degradation by Sphingobium chlorophenolicum ATCC 39723.J. Bacteriol.1907595–7600. 10.1128/JB.00489-08
69
HurstR.BaoY.JemthP.MannervikB.WilliamsonG. (1998). Phospholipid hydroperoxide glutathione peroxidase activity of human glutathione transferases.Biochem. J.332(Pt 1), 97–100. 10.1042/bj3320097
70
IslamS.RahmanI. A.IslamT.GhoshA. (2017). Genome-wide identification and expression analysis of glutathione S-transferase gene family in tomato: gaining an insight to their physiological and stress-specific roles.PLoS One12:e0187504. 10.1371/journal.pone.01875004
71
JakobssonP. J.MorgensternR.ManciniJ.Ford-HutchinsonA.PerssonB. (1999). Common structural features of MAPEG – a widespread superfamily of membrane associated proteins with highly divergent functions in eicosanoid and glutathione metabolism.Protein Sci.8689–692. 10.1110/ps.8.3.689
72
JakobyW. B. (1978). The glutathione S-transferases: a group of multifunctional detoxification proteins.Adv. Enzymol. Relat. Areas Mol. Biol.46383–414. 10.1002/9780470122914.ch6
73
JhaB.SharmaA.MishraA. (2011). Expression of SbGSTU (tau class glutathione S-transferase) gene isolated from Salicornia brachiata in tobacco for salt tolerance.Mol. Biol. Rep.384823–4832. 10.1007/s11033-010-0625-x
74
JiangH.-W.LiuM.-J.ChenI.-C.HuangC.-H.ChaoL.-Y.HsiehH.-L. (2010). A glutathione S-transferase regulated by light and hormones participates in the modulation of Arabidopsis seedling development.Plant Physiol.1541646–1658. 10.1104/pp.110.159152
75
KaoC. W.BakshiM.SherametiI.DongS.ReicheltM.OelmüllerR.et al (2016). A Chinese cabbage (Brassica campetris subsp. Chinensis) τ-type glutathione-S-transferase stimulates Arabidopsis development and primes against abiotic and biotic stress.Plant Mol. Biol.92643–659. 10.1007/s11103-016-0531-2
76
KepplerD. (1999). Export pumps for glutathione S-conjugates.Free Radic. Biol. Med.27985–991. 10.1016/s0891-5849(99)00171-9
77
KetleyJ. N.HabigW. H.JakobyW. B. (1975). Binding of nonsubstrate ligands to the glutathione S-transferases.J. Biol. Chem.2508670–8673.
78
KhanN.HuC.Amjad KhanW.HouX. (2018). Genome-wide identification, classification, and expression divergence of glutathione-transferase family in Brassica rapa under multiple hormone treatments.BioMed. Res. Int.20181–19. 10.1155/2018/6023457
79
KieferP. M.CopleyS. D. (2002). Characterization of the initial steps in the reductive dehalogenation catalyzed by tetrachlorohydroquinone dehalogenase †.Biochemistry411315–1322. 10.1021/bi0117504
80
KilianJ.WhiteheadD.HorakJ.WankeD.WeinlS.BatisticO.et al (2007). The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses: AtGenExpress global abiotic stress data set.Plant J.50347–363. 10.1111/j.1365-313X.2007.03052.x
81
KimJ.SuhH.KimS.KimK.AhnC.YimJ. (2006). Identification and characteristics of the structural gene for the Drosophila eye colour mutant sepia, encoding PDA synthase, a member of the Omega class glutathione S-transferases.Biochem. J.398451–460. 10.1042/BJ20060424
82
KitamuraS.AkitaY.IshizakaH.NarumiI.TanakaA. (2012). Molecular characterization of an anthocyanin-related glutathione S-transferase gene in cyclamen.J. Plant Physiol.169636–642. 10.1016/j.jplph.2011.12.011
83
KitamuraS.ShikazonoN.TanakaA. (2004). TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis.Plant J.37104–114. 10.1046/j.1365-313x.2003.01943.x
84
KrausP. (1980). Resolution, purification and some properties of three glutathione transferases from rat liver mitochondria.Hoppe-Seyler’s Z. Physiol. Chem.3619–15.
85
KwonY. S.RyuC.-M.LeeS.ParkH. B.HanK. S.LeeJ. H.et al (2010). Proteome analysis of Arabidopsis seedlings exposed to bacterial volatiles.Planta2321355–1370. 10.1007/s00425-010-1259-x
86
LallementP.-A.BrouwerB.KeechO.HeckerA.RouhierN. (2014). The still mysterious roles of cysteine-containing glutathione transferases in plants.Front. Pharmacol.5:192. 10.3389/fphar.2014.00192
87
LallementP.-A.MeuxE.GualbertoJ. M.DumarcayS.FavierF.DidierjeanC.et al (2015). Glutathionyl-hydroquinone reductases from poplar are plastidial proteins that deglutathionylate both reduced and oxidized glutathionylated quinones.FEBS Lett.58937–44. 10.1016/j.febslet.2014.11.021
88
LamoureuxG. L.ShimabukuroR. H.SwansonH. R.FrearD. S. (1970). Metabolism of 2-chloro-4-ethylamino-6-isopropylamino-s-triazine (atrazine) in excised sorghum leaf sections.J. Agric. Food Chem.1881–86. 10.1021/jf60167a029
89
LanT.YangZ.-L.YangX.LiuY.-J.WangX.-R.ZengQ.-Y. (2009). Extensive functional diversification of the populus glutathione S-transferase supergene family.Plant Cell213749–3766. 10.1105/tpc.109.070219
90
LarsenE. S.AlfenitoM. R.BriggsW. R.WalbotV. (2003). A carnation anthocyanin mutant is complemented by the glutathione S-transferases encoded by maize Bz2 and petunia An9.Plant Cell Rep.21900–904. 10.1007/s00299-002-0545-x
91
LedererB.BögerP. (2003). Binding and protection of porphyrins by glutathione S-transferases of Zea mays L.Biochim. Biophys. Acta (BBA) – Gen. Subjects1621226–233. 10.1016/S0304-4165(03)00073-4
92
LedererB.BögerP. (2005). A ligand function of glutathione S-transferase, Z. Naturforsch. C. 60166–171. 10.1515/znc-2005-3-403
93
LeviA. J.AriasI. M. (1969). Two hepatic cytoplasmic protein fractions, Y and Z, and their possible role in the hepatic uptake of bilirubin, sulfobromophthalein, and other anions.J. Clin. Invest.482156–2167. 10.1172/JCI106182
94
LieberherrD.WagnerU.DubuisP.-H.MétrauxJ.-P.MauchF. (2003). The rapid induction of glutathione S-transferases AtGSTF2 and AtGSTF6 by avirulent Pseudomonas syringae is the result of combined salicylic acid and ethylene signaling.Plant Cell Physiol.44750–757. 10.1093/pcp/pcg093
95
LitwackG.KettererB.AriasI. M. (1971). Ligandin: a hepatic protein which binds steroids, bilirubin, carcinogens and a number of exogenous organic anions.Nature234466–467. 10.1038/234466a0
96
LiuX.-F.LiJ.-Y. (2002). [Characterization of an ultra-violet inducible gene that encodes glutathione S-transferase in Arabidopsis thaliana].Yi Chuan Xue Bao29458–460.
97
LiuY.-J.HanX.-M.RenL.-L.YangH.-L.ZengQ.-Y. (2013). Functional divergence of the glutathione S-transferase supergene family in physcomitrella patens reveals complex patterns of large gene family evolution in land plants.Plant Physiol.161773–786. 10.1104/pp.112.205815
98
LvF.ZhouJ.ZengL.XingD. (2015). β-cyclocitral upregulates salicylic acid signalling to enhance excess light acclimation in Arabidopsis.J. Exp. Bot.664719–4732. 10.1093/jxb/erv231
99
MannervikB.AlinP.GuthenbergC.JenssonH.TahirM. K.WarholmM.et al (1985). Identification of three classes of cytosolic glutathione transferase common to several mammalian species: correlation between structural data and enzymatic properties.Proc. Natl. Acad. Sci. U.S.A.827202–7206. 10.1073/pnas.82.21.7202
100
MannervikB.DanielsonU. H. (1988). Glutathione transferases–structure and catalytic activity.CRC Crit. Rev. Biochem.23283–337.
101
MarrsK. A. (1996). The functions and regulation of glutathionE S-transferases in plants.Ann. Rev. Plant Physiol. Plant Mol. Biol.47127–158. 10.1146/annurev.arplant.47.1.127
102
MarrsK. A.AlfenitoM. R.LloydA. M.WalbotV. (1995). A glutathione S-transferase involved in vacuolar transfer encoded by the maize gene Bronze-2.Nature375397–400. 10.1038/375397a0
103
MarshM.ShoemarkD. K.JacobA.RobinsonC.CahillB.ZhouN.-Y.et al (2008). Structure of bacterial glutathione-S-transferase maleyl pyruvate isomerase and implications for mechanism of isomerisation.J. Mol. Biol.384165–177. 10.1016/j.jmb.2008.09.028
104
Martínez-MárquezA.Martínez-EstesoM. J.Vilella-AntónM. T.Sellés-MarchartS.Morante-CarrielJ. A.HurtadoE.et al (2017). A tau class glutathione-S-transferase is involved in trans-resveratrol transport out of grapevine cells.Front. Plant Sci.8:1457. 10.3389/fpls.2017.01457
105
MasaiE.IchimuraA.SatoY.MiyauchiK.KatayamaY.FukudaM. (2003). Roles of the enantioselective glutathione S-transferases in cleavage of beta-aryl ether.J. Bacteriol.1851768–1775. 10.1128/jb.185.6.1768-1775.2003
106
MashiyamaS. T.MalabananM. M.AkivaE.BhosleR.BranchM. C.HillerichB.et al (2014). Large-scale determination of sequence, structure, and function relationships in cytosolic glutathione transferases across the biosphere.PLoS Biol.12:e1001843. 10.1371/journal.pbio.1001843
107
MenonD.BoardP. G. (2013). A role for glutathione transferase omega 1 (GSTO1-1) in the glutathionylation cycle.J. Biol. Chem.28825769–25779. 10.1074/jbc.M113.487785
108
MeuxE.MorelM.LamantT.GérardinP.JacquotJ.-P.DumarçayS.et al (2013). New substrates and activity of Phanerochaete chrysosporium Omega glutathione transferases.Biochimie95336–346. 10.1016/j.biochi.2012.10.003
109
MeuxE.ProsperP.NgadinA.DidierjeanC.MorelM.DumarçayS.et al (2011). Glutathione transferases of Phanerochaete chrysosporium: S-glutathionyl-p-hydroquinone reductase belongs to a new structural class.J. Biol. Chem.2869162–9173. 10.1074/jbc.M110.194548
110
MomoseM.ItohY.UmemotoN.NakayamaM.OzekiY. (2013). Reverted glutathione S-transferase-like genes that influence flower color intensity of carnation (Dianthus caryophyllus L.) originated from excision of a transposable element.Breed. Sci.63435–440. 10.1270/jsbbs.63.435
111
MonticoloF.ColantuonoC.ChiusanoM. L. (2017). Shaping the evolutionary tree of green plants: evidence from the GST family.Sci. Rep.7:14363. 10.1038/s41598-017-14316-w
112
MoonsA. (2003). Osgstu3 and osgtu4, encoding tau class glutathione S-transferases, are heavy metal- and hypoxic stress-induced and differentially salt stress-responsive in rice roots 1.FEBS Lett.553427–432. 10.1016/S0014-5793(03)01077-9
113
MoonsA. (2005). “Regulatory and functional interactions of plant growth regulators and plant glutathione s-transferases (GSTs),” in Vitamins & Hormones, (Amsterdam: Elsevier), 155–202. 10.1016/S0083-6729(05)72005-7
114
MorelM.MeuxE.MathieuY.ThuillierA.ChibaniK.HarvengtL.et al (2013). Xenomic networks variability and adaptation traits in wood decaying fungi: fungal xenomic networks.Microb. Biotechnol.6248–263. 10.1111/1751-7915.12015
115
MuellerL. A.GoodmanC. D.SiladyR. A.WalbotV. (2000). AN9, a petunia glutathione S-transferase required for anthocyanin sequestration, is a flavonoid-binding protein.Plant Physiol.1231561–1570. 10.1104/pp.123.4.1561
116
MuellerS.HilbertB.DueckershoffK.RoitschT.KrischkeM.MuellerM. J.et al (2008). General detoxification and stress responses are mediated by oxidized lipids through TGA transcription factors in Arabidopsis.Plant Cell Online20768–785. 10.1105/tpc.107.054809
117
MunyampunduJ.-P.XuY.-P.CaiX.-Z. (2016). Phi class of glutathione S-transferase gene superfamily widely exists in nonplant taxonomic groups.Evol. Bioinform.12:EBO.S35909. 10.4137/EBO.S35909
118
NavrotN.CollinV.GualbertoJ.GelhayeE.HirasawaM.ReyP.et al (2006). Plant glutathione peroxidases are functional peroxiredoxins distributed in several subcellular compartments and regulated during biotic and abiotic stresses.Plant Physiol.1421364–1379. 10.1104/pp.106.089458
119
NeuefeindT.HuberR.DasenbrockH.PradeL.BieselerB. (1997a). Crystal structure of herbicide-detoxifying maize glutathione S-transferase-I in complex with lactoylglutathione: evidence for an induced-fit mechanism.J. Mol. Biol.274446–453. 10.1006/jmbi.1997.1402
120
NeuefeindT.HuberR.ReinemerP.KnäbleinJ.PradeL.MannK.et al (1997b). Cloning, sequencing, crystallization and X-ray structure of glutathione S-transferase-III from Zea mays var. mutin: a leading enzyme in detoxification of maize herbicides.J. Mol. Biol.274577–587. 10.1006/jmbi.1997.1401
121
NutricatiE.MiceliA.BlandoF.De BellisL. (2006). Characterization of two Arabidopsis thaliana glutathione S-transferases.Plant Cell Rep.25997–1005. 10.1007/s00299-006-0146-1
122
PantelidesI. S.TjamosS. E.PaplomatasE. J. (2010). Ethylene perception via ETR1 is required in Arabidopsis infection by Verticillium dahliae.Mol. Plant Pathol.11191–202. 10.1111/j.1364-3703.2009.00592.x
123
PégeotH.KohC. S.PetreB.MathiotS.DuplessisS.HeckerA.et al (2014). The poplar Phi class glutathione transferase: expression, activity and structure of GSTF1.Front. Plant Sci.5:712. 10.3389/fpls.2014.00712
124
PégeotH.MathiotS.PerrotT.GenseF.HeckerA.DidierjeanC.et al (2017). Structural plasticity among glutathione transferase Phi members: natural combination of catalytic residues confers dual biochemical activities.FEBS J.2842442–2463. 10.1111/febs.14138
125
PembleS. E.TaylorJ. B. (1992). An evolutionary perspective on glutathione transferases inferred from class-theta glutathione transferase cDNA sequences.Biochem. J.287(Pt 3), 957–963. 10.1042/bj2870957
126
PerperopoulouF.PouliouF.LabrouN. E. (2018). Recent advances in protein engineering and biotechnological applications of glutathione transferases.Crit. Rev. Biotechnol.38511–528. 10.1080/07388551.2017.1375890
127
PettersenE. F.GoddardT. D.HuangC. C.CouchG. S.GreenblattD. M.MengE. C.et al (2004). UCSF chimera–a visualization system for exploratory research and analysis.J. Comput. Chem.251605–1612. 10.1002/jcc.20084
128
Piślewska-BednarekM.NakanoR. T.HirumaK.PastorczykM.Sanchez-ValletA.Singkaravanit-OgawaS.et al (2018). Glutathione transferase U13 functions in pathogen-triggered glucosinolate metabolism.Plant Physiol.176538–551. 10.1104/pp.17.01455
129
PlomionC.AuryJ.-M.AmselemJ.LeroyT.MuratF.DuplessisS.et al (2018). Oak genome reveals facets of long lifespan.Nat. Plants4440–452. 10.1038/s41477-018-0172-3
130
PradeL.HuberR.BieselerB. (1998). Structures of herbicides in complex with their detoxifying enzyme glutathione S-transferase – explanations for the selectivity of the enzyme in plants.Structure61445–1452. 10.1016/S0969-2126(98)00143-9
131
RahantaniainaM.-S.LiS.Chatel-InnocentiG.TuzetA.MhamdiA.VanackerH.et al (2017). Glutathione oxidation in response to intracellular H2O2: key but overlapping roles for dehydroascorbate reductases.Plant Signal Behav.12:e1356531. 10.1080/15592324.2017.1356531
132
ReddyG. V. B.GoldM. H. (2001). Purification and characterization of glutathione conjugate reductase: a component of the tetrachlorohydroquinone reductive dehalogenase system from phanerochaete chrysosporium.Arch. Biochem. Biophys.391271–277. 10.1006/abbi.2001.2417
133
ReidR.GridleyK.KawamataY.ZhuY. (2013). Arsenite elicits anomalous sulfur starvation responses in barley.Plant Physiol.162401–409. 10.1104/pp.113.216937
134
ReinemerP.PradeL.HofP.NeuefeindT.HuberR.ZettlR.et al (1996). Three-dimensional structure of glutathione S-transferase from Arabidopsis thaliana at 2.2 Å resolution: structural characterization of herbicide-conjugating plant glutathione S-transferases and a novel active site architecture.J. Mol. Biol.255289–309. 10.1006/jmbi.1996.0024
135
RinaldiC.KohlerA.FreyP.DuchaussoyF.NingreN.CoulouxA.et al (2007). Transcript profiling of poplar leaves upon infection with compatible and incompatible strains of the foliar rust Melampsora larici-populina.Plant Physiol.144347–366. 10.1104/pp.106.094987
136
RouhierN.JacquotJ.-P. (2005). The plant multigenic family of thiol peroxidases.Free Radic. Biol. Med.381413–1421. 10.1016/j.freeradbiomed.2004.07.037
137
RouhierN.LemaireS. D.JacquotJ.-P. (2008). The role of glutathione in photosynthetic organisms: emerging functions for glutaredoxins and glutathionylation.Ann. Rev. Plant Biol.59143–166. 10.1146/annurev.arplant.59.032607.092811
138
RyuH. Y.KimS. Y.ParkH. M.YouJ. Y.KimB. H.LeeJ. S.et al (2009). Modulations of AtGSTF10 expression induce stress tolerance and BAK1-mediated cell death.Biochem. Biophys. Res. Commun.379417–422. 10.1016/j.bbrc.2008.11.156
139
SapplP. G.CarrollA. J.CliftonR.ListerR.WhelanJ.Harvey MillarA.et al (2009). The Arabidopsis glutathione transferase gene family displays complex stress regulation and co-silencing multiple genes results in altered metabolic sensitivity to oxidative stress: genomic and reverse genetic analysis of plant GSTs.Plant J.5853–68. 10.1111/j.1365-313X.2008.03761.x
140
SapplP. G.Oñate-SánchezL.SinghK. B.MillarA. H. (2004). Proteomic analysis of glutathione S -transferases of Arabidopsis thaliana reveals differential salicylic acid-induced expression of the plant-specific phi and tau classes.Plant Mol. Biol.54205–219. 10.1023/B:PLAN.0000028786.57439.b3
141
SchmidM.DavisonT. S.HenzS. R.PapeU. J.DemarM.VingronM.et al (2005). A gene expression map of Arabidopsis thaliana development.Nat. Genet.37501–506. 10.1038/ng1543
142
SchwartzM.DidierjeanC.HeckerA.GirardetJ.-M.Morel-RouhierM.GelhayeE.et al (2016). Crystal structure of Saccharomyces cerevisiae ECM4, a Xi-class glutathione transferase that reacts with glutathionyl-(hydro)quinones.PLoS One11:e0164678. 10.1371/journal.pone.0164678
143
ShahD. M.HironakaC. M.WiegandR. C.HardingE. I.KriviG. G.TiemeierD. C. (1986). Structural analysis of a maize gene coding for glutathione-S-transferase involved in herbicide detoxification.Plant Mol. Biol.6203–211. 10.1007/BF00015226
144
ShaoY.LvZ.LiC.ZhangW.DuanX.QiuQ.et al (2017). Molecular cloning and functional characterization of theta class glutathione S-transferase from Apostichopus japonicus.Fish Shellf. Immunol.6331–39. 10.1016/j.fsi.2017.02.004
145
SkopelitouK.MuletaA. W.PapageorgiouA. C.ChronopoulouE.LabrouN. E. (2015). Catalytic features and crystal structure of a tau class glutathione transferase from Glycine max specifically upregulated in response to soybean mosaic virus infections.Biochim. Biophys. Acta (BBA) – Proteins Proteom.1854166–177. 10.1016/j.bbapap.2014.11.008
146
SkopelitouK.MuletaA. W.PavliO.SkaracisG. N.FlemetakisE.PapageorgiouA. C.et al (2012). Overlapping protective roles for glutathione transferase gene family members in chemical and oxidative stress response in Agrobacterium tumefaciens.Funct. Integr. Genomics12157–172. 10.1007/s10142-011-0248-x
147
SmithA. P.DeRidderB. P.GuoW.-J.SeeleyE. H.RegnierF. E.GoldsbroughP. B. (2004). Proteomic analysis of Arabidopsis glutathione S-transferases from benoxacor- and copper-treated seedlings.J. Biol. Chem.27926098–26104. 10.1074/jbc.M402807200
148
SmithA. P.NourizadehS. D.PeerW. A.XuJ.BandyopadhyayA.MurphyA. S.et al (2003). Arabidopsis AtGSTF2 is regulated by ethylene and auxin, and encodes a glutathione S-transferase that interacts with flavonoids.Plant J.36433–442. 10.1046/j.1365-313x.2003.01890.x
149
SoranzoN.Sari GorlaM.MizziL.De TomaG.FrovaC. (2004). Organisation and structural evolution of the rice glutathione S-transferase gene family.Mol. Genet. Genomics271511–521. 10.1007/s00438-004-1006-8
150
SrivastavaD.VermaG.ChauhanA. S.PandeV.ChakrabartyD. (2019). Rice (Oryza sativa L.) tau class glutathione S -transferase (OsGSTU30) overexpression in Arabidopsis thaliana modulates a regulatory network leading to heavy metal and drought stress tolerance.Metallomics11375–389. 10.1039/c8mt00204e
151
StotzH. U.MuellerS.ZoellerM.MuellerM. J.BergerS. (2013). TGA transcription factors and jasmonate-independent COI1 signalling regulate specific plant responses to reactive oxylipins.J. Exp. Bot.64963–975. 10.1093/jxb/ers389
152
SuT.XuJ.LiY.LeiL.ZhaoL.YangH.et al (2011). Glutathione-indole-3-acetonitrile is required for camalexin biosynthesis in Arabidopsis thaliana.Plant Cell23364–380. 10.1105/tpc.110.079145
153
SunY.LiH.HuangJ.-R. (2012). Arabidopsis TT19 functions as a carrier to transport anthocyanin from the cytosol to tonoplasts.Mol. Plant5387–400. 10.1093/mp/ssr110
154
TangA. H.TuC. P. (1994). Biochemical characterization of Drosophila glutathione S-transferases D1 and D21.J. Biol. Chem.26927876–27884.
155
ThatcherL. F.KamphuisL. G.HaneJ. K.Oñate-SánchezL.SinghK. B. (2015). The Arabidopsis KH-domain RNA-binding protein ESR1 functions in components of jasmonate signalling, unlinking growth restraint and resistance to stress.PLoS One10:e0126978. 10.1371/journal.pone.0126978
156
ThomR.CumminsI.DixonD. P.EdwardsR.ColeD. J.LapthornA. J. (2002). Structure of a tau class glutathione S-transferase from wheat active in herbicide detoxification.Biochemistry417008–7020. 10.1021/bi015964x
157
ThomR.DixonD. P.EdwardsR.ColeD. J.LapthornA. J. (2001). The structure of a zeta class glutathione S-transferase from Arabidopsis thaliana: characterisation of a GST with novel active-site architecture and a putative role in tyrosine catabolism.J. Mol. Biol.308949–962. 10.1006/jmbi.2001.4638
158
TiwariV.PatelM. K.ChaturvediA. K.MishraA.JhaB. (2016). Functional characterization of the tau class glutathione-S-transferases gene (SbGSTU) promoter of salicornia brachiata under salinity and osmotic stress.PLoS One11:e0148494. 10.1371/journal.pone.0148494
159
TossounianM.-A.Van MolleI.WahniK.JacquesS.GevaertK.Van BreusegemF.et al (2018). Disulfide bond formation protects Arabidopsis thaliana glutathione transferase tau 23 from oxidative damage.Biochim. Biophys. Acta (BBA) – Gen. Subjects1862775–789. 10.1016/j.bbagen.2017.10.007
160
TripathiA.IndoliyaY.TiwariM.TiwariP.SrivastavaD.Kumar VermaP.et al (2014). Transformed yeast (Schizosaccharomyces pombe) overexpressing rice Tau class glutathione S-transferase (OsGSTU30 and OsGSTU41) shows enhanced resistance to hexavalent chromium.Metallomics61549–1557. 10.1039/C4MT00083H
161
Valenzuela-ChaviraI.Contreras-VergaraC. A.Arvizu-FloresA. A.Serrano-PosadaH.Lopez-ZavalaA. A.García-OrozcoK. D.et al (2017). Insights into ligand binding to a glutathione S-transferase from mango: structure, thermodynamics and kinetics.Biochimie13535–45. 10.1016/j.biochi.2017.01.005
162
WagnerU.EdwardsR.DixonD. P.MauchF. (2002). Probing the diversity of the Arabidopsis glutathione S-transferase gene family.Plant Mol. Biol.49515–532.
163
WentzellA. M.RoweH. C.HansenB. G.TicconiC.HalkierB. A.KliebensteinD. J. (2007). Linking metabolic QTLs with network and cis-eQTLs controlling biosynthetic pathways.PLoS Genet.3:e162. 10.1371/journal.pgen.0030162
164
WiegandR. C.ShahD. M.MozerT. J.HardingE. I.Diaz-CollierJ.SaundersC.et al (1986). Messenger RNA encoding a glutathione-S-transferase responsible for herbicide tolerance in maize is induced in response to safener treatment.Plant Mol. Biol.7235–243. 10.1007/BF00752897
165
WillettS. W.CopleyS. D. (1996). Identification and localization of a stable sulfenic acid in peroxide-treated tetrachlorohydroquinone dehalogenase using electrospray mass spectrometry.Chem. Biol.3851–857. 10.1016/S1074-5521(96)90071-X
166
XuJ.TianY.-S.XingX.-J.PengR.-H.ZhuB.GaoJ.-J.et al (2016). Over-expression of AtGSTU19 provides tolerance to salt, drought and methyl viologen stresses in Arabidopsis.Physiol. Plant.156164–175. 10.1111/ppl.12347
167
XuJ.ZhengA.-Q.XingX.-J.ChenL.FuX.-Y.PengR.-H.et al (2018). Transgenic Arabidopsis plants expressing grape glutathione S-Transferase gene (VvGSTF13) show enhanced tolerance to abiotic stress.Biochemistry83755–765. 10.1134/S0006297918060135
168
XunL.BelchikS. M.XunR.HuangY.ZhouH.SanchezE.et al (2010). S-Glutathionyl-(chloro)hydroquinone reductases: a novel class of glutathione transferases.Biochem. J.428419–427. 10.1042/BJ20091863
169
XunL.ToppE.OrserC. S. (1992). Purification and characterization of a tetrachloro-p-hydroquinone reductive dehalogenase from a Flavobacterium sp.J. Bacteriol.1748003–8007. 10.1128/jb.174.24.8003-8007.1992
170
YamamotoK.NagaokaS.BannoY.AsoY. (2009). Biochemical properties of an omega-class glutathione S-transferase of the silkmoth, Bombyx mori.Compar. Biochem. Physiol. Part C Toxicol. Pharmacol.149461–467. 10.1016/j.cbpc.2008.10.108
171
YamazakiM.ShibataM.NishiyamaY.SpringobK.KitayamaM.ShimadaN.et al (2008). Differential gene expression profiles of red and green forms of Perilla frutescens leading to comprehensive identification of anthocyanin biosynthetic genes: anthocyanin biosynthetic genes from Perilla.FEBS J.2753494–3502. 10.1111/j.1742-4658.2008.06496.x
172
YangG.XuZ.PengS.SunY.JiaC.ZhaiM. (2016). In planta characterization of a tau class glutathione S-transferase gene from Juglans regia (JrGSTTau1) involved in chilling tolerance.Plant Cell Rep.35681–692. 10.1007/s00299-015-1912-8
173
YangL.-P.FangY.-Y.AnC.-P.DongL.ZhangZ.-H.ChenH.et al (2013). C2-mediated decrease in DNA methylation, accumulation of siRNAs, and increase in expression for genes involved in defense pathways in plants infected with beet severe curly top virus.Plant J.73910–917. 10.1111/tpj.12081
174
YangQ.HanX.-M.GuJ.-K.LiuY.-J.YangM.-J.ZengQ.-Y. (2019). Functional and structural profiles of GST gene family from three Populus species reveal the sequence-function decoupling of orthologous genes.New Phytol.2211060–1073. 10.1111/nph.15430
175
ZakharyanR. A.Sampayo-ReyesA.HealyS. M.TsaprailisG.BoardP. G.LieblerD. C.et al (2001). Human monomethylarsonic acid (MMA(V)) reductase is a member of the glutathione-S-transferase superfamily.Chem. Res. Toxicol.141051–1057. 10.1021/tx010052h
176
ZettlR.SchellJ.PalmeK. (1994). Photoaffinity labeling of Arabidopsis thaliana plasma membrane vesicles by 5-azido-[7-3H]indole-3-acetic acid: identification of a glutathione S-transferase.Proc. Natl. Acad. Sci. U.S.A.91689–693. 10.1073/pnas.91.2.689
177
ZhaoJ. (2015). Flavonoid transport mechanisms: how to go, and with whom.Trends Plant Sci.20576–585. 10.1016/j.tplants.2015.06.007
Summary
Keywords
photosynthetic organisms, phylogeny, structure, glutathione transferases, ligandin property, secondary metabolism, xenobiotic detoxification
Citation
Sylvestre-Gonon E, Law SR, Schwartz M, Robe K, Keech O, Didierjean C, Dubos C, Rouhier N and Hecker A (2019) Functional, Structural and Biochemical Features of Plant Serinyl-Glutathione Transferases. Front. Plant Sci. 10:608. doi: 10.3389/fpls.2019.00608
Received
04 February 2019
Accepted
25 April 2019
Published
22 May 2019
Volume
10 - 2019
Edited by
Els Jm Van Damme, Ghent University, Belgium
Reviewed by
Yonghai Luo, Fujian Agriculture and Forestry University, China; Hsu-Liang Hsieh, National Taiwan University, Taiwan
Updates
Copyright
© 2019 Sylvestre-Gonon, Law, Schwartz, Robe, Keech, Didierjean, Dubos, Rouhier and Hecker.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nicolas Rouhier, nicolas.rouhier@univ-lorraine.fr Arnaud Hecker, arnaud.hecker@univ-lorraine.fr
This article was submitted to Plant Physiology, a section of the journal Frontiers in Plant Science
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.