- 1Departamento de Química Biológica, Facultad de Ciencias Exactas, Físicas y Naturales, Universidad Nacional de Córdoba, Córdoba, Argentina
- 2Instituto de Investigaciones Biológicas y Tecnológicas, Consejo Nacional de Investigaciones Científicas y Técnicas, Universidad Nacional de Córdoba, Córdoba, Argentina
- 3Departamento de Fundamentación Biológica, Facultad de Ciencias Agropecuarias, Universidad Nacional de Córdoba, Córdoba, Argentina
- 4Departamento de Química Biológica, Facultad de Ciencias Químicas, Universidad Nacional de Córdoba, Córdoba, Argentina
- 5Centro de Investigaciones en Química Biológica de Córdoba, Consejo Nacional de Investigaciones Científicas y Técnicas, Universidad Nacional de Córdoba, Córdoba, Argentina
Secreted phospholipases (sPLA2s) in plants are a growing group of enzymes that catalyze the hydrolysis of sn-2 glycerophospholipids to lysophospholipids and free fatty acids. Until today, around only 20 sPLA2s were reported from plants. This review discusses the newly acquired information on plant sPLA2s including molecular, biochemical, catalytic, and functional aspects. The comparative analysis also includes phylogenetic, evolutionary, and tridimensional structure. The observations with emphasis in Glycine max sPLA2 are compared with the available data reported for all plants sPLA2s and with those described for animals (mainly from pancreatic juice and venoms sources).
Introduction
For more than a century experiments were performed with sPLA2s enzymes, being used as lipid model enzymology and as paradigms for the formalism of interfacial catalysis (Dennis et al., 2011). The phospholipase A2 (PLA2, EC 3.1.1.4) superfamily is a broad and growing group of enzymes that stereo specifically catalyzes the cleavage at the sn-2 acyl ester bond from diacyl-phospholipid liberating lysophospholipid and free fatty acid. In plants, secreted PLA2 (sPLA2) represents one type of phospholipase A2 whose lipid products mediate a variety of cellular processes, including growth, development, defense, and stress responses (Stahl et al., 1998, 1999; Kim et al., 1999; Lee et al., 2003; Ryu, 2004; Mansfeld, 2009; Chen et al., 2011). Although numerous sPLA2 genes have been identified in plants, little is known about these enzymes in opposition to their insect, animal or human counterparts (Burke and Dennis, 2009). sPLA2 is best known from mammals where several sPLA2s have been identified in the last 25 years (Murakami et al., 2011). Moreover, many sPLA2s were found in sources as venoms from snakes, scorpions, bee, etc.; from microorganisms as bacteria and yeasts, as components of pancreatic juices, where it occurs abundantly and has a digestive role; arthritic synovial fluid; and in many different mammalian tissues (Valentin et al., 1999; Schaloske and Dennis, 2006; Burke and Dennis, 2009; Murakami et al., 2010, 2011). Additionally, for the first time, we have recently described the interfacial properties of purified recombinant sPLA2s from Streptomyces violaceoruber (Yunes Quartino et al., 2015) and from Glycine max (Mariani et al., 2012, 2015b), i.e., the optimal surface lipid packing conditions (interfacial quality) in which a sPLA2 can hydrolyze phospholipid in an organized membrane. This point, no less important for interfacial enzymes, was also addressed comparatively in the present review.
Glycine max (Soybean), in addition to being one of the most widely used oil crop grain in the world, possesses valuable contributions to health due to its high nutritional level. Lipids, proteins and other valuable bioactive components such as: phospholipids (known as lecithin), hormones, and antioxidants are present in soybean (Messina, 1999; Choi and Rhee, 2006). The industrial use of sPLA2s from animal pancreas extracts and microbes, especially in food production, has a long tradition (Guo et al., 2005; De Maria et al., 2007). One of the targets in the future may be the utilization of sPLA2 from plants for enzymatic processing to stereospecifically obtain lysoderivatives. This alternative has been recently recognized to satisfy food regulation requirements such as Kosher and Halal (Havinga, 2010). However, no sPLA2s from plants have now been yet available for industrial application (Mansfeld, 2009).
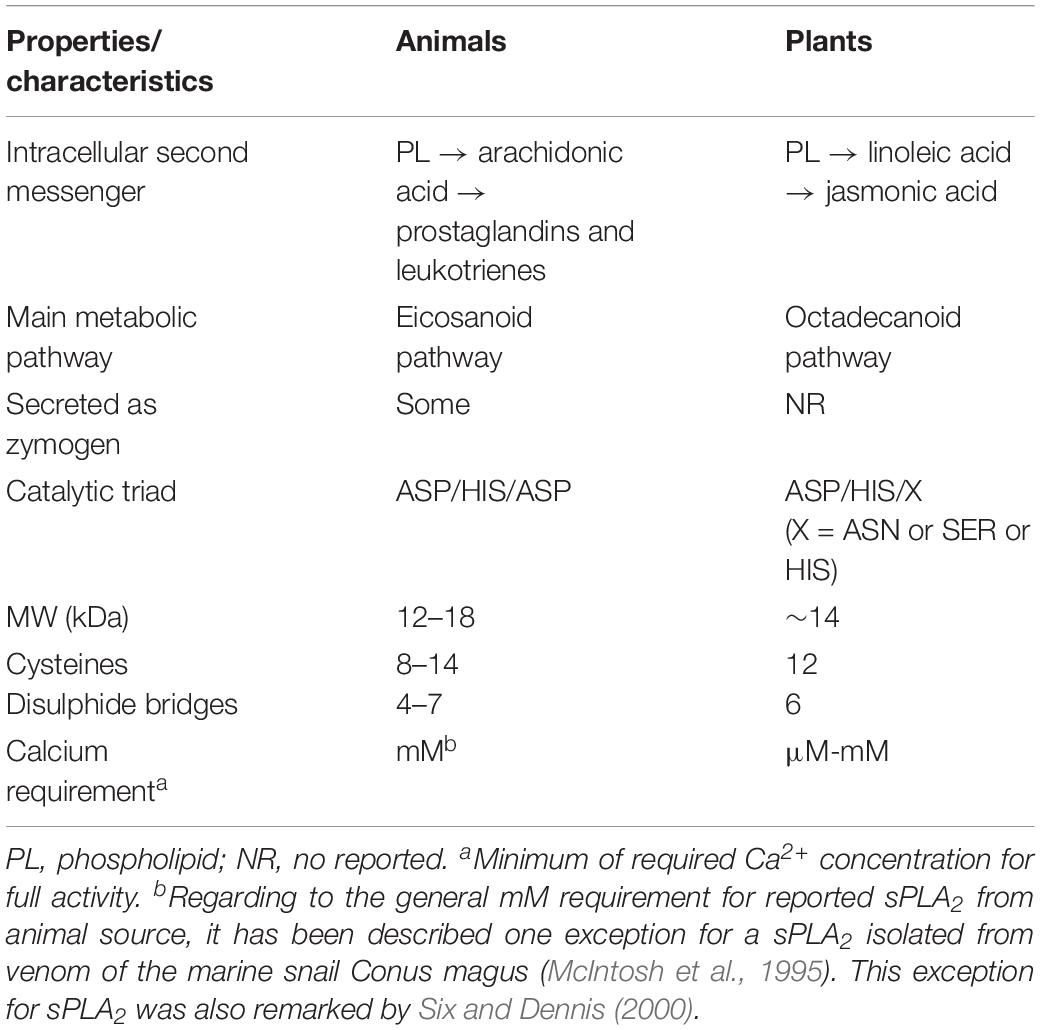
Secreted PLA2s are low MW calcium dependent enzymes (12–18 kDa) (Schaloske and Dennis, 2006). From a perusal revision of sequence data, almost all sPLA2s from plants and animals contain a signal sequence. So, in the general secretion way after removal of the N-terminal signal peptide in the endoplasmic reticulum (ER), they are secreted into the extracellular space in a either mature or pre-protein form (Fujikawa et al., 2005; Lee et al., 2005; Mansfeld et al., 2006). Although sPLA2s are recognized to be secreted proteins, a few of them were reported to act intracellularly prior or during secretion (Mounier et al., 2004; Shridas and Webb, 2014). Until now, the pre-protein form would be exclusive for animals (see Table 1).
Important common features shared for all sPLA2s are the presence of: (i) one HIS residue at the catalytic domain for nucleophilic attack at the sn-2 acyl ester bond of the glycerol backbone, (ii) requisite of calcium for full activity (μM-mM), and (iii) exceptionally heat-stable enzymes. sPLA2s also contain a domain, named PA2c, with the highly conserved Ca2+ binding loop (YGKYCGxxxxGC) and the active site motif (DACCxxHDxC), where the HIS/ASP pair was found to be highly well conserved in both animal and plants sPLA2s. At least, two characteristics are of great interest in the structure of all sPLA2s: the catalytic site and the interfacial recognition surface (IRS). All sPLA2s have the same architecture (about 55% of identity) at the catalytic site level (HIS-ASP) (Lee et al., 2005) but differ in the amino acid residues that conform the IRS region (Berg et al., 2001) sharing only 15% of identity in the amino acid sequence.
In the presence of reducing compounds such as β-mercaptoethanol or dithiothreitol (DTT) the activity is affected or abolished by disrupting the protein structure (reduction of disulfide bridges) (Stahl et al., 1998). They also show high resistance to organic solvents, acidic conditions and high temperatures (they are even more resistant in the presence of Ca2+). A common procedure to confirm the catalytic mechanism is by checking if the activity is chemically canceled by the alkylation of HIS localized in the catalytic triad HIS/ASP/X (where X may be either HIS, SER, or ASP) induced by p-bromophenacylbromide (BPB) (Minchiotti et al., 2008). A resume of the general characteristics comparing animals from plants sPLA2 is shown in Table 1.
Fatty acids produced by the hydrolysis carried out by sPLA2s, such as oleic (1:18) or arachidonic (4:20) acid, are sources of energy reserve. Furthermore, arachidonic acid can function as intracellular second messenger or as precursor of eicosanoids inflammation mediators, if is the extracellular product of the reaction catalyzed by secreted phospholipase as occurs for human synovial fluid (Baynes and Marek, 2004). The other product of the action of sPLA2, the lysophospholipid is important in cell signaling and remodeling or membrane perturbations (Khan et al., 1995). In contrast, in plants the jasmonic acid and its related compounds are important hormones involved in plant defense reaction against microbial pathogens, herbivores and UV light damaging as well as senescence mechano-transduction (Schaller, 2001).
In the past years, significant advances have been made toward understanding the role of these enzymes in normal cellular and tissue homeostasis or function particularly in mammals (Rhee and Bae, 1997; Assmann and Shimazaki, 1999; Williams, 1999; Liscovitch et al., 2000; Murakami et al., 2015) but, the more recent data reported for plant sPLA2s are rather scarce. Therefore, this review focuses on recently acquired information on all sPLA2 from plants reported until now with emphasis in GmsPLA2s identified in G. max (soybean), comparing them with the more relevant published data for several sPLA2s obtained from different sources. A comparative description with respect to the sequence characterization, biochemical, molecular, and functional aspects of sPLA2s enzymes was done.
Secretory Phospholipases A2 in Plants
In comparison with the animal sPLA2, the knowledge generated for sPLA2 from plants is still limited, even though when recombinant enzymes from plants have been recently expressed in Escherichia coli and yeast and characterized. Some studies about enzyme activities have been reported in more or less crude preparations (Moreau and Morgan, 1988; Mukherjee, 1990; Minchiotti et al., 2008; Murakami et al., 2011).
The first sPLA2 purified to homogeneity, sequenced and characterized from plants, was the sPLA2 from elm seed endosperm (Ulmus glabra) in 1998 (Stahl et al., 1998). Later in 1999, two cDNAs encoding sPLA2 (sPLA2-I and-II) were isolated from shoots of rice (Oryza sativa) and characterized (Stahl et al., 1999). cDNAs full sequences coding for putative sPLA2s were obtained from flowers of carnation (Dianthus caryophyllus) (Kim et al., 1999). These later clones from carnation and rice have not been further characterized to demonstrate that they encode functional enzymes. With progress in genome sequencing projects, more sPLA2s have been identified: in tomato (Lee et al., 2005) and outbreaks of castor bean (Ricinus communis) (Domingues et al., 2007). Four isoforms of sPLA2 from Arabidopsis thaliana have been also isolated, called AtsPLA2-α, -β, -γ, and -δ (Bahn et al., 2003; Lee et al., 2003, 2005; Mansfeld and Ulbrich-Hofmann, 2007; Seo et al., 2008), which have been expressed (Ryu et al., 2005; Mansfeld and Ulbrich-Hofmann, 2007) two isoforms have been studied in tobacco (Nicotiana tabacum) (Dhondt et al., 2000; Fujikawa et al., 2005, 2011) and orange (Citrus sinensis) (Liao and Burns, 2010). Three cDNA from durum wheat (Triticum durum) were isolated and two of them studied in detail (Verlotta et al., 2013; Verlotta and Trono, 2014). A novel sPLA2 from opium (Papaver somniferum) was purified and characterized (Jablonicka et al., 2016) and two sPLA2 from flax (Linum usitatissimum) were studied in detail (Gupta and Dash, 2017; Gupta et al., 2017). Moreover, one gene was reported for tomato (Lycopersicon esculentum) (Lee et al., 2005) and one gene for maize (Zea mays) found in UniProt and mentioned in (Mariani et al., 2012). From our laboratory, five G. max phospholipases A2 were reported (Mariani et al., 2012), and two of them (GmsPLA2-XIA-I and -XIB-II) were cloned, expressed in E. coli, further purified from inclusion bodies and the activity was evaluated using organized lipid systems such as mixed micelles and monomolecular films as substrates (Mariani et al., 2015b).
Table 2 summaries the different enzymes found in plants, their origin and source, GenBank accession numbers and the subgroup at which they belong to within the XI group of the PLA2 superfamily.
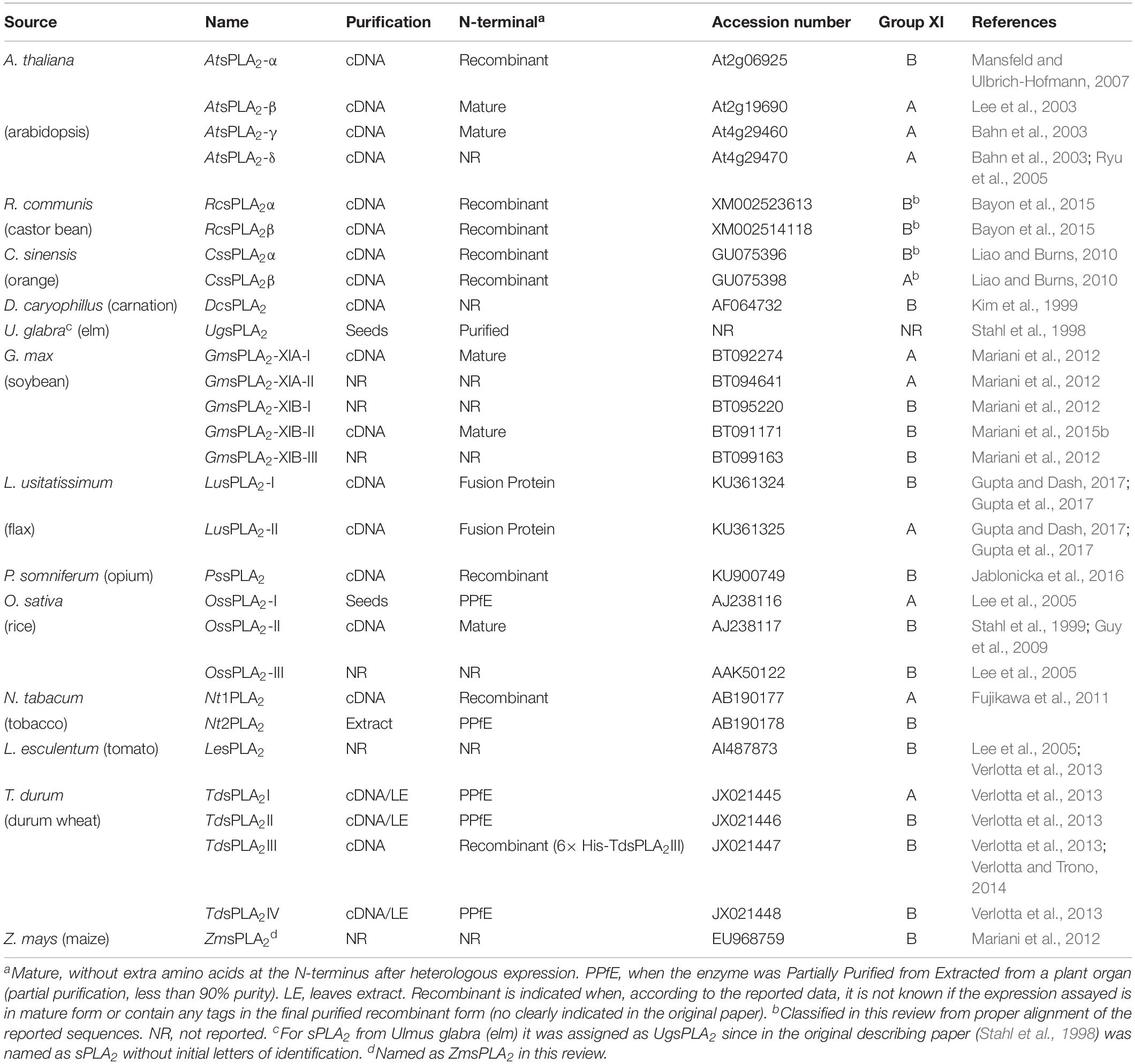
Table 2. sPLA2s from plants, accession numbers, N-terminus characteristics and purification/recombinant process applied.
Recombinant vs. Native sPla2s Proteins: Role of the Intact N-Terminal Preservation
Usually the N-terminus region of sPLA2 has an alpha helix domain which forms one wall of the channel through which the hydrophobic substrate entries as reported for groups I and II sPLA2s enzymes (according to Dennis, 1973b). Thus, in the case of pancreatic enzyme (group I), when the zymogen is converted into the active form by removing a short portion of the N-terminus, the remaining N-terminal helix is now able to be involved in the binding interfacial membrane (Scott et al., 1990). This would be affected by the extension of seven amino acids at the N-terminus in the zymogen (pro-enzyme) preventing the binding to lipid interfaces. Crystallographic evidence suggests that the zymogen has a more flexible N-terminus compared to the mature protein (van Deenen, 1971).
The effect of an extra amino acid on the N-terminus of pancreatic sPLA2 can be critical, for example, if it is of hydrophobic nature (van Scharrenburg et al., 1984). This was observed in the pioneering work of deHaas group, showing that the extension of an amino acid (doubling of the terminal ALA of the mature form) caused a decrease in enzyme catalysis to phosphatidylcholine (PC) short chain substrate presented as micelles or when the substrate was arranged as a lipid monolayer (Slotboom et al., 1977). Furthermore, in the case of porcine pancreatic enzyme, an absolute free amino terminal is required (Dijkstra et al., 1984).
In a recent work with a sPLA2 from group II of Crotalus atrox venom, the importance of a native N-terminus was also evident. By using chemically modified enzyme the authors concluded that N-terminal region plays a mechanistic role in catalysis and acts as a surface-active component of the complex interfacial catalytic site (Randolph and Heinrikson, 1982). This structural requirement is also found in other sPLA2 expressed in bacteria, such as human sPLA2 from synovial fluid (Marki and Hanulak, 1993). It was observed that, when expressing a sPLA2 in E. coli, the initial MET is not removed from the protein that had an ASN at position 1 in the sequence. This is because the bacterial aminopeptidase does not catalyze the removal of the initial MET if it is followed by ASN. The lipolytic activity of this protein was very low relative compared with the expressed correct N-terminus mature form (Othman et al., 1996). Similarly, another study reported that the protein with an extra MET at its N-terminus had the same pH optimum and prefered substrate compared to the one with native end (without MET), but the activity was drastically reduced (Marki and Hanulak, 1993). Bacterial aminopeptidases remove initial MET efficiently when the amino acid in position 2 of the mature sequence is little and without charge (such as ALA, GLY, SER), but fail when the residue is voluminous and charged as ASN (Hirel et al., 1989). Othman et al. (1996) have substituted the ASN by ALA to express the recombinant protein thus allowing the removal of the initial MET by the bacteria and avoiding a subsequent step of chemical or enzymatic cleavage.
A similar observation was made in sPLA2 mutants from Taiwan cobra (Anderson and Dufton, 1997). The addition of a MET at the N-terminus generates structural distortions, and it was postulated that affects the active site through hydrogen bonds network. Moreover, an extra MET decreases the activity with respect to the enzymes with native end (Chiou et al., 2008). Some reports suggest that the N-terminal helix of groups I and II sPLA2s acts as a regulatory domain that mediate the interfacial activation (Qin et al., 2005).
The correct design of the heterologous expression of the cloned enzyme is crucial because the recombinant protein must be generated with the correct native N-terminus, without any additional amino acid extension, since any modification or extension of the N-terminus in sPLA2 can severely alter the catalytic properties (van Scharrenburg et al., 1984; Othman et al., 1996). This is also valid for any additional N-terminal tag (such as HIS-Tag, frequently used in molecular biology protocols to express recombinant proteins). Both facts make the recombinant protein act as a zymogen like pre-protein.
In this sense, the sPLA2s obtained from G. max were expressed without N-terminal extension (Mariani et al., 2012, 2015b) by using the pHUE vector system that utilizes the ubiquitin fusion technique (Catanzariti et al., 2004), which allows easy purification and high yield of recombinant proteins (see Figure 1). The E. coli pHUE vector permits the expression of a particular protein as HIS-tagged ubiquitin fusion. Then, the HIS-tag-ubiquitin-sPLA2 fusion is further processed by the deubiquitylating enzyme used to cleave off the fusion to obtain the protein of interest free of any N-terminal extension (Figure 1).
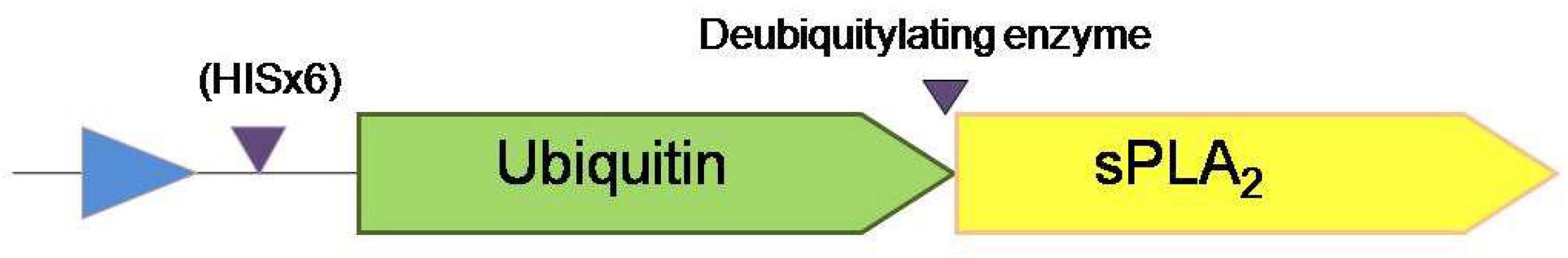
Figure 1. Schematic representation of the system applied for GmsPLA2s expression and purification. Triangles indicate the position where is the polyHIS-tag motif and the position where the deubiquitylating enzyme cut in order to release the protein with its native N-terminus without any extension.
In the particular case of the mature protein GmsPLA2-XIB-II, the LEU amino acid at the N-terminus was mutated to an ALA, to optimize the chance of obtaining the correct refolding as previously recommended (Kohler et al., 2006). In AtsPLA2-α, it was shown that an uncleaved signal peptide of the pre-processed forms produced a significant suppression of activity compared with the corresponding mature protein form (Ryu et al., 2005). Moreover, other sPLA2s from plants were expressed without the signal peptide (Mansfeld et al., 2006; Guy et al., 2009). In animals, a correct and functional sPLA2 from Bothrops diporus was produced without any extra extension at the N-terminus (Yunes Quartino et al., 2012). Using the ubiquitin/deubiquitinase system, in the latter case, it was clearly shown that the recombinant protein had the same interfacial catalytic profile when compared to the native one (Yunes Quartino et al., 2015).
As sPLA2 activity is very sensitive to N-terminus modifications, in Table 2 we include all sPLA2s from plants known until today and the process originally reported to obtain the final protein (either proteins purified from plant extracts, in a mature recombinant form or with an additional tag). It should be noted that not all reported information disclosed the sequence of phospholipase A2 either cloned or purified.
However, similar to its counterpart in animals, sPLA2s from plants have N-terminal signal peptides that were predicted to direct protein secretion into the extracellular or intracellular space (Bahn et al., 2003; Lee et al., 2003). It is noteworthy that some sPLA2s from plants have the sequences KTEL, KFEL, and KLEL at the C-terminal which are similar to the endoplasmic reticulum retention sequences KDEL and HDEL reported for animals (Matsushima et al., 2003). Even when this putative KxEL endoplasmic reticulum (ER) retention sequence (Pagny et al., 2000; Seo et al., 2008) is present in some plant sPLA2, the biochemical significance is still unknown (see Supplementary Figure S1).
GmsPla2s Gene Family, Classification, and Domain Structure
In G. max, five sPLA2s isoforms were identified (Mariani et al., 2012), named as GmsPLA2-XIA-I, GmsPLA2-XIA-II, GmsPLA2-XIB-I, GmsPLA2-XB-II, and GmsPLA2-XIB-III. Detailed information about the genes and proteins are shown in Table 3. As indicated above, the extension of the N-terminus of the mature protein is crucial for the activity, we show in Supplementary Figure S5 all the sequences of the sPLA2s of known plants with their signal sequence and their point of theoretical cut using the programs available online.
Moreover, the genes encoding for GmsPLA2-XIA-I and GmsPLA2-XIB-I are located in chromosome I, GmsPLA2-XIA-II y GmsPLA2-XIB-II are positioned in chromosome 7 and the gene of GmsPLA2-XIB-III is located in chromosome 8. Whereas GmsPLA2-XIB-I, GmsPLA2-XIB-II, and GmsPLA2-XIB-III possess three introns and four exons, the genes of GmsPLA2-XIA-I and GmsPLA2-XIA-II have two introns and three exons, respectively (see Supplementary Material in Mariani et al., 2012). These facts are indicative that during the course of evolution events of divergence and duplication might have occurred as it was suggested previously for AtsPLA2s (Lee et al., 2005).
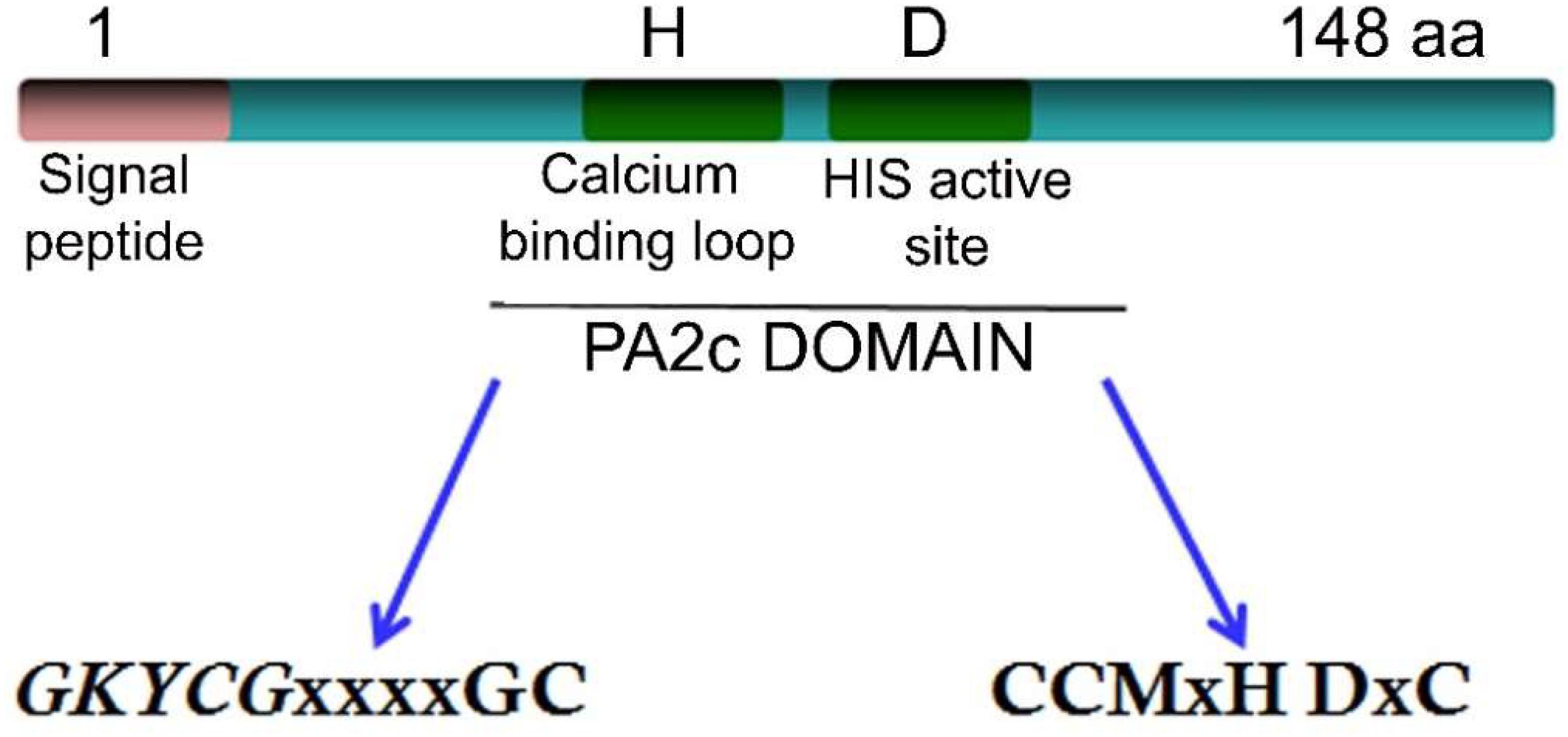
All sPLA2s sequences found in plants hold a PA2c (SMART accession number SM000851) domain that contains the highly conserved Ca2+-binding loop (YGKYCGxxxxGC) (see Figure 2). The active site motif (DACCxxHDxC) that holds the highly conserved HIS/ASP pair (Laigle et al., 1973) corresponds to position 49/50 for GmsPLA2-XIA-I, 47/48 for GmsPLA2-XIA-II and 62/63 for GmsPLA2-XIBs whereas for AtsPLA2α and AtsPLA2γ, it corresponds to the position 62/63 and 7/48, respectively (Mansfeld et al., 2006).
However, there is a dissimilarity that remains unclear in the HIS/ASP of the catalytic dyad in sPLA2s from plants compared with those found in animals (Mansfeld et al., 2006). It was proposed that water molecules assist in the Ca2+ coordination at the HIS48-ASP49 active site in bovine pancreatic bpsPLA2 (Bahnson, 2005), the roles of ASP99 in this sPLA2 (Kumar et al., 1994), and ASP64 in bee venom sPLA2 (West et al., 2013) were also claimed to take part in the hydroxyl-imidazole-carboxylate motif (Annand et al., 1996). However, for sPLA2 plant enzymes this important catalytic residue is replaced by an HIS or an ASN residue in enzymes from group XIA and by a SER or an ASN in those enzymes belonging to group XIB (Mansfeld et al., 2006) as shown in the alignment in Figure 3. Mansfeld et al. (2006) demonstrated that SER, ASN, or HIS in plant sPLA2s may fulfill the catalytic role assigned to ASP in animal’s sPLA2s (Mansfeld et al., 2006). Sequence alignment also reveals that, contrary to OssPLA2 isoforms, the ASP residue of the highly conserved HIS/ASP catalytic dyad of the animal counterpart is replaced by an HIS residue in the durum wheat TdsPLA2 isoform I, and by an ASN residue in all of the others durum wheat sPLA2 isoforms (Verlotta et al., 2013), see Figure 3 for more details of others sPLA2 from plants. Even though the comparison showed low homology among them within the overall amino acid sequences, both the catalytic site and the Ca2+ binding loop are highly conserved (Figure 3). Other relevant conserved residues within the Ca2+ binding loop are the two TYR and two GLY residues which are involved in the hydrogen bonding network reported for both animal and plant sPLA2s (Lee et al., 2005). A more perusal view of this domain offers additional information. The more conserved domain YGKYCG seems not to be exclusive, a change in the second TYR residue was observed for TdsPLA2-I changing to YGKFCG. Also, the following hydrophobic domain mainly formed by the LL pair may be VL, IL, IM, VS, IG, or VG (see Figure 3). However, the putative role of these differences on calcium affinity or phospholipase activity was not elucidate yet.
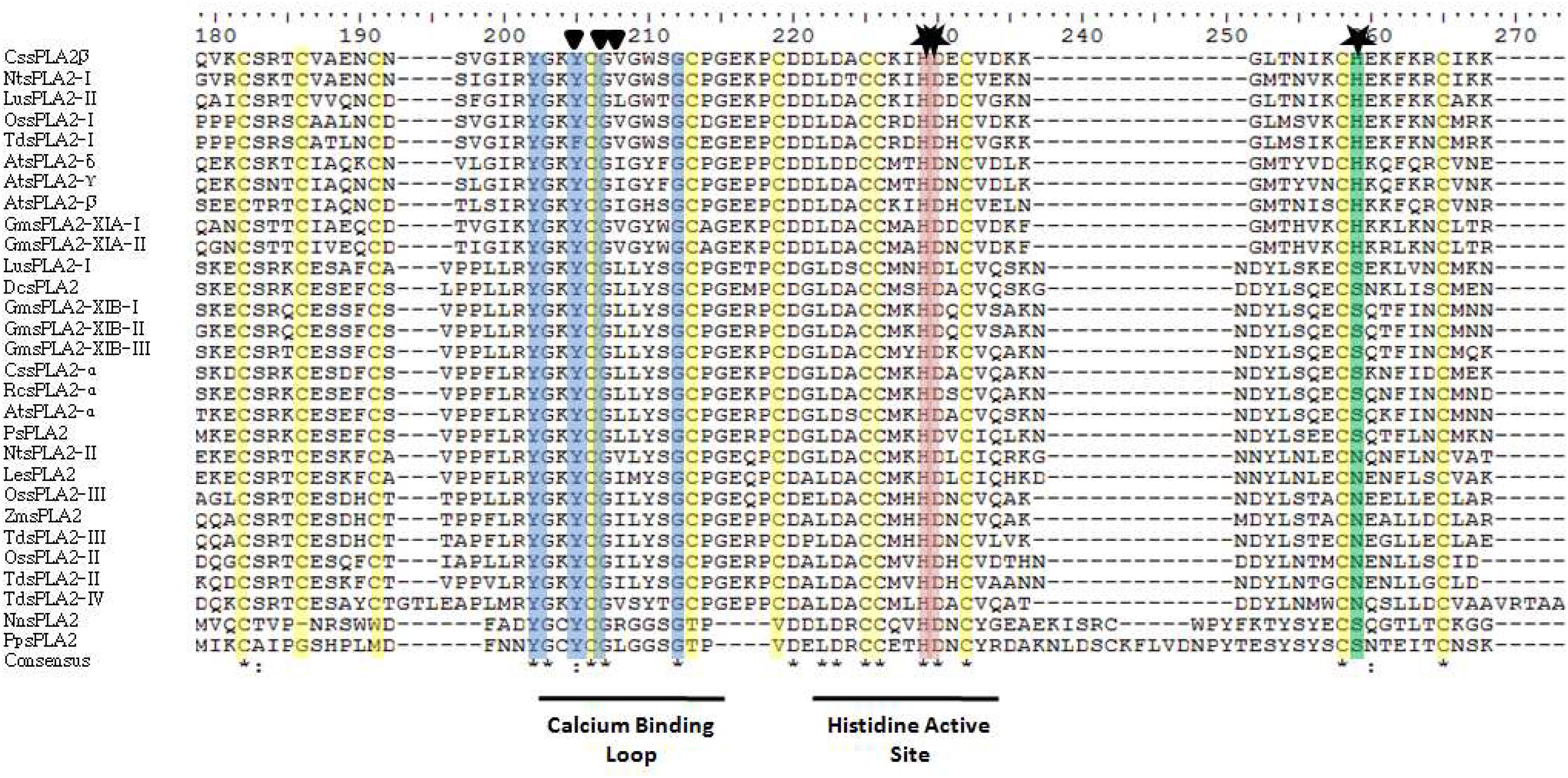
Figure 3. Sequence alignment of the sPLA2s reported from plants: analysis of the deduced amino acid sequences. The alignment was performed by using the Clustal X v2.1 program (http://www.clustal.org) and edited with jalview program. The triangles denote the amino acid residues involved in binding Ca2+ and stars denote amino acid residues putatively involved in catalysis. In yellow, CYS residues are marked. For abbreviation code of plants sPLA2 see Table 2. Two animals sPLA2 are included at the bottom of the figure for comparison: NnsPLA2 from Naja naja (cobra) venom and PpsPLA2 from pig pancreatic juice (Sus scrofa). The accession numbers of sPLA2 indicated in this figure are provided as Supplementary Appendix S1.
The mature proteins of both groups XIA and XIB contain 12 CYS residues (Figure 3) known to form six structural disulfide bonds that also are present in the same position as other known sPLA2s from plants (Mansfeld et al., 2006). CYS residues are essential for secreted sPLA2s and it has been shown to play a relevant role in the structural stability in mature sPLA2s (Six and Dennis, 2000; Mariani et al., 2015b).
The HIS residue (at position 49 in GmsPLA2-XIA-I, 47 in GmsPLA2-XIA-II and at 62 in GmsPLA2-XIB-II, -II, and -III) was suggested to play a crucial role in the nucleophilic attack at the sn-2 bond in the glycerol backbone of phospholipids for all sPLA2s (Six and Dennis, 2000; Berg et al., 2001; Burke and Dennis, 2009).
All plants sPLA2s are low MW enzymes (12–18 kDa) with the exception of CssPLA2β from Citrus that has an unexpected high MW (Table 4). The theoretical isoelectric points (pI) for each sPLA2 are shown also in Table 4. As it can be observed, four of the putative GmsPLA2s are rather acidic or neutral (GmsPLA2-XIA-I, GmsPLA2-XIB-I, -II, and -III) as reported for sPLA2s isolated from Bothrops diporus venom (de Haas et al., 1968; Daniele et al., 1997). Acidic sPLA2s were also reported for some enzymes found in the Crotalinae subfamily (dos Santos et al., 2011) and those found in rice (isoforms I and III) (Lee et al., 2005). On the other hand, the expected pI of GmsPLA2-XIA-II is slightly alkaline similar to those of all the sPLA2s found in Arabidopsis (Lee et al., 2005) and Papaver somniferum (Jablonicka et al., 2016); whereas other sPLA2s have a pI almost neutral as those found for carnation and tomato (Lee et al., 2005).
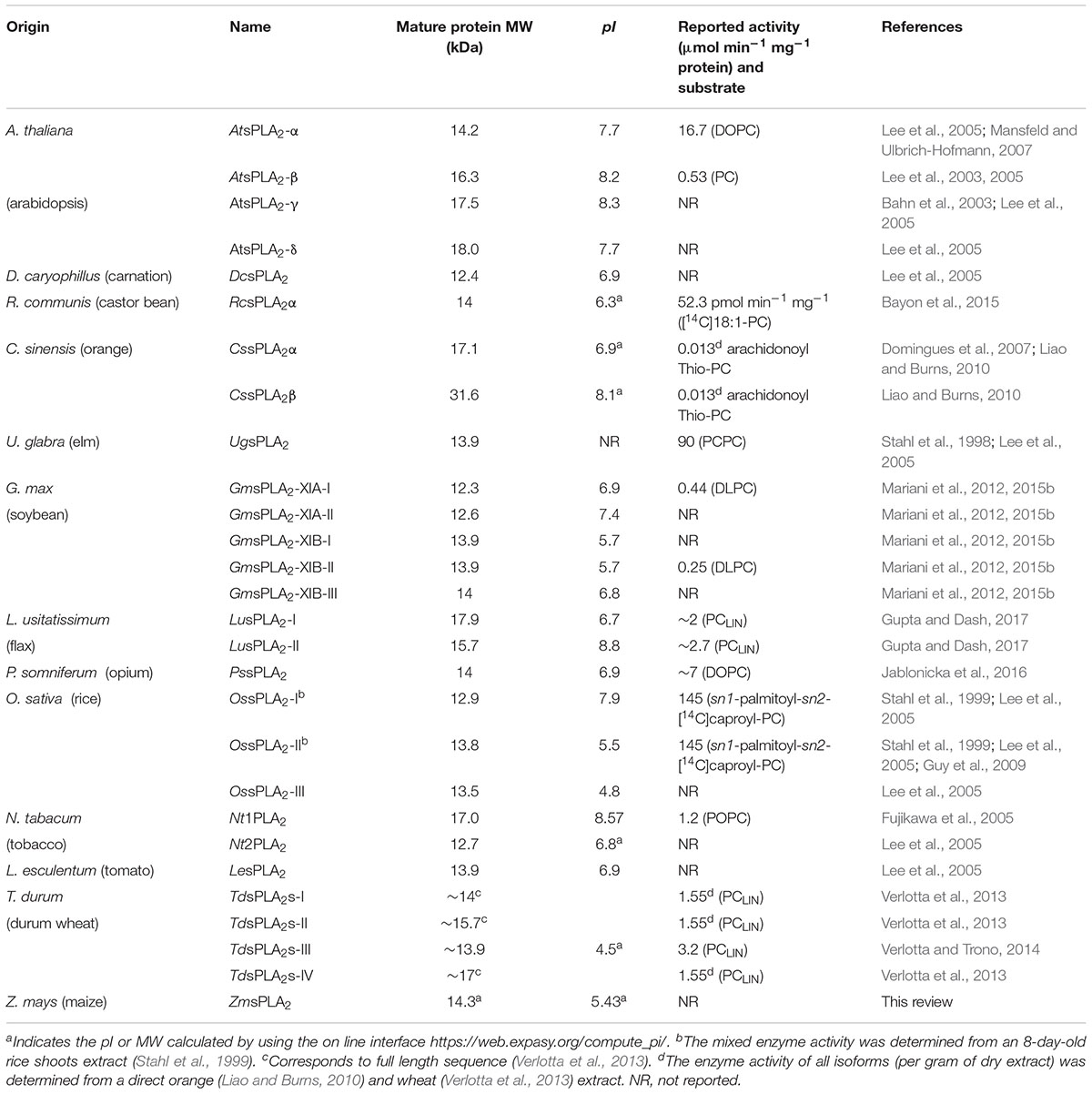
Table 4. Molecular weight, isoelectric point, and specific activity of different sPLA2s from plants.
The functional role of the diverse pIs found in different sPLA2s has not clearly been elucidated yet.
Another relevant domain information is that the enzymes from the different subgroups differ in the third Ca2+ coordinating amino acid, being a GLY residue in subgroup XIA or LEU residue in subgroup XIB (see Figure 3). The ASP located upstream in the sequence of the common HIS/ASP catalytic dyad found in animal sPLA2s does not correlate in the counterpart found in plants. Instead of this additional ASP residue, the plant enzymes that belong to group XIA contain an HIS residue, and the enzymes belonging to group XIB contain either a SER or an ASP residue (Mansfeld et al., 2006). The functional role of these differences with regard to the catalytic properties has not been completely elucidated yet.
GmsPla2s Classification in the sPla2 Superfamily
Secretory phospholipases in plants superfamily is composed of multiple members represented by multiple isoforms distinguishable by their structural, catalytic and physiological characteristics. sPLA2 are within the most populated group of PLA2 in nature which in turn is classified into 15 subgroups (Six and Dennis, 2000). In this context, the plant sPLA2s were classified into a separate group (group XI) (Meneghetti and Maggio, 2013), which, in turn, could be subdivided into two categories named XIA and XIB because of differences in MW and deviating sequences in the N- and C-terminal regions of the mature enzyme (Six and Dennis, 2000).
Figure 4 shows the phylogenetic classification into the two subgroups of all the sPLA2s from plants known until now. This way, GmsPLA2-XIA-I and -II are taking part of group XIA, which includes AtsPLA2-γ, AtsPLA2-β, AtsPLA2-δ, O. sativa isoform I, N. tabacum isoform I, T. durum isoform I, C. sinensis isoform β, and L. usitatissimum isoform II. Whereas two of the enzymes of G. max correspond to the subgroup XIA, three are grouped in the subgroup XIB (Mariani et al., 2012) named as GmsPLA2-XIB-I, -II, and -III together with AtsPLA2-α, O. sativa-II, -III, and -IV, D. caryophillus, N. tabacum isoform II, Z. maize, R. communis isoform α, P. somniferum, T. durum-II, -III, and -IV, L. esculentum, C. sinensis isoform α, and L. usitatissimum isoform I.
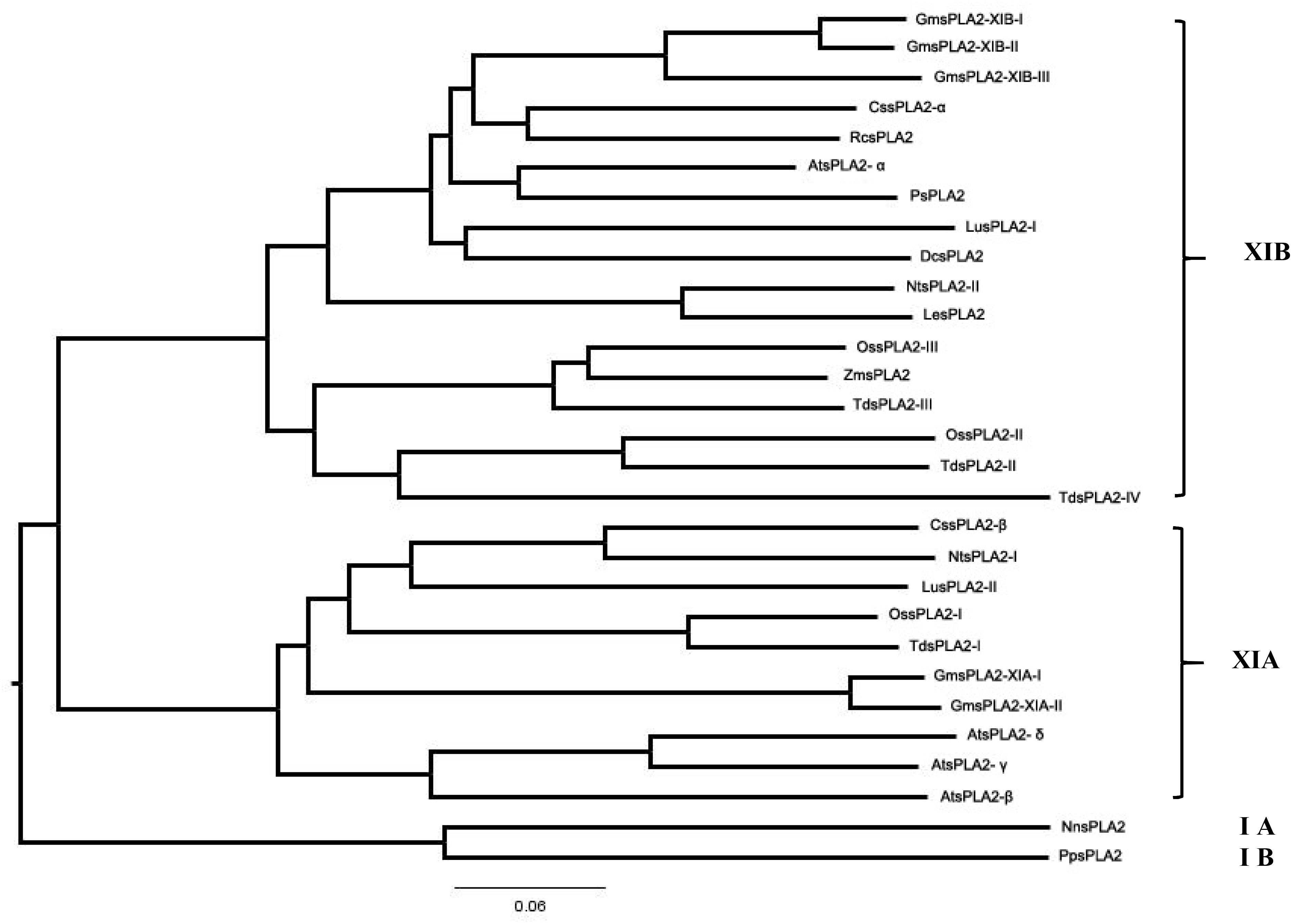
Figure 4. Phylogenetic tree of the deduced amino acid sequences of all reported plant sPLA2s. The program used to perform the sequence alignment was Clustal X (version 2.0) and to produce phylogenetic analysis the phylogenetic tree generation methods from the ClustalW2 package was used (on line tool http://www.ebi.ac.uk/Tools/phylogeny/clustalw2_phylogeny/). To draw the tree we used the Figtree v.1.4.4 program (http://tree.bio.ed.ac.uk/software/figtree/). The accession numbers of sPLA2 indicated in this figure are provided as Supplementary Appendix S1.
The data show a close evolutionary relationship among all sPLA2s from plants (see Figure 4). The highest level of similarity in amino acid sequences was observed between GmsPLA2-XIA-I and GmsPLA2-XIA-II, being of 95.5%, whereas between GmsPLA2-XIB-I and GmsPLA2-XIB-II the level of similarity is of 94.5% (Mariani et al., 2012) (see Supplementary Figure S2). Moreover, between LesPLA2 and NtsPLA2-II the level of similarity is of 90.4% and between TdsPLA2-I and OssPLA2-I, GmsPLA2-II and GmsPLA2-III, PsPLA2 and RcsPLA2-α and AtsPLA2-δ and AtsPLA2-γ the levels of similarity are of 89.9, 87.3, 83.7, and 82.7%, respectively.
Tridimensional Structure
Although the sPLA2 sequences from different sources differ significantly, the tridimensional structures have many features in common. There are more than 40 sPLA2s entries in the Protein Data Bank (PDB2) from all sources. Native and complex structures of sPLA2s simulated with mimic substrate have helped to identify the catalytically important residues involved in the active site (Pan et al., 2002).
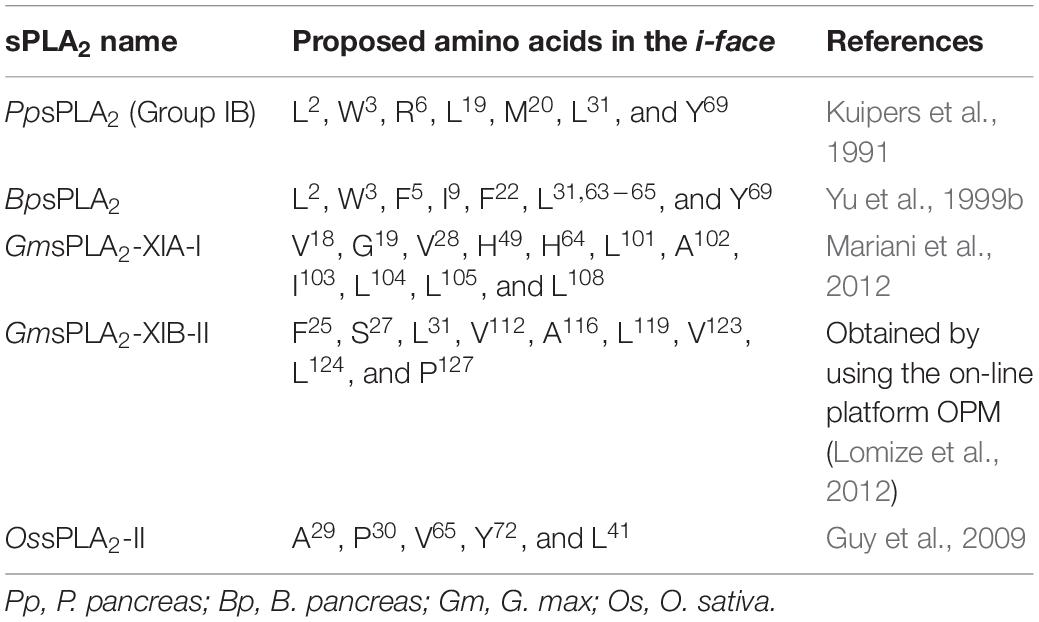
The tridimensional structure of many sPLA2s, such as porcine pancreas or bee venom (Dijkstra et al., 1984; Scott et al., 1990), has been elucidated by X-ray crystallography which revealed a common, rigid and highly conserved region with a similar tridimensional architecture compared with those from plants. The active site is not directly accessible to the aqueous phase and is within a rather local hydrophobic environment denoted as “i-face” that allows the interaction with the substrate in its monomeric form (Dijkstra et al., 1981). The putative residues involved in the “i-face” of some sPLA2 from plants are shown in Table 5.
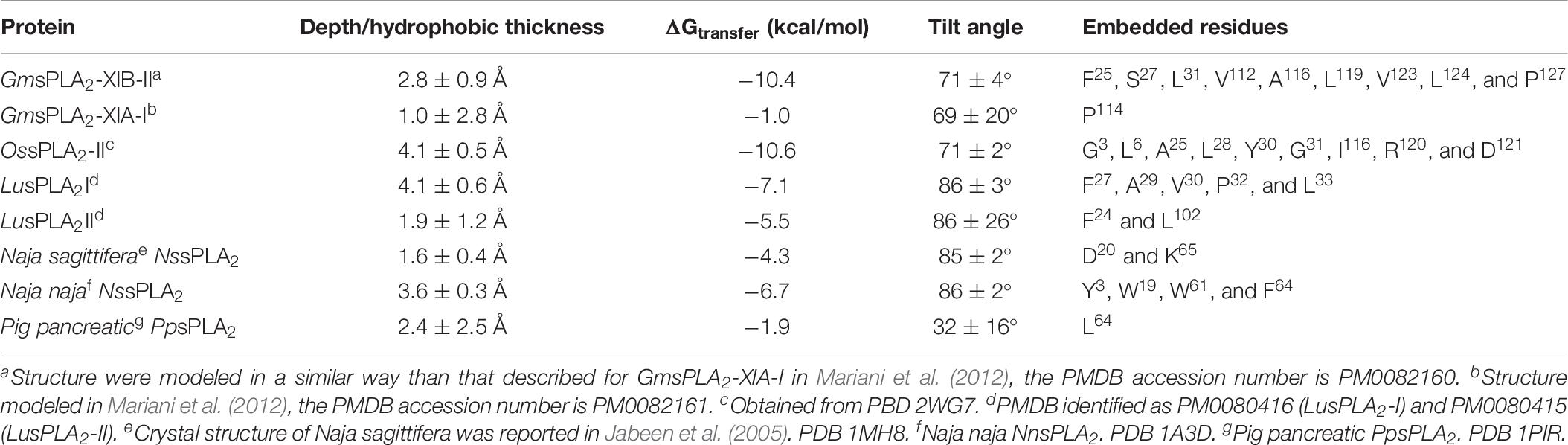
One of the first sPLA2 “i-face” identified was for the secreted pig pancreatic enzyme (Bai et al., 2008). In GmsPLA2-XIA-I the residues found in the putative “i-face” are VAL18, GLY19, VAL28, HIS49, HIS64, LEU101, ALA102, ILE103, LEU104, LEU105, and LEU108. Table 5 shows the putative amino acids proposed to be in contact with the membrane for different sPLA2s enzymes.
The binding of sPLA2 to the membrane is energetically favorable (Table 6) and, keeping in mind that most of the residues situated in the i-face are hydrophobic, the overall domain constitutes a hydrophobic environment that surrounds the active catalytic site. Hydrophobic side chains of the residues forming the “i-face” would be able to partition to the hydrophobic core, which allows the anchoring of the enzyme to the membrane, excluding water molecules in the region surrounding the active site and the diffusion of the substrate to the pocket of the active site to be hydrolyzed. The general molecular conformation proposed for plants sPLA2 is in agreement with the general vision proposed for secreted phospholipases of animal source (Scott et al., 1990).
Physically, the soluble sPLA2 protein must penetrate the phospholipid interface to exert its action. Therefore, the successful binding surface is located where the substrate is a prerequisite in the catalytic cycle, and this property can determine some specific characteristics of the enzyme activity. However, there are a limited number of charged residues in the flat topography of the “i-face” (see Table 5) that could modulate further interaction with the interface of the substrate in a way which has not been fully elucidated yet (Jain and Berg, 2006).
It is important to note that even when the energetic to membrane binding is favorable according to the available on-line calculation program (Lomize et al., 2012) used for some sPLA2s, the residues involved in the “i-face” differ for the same enzyme if a different approach is used instead (compare Tables 5, 6).
To date, only few structures corresponding to sPLA2s from plants were reported in the PDB or in the Protein Model Database (PMDB3) and correspond to O. sativa (rice) isoform II (PBD 2WG7), which belongs to the group XIB, and its tertiary structure was recently determined by X-ray crystallography to 2.0 Å resolution (Guy et al., 2009). Moreover, homology modeling and molecular dynamics were used to elucidate the structure of sPLA2 isoform α from Arabidopsis (Mansfeld et al., 2006) but its PDB is not available. The predicted models of LusPLA2s proteins were elucidated and submitted to PMDB identified as PM0080416 (LusPLA2-I) and PM0080415 (LusPLA2-II) (Gupta and Dash, 2017). The structure of GmsPLA2-XIA-I was modeled by using homology modeling and molecular dynamics (Mariani et al., 2012) and also GmsPLA2-XIB-II by using a similar methodology (see Figure 5, modeled structures in PDB format were not uploaded in the PDB). The data corresponding to Pig pancreatic (Sus scrofa), Naja naja (Indian cobra), Naja sagittifera (Andaman cobra venom) are also indicated in Table 6 for comparison in order to include sPLA2 able to hydrolyze aggregate lipids structured in a high packing organization, as it occurs with sPLA2 from cobra venom, or only at low packing as it certainly happens with sPLA2 from pig pancreas (see below and Table 10).
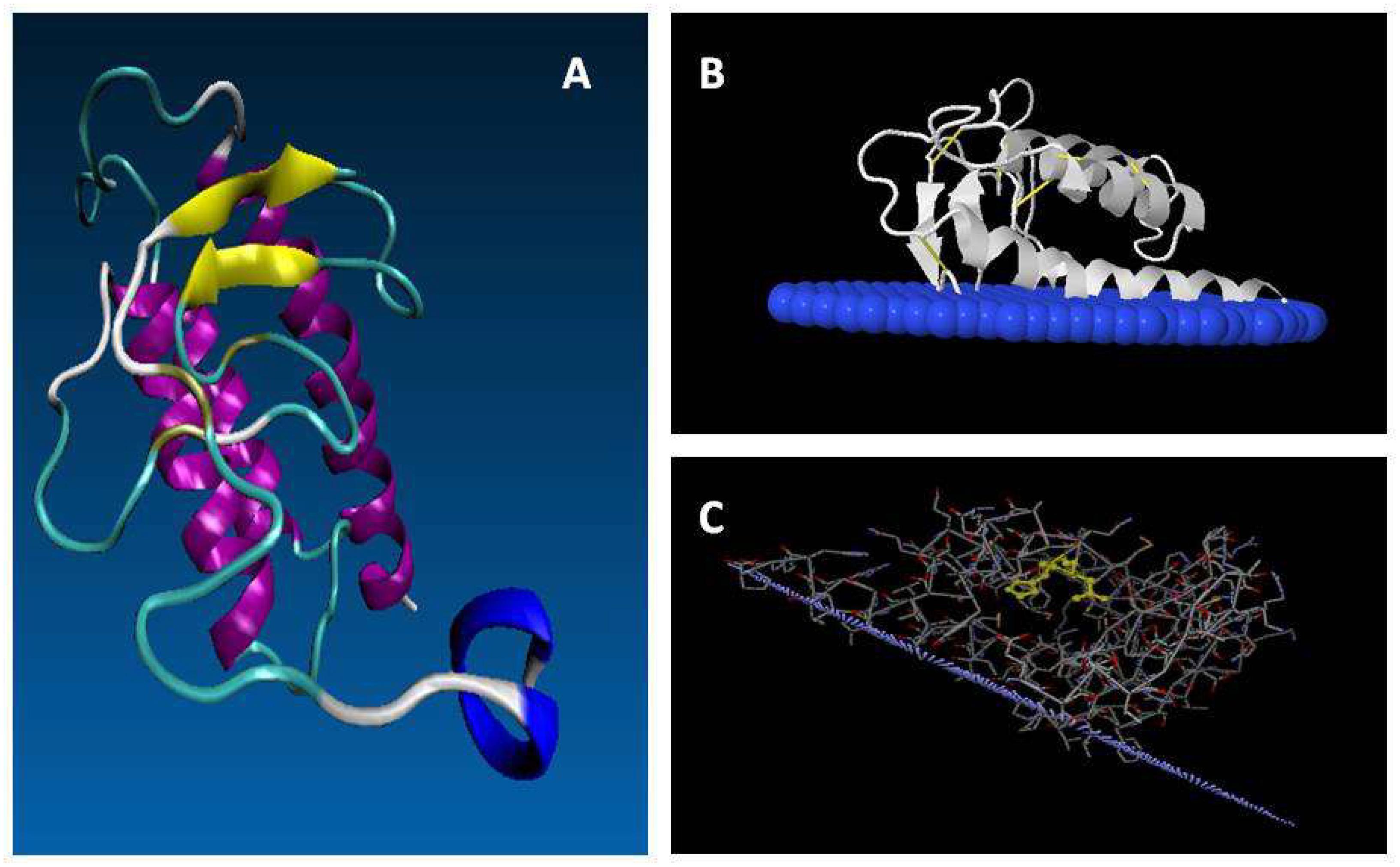
Figure 5. Putative mature structure of GmsPLA2-XIA-I. (A) Proposed structure from homology modeling of GmsPLA2-XIA-I (Mariani et al., 2012). Yellow: beta sheet strand; magenta: alpha-helix; blue: C-terminal; cyan: turns; white: coils. (B,C) Molecular simulation of the interaction between GmsPLA2-XIB-II with the membrane interface, simulated with the OPM (Orientation of Proteins in Membrane) database online service (opm.phar.umich.edu/; see Lomize et al., 2012). In panel (B) blue: interfacial membrane; white: protein. (C) Light purple represents the interfacial membrane; the sticks denote the protein amino acids with the H/D dyad highlighted in yellow.
The structure of rice sPLA2 shows that the half N-terminal chain contains mainly structured loops, including the conserved calcium binding loop domain together with two short anti-parallel β-strands. The half C-terminal is folded into three anti-parallel α-helix, in which two of them are highly conserved among others sPLA2s, containing the crucial catalytic HIS residue and the calcium binding/coordinating ASP residues (Guy et al., 2009). This overall general folded conformation seems to be shared by almost all known sPLA2 from plants. The complete putative mature structure of GmsPLA2-XIA-I protein was reported using homology modeling and molecular dynamics simulations (Mariani et al., 2012). The most mobile regions are the N- and C-terminal, followed by the loops in residues 74–85, 53–62, 34–37 that connect, respectively, the last two helices, the first with the second helix, and the last beta-sheet with the first helix (see Figure 5). As other sPLA2s in the family, the dominant secondary structure is the α-helix, with only a small portion of beta sheet with abundant regions containing turns and bends. The observations indicate that the terminal helix is rather a dynamic region and has three principal conformations: one fully helical, other with the last seven residues in coil, and the third one with a kink plus coil (Mariani et al., 2012). As noted before, this behavior can be attributed to a low number of hydrophobic contacts of this region, a high aqueous exposed area and the presence of a highly flexible GLY98 residue (Mariani et al., 2012).
The active site of the sPLA2 protein contains a crucial calcium ion cofactor commonly present in other plant sPLA2s (Mansfeld et al., 2006; Guy et al., 2009) that is important in the catalytic mechanism and is a requisite for full enzyme activity. The HIS-ASP pair constitutes the active center and the calcium binding loop (see Figure 3) is essential for the proper function of the enzyme (Scott et al., 1990). All sPLA2s catalyze the hydrolysis through the same mechanism: an abstraction of a proton from a water molecule followed by a nucleophilic attack on the sn-2 bond position of the diacylglycerophospholipids (Jorgensen et al., 1983; Berg et al., 2001). NMR structural studies of porcine pancreas sPLA2 show that the N-terminus is flexible with no defined structure in solution, unlike what it was evidenced by crystallography. It was hypothesized that this flexibility in solution would be related to the near null activity against monomeric substrate form [more unstructured unbound state (van den Berg et al., 1995)].
Enzymatic Properties of Plants sPla2s
Optimum Conditions for Plants sPLA2s Catalysis
The sPLA2s from N. tabacum and elm have optimum pH in the range of 8–10 and 8–9, respectively (Stahl et al., 1999; Fujikawa et al., 2005). In Arabidopsis the optimum pH ranges for the activities are pH 6–11, 6–7, 7–9, and 8–9 for AtsPLA2-α, -β, -γ, and -δ, respectively (Lee et al., 2005). Nevertheless, a similar situation was found for almost all the sPLA2s found in plants or animals. The pH optimum was at around 7 for GmsPLA2-XIA-I and -XIB-II (see Table 7), when using mixed micelles of DLPC:Triton X-100 as substrate in presence of calcium 10 mM. The optimum pH for pancreatic sPLA2 was reported to be 8 (de Haas et al., 1968; Fujikawa et al., 2005) similar to that reported for bee venom (Daniele et al., 1997). For human non-pancreatic PLA2 optimum pH is in between 8 and 10 (Kramer et al., 1989). However, it should be mentioned that different substrates (including different aggregation presentation of substrate) have been used to determine optimum pH for the different sPLA2s reported in the literature.
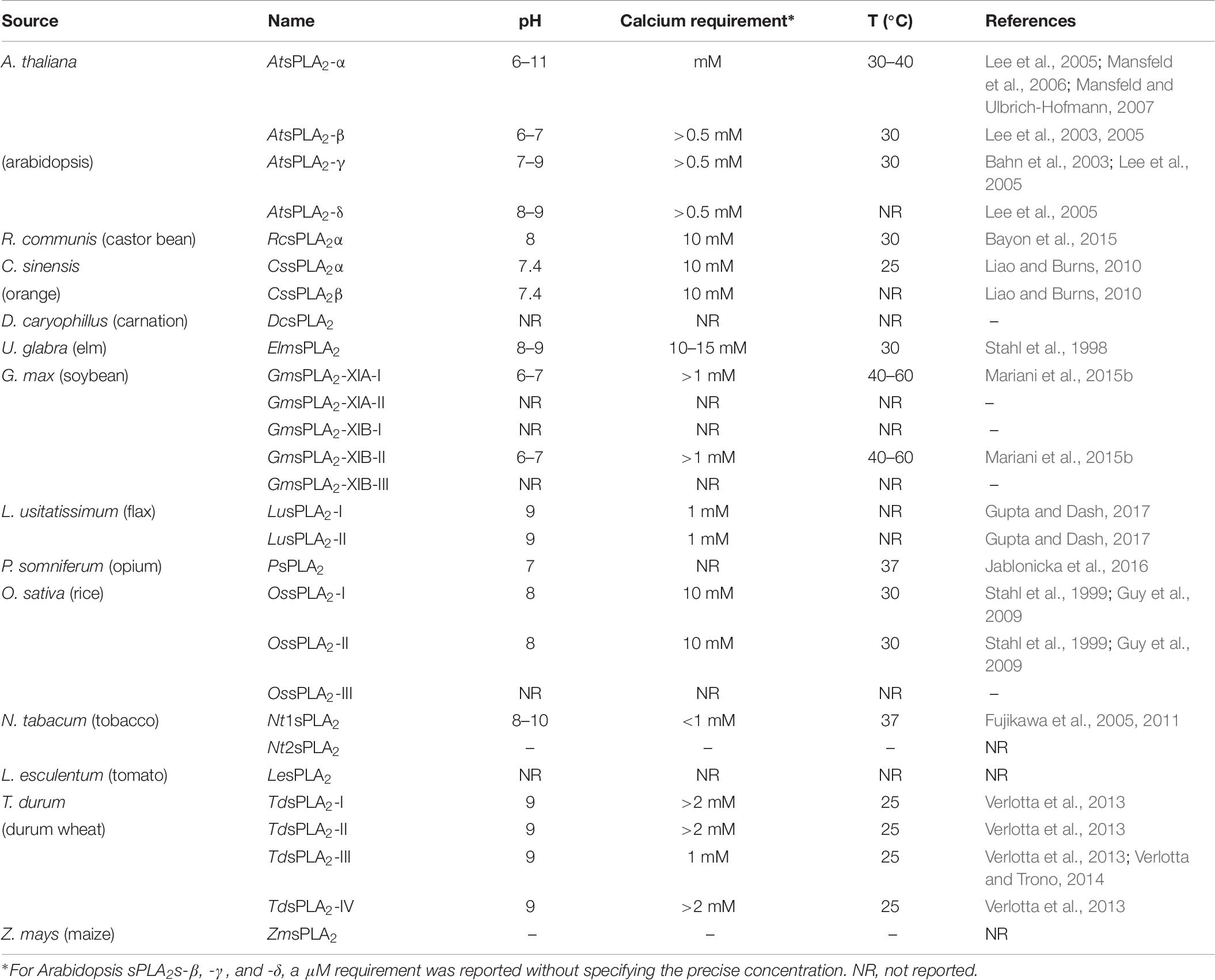
Table 7. Optimum requirements deduced for catalytic activity of the different sPLA2s found in plants.
Only few sPLA2s were investigated about the optimum temperature and stability. GmsPLA2s-XIA-I and -XIB-II demonstrate to be very stable when increasing the temperature (Mariani et al., 2015b) as previously determined by using an sPLA2 homogenate (Minchiotti et al., 2008). This proves that these enzymes are highly resistant to temperature denaturation due in part to the disulfide bridges that are postulated to be involved in the stability of sPLA2s (Berg et al., 2009; Murakami et al., 2010). Table 7 shows the optimal temperature reported for several sPLAs from plants.
The optimum calcium concentrations for activity of GmsPLA2-XIA-I and -XIB-II are in the micromolar range using DLPC:Triton X-100 mixed micelles as substrates (Table 7). This micromolar calcium requirement is rather unusual for sPLA2s enzymes that mostly possess millimolar requirement (Six and Dennis, 2000). Moreover, the same behavior was observed for the activities of AtsPLA2-β, -γ, and -δ (Lee et al., 2005). It is important to remark that none of these secreted enzymes (either from animals or plants) exhibit activity in absence of calcium. Particularly, for AtsPLA2-α the activity augmented as the calcium concentration increased up to 10 mM and for elm sPLA2 the range of calcium concentration for optimal activity was around 10–15 mM CaCl2 (Stahl et al., 1998; Lee et al., 2005; Mansfeld and Ulbrich-Hofmann, 2007). However, to achieve 50% of maximal enzyme activity a concentration of 0.5 mM CaCl2 was sufficient, at least, for these two latter enzymes. The maximal activity for sPLA2 from N. tabacum was detected above 1 mM CaCl2. This behavior is similar to that observed for the most animal sPLA2s, which require millimolar concentrations of Ca2+ and have no activity in the absence of this cation (Six and Dennis, 2000; Fujikawa et al., 2005). Even when it is evident the molecular differences among the enzymes in the sPLA2 family, the absolute requirement of Ca2+ for hydrolysis is indicative that all of them share a common mechanism for lipid hydrolysis. For durum wheat sPLA2 the activity continuously increased as Ca2+ concentration increased with a plateau close to 2–4 mM CaCl2, even though a 300 μM CaCl2 was sufficient to reach 50% of the maximal activity (Verlotta et al., 2013) (see Table 7).
The differences in the activity reported from many authors for plant sPLA2s is not easy to compare in absolute terms. Usually the reported activity values are informed as specific activities (μmol of hydrolyzed lipid.min–1.mg of protein–1) and this quantity may be affected by many factors. Among the main factors that can affect the sPLA2 activity can be mentioned (i) inherent deficiencies in the folding of recombinant enzymes, (ii) additional tags at the N-terminus, and (iii) the lack of standardization of substrate offered to the enzyme (lipid monolayers, micelles, SUVs, presence of detergents mixed with the lipid substrate, etc.). However, taking into account these precautions the activity of the reported enzymes could be compared although different substrates and systems were used in the assays (see Table 4).
Conformational Stability of sPLA2s
It is known that CYS residues are essential for the structural stability of sPLA2 and it has been shown to play an important role in the structural stability of the mature enzyme (Six and Dennis, 2000; Welker et al., 2011). In animals, sPLA2s contain between 10 and 16 CYS that have the potential to form 5–8 intramolecular disulfide bridges (Schaloske and Dennis, 2006). In contrast, all sPLA2s reported from plants have 12 CYS that can form 6 disulfide bridges (see Table 1). It is known that some sPLA2 from animals (especially type I and II), are rather stable upon heating compared with cytoplasmic cPLA2 (Mazereeuw-Hautier et al., 2000). Resistance to heating for sPLA2 from plants was reported for some enzymes indicating a similar behavior to that observed for animal sPLA2. The structural stability for durum wheat sPLA2 was demonstrated by the resistance to high temperatures (87% of the activity was retained after treatment of the crude leaf extract at 100°C for 15 min), see (Verlotta et al., 2013). Recombinant AtsPLA2α and AtsPLA2β retained 80–95% of their activities following 5 min treatment in boiling water (Lee et al., 2005), and a similar result was obtained for sPLA2 purified from elm seeds (Stahl et al., 1998). Moreover, for GmsPLA2-XIA-I and GmsPLA2-XIB-II preserved the activity after heating 5 min at 80°C (Mariani et al., 2015b).
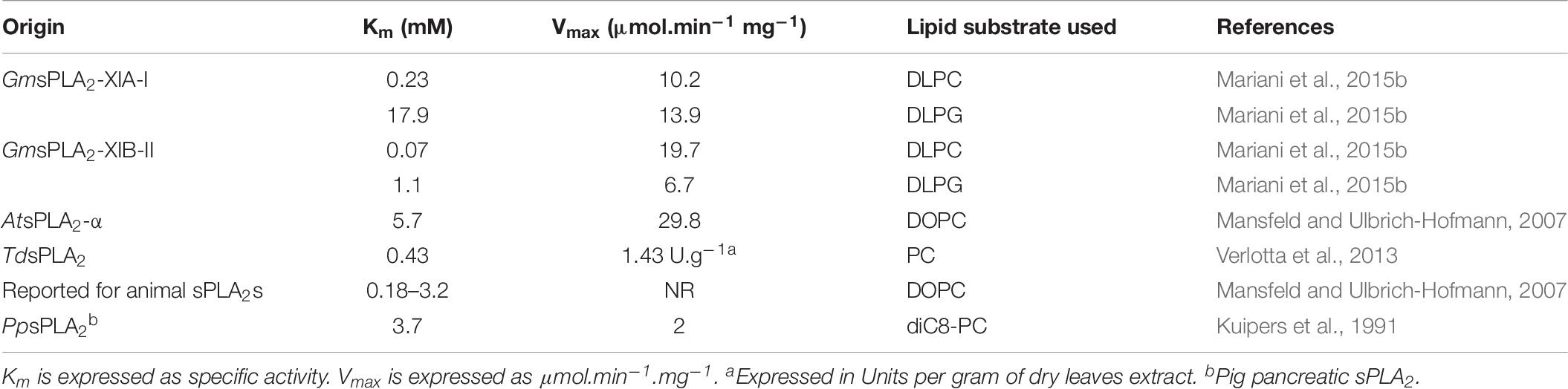
The main reason for the scarceness of information on recombinant plant sPLA2s may be attributed to the low expression yields obtained with the different protocols currently used and the strong propensity of the recombinant enzymes to aggregate (Mansfeld et al., 2006). The generally lower yields of the purified enzymes from inclusion bodies might be an indication for a higher fraction of misfolded and/or aggregated protein after the renaturation process. This may be the reason of different Vmax or specific activity values obtained when studying kinetic parameters in sPLA2 recombinant enzymes from plants (see Table 9).
In bee venom sPLA2 (BvsPLA2), it was reported that the formation of disulfide bonds is not essential for correct re-folding of the protein and an active enzyme form can be reobtained even from the completely denatured and reduced state (Welker et al., 2011). It is known that, in contrast to the seven disulfide bonds present in porcine pancreas enzyme (PpsPLA2), all five disulfide bonds of BvsPLA2 are essential for conformational stability and contribute to the activity (Welker et al., 2011). In the case of bacterial sPLA2 from Streptomyces violaceoruber, it possesses only two disulfide bridges (Sugiyama et al., 2002) which were sufficient to be active comparable to animal or plant sPLA2s (Yunes Quartino et al., 2015).
In sPLA2 from A. thaliana, the removal of disulfide bonds increased the proteolytic susceptibility of the native proteins whereas the stability decreased (Mansfeld et al., 2014). Regarding GmsPLA2s, it was demonstrated that the calcium ion also contributes to keep the protein folded in its native structure. This effect was observed by two independent assays using dynamic simulations and intrinsic fluorescence experiments (Mariani et al., 2012, 2015b).
The comparison of the data obtained on bovine pancreatic sPLA2, bee venom sPLA2, and porcine pancreatic sPLA2 with those obtained on sPLA2s from plants suggests that conserved disulfide bonds in those homologous proteins are important to keep the conformational architecture and stability. However, with the recompiled information, it is almost clear that not all the disulfide bridges are needed for the protein to be active, but are necessary for a protein correct folding.
Interfacial Catalysis Activation
Phospholipids are constituents of biological membranes, so a very important prerequisite step to perform the lipolytic action of sPLA2 is the interaction with the amphipathic nature of these interfaces; and in turn, determine the catalytic properties of the organized substrate (Jain and Berg, 2006). The interfacial binding step is crucial for enzymatic action of sPLA2, and it is mediated by a region of the protein often referred to as i-face (see above), also reported as IRS, the interfacial recognition site (Tatulian, 2001). The i-face or IRS is not a proper “site” or a flat face, it is rather a 3D domain with the confluence of several residues that crowns and precedes the catalytic site, giving an adequate environment for the catalysis, and also help keeping the enzyme attached to the membrane where the hydrolytic reaction takes place. The proper intimate contact of the i-face of sPLA2s with the interface is essential to provide the substrate access to the active site. Interfacial activation is a concept that means an adequate contact between the catalytic active site and the i-face modulating the catalytic activity (Scott et al., 1990; Tatulian et al., 2005; Jain and Berg, 2006; Winget et al., 2006). The binding and kinetic characteristics of interfacial catalysis by sPLA2 depend upon the organization and dynamics of the interface. The overall rate of catalytic turnover is not only determined by the kinetics at the interface, but also by the binding/desorption equilibrium kinetics of the enzyme with the interface (Ramirez and Jain, 1991). Hence, the hydrolysis of the organized substrate can occur in two extreme distinct modes: (i) in the scooting mode of catalysis, that requires that the enzyme remains bound at the interface between several catalytic turnover cycles and, (ii) in the pure hopping mode, where the binding and the desorption of the bound enzyme occur during each catalytic turnover cycle leading to a jumping mechanism (Jain et al., 2009) (see Supplementary Figure S4 for more details and a schematic description of both mechanism of lipids hydrolysis induced by sPLA2).
A few mode of interfacial catalysis for sPLA2s has been reported. Moreover, in plants, we were the only in studying the catalytic mode till today. The enzyme studied in order to determine the mode of catalysis was the sPLA2 from G. max (GmsPLA2-XIA-I) (Mariani et al., 2015b). Whereas pancreatic sPLA2 presents a scooting mode of catalysis when using anionic lipids (Berg et al., 1991), it presents a hopping mode of catalysis if the specific experimental conditions are changed to zwitterionic lipids (Scott et al., 1994). In our hands, GmsPLA2-XIA-I acts in the hopping mode against zwitterionic lipids (Mariani et al., 2015b).
Hydrolysis Using Micelles as Substrate Membrane Model System
There have been some reports in the literature regarding sPLA2 activity against different substrates and in different conditions. For some sPLA2, it has been demonstrated that the hydrolysis rate is sensitive to the surface charge density of the lipid aggregates (Volwerk et al., 1986). Several kinetics studies on pancreatic as well as snake venoms and plants phospholipases have been reported in which lipid phase transition, lipid membrane curvature, and composition may modulate the lipolysis (Wilschut et al., 1978; Bell and Biltonen, 1989; Bell et al., 1996; Leidy et al., 2004). However, it should not be forgotten, that sPLA2 has optimum of lipid packing for hydrolysis, i.e., that some enzymes have the ability to hydrolyze lipid in a low packing organization (low lateral pressure in lipid monolayers more compatible with micelles) but others also have optimum condition of hydrolysis at high lateral pressure in monolayers compatible with liposomes or biological membranes (Ramirez and Jain, 1991; Yunes Quartino et al., 2015).
Usually, short-chain zwitterionic phospholipids have been employed as substrates in single component systems (de Haas et al., 1971; Wells, 1972) or, for the case of long-chain phospholipids, they were mixed with neutral detergents (Dennis, 1973b; Yu et al., 1999a; Mansfeld and Ulbrich-Hofmann, 2007). Moreover, the activity is generally increased when the lipid substrate forms mixed micelles in presence of detergents (Dennis, 1973a; Dennis et al., 1981). The effect of enzyme immobilization on the sPLA2 kinetics was also reported (Madoery et al., 1999). Description and kinetics properties of sPLA2 from plants have been more frequent in their recombinant counterpart after appropriate expression, purification, and folding protocols (Bahn et al., 2003; Ryu et al., 2003; Fujikawa et al., 2005; Mansfeld and Ulbrich-Hofmann, 2007; Mariani et al., 2012) compared with their equivalent found in animals sPLA2s. The reason of this is due to the relative high amounts of the latter proteins found in their respective natural sources (venoms and pancreatic juice) and, therefore it allows an efficient purification of the mature forms of sPLA2. However, few studies using purified plant enzymes were reported from elm seeds (Stahl et al., 1998) and of G. max (Minchiotti et al., 2008).
Mammalian and plant enzymes differed in head group specificity. While some mammalian sPLA2s show high activity on anionic phospholipids (Ghomashchi et al., 1991; Bezzine et al., 2002), sPLA2s from plants preferred zwitterionic phospholipids (Mansfeld and Ulbrich-Hofmann, 2007; Mansfeld, 2009; Mariani et al., 2015b). In Table 8 we summarize the substrate lipid preference (head group or acyl chain) differences observed in some sPLA2s from plants reported in the literature (see also Table 9 additional kinetic data).
Table 9 shows the Km and Vmax values determined and reported for sPLA2s from different sources. As shown, we can infer that the values of Vmax could be sensitive to both the lipid substrate used in the assays and the interfacial quality of the surface in which the substrate is inserted.
Phospholipid Hydrolysis Using Langmuir Monolayers as Membrane Model System
The influence of substrate lipid packing on sPLA2s activities was studied for numerous authors using Langmuir-lipid monolayers performed at different surface pressures using almost exclusively sPLA2 from animal sources (Yunes Quartino et al., 2015). Moreover, to study the catalytic activity at the air-water interface the lipid monolayer technique in the “zero order” regime was used since the surface pressure is kept constant during the reaction (Panaiotov and Verger, 2000; Yunes Quartino et al., 2012) (see Supplementary Figure S3 for a schematic representation of this experimental system).
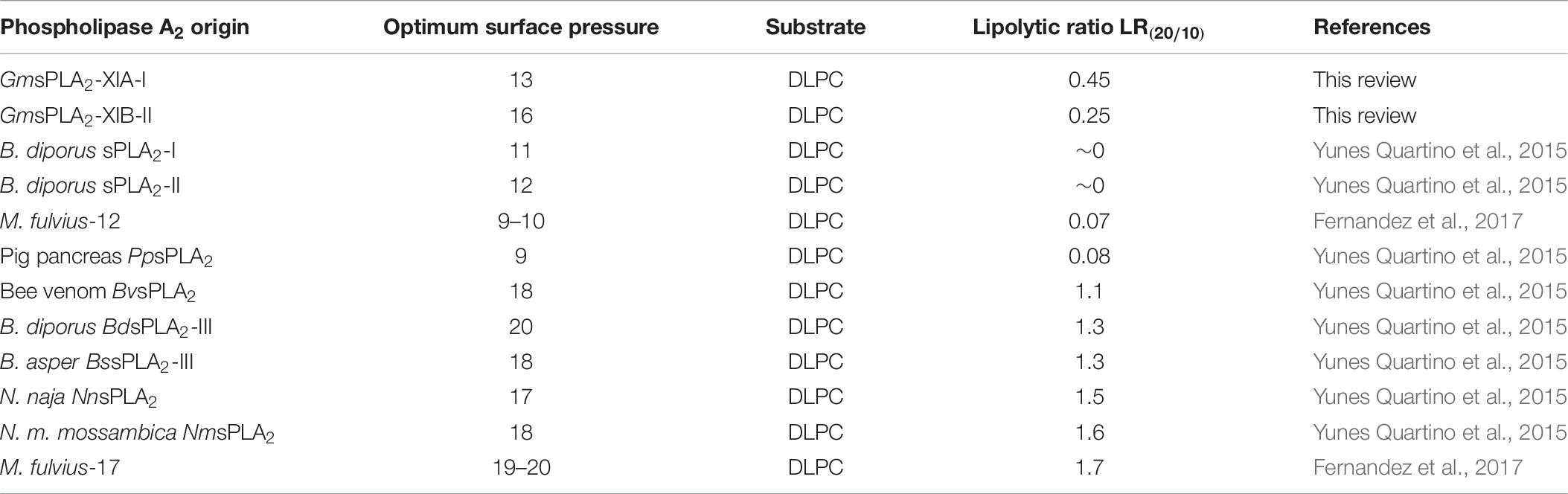
The optimum surface pressure of these enzymes to hydrolyze the lipids of the membranes differed with the origin of the sPLA2 (Ramirez and Jain, 1991; Mariani et al., 2015b; Yunes Quartino et al., 2015). GmsPLA2s were the first sPLA2s from plants to be studied with respect to their interfacial characteristics. Table 10 shows the optimum pressure determined for different sPLA2s. The optimum for plants GmsPLA2s seems to fall intermediate in between the values of “pancreatic like” enzymes that have high activity against micelles structured lipids rather bilayers (lipolytic ratio lower than 0.1) compared with toxic venom sPLA2s (lipolytic ratio higher than 1) that can hydrolyze intact cell membranes such as erythrocytes (Demel et al., 1975). Then, it may be concluded that sPLA2s from plants would have a more ubiquitous functionality, since they can be active in vitro against a rather wide range of curvature radio of structured lipid substrates (less sensitivity to the supramolecular organization).
Auxin Effect Over sPLA2 Activity
Studies of plant sPLA2s demonstrated that auxins play important roles in signal transduction regulating cellular processes and probably they are implicated in phospholipid signaling (Wang, 2001; Ryu et al., 2005; Scherer et al., 2010). At the cellular level, auxins control cell division, growth, extension, and differentiation (Davies, 1995). At the whole plant level, auxins play an essential role in processes such as apical dominance, lateral/adventitious root formation, tropisms, fruit set and development, vascular differentiation, and embryogenesis (Friml, 2003). A rapid increase in sPLA2 activity was first verified by treating isolated microsomes and cell cultures with auxins (Scherer and Andre, 1989; Scherer, 1990; Andre and Scherer, 1991; Scherer, 1992; Scherer and Andre, 1993; Scherer, 1996) and microsomes isolated from hypocotyls segments (Blanchet et al., 2008b). However, as the molecular mechanism of the putative effect of auxins over sPLA2s is unknown we have investigated whether these phytohormone have any direct effect over the enzyme by using simple in vitro assays.
Secretory phospholipases, like other lipolytic enzymes, are interfacial active proteins, since they access from water to the interface of the insoluble organized substrate to carry out the lipid hydrolysis. For this reason, the activity of the enzyme is directly modulated at the interface by the supramolecular organization of the substrate summarized in the concept of “interfacial quality” [e.g., the physical state of the lipids, proper lateral packing, modulation by non-substrate lipids, “membrane lateral defects,” among others (Verger et al., 1978; Daniele et al., 1996; Jain and Berg, 2006; Blanchet et al., 2008a; Campagnoli et al., 2008; Fico et al., 2008; De Tullio et al., 2013)].
The stimulation effect of auxins over recombinant sPLA2s from G. max is rather an interfacial effect. Despite porcine pancreas sPLA2 presents low identity with the known reported sPLA2s from plant sources, shows a significant similarity in the active site and calcium binding loop regions (Mansfeld and Ulbrich-Hofmann, 2007), making it an acceptable model for comparison. Using mixed micelles was determined that the effect of auxins on sPLA2 stimulation depends on the concentration of the phytohormones employed with an optimal effect around 100 μM [the maximum perturbing effect (Mariani et al., 2015a)]. The hypotheses states that a direct action over sPLA2 enzyme molecule or a synergic effect on the micelle surface doing more favorable the interface for lipolysis occurs. Both phytohormones IAA (indole 3-acetic acid) and IPA (indole 3-propionic acid) were active toward both type of sPLA2, either coming from plant or pancreatic sPLA2, suggesting that there is not a direct specific enzyme-phytohormone interaction involved. So, the effect of auxins can be attributable to changes in the interfacial quality of the organized substrate rather than a direct effect over the enzyme (Mariani et al., 2015a). The molecular details by which the particular mixed interfaces formed by auxins/phospholipids may modulate the sPLA2 activity, regardless of the enzyme origin, remain to be elucidated. However, to ascertain the interfacial hypothesis of auxins over the action of sPLA2 we further analyzed the surface properties of two auxins: IAA and IPA, i.e., the capability of these phytohormones to partition into lipid interfaces (Mariani et al., 2015a). Both IAA and IPA did not show any affinity toward lipid-clean interfaces (self-adsorption to water surface) but, very importantly, both auxins showed the ability to penetrate lipid interfaces forming stable and insoluble monolayers with phospholipids. This capability to form mixed lipid-auxin interfaces allowed the activation of two recombinants GmsPLA2s and pancreatic sPLA2 (Mariani et al., 2015a). The interfacial activation exerted by auxins was, regardless of sPLA2 source, supporting the theory that at the action is at lipid-auxin interface and not a direct effect over the enzyme (Mariani et al., 2015a).
Potential Industrial Application and Perspectives
The application of biotechnology, particularly enzymes in industrial processes, is continuously growing due to its minimal environmental impact, since they produce non-toxic waste substances and consume little energy (Warner, 2005). Natural and modified phospholipids have been extensively used in food industry, cosmetics, pharmaceuticals and agriculture (Guo et al., 2005). Therefore, in the production of these “modified phospholipids,” secreted phospholipases obtained mostly from microorganisms or mammals have been used by the industry either for refined oils, dairy products, baked goods and other health food industries (De Maria et al., 2007; Wang et al., 2012). As sPLA2 enzyme catalyze the stereospecific hydrolysis at the chiral carbon (sn-2) of glycerophospholipids converting them to lysoderivatives, the enzymatic bioconversion is the only selective pathway for obtaining sn-2-lysophospholipids. Lysophospholipids have a greater bioemulsifiers capability and have been applied in food and pharmaceutical industries (Stafford and Dennis, 1988). In this regard, most sPLA2s used are from animal pancreas (porcine or bovine) or venoms (bee, snake) since they are enzymes easily isolated in large quantities relatively to the low cost and they are commercially available (de Haas et al., 1968; De Maria et al., 2007). However, products of animal source, are rejected by many customers for religious reasons or risk of viral or prion contamination. Moreover, the use of enzymes from animal sources in processes for obtaining food additives may be incompatible with certain international regulations, which is not accepted in certain fields of application, since they do not meet the requirements of current international food standards. This is the reason why the industrial production of vegetable sPLA2s may become desirable. Nevertheless microbial sPLA2s are being accepted, sPLA2s from plant would be an advantage because its putative natural specificity (Lee et al., 2005; Mansfeld, 2009).
In the last decade, research has focused on the study of the still little known vegetable sPLA2s (Wang, 2001). Important advances have taken place in the identification, classification, biochemical characterization and functional analysis of plant sPLAs. Recent progress in understanding the biochemical and functional properties of plant sPLAs paves the way for approval of them for commercial use and various applications. Several sPLA2s have shown great potential as a target in the field of plant biotechnology, and molecular and catalytic diversity of plant sPLA2s shows that the phospholipases are of increasing value for biotechnology applications.
The possibility of using plant phospholipases in food processing would be an advantage, from the point of view of food regulations. Considering the large production of soybean in the world, it is of great interest to study the properties of its lipolytic enzymes in terms from of both agronomic and biotechnology point of views (Rönner, 2003; Hermida, 2005). Moreover, it should be noted that in the purification process of soybean oil, a byproduct named “gum” is a material enriched in phospholipids (about 65% of dry weight), which is usually used in animal’s food production or, after drying, it is sold as soybean lecithin. The hydrolytic products obtained by the action of sPLA2 over soybean lecithin, the lysophospholipids (lysoderivatives), are widely used as emulsifiers (Henderson et al., 1995; Dashiell, 2001).
Recently, sPLA2s were tested as catalysts for the synthesis of phospholipids with defined fatty acids by transesterification of lysophospholipids (Mansfeld, 2009). Furthermore, plant sPLA2s showed to be distinctive from animals due to differences in substrate selectivity regarding the polar head and the acyl chains of glycerophospholipids (Lee et al., 2005). The potential properties of plant sPLAs would open new horizons to the engineering of biocatalysts.
The plant sPLA2s is expected to have advantages over from animals regarding the performance or the incorporation of polyunsaturated fatty acids such as linoleic acid in egg PC for food production. Therefore, the processes for the production of phospholipids with fatty acids are not common and special performance requirements are desirable. Often, small differences in primary or 3D structure result in differences in the catalytic properties, which can be of great importance in biocatalytic applications. However, despite their enormous potential, plant enzymes have not been yet considered for industrial application. This could be attributed to the limited availability of these enzymes, recently discovered and characterized. Besides, these enzymes are much less abundant in the natural environment and no plant enzymes are available commercially.
Over 100 years, experiments with members of the sPLA2 superfamily have been carried out and kinetic and structural characterization established sPLA2 as an important model of interfacial enzymology. The future of this promising enzymes seems to be very exciting, leading to find out specific inhibitors of them, and further elucidating plants sPLA2’s roles in cellular processes, along with potential uses in the industry.
Author Contributions
MM and GF conceived the main idea, designed the general format of this manuscript, created the tables, and carried out the final corrections of this manuscript. MM prepared the figures and drafted this manuscript.
Funding
This work was supported, in part, by grants from the MinCyT Córdoba, CONICET, FONCYT PICT 2016-1010, and SeCyT-UNC. MM and GF are members of the Scientific Research Career (CIC) from the CONICET.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00861/full#supplementary-material
Footnotes
- ^ http://smart.embl-heidelberg.de/
- ^ https://www.rcsb.org/
- ^ http://srv00.recas.ba.infn.it/PMDB/main.php
References
Anderson, L. A., and Dufton, M. J. (1997). The action of Taiwan cobra venom on methionine enkephalin: a useful assay for oligopeptidase content. Toxicon 35, 1113–1123. doi: 10.1016/S0041-0101(97)00007-X
Andre, B., and Scherer, G. F. (1991). Stimulation by auxin of phospholipase A in membrane vesicles from an auxin-sensitive tissue is mediated by an auxin receptor. Planta 185, 209–214. doi: 10.1007/BF00194062
Annand, R. R., Kontoyianni, M., Penzotti, J. E., Dudler, T., Lybrand, T. P., and Gelb, M. H. (1996). Active site of bee venom phospholipase A2: the role of histidine-34, aspartate-64 and tyrosine-87. Biochemistry 35, 4591–4601. doi: 10.1021/bi9528412
Assmann, S. M., and Shimazaki, K. (1999). The multisensory guard cell. Stomatal responses to blue light and abscisic acid. Plant Physiol. 119, 809–816. doi: 10.1104/pp.119.3.809
Bahn, S. C., Lee, H. Y., Kim, H. J., Ryu, S. B., and Shin, J. S. (2003). Characterization of Arabidopsis secretory phospholipase A2-gamma cDNA and its enzymatic properties. FEBS Lett. 553, 113–118. doi: 10.1016/S0014-5793(03)00982-7
Bahnson, B. J. (2005). Structure, function and interfacial allosterism in phospholipase A2: insight from the anion-assisted dimer. Arch. Biochem. Biophys. 433, 96–106. doi: 10.1016/j.abb.2004.08.013
Bai, S., Jain, M. K., and Berg, O. G. (2008). Contiguous binding of decylsulfate on the interface-binding surface of pancreatic phospholipase A2. Biochemistry 47, 2899–2907. doi: 10.1021/bi702164n
Bayon, S., Chen, G., Weselake, R. J., and Browse, J. (2015). A small phospholipase A(2)-α from castor catalyzes the removal of hydroxy fatty acids from phosphatidylcholine in transgenic Arabidopsis Seeds. Plant Physiol. 167, 1259–1270. doi: 10.1104/pp.114.253641
Bell, J. D., and Biltonen, R. L. (1989). Thermodynamic and kinetic studies of the interaction of vesicular dipalmitoylphosphatidylcholine with Agkistrodon piscivorus piscivorus phospholipase A2. J. Biol. Chem. 264, 225–230.
Bell, J. D., Burnside, M., Owen, J. A., Royall, M. L., and Baker, M. L. (1996). Relationships between bilayer structure and phospholipase A2 activity: interactions among temperature, diacylglycerol, lysolecithin, palmitic acid, and dipalmitoylphosphatidylcholine. Biochemistry 35, 4945–4955. doi: 10.1021/bi952274i
Berg, O. G., Gelb, M. H., Tsai, M. D., and Jain, M. K. (2001). Interfacial enzymology: the secreted phospholipase A(2)-paradigm. Chem. Rev. 101, 2613–2654. doi: 10.1021/cr990139w
Berg, O. G., Yu, B. Z., and Jain, M. K. (2009). Thermodynamic reciprocity of the inhibitor binding to the active site and the interface binding region of IB phospholipase A2. Biochemistry 48, 3209–3218. doi: 10.1021/bi801244u
Berg, O. G., Yu, B. Z., Rogers, J., and Jain, M. K. (1991). Interfacial catalysis by phospholipase A2: determination of the interfacial kinetic rate constants. Biochemistry 30, 7283–7297. doi: 10.1021/bi00243a034
Bezzine, S., Bollinger, J. G., Singer, A. G., Veatch, S. L., Keller, S. L., and Gelb, M. H. (2002). On the binding preference of human groups IIA and X phospholipases A2 for membranes with anionic phospholipids. J. Biol. Chem. 277, 48523–48534. doi: 10.1074/jbc.M203137200
Blanchet, M. H., Le Good, J. A., Mesnard, D., Oorschot, V., Baflast, S., Minchiotti, G., et al. (2008a). Cripto recruits Furin and PACE4 and controls nodal trafficking during proteolytic maturation. EMBO J. 27, 2580–2591. doi: 10.1038/emboj.2008.174
Blanchet, M. H., Le Good, J. A., Oorschot, V., Baflast, S., Minchiotti, G., Klumperman, J., et al. (2008b). Cripto localizes nodal at the limiting membrane of early endosomes. Sci. Signal. 1:ra13. doi: 10.1126/scisignal.1165027
Burke, J. E., and Dennis, E. A. (2009). Phospholipase A2 structure/function, mechanism, and signaling. J. Lipid Res. 50(Suppl.), S237–S242. doi: 10.1194/jlr.R800033-JLR200
Campagnoli, M., Hansson, P., Dolcini, L., Caridi, G., Dagnino, M., Candiano, G., et al. (2008). Analbuminemia in a swedish male is caused by the kayseri mutation (c228_229delAT). Clin. Chim. Acta 396, 89–92. doi: 10.1016/j.cca.2008.06.008
Catanzariti, A. M., Soboleva, T. A., Jans, D. A., Board, P. G., and Baker, R. T. (2004). An efficient system for high-level expression and easy purification of authentic recombinant proteins. Protein Sci. 13, 1331–1339. doi: 10.1110/ps.04618904
Chen, G., Snyder, C. L., Greer, M. S., and Weselake, R. J. (2011). Biology and biochemistry of plant phospholipases. Crit. Rev. Plant Sci. 30, 239–258. doi: 10.1080/07352689.2011.572033
Chiou, Y. L., Cheng, Y. C., Kao, P. H., Wang, J. J., and Chang, L. S. (2008). Mutations on the N-terminal region abolish differentially the enzymatic activity, membrane-damaging activity and cytotoxicity of Taiwan cobra phospholipase A2. Toxicon 51, 270–279. doi: 10.1016/j.toxicon.2007.10.001
Choi, M. S., and Rhee, K. C. (2006). Production and processing of soybeans and nutrition and safety of isoflavone and other soy products for human health. J. Med. Food 9, 1–10. doi: 10.1089/jmf.2006.9.1
Daniele, J. J., Bianco, I. D., Delgado, C., Carrillo, D. B., and Fidelio, G. D. (1997). A new phospholipase A2 isoform isolated from Bothrops neuwiedii (Yarara chica) venom with novel kinetic and chromatographic properties. Toxicon 35, 1205–1215. doi: 10.1016/S0041-0101(97)00023-8
Daniele, J. J., Maggio, B., Bianco, I. D., Goni, F. M., Alonso, A., and Fidelio, G. D. (1996). Inhibition by gangliosides of Bacillus cereus phospholipase C activity against monolayers, micelles and bilayer vesicles. Eur. J. Biochem. 239, 105–110. doi: 10.1111/j.1432-1033.1996.0105u.x
Dashiell, G. (2001). Fuentes, métodos de procesos y usos comerciales de lecitina. Aceites Grasas 43, 197–204.
Davies, P. J. (1995). “The plant hormones: their nature, occurrence, and functions,” in Plant Hormones: Physiology, Biochemistry and Molecular Biology, ed. P. J. Davies (Boston, MA: Kluwer), 13–38.
de Haas, G. H., Bonsen, P. P., Pieterson, W. A., and van Deenen, L. L. (1971). Studies on phospholipase A and its zymogen from porcine pancreas. 3. Action of the enzyme on short-chain lecithins. Biochim. Biophys. Acta 239, 252–266. doi: 10.1016/0005-2760(71)901718
de Haas, G. H., Postema, N. M., Nieuwenhuizen, W., and van Deenen, L. L. (1968). Purification and properties of phospholipase A from porcine pancreas. Biochim. Biophys. Acta 159, 103–117. doi: 10.1016/0005-2744(68)90248-9
De Maria, L., Vind, J., Oxenboll, K. M., Svendsen, A., and Patkar, S. (2007). Phospholipases and their industrial applications. Appl. Microbiol. Biotechnol. 74, 290–300. doi: 10.1007/s00253-006-0775-x
De Tullio, L., Fanani, M. L., and Maggio, B. (2013). Surface mixing of products and substrate of PLA(2) in enzyme-free mixed monolayers reproduces enzyme-driven structural topography. Biochim. Biophys. Acta 1828, 2056–2063. doi: 10.1016/j.bbamem.2013.05.018
Demel, R. A., Geurts van Kessel, W. S., Zwaal, R. F., Roelofsen, B., and van Deenen, L. L. (1975). Relation between various phospholipase actions on human red cell membranes and the interfacial phospholipid pressure in monolayers. Biochim. Biophys. Acta 406, 97–107. doi: 10.1016/0005-2736(75)900450
Dennis, E. A. (1973a). Kinetic dependence of phospholipase A 2 activity on the detergent Triton X-100. J. Lipid Res. 14, 152–159.
Dennis, E. A. (1973b). Phospholipase A2 activity towards phosphatidylcholine in mixed micelles: surface dilution kinetics and the effect of thermotropic phase transitions. Arch. Biochem. Biophys. 158, 485–493. doi: 10.1016/0003-9861(73)905407
Dennis, E. A., Cao, J., Hsu, Y. H., Magrioti, V., and Kokotos, G. (2011). Phospholipase A2 enzymes: physical structure, biological function, disease implication, chemical inhibition, and therapeutic intervention. Chem. Rev. 111, 6130–6185. doi: 10.1021/cr200085w
Dennis, E. A., Darke, P. L., Deems, R. A., Kensil, C. R., and Pluckthun, A. (1981). Cobra venom phospholipase A2: a review of its action toward lipid/water interfaces. Mol. Cell. Biochem. 36, 37–45. doi: 10.1007/BF02354830
Dhondt, S., Geoffroy, P., Stelmach, B. A., Legrand, M., and Heitz, T. (2000). Soluble phospholipase A2 activity is induced before oxylipin accumulation in tobacco mosaic virus-infected tobacco leaves and is contributed by patatin-like enzymes. Plant J. 23, 431–440. doi: 10.1046/j.1365-313x.2000.00802.x
Dijkstra, B. W., Kalk, K. H., Drenth, J., de Haas, G. H., Egmond, M. R., and Slotboom, A. J. (1984). Role of the N-terminus in the interaction of pancreatic phospholipase A2 with aggregated substrates. Properties and crystal structure of transaminated phospholipase A2. Biochemistry 23, 2759–2766.
Dijkstra, B. W., Kalk, K. H., Hol, W. G., and Drenth, J. (1981). Structure of bovine pancreatic phospholipase A2 at 1.7A resolution. J. Mol. Biol. 147, 97–123. doi: 10.1016/0022-2836(81)900814
Domingues, S. J. S., de SouzaI, T. F., Soares, A. M. S., Jacinto, T., and Machado, O. L. T. (2007). Activation of phospholipase PLA2 actvity in Ricinus communis leaves in response to mechanical wounding. Braz. J. Plant Physiol. 19, 35–42.
dos Santos, J. I., Cintra-Francischinelli, M., Borges, R. J., Fernandes, C. A., Pizzo, P., Cintra, A. C., et al. (2011). Structural, functional, and bioinformatics studies reveal a new snake venom homologue phospholipase A class. Proteins 79, 61–78. doi: 10.1002/prot.22858
Fernandez, M. L., Quartino, P. Y., Arce-Bejarano, R., Fernandez, J., Camacho, L. F., Gutierrez, J. M., et al. (2017). Intravascular hemolysis induced by phospholipases A2 from the venom of the eastern coral snake, Micrurus fulvius: functional profiles of hemolytic and non-hemolytic isoforms. Toxicol. Lett. 286, 39–47. doi: 10.1016/j.toxlet.2017.11.037
Fico, A., Manganelli, G., Simeone, M., Guido, S., Minchiotti, G., and Filosa, S. (2008). High-throughput screening-compatible single-step protocol to differentiate embryonic stem cells in neurons. Stem Cells Dev. 17, 573–584. doi: 10.1089/scd.2007.0130
Friml, J. (2003). Auxin transport - shaping the plant. Curr. Opin. Plant Biol. 6, 7–12. doi: 10.1016/S1369526602000031
Fujikawa, R., Fujikawa, Y., Iijima, N., and Esaka, M. (2005). Molecular cloning, expression, and characterization of secretory phospholipase A2 in tobacco. Lipids 40, 901–908. doi: 10.1007/s11745-005-14509
Fujikawa, Y., Fujikawa, R., Iijima, N., and Esaka, M. (2011). Characterization of secretory phospholipase A(2) with phospholipase A(1) activity in tobacco, Nicotiana tabacum (L.). Lipids 47, 303–312. doi: 10.1007/s11745-011-36323
Ghomashchi, F., Yu, B. Z., Berg, O., Jain, M. K., and Gelb, M. H. (1991). Interfacial catalysis by phospholipase A2: substrate specificity in vesicles. Biochemistry 30, 7318–7329. doi: 10.1021/bi00243a037
Guo, Z., Vikbjerg, A. F., and Xu, X. (2005). Enzymatic modification of phospholipids for functional applications and human nutrition. Biotechnol. Adv. 23, 203–259. doi: 10.1016/j.biotechadv.2005.02.001
Gupta, P., and Dash, P. K. (2017). Molecular details of secretory phospholipase A2 from flax (Linum usitatissimum L.) provide insight into its structure and function. Sci. Rep. 7:11080. doi: 10.1038/s41598-017-109699
Gupta, P., Saini, R., and Dash, P. K. (2017). Origin and evolution of group XI secretory phospholipase A2 from flax (Linum usitatissimum) based on phylogenetic analysis of conserved domains. 3 Biotech. 7:216. doi: 10.1007/s13205-017-0790-x
Guy, J. E., Stahl, U., and Lindqvist, Y. (2009). Crystal structure of a class XIB phospholipase A2 (PLA2): rice (oryza sativa) isoform-2 pla2 and an octanoate complex. J. Biol. Chem. 284, 19371–19379. doi: 10.1074/jbc.M109.008466
Havinga, T. (2010). Regulating halal and kosher foods: different arrangements between state, industry and religious actors. Erasmus Law Rev. 3, 241–255. doi: 10.553/ELR221026712010003004004
Henderson, J., Atkinson, A. E., Lazarus, C. M., Hawes, C. R., Napier, R. M., Macdonald, H., et al. (1995). Stable expression of maize auxin-binding protein in insect cell lines. FEBS Lett. 371, 293–296. doi: 10.1016/0014-5793(95)00922-V
Hermida, R. (2005). Presente y futuro de la industria aceitera a nivel nacional e internacional. Aceites Grasas 58, 38–44.
Hirel, P. H., Schmitter, M. J., Dessen, P., Fayat, G., and Blanquet, S. (1989). Extent of N-terminal methionine excision from Escherichia coli proteins is governed by the side-chain length of the penultimate amino acid. Proc. Natl. Acad. Sci. U.S.A. 86, 8247–8251. doi: 10.1073/pnas.86.21.8247
Jabeen, T., Singh, N., Singh, R. K., Ethayathulla, A. S., Sharma, S., Srinivasan, A., et al. (2005). Crystal structure of a novel phospholipase A2 from Naja naja sagittifera with a strong anticoagulant activity. Toxicon 46, 865–875. doi: 10.1016/j.toxicon.2005.08.008
Jablonicka, V., Mansfeld, J., Heilmann, I., Oblozinsky, M., and Heilmann, M. (2016). Identification of a secretory phospholipase A2 from Papaver somniferum L. that transforms membrane phospholipids. Phytochemistry 129, 4–13. doi: 10.1016/j.phytochem.2016.07.010
Jain, E., Bairoch, A., Duvaud, S., Phan, I., Redaschi, N., Suzek, B. E., et al. (2009). Infrastructure for the life sciences: design and implementation of the UniProt website. BMC Bioinformatics 10:136. doi: 10.1186/1471-2105-10-136
Jain, M. K., and Berg, O. G. (2006). Coupling of the i-face and the active site of phospholipase A2 for interfacial activation. Curr. Opin. Chem. Biol. 10, 473–479. doi: 10.1016/j.cbpa.2006.08.015
Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W., and Klein, M. L. (1983). Comparison of single potential functions for simulating liquid water. J. Chem. Phys. 79, 2577–2637. doi: 10.1063/1.445869
Khan, W. A., Blobe, G. C., and Hannun, Y. A. (1995). Arachidonic acid and free fatty acids as second messengers and the role of protein kinase C. Cell Signal. 7, 171–184. doi: 10.1016/0898-6568(94)00089-T
Kim, J. Y., Chung, Y. S., Ok, S. H., Lee, S. G., Chung, W. I., Kim, I. Y., et al. (1999). Characterization of the full-length sequences of phospholipase A2 induced during flower development. Biochim. Biophys. Acta 1489, 389–392. doi: 10.1016/S0167-4781(99)001931
Kohler, G. A., Brenot, A., Haas-Stapleton, E., Agabian, N., Deva, R., and Nigam, S. (2006). Phospholipase A2 and phospholipase B activities in fungi. Biochim. Biophys. Acta 1761, 1391–1399. doi: 10.1016/j.bbalip.2006.09.011
Kramer, R. M., Hession, C., Johansen, B., Hayes, G., McGray, P., Chow, E. P., et al. (1989). Structure and properties of a human non-pancreatic phospholipase A2. J. Biol. Chem. 264, 5768–5775.
Kuipers, O. P., Vincent, M., Brochon, J. C., Verheij, H. M., de Haas, G. H., and Gallay, J. (1991). Insight into the conformational dynamics of specific regions of porcine pancreatic phospholipase A2 from a time-resolved fluorescence study of a genetically inserted single tryptophan residue. Biochemistry 30, 8771–8785. doi: 10.1021/bi00100a008
Kumar, A., Sekharudu, C., Ramakrishnan, B., Dupureur, C. M., Zhu, H., Tsai, M. D., et al. (1994). Structure and function of the catalytic site mutant Asp 99 Asn of phospholipase A2: absence of the conserved structural water. Protein Sci. 3, 2082–2088. doi: 10.1002/pro.5560031121
Laigle, J. L., Leger, H., Couraud, L., Guillard, J. M., Fontan, D., and Verger, P. (1973). Pulmonary fibroxanthoma in the child. Apropos of a new case. Pédiatrie 28, 755–763.
Lee, H. Y., Bahn, S. C., Kang, Y. M., Lee, K. H., Kim, H. J., Noh, E. K., et al. (2003). Secretory low molecular weight phospholipase A2 plays important roles in cell elongation and shoot gravitropism in Arabidopsis. Plant Cell 15, 1990–2002. doi: 10.1105/tpc.014423
Lee, H. Y., Bahn, S. C., Shin, J. S., Hwang, I., Back, K., Doelling, J. H., et al. (2005). Multiple forms of secretory phospholipase A2 in plants. Prog. Lipid Res. 44, 52–67. doi: 10.1016/j.plipres.2004.10.002
Leidy, C., Mouritsen, O. G., Jorgensen, K., and Peters, G. H. (2004). Evolution of a rippled membrane during phospholipase A2 hydrolysis studied by time-resolved AFM. Biophys. J. 87, 408–418. doi: 10.1529/biophysj.103.036103
Liao, H. L., and Burns, J. K. (2010). Light controls phospholipase A2alpha and beta gene expression in Citrus sinensis. J. Exp. Bot. 61, 2469–2478. doi: 10.1093/jxb/erq083
Liscovitch, M., Czarny, M., Fiucci, G., and Tang, X. (2000). Phospholipase D: molecular and cell biology of a novel gene family. Biochem. J. 345(Pt 3), 401–415. doi: 10.1042/bj3450401
Lomize, M. A., Pogozheva, I. D., Joo, H., Mosberg, H. I., and Lomize, A. L. (2012). OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 40, D370–D376. doi: 10.1093/nar/gkr703
Madoery, R. R., Gattone, C. G., and Fidelio, G. D. (1999). The effect of phospholipase A(2) immobilization upon calcium interaction: a kinetic study. J. Biochem. 126, 1060–1066. doi: 10.1093/oxfordjournals.jbchem.a022550
Mansfeld, J. (2009). Plant phospholipases A2: perspectives on biotechnological applications. Biotechnol. Lett. 31, 1373–1380. doi: 10.1007/s10529-009-00341
Mansfeld, J., Gebauer, S., Dathe, K., and Ulbrich-Hofmann, R. (2006). Secretory phospholipase A2 from Arabidopsis thaliana: insights into the three-dimensional structure and the amino acids involved in catalysis. Biochemistry 45, 5687–5694. doi: 10.1021/bi052563z
Mansfeld, J., Schopfel, M., Lorenz, J., Trutschel, T., Heilmann, I., Brandt, W., et al. (2014). Probing selected structural regions in the secreted phospholipase A(2) from Arabidopsis thaliana for their impact on stability and activity. Biochimie 101, 60–66. doi: 10.1016/j.biochi.2013.12.015
Mansfeld, J., and Ulbrich-Hofmann, R. (2007). Secretory phospholipase A2-alpha from Arabidopsis thaliana: functional parameters and substrate preference. Chem. Phys. Lipids 150, 156–166. doi: 10.1016/j.chemphyslip.2007.07.001
Mariani, M. E., Madoery, R. R., and Fidelio, G. D. (2015a). Auxins action on Glycine max secretory phospholipase A2 is mediated by the interfacial properties imposed by the phytohormones. Chem. Phys. Lipids 189, 1–6. doi: 10.1016/j.chemphyslip.2015.05.003
Mariani, M. E., Madoery, R. R., and Fidelio, G. D. (2015b). Kinetic characterization, optimum conditions for catalysis and substrate preference of secretory phospholipase A2 from Glycine max in model membrane systems. Biochimie 108, 48–58. doi: 10.1016/j.biochi.2014.10.016
Mariani, M. E., Villarreal, M. A., Cheung, F., Leiva, E. P., Madoery, R. R., and Fidelio, G. D. (2012). In silico and in vitro characterization of phospholipase A(2) isoforms from soybean (Glycine max). Biochimie 94, 2608–2619. doi: 10.1016/j.biochi.2012.07.021
Marki, F., and Hanulak, V. (1993). Recombinant human synovial fluid phospholipase A2 and N-terminal variant: kinetic parameters and response to inhibitors. J. Biochem. 113, 734–737. doi: 10.1093/oxfordjournals.jbchem.a124112
Matsushima, R., Kondo, M., Nishimura, M., and Hara-Nishimura, I. (2003). A novel ER-derived compartment, the ER body, selectively accumulates a beta-glucosidase with an ER-retention signal in Arabidopsis. Plant J. 33, 493–502. doi: 10.1046/j.1365-313X.2003.01636.x
Mazereeuw-Hautier, J., Redoules, D., Tarroux, R., Charveron, M., Salles, J. P., Simon, M. F., et al. (2000). Identification of pancreatic type I secreted phospholipase A2 in human epidermis and its determination by tape stripping. Br. J. Dermatol. 142, 424–431. doi: 10.1046/j.1365-2133.2000.03351.x
McIntosh, J. M., Ghomashchi, F., Gelb, M. H., Dooley, D. J., Stoehr, S. J., Giordani, A. B., et al. (1995). Conodipine-M, a novel phospholipase A2 isolated from the venom of the marine snail Conus magus. J. Biol. Chem. 270, 3518–3526. doi: 10.1046/j.1365-2133.2000.03351.x
Meneghetti, F., and Maggio, B. (2013). (E)-2-{[1-(3,11-Dimethyl-4-methyl-ene-10-oxo-1-phenyl-4,5,10,11-tetra-hydro-1H-be nzo[b]pyrazolo-[3,4-f][1,5]diazo-cin-5-yl)ethyl-idene]amino}-N-methyl-N-(3-methyl -1-phenyl-1H-pyrazol-5-yl)benzamide. Acta Crystallogr. Sect. E Struct. Rep. Online 69(Pt 10), o1583. doi: 10.1107/S1600536813025671
Messina, M. J. (1999). Legumes and soybeans: overview of their nutritional profiles and health effects. Am. J. Clin. Nutr. 70(3 Suppl.), 439S–450S. doi: 10.1093/ajcn/70.3.439s
Minchiotti, M., Scalambro, M. B., Vargas, L., Coronel, C., and Madoery, R. (2008). Isolation of phospholipase A2 from soybean (Glycine max) seeds: the study of its enzymatic properties. Enzyme Microb. Technol. 42, 389–394. doi: 10.1016/j.enzmictec.2007.11.015
Moreau, R. A., and Morgan, C. P. (1988). Photeolytic activation of a lipolytic enzyme activity in potato leaves. Plant Sci. 55, 205–211. doi: 10.1016/j.enzmictec.2007.11.015
Mounier, C. M., Ghomashchi, F., Lindsay, M. R., James, S., Singer, A. G., Parton, R. G., et al. (2004). Arachidonic acid release from mammalian cells transfected with human groups IIA and X secreted phospholipase A(2) occurs predominantly during the secretory process and with the involvement of cytosolic phospholipase A(2)-alpha. J. Biol. Chem. 279, 25024–25038. doi: 10.1074/jbc.M313019200
Mukherjee, A. B. (1990). Biochemistry, Molecular Biology and Physiology of Phospholipase A2 and its Regulatory Factors. New York, NY: Plenum Press.
Murakami, M., Sato, H., Miki, Y., Yamamoto, K., and Taketomi, Y. (2015). A new era of secreted phospholipase A(2). J. Lipid Res. 56, 1248–1261. doi: 10.1194/jlr.R058123
Murakami, M., Taketomi, Y., Miki, Y., Sato, H., Hirabayashi, T., and Yamamoto, K. (2010). Recent progress in phospholipase A research: from cells to animals to humans. Prog. Lipid Res. 50, 152–192. doi: 10.1016/j.plipres.2010.12.001
Murakami, M., Taketomi, Y., Sato, H., and Yamamoto, K. (2011). Secreted phospholipase A2 revisited. J. Biochem. 150, 233–255. doi: 10.1093/jb/mvr088
Narvaez-Vasquez, J., Florin-Christensen, J., and Ryan, C. A. (1999). Positional specificity of a phospholipase A activity induced by wounding, systemin, and oligosaccharide elicitors in tomato leaves. Plant Cell 11, 2249–2260. doi: 10.1105/tpc.11.11.2249
Othman, R., Baker, S., Li, Y., Worrall, A. F., and Wilton, D. C. (1996). Human non-pancreatic (group II) secreted phospholipase A2 expressed from a synthetic gene in Escherichia coli: characterisation of N-terminal mutants. Biochim. Biophys. Acta 1303, 92–102. doi: 10.1016/0005-2760(96)000835
Pagny, S., Cabanes-Macheteau, M., Gillikin, J. W., Leborgne-Castel, N., Lerouge, P., Boston, R. S., et al. (2000). Protein recycling from the Golgi apparatus to the endoplasmic reticulum in plants and its minor contribution to calreticulin retention. Plant Cell 12, 739–756. doi: 10.1105/tpc.12.5.739
Pan, Y. H., Yu, B. Z., Berg, O. G., Jain, M. K., and Bahnson, B. J. (2002). Crystal structure of phospholipase A2 complex with the hydrolysis products of platelet activating factor: equilibrium binding of fatty acid and lysophospholipid-ether at the active site may be mutually exclusive. Biochemistry 41, 14790–14800. doi: 10.1021/bi026922r
Panaiotov, I., and Verger, R. (2000). “Enzymatic reactions at interfaces: interfacial and temporal organization of enzymatic lipolysis,” in Physical Chemistry and Biological Interfaces, eds A. Baszkin and W. Norde (Boca Raton, FL: CRC Press), 359–400.
Qin, S., Pande, A. H., Nemec, K. N., He, X., and Tatulian, S. A. (2005). Evidence for the regulatory role of the N-terminal helix of secretory phospholipase A(2) from studies on native and chimeric proteins. J. Biol. Chem. 280, 36773–36783. doi: 10.1074/jbc.M506789200
Ramirez, F., and Jain, M. K. (1991). Phospholipase A2 at the bilayer interface. Proteins 9, 229–239. doi: 10.1002/prot.340090402
Randolph, A., and Heinrikson, R. L. (1982). Crotalus atrox phospholipase A2. Amino acid sequence and studies on the function of the NH2-terminal region. J. Biol. Chem. 257, 2155–2161.
Rhee, S. G., and Bae, Y. S. (1997). Regulation of phosphoinositide-specific phospholipase C isozymes. J. Biol. Chem. 272, 15045–15048.
Rönner, L. (2003). La Incorporación de Nuevas Tecnologías: el Caso de la Soja. Buenos Aires: Universidad de Buenos Aires.
Ryu, S. B. (2004). Phospholipid-derived signaling mediated by phospholipase A in plants. Trends Plant Sci. 9, 229–235. doi: 10.1016/j.tplants.2004.03.004
Ryu, S. B., Lee, H. Y., Doelling, J. H., and Palta, J. P. (2005). Characterization of a cDNA encoding Arabidopsis secretory phospholipase A2-alpha, an enzyme that generates bioactive lysophospholipids and free fatty acids. Biochim. Biophys. Acta 1736, 144–151. doi: 10.1016/j.bbalip.2005.08.005
Ryu, Y., Oh, Y., Yoon, J., Cho, W., and Baek, K. (2003). Molecular characterization of a gene encoding the Drosophila melanogaster phospholipase A2. Biochim. Biophys. Acta 1628, 206–210. doi: 10.1016/S0167-4781(03)00143-X
Schaller, F. (2001). Enzymes of the biosynthesis of octadecanoid-derived signalling molecules. J. Exp. Bot. 52, 11–23. doi: 10.1093/jxb/52.354.11
Schaloske, R. H., and Dennis, E. A. (2006). The phospholipase A2 superfamily and its group numbering system. Biochim. Biophys. Acta 1761, 1246–1259. doi: 10.1016/j.bbalip.2006.07.011
Scherer, G. F. (1990). Phospholipase A2 and phospholipid-activated protein kinase in plant signal transduction. Symp. Soc. Exp. Biol. 44, 257–270.
Scherer, G. F., and Andre, B. (1989). A rapid response to a plant hormone: auxin stimulates phospholipase A2 in vivo and in vitro. Biochem. Biophys. Res. Commun. 163, 111–117. doi: 10.1016/0006-291x(89)92106-2
Scherer, G. F., and Andre, B. (1993). Stimulation of phospholipase A 2 by auxin in microsomes from suspension-cultured soybean cells is receptormediated and influenced by nucleotides. Planta 191, 515–523.
Scherer, G. F., Ryu, S. B., Wang, X., Matos, A. R., and Heitz, T. (2010). Patatin-related phospholipase A: nomenclature, subfamilies and functions in plants. Trends Plant Sci. 15, 693–700. doi: 10.1016/j.tplants.2010.09.005
Scherer, G. F. E. (1992). Stimulation of growth and phospholipase A 2 by the peptides mastoparan and melittin and by the auxin 2,4-dichlorophenoxyacetic acid. Plant Growth Regul. 11, 153–157. doi: 10.1007/BF00024069
Scherer, G. F. E. (1996). Phospholipid signalling and lipid-derived second messengers in plants. Plant Growth Regul. 18, 125–133. doi: 10.1007/BF00028497
Scott, D. L., Mandel, A. M., Sigler, P. B., and Honig, B. (1994). The electrostatic basis for the interfacial binding of secretory phospholipases A2. Biophys. J. 67, 493–504. doi: 10.1016/S0006-3495(94)80546-6
Scott, D. L., White, S. P., Otwinowski, Z., Yuan, W., Gelb, M. H., and Sigler, P. B. (1990). Interfacial catalysis: the mechanism of phospholipase A2. Science 250, 1541–1546. doi: 10.1126/science.2274785
Seo, J., Lee, H. Y., Choi, H., Choi, Y., Lee, Y., Kim, Y. W., et al. (2008). Phospholipase A2beta mediates light-induced stomatal opening in Arabidopsis. J. Exp. Bot. 59, 3587–3594. doi: 10.1093/jxb/ern208
Shridas, P., and Webb, N. R. (2014). Diverse functions of secretory phospholipases A2. Adv. Vasc. Med. 2014:11. doi: 10.1155/2014/689815
Six, D. A., and Dennis, E. A. (2000). The expanding superfamily of phospholipase A(2) enzymes: classification and characterization. Biochim. Biophys. Acta 1488, 1–19. doi: 10.1016/s1388-1981(00)00105-0
Slotboom, A. J., van Dam-Mieras, M. C., and de Haas, G. H. (1977). Regulation of phospholipase A2 activity by different lipid-water interfaces. J. Biol. Chem. 252, 2948–2951.
Stafford, R. E., and Dennis, E. A. (1988). Lysophospholipids as Biosurfactants. Colloids Surf. 30, 47–64. doi: 10.1016/0166-6622(87)802032
Stahl, U., Ek, B., and Stymne, S. (1998). Purification and characterization of a low-molecular-weight phospholipase A2 from developing seeds of elm. Plant Physiol. 117, 197–205. doi: 10.2307/4278269
Stahl, U., Lee, M., Sjodahl, S., Archer, D., Cellini, F., Ek, B., et al. (1999). Plant low-molecular-weight phospholipase A2S (PLA2s) are structurally related to the animal secretory PLA2s and are present as a family of isoforms in rice (Oryza sativa). Plant Mol. Biol. 41, 481–490.
Sugiyama, M., Ohtani, K., Izuhara, M., Koike, T., Suzuki, K., Imamura, S., et al. (2002). A novel prokaryotic phospholipase A2. Characterization, gene cloning, and solution structure. J. Biol. Chem. 277, 20051–20058. doi: 10.1074/jbc.M200264200
Tatulian, S. A. (2001). Toward understanding interfacial activation of secretory phospholipase A2 (PLA2): membrane surface properties and membrane-induced structural changes in the enzyme contribute synergistically to PLA2 activation. Biophys. J. 80, 789–800. doi: 10.1016/S0006-3495(01)760584
Tatulian, S. A., Qin, S., Pande, A. H., and He, X. (2005). Positioning membrane proteins by novel protein engineering and biophysical approaches. J. Mol. Biol. 351, 939–947. doi: 10.1016/j.jmb.2005.06.080
Valentin, E., Ghomashchi, F., Gelb, M. H., Lazdunski, M., and Lambeau, G. (1999). On the diversity of secreted phospholipases A(2). Cloning, tissue distribution, and functional expression of two novel mouse group II enzymes. J. Biol. Chem. 274, 31195–31202. doi: 10.1074/jbc.274.44.31195
van Deenen, L. L. (1971). Chemistry of phospholipids in relation to biological membranes. Pure Appl. Chem. 25, 25–56. doi: 10.1351/pac197125010025
van den Berg, B., Tessari, M., Boelens, R., Dijkman, R., de Haas, G. H., Kaptein, R., et al. (1995). NMR structures of phospholipase A2 reveal conformational changes during interfacial activation. Nat. Struct. Biol. 2, 402–406. doi: 10.1038/nsb0595-402
van Scharrenburg, G. J., Jansen, E. H., Egmond, M. R., de Haas, G. H., and Slotboom, A. J. (1984). Structural importance of the amino-terminal residue of pancreatic phospholipase A2. Biochemistry 23, 6285–6294. doi: 10.1021/bi00320a059
Verger, R., Rietsch, J., Pattus, F., Ferrato, F., Pieroni, G., De Haas, G. H., et al. (1978). Studies of lipase and phospholipase A2 acting on lipid monolayers. Adv. Exp. Med. Biol. 101, 79–94. doi: 10.1002/bbpc.19780820923
Verlotta, A., Liberatore, M. T., Cattivelli, L., and Trono, D. (2013). Secretory Phospholipases A2 in durum wheat (Triticum durum Desf.): gene expression, enzymatic activity, and relation to drought stress adaptation. Int. J. Mol. Sci. 14, 5146–5169. doi: 10.3390/ijms14035146
Verlotta, A., and Trono, D. (2014). Expression, purification and refolding of active durum wheat (Triticum durum Desf.) secretory phospholipase A2 from inclusion bodies of Escherichia coli. Protein Expr. Purif. 101, 28–36. doi: 10.1016/j.pep.2014.05.009
Volwerk, J. J., Jost, P. C., de Haas, G. H., and Griffith, O. H. (1986). Activation of porcine pancreatic phospholipase A2 by the presence of negative charges at the lipid-water interface. Biochemistry 25, 1726–1733. doi: 10.1021/bi00355a042
Wang, G., Ryu, S., and Wang, X. (2012). “Plant phospholipases: an overview,” in Lipases and Phospholipases: Methods and Protocols, ed. G. Sandoval (New York, NY: Humana Press), 123–137.
Wang, X. (2001). Plant phospholipases. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 211–231. doi: 10.1146/annurev.arplant.52.1.211
Warner, S. (2005). Biotech takes on New Industries. Available at: https://www.the-scientist.com/biobusiness/biotech-takes-on-new-industries-49036Google Scholar
Welker, S., Markert, Y., Koditz, J., Mansfeld, J., and Ulbrich-Hofmann, R. (2011). Disulfide bonds of phospholipase A2 from bee venom yield discrete contributions to its conformational stability. Biochimie 93, 195–201. doi: 10.1016/j.biochi.2010.09.012
Wells, M. A. (1972). A kinetic study of the phospholipase A 2 (Crotalus adamanteus) catalyzed hydrolysis of 1,2-dibutyryl-sn-glycero-3-phosphorylcholine. Biochemistry 11, 1030–1041. doi: 10.1021/bi00756a013
West, N., Newcombe, R. G., Hughes, N., Mason, S., Maggio, B., Sufi, F., et al. (2013). A 3-day randomised clinical study investigating the efficacy of two toothpastes, designed to occlude dentine tubules, for the treatment of dentine hypersensitivity. J. Dent. 41, 187–194. doi: 10.1016/j.jdent.2012.11.007
Williams, R. L. (1999). Mammalian phosphoinositide-specific phospholipase C. Biochim. Biophys. Acta 1441, 255–267. doi: 10.1016/S1388-1981(99)00150-X
Wilschut, J. C., Regts, J., Westenberg, H., and Scherphof, G. (1978). Action of phospholipases A2 on phosphatidylcholine bilayers. Effects of the phase transition, bilayer curvature and structural defects. Biochim. Biophys. Acta 508, 185–196. doi: 10.1016/0005-2736(78)903243
Winget, J. M., Pan, Y. H., and Bahnson, B. J. (2006). The interfacial binding surface of phospholipase A2s. Biochim. Biophys. Acta 1761, 1260–1269. doi: 10.1016/j.bbalip.2006.08.002
Yu, B. Z., Berg, O. G., and Jain, M. K. (1999a). Hydrolysis of monodisperse phosphatidylcholines by phospholipase A2 occurs on vessel walls and air bubbles. Biochemistry 38, 10449–10456. doi: 10.1021/bi990194z
Yu, B. Z., Rogers, J., Tsai, M. D., Pidgeon, C., and Jain, M. K. (1999b). Contributions of residues of pancreatic phospholipase A2 to interfacial binding, catalysis, and activation. Biochemistry 38, 4875–4884. doi: 10.1021/bi982215f
Yunes Quartino, P. J., Barra, J. L., and Fidelio, G. D. (2012). Cloning and functional expression of secreted phospholipases A(2) from Bothrops diporus (Yarara Chica). Biochem. Biophys. Res. Commun. 427, 321–325. doi: 10.1016/j.bbrc.2012.09.051
Yunes Quartino, P. J., Portela, M., Lima, A., Duran, R., Lomonte, B., and Fidelio, G. D. (2015). A constant area monolayer method to assess optimal lipid packing for lipolysis tested with several secreted phospholipase A2. Biochim. Biophys. Acta 1848(10 Pt A), 2216–2224. doi: 10.1016/j.bbamem.2015.06.003
Keywords: secretory phospholipase A2, interfacial catalysis, auxin, Glycine max, phospholipase–membrane interaction
Citation: Mariani ME and Fidelio GD (2019) Secretory Phospholipases A2 in Plants. Front. Plant Sci. 10:861. doi: 10.3389/fpls.2019.00861
Received: 31 January 2019; Accepted: 14 June 2019;
Published: 10 July 2019.
Edited by:
Elisabeth Jamet, Université Toulouse III Paul Sabatier, FranceReviewed by:
Payal Gupta, Independent Researcher, Pune, IndiaStephen Beungtae Ryu, Korea Research Institute of Bioscience and Biotechnology (KRIBB), South Korea
Copyright © 2019 Mariani and Fidelio. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: María Elisa Mariani, emariani@unc.edu.ar; Gerardo Daniel Fidelio, gfidelio@unc.edu.ar; gerardo.fidelio@gmail.com
 María Elisa Mariani1,2,3*
María Elisa Mariani1,2,3* Gerardo Daniel Fidelio
Gerardo Daniel Fidelio