- 1Nottingham BBSRC Wheat Research Centre, Division of Plant and Cop Sciences, School of Biosciences, The University of Nottingham, Sutton Bonington Campus, Loughborough, Leicestershire, United Kingdom
- 2International Maize and Wheat Improvement Center (CIMMYT) Mexico, Mexico City, Mexico
- 3Division of Plant Sciences, University of Missouri, Columbia, MO, United States
The wild relatives of wheat provide an important source of genetic variation for wheat improvement. Much of the work in the past aimed at transferring genetic variation from wild relatives into wheat has relied on the exploitation of the ph1b mutant, located on the long arm of chromosome 5B. This mutation allows homologous recombination to occur between chromosomes from related but different genomes, e.g. between the chromosomes of wheat and related chromosomes from a wild relative resulting in the generation of interspecific recombinant chromosomes. However, the ph1b mutant also enables recombination to occur between the homologous genomes of wheat, e.g. A/B, A/D, B/D, resulting in the generation of wheat intergenomic recombinant chromosomes. In this work we report on the presence of wheat intergenomic recombinants in the genomic background of hexaploid wheat/Amblyopyrum muticum introgression lines. The transfer of genomic rearrangements involving the D-genome through pentaploid crosses provides a strategy by which the D-genome of wheat can be introgressed into durum wheat. Hence, a pentaploid crossing strategy was used to transfer D-genome segments, introgressed with either the A- and/or the B-genome, into the tetraploid background of two durum wheat genotypes Karim and Om Rabi 5 in either the presence or absence of different Am. muticum (2n = 2x = 14, TT) introgressions. Introgressions were monitored in backcross generations to the durum wheat parents via multi-color genomic in situ hybridization (mc-GISH). Tetraploid lines carrying homozygous D-genome introgressions, as well as simultaneous homozygous D- and T-genome introgressions, were developed. Introgression lines were characterized via Kompetitive Allele-Specific PCR (KASP) markers and multi-color fluorescence in situ hybridization (FISH). Results showed that new wheat sub-genomic translocations were generated at each generation in progeny that carried any Am. muticum chromosome introgression irrespective of the linkage group that the segment was derived from. The highest frequencies of homologous recombination were observed between the A- and the D-genomes. Results indicated that the genotype Karim had a higher tolerance to genomic rearrangements and T-genome introgressions compared to Om Rabi 5. This indicates the importance of the selection of the parental genotype when attempting to transfer/develop introgressions into durum wheat from pentaploid crosses.
Introduction
The most important cultivated Triticum species are hexaploid bread wheat (2n = 2x = 42; AABBDD, Triticum aestivum L. ssp. aestivum) and tetraploid durum wheat (2n = 2x = 28; AABB, Triticum turgidum L. ssp. durum). Tetraploid wheat arose 500,000 years ago from a cross between the wild ancestors of the A-genome, Triticum urartu Thum ex. Gandil (2n = 2x = 14; AuAu) (Feldman and Levy, 2005), and the B-genome from an Aegilops speltoides-like progenitor (Haider, 2013). After domestication, a spontaneous cross of tetraploid wheat as the female parent with the goat grass Ae. tauschii Coss. (2n = 2x = 14; DD) approximately 8,000 years ago gave rise to hexaploid bread wheat (Kihara, 1944; McFadden and Sears, 1944, Matsuoka and Nasuda, 2004). The addition of the D-genome to hexaploid wheat conferred baking characteristics and a wide climatic adaptation compared to durum wheat (Zohary et al., 1969) resulting in bread wheat becoming one of the most widely grown crops due to its high yields and nutritional and processing qualities (Shewry and Hey, 2015).
Despite the relatively small growing area (8%) and lower annual production compared to bread wheat, durum wheat remains a major crop in the Mediterranean basin where about 75% of the world’s durum wheat is produced (Li et al., 2013; Kabbaj et al., 2017) although Europe and North Africa are also the largest importers of durum wheat (Bonjean et al., 2016). According to data from the International Grain Council, durum wheat production has shown annual fluctuations, largely attributable to abiotic and biotic stresses, e.g., in the Mediterranean area, crops are often exposed to environmental stresses such as high temperature and drought during grain filling (Nazco et al., 2012). Breeding programs have greatly improved durum wheat yield and quality (Magallanes-López et al., 2017). However, the incorporation of new alleles into wheat germplasm is considered essential for the continued improvement of durum wheat productivity.
Wheat is related to a large number of other species, many of which are wild and uncultivated. These wild relatives provide a vast and largely untapped reservoir of genetic variation for agronomically important traits (Friebe et al., 1996; Jauhar and Chibbar, 1999; Qi et al., 2007; Schneider et al., 2008). The incorporation of these traits into wheat has the potential to increase the yield potential. For example, Ae. speltoides has been shown to be insect and disease resistant (Elek et al., 2014) and Thinopyrum bessarabicum salt tolerant (King et al., 1997).
Among the wild relatives of wheat, Am. muticum (2n = 2x = 14; TT) is an annual, native species of Turkey and Armenia (Kilian et al., 2013). This species has been reported to be resistant to environmental stresses (Iefimenko et al., 2015), powdery mildew (Eser, 1998), and leaf rust (Dundas et al., 2015). The introgression of Am. muticum into bread wheat is an ongoing project at the Wheat Research Centre (WRC) at the University of Nottingham (King et al., 2013; King et al., 2017) where 218 genome- wide bread wheat/Am. muticum introgressions have been developed covering the seven linkage groups of Am. muticum (King et al., 2017). Genomic in situ hybridization (GISH) analysis revealed that some of the introgression lines also contained intergenomic rearrangements between the A, B, and D sub-genomes of wheat. These intergenomic recombinants, and particularly those that involve the D-genome, can be transferred into durum wheat. Hybridization between bread and durum wheat leads to the production of a pentaploid hybrid (AABBD) with a chromosomal constitution of 2n = 5x = 35 (Kihara, 1924). Depending on the direction of the backcrosses, pentaploid hybrids have the potential to improve both bread wheat and durum (Eberhard et al., 2010; Martin et al., 2013; Kalous et al., 2015).
This paper describes the introgression of both wheat inter-genome rearrangements involving the D-genome and T-genome segments of Am. muticum present in hexaploid wheat/Am. muticum introgression (WMI) lines into two durum wheat genotypes using pentaploid crosses. The effect of the presence of the T-genome in the WMI lines, the efficiency of the crossing strategy as well as the choice of the durum wheat are discussed.
Materials and Methods
Plant Material
The self-fertilized or back-crossed seed of eight hexaploid wheat/Am. muticum introgression lines, designated as WMI (wheat/Am. muticum introgression) lines, were obtained from the Nottingham/BBSRC Wheat Research Centre (WRC) (King et al., 2017). The WMI lines were characterized by multi-color genomic in situ hybridization (mc-GISH) in the BC3 generation and shown to carry wheat inter-genomic rearrangements involving the D-genome. The genome rearrangements were designated by the letter of the genome involved (A, B, or D). An upper case letter designated the larger segment, a lower case letter the smaller segment. In the case of non centromeric translocations, the two letters were separated by a dash (e.g. A-d), whereas for centromeric translocations, a dot was used (e.g., A.D). Four of the WMI lines also carried one to three different large T-genome segments characterized using the Axiom® Wheat-Relative Genotyping Array (King et al., 2017) (Table1). Hence, the WMI lines were categorized into two groups, the G-1 lines without a T introgression/chromosome and the G-2 lines carrying a T introgression/chromosome. Four seeds of each line were germinated and screened for the presence of the D-genome introgression using mc-GISH. Lines that retained the introgressions were then used as the female parent in a pentaploid crossing strategy involving two durum wheat genotypes Karim and Om Rabi 5 (Figure 1).
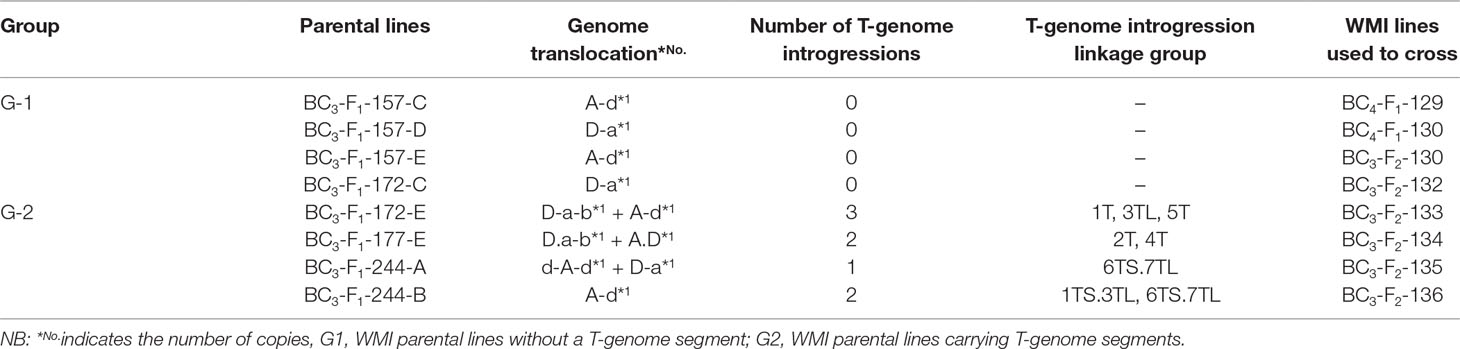
Table 1 Type and number of the D- genome and T-genome introgressions present in the parental introgression lines and the reference of the WMI lines used in the crosses.
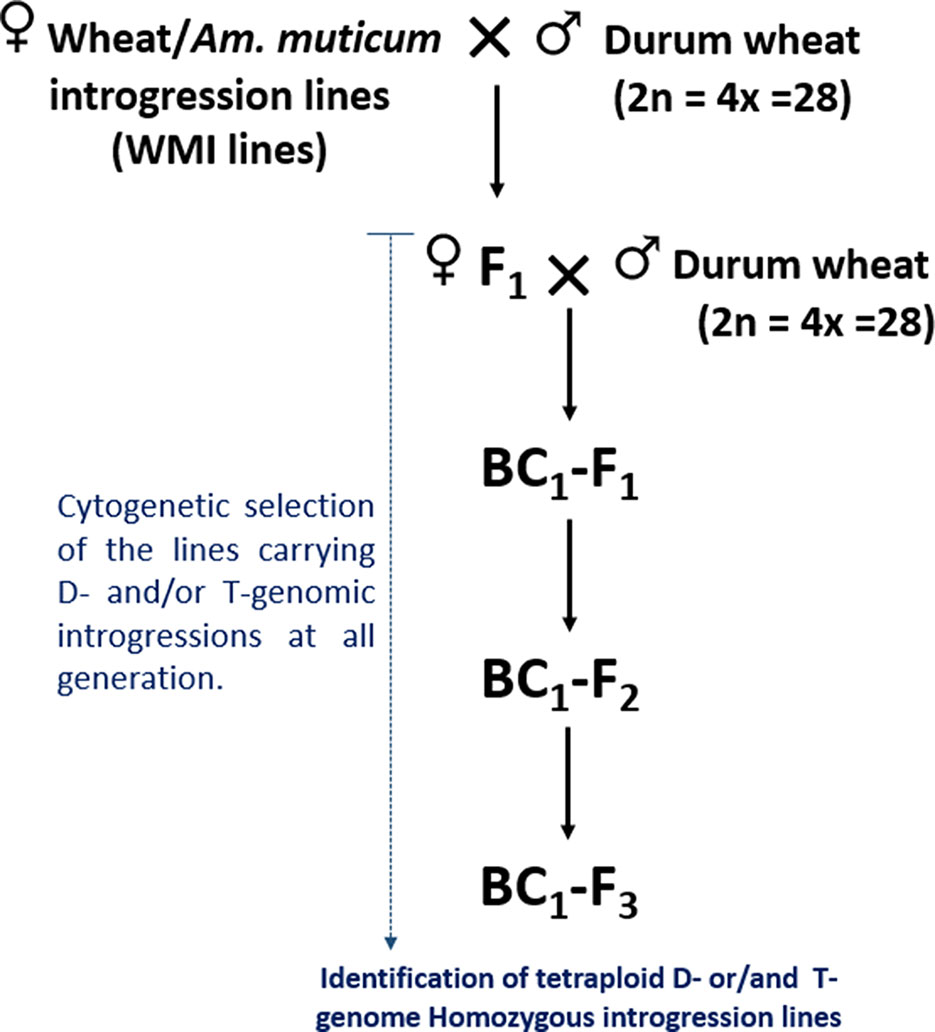
Figure 1 Crossing diagram for the introgression of the D- and T-genome segments identified in WMI lines into durum wheat.
Genomic In Situ Hybridization (GISH)
Slides of chromosome spreads were obtained as described in Kato et al. (2004) and King et al. (2017). Mc-GISH of the slides was conducted using the labeled total genomic DNA of the three putative progenitor species of wheat; T. urartu (A-genome), Ae. speltoides (B-genome) and Ae. tauschii (D-genome), as well as Am. muticum (T-genome). DNA was extracted from the young leaves using a CTAB method (Zhang et al., 2013) and labeled using the nick translation procedure (Luchniak et al., 2002). Slides were probed with T. urartu labeled with Chroma Tide Alexa Fluor 488-5-dUTP (Invitrogen; C11397; green), Ae. tauschii with Alexa Fluor 594-5-dUTP (Invitrogen; C11400; red), Am. muticum with Alexa Fluor 546 (Invitrogen; C11401; yellow) and the genomic DNA of Ae. speltoides fragmented to 300–500bp (using a heat block for 15 min at 110°C) used as a blocking DNA in a ratio of 1:1:2:30.
For the detection of T-genome introgression alone some of the lines were probed by single color GISH using the labeled Am. muticum genomic DNA with Chroma Tide Alexa Fluor 488-5-dUTP (Invitrogen; C11397; green) and the fragmented genomic DNA of wheat cv. Chinese Spring (300–500bp) as blocking DNA in a ratio of 1:50 per slide.
Slides were counterstained with 4’-6-diamidino-2- phenylindole (DAPI) and analyzed using a high throughput, fully automated Zeiss Axio Imager.Z2 upright epifluorescence microscope (Carl Zeiss Ltd, Oberkochen, Germany). Photographs were taken using a MetaSystems Coolcube 1 m CCD camera. Further slide analysis was carried out using an automated metaphase image capture software, Metafer4, and the ISIS software for image processing (Metasystems GmbH, Altlussheim, Germany).
Fluorescence in Situ Hybridization (FISH)
For multi-color fluorescence in situ hybridization (mc-FISH), two repetitive DNA sequences, pSc119.2 (McIntyre et al., 1990) and pAs1 (Rayburn and Gill, 1986), were labeled by nick translation with Alexa Fluor 488-5-dUTP (green) and Alexa Fluor 594-5-dUTP (red), respectively, and hybridized to the slides. Subsequent counterstaining and image capture were performed as described for GISH.
Genotyping With KASP™ Markers
Genomic DNA was isolated from leaf tissue of 10-day old seedlings in a 96-well plate as described by Thomson and Henry (1995). All lines showing T- and/or D-genome introgressions were genotyped alongside the two durum wheat genotypes, one Ae. tauschii accession P95-81.1.1-1 obtained from USDA and two bread wheat genotypes, Chinese Spring and Paragon, used as controls. The full set of Langdon disomic D-genome substitution lines (obtained from the USDA), were also used as control lines to verify the specificity of the Kompetitive Allele-Specific PCR (KASP) markers to the D-genome. In these lines a pair of D-genome chromosomes substitute a pair of either the A- or the B-genome chromosomes of the same linkage group (Joppa and Williams, 1988).
A total of 80 D-genome specific KASP™ markers (Grewal et al., 2019) of which 29 markers were polymorphic between wheat and Am. muticum, were used for simultaneous detection of the D- and T-genome introgression. For each KASP™ marker, two allele-specific forward primers and one common reverse primer were used (Supplementary Material). Genotyping reactions were performed in a ProFlex PCR system (Applied Biosystems by Life Technology) in a final volume of 5 µl with 1 ng genomic DNA, 2.5 µl KASP reaction mix (ROX), 0.068 µl primer mix and 2.43 µl nuclease free water. PCR conditions were set as 15 min at 94°C; 10 touchdown cycles of 10 s at 94°C, 1 min at 65–57°C (dropping 0.8°C per cycle); and 35 cycles of 10 s at 94°C, 1 min at 57°C.
Fluorescence detection of the reactions was performed using a QuantStudio 5 (Applied Biosystems) and the data analyzed using the QuantStudio™ Design and Analysis Software V1.5.0 (Applied Biosystems).
Results
The Development of Durum Wheat D- and/or T-Genome Introgression Lines
Only 23 seed, out of the 32 randomly selected from the eight WMI parental lines, germinated and reached maturity. Cytogenetic screening via mc-GISH showed that 15 of these lines had retained at least one copy of the D-genome introgression. Sixty-three crosses were made between these lines and the two durum genotypes to produce the F1 plants. A further 68 back-crosses were made between the F1 plants and durum wheat. The total number of crosses, percentage of crosses setting seed, number of seed produced and percentage germination are shown in Table 2.
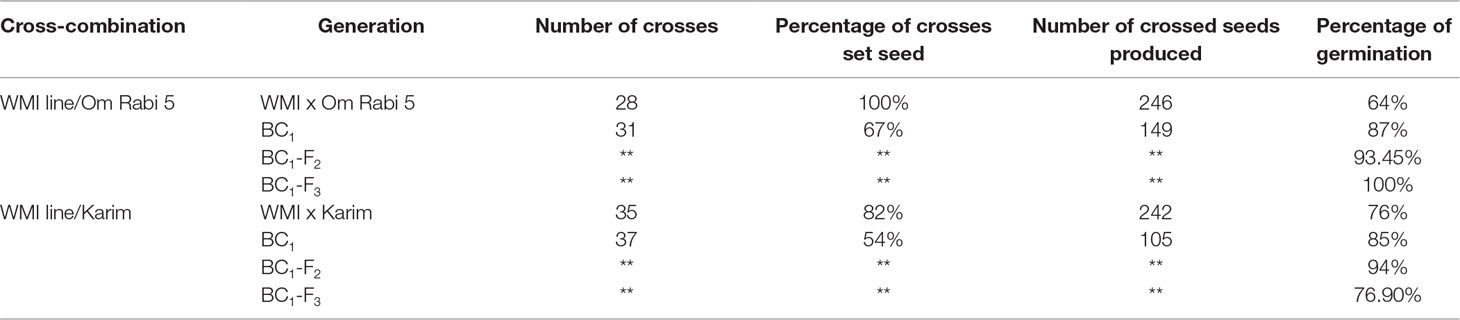
Table 2 Number of crosses, percentage of crosses setting seed, number of seeds produced and percentage of seed germination at every generation in the two cross-combination of the WMI lines and their subsequent backcross generations to both the Om Rabi 5 and Karim durum wheat genotypes.
Chromosome counts showed that 70% of the BC1-F2 generation and 88.4% of the BC1-F3 lines carrying D-genome introgressions had 28 chromosomes. Hence, backcrossing to the durum wheat parent had gradually decreased the average chromosome number through the loss of D-genome univalents.
Mc-GISH Analysis of the D-Genome Introgression Lines
Only lines carrying D-genome introgressions were selected using mc-GISH at every generation. The percentage of F1, BC1-F1, BC1-F2, and BC1-F3 carrying D-genome introgressions was 33.3, 35.5, 36.5, and 85, respectively, with the percentage retention slightly higher in the lines produced with Karim in all generations except the BC1-F3 (Table 3). The number of D-genome introgressions per line varied between one to three introgressions, with most lines carrying a single D-genome introgression. A higher number of lines carrying inter-genome introgressions were identified in the lines belonging to the G-2 plants, i.e. those in which the initial parental line had one to three introgressions of Am. muticum. New D-genome introgressions that were not present in the WMI parental lines were identified in all generations, with the highest number occurring in the BC1-F1 and BC1-F2 generations in Karim and Om Rabi 5 cross-combination, respectively (Table 3). However, these new introgressions occurred only in the G-2 group and mainly in the progeny from the crosses to Karim.
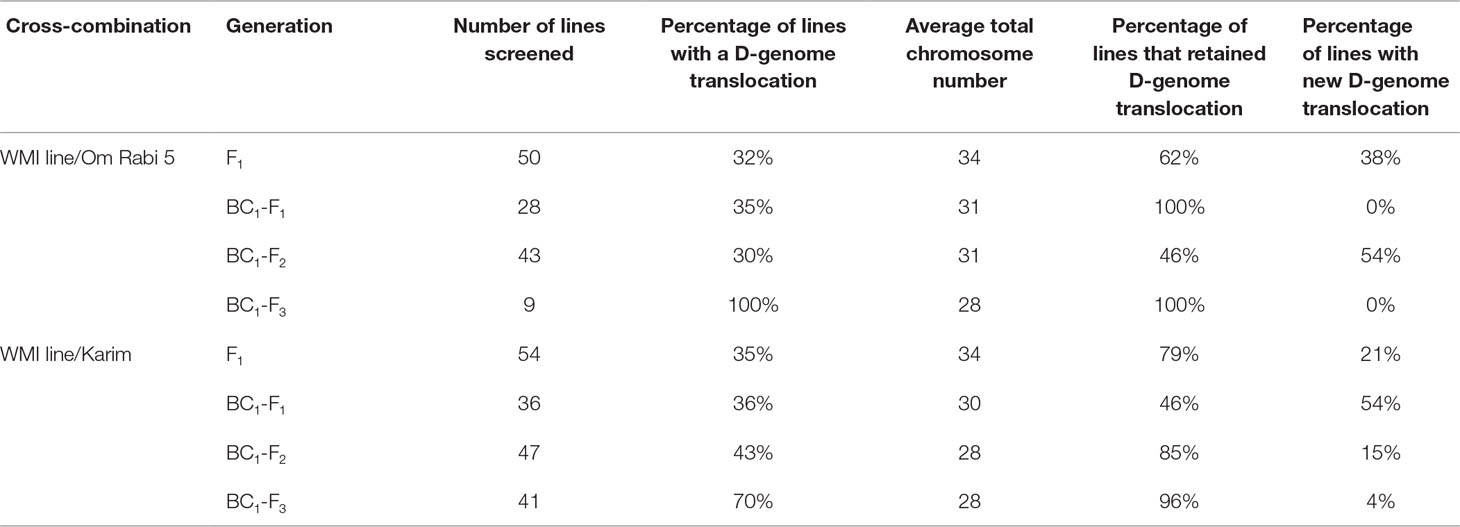
Table 3 Summary result table on the percentage of the retention and occurrence of new D-genome introgressions at the F1 and subsequent backcross generations of the WMI lines to Om Rabi 5 and Karim.
The most frequent introgressions identified initially in the WMI parental lines were D-introgressions into the A-genome, with recombination in the telomeric region (A-d or D-a introgressions—Table 4). The newly formed introgressions mainly involved either the D-genome with the A-genome or with both the A- and the B-genomes. Overall, a higher number of different AD (e.g., A-d, d-A-d, D-a, and A.D) and ABD (e.g., D.a-b, D-a-b, D.a-d, and B.a-d) recombinants were identified compared to BD (e.g., B.D and B-d) or AB (e.g., A.B and B-a) recombinants (Table 4). A-d recombinants, consisting of a small D-genome segment introgressed into either the long arm (LA) or the short arm (SA) of an A-chromosome, were retained the most between consecutive selfed generations.
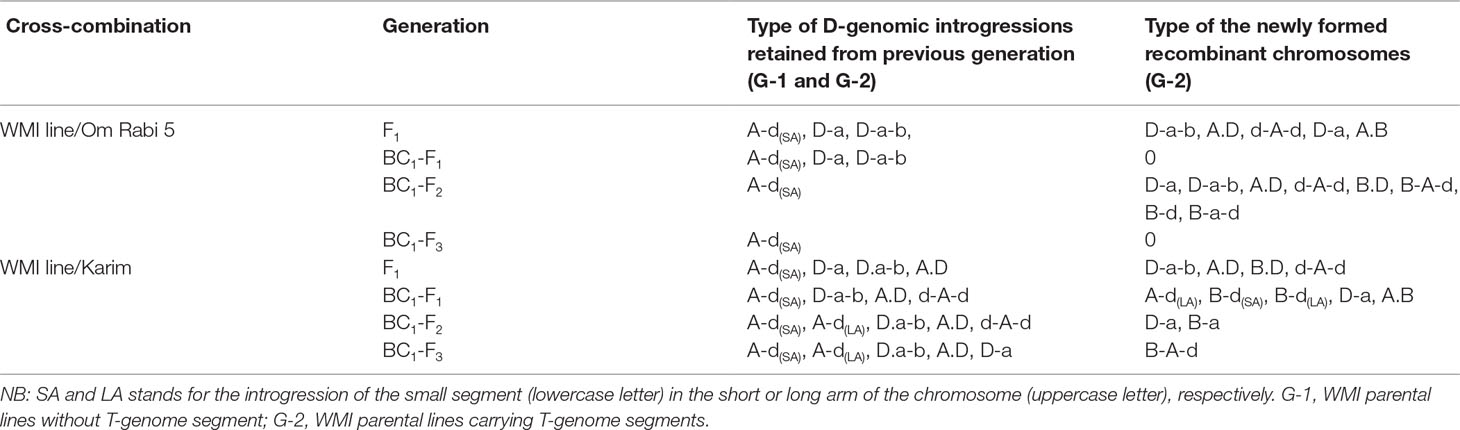
Table 4 Summary table of the introgressions identified and retained at the F1 and subsequent backcross generations of the WMI lines crossed to Om Rabi 5 and Karim genotypes and occurrence of new D-genome introgressions in the G-2 group.
The D-genome introgressions identified in the BC1-F2 progeny from Om Rabi 5 originated from only two BC1-F1 plants—a G-1 plant and a G-2 plant. Five progenies from the G-1 plant were found to contain a homozygous D-genome introgression of the A-d(SA) type. This introgression was initially identified in the parental hexaploid WMI line and hence, was successfully transferred into Om Rabi 5. Five of the seven progeny from the G-2 BC1-F1line showed the presence of either a single copy (3 lines) or two copies (2 lines) of a large T-genome introgression. One to three new D-genome introgressions were also identified in all seven lines screened.
The BC1-F2 Karim lines containing D-genome introgressions were progeny of the same G-2 WMI parental line (BC3-F2-134). The D-genome introgressions were all telomeric, recombined with an A-chromosome (A-d(SA) or A-d(LA)). Ninety-three percent of the lines carrying D-genome introgressions also contained at least one T-genome introgression or chromosome. Hence, simultaneous introgression of the D- and the T-genomes was identified in the tetraploid lines. Mc-GISH showed that the T-genome introgression was recombined with a B-genome chromosome near the telomere (T-b) and had substituted a B-genome chromosome. Homozygous tetraploid introgression lines for both the D- and the T-genomes were identified in the BC1-F3 generation in some of the selfed progeny of the tetraploid BC1-F2 lines.
Genotyping of the Introgression Lines
Five D-genome introgression lines (one in the Om Rabi 5 background and four in the Karim background) were isolated in the BC1-F3 generation. While the D-genome introgression in Om Rabi 5 background was present in the parental WMI line, the four D-genome introgressions into Karim were identified in four BC1-F1 lines which were derived from the same WMI line. A total of 80 KASP markers distributed across the seven linkage groups of the D- and T-genomes (except for the 1DS and 1TS arms) were used to characterise both the D-genome and T-genome introgressions in the progenies of the G2-WMI lines (Figure 2). A total of 16 D-genome introgression lines were genotyped (including at least two sister lines carrying each of the five introgressions described above).
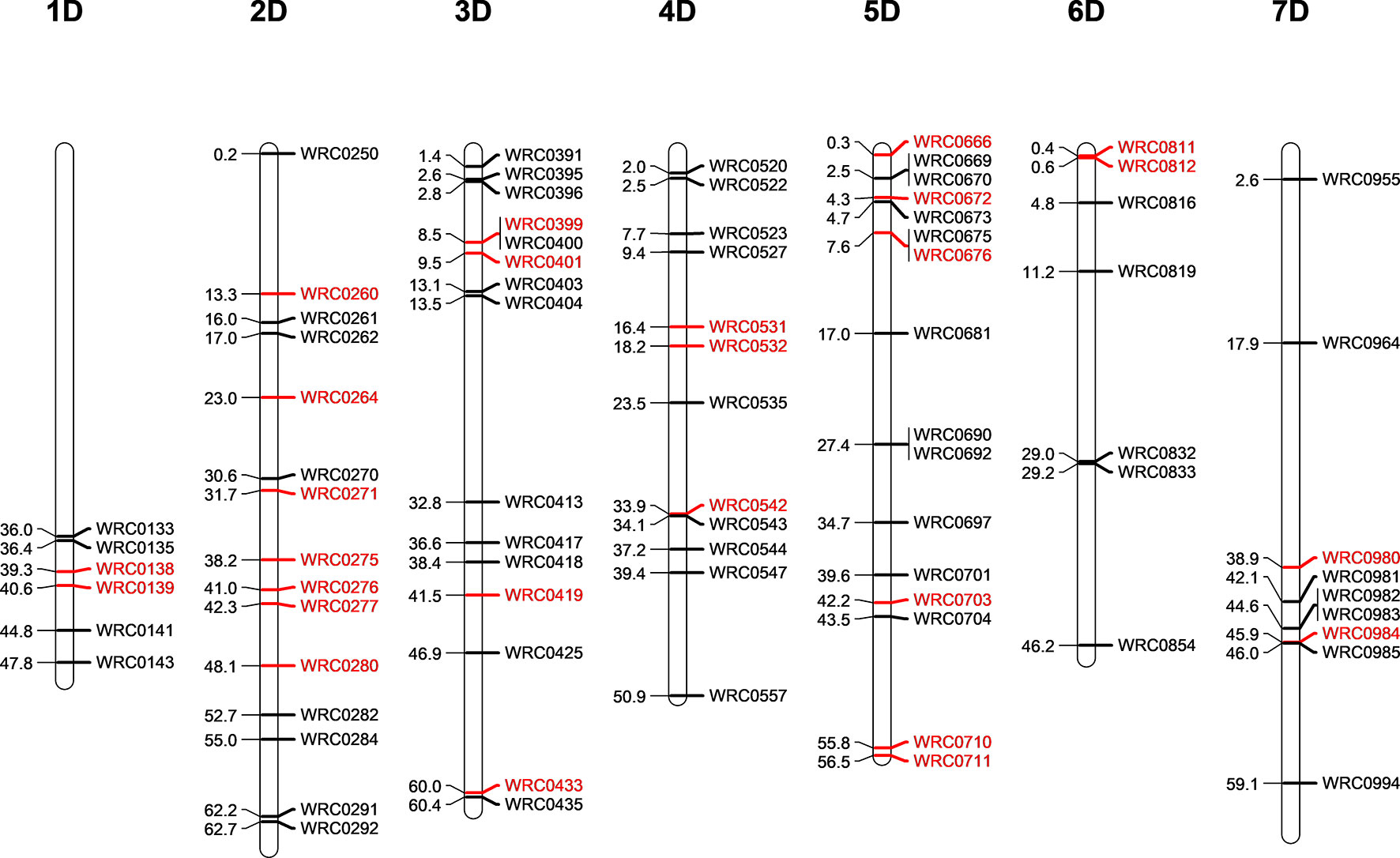
Figure 2 Physical position of the D-genome specific KASP markers on the seven linkage groups of the D-genome of wheat in bp×10−7. The D-specific KASP markers that are polymorphic between both wheat and the T-genome of Am. muticum (same linkage group as wheat) are highlighted in red.
Genotyping identified the D-genome introgression into Om Rabi 5 as the telomeric region of the 5DS chromosome arm via the amplification of the closely linked KASP markers WRC0669 and WRC0670 located at 24,574,003 and 24,971,617 bp (base pair) on wheat chromosome 5D (International Wheat Genome Sequercincy Consortium et al., 2018). However, the absence of amplification of marker WRC0666 (located at 3,031,923 bp) on chromosome 5D indicates that a deletion might have occurred in this region of the 5DS introgressed segment. The introgression was confirmed as homozygous by mc-GISH.
Three of the four D-introgressions into Karim were A-d(LA) introgressions. Two of these segments were characterized as the telomeric region of 1DL and one as the telomeric region of 6DL. KASP markers detected a small difference in segment size between the two 1DL introgressions. Only one marker, WRC0143, located at the telomeric region of 1DL amplified in the two BC1-F3-202-A and -B sister lines. However, the introgression in the BC1-F3-214, 215, 312, and 315 sister lines were shown to be larger due to the amplification of WRC0143 and WRC0141. The 6DL introgression was identified as a telomeric segment through amplification of marker WRC0854. The fourth introgression into Karim (A-d(SA)) was characterized as the very telomeric region of 2DS via the amplification of marker WRC0250.
The polymorphic wheat/Am. muticum KASP markers (highlighted in red in Figure 2) were able to detect the presence of T-genome introgressions in all the introgression lines from Karim. The two 6DL introgression lines had retained both the 2T and 4T Am. muticum introgressions, originally present in the WMI parental line. The remaining lines, however, had retained only the 4T introgression (mc-GISH showed that the large 4T introgression in all these lines had recombined with a B-genome chromosome). Combined analysis with genotyping and mc-GISH identified lines containing simultaneous homozygous introgressions of 4T and either 1DL or 6DL. The introgression remained heterozygous in the BC1-F3 line analyzed, although the 4T introgression was again homozygous. GISH analysis of the two tetraploid BC1-F3-324 sister lines showed that the 2T and 4T Am. muticum introgressions, identified via KASP, were both homozygous substituting two A- and two B-chromosomes. However, these two lines were both sterile and failed to produce seed (BC1-F4 seed was produced from the rest of the introgression lines).
Mc-FISH Characterization of the Introgression Lines
Mc-FISH based karyotyping of the introgression lines was used to identify the wheat chromosomes involved in the introgressions by comparison with the mc-FISH karyotype of Chinese Spring (Tang et al., 2014). Mc-FISH of the homozygous Om Rabi 5 5DS introgression identified it as being recombined with the short arm of chromosome 5A (Figures 3 A–C). Only two of the D-genome introgressions into Karim could be characterized as the 1DL introgression identified in the BC3-F2-202 sister lines and the 6DL introgression were too small to detect. The 1DL introgression identified in the BC1-F3-214, 215, 312, and 315 sister lines, however, was recombined with the long arm of chromosome 1A (Figures 3 D–F) and the 2DS introgression with the short arm of chromosome 2A (Figures 3 G–I). The B-genome introgression, recombined with the large 4T introgression, was also too small to detect. However, this single or homozygous 4T-b recombinant chromosome was found to have substituted either a single or a pair of 4B chromosomes (Figures 3 E–J).
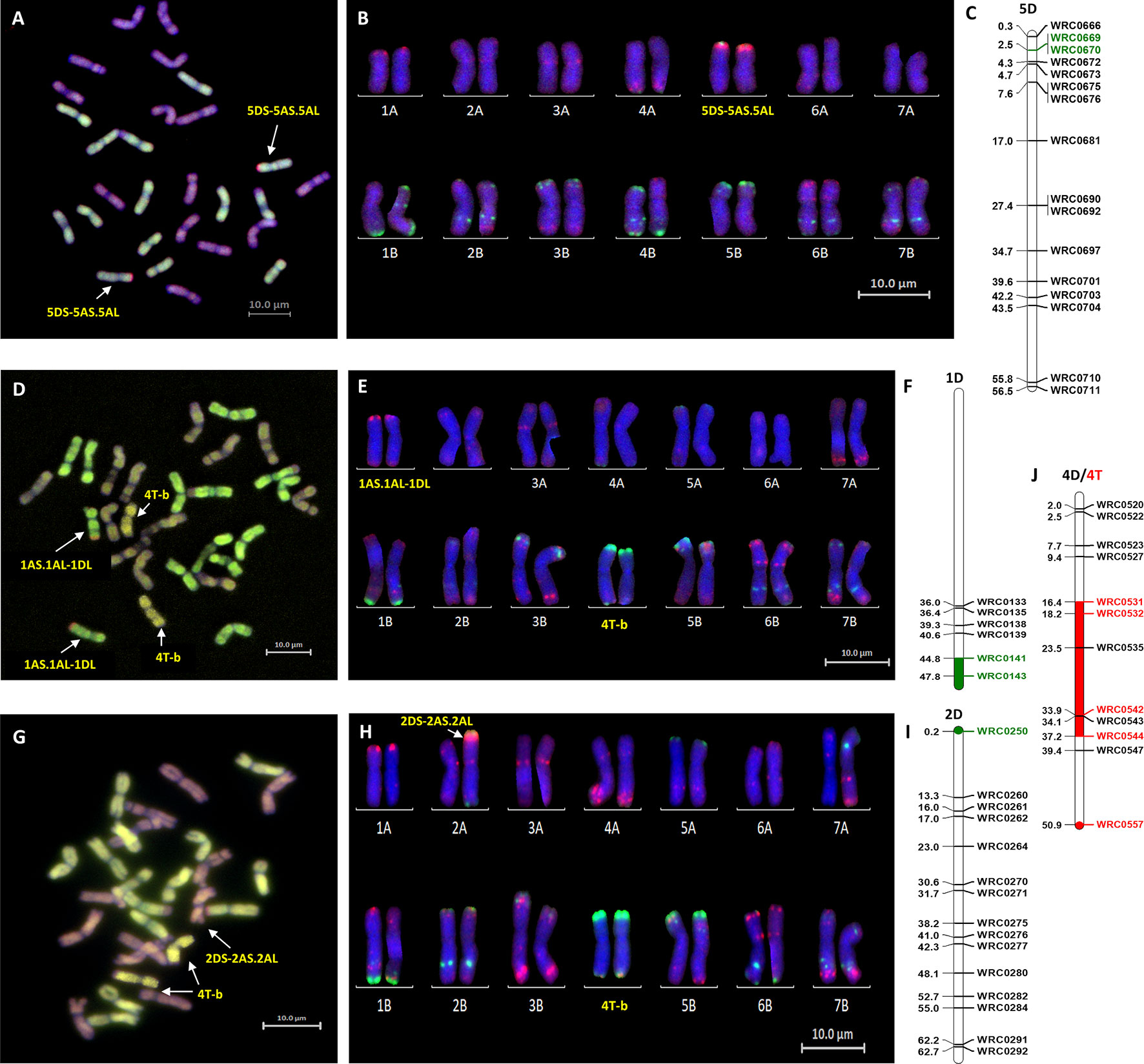
Figure 3 Molecular and cytogenetic characterisation of D-genome and T-genome introgression lines. (A, D, G) Mc-GISH showing the D-genome and T-genome introgression (A-genome in green, B-genome in purple, D-genome in red and T-genome in yellow), (B, E, H) mc-FISH based karyotype using the Oligo-pAs.1 (red) and Oligo-pSc119.2 (green) probes counterstained with DAPI (blue) (C,F, I) physical position (in bp×10−7) of the 5DS, 1DL and 2DS introgressions (green markers and region) using D-genome specific KASP markers showing the D-genome introgressions as (A, B, C) 5DS-5AS.5AL in the genomic backgroung of Om Rabi 5, and as (D, E, F) 1AS.1AL-1DL and (G, H, I) 2DS-2AS.2AL in the BC1-F3-315-E and BC1-F3-141-A lines, respectively, in the genomic background of Karim. (J) characterization of the T-genome introgression as a 4T chromosome recombined in its telomeric long arm with a small B-genome segment noted as 4T-b substituting the pair 4B chromosomes using wheat/T-genome polymorphic KASP markers (red marker and region) in both the BC1-F3-214-B and BC1-F3-141-A lines, respectively.
Discussion
Pentaploid crosses between bread and durum wheat have previously been shown to generate viable F1 seed that can be used in a backcrossing programme to either of the parents (Eberhard et al., 2010; Martin et al., 2013; Kalous et al., 2015). The presence of inter-genomic rearrangements in the hexaploid background of wheat/wild relative introgression lines [such as those identified in wheat/Am. muticum introgression lines by King et al. (2017)] can thus be used for the introgression of the D-genome of bread wheat into durum wheat. While the overall aim of the current programme was to introgress the D-genome, the crosses also had the potential to increase the genetic variability of the durum A- and B-genomes through recombination with their homologues of bread wheat.
In crosses involving parents of different ploidy levels, it has been shown that using the higher ploidy level genotype as the maternal parent is generally more successful in producing viable F1 progeny (Ramsey and Schemske, 1998; Kalous et al., 2015). In pentaploid wheat crosses, the hexaploid parent is usually used as the female parent (Padmanaban et al., 2017a; Padmanaban et al., 2017b) and thus, the hexaploid WMI lines were used here as the female parent. Viable F1 seeds were obtained with both durum wheat parents, Om Rabi 5 and Karim. However, a higher seed set was obtained in the F1 and the BC1 generations using Om Rabi 5 as compared to Karim suggesting a higher crossing compatibility of the Om Rabi 5 genotype in the crosses with the bread wheat used here. Other studies have highlighted the importance of the parental choice, as well as the direction of the cross, in the production of viable pentaploid hybrids (reviewed in Padmanaban et al., 2017b).
Mc-GISH was used to visualize the three genomes of wheat and thus, the presence of inter-genomic recombinant chromosomes within the wheat genome of the original introgression lines. This technique has been widely used for studying genome rearrangements, alien introgressions and the discrimination between different genomes in polyploid cereals (Schwarzacher et al., 1989; Schwarzacher et al., 1992; Schubert et al., 2001; Silva and Souza, 2013).
A higher number of lines carrying D-genome introgressions were distinguished in the progeny of Karim in all generations as compared to Om Rabi 5. This could indicate a higher tolerance of Karim to the presence of the D-genome introgressions emphasizing the importance of the durum parent selected for the work. However, a higher seed set with Om Rabi 5 does indicate that the choice of durum parent might not be straight forward. In addition to the D-genome introgressions present in the parental WMI lines, new wheat sub-genome introgressions involving the D- with either the A- and/or the B-genome and the A- with the B-genome were identified at all generations from the F1 to the BC1-F2. A mc-FISH karyotype [based on the karyotype for Chinese Spring developed by Tang et al. (2014)] was used to identify the wheat chromosomes present in the tetraploid BC1-F3 plants containing single or homozygous D-genome introgressions from both cross combinations. This showed the presence of a pair of 5B chromosomes in all lines. Since wild-type wheat (Paragon) was used to develop the WMI parental lines, instead of a ph1b mutant wheat (King et al., 2017), the inter-genomic rearrangements that occurred in the later generations were not due to the absence of the Ph1 gene. However, the presence of more than two genomes (A, B, D, and T) and unequal chromosome numbers in one cell could have promoted abnormal meiotic behavior leading to homologous paring. Wheat chromosomes in the selfed progeny of wheat/rye monosomic addition lines, such as 1R, 4R and 6R, show abnormal behavior at meiosis resulting in the elimination or the addition of some of the wheat chromosomes e.g., three 4A-chromosomes were observed in one of the progeny from a 7R monosomic addition line and chromosomes 5A and 4B were eliminated from some of the progeny of the 6R monosomic addition line in addition to alterations of the wheat chromosomes (Fu et al., 2012; Fu et al., 2013).
In the present study, new recombinant events occurred only in lines belonging to the G-2 group with at least one large T-genome introgression/chromosome present in the parental lines. Am. muticum is known to contain genes that promote pairing between homologous chromosomes/suppress the effect of the Ph1 gene in hybrids with allopolyploid wheat (Dvorak, 1972; Dover and Riley, 1972). Similarly, two major Ph1 suppressor loci, Su1-Ph1 and Su2-Ph1 were mapped on the distal end of the long arm of chromosomes 3S and 7S, respectively, in Ae. speltoides (Dvorak et al., 2006; Li et al., 2017). It may be possible that some of the introgressed segments from Am. muticum also carry a Ph1 suppressor gene. However, new introgressions were distinguished in the progeny of G-2 WMI lines carrying different introgressions of Am. muticum such as 2T, 4T, 6TS.7TL and 1TS.3TL. Hence, it is possible that the stress caused by the presence of Am. muticum introgression(s) might be one factor inducing recombination.
The new inter-genomic rearrangements were found to be made up of 80% D-genome with either the A- and/or the B-genome. The univalent state of the D-chromosomes in these lines may also have promoted the rearrangements. The A- and B-genomes have previously been shown to be more similar to the D-genome than they are to each other (Marcussen et al., 2014). Pairing is frequently observed between the A- and the D-genomes in wheat-rye hybrids denoting a much lower differentiation between these two genomes than between the A- and B- or B- and D-genomes, at least in the regions of high recombination in the distal chromosome regions (Naranjo et al., 1987; Marcussen et al., 2014). This is consistent with the high level of A-D recombinant chromosomes observed in the present study, especially in the telomeric regions of the chromosomes. For example, for the introgressions that could be identified with mc-FISH, analysis showed that the slightly larger 1DL introgression had recombined with the short arm of 1A and the 2DS introgression with the short arm of 2A.
Only the small telomeric D-genome introgressions were successfully transferred into the tetraploid background of both durum wheat varieties indicating that introgressions of a smaller size have a higher chance of being transmitted compared to larger D-genome introgressions. If the large D-genome segments do not have the ability for genetic compensation for the homologous A- or B- genome chromosome segments, it less likely they will be retained. The inter-genomic recombinant chromosomes that were present as additions were generally lost due to a lack of pairing at meiosis. For instance, the A-d translocation when present as a monosomic addition, in the tetraploid background with 29 chromosomes, was not retained after self-fertilisation. Whereas, the recombinant chromosomes that had substituted one of the wheat chromosomes had a higher rate of retention and transmission.
KASP marker analysis showed that the Am. muticum introgression in all the Karim D-genome introgression lines was a large 4T introgression previously confirmed as present in the WMI parental line, together with a 2T introgression, using the Axiom® Wheat-Relative Genotyping Array (King et al., 2017). The 4T introgression was highly retained in the progeny of Karim. Lines homozygous for both 4T and 1DL were identified in the BC1-F3 where FISH analysis showed that the pair of 4T recombinant chromosomes were substituting the pair of 4B-chromosomes. Under glasshouse conditions, these introgression lines were fertile with a normal spring wheat growth cycle and a durum wheat head type. Thus, the disomic 4T-b(4B) substitution did not affect fertility in these lines. Among the full set of Chinese Spring nullisomic–tetrasomic lines, only the 4B nullisomic tetrasomic line (N4BT4D) was completely male sterile (and had to be maintained as a monosomic tetrasomic line, M4BT4D) suggesting the presence an essential gene for male fertility on this chromosome (Sears, 1966). In addition, Endo and Gill (1996), failed to establish a homozygous deletion line for the short arm of chromosome 4B in a hexaploidy background, because plants were male sterile. However, Langdon durum 4D(4B) disomic substitution line is also fertile and can be selfed in the absence of the 4B chromosomes (Joppa and Williams, 1988). Hence, the 4D disomic substitution compensates for the absence of both copies of chromosome 4B at the tetraploid level but does not compensate when present as tetrasomic in the 4B nullisomic tetrasomic line at the hexaploid level. This can possibly be due to the interaction of several genes. Similar to the Langdon durum 4D(4B) disomic substitution line, the 4T-b introgression fully compensate for the absence of the male fertility gene, Ms1 (Driscoll, 1975), on chromosome 4B in durum wheat.
To our knowledge, this is the first study to transfer D-genome introgressions into either the A- or B-genomes, present in hexaploid wheat/wild relative introgression lines, into durum wheat. Advances in cytology and mc-GISH have made it possible to identify, characterize and track these genome rearrangements, together with wild relative introgressions, enabling their transfer via pentaploid crosses. Mc-GISH, however, is labour intensive and relatively low throughput. KASP markers, able to detect the presence of Am. muticum introgressions in wheat, have been developed at the WRC. Many of the KASP markers are wheat genome specific and those that are specific to the D-genome were used for the detection of the D-genome introgressions in the later generations. However, for future work, these markers will be used in the earlier generations such as the F1 and BC1-F1 with the mc-GISH analysis used for validation and chromosome counting in the later generations. The developed introgression lines can be of use in durum wheat breeding through marker assisted selection, to screen for several traits of interest such as disease resistance or agronomic traits. Once multiplied, D- and T-genome introgression lines as well as the KASP markers associated with the introgressed segments will be made freely available upon request from the GRU.
Data Availability
The raw genotyping data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
MO conducted the crossing program. MO and CY performed the in situ hybridization experiments. MO, SH-E, DS, and SA performed the genotyping analysis. MO, JK, IK, and SG wrote the manuscript. MO, AY, PG, PS, IK, and JK conceived the experimental design. All authors read and approved the final manuscript.
Funding
This study was supported by the Monsanto Beachell Borlaug International Scholarship program (2015), The Crop Trust and the Biotechnology and Biological Sciences Research Council [grant number BB/P016855/1] as part of the Designing Future Wheat programme (DFW). The funding body played no role in the design of the study and collection, analysis and interpretation of data and in writing the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
WMI, wheat/Am. muticum introgression; SA, short arm; LA, long arm.
Acknowledgments
Langdon D-genome disomic substitution lines were kindly provided by Justin Faris (USDA-ARS, Fargo).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01110/full#supplementary-material
Table S1 | Primer sequences for all KASP markers used in this study.
References
Bonjean, A. P., Angus, W. J., van Ginkel, M. (2016). The World Wheat Book: A History of Wheat Breeding Vol. 3. Paris: Lavoisier.
Dover, G. A., Riley, R. (1972). The prevention of pairing of homoeologous meiotic chromosomes of wheat by a genetic activity of supernumerary chromosomes of Aegilops. Nature 240, 159–161. doi: 10.1038/240159a0
Driscoll, C. J. (1975). Cytogenetic analysis of two chromosomal male-sterility mutants in hexaploidy wheat. Aus. J. Biol. Sci. 28, 413–416. doi: 10.1071/BI9750413
Dundas, I., Verlin, D., Islam, R. (2015). Chromosomal locations of stem and leaf rust resistance genes from Ae. caudata, Ae. searsii and Ae. mutica. Abstract retrieved from, BGRI Workshop, September 17–20, Sydney.
Dvorak, J. (1972). Genetic variability in Aegilops speltoides affecting homoeologous pairing in wheat. Can. 5. Genet. CytoL. 14, 371–380. doi: 10.1139/g72-046
Dvorak, J., Deal, K. R., Luo, M. C. (2006). Discovery and mapping of wheat Ph1 suppressors. Genetics 174, 17–27. doi: 10.1534/genetics.106.058115
Eberhard, F. S., Zhang, P., Lehmensiek, A., Hare, R. A., Simpfendorfer, S., Sutherland, W. M. (2010). Chromosome composition of an F2Triticum aestivum ¾ T. turgidum spp. Crop Pasture Sci. 61, 619–624. doi: 10.1071/CP10131
Elek, H., Smart, L., Ahmad, S., Anda, A., Werner, C. P., Pickett, J. A. (2014). A comparison of the levels of hydroxamic acids in Aegilops speltoides and a hexaploid wheat and effects on Rhopalosiphum padi behaviour and fecundity. Acta Biol. Hung. 65, 38–46. doi: 10.1556/ABiol.65.2014.1.4
Endo, T. R., Gill, B. S. (1996). The deletion stock of common wheat. Heredity. 87, 295–307. doi: 10.1093/oxfordjournals.jhered.a023003
Eser, V. (1998). Characterisation of powdery mildew resistant lines derived from crosses between Triticum aestivum and Aegilops speltoides and Ae. mutica. Euphytica 100, 269–272. doi: 10.1023/A:1018372726968
Feldman, M., Levy, A. A. (2005). Allopolyploidy: a shaping force in the evolution of wheat genomes. Cytogenet. Genome Res. 109, 250–258. doi: 10.1159/000082407
Friebe, B., Jiang, J., Raupp, W. J., McIntosh, R. A., Gill, B. S. (1996). Characterization of wheat alien translocations conferring resistance to diseases and pests: current status. Euphytica 71, 59–87. doi: 10.1007/BF00035277
Fu, S., Lv, Z., Qi, B., Guo, X., Li, J., Liu, B., et al. (2012). Molecular cytogenetic characterization of wheat-Thinopyrum elongatum addition, substitution and translocation lines with a novel source of resistance to wheat Fusarium head blight. J. Genet. Genom. 39, 103–110. doi: 10.1016/j.jgg.2011.11.008
Fu, S., Yang, M., Fei, Y., Tan, F., Ren, Z., Yan, B., et al. (2013). Alterations and abnormal mitosis of wheat chromosomes induced by wheat-rye monosomic addition lines. PLoS One 8, 70483. doi: 10.1371/journal.pone.0070483
Grewal, S., Hubbart-Edwards, S., Yang, C., Devi, U., Baker, L., Heath, J., et al. (2019). Rapid identification of homozygosity and site of wild relative introgressions in wheat through chromosome-specific KASP genotyping assays. Plant Biotechnology Journal. In Press.
Haider, N. (2013). The origin of the B-genome of bread wheat (Triticum aestivum L.). Russ. J. Genet. 49, 263–274. doi: 10.1134/S1022795413030071
Iefimenko, T. S., Fedak, Yu, G., Antonyuk, M. Z., Ternovska, T. K. (2015). Microsatellite analysis of chromosomes from the fifth homoeologous group in the introgressive Triticum aestivum/Amblyopyrum muticum wheat lines. Cytol. Genet. 49, 183–191. doi: 10.3103/S0095452715030056
International Wheat Genome Sequencing Consortium, Appels, R., Eversole, K., Feuillet, C., Keller, B., Rogers, J., et al. (2018). Shifting the limits in wheat research and breeding using a fully annotated reference sequence. Science 361, 7191. doi: 10.1126/science.aar7191
Jauhar, P. P., Chibbar, R. N. (1999). Chromosome-mediated and direct gene transfers in wheat. Genome 42, 570–583. doi: 10.1139/g99-045
Joppa, L. R., Williams, N. D. (1988). Langdon durum disomic substitution lines and aneuploid analysis in tetraploid wheat. Genome 30, 222–228. doi: 10.1139/g88-038
Kabbaj, H., Sall, A. T., Al-Abdallat, A., Geleta, M., Amri, A., Filali-Maltouf, A., et al. (2017). Genetic diversity within a global panel of durum wheat (Triticum durum) landraces and modern germplasm reveals the history of allele’s exchange. Front. Plant Sci. 8, 1277. doi: 10.3389/fpls.2017.01277
Kalous, J. R., Martin, J. M., Sherman, J. D., Heo, H. Y., Blake, N. K., Lanning, S. P., et al. (2015). Impact of the D genome and quantitative trait loci on quantitative traits in a spring durum by spring bread wheat cross. Theor. Appl. Genet. 128, 1799–1811. doi: 10.1007/s00122-015-2548-3
Kato, A., Lamb, J. C., Birchler, J. A. (2004). Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc. Natl Acad. Sci. U.S.A. 101, 13554–13559. doi: 10.1073/pnas.0403659101
Kihara, H. (1924). Cytologische und genetische Studien bei wichtigen Getreidearten mit besondere Rüchsicht auf das Verhalten der Chromosomen und die Sterilität in den Bastarden. Mem. Coll. Sci. Kyoto Imp. Univ. 1, 1–200.
Kihara, H. (1944). Discovery of the DD-analyser, one of the ancestors of Triticum vulgare (abstr) (in Japanese). Agric. Hortic. 19, 889–890. doi: 10.1016/j.jgg.2011.07.002
Kilian, B., Mammen, K., Millet, E., Sharma, R., Graner, A., Salamini, F., et al, (2013). “Aegilops,” in Wild crop relatives: genomic and breeding resources. Ed. Kole, Ch (Berlin, Heidelberg: Cereals. Springer), 1–76. doi: 10.1007/978-3-642-14228-4
King, I. P., Forster, B. P., Law, C. C., Cant, K. A., Orford, S. E., Gorham, J., et al. (1997). Introgression of salt-tolerance genes from Thinopyrum bessarabicum into wheat. New Phytol. 137, 75–81. doi: 10.1046/j.1469-8137.1997.00828.x
King, J., Armstead, I., Harper, J., Ramsey, L., Snape, J., Waugh, R., et al. (2013). Exploitation of interspecific diversity for monocot crop improvement. Heredity 110, 475–483. doi: 10.1038/hdy.2012.116
King, J., Grewal, S., Yang, C., Hubbart, S., Scholefield, D., Ashling, S., et al. (2017). A step change in the transfer of interspecific variation into wheat from Amblyopyrum muticum. Plant Biotech. J. 15, 217–226. doi: 10.1111/pbi.12606
Li, H., Deal, K. R., Luo, M.-C., Ji, W., Distelfeld, A., Dvorak, J. (2017). Introgression of the Aegilops speltoides Su1-Ph1 Suppressor into Wheat. Front. Plant Sci. 8, 2163. doi: 10.3389/fpls.2017.02163
Li, Y. F., Wu, Y., Hernandez-Espinosa, N., Peña, R. J. (2013). Heat and drought stress on durum wheat: responses of genotypes, yield, and quality parameters. J. Cereal Sci. 57, 398–404. doi: 10.1016/j.jcs.2013.01.005
Luchniak, P., Maluszynska, J., Olszewska, M. J. (2002). In situ fluorescent nick translation procedure for plant chromosomes.. Histochemistry 77, 15–19. doi: 10.1080/bih.77.1.15.19
Magallanes-López, A. M., Ammar, K., Morales-Dorantes, A., González-Santoyo, H., Crossa, J., Guzman, C. (2017). Grain quality traits of commercial durum wheat varieties and their relationships with drought stress and glutenins composition. J. Cereal Sci. 75, 1–9. doi: 10.1016/j.jcs.2017.03.005
Marcussen, T., Sandve, S. R., Heier, L., Spannagl, M., Pfeifer, M., The International Wheat Genome Sequencing Consortium, et al. (2014). Ancient hybridizations among the ancestral genomes of bread wheat. Science 345, 6194. doi: 10.1126/science.1250092
Martin, A., Simpfendorfer, S., Hare, R., Sutherland, M. (2013). Introgression of hexaploid sources of crown rot resistance into durum wheat. Euphytica 192, 463–470. doi: 10.1007/s10681-013-0890-6
Matsuoka, Y., Nasuda, S. (2004). Durum wheat as a candidate for the unknown female progenitor of bread wheat: An empirical study with a highly fertile F1 hybrid with Aegilops tauschii Coss. Theor. Appl. Genet. 109, 1710–1717. doi: 10.1007/s00122-004-1806-6
McFadden, E. S., Sears, E. R. (1944). The artificial synthesis of Triticum spelta. Rec. Genet. Soc. Am. 13, 26–27. doi: 10.4236/as.2014.514154
McIntyre, C. L., Pereira, S., Moran, L. B., Apples, R. (1990). New Secale cereale (rye) DNA derivatives for the detection of rye chromosome segments in wheat. Genome 33, 317–323. doi: 10.1139/g90-094
Naranjo, T., Roca, A., Goicoechea, P. G., Giraldez, R. (1987). Arm homoeology of wheat and rye chromosomes. Genome 29, 873–882. doi: 10.1139/g87-149
Nazco, R., Villegas, D., Ammar, K., Peña, R. J., Moragues, M., Royo, C. (2012). Can Mediterranean durum wheat landraces contribute to improved grain quality attributes in modern cultivars? Euphytica 185, 1–17. doi: 10.1007/s10681-011-0588-6
Padmanaban, S., Sutherland, M. W., Knight, N. L., Martin, A. (2017b). Genome inheritance in populations derived from hexaploid/tetraploid and tetraploid/hexaploid wheat crosses. Mol. Breed. 37, 48. doi: 10.1007/s11032-017-0647-3
Padmanaban, S., Zhang, P., Hare, R. A., Sutherland, M. W., Martin, A. (2017a). Pentaploid Wheat Hybrids: Applications, Characterisation, and Challenges. Front. Plant Sci. 8, 358. doi: 10.3389/fpls.2017.00358
Qi, L. L., Friebe, B., Zhang, P., Gill, B. S. (2007). Homoeologous recombination, chromosome engineering and crop improvement. Chromosome Res. 15, 3–19. doi: 10.1007/s10577-006-1108-8
Ramsey, J., Schemske, D. W. (1998). Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Syst. 29, 467–501. doi: 10.1146/annurev.ecolsys.29.1.467
Rayburn, A. L., Gill, B. S. (1986). Isolation of a D-genome specific repeated DNA sequence from Aegilops squarrosa. Plant. Mol. Biol. Rep. 4, 102–109. doi: 10.1007/BF02732107
Schneider, A., Molnár, I., Molnár-Láng, M. (2008). Utilisation of Aegilops (Goatgrass) species to widen the genetic diversity of cultivated wheat. Euphytica 163, 1–19. doi: 10.1007/s10681-007-9624-y
Schubert, I., Fransz, P. F., Fuchs, J., De Jong, J. H. (2001). Chromosome painting in plants. Methods Cell Sci. 23, 57. doi: 10.1023/A:1013137415093
Schwarzacher, T., Anamthawat-Jónsson, K., Harrison, G. E., Islam, A. K. M. R., Jia, J. Z., King, I. P., et al. (1992). Genomic in situ hybridization to identify alien chromosomes and chromosome segments in wheat. Theor. Appl. Genet. 84, 778–786. doi: 10.1007/BF00227384
Schwarzacher, T., Leitch, A., Bennett, M., Heslop-Harrison, J. (1989). In situ localization of parental genomes in a wide hybrid. Ann. Bot. 64, 315–324. doi: 10.1093/oxfordjournals.aob.a087847
Shewry, P. R., Hey, S. J. (2015). The contribution of wheat to human diet and health. Food Energy Secur. 4, 178–202. doi: 10.1002/fes3.64
Silva, G., Souza, M. (2013). Genomic in situ hybridization in plants. Genet. Mol. Res. 12, 2953–2965. doi: 10.4238/2013.August.12.11
Tang, Z. X., Yang, Z. J., Fu, S. L. (2014). Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J. Appl. Genet. 55, 313–318. doi: 10.1007/s13353-014-0215-z
Thomson, D., Henry, R. (1995). Single-step protocol for preparation of plant tissue for analysis by PCR. BioTechniques 19, 394–397, 400.
Zhang, H., Bian, Y., Gou, X., Zhu, B., Xu, C., Qi, B., et al. (2013). Persistent whole-chromosome aneuploidy is generally associated with nascent allohexaploid wheat. Proc. U.S.A. 110, 3447–3452. doi: 10.1073/pnas.1300153110
Keywords: durum wheat, pentaploid crosses, Amblyopyrum muticum, introgression, in situ hybridization, Kompetitive Allele-Specific PCR markers
Citation: Othmeni M, Grewal S, Hubbart-Edwards S, Yang C, Scholefield D, Ashling S, Yahyaoui A, Gustafson P, Singh PK, King IP and King J (2019) The Use of Pentaploid Crosses for the Introgression of Amblyopyrum muticum and D-Genome Chromosome Segments Into Durum Wheat. Front. Plant Sci. 10:1110. doi: 10.3389/fpls.2019.01110
Received: 14 June 2019; Accepted: 13 August 2019;
Published: 18 September 2019.
Edited by:
Agata Gadaleta, University of Bari Aldo Moro, ItalyReviewed by:
Daryl LaVerne Klindworth, Edward T. Schafer Agricultural Research Center, United StatesAssaf Distelfeld, Tel Aviv University, Israel
Peng Zhang, University of Sydney, Australia
Copyright © 2019 Othmeni, Grewal, Hubbart-Edwards, Yang, Scholefield, Ashling, Yahyaoui, Gustafson, Singh, King and King. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Julie King, SnVsaWUuS2luZ0Bub3R0aW5naGFtLmFjLnVr
 Manel Othmeni
Manel Othmeni Surbhi Grewal
Surbhi Grewal Stella Hubbart-Edwards
Stella Hubbart-Edwards Caiyun Yang
Caiyun Yang Duncan Scholefield
Duncan Scholefield Stephen Ashling
Stephen Ashling Amor Yahyaoui2
Amor Yahyaoui2 Perry Gustafson
Perry Gustafson Pawan K. Singh
Pawan K. Singh Ian P. King
Ian P. King Julie King
Julie King