Abstract
Plant hormones play central roles in plant growth, developmental processes, and plant response to biotic and abiotic stresses. On the one hand, plant hormones may allocate limited resources to the most serious stresses; on the other hand, the crosstalks among multiple plant hormone signaling regulate the balance between plant growth and defense. Many studies have reported the mechanism of crosstalks between jasmonic acid (JA) and other plant hormones in plant growth and stress responses. Based on these studies, this paper mainly reviews the crosstalks between JA and other plant hormone signaling in regulating the balance between plant growth and defense response. The suppressor proteins JASMONATE ZIM DOMAIN PROTEIN (JAZ) and MYC2 as the key components in the crosstalks are also highlighted in the review. We conclude that JA interacts with other hormone signaling pathways [such as auxin, ethylene (ET), abscisic acid (ABA), salicylic acid (SA), brassinosteroids (BRs), and gibberellin (GA)] to regulate plant growth, abiotic stress tolerance, and defense resistance against hemibiotrophic pathogens such as Magnaporthe oryzae and Pseudomonas syringae. Notably, JA may act as a core signal in the phytohormone signaling network.
Introduction
Under multiple environmental stresses, plant hormones allocate limited resources to respond to the most serious stress (Tian et al., 2003; Matyssek et al., 2005) and develop various signaling pathways (Pieterse et al., 2012; Sharma et al., 2013) to regulate the balance between plant growth and defense response (Tian et al., 2003; Matyssek et al., 2005). Understanding the similarities and differences of plant hormone signaling may be important in agricultural production.
Plant hormones are small endogenous signaling molecules, including gibberellin (GA), auxin (indole-3-acetic acid, IAA), cytokinin (CK), brassinosteroids (BRs), abscisic acid (ABA), ethylene (ET), jasmonic acid (JA), salicylic acid (SA), and strigolactone (SL). In recent decades, JA biosynthesis has been widely investigated in monocotyledons and dicotyledons, especially in Arabidopsis. In Arabidopsis, at least two pathways are responsible for JA biosynthesis, namely, the α-linolenic acid (18:3) initial octadecane pathway and hexadecatrienoic acid (16:3) initial hexadecane pathway (Engelberth et al., 2001; Wasternack, 2014; Kazan, 2015). In these pathways, the 18:3 and 16:3 unsaturated fatty acids are converted to 12-oxo-phytodienoic acid (12-OPDA) and deoxymethylated vegetable dienic acid (dn-OPDA) in the chloroplast, respectively. Then JA is formed from 12-OPDA and dn-OPDA through multiple β-oxidation in the peroxisome. Finally, different structures of JAs such as methyl jasmonate (MeJA). JA–isoleucine (JA–Ile) and 12-hydroxyjasmonic acid (12-OH-JA) are formed from JA in the cytoplasm. Among these JAs, JA–Ile is the biological active form of JA in plants (Wasternack and Strnad, 2016).
JA is widely distributed in plants as a natural plant growth regulator (Engelberth et al., 2001; Ghasemi Pirbalouti et al., 2014; Wasternack, 2014; Kazan, 2015; Ahmad et al., 2016; Wasternack and Strnad, 2016). The importance of the crosstalks between JA and other plant hormones in regulating plant stress responses has attracted extensive attention (Kazan, 2015; Ahmad et al., 2016; Wasternack and Strnad, 2016; Wasternack and Song, 2016). In this paper, the role of the crosstalks between JA and other plant hormone signaling in regulating plant stress responses as well as in the balance of plant growth and defense response is reviewed.
Core Components of JA Signaling
SKP1/CULLIN/F-Box (SCF)COI1 Complex
CORONATINE INSENSITIVE 1 (COI1) is assembled into SCF E3 ubiquitin ligase complex SCFCOI1 to be stabilized (Zhang et al., 2015). As an F-box protein and a component of SCFCOI1, COI1 plays an important role in recognizing JA signaling (Xie et al., 1998). In the JA signaling pathway, the F-box protein is recognized and combined with targeted proteins, and then the targeted proteins are degraded by 26S proteasome.
JASMONATE ZIM DOMAIN (JAZ) Protein: Inhibitor of JA Response
JAZ protein contains two conserved protein–protein interaction domains, ZIM and Jas (Thireault et al., 2015). The Jas domain of JAZ protein mediates the interaction between JAZ and COI1 or other transcription factors (TFs) (Gimenez-Ibanez et al., 2015), the ZIM (TIFY) domain of JAZ protein mediates JAZ dimerization and its interaction with NINJA, and NINJA recruits general transcriptional co-suppressor TPL through the conserved EAR domain. Moreover, it competes with MEDIATOR25 (MED25) to interact with MYCs (Zhang F et al., 2015).
MYC TFS
In contrast to the positive regulation of JAZ in plant growth, MYC TFs (MYC2, MYC3, and MYC4) negatively regulate gene expression in cell cycle, thereby inhibiting plant growth (Gasperini et al., 2015; Campos et al., 2016; Major et al., 2017). As the general switch of the JA signaling pathway, MYC2 is the most famous TF. Moreover, it participates in the crosstalks among JA, ABA, auxin, ET, GA, and other signaling pathways (Gangappa et al., 2013).
Roles of SCFCOI1 Complex, MYC2, and JAZ in JA Signaling Pathway
COI1 protein, JAZ, and MYC constitute the core signal transduction mechanism of JA signaling and have been proven to be the intersection of other signal transduction pathways under various stresses (Figure 1). JAZ and various TFs form specific JAZ/TFs that specifically regulate multiple downstream responses (Chini et al., 2016). The JAZ-MYC module increases the concentration of defense compounds to trigger defense response or inhibit plant growth against pathogen infection (Havko et al., 2016). In addition to the JAZ-MYC module, COI1-JAZ2-MYC2, 3,4-ANAC19,55,72 (Gimenez-Ibanez et al., 2017), and other specific JAZ-TF modules have been identified (Jin and Zhu, 2017; Mao et al., 2017). The interaction between MYCs and JAZs may involve other plant hormone signaling pathways such as ET-mediated cell division through ET RESPONSE FACTOR (ERF) TFs (Zhang F et al., 2015; Gimenez-Ibanez et al., 2017; Dubois et al., 2018).
Figure 1
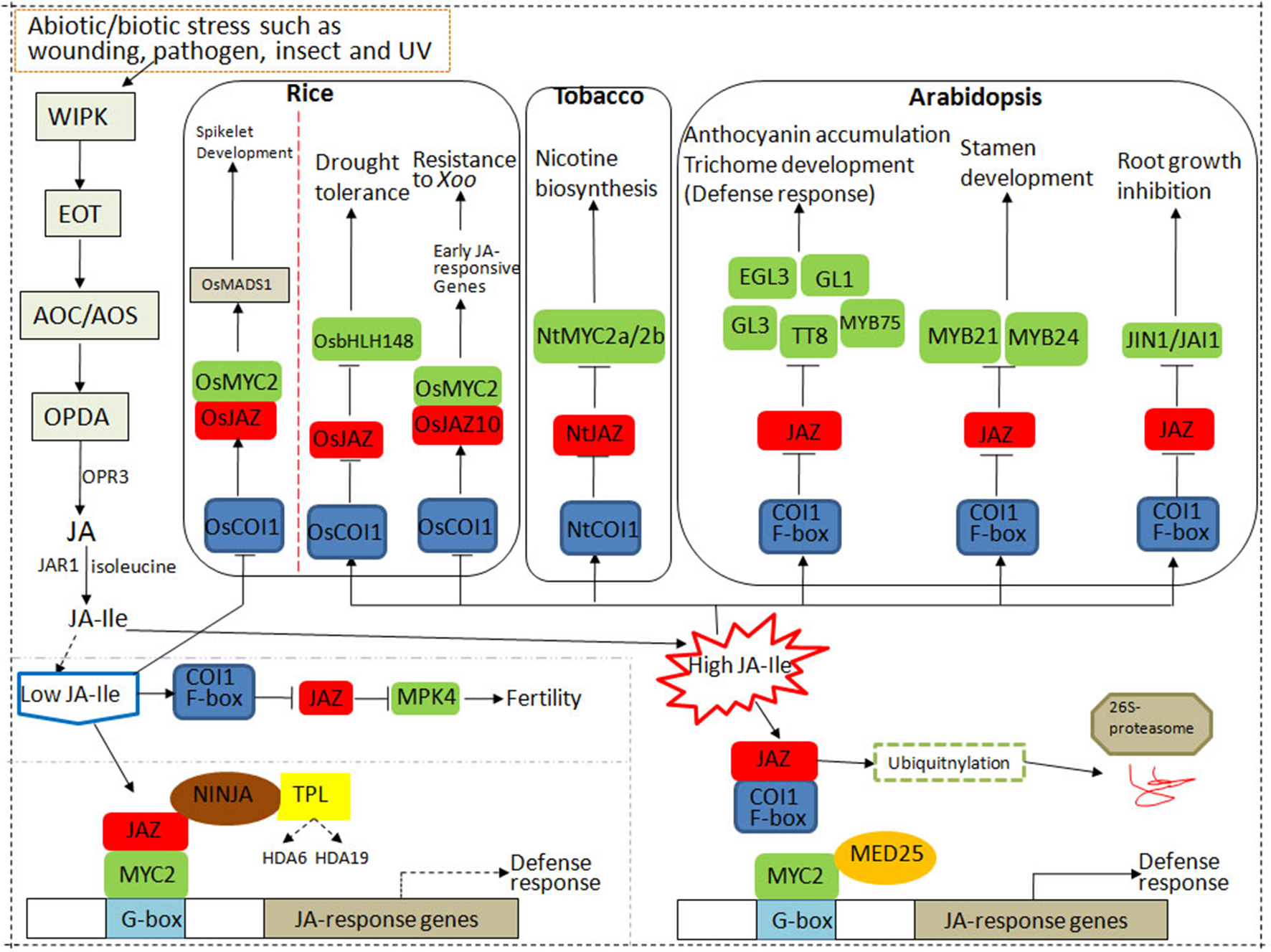
The core components of the jasmonic acid (JA) signaling pathway in rice, tobacco, and Arabidopsis. CORONATINE INSENSITIVE 1 (COI1) protein, JASMONATE ZIM DOMAIN PROTEIN (JAZ), and MYC constitute the core signal transduction mechanism of JA signaling. Under control conditions, the endogenous level of JA–isoleucine (Ile) is very low plants. JAZ repressors bind to MYC2 to inhibit its transcriptional activation on downstream genes. Under stress conditions, the endogenous level of JA–Ile is largely activated, which is perceived by JA receptor COI1. Then SKP1/CULLIN/F-box (SCF)COI1 binds to JAZs for ubiquitination and degradation through the 26S proteasome pathway, resulting in the release of the downstream transcription factors (TFs) such as MYCs and the activation of JA responses.
When JA–Ile content is high in rice, OsCOI1 interacts with OsJAZ to form a complex, JAZ protein is degraded, and the transcription of OsbHLH148 in the JA signal module is rapidly activated to induce drought tolerance. High levels of JA–Ile induce OsMYC2 to selectively bind to the G-box-like motif in the promoter region of OsJAZ10 to activate transcription activity, induce the expression of early JA-responsive genes, and thereby resist bacterial pathogens (Uji et al., 2016).
Generally, the endogenous level of biologically active JA (JA–Ile) is kept very low plants but can be rapidly activated in response to various stresses such as insect feeding or wounding. Then JA signaling is perceived by JA receptor COI1, an important component of SCFCOI1, which binds to JAZs for ubiquitination and degradation through the 26S proteasome pathway. The competitive binding and degradation of JAZ repressors can further release the downstream TFs such as MYCs, resulting in the activation of JA responses by MYCs (Gimenez-Ibanez et al., 2017; Dubois et al., 2018).
To sum up, the three main core components of JA signaling play an important role in plant growth, development, and response to biotic or abiotic stresses. When exogenous JA or extreme stress is applied, the concentration and application time may affect the transcriptional activities of different components in the JA signal module. Therefore, according to the JA metabolic pathway under stress conditions, corresponding stress-tolerance breeding may be developed to improve crop resistance in agriculture.
Crosstalks Between JA and Other Plant Hormone Signaling Pathways
The crosstalk between plant hormones is the core of plant stress response (He et al., 2017). JA does not work independently but acts in a complex signaling network combined with other plant hormone signaling pathways (Kazan, 2015; Ahmad et al., 2016; Wasternack and Strnad, 2016; Hu et al., 2017). As a core component of JA signaling, the role of JAZs-MYC2 in the crosstalks of plant hormone signaling pathways is highlighted in this review, especially in the crosstalks of JA–auxin, JA–ET, JA–ABA, JA–SA, JA–BR, and JA–GA signaling pathways (Pérez and Goossens, 2013; Gimenez-Ibanez et al., 2015; Chini et al., 2016; Goossens et al., 2016) (Figure 2).
Figure 2
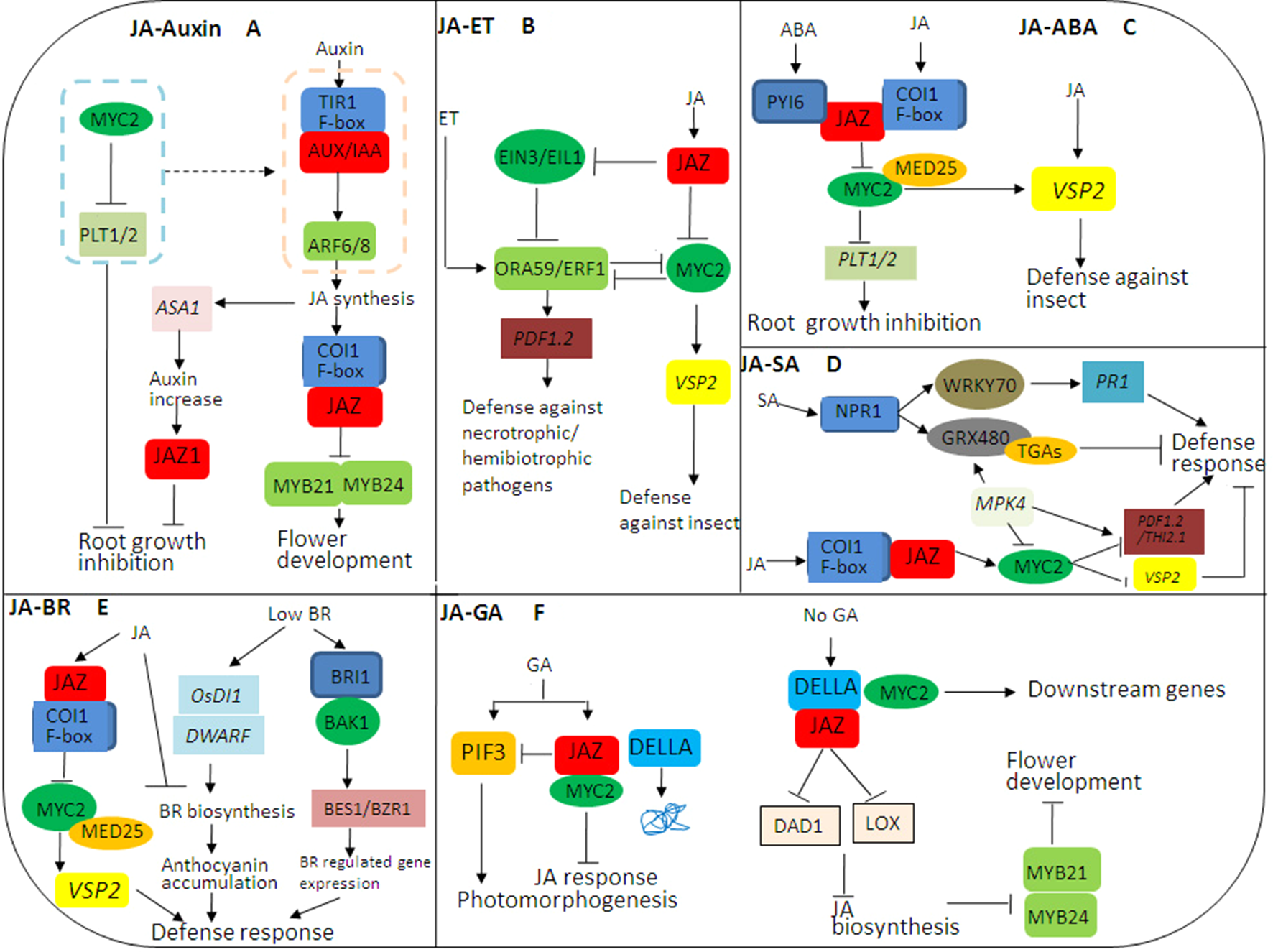
JASMONATE ZIM DOMAIN PROTEIN (JAZ)-mediated crosstalks among jasmonic acid (JA) hormone signaling pathways in plant growth and stress responses. (A) The complex crosstalk between JA and auxin signaling pathways. JA and auxin signaling pathways coordinately regulate flower development through modulation of JA, while JA and auxin antagonize root growth through JAZs-MYC2. (B) The complex crosstalk between JA and ethylene (ET) signaling pathways. JA and ET coordinately regulate plant stress responses through JAZs-MYC2 and EIN3/EIL1, especially in resisting necrotrophic or hemibiotrophic pathogens. (C) The complex crosstalk between JA and abscisic acid (ABA) signaling pathways. The crosstalk between PYRABACTIN RESISTANCE1-Like protein (PYL) and JAZ–MYC2 coordinates the balance between plant growth and defense resistance. (D) The complex crosstalk between JA and SA signaling pathways. SA initiates early defense-related gene expression in pathogen-infected plants, while JA induces late defense-related gene expression in pathogen-infected plants, mainly in the necrotrophic stage of necrotrophic or hemibiotrophic pathogens. (E) The complex crosstalk between JA and brassinosteroids (BR) signaling pathways. The crosstalk between JA and BR biosynthesis may be involved in the balance between plant growth and defense resistance. (F) The complex crosstalk between JA and GA signaling pathways. The JAZ-MYC2-DELLA-PIF signaling module being involved in the crosstalk between JA and GA signaling can be elucidated. In addition, many transcription factors (TFs) such as MYC3, MYC4, MYB21, and MYB24 can also interact with DELLAs, so there may be synergistic effect between JA and GA signaling.
JA–Auxin Crosstalk
JA and auxin signaling pathways coordinately regulate plant growth and development. COI1, MYC2, and JAZ, as the main core components, participate in the crosstalk of JA and auxin signaling pathways (Figure 2A). When plants are induced by exogenous auxin, the auxin–TIR–AUX/IAA–ARF signaling is activated, and JA synthesis is induced. On the one hand, the endogenous JA induces the expression of auxin synthase gene (ASA1) and auxin content, so JA regulates the biosynthesis of auxin and further regulates the expression of JAZ1 and root growth. On the other hand, JA induces the formation of a complex of COI1 and JAZ and leads to the degradation of JAZ, thereby activating the transcriptional activities of MYB21/MYB24 and inducing flower development. Notably, MYC2 inhibits the expressions of PLETHORAs (PTL1 and PTL2) and counteracts the auxin–TIR–AUX/IAA–ARF signaling, so as to regulate root growth (Chen et al., 2011). In addition, ARF6/ARF8 in the auxin signaling pathway regulates petal and stamen growth through modulation of endogenous JA level, and MYB21/MYB24 as downstream of JAZ in JA signaling pathways also coordinately regulate petal and stamen growth (Reeves et al., 2012). Thus, JA and auxin signaling pathways coordinately regulate flower development through modulation of JA, while JA and auxin antagonize root growth through JAZs-MYC2.
JA–ET Crosstalk
JA and ET antagonize or coordinately regulate plant stress response (Zhu, 2014; Zhu and Lee, 2015) (Figure 2B). ET INSENSITIVE3 (EIN3) and its homologue EIN3-like 1 (EIL1) in the ET signaling pathway as well as JAZs-MYC2 in the JA signaling pathway are involved in the crosstalk between JA and ET signaling pathways (Zhang et al., 2014). On the one hand, exogenous JA triggers the degradation of JAZ, and the release of MYC2 regulates the expression of ORA59/ERF1 and wound responsive gene VSP2, so as to resist herbivorous insects. On the other hand, JAZ inhibits the transcriptional activity of EIL2/EIN3 in the ET signaling pathway and activates downstream ORA59/ERF1 that targets the promoter of PLANT DEFENSIN 1.2 (PDF1.2) and induces its expression, thereby resisting the infection of necrotrophic pathogens and hemibiotrophic pathogens (Zhu et al., 2011). Generally, the JA signaling pathway synergistically crosstalks with the ET signaling pathway against necrotrophic pathogen attacks and activates the expression of defense proteins such as PDF1.2 through ERF1 and ORA59. Thus, JA and ET coordinately regulate plant stress responses through JAZs-MYC2 and EIN3/EIL1, especially in resisting necrotrophic or hemibiotrophic pathogens (Pieterse et al., 2012).
JA–ABA Crosstalk
ABA and JA signaling pathways coordinately regulate plant response to herbivorous insect feeding while antagonizing plant growth and development. JAZs-MYC2 participates in the crosstalk between JA and ABA signaling pathways, affecting plant growth and defense (Chen et al., 2011). ABA receptor PYRABACTIN RESISTANCE1-Like proteins (PYLs) regulate metabolic reprogramming in Arabidopsis thaliana and tobacco through the JA signaling pathway. Therefore, the crosstalk between ABA and JA signaling pathways can monitor elicitor-induced reprogramming of plant metabolism and growth (Per et al., 2018). ABA receptor PYL forms a complex with JAZ, which activates the transcriptional activated activity of MYC2. On the one hand, MYC2 activates the expression of JA responsive gene VSP2 under the mediation of MED25 to resist herbivorous insect feeding. On the other hand, MYC2 inhibits the expressions of PTL1 and PTL2 as well as root growth. Additionally, ABA initiates the degradation of JAZ12, which plays a specific role in the crosstalk between JA and ABA signaling pathways (Pauwels et al., 2015). Thus, the crosstalk between JA and ABA signaling, especially between PYL and JAZ-MYC2, coordinates the balance between plant growth and defense resistance (Figure 2C).
JA–SA Crosstalk
Generally, JA is widely involved in regulating disease resistance against necrotrophic pathogens, while SA mediates broad-spectrum resistance against biotrophic and hemibiotrophic pathogens (Fu et al., 2012). It has been shown that JA signaling can inhibit SA accumulation through modulation of multiple NAC TFs, such as ANAC019/055/072. Briefly, MYC2 directly binds to the promoters of these NACs and then activates their transcription. Then the activation of these NAC TFs further inhibits the expression of ISOCHORISMATE SYNTHASE 1 (ICS1) as an SA biosynthesis gene while triggering the expression of BENZOIC ACID/SA CARBOXYL METHYLTRANSFERASE 1 (BSMT1) as an SA methylation gene (Zheng et al., 2012). In addition, the crosstalk between JA and SA signaling pathways involves many components, including mitogen-activated protein kinase (MAPK) (Rodriguez et al., 2010); redox regulators glutathione (GRX) and thioredoxin (TRX) (Spoel and Loake, 2011), MYC2, TGAs, and PDF 1.2 (Gatz, 2013); and WRKY70 (Shim et al., 2013). In the presence of exogenous SA, NONEXPRESSOR OF PR GENES1 (NPR1) is activated to induce the transcriptional-activated activity of WRKY70, which promotes the expression of PR1 by binding to the promoter region of PR1 and inducing defense response. In the meanwhile, NPR1 polymers are monomerized by TRX through SA-induced redox state changes, and then monomers such as GRX480 are transported to the nucleus and specifically bind to TGAs, which also directly regulate the expression of PR1 (Leon-Reyes et al., 2009; Spoel et al., 2009; Fu et al., 2012; Zander et al., 2012; Gatz, 2013) (Figure 2D). Thus, the transformation between NPR1 polymer and monomer has a dual role in inhibiting and activating defense-related gene expression (Wasternack and Hause, 2013). Interestingly, the induction of GRXs can block TGA-mediated JA response gene expression, such as ORA59, further confirming SA–JA antagonism (Zander et al., 2012). MPK4 positively regulates GRX480 in the SA signaling pathway and negatively regulates MYC2 in the JA signaling pathway, which is necessary for JA responsive genes (PDF1.2 and THI2.1) (Wasternack and Hause, 2013). Therefore, MYC2 and its upstream MPK4 are involved in the crosstalk between JA and SA signaling pathways, which coordinately regulate plant disease resistance against necrotrophic or hemibiotrophic pathogens. SA initiates early defense-related gene expression in infected plants, while JA induces late defense-related gene expression in infected plants, mainly in the necrotrophic stage of necrotrophic or hemibiotrophic pathogens.
JA–BR Crosstalk
JA inhibits plant growth, while BR induces above-ground plant growth. The crosstalk between JA and BR signaling pathways is involved in the balance between plant growth and defense resistance. On the one hand, low concentration of BR induces the expression of OsDI1 and OsDWARF at the early and late stages of BR biosynthesis, respectively, and anthocyanin accumulation and activates defense response. On the other hand, high concentration of BR activates BR signaling cascades including BR receptor BRI1, BR-related kinase BAK1, and BR-related TFs to induce the expressions of downstream genes such as BES1 and BZR1, thereby regulating plant responses to abiotic stresses (Figure 2E). JA induces JAZ to bind COI1, and MYC2 activates the expression of VSP2 under the mediation of MED2, thereby resisting herbivorous insect feeding (Figure 2E). Notably, high concentration of BR inhibits endogenous biosynthesis of JA and BR, and JA also inhibits BR biosynthesis (Choudhary et al., 2012). Thus, the crosstalk between JA and BR biosynthesis may be involved in the balance between plant growth and defense resistance.
JA–GA Crosstalk
JA and GA signaling pathways coordinately and antagonistically regulate plant growth and defense response; however, plant defense response is exerted at the cost of inhibiting growth (Yang et al., 2012; Wasternack and Hause, 2013). The C-terminus of JAZs is necessary for the interaction between JAZs and MYC2 and between JAZs and DELLAs, so DELLAs can completely interact with JAZs (Hou et al., 2010). In the absence of GA, stable DELLA interacts with JAZ to release MYC2, resulting in the activation of MYC2 downstream genes. At the same time, DELLA interacts with JAZ to inhibit the expression of JA biosynthetic genes (DAD1 and LOX) and further inhibits JA biosynthesis as well as the activities of MYB21 and MYB24 (Figure 2F), thereby regulating stamen development (Song et al., 2011). GA induces the degradation of DELLA and the binding of JAZ to MYC2, so as to inhibit JA signaling. In addition, GA induces PIF3/PIF4 to regulate photomorphogenesis (Figure 2F) (Hou et al., 2013). Notably, JA delays GA-mediated degradation of DELLA, the della mutant is less sensitive to JA-inhibited plant growth inhibition, and AtJAZ9 inhibits the interaction between DELLA and PIF3 (Yang et al., 2012). Therefore, the molecular cascade involving the JAZ-MYC2-DELLA-PIF signaling module in the crosstalk between JA and GA signaling pathways can be elucidated. Moreover, many TFs such as MYC3, MYC4, MYB21, and MYB24 can also interact with DELLAs, so there may be a synergistic effect between JA and GA signaling pathways (Lyons et al., 2013).
JA Regulates Plant Response to Biotic/Abiotic Stresses
The role of JA in plant response to abiotic stresses has been extensively reported (Khan et al., 2012; Kazan, 2015; Ahmad et al., 2016; Sharma and Laxmi, 2016; Wasternack and Strnad, 2016; Per et al., 2018), such as heavy metals (Maksymiec et al., 2005), drought (Brossa et al., 2011), salt (Qiu et al., 2014; Zhao et al., 2014), heat (Clarke et al., 2009), and UV radiation (Liu et al., 2012). The role of JA in plant response to biotic stress is mainly to induce plant disease response against pathogen or insect (Campos et al., 2014). Under pathogen infection conditions, JA and JA–Ile can be rapidly induced (Chauvin et al., 2013; Fragoso et al., 2014), which induces almost all major secondary metabolites and protein expression involved in defense response, including alkaloids, terpenoids, phenylpropane, amino acid derivatives, anti-nutritional proteins, and some pathogen-related proteins (De et al., 2012; De et al., 2013). In this paper, the effects of JA on plant response to abiotic/biotic stresses including cold stress, drought stress, and fungal and bacterial diseases are discussed.
JA and Cold Stress
In order to adapt to extremely low temperatures, plants have evolved complex mechanisms by regulating physiological and biochemical processes, especially the modulation of stress-related gene expression. The INDUCER OF CBF EXPRESSION (ICE)-CBF transcriptional cascade signaling pathway plays a central role in plant cold stress response (Hu et al., 2017). ICE1 and ICE2, two basic helix–loop–helix (bHLH) TFs in A. thaliana, upregulate the expressions of CBFs through directly binding to CANNTG in the promoter region of CBFs. Rice grows at normal temperature. Under control conditions, JAZ1 and JAZ4 interact with ICE1 and ICE2 to inhibit the ICE-CBF signaling pathway (Figure 3). Under low-temperature conditions, the expressions of JA synthesis-related genes including allene oxide synthase1 (AOS1), DAD1, allene oxide cyclase (AOC), LOX2, and AOS1 are induced (Zhu, 2016; Hu et al., 2017), and bioactive JA–Ile is synthesized, thereby activating JA receptor COI1 to bind to JAZ1, resulting in the degradation of JAZ1 through 26S proteome after ubiquitination (Figure 3). Then, the ICE-CBF transcriptional regulation cascade signaling pathway is activated, and the expressions of cold-regulated genes are induced to improve plant cold tolerance. Moreover, JA–ABA crosstalk (Wang et al., 2016) and JA–GA crosstalk in plant response to cold stress have been extensively studied (Achard et al., 2008).
Figure 3
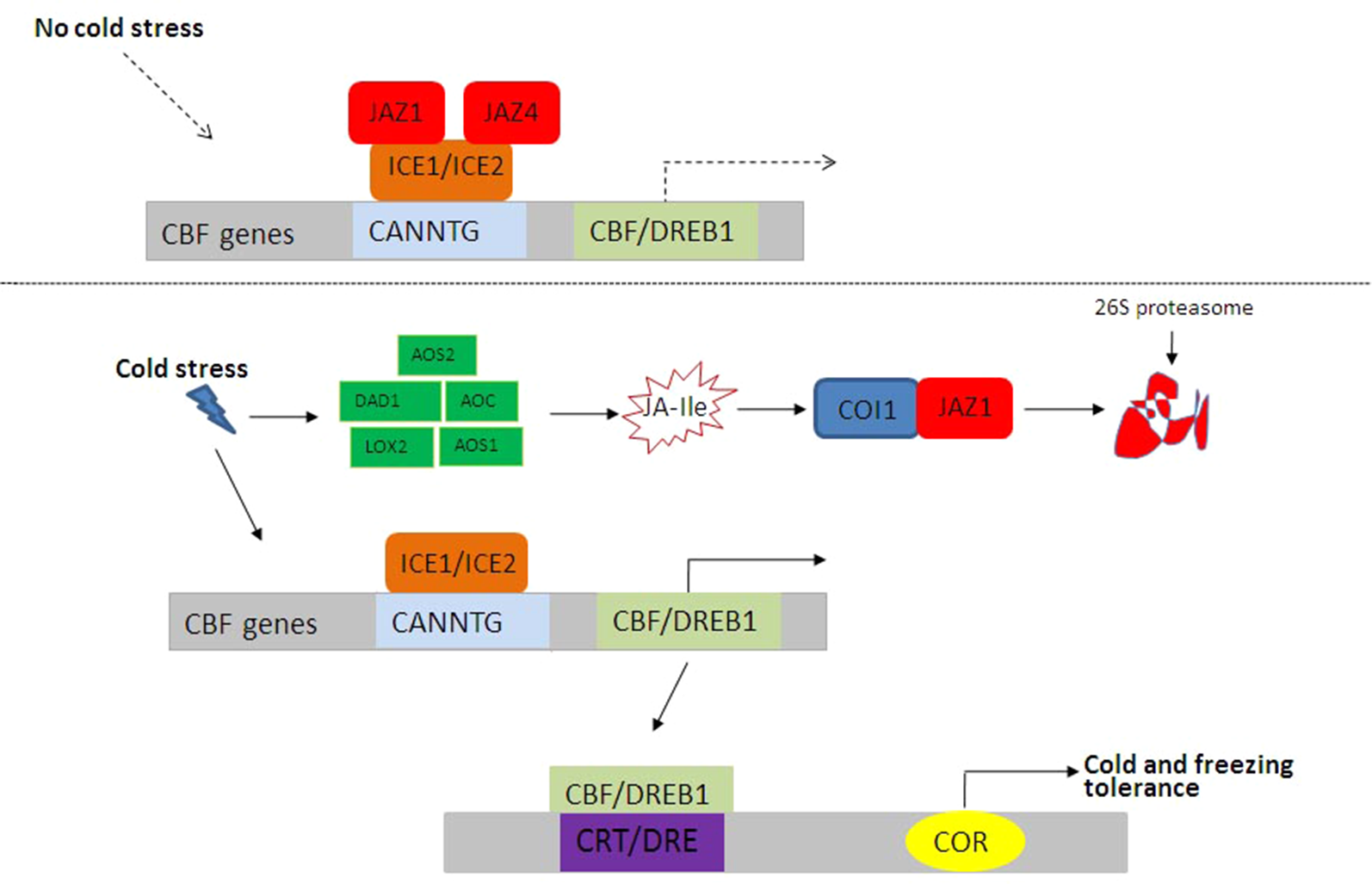
JA-mediated cold and freezing stress responses in plants. Under control conditions, JAZ1 and JAZ4 interact with ICE1 and ICE2 to inhibit the ICE–CBF signaling pathway. Under low-temperature conditions, bioactive JA–Ile and ICE–CBF pathways are activated, and the expressions of cold-regulated genes are induced to improve plant cold tolerance. CBF, C repeat binding factor; ICE, INDUCER OF CBF EXPRESSION; Ile, isoleucine; JA, jasmonic acid; JAZ, JASMONATE ZIM DOMAIN PROTEIN.
JA and Drought Stress
Drought stress response is a complex process in plants. Stomatal closure can reduce water loss and is a potential drought resistance mechanism of plants (Acharya and Assmann, 2009). JA and JA precursor 12-OPDA can promote stomatal closure in A. thaliana, and the increase of OPDA content is related to the decrease of stomatal aperture and improved drought resistance (Savchenko et al., 2014). 13-Lipoxygenase LOX6 is essential for the synthesis of 12-OPDA and plays an important role in plant drought tolerance. When drought stress is applied, LOX6 is synthesized to 12-OPDA, which promotes stomatal closure and improves drought tolerance in the presence or absence of ABA (Figure 4). In response to drought stress, the JA signaling pathway is activated, and OsbHLH148 interacts with OsJAZ1 to activate the expression of OsDREB1, thereby improving drought tolerance in rice (Figure 4) (Sarwat and Tuteja, 2017). Moreover, some antioxidant enzymes, including superoxide dismutase (SOD), peroxidase (POD), catalase (CAT), proline, and relative water content (RWC), are increased to enhance the ability of plants to cope with drought stress (Sarwat and Tuteja, 2017).
Figure 4
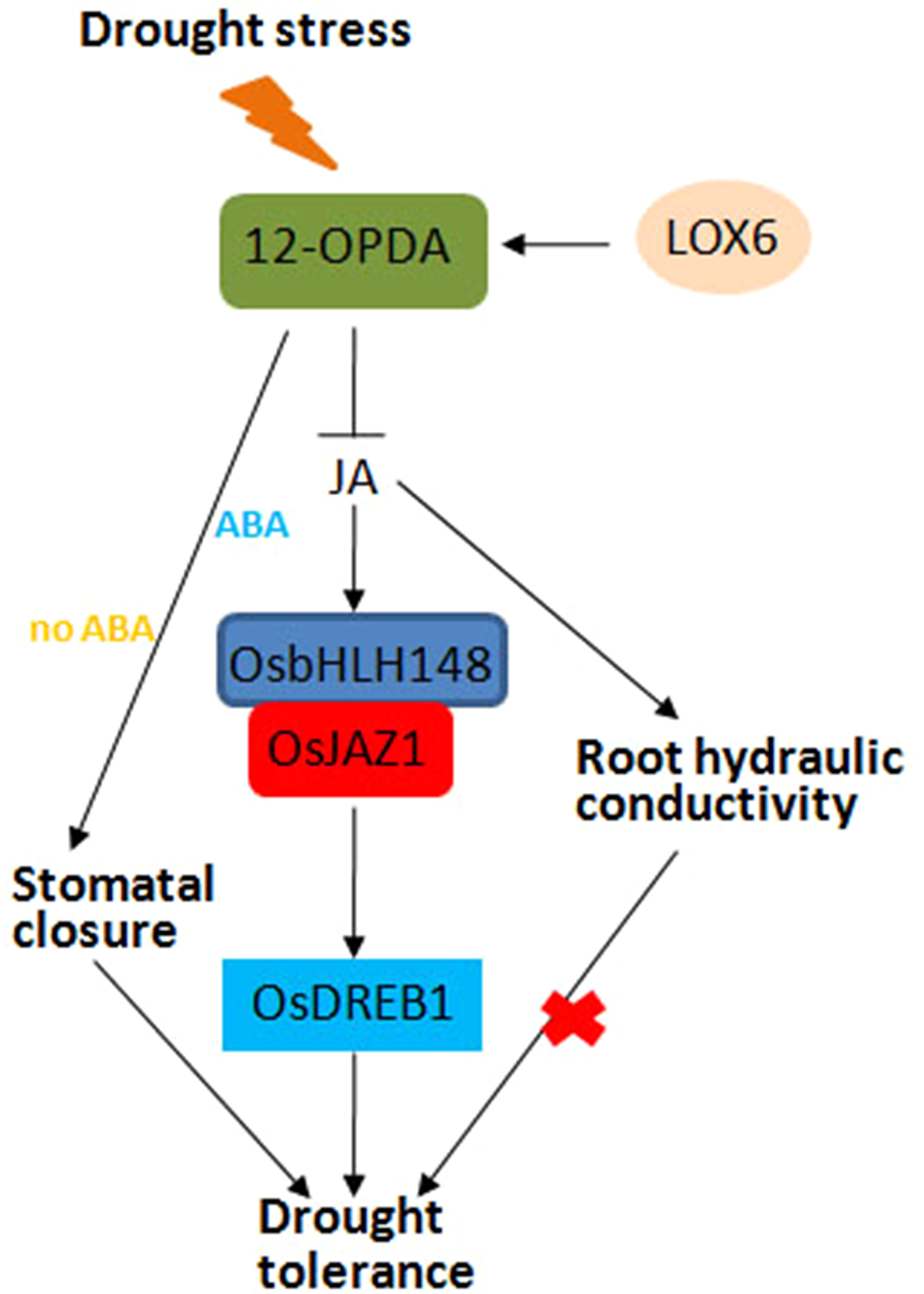
Jasmonic acid (JA)-mediated drought stress response in plants. In response to drought stress, the JA signaling pathway is activated; OsbHLH148 interacts with OsJAZ1 to activate the expression of OsDREB1, together with JA-mediated root hydraulic conductivity and stomatal closure, thereby improving drought tolerance in rice.
JA and Fungal Diseases
Hemibiotrophic and necrotrophic fungi have a wide host range, resulting to serious yield losses in many important crops (Mengiste, 2012; Pandey et al., 2016). JA plays an important role in inducing plant against necrotrophic and hemibiotrophic pathogen and herbivorous insect feeding (Hu et al., 2013; Okada et al., 2015; Pandey et al., 2016). Some hemibiotrophic fungi can metabolize JA produced by host plants. The antibiotic biosynthetic monooxygenase (Abm) formed by Magnaporthe grisea can convert JA into 12-OH-JA to weaken JA signaling and promote the colonization of Magnaporthe oryzae (Patkar et al., 2015; Zhang et al., 2017). When rice blast fungus is compatible with rice, it secretes Abm and inhibits JA activity and immune response (Figure 5A). When rice blast fungus and rice are incompatible, Abm secreted by rice blast fungus is degraded, resulting in the accumulation of MeJA and the activation of JA downstream response as well as immune response (Figure 5A). So far, the JA–MYC2–PDF1.2 module is widely involved in plant–fungi interaction (Zhang et al., 2017). With the effect on promoting pathogenesis, the expression of LOB DOMAIN-CONTAINING PROTEIN 20 (LBD20) is closely related to VSP2 and THIONIN 2.1 (Thi2.1) as well as MYC2 (Thireault et al., 2015). MED19a is an important member of the mediator co-activator complex in JA signaling, and it can be degraded by an oomycete effector protein HaRxL44 (Caillaud et al., 2013). We have to note that the underlying mechanism of the JA signaling pathway in plant–fungi interaction remains elusive and needs to be further investigated.
Figure 5
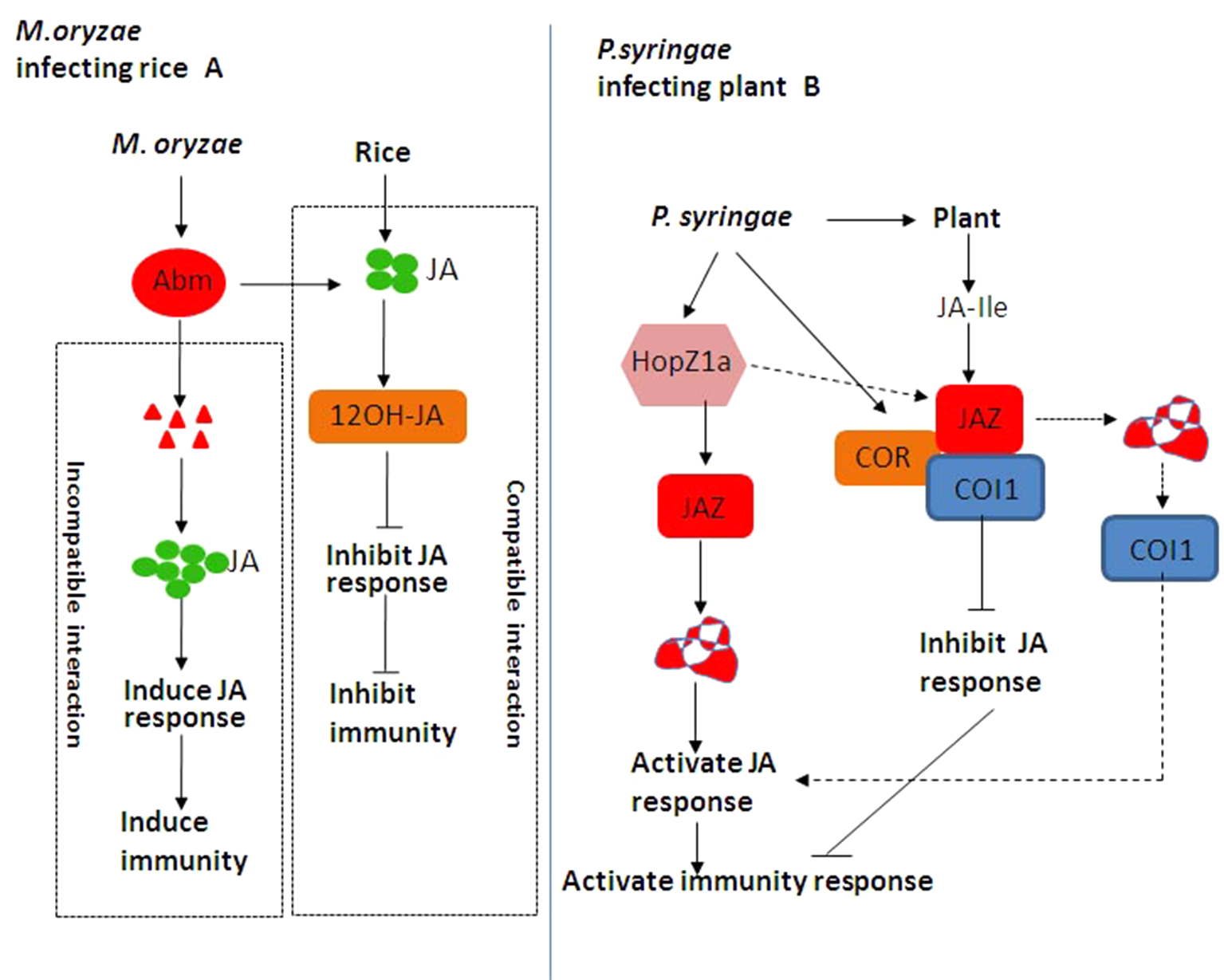
(A) Jasmonic acid (JA)-mediated disease resistance against Magnaporthe oryzae in rice. When rice blast fungus is compatible with rice, rice blast fungus secretes antibiotic biosynthetic monooxygenase (Abm) and inhibits JA activity and immune response. When rice blast fungus and rice are incompatible, Abm secreted by rice blast fungus is degraded, resulting in the accumulation of methyl jasmonate (MeJA) and the activation of JA downstream response as well as immune response. (B) JA-mediated disease resistance against Pseudomonas syringae in Arabidopsis. HopZ1a directly interacts with JASMONATE ZIM DOMAIN PROTEIN (JAZ) proteins and induces the acetylation of JAZ proteins, thereby activating the JA signaling pathway (Jiang et al., 2013). As one kind of functional JA analog, coronatine (COR) can induce CORONATINE INSENSITIVE 1 (COI1) to bind to JAZ proteins, thereby activating JA downstream response and plant immune response (Zhang F et al., 2015).
JA and Bacterial Diseases
Many pathogenic variants of hemibiotrophic bacteria Pseudomonas syringae can produce polyketide toxin coronatine (COR), AvrB, and HopZ1a. The most widely known example of JA-mediated plant–pathogen interaction is regulated by COR. COR is an active substance similar to JA–Ile in structure and function, with two moieties including coronamic acid and coronafacic acid (Bender et al., 1999). On the one hand, COR can promote bacterial infection through the modulation of pathogen-associated molecular pattern-triggered immunity (PTI)-activated stomatal closure and defense response (Sheard et al., 2010; Zhang L et al., 2015). On the other hand, COR can directly bind to the COI1–JAZ complex, and the activation of COR-mediated JA signaling pathway inhibits SA-mediated plant defense resistance against P. syringae infection (Zeng and He, 2010; Zhang L et al., 2015). In addition, COR has toxic functions by regulating secondary metabolites and inhibiting callose formation, which is independent of plant hormone antagonism (Millet et al., 2010; Geng et al., 2012; Yi et al., 2014). Therefore, JA–Ile mimics such as COR may be essential for the infection of some bacterial pathogens. COR can also enhance the interaction between COI1 and JAZ proteins (Zheng et al., 2012; Zhou et al., 2015; Zhang L et al., 2015).
AvrB regulates JA signaling through modulation of the COI1-dependent manner in Arabidopsis (He et al., 2004). In this case, the Arabidopsis protein RPM1-INTERACTING PROTEIN 4 (RIN4) appears to be involved (Cui et al., 2010; Zhou et al., 2015). AvrB interacts with RIN4 and triggers the plasma membrane-localized AHA1. Both AvrB and AHA1 promote the interaction between COI1 and JAZ, thereby regulating stomatal opening and plant defense response (Zhou et al., 2015). Unlike COR and AvrB, as an acetyl transferase, HopZ1a directly interacts with JAZ proteins and induces the acetylation of JAZ proteins, thereby activating the JA signaling pathway (Jiang et al., 2013) (Figure 5B).
Role of Crosstalks Between JA and Other Plant Hormones in Plant Growth and Defense Balance
The crosstalks between plant hormone signaling pathways promote the balance between plant growth and defense (Huot et al., 2014). In order to survive and reproduce, plants should not only maintain growth but also resist pathogen infection. Therefore, the balance between plant growth and defense resistance has important ecological, agricultural, and economic values. The JAZ-MYC module in the JA signaling pathway plays a central role in the balance by integrating TF complexes and plant metabolic pathways (Guo et al., 2018).
When plants are infected by pathogens, PTI as the first defense system of plants, is activated rapidly, followed by SA, JA, and other plant hormone signaling pathways. In the meanwhile, auxin, BR, and GA signaling pathways related to plant growth are inhibited (Boller and He, 2009; Dou and Zhou, 2012). The changes in the amount and composition of stress-related hormones promote plant defense response (Denancé et al., 2013; Xie et al., 2016). When plants are subjected to biotic stress, the transient PTI response and the subsequent SA, JA, GA, BR, and other plant hormone signaling pathways have a certain persistence (Pieterse et al., 2012). SA signaling is mainly involved in disease resistance against biotrophic pathogens, while JA signaling is mainly involved in disease resistance against necrotrophic pathogens or the necrotrophic stage of hemibiotrophic pathogens (Spoel et al., 2007). The crosstalk between GA and JA signaling pathways plays a major role in balancing plant growth and defense against biotic and abiotic stresses (Hou et al., 2010; Wild et al., 2012; Yang et al., 2012; Heinrich et al., 2013; Hou et al., 2013; Daviere and Achard, 2016). GA regulates many aspects of plant growth, and JA plays a major role in stress response. JA and BR coordinately regulate plant environmental stresses, while JA and BR antagonize plant growth (Checker et al., 2018). BR negatively regulates PTI response, because the inhibition of PTI-induced gene expression may lead to the decrease of BR biosynthesis (Jiménez-Góngora Tamara et al., 2015). In summary, the balance between plant growth and defense disease depends on the crosstalks between PTI, JA, SA, BR, and GA signaling pathways.
Prospect
In nature, plants are often subjected to various biotic and abiotic stresses. In response to these stresses, plants initiate a series of defense responses, PTI and effector-triggered immunity (ETI); among them are main defense responses (Katagiri and Tsuda, 2010). The plant hormone signaling network also plays an important role in the early regulation of plant defense response as well as plant–pathogen interaction. Hemibiotrophic fungi have a biotrophic stage, biotrophy-to-necrotrophy switch, and necrotrophic stage; the infected strategies are different at different stages, so it is necessary to further investigate whether plant hormones play the same or different roles at the whole stage of infection (Bhadauria et al., 2013; Chowdhury et al., 2017). Because plant hormones are natural and nontoxic in plants and the crosstalks among them regulate the balance between plant growth and defense resistance, it may have a broad application prospect to develop plant hormones as safe and environment-friendly elicitors by utilizing the crosstalks between plant hormone signaling pathways.
In recent decades, although the JA signaling pathway has been extensively investigated, the current understanding of its role in different environmental stresses is limited, due to the complex networks and crosstalk between multiple stresses and multiple signaling pathways. So far, the molecular mechanism of JA signaling in stress responses remains elusive. Compared with unidentified components, the identified components in plant hormone signaling pathways are limited. In addition, so many receptors and kinases exist in the cell membrane, and different environmental stresses may activate multiple enzymes, with a series of activation of secondary messengers such as Ca2+ and the reaction of kinase-TF-downstream genes. At present, there are still a lot of questions or gaps in understanding the crosstalks between JA and other hormones in plant stress responses, especially in the perception of multiple environmental signals. With the development of protein interaction omics, the complex protein interaction network may provide more clues to the understanding of complex stress signaling perception and protein complex-mediated plant hormone crosstalks.
The plant hormone signaling network is complex and changeable. Although “omics” methods have achieved global analyses of gene expression and protein spectrum changes to some extent, the data still fail to fully understand the dynamic spatial and temporal processes of plant hormone signaling networks in the balance between plant growth and defense resistance (Huot et al., 2014). Moreover, data in the lab may be largely different from those in the field, providing limited information for agriculture production. Therefore, it is necessary to comprehensively analyze plant hormone signaling networks during the whole developmental stages in the field; this will provide more values for crop breeding in the future. In addition, there are many questions that have remained unclear so far. For example, how do plant hormones regulate various plant responses to multiple environmental stress at the same time? How can plant hormones be effectively used to improve crop quality and stress tolerance in agricultural production? Therefore, we have to notice that the current understanding remains limited compared with unknown questions; further investigation will provide a novel insight into developing plant hormones for agricultural production by improving stress resistance and crop quality.
Funding
The work was supported by grants from the national Key Research and Development Program of China (2016YFD100600) and NSFC (31860483 and 31400073) and Program for Innovative Research Team (in Science and Technology, IRTSTYN) in the University of Yunnan Province and State Key Laboratory for Conservation and Utilization of Bio-Resources in Yunnan at Yunnan Agricultural University.
Statements
Author contributions
JY, GD, CL, LL, GH, YZ, and CW conceived the topic. GD, CL, and LL collected the data about jasmonate acid and crosstalking of JA and other plant hormones. GH, YZ, and CW collected the data about abiotic and biotic stresses. JY performed the network analysis and worked with other co-authors to complete the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1
AchardP.RenouJ. P.BerthomeR.HarberdN. P.GenschikP. (2008). Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr. Biol.18, 656–660. doi: 10.1016/j.cub.2008.04.034
2
AcharyaB. R.AssmannS. M. (2009). Hormone interactions in stomatal function. Plant Mol. Biol.69 (4), 451–462. doi: 10.1007/s11103-008-9427-0
3
AhmadP.RasoolS.GulA.SheikhS. A.AkramN. A.AshrafM.et al. (2016). Jasmonates: multifunctional roles in stress tolerance. Front. Plant Sci.7, 813. doi: 10.3389/fpls.2016.00813
4
BenderC. L.Alarcón-ChaidezF.GrossD. C. (1999). Pseudomonas syringae phytotoxins: mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol. Mol. Biol. Rev.63 (2), 266–292. doi: 1092-2172/99/$04.00+0
5
BhadauriaV.BannizaS.VandenbergA.SelvarajG.WeiY. (2013). Overexpression of a novel biotrophy-specific colletotrichum truncatum effector, ctnudix, in hemibiotrophic fungal phytopathogens causes incompatibility with their host plants. Eukaryot. Cell12 (1), 2–11. doi: 10.1128/EC.00192-12
6
BollerT.HeS. Y. (2009). Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science324 (5928), 742–744. doi: 10.1126/science.1171647
7
BrossaR.López-CarbonellMartaJubany-MaríT.AlegreL. (2011). Interplay between abscisic acid and jasmonic acid and its role in water-oxidative stress in wild-type, aba-deficient, ja-deficient, and ascorbate-deficient Arabidopsis plants. J. Plant Growth Regul.30 (3), 322–333. doi: 10.1007/s00344-011-9194-z
8
CaillaudM. C.AsaiS.RallapalliG.PiquerezS.FabroG.JonesJ. D. (2013). A downy mildew effector attenuates salicylic acid-triggered immunity in Arabidopsis by interacting with the host mediator complex. PLoS Biol.11 (12), e1001732. doi: 10.1371/journal.pbio.1001732
9
CamposM. L.KangJ. H.HoweG. A. (2014). Jasmonate-triggered plant immunity. J. Chem. Ecol.40 (7), 657–675. doi: 10.1007/s10886-014-0468-3
10
CamposM. L.YoshidaY.MajorI. T.DaltonD. O. F.WeraduwageS. M.FroehlichJ. E.et al. (2016). Rewiring of jasmonate and phytochrome b signalling uncouples plant growth-defense tradeoffs. Nat. Commun.7, 12570. doi: 10.1038/ncomms12570
11
ChauvinA.CaldelariD.WolfenderJ. L.FarmerE. E. (2013). Four 13-lipoxygenases contribute to rapid jasmonate synthesis in wounded Arabidopsis thaliana leaves: a role for lipoxygenase 6 in responses to long-distance wound signals. New Phytol.197 (2), 566–575. doi: 10.1111/nph.12029
12
CheckerV. G.KushwahaH. R.KumariP.YadavS. (2018). “Role of phytohormones in plant defense: signaling and cross talk,” in Molecular Aspects of Plant-Pathogen Interaction (Singapore: Springer), 159–184. doi: 10.1007/978-981-10-7371-7_7
13
ChenQ.SunJ.ZhaiQ.ZhouW.QiL.XuL.et al. (2011). The basic helix-loop-helix transcription factor myc2 directly represses plethora expression during jasmonate-mediated modulation of the root stem cell niche in arabidopsis. Plant Cell23 (9), 3335–3352. doi: 10.1105/tpc.111.089870
14
ChiniA.GimenezibanezS.GoossensA.SolanoR. (2016). Redundancy and specificity in jasmonate signalling. Curr. Opin. Plant Biol.33, 147–156. doi: 10.1016/j.pbi.2016.07.005
15
ChoudharyS. P.YuJ. Q.Yamaguchi-ShinozakiK.ShinozakiK.TranL. S. (2012). Benefits of brassinosteroid crosstalk. Trends Plant Sci.17 (10), 594–605. doi: 10.1016/j.tplants.2012.05.012
16
ChowdhuryS.BasuA.KunduS. (2017). Biotrophy-necrotrophy switch in pathogen evoke differential response in resistant and susceptible sesame involving multiple signaling pathways at different phases. Sci. Rep.7 (1), 17251. doi: 10.1038/s41598-017-17248-7
17
ClarkeS. M.CristescuS. M.MierschO.HarrenF. J.WasternackC.MurL. A. (2009). Jasmonates act with salicylic acid to confer basal thermotolerance in Arabidopsis thaliana. New Phytol.182 (1), 175–187. doi: 10.1111/j.1469-8137.2008.02735.x
18
CuiH.WangY.XueL.ChuJ.YanC.FuJ.et al. (2010). Pseudomonas syringae effector protein AvrB pertubs Arabidopsis hormone signaling by activating MAP kinase 4. Cell Host Microb.7, 164-175. doi: 10.1016/j.chom.2010.01.009
19
DaviereJ. M.AchardP. (2016). A pivotal role of DELLAs in regulating multiple hormone signals. Mol. Plant9 (1), 10–20. doi: 10.1016/j.molp.2015.09.011
20
DeGeyter N.GholamiA.GoormachtigS.GoossensA. (2012). Transcriptional machineries in jasmonate-elicited plant secondary metabolism. Trends Plant Sci.17 (6), 349–359. doi: 10.1016/j.tplants.2012.03.001
21
DeVleesschauwer. D.GheysenG.HöfteM. (2013). Hormone defense networking in rice: tales from a different world. Trends Plant Sci.18 (10), 555–565. doi: 10.1016/j.tplants.2013.07.002
22
DenancéN.Sánchez-ValletA.GoffnerD.MolinaA. (2013). Disease resistance or growth: the role of plant hormones in balancing immune responses and fitness costs. Front. Plant Sci.4, 155. doi: 10.3389/fpls.2013.00155
23
DouD.ZhouJ. M. (2012). Phytopathogen effectors subverting host immunity: different foes, similar battleground. Cell Host Microbe12 (4), 484–495. doi: 10.1016/j.chom.2012.09.003
24
DuboisM.Van den BroeckL.InzéD. (2018). The pivotal role of ethylene in plant growth. Trends Plant Sci.23 (4), 311–323. doi: 10.1016/j.tplants.2018.01.003
25
EngelberthJ.KochT.SchülerG.BachmannN.RechtenbachJ.BolandW. (2001). Ion channel-forming alamethicin is a potent elicitor of volatile biosynthesis and tendril coiling. Cross talk between jasmonate and salicylate signaling in lima bean. Plant Physiol.125 (1), 369–377. doi: 10.1104/pp.125.1.369
26
FragosoV.RotheE.BaldwinI. T.KimS. G. (2014). Root jasmonic acid synthesis and perception regulate folivore-induced shoot metabolites and increase Nicotiana attenuata resistance. New Phytol.202 (4), 1335–1345. doi: 10.1111/nph.12747
27
FuZ. Q.YanS.SalehA.WangW.RubleJ.OkaN.et al. (2012). Npr3 and npr4 are receptors for the immune signal salicylic acid in plants. Nature486 (7402), 3153. doi: 10.1038/nature11162
28
GangappaS. N.SrivastavaA. K.MauryaJ. P.RamH.ChattopadhyayS. (2013). Z-box binding transcription factors (zbfs): a new class of transcription factors in arabidopsis seedling development. Mol. Plant.6 (6), 1758–1768. doi: 10.1093/mp/sst140
29
GasperiniD.ChételatA.AcostaI. F.GoossensJ.PauwelsL.GoossensA.et al. (2015). Multilayered organization of jasmonate signalling in the regulation of root growth. PLoS Genet.11 (6), e1005300. doi: 10.1371/journal.pgen.1005300
30
GatzC. (2013). From pioneers to team players: TGA transcription factors provide a molecular link between different stress pathways. Mol. Plant-Microbe Interact.26 (2), 151–159. doi: 10.1094/MPMI-04-12-0078-IA
31
GengX.ChengJ.GangadharanA.MackeyD. (2012). The coronatine toxin of Pseudomonas syringae is a multifunctional suppressor of Arabidopsis defense. Plant Cell24 (11), 4763–4774. doi: 10.1105/tpc.112.105312
32
Ghasemi PirbaloutiA.SajjadiS. E.ParangK. (2014). A review (research and patents) on jasmonic acid and its derivatives. Arch. Pharm.347 (4), 229–239. doi: 10.1002/ardp.201300287
33
Gimenez-IbanezS.BoterM.OrtigosaA.García-CasadoG.SolanoR. (2015). Novel players fine-tune plant trade-offs. Essays Biochem.58, 83. doi: 10.1042/bse0580083
34
Gimenez-IbanezS.BoterM.ChiniA.LewseyM. G.et al. (2017). Jaz2 controls stomata dynamics during bacterial invasion. New Phytol.213 (3), 1378–1392. doi: 10.1111/nph.14354
35
GoossensJ.Fernández-CalvoPatriciaSchweizerF.GoossensA. (2016). Jasmonates: signal transduction components and their roles in environmental stress responses. Plant Mol. Biol.91 (6), 673–689. doi: 10.1007/s11103-016-0480-9
36
GuoQ.MajorI. T.HoweG. A. (2018). Resolution of growth–defense conflict: mechanistic insights from jasmonate signaling. Curr. Opin. Plant Biol.44, 72–81. doi: 10.1016/j.pbi.2018.02.009
37
HavkoN.MajorI.JewellJ.AttaranE.HoweG. (2016). Control of carbon assimilation and partitioning by jasmonate: an accounting of growth–defense tradeoffs. Plants5 (1), 7. doi: 10.3390/plants5010007
38
HeP.ChintamananiS.ChenZ.ZhuL.KunkelB. N.AlfanoJ. R.et al. (2004). Activation of a COI1-dependent pathway in Arabidopsis by Pseudomonas syringae type III effectors and coronatine. Plant J.37, 589–602. doi: 10.1111/j.1365-313X.2003.01986.x
39
HeX.JiangJ.WangC. Q.DeheshK. (2017). ORA59 and EIN3 interaction couples jasmonate-ethylene synergistic action to antagonistic salicylic acid regulation of PDF expression. J. Integr. Plant Biol.59 (4), 275–287. doi: 10.1111/jipb.12524
40
HeinrichM.HettenhausenC.LangeT.WuenscheH.FangJ.BaldwinI. T.et al. (2013). High levels of jasmonic acid antagonize the biosynthesis of gibberellins and inhibit the growth of Nicotiana attenuata stems. Plant J. Cell Mol. Biol.73 (4), 591–606. doi: 10.1111/tpj.12058
41
HouX.DingL.YuH. (2013). Crosstalk between GA and JA signaling mediates plant growth and defense. Plant Cell Rep.32 (7), 1067–1074. doi: 10.1007/s00299-013-1423-4
42
HouX.LeeL. Y. C.XiaK.YanY.YuH. (2010). Dellas modulate jasmonate signaling via competitive binding to jazs. Dev. Cell19 (6), 0–894. doi: 10.1016/j.devcel.2010.10.024
43
HuY.JiangL.WangF.YuD. (2013). Jasmonate regulates the inducer of CBF expression–C-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis. Plant Cell25 (8), 2907–2924. doi: 10.1105/tpc.113.112631
44
HuY.JiangY.HanX.WangH.PanJ.YuD. (2017). Jasmonate regulates leaf senescence and tolerance to cold stress: crosstalk with other phytohormones. J. Exp. Bot.68 (6), 1361–1369. doi: 10.1093/jxb/erx004
45
HuotB.YaoJ.MontgomeryB. L.HeS. Y. (2014). Growth-defense tradeoffs in plants: a balancing act to optimize fitness. Mol. Plant7 (8), 1267–1287. doi: 10.1093/mp/ssu049
46
JiangS.YaoJ.MaK. W.ZhouH.SongJ.HeS. Y.et al. (2013). Bacterial effector activates jasmonate signaling by directly targeting jaz transcriptional repressors. PLoS Pathog.9 (10), e1003715. doi: 10.1371/journal.ppat.1003715
47
Jiménez-Góngora TamaraS.-K.Lozano-Durán RosaK.CyrilZ. (2015). Flg22-triggered immunity negatively regulates key br biosynthetic genes. Front. Plant Sci.6, 981. doi: 10.3389/fpls.2015.00981
48
JinH.ZhuZ. (2017). Temporal and spatial view of jasmonate signaling. Trends Plant Sci.22 (6), 451–454. doi: 10.1016/j.tplants.2017.04.001
49
KatagiriF.TsudaK. (2010). Understanding the plant immune system. Mol. Plant-Microbe Interact.23 (12), 1531–1536. doi: 10.1094/mpmi-04-10-0099
50
KazanK. (2015). Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci.20 (4), 219–229. doi: 10.1016/j.tplants.2015.02.001
51
KhanN. A.NazarR.IqbalN.AnjumN. A. (2012). Phytohormones and abiotic stress tolerance in plants.Springer, Heidelberg, Dordrecht, London, New York: Springer Science and Business Media. doi: 10.1007/978-3-642-25829-9_3
52
Leon-ReyesA.SpoelS. H.De LangeE. S.AbeH.KobayashiM.TsudaS.et al. (2009). Ethylene modulates the role of NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 in cross talk between salicylate and jasmonate signaling. Plant Physiol.149, 1797–1809. doi: 10.1104/pp.108.133926
53
LiuX.ChiH.YueM.ZhangX.LiW.JiaE. (2012). The regulation of exogenous jasmonic acid on UV-b stress tolerance in wheat. J. Plant Growth Regul.31 (3), 436–447. doi: 10.1007/s00344-011-9253-5
54
LyonsR.MannersJ. M.KazanK. (2013). Jasmonate biosynthesis and signaling in monocots: a comparative overview. Plant Cell Rep.32, 815–827. doi: 10.1007/s00299-013-1400-y
55
MajorI. T.YoshidaY.CamposM. L.KapaliG.XinX. F.SugimotoK.et al. (2017). Regulation of growth–defense balance by the JASMONATE ZIM-DOMAIN (JAZ)-MYC transcriptional module. New Phytol.215 (4), 1533–1547. doi: 10.1111/nph.14638
56
MaksymiecW.WianowskaD.DawidowiczA. L.RadkiewiczS.MardarowiczM.KrupaZ. (2005). The level of jasmonic acid in Arabidopsis thaliana and Phaseolus coccineus plants under heavy metal stress. J. Plant Physiol.162 (12), 0–1346. doi: 10.1016/j.jplph.2005.01.013
57
MaoY. B.LiuY. Q.ChenD. Y.ChenF. Y.FangX.HongG. J.et al. (2017). Jasmonate response decay and defense metabolite accumulation contributes to age-regulated dynamics of plant insect resistance. Nat. Commun.8, 13925. doi: 10.1038/ncomms13925
58
MatyssekR.AgererR.ErnstD.MunchJ. C.OsswaldW.PretzschH.et al. (2005). The plant’s capacity in regulating resource demand. Plant Biol.7 (06), 560–580. doi: 10.1055/s-2005-872981
59
MengisteT. (2012). Plant immunity to necrotrophs. Annu. Rev. Phytopathol.50, 267–294. doi: 10.1146/annurev-phyto-081211-172955
60
MilletY. A.DannaC. H.ClayN. K.SongnuanW.SimonM. D.Werck-ReichhartD.et al. (2010). Innate immune responses activated in arabidopsis roots by microbe-associated molecular patterns. Plant Cell22 (3), 973–990. doi: 10.1105/tpc.109.069658
61
OkadaK.AbeH.ArimuraG. (2015). Jasmonates induce both defense responses and communication in monocotyledonous and dicotyledonous plants. Plant Cell Physiol.56, 16–27. doi: 10.1093/pcp/pcu158
62
PandeyD.RajendranS. R. C. K.GaurM.SajeeshP. K.KumarA. (2016). Plant defense signaling and responses against necrotrophic fungal pathogens. J. Plant Growth Regul.35 (4), 1159–1174. doi: 10.1007/s00344-016-9600-7
63
PatkarR. N.BenkeP. I.QuZ.ChenY. Y. C.YangF.SwarupS.et al. (2015). A fungal monooxygenase-derived jasmonate attenuates host innate immunity. Nat. Chem. Biol.11 (9), 733. doi: 10.1038/nchembio.1885
64
PauwelsL.RitterA.GoossensJ.DurandA. N.LiuH.GuY.et al. (2015). The ring e3 ligase keep on going modulates jasmonate zim-domain12 stability. Plant Physiol.169 (2), 1405–1417. doi: 10.1104/pp.15.00479
65
PerT. S.KhanM. I. R.AnjumN. A.MasoodA.HussainS. J.KhanN. A. (2018). Jasmonates in plants under abiotic stresses: crosstalk with other phytohormones matters. Environ. Exp. Bot.145, 104–120. doi: 10.1016/j.envexpbot.2017.11.004
66
PérezA. C.GoossensA. (2013). Jasmonate signalling: a copycat of auxin signalling? Plant Cell Environ.36 (12), 2071–2084. doi: 10.1111/pce.12121
67
PieterseC. M.Van der DoesD.ZamioudisC.Leon-ReyesA.Van WeesS. C. (2012). Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol.28, 489–521. doi: 10.1146/annurev-cellbio-092910-154055
68
QiuZ.GuoJ.ZhuA.ZhangL.ZhangM. (2014). Exogenous jasmonic acid can enhance tolerance of wheat seedlings to salt stress. Ecotoxicol. Environ. Saf.104, 202–208. doi: 10.1016/j.ecoenv.2014.03.014
69
ReevesP. H.EllisC. M.PloenseS. E.WuM. F.ReedJ. W. (2012). A regulatory network for coordinated flower maturation. PLoS Genet.8 (2), e1002506. doi: 10.1371/journal.pgen.1002506
70
RodriguezM. C.PetersenM.MundyJ. (2010). Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol.61 (1), 621–649. doi: 10.1146/annurev-arplant-042809-112252
71
SarwatM.TutejaN. (2017). Hormonal signaling to control stomatal movement during drought stress. Plant Gene11, 143–153. doi: 10.1016/j.plgene.2017.07.007
72
SavchenkoT.KollaV. A.WangC. Q.NasafiZ.HicksD. R.PhadungchobB.et al. (2014). Functional convergence of oxylipin and abscisic acid pathways controls stomatal closure in response to drought. Plant Physiol.164 (4), 1151–1160. doi: 10.4161/psb.28304
73
SharmaM.LaxmiA. (2016). Jasmonates: emerging players in controlling temperature stress tolerance. Front. Plant Sci.6, 1129. doi: 10.3389/fpls.2015.01129
74
SharmaR.De VleesschauwerD.SharmaM. K.RonaldP. C. (2013). Recent advances in dissecting stress-regulatory crosstalk in rice. Mol. Plant6 (2), 250–260. doi: 10.1093/mp/sss147
75
SheardL. B.TanX.MaoH.WithersJ.Ben-NissanG.HindsT. R.et al. (2010). Jasmonate perception by inositol-phosphate-potentiated coi1–jaz co-receptor. Nature468 (7322), 400–405. doi: 10.1038/nature09430
76
ShimJ. S.JungC.LeeS.MinK.LeeY. W.ChoiY.et al. (2013). Atmyb44 regulates wrky70 expression and modulates antagonistic interaction between salicylic acid and jasmonic acid signaling. Plant J.73 (3), 483–495. doi: 10.1111/tpj.12051
77
SongS.QiT.HuangH.RenQ.WuD.ChangC.et al. (2011). The jasmonate-zim domain proteins interact with the r2r3-myb transcription factors myb21 and myb24 to affect jasmonate-regulated stamen development in arabidopsis. Plant Cell23 (3), 1000–1013. doi: 10.1105/tpc.111.083089
78
SpoelS. H.JohnsonJ. S.DongX. (2007). Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc. Natl. Acad. Sci.104 (47), 18842–18847. doi: 10.1073/pnas.0708139104
79
SpoelS. H.LoakeG. J. (2011). Redox-based protein modifications: the missing link in plant immune signalling. Curr. Opin. Plant Biol.14 (4), 358–364. doi: 10.1016/j.pbi.2011.03.007
80
SpoelS. H.MouZ.TadaY.SpiveyN. W.GenschikP.DongX. (2009). Proteasome-mediated turnover of the transcription coactivator npr1 plays dual roles in regulating plant immunity. Cell137 (5), 0–872. doi: 10.1016/j.cell.2009.03.038
81
ThireaultC.ShyuC.YoshidaY.AubinB.CamposM. L.HoweG. A. (2015). Repression of jasmonate signaling by a non-TIFY JAZ protein in Arabidopsis. Plant J.82 (4), 669–679. doi: 10.1111/tpj.12841
82
TianD.TrawM. B.ChenJ. Q.KreitmanM.BergelsonJ. (2003). Fitness costs of R-gene-mediated resistance in arabidopsis thaliana. Nature423 (6935), 74–77. doi: 10.1038/nature01588
83
UjiY.TaniguchiS.TamaokiD.ShishidoH.AkimitsuK.GomiK. (2016). Overexpression of OsMYC2 results in the up-regulation of early JA-Rresponsive genes and bacterial blight resistance in Rice. Plant Cell Physiol.57 (9), 1814–1827. doi: 10.1093/pcp/pcw101
84
WangN.LiuM.GuoL.YangX.QiuD. (2016). A novel protein elicitor (PeBA1) from Bacillus amyloliquefaciens NC6 induces systemic resistance in tobacco. Int. J. Biol. Sci.12 (6), 757. doi: 10.7150/ijbs.14333
85
WasternackC. (2014). Action of jasmonates in plant stress responses and development—applied aspects. Biotechnol. Adv.32 (1), 31–39. doi: 10.1016/j.biotechadv.2013.09.009
86
WasternackC.HauseB. (2013). Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Annals of Botany111 (6), 1021–1058. doi: 10.1093/aob/mct067
87
WasternackC.SongS. (2016). Jasmonates: biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J. Exp. Bot.68 (6), 1303–1321. doi: 10.1093/jxb/erw443
88
WasternackC.StrnadM. (2016). Jasmonate signaling in plant stress responses and development–active and inactive compounds. New Biotechnol.33 (5), 604–613. doi: 10.1016/j.nbt.2015.11.001
89
WildM.DavièreJ. M.CheminantS.RegnaultT.BaumbergerN.HeintzD.et al. (2012). The Arabidopsis DELLA RGA-LIKE3 is a direct target of MYC2 and modulates jasmonate signaling responses. Plant Cell24 (8), 3307–3319. doi: 10.1105/tpc.112.101428
90
XieD. X.FeysB. F.JamesS.Nieto-RostroM.TurnerJ. G. (1998). COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science280 (5366), 1091–1094. doi: 10.1126/science.280.5366.1091
91
XieY.TanH.MaZ.HuangJ. (2016). DELLA proteins promote anthocyanin biosynthesis via sequestering MYBL2 and JAZ suppressors of the MYB/bHLH/WD40 complex in Arabidopsis thaliana. Mol. Plant9 (5), 711–721. doi: 10.1016/j.molp.2016.01.014
92
YangD. L.YaoJ.MeiC. S.TongX. H.ZengL. J.LiQ.et al. (2012). Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci.109 (19), E1192–E1200. doi: 10.1073/pnas.1201616109
93
YiS. Y.ShirasuK.MoonJ. S.LeeS. G.KwonS. Y. (2014). The activated sa and ja signaling pathways have an influence on flg22-triggered oxidative burst and callose deposition. PLoS One99 (2), e88951. doi: 10.1371/journal.pone.0088951
94
ZanderM.ChenS.ImkampeJ.ThurowC.GatzC. (2012). Repression of the arabidopsis thaliana jasmonic acid/ethylene-induced defense pathway by tga-interacting glutaredoxins depends on their c-terminal alwl motif. Mol. Plant5 (4), 831–840. doi: 10.1093/mp/ssr113
95
ZengW.HeS. Y. (2010). A prominent role of the flagellin receptor FLAGELLIN-SENSING2 in mediating stomatal response to Pseudomonas syringae pv tomato DC3000 in Arabidopsis. Plant Physiol.153 (3), 1188–1198. doi: 10.2307/25704949
96
ZhangL.YaoJ.WithersJ.XinX-F.BanerjeeR.FariduddinQ.et al. (2015). Host target modification as a strategy to counter pathogen hijacking of the jasmonate hormone receptor. Proc. Natl. Acad. Sci. U. S. A.112, 14354–14359. doi: 10.1073/pnas.1510745112
97
ZhangF.YaoJ.KeJ.ZhangL.LamV. Q.XinX. F.et al. (2015). Structural basis of jaz repression of myc transcription factors in jasmonate signalling. Nature525, 269–273. doi: 10.1038/nature14661
98
ZhangL.ZhangF.MelottoM.YaoJ.HeS. Y. (2017). Jasmonate signaling and manipulation by pathogens and insects. J. Exp. Bot.68 (6), 1371–1385. doi: 10.1093/jxb/erw478
99
ZhangX.ZhuZ.AnF.HaoD.LiP.SongJ.et al. (2014). Jasmonate-activated myc2 represses ethylene insensitive3 activity to antagonize ethylene-promoted apical hook formation in arabidopsis. Plant Cell26 (3), 1105–1117. doi: 10.1105/tpc.113.122002
100
ZhaoY.DongW.ZhangN.AiX.WangM.HuangZ.et al. (2014). A wheat allene oxide cyclase gene enhances salinity tolerance via jasmonate signaling. Plant Physiol.164 (2), 1068–1076. doi: 10.1104/pp.113.227595
101
ZhengX. Y.SpiveyN. W.ZengW.LiuP. P.FuZ. Q.KlessigD. F.et al. (2012). Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe11 (6), 587–596. doi: 10.1016/j.chom.2012.04.014
102
ZhouZ.WuY.YangY.DuM.ZhangX.GuoY.et al. (2015). An Arabidopsis plasma membrane proton ATPase modulates JA signaling and is exploited by the Pseudomonas syringae effector protein AvrB for stomatal invasion. Plant Cell27 (7), 2032–2041. doi: 10.1105/tpc.15.00466
103
ZhuZ.AnF.FengY.LiP.XueL.MuA.et al. (2011). Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci.108 (30), 12539–12544. doi: 10.1073/pnas.1103959108
104
ZhuZ. (2014). Molecular basis for jasmonate and ethylene signal interactions in Arabidopsis. J. Exp. Bot.65 (20), 5743–5748. doi: 10.1093/jxb/eru349
105
ZhuJ-K. (2016). Abiotic stress signaling and responses in plants. Cell167 (2), 313–324. doi: 10.1016/j.cell.2016.08.029
106
ZhuZ.LeeB. (2015). Friends or foes: new insights in jasmonate and ethylene co-actions. Plant Cell Physiol.56 (3), 414. doi: 10.1093/pcp/pcu171
Summary
Keywords
jasmonic acid, plant hormone, environmental stress, defense response, crosstalk
Citation
Yang J, Duan G, Li C, Liu L, Han G, Zhang Y and Wang C (2019) The Crosstalks Between Jasmonic Acid and Other Plant Hormone Signaling Highlight the Involvement of Jasmonic Acid as a Core Component in Plant Response to Biotic and Abiotic Stresses. Front. Plant Sci. 10:1349. doi: 10.3389/fpls.2019.01349
Received
06 March 2019
Accepted
27 September 2019
Published
18 October 2019
Volume
10 - 2019
Edited by
Zuhua He, Center for Excellence in Molecular Plant Sciences (CAS), China
Reviewed by
Dong-Lei Yang, Nanjing Agricultural University, China; Fengming Song, Zhejiang University, China
Updates
Copyright
© 2019 Yang, Duan, Li, Liu, Han, Zhang and Wang.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing Yang, yangjin-18@163.com
This article was submitted to Plant Physiology, a section of the journal Frontiers in Plant Science
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.