- 1Instutute of Agricultural Biotechnology, Jilin Academy of Agricultural Sciences, Changchun, China
- 2School of Basic Medicine, Guizhou University of Traditional Chinese Medicine, Guiyang, China
- 3Key Laboratory of Arable Land Conservation (Middle and Lower Reaches of Yangtze River), Ministry of Agriculture (MOA), Huazhong Agricultural University, Wuhan, China
- 4Microelement Research Center, College of Resources & Environment, Huazhong Agricultural University, Wuhan, China
Salinity and microbial pathogens are the major limiting factors for crop production. Although the manipulation of many genes could improve plant performance under either of these stresses, few genes have reported that could improve both pathogen resistance and saline-alkali stress tolerance. In this study, we identified a new chitinase gene CHITINASE 2 (LcCHI2) that encodes a class II chitinase from Leymus chinensis, which grows naturally on alkaline-sodic soil. Overexpression of LcCHI2 increased chitinase activity in transgenic plants. The transgenic tobacco and maize exhibited improved pathogen resistance and enhanced both neutral salt and alkaline salt stress tolerance. Overexpression of LcCHI2 reduced sodium (Na+) accumulation, malondialdehyde content and relative electrical conductivity in transgenic tobacco under salt stress. In addition, the transgenic tobacco showed diminished lesion against bacterial and fungal pathogen challenge, suggesting an improved disease resistance. Similar improved performance was also observed in LcCHI2-overexpressed maize under both pathogen and salt stresses. It is worth noting that this genetic manipulation does not impair the growth and yield of transgenic tobacco and maize under normal cultivation condition. Apparently, application of LcCHI2 provides a new train of thought for genetically engineering saline-alkali and pathogen resistant crops of both dicots and monocots.
Introduction
The biotic and abiotic stresses, such as pathogens and salinity, severely affected crop growth and agricultural productivity worldwide. According to incomplete statistics of UNESCO and FAO, 950 million ha (6.4%) of the world’s land area has saline-alkali soil, and about 10% of this area is found in China. The alkaline soils of China contains high levels of Na2CO3 and NaHCO3 (Wang et al., 1993). Only few species of plants could grow in such soils and they are sparsely distributed. Leymus chinensis, a perennial grass of family gramineae, grows naturally on alkaline soil, suggesting the existence of such a mechanism. Apparently, identification of functional gene(s) specifically responsible for stress tolerance would be a prerequisite of crop improvement through biotech breeding.
Chitinase is one of the important enzyme family that has been found to involve in such a mechanism of divergent biological functions. Chitinases (EC 3.2.1.14) are a family of glycosyl hydrolases (GHs) responsible for the hydrolysis of the chitin polymer (a β-1,4-linked N-acetylglucosamine), a structural component found in the cell walls of fungi, insects, a variety of crustaceans and nematode eggs (Kesari et al., 2015). Interestingly, although the chitin is not present in plants, different subgroups of chitinase genes were identified in plants (Kesari et al., 2015). Plant chitinase proteins are generally divided into six classes, I to VI (Patil et al., 2000). The classes III and V belong to the GH18 family, whereas class I, II, IV, and VI belong to the GH19 family (Patil et al., 2000). Both families exhibit diversity in their nucleic acid sequences, protein structures, substrate specificities, sensitivity to inhibitors and mechanisms of catalysis. Along with active chitinases, the plant genome also consists of a large number of sequences encoding catalytically inactive chitinases that are referred to as chitinase-like (CTL) proteins (Kesari et al., 2015). A few studies suggested that the likely substrates of plant CTL proteins may be arabinogalactan protein, chitooligosaccharides, N-acetylchitooligosaccharides and other GlcNAc-containing glycoproteins or glycolipids, but the substrates of most plant CTL proteins remain uncertain (van Hengel et al., 2001; Zhong et al., 2002; Sanchez-Rodriguez et al., 2012). The intrinsic diversities of plant chitinase imply that the chitinase/CTL genes likely have a broad function.
Because chitin is the major component of fungal cell walls, earlier studies on the role of chitinase genes focused extensively on its involvement in plant defense responses to fungal pathogen infection (Brogue et al., 1991; Verburg and Huynh, 1991). Chitinase gene expression in plant tissues is strongly induced by fungal pathogens and chitin oligosaccharides (Hong and Hwang, 2002; Seo et al., 2008). Interestingly, the expression of chitinase gene responded also to infection of viruses, bacteria and oomycetes, which do not have chitin or related structures in their cell wall (Hong and Hwang, 2006; van Loon et al., 2006). Accordingly, a number of plant chitinases are defined as classic pathogenesis-related proteins (PRs) including PR-3, -4, -8, and -11 families (van Loon et al., 2006). Overexpression of these PR proteins conferred resistance to plant disease in different plant species (Brogue et al., 1991; Nishizawa et al., 1999; Tabaeizadeh et al., 1999; Yamamoto et al., 2000; Takahashi et al., 2005; Shin et al., 2008).
Plants chitinase and CTL genes play a role not only in defense related processes but also in abiotic stress tolerance. In Arabidopsis thaliana, the AtPR3 gene which encodes a class II chitinase was induced by high salt (Seo et al., 2008). The pepper class II basic chitinase gene CaChi2 was induced by salt, drought and osmotic stresses (Hong and Hwang, 2002, 2006). The expression of ptch28 from Lycopersicon chilense plants induced by osmotic and abscisic acid (Chen et al., 1994). In addition to salt, osmotic and drought stresses, expression of the class II chitinase gene from Bermuda grass was reported to be up-regulated by cold stress and function as an antifreeze protein (Nakamura et al., 2008). Similarly, in winter rye (Secale cereal) and highbush blueberry, cold stress induced the chitinase like proteins, which showed antifreeze activity in vitro (Yeh et al., 2000; Kikuchi and Masuda, 2009). Interestingly, most of the reported chitinase genes that responsive to both biotic and abiotic stresses belonged to the class II family and homolog to PR3 proteins (Supplementary Figure S1). Furthermore, overexpression of these PR3 genes, such as AtPR3 and CaChi2, significantly increased the salt resistance of the transgenic plants (Hong and Hwang, 2006; Seo et al., 2008). In contrast, mutation of the AtCTL1 resulted in oversensitive to salt and drought stresses (Kwon et al., 2007).
Wildly grown plants have developed through evolution into adaptive mechanisms against the various environmental threatening. Apparently, identification of functional gene(s) specifically responsible for stress tolerance would be a prerequisite of crop improvement through biotech breeding. In a previous report, a chitinase like EST sequence was isolated from Leymus chinensis which was treated with Na2CO3 (Jin et al., 2006). Accordingly, we cloned a unique chitinase gene and designated it as LcCHI2, which belongs to class II chitinase gene family. In Leymus chinensis, expression of LcCHI2 was induced by salt stress leading to an increase in chitinase activity. Overexpression of LcCHI2 increased pathogen and salt stress resistance in both tobacco and maize. This study provides a novel train of thought in improvement of both pathogen resistance and salt stress tolerance, in particular alkaline salt stress, through overexpression of a single gene in dicots and monocots.
Materials and Methods
Cloning and Expression Analysis of LcCHI2
Total RNA was extracted from 4-week-old seedlings of Leymus chinensis treated with 100 mM Na2CO3 using RNAiso plus (TaKaRa, Japan) according to the instruction manual. Full-length cDNA of LcCHI2 sequence was obtained using a SMARTTM RACE cDNA Amplification Kit (Clontech, United States) for 3′ RACE with the primers 5′-CCGACCAGTTCCAATGGGGCT-3′ and 5′-GGCCACGCGTCGACTAGTACTTTT TTTTTTTTT TTTT-3′.
For expression analysis of LcCHI2, total RNA was extracted from 4-week-old Leymus chinensis harvested at different time points under various stress treatments. A 771 bp fragment of LcCHI2 cDNA was amplified in 30 cycles with the primers 5′-ATGGCGAGGTTTGCTGCCCTCG-3′ and 5′-CTAGCTAGCGAAGTTTCGCTGGGT G-3′. As an internal standard for cDNA levels, a 245 bp fragment of ACTIN cDNA (GenBank accession number AB181991) was amplified simultaneously in 28 cycles with primers 5′-GGACCTTGCTGGTCGTGACC-3′ and 5′-CCTCAGGGCACCTGAACCTT T-3′. DIG High Prime DNA Labeling and Detection Starter Kit I (Roche, United States) was used for Northern blot analysis. The specific LcCHI2 cDNA fragment was amplified with the primers 5′-ATGGCGAGGTTTGCTGCCC-3′ and 5′-CTAGCTAGCGAAGTTTCGCTGGG-3′ and then labeled with DIG-High Prime used for the hybridization probe.
Agrobacterium-Mediated Tobacco and Maize Transformation
The LcCHI2 gene was amplified with the primers 5′-ccggaattcATGGCGAGGTTTGCTGCCCTCG-3′ and 5′-cgcggatccCTAGCTAGCGAAGTTTCGCTGGGTG-3′. The PCR products were digested with EcoR I/BamH I and cloned into the pUC119-pCaMV35S-tNOS vector to generate pUC119-pCaMV35S::LcCHI2-tNOS. The fragment of pCaMV35S::LcCHI2-tNOS was collected by Hind III digestion and cloned into pBI121vector. The final construct map was shown in Supplementary Figure S1A. After sequencing the positive clones, the final construct pBI121-pCaMV35S::LcCHI2-tNOS were transformed into Agrobacterium strain EHA105. Transgenic tobacco (Nicotiana tabacum) plants were produced by using Agrobacterium-mediated transformation as described by the previously reported method (Hoekema et al., 1983). Transgenic plants were selected on MS medium containing 50 mg/ml kanamycin and 500 mg/ml carbenicillin.
For maize transformation, the sequence of LcCHI2 from pBI121-pCaMV35S::LcCHI2-tNOS was digested with SpeI/BamH I and cloned into the pTF101-pZmUBI-tNOS vector to generate pTF101-ZmUBI::LcCHI2-tNOS. The final construct map was shown in Supplementary Figure S1B. Immature zygotic embryos of HiII (B73 × A188) were infected with Agrobacterium strain EHA105 harboring the binary vector pTF101-ZmUBI::LcCHI2-tNOS. Transgenic plants were selected and produced by the previously reported method (Frame et al., 2002).
Chitinase Activity Assay
Chitinase activity was measured in 10 mM potassium phosphate buffer (pH 7.0) containing 0.1 mM 4-methylumbelliferyl-β-D-N, N′-diacetylchitobiose (Sigma, Germany) as a substrate by the previously reported method (O’Brien and Colwell, 1987). One unit of chitinase activity was defined as the amount of enzyme required to produce 1 μmol of 4-methylumbelliferone per minute.
Measurement of Malondialdehyde (MDA) Contents and Ion Leakage Ratio
Fresh leaves (0.3 g) were ground properly in 5 ml of 5 % trichloroacetic acid solution and centrifuged for 10 min at 3000 rpm. 2 ml of the supernatant was reacted with 2 ml 0.67% thiobarbituric acid and then it was heated for 30 min, at 100°C in a water bath and then immediately cooled on ice. After centrifugation for 10 min, at 3000 rpm, the absorbance of the supernatant was read at 450, 532, and 600 nm. The contents of MDA were calculated using the formula: MDA concentration (μmol/L) = 6.45 × (A532−A600) − 0.56 × A450).
Ion leakage ratio was measured as relative electrical conductivity parameter. 0.1 g leaves were sampled from different plants, rinsed briefly with deionized water and immediately placed into a tube with 10 mL of deionized water. Conductivity (I1) was measured using an electroconductivity meter (model 1054, VWR Scientific, Phoenix) after the tubes were placed at 22°C overnight. Then, the samples were heated at 100°C for 30 min and conductivity (I2) was measured again. Relative electrical conductivity was expressed as (I1/I2) × 100%.
Sodium Content
Seedings of wild type (WT) and T2 generation transgenic tobacco plants were cultured in nutrient solution for 1 month and then treated with 0 or 200 mM NaCl. After 6 days of treatment, the shoots and roots were oven dried, digested with H2SO4-H2O2. The digest was dissolved in deionized water and sodium content was estimated by flame spectrophotometer (M410, Waters Corporation, America).
Salt Stress Treatments
The plant seedlings of Leymus chinensis were grown in vermiculite with a light/dark cycle of 16 h/8 h at 25°C. They were irrigated with one-half-diluted Murashige and Skoog (MS) medium every 3 days. Four-week-old seedlings of Leymus chinensis were treated with 400 mM NaCl and 100 mM Na2CO3, respectively. Leaves of treated plants were harvested up to 0, 6, 12, and 24 h, respectively, by rapid freezing in liquid nitrogen and kept at −80°C for further analysis.
To determine abiotic stresses tolerance of the transgenic tobacco on disc, seeds of wild-type and T2 generation transgenic tobacco were sowed on MS medium containing 200 mM NaCl, 30 mM Na2CO3 or 500 mM sorbitol, respectively. For soil experiment, tobacco seeds germinated in soil cultured with a light/dark cycle of 16 h/8 h at 25°C. Thirty plants for each line were used for salt and drought stress treatment, respectively. For salt stress, nearly 3-week-old plants were irrigated for 25 days with either tap water or tap water containing 400 mM NaCl or 100 mM Na2CO3.
To determine abiotic stresses tolerance of the transgenic maize, the transgenic events with HiII background (CHI-Ox2, CHI-Ox5, and CHI-Ox7) were backcross to local commercial inbred line X923-1. The segregated positive and negative BC5F2 plants were grown in pots (10 × 10 cm) containing peat: vermiculite (5:1, v/v) medium in a greenhouse with a light/dark cycle of 16 h/8 h at 30°C. PCR analysis was used to screen for positive transformants, and 2-week-old seedlings were water with either tap water or tap water containing 200 mM NaCl or 50 mM Na2CO3 for 6 days. Photograph was recorded after each treatment by a camera.
Pathogen Response Assays of Transgenic Tobacco and Maize
For bacterial infection analysis, Pseudomonas tabaci (Wolf et Foster) Stevens was grown in NBY medium and bacterial cells were collected, washed and resuspended in 10 mM MgSO4. The density of bacterial cells was determined by measuring absorbance at OD600. Bacterial cells in suspension were infiltrated into fully expanded 6-week-old tobacco leaves using a 1 ml plastic syringe without a needle. After 5 days, the average lesion area for each independent transgenic line was calculated and compared with that of wild-type tobacco palnts.
For fungal resistance analysis, Alternaria alternata (Fries) Keissler was cultured on potato dextrose agar medium at 28°C. When the mycelia reached the edge of the plate, 0.5 cm diameter agar discs were excised from the edge of growing colonies using a cork borer and inverted onto the detached leaves from wild-type and transgenic tobacco plants. All leaves were placed on wet filter paper in Petri dishes and incubated at 28°C to permit normal disease development under high humidity. After 7 days, the average lesion area for each independent transgenic line was calculated and compared with that of wild-type tobacco plants.
Maize pathogen Exserohilum turcicum and curvularia lunata (Wakker) Boed were provided by Iinstitute of plant protection, Jilin academy of agricultural sciences. Exserohilum turcicum and curvularia lunata (Wakker) Boed were cultured on potato dextrose agar medium at 28°C. When the mycelia reached the edge of the plate, 0.6 cm diameter agar discs were excised from the edge of growing colonies using a cork borer and inverted onto the detached leaves from transgenic and null control maize plants. All leaves were placed on wet filter paper in Petri dishes and incubated at 28°C to permit normal disease development under high humidity. After 3 days, the average lesion area for each independent transgenic line was calculated.
Nucleotide and Protein Sequences Accession Numbers
The nucleotide and protein sequences reported in this paper have been deposited in the Genbank nucleotide database and protein database under the accession numbers GQ397277 and ACV20870.
Results
Cloning and Characterization of LcCHI2 From Leymus chinensis
According to the EST sequence information from literature (Jin et al., 2006), we cloned a full-length cDNA of the target chitinase gene by 3‘RACE technology from Leymus chinensis, and was designated as LcCHI2 (GenBank accession number GQ397277). Sequence analysis revealed that LcCHI2 contains a 771 bp open reading frame encoding a polypeptide of 256 amino acids. Pfam scan showed that the deduced protein is a class II chitinase belonging to the GH19 family (PF00182)1. To obtain more information of the LcCHI2 protein, a phylogenetic tree was constructed including LcCHI2 orthologes and all the class II chitinases of Arabidopsis. LcCHI2 belonged to a cluster including three pathogenesis-related protein 3 (HvPR3, TaPR3, and AtPR3) and two chitinases (ScCHI46, CaCHI2) (Supplementary Figure S2). LcCHI2 showed more than 95% homology to HvPR3, TaPR3 and ScCHI46 and 71% homology to CaCHI2 (Supplementary Figure S3A). Multiple protein alignment revealed two conserved chitinase family 19 signature domains in all the five orthologes (Supplementary Figure S3B). However, an N-terminal secretive signal peptide only found in LcCHI2 and its monocot orthologes (Supplementary Figure S3B).
Sodium Stress Induced the Expression of LcCHI2 and Chitinase Activity in Leymus chinensis
As the EST of LcCHI2 was isolated from alkaline-sodic stressed tissues, we measured the expression of LcCHI2 in Leymus chinensis under different abiotic conditions. A significantly induction of LcCHI2 transcripts was observed under 400 mM NaCl after 24 h treatment comparing with the control plant (Figure 1A). Similarly, the expression of LcCHI2 increased more than 2-fold under 100 mM Na2CO3 after 12 and 24 h treatment compared with the control plant (Figure 1A). Subsequently, the chitinase activities were determined under NaCl and Na2CO3 conditions after 24 h treatment. The results showed that chitinase activities increased by 4.3- and 2.6-fold after 24 h exposure to 400 mM NaCl and 100 mM Na2CO3, respectively (Figure 1B). In contrast, the expression of LcCHI2 and chitinase activity were not changed in Leymus chinensis under 20% PEG condition in 24 h (data not show).
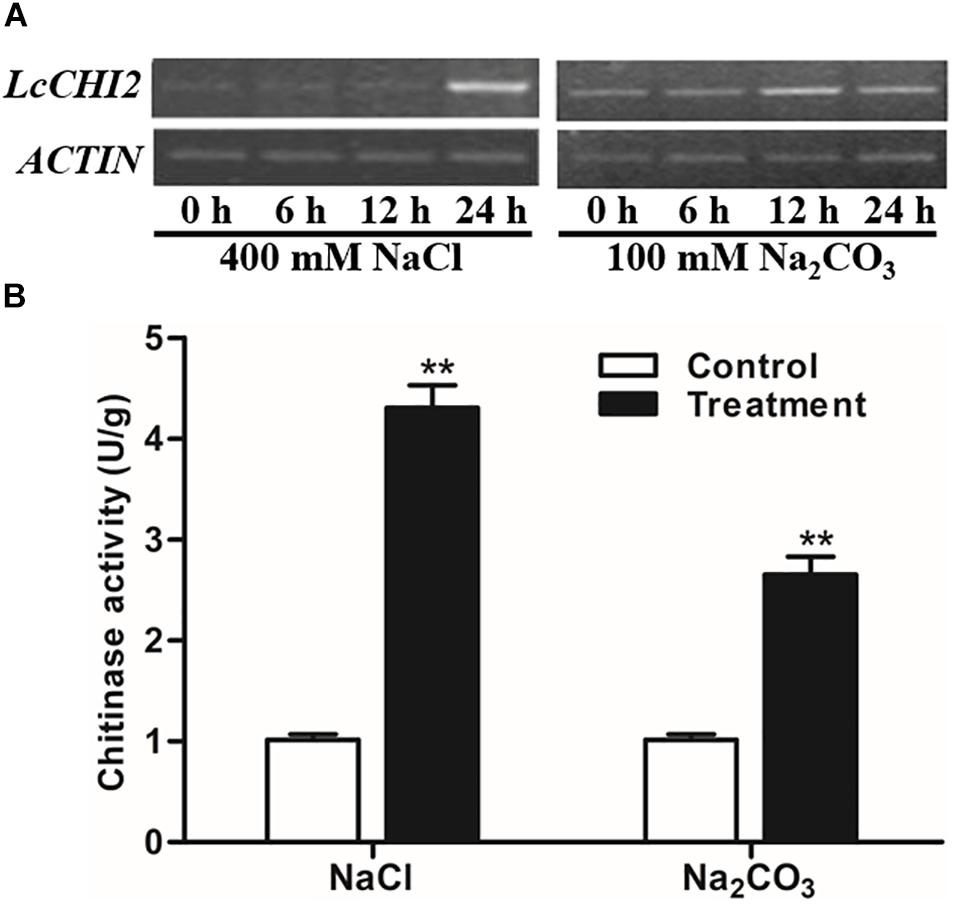
Figure 1. Regulation of LcCHI2 and chitinase activity under salt stress conditions in Leymus chinensis. (A) Expression analysis of the LcCHI2 after treatment with 400 mM NaCl and 100 mM Na2CO3 for the indicated times. Total RNA were extracted from 4-week-old seedlings of Leymus chinensis following treatments as indicated and reverse transcribed. The cDNA were used as templates for RT-PCR and the ACTIN gene was amplified as an internal control. The PCR products were examined by electrophoresis in 1% (w/v) agarose gel. (B) Chitinase activity assay under salt stress conditions in Leymus chinensis. Enzyme activity assays were carried out with the leaves of Leymus chinensis after treatment with 400 mM NaCl and 100 mM Na2CO3 for 24 h. Values are the mean ± SE obtained from three biological replicates. ** are significantly different at P < 0.01 (LSD test).
Overexpression of LcCHI2 in Tobacco Enhanced Plant Tolerance to Salt Stress
To further investigate the function of LcCHI2, transgenic tobacco plants that constitutively express the LcCHI2 gene under the control of the CaMV 35S promoter were developed by agrobacterium-mediated transformation. Twenty one independent positive transgenic lines were selected by kanamycin resistance. Three represented lines that showed high expression of LcCHI2 were selected for further analysis (Figure 2A). Moreover, chitinase activity increased by 1.5–1.7-fold over WT plants in the selected transgenic lines (Figure 2B). Comparing with the WT plants, the LcCHI2-overexpression plants exhibited no obvious phenotypic difference from the WTs under normal growth conditions (Supplementary Figure S4).
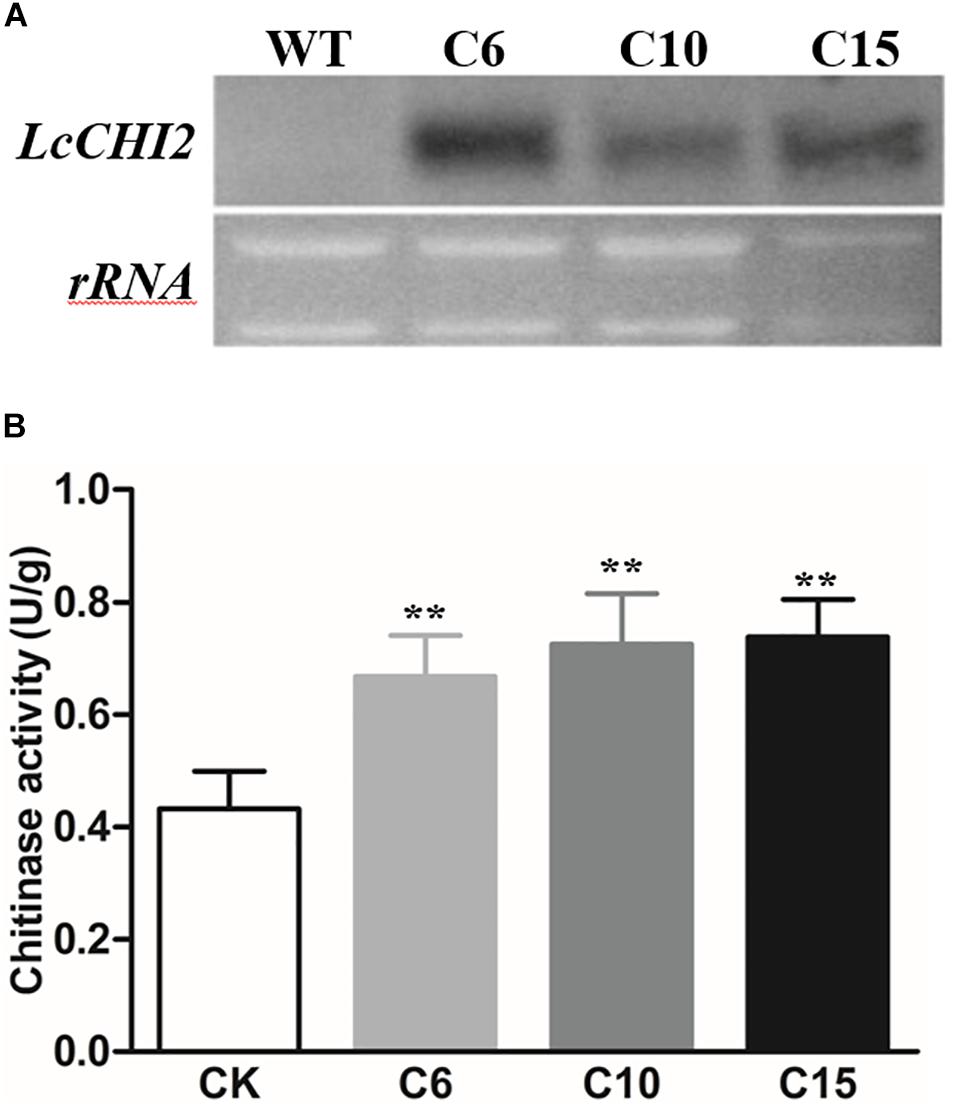
Figure 2. (A) Northern blot analysis of LcCHI2 transcripts in wild-type (WT) and transgenic tobacco plants (C6, C10, C15). A DIG-labeled LcCHI2 probe was used for hybridization. (B) Chitinase activity of WT and transgenic tobacco plants. Values are the mean ± SE obtained from three biological replicates. ** are significantly different at P < 0.01 (LSD test).
As the expression of LcCHI2 is induced by salt stress, phenotypes of the WT and LcCHI2-overexpression lines were analyzed on MS medium containing 200 mM NaCl, 30 mM Na2CO3 and 500 mM sorbitol, respectively. No difference was observed between WT and LcCHI2-overexpression plants growth on normal MS medium. However, the LcCHI2-overexpression plants survived well under salt stress (NaCl and Na2CO3) in comparison with WT plants, especially the C10 and C15 lines (Figure 3A). In contrast, the LcCHI2-overexpression plants only exhibited a minor effect for osmotic stress (Figure 3A). To further evaluate the abiotic resistance of LcCHI2-overexpression plants, soil growth plants were grown under salt and drought stress conditions. There were no observed differences in phenotypes between LcCHI2-overexpression and WT plants, when they were grown under normal and drought stress conditions (Supplementary Figure S5A). However, 6 days after watering 400 mM NaCl or 100 mM Na2CO3, the wild-type seedlings showed wilting phenotypes and ultimately died, whereas the transgenic tobacco plants continued to grow (Supplementary Figures S5B,C).
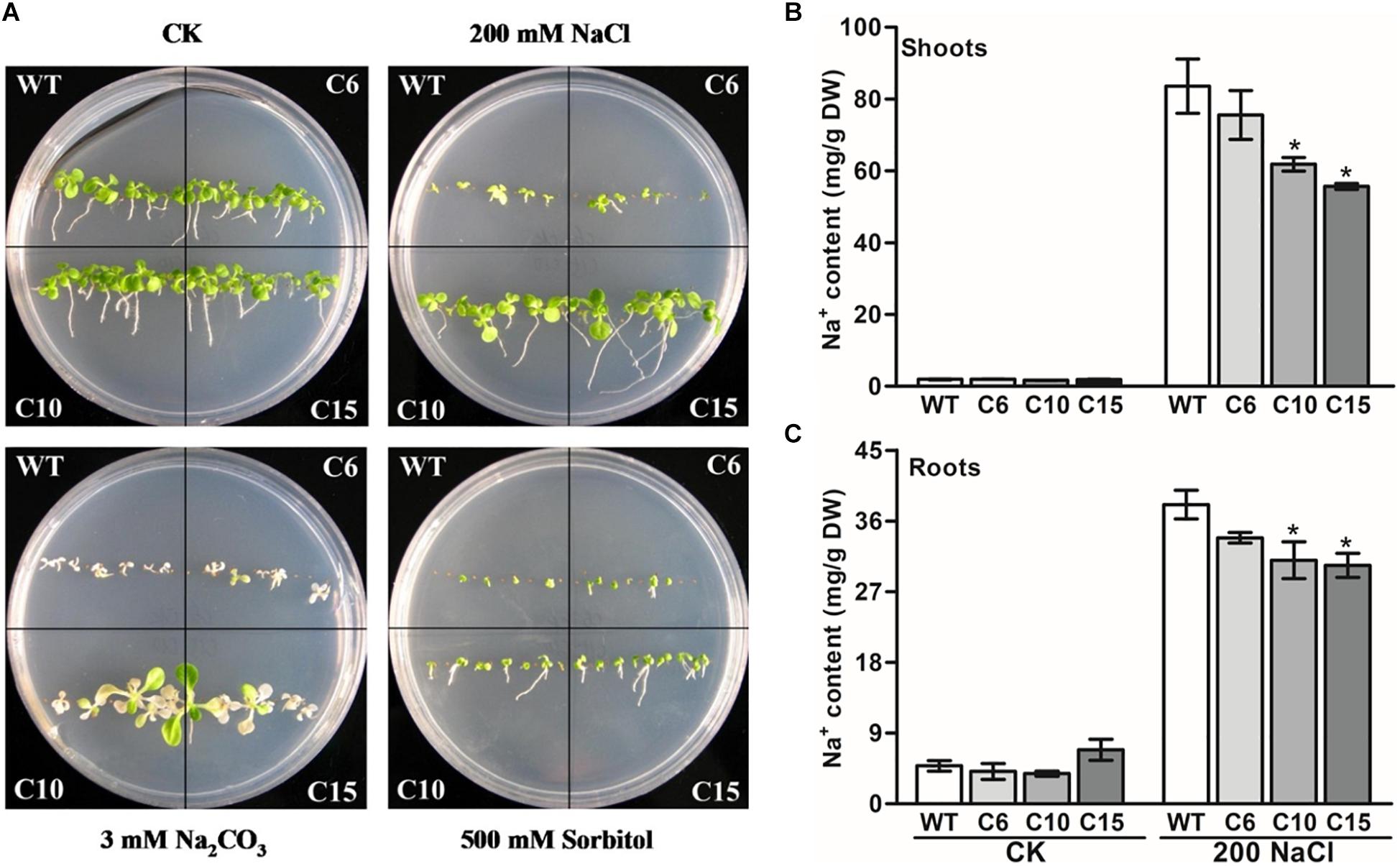
Figure 3. Abiotic stress tolerance assays of wild-type and LcCHI2-overexpressed plants in tobacco. (A) The WT and LcCHI2 overexpression (C6, C10 and C15) plants were grown on MS agar plates containing 200 mM NaCl, 30 mM Na2CO3, and 500 mM sorbitol for 30 days. (B, C) Na+ content of wild-type and LcCHI2 overexpression plants in shoots and roots. Values are the mean ± SE obtained from three biological replicates. * are significantly different at P < 0.05 (LSD test).
Because the overexpression of LcCHI2 increased salt stress tolerance other than osmotic or drought stresses, sodium contents were determined in the roots and shoots of WT and transgenic plants under normal and NaCl treatment conditions. As expected, NaCl treatment significantly increased Na+ content in the shoots and roots of WT and transgenic plants (Figures 3B,C). The overexpression of LcCHI2 decreased the Na+ content from 10 to 33% in the shoots and from 11 to 22% in the roots of transgenic lines compared with the corresponding WTs under NaCl treatment condition (Figures 3B,C). Meanwhile, the Na+ content showed a significantly decrease only in transgenic lines C10 and C15 other than line C6, which is coincident with the strong resistance to sodium stress of the transgenic lines C10 and C15 (Figure 3A).
Malondialdehyde (MDA) content and relative electrical conductivity are widely used as indicators for lipid peroxidation and the degree of plant cell injury under stress treatment, respectively. There are no obvious differences in MDA content and relative electrical conductivity between WT and LcCHI2 overexpression plants under normal growth condition. Although 24 h salt treatment significantly increased the MDA content and relative electrical conductivity in both WT and LcCHI2 overexpression plants, the transgenic lines showed a lower MDA content and relative electrical conductivity compared with WT under salt stress condition (Supplementary Figure S6).
Overexpression of LcCHI2 Conferred Pathogen Tolerance in Tobacco
As a homolog of PR3 proteins, LcCHI2 is presumed to be involve in pathogen resistance too. Thus, the LcCHI2-overexpression plants were inoculated with bacterial and fungal pathogens, respectively. Leaves of WT and LcCHI2-overexpression tobacco plants were inject-inoculated with spore suspensions of the bacterial pathogen Pseudomonas tabaci (Wolf et Foster) Stevens and symptom development was subsequently monitored for 5 days. The examined disease symptoms included chlorosis and necrosis expansion surrounding the primary infection sites. As shown in Figure 4, WT tobacco plants were more sensitive to bacterial infection than LcCHI2-overexpression tobacco plants in detached leaves, in which pathogen infection resulted in significantly reduced disease symptoms. The leaf lesion size of WT tobacco plants was 5- to 10-fold larger than that of LcCHI2-overexpression tobacco plants (Figures 4A,B). Resistance to the fungal pathogen Alternaria alternata (Fries) Keissler was also identified in LcCHI2-overexpression tobacco plants by a detached leaf inoculation test after 7 days inoculation. WT tobacco plants showed typical necrosis symptoms surrounded by chlorotic halos and extensive pathogen sporulation, while transgenic lines had significantly smaller lesion sizes than wild-type tobacco (Figures 4C,D). The enhanced resistance to Pseudomonas tabaci and Alternaria alternata in transgenic tobacco demonstrated that disease resistance conferred by LcCHI2 overexpression is effective against both bacterial and fungal pathogen.
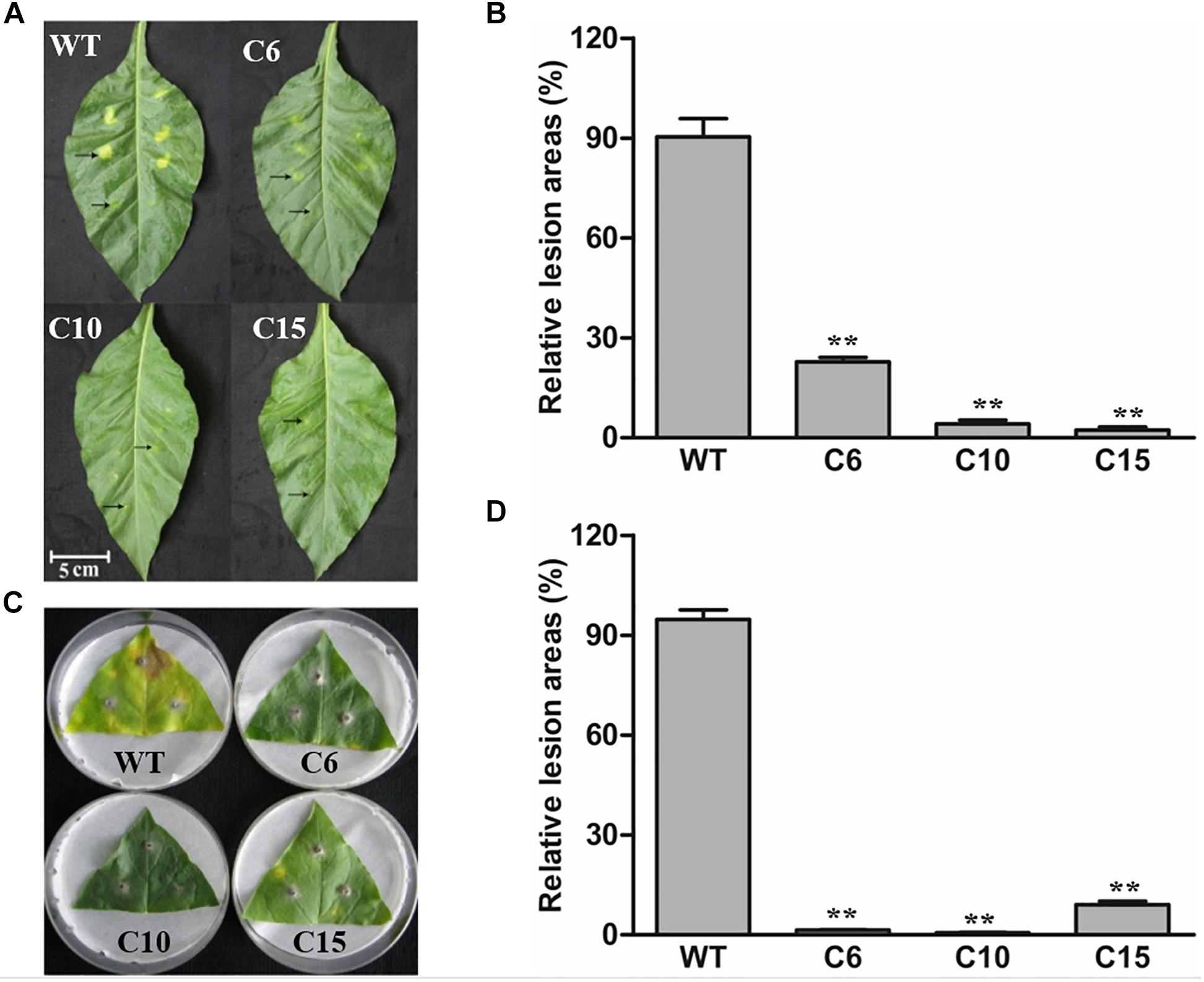
Figure 4. Pathogenesis analysis of LcCHI2-transgenic tobacco. (A, B) Resistance of transgenic tobacco to the bacterial pathogen Pseudomonas tabaci (Wolf and Foster) Stevens. Fully expanded leaves of tobacco plants were syringe-infiltrated with 0.1 ml solution of P. tabaci. Arrows in the upper line represent the sites inoculated with bacteria and arrows in the lower line represent the sites with mock-inoculated. The average lesion area of each independent transgenic line was calculated and their relative lesion areas are shown in columns after comparison with the average lesion area on wild-type tobacco. The photograph was taken 5 days after inoculation. (C, D) Responses of transgenic tobacco to the fungal pathogen Alternaria alternata (Fries) Keissler. Detached leaves were challenged with mycelia of A. alternata. The average lesion area of each independent transgenic line was calculated and their relative lesion areas are shown in columns after comparison with the average lesion area on wild-type tobacco. The photograph was taken 7 days after inoculation. Values are the mean ± SE obtained from three biological replicates. ** are significantly different at P < 0.01 (LSD test).
Overexpression of LcCHI2 in Maize Enhanced Tolerance to Salt and Pathogen Stresses
To determine whether the overexpression of the LcCHI2 gene could phenocopy the stress tolerances of transgenic tobacco in monocotyledons crops, we produced transgenic maize plants harboring the pTF101-pZmUBI-LcCHI2-tNOS construct. DNAs were extracted from leaves of T0 generation transgenic plants and use to detect the integration of target sequence into genomes of transgenic plants by PCR. The results showed that six transgenic events harbored both the selection marker and LcCHI2 genes (Supplementary Figure S7). Southern blot with bar gene as the probe showed that transgenic lines Ox2, Ox4, Ox7, and Ox10 were single copy while the transgenic lines Ox5 and Ox12 contained three copies of target gene (Supplementary Figure S8). Reverse-transcriptional PCR (RT-PCR) and chitinase activity assay indicated that three representative transgenic lines accumulated abundance LcCHI2 transcripts and enhanced chitinase activities compared to null plants with the absence of LcCHI2 (Figures 5A,B). Then these transgenic plants were grown in normal and alkaline soils in the greenhouse (Supplementary Figure S9). There are no significant different between the growth of transgenic (Ox) and their respective non-transgenic plants in normal soil (null). However, the transgenic plants were healthier than the corresponding null plants in alkaline soil. A significant increased plant height and SPAD value were observed in transgenic plants compared their null plants (Supplementary Figures S9B,C). To further evaluate the effects of divergent salt stress on the transgenic maize, 2 week-old plants were watered with 200 mM NaCl. As expected, no significant differences were observed between the growth of transgenic (Ox) and their respective null plants. In contrast, the overexpression plants showed resistance to NaCl other than the null plants after 6 days treatment (Figures 6A,B). Although NaCl treatments decreased the plant height and SPAD value in both overexpression and null plants, the magnitudes of the overexpression plants were significantly lower than null plants (Figure 6B).
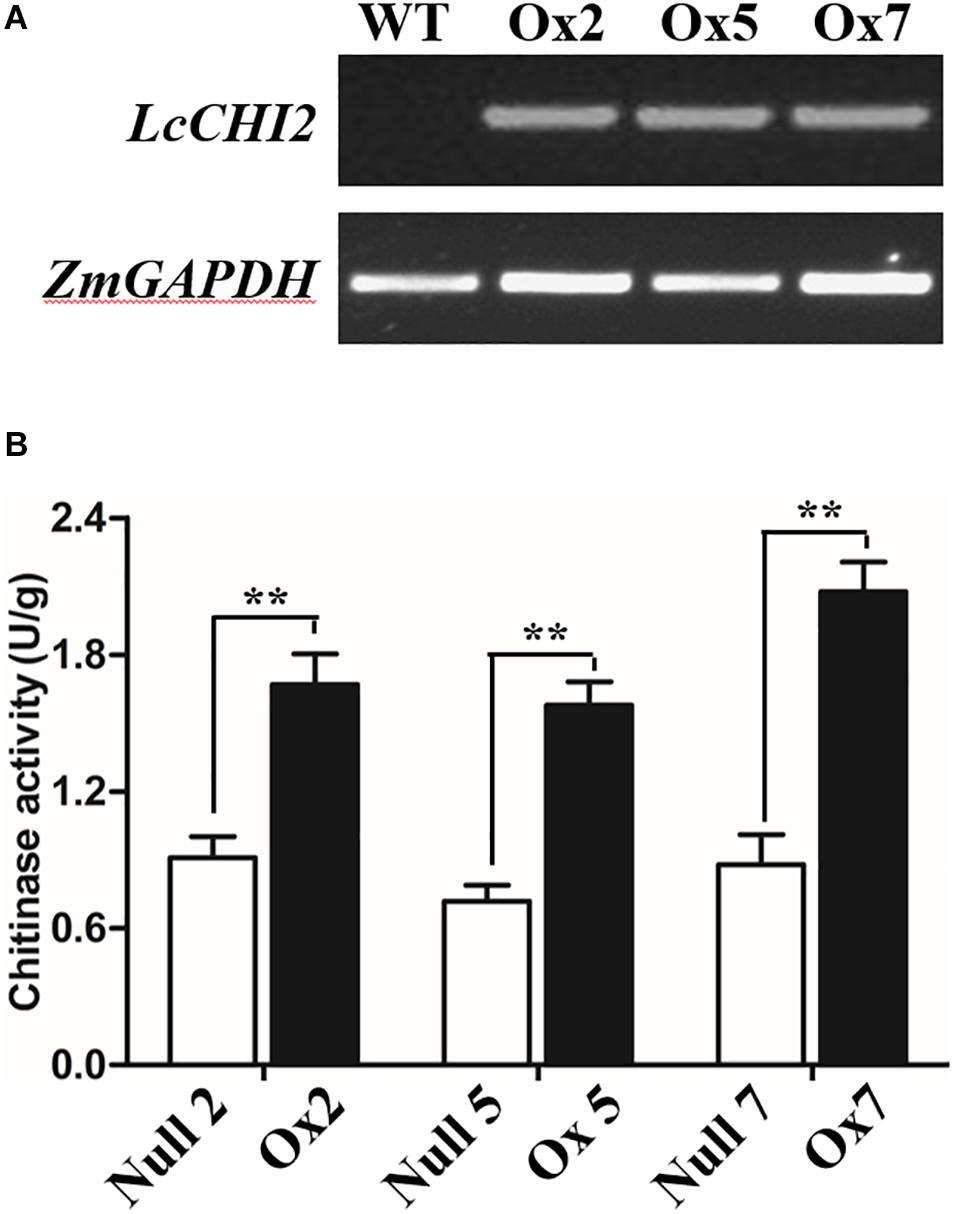
Figure 5. (A) RT-PCR analysis of LcCHI2 transcript in wild-type (WT) and transgenic maize plants (Ox2, Ox5, Ox7). Total RNA were extracted from 2-week-old seedlings of positive and null transgenic plants. The cDNA were used as templates for RT-PCR and the actin gene was amplified as an internal control. The PCR products were examined by electrophoresis in 1% (w/v) agarose gel. (B) Chitinase activity of null and transgenic maize plants. Values are the mean ± SE obtained from three biological replicates. ** are significantly different at P < 0.01 (LSD test).
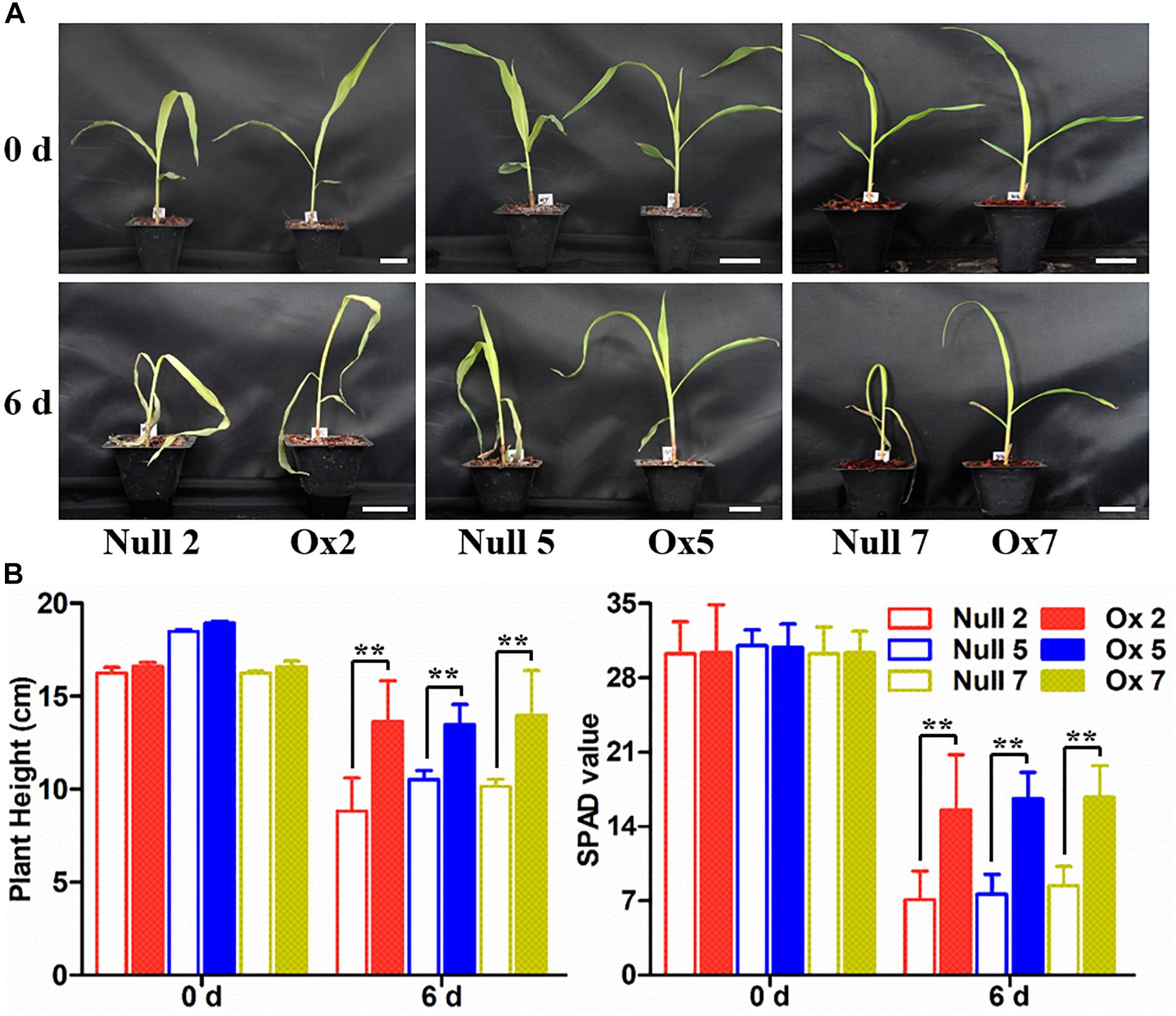
Figure 6. Salt stress tolerance assays of null and LcCHI2 overexpression plants in maize. (A) Soil growth null and LcCHI2 overexpression plants were watered with 200 mM NaCl for 6 days. (B) Plant height and SPAD value of null and LcCHI2 transgenic maize were measured every day. The transgenic events with HiII background were backcross to local commercial inbred line X923-1. The segregated positive and negative BC5F2 plants were used for the treatment. Values are the mean ± SE obtained from twenty biological replicates. ** are significantly different at P < 0.01 (LSD test).
Biotic resistances of the transgenic plants were also measured by infection the maize with two major fungal pathogens Exserohilum turcicum and curvularia lunata (Wakker) Boed, respectively. Typical necrosis symptoms were observed in the null plants while transgenic lines had significantly smaller lesion area after infection by Exserohilum turcicum or curvularia lunata (Wakker) Boed (Figures 7A–C). These results indicated that the overexpression of LcCHI2 in maize significantly inhibited the growth of fungal pathogens.
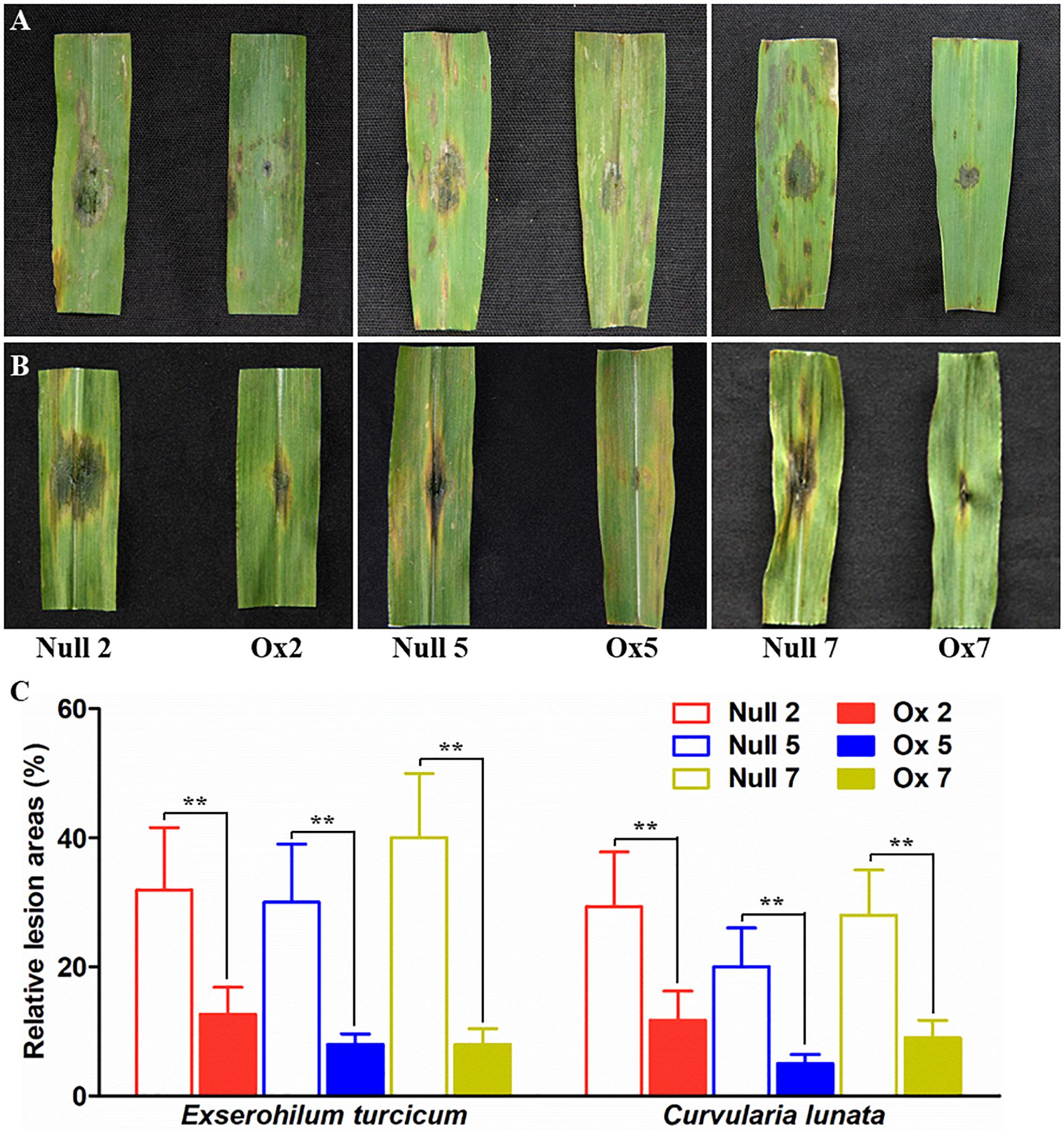
Figure 7. Pathogenesis analysis of null and LcCHI2-overexpressed maize. (A) Resistance of transgenic maize to fungal pathogen Exserohilum turcicum. (B) Responses of transgenic maize to the fungal pathogen curvularia lunata (Wakker) Boed. (C) The average lesion area of each independent transgenic line was calculated and their relative lesion areas are shown in columns after comparison with the average lesion area on wild-type tobacco. The transgenic events with HiII background were backcross to local commercial inbred line X923-1. The segregated positive and negative BC5F2 plants were used for the treatment. Values are the mean ± SE obtained from twenty biological replicates. ** are significantly different at P < 0.01 (LSD test).
To evaluate the effects of LcCHI2 overexpression in agronomic traits, the gene in the transgenic events with HiII background (CHI-Ox2, CHI-Ox5, and CHI-Ox7) were backcross to local commercial inbred line X923-1. The genetically segregated positive and negative BC5F2 plants were crossed, respectively, with Y822 to produce hybrid seeds. Agronomic traits were measured from the hybrid lines in two replicated experiments. The results (Supplementary Table S1) showed that all transgenic events exhibited statistically no negative impact such as cob weight, cob diameter, ear kernel weight and ear diameter, indicating that overexpression of LcCHI2 has no penalty on the growth and yield in the field conditions comparing with the controls.
Discussion
In this study, the full-length cDNA of a type II chitinase gene was cloned and named as LcCHI2, which was first identified from an EST library in Leymus Chinensis under Na2CO3 stress (Jin et al., 2006). The cDNA-deduced protein sequence analysis indicated that the LcCHI2 contains two conserved domains including the signature 1 (PS00773) and signature 2 (PS00774) of glycosyl hydrolase 19 family (Supplementary Figure S2). Protein cluster analysis indicates that LcCHI2 belongs to class II chitinase (Supplementary Figure S1). Several members of the class II chitinase proteins, such as AtPR3 and pcht28, were reported to have the typical chitinase activity and overexpression of those chitinase genes could increase chitinase activity in transgenic plants (Verburg and Huynh, 1991; Chen et al., 1994; Tabaeizadeh et al., 1999). In this study, a significantly increased chitinase activity was positively correlated with the up-regulation of LcCHI2 in Leymus Chinensis under both NaCl and Na2CO3 stress condition. Moreover, the transgenic tobacco and maize plants that overexpressed LcCHI2 also showed higher chitinase activity than control plants. Interestingly, protein alignment indicated that LcCHI2 and its homologs contain an N terminal secretary signal peptide (Supplementary Figure S3B). These results supported that LcCHI2 is a secreted active chitinase in Leymus Chinensis.
Different types of chitinases were induced by differential pathogen attack and have been characterized as pathogenesis-related (PR) proteins (van Loon et al., 2006; Kesari et al., 2015). Phylogenic analysis showed that LcCHI2 is a homolog of several reported PR3 proteins, including TaPR3, HvPR3, CaCHI2, pcht28, and AtPR3 (Supplementary Figure S3) (Verburg and Huynh, 1991; Tabaeizadeh et al., 1999; Hong and Hwang, 2002; Shin et al., 2008; Scheler et al., 2016). These PR3 proteins were proposed to be involved in plant defense responses. Plant chitinases were thought to degrade the major structural polysaccharide of fungal cell walls in the intercellular space to limit fungal growth (Brogue et al., 1991). The purified AtPR3 protein showed antifungal chitinase activity and inhibited the growth of Trichoderma reesei in vitro (Verburg and Huynh, 1991). The overexpression of chitinase resulted in enhanced resistance in tomato, rice, Italian ryegrass and grapevine to Verticillium dahlia, Magnaporthe grisea, Puccinia coronate, and Uncinula necator, respectively (Nishizawa et al., 1999; Tabaeizadeh et al., 1999; Yamamoto et al., 2000; Takahashi et al., 2005). Moreover, expression of a barley PR3 chitinase gene in transgenic wheat resulted in enhanced resistance to infection by Erysiphe graminis, Blumeria graminis, Pucinia recondita, and Fusarium graminearum (Oldach et al., 2001; Shin et al., 2008). To elucidate the defense response of LcCHI2, we investigated the biological functions of the LcCHI2 in transgenic tobacco and maize. The overexpression of LcCHI2 significantly enhanced resistance to multiple pathogens in tobacco and maize. The transgenic tobacco plants also showed resistance to a bacterial pathogen Pseudomonas tabaci Stevens, which do not have chitin or related structure in their cell wall. A similar phenomenon was observed in the transgenic Arabidopsis when overexpressing CaCHI2 gene (Hong and Hwang, 2006). In fact, chitinase expression in plant tissues is strongly induced by infection with viruses and bacteria in addition to fungal pathogen (van Loon et al., 2006). Therefore, the PR3 type protein may inhibit the pathogen invasion by an unknown mechanism other than directly degrading the chitin oligomers.
In addition to biotic stress, a number of cluster II chitinases were reported to be involved in developmental and various abiotic responses. The ScCHT46 showed 92% protein identify with LcCHI2 and encode a chitinase-antifreeze protein in winter rye (Yeh et al., 2000). The expression of ScCHT46 and its homologs in winter wheat, HvPR3, are responsive to cold and drought (Yeh et al., 2000). Moreover, it is reported that NaCl, drought and Mannitol induces the expression of several class II chitinases, including CaCHI2, pcht28, and AtPR3 (Chen et al., 1994; Hong and Hwang, 2002, 2006; Seo et al., 2008). The overexpression of CaCHI2 increased the osmotic stress in Arabidopsis (Hong and Hwang, 2006). In contrast, a mutation of AtPR3 affected the seeds geminating under high salt (Seo et al., 2008). Similarly, our results showed that the expression of LcCHI2 was up-regulated by Na+ stress conditions including treatment with NaCl and Na2CO3. Moreover, the activity of chitinase was increased under NaCl and Na2CO3 stresses in Leymus chinensis. Taking together, these results suggested that LcCHI2 play an important role in abiotic stress tolerance similar to its homologs in other plants.
Interestingly, the overexpression of LcCHI2 showed a significant tolerance to NaCl and Na2CO3 treatments in both tobacco and maize but not to the osmotic or drought stresses. These results implicated that the overexpression of LcCHI2 might assist to reduce Na+ ionic toxicity other than osmotic stress or improving water-use-efficiency. Coincidence with this assumption, the Na+ contents were decreased in the transgenic tobacco under NaCl treatment. Recently, it is proposed that the amount of the carboxyl groups on de-methesterified pectin could bind cations such as Na+ and sequester more Na+ in the cell wall, which may contribute to salt stress resistance in plants (An et al., 2014; de Lima et al., 2014; Byrt et al., 2018). However, we did not detect any significant difference in the pectin content between transgenic and WT plants (data not show). Instead, increased cellulose and decreased hemicelluloses were observed in the overexpression lines compared with WT plants under salt treatment condition (Supplementary Figure S10). This phenomenon is reminiscent of the Na+ sensitive mutant Atctl1, which affects the contents of cellulose and extractability of hemicelluloses (Hermans et al., 2011; Sanchez-Rodriguez et al., 2012). It is reported that mutation of AtCTL1 increased the Na+ influx rather than defects in Na+ efflux activity (Kwon et al., 2007). Therefore, the overexpression of the LcCHI2, a homolog of AtCTL1 (Supplementary Figure S2), may have an opposite effect and inhibit the Na+ influx, which could explain the deceased Na+ content in LcCHI2 transgenic plants.
Although cell wall metabolism is rationally correlated with salt stress, the exact mechanism is still largely unknown (Le Gall et al., 2015). Mutation of AtCES8 (cellulose synthase 8) or AtCSLD5 (cellulose synthase-like 5) resulted tolerance or hypersensitive to salt stress, respectively (Chen et al., 2005; Zhu et al., 2010). Moreover, mutation the chitinase-like gene AtCTL1 reduced the cellulose content and led to salt-sensitive of Arabidopsis (Kwon et al., 2007; Sanchez-Rodriguez et al., 2012). Compared with dicot, there has been no report on the modification of cell wall compound contributing to salt tolerance in grass. In this study, the overexpression of LiCHI2 in both tobacco and maize enhanced the salt and pathogen stress tolerances, suggesting similar roles of the chitinase in dicot and grass. Although the overall architectures of plant cell walls are similar in that they both consist of a network of cellulose fibers surrounded by a matrix of non-cellulosic polysaccharides, the types and abundance of non-cellulosic polysaccharides are significant different between grass and dicot cell walls (Vogel, 2008). It seems that cellulose represents the common component in plant cell walls and plays a pivotal role in regulating the salt stress. The changed cellulose content may regulate cross-linking and rigidity of the cell wall, which acts as a barrier for salt entrance or pathogen invasion.
Although a large number of alkali-responsive genes were reported from different plants, a few have been cloned and characterized (Zhao et al., 2016). Overexpression of the alkali-stress-inducable RMtATP6 and NADP-ME2 in rice and Arabidopsis enhanced the tolerance against osmotic, NaCl and Na2CO3 stresses (Zhang et al., 2006; Liu et al., 2007). Transgenic rice with PtNHA1 and PutNHX genes, which were cloned from an alkaline soil plant, increased tolerance of shoots to NaCl and roots to NaHCO3, respectively (Kobayashi et al., 2012). It is noted that all these studies were conducted in nutrient solution or agar plate in laboratory. In contrast, we presented the performance of the transgenic maize with natural alkaline soil or treated soil (Supplementary Figure S9). Moreover, the transgenic maize showed no penalty on growth and yield in field condition (Supplementary Table S1). Therefore, we proposed that overexpression of LcCHI2 represent a potential strategy for engineering biotic and alkaline resistance plants.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
XL, CW, and DH planned and designed the research, and wrote the manuscript. XL, YY, QL, and SD performed the tobacco experiments. XL, XJ, YJY, JG, NL, and YL performed the maize experiments. SH was in charge of the green house management.
Funding
This research was funded by the Science and Technology Innovation Program of Jilin Province (CXGC2017JQ014), the Ministry of Science and Technology if China (2016ZX08003005), the National Science Foundation of China (31772378 and 31771879), and Huazhong Agricultural University Scientific & Technological Self-innovation Foundation (2016RC005 and 2662017PY012).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Qiyun Li for the help in setting up the experiments of pathogen response assays of transgenic tobacco. We thank Dr. Bo Xu for the correction of the manuscript. This manuscript has been released as a pre-print at BiorXiv (Liu et al., 2019). We thank for Hongxiang Seed Industry Co., Ltd., provide maize commercial inbred line X923-1 and Y822.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00504/full#supplementary-material
FIGURE S1 | Construct maps of the recombinant plasmids for transformation in tobacco (A) and maize (B).
FIGURE S2 | Phylogenetic tree constructed with class II chitinase sequences retrieved by BLAST in the NCBI database.
FIGURE S3 | (A) Protein similarity of LcCHI2 with its homologs. (B) Alignment of deduced amino acid sequences of LcCHI2 with its homologs from other plant species.
FIGURE S4 | Morphology of the LcCHI2-overexpressing transgenic tobacco plants under normal growth condition.
FIGURE S5 | Drought (A), neutral salt (B), and alkaline salt (C) stress tolerance assays of wild-type and LcCHI2 overexpression tobacco plants grow in soil.
FIGURE S6 | MDA contents and relative electrical conductivity of wild-type and LcCHI2-transgenic tobacco plants under 200 mM NaCl treatment. ** are significantly different at P < 0.01 (LSD test).
FIGURE S7 | PCR detection of selection marker (A) and LcCHI2 genes (B) in transgenic maize plants.
FIGURE S8 | Southern blot of transgenic lines in maize.
FIGURE S9 | Growth performance (A), plant height (B), and SPAD value (C) of transgenic maize in normal and alkaline soils. Columns with different small letters are significantly different at P < 0.05.
FIGURE S10 | Cellulose and hemicelluloses contents in wild-type and LcCHI2-transgenic tobacco plants under 200 mM NaCl treatment.
TABLE S1 | Agronomic traits of transgenic maize grow in the field condition.
References
An, P., Li, X., Zheng, Y., Matsuura, A., Abe, J., Eneji, A. E., et al. (2014). Effects of NaCl on root growth and cell wall composition of two soya bean cultivars with contrasting salt tolerance. J. Agron. Crop Sci. 200, 212–218. doi: 10.1111/jac.12060
Brogue, K., Chet, I., Holliday, M., Cressman, R., Biddle, P., Knowlton, S., et al. (1991). Transgenic plants with enhanced resistance to the fungal pathogen Rhizoctonia solani. Science 254, 1194–1197. doi: 10.1126/science.254.5035.1194
Byrt, C. S., Munns, R., Burton, R. A., Gilliham, M., and Wege, S. (2018). Root cell wall solutions for crop plants in saline soils. Plant Sci. 269, 47–55. doi: 10.1016/j.plantsci.2017.12.012
Chen, R. D., Yu, L. X., Greer, A. F., Cheriti, H., and Tabaeizadeh, Z. (1994). Isolation of an osmotic stress-induced and abscisic-acid-induced gene encoding an acidic endochitinase from Lycopersicon-Chilense. Mol. Gen. Genet. 245, 195–202. doi: 10.1007/bf00283267
Chen, Z., Hong, X., Zhang, H., Wang, Y., Li, X., Zhu, J. K., et al. (2005). Disruption of the cellulose synthase gene, AtCesA8/IRX1, enhances drought and osmotic stress tolerance in Arabidopsis. Plant J. 43, 273–283. doi: 10.1111/j.1365-313X.2005.02452.x
de Lima, R. B., dos Santos, T. B., Vieira, L. G., de Lourdes Lucio Ferrarese, M., Ferrarese-Filho, O., Donatti, L., et al. (2014). Salt stress alters the cell wall polysaccharides and anatomy of coffee (Coffea arabica L.) leaf cells. Carbohydr. Polym. 112, 686–694. doi: 10.1016/j.carbpol.2014.06.042
Frame, B. R., Shou, H., Chikwamba, R. K., Zhang, Z., Xiang, C., Fonger, T. M., et al. (2002). Agrobacterium tumefaciens-mediated transformation of maize embryos using a standard binary vector system. Plant Physiol. 129, 13–22. doi: 10.1104/pp.000653
Hermans, C., Porco, S., Vandenbussche, F., Gille, S., De Pessemier, J., Van Der Straeten, D., et al. (2011). Dissecting the role of CHITINASE-LIKE1 in nitrate-dependent changes in root architecture. Plant Physiol. 157, 1313–1326. doi: 10.1104/pp.111.181461
Hoekema, A., Hirsh, P. R., Hooykaas, P. J., and Schilperoort, R. A. (1983). A binary plant vector strategy based on separation of vir-and T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature 303, 179–180. doi: 10.1038/303179a0
Hong, J. K., and Hwang, B. K. (2002). Induction by pathogen, salt and drought of a basic class II chitinase mRNA and its in situ localization in pepper (Capsicum annuum). Physiol. Plant. 114, 549–558. doi: 10.1034/j.1399-3054.2002.1140407.x
Hong, J. K., and Hwang, B. K. (2006). Promoter activation of pepper class II basic chitinase gene, CAChi2, and enhanced bacterial disease resistance and osmotic stress tolerance in the CAChi2-overexpressing Arabidopsis. Planta 223, 433–448. doi: 10.1007/s00425-005-0099-6
Jin, H., Plaha, P., Park, J. Y., Hong, C. P., Lee, I. S., Yang, Z. H., et al. (2006). Comparative EST profiles of leaf and root of Leymus chinensis, a xerophilous grass adapted to high pH sodic soil. Plant Sci. 170, 1081–1086. doi: 10.1016/j.plantsci.2006.01.002
Kesari, P., Patil, D. N., Kumar, P., Tomar, S., and Sharma, A. K. (2015). Structural and functional evolution of chitinase-like proteins from plants. Proteomics 15, 1693–1705. doi: 10.1002/pmic.201400421
Kikuchi, T., and Masuda, K. (2009). Class II chitinase accumulated in the bark tissue involves with the cold hardiness of shoot stems in highbush blueberry (Vaccinium corymbosum L.). Sci. Hortic. 120, 230–236. doi: 10.1016/j.scienta.2008.11.007
Kobayashi, S., Abe, N., Yoshida, K. T., Liu, S., and Takano, T. (2012). Molecular cloning and characterization of plasma membrane- and vacuolar-type Na(+)/H(+) antiporters of an alkaline-salt-tolerant monocot, Puccinellia tenuiflora. J. Plant Res. 125, 587–594. doi: 10.1007/s10265-012-0475-9
Kwon, Y., Kim, S. H., Jung, M. S., Kim, M. S., Oh, J. E., Ju, H. W., et al. (2007). Arabidopsis hot2 encodes an endochitinase-like protein that is essential for tolerance to heat, salt and drought stresses. Plant J. 49, 184–193. doi: 10.1111/j.1365-313X.2006.02950.x
Le Gall, H., Philippe, F., Domon, J. M., Gillet, F., Pelloux, J., and Rayon, C. (2015). Cell wall metabolism in response to abiotic stress. Plants (Basel) 4, 112–166. doi: 10.3390/plants4010112
Liu, S. K., Cheng, Y. X., Zhang, X. X., Guan, Q. J., Nishiuchi, S., Hase, K., et al. (2007). Expression of an NADP-malic enzyme gene in rice (Oryza sativa. L) is induced by environmental stresses; over-expression of the gene in Arabidopsis confers salt and osmotic stress tolerance. Plant Mol. Biol. 64, 49–58. doi: 10.1007/s11103-007-9133-3
Liu, X. G., Yu, Y., Liu, Q., Deng, S., Jin, X. B., Yin, Y., et al. (2019). A Na2CO3-responsive chitinase gene from Chinese wildrye improve pathogen resistance and saline-alkali stress tolerance in transgenic tobacco and maize. BioRxiv[Preprint]
Nakamura, T., Ishikawa, M., Nakatani, H., and Oda, A. (2008). Characterization of cold-responsive extracellular chitinase in bromegrass cell cultures and its relationship to antifreeze activity. Plant Physiol. 147, 391–401. doi: 10.1104/pp.106.081497
Nishizawa, Y., Nishio, Z., Nakazono, K., Soma, M., Nakajima, E., Ugaki, M., et al. (1999). Enhanced resistance to blast (Magnaporthe grisea) in transgenic Japonica rice by constitutive expression of rice chitinase. Theor. Appl. Genet. 99, 383–390. doi: 10.1007/s001220051248
O’Brien, M., and Colwell, R. R. (1987). A rapid test chitinase activity that uses 4-methylumbelliferyl-N-acetyl-β-D-glucosaminide. Appl. Environ. Microb. 53, 1718–1720. doi: 10.1128/aem.53.7.1718-1720.1987
Oldach, K. H., Becker, D., and Lorz, H. (2001). Heterologous expression of genes mediating enhanced fungal resistance in transgenic wheat. Mol. Plant Microbe Interact. 14, 832–838. doi: 10.1094/MPMI.2001.14.7.832
Patil, R. S., Ghormade, V., and Deshpande, M. V. (2000). Chitinolytic enzymes: an exploration. Enzyme Microb. Tech. 26, 473–483. doi: 10.1016/S0141-0229(00)00134-4
Sanchez-Rodriguez, C., Bauer, S., Hematy, K., Saxe, F., Ibanez, A. B., Vodermaier, V., et al. (2012). Chitinase-like1/pom-pom1 and its homolog CTL2 are glucan-interacting proteins important for cellulose biosynthesis in Arabidopsis. Plant Cell 24, 589–607. doi: 10.1105/tpc.111.094672
Scheler, B., Schnepf, V., Galgenmuller, C., Ranf, S., and Huckelhoven, R. (2016). Barley disease susceptibility factor RACB acts in epidermal cell polarity and positioning of the nucleus. J. Exp. Bot. 67, 3263–3275. doi: 10.1093/jxb/erw141
Seo, P. J., Lee, A. K., Xiang, F. N., and Park, C. M. (2008). Molecular and functional profiling of Arabidopsis pathogenesis-related genes: insights into their roles in salt response of seed germination. Plant Cell Physiol. 49, 334–344. doi: 10.1093/pcp/pcn011
Shin, S. Y., Mackintosh, C. A., Lewis, J., Heinen, S. J., Radmer, L., Dill-Macky, R., et al. (2008). Transgenic wheat expressing a barley class II chitinase gene has enhanced resistance against Fusarium graminearum. J. Exp. Bot. 59, 2371–2378. doi: 10.1093/jxb/ern103
Tabaeizadeh, Z., Agharbaoui, Z., Harrak, H., and Poysa, V. (1999). Transgenic tomato plants expressing a Lycopersicon chilense chitinase gene demonstrate improved resistance to Verticillium dahliae race 2. Plant Cell Rep. 19, 197–202. doi: 10.1007/s002990050733
Takahashi, W., Fujimori, M., Miura, Y., Komatsu, T., Nishizawa, Y., Hibi, T., et al. (2005). Increased resistance to crown rust disease in transgenic Italian ryegrass (Lolium multiflorum Lam.) expressing the rice chitinase gene. Plant Cell Rep. 23, 811–818. doi: 10.1007/s00299-004-0900-1
van Hengel, A. J., Tadesse, Z., Immerzeel, P., Schols, H., van Kammen, A., and de Vries, S. C. (2001). N-acetylglucosamine and glucosamine-containing arabinogalactan proteins control somatic embryogenesis. Plant Physiol. 125, 1880–1890. doi: 10.1104/pp.125.4.1880
van Loon, L. C., Rep, M., and Pieterse, C. M. (2006). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. doi: 10.1146/annurev.phyto.44.070505.143425
Verburg, J. G., and Huynh, Q. K. (1991). Purification and characterization of an antifungal chitinase from Arabidopsis thaliana. Plant Physiol. 95, 450–455. doi: 10.1104/pp.95.2.450
Vogel, J. (2008). Unique aspects of the grass cell wall. Curr. Opin. Plant Biol. 11, 301–307. doi: 10.1016/j.pbi.2008.03.002
Wang, Z. Q., Zhu, S. Q., Yu, R. P., Li, L., and Shan, G. Z. (1993). The Salinealkali Soil of China (In Chinese). Beijin: The Science Press.
Yamamoto, T., Iketani, H., Ieki, H., Nishizawa, Y., Notsuka, K., Hibi, T., et al. (2000). Transgenic grapevine plants expressing a rice chitinase with enhanced resistance to fungal pathogens. Plant Cell Rep. 19, 639–646. doi: 10.1007/s002999900174
Yeh, S., Moffatt, B. A., Griffith, M., Xiong, F., Yang, D. S., Wiseman, S. B., et al. (2000). Chitinase genes responsive to cold encode antifreeze proteins in winter cereals. Plant Physiol. 124, 1251–1264. doi: 10.1104/pp.124.3.1251
Zhang, X. X., Takano, T., and Liu, S. K. (2006). Identification of a mitochondrial ATP synthase small subunit gene (RMtATP6) expressed in response to salts and osmotic stresses in rice (Oryza sativa L.). J. Exp. Bot. 57, 193–200. doi: 10.1093/jxb/erj025
Zhao, Q., Suo, J. W., Chen, S. X., Jin, Y. D., Ma, X. L., Yin, Z. P., et al. (2016). Na2CO3-responsive mechanisms in halophyte Puccinellia tenuiflora roots revealed by physiological and proteomic analyses. Sci. Rep. 6:32717. doi: 10.1038/Srep32717
Zhong, R., Kays, S. J., Schroeder, B. P., and Ye, Z. H. (2002). Mutation of a chitinase-like gene causes ectopic deposition of lignin, aberrant cell shapes, and overproduction of ethylene. Plant Cell 14, 165–179. doi: 10.1105/tpc.010278
Keywords: chitinase, saline-alkali stress, biotic stress, Leymus chinensis, tobacco, maize
Citation: Liu X, Yu Y, Liu Q, Deng S, Jin X, Yin Y, Guo J, Li N, Liu Y, Han S, Wang C and Hao D (2020) A Na2CO3-Responsive Chitinase Gene From Leymus chinensis Improve Pathogen Resistance and Saline-Alkali Stress Tolerance in Transgenic Tobacco and Maize. Front. Plant Sci. 11:504. doi: 10.3389/fpls.2020.00504
Received: 02 September 2019; Accepted: 03 April 2020;
Published: 28 April 2020.
Edited by:
Zeng-Yu Wang, Qingdao Agricultural University, ChinaReviewed by:
Kan Wang, Iowa State University, United StatesWricha Tyagi, Central Agricultural University, India
Copyright © 2020 Liu, Yu, Liu, Deng, Jin, Yin, Guo, Li, Liu, Han, Wang and Hao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chuang Wang, Y2h1YW5nLndhbmdAbWFpbC5oemF1LmVkdS5jbg==; Dongyun Hao, ZHloYW9AY2phYXMuY29t
†These authors have contributed equally to this work
 Xiangguo Liu1†
Xiangguo Liu1† Ying Yu
Ying Yu Suren Deng
Suren Deng Chuang Wang
Chuang Wang