- 1Key Laboratory of Fujian Province for Agroecological Process and Safety Monitoring, College of Life Science, Fujian Agriculture and Forestry University, Fuzhou, China
- 2Key Laboratory for Genetics, Breeding and Multiple Utilization of Crops, Ministry of Education, College of Crop Sciences, Fujian Agriculture and Forestry University, Fuzhou, China
- 3Institute of Biotechnology and Genetic Engineering, The University of Agriculture Peshawar, Peshawar, Pakistan
In conventional tea plantations, a large amount of pruned material returns to the soil surface, putting a high quantity of polyphenols into the soil. The accumulation of active allelochemicals in the tea rhizosphere and subsequent shift in beneficial microbes may be the cause of acidification, soil sickness, and regeneration problem, which may be attributed to hindrance of plant growth, development, and low yield in long-term monoculture tea plantation. However, the role of pruning leaf litter in soil sickness under consecutive tea monoculture is unclear. Here, we investigated soil samples taken from conventional tea gardens of different ages (2, 15, and 30 years) and under the effect of regular pruning. Different approaches including liquid chromatography–mass spectrometry (LC-MS) analysis of the leaf litter, metagenomic study of root-associated bacterial communities, and in vitro interaction of polyphenols with selected bacteria were applied to understand the effect of leaf litter-derived polyphenols on the composition and structure of the tea rhizosphere microbial community. Our results indicated that each pruning practice returns a large amount of leaf litter to each tea garden. LC-MS results showed that leaf litter leads to the accumulation of various allelochemicals in the tea rhizosphere, including epigallocatechin gallate, epigallocatechin, epicatechin gallate, catechin, and epicatechin with increasing age of the tea plantation. Meanwhile, in the tea garden grown consecutively for 30 years (30-Y), the phenol oxidase and peroxidase activities increased significantly. Pyrosequencing identified Burkholderia and Pseudomonas as the dominant genera, while plant growth-promoting bacteria, especially Bacillus, Prevotella, and Sphingomonas, were significantly reduced in the long-term tea plantation. The qPCR results of 30-Y soil confirmed that the copy numbers of bacterial genes per gram of the rhizosphere soil were significantly reduced, while that of Pseudomonas increased significantly. In vitro study showed that the growth of catechin-degrading bacteria (e.g., Pseudomonas) increased and plant-promoting bacteria (e.g., Bacillus) decreased significantly with increasing concentration of these allelochemicals. Furthermore, in vitro interaction showed a 0.36-fold decrease in the pH of the broth after 72 h with the catechin degradation. In summary, the increase of Pseudomonas and Burkholderia in the 30-Y garden was found to be associated with the accumulation of catechin substrates. In response to the long-term monoculture of tea, the variable soil pH along with the litter distribution negatively affect the population of plant growth-promoting bacteria (e.g., Sphingomonas, Bacillus, and Prevotella). Current research suggests that the removal of pruned branches from tea gardens can prevent soil sickness and may lead to sustainable tea production.
Introduction
Plant soil feedbacks can alter the composition and structure of the soil microbial community and nutrient homeostasis as a result of all the interactions between plants and soil organisms, affecting soil fertility and plant growth (Kaur et al., 2009; Bever et al., 2010; van de Voorde et al., 2012; Baxendale et al., 2014). Soil disease or replanting disease is plant-soil negative feedback caused by consecutive planting of a single crop or its related species on the same field, resulting in a reduction in crop yield and quality (Muller, 1966; Huang et al., 2013; Zhao et al., 2015, 2016; Wu L. et al., 2016). Long-term monoculture not only impedes the growth and production of many annual crops, trees, and shrubs in orchards but also causes replanting and regeneration problems (Canals et al., 2005). Therefore, it is pertinent to understand the mechanisms underlying soil sickness associated with long-term monoculture practice, to explore the allelochemical interaction with soil microbiota, and to provide a solution for maintaining a sustainable agro-ecosystem.
Degradation of autotoxins or allelochemicals by microorganisms has been reported in previous studies (Bever et al., 2010; Weidenhamer et al., 2013). Several cinnamic and benzoic acids were initially detected in the Cecil Ap horizon soil, and after a few days, they were not detected in amended soil (Blum et al., 2000). Phenolics, being the most abundant plant metabolites, are thought to control the rates of soil organic matter decomposition and can be applied as a tool to evaluate the soil dynamics and ecosystem functioning (Min et al., 2015). Hence, the effects of soil microbiota are critical to the fate of plant phenolic compounds, including other potential allelochemicals found in the soil (Bever et al., 2010; Ehlers, 2011).
Polyphenols enter the soil in the form of leachates from above plant parts as plant residues and litter (Hättenschwiler and Vitousek, 2000). Contributions related to these input pathways, mainly underground flows, have a limited understanding. In most of the less disturbed terrestrial ecosystems, plant growth and net primary productivity depend on recycled nutrient availability, while external nutrient inputs contribute little to the total requirement. The rate-limiting steps in the nutrient cycle, such as climate, substrate quality (litter), and decomposing microbes, are decisive at influencing soil microbial mineralization (Heal, 1997). Polyphenols influence soil microbial activity and soil physicochemical properties, indicating interactions with nutrient cycling, thereby modifying the flux and pool of soil nutrients available to microbes and/or plants (Hättenschwiler and Vitousek, 2000). Several studies suggest that through specific mechanisms, soil macrofauna increases the biodegradation and mineralization of soil organic matter (Wardle and Lavelle, 1997) and high concentrations of polyphenols could limit such fauna in terms of activity and abundance (Neuhauser, 1978). Consequently, the direct effect of polyphenols on soil fauna is hard to explain owing to the complexity of soil food webs and the co-variability of other compounds.
Tea is an important economic crop, widely cultivated in Southeast Asia and China (Chen and Lin, 2015) and contains several phenolic compounds throughout the tea plant, especially in the leaves, accounting for 18–34% of the dry weight of the leaves (Wan, 2003). In tea plantation systems, the conventional management approach, especially pruning, is employed twice a year in terms of keeping tea bushes in the best shape, improving tea quality, increasing yield, and even inhibiting diseases and pests (Yilmaz et al., 2004; Maudu et al., 2010). The trimmed tea leaves and branches are rich in nutrients and are often amended into tea gardens to improve the soil physicochemical properties, soil organic matter, and nutrient availability (Weeraratna et al., 1977). However, some studies have demonstrated that the decomposition of accumulated residues and litter produces allelochemicals, mainly phenolic compounds, flavonoid, and alkaloids, which, in turn, inhibit the microbial activities in the rhizospheric soil. The indirect allelopathy of pruned tea leaves, especially polyphenols on soil sickness in long-term tea cropping systems, needs further consideration. The objectives of this study were to inspect the impact of tea litter and its polyphenols on (a) root-related bacterial communities in terms of structure and composition, (b) ratooning or regeneration problems related to soil sickness, and (c) growth and quality parameters of tea plants in response to continuous tea cultivation. Assuming that polyphenols represent a possible allelochemical substance, they regulate the tea soil bacterial community feedback processes, which leads to the progressive imbalance during long-term tea monocropping. In addition, changes in microbial community structure and composition play an important role in soil sickness as compared to the direct allelopathy.
Materials and Methods
Site Overview
Current experiment on tea plantation was carried out at Fujian Agriculture and Forestry University tea fields, Fuzhou City, Fujian Province, China (latitude: 26°0509.60′′ N; longitude: 119°1403.60′′ E) (27°43′ N, 118°72′ E). The area has a subtropical monsoon climate, with average annual temperature and rainfall (20–25°C, and around 900–1362 mm, respectively).
Soil Sampling
Rhizosphere soil along with roots was taken from different aged tea gardens (2, 15, and 30 years) and bulk soil (CK) from the same fields was used as control. The location, agronomic management practices, and environmental conditions of the tea gardens were the same. To overcome error began by spatial heterogeneity, five randomly taken soil samples from 15 sampling locations were combined into one replication, and three replications were carried out for each sample. In order to deliver soil samples to the laboratory, all soil samples were immediately stored in a sterile icebox (Lin et al., 2013). For the analysis of soil enzymes and microbes, a part of each soil sample and roots were stored at −80°C, and the remainder was air-dried for physicochemical attributes. With little modification in the method proposed by Edwards et al. (2015), we determined the microbial fauna in 2-Y, 30-Y, and CK soil. Shortly, the bacterial communities were obtained from tea root compartments, e.g., RS, rhizosphere (soil firmly adhered to the root surface); RP, rhizoplane (suite of microbes present on the root surface by sonication); and ES, endosphere (interior of the same plant roots after sonication).
Quality Parameters, Growth Index, and Yield Determination
Theophylline (TPY), theanine (TNN), and the total polyphenol (TPP) content of the uppermost leaves taken from three plants grown in each tea garden, i.e., 2-Y, 15-Y, and 30-Y, was determined by the method proposed by Peng et al. (2008) for quality determination. To examine the total chlorophyll content of the third leaf, eight plants were randomly selected from each garden and values were recorded using an instrument (SPAD-502 Plus). Further, using the US portable CO2 gas analyzer (CID-301), the net photosynthetic rate (Pn) of the third leaf was measured. Three replicates for each sample were taken to perform Pn. One hundred buds’ weight (fresh and dry) was estimated in grams for each of three biological replications.
Soil Enzyme Activities Analysis
With a slight modification, the activity of soil polyphenol oxidase and peroxidase was determined by the method previously adopted (Perucci et al., 2000; Saiya-Cork et al., 2002; Xu et al., 2015). Briefly, 1 g of soil was added to a 100-ml flask containing 10 ml of 1% pyrogallol before incubation at 300°C for 2 h, and then 35 ml of ether was added into the flask and then shaken for 15 min on a thermostat oscillator at 250°C. Moreover, peroxidase activity was determined using the same procedure along with the addition of 2 ml of a 0.5% H2O2 solution, and the final absorbance was recorded at 430 nm.
Leaf Litter Biomass Determination and Identification and Quantification of Allelochemicals
Standard Materials and Their Calibration Curves
In order to quantify and identify concentration of allelochemicals in leaf litter samples, ≥98% pure catechin, epigallocatechin (EGC), epicatechin gallate (ECG), epicatechin (EC), epigallocatechin gallate (EGCG), taxifolin (TF), and protocatechuic acid (PCA) were purchased as external standards from Cayman Chemical (1180 E. Ellsworth Road Ann Arbor, MI, United States). Formic acid and LC-MS grade methanol were also purchased from Sigma-Aldrich (St. Louis, MO, United States). For a stock solution preparation, 1 mg of every standard was suspended in 1 ml of 99.9% methanol and 0.1% by volume of formic acid. Serial dilutions were made using the same solvent as the stock solution, with a dilution range of 1.25–10 μg/ml in order to obtain a calibration curve. Each solution was distinctly injected with a 10-μl aliquot for HPLC-ESI-MS analysis. With all correlation coefficients > 0.998, excellent linearity was achieved within the calibration range (Supplementary Figures S1, S2).
Allelochemicals Determination From Leaf Litters
The leaf litter biomass that comes on the soil surface from each tea plant per pruning was determined in grams (Supplementary Figure S3). Leaf litters were collected randomly from three tea plants in each tea garden and were weighed by a digital balance. By adopting the method of Wang et al. (2013) with slight modifications, the allelochemicals concentration in leaf litter was determined by HPLC-ESI-MS. We took 5 g of the litter and fermented it for 3 days in 15 ml of a solution (containing 50% methanol and 0.1% acetic acid) at room temperature (25°C), and after three times sonication, it was vortexed at 150 rpm for 24 h. By vaporization at ultra-pressure, the methanol extract was air-dried and the crude extract was passed through a C-18 column (open column). After centrifugation at 4500 rpm for 10 min, the supernatant was then shifted to a sample collection vessel for liquid chromatography (LC).
The Chromatographic Conditions for Allelochemical Determination
Following this, HPLC–ESI–MS was carried out using a T3 RP-18 column (100 × 2.1 mm; 5 μm; Waters, Milford, MA, United States), eluted with buffer A (0.1% acetic acid) and buffer B (100% methanol) at a flow rate of 300 μl/min at 25°C. Initially, the column was eluted with 95% buffer B, followed by a linear increase in buffer A to 35% from 0 to 10 min, and further maintained in 90% buffer A until 10.50 min. Then, a linear increase in buffer B to 95% was maintained. Finally, the column was maintained in 95% buffer B for up to 19 min. The total time for running one sample was 19 min. The negative ionization mode was selected to perform mass spectrometry at a temperature of 100°C, and ion scans were carried out at low-energy collision (20 eV) using nitrogen as the collision gas. All the data from HPLC-ESI-MS were processed to determine the mean concentrations of the selected catechin compounds in each sample, by using Bruker Daltonics Data analysis software (version 4.0).
The Metagenomic Analysis of the Root-Associated Bacterial Communities
Soil DNA Extraction
Fresh soil samples were passed through a 2-mm mesh sieve and then 0.5 g was taken in 2-ml Eppendorf tubes for DNA extraction. DNA was extracted using a BioFast Soil Genomic DNA Extraction Kit (Bioer Technology Co., Ltd., Hanzhou, China) according to the manufacturer’s instructions. In order to estimate the quality and concentration of soil DNA, NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA, United States) was used. DNA was diluted to 1 ng/μl using sterile water accordingly to concentration.
PCR Amplification
Primers 515F/806R with the barcode were used in order to amplify the distinct region of 16S rRNA gene (16S V4). All PCR reactions were conducted with Phusion® High-Fidelity PCR Master Mix (New England Biolabs). PCR conditions were (95°C for 3 min, followed by 35 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 45 s, with a final extension at 72°C for 10 min) (GeneAmp 9700, ABI, United States). PCR reactions were carried out in triplicate in 20-μl mixture having 2 μl of 2.5 mM deoxyribonucleoside triphosphate (dNTPs), 4 μl of 5 × Fast Pfu buffer, 0.4 μl of Fast Pfu polymerase, 0.4 μl of every primer (5 μM), and template DNA (10 ng) (TransGen catalogue no. AP221–02). PCR products were purified with Qiagen Gel Extraction Kit (Qiagen, Germany).
Library Preparation and Illumina Hi Sequencing
Purified PCR products were sent to Novogene Bioinformatics Technology Co., Ltd., (Beijing, China) for high-throughput sequencing. Sequencing libraries were generated using TruSeq® DNA PCR-Free Sample Preparation Kit (Illumina, United States) following manufacturer’s recommendations and index codes were added. The library quality was assessed on the Qubit@ 2.0 Fluorometer (Thermo Scientific) and Agilent Bioanalyzer 2100 system. At last, the library was sequenced on an IlluminaHiSeq2500 platform and 250-bp paired-end reads were generated.
Processing and Analyzing of Data
Paired-End Reads Assembly and Quality Control
Paired-end reads was assigned to samples based on their unique barcode and truncated by cutting off the barcode and primer sequence. Paired-end reads were merged using FLASH (V1.2.7)1 (Magoč and Salzberg, 2011), and the splicing sequences were called raw tags. Quality filtering on the raw tags were performed under specific filtering conditions to obtain the high-quality clean tags (Bokulich et al., 2013), according to the QIIME(V1.7.0)2 (Caporaso et al., 2010). The tags were compared with the reference database(Unite Database)3 using UCHIME algorithm (UCHIME Algorithm4) (Edgar et al., 2011) to detect chimera sequences, and then the chimera sequences were removed (Haas et al., 2011).
OTU Cluster and Species Annotation
Sequence analysis was performed by Uparse software (Uparse v7.0.1001)5 (Edgar, 2013). Sequences with ≥97% similarity were assigned to the same OTUs. Representative sequence for each OTU was screened for further annotation. For each representative sequence, the Unite Database6 (Kõljalg et al., 2013). was used basing on Blast algorithm, which was calculated by QIIME software (Version 1.7.0)7 to annotate taxonomic information.
Heat Map and Graph Construction
Heat map was preceded by ggplot2 package in R software (Version 2.15.3).
Quantitative PCR Analysis of Total Bacteria in Ratooning Monoculture Tea Soil
A quantitative PCR (qPCR) assay was conducted to determine the size of bacterial population in the rhizosphere soil of different aged tea plantations. The Universal primer set Eub338/Eub518 was used to estimate the bacterial community size. qPCR was performed according to the method of Wu H. et al. (2016).
qPCR Analysis of Pseudomonas and Bacillus Population in Ratooning Monoculture Tea Soil
A qPCR assay using the primer sets Psf/Psr (Chen et al., 2017) was also conducted to determine the copy number of Pseudomonas in tea rhizosphere soil after different years of monoculture. However, for Bacillus genus, we used the method of Wu H. et al. (2016). The 15-μl PCR reaction contained 7.5 μl of 2 × SYBR green I Super Real Premix (TransGen, Beijing, China), 0.5 μl of each primer (10 μM), 1 μl of 20 ng/μl template DNA, and 5.5 μl of RNase-free H2O. Serial dilutions of plasmid DNA were set as standard curve.
In vitro Interactions of Allelochemicals With Selected Model Bacteria
Preparation of LB Medium
LB powder, 5 g, was mixed in 200 ml of ultrapure water in a 250-ml Erlenmeyer flask, covered the top with aluminum foil, and autoclaved at 121°C for 15 min.
For LB agar plates preparation, 3 g (1.5%) of bacteriological agar was added to 200 ml of LB liquid medium (5 g broth powder in 200 ml of ddH2O) in a 2500 ml Erlenmeyer flask and autoclaved at 121°C for 15 min. After autoclaving, when medium was cooled to ∼50°C, 200 μl of pimaricin antifungal (0.06 g/1 ml methanol) was added to 200 ml of media. The LB agar media was poured into the petri plates.
Preparation of Soil Serial Dilution
Five grams of soil was added to 45 ml of ddH2O in a 50-ml Erlenmeyer flask. The soil samples were diluted up to 10–3 times. Soil suspension (60 μl) of 10–2 and 10–3 dilutions was subjected to each hard-nutrient agar plate surface. The soil suspension was spread until it was equally distributed on hard agar plate. Last, the plates were incubated at 30°C for 24–72 h, and the colonies that show dark zone surrounding them were separated from a mixed bacterial community in the culture.
Sub-Culturing of Bacteria
Already prepared LB agar plates were used to get each purified bacterial colony. For this, divide the plates under sides by drawing equal numbers of squares to each plate and label it by giving specific numbers and then pick each colony carefully by sterilizing the pipette tip and attaching it to each label square. The plates were incubated at 30°C for 24–72 h again. These colonies represent each bacterial strain.
Enrichment and Preservation of Bacteria
The sterilized LB liquid broth was prepared and 5-ml broth solutions were distributed in sterilized 5-ml tubes. After that, one colony into each tube was suspended and incubated at 30°C for 72 h. To preserve each bacterium, put 1 ml of bacterial suspension in a sterilized Eppendorf tube and add an equal amount of glycerol (1:1) to it and preserve these colonies at −20°C for further use. DNA was extracted from each labeled bacterial suspension. The DNA were sent to the company for 16s RNA sequencing to identify key microbes.
The Influence of Identified Allelochemicals on the Growth of Selected Model Microbes
The LB Liquid broth was diluted six times and 5 ml was distributed in each glass tube; all tubes were sterilized at 121°C for 20 min. After autoclaving, the broth was amended with an appropriate amount of identified allelochemical stock solution that was filtered through a 0.22-μm filtration membrane. The final concentrations were 0, 1.25, 2.5, 5, and 10 μg/ml, respectively, for each allelochemical. The control was set in the same amount of ddH2O instead of allelochemical. Each treatment was repeated three times. Fifty microliters of each labeled bacteria was added to each tube, which has been already activated. All tubes were placed in a thermostatic shaker at 30°C and 200 rpm for 72 h. Further, 200 μl of bacterial culture was taken in a 96-hole enzyme label plate (Thermo Scientific Multiskan Mk3, Shanghai, China). To obtain the appropriate bacterial growth, the OD at 600 nm was regularly checked (Wu H. et al., 2016).
Biotransformation of Catechin and Its Effect on Soil pH
The LB liquid broth was diluted six times to minimize LB nutrient effect on bacterial growth and 5 ml was distributed in each glass tube. All the tubes were sterilized at 121°C for 20 min. After precooling, 50 μl of each labeled bacteria was added to each tube that has been already activated. The broth was amended with an appropriate amount of EC stock solution that was filtered through a 0.22-μm filtration membrane. After growing bacteria, an appropriate amount of broth was centrifuged at 13,000 rpm for 5 min at 4°C. The supernatant was taken for pH determination and for identification of their metabolites by LC-ESI-MS at 24, 48, 72, 96, and 120 h, respectively (Wang et al., 2013).
Statistical Analysis
All the experiments included three replications and repeated at least two times. The effects of the different treatments were analyzed by LSD test using SPSS 19.0, and the graphs were displayed using GraphPad Prism 5 software.
Results
The Performance of Tea Plants in Consecutively Ratooned Monoculture Soil
Tea plants grown in soil taken from different tea plantations (15-Y and 30-Y) showed weak growth, wilting, chlorosis, and regeneration obstacles compared to tea plants grown in the recently established tea garden (2-Y) soil (Figure 1). In addition, compared to the 2-Y garden, tea quality parameters, especially theophylline (TPY), total polyphenols (TPP), theanine (TNN), and total amino acids (TAA), were significantly lower in old gardens (15-Y and 30-Y) as shown (Figures 2A–D). Regarding physiological and growth parameters, continuous tea cropping significantly reduced net photosynthetic rate (Pn), new tea sprout’s length, and chlorophyll content of the third leaf (Figures 3A–D). Supplementary Table S1 shows the tea yield obtained from the tea gardens with different planting histories. In comparison with 2-Y gardens, the old gardens (15-Y and 30-Y) significantly reduced 100 buds fresh and dry weight (Supplementary Table S1). These results indicate that tea quality and productivity are negatively affected by long-term tea monoculture.

Figure 1. Replanting problems in continuous monoculture tea garden soil. 2-Y, 15-Y, and 30-Y indicate tea garden soil in which tea was planted continuously for different years (2, 15, and 30 years).
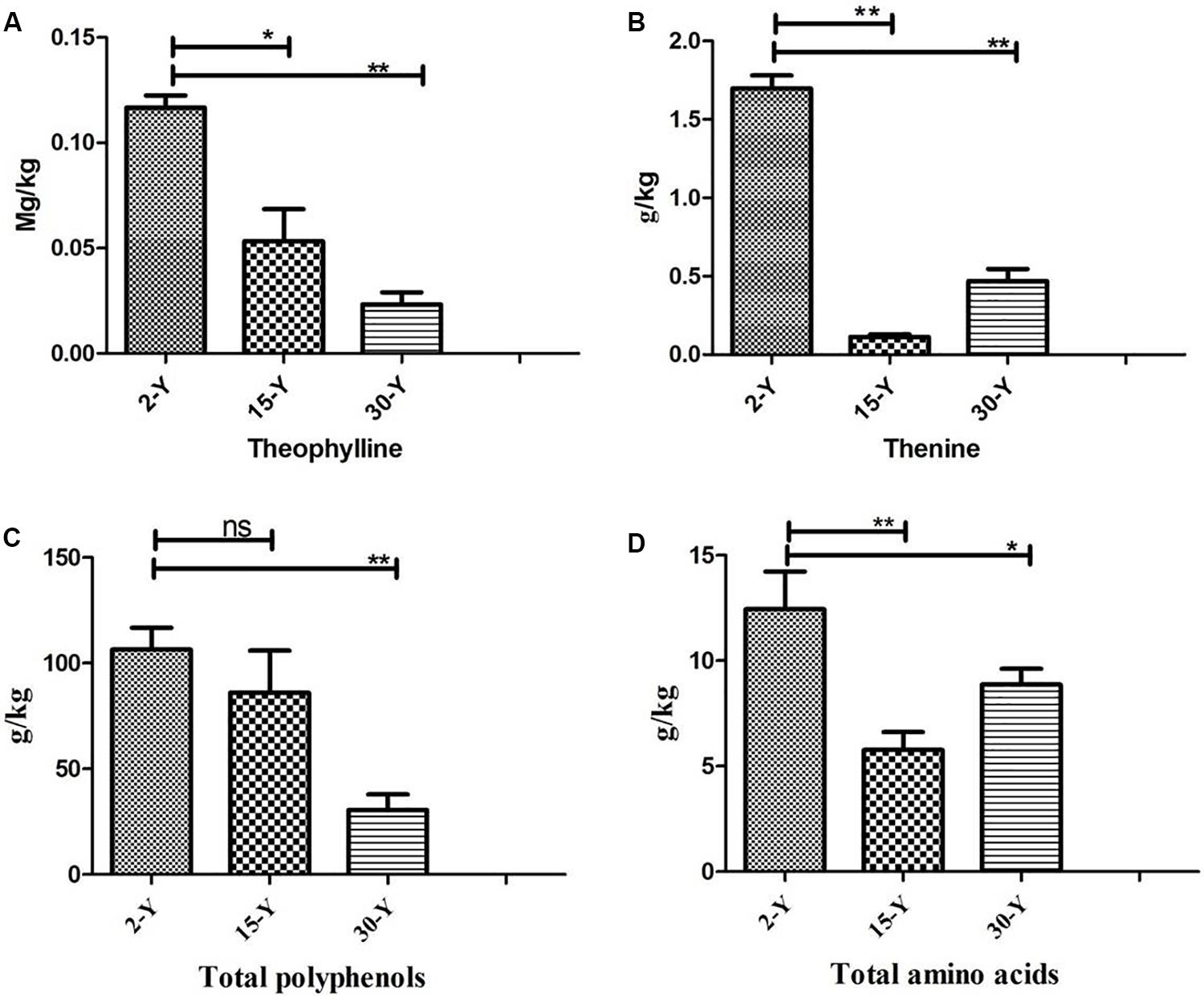
Figure 2. Quality parameters of tea leaves from tea plantations of different ages. (A) Theophylline, (B) theanine, (C) total polyphenols, and (D) total free amino acids. Stars in the column show significant differences (LSD test, P < 0.05, n = 3). 2-Y, 15-Y, and 30-Y indicate tea gardens planted continuously for different years (2, 15, and 30).
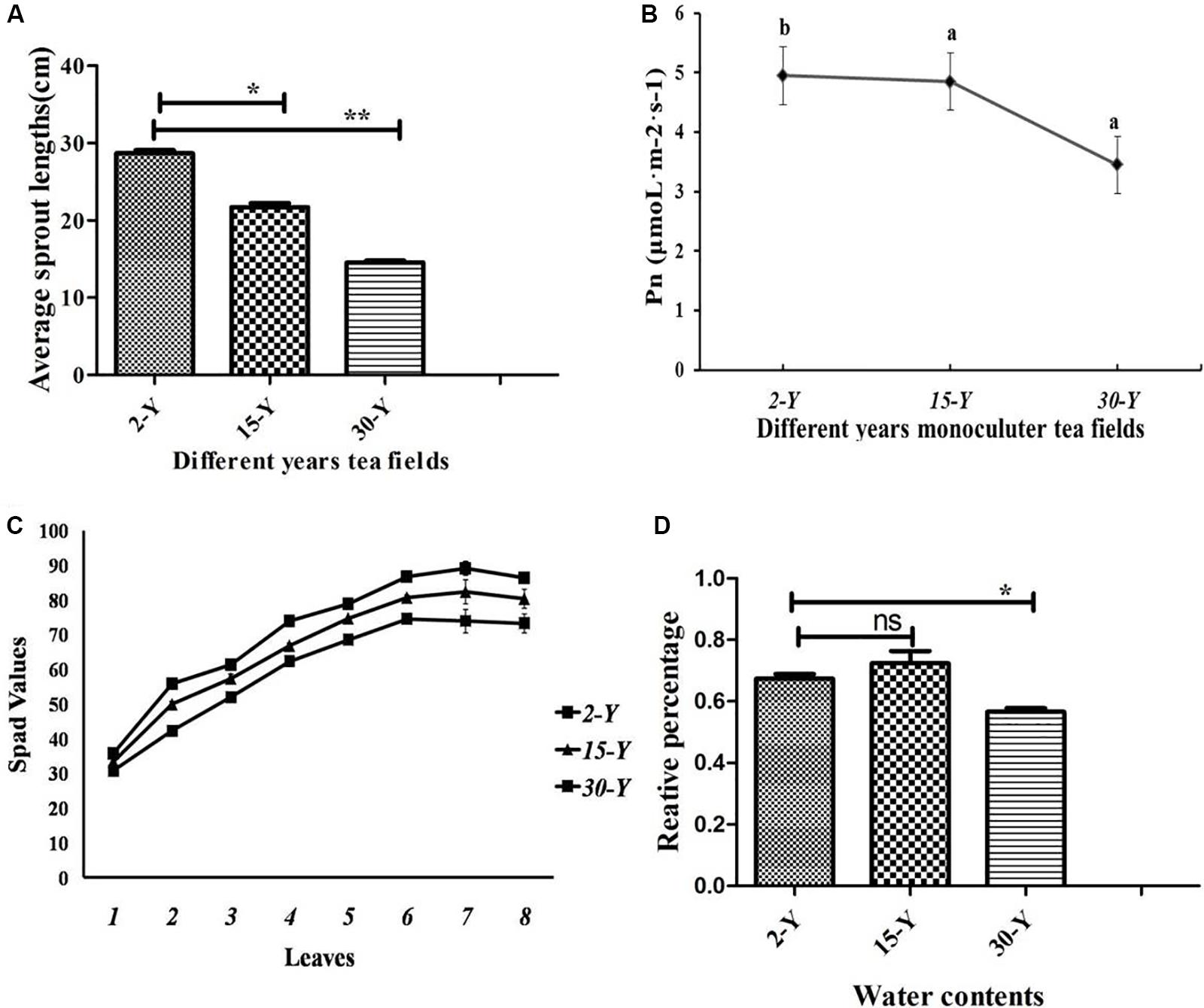
Figure 3. Physiological characteristics of tea leaves. (A) Length of new tea sprouts (P < 0.05, n = 5), (B) photosynthetic rate (Pn) of the third tea leaf in young sprout (P < 0.05, n = 3), (C) content of tea leaf chlorophyll with young sprouts from bottom to top (P < 0.05, n = 8), (D) water contents in five leaves. Stars in the column show significant differences (LSD test, P < 0.05, n = 5) under different fields. 2-Y, 15-Y, and 30-Y indicate tea gardens planted continuously for different years (2, 15, and 30).
Analysis of Soil Enzyme Activities in Different Age Monoculture Tea Plantations
The activity of phenol oxidase peroxidase were promoted by 13.78, 14.92, and 51.52% in 2-, 15-, and 30-Y tea plantations, respectively, as compared to CK. Likewise, the activity of peroxidase was sharply increased by 11.58, 10.12, and 17.88%, respectively, in 15- and 30-Y tea plantations as compared to CK (Supplementary Table S2).
Leaf Litter Biomass Input Into Tea Gardens and Analysis of Their Allelochemicals
Our results indicated that in each pruning turn, about 615, 838, and 969.67 g/plant leaf litters were returns into 2-Y, 15-Y, and 30-Y garden, respectively (Supplementary Figure S3). Five compounds including EGC, EGCG, EC, catechin (±C), and ECG, were identified in the leaf litters in both the newly planted tea gardens (2-Y) and the continuously monocultured tea gardens (15-Y and 30-Y) (Figure 4). Moreover, in the continuous monoculture tea gardens (15-Y and 30-Y tea gardens) the input of EGC, ±C, EC, EGCG, and ECG allelochemicals were high as compared to young tea plantations (2-Y).
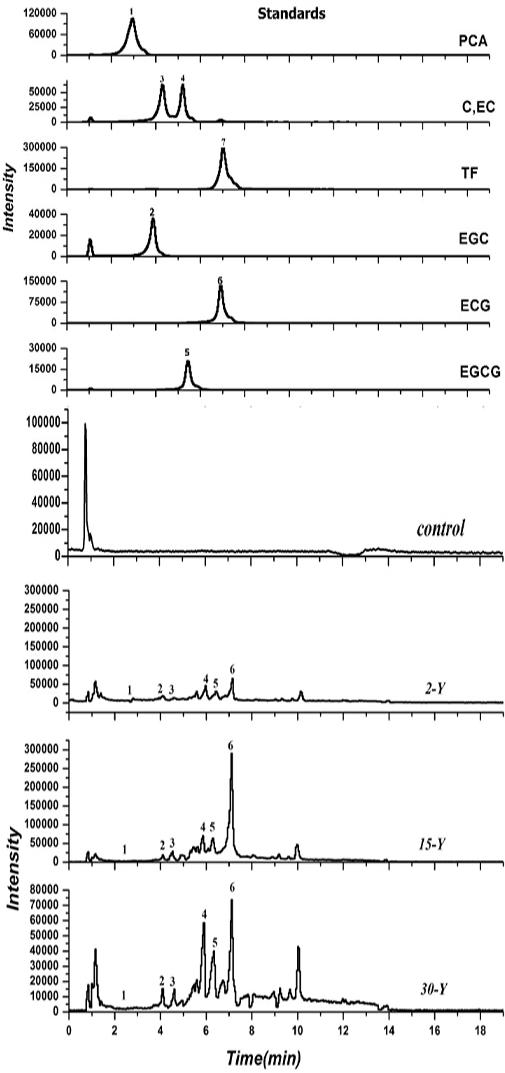
Figure 4. HPLC–ESI–MS spectra of catechins in leaf litters collected. (1) Represents protocatechuic acid (PCA) with a retention time of 3.08–3.12 min; (2) represents epigallocatechin (EGC) with a retention time of 4.07–4.11 min; (3) is catechin (±C) with a retention time of 4.52–4.55 min; (4) is epicatechin (EC) with a retention time of 5.42–5.44 min; (5) represents epigallocatechin-3-gallate (EGCG) with a retention time of 5.62–5.72 min; (6) represents epicatechin-3-gallate (ECG) with a retention time of 7.11–7.13 min; and (7) represents taxifolin (TF) with a retention time of 7.32–7.35 min. Control, 2-Y, 15-Y, and 30-Y indicate tea garden leaf litters planted continuously for different years (0, 2, 15, and 30).
Response of Bacterial Genera Under Continuous Monoculture Soil
Heat map analysis displayed that Acidobacteria (Candidatus_koribacter) and Proteobacteria (Rhodocyclus, Agrobacterium, Caulobacter, Hydrogenophaga, and Methylibium) were the dominant bacterial phyla in CK (Figure 5A). The genera (e.g., Pseudomonas, Burkholderia, Salinospora, and Helicobacter) were dominant in RS30, whereas Devosia, Coprococcus, Opitutus, Sphingomonas, Prevotella, Allobaculum, Bacteroides, and Oscillospira were dominant in RS2. In RP2, the dominant genera included Candidatus_solibacter, Opitutus, Devosia, Sphingomonas, and Rhodoplanes, while in RP30, the dominant groups found were of Flavobacterium, Novosphingobium, Polaromone, Pedobacter, Janthinobacterium, Paenibacillus, Escherichia, and Bifidobacterium, respectively. Similarly, Chitinophaga, Steroidobacter, Bradyrhizobium, and Methylibium were predominant in ES2, and Bradyrhizobium, Halomonas, and Bacillus were predominant in ES30. Stamp analysis verified that Salinispora, Pseudomonas, and Burkholderia were dominant genera in both rhizoplane (RP30) and rhizosphere (RS30) compartments of old tea garden (30-Y) as compared with Bulk soil (CK) and fresh soil tea plantation (2-Y) rhizosphere (RS2) and rhizoplane (RP2) (Figures 5B–D, respectively).
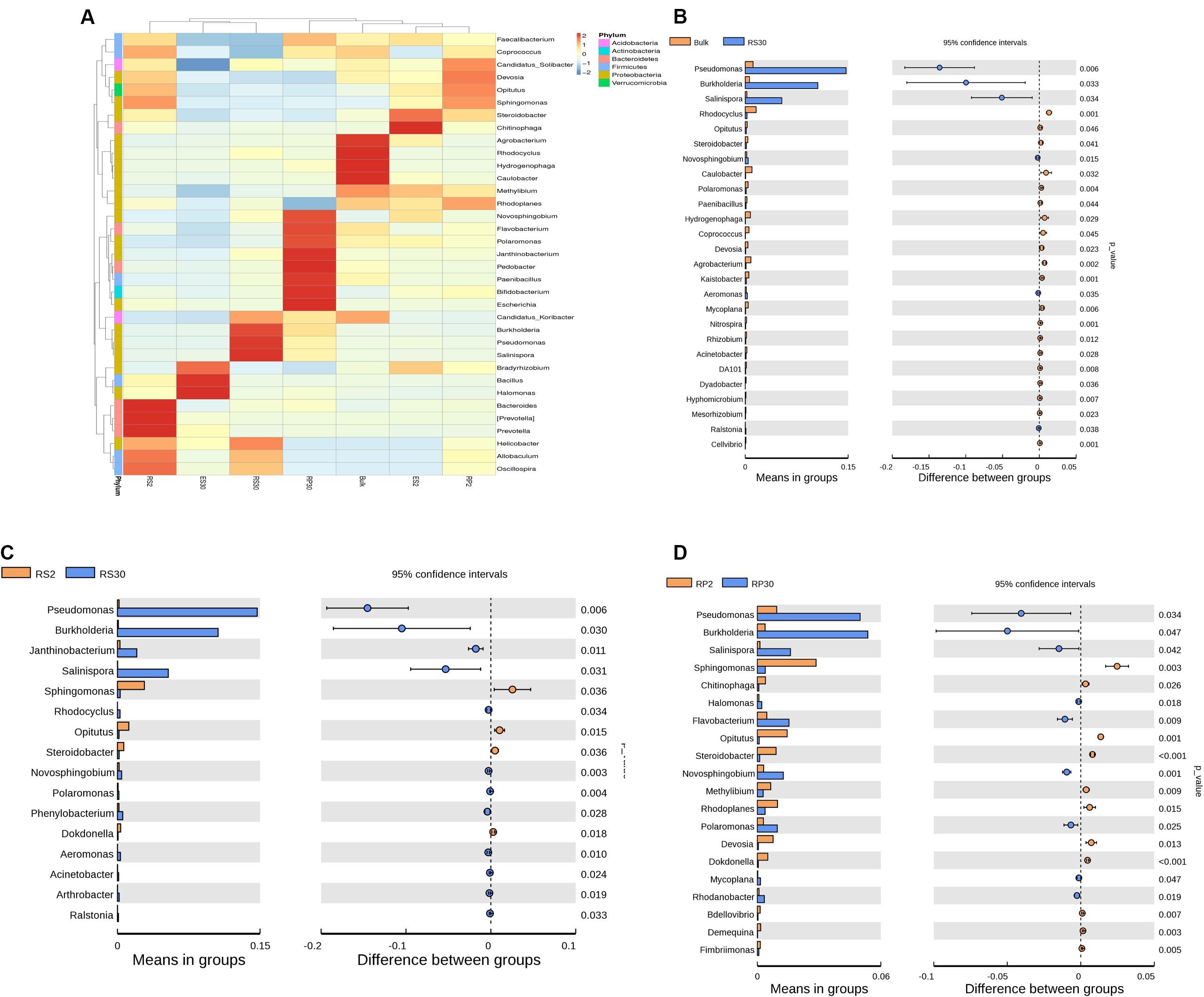
Figure 5. (A) Heat map showing the distribution of the 35 most abundant genera. Bulk soil (CK), rhizosphere (RS2 and RS30), rhizoplane (RP2 and RP30), and endosphere (ES2 and ES30) of tea gardens continuously cropped for 2 and 30 years, respectively (LSD test, P < 0.05, n = 3). (B) Error bar plots displaying significantly difference of most abundant genera among in Bulk (CK) and 30-Y rhizosphere (RS30) tea plantation (t-test, P < 0.05, n = 3). (C) Error bar plots displaying significant difference of most abundant genera in the 2-Y rhizosphere (RS2) and 30-Y rhizosphere (RS30) tea plantation (t-test, P < 0.05, n = 3). (D) Error bar plots displaying significant difference of most abundant genera in 2-Y rhizoplane (RP2) and 30-Y rhizoplane (RP30) tea plantation (t-test, P < 0.05, n = 3). The points explain differences among (“CK and RS30”), (“RS2 and RS30”), and (“RP2 and RP30”) (red, green, and blue bars, respectively); the values on the right-hand side display the P-values derived from the t-test error bar plots.
Abundance of Total Bacteria and Pseudomonas and Bacillus Genera by Colony-Forming Units’ qPCR Methods
Employing high-throughput sequencing, colony-forming units, and qPCR, we characterized the entire bacterial community composition and structure, including Pseudomonas and Bacillus in the CK, 2-Y, and 30-Y tea gardens. These results indicated that within 30-Y, the entire bacterial population and plant growth-promoting bacterial genus (Bacillus) were reduced compared to 2-Y and CK soil, while the catechin degradation bacteria (Pseudomonas) was increased (Figure 6).
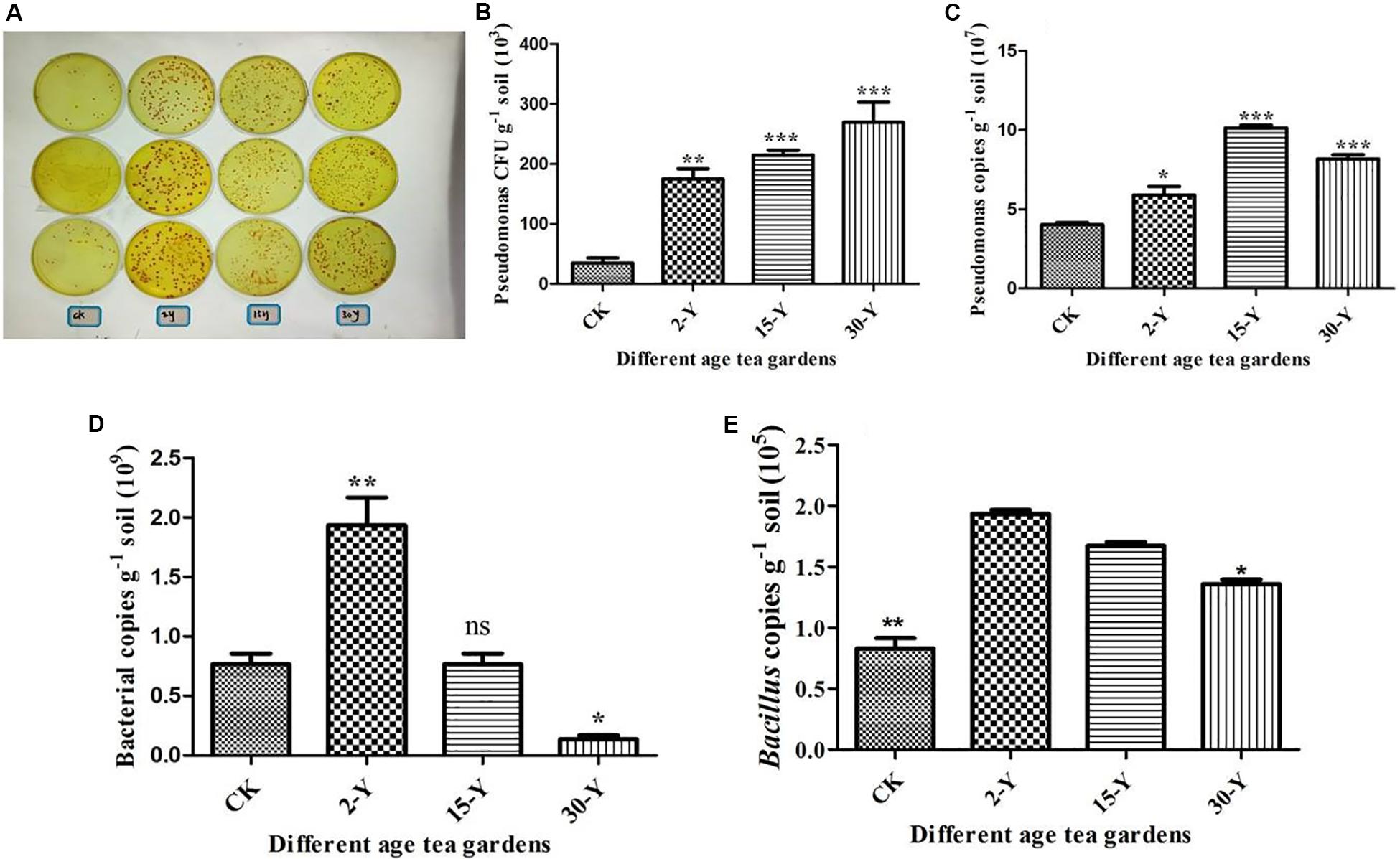
Figure 6. Abundance of total bacteria, Pseudomonas and Bacillus genera by qPCR analysis. (A) Pseudomonas populations. (B) CFU of Pseudomonas per gram of soil. (C) The contents of Pseudomonas genera in tea rhizosphere soils after different years of monoculture by qPCR analysis using the primer sets Psf/Psr (Tan and Ji, 2010). (D) The total bacterial contents by qPCR analysis using the primer set Eub338/Eub518. (E) The contents of Bacillus genera in tea rhizosphere soils after different years of monoculture by qPCR analysis (Wu H. et al., 2016). CK, 2Y, 15Y, and 30Y refer to bulk soil without planting any crop, newly planted 2-year garden, and replanted 15- and 30-year garden, respectively. Stars in the column show significant differences (LSD test, P < 0.05, n = 3).
In vitro Interactions of Different Types of Allelochemicals With Model Growth-Promoting Bacteria
Based on the HPLC-LC/MS identification of different types of active catechins allelochemicals in the leaf litter, a series of single and mixed allelochemicals with the final concentration gradients (0, 1.25, 2.5, 5, and 10 μg/ml) were set to identify the response of selected model growth-promoting bacteria such as Bacillus found in the tea garden. Our results determined that the different allelochemicals concentration significantly influenced the growth of selected growth-promoting bacteria. Moreover, as the concentration of these identified allelochemicals increased, the growth of Bacillus decreased (Figure 7).
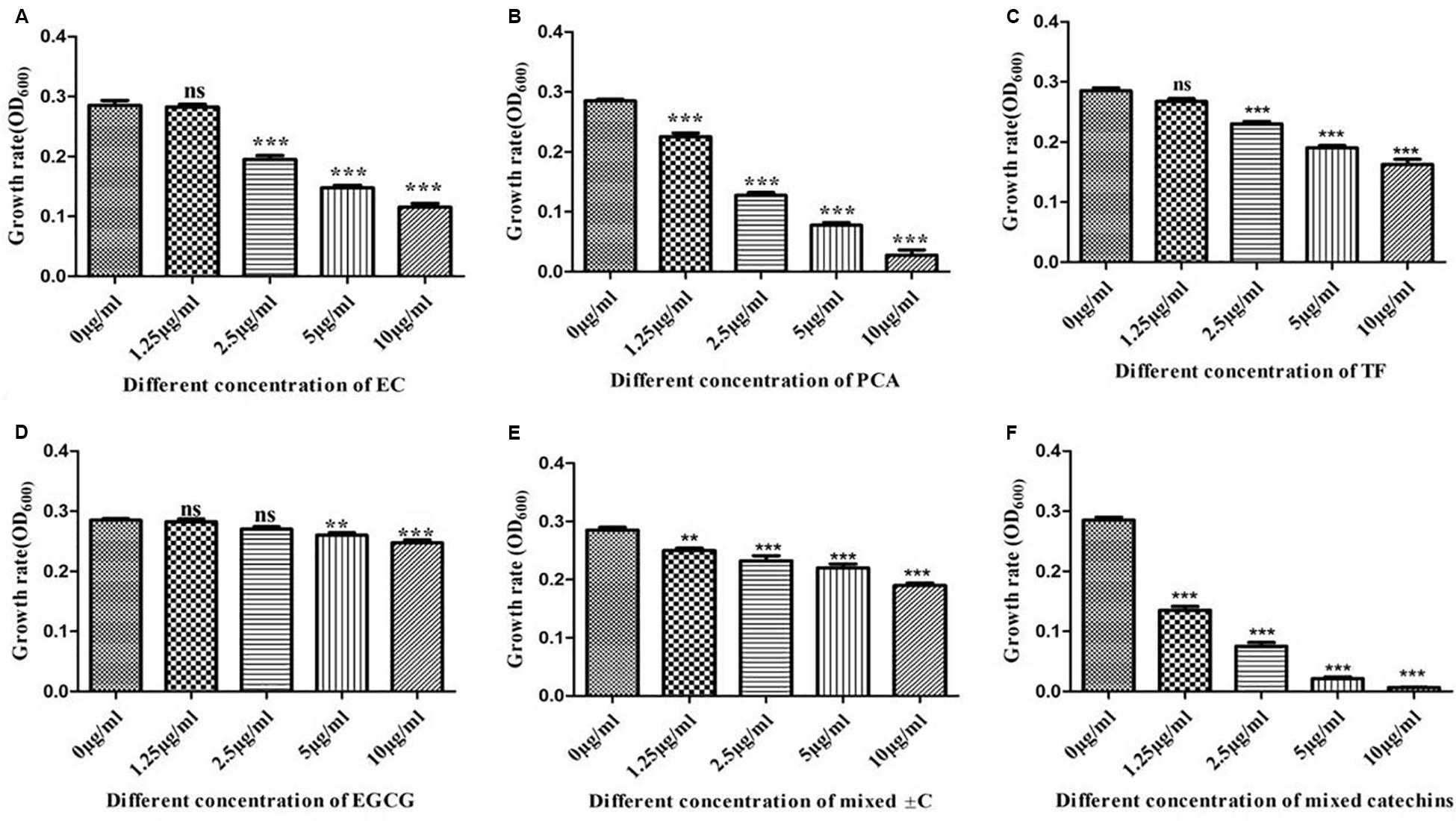
Figure 7. In vitro interactions of different types of allelochemicals with model growth-promoting bacteria Bacillus. (A) Epicatechin (EC), (B) protocatechuic acid (PCA), (C) taxifolin (TF), (D) epigallocatechin-3-gallate (EGCG), (E) catechin (±C), and (F) mix of all catechins. Stars in the column show significant differences (LSD test, P < 0.05, n = 4).
In vitro Interactions of Different Types of Allelochemicals With Model Catechins Degrading Bacteria
Similarly, a series of single and mixed allelochemicals with the final concentration gradients (0, 1.25, 2.5, 5, and 10 μg/ml) were also set to identify the response of catechin degrading bacteria, Pseudomonas spp., which was dominant in the 30-Y consecutively monoculture tea garden. The results showed that the different allelochemicals concentration have no significant effect on the growth of Pseudomonas spp. Low concentration at 1.25 μg/ml of ±C promoted the growth of Pseudomonas spp. However, the high concentration (10 μg/ml) of PCA and a mix of seven known allelochemicals significantly inhibit the growth of the selected model bacteria. It is therefore suggested that the dominant bacteria such as Pseudomonas used these leaf litters as a carbon substrate to some extent (Figure 8).
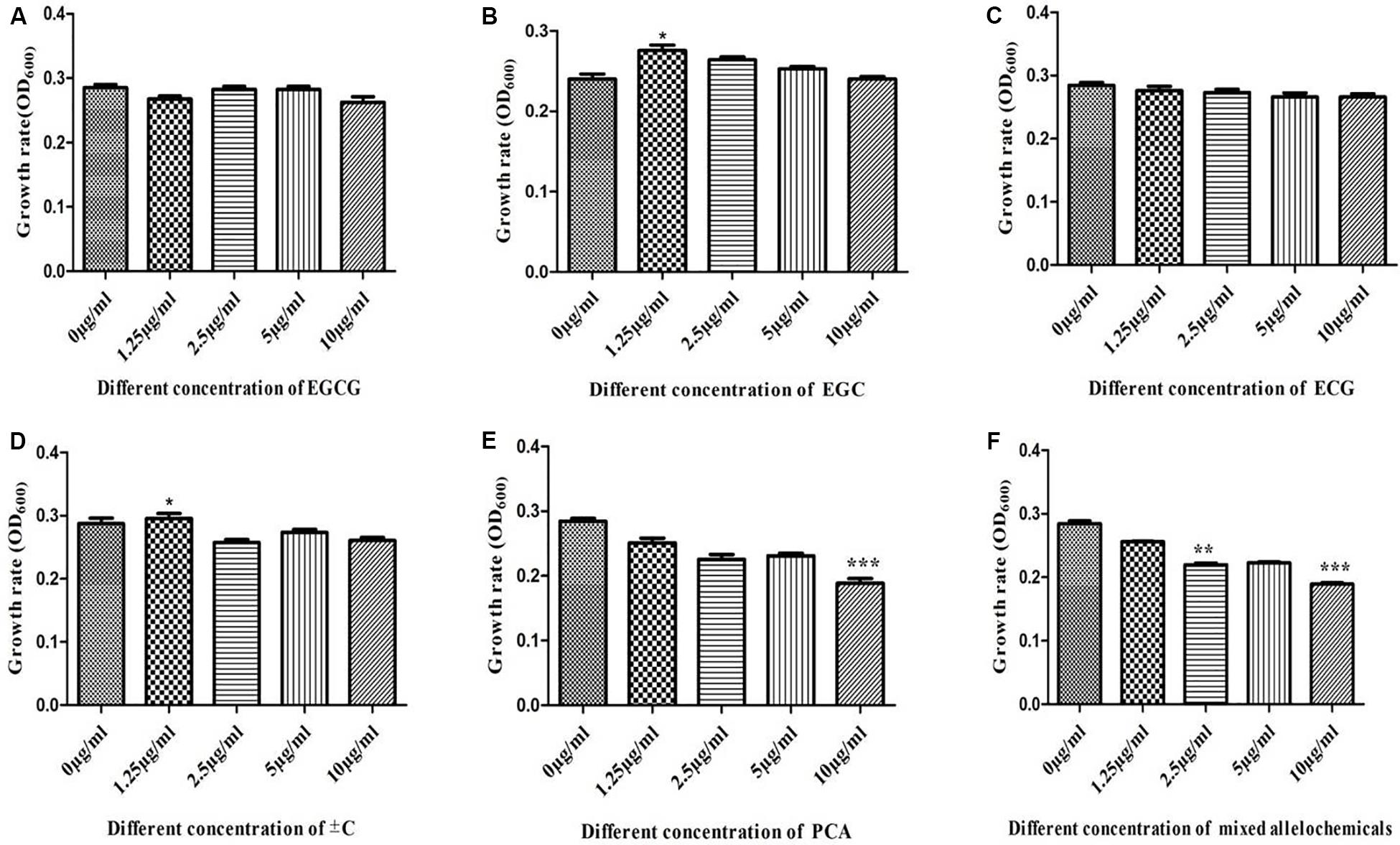
Figure 8. In vitro interactions of different types of allelochemicals with model catechins degrading bacteria Pseudomonas. (A) Epigallocatechin-3-gallate (EGCG), (B) epigallocatechin (EGC), (C) epicatechingallate (ECG), (D) catechin (±C), (E) protocatechuic acid (PCA), and (F) mix of all catechins. Stars in the column show significant differences (LSD test, P < 0.05, n = 4).
Biotransformation of Catechins and Their Effect on pH
In order to understand the role of the dominant bacterial populations in lowering the pH of the tea rhizosphere with the increasing tea planting age and consecutive monoculture problems, the dominant Pseudomonas was selected as a model bacterium. Epicatechin (EC) with the final concentration gradients 5 μg/ml and activated Pseudomonas bacterium (50 μl) was subjected to six times diluted LB medium. After 48 and 72 h, the media were purified and filtered for pH and metabolite analysis. The LC-MS analysis results identified the compound that was PCA after 72 h (Figure 9A). Moreover, results showed that the pH of the media drops to 0.36-fold after 72 h (Figure 9B). These results suggested that the dominant bacteria identified in 30-Y tea plantation used leaf litter. After using these substrates, the bacteria convert these allelochemicals into different types of acids like PCA, which may change the pH of the tea rhizosphere soil over time.
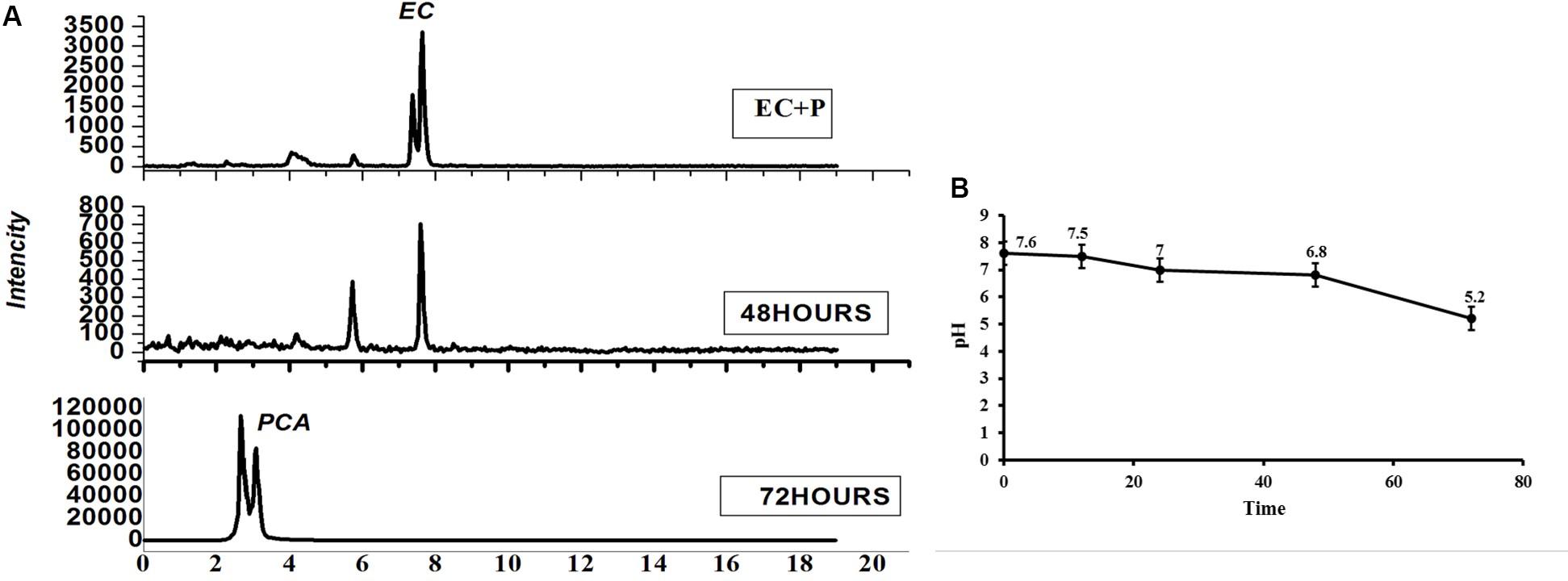
Figure 9. Impact of catechin degradation on pH. (A) Catechin degradation, (B) effect on pH. EC + P represents Epicatechin (EC) + Pseudomonas (P) and PCA represents protocatechuic acid.
Discussion
Our results indicated that long-term monoculture has an adverse effect on tea quality (TNN, TPY, TPP, and TAA content) and physiological and growth parameters of tea plants compared to freshly grown tea plants, which is consistent with previous findings (Arafat et al., 2017; Jiang et al., 2019). Similarly, previous studies have shown that tea production, as well as quality, declines dramatically with increasing cultivation time (Illukpitiya et al., 2004; Arafat et al., 2019). Owing to shifts in soil physicochemical and biological properties, a long-term tea monoculture can lead to a “soil-sickness” or “replanting disease,” which has an adverse impact on tea productivity (Owuor, 1996; Huang et al., 2006; Utkhede, 2006; Kamau, 2007; Qu and Wang, 2008). The underlying factors of soil sickness are usually considered to be nutrient imbalances, autotoxins generation, and/or shift in soil microbial community structure and diversity (Zhao et al., 2015). Plant-associated microbial communities are also considered as a second genome of the plant and are essential for soil fertility and plant health (Berendsen et al., 2012). The regulation of soil microbiota owing to allelochemicals interaction is closely associated with replanting disease in agricultural systems (Li et al., 2014; Liu et al., 2017).
Soil microbial biomass is commonly considered to be associated with soil fertility, including as a potential indicator of soil quality (Shengchun and Yinghua, 2003; Zhang and Zhang, 2003). Due to changes in soil management approaches, soil microbial dynamics shift faster than soil organic matter (Shengchun and Yinghua, 2003). Beneficial microbes, especially plant growth-promoting bacteria, are crucial for nutrient availability and plant growth (Acosta-Martínez et al., 2010; Cipollini et al., 2012; Huang et al., 2013). In the present study, we found Burkholderia and Pseudomonas as the dominant genera, while plant growth-promoting bacteria such as Prevotella, Bacillus, and Sphingomonas were significantly lower in the 30-Y tea garden. The qPCR results also confirmed that the bacterial density per gram of soil of 30-Y plantation was significantly lower compared to 2-Y. However, Pseudomonas population was selectively increased in 30-Y. It is generally believed that tea garden soils have an inhibitory effect on soil microorganisms due to high acidity and aluminum toxicity. We have previously reported that continuous planting of tea (30 years) has no significant effect on soil nutrients (N, P, and K). However, soil pH in continuous tea fields (30 years) was significantly lower than in the bulk and 2-year garden soil (2-Y) (Arafat et al., 2017).
Various effects of plant polyphenols have previously been observed under artificial laboratory conditions, indicating that they stimulate or inhibit (depending on the compound) the growth of the nitrifiers (Rice and Pancholy, 1973; Baldwin et al., 1983). Schimel et al. (1998), proving that plant polyphenols have a controlling effect on nutrient dynamics and species interaction in Alaskan taiga. The complexity of soil food webs, including the covariance of other compounds, makes it challenging to demonstrate the effects of polyphenols on soil biota directly. HPLC-LC/MS findings and in vitro interaction of the selected model growth-promoting bacteria with allelochemicals suggest that the various types of active catechins’ allelochemical concentration as they enhanced, in turn, significantly suppressed Bacillus growth (Figure 7). In vitro analysis of soil microbial density by catechin showed inhibition and microbe suppression (Inderjit et al., 2009). Catechin-degrading bacteria (Pseudomonas spp.) were greater in terms of abundance in the 30-Y tea garden and different concentrations of allelochemicals did not significantly influence the growth of Pseudomonas spp. (Figure 8). At a lower concentration (1.25 μg/ml of ±C), the growth of Pseudomonas was increased; on the other hand, a high frequency (10 μg/ml) of PCA and the combination of seven known allelochemicals significantly suppressed selected bacteria growth. These results suggest that dominant bacteria such as Pseudomonas utilized these catechins in leaf litter as a carbon substrate to a certain level. Wang et al. (2013) also showed that Pseudomonas can use catechins as substrate and biotransformed it into most toxic allelochemicals. We aimed to examine if the dominant bacterial groups associated with tea roots in the 30-Y tea garden has a role in soil acidification. EC with the final concentration of 5 μg/ml, which can activate the growth of Pseudomonas, was investigated for biotransformation and resulting metabolites. By LC-MS analysis, we detected compounds, especially PCA, and reduced the pH of the media (0.36 folds) after 72 h, indicating that abundant bacteria detected in the 30-Y garden converted the polyphenols in different types of acids (PCA, ferulic acids, etc.), which may lead to soil acidification.
Conclusion
Overall, long-term tea monocropping had a significant impact on the tea in terms of both quality and quantity, as well as soil regenerative capacity. We also studied the polyphenol interaction with microorganisms in tea residue and rhizosphere, which ultimately declined the plant growth-promoting bacteria. However, below a particular concentration gradient, no significant effect was observed on the growth of Pseudomonas. We found that Pseudomonas biotransformed polyphenols in various organic acids (PCA, TF, and EC) and changed soil pH, which may be the putative reason for altering the microbial structure and composition of tea rhizosphere during continuous tea planting. We recommend eliminating plant residues and trimmed materials from the tea plantation to avoid the accumulation of allelochemicals, which can delay soil acidification.
Data Availability Statement
The data set has been submitted to the NCBI-Sequence Read Archive with the SRA accession: PRJNA626070. Temporary Submission ID: SUB7295720.
Author Contributions
WL, SL, and YA conceived the study. YA and IU wrote the manuscript. YA, YJ, and ZC performed field sampling and lab experiments. YA, IU, TC, and HZ performed the statistical analyses. All authors discussed the results and commented on the manuscript.
Funding
This research was supported by the National Key Research and Development Plan 2017YFE0121800, the National Key Research and Development (R&D), China Plan 2016YFD0200900, Fujian–Taiwan Joint Innovative Centre for Germplasm Resources and Cultivation of Crop (grant no. 2015-75, Fujian 2011 Program, China), and 948 programs from the Ministry of Agriculture (2014-Z360) China.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00601/full#supplementary-material
Footnotes
- ^ http://ccb.jhu.edu/software/FLASH/
- ^ http://qiime.org/index.html
- ^ https://unite.ut.ee/
- ^ http://www.drive5.com/usearch/manual/uchime_algo.html
- ^ http://drive5.com/uparse/
- ^ https://unite.ut.ee/
- ^ http://qiime.org/scripts/assign_taxonomy.html
References
Acosta-Martínez, V., Burow, G., Zobeck, T., and Allen, V. (2010). Soil microbial communities and function in alternative systems to continuous cotton. Soil Sci. Soc. Am. J. 74, 1181–1192.
Arafat, Y., Tayyab, M., Khan, M. U., Chen, T., Amjad, H., Awais, S., et al. (2019). Long-term monoculture negatively regulates Fungal community composition and abundance of Tea orchards. Agronomy 9:466.
Arafat, Y., Wei, X., Jiang, Y., Chen, T., Saqib, H., Lin, S., et al. (2017). Spatial distribution patterns of root-associated bacterial communities mediated by root exudates in different aged ratooning tea monoculture systems. Int. J. Mol. Sci. 18:1727. doi: 10.3390/ijms18081727
Baldwin, I. T., Olson, R. K., and Reiners, W. A. (1983). Protein binding phenolics and the inhibition of nitrification in subalpine balsam fir soils. Soil Biol. Biochem. 15, 419–423.
Baxendale, C., Orwin, K. H., Poly, F., Pommier, T., and Bardgett, R. D. (2014). Are plant–soil feedback responses explained by plant traits? New Phytol. 204, 408–423. doi: 10.1111/nph.12915
Berendsen, R. L., Pieterse, C. M., and Bakker, P. A. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486.
Bever, J. D., Dickie, I. A., Facelli, E., Facelli, J. M., Klironomos, J., Moora, M., et al. (2010). Rooting theories of plant community ecology in microbial interactions. Trends Ecol. Evol. 25, 468–478. doi: 10.1016/j.tree.2010.05.004
Blum, U., Staman, K. L., Flint, L. J., and Shafer, S. R. (2000). Induction and/or selection of phenolic acid-utilizing bulk-soil and rhizosphere bacteria and their influence on phenolic acid phytotoxicity. J.Chem. Ecol. 26, 2059–2078.
Bokulich, N. A., Subramanian, S., Faith, J. J., Gevers, D., Gordon, J. I., Knight, R., et al. (2013). Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 10, 57–59. doi: 10.1038/nmeth.2276
Canals, R., Emeterio, L., and Peralta, J. (2005). Autotoxicity in Lolium rigidum: analyzing the role of chemically mediated interactions in annual plant populations. J. Theor. Biol. 235, 402–407. doi: 10.1016/j.jtbi.2005.01.020
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–356. doi: 10.1038/nmeth.f.303
Chen, J., Wu, L., Xiao, Z., Wu, Y., Wu, H., Qin, X., et al. (2017). Assessment of the Diversity of Pseudomonas spp. and Fusarium spp. in Radix pseudostellariae Rhizosphere under Monoculture by Combining DGGE and Quantitative PCR. Front. Microbiol. 8:1748. doi: 10.3389/fmicb.2017.01748
Chen, Z.-M., and Lin, Z. (2015). Tea and human health: biomedical functions of tea active components and current issues. J. Z. Univ. Sci. B 16, 87–102. doi: 10.1631/jzus.B1500207
Cipollini, D., Rigsby, C. M., and Barto, E. K. (2012). Microbes as targets and mediators of allelopathy in plants. J. Chem. Ecol. 38, 714–727. doi: 10.1007/s10886-012-0133-7
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27. 16, 2194–2200. doi: 10.1093/bioinformatics/btr381
Edwards, J., Johnson, C., Santos-Medellín, C., Lurie, E., Podishetty, N. K., Bhatnagar, S., et al. (2015). Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Scie. U.S.A. 112, E911–E920. doi: 10.1073/pnas.1414592112
Ehlers, B. K. (2011). Soil microorganisms alleviate the allelochemical effects of a thyme monoterpene on the performance of an associated grass species. PloS One 6:e26321. doi: 10.1371/journal.pone.0026321
Haas, B. J., Gevers, D., Earl, A. M., Feldgarden, M., Ward, D. V., Giannoukos, G., et al. (2011). Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21, 494–504. doi: 10.1101/gr.112730.110
Hättenschwiler, S., and Vitousek, P. M. (2000). The role of polyphenols in terrestrial ecosystem nutrient cycling. Trends Ecol. Evol. 15, 238–243. doi: 10.1016/s0169-5347(00)01861-9
Heal, O. (1997). “Plant litter quality and decomposition: an historical overview,” in Driven by Nature, Plant Litter Quality and Decomposition, eds G. Cadisch and K. E. Giller (Wallingford: CAB International), 3–30.
Huang, H., Chou, C., and Erickson, R. (2006). Soil sickness and its control. Allelopathy J. 18, 1–21.
Huang, L.-F., Song, L.-X., Xia, X.-J., Mao, W.-H., Shi, K., Zhou, Y.-H., et al. (2013). Plant-soil feedbacks and soil sickness: from mechanisms to application in agriculture. J. Chem. Ecol. 39, 232–242. doi: 10.1007/s10886-013-0244-9
Illukpitiya, P., Shanmugaratnam, N., and Kjosavik, D. J. (2004). Tea Agroecosystems in the Uva highlands of sri lanka. Mountain Res. Dev. 24, 52–60.
Inderjit, R. K., Surinder, K., and Callaway, R. M. (2009). Impact of (±)-catechin on soil microbial communities. Commun. Integrat. Biol. 2, 127–129. doi: 10.4161/cib.7744
Jiang, Y., Arafat, Y., Letuma, P., Ali, L., Tayyab, M., Waqas, M., et al. (2019). Restoration of long-term monoculture degraded tea orchard by green and goat manures applications system. Sustainability 11:1011.
Kamau, D. (2007). Confounding factors affecting the growth and production of ageing tea agro-ecosystems: a review. Tea 28, 26–40.
Kaur, H., Kaur, R., Kaur, S., and Baldwin, I. T. (2009). Taking ecological function seriously: soil microbial communities can obviate allelopathic effects of released metabolites. PloS One 4:e4700. doi: 10.1371/journal.pone.0004700
Kõljalg, U., Nilsson, R. H., Abarenkov, K., Tedersoo, L., Taylor, A. S. F., and Bahram, M. (2013). Towards a unified paradigm for sequence – based identification of fungi. Mol. Ecol. 22. 21, 5271–5277. doi: 10.1111/mec.12481
Li, X.-G., Ding, C.-F., Hua, K., Zhang, T.-L., Zhang, Y.-N., Zhao, L., et al. (2014). Soil sickness of peanuts is attributable to modifications in soil microbes induced by peanut root exudates rather than to direct allelopathy. Soil Biol. Biochem. 78, 149–159.
Lin, W., Wu, L., Lin, S., Zhang, A., Zhou, M., Lin, R., et al. (2013). Metaproteomic analysis of ratoon sugarcane rhizospheric soil. BMC Microbiol. 13:135. doi: 10.1186/1471-2180-13-135
Liu, J., Li, X., Jia, Z., Zhang, T., and Wang, X. (2017). Effect of benzoic acid on soil microbial communities associated with soilborne peanut diseases. Appl. Soil Ecol. 110, 34–42.
Magoč, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Maudu, M., Mudau, F., and Mariga, I. (2010). The effect of pruning on growth and chemical composition of cultivated bush tea (Athrixia phylicoides DC). J. Med. Plants Res. 4, 2353–2358.
Min, K., Freeman, C., Kang, H., and Choi, S.-U. (2015). The regulation by phenolic compounds of soil organic matter dynamics under a changing environment. Biomed Res. Int. 2015:825098. doi: 10.1155/2015/825098
Muller, C. H. (1966). The role of chemical inhibition (allelopathy) in vegetational composition. Bull. Torrey Bot. Club 93, 332–351.
Neuhauser, E. (1978). Phenolic content and palatability of leaves and wood to soil isopods and diplopods. Pedobiologia 18, 99–109.
Owuor, P. (1996). Problems of replanting tea or other crops on old tea lands: Possible role of allelochemicals: a review. Tea 17, 34–40.
Peng, L., Song, X., Shi, X., Li, J., and Ye, C. (2008). An improved HPLC method for simultaneous determination of phenolic compounds, purine alkaloids and theanine in Camellia species. J. Compos. Anal. 21, 559–563.
Perucci, P., Casucci, C., and Dumontet, S. (2000). An improved method to evaluate the o-diphenol oxidase activity of soil. Soil Biol. Biochem. 32, 1927–1933.
Qu, X. H., and Wang, J. (2008). Effect of amendments with different phenolic acids on soil microbial biomass, activity, and community diversity. Appl. Soil Ecol. 39, 172–179.
Rice, E. L., and Pancholy, S. K. (1973). Inhibition of nitrification by climax ecosystems. II. Additional evidence and possible role of tannins. Am. J. Bot. 60, 691–702.
Saiya-Cork, K., Sinsabaugh, R., and Zak, D. (2002). The effects of long term nitrogen deposition on extracellular enzyme activity in an Acer saccharum forest soil. Soil Biol. Biochem. 34, 1309–1315.
Schimel, J. P., Cates, R. G., and Ruess, R. (1998). The role of balsam poplar secondary chemicals in controlling soil nutrient dynamics through succession in the Alaskan taiga. Biogeochemistry 42, 221–234.
Shengchun, L. Z. Y. S. W., and Yinghua, W. J. P. (2003). Effects of different nitrogen fertilizers on the microbial biomass and the population of nitrifying-denitrifying bacteria in the rice rhizosphere. Soils 35, 490–494.
Tan, Y., and Ji, G. (2010). Bacterial community structure and dominant bacteria in activated sludge from a 70 C ultrasound-enhanced anaerobic reactor for treating carbazole-containing wastewater. Bioresour. Technol. 101, 174–180. doi: 10.1016/j.biortech.2009.08.044
Utkhede, R. (2006). Soil sickness, replant problem or replant disease and its integrated control. Allelopathy J. 18, 23–38.
van de Voorde, T. F., Ruijten, M., van der Putten, W. H., and Bezemer, T. M. (2012). Can the negative plant–soil feedback of Jacobaea vulgaris be explained by autotoxicity? Basic Appl. Ecol. 13, 533–541.
Wang, C.-M., Li, T.-C., Jhan, Y.-L., Weng, J.-H., and Chou, C.-H. (2013). The impact of microbial biotransformation of catechin in enhancing the allelopathic effects of Rhododendron formosanum. PLoS One 8:e85162. doi: 10.1371/journal.pone.0085162
Wardle, D., and Lavelle, P. (1997). “Linkages between soil biota, plant litter quality and decomposition,” in Driven by nature: Plant litter quality and decomposition, eds G. Cadisch and K. E. Giller (London: CAB International), 107–124.
Weeraratna, C., Watson, M., and Wettasingha, D. (1977). Effect of mineralization of tea prunings on some soil characteristics. Plant Soil 46, 93–99.
Weidenhamer, J. D., Li, M., Allman, J., Bergosh, R. G., and Posner, M. (2013). Evidence does not support a role for gallic acid in Phragmites australis invasion success. J. Chem. Ecolo. 39, 323–332. doi: 10.1007/s10886-013-0242-y
Wu, H., Wu, L., Wang, J., Zhu, Q., Lin, S., Xu, J., et al. (2016). Mixed phenolic acids mediated proliferation of pathogens Talaromyces helicus and Kosakonia sacchari in continuously monocultured Radix pseudostellariae rhizosphere soil. Front. Microbiol. 7:335. doi: 10.3389/fmicb.2016.00335
Wu, L., Chen, J., Wu, H., Wang, J., Wu, Y., Lin, S., et al. (2016). Effects of consecutive monoculture of Pseudostellaria heterophylla on soil fungal community as determined by pyrosequencing. Sci. Rep. 6:26601. doi: 10.1038/srep26601
Xu, W. H., Wang, Z., and Wu, F. (2015). The effect of D123 wheat as a companion crop on soil enzyme activities, microbial biomass and microbial communities in the rhizosphere of watermelon. Front. Microbiol. 6:899. doi: 10.3389/fmicb.2015.00899
Yilmaz, G., Kandemir, N., and Kinalioglu, K. (2004). Effects of different pruning intervals on fresh shoot yield and some quality properties of Tea (Camellia sinensis (L.) O. Kuntze) in Turkey. Pakistan J. Biol. Sci. 7, 1208–1212.
Zhang, H., and Zhang, G. (2003). Microbial biomass carbon and total organic carbon of soils as affected by rubber cultivation. Pedosphere 13, 353–357.
Zhao, Y., Wu, L., Chu, L., Yang, Y., Li, Z., Azeem, S., et al. (2015). Interaction of Pseudostellaria heterophylla with Fusarium oxysporum f. sp. heterophylla mediated by its root exudates in a consecutive monoculture system. Sci. Rep. 5:8197. doi: 10.1038/srep08197
Keywords: indirect allelopathy, catechins, soil sickness, monoculture problems, plant polyphenol
Citation: Arafat Y, Ud Din I, Tayyab M, Jiang Y, Chen T, Cai Z, Zhao H, Lin X, Lin W and Lin S (2020) Soil Sickness in Aged Tea Plantation Is Associated With a Shift in Microbial Communities as a Result of Plant Polyphenol Accumulation in the Tea Gardens. Front. Plant Sci. 11:601. doi: 10.3389/fpls.2020.00601
Received: 24 October 2019; Accepted: 20 April 2020;
Published: 28 May 2020.
Edited by:
Davide Neri, Marche Polytechnic University, ItalyReviewed by:
Hiroshi Noguchi, Nihon Pharmaceutical University, JapanDerek Stewart, The James Hutton Institute, United Kingdom
Copyright © 2020 Arafat, Ud Din, Tayyab, Jiang, Chen, Cai, Zhao, Lin, Lin and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenxiong Lin, bHd4QGZhZnUuZWR1LmNu; Sheng Lin, bHNqazE5NThAMTYzLmNvbQ==
 Yasir Arafat
Yasir Arafat Israr Ud Din
Israr Ud Din Muhammad Tayyab
Muhammad Tayyab Yuhang Jiang
Yuhang Jiang Ting Chen1,2
Ting Chen1,2 Xiangmin Lin
Xiangmin Lin Wenxiong Lin
Wenxiong Lin Sheng Lin
Sheng Lin