- 1College of Bioscience and Biotechnology, Hunan Agricultural University, Changsha, China
- 2State Key Laboratory of Hybrid Rice, Hunan Hybrid Rice Research Center, Changsha, China
- 3Hunan Academy of Agricultural Sciences, Changsha, China
“Easy-to-shatter” trait is a major cause of rice crop yield losses, emphasizing the economic value of developing elite rice cultivars with reduced seed shattering capable of achieving higher yields. In the present study, we describe the development of new indica rice lines that exhibit lower rates of seed shattering following the targeted CRISPR/Cas9-mediated editing of the qSH1 gene. We were able to identify qSH1 mutant T0 transgenic plants, with transgene-free homozygous mutants being obtained via segregation in the T1 generation. We then utilized two T2 transgene-free homozygous lines in order to assess the degree of seed shattering and major agronomic traits of these mutant lines and of wild-type rice plants (HR1128-WT). This approach revealed that qsh1 homozygous mutant lines exhibited significantly reduced seed shattering relative to HR1128-WT without any significant changes in other analyzed agronomic traits. We then used these mutant lines to develop new promising hybrid rice lines with intermediate seed shattering. Overall our results reveal that combining targeted gene editing via CRISPR/Cas9 with heterosis utilization approach can allow for the efficient development of novel promising hybrid rice cultivars that exhibit a intermediate of seed shattering, thereby ensuring better stability and improved rice yields.
Introduction
Rice (Oryza sativa L.) is a staple crop and one of the most important food sources in the world, being consumed by over half of the global population (Wang et al., 2013). In the wild, seed shattering is an advantageous behavior for rice. However, in a cultivated context, domesticated rice varieties that exhibit the “easy-to-shatter” trait can result in serious yield losses during the harvesting process (Doebley, 2006; Fuller et al., 2009). As such, there have been many efforts to breed rice varieties with an intermediate seed shattering phenotype in an effort to stabilize rice yields. A number of rice quantitative trait loci (QTLs) and genes associated with seed shattering have been cloned to date, such as SH-h, SH3, SH4, qSH1, SHAT1, and SSH1 (Li et al., 2006; Konishi et al., 2006; Lin et al., 2007; Ji et al., 2010; Lv et al., 2018; Jiang et al., 2019). Few of these genes, however, have been leveraged to develop elite rice varieties, particularly hybrid rice varieties. The qSH1 gene is an important QTL located on chromosome1 that is associated with seed shattering and that codes for a BEL1-type homeobox-containing protein. Loss of qSH1 expression in the abscission layer results in a significantly improved strong seed shattering phenotype (Konishi et al., 2006).
Hybrid rice plants offer yields that are 10–20% higher than those of conventional rice, and as a result these hybrid cultivars have been cultivated in over 40 countries throughout the world (Su et al., 2012; Zhou et al., 2016). At present, indica hybrids are the dominant crops used for commercial rice production, particularly in southern China. Such indica varietals, however, exhibit a greater propensity for the “easy-to-shatter” trait than do japanic rice cultivars. Previous reports suggest that certain hybrid rice varieties can suffer a 5.8–8.6% harvest yield loss owing to their susceptibility to seed shattering (Ye et al., 2012). As such, the development of genetically modified rice cultivars with an intermediate seed shattering phenotype has been a priority in recent years. Conventional breeding strategies require 5–10 years in order to develop novel seed shattering cultivars via mutagenesis and subsequent crossing and backcrossing. The current lack of robust germplasm resources associated with weaker seed shattering and the inefficient nature of such conventional breeding strategies have significantly constrained efforts to produce intermediate seed shattering rice cultivars to date. It is therefore essential that novel technologies be leveraged in an effort to reduce the propensity of rice plants to readily undergo seed shattering.
Many recent advances in plant genomic editing have been made through the use of tools such as sequence-specific nucleases (SSNs), including zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) 9 (CRISPR/Cas9) (Townsend et al., 2009; Li et al., 2012; Feng et al., 2013; Nekrasov et al., 2013; Baltes and Voytas, 2015; Hyun et al., 2015; Svitashev et al., 2015; Weeks et al., 2016). Of these tools, CRISPR/Cas9-mediated gene editing has been widely applied as a means of rapidly and reliably conducting genomic editing in important crops such as rice (Feng et al., 2013; Miao et al., 2013; Shan et al., 2013; Xu et al., 2014), maize (Liang et al., 2014; Feng et al., 2016; Zhu et al., 2016), tomato (Brooks et al., 2014; Ito et al., 2015), potato (Wang et al., 2015), wheat (Upadhyay et al., 2013; Shan et al., 2014; Wang et al., 2014), sorghum (Jiang et al., 2013), and soybean (Jacobs et al., 2015).
In the present study, we conducted the targeted mutagenesis of the qSH1 gene via a CRISPR/Cas9-mediated approach. This allowed us to develop qsh1 mutants on the HR1128 and Guangzhan63-4S (GZ63-4S) backgrounds, with both of these being parental lines of the commonly utilized the indica hybrid rice cultivar-GuangLiangYou1128 (GLY1128), which has many excellent agricultural traits, but with a strong seed shattering phenotype. Through gene editing approach, we were able to develop homozygous T1 mutant plants. These qsh1 mutants exhibited significant reductions in seed shattering activity without any changes in other major agronomic traits relative to wild-type plants under normal growth conditions. Furthermore, the seed shattering propensities of the S1 and S2 promising hybrid rice lines were significantly reduced following crossing with our qsh1 mutants with background of HR1128 and GZ63-4S.
Materials and Methods
Plant Materials and Growth
The HR-qsh1-ab, HR-qsh1-1, HR-qsh1-23, GZ-qsh1-ab, GZ-qsh1-1, and GZ-qsh1-23 mutants were produced via using the pYLCRISPR/Cas9-qsh1-Tab, T1, T23 vector to transform plants of the HR1128 and GZ63-4S rice varieties via Agrobacterium-mediated transformation. Plants were cultured at 28–35°C in a greenhouse in Changsha, or were grown in the transgenic crop planting field of the Hunan Hybrid Rice Research Centre in Changsha or Sanya during the standard rice-growing season.
Vector Construction
The pYLCRISPR/Cas9-qsh1-Tab, T1, T23 vector (pC-qsh1-Tab, T1, T23) was constructed as in previous reports (Ma et al., 2015). The gRNA expression vector (pYLgRNA-U3/U6a) and the Cas9 plant expression vector (pYLCRISPR/Cas9) were from Pro. Yao-guang Liu of the South China Agricultural University. For targeted mutagenesis of these rice plants, we selected a candidate target sequences composed of 19–20 bases upstream of the PAM motif (Figure 1A). We additionally obtained the target site sequence-containing primers qsh1-TaF/R, qsh1-TbF/R, qsh1-T1F/R, qsh1-T2F/R, and qsh1-T3F/R (Supplementary Table S1) from BLAST CRISPR-GE1 and NCBI2 as a means of ensuring that no off-target gene targeting occurred via this approach.
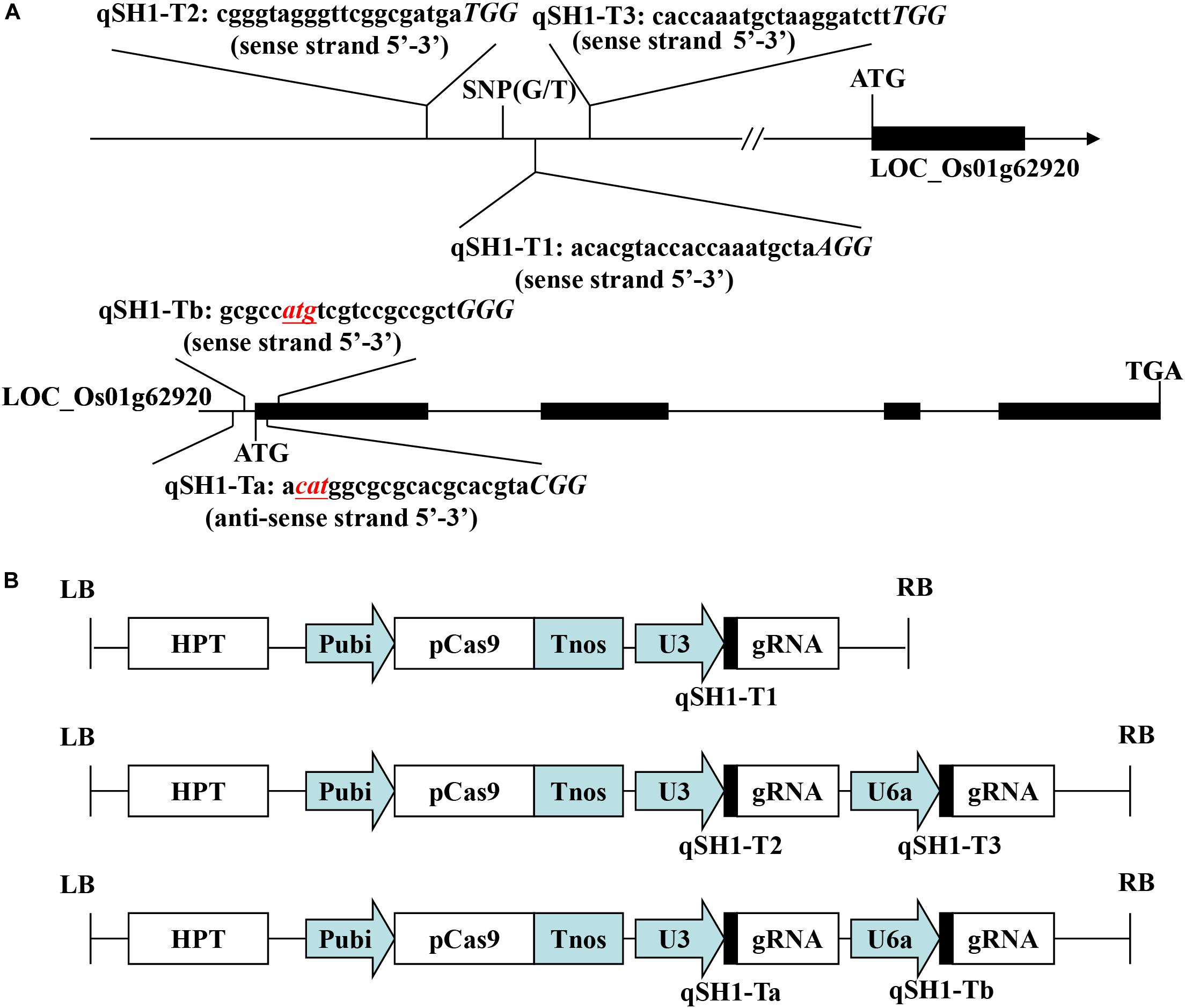
Figure 1. Target sites of the CRISPR/Cas9-qSH1-T1, T23, Tab vector. (A) Schematic of the qSH1 gene structure and target site. Exons and introns are indicated with black rectangles and black lines, respectively. Both the translation initiation codon (ATG) and the termination codon (TGA) are shown. The target site nucleotides are shown in lowercase, and the protospacer adjacent motif (PAM) site is capitalized and italicized. (B) A schematic presentation of the T-DNA structure in the CRISPR/Cas9-mediated genome editing construct. The expression of Cas9 is driven by the maize ubiquitin promoter (Pubi); the expression of the sgRNA scaffold is driven by the rice U3 or U6a small nuclear RNA promoter (OsU3 or OsU6a); the expression of hygromycin (HPT) is driven by two CaMV35S promoter (2 × 35S); Tnos, gene terminator; LB and RB, left border and right border, respectively.
For pC-qsh1-Tab vector preparation, the target site qsh1-Ta and qsh1-Tb sequence-containing chimeric primers were cloned into the sgRNA expression cassettes pYLsgRNA-U3 and pYLsgRNA-U6a, respectively, at a BsaI site (Ma et al., 2015). Synthesis of the gRNA expression cassette was achieved using the U-F/gRNA-R primers (Supplementary Table S1) for an initial round of overlapping PCR, followed by a second round using the site-specific B1’/B2 and B2’/BL primers (Supplementary Table S1). After this, the target sgRNA (qsh1-Ta-sgRNA and qsh1-Tb-sgRNA) expression cassettes were ligated into the pYLCRISPR/Cas9Pubi-H vector (Ma et al., 2015), yielding a pC-qsh1-Tab construct featuring a Cas9p expression cassette (Pubi:NLS:Cas9p:NLS:Tnos) and a hygromycin resistance cassette (2. 2 × P35S::HPT::T35STnos). This same approach was also used for pC-qsh1-T23 construct preparation.
For pC-qsh1-T1 vector preparation, the chimeric primers containing the qsh1-T1 target site sequence were cloned into the pYLsgRNA-U3 sgRNA cassette (Ma et al., 2015). Synthesis of the gRNA expression cassette was achieved using the U-F/gRNA-R primers (Supplementary Table S1) for an initial round of overlapping PCR, followed by a second round using the site-specific B1’/BL primers (Supplementary Table S1). The qsh1-T1-sgRNA constructs were then cloned into the pYLCRISPR/Cas9Pubi-H CRISPR/Cas9 Multi-targeting vector at a BsaI site. The pC-qsh1-T1 vector was prepared via ligation of qsh1-T1 sgRNA expression cassettes into the pYLCRISPR/Cas9Pubi-H vector. Primers used for PCR reactions are compiled in Supplementary Table S1.
Rice Transformation
The pC-qsh1-Tab or pC-qsh1-T1 or pC-qsh1-T23 binary vectors were introduced into the Agrobacterium tumefaciens EHA105. The parental indica hybrid rice varieties with stronger seed shattering (Oryza satva L. cv. HR1128 and Guangzan63-4S) were then transformed either as descried previously (Hiei et al., 2014) or by Wuhan Biorun biological technology Co., Ltd. After 4 weeks of rooting, regenerated plants were then transferred to plastic buckets and were grown in a greenhouse at 28°C/26°C (during the day and night, respectively).
Mutant Plants Identification and Characterization
In order to confirm the successful targeted mutagenesis of rice plants, genomic DNA (gDNA) was collected via the cetyltrimethyl ammonium bromide (CTAB) method from a minimum of five leaves from different tillers during the mature period of rice growth (Rowland and Nguyen, 1993). This isolated gDNA then served as a template for PCR amplification with KOD FX (TOYOBO, Osaka, Japan). Specific primers were then used to amplify the genomic regions that contained the CRISPR/Cas9 target sites (Supplementary Table S1), after which these PCR fragment were purified and subjected to sequencing. Transgenic plant sequences were compared to those of wild-type (WT) plants in order to identify any mutations located therein. When mutations were associated with a normal sequencing chromatogram, plants were considered to be homozygous. In contrast, mutations that exhibited superimposed sequencing chromatograms were considered to be heterozygous or biallelic. Samples with such superimposed sequence chromatograms were cloned into the pEASY-Blut vector (TransGen Biotech, Beijing, China) and 10 positive clones underwent sequencing as a means of determining the mutation genotype. The DNAMAN5.0 and MAGE4.0 software were used for sequence alignment analysis.
Analysis of Transgene-Free Plants
Plants of the T1 generation were used to identify transgene-free plants. For this analysis, HPT- and Cas9-specific PCR primers (Supplementary Table S1) and agarose gel electrophoresis were used to analyze transgenic plants, with pYLCRISPR/Cas9Pubi-H plasmids and WT plants serving as positive and negative controls, respectively. Plants that were negative for HPT- and Cas9- were considered to be transgene-free.
Analysis of Seed Shattering Phenotype
The propensity of individual lines to undergo seed shattering was analyzed by respective collecting three panicles from different main tillers of each plant once they were mature. A digital force gauge was then used to measure the breaking tensile strength (BTS) at which grains were detached from the pedicle when they were pulled by hand (Qin et al., 2010). This test was repeated for a total of five grains from the uppermost part of each panicle per plant, with three biological replicates per line being analyzed in this manner.
qSH1 Expression and qSH1 Amino Acid Sequence Analyses
Total RNA was isolated from panicles of HR1128-WT and transgene-free homozygous mutant plants using the RNAprep pure Plant Kit (TIANGEN, Beijing, China). Total RNA (1 μg) was reverse-transcribed by using One-Step gDNA Removal and cDNA Synthesis SuperMix (TRNSGEN, Beijing, China). qRT-PCR was performed with gene-specific primers (Supplementary Table S1) using TB Green Premix Ex Taq Ta (TAKARA, Kusatsu, Japan) and Roche LightCycler480 system. Ubiquitin was used an internal standard. Then the amino acid sequence of qSH1 was deduced and aligned for HR1128-WT and transgene-free homozygous mutant plants by using the MAGE4.0 and DNAMAN5.0 software.
Histological Analysis
At 40 days post-heading, panicles of HR1128-WT and transgene-free homozygous mutant plants were harvested for abscission zone (AZ) observation using an approach previously detailed by Ji et al. (2016), with samples being analyzed via light microscopy following Fast Green FCF and Safranine staining.
Agronomic Trait Characterization
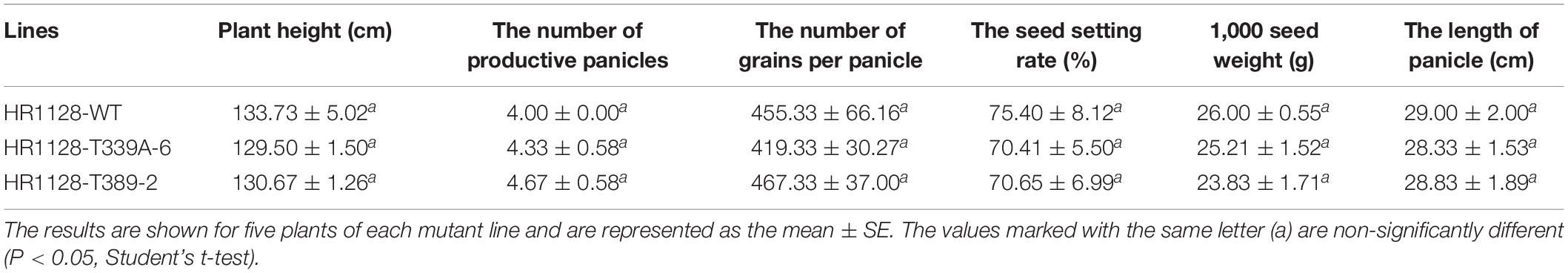
In order to assess the major agronomic traits of transgene-free homozygous mutant lines, wild-type, promising hybrid rice lines, and control-check, these plants were grown under standard conditions in fields located in Changsha or Sanya. Traits including plant height, number of productive panicles, number of grains per panicle, seed setting rates, the weight of 1,000 seeds, and the length of the mature panicle were all measured in three biological replicates per line.
Results
Site-Directed Mutagenesis of qSH1
In an effort to efficiently and specifically mutate the qSH1 gene in rice, we prepared three different CRISPR/Cas9 vectors including two containing two target sgRNAs (pC-qsh1-Tab and pC-qsh1-T23) and one containing a single-target sgRNA (pC-qsh1-T1). Both target-sites a and b (Figure 1A) contained the initiation codon and open reading frame of qSH1, while targets 1, 2, and 3 (Figure 1A) were all located near to a key SNP in the qSH1’s 5′ regulatory region that is associated with reduced seed shattering (Konishi et al., 2006). We then prepared three binary vectors (pC-qsh1-Tab, T1, T23; Figure 1B) based upon a previously described CRISPR/Cas9 vector (Ma et al., 2015). These three vectors were then introduced into the HR1128 and GZ63-4S rice varieties via Agrobacterium-mediated transformation. After transformation, site-specific PCR and Sanger sequencing were used to identify 7, 6, and 10 HR1128 mutants that had been recovered from 11, 7, 13 T0 transgenic HR1128 plants transformed using the pC-qsh1-Tab (63.64%, 7/11), T1 (85.71%, 6/7), and T23 (76.92%, 10/13) vectors, respectively. In addition, 6 GZ63-4S mutants were recovered from 12 T0 transgenic GZ63-4S plants that had undergone transformation with the pC-qsh1-Tab construct (50.00%, 6/12). These results revealed that these pC-qsh1-Tab, T1, and T23 constructs were all capable of rapidly generating qsh1-mutant rice plants.
Characterization of qSH1 Mutant Plants
We next selected 7 (pC-qsh1-Tab, HR1128), 6 (pC-qsh1-T1, HR1128), and 10 (pC-qsh1-T23, HR1128) T0 qsh1-mutant plants that were then used to analyze the target site mutation characterization following transformation. For this approach, we cloned the target-PCR fragments into the pEASY-Blut vector after which positive clones were sequenced. We were then able to classify mutant genotypes into three categories (Table 1 and Figure 2): (1) homozygotes (20.00 to 28.57% mutation efficiency; example: T339A at target-site a); (2) bi-allelic (42.86 to 100.00% mutation efficiency; example: T334 at target-site a); and (3) chimeras (10.00 to 28.57% mutation efficiency; example: T406 at target-site a). The most commonly detected mutations were bi-allelic, in line with previous studies (Zhang et al., 2014; Ma et al., 2015; Tang et al., 2017). Six different types of mutations were identified, including insertions, deletions, substitutions, insertions + substitutions, deletions + substitutions, and insertions + deletions (Table 1). The frequencies of insertions, deletions, and substitutions ranged from 21.05 to 58.33%, 21.05 to 73.68%, and 4.76 to 5.27%, respectively. Single nucleotide insertions were the most common form of mutation, accounting for 30.43% of all such mutations (Figure 3).
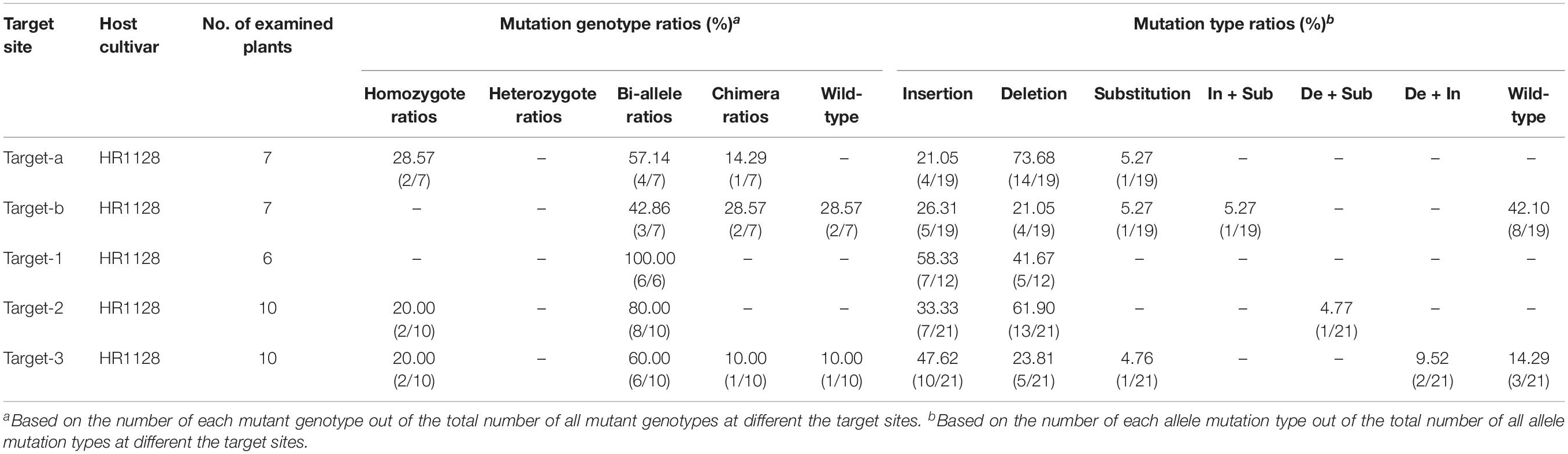
Table 1. Ratios of mutant genotype and mutation type at the different target sites in T0 mutant plants.

Figure 2. (A) is the mutant genotypes of pC-qsh1-T1 HR1128, (B) is the mutant genotypes of pC-qsh1-T23 HR1128, and (C) is the mutant genotypes of pC-qsh1-Tab HR1128. CRISPR/Cas9-induced qSH1 gene modification in rice. Nucleotide sequences at the target site in the 6, 10, and 7 T0 mutant rice plants. The recovered mutated alleles are shown below the wild-type sequence. The target site nucleotides are indicated using black lines. The PAM site is highlighted in blue. Insertions are represented using red letter, while deletions are shown using red hyphens. The initiation cordon is shown in black as an underlined and italicized sequence. WT corresponds to wild-type. The numbers on the right indicate the type of mutation and the number of nucleotides involved. “–,” “ +, ” “m” indicate the deletion, insertion, and substitution of the indicated number of nucleotides, respectively.
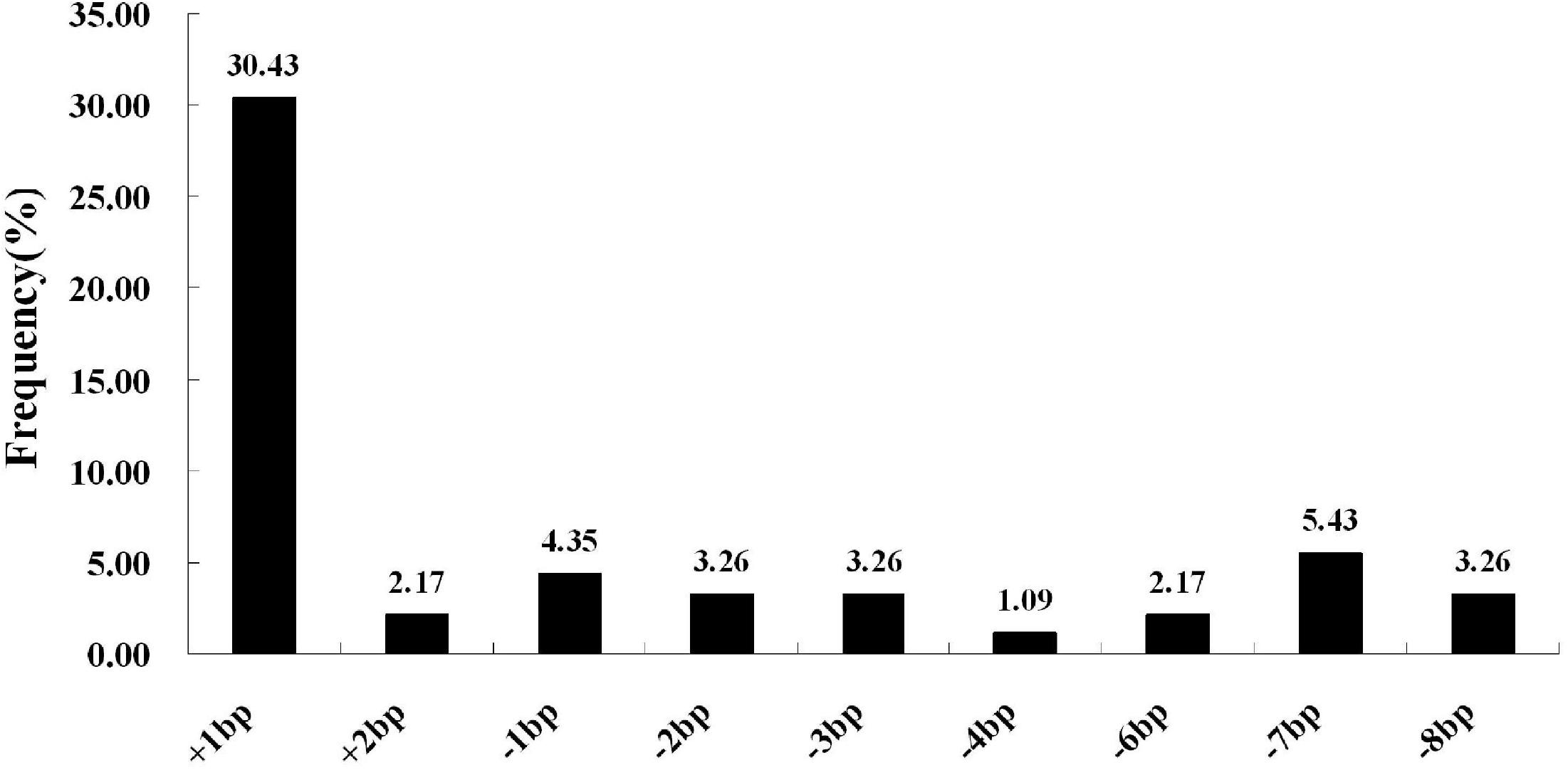
Figure 3. CRISPR/Cas9-induced mutation types and frequencies. The horizontal axis corresponds to different mutation types, whereas the vertical axis corresponds to the frequency at which each type was detected. Of all induced mutation types, single-nucleotide insertions were most frequently detected.
While powerful, the CRISPR/Cas9 technology is susceptible to the introduction of off-target mutations, as reported previously (Endo et al., 2014; Puchta, 2016). We therefore assessed the off-target efficiency of qSH1-Ta, qSH1-Tb, and qSH1-T1 at sites which have high sequence similarity (<5 mismatch bp) to our target sites. We examined four different potential off-target sites within the rice genome, but we detected no evidence of off-target events among 40 T1 plants (Supplementary Table S2). As such, our findings suggest that the sgRNAs used herein were able to achieve high mutagenic specificity.
Development of Transgene-Free Mutant T1 Rice Plants
We next sought to develop rice plants containing homozygous qSH1 mutations that were free of any transferred DNA (T-DNA) derived from the pC-qsh1-Tab, T1, or T23 constructs. In order to test for such plants, we utilized HPT- and Cas9-specific PCR primers (Supplementary Table S1) to amplify DNA from T1 populations derived from five T0 lines. T-DNA-free plants were those in which neither Cas9 nor HPT were detectable, and we were able to identify such plants at frequencies of 21.43 to 27.78% among analyzed plants (Supplementary Table S3 and Figure 4). This confirmed that T1 rice plants could be utilized to effectively develop transgene-free rice plants, as the pYLCRISPR/Cas9 vector and the qSH1 mutations are inherited independently of one another. We then selected three transgene-free plants bearing homozygous qSH1 mutations including plants with coding frameshifts, premature translational stops and altered 5′-UTR regions (HR1128-T339A-6, HR1128-T389-2, GZ63-4S-T622-6), and we then used these plants to generate the T2 population (Figure 4).
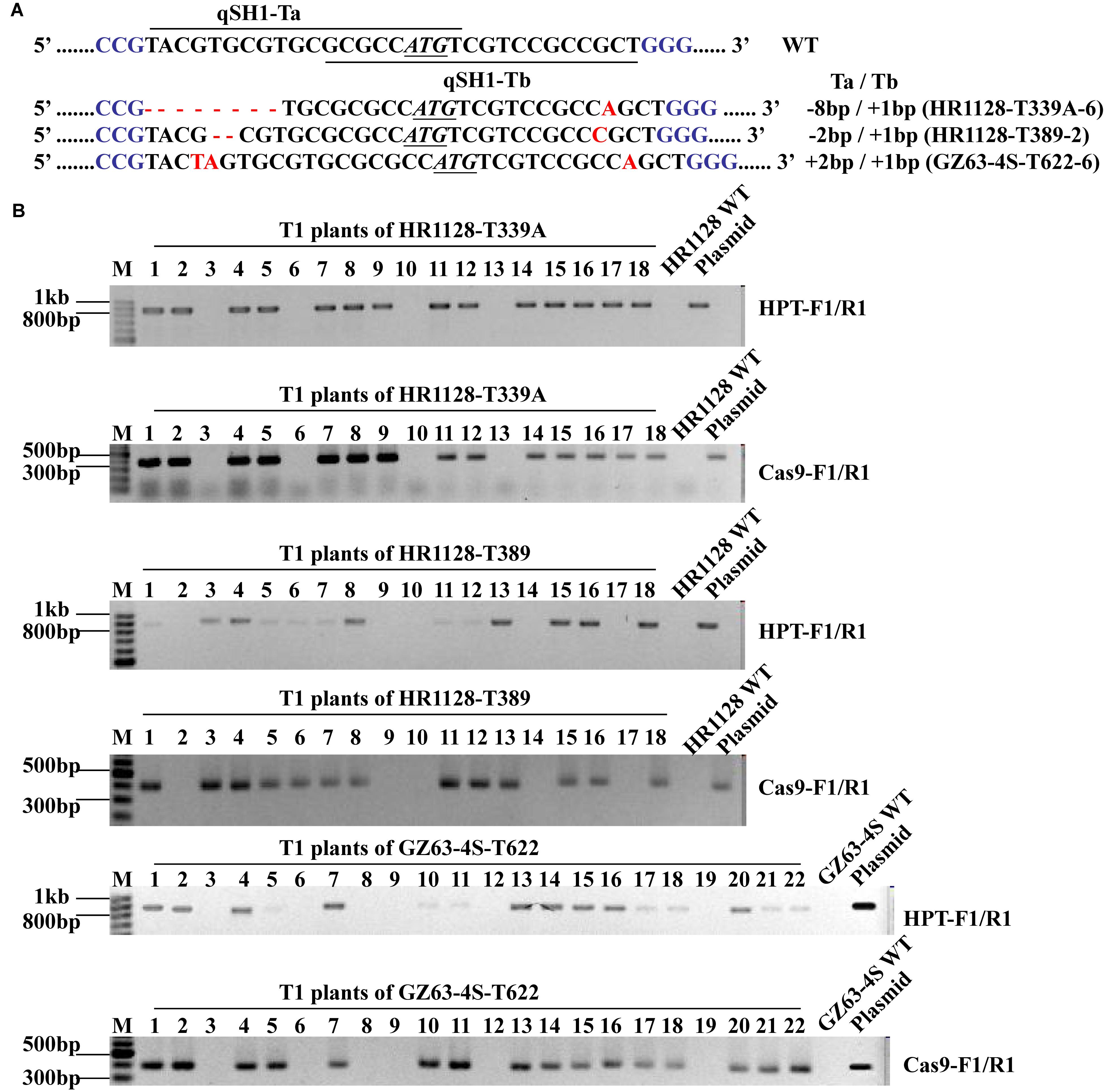
Figure 4. Mutation types and PCR-based identification of transgene-free in the T1 generation. (A) The mutation types of HR1128-T339A-6, HR1128-T389-2, and GZ63-4S-T622-6 (B). HPT and Cas9 gene-specific PCR amplification of HR1128-T339A, HR1128-T389, and GZ63-4S-T622 T1 generation plants.
T2 qSH1 Mutant Lines Exhibit Significantly Reduced Seed Shattering Without any Significant Changes in Major Agronomic Traits
We next measured the breaking tensile strength (BTS) of two T2 transgene-free homozygous mutant lines (HR1128-T339A-6, HR1128-T389-2) as a means of gauging the seed shattering behavior of these mutants relative to that of HR1128-WT. In this analysis, BTS values was negatively correlated with degree of shattering. We were able to detect significant differences in pulling strength when comparing HR1128-WT and mutant plant lines (P < 0.05; Figure 5). Specifically, we found that qSH1 mutants exhibited significantly higher BTS values relative to HR1128-WT, indicating that our CRISPR/Cas9-mediated editing of the qSH1 gene was able to significantly reduce the seed shattering of rice.
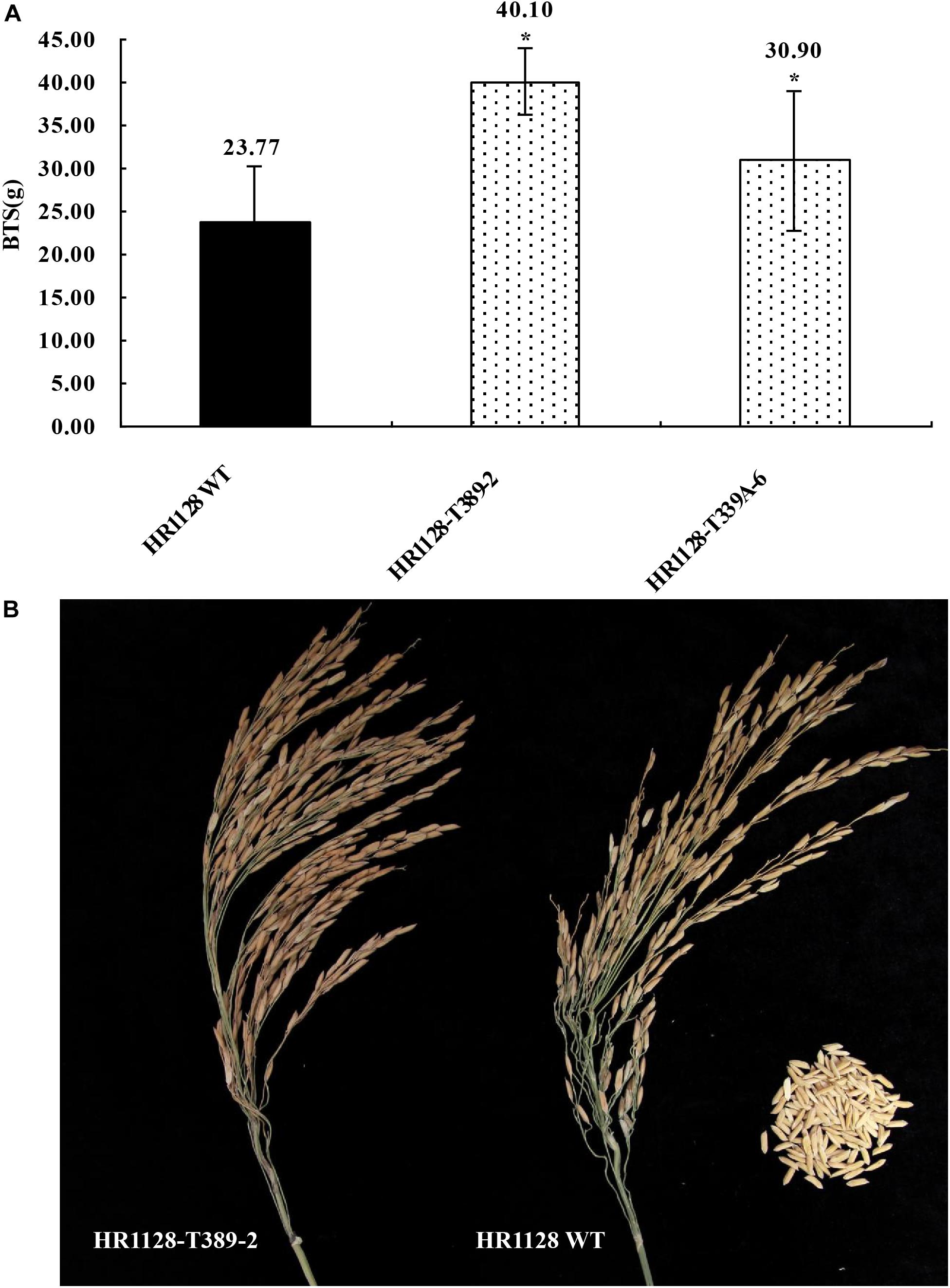
Figure 5. Characterization of seed shattering phenotypes in qsh1 transgene-free homozygous mutant lines. (A) The breaking tensile strength of qsh1 mutant lines (HR1128-T389-2, HR1128-T339A-6 mutant lines) and HR1128-WT line. *P < 0.05. (B) The seed shattering phenotype of qsh1 mutant lines (HR1128-T389-2 mutant lines) and HR1128-WT line.
In order to establish whether these qsh1 mutations had any impact on the major agronomic traits in mutant lines, we next analyzed both the HR1128-WT and the two transgene-free T2 mutant lines derived therefrom. When we measured plant height, number of productive panicles, number of grains per panicle, seed setting rate, weight of 1,000 seeds, and panicle length, we observed no significant differences in any of these traits between HR1128-WT and mutants under normal growth conditions (Table 2). As such, these findings indicate that the targeted mutagenesis of qSH1 has no impact on major agronomic traits when mutants are cultivated under standard conditions.
Analysis of the qSH1 Expression and qSH1 Amino Acid Sequence in Transgene-Free Homozygous Mutant Plants
We measured the relative expression of qSH1 in HR1128-WT and transgene-free homozygous mutant plants by quantitative real-time polymerase chain reaction (qRT-PCR) and found the expression of qSH1 was significantly reduced in HR1128-T389-2 and non-significantly different in HR1128-T339A-6, compared with the HR1128-WT (Figure 6). Next we deduced and aligned the qSH1 amino acid sequences from the HR1128-T339A-6, HR1128-T389-2, and HR1128-WT plants, revealing that each of the two mutant plants encoded the qSH1 protein that was only 231 amino acids in length, whereas the HR1128-WT qSH1 protein was 611 amino acids long (Figure 7). The amino acid sequences in these mutant proteins were also altered, emphasizing that these mutant alleles expressed truncated, disrupted and altered qSH1 proteins.
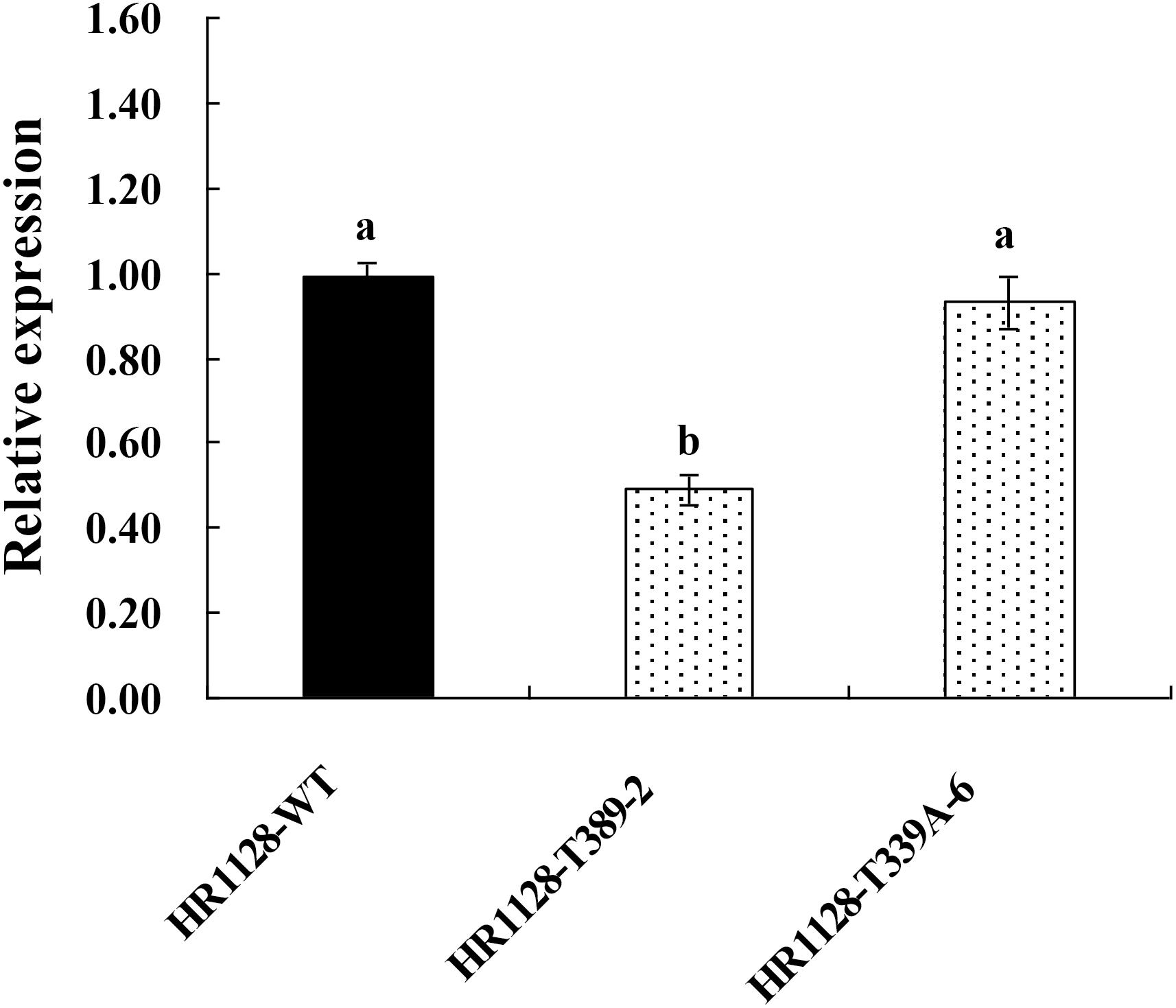
Figure 6. The expression of qSH1 gene in WT and mutant lines. qRT-PCR analysis of the transcription of qSH1 gene in WT line (HR1128-WT) and mutant lines (HR1128-T389-2, HR1128-T339A-6).
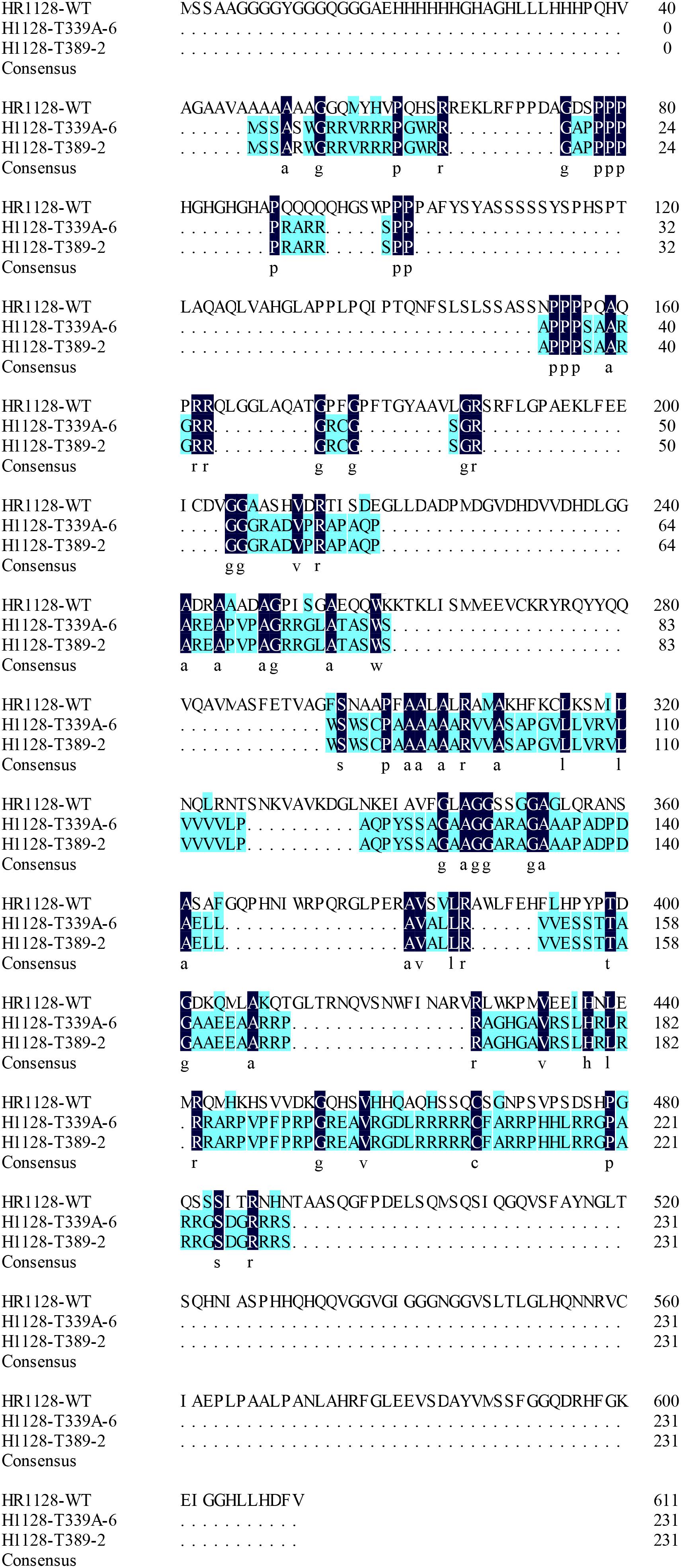
Figure 7. The amino aid sequence of qsh1 transgene-free homozygous mutant lines. Deduced qSH1 amino aid sequence alignment for the two transgene-free homozygous mutant lines (HR1128-T389-2, HR1128-T339A-6) and for WT line (HR1128-WT). Each of the mutant alleles codes for truncated and disrupted qSH1 proteins.
Analysis of the Abscission Zone in Mutant Lines
In order to assess abscission zone (AZ) morphology of our mutant lines, we next analyzed them via optical microscopy (Figure 8). We observed that the AZ between the pedicle and the spikelet at the rice seed base in these mutant lines was partially- developed (Figure 8), with a weaker seed shattering phenotype than that of HR1128-WT. In contrast, the AZ of HR1128-WT was well-developed. This indicates that the targeted mutagenesis of qSH1 resulted in a partial disruption of AZ formation, leading to the improved “easy-to-shatter” phenotype observed in these mutants.
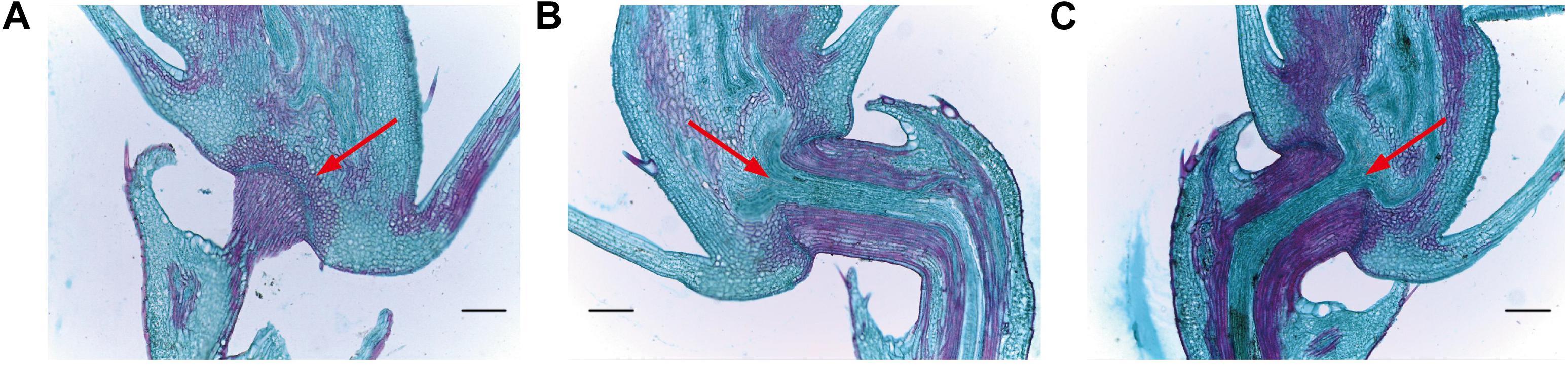
Figure 8. Longitudinal sections of the abscission zone in grain pedicle tissues. (A) Longitudinal abscission zone (AZ) sections from HR1128-WT. (B) Longitudinal abscission zone (AZ) sections from HR1128-T389-2. (C) Longitudinal abscission zone (AZ) sections from HR1128-T339A-6. Magnification time: ×100. Arrows indicate the AZ. Scale bar: 100 μm.
Development of New Promising Hybrid Rice Lines With Reduced Seed Shattering
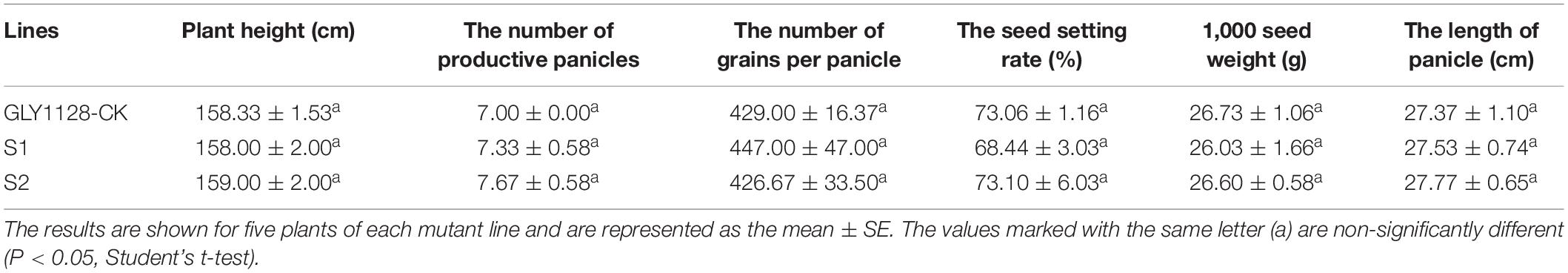
We next crossed the HR1128 mutants (HR1128-T339A-6, HR1128-T389-2) with the GZ63-4S mutant (GZ63-4S-T622-6) in order to generate two independent mutant hybrid rice lines (S1, S2). As a control, we additionally chose the commercial hybrid rice GLY1128 (by crossing HR1128-WT and GZ63-4S-WT). We then grew these mutant hybrid rice lines and GLY1128 line (CK) under normal conditions. When we analyzed the BTS values for these rice lines, we found that the S1 and S2 had significantly greater pulling strength values relative to the CK line (Figure 9), suggesting that the mutation of both parental lines was sufficient to improve the seed shattering of the resultant hybrid line (Table 3). Importantly, these new promising hybrid rice lines did not exhibit any significant morphological differences or changes in grain yield relative to control line (Table 3). These results together clearly demonstrate that CRISPR/Cas9-mediated mutagenesis of the qSH1 gene can be used to generate new promising hybrid rice lines with an intermediate seed shattering phenotype as a means of reducing yield losses during mechanized rice harvesting.
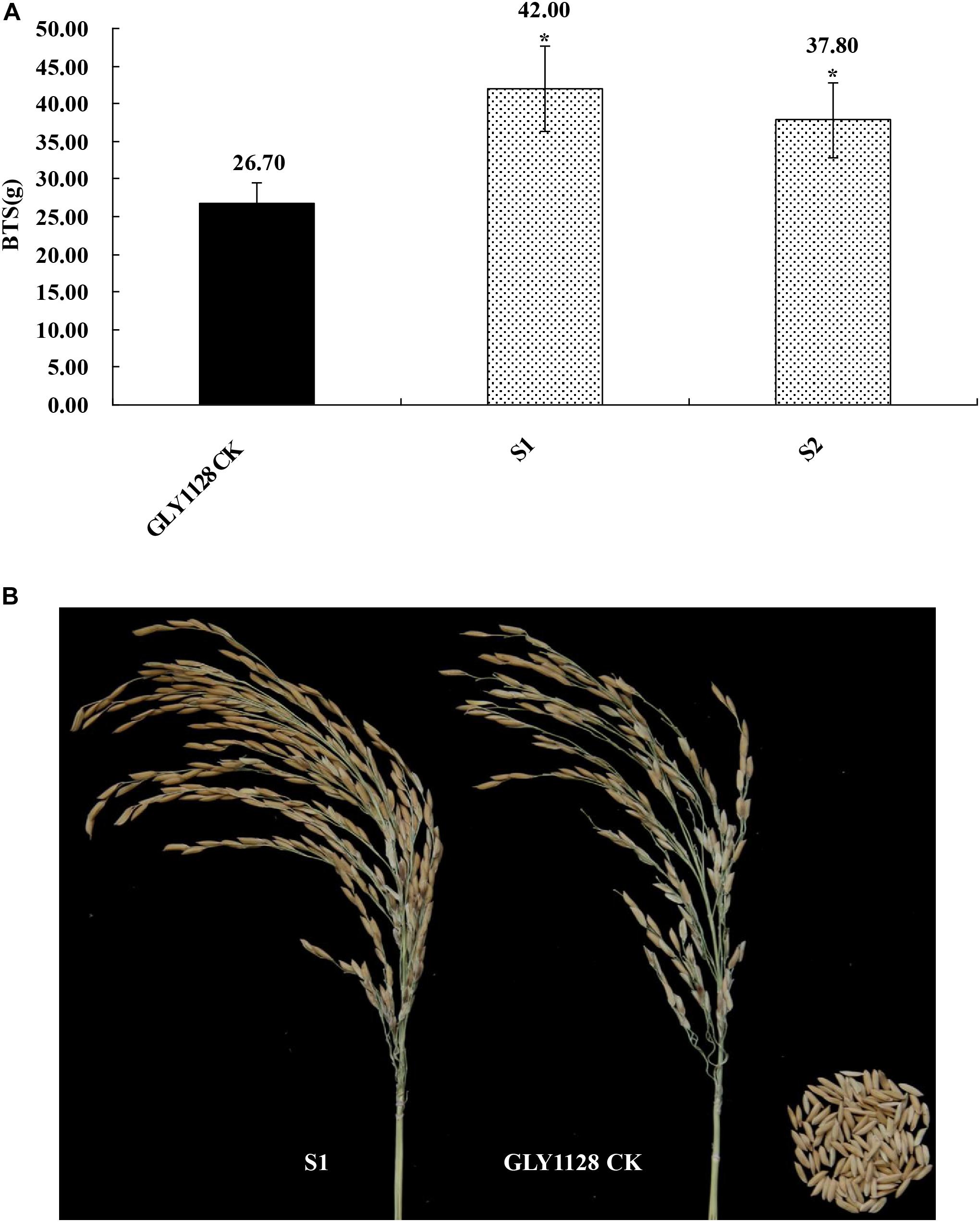
Figure 9. Characterization of the seed shattering phenotype in promising hybrid rice lines. (A) The breaking tensile strength of promising hybrid rice lines (S1, S2) and GLY1128 CK. *P < 0.05. (B) The seed shattering phenotype of the promising hybrid rice line (S1) and GLY1128 CK.
Discussion
In domesticated, rice seed shattering is disadvantageous as it results in significant losses during harvesting, leading to efforts to develop crop varieties with reduced seed shattering (Doebley, 2006). Despite these efforts, many rice varieties are still exhibit the undesirable “easy-to-shatter” phenotype, with this trait being particularly pronounced among indica subspecies. The qSH1 gene has been identified as a key regulator of seed shattering trait, with loss of expression of this gene at the mRNA level having been linked to reduced seed shattering. Meanwhile qSH1 functions downstream of SHAT1 and SH4 so as to maintain their expression in the AZ, leading to the promotion of AZ differentiation and showing stronger seed shattering (Konishi et al., 2006; Zhou et al., 2012). Previous efforts to develop intermediate seed shattering rice varieties have largely relied upon γ irradiation followed by crossing, backcrossing and marker assisted selection (MAS) during breeding, with no prior reports of the development of intermediate-shatter mutants produced via targeted genetic editing. More recently, CRISPR/Cas9-mediated gene editing has been used to genetically modify rice (Zhou et al., 2016; Wang et al., 2016; Tang et al., 2017; Zhang et al., 2019), maize (Feng et al., 2016), wheat (Puchta, 2016), and other crops. Few studies to date, however, have used this approach to edit the parental lines that are used to produce promising hybrid rice varieties. HR1128 is an elite indica restorer line with a lodging resistance, high seed setting rate and large panicle that has been used to generate many different hybrid rice varieties including GuangLiangYou1128 (GLY1128), LiangYou1128 (LY1128), and YLiangYou1128 (YLY1128). HR1128 and its derivatives lines, however, exhibit undesirably strong seed shattering phenotype. In an effort to overcome this limitation, we employed a CRISPR/Cas9 approach to knock-out the qSH1 gene in the parental lines of GLY1128, as the CRISPR/Cas9 technology allows for highly efficient mutagenesis without any sustained transgene insertion in progeny plants.
In the present study, we produced three transgene-free homozygous mutant lines (HR1128-T339A-6, HR1128-T389-2, GZ63-4S-T622-6) in the T2 generation. By further analyzing the qSH1 expression, qSH1 amino acid sequence and assessing the AZ of HR1128-T339A-6, HR1128-T389-2, and HR1128-WT lines, we found that qSH1 proteins were truncated, altered, and disrupted, and that AZ formation was partially disrupted in these mutants, and the HR1128-T389-2 showed significantly reduced qSH1 expression. Previous studies showed that mutations in cis-regulatory regions/elements often cause a change in the expression level and/or pattern of the genes (Li et al., 2017), thus the deletions in 5′-UTR region of HR1128-T389-2 may lead to qSH1 expression significantly reduced. Meanwhile, 1bp insertion in CDS possible activate the nonsense-mediated mRNA decay (NMD) pathway and the process of callus differentiation during transgenic tissue culture also may cause transposon activation, etc. can affect the mRNAs stability (Yin et al., 2007; Christopher and Rachel, 2012), as a result genes expression level of mutants have changed. In summary, we speculated that a loss-of-function of the qSH1 protein was a primary cause of incomplete AZ formation in these mutant plants, resulted in their improved strong seed shattering phenotype and reduced expression of qSH1 may promote for this. Then we produced two promising hybrid rice lines (S1, S2) with an intermediate seed shattering phenotype by crossing the HR1128 mutant line and the GZ63-4S mutant line. These S1 and S2 lines exhibited favorable agronomic traits, were transgene-free, and could be readily used for farming production if permitted by governmental policies. Overall, our results highlight the potential for the use of CRISPR/Cas9 technology to develop promising hybrid rice lines and parental varieties bearing targeted mutations capable of improving the “easy-to-shatter” trait, with qSH1 being a potentially ideal target for the accelerated development of intermediate seed shattering rice varieties.
Heterosis utilization has greatly benefited crop breeding efforts, and recent changes in rice breeding practices have significantly improved rice grain yields owing to the efficient use of heterosis (Wang et al., 2005; Cheng et al., 2007). In hybrid rice varieties (Oryza sativa), heterozygous first filial (F1) generation plants typically exhibit a 10–20% yield advantage over their parental lines (Cheng et al., 2007; Li et al., 2007; Luo et al., 2013), making heterosis an ideal property to leverage for rice breeding efforts. More recently, CRISPR/Cas9 technology has been developed and widely employed as a means of improving a range of crops. However, few studies to date, have combined CRISPR/Cas9 approach with heterosis utilization to produce new varieties, for example via editing the parental lines that are used to produce promising hybrid rice varieties. In the present study, we combined heterosis utilization with targeted gene editing as a means of rapidly producing new promising hybrid rice lines with an improved “easy-to-shatter” phenotype and beared desirable agronomic traits, suggesting that integrating both CRISPR/Cas9 approach and heterosis utilization may potentially represent a powerful, highly efficient, and green approach to the genetic improvement of rice and other hybrid crops breeding. At present the large panicle and large grain size traits are major reasons for high-yield in hybrid rice, however, these favorable agronomic traits easily lead to “easy-to-shatter” and cause serious yield losses. Therefore, reducing strong seed shattering being one beneficial approach to the achieving high rice yields, as a result this study offer a novel strategy and new materials for breeding new rice varieties with an intermediate seed shattering and desirable agronomic traits via combining CRISPR/Cas9 technology with heterosis utilization.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
DyY and MD designed the research work and drafted the manuscript. XS performed the experiments, analyzed the data, and drafted the manuscript. ZS and XW modified the drafted. YT, DoY, and GY participated in performing the experiments.
Funding
This work was supported by Entrustment Project of Hunan Province (Grant No. 2019TJ-Q08), by National Transgenic Projects of China (Grant No. 2014ZX0801001B and No. 2016ZX08001-001), by the National Natural Science Foundation of China (Grant No. 31771767 and No. 31801341), by Hunan Science and Technology Major Project (Grant No. 2018NK1010).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Yaoguang Liu (South China Agricultural University) provided the CRISPR/Cas9 editing vectors, pYLgRNA-U3, and pYLCRISPR/Cas9-MH, Prof. Kaijun Zhao (Chinese Academy of Agriculture Sciences) and Dr. Yan Peng (Hunan Agricultural University) for their suggestions for improvement of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00619/full#supplementary-material
Footnotes
References
Baltes, N., and Voytas, D. (2015). Enabling plant synthetic biology through genome engineering. Trends Biotechnol. 33, 120–131. doi: 10.1016/j.tibtech.2014.11.008
Brooks, C., Nekrasov, V., Lippman, Z., and Van Eck, J. (2014). Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol. 166, 1292–1297. doi: 10.1104/pp.114.247577
Cheng, S., Zhuang, J., Fan, Y., Du, J., and Cao, L. (2007). Progress in research and development on hybrid rice: a super-domesticate in China. Ann. Bot. 100, 959–966. doi: 10.1093/aob/mcm121
Christopher, J. S., and Rachel, G. (2012). Translation drives mRNA quality control. Nat. Struct. Mol. Biol. 19, 594–601. doi: 10.1038/nsmb.2301
Doebley, J. (2006). Plant science. unfallen grains: how ancient farmers turned weeds into crops. Science 312, 1318–1319. doi: 10.1126/science.1128836
Endo, M., Mikami, M., and Toki, S. (2014). Multigene knockout utilizing of-target mutations of the CRISPR/Cas9 system in rice. Plant Cell Physiol. 56, 41–47. doi: 10.1093/pcp/pcu154
Feng, C., Yuan, J., Wang, R., Liu, Y., Birchler, J., and Han, F. (2016). Efficient targeted genome modification in maize using CRISPR/Cas9 system. J. Genet. Genomics 43, 37–43. doi: 10.1016/j.jgg.2015.10.002
Feng, Z., Zhang, B., Ding, W., Liu, X., Yang, D., Wei, P., et al. (2013). Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 23, 1229–1232. doi: 10.1038/cr.2013.114
Fuller, D. Q., Qin, L., Zheng, Y. F., Zhao, Z. J., Chen, X., Hosoya, L. A., et al. (2009). The domestication process and domestication rate in rice: spikelet bases from the Lower Yangtze. Science 323, 1607–1610. doi: 10.1126/science.1166605
Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (2014). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacteriumand sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282. doi: 10.1046/j.1365-313X.1994.6020271.x
Hyun, Y., Kim, J., Cho, S., Choi, Y., Kim, J., and Coupland, G. (2015). Site-directed mutagenesis in Arabidopsis thaliana using dividing tissue-targeted RGEN of the CRISPR/Cas system to generate heritable null alleles. Planta 241, 271–284. doi: 10.1007/s00425-014-2180-5
Ito, Y., Nishizawa-Yokoi, A., Endo, M., Mikami, M., and Toki, S. (2015). CRISPR/Cas9-mediated mutagenesis of the RIN locus that regulates tomato fruit ripening. Biochem. Biophys. Res. Commun. 467, 76–82. doi: 10.1016/j.bbrc.2015.09.117
Jacobs, T., LaFayette, P., Schmitz, R., and Parrott, W. (2015). Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnol. 15:16. doi: 10.1186/s12896-015-0131-2
Ji, H., Chu, S., Jiang, W., Cho, Y., Hahn, J., Eun, M., et al. (2016). Characterization and mapping of a shattering mutant in rice that corresponds to a block of domestication genes. Genetics 173, 995–1005. doi: 10.1534/genetics.105.054031
Ji, H., Kim, S., Kim, Y., Kim, H., Eun, M., Jin, I., et al. (2010). Inactivation of the CTD phosphatase-like gene OsCPL1 enhances the development of the abscission layer and seed shattering in rice. Plant J. 61, 96–106. doi: 10.1145/1639950.1640004
Jiang, L., Ma, X., Zhao, S., Tang, Y., Liu, F., Gu, P., et al. (2019). The APETALA2-like transcription factor SUPERNUMERARY BRACT controls rice seed shattering and seed size. Plant Cell 31, 17–36. doi: 10.1105/tpc.18.00304
Jiang, W., Zhou, H., Bi, H., Fromm, M., Yang, B., and Weeks, D. (2013). Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 41:e188. doi: 10.1093/nar/gkt780
Konishi, S., Izawa, T., Lin, S. Y., Ebana, K., Fukuta, Y., Sasaki, T., et al. (2006). An SNP caused loss of seed shattering during rice domestication. Science 312, 1392–1396. doi: 10.1126/science.1126410
Li, C. B., Zhou, A. L., and Sang, T. (2006). Rice domestication by reducing shattering. Science 311, 1936–1939. doi: 10.1126/science.1123604
Li, S., Yang, D., and Zhu, Y. (2007). Characterization and use of male sterility in hybrid rice breeding. J. Integr. Plant Biol. 49, 791–804. doi: 10.1111/j.1744-7909.2007.00513.x
Li, T., Liu, B., Spalding, M. H., Weeks, D. P., and Yang, B. (2012). High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 30, 390–392. doi: 10.1038/nbt.2199
Li, X., Xie, Y., Zhu, Q., and Liu, Y. (2017). Targeted genome editing in genes and cis-regulatory regions improves qualitative and quantitative traits in crops. Mol. Plant 10, 1368–1370. doi: 10.1016/j.molp.2017.10.009
Liang, Z., Zhang, K., Chen, K., and Gao, C. (2014). Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J. Genet. Genomics 41, 63–68. doi: 10.1016/j.jgg.2013.12.001
Lin, Z., Griffith, M. E., Li, X., Zhu, Z., Tan, L., Fu, Y., et al. (2007). Origin of seed shattering in rice (Oryza sativa L.). Planta 226, 11–20. doi: 10.2307/23389646
Luo, D., Xu, H., Liu, Z., Guo, J., Li, H., Chen, L., et al. (2013). A detrimental mitochondrial-nuclear interaction causes cytoplasmic male sterility in rice. Nat. Genet. 45, 573–577. doi: 10.1038/ng.2570
Lv, S., Wu, W., Wang, M., Meyer, R. S., Ndjiondjop, M. N., Tan, L., et al. (2018). Genetic control of seed shattering during African rice domestication. Nat. Plants 4, 331–337. doi: 10.1038/s41477-018-0164-3
Ma, X., Zhang, Q., Zhu, Q., Liu, W., Chen, Y., Qiu, R., et al. (2015). A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8, 1274–1284. doi: 10.1016/j.molp.2015.04.007
Miao, J., Guo, D., Zhang, J., Huang, Q., Qin, G., Zhang, X., et al. (2013). Targeted mutagenesis in rice using CRISPR/Cas system. Cell Res. 23, 1233–1236. doi: 10.1038/cr.2013.123
Nekrasov, V., Staskawicz, B., and Weigel, D. (2013). Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31, 691–693. doi: 10.1038/nbt.2655
Puchta, H. (2016). Applying CRISPR/Cas for genome engineering in plants: the best is yet to come. Curr. Opin. Plant Biol. 36, 1–8. doi: 10.1016/j.pbi.2016.11.011
Qin, Y., Kim, S., Zhao, X., Jia, B., Lee, H., Kim, K., et al. (2010). Identification for quantitative trait loci controlling grain shattering in rice. Genes Genomics 32, 173–180. doi: 10.1007/s13258-009-0879-7
Rowland, L., and Nguyen, B. (1993). Use of polyethylene glycol for purifcation of DNA from leaf tissue of woody plants. Biotechniques 14, 734–736. doi: 10.1002/bip.360330513
Shan, Q., Wang, Y., Li, J., and Gao, C. (2014). Genome editing in rice and wheat using the CRISPR/Cas system. Nat. Protoc. 9, 2395–2410. doi: 10.1038/nprot.2014.157
Shan, Q., Wang, Y., Li, J., Zhang, Y., Chen, K., Liang, Z., et al. (2013). Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotech. 2013 31, 686–688. doi: 10.1038/nbt.2650
Su, N., Hu, M., Wu, D., Wu, F., Fei, G., Lan, Y., et al. (2012). Disruption of a rice pentatricopeptide repeat protein causes a seedling-specifc albino phenotype and its utilization to enhance seed purity in hybrid rice production. Plant Physiol. 159, 227–238. doi: 10.1104/pp.112.195081
Svitashev, S., Young, J., Schwartz, C., Gao, H., Falco, S., and Cigan, A. (2015). Targeted mutagenesis, precise gene editing, and site-specifc gene insertion in maize using Cas9 and guide RNA. Plant Physiol. 169, 931–945. doi: 10.1104/pp.15.00793
Tang, L., Mao, B., Li, Y., Lv, Q., Zhang, L. P., Chen, C., et al. (2017). Knockout of OsNramp5 using the Crispr/Cas9 system produces low Cd-accumulating indica rice without compromising yield. Sci. Rep. 7:14438. doi: 10.1038/s41598-017-14832-9
Townsend, J., Wright, D., Winfrey, R., Fu, F., Maeder, M., Joung, J., et al. (2009). High-frequency modifcation of plant genes using engineered zinc-fnger nucleases. Nature 459, 442–445. doi: 10.1038/nature07845
Upadhyay, S., Kumar, J., Alok, A., and Tuli, R. (2013). RNA-guided genome editing for target gene mutations in wheat. Genes Genomes Genetics 3, 2233–2238. doi: 10.1534/g3.113.008847
Wang, F., Wang, C., Liu, P., Lei, C., Hao, W., Gao, Y., et al. (2016). Enhanced Rice Blast Resistance by CRISPR/Cas9-Targeted Mutagenesis of the ERF Transcription Factor Gene OsERF922. PLoS One 11:e0154027. doi: 10.1371/journal.pone.0154027
Wang, N., Long, T., Yao, W., Xiong, L., Zhang, Q., and Wu, C. (2013). Mutant resources for the functional analysis of the rice genome. Mol. Plant 6, 596–604. doi: 10.1093/mp/sss142
Wang, S., Zhang, S., Wang, W., Xiong, X., Meng, F., and Cui, X. (2015). Efficient targeted mutagenesis in potato by the CRISPR/Cas9 system. Plant Cell Rep. 34, 1473–1476. doi: 10.1007/s00299-015-1816-7
Wang, Y., Cheng, X., Shan, Q., Zhang, Y., Liu, J., Gao, C., et al. (2014). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951. doi: 10.1038/nbt.2969
Wang, Y., Xue, Y., and Li, J. (2005). Towards molecular breeding and improvement of rice in China. Trends Plant Sci. 10, 610–614. doi: 10.1016/j.tplants.2005.10.008
Weeks, D., Spalding, M., and Yang, B. (2016). Use of designer nucleases for targeted gene and genome editing in plants. Plant Biotechnol. J. 14, 483–495. doi: 10.1111/pbi.12448
Xu, R., Li, H., Qin, R., Wang, L., Li, L., Wei, P., et al. (2014). Gene targeting using the Agrobacterium tumefaciensmediated CRISPR-Cas system in rice. Rice 7:5. doi: 10.1186/s12284-014-0005-6
Ye, X., Xu, L., Cong, S., Chuan, T., and Shen, S. (2012). Application evaluation on improved type of the super Rice‘Xieqingzao A/M9308’with medium shattering habit. Chinese Agricult. Sci. Bull. 28, 131–134. doi: 10.3969/j.issn.1000-6850.2012.21.024
Yin, L., Tao, Y., Zhao, K., Shao, J., Li, X., Liu, G., et al. (2007). Proteomic and transcriptomic analysis of rice mature seed-derived callus differentiation. Proteomics 7, 755–768. doi: 10.1002/pmic.200600611
Zhang, A., Liu, Y., Wang, F., Li, T., Chen, Z., Kong, D., et al. (2019). Enhanced rice salinity tolerance via CRISPR/Cas9-targeted mutagenesis of the OsRR22 gene. Mol. Breed. 39:47. doi: 10.1007/s11032-019-0954-y
Zhang, H., Zhang, J., Wei, P., Zhang, B., Gou, F., Feng, Z., et al. (2014). The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. 12, 797–807. doi: 10.1111/pbi.12200
Zhou, H., He, M., Li, J., Chen, L., Huang, Z., Zheng, S., et al. (2016). Development of commercial thermo-sensitive genic male sterile rice accelerates hybrid rice breeding using theCRISPR/Cas9-mediated TMS5 editing system. Sci Rep. 6:37395. doi: 10.1038/srep37395
Zhou, Y., Lu, D., Li, C., Luo, J., Zhu, B., Zhu, J., et al. (2012). Genetic control of seed shattering in rice by the APETALA2 transcription factor shattering Abortion1. Plant Cell 24, 1034–1048. doi: 10.1105/tpc.111.094383
Keywords: CRISPR/Cas9, heterosis utilization, qSH1, rice, seed shattering
Citation: Sheng X, Sun Z, Wang X, Tan Y, Yu D, Yuan G, Yuan D and Duan M (2020) Improvement of the Rice “Easy-to-Shatter” Trait via CRISPR/Cas9-Mediated Mutagenesis of the qSH1 Gene. Front. Plant Sci. 11:619. doi: 10.3389/fpls.2020.00619
Received: 25 February 2020; Accepted: 22 April 2020;
Published: 25 May 2020.
Edited by:
Zeng-Yu Wang, Qingdao Agricultural University, ChinaReviewed by:
Seonghoe Jang, World Vegetable Center Korea Office, South KoreaKeisuke Nagai, Nagoya University, Japan
Copyright © 2020 Sheng, Sun, Wang, Tan, Yu, Yuan, Yuan and Duan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dingyang Yuan, eXVhbmRpbmd5YW5nQGhocnJjLmFjLmNu; Meijuan Duan, ZHVhbm1laWp1YW5AMTYzLmNvbQ==
 Xiabing Sheng
Xiabing Sheng Zhizhong Sun
Zhizhong Sun Xuefeng Wang1
Xuefeng Wang1 Yanning Tan
Yanning Tan