- 1Department of Agricultural Chemistry, Institute of Environmentally Friendly Agriculture, College of Agriculture and Life Sciences, Chonnam National University, Gwangju, South Korea
- 2Department of Chemistry, Chonnam National University, Gwangju, South Korea
The over and repeated use of chemical bactericides to control plant bacterial diseases has resulted in unwanted effects, such as environmental pollution, residual toxicity, and resistance buildup in bacterial pathogens. Many previous studies have aimed to develop biological control agents to replace chemical bactericides. In this study, the antibacterial efficacy of the fermentation broth of Paenibacillus elgii JCK-5075 and its antibacterial compounds were evaluated against plant pathogenic bacteria, using both in vitro and in vivo bioassays. Pelgipeptins (PGPs) A, B, C, and D that were isolated from P. elgii JCK-5075 displayed broad-spectrum antibacterial activity against various plant pathogenic bacteria. The fermentation broth of P. elgii JCK-5075, at 5-fold dilution, effectively suppressed the development of tomato bacterial wilt, Kimchi cabbage soft rot, and red pepper bacterial leaf spot in pot experiments with control values of 81, 84, and 67%, respectively. PGP-A and C, at 200 μg/ml, were also found to markedly reduce the development of Kimchi cabbage bacterial soft rot by 75% and tomato bacterial wilt by 83%, respectively, and their disease control efficacy was comparable to that of oxolinic acid with control values of 81 and 85%, respectively. Additionally, the antibacterial activity of PGP-C was found to be directly correlated with membrane damage mechanisms. These results indicates that P. elgii JCK-5075 producing PGPs could be used as a biocontrol agent for the control of plant bacterial diseases. This is the first report on the in vitro and in vivo antibacterial activity of PGPs against bacterial plant pathogens.
Highlights
– We found that pelgipeptins isolated from P. elgii JCK-5075 showed very strong in vitro and in vivo antibacterial activity against plant pathogenic bacteria. Additionally, the antibacterial mechanism of pelgipeptin-C was elucidated to be membrance damage.
Introduction
Even though fungal diseases are more numerous and severe in plants, there are also several important pathogenic bacteria that can cause significant annual losses on a global scale, specifically in moist and hot climatic conditions (Egli and Sturm, 1981). The 10 most important bacterial plant pathogens, based on their scientific and economic importance are Pseudomonas syringae pathovars, Ralstonia solanacearum, Agrobacterium tumefaciens, Xanthomonas oryzae pv. oryzae, Xanthomonas campestris pathovars, Xanthomonas axonopodis pathovars, Erwinia amylovora, Xylella fastidiosa, Dickeya (dadantii and solani), and Pectobacterium carotovorum (Mansfield et al., 2012). Bacterial diseases in plants, however, are difficult to control for the following reasons: (1) bacterial pathogens under optimal environmental conditions can quickly increase their population sizes; (2) some soil-borne bacteria are an absence of effective fumigants for soil-borne bacteria; and (3) many pathogenic bacteria predominantly colonize the internal spaces of plants, while many of the standard control measures only target the plant surface (Sundin et al., 2016).
Over the last few decades, chemical control strategies have been applied to control plant diseases. However, the over and repeated use of several chemical pesticides, such as antibiotics and copper compounds, has caused pollution of the soil environment, a decrease in the population of beneficial microorganisms, residual toxicity on food commodities, and the occurrence of bactericide-resistant strains. In order to replace chemical pesticides, biological control agents, including bioactive metabolites produced by microorganisms or the microbial cells themselves, have previously been intensively studied (Roth et al., 1997; Kim et al., 2009; Lugtenberg and Kamilova, 2009; Sundin et al., 2016; Tsuda et al., 2016). The interest in biological control agents has increased, owing to their benefits, which include that they may potentially be self-sustaining as they may spread on their own after initial establishment; that they may require a reduced input of non-renewable resources; and that they may result in long-term disease suppression in the environment. The biological agents that have been used previously were mostly based on bacteria (90%) and fungi (10%) (Yuliar et al., 2015).
Bacteria produce a wide array of compounds to suppress their competitors and colonize new habitats, giving them a competitive advantage. Bacteria capable of thriving in extreme conditions are very likely to evolve special adaptations and produce unique molecules or compounds that may have beneficial uses. The use of beneficial bacteria is considered to be one of the most promising methods for more rational and safe crop-management practices (Fravel, 2005). Bacterial control agents have previously predominantly been from the genera of Pseudomonas, Bacillus, Streptomyces, and Paenibacillus (Shoda, 2000). Recently, many papers have reported the biocontrol potential of Paenibacillus spp. (Beatty and Jensen, 2002; Kim et al., 2009; Grady et al., 2016; Kumar et al., 2016). Paenibacillus species are a Gram variable, rod-shaped, endospore forming, and facultative anaerobes that were reclassified from Bacillus in 1993 by an extensive comparative analysis of their 16S ribosomal RNA gene sequences (Ash et al., 1993; Grady et al., 2016). Like other bacteria, the biocontrol capabilities of Paenibacillus spp. come from the promotion of plant growth, the induction of the plant’s own resistance, competition for nutrient and space conditions, antibiosis, and parasitism (Chung et al., 2000; Hong and Meng, 2003; He et al., 2007; Antonopoulos et al., 2008; Algam et al., 2010; Phi et al., 2010; Lee et al., 2012; Nguyen et al., 2015; Zhou et al., 2016). Paenibacillus species can be used in combination with other chemical products, as they have the capability to tolerate commercial fungicides or insecticides (Singh et al., 2009). Members of Paenibacillus are known to produce non-ribosomal lipopeptides with potent antimicrobial activities, such as polymyxin B by P. polymyxa, battacin by P. tianmuensis, pelgipeptins by P. ehimensis, and P. elgii produced PGPs (Khan et al., 2008; Wu et al., 2010; Ding et al., 2011; Shaheen et al., 2011; Qian et al., 2012; Huang et al., 2013).
As for the disease control efficacy of Paenibacillus species, the P. polymyxa strain KNUC265 was reported to protect pepper and tobacco from X. axonopodis and Erwinia carotovora (P. carotovorum), respectively, using bacterial volatiles and diffusible metabolites as elicitors of induced systemic resistance (Phi et al., 2010). P. polymyxa E681 was also shown to protect Arabidopsis thaliana against P. syringae via induced systemic resistance (Lee et al., 2012). Octapeptins and paenibacterin, that are cyclic lipopeptides produced by Paenibacillus, are active against both gram-negative and gram-positive bacteria (Huang and Yousef, 2014; Cochrane and Vederas, 2016). Fusaricidins are known to be active against many important phytopathogens and a variety of gram-positive bacteria (Kajimura and Kaneda, 1996). Additionally, pelgipeptin (PGP)-E that was isolated from P. elgii BC34-6, was reported to be active against both gram-negative and gram-positive bacteria (Kim et al., 2018). Recently, our research group reported that PGP-A, B, C, and D, that were produced by P. elgii JCK1400 strain, showed strong antifungal activity against plant pathogenic fungi and PGP-C was effective at controlling tomato gray mold and wheat leaf rust (Kim et al., 2019). However, the information on the disease control efficacy of the antibacterial metabolites produced by the Paenibacillus species, against plant bacterial diseases and their antibacterial action mechanisms, was limited.
During the discovery of the antimicrobial metabolites, we found that the PGP-A, B, C, and D, that were isolated from the P. elgii JCK-5075, were also highly active at inhibiting the cell growth of various phytopathogenic bacteria. To the best of our knowledge, there is no previous report on the disease control efficacy of PGPs against bacterial plant diseases and their mode of action in phytopathogenic bacteria. Therefore, the objectives of this study were as follows: (1) to examine the in vitro antibacterial activity of the fermentation filtrate of P. elgii JCK-5075 and the purified PGPs against 14 plant pathogenic bacteria; (2) to evaluate the biocontrol efficacy of the fermentation broth of the JCK-5075 and the purified PGPs against bacterial plant diseases, and (3) to elucidate the mode of action of PGP-C on plant pathogenic bacteria.
Materials and Methods
Culture Conditions for JCK-5075
Several bacterial strains were isolated from a soil sample which was collected from one red pepper land, Daejeon city, South Korea in 2015. Among the strains, one strain (JCK-5075) showed very strong in vitro antibacterial activity against R. solanacearum. The bacterial strain JCK-5075 was cultured on tryptic soy agar (TSA; Becton, Dickinson and Co., Sparks, MD, United States) medium at 30°C for 3 days and then stored at −80°C in 30% glycerin until use.
Identification of Antagonistic Bacteria
For the identification of JCK-5075, genomic DNA was extracted using the bacterial genomic DNA purification kit (ELPIS-Biotech, Daejeon, South Korea). The 16S rRNA gene was amplified by PCR using specific primer pair sets and sequencing (Genotech Co., Daejeon, South Korea). Cycles consisted of an initial denaturation at 95°C for 10 min, followed by 40 cycles (95°C for 30 s, 60°C for 30 s, 72°C for 40 s), and a final extension at 72°C for 10 min. The gene sequences obtained was compared with the database from GenBank using BioEdit version 5.0.9.1 (Kim et al., 2018). The phylogenetic trees were constructed by the neighbor-joining method in MEGA version 6.0 with 1000 bootstrap replicates.
Plants
The seeds of tomato (cultivar “Seokwnag;” Farm Hannong Co., Ltd., Seoul, South Korea), Kimchi cabbage (cultivar “Chunkwang;” Sakada Korea, Seoul, South Korea), and red pepper (cultivar “Josaengsintopgochu;” Non-gwoobio Co., Ltd., Suwon, South Korea), were sown in vinyl pots (6.0 cm diameter) (1 seed per pot) containing nursery soil (Punong nursery series #5; Seoulbio Co., Ltd., Gyeongju, South Korea), and kept in an incubation room with 12 h of daylight per day. The plants were transplanted to larger plastic pots (7.5 cm diameter) 24 h prior to the treatments.
Plant Pathogenic Bacteria
Fourteen pathogenic bacteria were used in this study: Acidovorax avenae subsp. cattleyae (bacterial brown spot of orchid), Acidovorax konjaci (bacterial black spot of cucumber), A. tumefaciens (crown gall of apple), Burkholderia glumae (bacterial panicle blight of rice), P. carotovorum subsp. carotovorum (bacterial soft rot of Kimchi cabbage), Pectobacterium chrysanthemi (bacterial soft rot of aloe), P. syringae pv. actinidiae (bacterial canker of kiwifruit), P. syringae pv. lachrymans (bacterial angular leaf spot of cucumber), Xanthomonas euvesicatoria (bacterial leaf spot of red pepper), Xanthomonas arboricola pv. pruni (bacterial spot of stone fruit), X. axonopodis pv. citri 24-20 (bacterial canker of citrus), X. oryzae pv. oryzae (bacterial blight of rice), R. solanacearum SL341 (bacterial wilt of tomato), and Clavibacter michiganensis subsp. michiganensis (bacterial canker of tomato). All plant pathogenic bacteria were isolated from infected plant tissues in South Korea by Dr. S. D. Lee of the National Academy of Agricultural Sciences, Prof. S. W. Lee of Dong-A University, and Prof. Y.-G. Ko of the Suncheon National University. The bacteria were all cultured on Tryptic Soy Agar (TSA) and Tryptic Soy Broth (TSB) medium at 30°C, except for Xanthomonas spp. which was incubated at 28°C.
Purification and Structural Determination of PGPs
The purification and structural determination of the antimicrobial substances from the culture supernatant of the P. elgii JCK-5075 was conducted as previously reported (Kim et al., 2019). Briefly, cell-free supernatant was obtained by centrifugation of the culture broth. Then, the supernatant was loaded onto a Diaion HP-20 column (IONTEC, South Korea). The fractions were eluted using a stepwise ethanol gradient (0, 20, 40, 60, 80, and 100%, v/v). Each fraction was concentrated by evaporation and further purification was performed using reverse-phase high-performance liquid chromatography (RP-HPLC; Shimadzu, Japan). The column used Shim-Pack C18 (20 × 250 mm) and the mobile phases were distilled water and acetonitrile (ACN), containing 0.05% trifluoroacetic acid (TFA). A linear gradient of 30–46% over 25 min was used with a flow rate of 10 ml/min. The purified substances were lyophilized, and their molecular masses were identified by liquid chromatography electrospray ionization mass spectrometry (LC-ESI-MS; API2000, AB SCIEX, United States), in positive mode.
In vitro Antibacterial Assay
The antibacterial assays of the culture filtrate of JCK-5075, crude substances (80% ethanol crude of Diaion HP-20), and PGPs, were conducted in a 96-well plate for microorganisms as previously reported, using the broth dilution method (Vu et al., 2017). Two-fold serial dilutions were performed with 10% of the culture supernatant, 100 μg ml–1 of crude substances, and 32 μg ml–1 of PGPs. Bacterial suspension (105 CFU/ml) was added to each well and the final volume of the solution in each well was 100 μl. Streptomycin sulfate and oxolinic acid were chosen as the positive controls. TSB was used as an untreated control for the culture filtrate and 1% methanol for crude substances and PGPs. The plates were incubated at 28 or 30°C for 1–2 days and the growth of the pathogenic bacteria were measured by optical density (OD) at 600 nm by Microplate Reader (iMark, Bio-Rad). Each run of the experiment contained three replicates, and the entire experiment was run in entirely twice. Minimum inhibitory concentration (MIC) values were at the lowest culture filtrate concentration and the inhibition rate was calculated as follows:
In vivo Antibacterial Assays
Preparation of the Samples
The biocontrol efficacy of the fermentation broth of the P. elgii JCK-5075 was evaluated against Kimchi cabbage soft rot, tomato bacterial wilt, and red pepper bacterial leaf spot. In addition, PGP-A and PGP-C were tested against Kimchi cabbage soft rot and tomato bacterial wilt, respectively. The fermentation broth of P. elgii JCK-5075 was diluted with distilled water at 5, 10, and 20-fold, and then Tween 20 was added to each dilution at a concentration of 250 μg/ml. PGP-A and C were dissolved in methanol and then diluted with distilled water at concentrations of 200, 100, and 50 μg/ml, followed by the addition of Tween 20 (250 μg/ml). The final concentrations of the methanol in the chemical solutions were 5%. A commercial bactericide, Ilpum (Oxolinic acid 20% WP; Dongbang Agro, South Korea), and Tween 20 solution (250 μg/ml) either with or without methanol (5%), were used as the positive and negative controls, respectively. Ilpum was diluted 1000-fold, as recommended by the manufacturer.
Kimchi Cabbage Soft Rot
The disease control efficacy of the fermentation broth of P. elgii JCK-5075 and the purified PGP-A against Kimchi cabbage soft rot was performed on the sixth-leaf stage of the Kimchi cabbage plants. Out of the four PGPs, PGP-A showed the strongest activity against P. carotovorum subsp. carotovorum with a MIC value of 16 μg/ml (Table 2). Therefore, PGP-A was chosen for an in vivo assay against bacterial soft rot of the Kimchi cabbage caused by P. carotovorum subsp. carotovorum. The plants were treated with 20 ml of the fermentation broth or PGP-A solutions by soil drench. After 24 h, the treated plants were inoculated with 20 ml of the P. carotovorum subsp. carotovorum cell suspension (107 CFU/ml, containing 10 mM MgCl2) by soil drench (Shrestha et al., 2009). The inoculated plants were incubated in the dark for 24 h at 30 ± 2°C and 100% RH, then kept in an incubation room with 12 h of daylight per day for 8 days. The disease severity (DS) was recorded on a scale ranging from 0 to 5 (Champoiseau et al., 2006). Where 0 = no symptom; 1 = one or two pencil-line streaks, 2 = more than two pencil-line streaks, 3 = leaf chlorosis or bleaching, 4 = leaf necrosis, and 5 = death of the plant.
Tomato Bacterial Wilt
The protective activity of the fermentation broth of the P. elgii JCK-5075 and the purified PGP-C against the tomato bacterial wilt was evaluated on the fourth-leaf stage tomato plants. R. solanacearum in the in vitro bioassay was sensitive to all four PGPs at 32 μg/ml, and the limited amounts of PGPs were obtained in this study. This allowed us to evaluate the biocontrol efficacy of the PGP-C alone against the tomato bacterial wilt in the pot experiments. The tomato plants were treated with 20 ml of the fermentation broth or PGP-C solutions by soil drench. After 24 h of treatment, the treated tomato plants were inoculated with 20 ml of the R. solanacearum cell suspension (108 CFU/ml, 10 mM MgCl2) by soil drench. The inoculated plants were kept in the dark for 24 h at 30 ± 2°C, and then transferred to an incubation room at 30 ± 2°C, with 75% RH for 10 days, with 12 h of daylight per day. The DS on the tomato plants was assessed on a scale of 0–4 as follows (Vu et al., 2017). 0 = no leaf symptoms, 1 = one leaf wilted, 2 = two or three leaves wilted, 3 = more than four leaves wilted, and 4 = a dead plant.
Red Pepper Bacterial Leaf Spot
The disease control efficacy of the P. elgii JCK-5075 fermentation broth against red pepper bacterial leaf spot was carried out using sixth-leaf stage plants. The 5 ml aliquots of each sample were applied onto the red pepper plants and then the treated plants were inoculated with 5 ml of the X. euvesicatoria cell suspension (107 CFU/ml) 24 h after the treatment by foliar spray. The inoculated plants were incubated in the dark at 25 ± 2°C and 100% RH for 24 h, they were then kept at 25 ± 2°C and 100% RH, for 16 days in a plastic cover with 12 h of daylight per day. The DS was recorded on a scale of 0–7 as follows (Abbasi et al., 2002). 0 = no symptom, 1 = symptomless, 2 = a few necrotic spots on a few leaflets, 3 = a few necrotic spots on many leaflets, 4 = many spots with coalescence on few leaf, 5 = many spots with coalescence on many leaflets, 6 = severe disease, and leaf defoliation, and 7 = a dead plant.
Calculation of the Control Value
The in vivo experiments were carried out with three replications and the entire experiment was repeated twice in entirely. The control value was calculated according to the following formula:
Calcein Leakage Assay
A calcein leakage assay was performed according to established protocols (Kim et al., 2018). Briefly, lipid mixtures consisting of 7:3 weight ratios of the phosphatidylcholine (POPC) and phosphatidylglycerol (POPG) were prepared from stock lipid solution in 100% methanol. The organic solvents were evaporated, and the resulting lipid film was lyophilized for 1 day. The dried lipid film was resuspended in 1 ml of dye buffer solution (70 mM calcein, 10 mM Tris–HCl, 0.1 mM EDTA, 150 mM NaCl, pH 7.4). Liposomes were prepared through 10 freeze-thaw cycles in liquid nitrogen, followed by an incubation in a 50°C water bath. Then, the suspensions were extruded through 100 nm pore-polycarbonate membranes 20 times. After extrusion, the calcein-entrapped large unilamellar vesicles (LUVs) were separated using gel filtration through a Sephadex G50 column and eluted with a buffer solution (10 mM Tris–HCl, 0.1 mM EDTA, 150 mM NaCl, pH 7.4). Calcein leakage from the LUVs was measured at 20°C by monitoring the fluorescence intensity at an excitation wavelength of 490 nm and an emission wavelength of 520 nm on a spectrophotometer (Cary 5000 UV-Vis-NIR). The peptide and lipid molar ratios ranged from 1:1 to 1:128. The liposome without the peptide was used as a negative control, and the Triton X-100 was used as a positive control. Each run of the experiment contained three replicates, and the entire experiment was run in entirely twice. The percentage of the calcein leakage was calculated according to the following formula:
Cytoplasmic Membrane Depolarization Assay
Cytoplasmic membrane depolarization activity of the PGP-C peptide was determined using a potential sensitive dye, DiSC3-(5), as previously described (Wu and Hancock, 1999; Papo and Shai, 2005; Kim et al., 2018). Briefly, X. oryzae pv. oryzae and R. solanacearum were grown at 30°C to mid-log phase (OD600 nm = 0.7) and were harvested by centrifugation. Cells were washed 3 times with buffer solution (5 mM HEPES, 20 mM glucose, pH 7.4) and gently resuspended to an OD600 nm of 0.15 in the same buffer solution. Then, the cell suspension was incubated with 2 μM of DiSC3-(5) at room temperature for 1 h. DiSC3-(5) was used with 2 mM stock in dimethylsulfoxide (DMSO). Membrane depolarization was monitored using a F-4500 FL fluorescence spectrophotometer (Hitachi, Japan), with filter wavelengths of 620 and 670 nm for excitation and emission, respectively. The peptide was treated after 100 s and the peptide-free cells were used as a negative control.
SYTOX Green Uptake Assay
The effect of the PGP-C peptide on the bacterial membrane permeabilization was evaluated using a SYTOX Green uptake assay, as previously described (Roth et al., 1997). Briefly, X. oryzae pv. oryzae and R. solanacearum were grown at 30°C to a mid-log phase (OD600 nm = 0.7), they were then harvested and washed three times with 1 × PBS buffer (pH 7.4). The cells were gently resuspended to an OD600 nm of 0.15 in the same buffer solution. Then, the cell suspension was incubated with 0.1 μM of SYTOX Green at room temperature for 16 h in the dark. SYTOX Green was used with 5 mM stock in DMSO. The uptake of the SYTOX Green was monitored using a F-4500 FL fluorescence spectrophotometer (Hitachi, Japan), with filter wavelengths of 480 and 520 nm for excitation and emissions, respectively. The peptide was treated after 100 s and the peptide-free cells were used as a negative control.
Statistical Analysis
The in vivo data were analyzed for homogeneity of variances using the SPSS statistical analysis software (version 23 software; IBM, Armonk, NY, United States). Data are expressed as means ± standard deviations of the replicates and were analyzed by one-way-analysis of variance (ANOVA) and the statistical differences among the treatments were determined according to Duncan’s multiple-range test (p < 0.05), among the means using SPSS statistical analysis software.
Results
Antagonistic Activity of the P. elgii JCK-5075 Against Plant Pathogenic Bacteria
The JCK-5075 strain was identified as P. elgii with accession number MT176528 by phylogenetic analysis (Supplementary Figure S1). Fermentation filtrate of the P. elgii JCK-5075 exhibited a range of strong antibacterial effects against the 14 plant pathogenic bacteria, with MIC values of 0.63–10% (Table 1). Among the 14 bacteria tested, X. arboricola pv. pruni was the most sensitive to the fermentation filtrate and ethanol crude substances of P. elgii JCK-5075, with MIC values of 0.31 and 3.13 μg/ml, respectively, followed by A. tumefaciens, X. euvesicatoria, X. axonopodis pv. citri, and X. oryzae pv. oryzae with MIC values of 0.63 and 6.25 μg/ml. A. avenae subsp. cattleyae, A. konjaci, and P. carotovorum subsp. carotovorum were also highly sensitive, with MIC values ranging from 1.25 to 2.5% for the fermentation filtrate, and from 12.5 to 25 μg/ml for the crude substances. In addition, the growth of five of the pathogenic bacteria including, B. glumae, C. michiganensis subsp. michiganensis, P. syringae pv. actinidiae, P. syringae pv. lachrymans, and R. solanacearum, were relatively resistant to the antibacterial metabolites of JCK-5075 with MIC values of 10% and 100 μg/ml for the fermentation filtrate and the crude substances, respectively. Oxolinic acid, a synthetic quinolone antibiotic, inhibited the growth of all the pathogenic bacteria tested with MIC values of 0.05–3.13 μg/ml. Out of the 14 plant pathogenic bacteria, four strains, A. avenae subsp. cattleyae, P. chrysanthemi, A. tumefaciens, X. euvesicatoria, were resistant to streptomycin sulfate but they were sensitive to the fermentation filtrate and crude substances of P. elgii JCK-5075 (Table 1).
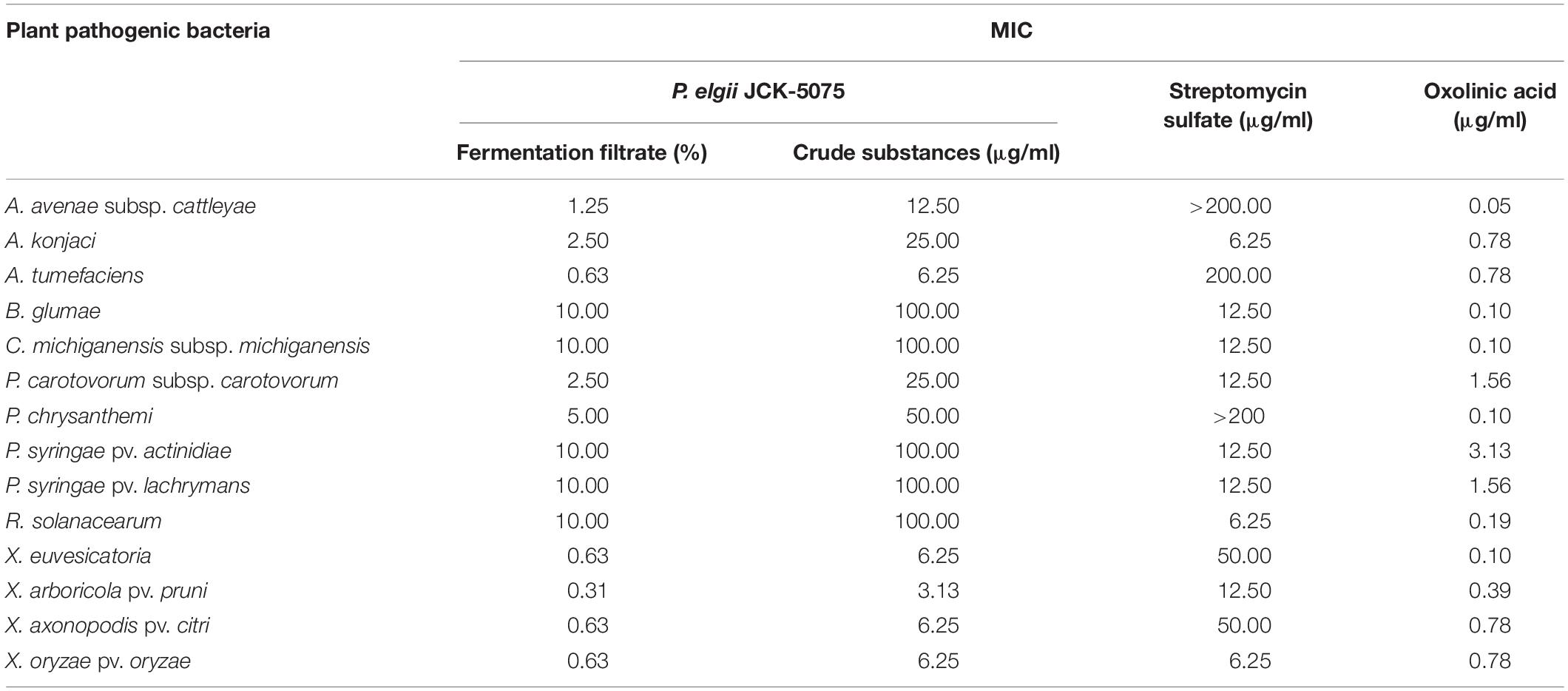
Table 1. Minimum inhibitory concentration (MIC) of the fermentation filtrate and the ethanol crude substances of Paenibacillus elgii JCK-5075 against 14 plant pathogenic bacteria.
Purification of the PGPs From the Fermentation Supernatant of Paenibacillus elgii JCK-5075
Purification of the PGPs from the culture broth was performed on a Diaion HP-20 column chromatography and preparative RP-HPLC. Of the eluted fractions, the 80% ethanol fraction contained PGPs including [M + H]+ ion peaks at m/z 1073.3, 1011.6, 1087.3, and 1087.2 Da, which corresponded to the masses of the PGP-A, B, C, and D, respectively. We further purified using preparative RP-HPLC to obtain pure PGPs. The purity was confirmed using LC-ESI-MS spectrometry (Supplementary Figure S2). The following amounts of the PGPs were obtained from the 1 L cultivation: 3.2 mg of PGP-A, 1.1 mg of PGP-B, 5.1 mg of PGP-C, and 1.4 mg of PGP-D.
Antibacterial Activity of PGPs Against Plant Pathogenic Bacteria
The four PGPs isolated from the fermentation broth of the P. elgii JCK-5075, exhibited strong antibacterial activity against a wide spectrum of plant pathogenic bacteria (Table 2). Among the four metabolites, PGP-A, C, and D presented similar antibacterial activity, while PGP-B was the least active. Xanthomonas species including X. euvesicatoria, X. arboricola pv. pruni, X. axonopodis pv. citri, and X. oryzae pv. oryzae were the most sensitive, with all the PGPs, with MIC values from 2 to 16 μg/ml. The B. glumae, C. michiganensis subsp. michiganensis, P. carotovorum subsp. carotovorum, P. chrysanthemi, P. syringae pv. actinidiae, P. syringae pv. Lachrymans, and R. solanacearum were relatively insensitive to the PGPs.
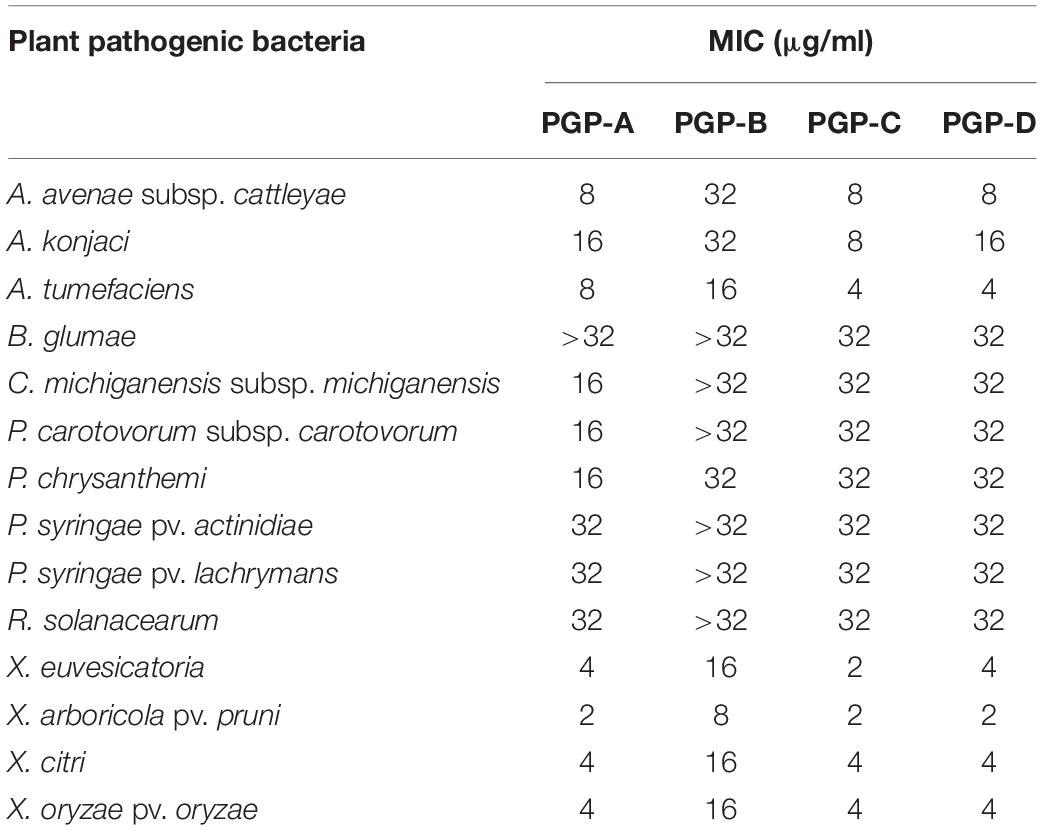
Table 2. The minimum inhibitory concentration (MIC) of the pelgipeptins (PGPs) isolated from Paenibacillus elgii JCK-5075 against 14 plant pathogenic bacteria.
Biocontrol Efficacy of the JCK-5075 and PGPs in Controlling Bacterial Plant Diseases
The fermentation broth of the P. elgii JCK-5075, suppressed the development of the tomato bacterial wilt, Kimchi cabbage soft rot, and red pepper bacterial leaf spot, in a dose-dependent manner (Figure 1). The control values of the fermentation broth of the P. elgii JCK-5075 against the tomato bacterial wilt were 81, 58, and 36%, at 5-, 10-, and 20-fold dilutions, respectively. In comparison, the commercial antibiotic Ilpum displayed a disease control efficacy of 86%, comparable to that of the 5-fold dilution of the fermentation broth of the JCK-5075. The fermentation broth of the JCK-5075 also reduced the development of the Kimchi cabbage soft rot, showing control values of 84, 60, and 29%, for the 5-, 10-, and 20-fold dilutions, respectively. The control value of the 5-fold dilution of the fermentation broth was not significantly different from that of Ilpum (89%). The control values of the fermentation broth of JCK-5075 against the red pepper bacterial leaf spot were 67, 51, and 37%, for the 5-, 10-, and 20-fold dilutions, respectively. The disease control efficacy (76%) of the Ilpum against the red pepper bacterial leaf spot was not significantly different from that of the 5-fold dilution of the fermentation broth of the JCK-5075.
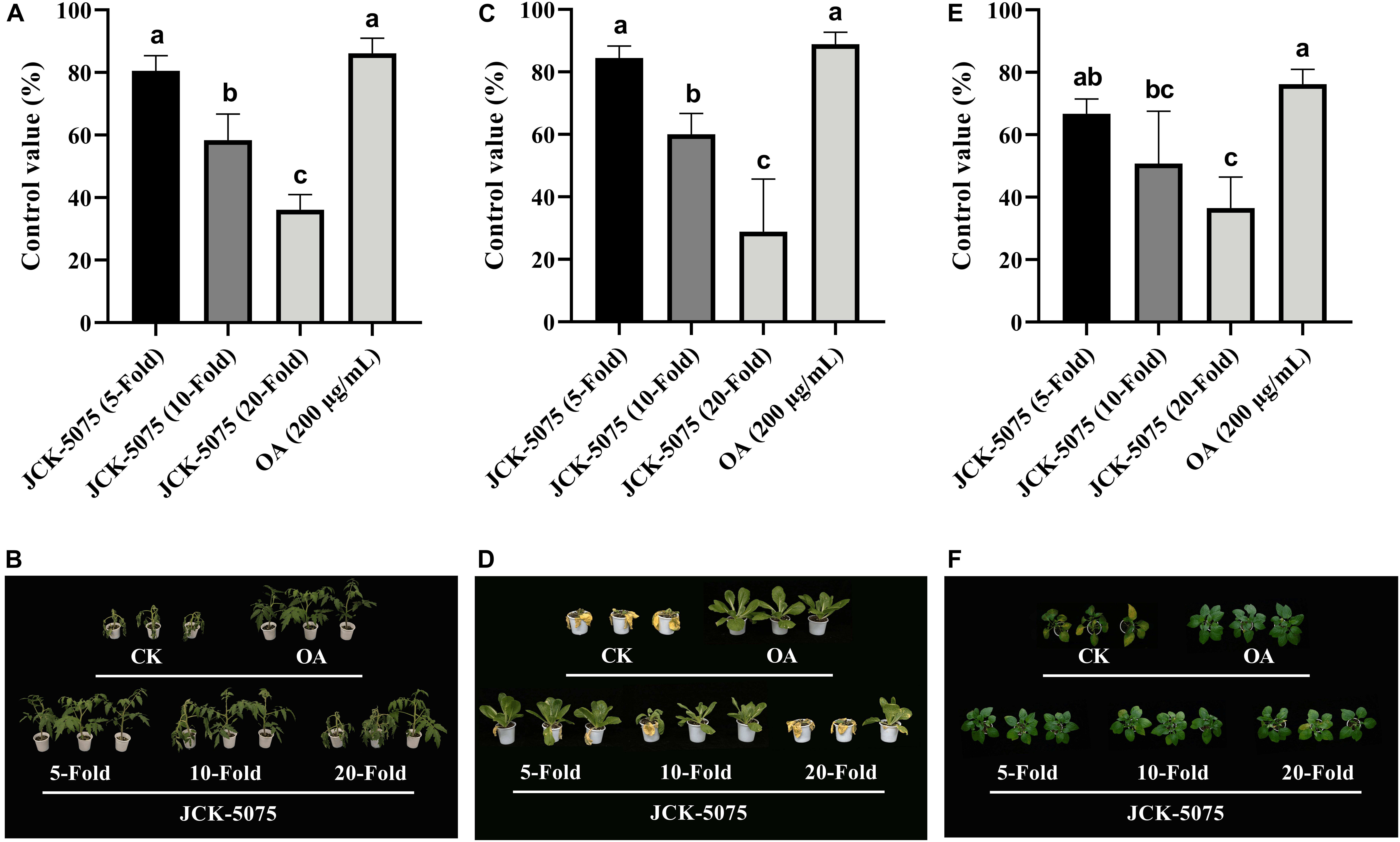
Figure 1. The biocontrol efficacy of the fermentation broth of Paenibacillus elgii JCK-5075 on tomato bacterial wilt (A,B), Kimchi cabbage bacterial soft rot (C,D), and red pepper bacterial leaf spot diseases (E,F). Each value represents the mean ± standard deviation (n = 3) of two runs, with three replicates each. Means followed by the same letters above the bars are not significantly different (p < 0.05) in a Duncan’s multiple range test. JCK-5075 ( 5-, 10-, and 20-fold) are 5-, 10-, and 20-fold dilutions of the fermentation broth of JCK-5075; OA, oxolinic acid (200 μg/ml).
PGP-A also effectively suppressed the development of the Kimchi cabbage soft rot with control values of 76, 53, and 29% for 200, 100, and 50 μg/ml (Figure 2). The control value of the 200 μg/ml of the PGP-A was comparable to that of the oxolinic acid (200 μg/ml). On the other hand, PGP-C reduced the tomato bacterial wilt by 83, 56, and 31%, for the 200, 100, and 50 μg/ml, respectively. The disease control efficacy of the 200 μg/ml PGP-C was comparable to that of the oxolinic acid (200 μg/ml).
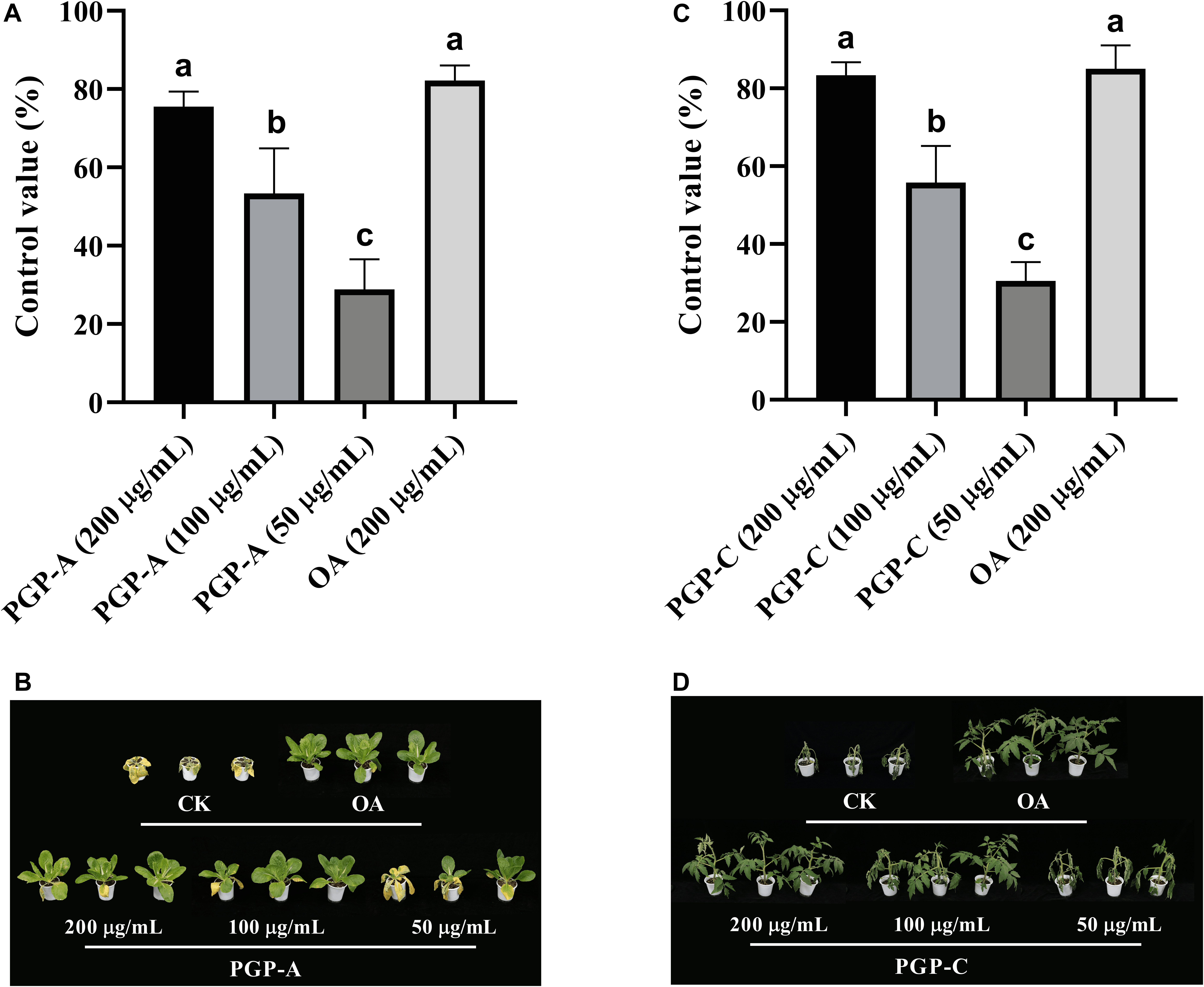
Figure 2. The disease-control efficacy of pelgipeptin (PGP)-A and C on Kimchi cabbage bacterial soft rot (A,B) and tomato bacterial wilt (C,D), respectively. Each value represents the mean ± standard deviation (n = 3) of two runs, with three replicates each. Means followed by the same letters above the bars are not significantly different (p < 0.05) in a Duncan’s multiple range test. OA, oxolinic acid (200 μg/ml).
Action Mechanisms of the PGP-C
The mechanisms of the antibacterial action of the PGP-C peptide were assessed by calcein leakage, membrane depolarization, and SYTOX-Green uptake. First, the calcein leakage assay was performed by mixing the PGP-C with 70 mM calcein encapsulated LUVs, at peptide:lipid molar ratios ranging from 1:128 to 1:1. Three measurements were averaged and plotted against the peptide:lipid ratios. PGP-C peptide gradually increased the calcein dye leakage by increasing the peptide ratio (Figure 3A). At a very low peptide:lipid ratio of 0.031 or less, the leakage did not exceed 10%, but after that the leakage gradually increased to reach 100% at 1.0 ratio. For the membrane depolarization and SYTOX-Green uptake experiments, we used a Xanthomonas strain (X. oryzae pv. oryzae), which was more sensitive to PGP peptides than other strains (Table 2). The potentiometric probe DiSC3-(5) is a carbocyanine with a short (C3) alkyl tail. This cationic dye accumulates on hyperpolarized membranes and is translocated into the lipid bilayer. When the cytoplasmic membrane is damaged, the membrane potential is dissipated and the DiSC3-(5) dye is released, causing an increase in fluoresce. The PGP-C with 2 × MIC (8 μg/ml) induced the complete depolarization of the cytoplasmic membrane within a few seconds (Figure 3B). We also applied a SYTOX-Green uptake assay to examine the ability of the PGP-C peptide to affect the bacterial membrane permeability, by monitoring the intracellular influx of the SYTOX-Green. The SYTOX-Green is a bright and high-affinity nucleic acid stain that easily penetrates the cells with damaged plasma membranes but does not cross the membranes of the living cells. The intensity of the fluorescence was immediately increased by the addition of the PGP-C (2 × MIC) (Figure 3C). Without the peptide, the fluorescence intensities in both the membrane depolarization and SYTOX-Green uptake experiments were not changed (Figures 3B,C), suggesting that the PGP-C specifically induced the increasing fluorescence by disrupting the membrane integrity.
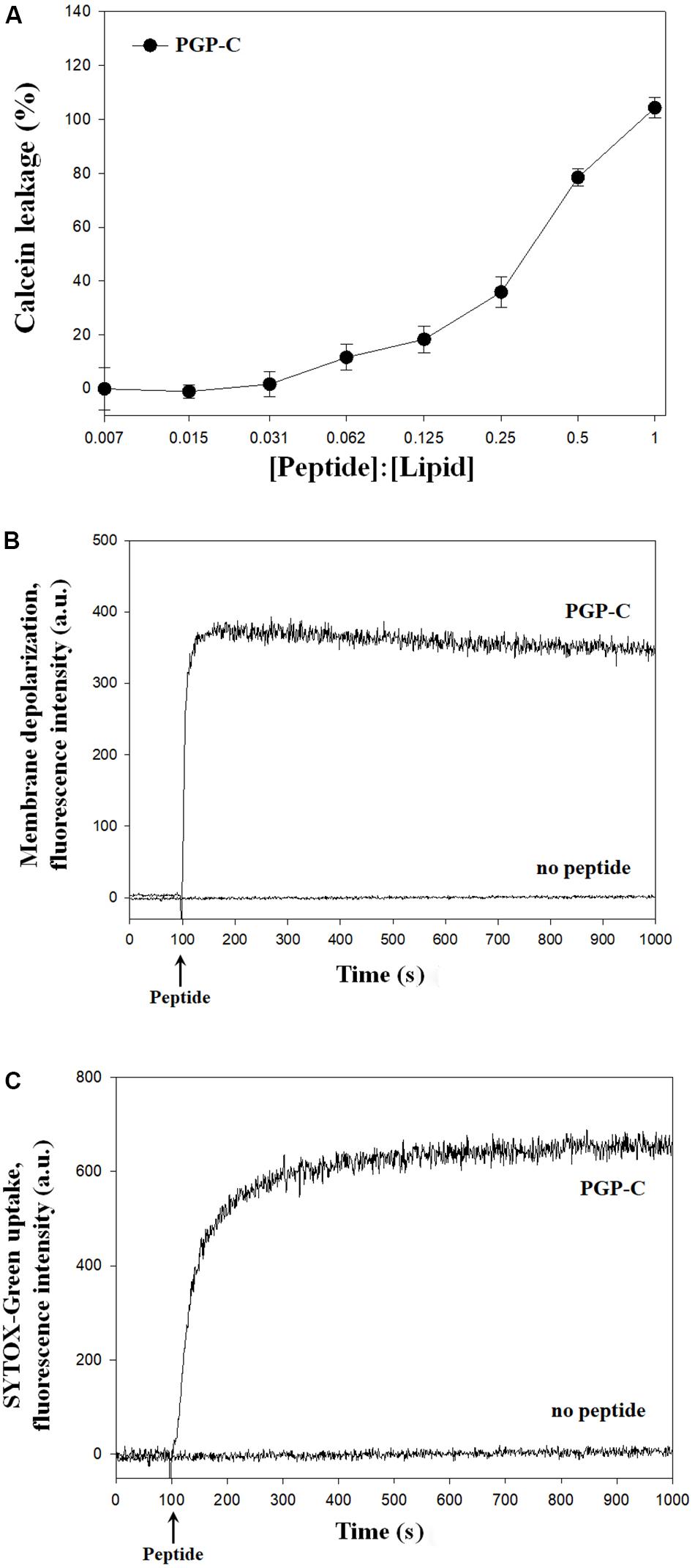
Figure 3. The mechanism studies of pelgipeptin (PGP)-C using calcein leakage (A), membrane depolarization (B), and SYTOX-Green uptake (C) experiments. For calcein leakage, PGP-C was added to calcein encapsulated large unilamellar vesicles (LUVs) at different peptide:lipid ratios. Each value represents the mean ± standard deviation (n = 3) of two runs, with three replicates each. The depolarization of the X. oryzae pv. oryzae cytoplasmic membrane was induced by the PGP-C peptide at 8 μg/ml concentration. The alterations of the cytoplasmic membrane of X. oryzae pv. oryzae by PGP-C (8 μg/ml) allowed the SYTOX Green probe to enter the cell and bind DNA, resulting in an increase of fluorescence.
Discussion
Many bacterial strains display their antagonistic activity both in vitro and in vivo, but only a few are successful for the control of plant diseases under field conditions (Shoda, 2000; Lugtenberg and Kamilova, 2009; Xu and Kim, 2014; Tsuda et al., 2016). The antimicrobial compounds produced by bacteria can be useful as biocontrol measures in agriculture, as demonstrated in several previous studies (Chung et al., 2000; Beatty and Jensen, 2002; Antonopoulos et al., 2008; Allard et al., 2014; Grady et al., 2016). Like the Bacillus spp., members of the Paenibacillus species produce a wide array of antimicrobial compounds, to compete with other microorganisms (Chung et al., 2000; Shaheen et al., 2011; Kai et al., 2013). Bacteria belonging to the Paenibacillus species were isolated from a variety of different environments, and their biological efficacy was different even within the same species; among the 25 strains of P. polymyxa isolated from ginseng roots, 15 strains showed strong activity against Phytophthora capsici, while 10 strains showed weak or no antimicrobial effects (Kim et al., 2009). In this study, P. elgii JCK-5075 presented strong antibacterial activity against various plant pathogenic bacteria. The fermentation filtrate of P. elgii JCK-5075 inhibited the growth of all 14 bacteria that were tested, with MIC values that ranged from 0.31 to 10%. The crude substances (80% ethanolic fraction of Diaion HP-20) also displayed antibacterial activity against all pathogenic bacteria, having a MIC range of 3.13–100 μg/ml (Table 1). Several studies reported on the disease control efficacy of Paenibacillus species against soil-borne diseases (Antonopoulos et al., 2008; Khan et al., 2008; Algam et al., 2010; Xu and Kim, 2014). To date, the information about the control of bacterial plant diseases using P. elgii species has been unavailable. Our results indicated the potential of P. elgii JCK-5075 for controlling plant diseases caused by pathogenic bacteria.
The well-known P. elgii strain may produce various antimicrobial compounds including PGPs, protocatechuic acid, and methyl 2,3-dihydroxybenzoate (Wu et al., 2010; Ding et al., 2011; Nguyen et al., 2015; Lee et al., 2017; Kim et al., 2018). PGPs, a class of cyclic lipopeptides, were first isolated from the fermentation broth of P. elgii B69 and displayed strong activity against human pathogenic bacteria in the range of 0.78–100 μg/ml, and PGP-B was the most active (Wu et al., 2010; Ding et al., 2011). PGP-E isolated by P. elgii BC34-6 also showed strong antibacterial activity, including methicillin-resistant Staphylococcus aureus with MICs of 2–8 μg/ml (Kim et al., 2018). In another report, PGP-D that was isolated from P. elgii AC13, showed the strongest antibacterial activity among the four types of PGP (Costa et al., 2019). In this study, 4 PGPs, namely A, B, C, and D, were isolated from the fermentation supernatant of P. elgii JCK-5075, showed strong in vitro antibacterial activity against 14 plant pathogenic bacteria, with MIC values of 2–32 μg/ml. Even though the antibacterial activity of the four chemicals varied depending on the test pathogens, PGP-A and C displayed the strongest activity, followed by PGP-D and B. Our data differ from the results of previous reports (Wu et al., 2010; Ding et al., 2011; Costa et al., 2019). This may be due to the different target bacteria, as the sensitivity of the bacteria to chemicals varies with the strains, even within one species.
The biological efficacy of the P. elgii JCK-5075 was confirmed herein, to control bacterial plant diseases in pot experiments. The fermentation broth of P. elgii JCK-5075, effectively controlled tomato bacterial wilt, Kimchi cabbage soft rot, and red pepper bacterial leaf spot. R. solanacearum, a causal agent of bacterial wilt in various crops, causes vascular disease, and is one of the most destructive soil-borne pathogens (Mansfield et al., 2012). Physical, chemical, and biological methods were applied to control this disease, and the biological methods have been of interest to researchers for decades. Algam et al. (2010) reported that the disease incidence of the tomato bacterial wilt was reduced by 82% when P. polymyxa MB02-1007 was applied (Algam et al., 2010). Similar results were obtained from the treatment with the fermentation broth of P. elgii JCK-5075 in our study, having a control value of 80.55% at a 5-fold dilution.
Pectobacterium carotovorum is also one of the most destructive soil-borne plant pathogens and causes soft rot to many economically important crops (Perombelon and Kelman, 1980; Charkowski, 2018). Several reports have previously presented the potential of Paenibacillus species as biological controls against the soft rot of Kimchi cabbage (Vanneste et al., 1998; Kyeremeh et al., 2000; Li et al., 2014; Tsuda et al., 2016). Shrestha et al. (2009) reported the biocontrol efficacy of Paenibacillus KPB3 against Kimchi cabbage soft rot, with control values of 58 and 62% in the greenhouse and field tests, respectively (Shrestha et al., 2009). In this study, the 5-fold dilution of the fermentation broth of P. elgii JCK-5075 displayed a control value of 84.45% in pot experiments.
Bacterial leaf spot disease occurs on red pepper and tomato worldwide. Bacterial leaf spot is a common disease on red pepper plants in Korea and the causal pathogen was identified as X. euvesicatoria. Repeated use of streptomycin and copper compounds has resulted in the spread of resistant strains (Marco and Stall, 1983; Araújo et al., 2012). The strain of X. euvesicatoria used in this study is resistant to streptomycin sulfate with a MIC value of 50 μg/ml. The fermentation broth of P. elgii JCK-5075, at a 5-fold dilution, displayed a control value of 67%, which was not significantly different to that of oxolinic acid. P. macerans was reported to control leaf spot disease on tomato, having a biocontrol efficacy of higher than 50% (Lanna Filho et al., 2010). None of the plants treated with the 5-fold dilutions of the fermentation broth of the P. elgii JCK-5075, showed any phytotoxic symptoms (Sarwar et al., 2018). To the best of our knowledge, this is the first report on the efficacy of P. elgii to control plant bacterial diseases.
Paenibacillus species were reported to protect crops from pathogen infections using mechanisms such as induced systemic resistance (Khan et al., 2008; Lee et al., 2012; Sang et al., 2014) or the production of antimicrobial substances (Hong and Meng, 2003; Shaheen et al., 2011; Huang et al., 2013). Bacterial lipopeptides have a wide range antimicrobial activities against pathogenic microorganisms and even some resistant strains, in both medicine and agriculture (Cochrane and Vederas, 2016). The lipopeptides produced by Bacillus amyloliquefaciens strain FJAT-2349, a mixture of iturin A, fengycin, and surfactin, could effectively control tomato bacterial wilt, with a biocontrol efficacy of 97.6% (Chen et al., 2019). Rice bakanae disease was reduced up to 80% with surfactin A purified from Bacillus strains AH-100 and NH217. In our study, PGPs (A and C), at 200 μg/ml, significantly reduced the disease severity in Kimchi cabbage soft rot and tomato bacterial wilt, with control values of 76 and 83%, respectively. Since their first isolations from the Paenibacillus spp., the PGPs have not been applied commercially.
Based on the membrane depolarization and SYTOX-Green uptake assay, PGP-C strongly affected the bacterial membranes of the X. oryzae pv. oryzae at concentrations of 8 μg/ml. However, at the same concentration, PGP-C caused a similar but substantially slower depolarization on the R. solanacearum cytoplasmic membrane (Supplementary Figure S3A). Moreover, the SYTOX-Green uptake experiment revealed that the PGP-C peptide cannot induce membrane permeabilization of R. solanacearum at the same concentration (Supplementary Figure S3B). The MIC of PGP-C, against R. solanacearum is much higher than that of X. oryzae pv. oryzae (32 vs. 4 μg/ml), indicating that the PGP-C is much more active toward X. oryzae pv. oryzae than R. solanacearum. PGP-C is not able to kill R. solanacearum at 8 μg/ml concentrations. These results suggest that the antibacterial activity of PGP-C is directly correlated with the membrane damage mechanisms by pore-formation or structural disruption of the bacterial membranes.
Conclusion
In this study, four members of the PGP family that were isolated from the fermentation supernatant of P. elgii JCK-5075, were found to have strong and broad-spectrum antibacterial activity against plant pathogenic bacteria. PGP-A and C also effectively controlled Kimchi cabbage soft rot and tomato bacterial wilt, respectively, and their disease control efficacy was comparable to that of a commercial bactericide, oxolinic acid. Additionally, PGP-C was found to impact on bacterial cell growth via membrane-active mechanisms. These results indicated that P. elgii JCK-5075 producing PGPs could be used as a biocontrol agent for the control of plant bacterial diseases. This is the first report on the in vitro and in vivo antibacterial activity of PGPs against bacterial plant pathogens.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
J-CK and CL designed the study. KL and JK designed and performed the experiments and analyzed the data. NY and BK performed the RNA isolation and identified species. J-CK, CL, KL, and JK wrote and revised the manuscript. All authors read and approved the final version of this manuscript.
Funding
This work was supported by the Cooperative Research Program for Agricultural Science and Technology Development (Project PJ015296042020), Rural Development Administration, South Korea.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The authors also declare that they will apply for one patent using the results of this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00775/full#supplementary-material
References
Abbasi, P. A., Soltani, N., Cuppels, D. A., and Lazarovits, G. (2002). Reduction of bacterial spot disease severity on tomato and pepper plants with foliar applications of ammonium lignosulfonate and potassium phosphate. Plant Dis. 86, 1232–1236. doi: 10.1094/pdis.2002.86.11.1232
Algam, S. A. E., Xie, G., Li, B., Yu, S., Su, T., and Larsen, J. (2010). Effects of Paenibacillus strains and chitosan on plant growth promotion and control of Ralstonia wilt in tomato. J. Plant Pathol. 92, 593–600. doi: 10.4454/jpp.v92i3.303
Allard, S., Enurah, A., Strain, E., Millner, P., Rideout, S. L., Brown, E. W., et al. (2014). In situ evaluation of Paenibacillus alvei in reducing carriage of Salmonella enterica serovar newport on whole tomato plants. Appl. Environ. Microbiol. 80, 3842–3849. doi: 10.1128/aem.00835-14
Antonopoulos, D. F., Tjamos, S. E., Antoniou, P. P., Rafeletos, P., and Tjamos, E. C. (2008). Effect of Paenibacillus alvei, strain K165, on the germination of Verticillium dahliae microsclerotia in planta. Biol. Control. 46, 166–170. doi: 10.1016/j.biocontrol.2008.05.003
Araújo, E., Pereira, R., Ferreira, M., Quezado-Duval, A., and Café-Filho, A. (2012). Sensitivity of xanthomonads causing tomato bacterial spot to copper and streptomycin and in vivo infra-specific competitive ability in Xanthomonas perforans resistant and sensitive to copper. J. Plant Pathol. 94, 79–87. doi: 10.4454/jpp.fa.2012.004
Ash, C., Priest, F. G., and Collins, M. D. (1993). Molecular identification of rRNA group 3 bacilli (Ash, Farrow, Wallbanks and Collins) using a PCR probe test. Anton. Van Lee 64, 253–260. doi: 10.1007/bf00873085
Beatty, P. H., and Jensen, S. E. (2002). Paenibacillus polymyxa produces fusaricidin-type antifungal antibiotics active against Leptosphaeria maculans, the causative agent of blackleg disease of canola. Can. J. Microbiol. 48, 159–169. doi: 10.1139/w02-002
Champoiseau, P., Daugrois, J. H., Girard, J. C., Royer, M., and Rott, P. (2006). Variation in albicidin biosynthesis genes and in pathogenicity of Xanthomonas albilineans, the sugarcane leaf scald pathogen. Phytopathology 96, 33–45. doi: 10.1094/phyto-96-0033
Charkowski, A. O. (2018). The changing face of bacterial soft-rot diseases. Annu. Rev. Phytopathol. 56, 269–288. doi: 10.1146/annurev-phyto-080417-045906
Chen, M. C., Wang, J. P., Zhu, Y. J., Liu, B., Yang, W. J., and Ruan, C. Q. (2019). Antibacterial activity against Ralstonia solanacearum of the lipopeptides secreted from the Bacillus amyloliquefaciens strain FJAT-2349. J. Appl. Microbiol. 126, 1519–1529. doi: 10.1111/jam.14213
Chung, Y. R., Kim, C. H., Hwang, I., and Chun, J. (2000). Paenibacillus koreensis sp. nov., a new species that produces an iturin-like antifungal compound. Int. J. Syst. Evol. Micro. 50, 1495–1500. doi: 10.1099/00207713-50-4-1495
Cochrane, S. A., and Vederas, J. C. (2016). Lipopeptides from Bacillus and Paenibacillus spp.: a gold mine of antibiotic candidates. Med. Res. Rev. 36, 4–31. doi: 10.1002/med.21321
Costa, R. A., Ortega, D. B., Fulgêncio, D. L., Costa, F. S., Araújo, T. F., and Barreto, C. C. (2019). Checkerboard testing method indicates synergic effect of pelgipeptins against multidrug resistant Klebsiella pneumoniae. Biotechnol. Res. Inn. 28:5. doi: 10.1016/j.biori.2018.12.001
Ding, R., Wu, X. C., Qian, C. D., Teng, Y., Li, O., Zhan, Z. J., et al. (2011). Isolation and identification of lipopeptide antibiotics from Paenibacillus elgii B69 with inhibitory activity against methicillin-resistant Staphylococcus aureus. J. Microbiol. 49, 942–949. doi: 10.1007/s12275-011-1153-7
Egli, T., and Sturm, E. (1981). “Bacterial plant diseases and their control,” in Insektizide Bakterizide Oomyceten-Fungizide / Biochemische und biologische Methoden Naturstoffe / Insecticides Bactericides Oomycete Fungicides / Biochemical and Biological Methods Natural Products, ed. R. Wegler (Berlin: Springer), 345–388.
Fravel, D. R. (2005). Commercialization and implementation of biocontrol. Annu. Rev. Phytopathol. 43, 337–359. doi: 10.1146/annurev.phyto.43.032904.092924
Grady, E. N., MacDonald, J., Liu, L., Richman, A., and Yuan, Z. C. (2016). Current knowledge and perspectives of Paenibacillus: a review. Microbiol. Cell. Fact. 15, 203–203. doi: 10.1186/s12934-016-0603-7
He, Z., Kisla, D., Zhang, L., Yuan, C., Green-Church, K. B., and Yousef, A. E. (2007). Isolation and identification of a Paenibacillus polymyxa strain that coproduces a novel lantibiotic and polymyxin. Appl. Environ. Microbiol. 73, 168–178. doi: 10.1128/aem.02023-06
Hong, T.-Y., and Meng, M. (2003). Biochemical characterization and antifungal activity of an endo-1,3-β-glucanase of Paenibacillus sp. isolated from garden soil. Appl. Microbiol. Biot. 61, 472–478. doi: 10.1007/s00253-003-1249-z
Huang, E., and Yousef, A. E. (2014). The lipopeptide antibiotic paenibacterin binds to the bacterial outer membrane and exerts bactericidal activity through cytoplasmic membrane damage. Appl. Environ. Microbiol. 80, 2700–2704. doi: 10.1128/aem.03775-13
Huang, Z., Hu, Y., Shou, L., and Song, M. (2013). Isolation and partial characterization of cyclic lipopeptide antibiotics produced by Paenibacillus ehimensis B7. BMC Microbiol. 13:87. doi: 10.1186/1471-2180-13-87
Kai, H., Yamashita, M., Takase, S., Hashimoto, M., Muramatsu, H., Nakamura, I., et al. (2013). KB425796-A, a novel antifungal antibiotic produced by Paenibacillus sp. 530603. J. Antibiot. 66:465. doi: 10.1038/ja.2013.63
Kajimura, Y., and Kaneda, M. (1996). Fusaricidin A, a new depsipeptide antibiotic produced by Bacillus polymyxa KT-8. J. Antibiot. 49, 129–135. doi: 10.7164/antibiotics.49.129
Khan, Z., Kim, S., Jeon, Y., Khan, H., Son, S., and Kim, Y. (2008). A plant growth promoting rhizobacterium, Paenibacillus polymyxa strain GBR-1, suppresses root-knot nematode. Bioresour. Technol. 99, 3016–3023. doi: 10.1016/j.biortech.2007.06.031
Kim, J., Il Kim, P., Bong, K. M., Il Kim, J., Shin, S. Y., Song, J., et al. (2018). Isolation and structural elucidation of pelgipeptin E, a novel pore-forming pelgipeptin analog from Paenibacillus elgii with low hemolytic activity. J. Antibiot. 71, 1008–1017. doi: 10.1038/s41429-018-0095-2
Kim, J., Le, K. D., Yu, N. H., Kim, J. I., Kim, J. C., and Lee, C. W. (2019). Structure and antifungal activity of pelgipeptins from Paenibacillus elgii against phytopathogenic fungi. Pestic. Biochem. Phys. 163, 154–163. doi: 10.1016/j.pestbp.2019.11.009
Kim, S. G., Khan, Z., Jeon, Y. H., and Kim, Y. H. (2009). Inhibitory effect of Paenibacillus polymyxa GBR-462 on Phytophthora capsici causing Phytophthora blight in Chili pepper. J. Phytopathol. 157, 329–337. doi: 10.1111/j.1439-0434.2008.01490.x
Kumar, S., Chauhan, P. S., Agrawal, L., Raj, R., Srivastava, A., Gupta, S., et al. (2016). Paenibacillus lentimorbus inoculation enhances tobacco growth and extenuates the virulence of Cucumber mosaic virus. PLoS One 11:e0149980. doi: 10.1371/journal.pone.0149980
Kyeremeh, A. G., Kikumoto, T., Chuang, D.-Y., Gunji, Y., Takahara, Y., and Ehara, Y. (2000). Biological control of soft rot of Chinese Cabbage using single and mixed treatments of bacteriocin-producing avirulent mutants of Erwinia carotovora subsp. carotovora. J. Gen. Plant Pathol. 66, 264–268. doi: 10.1007/pl00012957
Lanna Filho, R., Romeiro, R. D. S., and Alves, E. (2010). Bacterial spot and early blight biocontrol by epiphytic bacteria in tomato plants. Pesqui. Agropecu. Bras. 45, 1381–1387. doi: 10.1590/s0100-204x2010001200007
Lee, B., Farag, M. A., Park, H. B., Kloepper, J. W., Lee, S. H., and Ryu, C. M. (2012). Induced resistance by a long-chain bacterial volatile: elicitation of plant systemic defense by a C13 volatile produced by Paenibacillus polymyxa. PLoS One 7:e48744. doi: 10.1371/journal.pone.0048744
Lee, Y. S., Nguyen, X. H., Cho, J.-Y., Moon, J.-H., and Kim, K. Y. (2017). Isolation and antifungal activity of methyl 2,3-dihydroxybenzoate from Paenibacillus elgii HOA73. Microbiol. Pathog. 106, 139–145. doi: 10.1016/j.micpath.2016.01.007
Li, H. Y., Luo, Y., Zhang, X. S., Shi, W. L., Gong, Z. T., Shi, M., et al. (2014). Trichokonins from Trichoderma pseudokoningii SMF2 induce resistance against Gram-negative Pectobacterium carotovorum subsp. carotovorum in Chinese cabbage. FEMS Microbiol. Lett. 354, 75–82. doi: 10.1111/1574-6968.12427
Lugtenberg, B., and Kamilova, F. (2009). Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 63, 541–556. doi: 10.1146/annurev.micro.62.081307.162918
Mansfield, J., Genin, S., Magori, S., Citovsky, V., Sriariyanum, M., Ronald, P., et al. (2012). Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 13, 614–629. doi: 10.1111/j.1364-3703.2012.00804.x
Marco, G. M., and Stall, R. E. (1983). Control of bacterial spot of pepper initiated by strains of Xanthomonas campestris pv. vesicatoria that differ in sensitivity to copper. Plant Dis. 67:779. doi: 10.1094/pd-67-779
Nguyen, X. H., Naing, K. W., Lee, Y. S., Moon, J. H., Lee, J. H., and Kim, K. Y. (2015). Isolation and characteristics of protocatechuic acid from Paenibacillus elgii HOA73 against Botrytis cinerea on strawberry fruits. J. Basic Microbiol. 55, 625–634. doi: 10.1002/jobm.201400041
Papo, N., and Shai, Y. (2005). A molecular mechanism for lipopolysaccharide protection of Gram-negative bacteria from antimicrobial peptides. J. Biol. Chem. 280, 10378–10387. doi: 10.1074/jbc.m412865200
Perombelon, M. C. M., and Kelman, A. (1980). Ecology of the soft rot erwinias. Annu. Rev. Phytopathol. 18, 361–387. doi: 10.1146/annurev.py.18.090180.002045
Phi, Q. T., Park, Y. M., Seul, K. J., Ryu, C. M., Park, S. H., Kim, J. G., et al. (2010). Assessment of root-associated Paenibacillus polymyxa groups on growth promotion and induced systemic resistance in pepper. J. Microbiol. Biotechn. 20, 1605–1613. doi: 10.4014/jmb.1007.07014
Qian, C. D., Wu, X. C., Teng, Y., Zhao, W. P., Li, O., Fang, S. G., et al. (2012). Battacin (Octapeptin B5), a new cyclic lipopeptide antibiotic from Paenibacillus tianmuensis active against multidrug-resistant Gram-negative bacteria. Antimicrob. Agents. Chem. 56, 1458–1465. doi: 10.1128/aac.05580-11
Roth, B. L., Poot, M., Yue, S. T., and Millard, P. J. (1997). Bacterial viability and antibiotic susceptibility testing with SYTOX green nucleic acid stain. Appl. Environ. Microbiol. 63, 2421–2431. doi: 10.1128/aem.63.6.2421-2431.1997
Sang, M. K., Kim, E. N., Han, G. D., Kwack, M. S., Jeun, Y. C., and Kim, K. D. (2014). Priming-mediated systemic resistance in cucumber induced by Pseudomonas azotoformans GC-B19 and Paenibacillus elgii MM-B22 against Colletotrichum orbiculare. Phytopathology 104, 834–842. doi: 10.1094/phyto-11-13-0305-r
Sarwar, A., Hassan, M. N., Imran, M., Iqbal, M., Majeed, S., Brader, G., et al. (2018). Biocontrol activity of surfactin A purified from Bacillus NH-100 and NH-217 against rice bakanae disease. Microbiol. Res. 209, 1–13. doi: 10.1016/j.micres.2018.01.006
Shaheen, M., Li, J., Ross, A. C., Vederas, J. C., and Jensen, S. E. (2011). Paenibacillus polymyxa PKB1 produces variants of Polymyxin B-type antibiotics. Chem Biol. 18, 1640–1648. doi: 10.1016/j.chembiol.2011.09.017
Shoda, M. (2000). Bacterial control of plant diseases. J. Biosci. Bioeng. 89, 515–521. doi: 10.1016/s1389-1723(00)80049-3
Shrestha, A., Kim, E. C., Lim, C. K., Cho, S. Y., Hur, J. H., and Park, D. H. (2009). Biological control of soft rot on Chinese cabbage using beneficial bacterial agents in greenhouse and field. Korea. J. Pestic. Sci. 13, 325–331.
Singh, A. K., Ghodke, I., and Chhatpar, H. S. (2009). Pesticide tolerance of Paenibacillus sp. D1 and its chitinase. J. Environ. Manag. 91, 358–362. doi: 10.1016/j.jenvman.2009.09.001
Sundin, G. W., Castiblanco, L. F., Yuan, X., Zeng, Q., and Yang, C. H. (2016). Bacterial disease management: challenges, experience, innovation and future prospects: challenges in bacterial molecular plant pathology. Mol. Plant Pathol. 17, 1506–1518. doi: 10.1111/mpp.12436
Tsuda, K., Tsuji, G., Higashiyama, M., Ogiyama, H., Umemura, K., Mitomi, M., et al. (2016). Biological control of bacterial soft rot in Chinese cabbage by Lactobacillus plantarum strain BY under field conditions. Biol. Control. 100, 63–69. doi: 10.1016/j.biocontrol.2016.05.010
Vanneste, J., Cornish, D., Yu, J., and Voyle, M. (1998). A microcin produced by a strain of Erwinia herbicola is involved in biological control of fire blight and soft rot caused by Erwinia sp. Acta. Hortic. 513, 39–46. doi: 10.17660/actahortic.1998.513.3
Vu, T. T., Kim, H., Tran, V. K., Vu, H. D., Hoang, T. X., Han, J. W., et al. (2017). Antibacterial activity of tannins isolated from Sapium baccatum extract and use for control of tomato bacterial wilt. PLoS One 12:e0181499. doi: 10.1371/journal.pone.0181499
Wu, M., and Hancock, R. E. (1999). Interaction of the cyclic antimicrobial cationic peptide bactenecin with the outer and cytoplasmic membrane. J. Biol. Chem. 274, 29–35. doi: 10.1074/jbc.274.1.29
Wu, X. C., Shen, X. B., Ding, R., Qian, C. D., Fang, H. H., and Li, O. (2010). Isolation and partial characterization of antibiotics produced by Paenibacillus elgii B69. FEMS Microbiol. Lett. 310, 32–38. doi: 10.1111/j.1574-6968.2010.02040.x
Xu, S. J., and Kim, B. S. (2014). Biocontrol of fusarium crown and root rot and promotion of growth of tomato by Paenibacillus strains isolated from soil. Mycobiology 42, 158–166. doi: 10.5941/myco.2014.42.2.158
Yuliar, Nion, Y. A., and Toyota, K. (2015). Recent trends in control methods for bacterial wilt diseases caused by Ralstonia solanacearum. Microbes. Environ. 30, 1–11. doi: 10.1264/jsme2.me14144
Keywords: antibacterial activity, mode of action, Paenibacillus, pelgipeptins, plant pathogenic bacteria
Citation: Le KD, Kim J, Yu NH, Kim B, Lee CW and Kim J-C (2020) Biological Control of Tomato Bacterial Wilt, Kimchi Cabbage Soft Rot, and Red Pepper Bacterial Leaf Spot Using Paenibacillus elgii JCK-5075. Front. Plant Sci. 11:775. doi: 10.3389/fpls.2020.00775
Received: 16 March 2020; Accepted: 15 May 2020;
Published: 01 July 2020.
Edited by:
Dirk Albert Balmer, Syngenta, SwitzerlandReviewed by:
Kamal A. M. Abo-Elyousr, Assiut University, EgyptBruno Brito Lisboa, State University of Rio Grande do Sul, Brazil
Copyright © 2020 Le, Kim, Yu, Kim, Lee and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chul Won Lee, Y3dsZWVAam51LmFjLmty; Jin-Cheol Kim, a2ppbmNAam51LmFjLmty
†These authors have contributed equally to this work
 Khanh Duy Le
Khanh Duy Le Jueun Kim
Jueun Kim Nan Hee Yu
Nan Hee Yu Bora Kim
Bora Kim Chul Won Lee
Chul Won Lee Jin-Cheol Kim
Jin-Cheol Kim