- 1College of Life Sciences, Fujian Agriculture and Forestry University, Fuzhou, China
- 2Key Laboratory of Crop Ecology and Molecular Physiology, Fujian Agriculture and Forestry University, Fuzhou, China
- 3College of Crop Science, Fujian Agriculture and Forestry University, Fuzhou, China
- 4Wenxian Institute of Agricultural Sciences, Jiaozuo, China
- 5Fujian Provincial Key Laboratory of Agroecological Processing and Safety Monitoring, Fujian Agriculture and Forestry University, Fuzhou, China
Rehmannia glutinosa, a perennial medicinal plant, suffers from severe replant disease under consecutive monoculture. The rhizosphere microbiome is vital for soil suppressiveness to diseases and for plant health. Moreover, N-acyl homoserine lactone (AHL)-mediated quorum sensing (QS) regulates diverse behavior in rhizosphere-inhabiting and plant pathogenic bacteria. The dynamics of short-chain AHL-mediated QS bacteria driven by consecutive monoculture and its relationships with R. glutinosa replant disease were explored in this study. The screening of QS bacteria showed that 65 out of 200 strains (32.5%) randomly selected from newly planted soil of R. glutinosa were detected as QS bacteria, mainly consisting of Pseudomonas spp. (55.4%). By contrast, 34 out of 200 (17%) strains from the diseased replant soil were detected as QS bacteria, mainly consisting of Enterobacteriaceae (73.5%). Functional analysis showed most of the QS bacteria belonging to the Pseudomonas genus showed strong antagonistic activities against Fusarium oxysporum or Aspergillus flavus, two main causal agents of R. glutinosa root rot disease. However, the QS strains dominant in the replant soil caused severe wilt disease in the tissue culture seedlings of R. glutinosa. Microbial growth assays demonstrated a concentration-dependent inhibitory effect on the growth of beneficial QS bacteria (i.e., Pseudomonas brassicacearum) by a phenolic acid mixture identified in the root exudates of R. glutinosa, but the opposite was true for harmful QS bacteria (i.e., Enterobacter spp.). Furthermore, it was found that the population of quorum quenching (QQ) bacteria that could disrupt the beneficial P. brassicacearum SZ50 QS system was significantly higher in the replant soil than in the newly planted soil. Most of these QQ bacteria in the replant soil were detected as Acinetobacter spp. The growth of specific QQ bacteria could be promoted by a phenolic acid mixture at a ratio similar to that found in the R. glutinosa rhizosphere. Moreover, these quorum-quenching bacteria showed strong pathogenicity toward the tissue culture seedlings of R. glutinosa. In conclusion, consecutive monoculture of R. glutinosa contributed to the imbalance between beneficial and harmful short-chain AHL-mediated QS bacteria in the rhizosphere, which was mediated not only by specific root exudates but also by the QQ bacterial community.
Introduction
To ensure food security and meet market needs, the practice of consecutive monoculture is becoming popular in intensive agriculture. However, large-scale crop monoculture results in many problems including a loss of crop genetic diversity, an increase in disease incidence, a decline in crop quality and even the fragility of ecosystem functioning (Jacques and Jacques, 2012; Mariotte et al., 2018; Li et al., 2019). Consecutive monoculture problems, also known as replant problems or replant disease, are especially severe in the cultivation of medicinal herbs, such as Rehmannia glutinosa, Panax notoginseng, Pseudostellaria heterophylla, and Panax ginseng (Dong et al., 2018a; Luo et al., 2019; Wu et al., 2019; Zhang et al., 2019). It was reported that approximately 70% of medicinal herbs using tuberous roots were attacked by replant disease (Wu et al., 2016c). R. glutinosa, a member of the Scrophulariaceae family, is a traditional and famous Chinese medicinal herb with various pharmacological effects. However, 2-year consecutive monoculture of this plant on the same land led to a substantial increase in root rot disease and a serious reduction in tuberous root yield in the field trial conducted at our long-term orientation station. The consecutively- monocultured plants had unexpanded tuberous roots and large numbers of adventitious fibrous roots, which are of no commercial value (Wu et al., 2015, 2018b). In addition, consecutive monoculture of this plant resulted in a significant increase in the abundance of several soil-borne fungal pathogens (i.e., Aspergillus flavus and Fusarium oxysporum), which were frequently isolated from the consecutively monocultured soil and diseased plants and are pathogenic to R. glutinosa seedlings (Wu et al., 2015, 2016b, 2018a). Fields used for R. glutinosa production need to be planted with other crops for at least 15 years before they can be replanted again (Yang et al., 2011). Each year, replant disease causes a dramatic decrease in the area of the geo-authentic production zone (34° 48′ N to 35° 30′ N, 112° 02′ E to 113° 38′ E) (Zhang et al., 2019), an optimal production area with the most suitable soil and climate conditions for R. glutinosa cultivation. The quality of R. glutinosa tuberous roots cannot be assured when grown outside the geo-authentic production zone. Therefore, it is urgent to gain insight into the mechanisms underlying replant disease.
The plant microbiome is a key determinant of plant growth, development and health. The interactions between plants and soil microorganisms play key roles in maintaining soil quality and ecosystem sustainability (Kwak et al., 2018; Wang and Li, 2019). Plant roots influence soil microbial community assembly and alter the relative abundance of beneficial, harmful and neutral microorganisms, which in turn exert positive or negative effects on plant growth and resistance (Bakker et al., 2018; Stringlis et al., 2018; Zhalnina et al., 2018; Rolfe et al., 2019). Moreover, previous studies demonstrated that previous plants could affect the immunity and resistance of subsequent plant populations growing in the same soil through soil-borne microbial legacy (Bakker et al., 2018; Yuan et al., 2018; Kong et al., 2019). Therefore, recent research in the field of replant disease has increasingly focused on soil microbiome composition and function. Liu et al. (2019) indicated that consecutive soybean monoculture significantly decreased the fungal community diversity but increased the abundances of plant pathogens. Gao et al. (2019) found that consecutive sweet potato monoculture reduced the abundance of beneficial fungi such as Chaetomium but increased harmful fungi such as Verticillium, Fusarium, and Colletotrichum. Our previous study using barcoded pyrosequencing of 16S rDNA gene amplicons demonstrated that consecutive R. glutinosa monoculture modulated the rhizosphere microbiome with a reduction in the abundances of specific beneficial microorganisms and an increase in the harmful microorganisms (Wu et al., 2018b). Similar examples were also found for numerous medicinal plants including Panax quinquefolius, P. notoginseng, P. heterophylla and P. ginseng under a monoculture regime (Wu et al., 2016a; Zhao et al., 2017; Dong et al., 2018b; Jiang et al., 2019). Besides, a growing body of research has indicated that replant disease can be attributed to changes in the soil microbiome induced by phenolic allelochemicals, rather than their direct autotoxicity (Li et al., 2014; Wu et al., 2016a; Chen et al., 2017, 2018). Li et al. (2014) found that peanut root exudates could selectively stimulate or inhibit different microbial taxa, and the modifications in the soil microbiome mediated by phenolic acids led to the poor performance of the peanut plants. Our previous study found that the abundance of phenolic acids in R. glutinosa root exudates increased with the growth time of seedlings under sterile conditions but did not increase with the increasing years of monoculture under natural field conditions, suggesting that soil microbes might be involved in the degradation, utilization and conversion of root exudates (Wu et al., 2015).
The co-evolution between plants and associated microbial communities is common but complex in natural ecosystems (Zhang et al., 2017; Chagas et al., 2018). During coevolution with their host plant, microorganisms have evolved numerous strategies to talk with the hosts, intraspecific populations and other organisms for growth and survival (Badri et al., 2009; Venturi and Keel, 2016; Kan et al., 2017). Quorum sensing (QS) is a widespread phenomenon by which bacteria use intercellular communication to coordinate their behavior in response to environmental changes. Likewise, plant-associated bacteria utilize QS systems to sense the ecological niche, adapt to environmental stress, distribute their population under the existing conditions, and thereby influence the growth and health of host plants (Loh et al., 2002; Khan et al., 2019). N-acyl homoserine lactone (AHL)-mediated QS plays important roles in root-microbe interactions and the motility and colonization of rhizobacteria (Loh et al., 2002; von Bodman et al., 2003; Zhang et al., 2017). Plant root exudates not only provide nutrients and energy sources for root-associated microorganisms, but also select, attract or repel specific QS rhizobacteria (Bauer and Mathesius, 2004; Venturi and Keel, 2016; Chagas et al., 2018). Neal et al. (2012) found that 2,4-dihydroxy-7-methoxy-2H-1,4-benzoxazin-3(4H)-one (DIMBOA) released by maize roots could induce positive chemotaxis by Pseudomonas putida and attract these bacteria to the rhizosphere. Rosmarinic acid, a plant-derived compound that acts as a QS regulator agonist, activated the quorum sensing responses in Pseudomonas aeruginosa (Corral-Lugo et al., 2016). More interestingly, Schaefer et al. (2008) demonstrated that photosynthetic bacterium Rhodopseudomonas palustris had the ability to produce a novel QS signaling molecule, p-coumaroyl-homoserine lactone, by using a plant-derived aromatic acid, p-coumarate. In contrast, quorum quenching (QQ) refers to the process of interference in microbial QS (Dong et al., 2001; Uroz et al., 2009; Grandclement et al., 2016). Plant roots can also produce QS signal mimics or QS-interfering molecules to interfere with microbial QS systems (Teplitski et al., 2000; Zhang et al., 2017). Methyl gallate, a phenolic compound, was reported to inhibit both AHL synthesis and activity in Chromobacterium violaceum and to suppress biofilm formation and other QS-associated virulence factors in P. aeruginosa. In addition, microorganisms can utilize QQ strategies including the enzymatic degradation of QS molecules to prevent QS signaling and QS-regulated functions in plant-associated microorganisms.
Even though the rhizosphere microbiome plays crucial roles in maintaining soil health, the relationships between QS bacterial populations and replant disease are still unclear. A previous study on P. heterophylla replant disease indicated that the number of QS bacteria, all identified as Serratia marcescens that can rapidly cause wilt disease, significantly increased with the increasing years of monoculture. Moreover, it was found that P. heterophylla root exudates and root tuber extracts could significantly promote the growth of S. marcescens (Zhang et al., 2016). Our previous studies have demonstrated that consecutive R. glutinous monoculture led to soil microbiome dysbiosis, and phenolic acids in root exudates could significantly promote the mycelial growth and toxin production of pathogenic F. oxysporum (Wu et al., 2015, 2018a,b). However, little is known about the shifts in QS bacterial populations in the R. glutinous rhizosphere under consecutive monoculture, as well as the effects of phenolic acids in root exudates on the growth of specific QS bacteria. We hypothesized that consecutive R. glutinosa monoculture could restructure the short-chain AHL-mediated QS bacterial populations in the rhizosphere through the modulation of root exudates, with an increase in the abundance of harmful QS bacteria but a reduction in beneficial QS bacteria.
Materials and Methods
Field Experiment and Soil Sampling
The field experiment was conducted in Jiaozuo city, Henan Province (34°56′N, 112°58′E), the geo-authentic production zone. The mean annual precipitation in this region is 552 mm, and the mean annual temperature is 14.3°C. R. glutinosa cultivar “Wen 85-5” was used as the experimental material in this study. To ensure uniform soil and climate conditions among different treatments, a single field previously cultivated with wheat was divided into two parts for two cropping patterns: the newly planted (NP) part and the 2-year consecutively monocultured (CM) part. The soil pH of the tested field was 7.43, and the soil organic matter content was 12.52 g⋅kg–1. The contents of available nitrogen, phosphorus and potassium were 23.41, 51.36, and 223.89 mg⋅kg–1, respectively. The total nitrogen, phosphorus and potassium were 0.51, 1.46, and 6.98 g⋅kg–1, respectively. In brief, R. glutinosa in the NP plots was planted on April 15 in 2016 and harvested on October 30 in 2016. The CM plots were established in 2015; the plots were consecutively monocultured for 2 years (Wu et al., 2018b). Each treatment had three experimental repetitions (12 m2). All study plots had the same fertilization and water management during the whole experimental period.
The rhizosphere soil samples were collected from 5 random locations within each plot on July 15 in 2016. R. glutinosa tuberous roots were carefully excavated with a shovel, shaken to remove loosely attached soil, and then tightly attached soil on the tuberous roots was collected as rhizosphere soils. For bacterial isolation, the soil samples from three replicates of each treatment were combined into a composite sample. All soil samples were passed through a 2-mm sieve to remove plant residues, macrofauna and stones and then used immediately for bacterial isolation and soil total DNA extraction.
The Isolation and Identification of QS Bacteria From Rhizosphere Soil
The biosensor strain Chromobacterium violaceum CV026 (CV026), a mini-Tn5 mutant of C. violaceum ATCC31532 with kanamycin resistance (Sakr et al., 2013), was used to identify short-chain AHL-mediated QS bacteria (C4-C8-AHLs) (Remuzgo-Martínez et al., 2015). This strain cannot synthesize QS signal molecules but can sensitively respond to exogenous signal molecules (i.e., N-hexanoyl-l-homoserine lactone, C6-HSL) and produce purple violacein pigment (Hossain et al., 2017). The rhizosphere soils collected from healthy NP plants (denoted as NP soil) and the diseased CM plants with root rot disease symptoms (denoted as diseased soil, BT soil) were used to isolate short-chain AHL-mediated QS bacteria. Briefly, a serial dilution of soil suspension (10–1, 10–2, and 10–3) was perpared by using freshly collected soil. The soil suspension was spread on a plate containing Luria-Bertani (LB) medium and incubated at 37°C for 12 h. Then, 200 recognizable single colonies were randomly selected from an appropriate dilution (10–3 dilution) for each treatment because there were 200∼250 single colonies in this dilution on the LB agar plates. Each single colony was streaked and co-cultured with the biosensor CV026 on the LB plates at 30°C. The inoculation of a known non-QS bacterium and C6-HSL standard solution was used as a negative and positive control, respectively. The production of purple pigmentation by CV026 indicated that the co-cultured bacteria could produce QS signal molecules; these were identified as QS bacteria. The identified QS bacteria were stored at −80°C and were cultured overnight in LB medium for further use. The DNA of QS bacteria was extracted and then used for 16s rDNA amplification with the primer pair 357F (5′-CTCCTAGGGAGGCAGCAG-3′) and 1492r (5′-GGTTACCTTGTTACGACTT-3′) for Sanger sequencing. The sequence data have been submitted to GenBank (accession number MT355703∼MT355756).
Quantitative PCR (qPCR) of Pseudomonas sp. in Rhizosphere Soil
The abundance of Pseudomonas in NP and BT soils was determined via qPCR with the specific primer pairs Ps-for (5′-GGTCTGAGAGGATGATCAGT-3′) and Ps-rev (5′-TTAGCTCCACCTCGC GGC-3′) (Tan and Ji, 2010). The PCR reaction mixture and amplification conditions were set as described by Wu et al. (2015). Each treatment had three biological replicates.
Antagonistic Activity Assessment and Pathogenicity Test of QS Bacteria
The isolated QS bacteria were cultured overnight in LB medium at 37°C. The bacterial suspension was inoculated at the periphery of potato-dextrose-agar (PDA) plates and incubated for 2 days at 37°C. Then, the mycelium of A. flavus or F. oxysporum, two fungal causal agents of R. glutinosa root rot diseases (Wu et al., 2016b), was transferred to the center of the PDA plates and incubated again for several days to observe antagonistic activities of QS bacteria.
Simultaneously, QS bacteria were inoculated on Murashige-Skoog (MS) medium that was planted with tissue culture seedlings of R. glutinosa to assess the pathogenicity of QS bacteria. Firstly, seedlings shoots without roots were transferred to MS medium supplemented with 0.2 mg/L 6-benzyladenine, 0.2 mg/L indole-3- butyric acid and grown for 40 days. Then, the tissue culture seedlings inoculated with QS bacteria were cultured in a growth chamber at 25°C with a photoperiod of 16:8 h light/dark to observe the symptoms. Each treatment had three biological replicates.
The Effects of Phenolic Acid Mixture on the Growth of QS Bacteria
Based on the detection of the composition and total abundance of phenolic acids in the R. glutinosa rhizosphere (Wu et al., 2015), a phenolic acid mixture at the same ratio (molar ratio, protocatechuic acid : phthalic acid : p-hydroxybenzoic acid : vanillic acid : syringic acid : vanillin : ferulic acid : benzoic acid = 10 : 10 : 36 : 100 : 12 : 12 : 30 : 30) was applied to assess the effects of phenolic acids on the chemotactic response and growth of QS bacteria. The chemotaxis of QS bacteria was performed using the drop assay described by Li et al. (2012) with some modification. In detail, 500 μL of QS bacterial suspension was added into 20 mL of minimal medium (Rani et al., 1996) and poured into a petri plate. Subsequently, 0.1 g of phenolic acid powder mixture at the above-mentioned ratio was placed in the center of a petri plate and chemotactic response was observed via migrating rings after 24 h of incubation at 37°C. The growth response of QS bacteria to phenolic acids was assessed through the optic density (OD) value assay. Briefly, the stock solution of phenolic acids was filtered through 0.22 μm filters and then added to an 8-fold dilution of LB broth (1/8 LB) medium to prepare a series of solutions with different final concentrations (0, 30, 60, 120, 240, 480 μmol⋅L–1). Each treatment had three replicates. Thirty μL of QS bacteria was inoculated into 1/8 LB medium. Afer incubation at 200 rpm and 37°C for 8 h, the OD value at 600 nm (OD600) was detected using a plate reader (Thermo Scientific Multiskan MK3, Shanghai, China).
The Isolation of QQ Bacteria That Could Disrupt the Pseudomonas brassicacearum SZ50 QS System
The QS bacteria belonging to the Pseudomonas genus were widespread in NP soil in this study and showed a declining trend under consecutive R. glutinosa monoculture. Therefore, P. brassicacearum, a Pseudomonas baterium showing strong antagonistic activity against the fungal pathogen A. flavus, was selected as a target to isolate the corresponding QQ bacteria. In addition to NP and BT soils, another soil (denoted as consecutively cropped soil, CC soil) collected from 2-year CM plants, which had high numbers of adventitious fibrous roots but no root rot disease symptoms, was used for QQ bacterial isolation.
The biosensor strain CV026 was used to assess QQ activity against P. brassicacearum SZ50 and to isolate QQ bacteria from NP, CC and BT soils. Firstly, the P. brassicacearum SZ50 strain was cultured in LB medium for 16 h, and the QS signal molecules (AHL) were extracted using acidified ethyl acetate (supplemented with 1% acetic acid) according to the method of Anbazhagan et al. (2012) with modifications. AHL extracts were dissolved in HPLC-grade methanol and qualitatively detected by the well-diffusion assay using biosensor CV026 (Anbazhagan et al., 2012). Subsequently, the resulting AHL extracts were used for QQ bacterial isolation. In detail, the isolated strains were grown in sterile 96-well plates (plate No. 1) containing 300 μl of 1/2 TY broth (0.5% tryptone, 0.3% yeast extract, 15 mM KH2PO4 and 6 mM CaCl2) at 200 rpm and 37°C for 16 h. Then, 50 μL culture broths of the isolated strains were transferred into each well of sterile 96-well plates (plate No. 2) containing 50 μl of 1/2 TY broth supplemented with 1 μL of P. brassicacearum AHL extracts and incubated at 300 rpm and 30°C for 24 h. Wells containing 50 μL culture broth of a known QQ bacterium, 50 μl of 1/2 TY broth and 1 μl of C6-HSL were used as a positive control. Wells containing only 100 μl of 1/2 TY broth supplemented with 1 μl of AHL extracts or C6-HSL were both used as a negative control. After ultraviolet (UV) sterilization for 30 min, 50 μl of the sterilized culture broths were transferred into each well in new 96-well plates (plate No. 3) containing 100 μl of 1/2 TY broth, 50 μL of overnight-cultured CV026 and 0.18 μL kanamycin (50 μg⋅mL–1) and incubated at 300 rpm and 30°C for 24 h to observe the QQ activity of the isolated strains. No purple pigmentation produced by CV026 in plates No. 3 indicated that the corresponding bacteria in plate No. 1 could degrade P. brassicacearum AHL signal molecules and could be identified as QQ bacteria (Supplementary Figure S1). The QQ candidates were checked again through the above-mentioned method. Similarly, the molecular identification of QQ bacteria was performed by using 357F and 1492r as mentioned above. The sequence data have been submitted to GenBank (accession numbers MT355757∼MT355785).
The Functional Characterization of Specific QQ Bacteria
The effects of phenolic acid mixture on the growth of specific QQ bacteria and their pathogenicity test were carried out using the same method as described above for QS bacteria.
Statistical Analyses
Statistical analysis of data was performed by one-way analysis of variance (ANOVA) followed by the least significant difference (LSD) test (P < 0.05, n = 3) via Data Processing System (DPS) software (version 7.05, Zhejiang University, Hangzhou, China). Phylogenetic tree construction was performed with Molecular Evolutionary Genetics Analysis (MEGA) version 4.1.
Results
Changes in the Species Composition and Population Dynamics of Short-Chain AHL-Mediated QS Bacteria in the Rhizosphere Under Consecutive Monoculture
Compared with NP, CM plants had serious rot root disease and unexpanded tuberous roots. Moreover, 62% of consecutively- monocultured plants withered and died during sampling. The soil samples collected from the NP and diseased CM plants were used for QS bacterial isolation. Co-cultured with a non-QS bacterium, the biosensor strain CV026 did not produce purple pigmentation (Figure 1A). However, the positive control inoculated with the C6-HSL standard, the CV026 colony developed a deep purple color (Figure 1B). Based on this principle, the short-chain AHL-mediated QS bacteria were randomly isolated by biosensor CV026 from the newly planted (NP) soil and the diseased (BT) soil (Figure 1C). The results showed that 65 out of 200 strains (32.5%) randomly selected from the newly planted (NP) soil of R. glutinosa were detected as QS bacteria, mainly consisting of Pseudomonas spp. (55.4%). By contrast, 34 out of 200 (17%) strains from the diseased (BT) soil were detected as QS bacteria, mainly consisting of Enterobacteriaceae (73.5%), including Enterobacter spp. (61.8%) and Klebsiella spp. (11.8%). The phylogenetic trees of 16S rDNA genes of QS bacteria isolated from NP and BT soils are shown in Supplementary Figures S2, S3, respectively. Moreover, the number of QS bacteria belonging to Pseudomonas, Bacillus and Exiguobacterium was markedly higher in the NP soil than in the BT soil. However, the number of QS bacteria belonging to Enterobacter, Klebsiella and Pantoea increased under consecutive R. glutinosa monoculture (Figure 1D). Quantitative PCR confirmed that the abundance of the Pseudomonas genus was significantly higher in the NP soil (3.7 × 107 copies ⋅ g soil–1) than in the BT soil (1.1 × 107 copies ⋅ g soil–1) (Figure 2).
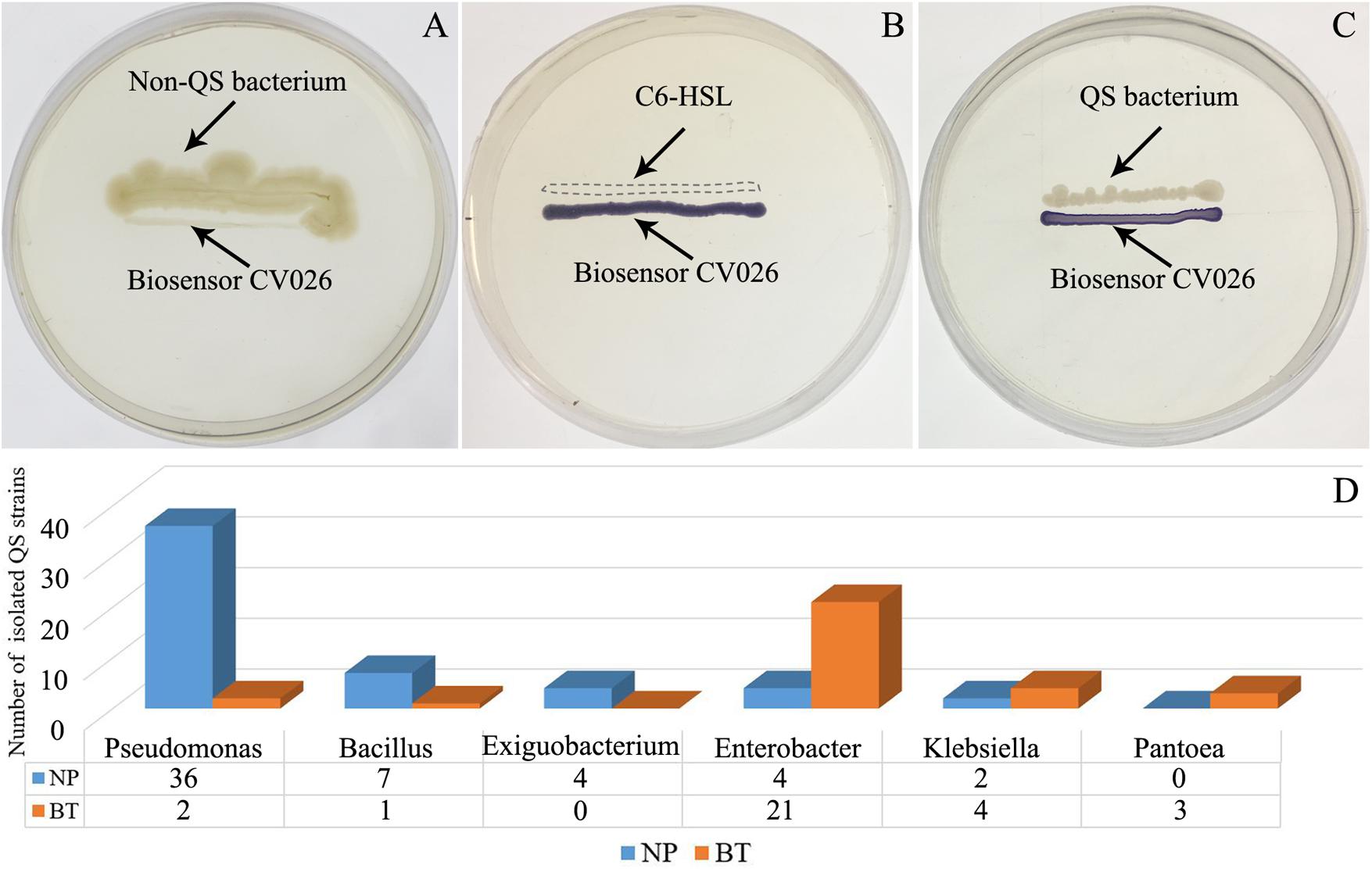
Figure 1. Screening of short-chain AHL-mediated QS bacteria by biosensor strain CV026 (A–C) and the dynamic changes of QS bacteria in the R. glutinosa rhizosphere under consecutive monoculture (D). (A) The biosensor strain CV026 did not produce the purple pigmentation when co-cultured with a known non-QS bacterium. (B) CV026 produced the purple pigmentation when inoculated with C6-HSL standard, as indicated by a dashed box. (C) The CV026 colony developed a purple color when co-cultured with a QS bacterium randomly isolated from the R. glutinosa rhizosphere.
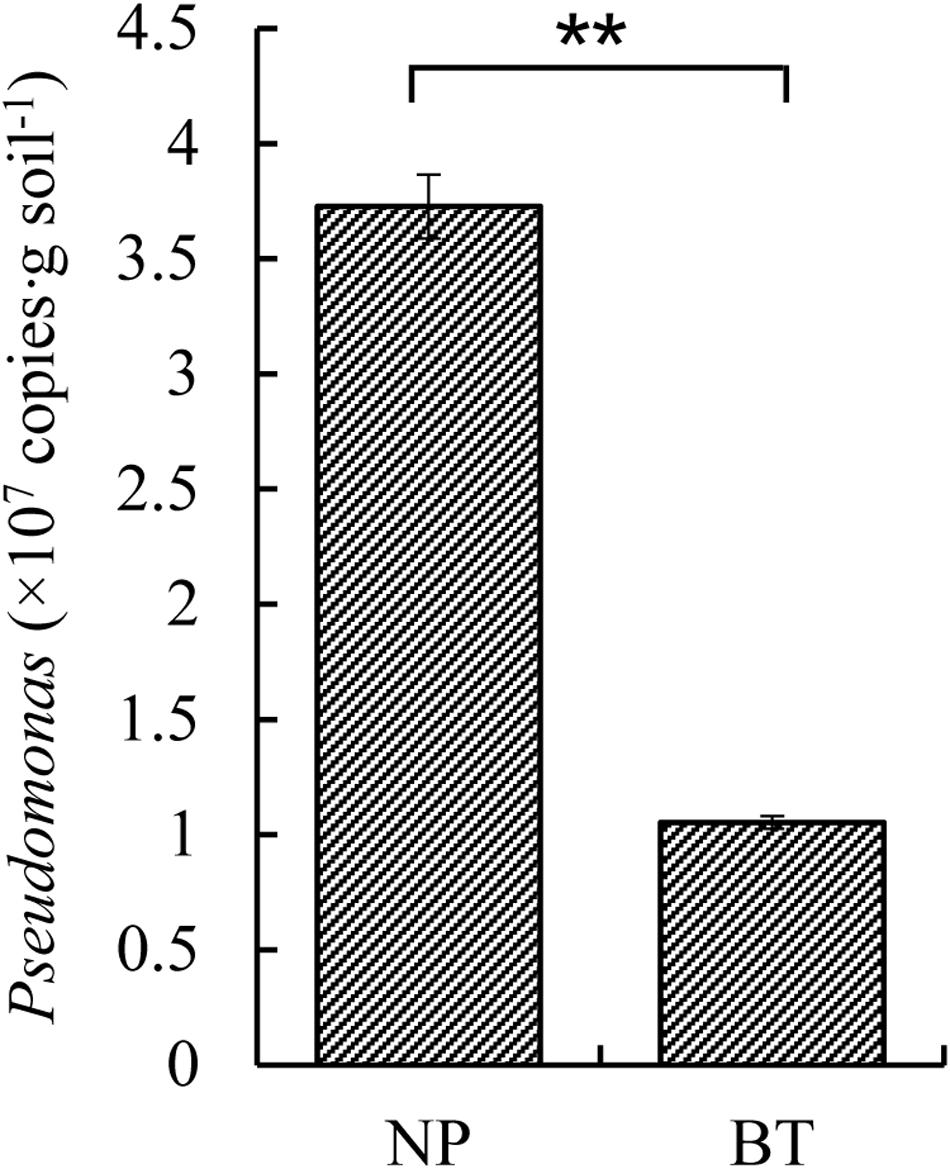
Figure 2. Quantitative PCR of Pseudomonas sp. in the newly planted (NP) soil and the diseased (BT) soil of R. glutinosa. ** indicates a statistically significant difference (P < 0.01, LSD test). Data are presented as means ± standard errors.
Antagonistic Activity Assessment and Pathogenicity Test of QS Bacteria
The antagonistic activity assessment showed that 33 out of 65 strains of QS bacteria (50.8%) in the NP soil had antagonistic activities against A. flavus or F. oxysporum, two fungal causal agents of R. glutinosa root rot diseases. In particular, most of the QS bacteria belonging to Pseudomonas (72.2%) had inhibitory effects on the growth of A. flavus. Further, several strains such as Pseudomonas spp. SZ88, SZ92, SZ95, SZ16, SZ90, and Bacillus sp. SZ93 possessed antagonistic activities against both fungal pathogens (Supplementary Table S1 and Figure 3). By contrast, 14 out of 34 strains of QS bacteria (41.2%) in the BT soil showed antagonistic activity against one of the two fungal pathogens. Except for BT7 with antagonistic activity against A. flavus, the others only showed antagonistic activity against F. oxysporum (Supplementary Table S1 and Figure 3). Morover, the dominant QS bacteria in the BT soil, such as Pantoea sp. BT3, Enterobacter sp. BT22, Enterobacter sp. BT23, and Enterobacter sp. BT56, were highly pathogenic to the tissue culture seedlings of R. glutinosa (Supplementary Figure S4).
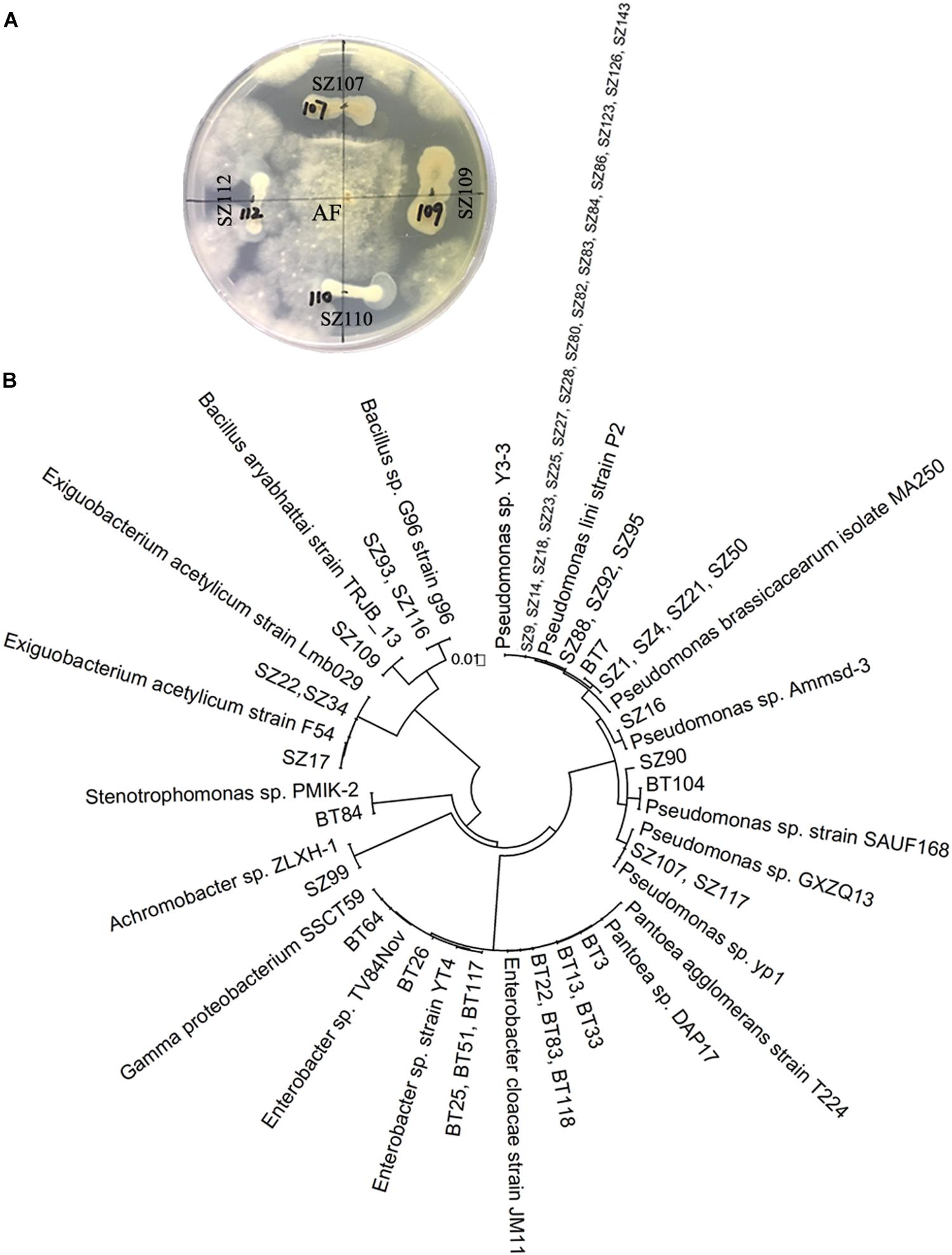
Figure 3. Antagonistic activity assessment of QS bacteria (A) and phylogenetic analysis of QS antagonists (B). (A) Strains SZ107 and SZ109, rather than SZ110 and SZ112, had antagonistic activities against A. flavus (AF). The prefixes “SZ” and “BT” indicate the QS bacteria isolated from the newly planted (NP) soil and the diseased (BT) soil, respectively.
Different Responses of Beneficial and Harmful QS Bacteria to R. glutinosa Root Exudates
Based on the phylogenetic trees of isolated QS bacteria (Supplementary Figures S2, S3), most of the dominant taxa were selected to test their chemotactic responses toward phenolic acids identified in root exudates of R. glutinosa through the drop assay. The results showed that most of the Enterobacter spp. and Klebsiella spp., frequently isolated from the BT soil, exhibited distinct chemotaxis toward the phenolic acid mixture. However, several QS strains including Pseudomonas sp. SZ90, Bacillus sp. 31, Bacillus sp.77, Exiguobacterium sp. SZ17 and Achromobacter sp. SZ99 showed no appreciable chemotaxis (Supplementary Figure S5).
Furthermore, the different responses of specific beneficial and harmful QS bacteria to the phenolic acid mixture were assessed through the OD value assay. The results showed that the phenolic acid mixture at the same ratio as detected in R. glutinosa rhizosphere soil had concentration-dependent inhibitory effects on the growth of beneficial QS bacteria including Pseudomonas spp. S9, SZ50, and SZ63, but not SZ8, that were dominant in the NP soil. However, the phenolic acid mixture significantly promoted the growth of harmful QS bacteria that were dominant in the BT soil including Pantoea sp. BT3, Enterobacter spp. BT22 and BT23, and Klebsiella sp. BT56, and the promotion effect increased as the concentration increased (Figure 4).
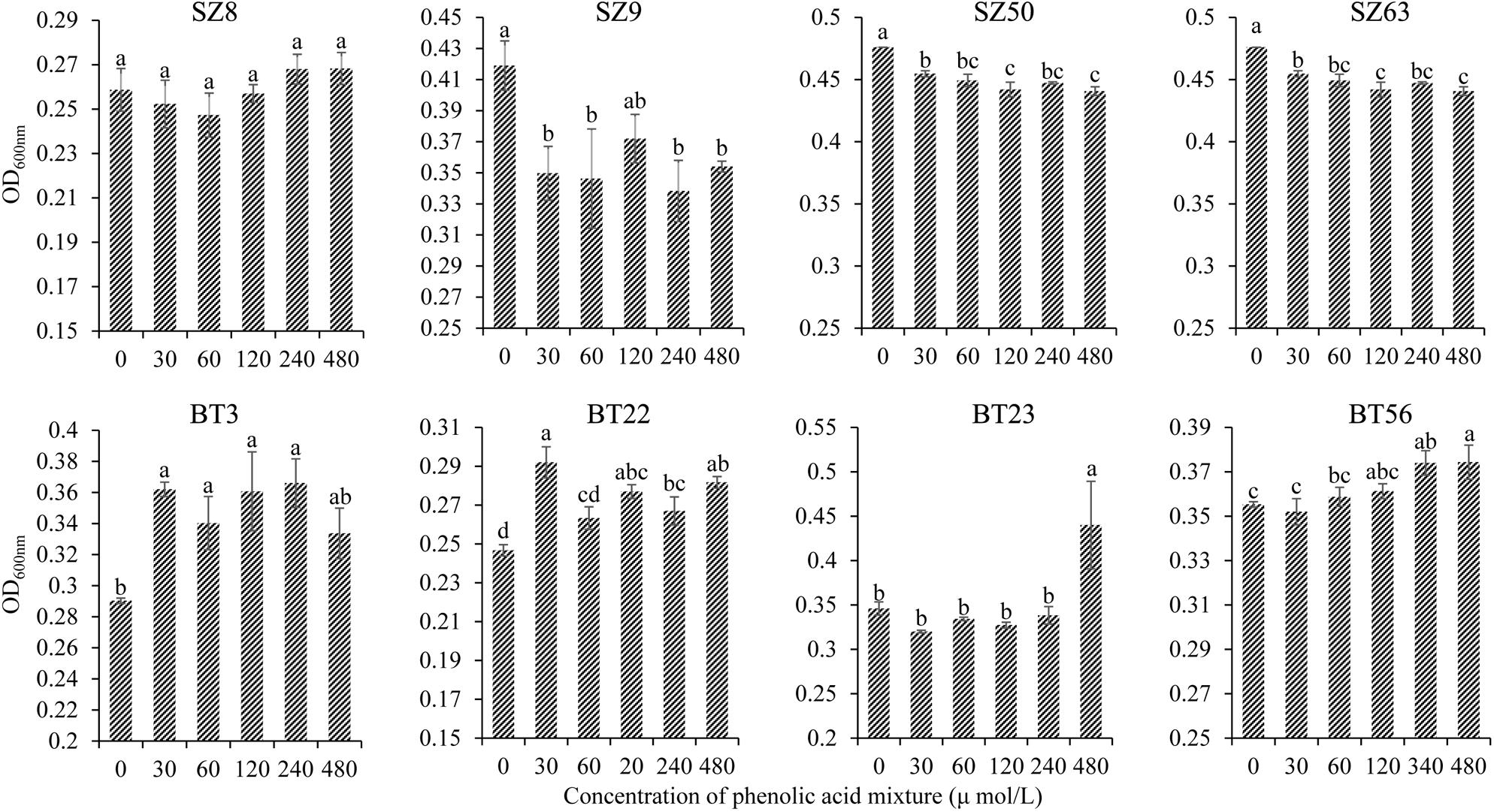
Figure 4. Effects of phenolic acid mixture on the growth of specific QS bacteria isolated from the newly planted (NP) soil and the diseased (BT) soil of R. glutinosa. The prefixes “SZ” and “BT” indicate the QS bacteria isolated from the NP soil and the BT soil, respectively. Strains S8, S9, SZ50, and SZ63 belong to the Pseudomonas genus. Strain BT3 belongs to the Pantoea genus, strains BT22 and BT23 belong to the Enterobacter genus and strain BT56 belongs to the Klebsiella genus. Data are presented using means ± standard errors. Different letters indicate different levels of significance (P < 0.05, LSD, n = 3).
Dynamic Change in Bacteria With Quorum Quenching Activity Against Beneficial QS Bacteria Under Consecutive Monoculture
The well-diffusion assay using biosensor CV026 demonstrated that QS signal molecules (AHLs) were successfully extracted from the culture broth of P. brassicacearum SZ50 strain (Supplementary Figure S6). Screening of QQ bacteria with enzymatic degradation activity against SZ50 AHLs showed that 8 out of 200 strains (4.0%) randomly selected from the NP soil were detected as QQ bacteria, consisting of Pseudomonas (37.5%), Exiguobacterium (12.5%), Achromobacter (12.5%), Acinetobacter (25.0%), and Stenotrophomonas (12.5%). However, 39 out of 200 strains (19.5%) and 23 out of 200 strains (11.5%) were identified as QQ bacteria in the CC soil and BT soil, respectively. Moreover, QQ bacteria in the consecutively monocultured soils mainly consisted of Acinetobacter spp., accounting for 87.2% (34 strains) in CC soil and 69.6% (16 strains) in BT soil (Figure 5). Further, several QQ bacteria belonging to Enterobacter spp. were detected in the consecutively monocultured soils, including Enterobacter sp. YQC23, Enterobacter sp. YQC24, and Enterobacter sp. YQC119. The phylogenetic trees of the 16S rDNA genes of QQ bacteria isolated from NP, CC, and BT soils are shown in Supplementary Figure S7.
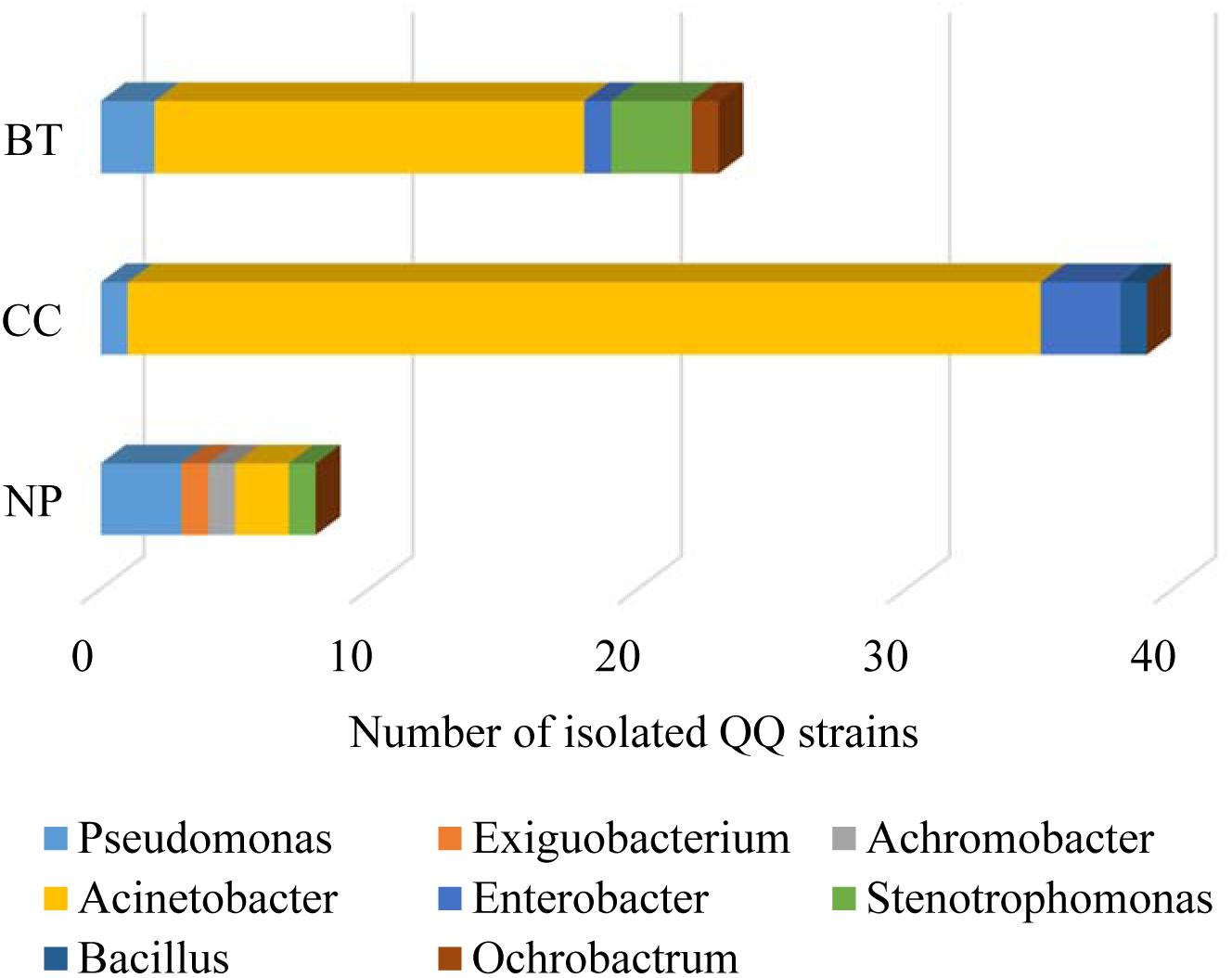
Figure 5. The species composition and population dynamics of rhizosphere bacteria with QQ activities against Pseudomonas SZ50 under consecutive R. glutinosa monoculture. NP, CC, and BT represent the newly planted soil, the 2-year consecutively cropped soil and the diseased soil, respectively.
Furthermore, the effects of phenolic acids on the growth of specific QQ bacteria isolated from the consecutively monocultured soil were tested by OD value assay. The results showed that the phenolic acid mixture at the same ratio as that detected in R. glutinosa rhizosphere soil could significantly increase the growth of specific QQ bacteria belonging to Acinetobacter spp. (i.e., YQC11, YQC12, and YQB98) and Enterobacter spp. (i.e., YQC23, YQC24, and YQC119) (Supplementary Figure S8), which were isolated from the consecutively monocultured soils. Furthermore, a pathogenicity test showed that these specific QQ bacteria isolated from the CC and BT soils could cause severe wilt disease in the tissue culture seedlings of R. glutinosa (Supplementary Figure S9).
Discussion
The collective genome of the rhizosphere microbial community is referred to as the second genome of the plant and plays crucial roles in plant growth and health (Berendsen et al., 2012). A growing body of research has demonstrated that replant disease is closely associated with changes in the rhizosphere microbiome (Franke-Whittle et al., 2015; Chen et al., 2017; Yu et al., 2019). Plant roots release variable but substantial amounts of compounds and can shape the rhizosphere microbial community structure (Hu et al., 2018; Stringlis et al., 2018; Voges et al., 2019). Soil microorganisms can actively respond to environmental changes, alter their physiological behavior including motility and chemotaxis, and even modulate the plant root exudation profile (Matilla et al., 2010; Gu et al., 2016; Zhalnina et al., 2018). Quorum sensing, a cell density-dependent type of intercellular communication, allows bacteria to monitor the surrounding environment, coordinate the behavior of populations, and mediate microbe-microbe and host-microbe interactions (Loh et al., 2002; Taga and Bassler, 2003; Chagas et al., 2018). AHL-mediated QS regulates diverse behavior involving both intra- and interspecies interactions in rhizosphere-inhabiting and plant pathogenic bacteria (von Bodman et al., 2003; Zaytseva et al., 2019).
In this study, the species composition and population dynamics of QS bacteria driven by consecutive monoculture of R. glutinosa were explored. The results showed that numerous bacteria in soil were detected as QS bacteria, accounting for 32.5% of all randomly selected strains in the NP soil and 17% in the diseased soil. This observation is consistent with the findings in previous studies that QS systems are widespread among the bacterial populations in the phytosphere (Veselova et al., 2003; Zhang et al., 2016). Molecular identification found that the QS bacteria isolated from the healthy NP soil mainly consisted of Pseudomonas spp. (accounting for 55.4%) and Bacillus spp. (accounting for 10.8%) (Figure 1D). Quantitative PCR confirmed the significant decline in Pseudomonas spp. in the R. glutinosa rhizosphere under consecutive monoculture (Figure 2). Moreover, more than half of the QS bacteria in the NP soil showed antagonistic activities against A. flavus or F. oxysporum, two fungal causal agents of R. glutinosa root rot diseases (Figure 3). Similar findings were reported in our previous study on the rhizosphere microbiome of R. glutinosa based on barcoded pyrosequencing (Wu et al., 2018b). Luo et al. (2019) indicated that P. notoginseng planting resulted in negative plant-soil feedback due to a decline in the beneficial rhizobacteria including the genera Pseudomonas and Bacillus in the rhizosphere. Pseudomonas spp. and Bacillus spp. have been widely proposed to play important roles in specific soil suppressiveness to plant pathogens (Mendes et al., 2011; Kyselková and Moënne-Loccoz, 2012; Shen et al., 2015; Zhou et al., 2019). By contrast, the QS bacteria isolated from the unhealthy BT soil mainly consisted of Enterobacter spp. (61.8%) and Klebsiella spp. (11.8%) (Figure 1D), and some of them were found to be pathogenic to R. glutinosa seedlings. Similarly, Lin et al. (2015) found that the abundance of Enterobacter spp. significantly increased in P. heterophylla rhizosphere soil with increasing years of monoculture. Köberl et al. (2017) indicated that F. oxysporum-infected banana plants harbored a higher abundance of Enterobacteriaceae known for their plant-degrading capacity. However, it should be noted that the culture-independent approaches in our previous study found that the relative abundance of Enterobacteriaceae was significantly higher in newly planted soil than in consecutively monocultured soil (Wu et al., 2018b), which might be due to the difference between culture-dependent and culture-independent assessments. Moreover, the CV026 reporter strain used in this study mainly detects short-chain AHLs (Remuzgo-Martínez et al., 2015). These results suggested that the imbalance between beneficial and harmful QS bacteria in the rhizosphere under consecutive monoculture might be an important factor for R. glutinosa replant disease. Further studies are needed to assess the effects of the synthetic multispecies community created using the isolated bacteria on the growth and development of tuberous roots of R. glutinosa in natural field sites.
Root exudates are the key determinant of rhizosphere microbiome assembly, acting as chemical attractants, repellants or antagonists of specific microorganisms in soil (el Zahar Haichar et al., 2014; Huang et al., 2014; Chagas et al., 2018; Stringlis et al., 2018). Hu et al. (2018) demonstrated that plant root exudates could modulate the rhizosphere microbiota and thereby affect the growth and defense of the next plant generation. Here, it was found that the phenolic acid mixture at the same ratio as that detected in R. glutinosa rhizosphere soil showed concentration-dependent inhibitory effects on the growth of several beneficial QS bacteria (i.e., Pseudomonas spp.) but promoted the growth of harmful QS bacteria (i.e., Enterobacter spp.) (Figure 4). This result is in line with previous findings that phenolic acids in P. heterophylla root exudates could selectively inhibit beneficial microorganisms (i.e., Pseudomonas spp.) and stimulate certain pathogenic bacteria and fungi (Wu et al., 2016c; Chen et al., 2017). Zhang et al. (2016) also demonstrated that both root exudates and tuberous root extracts of P. heterophylla could significantly promote the growth of S. marcescens, a QS bacterium that rapidly causes wilt disease of P. heterophylla. Numerous studies indicated that indirect allelopathy through modifications in soil microbiome induced by root exudates contributes to replant disease in agriculture and horticulture (Li et al., 2014; Chen et al., 2018). Besides, our findings indicated that the QS system of Pseudomonas sp. was disrupted by specific QQ bacteria in the soil via the enzymatic destruction of signal molecules. Moreover, the number of bacteria with QQ activity against P. brassicacearum SZ50 was considerably higher in the CC soil and BT soil than in the NP soil, and the number of QQ bacteria belonging to Acinetobacter spp. greatly increased in the rhizospehre soil under consecutive R. glutinosa monoculture (Figure 5). The AHL-degrading activity of Acinetobacter spp. has been widely documented in previous studies (Kang et al., 2004; Chan et al., 2011; Ochiai et al., 2013). The QS system is known to play a pivotal role in regulating diverse behavior and functions, such as rhizosphere colonization and competence and biocontrol activities (Loh et al., 2002; Venturi and Keel, 2016). Therefore, QS disruption might be another factor contributing to a decline in antagonistic Pseudomonas spp. in the R. glutinosa rhizosphere under consecutive monoculture.
In conclusion, consecutive monoculture of R. glutinosa resulted in negative soil-borne legacy effects including the build-up of potentially harmful short-chain AHL-mediated QS bacteria in the rhizosphere and the reduction of beneficial short-chain AHL-mediated QS bacteria, which was not only mediated by specific root exudates but also by QQ bacteria. The findings in this work indicated the importance of the rhizosphere microbiome in maintaining soil health and highlighted a link between short-chain AHL-mediated quorum sensing and R. glutinosa replant disease. However, further studies will be performed to investigate the roles of rhizosphere bacteria producing the long-chain AHLs in replant disease and the functions of QQ bacteria that target QS signaling of other strains. In addition, further work is needed to restore soil health and overcome the replant disease of R. glutinosa by quorum quenching.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
LW and WL conceived the study. LW and QL wrote the manuscript. LW, YW, QL, JW, BY, JC, and HW performed the experiments. LW, BY, and QL performed the statistical analyses. JC, ZZ, and CL were involved in the field management and soil sampling. All authors discussed the results and commented on the manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (Grant nos. 81973412 and 81573530), the Program for Innovative Research Team in Fujian Agriculture and Forestry University (Grant no. 712018009), the Natural Science Foundation of Fujian Province (Grant no. 2017J01803), the Outstanding Youth Scientific Fund of Fujian Agriculture and Forestry University (Grant no. XJQ201501), the National Key Research and Development Program of China (Grant no. 2017YFC1700705), and Fujian-Taiwan Joint Innovative Center for Germplasm Resources and Cultivation of Crop (FJ 2011 Program, no. 2015-75).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank National Natural Science Foundation of China and the Program for Innovative Research Team in Fujian Agriculture and Forestry University, the Natural Science Foundation of Fujian Province, the Outstanding Youth Scientific Fund of Fujian Agriculture and Forestry University for providing the funds used in this work.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00787/full#supplementary-material
References
Anbazhagan, D., Mansor, M., Yan, G. O. S., Yusof, M. Y. M., Hassan, H., and Sekaran, S. D. (2012). Detection of quorum sensing signal molecules and identification of an autoinducer synthase gene among biofilm forming clinical isolates of Acinetobacter spp. PLoS One 7:e36696. doi: 10.1371/journal.pone.0036696
Badri, D. V., Weir, T. L., van der Lelie, D., and Vivanco, J. M. (2009). Rhizosphere chemical dialogues: plant-microbe interactions. Curr. Opin. Biotech. 20, 642–650. doi: 10.1016/j.copbio.2009.09.014
Bakker, P. A., Pieterse, C. M., de Jonge, R., and Berendsen, R. L. (2018). The soil-borne legacy. Cell 172, 1178–1180. doi: 10.1016/j.cell.2018.02.024
Bauer, W. D., and Mathesius, U. (2004). Plant responses to bacterial quorum sensing signals. Curr. Opin. Plant Biol. 7, 429–433. doi: 10.1016/j.pbi.2004.05.008
Berendsen, R. L., Corné, Pieterse, M. J., and Bakker, P. A. H. M. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Chagas, F. O., Pessotti, R. C., Caraballo-Rodríguez, A. M., and Pupo, M. T. (2018). Chemical signaling involved in plant–microbe interactions. Chem. Soc. Rev. 47, 1652–1704. doi: 10.1039/c7cs00343a
Chan, K. G., Atkinson, S., Mathee, K., Sam, C. K., Chhabra, S. R., Cámara, M., et al. (2011). Characterization of N-acylhomoserine lactone-degrading bacteria associated with the Zingiber officinale (ginger) rhizosphere: co-existence of quorum quenching and quorum sensing in Acinetobacter and Burkholderia. BMC Microbiol. 11:51. doi: 10.1186/1471-2180-11-51
Chen, J., Wu, L. K., Xiao, Z. G., Wu, Y., Wu, H. M., Qin, X. J., et al. (2017). Assessment of the diversity of Pseudomonas spp. and Fusarium spp. in Radix pseudostellariae rhizosphere under monoculture by combining DGGE and quantitative PCR. Front. Microbiol. 8:1748. doi: 10.3389/fmicb.2017.01748
Chen, S. C., Yu, H. J., Zhou, X. G., and Wu, F. Z. (2018). Cucumber (Cucumis sativus L.) seedling rhizosphere Trichoderma and Fusarium spp. communities altered by vanillic acid. Front. Microbiol. 9:2195. doi: 10.3389/fmicb.2018.02195
Corral-Lugo, A., Daddaoua, A., Ortega, A., Espinosa-Urgel, M., and Krell, T. (2016). Rosmarinic acid is a homoserine lactone mimic produced by plants that activates a bacterial quorum-sensing regulator. Sci. Signal 9:ra1. doi: 10.1126/scisignal.aaa8271
Dong, L. L., Xu, J., Li, Y., Fang, H. L., Niu, W. H., Li, X. W., et al. (2018a). Manipulation of microbial community in the rhizosphere alleviates the replanting issues in Panax ginseng. Soil Biol. Biochem. 125, 64–74. doi: 10.1016/j.soilbio.2018.06.028
Dong, L. L., Xu, J., Zhang, L. J., Cheng, R. Y., Wei, G. F., Su, H., et al. (2018b). Rhizospheric microbial communities are driven by Panax ginseng at different growth stages and biocontrol bacteria alleviates replanting mortality. Acta Pharm. Sin. B 8, 272–282. doi: 10.1016/j.apsb.2017.12.011
Dong, Y., Wang, L., Xu, J., Zhang, H., Zhang, X., and Zhang, L. (2001). Quenching quorum-sensing-dependent bacterial infection by an N -acyl homoserine lactonase. Nature 411, 813–817. doi: 10.1038/35081101
el Zahar Haichar, F., Santaella, C., Heulin, T., and Achouak, W. (2014). Root exudates mediated interactions belowground. Soil Biol. Biochem. 77, 69–80. doi: 10.1016/j.soilbio.2014.06.017
Franke-Whittle, I. H., Manici, L. M., Insam, H., and Stres, B. (2015). Rhizosphere bacteria and fungi associated with plant growth in soils of three replanted apple orchards. Plant Soil 395, 317–333. doi: 10.1007/s11104-015-2562-x
Gao, Z. Y., Han, M. K., Hu, Y. Y., Li, Z. Q., Liu, C. F., Wang, X., et al. (2019). Effects of continuous cropping of sweet potato on the fungal community structure in rhizospheric soil. Front. Microbiol. 10:2269. doi: 10.3389/fmicb.2019.02269
Grandclement, C., Tannieres, M., Morera, S., Dessaux, Y., and Faure, D. (2016). Quorum quenching: role in nature and applied developments. FEMS Microb. Rev. 40, 86–116. doi: 10.1093/femsre/fuv038
Gu, Y., Wei, Z., Wang, X. Q., Friman, V. P., Huang, J. F., Wang, X. F., et al. (2016). Pathogen invasion indirectly changes the composition of soil microbiome via shifts in root exudation profile. Biol. Fert. Soils 52, 997–1005. doi: 10.1007/s00374-016-1136-2
Hossain, M. A., Lee, S. J., Park, N. H., Mechesso, A. F., Birhanu, B. T., Kang, J. W., et al. (2017). Impact of phenolic compounds in the acyl homoserine lactone-mediated quorum sensing regulatory pathways. Sci. Rep. 7, 1–16. doi: 10.1038/s41598-017-10997-5
Hu, L. F., Robert, C. A. M., Cadot, S., Zhang, X., Ye, M., Li, B., et al. (2018). Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat. Commun. 9:2738. doi: 10.1038/s41467-018-05122-7
Huang, X. F., Chaparro, J. M., Reardon, K. F., Zhang, R., Shen, Q. R., and Vivanco, J. M. (2014). Rhizosphere interactions: root exudates, microbes, and microbial communities. Botany 92, 267–275. doi: 10.1139/cjb-2013-0225
Jacques, P. J., and Jacques, J. R. (2012). Monocropping cultures into ruin: the loss of food varieties and cultural diversity. Sustainability 4, 2970–2997. doi: 10.3390/su4112970
Jiang, J. L., Yu, M., Hou, R. P., Li, L., Ren, X. M., Jiao, C. J., et al. (2019). Changes in the soil microbial community are associated with the occurrence of Panax quinquefolius L. root rot diseases. Plant Soil 438, 143–156. doi: 10.1007/s11104-018-03928-4
Kan, J. H., Fang, R. X., and Jia, Y. T. (2017). Interkingdom signaling in plant-microbe interactions. Sci. China Life Sci. 60, 785–796. doi: 10.1007/s11427-017-9092-3
Kang, B. R., Lee, J. H., Ko, S. J., Lee, Y. H., Cha, J. S., Cho, B. H., et al. (2004). Degradation of acyl-homoserine lactone molecules by Acinetobacter sp. strain C1010. Can. J. Microbiol. 50, 935–941. doi: 10.1139/w04-083
Khan, M., Bhargava, P., and Goel, R. (2019). “Quorum sensing molecules of rhizobacteria: a trigger for developing systemic resistance in plants,” in Plant Growth Promoting Rhizobacteria for Sustainable Stress Management, eds R. Z. Sayyed, N. K. Arora, and M. S. Reddy (Singapore: Springer), 117–138. doi: 10.1007/978-981-13-6536-2_7
Köberl, M., Dita, M., Martinuz, A., Staver, C., and Berg, G. (2017). Members of Gammaproteobacteria as indicator species of healthy banana plants on Fusarium wilt-infested fields in Central America. Sci. Rep. 7:45318. doi: 10.1038/srep45318
Kong, H. G., Song, G. C., and Ryu, C. M. (2019). Inheritance of seed and rhizosphere microbial communities through plant–soil feedback and soil memory. Environ. Microbiol. Rep. 11, 479–486. doi: 10.1111/1758-2229.12760
Kwak, M. J., Kong, H. G., Choi, K., Kwon, S. K., Song, J. Y., Lee, J., et al. (2018). Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat. Biotechnol. 36, 1100–1109. doi: 10.1038/nbt1118-1117
Kyselková, M., and Moënne-Loccoz, Y. (2012). “Pseudomonas and other microbes in disease-suppressive soils,” in Organic Fertilisation, Soil Quality and Human Health, ed. E. Lichtfouse (Dordrecht: Springer), 93–140. doi: 10.1007/978-94-007-4113-3_5
Li, P., Ma, L., Feng, Y. L., Mo, M. H., Yang, F. X., Dai, H. F., et al. (2012). Diversity and chemotaxis of soil bacteria with antifungal activity against Fusarium wilt of banana. J. Ind. Microbiol. Biot. 39, 1495–1505. doi: 10.1007/s10295-012-1163-4
Li, X. G., Ding, C. F., Hua, K., Zhang, T. L., Zhang, Y. N., Zhao, L., et al. (2014). Soil sickness of peanuts is attributable to modifications in soil microbes induced by peanut root exudates rather than to direct allelopathy. Soil Biol. Biochem. 78, 149–159. doi: 10.1016/j.soilbio.2014.07.019
Li, X. G., Panke-Buisse, K., Yao, X. D., Coleman-Derr, D., Ding, C. F., Wang, X. X., et al. (2019). Peanut plant growth was altered by monocropping-associated microbial enrichment of rhizosphere microbiome. Plant Soil 446, 655–669. doi: 10.1007/s11104-019-04379-1
Lin, S., Dai, L. Q., Chen, T., Li, Z. F., Zhang, Z. Y., and Lin, W. X. (2015). Screening and identification of harmful and beneficial microorganisms associated with replanting disease in rhizosphere soil of Pseudostellariae heterophylla. Int. J. Agric. Biol. 17, 458–466. doi: 10.17957/IJAB/17.3.14.224
Liu, J. J., Yao, Q., Li, Y. S., Zhang, W., Mi, G., Chen, X. L., et al. (2019). Continuous cropping of soybean alters the bulk and rhizospheric soil fungal communities in a Mollisol of Northeast PR China. Land Degrad. Dev. 30, 1725–1738. doi: 10.1002/ldr.3378
Loh, J., Pierson, E. A., Pierson, L. S. III, Stacey, G., and Chatterjee, A. (2002). Quorum sensing in plant-associated bacteria. Curr. Opin. Plant Biol. 5, 285–290. doi: 10.1016/S1369-5266(02)00274-1
Luo, L. F., Guo, C. W., Wang, L. T., Zhang, J. X., Deng, L. M., Luo, K. F., et al. (2019). Negative plant-soil feedback driven by re-assemblage of the rhizosphere microbiome with the growth of Panax notoginseng. Front. Microbiol. 10:1597. doi: 10.3389/fmicb.2019.01597
Mariotte, P., Mehrabi, Z., Bezemer, T. M., De Deyn, G. B., Kulmatiski, A., Drigo, B., et al. (2018). Plant–soil feedback: bridging natural and agricultural sciences. Trends Ecol. Evol. 33, 129–142. doi: 10.1016/j.tree.2017.11.005
Matilla, M. A., Ramos, J. L., Bakker, P. A. H. M., Doornbos, R., Badri, D. V., Vivanco, J. M., et al. (2010). Pseudomonas putida KT2440 causes induced systemic resistance and changes in Arabidopsis root exudation. Environ. Microbiol. Rep. 2, 381–388. doi: 10.1111/j.1758-2229.2009.00091.x
Mendes, R., Kruijt, M., De Bruijn, I., Dekkers, E., van der Voort, M., Schneider, J. H., et al. (2011). Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332, 1097–1100. doi: 10.1126/science.1203980
Neal, A. L., Ahmad, S., Gordon-Weeks, R., and Ton, J. (2012). Benzoxazinoids in root exudates of maize attract Pseudomonas putida to the rhizosphere. PLoS One 7:e35498. doi: 10.1371/journal.pone.0035498
Ochiai, S., Morohoshi, T., Kurabeishi, A., Shinozaki, M., Fujita, H., Sawada, I., et al. (2013). Production and degradation of N-acylhomoserine lactone quorum sensing signal molecules in bacteria isolated from activated sludge. Biosci. Biotechnol. Biochem. 77, 2436–2440. doi: 10.1271/bbb.130553
Rani, M., Prakash, D., Sobti, R. C., and Jain, R. K. (1996). Plasmid-mediated degradation of o-phthalate and salicylate by a Moraxella sp. Biochem. Biophys. Res. Commun. 220, 377–381. doi: 10.1006/bbrc.1996.0413
Remuzgo-Martínez, S., Lázaro-Díez, M., Mayer, C., Aranzamendi-Zaldumbide, M., Padilla, D., Calvo, J., et al. (2015). Biofilm formation and quorum-sensing-molecule production by clinical isolates of Serratia liquefaciens. Appl. Environ. Microb. 81, 3306–3315. doi: 10.1128/AEM.00088-15
Rolfe, S. A., Griffiths, J., and Ton, J. (2019). Crying out for help with root exudates: adaptive mechanisms by which stressed plants assemble health-promoting soil microbiomes. Curr. Opin. Microbiol. 49, 73–82. doi: 10.1016/j.mib.2019.10.003
Sakr, M. M., Aboshanab, K. M., Aboulwafa, M., and Hassouna, N. A. (2013). Characterization and ccomplete sequence of lactonase enzyme from Bacillus weihenstephanensis isolate P65 with potential activity against Acyl homoserine lactone signal molecules. Biomed. Res. Int. 2013:192589. doi: 10.1155/2013/192589
Schaefer, A. L., Greenberg, E. P., Oliver, C. M., Oda, Y., Huang, J. J., Bittan-Banin, G., et al. (2008). A new class of homoserine lactone quorum-sensing signals. Nature 454, 595–599. doi: 10.1038/nature07088
Shen, Z. Z., Ruan, Y. Z., Xue, C., Zhong, S. T., Li, R., and Shen, Q. R. (2015). Soils naturally suppressive to banana Fusarium wilt disease harbor unique bacterial communities. Plant Soil 393, 21–33. doi: 10.1007/s11104-015-2474-9
Stringlis, I. A., Yu, K., Feussner, K., de Jonge, R., Van Bentum, S., Van Verk, M. C., et al. (2018). MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc. Natl. Acad. Sci. U.S.A. 115, E5213–E5222. doi: 10.1073/pnas.1722335115
Taga, M. E., and Bassler, B. L. (2003). Chemical communication among bacteria. Proc. Natl. Acad. Sci. U.S.A. 84, 106–107. doi: 10.1086/598296
Tan, Y., and Ji, G. (2010). Bacterial community structure and dominant bacteria in activated sludge from a 70 degrees C ultrasound-enhanced anaerobic reactor for treating carbazole–containing wastewater. Bioresour. Technol. 101, 174–180. doi: 10.1016/j.biortech.2009.08.044
Teplitski, M., Robinson, J. B., and Bauer, W. D. (2000). Plants secrete substances that mimic bacterial N-acyl homoserine lactone signal activities and affect population density-dependent behaviors in associated bacteria. Mol. Plant Microbe Interact. 13, 637–648. doi: 10.1094/MPMI.2000.13.6.637
Uroz, S., Dessaux, Y., and Oger, P. (2009). Quorum sensing and quorum quenching: the yin and yang of bacterial communication. ChemBioChem 10, 205–216. doi: 10.1002/cbic.200800521
Venturi, V., and Keel, C. (2016). Signaling in the rhizosphere. Trends Plant Sci. 21, 187–198. doi: 10.1016/j.tplants.2016.01.005
Veselova, M., Kholmeckaya, M., Klein, S., Voronina, E., Lipasova, V., Metlitskaya, A., et al. (2003). Production of N-acylhomoserine lactone signal molecules by gram-negative soil-borne and plant-associated bacteria. Folia Microbiol. 48, 794–798. doi: 10.1007/bf02931516
Voges, M. J. E. E. E., Bai, Y., Schulze-Lefert, P., and Sattely, E. S. (2019). Plant-derived coumarins shape the composition of an Arabidopsis synthetic root microbiome. Proc. Natl. Acad. Sci. U.S.A. 116, 12558–12565. doi: 10.1073/pnas.1820691116
von Bodman, S. B., Bauer, W. D., and Coplin, D. L. (2003). Quorum sensing in plant-pathogenic bacteria. Annu. Rev. Phytopathol. 41, 455–482. doi: 10.1146/annurev.phyto.41.052002.095652
Wang, L., and Li, X. (2019). Steering soil microbiome to enhance soil system resilience. Crit. Rev. Microbiol. 45, 743–753. doi: 10.1080/1040841X.2019.1700906
Wu, H. M., Qin, X. J., Wang, J. Y., Wu, L. K., Chen, J., Fan, J. K., et al. (2019). Rhizosphere responses to environmental conditions in Radix pseudostellariae under continuous monoculture regimes. Agr. Ecosyst. Environ. 270, 19–31. doi: 10.1016/j.agee.2018.10.014
Wu, L. K., Chen, J., Khan, M. U., Wang, J. Y., Wu, H. M., Xiao, Z. G., et al. (2018a). Rhizosphere fungal community dynamics associated with Rehmannia glutinosa replant disease in a consecutive monoculture regime. Phytopathology 108, 1493–1500. doi: 10.1094/PHYTO-02-18-0038-R
Wu, L. K., Chen, J., Xiao, Z. G., Zhu, X. C., Wang, J. Y., Wu, H. M., et al. (2018b). Barcoded pyrosequencing reveals a shift in the bacterial community in the rhizosphere and rhizoplane of Rehmannia glutinosa under consecutive monoculture. Int. J. Mol. Sci. 19:850. doi: 10.3390/ijms19030850
Wu, L. K., Chen, J., Wu, H. M., Wang, J. Y., Wu, Y. H., Lin, S., et al. (2016a). Effects of consecutive monoculture of Pseudostellaria heterophylla on soil fungal community as determined by pyrosequencing. Sci. Rep. 6, 1–10. doi: 10.1038/srep26601
Wu, L. K., Wu, H. M., Chen, J., Wang, J. Y., and Lin, W. X. (2016b). Microbial community structure and its temporal changes in Rehmannia glutinosa rhizospheric soils monocultured for different years. Eur. J. Soil Biol. 72, 1–5. doi: 10.1016/j.ejsobi.2015.12.002
Wu, H. M., Wu, L. K., Wang, J. Y., Zhu, Q., Lin, S., Xu, J. H., et al. (2016c). Mixed phenolic acids mediated proliferation of pathogens Talaromyces helicus and Kosakonia sacchari in continuously monocultured Radix pseudostellariae rhizosphere soil. Front. Microbiol. 7:335. doi: 10.3389/fmicb.2016.00335
Wu, L. K., Wang, J. Y., Huang, W. M., Wu, H. M., Chen, J., Yang, Y. Q., et al. (2015). Plant-microbe rhizosphere interactions mediated by Rehmannia glutinosa root exudates under consecutive monoculture. Sci. Rep. 5:15871. doi: 10.1038/srep19101
Yang, Y. H., Chen, X. J., Chen, J. Y., Xu, H. X., Li, J., and Zhang, Z. Y. (2011). Differential miRNA expression in Rehmannia glutinosa plants subjected to continuous cropping. BMC Plant Biol. 11:53. doi: 10.1186/1471-2229-11-53
Yu, H. J., Chen, S. C., Zhang, X. X., Zhou, X. G., and Wu, F. Z. (2019). Rhizosphere bacterial community in watermelon-wheat intercropping was more stable than in watermelon monoculture system under Fusarium oxysporum f. sp. niveum invasion. Plant Soil 445, 369–381. doi: 10.1007/s11104-019-04321-5
Yuan, J., Zhao, J., Wen, T., Zhao, M. L., Li, R., Goossens, P., et al. (2018). Root exudates drive the soil-borne legacy of aboveground pathogen infection. Microbiome 6, 1–12. doi: 10.1186/s40168-018-0537-x
Zaytseva, Y. V., Sidorov, A. V., Marakaev, O. A., and Khmel, I. A. (2019). Plant-microbial interactions involving quorum sensing regulation. Microbiology 88, 523–533. doi: 10.1134/S0026261719040131
Zhalnina, K., Louie, K. B., Hao, Z., Mansoori, N., da Rocha, U. N., Shi, S. J., et al. (2018). Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 3, 470–480. doi: 10.1038/s41564-018-0129-3
Zhang, B., Weston, P. A., Gu, L., Zhang, B. Y., Li, M. J., Wang, F. Q., et al. (2019). Identification of phytotoxic metabolites released from Rehmannia glutinosa suggest their importance in the formation of its replant problem. Plant Soil 441, 439–454. doi: 10.1007/s11104-019-04136-4
Zhang, L. Y., Guo, Z. W., Gao, H. F., Peng, X. Q., Li, Y. Y., Sun, S. J., et al. (2016). Interaction of Pseudostellaria heterophylla with quorum sensing and quorum quenching bacteria mediated by root exudates in a consecutive monoculture system. J. Microbiol. Biotechn. 26, 2159–2170. doi: 10.4014/jmb.1607.07073
Zhang, R. F., Vivanco, J. M., and Shen, Q. R. (2017). The unseen rhizosphere root–soil–microbe interactions for crop production. Curr. Opin. Microbiol. 37, 8–14. doi: 10.1016/j.mib.2017.03.008
Zhao, J., Li, Y. L., Wang, B. Y., Huang, X. Q., Yang, L., Lan, T., et al. (2017). Comparative soil microbial communities and activities in adjacent Sanqi ginseng monoculture and maize-Sanqi ginseng systems. Appl. Soil Ecol. 120, 89–96. doi: 10.1016/j.apsoil.2017.08.002
Keywords: Rehmannia glutinosa, replant disease, quorum sensing, root exudate, quorum quenching
Citation: Li Q, Wu Y, Wang J, Yang B, Chen J, Wu H, Zhang Z, Lu C, Lin W and Wu L (2020) Linking Short-Chain N-Acyl Homoserine Lactone-Mediated Quorum Sensing and Replant Disease: A Case Study of Rehmannia glutinosa. Front. Plant Sci. 11:787. doi: 10.3389/fpls.2020.00787
Received: 22 February 2020; Accepted: 18 May 2020;
Published: 17 June 2020.
Edited by:
Xiangming Xu, NIAB EMR, United KingdomReviewed by:
Maria Ludovica Saccà, Council for Agricultural Research and Economics (CREA), ItalyUlrike Mathesius, Australian National University, Australia
Copyright © 2020 Li, Wu, Wang, Yang, Chen, Wu, Zhang, Lu, Lin and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenxiong Lin, bHd4QGZhZnUuZWR1LmNu; Linkun Wu, d3VsaW5rdW42MTlAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Qian Li
Qian Li Yanhong Wu
Yanhong Wu Juanying Wang
Juanying Wang Bo Yang
Bo Yang Jun Chen
Jun Chen Hongmiao Wu
Hongmiao Wu Zhongyi Zhang3
Zhongyi Zhang3 Wenxiong Lin
Wenxiong Lin Linkun Wu
Linkun Wu