- 1Rice Research Institute, Guangdong Academy of Agricultural Sciences, Guangzhou, China
- 2Guangdong Key Laboratory of New Technology in Rice Breeding, Guangzhou, China
Bacterial blight (BB) is an important constraint on achieving a high and stable rice grain yield. An increasing number of BB resistance (R) genes have been identified and cloned to increase the available options for rice disease resistance breeding. However, it is necessary to understand the distribution of R genes in rice varieties for rational distribution and breeding. Here, we genotyped five R genes, i.e. Xa4, Xa7, Xa21, Xa23, and Xa27, in seventy main cultivars from Guangdong Province, South China using the corresponding specific markers. Our results showed that 61 varieties carried Xa4, only three varieties carried Xa27, and Xa7, Xa21, or Xa23 was not detected in all tested varieties. Notably, only 33 varieties exhibited resistance to pathotype IV Xoo strains. These results indicate that Xa4 is no longer suitable for widespread use in rice breeding, although Xa4 is widely present in tested varieties. Remarkably, the strongly virulent BB strains of pathotype IX evolved quickly in southern China, and Xa23 was found to effectively confer resistance against the pathotype IX strains. Subsequently, we successfully bred two novel inbred rice varieties as also being restorer lines and two photoperiod- and thermo-sensitive genic male sterility (P/TGMS) lines using the broad-spectrum resistance gene Xa23 through marker-assisted selection (MAS) combined with phenotypic selection. All of the developed lines and derived hybrids exhibited enhanced resistance to BB with excellent yield performance. Our research may potentially facilitate both of the inbred and hybrid rice disease resistance breeding.
Introduction
Rice is one of the most important staple crops because it feeds nearly half of the global population. The grain production and quality of rice have to be increased and improved to guarantee global food security due to the rapid growth of the global population, global climate change, and other reasons (Tilman et al., 2001; Qian et al., 2016). However, rice grain yield and quality are frequently and severely threatened by devastating diseases caused by multiple pathogens, such as Xanthomonas oryzae pv. oryzae (Xoo) and Magnaporthe oryzae (Ke et al., 2017; Wang J. et al., 2018). Previous studies have shown that significant grain yield loss of more than 20% was caused by BB, especially in hybrid rice, and the degree of yield loss was affected by the stage of BB infection (Chukwu et al., 2019). Therefore, the breeding of resistant rice varieties is advocated as being the most cost-effective approach to overcome the major obstacle to achieving optimal yield. Consequently, the exploration of novel resistant germplasms and the utilization of excellent gene resources are crucial for the development of rice resistance to BB.
To date, approximately 45 resistance (R) genes conferring resistance against BB have been identified using diverse rice sources (Neelam et al., 2020). However, only eleven of these R genes, exhibiting multiple mechanisms of R-gene-mediated Xoo resistance, have been cloned and functionally analyzed (Song et al., 1995; Yoshimura et al., 1998; Iyer and McCouch, 2004; Sun et al., 2004; Gu et al., 2005; Chu et al., 2006; Liu et al., 2011; Tian et al., 2014; Hutin et al., 2015; Wang et al., 2015; Hu et al., 2017). Among these genes, Xa21, Xa23, and the un-cloned R gene Xa7 act in a dominant manner and confer the strongest resistance to BB with the broadest resistance spectrum (Zhang, 2009; Chukwu et al., 2019). Recently, many R genes for BB were successfully incorporated into both elite inbred varieties and parental lines of hybrid rice to control the disease using MAS (Chukwu et al., 2019; Jiang et al., 2020; Li W. et al., 2020). Notably, a few of those R genes, such as Xa3, Xa4, Xa7, and Xa21, have been widely utilized in rice resistance breeding since the 1980s (Deng Q. et al., 2006; Zhang, 2009; Chen et al., 2011; Luo et al., 2012). However, the resistance conferred by these genes has weakened and can be overcome by the new virulent BB strains due to pathogenic variation or evolution with consequent breakdown of rice resistance (Zhang, 2009; Chen et al., 2011; Wang et al., 2014). On the other hand, the R-gene-mediated resistance to Xoo in inbred rice or hybrid rice may be influenced by the rice genetic background and the incomplete dominance of R genes, thus hindering the application of these R genes for breeding of disease-resistant rice (Luo et al., 2012; Zeng L. et al., 2017). Remarkably, the resistance gene Xa23, a new executor R gene derived from wild rice (Oryza rufipogon), was found to effectively confer completely dominant and broad-spectrum resistance against BB of rice at all growth stages (Wang et al., 2015). Thus, CBB23, a near isogenic line (NIL) carrying the Xa23 resistance haplotype in the JG30 background, has been widely utilized as an important germplasm for disease resistance in rice breeding (Zhang, 2009; Wang et al., 2015).
There is high selection pressure due to the high cropping index in the two-season continuous-cropping rice area and the specific ecological conditions, such as high temperature with high humidity, in southern China, which facilitates pathogenic variation and results in epidemics of BB (Chen et al., 2018). Thus, the strongly virulent BB strains of pathotype V have replaced pathotype IV strains and have developed into the preponderant pathotype, and the more virulent pathotype IX has grown quickly in southern China (Chen et al., 2018). On the other hand, there have been few studies on the rationality of the distribution of rice varieties resistant to BB, whereas varieties carrying a single R gene are subjected to large-scale and long-term cultivation, resulting in breakdown of resistance (Kim, 2018; Chukwu et al., 2019). In the present study, we identified the distribution of the BB resistance genes of the main cultivars from Guangdong Province, China, for varieties that are rationally distributed and bred. We found that Xa4 was widely utilized, while other detected R genes were less frequently used for rice breeding in Guangdong. Based on this finding, the underlying objective of this study was to breed new rice varieties and novel P/TGMS lines using the broad-spectrum resistance gene Xa23, which conferred high resistance to all of the tested Xoo strains, including the strongly virulent BB strains of pathotype V and IX (Luo et al., 2013). Finally, two novel inbred rice varieties and two newly developed P/TGMS lines with significantly enhanced disease resistance to BB were successfully bred using MAS combined with phenotypic selection. The improved lines and the derived hybrid combinations with elite yield performance showed high resistance to the tested Xoo strains, and provided excellent paternal lines for production of inbred and hybrid rice with disease resistance to rice BB.
Materials and Methods
Plant Materials and Growth Conditions
Seventy main cultivars with a planting area of more than 6,000 ha in Guangdong Province in recent years, including fifty-one inbred lines, twelve restorer lines, six maintainer lines, and one P/TGMS line, were collected and planted for genotyping the rice bacterial blight resistance genes (Lu et al., 2019). The elite indica variety YueJinYinZhan (YJYZ) and the P/TGMS line YueJing1S (YJ1S) were used as recipient parents for disease resistance breeding in this study. XinHuangZhan (XHZ) and ‘V5400’, two indica type varieties harboring homozygous Xa23 and conferring high broad-spectrum resistance to rice bacterial blight, were used as the donors of Xa23. On the other hand, ‘V5400’, harboring homozygous Pi2 and conferring high resistance to rice blast, was used as the donor of Pi2 for novel P/TGMS-line breeding. The breeding schemes for the novel inbred lines and P/TGMS lines are illustrated in Figure 1. All of the materials were planted using normal field management practices in an area of 16.7 × 16.7 cm at the experimental fields of Guangdong Academy of Agricultural Sciences (GDAAS), Guangzhou city, Guangdong Province, China.
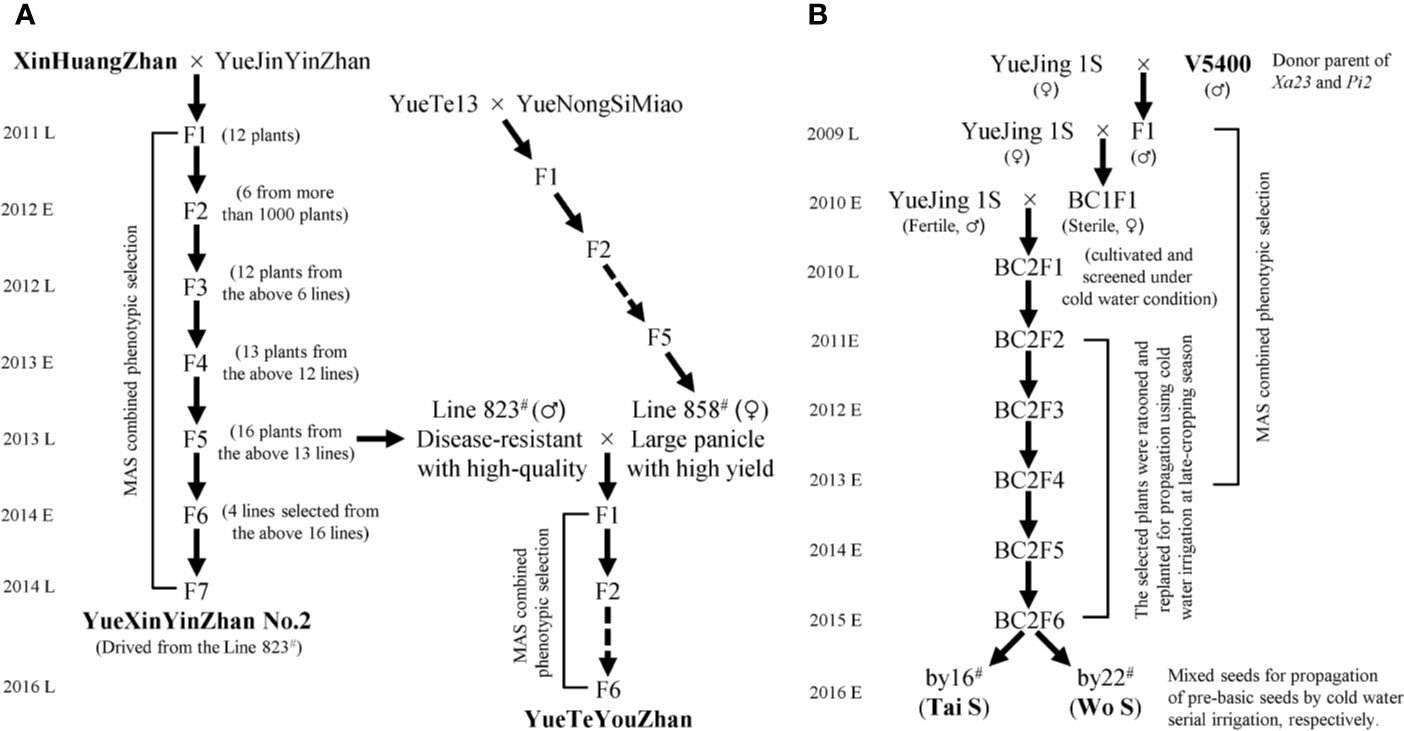
Figure 1 Flowchart for the development of disease-resistant varieties and P/TGMS lines. MAS, marker-assisted selection; E and L represent the early cropping season and later cropping season, respectively. (A) The pedigree breeding schemes for the inbred lines. (B) The backcross breeding schemes for the P/TGMS lines.
PCR-Based Genotyping and MAS Breeding
Plant genomic DNA was extracted from fresh leaves of the heading-stage plants using a previously described CTAB protocol (Liu et al., 1997). The molecular marker for genotyping Xa4 was the co-dominant simple sequence repeat (SSR) marker RM224, that for Xa7 was the sequence-tagged site (STS) marker STSP3, those for Xa21 were the co-dominant STS marker pTA248 and the optimized functional marker U1/I2, those for Xa23 were the SSR marker RM206 and the optimized marker P23, and that for Xa27 was the co-dominant marker XA27-Co (Chunwongse et al., 1993; Chen et al., 2000; Pai et al., 2003; Porter et al., 2003; Sun et al., 2003; Gu et al., 2005; Zhou et al., 2011). The markers RM206 and P23 were used for MAS breeding to develop novel rice bacterial blight-resistant varieties with Xa23. In addition, the SSR marker RM527 and the Indel marker S29742 were used for genotyping the rice blast resistance gene Pi2 and for MAS breeding (Deng Y. et al., 2006; Chen et al., 2008). Notably, MAS combining with phenotypic selection was used throughout the breeding process. PCR amplification was performed on a Bio-Rad C1000 Touch Thermal Cycler (USA) and the protocol for PCR amplification with the appropriate parameters was described previously (Wang et al., 2005). The PCR product for the marker RM206 was resolved on an 8% polyacrylamide gel, and other products were resolved on a 3.0% agarose gel in 1× TBE buffer. The primers used in this study are listed in Table S3.
Evaluation of Rice Bacterial Blight Resistance
Xoo strains were grown on PSA medium at 28°C for 2–3 days, and the bacterial cells were suspended in sterile water at an optical density of 0.5 (OD600). 161BYZ-2 and 13WH-D, the pathotype IV and IX Xoo strains, respectively, were used for artificial inoculation with bacterial blight separately through the leaf-clipping method as described previously (Lu et al., 2019). All plants were inoculated at the heading stage. Photos of disease symptoms were taken two weeks after inoculation, and lesion length was measured three weeks after inoculation to evaluate the level of rice bacterial blight resistance. The evaluation standard of bacterial blight resistance was referred to previous reference (Liu et al., 2016).
Fertility-Sterility Alteration Tested for the P/TGMS Lines
In 2016, the two P/TGMS lines, namely, Tai S and Wo S, were sown every 10 days from February 22nd to March 23rd in the early cropping season and from July 21st to August 20th in the later cropping season, respectively, in the experimental fields of GDAAS, Guangzhou city, Guangdong Province, China. YJ1S was used as the control. The dynamic pollen fertility of these P/TGMS lines was observed at several-day intervals from May 18th to June 11th during the early cropping season and September 25th to October 27th during the later cropping season in 2016. Male fertility conversion and multiplication of these P/TGMS lines were conducted using an artificial intelligence-based cold-water serial irrigation system. The cold water (20–22°C) irrigation treatment was started at the phase of differentiation of the secondary rachis branch and spikelet primordia (young panicle differentiation phase III) and was performed until the meiotic division of the pollen mother cell (phase VI).
Production Measurement and Statistical Analysis
Hybrid combinations were made through several cytoplasm male sterility (CMS) and P/TGMS lines crossed with the restorer lines, and the F1 hybrids were grown in the field in an area of 16.7 × 16.7 cm using normal field management practices. At maturity, all of the plants in each plot were harvested, and the grain yield of the hybrid combinations was measured according to a method described previously (Wang S. et al., 2018). Mean phenotypic values were compared using Student’s t test and the data were analyzed using Microsoft Excel 2013 as previously described (Wang S. et al., 2018).
Results
Identification of Bacterial Blight Resistance Genes in the Tested Cultivars
Understanding the distribution of disease resistance genes in rice cultivars is beneficial not only for rice breeding but also for the rational distribution of cultivars. In the present study, we collected seventy main cultivars from Guangdong Province, in the southern China rice region, and identified the distribution of the bacterial blight resistance genes using the corresponding specific markers, including Xa4, Xa7, Xa21, Xa23, and Xa27, which act in a dominant manner with broad-spectrum resistance against BB and have been widely utilized in rice resistance breeding. The detailed identification results are shown in Table S1 and Figure S1.
The results showed that 61 cultivars carried Xa4, accounting for 87.1%; and three cultivars, namely, Lvhuangzhan (LHZ), Qiu B, and Yuefeng B, carried Xa27, accounting for only 4.3% (Table S1). However, none of the tested cultivars carried Xa7, Xa21, or Xa23. On the other hand, 33 cultivars had moderate resistance or more to pathotype IV Xoo strains, and only two cultivars, namely, Huanglizhan (HLZ) and LHZ had moderate resistance to pathotype IX Xoo strains (Table S1). These results indicated that Xa4 had been widely used in rice breeding for BB resistance in Guangdong but was unable to meet the current requirements for rice disease resistance breeding. Two cultivars, HLZ and LHZ, have potential as excellent resistant germplasms for rice disease resistance breeding. The disease resistance mechanism of HLZ and LHZ should be further investigated to facilitate the subsequent marker-assisted selection.
Development of Elite Rice Varieties and Novel P/TGMS Lines
Based on the distribution of bacterial blight resistance genes and phenotypic results, it is necessary to explore disease-resistant germplasms or have excellent gene resources with broad-spectrum and high resistance to bacterial blight to improve or breed new varieties. First, we observed that XHZ, carrying the broad-spectrum and durable resistance gene Xa23 derived from CBB23, exhibited a typical hypersensitive disease resistance response to pathotype IV and also to pathotype IX Xoo strains (Figures 2 and 3A), which was consistent with the results of previous studies (Luo et al., 2013; Wang et al., 2015; Zeng L. et al., 2017). Then, we used XHZ as an important resistant donor for disease resistance improvement. As shown in the schematic (Figure 1A), XHZ harboring homozygous Xa23 was crossed with YJYZ, an excellent rice variety with high yield and high sensitivity to pathotype IX Xoo strains (Figures 2B, D). We used MAS combined with phenotypic selection for pedigree breeding and filtered out the high-quality and high resistance line 823# with homozygous Xa23 gene from the F5 generation in later cropping season of 2013. Finally, we bred a new excellent rice variety with high yield and high quality called YueXinYinZhan No.2 (YXYZ2, Figure 1A) from line 823#, and this variety was authorized in 2018 (Guangdong authorized variety no. 20180041). Subsequently, we crossed the line 823# with the large-panicle and high-yield intermediate material line 858#, which was derived from the high-quality and main cultivated rice variety YueNongSiMiao (YNSM). Then, we bred a new variety YueTeYouZhan (YTYZ) harboring homozygous Xa23 gene in later cropping season of 2016 (Figures 1A and 3A).

Figure 2 Disease reaction patterns of rice varieties to Xoo strains. (A, B) Leaves of rice varieties are presented to show the lesion patterns for the pathotype IV and IX Xoo strains, respectively. Pictures were taken on the 14th day after inoculation. Scale bars: 5 cm. (C, D) Statistical analysis of the lesion lengths of rice varieties inoculated with the pathotype IV and IX Xoo strains, respectively. Lesion length was measured on the 21st day after inoculation. Data are given as the means with SDs (n = 10). The uppercase and lowercase letters above the bars represent significant difference levels P < 0.01 and P < 0.05, respectively.
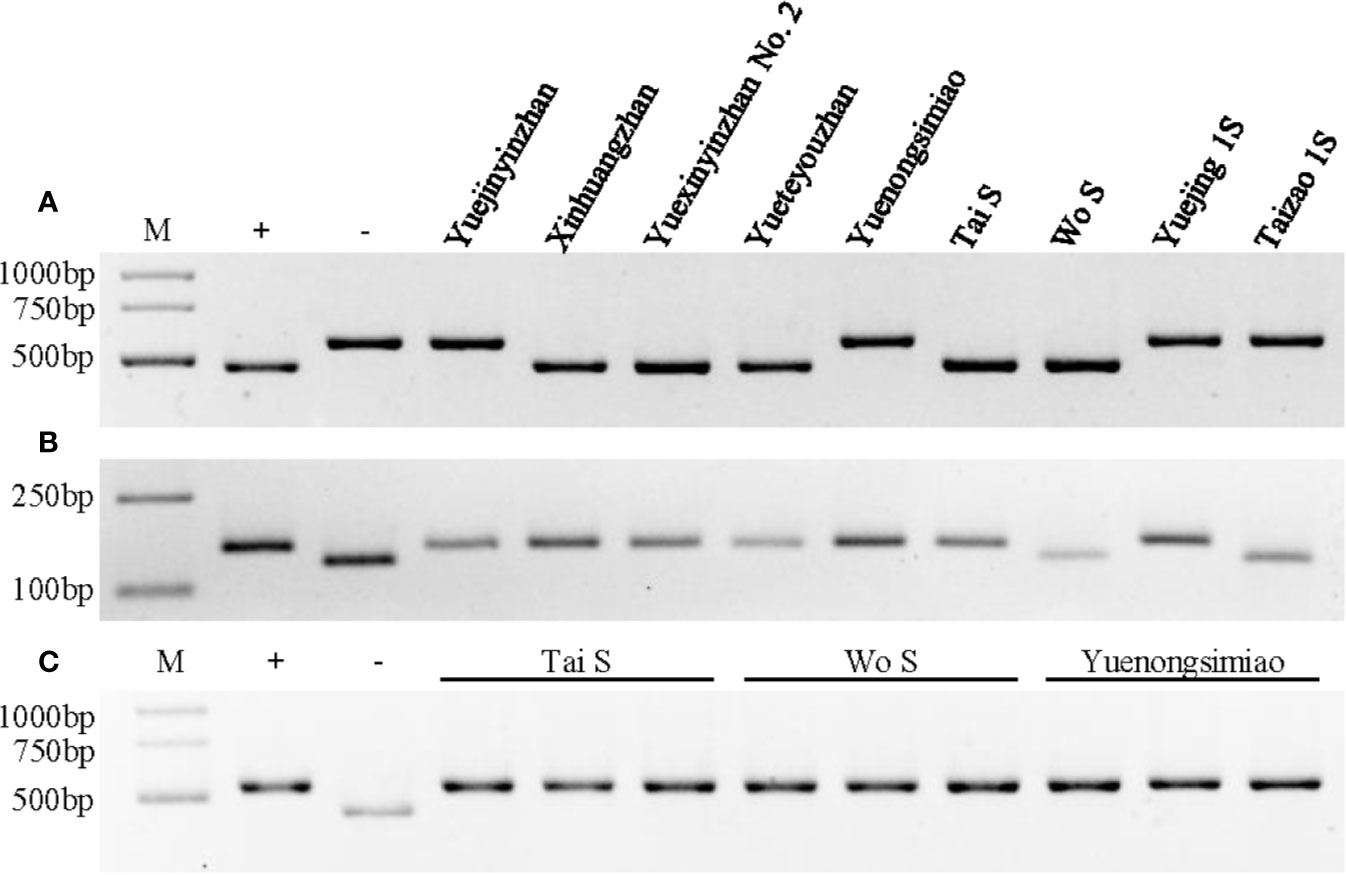
Figure 3 Genotype identification of Xa23, Xa4, and Pi2 in different parental lines. (A) Genotype identification of the Xa23 gene using the optimized marker P23. The positive (+) and negative control (−) were CBB23 and JG30, respectively. (B) Genotype identification of the Xa4 gene using the co-dominant SSR marker RM224. The positive (+) and negative control (−) were IR64 and IR24, respectively. (C) Genotype identification of the Pi2 gene using the Indel marker S29742. The positive (+) and negative control (−) were C101A51 and Lijiangxintuanheigu (LTH), respectively. M represents the DNA marker.
Previous results showed that Xa23 confers dominant bacterial blight resistance throughout the growth period and plays an important role in disease resistance breeding of hybrid rice (Wang et al., 2015). In this study, the multi-resistant germplasm “V5400”, harboring homozygous Xa23 and Pi2 genes, as the donor of resistance genes was used for backcrossing with the recipient parent YJ1S, a high-quality pollen-free P/TGMS line with high combination ability (Figure 1B). In the BC2F6 generation in early cropping season of 2015, we picked out two lines, by16# and by22#, showing excellent comprehensive agronomic traits and good outcrossing (Figure 1B). Genotyping analysis of resistance genes indicated that the two P/TGMS lines, tentatively named Tai S and Wo S, respectively, carried homozygous Xa23 and Pi2 (Figures 3A, C). Tai S exhibited similar heading date as YJ1S, while Wo S exhibited heading date approximately ten days later. In the field conditions, Tai S and Wo S exhibited complete male sterility until late October or early November, respectively, but exhibited conversion to male fertility and self-propagation under serial irrigation with cold water at temperatures below 23°C. These results indicated that the novel P/TGMS lines Tai S and Wo S had the characteristics of low critical sterility-inducing temperature and stable male sterility. Finally, Tai S and Wo S were released in 2018, and MAS combined with phenotypic selection was performed throughout the breeding process.
The Novel Varieties and P/TGMS Lines Exhibited High Resistance to Rice Bacterial Blight
To test whether YXYZ2 and YTYZ show improved the bacterial blight resistance, the pathotype IV and IX Xoo strains were used for inoculation. The results showed that YJYZ and YNSM exhibited moderate resistance and resistance to the pathotype IV Xoo strain with lesion lengths 3.46 ± 1.07 cm and 1.27 ± 0.52 cm, respectively (Figures 2A, C), high susceptibility to the pathotype IX Xoo strain with lesion lengths more than 24 cm (Figures 2B, D). Notably, the bacterial blight resistance of YXYZ2 and YTYZ to pathotype IV and IX Xoo strains was significantly higher than that of YJYZ and YNSM, and these varieties exhibited a typical hypersensitive disease resistance response with lesion lengths less than 0.2 cm as XHZ (Figure 2). Similarly, we used the pathotype IV and IX Xoo strains for inoculation to evaluate the bacterial blight resistance of the two novel P/TGMS lines (Figure S2). The results showed that Tai S and Wo S were highly resistant to the pathotype IV and IX Xoo strains, with lesion lengths ranging from 0.10 to 0.28 cm (Figure S2). However, the other widely applied sterile lines for two- or three-line hybrid rice breeding in China were susceptible to the pathotype IV Xoo strains with lesion lengths ranging from 5.39 to 23.80 cm (Figures S2A, C) and exhibited high susceptibility to the pathotype IX Xoo strains with lesion lengths more than 23.5 cm (Figures S2B, D). Meanwhile, we investigated the status of Xa4 in those improved lines and their parental lines (Figure 3B). The results showed that Xa4 was present in all of the parental lines and improved lines except Wo S, which indicated that the improved lines, YXYZ2, YTYZ, and Tai S, pyramided Xa23 and Xa4 at least (Figures 3A, B). The above results indicated that the broad-spectrum resistance gene Xa23 can be effectively applied to rice disease resistance breeding. In addition, the two P/TGMS lines Tai S and Wo S, which carried the homozygous Pi2 gene (Figure 3C), were resistant to rice blast, as determined by field resistance identification in a natural blast nursery for many years (Table S2). These results suggested that MAS coupled with excellent gene pyramiding is helpful for improving target traits rapidly and accelerating the breeding process.
Evaluation of Hybrids Derived From the Improved P/TGMS Lines or Cultivars
The development of hybrid rice has greatly contributed to global food security and continues to provide a fundamental guarantee for food supply (Yuan, 2017). In the field conditions, Tai S and Wo S exhibited complete male sterility, and the period of male sterility for the two P/TGMS lines was more than 150 days, as determined by sowing in stages and performing fertility identification in consecutive years. Furthermore, Tai S and Wo S exhibited high fertility restoration efficiency for multiplication under cold-water serial irrigation. Thus, we bred several hybrid combinations derived from Tai S and Wo S, namely, Tai S/YueYaSiMiao (YYSM) and Wo S/YYSM, Tai S/YueGuiZhan No.2 (YGZ2) and Wo S/YGZ2; these lines were planted, and the yields were evaluated in later cropping season of 2018 and early cropping season of 2019, respectively. The results showed that the yields of the above hybrids were significantly higher than that of the control variety Guangbayou 2168 (GBY2168) or Shenliangyou 58 xiangyouzhan (SLY58XYZ), increasing by 4.8–20.1% (Figure S5B). These results indicated that the two novel P/TGMS lines Tai S and Wo S are suitable for practical breeding of two-line hybrid rice.
Subsequently, we used two elite restorer lines, namely, HeHuangSiMiao (HHSM) and YGZ2, for hybridization with a series of P/TGMS lines and evaluated the resistance response of the derived hybrid combinations to bacterial blight (Figure S3). HHSM and YGZ2 exhibited moderate resistance to the pathotype IV Xoo strains with lesion lengths less than 5 cm (Figures 2A, C) and high susceptibility to the pathotype IX Xoo strains with lesion lengths more than 24 cm (Figures 2B, D). Compared with the results for hybrid combinations derived from other P/TGMS lines exhibiting susceptibility or high susceptibility to bacterial blight, the hybrid combinations derived from Tai S and Wo S showed a high-resistance or resistance response to bacterial blight, with lesion lengths ranging from 0.35 to 2.69 cm (Figure S3). Remarkably, the novel cultivars YXYZ2 and YTYZ are not only being excellent conventional rice varieties with high grain yield but also being elite restorer lines (Figure S5). The yield of the hybrid combinations, Tai S/YXYZ2 and Fa S/YTYZ, reached more than 9.4 t/ha in the trial conducted in early cropping season of 2018, which was significantly higher than that of the control variety Tianyou 3618 (TY3618), with an increase of more than 15% (Figure S5B). Next, we also evaluated the bacterial blight resistance of the hybrid combinations derived from YXYZ2 and YTYZ (Figure 4 and Figure S4). The hybrid combinations derived from YXYZ2 and YTYZ exhibited enhanced bacterial blight resistance compared with the other hybrids derived from the same sterile lines (Figure 4 and Figure S4). Bacterial lesion lengths of these hybrids ranged from 0.07 to 0.41 cm with the exception of the combination of Taizao 1S with YXYZ2, which were more than 1.2 cm (Figure 4 and Figure S4). These results indicated that the dominant resistance gene Xa23 could improve the bacterial blight resistance of hybrid and inbred rice, and the resistance of hybrid rice would probably be affected by the genetic background in the application of Xa23. Meanwhile, the results showed that the hybrids derived from Taizao 1S, without harboring Xa4 (Figure 3B), were more susceptible to the pathotype IV strains than those from Quan 93-11A (Figures 4A–D). Similarly, the hybrids derived from Shen 08S were significantly susceptible to the pathotype IV strains compared to those from Fulong S2 or Li 1S (Figures S3E, F).
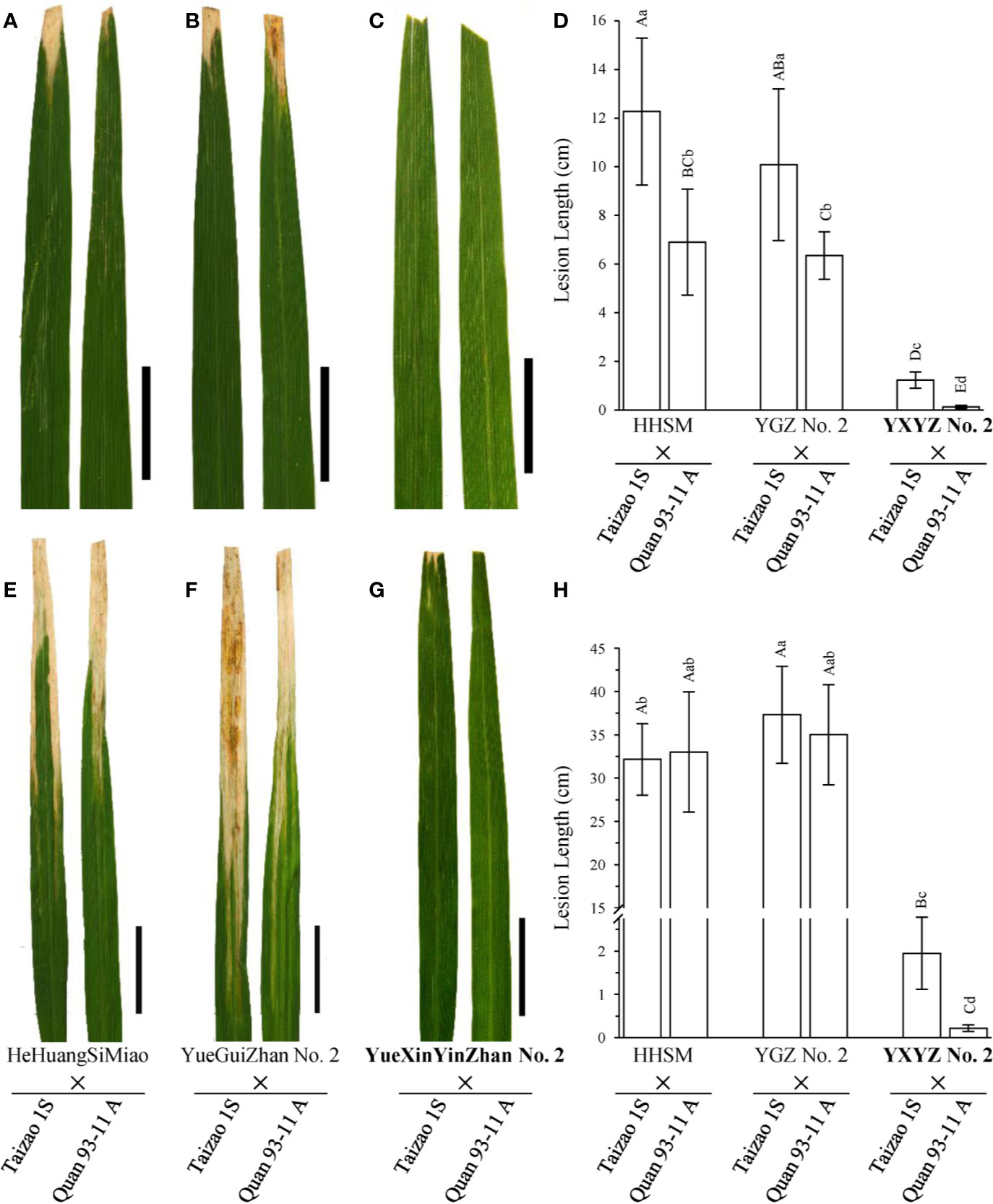
Figure 4 Comparison of the disease response of hybrid combinations of novel rice varieties. (A–C, E–G) Leaves of hybrids are presented to show the lesion patterns to the pathotype IV and IX Xoo strains, respectively. Pictures were taken on the 14th day after inoculation. Scale bars: 5 cm. (D, H) Statistical analysis of the lesion lengths of hybrids inoculated with the pathotype IV and IX Xoo strains, respectively. Lesion length was measured on the 21st day after inoculation. Data are given as the means with SDs (n = 10). The uppercase and lowercase letters above the bars represent significant difference levels P < 0.01 and P < 0.05, respectively.
Discussion
Rice plays a predominant role in the major strategies for demand-oriented national food security and achieving a guaranteed high and stable yield of rice is a fundamental goal (Zeng D. et al., 2017). However, for rice production and security, the major obstacle caused by multiple pathogens should be addressed (Ke et al., 2017). It is widely accepted that the utilization of broad-spectrum and durable resistance for rice improvement is the most cost-effective approach to preventing the loss of rice yield caused by various diseases (Khan et al., 2014; Li et al., 2017). Although an increasing number of bacterial blight resistance (R) genes have been identified and cloned in past decades, and main cultivars also showed resistant improvement to Xoo strains through the application of R genes (Vikal and Bhatia, 2017; Chukwu et al., 2019), with the continuous emergence of new strong pathogenic Xoo strains, the resistance of the main cultivars is at risk of loss.
In the present study, we genotyped and detected the distribution of a set of R genes, i.e. Xa4, Xa7, Xa21, Xa23, and Xa27, in seventy main cultivars from Guangdong Province using the corresponding specific markers. Among these detected genes, our results indicated that Xa4 was probably the most widely utilized R gene in rice breeding for BB resistance in Guangdong (Table S1). Meanwhile, it is worth noting that the maintainers, such as Guang 8B, Wufeng B, and Tianfeng B, corresponding to the widely applied CMS lines in rice hybrids harbored Xa4, which mediated a race-specific and durable resistance against Xoo at all stages of rice growth with dominant inheritance (Sun et al., 2003; Hu et al., 2017). These results indicated that Xa4 had also been widely utilized in rice hybrid breeding programs, which was consistent with the previous studies (Zhang et al., 2002; Sun et al., 2003; Hu et al., 2017). However, the resistance of Xa4 had weakened and even been overcome by the more virulent BB strains (Zhang et al., 2002; Chen et al., 2011). On the other hand, the strong virulent BB strains of pathotype V Xoo had replaced pathotype IV Xoo strains, and pathotype V had become the preponderant pathotype in South China; meanwhile, the new and the most virulent pathotype, pathotype IX, evolved quickly (Zeng et al., 2005; Chen et al., 2018). In the present study, our results showed that only two cultivars had moderate resistance to the strongly virulent pathotype IX Xoo strains (Table S1), which indicated that Xa4 has been unable to improve BB resistance in rice disease resistance breeding in Guangdong. Consequently, the exploration and utilization of the novel resistance resources play a pivotal role in rice resistance breeding for protection against the rapidly evolving Xoo races.
Bacterial blight races continue to evolve with artificial or natural selection and the utilization of BB resistance genes (Chukwu et al., 2019). As a result, the most virulent Xoo strains develop rapidly, and the preponderant Xoo pathotypes are also being frequently replaced (Zeng et al., 2005; Chen et al., 2018). Previous studies have indicated that Xa23, a transcription activator-like (TAL) effector dependent R gene, confers broad-spectrum resistance and provides strong resistance to all-natural Xoo strains tested so far at all growth stages of rice (Wang et al., 2015). Notably, Xa23 can trigger a strong hypersensitive response in crop plants (Vikal and Bhatia, 2017), and cultivars such as CBB23 and XHZ carrying the Xa23 gene exhibited high (grade 1) resistance to strains IV and V of Xoo from South China (Luo et al., 2013; Zeng L. et al., 2017). In the present study, our results showed that XHZ exhibited a typical hypersensitive disease resistance response to strain IV and also to strain IX of Xoo (Figure 2). Subsequently, we developed a series of highly disease-resistant intermediate materials and successfully bred two excellent rice varieties, namely, YXYZ2 and YTYZ, using XHZ as a resistant resource through MAS combined with phenotypic selection (Figure 1A). As expected, the two excellent varieties YXYZ2 and YTYZ exhibited typical hypersensitive disease resistance to the virulent Xoo pathotype IV or IX with high and stable grain production (Figure 2 and Figure S5A). In addition, the parental lines and derived hybrid combinations without harboring Xa23 resistance gene exhibited different responses to the pathotype IV Xoo strains and high susceptibility to the pathotype IX Xoo strains (Figures 2 and 4, Figures S3 and S4). These results indicated that the resistance conferred against pathotype IV Xoo strains was statistically significant, although this strain appears less aggressive.
Since the 1970s, the exploitation and utilization of heterosis in hybrid rice had led to a second leap in rice grain yield and to significant achievements (Qian et al., 2016; Yuan, 2017). However, breeding of disease-resistant and high-quality male sterile lines is a strategic problem that has long hindered the development and utilization of hybrid rice. Currently, the development of rice functional genomics is greatly accelerating the rational design and molecular breeding to enable new breakthroughs in rice production (Qian et al., 2016; Zeng D. et al., 2017). Nevertheless, improving and enhancing disease resistance is important for guaranteeing a high and stable yield of rice. Previous studies indicated that the rice genetic background would influence the R-gene-mediated resistance to Xoo in rice and, thus, hinder the utilization of a single R gene for disease resistance to BB in hybrid rice due to the heterozygous genetic background or incomplete dominance (Luo et al., 2012; Zeng L. et al., 2017). For recessive resistance genes, non-transgenic resistant varieties can be obtained quickly by gene editing, but these genes cannot be directly utilized in hybrid rice breeding (Kim et al., 2019; Li C. et al., 2020). Notably, Xa23 confers complete dominant bacterial blight resistance independently of genetic background during the whole growth period; thus, this gene has been widely adopted in rice breeding programs, and several Xa23-containing hybrid rice varieties have been released (Wang et al., 2014; Wang et al., 2015). On the other hand, the application of P/TGMS lines has effectively broadened the genetic diversity in sterile line breeding and is a breakthrough in the breeding of disease-resistant and high-quality male sterile lines (Ma and Yuan, 2015). Here, we bred two novel P/TGMS lines, namely, Tai S and Wo S, harboring homozygous Xa23 through backcrossing with MAS breeding (Figures 1B and 3A), and the derived hybrid combinations exhibited high resistance against the pathotype IV and IX Xoo strains as the two female parents (Figures S2 and S3). In addition, the hybrid combinations using YXYZ2 and YTYZ as restorer lines also exhibited high resistance to bacterial blight with high yield (Figure 4, Figures S4 and S5). These results indicated that Xa23 has wide application value due to its high and broad-spectrum resistance with complete dominant inheritance in inbred and hybrid rice resistance breeding. Remarkably, the Xa23 gene mediated resistance to Xoo would be probably influenced by the genetic background in hybrids (Figure 4 and Figure S3), indicating that Xa23 coupled with other resistance genes needs to be taken into consideration in future applications.
As mentioned above, we successfully bred two excellent cultivars and two novel P/TGMS lines harboring homozygous Xa23 and exhibiting high resistance against BB through conventional breeding aided by MAS. However, the Xoo or pathotypes keep evolving and overcoming the barrier presented by the R genes. Meanwhile, continuous planting of cultivars with a single major R gene imposes a strong selection on pathogen evolution and variation and results in resistance breakdown (Kim, 2018; Chukwu et al., 2019). Although the Xa23 locus confers high resistance to virtually all the Xoo races tested, it is possible that the Xa23-containing rice varieties would lose BB resistance if widely distributed and subjected to long-term planting and application (Wang et al., 2014; Wang et al., 2015). In other words, the vertical resistance controlled by a single resistance gene would be lost due to the arms race between plants and pathogens, as described by the Red Queen’s hypothesis. Fortunately, an increasing number of BB R genes that confer resistance against different Xoo strains via multiple mechanisms that have been identified and cloned (Chukwu et al., 2019). Recently, Xu et al. (2019) and Oliva et al. (2019) created new germplasms with robust and broad-spectrum resistance to BB using CRISPR/Cas9-mediated genome editing to introduce mutations in three effector-triggered susceptibility-associated SWEET genes, namely, Xa13/OsSWEET11, Xa25/OsSWEET13, and Xa41/OsSWEET14, successively. Therefore, pyramiding of multiple R genes is one of the more conducive and effective approaches to enhancing the horizontal resistance of rice varieties and achieving broad-spectrum and durable resistance (Luo et al., 2012; Suh et al., 2013; Pradhan et al., 2015; Luo et al., 2016; Oliva et al., 2019; Xu et al., 2019). In the present study, the improved lines, YXYZ2, YTYZ, and Tai S, pyramided Xa23 and Xa4 at least (Figures 3A, B), which could have been better than the Xa23 alone for disease resistance and durability, although Xa4 alone was unable to meet the current requirements for rice disease resistance breeding. Notably, pyramiding of multiple R genes in rice should not be accompanied by yield penalties or drawbacks for other traits (Antony et al., 2010; Li et al., 2012; Xu et al., 2019). In general, the exploitation of the new resistance resources and the molecular identification of novel genes for disease resistance are the key components for the rational design of rice resistance breeding.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
SW and XH designed the experiments and wrote the manuscript. SW, JX, and XH edited the manuscript. SW, WL, ZL, and DL performed the experiments. SW and XW analyzed the data. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Key R&D Program of China (2017YFD0100102), The National Natural Science Foundation of China (31471175), Science and Technology Programme of Guangdong Province, China (2015A020209043, 2017A070702006), Modern Agricultural Industry Technology System of Guangdong Province, China (2019KJ105), Joint Research on High Quality Rice Varieties (Yuecainong [2019]73), and the Special Fund for Science and Technology Innovation Strategy (Construction of High-level Academy of Agricultural Sciences) (Foundation of President of Guangdong Academy of Agricultural Sciences in China, 201924).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.555228/full#supplementary-material
References
Antony, G., Zhou, J., Huang, S., Li, T., Liu, B., White, F., et al. (2010). Rice xa13 Recessive Resistance to Bacterial Blight Is Defeated by Induction of the Disease Susceptibility Gene Os-11N3. Plant Cell 22, 3864.
Chen, S., Lin, X., Xu, C., Zhang, Q. (2000). Improvement of bacterial blight resistance of ‘Minghui 63’, an elite restorer line of hybrid rice, by molecular marker-assisted selection. Crop Sci. 40, 239–244.
Chen, H., Chen, Z., Ni, S., Zuo, S., Pan, X., Zhu, X. (2008). Pyramiding three genes with resistance to blast by marker assisted selection to improve rice blast resistance of Jin 23B. Chin. J. Rice Sci. 22, 23–27.
Chen, S., Liu, X., Zeng, L., Ouyang, D., Yang, J., Zhu, X. (2011). Genetic analysis and molecular mapping of a novel recessive gene xa34 (t) for resistance against Xanthomonas oryzae pv. oryzae. Theor. Appl. Genet. 122, 1331–1338.
Chen, S., Wang, C., Su, J., Feng, A., Zhu, X., Zeng, L. (2018). Differential Detection and Analysis of Pathotypes and Differentiation against Xanthomonas oryzae pv. oryzae in Southern China. Agric. Biotechnol. 7, 92–96.
Chu, Z., Fu, B., Yang, H., Xu, C., Li, Z., Sanchez, A., et al. (2006). Targeting xa13, a recessive gene for bacterial blight resistance in rice. Theor. Appl. Genet. 112, 455–461.
Chukwu, S. C., Rafii, M. Y., Ramlee, S. I., Ismail, S. I., Hasan, M. M., Oladosu, Y. A., et al. (2019). Bacterial leaf blight resistance in rice: a review of conventional breeding to molecular approach. Mol. Biol. Rep. 46, 1519–1532.
Chunwongse, J., Martin, G., Tanksley, S. (1993). Pre-germination genotypic screening using PCR amplification of half-seeds. Theor. Appl. Genet. 86, 694–698.
Deng, Q., Wang, S., Zheng, A., Zhang, H., Li, P. (2006). Breeding Rice Restorer Lines with High Resistance to Bacterial Blight by Using Molecular Marker-Assisted Selection. Rice Sci. 13, 22–28.
Deng, Y., Zhu, X., Shen, Y., He, Z. (2006). Genetic characterization and fine mapping of the blast resistance locus Pigm(t) tightly linked to Pi2 and Pi9 in a broad-spectrum resistant Chinese variety. Theor. Appl. Genet. 113, 705–713.
Gu, K., Yang, B., Tian, D., Wu, L., Wang, D., Sreekala, C., et al. (2005). R gene expression induced by a type-III effector triggers disease resistance in rice. Nature 435, 1122–1125.
Hu, K., Cao, J., Zhang, J., Xia, F., Ke, Y., Zhang, H., et al. (2017). Improvement of multiple agronomic traits by a disease resistance gene via cell wall reinforcement. Nat. Plants 3, 17009.
Hutin, M., Sabot, F., Ghesquière, A., Koebnik, R., Boris, S. (2015). A knowledge-based molecular screen uncovers a broad spectrum OsSWEET14 resistance allele to bacterial blight from wild rice. Plant J. 84, 694–703.
Iyer, A. S., McCouch, S. R. (2004). The Rice Bacterial Blight Resistance Gene xa5 Encodes a Novel Form of Disease Resistance. Mol. Plant-Microbe Interact. 17, 1348–1354.
Jiang, N., Yan, J., Liang, Y., Shi, Y., He, Z., Wu, Y., et al. (2020). Resistance Genes and their Interactions with Bacterial Blight/Leaf Streak Pathogens (Xanthomonas oryzae) in Rice (Oryza sativa L.)—an Updated Review. Rice 13, 3.
Ke, Y., Deng, H., Wang, S. (2017). Advances in understanding broad-spectrum resistance to pathogens in rice. Plant J. 90, 738–748.
Khan, M. A., Naeem, M., Iqbal, M. (2014). Breeding approaches for bacterial leaf blight resistance in rice (Oryza sativa L.), current status and future directions. Eur. J. Plant Pathol. 139, 27–37.
Kim, Y., Moon, H., Park, C.-J. (2019). CRISPR/Cas9-targeted mutagenesis of Os8N3 in rice to confer resistance to Xanthomonas oryzae pv. oryzae. Rice 12, 1–13.
Kim, S. (2018). Identification of novel recessive gene xa44(t) conferring resistance to bacterial blight races in rice by QTL linkage analysis using an SNP chip. Theor. Appl. Genet. 131, 2733–2743.
Li, T., Liu, B., Spalding, M. H., Weeks, D. P., Yang, B. (2012). High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 30, 390.
Li, W., Zhu, Z., Chern, M., Yin, J., Yang, C., Ran, L., et al. (2017). A natural allele of a transcription factor in rice confers broad-spectrum blast resistance. Cell 170, 114–126. e115.
Li, C., Li, W., Zhou, Z., Chen, H., Xie, C., Lin, Y. (2020). A new rice breeding method: CRISPR/Cas9 system editing of the Xa13 promoter to cultivate transgene-free bacterial blight-resistant rice. Plant Biotechnol. J. 18, 313–315.
Li, W., Deng, Y., Ning, Y., He, Z., Wang, G.-L. (2020). Exploiting Broad-Spectrum Disease Resistance in Crops: From Molecular Dissection to Breeding. Annu. Rev. Plant Biol. 71, 25.1–25.29.
Liu, K., Wang, J., Li, H., Xu, C., Liu, A., Li, X., et al. (1997). A genome-wide analysis of wide compatibility in rice and the precise location of the S5 locus in the molecular map[J]. Theor. Appl. Genet. 95, 809–814.
Liu, Q., Yuan, M., Zhou, Y. A. N., Li, X., Xiao, J., Wang, S. (2011). A paralog of the MtN3/saliva family recessively confers race-specific resistance to Xanthomonas oryzae in rice. Plant Cell Environ. 34, 1958–1969.
Liu, F., Liang, Q., Chen, W., Chen, Y., Liang, J., Chen, X. (2016). Analysis of Molecular Marker on Rice Germplasms of Bacterial Blight Resistant Gene Xa7 and Xa23. Mol. Plant Breed. 14, 935–940.
Lu, Z., He, X., Liu, W., Wang, L., Fu, W., Lu, D., et al. (2019). Distribution of a recessive blast resistant gene pi55 in the major rice varieties of Guangdong and germplasm creation. Mol. Plant Breed. 17, 6369-6377.
Luo, Y., Sangha, J. S., Wang, S., Li, Z., Yang, J., Yin, Z. (2012). Marker-assisted breeding of Xa4, Xa21 and Xa27 in the restorer lines of hybrid rice for broad-spectrum and enhanced disease resistance to bacterial blight. Mol. Breed. 30, 1601–1610.
Luo, S., Zhang, F., Chen, X., Jin, M., Zhou, Y., Li, Z. (2013). Evaluation of 26 Rice Cultivas (Lines) Resistance to Bacterial Blight at the Tillering and Booting Stages. J. Plant Genet. Resour. 14. 390–394.
Luo, Y., Ma, T., Zhang, A., Ong, K. H., Li, Z., Yang, J., et al. (2016). Marker-assisted breeding of the rice restorer line Wanhui 6725 for disease resistance, submergence tolerance and aromatic fragrance. Rice 9, 66.
Ma, G., Yuan, L. (2015). Hybrid rice achievements, development and prospect in China. J. Integr. Agric. 14, 197–205.
Neelam, K., Mahajan, R., Gupta, V., Bhatia, D., Gill, B. K., Komal, R., et al. (2020). High-resolution genetic mapping of a novel bacterial blight resistance gene xa-45(t) identified from Oryza glaberrima and transferred to Oryza sativa. Theor. Appl. Genet. 133, 689–705.
Oliva, R., Ji, C., Atienza-Grande, G., Huguet-Tapia, J. C., Perez-Quintero, A., Li, T., et al. (2019). Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat. Biotechnol. 37, 1344–1350.
Pai, H., Wang, C., Zhao, K., Zhang, Q., Fan, Y., Zhou, S., et al. (2003). Molecular Mapping by PCR-based Markers and Marker-assisted Selection of Xa 23, a Bacterial Blight Resistance Gene in Rice. Acta Agronom. Sin. 29, 501–507.
Porter, B., Chittoor, J., Yano, M., Sasaki, T., White, F. (2003). Development and mapping of markers linked to the rice bacterial blight resistance gene Xa7. Crop Sci. 43, 1484–1492.
Pradhan, S. K., Nayak, D. K., Mohanty, S., Behera, L., Barik, S. R., Pandit, E., et al. (2015). Pyramiding of three bacterial blight resistance genes for broad-spectrum resistance in deepwater rice variety, Jalmagna. Rice 8, 19.
Qian, Q., Guo, L., Smith, S. M., Li, J. (2016). Breeding high-yield superior quality hybrid super rice by rational design. Natl. Sci. Rev. 3, 283–294.
Song, W., Wang, G., Chen, L., Kim, H., Pi, L., Holsten, T., et al. (1995). A Receptor Kinase-Like Protein Encoded by the Rice Disease Resistance Gene, Xa21. Science 270, 1804.
Suh, J., Jeung, J., Noh, T., Cho, Y., Park, S., Park, H., et al. (2013). Development of breeding lines with three pyramided resistance genes that confer broad-spectrum bacterial blight resistance and their molecular analysis in rice. Rice 6, 5.
Sun, X., Yang, Z., Wang, S., Zhang, Q. (2003). Identification of a 47-kb DNA fragment containing Xa4, a locus for bacterial blight resistance in rice. Theor. Appl. Genet. 106, 683–687.
Sun, X., Cao, Y., Yang, Z., Xu, C., Li, X., Wang, S., et al. (2004). Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase-like protein. Plant J. 37, 517–527.
Tian, D., Wang, J., Zeng, X., Gu, K., Qiu, C., Yang, X., et al. (2014). The Rice TAL Effector–Dependent Resistance Protein XA10 Triggers Cell Death and Calcium Depletion in the Endoplasmic Reticulum. Plant Cell 26, 497.
Tilman, D., Fargione, J., Wolff, B., D’Antonio, C., Dobson, A., Howarth, R., et al. (2001). Forecasting agriculturally driven global environmental change. Science 292, 281–284.
Vikal, Y., Bhatia, D. (2017). “Genetics and genomics of bacterial blight resistance in rice” in Adv. Int. Rice Res. Ed. Li, J. (London: Intech publishers). 175–213.
Wang, C., Qi, H., Pan, H., Li, J., Fan, Y., Zhang, Q., et al. (2005). EST-Markers Flanking the Rice Bacterial Blight Resistance Gene Xa23 and Their Application in Marker-Assisted Selection. Sci. Agricult. Sin. 38, 1996–2001.
Wang, C., Fan, Y., Zheng, C., Qin, T., Zhang, X., Zhao, K. (2014). High-resolution genetic mapping of rice bacterial blight resistance gene Xa23. Mol. Genet. Genomics 289, 745–753.
Wang, C., Zhang, X., Fan, Y., Gao, Y., Zhu, Q., Zheng, C., et al. (2015). XA23 Is an Executor R Protein and Confers Broad-Spectrum Disease Resistance in Rice. Mol. Plant 8, 290–302.
Wang, J., Zhou, L., Shi, H., Chern, M., Yu, H., Yi, H., et al. (2018). A single transcription factor promotes both yield and immunity in rice. Science 361, 1026–1028.
Wang, S., Ma, B., Gao, Q., Jiang, G., Zhou, L., Tu, B., et al. (2018). Dissecting the genetic basis of heavy panicle hybrid rice uncovered Gn1a and GS3 as key genes. Theor. Appl. Genet. 131, 1391–1403.
Xu, Z., Xu, X., Gong, Q., Li, Z., Li, Y., Wang, S., et al. (2019). Engineering broad-spectrum bacterial blight resistance by simultaneously disrupting variable TALE-binding elements of multiple susceptibility genes in rice. Mol. Plant 12, 1434–1446.
Yoshimura, S., Yamanouchi, U., Katayose, Y., Toki, S., Wang, Z., Kono, I., et al. (1998). Expression of Xa1, a bacterial blight-resistance gene in rice, is induced by bacteriall inoculation. Proc. Natl. Acad. Sci. 95, 1663.
Zeng, L., Zhu, X., Yang, J., Wu, S., Chen, Z., Chen, S. (2005). A new pathotype of Xanthomonas oryzae pv. oryzae was found and tested for pathogenicity in Guangdong. Guangdong Agricult. Sci. 2, 58–59.
Zeng, D., Tian, Z., Rao, Y., Dong, G., Yang, Y., Huang, L., et al. (2017). Rational design of high-yield and superior-quality rice. Nat. Plants 3, 1–5.
Zeng, L., Wang, C., Feng, A., Shen, C., Su, J., Wang, W., et al. (2017). Resistance Analysis on Major Resistant Genes of Rice Bacterial Blight in Different Donors and Genetic Background. Agric. Biotechnol. 6, 11–65.
Zhang, Q., Wang, C., Zhao, K., Yang, W., Qiao, F., Zhou, Y., et al. (2002). Development of near-isogenic line CBB23 with a new resistance gene to bacterial blight in rice and its application. Chin. J. Rice Sci. 16, 206–210.
Zhang, Q. (2009). Genetics and Improvement of Bacterial Blight Resistance of Hybrid Rice in China. Rice Sci. 16, 83–92.
Keywords: rice, bacterial blight, distribution, Xa23, marker-assisted selection
Citation: Wang S, Liu W, Lu D, Lu Z, Wang X, Xue J and He X (2020) Distribution of Bacterial Blight Resistance Genes in the Main Cultivars and Application of Xa23 in Rice Breeding. Front. Plant Sci. 11:555228. doi: 10.3389/fpls.2020.555228
Received: 24 April 2020; Accepted: 18 August 2020;
Published: 31 August 2020.
Edited by:
Luigi Cattivelli, Council for Agricultural and Economics Research, ItalyReviewed by:
Sheshu Madhav Maganti, Indian Institute of Rice Research (ICAR), IndiaZhongchao Yin, Temasek Life Sciences Laboratory, Singapore
Andrea Volante, Council for Agricultural and Economics Research (CREA), Italy
Copyright © 2020 Wang, Liu, Lu, Lu, Wang, Xue and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiuying He, aGV4aXV5aW5nQGdkYWFzLmNu
†These authors have contributed equally to this work
 Shiguang Wang
Shiguang Wang Wei Liu1,2†
Wei Liu1,2† Jiao Xue
Jiao Xue