- Department of Plant Science and Technology, Chung-Ang University, Anseong, South Korea
Acidovorax citrulli (Ac) is a plant pathogenic bacterium that causes bacterial fruit blotch (BFB) in cucurbit crops. Despite its importance in the cucurbit industry, resistant cultivars/lines against BFB have not yet been identified. Therefore, there is a need to characterize the virulence factors/mechanisms in Ac to control the disease. Chorismate mutase, a key enzyme in the shikimate pathway, produces aromatic amino acids. Here, we report the functions of putative bifunctional chorismate mutase/prephenate dehydratase in Ac (CmpAc) determined by proteomic analysis and phenotypic assays. Ac strain lacking CmpAc, AcΔcmpAc(EV), were significantly less virulent on watermelon in the germinated-seed inoculation and leaf infiltration assays. Sequence analysis revealed that CmpAc possesses two distinct domains: chorismate mutase and prephenate dehydratase, indicating that CmpAc is a bifunctional protein. Auxotrophic assays demonstrated that CmpAc is required for the biosynthesis of phenylalanine, but not tyrosine. The comparative proteomic analysis revealed that CmpAc is mostly involved in cell wall/membrane/envelop biogenesis. Furthermore, AcΔcmpAc(EV) showed reduced twitching halo production and enhanced biofilm formation. In addition, AcΔcmpAc(EV) was less tolerant to osmotic stress but more tolerant to antibiotics (polymyxin B). Thus, our study provides new insights into the functions of a putative bifunctional protein related to virulence in Ac.
Introduction
Acidovorax citrulli (Ac), formerly known as A. avenae subsp. citrulli, is a gram-negative, rod-shaped, and seed-borne bacterium. Ac is the causal agent of bacterial fruit blotch (BFB) in watermelon, melon, and other cucurbit crops (Schaad et al., 1978; Willems et al., 1992). Watermelon seedlings infected by Ac show water-soaked lesions on cotyledons as initial symptoms, followed by the collapse of the emerging plant and subsequent wilting and dying of the seedlings. In case of the fruit, the infected watermelon shows dark, olive-like lesions. In the later stage, the water-soaked lesion spreads, leading to the ultimate decay and collapse of the fruit flesh by secondary infection or saprophytes (Latin and Hopkins, 1995). Despite the importance of this disease in watermelon production, lines/cultivars of watermelon resistant to Ac have not been identified. Thus, there is a limit to the efficient management of BFB, and Ac remains a major threat to the worldwide cucurbit crop industry (Burdman and Walcott, 2012). Although Ac strains isolated from rockmelon showed similar virulence on watermelon (O’Brien and Martin, 1999), Ac is generally known to be divided into two groups: group I strains are severely virulent on melon, and group II strains are mainly virulent on watermelon and moderately virulent on other Cucurbitaceae (Walcott et al., 2000).
Several virulence factors/mechanisms in Ac have been characterized. In the previous studies, Ac mutants showing impaired twitching motility and biofilm formation were less virulent on melon (Bahar et al., 2009). The type IV pili of Ac are also known to be involved in surface adhesion and virulence (Bahar et al., 2010). Deletion of aacR and aacI genes involved in quorum sensing in Ac showed reduced virulence but enhanced biofilm formation (Wang et al., 2016). In addition, Bahar et al. reported that a polar flagellum is involved in the full virulence in melon (Bahar et al., 2011). Similar to other gram-negative plant pathogenic bacteria, regulatory proteins for type III secretion system (T3SS), HrpG and HrpX, are also indispensable for the pathogenicity of Ac (Zhang et al., 2018). Recently, diverse type III effectors have been characterized in Ac (Jiwenez-Guerrero et al., 2020). In addition, functions of type II and VI secretion systems, quorum sensing mechanisms, and regulators related with ferric uptake have been also characterized regarding the virulence of Ac (Burdman and Walcott, 2012; Johnson and Walcott, 2013; Tian et al., 2015; Liu et al., 2019). However, other virulence factors/mechanisms related with biochemical pathways of Ac are still poorly understood.
Chorismate mutase is a critical enzyme in the shikimate pathway responsible for the synthesis of aromatic amino acids. This enzyme catalyzes chorismate to produce prephenate, which is an intermediate molecule for the biosynthesis of phenylalanine and tyrosine (Kast et al., 2000). Notably, many bacterial chorismate mutase are bifunctional proteins possessing two catalytic capacities for chorismate mutase and prephenate dehydratase (Duggleby et al., 1978; Ahmad et al., 1988; Huccetogullari et al., 2019). These bifunctional proteins are indispensable for phenylalanine production but not for tyrosine production because prephenate dehydratase is involved in phenylalanine but not tyrosine biosynthesis. The chorismate mutase of Mycobacterium tuberculosis plays a crucial role in the pathogenesis of tuberculosis (Khanapur et al., 2017). In addition, genes related to the shikimate pathway are involved in the production of toxoflavin, which is known to be a major virulence factor and promote tolerance to UV in Burkholderia glumae, the causative agent of bacterial panicle blight in rice (Karki and Ham, 2014). The secretome analysis from Xanthomonas citri. subsp. citri, a citrus canker pathogen, revealed chorismate mutase as a potential virulence factor (Ferreira et al., 2016). However, functions of chorismate mutase associated with virulence or other mechanisms have not been reported in Ac.
In this study, we report the functions of putative bifunctional chorismate mutase/prephenate dehydratase in the Ac strain KACC17005 whose genomic information was previously annotated (Park et al., 2017). Screening of the Ac Tn5-insertional library revealed a mutant that had lost virulence and contained a chorismate mutase gene disrupted by Tn5; the protein domain information indicates that it is a putative bifunctional chorismate mutase/prephenate dehydratase in Ac (CmpAc). To postulate mechanisms related to CmpAc, a comparative proteomic analysis combined with clusters of orthologous groups (COGs) were employed. Based on the proteomics, phenotypic changes in AcΔcmpAc were investigated.
Materials and Methods
Bacterial Strains and Growth Conditions
The Ac strain KACC17005 belonging to group II (Song et al., 2020) was used in this study as a wild-type strain. Ac strains were grown in TSB (Tryptic Soy Broth Soybean-Casein Digested, 30 g/L) or M9 (47.7 mM Na2HPO4·7H2O, 22 mM KH2PO4, 8.6 mM NaCl, 18.7 mM NH4Cl, 2 mM MgSO4, 0.1 mM CaCl2, 20 ml of 20% glucose in 1 L) at 28°C. The Escherichia coli strain DH5α was used for cloning genes and generating constructs. To identify genes disrupted by Tn5, the E. coli strain EC100D was used (Lucigen, Madison, WI, USA). E. coli strains were cultivated at 37°C in LB (Luria Bertani; 1% tryptone, 0.5% yeast extract, and 1% NaCl) with appropriate antibiotics. For selection, antibiotics were added to the media at the following final concentrations: kanamycin, 50 µg/ml; rifampicin, 50 µg/ml; gentamicin, 10 µg/ml; and ampicillin, 100 µg/ml.
Selection of AcΔcmpAc and Generation of the Complemented Strain
Bacterial strains and plasmids used in this study are listed in Supplementary Table 1. The knock out mutant of CmpAc was identified by screening the Tn5 insertional library generated with EZ-Tn5™ <R6Kγori/KAN-2> Insertion Kit (Lucigen, Middleton, WI, USA) as the manufacture’s protocol. Briefly, to construct the Tn5 insertional library, EZ-Tn5 transposome was introduced into the wild-type strain by electroporation using Bio Rad Micropulser™ (Bio-Rad, Hercules, CA, USA) under 2.5 kV. Tn5 insertional mutants were selected on TSA containing kanamycin and rifampicin as a selective marker. Approximately, 4,000 mutants were generated. Each colony was transferred into TSB containing kanamycin and rifampicin in 96-well cell plates, incubated for 2 days at 28°C and kept at −80°C. A germinated-seed inoculation assay was used for selecting virulence-deficient mutants. After extracting genomic DNA from the selected mutant, DNA fragments interrupted by Tn-5 were cloned using the manufacturer’s protocol (Lucigen). The interrupted regions were verified by Sanger sequencing using two primers (KAN-2F, 5′-acctacaacaaagctctcatcaacc-3′, and R6KAN-2R, 5′-ctaccctgtggaacacctactct-3′). The identified mutant was named as AcΔcmpAc. To generate the construct for the complemented strain, the open reading frame of cmpAc was amplified using cmpAc-specific primers: 5′-aagcttatgtcccaatcccc-3′, and 5′-ggatcctcagtggtggtggtggtggtgcggcgccaccg-3′. The amplicon was cloned into pGem-T easy vector (Promega, Madison, WI, USA) to produce pGem-CmpAc plasmid, which was confirmed by Sanger sequencing. The confirmed plasmid was digested with HindIII and BamHI, and the digested CmpAc construct was cloned again into pBBR1-MCS5 (Kovach et al., 1995), a broad host range vector containing LacZ promoter, creating pBBR1-CmpAc plasmid, which was then introduced into AcΔcmpAc cell by electroporation using Bio Rad Micropulser™ (Bio-Rad) under 2.5 kV. The transformant was selected on tryptic soy agar (TSA) with kanamycin and gentamycin and confirmed by polymerase chain reaction (PCR) using the cmpAc-specific primers used for cloning cmpAc amplicon, generating the complemented strain, AcΔcmpAc(CmpAc). To minimize the side effect caused by the pBBR1-MCS5 vector, the empty vector was introduced into Ac and AcΔcmpAc to create Ac(EV) and AcΔcmpAc(EV), respectively. The transformants were selected on TSA with gentamycin/rifampicin or gentamycin/rifampicin/kanamycin and confirmed by PCR using pBBR1-MCS5 specific primers: 5′-cagggttttcccagtcacga-3′ and 5′-atgcttccggctcgtatgtt-3′.
Pathogenicity Test
Citrullus lanatus var. vulgaris line SBA provided by Partner Seed Company (Gimje, Korea) was used for pathogenicity assay, which was carried out by two inoculation methods (the germinated-seed inoculation and the leaf infiltration methods). For germinated-seed inoculation, a previously reported protocol was used with slight modifications (Bahar et al., 2009). To improve the penetration rate, the seeds were germinated on moisturized-filter papers for 2 days. Ac strains were grown on TSA at 28°C, suspended in 10 mM MgCl2 to an OD600nm of 0.3, which corresponds to approximately 108 colony forming unit (CFU)/ml, and diluted (10−2) with the same buffer to obtain 106 CFU/ml. Ten germinated seeds were placed into 50-ml conical tubes containing 20 ml of bacterial suspension and incubated with moderate shaking (100 rpm) for 1 h at 22°C. The inoculated seeds were then sown in a 50-pot tray containing sterilized soil and grown in a growth chamber for 7 days. The disease severity of the inoculated watermelon was evaluated for 7 days. The disease severity was rated on a scale of 0–2 as follows: 0, no symptoms; 1, water-soaked region (Spot); 2, wilt. The disease index was then calculated using the following equation: Disease index = [Normal (numbers of plants)×0 + Spot (numbers of plants)×1 + Wilt (numbers of plants)×2]/Total (numbers of plants). For this assay, 10 biological replicates were examined, and seven independent experiments were carried out. For infiltration, the germinated seeds were grown in a growth chamber for 2 weeks until four true leaves stage. Ac strains were grown in the TSA plate and suspended in 10 mM MgCl2 to an OD600nm of 0.3 and diluted (10-3) with 10 mM MgCl2 to obtain 105 CFU/ml. The bacterial suspensions were infiltrated into first and second true leaves using 3-ml needleless syringes. For counting bacterial cell numbers, the infiltrated leaves were punched by cork-borers (0.4 cm in diameter), and two leaf disks were ground in 200 µL sterilized water using tissue grinders. The extracted Ac cells were serially diluted and dotted onto the antibiotics-containing TSA and incubated for 2 days at 28°C. Three biological replicates were employed, and seven independent experiments were carried out in this assay.
Auxotrophic Assay
The bacterial growth of Ac(EV), AcΔcmpAc(EV), and AcΔcmpAc(CmpAc) was evaluated in four different conditions (TSB, M9, M9 with one mM tyrosine, and M9 with one mM phenylalanine). The Ac strains were grown on TSA with appropriate antibiotics. The cultured bacterial cells were harvested and washed twice using sterilized water. After washing, the bacterial cells were adjusted to an OD600nm of 0.3, diluted (10−3) with TSB to obtain 105 CFU/ml, and grown in the shaking incubator at 28°C. Bacterial growth was measured using a spectrophotometer at OD600nm for 4 days at 12 h intervals. In minimal media, the bacterial suspension was adjusted to an OD600nm of 0.05 (approximately, 1.7×107 CFU/ml) and observed for 7 days at 24 h intervals. For this assay, three biological replicates for each strain were used, and six independent experiments were carried out.
For proteomic analysis, Ac and AcΔcmpAc were used, and a label-free shotgun comparative proteomics approach was carried out using a previously reported protocol (Park H. J. et al., 2020). Briefly, Ac strains were grown in TSB and harvested at an OD600 of 0.5 (approximately, 1.7×108 CFU/ml) by centrifugation. The harvested cells were disrupted by an Ultrasonic Processor (Colo Parmer, Vernon Hills, IL, USA). Total proteins were concentrated using trichloroacetic acid precipitation and digested by trypsin. After cleaning the tryptic-digested proteins using the Sep-Pak Vac 1cc tC18 cartridge (Waters, Milford, MA, USA), peptide concentration was determined using a BCA assay kit (Thermo Fisher Scientific, Waltham, MA, USA). Then, 1 μg of each sample (two strains with three biological replicates) was injected into a split-free nano LC (EASY-nLC II; Thermo Fisher Scientific, Bremen, Germany) combined with an LTQ Velos Pro instrument (Thermo Fisher Scientific) by the nanospray ion mode. Tryptic-digested peptides were loaded onto 7.5 cm of MAGIC C18AQ 200A (5 µm) (Michrom BioResources, Auburn, CA, USA) with 300 nl/min flow rate for 420 min under water/acetonitrile gradient using buffer A (water with 0.1% formic acid) and buffer B (100% acetonitrile with 0.1% formic acid). For the mass spectra, six data-dependent MS/MS scans with m/z in the 300–2,000 range of mass were used. Dynamic exclusion was allowed under one repeat, 0.5 min duration, and 3 min exclusion. The ion charge state selection was permitted for 2+ and 3+. From the full mass scan, six of the most intense ions were chosen.
The procedure for identifying and quantifying proteins and peptides was followed as reported previously except for the database (Park H. J. et al., 2020). The MS/MS spectra were identified by Thermo proteome discoverer (ver. 1.3.0.399) with a SEQUEST search algorithm. Genome information of Ac strain KACC17005 (Accession No. CP023687) was used from the National Center for Biotechnology Information. To improve the credibility, the target-decoy strategy was used (Elias and Gygi, 2007). Two missed cleavages were permitted, and the false discovery rate was 0.01. One hundred ppm precursor was set for mass accuracy, and the probability scores were at least 20. The identified proteins by the Thermo Proteome Discoverer were imported again into Scaffold 4 (Proteome Software, Portland, OR, USA), which was used for comparison. Peptide spectrum matches (PSMs) was used for comparative analyses (Choi et al., 2008). PSMs from the proteins were normalized against the total proteins in a sample. Three biological replicates were used, and the average of PSMs from the replicates was calculated for each protein and compared for the identification of proteins that are differentially abundant (over 2-fold) in Ac vs. AcΔcmpAc. The statistical analysis was performed using student’s t-test (P < 0.05). COG analysis was used for the categorization of the identified proteins (Tatusov et al., 2000).
Biofilm Formation
For biofilm formation assay, a previously established protocol was used with slight modification (Park et al., 2020). The Ac strains were grown for 24 h in TSA, washed, suspended in TSB to an OD600nm of 0.3, and diluted (10−3) with TSB (approximately, 105 CFU/ml). The bacterial suspension was incubated in 96-well polyvinyl chloride (PVC) plate at 28°C for 2 days. After incubation, the bacterial supernatant was removed carefully and washed using sterilized water. After washing, the remaining bacterial cells were stained with 0.1% crystal violet for 30 min, washed twice with sterilized water, suspended in 95% ethanol for 20 min, and measured the absorbance at 590 nm with a Spectramax 190 microplate reader (Molecular Devices, Sunnyvale, CA, USA); 20 biological replicates of each strain were assessed, and three independent experiments were carried out.
Twitching Motility Assay
The twitching motility assay was carried out as previously reported with slight modification (Turnbull and Whitchurch, 2014). The bacterial suspension was adjusted to an O.D.600nm of 0.3 and diluted (10−2) with sterilized water (approximately, 106 CFU/ml). Then, 5 μl of bacterial suspension was dotted onto TSA containing 0.5% agar and incubated at 28°C for 2 days. The twitching halos and colony diameter were evaluated by the stereoscopic microscope, a LEICA M205C (LEICA, Wetzlar, Germany). For this assay, five biological replicates of each strain were examined and five independent experiments were carried out.
Tolerance to Osmotic and Antibiotic Stresses
To investigate the effects of CmpAc in stress conditions, the strains were subjected to two stress conditions, namely, osmotic and antibiotic stress. For the osmotic stress, bacterial cell culture with OD600nm of 0.3 (approximately, 108 CFU/ml) was incubated in the presence of 2.5% NaCl at 28°C for 10 min with shaking (280 rpm) and were then serially diluted and dotted onto a TSA with appropriate antibiotics. After incubation for days, viable cell numbers were measured by a colony counting method. Tolerance against antibiotics was examined by supplementing the medium with 0.1 μg/ml polymyxin B at 28°C for two h with shaking (280 rpm). Water was used as a negative control in the assays. To calculate the survivability, the ratio of bacterial numbers in the control to stress conditions was calculated. For each assay, three biological replicates were evaluated, and at least seven independent experiments were carried out.
Statistical Analysis
The statistical significance was analyzed by one-way analysis of variance with Tukey HSDab using SPSS 12.0K software (SPSS, Inc., Chicago, IL, USA). A p-value of less than 0.05 was considered as a statistical difference.
Results
Identification of AcΔcmpAc Strain and Protein Sequence Analysis of CmpAc
A Tn5-insertional library in the background of Ac strain KACC17005 belonging to group II was screened to identify genes involved in the virulence of Ac. We found one mutant that did not cause disease on watermelon and confirmed that a gene, which was annotated as chorismate mutase (Accession No. ATG94418; Locus tag, CQB05_10545), was disrupted by Tn5. The deduced amino acids of ATG94418 revealed that the protein possesses two domains: chorismate mutase type II family (15-91 aa) and prephenate dehydratase family (92-366 aa) (Figure 1A) , indicating that ATG94418 encodes a putative bifunctional chorismate mutase/prephenate dehydratase protein. In agreement with our prediction, Zhang et al. reported that N-terminal and C-terminal of the P-protein, a bifunctional chorismate mutase/prephenate dehydratase protein, in E. coli are required for chorismate mutase and prephenate dehydratase activity, respectively (Zhang et al., 1998). In addition, CmpAc showed high homology with a putative chorismate mutase or prephenate dehydratase in other gram-negative bacteria (Figure 1B). ATG94418 showed 100% similarity with a putative prephenate dehydratase (Accession No. ABM33842) in Ac strain AAC00-1, 93% similarity with a putative chorismate mutase (Accession No. OGA85719) in Burkholderiales GWA2_64_37, and 92% similarity with a putative prephenate dehydratase (Accession No. TQK66165) in Nocardioides sp. SLBN-35. This suggests that the bifunctional chorismate mutase/prephenate dehydratase protein is conserved in the genus as well as in other bacteria. Therefore, ATG94418 was named as CmpAc (bifunctional chorismate mutase/prephenate dehydratase in Ac).
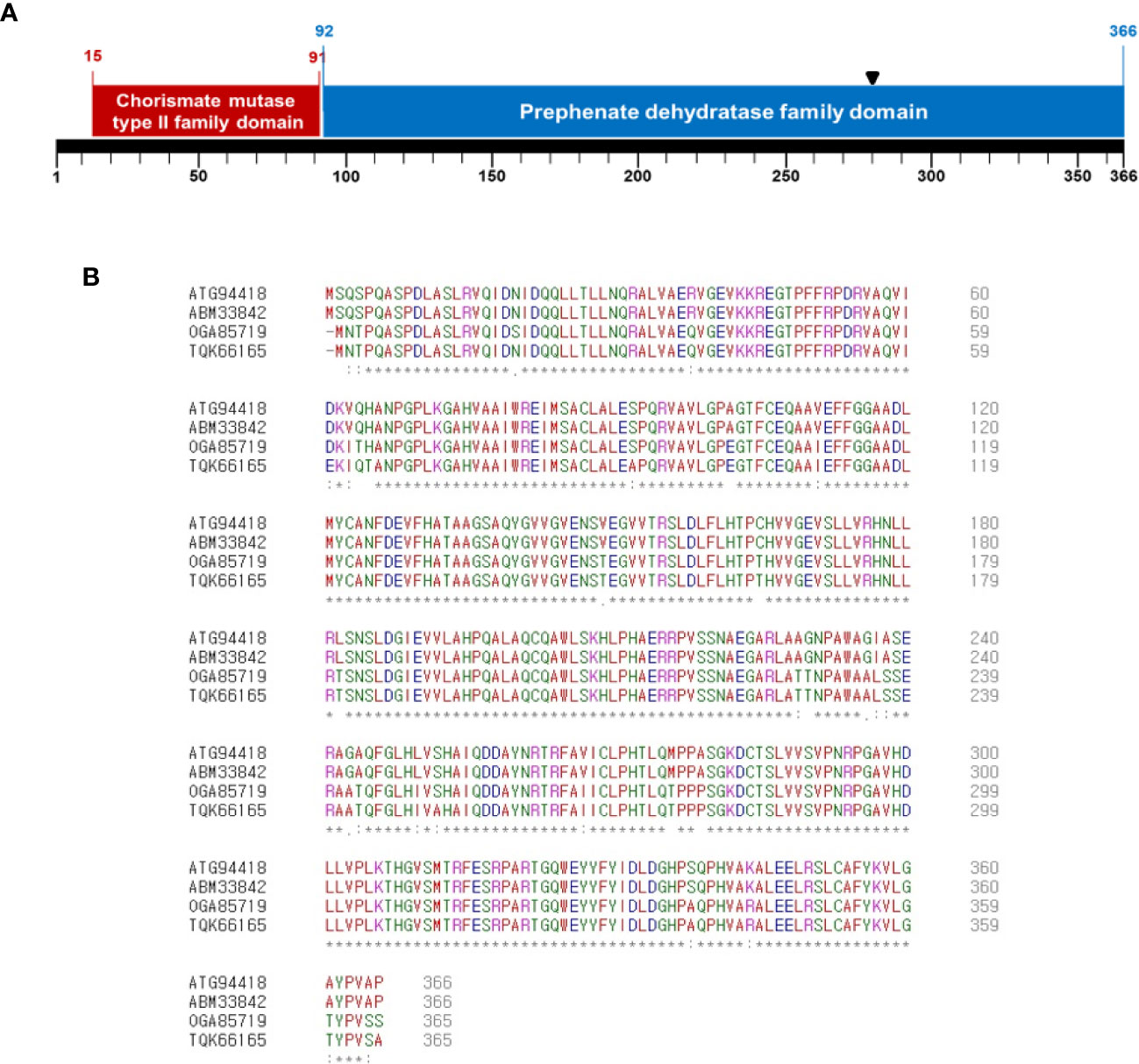
Figure 1 Sequence analysis of deduced amino acids of CmpAc. (A) Predicted domains in CmpAc. Red and blue boxes indicate chorismate mutase type II family and prephenate dehydratase family domains, respectively. A black arrow indicates the Tn5 insertional position. (B) Sequences alignment of CmpAc (ATG94418), prephenate dehydratase (ABM33842) from Ac strain AAC00-1, chorismate mutase (OGA85719) from Burkholderiales bacterium GWA2_64_37 (subsurface metagenome), and prephenate dehydratase (TQK66165) from Nocardioides sp. SLBN-35 using the Clustal Omega Program. The “*”, “:”, and “.” represent identical residues, conserved substitutions, and semi-conserved substitutions, respectively.
CmpAc Is Indispensable for Virulence of Ac
To identify whether CmpAc is involved in the virulence of Ac, pathogenicity assays using the seed-germinated inoculation and infiltration methods were conducted. Ac(EV) and AcΔcmpAc(EV), which are Ac and AcΔcmpAc strains carrying an empty vector, respectively, were used for the assay. In addition, we tested the virulence of the complemented strain, AcΔcmpAc(CmpAc), which is AcΔcmpAc carrying the CmpAc gene on pBBR1MCS5 vector. As shown in Figures 2A, B, seeds infected by AcΔcmpAc(EV) did not show any symptoms. However, Ac(EV) displayed typical wilt symptoms, and the complemented strain showed spots and some wilts. The disease severity by AcΔcmpAc(EV) was 0, while that by Ac(EV) was 2 at 7 days after inoculation (Figure 2C). The disease severity by AcΔcmpAc(CmpAc) was partially restored to the level of Ac(EV) at 1.4 (Figure 2C). The infiltration on leaves of watermelon showed similar patterns compared to the germinated-seed method (Figures 2D, E). Leaves infiltrated by Ac(EV) turned dark and wilted, but those by the mutant did not show any symptoms. AcΔcmpAc(CmpAc) showed similar results as Ac(EV). The bacterial population of AcΔcmpAc(EV) in infected leaves was markedly lower than those of Ac(EV) and AcΔcmpAc(CmpAc) at 2, 4, 6, and 8 days after inoculation. These data indicate that CmpAc is required for the virulence of Ac.
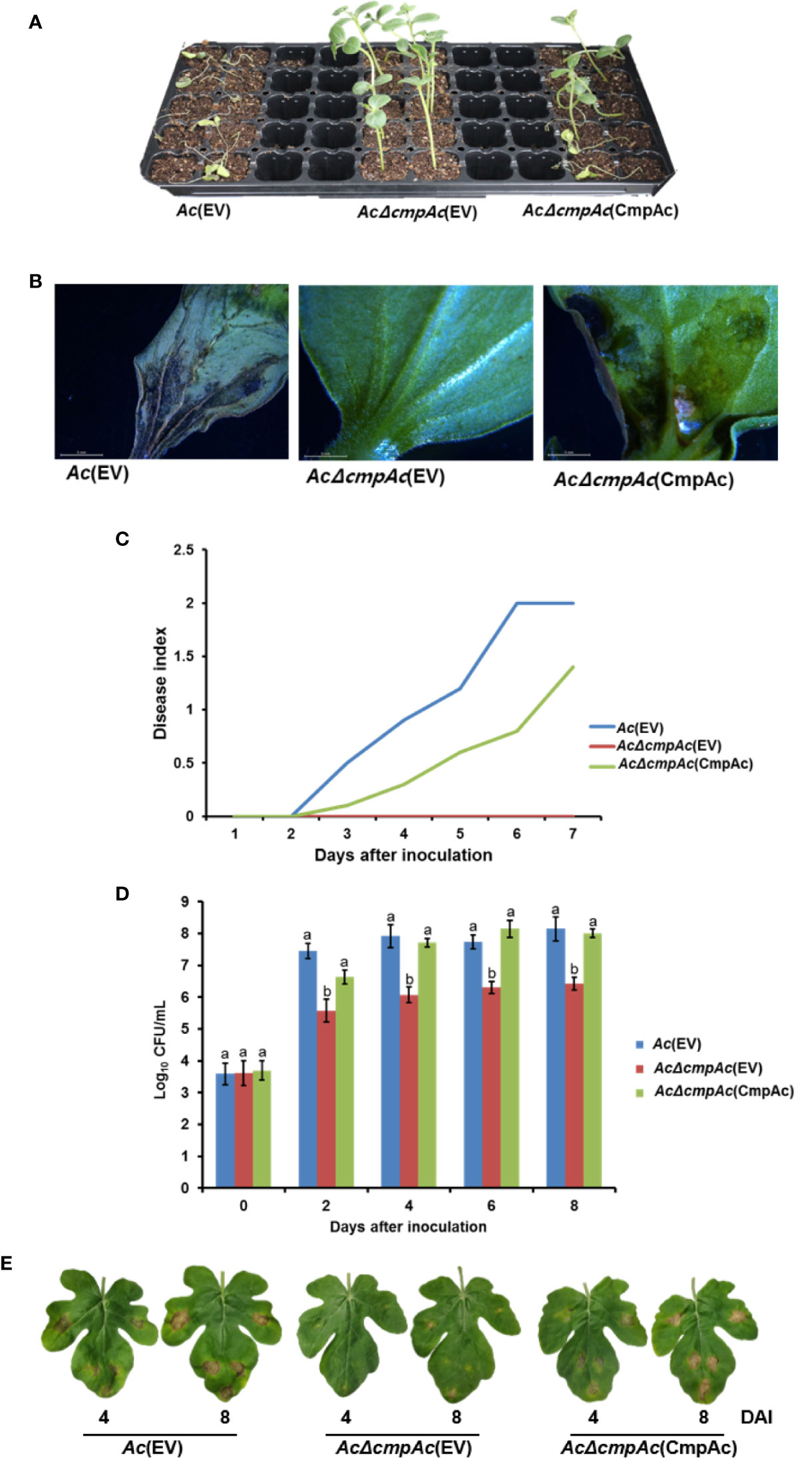
Figure 2 Pathogenicity assay for Ac(EV), AcΔcmpAc(EV), and AcΔcmpAc(CmpAc) using seed-germinated and infiltration inoculation method on watermelon. (A) The seed-germinated inoculation results in watermelon seedlings. The photographs were taken at 7 days after inoculation. (B) The midrib of infected leaves observed through a stereoscopic microscope. The observations were conducted 7 days after inoculation. (C) Disease index variation in the inoculated seedlings during 7 days after inoculation. The disease index is [Normal(numbers of plants)× 0+Spot(numbers of plants)×*1+Wilt(numbers of plants)×2]/Total(numbers of plants). (D) The bacterial population in the infected leaves for 8 days after inoculation determined by the colony counting method. The Ac strain suspension was adjusted to an O.D.600nm of 105 in 10 mM MgCl2, and then inoculated into first and second true leaves in 2-week-old watermelon seedlings using a needleless syringe. Different letters on error bars in the graph represent statistically significant differences by ANOVA (p < 0.05), Tukey HSDab. Error bars indicate standard errors of means. The graph was generated from seven independent experiments with three biological replicates. (E) The infected watermelon true leaf of each strain. The photographs were taken at 4 and 8 days after inoculation.
CmpAc Mutant Is a Phenylalanine Auxotroph
In the shikimate pathway, chorismate mutase is responsible for synthesizing both phenylalanine and tyrosine, while prephenate dehydratase is essential for phenylalanine, but not tyrosine. Therefore, the growth of Ac strains in the presence of phenylalanine or tyrosine was investigated (Figure 3). Firstly, the bacterial growth of the three strains was examined in a rich medium, and all three strains showed similar growth patterns (Figure 3A), indicating that CmpAc is not involved in the reproduction of Ac. In M9 medium, AcΔcmpAc(EV) did not grow without any amino acids (Figure 3B). On the other hand, Ac(EV) and AcΔcmpAc(CmpAc) grew well. In the presence of tyrosine, the growth of the three strains was comparable to those in M9 (Figure 3C). Strikingly, the growth of AcΔcmpAc(EV) was restored to the level of Ac(EV) in M9 with phenylalanine (Figure 3D). These results demonstrate that AcΔcmpAc(EV) is an auxotroph for phenylalanine, but not tyrosine.
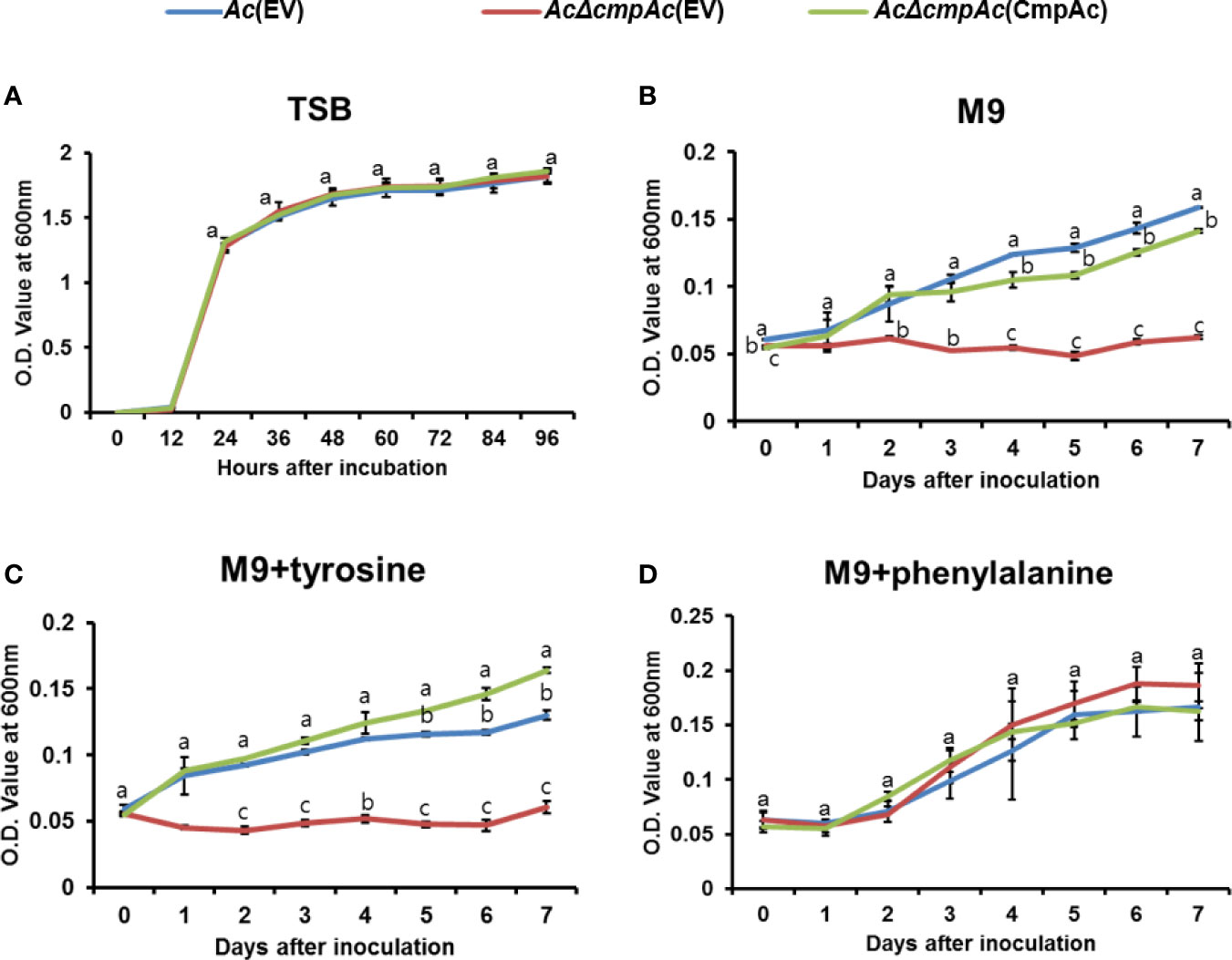
Figure 3 Auxotroph assay. (A) Bacterial growth measurement in (A) tryptic soy broth (TSB). (B) M9 medium without any amino acids. (C) M9 medium with 1 mM tyrosine. (D) M9 medium with 1 mM phenylalanine. The O.D. value at 600 nm (A) for 96 h with 12-h intervals and (B–D) for 7 days with 24-h intervals. Error bars indicate standard deviations. The different letters indicate statistically significant differences by ANOVA with Tukey HSDab (P < 0.05).
Proteomic Analysis
To further predict the biological or cellular processes affected by CmpAc, the comparative proteomic analysis combined with COG classification was carried out using Ac and AcΔcmpAc strains. Through LC-MS/MS, 913 and 872 proteins were commonly detected in three biological replicates of Ac and AcΔcmpAc, respectively (Supplementary Table 2), and these proteins were subjected to a comparative proteomic analysis. In the detected proteins, 53 proteins were only detected in Ac, and 63 proteins were uniquely found in AcΔcmpAc (Figure 4A). Additionally, 18 and 17 proteins were differentially (over 2-fold) abundant in Ac and AcΔcmpAc, respectively (Figure 4A and Supplementary Figure 1). These proteins were categorized using COG analysis. Proteins belonging to group E (Amino Acid metabolism and transport), I (Lipid metabolism), and J (Translation) were abundantly detected in Ac compared to that in AcΔcmpAc (Figure 4B and Supplementary Table 3). In addition, proteins categorized in group C (Energy production and conversion), M (Cell wall/membrane/envelop biogenesis), T (Signal transduction), and U (Intracellular trafficking and secretion) were highly found in AcΔcmpAc (Figure 4B and Supplementary Table 4). Among all the categories in COG, the group M including penicillin-binding protein showed the highest numbers of differentially abundant proteins. In addition, porins, ABC transporters, and methyl-accepting chemotaxis proteins were detected in the comparative proteomic analysis (Supplementary Tables 3, 4). These data suggest that CmpAc may be related to cell membrane/wall integrity.
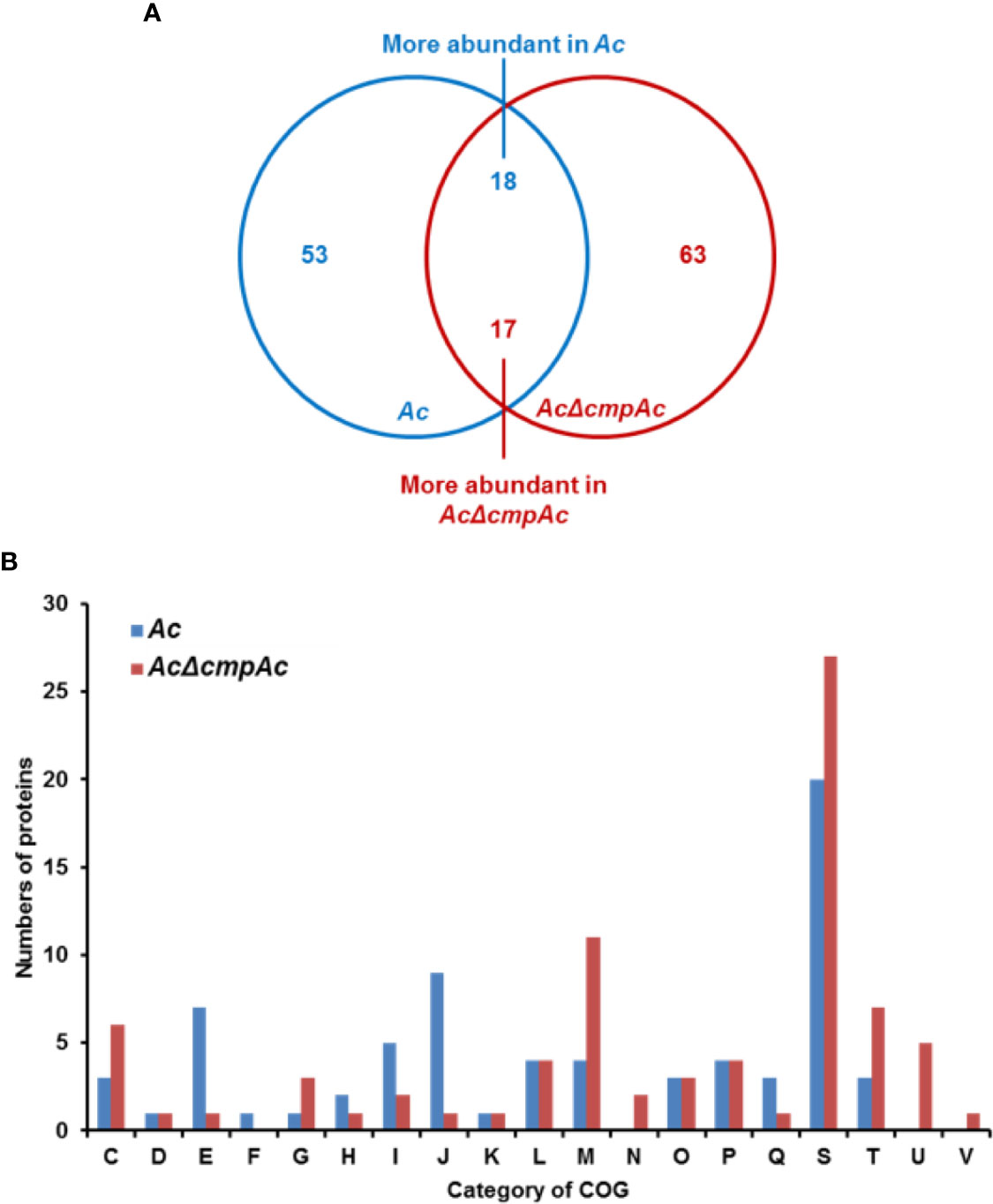
Figure 4 Comparative proteomic analysis in Ac and AcΔcmpAc. (A) The Venn diagram of 71 and 80 proteins whose abundance was altered. Fifty-three and 63 proteins were uniquely found in Ac and AcΔcmpAc, respectively. Eighteen and 17 proteins were differentially (>2 fold) enriched in Ac and AcΔcmpAc, respectively. (B) Clusters of orthologous groups classification of differentially abundant proteins (>2 fold) from the proteomic analysis. C, Energy production and conversion; D, Cell cycle control and mitosis; E, Amino acid metabolism and transport; F, Nucleotide metabolism and transport; G, Carbohydrate metabolism and transport; H, Coenzyme metabolism; I, Lipid metabolism; J, Translation; K, Transcription; L, Replication and repair; M, Cell wall/membrane/envelop biogenesis; N, Cell motility; O, Post-translational modification, protein turnover, chaperone functions; P, Inorganic ion transport and metabolism; Q, Secondary structure; S, Function unknown; T, Signal transduction; U, Intracellular trafficking and secretion; V, Defense mechanisms.
CmpAc Is Associated With Biofilm Formation
Our proteomics analysis revealed that the abundance of proteins related to the secretion of polysaccharides was altered. It is well known that the polysaccharide is one of the major components of biofilm in bacteria (Vu et al., 2009). Therefore, we hypothesized that CmpAc might have effects on biofilm formation. The biofilm formation of Ac strains was tested (Figure 5A) using a 96-well PVC microplate assay. Interestingly, the ability to form biofilm in AcΔcmpAc(EV) was enhanced (over 1.5-fold) compared to that in Ac(EV). In addition, AcΔcmpAc(CmpAc) showed a similar level of biofilm formation as Ac(EV). These data indicate that CmpAc is associated with biofilm formation in Ac.
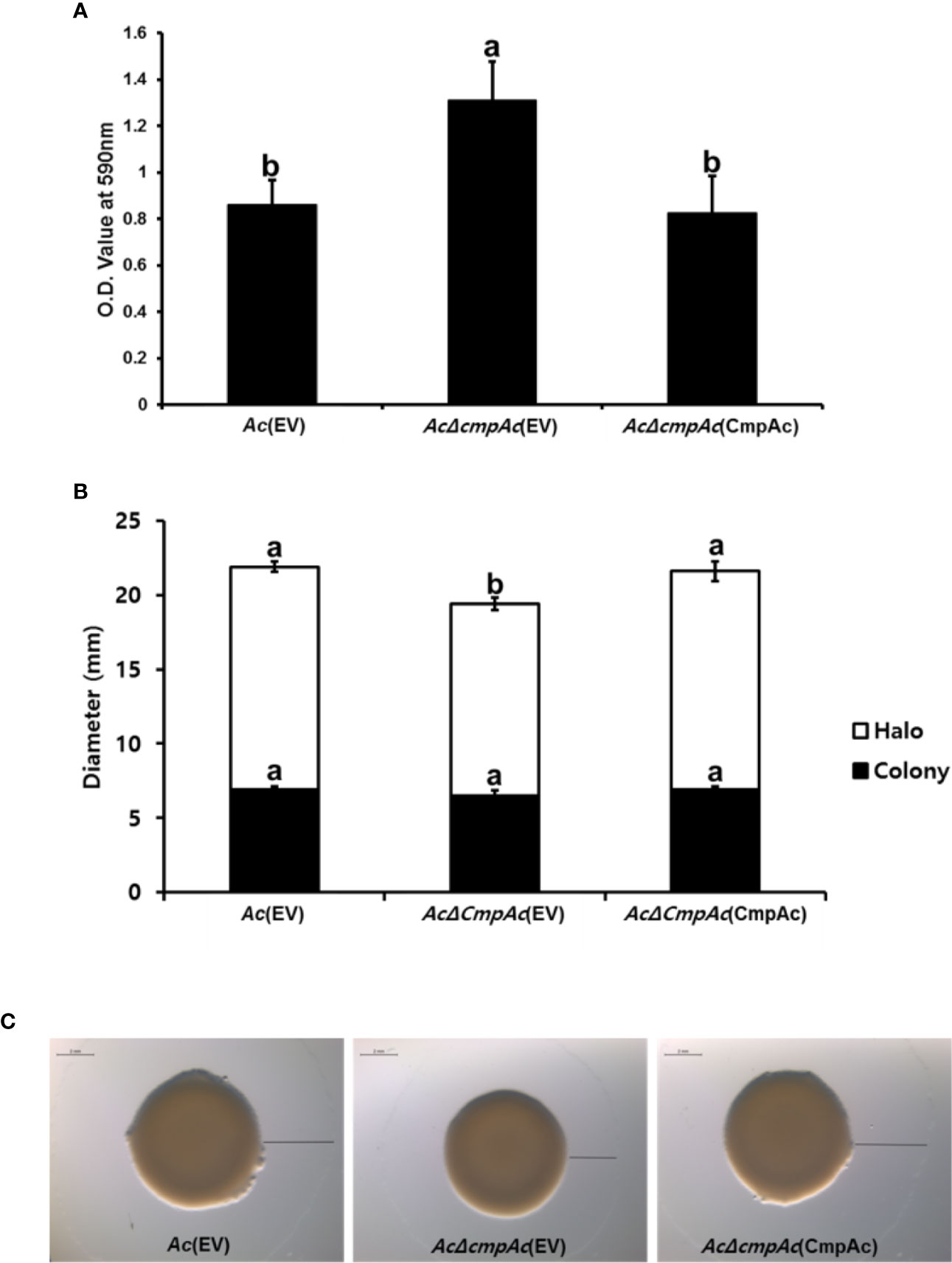
Figure 5 Biofilm formation and twitching motility halo production of Ac(EV), AcΔcmpAc(EV), and AcΔcmpAc(CmpAc). (A) Each strain was incubated in a polyvinyl chloride 96-well plate for 2 days. For the quantification, the biofilm was stained with 0.1% crystal violet. The stained biofilm was eluted using 95% ethanol, and the absorbance was measured by UV spectrophotometer at 590 nm. (B) Colony and halo production. The black bars indicate the colony diameter. The white bars show the diameter of the twitching halos produced. (C) Observation of colony and twitching halo production using a stereoscopic microscope. Black lines indicate the twitching halo diameter. Scale bar, 2 mm. Error bars indicate standard deviations. The different letters indicated statistically significant differences by ANOVA with Tukey HSDab (P < 0.05).
AcΔcmpAc(EV) Displayed Reduced Twitching Halos
Proteins associated with pili functions were found in the comparative proteomic analysis. Pili are known to be involved in the twitching motility in bacteria (Mattick, 2002). Thus, the twitching motility assay was conducted in TSA with 0.5% agar, and the diameters of bacterial colonies and twitching halos were measured (Figures 5B, C). The colony diameters of Ac(EV), AcΔcmpAc(EV), and AcΔcmpAc(CmpAc) were 6.9, 6.5, and 6.9 mm, respectively (Figures 5B, C), indicating that the expansion of the colony was not different in the three strains. The shapes of the marginal sides of colonies of the three strains were also similar (Data not shown). Interestingly, the twitching halos of AcΔcmpAc(EV) were reduced (12.9 mm) compared with those of Ac(EV) (15 mm). The diameter of the twitching halo in AcΔcmpAc(CmpAc) was comparable to that in Ac(EV). These results indicate that the functions of CmpAc are related to the production of twitching halos.
CmpAc Is Involved in Stress Tolerance
In the comparative proteomic analysis, the abundance of proteins related to cell wall/membrane/envelope biosynthesis was mostly affected. Previous studies reported that bacterial cell wall/membrane/envelope is associated with functions for protecting bacteria against the external environment (Hews et al., 2019; Park et al., 2019). Therefore, the tolerance of AcΔcmpAc(EV) against two external stresses was tested. Firstly, NaCl was used as an agent for osmotic stress (Figure 6A). In the presence of 2.5% NaCl, Ac(EV) showed 10.9% survivability, while AcΔcmpAc(EV) showed only 2.9% survivability. The survivability of AcΔcmpAc(CmpAc) was restored to the level of Ac(EV). Unexpectedly, in the supplementation of polymyxin B (0.1 µg/ml), which is a β–lactam antibiotics and has effects on membrane permeability, the survivability of AcΔcmpAc(EV) was enhanced (approximately, 1.5-fold) compared with that of Ac(EV) (Figure 6B). AcΔcmpAc(CmpAc) displayed comparable survivability to Ac(EV). These data indicate that CmpAc is associated with tolerance to environmental stresses.
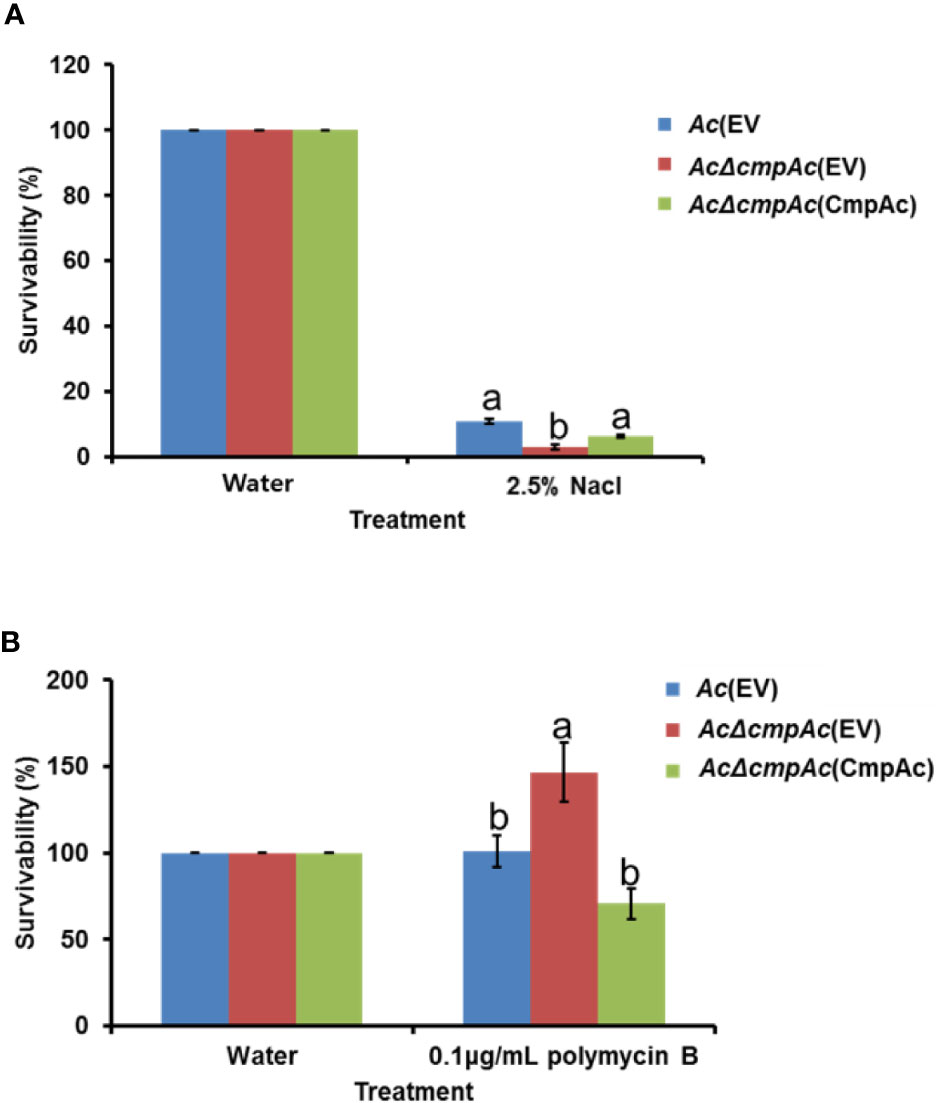
Figure 6 Tolerance assay for osmotic stress and antibiotics. Tolerance was determined in the presence of (A) 2.5% NaCl for 10 min or (B) 0.1 μg/ml polymyxin B for 2 h. The numbers of recovered bacterial cells were counted by the colony counting method. The survivability was calculated based on the number of recovered cells post water treatment (control) compared to that post the stress treatment. The different letters indicated statistically significant differences by ANOVA with Tukey HSDab (P < 0.05). Error bars indicate standard errors of means.
Discussion
It is generally known that the chorismate mutase gene is involved in virulence in both eukaryotic and prokaryotic microorganisms. For example, the inactivation of chorismate mutase in Ustilago maydis showed reduced virulence (Djamei and Kahmann, 2012). In contrast, the chorismate mutase knock out of X. oryzae pv. oryzae XKK.12 was more virulent than the wild-type strain in rice (Degrassi et al., 2010). In our study, we showed that the CmpAc is required for virulence of Ac using two different inoculation assays. But, there was no difference in bacterial growth in rich media, indicating that reduced virulence was not due to bacterial multiplication or reproduction. In addition to virulence, proteins required for the biosynthesis of aromatic amino acids are also involved in other mechanisms in bacteria. For example, aroA and aroB in the shikimate pathway are related to virulence, pigment production, and tolerance to UV stress in Burkholderia glumae (Karki and Ham, 2014). Felgner et al. reported that the auxotrophic mutant for aromatic amino acids in Salmonella enterica serovar Typhimurium showed the alterations in virulence, the lipid/amino acid metabolisms, sensitivity to penicillin, flagellin phase variation, motility and expression of the virulence-associated genes using transcriptomic and metabolic analyses as well as phenotypic assays (Felgner et al., 2016). In an agreement these studies, we also demonstrated that CmpAc is involved in virulence, phenylalanine biosynthesis, and other mechanisms, including biofilm formation, twitching motility, and stress tolerance by the comparative analysis and phenotypic observation. Thus, it suggests that enzymes, which are required for the biosynthesis of the primary metabolite, including CmpAc may be associated with diverse cellular functions in bacteria.
Chorismate mutase is a key enzyme of the shikimate pathway. Chorismate can be catalyzed by chorismate mutase to produce prephenate which is a branch point for the biosynthesis of tyrosine and phenylalanine (Kast et al., 2000). Prephenate is a substrate of two different enzymes: prephenate dehydratase and prephenate aminotransferase, which are responsible for phenylalanine and tyrosine, respectively (Tzin and Galili, 2010). Interestingly, some bacterial chorismate mutases possess two domains: chorismate mutase and prephenate dehydratase. These bifunctional proteins are involved in the production of phenylalanine, but not tyrosine because prephenate dehydratase is required for the production of phenylalanine (Duggleby et al., 1978; Zhang et al., 1998). In an agreement with these studies, our sequence analysis revealed that the chorismate mutase in Ac also contains two putative domains for chorismate mutase and prephenate dehydratase. We further demonstrated that CmpAc is indispensable for biosynthesis of phenylalanine, but not tyrosine in an auxotrophic assay. Interestingly, Ac strain KACC17005 possesses another putative chorismate mutase (ATG95797), containing only the chorismate mutase domain (Park et al., 2017). Although we did not test the functions of ATG95797 in this study, the protein may be responsible for the biosynthesis of tyrosine in the shikimate pathway in Ac.
Biofilm formation is one of the key elements for bacterial virulence (Davey and O’toole, 2000). Interestingly, AcΔcmpAc(EV) showed higher biofilm formation but was less virulent compared to other strains. According to a previous report, impaired capsular polysaccharides in Streptococcus pneumonia enhanced the biofilm formation but reduced the virulence (Qin et al., 2013). The proteomics analysis identified that an abundance of proteins involved in cell wall/membrane/envelop biogenesis was altered. In addition, proteins classified in the group U (intracellular trafficking and secretion) were detected only in AcΔcmpAc. These results implicate that impaired CmpAc in Ac may alter membrane integrity and/or secretion systems. Therefore, it can be postulated that the abnormal cell membranes may be weakened and are easily broken during the protein extraction steps, and hence, proteins related to cell wall/membrane/envelop biogenesis and intracellular trafficking and secretion were abundantly found in the comparative proteomic analysis. Similar to S. pneumonia (Qin et al., 2013), it is also hypothesized that the weakened cell membrane/wall may be responsible for the higher biofilm formation observed in AcΔcmpAc(EV) compared with other strains.
Biofilm formation is closely related to β-lactamase production in Pseudomonas aeruginosa (Heydari and Eftekhar, 2015), postulating that biofilm formation has a positive role for tolerance to β-lactams like polymyxin B. In addition, cell envelope synthesis and modification in Vibrio cholera also modulate tolerance to β-lactam antibiotics (Weaver et al., 2018). Similarly, in our study, AcΔcmpAc(EV) showed enhanced biofilm formation as well as tolerance to polymyxin B. Moreover, the mutant was less tolerant to osmotic stress compared with Ac(EV). In addition to the proteomic analysis, these results also support that CmpAc is associated with cell membrane/wall/envelope biosynthesis. Twitching motility rather than flagella-dependent motility is a crucial mechanism for bacterial movement in Ac (Bahar et al., 2010). It is also known that twitching motility in Ac is required for optimal virulence (Rosenberg et al., 2018). In X. oryzae pv. oryzae, the knockout mutant of outer membrane porin, has irregular shape twitching motility and an alteration in stress tolerance (Bae et al., 2018). Likewise, in this study, we showed that AcΔcmpAc(EV) reduced the production of twitching halo, which may contribute to the virulence of the group II Ac strain KACC17005.
In summary, we demonstrated that CmpAc, which is a putative bifunctional chorismate mutase/prephenate dehydratase, is related to the biosynthesis of phenylalanine, biofilm formation, twitching motility, and tolerance of osmotic stress and antibiotics, which may contribute to the virulence of Ac. Several enzymes, including chorismate mutase, which are involved in the production of amino acids, have been thought as potential targets for developing virulence inhibitors (Monti et al., 2016; Khanapur et al., 2017). Therefore, this study provides valuable information on an uncharacterized virulence factor, which could also be a potential target for anti-virulence reagent to control BFB.
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository with the dataset identifier PXD019444.
Author Contributions
S-WH conceived the study. S-WH and MK designed the experiments. MK, JL, and LH conducted the experiments. MK and S-WH analyzed the data and prepared the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Next-Generation BioGreen 21 Program (PJ01328901) of the Rural Development Administration and the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. NRF-2020R1A2C1013040), Republic of Korea (to S-WH). This research was supported by the Chung-Ang University Graduate Research Scholarship in 2020 (to LH).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.569552/full#supplementary-material
Supplementary Figure 1 | Differentially abundant (>2 fold) proteins detected in Ac and AcΔcmpAc strains. Normalized peptide spectral matches of proteins detected in AcΔcmpAc (Red) were comparted with these in Ac (Blue). Error bars indicate standard deviations. The abundance of all proteins was statistically different (P < 0.05).
References
Ahmad, S., Wilson, A. T., Jensen, R. A. (1988). Chorismate mutase-prephenate dehydratase from Acinetobacter calcoaceticus - purification, properties and immunological cross-reactivity. Eur. J. Biochem. 176, 69–79. doi: 10.1111/j.1432-1033.1988.tb14252.x
Bae, N., Park, H. J., Park, H., Kim, M., Han, S. W. (2018). Deciphering the functions of the outer membrane porin OprBXo involved in virulence, motility, exopolysaccharide production, biofilm formation and stress tolerance in Xanthomonas oryzae pv. oryzae. Mol. Plant Pathol. 19, 2527–2542. doi: 10.1111/mpp.12727
Bahar, O., Goffer, T., Burdman, S. (2009). Type IV pili are required for virulence, twitching motility, and biofilm formation of Acidovorax avenae subsp citrulli. Mol. Plant-Microbe Interact. 22, 909–920. doi: 10.1094/MPMI-22-8-0909
Bahar, O., De La Fuente, L., Burdman, S. (2010). Assessing adhesion, biofilm formation and motility of Acidovorax citrulli using microfluidic flow chambers. FEMS Microbiol. Lett. 312, 33–39. doi: 10.1111/j.1574-6968.2010.02094.x
Bahar, O., Levi, N., Burdman, S. (2011). The cucurbit pathogenic bacterium Acidovorax citrulli requires a polar flagellum for full virulence before and after host-tissue penetration. Mol. Plant-Microbe Interact. 24, 1040–1050. doi: 10.1094/MPMI-02-11-0041
Burdman, S., Walcott, R. R. (2012). Acidovorax citrulli: generating basic and applied knowledge to tackle a global threat to the cucurbit industry. Mol. Plant Pathol. 13, 805–815. doi: 10.1111/j.1364-3703.2012.00810.x
Choi, H., Fermin, D., Nesvizhskii, A. I. (2008). Significance analysis of spectral count data in label-free shotgun proteomics. Mol. Cell. Proteomics 7, 2373–2385. doi: 10.1074/mcp.M800203-MCP200
Davey, M. E., O’toole, G. A. (2000). Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64, 847–867. doi: 10.1128/MMBR.64.4.847-867.2000
Degrassi, G., Devescovi, G., Bigirimana, J., Venturi, V. (2010). Xanthomonas oryzae pv. oryzae XKK.12 contains an AroQgamma chorismate mutase that is involved in rice virulence. Phytopathology 100, 262–270. doi: 10.1094/PHYTO-100-3-0262
Djamei, A., Kahmann, R. (2012). Ustilago maydis: dissecting the molecular interface between pathogen and plant. PLoS Pathog. 8, e1002955. doi: 10.1371/journal.ppat.1002955
Duggleby, R. G., Sneddon, M. K., Morrison, J. F. (1978). Chorismate mutase-prephenate dehydratase from Escherichia coli - active-sites of a bifunctional enzyme. Biochemistry 17, 1548–1554. doi: 10.1021/bi00601a030
Elias, J. E., Gygi, S. P. (2007). Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214. doi: 10.1038/nmeth1019
Felgner, S., Frahm, M., Kocijancic, D., Rohde, M., Eckweiler, D., Bielecka, A., et al. (2016). aroA-deficient Salmonella enterica serovar Typhimurium is more than a metabolically attenuated mutant. Mbio 7, e1220–e1216. doi: 10.1128/mBio.01220-16
Ferreira, R. M., Moreira, L. M., Ferro, J. A., Soares, M. R. R., Laia, M. L., Varani, A. M., et al. (2016). Unravelling potential virulence factor candidates in Xanthomonas citri subsp citri by secretome analysis. Peerj 4, e1734. doi: 10.7717/peerj.1734
Hews, C. L., Cho, T., Rowley, G., Raivio, T. L. (2019). Maintaining integrity under stress: envelope stress response regulation of pathogenesis in gram-negative bacteria. Front. Cell. Infect. Microbiol. 9, 313. doi: 10.3389/fcimb.2019.00313
Heydari, S., Eftekhar, F. (2015). Biofilm formation and beta-lactamase production in burn isolates of Pseudomonas aeruginosa. Jundishapur J. Microbiol. 8, e15514. doi: 10.5812/jjm.15514
Huccetogullari, D., Luo, Z. W., Lee, S. Y. (2019). Metabolic engineering of microorganisms for production of aromatic compounds. Microbial. Cell Fact 18, 41. doi: 10.1186/s12934-019-1090-4
Jiwenez-Guerrero, I., Perez-Montano, F., Da Silva, G. M., Wagner, N., Shkedy, D., Zhao, M., et al. (2020). Show me your secret(ed) weapons: a multifaceted approach reveals a wide arsenal of type III-secreted effectors in the cucurbit pathogenic bacterium Acidovorax citrulli and novel effectors in the Acidovorax genus. Mol. Plant Pathol. 17, 17–37. doi: 10.1111/mpp.12877
Johnson, K. L., Walcott, R. R. (2013). Quorum sensing contributes to seed-to-seedling transmission of Acidovorax citrulli on watermelon. J. Phytopathol. 161, 562–573. doi: 10.1111/jph.12106
Karki, H. S., Ham, J. H. (2014). The roles of the shikimate pathway genes, aroA and aroB, in virulence, growth and UV tolerance of Burkholderia glumae strain 411gr-6. Mol. Plant Pathol. 15, 940–947. doi: 10.1111/mpp.12147
Kast, P., Grisostomi, C., Chen, I. A., Li, S. L., Krengel, U., Xue, Y. F., et al. (2000). A strategically positioned cation is crucial for efficient catalysis by chorismate mutase. J. Biol. Chem. 275, 36832–36838. doi: 10.1074/jbc.M006351200
Khanapur, M., Alvala, M., Prabhakar, M., Kumar, K. S., Edwin, R. K., Saranya, P. S. V. K. S., et al. (2017). Mycobacterium tuberculosis chorismate mutase: A potential target for TB. Bioorg. Med. Chem. 25, 1725–1736. doi: 10.1016/j.bmc.2017.02.001
Kovach, M. E., Elzer, P. H., Hill, D. S., Robertson, G. T., Farris, M. A., Roop, R. M., 2nd, et al. (1995). Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176. doi: 10.1016/0378-1119(95)00584-1
Latin, R. X., Hopkins, D. L. (1995). Bacterial fruit blotch of watermelon - the hypothetical exam question becomes reality. Plant Dis. 79, 761–765. doi: 10.1094/PD-79-0761
Liu, J., Tian, Y. L., Zhao, Y. Q., Zeng, R., Chen, B. H., Hu, B. S., et al. (2019). Ferric Uptake Regulator (FurA) is Required for Acidovorax citrulli Virulence on Watermelon. Phytopathology 109, 1997–2008. doi: 10.1094/PHYTO-05-19-0172-R
Mattick, J. S. (2002). Type IV pili and twitching motility. Annu. Rev. Microbiol. 56, 289–314. doi: 10.1146/annurev.micro.56.012302.160938
Monti, S. M., De Simone, G., D’ambrosio, K. (2016). L-histidinol dehydrogenase as a new target for old Diseases. Curr. Topics Med. Chem. 16, 2369–2378. doi: 10.2174/1568026616666160413140000
O’Brien, R. G., Martin, H. L. (1999). Bacterial blotch of melons caused by strains of Acidovorax avenae subsp citrulli. Aust. J. Exp. Agric. 39, 479–485. doi: 10.1071/EA98172
Park, H.-J., Seong, H. J., Sul, W. J., Oh, C.-S., Han, S.-W. (2017). Complete genome sequence of Acidovorax citrulli strain KACC17005, a causal agent for bacterial fruit blotch on watermelon. Korean J. Microbiol. 53, 340–341. doi: 10.7845/kjm.2017.7084
Park, H. J., Jung, B., Lee, J., Han, S. W. (2019). Functional characterization of a putative DNA methyltransferase, EadM, in Xanthomonas axonopodis pv. glycines by proteomic and phenotypic analyses. Sci. Rep. 9, 2446. doi: 10.1038/s41598-019-38650-3
Park, H., Do, E., Kim, M., Park, H. J., Lee, J., Han, S. W. (2020). A lysR-type transcriptional regulator LcrX is involved in virulence, biofilm formation, swimming motility, siderophore secretion, and growth in sugar sources in Xanthomonas axonopodis pv. glycines. Front. Plant Sci. 10, 1657. doi: 10.3389/fpls.2019.01657
Park, H. J., Lee, J., Kim, M., Han, S. W. (2020). Profiling differentially abundant proteins by overexpression of three putative methyltransferases in Xanthomonas axonopodis pv. glycines. Proteomics 20, 1900125. doi: 10.1002/pmic.201900125
Perez-Riverol, Y., Csordas, A., Bai, J. W., Bernal-Llinares, M., Hewapathirana, S., Kundu, D. J., et al. (2019). The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450. doi: 10.1093/nar/gky1106
Qin, L., Kida, Y., Imamura, Y., Kuwano, K., Watanabe, H. (2013). Impaired capsular polysaccharide is relevant to enhanced biofilm formation and lower virulence in Streptococcus pneumoniae. J. Infect. Chemother. 19, 261–271. doi: 10.1007/s10156-012-0495-3
Rosenberg, T., Salam, B. B., Burdman, S. (2018). Association between loss of type IV pilus synthesis ability and phenotypic variation in the cucurbit pathogenic bacterium Acidovorax citrulli. Mol. Plant-Microbe Interact. 31, 548–559. doi: 10.1094/MPMI-12-17-0324-R
Schaad, N. W., Sowell, G., Goth, R. W., Colwell, R. R., Webb, R. E. (1978). Pseudomonas pseudoalcaligenes subsp. citrulli subsp-Nov. Int. J. Syst. Bacteriol. 28, 117–125. doi: 10.1099/00207713-28-1-117
Song, Y. R., Hwang, I. S., Oh, C. S. (2020). natural variation in virulence of Acidovorax citrulli isolates that cause bacterial fruit blotch in watermelon, depending on infection routes. Plant Pathol. J. 36, 29–42. doi: 10.5423/PPJ.OA.10.2019.0254
Tatusov, R. L., Galperin, M. Y., Natale, D. A., Koonin, E. V. (2000). The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28, 33–36. doi: 10.1093/nar/28.1.33
Tian, Y. L., Zhao, Y. Q., Wu, X. R., Liu, F. Q., Hu, B. S., Walcott, R. R. (2015). The type VI protein secretion system contributes to biofilm formation and seed-to-seedling transmission of Acidovorax citrulli on melon. Mol. Plant Pathol. 16, 38–47. doi: 10.1111/mpp.12159
Turnbull, L., Whitchurch, C. B. (2014). Motility assay: twitching motility. Methods Mol. Biol. 1149, 73–86. doi: 10.1007/978-1-4939-0473-0_9
Tzin, V., Galili, G. (2010). New insights into the shikimate and aromatic amino acids biosynthesis pathways in plants. Mol. Plant 3, 956–972. doi: 10.1093/mp/ssq048
Vu, B., Chen, M., Crawford, R. J., Ivanova, E. P. (2009). Bacterial extracellular polysaccharides involved in biofilm formation. Molecules 14, 2535–2554. doi: 10.3390/molecules14072535
Walcott, R. R., Langston, D. B., Sanders, F. H., Gitaitis, R. D. (2000). Investigating intraspecific variation of Acidovorax avenae subsp citrulli using DNA fingerprinting and whole cell fatty acid analysis. Phytopathology 90, 191–196. doi: 10.1094/PHYTO.2000.90.2.191
Wang, T. L., Guan, W., Huang, Q., Yang, Y. W., Yan, W. R., Sun, B. X., et al. (2016). Quorum-sensing contributes to virulence, twitching motility, seed attachment and biofilm formation in the wild type strain Aac-5 of Acidovorax citrulli. Microbial. Pathogenesis 100, 133–140. doi: 10.1016/j.micpath.2016.08.039
Weaver, A. I., Murphy, S. G., Umans, B. D., Tallavajhala, S., Onyekwere, I., Wittels, S., et al. (2018). Genetic determinants of penicillin tolerance in Vibrio cholerae. Antimicrob. Agents Chemother. 62, e01326–e01318. doi: 10.1128/AAC.01326-18
Willems, A., Goor, M., Thielemans, S., Gillis, M., Kersters, K., Deley, J. (1992). Transfer of several phytopathogenic Pseudomonas species to Acidovorax as Acidovorax avenae subsp avenae subsp-nov, comb-nov, Acidovorax avenae subsp citrulli, Acidovorax avenae subsp cattleyae, and Acidovorax konjaci. Int. J. Syst. Bacteriol. 42, 107–119. doi: 10.1099/00207713-42-1-107
Zhang, S., Pohnert, G., Kongsaeree, P., Wilson, D. B., Clardy, J., Ganem, B. (1998). Chorismate mutase-prephenate dehydratase from Escherichia coli - study of catalytic and regulatory domains using genetically engineered proteins. J. Biol. Chem. 273, 6248–6253. doi: 10.1074/jbc.273.11.6248
Keywords: chorismate mutase/prephenate dehydratase, Acidovorax citrulli, virulence, proteomics, biofilm
Citation: Kim M, Lee J, Heo L and Han S-W (2020) Putative Bifunctional Chorismate Mutase/Prephenate Dehydratase Contributes to the Virulence of Acidovorax citrulli. Front. Plant Sci. 11:569552. doi: 10.3389/fpls.2020.569552
Received: 04 June 2020; Accepted: 09 September 2020;
Published: 25 September 2020.
Edited by:
Manuel Martinez, Polytechnic University of Madrid, SpainReviewed by:
Bhabesh Dutta, University of Georgia, United StatesDavid Mackey, The Ohio State University, United States
Copyright © 2020 Kim, Lee, Heo and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sang-Wook Han, c3doYW5AY2F1LmFjLmty
 Minyoung Kim
Minyoung Kim Jongchan Lee
Jongchan Lee Lynn Heo
Lynn Heo Sang-Wook Han
Sang-Wook Han