- Ministry of Agriculture, Key Laboratory of Molecular Biology of Crop Pathogens and Insects, Institute of Insect Sciences, Zhejiang University, Hangzhou, China
Plant-mediated interactions between plant viruses and their vectors are important determinants of the population dynamics of both types of organisms in the field. The whitefly Bemisia tabaci can establish mutualism with begomoviruses via their shared host plants. This mutualism is achieved by the interaction between virulence factors and their host proteins. While the virulence factor βC1 encoded by tomato yellow leaf curl China betasatellite (TYLCCNB), a subviral agent associated to the begomovirus tomato yellow leaf curl China virus (TYLCCNV), may interact with plant protein MYC2, thereby establishing the indirect mutualism between TYLCCNV and whitefly, whether other mechanisms are involved remains unknown. Here, we found the in vitro and in vivo interactions between βC1 and tobacco protein S-phase kinase associated protein 1 (NtSKP1). Silencing the expression of NtSKP1 enhanced the survival rate and fecundity of whiteflies on tobacco plants. NtSKP1 could activate the transcription of genes in jasmonic acid (JA) pathways by impairing the stabilization of JAZ1 protein. Moreover, βC1-NtSKP1 interaction could interfere JAZ1 degradation and attenuate the plant JA defense responses. These results revealed a novel mechanism underlying the better performance of whiteflies on TYLCCNV/TYLCCNB-infected plants.
Introduction
Plants are constantly exposed to environmental biotic factors, including plant pathogens and herbivores, both of which are propelled into complex interaction networks in microecosystems (Rzanny and Voigt, 2012; Franco et al., 2017; Noman et al., 2020). Thereinto, virus-plant-vector tripartite interactions can impact the population dynamics of all the three members. In some cases, this tripartite interaction can become a determining factor in the outbreak of diseases and pests (Islam et al., 2020; Noman et al., 2020). For instance, cucumber mosaic virus, a non-persistently transmitted-virus, alters the performance of its vector aphid, like survival, fecundity, wing dimorphism, and behavior via host plants to optimize viral spread (Ziebell et al., 2011; Carmo-Sousa et al., 2014; Shi et al., 2016; Wu et al., 2017; Tungadi et al., 2020). Soybean mosaic virus infection of soybean plants may reduce plant palatability for its aphid vectors, but increase aphid feeding preference for infected plants, thus promoting its transmission (Peñaflor et al., 2016; Li et al., 2018). Although the biological significance of virus-plant-vector tripartite interactions has been appreciated, molecular mechanisms underlying these complex interactions remain largely unknown.
The genus Begomovirus (family Geminiviridae) is the most populous genus in plant viruses (Brown et al., 2015; Zerbini et al., 2017). Based on genomic organization, begomovirus may be divided into monopartite and bipartite begomoviruses. The genome of bipartite begomoviruses contain two single-stranded circular DNA molecules (DNA-A and DNA-B), while monopartite begomoviruses have only a single genomic component similar to DNA-A of bipartite begomoviruses. Some monopartite begomoviruses were found to be associated with satellite DNA, such as alphasatellites, betasatellites, or deltasatellites (Nawaz-Ul-Rehman and Fauquet, 2009; Zhou, 2013; Lozano et al., 2016; Zerbini et al., 2017). Begomoviruses have long time been a major constraint in agricultural production in tropical and subtropical regions around the globe (Mansoor et al., 2006; Perefarres et al., 2012; Rojas et al., 2018). Begomoviruses are exclusively transmitted by whiteflies (Hemiptera: Aleyrodidae) of the Bemisia tabaci complex, the abundance and distribution of which are major determinants of virus epidemics (Hogenhout et al., 2008; Navas-Castillo et al., 2011; Gilbertson et al., 2015; Whitfield et al., 2015; Islam et al., 2018). The B. tabaci complex contains at least 44 cryptic species, among which the invasive Middle East Asia Minor 1 (MEAM1) can efficiently transmit many begomoviruses (De Barro et al., 2011; Liu et al., 2012; Lee et al., 2013; Wei et al., 2014; Kanakala and Ghanim, 2019). In recent years, the relationships among begomoviruses, plants, and whiteflies has been explored extensively, which could be positive, neutral, and negative (Luan et al., 2014; Wang et al., 2017). For example, tomato yellow leaf curl China virus (TYLCCNV) infection of tobacco plants could increase the fecundity and longevity of B. tabaci MEAM1 (Jiu et al., 2007; Zhang et al., 2012; Luan et al., 2013; Li et al., 2014), while having no influence on B. tabaci MEAM1 on tomato, or negative influence on B. tabaci Asia II 3 on tomato (Liu et al., 2009).
TYLCCNV is a monopartite begomovirus, which is associated with tomato yellow leaf curl China betasatellite (TYLCCNB, genus Betasatellite, family Tolecusatellitidae) (Adams et al., 2017). TYLCCNB only encodes one protein βC1 (Cui et al., 2004). In recent studies, βC1 from betasatellites was found to enhance the symptoms of the helper viruses and suppress multiple host defense pathways, including transcriptional gene silencing, post-transcriptional gene silencing, and jasmonic acid (JA) signaling (Yang et al., 2011; Zhou, 2013). For example, TYLCCNB βC1 was found to subvert JA signaling pathway in plants through interacting with the transcription factor MYC2 protein, thereby increasing the performance of whiteflies (Zhang et al., 2012; Li et al., 2014). However, whether other mechanisms are involved in the manipulation of plant defense against whiteflies by TYLCCNB βC1 remains obscure.
JA, one of the most intensively studied plant hormones, has emerged as a major signaling molecule that integrates information perceived at the plant-insect interface into broad spectrum defense responses (Erb et al., 2012; Verma et al., 2016; Berens et al., 2017). JA can promote the accumulation of metabolites like indole-glucosinolates, camalexin, the non-protein amino acid Nδ-acetylornithine and terpenes that are detrimental to phloem-feeding insects, like green peach aphid (GPA) and whitefly (Mewis et al., 2005; Goggin, 2007; Louis and Shah, 2013; Zhao et al., 2019). Foliage spray of methyl jasmonate (MeJA) can improve plant resistance to whiteflies (Zhang et al., 2012). The JA-mediated production of terpenoids reduce performance of whiteflies on tobacco plants (Luan et al., 2013). The COI1-JAZ-MYC signaling complex, which is constituted by coronatine insensitive 1 (COI1), jasmonate ZIM-domain (JAZ), and helix-loop-helix transcription factor MYC2, is a central regulatory module in JA signaling cascade. Stress responsive jasmonoyl-L-isoleucine (JA-Ile) directly promotes the interaction between JAZ and COI1, leading to ubiquitination and degradation of JAZ protein via 26S proteasome, thereby derepressing MYC-mediated transcriptional reprogramming (Pauwels and Goossens, 2011; Zhang et al., 2017). Recently, Jia et al. (2016) found that cotton leaf curl Multan betasatellite (CLCuMuB) βC1 can interact with tobacco S-phase kinase associated protein 1 (SKP1) to subvert plant ubiquitination, thus promoting virus infection and disease symptom induction. However, whether TYLCCNB βC1 can interact with plant SKP1, increase the stability of plant JAZ proteins and suppress plant resistance to whiteflies remain unknown.
In this study, TYLCCNB βC1 was used as a bait to search for binding proteins in tobacco by yeast two-hybrid screening. We found that βC1 interacted with Nicotiana tabacum SKP1 protein (NtSKP1) and verified the interaction via bimolecular fluorescence complementation (BiFC) and GST pull-down. Then, bioassays were conducted to examine how βC1-NtSKP1 interaction impacts plant defense against whiteflies. Furthermore, transcript level of genes in JA signaling pathway and contents of JAZ1 protein were analyzed to explore the contribution of NtSKP1 to plant defense responses against whiteflies.
Materials and Methods
Plants, Virus, Betasatellite, and Whitefly
Nicotiana tabacum var. NC89, N. benthamiana line 16c, and N. benthamiana line H2B-RFP were provided by the Institute of Biotechnology, Zhejiang University. All plants used were grown in climate chamber at 26 ± 2°C under 14 h light/10 h dark cycle with 60% humidity. Infectious clones of TYLCCNV (Y10, GenBank Accession No.: AJ319675.1) and TYLCCNB (AJ781200) were described previously (Cui et al., 2004). A population of whitefly (Middle East Asia Minor 1, MEAM1) was collected in Hangzhou, China and maintained on cotton (Gossypium hirsutum L. cv. Zhemian 1793) in climate chamber. The purity of the culture was monitored every three generations using PCR-restriction fragment length polymorphism (PCR-RFLP) and mtCO1 (GQ332577) sequencing analysis as described before (Qin et al., 2013).
Plasmid Construction and Transformation
Coding sequences of βC1 and NtSKP1 were cloned into pClone007 for sequencing (TSINGKE, China). Plant expression constructs (p2YC-βC1, p2YN-NtSKP1, pCambia1305-NtSKP1-GFP, pCambia1305-βC1-GFP, pBIN2mDNA1-NtSKP1) and prokaryotic expression recombinant protein constructs (pGEX-6P-1-GST-βC1, pMAL-c5x-MBP-NtSKP1) were constructed using restriction endonuclease and T4 DNA ligase. Target fragments and plasmids were digested by specific restriction endonucleases (Thermo Scientific, USA) at 37°C for 2 h. Then, fragments were ligated to plasmids using T4 DNA ligase (Thermo Scientific, USA) at 22°C for 10 min.
Recombinant plasmids used for expression in plant were transformed into Agrobacterium tumefaciens (EHA105) by electroporation. Recombinant plasmids used for prokaryotic expression were transformed into Escherichia coli (BL21) using heat-shock method.
Yeast Two Hybrid Assay
Forward primer (5′- CGCGGATCCATATGACTATCAAATACAA -3′) and reverse primer (5′-CCGCTCGAGTCATACATCTGAATTTG-3′) were used to amplify TYLCCNB βC1. Full-length of βC1 was amplified by PCR, digested by BamHI and XhoI, then cloned into yeast vector pGBKT7. Plasmid pGBKT7-βC1 and tobacco cDNA library were co-transformed into Saccharomyces cerevisiae Gold strain according to Yeastmaker Yeast Transformation System 2 Kit (Clontech, USA). Transformants were allowed to grow on synthetic medium -Leu/-Trp at 30°C for 3 days and then transferred to selective medium -Leu/-Trp/-His/-Ade X-a-Gal/AbA. Five days later, positive clones were transferred into fresh YPDA liquids (30°C, 250 rpm) for 24 h. cDNA library plasmids in yeast positive clones were extracted and transformed into E. coli (DH5α) for sequencing.
Verification of the interaction between βC1 and NtSKP1 was performed according to Yeastmaker Yeast Transformation System 2 Kit (Clontech, USA). Forward primer (5′-CATATGATGTCCTCCTCAAAG-3′) and reverse primer (5′- GAATTCTCACTCAAATGCCCA-3′) were used to amplify NtSKP1. Full-length of NtSKP1 was amplified by PCR, digested by NdeI and EcoRI, then cloned into pGADT7. Plasmid pGBKT7-βC1 and pGADT7-NtSKP1 were co-transformed into S. cerevisiae Gold strain. Transformants were grown on synthetic medium -Leu/-Trp at 30°C for 3 days and then transferred to selective medium -Leu/-Trp/-His/-Ade X-a-Gal/AbA.
Agrobacterium-Mediated Transient Expression
Agrobacterium containing plant expression constructs (p2YC-βC1, p2YN-NtSKP1, pCambia1305- NtSKP1-GFP, pCambia1305-βC1-GFP, pBIN2mDNA1-NtSKP1) were grown in LB liquid medium (50 μg/ml kanamycin, 50 μg/ml rifampicin) at 28°C, 200 rpm until OD600 reached 0.6. Fresh agrobacterium cultures were centrifuged (4,000 rpm,10 min, room temperature) and resuspended in infiltration buffer (10 mM MgCl2, 10 mM MES, and 200 μM acetosyringone, pH 5.6) until OD600 reached 1.0. Then the agrobacterium was kept at room temperature for at least 3 h. Suspension liquid was infiltrated into whole leaf using a needleless syringe.
Bimolecular Fluorescence Complementation (BiFC) Assay
Forward primer (5′-CCCTTAATTAACATGACTATCAAATACAACAA-3′) and reverse primer (5′-GGGACTAGTTACATCTGAATTTGTAAATA-3′) were used to amplify TYLCCNB βC1. Forward primer (5′-CCCTTAATTAACATGTCCTCCTCAAAGATGAT -3′) and reverse primer (5′-GGGACTAGTCTCAAATGCCCAAGCATTCT-3′) were used to amplify NtSKP1. Full-length of βC1 and NtSKP1 were digested by PacI and SpeI, then cloned into p2YC and p2YN vector respectively. Recombinant plasmids p2YC-βC1 and p2YN-NtSKP1 were transformed into agrobacterium respectively. Agrobacterium cultures containing p2YC-βC1 and p2YN-NtSKP1 were co-infiltrated into leaves of N. benthamiana line H2B-RFP. Next, 36-72 h after infiltration, RFP and YFP fluorescence were examined by Zeiss LSM710 confocal microscope.
GST Pull-Down Assay
Forward primer (5′-CGCGGATCCATGACTATCA AATACAA-3′) and reverse primer (5′-CCGCTCGAGTCATACATCTGAATTTG-3′) were used to amplify TYLCCNB βC1. Forward primer (5′-ATAAGAATGCGGCCGCATGTCCTCCTCAA-3′) and reverse primer (5′-CGCGGATCCTCACTCAAATGCC-3′) were used to amplify NtSKP1. Full-length of βC1 was digested by BamHI and XhoI, then cloned into vector pGEX-6P-1, and full-length of NtSKP1 was digested by NotI and BamHI, then cloned into vector pMal-c5x. Then, GST-βC1 and MBP-NtSKP1 fusion proteins were expressed in E. coli (BL21). MBP-NtSKP1 was purified by Amylose resin (NEB), eluted with Elution buffer (20 mM Tris-HCl, 200 mM NaCl, 1 mM DTT, 1 mM EDTA, 10 mM maltose, PH=7.4) and desalted by disposable PD-10 desalting columns (GE Healthcare). GST-βC1 was purified by Glutathione Sepharose 4 Fast Flow (GE Healthcare) and then used to pull down MBP-NtSKP1 in vitro for 2 h at 4°C. The beads were washed five times with ice-cold 1×PBS. Then washed beads were boiled with SDS loading buffer for 10 min. Proteins were separated by SDS-PAGE and detected by western blot with anti-MBP tag mouse monoclonal antibodies (Abcam, UK).
Phylogenetic Analysis
The amino acid sequences of SKP1 in different organisms were downloaded from NCBI database. These sequences were aligned by ClustalW. Phylogenetic analysis was conducted with MEGA 7.0 through neighbor-joining (NJ) method (1,000 replications).
Subcellular Localization Assay
Forward primer (5′-CGCGGATCCATGTCCTCCTCAAAG-3′) and reverse primer (5′-CGAGCTCTCACTCAAATGCCCA-3′) were used to amplify NtSKP1. Full-length of NtSKP1 was digested by BamHI and SacI, then cloned into the pCambia1305-GFP vector and transformed into agrobacterium. Agrobacterium clones expressing NtSKP1-GFP were infiltrated into leaves of N. benthamiana line H2B-RFP. Next, 36–72 h after infiltration, RFP and GFP fluorescence were observed by Zeiss LSM710 confocal microscope.
Virus Induced Gene Silencing (VIGS) Assay
About 300 bp fragment of NtSKP1 was amplified by forward primer (5′-CGCGGATCCCTACGATGTCCTCCTCAA-3′) and reverse primer (5′-TGCTCTAGATCAGAATCAAAGCCCTTA-3′). The fragment was digested by BamHI and XbaI, then cloned into the pBIN2mDNA1 vector, and the construct was transformed into agrobacterium. The method of VIGS was performed as previously described (Huang et al., 2011). After 15–20 days, total RNAs of tobacco plants were isolated to determine the silencing efficiency by qRT-PCR.
qRT-PCR and Data Analysis
Total RNAs of tobacco plants were isolated using TRIzolTM method. cDNA was synthesized using the PrimeScriptTM RT reagent Kit with gDNA Eraser (Takara, Dalian, China). qRT-PCRs were performed with the SYBR® Premix Ex TaqTM II (Takara, Dalian, China) using BIO-RAD CFX96 PCR System (Bio-Rad, California, USA). Glceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as internal control. The relative transcript levels of genes were calculated by the 2-ΔΔCT method. Nucleotide sequences of the gene-specific primers were as follows: NtSKP1 (forward primer, 5′-TGGCTGCCAACTACTTGAAC-3′; reverse primer, 5′-TCTCCTCTGGTGTCTTCCCT-3′); MYC2b (forward primer, 5′-ATCGGATGGGATGCTATGA-3′; reverse primer, 5′-GAAGCTGCTCTTGCGTGTA-3′); PDF1.2 (forward primer, 5′- GGAAATGGCAAACTCCATGCG-3′; reverse primer, 5′-ATCCTTCGGTCAGACAAACG-3′); GAPDH (forward primer, 5′-GCAGTGAACGACCCATTTATCTC-3′; reverse primer, 5′-AACCTTCTTGGCACCACCCT-3′).
Assessment of Plant Suitability to Whitefly
Newly emerged whiteflies (1–3 days) were used in bioassay. For the assessment of NtSKP1-silenced and control tobacco plants to whitefly, each cohort of five females and five males was released in a clip cage fixed on the abaxial surface of tobacco leaf. The number of live adults and total eggs laid were counted 7 days later. For the assessment of NtSKP1-overexpressed and control tobacco plants to whiteflies, number of live adults and total eggs whiteflies laid were counted 3 days after the release of whiteflies. Ten plants were used for each treatment in all experiments. The experiment was repeated three times.
Plant Protein Extraction and Western Blot of JAZ1
Total proteins of each leaf sample were extracted as previously described (Ryu et al., 2007). About 0.1 g of leaf samples were triturated and then lysed with buffer (20 mM Tris-HCl, pH 7.0, 250 mM sucrose, 25% glycerol, 20 mM KCl, 2 mM EDTA, 2.5 mM MgCl2, 30 mM β-mercaptoethanol, protease inhibitor cocktail, and 0.7% Triton X-100). Leaf samples in buffer were centrifuged for 30 min at 4°C. The supernatant was used for western blot analysis.
Protein extracts were boiled with SDS loading buffer for 10 min, and proteins were separated by SDS-PAGE and detected with anti-JAZ1 antibodies as previously described (Li P. et al., 2019).
Results
βC1 Interacts With NtSKP1 In Vivo and In Vitro
To further elucidate the mechanism underlying the plant-mediated indirect mutualism between TYLCCNV/TYLCCNB and whitefly, βC1 was used as a bait to screen the tobacco cDNA library by yeast two-hybrid system. NtSKP1, which is an indispensable subunit in plant Skp1-cullin-F-box (SCF) complex (Figure 1A), was captured. NtSKP1 comprises a 468 bp open reading frame (ORF), encoding a 155 aa protein (≈17.53 kDa).
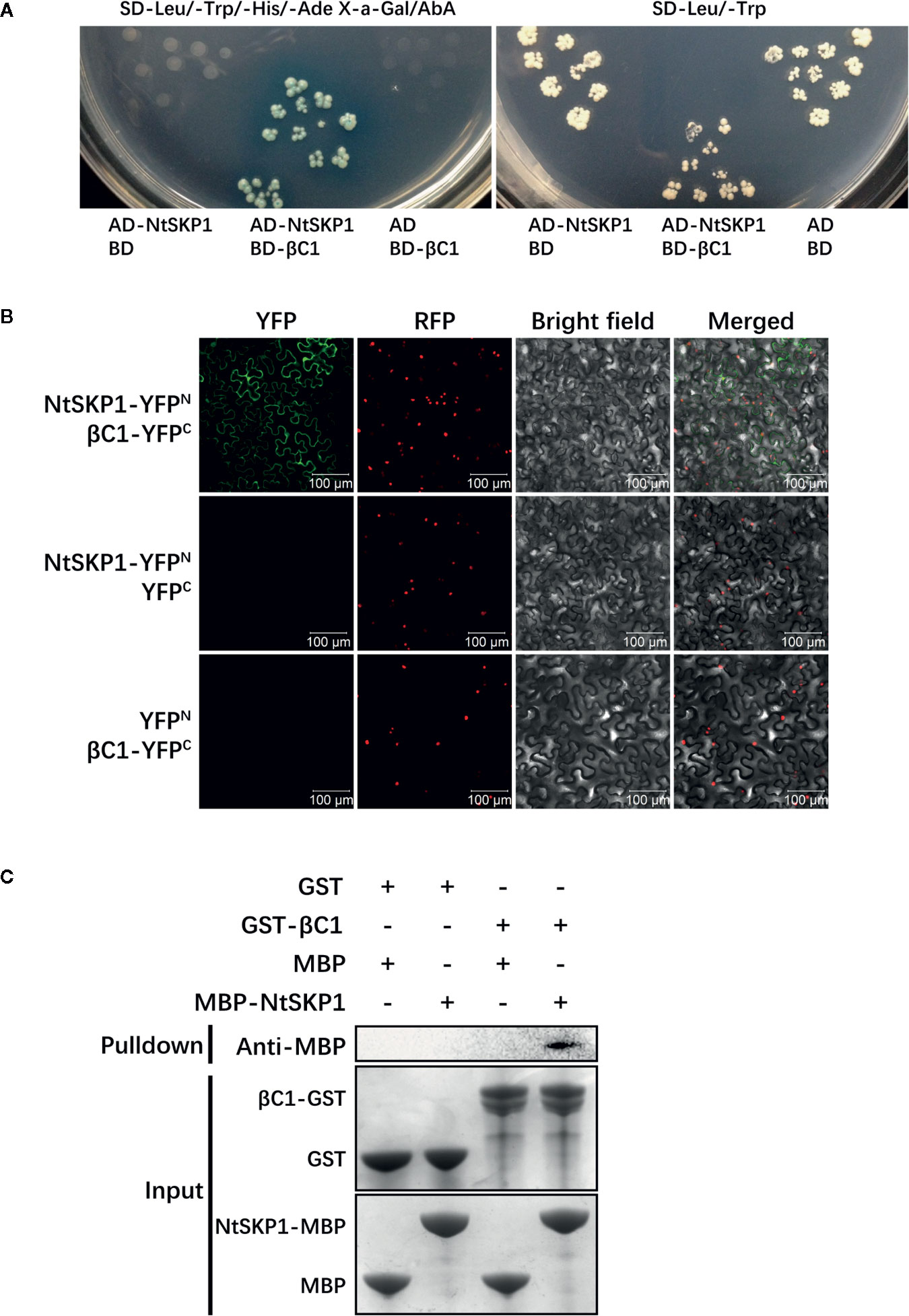
Figure 1 The interaction between βC1 and NtSKP1. (A) Interaction confirmation of βC1 and NtSBT1.7 via yeast two-hybrid system. Yeast strain Y2H Gold co-transformed indicated plasmids were grown on SD-Leu/-Trp/-His/-Ade X-a-Gal/AbA and SD-Leu/-Trp. Empty plasmids pGADT7/pGBKT7 were used as negative controls. (B) In vivo interaction confirmation of βC1 and NtSBT1.7 via BiFC analysis. βC1-YFPC and NtSKP1-YFPN were transiently co-expressed in N. benthamiana line H2B-RFP, of which nucleus were marked with RFP fusion protein. Photos were imaged at 48 h using a Zeiss LSM710 confocal microscope. Columns from left to right represent YFP fluorescence, RFP fluorescence, bright field, and YFP/RFP/bright field overlay. Empty plasmids were negative controls. Scale bars 100 μm. (C) In vitro interaction confirmation of βC1 and NtSBT1.7 via GST pull-down assay. Protein GST, βC1-GST, MBP, NtSKP1-MBP were expressed by prokaryotic expression, and purified by Glutathione agarose beads or Amylose resin. GST or GST-βC1 fusion proteins were used to pulldown MBP or MBP-NtSKP1 fusion proteins. Binding proteins were analyzed via SDS-PAGE and western blot assays using anti-MBP antibodies. At the start of Samples (Input) were stained by Coomassie blue solution. GST, MBP proteins were used as negative controls.
To verify the interaction between βC1 and NtSKP1, BiFC assay was performed in vivo. Agrobacterium containing βC1-YFPC or NtSKP1-YFPN were co-infiltrated into leaves of N. benthamiana (line H2B-RFP). Fluorescence confocal diagram showed strong YFP fluorescence in leaves expressing βC1-YFPC and NtSKP1-YFPN while no fluorescence was found in negative controls (Figure 1B), indicating the interaction between βC1 and NtSKP1 in plants.
Then, GST pull-down assay was performed to determine whether βC1 interacts with NtSKP1 in vitro. Fusion proteins GST-βC1 and MBP-NtSKP1 were expressed through prokaryotic expression. Figure 1C showed MBP-NtSKP1 could be captured by GST-βC1, and no signal was detected in negative controls.
Phylogenetic Analysis, Domain Analysis, and Subcellular Localization of NtSKP1
SKP1 commonly exists in eukaryotic organisms (Kong et al., 2004). Phylogenetic tree analysis showed that phylogenetically NtSKP1 was relatively closer to that in N. benthamiana and N. tomentosiformis than other plants, and it differed greatly from other organisms, such as microorganism or insects (Figure 2A). As a bridge between Cullin and F-box protein, SKP1 was composed of two typical conserved domains: tetramerisation domain and dimerisation domain (Figure 2B). The subcellular localization assay showed that protein NtSKP1 accumulated in nucleus and cytoplasm (Figure 2C).
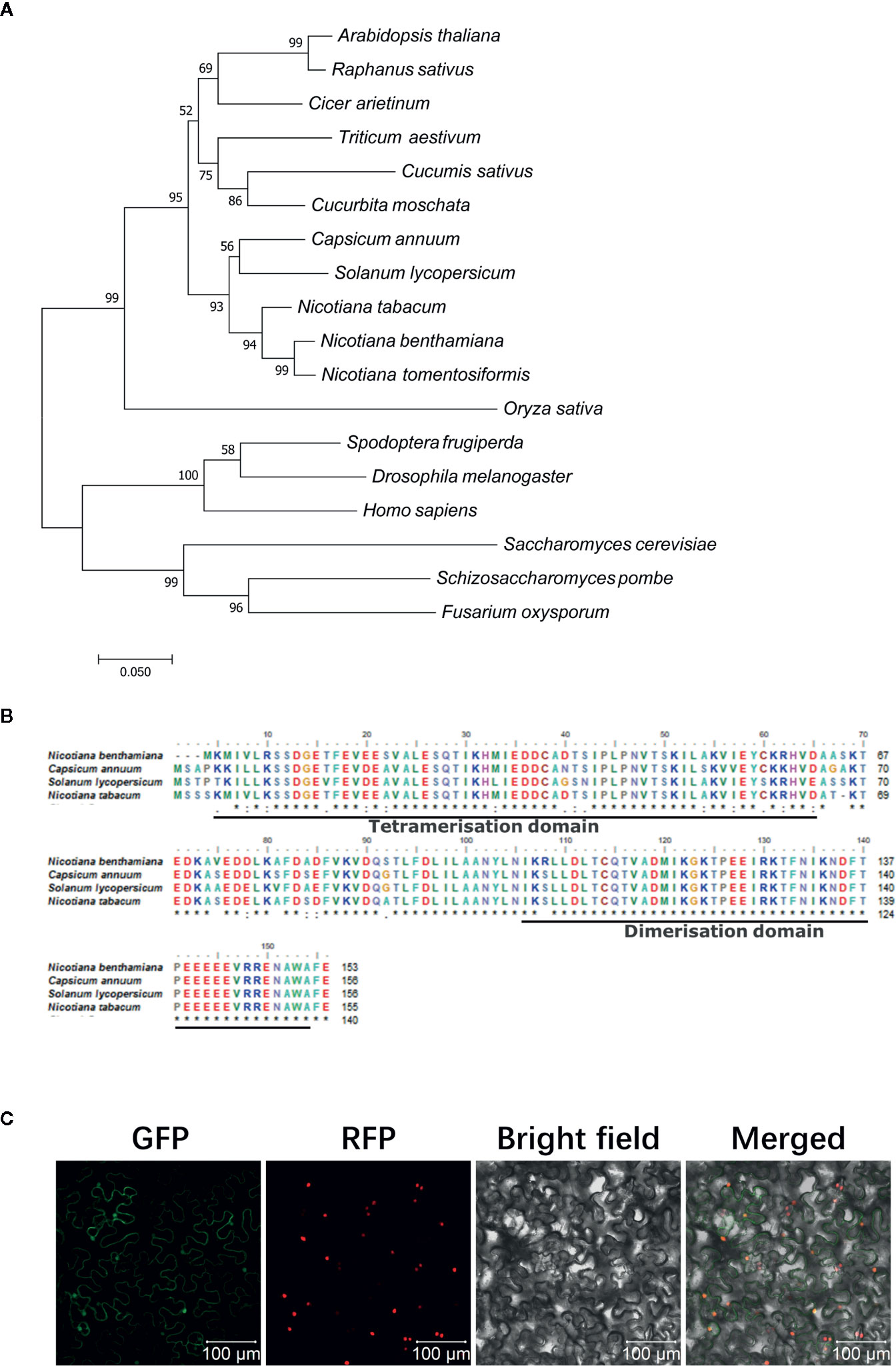
Figure 2 Phylogenetic analysis, sequence comparison, and subcellular localization of NtSKP1. (A) Phylogenetic tree analysis of SKP1. Accession numbers of the amino acid sequences included in the phylogenic tree as following as: Nicotiana benthamiana (AAO85510.1); common tobacco Nicotiana tabacum (XP_016435415.1); tobacco Nicotiana tomentosiformis (XP_009620406.1); radish Raphanus sativus (XP_018446355.1); chilly Capsicum annuum (AAX83944.1); tomato Solanum lycopersicum (ALA48968.1); Arabidopsis thaliana (NP_565123.1); wheat Triticum aestivum (AIJ50267.1); cucumber Cucumis sativus (XP_004146156.1); squash Cucurbita moschata (XP_022923340.1); Cicer arietinum (NP_001352203.1); rice Oryza sativa (AAT09201.1); fall armyworm Spodoptera frugiperda (AGP04177.1); fruit fly Drosophila melanogaster (NP_001284 755.1); human Homo sapiens (NP_008861.2); brewer’s yeast Saccharomyces cerevisiae (NP_010615.3); fission yeast Schizosaccharomyces pombe (BAB62325.1); Fusarium oxysporum (AAT85970.1). These sequences were aligned by ClustalW. Phylogenetic analysis was conducted with MEGA 7.0 through neighbor-joining (NJ) method (1,000 replications). (B) Amino acid sequence alignment and domain analysis of SKP1 proteins among Nicotiana benthamiana, Capsicum annuum, Solanum lycopersicum, and Nicotiana tabacum. (C) Subcellular localization of NtSKP1. Fusion protein NtSKP1-GFP were transiently expressed in N. benthamiana line H2B-RFP, of which nucleus were marked with RFP fusion protein. Photos were imaged at 48 h using a Zeiss LSM710 confocal microscope. Columns from left to right represent GFP fluorescence, RFP fluorescence, bright field, and GFP/RFP/bright field overlay. Scale bars 100 μm.
NtSKP1 Contributes to Plant Immunity Against Whitefly
To investigate the function of NtSKP1 in plant defense against whitefly, NtSKP1 was transiently expressed in tobacco plants (Figure 3A). The survival rate and fecundity of whiteflies had no significant differences between NtSKP1-overexpressed tobacco plants and control plants (Figures 3B, C). These results revealed that the overexpression of NtSKP1 did not affect plant defense response against whiteflies. However, when NtSKP1 was knocked down via VIGS (Figure 3D), the survival rate and fecundity of whiteflies on NtSKP1-silenced tobacco plants were significantly higher than control plants (Figures 3E, F). These results indicated that NtSKP1 positively regulated plant defenses against whiteflies.
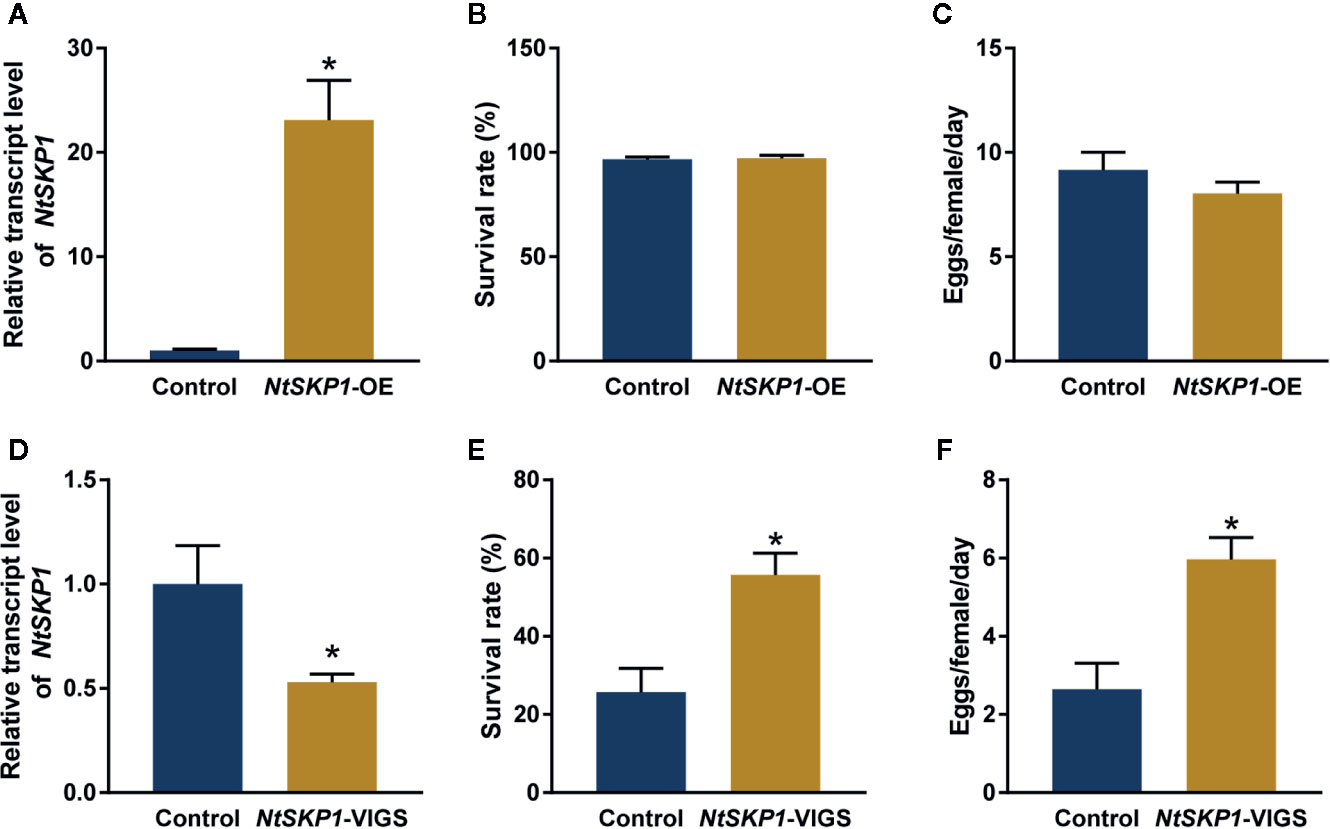
Figure 3 NtSKP1 regulates tobacco defense against whitefly. (A) Relative transcript levels of NtSKP1 in control and NtSKP1-overexpressed (NtSKP1-OE) tobacco plants. GFP or NtSKP1-GFP fusion proteins were transiently expressed in N. tabacum var. NC89. Total RNAs were extracted from each plant respectively at 48 h and subjected to qRT-PCR to quantify the mRNA levels of NtSKP1. GAPDH was used as the internal reference. Values are mean ± SEM, n = 5. (B) Survival rates of whiteflies on control and NtSKP1-overexpressed tobacco plants. GFP or NtSKP1-GFP fusion proteins were transiently expressed in N. tabacum var. NC89. Each cohort of five females and five males was released in a clip cage fixed on the abaxial surface of tobacco leaf at 48 h. The numbers of live adults were counted 3 days later. Values are mean ± SEM, n = 20. (C) Average numbers of eggs one day laid by per female whitefly on control and NtSKP1-overexpressed tobacco plants. GFP or NtSKP1-GFP fusion proteins were transiently expressed in N. tabacum var. NC89. Each cohort of five females and five males was released in a clip cage fixed on the abaxial surface of tobacco leaf at 48 h. The numbers of eggs were counted 3 days later. Values are mean ± SEM, n = 20. (D) Relative transcript levels of NtSKP1 in control and NtSKP1-silenced (NtSKP1-VIGS) tobacco plants. Gene silencing was achieved by VIGS assay. Total RNAs were extracted from each plant respectively and subjected to qRT-PCR to quantify the mRNA levels of NtSKP1. GAPDH was used as the internal reference. Values are mean ± SEM, n = 5. (E) Survival rates of whiteflies on control and NtSKP1-silenced tobacco plants. Gene silencing was achieved by VIGS assay. Each cohort of five females and five males was released in a clip cage fixed on the abaxial surface of tobacco leaf. The numbers of live adults were counted 7 days later. Values are mean ± SEM, n = 30. (F) Average numbers of eggs one day laid by per female whitefly on control and NtSKP1-silenced tobacco plants. Gene silencing was achieved by VIGS assay. Each cohort of five females and five males was released in a clip cage fixed on the abaxial surface of tobacco leaf. The numbers of eggs were counted 7 days later. Values are mean ± SEM, n = 30. Student’s t test was used for significant difference analysis (*P < 0.05).
NtSKP1 Modulates the Transcript Level of Genes in JA Signaling Pathway
SCF complex functions as a central regulator in JA signaling (Marino et al., 2012; Shu and Yang, 2017; Kelley, 2018). As JA functions in plant defense against whiteflies (Sun et al., 2017; Zhang et al., 2018), we wondered whether NtSKP1 impacted the JA signaling pathway in tobacco. Thus, transcript level of JA signaling pathway gene MYC2b and plant defensin gene PDF1.2 was measured by RT-qPCR. Our results showed that the transcript level of PDF1.2 was induced in NtSKP1-overexpressed plants (Figures 4A, B). In NtSKP1-silenced plants, the transcript level of MYC2b and PDF1.2 was down-regulated (Figures 4C, D). Taken together, these results showed that NtSKP1 participated in regulating JA signaling pathways, thereby promoting whitefly performance.
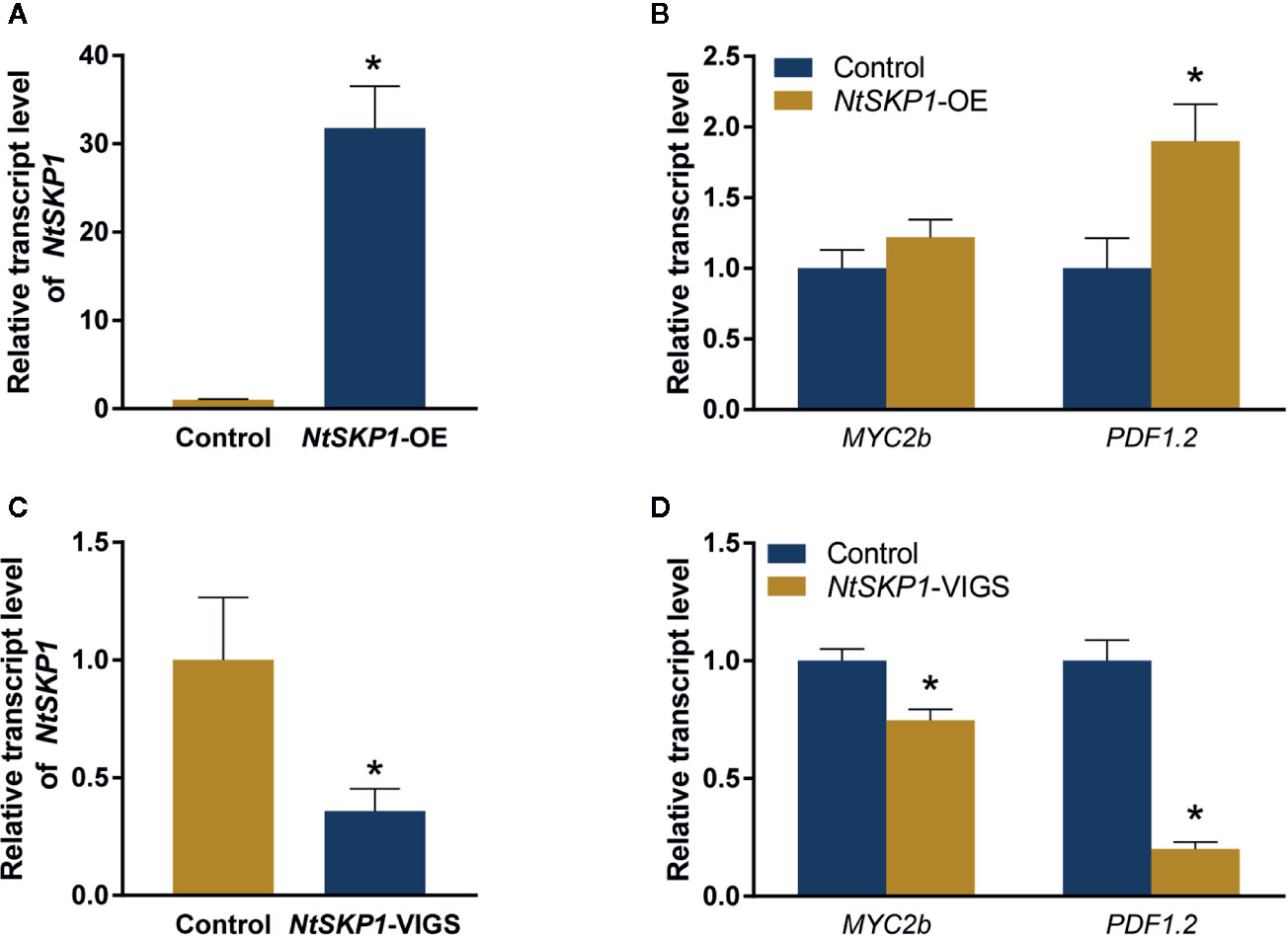
Figure 4 NtSKP1 affects the transcription of genes in JA signaling pathway. (A) Relative transcript levels of NtSKP1 in control and NtSKP1-overexpressed (NtSKP1-OE) tobacco plants. Values are mean ± SEM, n = 6. (B) Relative transcript levels of MYC2b and PDF1.2 in control tobaccos (Control) and NtSKP1-overexpressed (NtSKP1-OE) tobaccos. Values are mean ± SEM, n = 6. (C) Relative transcript levels of NtSKP1 in control and NtSKP1-silenced (NtSKP1-VIGS) tobacco plants. Values are mean ± SEM, n = 6. (D) Relative transcript levels of MYC2b and PDF1.2 in control tobaccos (Control) and NtSKP1-silenced (NtSKP1-VIGS) tobaccos. Values are mean ± SEM, n = 6. Total RNAs were extracted from each plant respectively and subjected to qRT-PCR to quantify the mRNA levels of target genes. GAPDH was used as the internal reference. Student’s t test was used for significant difference analysis (*P < 0.05).
βC1 Targets NtSKP1 to Regulate Plant Immunity Through Interfering With JAZ1 Degradation
JA-mediated responses are restrained by JAZ proteins, which are transcription repressors through binding to and repressing the activity of MYCs (Zhang et al., 2017). NtSKP1 is a component of SCF complex and plays an important role in ubiquitination-mediated protein degradation (Zeng et al., 2006; Shu and Yang, 2017; Zhou and Zeng, 2017; Serrano et al., 2018). We examined whether NtSKP1 took part in the turnover of JAZ1, thus modulating JA signaling pathway. Figure 5A showed that JAZ1 protein had higher accumulation in NtSKP1-silenced plant. In NtSKP1-overexpressed plant, the accumulation of JAZ1 was lower than control plant (Figure 5B). Similarly, JAZ1 was more stable in βC1-expressed plant (Figure 5C). Taken together, βC1 might interfere the degradation of JAZ1 through interacting with NtSKP1, thus suppressing the plant JA signaling pathway.

Figure 5 NtSKP1 and βC1 interfere the degradation of JAZ1 protein. (A) Western blot assay of JAZ1 protein in NtSKP1-silenced (NtSKP1-VIGS) tobacco plants. Gene silencing was achieved by VIGS assay. (B) Western blot assay of JAZ1 protein in NtSKP1-overexpressed (NtSKP1-OE) tobacco plants. GFP or NtSKP1-GFP fusion proteins were transiently expressed in N. tabacum var. NC89 for 48 h. (C) Western blot assay of JAZ1 protein in βC1-expressed (βC1-EX) tobacco plants. YFPC or βC1-YFPC fusion proteins were transiently expressed in N. tabacum var. NC89 for 48 h. Total plant protein extracts were analyzed via SDS-PAGE and western blot assays using anti-JAZ antibodies. Actin was the internal reference. Intensity was detected by image J.
Discussion
Betasatellites associated to begomoviruses have been shown to be determinants in virus disease symptom induction, virus movement, repression of plant defense, and other aspects of viral pathogenesis (Zhou, 2013). βC1 interacts with a number of host proteins to affect phytohormone, methylation, ubiquitin, and photosynthesis pathways in plants (Gnanasekaran et al., 2019; Yang et al., 2019). Here, we found that TYLCCNB βC1 interacted with NtSKP1 to disrupt the integrality of SCF complex, thereby suppressing JAZ1 degradation and the subsequent activation of JA signaling pathways (Figure 6).
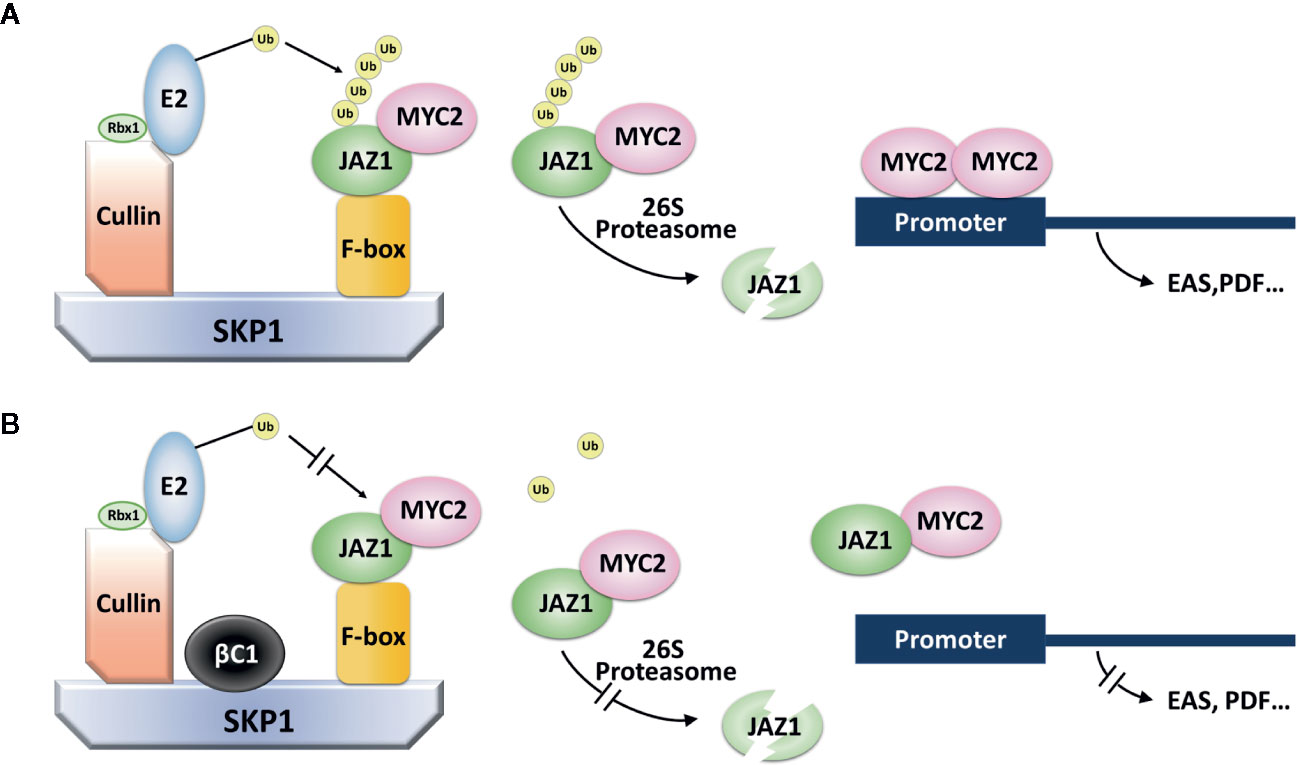
Figure 6 A model for the role of βC1 in regulating plant defense reaction. (A) The signaling pathway of SCF complex in plants. SCF complex is one kind of E3 ligase. E2-Ub complex can interact with E3 ligase via the help from Rbx1, making the E3 protein substrates (JAZ1) carry with polyubiquitin chain. Then, JAZ1-Ub chain is recognized by 26S proteasome, eventually degraded, releasing MYC2 transcription factor to activate genes expression in plant defense. (B) The pathway of SCF complex when plants were infected by TYLCCNV. βC1 can bind to SKP1 protein and interfere the normal function of SCF complex. JAZ1-MYC2 was stable and genes expression in plant defense were not activated.
SKP1 has been shown to be a target for several plant pathogen effectors, such as P7-2 from rice black streaked dwarf virus (Tao et al., 2017), βC1 from CLCuMuB (Jia et al., 2016), and P0 from brassica yellows virus (Li Y. et al., 2019). These pathogen effectors modulate plant immunity via damaging the integrity of SCF complex, thus establishing a successful infection. However, the role of SKP1 or SCF complex in plant defense against insects have not been proved yet. Kottapalli et al. (2006) found SKP1 was in eight tandem repeats on chromosome 9 and responsible for gall midge resistance phenotype. Hence, SKP1 may be a candidate gene in rice for gall midge insect resistance. In this study, we found the in vivo and in vitro interaction between TYLCCNB βC1 and NtSKP1 (Figure 1), and we confirmed that whiteflies performed better on NtSKP1-silenced tobacco plants (Figures 3E, F). Higher survival rate of whitefly adults and more offsprings on plant can in turn facilitate the spread of begomoviruses. However, the overexpression of NtSKP1 did not affect the survival rate and fecundity of whiteflies (Figures 3B, C). Perhaps the background expression of NtSKP1 is sufficient to achieve its function in plants, or JA signaling pathway have some mechanisms to alleviate or balance the overexpression of NtSKP1. These results showed a novel function of NtSKP1 in plant defense against insect herbivores, and the involvement of NtSKP1 in TYLCCNV/TYLCCNB-whitefly mutualism.
Previous studies have been conducted to investigate the role of JA in TYLCCNV/TYLCCNB-plant-whitefly tripartite interactions. TYLCCNV can inhibit the JA-biosynthesis genes FAD3 and FAD7, as well as JA-regulated gene PDF1.2. Also, it reduces the JA level through a SA-independent mechanism in tobacco plants (Zhang et al., 2012). TYLCCNB βC1 can interact with MYC2, a transcription factor in JA signaling pathway, to suppress the synthesis of tobacco terpenoids, thus improving the performance of whiteflies (Luan et al., 2013; Li et al., 2014). In this study, we found NtSKP1 contributed to JA signaling pathway and regulated the transcription of MYC2b and PDF1.2 (Fig 4). Furthermore, SCF complex, which consisted by SKP1, Cullin, and F-box proteins, is a kind of ubiquitination E3 ligase, and it plays prominent roles in the perception of many phytohormones including JA, gibberellin, ethylene, salicylic acid, and strigolactones (Yu et al., 2007; Kelley, 2018). SCF complex in Arabidopsis is essential for JA signaling, and it leads to the ubiquitination and degradation of JAZ protein via 26S proteasome to derepress MYC-mediated transcriptional reprogramming (Xu et al., 2002; Zhang et al., 2017). In this study, we verified NtSKP1 could impact the stabilization of JAZ1 protein (Figure 5), which might explain the role of SKP1 in JA signaling pathway. These results demonstrated that TYLCCNB βC1 might implement more than one strategy to suppress plant defense responses, and finally achieve the mutualism between TYLCCNV/TYLCCNB and whiteflies.
Taken together, our study uncovers a novel mechanism underlying the manipulation of whitefly-plant interactions by a begomovirus-associated betasatellite. The findings in this study not only add to our knowledge of βC1 function in modulating plant physiology, but also help to elucidate the nature of whitefly-plant interactions.
Data Availability Statement
All datasets presented in this study are included in the article/supplementary material.
Author Contributions
CZ designed and carried out the experiments, analyzed data, and wrote the first draft of the manuscript. Y-NS did the protein interaction verification assay and did the bioassays of the whitefly. J-JY and NC did the bioassays of the whitefly. L-LP, JZ, S-SL, and X-WW conceived the study and revised the manuscript.
Funding
Financial support for this study was provided by the National Natural Science Foundation of China (31930092, 31925033) and the Zhejiang Provincial Natural Science Foundation of China (LZ20C140003).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge the financial supports by the National Natural Science Foundation of China under Grant numbers 31930092 and 31925033 and the Zhejiang Provincial Natural Science Foundation of China under Grant numbers LZ20C140003. We also thank our lab engineer Gen-Hong Yan for his effort in plant growing.
References
Adams, M. J., Lefkowitz, E. J., King, A. M. Q., Harrach, B., Harrison, R. L., Knowles, N. J., et al. (2017). Changes to taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2017). Arch. Virol. 162, 2505–2538. doi: 10.1007/s00705-017-3358-5
Berens, M. L., Berry, H. M., Mine, A., Argueso, C. T., Tsuda, K. (2017). Evolution of hormone signaling networks in plant defense. Annu. Rev. Phytopathol. 55, 401. doi: 10.1146/annurev-phyto-080516-035544
Brown, J. K., Zerbini, F. M., Navas-Castillo, J., Moriones, E., Ramos-Sobrinho, R., Silva, J. C., et al. (2015). Revision of Begomovirus taxonomy based on pairwise sequence comparisons. Arch. Virol. 160, 1593–1619. doi: 10.1007/s00705-015-2398-y
Carmo-Sousa, M., Moreno, A., Garzo, E., Fereres, A. (2014). A non-persistently transmitted-virus induces a pull–push strategy in its aphid vector to optimize transmission and spread. Virus Res. 186, 38–46. doi: 10.1016/j.virusres.2013.12.012
Cui, X., Tao, X., Xie, Y., Fauquet, C. M., Zhou, X. (2004). A DNA beta associated with tomato yellow leaf curl China virus is required for symptom induction. J. Virol. 78, 13966–13974. doi: 10.1128/JVI.78.24.13966-13974.2004
De Barro, P. J., Liu, S. S., Boykin, L. M., Dinsdale, A. B. (2011). Bemisia tabaci: a statement of species status. Annu. Rev. Entomol. 56, 1–19. doi: 10.1146/annurev-ento-112408-085504
Erb, M., Meldau, S., Howe, G. A. (2012). Role of phytohormones in insect-specific plant reactions. Trends Plant Sci. 17, 250–259. doi: 10.1016/j.tplants.2012.01.003
Franco, F. P., Moura, D. S., Vivanco, J. M., Silva-Filho, M. C. (2017). Plant-insect-pathogen interactions: a naturally complex ménage à trois. Curr. Opin. Microbiol. 37, 54–60. doi: 10.1016/j.mib.2017.04.007
Gilbertson, R. L., Batuman, O., Webster, C. G., Adkins, S. (2015). Role of the insect supervectors Bemisia tabaci and Frankliniella occidentalis in the emergence and global spread of plant viruses. Annu. Rev. Virol. 2, 67–93. doi: 10.1146/annurev-virology-031413-085410
Gnanasekaran, P., Kishorekumar, R., Bhattacharyya, D., Kumar, R. V., Chakraborty, S. (2019). Multifaceted role of geminivirus associated betasatellite in pathogenesis: arms race between betasatellite and plant defense. Mol. Plant Pathol. 20, 1019–1033. doi: 10.1111/mpp.12800
Goggin, F. L. (2007). Plant-aphid interactions: molecular and ecological perspectives. Curr. Opin. Plant Biol. 10, 399–408. doi: 10.1016/j.pbi.2007.06.004
Hogenhout, S. A., Ammar, E. D., Whitfield, A. E., Redinbaugh, M. G. (2008). Insect vector interactions with persistently transmitted viruses. Annu. Rev. Phytopathol. 46, 327–359. doi: 10.1146/annurev.phyto.022508.092135
Huang, C. J., Zhang, T., Li, F. F., Zhang, X. Y., Zhou, X. P. (2011). Development and application of an efficient virus-induced gene silencing system in Nicotiana tabacum using geminivirus alphasatellite. J. Zhejiang Univ. Sci. B 12, 83–92. doi: 10.1631/jzus.b1000157
Islam, W., Akutse, K. S., Qasim, M., Khan, K. A., Ghramh, H. A., Idrees, A., et al. (2018). Bemisia tabaci-mediated facilitation in diversity of begomoviruses: evidence from recent molecular studies. Microb. Pathog. 123, 162–168. doi: 10.1016/j.micpath.2018.07.008
Islam, W., Noman, A., Naveed, H., Alamri, S. A., Hashem, M., Huang, Z., et al. (2020). Plant-insect vector-virus interactions under environmental change. Sci. Total Environ. 701, 135044. doi: 10.1016/j.scitotenv.2019.135044
Jia, Q., Liu, N., Xie, K., Dai, Y., Han, S., Zhao, X., et al. (2016). CLCuMuB βC1 subverts ubiquitination by interacting with NbSKP1s to enhance geminivirus infection in Nicotiana benthamiana. PLoS Pathog. 12, e1005668. doi: 10.1371/journal.ppat.1005668
Jiu, M., Zhou, X. P., Tong, L., Xu, J., Yang, X., Wan, F. H., et al. (2007). Vector-virus mutualism accelerates population increase of an invasive whitefly. PLoS One 2, e182. doi: 10.1371/journal.pone.0000182
Kanakala, S., Ghanim, M. (2019). Global genetic diversity and geographical distribution of Bemisia tabaci and its bacterial endosymbionts. PLoS One 14, e0213946. doi: 10.1371/journal.pone.0213946
Kelley, D. R. (2018). E3 ubiquitin ligases: key regulators of hormone signaling in plants. Mol. Cell. Proteomics 17, 1047–1054. doi: 10.1074/mcp.MR117.000476
Kong, H., Leebens-Mack, J., Ni, W., dePamphilis, C. W., Ma, H. (2004). Highly heterogeneous rates of evolution in the SKP1 gene family in plants and animals: functional and evolutionary implications. Mol. Biol. Evol. 21, 117–128. doi: 10.1093/molbev/msh001
Kottapalli, K. R., Sarla, N., Kikuchi, S. (2006). In silico insight into two rice chromosomal regions associated with submergence tolerance and resistance to bacterial leaf blight and gall midge. Biotechnol. Adv. 24, 561–589. doi: 10.1016/j.biotechadv.2006.05.003
Lee, W., Park, J., Lee, G. S., Lee, S., Akimoto, S. (2013). Taxonomic status of the Bemisia tabaci complex (Hemiptera: Aleyrodidae) and reassessment of the number of its constituent species. PLoS One 8, e63817. doi: 10.1371/journal.pone.0063817
Li, R., Weldegergis, B. T., Li, J., Jung, C., Qu, J., Sun, Y., et al. (2014). Virulence factors of geminivirus interact with MYC2 to subvert plant resistance and promote vector performance. Plant Cell 26, 4991–5008. doi: 10.1105/tpc.114.133181
Li, H., Liu, X., Liu, X., Michaud, J. P., Zhi, H., Li, K., et al. (2018). Host plant infection by soybean mosaic virus reduces the fitness of its vector, Aphis glycines (Hemiptera: Aphididae). J. Econ. Entomol. 111, 2017–2023. doi: 10.1093/jee/toy165
Li, P., Liu, C., Deng, W. H., Yao, D. M., Pan, L. L., Li, Y. Q., et al. (2019). Plant begomoviruses subvert ubiquitination to suppress plant defenses against insect vectors. PLoS Pathog. 15, e1007607. doi: 10.1371/journal.ppat.1007607
Li, Y. Y., Sun, Q., Zhao, T. Y., Xiang, H. Y., Zhang, X. Y., Wu, Z. Y., et al. (2019). Interaction between Brassica yellows virus silencing suppressor P0 and plant SKP1 facilitates stability of P0 in vivo against degradation by proteasome and autophagy pathways. New Phytol. 222, 1458–1473. doi: 10.1111/nph.15702
Liu, J., Zhao, H., Jiang, K., Zhou, X. P., Liu, S. S. (2009). Differential indirect effects of two plant viruses on an invasive and an indigenous whitefly vector: implications for competitive displacement. Ann. Appl. Biol. 155, 439–448. doi: 10.1111/j.1744-7348.2009.00366.x
Liu, S. S., Colvin, J., De Barro, P. J. (2012). Species concepts as applied to the whitefly Bemisia tabaci systematics: how many species are there? J. Integr. Agr. 11, 176–186. doi: 10.1016/s2095-3119(12)60002-1
Louis, J., Shah, J. (2013). Arabidopsis thaliana-Myzus persicae interaction: shaping the understanding of plant defense against phloem-feeding aphids. Front. Plant Sci. 4, 213. doi: 10.3389/fpls.2013.00213
Lozano, G., Trenado, H. P., Fiallo-Olive, E., Chirinos, D., Geraud-Pouey, F., Briddon, R. W., et al. (2016). Characterization of non-coding DNA satellites associated with sweepoviruses (Genus Begomovirus, Geminiviridae)-definition of a distinct class of begomovirus-associated satellites. Front. Microbiol. 7, 162. doi: 10.3389/fmicb.2016.00162
Luan, J. B., Yao, D. M., Zhang, T., Walling, L. L., Yang, M., Wang, Y. J., et al. (2013). Suppression of terpenoid synthesis in plants by a virus promotes its mutualism with vectors. Ecol. Lett. 16, 390–398. doi: 10.1111/ele.12055
Luan, J. B., Wang, X. W., Colvin, J., Liu, S. S. (2014). Plant-mediated whitefly-begomovirus interactions: research progress and future prospects. B. Entomol. Res. 104, 267–276. doi: 10.1017/s000748531400011x
Mansoor, S., Zafar, Y., Briddon, R. W. (2006). Geminivirus disease complexes: the threat is spreading. Trends Plant Sci. 11, 209–212. doi: 10.1016/j.tplants.2006.03.003
Marino, D., Peeters, N., Rivas, S. (2012). Ubiquitination during plant immune signaling. Plant Physiol. 160, 15–27. doi: 10.1104/pp.112.199281
Mewis, I., Appel, H. M., Hom, A., Raina, R., Schultz, J. C. (2005). Major signaling pathways modulate Arabidopsis glucosinolate accumulation and response to both phloem-feeding and chewing insects. Plant Physiol. 138, 1149–1162. doi: 10.1104/pp.104.053389
Navas-Castillo, J., Fiallo-Olive, E., Sanchez-Campos, S. (2011). Emerging virus diseases transmitted by whiteflies. Annu. Rev. Phytopathol. 49, 219–248. doi: 10.1146/annurev-phyto-072910-095235
Nawaz-Ul-Rehman, M. S., Fauquet, C. M. (2009). Evolution of geminiviruses and their satellites. FEBS Lett. 583, 1825–1832. doi: 10.1016/j.febslet.2009.05.045
Noman, A., Aqeel, M., Qasim, M., Haider, I., Lou, Y. (2020). Plant-insect-microbe interaction: a love triangle between enemies in ecosystem. Sci. Total Environ. 699, 134181. doi: 10.1016/j.scitotenv.2019.134181
Pauwels, L., Goossens, A. (2011). The JAZ Proteins: a crucial interface in the jasmonate signaling cascade. Plant Cell 23, 3089–3100. doi: 10.1105/tpc.111.089300
Peñaflor, M. F. G. V., Mauck, K. E., Alves, K. J., De Moraes, C. M., Mescher, M. C., Bell, J. R. (2016). Effects of single and mixed infections of bean pod mottle virus and soybean mosaic virus on host-plant chemistry and host-vector interactions. Funct. Ecol. 30, 1648–1659. doi: 10.1111/1365-2435.12649
Perefarres, F., Thierry, M., Becker, N., Lefeuvre, P., Reynaud, B., Delatte, H., et al. (2012). Biological invasions of geminiviruses: case study of TYLCV and Bemisia tabaci in Reunion Island. Viruses 4, 3665–3688. doi: 10.3390/v4123665
Qin, L., Wang, J., Bing, X. L., Liu, S. S. (2013). Identification of nine cryptic species of Bemisia tabaci (Hemiptera: Aleyrodidae) from China by using the mtCOI PCR-RFLP technique. Acta Entomol. Sin. 56, 186–194. in Chinese with English abstract. http://www.insect.org.cn/EN/Y2013/V56/I2/186.
Rojas, M. R., Macedo, M. A., Maliano, M. R., Soto-Aguilar, M., Souza, J. O., Briddon, R. W., et al. (2018). World management of Geminiviruses. Annu. Rev. Phytopathol. 56, 637–677. doi: 10.1146/annurev-phyto-080615-100327
Ryu, H., Kim, K., Cho, H., Park, J., Choe, S., Hwang, I. (2007). Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell 19, 2749–2762. doi: 10.1105/tpc.107.053728
Rzanny, M., Voigt, W. (2012). Complexity of multitrophic interactions in a grassland ecosystem depends on plant species diversity. J. Anim. Ecol. 81, 614–627. doi: 10.1111/j.1365-2656.2012.01951.x
Serrano, I., Campos, L., Rivas, S. (2018). Roles of E3 ubiquitin-ligases in nuclear protein homeostasis during plant stress responses. Front. Plant Sci. 9, 139. doi: 10.3389/fpls.2018.00139
Shi, X., Gao, Y., Yan, S., Tang, X., Zhou, X., Zhang, D., et al. (2016). Aphid performance changes with plant defense mediated by cucumber mosaic virus titer. Virol. J. 13, 70. doi: 10.1186/s12985-016-0524-4
Shu, K., Yang, W. (2017). E3 ubiquitin ligases: ubiquitous actors in plant development and abiotic stress responses. Plant Cell Physiol. 58, 1461–1476. doi: 10.1093/pcp/pcx071
Sun, Y. C., Pan, L. L., Ying, F. Z., Li, P., Wang, X. W., Liu, S. S. (2017). Jasmonic acid-related resistance in tomato mediates interactions between whitefly and whitefly-transmitted virus. Sci. Rep. 7, 566. doi: 10.1038/s41598-017-00692-w
Tao, T., Zhou, C. J., Wang, Q., Chen, X. R., Sun, Q., Zhao, T. Y., et al. (2017). Rice black streaked dwarf virus P7-2 forms a SCF complex through binding to Oryza sativa SKP1-like proteins, and interacts with GID2 involved in the gibberellin pathway. PLoS One 12, e0177518. doi: 10.1371/journal.pone.0177518
Tungadi, T., Donnelly, R., Qing, L., Iqbal, J., Murphy, A. M., Pate, A. E., et al. (2020). Cucumber mosaic virus 2b proteins inhibit virus-induced aphid resistance in tobacco. Mol. Plant Pathol. 21, 250–257. doi: 10.1111/mpp.12892
Verma, V., Ravindran, P., Kumar, P. P. (2016). Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 16, 86. doi: 10.1186/s12870-016-0771-y
Wang, X. W., Li, P., Naveed, H., Liu, S. (2017). Whitefly interactions with plants. Curr. Opin. Insect Sci. 19, 70–75. doi: 10.1016/j.cois.2017.02.001
Wei, J., Zhao, J. J., Zhang, T., Li, F. F., Ghanim, M., Zhou, X. P., et al. (2014). Specific cells in the primary salivary glands of the whitefly Bemisia tabaci control retention and transmission of begomoviruses. J. Virol. 88, 13460–13468. doi: 10.1128/JVI.02179-14
Whitfield, A. E., Falk, B. W., Rotenberg, D. (2015). Insect vector-mediated transmission of plant viruses. Virology 479-480, 278–289. doi: 10.1016/j.virol.2015.03.026
Wu, D., Qi, T., Li, W. X., Tian, H. X., Gao, H., Wang, J. J., et al. (2017). Viral effector protein manipulates host hormone signaling to attract insect vectors. Cell Res. 27, 402–415. doi: 10.1038/cr.2017.2
Xu, L., Liu, F., Lechner, E., Genschik, P., Crosby, W. L., Ma, H., et al. (2002). The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14, 1919–1935. doi: 10.1105/tpc.003368
Yang, X., Xie, Y., Raja, P., Li, S., Wolf, J. N., Shen, Q., et al. (2011). Suppression of methylation-mediated transcriptional gene silencing by betaC1-SAHH protein interaction during geminivirus-betasatellite infection. PLoS Pathog. 7, e1002329. doi: 10.1371/journal.ppat.1002329
Yang, X., Guo, W., Li, F., Sunter, G., Zhou, X. (2019). Geminivirus-associated betasatellites: exploiting chinks in the antiviral arsenal of plants. Trends Plant Sci. 24, 519–529. doi: 10.1016/j.tplants.2019.03.010
Yu, H., Wu, J., Xu, N., Peng, M. (2007). Roles of F-box proteins in plant hormone responses. Acta Bioch. Bioph. Sin. 39, 915–922. doi: 10.1111/j.1745-7270.2007.00358.x
Zeng, L. R., Vega-Sánchez, M. E., Zhu, T., Wang, G. L. (2006). Ubiquitination-mediated protein degradation and modification: an emerging theme in plant-microbe interactions. Cell Res. 16, 413–426. doi: 10.1038/sj.cr.7310053
Zerbini, F. M., Briddon, R. W., Idris, A., Martin, D. P., Moriones, E., Navas-Castillo, J., et al. (2017). ICTV virus taxonomy profile: Geminiviridae. J. Gen. Virol. 98, 131–133. doi: 10.1099/jgv.0.000738
Zhang, T., Luan, J. B., Qi, J. F., Huang, C. J., Li, M., Zhou, X. P., et al. (2012). Begomovirus-whitefly mutualism is achieved through repression of plant defences by a virus pathogenicity factor. Mol. Ecol. 21, 1294–1304. doi: 10.1111/j.1365-294X.2012.05457.x
Zhang, L., Zhang, F., Melotto, M., Yao, J., He, S. Y. (2017). Jasmonate signaling and manipulation by pathogens and insects. J. Exp. Bot. 68, 1371–1385. doi: 10.1093/jxb/erw478
Zhang, P. J., He, Y. C., Zhao, C., Ye, Z. H., Yu, X. P. (2018). Jasmonic acid-dependent defenses play a key role in defending tomato against Bemisia tabaci nymphs, but not adults. Front. Plant Sci. 9, 1065. doi: 10.3389/fpls.2018.01065
Zhao, P., Yao, X., Cai, C., Li, R., Du, J., Sun, Y. W., et al. (2019). Viruses mobilize plant immunity to deter nonvector insect herbivores. Sci. Adv. 5, eaav9801. doi: 10.1126/sciadv.aav9801
Zhou, B., Zeng, L. (2017). Conventional and unconventional ubiquitination in plant immunity. Mol. Plant Pathol. 18, 1313–1330. doi: 10.1111/mpp.12521
Zhou, X. P. (2013). Advances in understanding begomovirus satellites. Annu. Rev. Phytopathol. 51, 357–381. doi: 10.1146/annurev-phyto-082712-102234
Keywords: tripartite interaction, tomato yellow leaf curl China virus/tomato yellow leaf curl China betasatellite, βC1, whitefly, plant defense response
Citation: Zou C, Shu Y-N, Yang J-J, Pan L-L, Zhao J, Chen N, Liu S-S and Wang X-W (2020) Begomovirus-Associated Betasatellite Virulence Factor βC1 Attenuates Tobacco Defense to Whiteflies via Interacting With Plant SKP1. Front. Plant Sci. 11:574557. doi: 10.3389/fpls.2020.574557
Received: 20 June 2020; Accepted: 12 August 2020;
Published: 27 August 2020.
Edited by:
Rosa Lozano-Durán, Shanghai Institutes for Biological Sciences (CAS), ChinaReviewed by:
Murad Ghanim, Agricultural Research Organization (ARO), IsraelJesús Navas-Castillo, Institute of Subtropical and Mediterranean Horticulture La Mayora, Spain
Copyright © 2020 Zou, Shu, Yang, Pan, Zhao, Chen, Liu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao-Wei Wang, eHd3YW5nQHpqdS5lZHUuY24=
 Chi Zou
Chi Zou Yan-Ni Shu
Yan-Ni Shu Jing Zhao
Jing Zhao Shu-Sheng Liu
Shu-Sheng Liu Xiao-Wei Wang
Xiao-Wei Wang