- Department of Plant Pathology, University of Wisconsin–Madison, Madison, WI, United States
While fungal biotrophs are dependent on successfully suppressing/subverting host defenses during their interaction with live cells, necrotrophs, due to their lifestyle are often confronted with a suite of toxic metabolites. These include an assortment of plant defense compounds (PDCs) which can demonstrate broad antifungal activity. These PDCs can be either constitutively present in plant tissue or induced in response to infection, but are nevertheless an important obstacle which needs to be overcome for successful pathogenesis. Fungal necrotrophs have developed a number of strategies to achieve this goal, from the direct detoxification of these compounds through enzymatic catalysis and modification, to the active transport of various PDCs to achieve toxin sequestration and efflux. Studies have shown across multiple pathogens that the efficient detoxification of host PDCs is both critical for successful infection and often a determinant factor in pathogen host range. Here, we provide a broad and comparative overview of the various mechanisms for PDC detoxification which have been identified in both fungal necrotrophs and fungal pathogens which depend on detoxification during a necrotrophic phase of infection. Furthermore, the effect that these mechanisms have on fungal host range, metabolism, and disease control will be discussed.
Introduction
To successfully infect and colonize a host, phytopathogenic fungi must overcome a repertoire of plant defense compounds (PDCs) with broad antimicrobial properties. PDCs are often sub-classified as those which are induced in response to infection (phytoalexins) and/or those that exist as pre-formed antimicrobial compounds in plant tissue (phytoanticipins). These classifications are not mutually exclusive, as compounds may exist as both a phytoalexin and phytoanticipin within a single plant (Van Etten et al., 1994). The lifestyle of a pathogen can play a major role in how they interact with these compounds, as phytoalexin production is often dependent on the successful induction of plant defenses and phytoanticipin activity requires damage to plant tissue (Tiku, 2020). In the case of biotrophic pathogens that depend on the colonization of living tissue, it has been suggested that the maintenance of plant cellular integrity may prevent the release of phytoanticipins, thus subverting their role in plant defense (Glenn et al., 2002). Additionally, pathogens with greater host-specialization, as is seen in nearly all biotrophs, tend to present greater basal tolerance for co-evolved host antimicrobials and may be less dependent on specific detoxification mechanisms (Basse, 2005; Buxdorf et al., 2013). This may be why active detoxification of host PDCs by fungal biotrophs has only been noted in rare circumstances (Okmen et al., 2013).
Contrarily, necrotrophic and hemibiotrophic fungi, which induce cell death and compromise the integrity of plant tissues during colonization, do not benefit from this covertness and must actively detoxify host antimicrobials. This detoxification is facilitated by a number of mechanisms including metabolization of the compounds to less toxic derivatives and transporter-mediated efflux to maintain PDCs at sublethal thresholds. This review will serve to summarize the current state of knowledge surrounding the detoxification of plant PDCs during fungal necrotrophy and the implications this detoxification has on pathogen evolution, host range, and management.
Enzymatic Detoxification of Inducible PDCs
Inducible PDCs are defined in this review as low molecular weight antimicrobial compounds that can be produced/accumulated in response to pathogen invasion. This category includes all compounds classified as phytoalexins (i.e., induced in response to infection) and those which are both phytoalexins and phytoanticipins (i.e., compounds which exist in healthy plant tissue and additionally accumulate in response to infection). Examples of such compounds include the antimicrobials resveratrol and maackiain (Van Etten et al., 1994; Wang et al., 2013). While these compounds are often broadly mycotoxic, it has been observed for decades that pathogens of specific plants appear more tolerant of their PDC repertoire than non-pathogens (Delserone et al., 1999). This tolerance is a necessary trait of a pathogen that seeks to fully colonize host tissue and, in many cases, appears to result from its capacity to actively metabolize (detoxify) specific PDCs into less toxic derivatives (Table 1). Similar to the production of phytoalexins by the plant, this detoxification is typically inducible through fungal exposure to mycotoxic compounds or in response to specific cues (George and Van Etten, 2001; Pedras et al., 2004; Pedras et al., 2007; Chen et al., 2019). This stepwise induction follows: (1) pathogen recognition by the plant, (2) production and accumulation of PDCs at and around the site of infection, (3) recognition of PDCs by the pathogen, (4) production of detoxification enzymes. Some PDCs are not only antimicrobial themselves, but act as intermediates in the biosynthesis of other, often more mycotoxic compounds (Pedras et al., 2008; Dubrovina and Kiselev, 2017). Because of this, the metabolism of such compounds, including resveratrol and brassinin (discussed below), can serve to both actively protect the fungus and disrupt the production of other bioactive compounds. The following section discusses the enzymatic detoxification of inducible PDCs that have been observed or characterized in fungal necrotrophic/hemibiotrophic pathogens of plants.
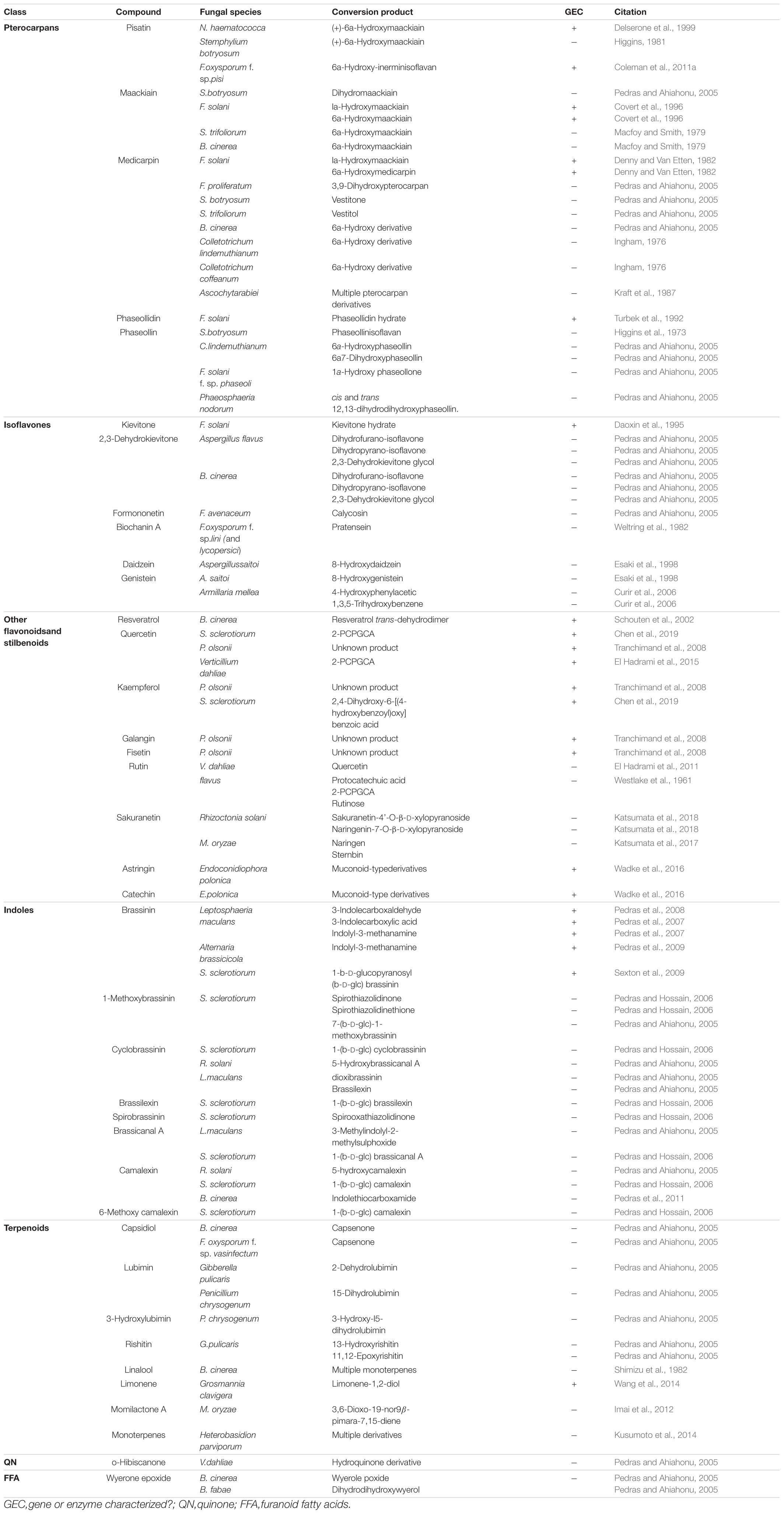
Pterocarpans
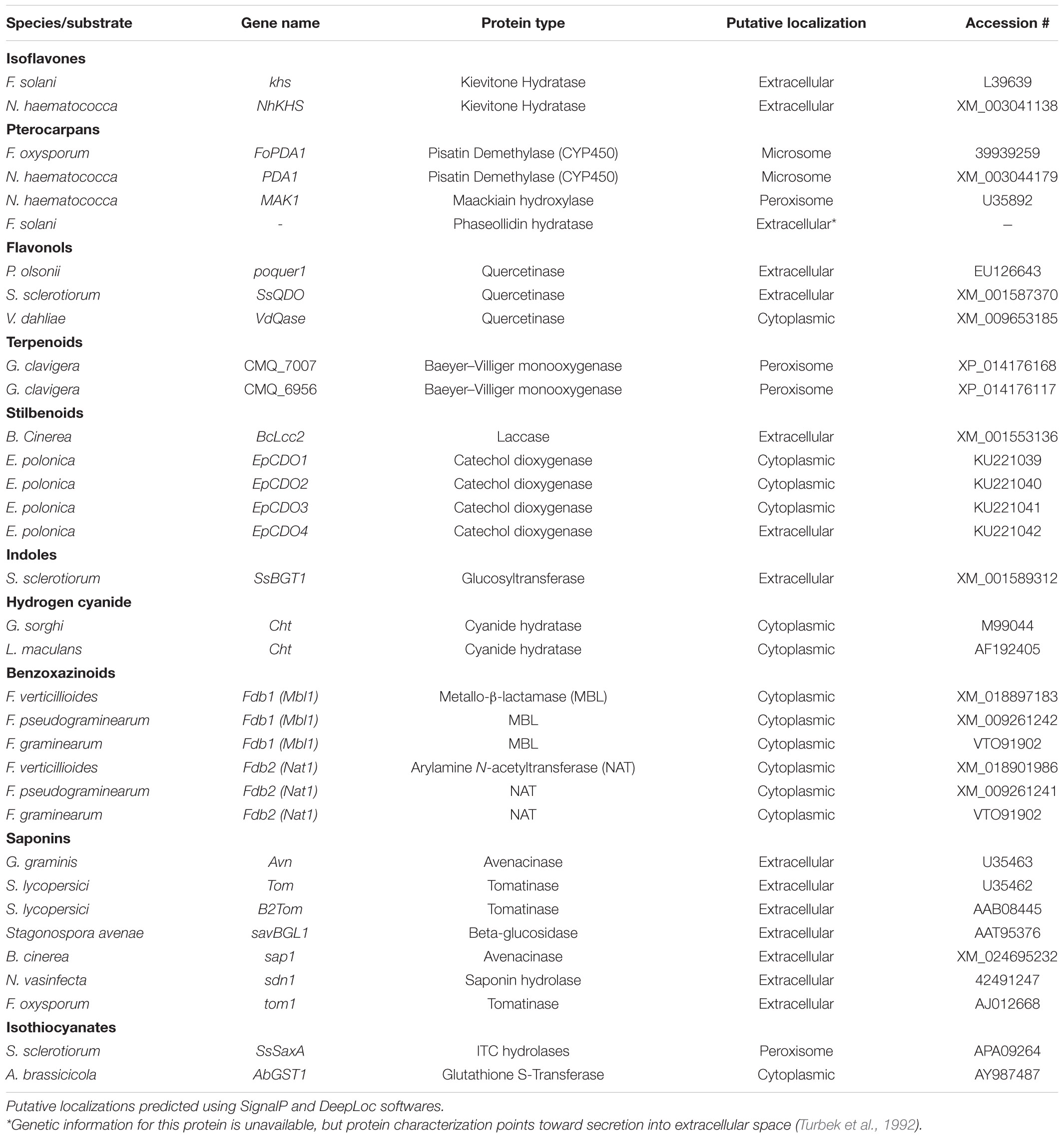
Pterocarpans are class of inducible PDC produced in the phenylpropanoid pathway that are primarily associated with legume defense against biotic stress (Dixon et al., 2002). Pisatin is an antifungal pterocarpan that is produced in pea pods of Pisum sativum during pathogen challenge (Perrin and Bottomley, 1961). As one of the earliest discovered PDCs, pisatin was initially credited with driving non-host resistance in P. sativum to fungal pathogens, as only fungi adapted to this host environment are able to cause disease (Delserone et al., 1999). Pisatin detoxification in Nectria haematococca was attributed to microsomal cytochrome P450s capable of efficiently demethylating pisatin to the less toxic (+)-6a-hydroxymaackiain, and termed pisatin demethylases (PDAs) (George et al., 1998; George and Van Etten, 2001). The gene encoding the primary PDA (PDA1) is located, along with several other pea pathogenicity genes (PEP genes), on a 1.6 Mb supernumerary chromosome which is dispensable for fungal survival in culture, but critical for pathogenicity on pea (Miao et al., 1991; Han et al., 2001). While the function of most of these PEP genes is still unknown a single gene, PEP5, was found to be a likely efflux transporter (discussed in section “Toxin Efflux”), capable of increasing virulence on pea in the absence of other detoxification mechanisms (Han et al., 2001). Although PDA genes have been primarily characterized in Fusarium spp., PDA activity has been noted in other fungal pathogens of pea and putative orthologs are induced during infection in some broad host range necrotrophs (Table 1 and Figure 1) (George and Van Etten, 2001).
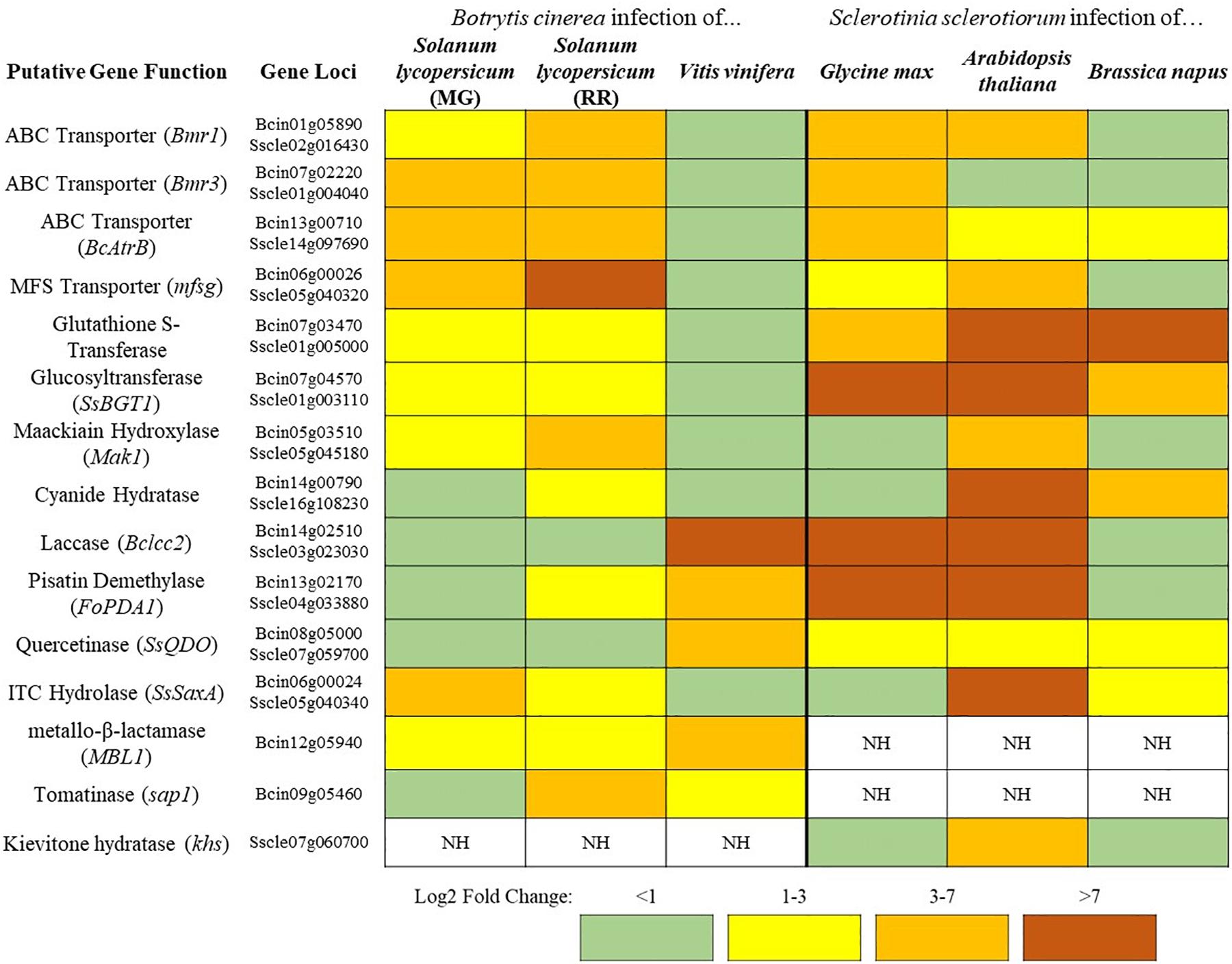
Figure 1. A sampling of Botrytis cinerea and Sclerotinia sclerotiorum genes putatively involved in detoxification of PDCs. Simplified differential gene expression values are presented for B. cinerea [infecting mature green (MG) and red ripe (RR) Solanum lycopersicum and Vitis vinifera] and S. sclerotiorum (infecting Glycine max, Arabidopsis thaliana, and Brassica napus) (Haile et al., 2017; Seifbarghi et al., 2017; Petrasch et al., 2019; Peyraud et al., 2019; Westrick et al., 2019). All samples were harvested at ∼24 h post inoculation and differential expression values were generated through comparison to in vitro culture controls. NH = no homolog found in the genome.
A closely related pterocarpan, maackiain, plays a similar role to pisatin in chickpea defense and its detoxification is also highly correlated with N. haematococca virulence (Lucy et al., 1988). Three loci involved in the degradation of maackiain, and the related medicarpin, have been identified (MAK1-3), but only MAK1 has been fully characterized as a hydroxylase capable of converting maackiain into 1α-hydroxy-maackiain (Miao and Van Etten, 1992; Enkerli et al., 1998). Similar to PDA1, MAK1, and MAK2 are on dispensable chromosomes associated with other putative virulence genes and MAK1 appears to share a chromosome with a putative PDA gene (PDA6-1) thought to be involved in pisatin tolerance (Covert et al., 1996). Disruption mutants of MAK1 have a reduced virulence and maackiain tolerant strains overexpressing MAK1 demonstrate increased aggression, confirming a previously noted correlation between maackiain detoxification and virulence (Enkerli et al., 1998). While it is assumed that a tolerance for maackiain is a prerequisite for pathogenicity on chickpea and the degradation of this pterocarpan has been observed in other chickpea pathogens, no other genes involved in maackiain degradation have been characterized (Birgit et al., 1989; Tenhaken et al., 1991).
Another pterocarpan, phaseollidin, is known to accumulate during infection of the common bean, Phaseolus vulgaris, along with the isoflavonol kievitone (discussed below), and is degraded by some pathogens of P. vulgaris (Van Etten et al., 1989; Turbek et al., 1992). Early evaluation of phaseollidin metabolism by the pathogen Fusarium solani suggested that a single enzyme was detoxifying both compounds through a hydration reaction (Kuhn and Smith, 1979). Later work determined that distinct proteins are responsible for kievitone and phaseollidin hydratase (PHase) activities, although the gene encoding the latter has never been characterized (Turbek et al., 1992). As phaseollidin is the precursor of other antifungal PDCs including phaseollin, PHase activity may serve to both detoxify an important PDC and suppress the formation of other antimicrobials (Dewick and Steele, 1982). This may also explain why PHase activity is accomplished by a secreted extracellular enzyme, while other pterocarpan detoxifying proteins are intracellular (Table 2).
Isoflavones
Isoflavones are a class of phenylpropanoids primarily known for their importance in legume defense to pathogens (Ahuja et al., 2012). Kievitone is a member of this class which accumulates, along with a range of other isoflavonoid phytoalexins, during fungal infection of P. vulgaris (Van Etten and Smith, 1975; Smith et al., 1984). Early studies determined that F. solani detoxified kievitone through a kievitone hydratase (KHase) reaction and a clear association between KHase activity and virulence on common bean has been readily observed (Kuhn and Smith, 1979; Smith et al., 1982). F. solani isolates/mutants lacking this activity were non-pathogenic, highlighting its critical importance during infection of P. vulgaris (Smith et al., 1984, 1982). The gene encoding kievitone hydratase was first cloned from F. solani f. sp. phaseoli and its function in converting kievitone to kievitone hydrate was confirmed through heterologous expression (Daoxin et al., 1995). Putative homologs of khs have been identified in other non-pathogenic Fusarium spp., suggesting that this gene is not a solely capable of granting virulence to P. vulgaris (Daoxin et al., 1995). More recently the gene encoding KHase in N. haematococca (NhKHS) was cloned and characterized through recombinant expression (Engleder et al., 2018). Purified NhKHS appears capable of acting on kievitone and several other flavonoid compounds, suggesting that this enzyme may be capable of detoxifying a wider range of PDCs than originally thought (Engleder et al., 2018). To date, khs deletion mutants have yet to be generated in any species, thus the involvement of other genes in this process cannot be excluded.
While no other isoflavone detoxification mechanisms have been specifically characterized, metabolic profiling has shown that other isoflavones, including the major isoflavones genistein and daidzein, can be metabolized by a number of other fungal pathogens (Table 1).
Other Flavonoids and Stilbenoids
Flavonol-type PDCs, such as quercetin and kaempferol, are found in a wide range of plants and demonstrate antimicrobial activity both in their free and glycosylated forms (Lygin et al., 2009; Sanzani et al., 2009; Chen et al., 2019). Quercetinase activity has been observed in a number of pathogens and quercetin dioxygenases (QDOs) responsible for this activity have been characterized in Penicillium olsonii, Sclerotinia sclerotiorum, and Verticillium dahlia (Tables 1, 2). While the enzyme from V. dahlia, VDQase, is putatively cytoplasmic, those found in the other two pathogens appear to be secreted, possibly as a dual mechanism that simultaneously detoxifies the compound and subvert the capacity of the host to produce glycosylated flavonols, such as rutin (Table 2). The detoxification of rutin requires the cleavage of the compounds sugar moiety through a proposed fungal rutinase, which subsequently releases the mycotoxic quercetin, thus implicating quercetinases in tolerance to both flavonols and their cognate flavonoid glycosides (El Hadrami et al., 2015). Interestingly, QDO activity has been primarily observed in broad-host range pathogens, whereas detoxification of other flavonoids, such as sakuranetin from rice, appears to utilize a distinct detoxification pathway in the more specialized pathogen Magnaporthe oryzae (Table 1).
Stilbenoids, another class of inducible PDC, help to modulate plant immunity to phytopathogens as well as UV and ozone stress (Chong et al., 2009). The most studied stilbenoid is the grapevine phytoalexin resveratrol, which has an important role in grapevine resistance to the bunch rot pathogen Botrytis cinerea. While resveratrol is suppressive to B. cinerea growth, it is well established that the pathogen is capable of oxidizing the phytoalexin in vitro and this activity has been attributed to a secreted laccase, Bclcc2 (Schouten et al., 2002). Counterintuitively, this laccase acts as a profungicide metabolizing resveratrol to a more toxic metabolite, and gene knockout mutants of Bclcc2 are both resistant to resveratrol and maintain full virulence on grapevine, leaving the role of this enzyme in pathogenesis unclear (Schouten et al., 2002). A more linear story of stilbenoid detoxification can be seen in the spruce pathogen Endoconidiophora polonica. After being deposited into a spruce tree by its bark-beetle vector, the host responds to invasion by producing antifungal compounds such as the stilbenoid astringin and the flavan-3-ol catechin. E. polonica detoxifies these compounds using a group of four catechol dioxygenases (CDOs), one of which is secreted and operates at the forefront of infection, and the other three of which operate intracellularly (Wadke et al., 2016) (Table 2).
Indoles
Indoles are a class of PDC known for their role in defense against biotic stress in cruciferous (brassica) vegetables (Pedras and Ahiahonu, 2005). Detoxification of indoles has been observed in broad-host range necrotrophs including Sclerotinia sclerotiorum and B. cinerea as well as the more specialized Brassicaceae pathogen Leptosphaeria maculans, albeit through seemingly distinct mechanisms (Table 1). A wide range of anti-fungal indoles have been characterized, yet only proteins with the capacity to degrade a single one, the phytoalexin brassinin, have been properly characterized (Table 1). Brassinin hydrolases (BHs), seen in the crucifer pathogens Alternaria brassicicola and L. maculans, transform brassinin to the less toxic indolyl-3-methanamine (Pedras et al., 2009). In addition to its BH activity, L. maculans also detoxifies brassinin through an inducible brassinin oxidase (BO) (Table 1) (Pedras et al., 2008). Both enzymes demonstrate a surprisingly narrow substrate range and are not only incapable of transforming related indoles, such as cyclobrassinin, but in many cases are competitively inhibited by them (Pedras et al., 2008, 2009). This inhibition is intriguing as brassinin is a metabolite required for the production of other antifungal indoles, meaning BH/BO activity may be achieving multiple goals simultaneously: (1) degrading antifungal brassinin in the host, (2) preventing the production of other antifungal indoles, and (3) preventing the production of BH/BO inhibitors.
Rather than oxidation/hydrolysis, S. sclerotiorum appears to detoxify brassinin using a rather uncommon glycosylation reaction in which brassinin is transformed through the conjugation of a glucose molecule. The enzyme responsible for this transformation, SsBGT1, appears to have a similar substrate specificity and induction pattern to other brassinin detoxifying enzymes, but inhibition by natural indoles has not been reported (Table 2) (Sexton et al., 2009). Metabolism of other indoles by fungal pathogens is summarized in Table 1.
Terpenes/Terpenoids
Unlike many of the above-described classes of PDC, which are formed as downstream products of the shikimate pathway, terpenoids are generated through either the mevalonate (MVA) or methylerythritol 4- phosphate (MEP) pathways in plants. Despite the broad range of terpenoids known to operate in chemical defense and the number of phytopathogens known to metabolize terpenoids in vitro, the only fungal enzymes characterized in terpenoid detoxification are from the pine pathogen Grosmannia clavigera (Table 1) (Wang et al., 2014). Multiple enzymes were involved in the metabolic utilization of monoterpenes, primarily the antifungal limonene, but only two Baeyer–Villiger monooxygenases were implicated specifically in enzymatic detoxification (Table 2) (Wang et al., 2014).
Enzymatic Detoxification of Non-Inducible PDCs
Non-inducible PDCs are defined in this review as low molecular weight antimicrobial compounds that are pre-formed in healthy plant tissue rather than being produced in response to pathogen invasion. This category includes many phytoanticipins, but excludes those which are both constitutively present in healthy tissue and are also induced in response to infection, examples of which include resveratrol and maackiain (Van Etten et al., 1994; Wang et al., 2013). Non-inducible PDCs are stored in either an active form or as inactive precursors which are activated in response to tissue damage (Tiku, 2020). As this damage is an inevitable result of fungal necrotrophy, these compounds provide protection that is less easily subverted by fungal effectors targeting plant defense pathways than phytoalexins might be. The following section will focus on the detoxification of important classes of non-inducible PDCs by fungal pathogens. A more complete summary of fungal metabolism of these compounds can be found in Table 3.
Hydrogen Cyanide
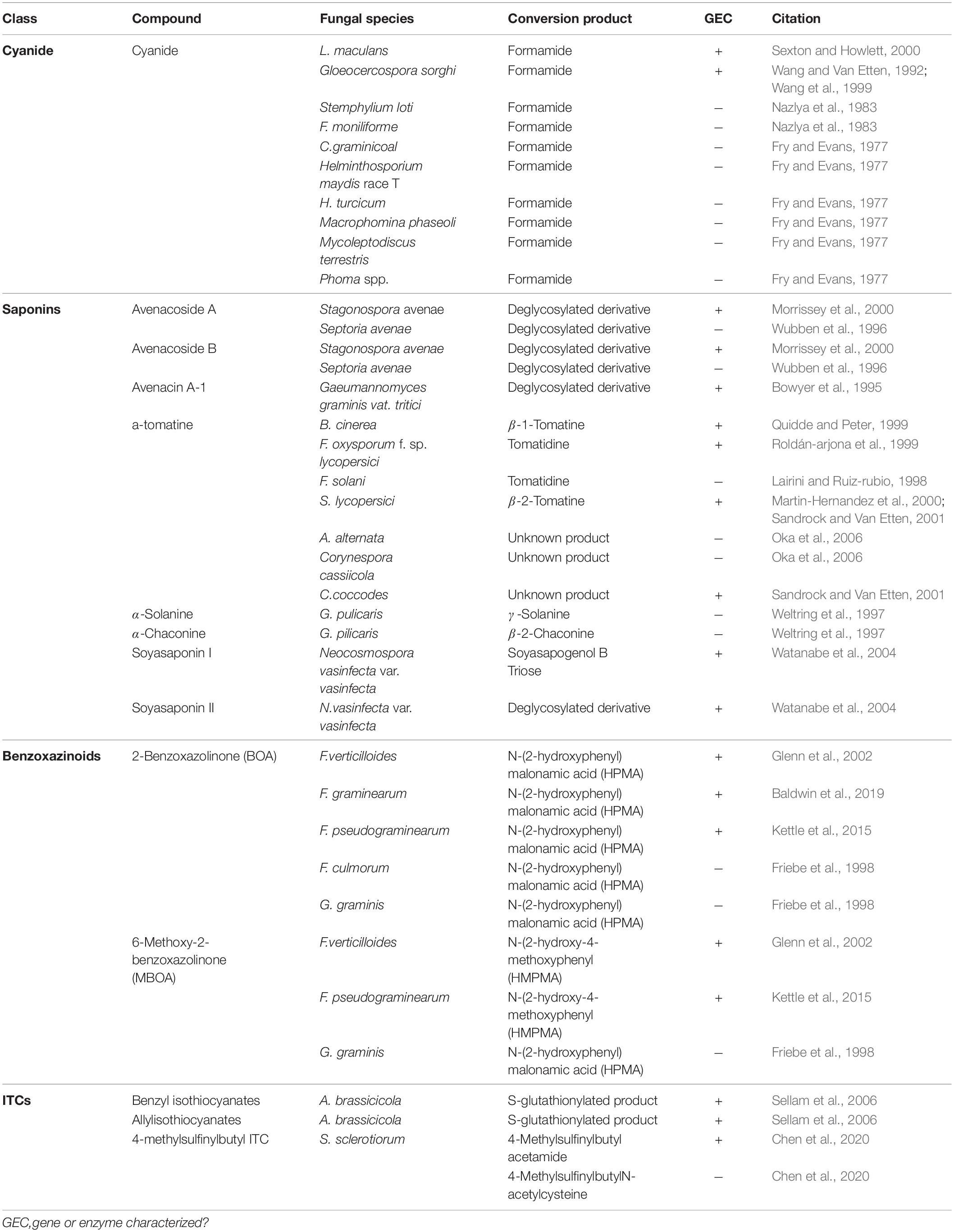
Cyanogenic plants are a broad group, including thousands of individual plant species and >120 plant families, that are characterized by the presence of cyanogenic glycosides in their tissue (Tiku, 2020). Tissue damage leads to the release of plant glycoside hydrolases which cleave the sugar moiety, converting these glycosides to cyanohydrins, which are subsequently transformed to hydrogen cyanide (HCN) by hydroxynitrile lyases (Tiku, 2020). Pathogens of cyanogenic plants are all capable of tolerating HCN and a single mechanism, cyanide hydration, appears to be an incredibly conserved detoxification mechanism used across fungal pathogens of these plants (Table 3) (Fry and Evans, 1977). Cyanide hydratases (CHT) responsible for this activity have been characterized in two pathogens, L. maculans and Gloeocercospora sorghi, both of which are putatively cytoplasmic proteins (Table 2). These CHTs efficiently convert HCN to formamide, and CHT knockouts in G. sorghi were far more sensitive to potassium cyanide, an inducer of CHT activity (Wang et al., 1999).
Saponins
Saponins are compounds containing a triterpenoid steroid, or steroidal glycoalkaloid bound to one or more sugar chains and have been utilized for hundreds of years as a surfactant, but have gained notoriety in recent decades for their antimicrobial properties (Osbourn, 1996). Prior to pathogen attack saponins are kept in an inactive form with multiple sugar chains bound to the alkaloid and are spatially separated from the hydrolytic enzymes needed to activate them. In response to tissue damage, this compartmentalization which maintains saponin precursors in vacuoles and hydrolytic enzymes in plastids breaks down, allowing the enzymes to cleave one or more sugars to generate bioactive saponins. Unlike most other phytoanticipins that are fully deglycosylated during activation, the antifungal activity of saponins are dependent on their sugar moieties (Osbourn, 1996).
Nearly all pathogens of saponin-containing plants depend on some detoxifying activity to tolerate these compounds during infection, and all characterized saponin detoxification enzymes operate through the cleavage of one or more sugar molecules from the compounds sugar chain (Osbourn, 1996) (Tables 2, 3). The first discovered of these enzymes, an avenacinase termed Avn, is used by the Take-All fungus Gaeumannomyces graminis to detoxify oat saponins and is seemingly a singular determinant in the pathogens capacity to infect oats (discussed further in section “The Role of PDC Tolerance in Fungal Host Range”) (Bowyer et al., 1995). Additional oat pathogens, Stagonospora avenae and Septoria avenae, encode multiple saponin hydrolases (SHs) which cleave distinct bonds in the sugar chains of these oat saponins, referred to as avenacosides, and are primarily present in oat-infecting isolates, but absent in those isolated from saponin-deficient wheat (Wubben et al., 1996; Morrissey et al., 2000) (Table 3).
While SHs were originally thought to be somewhat specific to oat pathogens, studies into the saponin detoxifying activity of the tomato pathogen Septoria lycopersici found that it utilized a secreted tomatinase enzyme (Tom) with homology to Avn from G. graminis (Osbourn et al., 1995). Tomatinases, named for their ability to degrade the important tomato saponin α-tomatine, are found in a wide range of tomato-infecting fungal pathogens (Tables 2, 3). The presence of avenacinases, tomatinases, and other SHs is highly correlated with the ability to infect saponin-containing plants, but as many pathogens contain multiple distinct SHs that are active during pathogenesis, the importance of individual enzymes in virulence is difficult to discern through simple gene disruption experiments (Quidde and Peter, 1999; Dufresne et al., 2000). Ectopic expression of the tomatinase-deficient pathogen N. haematococca MPVI with a β2-tomatinase gene from S. lycopersici allowed it to infect green tomato fruit, confirming that saponin detoxifying enzymes can act as singular determinant of pathogenicity on certain hosts (Sandrock and Van Etten, 2001). Although SHs appear to be primarily responsible for tolerance to plant saponins, evidence has also been presented to suggest that saponin degradation products may modulate plant immunity as well (discussed in section “Detoxification and Plant Defense Signaling”).
Benzoxazinoids
Benzoxazinoid-type phytoanticipins are indole-derived compounds common across cereal crops and are broadly toxic to pathogens and pests (Tiku, 2020). Similar to other phytoanticipins, benzoxazinoids are stores in a glycosylated form in plant vacuoles and are converted to their active form by plant glucosidases in response to tissue damage (Glenn and Bacon, 2009). Detoxification of these compounds has been primarily studied in cereal-infecting Fusarium spp. and relies on a two-step process to fully metabolize benzoxazolin-2(3H)-one (BOA) and 6-methoxybenzoxazolin-2(3H)-one (MBOA), the major antifungal benzoxazinoids found in maize and wheat (Table 3). The genes involved in this transformation are a metallo-β-lactamase referred to as MBL1, or Fdb1, and an arylamine N-acetyltransferase referred to at Nat1, or Fdb2 (Table 2). MBL1 catalyzes an initial conversion of BOA to 2-amino-phenoxazin-3-one (2-APO), which is subsequently converted to the negligibly mycotoxic N-(2-hydroxyphenyl) malonamic acid (HPMA) by Nat1. The specific requirement of benzoxazinoid detoxification in virulence is unclear, as genetic knockouts of these genes in multiple Fusarium spp. have resulted in conflicting results, with clear virulence defects observed in Fusarium pseudograminearum, but none in Fusarium graminearum or Fusarium verticillioides (Glenn et al., 2002; Kettle et al., 2015; Baldwin et al., 2019).
Isothiocyanates
Glucosinolates are amino acid derived plant compounds that are converted into toxic isothiocyanates (ITCs) in response to tissue damage. These compounds are found primarily in cruciferous plants, and ITCs are potent in suppressing both invasion by fungal/bacterial pathogens and herbivory by pests (Bednarek et al., 2009). The detoxification of ITCs has been well characterized in animals and is typically catalyzed by glutathione S-transferases (GSTs), a class of enzyme that can conjugate glutathione (GSH) to various xenobiotic substrates. The crucifer pathogen A. brassicicola detoxifies these compounds using a similar mechanism by producing a specific GST, AbGST1, in response to ITC exposure (Sellam et al., 2006). Other GSTs in A. brassicicola have also been characterized and appear to be important in pathogenesis, but their specific role in ITC detoxification is unclear (Calmes et al., 2015).
Evidence suggests that the broad-host range pathogen S. sclerotiorum also uses GST activity in the detoxification of ITCs to a minor extent, but the major detoxification mechanism (100-fold greater) appears to be through hydrolytic degradation. An ITC hydrolase, SsSaxA, is credited with this activity and is capable of efficiently degrading the aliphatic and aromatic ITCs of the host Arabidopsis thaliana (Chen et al., 2020).
Phenylpropanoid Derivatives
Derivatives of the phenylpropanoid pathway, including ferulic and cinnamic acid, are aromatic compounds necessary for the generation of structural lignin as well as other physiological processes (Mäkelä et al., 2015). While not typically classified as phytoanticipins due to their pivotal role in plant physiology, they share many characteristics including a constitutive presence in plant tissue and antifungal activity (Pacheco et al., 2018). It has been demonstrated that the accumulation of these compounds in plant tissue is important for soybean resistance to the broad host-range necrotroph S. sclerotiorum and the capacity to metabolize these compounds to some extent is a rather ubiquitous feature of filamentous fungi (Mäkelä et al., 2015; Ranjan et al., 2019). To our knowledge the mechanism for this metabolization of phenylpropanoid derivatives has not been characterized in any fungal species despite the necessity of such a process in colonizing plant tissue.
Toxin Efflux
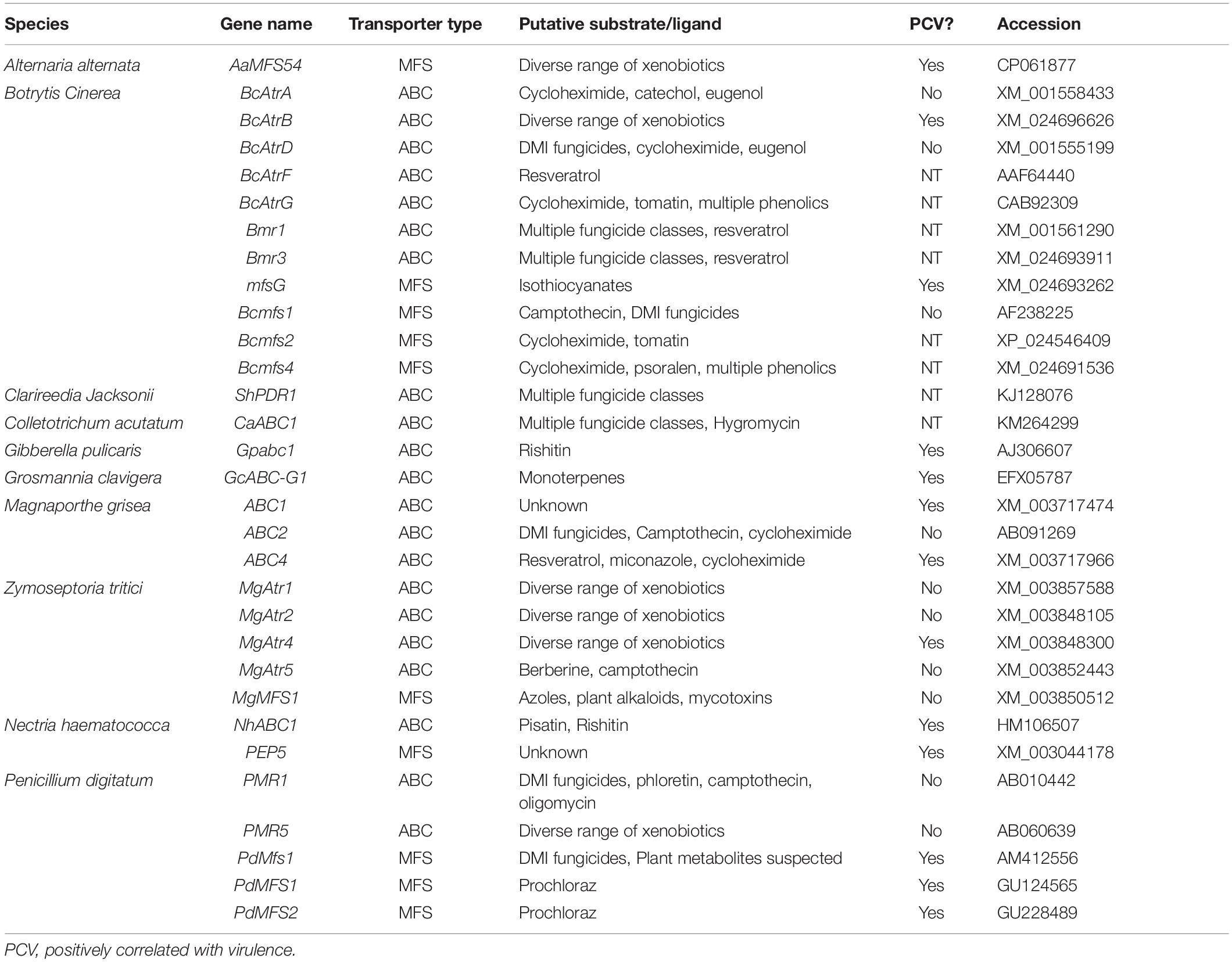
While degradative detoxification of PDCs has received a large amount of attention in recent years, it has long been known that some non-degradative mechanism of fungal tolerance to PDCs exists as well (Denny and Van Etten, 1983a, b). Despite that, it was not until the discovery of the first multidrug resistance (MDR) transporters in Saccharomyces cerevisiae, SNQ2 and PDR5 (Servos et al., 1993; Balzi et al., 1994), and the subsequent description of the first ATP-binding cassette (ABC) transporter in the rice pathogen M. oryzae that a true mechanism for this activity was described (Urban et al., 1999).
Contrary to the enzymatic detoxification of PDCs, toxin efflux operates through the activity of membrane bound substrate transport proteins that work to either export or sequester toxic compounds. Broadly, these proteins can be separated into ABC transporters that operate using energy from ATP hydrolysis and major facilitator superfamily (MFS) transporters which utilize a chemiosmotic gradient generated across the membrane to move substrate (Pao et al., 1998). As plant hosts typically possess a repertoire of chemically related PDCs, efflux proteins often have a broad substrate binding affinity and are thus referred to as pleiotropic and/or multidrug resistance proteins (PDR and/or MDR). These proteins are most often transcriptionally regulated and thus some insight into their substrate range can be gained through the accumulation of their RNA transcripts upon exposure to various compounds (Schoonbeek et al., 2001; Semighini et al., 2002; Lee et al., 2005; Gupta and Chattoo, 2008).
True to their name as pleiotropic drug resistance proteins, individual efflux transporters are often shown to be active against a range of chemically distinct compounds (Roohparvar et al., 2007). This broad range of activity has been functionally validated in multiple transporters within the wheat pathogen Zymoseptoria tritici, where ectopic expression of these transporters in yeast conferred increased tolerance to a variety of antifungal compounds. Surprisingly, though, genetic knockouts of these transporters often show no reduction in virulence (Table 4). Multiple theories have been discussed to address this, the most common being a functional redundancy between transporters with similar substrate ranges. Substrate redundancy across a range of toxic compounds has been observed in the broad host-range pathogen B. cinerea, the wheat pathogen Z. tritici, and the saprotroph Aspergillus nidulans (Semighini et al., 2002; Schoonbeek et al., 2003; Zwiers et al., 2003). This redundancy has been characterized in both plant and animal pathogens and often requires multiple transporters to be knocked out in a single mutant to see the expected increase in sensitivity to a suspected substrate (Sanglard et al., 1997; Hayashi et al., 2002).
The importance of a distinct transporter in infection is dependent on the hosts antimicrobial repertoire, so an additional explanation is that it may be necessary to screen multiple hosts before finding one in which a given transporter is pivotal to pathogenicity. In the case of the B. cinerea ABC transporter BcAtrB, knockout mutants showed no reduction in virulence on basil, but were compromised during infection of A. thaliana and grapevine (Schoonbeek et al., 2001, 2003; Stefanato et al., 2009).
Although there are certainly incidences in which the role of a putative efflux transporter is vague, many of those characterized in necrotrophic and hemibiotrophic pathogens have a clear role in virulence (Table 4). This role is often elucidated through a mixture of in vitro sensitivity and disease assays to establish a likely PDC substrate and quantify the importance during host colonization, but it’s typically difficult to prove causation between the loss of specific efflux activity and a reduction in virulence. This concern has been raised given that some transporters which contribute to MDR may do so as a side-effect of primary roles in lipid transport, plasma membrane integrity, and/or fungal development (Stergiopoulos et al., 2003; Gupta and Chattoo, 2008; Shahi and Moye-Rowley, 2009). An optimal method to establish this causation is by showing that efflux transporter mutants with a virulence defect on wild-type host plants are fully capable of infecting hosts deficient in the PDC substrate of interest. This has only been demonstrated in BcAtrB and mfsG from B. cinerea, which export the phytoalexin camalexin and ITC-type phytoanticipins, respectively. In both of these cases camalexin and ITC-deficient plants were generated in A. thaliana, an organism that benefits from genetic tractability, and thus, similar evidence remains elusive in other pathosystems (Stefanato et al., 2009; Vela-corcía et al., 2019).
While the importance of efflux in fungal tolerance to a number of PDCs is apparent, an inherent limitation to this mechanism is that it can only act on PDCs that target components within the fungal cell. Accordingly, there are a number of transporters with apparent activity against resveratrol, a plant stilbenoid thought to target cellular respiration, but none that act against plant saponins, which target the plasma membrane (Osbourn, 1996; Schoonbeek et al., 2001, 2003; Caruso et al., 2011; Adrian and Jeandet, 2012). Thus, in order to cope with a broader range of PDCs, efflux transporters often work in tandem with other detoxification mechanisms.
Working in Concert: Multiple Mechanisms of Toxin Tolerance
Cooperative relationships between efflux transporters and other detoxification mechanisms is expected, and have been described in multiple pathosystems, including G. clavigera (Wang et al., 2012, 2014), B. cinerea (Schouten et al., 2008), and N. haematococca (Han et al., 2001; Coleman et al., 2011b). In these relationships the efflux activities of the transporters activate very early during host colonization to avoid the accumulation of host PDCs in fungal hyphae, this is coupled with a subsequent activation of enzymes to detoxify compounds through chemical modification. Such a system has been observed on a broader scale by the fungal pathogen S. sclerotiorum during infection of soybean. A time-course transcriptomic analysis of this infection showed an enrichment of ABC and MFS transporters putatively involved in drug transport during early infection and a subsequent increase in genes putatively involved in degradative detoxification of xenobiotics during later timepoints (Westrick et al., 2019).
In addition to the broad use of detoxification enzymes, some fungi may also modify the infection court in a coordinated approach. B. cinerea has a repertoire of enzymes that are known to be involved in the degradation of saponins (Quidde and Peter, 1999). Interestingly, B. cinerea also acidifies host tissue during infection to facilitate disease, this increasingly acidic environment causes saponins to lose their antifungal activity (Kunz et al., 2006). Thus, the acidification of host tissue in B. cinerea may decrease the efficacy of plant saponins and allow for other degradative mechanisms to finish the job, through a similar coordinated relationship as that described above. This ability to reduce saponin efficacy through acidification may explain why the closely related S. sclerotiorum, a particularly prolific producer of oxalic acid, has no clear homolog of genes coding for some common saponin detoxifying enzymes tomatinase or avenacinase in its genome (Figure 1). Thus, S. sclerotiorum’s incredibly broad range may be owed to its ability to create an environment that renders plant chemical defenses powerless.
Another tool at the disposal of fungal pathogens noted in this review, is to simply deploy multiple enzymes that act in concert to deal with specific plant metabolites. It has been noted on multiple occasions that genes putatively involved in fungal detoxification are often conserved in gene clusters, similar to the secondary metabolite gene clusters found in most fungal genomes (Gluck-thaler and Slot, 2018). This has led to the assertion that these genes may be operating in tandem to efficiently metabolize host PDCs, but to date, only individual genes in these clusters have been credited with a role in detoxification, leaving the purpose of this gene clustering elusive (Glenn et al., 2016; Gluck-thaler and Slot, 2018).
Metabolism or Detoxification: Maybe Both?
Mechanisms for detoxifying PDC’s in phytopathogenic fungi are, at their core, responsible for overcoming plant immunity and facilitating the metabolism of host tissue. Given this fact it is tempting to draw a line between detoxification and metabolism, when in fact data suggests that they are far more interconnected than originally thought. An interesting example of this can be seen in the D-galacturonic acid (GalA) metabolic pathway, which is highly conserved in among pectin-degrading filamentous fungi (a category encompassing nearly all fungal necrotrophs and hemibiotrophs) (Martens-uzunova and Schaap, 2008). GalA is the primary constituent of pectin in plants and is degraded by fungal pathogens through a four step metabolic pathway into glycerol and pyruvate (Martens-uzunova and Schaap, 2008). While GalA is often considered a simple nutrient source for fungi, it is also capable of actively inhibiting the growth of the non-pathogenic yeast S. cerevisiae, indicating that the compound may have some antimicrobial properties (Huisjes et al., 2012). The components of the GalA metabolic pathway have been studied in multiple necrotrophic pathogens and individual gene knockouts of each component of this pathway have been shown to display reduced virulence on hosts rich in GalA (Martens-uzunova and Schaap, 2008; Zhang et al., 2011; Wei et al., 2020). Rather than a metabolic defect, this loss in virulence is due to the accumulation of suppressive amounts of GalA and its derivatives, compounds which could act as either carbon sources or toxins depending on the pathogen’s capacity to metabolize them (Zhang and van Kan, 2013).
Similar overlaps between metabolism and detoxification have been seen in the spruce pathogen E. polonica and the pine pathogen G. clavigera. Both pathogens must detoxify antimicrobials, primarily stilbenoids and terpenoids, induced by their beetle vectors at the site of infection (Franceschi et al., 2005; Keeling and Bohlmann, 2006). Rather than simply degrading these compounds, both E. polonica and G. clavigera have evolved multiple enzymes that facilitate the degradation and metabolic utilization of their respective PDCs (Wang et al., 2014; Wadke et al., 2016). G. clavigera is among the most aggressive fungal pathogens to pine trees in North America due its perceived tolerance and utilization of host monoterpenes to a greater degree than other pine ophiostomatoid fungi (Alamouti et al., 2011; Wang et al., 2014). The pathogen Armillaria mellea is capable of metabolizing the antifungal PDC genistein into multiple less toxic derivatives which can be utilized as carbon sources (Curir et al., 2006). Additionally, L. maculans can use formamide, the result of cyanide hydratase detoxification of HCN, as a sole carbon source (Sexton and Howlett, 2000).
The function of saponin hydrolase enzymes is another potential example of this crossover, they are used by many phytopathogenic fungi to detoxify plant saponins, a class of molecules typically consisting of a hydrophobic aglycone with appended sugar molecules. These enzymes reduce the antifungal activity of saponins through the removal of sugar molecules from their glycosyl chains, releasing glucose and/or other sugars in the process (Osbourn, 1996). While the metabolic utilization of these released sugars has not been specifically addressed to our knowledge, it would be expected if these sugars were used by the pathogen as a carbon source. Many of these enzymes belong to the glycoside hydrolase family 3 (GH3) family of proteins and are related to cellobiose degrading enzymes used to release sugars from plant cell walls during fungal necrotrophy (Osbourn et al., 1995).
The Role of PDC Tolerance in Fungal Host Range
Tolerance to specific plant defense compounds, be it through detoxification, efflux, or target modification, is an absolute necessity in a pathogen’s capacity to infect a host and is therefore a major determinant in its host range. The simplest example of this phenomenon is seen in the take-all pathogen G. graminis, which can infect a range of cereal crops. Varieties of G. graminis differ in their ability to infect oat and these differences appear to be largely defined by their capacity to detoxify saponins, which are found in most oats but are absent in many other cereal species (Bowyer et al., 1995). This detoxification activity was attributed to a specific enzyme (Avn) in the oat-infecting variety, G. graminis var. avenae, that effectively detoxifies the oat saponin Avenacin A-1. Avn acts as a singular determinant of pathogenicity on oats, with Avn-minus mutants losing the ability to infect oats but retaining virulence on saponin-deficient species including wheat (Bowyer et al., 1995). The potential for a similar host-range expansion can be seen from the S. lycopersici tomatinase, B2Tom, which efficiently hydrolyzes saponins found in green tomatoes (Table 2). When N. haematococca was transformed to express this protein, it gained the ability to infect green tomatoes, a feature that WT-strains lack (Sandrock and Van Etten, 2001). Therefore, the host range of some closely related fungi, or even strains of the same species, can differ due to minor changes to their detoxification repertoires.
Pisatin detoxification also appears to be important in the ability for Fusarium species to infect pea, as pisatin is one of its primary defense compounds induced during fungal infection and tolerance to pisatin appears critical for host tissue colonization (Cruickshank and Perrin, 1962; Delserone et al., 1999). The primary enzyme for pisatin degradation in F. solani is a pisatin demethylase (PDA), but unlike Avn in G. graminis, PDA-minus mutants do not fully lose pathogenicity, likely due to multiple mechanisms of tolerance to pisatin (Table 2) (Coleman et al., 2011b). Despite this, non-pathogenic strains of F. solani and the maize pathogen Cochliobolus heterostrophus which are not pea pathogens, gained pathogenicity through ectopic expression of PDA. This suggests that pisatin may be a critical driver of non-host resistance in pea and PDA expression is alarmingly enough to overcome this barrier (Schafer et al., 1989; Ciuffetti and Van Etten, 1996).
In the case of broad-host range pathogens detoxification is critically important as the fungus must maintain a repertoire of detoxification genes to tolerate the wide array of defense compounds being brought to bear in different hosts. These genes hold differential importance on distinct hosts as can be seen in the broad-host range pathogens S. sclerotiorum and B. cinerea (Figure 1). Both pathogens contain genes encoding putative cyanide hydratases for the detoxification of HCN and induction of this gene is likely regulated by the HCN content of the host, as cyanide hydratase expression has a dose-dependent relationship to cyanide content in other fungal systems (Barclay et al., 2002). Such a pattern is seen in the ITC detoxifying gene SsSaxA, which is only induced in the ITC containing crucifers A. thaliana and Brassica napus. The major toxin efflux transporters in B. cinerea are seemingly utilized to a much greater degree during infection of tomato than grapevine and inversely the laccase Bclcc2 which metabolizes the grapevine phytoalexin resveratrol is only induced during infection of grapevine (Figure 1). This transcriptional plasticity is a hallmark of broad-host range pathogens and the large number of detoxification mechanisms retained in their genome is likely a necessity for their lifestyle. This is similarly seen when comparing isolates of Alternaria alternata, a broad-host range necrotroph that retains tomatinase activity across a wide range of isolates regardless of their primary host, and Corynespora cassiicola, a comparatively narrow host range pathogen which has only retained tomatinase activity in specific tomato-infecting strains (Oka et al., 2006). Though it is convenient to utilize model plants to analyze virulence factors and pathogenic attributes of broad-host necrotrophs, considering the genomic plasticity seen in some of these pathogens, it is prudent to study their pathogenic development across multiple hosts.
An expansion of host range has been demonstrated through the horizontal gene transfer (HGT) of host-specific toxins in wheat pathogens, and a similar transfer of detoxification genes may have also allowed for the expansion of other pathogens to novel hosts (Mcdonald et al., 2019). Evidence suggests that phytopathogenic oomycetes gained cyanide hydratase genes through HGT from true fungi and benzoxazinoid detoxifying gene clusters appear to have spread through a similar mechanism from Fusarium spp. to species of Colletotrichum and Aspergillus (Richards et al., 2011; Glenn et al., 2016).
Detoxification and Plant Defense Signaling
In most plant–pathogen interactions the primary purpose of PDC detoxification appears to be the protection of the pathogen, but in some cases the metabolism of PDCs may play a larger role in subverting host defenses. This is highlighted by the tomatinase enzyme from S. lycopersici (Tom), which is known to hydrolyze the tomato phytoanticipin α-tomatine (Table 2) (Martin-Hernandez et al., 2000). An examination of tomatinase activity in Nicotiana benthamiana found evidence that the enzyme appears to suppress the host’s broader immune response in addition to its role in saponin detoxification (Bouarab et al., 2002). Indeed β2-tomatine, the hydrolysis product of α-tomatine, is capable of directly suppressing the induction of multiple plant defense genes (PR1, PR5, and AOS) during infection (Bouarab et al., 2002). β2-Tomatine also facilitated the growth of the bacterial pathogen Pseudomonas syringae pv. tabaci in planta (Bouarab et al., 2002). Although less well characterized, the Verticillium dahliae quercetinase enzyme, VDQase, is also suggested to manipulate the host phytohormone response to facilitate infection (Table 2) (El Hadrami et al., 2015).
These studies present an attractive avenue for future research, as the putative role of detoxification metabolites in actively facilitating infection has been largely overlooked. A potential example of this can be seen in the conversion of the flavanone sakuranetin into naringenin by the rice pathogen M. oryzae during infection (Katsumata et al., 2017). This process is assumed to be aimed at depleting the host of the antifungal sakuranetin, but naringenin is also a known inhibitor of lignin biosynthesis, revealing the possibility that its accumulation at the site of infection may suppress defensive lignification by the host (Deng et al., 2004). Moving into the future the effect of these detoxification metabolites on host physiology and defense must be evaluated as their role in pathogenesis may be as or more critical than the depletion of the initial PDC.
Manipulating Fungal Detoxification in Disease Control
Disruption of fungal detoxification activity can have a substantial effect on pathogenicity and subversion of this activity is potentially valuable in disease control across a wide range of crops. A novel approach to disease control through the subversion of fungal detoxification has been demonstrated with the usage synthetic compounds, termed paldoxins. Paldoxins, coined from “phytoalexin detoxification inhibitors,” are rationally designed compounds which inhibit the enzymatic detoxification mechanisms used by plant pathogens without the antifungal bioactivity seen in phytoalexins. This concept was originally demonstrated with the indoles cyclobrassinin and camalexin, which competitively bind to and inhibit brassinin degrading enzymes (BH and BO discussed in section “Enzymatic Detoxification of Inducible PDCs – Indoles”), but as phytoalexins themselves these compounds do not qualify as paldoxins. As opposed to other fungicides which are directly toxic to the pathogen itself, paldoxins would potentially allow for the plant to defend itself from invasion without the off target ecological effects of a broad-range fungicide (Pedras and Abdoli, 2017).
Inhibitors of ABC transporter-mediated efflux have also been proposed as a method to control fungal diseases, albeit in combination with chemical fungicides. Efflux is a major mechanism of derived fungal resistance to fungicides and by applying synthetic efflux inhibitors in combination with certain fungicides, it was shown that resistant isolates can be reverted to baseline levels of susceptibility, thus improving chemical control through fungicides (Reimann and Deising, 2005).
Reduced virulence is also achieved in a number of fungal pathogens through the silencing of pathogenicity and developmental factors (McLoughlin et al., 2018; Rosa et al., 2018). The usage of host-induced or spray-induced gene silencing (HIGS or SIGS) has never directly been used on genes involved in fungal detoxification, but as many studies demonstrated the critical importance many of these genes in host colonization, they may prove to be valuable targets for RNAi approaches (Bowyer et al., 1995; Vela-corcía et al., 2019).
Conclusion
In order to overcome both a plant’s induced and preformed PDCs during necrotrophic colonization, fungal pathogens utilize a number of distinct mechanisms (Figure 2). The earliest response to these compounds is the production of efflux transporters that work to export, or potentially sequester, toxic compounds below a lethal and/or inhibitory threshold. Secreted enzymes are then produced and metabolize PDCs in extracellular spaces, both to protect the pathogen from these compounds and, in some cases, prevent the production of other downstream antimicrobials. Many detoxification enzymes are intracellular as well, and the localization of these proteins either inside the fungal cell or in extracellular spaces can be a factor of the protein’s mechanism of action, the chemistry of its target substrate, and/or the PDC’s target in the fungus (i.e., plasma membrane vs. mitochondria) (Figure 2 and Table 2).
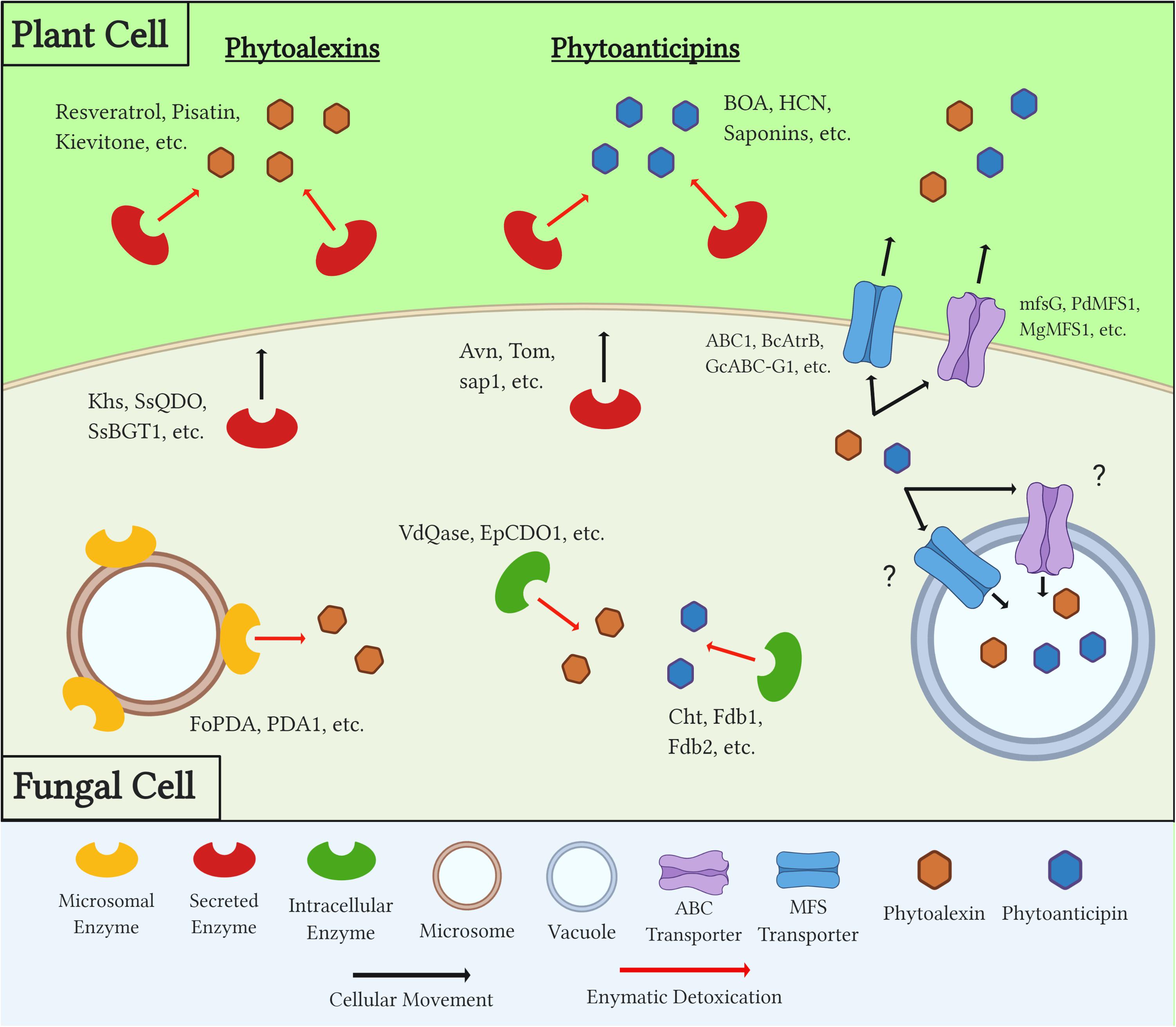
Figure 2. Diagram outlining generalized mechanisms of fungal tolerance for preformed and inducible plant defense compounds (PDCs). Examples of each class of PDC, detoxification enzyme, and transporter are included. Question marks denote a mechanism which is suspected but has yet to be confirmed through experimental evidence.
These detoxification mechanisms provide several benefits to the pathogen as they can quantitatively increase virulence and in some cases expand its host range to previously resistant plants. Additionally, when efficiently metabolized the formally toxic PDCs can serve as a valuable nutrient source during colonization. Overall, the range and redundancy of these mechanisms are evidence for the importance they play in most if not all necrotrophic/hemibiotrophic infections of plants, and a better understanding of these mechanisms will undoubtedly improve disease control strategies.
Author Contributions
NW wrote the manuscript. NW, MK, and DS revised the manuscript. All authors read and approved the manuscript.
Funding
This work was supported by the Wisconsin Soybean Marketing Board and the USDA National Institute of Food and Agriculture – National Sclerotinia Initiative (Award # 58-3060-8-023) to MK and DS.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Adrian, M., and Jeandet, P. (2012). Effects of resveratrol on the ultrastructure of Botrytis cinerea conidia and biological significance in Plant/Pathogen interactions. Fitoterapia 83, 1345–1350. doi: 10.1016/j.fitote.2012.04.004
Ahuja, I., Kissen, R., and Bones, A. M. (2012). Phytoalexins in defense against pathogens. Trends Plant Sci. 17, 73–90. doi: 10.1016/j.tplants.2011.11.002
Alamouti, S., Wang, V., Diguistini, S., Six, D. L., Bohlman, J., Hamelin, R. C., et al. (2011). Gene genealogies reveal cryptic species and host preferences for the pine fungal pathogen Grosmannia clavigera. Mol. Ecol. 20, 2581–2602. doi: 10.1111/j.1365-294x.2011.05109.x
Baldwin, T., Baldwin, S., Klos, K., Bregitzer, P., and Marshall, J. (2019). Deletion of the benzoxazinoid detoxification gene NAT1 in Fusarium graminearum reduces deoxynivalenol in spring wheat. PLoS One 14:e0214230. doi: 10.1371/journal.pone.0214230
Balzi, E., Wang, M., Leterme, S., Van Dyck, L., and Goffeau, A. (1994). PDR5, a novel yeast multidrug resistance conferring transporter controlled by the transcription regulator PDR1. J. Biol. Chem. 269, 2206–2214. doi: 10.1016/s0021-9258(17)42155-7
Barclay, M., Day, J. C., Thompson, I. P., Knowles, C. J., and Bailey, M. J. (2002). Substrate-regulated cyanide hydratase (Chy) gene expression in Fusarium solani: the potential of a transcription-based assay for monitoring the biotransformation of cyanide complexes. Environ. Microbiol. 4, 183–189. doi: 10.1046/j.1462-2920.2002.00284.x
Basse, C. W. (2005). Dissecting defense-related and developmental transcriptional responses of maize during ustilago maydis infection and subsequent tumor formation. Plant Physiol. 138, 1774–1784. doi: 10.1104/pp.105.061200
Bednarek, P., Pislewska-Bednarek, M., Svatos, A., Schneider, B., Doubsky, J., Mansurova, M., et al. (2009). A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323, 101–106. doi: 10.1126/science.1163732
Birgit, H., Arnemann, M., Schwenen, L., Stöckl, D., Bringmann, G., Jansen, J., et al. (1989). Degradation of the pterocarpan Phytoalexin (—)-Maackiain by ascochyta rabiei. Z. Naturforsc. 44c, 771–776. doi: 10.1515/znc-1989-9-1012
Bouarab, K., Melton, R., Peart, J., Baulcombe, D., and Osbourn, A. (2002). A saponin-detoxifying enzyme mediates suppression of plant defences. Nature 418, 889–892. doi: 10.1038/nature00950
Bowyer, P., Clarke, B. R., Lunness, P., Daniels, M. J., and Osbourn, A. E. (1995). Host range of a plant pathogenic fungus determined by a saponin detoxifying enzyme. Science 267, 371–374. doi: 10.1126/science.7824933
Buxdorf, K., Yaffe, H., Barda, O., and Levy, M. (2013). The effects of glucosinolates and their breakdown products on necrotrophic fungi. PLoS One 8:e70771. doi: 10.1371/journal.pone.0070771
Calmes, B., Morel-rouhier, M., Bataillé-simoneau, N., Gelhaye, E., Guillemette, T., and Simoneau, P. (2015). Characterization of glutathione transferases involved in the pathogenicity of Alternaria brassicicola. BMC Microbiol. 15:123. doi: 10.1186/s12866-015-0462-0
Caruso, F., Mendoza, L., Castro, P., Cotoras, M., Aguirre, M., Matsuhiro, B., et al. (2011). Antifungal activity of resveratrol against Botrytis cinerea is improved using 2-Furyl Derivatives. PLoS One 6:e25421. doi: 10.1371/journal.pone.0025421
Chen, J., Ullah, C., Reichelt, M., Beran, F., Yang, Z. L., Gershenzon, J., et al. (2020). The phytopathogenic fungus Sclerotinia sclerotiorum detoxifies plant glucosinolate hydrolysis products via an isothiocyanate hydrolase. Nat. Commun. 11, 1–12.
Chen, J., Ullah, C., Reichelt, M., Gershenzon, J., and Hammerbacher, A. (2019). Sclerotinia sclerotiorum circumvents flavonoid defenses by catabolizing flavonol glycosides and aglycones. Plant Physiol. 180, 1975–1987. doi: 10.1104/pp.19.00461
Chong, J., Poutaraud, A., and Hugueney, P. (2009). Metabolism and roles of stilbenes in plants. Plant Sci. 177, 143–155. doi: 10.1016/j.plantsci.2009.05.012
Ciuffetti, L. M., and Van Etten, H. D. (1996). Virulence of a pisatin demethylase-deficient nectria haematococca MPVI Isolate is increased by transformation with a pisatin demethylase Gene. Mol. Plant Microbe Interact. 9, 787–792. doi: 10.1094/mpmi-9-0787
Coleman, J. J., Wasmann, C. C., Usami, T., White, G. J., Esteban, D., Mccluskey, K., et al. (2011a). Characterization of the gene encoding pisatin demethylase (FoPDA 1) in Fusarium oxysporum. Mol. Plant Microbe Interact. 24, 1482–1491. doi: 10.1094/mpmi-05-11-0119
Coleman, J. J., White, G. J., Rodriguez-carres, M., and Van Etten, H. D. (2011b). An ABC transporter and a cytochrome P450 of nectria haematococca MPVI are virulence factors on pea and are the major tolerance mechanisms to the phytoalexin pisatin. Mol. Plant Microbe Interact. 24, 368–376. doi: 10.1094/mpmi-09-10-0198
Covert, S. F., Enkerli, J., Miao, V. P. W., and Van Etten, H. D. (1996). A gene for maackiain detoxification from a dispensable chromosome of nectria haematococca. Mol. General Genet. 251, 397–406. doi: 10.1007/bf02172367
Cruickshank, I. A. M., and Perrin, D. R. (1962). Studies on phytoalexins. Aust. J. Biol. Sci. 15, 147–159. doi: 10.1071/bi9620147
Curir, P., Dolci, M., Corea, G., Galeotti, F., and Lanzotti, V. (2006). The plant antifungal isoflavone genistein is metabolized by armillaria mellea vahl to give non- fungitoxic products. Plant Biosyst. 140, 156–162. doi: 10.1080/11263500600756363
Daoxin, L., Kuang-Ren, C., Smith, D. A., and Schardl, C. L. (1995). The Fusarium solani gene encoding kievitone hydratase, a secreted enzyme that catalyzes detoxification of a bean phytoalexin. Mol. Plant Microbe Interact. 8, 388–397. doi: 10.1094/mpmi-8-0388
Delserone, L. M., McCluskey, K., Matthews, D. E., and Van Etten, H. D. (1999). Pisatin demethylation by fungal pathogens and nonpathogens of pea: association with pisatin tolerance and virulence. Physiol. Mol. Plant Pathol. 55, 317–326. doi: 10.1006/pmpp.1999.0237
Deng, F., Aoki, M., and Yogo, Y. (2004). Effect of naringenin on the growth and lignin biosynthesis of gramineous plants. Weed Biol. Manag. 4, 49–55. doi: 10.1111/j.1445-6664.2003.00119.x
Denny, T. P., and Van Etten, H. D. (1982). Metabolism of the phytoalexins medicarpin and maackiain by Fusarium solani. Phytochemistry 21, 1023–1028. doi: 10.1016/s0031-9422(00)82409-7
Denny, T. P., and Van Etten, H. D. (1983a). Characterization of an inducible, nondegradative tolerance of nectria haematococca MP VI to phytoalexins. J. Gen. Microbiol. 129, 2903–2913. doi: 10.1099/00221287-129-9-2903
Denny, T. P., and Van Etten, H. D. (1983b). Tolerance of nectria haematococca MP VI to the phytoalexin pisatin in the absence of detoxification. J. Gen. Microbiol. 129, 2893–2901. doi: 10.1099/00221287-129-9-2893
Dewick, P. M., and Steele, M. (1982). Biosynthesis of the phytoalexin phaseollin in Phaseolus vulgaris. Phytochemistry 21, 1599–1603. doi: 10.1016/s0031-9422(82)85024-3
Dixon, R. A., Achnine, L., Kota, P., Liu, C., Srinivasa Reddy, M. S., and Wang, L. (2002). The Phenylpropanoid pathway and plant Defence — a genomics perspective. Mol. Plant Pathol. 3, 371–390. doi: 10.1046/j.1364-3703.2002.00131.x
Dubrovina, A. S., and Kiselev, K. V. (2017). Regulation of stilbene biosynthesis in plants. Planta 246, 597–623. doi: 10.1007/s00425-017-2730-8
Dufresne, M., Hugouvieux, V., Melton, R., and Osbourn, A. (2000). Effects of targeted replacement of the tomatinase gene on the interaction of septoria lycopersici with tomato plants. Mol. Plant Microbe Interact. 13, 1301–1311.
El Hadrami, A., Adam, L. R., and Daayf, F. (2011). Biocontrol treatments confer protection against verticillium dahliae infection of potato by inducing antimicrobial metabolites. Mol. Plant Microbe Interact. 24, 328–335. doi: 10.1094/mpmi-04-10-0098
El Hadrami, A., Islam, R., Adam, L. R., and Daayf, F. (2015). A Cupin domain-containing protein with a quercetinase activity (VdQase) regulates verticillium dahliae’s pathogenicity and contributes to counteracting host defenses. Front. Microbiol. 6:440. doi: 10.3389/fpls.2015.00440
Engleder, M., Horvat, M., Emmerstorfer-Augustin, A., Wriessnegger, T., Gabriel, S., Strohmeier, G., et al. (2018). Recombinant expression, purification and biochemical characterization of kievitone hydratase from nectria haematococca. PLoS One 13:e0192653. doi: 10.1371/journal.pone.0192653
Enkerli, J., Bhatt, G., and Covert, S. F. (1998). Maackiain detoxification contributes to the virulence of nectria haematococca MP VI on chickpea. Mol. Plant Microbe Interact. 11, 317–326. doi: 10.1094/mpmi.1998.11.4.317
Esaki, H., Onozaki, H., Morimitsu, Y., and Kawakishi, S. (1998). Potent antioxidative isoflavones isolated from soybeans fermented with Aspergillus saitoi. Biosci. Biotechnol. Biochem. 62, 740–746. doi: 10.1271/bbb.62.740
Franceschi, V. R., Krokene, P., Christiansen, E., and Krekling, T. (2005). Anatomical and chemical defenses of conifer bark against bark beetles and other pests. New Phytol. 167, 353–376. doi: 10.1111/j.1469-8137.2005.01436.x
Friebe, A., Vilich, V., Hennig, L., Kluge, M., and Sicker, D. (1998). Detoxification of benzoxazolinone allelochemicals from wheat by Gaeumannomyces graminis Var. Tritici, G. Graminis Var. Graminis, G. Graminis Var. Avenae, and Fusarium culmorum. Appl. Environ. Microbiol. 64, 2386–2391. doi: 10.1128/aem.64.7.2386-2391.1998
Fry, W. E., and Evans, P. H. (1977). Association of formamide hydrolyase with fungal pathogenicity to cyanogenic plants. Phytopathology 67, 1001–1006. doi: 10.1094/phyto-67-1001
George, H. L., Hirschi, K. D., and Van Etten, H. D. (1998). Biochemical properties of the products of cytochrome P450 Genes (PDA) encoding pisatin demethylase activity in nectria haematococca. Arch. Microbiol. 170, 147–154. doi: 10.1007/s002030050627
George, H. L., and Van Etten, H. D. (2001). Characterization of pisatin-inducible cytochrome P450s in fungal pathogens of pea that detoxify the pea phytoalexin pisatin. Fungal Genet. Biol. 33, 37–48. doi: 10.1006/fgbi.2001.1270
Glenn, A. E., and Bacon, C. W. (2009). FDB2 encodes a member of the arylamine N -acetyltransferase family and is necessary for biotransformation of benzoxazolinones by Fusarium verticillioides. J. Agric. Food Chem. 107, 657–671. doi: 10.1111/j.1365-2672.2009.04246.x
Glenn, A. E., Britton Davis, C., Gao, M., Gold, S. E., Mitchell, T. R., Proctor, R. H., et al. (2016). Two horizontally transferred xenobiotic resistance gene clusters associated with detoxification of benzoxazolinones by Fusarium Species. PLoS One 11:e0147486. doi: 10.1371/journal.pone.0147486
Glenn, A. E., Gold, S. E., and Bacon, C. W. (2002). Fdb1 and Fdb2, Fusarium verticillioides loci necessary for detoxification of preformed antimicrobials from corn. Mol. Plant Microbe Interact. 15, 91–101. doi: 10.1094/mpmi.2002.15.2.91
Gluck-thaler, E., and Slot, J. C. (2018). Specialized plant biochemistry drives gene clustering in fungi. ISME J. 12, 1694–1705. doi: 10.1038/s41396-018-0075-3
Gupta, A., and Chattoo, B. B. (2008). Functional analysis of a novel ABC transporter ABC4 from magnaporthe grisea. FEMS Microbiol. Lett. 278, 22–28. doi: 10.1111/j.1574-6968.2007.00937.x
Haile, Z. M., Pilati, S., Sonego, P., Malacarne, G., Vrhovsek, U., Engelen, K., et al. (2017). Molecular analysis of the early interaction between the grapevine flower and Botrytis cinerea reveals that prompt activation of specific host pathways leads to fungus quiescence. Plant Cell Environ. 40, 1409–1428. doi: 10.1111/pce.12937
Han, Y., Liu, X., Benny, U., Kistler, H. C., and Van Etten, H. D. (2001). Genes determining pathogenicity to pea are clustered on a supernumerary chromosome in the fungal plant pathogen Nectria Haematococca. Plant J. 25, 305–314. doi: 10.1046/j.1365-313x.2001.00969.x
Hayashi, K., Schoonbeek, H., and De Waard, M. A. (2002). Bcmfs1, a novel major facilitator superfamily transporter from Botrytis cinerea, provides tolerance towards the natural toxic compounds camptothecin and cercosporin and towards fungicides. Appl. Environ. Microbiol. 68, 4996–5004. doi: 10.1128/aem.68.10.4996-5004.2002
Higgins, J. (1981). Demethylation of the phytoalexin pisatin by Stemphylium botryosum. Can. J. Bot. 59, 1980–1981.
Higgins, V. J., Stoessl, A., and Heath, M. C. (1973). Conversion of phaseollin to phaseollinisoflavan by Stemphylium botryosum. Phytopathology 64, 105–107. doi: 10.1094/phyto-64-105
Huisjes, E. H., De Hulster, E., Van Dam, J. C., Pronk, J. T., and Van Maris, A. J. A. (2012). Galacturonic acid inhibits the growth of saccharomyces cerevisiae on galactose, xylose, arabinose. Appl. Environ. Microbiol. 78, 5052–5059. doi: 10.1128/aem.07617-11
Imai, T., Ohashi, Y., Mitsuhara, I., Seo, S., and Hasegawa, M. (2012). Identification of a degradation intermediate of the momilactone a rice phytoalexin by the rice blast fungus of the momilactone a rice phytoalexin by the rice blast fungus. Biosci. Biotechnol. Biochem. 76, 414–416. doi: 10.1271/bbb.110756
Ingham, J. L. (1976). Fungal modification of pterocarpan phytoalexins from melilotus alba and trifolium pratense. Phytochemistry 15, 1489–1495. doi: 10.1016/s0031-9422(00)88922-0
Katsumata, S., Hamana, K., Horie, K., Toshima, H., and Hasegawa, M. (2017). Identification of sternbin and naringenin as detoxified metabolites from the rice flavanone phytoalexin sakuranetin by Pyricularia Oryzae. Chem. Biodiv. 14:e1600240. doi: 10.1002/cbdv.201600240
Katsumata, S., Toshima, H., and Hasegawa, M. (2018). Xylosylated detoxification of the rice flavonoid phytoalexin sakuranetin by the rice sheath blight. Molecules 23:276. doi: 10.3390/molecules23020276
Keeling, C. I., and Bohlmann, J. (2006). Genes, enzymes and chemicals of terpenoid diversity in the constitutive and induced defence of conifers against insects and pathogens. New Phytol. 170, 657–675. doi: 10.1111/j.1469-8137.2006.01716.x
Kettle, A. J., Batley, J., Benfield, A. H., Manners, J. M., Kazan, K., and Gardiner, D. M. (2015). Degradation of the benzoxazolinone class of phytoalexins is important for virulence of Fusarium pseudograminearum towards wheat. Mol. Plant Pathol. 16, 946–962. doi: 10.1111/mpp.12250
Kraft, B., Schwenen, L., Stockl, D., and Barz, W. (1987). Degradation of the pterocarpan phytoalexin medicarpin by ascochyta rabiei. Arch. Microbiol. 147, 201–206. doi: 10.1007/bf00415285
Kuhn, P. J., and Smith, D. A. (1979). Isolation from Fusarium solani f. Sp. Phaseoli of an enzymic system responsible for kievitone and phaseollidin detoxification. Physiol. Plant Pathol. 14, 179–190. doi: 10.1016/0048-4059(79)90006-7
Kunz, C., Vandelle, E., Rolland, S., Poinssot, B., Bruel, C., Cimerman, A., et al. (2006). Characterization of a new, nonpathogenic mutant of Botrytis cinerea with impaired plant colonization capacity. New Phytol. 170, 537–550. doi: 10.1111/j.1469-8137.2006.01682.x
Kusumoto, N., Zhao, T., Swedjemark, G., Ashitani, T., Takahashi, K., and Borg-Karlson, A. K. (2014). Antifungal properties of terpenoids in picea abies against heterobasidion parviporum. For. Pathol. 44, 353–361.
Lairini, K., and Ruiz-rubio, M. (1998). Detoxification of α -Tomatine by Fusarium solani. Mycol. Res. 102, 1375–1380. doi: 10.1017/s095375629800656x
Lee, Y. J., Yamamoto, K., Hamamoto, H., Nakaune, R., and Hibi, T. (2005). A novel ABC transporter gene ABC2 involved in multidrug susceptibility but not pathogenicity in rice blast fungus, magnaporthe grisea. Pestic. Biochem. Physiol. 81, 13–23. doi: 10.1016/j.pestbp.2004.07.007
Lucy, M. C., Matthews, P. S., and Van Etten, H. D. (1988). Metabolic detoxification of the phytoalexins maackiain and medicarpin by nectria haematococca field isolates: relatioship to virulence on chickpea. Physiol. Mol. Plant Pathol. 33, 187–199. doi: 10.1016/0885-5765(88)90019-7
Lygin, A. V., Li, S., Vittal, R., Widholm, J. M., Hartman, G. L., and Lozovaya, V. V. (2009). The importance of phenolic metabolism to limit the growth of phakopsora pachyrhizi. Phytopathology 99, 1412–1420. doi: 10.1094/phyto-99-12-1412
Macfoy, C. A., and Smith, I. M. (1979). Phytoalexin production and degradation in relation to resistance of clover leaves to sclerofinia and Botrytis Spp. Physiol. Plant Pathol. 14, 99–111. doi: 10.1016/0048-4059(79)90029-8
Mäkelä, M. R., Marinović, M., Nousiainen, P., Liwanag, A. J. M., Benoit, I., Sipilä, J., et al. (2015). Aromatic metabolism of filamentous fungi in relation to the presence of aromatic compounds in plant biomass. Adv. Appl. Microbio. 91, 63–137. doi: 10.1016/bs.aambs.2014.12.001
Martens-uzunova, E. S., and Schaap, P. J. (2008). An evolutionary conserved D-Galacturonic acid metabolic pathway operates across filamentous fungi capable of pectin degradation an evolutionary conserved D -Galacturonic acid metabolic pathway operates across filamentous fungi capable of pectin degradation. Fungal Genet. Biol. 45, 1449–1457. doi: 10.1016/j.fgb.2008.08.002
Martin-Hernandez, A. M., Dufresne, M., Hugouvieux, V., Melton, R., and Osbourn, A. (2000). Effects of targeted replacement of the tomatinase gene on the interaction of septoria lycopersici with tomato plants. Mol. Plant Microbe Interact. 13, 1301–1311. doi: 10.1094/mpmi.2000.13.12.1301
Mcdonald, M. C., Taranto, A. P., Hill, E., Schwessinger, B., Liu, Z., Simpfendorfer, S., et al. (2019). Transposon-mediated horizontal transfer of the host-specific virulence protein toxa between three fungal wheat. mBio 10:e01515-19.
McLoughlin, A. G., Wytinck, N., Walker, P. L., Girard, I. J., Rashid, K. Y., De Kievit, T., et al. (2018). Identification and application of exogenous DsRNA confers plant protection against Sclerotinia sclerotiorum and Botrytis cinerea. Sci. Rep. 8:7320.
Miao, V. P., Covert, S. F., and Van Etten, H. D. (1991). A fungal gene for antibiotic resistance on a dispensable (“. B “) Chromosome. Science 254, 1773–1776. doi: 10.1126/science.1763326
Miao, V. P. W., and Van Etten, H. D. (1992). Three genes for metabolism of the phytoalexin maackiain in the plant pathogen nectria haematococca?: meiotic instability and relationship to a new gene for pisatin demethylase. Appl. Environ. Microbiol. 58, 801–808. doi: 10.1128/aem.58.3.801-808.1992
Morrissey, J. P., Wubben, J. P., and Osbourn, A. E. (2000). Stagonospora avenae secretes multiple enzymes that hydrolyze oat leaf saponins. Mol. Plant Microbe Interact. 13, 1041–1052. doi: 10.1094/mpmi.2000.13.10.1041
Nazlya, N., Knowlesa, C. J., Beardsmoreb, A. J., Naylorb, W. T., and Corcoranb, E. G. (1983). Detoxification of cyanide by immobilised fungi. J. Chem. Technol. Biotechnol. 33B, 119–126. doi: 10.1002/jctb.280330207
Oka, K., Okubo, A., and Kodama, M. (2006). Detoxification of a -tomatine by tomato pathogens Alternaria alternata tomato pathotype and corynespora cassiicola and its role in infection. Fungal Genet. Biol. 72, 152–158. doi: 10.1007/s10327-005-0262-8
Okmen, B., Etalo, D. W., Joosten, M. H. A. J., Bouwmeester, H. J., De Vos, R. C. H., and De Wit, P. J. G. M. (2013). Detoxification of a -tomatine by cladosporium fulvum is required for full virulence on tomato Er O. New Phytol. 198, 1203–1214. doi: 10.1111/nph.12208
Osbourn, A. (1996). Saponins and plant defence - a soap story. Trends Plant Sci. 1, 4–9. doi: 10.1016/s1360-1385(96)80016-1
Osbourn, A., Bowyer, P., Lunness, P., Clarke, B., and Daniels, M. (1995). Fungal pathogens of oat roots and tomato leaves employ closely related enzymes to detoxify different host plant saponins. Mol. Plant Microbe Interact. 8, 971–978. doi: 10.1094/mpmi-8-0971
Pacheco, T., Fassina, H., Ud, Z., Dias, I., Vinicius, J., and Rodrigues-filho, E. (2018). Bioorganic Chemistry conjugation of antifungal benzoic acid derivatives as a path for detoxi Fi Cation in Penicillium brasilianum, an endophyte from melia azedarach. Bioorg. Chem. 81, 367–372. doi: 10.1016/j.bioorg.2018.08.038
Pao, S. S., Paulsen, I. T., and Saier, M. H. (1998). Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62, 1–34.
Pedras, M. S. C., and Abdoli, A. (2017). Pathogen inactivation of cruciferous phytoalexins: detoxification reactions, enzymes and inhibitors. RSC Adv. 7, 23633–23646. doi: 10.1039/c7ra01574g
Pedras, M. S. C., and Ahiahonu, P. W. K. (2005). Metabolism and detoxification of phytoalexins and analogs by phytopathogenic fungi. Phytochemistry 66, 391–411. doi: 10.1016/j.phytochem.2004.12.032
Pedras, M. S. C., Ahiahonu, P. W. K., and Hossain, M. (2004). Detoxification of the cruciferous phytoalexin brassinin in Sclerotinia sclerotiorum requires an inducible glucosyltransferase. Phytochemistry 65, 2685–2694. doi: 10.1016/j.phytochem.2004.08.033
Pedras, M. S. C., Gadagi, R. S., Jha, M., and Sarma-mamillapalle, V. K. (2007). Detoxification of the phytoalexin brassinin by isolates of Leptosphaeria maculans pathogenic on brown mustard involves an inducible hydrolase. Phytochemistry 68, 1572–1578. doi: 10.1016/j.phytochem.2007.03.020
Pedras, M. S. C., and Hossain, M. (2006). Metabolism of crucifer phytoalexins in Sclerotinia sclerotiorum: detoxification of strongly antifungal compounds involves glucosylation †. Org. Biomol. Chem. 4, 2581–2590. doi: 10.1039/b604400j
Pedras, M. S. C., Hossain, S., and Snitynsky, R. B. (2011). Phytochemistry detoxification of cruciferous phytoalexins in Botrytis cinerea?: spontaneous dimerization of a camalexin metabolite. Phytochemistry 72, 199–206. doi: 10.1016/j.phytochem.2010.11.018
Pedras, M. S. C., Minic, Z., and Jha, M. (2008). Brassinin Oxidase, a fungal detoxifying enzyme to overcome a plant defense – purification, characterization and inhibition. FEBS J. 275, 3691–3705. doi: 10.1111/j.1742-4658.2008.06513.x
Pedras, M. S. C., Minic, Z., and Sarma-mamillapalle, V. K. (2009). Substrate specificity and inhibition of brassinin hydrolases, detoxifying enzymes from the plant pathogens Leptosphaeria maculans and Alternaria brassicicola. FEBS J. 276, 7412–7428. doi: 10.1111/j.1742-4658.2009.07457.x
Perrin, D. R., and Bottomley, W. (1961). Pisatin: an antifungal substance from pisum sativum L. Nature 191, 76–77. doi: 10.1038/191076a0
Petrasch, S., Silva, C. J., Mesquida-pesci, S. D., Gallegos, K., Van Den Abeele, C., Papin, V., et al. (2019). Infection strategies deployed by Botrytis cinerea, Fusarium Acuminatum, and rhizopus stolonifer as a function of tomato fruit ripening stage. Front. Plant Sci. 10:223. doi: 10.3389/fpls.2019.00223
Peyraud, R., Mbengue, M., Barbacci, A., and Raffaele, S. (2019). Intercellular cooperation in a fungal plant pathogen facilitates host colonization. Proc. Natl. Acad. Sci. U.S.A. 116, 3193–3201. doi: 10.1073/pnas.1811267116
Quidde, T., and Peter, B. (1999). Evidence for three different specific saponin-detoxifying activities in Botrytis cinerea and cloning and functional analysis of a gene coding for a putative avenacinase. Eur. J. Plant Pathol. 105, 273–283.
Ranjan, A., Westrick, N. M., Jain, S., Piotrowski, J. S., Ranjan, M., Kessens, R., et al. (2019). Resistance against Sclerotinia sclerotiorum in soybean involves a reprogramming of the phenylpropanoid pathway and Up-Regulation of Antifungal activity targeting ergosterol biosynthesis. Plant Biotechnol. J. 17, 1567–1581. doi: 10.1111/pbi.13082
Reimann, S., and Deising, H. B. (2005). Inhibition of efflux transporter-mediated fungicide resistance in pyrenophora tritici-repentis by a derivative of 4’-Hydroxyflavone and enhancement of fungicide activity. Appl. Environ. Microbiol. 71, 3269–3275. doi: 10.1128/aem.71.6.3269-3275.2005
Richards, T. A., Soanes, D. M., Jones, M. D. M., Vasieva, O., Leonard, G., Paszkiewics, K., et al. (2011). Horizontal gene transfer facilitated the evolution of plant parasitic mechanisms in the oomycetes. Proc. Natl. Acad. Sci. U.S.A. 108, 15258–15263. doi: 10.1073/pnas.1105100108
Roldán-arjona, T., Pérez-espinosa, A., and Ruiz-rubio, M. (1999). Tomatinase from Fusarium oxysporum f. Sp. lycopersici defines a new class of saponinases. Mol. Plant Microbe Interact. 12, 852–861. doi: 10.1094/mpmi.1999.12.10.852
Roohparvar, R., De Waard, M. A., Kema, G. H. J., and Zwiers, L. (2007). MgMfs1, a major facilitator superfamily transporter from the fungal wheat pathogen Mycosphaerella graminicola, is a strong protectant against natural toxic compounds and fungicides. Fungal Genet. Biol. 44, 378–388. doi: 10.1016/j.fgb.2006.09.007
Rosa, C., Kuo, Y., Wuriyanghan, H., and Falk, B. W. (2018). RNA interference mechanisms and applications in plant pathology. Annu. Rev. Phytopathol. 56, 581–610. doi: 10.1146/annurev-phyto-080417-050044
Sandrock, R. W., and Van Etten, H. D. (2001). The relevance of tomatinase activity in pathogens of tomato?: disruption of the B2 -Tomatinase Gene in Colletotrichum Coccodes and Septoria Lycopersici and Heterologous Expression of the Septoria Lycopersici B2 -Tomatinase in Nectria Haematococca, a pathogen of tomato fruit. Physiol. Mol. Plant Pathol. 58, 159–171. doi: 10.1006/pmpp.2001.0324
Sanglard, D., Ischer, F., Monod, M., and Billel, J. (1997). Cloning of candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology 143(Pt 2), 405–416. doi: 10.1099/00221287-143-2-405
Sanzani, S. M., De Girolamo, A., Schena, L., Solfrizzo, M., Ippolito, A., and Visconti, A. (2009). Control of Penicillium expansum and patulin accumulation on apples by quercetin and umbelliferone. Eur. Food Res. Technol. 228, 381–389. doi: 10.1007/s00217-008-0944-5
Schafer, W., Straney, D., Ciuffetti, L., Van Etten, H. D., and Yoder, O. C. (1989). One enzyme makes a fungal pathogen, but not a saprophyte, virulent on a new host plant. Science 246, 247–249. doi: 10.1126/science.246.4927.247
Schoonbeek, H., Del Sorbo, G., and de Waard, M. A. (2001). The ABC transporter BcatrB affects the sensitivity of Botrytis cinerea to the phytoalexin resveratrol and the fungicide fenpiclonil. Mol. Plant Microbe Interact. 14, 562–571. doi: 10.1094/mpmi.2001.14.4.562
Schoonbeek, H., Nistelrooy, J., and de Waard, M. (2003). Functional analysis of ABC transporter genes from Botrytis cinerea identifies BcatrB as a transporter of eugenol. Eur. J. Plant Pathol. 109, 1003–1011. doi: 10.1023/b:ejpp.0000003936.61182.14
Schouten, A., Maksimova, O., Cuesta-Arenas, Y., Van Den Berg, G., and Raaijmakers, J. M. (2008). Involvement of the ABC transporter BcAtrB and the Laccase BcLCC2 in defence of Botrytis cinerea against the broad-spectrum antibiotic 2,4- Diacetylphloroglucinol. Environ. Microbiol. 10, 1145–1157. doi: 10.1111/j.1462-2920.2007.01531.x
Schouten, A., Wagemakers, L., Stefanato, F. L., Van Der Kaaij, R. M., and Van Kan, J. A. L. (2002). Resveratrol acts as a natural profungicide and induces self-intoxication by a specific laccase. Mol. Microbiol. 43, 883–894. doi: 10.1046/j.1365-2958.2002.02801.x
Seifbarghi, S., Borhan, M. H., Wei, Y., Coutu, C., Robinson, S. J., and Hegedus, D. D. (2017). Changes in the Sclerotinia sclerotiorum transcriptome during infection of Brassica napus. BMC Genomics 18:266. doi: 10.1186/s12864-017-3642-5
Sellam, A., Poupard, P., and Simoneau, P. (2006). Molecular cloning of abgst1 encoding a glutathione transferase differentially expressed during exposure of Alternaria brassicicola to Isothiocyanates. FEMS Microbiol. Lett. 258, 241–249. doi: 10.1111/j.1574-6968.2006.00223.x
Semighini, C. P., Marins, M., Goldman, M. H. S., and Goldman, G. H. (2002). Quantitative analysis of the relative transcript levels of ABC transporter atr genes in Aspergillus nidulans by real-time reverse transcription-PCR assay. Appl. Environ. Microbiol. 68, 1351–1357. doi: 10.1128/aem.68.3.1351-1357.2002
Servos, J., Haase, E., and Brendel, M. (1993). Gene SNQ2 of saccharomyces cerevislae, which confers resistance to 4-Nitroquinoline-N-Oxide and other chemicals, encodes a 169 KDa protein homologous to ATP-Dependent permeases. MGG Mol. Gen. Genet. 236, 214–218. doi: 10.1007/bf00277115
Sexton, A. C., and Howlett, B. J. (2000). Characterisation of a cyanide hydratase gene in the phytopathogenic fungus Leptosphaeria maculans. Mol. Gen. Genet. 263, 463–470. doi: 10.1007/s004380051190
Sexton, A. C., Minic, Z., Cozijnsen, A. J., Pedras, M. S. C., and Howlett, B. J. (2009). Cloning, purification and characterisation of brassinin glucosyltransferase, a phytoalexin-detoxifying enzyme from the plant pathogen Sclerotinia sclerotiorum. Fungal Genet. Biol. 46, 201–209. doi: 10.1016/j.fgb.2008.10.014
Shahi, P., and Moye-Rowley, W. S. (2009). Coordinate control of lipid composition and drug transport activities is required for normal multidrug resistance in fungi. Biochim. Biophys. Acta 1794, 852–859. doi: 10.1016/j.bbapap.2008.12.012
Shimizu, J., Uehara, M., and Watanabe, M. (1982). Transformation of terpenoids in grape must by Botrytis cinerea. Agric. Biol. Chem. 46, 1339–1344. doi: 10.1271/bbb1961.46.1339
Smith, D. A., Harrer, J. M., and Cleveland, T. E. (1982). Relation between production of extracellular kievitone hydratase by isolates of Fusarium and their pathogenicity on Phaseolus vulgaris. Phytopathology 72, 1319–1323. doi: 10.1094/phyto-72-1319
Smith, D. A., Wheeler, H. E., Banks, S. W., and Cleveland, T. E. (1984). Association between lowered kievitone hydratase activity and reduced virulence to bean in Variants of Fusarium solani f. Sp. Phaseoli. Physiol. Plant Pathol. 25, 135–147. doi: 10.1016/0048-4059(84)90052-3
Stefanato, F. L., Abou-Mansour, E., Buchala, A., Kretschmer, M., Mosbach, A., Hahn, M., et al. (2009). The ABC transporter BcatrB from Botrytis cinerea exports camalexin and is a virulence factor on Arabidopsis thaliana. Plant J. 58, 499–510. doi: 10.1111/j.1365-313x.2009.03794.x
Stergiopoulos, I., Zwiers, L., and De Waard, M. A. (2003). The ABC transporter MgAtr4 Is a virulence factor of Mycosphaerella graminicola that affects colonization of substomatal cavities in wheat leaves. Mol. Plant Microbe Interact. 16, 689–698. doi: 10.1094/mpmi.2003.16.8.689
Tenhaken, R., Salmen, H. C., and Barz, W. (1991). Purification and characterization of pterocarpan hydroxylase, a flavoprotein monooxygenase from the fungus ascochyta rabid involved in pterocarpan phytoalexin metabolism. Arch. Microbiol. 155, 353–359.
Tiku, A. R. (2020). “Antimicrobial Compounds (Phytoanticipins and Phytoalexins) and their role in plant defense,” in Co-Evolution of Secondary Metabolites, eds J. M. Merillon and K. G. Ramawat (Berlin: Springer International Publishing), 845–868. doi: 10.1007/978-3-319-96397-6_63
Tranchimand, S., Ertel, G., Gaydou, V., Gaudin, C., Tron, T., and Iacazio, C. (2008). Biochemical and molecular characterization of a quercetinase from Penicillium olsonii. Biochimie 90, 781–789. doi: 10.1016/j.biochi.2007.12.004
Turbek, C. S., Smith, D. A., and Schardl, C. L. (1992). An extracellular enzyme from Fusarium solani f. Sp. phaseoli which catalyses hydration of the isoflavonoid phytoalexin, phaseollidin. FEMS Microbiol. Lett. 94, 187–190. doi: 10.1111/j.1574-6968.1992.tb05312.x
Urban, M., Bhargava, T., and Hamer, J. E. (1999). An ATP-Driven Efflux pump is a novel pathogenicity factor in rice blast disease. EMBO J. 18, 512–521. doi: 10.1093/emboj/18.3.512
Van Etten, H. D., Mansfield, J. W., Bailey, J. A., and Farmer, E. E. (1994). Two classes of plant antibiotics: phytoalexins versus “Phytoanticipins”. Plant Cell 6, 1191–1192. doi: 10.2307/3869817
Van Etten, H. D., Matthews, D. E., and Matthews, P. S. (1989). Phytoalexin detoxification?: importance for pathogenicity and practical implications. Annu. Rev. Phytopathol. 27, 143–164. doi: 10.1146/annurev.py.27.090189.001043
Van Etten, H. D., and Smith, D. A. (1975). Accumulation of antifungal isoflavonoids and in bean tissue infected with Fusarium solanif. Sp. Phaseoli. Physiol. Plant Pathol. 5, 225–237. doi: 10.1016/0048-4059(75)90089-2
Vela-corcía, D., Srivastava, D. A., Dafa-berger, A., Rotem, N., Barda, O., and Levy, M. (2019). MFS transporter from Botrytis cinerea provides tolerance to glucosinolate-breakdown products and is required for pathogenicity. Nat. Commun. 10:2886.
Wadke, N., Kandasamy, D., Vogel, H., Lah, L., Wing, B. D., Paetz, C., et al. (2016). The bark-beetle-associated fungus, endoconidiophora polonica, utilizes the phenolic defense compounds of its host as a carbon source. Plant Physiol. 171, 914–931.
Wang, L., Xu, M., Liu, C., Wang, J., Xi, H., Wu, B., et al. (2013). Resveratrols in grape berry skins and leaves in vitis germplasm. PLoS One 8:e0061642. doi: 10.1371/journal.pone.0061642
Wang, P., Sandrock, R. W., and Van Etten, H. D. (1999). Disruption of the cyanide hydratase gene in Gloeocercospora Sorghi Increases its sensitivity to the phytoanticipin cyanide but does not affect its pathogenicity on the cyanogenic plant sorghum. Fungal Genet. Biol. 28, 126–134.
Wang, P., and Van Etten, H. D. (1992). Cloning and properties of a cyanide hydratase gene from the phytopathogenic fungus Gloeocercospora sorghi. Biochem. Biophys. Res. Commun. 187, 1048–1054.
Wang, Y., Lim, L., Diguistini, S., Robertson, G., and Breuil, C. (2012). A specialized ABC efflux transporter GcABC-G1 confers monoterpene resistance to Grosmannia clavigera, a bark beetle-associated fungal pathogen of pine trees. New Phytol. 197, 886–898.
Wang, Y., Lim, L., Madilao, L., Lah, L., Bohlmann, J., and Breuil, C. (2014). Gene discovery for enzymes involved in limonene modification or utilization by the mountain pine beetle-associated pathogen Grosmannia clavigera. Appl. Environ. Microbiol. 80, 4566–4576.
Watanabe, M., Sumida, N., Yanai, K., and Murakami, T. (2004). A novel saponin hydrolase from Neocosmospora vasinfecta Var. Vasinfecta. Appl. Environ. Microbiol. 70, 865–872.
Wei, W., Pierre-pierre, N., Peng, H., Ellur, V., Vandemark, G. J., and Chen, W. (2020). The D-Galacturonic acid catabolic pathway genes differentially regulate virulence and salinity response in Sclerotinia sclerotiorum. Fungal Genet. Biol. 145:103482.
Weltring, K., Mackenbrock, K., and Barz, W. (1982). Demethylation, methylation and 3’-hydroxylation of isoflavones Bv Fusarium Fungi. Z. Naturforsc. 37, 570–574.
Weltring, K. M., Wessels, J., and Geyer, R. (1997). Metabolism of the potato saponins a-chaconine and a-solanine by Gibberella Pilicaris. Phytochemistry 46, 1005–1009.
Westlake, D. W. S., Roxburgh, J. M., and Talbot, G. (1961). Microbial production of carbon monoxide from flavonoids. Nature 189, 510–511.
Westrick, N. M., Ranjan, A., Jain, S., Grau, C. R., Smith, D. L., and Kabbage, M. (2019). Gene regulation of Sclerotinia sclerotiorum during infection of glycine max: on the road to pathogenesis. BMC Genomics 20:157. doi: 10.1186/s12864-019-5517-4
Wubben, J. P., Price, K. R., Daniels, M. J., and Osbourn, A. E. (1996). Detoxification of oat leaf saponins by septoria avenae. Phytopathology 86, 986–992.
Zhang, L., Thiewes, H., and Van Kan, J. A. L. (2011). The D -Galacturonic acid catabolic pathway in Botrytis cinerea. Fungal Genet. Biol. 48, 990–997.
Zhang, L., and van Kan, J. A. L. (2013). Botrytis cinerea mutants deficient in d-galacturonic acid catabolism have a perturbed virulence on Nicotiana benthamiana and Arabidopsis, but Not on Tomato. Mol. Plant Pathol. 14, 19–29.
Keywords: detoxification, necrotrophy, fungal pathogen, phytoalexin, phytoanticipin
Citation: Westrick NM, Smith DL and Kabbage M (2021) Disarming the Host: Detoxification of Plant Defense Compounds During Fungal Necrotrophy. Front. Plant Sci. 12:651716. doi: 10.3389/fpls.2021.651716
Received: 10 January 2021; Accepted: 26 March 2021;
Published: 30 April 2021.
Edited by:
Antonieta De Cal, Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (INIA), SpainReviewed by:
Morifumi Hasegawa, Ibaraki University, JapanChhana Ullah, Max Planck Institute for Chemical Ecology, Germany
Copyright © 2021 Westrick, Smith and Kabbage. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mehdi Kabbage, a2FiYmFnZUB3aXNjLmVkdQ==
 Nathaniel M. Westrick
Nathaniel M. Westrick Damon L. Smith
Damon L. Smith Mehdi Kabbage
Mehdi Kabbage