- 1Department of Agronomy, Iowa State University, Ames, IA, United States
- 2School of Plant and Environmental Sciences, Virginia Tech, Blacksburg, VA, United States
- 3Center for Quantitative Life Sciences and Department of Botany and Plant Pathology, Oregon State University, Corvallis, OR, United States
Phytophthora sojae is an oomycete that causes stem and root rot disease in soybean. P. sojae delivers many RxLR effector proteins, including Avr1b, into host cells to promote infection. We show here that Avr1b interacts with the soybean U-box protein, GmPUB1-1, in yeast two-hybrid, pull down, and bimolecular fluorescence complementation (BIFC) assays. GmPUB1-1, and a homeologous copy GmPUB1-2, are induced by infection and encode 403 amino acid proteins with U-Box domains at their N-termini. Non-synonymous mutations in the Avr1b C-terminus that abolish suppression of cell death also abolished the interaction of Avr1b with GmPUB1-1, while deletion of the GmPUB1-1 C-terminus, but not the U box, abolished the interaction. BIFC experiments suggested that the GmPUB1-1-Avr1b complex is targeted to the nucleus. In vitro ubiquitination assays demonstrated that GmPUB1-1 possesses E3 ligase activity. Silencing of the GmPUB1 genes in soybean cotyledons resulted in loss of recognition of Avr1b by gene products encoded by Rps1-b and Rps1-k. The recognition of Avr1k (which did not interact with GmPUB1-1) by Rps1-k plants was not, however, affected following GmPUB1-1 silencing. Furthermore, over-expression of GmPUB1-1 in particle bombardment experiments triggered cell death suggesting that GmPUB1 may be a positive regulator of effector-triggered immunity. In a yeast two-hybrid system, GmPUB1-1 also interacted with a number of other RxLR effectors including Avr1d, while Avr1b and Avr1d interacted with a number of other infection-induced GmPUB proteins, suggesting that the pathogen uses a multiplex of interactions of RxLR effectors with GmPUB proteins to modulate host immunity.
Introduction
Phytophthora root and stem rot disease caused by Phytophthora sojae is a destructive and the third most serious soybean disease in the U.S. (Tyler et al., 2007). In the U.S., the estimated average soybean annual yield suppression caused by this disease has been valued at $0.47 billion (Bandara et al., 2020). Losses to P. sojae have increased by 4-fold in the last 10 years (Wrather et al., 2001, 2003; Koenning and Wrather, 2010) as resistance genes have lost effectiveness. Soybean-P. sojae interactions have been extensively studied for understanding the signaling pathways involved in disease pathogenesis (Tyler, 2007, 2008).
Plants have evolved a number of strategies for combat disease-causing microorganisms (Duplan and Rivas, 2014) including effector-triggered immunity (ETI) (Gohre and Robatzek, 2008). ETI involves the direct or indirect recognition by R gene products of the presence of specific pathogen effectors. Effectors recognized by R proteins are termed avirulence (Avr) proteins. Rapid and localized cell death that limits pathogen growth, termed the hypersensitive response (HR), commonly accompanies ETI (Qutob et al., 2002; McDowell and Woffenden, 2003; Coll et al., 2011). Soybean R genes whose products recognize P. sojae Avr effectors and trigger Phytophthora resistance are known as Rps (Resistance to P. sojae) genes (Tyler and Gijzen, 2014). Soybean cultivars with Rps genes have proven to be an effective method to combat Phytophthora disease. One of the most durable Rps loci, Rps1-k, has been molecularly characterized. The Rps1-k locus carries two functional, highly similar coiled-coil nucleotide-binding and leucine-rich-repeat genes, Rps1-k-1 and Rps1-k-2 (Gao et al., 2005; Gao and Bhattacharyya, 2008). The Rps1-k locus enables soybean plants to recognize two genetically linked but dissimilar P. sojae effectors, Avr1b and Avr1k (Song et al., 2013), but it is unknown if recognition is mediated by distinct genes at the Rps1-k locus.
Degradation of ubiquitinated proteins through the 26S proteasome plays a major role in regulating defense responses (Goldberg, 2003; Vierstra, 2009; Marino et al., 2012; Duplan and Rivas, 2014). The formation of an isopeptide linkage between the C-terminal glycine residue of the ubiquitin peptide and the epsilon-amino groups of lysine residues of target proteins is mediated by a multi-enzyme cascade of three key enzymes: E1 (ubiquitin-activating enzyme), E2 (ubiquitin-conjugating enzyme), and E3 (ubiquitin-protein ligase) (Aravind and Koonin, 2000; Fang and Weissman, 2004). In the first step, ubiquitin is activated by E1 in an ATP-dependent manner. Then in the second step it is transferred to E2, to which it binds covalently. In the third step, E3 binds the E2-ubiquitin conjugate and the target protein and transfers the ubiquitin from E2 to a lysine residue on the target protein (Aravind and Koonin, 2000). The target specificity is determined by the E3 ubiquitin ligase. Compared to a relatively small number of E1 and E2 proteins, there are a very large number of E3 proteins with diverse specificities for E2 substrates and target proteins (Goldberg, 2003). Approximately 5% of Arabidopsis genes code for proteins of the ubiquitination pathway including two E1 proteins, at least 45 E2 proteins and more than 1,200 E3 proteins (Mazzucotelli et al., 2006; Yee and Goring, 2009). Depending on the structural features and mechanisms of action, E3 ligases are classified into four main sub-families: HECT (Homologous to the E6-AP Carboxy-Terminus), RING (Really Interesting New Gene), U-Box, and CRL (Cullin-RING ligases; Vierstra, 2009).
The Arabidopsis genome encodes 62 U-box proteins (Yee and Goring, 2009), while the soybean genome encodes more than 80 (Schmutz et al., 2010). Many of these U-Box proteins have been identified to be involved in self-incompatibility, hormone responses, biotic and abiotic stress responses (Monte et al., 2003; Stone et al., 2003; Yan et al., 2003; Cho et al., 2006, 2008; Liu et al., 2007; Samuel et al., 2008).
The importance of protein degradation in plant defenses was first established when SGT1 (Suppressor of G2 allele of SKP1), a component of the SCF complex for protein degradation, was demonstrated to be an essential component of R-gene mediated plant immunity (Azevedo et al., 2002). Subsequently, numerous examples of the integral role of ubiquitin-mediated degradation in the regulation of plant immunity, and its manipulation by pathogens, have been documented [reviewed in Trujillo and Shirasu (2010), Alcaide-Loridan and Jupin (2012), Marino et al. (2012), Duplan and Rivas (2014), Couto and Zipfel (2016), and Zhou and Zeng (2016)].
Plant U-Box (PUB) proteins with E3 Ub-ligase activity play an important role in regulating plant immune signaling, acting as positive or negative regulators. Positive regulators include tobacco NtCMPG1 and NtACRE276 (Gonzalez-Lamothe et al., 2006; Yang et al., 2006), the Arabidopsis ACRE276 homolog AtPUB17 (Yang et al., 2006; He et al., 2015), the Arabidopsis U-box proteins, MAC3A and MAC3B (Monaghan et al., 2009), pepper CaRING1 (Lee et al., 2011), Arabidopsis RGLG3 and RGLG4 (Zhang et al., 2012), and Arabidopsis SR1IP1 (Zhang et al., 2014). Negative regulators include rice SPL11 (Zeng et al., 2004; Shirsekar et al., 2014) and its Arabidopsis ortholog AtPUB13 (Li et al., 2012), the Arabidopsis U-box protein triplet, PUB22, PUB23, PUB24 (Trujillo et al., 2008), rice OsPUB51 (Park et al., 2012), and Arabidopsis ABD1 (Seo et al., 2014). In addition to U box protein genes with a demonstrated role in plant immunity, many others such as Arabidopsis CMPG1 homologs, AtPUB21 and AtPUB22 are induced by infection or elicitor treatment (Navarro et al., 2004).
Effector proteins produced by pathogens and mutualists also manipulate the ubiquitin-mediated degradation pathway, some by carrying E3 ligase activity and others by manipulating plant E3 ligases (Duplan and Rivas, 2014). Examples of effectors carrying E3 ligase activity include NopM (nodulation outer protein M) of Rhizobium sp. strain NGR234 (Xin et al., 2012), AvrPtoB of Pseudomonas syringae pv. tomato (Martin et al., 2003; Abramovitch et al., 2006; Rosebrock et al., 2007; Mathieu et al., 2014), and AvrPtoBB728a from Pseudomonas syringae pv. syringae B728a (homologous to AvrPtoB) (Chien et al., 2013). R. solanacearum GALA effectors (Angot et al., 2007) and Agrobacterium ASK1/ASK2 proteins (Anand et al., 2012) employ an F-box domain to form an active SCF-type E3 ubiquitin ligase required for virulence. Oomycete and fungal effectors without Ub-ligase related domains can target host E3 Ub-ligases. Avirulence effector AvrPiz-t from the rice blast fungus Magnaporthe oryzae suppresses the ubiquitin ligase activity of the rice RING E3 ubiquitin ligase APIP6 (Park et al., 2012), Avr3a from Phytophthora infestans stabilizes and inhibits the function of host U-box protein CMPG1 (Bos et al., 2010), and Avr1d from P. sojae binds to the U-Box of GmPUB13, acting as an E2 competitor (Lin et al., 2021).
Like other oomycete and fungal pathogens, P. sojae delivers a large collection of effector proteins into host cells during infection to promote colonization of soybean plants (Jiang et al., 2008; Wang et al., 2011; Jiang and Tyler, 2012). Avr1b is one such effector protein, conferring enhanced virulence when overexpressed in P. sojae (Shan et al., 2004; Dou et al., 2008). Although there is ample evidence suggesting involvement of U-Box proteins in the defense of rice, tobacco, tomato and Arabidopsis against invading pathogens (Zeng et al., 2004; Yang et al., 2006; Monaghan et al., 2009; Duplan and Rivas, 2014) very little is known about soybean E3 ligase proteins, except for a recently identified RING-finger protein with polyubiquitination activity (Du et al., 2010).
In the present study, a U-Box containing protein GmPUB1-1 with E3 ligase activity was identified by screening a soybean yeast two-hybrid library using Avr1b as a bait. Further yeast two-hybrid assays revealed multiplex interactions involving additional RxLR proteins and GmPUB proteins. Silencing and over-expression experiments suggest that GmPUB1-1 may be a positive regulator of effector-triggered immunity that is guarded against Avr1b-binding by proteins encoded by genes within the Rps1-b and Rps1-k loci.
Materials and Methods
Plant Materials, Transformation, and Growth Conditions
Seeds of soybean cultivars Williams and Williams 82 were sown in coarse vermiculite and soaked with 3 L of water immediately after sowing. The flats were kept in a Conviron Growth Chamber (16 h photoperiod, 200 μEs, 24°C day and 20°C night temperatures) for 7 days. On day 7, cotyledons were harvested for Agrobacterium rhizogenes-mediated transformation. The ideal cotyledons selected for the assay were ones that were medium green and not snapping completely into two halves. The 7-day old cotyledons were gently twisted off the plants and collected in Petri dishes with moist Whatman filter paper (10 cotyledons each plate). The surfaces of individual cotyledons were sterilized with alcohol wipes (North Safety Products, Cranston, RI). A shallow circular (4 mm in diameter) wound was made about 3 mm from the petiole end of the cotyledon by a razor blade. 20 μl of A. rhizogenes suspension (OD 0.3) in 10 mM MgSO4 was added to the wound. Plates were wrapped with Parafilm and incubated in a Conviron growth chamber (16 h photoperiod, 100 μEs, 24°C day and 20°C night temperatures).
A. rhizogenes strain K599 was transformed with pART27 GFP empty vector or pART27 GFP carrying a GmPUB1 inverted repeat by a freeze-thaw method. The transformants were selected on Luria-Bertani (LB) agar plates containing 50 μg/ml kanamycin and 100 μg/ml spectinomycin. Cultures for plant inoculation were prepared by inoculating 10 ml liquid LB broth (containing 50 μg/ml kanamycin and 100 μg/ml spectinomycin) with a single colony harboring the correct construct and growing the culture for two days at 28°C. Before inoculation onto the cotyledons, the cultures were centrifuged and resuspended in 10 mM MgSO4 for a final OD600 of 0.3.
P. sojae Infection Assays on Soybean Cotyledons
P. sojae strains CC5C (Sandhu et al., 2004), P6497 or P7063 (Förster et al., 1994) were grown on lima bean agar or V8 agar plates for 1 week in the dark at room temperature. Six days after inoculation of soybean cotyledons with A. rhizogenes cells, the non-petiole ends of the cotyledons were wounded lightly with a tip of a 1 ml pipette tip. A small piece (3 mm in diameter) of lima bean or V8 agar containing P. sojae mycelia was cut with a cork borer and placed onto the cut surfaces of the cotyledons. The cotyledons were incubated in the growth chamber (16 h photoperiod, 100 μEs, 24°C day and 20°C night temperatures) and observed every day for recording disease development.
The disease rating scale used in cotyledonary bioassays was as described by Park et al. (2002). Numbers 1–5 indicate different sizes of disease lesions. 0, no observable symptom development from the infection point; 1, 1–10%; 2, 10–25%; 3, 25–50%; 4, 50–75%; 5, 75–100% of the cotyledon area infected. Differences in average disease lesion ratings were statistically tested using the Wilcoxon Rank Sum Test.
For detection of GmPUB1 transcripts, cotyledons were collected 3, 6, 9, and 12 days post-transformation with A. rhizogenes for the time course experiment, and at 6 days for subsequent experiments. A column section was harvested by a cork borer (5 mm in diameter) at the non-petiole end (the site used for P. sojae inoculation) of the cotyledon. RNA was extracted from each column section for analysis by qRT-PCR.
Yeast Two-Hybrid Interaction Assays
Yeast two-hybrid interaction assays were carried out according to the manufacturer's protocol (Clontech Laboratories, Inc., Mountain View, CA). An unamplified prey cDNA library (>1.2 × 106 colony forming units) was generated in the pB42AD vector from poly (A)+ RNAs prepared from Williams 82 (Rps1-k) etiolated hypocotyls, 2 and 4 h following inoculation with the P. sojae isolate CC5C (Bhattacharyya and Ward, 1986; Sandhu et al., 2004). GmPUB1-1 fragments were identified from this prey soybean cDNA library by screening with Avr1b as bait. The GmPUB1-1 fragments were recovered in the prey vector pB42AD. This interaction was confirmed by introducing Avr1b wildtype and mutant alleles into the pLexA vector into a yeast strain containing the longest fragment, GmPUB1-171−397. Transformants were selected on SD/Gal/Raf/Xgal/BU salts (7 g/L Na2HP04, 7H2O; 3 g/L NaH2PO4, pH 7.0)/-His/-Trp/-Ura for screening β-galactosidase (LacZ) expression and SD/Gal/Raf/-His/-Trp/-Ura/-Leu plates for leucine auxotrophic selection. Full-length GmPUB1-1 and GmPUB1-2 genes were PCR amplified (Forward primer: 5′-ATATGGATCCGTATGGACGAAATTGAAATCCCTG-3′; reverse primer: 5′-TATCCTCGAGTCATGGATAGGAAGATAACAAAGGTAC-3′) from genomic DNA of the soybean cultivar Williams 82 and cloned into the BamHI-XhoI sites of the bait vector pLexA. pLexA_GmPUB1-1 and pLexA_GmPUB1-2 constructs were transformed into the yeast strain EGY48/pSH18-34 and selected on SD/-His/-Ura plates. An autoactivation assay was performed to test bait suitability. Because of the autonomous activation of GmPUB1-1 in the LexA system, only GmPUB1-2 was used as the bait for testing interactions of full length GmPUB proteins with Avr1b mutants. Avr1b genes carrying mutations in the W and Y motifs (Dou et al., 2008) and wild-type Avr1b were cloned into the pB42AD vector and introduced into yeast strains harboring the bait pLexA_GmPUB1-2. Double transformants were selected and screened for expression of LEU2 and LacZ reporter genes. Positive interactions were visualized as blue color on SD/Gal/Raf/X-gal/BU salts/-His/-Trp/-Ura plate from activation of LacZ gene and also from yeast growth on minimal medium lacking leucine due to complementation for the LEU2 gene.
For screening of GmPUB1-1, GmPUB1-2, and other PUB protein interactions with other Avr1b alleles and RxLR effectors, the effectors were cloned into pLAW10 and the PUB proteins were cloned into pLAW11. For reverse selection, the effectors were cloned into pLAW11 and the PUB proteins were cloned into pLAW10. pLAW10 and pLAW11 are Gateway™-compatible version of pGBKT7 and pGADT7, respectively (Yang et al., 2013). The genes were cloned into these vectors in a two-step process. First the genes were amplified with primers (listed in Supplementary Table 1) carrying attB sequences, then the fragments were cloned in pDONR207 via the Gateway™ BP reaction according to the manufacturer's protocol. Then the inserts were transferred into pLAW10 or pLAW11 using the Gateway™ LR reaction according to the manufacturer's protocol. pLAW10 constructs were introduced into yeast strain Y8930 (genotype: MATα, leu2-3,112 trp1-901 his3Δ200 ura3-52 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ cyh2R) using selection on –Trp medium with glucose as a carbon source. pLAW11 plasmids were introduced into Y8800 (genotype: MATaleu2-3,112 trp1-901 his3Δ200 ura3-52 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ cyh2R) using selection on –Leu medium with glucose as carbon source. To test for interactions, the bait- and prey-bearing strains were mated by co-inoculating 2 mL YEPD with each strain then incubating in a shaker at 240 rpm for overnight at 28°C. To select diploids, 40–80 μl of the mating cultures were plated on –Leu-Trp plates (glucose carbon course) and incubated for 40 h at 28°C. To test diploids for evidence of protein-protein interactions, single diploid colonies were inoculated into 2 mL –Leu-Trp liquid medium. After overnight incubation in a shaker with 240 rpm at 28°C, the cultures were diluted to an OD600 of 0.250 – 0.500 in a 96 well micro-titer plate. A 96-pin replicator was then used to transfer droplets from each well onto –Leu-Trp, -Leu-Trp-His, and -Leu-Trp-Ade plates. The plates were then incubated at 28°C for 3-5 days. GmPUB1-1 exhibited only very weak self-activation in pLAW10.
In vitro Pull-Down Assays Using Proteins Produced by in vitro Transcription-Translation
To produce Avr1b and GmPUB1-1 proteins by in vitro transcription-translation, the full-length Avr1b gene in pB42AD and GmPUB1-1 gene in pRSETA were used as the template for PCR amplification. Primers are presented in Supplementary Table 2. Forward primer with a Kozak consensus sequence and gene specific reverse primer were used in the first round of PCR. The resulting products were re-amplified with T7 promoter sequence containing forward primer and the same reverse primer. PCR products were translated using the wheat germ-based TNT® T7 quick for PCR system (Promega, Madison, WI) as recommended by the manufacturer's protocol. Expressed proteins were separated by SDS-PAGE and gel blotted for western analysis to assess the size and translational efficiency of individual proteins.
For pull-down assays, 90 μl TNT® reaction containing HA-Avr1b was diluted with 110 μl 1X PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4) pH 7.4. The total volume was incubated with 6 μl anti-HA agarose slurry (Pierce Technology Corporation, Holmdel, NJ) in a HandeeTM Mini-Spin Column (Pierce Technology Corporation, Holmdel, NJ) for 2 h at 4°C with gentle end-over-end rotation. The column was washed three times with PBS-T (0.15 M NaCl, 25 mM Tris-HCl, pH 7.2, 0.05% Tween-20) and the agarose was resuspended in 150 μl 1X PBS. 50 μl His-GmPUB1-1 TNT® reaction was added to the prepared column and incubated overnight at 4°C with end-over-end rotation. The columns were washed three times with 500 μl PBS-T buffer to remove any unbound proteins. Interacted GmPUB1 proteins were eluted with 25 μl non-reducing sample buffer (Pierce Technology Corporation, Holmdel, NJ) and boiling the slurry for 5 min. After adding 3 μl of β-mercaptoethanol, the samples were boiled again for 5 min and resolved on two 10% SDS-PAGE gels and detected with anti-His (Amersham Bioscience, PA) and anti-HA (Invitrogen, Carlsbad, CA) antibodies, respectively.
Bimolecular Fluorescence Complementation Transient Expression Assays in planta
For BiFC, the Avr1b ORF sequence was cloned into pSAT1-cEYFP-C1-B to generate the C-terminal in frame fusion with cEYFP (Forward primer: 5′-GACTAAGCTTCGATGCGTCTATCTTTTGTGC-3′; reverse primer: 5′-AGTCGGATCCTCACTGGTGGTGCTGGTGGTG-3'), whereas GmPUB1-1Δ71-397 was introduced into pSAT1-nEYFP-N1 to form the N-terminal in frame fusion with nEYFP (Forward primer: 5′-GACTAGATCTCGATGCAATCTTGGTGCACCCTC-3′; reverse primer: 5′-CAGGATCCCGGGTTCCTTTGCCCTCTCCTTAG-3′). Onion lamellas were placed inside Petri dishes containing moist filter papers and then bombarded with gold particles coated with each combination of plasmids: GmPUB1-1Δ71-397-YFPN/YFPC- Avr1b fusion plasmids or YFPN/YFPC as the negative control. Onion inner layer were bombarded at 1350 PSI Helium pressure and kept in dark at 22°C post-bombardment. The EGFP signal was detected 24 h after the bombardment with a fluorescence microscope (Carl Zeiss Axiostar Plus). The microscopic field was viewed under green isothiocyanate filters. Fluorescence was detected after 24 h with a fluorescence microscope (Carl Zeiss Axiostar Plus).
Expression and Purification of Recombinant Proteins in E. coli
The full-length wild type GmPUB1-1 and mutant GmPUB1-1 were cloned in the pRSET A (Invitrogen, Carlsbad, California) vector and used to produce 6XHis tagged fusion proteins. The mutant GmPUB1-1 was generated by substituting three highly conserved U-box domain amino acid residues (C12A, V23A, and W39Ile) previously shown to be critical for E3 ligase activity (Gonzalez-Lamothe et al., 2006; Yang et al., 2006). Plasmids were introduced into E. coli strain BL21(DE3)pLysS then the cells were induced with 1 mM isopropyl-β-D-thio-galactoside (IPTG) at OD 0.6. 16 h following induction at room temperature, cells were centrifuged and protein extraction done under native conditions using Ni-NTA resin (QIAGEN, Valencia, CA) following the manufacturer's protocol. The eluted proteins were dialyzed in 1X phosphate buffered saline pH 7.4.
In vitro Ubiquitination Assay
In vitro ubiquitination assays were conducted as described previously (Yang et al., 2006) with slight modifications. 1 μg of purified wild type and mutant His-GmPUB1 proteins, 50 ng of human recombinant E1 enzyme (Boston Biochem, Cambridge, MA), 150 ng of E2 UbcH5b (Boston Biochem, Cambridge, MA), 2 μg of HA-Ub (Boston Biochem, Cambridge, MA) were incubated in a final volume of 30 μl reaction buffer containing 50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 0.2 mM DTT, 3 mM ATP, 10 mM creatine phosphate and 0.1 unit of creatine phosphokinase (Sigma-Aldrich, St. Louis, MO). Reactions were incubated at 30°C for 2 h and stopped by boiling with 4X SDS-PAGE loading buffer for 5 min. 15 μl of each reaction was analyzed by electrophoresis on 10% SDS-PAGE and subjected to immunoblot analysis. Ubiquitination was detected with anti-HA antibody (Invitrogen, Carlsbad, CA) and anti-His (Amersham Bioscience, PA) antibody.
Western Blotting
Proteins were resolved in 10% polyacrylamide gel and then transferred to a nitrocellulose membrane (Whatman Inc. Florham Park, NJ). The membrane was blocked in PBS containing 0.1% Tween 20 and 5% fat-free dry milk for 1 h at room temperature followed by three washing with PBS/0.1% Tween-20. The membrane was incubated with primary antibodies (1:1500) and then with alkaline phosphatase conjugated secondary antibodies (1:1500). Specific proteins were visualized with AP conjugate substrate kit according to the manufacturer's instructions (Bio-rad, Hercules, CA).
RNAi Vector Construction
438 bp GmPUB1-1 fragments were PCR amplified with two sets of primers with different flanking restriction sites (Set 1: forward primer: 5′-GCATCTCGAGGCCTTTGCTACAATGCTGCT-3′ and reverse primer: 5′-GCATGGTACCCACTCTAGCATTGGCGGAAT-3′; Set 2: forward primer: 5′-GCATGGATCCGCCTTTGCTACAATGCTGCT-3′ and reverse primer: 5′-GCATATCGATCACTCTAGCATTGGCGGAAT-3′). The PCR products were ligated into the XhoI-KpnI and BamHI-ClaI sites of pHANNIBAL vector (Wesley et al., 2001) as an inverted repeat. A PDK (pyruvate orthophosphate dikinase) intron was placed between the two inverted repeats to stabilize the formation of a complementary hairpin structure (Wesley et al., 2001). The fragment containing the inverted repeats was sub-cloned as a NotI fragment into the pART27 GFP binary vector. The pART27 GFP vector was made by inserting a 35S:GFP from p35S-GFP vector (Clontech, Mountain View, CA) into the SacI site of pART27 (Wesley et al., 2001).
RT-PCR Assays of GmPUB1 Transcript Levels
Total RNA was extracted by TRIzol (Invitrogen, Carlsbad, California) or using a Qiagen RNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA) following the manufacturers' instructions. After treatment with RNase-free DNase (Invitrogen, Carlsbad, California), 2 μg RNA was incubated with 0.5 μg oligo(dT) at 70°C for 5 min then immediately transferred to ice. After denaturation, the following reagents were added to each tube: 5 μl M-MLV 5X reaction buffer, 5 μl 2.5 mM dNTPs, 20 units of RNase inhibitor (Invitrogen, Carlsbad, California), 200 units of M-MLV reverse transcriptase (Promega, Madison, WI) and RNase-free water to make up the volume to 25 μl. Reaction tubes were incubated at 30°C for 30 min and then at 42°C for 30 min.
For RT-PCR to detect GmPUB1 expression levels in silenced cotyledons at 3, 6, 9, and 12 days post-transformation with A. rhizogenes, 2 μl of 10X diluted first strand cDNA was used for specific gene amplification. GmPUB1 primer pair (forward primer: 5′-GGCTGCATTGAAGCTCATTGTGGAGCTC-3′, reverse primer: 5′-CGCCCTTCTGCACACCCACAAAGCTGATC-3′) was used to amplify both GmPUB1-1 and GmPUB1-2. The cycling parameters were 30 cycles of denaturing at 94°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 45 s; followed by 72°C for 10 min. The digested PCR products were separated in a 2% agarose gel and visualized under UV light. The soybean Actin gene was used as the internal control (forward primer: 5′-CCCTCAACCCAAAGGTCAACAG-3′, reverse primer: 5′-GGAATCTCTCTGCCCCAATTGTG-3′).
Quantitative RT-PCR assays of GmPUB1 transcript levels were conducted in a 7500 Fast Real-time PCR system (Applied Biosystems, Foster City, CA) using SYBR green. 20 ng of cDNA was used in each reaction and three technical replicates were performed. The primers used for analysis of GmPUB1 gene expression were designed based on the overlap region at the 3'end of GmPUB1-1 and GmPUB1-2. This region was not used in the RNAi construct. The sequence of the forward primer is: 5′-TCTAAGGGTCTCTCATGTGGC-3′ and the sequence of the reverse primer is 5′-GCCCCAACCTGCAACATTT-3′. GmCYP2 gene was used as a reference gene. The primers used to amplify GmCYP2 are: Forward primer 5′-CGGGACCAGTGTGCTTCTTCA-3′; reverse primer 5′-CCCCTCCACTACAAAGGCTCG-3′. The 2−ΔΔCt method was used to quantitate relative gene expression level.
Transcript Profiling of GmPUB Transcript Levels
Growth chamber-grown Williams soybean seedlings (7-day old) were inoculated in a slit in the hypocotyl with V8 broth-grown mycelia of P. sojae strain P6497, or hypocotyls were mock-inoculated by making an empty slit. After 12 h at 25°C in the light, the inoculated regions were harvested for RNA extraction. Total RNA was extracted from frozen and ground plant tissue using the Invitrogen ConcertTM Plant RNA Reagent and then pol(A)+ RNA was isolated using the Promega PolyATtract mRNA Isolation System III. Four independent biological replicates of 30 seedlings each were produced for mock and infected samples. The RNA samples were sequenced from the 3' end by Applied Biosystems (ABI) on their Solid™ platform. 22–27 million 25-nucleotide raw reads were obtained from each of the mock inoculated samples, and 55–66 million raw reads from each of the P. sojae-infected samples. Approximately 34–55% of the raw reads from each sample could be uniquely aligned to the soybean genome (Williams 82 version 1), and Reads Per Kilobase of transcript per Million mapped reads (RPKM) values were calculated for each soybean gene model for each sample using CuffLink. Differences in RPKM values between mock and infected samples were assessed using t-tests of log-transformed RPKM values, and using the multiple test correction of Benjamini and Hochberg (1995), using Excel software. For the GmPUB genes, RPKM values from closely related homeologs were averaged.
Particle Bombardment Assays of GmPUB1-1 Over-expression
For transient expression of GmPUB1-1 in bombardment assays, GmPUB1-1 was amplified using the primers 5′-AGATCCCGGGGGAGCAATGAGATATGGATGAAATTGAAATCCCTGCT-3′ and 5′-GCATGGGTACCTCATGGATAGGAAGATAACAAAGGT-3′ which introduced XmaI and KpnI sites upstream and downstream of the gene, respectively. These sites were then used to clone the gene into the bombardment vector pUC-SK (Dou et al., 2008). Bombardment assays and analysis of the resultant data were then conducted as described earlier (Kale and Tyler, 2011).
Results
Identification of a Soybean U-Box Protein That Interacts With Avr1b
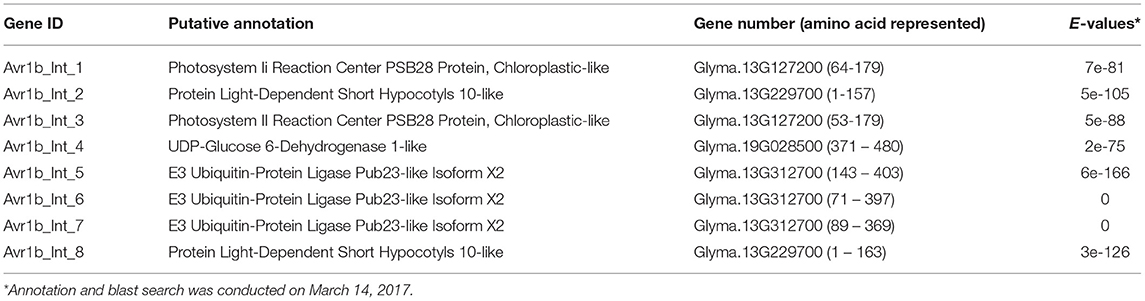
Screening of a yeast two-hybrid library prepared from P. sojae infected soybean hypocotyls using Avr1b as bait identified eight clones (Table 1). Among these eight Avr1b-interactors, three encoded portions of the same U-box protein, corresponding to gene model Glyma. 13G312700, which we named GmPUB1-1. In the yeast two-hybrid assay, full length GmPUB1-1 constructs produced positive interactions with Avr1b proteins encoded by Avr1b-1 genes from all four genotypes of P. sojae (Figure 1A). In addition to the GmPUB1-1 gene, the Glycine max genome contains a highly similar homeologous copy of GmPUB1-1, which we named GmPUB1-2 (Glyma.12g189000; https://phytozome-next.jgi.doe.gov/). GmPUB1-1 shares 94% amino acid identity with GmPUB1-2. Both the GmPUB1 genes encode proteins containing 403 amino acids with a U-Box domain located at the N-terminus. Based on the three fragments of GmPUB1-1 recovered in the initial yeast two-hybrid screen (Table 1), the minimal region of GmPUB1-1 required for binding of Avr1b lies between residues 143 and 369 and does not include the U-box.
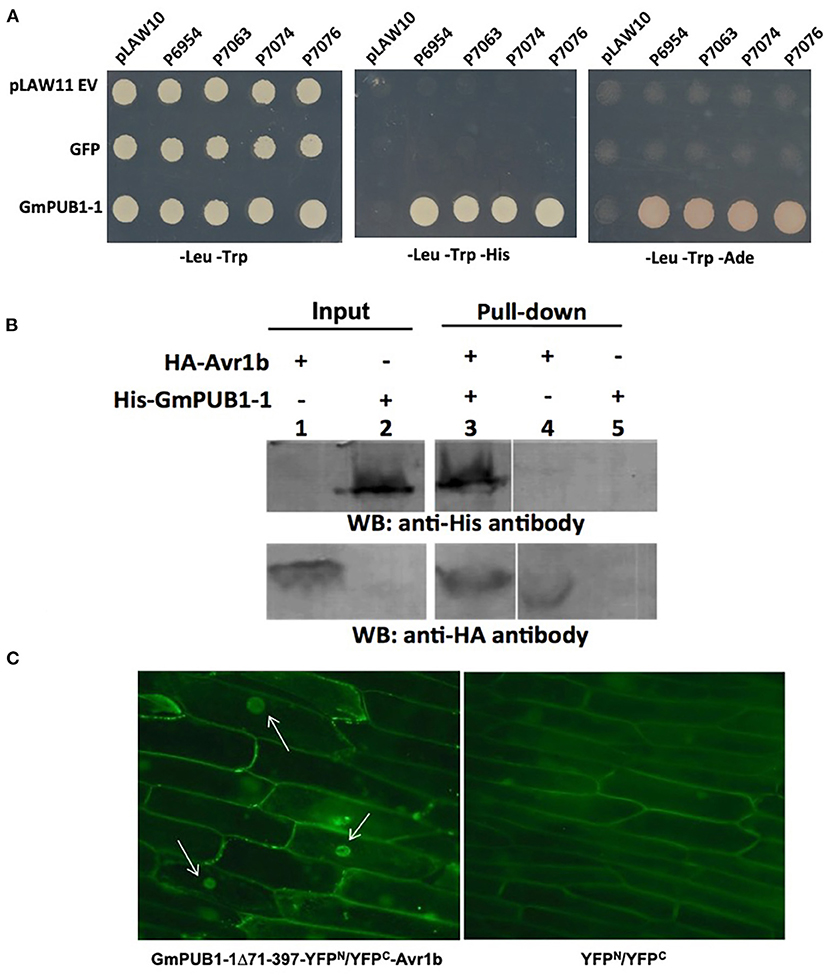
Figure 1. Specific interaction between GmPUB1-1 and Avr1b. (A) Interactions between GmPUB1-1 and four Avr1b alleles, detected by the yeast two-hybrid assay. Avr1b genes from P. sojae strains P6954 (genotype I, race 1), P7063 (genotype IV, race 6), P7074 (genotype III, race 17), or P7076 (genotype II, race 19) were cloned into bait plasmid pLAW10. GmPUB1-1 or GFP genes were cloned into prey vector pLAW11. pLAW10 or pLAW11 were used as empty vector (EV) controls, respectively. Haploids carrying the bait and prey plasmids were mated, and the diploids were tested for growth in the absence of histidine (-His) or adenine (-Ade) indicative of a positive interaction. (B) In vitro interaction of Avr1b with GmPUB1 proteins. Recombinant fusion proteins HA-Avr1b and His GmPUB1-1 were expressed via TNT® T7-quick-for-PCR DNA system (Promega, Madison, WI). Aliquots of HA-Avr1b (lane 1) or His-GmPUB1-1 (lane 2) proteins used in the pull-down assay were blotted onto membrane and probed with anti-HA and anti-His antibodies, respectively. His-GmPUB1-1 was incubated with HA-Avr1b that was immobilized onto anti-HA agarose beads overnight at 4°C. The bound proteins were eluted, resolved by SDS-PAGE and blotted onto membrane and then probed with anti-His and anti-HA antibodies, respectively (lane 3). HA-Avr1b (lane 4) or His-GmPUB1-1 (lane 5) alone were used as negative controls. Predicted sizes of the recombinant proteins are: HA-Avr1b, ~18.6 kDa; His-GmPUB1-1, ~48 kDa. (C) BiFC analysis showing in planta interaction between GmPUB1-1 and Avr1b. (Left) Nuclear fluorescence (arrows) observed from complementation of the C-terminus of YFP fused to Avr1b (Avr1b-C-EYFP) with the N-terminus of YFP fused to GmPUB1-171−397 (GmPUB1-171−397-N-EYFP) after bombardment into onion bulb epidermal cells. (Right) No fluorescence was detected in controls with C-EYFP and N-EYFP alone.
In vitro Pull-Down Assay Confirms the Interaction Between Avr1b and GmPUB1-1
To verify the protein interaction observed in the yeast two-hybrid experiment, we assayed the interaction between Avr1b and GmPUB1-1 in vitro by conducting a pull-down assay. His-tagged GmPUB1-1 protein was incubated with HA-tagged Avr1b protein immobilized on HA beads. Unbound proteins were washed away using PBS-T buffer. Associated proteins were then eluted, separated by SDS-PAGE, and immunoblotted with anti-His and anti-HA antibodies. Figure 1B shows that His-GmPUB1-1 was pulled down by HA-Avr1b (lane 3). However, when HA-Avr1b was omitted, His-GmPUB1-1 did not appear in the pull-down fraction (lane 5), indicating the specificity of the assay. This result therefore provides in vitro confirmation of the interaction between GmPUB1-1 and Avr1b.
Complexes of Avr1b and GmPUB1-1 Become Predominantly Localized to the Nuclei in planta
Bimolecular fluorescence complementation (BiFC) assays were conducted to examine the interaction of Avr1b and GmPUB1-1 in planta. Onion cells were bombarded with two plasmids carrying either Avr1b or GmPUB1-1 fused to split yellow fluorescence protein; i.e., in one construct Avr1b was fused to the C-terminal half of YFP and in the other construct, GmPUB1-171−397 (lacking the entire U-box domain) was fused to the N-terminal half of YFP. Assuming Avr1b and GmPUB1-171−397 interacted in planta, the interaction would bring the two halves of YFP into close proximity to form a functional fluorophore. Empty vector combinations (half of YFP not fused to an interaction partner) were used as the negative control. As shown in Figure 1C, bright fluorescence was detected in nuclei with weaker fluorescence in the cytoplasm, consistent with Avr1b interacting with GmPUB1-1 in plant cells, and with the complex being predominantly targeted to nuclei. No fluorescence was detected in the negative control (Figure 1C).
Mutations in the C-Terminal W-Y Domain of Avr1b Result in Loss of Interaction With GmPUBs
More than half of the Phytophthora effector proteins share conserved W and Y motifs in their C-terminal domains, which form a conserved structural fold (Win et al., 2012b). Different amino acid substitutions in the C-terminal W and Y motifs of Avr1b abolish recognition of the protein by Rps1-b plants or the ability of Avr1b to suppress cell death in soybean lines lacking Rps1-b (Shan et al., 2004; Dou et al., 2008). To test whether these two motifs might also be involved in interaction with GmPUB1 proteins, a yeast two-hybrid assay was conducted to test the interaction of the mutant proteins with GmPUB1-1. Constructs encoding Avr1b point mutants in the W and Y motifs (Figure 2A) were cloned into the “bait” vector pLexA. For the “prey” plasmid, we used the C-terminal fragment of GmPUB1-1 (GmPUB1-171−397 lacking the U box domain) that was recovered in the original yeast two-hybrid screen (Figure 2B). The “bait” and “prey” plasmids were introduced into the yeast strain EGY48/pSH18-34. This yeast strain has two reporter genes, LEU2 and LacZ. Protein interaction results in β-galactosidase (LacZ) activity and growth in medium lacking leucine (Leu2). Avr1b mutants W3, W4, and Y1 did not interact with GmPUB1- 171−397 (Figure 2C); whereas the wild type and mutants W2, W5 (weakly) and W6 did interact. In order to examine interactions between full-length GmPUB1s and Avr1b, we cloned GmPUB1-1 into the bait vector, pLexA. However, GmPUB1-1 displayed strong self-activation, so reliable results could not be obtained. GmPUB1-2 did not display self-activation; and therefore, considering the high identity (94%) between GmPUB1-1 and GmPUB1-2, we tested the interactions of full length GmPUB1-2 with the Avr1b wild type and mutants. GmPUB1-2 reproducibly interacted with Avr1b and some of its mutants, albeit more weakly than GmPUB1-1 (Figure 2C). Avr1b wild type and mutant W6 preserved some ability to interact with GmPUB1-2 but interaction with W2 was especially weak, and no interaction with W5 could be detected (Figure 2C). In summary, mutations such as W3 and W4 that abolish the ability of Avr1b to suppress cell death also abolished the ability to interact with GmPUB1-1; whereas mutations such as W2 that abolish recognition of Avr1b by Rps1-b, but not cell death suppression, did not abolish the GmPUB1-1 interaction. Mutants W5 and Y1 showed partial departure from this pattern: W5 abolished cell death suppression but only weakened the interaction with GmPUB1-1, while Y1 abolished the interaction with GmPUB1-1 but only weakened cell death suppression. All mutations that abolished the interaction also abolished recognition by Rps1-b-containing soybean lines. However, some mutations that abolished recognition by Rps1-b-containing lines did not abolish the interaction with GmPUB1-1. These results suggest that the interaction is necessary but not sufficient for recognition by Rps1-b plants. Overall, the results from the mutation analysis support that the interaction between Avr1b and GmPUB1-1 is relevant to the functioning of Avr1b inside soybean cells.
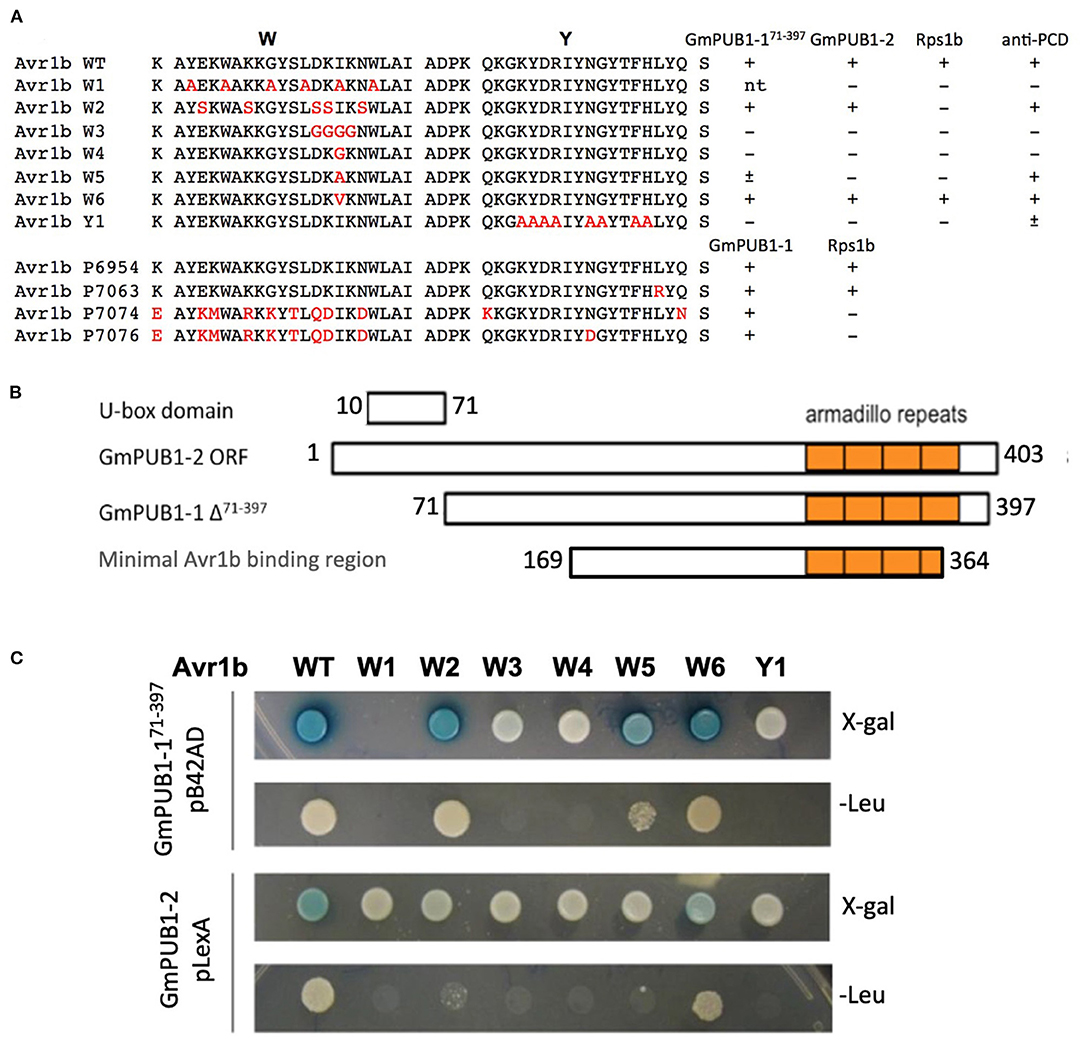
Figure 2. Interactions of GmPUB1s with Avr1b and mutant Avr1b proteins. (A) Sequences of W and Y motifs of Avr1b, Avr1b mutants, Avr1b variants and their interaction with GmPUB1s and PCD responses in an Rps1-b containing line. Sequence differences are in red. +, interaction; -, no interaction; ±, partially active; n.t., not tested. Anti-PCD activity was cited from Dou et al. (2008), indicating the ability of Avr1b to suppress pro-apoptopic protein BAX-induced programmed cell death (PCD) in soybean leaves containing Rps1-b. (B) Schematic representation of protein fragment of GmPUB1s used in yeast two-hybrid assay, and the minimal region required for Avr1b binding, based on the fragment of GmPUB1-1 recovered in the original two-hybrid screen (Table 1). (C) Yeast two-hybrid assays for in vivo interactions of GmPUB1 with Avr1b and mutant Avr1b proteins. Yeast strain EGY48/pSH18-34 containing combinations of truncated GmPUB1-1 in the prey vector pB42AD and C-terminal mutated forms of Avr1b in the bait vector pLexA, or full length GmPUB1-2 in the bait vector pLexA and Avr1b mutants in the prey vector pB42AD were grown on SD/Gal/Raf/X-gal/BU/-His/-Trp/-Ura plate and SD/Gal/Raf/-His/-Trp/- Ura/-Leu plate for 2 days to allow detection of LacZ and Leu2 gene expression respectively. Blue color in X-gal containing plate and colony growth on minimum medium lacking leucine suggest the presence of protein-protein interactions.
GmPUB1-1 Has Ubiquitin-E3 Ligase Activity
To determine whether GmPUB1-1 protein is a functional E3 ubiquitin ligase, full-length wild type GmPUB1-1 protein was expressed in E. coli as a fusion protein with a His tag. As a negative control, we also expressed mutant GmPUB1-1m3 protein, with three substitution mutations of highly conserved U-box domain amino acids, Cys-12 to Ala, Val-23 to Ile, and Trp-39 to Ala. The purified proteins were used for in vitro ubiquitination assays. His-tagged GmPUB1-1 and GmPUB1-1m3 were incubated with or without HA-Ub, ATP, E1 protein and E2 protein for 2 hours followed by western blot analysis with anti-HA and anti-His antibodies. In the presence of E1, E2, Ub, ATP and GmPUB1-1, ubiquitination of E. coli proteins (lane 6 in Figure 3) was detected with a monoclonal anti-HA antibody. No clear protein ubiquitination was detected in the absence of ATP, Ub, E1, E2 or E3 (lanes 1–5, Figure 3). Mutation of the conserved U-Box domain residues in GmPUB1-1m3 abolished the E3 activity (lane 7, Figure 3; Gonzalez-Lamothe et al., 2006; Yang et al., 2006). These results indicate that the U-Box domain of GmPUB1-1 protein possesses E3 ligase activity.
Figure 3. E3 Ubiquitin ligase activity of GmPUB1-1. In vitro ubiquitination assay with E. coli expressed His-GmPUB1-1 was performed in presence of combination of components as listed on top of the Figure. As a negative control for the E3 activity, a mutated version of His-GmPUB1-1 was used (three conserved amino acid residues Cys-12, Val23 and Trp-39 mutated to Ala, Ile and Ala respectively, in U-box domain). The immunoblots were probed with anti-HA antibody (top) to detect ubiquitinated E. coli proteins. Anti-His antibody (bottom) was used to detect His-GmPUB1-1 (~ 48 kDa) and its mutated version. Numbers on the left are molecular weight markers in kDa.
GmPUB1 Silencing Compromises Rps1-b- and Rps1-k-Mediated P. sojae Resistance Triggered by Avr1b
To investigate if the two GmPUB1 genes are required for P. sojae resistance in soybean, we silenced GmPUB1-1 and GmPUB1-2 jointly using a dsRNA construct in an Agrobacterium rhizogenes cotyledon assay developed by Subramanian et al. (2005). It was previously reported that silencing of the isoflavone synthase (IFS) gene occurred systemically in soybean cotyledons distal to A. rhizogenes-inoculation sites, and the most effective silencing period was between 5 and 7 days following transformation (Subramanian et al., 2005). We produced the GmPUB1 RNAi constructs by cloning a 438 bp DNA fragment of the GmPUB1-1 gene as inverted repeats separated by an intron in the pART27 vector (Wesley et al., 2001). The GmPUB1-1 438 bp fragment was highly similar to GmPUB1-2, and was expected to silence both genes. Soybean cotyledons were transformed with A. rhizogenes strain K599 carrying the empty vector pART27 GFP or the GmPUB1-1 RNAi construct. Cotyledon tissues distal to the A. rhizogenes-inoculation sites were collected 3, 6, 9, 12 days following transformation. RT-PCR assays revealed that GmPUB1 silencing was maximal at around 6 days (Supplementary Figure 1). To determine the effect of GmPUB1 silencing on resistance to P. sojae, on day 6 an agar pulp containing P. sojae mycelia was placed on wounded sites distal to the A. rhizogenes-inoculation sites and symptoms were recorded every 24 h period following infection. When susceptible cultivar Williams was inoculated with P. sojae strain P6497 (race 2), which does not express Avr1b, there was no obvious difference in the susceptibility of the cotyledons in which GmPUB1 genes were silenced (Figures 4A,B). Dark brown, water-soaked lesions spread to the entire cotyledon at the same rate in each case. Avr1b is recognized by the products of two Rps genes, Rps1-b, and Rps1-k. When cultivars L77-1863 or Williams 82 containing Rps1-b or Rps1-k respectively in the Williams background were used, silencing of the GmPUB1 genes in both genotypes caused loss of resistance to P. sojae strain P7063 (race 6), which expresses Avr1b (Figures 4C–F). However, the effect of silencing was specific to recognition of Avr1b; when Williams 82 (Rps1-k) was inoculated with P. sojae P6497, which expresses a different effector recognized by Rps1-k plants, namely Avr1k, silencing of GmPUB1 genes did not result in loss of resistance (Figures 4F,G). Furthermore, when P6497 was used to inoculate soybean cultivars containing Rps1-d, Rps3-a, or Rps4 and silenced for GmPUB1 genes, no loss of recognition was observed (Figure 5). Thus, silencing of GmPUB1 genes only abolished resistance triggered by Avr1b.
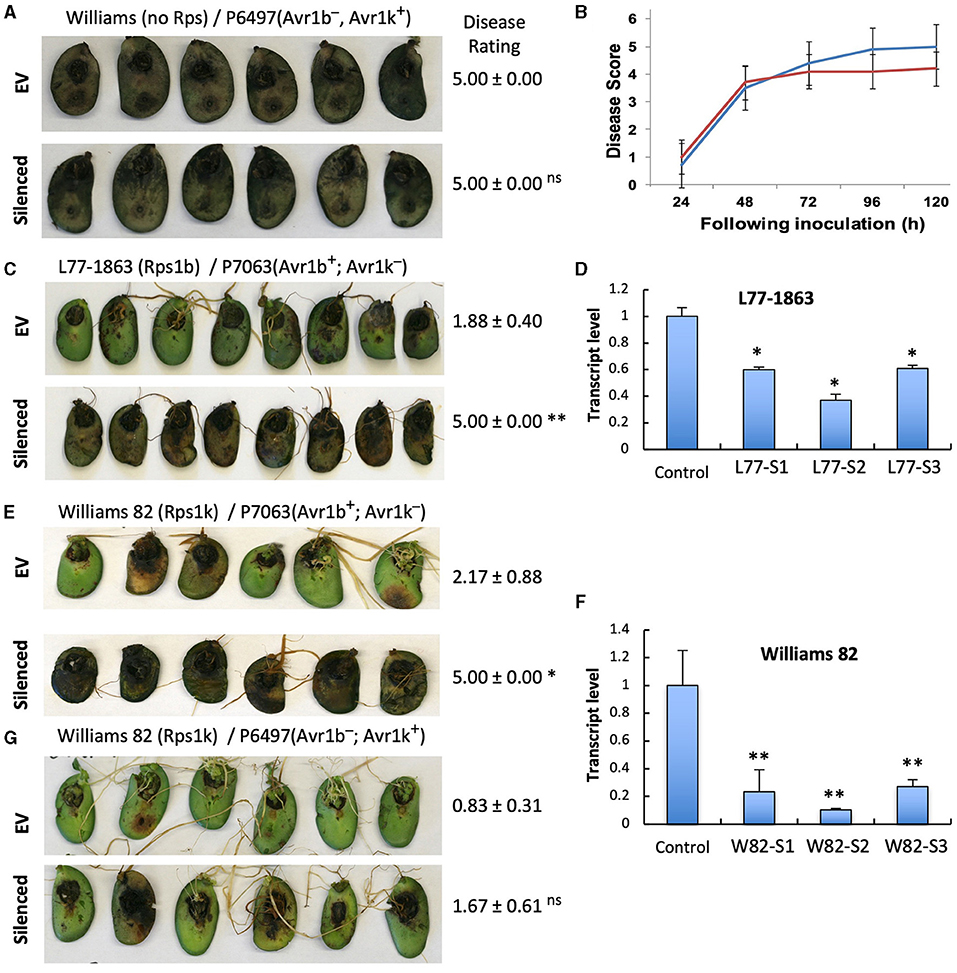
Figure 4. Silencing of GmPUB1 genes resulted in specific loss of Avr1b recognition by products of the Rps1-b and Rps1-k loci. (A,C,E,G) Soybean cotyledons carrying Rps1-b (C), Rps1-k (E,G), or no Rps gene (A) were inoculated with A. rhizogenes harboring the GmPUB1-1:RNAi vector or pART27GFP empty vector. Six days later, the cotyledons were inoculated with mycelia of P. sojae strains P6497 (A,G) or P7063 (C,E). Cotyledons were photographed 3 days after P. sojae inoculation and the disease rating scored as described in the Methods. True disease lesions showed diffuse brown margins. HR lesions were smaller and showed sharp black margins. Disease rating means and standard errors are shown. Significance of the differences between empty vector (EV) and silenced cotyledons were determined by the Wilcoxon Rank Sum Test; *p < 0.05, **p < 0.01, ns = not significant p > 0.05. GmPUB1-1 and GmPUB1-2 sequences are near-identical and expected to be co-silenced. (B) Disease development over time following inoculation with P. sojae CC5C. Means and standard errors were calculated from 20 infected cotyledons. Red line represents the GmPUB1 silenced cotyledons and blue line represents the empty vector transformed cotyledons. (D,F) Transcript levels of GmPUB1 (GmPUB1-1 and GmPUB1-2 combined) were measured by qRT-PCR in three individual soybean cotyledons of L77-1863 (D) or Williams 82 (F) inoculated with A. rhizogenes harboring the GmPUB1-1:RNAi vector. Error bars represent standard errors from three technical replicates. * and ** indicate p < 0.01 and 0.001, respectively, for the difference with the control.
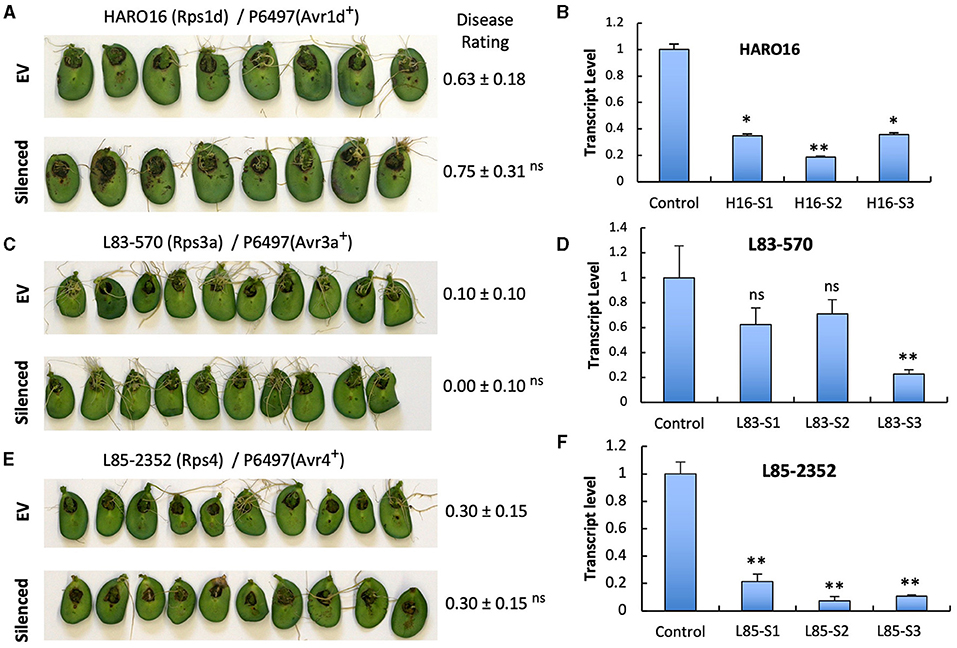
Figure 5. Silencing of GmPUB1 genes does not affect effector recognition by products of the Rps1-d, Rps3-a, and Rps4 genes. (A,C,E) Soybean cotyledons carrying Rps1-a (A), Rps3-a (C), or Rps4 (E) were inoculated with A. rhizogenes harboring the GmPUB1-1:RNAi vector or pART27GFP empty vector. Six days later, the cotyledons were inoculated with mycelia of P. sojae strains P6497. Cotyledons were photographed 3 days after P. sojae inoculation and disease levels were assessed as in Figure 4. (B,D,F) Transcript levels of GmPUB1 (GmPUB1-1 and GmPUB1-2 combined) were measured by qRT-PCR in three individual soybean cotyledons of HARO16 (B), L83-570 (D), or L85-2352 (F) inoculated with A. rhizogenes harboring the GmPUB1-1:RNAi vector. Error bars represent standard errors from three technical replicates. ns, * or ** indicate p > 0.05, p < 0.01 or p < 0.001, respectively, for the difference with the control.
To determine if GmPUB1-1 may be a positive or negative regulator of cell death, a double-barrel-bombardment transient assay (Kale and Tyler, 2011) was conducted using the reporter gene GUS. By using a control bombardment side-by-side with the test (i.e., GmPUB1-1) bombardment, this assay can directly measure cell death triggered by a gene of interest. It was observed that transient expression of GmPUB1-1 caused significant (p < 0.001) cell death (Supplementary Figure 2) suggesting that GmPUB1-1 might be a positive regulator of cell death.
GmPUB Family Proteins Interact With Multiple P. sojae RxLR Effectors
Phylogenetic analysis (Figure 6A) revealed that GmPUB1-1 and GmPUB1-2 share close similarity with Arabidopsis U-box proteins, AtPUB22, AtPUB23, and AtPUB24 (Trujillo et al., 2008). GmPUB1-1 and GmPUB1-2 belong to a clade of U box proteins containing 17 GmPUB proteins (including seven pairs of homeologs), as well as U box proteins from Arabidopsis (AtPUB20-24), Nicotiana (CMPG1) and Capsicum (CaPUB1) (Figure 6A). Many of the GmPUB genes are strongly up-regulated during P. sojae infection (Figure 6A; Supplementary Table 3). To determine if Avr1b could interact with other U-box proteins in the same clade as GmPUB1-1, and determine if other RxLR effectors could interact with GmPUB1-1, we conducted the yeast two-hybrid assay to screen 15 RxLR effectors (including Avr1b and six other Avr effectors) against 6 representative GmPUB proteins (including GmPUB1-1 and GmPUB1-2) as well as the three AtPUB proteins most closely related to GmPUB1-1.
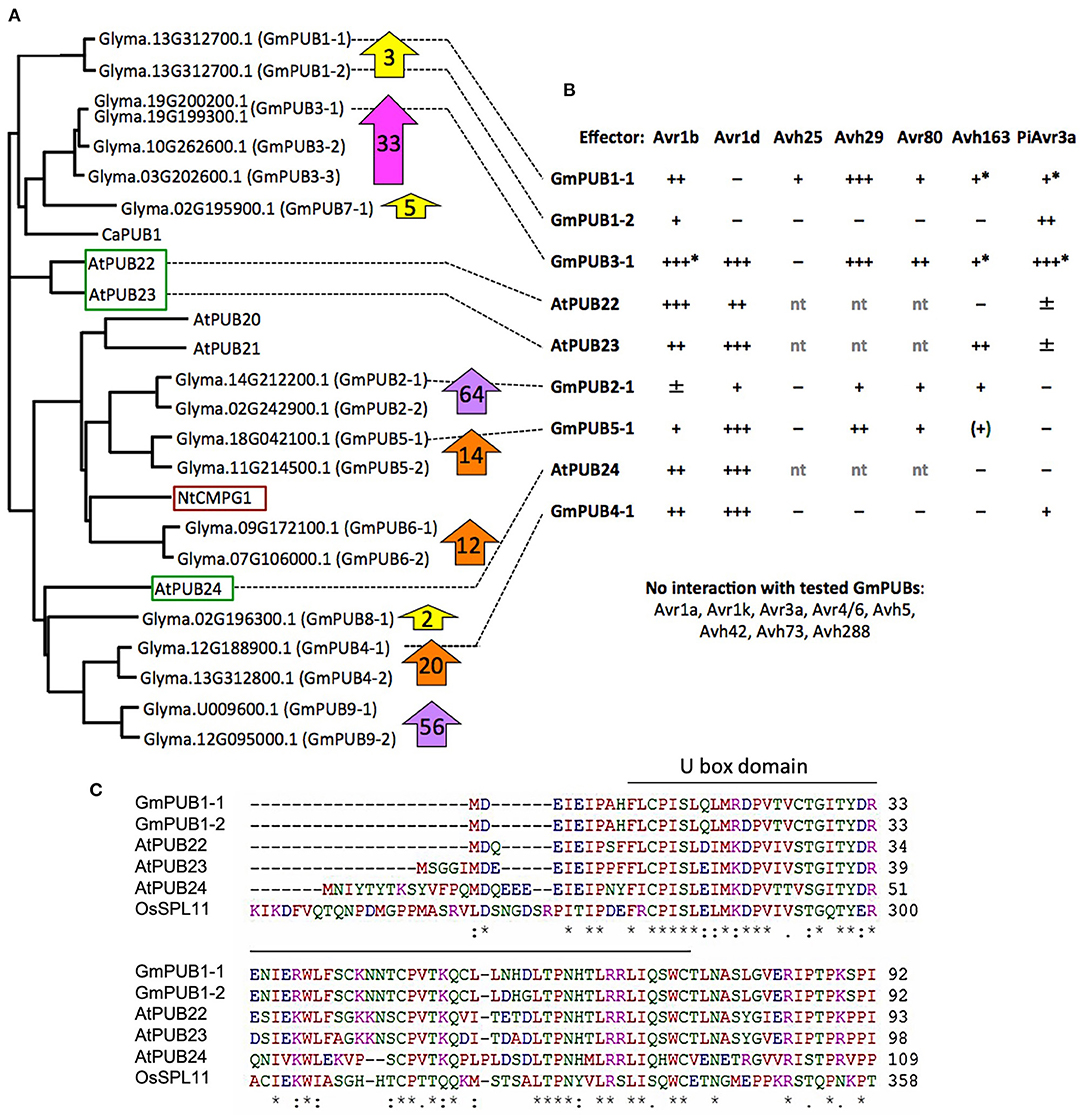
Figure 6. Interaction of GmPUB1-1 and paralogs with multiple RxLR effectors. (A) Distance tree of selected soybean and Arabidopsis PUBs, including known regulators of defense (red box, positive regulator; green box, negative regulators). Glyma, soybean (Williams 82 assembly v2 annotation 1); At, Arabidopsis; Nt, Nicotiana tabacum; Ca, Capsicum annuum. ClustalW was used for the sequence alignment and Phylip was used to create an unrooted UPGMA tree. Block arrows indicate fold change in soybean transcript levels following P. sojae infection (Supplementary Table 3). Levels for closely related genes are combined. (B) Interactions of PUBs shown in (A) with RxLR effectors. Most assays were done with pLAW10 (bait vector) and pLAW11 (prey vector). +++, strong interaction (growth on –Ade and –His); ++, medium (growth only on –His); +, weak (limited growth on –His); ±, very weak (micro-colonies on –His); –, negative. nt, not tested; *confirmed with bait-prey swap; (+) interaction observed only with effector as bait, but not with effector as prey. (C) Comparison of selected U-Box domain sequences from soybean, Arabidopsis and rice. Amino acids with a single color are clusters of amino acids with similar physiochemical properties. Amino acids that are identical in all six proteins are marked with asterisk. Conserved and semi- conserved substitutions are marked with colon and dot.
Five effectors (Avr1b, Avr1d, Avh29, Avh80, and Avh163) showed medium to strong interactions in the yeast two-hybrid assay with different subsets of GmPUBs 1, 3, 4, and 5, as well as with AtPUBs 22, 23, and 24 (Figure 6B; Supplementary Figure 3). Upon alignment of the protein sequences, the U-box domains of the GmPUB1 proteins were found to share 77 and 73% identity with the Arabidopsis PUB22 and PUB23 proteins, while PUB24 showed 66% identity (Figure 6C). Avr1b interacted most strongly with GmPUB3-1 and AtPUB22, while interactions with GmPUB4-1, AtPUB23, and AtPUB24 were comparable to its somewhat weaker interaction with GmPUB1-1. Avr1d interacted strongly with GmPUB3-1, GmPUB4-1, GmPUB5-1, AtPUB23, and AtPUB24, but not at all with GmPUB1-1 or GmPUB1-2. Avh29 interacted equally strongly with GmPUB1-1 and GmPUB3-1. P. infestans effector PiAvr3a, which is a homolog of Avr1b that interacts with Nicotiana CMPG1 (Bos et al., 2010), also showed interactions with several GmPUBs but not AtPUBs (Figure 6B). GmPUB3-1 interacted strongly with all of the RxLR effectors, except Avh80 which showed a moderate interaction. GmPUB1-2 showed only a weak interaction with Avr1b and a moderate interaction with PiAvr3a. These results suggest that GmPUBs are targeted by multiple P. sojae RxLR effectors, presumably to interfere with regulation of defense responses.
Discussion
In this study, cDNA clones encoding an Avr1b-interacting protein fragment, GmPUB1-171−397, were identified by screening a yeast two-hybrid library generated from P. sojae-infected etiolated soybean hypocotyls. The gene encoding the interactor, named GmPUB1-1, corresponded to soybean gene Glyma.13G312700. The homeologous gene GmPUB1-2 (Glyma.12g189000) encoded a protein with 94% identity to GmPUB1-1, which also interacted with Avr1b in the yeast two-hybrid assay, albeit more weakly (Figure 1A). Pull-down (Figure 1B) and BIFC (Figure 1C) assays supported that GmPUB1-1 is most likely a bona fide target of the P. sojae effector protein, Avr1b.
Full length GmPUB1-1 and GmPUB1-2 interact with Avr1b, but so does the original interacting cDNA, GmPUB1-171−397, which entirely lacks the U-box. This suggests that the N-terminal U-Box region is not required for the interactions of GmPUB1-1 or GmPUB1-2 with Avr1b. This contrasts with the binding of P. sojae Avr1d to the U-Box of GmPUB13 (Lin et al., 2021). U-Box domains are known to bind E2 (ubiquitin conjugating enzyme) and to be responsible for E3 ligase activity, while C-terminal domains are known to bind substrates targeted for ubiquitination (Azevedo et al., 2001). The weaker binding by GmPUB1-2 compared to GmPUB1-1, despite the 94% amino acid sequence identity between the two, suggests that the some of the polymorphic residues may contribute to the Avr1b interaction.
More than half of all oomycete effectors, including Avr1b, contain the conserved W and Y C-terminal motifs (Jiang et al., 2008) that comprise a common structural fold (Win et al., 2012b). Investigation of the Avr1b mutants (Figure 3) suggested that both the W and Y domains were required for interaction of Avr1b with GmPUBs. Furthermore, the same domains of Avr1b are most likely involved in both the interaction with GmPUB proteins and suppression of programmed cell death (Dou et al., 2008). Mutations in the W and Y motifs of Avr1b abolish Avr1b's ability to suppress cell death triggered by BAX (Dou et al., 2008) (Figure 2A). The pattern of responses to different W and Y motif mutations closely matched the pattern observed for recognition for suppression of BAX-triggered PCD by Avr1b (Figure 2A). The similarity of the yeast two-hybrid interaction patterns and anti-PCD activities of the Avr1b mutants supports that (i) the interactions of the GmPUBs with Avr1b are most likely biologically relevant and (ii) one or more of the GmPUBs may mediate the ability of Avr1b to suppress defense-related PCD.
Another study (Bos et al., 2010) has also reported that the N-terminal signal peptide and RxLR domain of Avr3a effector from P. infestans are not needed and that C-terminal domain with conserved W and Y motifs is sufficient for its interaction with the plant U-Box protein CMPG1 (R protein) and suppression of cell death. It was also reported that W and Y domains of effector proteins are important for interactions with R proteins and effector-related activities (Win et al., 2007). Lin et al. (2021) showed that P. sojae effector Avr1d binds to the U-box domain of GmPUB13 via its W, Y, and L domains. By solving the crystal structure of the Avr1d-GmPUB13 PUB domain complex Lin et al. (2021) showed that Avr1d occupies the binding site for E2 ubiquitin conjugating enzyme on GmPUB13.
Our BIFC study suggested that the complex of Avr1b and GmPUB1-171−397 mostly localizes to nuclei (Figure 1C). Nuclear localization is required for Avr1b's avirulence activity mediated by Rps1-b and for suppression of effector-triggered PCD (Dou et al., 2008). Our data do not indicate whether Avr1b and GmPUB1-1 form a complex before moving into the nuclei or whether Avr1b moves into the nucleus before interacting with GmPUB1-1 located in the nuclei.
We confirmed that the U-box domain of GmPUB1-1 possesses ubiquitination activity and presumably therefore is involved in the 26S proteasome degradation pathway. In our assays, GmPUB1-1 was able to conjugate ubiquitin moieties to E. coli proteins. Furthermore, mutation of the conserved Cys-12, Val-23, and Trp-39 residues within the U-Box domain completely abolished ubiquitin ligase activity (Figure 3). Similar in vitro ubiquitin E3 ligase activity has been observed in a number of other U-Box proteins (Jiang et al., 2001; Murata et al., 2001; Ohi et al., 2005). Mutations in highly conserved amino acid residues in the U-box central core domain, such as valine, can abolish the E3 ligase activity of U-Box proteins by disrupting their tertiary structures (Ohi et al., 2003).
Several U-box proteins have been identified as positive or negative regulators of plant defense. In rice, mutations in the U-Box Spl11 protein result in spontaneous cell death and enhanced disease resistance (Zeng et al., 2004). In N. benthamiana and tomato, the U-box protein CMPG1 is a positive regulator required for the full hypersensitive response (Gonzalez-Lamothe et al., 2006). Also, CMPG1 is targeted by the P. infestans effector Avr3a (Bos et al., 2010). Another positive regulator is AtPUB17 and its Nicotiana ortholog ACRE276 (Yang et al., 2006). The rice pathogen Magnaporthe oryzae effector AvrPi-zt targets a rice RING E3 ligase (Park et al., 2012).
In Arabidopsis, AtPUB13 (Li et al., 2012) and a closely related trio of U-box proteins, AtPUB22, AtPUB23 and AtPUB24 (Trujillo et al., 2008) negatively regulate resistance against H. arabidopsidis and the bacterial pathogen Pseudomonas syringae; over-expression results in increased susceptibility, while the triple mutant for the U-box trio is more resistant (Trujillo et al., 2008). Using a yeast two-hybrid system, a subunit of the exocyst complex, Exo70B2, has been shown to be a substrate of AtPUB22 that mediates its ubiquitination and degradation via the 26S proteasome (Stegmann et al., 2012). Degradation of Exo70B2 leads to reduction in the transport of toxic chemicals such as ROS to the infection sites and attenuation of PAMP-triggered immunity responses.
The phylogenetic tree (Figure 6A) placed the GmPUB1 proteins close to the triplet of Arabidopsis U-Box E3 ligases PUB22, PUB23, PUB24, which are negative regulators of PAMP-triggered immunity (Trujillo et al., 2008). However, in our study it was observed that over-expression of GmPUB1-1 causes cell death (Supplementary Figure 2) suggesting that GmPUB1-1 might be a positive regulator of cell death.
Joint silencing of GmPUB1-1 and GmPUB1-2 in soybean cotyledons resulted in loss of the P. sojae resistance normally mediated by the recognition of Avr1b by the products of genes at the Rps1-b and Rps1-k loci. On the other hand, P. sojae resistance mediated by the recognition of Avr1k by the products of genes at the Rps1-k locus was not affected by silencing of the GmPUB1 genes. Avr1k, which has a completely different amino acid sequence than Avr1b, did not interact in the yeast two-hybrid assay with GmPUB1-1, GmPUB1-2, or any of the other tested GmPUB proteins. These results suggest that at least one of the GmPUB1 proteins is essential for recognition of Avr1b by proteins encoded by genes at the Rps1-b and Rps1-k loci. Since recognition of Avr1k was unaffected by the silencing of the GmPUB1 genes, and Avr1k has a different amino acid sequence than Avr1b, we speculate (Figure 7) that genes at the Rps1-k locus encode at least two different proteins, one able to recognize Avr1b dependent on GmPUB1 (Rps1-bk) and one able to recognize Avr1k independent of GmPUB1 (Rps1-kk). We further speculate that the Rps1-b locus, which is allelic to the Rps1-k locus, encodes a protein similar or identical to Rps1-bk, namely Rps1-bb. The Rps1-b locus presumably does not contain a functional allele encoding a protein similar or identical to the Rps1-kk protein. The Rps1-k locus indeed contains two highly similar functional CC-NB-LRR type Phytophthora resistance genes (Gao et al., 2005; Gao and Bhattacharyya, 2008), though it is currently unknown if they differentially recognize Avr1b and Avr1k.
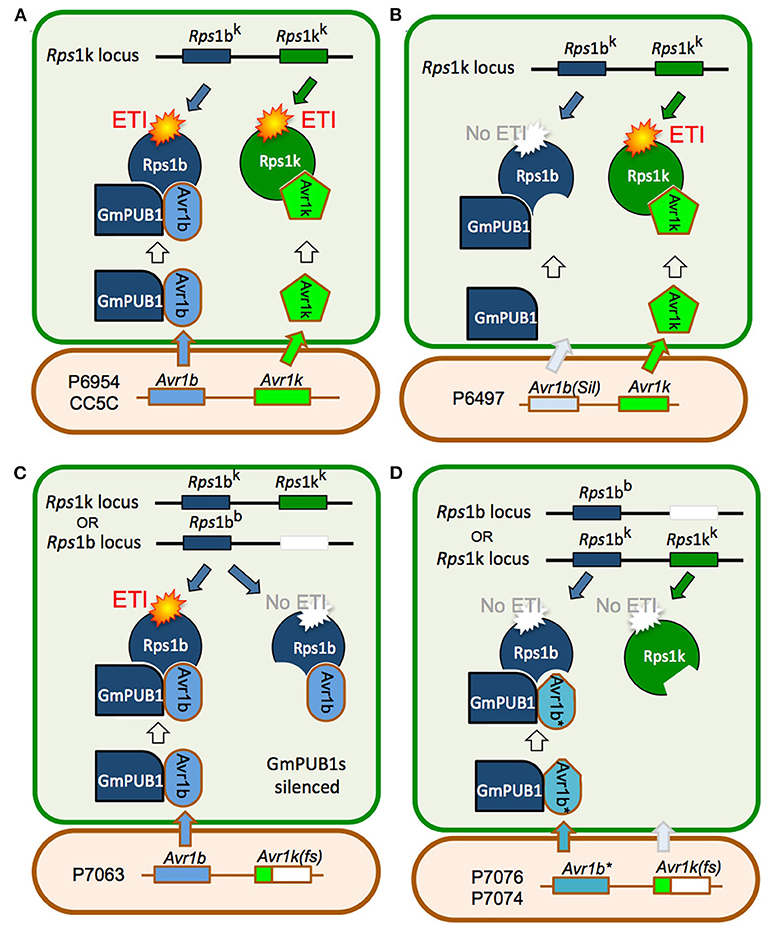
Figure 7. Model for the role of GmPUB1 proteins in recognition of Avr1b by plants carrying Rps1-b and Rps1-k. (A) P. sojae isolates of race 1 such as P6954 (genotype I) and CC5C, produce two completely different RxLR effectors, Avr1b and Avr1k, encoded by closely linked genes. The Rps1-k locus is proposed to encode two distinct proteins, Rps1-bk and Rps1-kk, responsible for recognition of Avr1b and Avr1k, respectively, triggering ETI. Recognition of Avr1b, but not Avr1k, requires the presence of GmPUB1 (either to form a physical complex as diagrammed, or to modify Avr1b). (B) When Avr1b is silenced, either experimentally or, naturally as in the case of P. sojae strain P6497 (race 2, genotype I), recognition by plants containing Rps1-k still occurs (via Rps1-kk), but plants containing Rps1-b cannot recognize the P. sojae strain (not diagrammed). (C) The soybean locus Rps1-b is proposed to encode an Rps1-bb protein (functionally identical to Rps1-bk) but no Rps1-kk protein. When the soybean GmPUB1 genes are silenced, Avr1b cannot be recognized by plants containing either Rps1-b or Rps1-k. If the P. sojae strain lacks Avr1k, as in the case of P7063 (race 6, genotype IV), silencing of GmPUB1 genes makes both Rps1-b and Rps1-k plants susceptible. (D) Some P. sojae strains such as P7076 (race 19, genotype II) or P7074 (race 17, genotype III) produce a variant Avr1b* protein that can bind to GmPUB1-1 but cannot be recognized either by plants carrying Rps1-b or Rps1-k. Since these strains also lack Avr1k, they can successfully infect plants carrying either Rps1-b or Rps1-k irrespective of the presence of GmPUB1 proteins.
One possible mechanistic explanation for the requirement for GmPUB1 for recognition of Avr1b is that the Rps1-bb and Rps1-bk proteins guard the GmPUB1 proteins, and that binding of GmPUB1 proteins by Avr1b disrupts the binding of GmPUB1 to the Rps1b proteins, triggering defense signaling by the Rps1b proteins. However, several lines of evidence suggest that this conventional guardee scenario is not correct. First, GmPUB1-1 and GmPUB1-2 bound to all alleles of Avr1b in yeast two-hybrid assays, even alleles (from P7076 and P7074) that do not trigger Rps1-b-mediated resistance. Second, in other gene-for-gene systems, Avr effector proteins that interact directly with the cognate R protein, such as AvrPi-ta (Jia et al., 2000), AvrL567 (Dodds et al., 2006), and AvrPi-tz (Park et al., 2012), typically show high levels of sequence polymorphism in alleles that evade R protein detection. In contrast, Avr effectors that bind R-protein-guarded targets typically do not show high levels of sequence polymorphisms (they may be deleted or silenced in virulent strains of the pathogen). The high levels of sequence polymorphisms shown by Avr1b are thus more typical of proteins that interact directly with an R protein. Third, current models of R protein-guardee interactions propose that the guardee (here GmPUB1) holds the R protein (i.e., Rps1b) in an inactive state; when an effector such as Avr1b binds to the guardee disrupting its interaction with the R protein, the R protein is released into an active state and signals the activation of ETI. However, when we silenced the GmPUB1 genes in the background of Rps1-b, the cotyledons became fully susceptible. Therefore, we propose a modified guard model, shown in Figure 7, in which Rps1-b is activated only when it recognizes avirulence alleles of Avr1b (from P6954 and P7063) in the context of GmPUB1. A variety of related models are also possible: for example, perhaps Rps1-b proteins can only recognize Avr1b proteins that have been ubiquitinated by GmPUB1; this would make GmPUB1 an “effector helper” protein as described by (Win et al., 2012a) rather than a guardee.
Despite extensive characterization of plant proteins and processes targeted by oomycete RxLR effectors, a systematic understanding of how the collection of RxLR effectors from an oomycete pathogen impact the immunity of its host remains poorly understood. The findings presented here suggest that there may be an extensive network of interactions involving multiple RxLR effectors and multiple plant U-Box E3 ligases (Supplementary Figure 3). Given the large numbers of E3 ligases encoded in plant genomes, and the complex sets of processes regulated by these proteins, the cumulative effect of numerous effector-E3 ligase interactions could profoundly re-program cell physiology. A major goal for the future research will be to finish mapping the network of effector E3 ligase interactions. This should encompass all families of E3 ligases on the host side and crinkler effectors as well as RxLR effectors on the pathogen side.
Data Availability Statement
The original contributions (GSE182773) presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
SL characterized the interaction of Avr1b with GmPUB1 genes and wrote the first draft of the manuscript. RH designed and executed the Y2H experiments. HW designed and executed the cotyledon experiments and contributed to writing the manuscript. NP conducted the western blot analysis for determining the function of GmPUB1-1 protein. HB screened the yeast 2-hybrid library. CL conducted the double-barreled particle bombardment experiment to determine the function of GmPUB1. EP executed the cotyledon experiments. LZ designed and executed transcriptome experiments. HG constructed the yeast 2-hybrid library. BT planned the project, designed the experiments, analyzed the data, and wrote the manuscript. MB contributed to conception and design of the experiments and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported in part by grants to MB from the Iowa Soybean Association and Agronomy Department, Iowa State University, in part by grants to BT from the National Science Foundation (IOS #0744875), from the Agriculture and Food Research Initiative of the USDA National Institute of Food and Agriculture (#2007-35319-18100 and #2010-65110-20764), and by support from Oregon State University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Matthew Dyer (Applied Biosystems) for transcriptome sequencing and data processing, Shiv Kale (Virginia Tech) for advice on particle bombardment assays, Felipe Arredondo, Samantha Taylor, and Madeleine Duquette for technical assistance, Ryan Anderson (Virginia Tech) and John McDowell (Virginia Tech) for useful suggestions, and the ISU DNA Facility, the Core Laboratory Facility at Virginia Tech, and the Center for Genome Research and Biocomputing at Oregon State University for DNA sequencing and synthesis of oligonucleotides.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.725571/full#supplementary-material
Supplementary Figure 1. RT-PCR analysis of the GmPUB1 transcript levels following transformation of soybean cotyledons with A. rhizogenes harboring the GmPUB1 RNAi vector. Soybean Williams 82 cotyledonary tissues, different days following transformation with A. rhizogenes harboring the GmPUB1 RNAi vector, were harvested for preparation of total RNAs that were subjected to RT-PCR. Reduced GmPUB1 mRNA accumulation was observed on day 6 following transformation. Soybean Actin gene was used as the internal control to normalize the total RNAs used in RT-PCR analysis.
Supplementary Figure 2. Over-expression of GmPUB1-1 triggers cell death. Cell death was expressed as percentage survival of GUS-positive cells relative to a parallel empty vector control, using a double-barrel bombardment assay (Kale and Tyler, 2011). Lower percentages of surviving GUS-positive cells indicate increased cell death. The data means and standard errors were calculated from 7 to 8 bombardments per replicate; three replicates were conducted on Williams (rps) leaves, and one each on L77-1863 (Rps1-b) and Williams 82 (Rps1-k) leaves. Cell death due to GmPUB1-1 over-expression was significant (**p < 0.001) compared to empty vector for all three experiments, based on the Wilcoxon signed-ranks test (Kale and Tyler, 2011). There were no significant differences among soybean lines, based on the Wilcoxon rank sum test (p > 0.05) (Kale and Tyler, 2011).
Supplementary Figure 3. Yeast two-hybrid assays for interactions between RxLR effectors and PUB proteins. RxLR effector genes (lacking signal peptide coding regions) were cloned into pLAW10 and introduced into Y8930. GmPUB and AtPUB genes were cloned into pLAW11 and introduced into Y8800. The RxLR-containing strains were then mated with PUB-containing strains and plated on the indicated SD drop-out media. Strong interactions produce growth on –LTA while weak to medium interactions produce moderate or strong growth on –LTH plates, respectively.
Supplementary Table 1. Primers used for cloning soybean GmPUB genes and P. sojae effector genes into pLAW10 and pLAW11 plasmids (Spreadsheet).
Supplementary Table 2. The nucleotide sequences of primers used in pull down assay.
Supplementary Table 3. GmPUB gene expression levels during infection.
References
Abramovitch, R. B., Janjusevic, R., Stebbins, C. E., and Martin, G. B. (2006). Type III effector AvrPtoB requires intrinsic E3 ubiquitin ligase activity to suppress plant cell death and immunity. Proc. Natl. Acad. Sci. U. S. A. 103, 2851–2856. doi: 10.1073/pnas.0507892103
Alcaide-Loridan, C., and Jupin, I. (2012). Ubiquitin and plant viruses, let's play together! Plant Physiol. 160, 72–82. doi: 10.1104/pp.112.201905
Anand, A., Rojas, C. M., Tang, Y., and Mysore, K. S. (2012). Several components of SKP1/Cullin/F-box E3 ubiquitin ligase complex and associated factors play a role in Agrobacterium-mediated plant transformation. New Phytol. 195, 203–216. doi: 10.1111/j.1469-8137.2012.04133.x
Angot, A., Vergunst, A., Genin, S., and Peeters, N. (2007). Exploitation of eukaryotic ubiquitin signaling pathways by effectors translocated by bacterial type III and type IV secretion systems. PLoS Pathog. 3:e3. doi: 10.1371/journal.ppat.0030003
Aravind, L., and Koonin, E. V. (2000). The U box is a modified RING finger - a common domain in ubiquitination. Curr. Biol. 10, R132–R134. doi: 10.1016/S0960-9822(00)00398-5
Azevedo, C., Sadanandom, A., Kitagawa, K., Freialdenhoven, A., Shirasu, K., and Schulze-Lefert, P. (2002). The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295, 2073–2076. doi: 10.1126/science.1067554
Azevedo, C., Santos-Rosa, M. J., and Shirasu, K. (2001). The U-box protein family in plants. Trends Plant Sci. 6, 354–358. doi: 10.1016/S1360-1385(01)01960-4
Bandara, A. Y., Weerasooriya, D. K., Conley, S. P., Bradley, C. A., Allen, T. W., and Esker, P. D. (2020). Modeling the relationship between estimated fungicide use and disease-associated yield losses of soybean in the United States I: foliar fungicides vs foliar diseases. PLoS ONE 15:e0234390. doi: 10.1371/journal.pone.0234390
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate - a practical and powerful approach to multiple testing. J. R Stat. Soc. Ser. B 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Bhattacharyya, M. K., and Ward, E. W. B. (1986). Resistance, susceptibility and accumulation of glyceollin I-III in soybeans inoculated with Phytophthora megasperma f. sp. glycinea. Physiol. Mol. Plant Pathol. 29, 227–237. doi: 10.1016/S0048-4059(86)80023-6
Bos, J. I., Armstrong, M. R., Gilroy, E. M., Boevink, P. C., Hein, I., Taylor, R. M., et al. (2010). Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proc. Natl. Acad. Sci. U. S. A. 107, 9909–9914. doi: 10.1073/pnas.0914408107
Chien, C. F., Mathieu, J., Hsu, C. H., Boyle, P., Martin, G. B., and Lin, N. C. (2013). Nonhost resistance of tomato to the bean pathogen Pseudomonas syringae pv. syringae B728a is due to a defective E3 ubiquitin ligase domain in avrptobb728a. Mol. Plant Microbe Interact. 26, 387–397. doi: 10.1094/MPMI-08-12-0190-R
Cho, S. K., Chung, H. S., Ryu, M. Y., Park, M. J., Lee, M. M., Bahk, Y. Y., et al. (2006). Heterologous expression and molecular and cellular characterization of CaPUB1 encoding a hot pepper U-Box E3 ubiquitin ligase homolog. Plant Physiol. 142, 1664–1682. doi: 10.1104/pp.106.087965
Cho, S. K., Ryu, M. Y., Song, C., Kwak, J. M., and Kim, W. T. (2008). Arabidopsis PUB22 and PUB23 are homologous U-Box E3 ubiquitin ligases that play combinatory roles in response to drought stress. Plant Cell 20, 1899–1914. doi: 10.1105/tpc.108.060699
Coll, N. S., Epple, P., and Dangl, J. L. (2011). Programmed cell death in the plant immune system. Cell Death Differ. 18, 1247–1256. doi: 10.1038/cdd.2011.37
Couto, D., and Zipfel, C. (2016). Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 16, 537–552. doi: 10.1038/nri.2016.77
Dodds, P. N., Lawrence, G. J., Catanzariti, A. M., Teh, T., Wang, C. I., Ayliffe, M. A., et al. (2006). Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl. Acad. Sci. U. S. A. 103, 8888–8893. doi: 10.1073/pnas.0602577103
Dou, D., Kale, S. D., Wang, X., Chen, Y., Wang, Q., Wang, X., et al. (2008). Carboxy-terminal motifs common to many oomycete RXLR effectors are required for avirulence and suppression of BAX-mediated programmed cell death by Phytophthora sojae effector Avr1b. Plant Cell 20, 1118–1133. doi: 10.1105/tpc.107.057067
Du, Q. L., Cui, W. Z., Zhang, C. H., and Yu, D. Y. (2010). GmRFP1 encodes a previously unknown RING-type E3 ubiquitin ligase in Soybean (Glycine max). Mol. Biol. Rep. 37, 685–693. doi: 10.1007/s11033-009-9535-1
Duplan, V., and Rivas, S. (2014). E3 ubiquitin-ligases and their target proteins during the regulation of plant innate immunity. Front. Plant Sci. 5:42. doi: 10.3389/fpls.2014.00042
Fang, S., and Weissman, A. M. (2004). A field guide to ubiquitylation. Cell Mol. Life Sci. 61, 1546–1561. doi: 10.1007/s00018-004-4129-5
Förster, H., Tyler, B. M., and Coffey, M. D. (1994). Phytophthora sojae races have arisen by clonal evolution and by rare outcrosses. Mol. Plant-Microbe Interact. 7, 780–791. doi: 10.1094/MPMI-7-0780
Gao, H., and Bhattacharyya, M. K. (2008). The soybean-Phytophthora resistance locus Rps1-k encompasses coiled coil-nucleotide binding-leucine rich repeat-like genes and repetitive sequences. BMC Plant Biol. 8:29. doi: 10.1186/1471-2229-8-29
Gao, H., Narayanan, N. N., Ellison, L., and Bhattacharyya, M. K. (2005). Two classes of highly similar coiled coil-nucleotide binding-leucine rich repeat genes isolated from the Rps1-k locus encode Phytophthora resistance in soybean. Mol. Plant Microbe Interact. 18, 1035–1045. doi: 10.1094/MPMI-18-1035
Gohre, V., and Robatzek, S. (2008). Breaking the barriers: microbial effector molecules subvert plant immunity. Annu. Rev. Phytopathol. 46, 189–215. doi: 10.1146/annurev.phyto.46.120407.110050
Goldberg, A. L. (2003). Protein degradation and protection against misfolded or damaged proteins. Nature 426, 895–899. doi: 10.1038/nature02263
Gonzalez-Lamothe, R., Tsitsigiannis, D. I., Ludwig, A. A., Panicot, M., Shirasu, K., and Jones, J. D. (2006). The U-box protein CMPG1 is required for efficient activation of defense mechanisms triggered by multiple resistance genes in tobacco and tomato. Plant Cell 18, 1067–1083. doi: 10.1105/tpc.106.040998
He, Q., McLellan, H., Boevink, P. C., Sadanandom, A., Xie, C., Birch, P. R., et al. (2015). U-box E3 ubiquitin ligase PUB17 acts in the nucleus to promote specific immune pathways triggered by Phytophthora infestans. J. Exp. Bot. 66, 3189–3199. doi: 10.1093/jxb/erv128
Jia, Y., McAdams, S. A., Bryan, G. T., Hershey, H. P., and Valent, B. (2000). Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19, 4004–4014. doi: 10.1093/emboj/19.15.4004
Jiang, J., Ballinger, C. A., Wu, Y., Dai, Q., Cyr, D. M., Hohfeld, J., et al. (2001). CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J. Biol. Chem. 276, 42938–42944. doi: 10.1074/jbc.M101968200
Jiang, R. H. Y., Tripathy, S., Govers, F., and Tyler, B. M. (2008). RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving super-family with more than 700 members. Proc. Natl. Acad. Sci. U. S. A. 105, 4874–4879. doi: 10.1073/pnas.0709303105
Jiang, R. H. Y., and Tyler, B. M. (2012). Mechanisms and evolution of virulence in oomycetes. Ann. Rev. Phytopath. 50, 295–318. doi: 10.1146/annurev-phyto-081211-172912
Kale, S. D., and Tyler, B. M. (2011). “Assaying effector function in planta using double-barreled particle bombardment,” in Methods in Molecular Biology. The Plant Immune Response, ed J. M. McDowell (Totowa, NJ: Humana), 153–172. doi: 10.1007/978-1-61737-998-7_13
Koenning, S. R., and Wrather, J. A. (2010). Suppression of soybean yield potential in the continental United States by plant diseases from 2006 to 2009. Plant Health Prog. doi: 10.1094/PHP-2010-1122-01-RS. [Epub ahead of print].
Lee, D. H., Choi, H. W., and Hwang, B. K. (2011). The pepper E3 ubiquitin ligase RING1 gene, CaRING1, is required for cell death and the salicylic acid-dependent defense response. Plant Physiol. 156, 2011–2025. doi: 10.1104/pp.111.177568
Li, W., Ahn, I. P., Ning, Y., Park, C. H., Zeng, L., Whitehill, J. G., et al. (2012). The U-Box/ARM E3 ligase PUB13 regulates cell death, defense, and flowering time in Arabidopsis. Plant Physiol. 159, 239–250. doi: 10.1104/pp.111.192617
Lin, Y., Hu, Q., Zhou, J., Yin, W., Yao, D., Shao, Y., et al. (2021). Phytophthora sojae effector Avr1d functions as an E2 competitor and inhibits ubiquitination activity of GmPub13 to facilitate infection. Proc. Natl. Acad. Sci. U. S. A. 118:312. doi: 10.1073/pnas.2018312118
Liu, P., Sherman-Broyles, S., Nasrallah, M. E., and Nasrallah, J. B. (2007). A cryptic modifier causing transient self-incompatibility in Arabidopsis thaliana. Curr. Biol. 17, 734–740. doi: 10.1016/j.cub.2007.03.022
Marino, D., Peeters, N., and Rivas, S. (2012). Ubiquitination during plant immune signaling. Plant Physiol. 160, 15–27. doi: 10.1104/pp.112.199281
Martin, G. B., Bogdanove, A. J., and Sessa, G. (2003). Understanding the functions of plant disease resistance proteins. Annu. Rev. Plant Biol. 54, 23–61. doi: 10.1146/annurev.arplant.54.031902.135035
Mathieu, J., Schwizer, S., and Martin, G. B. (2014). Pto kinase binds two domains of AvrPtoB and its proximity to the effector E3 ligase determines if it evades degradation and activates plant immunity. PLoS Pathog. 10:e1004227. doi: 10.1371/journal.ppat.1004227
Mazzucotelli, E., Belloni, S., Marone, D., De Leonardis, A., Guerra, D., Di Fonzo, N., et al. (2006). The e3 ubiquitin ligase gene family in plants: regulation by degradation. Curr. Genom. 7, 509–522. doi: 10.2174/138920206779315728
McDowell, J. M., and Woffenden, B. J. (2003). Plant disease resistance genes: recent insights and potential applications. Trends Biotechnol. 21, 178–183. doi: 10.1016/S0167-7799(03)00053-2
Monaghan, J., Xu, F., Gao, M., Zhao, Q., Palma, K., Long, C., et al. (2009). Two Prp19-like U-box proteins in the MOS4-associated complex play redundant roles in plant innate immunity. PLoS Pathog. 5:e1000526. doi: 10.1371/journal.ppat.1000526
Monte, E., Amador, V., Russo, E., Martinez-Garcia, J., and Prat, S. (2003). PHOR1: a U-Box GA signaling component with a role in proteasome degradation? J. Plant Growth Regul. 22, 152–162. doi: 10.1007/s00344-003-0029-4
Murata, S., Minami, Y., Minami, M., Chiba, T., and Tanaka, K. (2001). CHIP is a chaperone-dependent E3 ligase that ubiquitylates unfolded protein. EMBO Rep. 2, 1133–1138. doi: 10.1093/embo-reports/kve246
Navarro, L., Zipfel, C., Rowland, O., Keller, I., Robatzek, S., Boller, T., et al. (2004). The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol. 135, 1113–1128. doi: 10.1104/pp.103.036749
Ohi, M. D., Vander Kooi, C. W., Rosenberg, J. A., Chazin, W. J., and Gould, K. L. (2003). Structural insights into the U-box, a domain associated with multi-ubiquitination. Nat. Struct. Biol. 10, 250–255. doi: 10.1038/nsb906
Ohi, M. D., Vander Kooi, C. W., Rosenberg, J. A., Ren, L., Hirsch, J. P., Chazin, W. J., et al. (2005). Structural and functional analysis of essential pre-mRNA splicing factor Prp19p. Mol. Cell Biol. 25, 451–460. doi: 10.1128/MCB.25.1.451-460.2005
Park, C. H., Chen, S., Shirsekar, G., Zhou, B., Khang, C. H., Songkumarn, P., et al. (2012). The Magnaporthe oryzae effector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern-triggered immunity in rice. Plant Cell 24, 4748–4762. doi: 10.1105/tpc.112.105429
Park, D. S., Landini, S., Graham, M. Y., and Graham, T. L. (2002). Induced distal defence potentiation against Phytophthora sojae in soybean. Physiol. Mol. Plant Pathol. 60, 293–310. doi: 10.1016/S0885-5765(02)90409-1
Qutob, D., Kamoun, S., and Gijzen, M. (2002). Expression of a Phytophthora sojae necrosis-inducing protein occurs during transition from biotrophy to necrotrophy. Plant J. 32, 361–373. doi: 10.1046/j.1365-313X.2002.01439.x
Rosebrock, T. R., Zeng, L., Brady, J. J., Abramovitch, R. B., Xiao, F., and Martin, G. B. (2007). A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature 448, 370–374. doi: 10.1038/nature05966
Samuel, M. A., Mudgil, Y., Salt, J. N., Delmas, F., Ramachandran, S., Chilelli, A., et al. (2008). Interactions between the S-domain receptor kinases and AtPUB-ARM E3 ubiquitin ligases suggest a conserved signaling pathway in Arabidopsis. Plant Physiol. 147, 2084–2095. doi: 10.1104/pp.108.123380
Sandhu, D., Gao, H., Cianzio, S., and Bhattacharyya, M. K. (2004). Deletion of a disease resistance nucleotide-binding-site leucine-rich- repeat-like sequence is associated with the loss of the Phytophthora resistance gene Rps4 in soybean. Genetics 168, 2157–2167. doi: 10.1534/genetics.104.032037
Schmutz, J., Cannon, S. B., Schlueter, J., Ma, J., Mitros, T., Nelson, W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183. doi: 10.1038/nature08670
Seo, K. I., Lee, J. H., Nezames, C. D., Zhong, S., Song, E., Byun, M. O., et al. (2014). ABD1 is an Arabidopsis DCAF substrate receptor for CUL4-DDB1-based E3 ligases that acts as a negative regulator of abscisic acid signaling. Plant Cell 26, 695–711. doi: 10.1105/tpc.113.119974
Shan, W., Cao, M., Leung, D., and Tyler, B. M. (2004). The Avr1b locus of Phytophthora sojae encodes an elicitor and a regulator required for avirulence on soybean plants carrying resistance gene Rps1b. Mol. Plant Microbe Interact. 17, 394–403. doi: 10.1094/MPMI.2004.17.4.394
Shirsekar, G. S., Vega-Sanchez, M. E., Bordeos, A., Baraoidan, M., Swisshelm, A., Fan, J., et al. (2014). Identification and characterization of suppressor mutants of spl11- mediated cell death in rice. Mol. Plant Microbe Interact. 27, 528–536. doi: 10.1094/MPMI-08-13-0259-R
Song, T.-Q., Kale, S. D., Arredondo, F. D., Shen, D.-Y., Su, L.-M., Liu, L., et al. (2013). Two RxLR avirulence genes in Phytophthora sojae determine soybean Rps1k-mediated disease resistance. Mol. Plant-Microbe Interact. 26:711–720. doi: 10.1094/MPMI-12-12-0289-R
Stegmann, M., Anderson, R. G., Ichimura, K., Pecenkova, T., Reuter, P., Zarsky, V., et al. (2012). The ubiquitin ligase PUB22 targets a subunit of the exocyst complex required for PAMP-triggered responses in Arabidopsis. Plant Cell 24, 4703–4716. doi: 10.1105/tpc.112.104463
Stone, S. L., Anderson, E. M., Mullen, R. T., and Goring, D. R. (2003). ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible Brassica pollen. Plant Cell 15, 885–898. doi: 10.1105/tpc.009845
Subramanian, S., Graham, M. Y., Yu, O., and Graham, T. L. (2005). RNA interference of soybean isoflavone synthase genes leads to silencing in tissues distal to the transformation site and to enhanced susceptibility to Phytophthora sojae. Plant Physiol. 137, 1345–1353. doi: 10.1104/pp.104.057257
Trujillo, M., Ichimura, K., Casais, C., and Shirasu, K. (2008). Negative regulation of PAMP-triggered immunity by an E3 ubiquitin ligase triplet in Arabidopsis. Curr. Biol. 18, 1396–1401. doi: 10.1016/j.cub.2008.07.085
Trujillo, M., and Shirasu, K. (2010). Ubiquitination in plant immunity. Curr. Opin. Plant Biol. 13, 402–408. doi: 10.1016/j.pbi.2010.04.002
Tyler, B. M. (2007). Phytophthora sojae: root rot pathogen of soybean and model oomycete. Mol. Plant Pathol. 8, 1–8. doi: 10.1111/j.1364-3703.2006.00373.x
Tyler, B. M. (2008). “Genomics of fungal- and oomycete-soybean interactions,” in Soybean Genomics, ed G. Stacey (New York, NY: Springer), 243–267. doi: 10.1007/978-0-387-72299-3_14
Tyler, B. M., and Gijzen, M. J. (2014). “The Phytophthora sojae genome sequence: foundation for a revolution,” in Genomics of Plant-Associated Fungi and Oomycetes: Dicot Pathogens, eds R. A. Dean, A. Lichens-Park, and C. Kole (Heidelberg: Springer), 133–157. doi: 10.1007/978-3-662-44056-8_7
Tyler, B. M., Jiang, R. H. Y., Zhou, L., Tripathy, S., Dou, D., Torto-Alalibo, T., et al. (2007). “Functional genomics and bioinformatics of the Phytophthora sojae-soybean interaction,” in The Genomics of Disease, eds P. Gustafson, G. Stacey, and J. Taylor (New York, NY: Kluwer Academic/Plenum Publisher), 67–78. doi: 10.1007/978-0-387-76723-9_6
Vierstra, R. D. (2009). The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10, 385–397. doi: 10.1038/nrm2688
Wang, Q., Han, C., Ferreira, A. O., Yu, X., Ye, W., Tripathy, S., et al. (2011). Transcriptional programming and functional interactions within the Phytophthora sojae RXLR effector repertoire. Plant Cell 23, 2064–2086. doi: 10.1105/tpc.111.086082
Wesley, S. V., Helliwell, C. A., Smith, N. A., Wang, M. B., Rouse, D. T., Liu, Q., et al. (2001). Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27, 581–590. doi: 10.1046/j.1365-313X.2001.01105.x
Win, J., Chaparro-Garcia, A., Belhaj, K., Saunders, D. G., Yoshida, K., Dong, S., et al. (2012a). Effector biology of plant-associated organisms: concepts and perspectives. Cold Spring Harb. Symp. Quant. Biol. 77, 235–247. doi: 10.1101/sqb.2012.77.015933
Win, J., Krasileva, K. V., Kamoun, S., Shirasu, K., Staskawicz, B. J., and Banfield, M. J. (2012b). Sequence divergent RXLR effectors share a structural fold conserved across plant pathogenic oomycete species. PLoS Pathog. 8:e1002400. doi: 10.1371/journal.ppat.1002400
Win, J., Morgan, W., Bos, J., Krasileva, K. V., Cano, L. M., Chaparro-Garcia, A., et al. (2007). Adaptive evolution has targeted the C-terminal domain of the RXLR effectors of plant pathogenic oomycetes. Plant Cell 19, 2349–2369. doi: 10.1105/tpc.107.051037
Wrather, J. A., Anderson, T. R., Arsyad, D. M., Tan, Y., Ploper, L. D., Porta-Puglia, A., et al. (2001). Soybean disease loss estimates for the top 10 soybean-producing countries in 1998. Canada J. Plant Pathol. 23, 115–121. doi: 10.1080/07060660109506918
Wrather, J. A., Koenning, S. R., and Anderson, T. R. (2003). Effect of diseases on soybean yields in the United States and Ontario (1999 to 2002). Plant Health Progr. 4:325. doi: 10.1094/PHP-2003-0325-01-RV
Xin, D. W., Liao, S., Xie, Z. P., Hann, D. R., Steinle, L., Boller, T., et al. (2012). Functional analysis of NopM, a novel E3 ubiquitin ligase (NEL) domain effector of Rhizobium sp. strain NGR234. PLoS Pathog. 8:e1002707. doi: 10.1371/journal.ppat.1002707
Yan, J., Wang, J., Li, Q., Hwang, J. R., Patterson, C., and Zhang, H. (2003). AtCHIP, a U-box-containing E3 ubiquitin ligase, plays a critical role in temperature stress tolerance in Arabidopsis. Plant Physiol. 132, 861–869. doi: 10.1104/pp.103.020800
Yang, B., Ruan, R., Cantu, D., Wang, X., Ji, W., Ronald, P. C., et al. (2013). A comparative approach expands the protein-protein interaction node of the immune receptor XA21 in wheat and rice. Genome 56, 315–326. doi: 10.1139/gen-2013-0032
Yang, C.-W., Gonzalez-Lamothe, R., Ewan, R. A., Rowland, O., Yoshioka, H., Shenton, M., et al. (2006). The E3 ubiquitin ligase activity of arabidopsis PLANT U-BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. Plant Cell 18, 1084–1098. doi: 10.1105/tpc.105.039198
Yee, D., and Goring, D. R. (2009). The diversity of plant U-box E3 ubiquitin ligases: from upstream activators to downstream target substrates. J. Exp. Bot. 60, 1109–1121. doi: 10.1093/jxb/ern369
Zeng, L. R., Qu, S., Bordeos, A., Yang, C., Baraoidan, M., Yan, H., et al. (2004). Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 16, 2795–2808. doi: 10.1105/tpc.104.025171
Zhang, L., Du, L., Shen, C., Yang, Y., and Poovaiah, B. W. (2014). Regulation of plant immunity through ubiquitin-mediated modulation of Ca(2+) -calmodulin-AtSR1/CAMTA3 signaling. Plant J. 78, 269–281. doi: 10.1111/tpj.12473
Zhang, X., Wu, Q., Ren, J., Qian, W., He, S., Huang, K., et al. (2012). Two novel RING-type ubiquitin ligases, RGLG3 and RGLG4, are essential for jasmonate-mediated responses in Arabidopsis. Plant Physiol. 160, 808–822. doi: 10.1104/pp.112.203422
Keywords: soybean U-box protein, E3 ubiquitin-ligase, Phytophthora sojae, oomycete, RXLR effector, plant immunity
Citation: Li S, Hanlon R, Wise H, Pal N, Brar H, Liao C, Gao H, Perez E, Zhou L, Tyler BM and Bhattacharyya MK (2021) Interaction of Phytophthora sojae Effector Avr1b With E3 Ubiquitin Ligase GmPUB1 Is Required for Recognition by Soybeans Carrying Phytophthora Resistance Rps1-b and Rps1-k Genes. Front. Plant Sci. 12:725571. doi: 10.3389/fpls.2021.725571
Received: 15 June 2021; Accepted: 30 August 2021;
Published: 06 October 2021.
Edited by:
Aardra Kachroo, University of Kentucky, United StatesReviewed by:
Xiao-Ren Chen, Yangzhou University, ChinaWolfgang Moeder, University of Toronto, Canada
Copyright © 2021 Li, Hanlon, Wise, Pal, Brar, Liao, Gao, Perez, Zhou, Tyler and Bhattacharyya. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Madan K. Bhattacharyya, bWJoYXR0YWNAaWFzdGF0ZS5lZHU=; Brett M. Tyler, YnJldHQudHlsZXJAb3JlZ29uc3RhdGUuZWR1
†These authors have contributed equally to this work
‡Present address: Shan Li, Discover Financial Services, Riverwoods, IL, United States
Regina Hanlon, Department of Plant Pathology, Physiology and Weed Science, Virginia Tech, Blacksburg, VA, United States
Hua Wise, LynnTech, Inc., College Station, TX, United States
Narinder Pal, North Central Regional Plant Introduction Station, U.S. Department of Agriculture-Agricultural Research Service (USDA-ARS), Ames, IA, United States
Hargeet Brar, ACGT Inc., Wheeling, IL, United States
Chunyu Liao, Helmholtz Institute for RNA-based Infection Research (HIRI), Helmholtz-Centre for Infection Research (HZI), Würzburg, Germany
Hongyu Gao, Indiana University School of Medicine, R3 C420, Medical and Molecular Genetics (MMGE), Indianapolis, IN, United States
Eli Perez, Department of Pathology, University of Iowa Medical Research Center, Iowa City, IA, United States
 Shan Li1†‡
Shan Li1†‡ Regina Hanlon
Regina Hanlon Chunyu Liao
Chunyu Liao Brett M. Tyler
Brett M. Tyler Madan K. Bhattacharyya
Madan K. Bhattacharyya