- 1 Laboratorio Nacional de Genómica para la Biodiversidad, Centro de Investigación y de Estudios Avanzados del Instituto Politécnico Nacional, Irapuato, Guanajuato, México
- 2 College of Life Science, Northwest A&F University, Yangling, Shaanxi, China
A commentary on
Target genes of the MADS transcription factor SEPALLATA3: integration of developmental and hormonal pathways in the Arabidopsis flower
by Kaufmann, K., Muino, J. M., Jauregui, R., Airoldi, C. A., Smaczniak, C., Krajewski, P., and Angenent, G. C. (2009). PLoS Biol. 7, e1000090. doi: 10.1371/journal.pbio.1000090
Global identification of targets of the Arabidopsis MADS domain protein AGAMOUS-Like15
by Zheng, Y., Ren, N., Wang, H., Stromberg, A. J., and Perry, S. E. (2009). Plant Cell 21, 2563–2577.
Orchestration of floral initiation by APETALA1
by Kaufmann, K., Wellmer, F., Muino, J. M., Ferrier, T., Wuest, S. E., Kumar, V., Serrano-Mislata, A., Madueno, F., Krajewski, P., Meyerowitz, E. M., Angenent, G. C., and Riechmann, J. L. (2010). Science 328, 85–89.
Flowering locus C (FLC) regulates development pathways throughout the life cycle of Arabidopsis
by Deng, W., Ying, H., Helliwell, C. A., Taylor, J. M., Peacock, W. J., and Dennis, E. S. (2011). Proc. Natl. Acad. Sci. U.S.A. 108, 6680–6685.
MADS domain transcription factors are key regulators in yeast, animals, and plants. They guide important developmental processes like heart and muscle development in mammals, mating in yeast, and flower development in plants (Messenguy and Dubois, 2003). The number of MADS genes varies greatly among taxa, with yeast (4) and animals (5) having modest numbers. MADS genes have undergone significant duplication in plant lineages, ranging from 1 gene in green algae, to over 20 in mosses, to more than 100 in angiosperms (Gramzow and Theissen, 2010). MADS domain proteins function in multimeric complexes (Immink et al., 2010) and bind to CArG-boxes, which are small 8–10 bp DNA sequences composed of 6–8 A or Ts flanked by C and G bases (de Folter and Angenent, 2006). The Arabidopsis genome contains an estimated 340,000 potential MADS binding sites (de Folter and Angenent, 2006), leading to the intriguing question: What is the biological significance of the large number of potential MADS binding sites, and how many target genes do MADS proteins really regulate?
Recently, several studies reported on the genome-wide identification of target genes of MADS proteins using chromatin immunoprecipitation (ChIP) followed by sequencing (ChIP-seq) or microarray analysis (ChIP–chip; Kaufmann et al., 2009, 2010; Zheng et al., 2009; Deng et al., 2011). Based on these studies, it appears that MADS proteins may directly regulate thousands of target genes (Figure 1A), and may serve to integrate different biological processes.
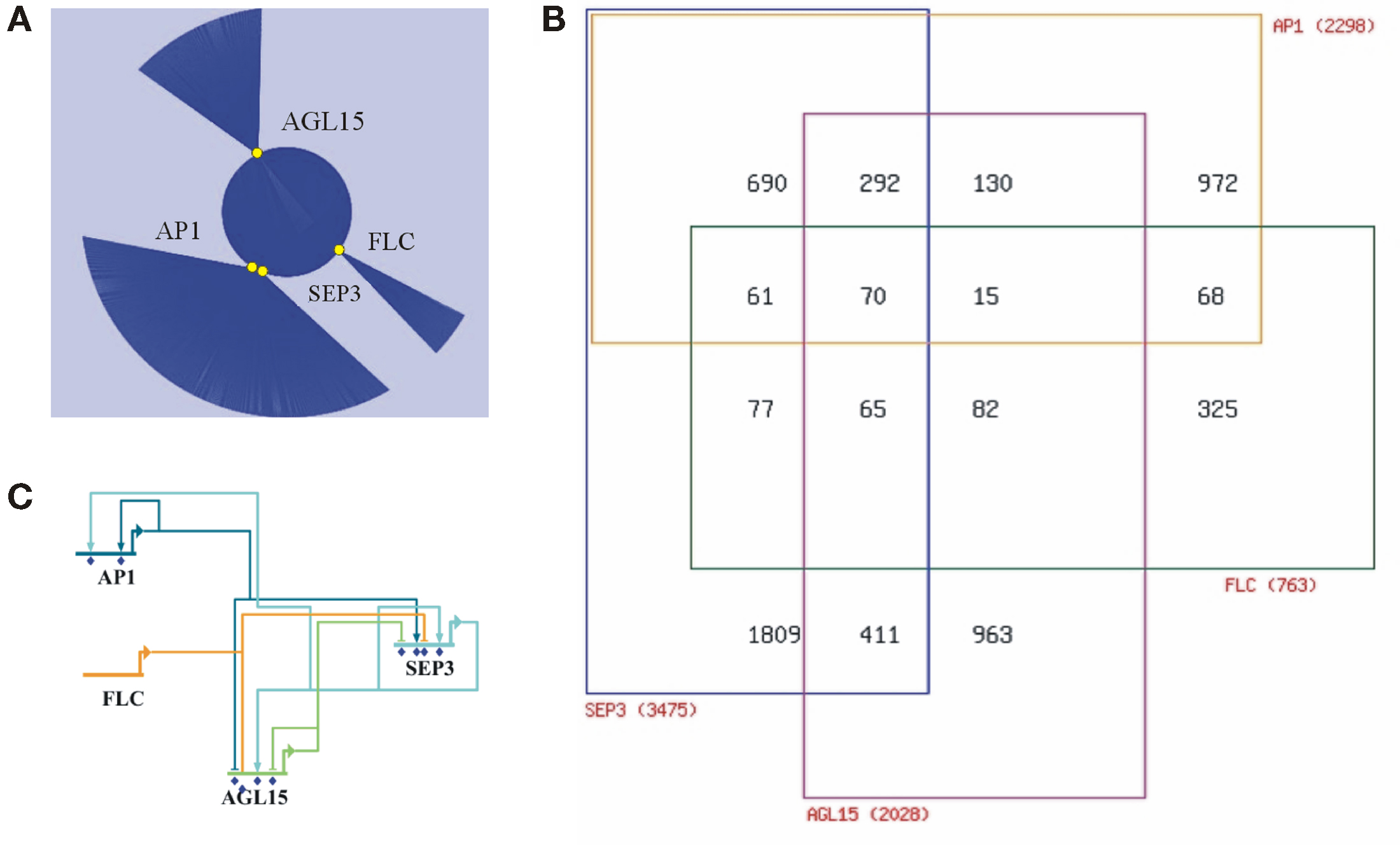
Figure 1. Integrative analysis of the reported target genes of the MADS proteins SEP3, AP1, AGL15, and FLC. (A) Network representation (Cytoscape) of the reported target genes (represented by blue lines) of the four MADS proteins (yellow circles). (B) Venn diagram depicting the numbers of shared target genes. The four MADS proteins share 70 target genes. (C) Wire diagram (BioTapestry) showing the complex regulatory network between the four MADS proteins themselves. MADS genes are MADS targets and autoregulatory feedback loops are present.
The first study reported on the identification of target genes of the floral homeotic protein SEPALLATA3 (SEP3), necessary for correct development of all floral organs (Kaufmann et al., 2009). SEP3 is known to form a heterodimer with AG, essential for stamen and carpel development. ChIP-seq was performed using antibodies against SEP3 in wild type and agamous (ag-1) inflorescence tissues, resulting in 4282 and 2828 peaks (binding events) respectively, corresponding to 3475 and 2424 putative target genes. In this study the authors were also interested in identifying target genes associated with perianth development. To accomplish this goal, the authors used an agamous loss-of-function mutant, which fails to produce stamens and carpels and instead produces an indeterminate set of sepals and petals. Most peaks, clearly enriched for CArG-box sequences, are present in upstream promoter regions close to the transcriptional start site, though some are also found in introns and 3′-UTR regions. This distribution of binding peaks is consistent for all four studies. Based on public expression data, over 60% of the putative SEP3 targets show differential expression patterns during wild type floral development. The putative targets are enriched for transcription factors of different families and, interestingly, for hormonal signaling pathways (auxin, brassinosteroids, and gibberellins). The authors suggest that SEP3 acts as a transcriptional activator involved in directing growth-related and hormonal pathways in a combinatorial fashion with other MADS proteins and possibly also with non-MADS transcription factors (e.g., TCP and bHLH; Kaufmann et al., 2009; Dornelas et al., 2011).
The second study focused on the floral repressor and somatic embryogenesis promoter AGL15. ChIP–chip with an AGL15-specific antibody in wildtype Arabidopsis embryogenic cultures identified 2028 binding sites (Zheng et al., 2009). As for SEP3, AGL15 targets were also enriched for transcription factors and components of gibberellic acid and auxin signaling pathways. Transcript profiling comparisons among wild type, an AGL15 overexpressor line, and the agl15 agl18 mutant, demonstrated that over 20% of the targets are differentially expressed. Interestingly, 40% of the total down-regulated and 23% of up-regulated genes are directly bound by AGL15. A working model for embryogenesis is presented that places AGL15/AGL18 in a highly connected network linking auxin and gibberellic acid/abscisic acid signaling pathways, and embryo B3 domain genes (Zheng et al., 2009).
The third study used an APETALA1 (AP1) specific antibody in an ap1 cal meristematic tissue containing an inducible AP1:GR transgene. AP1 is essential for sepal and petal formation, and it shares with CAL overlapping functions in floral meristem determination. Target gene identification using ChIP-seq suggests that AP1 orchestrates floral initiation (Kaufmann et al., 2010). In total 1942 peaks were identified and after cis motif analysis this resulted in the identification of 2298 putative targets. High-confidence targets were enriched in transcription factors, including those known to regulate AP1 itself. As with the previous studies, signaling components of the gibberellin pathway were found. Transcript profiling revealed that around 44% of the putative targets respond in a moderate way to AP1 activation, while around 11% show a robust change (at least 1.8-fold with p-value < 0.05). The authors speculate that weak transcriptional changes could be explained by a “fine-tuning” regulatory mechanism. It appears that AP1 predominantly acts as a transcriptional repressor during the establishment of the floral meristem identity. Interestingly, around 64% of the targets overlap with the SEP3 targets (Figure 1B), indicating that AP1/SEP3 function mainly as heterodimers to promote floral initiation during early flower development. The authors concluded that AP1 acts as a true hub in the regulatory network that mediates the switch from floral induction to flower formation (Kaufmann et al., 2010).
Most recently, a ChIP-seq study identified 505 binding sites corresponding to 786 potential targets genes for the MADS protein flowering locus C (FLC; Deng et al., 2011). FLC is a flowering repressor down-regulated by vernalization. An FLC specific antibody was used in wild type seedling tissue. Once more, targets were enriched for transcription factors and for hormone response pathways (abscisic acid, jasmonate, ethylene, auxin, and gibberellin). A small-scale gene expression study showed that over 50% of the putative targets are differentially expressed in the flc mutant. FLC regulates genes involved in many different pathways and seems to be important during the complete life cycle of the plant, like juvenile-to-adult transition, flowering initiation, and floral morphogenesis. Surprisingly, FLC shares 28% of the AP1 and 36% of the SEP3 targets (Deng et al., 2011; Figure 1B).
Taken together, the reviewed studies demonstrate that plant MADS proteins may have thousands of target genes. So far, similar studies of plant transcription factors belonging to different families (e.g., LEAFY, APETALA2, BRI1-EMS-SUPPRESSOR1, PHYTOCHROME INTERACTING FACTOR 3-LIKE5, and GLABRA1/3), also show equivalent numbers of target genes (Lee et al., 2007; Morohashi and Grotewold, 2009; Oh et al., 2009; Yant et al., 2010; Moyroud et al., 2011; Winter et al., 2011; Yu et al., 2011). Nevertheless, considering that there are numerous potential CArG-boxes in the genome, and that there are 39 MADS type II genes which might have thousands of targets each, even considering some degree of overlap, it is tempting to question whether this gene family could control a major part of the Arabidopsis genome (and perhaps true for angiosperm genomes in general). Moreover, target analysis revealed that MADS genes are MADS targets, and reciprocal analysis showed a complex relationship among the assayed MADS (Figure 1C).
The wide range of targets may not be exclusive to the plant kingdom. Recently, in mammalian cell lines, it was reported that the MADS proteins SRF and MEF2 have also over 1000 target genes (Schlesinger et al., 2011; Sullivan et al., 2011). Binding events are also enriched in CArG-boxes and targets are involved in cytoskeleton, heart, and muscle development. Interestingly, the percentage of targets showing transcriptional alterations in knock-down backgrounds can be as low as 10% due to the combinatorial nature of gene regulation, with predominant activating regulators compensating for each other. Like plant MADS proteins, mammalian MADS proteins can show a large overlap of targets (up to almost 50%; Schlesinger et al., 2011). In general, besides target genes being redundantly regulated by different transcription factors (e.g., Figure 1C), other reasons for the discrepancy between the genes identified as direct targets, versus genes that are misregulated in mutants for the same transcription factor, could be that transcription only occurs under certain environmental conditions or at developmental time points not sampled by the gene expression experiments. A transcription factor for instance is bound to a target site, but requires an interacting partner to be able to transcriptionally regulate the target gene. On the other hand, given a certain binding event, many times it is difficult to point out which gene is the real target, the upstream, or the downstream gene? This causes false-positives. Probably, the explanation for the “binding event – transcriptionally regulated gene” gap is a combination of all these possibilities, resulting in relatively low frequencies of transcriptionally altered target genes.
In summary, all the findings show that the MADS proteins function in a combinatorial nature as “master” regulators in highly complex regulatory networks. The picture that emerges from these interesting studies depicts MADS domain transcription factors as musical directors performing simultaneous living symphonies. Numerous musicians follow their instructions, and their coordinated sounds and silences (at the exact time and intensity) together weave exquisite melodies, just as our MADS directors coordinate the development of exquisite developmental programs in nature, where, as in a symphony, silence is not the same as “doing nothing.”
References
de Folter, S., and Angenent, G. C. (2006). Trans meets cis in MADS science. Trends Plant Sci. 11, 224–231.
Deng, W., Ying, H., Helliwell, C. A., Taylor, J. M., Peacock, W. J., and Dennis, E. S. (2011). Flowering locus C (FLC) regulates development pathways throughout the life cycle of Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 6680–6685.
Dornelas, M. C., Patreze, C. M., Angenent, G. C., and Immink, R. G. (2011). MADS: the missing link between identity and growth? Trends Plant Sci. 16, 89–97.
Gramzow, L., and Theissen, G. (2010). A Hitchhiker’s guide to the MADS world of plants. Genome Biol. 11, 214.
Immink, R. G., Kaufmann, K., and Angenent, G. C. (2010). The “ABC” of MADS domain protein behaviour and interactions. Semin. Cell Dev. Biol. 21, 87–93.
Kaufmann, K., Muino, J. M., Jauregui, R., Airoldi, C. A., Smaczniak, C., Krajewski, P., and Angenent, G. C. (2009). Target genes of the MADS transcription factor sepallata3: integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol. 7, e1000090.
Kaufmann, K., Wellmer, F., Muino, J. M., Ferrier, T., Wuest, S. E., Kumar, V., Serrano-Mislata, A., Madueno, F., Krajewski, P., Meyerowitz, E. M., Angenent, G. C., and Riechmann, J. L. (2010). Orchestration of floral initiation by apetala1. Science 328, 85–89.
Lee, J., He, K., Stolc, V., Lee, H., Figueroa, P., Gao, Y., Tongprasit, W., Zhao, H., Lee, I., and Deng, X. W. (2007). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19, 731–749.
Messenguy, F., and Dubois, E. (2003). Role of MADS box proteins and their cofactors in combinatorial control of gene expression and cell development. Gene 316, 1–21.
Morohashi, K., and Grotewold, E. (2009). A systems approach reveals regulatory circuitry for Arabidopsis trichome initiation by the GL3 and GL1 selectors. PLoS Genet. 5, e1000396.
Moyroud, E., Minguet, E. G., Ott, F., Yant, L., Pose, D., Monniaux, M., Blanchet, S., Bastien, O., Thevenon, E., Weigel, D., Schmid, M., and Parcy, F. (2011). Prediction of regulatory interactions from genome sequences using a biophysical model for the Arabidopsis leafy transcription factor. Plant Cell 23, 1293–1306.
Oh, E., Kang, H., Yamaguchi, S., Park, J., Lee, D., Kamiya, Y., and Choi, G. (2009). Genome-wide analysis of genes targeted by phytochrome interacting factor 3-like5 during seed germination in Arabidopsis. Plant Cell 21, 403–419.
Schlesinger, J., Schueler, M., Grunert, M., Fischer, J. J., Zhang, Q., Krueger, T., Lange, M., Tonjes, M., Dunkel, I., and Sperling, S. R. (2011). The cardiac transcription network modulated by Gata4, Mef2a, Nkx2.5, Srf, histone modifications, and microRNAs. PLoS Genet. 7, e1001313.
Sullivan, A. L., Benner, C., Heinz, S., Huang, W., Xie, L., Miano, J. M., and Glass, C. K. (2011). Serum response factor utilizes distinct promoter- and enhancer-based mechanisms to regulate cytoskeletal gene expression in macrophages. Mol. Cell. Biol. 31, 861–875.
Winter, C. M., Austin, R. S., Blanvillain-Baufumé, S., Reback, M. A., Monniaux, M., Wu, M.-F., Sang, Y., Yamaguchi, A., Yamaguchi, N., Parker, J. E., Parcy, F., Jensen, S. T., Li, H., and Wagner, D. (2011). Leafy target genes reveal floral regulatory logic, cis motifs, and a link to biotic stimulus response. Dev. Cell 20, 430–443.
Yant, L., Mathieu, J., Dinh, T. T., Ott, F., Lanz, C., Wollmann, H., Chen, X., and Schmid, M. (2010). Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor apetala2. Plant Cell 22, 2156–2170.
Yu, X., Li, L., Zola, J., Aluru, M., Ye, H., Foudree, A., Guo, H., Anderson, S., Aluru, S., Liu, P., Rodermel, S., and Yin, Y. (2011). A brassinosteroid transcriptional network revealed by genome-wide identification of BES1 target genes in Arabidopsis thaliana. Plant J. 65, 634–646.
Citation: Marsch-Martínez N, Wu W and de Folter S (2011) The MADS symphonies of transcriptional regulation. Front. Plant Sci. 2:26. doi: 10.3389/fpls.2011.00026
Received: 16 May 2011;
Accepted: 19 June 2011;
Published online: 01 July 2011.
Copyright: © 2011 Marsch-Martínez, Wu and de Folter. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence:c2Rmb2x0ZXJAaXJhLmNpbnZlc3Rhdi5teA==


