- Graduate School of Agricultural Science, Tohoku University, Sendai, Japan
Inorganic ammonium ions are assimilated by a coupled reaction of glutamine synthetase and glutamate synthase (GOGAT). In rice, three genes encoding either ferredoxin (Fd)-GOGAT, NADH-GOGAT1, or NADH-GOGAT2, have been identified. OsNADH-GOGAT2, a newly identified gene, was expressed mainly in fully expanded leaf blades and leaf sheaths. Although the distinct expression profile to OsNADH-GOGAT1, which is mainly detected in root tips, developing leaf blades, and grains, was shown in our previous studies, physiological role of NADH-GOGAT2 is not yet known. Here, we isolated retrotransposon mediated-knockout mutants lacking OsNADH-GOGAT2. In rice grown under paddy field conditions, disruption of the OsNADH-GOGAT2 gene caused a remarkable decrease in spikelet number per panicle associated with a reductions in yield and whole plant biomass, when compared with wild-type (WT) plants. The total nitrogen contents in the senescing leaf blade of the mutants were approximately a half of the WT plants. Expression of this gene was mainly detected in phloem companion cells and phloem parenchyma cells associated with large vascular bundles in fully expanded leaf blades, when the promoter region fused with a β-glucuronidase gene was introduced into the WT rice. These results suggest that the NADH-GOGAT2 is important in the process of glutamine generation in senescing leaves for the remobilization of leaf nitrogen through phloem to the panicle during natural senescence. These results also indicate that other GOGATs, i.e., NADH-GOGAT1 and ferredoxin-GOGAT are not able to compensate the function of NADH-GOGAT2.
Introduction
In rice (Oryza sativa L.) plants, approximately 80% of the total nitrogen in the panicle arises from remobilization through the phloem from senescing organs (Mae and Ohira, 1981). The major forms of nitrogen in the phloem sap are glutamine and asparagine (Hayashi and Chino, 1990). Synthesis of glutamine is the essential step for nitrogen remobilization in rice, as asparagine is synthesized from glutamine (Ireland and Lea, 1999). Biosynthesis of glutamine occurs via glutamine synthetase (GS, EC 6.3.1.2; Ireland and Lea, 1999). The reaction of GS is coupled with a glutamate synthase (GOGAT), which generates two molecules of glutamate. One molecule of glutamate is cycled back to the GS reaction as substrate with the other molecule exported or used to produce other amino acids (Ireland and Lea, 1999). In rice, there are three genes for cytosolic GS (OsGS1;1, OsGS1;2, and OsGS1;3) and one gene for chloroplastic GS (OsGS2; Tabuchi et al., 2005). There are two types of GOGAT using either reduced ferredoxin (Fd-GOGAT, EC 1.4.7.1) or NADH (NADH-GOGAT, EC 1.4.1.14) as an electron donor. In rice plants, NADH-GOGAT is coded for by two genes, OsNADH-GOGAT1 and OsNADH-GOGAT2, while one gene codes for Fd-GOGAT (Tabuchi et al., 2007). Occurrence of a pseudo-gene for rice Fd-GOGAT has been reported by Zhao and Shi (2006). The major function of GS2 and Fd-GOGAT is in photorespiratory nitrogen metabolisms (Lea and Miflin, 2003). The three GS1 and two NADH-GOGAT species appear to be more important for the normal growth and development (Yamaya and Oaks, 2004), since a mutant lacking either gene for GS2 or Fd-GOGAT was able to grow normally under non-photorespiratory conditions (Kendall et al., 1986; Wallsgrove et al., 1987).
Reverse genetic approaches, together with cell type and age-specific gene expression analysis, are powerful for elucidating gene function in nitrogen metabolisms. For example, our previous results using retrotransposon mediated-knockout mutants clearly show that one of the cytosolic GS species, GS1;1, is important for normal growth and grain filling in rice (Tabuchi et al., 2005). Expression of OsGS1;1 was mainly detected in senescing leaf blades of rice (Tabuchi et al., 2007) with the GS1 protein specifically located in phloem companion cells and parenchyma cells of vascular bundles of leaf blade (Sakurai et al., 1996). Metabolomics data reveal that the GS1;1 possesses a crucial role in coordinating metabolic balance in rice (Kusano et al., 2011). Similar results were also obtained with maize plants, where two cytosolic GS species are involved in the control of grain production (Martin et al., 2006). Furthermore, rice mutants lacking OsNADH-GOGAT1 exhibit a reduced number of active tillers, when grown in a paddy field until harvest (Tamura et al., 2010). Rice NADH-GOGAT protein was mainly detected in vascular parenchyma cells of developing leaves and grains (Hayakawa et al., 1994) and two cell layers of the root surface following the supply of  (Ishiyama et al., 1998). Expression of the OsNADH-GOGAT1 gene in those root cell types (Ishiyama et al., 2003) as well as young leaf blade and grains during development was confirmed in our recent study (Tabuchi et al., 2007). In addition,
(Ishiyama et al., 1998). Expression of the OsNADH-GOGAT1 gene in those root cell types (Ishiyama et al., 2003) as well as young leaf blade and grains during development was confirmed in our recent study (Tabuchi et al., 2007). In addition,  -induced expression of OsGS1;2 in the surface cells of rice roots was confirmed (Ishiyama et al., 2004). These results strongly suggest that NADH-GOGAT1, together with GS1;2, is important in the primary assimilation of
-induced expression of OsGS1;2 in the surface cells of rice roots was confirmed (Ishiyama et al., 2004). These results strongly suggest that NADH-GOGAT1, together with GS1;2, is important in the primary assimilation of  in roots. NADH-GOGAT1 is also active in the reutilization of glutamine remobilized from senescing organs to developing organs such as grains (Tabuchi et al., 2007).
in roots. NADH-GOGAT1 is also active in the reutilization of glutamine remobilized from senescing organs to developing organs such as grains (Tabuchi et al., 2007).
The newly identified OsNADH-GOGAT2 gene was mainly expressed in the fully expanded leaf blades and sheathes at vegetative stage (Tabuchi et al., 2007). However, the physiological function of this gene in rice remains unknown. Occurrence of a single gene for NADH-GOGAT in Arabidopsis thaliana (Kaul et al., 2000) caused some difficulties in understanding the function of rice NADH-GOGAT2. To characterize its function, a reverse genetics approach is adopted and phenotypes of the knockout mutants grown in field conditions are characterized in the current study. Here, we discuss possible function of NADH-GOGAT2 in rice plants.
Materials and Methods
Plant Materials and Growth Conditions
The Project for Rice Genome Research, supported by the Ministry of Agriculture, Forestry, and Fisheries of Japan (mutant panel: http//tos.nias.affrc.go.jo/∼miyao/pub/tos17/) kindly provided 20 seeds each from three lines (ND2034, NC0418, and NG3002) of rice (Oryza sativa L. cv. Nipponbare) in which the retrotransposon Tos17 could potentially have been inserted into OsNADH-GOGAT2 gene. For identification of these knockout mutants, border sequences of Tos17 were surveyed against the OsNADH-GOGAT2 sequence (accession number; AB274818, Tabuchi et al., 2007) using the BLAST program (Altschul et al., 1990). A Nipponbare wild-type (WT) was also used. Seeds were germinated in distilled water at 30°C for 48 h in the dark. Each germinated seed was planted on a synthetic culture soil (Mitsui-Toatsu No.3, Tokyo, Japan) in a small container. Twenty days later, one seedling of each of these mutants and the WT were transplanted into a 1.3-l plastic pot with 1.0 g of slow-release fertilizer (N, 16%; P, 16%; K, 16%: Coop Chemical Co., Tokyo, Japan) and grown in a greenhouse with irrigation, as described by Obara et al. (2001). For determination of the insertion position of Tos17 as well as the genotyping, 2 g fresh weight of leaf blade from a vegetative tiller was harvested when the 10th leaf was fully expanded on the main stem, and immediately frozen in liquid nitrogen. For determination of mRNA expression of OsNADH-GOGAT2, the seedling of WT and Tos17-inserted homozygous lines were grown on a synthetic cultured soil in green house for 22 days, and its third leaf blade was harvested for reverse transcription polymerase chain reaction (RT-PCR) analysis. For quantitative real-time PCR, seedlings were transplanted into1.3 l pots as described above and further grown until the flag leaf was fully expanded. A part of the flag leaf of these lines was used for extraction of RNA. Under field conditions, seeds of WT and three mutant lines were first germinated, planted on a synthetic culture soil, and transplanted into paddy field in Kashimadai, Miyagi, Japan, until the ripening stage and were harvested in 2008 and 2009, as described by Obara et al. (2004). When total nitrogen contents in the 6th and 10th leaf blade of the OsNADH-GOGAT2 mutants and WT were determined, all lines were first grown in one-fourth strength hydroponic solution (Mae and Ohira, 1981) for 39 days and half strength hydroponic solution for 11 days, and then transferred to the 1/10 strength solution for 5 days. Under these conditions, the 10th leaf blade was just fully expanded in all lines. Transgenic rice seeds at T1 generation having OsNADH-GOGAT2 promoter::β-glucuronidase (GUS) was germinated, planted on a synthetic culture soil, and grown in a green house for 26 days after the sowing.
Screening for the OsNADH-GOGAT2 Mutants from Mutants Panel
Rice lines (ND2034, NC0418, and NG3002) were screened by searching the flanking sequence database of the mutant panel. Genomic DNA was prepared from leaf blades of these mutants and WT plants and used for the determination of genotype and position of Tos17 which inserted into OsNADH-GOGAT2 gene. For PCR amplification and sequencing, we used Tos17-specific primers (T17F-1 and T17R-1) as described by Tabuchi et al. (2005). OsNADH-GOGAT2 gene specific primers used were as follows; forward primer, 5′-ATTGGGAATACGTGAGACGC-3′ and reverse primer, 5′-AGGAGGAACATCGACCACAC-3′ for ND2034 and NC0418, forward primer, 5′-GCTCTTGGATCAATGGGAAA-3′ and reverse primer, 5′-CTGTTTTAAGCCCATCCTGC-3′ for NG3002.
Promoter::GUS Construction and Plant Transformation
An approximately 1.3 kb sequence (from −1302 to +4) upstream of the ATG start codon (position +1) of OsNADH-GOGAT2 was amplified from genomic DNA by the following gene specific primer pairs containing attB1 or attB2 site in the 5′ end; forward primer, 5′-AAAAAGCAGGCTGGCTTTGCGGTTTGCGATCAACA-3′ and reverse primer, 5′-AGAAAGCTGGGTACATTGCCCGCGCGGGAACGAAT-3′. This fragment was first recombined into the corresponding sites of pDONR221 entry vector by the BP recombination reaction (Invitrogen Corp., Carlsbad, CA, USA), and then subsequently recombined into the pGWB3 binary vector by performing the LR recombination reaction (Invitrogen Corp.). Agrobacterium-mediated transformation into Nipponbare genome was carried out as described by Kojima et al. (2000).
GUS Staining
A fully expanded fourth leaf blade was harvested from 26-day-old transgenic seedlings. The leaf was cut at the middle part of the blade into 1 mm-thick cross-sections, and then GUS staining was carried out as described by Kojima et al. (2000). A basal part of node was also harvested from the same seedling. The basal part was cut into longitudinal-sections and stained. After staining, the sections were washed with distilled water, embedded in paraffin, and deparaffinized as described by Hayakawa et al. (1994). Light (Leica Microsystems, Tokyo, Japan) or a stereo microscopy (Leica Microsystems) was used for observation of GUS staining.
RT–PCR and Quantitative Real-Time PCR
Total RNA was extracted from leaf blades by using RNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). The first-strand cDNA was synthesized using PrimeScript_RT reagent Kit (Takara Bio Inc., Otsu, Japan). RT-PCR analysis of for Tos17 insertion lines was carried out with specific primers of OsNADH-GOGAT2 gene as follows; forward primer, 5′-ATTGGGAATACGTGAGACGC-3′ and reverse primer, 5′-CTGTTTTAAGCCCATCCTGC-3′. Forward primer, 5′-CTTCATAGGAATGGAAGCTGCGGGTA-3′ and reverse primer, 5′-CGACCACCTTGATCTTCATGCTGCTA-3′ were used for the detection of actin mRNA as described by Sonoda et al. (2003). PCR amplification and electrophoresis were carried out as described by Tabuchi et al. (2005). Quantitative real-time PCR analysis of OsNADH-GOGAT2 and OsNADH-GOGAT1 were carried out using gene specific primers as described by Tabuchi et al. (2007). Primers for OsFd-GOGAT and GAPDH were used as described by Tamura et al. (2010). Primers for OsGS2 were used as follows; forward primer, 5′-AGAACTTGGACGATGAATCGGGGC-3′ and reverse primer, 5′-GAGGGAAGGACGCAGGACTGAAGA-3′. Primers for OsGS1;1 and OsGS1;2 were used as described by Ishiyama et al. (2004). The PCR products were detected by SYBR Premix Ex-Taq™ II (Takara Bio Inc.,) by using Light Cycler 480 (Roche Diagnostics Corp., Tokyo, Japan).
Total Nitrogen
Total nitrogen content in rice leaf blade was determined by Elemental Analyzer (FLASH2000, Thermo Fisher Scientific, Waltham, MA, USA) according to the protocol of the manufacture. Acetanilide was used as a standard.
Field Trait and Yield
Field traits and yield components were evaluated by observing the ripening stage of WT and three lines of NADH-GOGAT2 mutant grown in paddy fields in 2008 and 2009, as described by Obara et al. (2004). Phenotypic measurements were performed as described by Tamura et al. (2010).
Statistics
Data were analyzed with Excel (Microsoft) and SigmaStat (Aspire Software International, Ashburn, VA, USA). Data were compared using one-way analysis of variance (ANOVA) followed by Holm–Sidak post hoc tests with P < 0.05 altered as necessary for multiple comparisons using the Holm correction.
Results
Isolation of Mutants Lacking NADH-GOGAT2
Three OsNADH-GOGAT2 knockout lines (ND2034, NC0418, and NG3002) were screened by searching the flanking sequence database (Miyao et al., 2003) of the mutant panel of the Project for Rice Genome Research, where mutant lines, caused by the random insertion of the endogenous retrotransposon Tos17 into the rice genome (Hirochika et al., 1996), were collected. In line ND2034, Tos17 was inserted into exon-9 of the OsNADH-GOGAT2 gene from +2926 to +2922 where the translation start is +1 (Figure 1A). The apparent abnormal order of nucleotide number was caused by the addition of identical 5 bp sequences (5′-GGATG-3′) at the right and left borders of Tos17 in the ND2034 line, as seen in our previous studies on OsGS1;1 mutants (Tabuchi et al., 2005). In line NC0418, Tos17 was inserted into exon-10 from +3058 to +3054 with the addition of 5′-GGTGC-3′. Tos17 was inserted into exon-12 from +3828 to +3824 with the addition of 5′-CATGA-3′ in line ND2034. Genotypic analyses confirmed that Tos17 was homozygously inserted into each corresponding exon of the OsNADH-GOGAT2 gene in these three lines (results not shown).
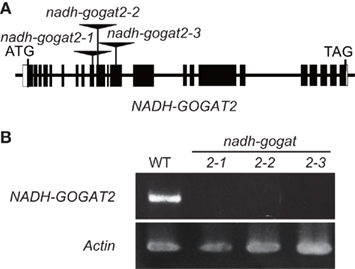
Figure 1. Isolation of OsNADH-GOGAT2knockout mutants of rice. (A) Diagram of the insertion position of retrotransposon Tos17 in OsNADH-GOGAT2. Exons are indicated as boxed regions whereas lines represent introns and the 5′- and 3′-untranscribed region. The open box corresponds to untranslated sequences. (B) Confirmation of no expression of mRNA for OsNADH-GOGAT2 in three knockout lines with RT-PCR. The seedling of wild-type (WT) and Tos17-inserted homozygous lines were grown on a synthetic cultured soil in green house for 22 days, and its third leaf blade was harvested for RT-PCR analysis.
Reverse transcription polymerase chain reaction showed that there was no detectable mRNA for OsNADH-GOGAT2 in the fully expanded third leaf blades of three mutants at 22-day-old seedlings, while OsNADH-GOGAT2 mRNA was actively accumulated in the third leaf blade of WT rice (Figure 1B). Three knockout mutant lines, ND2034, NC0418, and NG3002, are referred to as nadh-gogat2-1, nadh-gogat2-2, and nadh-gogat2-3, respectively. Real-time PCR indicated that these three lines expressed identical amounts of mRNAs for OsNADH-GOGAT1, OsFd-GOGAT, OsGS2, OsGS1;1, and OsGS1;2 in the flag leaf blade, when these mutants and WT rice were grown for 127 days (Figure 2). Since OsGS1;3 mRNA was not detected in leaf blade (Tabuchi et al., 2007), its content was not referred to this result. Thus, we succeeded in obtaining three independent mutant lines specifically lacking OsNADH-GOGAT2.
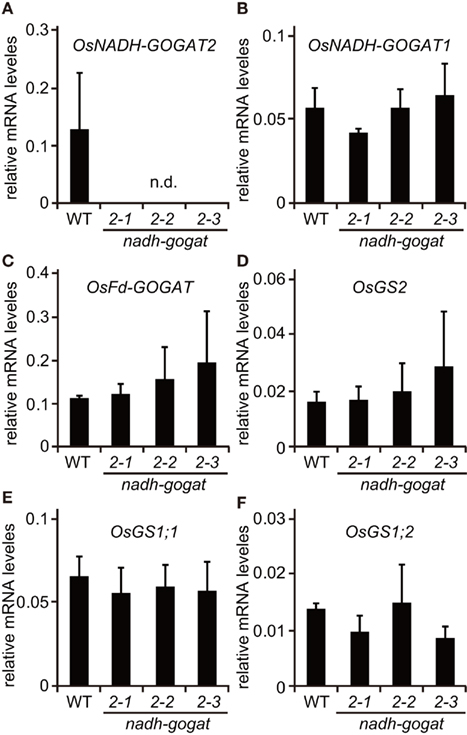
Figure 2. Real-time PCR detection of mRNAs for OsNADH-GOGAT2(A), OsNADH-GOGAT1 (B), OsFd-GOGAT (C), OsGS2 (D), OsGS1;1 (E), and OsGS1;2 (F) from a flag leaf of wild-type (WT) and Tos17-inserted homozygous lines grown with soil in a green house. Three knockout lines, ND2034, NC0418, and NG3002, were indicated as in nadh-gogat2-1, 2-2, and 2-3 in this figure, respectively. mRNA contents were normalized using actin mRNA. Means of independent triplicate samples and SD are indicated.
Severe Reduction in Productivity Via Decrease of Spikelet Number in OsNADH-GOGAT2 Mutants Grown till the Harvest in Paddy Field
Field tests were carried out in 2008 and 2009. The phenotypic characteristics were basically identical in the independent cultivations during couple years. Here, we show results obtained in 2008 (Figures 3 and 4) and those in 2009 are presented as a Figure A1 in Appendix. Three lines of the knockout mutants were able to grow well during the vegetative stage, similar to the growth of the WT rice in the paddy field. However, at the harvest, these three mutants exhibited a slight, but significant reduction in plant height (Figure 3A). Total shoot dry weight was decreased approximately 35–39% in the mutant lines, compared to the WT (Figure 3B). Severe decreases (approximately 43–47%) in panicle dry weight were observed in these mutants (Figure 3C). Along with the decrease in dry mass production, total yield, and ripened spikelet number were also severely reduced in the mutants (Figures 4A,B). Rice yield is determined by the product of the panicle number, spikelet number per panicle, proportion of well ripened grains, and thousand-grain weight (Mae, 1997; Sakamoto and Matsuoka, 2008). When the yield components of NADH-GOGAT2 mutants were investigated by comparing those of the WT, spikelet number per panicle was also significantly reduced (approximately 26–39% reduction) in the mutants (Figure 4E). The proportion of well ripened grains was also decreased by approximately 5–25% (Figure 4F). Panicle number (Figure 4C) and thousand spikelet weight (Figure 4D) were, however, identical between WT and mutant plants. Thus, OsNADH-GOGAT2 gene knockout caused severe reduction in rice yield under conditions grown in paddy field.
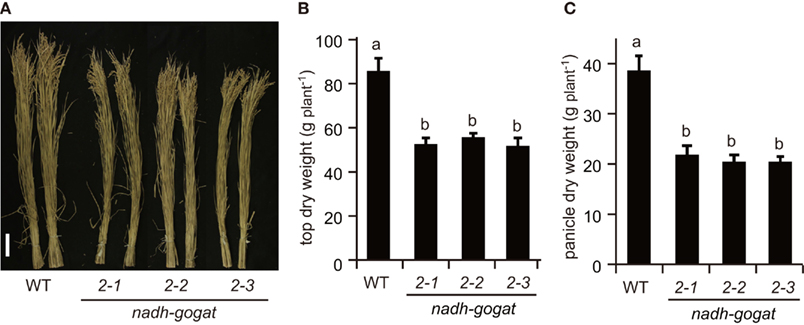
Figure 3. Phenotypic characterization of OsNADH-GOGAT2knockout mutants at ripening stage grown in paddy field in 2008. (A) Phenotype at harvest. Names of three mutant lines were the same as described in Figure 2. Scale bar is 10 cm. (B) Total dry weight of top part (above ground) per plant. (C) Total dry weight of panicles per plant. Means of three independent samples and SD are indicated in (B,C). Significant differences in these three lines by one-way ANOVA are indicated by lower-case letters (A,B) on error bars. Different letters indicate significant differences (P < 0.05) in Holm–Sidak corrected post hoc tests following an ANOVA.
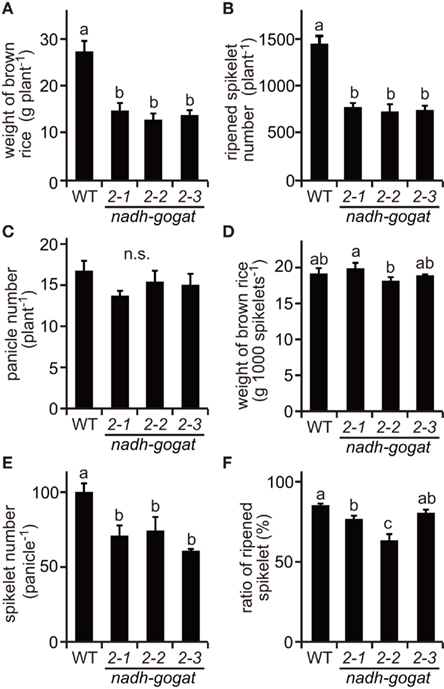
Figure 4. Productivity of OsNADH-GOGAT2knockout mutants cultivated in paddy field in 2008. (A) Total weight of brown rice per plant. Names of three mutant lines were the same as described in Figure 2. (B) Total ripened spikelet number per plant. (C) Total panicle number per plant. (D) 1,000 brown rice weight. (E) Spikelet number per panicle. (F) Proportion of well ripened grains. Means of three independent samples and SD are indicated. Significant differences in these three lines by one-way ANOVA are indicated by lower-case letters (a, ab, b, and c). No significant difference is indicated as in n.s. Different letters indicate significant differences (P < 0.05) in Holm–Sidak corrected post hoc tests following an ANOVA.
Total nitrogen contents in leaf blades were determined, all mutant lines as well as WT were grown hydroponically for 55 days, when the 10th leaf blade was just fully expanded. While nitrogen contents in the 10th leaf blade (the youngest fully expanded leaf) were similar among all mutant lines and WT tested, however, nitrogen contents in the senescing sixth leaf blade of the mutants were approximately a half, when compared with WT (Figure 5).
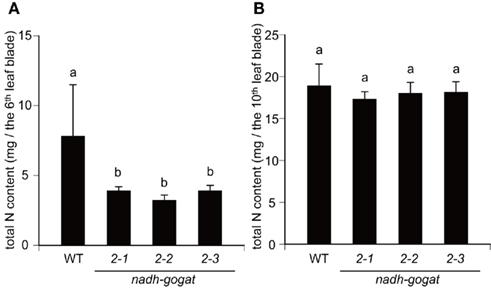
Figure 5. Total nitrogen contents in the 6th and 10th leaf blade of the OsNADH-GOGAT2 mutants hydroponically grown for 55 days. Under these growth conditions, the 10th leaf blade was just fully expanded in all line of the mutants and WT. (A) Nitrogen contents in sixth leaves. (B) Nitrogen contents in 10th leaves. Means of five independent samples and SD are indicated. Different letters indicate significant differences (P < 0.05) in Holm–Sidak corrected post hoc tests following an ANOVA.
Vascular Bundle-Specific Expression of OsNADH-GOGAT2 Gene in Fully Expanded Leaf Blade of Rice
In order to analyze tissue-specific expression of the OsNADH-GOGAT2 gene, the apparent promoter region of this gene (−1302 to +4 bp from the translational initiation site) was fused with GUS reporter gene and this chimeric gene was introduced into the WT rice by using Agrobacterium-mediated transformation (Kojima et al., 2000). Transgenic T1 seeds were germinated and grown for 26 days. Fully expanded fourth leaf blades were harvested and the GUS activity visualized by staining. GUS activity was mainly detected in the phloem parenchyma cells and phloem companion cells of large vascular bundles, when cross-sections of the leaves were stained (Figures 6A,B). GUS activity was also detected in small vascular bundles (Figure 6C). In contrast, there was no detectable GUS activity in other cell types, such as mesophyll or epidermal cells. When a longitudinal-section from the leaf base was stained, GUS activity was mainly detected in large vascular bundle and nodal plexus vascular bundle (Figures 6D,E).
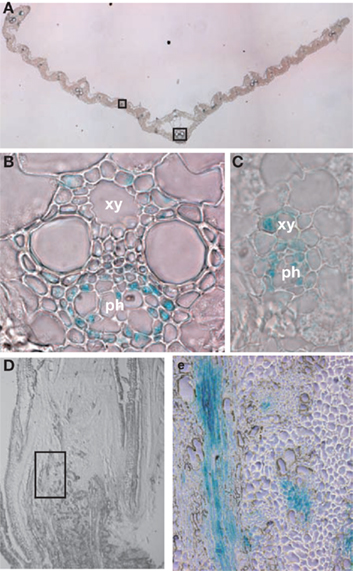
Figure 6. Cell-specific expression of OsNADH-GOGAT2. An approximately 1.3 kb sequence (from −1302 to +4) upstream of the ATG start codon (position +1) of OsNADH-GOGAT2 was fused with GUS structural gene and this chimeric gene was introduced into wild-type (WT) rice. The transformants at T1 generation were grown for 26 days and fully expanded fourth leaf blade were cut into 1 mm-thick cross-sections, and then GUS activity was visualized by staining. Longitudinal-sections were also prepared from a basal node area. (A) GUS activity in cross-section of fully expanded leaf blade. (B) Enlarged view of a large vascular bundle. (C) Enlarged view of a small vascular bundle. (D) Longitudinal-sections of a basal node area. (E) Enlarged view of a large vascular bundle and nodal plexus vascular bundle area. ph, phloem element; xy, xylem vessel element.
Discussion
Unlike Arabidopsis thaliana, there are two genes for NADH-GOGAT, i.e., OsNADH-GOGAT1 and OsNADH-GOGAT2, in rice (Tabuchi et al., 2007). Cellular localization (Hayakawa et al., 1994; Ishiyama et al., 1998) as well as tissue-specific expression of OsNADH-GOGAT1 gene (Ishiyama et al., 2003; Tabuchi et al., 2007) has been extensively studied in rice plants. On the other hand, there exists limited information on NADH-GOGAT2 in rice. It is only known that the OsNADH-GOGAT2 gene is mainly expressed in fully expanded leaf blades and leaf sheaths at vegetative stage (Tabuchi et al., 2007). Since fully expanded leaves contain relatively low activity, as well as protein, of total NADH-GOGAT compared to the developing young leaves (Yamaya et al., 1992), efforts have not actively been made to understand any function of NADH-GOGAT2 in rice. Reverse genetic approaches are a powerful tool for characterizing the function of gene products in plants including rice. For example, our previous work with knockout mutants showed that GS1;1 is important for normal growth and grain filling (Tabuchi et al., 2005). NADH-GOGAT1 was shown to be important in the development of active tillers, when OsNADH-GOGAT1 knockout mutants were characterized (Tamura et al., 2010). Using the identical approach, we have successively extended our fundamental understanding of the physiological function of the newly identified NADH-GOGAT2 in rice. Three independent lines, mutated by the random insertion of an endogenous retrotransposon Tos17 into the OsNADH-GOGAT2 structural gene, were screened from the Mutant Panel of the Project for Rice Genome Research, supported by the Ministry of Agriculture, Forestry, and Fisheries of Japan. These mutants are genetically stable with the homozygous progeny exhibiting the identical phenotype, i.e., severe reduction in total biomass production and panicle weight. Reductions in plant panicle weight were mainly caused by the reductions in spikelet number per panicle and ripened spikelet number per plant. Our previous study with mutants lacking OsNADH-GOGAT1 also demonstrated severe reductions in productivity. However, the OsNADH-GOGAT1 mutants mainly caused reductions in panicle numbers per plant. Rice yield is defined as the product of panicle number, spikelet number per panicle, proportion of well ripened gains, and thousand-grain weight (Mae, 1997; Sakamoto and Matsuoka, 2008). Knockout of OsNADH-GOGAT1 affected the panicle number, while OsNADH-GOGAT2 knockout resulted in a reduction in spikelet numbers, suggesting that these two NADH-GOGATs have discrete functions in rice. Development of tillers and spikelets in rice is suppressed by nitrogen deficient conditions during the early stages of reproductive growth (Mae, 1997). OsNADH-GOGAT2 knockout may have resulted in conditions similar to nitrogen deficiency, in turn causing the decrease in spikelet numbers in the mutant rice. As seen in Figure 3, severe decreases in shoot dry weight and panicle dry weight simply suggested that total nitrogen contents per one plant was severely reduced in the knockout mutants. Indeed, total nitrogen contents in the senescing leaf blade of the mutants were approximately a half, when compared with WT (Figure 5). Thus, the less content of nitrogen in senescing leaf blade in the mutants was probably caused by the shortage of total nitrogen in whole plant body. Comparison of metabolite profiling and remobilization analysis with 15N-palse-chase labeling between the mutants and WT rice will be conducted in near future. Since expression of OsNADH-GOGAT1 and OsFd-GOGAT genes in three mutants unaltered compared with the WT, these GOGATs were not able to compensate for NADH-GOGAT2 function in rice.
The current study with transgenic rice expressing the OsNADH-GOGAT2 promoter-GUS reporter gene clearly showed that this gene was specifically expressed in vascular tissues of the fully expanded fourth leaf blade of the 26-day-old transgenic seedlings grown on a synthetic culture soil (Figure 6). Rice seedlings at this stage are autotrophic, since endosperm of germinated seed had completely been consumed (Tabuchi et al., 2005). Within large vascular bundles, strong signals were detected in phloem companion cells and phloem parenchyma cells. These cells also contain GS1 protein (Sakurai et al., 1996) and recent studies with real-time PCR detection strongly suggest that this GS1 is the GS1;1 (Tabuchi et al., 2007). According to our previous studies, the fully expanded leaf blades contain 16–24% of total NADH-GOGAT activity on a fresh weight basis, when compared to the maximum activity detected in the unexpanded leaf blade (Yamaya et al., 1992). Since major species of mRNA for NADH-GOGAT accumulated in the fully expanded leaf blades at vegetative stage of rice grown on a synthetic culture soil was the OsNADH-GOGAT2 mRNA (Tabuchi et al., 2007), the activity detected in the fully expanded leaf blades would probably caused by NADH-GOGAT2. The identical localization of OsNADH-GOGAT2 promoter-GUS gene and GS1;1 protein in fully expanded leaf blade probably implies that these two enzymes are responsible for the generation of glutamine for remobilization of nitrogen during senescence. Glutamine synthesis in senescing organs is an essential step for the dynamic recycling of nitrogen with asparagine derived from glutamine (Tobin and Yamaya, 2001). The current study suggests that NADH-GOGAT2 provides glutamate to GS1;1 in phloem companion cells and phloem parenchyma cells in senescing leaves for the long distant transport of nitrogen in rice.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Dr. Louis J. Irving, University of Tsukuba, Japan for helpful comments and critical reading of the manuscript. We thank Drs. H. Hirochika and A. Miyao, The Project for Rice Genome Research, National Institute of Agrobiological Sciences, Tsukuba, Japan, for providing seeds of Tos17-inserted rice mutants. The pGWB3 vector used in this work was kindly provided by Dr. T. Nakagawa (Shimane University). We also thank Ms. K. Matsuoka, Mr. T. Honda, and Miss A. Matsuzaki for technical assistance. This work was supported in part by a grant from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Genomics for Agricultural Innovation, IPG-0008), in part by Grants-in-Aid for Scientific Research (A) from the JSPS (19208007 and 22248005 to Tomoyuki Yamaya), and in part by a Grant-in-Aid for Scientific Research on Innovative Area (22119003 to Tomoyuki Yamaya) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410.
Hayakawa, T., Nakamura, T., Hattori, F., Mae, T., Ojima, K., and Yamaya, T. (1994). Cellular localization of NADH-dependent glutamate-synthase protein in vascular bundles of unexpanded leaf blades and young grains of rice plants. Planta 193, 455–460.
Hayashi, H., and Chino, M. (1990). Chemical composition of phloem sap from the upper most internode of the rice plant. Plant Cell Physiol. 31, 247–251.
Hirochika, H., Sugimoto, K., Otsuki, Y., Tsugawa, H., and Kanda, M. (1996). Retrotransposon of rice involved in mutations induced by tissue culture. Proc. Natl. Acad. Sci. U.S.A. 93, 7783–7788.
Ireland, R. J., and Lea, P. J. (1999). “The enzymes of glutamine, glutamate, asparagine and aspartate metabolisms,” in Plant Amino Acids. Biochemistry and Biotechnology, ed. B. K. Singh (New York: Marcel Dekker Inc.), 49–109.
Ishiyama, K., Hayakawa, T., and Yamaya, T. (1998). Expression of NADH-dependent glutamate synthase protein in the epidermis and exodermis of rice roots in response to the supply of nitrogen. Planta 204, 288–294.
Ishiyama, K., Inoue, E., Tabuchi, M., Yamaya, T., and Takahashi, H. (2004). Biochemical background and compartmentalized functions of cytosolic glutamine synthetase for active ammonium assimilation in rice roots. Plant Cell Physiol. 45, 1640–1647.
Ishiyama, K., Kojima, S., Takahashi, H., Hayakawa, T., and Yamaya, T. (2003). Cell type distinct accumulations of mRNA and protein for NADH-dependent glutamate synthase in rice roots in response to the supply of NH4+. Plant Physiol. Biochem. 41, 643–647.
Kaul, S., Koo, H. L., Jenkins, J., Rizzo, M., Rooney, T., Tallon, L. J., Feldblyum, T., Nierman, W., Benito, M. I., Lin, X. Y., Town, C. D., Venter, J. C., Fraser, C. M., Tabata, S., Nakamura, Y., Kaneko, T., Sato, S., Asamizu, E., Kato, T., Kotani, H., Sasamoto, S., Ecker, J. R., Theologis, A., Federspiel, N. A., Palm, C. J., Osborne, B. I., Shinn, P., Conway, A. B., Vysotskaia, V. S., Dewar, K., Conn, L., Lenz, C. A., Kim, C. J., Hansen, N. F., Liu, S. X., Buehler, E., Altafi, H., Sakano, H., Dunn, P., Lam, B., Pham, P. K., Chao, Q., Nguyen, M., Yu, G. X., Chen, H. M., Southwick, A., Lee, J. M., Miranda, M., Toriumi, M. J., Davis, R. W., Wambutt, R., Murphy, G., Dusterhoft, A., Stiekema, W., Pohl, T., Entian, K. D., Terryn, N., Volckaert, G., Salanoubat, M., Choisne, N., Rieger, M., Ansorge, W., Unseld, M., Fartmann, B., Valle, G., Artiguenave, F., Weissenbach, J., Quetier, F., Wilson, R. K., de la Bastide, M., Sekhon, M., Huang, E., Spiegel, L., Gnoj, L., Pepin, K., Murray, J., Johnson, D., Habermann, K., Dedhia, N., Parnell, L., Preston, R., Hillier, L., Chen, E., Marra, M., Martienssen, R., McCombie, W. R., Mayer, K., White, O., Bevan, M., Lemcke, K., Creasy, T. H., Bielke, C., Haas, B., Haase, D., Maiti, R., Rudd, S., Peterson, J., Schoof, H., Frishman, D., Morgenstern, B., Zaccaria, P., Ermolaeva, M., Pertea, M., Quackenbush, J., Volfovsky, N., Wu, D. Y., Lowe, T. M., Salzberg, S. L., Mewes, H. W., Rounsley, S., Bush, D., Subramaniam, S., Levin, I., Norris, S., Schmidt, R., Acarkan, A., Bancroft, I., Brennicke, A., Eisen, J. A., Bureau, T., Legault, B. A., Le, Q. H., Agrawal, N., Yu, Z., Copenhaver, G. P., Luo, S., Pikaard, C. S., Preuss, D., Paulsen, I. T., Sussman, M., Britt, A. B., Selinger, D. A., Pandey, R., Mount, D. W., Chandler, V. L., Jorgensen, R. A., Pikaard, C., Juergens, G., Meyerowitz, E. M., Dangl, J., Jones, J. D. G., Chen, M., Chory, J., and Somerville, M. C. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815.
Kendall, A. C., Wallsgrove, R. M., Hall, N. P., Turner, J. C., and Lea, P. J. (1986). Carbon and nitrogen metabolism in barley (Hordeum vulgare L.) mutants lacking ferredoxin-dependent glutamate synthase. Planta 168, 316–323.
Kojima, S., Kimura, M., Nozaki, Y., and Yamaya, T. (2000). Analysis of a promoter for the NADH-glutamate synthase gene in rice (Oryza sativa L.): cell type-specific expression in developing organs of transgenic rice plants. Aust. J. Plant Physiol. 27, 787–793.
Kusano, M., Tabuchi, M., Fukushima, A., Funayama, K., Diaz, C., Kobayashi, M., Hayashi, N., Tsuchiya, Y. N., Takahashi, H., Kamata, A., Yamaya, T., and Saito, K. (2011). Metabolomics data reveal a crucial role of cytosolic glutamine synthetase 1;1 in coordinating metabolic balance in rice. Plant J. 66, 456–466.
Lea, P. J., and Miflin, B. J. (2003). Glutamate synthase and the synthesis of glutamate in plants. Plant Physiol. Biochem. 41, 555–564.
Mae, T. (1997). Physiological nitrogen efficiency in rice: nitrogen utilization, photosynthesis, and yield potential. Plant Soil 196, 201–210.
Mae, T., and Ohira, K. (1981). The remobilization of nitrogen related to leaf growth and senescence in rice plants (Oryza sativa L.). Plant Cell Physiol. 22, 1067–1074.
Martin, A., Lee, J., Kichey, T., Gerentes, D., Zivy, M., Tatout, C., Dubois, F., Balliau, T., Volt, B., Davanture, M., Tercé-Laforgue, T., Quilleré, I., Coque, M., Gallais, A., Gonzalez-Moro, M.-B., Bethencourt, L., Habash, D. Z., Lea, P. J., Charcosset, A., Perez, P., Murigneux, A., Sakakibara, H., Edwards, K. J., and Hirel, B. (2006). Two cytosolic glutamine synthetase isoforms of maize are specifically involved in the control of grain production. Plant Cell 18, 3252–3274.
Miyao, A., Tanaka, K., Murata, K., Sawaki, H., Takeda, S., Abe, K., Shinozaki, Y., Onosato, K., and Hirochika, H. (2003). Target site specificity of the Tos17 retrotransposon shows a preference for insertion in retrotransposon-rich regions of the genome. Plant Cell 15, 1771–1780.
Obara, M., Kajiura, M., Fukuta, Y., Yano, M., Hayashi, M., and Yamaya, T. (2001). Mapping of QTLs associated with cytosolic glutamine synthetase and NADH-glutamate synthase in rice (Oryza sativa L.). J. Exp. Bot. 52, 1209–1217.
Obara, M., Sato, T., Sasaki, S., Kashiba, K., Nakamura, I., Ebitani, T., Yano, M., and Yamaya, T. (2004). Identification and characterization of QTL on chromosome 2 for cytosolic glutamine synthetase content and panicle number in rice (Oryza sativa L.). Theor. Appl. Genet. 110, 1–11.
Sakamoto, T., and Matsuoka, M. (2008). Identifying and exploiting grain yield genes in rice. Curr. Opin. Plant Biol. 11, 209–214.
Sakurai, N., Hayakawa, T., Nakamura, T., and Yamaya, T. (1996). Changes in the cellular localization of cytosolic glutamine synthetase protein in vascular bundles of rice leaves at various stages of development. Planta 200, 306–311.
Sonoda, Y., Ikeda, A., Saiki, S., von Wirén, N., Yamaya, T., and Yamaguchi, J. (2003). Distinct expression and function of three ammonium transporter genes (OsAMT1;1-1;3) in rice. Plant Cell Physiol. 44, 726–734.
Tabuchi, M., Abiko, T., and Yamaya, T. (2007). Assimilation of ammonium ions and reutilization of nitrogen in rice (Oryza sativa L.). J. Exp. Bot. 58, 2319–2327.
Tabuchi, M., Sugiyama, K., Ishiyama, K., Inoue, E., Sato, T., Takahashi, H., and Yamaya, T. (2005). Severe reduction in growth rate and grain filling of rice mutants lacking OsGS1;1, a cytosolic glutamine synthetase1;1. Plant J. 42, 641–651.
Tamura, W., Hidaka, Y., Tabuchi, M., Kojima, S., Hayakawa, T., Sato, T., Obara, M., Kojima, K., Sakakibara, H., and Yamaya, T. (2010). Reverse genetic approach to characterize a function of NADH-glutamate synthase1 in rice plants. Amino Acids 39, 1003–1012.
Tobin, A. K., and Yamaya, T. (2001). Cellular compartmentation of ammonium assimilation in rice and barley. J. Exp. Bot. 52, 591–604.
Wallsgrove, R. M., Turner, J. C., Hall, N. P., Kendall, A. C., and Bright, S. W. J. (1987). Barley mutants lacking chloroplast glutamate synthetase – biochemical and genetic analysis. Plant Physiol. 83, 155–158.
Yamaya, T., Hayakawa, T., Tanasawa, K., Kamachi, K., Mae, T., and Ojima, K. (1992). Tissue distribution of glutamate synthase and glutamine synthetase in rice leaves. Occurrence of NADH-dependent glutamate synthase protein and activity in the unexpanded, nongreen leaf blades. Plant Physiol. 100, 1427–1432.
Yamaya, T., and Oaks, A. (2004). “Metabolic regulation of ammonium uptake and assimilation,” in Nitrogen Acquisition and Assimilation in Higher Plants, eds S. Amâncio and I. Stulen (Dordrecht: Kluwer Academic Publishers), 35–63.
Zhao, X.-Q., and Shi, W.-M. (2006). Expression analysis of the glutamine synthetase and glutamate synthase gene families in young rice (Oryza sativa) seedlings. Plant Sci. 170, 748–754.
Appendix
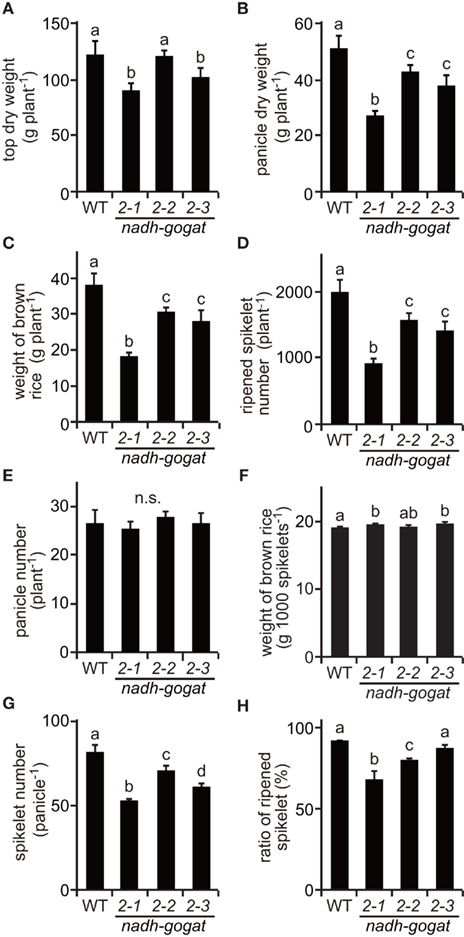
Figure A1. Phenotypic characterization of the mutants at ripening stage cultivated in 2009. (A) Total dry weight of top part (above ground) per plant. (B) Total dry weight of panicles per plant. (C) Total weight of brown rice per plant. Names of three mutant lines were the same as described in Figure 2. (D) Total ripened spikelet number per plant. (E) Total panicle number per plant. (F) 1,000 brown rice weight. (G) Spikelet number per panicle. (H) Proportion of well ripened grains. Means of three independent samples and SD are indicated. Significant differences in these three lines by one-way ANOVA are indicated by lower-case letters (a, ab, b and c). No significant difference is indicated as in n.s. Different letters indicate significant differences (P < 0.05) in Holm–Sidak corrected post hoc tests following an ANOVA.
Keywords: NADH-glutamate synthase2, knockout mutant, nitrogen remobilization, retrotransposon, rice, spikelet number
Citation: Tamura W, Kojima S, Toyokawa A, Watanabe H, Tabuchi-Kobayashi M, Hayakawa T and Yamaya T (2011) Disruption of a novel NADH-glutamate synthase2 gene caused marked reduction in spikelet number of rice. Front. Plant Sci. 2:57. doi: 10.3389/fpls.2011.00057
Received: 04 July 2011;
Accepted: 09 September 2011;
Published online: 28 September 2011.
Edited by:
Lixing Yuan, China Agricultural University, ChinaReviewed by:
Miyako Kusano, RIKEN Plant Science Center, JapanGuohua Xu, Nanjing Agricultural University, China
Copyright: © 2011 Tamura, Kojima, Toyokawa, Watanabe, Tabuchi-Kobayashi, Hayakawa and Yamaya. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Tomoyuki Yamaya, Graduate School of Agricultural Science, Tohoku University, 1-1 Tsutsumidori-Amamiyamachi, Aoba-ku, Sendai 981-8555, Japan. e-mail:dHlhbWF5YUBiaW9jaGVtLnRvaG9rdS5hYy5qcA==
 Wataru Tamura
Wataru Tamura