- Institute of Molecular Physiology and Biotechnology of Plants, University of Bonn, Bonn, Germany
Aldehyde dehydrogenases (ALDHs) are a family of enzymes which catalyze the oxidation of reactive aldehydes to their corresponding carboxylic acids. Here we summarize molecular genetic and biochemical analyses of selected Arabidopsis ALDH genes. Aldehyde molecules are very reactive and are involved in many metabolic processes but when they accumulate in excess they become toxic. Thus activity of aldehyde dehydrogenases is important in regulating the homeostasis of aldehydes. Overexpression of some ALDH genes demonstrated an improved abiotic stress tolerance. Despite the fact that several reports are available describing a role for specific ALDHs, their precise physiological roles are often still unclear. Therefore a number of genetic and biochemical tools have been generated to address the function with an emphasis on stress-related ALDHs. ALDHs exert their functions in different cellular compartments and often in a developmental and tissue specific manner. To investigate substrate specificity, catalytic efficiencies have been determined using a range of substrates varying in carbon chain length and degree of carbon oxidation. Mutational approaches identified amino acid residues critical for coenzyme usage and enzyme activities.
Introduction
Aldehyde dehydrogenases (ALDHs) represent an evolutionary conserved protein superfamily of NAD(P)+-dependent enzymes that metabolize a wide range of endogenous and exogenous aliphatic and aromatic aldehydes, which are toxic, if their steady state level is not strictly regulated. A steadily increasing number of ALDHs has been identified in almost all taxa (Sophos and Vasiliou, 2003), but a comprehensive structural and functional analysis has so far only be performed on the human ALDHs (Marchitti et al., 2008). Although the completion of a number of genome projects in recent years has led to the identification of ALDH protein superfamilies of some lower and higher plant species, as Physcomitrella patens, Arabidopsis, maize, and rice (Kirch et al., 2004; Wood and Duff, 2009; Jimenez-Lopez et al., 2010; Kotchoni et al., 2010), their physiological roles are largely unclear. The interest in studying ALDHs is triggered by available data that indicate essential functions of ALDHs in growth, development, and stress adaptation. The maize mitochondrial ALDH2B2 has been identified as the nuclear restorer of cytoplasmic male sterility (Cui et al., 1996; Liu et al., 2001). The rice mitochondrial ALDH family 2 probably is essential for the detoxification of acetaldehyde during reaeration after submergence (Tsuji et al., 2003), whereas OsALDH7B6 is necessary for seed maturation and maintenance of seed viability by detoxifying aldehydes generated by lipid peroxidation (Shin et al., 2009). Nair et al. (2004) showed that the reduced epidermal fluorescence1 (ref1) mutant of Arabidopsis is caused by a mutation in the ALDH2C4 gene (At3g24503) and that ALDH2C4 plays an important role in the phenyl-propanoid pathway, being involved in ferulic acid and sinapic acid biosynthesis. The Arabidopsis ALDH2B4 may play a role in the pyruvate dehydrogenase bypass pathway (Wei et al., 2009).
The activities of aldehyde dehydrogenases seem to be crucial to regulate the imbalanced accumulation of toxic aldehydes in plants exposed to stress conditions. Different plant ALDH genes have been reported to respond to environmental stress conditions such as dehydration, salinity, or excessive light. Overexpression of selected ALDH genes confers enhanced stress tolerance to transgenic plants, which demonstrates the importance of ALDH proteins in stress responses (Deuschle et al., 2001, 2004; Kirch et al., 2001, 2005; Bouché et al., 2003; Sunkar et al., 2003; Kotchoni et al., 2006; Rodrigues et al., 2006).
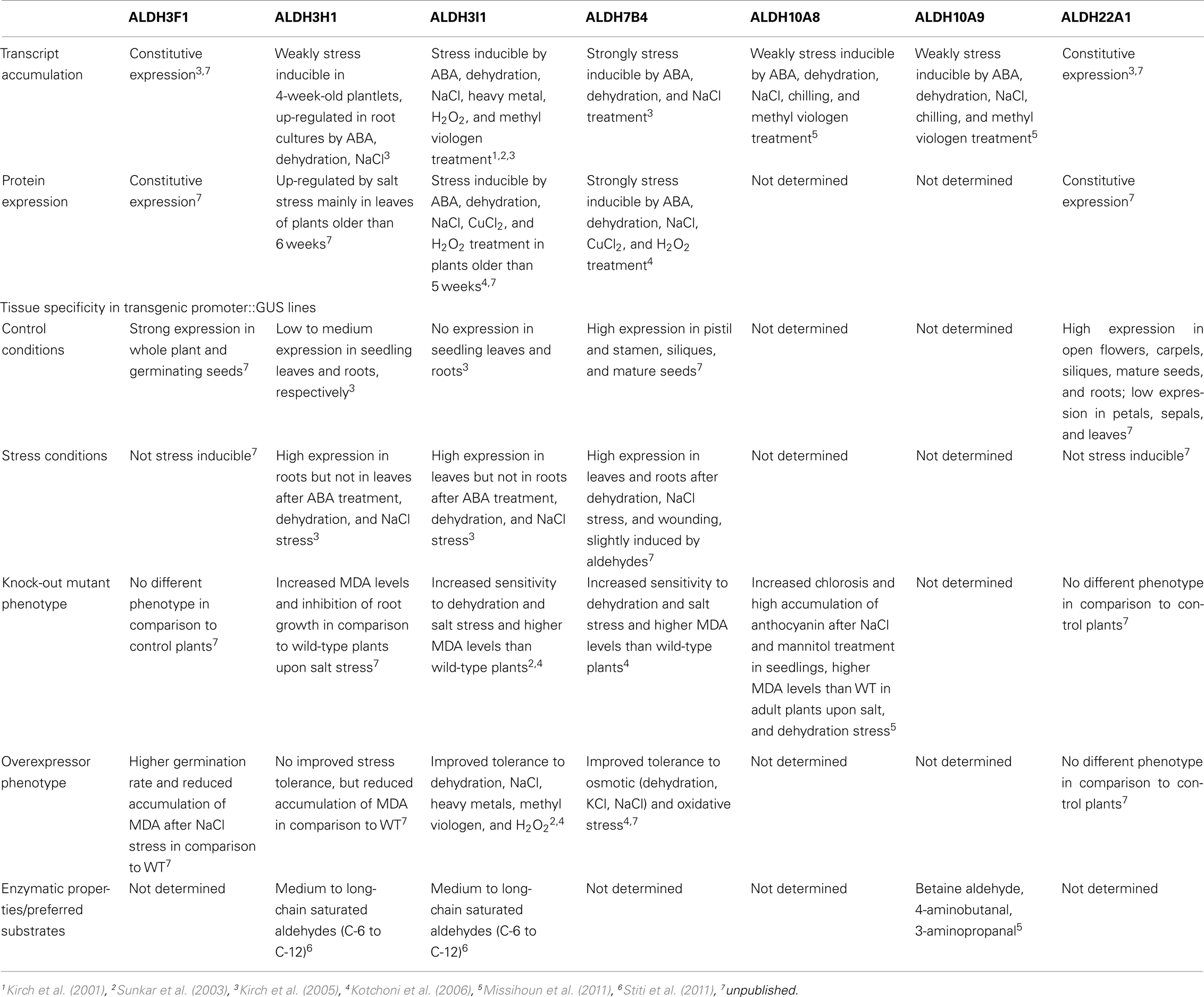
The Arabidopsis thaliana genome encodes 14 genes belonging to nine different aldehyde dehydrogenase families ranging from substrate specific to variable substrate ALDH proteins (Kirch et al., 2004; Table 1). In this review we summarize the current knowledge on selected aldehyde dehydrogenases, which have been studied in our laboratory, using genomic, and biochemical tools. The focus has been on the functional analysis of selected members of different Arabidopsis ALDH families, implicated in abiotic stress tolerance. A platform encompassing genetic and molecular tools was developed for Arabidopsis ALDH genes with a potential role in osmotic stress adaptation and novel ALDHs. Functional genomics tools have been generated for the stress-related family 3 and family 7 ALDH genes (ALDH3I1, ALDH3H1, ALDH3F1, and ALDH7B4), the betaine aldehyde dehydrogenase (BADH) homologues ALDH10A8 and ALDH10A9, and the novel plant specific gene ALDH22A1.
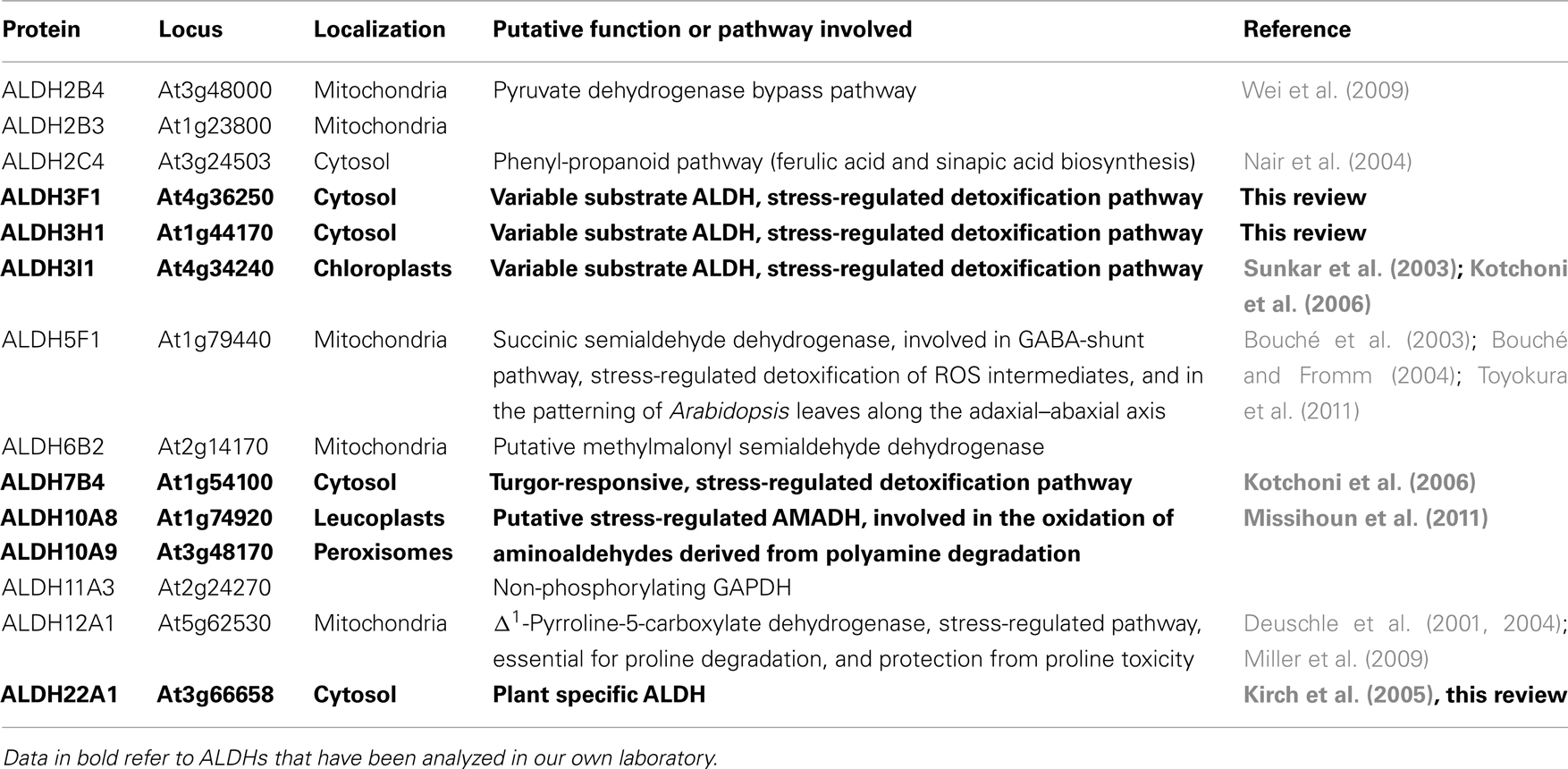
Table 1. Localization and putative physiological functions of the Arabidopsis thaliana ALDH protein superfamily.
Molecular, Biochemical, and Physiological Features of Arabidopsis Aldehyde Dehydrogenases
Subcellular Localization and Expression Patterns
Aldehyde dehydrogenases genes were analyzed with regard to expression patterns, subcellular localization, and biochemical properties. Results of these studies are summarized in Tables 1 and 2. GFP reporter gene analysis demonstrates that ALDH proteins are targeted to different cellular compartments (Figure 1). ALDH3I1 proteins are targeted to the chloroplasts, while ALDH3H1, ALDH3F1, ALDH22A1, and ALDH7B4 are found in the cytosol (Kuhns, 2005; Kotchoni et al., 2006; Ditzer, unpublished results). The putative BADH proteins, ALDH10A8 and ALDH10A9 are targeted to leucoplasts and peroxisomes, respectively (Missihoun et al., 2011), which is in accordance to the localization of BADH proteins from other plant species (Weretilnyk and Hanson, 1990; Nakamura et al., 1997, 2001; Shirasawa et al., 2006; Fitzgerald et al., 2009). The differential localization of ALDH enzymes implies a functional specialization and suggests that different compartments may require ALDH proteins with specific biochemical properties. The fact that several ALDHs proteins are conserved and ubiquitously present in plant cells emphasizes their biological importance, although their function is often still unclear.
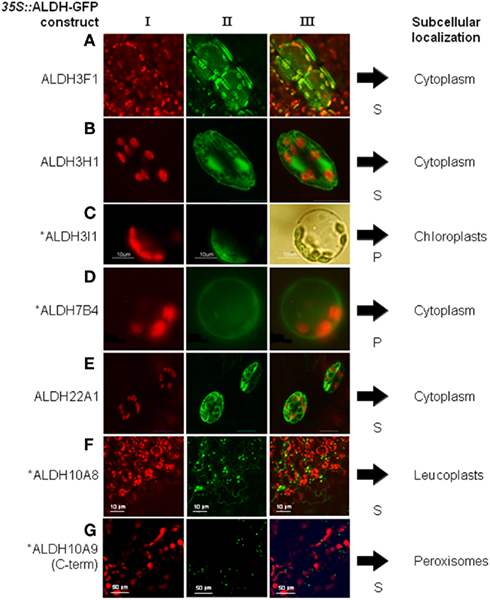
Figure 1. Overview of subcellular localizations of Arabidopsis ALDH-GFP fusion proteins. 35S::ALDH-GFP fusion proteins were expressed in Arabidopsis and viewed under fluorescence light with appropriate filters. (I) shows chlorophyll auto-fluorescence, (II) the GFP-signal, and (III) the merged signals. For ALDH3I1 the transformed protoplast in (C) is shown in bright field to visualize the chloroplasts. S, stable expression after Agrobacterium-mediated transformation; P, transient expression in protoplasts. *GFP-data for ALDH3I1, ALDH7B4, ALDH10A8, and ALDH10A9 have been published (Kotchoni et al., 2006; Missihoun et al., 2011). (A) gene ALDH3F1, (B) gene ALDH3H1, (C) gene ALDH3I1, (D) ALDH7B4, (E) ALDH22A1, (F) ALDH10A8, (G) ALDH10A9.
To elucidate the function of ALDH genes expression was analyzed on the transcript level and by examining transgenic promoter-GUS Arabidopsis plants. ALDH genes are expressed in a tissue specific manner. GUS expression in ALDH3H1 and ALDH22A1 transgenic plants is much stronger in roots than in leaves. ALDH3F1::GUS plants display a very strong expression in the whole plant and in germinating seeds (Kirch et al., 2005; Ditzer, unpublished results). A high GUS activity was detected in pistil and stamen, siliques and mature seeds of ALDH7B4::GUS transgenic plants (Missihoun, 2011). Plants carrying the ALDH22A1 promoter-GUS-fusion showed constitutive GUS expression in all tissues but with variable intensities (Figure 2A). GUS expression was high in siliques, flowers, roots, and relatively low in leaves under control and stress conditions.
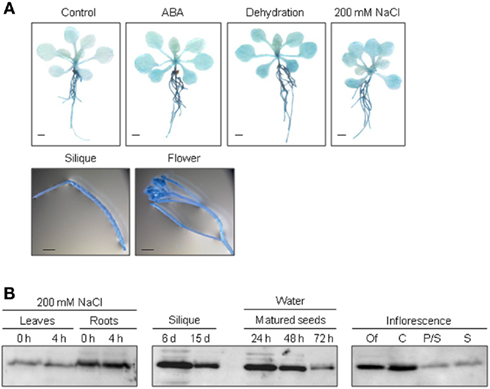
Figure 2. Tissue specific expression of ALDH22A1 in Arabidopsis. (A) ALDH22A1::GUS expression in transgenic Arabidopsis plants: 4-week-old (approximately eight rosette leaves) transgenic Arabidopsis seedlings were subjected to ABA (100 μM), dehydration or salt stress (200 mM NaCl) for a period of 4 h and analyzed for in situ GUS activity. Bars represent 1 mm. (B) Total proteins (20 μg) from leaves and roots of salt-stressed and control plants as well as from different flower tissues were separated on a 12% (w/v) SDS-PAGE and blotted onto a nitrocellulose membrane. Immunodetection was performed using the polyclonal anti-ALDH22A1 antiserum. S, stamina; P/S, petals and sepals; C, carpels; Of, open flowers.
Transcript accumulation analyses demonstrated that five of the analyzed Arabidopsis ALDH genes (ALDH3H1, ALDH3I1, ALDH7B4, ALDH10A8, and ALDH10A9) respond to ABA treatment and abiotic stress conditions such as salt and dehydration, while ALDH3F1 and ALDH22A1 are constitutively expressed and appear to be not related to stress (Kirch et al., 2001, 2005; Sunkar et al., 2003; Missihoun et al., 2011).
Stress induces ALDH genes differentially (Kirch et al., 2005). ALDH3I1, ALDH3H1, ALDH7B4 promoters are induced in response to abiotic stress conditions (Table 2). ALDH3I1::GUS plants display a high expression in leaves but not in roots after ABA treatment, dehydration, or NaCl stress, while the situation is vice versa for ALDH3H1 (Kirch et al., 2005). The ALDH7B4 promoter shows the strongest stress-inducibility, and high GUS expression was found in all tissues in response to ABA, dehydration, NaCl, and wounding. In addition, the ALDH7B4 promoter is also slightly inducible by aldehydes (Höller, 2010; Missihoun, 2011), which may point to a feedback regulation in some metabolic conditions.
To identify possible posttranscriptional regulation steps ALDH protein accumulation was analyzed with specific antibodies. The protein blot analyses reveal a developmental-specific expression pattern for two of the analyzed ALDH proteins. The ALDH3I1 protein is stress inducible in plants older than 5 weeks (approximately 8–10 rosette leaves; Kotchoni et al., 2006; Schmidt, 2006). In contrast, ALDH3H1 accumulates in response to salt stress in 7-week-old (approximately 12–14 rosette leaves) plants and on a lower level in 6-week-old plants (approximately 10–12 rosette leaves; Schmidt, 2006). ALDH3F1 is stress inducible only in young plants at a very low level. ALDH7B4 shows a strong stress response irrespective of age (Kotchoni et al., 2006; Schmidt, 2006). ALDH22A1 proteins accumulate in all tissues but to different amounts. Protein expression is higher in open flowers, carpels, siliques, mature seeds, and roots than in petals/sepals, stamina, and leaves (Figure 2B).
The comparison between transcript expression and protein accumulation as well as the promoter-GUS analyses indicate that ALDH gene expression is predominantly regulated transcriptionally.
Functional Significance of ALDHs
The stress inducible ALDH proteins are predicted to be important for the mechanisms of stress adjustment and long-term adaptation. This hypothesis was investigated by analyzing the stress-performance of transgenic plants overexpressing ALDH genes and ALDH-knock-out lines. Transgenic Arabidopsis plants overexpressing ALDH3F1, ALDH3I1, or ALDH7B4 genes are more tolerant to salt (NaCl and/or KCl) dehydration and oxidative stress. They show higher germination rates and reduced accumulation of malondialdehyde (MDA) in comparison to wild-type plants when challenged with salt, drought, or oxidative stress (Sunkar et al., 2003; Kotchoni et al., 2006; Martens, 2009). ALDH gene overexpressor phenotypes in transgenic plants are in agreement with physiological and molecular analyses of corresponding ALDH T-DNA single and double insertion mutants, which generally reveal an increased sensitivity to dehydration and salt stress and accumulate higher MDA levels than wild-type plants (Figure 3; Sunkar et al., 2003; Kotchoni et al., 2006; Missihoun, 2011; Missihoun et al., 2011). Plants overexpressing ALDH3H1 do not display improved stress tolerance, although accumulation of MDA under stress conditions was reduced in comparison to wild-type plants (Missihoun, 2011). Thus, this gene may have another function.
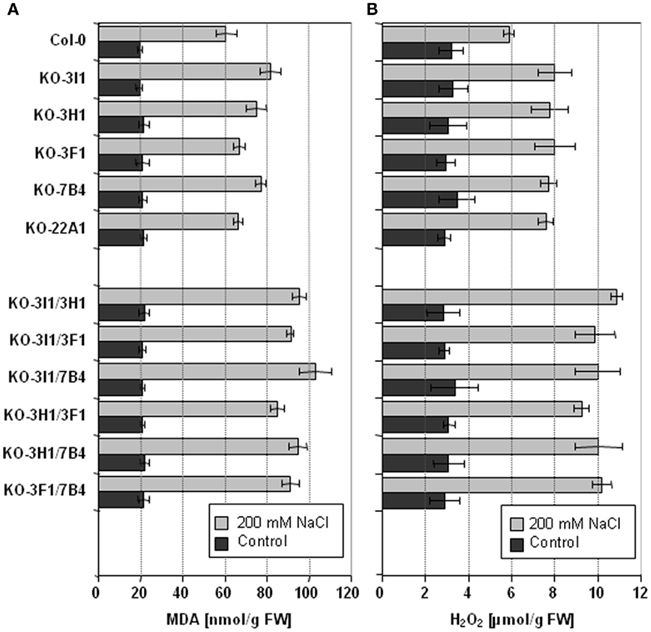
Figure 3. Lipid peroxidation and H2O2 accumulation in Arabidopsis ALDHT-DNA knock-out lines. Lipid peroxidation levels (A) and H2O2 accumulation (B) of 4-week-old, soil-grown Arabidopsis T-DNA knock-out plants that were irrigated with 200 mM NaCl in tap water every second day for 7 days. Control plants were treated with tap water for the same period. Lipid peroxidation was determined as the amount of malondialdehyde (MDA) accumulating as an end product of lipid peroxidation (Sunkar et al., 2003). Quantitative H2O2 measurements were performed according to Kotchoni et al. (2006). The data represent mean values (±SD) of three replicate experiments.
The stress tolerant phenotype of transgenic Arabidopsis plants overexpressing either ALDH3I1 or ALDH7B4 is further supported by experiments with transgenic tobacco plants (Raza, 2010). Results regarding the protective role of ALDH7B4 are confirmed by Rodrigues et al. (2006), who report that Arabidopsis and tobacco plants ectopically expressing a soybean ALDH7 gene also show enhanced tolerance to abiotic stress.
ALDH10A8 and ALDH10A9: Betaine Aldehyde Dehydrogenase Orthologous Genes?
Glycine betaine is a protective osmolyte that is synthesized from choline in response to stress by a two-step reaction catalyzed by choline monooxygenase (EC 1.14.15.7) and betaine aldehyde dehydrogenase (BADH; EC 1.2.1.8). A. thaliana possesses two BADH orthologs, ALDH10A8 and ALDH10A9 (Sakamoto and Murata, 2002; Kirch et al., 2004) but Arabidopsis, like rice, belongs to those species that do not accumulate detectable amounts of glycine betaine under stress conditions (Rhodes and Hanson, 1993). The function of the two Arabidopsis genes, ALDH10A8 and ALDH10A9, are therefore of particular interest. Xing and Rajashekar (2001) reported the detection of endogenous glycine betaine in Arabidopsis using nuclear magnetic resonance (1H NMR). About 3 μmol. g−1 dry weight of glycine betaine were found to accumulate in cold acclimated seedlings. The accumulation of glycine betaine was shown to correlate with the freezing tolerance of leaf tissues. No other report indicated endogenous glycine betaine accumulation in Arabidopsis. This confirms the widely accepted observation that Arabidopsis may only contain none or hardly measurable amounts of glycine betaine. In comparison naturally glycine betaine accumulating species such as spinach accumulate more than 10 μmol glycine betaine. g−1 fresh weight in response to salt stress (Di Martino et al., 2003). Interestingly, when Arabidopsis plants have been transformed with BADH genes isolated from naturally accumulating plant species, glycine betaine was detected and osmotic stress tolerance has been reported (Fitzgerald et al., 2009).
The absence of a functional choline monooxygenase was proposed to explain the lack of glycine betaine in some plant species (Nuccio et al., 1998). For instance, recombinant protein corresponding to the Arabidopsis choline monooxygenase-like gene did not complement Escherichia coli strains deficient in choline dehydrogenase (Hibino et al., 2002). The absence of glycine betaine in some plant species could also be due to choline availability (Peel et al., 2010). So far, the reasons for the non-accumulation of glycine betaine in some plant species are still elusive.
Variation seems to exist among betaine accumulating plants with respect to the subcellular localization of glycine betaine and the biosynthetic enzymes (Fitzgerald et al., 2010; Chen and Murata, 2011). Glycine betaine was found to accumulate in spinach chloroplasts and accordingly both, choline monooxygenase and betaine aldehyde dehydrogenase activities were detected in chloroplasts (Robinson and Jones, 1986). In other glycine betaine accumulators such as mangrove and barley no choline monooxygenase and betaine aldehyde dehydrogenase activities were found in chloroplasts (Hibino et al., 2001; Fujiwara et al., 2008). The BADH isoforms in barley, BBD1 and BBD2, are peroxisomal and cytosolic proteins, respectively (Nakamura et al., 2001; Fujiwara et al., 2008). In Arabidopsis, the ALDH10A8 and ALDH10A9 proteins are targeted to leucoplasts and peroxisomes, respectively (Missihoun et al., 2011). These observations indicate that glycine betaine may be synthesized in other cellular compartments than chloroplasts or other unknown proteins may be involved in its biosynthesis (Chen and Murata, 2011). Alternatively, BADH genes may have other functions. The latter hypothesis is supported by the recent discovery that the fgr mutation associated with the fragrance phenotype in rice resides within the OsBADH2 gene isoform (Bradbury et al., 2008). This mutation results in a non-functional BADH protein that cannot oxidize 4-aminobutanal to γ-aminobutyric acid (GABA), leading to the accumulation of 2-acetyl-1-pyrroline, responsible for the fragrance. The functional analysis of BADH proteins in naturally non-accumulating glycine betaine species may lead to the discovery of alternative mechanism(s) used by those plants to compensate for the lack of glycine betaine. This was addressed with biochemical and genetic tools that we established for Arabidopsis.
We recently reported on the molecular and functional analysis of the Arabidopsis ALDH10A8 (At1g74920) and ALDH10A9 (At3g48170) genes, which are differentially regulated (Missihoun et al., 2011). Generally ALDH10A8 is expressed at higher levels than ALDH10A9. Both genes are responsive to ABA treatment and to a lesser extent to NaCl, dehydration, chilling, and methyl viologen. A T-DNA insertion mutant for the ALDH10A8 gene was found to be sensitive to drought and salinity, indicating that the ALDH10A8 gene product is functional in planta, likely in a pathway different from glycine betaine synthesis. Our data point to a possible involvement in the oxidation of aminoaldehydes derived from the degradation of polyamines.
ALDH10A9 recombinant protein is able to metabolize betaine aldehyde as well as the two aminoaldehydes, 4-aminobutanal and 3-aminopropanal, implying that ALDH10A9 might be involved in the polyamine metabolism (Missihoun et al., 2011). As there was no mutant for ALDH10A9 available, this hypothesis was not fully investigated. Only recently, a T-DNA mutant line (SK17639; Robinson et al., 2009) has become available and the analysis of this line should shed some light on the function of this gene. Evidence supporting the involvement of plant aminoaldehyde dehydrogenases (AMADH) in the terminal catabolism of polyamines has been reported. Petrivalsky et al. (2007 found that the activity of pea AMADH increased during wound healing of injured etiolated seedlings, which was spatially correlated with lignification, a physiological process that involves both copper amine oxidase and polyamine oxidase activities. Likewise, simultaneous increase of diamine oxidase activity and the production of GABA from 4-aminobutanal was also reported in soybean (Xing et al., 2007). This resulted in an increase of the GABA contents in roots of soybean [Glycine max (L.) Merr.] grown on salt. The GABA content derived from the activity of AMADH proteins was estimated to be about 39% of the total GABA pool in soybean (Xing et al., 2007). This suggests that the production of GABA from the polyamine catabolism-derived aminoaldehydes under adverse conditions is a general feature shared by both monocots and dicots. We propose that AMADH may link the polyamine metabolism and the tricarboxylic acid cycle via the GABA-shunt (Figure 4). The metabolic AMADH activity can be responsible for the detoxification of toxic aminoaldehydes (Tylichová et al., 2007). Based on sequence similarity, ALDH10A8 and ALDH10A9 represent the best candidates for AMADH in Arabidopsis. Whether ALDH10A8 and ALDH10A9 function in vivo in this pathway remains to be demonstrated.
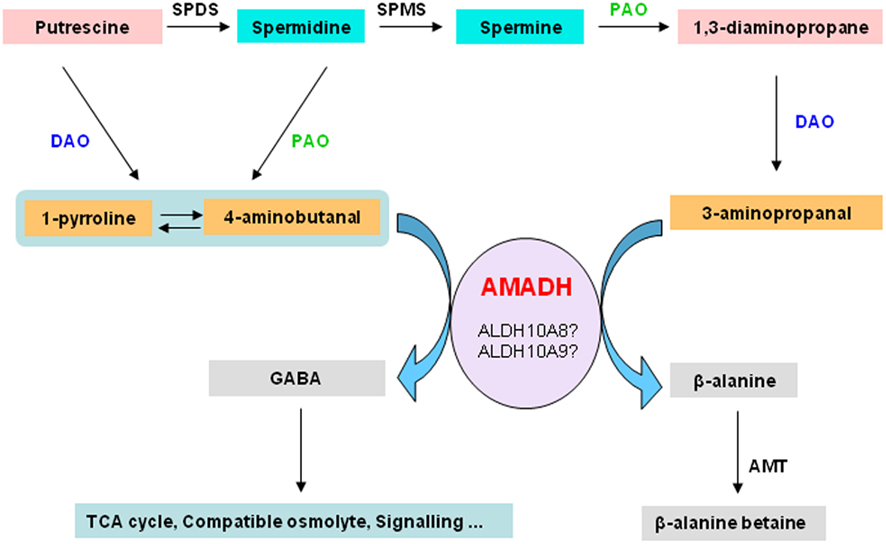
Figure 4. Aminoaldehyde dehydrogenases and the degradation of polyamines. The terminal catabolism of diamine putrescine and polyamines, spermidine and spermine, involves diamine oxidase and polyamine oxidase, respectively. This generates aminoaldehydes such as 4-aminobutanal and 3-aminopropanal, which are further converted by aminoaldehyde dehydrogenases to GABA and β-alanine or β-alanine betaine. GABA can act as a signal compound. It can also be funneled to the TCA cycle or accumulated as compatible osmolyte. Beta-alanine–betaine accumulates in certain members of the Plumbaginaceae as compatible osmolyte. SPDS, spermidine synthase; AMADH, aminoaldehyde dehydrogenase; SPMS, spermine synthase; PAO, polyamine oxidase; DAO, diamine oxidase; GABA, γ-aminobutyric acid; TCA, tricarboxylic acid; AMT, β-alanine methyl transferase.
As reported by Xing et al. 2007) and illustrated in the Figure 4, GABA can be produced from the oxidation of 4-aminobutanal by AMADH. Several functions have been attributed to GABA in plants, ranging from acting as a compatible solute to a signal molecule (Bouché and Fromm, 2004). Although GABA is believed to derive mainly from the decarboxylation of glutamic acid (Shelp et al., 1999), the question as to how cells control the GABA biosynthesis to match the physiological need is unclear. A complex regulatory mechanism may take place to adjust the GABA levels according to the cellular requirement. The site of GABA synthesis may be an important factor. Thus, GABA produced in the cytosol by decarboxylation of glutamic acid may not play the same role as the one generated by AMADHs in peroxisomes or in plastids. AMADHs also oxidise 3-aminopropanal to beta-alanine (Figure 4). Beta-alanine is a non-proteinic amino acid involved in the biosynthesis of pantothenate, a precursor of coenzyme A. It has also been associated to thermotolerance in transgenic tobacco plants (Fouad and Rathinasabapathi, 2006; Fouad and Altpeter, 2009). Beta-alanine can be trimethylated to beta-alanine–betaine, which accumulates as compatible solutes in certain members of the Plumbaginaceae. It is worth validating the existence of enzymatically active AMADHs in Arabidopsis and to establish unequivocally the pivotal role of AMADHs in linking polyamine metabolism to the energy metabolism in plant cells. This will facilitate future investigations of the physiological importance of GABA in plants and help uncover subtle details related to its functions.
Biochemical Analysis and Coenzyme Affinity Studies of ALDH3H1 and ALDH3I1
Research on Arabidopsis ALDHs was started after it was discovered that Cp-ALDH isolated from the desiccation tolerant plant Craterostigma plantagineum is responsive to dehydration and may contribute to detoxify chloroplasts when aldehydes accumulated under stress conditions (Kirch et al., 2001). ALDH3I1 and ALDH3H1 from Arabidopsis are the closest homologues to Cp-ALDH (Cp-ALDH shares 81% and 80% amino acid homologies with ALDH3H1 and ALDH3I1, respectively; ALDH3I1 and ALDH3H1 have 79% homology). Cp-ALDH and ALDH3I1 show a similar tissue specific expression, are targeted to the chloroplast, use NADP+ as coenzyme in enzymatic reactions and contain a conserved valine in their coenzyme binding site. The cytosolic ALDH3H1 has an isoleucine residue instead of the valine and only uses NAD+ as coenzyme. Overexpressing either Cp-ALDH or ALDH3I1 in Arabidopsis leads to improved stress tolerance, which was not observed in plants overexpressing ALDH3H1 (Table 2). We addressed the question how the different subcellular localizations and the single amino acid difference determine substrate specificities and coenzyme affinities. These biochemical properties may be important in the observed stress tolerance. Enzymatic activities of recombinant ALDH3H1 and ALDH3I1 proteins were analyzed to identify their substrate requirements. Both display a preference for medium to long-chain saturated aldehydes with dodecanal as most effective substrate (Table 2).
Structures of several ALDHs revealed that they bind the coenzyme in an atypical five stranded open α/β Rossmann fold (Figure 5A; Liuetal et al., 1997). Different amino acids are important for coenzyme binding and influence coenzyme specificity. Most importantly a lysine residue interacts with the adenine ribose of NAD+ or the 2′-phosphate of NADP+ (Perozich et al., 2000). A glutamic acid residue occupies a central position in the coenzyme binding site and coordinates the adenine ribose 2′- and 3′-hydroxyls of the adenine ribose of NAD+, while repelling the 2′-phosphate of the ribose of adenosine in NADP+ (Perozich et al., 2000, Stiti unpublished). Thus, space in the opposite side of the coenzyme binding cleft is required to keep the interacting NADP+ molecule in an active conformation.
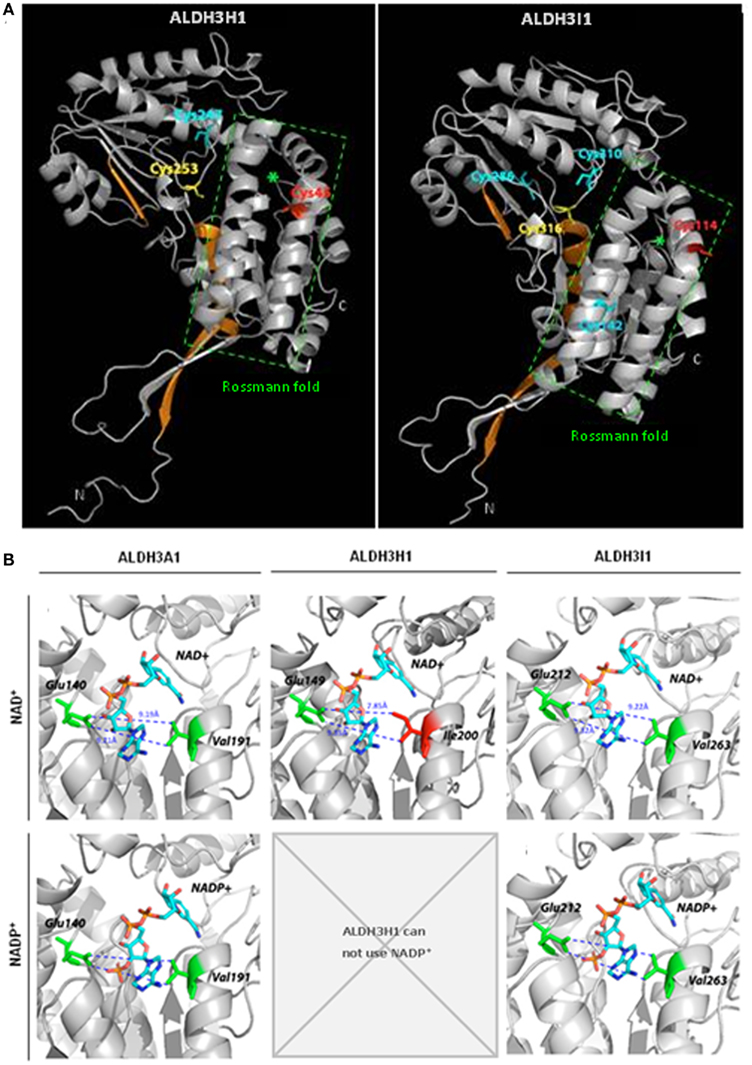
Figure 5. Predicted structures of Arabidopsis ALDH3H1 and ALDH3I1 and models of coenzyme binding. (A) Ribbon diagrams of the predicted structures of Arabidopsis thaliana ALDH3H1 and ALDH3I1 monomeric subunits. Green asterisks denote the central helices of the coenzyme binding Rossmann fold domain. Catalytic cysteine residues are depicted in yellow, redox-sensitive cysteines in red, other cysteines in cyan (Stiti et al., 2011). Regions highlighted in orange indicate helix αD and sheets β12 as well as β13 involved in hydrogen bond-mediated homodimerization in the functional native homologous ALDH3A1 protein. (B) Location of NAD+ and NADP+ and amino acid residues in the coenzyme binding cleft of the previously reported structure of ALDH3A1 from Rattus norvegicus and the predicted structures of ALDH3H1 and ALDH3I1. The predicted structures of ALDH3H1 and ALDH3I1 were built using the crystal structure of Rattus norvegicus ALDH3A1 as template. (This figure was redrawn from Stiti et al., 2011, permission to reproduce the figure was obtained from Portland press).
Comparison of the amino acid sequences showed that the NAD+-specific ALDH3H1 has an isoleucine instead of a valine in motif 4 of the coenzyme binding cleft. The isoleucine has a large side chain due to an additional methyl group compared to the valine occupying this position in ALDH3I1. A valine residue is highly conserved in this position in all family 3 ALDHs except for ALDH3H1 and ALDH3F1. Therefore it was tested in a site-directed mutagenesis approach whether the unusual isoleucine is the reason for the inability to use NADP+. The space occupied by isoleucine may not allow to accommodate the 2′-phosphate of NADP+ while interaction with NAD+ is not affected. Thus the isoleucine-200 was substituted by residues with a shorter side chain, valine, or glycine (Stiti et al. 2011). For the generated mutated enzyme ALDH3H1Ile200Val, the cofactor binding cleft is enlarged by 1.4 Å compared to the wild-type enzyme (7.85 Å). The distance from valine-200 to glutamic acid-149 is approximately 9.23 Å as determined by structure modeling. But the width of the cleft is similar to ALDH3I1 (9.22 Å) or the rat ALDH3A1 (9.19 Å; Figure 5B). The latter two enzymes can use either NAD+ or less effectively NADP+. The ALDH3H1Ile200Val mutant is able to use NADP+ as coenzyme with a KmNADP+ in the range comparable to the wild-type ALDH3I1 with hexanal as a substrate (KmNADP+ = 2300 μM for ALDH3H1Ile200Val and KmNADP+ = 1868 μM for ALDH3I1). A minor increase was observed in KmNAD+ and no significant change of Vmax was observed. Similarly to the wild-type enzyme, ALDH3H1Ile200Val prefers NAD+.
Substitution of isoleucine-200 by glycine should enlarge the cleft to reach a width of 11.63 Å, which makes the NAD+ binding weaker and at the same time increases the affinity for NADP+ (KmNAD+ = 3218μM vs. KmNADP+ = 1817 μM). This demonstrates that the enlargement of the coenzyme binding site resulted in a shift of specificity from NAD+ to NADP+, which confirms that the distance is critical between the residues occupying the positions 200 and 149 for coenzyme affinity. Substituting isoleucine by glycine provides the space necessary to accommodate NADP+, but the larger space possibly makes the cleft too wide for correct binding of NAD+. The isoleucine side chain may force the adenine ribose of NAD+ to allow interaction with the enzyme surface inside the coenzyme binding pocket. Point mutations in enzymatically critical positions alter coenzyme affinities and may be an evolutionary adaptation to the requirements of the physiology needed for the function of the ALDH enzymes.
This review demonstrates that despite strict sequence conservations among ALDH enzymes a few changes in gene sequences lead to diversity in enzymatic properties, subcellular localization, expression patterns, and contribution to stress tolerance. A high degree of sequence conservation of ALDH genes between plant species suggests that these alterations must have been positively selected during evolution for the specific role of individual ALDH genes.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was funded by the DFG grant BA 712/8-1 within the framework of the Arabidopsis Functional Genomics Network (AFGN). We thank Dr. Andrea Ditzer and Tobias Dieckmann, IMBIO University of Bonn for providing some experimental support.
References
Bouché, N., Fait, A., Bouchez, D., Møller, S. G., and Fromm, H. (2003). Mitochondrial succinic-semialdehyde dehydrogenase of the gamma-aminobutyrate shunt is required to restrict levels of reactive oxygen intermediates in plants. Proc. Natl. Acad. Sci. U.S.A. 100, 6843–6849.
Bradbury, L. M. E., Gillies, S. A., Brushett, D., Waters, D. L. E., and Henry, R. J. (2008). Inactivation of an aminoaldehyde dehydrogenase is responsible for fragrance in rice. Plant Mol. Biol. 68, 439–449.
Chen, T. H. H., and Murata, N. (2011). Glycine betaine protects plants against abiotic stress, mechanisms and biotechnological applications. Plant Cell Environ. 34, 1–20.
Cui, X., Wise, R. P., and Schnable, P. S. (1996). The RF2 nuclear restorer gene of male-sterile, T-cytoplasm maize. Science 272, 1334–1336.
Deuschle, K., Funck, D., Forlani, G., Stransky, H., Biehl, A., Leister, D., van der Graaff, E., Kunze, R., and Frommer, W. B. (2004). The role of Δ1-pyrroline-5-carboxylate dehydro-genase in proline degradation. Plant Cell 16, 3413–3425.
Deuschle, K., Funck, D., Hellmann, H., Däschner, K., Binder, S., and Frommer, W. B. (2001). A nuclear gene encoding mitochondrial Delta-pyrroline-5-carboxylate dehydrogenase and its potential role in protection from proline toxicity. Plant J. 27, 345–356.
Di Martino, C., Delfine, S., Pizzuto, R., Loreto, F., and Fuggi, A. (2003). Free amino acids and glycine betaine in leaf osmoregulation of spinach responding to increasing salt stress. New Phytologist 158, 455–463.
Fitzgerald, T. L., Waters, D. L. E., Brooks, L. O., and Henry, R. J. (2010). Fragrance in rice (Oryza sativa L.) is associated with reduced yield under salt treatment. Environ. Exp. Bot. 68, 292–300.
Fitzgerald, T. L., Waters, D. L. E., and Henry, R. J. (2009). Betaine aldehyde dehydrogenase in plants. Plant Biol. 11, 119–130.
Fouad, W. M., and Altpeter, F. (2009). Transplastomic expression of bacterial L-aspartate-alpha-decarboxylase enhances photosynthesis and biomass production in response to high temperature stress. Transgenic Res. 18, 707–718.
Fouad, W. M., and Rathinasabapathi, B. (2006). Expression of bacterial L-aspartate-alpha-decarboxylase in tobacco increases beta-alanine and pantothenate levels and improves thermotolerance. Plant Mol. Biol. 60, 495–505.
Fujiwara, T., Hori, K., Ozaki, K., Yokota, Y., Mitsuya, S., Ichiyanagi, T., Hattori, T., and Takabe, T. (2008). Enzymatic characterization of peroxisomal and cytosolic betaine aldehyde dehydrogenases in barley. Physiol. Plant 134, 22–30.
Hibino, T., Meng, Y. L., Kawamitsu, Y., Uehara, N., Matsuda, N., Tanaka, Y., Ishikawa, H., Baba, S., Takabe, T., Wada, K., Ishii, T., and Takabe, T. (2001). Molecular cloning and functional characterization of two kinds of betaine-aldehyde dehydrogenase in betaine-accumulating mangrove Avicennia marina (Forsk.) Vierh. Plant Mol. Biol. 45, 353–363.
Hibino, T., Waditee, R., Araki, E., Ishikawa, H., Aoki, K., Tanaka, Y., and Takabe, T. (2002). Functional characterization of choline monooxygenase, an enzyme for betaine synthesis in plants. J. Biol. Chem. 277, 41352–41360.
Höller, S. (2010). Untersuchungen zur Regulation des Gens ALDH7B4 aus Arabidopsis thaliana. Diploma thesis, University of Bonn, Bonn.
Jimenez-Lopez, J. C., Gachomo, E. W., Seufferheld, M. J., and Kotchoni, S. O. (2010). The maize ALDH protein superfamily: linking structural features to functional specificity. BMC Struct. Biol. 10, 43. doi: 10.1186/1472-6807-10-43
Kirch, H. H., Bartels, D., Wei, Y., Schnable, P. S., and Wood, A. J. (2004). The ALDH gene superfamily of Arabidopsis. Trends Plant Sci. 9, 371–377.
Kirch, H. H., Nair, A., and Bartels, D. (2001). Novel ABA- and dehydration-inducible aldehyde dehydrogenase genes isolated from the resurrection plant Craterostigma plantagineum and Arabidopsis thaliana. Plant J. 28, 555–567.
Kirch, H. H., Schlingensiepen, S., Kotchoni, S., Ramanjulu, S., and Bartels, D. (2005). Detailed expression analysis of selected genes of the aldehyde dehydrogenase (ALDH) gene superfamily in Arabidopsis thaliana. Plant Mol. Biol. 57, 315–332.
Kotchoni, S. O., Jimenez-Lopez, J. C., Gao, D., Edwards, V., Gachomo, E. W., Margam, V. M., and Seufferheld, M. J. (2010). Modeling-dependent protein characterization of the rice aldehyde dehydrogenase (ALDH) superfamily reveals distinct functional and structural features. PLoS ONE 5, e11516. doi: 10.1371/journal.pone.0011516
Kotchoni, S. O., Kuhns, C., Ditzer, D., Kirch, H. H., and Bartels, D. (2006). Over-expression of different aldehyde dehydrogenase genes in Arabidopsis thaliana confers tolerance to abiotic stress and protects plants against lipid peroxidation and oxidative stress. Plant Cell Environ. 29, 1033–1048.
Kuhns, C. (2005). Subzelluläre Lokalisierung Von Aldheyddehydrogenasen aus Arabidopsis thaliana Mittels GFP-Markierung. Diploma thesis, University of Bonn, Bonn.
Liu, F., Cui, X., Horner, H. T., Weiner, H., and Schnable, P. S. (2001). Mitochondrial aldehyde dehydrogenase activity is required for male fertility in maize (Zea mays L.). Plant Cell 13, 1063–1078.
Liu, Z. J., Sun, Y. J., Rose, J., Chung, Y.-J., Hsiao, C. D., Chang, W. R., Kuo, I., Perozich, J., Lindahl, R., Hempel, J., and Wang, B. C. (1997). The first structure of an aldehyde dehydrogenase reveals novel interactions between NAD and the Rossmann fold. Nat. Struct. Biol. 4, 317–326
Marchitti, S. A., Brocker, C., Stagos, D., and Vasiliou, V. (2008). Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert Opin. Drug Metab. Toxicol. 4, 697–720.
Martens, M. (2009). Generierung von Aldehyd Dehydrogenase Überexpressions-Konstrukten und deren Transformation in Arabidopsis thaliana zur Steigerung der Toleranz gegen abiotischen Stressfaktoren. Diploma thesis, University of Bonn, Bonn.
Miller, G., Honig, A., Stein, H., Suzuki, N., Mittler, R., and Zilberstein, A. (2009). Unraveling Δ1-pyrroline-5-carboxylate-proline cycle in plants by uncoupled expression of proline oxidation enzymes. J. Biol. Chem. 284, 26482–26492.
Missihoun, T. D. (2011). Characterisation of Selected Arabidopsis Aldehyde Dehydrogenase Genes: Role in Plant Stress Physiology and Regulation of Gene Expression. Ph.D. thesis, University of Bonn, Bonn.
Missihoun, T. D., Schmitz, J., Klug, R., Kirch, H. H., and Bartels, D. (2011). Betaine aldehyde dehydrogenase genes from Arabidopsis with different sub-cellular localization affect stress responses. Planta 233, 369–382.
Nair, R. B., Bastress, K. L., Ruegger, M. O., Denault, J. W., and Chapple, C. (2004). The Arabidopsis thaliana REDUCED EPIDERMAL FLUORESCENCE1 gene encodes an aldehyde dehydrogenase involved in ferulic acid and sinapic acid biosynthesis. Plant Cell 16, 544–554.
Nakamura, T., Nomura, M., Mori, H., Jagendorf, A., Ueda, A., and Takabe, T. (2001). An isozyme of betaine aldehyde dehydrogenase in barley. Plant Cell Physiol. 42, 1088–1092.
Nakamura, T., Yokota, S., Muramoto, Y., Tsutsui, K., Oguri, Y., Fukui, K., and Takabe, T. (1997). Expression of a betaine aldehyde dehydrogenase gene in rice, a glycine betaine nonaccumulator, and possible localization of its protein in peroxisomes. Plant J. 11, 1115–1120.
Nuccio, M. L., Russell, B. L., Nolte, K. D., Rathinasabapathi, B., Gage, D. A., and Hanson, A. D. (1998). The endogenous choline supply limits glycine betaine synthesis in transgenic tobacco expressing choline monooxygenase. Plant J. 16, 487–496.
Peel, G. J., Mickelbart, M. V., and Rhodes, D. (2010). Choline metabolism in glycinebetaine accumulating and non-accumulating near-isogenic lines of Zea mays and Sorghum bicolor. Phytochemistry 71, 404–414.
Perozich, J., Kuo, I., Wang, B. C., Boesch, J. S., Lindahl, R., and Hempel, J. (2000). Shifting the NAD/NADP preference in class 3 aldehyde dehydrogenase. Eur. J. Biochem. 267, 6197–6203.
Petrivalsky, M., Brauner, F., Luhova, L., Gagneul, D., and Sebela, M. (2007). Aminoaldehyde dehydrogenase activity during wound healing of mechanically injured pea seedlings. J. Plant Physiol. 164, 1410–1418.
Raza, H. (2010). Functional Characterization of Transgenic Arabidopsis Thaliana Plants Co-Overexpressing Aldehyde Dehydrogenases and Genes for Soluble Osmolytes. Ph.D. thesis, University of Bonn, Bonn.
Rhodes, D., and Hanson, A. D. (1993). Quaternary ammonium and tertiary sulfonium compounds in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 44, 357–384.
Robinson, S. J., Tang, L. H., Mooney, B. A., McKay, S. J., Clarke, W. E., Links, M. G., Karcz, S., Regan, S., Wu, Y. Y., Gruber, M. Y., Cui, D., Yu, M., and Parkin, I. A. (2009). An archived activation tagged population of Arabidopsis thaliana to facilitate forward genetics approaches. BMC Plant Biol. 9, 101. doi: 10.1186/1471-2229-9-101
Robinson, S. P., and Jones, G. P. (1986). Accumulation of glycine betaine in chloroplasts provides osmotic adjustment during salt stress. Australian J. Plant Physiol. 13, 659–668.
Rodrigues, S. M., Andrade, M. O., Soares Gomes, A. P., DaMatta, F. M., Baracat-Pereirea, M. C., and Fontes, E. P. B. (2006). Arabidopsis and tobacco plants ectopically expressing the soybean antiquitin-like ALDH7 gene display enhanced tolerance to drought, salinity, and oxidative stress. J. Exp. Bot. 57, 1909–1918.
Sakamoto, A., and Murata, N. (2002). The role of glycine betaine in the protection of plants from stress: clues from transgenic plants. Plant Cell Environ. 25, 163–171.
Schmidt, S. M. (2006). Biochemische Untersuchungen Von Aldehyd-Dehydrogenasen in Arabidopsis thaliana. Diploma thesis, University of Bonn, Bonn.
Shelp, B. J., Bown, A. W., and McLean, M. D. (1999). Metabolism and functions of gamma-aminobutyric acid. Trends Plant Sci. 4, 446–452.
Shin, J. H., Kim, S. R., and An, G. (2009). Rice aldehyde dehydrogenase7 is needed for seed maturation and viability. Plant Physiol. 149, 905–915.
Shirasawa, K., Takabe, T., Takabe, T., and Kishitani, S. (2006). Accumulationof glycine betaine in rice plants that overexpress choline monooxygenase from spinach and evaluation of their tolerance to abiotic stress. Ann. Bot. 98, 565–571.
Sophos, N. A., and Vasiliou, V. (2003). Aldehyde dehydrogenase gene superfamily: the 2002 update. Chem. Biol. Interact. 143–144, 5–22.
Stiti, N., Adewale, I. O., Petersen, J., Bartels, D., and Kirch, H. H. (2011). Engineering the nucleotide coenzyme specificity and sulfhydryl redox sensitivity of two stress-responsive aldehyde dehydrogenase isoenzymes of Arabidopsis thaliana. Biochem. J. 434, 459–471.
Sunkar, R., Bartels, D., and Kirch, H. H. (2003). Overexpression of a stress-inducible aldehyde dehydrogenase gene from Arabidopsis thaliana in transgenic plants improves stress tolerance. Plant J. 35, 452–464.
Toyokura, K., Watanabe, K., Oiwaka, A., Kusano, M., Tameshige, T., Tatematsu, K., Matsumoto, N., Tsugeki, R., Saito, K., and Okada, K. (2011). Succinic semialdehyde dehydrogenase is involved in the robust patterning of Arabidopsis leaves along the adaxial-abaxial axis. Plant Cell Physiol. 52, 1340–1353.
Tsuji, H., Meguro, N., Suzuki, Y., Tsutsumi, N., Hirai, A., and Nakazono, M. (2003). Induction of mitochondrial aldehyde dehydrogenase by submergence facilitates oxidation of acetaldehyde during reaeration in rice. FEBS Lett. 546, 369–373.
Tylichová, M., Kopecný, D., Snégaroff, J., and Šebela, M. (2007). Aminoaldehyde dehydrogenases: has the time now come for new interesting discoveries? Curr. Topics Plant Biol. 8, 45–70.
Wei, Y., Lin, M., Oliver, D. J., and Schnable, P. S. (2009). The roles of aldehyde dehydrogenases (ALDHs) in the PDH bypass of Arabidopsis. BMC Biochem. 10, 7. doi: 10.1186/1471-2091-10-7
Weretilnyk, E. A., and Hanson, A. D. (1990). Molecular cloning of a plant betaine-aldehyde dehydrogenase, an enzyme implicated in adaptation to salinity and drought. Proc. Natl. Acad. Sci. U.S.A. 87, 2745–2749.
Wood, A. J., and Duff, R. J. (2009). The aldehyde dehydrogenase (ALDH) gene superfamily of the moss Physcomitrella patens and the algae Chlamydomonas reinhardtii and Ostreococcus tauri. Bryologist 112, 1–11.
Xing, S. G., Jun, Y. B., Hau, Z. W., and Liang, L. Y. (2007). Higher accumulation of γ-aminobutyric acid induced by salt stress through stimulating the activity of diamine oxidases in Glycine max (L.) Merr. roots. Plant Physiol. Biochem. 45, 560–566.
Keywords: aldehyde dehydrogenases, aminoaldehyde dehydrogenases, betaine aldehyde dehydrogenases, coenzyme binding, enzyme activities, stress tolerance
Citation: Stiti N, Missihoun TD, Kotchoni SO, Kirch H-H and Bartels D (2011) Aldehyde dehydrogenases in Arabidopsis thaliana: biochemical requirements, metabolic pathways, and functional analysis. Front. Plant Sci. 2:65. doi: 10.3389/fpls.2011.00065
Received: 23 August 2011; Accepted: 23 September 2011;
Published online: 13 October 2011.
Edited by:
Andreas P. M. Weber, University of Duesseldorf, GermanyReviewed by:
Andrew Wood, Southern Illinois University, USADavid Gagneul, University of Lille 1, France
Copyright: © 2011 Stiti, Missihoun, Kotchoni, Kirch and Bartels. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Dorothea Bartels, Institute of Molecular Physiology and Biotechnology of Plants, University of Bonn, Kirschallee 1, D-53115 Bonn, Germany. e-mail:ZGJhcnRlbHNAdW5pLWJvbm4uZGU=
†Present address: Simeon O. Kotchoni, Department of Biology, Rutgers University, 315 Penn Street, Camden, NJ 08102, USA.
‡Naim Stiti and Tagnon D. Missihoun have contributed equally to this work.