- Energy Biosciences Institute, University of California Berkeley, Berkeley, CA, USA
Plant cell walls are composed of structurally diverse polymers, many of which are O-acetylated. How plants O-acetylate wall polymers and what its function is remained elusive until recently, when two protein families were identified in the model plant Arabidopsis that are involved in the O-acetylation of wall polysaccharides – the reduced wall acetylation (RWA) and the trichome birefringence-like (TBL) proteins. This review discusses the role of these two protein families in polysaccharide O-acetylation and outlines the differences and similarities of polymer acetylation mechanisms in plants, fungi, bacteria, and mammals. Members of the TBL protein family had been shown to impact pathogen resistance, freezing tolerance, and cellulose biosynthesis. The connection of TBLs to polysaccharide O-acetylation thus gives crucial leads into the biological function of wall polymer O-acetylation. From a biotechnological point understanding the O-acetylation mechanism is important as acetyl-substituents inhibit the enzymatic degradation of wall polymers and released acetate can be a potent inhibitor in microbial fermentations, thus impacting the economic viability of, e.g., lignocellulosic based biofuel production.
Introduction
The cells of higher plants are encased in a wall consisting of multiple, structurally complex, highly substituted polymers. One of the substituents found on many of these polymers is O-linked acetate. How the O-acetyl-substituent is transferred to the wall polymers and its role in the life cycle of a plant is not known. From an industrial perspective O-acetyl-groups on plant polymers contribute significantly to their viscosity and gelation properties and hence applicability, e.g., in the food industry (Rombouts and Thibault, 1986; Huang et al., 2002). Recently, O-acetylation of cell wall polysaccharides has received increased attention, as wall polymers, the primary constituent of plant biomass, can be used as a renewable source for the production of biofuels and other commodity chemicals (Pauly and Keegstra, 2008; Carroll and Somerville, 2009). During the processing of plant biomass acetylation can reduce saccharification yields, if not removed by pre-treatments (Selig et al., 2009). Even if acetate is released during processing it inhibits some sugar-to-ethanol fermenting organisms (Helle et al., 2003). Techno-economic models predict that a 20% reduction in biomass O-acetylation could result in a 10% reduction in ethanol price (Klein-Marcuschamer et al., 2010). Hence, a major goal in plant research in the biofuel area is to try to reduce O-acetylation in the walls of plant feedstocks.
Many Plant Cell Wall Polymers are O-Acetylated
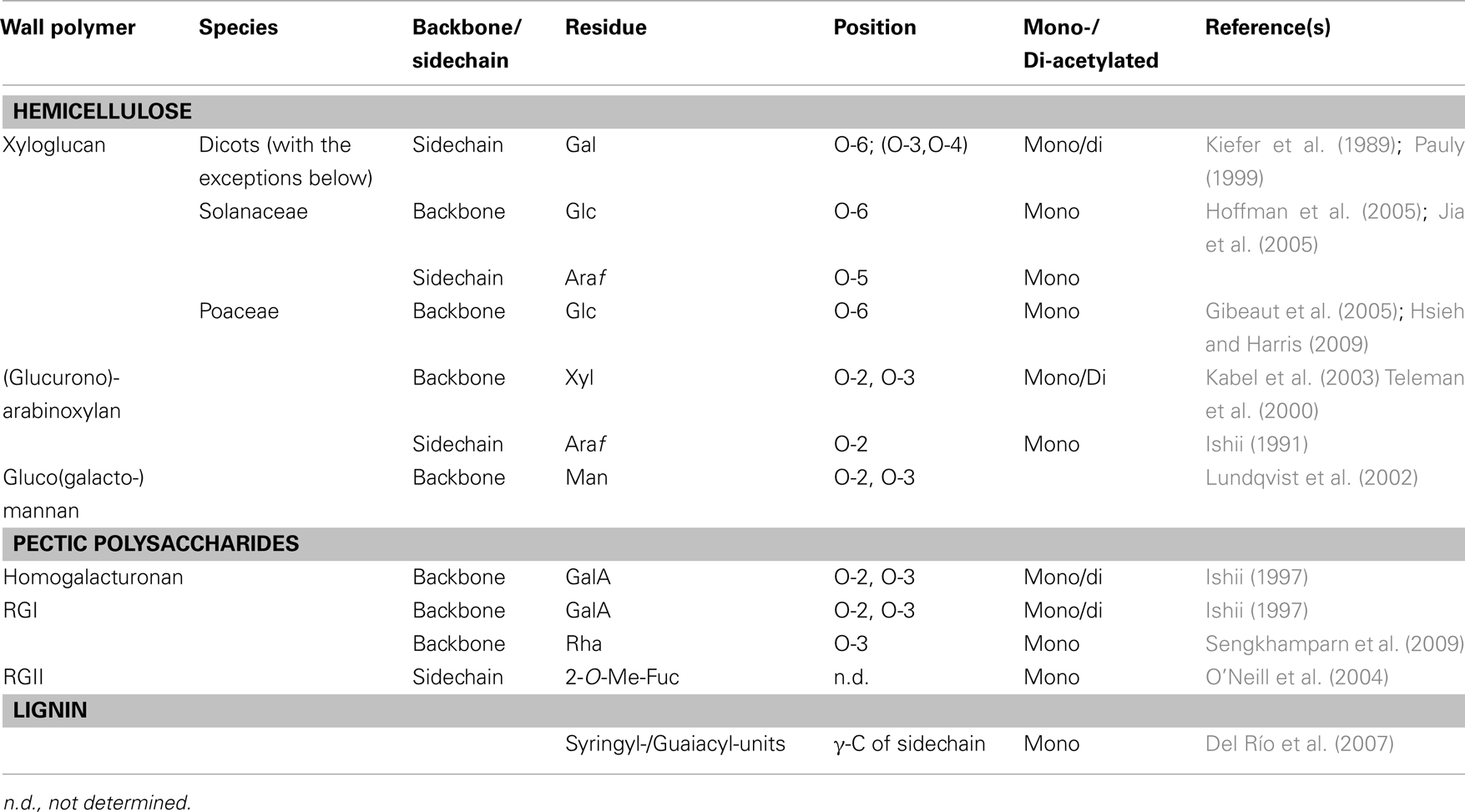
O-acetyl-substituents are present on nearly all wall polymers with the exception of cellulose, β-1,3-1,4-linked glucan in grasses, and the wall glycoproteins. In particular the various hemicelluloses, the pectic polysaccharides and the polyphenol lignin can be O-acetylated (Table 1 and references therein). The degree of O-acetylation can vary depending on species, tissue type, and developmental state as has been observed with any other wall substituent (Pauly and Keegstra, 2010). In the dicot hemicellulose xyloglucan (XyG) only the sidechain galactosyl-residue is O-acetylated (Pauly, 1999; Scheller and Ulvskov, 2010; Gille et al., 2011b). In contrast, in members of the Solanaceae and the Poaceae the glucosyl-residues of the XyG backbone are O-acetylated (Gibeaut et al., 2005; Jia et al., 2005). Presumably, polymer backbone O-acetylation has a larger effect on the polymer properties such as conformation and hydrophobicity, while on the sidechain O-acetates might present just an alternative substituent. Other polymers, whose glycan-backbones can be O-acetylated, include arabinoxylan (Bastawde, 1992; Van Dongen et al., 2011), glucomannans present in softwood (Lundqvist et al., 2002) or in storage polymers (Gille et al., 2011a), as well as the pectic polysaccharides homogalacturonan (Liners et al., 1994) and rhamnogalacturonan I (Schols and Voragen, 1994). Lignin can also be O-acetylated; up to 58% of the phenolic units were found to be O-acetylated in species such as Kenaf (Del Río et al., 2008).
The position of the O-acetyl-group has been identified in nearly all of the polymers (Table 1). Some monosaccharides present in wall polymers can be substituted with two O-acetyl-groups such as XyG, arabinoxylan, and some of the pectic polysaccharides. It should be noted that O-acetyl-groups can migrate in aqueous solutions to non-substituted hydroxyl-groups within the same glycosyl-residue by a chemical, non-protein mediated mechanism forming an equilibrium of acetylation on the various positions, e.g., O-2 and O-3 on xylosyl-residues in xylan (Kabel et al., 2003). Migration has also been observed on the galactosyl-residue in XyG (Pauly, 1999).
Mechanisms of Cell Wall Polymer O-Acetylation: Comparison to Other Organisms
Despite the abundance of O-acetyl-substituents on various wall polymers virtually nothing was known about the molecular mechanism of O-acetylation in plants until recently. More information had been amassed from the polysaccharide O-acetylation systems in bacteria, fungi, and mammals. In the fungal pathogen Cryptococcus neoformans the mannosyl-residues of the capsular polysaccharide glucuronoxylomannan can be O-acetylated (Cherniak and Sundstrom, 1994). The responsible protein, CnCas1p, has been identified as essential for the O-acetylation of this capsular polysaccharide (Janbon et al., 2001). CnCas1p contains 12 transmembrane domains as well as a 347 aa loop predicted to face the Golgi lumen between the first and second transmembrane domain from the N-terminus (Figure 1). Orthologs to CnCas1p were also identified in the human and drosophila genomes (Janbon et al., 2001). The human ortholog HsCasD1 has a very similar protein structure; multiple (13) transmembrane domains and a large loop, also predicted to face a membrane enclosed lumen (Arming et al., 2011; Figure 1). Ectopic expression of HsCasD1 leads to an increase in the O-acetylation of sialic acids and subcellular localization experiments located the protein to the Golgi apparatus (Arming et al., 2011). In bacteria such as Staphylococcus, Campylobacter, Helicobacter, Neisseria, and Bacillus the muramyl-residue in the wall polymer peptidoglycan is O-acetylated (Clarke and Dupont, 1992; Moynihan and Clarke, 2010). In Gram-positive bacteria O-acetylation of peptidoglycan is facilitated by OatA (Bera et al., 2005; Aubry et al., 2011; Bernard et al., 2011). Similar to the fungal and human Cas1 proteins, also the bacterial OatA proteins consist of multiple transmembrane domains and a large loop (Bernard et al., 2011; see LpOatA in Figure 1). In contrast, in Gram-negative bacteria such as Neisseria gonorrhoeae or Bacillus anthracis, the peptidoglycan acetylation is facilitated by two separate proteins – PatA and PatB (Moynihan and Clarke, 2010; Laaberki et al., 2011). While PatA represents a protein with multiple transmembrane domains, PatB is a membrane bound periplasmic protein reminiscent of the loop present in the Cas1 proteins in other organisms (Figure 1). Another O-acetylated bacterial polysaccharide is alginate – a viscous exopolysaccharide produced by certain bacteria such as the human pathogen Pseudomonas aeruginosa (Franklin and Ohman, 1996). Here, three proteins were shown to be required for alginate acetylation; PaAlgI, PaAlgJ, and PaAlgF (Franklin and Ohman, 1993, 1996; Shinabarger et al., 1993; Figure 1). The PaAlgI protein contains multiple transmembrane domains while the PaAlgJ protein is a membrane-anchored protein with one transmembrane domain and a large loop facing the extracellular space (Figure 1). The function of the periplasmic PaAlgF protein is unknown but it is thought to interact with PaAlgI and PaAlgJ in the alginate biosynthetic complex (Franklin and Ohman, 2002).
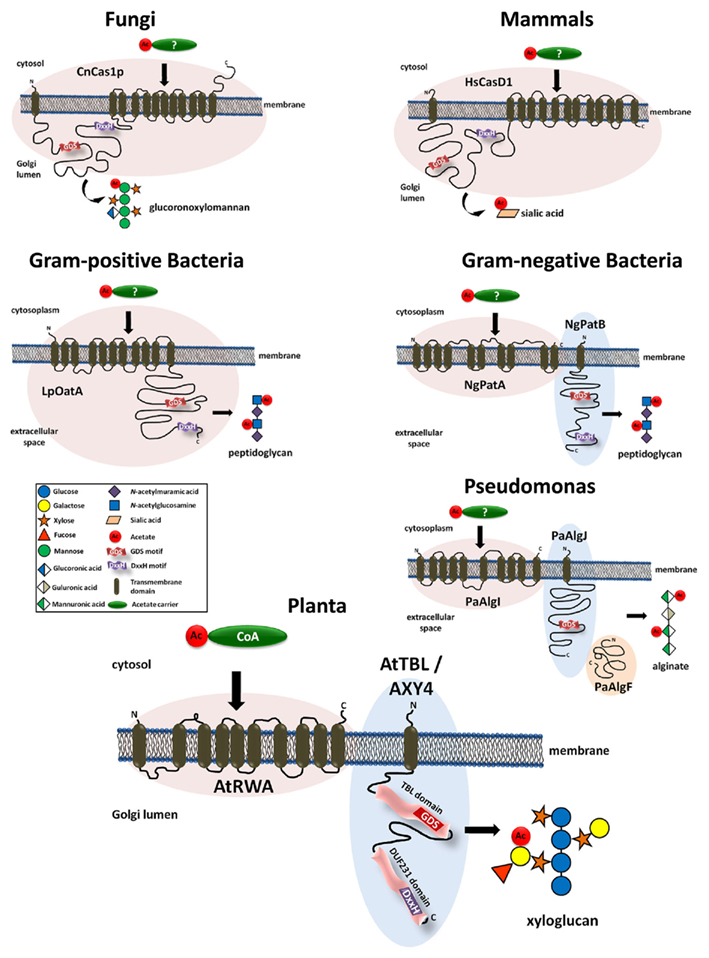
Figure 1. Models of proteins involved in the O-acetylation of carbohydrates in various organisms. Presented are proteins that have been shown to impact O-acetylation of glucuronoxylomannan in Cryptococcus neoformans (Fungi), sialic acid in humans (Mammals), peptidoglycan in Gram-positive and Gram-negative bacteria, alginate in Pseudomonas aeruginosa, and xyloglucan in Arabidopsis thaliana (Plants). The structural organization of the proteins and protein complexes seems to be conserved among those organisms. All systems have in common a multi-transmembrane domain, proposed to be involved in the translocation of an acetyl-carrier across a membrane. In addition, they contain a large globular domain with highly conserved GDS and DxxH peptide motifs (see also Figure 2) representing the presumed acetyltransferase. While in fungi, mammals, and Gram-positive bacteria this mechanism is realized in a single protein, in Gram-negative bacteria and plants this mechanism is split into two proteins. Alginate O-acetylation in Pseudomonas seems to be a special case as no DxxH motif can be found and three proteins were shown to be essential.
Recent findings indicate that plants harbor a similar O-acetylation system. Based on protein homology to the CnCas1p protein from Cryptococcus, a family of four proteins has been identified in Arabidopsis that lead to a reduced cell wall acetylation phenotype (rwa-mutants; Manabe et al., 2011; Lee et al., 2011). RWA consists of 10 transmembrane domains but lacks the extended loop found in CnCas1p (Figure 1). Mutants affected only in RWA2 where shown to have an overall reduction of 20% in wall polymer acetylation levels with multiple polymers being affected including pectin, XyG, and xylan (Manabe et al., 2011).
A second protein involved in plant polysaccharide O-acetylation was identified using a forward genetic screen for mutants with altered xyloglucan structures (axy; Gille et al., 2011b). Knock-out mutants of the identified gene AXY4 lack O-acetyl-substituents on XyG completely in non-seed tissues, while the O-acetylation level of other polysaccharides is not affected. These findings indicate that unlike RWA, AXY4 acts in a polysaccharide specific manner as a likely XyG specific O-acetyltransferase. The O-acetyltransferase activity of AXY4 and the position, to which the O-acetyl-group is transferred has yet to be experimentally demonstrated. However, overexpression of the AXY4 gene leads not only to an increase in XyG acetylation but there is also evidence that one galactosyl-unit of XyG becomes di-acetylated (Gille et al., 2011b). Hence, AXY4 is sufficient to add multiple acetyl-substituents to the same galactosyl-residue. AXY4 could accomplish this by adding acetyl-substituents onto two different positions. Alternatively, AXY4 adds the acetyl-group to a single specific position. The acetyl-group is then distributed through chemical migration to the other position, leaving the initial O-acetylation site open for a further addition of a second O-acetate. However, overexpression of AXY4 did not lead to completely acetylated XyG, suggesting that the acetylated polymer can be further modified by acetylesterases, which remove the acetate, presumably after deposition in the apoplastic space (Gille et al., 2011b).
A homolog of AXY4, a second putative XyG O-acetyltransferase termed AXY4L for AXY4like, was identified as acting on XyG specifically in seeds (Gille et al., 2011b). AXY4 and AXY4L are both members of a large, plant-specific protein family annotated as trichome birefringence-like (TBL; Bischoff et al., 2010a). In Arabidopsis this family includes 46 members (TBR, TBL1-45). Based on the data for AXY4/TBL27 and AXY4L/TBL22 also other members of the TBL family are proposed to encode additional wall polysaccharide specific O-acetyltransferases (Gille et al., 2011b). All TBL proteins consist of one N-terminal transmembrane domain and two highly conserved plant-specific domains; a TBL domain and a DUF231 domain (Bischoff et al., 2010a). Both domains harbor conserved motifs; a Gly-Asp-Ser (GDS) peptide in the TBL domain and an Asp-x-x-His (DxxH) motif in the C-terminal DUF231 domain (Figure 1). These two motifs were shown to be conserved in esterases and lipases, including a fungal rhamnogalacturonan acetylesterase and seem to infer catalytic activity (Molgaard et al., 2000; Akoh et al., 2004; Bischoff et al., 2010a,b). One of the two domains could be involved in binding a particular polymer while the other domain facilitates the binding of the acetyl-donor substrate and transfer of the acetate group to the polymer. This proposed mechanism might require an esterase-like activity in order to hydrolyze the acetate from the donor substrate (Gille et al., 2011b). Sequence comparisons of selected TBL family proteins to the O-acetylation systems described in the various organisms above (Figure 1) indicates that despite significant sequence diversity both motifs, the GDS and the DxxH motif, are present in the large loops of nearly all O-acetylation systems (Figure 2). Solely in the Pseudomonas alginate O-acetylation system only a GDS-motif can be found, but not the DxxH motif, neither in PaAlgJ nor PaAlgF.
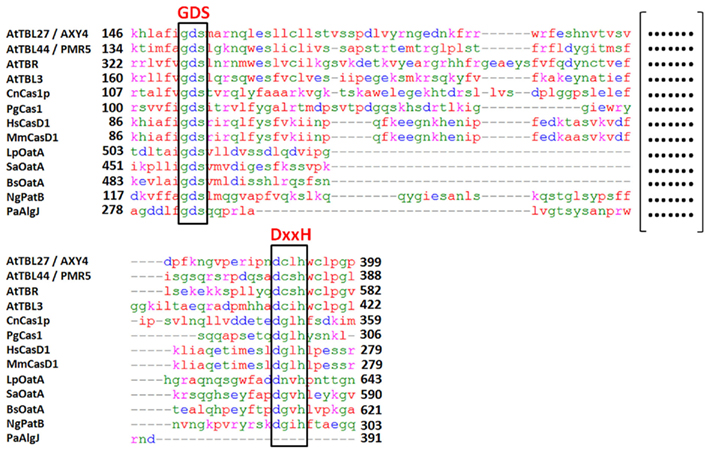
Figure 2. Protein sequence alignment of domains containing the GDS and DxxH motives of proteins displayed in Figure 1. Alignment was performed using Kalign 2.0 (Lassmann and Sonnhammer, 2005). (Format: ClustalW, Gap open penalty: 30, Gap extension penalty: 0.2, Terminal Gap penalties: 0.45, Bonus score: 0). Proteins were trimmed to the amino acid residues given in bold numbers for the alignment. At, Arabidopsis thaliana; Cn, Cryptococcus neoformans; Pg, Puccinia graminis; Hs, Homo sapiens; Mm, Mus musculus; Lp, Lactobacillus plantarum; Sa, Staphylococcus aureus; Bs, Bacillus subtilis; Ng, Neisseria gonorrhoeae; Pa, Pseudomonas aeruginosa. (At_AXY4 – AT2G70230, At_PMR5 – AT5G58600, At_TBR – AT5G06700, At_TBL3 – AT5G01360, Cn_Cas1p – AF355592, Pg_Cas1 – XP_003331718, Hs_Cas1 – NP_075051, Mm_Cas1 – NP_663373, Lp_OatA – NP_784589, Sa_OatA – ZP_06313146, Bs_OatA – NP_390592, Ng_PatB – YP_207683, Pa_AlgJ – AAB09782).
It thus appears that the O-acetylation mechanism is highly conserved among organisms and kingdoms. However, while in some organisms the responsible protein remains conserved as a single protein (mammals, fungi, Gram-positive bacteria) in other organisms this protein is split into two (Gram-negative bacteria, plants) or three proteins (Pseudomonas). Such a split allows the diversification of the proteins. For example, in plants there are four RWA and 46 TBL proteins perhaps reflecting the diversity of acetylated substrates and specificity in the various tissues and/or developmental stages. It has been proposed that the proteins harboring the multiple transmembrane domains such as PatA and AlgI are involved in translocation of an acetyl-donor molecule across the membrane (Franklin and Ohman, 2002), while the loop proteins PatB and AlgJ can be considered putative O-acetyltransferases that transfer the acetyl-group to the polysaccharide. By analogy in plants RWA would be an acetyl-donor translocator and the TBL proteins would represent the polysaccharide specific O-acetyltransferases (Gille et al., 2011b; Manabe et al., 2011). Analysis of the XyG O-acetylation status in an rwa/axy4 (tbl27) double mutant does not exclude the possibility that both proteins act in the same pathway (Gille et al., 2011b). In mammals and fungi both functions of translocation and transfer are combined into a single protein (Figure 1). Further research is needed to ascertain the function of the two protein domains (putative translocator and putative transferase) as well as the role of the two motifs (GDS and DxxH). Moreover, there might be other proteins involved in the O-acetylation mechanism. For example, knocking out all four RWA proteins in Arabidopsis in a RWA1/2/3/4 quadruple mutant does not eliminate wall polysaccharide O-acetylation but leads only to a 40% reduction in xylan acetylation in the stem (Lee et al., 2011) demonstrating that other hitherto unknown proteins can substitute for the loss of the presumed translocator RWA.
There are several pieces of evidence that O-acetylation of wall polysaccharides in plants occurs in the endomembrane system, most likely the Golgi apparatus, where non-cellulosic polysaccharides are synthesized (Moore et al., 1986; Scheible and Pauly, 2004). First, O-acetylated XyG can be isolated from intact microsomes of Arabidopsis confirming that O-acetylation occurs prior to deposition of the wall polysaccharide in the apoplast (Obel et al., 2009). Second, when microsomal preparations of a potato cell suspension cultures are fed with radio-labeled acetyl-CoA, the radio-labeled acetate can be found in an esterified form on several polysaccharides including XyG and pectin (Pauly and Scheller, 2000). Third, RWA and AXY4/TBL27 have been localized to the endomembrane system and their loop containing the GDS and DxxH motives are predicted to face the lumen (Gille et al., 2011b; Lee et al., 2011; Manabe et al., 2011).
The microsome experiments in plants indicate that acetyl-CoA is an acetyl-donor for the acetylation of wall polysaccharides. Other additional acetyl-donors or acetyl-intermediates cannot be excluded. Acetyl-CoA is also the donor substrate for acetylation of sialic acids in the mammalian system (Vandamme-Feldhaus and Schauer, 1998). The nature of the O-acetyl-donor in the other systems has not been identified but acetyl-CoA is also a possibility there (Franklin and Ohman, 2002; Laaberki et al., 2011). Is acetyl-CoA translocated to act as a donor substrate for the O-acetyltransferases? This seems unlikely in bacteria as acetyl-CoA would be translocated into the extracellular space and thus lost in the culture medium. It seems thus likely that the acetyl-group is transferred to an intermediate preferably one that remains tightly bound to the membrane, potentially the translocator protein itself. If one hypothesizes similar O-acetylation mechanisms in plants/human/fungi, where O-acetylation occurs in the Golgi, the possibility of translocation of acetyl-CoA seems also improbable. However, future experimental evidence is needed to elucidate how and in what form the acetyl-group is translocated and transferred to the polysaccharide.
Biological Functions of Plant Wall Polymer O-Acetylation
The biological function of O-acetyl-substituents in plant development and morphology is not known. However, surveys of O-acetylation in various plant tissues and during plant development indicate that the degree of O-acetylation on wall polymers is modified by the plant (Liners et al., 1994; Pauly et al., 2001; Obel et al., 2009) suggesting an important functional role in the plant. Since in vitro polymer experiments indicate that O-acetylation impacts the rheological properties and hinders enzymatic breakdown of the polysaccharide presumably through steric hindrance and/or conformational changes of the polymer (Biely et al., 1986), it is possible that fine-tuning the acetylation level of wall polymers impacts non-covalent interaction of wall polymers and hinders degradation of the polysaccharide by invading pathogens. Adding O-acetyl-substituents (C2-unit) to wall polysaccharides instead of additional pentosyl- or hexosyl-units (C5- and C6-units, respectively) does not only increase the structural complexity of the polysaccharide but it is also energetically favorable as less carbon is deposited in the wall. It seems such a strategy has been pursued in the grass XyG, where compared to the structure of the dicot XyG a specific xylosyl-residue has been regularly replaced with an O-acetyl-group (Gibeaut et al., 2005; Jia et al., 2005).
The recent discoveries of the first plant mutants affected in wall polysaccharide O-acetylation give more insights in the biological function of this substituent. The rwa2 single mutant exhibiting a 20% reduction in overall cell wall acetylation has a wild type like growth and morphology when grown under laboratory conditions (Manabe et al., 2011), while the RWA quadruple mutant exhibits collapsed xylem vessels (Lee et al., 2011). Consistent with observations made with mutants affected in the substitution pattern of XyG (Madson et al., 2003; Perrin et al., 2003; Cavalier et al., 2008; Günl et al., 2011) the axy4 mutant containing non-acetylated XyG exhibits no obvious growth or developmental phenotypes. In fact, the Arabidopsis ecotype Ty-0 containing only non-acetylated XyG due to point mutations in AXY4 apparently thrives in its habitat, Taynuilt in northern Scotland (Gille et al., 2011b). Apparently, the lack of XyG O-acetylation does not impact the fitness of the plant and does not present a selective disadvantage under the environmental conditions at this location. Interestingly, Ty-0 has been shown to be highly susceptible to the fungal pathogen Fusarium oxysporum (Diener and Ausubel, 2005). Although the mapping of one of the QTLs in this study led to a wall-associated kinase-like kinase 22 (WAKL22) there is a possibility that XyG O-acetylation may play a role in the susceptibility of this pathogen. Evidence for a potential role of wall O-acetylation in pathogen defense is provided by rwa2, which was shown to have an increased resistance toward the necrotrophic fungal pathogen Botrytis cinerea (Manabe et al., 2011). Moreover, if one assumes that all TBL proteins are involved in wall polysaccharide O-acetylation, a mutant in one of its members, PMR5, exhibits increased resistance to powdery mildew, Erysiphe spec (Vogel et al., 2004). Indeed, the cell wall of pmr5 exhibits a decrease in esterification as demonstrated by Fourier transform infrared (FTIR) analysis. If and which polysaccharide contains an alteration in O-acetylation has yet to be demonstrated in pmr5. Other TBL mutants also show significant phenotypes. The original mutant after which this class of genes was named, trichome birefringence (tbr), exhibits a defect in cellulose crystallinity in its trichomes (Potikha and Delmer, 1995; Bischoff et al., 2010a). The tbr-mutant and a mutant in its homolog TBL3 show alterations in their pectin esterification (Bischoff et al., 2010a). Another mutant affected in the expression of a gene in the TBL gene family is the eskimo1 mutant that displays strong freezing tolerance in the absence of cold acclimation (Xin et al., 2007). More detailed analysis of the mutant indicated collapsed xylem and alterations of the wall structure (Lefebvre et al., 2011). Although the precise nature of the wall structural change in eskimo1 has not been elucidated it is tempting to speculate that it could be O-acetylation of a particular polysaccharide. Bridging the gap between those observed mutant phenotypes and the proposed O-acetyltransferases will be vital in understanding the function of O-acetylation in the life cycle of plants.
Homologs of RWA and TBLs can be found in crop plants (Gille et al., 2011b) and the recent finding that these genes are involved in the O-acetylation of wall polysaccharides opens the door to identify or generate crop plants whose biomass contains lower amounts of acetate. This can be achieved through genetic engineering of repressing O-acetylation or marker assisted breeding. The resulting plant biomass should represent an improved feedstock for the emerging lignocellulosic based biofuel industry.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by Award OO0G01 from the Energy Biosciences Institute.
References
Akoh, C. C., Lee, G.-C., Liaw, Y.-C., Huang, T.-H., and Shaw, J.-F. (2004). GDSL family of serine esterases/lipases. Prog. Lipid Res. 43, 534–552.
Arming, S., Wipfler, D., Mayr, J., Merling, A., Vilas, U., Schauer, R., Schwartz-Albiez, R., and Vlasak, R. (2011). The human Cas1 protein: a sialic acid-specific O-acetyltransferase? Glycobiology 21, 553–564.
Aubry, C., Goulard, C., Nahori, M.-A., Cayet, N., Decalf, J., Sachse, M., Boneca, I. G., Cossart, P., and Dussurget, O. (2011). OatA, a peptidoglycan O-acetyltransferase involved in Listeria monocytogenes immune escape, is critical for virulence. J. Infect. Dis. 204, 731–740.
Bastawde, K. (1992). Xylan structure, microbial xylanases, and their mode of action. World J. Microbiol. Biotechnol. 8, 353–368.
Bera, A., Herbert, S., Jakob, A., Vollmer, W., and Götz, F. (2005). Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol. Microbiol. 55, 778–787.
Bernard, E., Rolain, T., Courtin, P., Guillot, A., Langella, P., Hols, P., and Chapot-Chartier, M.-P. (2011). Characterization of O-acetylation of N-acetylglucosamine: a novel structural variation of bacterial peptidoglycan. J. Biol. Chem. 286, 23950–23958.
Biely, P., Mackenzie, C., Puls, J., and Schneider, H. (1986). Cooperativity of esterases and xylanases in the enzymatic degradation of acetyl xylan. Biotechnology 4, 731–733.
Bischoff, V., Nita, S., Neumetzler, L., Schindelasch, D., Urbain, A., Eshed, R., Persson, S., Delmer, D., and Scheible, W. R. (2010a). Trichome birefringence and its homolog AT5G01360 encode plant-specific DUF231 proteins required for cellulose biosynthesis in Arabidopsis. Plant Physiol. 153, 590–602.
Bischoff, V., Selbig, J., and Scheible, W. R. (2010b). Involvement of TBL/DUF231 proteins into cell wall biology. Plant Signal. Behav. 5, 1057–1059.
Cavalier, D. M., Lerouxel, O., Neumetzler, L., Yamauchi, K., Reinecke, A., Freshour, G., Zabotina, O. A., Hahn, M. G., Burgert, I., Pauly, M., Raikhel, N. V., and Keegstra, K. (2008). Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. Plant Cell 20, 1519–1537.
Cherniak, R., and Sundstrom, J. (1994). Polysaccharide antigens of the capsule of Cryptococcus neoformans. Infect. Immun. 62, 1507–1512.
Clarke, A. J., and Dupont, C. (1992). O-acetylated peptidoglycan: its occurrence, pathobiological significance, and biosynthesis. Can. J. Microbiol. 38, 85–91.
Del Río, J. C., Marques, G., Rencoret, J., Martínez, Á. T., and Gutiérrez, A. (2007). Occurrence of naturally acetylated lignin units. J. Agric. Food Chem. 55, 5461–5468.
Del Río, J. C., Rencoret, J., Marques, G., Gutiérrez, A., Ibarra, D., Santos, J. I., Jiménez-Barbero, J., Zhang, L., and Martínez, Á. T. (2008). Highly acylated (acetylated and/or p-coumaroylated) native lignins from diverse herbaceous plants. J. Agric. Food Chem. 56, 9525–9534.
Diener, A. C., and Ausubel, F. M. (2005). Resistance to Fusarium oxysporum 1, a dominant Arabidopsis disease-resistance gene, is not race specific. Genetics 171, 305–321.
Franklin, M., and Ohman, D. (1993). Identification of algf in the alginate biosynthetic gene-cluster of Pseudomonas aeruginosa which is required for alginate acetylation. J. Bacteriol. 175, 5057–5065.
Franklin, M., and Ohman, D. (1996). Identification of algI and algJ in the Pseudomonas aeruginosa alginate biosynthetic gene cluster which are required for alginate O acetylation. J. Bacteriol. 178, 2186–2195.
Franklin, M. J., and Ohman, D. E. (2002). Mutant analysis and cellular localization of the AlgI, AlgJ, and AlgF proteins required for O acetylation of alginate in Pseudomonas aeruginosa. J. Bacteriol. 184, 3000–3007.
Gibeaut, D. M., Pauly, M., Bacic, A., and Fincher, G. B. (2005). Changes in cell wall polysaccharides in developing barley (Hordeum vulgare) coleoptiles. Planta 221, 729–738.
Gille, S., Cheng, K., Skinner, M. E., Liepman, A. H., Wilkerson, C. G., and Pauly, M. (2011a). Deep sequencing of voodoo lily (amorphophallus konjac): an approach to identify relevant genes involved in the synthesis of the hemicellulose glucomannan. Planta 234, 515–526.
Gille, S., de Souza, A., Xiong, G., Benz, M., Cheng, K., Schultink, A., Reca, I. B., and Pauly, M. (2011b). O-acetylation of Arabidopsis hemicellulose xyloglucan requires AXY4 or AXY4L, proteins with a TBL and DUF231 domain. Plant Cell 23, 4041–4053.
Günl, M., Neumetzler, L., Kraemer, F., de Souza, A., Schultink, A., Pena, M., York, W. S., and Pauly, M. (2011). AXY8 encodes a fucosidase, underpinning the importance of apoplastic metabolism on the fine structure of plant cell wall polysaccharides. Plant Cell 23, 4025–4040.
Helle, S., Cameron, D., Lam, J., and White, B. (2003). Effect of inhibitory compounds found in biomass hydrolysates on growth and xylose fermentation by a genetically engineered strain of S. cerevisiae. Enzyme Microb. Technol. 33, 786–792.
Hoffman, M., Jia, Z., Pena, M., Cash, M., Harper, A., Blackburn, A., Darvill, A., and YORK, W. (2005). Structural analysis of xyloglucans in the primary cell walls of plants in the subclass Asteridae. Carbohydr. Res. 340, 1826–1840.
Hsieh, Y. S. Y., and Harris, P. J. (2009). Xyloglucans of monocotyledons have diverse structures. Mol. Plant 2, 943–965.
Huang, L., Takahashi, R., Kobayashi, S., Kawase, T., and Nishinari, K. (2002). Gelation behavior of native and acetylated konjac glucomannan. Biomacromolecules 3, 1296–1303.
Ishii, T. (1991). Acetylation at O-2 of arabinofuranose residues in feruloylated arabinoxylan from bamboo shoot cell-walls. Phytochemistry 30, 2317–2320.
Ishii, T. (1997). O-acetylated oligosaccharides from pectins of potato tuber cell walls. Plant Physiol. 113, 1265–1272.
Janbon, G., Himmelreich, U., Moyrand, F., Improvisi, L., and Dromer, F. (2001). Cas1p is a membrane protein necessary for the O-acetylation of the Cryptococcus neoformans capsular polysaccharide. Mol. Microbiol. 42, 453–467.
Jia, Z., Cash, M., Darvill, A., and York, W. (2005). NMR characterization of endogenously O-acetylated oligosaccharides isolated from tomato (Lycopersicon esculentum) xyloglucan. Carbohydr. Res. 340, 1818–1825.
Kabel, M. A., de Waard, P., Schols, H. A., and Voragen, A. G. J. (2003). Location of O-acetyl substituents in xylo-oligosaccharides obtained from hydrothermally treated eucalyptus wood. Carbohydr. Res. 338, 69–77.
Kiefer, L., York, W., Darvill, A., and Albersheim, P. (1989). Structure of plant-cell walls.27. Xyloglucan isolated from suspension-cultured sycamore cell-walls is o-acetylated. Phytochemistry 28, 2105–2107.
Klein-Marcuschamer, D., Oleskowicz-Popiel, P., Simmons, B. A., and Blanch, H. W. (2010). Technoeconomic analysis of biofuels, A wiki-based platform for lignocellulosic biorefineries. Biomass Bioenergy 34, 1914–1921.
Laaberki, M. H., Pfeffer, J., Clarke, A. J., and Dworkin, J. (2011). O-acetylation of peptidoglycan is required for proper cell separation and s-layer anchoring in Bacillus anthracis. J. Biol. Chem. 286, 5278–5288.
Lassmann, T., and Sonnhammer, E. L. L. (2005). Kalign – an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics 6, 298. doi:10.1186/1471-2105-6-298
Lee, C., Teng, Q., Zhong, R., and Ye, Z.-H. (2011). The four Arabidopsis reduced wall acetylation genes are expressed in secondary wall-containing cells and required for the acetylation of xylan. Plant Cell Physiol. 52, 1289–1301.
Lefebvre, V., Fortabat, M. N., Ducamp, A., North, H. M., Maia-Grondard, A., Trouverie, J., Boursiac, Y., Mouille, G., and Durand-Tardif, M. (2011). ESKIMO1 disruption in Arabidopsis alters vascular tissue and impairs water transport. PLoS ONE 6, e16645. doi:10.1371/journal.pone.0016645
Liners, F., Gaspar, T., and Vancutsem, P. (1994). Acetyl-esterification and methyl-esterification of pectins of friable and compact sugar-beet calli – consequences for intercellular-adhesion. Planta 192, 545–556.
Lundqvist, J., Teleman, A., Junel, L., Zacchi, G., Dahlman, O., Tjerneld, F., and Stålbrand, H. (2002). Isolation and characterization of galactoglucomannan from spruce (Picea abies). Carbohydr. Polym. 48, 29–39.
Madson, M., Dunand, C., Li, X., Verma, R., Vanzin, G., Calplan, J., Shoue, D., Carpita, N., and Reiter, W. (2003). The MUR3 gene of Arabidopsis encodes a xyloglucan galactosyltransferase that is evolutionarily related to animal exostosins. Plant Cell 15, 1662–1670.
Manabe, Y., Nafisi, M., Verhertbruggen, Y., Orfila, C., Gille, S., Rautengarten, C., Cherk, C., Marcus, S. E., Somerville, S., Pauly, M., Knox, J. P., Sakuragi, Y., and Scheller, H. V. (2011). Loss-of-function mutation of reduced wall acetylation2 in Arabidopsis leads to reduced cell wall acetylation and increased resistance to Botrytis cinerea. Plant Physiol. 155, 1068–1078.
Molgaard, A., Kauppinen, S., and Larsen, S. (2000). Rhamnogalacturonan acetylesterase elucidates the structure and function of a new family of hydrolases. Structure 8, 373–383.
Moore, P., Darvill, A., Albersheim, P., and Staehelin, L. (1986). Immunogold localization of xyloglucan and rhamnogalacturonan-i in the cell-walls of suspension-cultured sycamore cells. Plant Physiol. 82, 787–794.
Moynihan, P. J., and Clarke, A. J. (2010). O-acetylation of peptidoglycan in Gram-negative bacteria: identification and characterization of peptidoglycan O-acetyltransferase in Neisseria gonorrhoeae. J. Biol. Chem. 285, 13264–13273.
Obel, N., Erben, V., Schwarz, T., Kuhnel, S., Fodor, A., and Pauly, M. (2009). Microanalysis of plant cell wall polysaccharides. Mol. Plant 2, 922–932.
O’Neill, M. A., Ishii, T., Albersheim, P., and Darvill, A. G. (2004). Rhamnogalacturonan II: structure and function of a borate cross-linked cell wall pectic polysaccharide. Annu. Rev. Plant Biol. 55, 109–139.
Pauly, M. (1999). Development of Analytical Tools to Study Plant Cell Wall Xyloglucan, Ph.D. thesis, Shaker Verlag, Aachen.
Pauly, M., Eberhard, S., Albersheim, P., Darvill, A., and York, W. (2001). Effects of the mur1 mutation on xyloglucans produced by suspension-cultured Arabidopsis thaliana cells. Planta 214, 67–74.
Pauly, M., and Keegstra, K. (2008). Cell-wall carbohydrates and their modification as a resource for biofuels. Plant J. 54, 559–568.
Pauly, M., and Keegstra, K. (2010). Plant cell wall polymers as precursors for biofuels. Curr. Opin. Plant Biol. 13, 304–311.
Pauly, M., and Scheller, H. (2000). O-acetylation of plant cell wall polysaccharides: identification and partial characterization of a rhamnogalacturonan O-acetyl-transferase from potato suspension-cultured cells. Planta 210, 659–667.
Perrin, R., Jia, Z., Wagner, T., O’Neill, M., Sarria, R., York, W., Raikhel, N., and Keegstra, K. (2003). Analysis of xyloglucan fucosylation in Arabidopsis. Plant Physiol. 132, 768–778.
Potikha, T., and Delmer, D. (1995). A mutant of Arabidopsis thaliana displaying altered patterns of cellulose deposition. Plant J. 7, 453–460.
Rombouts, F., and Thibault, J. (1986). Sugar-beet pectins – chemical structure and gelation through oxidative coupling. ACS Symp. Ser. 310, 49–60.
Scheible, W.-R., and Pauly, M. (2004). Glycosyltransferases and cell wall biosynthesis: novel players and insights. Curr. Opin. Plant Biol. 7, 285–295.
Schols, H., and Voragen, A. (1994). Hairy (ramified) regions of pectins occurrence of pectic hairy regions in various plant-cell wall materials and their degradability by rhamnogalacturonase. Carbohydr. Res. 256, 83–95.
Selig, M., Adney, W., and Himmel, M. (2009). The impact of cell wall acetylation on corn stover hydrolysis by cellulolytic and xylanolytic enzymes. Cellulose 16, 711–722.
Sengkhamparn, N., Bakx, E. J., Verhoef, R., Schols, H. A., Sajjaanantakul, T., and Voragen, A. G. J. (2009). Okra pectin contains an unusual substitution of its rhamnosyl residues with acetyl and alpha-linked galactosyl groups. Carbohydr. Res. 344, 1842–1851.
Shinabarger, D., May, T., Boyd, A., Ghosh, M., and Chakrabarty, A. (1993). Nucleotide-sequence and expression of the Pseudomonas aeruginosa algf gene controlling acetylation of alginate. Mol. Microbiol. 9, 1027–1035.
Teleman, A., Lundqvist, J., Tjerneld, F., Stålbrand, H., and Dahlman, O. (2000). Characterization of acetylated 4-O-methylglucuronoxylan isolated from aspen employing 1H and 13C NMR spectroscopy. Carbohydr. Res. 329, 807–815.
Van Dongen, F., Van Eylen, D., and Kabel, M. (2011). Characterization of substituents in xylans from corn cobs and stover. Carbohydr. Polym. 86, 722–731.
Vandamme-Feldhaus, V., and Schauer, R. (1998). Characterization of the enzymatic 7-O-acetylation of sialic acids and evidence for enzymatic O-acetyl migration from C-7 to C-9 in bovine submandibular gland. J. Biochem. 124, 111–121.
Vogel, J., Raab, T., Somerville, C., and Somerville, S. (2004). Mutations in PMR5 result in powdery mildew resistance and altered cell wall composition. Plant J. 40, 968–978.
Keywords: cell wall, polysaccharides, O-acetylation, acetyltransferase
Citation: Gille S and Pauly M (2012) O-acetylation of plant cell wall polysaccharides. Front. Plant Sci. 3:12. doi: 10.3389/fpls.2012.00012
Received: 13 December 2011;
Paper pending published: 30 December 2011;
Accepted: 12 January 2012;
Published online: 31 January 2012.
Edited by:
Jose Manuel Estevez, University of Buenos Aires and CONICET, ArgentinaCopyright: © 2012 Gille and Pauly. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Markus Pauly, Calvin Lab, Energy Biosciences Institute, University of California Berkeley, Berkeley, CA 94720, USA. e-mail:bXBhdWx5NjlAYmVya2VsZXkuZWR1