- 1 Department of Cell Biology and Molecular Genetics, Maryland Agricultural Experiment Station, University of Maryland, College Park, MD, USA
- 2 Departmento de Bioquímica, Biología Celular y Molecular de Plantas, Estación Experimental del Zaidín, Consejo Superior de Investigaciones Científicas, Granada, Spain
All organisms have evolved strategies to regulate ion and pH homeostasis in response to developmental and environmental cues. One strategy is mediated by monovalent cation–proton antiporters (CPA) that are classified in two superfamilies. Many CPA1 genes from bacteria, fungi, metazoa, and plants have been functionally characterized; though roles of plant CPA2 genes encoding K+-efflux antiporter (KEA) and cation/H+ exchanger (CHX) families are largely unknown. Phylogenetic analysis showed that three clades of the CPA1 Na+–H+ exchanger (NHX) family have been conserved from single-celled algae to Arabidopsis. These are (i) plasma membrane-bound SOS1/AtNHX7 that share ancestry with prokaryote NhaP, (ii) endosomal AtNHX5/6 that is part of the eukaryote Intracellular-NHE clade, and (iii) a vacuolar NHX clade (AtNHX1–4) specific to plants. Early diversification of KEA genes possibly from an ancestral cyanobacterium gene is suggested by three types seen in all plants. Intriguingly, CHX genes diversified from three to four members in one subclade of early land plants to 28 genes in eight subclades of Arabidopsis. Homologs from Spirogyra or Physcomitrella share high similarity with AtCHX20, suggesting that guard cell-specific AtCHX20 and its closest relatives are founders of the family, and pollen-expressed CHX genes appeared later in monocots and early eudicots. AtCHX proteins mediate K+ transport and pH homeostasis, and have been localized to intracellular and plasma membrane. Thus KEA genes are conserved from green algae to angiosperms, and their presence in red algae and secondary endosymbionts suggest a role in plastids. In contrast, AtNHX1–4 subtype evolved in plant cells to handle ion homeostasis of vacuoles. The great diversity of CHX genes in land plants compared to metazoa, fungi, or algae would imply a significant role of ion and pH homeostasis at dynamic endomembranes in the vegetative and reproductive success of flowering plants.
Introduction
Cells have evolved mechanisms to regulate ion and pH homeostasis in order to respond to developmental cues and to adapt to a constantly changing environment. In prokaryotes, this feat is accomplished with a diverse array of transporters at the plasma membrane. In eukaryotes, cells have increased in size and contain more intracellular membranes, including organelles and various compartments of the secretory and endocytic pathways. Consequently, the number of ion transporters localized to endomembranes has increased considerably. As plant ancestors initially living in an aquatic or marine environment colonized land, we wondered how transport systems required for growth, reproduction, and adaptation evolved in their structure and function. The genomes of diverse plants have been sequenced in recent years offering the first opportunity to understand how plant transporters evolved over one billion years. Here we focus on the monovalent cation/proton antiporter (CPA) superfamily whose members are thought to regulate cation and pH homeostasis (Brett et al., 2005a) by exchanging Na+, Li+, or K+ for H+. Prokaryote and eukaryote CPA members fall into two major families designated as CPA1 (2.A.36) and CPA2 (2.A.37; Saier, 2000; Brett et al., 2005a). In plants, CPA1 includes the well-studied Na+–H+ exchanger (NHX) family, and the relatively obscure CPA2 family that includes K+ efflux Antiporter (KEA) and Cation–H+ exchanger (CHX) families. The purpose of this study is to understand the evolutionary relationship of NHX, KEA, and CHX homologs from algae to flowering plants, to use comparative biology to infer functions of uncharacterized plant genes (Chang et al., 2004), and hopefully reveal new model systems to determine functions of KEA and CHX genes which remain largely uncharacterized.
Materials and Methods
Organisms
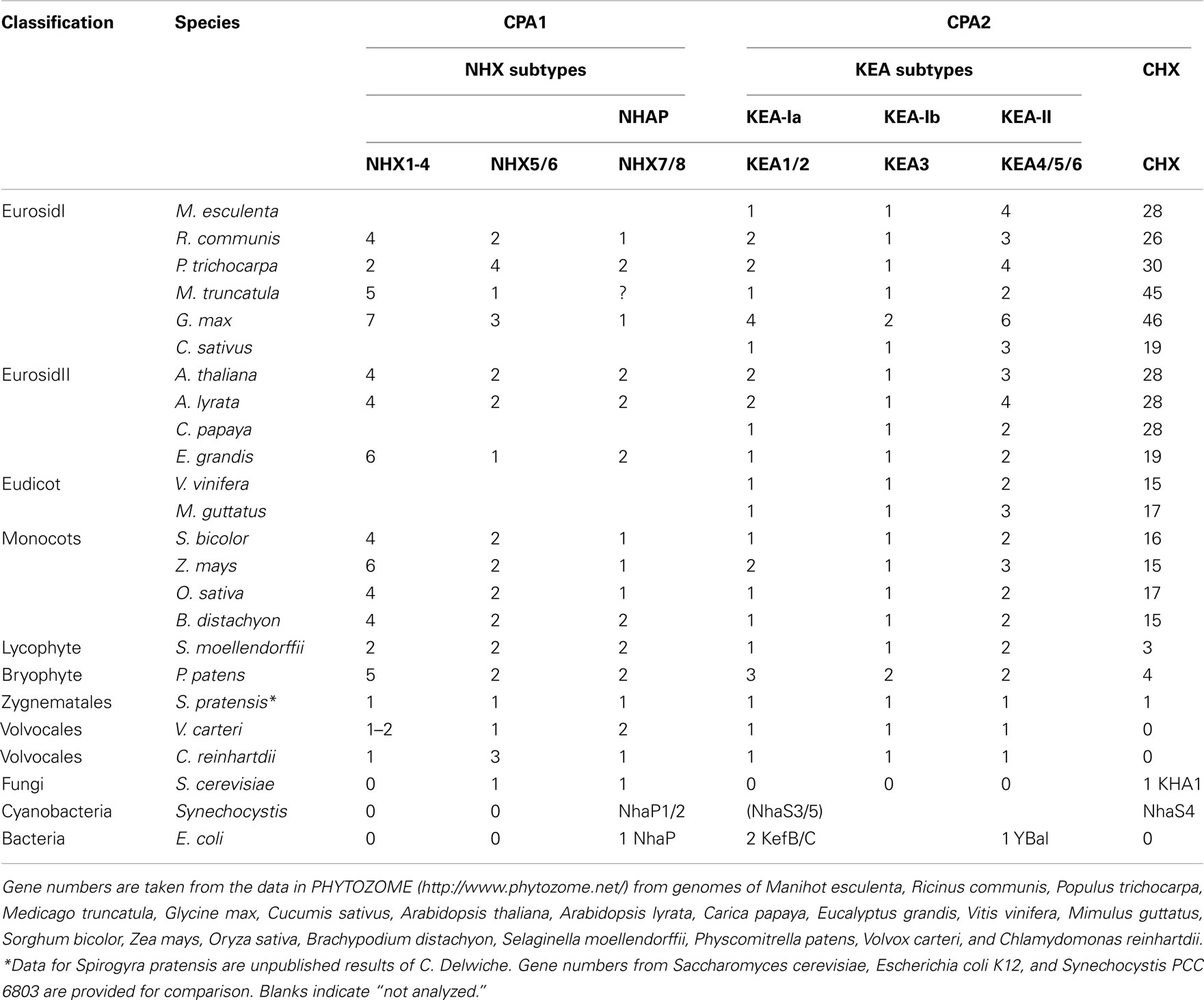
Plant genomes that have been completely sequenced and available in Phytozome were used. The list of plant species and their abbreviated names in parenthesis include Manihot esculenta (Mes), Ricinus communis (Rco), Populus trichocarpa (Ptr), Medicago truncatula (Mtr), Glycine max (Gma), Cucumis sativus (Csa), Arabidopsis thaliana (Ath), Arabidopsis lyrata (Aly), Carica papaya (Cpa), Eucalyptus grandis (Egr), Vitis vinifera (Vvi), Mimulus guttatus (Mgu), Sorghum bicolor (Sbi), Zea mays (Zma), Oryza sativa (Osa), Brachypodium distachyon (Bdi), Selaginella moellendorffii (Smo or club moss), Physcomitrella patens (Ppa or moss), Volvox carteri (Vca), and Chlamydomonas reinhartdii (Cre).
Collecting Sequences from Plants
Deduced protein sequences from diverse plants were initially collected from Phytozome1 by Blast analyses using NHX1, NHX5, NHX7, KEA1, KEA3, KEA4, or CHX17 from Arabidopsis thaliana. The sequences of each plant species were then compared with Arabidopsis protein by alignment with Clustal W or MUSCLE (Edgar, 2004) and analyzed with T-coffee (Notredame et al., 2000) or MEGA5 (Tamura et al., 2011). Abbreviated names were given to each gene according to the nomenclature used for A. thaliana (Maser et al., 2001). Blast analyses with Arabidopsis genes also recovered predicted proteins from an EST sequencing project of Charophyceae (Timme et al., 2012); though many transcripts were truncated and were not used in phylogenetic analysis. Other sequences from bacteria, cyanobacteria, fungi, protist, or metazoa were obtained by BLAST from JGI2 or NCBI3.
Alignment and Constructing Trees
RAxML
Sequences were assembled using Clustal X2 program (Larkin et al., 2007) to an original fasta file (fa.), and then aligned by Muscle software (Edgar, 2004). Alignment was exported and saved as fasta (afa), phylip (phy), or MSF format. RAxML was used for phylogenetic analysis and MSF was read by Genedoc (Nicholas et al., 19974) for editing. After alignment with Clustal X2 or MUSCLE, sequences that included the Pfam domain (00999) Na/H exchanger domain were retained for further analysis. Short sequences or those lacking >30% of the transmembrane domain of ~400 residues were dropped, though they resemble members of the family. When sequences outside the conserved domain diverged significantly, the N-terminus and C-tail were trimmed to maximize alignment (e.g., Figure 1). We used maximum-likelihood and tested phylogeny with a bootstrap of 100–500, JTT model (Jones et al., 1992), to construct trees with either RAxML (Stamatakis, 2006) or MEGA5 (Tamura et al., 2011). The bestTree file generated from the program gives the evolutionary value whereas the bipartition file informs the statistical value.
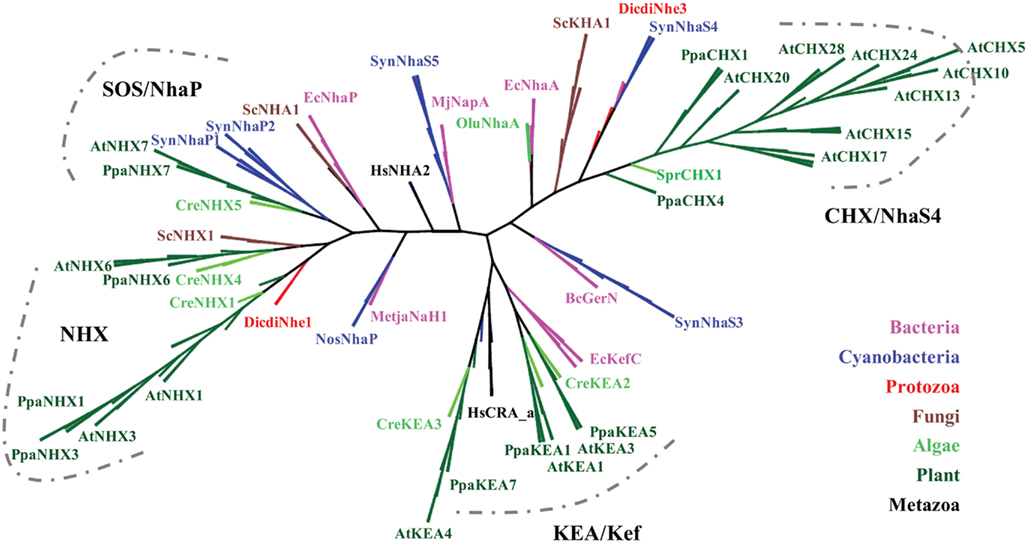
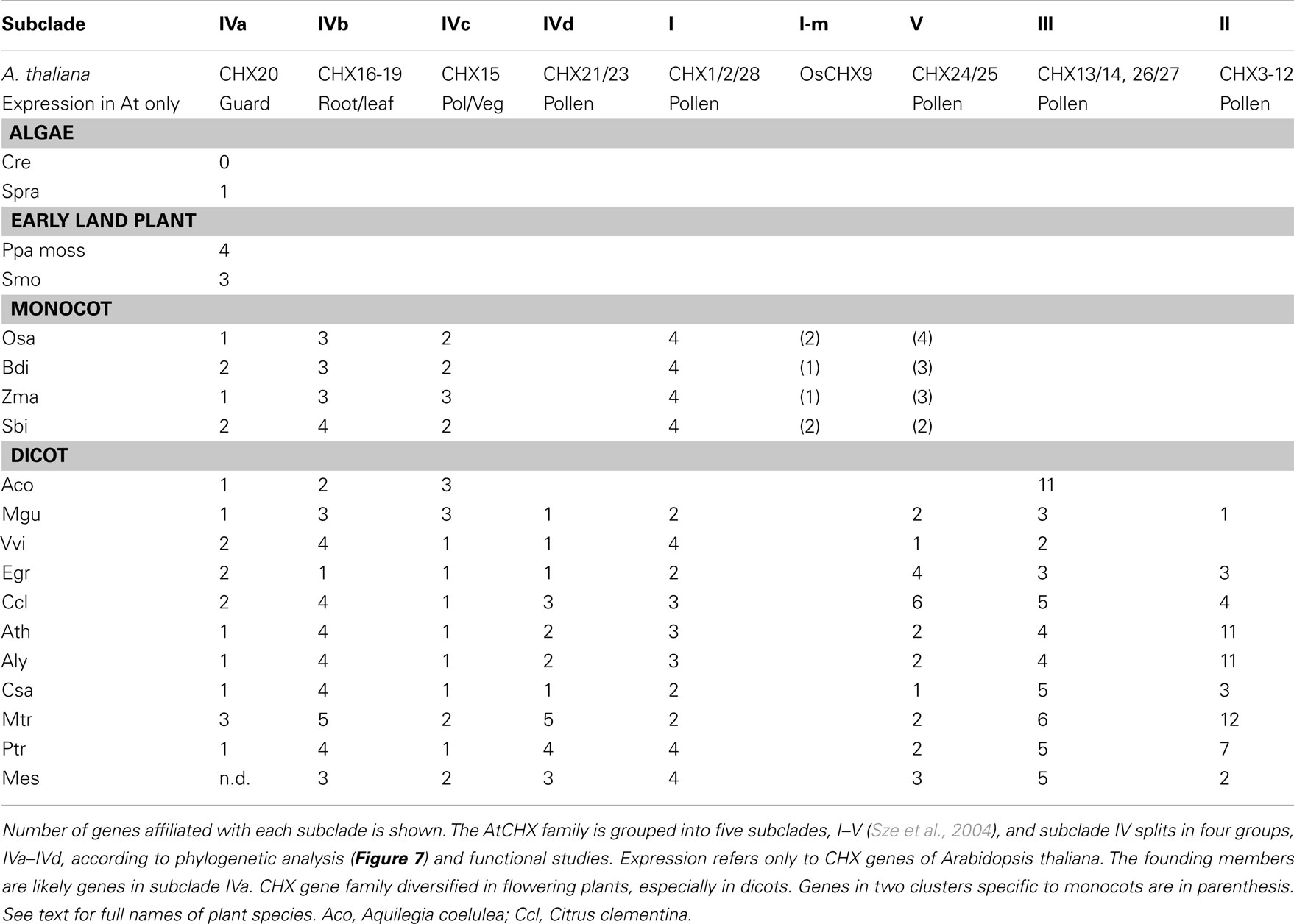
Figure 1. Protein phylogeny of cation–proton antiporters shows the evolutionary history of CPA1 and CPA2 families in plants. The conserved Pfam 00999 domains from diverse organisms were aligned by MUSCLE and the evolutionary history of the sequences were determined by RAxML, maximum likelihood using the Jones–Taylor–Thornton (JTT) model with a bootstrap of 500. (Final ML optimization likelihood = −104241.644676). Both N and C ends were trimmed, so aligned TM domain for proteins was ~400 residues, corresponding to AtCHX17 residue 33–426. Organisms are color-coded as follows: bacteria (magenta), Cyanobacteria (dark-blue), protist: Dictyostelium (red), fungi (brown), algae (green), and plants (dark green). NhaP and NhaA genes from ancestral bacteria likely gave rise to eukaryote CPA1 (NhaP and NHX) and CPA2 (KEA and CHX) families, respectively. Abbreviated protein names and organisms are defined in Table S1 in Supplementary Material. Unrooted rectangular tree is shown in Figure S1 in Supplementary Material.
Results
We first examined the evolutionary origin of plant cation–proton antiporters (CPA1 and CPA2) using Arabidopsis and rice as representatives of flowering plants. All these sequences had a conserved Na/H exchanger domain (Pfam00999) of about 400 residues. Figure 1 shows three distinct branches which are labeled as CPA1 (NHX and NHAP), CPA2 with KEA, and with CHX families.
Monovalent Cation Proton Antiporter-1
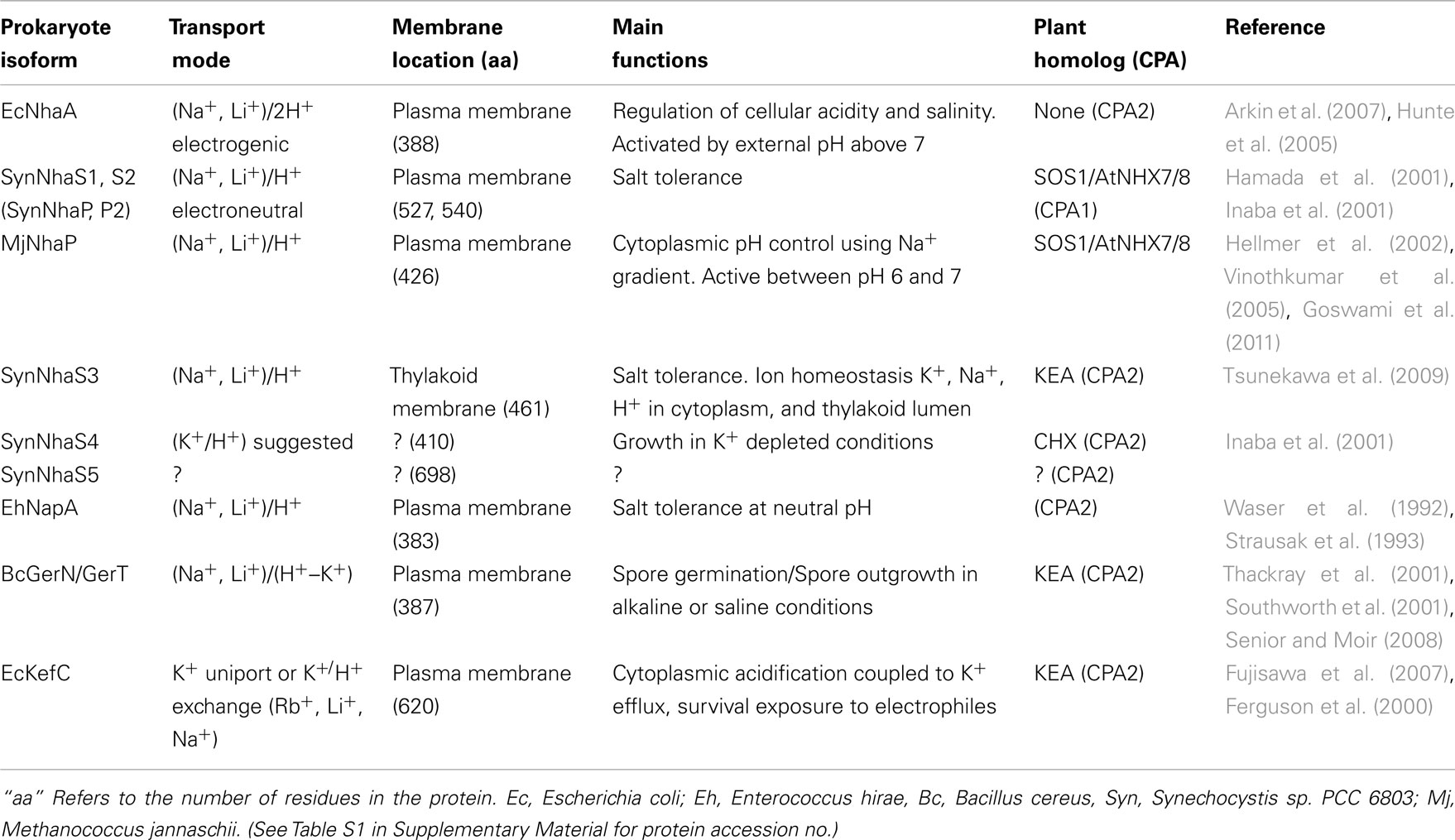
Several prokaryotic and archaea NhaP genes are located at the base of the CPA1 branch supporting the idea that an ancestral NhaP gene gave rise to eukaryote CPA1 genes (Figures 1 and 2). This family of transporters is predicted to have 10–12 membrane-spanning domains, and bacterial NhaP mediates electroneutral Na+/H+ exchange (see Table 1). CPA1-type transporters are found in all kingdoms, including archaea, bacteria, fungi, plants, and metazoa (Brett et al., 2005a). Our analysis shows that ancestral NhaP evolved and diverged to give two distinct types: NHX and NHAP (Figure 2).
Figure 2. Diversification of CPA1 genes into three clades preceded the evolution of early land plants. The C-tails were trimmed to maximize alignment, so AtNHX1 sequence included residues 1–448. Tree was generated with maximum likelihood of RAxML using the Jones–Taylor–Thornton (JTT) model and tested by bootstrap of 500. (Final ML optimization likelihood = −30244.238457). Single-celled alga (Cre), moss (Ppa), and club moss (Smo) NHX members were present in each of three clades, NhaP/SOS1, endosomal AtNHX5/6, and vacuolar AtNHX1–4. Organisms and proteins are defined in Table S2 in Supplementary Material, and color-coded as bacteria (magenta), Cyanobacteria (dark-blue), fungi (brown), protist (red), green algae (green), early land plants, and angiosperms (dark green). The “vacuolar” AtNHX1–4 clade is specific to plants.
NHAP/SOS1 clade
Phylogenetic analysis showed that plant members of Na+/H+ antiporter (NHAP) clade arose from ancestral NhaP1 and NhaP2 genes of a cyanobacterium (Figures 1 and 2). The best characterized plant NHAP is SOS1/AtNHX7 which mediates electroneutral Na+/H+ exchange (Qiu et al., 2003). Although AtSOS1 and AtNHX8 were annotated previously as NHX, our analysis places this group within NHAP clade consistent with results of Brett et al. (2005a). AtSOS1 and its homolog, AtNHX8, are localized to the plasma membrane (Qiu et al., 2003; An et al., 2007). Intriguingly, plant NHAP, such as SOS1/NHX7, has a particularly long C-terminal tail compared to other plant NHX. Recent studies show that this long C-tail interacts with other regulatory proteins (Katiyar-Agarwal et al., 2006; Quintero et al., 2011).
Arabidopsis NHAP or AtSOS1 has been extensively studied since the discovery that sos1 mutant is salt-overly sensitive (Wu et al., 1996; Quintero et al., 2002; Zhou et al., 2006; Fraile-Escanciano et al., 2010; see Table 2). SOS1 was shown to be associated with salt stress response (Shi et al., 2000, 2002) by mediating extrusion of Na+ (Qiu et al., 2003; Olias et al., 2009). It was shown that SOS1 had a physiological role in protecting plasma membrane K+ transporters from Na+ inhibition (Qi and Spalding, 2004). Furthermore, interaction of AtSOS1 with radical-induced cell death (RCD1) prevented RCD1, which is important for oxidative stress response, from nuclear localization (Katiyar-Agarwal et al., 2006). This relationship suggested a role of SOS1 in signal transduction. Analysis of sos1 mutant revealed the consequence of Na+ accumulation which affected membrane trafficking, vacuolar biogenesis, and pH homeostasis in roots (Oh et al., 2010). Interestingly, cation specificity for AtSOS1 and AtNHX8 is limited to Na+ and Li+ (Qiu et al., 2003; An et al., 2007) consistent with the substrate specificity of prokaryotic NhaP (Table 1). Results indicate that the major role of plant SOS1 transporters, like other NHAP members, is to extrude Na+ and confer tolerance to salt stress.
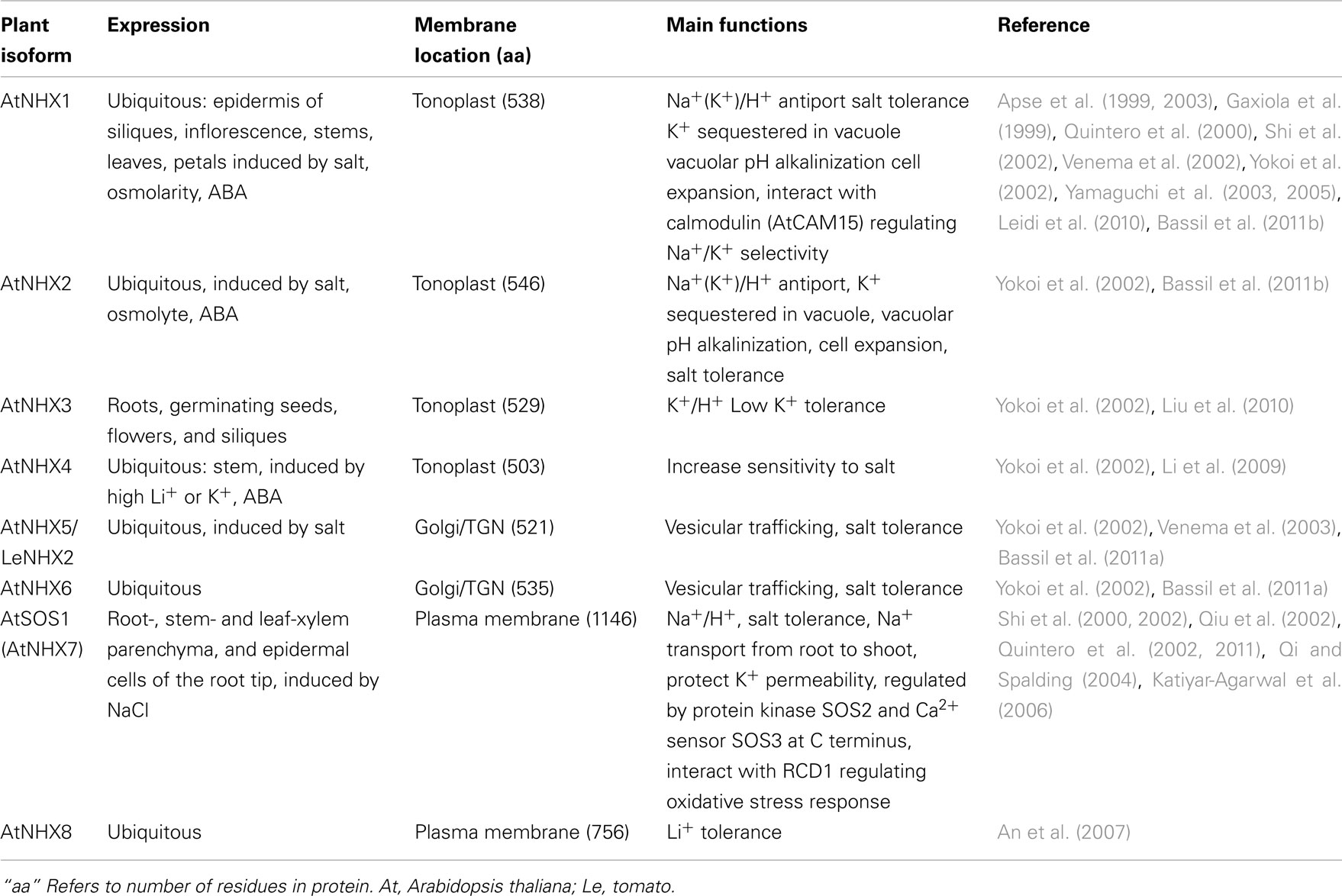
Two NHX clades
The diversification of NHAP to eukaryote intracellular-NHE (IC-NHE) and PM-NHE occurred early in evolution most likely when primitive eukaryote cells appeared (Brett et al., 2005a; Figure 1). Plant NHX members (excluding SOS1) belong to the IC-NHE family. NHX genes are present in single-celled algae like Chlamydomonas, early land plants, like Physcomitrella patens, as well as Arabidopsis thaliana (Figure 2). Furthermore, plant NHX has diversified into two clades that cluster either with Arabidopsis NHX5/6 or with AtNHX1–4. Consistent with the phylogenetic analysis, NHX proteins have been localized to intracellular membranes. In flowering plants, AtNHX5/6 is localized to Golgi or TGN, and AtNHX1 is at the vacuolar membrane (see Table 2 for references). The functions and localization of plant NHX and NhaP/SOS1 proteins are summarized in Table 2. These findings support the idea that NHX genes evolved as endomembranes appeared in early eukaryote and plant cells.
Initial studies showed that vacuolar NHX1 mediated electroneutral Na+/H+ exchange (Darley et al., 2000), though later studies broadened cation specificity to K+ in plants (Table 2). Vacuolar NHX was associated with salt stress responses and pH regulation (reviewed by Rodriguez-Rosales et al., 2009). InNHX1/2 of Japanese morning glory was shown to alkalinize vacuolar pH causing purple buds to turn into blue flowers (Yamaguchi et al., 2001; Ohnishi et al., 2005). The increase in vacuolar pH was accompanied by K+ accumulation indicating that NHX1 mediated K+/H+ exchange in Ipomea tricolor (Yoshida et al., 2009). Cation specificity was directly shown using proteoliposome where AtNHX1 catalyzed Na+/H+ and K+/H+ exchange with similar affinity (Venema et al., 2002). Moreover, silencing LeNHX2 resulted in tomato plants that accumulated less K+ and were susceptible to salt stress (Rodriguez-Rosales et al., 2008). In contrast, over-expression of AtNHX1 in tomato resulted in enhanced K+ accumulation and resistance to salt (Leidi et al., 2010). Finally, nhx1/nhx2 double mutants displayed reduced K+ concentration and pH in vacuoles, supporting a role of AtNHX1 and AtNHX2 in mediating H+ efflux coupled to K+ uptake. Thus regulating intravacuolar (K+) and pH are essential to cell expansion and flower development (Bassil et al., 2011b). There is increasing acceptance that plant NHX has a major role in pH and K+ homeostasis in plant cells where salt tolerance is conferred by enrichment of intracellular K+ pool and sequestration of Na+ in vacuole.
Interestingly, endosomal NHXs, such as LeNHX2, AtNHX5, and AtNHX6 were able to confer salt tolerance (Yokoi et al., 2002; Rodriguez-Rosales et al., 2008). Double mutants of nhx5/nhx6 were hypersensitive to salt (Bassil et al., 2011a). In previous studies of yeast, evidence emerged that ScNHX1 affected endosomal pH and K+ homeostasis that influenced protein sorting and membrane trafficking (Bowers et al., 2000; Ali et al., 2004; Brett et al., 2005b; Wagner et al., 2006). A recent study with plants suggested that regulation of endosomal pH and K+ homeostasis by AtNHX5 and AtNHX6 had a role in sorting of vacuolar protein and cellular stress responses (Bassil et al., 2011a), consistent with the phylogenetic analysis showing ScNHX1 and AtNHX5/6 are in the same clade (Figures 1 and 2).
Curiously, budding yeast (and probably most fungi) has only one endosomal NHX1 localized to endosome/prevacuolar compartment (PVC), and does not have genes related to “plant vacuolar-type” AtNHX1–4. However, Dictyostelium Nhe1 and Nhe2 are at the base of the plant vacuolar NHX clade (Figure 2), suggesting plant vacuolar NHX arose from a gene in an ancestral protozoa possibly with contractile vacuoles for osmoregulation (Allen and Naitoh, 2002; Gerisch et al., 2002; Uchikawa et al., 2011). The plant-specific AtNHX1–4 type has apparently undergone gene duplications (Figure 2) underscoring the significance and diversity of vacuole functions in multicellular land plants.
Monovalent Cation Proton Antiporter-2
Members of CPA2 superfamily are found in bacteria, fungi, and plants; however, they are rare in metazoa (Brett et al., 2005a). They are predicted to have 8–14 membrane-spanning domains with a Pfam00999 domain for Na+(K+)/H+ exchange. Interestingly, substrate and transport mode of CPA2 from bacteria vary among family members. The general transport mode of CPA2 is 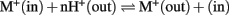 for the carrier-mediated mode, whereas some members may catalyze
for the carrier-mediated mode, whereas some members may catalyze 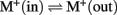 in the channel-mediated mode (Saier, 2000). The CPA2 superfamily consists of three families named here as NHA/NhaA, KEA/Kef, and CHX/NhaS4 based on phylogenetic analyses and prokaryote ancestry.
in the channel-mediated mode (Saier, 2000). The CPA2 superfamily consists of three families named here as NHA/NhaA, KEA/Kef, and CHX/NhaS4 based on phylogenetic analyses and prokaryote ancestry.
NHA clade
A eukaryote NHA clade was previously classified within CPA2 (Brett et al., 2005a), though they seem related to the NHAP clade in CPA1 (Figure 1). Eukaryote NHA, like NhaP, is localized at the plasma membrane (Kinclova et al., 2001). Functional studies of yeast show that plasma membrane ScNHA1 mediates both Na+/H+ and K+/H+ exchange (Banuelos et al., 1998; Sychrova et al., 1999; Ohgaki et al., 2005). Human NHA2 is localized to the PM and its expression in yeast conferred tolerance to Na+ and Li+, but not to K+ (Xiang et al., 2007), suggesting it mediates Na+ transport similar to NhaP or NHX of CPA1. Our phylogenetic analysis suggests that eukaryote NHA is likely evolved from an NhaP gene of an ancestral bacterium (Figure 1). Unlike NhaP, E. coli NhaA is active at pH 8 and inactive at pH 6 (Taglicht et al., 1991; see Table 1). EcNhaA mediated exchange of 2H+ for 1Na+ or 1Li+ (Padan, 2008). Intriguingly, only tiny marine green alga, such as Ostreococcus lucimarinus and Micromonas pusilla (Chlorophyta) have a protein homologous to bacterial NhaA (Figure 1; Table S1 in Supplementary Material). NHA members are thus found in bacteria, animals, and fungi (Brett et al., 2005a), but not in early land plants or flowering plants (Figure 1).
KEA family
Genes encoding putative KEA from higher plants were first classified in Arabidopsis thaliana (Maser et al., 2001); thus AtKEA1–AtKEA6 members are used as a reference. KEA-like transporters from bacteria have been extensively studied, and are characterized by an N-terminal Na_H exchanger domain (Pfam PF00999), and a C-terminal KTN NAD(H)-binding domain, also called TrkA_N domain (Pfam PF02254; Figure 4). For example, Kch, TrkA, Ybal, and KefB or KefC are K+ channels or transporters with a C-terminal KTN domain (Choe, 2002). The minimal functional unit of the KTN domain is a dimeric molecule connected by a flexible hinge. Hinge motion can be physically coupled to transmembrane loops to control K+ flux, providing a mechanism of gating control (Choe, 2002). The domain contains the typical Rossman fold GXGXXG…D Glycine motif involved in NAD binding, indicating metabolic control of transporter function (Jiang et al., 2001; Roosild et al., 2002).
Divergence of two KEA clades. To understand the evolutionary origin of higher plant KEA, we searched and analyzed homologs in prokaryotes, protists, metazoa, and plants. AtKEA genes diverged into two main branches with AtKEA1–3 in clade I and AtKEA4–6 in clade II. Clade I consists of the only eukaryote-encoded proteins with a complete C-terminal KTN domain (Figures 3 and 4), and are closely related to EcKefB and EcKefC-like proteins from bacteria, including cyanobacterium Gloeobacter violaceus (Figure 3). Red algae as well as secondary photosynthetic organisms of the Kingdom Chromista contain sequences from clade I exclusively (Figure 3; Table S3 in Supplementary Material). In contrast, proteins of clade II lack a KTN domain (Figures 3, 4, and 6; Table S3 in Supplementary Material). Phylogenetic analysis suggests AtKEA4–6 arose from an ancestral cyanobacterium judging by the presence of related proteins in unicellular and filamentous cyanobacteria (Figure 3). Clade II genes are also present in parasitic organisms of the phylum Apicomplexa. These organisms contain a high number of plant and bacterial-like proteins (Abrahamsen et al., 2004) possibly due to apicoplast (plastid) resulting from secondary endosymbiont of an ancestral green or red alga (Keeling, 2004). A clade II protein is found in the oomycete, Phytophthora infestans, possibly also related to the presence of a plastid in a common photosynthetic ancestor of the chromista (Tyler et al., 2006). Oddly, AtKEA4–6 like proteins are also found in Trichoplax adhaerens (a Placozoa), Ciona intestinalis, a basal Chordata, and in higher Chordata, such as mammals, amphibians, and fish (Figure 3, Table S3 in Supplementary Material). These AtKEA4–6 like proteins contain a trans membrane coiled coil protein 3 (TMCO3) and also lack the KTN domain.
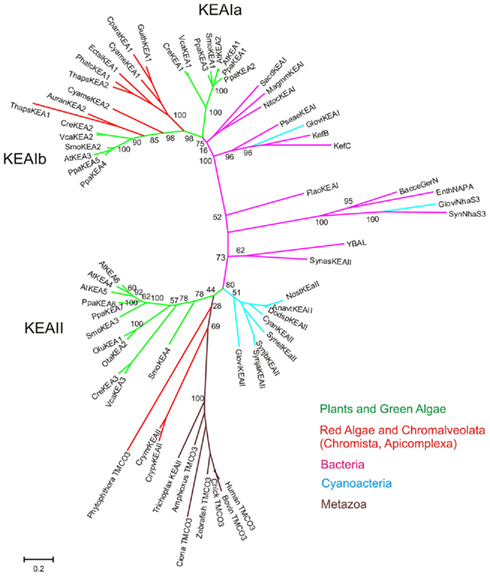
Figure 3. Phylogenetic tree shows plant KEAs originated from ancestral genes in Cyanobacteria. The conserved Pfam 00999 domains in KEA sequences from diverse organisms were aligned by MUSCLE and the evolutionary history of the genes were determined by maximum likelihood using the Jones–Taylor–Thornton (JTT) model with a Bootstrap of 100. The percentage of trees in which the associated sequences clustered together is shown next to the branches. The analysis involved 66 amino acid sequences. All positions containing gaps and missing data were eliminated, resulting in a total of 307 positions in the final dataset. Branch colors refer to bacteria (pink), Cyanobacteria (blue), red alga (red), metazoa (brown), and plants (green). Organisms and accession numbers of protein sequences are listed in Table S3 in Supplementary Material.

Figure 4. Alignment of AtKEA sequences with their bacterial homologs – AtKEA sequences were aligned using MUSCLE (Edgar, 2004) with their closest prokaryotic homologs: E. coli KefC and Cyanobacterial sequence SynjbKEAII (Q2JI45 Table S3 in Supplementary Material). The soluble N-terminal domain of AtKEA1 was removed. Conserved residues are shown in orange. Approximate positions of possible transmembrane helices are shown above the alignment. Secondary structure of the KTN domain is indicated as well as residues that were shown to be involved in Glutathione-binding in the KefC KTN structure. A short cytoplasmic regulatory loop in the E. coli KefC sequence as well as the conserved residues in the Arabidopsis sequences are shown in magenta. The residues corresponding to the proton binding residues in the structure of the bacterial NhaA sequence are shown in red. The Rossman fold glycine motif is shown in green. Below the alignment the two specific clade Ib inserts in different plants are shown.
These results would suggest that KEA genes were acquired by primary endosymbiosis of a cyanobacterium, and by secondary endosymbiosis in the Archaeplastida ancestor of red and green algae (Adl et al., 2005; Bowman et al., 2007). Furthermore, it appears the divergence of clade I and clade II KEAs occurred early as prokaryotes diversified perhaps before eukaryote cells evolved.
Intriguingly, KEA genes from clade I are found in a red alga (C merolae) and in chromista (Figure 3). Chromista possibly became photosynthetic by endosymbiosis with a unicellular red alga, as genes related to KEA sequences in C. merolae are also present in these species. In chromista of the cryptomad subgroup, a KEA gene constitutes one of three transport genes that are maintained in the highly reduced nucleomorph genome (Douglas et al., 2001). Nucleomorphs are the remnant nuclei of algal endosymbionts that were engulfed by non-photosynthetic eukaryotes (Tanifuji et al., 2011). This observation suggests that KEAs play important roles in these organisms, such as ion transport across one of four chloroplast membranes. Its role is apparently not in photosynthesis, as KEA-like gene is conserved in the nucleomorph of a non-photosynthetic Cryptomonas paramecium, but not in nucleomorphs of green algal origin (Tanifuji et al., 2011).
Evolution of KEA in plants. Protein phylogenetic analyses showed that all plants, from unicellular algae to flowering plants, had three types of KEAs classified as clades Ia, Ib, and II (Figure 5). Green algae, like C. reinhartdii and V. carteri, contain only one gene in each group. In other plants, diversification occurred in clades KEA-Ia, KEA-II, or both.
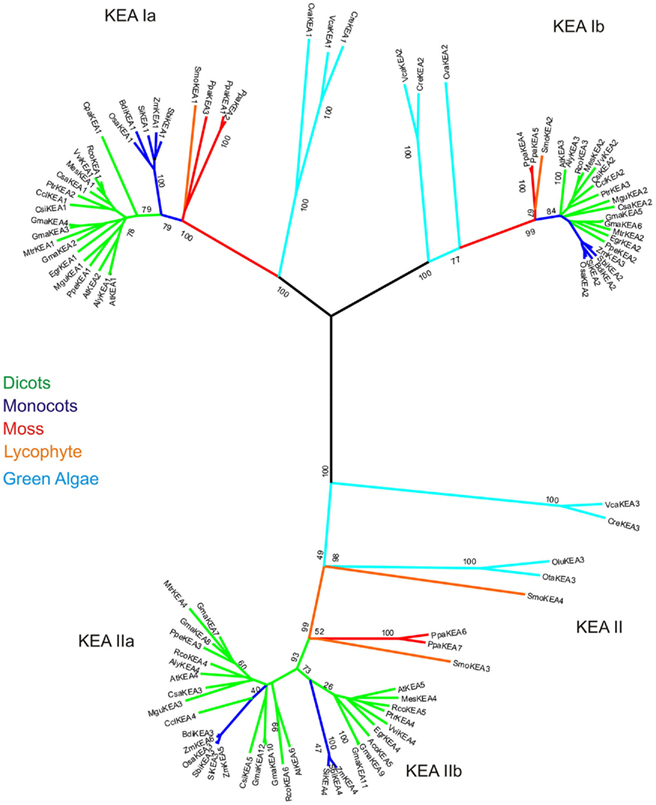
Figure 5. K+-efflux antiporter diversified into three types in early photosynthetic eukaryotes. The analysis involved 97 amino acid sequences. The tree includes all proteins found in A. thaliana, G. max, O. sativa, Z. mays, S. moellendorffii, P. patens, C. reinhardtii, and V. carteri, as well as one representative sequence for each group from the remaining plant species listed in Table S3 in Supplementary Material. All positions containing gaps and missing data were eliminated. There were a total of 152 (335 excluding incomplete sequences GmaKEA11 and RcoKEA6) positions in the final dataset. Evolutionary analyses were conducted in MEGA5 (Tamura et al., 2011). Branch colors refer to dicots (green), monocot (dark-blue), early land plants (orange–red), and green algae (light blue). Species and accession numbers of protein sequences are listed in Table S3 in Supplementary Material.
Clade Ia KEAs possess a long N-terminal domain. Phylogenetic analysis showed that plant KEAs in clade I diverged into two types that differ in their protein organization. Within clade Ia, all plant sequences from moss to monocot and dicots have an long hydrophilic N-terminal domain of ~570–770 amino acids (Figure 6) that is often missed in preliminary gene annotations. This domain is typically rich in negatively charged glutamate residues, and predicted coiled coil structures (Figure S4 in Supplementary Material). The role of this unusual extra sequence is unknown (Rose et al., 2004). The N-terminal domain of green algae Cre and Cva share some similarity (46%) and are slightly shorter than that of higher plants (Figure 6). The absence of this region in prokaryotes would indicate that the N-terminal domain is a recent acquisition that gained increasing relevance in land plants.
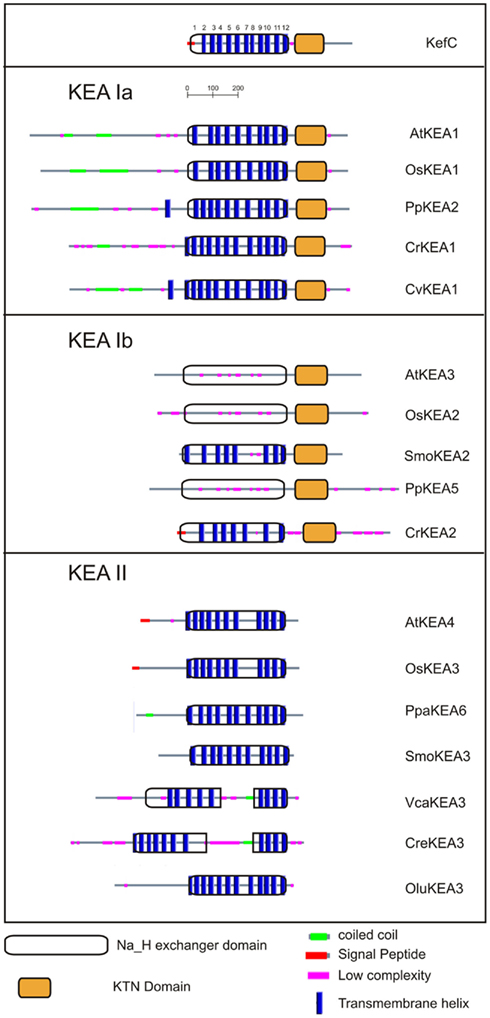
Figure 6. Distinct protein domains of three KEA clades. The relative length and position of the KTN and Na_H exchanger domain in the three KEA clades is shown graphically using information obtained with the Simple Modular Architecture Research Tool (Smart, http://smart.embl-heidelberg.de/). All plant clade I proteins contain a KTN domain. In the clade II group this domain is lost, making it the shortest KEA proteins. The clade Ia proteins all have acquired a long N-terminal domain, typically containing predicted coiled coil structures. The C-terminus of the clade Ib proteins is slightly longer as compared to the other sequences. Some green algal clade II proteins have a split Na_H domain, which might be a result of erroneous gene predictions. The transmembrane helix predictions are very variable, and especially unsuccessful for the clade Ib group of proteins. The transmembrane helices shown in Figure 2 are numbered from 1 to 12 in KefC.
In contrast, proteins in clade Ib, such as AtKEA3, do not harbor the N-terminal extension, instead they have gained specific insertions between predicted membrane helices 4 and 5 and in the Rossman fold glycine motif of the KTN domain (Figures 4 and 6). Although all hydrophobic regions are conserved in the clade Ib proteins, transmembrane helices are poorly identified by transmembrane prediction programs (Figure 6). In general, only one gene of this type is present per plant, except in G. max and P. patens, where two genes are present (Table 4; Table S3 in Supplementary Material; Figure 5).
Recent diversification of clade II KEAs. Genes encoding AtKEA4–6 have a hydrophilic N-terminal domain of ~157–235 residues that is predicted to include a signal peptide (Figures 4 and 6). Green algal sequences found in V. carteri and C. reinhartdii have an anomalous Na_H exchanger domain that is interrupted by a similar sequence in both species (Figure 5). Interestingly, clade KEA-II has diversified further in angiosperms where AtKEA5 is clearly separate from AtKEA4 and AtKEA6 indicating this occurred recently after the evolution of early land plants.
Function of KEA. Nearly nothing is known about plant KEA function, so bacterial homologs could provide insights. KefB and KefC in E. coli are glutathione regulated K+ efflux transporters, that are important for bacterial survival during exposure to toxic metabolites (Booth, 2003). Electrophiles react with glutathione to form glutathione adducts, thereby releasing KefC and KefB inhibition by GSH, and eliciting K+ efflux and H+ uptake. Cytoplasmic acidification is suggested to prevent DNA damage (Ferguson et al., 2000; Miller et al., 2000). Reduced GSH stabilizes the association of the two KTN domains, whilst the larger adducts disrupt this interaction (Roosild et al., 2010). Based on mutagenesis studies, a short central hydrophilic regulatory loop was proposed to interact with the KTN domain. This regulatory loop is partially conserved in the plant KEA1–3 sequences, but the residues forming the glutathione-binding pocket are less conserved in the plant sequences (Figure 2), indicating that plant KEA1–3 are regulated in a different way (Roosild et al., 2010). Although originally proposed to function as K+ channels, recent studies indicate KefC has K+/H+ antiport activity (Fujisawa et al., 2007).
Recently, AtKEA1 and AtKEA3 peptides were detected by mass spectroscopy in Arabidopsis chloroplast preparations (Zybailov et al., 2008). Moreover, localization to thylakoid membrane of SynNha3 (Tsunekawa et al., 2009), a Synechocystis protein closely related to KEA-I and KEA-II proteins (Figure 1), would suggest a role of plant KEA in chloroplast ion homeostasis. So far only one cyanobacterium, Gloeobacter violaceus, has a gene encoding a KEA-I homolog (Figure 3). Perhaps ancestral primary plastids originated just after the separation of primitive cyanobacterium, like Gloeobacter, from present-day lineages (Criscuolo and Gribaldo, 2011). In Gloeobacter, thylakoids are absent and photosystems reside at the plasma membrane (Vothknecht and Westhoff, 2001). We hypothesize that the long N-terminal sequence of AtKEA1/2 has a role in chloroplast-specific processes in plant (Vothknecht and Westhoff, 2001). These and other possibilities need to be tested.
Evolutionary origin of CHX clade in plants
The CHX clade is populated by genes from higher plants, but none from metazoa so far. Nearly all deduced proteins from plants have ~800 residues (Table S4 in Supplementary Material) that consist of a hydrophobic Na+/H+ exchanger domain of ~400 residues at the amino terminus, and a carboxyl hydrophilic domain of 300–400 residues. The long hydrophilic C-tail does not contain any known conserved motifs, so its role is a complete mystery. NhaS4 from the cyanobacterium Synechocystis PCC 6803 encodes a much shorter protein of 410 residues, though the predicted protein from Synechococcus elongatus PCC 6301 has 715 residues.
Based on analyses using the TM domain alone, we show that the present-day NhaS4 from cyanobacteria, shares the highest similarity with AtCHX16–CHX20 (Figure 1; Sze et al., 2004). NhaS4 gene is found in single-celled Synechocystis PCC 6803 (Inaba et al., 2001) as well as filamentous Anabaena variabilis (Ava1632). Many non-cyanobacteria do not have genes that cluster with the CHX/NhaS4 clade. One exception is MyxxaNhaS4 (Table S1 in Supplementary Material) from the gliding bacterium Myxococcus xanthus (a member of Myxococcales) which form fruiting body in response to nutrient starvation (Mauriello et al., 2010). Intriguingly, the mechanisms of motility and establishment of cell polarity in M. xanthus and eukaryotes share several similarities (Leonardy et al., 2010). Although genes from metazoa are absent from this clade, Dictyostelium discoideum (a protist and slime mold) has two members, Nhe3 and Nhe4, of unknown function that cluster with NhaS4 (Figure 1). The function and membrane location of cyanobacterial NhaS4 are still unclear. Interestingly, all fungi examined have a single gene KHA1 that encodes a homolog of Synechocystis NhaS4 as seen in Figure 1. Fungi include unicellular Saccharomyces cerevisiae, and filamentous fungi such as Neurospora crassa, Aspergillus oryzae, and Ustilago maydis (Figure 1; Table S1 in Supplementary Material). Mutants of ScKHA1 have served as a useful heterologous system to test functions of Arabidopsis CHX16–20 (Chanroj et al., 2011; Table 3).
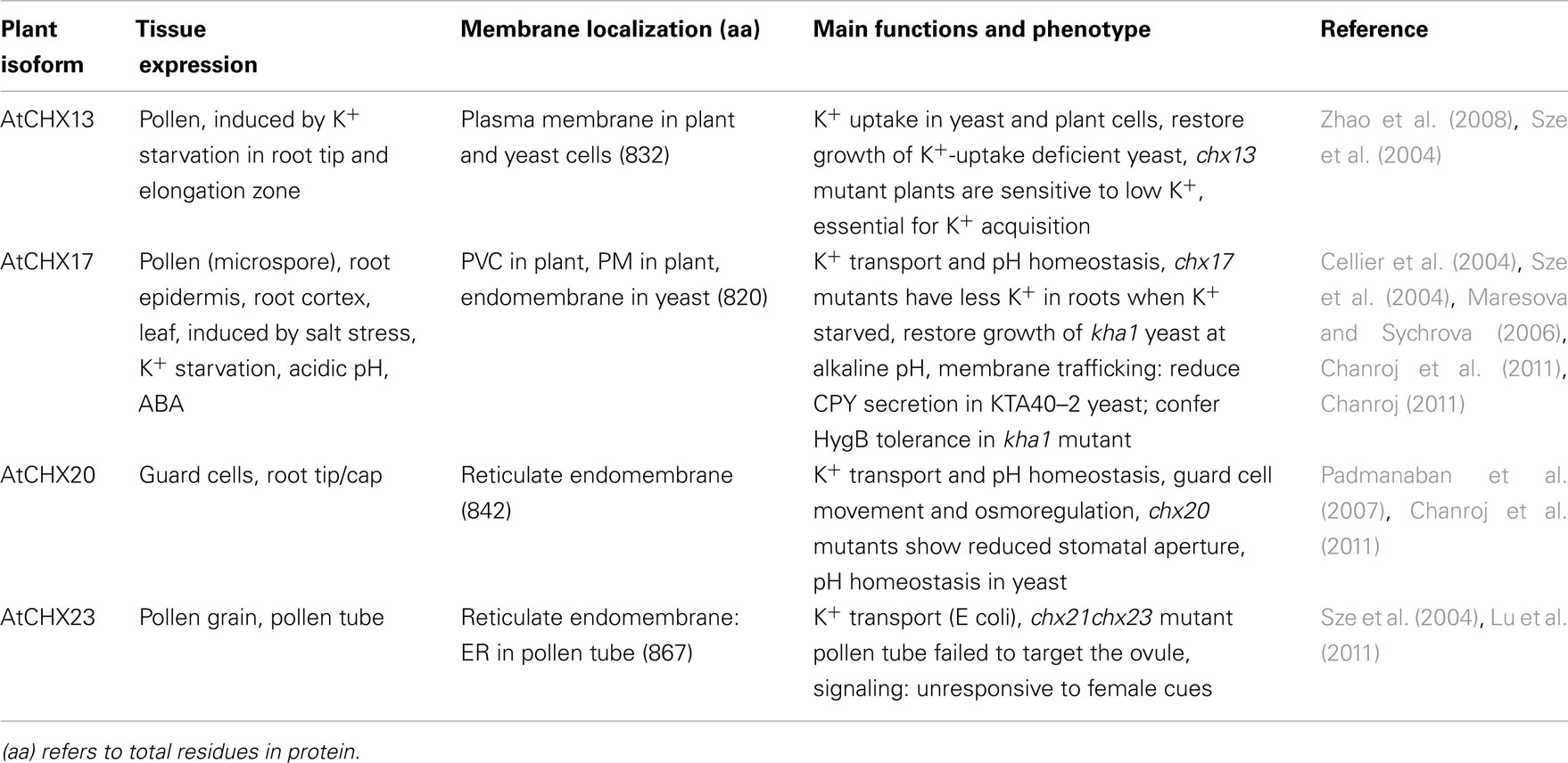
Comparative analyses of full-length protein sequences revealed additional insights on the origin of plant CHX members. One CHX homolog was found in Spirogyra pratensis, a Zygnematales, though none was detected in Chlamydomonas reinhartdii, Volvox carteri (Figures 1 and 7), or red algae (not shown). Early land plants, like Physcomitrella and Selaginella, had four and three genes, respectively encoding CHX homologs (Figure 1). Interestingly, all these proteins cluster in one subclade closest to Arabidopsis CHX20 (Figure 7) and CHX16–19. Results suggest that AtCHX16–CHX20 genes arose early in CHX gene family diversification. In Arabidopsis, these genes are expressed in leaves and roots, especially in dermal tissues, such as root epidermis (Cellier et al., 2004; Chanroj, 2011), root cap, and guard cells (Padmanaban et al., 2007; see Table 3).
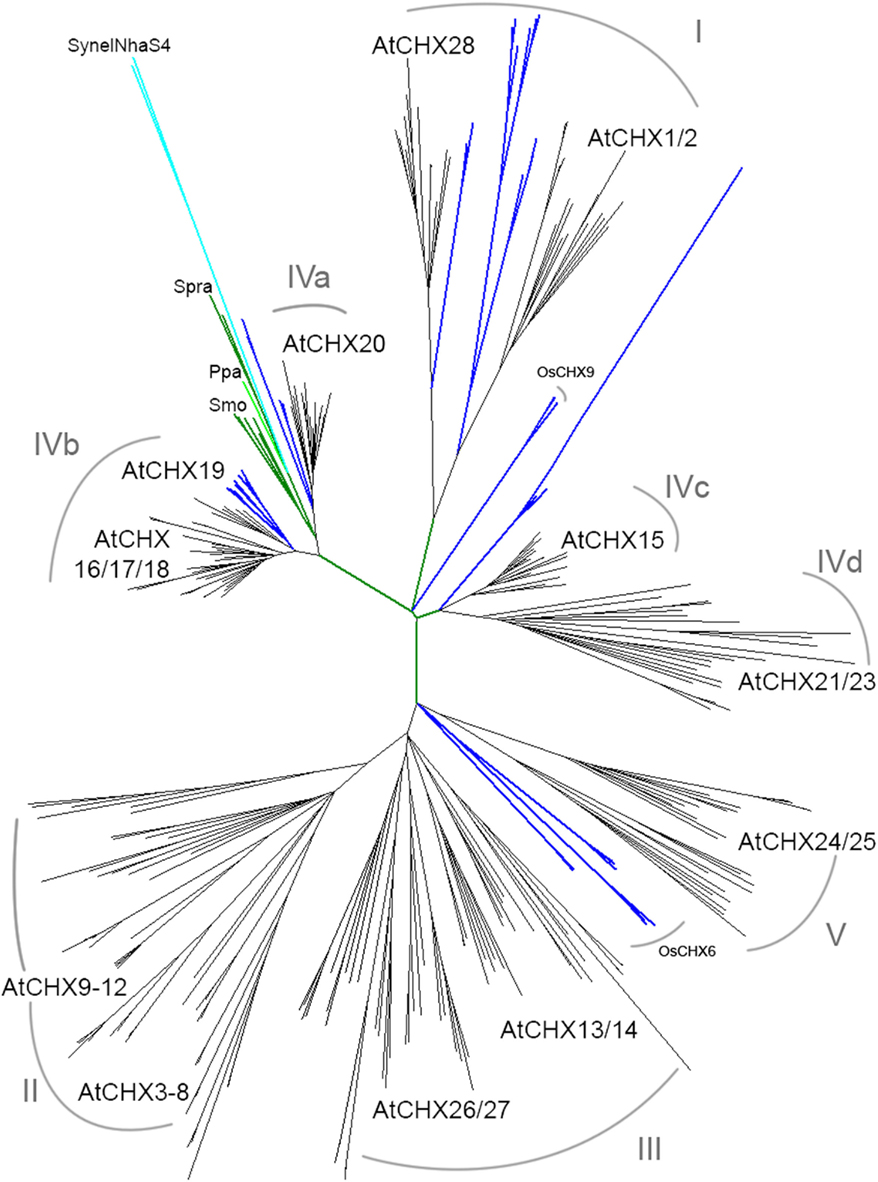
Figure 7. CHX homologs diversified from moss to flowering plants. Full-length CHX proteins from 15 species were aligned and then analyzed by maximum likelihood. Spirogyra (Spra), moss (Ppa), and club moss (Smo; green) CHXs clustered with AtCHX20 (subclade IVa). Both monocot (blue) and dicot (black) plants had orthologs of AtCHX20, AtCHX16–19 (IVb), AtCHX28 (I), AtCHX1/2 (I), and AtCHX15 (IVc). Corn, sorghum, or rice had two to four genes encoding OsCHX6 homologs in a monocot cluster near AtCHX24/25. Most dicots had additional CHX homologs that clustered with AtCHX24/25 (V), 26/27, and 13/14 (III). AtCHX3–12 proteins (subclade II) are specific to A. thaliana and A. lyrata. A cluster of CHX resulting from multiple gene duplications is specific to Medicago truncatula (Mtr). See summary in Table 5. Species, protein accession numbers and protein properties are described in Table S4 in Supplementary Material, and unrooted rectangular tree is in Figure S4 in Supplementary Material.
Multiple subclades within the CHX family. Analysis of CHX homologs from 14 higher plants unveiled the diversification of this family in flowering plants (Figure 7). In general, CHX genes diverged less in monocots than in dicots. First, the average number of CHX genes (16) per monocot genome is less than that seen in many dicots (>26; Table 4). Second, Figure 7 shows that CHX members in Arabidopsis and other dicots can be separated into eight subclades, and five of these are conserved in monocots (Table 5). For example, orthologs of AtCHX20 and AtCHX16–CHX19 were evident in rice, corn, and sorghum, as shown by the blue branches in Figure 7 (Table S4 in Supplementary Material). Furthermore, monocot orthologs of AtCHX15, AtCHX1/2, and AtCHX28 were suggested by their close association with these subclades. Many of these genes in Arabidopsis are preferentially expressed in pollen (Sze et al., 2004; Table 5). Oddly, homologs of AtCHX21/23 were not obvious in monocot genomes. However, rice had four other genes (OsCHX5–8) that clustered near AtCHX24/25, though none were closely related to 16 additional Arabidopsis gene products, CHX3–12, 13/14, or 26/27. Three to four CHX genes from other monocot plants including Brachypodium, a model of an energy grass, sorghum, and corn also clustered with rice OsCHX5–8, suggesting these represent a monocot subclade of CHX homologs (Figure 7).
In contrast, some dicot plants shared additional genes that are potential orthologs of AtCHX24/25, 13/14, and 26/27. Other dicot CHXs from poplar, Citrus clementina, Mimulus, and Manihot esculenta (cassava) formed clusters that were separate from AtCHX13/14 or 24/25, 26/27 subclades, suggesting these genes had diverged later in speciation. Notably, AtCHX3–8 and AtCHX9–12 proteins were in general specific to Arabidopsis genus, indicating diversification of CHX genes during Arabidopsis speciation. Medicago truncatula also had a cluster of 12 genes that could be a result of additional duplications during speciation.
Functions of plant CHX proteins. Functional studies of plant CHX proteins are just beginning and so far limited to Arabidopsis thaliana. According to results from the founding members of the family, AtCHX16–AtCHX20, these proteins are implicated in K+ and pH homeostasis of dynamic endomembranes. First, expression of AtCHX17 or its homologs CHX16–CHX20, rescued alkaline-sensitive growth phenotype of the host yeast strain (Chanroj et al., 2011). Second, CHX20 caused acidification and alkalization of the cytosol and vacuole of yeast, respectively; although CHX17 did not alter cytoplasmic or vacuolar pH (Chanroj et al., 2011). Third, AtCHX17 restored growth of bacteria defective in several K+ uptake systems, and tracer studies demonstrated that CHX17, CHX20, and CHX23 mediated monovalent cation transport with a preference for K+ over Na+ (Table 3). Thus CHXs, including plasma membrane-localized CHX13, transport K+ (Zhao et al., 2008), though their ability to restore yeast growth depends on different external pH’s. Interestingly, these findings of plant CHX are consistent with mutant studies that suggest Synechocystis NhaS4 probably transported K+, but not Na+ (Inaba et al., 2001).
The association of AtCHX proteins with endomembranes and their role in pH and cation homeostasis would suggest they perform important roles in membrane trafficking events that affect protein and cargo sorting. Two lines of evidence support this idea. First, AtCHX17, AtCHX18, or AtCHX19, but not CHX20, conferred tolerance to hygromycin B in yeast lacking multiple cation-handling mechanisms (Chanroj et al., 2011). Sensitivity to aminoglycosides in yeast has been related to altered intracellular pH homeostasis and membrane trafficking. Second, Chanroj et al. (2011) showed that yeast mutants expressing CHX17 secreted less vacuolar-destined carboxypeptidase Y (CPY) to the external medium similar to cells expressing NHX1. Thus cation and pH homeostasis in endosomes, such as PVC, appeared to influence sorting and trafficking of proteins in the endomembrane system. Based on these findings and analysis of Atchx20 mutants (Padmanaban et al., 2007), we propose that CHX20 affects endomembrane dynamics and osmoregulation needed for guard cell swelling and stomatal opening.
The multiplicity of CHX genes in Arabidopsis pollen and possibly other plants is surprising and not understood. The first study on pollen CHX proteins showed that AtCHX21 and AtCHX23 are functionally related and that they are involved in either the reception or the transduction of female signals that target pollen tube to the ovule (Lu et al., 2011). Pollen carrying double mutant chx21chx23 developed into mature grains, extended a tube into the transmitting tract, however the tube failed to find the egg (Lu et al., 2011). Pollen tube elongates by tip growth and failure to shift the axis of polarity in the double mutant suggests that localized cation and pH homeostasis by CHX has a role in the establishment, or maintenance of polarity or both. Homologs of Arabidopsis CHX21 and CHX23 are present in dicot, but not in monocot, plants (Figure 7; Table 5; Table S4 in Supplementary Material). Whether they influence pollen tube guidance of dicots in general or have other roles in reproductive fitness will need to be investigated.
Discussion
Origin of Plant KEA and CHX Genes from Cyanobacteria
Our phylogenetic analyses confirm that plant KEA and CHX genes of the CPA2 superfamily most likely originated from ancestral NhaA/NapA genes of prokaryotes (Chang et al., 2004; Brett et al., 2005a) of cyanobacterial origin. Although many bacteria, such as E. coli and K. pneumonia possess both NhaP and NhaA genes, CPA2 genes related to KEA/KefB/C are detected in a subset of bacteria, including Cyanobacteria Gloeobacter and Anabaena variabilis (Figure 3). Oddly, KEA or Kef-like genes have not been found in any fungi yet. These results suggest that plant CPA2 genes were derived from a cyanobacterium after it formed an endosymbiotic relationship with a primitive non-photosynthetic eukaryote cell. Gloeobacter violaceus may be a descendant of that primitive cyanobacterium (Criscuolo and Gribaldo, 2011), as it has genes encoding proteins homologous to NhaP, NhaS3, NhaS4, and KefB (KEA1/2). Similarly, NhaS4 from predominantly Cyanobacteria is highly homologous to plant CHX (Figure 1). However, NhaS4 also shares high similarity with fungi KHA1 and Nhe3/4 from a protist Dictyostelium discoideum.
Our analysis suggests that KEA transporters in eukaryotes serve functions preferentially in plant cells, and possibly in chloroplasts or plastids. First, we find that all plants sequenced to date possess three subtypes of KEA (Table 4). Green algae, Chlamydomonas and Volvox, have one KEA in each subtype, while higher plant KEAs diversified in subtype KEA1/2 or KEA4–6 (Figure 3). Second, KEAs are found mainly in plastid-containing organisms, including red algae and secondary endosymbiont like Chromista, but are not detected in fungi. Third, peptide sequences of KEA-Ia subtype were identified in proteomic analysis of isolated Arabidopsis plastids (Zybailov et al., 2008). Fourth, a functional study showed that Synechocystis NhaS3 was localized to thylakoid membranes and that it conferred tolerance to high Na and low K+ when expressed in a salt-sensitive E. coli (Inaba et al., 2001; Tsunekawa et al., 2009). As NhaS3 is related to KefB and KEAs, plant KEA-I subtype may participate in monovalent cation and pH homeostasis of thylakoid membranes in chloroplasts. Plant KEA-Ia proteins are distinguished by a long N-terminal domain. The roles of this unusual domain and of KEA subtypes in plants need to be determined.
Vacuolar NHX1–4 Clade is Specific to Plants
The finding that single-celled algae and early land plants have homologs of plasma membrane NhaP/SOS1, endosomal AtNHX5/6, and vacuolar AtNHX1–4 suggest these genes were present in ancestral algae before plants colonized land (Table 4). Furthermore, their roles are conserved and fundamental to plant cells, as the three NHX subtypes have persisted from single-celled algae to flowering plants. Endosomal AtNHX5/6 is part of the IC-NHE clade found in all eukaryote, including protist, fungi, and metazoa (Brett et al., 2005a). It is noteworthy that yeast NHX1 is localized to PVC and shares more similarity to endosomal NHX5/6 than to plant vacuolar NHX1. Importantly, our analysis extends a previous study (Brett et al., 2005a) that a clade of “vacuolar” AtNHX1–4 is specific to plants as seen from Chlamydomonas and moss to rice and Arabidopsis (Figure 2). Importantly, gene diversification of the “plant vacuolar NHX” in monocots and dicots (Table 4) underscores the diverse and critical roles vacuoles play in plant life (Martinoia et al., 2007; Muntz, 2007).
Diversification of CHX Gene Family during Plant Evolution
The multiplicity of the CHX gene family as plants colonized terrestrial habitats is striking. Phylogenetic analysis demonstrated this gene family in plants was founded by members similar to AtCHX20 and AtCHX16–19. First, cyanobacterium SynNhaS4 shares highest similarity with AtCHX17–20. Second, one CHX from a Charophyte, Spirogyra, is homologous to AtCHX20. Third, CHX genes from Selaginella and Physcomitrella encode proteins that are most homologous to AtCHX20 and AtCHX16–19, and fourth all flowering plants sequenced so far have CHX genes orthologs to guard cell-specific AtCHX20 and AtCHX16–19 (Figure 7). Thus a CHX member in Spirogyra, an alga that is closely related to land plants (Karol et al., 2001), is conserved in early land plants. In Arabidopsis, AtCHX20 is preferentially expressed in guard cells (Padmanaban et al., 2007), and CHX16–19 are expressed in various root cells, leaf hydathodes, as well as at the micropylar end of developing seeds (Chanroj, 2011). It would be interesting to determine if one or more of moss CHX genes are expressed in cells forming leaf pores and/or stomata which first appeared in mosses as they developed specialized epidermal cells to prevent desiccation (Freeman, 2008; Peterson et al., 2010).
Analysis of higher plants showed several monocot CHXs are orthologous to AtCHX15, AtCHX1/2, and AtCHX28. Thus these genes were present before the diversification of dicots and monocots. As these Arabidopsis genes are expressed in pollen (Sze et al., 20045), we suggest CHX genes evolved as plants developed pollen grains to protect and transfer sperms for successful reproduction on land. Several rice CHX genes are expressed in developing anthers when pollen is at the bicellular and tricellular stages (Fujita et al., 20106), supporting this idea. In general, monocots have less CHX genes than dicots encoding proteins homologous to pollen-expressed AtCHXs (Table 4). Thus CHX genes further diversified as dicot plants evolved as seen in AtCHX3–12 (Sze et al., 2004) from Arabidopsis thaliana or A. lyrata. Several homologs exist in Medicago truncatula in a sister clade to pollen-expressed Arabidopsis CHX. Appearance of pollen is an important innovation for successful reproduction on land (Freeman, 2008). Whether CHX homologs are related to development of monocolpate versus tricolpate pollen seen in monocots and dicots, respectively, remains to be seen.
Structural and Functional Innovations for Survival on Land
Based on functional studies of CHXs and phylogenetic results, we propose a simple model. The emergence of CHX genes in early land plants and their multiplicity in flowering plants, but not in higher metazoa, suggest CHX function is tied to land plant-specific cellular processes. Their roles in guard cell movement (Padmanaban et al., 2007) and pollen tube guidance (Lu et al., 2011) would support this idea. Based on these two studies, it is obvious that endomembrane-associated NHX members cannot substitute for CHX functions (Chanroj et al., 2011), even though vacuolar and endosomal NHX homologs are expressed in most cell types. Studies showed that AtCHX17 and AtCHX20 are associated with dynamic endomembranes of the endocytic and exocytic pathways suggesting roles in osmoregulation, remodeling of membranes as well as in secretion. Land plants secrete a variety of compounds for growth, development, and defense, including signaling molecules, soluble proteins, membrane complexes, extracellular and wall components. We propose that CHX transporters modulate the cation and pH environment of diverse intracellular compartments for protein activities within the lumen and/or for directed vesicle trafficking to deliver cargo in flowering plants to designated locations.
Summary
Based on the prevalence of CPA1 genes in all organisms, we suggest that plant NHX gene family evolved to serve fundamental needs for cation and pH homeostasis in all eukaryote cells. NhaP/SOS1 regulates cation/H+ fluxes at the plasma membrane as in prokaryotes, and endosomal NHX appeared later to modulate homeostasis and support functions of multiple intracellular compartments in primitive eukaryote cells. Further diversification of plant-specific vacuolar AtNHX1–4 type genes occurred in algal ancestors and in multicellular plants as vacuolar functions increased. In contrast, plant CPA2 genes likely originated from an ancestral cyanobacterium, where NhaS3- or NhaS5-like genes were transferred to the nuclear genome of ancestral plant cells. Three subtypes of KEA proteins are conserved from algae to flowering plants. Although their functions are still unknown, the evolutionary history would implicate a role of some KEA in ion homeostasis of plastids. A NhaS4-like gene of an ancient cyanobacterium is a likely progenitor of the CHX gene family which has multiplied and diversified specifically in flowering plants. Their functions in guard cell movement and pollen tube guidance determined so far would indicate they evolved to support vegetative and reproductive innovations required by plants to adapt and survive on land.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The sequence data encoding CHX homologs from many plants were produced by the US Department of Energy Joint Genome Institute (www.Phytozome.net). This work was support in part by National Science Foundation Grant IBN0209788 and US Department of Energy Grant BES DEFG0207ER15883 to Heven Sze, grant BIO2008-01691 from Spanish Plan Nacional I + D + I to Kees Venema, and a Royal Thai Government Graduate Fellowship to Salil Chanroj. Work of CFD was supported by NSF grants #MCB-0523719 and DEB1036506.
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/Plant_Physiology/10.3389/fpls.2012.00025/abstract
Table S1. Cation–proton antiporters 1 and CPA2 genes and proteins from prokaryotes, algae, protist, metazoa, and plants used in Figure 1.
Table S2. Gene descriptions of selected CPA1 proteins used in Figure 2.
Table S3. Gene descriptions of selected KEA/Kef proteins in Figure 3.
Table S4. Description of CHX genes and proteins from Figure 7.
Table S5. Xls. Gene descriptions of selected KEA/Kef proteins in Figure 3 with hyperlinks.
Figure S1. Phylogenetic tree showing CPA1 and CPA2 proteins used in Figure 1.
Figure S2. Tree showing all CPA1 and NHX proteins used in analysis of Figure 2.
Figure S3. Tree of KEA/Kef proteins used in Figure 3.
Figure S4. Alignment of the N-terminal domain of KEA clade Ia sequences. The N-terminal domain of the clade Ia protein sequences found in the different plant taxa were aligned by MUSCLE (Edgar, 2004). Conserved residues are shown in orange. Regions of predicted coiled coil structures are underlined in blue. The database references for the used species are listed in Table S3.
Figure S5. Tree showing all CHX proteins used in analysis of Figure 7.
Footnotes
References
Abrahamsen, M. S., Templeton, T. J., Enomoto, S., Abrahante, J. E., Zhu, G., Lancto, C. A., Deng, M., Liu, C., Widmer, G., Tzipori, S., Buck, G. A., Xu, P., Bankier, A. T., Dear, P. H., Konfortov, B. A., Spriggs, H. F., Iyer, L., Anantharaman, V., Aravind, L., and Kapur, V. (2004). Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 304, 441–445.
Adl, S. M., Simpson, A. G., Farmer, M. A., Andersen, R. A., Anderson, O. R., Barta, J. R., Bowser, S. S., Brugerolle, G., Fensome, R. A., Fredericq, S., James, T. Y., Karpov, S., Kugrens, P., Krug, J., Lane, C. E., Lewis, L. A., Lodge, J., Lynn, D. H., Mann, D. G., McCourt, R. M., Mendoza, L., Moestrup, O., Mozley-Standridge, S. E., Nerad, T. A., Shearer, C. A., Smirnov, A. V., Spiegel, F. W., and Taylor, M. F. (2005). The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol. 52, 399–451.
Ali, R., Brett, C. L., Mukherjee, S., and Rao, R. (2004). Inhibition of sodium/proton exchange by a Rab-GTPase-activating protein regulates endosomal traffic in yeast. J. Biol. Chem. 279, 4498–4506.
Allen, R. D., and Naitoh, Y. (2002). Osmoregulation and contractile vacuoles of protozoa. Int. Rev. Cytol. 215, 351–394.
An, R., Chen, Q. J., Chai, M. F., Lu, P. L., Su, Z., Qin, Z. X., Chen, J., and Wang, X. C. (2007). AtNHX8, a member of the monovalent cation: proton antiporter-1 family in Arabidopsis thaliana, encodes a putative Li/H antiporter. Plant J. 49, 718–728.
Apse, M. P., Aharon, G. S., Snedden, W. A., and Blumwald, E. (1999). Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285, 1256–1258.
Apse, M. P., Sottosanto, J. B., and Blumwald, E. (2003). Vacuolar cation/H+ exchange, ion homeostasis, and leaf development are altered in a T-DNA insertional mutant of AtNHX1, the Arabidopsis vacuolar Na+/H+ antiporter. Plant J. 36, 229–239.
Arkin, I. T., Xu, H., Jensen, M. O., Arbely, E., Bennett, E. R., Bowers, K. J., Chow, E., Dror, R. O., Eastwood, M. P., Flitman-Tene, R., Gregersen, B. A., Klepeis, J. L., Kolossvary, I., Shan, Y., and Shaw, D. E. (2007). Mechanism of Na+/H+ antiporting. Science 317, 799–803.
Banuelos, M. A., Sychrova, H., Bleykasten-Grosshans, C., Souciet, J. L., and Potier, S. (1998). The Nha1 antiporter of Saccharomyces cerevisiae mediates sodium and potassium efflux. Microbiology 144(Pt 10), 2749–2758.
Bassil, E., Ohto, M. A., Esumi, T., Tajima, H., Zhu, Z., Cagnac, O., Belmonte, M., Peleg, Z., Yamaguchi, T., and Blumwald, E. (2011a). The Arabidopsis intracellular Na+/H+ antiporters NHX5 and NHX6 are endosome associated and necessary for plant growth and development. Plant Cell 23, 224–239.
Bassil, E., Tajima, H., Liang, Y. C., Ohto, M. A., Ushijima, K., Nakano, R., Esumi, T., Coku, A., Belmonte, M., and Blumwald, E. (2011b). The Arabidopsis Na+/H+ antiporters NHX1 and NHX2 control vacuolar pH and K+ homeostasis to regulate growth, flower development, and reproduction. Plant Cell 23, 3482–3497.
Bowers, K., Levi, B. P., Patel, F. I., and Stevens, T. H. (2000). The sodium/proton exchanger Nhx1p is required for endosomal protein trafficking in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 11, 4277–4294.
Bowman, J. L., Floyd, S. K., and Sakakibara, K. (2007). Green genes-comparative genomics of the green branch of life. Cell 129, 229–234.
Brett, C. L., Donowitz, M., and Rao, R. (2005a). Evolutionary origins of eukaryotic sodium/proton exchangers. Am. J. Physiol. Cell Physiol. 288, C223–C239.
Brett, C. L., Tukaye, D. N., Mukherjee, S., and Rao, R. (2005b). The yeast endosomal Na+K+/H+ exchanger Nhx1 regulates cellular pH to control vesicle trafficking. Mol. Biol. Cell 16, 1396–1405.
Cellier, F., Conejero, G., Ricaud, L., Luu, D. T., Lepetit, M., Gosti, F., and Casse, F. (2004). Characterization of AtCHX17, a member of the cation/H exchangers, CHX family, from Arabidopsis thaliana suggests a role in K homeostasis. Plant J. 39, 834–846.
Chang, A. B., Lin, R., Keith Studley, W., Tran, C. V., and Saier, M. H. Jr. (2004). Phylogeny as a guide to structure and function of membrane transport proteins. Mol. Membr. Biol. 21, 171–181.
Chanroj, S. (2011). Plant-Specific K+ Transporters with Distinct Properties and Their Emerging Roles in Endomembrane Trafficking. Ph.D. Dissertation, University of Maryland, College Park.
Chanroj, S., Lu, Y., Padmanaban, S., Nanatani, K., Uozumi, N., Rao, R., and Sze, H. (2011). Plant-specific cation/H+ exchanger 17 and its homologs are endomembrane K+ transporters with roles in protein sorting. J. Biol. Chem. 286, 33931–33941.
Criscuolo, A., and Gribaldo, S. (2011). Large-scale phylogenomic analyses indicate a deep origin of primary plastids within Cyanobacteria. Mol. Biol. Evol. 28, 3019–3032.
Darley, C. P., van Wuytswinkel, O. C., van der Woude, K., Mager, W. H., and de Boer, A. H. (2000). Arabidopsis thaliana and Saccharomyces cerevisiae NHX1 genes encode amiloride sensitive electroneutral Na+/H+ exchangers. Biochem. J. 351, 241–249.
Douglas, S., Zauner, S., Fraunholz, M., Beaton, M., Penny, S., Deng, L. T., Wu, X., Reith, M., Cavalier-Smith, T., and Maier, U. G. (2001). The highly reduced genome of an enslaved algal nucleus. Nature 410, 1091–1096.
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797.
Ferguson, G. P., Battista, J. R., Lee, A. T., and Booth, I. R. (2000). Protection of the DNA during the exposure of Escherichia coli cells to a toxic metabolite: the role of the KefB and KefC potassium channels. Mol. Microbiol. 35, 113–122.
Fraile-Escanciano, A., Kamisugi, Y., Cuming, A. C., Rodriguez-Navarro, A., and Benito, B. (2010). The SOS1 transporter of Physcomitrella patens mediates sodium efflux in planta. New Phytol. 188, 750–761.
Freeman, S. (2008). “Green plants,” in Biological Sciences, 3rd Edn. San Francisco, CA: Pearson Education, 626–663.
Fujisawa, M., Ito, M., and Krulwich, T. A. (2007). Three two-component transporters with channel-like properties have monovalent cation/proton antiport activity. Proc. Natl. Acad. Sci. U.S.A. 104, 13289–13294.
Fujita, M., Horiuchi, Y., Ueda, Y., Mizuta, Y., Kubo, T., Yano, K., Yamaki, S., Tsuda, K., Nagata, T., Niihama, M., Kato, H., Kikuchi, S., Hamada, K., Mochizuki, T., Ishimizu, T., Iwai, H., Tsutsumi, N., and Kurata, N. (2010). Rice expression atlas in reproductive development. Plant Cell Physiol. 51, 2060–2081.
Gaxiola, R. A., Rao, R., Sherman, A., Grisafi, P., Alper, S. L., and Fink, G. R. (1999). The Arabidopsis thaliana proton transporters, AtNhx1 and Avp1, can function in cation detoxification in yeast. Proc. Natl. Acad. Sci. U.S.A. 96, 1480–1485.
Gerisch, G., Heuser, J., and Clarke, M. (2002). Tubular-vesicular transformation in the contractile vacuole system of Dictyostelium. Cell Biol. Int. 26, 845–852.
Goswami, P., Paulino, C., Hizlan, D., Vonck, J., Yildiz, O., and Kuhlbrandt, W. (2011). Structure of the archaeal Na+/H+ antiporter NhaP1 and functional role of transmembrane helix 1. EMBO J. 30, 439–449.
Hamada, A., Hibino, T., Nakamura, T., and Takabe, T. (2001). Na+/H+ antiporter from Synechocystis species PCC 6803, homologous to SOS1, contains an aspartic residue and long C-terminal tail important for the carrier activity. Plant Physiol. 125, 437–446.
Hellmer, J., Patzold, R., and Zeilinger, C. (2002). Identification of a pH regulated Na(+)/H(+) antiporter of Methanococcus jannaschii. FEBS Lett. 527, 245–249.
Hunte, C., Screpanti, E., Venturi, M., Rimon, A., Padan, E., and Michel, H. (2005). Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH. Nature 435, 1197–1202.
Inaba, M., Sakamoto, A., and Murata, N. (2001). Functional expression in Escherichia coli of low-affinity and high-affinity Na(+) (Li(+))/H(+) antiporters of Synechocystis. J. Bacteriol. 183, 1376–1384.
Jiang, Y., Pico, A., Cadene, M., Chait, B. T., and MacKinnon, R. (2001). Structure of the RCK domain from the E. coli K+ channel and demonstration of its presence in the human BK channel. Neuron 29, 593–601.
Jones, D. T., Taylor, W. R., and Thornton, J. M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8, 275–282.
Karol, K. G., McCourt, R. M., Cimino, M. T., and Delwiche, C. F. (2001). The closest living relatives of land plants. Science 294, 2351–2353.
Katiyar-Agarwal, S., Zhu, J., Kim, K., Agarwal, M., Fu, X., Huang, A., and Zhu, J. K. (2006). The plasma membrane Na+/H+ antiporter SOS1 interacts with RCD1 and functions in oxidative stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103, 18816–18821.
Keeling, P. J. (2004). Diversity and evolutionary history of plastids and their hosts. Am. J. Bot. 91, 1481–1493.
Kinclova, O., Ramos, J., Potier, S., and Sychrova, H. (2001). Functional study of the Saccharomyces cerevisiae Nha1p C-terminus. Mol. Microbiol. 40, 656–668.
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., Valentin, F., Wallace, I. M., Wilm, A., Lopez, R., Thompson, J. D., Gibson, T. J., and Higgins, D. G. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948.
Leidi, E. O., Barragan, V., Rubio, L., El-Hamdaoui, A., Ruiz, M. T., Cubero, B., Fernandez, J. A., Bressan, R. A., Hasegawa, P. M., Quintero, F. J., and Pardo, J. M. (2010). The AtNHX1 exchanger mediates potassium compartmentation in vacuoles of transgenic tomato. Plant J. 61, 495–506.
Leonardy, S., Miertzschke, M., Bulyha, I., Sperling, E., Wittinghofer, A., and Søgaard-Andersen, L. (2010). Regulation of dynamic polarity switching in bacteria by a Ras-like G-protein and its cognate GAP. EMBO J. 29, 2276–2289.
Li, H. T., Liu, H., Gao, X. S., and Zhang, H. X. (2009). Knock-out of Arabidopsis AtNHX4 gene enhances tolerance to salt stress. Biochem. Biophys. Res. Commun. 382, 637–641.
Liu, H., Tang, R. J., Zhang, Y., Wang, C. T., Lv, Q. D., Gao, X. S., Li, W. B., and Zhang, H. X. (2010). AtNHX3 is a vacuolar K+/H+ antiporter required for low-potassium tolerance in Arabidopsis thaliana. Plant Cell Environ. 33, 1989–1999.
Lu, Y., Chanroj, S., Zulkifli, L., Johnson, M. A., Uozumi, N., Cheung, A., and Sze, H. (2011). Pollen tubes lacking a pair of K+ transporters fail to target ovules in Arabidopsis. Plant Cell 23, 81–93.
Maresova, L., and Sychrova, H. (2006). Arabidopsis thaliana CHX17 gene complements the kha1 deletion phenotypes in Saccharomyces cerevisiae. Yeast 23, 1167–1171.
Martinoia, E., Maeshima, M., and Neuhaus, H. E. (2007). Vacuolar transporters and their essential role in plant metabolism. J. Exp. Bot. 58, 83–102.
Maser, P., Thomine, S., Schroeder, J. I., Ward, J. M., Hirschi, K., Sze, H., Talke, I. N., Amtmann, A., Maathuis, F. J., Sanders, D., Harper, J. F., Tchieu, J., Gribskov, M., Persans, M. W., Salt, D. E., Kim, S. A., and Guerinot, M. L. (2001). Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 126, 1646–1667.
Mauriello, E. M., Mignot, T., Yang, Z., and Zusman, D. R. (2010). Gliding motility revisited: how do the myxobacteria move without flagella? Microbiol. Mol. Biol. Rev. 74, 229–249.
Miller, S., Ness, L. S., Wood, C. M., Fox, B. C., and Booth, I. R. (2000). Identification of an ancillary protein, YabF, required for activity of the KefC glutathione-gated potassium efflux system in Escherichia coli. J. Bacteriol. 182, 6536–6540.
Nicholas, K. B., Nicholas, H. B., and Deerfield, D. W. (1997). GeneDoc: analysis and visualization of genetic variation. EMBNEW NEWS 4, 14.
Notredame, C., Higgins, D. G., and Heringa, J. (2000). T-coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302, 205–217.
Oh, D. H., Lee, S. Y., Bressan, R. A., Yun, D. J., and Bohnert, H. J. (2010). Intracellular consequences of SOS1 deficiency during salt stress. J. Exp. Bot. 61, 1205–1213.
Ohgaki, R., Nakamura, N., Mitsui, K., and Kanazawa, H. (2005). Characterization of the ion transport activity of the budding yeast Na+/H+ antiporter, Nha1p, using isolated secretory vesicles. Biochim. Biophys. Acta 1712, 185–196.
Ohnishi, M., Fukada-Tanaka, S., Hoshino, A., Takada, J., Inagaki, Y., and Iida, S. (2005). Characterization of a novel Na+/H+ antiporter gene InNHX2 and comparison of InNHX2 with InNHX1, which is responsible for blue flower coloration by increasing the vacuolar pH in the Japanese morning glory. Plant Cell Physiol. 46, 259–267.
Olias, R., Eljakaoui, Z., Li, J., De Morales, P. A., Marin-Manzano, M. C., Pardo, J. M., and Belver, A. (2009). The plasma membrane Na+/H+ antiporter SOS1 is essential for salt tolerance in tomato and affects the partitioning of Na+ between plant organs. Plant Cell Environ. 32, 904–916.
Padan, E. (2008). The enlightening encounter between structure and function in the NhaA Na+-H+ antiporter. Trends Biochem. Sci. 33, 435–443.
Padmanaban, S., Chanroj, S., Kwak, J. M., Li, X., Ward, J. M., and Sze, H. (2007). Participation of endomembrane cation/H+ exchanger AtCHX20 in osmoregulation of guard cells. Plant Physiol. 144, 82–93.
Peterson, K. M., Rychel, A. L., and Torii, K. U. (2010). Out of the mouths of plants: the molecular basis of the evolution and diversity of stomatal development. Plant Cell 22, 296–306.
Qi, Z., and Spalding, E. P. (2004). Protection of plasma membrane K+ transport by the salt overly sensitive1 Na+-H+ antiporter during salinity stress. Plant Physiol. 136, 2548–2555.
Qiu, Q. S., Barkla, B. J., Vera-Estrella, R., Zhu, J. K., and Schumaker, K. S. (2003). Na+/H+ exchange activity in the plasma membrane of Arabidopsis. Plant Physiol. 132, 1041–1052.
Qiu, Q. S., Guo, Y., Dietrich, M. A., Schumaker, K. S., and Zhu, J. K. (2002). Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. U.S.A. 99, 8436–8441.
Quintero, F. J., Blatt, M. R., and Pardo, J. M. (2000). Functional conservation between yeast and plant endosomal Na(+)/H(+) antiporters. FEBS Lett. 471, 224–228.
Quintero, F. J., Martinez-Atienza, J., Villalta, I., Jiang, X., Kim, W. Y., Ali, Z., Fujii, H., Mendoza, I., Yun, D. J., Zhu, J. K., and Pardo, J. M. (2011). Activation of the plasma membrane Na/H antiporter Salt-Overly-Sensitive 1 (SOS1) by phosphorylation of an auto-inhibitory C-terminal domain. Proc. Natl. Acad. Sci. U.S.A. 108, 2611–2616.
Quintero, F. J., Ohta, M., Shi, H. Z., Zhu, J. K., and Pardo, J. M. (2002). Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc. Natl. Acad. Sci. U.S.A. 99, 9061–9066.
Rodriguez-Rosales, M. P., Galvez, F. J., Huertas, R., Aranda, M. N., Baghour, M., Cagnac, O., and Venema, K. (2009). Plant NHX cation/proton antiporters. Plant Signal. Behav. 4, 265–276.
Rodriguez-Rosales, M. P., Jiang, X., Galvez, F. J., Aranda, M. N., Cubero, B., and Venema, K. (2008). Overexpression of the tomato K+/H+ antiporter LeNHX2 confers salt tolerance by improving potassium compartmentalization. New Phytol. 179, 366–377.
Roosild, T. P., Castronovo, S., Healy, J., Miller, S., Pliotas, C., Rasmussen, T., Bartlett, W., Conway, S. J., and Booth, I. R. (2010). Mechanism of ligand-gated potassium efflux in bacterial pathogens. Proc. Natl. Acad. Sci. U.S.A. 107, 19784–19789.
Roosild, T. P., Miller, S., Booth, I. R., and Choe, S. (2002). A mechanism of regulating transmembrane potassium flux through a ligand-mediated conformational switch. Cell 109, 781–791.
Rose, A., Manikantan, S., Schraegle, S. J., Maloy, M. A., Stahlberg, E. A., and Meier, I. (2004). Genome-wide identification of Arabidopsis coiled-coil proteins and establishment of the ARABI-COIL database. Plant Physiol. 134, 927–939.
Saier, M. H. Jr. (2000). A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol. Mol. Biol. Rev. 64, 354–411.
Senior, A., and Moir, A. (2008). The Bacillus cereus GerN and GerT protein homologs have distinct roles in spore germination and outgrowth, respectively. J. Bacteriol. 190, 6148–6152.
Shi, H., Ishitani, M., Kim, C., and Zhu, J. K. (2000). The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. U.S.A. 97, 6896–6901.
Shi, H., Quintero, F. J., Pardo, J. M., and Zhu, J. K. (2002). The putative plasma membrane Na(+)/H(+) antiporter SOS1 controls long-distance Na(+) transport in plants. Plant Cell 14, 465–477.
Southworth, T. W., Guffanti, A. A., Moir, A., and Krulwich, T. A. (2001). GerN, an endospore germination protein of Bacillus cereus, is an Na(+)/H(+)-K(+) antiporter. J. Bacteriol. 183, 5896–5903.
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690.
Strausak, D., Waser, M., and Solioz, M. (1993). Functional expression of the Enterococcus hirae NaH-antiporter in Escherichia coli. J. Biol. Chem. 268, 26334–26337.
Sychrova, H., Ramirez, J., and Pena, A. (1999). Involvement of Nha1 antiporter in regulation of intracellular pH in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 171, 167–172.
Sze, H., Padmanaban, S., Cellier, F., Honys, D., Cheng, N. H., Bock, K. W., Conejero, G., Li, X., Twell, D., Ward, J. M., and Hirschi, K. D. (2004). Expression patterns of a novel AtCHX gene family highlight potential roles in osmotic adjustment and K+ homeostasis in pollen development. Plant Physiol. 136, 2532–2547.
Taglicht, D., Padan, E., and Schuldiner, S. (1991). Overproduction and purification of a functional Na+/H+ antiporter coded by nhaA (ant) from Escherichia coli. J. Biol. Chem. 266, 11289–11294.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739.
Tanifuji, G., Onodera, N. T., Wheeler, T. J., Dlutek, M., Donaher, N., and Archibald, J. M. (2011). Complete nucleomorph genome sequence of the nonphotosynthetic alga Cryptomonas paramecium reveals a core nucleomorph gene set. Genome Biol. Evol. 3, 44–54.
Thackray, P. D., Behravan, J., Southworth, T. W., and Moir, A. (2001). GerN, an antiporter homologue important in germination of Bacillus cereus endospores. J. Bacteriol. 183, 476–482.
Timme, R. E., Bachvaroff, T. R., and Delwiche, C. F. (2012). Broad phylogenomic sampling and the sister lineage of land plants. PLoS ONE 7, e29696.
Tsunekawa, K., Shijuku, T., Hayashimoto, M., Kojima, Y., Onai, K., Morishita, M., Ishiura, M., Kuroda, T., Nakamura, T., Kobayashi, H., Sato, M., Toyooka, K., Matsuoka, K., Omata, T., and Uozumi, N. (2009). Identification and characterization of the Na+/H+ antiporter Nhas3 from the thylakoid membrane of Synechocystis sp. PCC 6803. J. Biol. Chem. 284, 16513–16521.
Tyler, B. M., Tripathy, S., Zhang, X., Dehal, P., Jiang, R. H., Aerts, A., Arredondo, F. D., Baxter, L., Bensasson, D., Beynon, J. L., Chapman, J., Damasceno, C. M., Dorrance, A. E., Dou, D., Dickerman, A. W., Dubchak, I. L., Garbelotto, M., Gijzen, M., Gordon, S. G., Govers, F., Grunwald, N. J., Huang, W., Ivors, K. L., Jones, R. W., Kamoun, S., Krampis, K., Lamour, K. H., Lee, M. K., McDonald, W. H., Medina, M., Meijer, H. J., Nordberg, E. K., Maclean, D. J., Ospina-Giraldo, M. D., Morris, P. F., Phuntumart, V., Putnam, N. H., Rash, S., Rose, J. K., Sakihama, Y., Salamov, A. A., Savidor, A., Scheuring, C. F., Smith, B. M., Sobral, B. W., Terry, A., Torto-Alalibo, T. A., Win, J., Xu, Z., Zhang, H., Grigoriev, I. V., Rokhsar, D. S., and Boore, J. L. (2006). Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science 313, 1261–1266.
Uchikawa, T., Yamamoto, A., and Inouye, K. (2011). Origin and function of the stalk-cell vacuole in Dictyostelium. Dev. Biol. 352, 48–57.
Venema, K., Belver, A., Marin-Manzano, M. C., Rodriguez-Rosales, M. P., and Donaire, J. P. (2003). A novel intracellular K+/H+ antiporter related to Na+/H+ antiporters is important for K+ ion homeostasis in plants. J. Biol. Chem. 278, 22453–22459.
Venema, K., Quintero, F. J., Pardo, J. M., and Donaire, J. P. (2002). The arabidopsis Na+/H+ exchanger AtNHX1 catalyzes low affinity Na+ and K+ transport in reconstituted liposomes. J. Biol. Chem. 277, 2413–2418.
Vinothkumar, K. R., Smits, S. H., and Kuhlbrandt, W. (2005). pH-induced structural change in a sodium/proton antiporter from Methanococcus jannaschii. EMBO J. 24, 2720–2729.
Vothknecht, U. C., and Westhoff, P. (2001). Biogenesis and origin of thylakoid membranes. Biochim. Biophys. Acta 1541, 91–101.
Wagner, M. C., Molnar, E. E., Molitoris, B. A., and Goebl, M. G. (2006). Loss of the homotypic fusion and vacuole protein sorting or golgi-associated retrograde protein vesicle tethering complexes results in gentamicin sensitivity in the yeast Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 50, 587–595.
Waser, M., Hess-Bienz, D., Davies, K., and Solioz, M. (1992). Cloning and disruption of a putative NaH-antiporter gene of Enterococcus hirae. J. Biol. Chem. 267, 5396–5400.
Wu, S. J., Ding, L., and Zhu, J. K. (1996). SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell 8, 617–627.
Xiang, M., Feng, M., Muend, S., and Rao, R. (2007). A human Na+/H+ antiporter sharing evolutionary origins with bacterial NhaA may be a candidate gene for essential hypertension. Proc. Natl. Acad. Sci. U.S.A. 104, 18677–18681.
Yamaguchi, T., Aharon, G. S., Sottosanto, J. B., and Blumwald, E. (2005). Vacuolar Na+/H+ antiporter cation selectivity is regulated by calmodulin from within the vacuole in a Ca2+- and pH-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 102, 16107–16112.
Yamaguchi, T., Apse, M. P., Shi, H. Z., and Blumwald, E. (2003). Topological analysis of a plant vacuolar Na+/H+ antiporter reveals a luminal C terminus that regulates antiporter cation selectivity. Proc. Natl. Acad. Sci. U.S.A. 100, 12510–12515.
Yamaguchi, T., Fukada-Tanaka, S., Inagaki, Y., Saito, N., Yonekura-Sakakibara, K., Tanaka, Y., Kusumi, T., and Iida, S. (2001). Genes encoding the vacuolar Na+/H+ exchanger and flower coloration. Plant Cell Physiol. 42, 451–461.
Yokoi, S., Quintero, F. J., Cubero, B., Ruiz, M. T., Bressan, R. A., Hasegawa, P. M., and Pardo, J. M. (2002). Differential expression and function of Arabidopsis thaliana NHX Na+/H+ antiporters in the salt stress response. Plant J. 30, 529–539.
Yoshida, K., Miki, N., Momonoi, K., Kawachi, M., Katou, K., Okazaki, Y., Uozumi, N., Maeshima, M., and Kondo, T. (2009). Synchrony between flower opening and petal-color change from red to blue in morning glory, Ipomoea tricolor cv. Heavenly Blue. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 85, 187–197.
Zhao, J., Cheng, N. H., Motes, C. M., Blancaflor, E. B., Moore, M., Gonzales, N., Padmanaban, S., Sze, H., Ward, J. M., and Hirschi, K. D. (2008). AtCHX13 is a plasma membrane K+ transporter. Plant Physiol. 148, 796–807.
Zhou, G. A., Jiang, Y., Yang, Q., Wang, J. F., Huang, J., and Zhang, H. S. (2006). Isolation and characterization of a new Na+/H+ antiporter gene OsNHA1 from rice (Oryza sativa L.). DNA Seq. 17, 24–30.
Keywords: cation homeostasis, pH homeostasis, dynamic endomembrane, secretory system, protein, cargo sorting
Citation: Chanroj S, Wang G, Venema K, Zhang MW, Delwiche CF and Sze H (2012) Conserved and diversified gene families of monovalent cation/H+ antiporters from algae to flowering plants. Front. Plant Sci. 3:25. doi: 10.3389/fpls.2012.00025
Received: 20 October 2011; Accepted: 21 January 2012;
Published online: 14 February 2012.
Edited by:
Markus Geisler, University of Fribourg, SwitzerlandReviewed by:
Eduardo Blumwald, University of California Davis, USAWei-Hua Tang, Chinese Academy of Sciences, China
Enrico Martinoia, Universitaet Zuerich, Switzerland
Copyright: © 2012 Chanroj, Wang, Venema, Zhang, Delwiche and Sze. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Heven Sze, Department of Cell Biology and Molecular Genetics, Maryland Agricultural Experiment Station, University of Maryland, Bioscience Research Building # 413, College Park, MD 20742, USA. e-mail:aHN6ZUB1bWQuZWR1
 Salil Chanroj1
Salil Chanroj1