- 1 Centre of Microbial and Plant Genetics, Katholieke Universiteit Leuven, Heverlee, Belgium
- 2 Scientia Terrae Research Institute, Sint-Katelijne-Waver, Belgium
- 3 Laboratory for Process Microbial Ecology and Bioinspirational Management, Consortium for Industrial Microbiology and Biotechnology (CIMB), Department of Microbial and Molecular Systems, Katholieke Universiteit Leuven Association, Sint-Katelijne-Waver, Belgium
In this study, the molecular basis of the induced systemic resistance (ISR) in Arabidopsis thaliana by the biocontrol fungus Trichoderma hamatum T382 against the phytopathogen Botrytis cinerea B05-10 was unraveled by microarray analysis both before (ISR-prime) and after (ISR-boost) additional pathogen inoculation. The observed high numbers of differentially expressed genes allowed us to classify them according to the biological pathways in which they are involved. By focusing on pathways instead of genes, a holistic picture of the mechanisms underlying ISR emerged. In general, a close resemblance is observed between ISR-prime and systemic acquired resistance, the systemic defense response that is triggered in plants upon pathogen infection leading to increased resistance toward secondary infections. Treatment with T. hamatum T382 primes the plant (ISR-prime), resulting in an accelerated activation of the defense response against B. cinerea during ISR-boost and a subsequent moderation of the B. cinerea induced defense response. Microarray results were validated for representative genes by qRT-PCR. The involvement of various defense-related pathways was confirmed by phenotypic analysis of mutants affected in these pathways, thereby proving the validity of our approach. Combined with additional anthocyanin analysis data these results all point to the involvement of the phenylpropanoid pathway in T. hamatum T382-induced ISR.
Introduction
Some types of soil can suppress the symptoms of plant diseases (Craft and Nelson, 1996). Research showed that the observed increased resistance in these plants is the result of the presence of biocontrol agents (BCAs) in the soil. BCAs typically exercise their protective effect by direct interaction with pathogens (Punja and Utkhede, 2003). Surprisingly, some BCAs were also found to be also effective against pathogens through indirect interactions (De Meyer et al., 1998; Krause et al., 2003; Horst et al., 2005; Ongena et al., 2005) by means of the activation of a part of the plant’s immune system, called induced systemic resistance (ISR; Harman et al., 2004; Conrath, 2011). This is in contrast to the more intensively studied systemic acquired resistance (SAR), which is induced by pathogens (Ryals et al., 1996).
According to the classical vision on SAR in Arabidopsis thaliana, it is dependent on the salicylic acid (SA) signaling pathway (Métraux et al., 1990; Gaffney et al., 1993) and on NPR1-mediated induction (Delaney et al., 1995; Kinkema et al., 2000) of pathogenesis-related protein (PR) genes PR1, PR2, and PR5 (van Loon, 1997). It has been shown that SAR leads to increased resistance toward secondary infections by a broad spectrum of plant pathogens (Ryals et al., 1996). However, not all pathogens induce SAR in the plant. The necrotrophic pathogen Botrytis cinerea, for example, does not trigger SAR during infection (Govrin and Levine, 2002) but rather NPR1-independent defense responses controlled by the ethylene (Et) and jasmonic acid (JA) signaling pathways (Thomma et al., 1998; Zimmerli et al., 2001; Glazebrook, 2005).
Induced systemic resistance, on the other hand, is thought to be regulated by the Et- and JA-signaling pathway with mediation of NPR1 but without induction of PR1, PR2, and PR5 (Pieterse et al., 1996, 2009). However, this traditional view on ISR appears to be more complex (Korolev et al., 2008; Niu et al., 2011), a notion that is confirmed by the results of our present study. The nature and composition of ISR strongly depends on the tripartite combination plant-BCA-pathogen (Duijff et al., 1998; Tjamos et al., 2005) and the overlap between SAR and ISR could be much larger than only through the reported key marker NPR1 (Pieterse et al., 1998). In this manuscript, a clear distinction is made between ISR that is induced by BCAs before (ISR-prime) and after (ISR-boost) additional inoculation with a pathogen, a distinction earlier observed by others (van Wees et al., 1999; Attitalla et al., 2001; Harman et al., 2004; Verhagen et al., 2004). In 2006, a consortium of different research groups defined the concept of priming in plant defense as follows: various treatments, like inoculation with pathogens or BCAs, are said to prime plants or – in other words – prepare the plants’ defense system to respond to stresses more quickly and aggressively (Prime-A-Plant Group et al., 2006). Based on this definition, we chose the terms ISR-prime for the actual process of priming of the plant by the BCA and ISR-boost for the subsequent boost of the defense response upon pathogen inoculation.
So far, whole genome analysis of ISR was predominantly focused on bacterial BCAs (Cartieaux et al., 2003, 2008; Verhagen et al., 2004; Wang et al., 2005; Pozo et al., 2008; Van Oosten et al., 2008) and ISR-related microarray data for fungal BCAs are limited. Moreover, the few reported data mostly focus on ISR-prime in tomato (Alfano et al., 2007; Jiang et al., 2009) or on transcriptional changes in the fungus itself (Chacón et al., 2007; Samolski et al., 2009; Lorito et al., 2010; Rubio et al., 2012). In the same context, but not on whole genome level, Brotman et al. (2012) very recently reported on their study in which expression of 137 A. thaliana genes was monitored by qRT-PCR analysis during the ISR that is induced by Trichoderma asperelloides T203 against Pseudomonas syringae.
For this reason, we initiated a large-scale analysis of the ISR induced by the biocontrol fungus Trichoderma hamatum T382 against Botrytis cinerea in A. thaliana both before (ISR-prime) and after (ISR-boost) additional inoculation with the pathogen. Trichoderma spp. are known to raise resistance against pathogens by the induction of ISR in the plant (Shoresh et al., 2005; Segarra et al., 2009; Tucci et al., 2011). This effect was shown for mono- and dicotyledons and for different types of pathogens (fungi, bacteria, and viruses; Harman et al., 2004). Comparison of more than 500 micro-organisms, isolated from several types of soil, showed that treatment with T. hamatum T382 causes the strongest decrease of the symptoms of foliar infections (Krause et al., 2003), supporting our choice for this BCA in the present study. Further we opted for the necrotrophic broad spectrum pathogen B. cinerea (review by van Kan, 2006), since it only infects the above-ground parts of the plant and as a result it is a suitable pathogen to study ISR-effects induced by soil BCAs. Finally, we chose A. thaliana as a model plant because of the availability of the complete genome sequence, annotation, advanced techniques and mutants. Up to now, the activity of T. hamatum T382 against B. cinerea was shown in begonia (Horst et al., 2005) and geranium (Olson and Benson, 2007), but not yet in A. thaliana. Additionally, little is known about the ISR that is induced in A. thaliana by Trichoderma spp., at least at the level of gene expression.
In this study we started to phenotypically demonstrate the ISR-effect of T. hamatum T382 against B. cinerea in A. thaliana. This allowed us to use this model for an in-depth transcriptomic analysis of both ISR-prime and ISR-boost mechanisms induced by T. hamatum T382 before and after B. cinerea inoculation. In addition, this study pioneered the large-scale characterization of the ISR-boost that is induced by a fungal BCA. The involvement of different defense-related pathways identified in this transcriptomic ISR study was validated using phenotypic analysis of A. thaliana disease signaling mutants.
Results
Trichoderma hamatum T382 suppresses disease symptoms in A. thaliana infected with B. cinerea
As a basis for our study on the ISR-effect of T. hamatum T382 we provided proof-of-principle that T. hamatum T382 is effectively able to induce ISR and subsequently to reduce the symptoms of B. cinerea infection in A. thaliana (Figure 1A). To this end, 3-weeks old A. thaliana plants were treated with T. hamatum T382 by pipetting a spore suspension directly onto the roots, and their susceptibility to subsequent B. cinerea leaf infection was evaluated as compared to mock-treated plants. A preliminary experiment, in which T. hamatum T382 was administered on different time points ranging from 4 to 20 days before B. cinerea inoculation, indicated that a T. hamatum T382 treatment 6 days before B. cinerea inoculation resulted in the highest reduction of disease symptoms (results not shown). These conditions were applied in an extended disease experiment consisting of 12 repeats comprising a total of 2000 plants in which B. cinerea infection was scored symptomatically on a daily basis. By comparing the average lesion diameters of T. hamatum T382 and mock-treated plants using a two-sample one-sided Student’s t-test (Figure 1B) it was shown that treatment of A. thaliana with T. hamatum T382 resulted in a statistically significant (p < 0.001) reduction of the disease symptoms of on average 60% on 2, 3, and 4 days after B. cinerea inoculation. Moreover, the results of these disease assays were highly reproducible making the model A. thaliana – T. hamatum T382 – B. cinerea suitable for further transcriptomic analysis of ISR.
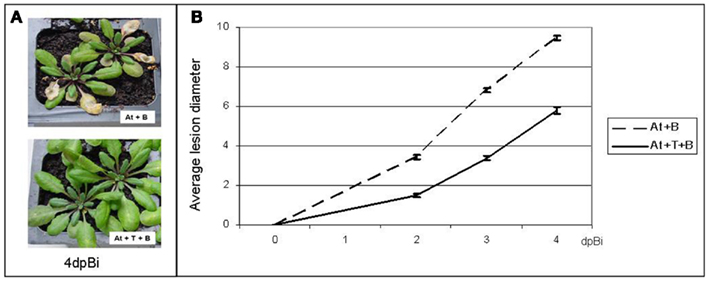
Figure 1. Analysis of the ISR-effect of T. hamatum T382 (T) in A. thaliana (At) against B. cinerea (B). (A) Visual assessment of disease symptoms in mock-treated control plants (top) and in plants treated with T. hamatum T382 (bottom), on 4 days post-inoculation (dpBi) with B. cinerea. (B). Quantitative analysis of disease symptoms, calculated as average lesion diameters on 2, 3, and 4 days post-inoculation (dpBi) with B. cinerea on plants treated with T. hamatum T382 (solid line) and mock-treated control plants (dashed line). The disease assay was repeated 12 times comprising a total of 2000 plants. Confidence intervals (95%) are shown.
Gene-specific Determination by qRT-PCR of Relevant Time Points for a Further ISR Transcriptome Study
To determine relevant time points for a transcriptomic analysis of ISR, known marker genes were selected for the main defense-related pathways and their expression was monitored by qRT-PCR. We chose PDF1.2a for the combined JA- and Et-mediated pathway and PR1 for the SA-mediated pathway. For ISR-prime analysis, pools of leaves were collected from both T. hamatum T382 and mock-treated A. thaliana Col-0 plants (At + T vs. At) and the expression of the marker genes was examined on six consecutive days after T. hamatum T382 treatment. For ISR-boost analysis, the same two sets of plants were subsequently inoculated with B. cinerea and pools of systemic leaves were analyzed for marker gene expression on four consecutive days after B. cinerea inoculation (At + T + B vs. At + B).
During ISR-prime, a statistically significant (p < 0.01) induction was shown for PR1 on the first 3 days post-T. hamatum T382 inoculation (dpTi; Figure 2A). Two other SA-markers, PR2 and PR5, showed a similar induction pattern as PR1 (results not shown), while the expression of the Et/JA-marker PDF1.2a was not affected (p < 0.01; Figure 2B). After 3 dpTi, expression levels decreased until basal levels were reached at 5 and 6 dpTi. Based on these results we decided to use the samples taken on 2 dpTi in the subsequent transcriptomic analysis of ISR-prime. Interestingly, the changes in the expression of defense-related genes during ISR-prime coincided with the explosive growth of T. hamatum T382 in the soil and on the roots of the plants, which was observed both by qPCR using specific primers and plating on a selective medium. Both techniques showed an increase from 4 × 105 CFU/g to 108 CFU/g from 1 h to 3 days after T. hamatum T382 application in the soil. On the roots, T. hamatum T382 increased from 4 × 104 CFU/g to 4 × 108 CFU/g in the same time period.
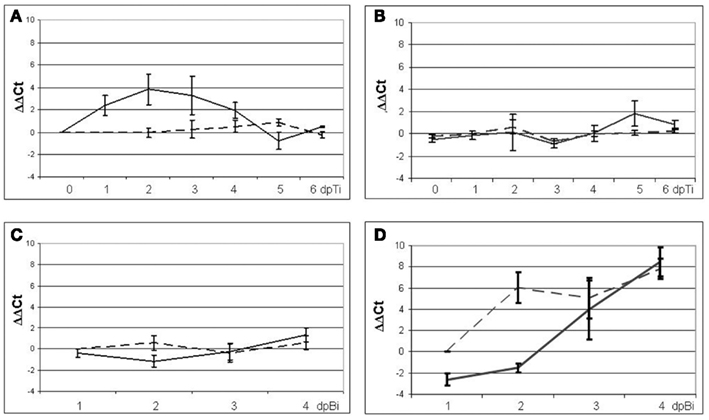
Figure 2. Analysis of expression of the marker genes PR1 and PDF1.2a in mock-treated control plants (dashed lines) and plants treated with T. hamatum T382 (solid lines) using qRT-PCR. For ISR-prime gene expression analysis of PR1(A) and PDF1.2a (B) was done in six samples (biological replicates) collected on six consecutive days after T. hamatum T382 administration. For ISR-boost gene expression analysis of PR1 (C) and PDF1.2a (D) was done in five samples (biological replicates) collected on four consecutive days after B. cinerea inoculation.
During ISR-boost, the expression of PR1 was unaltered (Figure 2C) as was the case for the two other SA-responsive markers PR2 and PR5 (results not shown). Interestingly, PDF1.2a showed a six-fold downregulation in T. hamatum T382 treated plants on 1 day post-B. cinerea inoculation (dpBi) whereas no effect was observed in mock-treated control plants (Figure 2D). On 2 dpBi, however, the PDF1.2a expression levels increased up to 60-fold in control plants while in T. hamatum T382 treated plants still a slight downregulation (2.86-fold) was observed. Thus, during ISR-boost, we could not observe the strong induction of PDF1.2a on 2 dpBi, which is characteristic during the defense response induced by B. cinerea in A. thaliana (Manners et al., 1998). Based on these findings 1 and 2 dpBi were chosen as relevant time points for ISR-boost.
Transcriptome Analysis of A. thaliana after Interaction with T. hamatum T382 and/or B. cinerea Using Microarrays
For both ISR-prime and ISR-boost, plant samples were collected at the above-selected time points from plants that showed a clear ISR-effect (defined as 40–80% reduction in disease index in the subsequent disease assay) and from their corresponding control plants. Three independent biological replicates were performed, each with a dye swap. Furthermore, control samples of the ISR-boost experiment (systemic leaves from A. thaliana Col-0 plants that were inoculated with B. cinerea without pretreatment with T. hamatum T382) were compared to those from untreated and uninfected plants (At + B vs. At) to characterize the defense response induced by B. cinerea (BIDR).
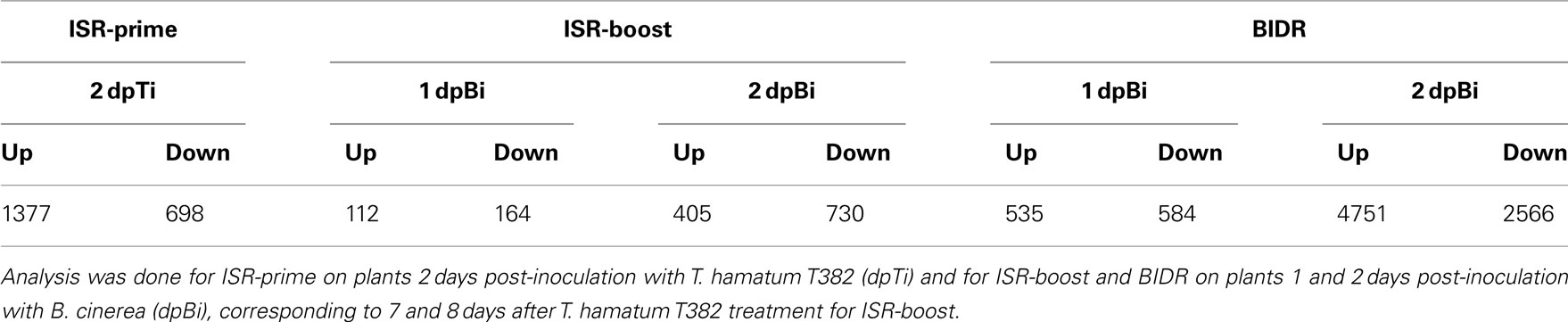
From all selected plant samples high quality mRNA was isolated, labeled, and hybridized on Agilent 4-pack Arabidopsis microarrays. Normalized microarray data were used to identify genes that are differentially expressed during ISR (either prime or boost) and BIDR. Comparing plants treated with T. hamatum T382 with mock-treated controls (At + T vs. At), we identified 2075 genes that are differentially expressed during ISR-prime as summarized in Table 1 (for a complete list see Table S1 in Supplementary Material). Furthermore, the microarray analysis revealed 276 and 1135 annotated genes that were differentially expressed during ISR-boost (Table 1, Table S1 in Supplementary Material) on 1 and 2 dpBi, respectively. The analysis of ISR-boost consisted of a comparison of plants treated with T. hamatum T382 and consequently inoculated with B. cinerea and mock-treated controls consequently inoculated with B. cinerea (At + B + T vs. At + B). For BIDR, comparison of these mock-treated plants that were consequently inoculated with B. cinerea and mock-treated plants without subsequent B. cinerea inoculation (At + B vs. At) resulted in the identification of 1119 and 7317 annotated genes that were differentially expressed on 1 and 2 dpBi, respectively (Table 1, Table S1 in Supplementary Material).
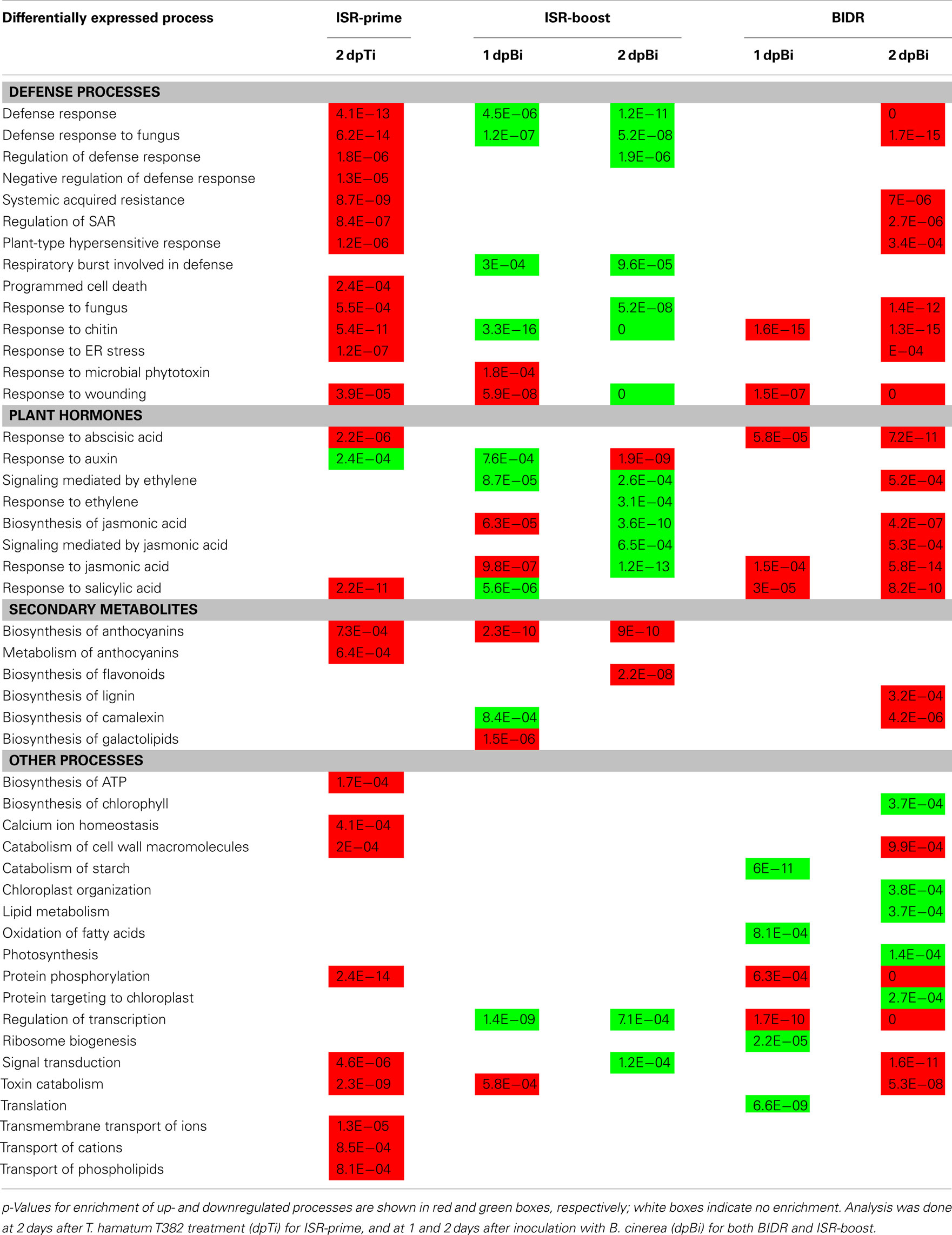
The high number of differentially expressed (DE) genes allowed us to examine the underlying mechanism of ISR at the level of biological processes and pathways leading to a more holistic view on ISR. As shown in Table 2, classification of the DE genes based on the standard Gene Ontology annotation (Gene Ontology Consortium, 2000), led to the identification of biological processes that were significantly (p < 0.001) enriched in the sets of DE genes as compared to the complete A. thaliana genome, according to the method described by Tavazoie et al. (1999). The main observations are discussed below for both ISR-prime and ISR-boost and compared to BIDR. A concise overview of these results is shown in Table 2 while the complete list of up- and downregulated processes can be found Table S2 in Supplementary Material.
Biological Processes Involved in ISR-prime
With respect to upregulated processes a striking analogy between ISR-prime and BIDR (on 2 dpBi; Table 2) was observed including both defense responses such as “defense response to fungus,” “plant-type hypersensitive response,” or “response to chitin” and defense-related plant hormone responses like “response to SA” and “response to abscisic acid” (ABA). Nevertheless, still a number of differences between ISR-prime and BIDR could be observed. For instance, ISR-prime was characterized by the induction of “negative regulation of defense response” while this was not the case for BIDR. On the other hand, ISR-prime was not characterized by the induction of the Et- and JA-signaling pathways. Furthermore, anthocyanins were the main secondary metabolites formed during ISR-prime while defense during BIDR relied mainly on the production of camalexin. Specific for ISR-prime is also the stimulation of the transport of a variety of compounds in the plant, e.g. phospholipids and ions.
Remarkably, when looking at the downregulated processes, the correspondence between ISR-prime and BIDR was very low (Table 2). Where BIDR negatively affected the general metabolism of the plant (e.g. photosynthesis, translation, lipid metabolism), this phenomenon was not observed during ISR-prime.
Biological Processes Involved in ISR-Boost as Compared to BIDR
It is important to stress here that, in order to characterize ISR-boost, we compared plants treated with T. hamatum T382 and subsequently inoculated with B. cinerea with mock-treated control plants subsequently inoculated with B. cinerea (At + T + B vs. At + B). As such the results for ISR-boost represent the biological processes that are specifically affected by the pretreatment with T. hamatum T382 and that occur on top of the B. cinerea induced defense response (BIDR).
On the first day after B. cinerea inoculation, BIDR affected a limited number of defense processes (Table 2) like “response to chitin” and “response to wounding.” Several defense-related plant hormone responses were induced, e.g. responses to JA, SA, and ABA (Table 2) while general metabolic processes, like translation, were downregulated. In contrast to BIDR, defense-related processes and hormone pathways were activated more rapidly (primed) during ISR-boost, such as “biosynthesis of JA” and “response to microbial phytotoxin,” or more strongly, such as “response to JA” and “response to wounding” (Table 2). Production of secondary metabolites such as galactolipids and anthocyanins were specifically induced during ISR-boost. Interestingly, defense–related ROS-production as reflected by the biological process “respiratory burst involved in the defense response” is specifically downregulated during ISR-boost.
On the second day after B. cinerea inoculation, the strong induction of defense processes that characterized BIDR was not or to a less extent observed during ISR-boost, indicating that the T. hamatum T382 treatment restrained the Botrytis-induced defense response (Table 2). Many defense-related processes showed an opposite behavior in ISR-boost as compared to BIDR such as “defense response to fungus,” “regulation of defense response,” and “response to chitin.” Similar observations can be made for the defense-related hormone pathways such as signaling mediated by JA and Et. However, in line with the situation on the first day after B. cinerea inoculation, the phenylpropanoid pathway–based biosynthesis of anthocyanins, as well as flavonoids, were specifically induced while “respiratory burst involved in the defense response” was specifically downregulated during ISR-boost. Again, general metabolic processes, like photosynthesis, were downregulated during BIDR.
Validation of Microarray Results
The above-mentioned microarray results were validated by qRT-PCR and GUS-staining.
Validation of microarray results using qRT-PCR
Because of the high number of differentially expressed (DE) genes during ISR, a set of 26 genes was selected representing the different ISR-related pathways identified in our transcriptome analysis (Table 3) and their expression levels were measured by qRT-PCR (primers shown in Table S3 in Supplementary Material) under the same conditions and at the same time points as used in the microarray analysis. To this end, additional independent biological samples were tested. As can be concluded from Table 3, the resulting qRT-PCR data are generally in line with the normalized log ratios from the microarrays, thereby confirming the observed gene expression patterns.
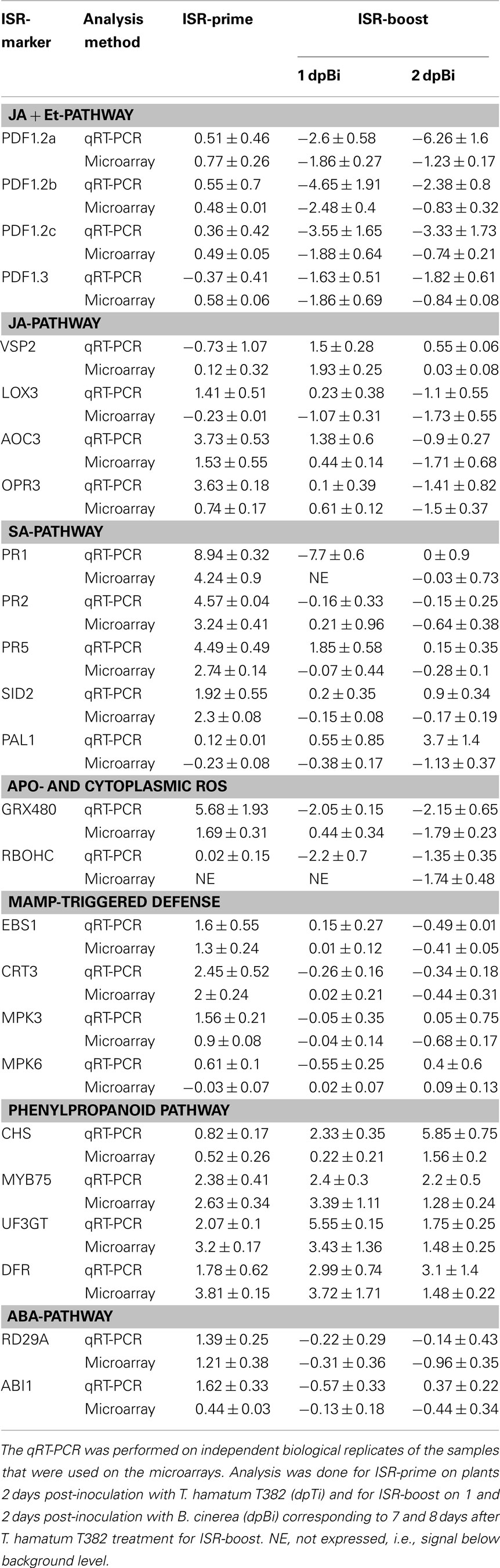
Table 3. Comparison between the results of the microarrays (shown as log2 ratio) and the qRT-PCR (shown as log2 ratio) for various ISR-markers.
Validation of microarray results using GUS-staining
The microarray results were further validated in planta using a histochemical staining approach. We therefore made use of promoter-GUS lines corresponding to PDF1.2a, known as well-established markers for the Et/JA-response pathway (Penninckx et al., 1998; Zimmerli et al., 2000), and PR1 which is considered a marker for the SA-mediated pathway (Shah, 2003). In the present study, we evaluated the expression of these marker genes after treatment with T. hamatum T382 and additional B. cinerea inoculation. The results of the GUS-staining for both PDF1.2a and PR1 on different time points in the ISR-process (Figure 3) confirmed both the microarray results and the results of the preliminary qRT-PCR analysis (Figure 2). For ISR-prime, both the strong induction of PR1 and the absence of the induction of PDF1.2a after T. hamatum T382 treatment were clearly confirmed by the GUS-staining. During ISR-boost, no expression of PR1 could be detected in any of the samples. Conform to its well-established role as a marker of B. cinerea induced defense (Manners et al., 1998) a strong induction of PDF1.2a was observed upon B. cinerea inoculation, at least in plants that were not pretreated with T. hamatum T382. Interestingly, in plants primed with T. hamatum T382 this strong induction of PDF1.2a was not observed, again confirming our microarray results.
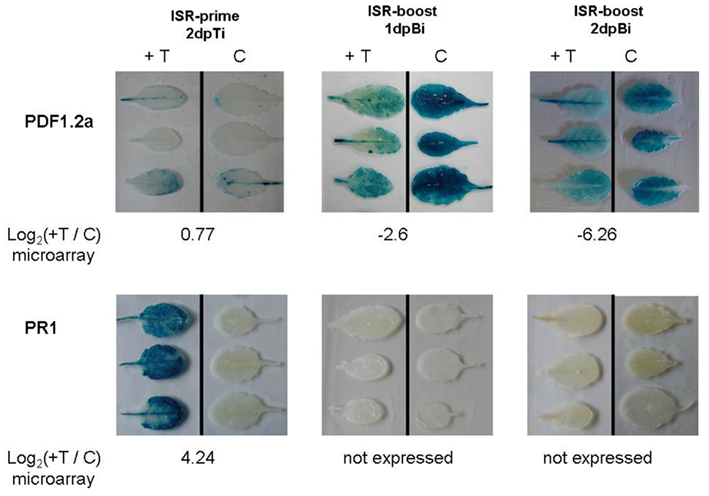
Figure 3. Effect of T. hamatum T382 treatment on the expression of the GUS-gene controlled by the promoter of the plant defensin PDF1.2a and the pathogenesis-related protein PR1. GUS-staining was done on leaves of transgenic lines expressing the different promoter-GUS fusions. For each of these genes results are shown for T. hamatum T382 treated plants (+T) vs. mock-treated control plants (C), during ISR-prime (left column) and ISR-boost at 1 day (middle column) and 2 days (right column) post-inoculation with B. cinerea (dpBi). Corresponding microarray results (log2 ratios) are shown below each figure.
Confirmation of the Involvement of Various Defense-Related Pathways in ISR
The involvement of the above-mentioned pathways that were identified in this study as contributing to ISR was further confirmed by mutant evaluation and anthocyanin analysis.
Confirmation of the involvement of defense-related pathways in ISR using mutants
We investigated the ISR-effect of T. hamatum T382 on B. cinerea infection in different A. thaliana mutants as compared to wild-type plants. More specifically, mutants or transformants were selected that are affected in a specific defense-related pathway including npr1, sid2, and NahG for the SA-pathway (Cao et al., 1994; Delaney et al., 1994), ein2 and etr1 for the Et-pathway (Guzmán and Ecker, 1990; Chang et al., 1993) and myc2 for the JA-pathway (Berger et al., 1996), or in defense-related mechanisms such as tt, chs, and f3h, all carrying mutations in the phenylpropanoid pathway (Teng et al., 2005). Additionally the gene encoding the respiratory burst oxidase RBOHD (Torres et al., 1998) is inactivated in the rbohD mutant affecting defense-related ROS-production (Mersmann et al., 2010) while the vtc1 mutant is deficient in ascorbic acid, an important ROS scavenger (Conklin et al., 2000), resulting in a SAR-phenotype (Pastori et al., 2003; Barth et al., 2004; Mukherjee et al., 2010) with increased levels of SA, PR1, PR2, and PR5 (thus also similar to ISR-prime).
More specifically, the SA-signaling mutants sid2 and npr1 are impaired in biosynthesis and perception of SA, respectively, while in the transgenic NahG-line SA-accumulation is impossible through transformation of SA to catechol. In our experiments NahG showed a basal susceptibility toward B. cinerea that was equal (2 dpi) or significantly higher (3 and 4 dpi; p < 0.001) to that of wild-type (wt) Col-0 and npr1 showed increased susceptibility (p < 0.01 on 2 dpi and p < 0.001 on 3 and 4 dpi; Figure 4), confirming the results of earlier reports on both equal (Ferrari et al., 2003; Kishimoto et al., 2006; Veronese et al., 2006) and significantly increased susceptibility of these mutants toward B. cinerea infection (Zimmerli et al., 2001; Govrin and Levine, 2002). The sid2 mutants showed significantly increased lesion development compared to wild-type (p < 0.001), in contrast to an earlier report (Ferrari et al., 2003).
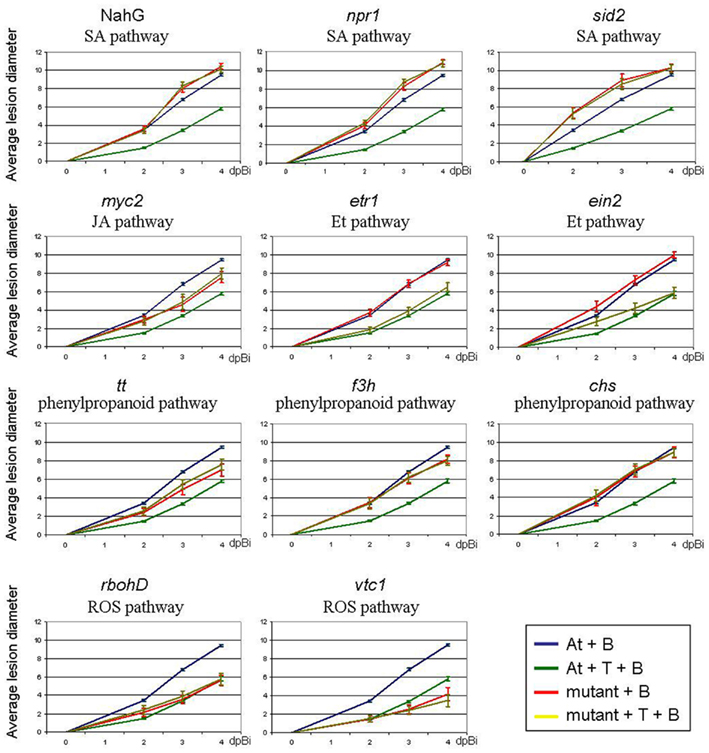
Figure 4. Effect of T. hamatum T382 treatment on suppression of B. cinerea disease in A. thaliana mutants (npr1, sid2, myc2, etr1, ein2, tt, chs, f3h, rbohD, vtc1) or transformants (NahG), affected in different disease-related pathways. Affected pathways are indicated below the mutant name. Quantitative analysis of the ISR-effect of T. hamatum T382 (T) against B. cinerea infection. Comparison of disease symptoms, calculated as average lesion diameters and measured on 2, 3, and 4 days post-inoculation (dpBi) with B. cinerea of mock-treated wild-type (blue) or mutant plants (red) and of T. hamatum T382 treated wild-type (dark green) and mutant plants (light green). Disease assays were repeated four to six times comprising a total of 100–250 plants depending on the mutant used. Confidence intervals (95%) are shown.
The transcription factor (TF) MYC2 acts as both activator and repressor of distinct JA-responsive genes in Arabidopsis (Lorenzo et al., 2004). The corresponding myc2 mutants showed significantly increased resistance (p < 0.01) to B. cinerea on all time points, in conformity with earlier findings (Lorenzo et al., 2004).
Mutations of the Et-receptor ETR1 are known to result in loss of sensitivity to Et (Schaller and Bleecker, 1995) and in some studies reported to increase sensitivity toward B. cinerea infections (Zimmerli et al., 2001; Kishimoto et al., 2006; Lloyd et al., 2011). The latter could not be observed in our experiments. However, variations in the sensitivity of different mutants to B. cinerea infection have been regularly reported and have been attributed to the use of different strains of this pathogen in the different studies (Govrin and Levine, 2002; Rowe et al., 2010). Indeed, the B. cinerea strain used in this study (B05-10) differs from these used in the studies that showed increased sensitivity toward B. cinerea in the etr1 mutant. When using ein2, another mutant affected in Et-signaling we do observe increased sensitivity toward B. cinerea on 2 dpi (p = 0.0001) and 4 dpi (p < 0.05). EIN2 which encodes a signal transducer that interacts with ETR1 (Alonso et al., 1999), is the only gene of the Et-pathway whose loss-of-function mutations lead to complete ethylene insensitivity (Alonso et al., 1999) and block the induction of the marker gene PDF1.2a (Penninckx et al., 1996) and therefore might explain the more pronounced phenotypic effects, including the influence on B. cinerea sensitivity in ein2 as compared to etr1.
The transparent testa (tt) mutant shows reduced anthocyanin content whereas f3h and chs are characterized by deletions of the flavanone 3-hydroxylase and the chalcone synthase genes, respectively. Both genes encode enzymes that catalyze different steps in the biosynthesis of flavonoids and anthocyanins. The relation of these mutants to B. cinerea resistance has to our knowledge not yet been reported. Our results indicate that these mutants display similar or significantly decreased sensitivity toward B. cinerea infections compared to wt (p < 0.001 for tt and for f3h on 4 dpi; p < 0.05 for chs on 4 dpi). Additionally, both vtc1 and rbohD mutants exhibited significantly increased resistance to B. cinerea (p < 0.001). These results are in accordance with former observations of reduced fungal biomass on rbohD mutants compared to wild-type A. thaliana Col-0 during infection with Alternaria brassicicola (Pogány et al., 2009), a necrotrophic fungal pathogen like B. cinerea.
As shown in Figure 4 the suppressive effect on B. cinerea disease, as earlier observed in wild-type A. thaliana plants pretreated with T. hamatum T382, was not detected in most of these mutants, indicating that the corresponding genes (and pathways) play an important role in this interaction. Indeed, mutants corresponding to key genes in SA- or JA-mediated signaling, or anthocyanin production did not display the T. hamatum T382-induced ISR against B. cinerea. Also in vtc1 and rbohD mutants, additional T. hamatum T382 treatment did not further increase the resistance that was generated by the mutation. More specifically, we could demonstrate that the regulatory protein NPR1 which (nuclear localization) was demonstrated to be essential for SA-mediated defense-gene expression during SAR (Wang et al., 2006), is also a key element in the T. hamatum T382-induced ISR. This protein has recently been shown to be implicated in ISR (Segarra et al., 2009), albeit operating from the cytosol (Stein et al., 2008). The significant divergence of expression of the SID2 gene earlier observed (Table S1 in Supplementary Material) during the Botrytis-induced defense response (not affected) and T. hamatum T382-induced ISR (strongly induced), indicated an important role of SID2 in the former. This was clearly confirmed by the results on the sid2 mutants (Figure 4). These results are not in accordance with the findings of Segarra et al. (2009) who observed a preserved ISR-effect in sid2 mutants treated with another biocontrol Trichoderma strain.
In contrast to the results obtained with the other mutants, the ISR-effect was preserved in etr1 and ein2 mutants (p < 0.001) affected in Et-signaling (Cancel and Larsen, 2002) and no difference was observed in ISR-effect in these mutants as compared to wild-type plants, indicating that the Et-pathway, or at least that part in which ETR1 and EIN2 are involved, is not essential in the T. hamatum T382-induced ISR against B. cinerea in A. thaliana. This finding clearly corresponds to our microarray results which indicate that the Et-signaling pathway, despite of being strongly induced during BIDR, remains unaffected during ISR-prime or is even downregulated during ISR-boost on 1 dpBi.
Confirmation of the involvement of the phenylpropanoid pathway in ISR using anthocyanin measurements
In order to confirm the observed upregulation of the biological process “anthocyanin biosynthesis” during both ISR-prime and ISR-boost, anthocyanin concentrations were compared between ISR-primed and ISR-boosted plants and their corresponding controls at several time points after T. hamatum T382 treatment or B. cinerea inoculation. In agreement with the results of the microarray analysis, we observed a significant increase in anthocyanin content between ISR-primed plants and mock-treated controls and between ISR-boosted plants and B. cinerea infected controls (Figure 5).
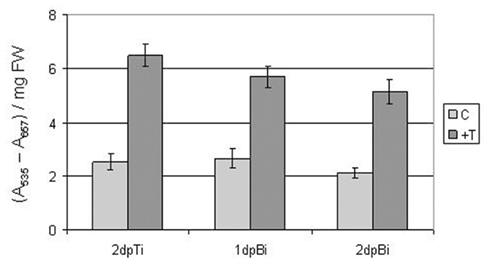
Figure 5. Quantification of anthocyanin levels (expressed as corrected A535 per milligram fresh weight) in ISR-primed and mock-treated control plants at 2 days post-treatment with T. hamatum T382 (dpTi) and in ISR-boosted and Botrytis inoculated control plants at 1 and 2 days post-inoculation with B. cinerea (dpBi). The values are means ± SE of six measurements of four plants each. FW, fresh weight.
Discussion
In this study we demonstrated that application of T. hamatum T382 to the roots of A. thaliana results in an increased resistance to subsequent leaf infections by the necrotrophic pathogen B. cinerea, characterized by a significant reduction in symptom development. This tripartite model allowed a genome-wide analysis of ISR-related gene expression using microarrays. More specifically, gene expression was characterized both before (ISR-prime) and after (ISR-boost) additional B. cinerea inoculation in T. hamatum T382-pretreated plants as compared to mock-pretreated controls. To allow further comparison with the regular defense response induced by this pathogen, further mentioned as BIDR, we also compared B. cinerea infected plants without T. hamatum T382-pretreatment with uninfected controls. Aiming at a holistic view on ISR, we classified the ISR- and BIDR-related genes into the standard biological processes, as defined by the Gene Ontology Consortium (2000), that were significantly induced or downregulated (Table 2). However, since (i) not all relevant biological processes are represented in Gene Ontology (e.g. MAMP-triggered defense is as such not present), and (ii) the analysis of enriched biological processes might miss small differences between ISR and BIDR, we opted to perform an additional reverse analysis process by starting from the structure of a pathway and identifying the differentially expressed genes in that pathway. We focused on pathways with a relatively well-known structure on different reported levels of the plant’s defense response including (i) early processes related to MAMP-triggered defense and the subsequent production of reactive oxygen species (ROS; Figure 6A), (ii) downstream signaling [e.g. mediated by SA (Figure 6B) and JA (Figure 6C)] leading to the production of defense-related components (e.g. different types of PR proteins), and (iii) general stress responses such as those occurring via the phenyl propanoid pathway (Figure 6D). The induction or downregulation of genes was visualized in Figure 6 by red and green boxes, respectively, allowing visual comparison of general gene modulation during ISR-prime, ISR-boost and BIDR. By combining this overview with results shown earlier in this manuscript the following overall conclusions can be drawn.
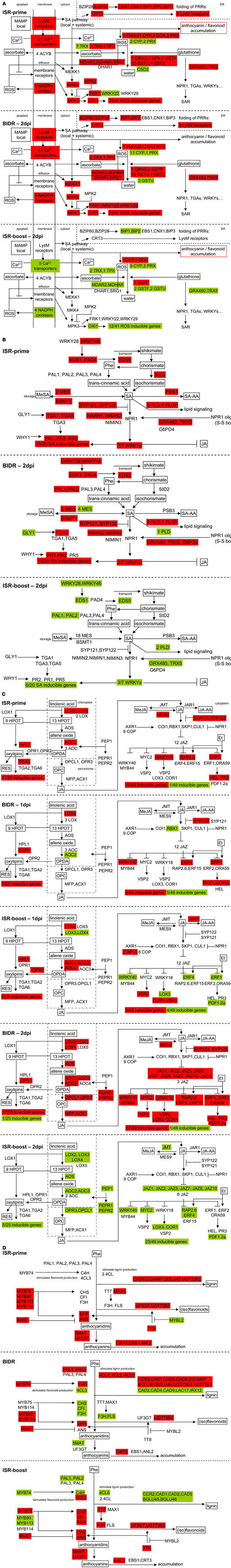
Figure 6. Overview of modulation of expression of genes in four defense-related pathways as a result of ISR and BIDR. Selected pathways include those involved in MAMP-triggered defense and subsequent ROS-production (A) (based on Asai et al., 2002; Kaupp and Seifert, 2002; Mittler et al., 2004; Wan et al., 2004; Kotchoni and Gachomo, 2006; Pitzschke et al., 2006; Tsuda et al., 2008; Van Breusegem et al., 2008; Li et al., 2009; Christensen et al., 2010; Foyer and Noctor, 2011; Ranf et al., 2011), those mediated by SA (B) (based on Shah, 2003; Dong, 2004; Katagiri, 2004; Glazebrook, 2005; Wang et al., 2006; Krinke et al., 2007; van Leeuwen et al., 2007), or JA (C) (based on Sasaki et al., 2001; Stintzi et al., 2001; Glazebrook et al., 2003; Delker et al., 2006; Ndamukong et al., 2007; van Leeuwen et al., 2007; Wasternack, 2007; Chico et al., 2008; Koornneef et al., 2008; Mueller et al., 2008; Lee et al., 2009; Fonseca et al., 2010) and the phenylpropanoid pathway leading to the production of lignin, flavonoids, and anthocyanins (D) (based on Winkel-Shirley, 2002; Routaboul et al., 2006; Ferrer et al., 2008). For each pathway except for the JA-pathway three panels can be distinguished: Upper panel: overview of modulated gene expression during ISR-prime in plants 2 days post-inoculation with T. hamatum T382 vs. mock-treated control plants. Middle panel: overview of modulated gene expression during BIDR in plants 2 days post-inoculation with B. cinerea vs. mock-treated control plants. Lower panel: overview of modulated gene expression during ISR-boost in plants pretreated with T. hamatum T382 vs. mock-treated control plants, both 2 days post-inoculation with B. cinerea. For the JA-pathway five panels can be distinguished because both 1 and 2 days post-inoculation with B. cinerea are shown for ISR-boost and BIDR. Up- and downregulated genes are shown in red and green respectively. Gene names are in conformity with TAIR annotation.
Trichoderma hamatum T382 Induces a MAMP-Triggered Defense Reaction in the Plant
In contrast to what was generally observed for BCOs of bacterial origin (Verhagen et al., 2004; Cartieaux et al., 2008) which are believed not to significantly alter gene expression upon treatment, addition of Trichoderma hamatum T382 to the roots of the plant triggers a clear and pronounced defense response in the leaves on the second day after the treatment. Our finding is supported by the recent observation that Bacillus cereus AR156 treatment to the roots of A. thaliana activates expression of defense-related genes PR1, PR2, PR5, and PDF1.2 in the leaves (Niu et al., 2011) thereby inducing ISR against Pseudomonas syringae pv. tomato DC3000. Furthermore, a striking analogy can be observed between the biological processes that are induced during ISR-prime and BIDR. Indeed, based on the results for the enriched biological processes (Table 2) and genes (Table S4 in Supplementary Material; Figure 6A) it can be concluded that treatment with T. hamatum T382 results in a microbe-associated molecular pattern (MAMP)-induced defense response in A. thaliana leaves. The importance of MAMP-triggered immunity in the plant response to Trichoderma spp. has been described by Lorito et al. (2010). According to this review, Trichoderma-induced ISR includes the increase of the plant’s basic immunity or MAMP-triggered immunity upon the detection of a variety of MAMPs that are produced by the Trichoderma spp. This suggests that MAMP-triggered immunity by Trichoderma spp. at the roots can affect MAMP-triggered immunity in the leaves.
The MAMPs by which the plant recognizes T. hamatum T382, could be chitin-related because we observe a clear induction of “response to chitin” (Table 2) and several chitinases (Table S4 in Supplementary Material). However, it is not clear whether recognition of chitin from T. hamatum T382 at the roots would also be reflected by similar alterations in gene expression in systemic tissues such as leaves. In our results on BIDR we also see a clear induction of “response to chitin” and the MAMP-defense pathway in systemic leaves (Table 2). Nevertheless, MAMP recognition is known to result in SID2-dependent accumulation of SA and concomitant activation of the SA-signaling pathway both in local and systemic leaves, leading to increased resistance against subsequent pathogen infections (Mishina and Zeier, 2007; Tsuda et al., 2008), as reflected in our data by the induction of “response to SA” (Table 2). Additionally, Figure 6B clearly shows that during ISR-prime SA is indeed produced via isochorismate (through the actions of SID2) instead of via phenylalanine (through the actions of PAL1–4), which is the main source of SA during BIDR. The MAMP-triggered SA-signal activates ER-localized proteins that are involved in the folding of defense-related proteins (Li et al., 2009; Christensen et al., 2010) through the actions of TF BZIP60 (Iwata et al., 2008), which are all also upregulated in our data (Figure 6A; Table S4 in Supplementary Material). The latter is also the case for the LysM domain containing proteins, which are MAMP recognition receptors (PRRs; Li et al., 2009; Table S4 in Supplementary Material). Activation of PRRs leads to the onset of (i) the phenylpropanoid pathway leading to the accumulation of anthocyanins as reflected by the induction of “biosynthesis of anthocyanins” and “metabolism of anthocyanins” (Table 2; Figure 6D), (ii) a MAPK signaling cascade indicated by the induction of, e.g. MKK4 and MPK3 (Asai et al., 2002; Wan et al., 2004; Figure 6A; Table S4 in Supplementary Material), (iii) cell wall reinforcement, as reflected by the induction of PEN3 (Table S4 in Supplementary Material), and (iv) an increased Ca2+ influx in the cell by ion channels in the plasma membrane that are regulated by glutamate binding (Ranf et al., 2011), as reflected by the induction of “calcium ion homeostasis” and several Ca2+ transporters (Table 2; Table S4 in Supplementary Material). Ultimately Ca2+ regulates the channel activity of cyclic nucleotide-gated channels (Kaupp and Seifert, 2002), of which several are upregulated in our experiments, leading to the induction of the hypersensitive response (Moeder and Yoshioka, 2008) and the concomitant programmed cell death. Both processes are included in the list of upregulated biological processes during ISR-prime. In addition to Trichoderma-derived MAMPs inducing the defense response during ISR-prime, pectin could be involved in priming too. Indeed, our data demonstrate also upregulation of wall-associated kinases, which are known to interact with pectin released from the plant cell wall and to subsequently activate the MAPK cascade transferring the signal to the nucleus of the cell (Ringli, 2010). Furthermore, it has been demonstrated that MAMP-perception also results in systemic acquired resistance (SAR) in A. thaliana (Mishina and Zeier, 2007), which is reflected in our data by the induction of “systemic acquired resistance” and “regulation of SAR” during ISR-prime (Table 2).
In our data we also observe an induction of genes involved in the “negative regulation of defense response” (Table 2). This might be explained by the fact that T. hamatum T382 is recognized by the plant as a “beneficial invader.” For instance, we observe a clear induction of both PYK10 and BGLU18, which encode proteins accumulating in ER bodies (Table S4 in Supplementary Material). The latter are recently discovered ER compartments reportedly linked to wounding and defense responses (Ogasawara et al., 2009). PYK10, for example, is proposed to be involved in maintaining the interaction between the beneficial endophytic fungus Piriformospora indica and A. thaliana by repressing the defense response (Sherameti et al., 2008). Additionally, both the gene encoding protein kinase OXI1 and the gene encoding monodehydroascorbate reductase MDHAR, two enzymes which are also linked to the interaction with this endophyte (Vadassery et al., 2009; Camehl et al., 2011), are upregulated by T. hamatum T382 (Table S4 in Supplementary Material). Interestingly, mutant analysis showed that the production of ascorbate by MDHAR keeps the interaction between plant and endophyte in a mutualistic state whereas the OXI1 pathway seems to control plant growth promotion by the endophyte. Our observation of PYK10, MDHAR, and OXI1 induction during ISR-prime could support a similar role. However, it should be noted again that, in contrast to the results from the P. indica – A. thaliana interaction, our study did not focus on gene expression in roots but in systemic leaves.
In the next sections we will zoom in on the different defense-related pathways and discuss their involvement in ISR in more detail.
The SA-pathway as a first key player in T. hamatum T382-induced ISR
While some reported studies on ISR conclude that in general it would occur independently of SA-signaling (van Loon et al., 1998; Yan et al., 2002; De Vleesschauwer et al., 2008; Segarra et al., 2009), others have contradicted this generalization both in A. thaliana (Tjamos et al., 2005; Hossain et al., 2007; Conn et al., 2008; Niu et al., 2011) and in other plants (De Meyer et al., 1999; Audenaert et al., 2002; Schuhegger et al., 2006; Alfano et al., 2007). Triggering of this pathway is reported for inducers of SAR including exogenous application of SA (Delaney et al., 1995), and plant defense activators, such as benzo-(1,2,3)-thiadiazole-7-carbothioic acid S-methyl ester (BTH; Lawton et al., 1996), β-aminobutyric acid (BABA; Zimmerli et al., 2000; Ton and Mauch-Mani, 2004) and thiamin (Ahn et al., 2005). SAR is a systemic defense response that is dependent on the SA-pathway (Ward et al., 1991; Uknes et al., 1992; Delaney et al., 1994) and leads via the actions of NPR1 (Cao et al., 1994, 1997) and WRKY TFs (Jaskiewicz et al., 2011) to the activation of pathogenesis-related proteins (PRs) such PR1, PR2, and PR5 (Ward et al., 1991; Uknes et al., 1992). Subsequent pathogen challenge on plants that display SAR leads to augmented defense-related responses in the whole plant resulting in a long-lasting and increased resistance of the plant toward a broad-spectrum of pathogens (Ryals et al., 1996). As discussed previously, we observe a strong induction of “systemic acquired resistance” and “regulation of SAR” during ISR-prime (Table 2). The similarity between SAR and ISR-prime is further highlighted by the induction of the entire SA-pathway during ISR-prime including WRKY6, WRKY53, PR1, PR2, and PR5 (Figure 6B) and by the analysis of NahG, npr1, and sid2 mutants (Figure 4). Previous detailed transcriptome analysis of SAR led to the identification of the PR1-regulon, consisting of 30 genes that were coexpressed with the SAR-marker PR1 in SAR-related conditions (Maleck et al., 2000). Interestingly, in the present study this regulon was also significantly induced during ISR-prime (p = 1.1 × 10−15; Table S4 in Supplementary Material).
Thus, according to our data on both gene expression and mutants, the SA-pathway is indeed a key player in the induction of ISR(-prime) as is the case in SAR. Remarkably, we observe that during ISR-prime SA was synthesized from chorismate rather than via the phenylalanine pathway, the route that is operative during the BIDR (Figure 6B). The active role of the chorismate pathway in ISR was further confirmed by the results on the sid2 mutant (Figure 4). Although the phenylalanine pathway is generally considered the main route of SA-synthesis, the chorismate way is postulated as an important alternative route for the production of SA required for defense responses (Strawn et al., 2007) such as MAMP-triggered defense and SAR (Nawrath and Métraux, 1999). Additionally, our results on the npr1 mutant (Figure 4) confirmed the role of TF NPR1 as a mediator of ISR (Pieterse et al., 1998). However, in this study on ISR triggered by bacterial BCOs, NPR1 was demonstrated to involve Et- and JA-signaling but not SA, whereas SA-dependent activation of NPR1 is considered characteristic for SAR (Cao et al., 1997). Although in our data NPR1 is not induced at the transcriptional level during ISR-prime we did observe an upregulation of GRX480 and several genes encoding thioredoxins, reflecting the activation of NPR1 in the cytoplasm (Cao et al., 1997; Ndamukong et al., 2007).
The JA-pathway as a second important player in T. hamatum T382-induced ISR
Both gene expression and mutant analysis data confirmed the involvement of the JA-pathway in ISR (Table 2; Figures 4 and 6C), which supports earlier findings (Pieterse et al., 1998; Niu et al., 2011). More detailed analysis in our study indicates that the pathway is not induced during ISR-prime (Figure 6C). On the first day of ISR-boost, however, a clear but transient (reinforcement of the) activation of the “biosynthesis of JA” and “response to JA” is observed (as a result of the priming of the JA-pathway by the ISR) suggesting a role of this pathway in countering the B. cinerea infection during the ISR-boosted defense response as was described for bacterial BCOs (Pieterse et al., 1998) and for hexanoic acid (Kravchuk et al., 2011). Furthermore, while the transcription factor MYC2 is regarded a marker gene of the ISR induced by rhizobacteria (Pozo et al., 2008), in our study its expression was not significantly affected during the ISR-prime neither during ISR-boost. However, our mutant analysis clearly showed absence of ISR in myc2 mutants (Figure 4), thereby confirming that it is essential for ISR.
The Et-pathway shows no or minimal involvement in T. hamatum T382-induced ISR
Our observations on a limited role of the Et-pathway in T. hamatum T382-induced ISR, differs from earlier findings in rhizobacteria-mediated ISR in which both Et and JA are postulated to play key roles (Pieterse et al., 1998). Several reasons for this discrepancy can be given. First, rhizobacteria and the fungus T. hamatum T382 do not necessarily use the same working mechanism to trigger ISR in plants. In this aspect our observed induction of “response to chitin” and several chitinases as a consequence of recognition by the plant of fungal (T382) chitin-like MAMPs might be explanatory. Second, since in rhizobacteria-mediated ISR Et is proposed to act downstream of JA (Pieterse et al., 1998) and our results do show a clear induction of JA-related pathways during T. hamatum T382-mediated ISR, it can not be excluded that in the latter Et-induction occurs outside the time frame analyzed in the present study. Third, it has been shown that plants undergoing rhizobacteria-mediated ISR do not display induction of JA/Et-responsive genes during ISR-prime (Pieterse et al., 1998). Therefore, the unaltered expression of JA-Et markers (like PDF1.2) during ISR-prime does not exclude that the Et-pathway is not involved in the induction of T. hamatum T382-mediated ISR. The unaltered ISR-effect observed in the etr1 and ein2 mutants (Figure 4) only indicates that the Et-pathway, or at least the part in which ETR1 and EIN2 are involved, does not play an important role in T. hamatum T382-mediated ISR.
The phenylpropanoid pathway is involved in T. hamatum T382-induced ISR
In accordance with the results on gene expression (Table 2), anthocyanin measurements (Figure 5) and the tt, chs, and f3h mutant (Figure 4) studies, an overview of ISR-modulated expression of genes involved in the phenylpropanoid pathway showed that this pathway is clearly involved in ISR, and that mainly the final branch of the pathway, leading to the production of anthocyanins, is activated (Figure 6D). The latter is characterized by absence of induction of enzymes that catalyze the first steps of the phenylpropanoid pathway like the PAL proteins, MYB74, C4H, and 4CL2 during ISR-prime (Figure 6D). These findings for Trichoderma-induced priming are very similar to the results of Shetty et al. (2011). They found that silicon-induced priming of roses is characterized by upregulation of CHS expression and subsequent elevated levels of phenolic acids and flavonoids in response to infection by rose powdery mildew.
In line with the observation that T. hamatum T382-induced ISR-prime closely resembles MAMP-triggered immunity and the resulting SAR, it can be concluded that despite overall similarities with BIDR, ISR-prime can be distinguished by the much more pronounced induction of the SA-pathway (Figure 6B), production of SA via isochorismate instead of phenylalanine (Figure 6B), the absence of any involvement of the JA- and the Et-pathway (Table 2), and the induction of the phenylpropanoid pathway instead of the camalexin pathway as the main source of secondary metabolites (Table 2; Figure 6D).
The most remarkable difference between ISR and BIDR/SAR is the absence in ISR of downregulation of general processes like photosynthesis and translation (Table 2). The latter might explain why ISR-prime as fully fledged defense reaction is much better tolerated by the plant than pathogen-induced defense responses. A negative correlation between SAR and plant growth and metabolism has been frequently reported in literature (Cohen and Kuć, 1981; Heil et al., 2000; Latunde-Dada and Lucas, 2001; Louws et al., 2001; Bolton, 2009). In contrast, we never observed such negative effects in any of our experiments on T. hamatum T382-induced ISR.
Trichoderma hamatum T382 Primes the Plant to Respond more Quickly to Pathogen Infection
On the first day after B. cinerea inoculation, ISR-boost was characterized by a transient activation of JA-biosynthesis which was not observed during BIDR and reinforcement of the BIDR-related induction of JA-response (Table 2). Additionally, the induction of the defense-related process “response to wounding,” which is linked to the JA-pathway, was also reinforced by the ISR. Furthermore, during ISR-boost, the defense-related process “response microbial phytotoxin” and the biosynthesis of secondary metabolites (e.g. anthocyanins, galactolipids) were induced, whereas during BIDR these defense responses were not yet activated on the first day after B. cinerea inoculation. These findings correspond to the current view that (i) ISR primes the plant to react faster to subsequent pathogen infections (Prime-A-Plant Group et al., 2006; Pozo et al., 2008; De Vleesschauwer et al., 2009) and that (ii) Trichoderma spp. are able to activate ISR that leads to such primed responses (Lorito et al., 2010).
On the second day after B. cinerea inoculation the production of secondary metabolites (e.g. anthocyanins and flavonoids; Figure 6D; Table 2), was still reinforced. With respect to anthocyanins and flavonoids many different in planta functions have been proposed including their role as antioxidants or protectants against different types of abiotic (e.g. UV) and biotic stress. Regarding the latter, it is suggested that both compounds accumulate around fungal infection sites to protect host cells from oxidative damage as a result of the defense-related ROS-production (Hipskind et al., 1996; Kangatharalingam et al., 2002; Treutter, 2006). Furthermore, anthocyanins have been reported to play a role in attenuating defense-related ROS-production (Figueroa-Balderas et al., 2006; Senthil-Kumar and Mysore, 2010), while some flavonoids have direct antifungal activity (Treutter, 2006; Buer et al., 2010). Like flavonoids, galactolipids are also known to display antifungal effects. For instance, esters formed between jasmonates and galactolipids can inhibit the growth of Botrytis cinerea (Kourtchenko et al., 2007). The enhanced (“primed”) production of these antifungal compounds during ISR-boost might give a first explanation for the observed increased resistance toward B. cinerea resulting from T382-induced ISR.
Trichoderma hamatum T382 Restrains the Defense Response after B. cinerea Inoculation
On the second day after B. cinerea inoculation, we observed a clear moderation of BIDR during ISR-boost (Table 2), possibly as a result of the priming and the subsequent increased inhibition of B. cinerea proliferation (and concomitant BIDR-triggering) in T. hamatum T382 treated plants. The major induction of the defense system during BIDR was thus restrained during ISR-boost, as was the case for various defense processes (e.g. “defense response to fungus,” “response to chitin,”…), JA-, Et-, and ABA-responses, SA- and JA-mediated signaling and the production of camalexin. Such moderation of the normal defense response against pathogens has been previously observed for other BCAs, from both fungal (Wen et al., 2005) and bacterial (Cartieaux et al., 2008) origin. When studying the respective pathways in more detail, we observed the same sets of genes being upregulated during BIDR and downregulated during ISR-boost, thereby confirming the restraining of the BIDR (Figure 6). It is possible that during ISR-prime defensive proteins or their respective transcripts have accumulated so that the levels of these compounds at the moment of Botrytis inoculation are already higher in T. hamatum T382-treated plants than in control plants. This would leave out the need for a further strong induction of the defense response at the level of gene expression and, hence, would result in the observed restraining of the defense response during ISR-boost.
The induction of compounds with an antioxidant activity (e.g. anthocyanins, flavonoids), together with the observed restraining of the ROS response during ISR-boost (Figure 6A), might give a reasonable second explanation for the T382-induced ISR efficient against B. cinerea. The importance of ROS response in ISR is also reflected in our results for the vtc1 and rbohD mutant studies (Figure 4). Both mutants display increased resistance to B. cinerea infection and additional T. hamatum T382 treatment did not further boost this enhanced resistance. However, it should be noted here that the absence of an ISR-effect in mutants with already increased resistance to pathogens (such as B. cinerea) can be related to the fact that an additional boost of defense-related compounds by the biocontrol organism might not be possible in the plant. Different findings support the general idea that infection by B. cinerea is favored by ROS-production in the plant. First, as for other necrotrophic pathogens, infection and colonization of the plant will be promoted by necrosis-inducing components such as ROS (Govrin and Levine, 2000; Glazebrook, 2005). It has indeed been shown that B. cinerea virulence correlates with the intensity by which the plant produces ROS as defense reaction (Temme and Tudzynski, 2009). Secondly, it is known that this pathogen is actively secreting compounds to elicit an oxidative burst and subsequent programmed cell death (Govrin et al., 2006). Additionally, it has been demonstrated that B. cinerea infection can be suppressed by spraying antioxidants on plants (Elad, 1992).
In view of the very recent report by Brotman et al. (2012), it is worth here to briefly compare our results with their findings. In their study analysis of gene expression in A. thaliana leaves was done on a restricted set of 137 genes during the ISR induced by another Trichoderma species, T. asperelloides T203, and effective against the bacterial pathogen Pseudomonas syringae. During ISR-prime induced by T. asperelloides T203 they identified 15 up- and 2 downregulated genes. Four of these upregulated genes are also induced during T. hamatum T382-induced ISR-prime, namely WRKY40, WRKY55, TAT3, and PR5. The first one is a gene that is induced by SA-signaling, encoding a transcription factor that interacts with WRKY18 to form a negative feedback loop during MAMP-triggered defense (Pandey et al., 2010). Remarkably, the WRKY18 gene is also induced during T. hamatum T382-ISR-prime. The TAT3 gene is controlled by the JA-signaling pathway in which NPR1 acts as a positive regulator. This induction is linked to the activation of ROS signaling by NPR1 (Brosché and Kangasjärvi, 2012) upon activation of SA-signaling during the MAMP-response. Overall the similarities in ISR-prime between our results and these of Brotman et al. (2012) are low. A possible explanation for this discrepancy might be the fact that both studies characterize ISR-prime at different time points after Trichoderma treatment (4 dpTi in Brotman et al., 2012 vs. 2 dpTi in our study). As shown in Figure 2A, the induced response during ISR-prime triggered by T. hamatum T382 is transient, peaking in the first few days after Trichoderma treatment and then decreasing again until basal expression levels are reached.
Comparing our results on ISR-boost with their data is even more complex. First of all, as ISR-boost in both studies is initiated by different types of pathogen (fungal vs. bacterial) with different infection strategies, comparison of induced responses are expected to also significantly differ. Furthermore their study is restricted to only a comparison between the tripartite interaction (A. thaliana – T. asperelloides T203 – P. syringae) with untreated and mock-inoculated control plants, one can not rule out the effects of the pathogen (P. syringae) infection, as is the case in our study (BIDR). Nevertheless, some overlap in differentially expressed genes can be identified in both studies such as the induction of LTP4 and LOX2, and a downregulation of WRKY40. The LTP4 gene encodes a lipid transfer protein, a member of a family 14 of pathogenesis-related peptides (PR14) with reported in vitro antimicrobial activity (Sels et al., 2008). The JA-responsive LOX2 gene encodes a lipoxygenase that is required for JA production (Bell et al., 1995) during pathogen infection (Spoel et al., 2003). Enhanced expression of this gene also takes place during rhizobacteria-induced ISR-boost (Pineda et al., 2012).
Conclusion
In this study we demonstrated that application of T. hamatum T382 to the roots of A. thaliana results in an increased resistance to subsequent leaf infections by the necrotrophic pathogen B. cinerea, characterized by a significant reduction in symptom development. Further analysis of this ISR through an extended transcriptome study, more specifically of the responses induced before (ISR-prime) and after addition (ISR-boost) of B. cinerea, and comparison with the more intensively studied pathogen-induced responses (like SAR and MAMP-triggered defense) led to the following general conclusions.
During ISR-prime T. hamatum T382 evokes a fully fledged MAMP-triggered defense response in Arabidopsis leaves on the second day after the treatment that leads to a SAR-like response and that prepares the plant to react more quickly to subsequent pathogen inoculation. Both the SA- and NPR1 were identified as important players in the signaling of this ISR-response. However, in contrast to pathogen-induced defense responses like SAR and BIDR (Cohen and Kuć, 1981; Heil et al., 2000; Latunde-Dada and Lucas, 2001; Louws et al., 2001; Bolton, 2009), the T. hamatum T382-induced ISR-response did not negatively affect translation and photosynthesis, processes that are essential for growth and survival of the plant. This could explain why ISR is relatively well-tolerated by the plant and does not cause any visually observable negative effects on plant growth previously reported for SAR.
The primed defense response that is mounted in T. hamatum T382-treated plants upon pathogen inoculation is characterized by the faster induction of defense processes, JA-synthesis and JA-response, and the production of several secondary metabolites like anthocyanins, flavonoids and galactolipids. The observed induction of the JA-pathway is transient since at a later stage, the Botrytis-induced defense responses are in general reduced as compared to those in plants not pretreated with T. hamatum T382 (BIDR), although this is not the case for the production of anthocyanins and flavonoids. Interestingly, the restrained induction of ROS together with an increased production of antioxidants (like anthocyanins and flavonoids) during ISR-boost could explain a reduction in plant cell necrosis, a process which is considered favorable for necrotrophic pathogens such as B. cinerea, and as such for the observed decrease in B. cinerea-caused disease symptoms. However, the role of anthocyanins and flavonoids in ISR could extend a mere antioxidant activity since they are also known to display direct antimicrobial properties (Buer et al., 2010). Furthermore, the induction of both JA- and galactolipid biosynthesis during ISR-boost might point to a second route for producing antifungal compounds to neutralize the B. cinerea infection (Kourtchenko et al., 2007).
Materials and Methods
Biological Materials
Cultivation and spore harvesting of Trichoderma hamatum strain T382 (kindly provided by Tom De Ceuster, DCM, Sint-Katelijne-Waver, Belgium) and Botrytis cinerea strain B05-10 (kindly provided by Rudi Aerts, Katholieke Hogeschool Kempen, Geel, Belgium) was performed as described previously (Broekaert et al., 1990). Arabidopsis thaliana Col-0 plants were obtained from the European Arabidopsis thaliana stock centre (NASC). The Arabidopsis thaliana mutants sid2, npr1, myc2, ein2, etr1, chs, f3h, rbohd, vtc1, and tt were kindly provided by Prof. U. Conrath (RWTH Aachen University, Aachen, Germany), Prof. X. Dong (Duke University, Durham, NC, USA), Prof. S. Berger (Institut für Pflanzenbiochemie, Halle/Saale, Germany), Prof. F. Ausubel (Massachusetts General Hospital, Boston, MA, USA), Prof. J. Glazebrook (University of Maryland, Maryland, MD, USA) and the Arabidopsis Biological Resource Center, Columbus, OH, USA (accession numbers CS237 for etr1, CS3071 for ein2, N3130 for tt, N653439 for f3h, N671192 for chs, N671557 for rbohD and N8326 for vtc1), respectively. The transgenic Arabidopsis thaliana line containing the NahG gene was obtained from J. Ryals (Ciba Geigy, Research Triangle Park, NC, USA). The transgenic lines carrying PDF-promoter-GUS constructs were developed in house (De Coninck et al., 2010); the constructs contain the GUS-gene linked to the promoter fragments (1.25 kb) upstream of the predicted start codon of the PDF1.2a (At5g44420) and PR1 (At2g14610) genes.
Disease Assays
Arabidopsis thaliana Col-0 plants were grown in untreated and unsterilized soil (“DCM potgrond voor Zaaien and Stekken”, DCM, Sint-Katelijne-Waver, Belgium) in a growth chamber with 21°C daytime temperature, 18°C night-time temperature, 75% humidity and a 12-h day-light cycle with a light intensity of approximately 120 μmol/m2 s. Three weeks old plants were treated with Trichoderma hamatum T382 by pipetting a 50 μl spore suspension (2 × 107/ml) directly onto the roots. Six days later, these and an equal number of mock-treated plants were inoculated with Botrytis cinerea B05-10 as described previously (Thomma et al., 1998). Briefly, a 5 μl drop of a Botrytis cinerea spore suspension (5 × 105/ml in 1/2 PDB) was inoculated onto two leaves per plant. Disease symptoms were scored by measuring the diameters of the necrotic lesions on various days after B. cinerea inoculation. The disease assay was repeated 12 times with a total of 2000 plants. Each day, lesions diameters were measured and the average lesion diameters of treated and untreated plants were compared using a two-sample one-sided Student’s t-test (alternative hypothesis average lesion diameter of treated plants < average lesion diameter of untreated plants) implemented in R (R Development Core Team, 2011).
Determination of the Level of Trichoderma hamatum T382
Trichoderma hamatum T382 density was determined by (i) dilution plate enumeration on Trichoderma selective medium and (ii) qPCR using Trichoderma hamatum T382 specific primers as previously described (Lievens et al., 2007).
In short, 10 g of each root sample was washed intensely in 100 ml phosphate buffer and 10 g of each soil sample was mixed with 90 ml phosphate buffer in a blender for 30 s at high speed. Next, a 10-fold dilution series was prepared and 100 μl of each dilution was plated and spread on a Trichoderma selective medium (Chung and Hoitink, 1990) in triplicate. After 5 days of incubation at 25°C in the dark, the colonies were counted.
In parallel, genomic DNA was extracted from 0.5 ml washed root or 0.5 g soil sample using Mo Bio Ultraclean Soil DNA Isolation kit according to the manufacturer’s specifications (Mo Bio Laboratories, Solana Beach, CA, USA). DNA extracts were diluted 10-fold and stored at −20°C. To specifically detect Trichoderma hamatum T382, qPCR amplification was performed in a total volume of 25 ml using the intercalating dye SYBR1 Green I on a SmartCyclerII1 instrument (Cepheid, Sunnyvale, CA, USA). Each reaction mixture contained 2 ml of the target DNA extract, 12.5 ml of the QuantiTectTM SYBR1 Green PCR Master Mix (Qiagen, Inc., Valencia, CA, USA), 0.625 ml of each primer (20 mM), and 9.25 ml sterile distilled water. Sequences of the Trichoderma hamatum T382 specific primers are shown in Table S3 in Supplementary Material. Thermal cycling conditions consisted of 10 min at 95°C followed by 45 amplification cycles of 15 s at 95°C, 30 s at 60°C, and 30 s at 72°C with a final 2-min elongation step at 72°C. Fluorescence was detected at the end of the elongation phase of each cycle. To evaluate amplification specificity, melt curve analysis was performed at the end of the PCR run. A melt curve profile was obtained by slowly heating the mixture from 60 to 95°C at 0.2°C/s with continuous measurement of fluorescence. Standard curves were generated by plotting the threshold cycle (Ct) of a 10-fold dilution series of standard DNA against the logarithm of the concentration. The regression line was used to calculate the DNA concentration of Trichoderma hamatum T382 in the samples via the obtained Ct-values (Brouwer et al., 2003; Lievens et al., 2006).
qRT-PCR
Primers were developed using Primer3 software (Rozen and Skaletsky, 2000), primer sequences are shown in Table S3 in Supplementary Material. Before qRT-PCR the ideal annealing temperature of each primer was determined in a regular PCR. Six different sets of leaves from mock-treated and T. hamatum T382 treated were collected daily and used for qRT-PCR to study gene expression during ISR-prime. For ISR-boost systemic leaves were collected from five of the same sets of mock-treated and Trichoderma hamatum T382-treated plants after additional Botrytis cinerea inoculation. For qRT-PCR validation of the microarray results, an additional biological repeat was used, for which samples were collected in the same manner. RNA extraction, DNase treatment and reverse transcription were done as described previously (Mirouze et al., 2006). The qRT-PCR analysis was carried out using the StepOnePlus System and Power SYBR Green PCR Master Mix (Applied Biosystems). The PCR parameters were: 10 min at 95°C, 40 cycles of amplification (10 s at 95°C, 10 s at 58°C, 10 s at 72°C) and a melting curve stage (15 s at 95°C, 1 min at 60°C increased to 95°C with steps of 0.3°C). Melt curve analysis was performed as described in the previous section. Elongation factor 1α (EF1α; At5g60390) was used as a reference gene (Becher et al., 2004). Transcript levels were normalized to the respective transcript level of EF1α. Relative log2 induction ratios of treated samples compared with the mock treatment were calculated based on the ΔΔCt method (Livak and Schmittgen, 2001).
Microarrays
Samples from three independent sets of both ISR-primed (At + T) and ISR-boosted (At + B + T) plants, and from the corresponding control plants (At and At + B) were used for microarray analysis. For all samples a dye swap was performed. Three biological replicates were included to assess technical and biological variation. RNA was extracted using a combination of Trizol® Reagent (Invitrogen Life Technologies, Paisley, UK) and a Qiagen RNeasy kit (Qiagen, Hilden, Germany). RNA quality control, labeling, hybridizations and imaging were performed at the MicroArray Facility (VIB, Leuven) according to the protocols specified on the web site (http://www.microarrays.be). Agilent Arabidopsis 4-pack microarrays (Agilent Technologies, Palo Alto, CA, USA) were used, normalization was done with the accompanying software (Agilent Technologies, Palo Alto, CA, USA). The correlations among the replicates (0.98–0.99 among technical replicates and 0.92 among dye swaps) and the scatter plots confirmed the high quality and reproducibility of the microarray experiments. Data from different hybridizations were centered and scaled using quantile normalization, implemented in R (R Development Core Team, 2011). For ISR-prime, ISR-boost and BIDR differentially expressed genes, defined as genes of which the expression level is significantly modified (raised or reduced) in a specific condition as compared to a control treatment were selected using the (adapted) t-test developed by Tusher et al. (2001). Analysis of enrichment of gene ontology (GO) terms was performed on the five sets of differentially expressed (DE) genes that were produced (ISR-prime, BIDR on 1 and 2 dpBi, ISR-boost on 1 and 2 dpBi) as described previously (Tavazoie et al., 1999). In short, each gene in the DE gene sets was attributed its corresponding GO terms (Gene Ontology Consortium, 2000) from the biological process ontology using the GO annotation on the TAIR website (Lamesch et al., 2012). Next, the hypergeometric probability statistic was used to calculate the probability that at random each GO term would have the observed number of instances among the DE gene sets as follows:
where m is the total number of DE genes in a specific set, N is the total number of genes in the genome for which GO annotation is available, n is the total number of genes that are annotated with a specific GO term and k is the number of genes which belong to the set of DE genes and are annotated with that GO term. Hypergeometric probability calculations were implemented in R (R Development Core Team, 2011).
GUS-Staining
Histochemical GUS-staining was performed as described (De Bondt et al., 1994) except that tissues soaked in substrate buffer were vacuum infiltrated for 5 min. prior to overnight incubation at 37°C.
Anthocyanin Analysis
Anthocyanin content was determined using a procedure modified from that of Neff and Chory (1998). For six replicates of four plants each, leafs of both ISR-primed (At + T) and ISR-boosted (At + B + T) plants, and from the corresponding control plants (At and At + B) were weighed, powdered in liquid nitrogen, and total plant pigments were extracted 48 h at 4°C with 0.6 ml methanol containing 1% HCl (w/v). After centrifugation (14000 rpm, 5 min), anthocyanin was extracted with 0.6 ml of chloroform. The absorbance of the aqueous phase was measured at 535 and 657 nm (A535–A657) and corrected for weight.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Prof. U. Conrath, Dr. K. Thevissen, and Dr. G. Beckers for critical reading of the manuscript. This research was funded by a postdoctoral fellowship from IWT-Vlaanderen to Janick Mathys and Barbara De Coninck and from KU Leuven (PDM) to Mieke Vanhaecke; a doctoral fellowship from IWT-Vlaanderen to Kaat De Cremer and Pieter Timmermans; and project funding by KULeuven (GOA/2008/11) and FWO Vlaanderen (G.0414.09N and Krediet aan navorsers 1.5.141.09N). The authors acknowledge DCM (Sint-Kathelijne-Waver, Belgium) for providing the necessary material and knowledge concerning T. hamatum T382.
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/Plant-Microbe_Interaction/10.3389/fpls.2012.00108/abstract
Table S1 List of genes and fold changes that are differentially expressed during ISR-prime, ISR-boost and BIDR.
Table S2 Complete overview of enriched biological processes.
Table S3 Sequences of all primers used in qRT-PCR analyses.
Table S4 List of At numbers, names, log ratios, and standard errors of genes that are differentially expressed during ISR-prime, ISR-boost and BIDR for the biological processes: MAMP-triggered defense, SA-pathway, JA-pathway, phenylpropanoid pathway, and PR1-regulon.
References
Ahn, I. P., Kim, S., and Lee, Y. H. (2005). Vitamin B1 functions as an activator of plant disease resistance. Plant Physiol. 138, 1505–1515.
Alfano, G., Ivey, M. L. L., Cakir, C., Bos, J. I. B., Miller, S. A., Madden, L. V., Kamoun, S., and Hoitink, H. A. J. (2007). Systemic modulation of gene expression in tomato by Trichoderma hamatum 382. Phytopathology 97, 429–437.
Alonso, J. M., Hirayama, T., Roman, G., Nourizadeh, S., and Ecker, J. R. (1999). EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284, 2148–2152.
Asai, T., Tena, G., Plotnikova, J., Willmann, M. R., Chiu, W. L., Gomez-Gomez, L., Boller, T., Ausubel, F. M., and Sheen, J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415, 977–983.
Attitalla, I. H., Johnson, P., Brishammar, S., and Quintanilla, P. (2001). Systemic resistance to Fusarium wilt in tomato induced by Phytophthora cryptogea. J. Phytopathol. 149, 373–380.
Audenaert, K., De Meyer, G. B., and Höfte, M. M. (2002). Abscisic acid determines basal susceptibility of tomato to Botrytis cinerea and suppresses salicylic acid-dependent signaling mechanisms. Plant Physiol. 128, 491–501.
Barth, C., Moeder, W., Klessig, D. F., and Conklin, P. L. (2004). The timing of senescence and response to pathogens is altered in ascorbate-deficient mutant vitamin C-1. Plant Physiol. 134, 178–192.
Becher, M., Talke, I. N., Krall, L., and Krämer, U. (2004). Cross-species microarray transcript profiling reveals high constitutive expression of metal homeostasis genes in shoots of the zinc hyperaccumulator Arabidopsis halleri. Plant J. 37, 251–268.
Bell, E., Creelman, R. A., and Mullet, J. E. (1995). A chloroplast lipoxygenase is required for wound-induced accumulation of jasmonic acid in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 92, 8675–8679.
Berger, S., Bell, E., and Mullet, J. E. (1996). Two methyl jasmonate-insensitive mutants show altered expression of AtVsp in response to methyl jasmonate and wounding. Plant Physiol. 111, 525–531.
Bolton, M. D. (2009). Primary metabolism and plant defense – fuel for the fire. Mol. Plant Microbe Interact. 22, 487–497.
Broekaert, W. F., Terras, F. R., Cammue, B. P. A., and Vanderleyden, J. (1990). An automated quantitative assay for fungal growth inhibition. FEMS Microbiol. Lett. 69, 55–60.
Brosché, M., and Kangasjärvi, J. (2012). Low antioxidant concentrations impact on multiple signalling pathways in Arabidopsis thaliana partly through NPR1. J. Exp. Bot. 63, 1849–1861.
Brotman, Y., Lisec, J., Méret, M., Chet, I., Willmitzer, L., and Viterbo, A. (2012). Transcript and metabolite analysis of the Trichoderma-induced systemic resistance response to Pseudomonas syringae in Arabidopsis thaliana. Microbiology 158(Pt 1), 139–146.
Brouwer, M., Lievens, B., Van Hemelrijck, W., Van den Ackerveken, G., Cammue, B. P. A., and Thomma, B. P. H. J. (2003). Quantification of disease progression of several microbial pathogens on Arabidopsis thaliana using real-time fluorescence PCR. FEMS Microbiol. Lett. 228, 241–248.
Buer, C. S., Imin, N., and Djordjevic, M. A. (2010). Flavonoids: new roles for old molecules. J. Integr. Plant Biol. 52, 98–111.
Camehl, I., Drzewiecki, C., Vadassery, J., Shahollari, B., Sherameti, I., Forzani, C., Munnik, T., Hirt, H., and Oelmüller, R. (2011). The OXI1 kinase pathway mediates Piriformospora indica-induced growth promotion in Arabidopsis. PLoS Pathog. 7, e1002051.
Cancel, J. D., and Larsen, P. B. (2002). Loss-of-function mutations in the ethylene receptor ETR1 cause enhanced sensitivity and exaggerated response to ethylene in Arabidopsis. Plant Physiol. 129, 1557–1567.
Cao, H., Bowling, S. A., Gordon, A. S., and Dong, X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6, 1583–1592.
Cao, H., Glazebrook, J., Clarke, J. D., Volko, S., and Dong, X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88, 57–63.
Cartieaux, F., Contesto, C., Gallou, A., Desbrosses, G., Kopka, J., Taconnat, L., Renou, J. P., and Touraine, B. (2008). Simultaneous interaction of Arabidopsis thaliana with Bradyrhizobium sp. strain ORS278 and Pseudomonas syringae pv. tomato DC3000 leads to complex transcriptome changes. Mol. Plant Microbe Interact. 21, 244–259.
Cartieaux, F., Thibaud, M. C., Zimmerli, L., Lessard, P., Sarrobert, C., and David, P. (2003). Transcriptome analysis of Arabidopsis colonized by a plant-growth promoting rhizobacterium reveals a general effect on disease resistance. Plant J. 36, 177–188.
Chacón, M. R., Rodríguez-Galán, O., Benítez, T., Sousa, S., Rey, M., Llobell, A., and Delgado-Jarana, J. (2007). Microscopic and transcriptome analyses of early colonization of tomato roots by Trichoderma harzianum. Int. Microbiol. 10, 19–27.
Chang, C., Kwok, S. F., Bleecker, A. B., and Meyerowitz, E. M. (1993). Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science 262, 539–544.
Chico, J. M., Chini, A., Fonseca, S., and Solano, R. (2008). JAZ repressors set the rhythm in jasmonate signaling. Curr. Opin. Plant Biol. 11, 486–494.
Christensen, A., Svensson, K., Thelin, L., Zhang, W., Tintor, N., Prins, D., Funke, N., Michalak, M., Schulze-Lefert, P., Saijo, Y., Sommarin, M., Widell, S., and Persson, S. (2010). Higher plant calreticulins have acquired specialized functions in Arabidopsis. PLoS ONE 5, e11342.
Chung, Y. R., and Hoitink, H. A. J. (1990). Interactions between thermophylic fungi and Trichoderma hamatum in suppression of Rhizoctonia damping-off in a bark-amended container medium. Phytopathology 80, 73–77.
Cohen, Y., and Kuć, J. (1981). Evaluation of systemic resistance to blue mold induced in tobacco leaves by prior stem inoculation with Peronospora hysoscyami f. sp. tabacina. Phytopathology 71, 783–787.
Conklin, P. L., Saracco, S. A., Norris, S. R., and Last, R. L. (2000). Identification of ascorbic acid-deficient Arabidopsis thaliana mutants. Genetics 154, 847–856.
Conn, V. M., Walker, A. R., and Franco, C. M. (2008). Endophytic actinobacteria induce defense pathways in Arabidopsis thaliana. Mol. Plant Microbe Interact. 21, 208–218.
Craft, C. M., and Nelson, E. B. (1996). Microbial properties of composts that suppress damping-off and root rot of creeping bentgrass caused by Pythium graminicola. Appl. Environ. Microbiol. 62, 1550–1557.
De Bondt, A., Eggermont, K., Druart, P., Vil, M., Goderis, I., Vanderleyden, J., and Broekaert, W. F. (1994). Agrobacterium-mediated transformation of apple (Malus × domestica Bork.): an assessment of factors affecting gene transfer efficiency during early transformation steps. Plant Cell Rep. 13, 587–593.
De Coninck, B. M., Sels, J., Venmans, E., Thys, W., Goderis, I. J., Carron, D., Delaure, S. L., Cammue, B. P., De Bolle, M. F., and Mathys, J. (2010). Arabidopsis thaliana plant defensin AtPDF1.1 is involved in the plant response to biotic stress. New Phytol. 187, 1075–1088.
De Meyer, G., Bigirimana, J., Elad, Y., and Höfte, M. (1998). Induced systemic resistance in Trichoderma harzianum T39 biocontrol of Botrytis cinerea. Eur. J. Plant Pathol. 104, 279–286.
De Meyer, G., Capieau, K., Audenaert, K., Buchala, A., Métraux, J. P., and Höfte, M. (1999). Nanogram amounts of salicylic acid produced by the rhizobacterium Pseudomonas aeruginosa 7NSK2 activate the systemic acquired resistance pathway in bean. Mol. Plant Microbe Interact. 12, 450–458.
De Vleesschauwer, D., Chernin, L., and Höfte, M. M. (2009). Differential effectiveness of Serratia plymuthica IC1270-induced systemic resistance against hemibiotrophic and necrotrophic leaf pathogens in rice. BMC Plant Biol. 9, 9.
De Vleesschauwer, D., Djavaheri, M., Bakker, P. A. H. M., and Höfte, M. (2008). Pseudomonas fluorescens WCS374r-induced qystemic resistance in rice against Magnaporthe oryzae is based on pseudobactin-mediated priming for a salicylic acid-repressible multifaceted defense response. Plant Physiol. 148, 1996–2012.
Delaney, T. P., Friedrich, L., and Ryals, J. A. (1995). Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. U.S.A. 92, 6602–6606.
Delaney, T. P., Uknes, S., Vernooij, B., Friedrich, L., Weymann, K., Negrotto, D., Gaffney, T., Gut-Rella, M., Kessmann, H., Ward, E., and Ryals, J. (1994). A central role of salicylic acid in plant disease resistance. Science 266, 1247–1250.
Delker, C., Stenzel, I., Hause, B., Miersch, O., Feussner, I., and Wasternack, C. (2006). Jasmonate biosynthesis in Arabidopsis thaliana – enzymes, products, regulation. Plant Biol. (Stuttg.) 8, 297–306.
Duijff, B. J., Pouhair, D., Olivain, C., Alabouvette, C., and Lemanceau, P. (1998). Implication of systemic induced resistance in the suppression of fusarium wilt of tomato by Pseudomonas fluorescens WCS417r and by nonpathogenic Fusarium oxysporum Fo47. Eur. J. Plant Pathol. 104, 903–910.
Elad, Y. (1992). The use of antioxidants (free radical scavengers) to control grey and white moulds in various crops. Plant Pathol. 41, 417–426.
Ferrari, S., Plotnikova, J. M., De Lorenzo, G., and Ausubel, F. M. (2003). Arabidopsis local resistance to Botrytis cinerea involves salicylic acid and camalexin and requires EDS4 and PAD2, but not SID2, EDS5 or PAD4. Plant J. 35, 193–205.
Ferrer, J. L., Austin, M. B., Stewart, C. Jr., and Noel, J. P. (2008). Structure and function of enzymes involved in the biosynthesis of phenylpropanoids. Plant Physiol. Biochem. 46, 356–370.
Figueroa-Balderas, R. E., García-Ponce, B., and Rocha-Sosa, M. (2006). Hormonal and stress induction of the gene encoding common bean acetyl-coenzyme A carboxylase. Plant Physiol. 142, 609–619.
Fonseca, J. P., Menossi, M., Thibaud-Nissen, F., and Town, C. D. (2010). Functional analysis of a TGA factor-binding site located in the promoter region controlling salicylic acid-induced NIMIN-1 expression in Arabidopsis. Genet. Mol. Res. 9, 167–175.
Foyer, C. H., and Noctor, G. (2011). Ascorbate and glutathione: the heart of the redox hub. Plant Physiol. 155, 2–18.
Gaffney, T., Friedrich, L., Vernooij, B., Negrotto, D., Nye, G., Uknes, S., Ward, E., Kessmann, H., and Ryals, J. (1993). Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261, 754–756.
Gene Ontology Consortium. (2000). Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29.
Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227.
Glazebrook, J., Chen, W., Estes, B., Chang, H. S., Nawrath, C., Métraux, J. P., Zhu, T., and Katagiri, F. (2003). Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping. Plant J. 34, 217–228.
Govrin, E. M., and Levine, A. (2000). The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr. Biol. 10, 751–757.
Govrin, E. M., and Levine, A. (2002). Infection of A. thaliana with a necrotrophic pathogen, Botrytis cinerea, elicits various defense responses but does not induce systemic acquired resistance (SAR). Plant Mol. Biol. 48, 267–276.
Govrin, E. M., Rachmilevitch, S., Tiwari, B. S., Solomon, M., and Levine, A. (2006). An elicitor from Botrytis cinerea induces the hypersensitive response in Arabidopsis thaliana and other plants and promotes the gray mold disease. Phytopathology 96, 299–307.
Guzmán, P., and Ecker, J. R. (1990). Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2, 513–523.
Harman, G. E., Howell, C. R., Viterbo, A., Chet, I., and Lorito, M. (2004). Trichoderma species-opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol. 2, 1–14.
Heil, M., Hilpert, A., Kaiser, W., and Linsenmair, K. E. (2000). Reduced growth and seed set following chemical induction of pathogen defence: does systemic acquired resistance (SAR) incur allocation costs? J. Ecol. 88, 645–654.
Hipskind, J., Wood, K., and Nicholson, R. L. (1996). Localized stimulation of anthocyanin accumulation and delineation of pathogen ingress in maize genetically resistant to Bipolaris maydis race O. Physiol. Mol. Plant Pathol. 49, 247–256.
Horst, L., Locke, J. C., Krause, C. R., McMahon, R. W., Madden, L. V., and Hoitink, H. A. (2005). Suppression of Botrytis blight of begonia by Trichoderma hamatum 382 in peat and compost-amended potting mixes. Plant Dis. 89, 1195–1200.
Hossain, M. D. M., Sultana, F., Kubota, M., Koyama, H., and Hyakumachi, M. (2007). The plant growth-promoting fungus Penicillium simplicissimum GP17-2 induces resistance in Arabidopsis thaliana by activation of multiple defense signals. Plant Cell Physiol. 48, 1724–1736.
Iwata, Y., Fedoroff, N. V., and Koizumi, N. (2008). Arabidopsis bZIP60 is a proteolysis-activated transcription factor involved in the endoplasmic reticulum stress response. Plant Cell 20, 3107–3121.
Jaskiewicz, M., Conrath, U., and Peterhänsel, C. (2011). Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO Rep. 12, 50–55.
Jiang, F., Zheng, X., and Chen, J. (2009). Microarray analysis of gene expression profile induced by the biocontrol yeast Cryptococcus laurentii in cherry tomato fruit. Gene 430, 12–16.
Kangatharalingam, N., Pierce, M. L., Bayles, M. B., and Essenberg, M. (2002). Epidermal anthocyanin production as an indicator of bacterial blight resistance in cotton. Physiol. Mol. Plant Pathol. 61, 189–195.
Katagiri, F. (2004). A global view of defense gene expression regulation – a highly interconnected signaling network. Curr. Opin. Plant Biol. 7, 506–511.
Kaupp, U. B., and Seifert, R. (2002). Cyclic nucleotide-gated ion channels. Physiol. Rev. 82, 769–824.
Kinkema, M., Fan, W., and Dong, X. (2000). Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12, 2339–2350.
Kishimoto, K., Matsui, K., Ozawa, R., and Takabayashi, J. (2006). ETR1-, JAR1- and PAD2-dependent signaling pathways are involved in C6-aldehyde-induced defense responses of Arabidopsis. Plant Sci. 171, 415–423.
Koornneef, A., Leon-Reyes, A., Ritsema, T., Verhage, A., Den Otter, F. C., Van Loon, L. C., and Pieterse, C. M. (2008). Kinetics of salicylate-mediated suppression of jasmonate signaling reveal a role for redox modulation. Plant Physiol. 147, 1358–1368.
Korolev, N., Rav David, D., and Elad, Y. (2008). The role of phytohormones in basal resistance and Trichoderma-induced systemic resistance to Botrytis cinerea in Arabidopsis thaliana. BioControl 53, 667–683.
Kotchoni, S. O., and Gachomo, E. W. (2006). The reactive oxygen species network pathways: an essential prerequisite for perception of pathogen attack and the acquired disease resistance in plants. J. Biosci. 31, 389–404.
Kourtchenko, O., Andersson, M. X., Hamberg, M., Brunnström, A., Göbel, C., McPhail, K. L., Gerwick, W. H., Feussner, I., and Ellerström, M. (2007). Oxo-phytodienoic acid-containing galactolipids in Arabidopsis: jasmonate signaling dependence. Plant Physiol. 145, 1658–1669.
Krause, M. S., De Ceuster, T. J. J., Tiquia, S. M., Michel, F. C. Jr., Madden, L. V., and Hoitink, H. A. J. (2003). Isolation and characterization of rhizobacteria from composts that suppress the severity of bacterial leaf spot of radish. Phytopathology 93, 1292–1130.
Kravchuk, Z., Vicedo, B., Flors, V., Camañes, G., González-Bosch, C., and García-Agustín, P. (2011). Priming for JA-dependent defenses using hexanoic acid is an effective mechanism to protect Arabidopsis against B. cinerea. J. Plant Physiol. 168, 359–366.
Krinke, O., Ruelland, E., Valentová, O., Vergnolle, C., Renou, J. P., Taconnat, L., Flemr, M., Burketová, L., and Zachowski, A. (2007). Phosphatidylinositol 4-kinase activation is an early response to salicylic acid in Arabidopsis suspension cells. Plant Physiol. 144, 1347–1359.
Lamesch, P., Berardini, T. Z., Li, D., Swarbreck, D., Wilks, C., Sasidharan, R., Muller, R., Dreher, K., Alexander, D. L., Garcia-Hernandez, M., Karthikeyan, A. S., Lee, C. H., Nelson, W. D., Ploetz, L., Singh, S., Wensel, A., and Huala, E. (2012). The Arabidopsis information resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40, D1202–D1210.
Latunde-Dada, A. O., and Lucas, J. A. (2001). The plant defence activator acibenzolar-S-methyl primes cowpea (Vigna unguiculata (L.) Walp.) seedlings for rapid induction of resistance. Physiol. Mol. Plant Pathol. 58, 199–208.
Lawton, K. A., Friedrich, L., Hunt, M., Weymann, K., Delaney, T., Kessmann, H., Staub, T., and Ryals, J. (1996). Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. Plant J. 10, 71–82.
Lee, D. S., Kim, B. K., Kwon, S. J., Jin, H. C., and Park, O. K. (2009). Arabidopsis GDSL lipase 2 plays a role in pathogen defense via negative regulation of auxin signaling. Biochem. Biophys. Res. Commun. 379, 1038–1042.
Li, J., Zhao-Hui, C., Batoux, M., Nekrasov, V., Roux, M., Chinchilla, D., Zipfel, C., and Jones, J. D. (2009). Specific ER quality control components required for biogenesis of the plant innate immune receptor EFR. Proc. Natl. Acad. Sci. U.S.A. 106, 15973–15978.
Lievens, B., Brouwer, M., Vanachter, A. C. R. C., Cammue, B. P. A., and Thomma, B. P. H. J. (2006). Real-time PCR for detection and quantification of fungal and oomycete tomato pathogens in plant and soil samples. Plant Sci. 171, 155–165.
Lievens, B., Claes, L., Vanachter, A. C. R. C., Krause, M. S., Cammue, B. P. A., and Thomma, B. P. H. J. (2007). Assessing populations of a disease suppressive microorganism and plant pathogen using DNA arrays. Plant Sci. 172, 505–515.
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta c(t)) method. Methods 25, 402–428.
Lloyd, A. J., William Allwood, J., Winder, C. L., Dunn, W. B., Heald, J. K., Cristescu, S. M., Sivakumaran, A., Harren, F. J., Mulema, J., Denby, K., Goodacre, R., Smith, A. R., and Mur, L. A. (2011). Metabolomic approaches reveal that cell wall modifications play a major role in ethylene-mediated resistance against Botrytis cinerea. Plant J. 67, 852–868.
Lorenzo, O., Chico, J. M., Sanchez-Serrano, J. J., and Solano, R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16, 1938–1950.
Lorito, M., Woo, S. L., Harman, G. E., and Monte, E. (2010). Translational research on Trichoderma: from omics to the field. Annu. Rev. Phytopathol. 48, 395–417.
Louws, F. J., Wilson, M., Cuppels, D. A., Jones, J. B., Shoemaker, P. B., Sahin, F., and Miller, S. A. (2001). Field control of bacterial spot of tomato and pepper and bacterial speck of tomato using a plant activator. Plant Dis. 85, 481–488.
Maleck, K., Levine, A., Eulgem, T., Morgan, A., Schmid, J., Lawton, K. A., Dangl, J. L., and Dietrich, R. A. (2000). The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat. Genet. 26, 403–410.
Manners, J. M., Penninckx, I. A., Vermaere, K., Kazan, K., Brown, R. L., Morgan, A., Maclean, D. J., Curtis, M. D., Cammue, B. P., and Broekaert, W. F. (1998). The promoter of the plant defensin gene PDF1.2 from Arabidopsis is systemically activated by fungal pathogens and responds to methyl jasmonate but not to salicylic acid. Plant Mol. Biol. 38, 1071–1080.
Mersmann, S., Bourdais, G., Rietz, S., and Robatzek, S. (2010). Ethylene signaling regulates accumulation of the FLS2 receptor and is required for the oxidative burst contributing to plant immunity. Plant Physiol. 154, 391–400.
Métraux, J.-P., Signer, H., Ryals, J., Ward, E., Wyss-Benz, M., Gaudin, J., Raschdorf, K., Schmid, E., Blum, W., and Inverardi, B. (1990). Increase in salicylic acid at the onset of systemic acquired resistance in cucumber. Science 250, 1004–1006.
Mirouze, M., Sels, J., Richard, O., Czernic, P., Loubet, S., Jacquier, A., François, I. E. J., Cammue, B. P. A., Lebrun, M., Berthomieu, P., and Marquès, L. (2006). A putative novel role for plant defensins: a defensin from the zinc hyper-accumulating plant, Arabidopsis halleri, confers zinc tolerance. Plant J. 47, 329–342.
Mishina, T. E., and Zeier, J. (2007). Pathogen-associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. Plant J. 50, 500–513.
Mittler, R., Vanderauwera, S., Gollery, M., and Van Breusegem, F. (2004). The reactive oxygen gene network of plants. Trends Plant Sci. 9, 490–498.
Moeder, W., and Yoshioka, K. (2008). Lesion mimic mutants: a classical, yet still fundamental approach to study programmed cell death. Plant Signal. Behav. 3, 764–767.
Mueller, S., Hilbert, B., Dueckershoff, K., Roitsch, T., Krischke, M., Muelle, M. J., and Berger, S. (2008). General detoxification and stress responses are mediated by oxidized lipids through TGA transcription factors in Arabidopsis. Plant Cell 20, 768–785.
Mukherjee, M., Larrimore, K. E., Ahmed, N. J., Bedick, T. S., Barghouthi, N. T., Traw, M. B., and Barth, C. (2010). Ascorbic acid deficiency in Arabidopsis induces constitutive priming that is dependent on hydrogen peroxide, salicylic acid, and the NPR1 gene. Mol. Plant Microbe Interact. 23, 340–351.
Nawrath, C., and Métraux, J. P. (1999). Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11, 1393–1404.
Ndamukong, I., Abdallat, A. A., Thurow, C., Fode, B., Zander, M., Weigel, R., and Gatz, C. (2007). SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF1.2 transcription. Plant J. 50, 128–139.
Neff, M. M., and Chory, J. (1998). Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol. 118, 27–35.
Niu, D. D., Liu, H. X., Jiang, C. H., Wang, Y. P., Wang, Q. Y., Jin, H. L., and Guo, J. H. (2011). The plant growth-promoting rhizobacterium Bacillus cereus AR156 induces systemic resistance in Arabidopsis thaliana by simultaneously activating salicylate- and jasmonate/ethylene-dependent signaling pathways. Mol. Plant Microbe Interact. 24, 533–542.
Ogasawara, K., Yamada, K., Christeller, J. T., Kondo, M., Hatsugai, N., Hara-Nishimura, I., and Nishimura, M. (2009). Constitutive and inducible ER bodies of Arabidopsis thaliana accumulate distinct beta-glucosidases. Plant Cell Physiol. 50, 480–488.
Olson, H. A., and Benson, D. M. (2007). Induced systemic resistance and the role of binucleate Rhizoctonia and Trichoderma hamatum 382 in biocontrol of Botrytis blight in geranium. Biol. Control 42, 233–241.
Ongena, M., Jourdan, E., Schafer, M., Kech, C., Budzikiewicz, H., Luxen, A., and Thonart, P. (2005). Isolation of an N-alkylated benzylamine derivative from Pseudomonas putida BTP1 as elicitor of induced systemic resistance in bean. Mol. Plant Microbe Interact. 18, 562–569.
Pandey, S. P., Roccaro, M., Schön, M., Logemann, E., and Somssich, I. E. (2010). Transcriptional reprogramming regulated by WRKY18 and WRKY40 facilitates powdery mildew infection of Arabidopsis. Plant J. 64, 912–923.
Pastori, G. M., Kiddle, G., Antoniw, J., Bernard, S., Veljovic-Jovanovic, S., Verrier, P. J., Noctor, G., and Foyer, C. H. (2003). Leaf vitamin C contents modulate plant defense transcripts and regulate genes that control development through hormone signaling. Plant Cell 15, 939–951.
Penninckx, I. A., Eggermont, K., Terras, F. R., Thomma, B. P., De Samblanx, G. W., Buchala, A., Métraux, J. P., Manners, J. M., and Broekaert, W. F. (1996). Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 8, 2309–2323.
Penninckx, I. A., Thomma, B. P., Buchala, A., Métraux, J. P., and Broekaert, W. F. (1998). Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10, 2103–2113.
Pieterse, C. M., Leon-Reyes, A., Van der Ent, S., and Van Wees, S. C. (2009). Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5, 308–316.
Pieterse, C. M., van Wees, S. C., Hoffland, E., van Pelt, J. A., and van Loon, L. C. (1996). Systemic resistance in Arabidopsis induced by biocontrol bacteria is independent of salicylic acid accumulation and pathogenesis-related gene expression. Plant Cell 8, 1225–1237.
Pieterse, C. M. J., van Wees, S. C. M., van Pelt, J. A., Knoester, M., Laan, R., Gerrits, H., Weisbeek, P. J., and van Loon, L. C. (1998). A novel signalling pathway controlling induced systemic resistance in A. thaliana. Plant Cell 10, 1571–1580.
Pineda, A., Zheng, S. J., van Loon, J. J., and Dicke, M. (2012). Rhizobacteria modify plant-aphid interactions: a case of induced systemic susceptibility. Plant Biol. (Stuttg). 14(Suppl. 1), 83–90.
Pitzschke, A., Forzani, C., and Hirt, H. (2006). Reactive oxygen species signaling in plants. Antioxid. Redox Signal. 8, 1757–1764.
Pogány, M., von Rad, U., Grün, S., Dongó, A., Pintye, A., Simoneau, P., Bahnweg, G., Kiss, L., Barna, B., and Durner, J. (2009). Dual roles of reactive oxygen species and NADPH oxidase RBOHD in an Arabidopsis-alternaria pathosystem. Plant Physiol. 151, 1459–1475.
Pozo, M. J., Van der Ent, S., Van Loon, L. C., and Pieterse, C. M. J. (2008). The transcription factor MYC2 is involved in priming for enhanced defense during rhizobacteria-induced systemic resistance in Arabidopsis. New Phytol. 180, 511–523.
Prime-A-Plant GroupConrath, U., Beckers, G. J., Flors, V., García-Agustín, P., Jakab, G., Mauch, F., Newman, M. A., Pieterse, C. M., Poinssot, B., Pozo, M. J., Pugin, A., Schaffrath, U., Ton, J., Wendehenne, D., Zimmerli, L., and Mauch-Mani, B. (2006). Priming: getting ready for battle. Mol. Plant Microbe Interact. 19, 1062–1071.
Punja, Z. K., and Utkhede, R. S. (2003). Using fungi and yeasts to manage vegetable crop diseases. Trends Biotechnol. 21, 400–407.
R Development Core Team. (2011). R: A Language and Environment for Statistical Computing, Vienna: R Foundation for Statistical Computing. ISBN 3-900051-07–0.
Ranf, S., Eschen-Lippold, L., Pecher, P., Lee, J., and Scheel, D. (2011). Interplay between calcium signalling and early signalling elements during defence responses to microbe- or damage-associated molecular patterns. Plant J. 68, 100–113.
Ringli, C. (2010). Monitoring the outside: cell wall-sensing mechanisms. Plant Physiol. 153, 1445–1452.
Routaboul, J. M., Kerhoas, L., Debeaujon, I., Pourcel, L., Caboche, M., Einhorn, J., and Lepiniec, L. (2006). Flavonoid diversity and biosynthesis in seed of Arabidopsis thaliana. Planta 224, 96–107.
Rowe, H. C., Walley, J. W., Corwin, J., Chan, E. K., Dehesh, K., and Kliebenstein, D. J. (2010). Deficiencies in jasmonate-mediated plant defense reveal quantitative variation in Botrytis cinerea pathogenesis. PLoS Pathog. 6, e1000861.
Rozen, S., and Skaletsky, H. (2000). Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132, 365–386.
Rubio, M. B., Domínguez, S., Monte, E., and Hermosa, R. (2012). Comparative study of Trichoderma gene expression in interactions with tomato plants using high-density oligonucleotide microarrays. Microbiology 158(Pt 1), 119–128.
Ryals, J. A., Neuenschwander, U. H., Willits, M. G., Molina, A., Steiner, H. Y., and Hunt, M. D. (1996). Systemic acquired resistance. Plant Cell 8, 1809–1819.
Samolski, I., de Luis, A., Vizcaíno, J. A., Monte, E., and Suárez, M. B. (2009). Gene expression analysis of the biocontrol fungus Trichoderma harzianum in the presence of tomato plants, chitin, or glucose using a high-density oligonucleotide microarray. BMC Microbiol. 9, 217.
Sasaki, Y., Asamizu, E., Shibata, D., Nakamura, Y., Kaneko, T., Awai, K., Amagai, M., Kuwata, C., Tsugane, T., Masuda, T., Shimada, H., Takamiya, K., Ohta, H., and Tabata, S. (2001). Monitoring of methyl jasmonate-responsive genes in Arabidopsis by cDNA macroarray: self-activation of jasmonic acid biosynthesis and crosstalk with other phytohormone signaling pathways. DNA Res. 8, 153–161.
Schaller, G. E., and Bleecker, A. B. (1995). Ethylene-binding sites generated in yeast expressing the Arabidopsis ETR1 gene. Science 270, 1809–1811.
Schuhegger, R., Ihring, A., Gantner, S., Bahnweg, G., Knappe, C., Vogg, G., Hutzler, P., Schmid, M., Van Breusegem, F., Eberl, L., Hartmann, A., and Langebartels, C. (2006). Induction of systemic resistance in tomato by N-acyl-L-homoserine lactone-producing rhizosphere bacteria. Plant Cell Environ. 29, 909–918.
Segarra, G., Van der Ent, S., Trillas, I., and Pieterse, C. M. J. (2009). Source MYB72, a node of convergence in induced systemic resistance triggered by a fungal and a bacterial beneficial microbe. Plant Biol. 11, 90–96.
Sels, J., Mathys, J., De Coninck, B. M., Cammue, B. P., and De Bolle, M. F. (2008). Plant pathogenesis-related (PR) proteins: a focus on PR peptides. Plant Physiol. Biochem. 46, 941–950.
Senthil-Kumar, M., and Mysore, K. S. (2010). Assessing functional role of three water deficit stress-induced genes in nonhost disease resistance using virus-induced gene silencing in Nicotiana benthamiana. Plant Signal. Behav. 5, 586–590.
Sherameti, I., Venus, Y., Drzewiecki, C., Tripathi, S., Dan, V. M., Nitz, I., Varma, A., Grundler, F. M., and Oelmüller, R. (2008). PYK10, a beta-glucosidase located in the endoplasmatic reticulum, is crucial for the beneficial interaction between Arabidopsis thaliana and the endophytic fungus Piriformospora indica. Plant J. 54, 428–439.
Shetty, R., Fretté, X., Jensen, B., Shetty, N. P., Jensen, J. D., Jørgensen, H. J., Newman, M. A., and Christensen, L. P. (2011). Silicon-induced changes in antifungal phenolic acids, flavonoids and key phenylpropanoid pathway genes during the interaction between miniature roses and the biotrophic pathogen Podosphaera pannosa. Plant Physiol. 157, 2194–2205.
Shoresh, M., Yedidia, I., and Chet, I. (2005). Involvement of jasmonic acid/ethylene signaling pathway in the systemic resistance induced in cucumber by Trichoderma asperellum T203. Phytopathology 95, 76–84.
Spoel, S. H., Koornneef, A., Claessens, S. M., Korzelius, J. P., Van Pelt, J. A., Mueller, M. J., Buchala, A. J., Métraux, J. P., Brown, R., Kazan, K., Van Loon, L. C., Dong, X., and Pieterse, C. M. (2003). NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15, 760–770.
Stein, E., Molitor, A., Kogel, K. H., and Waller, F. (2008). Systemic resistance in Arabidopsis conferred by the mycorrhizal fungus Piriformospora indica requires jasmonic acid signaling and the cytoplasmic function of NPR1. Plant Cell Physiol. 49, 1747–1751.
Stintzi, A., Weber, H., Reymond, P., Browse, J., and Farmer, E. E. (2001). Plant defense in the absence of jasmonic acid: the role of cyclopentenones. Proc. Natl. Acad. Sci. U.S.A. 98, 12837–12842.
Strawn, M. A., Marr, S. K., Inoue, K., Inada, N., Zubieta, C., and Wildermuth, M. C. (2007). Arabidopsis isochorismate synthase functional in pathogen-induced salicylate biosynthesis exhibits properties consistent with a role in diverse stress responses. J. Biol. Chem. 282, 5919–5933.
Tavazoie, S., Hughes, J. D., Campbell, M. J., Cho, R. J., and Church, G. M. (1999). Systematic determination of genetic network architecture. Nat. Genet. 22, 281–285.
Temme, N., and Tudzynski, P. (2009). Does Botrytis cinerea ignore H2O2-induced oxidative stress during infection? Characterization of the Botrytis activator protein 1. Mol. Plant Microbe Interact. 22, 987–998.
Teng, S., Keurentjes, J., Bentsink, L., Koornneef, M., and Smeekens, S. (2005). Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol. 139, 1840–1852.
Thomma, B. P. H. J., Eggermont, K., Penninckx, I. A. M. A., Mauch-Mani, B., Vogelsang, R., Cammue, B. P. A., and Broekaert, W. F. (1998). Separate jasmonate-dependent and salicylate-dependent defense response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. U.S.A. 95, 15107–15111.
Tjamos, S. E., Flemetakis, E., Paplomatas, E. J., and Katinakis, P. (2005). Induction of resistance to Verticillium dahliae in Arabidopsis thaliana by the biocontrol agent K-165 and pathogenesis-related proteins gene expression. Mol. Plant Microbe Interact. 18, 555–561.
Ton, J., and Mauch-Mani, B. (2004). Beta-amino-butyric acid-induced resistance against necrotrophic pathogens is based on ABA-dependent priming for callose. Plant J. 38, 119–130.
Torres, M. A., Onouchi, H., Hamada, S., Machida, C., Hammond-Kosack, K. E., and Jones, J. D. (1998). Six Arabidopsis thaliana homologues of the human respiratory burst oxidase (gp91phox). Plant J. 14, 365–370.
Treutter, D. (2006). Significance of flavonoids in plant resistance: a review. Environ. Chem. Lett. 4, 147–157.
Tsuda, K., Sato, M., Glazebrook, J., Cohen, J. D., and Katagiri, F. (2008). Interplay between MAMP-triggered and SA-mediated defense responses. Plant J. 53, 763–775.
Tucci, M., Ruocco, M., De Masi, L., De Palma, M., and Lorito, M. (2011). The beneficial effect of Trichoderma spp. on tomato is modulated by the plant genotype. Mol. Plant Pathol. 12, 341–354.
Tusher, V. G., Tibshirani, R., and Chu, G. (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U.S.A. 98, 5116–5121.
Uknes, S., Mauch-Mani, B., Moyer, M., Potter, S., Williams, S., Dincher, S., Chandler, D., Slusarenko, A., Ward, E., and Ryals, J. (1992). Acquired resistance in Arabidopsis. Plant Cell 4, 645–656.
Vadassery, J., Tripathi, S., Prasad, R., Varma, A., and Oelmüller, R. (2009). Monodehydroascorbate reductase 2 and dehydroascorbate reductase 5 are crucial for a mutualistic interaction between Piriformospora indica and Arabidopsis. J. Plant Physiol. 166, 1263–1274.
Van Breusegem, F., Bailey-Serres, J., and Mittler, R. (2008). Unraveling the tapestry of networks involving reactive oxygen species in plants. Plant Physiol. 147, 978–984.
van Kan, J. A. (2006). Licensed to kill: the lifestyle of a necrotrophic plant pathogen. Trends Plant Sci. 11, 247–253.
van Leeuwen, H., Kliebenstein, D. J., West, M. A., Kim, K., van Poecke, R., Katagiri, F., Michelmore, R. W., Doerge, R. W., and St Clair, D. A. (2007). Natural variation among Arabidopsis thaliana accessions for transcriptome response to exogenous salicylic acid. Plant Cell 19, 2099–2110.
van Loon, L. C. (1997). Induced resistance in plants and the role of pathogenesis-related proteins. Eur. J. Plant Pathol. 103, 753–765.
van Loon, L. C., Bakker, P. A. H. M., and Pieterse, C. M. J. (1998). Systemic resistance induced by rhizosphere bacteria. Annu. Rev. Phytopathol. 36, 453–483.
Van Oosten, V. R., Bodenhausen, N., Reymond, P., Van Pelt, J. A., Van Loon, L. C., Dicke, M., and Pieterse, C. M. (2008). Differential effectiveness of microbially induced resistance against herbivorous insects in Arabidopsis. Mol. Plant Microbe Interact. 21, 919–930.
van Wees, S. C., Luijendijk, M., Smoorenburg, I., van Loon, L. C., and Pieterse, C. M. (1999). Rhizobacteria-mediated induced systemic resistance (ISR) in A. thaliana is not associated with a direct effect on expression of known defense-related genes but stimulates the expression of the jasmonate-inducible gene Atvsp upon challenge. Plant Mol. Biol. 41, 537–549.
Verhagen, B. W. M., Glazebrook, J., Zhu, T., Chang, H., van Loon, L. C., and Pieterse, C. M. J. (2004). The transcriptome of rhizobacteria-induced systemic resistance in Arabidopsis. Mol. Plant Microbe Interact. 17, 895–908.
Veronese, P., Nakagami, H., Bluhm, B., AbuQamar, S., Chen, X., Salmeron, J., Dietrich, R. A., Hirt, H., and Mengiste, T. (2006). The membrane anchored Botrytis-induced kinase1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 18, 257–273.
Wan, J., Zhang, S., and Stacey, G. (2004). Activation of a mitogen-activated protein kinase pathway in Arabidopsis by chitin. Mol. Plant Pathol. 5, 125–135.
Wang, D., Amornsiripanitch, N., and Dong, X. (2006). A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2, 1042–1050.
Wang, Y., Ohara, Y., Nakayashiki, H., Tosa, Y., and Mayama, S. (2005). Microarray analysis of the gene expression profile induced by the endophytic plant growth-promoting rhizobacteria, Pseudomonas fluorescens FPT9601-T5 in Arabidopsis. Mol. Plant Microbe Interact. 18, 385–396.
Ward, E. R., Uknes, S. J., Williams, S. C., Dincher, S. S., Wiederhold, D. L., Alexander, D. C., Ahl-Goy, P., Metraux, J. P., and Ryals, J. A. (1991). Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell 3, 1085–1094.
Wasternack, C. (2007). Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. 100, 681–697.
Wen, K., Seguin, P., St Arnaud, M., and Jabaji-Hare, S. (2005). Real-Time Quantitative RT-PCR of defense-associated gene transcripts of Rhizoctonia solani-infected bean seedlings in response to inoculation with a nonpathogenic binucleate Rhizoctonia isolate. Phytopathology 95, 345–353.
Winkel-Shirley, B. (2002). Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 5, 218–223.
Yan, Z., Reddy, M. S., Ryu, C.-M., McInroy, J. A., Wilson, M., and Kloepper, J. W. (2002). Induced systemic protection against tomato late blight elicited by plant growth-promoting rhizobacteria. Phytopathology 92, 1329–1333.
Zimmerli, L., Jakab, G., Metraux, J. P., and Mauch-Mani, B. (2000). Potentiation of pathogen-specific defense mechanisms in Arabidopsis by beta -aminobutyric acid. Proc. Natl. Acad. Sci. U.S.A. 97, 12920–12925.
Keywords: induced systemic resistance, microarrays, Arabidopsis thaliana, Trichoderma hamatum T382, Botrytis cinerea
Citation: Mathys J, De Cremer K, Timmermans P, Van Kerckhove S, Lievens B, Vanhaecke M, Cammue BPA and De Coninck B (2012) Genome-wide characterization of ISR induced in Arabidopsis thaliana by Trichoderma hamatum T382 against Botrytis cinerea infection. Front. Plant Sci. 3:108. doi: 10.3389/fpls.2012.00108
Received: 13 March 2012; Accepted: 07 May 2012; Published online: 29 May 2012.
Edited by:
Corné M. J. Pieterse, Utrecht University, NetherlandsReviewed by:
Christos Zamioudis, Utrecht University, NetherlandsMonica Höfte, Ghent University, Belgium
Copyright: © 2012 Mathys, De Cremer, Timmermans, Van Kerckhove, Lievens, Vanhaecke, Cammue and De Coninck. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Bruno P. A. Cammue, Centre of Microbial and Plant Genetics, Katholieke Universiteit Leuven, Kasteelpark Arenberg 20, 3001 Heverlee, Belgium. e-mail:YnJ1bm8uY2FtbXVlQGJpdy5rdWxldXZlbi5iZQ==



 Stefan Van Kerckhove2
Stefan Van Kerckhove2