- 1 Department of Plant Biology, Cornell University, Ithaca, NY, USA
- 2 Horticultural Sciences Department, University of Florida, Gainesville, FL, USA
The embryo and endosperm are the products of double fertilization and comprise the clonally distinct products of angiosperm seed development. Recessive mutations in the maize gene discolored1 (dsc1) condition inviable seed that are defective in both embryo and endosperm development. Here, detailed phenotypic analyses illustrate that discolored mutant kernels are able to establish, but fail to maintain, differentiated embryo, and endosperm structures. Development of the discolored mutant embryo and endosperm is normal albeit delayed, prior to the abortion and subsequent degeneration of all differentiated kernel structures. Using a genomic fragment that was previously isolated by transposon tagging, the full length dsc1 transcript is identified and shown to encode an ADP-ribosylation factor-GTPase activating protein (ARF-GAP) that co-localizes with the trans-Golgi network/early endosomes and the plasma membrane during transient expression assays in N. benthamiana leaves. DSC1 function during endomembrane trafficking and the maintenance of maize kernel differentiation is discussed.
Introduction
The maize kernel is a single-seeded fruit composed of the triploid endosperm, the diploid embryo, and the maternally derived pericarp, pedicel, and placenta. At maturity the maize embryo comprises a primary root axis and a relatively precocious shoot axis that may develop up to six leaf primordia. During germination the endosperm nurtures the embryo as it matures into a seedling. Development of the clonally distinct endosperm and embryo ensues following double fertilization of the female gametophyte. The fertilized central cell gives rise to the triploid endosperm, which becomes cellularized 4 days after pollination (DAP; Olsen, 2001). Four distinct structures differentiate to form the maize endosperm, and include: the basal endosperm transfer layer (BETL); the embryo surrounding region (ESR); the starchy endosperm; and the aleurone. Developing at the base of the kernel, the BETL facilitates the transfer of maternally derived nutrients and photosynthates from the placenta into the developing endosperm (Brink and Cooper, 1947; Kiesselbach and Walker, 1952). A distinguishing feature of the BETL is cell wall projections that increase the surface area of the plasma membrane, and thus facilitate the transport function of these cells (Thompson et al., 2001). The ESR forms around the earliest-staged embryo (the proembryo) and has a predicted signaling function between the embryo and the early endosperm compartments but is not present in the mature endosperm (Schel et al., 1984; Opsahl-Ferstad et al., 1997). Functioning as energy reserves, starchy endosperm cells accumulate starch and proteins that will nurture development of the germinating seedling (Duvik, 1961). Cuboidal-shaped aleurone cells form a single-cell layer that encompasses the starchy endosperm; during germination the aleurone functions to digest the stored energy reserves in the kernel (Becraft and Yi, 2011). Aleurone cells are clonally related to the starchy endosperm and can re-differentiate into starchy endosperm in the absence of an as yet unidentified signal that is required to maintain aleurone-specific cell fate (Becraft and Asuncion-Crabb, 2000).
The diploid embryo is derived from the fertilized egg cell, and at maturity is composed of the shoot and root apical meristems, the scutellum, five to six foliar leaves protected by the sheathing coleoptile, and a primary root protected by the sheathing coleorhiza (Abbe and Stein, 1954). Landmark events during embryo development (Kaplan and Cooke, 1997) include formation of the pre-meristematic embryo proper, the establishment of the shoot apical meristem (SAM), and the elaboration of three distinct varieties of shoot lateral organs (the scutellum, the coleoptile, and foliar leaves). Historically, these key events in maize embryo development have been described in nine discrete stages (Abbe and Stein, 1954) and include: (1) the proembryo, before the meristem is formed; (2) the transition stage, when the SAM is established; (3) the coleoptile stage, after the SAM has formed and initiates the coleoptile; and (4) stages L1-L6, wherein the SAM initiates up to six foliar leaves (Abbe and Stein, 1954; Poethig et al., 1986).
Defective kernel (dek) mutations condition defects in both embryo and endosperm development, and thus are useful genetic tools to study kernel development (Neuffer and Sheridan, 1980; Scanlon et al., 1994). Several dek mutants have shared defects in embryonic epidermal patterning and in the maintenance of the endosperm aleurone layer (Becraft et al., 1996, 2002; Becraft and Asuncion-Crabb, 2000; Kessler et al., 2002; Lid et al., 2002; Shen et al., 2003). Several genes underlying these dek mutations are implicated to function in cell-to-cell signaling during endosperm differentiation. Dek1 encodes a trans-membrane domain protein with a cytoplasmic calpain-like protease (Lid et al., 2002), and cr4 encodes a tumor necrosis factor-like receptor kinase (Becraft et al., 1996); both DEK1 and CR4 are required to maintain aleurone cell fate. In contrast, sal1 functions to inhibit aleurone cell fate during kernel development and encodes an E class vacuolar sorting protein (Shen et al., 2003), which implicates an essential role for endomembrane trafficking in endosperm differentiation.
Endomembrane vesicle trafficking involves the intracellular and intercellular transport of cellular cargo, including proteins, cell wall pectins, structural sterols, receptors, lipids, and signaling molecules, from one membrane-bound compartment to another (Cosgrove, 1997; Takai et al., 2001; Samaj et al., 2005). Vesicle trafficking is regulated in part by the activity of ARF-GTPases, which cycle between active and inactive forms that correlate with vesicle formation and dissociation, respectively (Nie and Randazzo, 2006). Active ARF-GTPases associate with GTP and are membrane-bound during vesicle formation; inactive ARF-GTPases associate with GDP in the cytosol and function during vesicle dissociation. These cyclic activities of ARF-GTPases are regulated by ADP-ribosylation factor-guanine exchange factors (ARF-GEFs) that catalyze the exchange of GDP for GTP, and by ADP-ribosylation factor-GTPase activating proteins (ARF-GAPs) that catalyze the subsequent hydrolysis of GTP-bound ARFs (Chardin et al., 1996; Scheffzek et al., 1998; Goldberg, 1999). In Arabidopsis, endomembrane cycling of the PINFORMED (PIN) family of auxin efflux proteins requires the activities of both the ARF-GEF GNOM/EMB30 (GN) and the ARF-GAP vascular network defective3 (VAN3)/SCARFACE (SFC; Geldner et al., 2003; Koizumi et al., 2005; Sieburth et al., 2006; Naramoto et al., 2010). Mutations in GN and VAN3/SFC give rise to mutants that have defects in embryo development or vascular differentiation, respectively (Geldner et al., 2003; Koizumi et al., 2005; Sieburth et al., 2006). ARF-GAP domain1 (AGD1), a second ACAP-type ARF-GAP characterized in Arabidopsis, functions in signaling pathways that remodel the actin cytoskeleton and direct membrane trafficking to maintain polarized root hair growth (Yoo et al., 2008, 2012). The characterization of these ARF-GAPs in Arabidopsis indicates that there is a wide diversity of ACAP-type ARF-GAP function and cargo specificity.
Previously, the maize dek mutation discolored1 (dsc1) was identified in a Mutator (Mu) transposon-mutagenized population and named for the shrunken, brown phenotype of homozygous mutant kernels (Scanlon et al., 1994). Recessive mutations in DSC1 condition inviable kernel phenotypes, and transposon-tagging identified a Mu1-inserted genomic DNA fragment from the 5′ UTR region of the dsc1-Reference (dsc1-R) mutation (AF006498; Scanlon and Myers, 1998). Herein, detailed phenotypic analyses reveal that dsc1 mutant kernels are developmentally delayed but undergo differentiation of embryo and endosperm structures, prior to kernel abortion and tissue degeneration. The full length dsc1 transcript encodes a predicted ARF-GAP protein and accumulates in kernels harvested after 6 DAP and in seedling roots and shoots. Transient expression assays in N. benthamiana leaf tissue show that YFP-tagged DSC1 proteins co-localize with the trans-Golgi network/early endosomes and with the plasma membrane. Taken together, these data reveal that DSC1 functions in endomembrane trafficking between the trans-Golgi and the plasma membrane, and is required for the maintenance of differentiated cell types in the maize kernel.
Materials and Methods
Plant Materials
The dsc1-R mutation arose from a Mu transposon line (Scanlon et al., 1994) and was introgressed into a B73 background for at least six generations before harvesting kernels used in phenotypic and gene expression analyses. The dsc1-C06, dsc1-H02, and dsc1-B09 alleles were identified after screening the trait utility system for corn (TUSC), a Mu transposon-mutagenized population (Meeley and Briggs, 1995). Primers for this screen can be found in Table A1 in Appendix.
Histological Analyses and In Situ Hybridizations
For histological analyses, wild type and discolored mutant kernels were harvested 6 DAP to 20 DAP and fixed overnight in FAA (37% formaldehyde: ethanol: glacial acetic acid: water at 10:50:5:35). The kernels were dehydrated in an ethanol/tert-butyl alcohol series, embedded in paraplast, and 10 μm thick sections were stained with either safranin O-fast green or safranin O-orange G as previously described (Ruzin, 1999). For in situ hybridizations, wild type and discolored mutant kernels were harvested 6 DAP to 20 DAP, fixed in FAA, dehydrated, embedded in paraplast, sectioned, and hybridized with gene specific probes as previously described (Jackson, 1991). Primers used to make probes can be found in Table A1 in Appendix. All samples were imaged using the Zeiss Axio Imager Z1-Apotome microscope (Thornwood, New York) and Zeiss Axiovision release 4.6 software.
Identification of the Full Length dsc1 Transcript and Gene Expression Analysis
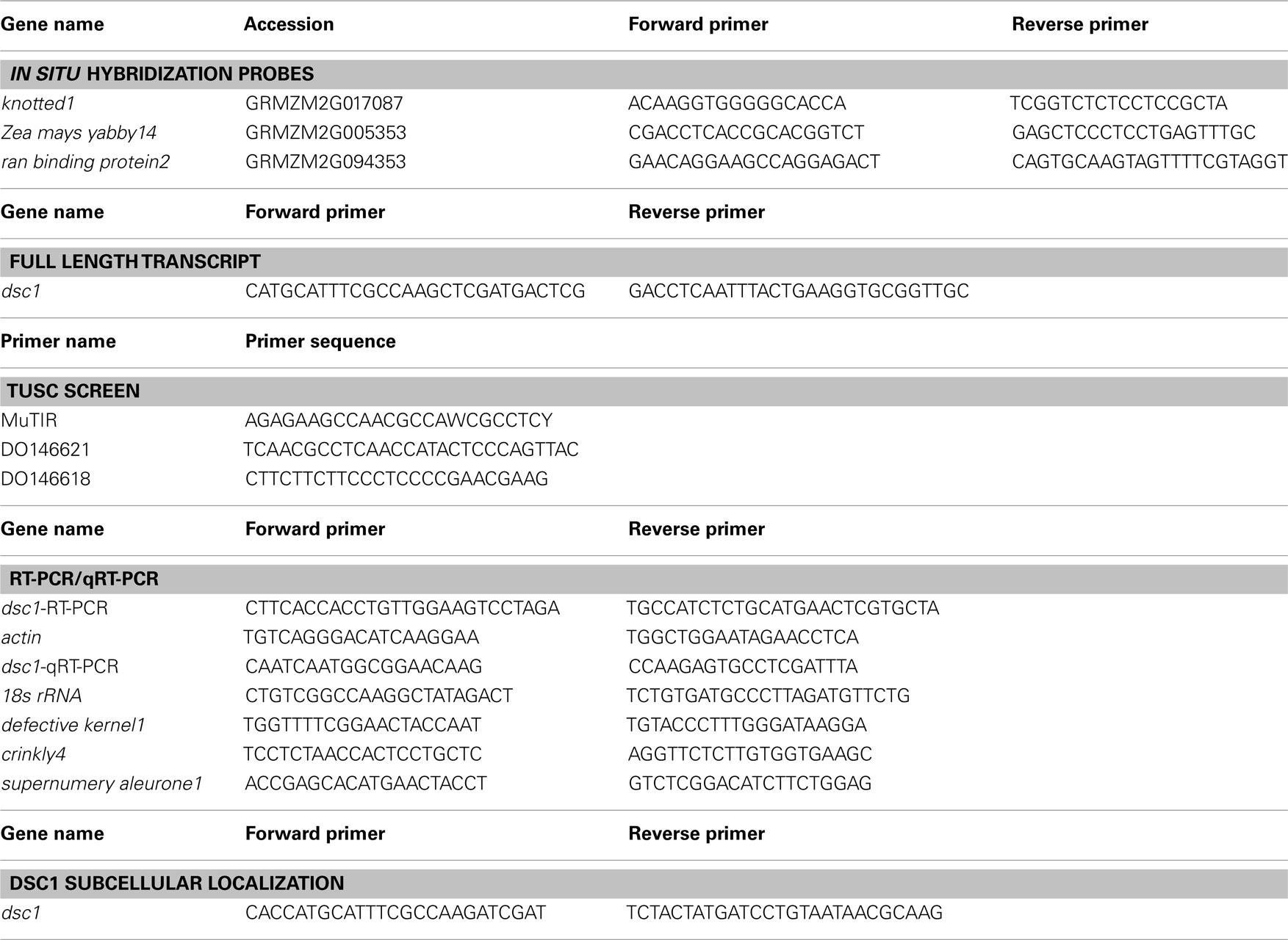
The Invitrogen Superscript III One Step RT-PCR Platinum Taq HiFi kit was used to clone dsc1. Briefly, tissue segments including the vegetative SAM were cut from 10 day old B73 seedlings. Total RNA was isolated using the RNeasy Plant Mini Kit (Qiagen) and poly(A) RNA was isolated by the Oligotex mRNA mini kit (Qiagen). Primers, which can be found in Table A1 in Appendix, were designed in the 5′ and 3′ ends of the predicted full length transcript and RT-PCR was performed to generate the full length dsc1 transcript. For analysis of gene expression using RT-PCR and quantitative RT-PCR, total RNA was isolated from harvested wild type kernels (6 DAP, 8 DAP, 12 DAP, 14 DAP, 16 DAP, 18 DAP) and discolored mutant kernels (16 DAP). Dissected embryo and endosperm tissue was flash frozen in liquid nitrogen and ground in SDS extraction buffer as previously described (Prescott and Martin, 1986) with some modifications. Following the five minute incubation on ice with chloroform/isoamyl alcohol (24:1), samples were centrifuged for 10 min at 4°C. After transferring the aqueous phase to a new tube, 1 mL of TRIzol (Invitrogen) was used to extract RNA following the manufacturer’s protocol. Total RNA was extracted from whole 14 day old seedlings and from the upper third of the emerging leaf blade from 14 day old seedlings grown on soil, and roots from 14 day old seedlings grown on 0.02% agar using TRIzol (Invitrogen) according to the manufacturer’s protocol. Superscript III (Invitrogen) was used to synthesize cDNA from 1 μg of RNA treated with DNaseI (Invitrogen). SYBR-green (Quanta) methodology combined with gene specific primers (Table A1 in Appendix) as described in (Zhang et al., 2007) was used in the quantitative RT-PCR analysis. Relative gene expression normalized to 18 s rRNA was determined using the 2−ΔΔCT method as described in Livak and Schmittgen (2001).
Transient Expression Assays
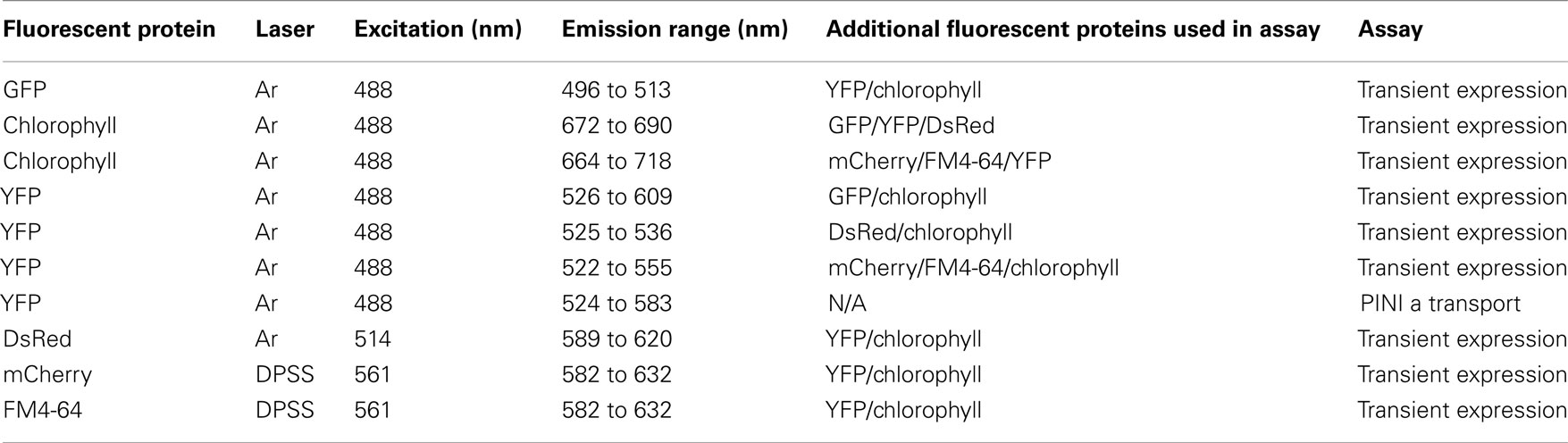
The Gateway Recombination Cloning System (Invitrogen) was used to clone the DSC1 ORF into the pEarleyGate104 destination vector as described (Earley et al., 2006). Details about the organelle marker constructs are previously described (Kohler et al., 1997; Boevink et al., 1998; Reisen and Hanson, 2007; Geldner et al., 2009). Electroporation was used to transform agrobacterium strain C58C1 with the N-terminal 35S-YFP<DSC1 fusion construct, the control construct (empty pEarleyGate104 vector), and the organelle marker constructs (35S-ERD2<GFP, 35S35SAMV-COXIV<GFP, 35S-DsRed<CAT, UBQ10-mCherry<VTI12, UBQ10-mCherry<Got1p; Kohler et al., 1997; Boevink et al., 1998; Reisen and Hanson, 2007; Geldner et al., 2009). Transformants from individual construct lines were grown overnight at 28°C in 2 mL of LB medium containing 50 μg/mL kanamycin and 5 μg/mL tetracycline. After centrifugation, the cells were resuspended in 10 mM MgCl2 to an OD of 0.5 and incubated at room temperature for 2–4 h. For localization of YFP-tagged DSC1 and the control construct 35S-YFP, N. benthamiana leaves were infiltrated as described in Goodin et al. (2002) and observed using epifluorescence microscopy between 48 and 72 h later. Images were obtained on the Zeiss Axio Imager Z1-Apotome microscope (Thornwood, New York) and Zeiss Axiovision release 4.6 software. For co-localization assays, equal parts of the suspensions transformed with each one of the five organelle markers were individually mixed with the suspension transformed with YFP-tagged DSC1 prior to incubation at room temperature. N. benthamiana leaves were infiltrated as described in Goodin et al. (2002) and observed using confocal microscopy between 48 and 72 h later. Imaging of fluorescent proteins was performed using a Leica TCS-SP5 confocal microscope (Leica Microsystems, Exton, PA, USA) using either 10× or 40× objectives (NA 0.4 or 0.85, respectively). Images were obtained sequentially to separate signal from the two channels and were later superimposed. Time lapse series were collected non-sequentially. All images were taken using either a blue argon ion laser (Ar) or a diode pumped solid state laser (DPSS). Excitation and emission parameters are presented in Table A2 in Appendix. Leica LAS-AF software (version 1.8.2) was used to process all images.
PIN1a Transport Assays
Plants heterozygous for dsc1-R were crossed with ZmPIN1a∼YFP transgenic individuals (Gallavotti et al., 2008). The resulting progeny were planted, screened for the dsc1-R allele and YFP, grown to maturity, and self-pollinated. Ears were harvested the same day kernels were removed for live imaging. Embryos were dissected and put on culture media as described in Scanlon et al. (1997). A BFA (Sigma-Aldrich) stock solution was diluted in DMSO and added to liquid culture media to make a final concentration of 100 μM BFA. Mock treatments were made by adding the same amount of DMSO (minus BFA) to liquid culture media. Harvested embryos were incubated in culture media containing BFA or the mock treatment for at least 4 h before confocal microscopy image analysis. Images were collected non-sequentially as described above.
Results
DSC1 is Required to Maintain Differentiation of Embryo and Endosperm Structures
Self-pollinated plants heterozygous for the dsc1-Rmutation segregate mutant kernels with aberrations in both embryo and endosperm development (Scanlon et al., 1994; Scanlon and Myers, 1998). Wild type kernels are yellow at 12 DAP, where upon discolored mutant kernels are white and smaller than wild type siblings (Figure 1A). Embryo structures are not discernible in mutant kernels dissected after 18 DAP, and the reduced endosperm development fails to fill the kernel space (Figures 1B,C; Scanlon et al., 1994; Scanlon and Myers, 1998). At maturity, all discolored mutant kernels are brown, misshapen, and embryo lethal (Figure 1D).
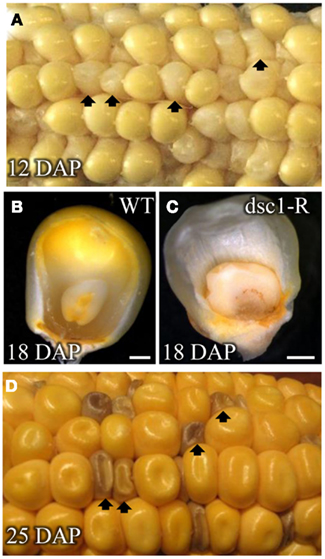
Figure 1. dscl-R is a defective kernel mutation. Self-pollinated ear of a Dscl/dscl-R heterozygous plant harvested at (A) 12 DAP segregating dscl-R mutant kernels. Frontal view of (B) non-mutant sibling and (C) dscl-R mutant kernels harvested at 18 DAR The outer layer is removed to reveal the embryo and endosperm inside of the pericarp. (D) Ear harvested at 25 DAP segregating dscl-R mutant kernels. Arrows denote dscl-R mutant kernels. DAP, days after pollination. Scale bars represent 1 mm.
Detailed phenotypic analyses of discolored mutant kernels harvested at different time-points following pollination show that development of both the embryo and endosperm is normal, albeit delayed, before eventual kernel abortion and tissue disintegration. For example, whereas late proembryo-staged embryos are harvested from wild type embryos at 8 DAP (Figures 2A,B), mutant embryos harvested from the same ear comprise far fewer cells than wild type siblings and are still encased within the ESR (Figures 2G,H). Wild type embryos harvested at 12 DAP are in stage L2, having already elaborated the scutellum, the coleoptile, and the first foliar leaf (Figures 2C,D). In contrast, 12 DAP discolored mutant sibling embryos are stalled at the early transition stage (Figures 2I,J). Three foliar leaf primordia (i.e., stage L3) are initiated in wild type embryos harvested at 16 DAP (Figures 2E,F), where upon discolored mutant embryos exhibit a variable range of phenotypes, including embryos retarded at stage L1(Figures 2K,L) and others in which identification of embryo developmental stage is impossible owing to tissue degeneration (Figures 2M–P). In agreement with the mutant embryo phenotypes described above, molecular markers for shoot meristem maintenance [knotted1(kn1)], vasculature development [ran binding protein2(ranbp2)], and scutellum initiation [Zea mays yabby14(zyb14)] showed normal transcript accumulation in transition-staged discolored mutant embryos (Figures 3A–F; Smith et al., 1995; Juarez et al., 2004). Likewise, degenerated later-staged discolored mutant embryos do not accumulate kn1or zyb14 transcripts after 16 DAP (Figures 3G,H).
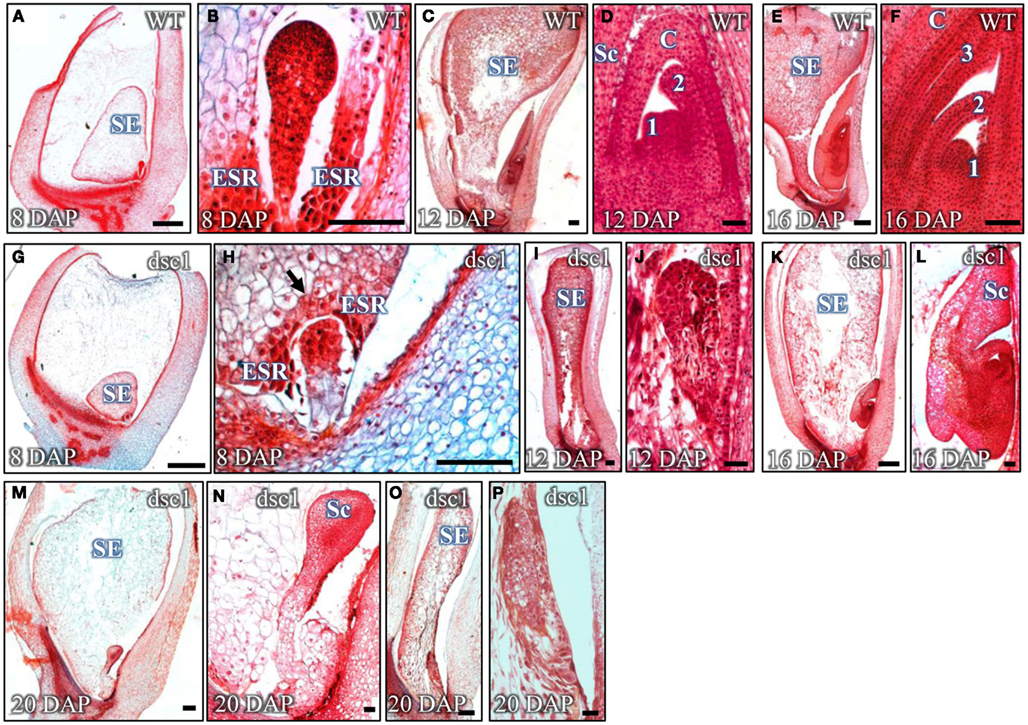
Figure 2. Embryogenesis is delayed in dscl-R mutant embryos prior to degeneration. Sagittal sections of (A–F) non-mutant and (G–L) dscl-R mutant embryos harvested at 8 DAP (A,B,G,H), 12 DAP (C,D,I,J), and 16 DAP (E,F,K,L) stained with Safranin-0 and Fast Green. dscl-R mutants harvested at 20 DAP are degenerate (M–P). Images of embryos in (B,D,F,H,J,L,N,P) are magnified from the whole kernel images in (A,C,E,G,I,K,M,O), respectively. DAP, days after pollination; ESR, embryo surrounding region; SE, starchy endosperm; Sc, scutellum; C, coleoptile; 3, foliar leaf 3; 2, foliar leaf 2; 1, foliar leaf 1. Arrow points to intact ESR in dscl mutant kernel. Scale bars in (A,C,E,G,I,K,M,O) represent 500 um and in (B,D,F,H,J,L,N,P) represent 100 um.
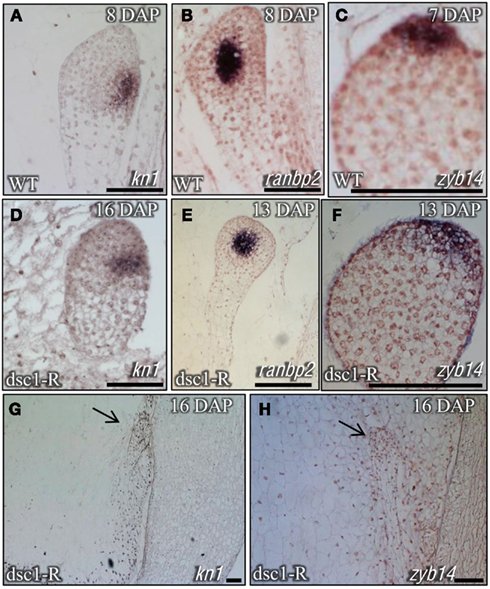
Figure 3. Transcriptional markers illustrate that dscl-R mutants develop a shoot meristem, vasculature, and initiate lateral organs. Severe dscl-R mutants are not able to maintain a meristem and lateral organ initiation. Sagittal sections of (A–C) non-mutant and (D–H) dscl-R mutant embryos probed with knotted 1 (kn1; A,D,G), ran binding protein2 (ranbp2; B,E), and Zea mays yabby14 (zyb14; C,F,H). (A–F) Non-mutant and dscl-R mutant embryos were harvested at different time-points to show the same stage of embryo development. DAP, days after pollination. Scale bars represent 100 um.
Similar to the development discolored mutant embryos, the differentiation of aleurone, BETL, and starchy endosperm cell types is also delayed and eventually aborted in discolored mutant kernels. For example, all these endosperm-specific structures are fully differentiated in 8 DAP wild type kernels. Aleurone cells assume their distinctive cuboidal shape within a single layer surrounding the perimeter of the starchy endosperm (Figure 4A), and three layers of highly extended BETL cells develop in the base of the endosperm, immediately juxtaposed to the maternally derived placenta (Figure 4B). In contrast, differentiated aleurone cells are not identified in the discolored endosperm at 8 DAP (Figure 4E), and just a single layer of BETL cells form at the base of the mutant endosperm (Figure 4F). By 16 DAP however, both discolored mutant and wild type sibling kernels have fully differentiated endosperm structures, including an anatomically distinct aleurone layer surrounding the enlarged, vacuolated cells of the starchy endosperm, and a BETL comprising three cell layers (Figures 4C,D,G,H). Finally, discolored mutant endosperm development is again aberrant by 20 DAP, wherein undifferentiated cells are observed within the mutant aleurone and BETL layers and other cells are degenerated and of non-descript identity (Figures 4I–L).
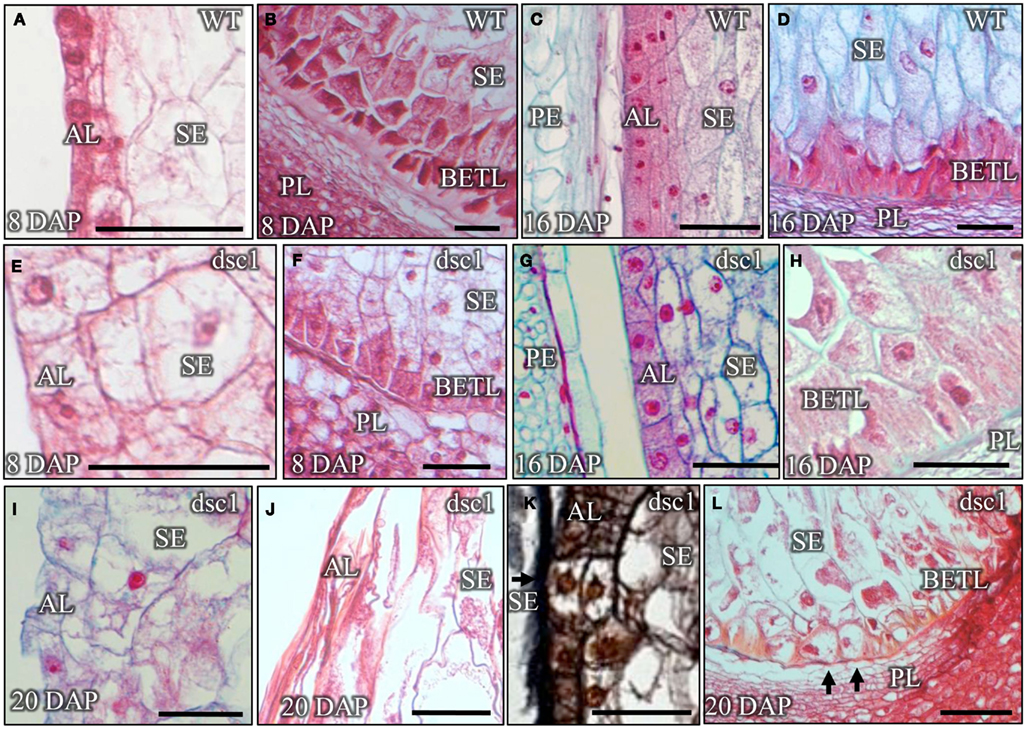
Figure 4. Development of endosperm tissues is delayed in dscl-R mutant kernels, prior to eventual tissue degeneration. Sagittal sections of (A–D) non-mutant and (E–L) dscl-R mutant kernels harvested at 8 DAP (A,B,E,F), 16DAP (C,D,G,H), and 20 DAP (I,L). (I) The lack of staining due to the presence of dense cytoplasm and the round cell morphology indicate that the aleurone layer has not fully differentiated in this 20 DAP kernel. (J) The aleurone and the starchy endosperm cells in this 20 DAP kernel have already degenerated. (K) The arrow points to undifferentiated cells in the aleurone cell layer in this kernel harvested 20 DAP. (L) The arrows point to misshapen basal endosperm transfer layer cells that lack cell wall ingrowths at the base of this 20 DAP kernel. All sections were stained with Safranin-0 and Fast Green for the exception of (K), which is stained with Safranin-O and Orange G. AL, aleurone; BETL, basal endosperm transfer layer; DAP, days after pollination; PE, pericarp; PL, placenta; SE, starchy endosperm. Scale bars represent 100 μm.
dsc1 Encodes an ADP-Ribosylation Factor-GTPase Activating Protein
Previously, a 3,808 bp Mu1-inserted 5′ genomic fragment of the dsc1 locus on chromosome 4S was identified by transposon-tagging analysis of the dsc1-R mutation (Scanlon and Myers, 1998). Three additional Mu-insertion alleles of dsc1 (dsc1-H02, dsc1-C06, and dsc1-B09) obtained from the TUSC (Meeley and Briggs, 1995) failed to complement the dsc1-R mutation (Figure 5A), and thus provided further confirmation that the dsc1 locus has been cloned. Alignment of the ∼2.4 KB genomic DNA fragment of the dsc1 clone to the sequenced maize genome (MaizeSequence.org Release 5b.60; Schnable et al., 2009) identified the predicted full length gene, the 2.472 KB full length transcript, the boundaries of the 17 predicted introns, and the 823 amino acid sequence of the predicted dsc1 gene product. The sequence of the dsc1 cDNA was confirmed by RT-PCR using primers anchored within the dsc1-R transposon-inserted genomic DNA fragment described above (Figures 5A,B). PCR analyses utilizing dsc1gene-specific primers (Table A1 in Appendix) and a primer targeted to the Mu transposon termini (MuTIR; Table A1 in Appendix) identified the Mu-insertion sites of the four dsc1 mutant alleles. As indicated in Figure 5A, the dsc1-R and dsc1-H02 alleles harbor 5′ Mu transposon insertions located 233 bp and 283 bp upstream of the dsc1start codon, respectively. In contrast, the dsc1-C06 and dsc1-B09 alleles contain Mu-insertions within the first intron, 284 bp and 250 bp downstream from the DSC1 start codon. Notably, no exon insertions were identified among our dsc1 mutant alleles. The predicted 823 amino acids of the DSC1 protein encode a putative ARF-GAP protein comprising a BIN-amphiphysin-RVS (BAR) domain, a pleckstrin homology motif, and two ankyrin repeats, in addition to an ARF-GAP domain (Figure 5B). Both the BAR and pleckstrin homology domains are implicated in membrane interactions (Hurley, 2006), whereas ankyrin repeats are known to function in protein-protein interactions (Inoue and Randazzo, 2007). ARF-GAPs comprise a highly conserved group of proteins within the eukaryotes, and function during the regulation of vesicle trafficking (Vernoud et al., 2003; Jiang and Ramachandran, 2006; Inoue and Randazzo, 2007). Taken together, the DSC1 protein is predicted to function during endomembrane trafficking in maize.
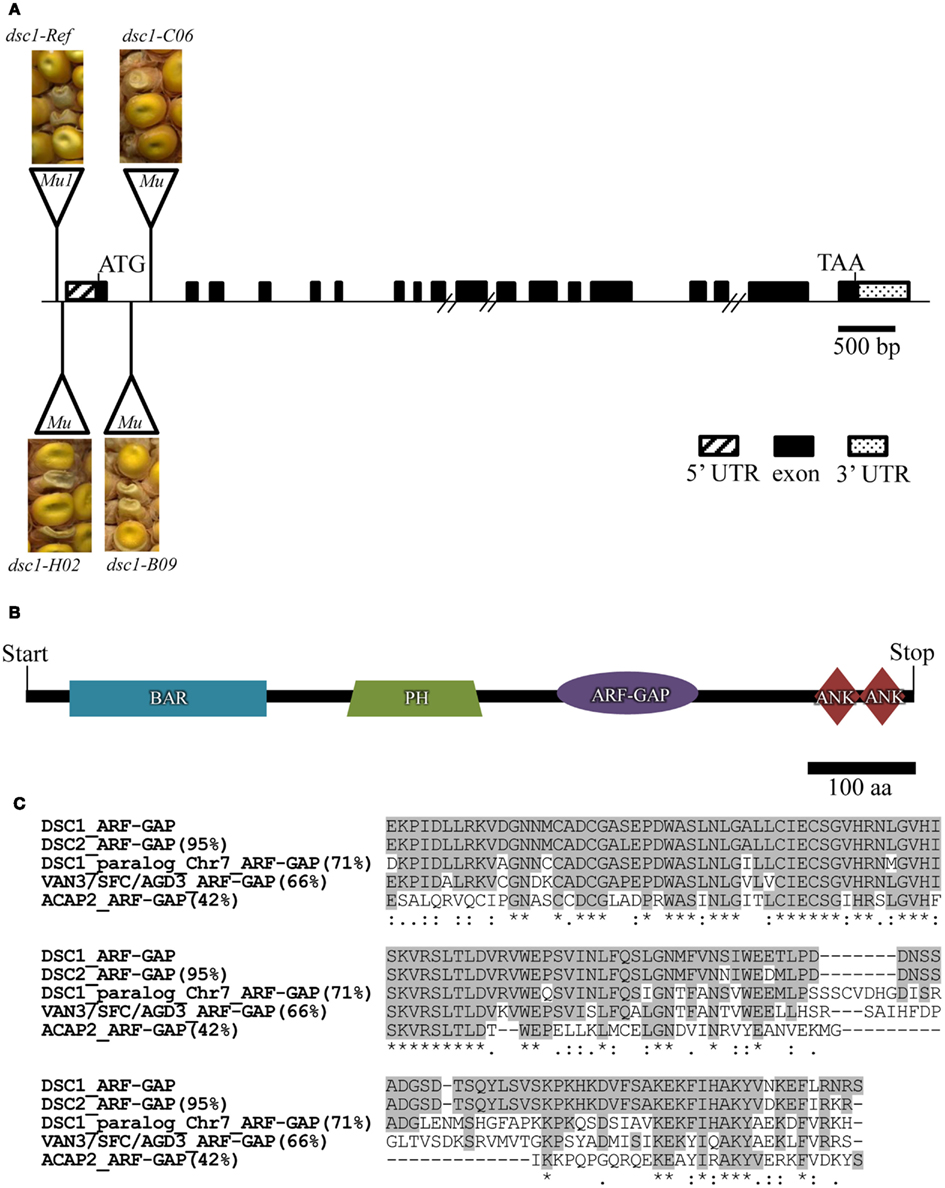
Figure 5. dscl encodes an ARF-GAP. (A) The full length dscl transcript is 3.194 kb. Positions of Mu transposon insertions in individual dscl mutant alleles with images of their kernel phenotypes from failed complementation crosses are indicated. Hash marks denote unknown lengths of genomic fragments. (B) The DSC1 protein comprises 823 amino acids and is composed of a BAR, a PH, an ARF-GAP, and two ANKYRIN domains. (C) ClustalW mediated amino acid alignment of the ARF-GAP of DSC 1 with that of the two maize paralogs on chromosomes 1 (DSC2, GRMZM5G872204) and 7 (GRMZM2G059225), the Arabidopsis protein VAN3/SFC/AGD3 (NP196834), and ACAP2 (AAH60767) from Homo sapiens. Amino acid percent identity to DSC1 ARF-GAP domain is in parentheses.
A total of 43 predicted maize genes encode an ARF-GAP domain, including dsc1 and a close paralog (95% nucleotide identity/98% amino acid identity) located on chromosome 1 (GRMZM5G872204, designated here as dsc2). In adherence to the mammalian classification system, nine predicted maize ARF-GAP proteins (including DSC1 and DSC2) belong to the ACAP subgroup, comprising ARF-GAPs that function during post-Golgi transport (Jackson et al., 2000; Miura et al., 2002; Nie et al., 2003; Randazzo and Hirsch, 2004). The ARF-GAP domain of DSC1 is homologous (66% identity) to that of the Arabidopsis protein VAN3 (Koizumi et al., 2005; Sieburth et al., 2006), which regulates endomembrane trafficking of the auxin efflux protein PIN1, although yet another maize ARF-GAP (GRMZM2G059225) is more similar to VAN3 (69% identity). ClustalW alignments of the ARF-GAP domains of DSC1, DSC2, VAN3, and a human ACAP-type ARF-GAP (ACAP2) are shown in Figure 5C.
RT-PCR identified dsc1 transcript accumulation during multiple stages of kernel development (6DAP, 8 DAP, 12 DAP, 14 DAP, 16 DAP, and 18 DAP) and in 14-day-old seedling shoots and roots, but not in the fully differentiated distal tips of emerged seedling leaves (Figure 6A). Notably, dsc1 transcript accumulation in 12 DAP, early transition-staged discolored mutant kernels is reduced to less than 0.3 times the level found in wild-type transition-staged seed harvested at 8 DAP (Figure 6B). Furthermore, transcripts of the kernel patterning genes dek1 and cr4 both accumulate to less than 0.4 fold the level of wild type in transition-staged discolored mutants, although accumulation of sal1 is not significantly altered by the dsc1-R mutation (Figure 6B).
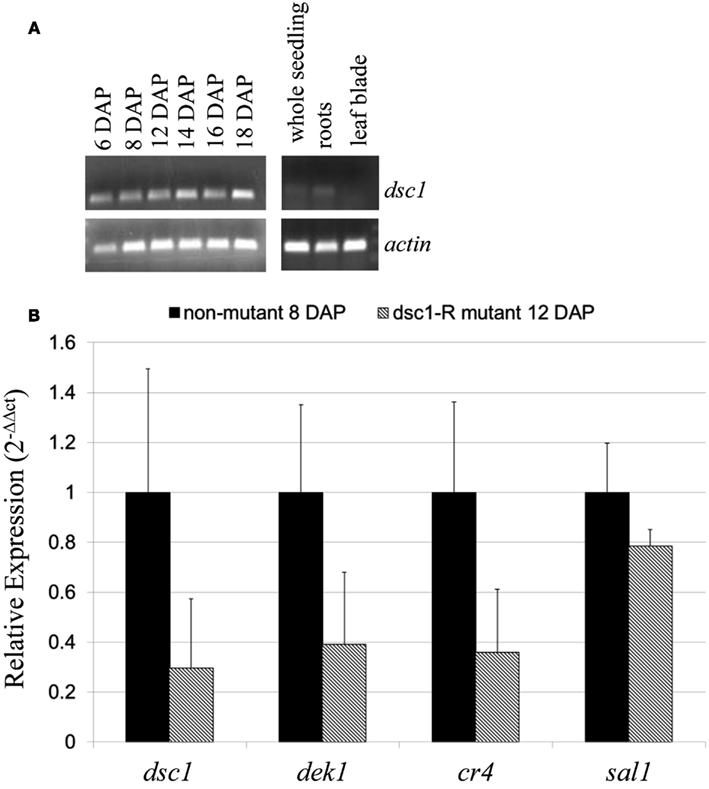
Figure 6. Transcript accumulation of dscl. (A) RT-PCR of dscl transcript accumulation in developing maize kernels (6–18 DAP), 14-day-old seedlings, 14-day-old roots, and leaf blade tips. (B) qRT-PCR analysis of dscl, dekl, cr4, and sall in 8 DAP non-mutant and dscl-R mutant transition-staged kernels. Accumulation was normalized to 18S rRNA. Each experiment utilized three biological replicates. Error bars denote SE. Primers were designed to the 3′ end of the dscl transcript and can be found in Table A1 in Appendix.
DSC1 Co-Localizes with the Trans-Golgi Network/Early Endosomes and the Plasma Membrane
Yellow fluorescent protein (YFP)-tagged DSC1 constructs were generated to determine the subcellular localization of DSC1 in the N. benthamiana leaf (Earley et al., 2006). Epifluorescence and confocal microscopic imaging of infiltrated transgenic leaf sectors reveals35S-YFP<DSC1 accumulation within distinct intracellular compartments, and as foci at the plasma membrane (Figures 7A,D,G,J,M; Figures A1A,C and A2A,D,G in Appendix). 35S-YFP<DSC1 labeled bodies also actively move around the cell, as evidenced by time series taken of cells to illustrate the intracellular movement of 35S-YFP-tagged DSC1 (Movie S1 in Supplementary Material). In contrast, N. benthamiana leaves expressing control 35S-YFP construct that lacked the DSC1 open reading frame show fluorescent signal within the nucleus, or dispersed within the cytoplasm (Figures A1E,G in Appendix).
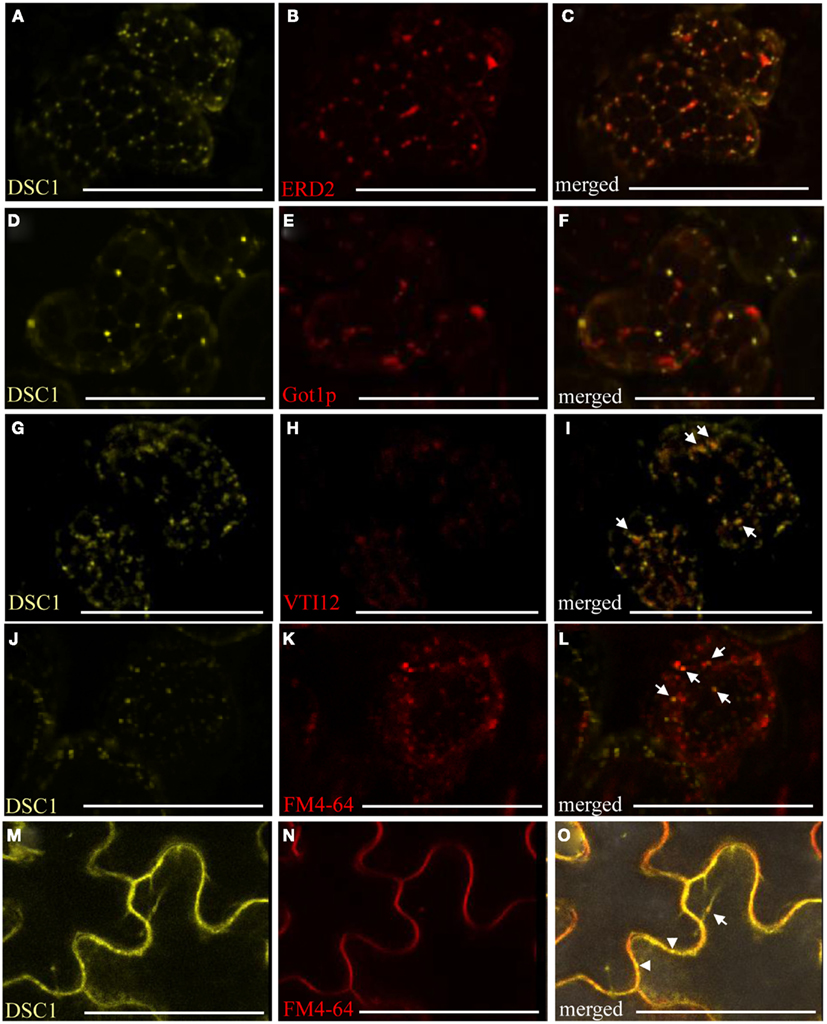
Figure 7. YFP-tagged DSC 1 in transient expression assays in N. benthamiana leaves. (A,D) 35S-YFP<DSC1 bodies do not co-localize with (B) cis-Golgi marker 35S-ERD2<GFP [(C), merged], or (E) the Golgi marker UBQ10-mCherry<Gotlp [(F), merged]. (G,J,M) 35S-YFP<DSC1 bodies do co-localize with some of the intracellular compartments labeled with (H) the trans-Golgi/early endosomal marker UBQ10-mCherry<VTI12 [(I), merged] and (K,N) the endocytic tracer FM4-64 in mesophyll and epidermal cells [(L,O), merged]. Arrows point to compartments that co-localize with YFP-tagged DSC1. Arrowheads point to YFP-tagged DSC1 compartments at the plasma membrane. Scale bars represent 50 urn.
The lipophilic, styryl dye FM4-64 (Invitrogen) fluoresces in hydrophobic environments, and is commonly utilized to trace endocytotic activities in cells (Bolte et al., 2004). Once internalized FM4-64 labeled membranes successively co-localize with the trans-Golgi network/early endosomes, prevacuolar compartments, and ultimately with the tonoplast (vacuolar membrane; Bolte et al., 2004; Geldner et al., 2009). Co-localization assays with several organelle-specific markers and the endocytic tracer FM4-64 were performed to identify more precisely the intracellular localization of YFP-tagged DSC1 protein. These studies confirmed that 35S-YFP<DSC1does not co-localize with the mitochondrial marker 35S35SAMV-COXIV<GFP, the peroxisome marker 35S-DsRed<catalase, auto-fluorescent chloroplasts, the cis-Golgi marker 35S-ERD2<GFP; or with the Golgi stacks marker UBQ10-mCherry<Got1p (Figures A2A–I in Appendix; Figures 7A–F; Kohler et al., 1997; Boevink et al., 1998; Reisen and Hanson, 2007; Geldner et al., 2009). However, in some instances35S-YFP<DSC1 labeled bodies localize next to the cis-Golgi and Golgi stacks markers (Figures 7A–F). Recurrently, 35S-YFP<DSC1 labeled bodies co-localize with the trans-Golgi network/early endosome marker UBQ10-mCherry<VTI12 (Figures 7G–I), in addition toFM4-64 labeled intracellular compartments and the plasma membrane (Figures 7J–O; Geldner et al., 2009). Taken together, the cellular mobility of YFP-tagged DSC1 labeled compartments and their co-localization with both FM4-64 and a marker for the trans-Golgi network/early endosome illustrate that DSC1 functions in endomembrane trafficking.
ZmPIN1a Endomembrane Transport is not Disrupted in Discolored Mutant Embryos
In Arabidopsis, the ARF-GAP VAN3 functions during the transport of PIN1 from the plasma membrane to the recycling endosome (Sieburth et al., 2006). YFP-tagged ZmPIN1a localization was observed in wild type and discolored mutant embryos to determine if the endomembrane transport of ZmPIN1a is disrupted by the dsc1-R mutation. ZmPIN1a<YFP localizes to the plasma membrane in both mutant and non-mutant 14 DAP embryos (Figures 8A–D). After treatment with Brefeldin A, which blocks vesicle cycling from the endosome to the plasma membrane (Steinmann et al., 1999; Geldner et al., 2001), ZmPIN1a<YFP accumulates in endosomal compartments in both wild type and discolored mutant embryos and no differences in size or number of ZmPIN1a<YFP-tagged endosomal compartments are observed (Figures 8E,F).
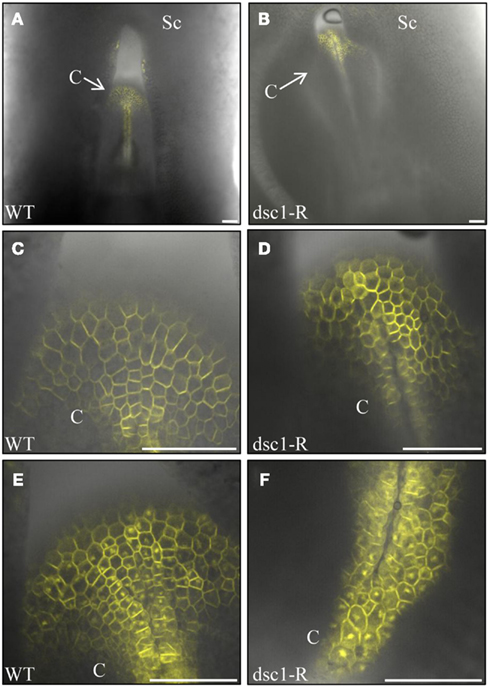
Figure 8. PINFORMEDla (PINla) endomembrane transport is not disrupted in dscl-R mutant embryos. (A) YFP-tagged PPSTla preferentially localizes at the plasma membrane in mock-treated (A,C) non-mutant and (B,D) dscl-R mutant embryos. The coleoptiles imaged in (C,D) are magnified from (A,B), respectively. YFP-tagged PIN la accumulates internally after Brefeldin A (BFA) treatment of embryos in both (E) non-mutant and (F) dscl-R mutant embryos. No difference in the shape or number of BFA bodies in non-mutant and dscl mutant embryos is observed. DAP, days after pollination; C, coleoptile; Sc, scutellum; BFA, Brefeldin A. Scale bars represent 100 urn.
Discussion
Defective kernel mutants are phenotypically variable (Sheridan and Neuffer, 1980; Scanlon et al., 1994), such that the wide range in developmental progression observed in the dsc1-R homozygous mutant endosperm and embryo is not unusual, even after more than six introgressions into the B73 inbred background. Analyses of dsc1 transcript accumulation in equivalently staged discolored mutant and wild type kernels reveal that dsc1 transcripts are reduced in dsc1-Rmutants. Another previously reported phenotype observed in maize dek mutants is the relatively normal, albeit retarded, embryo, and endosperm development of the early-staged kernel, followed by kernel abortion and reabsorption of most or all kernel tissues (Clark and Sheridan, 1988; Scanlon et al., 1997; Fu et al., 2002). In this way, discolored mutant embryos initiate a SAM, vascular tissue, and a scutellum, whereas some mutant embryos progress to the coleoptilar stage or stage L1 before embryo structures completely degenerate (Figures 2G–P). Likewise, the discolored mutant endosperm forms at least some fully differentiated BETL and aleurone cell types prior to kernel abortion. The delay in discolored mutant endosperm development is evidenced by the delay in forming the aleurone cell layer and the presence of only one file of BETL cells (Figures 4E,F). dsc1and dsc2display similar transcript accumulation patterns in previous transcriptomic analyses performed on mixed maize tissues (Sen et al., 2009; Sekhon et al., 2011); partial genetic redundancy of these paralogous ACAP/ARF-GAP proteins early in kernel development may explain why discolored mutant kernels can progress, albeit at a retarded rate, through at least the first stages of kernel development. Perhaps the most interesting and informative aspect of the discolored mutant phenotype is the appearance of undifferentiated cells in the aleurone cell layer, prior to kernel abortion, and tissue degeneration (Figure 4K). This represents a mosaic phenotype and a failure to complete differentiation of the aleurone layer. This striking phenotype reaffirms the conclusions drawn from previous mosaic analyses of maize kernel development (Becraft and Asuncion-Crabb, 2000), in that determination of endosperm cell fate requires a differentiation signal that must be maintained until late stages in kernel development. Our data also implicate a role for DSC1 in the maintenance of endosperm cell fate.
In a heterologous transient expression system, the fluorescently tagged DSC1 protein exhibited intracellular motility and localized to the plasma membrane as well as the trans-Golgi network/early endosomes of N. benthamiana leaves (Figures 7G–O; Movie 1). These data suggest that the DSC1 ARF-GAP protein regulates endomembrane transport of an unknown cargo between the plasma membrane and the trans-Golgi network/early endosomes. Additional co-localization assays could be performed to determine if DSC1 co-localizes with late endosomes or prevacuolar compartments. In addition, localization of ZmPINa<YFP in maize embryos before and after Brefeldin A treatment reveals that DSC1 is not required for trafficking of this auxin efflux transporter from the plasma membrane to endosomal compartments. Taken together, these data suggest that the DSC1 ARF-GAP regulates transport of an unidentified cargo that is required to maintain differentiation of maize kernel tissues. Interestingly, genetic data suggest that SAL1, a class E vacuolar sorting protein, is antagonistic to DEK and CR4, two maize proteins that were initially identified in an intercellular signaling pathway required for the maintenance of endosperm cell fate (Becraft and Asuncion-Crabb, 2000; Becraft et al., 2002; Lid et al., 2002; Shen et al., 2003). However, no direct interaction has yet been demonstrated. We show that DSC1 function is required for normal accumulation of dek1and cr4 mRNA, although transcript accumulation of sal1 is not significantly disrupted in discolored kernels (Figure 6B). These data suggest that DSC1 either functions downstream of SAL1, or in an unrelated signaling pathway. Likewise, these data are consistent with DSC1 function upstream of the CR4/DEK1 signal transduction pathway, however no genetic or biochemical evidence of this interaction is currently available. Our analyses of DSC1 further implicate a role for endomembrane trafficking during maize kernel development, and future studies will investigate the identity of the specific cargo implicated during DSC1 function and its specific role in maize kernel development.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Robert Meeley at Pioneer Hi-Bred International, Inc. for providing us with the TUSC alleles, Maureen Hanson at Cornell University for providing us with the mitochondria, peroxisome, and cis-Golgi markers for our transient expression assays, and David Jackson at the Cold Spring Harbor Laboratory for providing us with the ZmPIN1a∼YFP transgenic line. We also thank the Plant Cell Imaging Center at the Boyce Thompson Institute for Plant Research (Ithaca, New York) for use of the Leica TCS-SP5 confocal microscope, which was purchased with funds provided by the National Science Foundation Major Research Instrumentation Program (NSF DBI-0618969).
Supplementary Material
The Movie S1 for this article can be found online at http://www.frontiersin.org/Plant_Evolution_and_Development/10.3389/fpls.2012.00115/abstract
References
Abbe, E. C., and Stein, O. L. (1954). The growth of the shoot apex in maize: embyrogeny. Am. J. Bot. 41, 285–293.
Becraft, P. W., and Asuncion-Crabb, Y. (2000). Positional cues specify and maintain aleurone cell fate in maize endosperm development.
Becraft, P. W., Li, K., Dey, N., and Asuncion-Crabb, Y. (2002). The maize dek1 gene functions in embryonic pattern formation and cell fate specification. Development 129, 5217–5225.
Becraft, P. W., Stinard, P. S., and McCarty, D. R. (1996). CRINKLY4: a TNFR-like receptor kinase involved in maize epidermal differentiation. Science 273, 1406–1409.
Becraft, P. W., and Yi, G. (2011). Regulation of aleurone development in cereal grains. J. Exp. Bot. 62, 1669–1675.
Boevink, P., Opraka, K., Santa Cruz, S., Martin, B., Betteridge, A., and Hawes, C. (1998). Stacks on tracks: the plant Golgi apparatus traffics on an actin/ER network. Plant J. 15, 441–447.
Bolte, S., Talbot, C., Boutted, Y., Catrice, O., Read, N. D., and Satiat-Jeunemaitre, B. (2004). FM-dyes as experimental probes for dissecting vesicle trafficking in living plant cells. J. Microsc. 214, 159–173.
Chardin, P., Paris, S., Antonny, B., Robinueau, S., Beroud-Dufour, S., Jackson, C. L., and Charbe, M. (1996). A human exchange factor for ARF contains Sec7 and pleckstrin-homology domains. Nature 384, 481–484.
Clark, J. K., and Sheridan, W. F. (1988). Characterization of the two maize embryo-lethal defective kernel mutants rgh*-1210 and fl*-1253B: effects on embryo and gametophyte development. Genetics 120, 279–290.
Cosgrove, D. J. (1997). Assembly, and enlargement of the primary cell wall in plants. Annu. Rev. Dev. Biol. 13, 171–201.
Earley, K. W., Haag, J. R., Pontes, O., Opper, K., Juehne, T., Song, K., and Pikaard, C. S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45, 616–629.
Fu, S., Meeley, R., and Scanlon, M. J. (2002). empty pericarp2 Encodes a negative regulator of the heat shock response and is required for maize embryogenesis. Plant Cell 14, 3119–3132.
Gallavotti, A., Yang, Y., Schmidt, R. J., and Jackson, D. (2008). The relationship between auxin transport and maize branching. Plant Physiol. 147, 1913–1923.
Geldner, N., Anders, N., Wolters, H., Keicher, J., Kornberger, W., Muller, P., Delbarre, A., Ueda, T., Nakano, A., and Jurgens, G. (2003). The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 11, 219–230.
Geldner, N., Devervaud-Tendon, V., Hyman, D. L., Mayer, U., Stierhof, Y-D., and Chory, J. (2009). Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J. 59, 169–178.
Geldner, N., Friml, J., Stierhofm, Y-D., Jurgensm, G., and Palme, K. (2001). Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413, 425–428.
Goldberg, J. (1999). Structural and functional analysis of the ARF1-ARFGAP complex reveals a role for coatmer in GTP hydrolysis. Cell 96, 893–902.
Goodin, M. M., Dietzgen, R. G., Schichnes, D., Ruzin, S., and Jackson, A. O. (2002). pGD vectors: versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J. 31, 375–383.
Inoue, H., and Randazzo, P. A. (2007). Arf GAPs and their interacting proteins. Traffic 8, 1465–1475.
Jackson, D. (1991). “In situ hybridization in plants,” in Molecular Plant Pathology: A Practical Approach, eds D. J. Bowles, S. J. Gurr, and M. McPherson (Oxford, UK: Oxford University Press), 163–174.
Jackson, T. R., Brown, F. D., Nie, Z., Miura, K., Foroni, L., Sun, J., Hsu, V. W., Donaldson, J. G., and Randazzo, P. A. (2000). ACAPs are Arf6 GTPase-activating proteins that function in the cell periphery. J. Cell Biol. 151, 627–638.
Jiang, S-Y., and Ramachandran, S. (2006). Comparative and evolutionary analysis of genes encoding small GTPases and their activating proteins in eukaryotic genomes. Physiol. Genomics 24, 235–251.
Juarez, M. T., Twigg, R. W., and Timmermans, M. C. P. (2004). Specification of adaxial cell fate during maize leaf development. Development 131, 4533–4544.
Kaplan, D. R., and Cooke, T. J. (1997). Fundamental concepts in the embryogenesis of dicotyledons: a morphological interpretation of embryo mutants. Plant Cell 9, 1903–1919.
Kessler, S., Seiki, S., and Sinha, N. (2002). Xcl1 causes delayed oblique periclinal cell division in developing maize leaves, leading to cellular differentiation by lineage instead of position. Development 129, 1859–1869.
Kiesselbach, T. A., and Walker, E. R. (1952). Structure of certain specialized tissue in the kernel of corn. Am. J. Bot. 39, 561–569.
Kohler, R. H., Zipfel, W. R., Webb, W. W., and Hanson, M. R. (1997). The green fluorescent protein as a marker to visualize plant mitochondria in vivo. Plant J. 11, 613–621.
Koizumi, K., Naramoto, S., Sawa, S., Yahara, N., Uedo, T., Nakano, A., Suglyama, M., and Fukuda, H. (2005). VAN3 ARF-GAP-mediated vesicle transport is involved in leaf vascular network formation. Development 132, 1699–1711.
Lid, S. E., Gruis, D., Jung, R., Lorentzen, J. A., Ananiev, E., Chamberlin, M., Niu, X., Meeley, R., Nichols, S. E., and Olsen, O-A. (2002). The defective kernel1 (dek1) gene required for aleurone cell development in the endosperm of maize grains encodes a membrane protein of the calpain gene superfamily. Proc. Natl. Acad. Sci. U.S.A. 99, 5460–5465.
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408.
Meeley, R., and Briggs, S. (1995). Reverse genetics for maize. Maize Genet. Coop. News Lett. 69, 67–82.
Miura, K., Jacques, K. M., Stauffer, S., Kubosaki, A., Zhu, K., Hirsch, D. S., Resau, J., Zheng, Y., and Randazzo, P. A. (2002). ARAP1: a point of convergence for Arf and Pho signaling. Mol. Cell 9, 109–119.
Naramoto, S., Klein-Vehn, J., Robert, S., Fujimoto, M., Dainobu, T., Paciorek, T., Ueda, T., Nakano, A., Van Montagu, M. C., Fukuda, H., and Friml, J. (2010). ADP-ribosylation facotre machinery mediates endocytosis in plant cells. Proc. Natl. Acad. Sci. U.S.A. 107, 21890–21895.
Neuffer, M. G., and Sheridan, W. F. (1980). Defective kernel mutants of maize. I. Genetic and lethality studies. Genetics 95, 929–944.
Nie, Z., Boehm, M., Boja, E. S., Vass, W. C., Bonifacino, J. S., Fales, H. M., and Randazzo, P. A. (2003). Specific regulation of the adaptor protein complex AP-3 by the Arf GAP AGAP1. Dev. Cell 5, 455–463.
Olsen, O.-A. (2001). Endosperm development: cellularization and cell fate specification. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 233–267.
Opsahl-Ferstad, H-G., Le Deunff, E., Dumas, C., and Rogowsky, P. M. (1997). ZmEsr, a novel endosperm-specific gene expressed in a restricted region around the maize embryo. Plant J. 12, 235–246.
Poethig, R. S., Coe, E. H. Jr., and Johri, M. M. (1986). Cell lineage patterns in maize embryogenesis: A clonal analysis. Dev. Biol. 117, 392–404.
Prescott, A., and Martin, C. (1986). A rapid method for the quantitative assessment of levels of specific mRNAs in plants. Plant Mol. Biol. Rep. 4, 219–224.
Randazzo, P. A., and Hirsch, D. S. (2004). Arf GAPs: multifunctional proteins that regulate membrane traffic and actin remodeling. Cell. Signal. 16, 401–413.
Reisen, D., and Hanson, M. R. (2007). Association of six YFP-myosin XI-tail fusions with mobile plant cell organelles. BMC Plant Biol. 7, 6. doi:10.1186/1471-2229-7-6
Samaj, J., Readm, N. D., Volkmann, D., Menzel, D., and Baluska, F. (2005). The endocytic network in plants. Trends Cell Biol. 15, 425–433.
Scanlon, M. J., and Myers, A. M. (1998). Phenotypic analysis and molecular cloning of discolored-1 (dsc1) a maize gene required for early kernel development. Plant Mol. Biol. 37, 483–493.
Scanlon, M. J., Myers, A. M., Schneeberger, R. G., and Freeling, M. (1997). The maize gene empty pericarp-2 is required for progression beyond early stages of embryogenesis. Plant J. 12, 901–909.
Scanlon, M. J., Stinard, P. S., James, M. G., Myers, A. M., and Robertson, D. S. (1994). Genetic analysis of 63 mutations affecting maize kernel development isolated from Mutator stocks. Genetics 136, 281–294.
Scheffzek, K., Ahmadian, M. R., and Wittinghofer, A. (1998). GTPase-activating proteins: helping hands to complement an active site. Trends Biochem. Sci. 23, 257–262.
Schel, J. H. N., Kieft, H., and Van Lammeren, A. A. M. (1984). Interactions between embryo and endosperm during early developmental stages of maize caryopses (Zea mays). Can. J. Bot. 62, 2842–2853.
Schnable, P. S., Ware, D., Fulton, R. S., Stein, J. C., Wei, F., Pasternak, S., Liang, C., Zhang, J., Fulton, L., Graves, T. A., Minx, P., Reily, A. D., Courtney, L., Kruchowski, S. S., Tomlinson, C., Strong, C., Delehaunty, K., Fronick, C., Courtney, B., Rock, S. M., Belter, E., Du, F., Kim, K., Abbott, R. M., Cotton, M., Levy, A., Marchetto, P., Ochoa, K., Jackson, S. M., Gillam, B., Chen, W., Yan, L., Higginbotham, J., Cardenas, M., Waligorski, J., Applebaum, E., Phelps, L., Falcone, J., Kanchi, K., Thane, T., Scimone, A., Thane, N., Henke, J., Wang, T., Ruppert, J., Shah, N., Rotter, K., Hodges, J., Ingenthron, E., Cordes, M., Kohlberg, S., Sgro, J., Delgado, B., Mead, K., Chinwalla, A., Leonard, S., Crouse, K., Collura, K., Kudrna, D., Currie, J., He, R., Angelova, A., Rajasekar, S., Mueller, T., Lomeli, R., Scara, G., Ko, A., Delaney, K., Wissotski, M., Lopez, G., Campos, D., Braidotti, M., Ashley, E., Golser, W., Kim, H., Lee, S., Lin, J., Dujmic, Z., Kim, W., Talag, J., Zuccolo, A., Fan, C., Sebastian, A., Kramer, M., Spiegel, L., Nascimento, L., Zutavern, T., Miller, B., Ambroise, C., Muller, S., Spooner, W., Narechania, A., Ren, L., Wei, S., Kumari, S., Faga, B., Levy, M. J., McMahan, L., Van Buren, P., Vaughn, M. W., Ying, K., Yeh, C. T., Emrich, S. J., Jia, Y., Kalyanaraman, A., Hsia, A. P., Barbazuk, W. B., Baucom, R. S., Brutnell, T. P., Carpita, N. C., Chaparro, C., Chia, J. M., Deragon, J. M., Estill, J. C., Fu, Y., Jeddeloh, J. A., Han, Y., Lee, H., Li, P., Lisch, D. R., Liu, S., Liu, Z., Nagel, D. H., McCann, M. C., SanMiguel, P., Myers, A. M., Nettleton, D., Nguyen, J., Penning, B. W., Ponnala, L., Schneider, K. L., Schwartz, D. C., Sharma, A., Soderlund, C., Springer, N. M., Sun, Q., Wang, H., Waterman, M., Westerman, R., Wolfgruber, T. K., Yang, L., Yu, Y., Zhang, L., Zhou, S., Zhu, Q., Bennetzen, J. L., Dawe, R. K., Jiang, J., Jiang, N., Presting, G. G., Wessler, S. R., Aluru, S., Martienssen, R. A., Clifton, S. W., McCombie, W. R., Wing, R. A., and Wilson, R. K. (2009). The B73 maize genome: complexity, diversity, and dynamics. Science 326, 1112–1115.
Sekhon, R. S., Haining, L., Childs, K. L., Hansey, C. N., Buell, C. R., de Leon, N., and Kaeppler, S. M. (2011). Genome-wide atlas of transcription during maize development. Plant J. 66, 553–563.
Sen, T. Z., Andorf, C. M., Schaeffer, M. L., Harper, L. C., Sparks, M. E., Duvick, J., Brendel, V. P., Cannon, E., Campbell, D. A., and Lawrence, C. J. (2009). MaizeGDB becomes ‘sequence-centric’. Database (Oxford) 2009:bap020.
Shen, B., Li, C., Min, Z., Meeley, R. B., Tarczynski, M. C., and Olsen, O-A. (2003). sal1 determines the number of aleurone cell layers in maize endosperm and encodes a class E vauolar sorting protein. Proc. Natl. Acad. Sci. of U.S.A. 100, 6552–6557.
Sheridan, W. F., and Nueffer, M. G. (1980). Defective kernel mutants of maize II. Morphological and embryo culture studies. Genetics 95, 945–960.
Sieburth, L. E., Muday, G. K., King, E. J., Benton, G., Kim, S., Metcalf, K. E., Myers, L., Seamen, E., and Van Norman, J. M. (2006). SCARFACE encodes an ARF-GAP that is required form normal auxin efflux and vein patterning in Arabidopsis. Plant Cell 18, 1396–1411.
Smith, L. G., Jackson, D., and Hake, S. (1995). Expression of knotted1 marks shoot meristem formation during maize embryogenesis. Dev. Genet. 16, 344–348.
Steinmann, T., Geldner, N., Grebe, M., Mangold, S., Jackson, C. L., Paris, S., Galweiler, L., Palme, K., and Jurgens, G. (1999). Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science 286, 316–318.
Takai, Y., Sasaki, T., and Matozaki, T. (2001). Small GTP-binding proteins. Physiol. Rev. 81, 153–208.
Thompson, R. D., Hueros, G., Becker, H., and Maitz, M. (2001). Development and functions of seed transfer cells. Plant Sci. 160, 775–783.
Vernoud, V., Horton, A. C., Yang, Z., and Nielsen, E. (2003). Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol. 131, 1191–1208.
Yoo, C.-M., Quan, L., Cannon, A. E., Wen, J., and Blancaflor, E. B. (2012). AGD1, a class 1 ARF-GAP, acts in common signaling pathways with phosphoinositide metabolism and the actin cytoskeleton in controlling Arabidopsis root hair polarity. Plant J. 69, 1064–1076.
Yoo, C.-M., Wen, J., Motes, C. M., Sparks, J. A., and Blancaflor, E. B. (2008). A class I ADP-ribosylation factor GTPase-activating protein is critical for maintaining direction root hair growth in Arabidopsis. Plant Physiol. 147, 1659–1674.
Zhang, X., Madi, S., Borsuk, L., Nettleton, D., Elshire, R. J., Buckner, B., Janick-Buckner, D., Beck, J., Timmermans, M., Schnable, P. S., and Scanlon, M. J. (2007). Laser microdissection of narrow sheath mutant maize uncovers novel gene expression in the shoot apical meristem. PLoS Genet. 3, e101. doi:10.1371/journal.pgen.0030101
Appendix
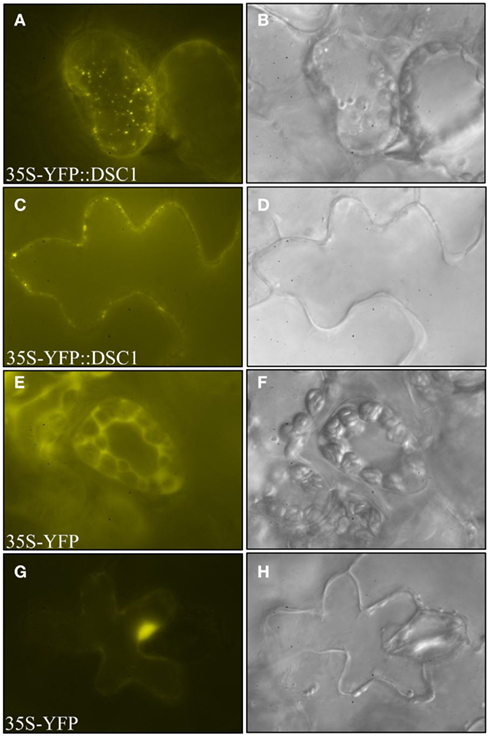
Figure A1. Subcellular localization of YFP-tagged DSC1 and 35S-YFP control construct. DSC1 localizes to punctate structures in (A) mesophyll and (C) epidermal cells. The control construct has nucleo-cytoplasmic localization in (E) mesophyll and (G) epidermal cells. (B,D,F,H) Bright field images were taken of the cells located directly to the left. Images were obtained using epifluorescence microscopy.
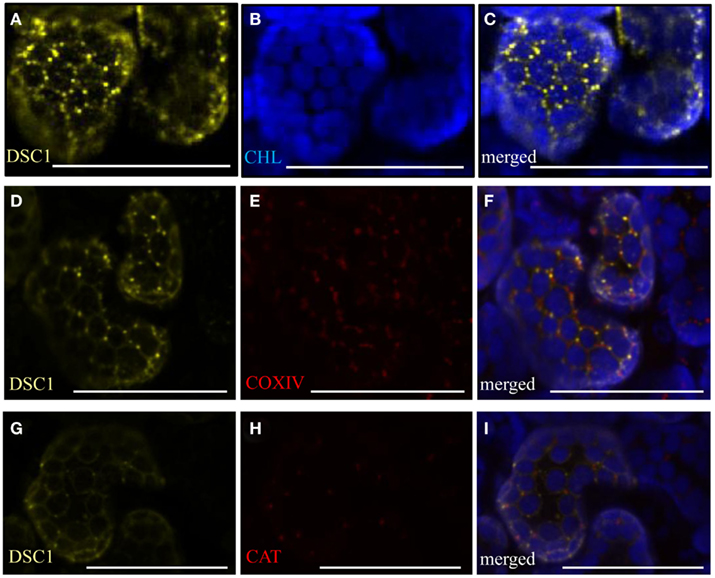
Figure A2. YFP-tagged DSC1 transient expression assays in N. benthamiana. (A,D,G) 35S-YFP<DSC1 bodies do not co-localize with (B) chlorophyll auto-fluorescence [(C), merged], (E) the mitochondria marker 35S35SAMV-COXIV<GFP [(F), merged], or (H) the peroxisome marker 35S-DsRed<catalase [(I), merged]. Chlorophyll auto fluorescence is shown in each of the merged images (C,F,I). Scale bars represent 50 μm. CHL, chlorophyll.
Keywords: maize, embryo, endosperm, dsc1, ADP-ribosylation factor-GTPase activating protein (ARF-GAP), endomembrane trafficking
Citation: Takacs EM, Suzuki M and Scanlon MJ (2012) Discolored1 (DSC1) is an ADP-ribosylation factor-GTPase activating protein required to maintain differentiation of maize kernel structures. Front. Plant Sci. 3:115. doi: 10.3389/fpls.2012.00115
Received: 09 February 2012; Accepted: 14 May 2012;
Published online: 31 May 2012.
Edited by:
Kimberly Lynne Gallagher, University of Pennsylvania, USAReviewed by:
Philip W. Becraft, Iowa State University, USAA. Mark Settles, University of Florida, USA
Sarah Liljegren, University of Mississippi, USA
Copyright: © 2012 Takacs, Suzuki and Scanlon. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Michael J. Scanlon, Department of Plant Biology, Cornell University, 412 Mann Library, Ithaca, NY 14853, USA. e-mail:bWpzMjk4QGNvcm5lbGwuZWR1

 Masaharu Suzuki2
Masaharu Suzuki2