- 1 Istituto di Genomica Applicata, Udine, Italy
- 2 CNR, Istituto di Genetica Vegetale, Sezione di Firenze, Firenze, Italy
- 3 Dipartimento di Scienze Agrarie e Ambientali, Università di Udine, Udine, Italy
Next generation sequencing (NGS) instruments produce an unprecedented amount of sequence data at contained costs. This gives researchers the possibility of designing studies with adequate power to identify rare variants at a fraction of the economic and labor resources required by individual Sanger sequencing. As of today, few research groups working in plant sciences have exploited this potentiality, showing that pooled NGS provides results in excellent agreement with those obtained by individual Sanger sequencing. The aim of this review is to convey to the reader the general ideas underlying the use of pooled NGS for the identification of rare variants. To facilitate a thorough understanding of the possibilities of the method, we will explain in detail the possible experimental and analytical approaches and discuss their advantages and disadvantages. We will show that information on allele frequency obtained by pooled NGS can be used to accurately compute basic population genetics indexes such as allele frequency, nucleotide diversity, and Tajima’s D. Finally, we will discuss applications and future perspectives of the multiplexed NGS approach.
Introduction
Progress in genetics and genomics has the potential to greatly benefit plant sciences in many aspects, from crop improvement (Varshney et al., 2005) to forest trees management (Neale and Kremer, 2011; Harfouche et al., 2012). One of the main advances that researchers are expecting is the elucidation of the genetic contribution to complex phenotypes. Despite the efforts devoted to this aim and the progress in knowledge that such efforts produced, the genetic contribution to complex phenotypes still remains largely unexplored, and a substantial proportion of the heritability of complex traits is not explained by identified polymorphisms. Several researchers addressed the problem of “missing heritability,” and suggested several determinants of complex traits, including rare variants (Eichler et al., 2010).
In the last decade, researchers focused on genetic association studies, according to the hypothesis that most of the genetic variance was due to common variants with moderate effect (Lander, 1996; Risch and Merikangas, 1996). Association studies allowed the identification of several polymorphisms, cumulatively explaining usually less than 50% of the heritability. This prompted researchers to acknowledge that the common variant/common disease hypothesis could not explain a large proportion of genetic variance, and raised again the interest for the study of rare variants (Bodmer and Bonilla, 2008).
In plant science, the identification of variants affecting complex traits has practical implications for the development of breeding programs. Researchers working on experimental organisms learnt how to induce mutations in DNA using ionizing radiations even before the DNA structure became actually known (Muller, 1927); this gave rise to the reverse genetic approach to study gene function. One of the best-known evolutions of traditional mutagenesis in plant science is TILLING, in which traditional chemical mutagenesis is followed by a high-throughput screening for point mutations (McCallum et al., 2000). TILLING has then been adapted to the discovery of polymorphisms in natural populations and termed ecotilling (Comai et al., 2004). Ecotilling allows the effective screening of large samples and the identification of rare genetic variants at reduced costs, by reducing the amount of sequencing required. As an alternative, several researchers focused on individual Sanger sequencing of large samples, although with relatively large investments in labor and reagents (Marroni et al., 2011b).
The advent of next generation sequencing (NGS), also referred to as deep sequencing and massively parallel sequencing, revolutionized the fields of genetics and genomics (Mardis, 2008). NGS enables researchers to obtain large amount of sequence data at relatively low cost so that a single research group can easily obtain the entire genome of a human individual (Wheeler et al., 2008).
With a shift of perspective, the same amount of sequencing data can be used to obtain sequence information in a limited number of PCR amplicons for a large number of subjects (Ingman and Gyllensten, 2009; Out et al., 2009). Plant scientists just discovered the potential of this approach and deemed it as a potential evolution of TILLING and ecotilling (Marroni et al., 2011a; Tsai et al., 2011). As of today, four studies have faced the problem of identifying variants in candidate regions in large plant populations via NGS (Maughan et al., 2010; Kharabian-Masouleh et al., 2011; Marroni et al., 2011a; Tsai et al., 2011). We will review the four studies to give a perspective of the different options that are currently available to perform the screening for rare variants, and emphasize the possible contribution of this approach to plant genetics and genomics in the next few years.
Prolog: The Origins of Pooled Multiplexed NGS
Next generation sequencing is considered the ideal tool to perform follow-up sequencing of interesting regions identified by genome-wide association or linkage studies. This has been made possible by the use of DNA barcoding in NGS experiments (Parameswaran et al., 2007; Craig et al., 2008). DNA barcoding also referred to as multiplexing allows the sequencing of target regions in several subjects retaining the individual information. Commercial kits to perform multiplexed NGS experiments are now available, opening the way to several application studies. Plant scientists also took advantage of this technology to sequence plant chloroplasts in multiplex (Cronn et al., 2008).
The first experiments of multiplex sequencing had limitations imposed by the relatively low throughput of the newly released next generation sequencers. As the throughput of next generation instruments increased, researchers became interested in applying NGS to the screening of candidate genes in large samples. This prompted several research groups to perform NGS experiments on pooled DNA (Ingman and Gyllensten, 2009; Out et al., 2009). Out and collaborators sequenced the candidate gene MUTYH in 287 cancer patients and compared results of multiplexed NGS with individual Sanger sequencing. Using variants identified by Sanger as a gold standard they identified 2 false negatives, 4 false positives, and 15 true positives. In addition, they showed that the frequency of the variants identified by multiplexed NGS and individual Sanger sequencing were strongly correlated. Ingman and Gyllensten (2009) obtained a similar result. They used NGS to sequence the gene TSCOT in pooled DNA of 96 patients. They selected 16 identified polymorphisms for which Taqman assays were available. They then compared allele frequencies estimated by pooled NGS and by Taqman assays and observed a strong correlation.
In summary, the above-mentioned studies provided the basis for multiplexed NGS (Parameswaran et al., 2007; Craig et al., 2008) and pooled NGS (Ingman and Gyllensten, 2009; Out et al., 2009). Plant scientists seeking to identify carriers of rare variants took inspiration from work published in human genetics. They combined multiplexing and pooling in a single experiment with the aim to minimize the amount of work needed for the discovery of rare variants and the identification of the carriers of such variants (Marroni et al., 2011a; Tsai et al., 2011).
The Quest for Rare Variants
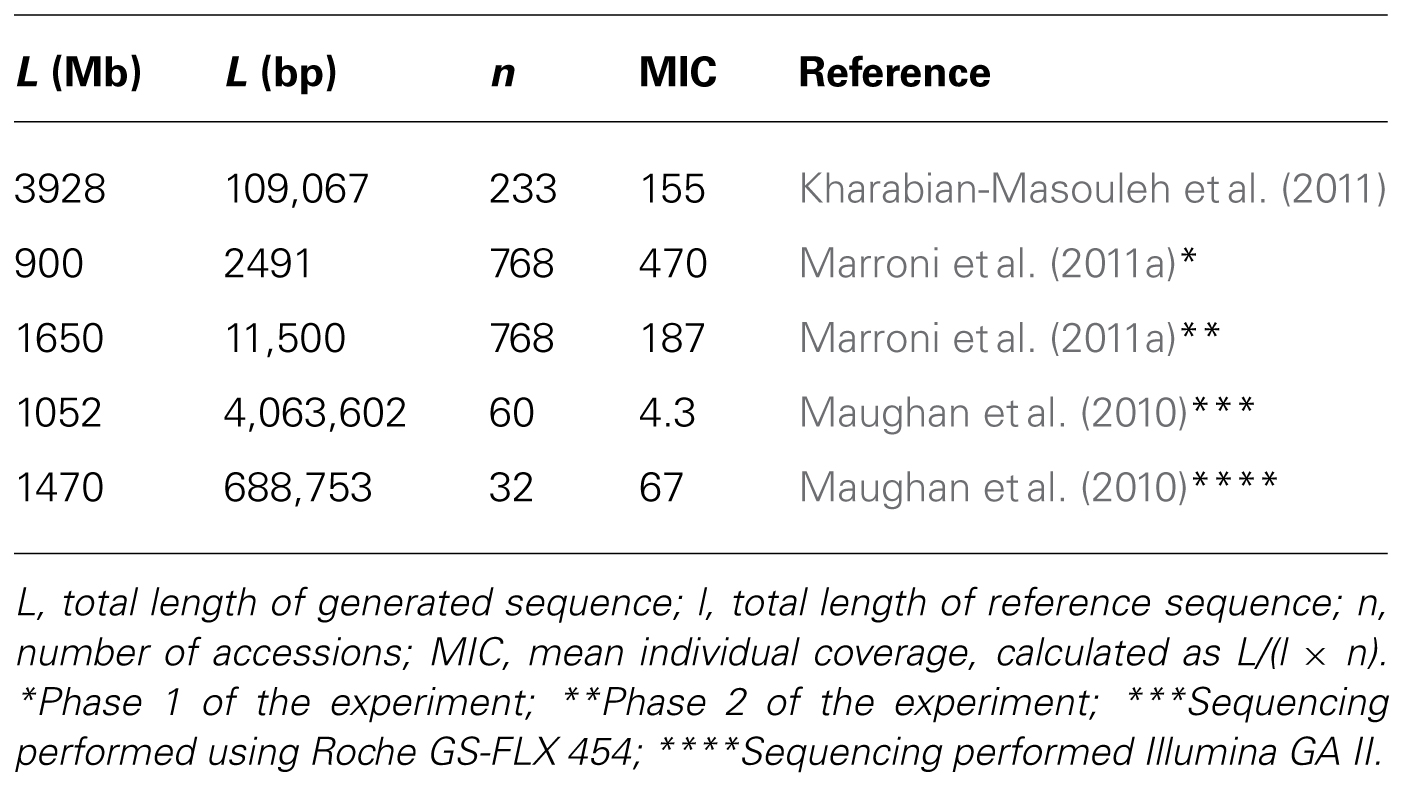
Although there is no universal definition of rare variant, a frequency threshold of 1% is usually considered a reasonable boundary between rare and common variants (Bodmer and Bonilla, 2008). The use of pooled NGS to identify rare variants requires the screening of a large number of individuals for mutations. According to the normal approximation to the binomial distribution, a sample size of about 150 chromosomes (75 individuals for diploid species) is required to have a 95% probability of sampling a variant with a 1% frequency. Studies explicitly aiming at the identification of rare variants (Marroni et al., 2011a; Tsai et al., 2011) require larger sample size than studies not focusing on rare variants (Maughan et al., 2010; Kharabian-Masouleh et al., 2011). Marroni and colleagues investigated 768 poplar (Populus nigra) accessions and Tsai and colleagues sequenced 768 rice (Oryza sativa) accessions and 768 wheat (Triticum durum) accessions, Kharabian-Masouleh and colleagues sequenced 233 rice accessions, and Maughan and colleagues sequenced 60 Arabidopsis thaliana accessions.
Given the above definition, a rare variant might be present in one or a few out of hundreds of sequenced chromosomes. Experiments focusing on the identification of rare variants should therefore be carefully designed to increase researchers’ ability to discriminate mutations from sequencing errors or other kinds of bias. All the reviewed studies selected NGS of pooled libraries as the best option for SNP discovery in large samples. However, each research group applied different variations to the study design according to the research question they were trying to answer. Different approaches have been used regarding (a) selection of the target region, (b) DNA amplification, (c) pooling, and (d) multiplexing. We summarize in Table 1 the different modifications that each group brought to the basic experimental design.
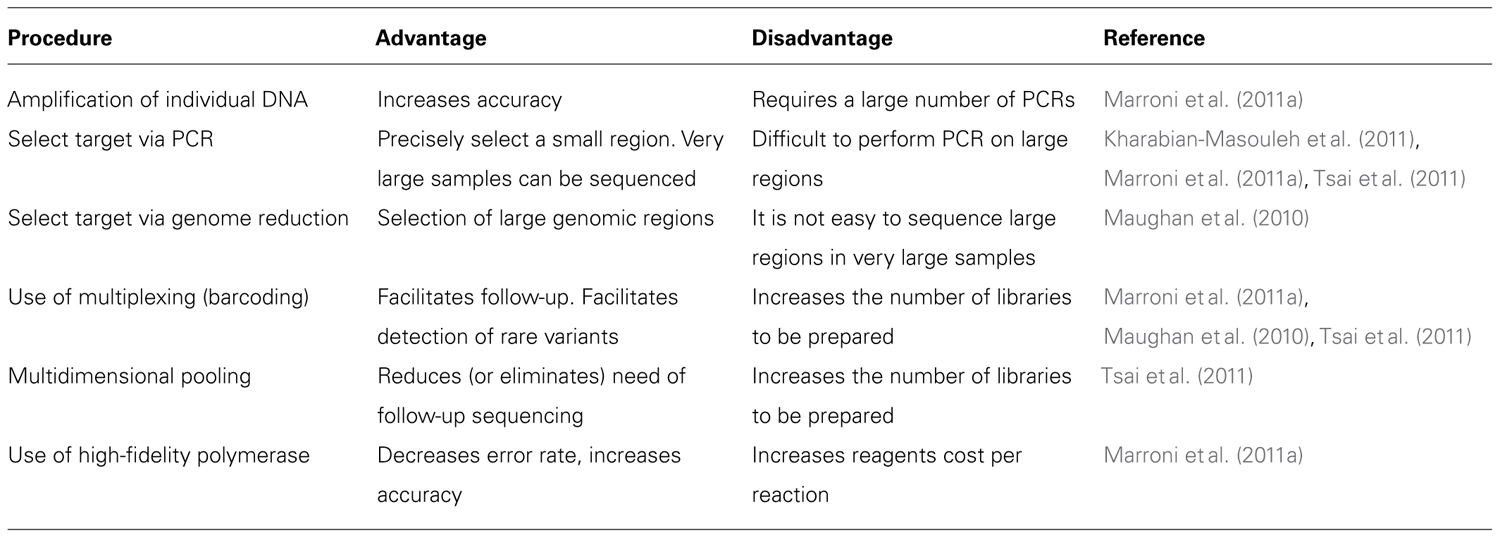
TABLE 1. List of different procedures applied to the basic pooled NGS approach with the respective advantages and disadvantages.
The reviewed studies used outcrossing species, such as P. nigra, and self-pollinating species, such as T. durum and O. sativa. From a technical point of view, identification of rare variants can be performed in the same way in self-pollinating and in outcrossing species. However, if the interest lies in loss-of-function variants likely to affect fitness, the study design might differ. Loss-of-function variants are less likely to occur in self-pollinating species, in which such mutations have a high probability to be observed in homozygous state. Studies aimed at the identification of loss-of-function variants in self-pollinating species are performed on mutant populations (Tsai et al., 2011), while loss-of-function variants have been successfully identified in natural populations of outcrossing species (Marroni et al., 2011a).
Selection of the Target Region
Identification of rare variants is at present not feasible at a genome-wide scale; researchers need to enrich their libraries with DNA fragments originating from the genomic regions of interest. The length of the selected region has a crucial role in the choice of the enrichment method. Studies focusing on small region used PCR amplification (Marroni et al., 2011a; Tsai et al., 2011) or long-range PCR (Kharabian-Masouleh et al., 2011). If the selected regions are larger than a few hundred Kb, then PCR is not the optimal method. One possible alternative is the “reduced representation libraries” or “genome reduction” approach, consisting in the use of restriction endonucleases to randomly select a given number of genomic regions (Maughan et al., 2010).
PCR amplification is better suited when the interest is focused on a limited number of candidate genes to be screened in a very large number of accessions. Marroni and colleagues used PCR amplification to select candidate genes spanning less than 50 Kb of the genome, and sequenced them in 768 accessions. Genome reduction is better suited to survey relatively large genomic regions in a more limited number of accessions. Maughan and collaborators sequenced a target region spanning about 4 Mb in 60 individuals using Roche GS-FLX 454 and a region spanning about 700 Kb using Illumina GA II.
Pooling Strategies, Target Enrichment, and DNA Amplification
Illumina NGS instruments allow the generation of a large amount of sequence data in a single run. Each run is divided into 8 or 16 independent lanes of a flow cell. At the time the studies were performed, each lane could generate “only” one billion base pairs. Sequencing of a region spanning 50 Kb in a single individual in a sequencing lane would have resulted in an average coverage of 20,000-fold. Sequencing pooled DNA of 20 individuals in a single lane would have resulted in a 1000-fold individual coverage. In order to save sequencing resources, sequencing of pooled samples in a single lane was required (Maughan et al., 2010; Kharabian-Masouleh et al., 2011; Marroni et al., 2011a; Tsai et al., 2011). The need for analyzing multiple samples in a single lane is even stronger today, given that the new next generation sequencers can easily generate more than 10 Gb per lane.
The approaches chosen to design the pooling strategies vary. Some researchers minimized the number of PCRs, and amplified pooled DNA (Kharabian-Masouleh et al., 2011; Tsai et al., 2011). However, DNA samples from different individuals may not be amplified with similar efficiency in PCRs, creating random biases (Kharabian-Masouleh et al., 2011). To avoid PCR bias, other researchers performed individual PCRs with the aim of obtaining less bias in the abundance of PCR products for each individual (Marroni et al., 2011a). Amplification of pooled DNA reduces costs, while individual amplification aims at increasing accuracy. Finally, when the population size is not prohibitively large, it is possible to uniquely tag each accession with a barcode (Maughan et al., 2010); we refer to this approach as multiplex sequencing in contrast to pooled multiplex sequencing, where each barcode tags a pool of individuals.
In pooled multiplex sequencing, the size of each pool is a critical issue. Having a large number of accessions in each pool allows saving sequencing resources and minimizes efforts for library preparation. On the other hand, having large pools makes identification of singleton variants difficult, because of the decrease in signal-to-noise ratio. In addition, many studies aim at the follow-up and phenotypic characterization of subjects carrying a given mutation. In such cases, all the accessions composing the pool have to undergo individual sequencing. The larger the pool size, the larger the number of subjects that needs to be sequenced for follow-up (Figure 1). The reviewed studies chose a pool size of 64–96 subjects, corresponding to 128–192 chromosomes. Thus, a mutation occurring only once in the pool would have a frequency of 0.5–0.8%, which can be distinguished from sequencing and amplification errors (Marroni et al., 2011a; Tsai et al., 2011).
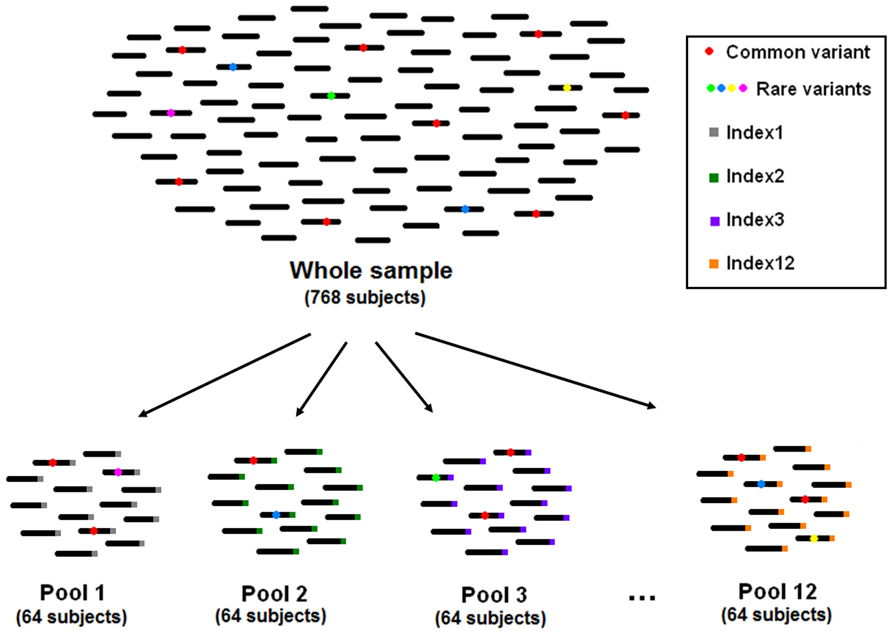
FIGURE 1. The pooling strategy used by Marroni et al., (2011a)
A final option in pooling is multidimensionality. The plain (unidimensional) pooling strategy implies that the n accessions to be sequenced are divided into p pools, each including s accessions, where n = p × s. Multidimensional pooling consists in performing more than one pooling layer. Tsai and colleagues performed a bidimensional pooling by deploying 8 accessions in each of the 96 wells of a plate, and then created 8 row pools (64 subjects each) and 12 column pools (96 subjects each). Each accession was thus sequenced twice; the total number of required libraries was 20. The increase in library preparation effort was compensated by the advantage that follow-up of a variant appearing only once in the whole sample required to sequence only eight subjects. Tsai and colleagues also performed three-dimensional pooling (Figure 2). Accessions were deployed in 12 plates of 64 wells (one accession per plate). Pooling was performed by (1) pooling all the accessions in a plate, thus creating 12 pools of size 64; (2) pooling over columns, creating 16 pools of 48 individuals each; and (3) pooling over rows, creating 16 pools of 48 individuals each. The total number of libraries required by this pooling scheme was 44. The increase in the number of required libraries was balanced by the fact that no follow-up sequencing was needed, since each accession was univocally defined by its coordinates in the three pooling schemes.
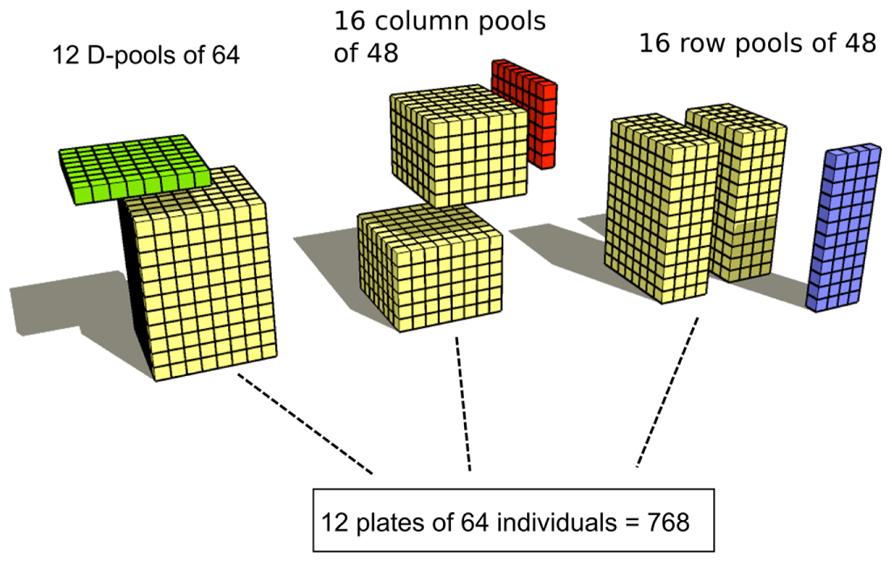
FIGURE 2. The three-dimensional pooling strategy used by (Tsai et al. (2011, reprinted with permission).
Multiplexing
Once PCR has been performed for each selected amplicon, PCR products of the different amplicons are pooled in an equimolar amount for library preparation. Commercially available kits allow tagging of each library with a short DNA sequence as a barcode to perform multiplexed experiments. This enables the sequencing of several libraries in a single lane, thus reducing the number of lanes without losing ability of discriminating between libraries. Some authors used this feature in combination with pooling (Marroni et al., 2011a; Tsai et al., 2011). The use of multiplexing increases the number of libraries that need to be prepared, but facilitates the identification of carriers of an interesting variant. In Figure 1, we show how a sample of 768 accessions can be divided into 12 pools of 64 subjects each, by preparing 12 libraries and tagging each library with a unique DNA barcode (index). When a variant of interest is identified, researchers often need to identify the carrier in order to analyze the effects of the variant on the phenotype. The use of multiplexing drastically reduces the amount of follow-up sequencing needed to identify the mutation carrier. The combined use of multidimensional pooling and multiplexing results in a greater number of libraries, but also in a further reduction of the required follow-up sequencing.
Tsai and collaborators, using three-dimensional pooling and multiplexing, were able to identify the carriers of a given mutation without the need of performing additional individual sequencing.
In one of the reviewed studies, there was no interest in the exact identification of the carriers of interesting variants and researchers performed pooled sequencing without the use of multiplexing (Kharabian-Masouleh et al., 2011).
When the number of individuals is limited, it is possible to construct a library for each individual and use a DNA barcode to uniquely tag each sequenced accession. Maughan and colleagues used 32 barcodes to tag a total of 60 A. thaliana accessions sequenced in two sequencing lanes. This required the preparation of 60 different libraries, but allowed the exact identification of each accession.
Sequencing Depth
The number of sequencing reads produced in NGS experiments is impressive and constantly increasing. Kharabian-Masouleh and collaborators generated a total of almost 4 Gb of sequence, while Marroni and colleagues generated about 2.5 Gb. Maughan and colleagues generated a total of 1 Gb using 454 and 1.5 Gb using Illumina GAII. No explicit information on sequencing data is available for the work of Tsai and colleagues, but indirect evidence suggests that they generated a comparable amount of sequence data.
In all the studies, the amount of generated reads resulted in a very high coverage (the number of times any position of the target region is sequenced). In each pool, coverage can be calculated as the total length of generated sequence L, divided by the length of the target region, l. In pooled NGS experiments, an important measure is the Mean Individual Coverage (MIC), expressing the average number of times that each accession is sequenced. MIC is given by L/(l × n), where n is the number of accessions in the pool.
It is often useful to know the value of MIC at each base position, also referred to as Mean Individual Base Coverage (MIBC). MIBC is calculated as R/n where R is the number of reads covering the considered base. MIBC is essential to filter positions with excessively high or low coverage, while MIC is useful to give a general idea of the coverage of the experiment.
Pooled experiments aiming at the identification of rare variants require a high MIC. In bidimensional pooling experiments, Tsai and colleagues aimed at obtaining an MIC of about 75× (30× per row and 45× per column). Unidimensional pooling requires even higher coverage; Kharabian-Masouleh and colleagues reported an MIC ranging 60–172×, while Marroni and colleagues reported an MIC ranging 100–350×, depending on the considered amplicon. Maughan and colleagues obtained an MIC of 4.3× with 454 data and 67× for Illumina GAII data (Table 2).
Three of the reviewed studies (Maughan et al., 2010; Kharabian-Masouleh et al., 2011; Marroni et al., 2011a) provided sufficient details to investigate how MIC, calculated as L/(l × n), is affected by the variation of (1) the total amount of generated sequence (L); (2) the length of the reference “genome” (or targeted region, l); and (3) the number of subjects present in the pool (n).
The differences in MIC between studies in spite of the similar amount of generated sequence reflect different experimental designs (Table 2). The two extremes are the works of Marroni and colleagues and that of Maughan and colleagues. The first explicitly aimed at the identification of rare variants in a very large number of accessions (large n) and focused on a small region (small l). The work of Maughan and colleagues aimed at discovering (common) variants in a large proportion of the genome (very large l) in a moderately sized sample (small n).
Dealing with Sequencing Errors
The search for rare variants is hampered by errors in the sequencing process. Errors might be introduced by inaccuracy of the DNA polymerase during amplification or by mistakes in the identification of the incorporated nucleotide in the sequencing by synthesis process used by most NGS instruments. Most researchers decide to use high-fidelity polymerases to reduce the error rate of the amplification step (Kharabian-Masouleh et al., 2011; Marroni et al., 2011a). In Section “Variants Identification and Validation,” we discuss further the improvement due to the use of a high-fidelity polymerase according to the results reported by Marroni and colleagues.
Another source of errors is the sequencing by synthesis process itself. Such error is low but not negligible and is reported to be in the range of 0.5–1% (Kharabian-Masouleh et al., 2011). In multiplexed experiments, errors in the sequencing step will affect both the sequencing reads, potentially causing false-positive findings, and the DNA barcode (index), causing the loss of reads for which the barcode cannot be unambiguously determined. Marroni and colleagues reported that about 4% of the reads contained at least one error in the barcode.
Bioinformatics
In NGS experiments involving one single individual, researchers can record three genotypic states: (1) the subject does not carry the variant (no sequencing read carry the variant); (2) the subject is heterozygous for the variant (roughly 50% of the reads carry the variant); and (3) the subject is homozygous carrier of the variant (all the reads carry the variant). These thresholds, with reasonable approximations and/or with the aid of statistical models, are used to determine genotype of individuals in NGS experiments. The basic assumption of the approach is that the number of reads carrying a variant is roughly proportional to the number of chromosomes carrying the same variant. It is possible to apply the same concept to experiments involving the sequencing of several individuals by assuming that the frequency of the variant in the population is roughly proportional to the fraction of reads carrying the variant itself.
However, the identification of variants in pooled samples poses additional challenges. In particular, it requires optimizing SNP detection, so that rare variants can be discriminated from errors. This might require adapting existing algorithms for variant calling to pooled samples (Marroni et al., 2011a) or developing completely novel algorithms (Tsai et al., 2011), although some researchers prefer to rely on the use of commercially available software packages (Kharabian-Masouleh et al., 2011).
The first step in the detection of variants in pooled NGS experiments is the alignment to a reference sequence. Several software packages are available to perform this task. Kharabian-Masouleh and colleagues used a commercial software package, CLCbio1 Marroni and colleagues used a free version of the commercial aligner Novoalign2 and Tsai and colleagues used the freely available BWA aligner (Li and Durbin, 2009). The second step is the variant calling itself. Again, researchers may use several available tools. Kharabian-Masouleh and colleagues relied on CLCbio, Marroni and coworkers used Varscan (Koboldt et al., 2009), while Tsai and colleagues used CAMBa (Missirian et al., 2011).
To obtain reliable results, all authors imposed limitations regarding base coverage and the proportion of reads carrying the variant. Bases with coverage below a defined threshold are discarded because they do not allow confident assignment of genotype. In addition, variants occurring with low frequency cannot be confidently distinguished from errors. Such step is essential to perform variant calling in pooled samples, and can be easily performed with commercially available software (Kharabian-Masouleh et al., 2011). However, several authors acknowledge that additional improvements are needed. Such improvements often require to develop custom algorithms or at least to perform custom operations on sequencing data. Tsai and colleagues used CAMBa (Missirian et al., 2011), a pipeline optimized to work with multidimensional pooling strategies. It takes into consideration the pooling depth, the expected frequency and type of mutations, the probability of heterozygosis, the sequencing quality of the call, and the frequency of forward and reverse reads. Marroni and colleagues used additional filtering steps to improve variant calling accuracy. Relying on the observation that false-positive SNPs can arise at the 5′ and 3′ ends of Illumina reads due to the higher error rate of the sequencing process or the presence of small insertions/deletions near the read ends, they introduced two additional filtering steps. They (1) analyzed the forward and reverse strands separately and required that each SNP was identified in both strands and (2) partitioned each read into three segments of equal length, and required that each SNP was identified in each of the segments of the read.
Variants Identification and Validation
Two of the reviewed studies compared variants identified by pooled multiplexed NGS with those identified in the same accessions by individual screening with an established protocol used as a gold standard ( Marroni et al., 2011a; Tsai et al., 2011). Both studies showed excellent agreement between pooled NGS and individual Sanger sequencing. Tsai and colleagues tested in total 84 variants passing an ad hoc probability threshold. Of them, only one was identified as a false positive. Marroni and colleagues tested 137 variants passing an ad hoc frequency threshold, and six of them were false positives.
Validation of the identified variants showed that pooled NGS reliably identified variants with frequency lower than 1%. Marroni and coworkers performed an ROC analysis comparing pooled NGS and individual Sanger sequencing on a randomly chosen subset, and used this analysis to choose the variant frequency threshold maximizing true positives while minimizing false positives. Their results showed that the optimal frequency threshold was around 0.4%. Tsai and colleagues also identified variants with frequencies below the 1% threshold. Kharabian-Masouleh and colleagues reported for the Illumina GA II sequencer an error rate in the range 0.5–1%. The studies from Marroni and Tsai showed that the achievement of very high MIC and the development of specific algorithms for variant calling allowed to overcome this limit and to effectively identify rare variants. An additional experiment performed by Marroni and coworkers showed the effect of polymerase accuracy on the error rate of the sequencing process. The authors used a high-fidelity polymerase to amplify one of the studied amplicons, while the remaining amplicons were amplified by a standard polymerase (Marroni et al., 2011a). To assess differences due to the use of different polymerases, they measured the proportion of polymorphic positions in each amplicon as a function of the frequency threshold applied for variant calling (Figure 3). At very low frequency thresholds, the proportion of bases carrying a putative SNP was between 0.9 and 1.0. Given that the actual proportion of polymorphic positions in the studied genes was less than 5%, most of such putative SNPs were false positives. Increasing the frequency threshold for variant calling caused a decrease in the proportion of false positives. These results showed that the use of a high-fidelity polymerase sensibly decreased false positives, thus suggesting that a significant amount of errors in NGS of pooled amplicons were due to DNA polymerase errors. In conclusion, the use of a high-fidelity polymerase can sensibly decrease the error rate and the optimal variant frequency threshold, and increase sensitivity and specificity of the method. However, at relatively high frequency thresholds, the performance of the standard polymerase is comparable to that of high-fidelity polymerase. Thus, the authors suggested the use of high-fidelity polymerase for amplification of the target genes only when rare variants need to be identified. The observation that DNA polymerase errors are responsible for the identification of potential false positives have been put forward also by Tsai and colleagues. In their work, DNA polymerase errors gave rise to “orphan” mutations, i.e., mutations that in bidimensional or three-dimensional pooling were identified only in one pool. Tsai and colleagues were able to decrease the occurrence of orphans by increasing the amount of DNA template.
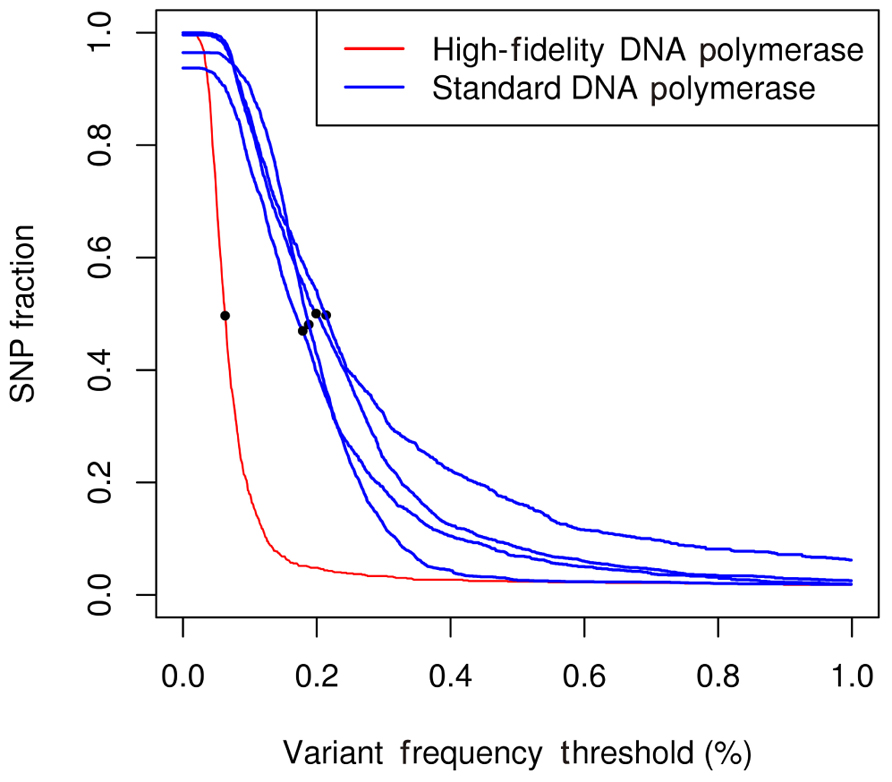
FIGURE 3. Number of called SNP per nucleotide as a function of the frequency set as the threshold for the five genes studied in the experiment. The dots represent the frequency threshold at which the proportion of bases identified as polymorphic is 50% of the maximum value. Blue lines represent amplicons sequenced using a standard polymerase. Red line represents amplicons sequenced using a high-fidelity polymerase.
A number of studies in human and plant genetics suggest that pooled multiplexed NGS gives reliable estimates of SNP allele frequency (Ingman and Gyllensten, 2009; Out et al., 2009; Marroni et al., 2011a). Several population genetics parameters can be estimated using information on allele frequency and sample size. One of such parameters is nucleotide diversity, that can be measured as the sum of the unbiased heterozygosity of segregating sites (Tajima, 1989), averaged over all nucleotides. The work by Marroni and colleagues showed that pooled multiplexed NGS gives reliable estimates of nucleotide diversity, showing strong correlation with estimates obtained with individual Sanger sequencing (Figure 4). Another parameter that can be estimated using pooled multiplexed NGS is Tajima’s D, a statistic used to test for departure from neutral evolution (Tajima, 1989). Negative values of D indicate negative selection, while positive values of D are suggestive of balancing selection.
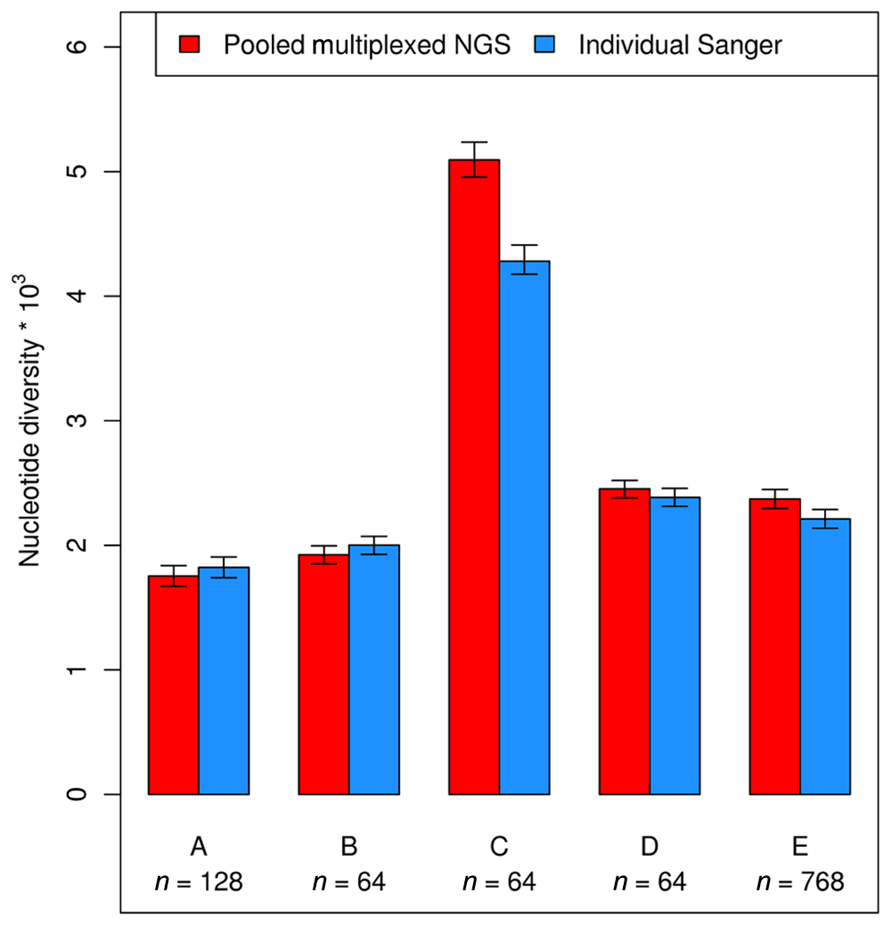
FIGURE 4. Nucleotide diversity calculated using allele frequency estimated by pooled multiplexed NGS (red bars) and by individual Sanger sequencing (blue bars).
Future Perspectives
With the ever increasing sequence output of next generation sequencers, it is becoming feasible to sequence megabases of target regions at high coverage in a large number of subjects. In such an experiment, the PCR amplification would become the bottleneck (Mamanova et al., 2010). An alternative approach is the use of hybridization-based target enrichment methods (Gnirke et al., 2009), which can target regions from few to hundreds of megabases (the size of human exome). However, commercial products based on hybridization can sensibly increase sequencing costs. An alternative method is the use of a set of restriction enzymes to fragment DNA, followed by targeted circularization and amplification of the target regions (Dahl et al., 2007). The commercial application of the latter approach is less expensive than hybridization-based applications and can target regions from less than 1 Mb to hundreds of megabases, but is at present only available for human genome. Adapting the technology to other organisms is feasible, but it would require the optimization of a different set of restriction enzymes for DNA fragmentation. A similar approach has already been implemented to genotype an A. thaliana mapping population (Maughan et al., 2010). The work by Maughan and coworkers confirmed that the construction of reduced representation libraries is a valid and cost-effective approach to target selection in plants.
Another interesting option is the use of hybridization-based target enrichment methods on pooled multiplexed samples. The advantage of this approach would be that only one hybridization experiment is needed to perform target enrichment in several subjects. This approach has already been successfully applied for the identification of mutations in humans (Cummings et al., 2010; Kenny et al., 2011) and in rats (Nijman et al., 2010). The possibility of performing the hybridization step on pooled multiplexed samples can sensibly reduce costs and labor and is an interesting opportunity for plant genomics experiments.
Researchers in human genetics can target the whole human genome by exon capture (Hodges et al., 2007). Several companies offer commercial kits to perform whole exome enrichment in human subjects. This option is extremely appealing since allows to obtain sequence information on the entire coding portion of the human genome at a reduced cost. The cost of sequencing a human exome is already well below 1000 if only sequencing costs are considered (Mertes et al., 2011).
As of today, the saving in sequencing costs obtained using target enrichment is greater than the increase in costs of library preparation due to enrichment protocols (Mamanova et al., 2010). Given the steady decrease of sequencing costs, it is possible that in the future whole-genome sequencing of large samples will be more efficient than sequencing targeted regions. However, at present, only large research consortia can undertake whole-genome resequencing in sample sizes larger than 100 subjects. This is the case of the 1000 Genomes Project, in which nearly 200 subjects underwent low-coverage (<6×) genome sequencing and nearly 700 subjects underwent high coverage sequencing of slightly less than 1000 genes (The 1000 Genomes Project Consortium, 2010), and of the 1001 Genomes, a project aiming at surveying genetic variation in A. thaliana (Weigel and Mott, 2009), which produced so far complete genome sequence of several A. thaliana strains. Although we hypothesize that a long-term aim will be to obtain high coverage sequence of whole genome of large samples, we foresee that in the near future the search for rare variants will still be driven by the use of targeted resequencing.
Conclusion
Pooled NGS of target regions is an efficient method to screen large populations for polymorphisms. It has been applied with excellent results to mainly self-pollinating crops such as wheat and rice (Kharabian-Masouleh et al., 2011; Tsai et al., 2011) and to outcrossing trees (Marroni et al., 2011a).
The main interest of pooled NGS of target regions is the identification of variants with a functional role (Marroni et al., 2011a; Tsai et al., 2011). Marroni and colleagues identified a mutation causing a premature stop codon in HCT1, a gene involved in lignin production. Phenotype of mutation carriers is now being thoroughly investigated, and experimental crosses are being carried out to extensively evaluate the potential impact of the mutation. If the identified mutation has a functional role, then it will be of importance for improving poplar breeding programs.
The first studies using multiplexed pooled NGS were performed in humans (Ingman and Gyllensten, 2009; Out et al., 2009) and reported a good correlation between the proportion of reads carrying a polymorphism and the polymorphism frequency. The ability to accurately estimate allele frequency enables researchers to accurately estimate several population genetics parameters, such as nucleotide diversity and Tajima’s D. One of the reviewed studies used pooled multiplexed NGS to obtain accurate estimates of nucleotide diversity and Tajima’s D (Marroni et al., 2011a). Comparison of nucleotide diversity estimated by pooled multiplexed NGS with estimates obtained by individual Sanger sequencing showed a strong agreement between the two approaches (Figure 4).
Thus, pooled multiplexed NGS can be successfully used to accurately estimate population genetics parameters. The approach is also naturally suited for projects investigating the genetic structure of different populations. In projects investigating the genetic background of two or more populations, pooling of individuals of each population would allow saving resources without loss of information.
Finally, the ability to contrast allele frequencies between two samples will also greatly facilitate pooled association mapping, a method already proposed to identify association between polymorphisms and qualitative traits (Sham et al., 2002).
We foresee that the increase of next generation sequencers throughput, the availability of cost-effective targeting methods and the improvement of multiplexing capabilities will lead to a further improvement of performance, making it feasible to genotype thousands of variants in large numbers of individuals in a single sequencing run retaining individual information.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the European Community’s Seventh Framework Program (FP7/2007-2013) under grant agreement number 211917 (ENERGYPOPLAR). Figure 2 was kindly provided by Professor Luca Comai.
Footnotes
References
Bodmer, W., and Bonilla, C. (2008). Common and rare variants in multifactorial susceptibility to common diseases. Nat. Genet. 40, 695–701.
Comai, L., Young, K., Till, B. J., Reynolds, S. H., Greene, E. A., Codomo, C. A., Enns, L. C., Johnson, J. E., Burtner, C., Odden, A. R., and Henikoff, S. (2004). Efficient discovery of DNA polymorphisms in natural populations by Ecotilling. Plant J. 37, 778–786.
Craig, D. W., Pearson, J. V., Szelinger, S., Sekar, A., Redman, M., Corneveaux, J. J., Pawlowski, T. L., Laub, T., Nunn, G., Stephan, D. A., Homer, N., and Huentelman, M. J. (2008). Identification of genetic variants using bar-coded multiplexed sequencing. Nat. Methods 5, 887–893.
Cronn, R., Liston, A., Parks, M., Gernandt, D. S., Shen, R., and Mockler, T. (2008). Multiplex sequencing of plant chloroplast genomes using Solexa sequencing-by-synthesis technology. Nucleic Acids Res. 36, e122.
Cummings, N., King, R., Rickers, A., Kaspi, A., Lunke, S., Haviv, I., and Jowett, J. B. M. (2010). Combining target enrichment with barcode multiplexing for high throughput SNP discovery. BMC Genomics 11, 641. doi: 10.1186/1471-2164-11-641
Dahl, F., Stenberg, J., Fredriksson, S., Welch, K., Zhang, M., Nilsson, M., Bicknell, D., Bodmer, W. F., Davis, R. W., and Ji, H. (2007). Multigene amplification and massively parallel sequencing for cancer mutation discovery. Proc. Natl. Acad. Sci. U.S.A. 104, 9387–9392.
Eichler, E. E., Flint, J., Gibson, G., Kong, A., Leal, S. M., Moore, J. H., and Nadeau, J. H. (2010). Missing heritability and strategies for finding the underlying causes of complex disease. Nat. Rev. Genet. 11, 446–450.
Gnirke, A., Melnikov, A., Maguire, J., Rogov, P., LeProust, E. M., Brockman, W., Fennell, T., Giannoukos, G., Fisher, S., Russ, C., Gabriel, S., Jaffe, D. B., Lander, E. S., and Nusbaum, C. (2009). Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat. Biotechnol. 27, 182–189.
Harfouche, A., Meilan, R., Kirst, M., Morgante, M., Boerjan, W., Sabatti, M., and Scarascia Mugnozza, G. (2012). Accelerating the domestication of forest trees in a changing world. Trends Plant Sci. 17, 64–72.
Hodges, E., Xuan, Z., Balija, V., Kramer, M., Molla, M. N., Smith, S. W., Middle, C. M., Rodesch, M. J., Albert, T. J., Hannon, G. J., and McCombie, W. R. (2007). Genome-wide in situ exon capture for selective resequencing. Nat. Genet. 39, 1522–1527.
Ingman, M., and Gyllensten, U. (2009). SNP frequency estimation using massively parallel sequencing of pooled DNA. Eur. J. Hum. Genet. 17, 383–386.
Kenny, E. M., Cormican, P., Gilks, W. P., Gates, A. S., O’Dushlaine, C. T., Pinto, C., Corvin, A. P., Gill, M., and Morris, D. W. (2011). Multiplex target enrichment using DNA indexing for ultra-high throughput SNP detection. DNA Res. 18, 31–38.
Kharabian-Masouleh, A., Waters, D. L. E., Reinke, R. F., and Henry, R. J. (2011). Discovery of polymorphisms in starch-related genes in rice germplasm by amplification of pooled DNA and deeply parallel sequencing. Plant Biotechnol. J. 9, 1074–1085.
Koboldt, D. C., Chen, K., Wylie, T., Larson, D. E., McLellan, M. D., Mardis, E. R., Weinstock, G. M., Wilson, R. K., and Ding, L. (2009). VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 25, 2283–2285.
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760.
Mamanova, L., Coffey, A. J., Scott, C. E., Kozarewa, I., Turner, E. H., Kumar, A., Howard, E., Shendure, J., and Turner, D. J. (2010). Target-enrichment strategies for next-generation sequencing. Nat. Methods 7, 111–118.
Mardis, E. R. (2008). The impact of next-generation sequencing technology on genetics. Trends Genet. 24, 133–141.
Marroni, F., Pinosio, S., Di Centa, E., Jurman, I., Boerjan, W., Felice, N., Cattonaro, F., and Morgante, M. (2011a). Large scale detection of rare variants via pooled multiplexed next generation sequencing: towards next generation Ecotilling. Plant J. 67, 736–745.
Marroni, F., Pinosio, S., Zaina, G., Fogolari, F., Felice, N., Cattonaro, F., and Morgante, M. (2011b). Nucleotide diversity and linkage disequilibrium in Populus nigra cinnamyl alcohol dehydrogenase (CAD4) gene. Tree Genet. Genomes 7, 1011–1023.
Maughan, P. J., Yourstone, S. M., Byers, R. L., Smith, S. M., and Udall, J. A. (2010). Single-nucleotide polymorphism genotyping in mapping populations via genomic reduction and next-generation sequencing: proof of concept. Plant Genome J. 3, 166.
McCallum, C. M., Comai, L., Greene, E. A., and Henikoff, S. (2000). Targeting induced local lesions IN genomes (TILLING) for plant functional genomics. Plant Physiol. 123, 439–442.
Mertes, F., Elsharawy, A., Sauer, S., van Helvoort, J. M., van der Zaag, P. J., Franke, A., Nilsson, M., Lehrach, H., and Brookes, A. J. (2011). Targeted enrichment of genomic DNA regions for next-generation sequencing. Brief. Funct.Genomics 10, 374–386.
Missirian, V., Comai, L., and Filkov, V. (2011). Statistical mutation calling from sequenced overlapping DNA pools in TILLING experiments. BMC Bioinformatics 12, 287. doi: 10.1186/1471-2105-12-287
Neale, D. B., and Kremer, A. (2011). Forest tree genomics: growing resources and applications. Nat. Rev. Genet. 12, 111–122.
Nijman, I. J., Mokry, M., van Boxtel, R., Toonen, P., de Bruijn, E., and Cuppen, E. (2010). Mutation discovery by targeted genomic enrichment of multiplexed barcoded samples. Nat. Methods 7, 913–915.
Out, A. A., van Minderhout, I. J., Goeman, J. J., Ariyurek, Y., Ossowski, S., Schneeberger, K., Weigel, D., van Galen, M., Taschner, P. E, Tops, C. M., Breuning, M. H., van Ommen, G. J., den Dunnen, J. T., Devilee, P., and Hes, F. J. (2009). Deep sequencing to reveal new variants in pooled DNA samples. Hum. Mutat. 30, 1703–1712.
Parameswaran, P., Jalili, R., Tao, L., Shokralla, S., Gharizadeh, B., Ronaghi, M., and Fire, A. Z. (2007). A pyrosequencing-tailored nucleotide barcode design unveils opportunities for large-scale sample multiplexing. Nucleic Acids Res. 35, e130.
Risch, N., and Merikangas, K. (1996). The future of genetic studies of complex human diseases. Science 273, 1516–1517.
Sham, P., Bader, J. S., Craig, I., O’Donovan, M., and Owen, M. (2002). DNA Pooling: a tool for large-scale association studies. Nat. Rev. Genet. 3, 862–871.
Tajima, F. (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595.
The 1000 Genomes Project Consortium (2010). A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073.
Tsai, H., Howell, T., Nitcher, R., Missirian, V., Watson, B., Ngo, K. J., Lieberman, M., Fass, J., Uauy, C., Tran, R. K., Khan, A. A., Filkov, V., Tai, T. H., Dubcovsky, J., and Comai, L. (2011). Discovery of rare mutations in populations: TILLING by sequencing. Plant Physiol. 156, 1257–1268.
Varshney, R. K., Graner, A., and Sorrells, M. E. (2005). Genomics-assisted breeding for crop improvement. Trends Plant Sci. 10, 621–630.
Weigel, D., and Mott, R. (2009). The 1001 genomes project for Arabidopsis thaliana. Genome Biol. 10, 107.
Wheeler, D. A., Srinivasan, M., Egholm, M., Shen, Y., Chen, L., McGuire, A., He, W., Chen, Y.-J., Makhijani, V., Roth, G. T., Gomes, X., Tartaro, K., Niazi, F., Turcotte, C. L., Irzyk, G. P., Lupski, J. R., Chinault, C., Song, X. Z., Liu, Y., Yuan, Y., Nazareth, L., Qin, X., Muzny, D. M., Margulies, M., Weinstock, G. M., Gibbs, R. A., and Rothberg, J. M. (2008). The complete genome of an individual by massively parallel DNA sequencing. Nature 452, 872–876.
Keywords: rare variants, next generation sequencing, plants, multiplex, barcode, pool, polymerase, nucleotide diversity
Citation: Marroni F, Pinosio S and Morgante M (2012) The quest for rare variants: pooled multiplexed next generation sequencing in plants. Front. Plant Sci. 3:133. doi: 10.3389/fpls.2012.00133
Received: 07 March 2012; Accepted: 04 June 2012;
Published online: 28 June 2012.
Edited by:
Patrick J. Krysan, University of Wisconsin-Madison, USAReviewed by:
Vagner Benedito, West Virginia University, USAHaiquan Li, National University of Singapore, Singapore
Copyright: © 2012 Marroni, Pinosio and Morgante. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Fabio Marroni, Istituto di Genomica Applicata, Via J. Linussio 51, 33100 Udine, Italy. e-mail:bWFycm9uaUBhcHBsaWVkZ2Vub21pY3Mub3Jn