- Department of Biological Chemistry, John Innes Centre, Norwich Research Park, Norwich, UK
Mono ADP-ribosyltransferases (mADP-RTs) are a family of enzymes that cleave NAD+ and covalently attach the ADP-ribosyl moiety to target proteins. mADP-RTs are well established as important virulence factors of bacteria that infect mammals. Cholera toxin, pertussis toxin, and diphtheria toxin are three of the best-known examples of mADP-RTs. They modify host target proteins in order to promote infection and/or killing of the host cell. Despite low sequence similarity at the primary amino acid level, mADP-RTs share a conserved core catalytic fold and structural biology has made important contributions to elucidating how mADP-RTs modify mammalian host targets. Recently, mADP-RTs were shown to be present in plant pathogenic bacteria, suggesting that mADP-RTs are also important virulence factors of plant pathogens. Crystal structures of plant pathogenic bacterial mADP-RTs are also now available. Here we review the structure/function of mADP-RTs from pathogens of mammals and plants, highlighting both commonalities and differences.
A Common Fold Mediates Merciless Killing or Subtle Manipulation
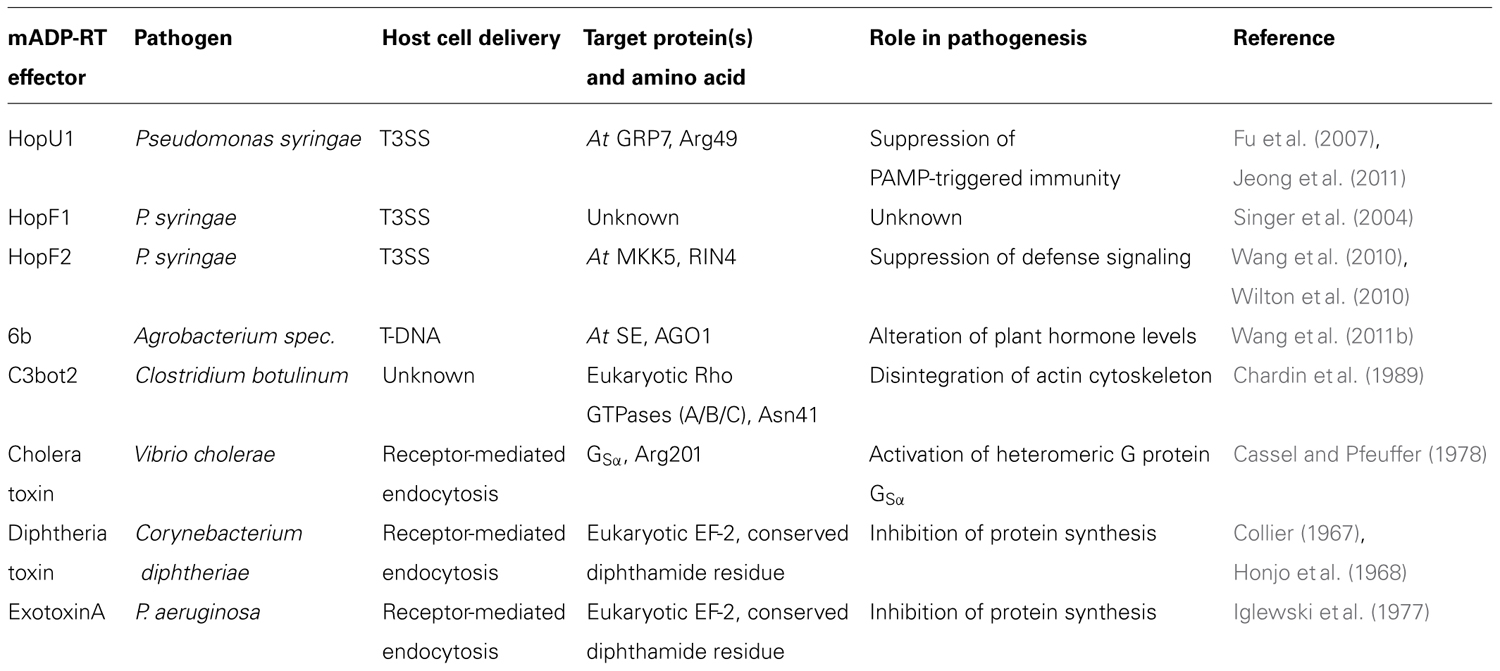
Several important pathogenic bacteria use secreted proteins, frequently termed “effectors,” to modify the physiology of their host cells to promote infection. One family of effectors encodes a mono-ADP-ribosyltransferase (mADP-RT) enzymatic activity. The manipulating activities of mADP-RTs often culminate in killing the infected host cell and therefore many pathogen secreted mADP-RTs are referred to as toxins (Holbourn et al., 2006). ADP-ribosylating toxins typically comprise several distinct domains that mediate attachment to host cell receptors, translocation into the host cell and mADP-ribosylation (Figure 1; Deng and Barbieri, 2008). Some mADP-RTs from Gram-negative bacteria rely on host cell delivery by the bacterial type three secretion system (T3SS) and consist only of the catalytic domain preceded by a type three secretion signal and host cell targeting domains (Deng and Barbieri, 2008). The catalytic domain mediates cleavage of an ADP-ribose moiety from NAD+ and its subsequent transfer onto an amino acid of the target protein (often Cys, Arg, Asn, or Diphthamide – a modified form of His). In most cases mADP-ribosylation of target proteins leads to their inactivation. Transfer of an ADP-ribose moiety onto the GSα subunit of heteromeric G proteins by cholera toxin (CT) results in constitutive production of cAMP and leads to the severe diarrhea associated with infection by Vibrio cholerae (Cassel and Pfeuffer, 1978; Vanden Broeck et al., 2007). Diphtheria toxin (DT) from Corynebacterium diphtheriae mADP-ribosylates eukaryotic elongation factor-2, leading to its inactivation and interference with mRNA translation that can have detrimental effects in the host organism (Collier, 1967; Honjo et al., 1968; Collier, 2001).
FIGURE 1. Functional domains of mADP-RTs discussed in this review. T3S, type 3 secretion peptide; mADP-RT, mono-ADP-ribosylating catalytic domain; binding, host cell receptor binding; translocation, host cell translocation.
Despite a low level of sequence similarity, the catalytic domains of most mADP-RTs share a common core αβ fold (Holbourn et al., 2006). A series of anti-parallel β-strands provides structural support for the active site. This active site comprises a hydrophobic NAD+ binding cleft and a conserved Glu residue that promotes cleavage of NAD+ at the anomeric carbon of the nicotinamide ribose and subsequent transfer of the mADP-ribose moiety onto the target amino acid. The α helices and loops connecting the β-strands are structurally less conserved but make important contributions to the NAD+ binding cleft and mediate substrate binding. The enzymatic activity of several mADP-RTs is regulated by an active site loop located adjacent to the catalytic cleft. This loop can undergo significant re-orientation to allow access to the active site upon substrate binding (Bell and Eisenberg, 1996; Sun et al., 2004; O’Neal et al., 2005).
Most mADP-RTs retain three structurally conserved features (Holbourn et al., 2006): (1) The arom-H/R motif is composed of an aromatic amino acid followed by His or Arg. This motif either contributes to NAD+ binding or supports the structural integrity of the NAD+ binding site; (2) The ARTT loop contains the conserved catalytic Glu residue required for NAD+ cleavage and transferase activity. The catalytic Glu is often part of a Q/E-X-E motif in which the presence of Glu or Gln two residues upstream appears to determine substrate specificity for either Arg or Asn mADP-ribosylation; and (3) The STS motif that maintains the structure of the active site through hydrogen bonds with the catalytic Glu, and other conserved residues of the NAD+ binding site. However, the STS motif is less conserved in DT and related structures.
Dependent on the conservation of additional motifs that form the active site, mADP-RTs can be annotated as DT- or CT-like in their folds. Intriguingly, both groups are not limited to bacterial toxins. The DT group also includes the mammalian poly-ADP-ribosyltransferases (PARPs) that regulate diverse cellular mechanisms including apoptosis, DNA repair, and intracellular transport (Schreiber et al., 2006). The CT group is also found in a class of eukaryotic extracellular enzymes, the Ecto-ARTs (Di Girolamo et al., 2005). Plants possess PARPs, but examples of mADP-RTs in plants have yet to be found (Wang et al., 2011a; Lamb et al., 2012). Recent structural studies on virulence effectors of plant pathogenic bacteria have revealed examples of both DT- and CT-like folds in several bacterial effector proteins, and most of these effectors have mADP-RT activity. In contrast to the fatal consequences associated with delivery of mADP-ribosylating toxins to host cells, but consistent with the (hemi-) biotrophic nature of plant pathogenic bacteria, these effectors appear to induce more subtle modifications to plant innate immunity pathways or the physiology of the infected cell. Molecular targets of selected mADP-RTs are shown in Table 1.
HopU1: Targeting RNA-Binding Proteins
The effector HopU1 from Pseudomonas syringae strain DC3000 is delivered through the T3SS into host cells where it interferes with plant immune responses (Fu et al., 2007). HopU1 is an active mADP-RT and several RNA binding proteins have been identified as in vitro targets of this effector. One of these RNA binding proteins, GRP7, regulates circadian mRNA oscillations and is also required for immunity towards P. syringae DC3000 (Heintzen et al., 1997; Fu et al., 2007).
Recently, the crystal structure of HopU1 has been determined (Jeong et al., 2011). This showed that the HopU1 catalytic subunit shares structural homology to mADP-RTs of the CT-class. For example, 86 Cα atoms of HopU1 can be superimposed onto the structure of the Clostridium botulinum mADP-RT C3bot2 with a root mean square deviation of 2.12Å. The two opposing β sheets, that support the ARTT loop with the catalytic Glu and the STS motif (STT in HopU1), align well with the corresponding β sheets of C3bot2 (Figure 2A). In both structures the first Ser of the STS motif forms hydrogen bonds with the catalytic Glu.
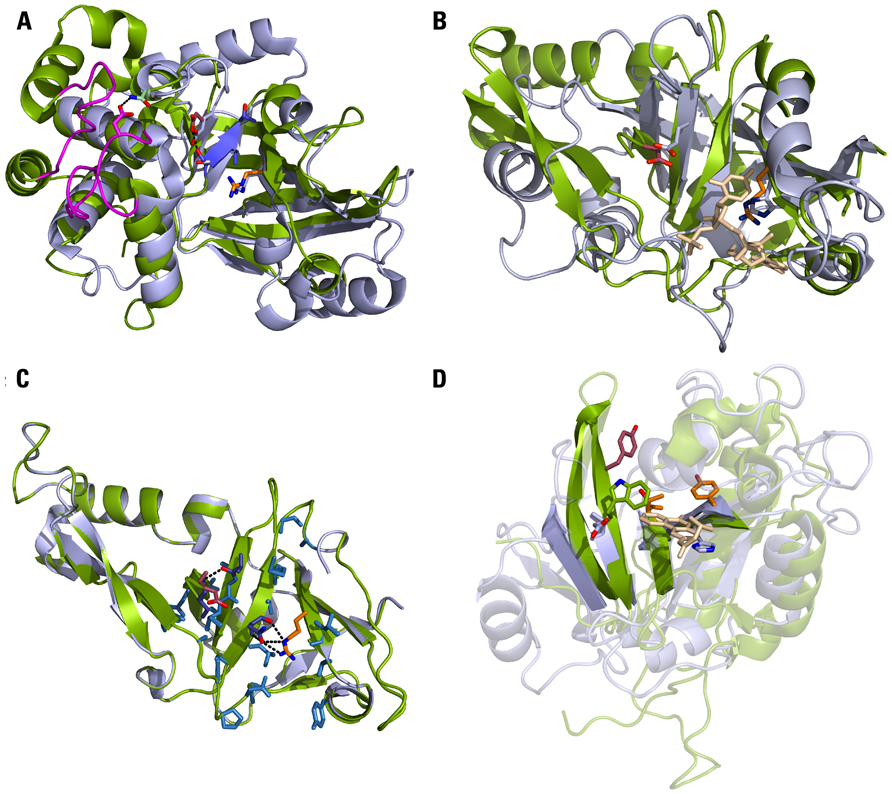
FIGURE 2. Plant and animal pathogen mADP-RTs share the same protein fold. (A) HopU1 structure (green) superimposed on the catalytic subunit of C3bot2 (light blue). Highlighted in the HopU1 structure are the conserved Arg of the arom-H/S motif (orange), the STS motif (blue) and the catalytic Glu (dark red). The corresponding amino acids of C3bot2 are shown as sticks. The two loops that mediate binding to GRP7 are shown in magenta. PDB: 3U0J (HopU1), 1R45 (C3bot2). (B) HopF1 structure (green) superimposed on the catalytic subunit of DT (light blue) in complex with the NAD+ analog adenylyl 3′,5′ uridine 3′ monophosphate (beige). Highlighted in the HopF1 structure are the conserved Arg of the arom-H/S motif (orange) and the catalytic Asp (dark red). The corresponding amino acids of DT are shown as sticks. PDB: 1S21 (HopF1), 1DDT (DT). (C) Homology model of HopF2 (green) based on the HopF1 structure (light blue). Amino acids colored in blue contribute to the active site and are conserved in both effectors. These also include the catalytic Asp174/175 (dark red) and Arg 72/71 (orange) as well as Asn135/134 and Ala187/188 (purple) that form hydrogen bonds with Arg and Asp, respectively. (D) Agrobacterium vitis 6b structure (green) superimposed on the catalytic subunit of Exotoxin A (light blue) in complex with the inhibitor PJ34 (beige). Highlighted is the conserved arrangement of b-strands with 6b residues Tyr66 and Thr93 (orange) as well as the proposed catalytic Tyr153 (red). The catalytic Glu553 and His440 of Exotoxin A are shown as sticks. The amino acid of 6b that is located closest to the position of the catalytic Glu of Exotoxin A is Trp155 (green). PDB: 3AQ2 (6b), 1XK9 (Exotoxin A).
Whereas the catalytic subunits of mADP-RTs are structurally conserved, the regions mediating substrate binding can adopt more divergent conformations. The structure of HopU1 revealed two protruding loops that are not conserved in other mADP-RTs (Jeong et al., 2011). Ala substitutions in both of these loops were shown to affect GRP7 binding in vitro and abolish mADP-ribosylation of GRP7, suggesting they are essential for substrate recognition. Two Arg residues located in the GRP7 RNA binding domain have been previously identified as potential HopU1 target amino acids (Fu et al., 2007). Using a proteomics approach Jeong et al. (2011) further demonstrate that at least one residue, Arg49, is mADP-ribosylated by HopU1 in vitro. This is consistent with the close proximity of the putative HopU1 active site and Arg47/Arg49 in a structural model of the HopU1/GRP7 RNA binding domain complex. Interestingly, GRP7 Arg49 makes direct contact to RNA (Schöning et al., 2007). Therefore, HopU1 specifically targets an amino acid of GRP7 that is essential for RNA binding.
HopF1: In Search of a Target
Structural homology to mADP-RTs has also been observed for other T3SS effectors from P. syringae. The crystal structure of HopF1 (formerly AvrPphF) from the bean-infecting P. syringae pathovar phaseolicola reveals structural similarity to DT (Figure 2B; Singer et al., 2004). However, to date, no mADP-RT activity has been shown for HopF1 in vitro, or in a plant cell extract (Singer et al., 2004). One explanation for this lack of activity could be changes in the amino acid composition at key positions compared to DT. This includes the catalytic Glu (Glu148, DT-numbering), which is an Asp in HopF1 (Asp174) and His21, part of the arom-H/S motif in DT (Bennett and Eisenberg, 1994), which is replaced by an Arg in HopF1 (Arg72). Interestingly, the side chains of each of these residues occupy almost identical positions in the active sites (Figure 2B). However, in the case of the DT(Glu148)/HopF1(Asp174) substitution, the residues originate from different secondary structure elements (β7 in DT and β6 in HopF1).
Despite the lack of mADP-RT activity under the conditions tested, Arg72 and Asp174 are still required for the virulence and avirulence activities of HopF1. Whilst wild type HopF1 enhances growth of P. syringae on the susceptible bean cultivar Tendergreen ~eightfold, Ala substitutions of either Arg72 or Asp174 completely abolish this virulence activity (Singer et al., 2004). In bean cultivar Red Mexican the R1 disease resistance gene confers recognition of HopF1 (Tsiamis et al., 2000). This recognition event requires both Arg72 and Asp174 as P. syringae expressing the corresponding Ala substitutions evades recognition on Red Mexican (Singer et al., 2004). Therefore, even though HopF1 does not exhibit mADP-RT activity under the conditions tested, the conserved cleft forming the putative active site is essential for both HopF1’s virulence activity and recognition on resistant host plants. The side chains of both Arg72 and Asp174 form hydrogen bonds to other residues located within the cleft and these intramolecular interactions would be impaired by Ala substitutions, suggesting that the structural integrity of the cavity is required for HopF1’s virulence function and recognition by R1. It is conceivable that the interface supported by Arg72 and Asp174 forms a binding site for virulence target/s of HopF1. As the same binding site is required for HopF1 recognition on resistant bean cultivars this recognition event might be mediated by a “decoy” protein mimicking a virulence target (van der Hoorn and Kamoun, 2008). In this scenario modification of the decoy protein would trigger defense gene activation by the R1 resistance protein.
HopF2: Targeting Kinase Cascades
In contrast to HopF1, the sequence related P. syringae strain DC3000 effector HopF2 is known to be an active mADP-RT (Wang et al., 2010). HopF2 contributes to virulence of P. syringae as it enhances growth of the bacteria in Arabidopsis and tomato (Robert-Seilaniantz et al., 2006; Wilton et al., 2010). When delivered via the T3SS of a non-pathogenic Pseudomonas strain, HopF2 suppresses MAMP-triggered MAP-kinase activation and direct interaction of HopF2 with several MAP-kinase-kinases (MKKs) has been shown in vitro and in planta (Wang et al., 2010). Using biotin-labeled NAD+ as co-factor, HopF2 is able to transfer biotin-ADP-ribose onto MKK5 and this impairs phosphorylation of MPK6 (Wang et al., 2010). Interestingly, HopF2 appears to modify the constitutively active MKK5DD mutant more efficiently than the wild type form, suggesting that the effector may preferentially target activated MKKs. ADP-ribosylation of MKK5 is dependent on a conserved Arg in the C-terminus of MKK5 and an Ala substitution of this residue largely impairs MKK5-mediated activation of the defense marker gene FRK1. Hence, one virulence activity of HopF2 is to interfere with MKK signaling to suppress MAMP-triggered immunity (Wang et al., 2010).
When over-expressed in Arabidopsis, HopF2 also interferes with activation of the resistance protein RPS2 by its cognate P. syringae effector AvrRpt2 (Wilton et al., 2010). AvrRpt2 is a Cys protease that cleaves Arabidopsis RIN4 and the disappearance of RIN4 triggers RPS2 activation (Axtell and Staskawicz, 2003; Mackey et al., 2003). HopF2 interferes with AvrRpt2-mediated cleavage of RIN4 and in vitro assays show that HopF2 binds to and mADP-ribosylates RIN4 (Wang et al., 2010; Wilton et al., 2010). Therefore, a possible mechanism of HopF2 interference with RIN4 cleavage is mADP-ribosylation of an amino acid in the RIN4 peptide that is cleaved by AvrRpt2 (Day et al., 2005; Kim et al., 2005).
HopF2 elicits a hypersensitive response in the non-host plant Nicotiana tabacum (Robert-Seilaniantz et al., 2006). In striking accordance to the requirement of Arg72 and Asp174 residues for HopF1 virulence function and recognition, the corresponding two amino acids of HopF2 (Arg71 and Asp175) are essential for both the virulence and avirulence activities of HopF2 (Wang et al., 2010). HopF2 has 48% amino acid identity and 92% amino acid similarity to HopF1. The level of sequence conservation between the two effectors allows building of a reliable homology model of HopF2 based on the HopF1 structure (Figure 2C). According to this homology model, amino acids forming the putative active site, including the Arg/Asp pair critical for effector virulence and recognition, are conserved in both effectors. The enzymatic activity of HopF2 suggests that replacement of the catalytic Glu residue, which is highly conserved in ADP-ribosylating toxins, by Asp174 in HopF1 does not explain why HopF1 is enzymatically inactive. Based on the structural conservation of the active site in both effectors and the requirement of corresponding amino acids for virulence and avirulence functions it would be surprising if only HopF2 but not HopF1 functions as a mADP-RT. Hence, it would be informative to re-test whether HopF1 has mADP-RT function under conditions used for HopF2.
Based on the finding that Ala substitutions of HopF2 Arg71 or Asp175 abolish MKK5 binding, Wang and co-workers (Wang et al., 2010) suggest that Arg71/Asp175 are more likely to be involved in substrate binding than in catalysis. However, both functions are likely to depend on the structural integrity of the active site and loss of hydrogen bonding mediated by Arg71 and Asp175 might affect its overall conformation. In a similar manner, substitutions of the conserved Glu 233/235 in the HopU1 ARTT loop impair not only mADP-ribosylating activity, but also decrease binding to GRP7 (Jeong et al., 2011).
Agrobacterium 6b: A Divergent mADP-RT
Plant pathogen effectors adopting the mADP-RT core fold are not limited to P. syringae. The Agrobacterium 6b protein, encoded on the Ti plasmids of A. tumefaciens and A. vitis, shows structural homology to mADP-ribosylating toxins such as Exotoxin A from P. aeruginosa (Wang et al., 2011b). Although the 6b protein is dispensable for crown gall formation in a natural infection, ectopic expression of 6b in several host species is sufficient to induce tumors, probably by altering auxin and cytokinin physiology of the host cell (Hooykaas et al., 1988; Tinland et al., 1989). Further, ectopic expression of 6b in Arabidopsis leads to formation of serrated leaves, a phenotype also observed in mutants deficient in microRNA (miRNA) metabolism (Wang et al., 2011b). Indeed, the accumulation of several miRNAs is altered in plants expressing 6b and the reduced levels of one particular miRNA, miR319, could provide a direct link to activation of auxin signaling (Navarro et al., 2006). 6b interacts with two proteins involved in miRNA processing, SE and AGO1, in vitro and in plant cells (Kidner and Martienssen, 2004; Yang et al., 2006; Wang et al., 2011b). Thus, manipulation of the miRNA processing machinery, to alter hormone levels in host cells, appears to be one virulence mechanism of 6b proteins.
Despite the lack of sequence similarity, the structure of 6b can be superimposed on Exotoxin A and CT with a root mean square deviation of <4Å (Figure 2D; Wang et al., 2011b). Although the overall position of the central β-sheets in both proteins is conserved, dramatic changes can be observed at amino acids forming the putative active site of 6b (Wang et al., 2011b). There is a notable lack of residues in 6b that could functionally substitute for the Arg/His of the arom-H/S motif and the catalytic Glu in CT/Exotoxin A. Remarkably, the authors still provide evidence that 6b is an active mADP-RT: (1) the morphological phenotype induced by 6b expression in Arabidopsis can be rescued by application of the less-hydrolysable NAD+ analog TAD; (2) in the presence of the putative target, SE, 6b is able to hydrolyze the NAD+ analog ε-NAD+ in vitro and this activity is dependent on three Tyr and one Thr residue that contribute to the active site; (3) the hydrolytic activity of 6b is enhanced >20-fold in presence of an Arabidopsis ARF protein. This observation is in striking accordance with activation of CT by human ARF6-GTP (O’Neal et al., 2005). ARF6-GTP binding to CT leads to a conformational change in the active site loop rendering the catalytic cleft accessible to NAD+ and the substrate. A similar active site loop occluding the NAD+ binding site in absence of ARF is found in the 6b structures. Thus, it appears that an ARF-GTP-dependent activation mechanism has been conserved in mADP-RTs of mammalian and plant pathogens.
How do 6b proteins catalyze NAD+ hydrolysis without the conserved catalytic Glu? Although the detailed reaction mechanisms of most mADP-RTs remain to be elucidated, the conserved Glu is generally assumed to play a critical role in stabilizing the bound NAD+ molecule in a transition state that renders the anomeric carbon of the nicotinamide ribose more vulnerable to nucleophilic attack by the substrate (Holbourn et al., 2006). It has been proposed that Tyr153 might perform the same function in 6b (Wang et al., 2011b). Consistent with this, the 6B Tyr153Ala mutant is impaired in ε-NAD+ hydrolysis. However, it is debatable whether amino acids less electronegative than Glu or Asp would be able to stabilize the NAD+ transition state. It is equally plausible that Tyr153, together with Tyr66 and Tyr121, contributes to the structural integrity of the active site.
An alternative explanation for retention of enzymatic activity in absence of a catalytic Glu is suggested by research on mammalian PARPs. Human PARP10 and PARP14, which both lack the conserved Glu in the catalytic core motif, do not function as PARPs (Kleine et al., 2008). However, both enzymes show mADP-ribosylating activity. Based on their findings, Kleine et al. (2008) proposed an alternative substrate-assisted catalytic mechanism where the catalytic Glu is provided not by the enzyme but by the substrate. It is conceivable that the enzymatic activity of Agrobacterium 6b effectors relies on a similar mechanism.
The work reviewed here suggests that several plant pathogenic bacteria evolved host-targeted enzymes with mADP-RT activity to manipulate the physiology and immune system in infected host cells. Notably, mADP-RTs from plant pathogens appear to show greater structural diversity than secreted mADP-RTs from mammalian pathogens, or the eukaryotic Ecto-ARTs. How some of these effectors retain enzymatic activity despite considerable changes in the active site is an intriguing question. The approaches summarized here need to be extended to include mADP-RT structures in the presence of non-hydrolysable NAD+ analogs and complemented with more sophisticated enzymatic analysis. Elucidating how this structurally diverged group of host-targeted effectors catalyzes transfer of mADP-ribose onto target proteins, and defining the their target specificity, will not only provide new insights into manipulation of plant immunity but also extend our functional understanding of mADP-RTs.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We apologize to colleagues whose work could not be included in this review due to space constraints. Lennart Wirthmueller is supported by a FEBS long-term fellowship. Mark J. Banfield acknowledges the BBSRC (grant BB/J004553/1), the John Innes Foundation, and the Gatsby Charitable Foundation for funding.
References
Axtell, M. J., and Staskawicz, B. J. (2003). Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112, 369–377.
Bell, C. E., and Eisenberg, D. (1996). Crystal structure of diphtheria toxin bound to nicotinamide adenine dinucleotide. Biochemistry 35, 1137–1149.
Bennett, M. J., and Eisenberg, D. (1994). Refined structure of monomeric diphtheria toxin at 2.3Å resolution. Protein Sci. 3, 1464–1475.
Cassel, D., and Pfeuffer, T. (1978). Mechanism of cholera toxin action: covalent modification of the guanyl nucleotide-binding protein of the adenylate cyclase system. Proc. Natl. Acad. Sci. U.S.A. 75, 2669–2673.
Chardin, P., Boquet, P., Madaule, P., Popoff, M. R., Rubin, E. J., and Gill, D. M. (1989). The mammalian G protein rhoC is ADP-ribosylated by Clostridium botulinum exoenzyme C3 and affects actin microfilaments in Vero cells. EMBO J. 8, 1087–1092.
Collier, R. J. (1967). Effect of diphtheria toxin on protein synthesis: inactivation of one of the transfer factors. J. Mol. Biol. 25, 83–98.
Collier, R. J. (2001). Understanding the mode of action of diphtheria toxin: a perspective on progress during the 20th century. Toxicon 39, 1793–1803.
Day, B., Dahlbeck, D., Huang, J., Chisholm, S. T., Li, D., and Staskawicz, B. J. (2005). Molecular basis for the RIN4 negative regulation of RPS2 disease resistance. Plant Cell 17, 1295–1305.
Deng, Q., and Barbieri, J. T. (2008). Molecular mechanisms of the cytotoxicity of ADP-ribosylating toxins. Annu. Rev. Microbiol. 62, 271–288.
Di Girolamo, M., Dani, N., Stilla, A., and Corda, D. (2005). Physiological relevance of the endogenous mono-(ADP-ribosyl)ation of cellular proteins. FEBS J. 272, 4565–4575.
Fu, Z. Q., Guo, M., Jeong, B. R., Tian, F., Elthon, T. E., Cerny, R. L., Staiger, D., and Alfano, J. R. (2007). A type III effector ADP-ribosylates RNA-binding proteins and quells plant immunity. Nature 447, 284–288.
Heintzen, C., Nater, M., Apel, K., and Staiger, D. (1997). AtGRP7, a nuclear RNA-binding protein as a component of a circadian-regulated negative feedback loop in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 94, 8515–8520.
Holbourn, K. P., Shone, C. C., and Acharya, K. R. (2006). A family of killer toxins. Exploring the mechanism of ADP-ribosylating toxins. FEBS J. 273, 4579–4593.
Honjo, T., Nishizuka, Y., and Hayaishi, O. (1968). Diphtheria toxin-dependent adenosine diphosphate ribosylation of aminoacyl transferase II and inhibition of protein synthesis. J. Biol. Chem. 243, 3553–3555.
Hooykaas, P. J., Dulk-Ras, H., and Schilperoort, R. A. (1988). The Agrobacterium tumefaciens T-DNA gene 6b is an onc gene. Plant Mol. Biol. 11, 791–794.
Iglewski, B. H., Liu, P. V., and Kabat, D. (1977). Mechanism of action of Pseudomonas aeruginosa exotoxin: adenosine diphosphate-ribosylation of mammalian elongation factor 2 in vitro and in vivo. Infect. Immun. 15, 138–144.
Jeong, B. R., Lin, Y., Joe, A., Guo, M., Korneli, C., Yang, H., Wang, P., Yu, M., Cerny, R. L., Staiger, D., Alfano, J. R., and Xu, Y. (2011). Structure function analysis of an ADP-ribosyltransferase type III effector and its RNA-binding target in plant immunity. J. Biol. Chem. 286, 43272–43281.
Kidner, C. A., and Martienssen, R. A. (2004). Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 428, 81–84.
Kim, H. S., Desveaux, D., Singer, A. U., Patel, P., Sondek, J., and Dangl, J. L. (2005). The Pseudomonas syringae effector AvrRpt2 cleaves its C-terminally acylated target, RIN4, from Arabidopsis membranes to block RPM1 activation. Proc. Natl. Acad. Sci. U.S.A. 102, 6496–6501.
Kleine, H., Poreba, E., Lesniewicz, K., Hassa, P. O., Hottiger, M. O., Litchfield, D. W., Shilton, B. H., and Lüscher, B. (2008). Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol. Cell 32, 57–69.
Lamb, R. S., Citarelli, M., and Teotia, S. (2012). Functions of the poly-ADP-ribose- polymerase superfamily in plants. Cell. Mol. Life Sci. 69, 175–189.
Mackey, D., Belkhadir, Y., Alonso, J. M., Ecker, J. R., and Dangl J. L. (2003). Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112, 379–389.
Navarro, L., Dunoyer, P., Jay, F., Arnold, B., Dharmasiri, N., Estelle, M., Voinnet, O., and Jones, J. D. (2006). A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312, 436–439.
O’Neal, C. J., Jobling, M. G., Holmes, R. K., and Hol, W. G. (2005). Structural basis for the activation of cholera toxin by human ARF6-GTP. Science 309, 1093–1096.
Robert-Seilaniantz, A., Shan, L., Zhou, J. M., and Tang, X. (2006). The Pseudomonas syringae pv. tomato DC3000 type III effector HopF2 has a putative myristoylation site required for its avirulence and virulence functions. Mol. Plant Microbe Interact. 19, 130–138.
Schöning, J. C., Streitner, C., Page, D. R., Hennig, S., Uchida, K., Wolf, E., Furuya, M., and Staiger, D. (2007). Auto-regulation of the circadian slave oscillator component AtGRP7 and regulation of its targets is impaired by a single RNA recognition motif point mutation. Plant J. 52, 1119–1130.
Schreiber, V., Dantzer, F., Ame, J. C., and de Murcia, G. (2006). Poly-(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 7, 517–28.
Sun, J., Maresso, A. W., Kim, J. J. P., and Barbieri, J. T. (2004). How bacterial ADP-ribosylating toxins recognize substrates. Nat. Struct. Mol. Biol. 11, 868–876.
Singer, A. U., Desveaux, D., Betts, L., Chang, J. H., Nimchuk, Z., Grant, S. R., Dangl, J. L., and Sondek, J. (2004). Crystal structures of the type III effector protein AvrPphF and its chaperone reveal residues required for plant pathogenesis. Structure 12, 1669–1681.
Tinland, B., Huss, B., Paulus, F., Bonnard, G., and Otten, L. (1989). Agrobacterium tumefaciens 6b genes are strain-specific and affect the activity of auxin as well as cytokinin genes. Mol. Gen. Genet. 219, 217–224.
Tsiamis, G., Mansfield, J. W., Hockenhull, R., Jackson, R. W., Sesma, A., Athanassopoulos, E., Bennett, M. A., Stevens, C., Vivian, A., Taylor, J. D., and Murillo, J. (2000). Cultivar-specific avirulence and virulence functions assigned to avrPphF in Pseudomonas syringae pv. phaseolicola, the cause of bean halo-blight disease. EMBO J. 19, 3204–3214.
Vanden Broeck, D., Horvath, C., and De Wolf, M. J. (2007). Vibrio cholerae: cholera toxin. Int. J. Biochem. Cell Biol. 39, 1771–1775.
van der Hoorn, R. A., and Kamoun, S. (2008). From guard to decoy: a new model for perception of plant pathogen effectors. Plant Cell 20, 2009–2017.
Wang, H., Liang, Q., Cao, K., and Ge, X. (2011a). Endogenous protein mono-ADP-ribosylation in Arabidopsis thaliana. Planta 233, 1287–1292.
Wang, M., Soyano, T., Machida, S., Yang, J. Y., Jung, C., Chua, N. H., and Yuan, Y. A. (2011b). Molecular insights into plant cell proliferation disturbance by Agrobacterium protein 6b. Genes Dev. 64–76.
Wang, Y., Li, J., Hou, S., Wang, X., Li, Y., Ren, D., Chen, S., Tang, X., and Zhou, J. M. (2010). A Pseudomonas syringae ADP-ribosyltransferase inhibits Arabidopsis mitogen-activated protein kinase kinases. Plant Cell 22, 2033–2044.
Wilton, M., Rajagopal, S., Elmore, J., Felsensteiner, C., Coaker, G., and Desvaux, D. (2010). The type III effector HopF2 Pto targets Arabidopsis RIN4 protein to promote Pseudomonas syringae virulence, Proc. Natl. Acad. Sci. U.S.A. 107, 2349–2354
Keywords: pathogen effector, mono-ADP-ribosyltransferase, crystal structure, plant innate immunity
Citation: Wirthmueller L and Banfield MJ (2012) mADP-RTs: versatile virulence factors from bacterial pathogens of plants and mammals. Front. Plant Sci. 3:142. doi: 10.3389/fpls.2012.00142
Received: 30 April 2012; Paper pending published: 20 May 2012;
Accepted: 12 June 2012; Published online: 27 June 2012.
Edited by:
Jacqueline Monaghan, The Sainsbury Laboratory, UKReviewed by:
Christian Lindermayr, Helmholtz Zentrum München – German Research Center for Environmental Health, GermanyJesus V. Jorrin Novo, University of Cordoba, Spain
Copyright: © 2012 Wirthmueller and Banfield. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Lennart Wirthmueller and Mark J. Banfield, Department of Biological Chemistry, John Innes Centre, Norwich Research Park, Norwich, NR4 7UH, UK. e-mail:bGVubmFydC53aXJ0aG11ZWxsZXJAamljLmFjLnVr;bWFyay5iYW5maWVsZEBqaWMuYWMudWs=