- 1 Department of Plant Biology, Cornell University, Ithaca, NY, USA
- 2 USDA-ARS, Robert W. Holley Center for Agriculture and Health, Ithaca, NY, USA
- 3 Institute for Biotechnology and Life Science Technologies, Cornell University, Ithaca, NY, USA
N-glycosylation is one of the most common and complex post-translational modifications of eukaryotic proteins and one that has numerous roles, such as modulating protein stability, sorting, folding, enzyme activity, and ligand interactions. In plants, the functional significance of N-glycosylation is typically obscure, although it is a feature of most secreted proteins and so is potentially of considerable interest to plant cell wall biologists. While analytical pipelines have been established to characterize yeast, mammalian, and bacterial N-glycoproteomes, such large-scale approaches for the study of plant glycoproteins have yet to be reported. Indeed, the N-glycans that decorate plant and mammalian or yeast proteins are structurally distinct and so modification of existing analytical approaches are needed to tackle plant N-glycoproteomes. In this review, we summarize a range of existing technologies for large-scale N-glycoprotein analysis and highlight promising future approaches that may provide a better understanding of the plant N-glycoproteome, and therefore the cell wall proteome and other proteins associated with the secretory pathway.
Introduction
Characterization of the detailed structures of N-glycoproteins can provide valuable insights into basics aspects of cell and organismal biology and is also becoming increasingly important for pharmaceutical development and production (De Marchis et al., 2011; Xu et al., 2011; Yang et al., 2012). In plants, N-glycosylation is a common feature of secreted proteins and there is considerable interest in developing more advanced analytical platforms to characterize N-glycosylation in order to provide new insights in the plant cell wall proteome (Minic et al., 2007; Marino et al., 2010; Zhang et al., 2010; Catala et al., 2011; Ruiz-May et al., 2012). However, structural elucidation of glycans, glycopeptides, and glycoproteins is notoriously difficult as glycans are structurally complex, being branched, and containing a variety of types of intersaccharide linkages, and heterogeneous, with a wide range of abundance between glycoforms and occupancy of glycosylated sites. Ideally, a comprehensive analysis of a plant glycoprotein involves the identification of glycosylated peptides, determination of the location of the glycosylation sites, and elucidation of the glycan structure (Fitchette et al., 2007), each of which presents a specific set of technical challenges.
In the past decade, biological mass spectrometry (MS) has become established as a core analytical tool for proteome profiling (Thelen and Peck, 2007; Yates et al., 2009; Ytterberg and Jensen, 2010; Song et al., 2011) as well as characterization of protein glycosylation patterns and glycan analysis (Harvey, 2005; Park and Lebrilla, 2005). However, while advanced tandem MS (MS/MS) technologies provide an effective means for glycopeptide amino acid sequencing and obtaining structural information from oligosaccharides (e.g., sequence, linkages, and branching), the high degree of complexity and variability in glycoprotein populations from any eukaryotic source still poses significant analytical obstacles. Sample preparation that allows enrichment of the targeted glycoproteins/glycopeptides is therefore often an important step as it increases the concentration of the desired analyte while reducing sample complexity. A second technical hurdle is the hydrophilic nature of glycans and their relatively larger masses compared with the native tryptic peptides to which they are attached. This often results in poor retention on the reverse phase chromatographic materials that are typically used in peptide separations, reduced ionization efficiency and larger than optimal m/z detection range in MS analysis, all of which can contribute to poor sensitivity (An et al., 2009; Ytterberg and Jensen, 2010). Furthermore, the labile nature of the glycan–peptide bond often causes the neutral loss of individual carbohydrates and few fragmented ions from the peptide during the collision induced dissociation (CID), yielding little or no information for peptide identification, glycol-site determination, and full glycan sequence. As a result, deglycosylation is often required and the free peptide and glycan are subjected separately to tandem MS (MS/MS) analysis. Finally, due to the high glycan heterogeneity, determination of the glycan structure through bioinformatics tool-based database searching is often difficult and requires time-consuming manual interpretation.
To overcome these analytical challenges, many new strategies have been developed and implemented over the past decade involving the development of three major tools: (1) selective enrichment technologies for glycoproteins, glycopeptides, and released glycans, and improvements in separation science including the miniaturization of chromatographic formats, capillary electrophoresis (CE), and ion mobility technologies; (2) enhanced MS technology and development of more effective analysis workflows; and (3) newly developed software packages and algorithms for interpretation of MS fragmentation spectra. Most of these technologies have been initially developed to study mammalian glycoproteins and there are only a few reported examples of their successful application to characterize plant N-glycosylation (Fitchette et al., 2007; Bardor et al., 2009; Zhang et al., 2012), which is structurally distinct from that of mammalian or yeast proteins (Ruiz-May et al., 2012). Key structural differences, such as the absence of non-reducing terminal sialic acid in plant N-glycans and the presence of α-1,3-fucose (Fuc) in the innermost GlcNAc residue necessitate tailoring of existing analytical workflows.
In this review we present some of the analytical platforms or strategies, including new developments in MS instrumentation, which can be used for the systematic characterization of N-glycoproteins, with particular reference to the challenges of studying those from plants, as well as a perspective of future developments in the field. To our knowledge there are no published reports describing large- or even intermediate-scale analysis of plant protein glycosylation together with associated N-glycans analysis. However, given the new and emerging analytical pipelines we are soon likely to see a dramatic expansion in the identification and functional characterization of plant N-glycoproteomes, which in turn will be of great value for researchers studying the plant cell wall and associated secretory pathways.
Glycoprotein Detection
A summary of a typical workflow for characterizing N-glycoproteins/larger scale N-glycoproteomes is shown in Figure 1. A valuable first step is to confirm the presence of glycoproteins in a crude extract using 1D or 2D gel-based electrophoretic separation, followed by immunoblot analysis or direct visualization using a commercially available sugar detection stain (Fitchette et al., 2007; Bardor et al., 2009). Antibodies to horseradish peroxidase (HRP) can recognize glycoproteins with an epitope associated with β1→2 xylose and/or α1→3 fucose structures on complex-type N-glycans (Fitchette et al., 2007; Bardor et al., 2009; Figure 2). They are therefore useful for detecting plant N-glycoproteins since plant complex-type glycans carry β1→2 xylose and/or α1→3 core fucose structures (Strasser et al., 2008). This is a relatively straightforward technique, but does not distinguish between different types of N-glycans, or between N- and O-linked glycans. The presence of glycoproteins can also be suggested by the presence of smeared bands on SDS-PAGE gels (Zhang et al., 2010; Catala et al., 2011), resulting from the presence of heterogonous glycans. Another practical, but sometimes less informative means to assess the presence of glycoproteins in a sample is by comparison of changes in the electrophoretic mobility of the constituent proteins following specific deglycosylation steps (Fitchette et al., 2007; Bardor et al., 2009). It is important to note that that the enzyme PNGase-F, which is typically use to deglycosylate mammalian proteins does not process N-glycans that contain an α-1,3-fucose residue that is a common component of plant N-glycans, although enzymes such as PNGase-A can be used as an alternative (Ytterberg and Jensen, 2010).
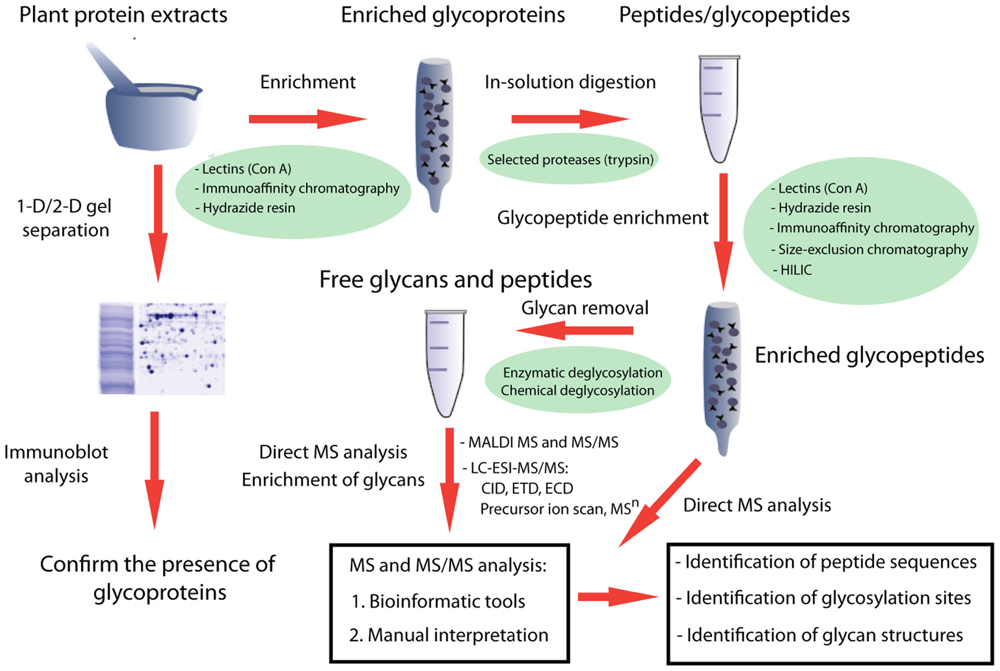
FIGURE 1. A schematic overview of experimental approaches for the systematic characterization of glycoproteins by mass spectrometry-based proteomic analysis. Regardless of the sample sources, the general workflow includes three major steps: enrichment of glycoproteins, glycopeptides, and released glycans; mass spectrometry analysis of glycopeptides, glycans, and peptides; and interpretation of MS fragmentation spectra, allowing identification of core amino acid sequences of the glycopeptides, glycosylation sites, and potential glycan structures.
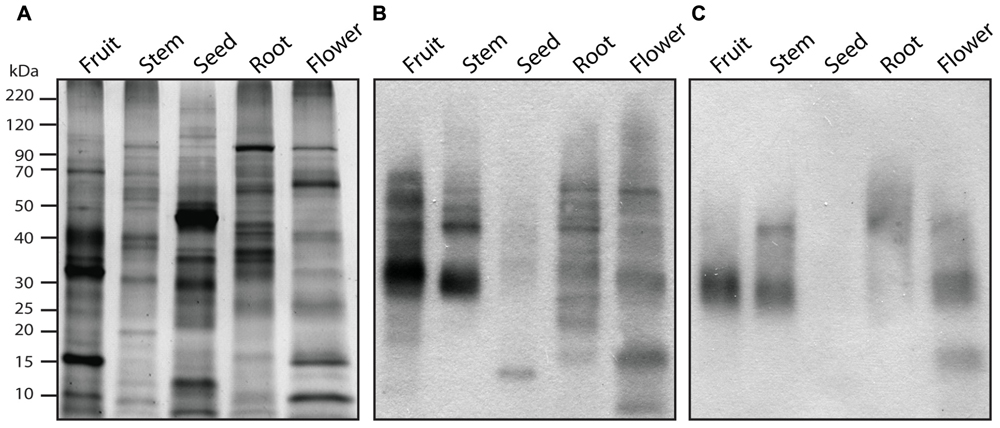
FIGURE 2. Organ-specific immunodetection of glycan epitopes. Protein extracts from different tomato organs were fractionated 12% polyacrylamide gels and visualized by SYPRO Ruby protein staining (A) or transferred onto nitrocellulose membrane for immunoblotting using antisera that recognize β-1,2-xylose (B) or α-1,3-fucose (C) residues.
Glycoprotein/Glycopeptide Enrichment and Preparation
Depending on the protein complexity in the initial samples, the analysis of glycoproteins then often proceeds with the enrichment of glycoproteins or glycopeptides by specific lectins or immunoaffinity chromatography (Rudiger and Gabius, 2001) to remove non-glycosylated forms. Lectin chromatography using Concanavalin A (Con A) has been reported for the enrichment of N-glycoproteins from diverse sources including human serum, Caenorhabditis elegans plasma membrane glycoproteins (Bunkenborg et al., 2004; Fan et al., 2004), and plant tissues (Minic et al., 2007; Zhang et al., 2010; Catala et al., 2011). Various plants lectins are available with affinities for different sugars (Gabius et al., 2004) and such lectins have been used sequentially (Yamamoto et al., 1995, 1998), in parallel (Yang et al., 2006; Lee et al., 2009; Zielinska et al., 2010), and as mixtures (Li et al., 2004) to increase the range of identified glycoproteins. When working with limited amounts of sample it is often necessary to miniaturize the chromatography. Both silica-based and monolithic materials incorporating a lectin-containing stationary phase are now commercially available that can be packed into micro or capillary columns, greatly increasing the number of glycoproteins that can be detected in low concentrations compared with standard column formats (Madera et al., 2006; Feng et al., 2009).
Once an enriched glycoprotein sample has been prepared, a typical next step is to fragment the parent protein backbone and a potential technical challenge is that the glycan moiety may spatially interfere with proteolysis, particularly near the glycosylation site. This can result in missed cleavages, yielding glycosylated polypeptides that are too large to be detected by some MS instruments. However, the use of less specific proteases, such as pepsin, thermolysin, Proteinase K, either alone or in combination, can reduce this problem (Chen et al., 2009). Nevertheless, no matter what the digestion procedure, the degree of heterogeneity of the resulting mixture of peptides and glycopeptides can complicate the subsequent MS analysis as glycopeptides often ionize less efficiently than non-glycopeptides and may be less abundant than their unmodified counterparts. Therefore, in some cases, deglycosylation prior to or concurrent with enzymatic digestion is desirable (see below). If deglycosylation is not employed, an additional enrichment of glycopeptides following glycoprotein digestion is often implemented to improve specificity. An alternative chemical approach for glycoprotein/glycopeptides enrichment is the use of hydrazine resin. N-glycans bind covalently to this matrix after oxidation with periodic acid and can then be released with PNGase A, although some information related to the N-glycan structure is lost (Zhang et al., 2003). Other alternative chemical methods involve graphitized carbon (Larsen et al., 2005) or boronic acid-functionalized beads, since boric acid is able to form diesters with glycans that have cis-diol groups (Sparbier et al., 2005).
Several such enrichment methods have been successfully applied in large-scale N-glycoproteome analyses, such as the combined use of lectin (Con A) affinity chromatography and subsequent hydrophilic interaction chromatography (HILIC) enrichment of tryptic glycopeptides (Calvano et al., 2008; Selby et al., 2008), and size-exclusion chromatography has also been used to enrichment for tryptic glycopeptides (Alvarez-Manilla et al., 2006).
MS Analysis
Once enriched glycopeptide samples have been isolated, they are then commonly subjected to MS/MS analysis, or to a deglycosylation step for simplifying MS analysis of individual peptides and glycans. However, to overcome the relatively poor sensitivity resulting from the hydrophilic nature of glycans, online nano-scale LC enrichment coupled with nano-electrospray ionization tandem MS (nanoESI-MS/MS; Bahr et al., 1997; Sandra et al., 2004; Zhang and Van Pelt, 2004) or additional derivatization (Zaia, 2004) is often used to improve glycopeptide ionization efficiency. Alternatively, matrix assisted laser desorption/ionization (MALDI) MS is used extensively for direct analysis of the released glycan classes or followed by glycan derivatization, as has been reviewed in detail (Zaia, 2010; Antonopoulos et al., 2011). Nano-scale liquid chromatography electrospray ionization (nanoLC-ESI) allows for low concentration of analytes, improves ionization efficiency and allows for low flow rates, which in turn makes it possible to use extended analysis times for MS/MS and multi-stage fragmentation (MSn) oligosaccharide analyses (Bahr et al., 1997). Traditionally, low-energy CID MS/MS has been the method of choice for peptide and glycopeptides sequencing operating in either automated data-dependent acquisition mode, or manual selection for MS/MS through infusion analysis. Another unique feature of CID MS/MS analysis of glycopeptides or glycans is the production of oxonium ions, such as m/z 163 (Hex + H)+, m/z 204 (HexNAc + H)+, and m/z 366 (Hex-HexNAc + H)+, which can be used as diagnostic marker ions, indicating the presence of specific sugar compositions. The oxonium ions are often monitored in precursor ion scanning mode for highly selective detection of glycopeptides in digestion mixture and subjected to MS/MS and MS/MS/MS of the selectively detected glycopeptides ions, yielding complete peptide and glycan sequences (Sandra et al., 2004; Zhang and Williamson, 2005). It should be note that highly selective precursor ion scanning for sugar oxonium ions also allows direct analysis of relatively less complex protein digests without further enrichment of glycopeptides (Sandra et al., 2004; Zhang and Williamson, 2005).
However, low-energy CID MS/MS can be rendered less effective by the labile nature of the glycosidic bond and often fails to produce fragment ions that can be used to determine both the peptide sequence and glycosylation site. In this regard, two new dissociation technologies, electron capture dissociation (ECD; Zubarev et al., 1998) and electron transfer dissociation (ETD; Syka et al., 2004), an analogue of ECD, provide valuable alternative approaches. Both ECD and ETD are non-ergodic fragmentation techniques that induce backbone fragmentation at N-Cα bonds and create complementary c- and z-type ion series, yielding information that is highly complementary to conventional CID fragmentation (Zubarev et al., 1998; Syka et al., 2004; Coon et al., 2005). ETD MS is becoming more widely accessible as it enables the incorporation of ECD-type fragmentation in more common RF ion trap mass spectrometers, thus eliminating the need for highly sophisticated higher-cost FTICR MS instruments. ETD cleaves the peptide backbone in a sequence-independent manner, leaving the glycan preserved on the peptide and is increasingly recognized as an important alternative dissociation technique to CID for glycosylation analysis (Catalina et al., 2007; Khidekel et al., 2007; Wuhrer et al., 2007b). With the combination of CID and ETD fragmentation available in single ion trap instruments, it is now possible to analyze intact glycopeptides and determine both peptide and glycan sequence information (Wuhrer et al., 2007a). To some degree the development of these non-ergodic fragmentation methods, which can be used to structurally characterize much larger analytes, has reduced the need to ensure that the proteolytic digestion proceeds to the point where the mass contribution of the peptidyl moiety is minimal. Wu et al. (2007) have successfully demonstrated the use of endoproteinase Lys-C to study the glycosylation of several model proteins using a combination of ETD and CID. This approach reduces the number of non-glycosylated peptides, thereby facilitating the characterization of the glycopeptides and reducing the need for additional chromatographic steps.
The development of ion mobility spectrometry (IMS) has extended the ability to characterize glycoprotein/glycopeptides to the realm of isomeric and conformational structure (Plasencia et al., 2008). For example, Olivova et al. (2008), using the high resolution provided by a recently developed quadrupole ion-mobility time-of-flight instrument, was able to determine the total glycosylation profile of a monoclonal antibody. The IMS function was used to resolve the light and heavy chains in the gas phase allowing the accurate mass measurement of each. Furthermore, the innovative dual-collision cell design of this instrument was leveraged in a two step fragmentation process to provide a detailed characterization of the glycan structures and the determination of the glycosylation site.
Other than direct MS analysis of enriched glycopeptides, deglycosylation steps can also be performed to gain additional information about peptide sequence and complete glycan structure. Three enzymes are commonly used for enzymatic deglycosylation. PNGase F, an amidase that hydrolyzes the amide bond between the Asn residue on the peptide backbone and the innermost HexNAc unit, is active on high mannose and complex plant N-glycans, except those with an α-1,3-fucose residue linked to the innermost HexNAc (Fitchette et al., 2007; Bardor et al., 2009). In this case, PNGase A can be used to release glycans from relatively short glycopeptides (Fitchette et al., 2007; Bardor et al., 2009). Alternatively, endoglycosidase H (Endo H) cleaves the glycosidic bond between the two GlcNAc residues on the high-mannose-type N-glycans of plant glycoproteins (Harvey, 2005). While these deglycosylation steps are time consuming it has been shown that pressure cycling and exposure to microwaves can greatly reduce deglycosylation times (Chen et al., 2009). Chemical deglycosylation can also be used, although enzymatic deglycosylation is generally favored as chemical approaches often introduce unexpected modifications of peptide side chains (Fitchette et al., 2007; Bardor et al., 2009).
After deglycosylation, LC-MS/MS is used to identify the amino acid sequence of the deglycosylated peptide. Alternatively, the released glycans can be further purified and analyzed by MALDI-TOF or LC-ESI-MS/MS analysis to identify the glycan structure (An et al., 2009). In practice, MS/MS spectra derived from the glycosidic bond fragment ions, along with cross-ring fragmentation of glycans and treatments with different exo-hexosidases are required for complete structural elucidation (Zaia, 2004). In the past, per-derivation, such as permethylation, of glycans has often been used to improve sensitivity and has yielded more structurally relevant fragment ions during MS/MS. However, as new MS technologies are constantly being developed, the sensitivity improvements provided by derivatization has only been marginal and insufficient to compensate for the more complex sample preparation and the fact that derivatization makes it difficult to employ glycosidase treatment to determine linkages and identify saccharide units (Kang et al., 2007). Another technique, called isotope coded glycosylation site-specific tagging (IGOT), has been used for the large-scale identification of N-linked glycoproteins from complex biological samples of C. elegans (Kaji et al., 2003, 2007) and mouse (Zielinska et al., 2010). This approach involves protein tryptic digestion and enrichment of resulting glycopeptides followed by conversion of glycosylated Asn residues into 18O-labeled aspartic acids by PNGase F digestion in 18O water. The subsequent 3 Da mass difference created by this process is readily identified by LC-MS/MS analysis, indicating N-linked glycosylation sites. However, a potential pitfall of this approach is that a significantly high false positive rate (~60%) has been reported (Angel et al., 2007), apparently due to the fact that the trypsin used for proteolysis was still active, leading to the incorporation of 18O into the C-termini of the peptides during the deglycosylation step.
Capillary electrophoresis has been used to analyze the heterogeneity of isolated glycoproteins with MS detection (Balaguer and Neususs, 2006; Thakur et al., 2009; Szabo et al., 2010) and the incorporation of microfluidic devices can also greatly facilitate the analysis by increasing sensitivity and reducing analysis times (Zhuang et al., 2007, 2011). While these CE/MS methods have proven useful as a means to characterize glycoprotein heterogeneity and to investigate the structure of glycans at the N- and O-glycosylation sites of recombinant glycoproteins (Zaia, 2010), they have not yet proven sufficiently robust for the analysis of complex glycoprotein mixtures.
Quantitative N-Glycoprotein Analysis by MS
In principle quantitative glycoproteomics experiments may be carried out by the same assortment of techniques that have been developed and are currently in use for labeling and label-free approaches in other proteomics disciplines. There have been several excellent reviews (Bindschedler and Cramer, 2011; Matros et al., 2011, Pan et al., 2011) and the reader is referred to these for detailed descriptions of the various experimental strategies employed and methodologies in use.
Historically, the quantification of proteoglycans has been difficult as it involves both the structural elucidation of glycan and peptide moieties. While structural characterization of peptides is well developed and is facilitated by the existence of complete genomic databases for a growing number of species, the structural characterization of the glycan moiety remains a challenge as they are often exceedingly heterogeneous and chemically complex, with differing chemical configurations and abundant isoforms. Furthermore, glycans are frequently branched and contain an assortment of glycan linkages (Zaia, 2004). These complications combine in such a way that quantitative experiments must be carried out in different tiers with each providing a different level of quantitative information. The first tier involves determining glycosylation sites and their degree of occupancy. For N-linked glycans potential glycosylation sites can be recognized due to the presence of the consensus sequence Asn-X-Ser/Thr using any of several open access programs such as Net Glyc (http://www.cbs.dtu.dk/services/NetOGlyc/). However, not all potential sites are occupied and those that are occupied are often only partially so but by a variety of differing glycans. Determining the number and types of glycoforms and their relative proportion present at each site represents the second tier of quantifications. The third tier of glycoprotein quantification involves comparing the state of glycosylation between different samples, i.e., control verses treated, control verses diseased, etc. Of the methods available, only MS allows for quantitative characterization at all three tiers.
Two-dimensional gel electrophoresis with glycan-specific staining has been the traditional method used to identify and quantify glycoproteins (Bardor et al., 2009). When coupled with MS of tryptic digests of excised spots it can provide quantitative characterization on all three levels. However, the heterogeneity and structural complexity of the glycans has important consequences on a protein's mobility which complicate the interpretation of the electropherograms. Thus, LC-MS has emerged as the preeminent tool for the identification and quantitative characterization of glycoproteins and glycopeptides. The development of novel hybrid mass spectrometers incorporating ion mobility cells, high mass accuracy and resolution, improved sensitivity, and scanning capabilities has allowed detailed glycoprotein structural characterization, with accurate empirical information concerning the position and degree of occupancy of the glycosylation sites as well as the composition, structure, and distribution of glycoforms.
In the discovery phase of research, most quantitative MS strategies involve either a labeling approach (Gygi et al., 1999; Kaji et al., 2003; Zhang et al., 2003; Ross et al., 2004; Aggarwal et al., 2006; Atwood et al., 2008; Haqqani et al., 2008) or a label-free strategy (Hill et al., 2009; Rebecchi et al., 2009; Zhang et al., 2012). These non-targeted approaches allow one to quickly survey complex proteomes to determine the features that vary between samples and more precise quantitative information can be obtained through a targeted approach involving specific reaction monitoring (SRM) as recently described by Zhang et al. (2012). New developments involving data independent acquisition (DIA) using approaches such as MSE or SWATH (Levin et al., 2011; Gillet et al., 2012; Hopfgartner et al., 2012) could potentially simplify these procedures further by allowing the discovery and targeted data to be collected simultaneously greatly reducing the instrument time required.
Bioinformatic Tools
One of the biggest challenges in the field of glycoproteomics is the development of bioinformatic tools for glycan analysis. As a consequence of the complex fragmentation behaviors of glycopeptides and glycans, which tend to vary under different CID conditions utilized by various MS instruments, and the high level of glycan heterogeneity, there are few tools for direct large-scale identification of glycoproteins through database searching. As a result, tedious manual data interpretation represents the major bottle-neck for glycoprotein characterization. However, several computer programs have been developed, such as GlysodeIQ (Joshi et al., 2004), GlyMod (Cooper et al., 2001), GlycoMiner (Ozohanics et al., 2008), and Peptoonist (Goldberg et al., 2007), to aid identification of intact N-linked glycopeptides and oligosaccharides, as well as CartoonistTwo (Goldberg et al., 2006) for automatic determination of the topology of O-glycan without linkage information from MS/MS spectra. Nevertheless, there is an unquestionable need for improved bioinformatic tools to keep pace with the rapid advances in MS technologies and strategies for sample preparation, such as software for the automatic determination of N-linked glycopeptides.
Discussion and Perspectives
As described above, the characterization of the plant N-glycoproteome is methodologically challenging due to the extreme structural heterogeneity and the often low concentration of several glycoforms attached to each glycopeptide residing in the total peptide pool. However, several approaches have been established to study specific population of glycoproteins from different tissues. Until now, there had not been a single enrichment approach that permits the analysis of the complete population of glycoprotein in a specific tissue, but lectin affinity chromatography is an effective option because it permits the purification and analysis of native glycoproteins, allowing the characterization of endogenous glycoforms without any modification of the N-glycans structure. Moreover, the availability of several lectins with different ligand affinities allows broader coverage of the N-glycoproteome, as has been described in mammals (Zielinska et al., 2010).
As far as we are aware, there have been no such reports describing the application of large-scale enrichment techniques in combination with cutting-edge MS/MS methods for the direct analysis of intact plant glycopeptides to gain information of both glycan and peptide moiety. However, the establishment of new MS platforms, such as the use of CID and ETD fragmentation in a single ion trap instrument for simultaneous analysis of the sugar and protein components of glycopeptides (Wuhrer et al., 2007a) is likely to revolutionize the study of plant glycoproteins. Rapid progress will depend on other technical advances such as solving the problems of the recalcitrance of plant proteins to deglycosylation by standard PNGase F treatments and the development of new, robust bioinformatics tools.
Taken together, such developments hold great promise for plant researchers in many fields, including those interested in the plant cell wall. Increasing numbers of wall localized proteins are being identified (Ruiz-May et al., 2012) and yet remarkably little is known about the functional significance of post-translational modifications such as glycosylation. New analytical pipelines such as those described here, perhaps coupled with other techniques that will likely result in glycoprotein enrichment (e.g., Parsons et al., 2012), hold great potential in this regard. This represents an important new frontier in plant cell wall biology and it can be argued that the advent of the plant N-glycoproteome starts now.
Conflict of Interest Statement:
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Funding to Theodore W. Thannhauser was provided by ARS CRIS Project No. 1907-21000-024-00D and by NSF DBI-0606596. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. The USDA is an equal opportunity provider and employer.
References
Aggarwal, K., Choe, L. H., and Lee, K. H. (2006). Shotgun proteomics using the iTRAQ isobaric tags. Brief. Funct. Genomic. Proteomic. 5, 112–120.
Alvarez-Manilla, G., Atwood, J. III, Guo, Y., Warren, N. L., Orlando, R., and Pierce, M. (2006). Tools for glycoproteomic analysis: size exclusion chromatography facilitates identification of tryptic glycopeptides with N-linked glycosylation sites. J. Proteome Res. 5, 701–708.
An, H. J., Froehlich, J. W., and Lebrilla, C. B. (2009). Determination of glycosylation sites and site-specific heterogeneity in glycoproteins. Curr. Opin. Chem. Biol. 13, 421–426.
Angel, P. M., Lim, J. M., Wells, L., Bergmann, C., and Orlando, R. (2007). A potential pitfall in 18O-based N-linked glycosylation site mapping. Rapid Commun. Mass Spectrom. 21, 674–682.
Antonopoulos, A., North, S. J., Haslam, S. M., and Dell, A. (2011). Glycosylation of mouse and human immune cells: insights emerging from N-glycomics analyses. Biochem. Soc. Trans. 39, 1334–1340.
Atwood, J. A. III, Cheng, L., Alvarez-Manilla, G., Warren, N. L., York, W. S., and Orlando, R. (2008). Quantitation by isobaric labeling: applications to glycomics. J. Proteome Res. 7, 367–374.
Bahr, U., Pfenninger, A., Karas, M., and Stahl, B. (1997). High-sensitivity analysis of neutral underivatized oligosaccharides by nanoelectrospray mass spectrometry. Anal. Chem. 69, 4530–4535.
Balaguer, E., and Neususs, C. (2006). Glycoprotein characterization combining intact protein and glycan analysis by capillary electrophoresis-electrospray ionization-mass spectrometry. Anal. Chem. 78, 5384–5393.
Bardor, M., Cabrera, G., Stadlmann, J., Lerouge, P., Cremata, J. A., Gomord, V., and Fitchette, A. C. (2009). N-glycosylation of plant recombinant pharmaceuticals. Methods Mol. Biol. 483, 239–264.
Bunkenborg, J., Pilch, B. J., Podtelejnikov, A. V., and Wiśniewski, J. R. (2004). Screening for N-glycosylated proteins by liquid chromatography mass spectrometry. Proteomics 4, 454–465.
Calvano, C. D., Zambonin, C. G., and Jensen, O. N. (2008). Assessment of lectin and HILIC based enrichment protocols for characterization of serum glycoproteins by mass spectrometry. J. Proteomics 71, 304–317.
Catala, C., Howe, K. J., Hucko, S., Rose, J. K., and Thannhauser, T. W. (2011). Towards characterization of the glycoproteome of tomato (Solanum lycopersicum) fruit using Concanavalin A lectin affinity chromatography and LC-MALDI-MS/MS analysis. Proteomics 11, 1530–1544.
Catalina, M. I., Koeleman, C. A., Deelder, A. M., and Wuhrer, M. (2007). Electron transfer dissociation of N-glycopeptides: loss of the entire N-glycosylated asparagine side chain. Rapid Commun. Mass Spectrom. 21, 1053–1061.
Chen, R., Jiang, X., Sun, D., Han, G., Wang, F., Ye, M., Wang, L., and Zou, H. (2009). Glycoproteomics analysis of human liver tissue by combination of multiple enzyme digestion and hydrazide chemistry. J. Proteome Res. 8, 651–661.
Coon, J. J., Ueberheide, B., Syka, J. E., Dryhurst, D. D., Ausio, J., Shabanowitz, J., and Hunt, D. F. (2005). Protein identification using sequential ion/ion reactions and tandem mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 102, 9463–9468.
Cooper, C. A., Gasteiger, E., and Packer, N. (2001). GlycoMod – a software tool for determining glycosylation compositions from mass spectrometric data. Proteomics 1, 340–349.
De Marchis, F., Balducci, C., Pompa, A., Riise Stensland, H. M., Guaragno, M., Pagiotti, R., Menghini, A. R., Persichetti, E., Beccari, T., and Bellucci, M. (2011). Human alpha-mannosidase produced in transgenic tobacco plants is processed in human alpha-mannosidosis cell lines. Plant Biotechnol. J. 9, 1061–1073.
Fan, X., She, Y. M., Bagshaw, R. D., Callahan, J. W., Schachter, H., and Mahuran, D. J. (2004). A method for proteomic identification of membrane-bound proteins containing Asn-linked oligosaccharides. Anal. Biochem. 332, 178–186.
Feng, S., Yang, N., Pennathur, S., Goodison, S., and Lubman, D. M. (2009). Enrichment of glycoproteins using nanoscale chelating concanavalin A monolithic capillary chromatography. Anal. Chem. 81, 3776–3783.
Fitchette, A. C., Dinh, O. T., Faye, L., and Bardor, M. (2007). Plant proteomics and glycosylation. Methods Mol. Biol. 355, 317–342.
Gabius, H. J., Siebert, H. C., André, S., Jiménez-Barbero, J., and Rüdiger, H. (2004). Chemical biology of the sugar code. Chembiochem 5, 740–764.
Gillet, L. C., Navarro, P., Tate, S., Röst, H., Selevsek, N., Reiter, L., Bonner, R., and Aebersold, R. (2012). Targeted data extraction of the MS/MS spectra generated by data independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics 11, O111.016717.
Gygi, S. P., Rist, B., Gerber, S. A., Turecek, F., Gelb, M. H., and Aebersold, R. (1999). Quantitative analysis of mixtures using isotope-coded affinity tags. Nat. Biotechnol. 17, 994–999.
Goldberg, D., Bern, M., Li, B., and Lebrilla, C. B. (2006). Automatic determination of O-glycan structure from fragmentation spectra. J. Proteome Res. 5, 1429–1434.
Goldberg, D., Bern, M., Parry, S., Sutton-Smith, M., Panico, M., Morris, H. R., and Dell, A. (2007). Automated N-glycopeptide identification using a combination of single- and tandem-MS. J. Proteome Res. 6, 3995–4005.
Haqqani, A. S., Kelly, J. F., and Stanimirovic, D. B. (2008). Quantitative protein profiling by mass spectrometry using isotope-coded affinity tags. Methods Mol. Biol. 439, 225–240.
Harvey, D. J. (2005). Proteomic analysis of glycosylation: structural determination of N- and O-linked glycans by mass spectrometry. Exp. Rev. Proteomics 2, 87–101.
Hill, J. J., Tremblay, T.-L., Cantin, C., O'Connor-McCourt, M., Kelly, J. F., and Lenferink, A. E. (2009). Glycoproteomic analysis of two mouse mammary cell lines during transforming growth factor (TGF)-β induced epithelial to mesenchymal transition. Proteome Sci. 7, 2. doi: 10.1186/1477-5956-7-2
Hopfgartner, G., Tonoli, D., and Varesio, E. (2012). High-resolution mass spectrometry for integrated qualitative and quantitative analysis of pharmaceuticals in biological matrices. Anal. Bioanal. Chem. 402, 2587–2596.
Joshi, H. J., Harrison, M. J., Schulz, B. L., Cooper, C. A., Packer, N. H., and Karlsson, N. G. (2004). Development of a mass fingerprinting tool for automated interpretation of oligosaccharide fragmentation data. Proteomics 4, 1650–1664.
Kaji, H., Kamiie, J., Kawakami, H., Kido, K., Yamauchi, Y., Shinkawa, T., Taoka. M., Takahashi, N., and Isobe, T. (2007). Proteomics reveals N-linked glycoprotein diversity in Caenorhabditis elegans and suggests an atypical translocation mechanism for integral membrane proteins. Mol. Cell Proteomics 6, 2100–2109.
Kaji, H., Saito, H., Yamauchi, Y., Shinkawa, T., Taoka, M., Hirabayashi, J., Kasai, K.-I., Takahashi, N., and Isobe, T. (2003). Lectin affinity capture, isotope-coded tagging and mass spectrometry to identify N-linked glycoproteins. Nat. Biotechnol. 21, 667–672.
Kang, P., Mechref, Y., Kyselova, Z., Goetz, J. A., and Novotny, M. V. (2007). Comparative glycomic mapping through quantitative permethylation and stable-isotope labeling. Anal. Chem. 79, 6064–6073.
Khidekel, N., Ficarro, S. B., Clark, P. M., Bryan, M. C., Swaney, D. L., Rexach, J. E., Sun, Y. E., Coon, J. J., Peters, E. C., and Hsieh-Wilson, L. C. (2007). Probing the dynamics of O-GlcNAc glycosylation in the brain using quantitative proteomics. Nat. Chem. Biol. 3, 339–348.
Larsen, M. R., Hojrup, P., and Roepstorff, P. (2005). Characterization of gel-separated glycoproteins using two-step proteolytic digestion combined with sequential microcolumns and mass spectrometry. Mol. Cell Proteomics 4, 107–119.
Lee, A., Kolarich, D., Haynes, P. A., Jensen, P. H., Baker, M. S., and Packer, N. H. (2009). Rat liver membrane glycoproteome: enrichment by phase partitioning and glycoprotein capture. J. Proteome Res. 8, 770–781.
Levin, Y., Hradetzky, E., and Bahn, S. (2011). Quantification of proteins using data-independent analysis (MSE) in simple andcomplex samples: a systematic evaluation. Proteomics 11, 3273–3287.
Li, L., Wang, L., Zhang, W., Tang, B., Zhang, J., Song, H., Yao, D., Tang, Y., Chen, X., Yang, Z., Wang, G., Li, X., Zhao, J., Ding, H., Reed, E., and Li, Q. Q. (2004). Correlation of serum VEGF levels with clinical stage, therapy efficacy, tumor metastasis and patient survival in ovarian cancer. Anticancer Res. 24, 1973–1979.
Madera, M., Mechref, Y., Klouckova, I., and Novotny, M. V. (2006). Semiautomated high-sensitivity profiling of human blood serum glycoproteins through lectin preconcentration and multidimensional chromatography/tandem mass spectrometry. J. Proteome Res. 5, 2348–2363.
Marino, K., Bones, J., Kattla, J. J., and Rudd, P. M. (2010). A systematic approach to protein glycosylation analysis: a path through the maze. Nat. Chem. Biol. 6, 713–723.
Matros, A., Kaspar, S., Witzel, K., and Mock, H. P. (2011). Recent progress in liquid chromatography-based separation and label-free quantitative plant proteomics. Phytochemistry 72, 963–974.
Minic, Z., Jamet, E., Négroni, L., der Garabedian, P. A., Zivy, M., and Jouanin, L. (2007). A sub-proteome of Arabidopsis thaliana mature stems trapped on Concanavalin A is enriched in cell wall glycoside hydrolases. J. Exp. Bot. 58, 2503–2512.
Olivova, P., Chen, W., Chakraborty, A. B., and Gebler, J. C. (2008). Determination of N-glycosylation sites and site heterogeneity in a monoclonal antibody by electrospray quadrupole ion-mobility time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 22, 29–40.
Ozohanics, O., Krenyacz, J., Ludányi, K., Pollreisz, F., Vékey, K., and Drahos, L. (2008). GlycoMiner: a new software tool to elucidate glycopeptide composition. Rapid Commun. Mass Spectrom. 22, 3245–3254.
Pan, S., Chen, R., Aebersold, R., and Brentnall, T. A. (2011). Mass spectrometry based glycoproteomics-from a proteomics perspective. Mol. Cell Proteomics 10, R110.003251.
Park, Y., and Lebrilla, C. B. (2005). Application of Fourier transform ion cyclotron resonance mass spectrometry to oligosaccharides. Mass Spectrom. Rev. 24, 232–264.
Parsons, H. T., Christiansen, K., Knierim, B., Carroll, A., Ito, J., Batth, T. S., Smith-Moritz, A. M., Morrison, S., McInerney, P., Hadi, M. Z., Auer, M., Mukhopadhyay, A., Petzold, C. J., Scheller, H. V., Loqué, D., and Heazlewood, J. L. (2012). Isolation and proteomic characterization of the Arabidopsis Golgi defines functional and novel components involved in plant cell wall biosynthesis. Plant Physiol. 159, 12–26.
Plasencia, M. D., Isailovic, D., Merenbloom, S. I., Mechref, Y., Novotny, M. V., and Clemmer, D. E. (2008). Resolving and assigning N-linked glycan structural isomers from ovalbumin by IMS-MS. J. Am. Soc. Mass Spectrom. 19, 1706–1715.
Rebecchi, K. R., Wenke, J. L., Go, E. P., and Desaire, H. (2009). Label-free quantitation: a new glycoproteomics approach. J. Am. Soc. Mass Spectrom. 20, 1048–1059.
Ross, P. L., Huang, Y. N., Marchese, J. N., Williamson, B., Parker, K., Hattan, S., Khainovski, N., Pillai, S., Dey, S., Daniels, S., Purkayastha, S., Juhasz, P., Martin, S., Bartlet-Jones, M., He, F., Jacobson, A., and Pappin, D. J. (2004). Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell Proteomics 3, 1154–1169.
Rudiger, H., and Gabius, H. J. (2001). Plant lectins: occurrence, biochemistry, functions and applications. Glycoconj. J. 18, 589–613.
Ruiz-May, E., Kim, S. J., Brandizzi, F., and Rose, J. K. C. (2012). The secreted plant N-glycoproteome and associated secretory pathways. Front. Plant Sci. 3:117. doi: 10.3389/fpls.2012.00117
Sandra, K., Devreese, B., Van Beeumen, J., Stals, I., and Claeyssens, M. (2004). The Q-Trap mass spectrometer, a novel tool in the study of protein glycosylation. J. Am. Soc. Mass Spectrom. 15, 413–423.
Selby, D. S., Larsen, M. R., Calvano, C. D., and Jensen, O. N. (2008). Identification and characterization of N-glycosylated proteins using proteomics. Methods Mol. Biol. 484, 263–276.
Song, W., Henquet, M. G., Mentink, R. A., van Dijk, A. J., Cordewener, J. H., Bosch, D., America, A. H., and van der Krol, A. R. (2011). N-glycoproteomics in plants: perspectives and challenges. J. Proteomics 74, 1463–1474.
Sparbier, K., Koch, S., Kessler, I., Wenzel, T., and Kostrzewa, M. (2005). Selective isolation of glycoproteins and glycopeptides for MALDI-TOF MS detection supported by magnetic particles. J. Biomol. Tech. 16, 407–413.
Strasser, R., Stadlmann, J., Schähs, M., Stiegler, G., Quendler, H., Mach, L., Glössl, J., Weterings, K., Pabst, M., and Steinkellner, H. (2008). Generation of glyco-engineered Nicotiana benthamiana for the production of monoclonal antibodies with a homogeneous human-like N-glycan structure. Plant Biotechnol. J. 6, 392–402.
Syka, J. E., Coon, J. J., Schroeder, M. J., Shabanowitz, J., and Hunt, D. F. (2004). Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 101, 9528–9533.
Szabo, Z., Guttman, A., Rejtar, T., and Karger, B. L. (2010). Improved sample preparation method for glycan analysis of glycoproteins by CE-LIF and CE-MS. Electrophoresis 31, 1389–1395.
Thakur, D., Rejtar, T., Karger, B. L., Washburn, N. J., Bosques, C. J., Gunay, N. S., Shriver, Z., and Venkataraman, G. (2009). Profiling the glycoforms of the intact alpha subunit of recombinant human chorionic gonadotropin by high-resolution capillary electrophoresis-mass spectrometry. Anal. Chem. 81, 8900–8907.
Thelen, J. J., and Peck, S. C. (2007). Quantitative proteomics in plants: choices in abundance. Plant Cell 19, 3339–3346.
Wu, S. L., Huhmer, A. F., Hao, Z., and Karger, B. L. (2007). On-line LC-MS approach combining collision-induced dissociation (CID), electron-transfer dissociation (ETD), and CID of an isolated charge-reduced species for the trace-level characterization of proteins with post-translational modifications. J. Proteome Res. 6, 4230–4244.
Wuhrer, M., Catalina, M. I., Deelder, A. M., and Hokke, C. H. (2007a). Glycoproteomics based on tandem mass spectrometry of glycopeptides. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 849, 115–128.
Wuhrer, M., Stam, J. C., van de Geijn, F. E., Koeleman, C. A., Verrips, C. T., Dolhain, R. J., Hokke, C. H., and Deelder, A. M. (2007b). Glycosylation profiling of immunoglobulin G (IgG) subclasses from human serum. Proteomics 7, 4070–4781.
Xu, J., Ge, X., and Dolan, M. C. (2011). Towards high-yield production of pharmaceutical proteins with plant cell suspension cultures. Biotechnol. Adv. 29, 278–299.
Yamamoto, K., Tsuji, T., and Osawa, T. (1995). Analysis of asparagine-linked oligosaccharides by sequential lectin affinity chromatography. Mol. Biotechnol. 3, 25–36.
Yamamoto, K., Tsuji, T., and Osawa, T. (1998). Analysis of asparagine-linked oligosaccharides by sequential lectin-affinity chromatography. Methods Mol. Biol. 76, 35–51.
Yang, Z., Drew, D. P., Jørgensen, B., Mandel, U., Bach, S. S., Ulvskov, P., Levery, S. B., Bennett, E. P., Clausen, H., and Petersen, B. L. (2012). Engineering mammalian mucin-type O-glycosylation in plants. J. Biol. Chem. 287, 11911–11912
Yang, Z., Harris, L. E., Palmer-Toy, D. E., and Hancock, W. S. (2006). Multilectin affinity chromatography for characterization of multiple glycoprotein biomarker candidates in serum from breast cancer patients. Clin. Chem. 52, 1897–1905.
Yates, J. R., Ruse, C. I., and Nakorchevsky, A. (2009). Proteomics by mass spectrometry: approaches, advances, and applications. Annu. Rev. Biomed. Eng. 11, 49–79.
Ytterberg, A. J., and Jensen, O. N. (2010). Modification-specific proteomics in plant biology. J. Proteomics 73, 2249–2266.
Zhang, H., Li, X. J., Martin, D. B., and Aebersold, R. (2003). Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat. Biotechnol. 21, 660–666.
Zhang, S., Sherwood, R. W., Yang, Y., Fish, T., Chen, W., McCardle, J. A., Jones, R, M., Yusibov, V., May, E. R., Rose, J. K., and Thannhauser, T. W. (2012). Comparative characterization of the glycosylation profiles of an influenza hemagglutinin produced in plant and insect hosts. Proteomics 12, 1269–1288.
Zhang, S., and Van Pelt, C. K. (2004). Chip-based nanoelectrospray mass spectrometry for protein characterization. Exp. Rev. Proteomics 1, 449–468.
Zhang, S., and Williamson, B. L. (2005). Characterization of protein glycosylation using chip-based nanoelectrospray with precursor ion scanning quadrupole linear ion trap mass spectrometry. J. Biomol. Tech. 16, 209–219.
Zhang, Y., Giboulot, A., Zivy, M., Valot, B., Jamet, E., and Albenne, C. (2010). Combining various strategies to increase the coverage of the plant cell wall glycoproteome. Phytochemistry 72, 1109–1123.
Zhuang, Z., Mitra, I., Hussein, A., Novotny, M. V., Mechref, Y., and Jacobson, S. C. (2011). Microchip electrophoresis of N-glycans on serpentine separation channels with asymmetrically tapered turns. Electrophoresis 32, 246–253.
Zhuang, Z., Starkey, J. A., Mechref, Y., Novotny, M. V., and Jacobson, S. C. (2007). Electrophoretic analysis of N-glycans on microfluidic devices. Anal. Chem. 79, 7170–7175.
Zielinska, D. F., Gnad, F., Wiśniewski, J. R., and Mann, M. (2010). Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell 141, 897–907.
Keywords: glycoproteins, N-glycans, glycopeptides, lectins, mass spectrometry
Citation: Ruiz-May E, Thannhauser TW, Zhang S and Rose JKC (2012) Analytical technologies for identification and characterization of the plant N-glycoproteome. Front. Plant Sci. 3:150. doi: 10.3389/fpls.2012.00150
Received: 30 March 2012; Accepted: 18 June 2012;
Published online: 04 July 2012.
Edited by:
Seth DeBolt, University of Kentucky, USACopyright: © 2012 Ruiz-May, Thannhauser, Zhang and Rose. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Jocelyn K. C. Rose, Department of Plant Biology, Cornell University, 412 Mann Library Building, Ithaca, NY 14853, USA. e-mail:anIyODZAY29ybmVsbC5lZHU=

 Theodore W. Thannhauser2
Theodore W. Thannhauser2 and
and