- 1Bio Research Laboratory, Toyota Motor Corporation, Toyota, Aichi, Japan
- 2Laboratory of Crop Science, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Bunkyo-ku, Tokyo, Japan
- 3NARO Agricultural Research Center, Joetsu, Niigata, Japan
- 4Center for Gene Research, Nagoya University, Chikusa-ku, Nagoya, Japan
Although sucrose plays a role in sugar sensing and its signaling pathway, little is known about the regulatory mechanisms of the expressions of plant sucrose-related genes. Our previous study on the expression of the sucrose phosphate synthase gene family in rice (OsSPSs) suggested the involvement of sucrose sensing and/or circadian rhythm in the transcriptional regulation of OsSPS. To examine whether the promoters of OsSPSs can be controlled by sugars and circadian clock, we produced transgenic rice plants harboring a promoter–luciferase construct for OsSPS1 or OsSPS11 and analyzed the changes in the promoter activities by monitoring bioluminescence from intact transgenic plants in real-time. Transgenic plants fed sucrose, glucose, or mannitol under continuous light conditions showed no changes in bioluminescence intensity; meanwhile, the addition of sucrose increased the concentration of sucrose in the plants, and the mRNA levels of OsSPS remained constant. These results suggest that these OsSPS promoters may not be regulated by sucrose levels in the tissues. Next, we investigated the changes in the promoter activities under 12-h light/12-h dark cycles and continuous light conditions. Under the light–dark cycle, both OsSPS1 and OsSPS11 promoter activities were low in the dark and increased rapidly after the beginning of the light period. When the transgenic rice plants were moved to the continuous light condition, both POsSPS1::LUC and POsSPS11::LUC reporter plants exhibited circadian bioluminescence rhythms; bioluminescence peaked during the subjective day with a 27-h period: in the early morning as for OsSPS1 promoter and midday for OsSPS11 promoter. These results indicate that these OsSPS promoters are controlled by both light illumination and circadian clock and that the regulatory mechanism of promoter activity differs between the two OsSPS genes.
Introduction
Sucrose is the major photosynthetic product and plays a central role in plant metabolism. Many plant species utilize sucrose as the main sugar for translocation of carbohydrate form source leaves to sink tissues. Sucrose is also reported to be a signal molecule that regulates gene expression in plants (Chiou and Bush, 1998; Vaughn et al., 2002; Ransom-Hodgkins et al., 2003). However, there are fewer reports about sucrose-mediated signaling pathways than glucose-mediated ones (see Rolland et al., 2006 for review).
Sucrose phosphate synthase (SPS, EC 2.3.1.14) catalyzes the conversion of fructose-6-phosphate and UDP-glucose into sucrose-6-phosphate and is known to be the major rate-limiting enzyme in sucrose biosynthesis in plants (Winter and Huber, 2000; Lunn and MacRae, 2003). SPS activity is regulated by allosteric effectors, glucose-6-phosphate (activator) and inorganic phosphate (inhibitor), and by environmental factors such as light and osmotic stress via the phosphorylation of several serine residues. In particular, light–dark modulation of SPS activity via reversible phosphorylation is well established in several plant species (see Winter and Huber, 2000, for review). In addition, the expression of SPS genes can be regulated by light and cold stress at the transcriptional level (Chávez-Bárcenas et al., 2000; Lutfiyya et al., 2007; Okamura et al., 2011).
It has been shown that plants have multiple forms of SPS, and that plant SPS genes are clustered into four groups (groups A, B, C, and D), based on their amino acid sequences (Castleden et al., 2004; Lutfiyya et al., 2007). Alignment of SPS sequences indicates that all of plant SPS proteins examined possess the phosphorylation site involved in light–dark regulation. The group-D SPS proteins, which can be found only in grass species including rice, are smaller than those from the other groups, lacking two phosphorylation sites involved in 14-3-3 protein binding and osmotic regulation. Rice has five SPS genes, OsSPSs, classified into four groups: OsSPS8, OsSPS1, OsSPS11, and OsSPS2 and OsSPS6 in groups A, B, C, and D, respectively (Castleden et al., 2004; Okamura et al., 2011). Although each OsSPS may have different roles in rice plants, little is known about the physiological significance of each OsSPS gene except for OsSPS1. For example, the suppression of OsSPS1 shortens the plant length of rice seedlings (Hirose et al., 2012), and the locus of OsSPS1 appears to coincide with the quantitative trait locus (QTL) for plant height (Ishimaru et al., 2004). Previously, we measured the mRNA levels of five OsSPSs by real-time quantitative RT-PCR with respect to developmental stages, tissues, diurnal changes, and circadian rhythm. We revealed differential expression patterns in the rice SPS gene family and part of the complex mechanisms underlying their transcriptional controls (Okamura et al., 2011). We found, in rice leaves, that the expressions of all OsSPSs tend to be higher at night than during the day, that OsSPS1 and OsSPS6 mRNA levels are negatively correlated with sucrose concentration, and that all OsSPSs except for OsSPS11 exhibit circadian rhythms. These results suggested that the mechanisms of transcriptional control differ between OsSPS1 and OsSPS11; the transcription of OsSPS1 could be regulated by sugar levels and/or circadian clock and that of OsSPS11 might not.
In this study, we performed promoter–reporter assays for OsSPS1 and OsSPS11 in vivo using an automated bioluminescence-monitoring apparatus to investigate the potential regulation of the promoter activities of OsSPSs by sugars and/or circadian clock. The promoter activities of transgenic rice plants carrying a promoter–luciferase reporter for OsSPS1 or OsSPS11 were measured in real-time under sugar-fed conditions, light–dark cycles, and continuous light. We found that the promoter activities of OsSPS1 and OsSPS11 are unaffected by an exogenous supply of sucrose or glucose but are regulated by circadian clock and light.
Materials and Methods
Growth Conditions of Rice Plants and Sampling
Surface-sterilized seeds (Oryza sativa L. cv. Nipponbare) were grown for 14 days in Milli-Q water (Millipore, Tokyo, Japan) under 18-h light/6-h dark cycles. The light intensity in the light period was 80 μmol m-2 s-1 emitted by halogen lamps. The temperature was maintained at 30°C in the light period and 25°C in the dark period. For RNA analysis and the measurement of sugar contents, detached aerial parts of plants were treated with each sugar, immediately frozen in liquid nitrogen, and stored at -80°C.
PSPS1::LUC and PSPS11::LUC Reporter Constructs
MultiSite Gateway cloning technology (Life Technologies, Tokyo, Japan) was employed to obtain promoter–reporter constructs containing either OsSPS1 (Os01g0919400) or OsSPS11 (Os11g0236100) promoters, modified firefly luciferase gene (LUC+) as a reporter gene, and nopaline synthase terminator (TNOS). PCR amplification of the elements required for the constructs was carried out with a high-fidelity DNA polymerase, PrimeSTAR GXL (Takara Bio, Shiga, Japan), according to the manufacturer’s instructions. The primers used to make the constructs are listed in Table 1.
A destination vector, pIG-R4R3, was constructed by introducing a Gateway cassette comprising attR4, ccdB, chloramphenicol resistance gene (CmR), and attR3 into the HindIII site of an ordinary binary vector, pIG121-Hm (AB489142). Entry clones harboring SPS promoters, the reporter, and the terminator were prepared as follows. The promoter regions of OsSPS1 and OsSPS11, PSPS1 and PSPS11, respectively, were amplified from genomic DNA from a japonica rice cultivar, Nipponbare, as a template. PSPS1 and PSPS11 cover the genome region from -2,415 to +6 and -2,380 to +6 nucleotides from the translation start point, respectively. These two promoter DNAs were cloned into the pDONR P4-P1R vector (Invitrogen, Tokyo, Japan) using BP clonase II (Life Technologies).
A modified gene for firefly luciferase, LUC+, was amplified from the pSP-luc+NF Fusion Vector (Promega, Tokyo, Japan) and cloned into the pDONR221 vector (Life Technologies). Nopaline synthase terminator was amplified from binary vector pBI121 (AF485783; Jefferson et al., 1987) and cloned into the pDONR P2R-P3 vector (Invitrogen).
Finally, these entry clones were assembled into the pIG-R4R3 destination vector using LR Clonase II Plus (Life Technologies) and designated either pSPS1 or pSPS11 vector, in which the initial two amino acid residues of SPSs were fused in frame with luciferase.
Generation of Transgenic Rice Plants
Five-week-old calli derived from a mature rice seed (cv. Nipponbare) were co-cultivated with Agrobacterium tumefaciens strain EHA101 harboring either pSPS1 or pSPS11 plasmid for 3 days in N6 (Chu et al., 1975) co-cultivation medium supplemented with 3% (w/v) sucrose, 1% (w/v) glucose, 0.03% (w/v) casamino acids, 2 mg L-1 2,4-dichlorophenoxyacetic acid (2,4-D), 10 mg L-1 acetosyringone, and 0.4% (w/v) gelrite at 28°C in the dark. The calli were then transferred to N6 selection medium supplemented with 3% (w/v) sucrose, 1% (w/v) glucose, 0.03% (w/v) casamino acids, 2 mg L-1 2,4-D, 0.4% (w/v) gelrite, 500 mg L-1 carbenicillin, and 25 mg L-1 hygromycin B as selective antibiotics for 4 weeks (subcultured every 2 weeks) at 30°C under dark conditions. The resistant calli were transferred to MS (Murashige and Skoog, 1962) regeneration medium supplemented with 3% (w/v) sucrose, 3% (w/v) sorbitol, 2% (w/v) casamino acids, 2 mg L-1 kinetin, 2 μg L-1 1-naphthylacetic acid, 0.4% (w/v) gelrite, and 25 mg L-1 hygromycin B for 4 weeks (subcultured every 2 weeks) to regenerate the aerial parts of rice and then transferred to MS regeneration hormone-free medium for 2 weeks to regenerate the roots at 30°C under continuous light conditions. Regenerated plants (T0 plants) were transplanted into sterilized soil and grown in a greenhouse at 30°C under 12-h light/12-h dark cycles. Self-pollinated heterozygous progenies (T1 seeds and T2 plants) were used for the experiment. Several independent lines were established for POsSPS1::LUC and POsSPS11::LUC reporter plants, and all lines exhibited bioluminescence in preliminary assay for luciferase activity. For further experiments, we selected two lines each, the line #8 and #10 as POsSPS1::LUC reporter plants, and the line #10 and #18 as POsSPS11::LUC reporter plants, judging from the abundance of seeds.
Measurement of Bioluminescence of POsSPS1::LUC and POsSPS11::LUC Transgenic Plants
Plants were grown for 14 days in 100 μM D-luciferin-K (Biosynth, Naperville, IL, USA) dissolved in Milli-Q water under 18-h light/6-h dark cycles, and transferred to continuous light at 30°C. The following day, the aerial parts of each plant were detached, rolled, and transferred to 35-mm petri dishes (BD Biosciences, Tokyo, Japan) containing 3.5 mL of 100 μM D-luciferin. The bioluminescence from each petri dish was subsequently measured using an automated bioluminescence-monitoring apparatus (Okamoto, Onai, and Ishiura, unpublished; model LL04-1; Churitsu Electric Corp., Nagoya, Japan) at 30°C under continuous light or 12-h light/12-h dark cycle conditions. The light intensity was 80 μmol m-2 s-1 emitted from white fluorescent lamps. To examine the effects of exogenous sugar on the reporter expressions, each sugar was added to the D-luciferin water in each petri dish and the bioluminescence was measured in continuous light condition without resetting the circadian clock. To measure circadian bioluminescence rhythms, each petri dish was exposed to an additional three 12-h light/12-h dark cycles to reset the clock and the bioluminescence was measured in continuous light condition. We analyzed the bioluminescence data using a commercially available bioluminescence-analyzing software (Onai, Shiraki, and Ishiura, unpublished; SL00-01; Churitsu Electric Corp.) based on RAP software (Okamoto et al., 2005). The phase of rhythms was represented as circadian time (CT) calculated by dividing the peak-phase value by the period length and multiplying by 24.
Real-Time Quantitative RT-PCR
Total RNA was extracted from sugar-treated wild-type (non-transgenic) plants as described by Okamura et al. (2011). Real-time quantitative RT-PCR analysis was performed using sets of gene-specific primers for OsSPS1 or OsSPS11 (Table 1; Okamura et al., 2011). Total RNA (10 ng) was used for one-step real-time quantitative PCR using the One Step SYBR PrimeScript PLUS RT-PCR Kit (Takara Bio, Shiga, Japan) and SMART Cycler II System (Cepehid Inc., CA, USA) according to the manufacturers’ instructions. The results were normalized according to the transcript levels of a rice polyubiquitin gene, RUBIQ1 (Wang et al., 2000).
Determination of Soluble Sugar Content
Soluble sugars were extracted from sugar-treated non-transgenic plants with 80% (v/v) ethanol and determined enzymatically as described by Okamura et al. (2011).
Results
Effects of Sugars on the Promoter Activities of OsSPS1 and OsSPS11
Several independent lines of transgenic rice plants carrying a promoter–luciferase reporter for OsSPS1 or OsSPS11 (POsSPS1::LUC or POsSPS11::LUC) were established. When D-luciferin was added to the medium, all reporter plants used exhibited bioluminescence and we found no significant differences among lines (data not shown). Therefore, both promoter sequences, POsSPS1 and POsSPS11, were functional in the rice cells.
To examine whether the promoter activities of OsSPS1 and OsSPS11 are regulated by sugars, we measured the bioluminescence of the POsSPS1::LUC and POsSPS11::LUC reporter plants after exogenously supplying 5% (w/v) sucrose, 5% (w/v) glucose, or 5% (w/v) mannitol. In both of the reporter plants, bioluminescence levels did not fluctuate significantly during the 48-h continuous light, even without the sugar treatments (0%). We did not observed significant changes in bioluminescence with any treatment (Figures 1A,B).
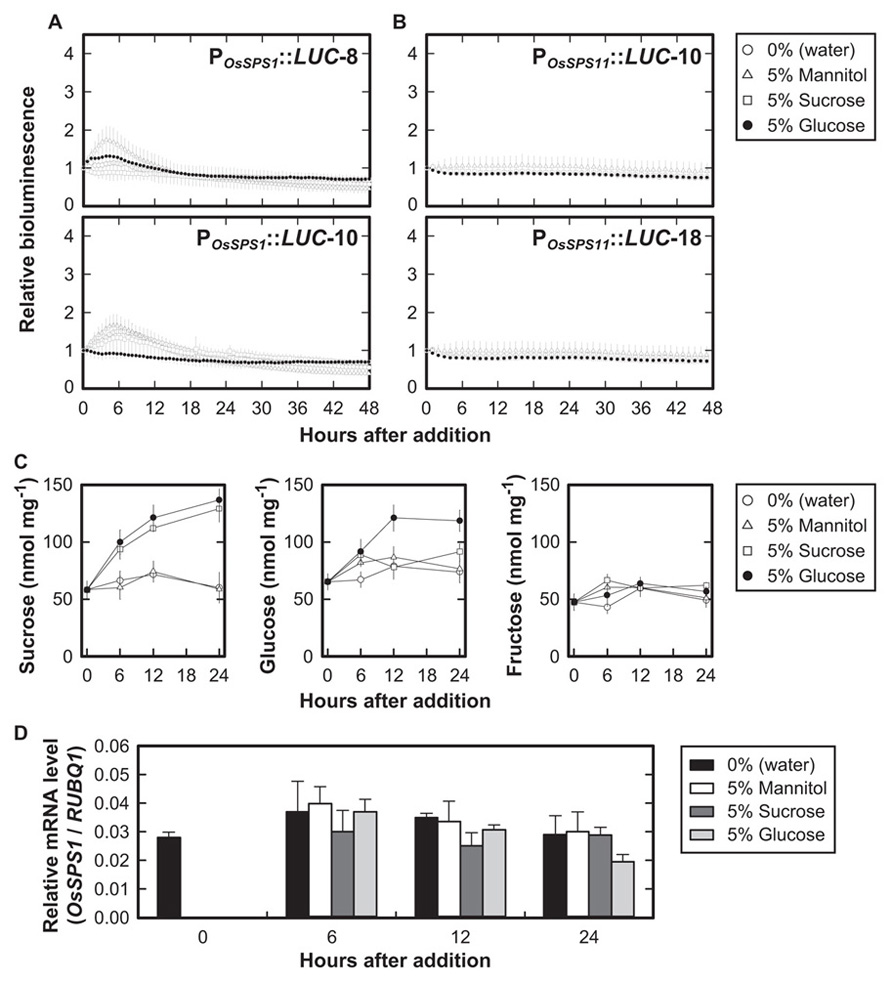
FIGURE 1. Promoter activities of OsSPS1 and OsSPS11 with the addition of sugars in continuous light. Fifteen-day-old plants were exposed to sugars added to the medium 24 h after the onset of illumination. (A) Bioluminescence of POsSPS1::LUC reporter plants without resetting the circadian clock. Data points indicate the mean ±SD of four independent plants in each line. We obtained essentially the same results from three independent experiments. (B) Bioluminescence of POsSPS11::LUC reporter plants. Data points indicate the mean ±SD of five independent plants in each line. We obtained essentially the same results from three independent experiments. (C) Endogenous sugar contents of wild-type (non-transgenic) plants after an exogenous supply of 5% sucrose, 5% glucose, 5% mannitol, or no sugar (0%, water). The y-axis represents sugar contents (nmol) per fresh weights of the plants (mg). Error bars indicate the SD of three independent plants. We obtained essentially the same results from three independent experiments. (D) mRNA levels of OsSPS1 in non-transgenic plants after an exogenous supply of 5% sucrose, 5% glucose, 5% mannitol, or water. Values were normalized to the expression level of a rice polyubiquitin gene (RUBIQ1). Note that the data at time 0 indicate the relative expression levels before the supply of sugars. Error bars indicate the standard errors of three independent plants. We obtained essentially the same results from three independent experiments.
To confirm if exogenous sugars added to the medium were successfully taken up into the plant cells, we examined concentrations of sucrose, glucose, and fructose in the plants after the addition of sugars (Figure 1C). When 5% sucrose was added to the medium, the amount of sucrose within the tissues increased 2.5-fold for 24 h whereas glucose and fructose levels remained unchanged. The addition of 5% glucose increased the amount of glucose twofold after 12 h of treatment. The amount of sucrose also increased twofold compared to the control after 6 h of treatment, but fructose levels remained unchanged. No changes in the amounts of these sugars were observed with the addition of 5% mannitol.
We also examined the mRNA levels of OsSPS1 and OsSPS11 during the sugar treatments and found no significant differences between OsSPS1 mRNA levels at any time point (Figure 1D). The result was consistent with those from POsSPS1::LUC reporter plants. In contrast, we did not detect any significant amounts of OsSPS11 mRNA under these experimental conditions, suggesting low levels of OsSPS11 mRNA.
These results indicated that sucrose and glucose did not affect the promoter activities of OsSPS1 or OsSPS11 even though exogenous sugars in the medium were taken up into the cells.
Diurnal Changes in the Promoter Activities of OsSPS1 and OsSPS11
Previously, we found that OsSPS1 and OsSPS11 mRNA levels changed within a day–night cycle (Okamura et al., 2011). We measured the diurnal changes of the promoter activities of OsSPS1 and OsSPS11 according to bioluminescence from the reporter plants under 12-h light/12-h dark cycles (Figure 2). The promoter activities of both OsSPS1 and OsSPS11 were low during the dark period and increased rapidly after the onset of the light period. While the promoter activity of OsSPS1 decreased gradually during the day and remained low throughout the night (Figure 2A), the OsSPS11 promoter remained active throughout the day and became inactive immediately after the onset of the dark period (Figure 2B). These results indicated that the promoter activities of both OsSPSs were controlled by light but not in the same manner.
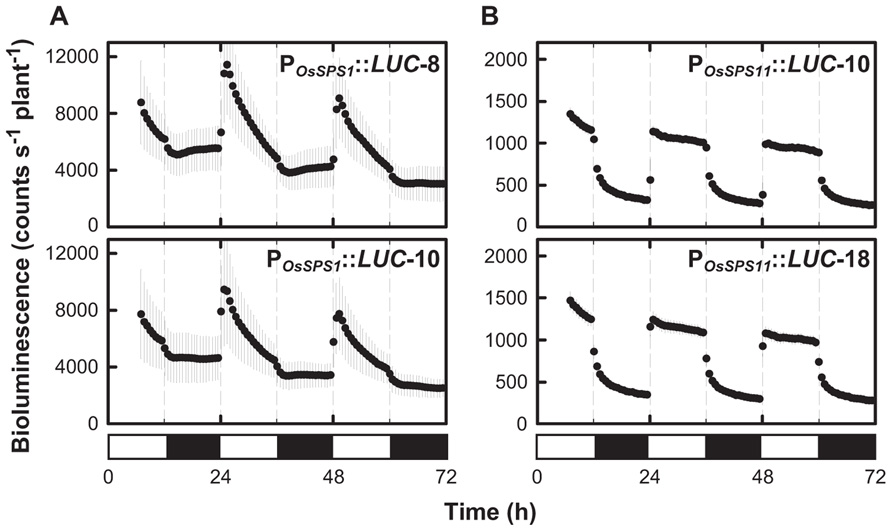
FIGURE 2. Diurnal patterns of the promoter activities of OsSPS1 and OsSPS11 under light/dark cycles. The bioluminescence of POsSPS1::LUC (A) and POsSPS11::LUC (B) reporter plants was measured under 12-h light/12-h dark cycles at 30°C. Data points indicate the mean ±SD from 6 to 8 independent plants in each line. We obtained essentially the same results from three independent experiments. White and black bars represent light and dark periods, respectively.
Regulation of the Promoter Activities of OsSPS1 and OsSPS11 by Circadian Clock
Circadian rhythms are endogenous daily fluctuations in physiological activities sustained under constant conditions with a periodicity of nearly 1 day. As shown in Figure 1, neither POsSPS1::LUC nor POsSPS11::LUC reporter plants exhibited a significant fluctuation in bioluminescence under continuous light conditions. However, the sugar-feeding experiments were conducted without a pretreatment to synchronize the circadian clock of measured plants, which is a general procedure for measuring circadian rhythms. In fact, when the reporter plants were exposed to three 12-h light/12-h dark cycles prior to the transfer to continuous light conditions, a clear circadian rhythm in bioluminescence was observed in both POsSPS1::LUC and POsSPS11::LUC reporter plants (Figure 3). The period lengths, peak phases, and amplitudes of rhythms are shown in Table 2. Both reporters exhibited circadian bioluminescence rhythms for at least 5 days with period lengths of approximately 27 h. However, these two reporters exhibited different rhythm profiles with respect to their peak phases and amplitudes. The rhythms of the POsSPS1::LUC reporter plants peaked in the early morning (around CT1), whereas those of POsSPS11::LUC reporter plants peaked in midday (around CT9). The rhythms of the POsSPS11::LUC reporter plants were weak; their amplitudes were quarter those of POsSPS1::LUC reporter plants (0.13 and 0.10 for POsSPS1::LUC reporter and 0.03 and 0.02 for POsSPS11::LUC reporter). These results indicated that the promoter activities of both OsSPSs were controlled by the circadian clock but their regulations by the clock were distinct.
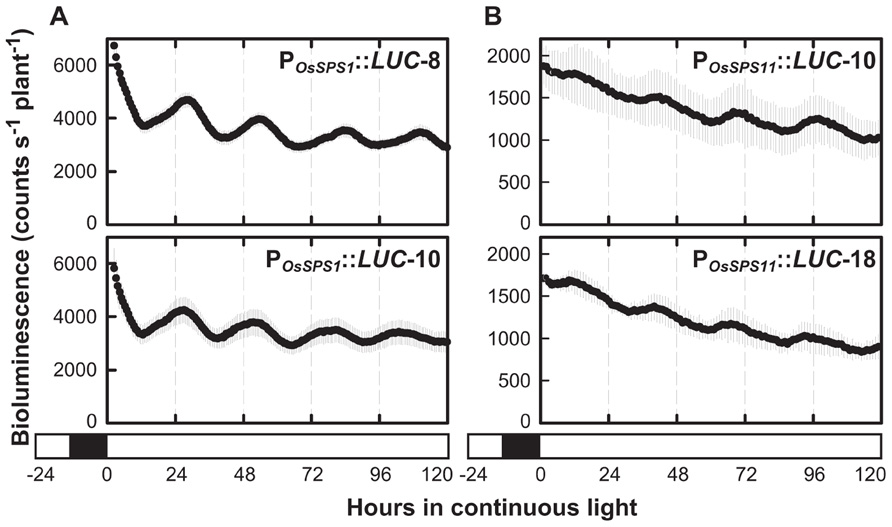
FIGURE 3. Bioluminescence rhythms of POsSPS1::LUC (A) and POsSPS11::LUC (B) reporter plants in continuous light. Fourteen-day-old plants were exposed to three 12-h light/12-h dark cycles to reset the circadian clock and transferred to continuous light conditions at 30°C for bioluminescence measurement. Data points indicate the mean ±SD from three independent plants in each line. The results are summarized in Table 2. We obtained essentially the same results from three independent experiments. White and black bars represent light and dark periods, respectively.
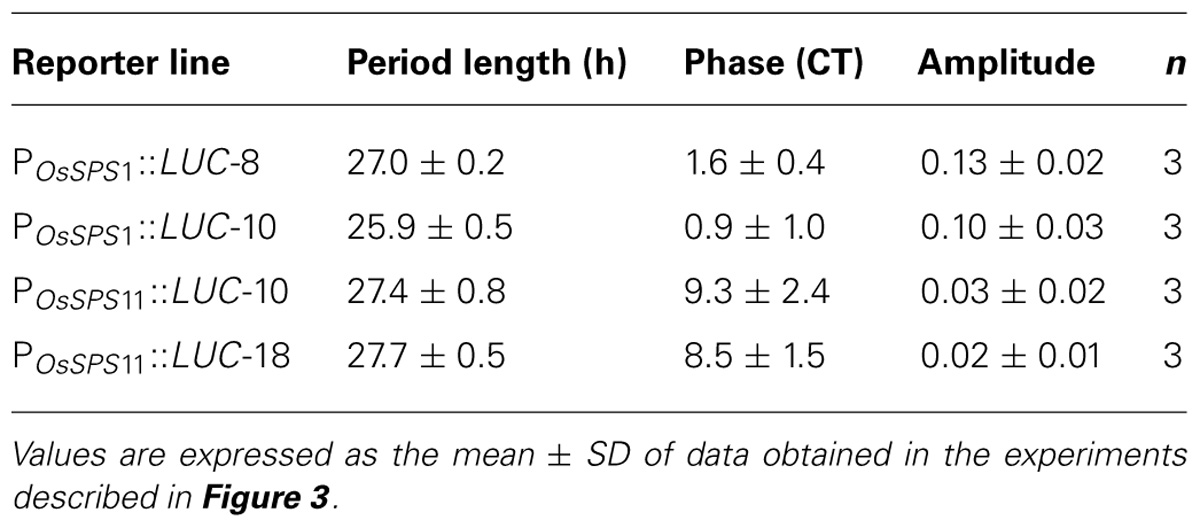
TABLE 2. Bioluminescence rhythms of POsSPS1::LUC and POsSPS11::LUC reporter plants at 30°C under continuous light conditions.
Discussion
Since sucrose is a signaling molecule as well as the major photoassimilate in plants, the transcriptional regulation of genes related to the synthesis, transport, and metabolism of sucrose may play roles in sugar sensing and its signaling pathway. However, little is known about the regulatory mechanisms of the expressions of plant sucrose-related genes except for a gene for a proton–sucrose symporter in sugar beet (Vaughn et al., 2002; Ransom-Hodgkins et al., 2003). In the present study, we examined whether genes for SPS can be transcriptionally regulated by sugars in rice plants using promoter–reporter assays for OsSPS1 and OsSPS11. However, neither sucrose nor glucose influenced the activities of the OsSPS1 and OsSPS11 promoters (Figures 1A,B). In addition, the sugar treatments did not alter OsSPS1 mRNA levels in rice plants (Figure 1D). These results suggest that the expressions of OsSPS1 and OsSPS11 are not regulated by sugar levels in rice plants. There is a possibility that the exogenously supplied sucrose is hydrolyzed by invertases in the apoplast and then taken up into cells in the form of glucose and fructose. If so, it would not be surprising that the effect of sucrose on gene expression looks similar to that of glucose.
It should be noted that feeding sucrose to rice plants induced an increase of sucrose level within the tissues whereas glucose and fructose levels remained unchanged (Figure 1C). It is also surprising that glucose feeding induced the accumulation of sucrose as well as glucose. Sucrose feeding in Arabidopsis thaliana results in the accumulation of glucose and fructose as well as sucrose (Mita et al., 1997; Sokolov et al., 1998); moreover, sucrose does not accumulate as a result of glucose feeding (Yonekura, Aoki, Hirose, and Onai, unpublished). These differential outcomes of sugar feeding between rice and Arabidopsis suggest that caution should be taken when assuming that these plants have the same regulation of sugar metabolism through sugar sensing and signaling.
We also investigated the involvement of light and circadian rhythms in the transcriptional regulation of the two OsSPSs. When bioluminescence from OsSPS1- and OsSPS11-transgenic plants was measured under a light–dark cycle, OsSPS1 and OsSPS11 promoter activities were low in the dark and increased rapidly after the beginning of the light period (Figure 2). These results indicate that light is an important cue for the expressions of the two OsSPSs at the transcriptional level as well as the post-translational regulation mechanism of SPS protein, which is well-established in several plant species (Winter and Huber, 2000). OsSPS1 is reported to possess a light-responsive element in its promoter region and that its expression is positively regulated by light (Argüello-Astorga and Herrera-Esstrella, 1996; Chávez-Bárcenas et al., 2000). In the OsSPS11 promoter sequence, as well as OsSPS1, we found putative light-responsive cis-acting regulatory elements such as GATA-box and I-box core sequences, which were previously identified in light-regulated genes (Higo et al., 1999; Prestridge, 1991; Terzaghi and Cashmore, 1995). The existence of these cis-elements corroborates the present experimental data. However, another interpretation could be given for the diurnal pattern of promoter activities: light illumination may influence indirectly to the promoter activities of OsSPS1 and OsSPS11. It is generally known in plants that many metabolites fluctuate in response to light–dark transition. The OsSPS promoters could be regulated by metabolic changes. Even though sugar levels in tissues did not affect the activities of the two OsSPS promoters (Figure 1), the sugar levels measured with whole-tissue extracts may not reflect the levels within cells where the OsSPSs are able to be active.
The diurnal changes in the promoter activities of OsSPS1 and OsSPS11 (Figure 2) are inconsistent with our previous data on the day–night accumulation patterns of their mRNAs (Okamura et al., 2011; see Introduction). We can attribute this inconsistency in OsSPS expression to post-transcriptional regulation, which influences the stability of mRNAs. For example, in CIRCADIAN CLOCK ASSOCIATED1 (CCA1), one of the clock genes in Arabidopsis, the mRNA levels in the PCaMV35S::CCA1 plants are constantly high in the dark; meanwhile, in the light, they rapidly decline to about 40% of the dark level. The rapid decrease in CCA1 transcript level is attributable to the light-modulated stability of the mRNA (Yakir et al., 2007). Likewise, the two OsSPS mRNAs might be unstable and degraded immediately in the light, although the transcription of the genes is activated by light. Therefore, it can be speculated that a decrease in the pool size of the transcripts during the daytime enables the sucrose biosynthesis system to respond quickly to rapidly changing environments, including light condition, by the de novo transcription of the gene and/or post-translational regulation of the enzyme. In this study, we only used the 5′-untranslated region (UTR). Since Newman et al. (1993) reported that 3′-UTR sequences contribute to mRNA stability like downstream elements, the lack of a 3′-UTR might account for the differences between the diurnal changes in promoter activity and RNA accumulation.
In the sugar-feeding experiments (Figure 1), both POsSPS1::LUC and POsSPS11::LUC reporter plants did not exhibit circadian rhythms in bioluminescence even under constant condition. However, clear circadian rhythms became detectable when bioluminescence was monitored under constant condition after the reporter plants were exposed to three 12-h light/12-h dark cycles (Figure 3). Apparently, the circadian rhythms of promoter activities were masked in the sugar-feeding experiments, probably because the rhythms of four individual plants used were not fully synchronized without the pretreatment before measurements. Although the reason why no circadian rhythms could be detectable without pretreatment still remains unclear, the different results between the two experiments indicate that the pretreatment for synchronization is important to assess the existence of circadian rhythm.
By monitoring bioluminescence in real-time, we found that the decay pattern of promoter activity during the dark period differed between OsSPS1 and OsSPS11 (Figure 2). We also discovered that both OsSPS1 and OsSPS11 promoters are controlled by the circadian clock at least under constant light condition, but their phases and amplitudes were distinct (Figure 3). These results clearly show that OsSPS1 and OsSPS11 are regulated differently at the transcriptional level, implying multiplicity in the transcriptional control mechanisms of the five OsSPSs. Further studies at the both transcriptional and post-transcriptional levels are necessary to fully elucidate the complex regulatory mechanisms of the expressions of OsSPSs and characterize the roles of the five OsSPSs in sucrose metabolism and assimilate partitioning in rice plants.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to express our gratitude to Shiori Suzuki and Kiiko Takatsuto for their excellent technical assistance. Masahiro Ishiura was supported by SENTAN, JST. Masahiro Ishiura and Kiyoshi Onai were supported by grants from MEXT. Masaki Okamura is a recipient of a fellowship from the Japan Society for the Promotion of Science.
References
Argüello-Astorga, G. R., and Herrera-Esstrella, L. R. (1996). Ancestral multipartite units in light-responsive plant promoters have structural features correlating with specific phototransduction pathways. Plant Physiol. 112, 1151–1166.
Castleden, C. K., Aoki, N., Gillespie, V. J., MacRae, E. A., Quick, P., Buchner, P., et al. (2004). Evolution and function of the sucrose-phosphate synthase gene families in wheat and other grasses. Plant Physiol. 135, 1753–1764.
Chávez-Bárcenas, A. T., Valdez-Alarcón, J. J., Martínez-Trujillo, M., Chen, L., Xoconostle-Cázares, B., Lucas, W. J., et al. (2000). Tissue-specific and developmental pattern of expression of the rice sps1 gene. Plant Physiol. 124, 641–653.
Chiou, T. J., and Bush, D. R. (1998). Sucrose is a signal molecule in assimilate partitioning. Proc. Natl. Acad. Sci. U.S.A. 95, 4784–4788.
Chu, C. C., Wang, C. C., Sun, C. S., Hsu, K. C., Yin, K. C., Chu, C. Y., et al. (1975). Establishment of an efficient medium for anther culture of rice through comparative experiments on the nitrogen sources. Sci. Sinica 18, 659–668.
Higo, K., Ugawa, Y., Iwamoto, M., and Korenaga, T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res. 27, 297–300.
Hirose, T., Mizutani, R., Mitsui, T., and Terao, T. (2012). A chemically inducible gene expression system and its application to inducible gene suppression in rice. Plant Prod. Sci. 15, 73–78.
Ishimaru, K., Ono, K., and Kashiwagi, T. (2004). Identification of a new gene controlling plant height in rice using the candidate-gene approach. Planta 218, 388–395.
Jefferson, R. A., Kavanagh, T. A., and Bevan, M. W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907.
Lunn, J. E., and MacRae, E. (2003). New complexities in the synthesis of sucrose. Curr. Opin. Plant Biol. 6, 208–214.
Lutfiyya, L. L., Xu, N., D’Ordine, R. L., Morrell, J. A., Miller, P. W., and Duff, S. M. G. (2007). Phylogenic and expression analysis of sucrose phosphate synthase isozymes in plants. J. Plant Physiol. 164, 923–993.
Mita, S., Murano, N., Akaike, M., and Nakamura, K. (1997). Mutants of Arabidopsis thaliana with pleiotropic effects on the expression of the gene for β-amylase and on the accumulation of anthocyanin that are inducible by sugars. Plant J. 11, 841–851.
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15, 473–497.
Newman, T. C., Ohme-Takagi, M., Taylor, C. B., and Green, P. J. (1993). DST sequences, highly conserved among plant SAUR genes, target reporter transcripts for rapid decay in tobacco. Plant Cell 5, 701–714.
Okamoto, K., Onai, K., and Ishiura, M. (2005). RAP, an integrated program for monitoring bioluminescence and analyzing circadian bioluminescence rhythms in real time. Anal. Biochem. 340, 193–200.
Okamura, M., Aoki, N., Hirose, T., Yonekura, M., Ohto, C., and Ohsugi, R. (2011). Tissue specificity and diurnal change in gene expression of the sucrose phosphate synthase gene family in rice. Plant Sci. 181, 159–166.
Prestridge, D. S. (1991). SIGNAL SCAN: a computer program that scans DNA sequences for eukaryotic transcriptional elements. Comput. Appl. Biosci. 7, 203–206.
Ransom-Hodgkins, W. D., Vaughn, M. W., and Bush, D. R. (2003). Protein phosphorylation plays a key role in sucrose-mediated transcriptional regulation of a phloem-specific proton–sucrose symporter. Planta 217, 483–489.
Rolland, F., Baena-Gozalez, E., and Sheen, J. (2006). Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu. Rev. Plant Biol. 57, 675–709.
Sokolov, L. N., Déjardin, A., and Kleczkowski, L. A. (1998). Sugars and light/dark exposure trigger differential regulation of ADP-glucose pyrophosphorylase gene in Arabidopsis thaliana (thale cress). Biochem. J. 336, 681–687.
Terzaghi, W. B., and Cashmore, A. R. (1995). Light-regulated transcription. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46, 445–474.
Vaughn, M. W., Harrington, G. N., and Bush, D. R. (2002). Sucrose-mediated transcriptional regulation of sucrose symporter activity in the phloem. Proc. Natl. Acad. Sci. U.S.A. 99, 10876–10880.
Wang, J., Jiang, J., and Oard, J. H. (2000). Structure, expression and promoter activity of two polyubiquitin genes from rice. Plant Sci. 156, 201–211.
Winter, H., and Huber, S. C. (2000). Regulation of sucrose metabolism in higher plants: localization and regulation of activity of key enzymes. Crit. Rev. Plant Sci. 19, 31–67.
Keywords: sucrose phosphate synthase, promoter–luciferase reporter, transcriptional regulation, circadian clock, sugar sensing, rice
Citation: Yonekura M, Aoki N, Hirose T, Onai K, Ishiura M, Okamura M, Ohsugi R and Ohto C (2013) The promoter activities of sucrose phosphate synthase genes in rice, OsSPS1 and OsSPS11, are controlled by light and circadian clock, but not by sucrose. Front. Plant Sci. 4:31. doi: 10.3389/fpls.2013.00031
Received: 29 November 2012; Accepted: 09 February 2013;
Published online: 01 March 2013.
Edited by:
Hanjo A. Hellmann, Washington State University, USAReviewed by:
Woei-Jiun Guo, National Cheng Kung University, TaiwanAnke Reinders, University of Minnesota, USA
Copyright: © 2013 Yonekura, Aoki, Hirose, Onai, Ishiura, Okamura, Ohsugi and Ohto. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Naohiro Aoki, Laboratory of Crop Science, Graduate School of Agricultural and Life Sciences, The University of Tokyo, 1-1-1 Yayoi, Bunkyo-ku, Tokyo, 113-8657, Japan. e-mail:YWFva2luQG1haWwuZWNjLnUtdG9reW8uYWMuanA=




