- 1Department of Genetics, University of Pretoria, Pretoria, South Africa
- 2Forestry and Agricultural Biotechnology Institute, University of Pretoria, Pretoria, South Africa
- 3Department of Plant Science, University of Pretoria, Pretoria, South Africa
Two important role players in plant defence response are the phytohormones salicylic acid (SA) and jasmonic acid (JA); both of which have been well described in model species such as Arabidopsis thaliana. Several pathogenesis related (PR) genes have previously been used as indicators of the onset of SA and JA signaling in Arabidopsis. This information is lacking in tree genera such as Eucalyptus. The aim of this study was to characterize the transcriptional response of PR genes (EgrPR2, EgrPR3, EgrPR4, EgrPR5, and EgrLOX) identified in Eucalyptus grandis to SA and methyl jasmonate (MeJA) treatment as well as to qualify them as diagnostic for the two signaling pathways. Using the genome sequence of E. grandis, we identified candidate Eucalyptus orthologs EgrPR2, EgrPR3, EgrPR4, EgrPR5, and EgrLOX based on a co-phylogenetic approach. The expression of these genes was investigated after various doses of SA and MeJA (a derivative of JA) treatment as well as at various time points. The transcript levels of EgrPR2 were decreased in response to high concentrations of MeJA whereas the expression of EgrPR3 and EgrLOX declined as the concentrations of SA treatment increased, suggesting an antagonistic relationship between SA and MeJA. Our results support EgrPR2 as potentially diagnostic for SA and EgrPR3, EgrPR4, and EgrLOX as indicators of MeJA signaling. To further validate the diagnostic potential of the PR genes we challenged E. grandis clones with the fungal necrotrophic pathogen Chrysoporthe austroafricana. The tolerant clone showed high induction of EgrPR2 and decreased transcript abundance of EgrPR4. Pre-treatment of the susceptible genotype with 5 mM SA resulted in lesion lengths comparable to the tolerant genotype after artificial inoculation with C. austroafricana. Thus expression profiling of EgrPR2 and EgrPR4 genes could serve as a useful diagnostic approach to determine which of the two signaling pathways are activated against various pathogens in Eucalyptus.
Introduction
The defence mechanisms that are employed by plants to deter pathogens have been well-studied in various model organisms such as Arabidopsis thaliana. These model systems have created a foundation for understanding general host responses to pathogens. Following the plants perception of an invading pathogen, a plethora of defences responses are activated. Among these responses is the activation of various phytohormone signaling molecules including salicylic acid (SA), jasmonic acid (JA), ethylene (ET), abscisic acid (ABA), auxin, cytokinins (CK), gibberelins (GA), and brassinosteriods (BR). In particular, the phytohormones SA and JA have been extensively investigated in various pathosystems. These studies have shown that biotrophic pathogens are impeded by the activation of the SA pathway whereas necrotrophic pathogens are targeted by induction of JA and ET signaling pathways (Glazebrook, 2005). Each of these signaling cascades has been shown to involve the activation of certain signature defence genes, e.g., Pathogenesis Related (PR) genes, which can be representative of the induction of a pathway (Reymond and Farmer, 1998).
Stimulation of the SA pathway can be represented by an increase in the expression levels of PR1, PR2, and PR5 defence genes (Kunkel and Brooks, 2002; Delaure et al., 2008). Arabidopsis SA signaling mutants npr1, nim1, and sai1 as well as plants expressing the bacterial salicylate hydroxylase (nahG) are all impaired in their ability to induce expression of the PR1, PR2, and PR5 thereby indicating that these PR candidates can be used as a measure of SA signaling induction (Cao et al., 1994; Delaney et al., 1995; Shah et al., 1997). In the case of eds and pad mutants, there is a lack of SA signaling thereby allowing for increase in JA signaling due to the lack of antagonism by SA (Zhou et al., 1998; Gupta et al., 2000; Nawrath et al., 2002; Glazebrook et al., 2003). Transgenic plants over-expressing these SA signature defence genes have also been shown to result in increased resistance against pathogens such as Phytophthora parasitica and Alternaria alternata (Alexander et al., 1993; Jach et al., 1995). Induction of a derivative of JA, MeJA, can be represented in Arabidopsis by an increase in the expression levels of PR3, PR4, Vegetative Storage Protein (VSP), and Lipoxygenase (LOX). Over-expression of these proteins has also been shown to confer resistance to Phytophthora nicotianae and Rhizoctonia solani (Boter et al., 2004; Mishina and Zeier, 2007; Kusajima et al., 2010). Mutants of the JA signaling pathway in Arabidopsis, e.g., fad3/7/8, coi1, and jar1 have been shown to inhibit the expression of PR3, PR4, VSP, and LOX and thus increase the susceptibility of the mutant lines to numerous pathogens (Staswick et al., 1998; Vijayan et al., 1998; Norman-Setterblad et al., 2000). Additional JA mutants, mpk4 and ssi2, display increased levels of PR1, PR2, and PR5 whilst impaired in JA defence gene expression, thereby indicating that these mutants are involved in JA and SA antagonism (Petersen et al., 2000; Kachroo et al., 2001; Shah et al., 2001). Consequently PR3, PR4, and LOX defence genes can be used as indicators for the onset of JA signaling. One can thus refer to PR2 and PR5 as signature defence response genes for SA and PR3, PR4, and LOX as signature defence response genes for JA. Although there have been significant advances in the understanding of plant defences in model systems, signature defence genes associated with SA and JA in woody plants such as Eucalyptus has not been extensively explored.
Eucalyptus species and hybrid clones are commercially planted because of their valuable wood and fiber properties which have been exploited by the pulp and paper industry. Due to the importance and value associated with this genus of hardwood trees, the initiative to sequence the genome of Eucalyptus grandis was undertaken by the US Department of Energy (DOE—Joint Genome Institute) in 2008. Currently, the first annotated version of the genome, released in January 2011, is available through Phytozome v7.0 and consists of 4952 scaffolds including 11 linkage groups/chromosomal assemblies (Phytozome, 2010). This resource provides a useful platform for elucidating various physiological aspects of Eucalyptus, such as their responses to biotic and abiotic factors. Although Eucalyptus trees are generally disease tolerant, they can and do succumb to diseases caused by a wide range of pathogens (Wingfield et al., 2008). A stepping stone for improving our understanding of Eucalyptus responses would be to identify genes associated with the SA and JA signaling pathways in these trees. The first aim of this study was to identify Eucalyptus orthologs of signature defence genes specific for the SA (PR2 and PR5) and JA (PR3, PR4, and LOX) signaling pathways using sequence information from other plant species and the E. grandis genome sequence. Secondly we aimed to characterize the expression profiles of the putative orthologs using reverse transcriptase quantitative PCR (RT-qPCR). Transcript profiling that was conducted under mock induction of the signaling pathways revealed dose-dependent induction of the orthologous signature defence genes, as well as key time points for their expression. Furthermore, the orthologous genes were found to corroborate the antagonistic relationship observed between SA and JA in Arabidopsis. The ability of these putative signature defence genes to respond to fungal infection by Chrysoporthe austroafricana was examined in tolerant (TAG5) and susceptible (ZG14) E. grandis genotypes (Van Heerden et al., 2005). Expression profiling of these signature genes revealed the possible involvement of SA in defence against C. austroafricana.
Materials and Methods
Plant Material
Disease free E. grandis (Clone A, Mondi Tree Improvement Research) plantlets were propagated in vitro and following rooting the plantlets were transferred to Jiffy pots and grown at 25–28°C under long day (16 h) conditions under light intensity of 300–500 lum/sqf. Potted cuttings of E. grandis clonal genotypes, ZG14 and TAG5 (Mondi) with a stem diameter of 1 cm, were subsequently used for the infection trial with C. austroafricana and kept under the same conditions as stated above.
Phylogenetic Identification of Putative Orthologs for Signature Defence Genes Associated with SA and MeJA
The Arabidopsis thaliana amino acid sequences of the genes of interest were obtained from The Arabidopsis Information Resource (TAIR, version 10) (https://www.arabidopsis.org). A BLASTP similarity search was conducted against the predicted E. grandis proteome (first ab initio and homology-based annotation) using the amino acid sequence as a query. This analysis was performed in Phytozome v7.0 (www.phytozome.net) and predicted E. grandis transcripts with e-values <10−50 were downloaded. Putative Populus trichocarpa orthologs of the gene of interest were retrieved from NCBI and added to the analysis using the same BLAST parameters. Aligned sequences were imported into MEGA v5.01 (Tamura et al., 2011) for the construction of a neighbor joining (NJ) tree. Confidence in the clades was substantiated by a bootstrap value calculated after 10,000 permutations. For the maximum likelihood (ML) analysis, the aligned sequences were assessed using Prottest 3.0 (Abascal et al., 2005) and PhyML 3.0 (Guindon and Gascuel, 2003) was used to perform the ML analysis using the parameters of the best model obtained from the Prottest results. Confidence in the clades was substantiated by a bootstrap value calculated after 1000 permutations. Furthermore the expression pattern of the selected gene model across different tissues was assessed on the Eucalyptus Genome Integrative Explorer (EucGenIE, http://eucgenie.bi.up.ac.za, Mizrachi et al., 2010). Following the identification of putative orthologs in E. grandis based on the expression data and NJ and ML trees, primers were designed and verified in Phytozome v7.0 using a BLASTN similarity search against the E. grandis genome (Table 1). Eucalyptus orthologs for PR1a (AT2G14610), VSP1 (AT5G24780), and PDF1.2 (AT5G44420) could not be identified based on the phylogenetic approach and were thus not assessed further.
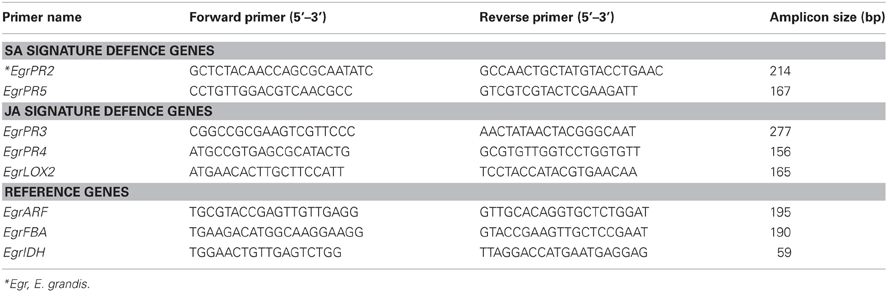
Table 1. Primer sequence of Eucalyptus target signature defence genes and reference genes assessed using RT-qPCR.
Dose Response of Putative Orthologous Signature Defence Genes for SA and MeJA Signature Defence Genes
SA and MeJA phytohormones were administered to E. grandis (clone A) plantlets by spraying the aerial portions with varying concentrations of the inducers until run-off. The following inducer concentrations were assessed: 25 μM, 50 μM, 100 μM, 250 μM, 500 μM, 1 mM, and 5 mM. Sodium salicylate (Riedel-de Haen, Seelze, Germany) was used to prepare the SA solutions (adjusted to pH 7.0 with NaOH solution) with the addition of 0.1% Tween® 20 (Sigma-Aldrich, Missouri, USA). MeJA (methyl jasmonate 95%, Sigma-Aldrich) was prepared with the addition of 0.1% ethanol (100%) as well as 0.1% Tween® 20 (Sigma-Aldrich). Control plants for SA treatment were sprayed with distilled water containing 0.1% Tween® 20. The control plants for the MeJA treatment were sprayed with distilled water containing 0.1% Tween® 20 and 0.1% ethanol. Aerial parts of the plantlets were harvested 24 h post-treatment (hpt). Three biological replicates of consisting of five plants each was harvested for the control and treated samples.
Investigation of the Expression Profiles of Putative E. grandis Orthologs Over a Time Course
Phytohormones, SA and MeJA, were administered to E. grandis (clone A) plantlets as described in the previous section. A single concentration selected from the dose response experiment for SA and MeJA was assessed at the following time points: 6, 12, 24, and 48 hpt. Controls were harvested at each individual time point as well as at time zero which refers to the time prior to the application of inducers. Three biological replicates consisting of five plants each was harvested for the control and treated samples at the different time points.
Infection Trial with Chrysoporthe Austroafricana
Ramets of two E. grandis clones, TAG5 and ZG14 trees, with an approximate stem diameter of 1 cm were inoculated with the fungus C. austroafricana CMW2113 as previously described (Roux et al., 2003). Lesion lengths were recorded and plant material (stem tissue, 1 cm above and below the lesion) was harvested at 48 h post-inoculation, the earliest time point at which confirmation of infection was observed, as well as 2 and 6 weeks post-inoculation (wpi). Three biological replicates consisting of three trees each was harvested for the control and inoculated samples. Re-isolation of the fungus was performed by excising a piece from the periphery of the lesion after 6 weeks and placing the block on 2% Malt Extract Agar (Merck, Gauteng, South Africa). Confirmation of infection by C. austroafricana was done by observing the culture morphology after 5 days.
RNA Extraction and First Strand cDNA Synthesis
Total RNA was extracted from the plant powder using a modified cetyl-trimethyl-ammonium-bromide (CTAB) extraction protocol (Zeng and Yang, 2002). Extracted samples were treated with RNase-free DNaseI enzyme (Qiagen Inc, Valencia, CA) and subsequently column purified using the RNeasy® MinElute Kit (Qiagen Inc) as per the manufacturer's instructions. Purified RNA (1 μg) was used as the template for reverse transcription using Improm II reverse transcriptase enzyme (Promega, Wisconsin, USA).
Reverse Transcriptase Quantitative PCR (RT-qPCR) Analysis
Reverse transcriptase quantitative PCR was performed according to the Minimum Information for Publication of Quantitative Real-Time PCR Experiments guidelines (MIQE) (Bustin et al., 2009). For each target, three biological replicates and three technical replicates per biological replicate was performed. The LightCycler® 480 SYBR Green I Master Mix (2× concentration) kit (Roche, Mannheim, Germany) was used to perform the RT-qPCR experiments on the LightCycler® 480 Real-Time PCR system (Roche Diagnostics, GmBH, Basa, Switzerland) according to the manufactures instructions. Reactions were set up in 11 μl volumes containing: 1 μl (1:10 diluted cDNA template), 5 μl LightCycler® 480 SYBR Green I Master Mix, 0.5 μM of each primer, and water to make up the total volume. For each primer pair, a negative no template control was included. Samples were normalized to a combination of the following reference genes: ADP ribosylation factor (EgrARF), Fructose bisphosphate aldolase (EgrFBA), and NADP-isocitrate dehydrogenase (EgrIDH, Boava et al., 2010). Relative quantification and normalization was performed using qBASEplus v1.0 (Hellemans et al., 2007). The datasets were tested for normality using the Shapiro–Wilk's test with the statistical software package Analyse-it® (Analyse-it Software, Ltd., Leeds, UK). The pairwise comparison Kruskal–Wallis test (p < 0.05) was applied to investigate significant differential expression unless otherwise stated.
Results
Phylogenetic Identification of Putative Orthologs for PR Genes Associated with the Salicylic Acid and Jasmonic Acid Signaling Pathways in Eucalyptus Grandis
Putative orthologs of defence genes that are known to be responsive to the SA and JA signaling pathways from Arabidopsis were identified in E. grandis using BLAST algorithms and phylogenetic analyses (Table 2). All of the genes, except for EgrPR2, had predicted transcripts that were congruent with the annotated sequence of E. grandis located on Phytozome v7.0. Further investigation into EgrPR2 revealed a region on scaffold 1:33791675_33792649 that had the highest similarity to the Arabidopsis candidate. Therefore an ab initio prediction of this region was performed using GeneMark (designated GM_Egrandis_V1_Scaffold1) and the result of this was included in the phylogenetic tree. The Arabidopsis PR2 gene formed a clade with GM_Egrandis_V1_Scaffold1 that was accompanied by a strong bootstrap statistical support in the ML phylogenetic tree (Results not shown) and the GeneMark predicted gene model therefore was selected as the putative ortholog (Table 2).
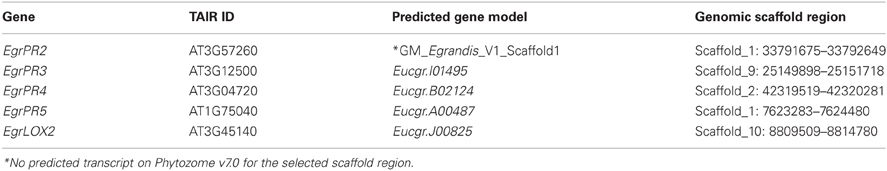
Table 2. Predicted gene models and corresponding genomic scaffold regions selected as putative orthologs for the SA and MeJA defence signature genes in E. grandis.
Expression Profiling of the Putative Orthologous EgrPR Genes at Various Concentrations of SA and JA Reveals Dose-Specific Induction
Following the identification of putative orthologs of signature defence genes of the SA and JA signaling pathways, we investigated the expression profile of the candidates under various doses of phytohormone application. Putative orthologous defence signature genes for the SA pathway, EgrPR2 and EgrPR5 both displayed increased transcript abundance at 25 μM SA (Figure 1A). Although both targets had increased transcript abundance at 25 μM, EgrPR2 had a much higher increase (16-fold compared to the control) at 5 mM and therefore this concentration was used for further experiments. Putative orthologs for the following candidates, EgrPR3, EgrPR4, and EgrLOX2 were profiled as signature defence genes of the JA pathway. EgrPR3 and EgrPR4 exhibited increased transcript abundance at a common concentration of 100 μM. EgrPR3 was also significantly increased at 25 μM and 5 mM but the fold change was lower than at 100 μM for EgrPR4 (Figure 1B). Although the expression of EgrLOX2 was significantly induced at 1 mM, it was decided to proceed with 100 μM for further experiments as both EgrPR3 and EgrPR4 exhibited significant differential expression at this concentration.
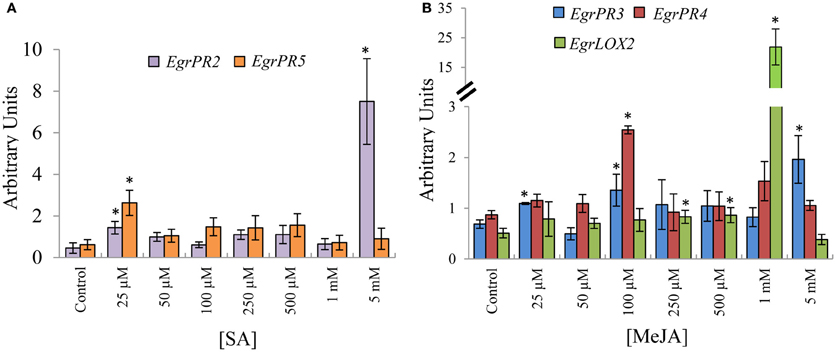
Figure 1. Relative transcript abundance of EgrPR signature defence genes under hormone treatment. (A) Putative SA signature defence genes following normalization with EgrARF and EgrFBA. (B) Putative MeJA signature defence genes following normalization with EgrARF and EgrIDH. The y-axis represents the relative transcript abundance ratios expressed in arbitrary units. The x-axis shows the respective concentration range that was applied to the aerial parts of E. grandis plants. Error bars show the standard error of the mean of the biological replicates (n = 3) sampled after 24 hpt. Significance, indicated by *, is relative to the control for each target and was calculated by the Kruskal–Wallis test (p < 0.05).
Expression of EgrPR Defence Genes Validate SA–JA Antagonism in E. grandis
To investigate the hypothesis that SA and JA display an antagonistic relationship, the candidates were assessed by profiling the SA defence signature genes in material induced with MeJA and vice versa. The antagonistic relationship between SA and JA was clearly validated to occur in E. grandis in tissue treated with the phytohormone at selected concentrations. EgrPR2 was suppressed at higher concentrations of MeJA relative to the control (Figure 2A). EgrPR3 expression was reduced at 100 μM, 250 μM, 1 mM, and 5 mM whereas EgrLOX2 was significantly lower at 100 μM, 1 mM, and 5 mM SA. EgrPR4 had higher abundance at 25 μM SA and was not repressed at any of the other concentrations (Figure 2B).
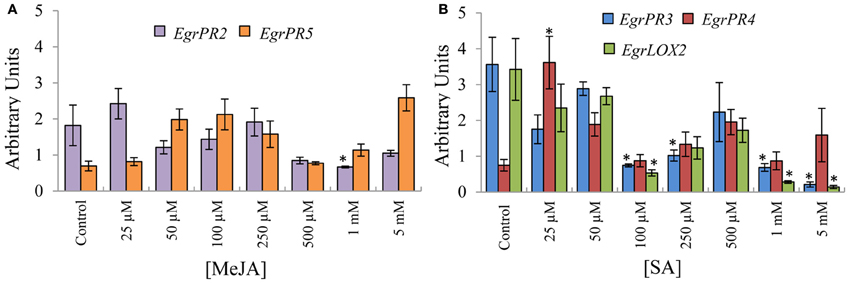
Figure 2. Relative transcript abundance of the putative orthologs for the EgrPR defence genes in tissue treated with the opposite phytohormone. The y-axis represents the relative expression ratios expressed in arbitrary units. Putative SA signature defence genes (A) were normalized with EguIDH and EgrARF whereas the putative MeJA signature defence genes (B) were normalized with EgrARF and EgrFBA. The x-axis represents the concentration range for the applied inducer. Error bars show the standard error of the mean of the biological replicates (n = 3). Significance, indicated by *, is relative to the control in each graph and was calculated by the Kruskal–Wallis test (p < 0.05).
Time-Dependent Expression of Putative EgrPR Genes Identifies Key Points of Induction
To investigate the expression profile of the suite of signature defence genes over a time course, 100 μM MeJA and 5 mM SA was applied to aerial portions of the E. grandis (clone A) tissue culture plants and the harvested material was profiled over various time points. The relative expression values for each time point was compared to the T = 0 control as well as the time specific control using the Kruskal–Wallis test (Note that the significance indicated on the graphs is only in relation to the time specific control). The T = 0 control was included in the experiment to indicate the basal level of gene expression prior to any treatment. Transcript abundance of the SA signature defence gene candidate, EgrPR2 was significantly increased at 12, 24, and 48 hpt with a drastic peak at 24 hpt followed by a decline at 48 hpt (Figure 3A). EgrPR5 displayed a gradual increase in expression from 6 to 48 hpt, with the expression of the target showing statistical significance all the time points except 12 hpt (Figure 3B). Signature defence genes for JA, EgrPR3, EgrPR4, and EgrLOX2 all displayed altered levels of expression at the various time points (Figures 3C–E). EgrPR4 transcript levels increased progressively from 6 to 48 hpt, with all the time points being statistically significant (Figure 3D). Notably the level at which EgrPR2 and EgrPR4 are expressed at 24 hpt was approximately the same level as was observed in the dose response experiment, thereby indicating reproducibility of the results.
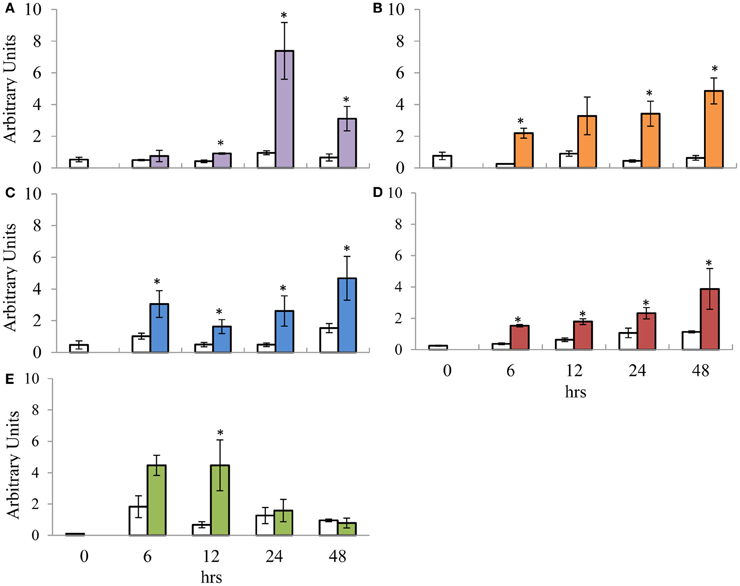
Figure 3. Relative transcript abundance of putative orthologs for SA and MeJA signature defence genes assessed during the time course trial. (A) EgrPR2; (B) EgrPR5; (C) EgrPR3; (D) EgrPR4; (E) EgrLOX2. The y-axis represents the relative expression ratios expressed in arbitrary units. The x-axis represents the time course (h) post-treatment with 5 mM SA (A and B) and 100 μM MeJA (C–E). Samples were normalized with EgrARF and EgrFBA. Error bars are show the standard error of the mean of the biological replicates (n = 3). White boxes represent the control samples whereas the colored boxes represent the treated samples. Significance between the control and treated samples is indicated by * at a specific time point and was calculated by the Kruskal–Wallis test (p < 0.05).
Expression Profiling of the Putative Orthologous Defence Genes During Infection by a Pathogen Qualifies the Potential of the Candidates to be Diagnostic of SA and MeJA and Implicates SA in Defence Against C. austroafricana
The potential of these defence signature genes to be used as diagnostic markers was investigated under pathogen stress by employing the E. grandis—C. austroafricana pathosystem. Using the Kruskal–Wallis statistic test a significant difference (p = 0.0295) was observed between the lesion lengths of TAG5 (4.8 ± 2.1 cm) and ZG14 (8.2 ± 3 cm) at 6 wpi whereas no significance was observed at 48 h and 2 wpi. In TAG5, the SA signature gene EgrPR2 showed significant differential expression at 2 and 6 wkpi (Figure 4A). In TAG5, the JA signature genes, EgrPR4 significantly decreased at 2 wpi and increased once again at 6 wpi. Despite significant up-regulation of EgrPR4 at 6 wpi in TAG5 compared to its control, the level to which it was induced was lower than EgrPR2 levels (Figure 4B). In ZG14, the level of expression of EgrPR2 was only significantly up-regulated at 6 wpi (Figure 4A) whereas the expression of EgrPR4 transcripts was found to be significantly up-regulated at 2 and 6 wpi (Figure 4B). The pre-treatment of the susceptible genotype of Eucalytpus with 5 mM SA, prior to manual inoculation with C. austroafricana, resulted in a smaller lesion lengths (5 ± 0.5 cm) compared to the untreated plants (7 ± 0.6 cm) at 5 wpi (One-Way ANOVA, p < 0.05). These lesion lengths were comparable to lesions found on the tolerant genotype (4.8 ± 0.4 cm).
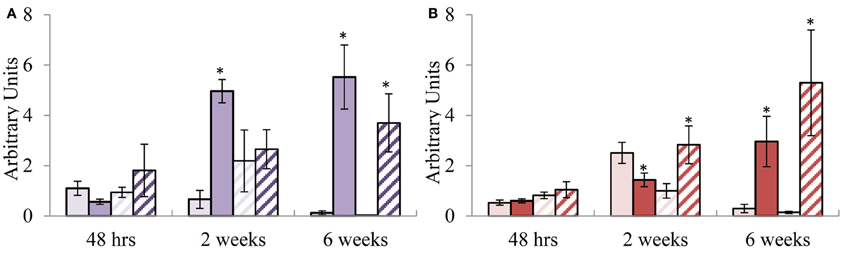
Figure 4. Relative transcript abundance of putative orthologs for EgrPR signature defence genes during infection with C. austroafricana. (A) EgrPR2; (B) EgrPR4. The y-axis represents the relative expression ratios expressed in arbitrary units. The x-axis represents the time points post-inoculation at which the samples were analyzed. Error bars show the standard error of the mean of the biological replicates (n = 3). Samples were normalized to EgrARF and EgrFBA. Light and dark solid boxes represent the TAG5 control and inoculated samples respectively whereas the light and dark striped boxes represent the ZG14 control and inoculated samples respectively. Significance, indicated by *, is relative to the control and was calculated by the Student's t-test (p < 0.05).
Discussion
PR genes have been shown to be indicators of the SA and MeJA signaling pathways and can be termed signatures of these pathways. This study aimed to identify orthologs of signature defence genes for the SA and MeJA signaling pathways from A. thaliana in E. grandis using sequence similarity and phylogenetic analysis. Phylogenetics provides a solid starting point for selecting candidates to investigate, however it does not provide definitive evidence that the selected gene is the true functional ortholog (Chen et al., 2007). Eucalyptus, Populus, and Arabidopsis share an ancient hexaploidization event and therefore on average there should be three genes in each species relative to the ancestor (Jaillon et al., 2007). These genes may have undergone various gene loss and/or duplication events which have changed this number for many genes and gene families thereby possibly creating multiple functional orthologs. The putative orthologous signature defence genes identified here provide suitable candidates for further investigation in complementation and functional studies to better understand the role of these genes in E. grandis.
Orthologs for EgrPR Signature Defence Genes Exhibit Dose-Specific Induction and Pathway Specificity for EgrPR2, EgrPR4, and EgrLOX
Based on the premise that the candidates identified through phylogeny were defence signature genes for SA and JA, we subsequently investigated the expression of these targets under various doses of phytohormone treatment. The concentrations used in this study were based on experiments conducted in A. thaliana and on the level of the phytohormone following a pathogen challenge in other model organisms (Rasmussen et al., 1991; Jung et al., 2007). The transcript abundance levels of the putative SA signature defence genes, EgrPR2 and EgrPR5 were increased (Figure 1A) by application of the inducer which was consistent with literature in Arabidopsis (Reymond and Farmer, 1998; Kunkel and Brooks, 2002; Delaure et al., 2008). Furthermore, when these genes were evaluated for their specificity to the SA pathway, it was clearly demonstrated that expression levels of EgrPR2 was suppressed by higher concentrations of MeJA (Figure 2A). These results suggest that in E. grandis EgrPR2 could serve as a diagnostic signature gene for the SA pathway. Expression of the MeJA defence signature genes, EgrPR3 and EgrLOX2 were significantly differentially regulated at varying concentrations of this phytohormone. These signature genes were additionally repressed at high concentrations of SA treatment confirming the suppressive effect of SA on MeJA responses. Transcripts of EgrPR4 were found to be up-regulated by the application of MeJA, but showed no differential expression under SA treatment other than at 25 μM. Nonetheless, EgrPR4 can be utilized as a defence signature for MeJA as expression levels of this gene were significantly altered upon application of that phytohormone. The data suggests that the known antagonistic relationship between MeJA and SA in Arabidopsis may also occur in Eucalyptus. All of the MeJA responsive defence signature genes profiled in this study were found to be diagnostic to the MeJA pathway in E. grandis and could serve as suitable markers for the pathway. Although SA and JA predominantly have an antagonistic relationship (Pieterse et al., 2009), there have been situations whereby these pathways act synergistically (Mur et al., 2006; Lazniewska et al., 2010). The outcome of the interaction between SA and JA seems to be largely dependent of the timing of activation and the concentration of the phytohormones (Mur et al., 2006).
Time Dependent Expression Profiles Suggest that MeJA and SA Signature Defence Genes in E. grandis are Differentially Regulated as Early as 6 hpt
To further elucidate the expression profiles of the Eucalyptus signature defence genes, we investigated the response of the candidates over time. The time at which a host's defences are activated has a crucial role in determining the outcome of a pathogen interaction. Susceptibility may not only be due to the lack of required artillery (e.g., defence genes), but also to the delayed activation of the genes required to curb the pathogen (Loon, 2009). Elucidating the time dependent expression profiles of the putative orthologous signature genes under mock induction of the signaling pathways would provide a glimpse into how the genes would respond under pathogen conditions. SA hormone levels in tobacco plants infected with tobacco mosaic virus (Malamy et al., 1990, 1996) parallels the expression profile observed for EgrPR2 in this study (Figure 3A) under application of SA in E. grandis. In contrast to EgrPR2, EgrPR5 was shown to gradually increase over the time points with a maximum expression level detected at 48 hpt (Figure 3B). This suggests that the signature defence genes identified in this study respond to mock induction of the signaling pathway in a similar manner as they would under pathogen incursion. Tobacco plants that have been treated with exogenous MeJA displayed time course patterns similar to that found in E. grandis for LOX and PR3 (Bell and Mullet, 1993). In E. grandis, EgrLOX2 transcript levels were significantly up-regulated as early as 12 hpt followed by a decline at 24 hpt (Figure 3E). This could indicate a possible role for EgrLOX2 in the early stages of defence activation in a host as this gene is involved in jasmonate biosynthesis. EgrPR3 displayed a similar profile with the level of transcripts increasing from 6 to 48 hpt in E. grandis (Figure 3C) compared to increasing levels from 8 to 24 h post-MeJA treatment in tobacco (Rickauer et al., 1997). A microarray time course study in which Arabidopsis plants were treated with MeJA revealed that EgrPR4 transcripts began to increase as early as 1 h then slowly declined by 24 h (Jung et al., 2007). Conversely in E. grandis, EgrPR4 increased from 6 hpt with the maximum expression level detected at 48 hpt (Figure 3D). Although the time points differ between the two organisms, the general trend of expression remains the same. The observed increase in the transcript levels of EgrPR3 and EgrPR4 over time could also be due to the role of these proteins in the host during defence. Both of these genes encode for products that target and alter the cell wall composition of a fungal pathogen and during infection an increase in expression would be beneficial in preventing the spread of the pathogen (Selitrennikoff, 2001).
Pathogenicity Experiments Conducted with C. austroafricana Establishes the Diagnostic Potential of the EgrPR Signature Defence Genes and Elucidates the Importance of SA in Defence Against this Pathogen
In Arabidopsis, the involvement of a specific signaling pathway during an interaction with a pathogen can be elucidated by the diagnostic ability of the assigned signature genes. This study examined the diagnostic potential of the putative orthologous signature genes for SA and MeJA found in Eucalyptus upon infection with C. austroafricana. It was found that at 2 wpi there was no substantial difference in lesion length between TAG5 and ZG14 whilst at 6 wpi there was a significant lesion difference, suggesting that during the initial 2 weeks following infection the tolerant host was able to initiate a certain response to curb the spread of the disease. Interestingly, the signature defence gene expression profiles that were observed in the two hosts suggest a probable role of SA in the tolerance mechanism of TAG5. In the incompatible interaction (TAG5 and C. austroafricana), at 2 and 6 wpi, EgrPR2 transcripts were considerably up-regulated compared to the control, whereas up-regulation only occurred at 6 wpi in the compatible interaction (ZG14 and C. austroafricana) and to a lower lever (Figure 4A). In addition, the level of MeJA signaling at 2 wpi was lower in the incompatible interaction compared to the compatible interaction as indicated by the expression levels of EgrPR4 (Figure 4B). The antagonistic relationship between SA and MeJA evidently occurs within these hosts at this time point and could possibly have a key role in determining the outcome of the interaction with C. austroafricana. From other plant species, PR2 is known to encode for the β-1, 3-glucanase enzyme which facilitates the enzymatic degradation of the glucan component of fungal cell walls (Theis and Stah, 2004). In TAG5, the elevated level of EgrPR2 could contribute to confining the spread of C. austroafricana by hydrolyzing the β-1, 3-glucan component of the cell wall. In the review by Selitrennikoff (2001), it's hypothesized that this particular glucan component maybe abundant in the hypal apex of a growing fungus and degradation of the β-1, 3-glucan may lead to a loss in rigidity of the cell wall thereby resulting in cell lysis and eventual cell death. EgrPR2 was significantly up-regulated at the later time points (2 and 6 wpi) but not at the early time point of 48 hpi (Figure 4A) suggesting that the lack of an early response could be a partial reason as to why TAG5 is tolerant but not fully resistant against C. austroafricana. Based on the premise that SA may facilitate tolerance, ZG14 plants were sprayed with 5 mM SA to determine if this hormone would increase the tolerance of this host. A significant reduction in the lesion lengths of ZG14 treated with SA was observed and the lesions were of similar length to that seen in the tolerant TAG5 plants. Induction of systemic resistance in E. urophylla upon application of 5 mM SA has been previously documented (Ran et al., 2005). EgrPR4 encodes a hevein-like protein which acts like a chitin binding protein by targeting the β -chitin component of the cell wall. These proteins migrate to the cell walls of an invading fungus and disrupt the formation of the septa and hyphal tips (Selitrennikoff, 2001; Theis and Stah, 2004). In ZG14, EgrPR4 was elevated at 2 wpi however the host was still susceptible to C. austroafricana. A possible explanation for this is that the level to which this gene is expressed was not high enough to curb the pathogen. Timing of defence gene expression is crucial in a pathogen interaction and the lack of significant EgrPR4 expression at 48 h in TAG5 or in ZG14, may contribute to the ability of C. austroafricana to proliferate within these hosts during the initial 2 weeks of infection.
Our results suggest that EgrPR2 and EgrPR4 were diagnostic of SA and MeJA signaling pathways respectively against C. austroafricana as SA was recognized as playing a role in enhancing tolerance against the pathogen in Eucalyptus. It is possible that other signaling pathways may have a role in contributing to resistance in this interaction. The involvement of SA in facilitating a defence response to a necrotrophic pathogen is in contrast to the published literature from Arabidopsis which implicates the involvement of the MeJA pathway (Glazebrook, 2005). In spite of this, there have been studies that have shown that SA could also assist in impeding necrotrophic pathogens (Ferrari et al., 2003; Azaiez et al., 2009). It may also be possible that in tree species the roles of SA and MeJA in pathogen defence could differ from what is known in Arabidopsis.
This study provides a first step toward understanding hormone mediated defence responses of Eucalyptus trees. It is envisaged that expression profiling of the diagnostic markers, EgrPR2 and EgrPR4, can be adopted as a tool to determine which of the two major defence pathways are active against different pathogens in Eucalyptus in future.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Mondi, Sappi South Africa and the National Research Foundation (NRF) for funding. Opinions expressed and conclusions arrived at are those of the authors and are not necessarily to be attributed to the NRF.
References
Abascal, F., Zardoya, R., and Posada, D. (2005). ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21, 2104–2105.
Alexander, D., Goodman, R. M., Gut-Rella, M., Glascock, C., Weymann, K., Friedrich, L., et al. (1993). Increased tolerance to two oomycete pathogens in transgenic tobacco expressing pathogenesis-related protein 1a. Proc. Natl. Acad. Sci. U.S.A. 90, 7327–7331.
Azaiez, A., Boyle, B., Levée, V., and Séguin, A. (2009). Transcriptome profiling in hybrid poplar following interactions with Melampsora rust fungi. Mol. Plant Microbe Interact. 22, 190–200.
Bell, E., and Mullet, J. E. (1993). Characterization of an Arabidopsis lipoxygenase gene responsive to methyl jasmonate and wounding. Plant Physiol. 103, 1133–1137.
Boava, L. P., Laia, M. L., Jacob, T. R., Dabbas, K. M., Goncalves, J. F., Ferro, J. A., et al. (2010). Selection of endogenous genes for gene expression studies in Eucalyptus under biotic (Puccinia psidii) and abiotic (acibenzolar-S-methyl) stresses using RT-qPCR. BMC Res. Notes 3:43. doi: 10.1186/1756-0500-3-43
Boter, M., Ruız-Rivero, O., Abdeen, A., and Prat, S. (2004). Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 18, 1577–1591.
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622.
Cao, H., Bowling, S. A., Gordon, A. S., and Dong, X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6, 1583–1592.
Chen, F., Mackey, A. J., Vermunt, J. K., and Roos, D. S. (2007). Assessing performance of orthology detection strategies applied to eukaryotic genomes. PLoS ONE 2:e383. doi: 10.1371/journal.pone.0000383
Delaney, T. P., Friedrich, L., and Ryals, J. A. (1995). Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. U.S.A. 92, 6602–6606.
Delaure, S. L., Hemelrijck, W. V., Bolle, M. F. C. D., Cammue, B. P. A., and De Coninck, B. M. A. (2008). Building up plant defenses by breaking down proteins. Plant Sci. 174, 375–385.
Ferrari, S., Plotnikova, J. M., Lorenzo, G. D., and Ausubel, F. M. (2003). Arabidopsis local resistance to Botrytis cinerea involves salicylic acid and camalexin and requires EDS4 and PAD2 but not SID2, EDS5 or PAD4. Plant J. 35, 193–205.
Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227.
Glazebrook, J., Chen, W., Estes, B., Chang, H.-S., Nawrath, C., Metraux, J.-P., et al. (2003). Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression profiling. Plant J. 34, 217–228.
Guindon, S., and Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704.
Gupta, V., Willits, M. G., and Glazebrook, J. (2000). Arabidopsis thaliana EDS4 contributes to Salicylic Acid (SA)-dependent expression of defense responses: evidence for inhibition of jasmonic acid signaling by SA. Mol. Plant Microbe Interact. 13, 503–511.
Hellemans, J., Mortier, G., Paepe, A. D., Speleman, F., and Vandesompele, J. (2007). qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8:R19. doi: 10.1186/gb-2007-8-2-r19
Jach, G., Gornhardt, B., Mundy, J., Logemann, J., Pinsdorf, E., Leah, R., et al. (1995). Enhanced quantitative resistance against fungal disease by combinatorial expression of different barley antifungal proteins in transgenic tobacco. Plant J. 8, 97–109.
Jaillon, O., Aury, J.-M., Noel, B., Policriti, A., Clepet, C., Casagrande, A., et al. (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463–468.
Jung, C., Lyou, S. H., Yeu, S., Kim, M. A., Rhee, S., Kim, M., et al. (2007). Microarray-based screening of jasmonate-responsive genes in Arabidopsis thaliana. Plant Cell Rep. 26, 1053–1063.
Kachroo, P., Shanklin, J., Shah, J., Whittle, E. J., and Klessig, D. F. (2001). A fatty acid desaturase modulates the activation of defense signaling pathways in plants. Proc. Natl. Acad. Sci. U.S.A. 98, 9448–9453.
Kunkel, B. N., and Brooks, D. M. (2002). Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 5, 325–331.
Kusajima, M., Yasuda, M., Kawashima, A., Nojiri, H., Yamane, H., Nakajima, M., et al. (2010). Suppressive effect of abscisic acid on systemic acquired resistance in tobacco plants. J. Gen. Plant Pathol. 76, 161–167.
Lazniewska, J., Macioszek, V. K., Lawrence, C. B., and Kononowicz, A. K. (2010). Fight to the death: Arabidopsis thaliana defense response to fungal necrotrophic pathogens. Acta Physiologiae Plantarum 32, 1–10.
Loon, L. C. V. (2009). Advances in Botanical Research: Plant Innate Immunity. San Diego, CA: Academic Press.
Malamy, J., Carr, J. P., Klessig, D. F., and Raskin, I. (1990). Salicylic acid: a likely endogenous signal in the resistance response of tobacco to viral infection. Science 250, 1002–1004.
Malamy, J., Sanchez-Casas, P., Hennig, J., Guo, A., and Klessig, D. F. (1996). Dissection of the Salicylic acid signalling pathway in tobacco. Mol. Plant Microbe Interact. 9, 474–482.
Mishina, T. E., and Zeier, J. (2007). Pathogen-associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. Plant J. 50, 500–513.
Mizrachi, E., Hefer, C. A., Ranik, M., Joubert, F., and Myburg, A. A. (2010). De novo assembled expressed gene catalog of a fast-growing Eucalyptus tree produced by Illumina mRNA-Seq. BMC Genomics 11:681. doi: 10.1186/1471-2164-11-681
Mur, L. A. J., Kenton, P., Atzorn, R., Miersch, O., and Wasternack, C. (2006). The outcomes of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol. 140, 249–262.
Nawrath, C., Heck, S., Parinthawong, N., and Métraux, J.-P. (2002). EDS5, an essential component of salicylic acid–dependent signaling for disease resistance in Arabidopsis, is a nember of the MATE transporter family. Plant Cell 14, 275–286.
Norman-Setterblad, C., Vidal, S., and Palva, E. T. (2000). Interacting signal pathways control defense gene expression in Arabidopsis in response to cell wall-degrading enzymes from Erwinia carotovora. Mol. Plant Microbe Interact. 13, 430–438.
Petersen, M., Brodersen, P., Naested, H., Andreasson, E., Lindhart, U., Johansen, B., et al. (2000). Arabidopsis MAP Kinase 4 negatively regulates systemic acquired resistance. Cell 103, 1111–1120.
Phytozome. (2010). Eucalyptus Grandis Genome Project 2010. Accessed March 4, 2011. Available online at: http://www.phytozome.net/eucalyptus
Pieterse, C. M. J., Leon-Reyes, A., Ent, S. V. D., and Wees, S. C. M. V. (2009). Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5, 308–316.
Ran, L. X., Li, Z. N., Wu, G. J., Loon, L. C. V., and Bakker, P. A. H. M. (2005). Induction of systemic resistance against bacterial wilt in Eucalyptus urophylla by fluorescent Pseudomonas spp. Eur. J. Plant Pathol. 113, 59–70.
Rasmussen, J. B., Hammerschmidt, R., and Zook, M. N. (1991). Systemic induction of salicylic acid accumulation in cucumber after inoculation with Pseudomonas syringae pv syringae. Plant Physiol. 97, 1342–1347.
Reymond, P., and Farmer, E. E. (1998). Jasmonate and salicylate as global signals for defense gene expression Curr. Opin. Plant Biol. 1, 404–411.
Rickauer, M., Brodschelm, W., Bottin, A., Veronesi, C., Grimal, H., and Esquerre-Tugaye, M. T. (1997). The jasmonate pathway is involved differentially in the regulation of different defence responses in tobacco cells. Planta 202, 155–162.
Roux, J., Myburg, H., Wingfield, B. D., and Wingfield, M. J. (2003). Biological and phylogenetic analyses suggest that two Cryphonectria spp.cause cankers of Eucalyptus in Africa. Plant Dis. 87, 1329–1332.
Shah, J., Kachroo, P., Nandi, A., and Klessig, D. F. (2001). A recessive mutation in the Arabidopsis SSI2 gene confers SA- and NPR1-independent expression of PR genes and resistance against bacterial and oomycete pathogens. Plant J. 25, 563–574.
Shah, J., Tsui, F., and Klessig, D. F. (1997). Characterization of a salicylic acid–insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 Gene. Mol. Plant Microbe Interact. 10, 69–78.
Staswick, P. E., Yuen, G. Y., and Lehman, C. C. (1998). Jasmonate signaling mutants of Arabidopsis are susceptible to the soil fungus Pythium irregulare. Plant J. 15, 747–754.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739.
Theis, T., and Stah, U. (2004). Antifungal proteins: targets, mechanisms and prospective applications. Cell. Mol. Life Sci. 61, 437–455.
Van Heerden, S. W., Amerson, H. V., Preisig, O., Wingfield, B. D., and Wingfield, M. J. (2005). Relative pathogenicity of Cryphonectria cubensis on Eucalyptus clones differing in their resistance to C. cubensis. Plant Dis. 89, 659–662.
Vijayan, P., Shockey, J., Levesque, C. A., Cook, R. J., and Browse, J. (1998). A role for jasmonate in pathogen defense of Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 95, 7209–7214.
Wingfield, M. J., Slippers, B., Hurley, B. P., Coutinho, T. A., Wingfield, B. D., and Roux, J. (2008). Eucalypt pests and diseases: growing threats to plantation productivity. South. For. 70, 139–144.
Zeng, Y., and Yang, T. (2002). RNA isolation from highly viscous samples rich in polyphenols and polysaccharides. Plant Mol. Biol. Rep. 20, 417a–417e.
Keywords: Eucalyptus, salicylic acid, methyl jasmonate, PR genes, defence, PR2, PR4
Citation: Naidoo R, Ferreira L, Berger DK, Myburg AA and Naidoo S (2013) The identification and differential expression of Eucalyptus grandis pathogenesis-related genes in response to salicylic acid and methyl jasmonate. Front. Plant Sci. 4:43. doi: 10.3389/fpls.2013.00043
Received: 14 December 2012; Accepted: 19 February 2013;
Published online: 06 March 2013.
Edited by:
Sergi Munné-Bosch, University of Barcelona, SpainReviewed by:
Jin-Long Qiu, Chinese Academy of Sciences, ChinaMaria Elizabeth Abreu, Universidade Federal do Rio de Janeiro, Brazil
Copyright © 2013 Naidoo, Ferreira, Berger, Myburg and Naidoo. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Sanushka Naidoo, Department of Genetics, University of Pretoria, Private bag X20, Pretoria 0028, South Africa. e-mail:c2FudXNoa2EubmFpZG9vQGZhYmkudXAuYWMuemE=



