- 1Department of Plant Biology, Michigan State University, East Lansing, MI, USA
- 2DOE Great Lakes Bioenergy Research Center, Michigan State University, East Lansing, MI, USA
- 3Department of Biochemistry and Molecular Biology, Michigan State University, East Lansing, MI, USA
The exact biochemical steps of xylan backbone synthesis remain elusive. In Arabidopsis, three non-redundant genes from two glycosyltransferase (GT) families, IRX9 and IRX14 from GT43 and IRX10 from GT47, are candidates for forming the xylan backbone. In other plants, evidence exists that different tissues express these three genes at widely different levels, which suggests that diversity in the makeup of the xylan synthase complex exists. Recently we have profiled the transcripts present in the developing mucilaginous tissue of psyllium (Plantago ovata Forsk). This tissue was found to have high expression levels of an IRX10 homolog, but very low levels of the two GT43 family members. This contrasts with recent wheat endosperm tissue profiling that found a relatively high abundance of the GT43 family members. We have performed an in-depth analysis of all GTs genes expressed in four developmental stages of the psyllium mucilagenous layer and in a single stage of the psyllium stem using RNA-Seq. This analysis revealed several IRX10 homologs, an expansion in GT61 (homologs of At3g18170/At3g18180), and several GTs from other GT families that are highly abundant and specifically expressed in the mucilaginous tissue. Our current hypothesis is that the four IRX10 genes present in the mucilagenous tissues have evolved to function without the GT43 genes. These four genes represent some of the most divergent IRX10 genes identified to date. Conversely, those present in the psyllium stem are very similar to those in other eudicots. This suggests these genes are under selective pressure, likely due to the synthesis of the various xylan structures present in mucilage that has a different biochemical role than that present in secondary walls. The numerous GT61 family members also show a wide sequence diversity and may be responsible for the larger number of side chain structures present in the psyllium mucilage.
Introduction
A number of plants have seeds that produce mucilage that aids in hydration, dispersal and germination. The composition of mucilage varies considerably across species. As examples, Arabidopsis thaliana uses primarily pectin (Goto, 1985; Western et al., 2000) while flax utilizes a mixture of both pectin and arabinoxylan (Naran et al., 2008). Psyllium (Plantago ovata Forsk) mucilage is composed predominantly of complex heteroxylan (Edwards et al., 2003; Fischer et al., 2004; Guo et al., 2008) and, as such, presents an opportunity to discover genes involved in xylan production. The mucilage of psyllium is produced in a single cell tissue layer that is relatively easy to dissect from the developing seed. The mucilage produced by this tissue forms a large part of the tissue's dry mass and the ratio of xylan to cellulose is much higher than that found in secondary cell walls and thus represents an opportunity to distinguish genes involved in xylan formation from those involved in secondary cell wall biosynthesis. We have investigated this tissue, using transcriptional profiling, to determine which genes are highly expressed during mucilage formation. Using this approach we identified a previously uncharacterized component of the xylan synthases, IRX15 (Jensen et al., 2011).
Currently, a number of genes that affect xylan biosynthesis have been identified. In a few cases, the biochemical activities of these genes have been demonstrated; specifically, the addition of glucuronic acid side chain (GUX1, GUX2, GUX4; Lee et al., 2012a; Rennie et al., 2012) and the o-methylation of the glucuronic acid (GXMT1; Lee et al., 2012b; Urbanowicz et al., 2012). Three complementation groups of putative glycosyltransferase (GT) genes have been implicated in the synthesis of the β-(1,4)-linked xylose backbone of xylan. Each of these three complementation groups consist of two genes, one gene with secondary cell wall expression pattern, named IREGULAR XYLEM (IRX) 9, IRX10 and IRX14, respectively, and one gene with much lower expression level and a more general expression pattern, named as their redundant homolog but with the suffix “LIKE” abbreviated L, e.g., IRX9-L. The four genes IRX9(-L) and IRX14(-L) are members of the GT family 43 (GT43) while the IRX10(-L)genes are members of the GT47 family (Brown et al., 2005, 2009; Persson et al., 2005; Peña et al., 2007; Wu et al., 2009, 2010; Lee et al., 2010). Our finding that IRX15, and its redundant homolog IRX15-L, also affects xylan chain length indicates further complexity of the xylan synthase (Brown et al., 2011; Jensen et al., 2011). Recently, a study performed in wheat endosperm has shown that, in contrast to Arabidopsis and psyllium, IRX15 is not expressed at high levels in the endosperm tissue, but homologs of IRX9, IRX14 and IRX10 are highly expressed (Pellny et al., 2012). This result indicates that variation is possible in the makeup of the xylan synthase. It would appear that the synthesis of xylan in wheat endosperm does not require IRX15. Our previous results demonstrate that the xylan synthase responsible for complex heteroxylan biosynthesis in psyllium does not require IRX9 or IRX14, as these were found to be expressed at very low levels in this tissue. A homolog of IRX10 was, on the other hand, found to be abundantly expressed (Jensen et al., 2011). These indications of diversity in the xylan synthase seem to suggest that the one constant in xylan synthesis is IRX10. If IRX10 is primarily responsible for the synthesis of the xylan backbone it would be expected that the xylan synthase from the psyllium mucilaginous layer (ML) would express an IRX10 gene with different properties than found in tissues containing both GT47 and GT43 family members. Additionally, one would expect to find GTs responsible for the larger variety of xylan side chains found in the psyllium mucilage. We present in this study an examination of the IRX10 genes present in the ML, as well as stem tissue, and we examine other highly abundant transcripts in the ML encoding proteins likely involved in xylan biosynthesis.
Materials and Methods
Plant Growth, Cell Wall Analysis, and RNA-SEQ
Psyllium (Indian, Plantago ovata, Sand Mountain Herbs, AL, USA) and Arabidopsis (Col-0) plants were grown as previously described (Jensen et al., 2011).
Toluidine blue staining of psyllium inflorescence, stem top half and stem bottom half was performed on free-hand sections of fresh material. Sequential extraction of cell wall material from leaves, inflorescence, stem top half and stem bottom half and subsequent neutral monosaccharide analysis of the 1 M KOH fraction was performed as described in Jensen et al. (2011).
Whole stems from 3-month-old psyllium plants were used for the preparation of total RNA extraction using Trizol reagent (15596-026; Invitrogen, http://www.invitrogen.com/). Of the crude RNA preparation 20 μg was subjected to additional purification using the RNeasy Micro Kit (74004; Qiagen, http://www.qiagen.com/) with DNase treatment (79254; Qiagen, http://www.qiagen.com/) as per manufacture's protocol. The subsequent cDNA library and high-throughput cDNA sequencing (RNA-Seq) was performed as described in Jensen et al. (2011). The RNA-Seq datasets were deposited at NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) with the following accessions: 6 DPA, SRX096079; 8 DPA, SRX027102; 10 DPA, SRX096080; 12 DPA, SRX027103; stems 10 weeks, SRX027101.
Assembly of 454 ESTs and Database Construction
The five datasets of 454 ESTs were assembled collectively using the CLC Genomics Workbench version 4.7.2 (CLC bio, Cambridge, MA, USA) and the De-novo assembly algorithm (Parameters: Similarity 0.8; Length fraction 0.5; Insertion cost 3; Deletion cost 3; Mismatch cost 2). Unique counts were generated by aligning ESTs to the assembled contigs using the RNA-Seq Analysis algorithm for non-annotated sequences. (Parameters: Similarity 0.8; Length fraction 0.9). The assembled sequence contigs were annotated using TBLASTN (Altschul et al., 1997) against the TAIR 9 annotation of the Arabidopsis genome. The annotations were subsequently expanded with the following information: Arabidopsis gene family assignments from the Carbohydrate Active enZyme (CAZy) database (Cantarel et al., 2009; http://www.cazy.org; update 2012-05-31) were labeled e.g., “Glycosyltransferase Family 47″ or “Glycoside Hydrolase Family 19″; Arabidopsis proteins not included in CAZy but recently proposed to also encode GTs (Nikolovski et al., 2012) were labeled GT and the respective family name, eg. “Glycosyltransferase Family GT14R”; members of the nucleotide sugar transporter/triose phosphate translocators family in Arabidopsis (Ward, 2001) were added the label “NST/TPT family”; and transcription factors in the Database of Arabidopsis Transcription Factors (DATF; Guo et al., 2005; http://datf.cbi.pku.edu.cn/) were added the label “Transcription Factor”; genes co-expressed with IRX10 (r > 0.5; 184 genes) and with secondary cell wall CESA4, CESA7 and CESA8 (r > 0.5; 227 genes) (GeneCAT database; http://genecat.mpg.de/cgi-bin/Ainitiator.py; Mutwil et al., 2008) were added the label “AtIRX10 Co-expression” and “At SCW CESA Co-expression,” respectively. Contig name, DNA sequence, annotation and expression information were stored in an Oracle relational database that is located at http://glbrc.bch.msu.edu/psyllium. The database can be queried using keywords that search contig annotation, including the added annotations mentioned above, while the contig sequence information can be analyzed using BLAST (Altschul et al., 1997) and query sequences, either DNA or protein, provided by the user. Information about each contig, such as DNA sequence, EST coverage and BLAST report against TAIR9, can be retrieved by clicking on the contig ID numbers and the “T” icon associated with each contig. Access to the individual contig data facilitates manual analysis for artifact assembly, such as ESTs from different genes grouped into the same contig or the identification of multiple contigs originating from the same transcript. Finally, a micro array viewer based on a gene expression map of Arabidopsis development (Schmid et al., 2005) is provided for each contig by clicking on the associated AGI.
Identifying Genes of Interest
Because of sequencing errors, ESTs from one gene were in some cases assembled into two or more individual contigs. In the cases of PoIRX10_1 to _4 and PoGT61_1 to _7 the complete cDNA sequences were determined by cDNA cloning and Sanger sequencing. Four independent clones were sequenced in each case. PoIRX10_2 is not full length. The verified cDNA sequences were deposited at NCBI GenBank (http://www.ncbi.nlm.nih.gov/genbank) with the following accessions KC832826 to KC832829 (PoIRX10_1 to _4) and KC894060 to KC894066 (PoGT61_1 to _7).
Phylogenetic Analysis
Phylogenetic trees were calculated by the use of MEGA 5.05 (Tamura et al., 2011), using the built-in ClustalW (Larkin et al., 2007) sequence alignment program, the Maximum Likelihood algorithm (Nei and Kumar, 2000), using the Poisson substitution model and bootstrapping based on 500 trees (Felsenstein, 1985). The phylogenetic analysis of GT61 members was based on protein sequences only. The phylogenetic analysis of GT47 members was based on cDNA sequences. First cDNA sequences were loaded in the MEGA program, then translated into protein sequences and aligned using the built-in ClustalW function (File S2; Larkin et al., 2007). The resulting codon based cDNA alignment was then used for phylogenetic analysis. Codon positions included were first, second, third, and non-coding.
Protein sequences were obtained from the Phytozome v8.0 database (Goodstein et al., 2012; http://www.phytozome.net/). For poplar (Populus trichocarpa, annotation v3.0) the genes Potri015G107200 and Potri015G116700 were not included in the analysis as these represent partial sequences. GT family 61 proteins from Arabidopsis and rice (Oryza sativa Japonica Group) were obtained from the CAZy database. In Brachypodium distachyon, all proteins annotated as GT family 61 proteins based on the recent genome annotation (International Brachypodium Initiative, 2010) were included.
Determining Degree of Cell Wall Acetylation
Ground plant material of Arabidopsis lower stem, dissected mucilaginous layers (8–10 DPA), psyllium husk (Now Foods, www.nowfoods.com), and whole psyllium seeds were washed three times with 70% ethanol, three times with 1:1 methanol-chloroform, and two times with acetone to obtain alcohol insoluble residue (AIR). Acetyl groups from the alcohol insoluble residue were then released by alkaline hydrolysis by treating with 1 M KOH at room temperature for 5 min and then neutralized with an equal amount of HCl. The amount of freed acetic acid in solution was then subsequently determined using the K-ACETRM acetic acid quantification kit from Megazyme (www.megazyme.com).
Results and Discussion
Transcript Profiling of Psyllium Stem Tissue, Assembly of ESTs and Assignment of Functional Annotation
In order to compare xylan biosynthesis in the ML with xylan formation in other tissues of psyllium we first determined the neutral monosaccharide composition for different aerial parts of the plant (Figure 1A). The psyllium stem and inflorescence yielded the highest levels of xylose, which were at levels comparable to Arabidopsis stem. Given glucose levels are low in these tissues, the high levels of xylose likely result from xylan as opposed to xyloglucan. Anatomical investigation by hand sectioning and toluidine blue staining verified the presence of secondary cell wall formation in both inflorescence and stem (Figures 1B–D). Subsequently, a series of sequential extractions, using CDTA, Na2CO3 and KOH, were performed and the xylan enriched 1 M KOH fraction was subjected to neutral monosaccharide composition analysis (Figure 1E). Only minor differences were found in the monosaccharide profiles between Arabidopsis lower stem, psyllium inflorescence and psyllium stem samples. Based on these analyses we chose to profile the transcriptome of psyllium stem.
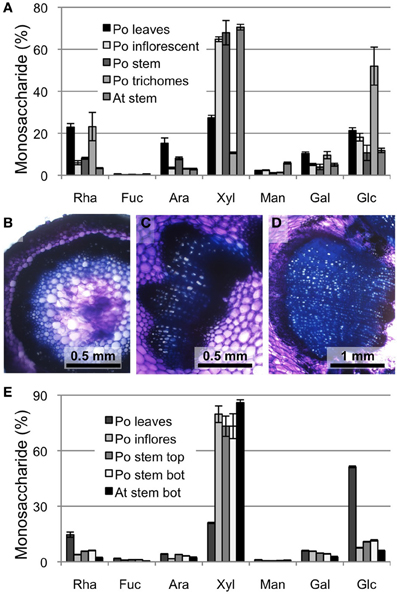
Figure 1. Cell wall analysis of psyllium aerial tissues. (A) Neutral monosaccharide composition of total cell walls from various tissues from psyllium (Po) leaves, inflorescent, stem and trichomes and Arabidopsis (At) stem. (B–D) Toluidine blue staining of free-hand sections of psyllium inflorescence (B), stem top (C) and stem bottom (D). (E) Neutral monosaccharide composition of 1 M KOH extractions of various tissues from psyllium leaves, inflorescent, stem top and stem bottom, and Arabidopsis stems bottom. The selected tissues were subjected to sequential extractions with CDTA, Na2CO3 and 1 M KOH.
The sequence data from the psyllium stem RNA-Seq experiment was added to four previous RNA-Seq datasets from psyllium ML (Jensen et al., 2011). This dataset of approximately 1 million ESTs was assembled into transcript models (contigs; Table S1 in Supplementary Material), annotated and stored in an Oracle relational database that is located at http://glbrc.bch.msu.edu/psyllium.
Overview of Glycosyltransferases Highly Expressed in Psyllium Mucilaginous Layers
Assembly and annotation of the five RNA-Seq datasets from psyllium resulted in identification of 634 contigs encoding putative GTs. The top 50 transcripts from this set are listed in Table 1 ranked by expression in the ML at 10 days post anthesis (DPA) stage. The most abundant transcripts encoding putative GTs (1000 ppm or higher in at least one of the four ML stages) are homologs of IRX10(-L) (GT47), GUX5 (GT8; Mortimer et al., 2010), RGP1/UAM (GT75; Konishi et al., 2007), and AT3G18170/AT3G18180 (GT61), and are likely involved in complex heteroxylan biosynthesis. Most of these highly abundant ML transcripts are not found in the stem transcriptome (Table 1). Multiple homologous genes related to AT3G18170/AT3G18180 and IRX10(-L) are present in psyllium. These two gene families were investigated in further detail.
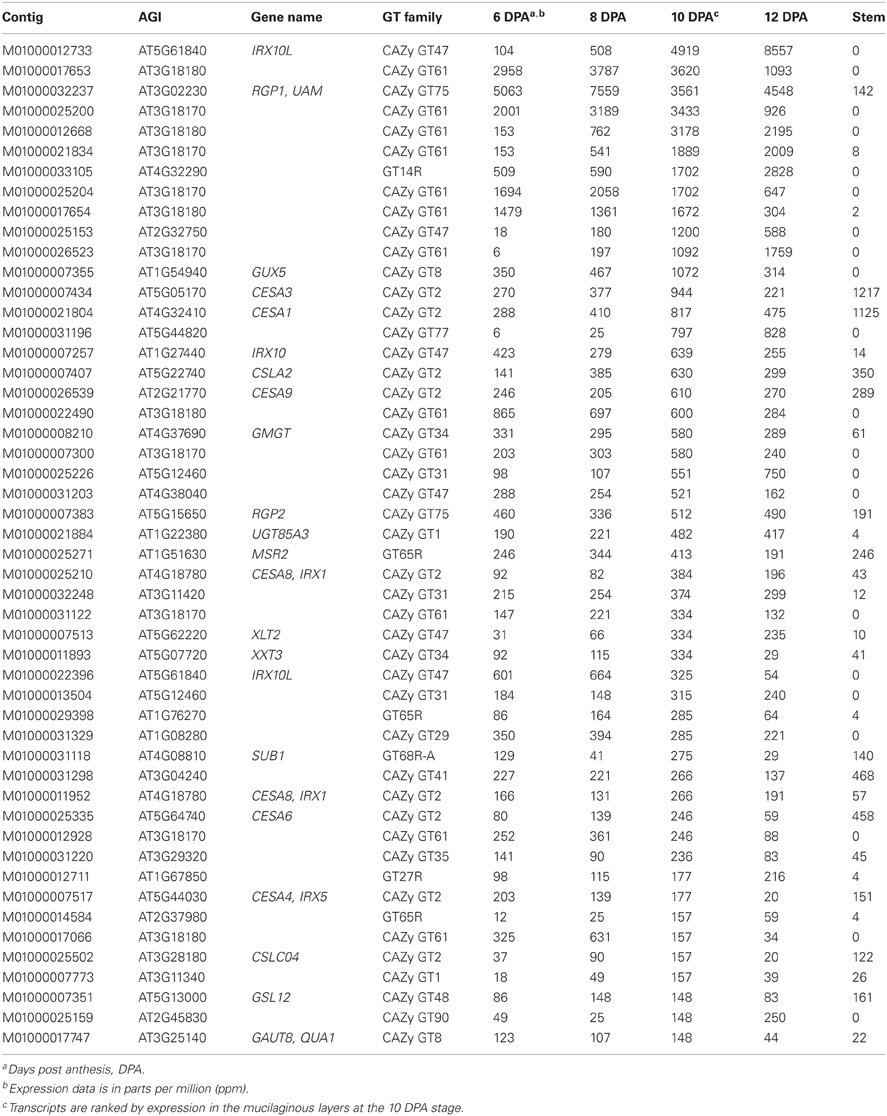
Table 1. The 50 most abundant transcripts expressed in psyllium mucilaginous layers encoding putative glycosyltransferases.
A significant level of primary cell wall biosynthesis is evident in the ML. Homologs of CESA1 and CESA3 (Arioli et al., 1998; Desprez et al., 2007; Persson et al., 2007) are found expressed in the range of 200 to 1000 ppm, while expression of putative xyloglucan GTs are found in the range of 50 to 350 ppm; e.g., homologs of CSLC4 (Cocuron et al., 2007), XLT2 (Jensen et al., 2012) and XXT3 (Vuttipongchaikij et al., 2012) (Table 1). A homolog of GAUT1 (Sterling et al., 2006) is found to be expressed at 79 ppm at 10 DPA, providing evidence for homogalacturonan synthesis. A homolog of the callose synthase, GSL12, is most abundant at 8 to 10 DAP (148 ppm) in the ML, indicating that cell division is taking place (Chen et al., 2009). Some level of secondary cell wall biosynthesis also appears to be present. Transcripts with homology to secondary cell wall CESA8 (IRX1) and CESA4 (IRX5) (Turner and Somerville, 1997; Persson et al., 2005) are found at a similar abundance as the GTs involved in xyloglucan biosynthesis. Transcripts with homology to CESA2, CESA5 and CESA9 are present in the ML transcriptome, especially abundant are transcripts with homology to CESA9. These three CESA proteins have been found to play important roles in Arabidopsis seed coat development, namely in mucilage attachment (CESA5) and formation of a secondary cell wall that reinforces the columella and radial wall (Mendu et al., 2011).
Evidence of mannan biosynthesis is indicated by the presence of CSLA2 (Dhugga et al., 2004; Goubet et al., 2009), MSR2 (Wang et al., 2012) and galactomannan galactosyltransferase (GMGT) (Edwards et al., 1999) homologs that have expression levels as high as 630 ppm (CSLA2 homolog, 10 DPA; Table 1). This finding is likely a result of endosperm tissue contamination in the dissected ML. The endosperm stores large amounts of mannan (Jensen et al., 2011) and given the attachment of the endosperm to the ML it is difficult to obtain ML tissue completely devoid of endosperm.
Out of the 50 most abundant transcripts shown in Table 1 there are 14 putative GT transcripts that cannot readily be assigned a function or to a pathway. Notably, many of these abundant transcripts have no expression in the stem transcriptome, as is seen for transcripts likely involved in heteroxylan biosynthesis (GT8, GT47, GT61, and GT75). This is in contrast to GTs involved in primary and secondary cell wall biosynthesis which reach expression levels in the stem of approximately 50 ppm or higher. The ML specific GTs without an assigned function therefore represent GTs possibly involved in complex heteroxylan synthesis in the psyllium ML, though involvement in other pathways unrelated to xylan syntheis is also possible.
Psyllium Stem Xylan Biosynthesis is Similar to Arabidopsis
All the transcripts identified encoding proteins homologous to IRX9(-L), IRX10(-L), IRX14(-L) and IRX15(-L) are listed in Table 2. This group of transcripts, with the exception of some IRX10(-L) and IRX15(-L) transcripts, had low expression or were not found in the ML. In the stem, the expression of these xylan specific genes was found to be unexpectedly low (100 ppm or lower). It appears, however, that this tissue is principally engaged in primary rather than secondary cell wall biosynthesis. When examining the expression of both the primary and secondary cell wall CESAs in the stem, the primary CESAs were found at levels as high as 1217 ppm (CESA3; Table 1) while the secondary CESAs were found at 10 fold lower levels. The expression of IRX9(-L), IRX10(-L), IRX14(-L), and IRX15(-L) in the stem therefore matches the level of secondary cell wall formation in this tissue. Therefore, it appears that psyllium has a similar complement of GTs found to be responsible for xylan synthesis as in Arabidopsis and that these genes are expressed at comparable levels in the psyllium stem.
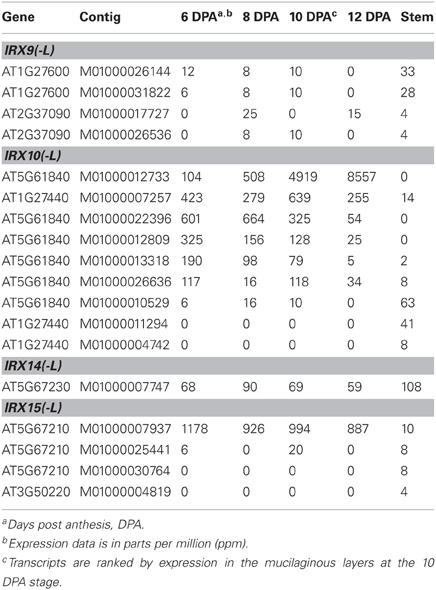
Table 2. All transcripts from psyllium stem and mucilaginous layers encoding proteins homologous to Arabidopsis IRX9(-L), IRX10(-L), IRX14(-L), and IRX15(-L).
Four Homologs of Arabidopsis IRX10 are Highly Expressed in Psyllium Mucilaginous Layers
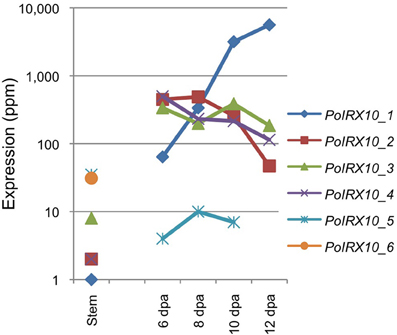
Transcripts encoding proteins homologous to IRX10(-L) show tissue specific distributions (Table 2), with transcripts present at high levels in the ML showing little or no expression in the stem, and vice versa. The presence of these two categories of IRX10(-L) transcripts led us to consider that at least two different genes with homology to IRX10(-L) are present in psyllium. We therefore manually examined a total of 12 IRX10(-L) contigs and found evidence of six unique IRX10(-L) genes in psyllium, named Plantago ovata IRX10 1 to 6 (PoIRX10_1 to _6). Four of these, those showing abundant expression in the ML (PoIRX10_1 to _4), were cloned from cDNA and sequenced. Analysis of the deduced amino acid sequence of PoIRX10_1, PoIRX10_3, and PoIRX10_4 for transmembrane domains as predicted by the TMHMM Server v. 2.0 (Krogh et al., 2001; http://www.cbs.dtu.dk/services/TMHMM/) resulted in a high score for a single N-terminal transmembrane domain for PoIRX10_1, an intermediate score for PoIRX10_4, and a very low score for PoIRX10_3 (File S1). The PoIRX10_2 cDNA sequence is missing the 5' end and was not analyzed.
The expression of PoIRX10_1 to _6 is shown in Figure 2. The expression profiles for PoIRX10_1 to _4 were generated by mapping the RNA-Seq data to the sequences obtained from the cDNA clones. The expression profile for PoIRX10_1 shows strong induction in the ML and reached maximum levels at 12 DPA, while PoIRX10_2 to _4 show a flat or a decreasing expression pattern over the four ML stages. PoIRX10_6 is not detected in the ML but is present in stem together with PoIRX10_5. The PoIRX10_5 is found in the ML but at a 10 fold lower level than PoIRX10_1 to _4.
PoIRX10_1, _2 and _4 Represent Some of the Most Divergent IRX10 Proteins Yet Identified
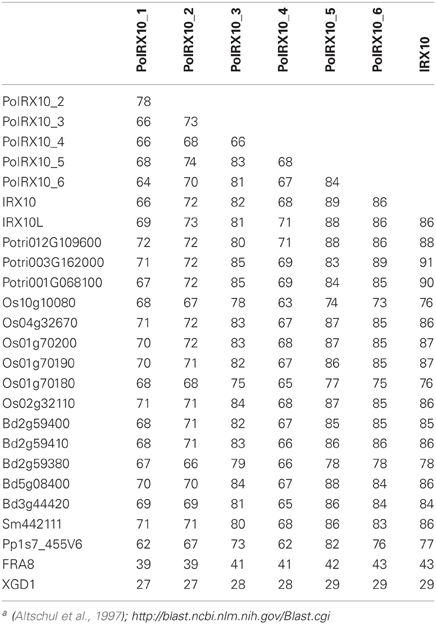
An examination of homologs of IRX10 from various higher plants showed a high degree of sequence conservation among these proteins. To obtain a broader view of this, we collected all IRX10 homologs from six different plant species with extensive phylogenetic diversity, all with fully sequenced and annotated genomes. This resulted in 18 IRX10 homologs from Physcomitrella patens (1), Selaginella moellendorffii (2), Arabidopsis thaliana (3), Populus trichocarpa (4), Brachypodium distachyon (5) and Oryza sativa (6). Table 3 shows the pair-wise amino acid maximum identity scores using the BLAST algorithm (Altschul et al., 1997; http://blast.ncbi.nlm.nih.gov/Blast.cgi) for these 18 IRX10 proteins compared against Arabidopsis IRX10 (AtIRX10) and the six PoIRX10. Arabidopsis FRA8 and XGD1 were included for comparison of more distantly related genes. FRA8 is the closest homolog to the IRX10(-L) genes in Arabidopsis (Zhong et al., 2005) and XGD1 is a xylosyltransferase from GT47 subgroup D (Jensen et al., 2008). The remaining of the pair-wise matrix is shown Table S2 in Supplementary Material. Eudicot sequences, including PoIRX10_3, _5 and _6, share 81–91% identity with AtIRX10, while monocot sequences show 76–87% identity with AtIRX10. Remarkably, the evolutionarily more distant SmIRX10 and PpIRX10 follow a similar trend with 86% and 77% identity, respectively, to AtIRX10. This conservation is also observed when comparing SmIRX10 and PpIRX10 to the remaining sequences from poplar, B. distachyon and rice; here SmIRX10 shows 77–88% identity, while PpIRX10 shows 68 to 80% identity (Table S2 in Supplementary material). The difference in identity between PoIRX10_1, _2 and _4 and mono- and dicot IRX10s is similar or lower than the difference in identity between higher plants and PpIRX10. Thus, IRX10 proteins show a high degree of conservation over the phylogenetic distance from P. patens to higher plants, while three of the four ML PoIRX10 proteins show notably less conservation, with PoIRX10_4 being the most divergent.
A phylogenetic tree of the 24 IRX10 proteins, FRA8 and XGD1 is shown in Figure 3A. The phylogenetic analysis was performed on a codon based cDNA sequence alignment. This approach is beneficial when performing phylogenetic analysis of conserved proteins with many synonymous mutations. The tree identifies two major clades rooted by PpIRX10. Eudicot IRX10 sequences make up one of the major clades, while the other clade contains monocot IRX10 sequences and SmIRX10. Of the six psyllium proteins, PoIRX10_6 is grouped with AtIRX10 and two of the three poplar IRX10 proteins, while PoIRX10_1 to _5 form a separate group. The phylogenetic analysis therefore suggests that the expansion of PoIRX10 proteins has taken place after the separation of monocots and dicots.
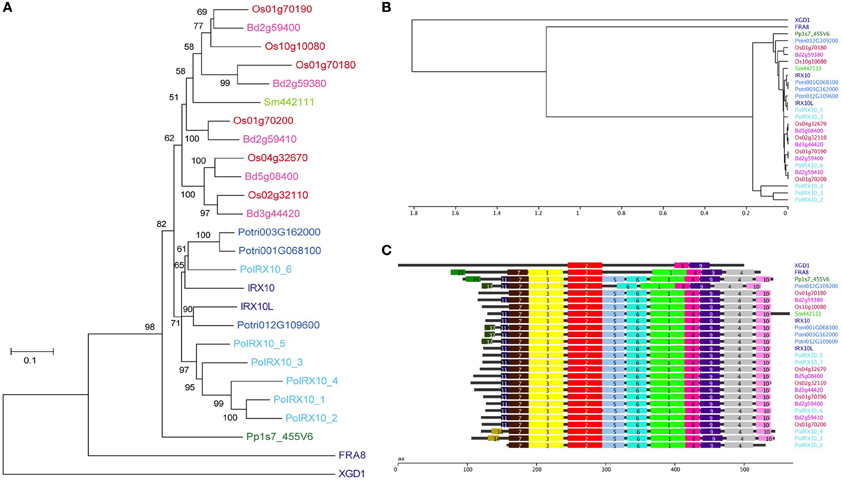
Figure 3. Phylogenetic and motif analysis of IRX10 homologs in psyllium. (A) Phylogenetic analysis of IRX10 homologs in psyllium (light blue) and six other plants; Brachypodium distachyon (pink), rice (red), Arabidopsis (dark blue), poplar (blue), Selaginella moellendorffii (light green) and Physcomitrella patens (green). (B, C) Hierarchical clustering (B) of motif analysis (C) generated using the interactive feature in the SALAD database (http://salad.dna.affrc.go.jp/CGViewer/en/cgv_upload.html). Both graphs are provided in File S3 including bootstrap values from the hierarchical clustering.
Evaluation of evolutionarily conserved protein domains are a powerful method for predicting protein function and are collected in a number of searchable databases, e.g., Pfam (Punta et al., 2012) and InterPro (Hunter et al., 2009). The algorithm behind the SALAD database uses patterns of evolutionarily conserved motifs to determine relatedness (Mihara et al., 2010; http://salad.dna.affrc.go.jp/salad/en/). As with other protein domain predicting methods, this approach emphasizes conserved protein function rather than phylogenetic relationships. In Figure 3B the 26 proteins from Figure 3A are depicted in a SALAD dendrogram. It shows that IRX10 proteins ranging in phylogenetic distance from P. patens to Arabidopsis are tightly clustered while PoIRX10_1, _2 and _4 form a distinct group. Notably, this psyllium specific clade consists of PoIRX10 proteins exclusively expressed in the ML. The SALAD motif structure (Figure 3C), used to construct the dendrogram, is conserved across the majority of IRX10 proteins. A few exceptions exist such as motif 5 is absent in the poplar gene Potri012G109200, motif 10 is absent in PoIRX10_2 and there is some motif variation in the N-terminus involving motif 11, 12, 14, and 15. In FRA8 motif 5, 6, and 10 are absent; while in XGD1 most of the motifs found in the IRX10 proteins are absent. This indicates that PoIRX10_1, _2 and _4 have conserved the motif structure despite their more divergent protein sequences and suggests they have conserved protein function with the IRX10 proteins found in the other plant species.
Similarities in Xylan Side Chain Decorations Between Psyllium and Grasses are Likely the Result of Convergent Evolution
The psyllium database contains 18 contigs encoding proteins with close homology to AT3G18170 and AT3G18180. Many of these contigs represented partial transcripts and were assembled into full transcripts by manual inspection. These efforts yielded evidence for the presence of nine unique GT61 genes in psyllium, seven of which were cloned from cDNA and named Plantago ovata GT61 1 to 7 (PoGT61_1 to _7).
The expression profiles of PoGT61_1 to _7 in psyllium stem and ML are depicted in Figure 4. These expression levels were similarly high as those of the PoIRX10_1 to _4 genes in the ML and show either induction or flat to decreasing levels of expression during ML development. These proteins are therefore likely candidates for GT activities that form the side chain decorations on the ML complex heteroxylan.
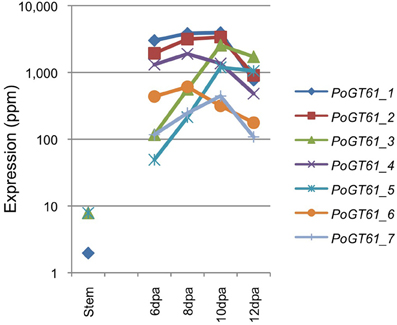
Figure 4. Expression levels of glycosyltransferase family 61 genes in psyllium in stem and mucilaginous layers.
Figure 5 presents a phylogenetic tree of PoGT61_1 to _7 and all GT61 proteins identified in Arabidopsis, rice and B. distachyon (ClustalW alignment in File S4). The phylogenetic tree shows that the large diversification in grasses of this family is unrelated to the diversification found in psyllium. Therefore, the similar modifications of the xylan backbone found in psyllium ML and grasses are likely the results of convergent evolution.
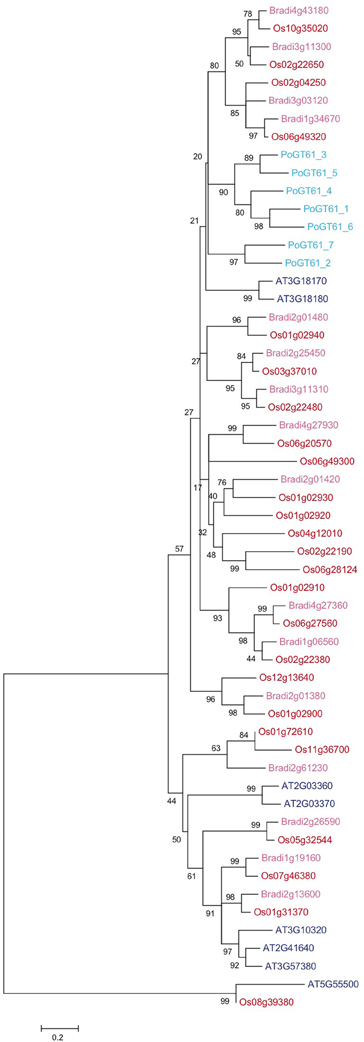
Figure 5. Phylogenetic analysis of glycosyltransferase family 61 proteins from psyllium, Arabidopsis, rice and Brachypodium distachyon. Seven cDNAs displaying homology to At3g18170 and At3g18180 were cloned from psyllium mucilaginous layers and their full-length protein sequences deduced. A few transcripts encoding protein sequences homologous to some of the other six GT61 proteins in Arabidopsis were identified in the mucilagionous layers but these were expressed at negligible levels (<10 pm) and were not included in this analysis. The seven GT61 proteins highly expressed in psyllium mucilaginous layers (light blue) were aligned with all glycosyltransferase family 61 proteins from Arabidopsis (dark blue), rice (red) and Brachypodium distachyon (pink).
Possible Function of the Numerous Putative Glycosyltransferases Highly Expressed in Psyllium Mucilaginous Layer
The structure of the xylan-based mucilage from the Plantago genus (ovata F., major L., asiatica L.) is highly complex (Samuelsen et al., 1999; Fischer et al., 2004; Yin et al., 2012). In the work of Guo et al. (2008) psyllium husk was fractionated using hot water and successive rounds of increasing concentrations of NaOH. This resulted in three fractions collectively accounting for 90% of the husk mass and predominantly consisting of Ara (15–25%) and Xyl (65–70%). Two of these fractions also yielded approximately 15% uronic acid. Each of the three fractions showed related but distinct glycosyl-linkage compositions providing evidence for the presence of extensively branched xylans in all three fractions. In all fractions, the branching appears to consist of single xylose residues, single arabinose residues and side chains of two to three sugars containing different combinations of xylose, arabinose, galactose, and mannose (Guo et al., 2008). An abundant side chain of α-Araf-(1→3)-β-Xylp-(1→3)-Araf present in the non-acidic fraction has been isolated and structurally characterized by NMR (Fischer et al., 2004). It therefore appears that the mucilage of P. ovata F. consist of several species of complex heteroxylans that have different structural compositions and physical characteristics.
The multiple side chains found in the psyllium mucilage are consistent with finding numerous GTs highly expressed in the ML. The identification of four different and abundantly expressed PoIRX10 genes is noteworthy. This may indicate that there are several heteroxylan subspecies being produced in the tissue and that each PoIRX10 protein is involved in making separate xylans by interacting with different decorating GTs. Alternatively, the four PoIRX10 proteins could form one or more complexes necessary to form the β-(1,4)-xylan backbone. Thirdly, some of the PoIRX10 proteins may be backbone decorating GTs and not involved in backbone synthesis (Table S3 in Supplementary Material). It seems likely, however, that at least one of the ML specific PoIRX10 proteins constitutes the xylan synthase in this tissue, hence forming a xylan synthase activity different than that found in Arabidopsis and other eudicots.
Several GTs from rice and wheat have been implicated in xylan side-chain formation. TaXAT2, OsXAT2 and OsXAT3 are xylan α-(1,3)-arabinosyltransferases, transferring single arabinofuranose onto the xylan backbone at the O-3 position (Anders et al., 2012). While XAX1 from rice is likely a β-(1,2)-xylosyltransferase involved in making the side-chain β-Xylp-(1→2)-α-Araf-(1→3) (Chiniquy et al., 2012). These proteins all group together in the same phylogenetic subgroup as Arabidopsis AT3G18170 and AT3G18180 and PoGT61_1 to _7. Therefore, it seems likely that the seven PoGT61 proteins are arabinosyl- and xylosyltransferases involved in forming the large diversity of xylan side-chains found in psyllium heteroxylan (Table S3 in Supplementary Material).
Small amounts of rhamnose, glucose, glucuronic acid, galactose, and mannose have been identified in psyllium husk and proposed to be side chain decorations (Fischer et al., 2004; Guo et al., 2008; Yin et al., 2012). Additional GTs, from families other than GT61, may be involved in forming these side chains in psyllium heteroxylan. Two such candidates are the transcripts homologous to AT4G32290 and AT2G32750 (Table S3 in Supplementary Material). AT4G32290 is a member of the GT14R family (Nikolovski et al., 2012). None of the members in this family have been characterized apart from having Golgi localization. The homologous transcript in psyllium is highly abundant in the ML. AT2G32750 is homologous to Arabidopsis MUR3 (Madson et al., 2003) and RLXT2 (Jensen et al., 2012), both of which transfer galactose onto xylose as part of xyloglucan biosynthesis. The homologous protein in psyllium is copiously expressed in the ML and could possibly transfer galactose onto xylose in psyllium heteroxylan.
Putative Nucleotide Sugar Transporters are Highly Expressed in Psyllium Mucilaginous Layers
Golgi transport proteins for UDP-galactose, UDP-galactose/glucose and GDP-mannose have been identified in Arabidopsis (Reyes and Orellana, 2008; Handford et al., 2012) and rice (Seino et al., 2010) and are members of the NST/TPT superfamily (Ward, 2001). Proteins transporting other UDP-sugars have been proposed to also be members of this superfamily (Ward, 2001; Reyes and Orellana, 2008). Several UDP-sugar transporters are likely to be expressed in the ML in order to supply UDP-xylose and UDP-arabinofuranose to Golgi localized enzymes for the biosynthesis of complex heteroxylan. The UDP-arabinopyranose mutase (UAM) interconverts UDP-arabinofuranose and UDP-arabinopyranose (Konishi et al., 2007) and is located in the cytosol (Bar-Peled and O'Neill, 2011). The synthesis of arabinoxylan occurs in the Golgi and requires UDP-arabinofuranose, which appears to be uniquely produced by this mutase (Rautengarten et al., 2011). It is therefore necessary for UDP-arabinofuranose to be transported across the Golgi membrane in order for it to be incorporated into cell wall carbohydrates such as heteroxylan. Approximately 40% of the neutral sugar content of the ML cell wall is arabinose, likely requiring higher amounts of UDP-arabinofuranose import into ML Golgi. It is therefore likely that transcript levels for the UDP-arabinofuranose transporter would be high in the ML.
The enzyme UDP-xylose epimerase 1 (UXE1/MUR4), that interconverts UDP-xylose and UDP-arabinopyranose, has been found to be Golgi localized in Arabidopsis (Burget et al., 2003). Two contigs in the psyllium ML (M01000013775 and M01000025234) match sequences at the N-terminal and C-terminal of UXE1 and together may represent the full-length transcript of a psyllium UXE1 homolog. This homolog shares 81% amino acid sequence identity with UXE1 in the N-terminal where both proteins have transmembrane domains as predicted by the TMHMM Server v. 2.0. The psyllium UXE1 homolog is therefore likely a Golgi localized protein. To provide UDP-xylose for UXE1, psyllium appears to express two isoforms of UDP-xylose synthase (UXS) at comparable levels in the ML (1000–6000 ppm) with just one of the two having a predicted transmembrane domain. Psyllium therefore appears to have the capacity to produce UDP-xylose in the cytosol, as well as in the Golgi. Finally, the substrate for UXS, UDP-glucuronic acid, is usually synthesized by the enzyme UDP-glucose 6-dehydrogenase (UGD) from UDP-glucose. The subcellular localization of this enzyme in psyllium ML could not be inferred as the contig lacks the N-terminal sequences, which would contain the transmembrane domain. The putative subcellular localization of these UDP-sugar interconverting activities in psyllium ML present several possible routes for the supply of UDP-xylose needed for xylan biosynthesis. The UDP-sugars imported into Golgi may be UDP-glucose, UDP-glucuronic acid or UDP-xylose. Furthermore, as the psyllium ML UXE activity appears to be exclusively Golgi localized, UDP-arabinopyranose needs to be exported from Golgi to the cytosol in order to be converted to UDP-arabinofuranose by UMA. Hence, psyllium transporters of these UDP-sugars, as well as UDP-arabinofuranose as mentioned above, are likely to be expressed at elevated levels in the ML.
The psyllium database holds a total of 50 contigs encoding proteins with close homology to the Arabidopsis NST/TPT Family. Homologs of characterized proteins such as ATUTR3 (Reyes et al., 2010) and GONST1 (Baldwin et al., 2001) are found at nearly undetectable levels in the ML, while the most abundant transcripts reach expression levels as high as 2000 ppm. When ranked by abundance in the ML 10 DPA stage, three of the top four transcripts show homology to Arabidopsis NST proteins AT5G25400, AT1G21070 and AT4G32390, all of which are in an uncharacterized branch of the NST/TPT superfamily. Several members of this branch, including AT4G32390, have been found to be localized in the Golgi (Nikolovski et al., 2012). The second most abundant transcript has closest homology to AT1G06890. This protein has also been found in Golgi (Nikolovski et al., 2012) and is related to GONST4 and GONST5 (Handford et al., 2004). These highly abundant NST transcripts might therefore be candidates for encoding UDP-glucose, UDP-glucuronic acid, UDP-xylose, UDP-arabinopyranose, or UDP-arabinofuranose transport proteins.
Additional Genes Possibly Involved in Xylan Biosynthesis in Psyllium Mucilaginous Layers
Identifying an Arabidopsis gene with secondary cell wall expression and with a close homolog highly expressed in psyllium ML may indicate that such a gene is involved in xylan biosynthesis in Arabidopsis and psyllium ML, as has proved to be the case for the Arabidopsis IRX15(-L) proteins (Jensen et al., 2011). Table 4 shows the 12 most abundant transcripts in psyllium that show a similar expression pattern. These genes are likely involved in complex heteroxylan biosynthesis or in secondary cell wall formation associated with the psyllium ML.
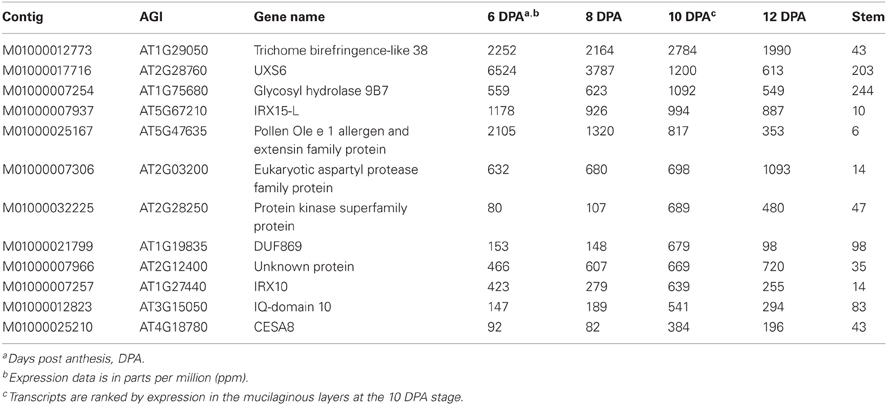
Table 4. The 12 most abundant transcripts expressed in psyllium mucilaginous layers where the closest homolog in Arabidopsis is co-expressed with IRX10 or CESA4, CESA7 and CESA8.
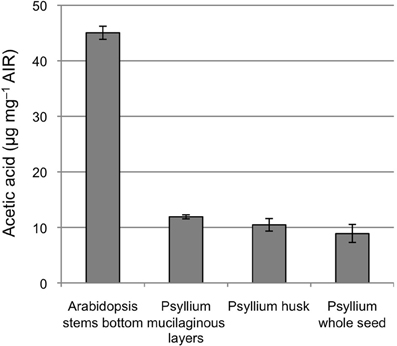
The top member is homologous to Arabidopsis TBR38 and contains a domain of unknown function (DUF) 231. The DUF231 proteins constitute a 46-member protein family in Arabidopsis (Bischoff et al., 2010) in which the genes AXY4 (TBL27) and AXY4-Like (TBL22) have been shown to be involved in acetylation of xyloglucan (Gille et al., 2011) and ESK1 (TBL29) have been shown to be involved in acetylation of secondary cell wall xylan (Xiong et al., 2013). The other members of this family have been proposed to also be acetyltransferases specific for xyloglucan or other cell wall polymers, e.g., pectins and xylan (Oikawa et al., 2010; Gille and Pauly, 2012). TBR38 is part of an uncharacterized subclade of the TBR protein family. Given that the psyllium homolog has a much higher expression in the ML than the secondary cell wall CESA proteins it is likely involved in complex heteroxylan biosynthesis rather than secondary cell wall formation in this tissue. The level of cell wall acetylation in dissected psyllium ML is 12 μg acetic acid per milligram alcohol insoluble residue, approximately 4 fold lower that found in the alcohol insoluble residue of Arabidopsis lower stem (Figure 6). The acetic acid content in the psyllium ML corresponds to one acetic acid group for every 25 pentose sugars, assuming the cell wall material from the ML consist of 100% pentose sugars. Glucuronoxylan from aspen wood has been found to have an average degree of xylose backbone acetylation of approximately 60% (Teleman et al., 2000), while a degree of acetylation of approximately 50% has been found for arabinoxylan from corncobs and corn stover (Dongen et al., 2011). These findings may indicate that TBR38 and its psyllium homolog could function as xylan specific acetyltransferases.
Another candidate gene possibly involved in xylan formation in psyllium ML is a homolog of At5g47635 encoding a Pollen Ole e 1 allergen and extensin family protein. The work of Tan et al. (2013) identified and characterized two isoforms of a highly glycosylated AGP, named ARABINOXYLAN PECTIN ARABINOGALACTAN PROTEIN1 (APAP1). The authors identified two individual xylan oligomers attached as separate side chains of the APAP1 carbohydrate branch structure and so provide a link between AGP and xylan. Though other possibilities exist, the high expression of an extensin protein in psyllium ML and the secondary cell wall expression pattern of the closest Arabidopsis homolog may suggest that this extensin homolog functions by cross-linking mucilaginous heteroxylan into a bigger covalent network in the mucilage wall.
Homologs of Several Secondary Cell Wall Transcription Factors are Highly Expressed in Psyllium Mucilaginous Layers
The most abundant psyllium transcripts encoding putative transcription factors reach levels of 1000 ppm in the ML (Table 5). Many of these transcripts show closest homology to Arabidopsis genes that are highly expressed throughout the Arabidopsis plant, including seed development, while another set of these transcripts have close homology to Arabidopsis genes that are specifically expressed during seed development, such as MYB61, NARS1, AT3G51880 and AT5G67480. Both MYB61 and NARS1 have been shown to play roles in seed coat development in Arabidopsis. Knockout mutants of MYB61 have reduced mucilage deposition and extrusion (Penfield et al., 2001), while NARS1 is expressed in the outer integument of the Arabidopsis seed where it regulates the degeneration of this tissue (Kunieda et al., 2008).
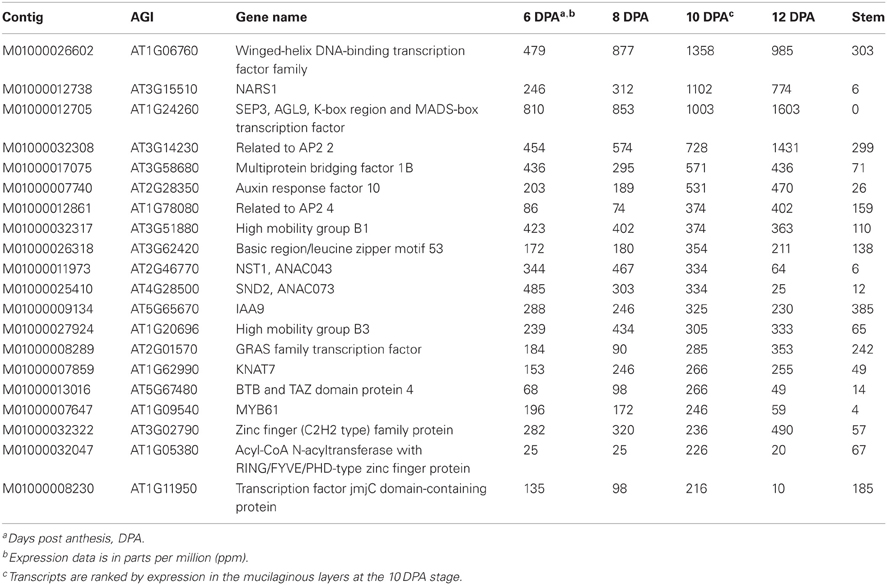
Table 5. The 20 most abundant transcripts expressed in psyllium mucilaginous layers encoding putative transcription factors.
A third category of transcripts consists of ones with closest homology to Arabidopsis transcription factors involved in secondary cell wall formation, namely NST1, SND2, and KNAT7. All three transcription factors are potent regulators of secondary cell wall formation in Arabidopsis. NST1 was identified as a regulator of secondary wall thickening in anther endothecium (Mitsuda et al., 2005) and was later found to act redundantly with SND1 as a master regulator of secondary wall synthesis in fiber cells of Arabidopsis stem (Zhong et al., 2007). Furthermore, in protoplast transactivation assays, NST1 directly activates MYB46, SND3, MYB103, and KNAT7 (Zhong et al., 2008, 2010). Overexpression of SND2 also leads to increased secondary wall thickening in Arabidopsis stem fiber cells (Zhong et al., 2008) and upregulation of, among other genes, MYB103 and SND1 (Hussey et al., 2011). KNAT7 loss-of-function mutants display IRX phenotypes (Brown et al., 2005) and Arabidopsis plants transformed with dominant repression constructs of KNAT7 lead to a moderate decrease of secondary cell wall thickening in Arabidopsis stems (Zhong et al., 2008). The KNAT7 protein has been shown to interact with OFP4 and both act as repressor proteins in protoplast transactivation assays and in planta (Li et al., 2011, 2012). The homologs of NST1, SND2, and KNAT7 found in psyllium ML may or may not be true orthologs to the Arabidopsis genes. However, it is striking to find several abundantly expressed homologs of transcription factors that have been implicated in secondary cell wall formation, a process involving extensive biosynthesis of xylan. The highly elevated expression levels of homologs NST1, SND2 and KNAT7 in the psyllium ML may therefore suggest that they are involved in regulating xylan biosynthesis in this tissue. Such regulatory circuit(s) in the psyllium ML may have evolved from the secondary cell wall regulatory cascade. It should be noted that this it is only a partial set of the transcriptional regulatory network controlling secondary cell wall formation in Arabidopsis (Demura and Ye, 2010) that may be detected in the psyllium ML. Of the proven downstream targets for NST1 and SND2, it is only a homolog of KNAT7 that is found highly expressed in the psyllium ML. Homologs of proven targets of NST1 in Arabidopsis, such as SND1, MYB46, and MYB103, are not detected, while a homolog of SND3 is detected but at low levels of approximately 50 ppm. When over-expressed, NST1 will induce abundant secondary cell wall formation in Arabidopsis mesophyll leaf cells (Zhong et al., 2007). If the psyllium homolog of NST1 found in the ML is functionally othologous to Arabidopsis NST1, it appears that branches of the NST1 transcriptional cascade that leads to cellulose and lignin deposition, rather than xylan formation, has been specifically suppressed in the psyllium ML.
Lessons Learned from Xylan Biosynthesis in Psyllium Mucilaginous Layers May Prove Valuable for Biofuels Research and Biotechnology
The study of tissues having cell walls with unusual composition may provide valuable insights into manipulating plant cell walls for improved characteristics as biofuel feedstocks, such as improved digestibility, higher biomass, and altered composition of lignin, cellulose, and hemicellulose. It seems plausible that the diverse cell walls found in many highly specialized tissues, for instance in many seeds, are derived from existing cell wall biosynthetic pathways and so provide examples of cell wall alterations which provide new characteristics.
This study provides evidence for biosynthetic enzymes, sugar-nucleotide transporters and transcription factors as likely candidates involved in xylan biosynthesis. These new targets may serve as novel entry points to manipulate xylan deposition and structure. To date, it has not been possible to reconstitute the xylan synthase activity from known components. This has limited our ability to assign roles for the genes shown to be components of the synthase by genetic methods. The four cloned PoIRX10 from the ML may constitute a simpler xylan synthase, as it has a reduced set of components, suggesting that it may be more tractable than xylan synthases from systems such as Arabidopsis. If so, the psyllium IRX10 genes would offer a tool for future research in understanding and manipulating xylan formation.
The seven cloned PoGT61 sequences may prove useful in altering xylan branch structures in cell walls of both monocot and eudicot crops for improved biofuel traits such as digestibility. Finally, identification of direct transcriptional regulators of xylan biosynthetic genes, such as IRX10, is likely to identify more genes involved in xylan biosynthesis which could constitute key points of regulating xylan biosynthesis.
Full access has been provided to the RNA-Seq data from psyllium through a user-friendly web interface. The database features several custom made tools facilitating further analysis and may provide a valuable resource for the research community in other areas than xylan biosynthesis, such as mucilage development.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the DOE Great Lakes Bioenergy Research Center (DOE BER Office of Science DE-FC02-07ER64494). We thank Christa Pennacchio and Erika Linquist of the US Department of Energy Joint Genome Institute for high-throughput cDNA sequencing, which was supported by the Office of Science of the US Department of Energy under Contract No. (DE-AC02-05CH11231). We also thank Nick Thrower for providing the bioinformatic expertise clustering of the cDNA RNA-Seq libraries.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/Plant_Biotechnology/10.3389/fpls.2013.00183/abstract
File S1. Transmembrane domain predictions for PoIRX10_1, PoIRX10_3, and PoIRX10_4.
File S2. Protein sequence alignment of IRX10 proteins.
File S3. SALAD dendrogram and motif alignment of IRX10 proteins.
File S4. Protein sequence alignment of GT61 proteins.
Table S1. 454 FLX sequencing and assembly of sequence reads.
Table S2. Pairwise amino acid maximum identity scores using BLAST.
Table S3. Glycosyl linkages identified in psyllium mucilaginous heteroxylan and putative glycosyltransferases identified in this study that are hypothesized to form these linkages.
Abbreviations
GT, glycosyltransferase; DPA, days post anthesis; ML, mucilaginous layer.
References
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Anders, N., Wilkinson, M. D., Lovegrove, A., Freeman, J., Tryfona, T., Pellny, T. K., et al. (2012). Glycosyl transferases in family 61 mediate arabinofuranosyl transfer onto xylan in grasses. Proc. Natl. Acad. Sci. U.S.A. 109, 989–993. doi: 10.1073/pnas.1115858109
Arioli, T., Peng, L., Betzner, A. S., Burn, J., Wittke, W., Herth, W., et al. (1998). Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279, 717–720. doi: 10.1126/science.279.5351.717
Baldwin, T. C., Handford, M. G., Yuseff, M. I., Orellana, A., and Dupree, P. (2001). Identification and characterization of GONST1, a golgi-localized GDP-mannose transporter in Arabidopsis. Plant Cell 13, 2283–2295. doi: 10.1105/tpc.010247
Bar-Peled, M., and O'Neill, M. A. (2011). Plant nucleotide sugar formation, interconversion, and salvage by sugar recycling. Annu. Rev. Plant Biol. 62, 127–155. doi: 10.1146/annurev-arplant-042110-103918
Bischoff, V., Nita, S., Neumetzler, L., Schindelasch, D., Urbain, A., Eshed, R., et al. (2010). TRICHOME BIREFRINGENCE and Its homolog AT5G01360 encode plant-specific DUF231 proteins required for cellulose biosynthesis in Arabidopsis. Plant Physiol. 153, 590–602. doi: 10.1104/pp.110.153320
Brown, D., Wightman, R., Zhang, Z., Gomez, L. D., Atanassov, I., Bukowski, J.-P., et al. (2011). Arabidopsis genes IRREGULAR XYLEM (IRX15) and IRX15L encode DUF579-containing proteins that are essential for normal xylan deposition in the secondary cell wall. Plant J. 66, 401–413. doi: 10.1111/j.1365-313X.2011.04501.x
Brown, D. M., Zeef, L. A. H., Ellis, J., Goodacre, R., and Turner, S. R. (2005). Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17, 2281–2295. doi: 10.1105/tpc.105.031542
Brown, D. M., Zhang, Z., Stephens, E., Dupree, P., and Turner, S. R. (2009). Characterization of IRX10 and IRX10-like reveals an essential role in glucuronoxylan biosynthesis in Arabidopsis. Plant J. 57, 732–746. doi: 10.1111/j.1365-313X.2008.03729.x
Burget, E. G., Verma, R., Mølhøj, M., and Reiter, W.-D. (2003). The biosynthesis of L-arabinose in plants: molecular cloning and characterization of a Golgi-localized UDP-D-xylose 4-epimerase encoded by the MUR4 gene of Arabidopsis. Plant Cell 15, 523–531. doi: 10.1105/tpc.008425
Cantarel, B. L., Coutinho, P. M., Rancurel, C., Bernard, T., Lombard, V., and Henrissat, B. (2009). The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238. doi: 10.1093/nar/gkn663
Chen, X.-Y., Liu, L., Lee, E., Han, X., Rim, Y., Chu, H., et al. (2009). The Arabidopsis callose synthase gene GSL8 is required for cytokinesis and cell patterning. Plant Physiol. 150, 105–113. doi: 10.1104/pp.108.133918
Chiniquy, D., Sharma, V., Schultink, A., Baidoo, E. E., Rautengarten, C., Cheng, K., et al. (2012). XAX1 from glycosyltransferase family 61 mediates xylosyltransfer to rice xylan. Proc. Natl. Acad. Sci. U.S.A. 109, 17117–17122. doi: 10.1073/pnas.1202079109
Cocuron, J.-C., Lerouxel, O., Drakakaki, G., Alonso, A. P., Liepman, A. H., Keegstra, K., et al. (2007). A gene from the cellulose synthase-like C family encodes a beta-1, 4 glucan synthase. Proc. Natl. Acad. Sci. U.S.A. 104, 8550–8555. doi: 10.1073/pnas.0703133104
Demura, T., and Ye, Z.-H. (2010). Regulation of plant biomass production. Curr. Opin. Plant Biol. 13, 299–304. doi: 10.1016/j.pbi.2010.03.002
Desprez, T., Juraniec, M., Crowell, E. F., Jouy, H., Pochylova, Z., Parcy, F., et al. (2007). Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 104, 15572–15577. doi: 10.1073/pnas.0706569104
Dhugga, K., Barreiro, R., Whitten, B., Stecca, K., Hazebroek, J., Randhawa, G., et al. (2004). Guar seed beta-mannan synthase is a member of the cellulose synthase super gene family. Science 303, 363–366. doi: 10.1126/science.1090908
Dongen, F. E. M. V., Eylen, D. V., and Kabel, M. A. (2011). Characterization of substituents in xylans from corn cobs and stover. Carbohydr. Polym. 86, 722–731. doi: 10.1016/j.carbpol.2011.05.007
Edwards, M. E., Dickson, C. A., Chengappa, S., Sidebottom, C., Gidley, M. J., and Reid, J. S. (1999). Molecular characterisation of a membrane-bound galactosyltransferase of plant cell wall matrix polysaccharide biosynthesis. Plant J. 19, 691–697. doi: 10.1046/j.1365-313x.1999.00566.x
Edwards, S., Chaplin, M. F., Blackwood, A. D., and Dettmar, P. W. (2003). Primary structure of arabinoxylans of ispaghula husk and wheat bran. Proc. Nutr. Soc. 62, 217–222. doi: 10.1079/PNS2003202
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.2307/2408678
Fischer, M. H., Yu, N., Gray, G. R., Ralph, J., Anderson, L., and Marlett, J. A. (2004). The gel-forming polysaccharide of psyllium husk (Plantago ovata Forsk). Carbohydr. Res. 339, 2009–2017. doi: 10.1016/j.carres.2004.05.023
Gille, S., De Souza, A., Xiong, G., Benz, M., Cheng, K., Schultink, A., et al. (2011). O-acetylation of Arabidopsis hemicellulose xyloglucan requires AXY4 or AXY4L, proteins with a TBL and DUF231 domain. Plant Cell 23, 4041–4053. doi: 10.1105/tpc.111.091728
Gille, S., and Pauly, M. (2012). O-acetylation of plant cell wall polysaccharides. Front. Plant Sci. 3:12. doi: 10.3389/fpls.2012.00012
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Goto, N. (1985). A mucilage polysaccharide secreted from testa of Arabidopsis thaliana. Arab. Inf. Ser. 22, 143.
Goubet, F., Barton, C. J., Mortimer, J. C., Yu, X., Zhang, Z., Miles, G. P., et al. (2009). Cell wall glucomannan in Arabidopsis is synthesised by CSLA glycosyltransferases, and influences the progression of embryogenesis. Plant J. 60, 527–538. doi: 10.1111/j.1365-313X.2009.03977.x
Guo, A., He, K., Liu, D., Bai, S., Gu, X., Wei, L., et al. (2005). DATF: a database of Arabidopsis transcription factors. Bioinformatics 21, 2568–2569. doi: 10.1093/bioinformatics/bti334
Guo, Q., Cui, S. W., Wang, Q., and Young, J. C. (2008). Fractionation and physicochemical characterization of psyllium gum. Carbohydr. Polym. 73, 35–43. doi: 10.1016/j.carbpol.2007.11.001
Handford, M., Rodríguez-Furlán, C., Marchant, L., Segura, M., Gómez, D., Alvarez-Buylla, E., et al. (2012). Arabidopsis thaliana AtUTr7 encodes a golgi-localized UDP-glucose/UDP-galactose transporter that affects lateral root emergence. Mol. Plant 5, 1263–1280. doi: 10.1093/mp/sss074
Handford, M. G., Sicilia, F., Brandizzi, F., Chung, J. H., and Dupree, P. (2004). Arabidopsis thaliana expresses multiple Golgi-localised nucleotide-sugar transporters related to GONST1. Mol. Genet. Genom. 272, 397–410. doi: 10.1007/s00438-004-1071-z
Hunter, S., Apweiler, R., Attwood, T. K., Bairoch, A., Bateman, A., Binns, D., et al. (2009). InterPro: the integrative protein signature database. Nucleic Acids Res. 37, D211–D215. doi: 10.1093/nar/gkn785
Hussey, S. G., Mizrachi, E., Spokevicius, A. V., Bossinger, G., Berger, D. K., and Myburg, A. A. (2011). SND2, a NAC transcription factor gene, regulates genes involved in secondary cell wall development in Arabidopsis fibres and increases fibre cell area in Eucalyptus. BMC Plant Biol. 11:173. doi: 10.1186/1471-2229-11-173
International Brachypodium Initiative. (2010). Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463, 763–768. doi: 10.1038/nature08747
Jensen, J. K., Schultink, A., Keegstra, K., Wilkerson, C. G., and Pauly, M. (2012). RNA-Seq analysis of developing nasturtium seeds (Tropaeolum majus): identification and characterization of an additional galactosyltransferase involved in xyloglucan biosynthesis. Mol. Plant 5, 984–992. doi: 10.1093/mp/sss032
Jensen, J. K., Kim, H., Cocuron, J.-C., Orler, R., Ralph, J., and Wilkerson, C. G. (2011). The DUF579 domain containing proteins IRX15 and IRX15-L affect xylan synthesis in Arabidopsis. Plant J. 66, 387–400. doi: 10.1111/j.1365-313X.2010.04475.x
Jensen, J. K., Sørensen, S. O., Harholt, J., Geshi, N., Sakuragi, Y., Møller, I., et al. (2008). Identification of a xylogalacturonan xylosyltransferase involved in pectin biosynthesis in Arabidopsis. Plant Cell 20, 1289–1302. doi: 10.1105/tpc.107.050906
Konishi, T., Takeda, T., Miyazaki, Y., Ohnishi-Kameyama, M., Hayashi, T., O'Neill, M. A., et al. (2007). A plant mutase that interconverts UDP-arabinofuranose and UDP-arabinopyranose. Glycobiology 17, 345–354. doi: 10.1093/glycob/cwl081
Krogh, A., Larsson, B., Von Heijne, G., and Sonnhammer, E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
Kunieda, T., Mitsuda, N., Ohme-Takagi, M., Takeda, S., Aida, M., Tasaka, M., et al. (2008). NAC family proteins NARS1/NAC2 and NARS2/NAM in the outer integument regulate embryogenesis in Arabidopsis. Plant Cell 20, 2631–2642. doi: 10.1105/tpc.108.060160
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., Mcgettigan, P. A., Mcwilliam, H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Lee, C., Teng, Q., Huang, W., Zhong, R., and Ye, Z.-H. (2010). The Arabidopsis family GT43 glycosyltransferases form two functionally nonredundant groups essential for the elongation of glucuronoxylan backbone. Plant Physiol. 153, 526–541. doi: 10.1104/pp.110.155309
Lee, C., Teng, Q., Zhong, R., and Ye, Z.-H. (2012a). Arabidopsis GUX proteins are glucuronyltransferases responsible for the addition of glucuronic acid side chains onto xylan. Plant Cell Physiol. 53, 1204–1216. doi: 10.1093/pcp/pcs064
Lee, C., Teng, Q., Zhong, R., Yuan, Y., Haghighat, M., and Ye, Z.-H. (2012b). Three Arabidopsis DUF579 domain-containing GXM proteins are methyltransferases catalyzing 4-O-methylation of glucuronic acid on xylan. Plant Cell Physiol. 53, 1934–1949. doi: 10.1093/pcp/pcs138
Li, E., Bhargava, A., Qiang, W., Friedmann, M. C., Forneris, N., Savidge, R. A., et al. (2012). The class II KNOX gene KNAT7 negatively regulates secondary wall formation in Arabidopsis and is functionally conserved in Populus. New Phytol. 194, 102–115. doi: 10.1111/j.1469-8137.2011.04016.x
Li, E., Wang, S., Liu, Y., Chen, J.-G., and Douglas, C. J. (2011). OVATE FAMILY PROTEIN4 (OFP4) interaction with KNAT7 regulates secondary cell wall formation in Arabidopsis thaliana. Plant J. 67, 328–341. doi: 10.1111/j.1365-313X.2011.04595.x
Madson, M., Dunand, C., Li, X., Verma, R., Vanzin, G. F., Caplan, J., et al. (2003). The MUR3 gene of Arabidopsis encodes a xyloglucan galactosyltransferase that is evolutionarily related to animal exostosins. Plant Cell. 15, 1662–1670. doi: 10.1105/tpc.009837
Mendu, V., Stork, J., Harris, D., and Debolt, S. (2011). Cellulose synthesis in two secondary cell wall processes in a single cell type. Plant Signal. Behav. 6, 1638–1643. doi: 10.4161/psb.6.11.17709
Mihara, M., Itoh, T., and Izawa, T. (2010). SALAD database: a motif-based database of protein annotations for plant comparative genomics. Nucleic Acids Res. 38, D835–D842. doi: 10.1093/nar/gkp831
Mitsuda, N., Seki, M., Shinozaki, K., and Ohme-Takagi, M. (2005). The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17, 2993–3006. doi: 10.1105/tpc.105.036004
Mortimer, J. C., Miles, G. P., Brown, D. M., Zhang, Z., Segura, M. P., Weimar, T., et al. (2010). Absence of branches from xylan in Arabidopsis gux mutants reveals potential for simplification of lignocellulosic biomass. Proc. Natl. Acad. Sci. U.S.A. 107, 17409–17414. doi: 10.1073/pnas.1005456107
Mutwil, M., Obro, J., Willats, W. G. T., and Persson, S. (2008). GeneCAT–novel webtools that combine BLAST and co-expression analyses. Nucleic Acids Res. 36, W320–W326. doi: 10.1093/nar/gkn292
Naran, R., Chen, G., and Carpita, N. C. (2008). Novel rhamnogalacturonan I and arabinoxylan polysaccharides of flax seed mucilage. Plant Physiol. 148, 132–141. doi: 10.1104/pp.108.123513
Nei, M., and Kumar, S. (2000). Molecular Evolution and Phylogenetics. New York, NY: Oxford University Press. doi: 10.1186/1471-2148-12-219
Nikolovski, N., Rubtsov, D., Segura, M. P., Miles, G. P., Stevens, T. J., Dunkley, T. P., et al. (2012). Putative glycosyltransferases and other plant Golgi apparatus proteins are revealed by LOPIT proteomics. Plant Physiol. 160, 1037–1051. doi: 10.1104/pp.112.204263
Oikawa, A., Joshi, H. J., Rennie, E. A., Ebert, B., Manisseri, C., Heazlewood, J. L., et al. (2010). An integrative approach to the identification of Arabidopsis and rice genes involved in xylan and secondary wall development. PLoS ONE 5:e15481. doi: 10.1371/journal.pone.0015481
Peña, M. J., Zhong, R., Zhou, G.-K., Richardson, E. A., O'Neill, M. A., Darvill, A. G., et al. (2007). Arabidopsis irregular xylem8 and irregular xylem9: implications for the complexity of glucuronoxylan biosynthesis. Plant Cell 19, 549–563. doi: 10.1105/tpc.106.049320
Pellny, T. K., Lovegrove, A., Freeman, J., Tosi, P., Love, C. G., Knox, J. P., et al. (2012). Cell walls of developing wheat starchy endosperm: comparison of composition and RNA-Seq transcriptome. Plant Physiol. 158, 612–627. doi: 10.1104/pp.111.189191
Penfield, S., Meissner, R. C., Shoue, D. A., Carpita, N. C., and Bevan, M. W. (2001). MYB61 is required for mucilage deposition and extrusion in the Arabidopsis seed coat. Plant Cell 13, 2777–2791. doi: 10.1105/tpc.010265
Persson, S., Paredez, A., Carroll, A., Palsdottir, H., Doblin, M., Poindexter, P., et al. (2007). Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 15566–15571. doi: 10.1073/pnas.0706592104
Persson, S., Wei, H., Milne, J., Page, G. P., and Somerville, C. R. (2005). Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc. Natl. Acad. Sci. U.S.A. 102, 8633–8638. doi: 10.1073/pnas.0503392102
Punta, M., Coggill, P. C., Eberhardt, R. Y., Mistry, J., Tate, J., Boursnell, C., et al. (2012). The Pfam protein families database. Nucleic Acids Res. 40, D290–D301. doi: 10.1093/nar/gkr1065
Rautengarten, C., Ebert, B., Herter, T., Petzold, C. J., Ishii, T., Mukhopadhyay, A., et al. (2011). The interconversion of UDP-Arabinopyranose and UDP-Arabinofuranose is indispensable for plant development in Arabidopsis. Plant Cell 23, 1373–1390. doi: 10.1105/tpc.111.083931
Rennie, E. A., Hansen, S. F., Baidoo, E. E. K., Hadi, M. Z., Keasling, J. D., and Scheller, H. V. (2012). Three members of the Arabidopsis glycosyltransferase family 8 are xylan glucuronosyltransferases. Plant Physiol. 159, 1408–1417. doi: 10.1104/pp.112.200964
Reyes, F., León, G., Donoso, M., Brandizzí, F., Weber, A. P. M., and Orellana, A. (2010). The nucleotide sugar transporters AtUTr1 and AtUTr3 are required for the incorporation of UDP-glucose into the endoplasmic reticulum, are essential for pollen development and are needed for embryo sac progress in Arabidopsis thaliana. Plant J. 61, 423–435. doi: 10.1111/j.1365-313X.2009.04066.x
Reyes, F., and Orellana, A. (2008). Golgi transporters: opening the gate to cell wall polysaccharide biosynthesis. Curr. Opin. Plant Biol. 11, 244–251. doi: 10.1016/j.pbi.2008.03.008
Samuelsen, A. B., Cohen, E. H., Paulsen, B. S., Brüll, L. P., and Thomas-Oates, J. E. (1999). Structural studies of a heteroxylan from Plantago major L. seeds by partial hydrolysis, HPAEC-PAD, methylation and GC-MS, ESMS and ESMS/MS. Carbohydr. Res. 315, 312–318. doi: 10.1016/S0008-6215(99)00038-5
Schmid, M., Davison, T. S., Henz, S. R., Pape, U. J., Demar, M., Vingron, M., et al. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37, 501–506. doi: 10.1038/ng1543
Seino, J., Ishii, K., Nakano, T., Ishida, N., Tsujimoto, M., Hashimoto, Y., et al. (2010). Characterization of rice nucleotide sugar transporters capable of transporting UDP-galactose and UDP-glucose. J. Biochem. 148, 35–46. doi: 10.1093/jb/mvq031
Sterling, J. D., Atmodjo, M. A., Inwood, S. E., Kumar Kolli, V. S., Quigley, H. F., Hahn, M. G., et al. (2006). Functional identification of an Arabidopsis pectin biosynthetic homogalacturonan galacturonosyltransferase. Proc. Natl. Acad. Sci. U.S.A. 103, 5236–5241. doi: 10.1073/pnas.0600120103
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tan, L., Eberhard, S., Pattathil, S., Warder, C., Glushka, J., Yuan, C., et al. (2013). An Arabidopsis cell wall proteoglycan consists of pectin and arabinoxylan covalently linked to an arabinogalactan protein. Plant Cell 25, 270–287. doi: 10.1105/tpc.112.107334
Teleman, A., Lundqvist, J., Tjerneld, F., Stålbrand, H., and Dahlman, O. (2000). Characterization of acetylated 4-O-methylglucuronoxylan isolated from aspen employing 1H and 13C NMR spectroscopy. Carbohydr. Res. 329, 807–815. doi: 10.1016/S0008-6215(00)00249-4
Turner, S. R., and Somerville, C. R. (1997). Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell 9, 689–701. doi: 10.1105/tpc.9.5.689
Urbanowicz, B. R., Peña, M. J., Ratnaparkhe, S., Avci, U., Backe, J., Steet, H. F., et al. (2012). 4-O-methylation of glucuronic acid in Arabidopsis glucuronoxylan is catalyzed by a domain of unknown function family 579 protein. Proc. Natl. Acad. Sci. U.S.A. 109, 14253–14258. doi: 10.1073/pnas.1208097109
Vuttipongchaikij, S., Brocklehurst, D., Steele-King, C., Ashford, D. A., Gomez, L. D., and Mcqueen-Mason, S. J. (2012). Arabidopsis GT34 family contains five xyloglucan α-1, 6-xylosyltransferases. New Phytol 195, 585–595. doi: 10.1111/j.1469-8137.2012.04196.x
Wang, Y., Alonso, A. P., Wilkerson, C. G., and Keegstra, K. (2012). Deep EST profiling of developing fenugreek endosperm to investigate galactomannan biosynthesis and its regulation. Plant Mol. Biol. 79, 243–258. doi: 10.1007/s11103-012-9909-y
Ward, J. M. (2001). Identification of novel families of membrane proteins from the model plant Arabidopsis thaliana. Bioinformatics 17, 560–563. doi: 10.1093/bioinformatics/17.6.560
Western, T. L., Skinner, D. J., and Haughn, G. W. (2000). Differentiation of mucilage secretory cells of the Arabidopsis seed coat. Plant Physiol. 122, 345–356. doi: 10.1104/pp.122.2.345
Wu, A.-M., Hornblad, E., Voxeur, A., Gerber, L., Rihouey, C., Lerouge, P., et al. (2010). Analysis of the Arabidopsis IRX9/IRX9-L and IRX14/IRX14-L pairs of glycosyltransferase genes reveals critical contributions to biosynthesis of the hemicellulose glucuronoxylan. Plant Physiol. 153, 542–554. doi: 10.1104/pp.110.154971
Wu, A.-M., Rihouey, C., Seveno, M., Hörnblad, E., Singh, S. K., Matsunaga, T., et al. (2009). The Arabidopsis IRX10 and IRX10-LIKE glycosyltransferases are critical for glucuronoxylan biosynthesis during secondary cell wall formation. Plant J. 57, 718–731. doi: 10.1111/j.1365-313X.2008.03724.x
Xiong, G., Cheng, K., and Pauly, M. (2013). Xylan O-acetylation impacts xylem development and enzymatic recalcitrance as indicated by the Arabidopsis mutant tbl29. Mol. Plant. doi: 10.1093/mp/sst014. [Epub ahead of print].
Yin, J.-Y., Lin, H.-X., Nie, S.-P., Cui, S. W., and Xie, M.-Y. (2012). Methylation and 2D NMR analysis of arabinoxylan from the seeds of Plantago asiatica L. doi: 10.1016/j.carbpol.2012.02.025
Zhong, R., Lee, C., and Ye, Z.-H. (2010). Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis. Mol. Plant 3, 1087–1103. doi: 10.1093/mp/ssq062
Zhong, R., Lee, C., Zhou, J., Mccarthy, R. L., and Ye, Z.-H. (2008). A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20, 2763–2782. doi: 10.1105/tpc.108.061325
Zhong, R., Peña, M. J., Zhou, G.-K., Nairn, C. J., Wood-Jones, A., Richardson, E. A., et al. (2005). Arabidopsis fragile fiber8, which encodes a putative glucuronyltransferase, is essential for normal secondary wall synthesis. Plant Cell 17, 3390–3408. doi: 10.1105/tpc.105.035501
Keywords: xylan, psyllium, secondary cell wall, irx, glycosyltransferase, mucilage
Citation: Jensen JK, Johnson N and Wilkerson CG (2013) Discovery of diversity in xylan biosynthetic genes by transcriptional profiling of a heteroxylan containing mucilaginous tissue. Front. Plant Sci. 4:183. doi: 10.3389/fpls.2013.00183
Received: 26 February 2013; Accepted: 20 May 2013;
Published online: 07 June 2013.
Edited by:
Samuel P Hazen, University of Massachusetts, USAReviewed by:
Staffan Persson, Max-Planck Gesellschaft, GermanyRowan Mitchell, Rothamsted Research, UK
Copyright © 2013 Jensen, Johnson and Wilkerson. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Curtis G. Wilkerson, Department of Plant Biology, Michigan State University, 612 Wilson Rd., Room 122, East Lansing, MI 48824-1312 USA e-mail:d2lsa2VyMTNAbXN1LmVkdQ==

 Nathan Johnson1,2
Nathan Johnson1,2