Abstract
In plants, a wide frame of physiological processes are regulated in liaison by both, nitric oxide (NO) and hormones. Such overlapping roles raise the question of how the cross-talk between NO and hormones trigger common physiological responses. In general, NO has been largely accepted as a signaling molecule that works in different processes. Among the most relevant ways NO and the NO-derived reactive species can accomplish their biological functions it is worthy to mention post-translational protein modifications. In the last years, S-nitrosylation has been the most studied NO-dependent regulatory mechanism. Briefly, S-nitrosylation is a redox-based mechanism for cysteine residue modification and is being recognized as a ubiquitous regulatory reaction comparable to phosphorylation. Therefore, it is emerging as a crucial mechanism for the transduction of NO bioactivity in plants and animals. In this mini-review, we provide an overview on S-nitrosylation of target proteins related to hormone networks in plants.
INTRODUCTION
Nitric oxide (NO) is a free-radical product of cell metabolism, being nitrate reductase the best characterized enzymatic pathway for NO production in plants. However, other reductive and oxidative routes have been also described (Lamattina and Polacco, 2007). It functions as a ubiquitous signal involved in diverse physiological processes and it is frequently implicated in multiple cell signaling events under the control of phytohormones including growth, development, and stress responses. Nevertheless, in most cases the molecular mechanisms underlying NO action in the plant cell are still undeciphered. The overlapping roles between plant hormones and NO raise the question of how both molecules may act in coordination. In general, regulatory effects of NO are mediated through protein modifications, including tyrosine nitration, metal nitrosylation, and S-nitrosylation of cysteines. Thus, the identification of NO primary targets has provided new opportunities to link NO reactivity and biological processes. In this review, we highlight the progress brought by the identification of S-nitrosylated target proteins related to stress and growth-promoting plant hormones. Our focus is the broad role of this post-translational modification that allows NO to modulate plant hormone homeostasis as well as signaling pathways. However, the participation of NO beyond its action through S-nitrosylation in hormone-regulated processes is out of the scope of this work and it is widespreadly covered in recent reviews by Simontacchi et al. (2013) and Astier and Lindermayr (2012).
S-NITROSYLATION AS AN EMERGING POST-TRANSLATIONAL MODIFICATION OF PLANT PROTEINS
S-nitrosylation is the reversible binding of a NO moiety to a reactive cysteine residue of a target protein to form an S-nitrosothiol (SNO; Stamler et al., 2001). It is recognized as a reversible and ubiquitous regulatory reaction. Thus, like in animals, this redox-based post-translational mechanism is also crucial for the transduction of NO bioactivity in many plant cellular responses (Hess et al., 2005). At first, protein S-nitrosylation was thought to be controlled mainly through the regulation of NO biosynthesis. However, in mammals it has been postulated as a short-range NO post-translational mechanism limited to proximity of NO sources (Martinez-Ruiz et al., 2013). In addition to the enzymatic NO-producing enzymes, it is important to consider that both, favorable environment to S-nitrosylating agent formation as well as transnitrosylating reactions could promote the expansion of the S-nitrosylation range of action (Martinez-Ruiz et al., 2013). The SNO turnover could also provide an alternative mechanism to control protein S-nitrosylation in the cell. Given the labile nature of this post-translational modification, it was conceived initially as a spontaneous and non-regulated process. However, different denitrosylase enzymes have been described, which directly mediate denitrosylation or govern the cellular equilibrium between protein and low-molecular weight SNOs. Two main enzymatic systems have emerged as physiologically relevant denitrosylases: the glutathione/S-nitrosoglutathione reductase (GSH/GSNOR) and the thioredoxin/thioredoxin reductase (Trx/TrxR; Benhar et al., 2009). S-nitrosylation of the major intracellular antioxidant tripeptide GSH forms S-nitrosoglutathione (GSNO) that functions as a mobile reservoir of NO. Consequently, the enzyme GSNOR or GSNOR1 in Arabidopsis does not display a direct denitrosylase activity but controls intracellular levels of both, GSNO and SNO affecting the global level of S-nitrosylation (Feechan et al., 2005; Malik et al., 2011). On the other side, the mechanism described in animals for Trx denitrosylation involves direct interaction with SNO-proteins by formation of an intermolecular disulphide intermediate in which Trx is covalently linked to the substrate protein through a disulphide bridge, or transnitrosylation in which Trx is transiently S-nitrosylated (Benhar et al., 2009). Trx have been also described in the denitrosylation process taking part in hormonal signaling in plants (Tada et al., 2008). Therefore, it appears that the balance between S-nitrosylation/denitrosylation is critical for the precise transduction of NO signal.
S-glutathionylation is the post-translational modification of protein cysteine residues by the addition of GSH (Martinez-Ruiz and Lamas, 2007). The integrative interplay between protein S-glutathionylation and S-nitrosylation could be recognized as another crucial network for post-translational modification of certain proteins. Although the S-glutathionylation of proteins has been generally described more than 20 years ago, the identification of protein targets for this modification remains rather unexplored. Interestingly, for some mammal proteins involved in clinical disorders such as cardiovascular disease and diabetes among others, S-nitrosylation has been described as an intermediate for more stable modifications like S-glutathionylation (Martinez-Ruiz and Lamas, 2007). In summary, S-nitrosylation is crucial for NO signal transduction pathway but it should also be noted that other related-S-nitrosylation regulators can converge in NO-mediated protein functionality in plants.
S-NITROSYLATION OF TARGET PROTEINS LINKED TO STRESS PHYTOHORMONES
Salicylic acid (SA) and ethylene (ET) are key signaling molecules for plants in the resistance to biotic stress (Fujita et al., 2006; Loake and Grant, 2007). NO has an essential role in restriction of pathogen attack by induction of the defense response and programed host cell death (reviewed by Mur et al., 2013). Thus, NO bioactivity may exert a role on SA and ET hormone signaling pathways.
In Arabidopsis, one of the first comprehensive proteomic studies allowed the identification of more than 100 S-nitrosylated proteins (Lindermayr et al., 2005). Interestingly, one of the identified S-nitrosylated proteins corresponded to a methionine adenosyltransferase (MAT) which catalyzes the synthesis of S-adenosylmethionine (SAM), a substrate for ET biosynthesis. Later on, Lindermayr et al. (2006) provided the first detailed molecular characterization of an S-nitrosylated target protein in plants. This study describes the S-nitrosylation of Cys-114 residue of the MAT1 isoform and the consequently inhibition of its activity. The enzymes S-adenosylhomocysteinase and cobalamin-independent methionine synthase are also part of the methylmethionine cycle and both enzymes have been found to be S-nitrosylated in proteomic analysis in Arabidopsis and Kalanchoe pinnata plants (Lindermayr et al., 2005; Abat et al., 2008). Activation/inactivation of these enzymes controls the SAM pool impacting in ET biosynthesis. All these evidences point out a multi-step control of ET biosynthesis by S-nitrosylation and opened the possibility to elucidate new mechanisms of NO and ET cross-talk (Figure 1A).
FIGURE 1
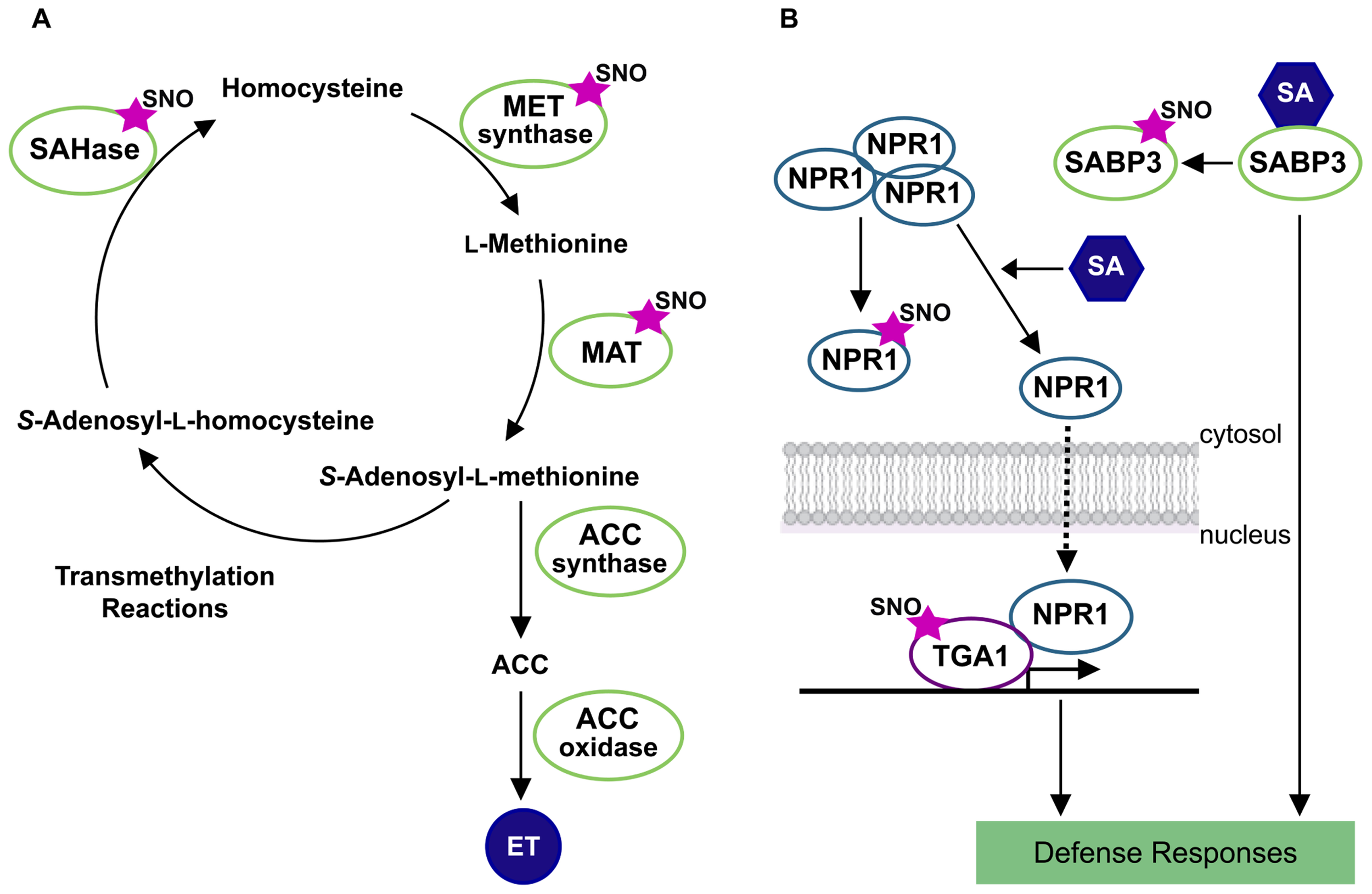
S-nitrosylation of target proteins in ethylene biosynthesis and salicylic acid network. The figure shows a schematic representation of methylmethionine cycle in the ethylene (ET) synthesis (A) and salicylic acid (SA) signaling networks (B). Protein S-nitrosothiols are represented by an SNO mark. References to physiological processes regulated by hormones, and subcellular localizations in the cell are also indicated. SAHase, adenosylhomocysteinase; MET synthase, cobalamin-independent methionine synthase; MAT, methionine adenosyltransferase; ACC, aminocyclopropane-1-carboxylic acid; SABP3, salicylic acid binding protein 3; NPR1, non-expresser of pathogenesis-related gene1 protein; TGA1, transcription factor TGA1.
Salicylic acid is synthesized by plants in response to pathogen infection and is essential to the establishment of resistance mechanisms, including host cell death and systemic acquired resistance. Mutations in AtGSNOR1 showed a pivotal role in the GSNO turnover, influencing cellular SNO levels under both, basal conditions and attempted microbial attack (Feechan et al., 2005). Interestingly, in the absence of AtGSNOR1 both SA biosynthesis and signaling are affected, suggesting that S-nitrosylation may control at least, two nodes of the SA-signaling network. GSNOR1 regulates the S-nitrosylation extent of non-expresser of pathogenesis-related gene1 (NPR1) and SA binding protein 3 (SABP3; Tada et al., 2008; Wang et al., 2009). S-nitrosylation of SABP3 is triggered during bacterial infection and suppresses SA binding capacity and carbonic anhydrase (CA) activity (Wang et al., 2009). Since, CA activity is required for the establishment of plant disease resistance, its inhibition by S-nitrosylation during late infection stages could contribute to a negative feedback loop which could be crucial for the proper modulation of SA-dependent plant defense mechanism (Figure 1B).
S-nitrosylation also exerts a key redox control of systemic acquired resistance in plants through targeting NPR1/TGA1 system. The SA NPR1-dependent signaling mechanism is mediated by redox changes that lead to reduction of NPR1 cysteines. This event switches NPR1 from cytosolic, disulfide-bound oligomers, to active monomers that are subsequently translocated into the nucleus and interacts with the TGA class of basic leucine zipper transcription factors. The result is an enhanced binding activity of TGA1 to the promoter region of pathogenesis-related (PR) genes, stimulating SA-dependent immune defense (Vlot et al., 2009). Upon pathogen attack, SA induces Trx which facilitates NPR1 monomerization, nuclear translocation, and activation of PR genes (Tada et al., 2008). Additionally, Tada et al. (2008) demonstrated that NPR1 is an S-nitrosylated protein. Notably, TGA1 is regulated by S-nitrosylation and S-glutathionylation improving TGA1 binding activity to PR1 promoter region (Lindermayr et al., 2010). However, it has not been demonstrated which type of modification, S-nitrosylation, S-glutathionylation, and/or both, is responsible for such protein–DNA binding activity (Figure 1B).
Concluding, plant immunity is regulated by a precise redox balance between the opposing actions of distinct redox-signals that catalyze NPR1 oligomer–monomer switch and NPR1/TGA1 interaction through transient redox fluctuations that includes S-nitrosylation and S-glutathionylation. Moreover, in the cytosol NPR1 also contributes to the suppression of jasmonic acid (JA)-dependent responses (Spoel et al., 2003), evidencing S-nitrosylation as a mediator of the integrative hormonal regulation network for guarantee immunity in plants.
Meanwhile, abscisic acid (ABA) is the major player mediating adaptation of plants to drought stress. ABA induces stomatal closure and inhibits stomatal opening by facilitating osmotic solute loss to reduce guard cell turgor. These events take place through a complex signaling network that involves multiple components including Ca2+, K+, IP3, MAPK, and H2O2 (Fan et al., 2004). NO enhances plant tolerance to drought and it contributes to stomatal closure evoked by ABA. Mechanistically, NO regulates inward-rectifying K+ channels through its action on Ca2+ release from intercellular stores. Alternative pathways have been also indicated for NO action on the outward-rectifying K+ channels, which are Ca2+ insensitive. It is probable that NO directly modifies the K+ channel at the guard cell plasma membrane or a closely associated regulatory protein through S-nitrosylation (Sokolovski and Blatt, 2004). However, the physiological significance of this regulation remains unexplored.
TARGETS FOR PROTEIN S-NITROSYLATION IN SIGNALING PATHWAYS OF GROWTH-PROMOTING PHYTOHORMONES AUXINS AND CYTOKININS
Auxins and cytokinins (CKs) are critical regulators of cell division, expansion, and differentiation.Relatively recent breakthroughs were found by comparing functions of NO and the well-known growth-promoting hormones (reviewed by Mur et al., 2013). There are several examples of NO and auxin overlapping effects during shoot and root organogenesis such as, NO mediation of auxin-induced adventitious and lateral roots (Pagnussat et al., 2002; Correa-Aragunde et al., 2004), root hair formation (Lombardo et al., 2006), and adventitious root formation (Pagnussat et al., 2003). NO stimulates the activation of cell division and embryogenic cell formation in leaf protoplast in the presence of auxin (Otvos et al., 2005). Copper-induced morphological responses are also mediated by auxin and NO in Arabidopsis seedlings (Peto et al., 2011). All these previous evidences led to investigate the possible interplay between these two signal molecules. Briefly, in the case of auxin, its perception is mediated by the F-box protein TIR1 (transport inhibitor response1) and the related proteins, AUXIN SIGNALING F-BOX proteins (AFBs; Dharmasiri et al., 2005; Kepinski and Leyser, 2005). Auxin binding stabilizes the interaction between TIR1/AFBs and the transcriptional repressor proteins, auxin/indole-3-acetic acid (Aux/IAA) causing a rapid proteasomal degradation of them (Gray et al., 2001). Then, Aux/IAA degradation results in the activation of transcriptional responses with the concomitant impact in plant growth and development (Tan et al., 2007). In an attempt to study the possible mechanism by which NO might regulate auxin signaling, S-nitrosylation of auxin receptor was analyzed. S-nitrosylation of TIR1 was demonstrated by Terrile et al. (2012). This redox-based modification enhances the efficiency by which TIR1 interacts with Aux/IAAs facilitating their degradation and modulating auxin signaling during root growth in Arabidopsis seedlings (Figure 2). Particularly, Cys-140 is a critical residue for TIR1–Aux/IAA interaction and TIR1 function. S-nitrosylation of TIR1 represents an efficient mechanism by which NO might enhance sensitivity and/or ligand selectivity. Furthermore, NO modulation of auxin signaling is more complex since a combinatorial TIR1/AFB–Aux/IAA co-receptor system could be assembled, contributing to the versatility of auxin response (Calderon Villalobos et al., 2012). However, cellular effectors of denitrosylation remain to be explored. Recently, Correa-Aragunde et al. (2013) described a new convergence where auxins are thought to influence S-nitrosylation/denitrosylation balance in Arabidopsis roots. The antioxidant enzyme, APX1 is an S-nitrosylation target and auxin induces denitrosylation and partial inhibition of its activity (Correa-Aragunde et al., 2013). These authors postulated that an auxin-regulated balance of APX1 S-nitrosylation/denitrosylation state contributes to a fine-tuned control of reactive oxygen species (ROS) that finally impacts on root architecture and development. Recent studies have pointed out the correlation between ROS and auxin homeostasis in signal transduction during plant development and stress response (Tognetti et al., 2012). In this direction, Bashandy et al. (2010) also highlighted the intercellular redox status as a critical parameter determining plant development through modulation of auxin signaling, transport, and homeostasis. Although our knowledge about auxin and NO is currently being born, most probably S-nitrosylation/denitrosylation is of great impact throughout to interlink these two molecules along plant lifecycle.
FIGURE 2
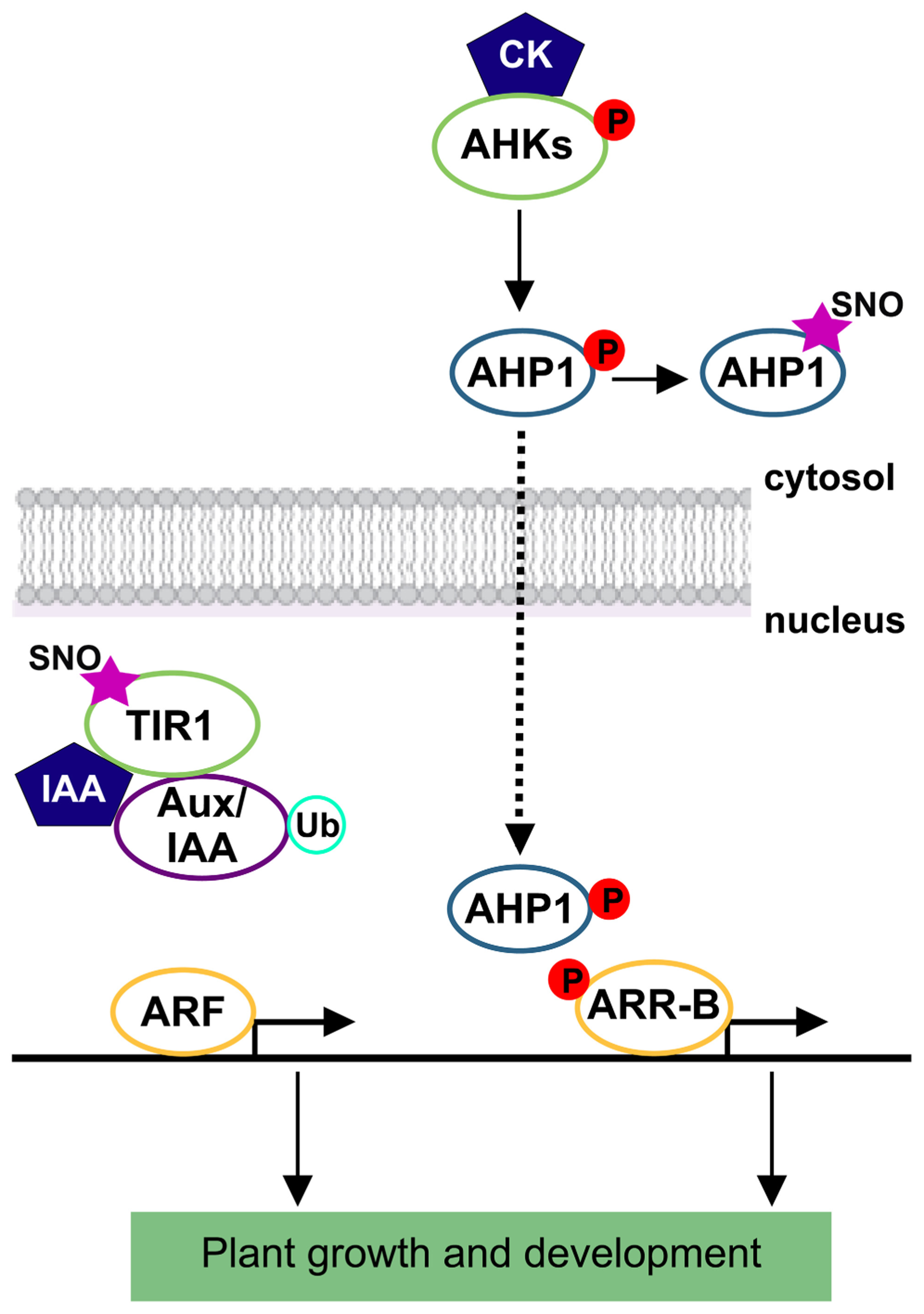
Auxin and cytokinin signaling pathways are under protein S-nitrosylation influence. Overview of indole-acetic acid (IAA) and cytokinin (CK) signaling pathways. S-nitrosothiols are represented by an SNO mark. Protein phosphorylation is represented by a P letter. References to physiological processes regulated by hormones, and subcellular localizations are also indicated. TIR1, transport inhibitor response 1 protein; Aux/IAA, auxin/indole-3-acetic acid protein; ARF, auxin response factor; Ub, ubiquitin; AHKs, hybrid histidine protein kinases; AHP1, histidine phosphotransfer protein 1; ARR-B, primary response regulator type B.
Plant hormones CKs are well known for their ability to promote cell division and they are associated with growth and development, including lateral root formation and nodulation in legumes (Gonzalez-Rizzo et al., 2006; Murray et al., 2007; Tirichine et al., 2007), circadian rhythms (Salome et al., 2006), and shoot and root development (Werner and Schmulling, 2009). Recently, NO-mediated CK functions have been associated to cell proliferation and meristem maintenance in Arabidopsis (Shen et al., 2013). CKs are perceived and mediated by a multi-step two-component circuit through a histidine and aspartate phosphorelay (Muller and Sheen, 2007). CKs regulate their signals through a variety of mechanisms, such as modulating transcription, controlling phosphorelay and regulating protein localization and stability (To and Kieber, 2008). In a recent report, Feng et al. (2013) demonstrated that NO represses CK signaling by inhibiting the phosphorelay activity through S-nitrosylation. Interestingly, the authors showed that NO-overproducing mutants, nox-1 (NO overproducer1) and gsnor1–3 do not respond to CK-induced shoot regeneration in Arabidopsis explants. Moreover, gsnor1–3 has a substantial reduction on the expression of the primary response regulator genes (ARRs) for CK signaling. Centrally, by the use of an in vivo biotin-switch assay, it was demonstrated that the histidine phosphotransfer protein AHP1 is in plantaS-nitrosylated under normal growth conditions. Cys-115 was proposed as an S-nitrosylated residue. Comprehensively, AHP1 S-nitrosylation compromises CK action revealing again, a mechanism through which CK signaling components perceive and integrate a redox signal in the regulation of plant growth and development (Figure 2). Although several lines of evidence support the involvement of NO in CK signaling (Carimi et al., 2005; Tun et al., 2008), other works claim an opposite effect of NO in CK action (Werner et al., 2003; Riefler et al., 2006; Xiao-Ping and Xi-Gui, 2006). Much more recently, a direct interaction between NO and CK has been also described (Liu et al., 2013). In summary, NO roles could be of the most varied because in addition to its own action it meets specific cellular functions according to the target molecules amending within the routes of hormonal regulation in plant cells.
CONCLUDING REMARKS AND PERSPECTIVES
NO is a fascinating molecule with remarkable feats and properties to modulate signaling pathways in biological systems. The bioactivity of NO is high enough for it to occur in a wide variety of biochemical circumstances. S-nitrosylation/denitrosylation is currently accepted as critical redox-mediated regulation processes in plant cells. Certainly, S-nitrosylation could be a possible mechanism by which NO impacts on plant hormonal regulation by modulating hormone biosynthesis, perception, transport, and/or degradation. Clearly, multiple layers of interactions may be involved in the plant hormones and NO cross-talks, depending on complex biological and biochemical scenarios in cells. However,nowadays fragmented studies on its in vivo function hamper our thorough understanding on hormone–NO cross-talking. Probably, high-throughput genetic and protein-based approaches in combination with a deeper understanding on the basic structure/function relationships of NO generating systems will shed light on this scientific riddle.
Statements
Acknowledgments
Ramiro París, María C. Terrile, and Claudia A. Casalongué are researchers from CONICET. María J. Iglesias is postgraduate fellow from the same Institution. Work in the lab of Claudia A. Casalongué is supported by grants from CONICET, ANPCyT, and Universidad Nacional de Mar del Plata.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1
Abat J. K. Mattoo A. K. Deswal R. (2008) S-nitrosylated proteins of a medicinal CAM plant Kalanchoe pinnata – ribulose-1,5-bisphosphate carboxylase/oxygenase activity targeted for inhibition.FEBS J.2752862–2872.10.1111/j.1742-4658.2008.06425.x
2
Astier J. Lindermayr C. (2012) Nitric oxide-dependent posttranslational modification in plants: an update.Int. J. Mol. Sci.1315193–15208.10.3390/ijms131115193
3
Bashandy T. Guilleminot J. Vernoux T. Caparros-Ruiz D. Ljung K. Meyer Y. (2010) Interplay between the NADP-linked thioredoxin and glutathione systems in Arabidopsis auxin signaling.Plant Cell22376–391.10.1105/tpc.109.071225
4
Benhar M. Forrester M. T. Stamler J. S. (2009) Protein denitrosylation: enzymatic mechanisms and cellular functions.Nat. Rev. Mol. Cell Biol.10721–732.10.1038/nrm2764
5
Calderon Villalobos L. I. Lee S. De Oliveira C. Ivetac A. Brandt W. Armitage L. (2012) A combinatorial TIR1/AFB–Aux/IAA co-receptor system for differential sensing of auxin.Nat. Chem. Biol.8477–485.10.1038/nchembio.926
6
Carimi F. Zottini M. Costa A. Cattelan I. De Michele R. Terzi M. (2005) NO signalling in cytokinin-induced programmed cell death.Plant Cell Environ.281171–1178.10.1111/j.1365-3040.2005.01355.x
7
Correa-Aragunde N. Foresi N. Delledonne M. Lamattina L. (2013) Auxin induces redox regulation of ascorbate peroxidase activity by S-nitrosylation/denitrosylation balance resulting in changes of root growth pattern in Arabidopsis.J. Exp. Bot.10.1093/jxb/ert172
8
Correa-Aragunde N. Graziano M. Lamattina L. (2004) Nitric oxide plays a central role in determining lateral root development in tomato.Planta218900–905.10.1007/s00425-003-1172-7
9
Dharmasiri N. Dharmasiri S. Estelle M. (2005) The F-box protein TIR1 is an auxin receptor.Nature435441–445.10.1038/nature03543
10
Fan L. M. Zhao Z. Assmann S. M. (2004) Guard cells: a dynamic signaling model.Curr. Opin. Plant Biol.7537–546. 10.1016/j.pbi.2004.07.009
11
Feechan A. Kwon E. Yun B. W. Wang Y. Pallas J. A. Loake G. J. (2005) A central role for S-nitrosothiols in plant disease resistance.Proc. Natl. Acad. Sci. U.S.A.1028054–8059.10.1073/pnas.0501456102
12
Feng J. Wang C. Chen Q. Chen H. Ren B. Li X. (2013) S-nitrosylation of phosphotransfer proteins represses cytokinin signaling.Nat. Commun.41529 10.1038/ncomms2541
13
Fujita M. Fujita Y. Noutoshi Y. Takahashi F. Narusaka Y. Yamaguchi-Shinozaki K. (2006) Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks.Curr. Opin. Plant Biol.9436–442.10.1016/j.pbi.2006.05.014
14
Gonzalez-Rizzo S. Crespi M. Frugier F. (2006) The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti.Plant Cell182680–2693.10.1105/tpc.106.043778
15
Gray W. M. Kepinski S. Rouse D. Leyser O. Estelle M. (2001) Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins.Nature414271–276.10.1038/35104500
16
Hess D. T. Matsumoto A. Kim S. O. Marshall H. E. Stamler J. S. (2005) Protein S-nitrosylation: purview and parameters.Nat. Rev. Mol. Cell Biol.6150–166.10.1038/nrm1569
17
Kepinski S. Leyser O. (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor.Nature435446–451.10.1038/nature03542
18
Lamattina L. Polacco J. C. (2007) Nitric Oxide in Plant Growth, Development and Stress Physiology.Berlin: Springer-Verlag.10.1007/11563280
19
Lindermayr C. Saalbach G. Bahnweg G. Durner J. (2006) Differential inhibition of Arabidopsis methionine adenosyltransferases by protein S-nitrosylation.J. Biol. Chem.2814285–4291.10.1074/jbc.M511635200
20
Lindermayr C. Saalbach G. Durner J. (2005) Proteomic identification of S-nitrosylated proteins in Arabidopsis.Plant Physiol.137921–930.10.1104/pp.104.058719
21
Lindermayr C. Sell S. Muller B. Leister D. Durner J. (2010) Redox regulation of the NPR1–TGA1 system of Arabidopsis thaliana by nitric oxide.Plant Cell222894–2907.10.1105/tpc.109.066464
22
Liu W. Z. Kong D. D. Gu X. X. Gao H. B. Wang J. Z. Xia M. (2013) Cytokinins can act as suppressors of nitric oxide in Arabidopsis.Proc. Natl. Acad. Sci. U.S.A.1101548–1553.10.1073/pnas.1213235110
23
Loake G. Grant M. (2007) Salicylic acid in plant defence – the players and protagonists.Curr. Opin. Plant Biol.10466–472.10.1016/j.pbi.2007.08.008
24
Lombardo M. C. Graziano M. Polacco J. Lamattina L. (2006) Nitric oxide functions as a positive regulator of root hair development.Plant Signal. Behav.128–33.10.4161/psb.1.1.2398
25
Malik S. I. Hussain A. Yun B. W. Spoel S. H. Loake G. J. (2011) GSNOR-mediated de-nitrosylation in the plant defence response.Plant Sci.181540–544.10.1016/j.plantsci.2011.04.004
26
Martinez-Ruiz A. Araujo I. M. Izquierdo-Alvarez A. Hernansanz-Agustin P. Lamas S. Serrador J. M. (2013) Specificity in S-nitrosylation: a short-range mechanism for NO signaling? Antioxid.Redox Signal.10.1089/ars.2012.5066 [Epub ahead of print].
27
Martinez-Ruiz A. Lamas S. (2007) Signalling by NO-induced protein S-nitrosylation and S-glutathionylation: convergences and divergences.Cardiovasc. Res.75220–228.10.1016/j.cardiores.2007.03.016
28
Muller B. Sheen J. (2007) Arabidopsis cytokinin signaling pathway.Sci. STKE2007cm5 10.1126/stke.4072007cm5
29
Mur L. A. Mandon J. Persijn S. Cristescu S. M. Moshkov I. E. Novikova G. V. (2013) Nitric oxide in plants: an assessment of the current state of knowledge.AoB Plants5pls052 10.1093/aobpla/pls052
30
Murray J. D. Karas B. J. Sato S. Tabata S. Amyot L. Szczyglowski K. (2007) A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis.Science315101–104.10.1126/science.1132514
31
Otvos K. Pasternak T. P. Miskolczi P. Domoki M. Dorjgotov D. Szucs A. (2005) Nitric oxide is required for, and promotes auxin-mediated activation of, cell division and embryogenic cell formation but does not influence cell cycle progression in alfalfa cell cultures.Plant J.43849–860.10.1111/j.1365-313X.2005.02494.x
32
Pagnussat G. C. Lanteri M. L. Lamattina L. (2003) Nitric oxide and cyclic GMP are messengers in the indole acetic acid-induced adventitious rooting process.Plant Physiol.1321241–1248.10.1104/pp.103.022228
33
Pagnussat G. C. Simontacchi M. Puntarulo S. Lamattina L. (2002) Nitric oxide is required for root organogenesis.Plant Physiol.129954–956.10.1104/pp.004036
34
Peto A. Lehotai N. Lozano-Juste J. Leon J. Tari I. Erdei L. (2011) Involvement of nitric oxide and auxin in signal transduction of copper-induced morphological responses in Arabidopsis seedlings.Ann. Bot.108449–457.10.1093/aob/mBR176
35
Riefler M. Novak O. Strnad M. Schmulling T. (2006) Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism.Plant Cell1840–54.10.1105/tpc.105.037796
36
Salome P. A. To J. P. Kieber J. J. McClung C. R. (2006) Arabidopsis response regulators ARR3 and ARR4 play cytokinin-independent roles in the control of circadian period.Plant Cell1855–69.10.1105/tpc.105.037994
37
Shen Q. Wang Y. T. Tian H. Guo F. Q. (2013) Nitric oxide mediates cytokinin functions in cell proliferation and meristem maintenance in Arabidopsis.Mol. Plant. 10.1093/mp/sss148 [Epub ahead of print]
38
Simontacchi M. Garcia-Mata C. Bartoli C. G. Santa-Maria G. E. Lamattina L. (2013) Nitric oxide as a key component in hormone-regulated processes.Plant Cell Rep.32853–866.10.1007/s00299-013-1434-1
39
Sokolovski S. Blatt M. R. (2004) Nitric oxide block of outward-rectifying K+ channels indicates direct control by protein nitrosylation in guard cells.Plant Physiol.1364275–4284.10.1104/pp.104.050344
40
Spoel S. H. Koornneef A. Claessens S. M. C. Korzelius J. P. Van Pelt J. A. Mueller M. J. (2003) NPR1 Modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol.Plant Cell15760–770. 10.1105/tpc.009159
41
Stamler J. S. Lamas S. Fang F. C. (2001) Nitrosylation.the prototypic redox-based signaling mechanism. Cell106675–683.10.1016/S0092-8674(01)00495-0
42
Tada Y. Spoel S. H. Pajerowska-Mukhtar K. Mou Z. Song J. Wang C. (2008) Plant immunity requires conformational charges of NPR1 via S-nitrosylation and thioredoxins.Science321952–956.10.1126/science.1156970
43
Tan X. Calderon-Villalobos L. I. Sharon M. Zheng C. Robinson C. V. Estelle M. (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase.Nature446640–645.10.1038/nature05731
44
Terrile M. C. Paris R. Calderon-Villalobos L. I. Iglesias M. J. Lamattina L. Estelle M. (2012) Nitric oxide influences auxin signaling through S-nitrosylation of the Arabidopsis TRANSPORT INHIBITOR RESPONSE 1 auxin receptor.Plant J.70492–500.10.1111/j.1365-313X.2011.04885.x
45
Tirichine L. Sandal N. Madsen L. H. Radutoiu S. Albrektsen A. S. Sato S. (2007) A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis.Science315104–107.10.1126/science.1132397
46
To J. P. Kieber J. J. (2008) Cytokinin signaling: two-components and more.Trends Plant Sci.1385–92.10.1016/j.tplants.2007.11.005
47
Tognetti V. B. Muhlenbock P Van Breusegem F. (2012) Stress homeostasis – the redox and auxin perspective.Plant Cell Environ.35321–333.10.1111/j.1365-3040.2011.02324.x
48
Tun N. N. Livaja M. Kieber J. J. Scherer G. F. (2008) Zeatin-induced nitric oxide (NO) biosynthesis in Arabidopsis thaliana mutants of NO biosynthesis and of two-component signaling genes.New Phytol.178515–531.10.1111/j.1469-8137.2008.02383.x
49
Vlot A. C. Dempsey D. A. Klessig D. F. (2009) Salicylic acid, a multifaceted hormone to combat disease.Annu. Rev. Phytopathol.47177–206.10.1146/annurev.phyto.050908.135202
50
Wang Y. Q. Feechan A. Yun B. W. Shafiei R. Hofmann A. Taylor P. (2009) S-nitrosylation of AtSABP3 antagonizes the expression of plant immunity.J. Biol. Chem.2842131–2137.10.1074/jbc.M806782200
51
Werner T. Motyka V. Laucou V. Smets R. Van Onckelen H. Schmulling T. (2003) Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity.Plant Cell152532–2550.10.1105/tpc.014928
52
Werner T. Schmulling T. (2009) Cytokinin action in plant development.Curr. Opin. Plant Biol.12527–538.10.1016/j.pbi.2009.07.002
53
Xiao-Ping S. Xi-Gui S. (2006) Cytokinin- and auxin-induced stomatal opening is related to the change of nitric oxide levels in guard cells in broad bean.Physiol. Plant.128569–579.10.1111/j.1399-3054.2006.00782.x
Summary
Keywords
nitric oxide, phytohormones, redox mechanism, signaling, S-nitrosylation
Citation
Parí R, Iglesias MJ, Terrile MC and Casalongué CA (2013) Functions of S-nitrosylation in plant hormone networks. Front. Plant Sci. 4:294. doi: 10.3389/fpls.2013.00294
Received
30 April 2013
Accepted
15 July 2013
Published
01 August 2013
Volume
4 - 2013
Edited by
Emmanuel Baudouin, Université Pierre et Marie Curie - Paris 6, France
Reviewed by
Georgia Tanou, Aristotle University of Thessaloniki, Greece; Christian Lindermayr, Helmholtz Zentrum München - German Research Center for Environmental Health, Germany
Copyright
© París, Iglesias, Terrile and Casalongué.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Claudia A. Casalongué, Instituto de Investigaciones Biológicas, Facultad de Ciencias Exactas y Naturales, Unidade Ejecutora-Consejo Nacional de Investigaciones Cientïficas y Técnicas - Universidad Nacional de Mar del Plata, Funes 3250, CC 1245, 7600 Mar del Plata, Argentina e-mail: casalong@mdp.edu.ar
This article was submitted to Frontiers in Plant Physiology, a specialty of Frontiers in Plant Science.
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.