- Laboratório de Biologia Molecular da Assimilação do Azoto, Instituto de Biologia Molecular e Celular, Universidade do Porto, Porto, Portugal
Nitric oxide (NO) is emerging as an important regulatory player in the Rhizobium-legume symbiosis. The occurrence of NO during several steps of the symbiotic interaction suggests an important, but yet unknown, signaling role of this molecule for root nodule formation and functioning. The identification of the molecular targets of NO is key for the assembly of the signal transduction cascade that will ultimately help to unravel NO function. We have recently shown that the key nitrogen assimilatory enzyme glutamine synthetase (GS) is a molecular target of NO in root nodules of Medicago truncatula, being post-translationally regulated by tyrosine nitration in relation to nitrogen fixation. In functional nodules of M. truncatula NO formation has been located in the bacteroid containing cells of the fixation zone, where the ammonium generated by bacterial nitrogenase is released to the plant cytosol and assimilated into the organic pools by plant GS. We propose that the NO-mediated GS post-translational inactivation is connected to nitrogenase inhibition induced by NO and is related to metabolite channeling to boost the nodule antioxidant defenses. Glutamate, a substrate for GS activity is also the precursor for the synthesis of glutathione (GSH), which is highly abundant in root nodules of several plant species and known to play a major role in the antioxidant defense participating in the ascorbate/GSH cycle. Existing evidence suggests that upon NO-mediated GS inhibition, glutamate could be channeled for the synthesis of GSH. According to this hypothesis, GS would be involved in the NO-signaling responses in root nodules and the NO-signaling events would meet the nodule metabolic pathways to provide an adaptive response to the inhibition of symbiotic nitrogen fixation by reactive nitrogen species.
Introduction
Leguminous plants associated with symbiotic bacteria of the family Rhizobiaceae are able to grow under nitrogen-limiting conditions. Key to this achievement is the bacterial ability to reduce atmospheric nitrogen in a functional symbiotic interaction, in which ammonia is provided to the plant and assimilated into organic composition by the plant enzyme glutamine synthetase (GS; EC 6.3.1.2). The establishment of this symbiosis requires a constant fine-tuned signal exchange between plant and bacteria culminating with the formation of a novel organ, the root nodule, which provide an environment suitable for bacterial nitrogen fixation (Oldroyd et al., 2011). Symbiotic nitrogen fixation is of particular agricultural and ecological importance, as it constitutes one of the largest contributions to biologically available nitrogen in the biosphere. Therefore, the identification of the regulatory signaling network underlying the symbiotic interaction is of utmost importance and has been the subject of intense research (for recent reviews, see Oldroyd et al., 2011; Udvardi and Poole, 2013). In recent years, nitric oxide (NO), widely recognized as an endogenous signaling molecule, emerged as an important player in the legume–rhizobium interaction, but its mechanisms of action are still far from being understood (Besson-Bard et al., 2008; Neill et al., 2008; Meilhoc et al., 2011; Puppo et al., 2013). To unravel the signal transduction cascade and ultimately NO function, it is necessary to identify its molecular targets. We have recently shown that GS, a key enzyme for nodule functioning, is a molecular target of NO, being post-translationally regulated by tyrosine nitration in relation to active nitrogen fixation (Melo et al., 2011). In functional nodules of Medicago truncatula NO production has been located in the bacteroid containing cells of the nodule fixation zone (Baudouin et al., 2006), where glutamine synthetase is highly abundant. The location of the enzyme at the sites of NO production together with its position at the center of the complex matrix of nitrogen metabolism conveys an important role of the enzyme at the crossroads of signaling events. We propose that the regulation of glutamine synthetase by NO is related to metabolite channeling to boost the nodule antioxidant defenses, linking NO signaling with nitrogen metabolism. This article discusses this hypothesis in view of the existing evidence supporting a role of glutamine synthetase in the NO signaling cascade in root nodules.
Evidence for a Signaling Role of No in the Symbiotic Interaction
The formation of NO and its involvement in the legume-rhizobia symbiosis has been the subject of much research in the last few years. It is now well established that the molecule is produced in root nodules and is important both for nodule development and functioning (Meilhoc et al., 2011; Wang and Ruby, 2011; Puppo et al., 2013). Nodule formation is highly complex and involves a progression of temporally and spatially regulated events, which require extensive recognition and signaling by both partners. The first signal is plant-released flavonoids and related compounds, which elicit synthesis of lipochito-oligosaccharides (Nod factors) by rhizobia. Nod factors induce cell division in the inner root cortex and the formation of a nodule primordium. In parallel, bacteria enter the root hairs via infection threads, are released to the plant cells by endocytosis and remain surrounded by a plant-derived symbiosome membrane (Oldroyd et al., 2011). As the nodule primordia continue to grow, new plant cells are continuously being infected and fully developed legume nodules contain a large central tissue harboring thousands of nitrogen fixing bacteria. The fixed nitrogen is exported as ammonium to the plant cytosol where it is assimilated into organic compounds by plant GS. In exchange for reduced nitrogen from the bacteria, the plant provides rhizobia with reduced carbon and all the essential nutrients required for bacterial metabolism. As nitrogenase is strongly inhibited by oxygen, nitrogen fixation is made possible by the microaerophilic conditions prevailing in the nodule, where the oxygen concentration is controlled by a variable-permeability barrier in the nodule parenchyma and by leghemoglobin, an oxygen-binding plant protein regulating and delivering oxygen to the infected cells (Udvardi and Poole, 2013). As such, the process of nodulation involves infection, development and metabolic processes and the signals exchanged between the two partners will encompass very different physiological contexts. NO is known to be involved in physiological processes ranging from biotic and abiotic stress responses, to normal plant growth and development (Besson-Bard et al., 2008). A number of reports document that NO is involved in the signaling network in root nodules, both at the early steps of plant–bacteria interaction and at later stages in mature nitrogen-fixing nodules, suggesting distinct roles of the molecule at different steps of the symbiosis. This subject has been reviewed comprehensively elsewhere (Meilhoc et al., 2011; Wang and Ruby, 2011; Puppo et al., 2013). Here we recapitulate very briefly the recent disclosures obtained using the model legume Medicago truncatula and its symbiotic partner Sinorhizobium meliloti.
During early steps of the M. truncatula–S. meliloti interaction, NO has been detected both at the infection sites and in the nodule primordia, suggesting an involvement of NO in both bacterial infection and nodule organogenesis. Evidence for an important role of the molecule in nodule formation was given by the finding that NO depletion resulted in a significant delay in nodule appearance and provoked the down regulation of genes involved in nodule development (del Giudice et al., 2011). In fully developed root nodules NO has been located exclusively in the infected cells and appears to be confined to the nodule fixation zone, pointing to an involvement of the molecule in root nodule metabolism (Baudouin et al., 2006; Horchani et al., 2011). A metabolic function for NO in providing a significant energy input in mature nitrogen-fixing nodules through the nitrate-NO respiration process has been recently highlighted (Horchani et al., 2011). NO has also been shown to modulate the expression of a wide number of genes both from S. meliloti (Meilhoc et al., 2010) and M. truncatula (Ferrarini et al., 2008). Many of the NO-responsive M. truncatula genes are involved in nodule development and functioning, with a significant number of the NO-responsive genes being involved in primary metabolism, further supporting a signaling role of NO in the nodule metabolic pathways (Ferrarini et al., 2008). More recently, NO production has also been associated with nodule senescence. Using both genetic and pharmacological approaches, it was shown that NO accumulation in aging nodules of M. truncatula has deleterious effects on the symbiosis by inhibiting nitrogen fixation and activating nodule senescence, whereas a decrease in NO levels leads to a delay in nodule senescence (Cam et al., 2012).
The origin of NO in plants is still not clearly understood, and in root nodules the picture is even more complex because the source of NO is probably variable at different stages of the symbiotic interaction and can arise from both symbiotic partners (Meilhoc et al., 2011). Several routes capable of yielding NO in root nodules have been described: NO synthase (NOS)-like activity converting arginine to citrulline and NO (Cueto et al., 1996; Baudouin et al., 2006; Leach et al., 2010), and nitrate reductase and the electron transfer chains from both plants and bacteria (Mesa et al., 2004; Meakin et al., 2007; Gupta et al., 2011a; Horchani et al., 2011).
Nitric oxide can signal fundamental physiological processes by changing both gene expression and protein function and a major step towards understanding the mechanisms regulated by NO during the symbiosis relies on the identification of its molecular targets. This task is made difficult, because the physiological contexts underlying discrete symbiotic stages are highly variable, ranging from infection, to development and senescence and thus the molecular targets of NO are expected to vary at different stages of the symbiotic interaction. While considerable effort is being put forward to identify the molecular targets of NO using large scale approaches, either by proteomics (Cecconi et al., 2009; Chaki et al., 2009; Lozano-Juste et al., 2011) or transcriptomics (Ferrarini et al., 2008; De Michele et al., 2009; Boscari et al., 2013), GS was identified as a molecular target of NO by a simple biochemical approach (Melo et al., 2011).
Evidence for a Crucial Role of the Nodule Antioxidant Responses in Nitrogen Fixation
Whilst it is now evident that NO is required for nodule functioning, paradoxically it is also clear that it is a potent inhibitor of nitrogenase activity (Trinchant and Rigaud, 1982; Kato et al., 2010). The involvement of NO in nitrogenase inactivation has been demonstrated in soybean and Lotus after nitrate supply (Kanayama et al., 1990; Meakin et al., 2007; Kato et al., 2010). In Lotus japonicus, the artificial application of the NO donor sodium nitroprusside (SNP) decreased nitrogen fixation, whereas the application of a NO scavenger (cPTIO) had the opposite effect (Shimoda et al., 2009; Kato et al., 2010). Thus, the NO concentration inside the nodule needs to be maintained at levels compatible with nitrogenase activity, but still be sufficient to achieve its signaling function. This implies a balance between NO production and detoxification. The plant antioxidant responses are therefore of crucial importance to maintain nodule functioning (Pauly et al., 2006; Becana et al., 2010; Sanchez et al., 2011). Most of the antioxidants in legume nodules are also present in other plant organs or tissues, but the concentrations in nodules are generally higher, denoting a connection between N2 fixation and the antioxidant response (Puppo et al., 2013). The data published to date indicate that hemoglobins (Hbs) and the GSH/ascorbate pathway constitute the chief antioxidant mechanisms in root nodules (Becana et al., 2010) and will be considered separately.
Hemoglobins
The levels of NO inside the nodule appear to be controlled by Hbs, which are able to scavenge NO, and in this way may protect nitrogenase from inactivation. In legumes, three types of Hb have been described: symbiotic Hb (Lb), non-symbiotic Hb (nsHb) and truncated Hb (trHb; Bustos-Sanmamed et al., 2011). The nsHbs are subdivided into nsHb-1s (class 1 nsHbs), which have a very high affinity for O2, and nsHb-2s (class 2 nsHbs), which have lower affinity for O2 and are similar to the sHbs (Gupta et al., 2011b). The first evidence of NO binding to Hb was given by the detection of nitroso-leghemoglobin complexes (LbNO) in nodules of soybean and Lotus (Kanayama et al., 1990; Mathieu et al., 1998; Meakin et al., 2007; Sanchez et al., 2010). Later, this NO-scavenging function has also been attributed to non-symbiotic class 1 Hbs (nsHb1) in Lotus japonicus (Shimoda et al., 2009) and more recently the three types of Hb were found to be expressed in nodules of Lotus japonicus, suggesting complementary roles of the different types of Hb for root nodule formation and/or functioning (Bustos-Sanmamed et al., 2011). Because class 1 nsHbs have an extreme affinity for O2, it is unlikely that they function as O2 transporters, stores, or sensors, therefore they have been supposed to play the role of NO scavenger in NO detoxifying pathways (Gupta et al., 2011b; Igamberdiev et al., 2011). These proteins are induced upon symbiotic infection, accumulate in nitrogen fixing nodules and their overexpression enhances symbiotic N2 fixation, further supporting a role in NO quenching in root nodules (Shimoda et al., 2009). An NO scavenging role has also been attributed to the flavohemoprotein Hmp of the bacterial partner (Meilhoc et al., 2010). Indeed, using S. meliloti hmp mutant strains and Hmp overexpressing strains, it was recently shown that this protein can modulate the levels of NO inside the nodules (Cam et al., 2012). A direct relationship between NO scavenging by Hbs and nitrogen fixation is reinforced by the fact that the over-expression of either plant ns-Hb1 in the plant partner (Nagata et al., 2008; Shimoda et al., 2009) or bacterial Hbs in the rhizobial partner (Ramirez et al., 1999; Cam et al., 2012) lead to enhanced symbiotic N2 fixation, whereas this process is impaired in rhizobial hmp- mutants in M. truncatula (Meilhoc et al., 2010; Cam et al., 2012). All together, the available data suggest that both the plant and the bacterial Hbs are involved in the signaling responses to NO and are important for N metabolism in root nodules.
GSH/Ascorbate Cycle
The GSH/ascorbate pathway provides one of the main antioxidant mechanisms in plants and several lines of evidence indicate that this pathway is a major contributor to the antioxidant defenses in nodules (reviewed in Matamoros et al., 2003; Pauly et al., 2006; Becana et al., 2010; Puppo et al., 2013). In legume root nodules there is a close positive correlation between nitrogenase activity, ascorbate and glutathione (GSH)/homoglutathione content (Dalton et al., 1993; Matamoros et al., 2003; El Msehli et al., 2011). The thiol tripeptides GSH and hGSH are known to be at high concentrations in nodules and to play key roles in both nodule formation and functioning (Frendo et al., 2005; Pauly et al., 2006; El Msehli et al., 2011). The substrates for GSH and hGSH synthesis are glutamate and cysteine and the pathway involves two ATP-dependent steps. In the first reaction, γ-glutamyl-cysteine synthetase (γ ECS; EC 6.3.2.2) catalyses the formation of γ-glutamylcysteine, and in the second reaction, glycine or β-alanine is added to the C-terminal site of γ-glutamylcysteine by GSH synthetase (GSHS; EC 6.3.2.3) or hGSH synthetase (hGSHS), respectively (Frendo et al., 1999, 2001). Recently, it was shown that GSHS and hGSHS follow a tissue-specific pattern of expression in the nodules of M. truncatula, pointing to a tissue-specific differential regulation of GSH and hGSH synthesis in M. truncatula (El Msehli et al., 2011). The importance of (h)GSH for nitrogen fixation was recently evidenced by studies in transgenic nodules with decreased or increased (h)GSH content in the nitrogen-fixing zone. These studies showed that the concentration of (h)GSH regulates nitrogen fixation efficiency and that a deficiency in (h)GSH impairs nodule growth (El Msehli et al., 2011).
Glutathione can readily react with NO to form S-nitrosoglutathione (GSNO) and may play an important role in regulating NO bioactivity. While the half-life of NO in biological systems is only a few seconds, GSNO is relatively stable and thought to function as a NO reservoir, since it can release NO or function as a transnitrosylating agent. The key enzyme regulating GSNO pools is S-nitrosoglutathione reductase (GSNOR), reducing GSNO to ultimately produce glutathione disulfide (GSSG), which can be reduced by glutathione reductase (GR) to re-enter the GSH pool and ammonia, which can be re-assimilated by GS (Liu et al., 2001; Lamotte et al., 2005).
Interestingly, it was reported that GSH is produced in response to elevated NO in roots of M. truncatula (Innocenti et al., 2007). As GS activity is inhibited by NO and one of its substrates, glutamate is also a substrate for (h)GSH synthesis, we proposed that upon NO-induced inhibition of GS, glutamate could be channeled to the synthesis of (h)GSH, contributing in this way, to the nodule antioxidant defenses and to the protection of nitrogenase from inactivation by NO. This aspect will be further discussed in the last section of this article.
Evidence for the Regulation of Glutamine Synthetase Activity by No
Glutamine synthetase is abundantly present in root nodules where it plays a pivotal role in the assimilation of the ammonium released by nitrogen fixation. The enzyme catalyses the ATP-dependent condensation of ammonium with glutamate to yield glutamine, which can be directly exported from the nodules or used to synthetize asparagine, the main nitrogen export compound in indeterminate nodules (Vance, 2008). In the model legume M. truncatula GS is encoded by four expressed genes, two (MtGS1a and MtGS1b) encoding cytosolic isoenzymes, and two (MtGS2a and MtGS2b) encoding plastid located isoenzymes (Stanford et al., 1993; Carvalho et al., 2000a,b; Melo et al., 2003; Seabra et al., 2010), the latter of which is exclusively expressed in the seeds and is unique to M. truncatula and closely related species (Seabra et al., 2010). The other three GS genes are expressed in root nodules, but MtGS1a is highly up regulated, accounting for the production of over 90% of the total nodule GS activity, and encodes the isoenzyme responsible for the assimilation of the ammonia released by nitrogen fixation (Carvalho et al., 2000a). We have previously shown that MtGS1a is abundantly present in the infected cells of the nodule fixation zone (Carvalho et al., 2000a), coinciding with the major site of NO formation in this model species (Baudouin et al., 2006; Horchani et al., 2011). The enzyme is thus in vivo accessible to the oxidative effects induced by this reactive compound and it was shown to be a molecular target of NO in root nodules (Melo et al., 2011). In vitro studies using purified recombinant enzymes, demonstrated that the M. truncatula nodule enzyme MtGS1a is subjected to tyrosine nitration and that this modification provokes a total loss of enzyme activity (Melo et al., 2011). It is noteworthy that the plastid located GS isoenzyme, MtGS2a, which is also expressed in root nodules but at considerably lower levels, is also affected by NO, but by a different mechanism, cysteine nitrosylation. The finding that two isoenzymes that share a high degree of sequence homology and a remarkably conserved active site fold are differentially modified by NO, strengthens the idea that the NO signaling effects are specific under different physiological contexts. In addition to a differential sensitivity of individual GS isoenzymes to NO, the differential localization of the isoenzymes in specific organelles and/or plant tissues is likely to be implied in the NO-mediated regulation of GS activity. Future studies should address the regulation of the plastid located GS isoenzyme by S-nitrosylation. The enzyme is also expressed in the infected cells of root nodules and its expression is positively correlated with active nitrogen fixation (Melo et al., 2003). Here, we will focus on the regulation of MtGS1a by NO, because it is the M. truncatula GS isoenzyme responsible for the assimilation of the ammonium released by bacterial nitrogenase.
Mechanistic of MtGS1a Inactivation by Tyrosine Nitration
Protein tyrosine nitration is a post-translational modification (PTM) mediated by reactive nitrogen species (RNS), resulting from the addition of a nitro (–NO2) group to one of two equivalent ortho carbons in the aromatic ring of tyrosine residues (Radi, 2004). The incorporation of a nitro group (–NO2) into protein tyrosines can lead to profound structural and functional changes, the most common being loss of function (Radi, 2013). This PTM has been best studied in animals and it is a relatively new area of research in higher plants. A number of nitrated proteins have been identified in plants by proteomic approaches (Cecconi et al., 2009; Chaki et al., 2009; Lozano-Juste et al., 2011; Begara-Morales et al., 2013), however the functional effects of nitration on specific proteins are Known only for a few plant proteins (Lozano-Juste et al., 2011; Corpas et al., 2013) and the physiological significance of this PTM remains largely unknown. The tyrosine nitration of MtGS1a has become a good case study on how nitration of tyrosines can promote conformational changes leading to a loss of function. Furthermore, the nitration of MtGS1a in root nodules negatively correlates with active nitrogen fixation, strongly suggesting that the nitration of the enzyme is physiologically relevant for root nodule functioning, an aspect that will be further discussed in the next section.
Tyrosine nitration is considered a selective process, and typically only one or two of the tyrosine residues present in a protein become preferentially nitrated, depending on the structural environment (Abello et al., 2009). By site-directed mutagenesis it was shown that at least two of the 19 tyrosine residues of MtGS1a are prone to nitration, as the substitution of either Tyr 167 or Tyr 263 to phenylalanine reduced by half the protein anti-nitrotyrosine immunoreactivity. However, only mutation on Tyr 167 results in a significant reduction in the NO-mediated inhibitory effect, thus indicating that it is the relevant regulatory site (Melo et al., 2011). Since the three-dimensional structure of MtGS1a is available (Seabra et al., 2009), it was possible to enlighten the structural basis by which the nitration of Tyr167 leads to enzyme inactivation. An analysis of the structural environment of Tyr167 revealed that this residue is located in a solvent-accessible loop, close to the enzyme active site and in close proximity to a basic residue (Lys-137). In the M. truncatula enzyme, Tyr-167 establishes a hydrogen bond with Lys-137, and the nitration of this residue could prevent the formation of this bond, which appears to be important to maintain a correct conformation of the active site and is expected to interfere with the catalytic activity of the enzyme (Figure 1). Thus, the mechanism of MtGS1a inactivation by tyrosine nitration can be elucidated in structural terms. A rare example, since protein tyrosine nitration is a non-enzymatic mechanism based on free radical reactions and its selectivity for target residues in proteins is far from obvious.
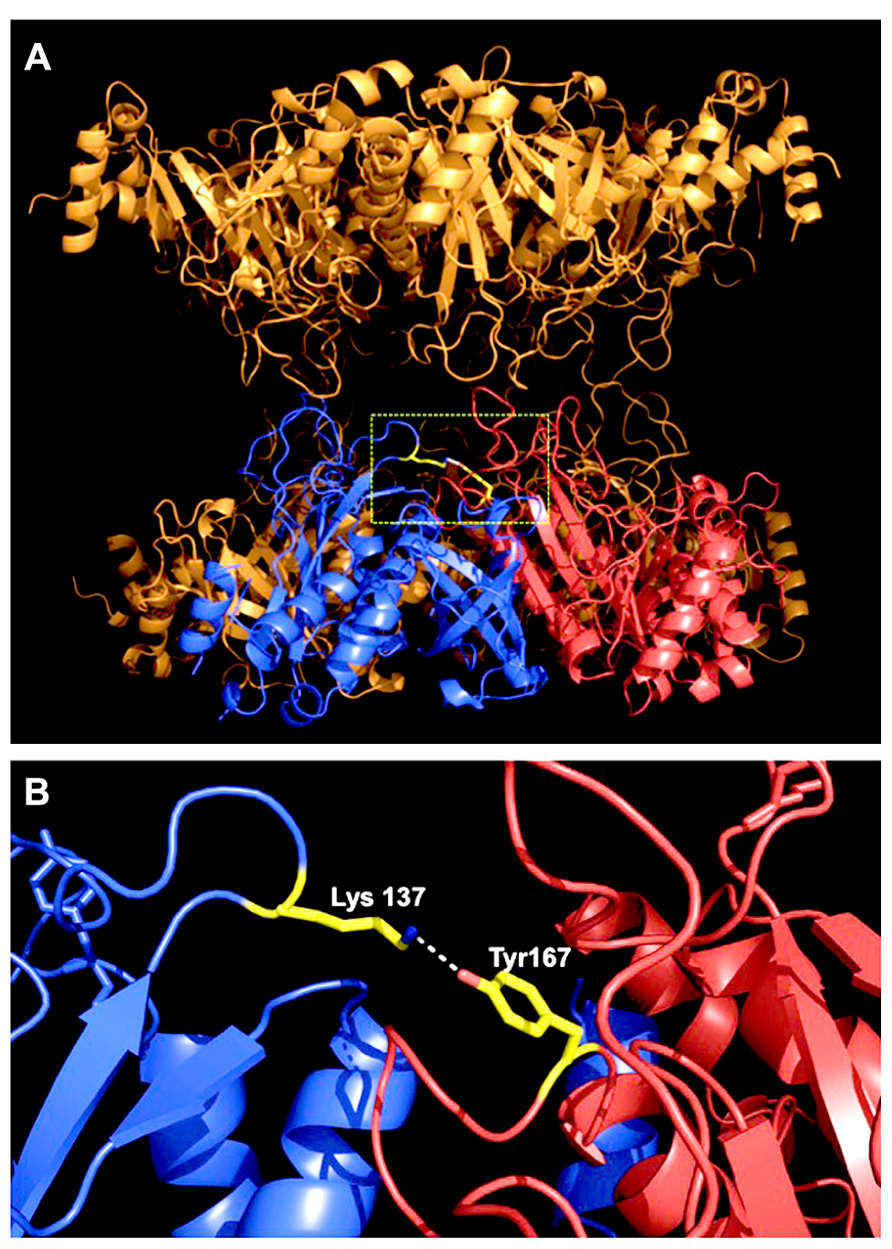
FIGURE 1. Location of the regulatory nitration site within the three-dimensional structure of MtGS1a. (A) Side-view of the MtGS1a molecule, which is a decamer composed of two stacked (face-to-face) pentameric rings, with 10 active sites formed between the C-terminal domain of one subunit and the N-terminal domain of the other subunit within a pentameric ring (Seabra et al., 2009). The position of Tyr167 is shown in yellow, in a solvent-accessible loop at the interface between two neighboring subunits, which are colored blue and red. (B) Arrangement of two neighboring subunits, highlighting the position of tyrosine 167 of the subunit labeled in red, close to the enzyme active site, and establishing an hydrogen bound with Lys-137 of the neighboring subunit, which is presented in blue.
Physiological Significance of GS Nitration for Root Nodule Functioning
Glutamine synthetase in conjunction with NADH-glutamate synthase (NADH-GOGAT, EC 1.4.1.14) operates the GOGAT cycle leading to the synthesis of glutamine and glutamate, which then serve as nitrogen donors for the biosynthesis of essentially all nitrogenous compounds. In temperate legumes, fixed nitrogen is exported from the nodules to the rest of the plant mainly as asparagine, which is synthesized by the concerted action of two additional enzymes, aspartate aminotransferase (AAT, EC 2.6.1.1) and asparagine synthetase (AS, EC 6.3.5.4). Being the first enzyme of the pathway, GS is placed in a key position to play a regulatory role in the nitrogen assimilatory pathways in nodules. The finding that it is a molecular target of NO, is thus particularly interesting. The formation of NO by plants is necessarily closely linked to nitrogen metabolism, since it is produced from inorganic (reduction of nitrate via nitrite; Horchani et al., 2011), or organic nitrogen sources like arginine via NOS-like activity (Cueto et al., 1996; Baudouin et al., 2006) and potentially polyamines, via a yet non-identified polyamine oxidation pathway (Gupta et al., 2011a; Meilhoc et al., 2011). Research in nodules of M. truncatula established that NO accumulation is not a by-product of symbiotic nitrogen fixation (Baudouin et al., 2006), however any of the NO generating pathways that have been described will require adequate nitrogen supply at the sites of NO production. GS is a key enzyme in nitrogen metabolism and in addition to its vital role in primary N assimilation is also crucial in N recycling in plants. Therefore, the regulation of GS by NO establishes a connection between NO signaling and N metabolism.
Nitric oxide is a strong inhibitor of nitrogenase activity, and it seems reasonable that the same signaling molecule also inhibits GS, the enzyme that uses the product of nitrogenase activity as a substrate. Indeed, in root nodules of M. truncatula it was shown that GS is inactivated by tyrosine nitration in planta and that the GS nitration status is positively correlated with the inhibition of nitrogen fixation (Melo et al., 2011). The GS nitration status was quantified in planta in situations where nitrogen fixation is impaired and NO is known to be produced, namely in ineffective nodules, induced either by nifH- or fixJ-rhizobial strains, as well as in nodules fed with nitrate or treated with the NO donor SNP. A direct relationship could be established between increased GS nitration, reduced nodule GS activity and reduced nitrogen fixation activity, strongly suggesting that GS is post-translationally inactivated by NO-mediated nitration in response to lower nitrogen fixation rates (Melo et al., 2011). NO concentration is expected to raise in root nodules following nitrate application (Kanayama et al., 1990; Kato et al., 2010), however in ineffective nodules NO production appears to be unaffected (Baudouin et al., 2006) and thus it seems that the regulation of GS activity by tyrosine nitration is a specific process associated with nitrogen fixation rather than a general effect resulting from increased NO levels inside the nodule.
Additional evidence for a specific regulation of GS by NO in root nodules is given by recent studies using S. meliloti strains carrying a mutation in the gene encoding flavohemoglobin (hmp), which is involved in NO degradation and leads to increased NO content inside the nodules(Meilhoc et al., 2010; Cam et al., 2012). Quantification of GS nitration in hmp- mutant nodules revealed a considerable increase in GS nitration in relation to wild type nodules, with a concomitant decrease in GS activity (H. Carvalho, unpublished results). As it has been shown that nodules formed by the hmp- mutant rhizobium suffer a premature senescence induced by NO (Cam et al., 2012), it is tempting to speculate that the NO-induced GS inhibition could be associated with this premature nodule senescence. This idea is supported by the finding that the application of the GS inhibitor phosphinothricin (PPT) to root nodules promotes nodule senescence (Seabra et al., 2012).
The finding that the root enzymes appear to respond differently to NO also supports a specific role of GS in the NO signaling response in root nodules. Following nitrate supply, the GS nitration status was found to be unaffected in roots but increased in root nodules (Melo et al., 2011). The total amount of nitrated proteins, which was quantified by direct ELISA using a specific anti-nitrotyrosine antibody, increases in both the roots and the nodules following nitrate supply, but GS does not appear to be among these proteins in the roots (H. Carvalho, unpublished results). It is noteworthy that in M. truncatula roots, GS is mainly composed of a different cytosolic isoenzyme, MtGS1b, which is largely located in the root cortex, whereas MtGS1a is confined to the root vascular tissues (Carvalho et al., 2000b). It is probable that both the formation of NO at the sites of expression of each individual GS isoenzyme and the differential sensitivity of the two isoenzymes to NO account for the differential regulation of GS in roots and root nodules. This is in agreement with the general idea that the effects of NO are not simply a consequence of the amount of NO produced but, are determined by the local environment in which NO is released and the nature of the generated RNS, which in turn will be dependent of the cellular redox state, the bioavailability of NO-generating enzyme substrates, the nature and proximity of molecular targets and of NO-metabolizing proteins.
Taken together, the available information supports a role of NO in mediating GS activity in root nodules as a function of the nitrogen fixation status, rather than this being a consequence of a general increase in NO concentration inside the nodule.
Proposed Model for the Involvement of GS in the No Signaling Pathway in Root Nodules
We hypothesize that the inactivation of GS by tyrosine nitration is an NO-mediated regulatory process important for nodule functioning. In view of the overall available evidence, which was described in the previous sections, we propose a model to explain the involvement of GS in the NO signaling pathway in root nodules (Figure 2). According to this model, the inhibition of GS activity by tyrosine nitration would be directly related to the NO-induced nitrogenase inhibition. In view of the fact that elevated levels of NO in root nodules lead to decreased production of ammonium for GS assimilation, the enzyme would be shut down by post-translational inactivation through tyrosine nitration in response to the signal NO, the same signal that shuts down nitrogenase. This way, NO would be placed as a regulatory molecule coordinating N fixation and assimilation in root nodules.
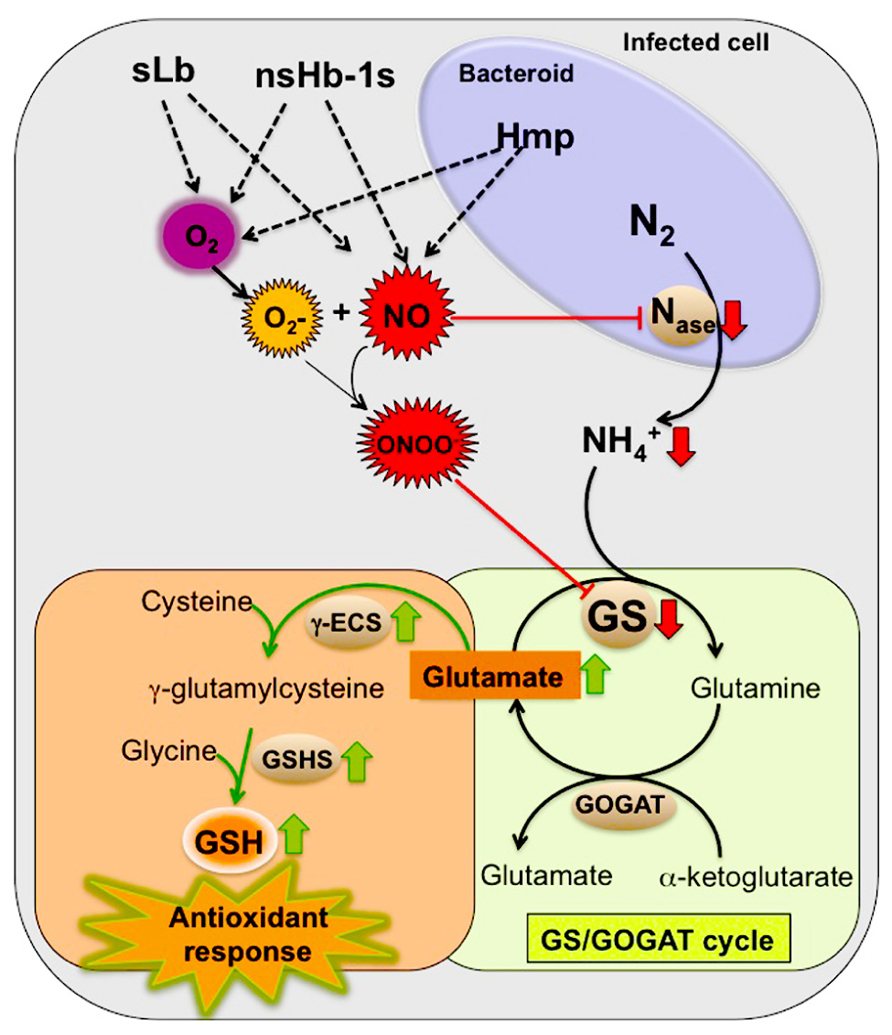
FIGURE 2. Proposed model for the involvement of GS in the NO signaling events in root nodules by contributing to the nodule antioxidant responses. Red arrows indicate down regulation by NO and the green arrows indicate up regulation by NO. Enzymes: nitrogenase (Nase), glutamine synthetase (GS), glutamate synthase (GOGAT), γ-glutamylcysteine synthetase (γ-ECS) and glutathione synthetase (GSHS). Hemoglobins: symbiotic leghemoglobin (sLb), class 1 non-symbiotic hemoglobins (nsHb-1s), rhizobial flavohemoprotein (Hmp).
We further propose that the NO-induced GS inhibition is involved in the nodule antioxidant response to NO and related RNS. Glutamate, a substrate for GS activity is also the precursor for the synthesis of GSH, which as described in a previous section, is known to be highly abundant in root nodules of several plant species and to play a major role in the antioxidant defense participating in the GSH/ascorbate cycle (Matamoros et al., 1999b, 2003; Becana et al., 2010). Upon NO-mediated GS inhibition, glutamate could be channeled for the synthesis of GSH contributing to neutralize the deleterious effects of RNS. This idea is supported by the finding that the synthesis of the two enzymes involved in GSH production from glutamate, γ-glutamylcysteine synthetase (γ-ECS) and GSHS is up regulated by NO in M. truncatula, correlating with the accumulation of the end product GSH (Innocenti et al., 2007). According to this theory, GS would be involved in the NO signaling pathway, functioning both as a sensor of increased levels of NO inside the nodules and as an activator, by forcing the N metabolic pathways to shift from primary N assimilation to the synthesis of GSH. This, in turn, would boost the nodule antioxidant responses and adjust the levels of NO inside the nodule. Since GSNO, formed by the reaction of NO with GSH, is thought to function as a mobile reservoir of NO bioactivity, GS would play an additional share in the NO signaling cascade by contributing to storage of the signaling molecule in the form of GSNO. NO release from GSNO would then be controlled by the enzyme GSNOR (Leterrier et al., 2011).
The proposed model also predicts that Hbs are important players in the process by regulating the levels of both O2 and NO, which may compete for binding sites, controlling in this way, the formation of peroxynitrite (ONOO–). Peroxynitrite is probably the main nitrating agent in vivo and is formed rapidly in the reaction of the superoxide anion () with NO (Abello et al., 2009; Arasimowicz-Jelonek and Floryszak-Wieczorek, 2011). As discussed before, it is documented that at least three types of Hbs have the capacity to scavenge NO, contributing in this way to modulate NO bioactivity and protecting nitrogenase from inactivation (Kanayama et al., 1990; Herold and Puppo, 2005; Meakin et al., 2007; Sanchez et al., 2010). We thus anticipate the participation from the plant side, of leghemoglobin and non-symbiotic Hb, pointing to class1 nsHb as the best candidates, and from the bacterial side the flavohemoprotein Hmp.
According to the proposed model, the NO-signaling events would meet the nodule metabolic pathways to provide an adaptive response to the inhibition of symbiotic nitrogen fixation by RNS.
Conclusion
Post-translational nitration of key enzymes and the subsequent alteration of their catalytic properties may represent a new level of regulation of primary metabolism. Here we propose that the key nitrogen metabolic enzyme, glutamine synthetase is involved in the NO signaling pathways in root nodules by shifting primary N assimilation to the production of GSH in response to increased NO. For a signaling molecule to be effective, it needs to be produced quickly on demand, induce defined effects within a cell and to be removed rapidly and effectively when it is no longer required. According to the proposed hypothesis, GS would be involved in NO sensing and removal and also in NO storage by controlling GSNO pools. This mechanism would be important, on one hand to coordinate N-fixation and assimilation in the nodules and on the other hand, to boost the antioxidant defenses of the nodule in response to NO. The proposed model conveys an important role for the enzyme at the crossroads of signaling events, connecting nitrogen metabolism to NO production, storage and detoxification.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge Pedro Pereira for the structural analysis of MtGS1a and Claude Bruand and Eliane Meilhoc for kindly providing the flavohemoprotein hmp- rhizobium mutant strain. We would like to thank Nuno Leitão and Ana Campilho for helpful discussions and for critically revising this manuscript. We acknowledge support from community funds (FEDER) through the Operational Competitiveness Program COMPETE and from national funds through Fundação para a Ciência e a Tecnologia (FCT) under the projects FCOMP-01-0124-FEDER-028335 (PTDC/BIA-PLA/2291/2012) and FCOMP-01-0124-FEDER-008678 (PTDC/AGR-AAM/099577/2008).
References
Abello, N., Kerstjens, H. A., Postma, D. S., and Bischoff, R. (2009). Protein tyrosine nitration: selectivity, physicochemical and biological consequences, denitration, and proteomics methods for the identification of tyrosine-nitrated proteins. J. Proteome Res. 8, 3222–3238. doi: 10.1021/pr900039c
Arasimowicz-Jelonek, M., and Floryszak-Wieczorek, J. (2011). Understanding the fate of peroxynitrite in plant cells – from physiology to pathophysiology. Phytochemistry 72, 681–688. doi: 10.1016/j.phytochem.2011.02.025
Baudouin, E., Pieuchot, L., Engler, G., Pauly, N., and Puppo, A. (2006). Nitric oxide is formed in Medicago truncatula-Sinorhizobium meliloti functional nodules. Mol. Plant Microbe Interact. 19, 970–975. doi: 10.1094/MPMI-19-0970
Becana, M., Matamoros, M. A., Udvardi, M., and Dalton, D. A. (2010). Recent insights into antioxidant defenses of legume root nodules. New Phytol. 188, 960–976. doi: 10.1111/j.1469-8137.2010.03512.x
Begara-Morales, J. C., Chaki, M., Sanchez-Calvo, B., Mata-Perez, C., Leterrier, M., Palma, J. M., et al. (2013). Protein tyrosine nitration in pea roots during development and senescence. J. Exp. Bot. 64, 1121–1134. doi: 10.1093/jxb/ert006
Besson-Bard, A., Pugin, A., and Wendehenne, D. (2008). New insights into nitric oxide signaling in plants. Annu. Rev. Plant Biol. 59, 21–39. doi: 10.1146/annurev.arplant.59.032607.092830
Boscari, A., Del Giudice, J., Ferrarini, A., Venturini, L., Zaffini, A. L., Delledonne, M., et al. (2013). Expression dynamics of the Medicago truncatula transcriptome during the symbiotic interaction with Sinorhizobium meliloti: which role for nitric oxide? Plant Physiol. 161, 425–439. doi: 10.1104/pp.112.208538
Bustos-Sanmamed, P., Tovar-Mendez, A., Crespi, M., Sato, S., Tabata, S., and Becana, M. (2011). Regulation of nonsymbiotic and truncated hemoglobin genes of Lotus japonicus in plant organs and in response to nitric oxide and hormones. New Phytol. 189, 765–776. doi: 10.1111/j.1469-8137.2010.03527.x
Cam, Y., Pierre, O., Boncompagni, E., Herouart, D., Meilhoc, E., and Bruand, C. (2012). Nitric oxide (NO): a key player in the senescence of Medicago truncatula root nodules. New Phytol. 196, 548–560. doi: 10.1111/j.1469-8137.2012.04282.x
Carvalho, H., Lescure, N., de Billy, F., Chabaud, M., Lima, L., Salema, R., et al. (2000a). Cellular expression and regulation of the Medicago truncatula cytosolic glutamine synthetase genes in root nodules. Plant Mol. Biol. 42, 741–756. doi: 10.1023/A:1006304003770
Carvalho, H., Lima, L., Lescure, N., Camut, S., Salema, R., and Cullimore, J. (2000b). Differential expression of the two cytosolic glutamine synthetase genes in various organs of Medicago truncatula. Plant Sci. 159, 301–312. doi: 10.1016/S0168-9452(00)00360-5
Cecconi, D., Orzetti, S., Vandelle, E., Rinalducci, S., Zolla, L., and Delledonne, M. (2009). Protein nitration during defense response in Arabidopsis thaliana. Electrophoresis 30, 2460–2468. doi: 10.1002/elps.200800826
Chaki, M., Valderrama, R., Fernandez-Ocana, A. M., Carreras, A., Lopez-Jaramillo, J., Luque, F., et al. (2009). Protein targets of tyrosine nitration in sunflower (Helianthus annuus L.) hypocotyls. J. Exp. Bot. 60, 4221–4234. doi: 10.1093/jxb/erp263
Corpas, F. J., Palma, J. M., Del Rio, L. A., and Barroso, J. B. (2013). Protein tyrosine nitration in higher plants grown under natural and stress conditions. Front. Plant Sci. 4:29. doi: 10.3389/fpls.2013.00029
Cueto, M., Hernandez-Perera, O., Martin, R., Bentura, M. L., Rodrigo, J., Lamas, S., et al. (1996). Presence of nitric oxide synthase activity in roots and nodules of Lupinus albus. FEBS Lett. 398, 159–164. doi: 10.1016/S0014-5793(96)01232-X
Dalton, D. A., Langeberg, L., and Treneman, N. C. (1993). Correlations between the ascorbate-glutathione pathway and effectiveness in legume root nodules. Physiol. Plant. 84, 365–371. doi: 10.1111/j.1399-3054.1993.tb01743.x
del Giudice, J., Cam, Y., Damiani, I., Fung-Chat, F., Meilhoc, E., Bruand, C., et al. (2011). Nitric oxide is required for an optimal establishment of the Medicago truncatula-Sinorhizobium meliloti symbiosis. New Phytol. 191, 405–417. doi: 10.1111/j.1469-8137.2011.03693.x
De Michele, R., Formentin, E., Todesco, M., Toppo, S., Carimi, F., Zottini, M., et al. (2009). Transcriptome analysis of Medicago truncatula leaf senescence: similarities and differences in metabolic and transcriptional regulations as compared with Arabidopsis, nodule senescence and nitric oxide signalling. New Phytol. 181, 563–575. doi: 10.1111/j.1469-8137.2008.02684.x
El Msehli, S., Lambert, A., Baldacci-Cresp, F., Hopkins, J., Boncompagni, E., Smiti, S. A., et al. (2011). Crucial role of (homo)glutathione in nitrogen fixation in Medicago truncatula nodules. New Phytol. 192, 496–506. doi: 10.1111/j.1469-8137.2011.03810.x
Ferrarini, A., De Stefano, M., Baudouin, E., Pucciariello, C., Polverari, A., Puppo, A., et al. (2008). Expression of Medicago truncatula genes responsive to nitric oxide in pathogenic and symbiotic conditions. Mol. Plant Microbe Interact. 21, 781–790. doi: 10.1094/MPMI-21-6-0781
Frendo, P., Gallesi, D., Turnbullet, R., Van de Sype, G., Herouart, D., and Puppo, A. (1999). Localisation of glutathione and homoglutathione in Medicago truncatula is correlated to a differential expression of genes involved in their synthesis. Plant J. 17, 215–219. doi: 10.1046/j.1365-313X.1999.00367.x
Frendo, P., Harrison, J., Norman, C., Hernandez Jimenez, M. J., Van de Sype, G., Gilabert, A., et al. (2005). Glutathione and homoglutathione play a critical role in the nodulation process of Medicago truncatula. Mol. Plant Microbe Interact. 18, 254–259. doi: 10.1094/MPMI-18-0254
Frendo, P., Jimenez, M. J., Mathieu, C., Duret, L., Gallesi, D., Van de Sype, G., et al. (2001). A Medicago truncatula homoglutathione synthetase is derived from glutathione synthetase by gene duplication. Plant Physiol. 126, 1706–1715. doi: 10.1104/pp.126.4.1706
Gupta, K. J., Fernie, A. R., Kaiser, W. M., and van Dongen, J. T. (2011a). On the origins of nitric oxide. Trends Plant Sci. 16, 160–168. doi: 10.1016/j.tplants.2010.11.007
Gupta, K. J., Hebelstrup, K. H., Mur, L. A., and Igamberdiev, A. U. (2011b). Plant hemoglobins: important players at the crossroads between oxygen and nitric oxide. FEBS Lett. 585, 3843–3849. doi: 10.1016/j.febslet.2011.10.036
Herold, S., and Puppo, A. (2005). Oxyleghemoglobin scavenges nitrogen monoxide and peroxynitrite: a possible role in functioning nodules? J. Biol. Inorg. Chem. 10, 935–945. doi: 10.1007/s00775-005-0046-9
Horchani, F., Prevot, M., Boscari, A., Evangelisti, E., Meilhoc, E., Bruand, C., et al. (2011). Both plant and bacterial nitrate reductases contribute to nitric oxide production in Medicago truncatula nitrogen-fixing nodules. Plant Physiol. 155, 1023–1036. doi: 10.1104/pp.110.166140
Igamberdiev, A. U., Bykova, N. V., and Hill, R. D. (2011). Structural and functional properties of class 1 plant hemoglobins. IUBMB Life 63, 146–152. doi: 10.1002/iub.439
Innocenti, G., Pucciariello, C., Le Gleuher, M., Hopkins, J., de Stefano, M., Delledonne, M., et al. (2007). Glutathione synthesis is regulated by nitric oxide in Medicago truncatula roots. Planta 225, 1597–1602. doi: 10.1007/s00425-006-0461-3
Kanayama, Y., Watanabe, I., and Yamamoto, I. (1990). Inhibition of nitrogen fixation in soybean plants supplied with nitrate. I. Nitrite accumulation and formation of nitrosylleghemoglobin in nodules. Plant Cell Physiol. 31, 341–346.
Kato, K., Kanahama, K., and Kanayama, Y. (2010). Involvement of nitric oxide in the inhibition of nitrogenase activity by nitrate in Lotus root nodules. J. Plant Physiol. 167, 238–241. doi: 10.1016/j.jplph.2009.08.006
Lamotte, O., Courtois, C., Barnavon, L., Pugin, A., and Wendehenne, D. (2005). Nitric oxide in plants: the biosynthesis and cell signalling properties of a fascinating molecule. Planta 221, 1–4. doi: 10.1007/s00425-005-1494-8
Leach, J., Keyster, M., Du Plessis, M., and Ludidi, N. (2010). Nitric oxide synthase activity is required for development of functional nodules in soybean. J. Plant Physiol. 167, 1584–1591. doi: 10.1016/j.jplph.2010.06.019
Leterrier, M., Chaki, M., Airaki, M., Valderrama, R., Palma, J. M., Barroso, J. B., et al. (2011). Function of S-nitrosoglutathione reductase (GSNOR) in plant development and under biotic/abiotic stress. Plant Signal. Behav. 6, 789–793. doi: 10.4161/psb.6.6.15161
Liu, L., Hausladen, A., Zeng, M., Que, L., Heitman, J., and Stamler, J. S. (2001). A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature 410, 490–494. doi: 10.1038/35068596
Lozano-Juste, J., Colom-Moreno, R., and Leon, J. (2011). In vivo protein tyrosine nitration in Arabidopsis thaliana. J. Exp. Bot. 62, 3501–3517. doi: 10.1093/jxb/err042
Matamoros, M. A., Dalton, D. A., Ramos, J., Clemente, M. R., Rubio, M. C., and Becana, M. (2003). Biochemistry and molecular biology of antioxidants in the rhizobia-legume symbiosis. Plant Physiol. 133, 499–509. doi: 10.1104/pp.103.025619
Matamoros, M. A., Moran, J. F., Iturbe-Ormaetxe, I., Rubio, M. C., and Becana, M. (1999b). Glutathione and homoglutathione synthesis in legume root nodules. Plant Physiol. 121, 879–888. doi: 10.1104/pp.121.3.879
Mathieu, C., Moreau, S., Frendo, P., Puppo, A., and Davies, M. J. (1998). Direct detection of radicals in intact soybean nodules: presence of nitric oxide-leghemoglobin complexes. Free Radic. Biol. Med. 24, 1242–1249. doi: 10.1016/S0891-5849(97)00440-1
Meakin, G. E., Bueno, E., Jepson, B., Bedmar, E. J., Richardson, D. J., and Delgado, M. J. (2007). The contribution of bacteroidal nitrate and nitrite reduction to the formation of nitrosylleghaemoglobin complexes in soybean root nodules. Microbiology 153, 411–419. doi: 10.1099/mic.0.2006/000059-0
Meilhoc, E., Boscari, A., Bruand, C., Puppo, A., and Brouquisse, R. (2011). Nitric oxide in legume-rhizobium symbiosis. Plant Sci. 181, 573–581. doi: 10.1016/j.plantsci.2011.04.007
Meilhoc, E., Cam, Y., Skapski, A., and Bruand, C. (2010). The response to nitric oxide of the nitrogen-fixing symbiont Sinorhizobium meliloti. Mol. Plant Microbe Interact. 23, 748–759. doi: 10.1094/MPMI-23-6-0748
Melo, P. M., Lima, L. M., Santos, I. M., Carvalho, H. G., and Cullimore, J. V. (2003). Expression of the plastid-located glutamine synthetase of Medicago truncatula. Accumulation of the precursor in root nodules reveals an in vivo control at the level of protein import into plastids. Plant Physiol. 132, 390–399. doi: 10.1104/pp.102.016675
Melo, P. M., Silva, L. S., Ribeiro, I., Seabra, A. R., and Carvalho, H. G. (2011). Glutamine synthetase is a molecular target of nitric oxide in root nodules of Medicago truncatula and is regulated by tyrosine nitration. Plant Physiol. 157, 1505–1517. doi: 10.1104/pp.111.186056
Mesa, S., Alche, J. D., Bedmar, E., and Delgado, M. J. (2004). Expression of nir, nor and nos denitrification genes from Bradyrhizobium japonicum in soybean root nodules. Physiol. Plant. 120, 205–211. doi: 10.1111/j.0031-9317.2004.0211.x
Nagata, M., Murakami, E., Shimoda, Y., Shimoda-Sasakura, F., Kucho, K., Suzuki, A., et al. (2008). Expression of a class 1 hemoglobin gene and production of nitric oxide in response to symbiotic and pathogenic bacteria in Lotus japonicus. Mol. Plant Microbe Interact. 21, 1175–1183. doi: 10.1094/MPMI-21-9-1175
Neill, S., Bright, J., Desikan, R., Hancock, J., Harrison, J., and Wilson, I. (2008). Nitric oxide evolution and perception. J. Exp. Bot. 59, 25–35. doi: 10.1093/jxb/erm218
Oldroyd, G. E. D., Murray, J. D., Poole, P. S., and Downie, J. A. (2011). The rules of engagement in the legume-rhizobial symbiosis. Annu. Rev. Genet. 45, 119–144. doi: 10.1146/annurev-genet-110410-132549
Pauly, N., Pucciariello, C., Mandon, K., Innocenti, G., Jamet, A., Baudouin, E., et al. (2006). Reactive oxygen and nitrogen species and glutathione: key players in the legume-rhizobium symbiosis. J. Exp. Bot. 57, 1769–1776. doi: 10.1093/jxb/erj184
Puppo, A., Pauly, N., Boscari, A., Mandon, K., and Brouquisse, R. (2013). Hydrogen peroxide and nitric oxide: key regulators of the legume-rhizobium and mycorrhizal symbioses. Antioxid. Redox Signal. 18, 2202–2219. doi: 10.1089/ars.2012.5136
Radi, R. (2004). Nitric oxide, oxidants, and protein tyrosine nitration. Proc. Natl. Acad. Sci. U.S.A. 101, 4003–4008. doi: 10.1073/pnas.0307446101
Radi, R. (2013). Protein tyrosine nitration: biochemical mechanisms and structural basis of functional effects. Acc. Chem. Res. 46, 550–559. doi: 10.1021/ar300234c
Ramirez, M., Valderrama, B., Arredondo-Peter, R., Soberon, M., Mora, J., and Hernandez, G. (1999). Rhizobium etli genetically engineered for the heterologous expression of Vitreoscilla sp. hemoglobin: effects on free-living and symbiosis. Plant Microbe Interact. 12, 1008–1015. doi: 10.1094/MPMI.1999.12.11.1008
Sanchez, C., Cabrera, J. J., Gates, A. J., Bedmar, E. J., Richardson, D. J., and Delgado, M. J. (2011). Nitric oxide detoxification in the rhizobia-legume symbiosis. Biochem. Soc. Trans. 39, 184–188. doi: 10.1042/BST0390184
Sanchez, C., Gates, A. J., Meakin, G. E., Uchiumi, T., Girard, L., Richardson, D. J., et al. (2010). Production of nitric oxide and nitrosylleghemoglobin complexes in soybean nodules in response to flooding. Mol. Plant Microbe Interact. 23, 702–711. doi: 10.1094/MPMI-23-5-0702
Seabra, A. R., Carvalho, H., and Pereira, P. J. (2009). Crystallization and preliminary crystallographic characterization of glutamine synthetase from Medicago truncatula. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 65, 1309–1312. doi: 10.1107/S1744309109047381
Seabra, A. R., Pereira, P. A., Becker, J. D., and Carvalho, H. G. (2012). Inhibition of glutamine synthetase by phosphinothricin leads to transcriptome reprograming in root nodules of Medicago truncatula. Mol. Plant Microbe Interact. 25, 976–992. doi: 10.1094/MPMI-12-11-0322
Seabra, A. R., Vieira, C. P., Cullimore, J. V., and Carvalho, H. G. (2010). Medicago truncatula contains a second gene encoding a plastid located glutamine synthetase exclusively expressed in developing seeds. BMC Plant Biol. 10:183. doi: 10.1186/1471-2229-10-183
Shimoda, Y., Shimoda-Sasakura, F., Kucho, K., Kanamori, N., Nagata, M., Suzuki, A., et al. (2009). Overexpression of class 1 plant hemoglobin genes enhances symbiotic nitrogen fixation activity between Mesorhizobium loti and Lotus japonicus. Plant J. 57, 254–263. doi: 10.1111/j.1365-313X.2008.03689.x
Stanford, A. C., Larsen, K., Barker, D. G., and Cullimore, J. V. (1993). Differential expression within the glutamine synthetase gene family of the model legume Medicago truncatula. Plant Physiol. 103, 73–81. doi: 10.1104/pp.103.1.73
Trinchant, J. C., and Rigaud, J. (1982). Nitrite and nitric oxide as inhibitors of nitrogenase from soybean bacteroids. Appl. Environ. Microbiol. 44, 1385–1388.
Udvardi, M., and Poole, P. S. (2013). Transport and metabolism in legume-rhizobia symbioses. Annu. Rev. Plant Biol. 64, 781–805. doi: 10.1146/annurev-arplant-050312-120235
Vance, C. P. (2008). “Carbon and nitrogen metabolism in legume nodules,” in Nitrogen-Fixing Leguminous Symbioses, Vol. 7, eds M. J. Dilworth, E. K. James, J. I. Sprent, and W. E. Newton (Dordrecht, The Netherlands: Springer), 293–320.
Keywords: root nodules, nitrogen fixation, glutamine synthetase, tyrosine nitration, nitric oxide, Medicago truncatula
Citation: Silva L and Carvalho H (2013) Possible role of glutamine synthetase in the NO signaling response in root nodules by contributing to the antioxidant defenses. Front. Plant Sci. 4:372. doi: 10.3389/fpls.2013.00372
Received: 14 June 2013; Accepted: 31 August 2013;
Published online: 19 September 2013.
Edited by:
Emmanuel Baudouin, Université Pierre et Marie Curie-Paris 6, FranceReviewed by:
Vasileios Fotopoulos, Cyprus University of Technology, CyprusThomas Ott, University of Munich, Germany
Copyright © 2013 Silva and Carvalho. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Helena Carvalho, Laboratório de Biologia Molecular da Assimilação do Azoto, Instituto de Biologia Molecular e Celular, Universidade do Porto, Rua do Campo Alegre 823, 4150-180 Porto, Portugal e-mail:bWhjYXJ2YWxAaWJtYy51cC5wdA==

