- Departamento de Bioquímica, Biología Celular y Molecular de Plantas, Estación Experimental del Zaidín, Consejo Superior de Investigaciones Científicas, Granada, Spain
Abiotic stress is one of the main problems affecting agricultural losses, and understanding the mechanisms behind plant tolerance and stress response will help us to develop new means of strengthening fruitful agronomy. The mechanisms of plant stress response are complex. Data obtained by experimental procedures are sometimes contradictory, depending on the species, strength, and timing applied. In recent years nitric oxide has been identified as a key signaling molecule involved in most plant responses to abiotic stress, either indirectly through gene activation or interaction with reactive oxygen species and hormones; or else directly, as a result of modifying enzyme activities mainly by nitration and S-nitrosylation. While the functional relevance of the S-nitrosylation of certain proteins has been assessed in response to biotic stress, it has yet to be characterized under abiotic stress. Here, we review initial works about S-nitrosylation in response to abiotic stress to conclude with a brief overview, and discuss further perspectives to obtain a clear outlook of the relevance of S-nitrosylation in plant response to abiotic stress.
Introduction
During their life, plants are continuously exposed to extremes in environmental conditions, in particular abiotic stress sources such as drought, high or low temperatures, high salinity, heavy metal exposure, or herbicides affecting plant development and production (Mittler, 2006; Sreenivasulu et al., 2007). Over 90% of the world’s arable lands are reportedly exposed to major environmental stresses (Pinho dos Reis et al., 2012). Plants use complex recognition and response mechanisms to protect themselves from environment-related changes (Yamaguchi-Shinozaki and Shinozaki, 2006). The effects of abiotic stress may be general or non-specific, such as growth inhibition, electrolyte leakage, and excess of reactive oxygen species (ROS), all of which could lead to cell death. Each type of stress, however, induces specific responses involved in plant acclimation to the particular stress (Kreps et al., 2002; Rizhsky et al., 2004). Therefore, determining the mechanism underlying plant stress tolerance and adaptation is a high priority to ensure plant fitness over a wider range of environmental conditions.
Nitric oxide (NO) is a ubiquitous inter- and intracellular signaling molecule found to be involved in a myriad of cellular functions in plants (Neill et al., 2008b; Mur et al., 2013). Having gained knowledge about its high reactivity and its ambivalent effect, depending on the rate/place of production, there is a need to explore how NO develops all these functions. NO regulates different processes by inducing gene transcription or activating secondary messengers (Besson-Bard et al., 2008; Palmieri et al., 2008; Gaupels et al., 2011). Moreover, NO controls diverse biological processes by directly altering proteins (Martinez-Ruiz et al., 2011). It is able to regulate enzyme activity through covalent post-translational modifications (PTMs) joining metal centers of the proteins and NO tends to affect cysteine and tyrosine residues of the proteins (S-nitrosylation and nitration, respectively) changing their activity, location, or aggregation state (Souza et al., 2008; Martinez-Ruiz et al., 2011). Protein S-nitrosylation, the incorporation of a nitroso group to a Cys thiol, has been established as a significant route through which NO transmits its global cellular influence, and as a broad-based mechanism for the post-translational regulation of most or all classes of proteins (Stamler et al., 2001; Lindermayr and Durner, 2009; Astier et al., 2011). In this review we will focus on the current state of knowledge regarding S-nitrosylation in plants grown under abiotic stress and the elucidation of the function of NO as a signaling mechanism in plant response to environmental modifications.
NO and Abiotic Stress
Klepper (1979) demonstrated by the end of 1970s that herbicides treated soybean leaves release NOx (thought to be mainly NO). Then, in the 1990s, new works evidenced plant NO production in response to abiotic stress (Leshem and Haramaty, 1996; Leshem et al., 1998). Notwithstanding, the first confirmations presenting NO as a key signaling molecule were achieved in response to biotic stress (Delledonne et al., 1998; Durner et al., 1998). In parallel a number of reports on exogenous NO effects in plants were shown (Lamattina et al., 2003). From the very beginning the dual effects of NO as a promoter and inhibitor were seen to depend mainly on its concentration (Leshem and Haramaty, 1996; Beligni and Lamattina, 1999). Currently, many examples of the effect of NO donors on plant biology highlight the protective role of NO against abiotic stresses such as salinity, drought, heavy metals, or UV-radiation (Corpas et al., 2006; Tossi et al., 2012a). Special caution needs to be taken with results obtained with NO donors (Murgia et al., 2004), as each one has specific chemical characteristics that are determinant for the timing of NO release, in addition to features such as the pH of the media, temperature, or light (Ramamurthi and Lewis, 1997; Lamattina et al., 2003). It is therefore difficult to derive physiological conclusions with NO donor effects on plants, as it is hard to measure the real concentration of this molecule in the cell and identify specific targets for each one. Still, the pharmacological approach seems necessary until the mechanism of NO generation in plants can be better defined (Gupta et al., 2011; Mur et al., 2011). Some mutant plants with altered endogenous NO production have been described, involving different NO production pathways or inducing NO production indirectly (Mur et al., 2013). Thus, the use of a combined approach, genetic and pharmacological, would shed light on the functional relevance of NO in response to abiotic stress.
In the past decade, a number of articles have addressed plant endogenous NO production/reduction in a wide range of species, in response to different abiotic stresses (Gould et al., 2003; Corpas et al., 2006; Tossi et al., 2012a). Such studies entail technical problems in assessing the precise location and amount of NO in planta (Mur et al., 2013), and the timing and concentration of the stress. It is lacking: are key factors in NO production. Thus, one single stress type could produce contrary effects depending on the application time and strength of the stress. Meanwhile, there are non-technical problems due to the idiosyncrasy of plants, at least seven sources of NO have been described (Gupta et al., 2011), and more than one source could be involved in the response of a certain stress. Contradictory effects might also depend on the species, tissue analyzed, and developmental stage of the plant.
Heavy metals, and specifically Cd provide a clear example of this diversity of NO-timing and effects (see Bartha et al., 2005; Barroso et al., 2006; Rodriguez-Serrano et al., 2006, 2009; Besson-Bard et al., 2009; De Michele et al., 2009). It appears that initial peaks of NO in response to Cd could have to do with signaling functions involved in iron homeostasis and root growth (Besson-Bard et al., 2009).When Cd treatment is in excess (150 μM), NO may be related with programmed cell death (PCD) induction (De Michele et al., 2009), whereas in long-term Cd treatment (50 μM), NO is associated with an induced senescence process stemming from an excess of ROS and ethylene (McCarthy et al., 2001; Romero-Puertas et al., 2002; Rodriguez-Serrano et al., 2009).
Drought is one of the main stresses affecting crop production, and again the role of NO in plant response is not clear (see Leshem and Haramaty, 1996; Magalhaes et al., 2000; Zhao et al., 2001; Garcia-Mata and Lamattina, 2002; Gould et al., 2003; Zhang et al., 2007). An important point is that stomata closure/aperture is essential during drought stress, a process controlled by abscisic acid (ABA), and NO is needed during the ABA-induced stomatal closure of turgid leaves, but there is not such necessity under conditions of rapid dehydration. NO could therefore be involved in the fine tuning of stomata closure in turgid leaves that occurs in response to oscillations in the environment (Garcia-Mata and Lamattina, 2002; Neill et al., 2008a; Wilson et al., 2009).
The literature describes NO responses to other abiotic stresses such as extreme temperatures, salinity, mechanical damage, UV-B, ozone, or herbicides (Gould et al., 2003; Corpas et al., 2006; Neill et al., 2008a; Molassiotis et al., 2010; Tossi et al., 2012b; Sehrawat et al., 2013; Xie et al., 2013). Although much work remains to define the physiological function of this molecule in response to abiotic stress, progress is underway. The NO signaling mechanisms has been building up beginning with transcriptomic analysis (Besson-Bard et al., 2008). NO has also been shown to mediate in different hormone-regulated processes in plants such as salicylic acid (SA), ABA, auxins, ethylene, or DELLAs, and a cross-talk between NO and hormones has been described in response to environmental fluctuations that may involve second messengers such as Ca or kinases (Lamattina et al., 2003; Simontacchi et al., 2013). The existence of a feedback mechanism between NO and ROS has been demonstrated, and ROS/NO balance is an important factor for the fate of the cells, especially in response to abiotic stress in the context of antioxidant systems and ROS production (Neill et al., 2003, 2008b; Rodriguez-Serrano et al., 2009). A further line of study focuses on direct NO-dependent protein regulation, mainly through S-nitrosylation and nitration (Astier et al., 2011; Vandelle and Delledonne, 2011).
S-Nitrosylation Under Abiotic Stress
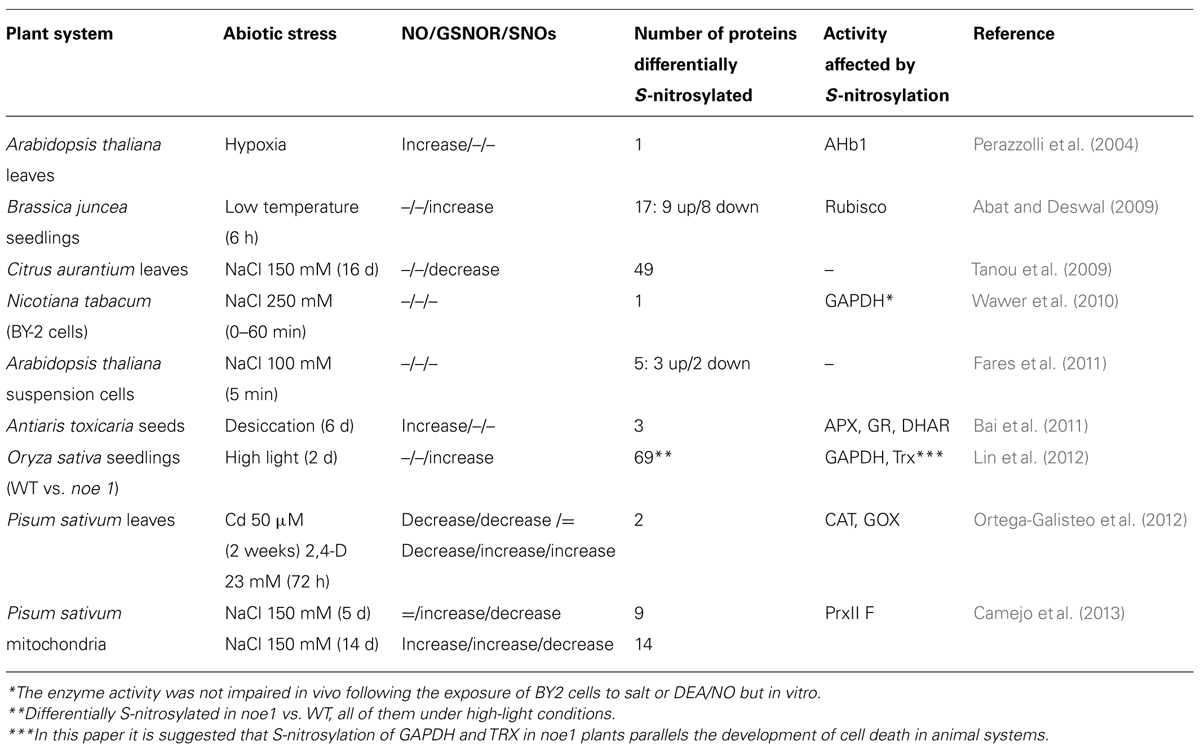
A key feature of NO biology is the PTM of cysteine thiol to form nitrosothiols (S-nitrosylation; Stamler et al., 2001). The development of biotin-switch (BST) technology that overcomes the sensitivity of the nitrosothiol group ensures more rapid entry to the world of NO biology. This elegant approach, formulated in the Snyder laboratory, facilitates the identification of S-nitrosylated proteins in situ as well as in vitro (Jaffrey and Snyder, 2001). The BST is currently the most commonly used method, though other promising approaches that are based on the BST have been developed (Seth and Stamler, 2011). Both approaches to detect S-nitrosylation have been used in plants grown under abiotic stress, proteome-wide scale and analysis of specific proteins known to be involved in the response of the plant to the mentioned stress (Table 1; Astier et al., 2011). Basically, nitrosylating agents (mainly S-nitrosoglutathione; GSNO) were used in proteome-wide scale analysis to increase the number of S-nitrosylated proteins before facing the proteomic study (Lindermayr et al., 2005; Abat et al., 2008; Abat and Deswal, 2009; Palmieri et al., 2010; Ortega-Galisteo et al., 2012; Begara-Morales et al., 2013; Kato et al., 2013). Other studies have been made in plants grown under stress to pathogen challenge (Romero-Puertas et al., 2008; Maldonado-Alconada et al., 2011) and later on under abiotic stress (Abat and Deswal, 2009; Tanou et al., 2009; Fares et al., 2011; Lin et al., 2012; Camejo et al., 2013). Salinity is the best characterized abiotic stress with regard to S-nitrosylation. Forty-nine proteins differentially S-nitrosylated were found in Citrus aurantium leaves under salt stress. Interestingly, a link between S-nitrosylation and oxidative damages (carbonylation) was detected, somehow involved in the prevention of protein loss of function by carbonylation, especially under stress conditions (Tanou et al., 2009). Brief salt stress in Arabidopsis cell suspensions showed that NaCl modified the S-nitrosylation level of a small proportion of endogenously S-nitrosylated proteins (around 10%), suggesting that salt stress induced minor modulations of the S-nitrosylation pattern rather than major changes (Wawer et al., 2010; Fares et al., 2011). Interestingly, this lab adapted the method for detection of endogenous S-nitrosylated Cys (Fares et al., 2011). Deeper analysis in isolated organelles could contribute to finding new targets as yet undetected in total extracts; along this lines, a proteomic study was done in mitochondria from pea plants subjected to salt stress (Camejo et al., 2013). A reduction of the S-nitrosylation pattern was reported in both short and long-term salt treatment being greater in the latter (Camejo et al., 2013). During salt treatment, proteins from respiratory and photorespiratory pathways and, significantly, antioxidant enzymes changed their S-nitrosylation pattern (Fares et al., 2011; Camejo et al., 2013). Changes in the S-nitrosoproteome were studied under low temperature as well, with nine spots induced and eight spots reduced differentially identified as plant defense-related, photosynthetic, glycolytic, and signaling-associated mechanisms (Abat and Deswal, 2009). Another noteworthy study looked at the S-nitrosoproteome under high-light conditions from wild type (WT) and noe1 (NO excess1) mutant rice, and showed 48 proteins differentially S-nitrosylated in the mutant, 10% related to environmental adaptation and 14% to redox homeostasis (Lin et al., 2012). Indeed, noe1 mutants have increased H2O2 and NO levels and display NO-dependent PCD under high-light conditions. The authors found glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and thioredoxin (Trx) S-nitrosylated in noe1 mutants but not in WT, suggesting a relation of these proteins with the control of light-mediated leaf cell death in rice (Lin et al., 2012), reportedly involved in cell death in animals.
S-nitrosylation pattern of particular proteins involved in a specific stress have been studied in parallel. The first protein identified as undergoing S-nitrosylation was hemoglobin AHb1 from Arabidopsis thaliana, which reduces NO emission under hypoxic stress through the production of S-nitrosohemoglobin (Perazzolli et al., 2004). Then, GAPDH showed a transient increase in its S-nitrosylation level in a tobacco cell culture in response to salt stress. However, further analysis is needed to explore the physiological relevance of this change over the treatment period, as its interaction with the osmotic stress-activated protein kinase (NtOSAK) was not affected (Wawer et al., 2010). Antioxidant enzymes from the Asc-Glu cycle, ascorbate peroxidase, glutathione reductase, and dehydroascorbate reductase (APX, GR, and DHAR, respectively) have reduced their S-nitrosylated pattern in response to seed desiccation, thereby suggesting a regulation of antioxidant enzyme activities to stabilize H2O2 accumulation at an appropriate concentration and increasing seed tolerance to dehydration (Bai et al., 2011). S-nitrosylation level of the peroxisomal protein glycolate oxidase decreased under cadmium and 2,4-dichlorophenoxyacetic acid (2,4-D) treatments while no differences were found under 2,4-D in S-nitrosylation level of CAT. Also, a reduction of S-nitrosylated CAT under Cd treatment was observed but similar to the changes occurred in the total amount of this protein under Cd stress. These results point to a regulation of H2O2 level under these stress conditions by NO through the control of ROS sources and antioxidant defenses (Ortega-Galisteo et al., 2012). Additionally, phytochelatins (PCs) with a specific nitrosylation signature were found in Arabidopsis cells treated with Cd, suggesting an interference with the capacity of PC to chelate the metal (De Michele et al., 2009).
Conclusions and Perspectives
The identification of a number of plant proteins that change their S-nitrosylation pattern under abiotic stress is the starting point for the functional and biochemical characterization of S-nitrosylation in plants under such pathophysiological conditions (Figure 1). While the physiological relevance of S-nitrosylation have been shown with various proteins during plant–pathogen interactions (Spoel and Loake, 2011; Yu et al., 2012) this process has been poorly investigated under abiotic stress. Redox and oxygen metabolism-related proteins comprise an interesting group of targets of S-nitrosylation under abiotic stress, as these enzymes and the redox state of the cell play a key role in plant responses to environmental changes. A future challenge will be to unravel the NO-dependent control of these proteins, as a fine-tune regulation may exists in the NO/ROS balance to define the fate of the cell, especially under abiotic stress. Likewise important is the determination of NO-dependent interaction with hormones, especially those linked to plant responses to abiotic stress such as ABA (Roychoudhury et al., 2013), given that a link has been found between S-nitrosylation and auxins (Terrile et al., 2012). The regulation by S-nitrosylation of other protein PTMs that play a crucial role in cellular signaling, including phosphorylation, acetylation, or ubiquitylation, is an intringuing topic (Hess and Stamler, 2012). In this sense, initial data support the idea that S-nitrosylation could interfere with protein carbonylation under abiotic stress (Tanou et al., 2012).
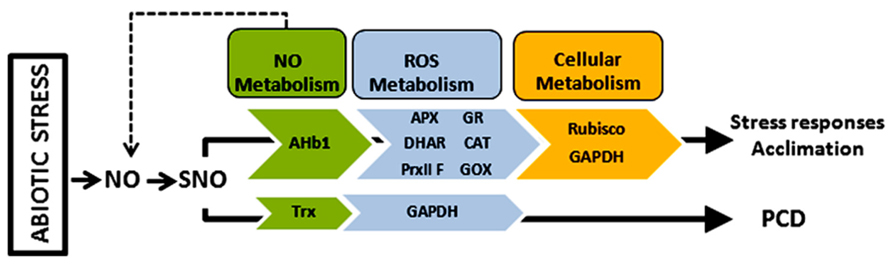
FIGURE 1. S-nitrosylated proteins under abiotic stress. Proteins that change their S-nitrosylation pattern and activity in response to abiotic stress are related to NO, ROS, and cellular metabolism. AHb1, haemoglobin 1; APX, ascorbate peroxidase; CAT, catalase; DHAR, dehydroascobate reductase; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; GOX, glycolate oxidase; Trx, thioredoxin. It is lacking: PrxII F, peroxiredoxin II F.
Two important open questions regarding NO production can be summed up as “how and where”; especially in response to abiotic stress where the adverse condition is not as localized as in response to pathogens. The regulation of NO levels, and particularly its degradation, would be another topic of debate calling for study. Hemoglobin AHb1 has been shown to detoxify NO during hypoxic stress (Perazzolli et al., 2004), but its involvement in different abiotic stresses is still unknown. GSNO reductase (GSNOR) can control levels of GSNO, indirectly regulating S-nitrosylation-dependent signaling, though its participation in abiotic stress must be further explored. Finally, elucidation of the cellular distribution and characterization of nitrosylases and de-nitrosylases that may or may not involve Trx system is a matter essential for our understanding of the full scope of S-nitrosylation in plants under physiological and pathophysiological conditions.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
María C. Romero-Puertas and Luisa M. Sandalio lab is supported by ERDF-cofinanced grant BIO2012-36742 from MICINN and Fundación Ramón Areces (Ayuda CIVP16A1840; http://www.fundacionareces.es), Spain. The authors apologize to the scientists that are not cited because of space limitation.
References
Abat, J. K., and Deswal, R. (2009). Differential modulation of S-nitrosoproteome of Brassica juncea by low temperature: change in S-nitrosylation of Rubisco is responsible for the inactivation of its carboxylase activity. Proteomics 9, 4368–4380. doi: 10.1002/pmic.200800985
Abat, J. K., Mattoo, A. K., and Deswal, R. (2008). S-nitrosylated proteins of a medicinal CAM plant Kalanchoe pinnata- ribulose-1,5-bisphosphate carboxylase/oxygenase activity targeted for inhibition. FEBS J. 275, 2862–2872. doi: 10.1111/j.1742-4658.2008.06425.x
Astier, J., Kulik, A., Koen, E., Besson-Bard, A., Bourque, S., Jeandroz, S., et al. (2011). Protein S-nitrosylation: what’s going on in plants? Free Radic. Biol. Med. 53, 1101–1110. doi: 10.1016/j.freeradbiomed.2012.06.032
Bai, X. G., Yang, L. M., Tian, M. H., Chen, J. H., Shi, J. S., Yang, Y. P., et al. (2011). Nitric oxide enhances desiccation tolerance of recalcitrant Antiaris toxicaria seeds via protein S-nitrosylation and carbonylation. PLoS ONE 6:e20714. doi: 10.1371/journal.pone.0020714
Barroso, J. B., Corpas, F. J., Carreras, A., Rodriguez-Serrano, M., Esteban, F. J., Fernandez-Ocana, A., et al. (2006). Localization of S-nitrosoglutathione and expression of S-nitrosoglutathione reductase in pea plants under cadmium stress. J. Exp. Bot. 57, 1785–1793. doi: 10.1093/jxb/erj175
Bartha, B., Kolbert, Z., and Erdei, L. (2005). Nitric oxide production induced by heavy metals in Brassica juncea L. Czern. and Pisum sativum L. Acta Biol. Szeged. 49, 9–12.
Begara-Morales, J. C., Lopez-Jaramillo, F. J., Sanchez-Calvo, B., Carreras, A., Ortega-Munoz, M., Santoyo-Gonzalez, F., et al. (2013). Vinyl sulfone silica: application of an open preactivated support to the study of transnitrosylation of plant proteins by S-nitrosoglutathione. BMC Plant Biol. 13:61. doi: 10.1186/1471-2229-13-61
Beligni, M., and Lamattina, L. (1999). Is nitric oxide toxic or protective? Trends Plant Sci. 4, 299–300. doi: 10.1016/S1360-1385(99)01451-X
Besson-Bard, A., Gravot, A., Richaud, P., Auroy, P., Duc, C., Gaymard, F., et al. (2009). Nitric oxide contributes to cadmium toxicity in Arabidopsis by promoting cadmium accumulation in roots and by up-regulating genes related to iron uptake. Plant Physiol. 149, 1302–1315. doi: 10.1104/pp.108.133348
Besson-Bard, A., Pugin, A., and Wendehenne, D. (2008). New insights into nitric oxide signaling in plants. Annu. Rev. Plant Biol. 59, 21–39. doi: 10.1146/annurev.arplant.59.032607.092830
Camejo, D., Romero-Puertas, M. D. C., Rodriguez-Serrano, M., Sandalio, L. M., Lazaro, J. J., Jimenez, A., et al. (2013). Salinity-induced changes in S-nitrosylation of pea mitochondrial proteins. J. Proteomics 79, 87–99. doi: 10.1016/j.jprot.2012.12.003
Corpas, F. J., Barroso, J. B., Carreras, A., Valderrama, R., Palma, J. M., and Del Rio, L. A. (2006). “Nitrosative stress in plants: a new approach to understand the role of NO in abiotic stress,” in Nitric Oxide in Plant Growth, Development and Stress Physiology, eds L. Lamattina and J. C. Polacco (Berlin: Springer-Verlag), 187–205.
Delledonne, M., Xia, Y., Dixon, R. A., and Lamb, C. (1998). Nitric oxide functions as a signal in plant disease resistance. Nature 394, 585–588. doi: 10.1038/29087
De Michele, R., Vurro, E., Rigo, C., Costa, A., Elviri, L., Di Valentin, M., et al. (2009). Nitric oxide is involved in cadmium-induced programmed cell death in Arabidopsis suspension cultures. Plant Physiol. 150, 217–228. doi: 10.1104/pp.108.133397
Durner, J., Wendehenne, D., and Klessig, D. F. (1998). Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc. Natl. Acad. Sci. U.S.A. 95, 10328–10333. doi: 10.1073/pnas.95.17.10328
Fares, A., Rossignol, M., and Peltier, J. B. (2011). Proteomics investigation of endogenous S-nitrosylation in Arabidopsis. Biochem. Biophys. Res. Commun. 416, 331–336. doi: 10.1016/j.bbrc.2011.11.036
Garcia-Mata, C., and Lamattina, L. (2002). Nitric oxide and abscisic acid cross talk in guard cells. Plant Physiol. 128, 790–792. doi: 10.1104/pp.011020
Gaupels, F., Kuruthukulangarakoola, G. T., and Durner, J. (2011). Upstream and downstream signals of nitric oxide in pathogen defence. Curr. Opin. Plant Biol. 14, 707–714. doi: 10.1016/j.pbi.2011.07.005
Gould, K. S., Lamotte, O., Klinguer, A., Pugin, A., and Wendehenne, D. (2003). Nitric oxide production in tobacco leaf cells: a generalized stress response? Plant Cell Environ. 26, 1851–1862. doi: 10.1046/j.1365-3040.2003.01101.x
Gupta, K. J., Fernie, A. R., Kaiser, W. M., and Van Dongen, J. T. (2011). On the origins of nitric oxide. Trends Plant Sci. 16, 160–168. doi: 10.1016/j.tplants.2010.11.007
Hess, D. T., and Stamler, J. S. (2012). Regulation by S-nitrosylation of protein post-translational modification. J. Biol. Chem. 287, 4411–4418. doi: 10.1074/jbc.R111.285742
Jaffrey, S. R., and Snyder, S. H. (2001). The biotin switch method for the detection of S-nitrosylated proteins. Sci. STKE 2001, PL1. doi: 10.1126/stke.2001.86.pl1
Kato, H., Takemoto, D., and Kawakita, K. (2013). Proteomic analysis of S-nitrosylated proteins in potato plant. Physiol. Plant. 148, 371–386. doi: 10.1111/j.1399-3054.2012.01684.x
Klepper, L. A. (1979). Effects of certain herbicides and their combinations on nitrate and nitrite reduction. Plant Physiol. 64, 273–275. doi: 10.1104/pp.64.2.273
Kreps, J. A., Wu, Y., Chang, H.-S., Zhu, T., Wang, X., and Harper, J. F. (2002). Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 130, 2129–2141. doi: 10.1104/pp.008532
Lamattina, L., Garcia-Mata, C., Graziano, M., and Pagnussat, G. (2003). Nitric oxide: the versatility of an extensive signal molecule. Annu. Rev. Plant Biol. 54, 109–136. doi: 10.1146/annurev.arplant.54.031902.134752
Leshem, Y. Y., and Haramaty, E. (1996). The characterization and contrasting effects of the nitric oxide free radical in vegetative stress and senescence of Pisum sativum Linn. foliage. J. Plant Physiol. 148, 258–263. doi: 10.1016/S0176-1617(96)80251-3
Leshem, Y. Y., Wills, R. B. H., and Ku, V. V. (1998). Evidence for the function of the free radical gas – nitric oxide (NO) – as an endogenous maturation and senescence regulating factor in higher plants. Plant Physiol. Biochem. 36, 825–833. doi: 10.1016/S0981-9428(99)80020-5
Lin, A. H., Wang, Y. Q., Tang, J. Y., Xue, P., Li, C. L., Liu, L. C., et al. (2012). Nitric oxide and protein S-nitrosylation are integral to hydrogen peroxide-induced leaf cell death in rice. Plant Physiol. 158, 451–464. doi: 10.1104/pp.111.184531
Lindermayr, C., and Durner, J. (2009). S-Nitrosylation in plants: pattern and function. J. Proteomics 73, 1–9. doi: 10.1016/j.jprot.2009.07.002
Lindermayr, C., Saalbach, G., and Durner, J. (2005). Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol. 137, 921–930. doi: 10.1104/pp.104.058719
Magalhaes, J., Monte, D. C., and Durzan, D. (2000). Nitric oxide and ethylene emission in Arabidopsis thaliana. Physiol. Mol. Biol. Plants 2, 117–127.
Maldonado-Alconada, A. M., Echevarria-Zomeno, S., Lindermayr, C., Redondo-Lopez, I., Durner, J., and Jorrin-Novo, J. V. (2011). Proteomic analysis of Arabidopsis protein S-nitrosylation in response to inoculation with Pseudomonas syringae. Acta Physiol. Plant. 33, 1493–1514. doi: 10.1007/s11738-010-0688-2
Martinez-Ruiz, A., Cadenas, S., and Lamas, S. (2011). Nitric oxide signaling: classical, less classical, and nonclassical mechanisms. Free Radic. Biol. Med. 51, 17-29. doi: 10.1016/j.freeradbiomed.2011.04.010
McCarthy, I., Romero-Puertas, M. C., Palma, J. M., Sandalio, L. M., Corpas, F. J., Gomez, M., et al. (2001). Cadmium induces senescence symptoms in leaf peroxisomes of pea plants. Plant Cell Environ. 24, 1065–1073. doi: 10.1046/j.1365-3040.2001.00750.x
Mittler, R. (2006). Abiotic stress, the field environment and stress combination. Trends Plant Sci. 11, 15–19. doi: 10.1016/j.tplants.2005.11.002
Molassiotis, A., Tanou, G., and Diamantidis, G. (2010). NO says more than ‘YES’ to salt tolerance: salt priming and systemic nitric oxide signaling in plants. Plant Signal. Behav. 5, 209–212. doi: 10.4161/psb.5.3.10738
Mur, L. A. J., Mandon, J., Cristescu, S. M., Harren, F. J. M., and Prats, E. (2011). Methods of nitric oxide detection in plants: a commentary. Plant Sci. 181, 509–519. doi: 10.1016/j.plantsci.2011.04.003
Mur, L. A. J., Mandon, J., Persijn, S., Cristescu, S. M., Moshkov, I. E., Novikova, G. V., et al. (2013). Nitric oxide in plants: an assessment of the current state of knowledge. AoB Plants 5, pls052. doi: 10.1093/aobpla/pls052
Murgia, I., De Pinto, M. C., Delledonne, M., Soave, C., and De Gara, L. (2004). Comparative effects of various nitric oxide donors on ferritin regulation, programmed cell death and cell redox state in plant cells. J. Plant Physiol. 161, 777–783. doi: 10.1016/j.jplph.2003.12.004
Neill, S., Barros, R., Bright, J., Desikan, R., Hancock, J., Harrison, J., et al. (2008a). Nitric oxide, stomatal closure, and abiotic stress. J. Exp. Bot. 59, 165–176. doi: 10.1093/jxb/erm293
Neill, S., Bright, J., Desikan, R., Hancock, J., Harrison, J., and Wilson, I. (2008b). Nitric oxide evolution and perception. J. Exp. Bot. 59, 25–35. doi: 10.1093/jxb/erm218
Neill, S. J., Desikan, R., and Hancock, J. T. (2003). Nitric oxide signaling in plant. New Phytol. 159, 11–35. doi: 10.1046/j.1469-8137.2003.00804.x
Ortega-Galisteo, A. P., Rodriguez-Serrano, M., Pazmino, D. M., Gupta, D. K., Sandalio, L. M., and Romero-Puertas, M. C. (2012). S-Nitrosylated proteins in pea (Pisum sativum L.) leaf peroxisomes: changes under abiotic stress. J. Exp. Bot. 63, 2089–2103. doi: 10.1093/jxb/err414
Palmieri, M. C., Lindermayr, C., Bauwe, H., Steinhauser, C., and Durner, J. (2010). Regulation of plant glycine decarboxylase by S-nitrosylation and glutathionylation. Plant Physiol. 152, 1514–1528. doi: 10.1104/pp.109.152579
Palmieri, M. C., Sell, S., Huang, X., Scherf, M., Werner, T., Durner, J., et al. (2008). Nitric oxide-responsive genes and promoters in Arabidopsis thaliana: a bioinformatics approach. J. Exp. Bot. 59, 177–186. doi: 10.1093/jxb/erm345
Perazzolli, M., Dominici, P., Romero-Puertas, M. C., Zago, E., Zeier, A., Sonoda, M., et al. (2004). Arabidopsis nonsymbiotic hemoglobin AHb1 modulates nitric oxide bioactivity. Plant Cell 16, 2785–2794. doi: 10.1105/tpc.104.025379
Pinho dos Reis, S., Medeiros, L. A., and De Souza, C. R. B. (2012). Recent molecular advances on downstream plant responses to abiotic stress. Int. J. Mol. Sci. 13, 8628–8647. doi: 10.3390/ijms13078628
Ramamurthi, A., and Lewis, R. S. (1997). Measurements and modeling of nitric oxide release rates for nitric oxide donors. Chem. Res. Toxicol. 10, 408–413. doi: 10.1021/tx960183w
Rizhsky, L., Liang, H., Shuman, J., Shulaev, V., Davletova, S., and Mittler, R. (2004). When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiol. 134, 1683–1696. doi: 10.1104/pp.103.033431
Rodriguez-Serrano, M., Romero-Puertas, M. C., Pazmino, D. M., Testillano, P. S., Risueno, M. C., Del Rio, L. A., et al. (2009). Cellular response of pea plants to cadmium toxicity: cross talk between reactive oxygen species, nitric oxide, and calcium. Plant Physiol. 150, 229–243. doi: 10.1104/pp.108.131524
Rodriguez-Serrano, M., Romero-Puertas, M. C., Zabalza, A., Corpas, F. J., Gomez, M., Del Rio, L. A., et al. (2006). Cadmium effect on oxidative metabolism of pea (Pisum sativum L.) roots. Imaging of reactive oxygen species and nitric oxide accumulation in vivo. Plant Cell Environ. 29, 1532–1544. doi: 10.1111/j.1365-3040.2006.01531.x
Romero-Puertas, M. C., Campostrini, N., Matte, A., Righetti, P. G., Perazzolli, M., Zolla, L., et al. (2008). Proteomic analysis of S-nitrosylated proteins in Arabidopsis thaliana undergoing hypersensitive response. Proteomics 8, 1459–1469. doi: 10.1002/pmic.200700536
Romero-Puertas, M. C., Palma, J. M., Gomez, M., Del Rio, L. A., and Sandalio, L. M. (2002). Cadmium causes the oxidative modification of proteins in pea plants. Plant Cell Environ. 25, 677–686. doi: 10.1046/j.1365-3040.2002.00850.x
Roychoudhury, A., Paul, S., and Basu, S. (2013). Cross-talk between abscisic acid-dependent and abscisic acid-independent pathways during abiotic stress. Plant Cell Rep. 32, 985–1006. doi: 10.1007/s00299-013-1414-5
Sehrawat, A., Gupta, R., and Deswal, R. (2013). Nitric oxide-cold stress signalling cross-talk, evolution of a novel regulatory mechanism. Proteomics 13, 1816–1835. doi: 10.1002/pmic.201200445
Seth, D., and Stamler, J. S. (2011). The SNO-proteome: causation and classifications. Curr. Opin. Chem. Biol. 15, 129–136. doi: 10.1016/j.cbpa.2010.10.012
Simontacchi, M., Garcia-Mata, C., Bartoli, C. G., Santa-Marïa, G. E., and Lamattina, L. (2013). Nitric oxide as a key component in hormone-regulated processes. Plant Cell Rep. 32, 853–866. doi: 10.1007/s00299-013-1434-1
Souza, J. M., Peluffo, G., and Radi, R. (2008). Protein tyrosine nitration–functional alteration or just a biomarker? Free Radic. Biol. Med. 45, 357–366. doi: 10.1016/j.freeradbiomed.2008.04.010
Spoel, S. H., and Loake, G. J. (2011). Redox-based protein modifications: the missing link in plant immune signalling. Curr. Opin. Plant Biol. 14, 358–364. doi: 10.1016/j.pbi.2011.03.007
Sreenivasulu, N., Sopory, S. K., and Kavi Kishor, P. B. (2007). Deciphering the regulatory mechanisms of abiotic stress tolerance in plants by genomic approaches. Gene 388:1–13. doi: 10.1016/j.gene.2006.10.009
Stamler, J. S., Lamas, S., and Fang, F. C. (2001). Nitrosylation. the prototypic redox-based signaling mechanism. Cell 106, 675–683. doi: 10.1016/S0092-8674(01)00495-0
Tanou, G., Filippou, P., Belghazi, M., Job, D., Diamantidis, G., Fotopoulos, V., et al. (2012). Oxidative and nitrosative-based signaling and associated post-translational modifications orchestrate the acclimation of citrus plants to salinity stress. Plant J. 72, 585–599. doi: 10.1111/j.1365-313X.2012.05100.x
Tanou, G., Job, C., Rajjou, L., Arc, E., Belghazi, M., Diamantidis, G., et al. (2009). Proteomics reveals the overlapping roles of hydrogen peroxide and nitric oxide in the acclimation of citrus plants to salinity. Plant J. 60, 795–804. doi: 10.1111/j.1365-313X.2009.04000.x
Terrile, M. C., Paris, R., Calderon-Villalobos, L. I. A., Iglesias, M. J., Lamattina, L., Estelle, M., et al. (2012). Nitric oxide influences auxin signaling through S-nitrosylation of the Arabidopsis TRANSPORT INHIBITOR RESPONSE 1 auxin receptor. Plant J. 70, 492–500. doi: 10.1111/j.1365-313X.2011.04885.x
Tossi, V., Amenta, M., Lamattina, L., and Cassia, R. (2012a). Nitric oxide enhances plant ultraviolet-B protection up-regulating gene expression of the phenylpropanoid biosynthetic pathway. Plant Cell Environ. 34, 909–921. doi: 10.1111/j.1365-3040.2011.02289.x
Tossi, V., Cassia, R., Bruzzone, S., Zocchi, E., and Lamattina, L. (2012b). ABA says NO to UV-B: a universal response? Trends in Plant Sci. 17, 510–517. doi: 10.1016/j.tplants.2012.05.007
Vandelle, E., and Delledonne, M. (2011). Peroxynitrite formation and function in plants. Plant Sci. 181, 534–539. doi: 10.1016/j.plantsci.2011.05.002
Wawer, I., Bucholc, M., Astier, J., Anielska-Mazur, A., Dahan, J., Kulik, A., et al. (2010). Regulation of Nicotiana tabacum osmotic stress-activated protein kinase and its cellular partner GAPDH by nitric oxide in response to salinity. Biochem. J. 429, 73–83. doi: 10.1042/BJ20100492
Wilson, I. D., Ribeiro, D. M., Bright, J., Confraria, A., Harrison, J., Barros, R. S., et al. (2009). Role of nitric oxide in regulating stomatal apertures. Plant Signal. Behav. 4, 467–469. doi: 10.4161/psb.4.5.8545
Xie, Y., Mao, Y., Lai, D., Zhang, W., Zheng, W., and Shen, W. (2013). Roles of NIA/NR/NOA1-dependent nitric oxide production and HY1 expression in the modulation of Arabidopsis salt tolerance. J. Exp. Bot. 64, 3045–3060. doi: 10.1093/jxb/ert149
Yamaguchi-Shinozaki, K., and Shinozaki, K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57, 781–803. doi: 10.1146/annurev.arplant.57.032905.105444
Yu, M. D., Yun, B. W., Spoel, S. H., and Loake, G. J. (2012). A sleigh ride through the SNO: regulation of plant immune function by protein S-nitrosylation. Curr. Opin. Plant Biol. 15, 424–430. doi: 10.1016/j.pbi.2012.03.005
Keywords: abiotic stress, nitric oxide, plant, post-translational modifications, S-nitrosylation
Citation: Romero-Puertas MC, Rodrïguez-Serrano M and Sandalio LM (2013) Protein S-nitrosylation in plants under abiotic stress: an overview. Front. Plant Sci. 4:373. doi: 10.3389/fpls.2013.00373
Received: 02 July 2013; Accepted: 31 August 2013;
Published online: 20 September 2013.
Edited by:
Emmanuel Baudouin, Université Pierre et Marie Curie, FranceReviewed by:
Francisca Sevilla, Agencia Estatal Consejo Superior de Investigaciones Científicas, SpainDiana Bellin, University of Verona, Italy
Copyright © 2013 Romero-Puertas, Rodríguez-Serrano and Sandalio. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: María C. Romero-Puertas, Departamento de Bioquímica, Biología Celular y Molecular de Plantas, Estación Experimental del Zaidín, Consejo Superior de Investigaciones Cientïficas, Calle Profesor Albareda 1, 18008 Granada, Spain e-mail:bWFyaWEucm9tZXJvQGVlei5jc2ljLmVz