- 1Laboratory of Environmental Biotechnology, Graduate School of Agricultural Science, Tohoku University, Sendai, Japan
- 2Faculty of Agriculture, Kyushu University, Fukuoka, Japan
Introduction
Genomic organization and gene expression system of plant mitochondria are distinct from those of other eukaryotes, including animals, even though of chloroplast, another organelle in plants that contain its own genomes. Recent research has revealed the significance of the control of gene expression at the RNA level, including the formation of the 5′- and 3′-ends of mitochondrial mRNA. Although the details of gene expression in mitochondria are not the same as in chloroplasts, huge numbers of nuclear-encoded pentatricopeptide repeat (PPR) proteins are transported in both mitochondria and chloroplasts and control their gene expression. (See other articles in this issue.)
CMS/Rf System
Research on mitochondrial gene expression in plants has focused on understanding cytoplasmic male sterility (CMS), which is caused by the incompatibility between the mitochondrial and nuclear genomes. The mitochondrial genome allows active recombination of genes to occur and easily generates new protein-coding genes. Expression of such a gene sometimes causes male sterility, and consequently the gene is called a CMS-associated gene. Pollen fertility can be restored by suppressing the expression of the CMS-associated gene with a nuclear-encoded fertility restorer (Rf) gene. The CMS/Rf system is useful in agriculture because it enables easy crossbreeding of varieties to produce hybrid seeds. It is also an excellent model in which to study the nuclear control of mitochondrial gene expression. Extensive research has been conducted to identify CMS-associated genes and Rf genes.
One organism that has been studied for this purpose is BT-CMS rice. The BT-CMS/Rf system contains the mitochondrial CMS gene orf79 and the nuclear-encoded Rf1a gene, which codes for a PPR protein (Kazama and Toriyama, 2003). The orf79 gene is co-transcribed with its upstream gene, atp6. The protein of Rf1a promotes the cleavage of atp6–orf79 co-transcribed mRNAs. The cleavage prevents the translation of orf79 (Kazama et al., 2008).
A similar system exists in a Brassica CMS of Ogura. The mitochondrial genome contains the CMS-associated gene orf138, which is co-transcribed as orf138–atp8 (Bonhomme et al., 1992). The amount and processing pattern of the co-transcribed mRNA is not affected by the presence or absence of the Rf gene, which codes for a PPR protein (Koizuka et al., 2003). The RFo/PPR-B protein is known to be associated with the orf138 gene containing RNA, suggesting that its function is direct suppression of the translation (Uyttewaal et al., 2008a). In the cases of both rice and the Brassica CMS, the translational step seems to be the critical step in the CMS/Rf system.
Translation in Plant Mitochondria
Hardly anything is known about the translational control of gene expression in plant mitochondria. Regarding the cis-regulatory element, the mitochondrial mRNAs do not follow the Shine-Dalgarno sequence that exists in prokaryotic organisms. An early informatics study found three conserved sequence blocks in the 5′ untranslated region (UTR) of mitochondrial RNAs (Pring et al., 1992): block I (GGGAGCAGAG), block II (AGUCUCCCUUUC), and block III [GU (n) CGUUGG]. These blocks generally occur within 100 bases of the 5′ flanking region of the start codons, suggesting that they are involved in mitochondrial translation. However, their functionality has not been evaluated, due to the lack of experimental techniques to study mitochondrial translation.
Recent advances in genetic studies have revealed, although sporadically, the protein factors that are involved in plant mitochondrial translation. The silencing of the nuclear-encoded rps10 gene, which codes for the mitochondrial ribosomal protein S10, has induced differential translations of mitochondrial transcripts, including over-expression of ribosomal proteins and down-regulation of oxidative phosphorylation subunits (Kwasniak et al., 2013). The PPR protein of the MPPR6 gene in maize has been shown to interact with the 5′ UTR of the rps3 mRNA, encoding mitochondrial ribosomal protein S3. This protein may also be involved in 5′ maturation and translational initiation of the rps3 mRNA. The loss of MPPR6 results, consequently, in a considerable reduction of mitochondrial translation (Manavski et al., 2012). The loss of translation activity induces general down-regulation of mitochondrial RNA, in contrast with the silencing of the rps10 gene (Kwasniak et al., 2013). Another PPR protein of the PPR336 gene has been shown to associate with polysomes in the mitochondria. The mutant plant has unusual polysomal profiles, suggesting that PPR336 could be involved in translation (Uyttewaal et al., 2008b), although the actual mechanism has not been elucidated.
Putative Role of 5′-End Heterogeneity of mRNA for Translation in Plant Mitochondria
Forner et al. (2007) reported that Arabidopsis mitochondrial RNAs tend to have heterogeneous 5′-ends but uniform 3′-ends. We conducted an analysis using circularized (CR) reverse transcriptase (RT) PCR to determine whether the heterogeneity of the 5′-end of mRNA is involved in translational efficiency. We compared the 5′-ends of mRNA derived from total mitochondrial RNA with those of mRNA during translation. The RNAs during translation (i.e., the RNAs associated with polysomes) were fractionated by centrifuging at 100,000 × g (Figure 1A). The purity of the mitochondria and the enrichment of ribosomes in the polysomal fraction used this study were verified by the western and northern blot analyses (Figure 1B). Next, the RNA termini were determined by CR-RT-PCR for several genes in rice mitochondria (atp1, atp6, atp8, and atp9; Figure 1).
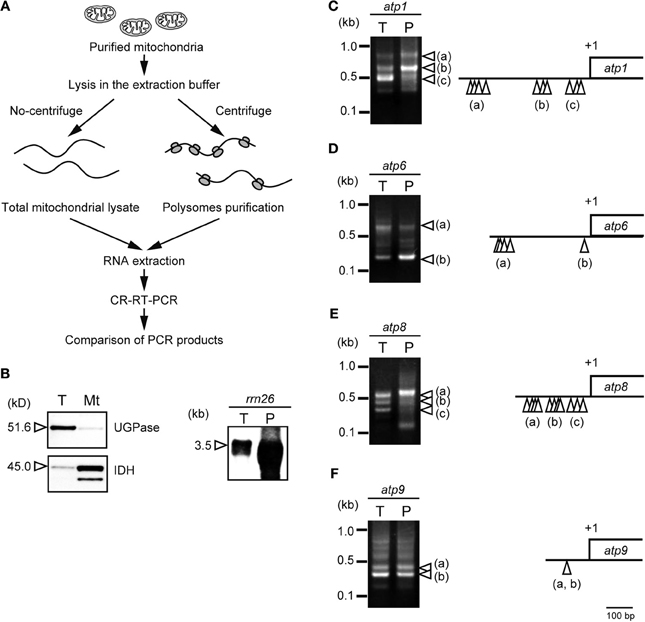
Figure 1. Heterogeneity in the 5′-end of mRNA and its involvement in translational efficiency in plant mitochondria. (A) Overview of the CR-RT-PCR analysis compared mitochondrial total RNA with the polysomal RNA. The rice mitochondria were purified as described (Kazama et al., 2008). Purified mitochondria were homogenized in extraction buffer [0.1 M Tris-HCl (pH 7.5), 0.2 M KCl, 35 mM MgCl2, 1% triton X-100, 0.5 mg/ml heparin, 3 mM DTT, 0.5 mg/ml chloramphenicol, 25 mM EGTA, 0.2 M sucrose, 2% polyoxyethylene-10-tridecyl ether]. The suspension was centrifuged for 5 min at 20,000 × g and 4°C, twice to remove cell debris. The cell debris was further removed by centrifugation for 30 min at 30,000 × g and 4°C, twice. The resultant supernatant was transferred to a new tube and centrifuged for 3 hours at 100,000 × g and 4°C in TLA100 rotor (Beckman). The precipitate was used for polysomal fraction. (B) Purity of the isolated mitochondria was examined by a cytosolic marker protein of (UGPase, UDP-glucose pyrophosphorylase) and a mitochondrial marker protein (IDH, isocitrate dehydrogenase) (left panel). The western blot analyses were performed using total protein (T) and isolated mitochondria (Mt) using the specific antibodies (Agrisera). The enrichment of ribosomes in the polysomal fraction was also assessed by northern blot analysis in total mitochondrial RNA (T) and polysomal mitochondrial RNA (P) using rrn26 probe (right panel). The images of CR-RT-PCR were shown for mRNAs of atp1 (C), atp6 (D), atp8 (E), and atp9 (F). The 5′ terminal of each transcript is indicated by white arrowheads in the CR-RT-PCR images mapped on the schematic gene structure (right panel).
The CR-RT-PCR analysis revealed the three major signals for atp1 (Figure 1C; a, b, and c). Sequence analysis revealed that the 3′-ends are uniform (+141/+142 relative to the stop codon). The multiple 5′-ends were mapped at −390 to −453 relative to the start codon (Figure 1C; a), −167 to −207 (Figure 1C; b), and −34 to −78 (Figure 1C; c). Interestingly, the CR-RT-PCR profile was different in the polysome-associated RNA. The atp1-b RNA was most enriched in the polysomal fraction, whereas the atp1-c RNA was predominant in the total mitochondrial RNA, suggesting that the heterogenity of the 5′-end could be involved in the translational efficiency of plant mitochondrial RNA.
Similar observations have been made in other mRNAs. The atp6 RNA accumulated in two forms (Figure 1D; a and b). Their 5′-ends were mapped at around −300 for atp6-a and –24/–23 for atp6-b, relative to the start codon. The 3′-ends of all cDNAs for atp6 have been mapped in the dense region (+27 to +29, relative to the stop codon). Polysomal analysis revealed that the shorter form of RNA (atp6-b) seems to be more enriched in polysome than the longer one, as shown by the distinct CR-RT-PCR profiles of total RNA vs. polysomal RNA. The atp8 RNA was shown to accumulate in three different forms with the 5′-ends at −240 to −199, −171 to −113, and −76 to −19 (Figure 1E; a, b, and c, respectively). The 3′-ends were mapped at the same position in all the RNA species (+119/+121, relative to the stop codon). The longest RNA (Figure 1E; a) was concentrated in the polysomal fraction. The atp9 contains two major isoforms with identical 5′ termini (−85/−84, relative to the start codon) but with different 3′ termini (+110/+111 and +6/+9) (Figure 1F; a and b, respectively). Polysome analysis suggested that the different 3′-end status is not involved in translation.
Together, these results suggested that the heterogeneity of 5′-ends could be involved in translational efficiency in plant mitochondria. Preliminary in silico searches have failed to find conserved motifs within the putative translational active RNAs.
Mitochondrial Translation in Yeasts and Humans
The status of the 5′ UTR differs among different species. As in plants, the mitochondrial mRNAs of the yeast Saccharomyces cerevisiae possess characteristic 5′ and 3′ UTRs. The S. cerevisiae would be regarded as the best system studying mitochondrial translation. Currently, tens of the translational activators have been identified for the several mitochondrial transcripts (Herrmann et al., 2013). For instance, PET309, which is a membrane-bound PPR protein, acts on the 5′ UTR of the cox1 mRNA to activate translation and is required to stabilize the precursor of cox1 RNAs (Manthey and McEwen, 1995). A series of mutations in PPR motifs within PET309 revealed that the PPR motifs are necessary for cox1 mRNA translation, but not for stabilization (Tavares-Carreón et al., 2008). Thus, the PPR motifs of PET309 may induce a particular RNA conformation to attract and/or interact with the translational machinery. This evidence indicates that mitochondrial translation in S. cerevisiae is mainly controlled by the gene-specific translation activator through its association with the 5′ UTR in mRNA (Gruschke and Ott, 2013; Herbert et al., 2013).
Human mitochondrial genes are transcribed using three promoters. The RNAs are subsequently processed and polyadenylated to generate the stop codons (Rorbach and Minczuk, 2012). Mapping of the 5′-end of human mitochondrial mRNAs revealed that mRNAs start directly at or very near the start codon (Montoya et al., 1981). Thus, their mRNAs lack the ribosome-binding site at the 5′ UTR. The analysis of secondary structure at the 5′-ends indicated that the 5′-ends of all mRNA are highly unstructured (Jones et al., 2008). The mechanism of human mitochondrial translation is poorly understood (Koc and Koc, 2013).
Plant mitochondrial RNAs have long 5′ UTRs with no obvious conserved motifs, suggesting that gene-specific translational regulation occurs, as in S. cerevisiae. PPR proteins are believed to play a pivotal role in the translation regulation via the 5′ UTR in plant mitochondria.
Perspective
Translation is a critical step that determines the final level of protein production. Recent research has suggested that mitochondrial gene expression is important in various plant phenomena, such as the pollen production, stress response, germination, and metabolite synthesis.
It is critical to develop techniques to analyze the mitochondrial translational system. The recently developed “genome editing” technology, which has been used to study the human mitochondrial genome, may be applicable to studies of transformation in plant mitochondria (Bacman et al., 2013). Alternatively, incorporation of exogenous RNA into mitochondria using the import pathway for tRNA or via PNPase may facilitate analysis of the cis-regulatory element (Wang et al., 2010; Mahato et al., 2011). The tRNA import pathway is applicable for plant as already shown (Sieber et al., 2011; Val et al., 2011). These approaches will facilitate elucidation of the plant mitochondrial translational system and understanding of the diverse methods of mitochondrial translation among different organisms.
Acknowledgments
This work was supported by Grants-in-Aid 22681028 and 22380008 (Takahiro Nakamura), and 2338002 (Kinya Toriyama) from the Ministry of Education, Culture, Sports, Science, and Technology; the Adaptable and Seamless Technology Transfer Program through Target-driven R&D, JST (Takahiro Nakamura); and Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry (Kinya Toriyama). Yusuke Yagi is a recipient of JSPS post-doctoral fellowship.
References
Bacman, S. R., Williams, S. L., Pinto, M., Peralta, S., and Moraes, C. T. (2013). Specific elimination of mutant mitochondrial genomes in patient-derived cells by mitoTALENs. Nat. Med. 19, 1111–1113. doi: 10.1038/nm.3261
Bonhomme, S., Budar, F., Lancelin, D., Small, I., Defrance, M. C., and Pelletier, G. (1992). Sequence and transcript analysis of the Nco2.5 Ogura-specific fragment correlated with cytoplasmic male sterility in Brassica cybrids. Mol. Gen. Genet. 235, 340–348. doi: 10.1007/BF00279379
Forner, J., Weber, B., Thuss, S., Wildum, S., and Binder, S. (2007). Mapping of mitochondrial mRNA termini in Arabidopsis thaliana: t-elements contribute to 5′ and 3′ end formation. Nucleic Acids Res. 35, 3676–3692. doi: 10.1093/nar/gkm270
Gruschke, S., and Ott, M. (2013). “Mechanisms and control of protein synthesis in yeast mitochondria,” in Translation in Mitochondria and Other Organelles, ed A.-M. Duchêne (Berlin; Heidelberg: Springer), 109–131 doi: 10.1007/978-3-642-39426-3_5
Herbert, C. J., Golik, P., and Bonnefoy, N. (2013). Yeast PPR proteins, watchdogs of mitochondrial gene expression. RNA Biol. 10, 1477–1494. doi: 10.4161/rna.25392
Herrmann, J. M., Woellhaf, M. W., and Bonnefoy, N. (2013). Control of protein synthesis in yeast mitochondria: the concept of translational activators. Biochim. Biophys. Acta 1833, 286–294. doi: 10.1016/j.bbamcr.2012.03.007
Jones, C. N., Wilkinson, K. A., Hung, K. T., Weeks, K. M., and Spremulli, L. L. (2008). Lack of secondary structure characterizes the 5′ ends of mammalian mitochondrial mRNAs. RNA 14, 862–871. doi: 10.1261/rna.909208
Kazama, T., Nakamura, T., Watanabe, M., Sugita, M., and Toriyama, K. (2008). Suppression mechanism of mitochondrial ORF79 accumulation by Rf1 protein in BT-type cytoplasmic male sterile rice. Plant J. 55, 619–628. doi: 10.1111/j.1365-313X.2008.03529.x
Kazama, T., and Toriyama, K. (2003). A pentatricopeptide repeat-containing gene that promotes the processing of aberrant atp6 RNA of cytoplasmic male-sterile rice. FEBS Lett. 544, 99–102. doi: 10.1016/S0014-5793(03)00480-0
Koc, E., and Koc, H. (2013). “Mechanism and regulation of protein synthesis in mammalian mitochondria,” in Translation in Mitochondria and Other Organelles, ed A.-M. Duchêne (Berlin; Heidelberg: Springer), 29–53. doi: 10.1007/978-3-642-39426-3_2
Koizuka, N., Imai, R., Fujimoto, H., Hayakawa, T., Kimura, Y., Kohno-Murase, J., et al. (2003). Genetic characterization of a pentatricopeptide repeat protein gene, orf687, that restores fertility in the cytoplasmic male-sterile Kosena radish. Plant J. 34, 407–415. doi: 10.1046/j.1365-313X.2003.01735.x
Kwasniak, M., Majewski, P., Skibior, R., Adamowicz, A., Czarna, M., Sliwinska, E., et al. (2013). Silencing of the nuclear RPS10 gene encoding mitochondrial ribosomal protein alters translation in Arabidopsis mitochondria. Plant Cell 25, 1855–1867. doi: 10.1105/tpc.113.111294
Mahato, B., Jash, S., and Adhya, S. (2011). RNA-mediated restoration of mitochondrial function in cells harboring a Kearns Sayre Syndrome mutation. Mitochondrion 11, 564–574. doi: 10.1016/j.mito.2011.03.006
Manavski, N., Guyon, V., Meurer, J., Wienand, U., and Brettschneider, R. (2012). An essential pentatricopeptide repeat protein facilitates 5′ maturation and translation initiation of rps3 mRNA in maize mitochondria. Plant Cell 24, 3087–3105. doi: 10.1105/tpc.112.099051
Manthey, G. M., and McEwen, J. E. (1995). The product of the nuclear gene PET309 is required for translation of mature mRNA and stability or production of intron-containing RNAs derived from the mitochondrial COX1 locus of Saccharomyces cerevisiae. EMBO J. 14, 4031–4043.
Montoya, J., Ojala, D., and Attardi, G. (1981). Distinctive features of the 5′-terminal sequences of the human mitochondrial mRNAs. Nature 290, 465–470. doi: 10.1038/290465a0
Pring, D. R., Mullen, J. A., and Kempken, F. (1992). Conserved sequence blocks 5′ to start codons of plant mitochondrial genes. Plant Mol. Biol. 19, 313–317. doi: 10.1007/BF00027353
Rorbach, J., and Minczuk, M. (2012). The post-transcriptional life of mammalian mitochondrial RNA. Biochem. J. 444, 357–373. doi: 10.1042/BJ20112208
Sieber, F., Placido, A., El Farouk-Ameqrane, S., Duchene, A. M., and Marechal-Drouard, L. (2011). A protein shuttle system to target RNA into mitochondria. Nucleic Acids Res. 39:e96. doi: 10.1093/nar/gkr380
Tavares-Carreón, F., Camacho-Villasana, Y., Zamudio-Ochoa, A., Shingú-Vázquez, M., Torres-Larios, A., and Pérez-Martinez, X. (2008). The pentatricopeptide repeats present in Pet309 are necessary for translation but not for stability of the mitochondrial COX1 mRNA in yeast. J. Biol. Chem. 283, 1472–1479. doi: 10.1074/jbc.M708437200
Uyttewaal, M., Arnal, N., Quadrado, M., Martin-Canadell, A., Vrielynck, N., Hiard, S., et al. (2008a). Characterization of Raphanus sativus pentatricopeptide repeat proteins encoded by the fertility restorer locus for Ogura cytoplasmic male sterility. Plant Cell 20, 3331–3345. doi: 10.1105/tpc.107.057208
Uyttewaal, M., Mireau, H., Rurek, M., Hammani, K., Arnal, N., Quadrado, M., et al. (2008b). PPR336 is associated with polysomes in plant mitochondria. J. Mol. Biol. 375, 626–636. doi: 10.1016/j.jmb.2007.11.011
Val, R., Wyszko, E., Valentin, C., Szymanski, M., Cosset, A., Alioua, M., et al. (2011). Organelle trafficking of chimeric ribozymes and genetic manipulation of mitochondria. Nucleic Acids Res. 39, 9262–9274. doi: 10.1093/nar/gkr580
Keywords: plants, mitochondria, translation, RNA processing, protein-RNA interaction
Citation: Kazama T, Yagi Y, Toriyama K and Nakamura T (2013) Heterogeneity of the 5′-end in plant mRNA may be involved in mitochondrial translation. Front. Plant Sci. 4:517. doi: 10.3389/fpls.2013.00517
Received: 14 November 2013; Accepted: 02 December 2013;
Published online: 17 December 2013.
Edited by:
Philippe Giegé, Centre National de la Recherche Scientifique, FranceReviewed by:
Thalia Salinas, Centre National de la Recheche Scientifique associé à l'Université de Strasbourg, FranceCopyright © 2013 Kazama, Yagi, Toriyama and Nakamura. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence:dG5ha2FAYWdyLmt5dXNodS11LmFjLmpw
 Tomohiko Kazama
Tomohiko Kazama Yusuke Yagi
Yusuke Yagi Kinya Toriyama1
Kinya Toriyama1 Takahiro Nakamura
Takahiro Nakamura