- Institut de Biologie Moléculaire des Plantes, UPR2357 CNRS, Université de Strasbourg, Strasbourg, France
Polyubiquitin chain deposition on a target protein frequently leads to proteasome-mediated degradation whereas monoubiquitination modifies target protein property and function independent of proteolysis. Histone monoubiquitination occurs in chromatin and is in nowadays recognized as one critical type of epigenetic marks in eukaryotes. While H2A monoubiquitination (H2Aub1) is generally associated with transcription repression mediated by the Polycomb pathway, H2Bub1 is involved in transcription activation. H2Aub1 and H2Bub1 levels are dynamically regulated via deposition and removal by specific enzymes. We review knows and unknowns of dynamic regulation of H2Aub1 and H2Bub1 deposition and removal in plants and highlight the underlying crucial functions in gene transcription, cell proliferation/differentiation, and plant growth and development. We also discuss crosstalks existing between H2Aub1 or H2Bub1 and different histone methylations for an ample mechanistic understanding.
Introduction
Ubiquitin (Ub) and Ub-like (e.g., SUMO) proteins constitute a family of modifiers that are linked covalently to target proteins. Although ubiquitination (also called ubiquitylation or ubiquitinylation) first came to light in the context of protein destruction, it is now clear that ubiquitination can also carry out proteolysis-independent functions. Ubiquitination can alter biochemical, molecular and/or subcellular localization activities of a target protein. The first ubiquitinated protein to be described was histone H2A in calf thymus, a finding dated more than 36 years ago (Goldknopf et al., 1975; Hunt and Dayhoff, 1977). Yet, only more recently have the underlying mechanisms and regulatory functions of histone ubiquitination begun to emerge (reviewed in Zhang, 2003; Shilatifard, 2006; Weake and Workman, 2008; Braun and Madhani, 2012; Pinder et al., 2013). Histones are highly alkaline proteins, found in the nuclei of eukaryotic cell, which package and order the DNA into structural units named nucleosomes. A nucleosome is composed of roughly 146 bp of DNA wrapping around the histone octamer comprising two molecules each of the four core histones H2A, H2B, H3, and H4 (Luger et al., 1997). Histone monoubiquitination together with other types of posttranslational modifications, e.g., acetylation, methylation, phosphorylation, and SUMOylation, can modulate nucleosome/chromatin structure and DNA accessibility and thus regulate diverse DNA-dependent processes, such as genome replication, repair, and transcription (Zhang, 2003; Shilatifard, 2006; Weake and Workman, 2008; Braun and Madhani, 2012; Pinder et al., 2013).
Ubiquitination occurs via conjugation of the C-terminal residue of Ub to the side chain of a lysine (K) residue of the substrate/acceptor protein, a reaction involving three coordinated enzymatic activities (reviewed in Hershko and Ciechanover, 1998). Ub is first activated by an ATP-dependent reaction involving the Ub-activating enzyme E1, then conjugated to the active site cysteine residue of the Ub-conjugating (UBC) enzyme E2, and finally transferred to the target K residue of the substrate protein by the Ub-protein isopeptide ligase E3. Most organisms have only one E1, but dozens of different E2 and hundreds up to thousands of different E3 enzymes, providing the need in coping with effective substrate specificity (Hua and Vierstra, 2011; Braun and Madhani, 2012). Identification and characterization of E3s and some E2s involved in histone ubiquitination had been a key for understanding biological functions of histone ubiquitination in various organisms. Because of its suitability for genomics, genetics, and cellular and molecular biological approaches, Arabidopsis thaliana is an ideal model to investigate histone ubiquitination functions. In this review, we focus on this reference plant to expose current progress made on ubiquitination of different types of histones.
H2B Monoubiquitination in Arabidopsis
Genome-Wide Distribution of H2Bub1
Monoubiquitinated H2B (H2Bub1) was first discovered in mouse cells and was estimated to represent about 1–2% of total cellular H2B (West and Bonner, 1980). Later, H2Bub1 was detected widely throughout eukaryotes spanning from yeast to humans and plants (Zhang, 2003; Shilatifard, 2006; Sridhar et al., 2007; Zhang et al., 2007a; Weake and Workman, 2008). The ubiquitination site is mapped to a highly conserved K residue, H2BK123 in budding yeast, H2BK119 in fission yeast, H2BK120 in humans, and H2BK143 in Arabidopsis. Genome-wide analysis revealed that in Arabidopsis as in animals H2Bub1 is associated with active genes distributed throughout the genome and marks chromatin regions notably in combination with histone H3 trimethylated on K4 (H3K4me3) and/or with H3K36me3 (Roudier et al., 2011). During early photomorphogenesis, gene upregulation was found to be associated with H2Bub1 enrichment whereas gene downregulation did not show detectable correlation with any H2Bub1 level changes (Bourbousse et al., 2012). In general, H2Bub1 is considered to represent an active chromatin mark broadly involved in genome transcription regulation.
Enzymes Involved in Regulation of H2Bub1 Levels
The budding yeast Rad6 (radiation sensitivity protein 6) was the first factor identified and shown to work as an E2 enzyme involved in catalyzing H2Bub1 formation both in vitro and in vivo (Robzyk et al., 2000). It contains a highly conserved catalytic UBC domain of approximately 150 amino acids in length with an active-site cysteine for linking Ub. The E3 enzyme working together with Rad6 in catalyzing H2Bub1 formation in budding yeast is Bre1 (Brefeldin-A sensitivity protein 1), which contains a C3HC4-type RING finger domain typical for all E3s (Hwang et al., 2003; Wood et al., 2003). The depletion of either Rad6 or Bre1 eliminates genome-wide H2Bub1 and causes yeast cell growth defects (Robzyk et al., 2000; Hwang et al., 2003; Wood et al., 2003). Human contains at least two homologs of Rad6, namely hHR6A and hHR6B, and two homologs of Bre1, namely RNF20/hBRE1A and RNF40/hBRE1B (Kim et al., 2005; Zhu et al., 2005). In Arabidopsis, three homologs of Rad6, namely UBC1, UBC2, and UBC3, were identified and UBC1 and UBC2 but not UBC3 were shown to be redundantly responsible for H2Bub1 formation in planta (Cao et al., 2008; Gu et al., 2009; Xu et al., 2009). The two Bre1 homologs HUB1 (HISTONE MONOUBIQUITINATION 1) and HUB2 work non-redundantly, possibly as a hetero-tetramer composed of two copies of HUB1 and two copies of HUB2, in catalyzing H2Bub1 formation in Arabidopsis (Fleury et al., 2007; Liu et al., 2007; Cao et al., 2008). H2Bub1 levels are drastically reduced or undetectable in Western blot analysis in the loss-of-function hub1 and hub2 single mutants as well as in the hub1 hub2 and ubc1 ubc2 double mutants, but are unaffected in the ubc1, ubc2, and ubc3 single mutants or in the ubc1 ubc3 and ubc2 ubc3 double mutants (Cao et al., 2008; Gu et al., 2009; Xu et al., 2009).
H2Bub1 levels are also regulated by deubiquitination enzymes. Two Ub-specific proteases, Ubp8 and Ubp10, are involved in deubiquitination of H2Bub1 in budding yeast. Strikingly, while Ubp8 acts as a component of the SAGA (Spt-Ada-Gcn5-acetyltransferase) complex specifically in H2Bub1 deubiquitination in transcription activation, Ubp10 functions independently of SAGA and primarily acts in Sir-mediated silencing of telomeric and rDNA regions (reviewed in Weake and Workman, 2008). In human, USP22 acts as Ubp8 ortholog in a SAGA complex in H2Bub1 deubiquitination (Weake and Workman, 2008). In Arabidopsis, although a SAGA complex remains uncharacterized so far, the Ub protease UBP26/SUP32 has been shown to deubiquitinate H2Bub1 involved in both heterochromatic silencing (Sridhar et al., 2007) and transcription activation of the FLC (FLOWERING LOCUS C) gene (Schmitz et al., 2009). More recently, the otubain-like deubiquitinase OTLD1 was reported as implicated in deubiquitination of H2BUb1 and repression of At5g39160, a gene of unknown function (Krichevsky et al., 2011).
Role of H2Bub1 in Flowering Time Regulation
The timing of flowering is critical for the reproductive success of plants. As compared to wild type, the hub1 and hub2 single mutants as well as the hub1 hub2 and ubc1 ubc2 double mutants exhibit an early flowering phenotype whereas but the ubc1, ubc2, and ubc3 single mutants and the ubc1 ubc3 and ubc2 ubc3 double mutants have a normal phenotype (Cao et al., 2008; Gu et al., 2009; Xu et al., 2009). This early flowering phenotype is detectable under both long-day and short-day photoperiod plant growth conditions. Molecular analyses of the mutants indicate that H2Bub1 controls flowering time primarily through transcriptional activation of FLC (Figure 1). FLC encodes a key transcription repressor involved in both the autonomous/developmental and vernalization flowering pathways, and its active transcription is associated with several histone marks, e.g., H3K4me3, H3K36me2/3 and H2Bub1 (reviewed in Berr et al., 2011). In the early flowering mutants hub1, hub2, hub1 hub2, and ubc1 ubc2, FLC expression levels are reduced and the FLC chromatin shows reduced H2Bub1 levels (Cao et al., 2008; Gu et al., 2009). The loss-of-function mutant ubp26/sup32 showed also an early flowering phenotype and reduced FLC expression but an elevated level of H2Bub1 in the FLC chromatin (Schmitz et al., 2009), indicating that not only H2Bub1 formation but also H2Bub1 removal are necessary for FLC transcription. Accompanying H2Bub1 reduction compromised levels of H3K4me3 and to a less extent H3K36me2 were detected at FLC in hub1 and ubc1 ubc2 (Cao et al., 2008), and reduced level of H3K36me3 but elevated level of H3K27me3 was observed at FLC in ubp26/sup32 (Schmitz et al., 2009). On parallels to the knowledge in yeast, it was proposed that the UBC-HUB-mediated H2Bub1 formation is necessary for H3K4me3 deposition at transcription initiation whereas UBP26/SUP32-mediated H2Bub1 removal is required for H3K36me3 deposition during transcription elongation (Cao et al., 2008; Schmitz et al., 2009). Nonetheless, this hierarchy of histone modifications needs to be cautioned because multiple factors are involved in H3K4me3 and H3K36me2/3 depositions and the SDG8 (SET DOMAIN GROUP 8)-mediated H3K36me2/3 deposition remarkably override H3K4me2/3 deposition in FLC transcription (Yao and Shen, 2011; Shafiq et al., 2014). Besides FLC, Arabidopsis has five FLC paralogs, namely MAF1 (MADS AFFECTING FLOWERING 1), MAF2, MAF3, MAF4 and MAF5. Some MAFs are also downregulated in the early flowering mutants hub1, hub2, hub1 hub2, ubc1 ubc2, and ubp26/sup32 (Cao et al., 2008; Gu et al., 2009; Schmitz et al., 2009; Xu et al., 2009). Thus, H2Bub1 may also regulate flowering time through control of MAF gene expression under some plant growth conditions.
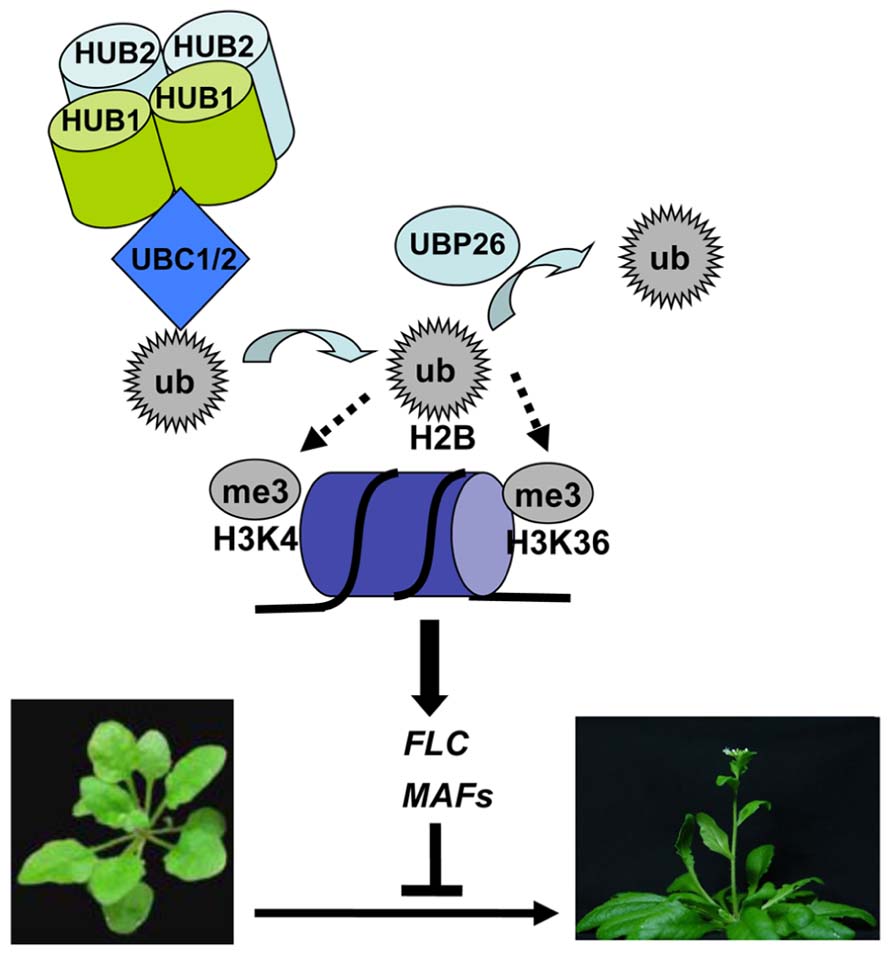
FIGURE 1. A proposed model for deposition and removal of histone H2B monoubiquitination in transcriptional activation of FLC and MAFs in flowering time regulation. In this model, HUB1 and HUB2 form a heterotetramer and recruit UBC1 or UBC2 to FLC/MAFs chromatin, leading to transfer of a ubiquitin (ub) monomer from UBC1 or UBC2 onto H2B. H2Bub1 formation enhances H3K4me3 deposition by methyltransferases, together promoting transcription initiation. UBP26 removes ubiquitin on H2B, favoring H3K36me3 deposition in promoting transcription elongation. Active transcription of FLC/MAFs represses Arabidopsis flowering, a transition from vegetative to reproductive plant development.
H2Bub1 Function in other Processes
In addition to flowering, many other processes also involve H2Bub1 as evidenced by studies of the Arabidopsis hub1 and hub2 mutants. The hub mutants display reduced seed dormancy associated with reduced expression of several dormancy-related genes, including DOG1 (DELAY OF GERMINATION 1), ATS2 (ACYLTRANSFERASE 2), NCED9 (NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 9), PER1 (CYSTEINE PEROXIREDOXIN 1), and CYP707A2 (Liu et al., 2007). At vegetative growth stages, the hub mutants exhibit pale leaf coloration, modified leaf shape, reduced rosette biomass, and inhibited root growth (Fleury et al., 2007). Cell cycle genes, particularly some key regulators of the G2-to-M transition, are downregulated, which could largely explain the plant growth defects of the hub mutants (Fleury et al., 2007). A more recent study shows that several circadian clock genes, including CCA1 (CIRCADIAN CLOCK ASSOCIATED 1), ELF4 (EARLY FLOWERING 4) and TOC1 (TIMING OF CAB EXPRESSION 1), are downregulated and their chromatin regions contain lower levels of H2Bub1 in the hub mutants, suggesting that H2Bub1 may contribute to the regulation of plant growth fitness to environment through expression modulation of some circadian clock genes (Himanen et al., 2012). It is worth to note that SDG2-mediated H3K4me3 deposition is also required for expression of several circadian clock genes (e.g., CCA1, TOC1) and the hub mutants exhibit reduced levels of H3K4me3 in chromatin regions of the circadian clock genes (Himanen et al., 2012; Malapeira et al., 2012).
During photomorphogenesis, hundreds of genes show upregulation associated with H2Bub1 enrichment in their chromatin in response to light exposure (Bourbousse et al., 2012). Strikingly, over 50% of these genes gain H2Bub1 enrichment upon the 1 h of illumination, illustrating the highly dynamic nature of H2Bub1 deposition during a likely cell division-independent genome regrogramming process. In contrast to the above discussed cases, in this study the H2Bub1 changes is neither accompanied by any detectable changes of H3K36me3 nor required for H3K4me3 enrichment following six hours of light exposure (Bourbousse et al., 2012). In line with the function of H2Bub1 in gene activation in response to light, the hub1-3 mutant seedlings are overly light sensitive, exhibiting a photobleaching phenotype (Bourbousse et al., 2012).
The hub1 mutants also show increased susceptibility to the necrotrophic fungal pathogens Botrytis cinerea and Alternaria brassicicola (Dhawan et al., 2009). Precise role of H2Bub1 in plant defense against pathogens still remains largely unclear. Structure defects, e.g., thinner cell walls and altered surface cutin and wax compositions, together with impaired induction of some defense genes might have partly contributed to the increased susceptibility to pathogen infection in the hub mutant plants (Dhawan et al., 2009; Ménard et al., 2014). It is worthy noting that the sdg8 mutants impaired in H3K36me3 deposition also display reduced resistance to necrotrophic fungal pathogen infection (Berr et al., 2010, 2012; Palma et al., 2010). It will be interesting to study in future research whether a trans-histone crosstalk between H2Bub1 and H3K36me3 acts on transcription induction in plant response to pathogens.
Mechanisms of H2Bub1 in Transcription Regulation
So far only limited information is available concerning how H2Bub1 enzymes are recruited to the target chromatin. The evolutionarily conserved PAF1 (Polymerase Associated Factor 1) complex interacts with Pol II (RNA polymerase II) and plays a role as a “platform” for association of enzymes involved in H2bub1, H3K4me3, and H3K36me2/3 deposition, linking histone modifications with active transcription (Shilatifard, 2006; Weake and Workman, 2008; Berr et al., 2011; Braun and Madhani, 2012). A direct interaction between PAF1 complex and Rad6-Bre1 has been detected and shown as required for catalyzing H2Bub1 formation (Xiao et al., 2005). As in yeast and animals, deletion or knockdown of PAF1 components markedly reduces H2Bub1 in Arabidopsis (Schmitz et al., 2009). Genetic analysis shows that HUB2 and ELF8 encoding a PAF1 subunit act in a same floral-repression pathway in Arabidopsis flowering time regulation (Gu et al., 2009). Although physical interaction between UBC-HUB and PAF1 needs future investigation, interactions were observed between UBC and HUB (Cao et al., 2008) and between HUB and MED21 (mediator complex subunit 21), a subunit of the evolutionarily conserved Mediator complex (Dhawan et al., 2009). Mediator complex is associated with both general transcription factors and Pol II and is essential for activator-dependent transcription in all eukaryotes (for a recent review, see Carlsten et al., 2013). Nevertheless, the aforementioned interactors are generally involved in Pol II transcribed genes and thus cannot fully explain why UBC-HUB targets some but not all active genes. It is reasonable to speculate that UBC-HUB recruitment might also involve some gene-specific yet uncharacterized factors.
The next question is how H2Bub1 affects transcription. In yeast and animals, H2Bub1 can promote transcription elongation by enhancing the recruitment of RNA Pol II and by facilitating nucleosome removal through interplay with FACT (facilitates chromatin transcription), an evolutionarily conserved histone chaperone complex (Pavri et al., 2006; Tanny et al., 2007). FACT acts on displacement of H2A/H2B dimer from a nucleosome core, facilitating transcription elongation on chromatin template. In Arabidopsis, FACT genetically interacts with HUB1 and plays critical roles in multiple plant developmental processes (Lolas et al., 2010). Yet its precise interplay with H2Bub1 in transcription regulation needs future investigations.
Alternatively or additionally, H2Bub1 may regulate transcription indirectly through crosstalk with H3K4me3 and H3K36me2/3 (Shilatifard, 2006; Weake and Workman, 2008; Berr et al., 2011; Braun and Madhani, 2012). In line with this idea, lack of H2Bub1 in Arabidopsis impairs H3K4me3 and H3K36me2 formation in chromatin at FLC and clock genes (Cao et al., 2008; Himanen et al., 2012), and elevated H2Bub1 inhibits H3K36me3 formation in the FLC chromatin (Schmitz et al., 2009). Nevertheless, in contrast to the requirement of H2Bub1 for genome-wide H3K4me3 formation in yeast, lack of H2Bub1 in Arabidopsis barely affects global H3K4me2/3 and H3K36me2/3 levels, as evidenced by Western blot analysis (Cao et al., 2008; Dhawan et al., 2009; Gu et al., 2009) as well as by ChIP (chromatin immunoprecipitation) analysis of light responsive genes during photomorphogenesis (Bourbousse et al., 2012). It is currently unclear to which extent applies the crosstalk of H2Bub1 with H3K4me2/3 and H3K36me2/3 in Arabidopsis gene transcription regulation and what are the molecular mechanisms underlying the crosstalk.
Finally, while H2Bub1 is generally associated with active gene transcription, it can also regulate transcription repression in a chromatin context-dependent manner. The ubp26/sup32 mutant shows release of transgene and transposon silencing (Sridhar et al., 2007) as well as elevated expression of PHE1 (PHERES1) associated with seed developmental defects (Luo et al., 2008). It has been shown that the silencing release is accompanied by reduction of H3K9me2 and of siRNA-mediated DNA methylation and the PHE1 expression elevation is associated with a reduced level of H3K27me3. Nevertheless, whether these changes of repressive marks are directly linked with H2Bub1 still need to be investigated.
H2A Monoubiquitination in Arabidopsis
Presence of H2Aub1
In contrast to H2Bub1, H2Aub1 has not been found in yeast and has been generally implicated in transcription repression in animal cells (Weake and Workman, 2008; Braun and Madhani, 2012). Albeit its early discovery and high abundance (about 5–15% of the total H2A) in animal cells (Goldknopf et al., 1975; Hunt and Dayhoff, 1977; Zhang, 2003), H2Aub1 function has only more recently begun to be elucidated, thanking to the first identification of the human PRC1 (Polycomb repressive complex 1) component Ring1B (also known as Ring2 and RNF2) as a E3 involved in catalyzing H2Aub1 formation (Wang et al., 2004). In Arabidopsis, H2Aub1 was undetectable in a large-scale analysis of histone post-translational modifications by mass spectrometry (Sridhar et al., 2007; Zhang et al., 2007a) and had been thought for a long time to be non-existent (Weake and Workman, 2008). However, five PRC1-like RING-finger proteins, namely AtRING1a, AtRING1b, AtBMI1a, AtBMI1b, and AtBMI1c, have been identified in Arabidopsis (Sanchez-Pulido et al., 2008; Xu and Shen, 2008). More recent immunodetection and in vitro enzyme activity assays have revealed that these RING-finger proteins are effectively involved in catalyzing H2Aub1 formation in Arabidopsis (Bratzel et al., 2010; Li et al., 2011; Yang et al., 2013).
PRC2 and PRC1 in H2Aub1 Deposition
Polycomb group (PcG) proteins, first identified in Drosophila as repressors of homeotic (Hox) genes, are nowadays known to act in multiprotein complexes in transcription repression of a large number of genes in many multicellular organisms including plants (Bemer and Grossniklaus, 2012; Molitor and Shen, 2013; Schwartz and Pirrotta, 2013; Simon and Kingston, 2013). The most intensively studied complexes are PRC1 and PRC2. In Drosophila, PRC2 is composed of four core subunits, namely Ez (Enhancer of zeste), Suz12 (Suppressor of zeste 12), Esc (Extra sex combs) and N55 (a 55 kDa WD40 repeat protein), and PRC1 also contains four main subunits, namely Pc (Polycomb), Ph (Polyhomeotic), Psc (Posterior sex combs) and Ring1 (also known as dRing). In mammals, alternate subunit compositions create larger families of related PRC2-type and PRC1-type complexes (Schwartz and Pirrotta, 2013; Simon and Kingston, 2013). Nevertheless, defined biochemical activities of PRC2 and PRC1 are conserved from flies to humans. The classical model proposes a sequential mode of action of the two complexes: PRC2 catalyzes H3K27me3 formation, and PRC1 recognizes the H3K27me3 mark and further mediates downstream H2Aub1 deposition. The PRC1 components, acting as E3 ligases in H2Aub1 formation, are RING-finger proteins: Ring1 in Drosophila and Ring1A and Ring1B in human (Braun and Madhani, 2012; Schwartz and Pirrotta, 2013).
In Arabidopsis, the four PRC2 core components are highly conserved (Figure 2) and encoded by small gene families, and their function in H3K27me3 deposition and transcription repression have been intensively studied (Bemer and Grossniklaus, 2012). In contrast, PRC1 compositions are drastically diverged in plants as compared to animals (Molitor and Shen, 2013). No sequence homologue of Ph could be identified in plants so far. LHP1 (LIKE HETEROCHROMATIN PROTEIN 1), also known as TFL2 (TERMINAL FLOWER 2), binds H3K27me3 and may play a Pc-like function (Turck et al., 2007; Zhang et al., 2007b). This remarkably differs from the distinct roles of HP1 and Pc in animals, where HP1 binds H3K9me3 involved in hetereochromatin formation whereas Pc binds H3K27me3 involved in PRC1-mediated silencing in euchromatin. The best conservations found about PRC1 core components are from RING-finger proteins structured by a RING domain at N-terminus and a Ub-like RAWUL domain at C-terminus (Sanchez-Pulido et al., 2008; Xu and Shen, 2008). These RING-finger proteins can be classified into two phylogenic groups: the first group comprises Drosophila Ring1, human Ring1A and Ring1B, and Arabidopsis AtRING1a and AtRING1b; the second group comprises Drosophila Psc, human Bmi1, and Arabidopsis AtBMI1a, AtBMI1b, and AtBMI1c. Consistent with their sequence conservation, AtRING1a, AtRING1b, AtBMI1a, and AtBMI1b each can ubiquitinate H2A in vitro, and loss of function of AtBMI1a and AtBMI1b causes H2Aub1 reduction in planta (Bratzel et al., 2010; Yang et al., 2013).
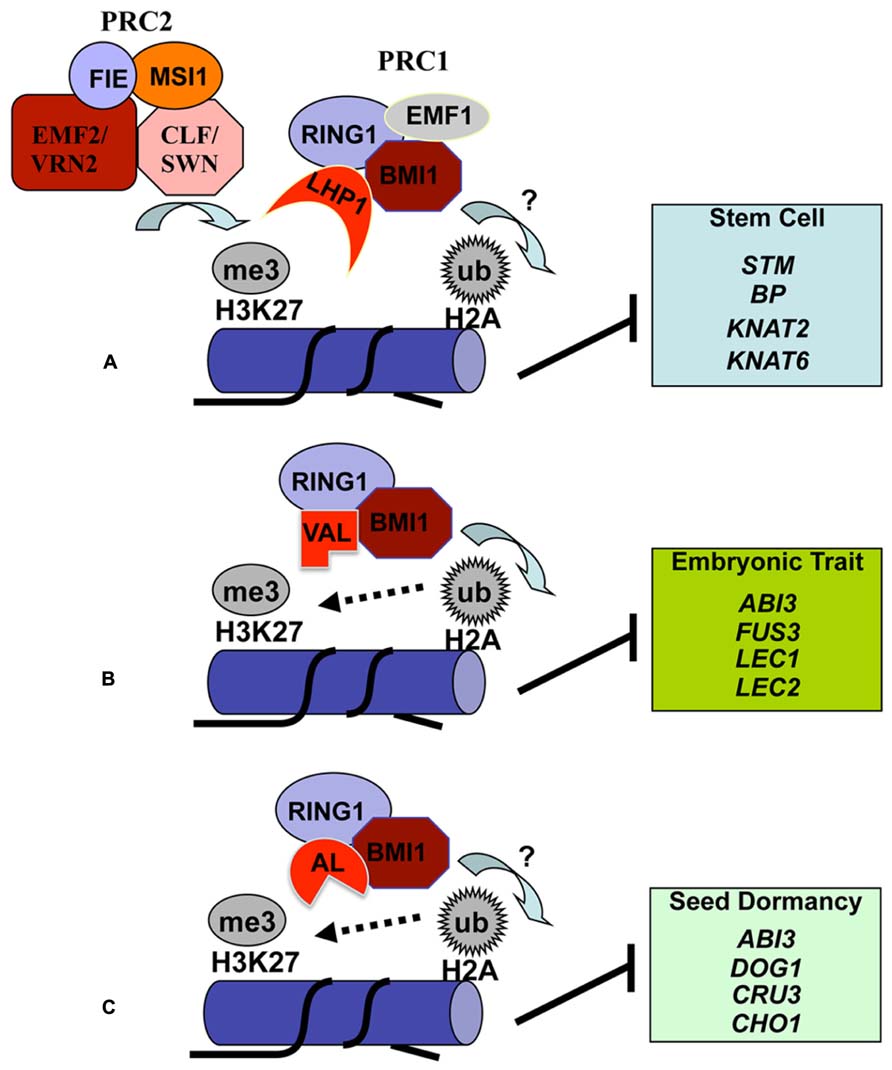
FIGURE 2. Proposed models for histone H2A monoubiquitination deposition in transcriptional repression of varied target genes. The Arabidopsis PRC1-like RING-finger proteins AtRING1a/b (RING1) and AtBMI1a/b/c (BMI1) have the E3 ligase activity in catalyzing H2A monoubiquitination (H2Aub1). Comparable to the classical model of sequential PRC2 then PRC1 action in Polycomb silencing in animal cells, the Arabidopsis PRC1-like protein LHP1 binds H3K27me3 pre-deposited by the evolutionarily conserved PRC2 complexes and recruits RING1, BMI1 and possibly also EMF1 through protein–protein interactions (A). This combinatorial action by PRC2 then PRC1 likely plays a broad role in suppression of numerous genes, including the key stem cell regulatory KNOX genes that need to be stably repressed during lateral organ development. The transcription factor VAL is involved in recruitment of BMI1 and RING1 in suppression of embryonic trait genes in somatic cells (B). AL proteins bind BMI1 and RING1 and play important roles in suppression of several key seed dormancy regulatory genes to promote germination (C). H3K27me3 deposition at embryonic/seed genes is enhanced by VAL/AL-PRC1 (B,C), unraveling a non-canonical crosstalk between H3K27me3 and H2Aub1. The question marks indicate that H2Aub1 deposition in the specified target gene chromatin still requires future investigation.
Role of PRC1-Like Ring-Finger Proteins in Stem Cell Maintenance
Plant growth and development largely depend on stem cells located in SAM (shoot apical meristem) and RAM (root apical meristem), whose activities are fine-tuned by multiple families of chromatin factors (Sang et al., 2009; Shen and Xu, 2009). The first uncovered biological role of the Arabidopsis PRC1-like RING-finger proteins are on the regulation of SAM activity (Xu and Shen, 2008). While the single loss-of-function mutants Atring1a and Atring1b have a normal phenotype, the double mutant Atring1a Atring1b exhibits enlarged SAM, fasciated stem, and ectopic-meristem formation in cotyledons and leaves. This indicates that AtRING1a and AtRING1b play a redundant role in stable repression of stem cell activity to allow appropriate lateral organ differentiation. The balances between stem cell maintenance and cell differentiation for organ formation are controlled by specific transcription factors, including KNOX (Class I KNOTTED1-like homeobox) proteins. Strikingly, several KNOX genes, e.g., STM (SHOOT-MERISTEMLESS), BP (BREVIPEDICELLUS)/KNAT1, KNAT2 and KNAT6, are upregulated in Atring1a Atring1b (Xu and Shen, 2008). Ectopic expression of KNOX genes colocalizes with and precedes ectopic meristem formation. It has been proposed that AtRING1a/b acts as a crucial PRC1 component in conjunction with PRC2 in repression of KNOX genes to promote lateral organ formation in the SAM (Figure 2A).
Role of PRC1-Like Ring-Finger Proteins in Embryonic Cell Fate Determinacy
Further characterization of the ectopic meristem structures observed in Atring1a Atring1b unravels that these callus structures exhibit embryonic traits (Chen et al., 2010). The Atbmi1a Atbmi1b mutant also displays derepression of embryonic traits (Bratzel et al., 2010; Chen et al., 2010). Embryonic callus formation has been observed broadly in somatic tissues of cotyledons, leaves, shoots and roots of the mutant plants. Treatment with an auxin transport inhibitor can inhibit embryonic callus formation in Atring1a Atring1b, indicating that a normal auxin gradient is required for somatic embryo formation in the mutant (Chen et al., 2010). Both Atring1a Atring1b and Atbmi1a Atbmi1b mutants exhibit elevated expression of several key embryonic regulatory genes, including ABI3 (ABSCISIC ACID INSENSITIVE 3), AGL15 (AGAMOUS LIKE 15), BBM (BABYBOOM), FUS3 (FUSCA 3), LEC1 (LEAFY COTYLEDON 1), and LEC2 (Bratzel et al., 2010; Chen et al., 2010). It is likely that derepression of these regulatory genes together with KNOX has contributed to the ectopic meristem and embryonic callus formation in somatic tissues of the Atring1a Atring1b and Atbmi1a Atbmi1b mutants (Figure 2B). The VAL (VP1/ABI3-LIKE) transcription factors can physically interact with AtBMI1 proteins and the val1 val2 mutant exhibits comparable phenotype to Atbmi1a Atbmi1b, suggesting that VAL and AtBMI1 proteins may form complexes in repression of embryonic regulatory genes during vegetative development (Yang et al., 2013). Notably, loss of VAL or AtBMI1 causes H2Aub1 reduction in chromatin regions at ABI3, BBM, FUS3 and LEC1 but not STM (Yang et al., 2013). Future investigation is necessary to clarify whether AtBMI1 and AtRING1 proteins repress KNOX transcription via H2Aub1 deposition or other independent chromatin remodeling mechanisms.
Role of PRC1-Like Ring-Finger Proteins in Seed Germination
Seed germination defines the entry into a new generation of the plant life cycle. It is generally accepted that the process of germination starts with water uptake followed by seed coat rupture and is completed following radicle protrusion (Bentsink and Koornneef, 2008). During the very early phase, the embryonic growth program remains latent and can be reinstated in response to unfavorable environmental cues. With the attainment of photosynthetic competence, the irreversible transition to autotrophic growth is accomplished and embryonic program is stably suppressed. A recent study (Molitor et al., 2014) has identified the Arabidopsis PHD-domain H3K4me3-binding AL (ALFIN1-like) proteins as interactors of AtBMI1 and AtRING1 proteins and has demonstrated a crucial function of chromatin state switch in establishment of seed developmental gene repression during seed germination (Figure 2C). Loss of AL6 and AL7 as well as loss of AtBMI1a and AtBMI1b retards seed germination and causes transcriptional derepression and a delayed chromatin state switch from H3K4me3 to H3K27me3 enrichment of seed developmental genes, including ABI3 and DOG1. The germination delay phenotype of the al6 al7 and Atbmi1a Atbmi1b mutants is more pronounced under osmotic stress (Molitor et al., 2014), suggesting that AL PHD-PRC1 complexes may participate in regulation of seed germination in response to environmental cues.
Role of PRC1-Like Ring-Finger Proteins in other Processes
AtBMI1a and AtBMI1b, also named DRIP1 (DREB2A-INTERACTING PROTEIN 1) and DRIP2, had been reported first as E3 ligases involved in ubiquitination of DREB2A (DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2A), a transcription factor controlling water deficit-inducible gene expression (Qin et al., 2008). The drip1 drip2 mutant shows enhanced expression of water deficit-inducible genes and more tolerance to drought (Qin et al., 2008). Overexpression of AtBMI1c accelerates flowering time, which is associated with reduction of FLC expression (Li et al., 2011). In addition to SAM maintenance defects and derepression of embryonic traits, the Atring1a Atring1b mutant also displays homeotic conversions of floral tissues (Xu and Shen, 2008). Therefore, more precise functions and underlying molecular mechanisms for the PRC1-like RING-finger proteins are still waiting to be uncovered during plant development and in plant response to environmental changes.
Mechanisms of PRC1-Like Ring-Finger Proteins in Transcription Repression
H2Aub1 function in plants is primarily evidenced through investigation of roles of the Arabidopsis PRC1-like RING-finger proteins (Xu and Shen, 2008; Bratzel et al., 2010; Chen et al., 2010; Li et al., 2011; Yang et al., 2013). Although these RING-finger proteins act nicely in vitro as E3 ligases, their in vivo functions in H2Aub1 deposition are still poorly documented. H2Aub1 level in Arabidopsis seems very low because large-scale analyses of either the histone-enriched or the Ub-affinity-purified protein preparations fail to detect H2Aub1 (Maor et al., 2007; Sridhar et al., 2007; Zhang et al., 2007a; Manzano et al., 2008; Saracco et al., 2009). H2Aub1 has been detected only by using specific antibodies, and in this case AtBMI1 genes have been shown to act as positive regulators for H2Aub1 deposition in Arabidopsis plants (Bratzel et al., 2010; Li et al., 2011; Yang et al., 2013). It is unknown whether any deubiquitinases might cause low levels of H2Aub1 in Arabidopsis. In animal cells, several deubiquitinases are characterized as specific for H2Aub1 (Weake and Workman, 2008; Simon and Kingston, 2013). Future characterization of Arabidopsis H2Aub1 deubiquitinases may provide useful information regarding regulatory mechanisms of H2Aub1 dynamics.
AtRING1 and AtBMI1 proteins physically interact each other and with the H3K27me3-binding protein LHP1 (Xu and Shen, 2008; Bratzel et al., 2010; Chen et al., 2010), providing a possible recruitment mechanism similar to the classical sequential PRC2 then PRC1 silencing pathway in animal cells. However, the Atring1a Atring1b, Atbmi1a Atbmi1b, or Atbmi1a Atbmi1b Atbmi1c mutant exhibits much more severe phenotypic defects than the lhp1 mutant does, and lhp1 enhances the Atring1a Atring1b mutant defects. It is thus apparent that AtRING1 and AtBMI1 proteins also act independently from LHP1. Recent identification of the transcriptional regulator VAL as AtBMI1-binding protein and of AL as AtRING1 and AtBMI1 interactor provides some novel insight about recruitment mechanisms (Yang et al., 2013; Molitor et al., 2014). It is particular intriguing that loss of AtBMI1 impairs H3K27me3 enrichment at seed developmental genes during seed germination and vegetative growth (Yang et al., 2013; Molitor et al., 2014). It has also been reported that loss of LHP1 impairs H3K27me3 enrichment at flower gene loci in roots (Derkacheva et al., 2013). These recent findings challenge the classic hierarchical paradigm where PRC2-mediated H3K27me3 deposition precedes PRC1 recruitment (Figure 2). It is obvious that future investigations are necessary to better understand the composition and function of different PRC1-like complexes in Arabidopsis.
Conclusions and Perspectives
Studies over the last few years in the model plant Arabidopsis have greatly advanced our knowledge about the roles of H2Aub1 and H2Bub1 in transcription regulation in plant growth and development. In view of additional functions described in animal cells for both H2Aub1 and H2Bub1 in DNA damage repair (Bergink et al., 2006; Marteijn et al., 2009; Chernikova et al., 2010; Ginjala et al., 2011; Moyal et al., 2011; Nakamura et al., 2011), it is anticipated that more roles of H2Aub1 and H2Bub1 in plant response to environmental stresses are waiting to be uncovered. Mutagenesis of enzymes involved in H2Aub1 and H2Bub1 deposition or removal is required to address the question whether these enzymes effectively exert their biological functions via H2Aub1 and H2Bub1. Identification and characterization of factors associated with these different enzymes will be essential for understanding molecular mechanisms of their recruitment and function at specific targets within the genome. We need to know whether and how their function is spatially and temporally integrated with plant development. Genome-wide tools need to be further explored to provide a global view of links among enzyme or associated factor binding, H2Aub1/H2Bub1 enrichment, H3 methylation, and Pol II occupation. Crosstalks between H2Aub1 or H2Bub1 and different H3 methylations need to be addressed for chromatin context specificity.
In addition to H2Aub1 and H2Bub1, ubiquitinated H1, H3, and H4 are also found in Arabidopsis (Maor et al., 2007; Manzano et al., 2008; Saracco et al., 2009). H3 ubiquitination catalyzed by Rtt101-Mms1 in yeast and by Cul4-DDB1 in human has been recently shown to play an important role in the histone chaperone Asf1-mediated nucleosome assembly (Han et al., 2013). Arabidopsis contains a conserved family of CULLINs and CUL4-DDB1 complexes are reported (Shen et al., 2002; Hua and Vierstra, 2011). The Asf1 homologues in Arabidopsis are also identified (Zhu et al., 2011). It remains to be investigated whether CUL4-DDB and AtASF1 collaboratively act on nucleosome assembly via H3 ubiquitination in epigenetic regulation in Arabidopsis.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The work in Wen-Hui Shen laboratory was supported by the French “Centre National de la Recherche Scientifique” (CNRS) and in part by the French “Agence Nationale de la Recherche” (ANR-08-BLAN-0200-CSD7; ANR-12-BSV2-0013-02). Jing Feng received a fellowship from the China Scholarship Council of the Ministry of Education of P. R. China.
References
Bemer, M., and Grossniklaus, U. (2012). Dynamic regulation of Polycomb group activity during plant development. Curr. Opin. Plant Biol. 15, 523–529. doi: 10.1016/j.pbi.2012.09.006
Bentsink, L., and Koornneef, M. (2008). Seed dormancy and germination. Arabidopsis Book 6:e0119. doi: 10.1199/tab.0119
Bergink, S., Salomons, F. A., Hoogstraten, D., Groothuis, T. A. M., De Waard, H., Wu, J., et al. (2006). DNA damage triggers nucleotide excision repair-dependent monoubiquitylation of histone H2A. Genes Dev. 20, 1343–1352. doi: 10.1101/gad.373706
Berr, A., McCallum, E. J., Alioua, A., Heintz, D., Heitz, T., and Shen, W.-H. (2010). Arabidopsis histone methyltransferase SET DOMAIN GROUP 8 mediates induction of the jasmonate/ethylene pathway genes in plant defense response to necrotrophic fungi. Plant Physiol. 154, 1403–1414. doi: 10.1105/tpc.110.079962
Berr, A., Ménard, R., Heitz, T., and Shen, W.-H. (2012). Chromatin modification and remodelling: a regulatory landscape for the control of Arabidopsis defence responses upon pathogen attack. Cell. Microbiol. 14, 829–839. doi: 10.1111/j.1462-5822.2012.01785.x
Berr, A., Shafiq, S., and Shen, W.-H. (2011). Histone modifications in transcriptional activation during plant development. Biochim. Biophys. Acta 1809, 567–576. doi: 10.1016/j.bbagrm.2011.07.001
Bourbousse, C., Ahmed, I., Roudier, F., Zabulon, G., Blondet, E., Balzergue, S., et al. (2012). Histone H2B monoubiquitination facilitates the rapid modulation of gene expression during Arabidopsis photomorphogenesis. PLoS Genet. 8:e1002825. doi: 10.1371/journal.pgen.1002825
Bratzel, F., López-Torrejón, G., Koch, M., Del Pozo, J. C., and Calonje, M. (2010). Keeping cell identity in Arabidopsis requires PRC1 RING-finger homologs that catalyze H2A monoubiquitination. Curr. Biol. 20, 1853–1859. doi: 10.1016/j.cub.2010.09.046
Braun, S., and Madhani, H. D. (2012). Shaping the landscape: mechanistic consequences of ubiquitin modification of chromatin. EMBO Rep. 13, 619–630. doi: 10.1038/embor.2012.78
Cao, Y., Dai, Y., Cui, S., and Ma, L. (2008). Histone H2B monoubiquitination in the chromatin of FLOWERING LOCUS C regulates flowering time in Arabidopsis. Plant Cell 20, 2586–2602. doi: 10.1105/tpc.108.062760
Carlsten, J. O. P., Zhu, X., and Gustafsson, C. M. (2013). The multitalented mediator complex. Trends Biochem. Sci. 38, 531–537. doi: 10.1016/j.tibs.2013.08.007
Chen, D., Molitor, A., Liu, C., and Shen, W.-H. (2010). The Arabidopsis PRC1-like RING-finger proteins are necessary for repression of embryonic traits during vegetative growth. Cell Res. 20, 1332–1344. doi: 10.1038/cr.2010.151
Chernikova, S. B., Dorth, J. A., Razorenova, O. V., Game, J. C., and Brown, J. M. (2010). Deficiency in Bre1 impairs homologous recombination repair and cell cycle checkpoint response to radiation damage in mammalian cells. Radiat. Res. 174, 558–565. doi: 10.1667/RR2184.1
Derkacheva, M., Steinbach, Y., Wildhaber, T., Mozgova, I., Mahrez, W., Nanni, P., et al. (2013). Arabidopsis MSI1 connects LHP1 to PRC2 complexes. EMBO J. 32, 2073–2085. doi: 10.1038/emboj.2013.145
Dhawan, R., Luo, H., Foerster, A. M., Abuqamar, S., Du, H.-N., Briggs, S. D., et al. (2009). HISTONE MONOUBIQUITINATION1 interacts with a subunit of the Mediator complex and regulates defense against necrotrophic fungal pathogens in Arabidopsis. Plant Cell 21, 1000–1019. doi: 10.1105/tpc.108.062364
Fleury, D., Himanen, K., Cnops, G., Nelissen, H., Boccardi, T. M., Maere, S., et al. (2007). The Arabidopsis thaliana homolog of yeast BRE1 has a function in cell cycle regulation during early leaf and root growth. Plant Cell 19, 417–432. doi: 10.1105/tpc.106.041319
Ginjala, V., Nacerddine, K., Kulkarni, A., Oza, J., Hill, S. J., Yao, M., et al. (2011). BMI1 is recruited to DNA breaks and contributes to DNA damage-induced H2A ubiquitination and repair. Mol. Cell. Biol. 31, 1972–1982. doi: 10.1128/MCB.00981-10
Goldknopf, I. L., Taylor, C. W., Baum, R. M., Yeoman, L. C., Olson, M. O., Prestayko, A. W., et al. (1975). Isolation and characterization of protein A24, a “histone-like” non-histone chromosomal protein. J. Biol. Chem. 250, 7182–7187.
Gu, X., Jiang, D., Wang, Y., Bachmair, A., and He, Y. (2009). Repression of the floral transition via histone H2B monoubiquitination. Plant J. 57, 522–533. doi: 10.1111/j.1365-313X.2008.03709.x
Han, J., Zhang, H., Zhang, H., Wang, Z., Zhou, H., and Zhang, Z. (2013). A Cul4 E3 ubiquitin ligase regulates histone hand-off during nucleosome assembly. Cell 155, 817–829. doi: 10.1016/j.cell.2013.10.014
Hershko, A., and Ciechanover, A. (1998). The ubiquitin system. Annu. Rev. Biochem. 67, 425–479. doi: 10.1146/annurev.biochem.67.1.425
Himanen, K., Woloszynska, M., Boccardi, T. M., De Groeve, S., Nelissen, H., Bruno, L., et al. (2012). Histone H2B monoubiquitination is required to reach maximal transcript levels of circadian clock genes in Arabidopsis. Plant J. 72, 249–260. doi: 10.1111/j.1365-313X.2012.05071.x
Hua, Z., and Vierstra, R. D. (2011). The Cullin-RING ubiquitin-protein ligases. Annu. Rev. Plant Biol. 62, 299–334. doi: 10.1146/annurev-arplant-042809-112256
Hunt, L. T., and Dayhoff, M. O. (1977). Amino-terminal sequence identity of ubiquitin and the nonhistone component of nuclear protein A24. Biochem. Biophys. Res. Commun. 74, 650–655. doi: 10.1016/0006-291X(77)90352-7
Hwang, W. W., Venkatasubrahmanyam, S., Ianculescu, A. G., Tong, A., Boone, C., and Madhani, H. D. (2003). A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol. Cell 11, 261–266. doi: 10.1016/S1097-2765(02)00826-2
Kim, J., Hake, S. B., and Roeder, R. G. (2005). The human homolog of yeast BRE1 functions as a transcriptional coactivator through direct activator interactions. Mol. Cell 20, 759–770. doi: 10.1016/j.molcel.2005.11.012
Krichevsky, A., Zaltsman, A., Lacroix, B., and Citovsky, V. (2011). Involvement of KDM1C histone demethylase–OTLD1 otubain-like histone deubiquitinase complexes in plant gene repression. Proc. Natl. Acad. Sci. U.S.A. 108, 11157–11162. doi: 10.1073/pnas.1014030108
Li, W., Wang, Z., Li, J., Yang, H., Cui, S., Wang, X., et al. (2011). Overexpression of AtBMI1C, a Polycomb group protein gene, accelerates flowering in Arabidopsis. PLoS ONE 6:e21364. doi: 10.1371/journal.pone.0021364
Liu, Y., Koornneef, M., and Soppe, W. J. J. (2007). The absence of histone H2B monoubiquitination in the Arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy. Plant Cell 19, 433–444. doi: 10.1105/tpc.106.049221
Lolas, I. B., Himanen, K., Grønlund, J. T., Lynggaard, C., Houben, A., Melzer, M., et al. (2010). The transcript elongation factor FACT affects Arabidopsis vegetative and reproductive development and genetically interacts with HUB1/2. Plant J. 61, 686–697. doi: 10.1111/j.1365-313X.2009.04096.x
Luger, K., Mader, A. W., Richmond, R. K., Sargent, D. F., and Richmond, T. J. (1997). Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260. doi: 10.1038/38444
Luo, M., Luo, M.-Z., Buzas, D., Finnegan, J., Helliwell, C., Dennis, E. S., et al. (2008). UBIQUITIN-SPECIFIC PROTEASE 26 is required for seed development and the repression of PHERES1 in Arabidopsis. Genetics 180, 229–236. doi: 10.1534/genetics.108.091736
Malapeira, J., Khaitova, L. C., and Mas, P. (2012). Ordered changes in histone modifications at the core of the Arabidopsis circadian clock. Proc. Natl. Acad. Sci. U.S.A. 109, 21540–21545. doi: 10.1073/pnas.1217022110
Manzano, C., Abraham, Z., López-Torrejón, G., and Pozo, J. (2008). Identification of ubiquitinated proteins in Arabidopsis. Plant Mol. Biol. 68, 145–158. doi: 10.1007/s11103-008-9358-9
Maor, R., Jones, A., Nühse, T. S., Studholme, D. J., Peck, S. C., and Shirasu, K. (2007). Multidimensional protein identification technology (MudPIT) analysis of ubiquitinated proteins in plants. Mol. Cell. Proteomics 6, 601–610. doi: 10.1074/mcp.M600408-MCP200
Marteijn, J. A., Simon B. J., Mailand N., Lans H., Schwertman P., Gourdin A. M., et al. (2009). Nucleotide excision repair–induced H2A ubiquitination is dependent on MDC1 and RNF8 and reveals a universal DNA damage response. J. Cell Biol. 186, 835–847. doi: 10.1083/jcb.200902150
Ménard, R., Verdier G., Ors M., Erhardt M., Beisson F., and Shen W. H. (2014). Histone H2B monoubiquitination is involved in the regulation of cutin and wax composition in Arabidopsis thaliana. Plant Cell Physiol. 55, 455–466. doi: 10.1093/pcp/pct182
Molitor, A. M., Bu, Z., Yu, Y., and Shen, W. H. (2014). Arabidopsis AL PHD-PRC1 complexes promote seed germination through H3K4me3-to-H3K27me3 chromatin state switch in repression of seed developmental genes. PloS Genet. 10:e1004091. doi: 10.1371/journal.pgen.1004091
Molitor, A., and Shen, W.-H. (2013). The Polycomb complex PRC1: composition and function in plants. J. Genet. Genomics 40, 231–238. doi: 10.1016/j.jgg.2012.12.005
Moyal, L., Lerenthal, Y., Gana-Weisz, M., Mass, G., So, S., Wang, S.-Y., et al. (2011). Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol. Cell 41, 529–542. doi: 10.1016/j.molcel.2011.02.015
Nakamura, K., Kato, A., Kobayashi, J., Yanagihara, H., Sakamoto, S., Oliveira, D. V., et al. (2011). Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol. Cell 41, 515–528. doi: 10.1016/j.molcel.2011.02.002
Palma, K., Thorgrimsen, S., Malinovsky, F. G., Fiil, B. K., Nielsen, H. B., Brodersen, P., et al. (2010). Autoimmunity in Arabidopsis acd11 is mediated by epigenetic regulation of an immune receptor. PLoS Pathog. 6:e1001137. doi: 10.1371/journal.ppat.1001137
Pavri, R., Zhu, B., Li, G., Trojer, P., Mandal, S., Shilatifard, A., et al. (2006). Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125, 703–717. doi: 10.1016/j.cell.2006.04.029
Pinder, J. B., Attwood, K. M., and Dellaire, G. (2013). Reading, writing, and repair: the role of ubiquitin and the ubiquitin-like proteins in DNA damage signaling and repair. Front. Genet. 4:45. doi: 10.3389/fgene.2013.00045
Qin, F., Sakuma, Y., Tran, L.-S. P., Maruyama, K., Kidokoro, S., Fujita, Y., et al. (2008). Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress–responsive gene expression. Plant Cell 20, 1693–1707. doi: 10.1105/tpc.107.057380
Robzyk, K., Recht, J., and Osley, M. A. (2000). Rad6-dependent ubiquitination of histone H2B in yeast. Science 287, 501–504. doi: 10.1126/science.287.5452.501
Roudier, F., Ahmed, I., Bérard, C., Sarazin, A., Mary-Huard, T., Cortijo, S., et al. (2011). Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 30, 1928–1938. doi: 10.1038/emboj.2011.103
Sanchez-Pulido, L., Devos, D., Sung, Z., and Calonje, M. (2008). RAWUL: a new ubiquitin-like domain in PRC1 RING finger proteins that unveils putative plant and worm PRC1 orthologs. BMC Genomics 9:308. doi: 10.1186/1471-2164-9-308
Sang, Y., Wu, M.-F., and Wagner, D. (2009). The stem cell–chromatin connection. Semin. Cell Dev. Biol. 20, 1143–1148. doi: 10.1016/j.semcdb.2009.09.006
Saracco, S. A., Hansson, M., Scalf, M., Walker, J. M., Smith, L. M., and Vierstra, R. D. (2009). Tandem affinity purification and mass spectrometric analysis of ubiquitylated proteins in Arabidopsis. Plant J. 59, 344–358. doi: 10.1111/j.1365-313X.2009.03862.x
Schmitz, R. J., Tamada, Y., Doyle, M. R., Zhang, X., and Amasino, R. M. (2009). Histone H2B deubiquitination is required for transcriptional activation of FLOWERING LOCUS C and for proper control of flowering in Arabidopsis. Plant Physiol. 149, 1196–1204. doi: 10.1104/pp.108.131508
Schwartz, Y. B., and Pirrotta, V. (2013). A new world of Polycombs: unexpected partnerships and emerging functions. Nat. Rev. Genet. 14, 853–864. doi: 10.1038/nrg3603
Shafiq, S., Berr, A., and Shen, W.-H. (2014). Combinatorial functions of diverse histone methylations in Arabidopsis thaliana flowering time regulation. New Phytol. 201, 312–322. doi: 10.1111/nph.12493
Shen, W.-H., Parmentier, Y., Hellmann, H., Lechner, E., Dong, A., Masson, J., et al. (2002). Null mutation of AtCUL1 causes arrest in early embryogenesis in Arabidopsis. Mol. Biol. Cell 13, 1916–1928. doi: 10.1091/mbc.E02-02-0077
Shen, W.-H., and Xu, L. (2009). Chromatin remodeling in stem cell maintenance in Arabidopsis thaliana. Mol. Plant 2, 600–609. doi: 10.1093/mp/ssp022
Shilatifard, A. (2006). Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 75, 243–269. doi: 10.1146/annurev.biochem.75.103004.142422
Simon, J. A., and Kingston, R. E. (2013). Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell 49, 808–824. doi: 10.1016/j.molcel.2013.02.013
Sridhar, V. V., Kapoor, A., Zhang, K., Zhu, J., Zhou, T., Hasegawa, P. M., et al. (2007). Control of DNA methylation and heterochromatic silencing by histone H2B deubiquitination. Nature 447, 735–738. doi: 10.1038/nature05864
Tanny, J. C., Erdjument-Bromage, H., Tempst, P., and Allis, C. D. (2007). Ubiquitylation of histone H2B controls RNA polymerase II transcription elongation independently of histone H3 methylation. Genes Dev. 21, 835–847. doi: 10.1101/gad.1516207
Turck, F., Roudier, F., Farrona, S., Martin-Magniette, M., Guillaume, E., Buisine, N., et al. (2007). Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 3:e86. doi: 10.1371/journal.pgen.0030086
Wang, L., Brown, J. L., Cao, R., Zhang, Y., Kassis, J. A., and Jones, R. S. (2004). Hierarchical recruitment of polycomb group silencing complexes. Mol. Cell 14, 637–646. doi: 10.1016/j.molcel.2004.05.009
Weake, V. M., and Workman, J. L. (2008). Histone ubiquitination: triggering gene activity. Mol. Cell 29, 653–663. doi: 10.1016/j.molcel.2008.02.014
West, M. H. P., and Bonner, W. M. (1980). Histone 2B can be modified by the attachment of ubiquitin. Nucleic Acids Res. 8, 4671–4680. doi: 10.1093/nar/8.20.4671
Wood, A., Krogan, N. J., Dover, J., Schneider, J., Heidt, J., Boateng, M. A., et al. (2003). Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell 11, 267–274. doi: 10.1016/S1097-2765(02)00802-X
Xiao, T., Kao, C.-F., Krogan, N. J., Sun, Z.-W., Greenblatt, J. F., Osley, M. A., et al. (2005). Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol. Cell. Biol. 25, 637–651. doi: 10.1128/MCB.25.2.637-651.2005
Xu, L., Ménard, R., Berr, A., Fuchs, J., Cognat, V., Meyer, D., et al. (2009). The E2 ubiquitin-conjugating enzymes, AtUBC1 and AtUBC2, play redundant roles and are involved in activation of FLC expression and repression of flowering in Arabidopsis thaliana. Plant J. 57, 279–288. doi: 10.1111/j.1365-313X.2008.03684.x
Xu, L., and Shen, W.-H. (2008). Polycomb silencing of KNOX genes confines shoot stem cell niches in Arabidopsis. Curr. Biol. 18, 1966–1971. doi: 10.1016/j.cub.2008.11.019
Yang, C., Bratzel, F., Hohmann, N., Koch, M., Turck, F., and Calonje, M. (2013). VAL- and AtBMI1-mediated H2Aub initiate the switch from embryonic to postgerminative growth in Arabidopsis. Curr. Biol. 23, 1324–1329. doi: 10.1016/j.cub.2013.05.050
Yao, X. Z., and Shen, W.-H. (2011). Crucial function of histone lysine methylation in plant reproduction. Chin. Sci. Bull. 56, 3493–3499. doi: 10.1007/s11434-011-4814-3
Zhang, K., Sridhar, V. V., Zhu, J., Kapoor, A., and Zhu, J.-K. (2007a). Distinctive core histone post-translational modification patterns in Arabidopsis thaliana. PLoS ONE 2:e1210. doi: 10.1371/journal.pone.0001210
Zhang, X., Germann, S., Blus, B. J., Khorasanizadeh, S., Gaudin, V., and Jacobsen, S. E. (2007b). The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat. Struct. Mol. Biol. 14, 869–871. doi: 10.1038/nsmb1283
Zhang, Y. (2003). Transcriptional regulation by histone ubiquitination and deubiquitination. Genes Dev. 17, 2733–2740. doi: 10.1101/gad.1156403
Zhu, B., Zheng, Y., Pham, A.-D., Mandal, S. S., Erdjument-Bromage, H., Tempst, P., et al. (2005). Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol. Cell 20, 601–611. doi: 10.1016/j.molcel.2005.09.025
Keywords: chromatin, epigenetics, ubiquitin, histone monoubiquitination, transcription regulation, plant development, Arabidopsis thaliana
Citation: Feng J and Shen W-H (2014) Dynamic regulation and function of histone monoubiquitination in plants. Front. Plant Sci. 5:83. doi: 10.3389/fpls.2014.00083
Received: 02 January 2014; Paper pending published: 24 January 2014;
Accepted: 22 February 2014; Published online: 13 March 2014.
Edited by:
Hongyong Fu, Institute of Plant and Microbial Biology – Academia Sinica, TaiwanReviewed by:
Keqiang Wu, National Taiwan University, TaiwanHongyong Fu, Institute of Plant and Microbial Biology – Academia Sinica, Taiwan
Copyright © 2014 Feng and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wen-Hui Shen, Institut de Biologie Moléculaire des Plantes, UPR2357 CNRS, Université de Strasbourg, 12 rue du Général Zimmer, 67084 Strasbourg Cedex, France e-mail:d2VuLWh1aS5zaGVuQGlibXAtY25ycy51bmlzdHJhLmZy
 Jing Feng
Jing Feng Wen-Hui Shen
Wen-Hui Shen