- Commonwealth Scientific and Industrial Research Organisation, Plant Industry, Cotton Biotechnology, Canberra, ACT, Australia
MYBMIXTA-like (MML) transcription factors form the subgroup 9 of R2R3-MYBs (Stracke et al., 2001) whose first characterized member was MIXTA from Antirrhinum majus. Various MML genes have been shown to be important regulators of epidermal cell differentiation in different plant species, including specifying cell shape in petals, vegetative trichome initiation and branching and seed fiber initiation (Martin et al., 2002; Machado et al., 2009; Walford et al., 2011). Indeed, the conical cells of petals look very much like young fibers shortly after they protrude from the epidermal surface of the cotton seed and begin to elongate, so it is likely there is some commonality in cellular regulation between the different tissue types. In tetraploid cotton, Gossypium hirsutum L. (Gh), the silencing or over-expression of two MML genes (GhMYB25 and GhMYB25Like) expressed predominantly in the ovule epidermis during fiber initiation affect the initiation or timing of expansion of fiber initials (Machado et al., 2009; Walford et al., 2011). Based on silencing phenotypes and gene expression data, GhMYB25Like may be one of the most upstream genes in a regulatory cascade currently known to involve GhMYB25, GhMYB109 (an R2R3 MYB from subgroup 15, Pu et al., 2008) and other types of transcription factors, such as the homeodomain leucine zipper (HDzip) factor GhHD-1, that may act in a protein complex with GhMYB25 (Zhang et al., 2010; Walford et al., 2012). The recent release of the genome sequence from the diploid G. raimondii (Gr), an extant species most closely related to the D-genome progenitor of tetraploid cotton, may help draw a more complete picture about the evolution of the MML gene subgroup in cotton and their apparent expansion and recruitment to specialized functions in epidermal seed fiber development.
Genomic Organization and Phylogeny of Cotton MMLs
The Gr genome contains over 200 R2R3 MYBs (Paterson et al., 2012), but 10 (GrMML1-10, Figure 1A) cluster with MIXTA, and all of these contain the signature protein motif AQWESARxxAExRLxRES previously indicated to be unique to subgroup 9 (Stracke et al., 2001). This number is considerably greater than the three MML genes found in Arabidopsis: AtMYB16, proposed to control the shape of petal epidermal cells (Baumann et al., 2007), AtMYB17 a putative regulator of early inflorescence development and seed germination (Zhang et al., 2009) and AtMYB106/NOK a negative regulator of trichome branching (Jakoby et al., 2008). The MML factors are distinct from the AtGL1-like MYBs (AtMYB0, AtMYB23, AtMYB66) of subgroup 15, involved in trichome and root hair initiation and development, that are often suggested as the likely models for regulators of seed fiber development in cotton. Subgroup 15 in Gr appears to have only one member, Gorai.012G061800/GrMYB109 (Paterson et al., 2012) that is a homolog of GhMYB109 with a demonstrated role in fiber elongation rather than initiation (Pu et al., 2008). Gr has very close homologs of GhMYB25 (Gorai.012G186500, GrMML7) and GhMYB25Like (Gorai.008G179600, GrMML3) as well as eight other novel MML genes, each with a pair of very close homologs in the A- and D-genomes of tetraploid Gh (Figure 1A, Supplementary data 1). The 10 MML proteins fall into 4 distinct clades supported by elevated bootstrap scores. One of these, Sg9-2, includes GrMML3/GhMYB25Like, GrMML7/GhMYB25, Gr/GhMML9 and 10, but no Arabidopsis or Antirrhinum MML MYBs (Figure 1A). In the Gr genome the 10 MML genes are distributed across 6 of the 13 diploid cotton chromosomes. However, on chromosomes Chr08 and Chr13 there are clusters of two (GrMML3 and 4 and a fragment of another MML gene) and three genes (GrMML8, 9, and 10), respectively, in tandem arrangements (Paterson et al., 2012; Figure 1B). This suggests that they may have evolved from gene duplications of ancestral MML genes and subsequently evolved new functions and in some cases different expression patterns.
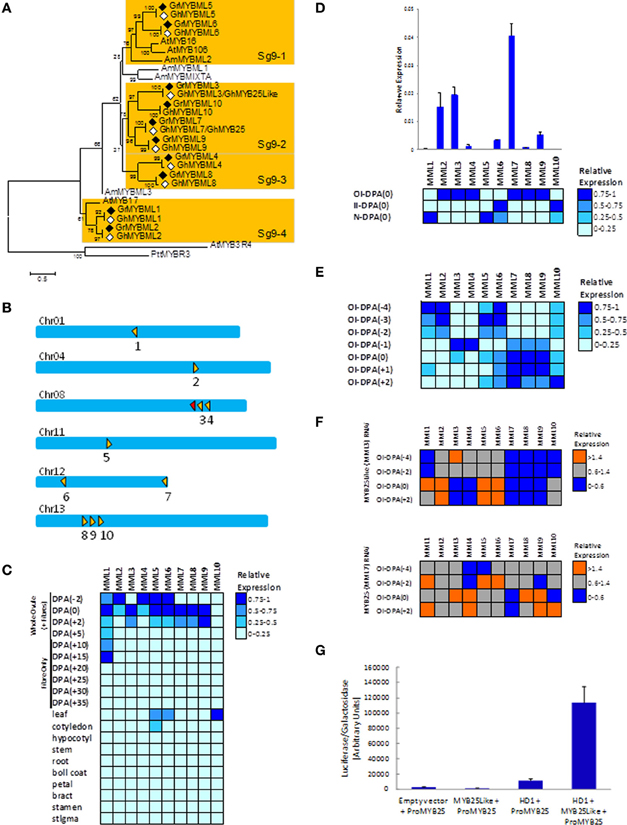
Figure 1. Phylogenetic analysis, chromosomal location, gene expression of the cotton MML genes in wild type and transgenic G. hirsutum, and transactivation of the GhMML3 promoter in cotton protoplasts. (A) Phylogenetic analysis of the G. raimondii (Gr, filled diamonds) MML proteins and their putative G. hirsutum (Gh, empty diamonds) D-genome homologs. Four clades (Sg9-1 to 4) are indicated by shading. The rooted Neighbour–Joining tree was obtained in MEGA 5.0 with Clustal W alignment using the full length amino acid sequences (details in Supplementary data 1). Arabidopsis thaliana (At) MYB3R4 and Populus tremula × P. Tremuloides (Ptt) MYBR3 are R1R2R3-MYBs used as outgroups and the other AtMYB and Anthirrinum majus (Am) sequences are landmarks of Subgroup 9. (B) Schematic of the chromosomal distribution of the 10 GrMML genes indicated by the filled triangles, while the unfilled triangle represents a fragment of a MML gene. The directions of the triangle indicate the coding strand of the transcripts and the numbers under the triangle the particular GrMML gene. Adjacent triangles are tandemly arranged genes. (Chromosomes are only approximately to scale). The heat maps visualize the transcript level differences between the G. hirsutum homologues of the GrMML genes in: (C) cotton fibers, ovules and selected other plant organs, (D) three dissected tissues (OI, II and N) from wild type (G. hirsutum) ovules collected the day of anthesis [DPA(0)], (E) dissected OI of wild type ovules collected from 4 days before anthesis [DPA(−4)] to 2 days after anthesis [DPA(+2)], (F) OI dissected from G. hirsutum ovules silenced by RNAi for GhMYB25Like (ie., MYBML3) and GhMYB25 (i.e., MYBML7) and their respective controls (ie., null segregant plants) collected at DPA(−4), (−2), (0), and (+2) (F). Heat maps were made using Expander software based on gene expression relative to the cotton ubiquitin gene and normalized for each MML gene and separate experiment (details in Supplementary data 2). Primers used detect both the A- and D-genome homoeologues of each MML gene. (G) Transactivation assay of the GhMML3/GhMYB25 promoter-Luciferase reporter by GhMYB25Like and/or GhHD-1 in cotton cotyledon protoplasts (details in Supplementary data 3).
MML Gene Expression During the Early Events of Fiber Initiation and Development
The 10 MML genes are nearly all expressed predominantly in early fiber development in whole ovules/seeds of tetraploid cotton from −2 to +2 day post anthesis (DPA) (Figure 1C) with little expression in other cotton tissues. A few (GhMML5, 6, 10) are also expressed in leaves and/or cotyledons which are rich in either hair or glandular trichomes that are structurally related to seed fibers and share some common regulators like GhHD-1 (Walford et al., 2012). MML gene expression levels were also investigated using hand-microdissected tissues (Bedon et al., 2013) from whole ovules and young seeds to focus just on the fiber initiation process occurring in the epidermis. This circumvents the transcript dilution or confounding with expression from the other layers of ovule tissues. Six of the GhMML genes were preferentially expressed at 0 DPA in outer-integuments (OI; GhMML2, 3, 4, 7, 8, and 9), two in inner-integument (II; GhMML6 and 10) and two in the nucellus (N; GhMML1 and 5) (heat map in Figure 1D). Based on their differences in relative expression levels in the OI, three groups can be classified as having; high (GhMML2, 3 and 7), low to medium (GhMML 4, 6, 8, and 9), and very low (GhMML1, 5, and 10) expression (chart in Figure 1D). Further expression analysis in the OI was performed from −4 to +2 DPA which covered the stages of ovule epidermal cell differentiation (−4 to −1 DPA), fiber initiation (i.e., ballooning of fiber initials above the epidermis surface at 0 DPA), and early fiber elongation (+1 to +2 DPA) (Figure 1E). There are four different patterns with some GhMML genes having their highest expression from −4 to −2 DPA (GhMML1, 2, 5, 6); genes peaking at −1 DPA (GhMML3 and 4); genes peaking at 0 to +1 DPA (GhMML7, 8, 9); and one member with highest expression at +2 DPA (GhMML10) (Figure 1E). These expression patterns support the specialization of different sets of MML genes for specific aspects of epidermal cell differentiation, although they may still have some roles in other tissues.
MML Regulatory Cascades and Interactions
To unravel the potential transcriptional networks among the different GhMML members their expression levels were assessed in dissected OI from transgenic tetraploid cotton silenced (through RNAi) for GhMML3/GhMYB25Like (Walford et al., 2011) or GhMML7/GhMYB25 (Machado et al., 2009) compared to the transcript levels from the corresponding null segregant plants as controls (Figure 1F, Supplementary data 2). In the GhMYB25Like silenced plants Figure 1F (upper panel), the transcript level of GhMYB25Like/GhMML3 was decreased at 0 and +2 DPA, as previously reported (Walford et al., 2011). Transcripts of GhMML7/GhMYB25, GhMML8, and 9 were completely abolished at 0 and +2 DPA, and GhMML4 was decreased to a lesser extent, indicating that they may all be downstream of GhMYB25-like. A significant increase in GhMML2 and 6 transcripts were observed at 0 and +2 DPA (Figure 1F), suggesting that they may be repressed by GhMYB25-like. A slight decrease in GhMML1 at −4 DPA was also seen (see chart representation in Supplementary data 2). In the GhMYB25 silenced plants (Figure 1F- lower panel), the transcript levels of GhMML7/GhMYB25 was decreased at 0 and +2 DPA as previously reported by Machado et al. (2009). Transcript levels of GhMML3/GhMYB25Like, GhMML4 and 8, and to a lesser extent GhMML9, were increased at 0 DPA (Supplementary data 2), suggesting there may be some feedback regulation of GhMYB25Like by GhMYB25. The direct requirement of GhMYB25Like/GhMML3 for expression of GhMML7/GhMYB25 was assessed using a transient assay in cotton protoplasts (Figure 1G, Supplementary data 3). Transactivation of the GhMYB25 promoter, fused to the luciferase reporter, did occur in the presence of GhMYB25Like, but only when co-expressed with the HDzip factor GhHD-1, recently shown to be involved in fiber initiation (Walford et al., 2012), so the two may be involved in a complex to activate GhMYB25.
Conclusion
The MYBMIXTA-like subgroup appears to have expanded in cotton compared to non-fiber plants, probably following a cotton specific triplication as reported in Paterson et al. (2012), thus contributing to the evolution of seed fiber. The spatio-temporal expression study of the different GhMML genes reported here distinguishes them according to the plant organs and/or ovule tissues in which they are expressed and the timing of their expression during the early stages of seed fiber development. The newly described GhMML2 is one of the more highly expressed MML genes in the OI along with GhMML7/GhMYB25 and GhMML3/GhMYB25Like which have already been shown to be involved in seed fiber initiation (Machado et al., 2009; Walford et al., 2011), but GhMML2 shows a different pattern of expression with highest transcript accumulation before the fiber initiation step. GhMML2, together with GhMML6, transcripts are increased rather than decreased at 0 and +2 DPA in GhMYB25Like RNAi plants, suggesting a molecular function different to GhMML7, 8, and 9 that are down-regulated in this transgenic background. The differences in GhMML expression in OI in the two MMLs silenced plants highlights a possible hierarchical network between MMLs; with MML3/GhMYB25-like activating the transcription of MML7/GhMYB25, MML8, and MML9 and perhaps being involved in the repression of MML2 and MML6 at 0 and +2 DPA as the fibers begin to expand and elongate. MML7/GhMYB25 might be involved in a feedback loop to transcriptionally activate MML3, 4, 8, and 9 at 0 DPA. We were able to show that MML3/GhMYB25Like is able to transactivate the MML7/MYB25 promoter, but only when associated with GhHD-1. Such results suggest the presence of a transcriptional complexes, involving MML(s) and GhHD-1, necessary for fiber development that are similar to the sorts of complexes seen in Arabidopsis trichome development (Ishida et al., 2008), although clearly involving different factors. Here, we identified some potential new players in fiber development that belong to the same phylogenetic group of MYBs and suggest a very subtle networking involving both protein-protein and protein-DNA interactions. The next steps involve the functional characterization of these novel cotton MML genes by silencing and over-expression in transgenic cotton and the study of their interactions in both cotton protoplasts and yeast to confirm their roles in generating the fibers of commercially important cotton species.
Author Contributions
Phylogeny analysis (Figure 1A): Frank Bedon. Genomic organization of MMLs (Figure 1B): Danny J. Llewellyn. Gene expression studies (Figures 1D–F): Frank Bedon and Lisa Ziolkowski, (Figure 1C): Sally A. Walford. Transient assays in protoplast (Figure 1G): Frank Bedon and Lisa Ziolkowski. Wrote the paper: Frank Bedon, Sally A. Walford, Elizabeth S. Dennis and Danny J. Llewellyn.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported in part by funding from the Monsanto Company (Frank Bedon, Lisa Ziolkowski), as well as Cotton Breeding Australia, a joint venture between CSIRO and Cotton Seed Distributors (Sally A. Walford, Danny J. Llewellyn).
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2014.00179/full
Supplementary data 1. Sequence information.
Supplementary data 2. Gene expression data.
Supplementary data 3. Transient assays.
References
Baumann, K., Perez-Rodriguez, M., Bradley, D., Venail, J., Bailey, P., Jin, H., et al. (2007). Control of cell and petal morphogenesis by R2R3 MYB transcription factors. Development 134, 1691–1701. doi: 10.1242/dev.02836
Bedon, F., Ziolkowski, L., Osabe, K., Venables, I., Machado, A., and Llewellyn, D. J. (2013). Separation of integument and nucellar tissues from cotton ovules (Gossypium hirsutum L.) for both high- and low- throughput molecular applications. Biotechniques 54, 44–46. doi: 10.2144/000113969
Ishida, T., Kurata, T., Okada, K., and Wada, T. (2008). A genetic regulatory network in the development of trichomes and root hairs. Annu. Rev. Plant Biol. 59, 365–386. doi: 10.1146/annurev.arplant.59.032607.092949
Jakoby, M. J., Falkenhan, D., Mader, M. T., Brininstool, G., Wischnitzki, E., Platz, N., et al. (2008). Transcriptional profiling of mature Arabidopsis trichomes reveals that NOECK encodes the MIXTA-like transcriptional regulator MYB106. Plant Physiol. 148, 1583–1602. doi: 10.1104/pp.108.126979
Machado, A., Wu, Y., Yang, Y., Llewellyn, D. J., and Dennis, E. S. (2009). The MYB transcription factor GhMYB25 regulates early fibre and trichome development. Plant J. 59, 52–62. doi: 10.1111/j.1365-313X.2009.03847.x
Martin, C., Bhatt, K., Baumann, K., Jin, H., Zachgo, S., Roberts, K., et al. (2002). The mechanics of cell fate determination in petals. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357, 809–813. doi: 10.1098/rstb.2002.1089
Paterson, A. H., Wendel, J. F., Gundlach, H., Guo, H., Jenkins, J., Jin, D., et al. (2012). Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492, 423–427. doi: 10.1038/nature11798
Pu, L., Li, Q., Fan, X., Yang, W., and Xue, Y. (2008). The R2R3 MYB transcription factor GhMYB109 is required for cotton fibre development. Genetics 180, 811–820. doi: 10.1534/genetics.108.093070
Stracke, R., Werber, M., and Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4, 447–456. doi: 10.1016/S1369-5266(00)00199-0
Walford, S. A., Wu, Y. R., Llewellyn, D. J., and Dennis, E. S. (2011). GhMYB25-like: a key factor in early cotton fibre development. Plant J. 65, 785–797. doi: 10.1111/j.1365-313X.2010.04464.x
Walford, S. A., Wu, Y., Llewellyn, D. J., and Dennis, E. S. (2012). Epidermal cell differentiation in cotton mediated by the homeodomain leucine zipper gene, GhHD-1. Plant J. 71, 464–478. doi: 10.1111/j.1365-313X.2012.05003.x
Zhang, F., Zuo, K., Zhang, J., Liu, X., Zhang, L., Sun, X., et al. (2010). An L1 box binding protein, GbML1, interacts with GbMYB25 to control cotton fibre development. J. Exp. Bot. 61, 3599–3613. doi: 10.1093/jxb/erq173
Keywords: Gossypium, ovule, protoplast, transient assay, phylogeny, transcription, HDzip, fiber initiation
Citation: Bedon F, Ziolkowski L, Walford SA, Dennis ES and Llewellyn DJ (2014) Members of the MYBMIXTA-like transcription factors may orchestrate the initiation of fiber development in cotton seeds. Front. Plant Sci. 5:179. doi: 10.3389/fpls.2014.00179
Received: 21 February 2014; Accepted: 14 April 2014;
Published online: 01 May 2014.
Edited by:
John Schiefelbein, University of Michigan, USAReviewed by:
Xiaoya Chen, Shanghai Institutie of Plant Physiology and Ecology, ChinaMingxiong Pang, Texas Tech University, USA
Copyright © 2014 Bedon, Ziolkowski, Walford, Dennis and Llewellyn. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence:ZGFubnkubGxld2VsbHluQGNzaXJvLmF1
†Present Address: Frank Bedon, Department of Botany, La Trobe University, Melbourne, VIC, Australia
 Frank Bedon
Frank Bedon Lisa Ziolkowski
Lisa Ziolkowski Sally A. Walford
Sally A. Walford Elizabeth S. Dennis
Elizabeth S. Dennis Danny J. Llewellyn
Danny J. Llewellyn