- 1NARO Institute of Crop Science, Tsukuba, Japan
- 2Agrogenomics Research Center, National Institute of Agrobiological Sciences, Tsukuba, Japan
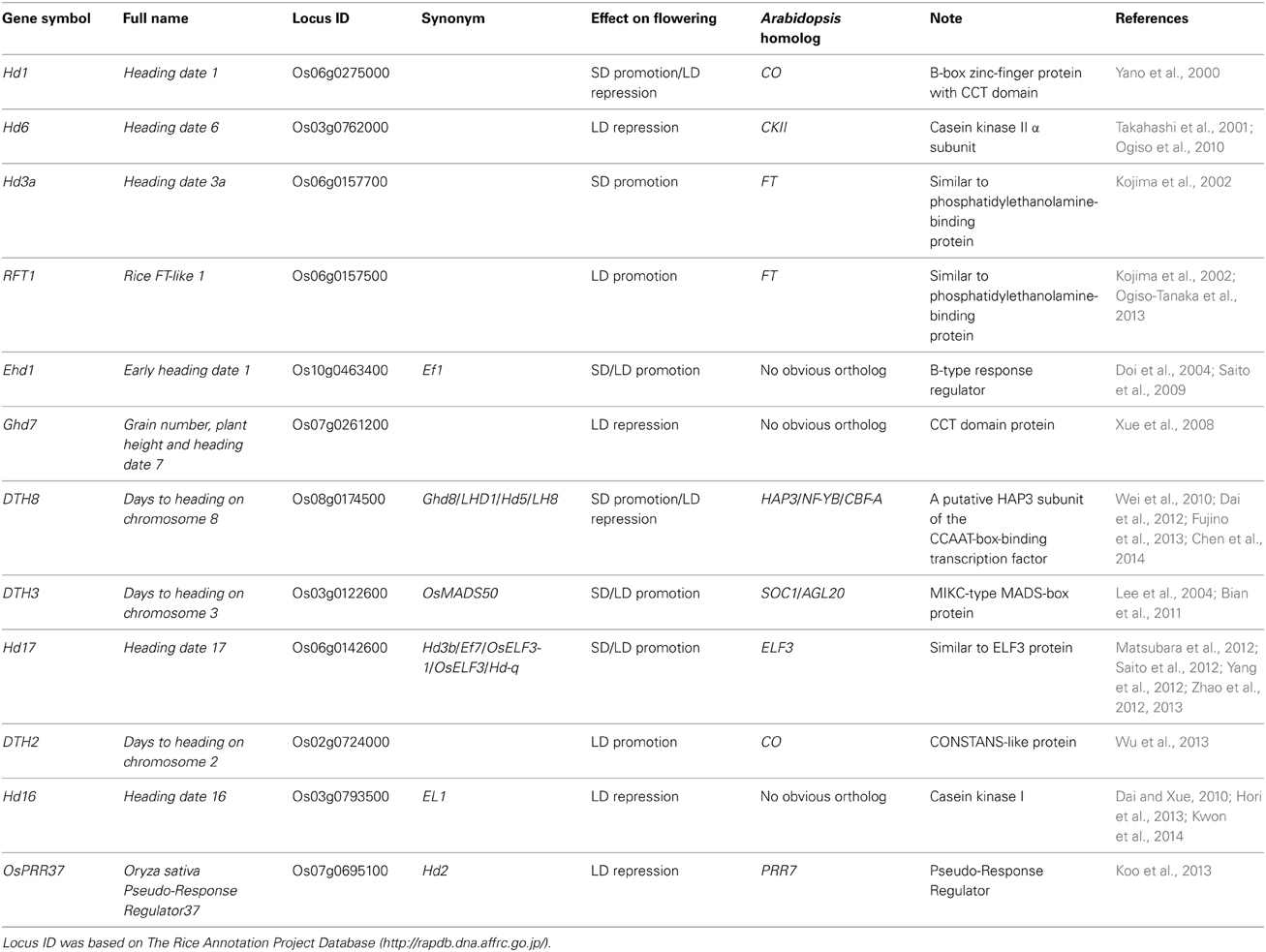
Flowering time in rice (Oryza sativa L.) is determined primarily by daylength (photoperiod), and natural variation in flowering time is due to quantitative trait loci involved in photoperiodic flowering. To date, genetic analysis of natural variants in rice flowering time has resulted in the positional cloning of at least 12 quantitative trait genes (QTGs), including our recently cloned QTGs, Hd17, and Hd16. The QTGs have been assigned to specific photoperiodic flowering pathways. Among them, 9 have homologs in the Arabidopsis genome, whereas it was evident that there are differences in the pathways between rice and Arabidopsis, such that the rice Ghd7–Ehd1–Hd3a/RFT1 pathway modulated by Hd16 is not present in Arabidopsis. In this review, we describe QTGs underlying natural variation in rice flowering time. Additionally, we discuss the implications of the variation in adaptive divergence and its importance in rice breeding.
Introduction
Photoperiodic flowering is one of the most important responses of plants to their environment (Thomas and Vince-Prue, 1997). In the last two decades, molecular genetics demonstrated that external light signals perceived by photoreceptors induce florigens (flowering signals). This process is regulated in part by the circadian clock, and promotes flowering in response to favorable daylength in both monocots and eudicots (reviewed by Andrés and Coupland, 2012).
Rice is a short-day (SD) plant, i.e., flowering is accelerated under SD conditions. Natural variation in rice flowering time is generated mainly by quantitative trait genes (QTGs) involved in photoperiod pathways, unlike in other cereals (such as wheat and barley) and Arabidopsis thaliana, which also need to be subjected to low temperature (vernalization requirement) (reviewed by Greenup et al., 2009; Tsuji et al., 2011; Bentley et al., 2013; Brambilla and Fornara, 2013; Itoh and Izawa, 2013).
To date, at least 12 QTGs, which belong to the two independent flowering pathways, have been mapped and cloned through quantitative trait locus (QTL) analysis of natural variation in flowering time (Figure 1 and Table 1). In this review, we describe the molecular basis of QTGs underlying natural variation in rice flowering time and discuss the implications on adaptive divergence and consequences for breeding.
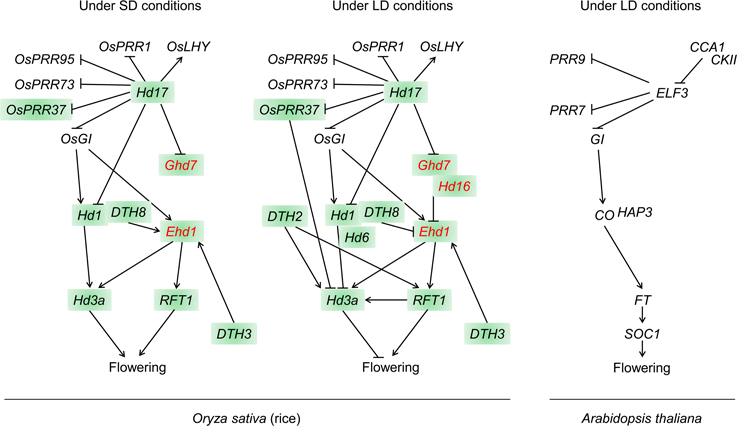
Figure 1. Schematic representation of roles of QTGs within the photoperiodic flowering pathways in rice and Arabidopsis thaliana. QTGs that are marked in red are not present in Arabidopsis. QTGs that were cloned by QTL analysis of natural variation are highlighted. Arrow heads, up-regulation; Bars, down-regulation.
Molecular basis of Natural Variation in Rice Flowering
A genetic pathway resembling that the photoperiod pathway in Arabidopsis [a long-day (LD) plant] is conserved in rice. Hd1 [a CONSTANS (CO) homolog in rice] was the first flowering time QTG cloned from natural rice variants. Hd1 promotes flowering under SD conditions and represses it under LD conditions (Yano et al., 2000) (Figure 1). By contrast, the Arabidopsis CO gene promotes flowering under LD conditions (Putterill et al., 1995). The daylength-dependent conversion of Hd1 activity between flowering activator and flowering repressor is caused by phytochrome-mediated signaling (e.g., Ishikawa et al., 2011). The repression of flowering by Hd1 under LD conditions is enhanced by the kinase activity of Hd6, which encodes the α-subunit of casein kinase II (Takahashi et al., 2001; Ogiso et al., 2010). Hd6 enhances the Hd1 repressor function under LD conditions through the phosphorylation of an unknown protein (Ogiso et al., 2010).
Hd1 regulates Hd3a, a rice homolog of Arabidopsis FLOWERING LOCUS T (FT) (Kojima et al., 2002) (Figure 1). Tamaki et al. (2007) demonstrated that Hd3a functions as a florigen. Another florigen gene, RFT1, is a tandemly duplicated paralog of Hd3a (Kojima et al., 2002; Ogiso-Tanaka et al., 2013). Komiya et al. (2008, 2009) found that RFT1 expression increases under LD conditions, indicating that RFT1 is a LD-specific florigen. More recently, Ogiso-Tanaka et al. (2013) demonstrated that functional defects, which were caused by sequence polymorphisms in the regulatory and coding regions of RFT1, contribute to late flowering under LD conditions in an indica cultivar.
In Arabidopsis, the CO–FT pathway is regulated by GIGANTEA (GI), which is a component of the circadian clock (Fowler et al., 1999; Park et al., 1999) (Figure 1). Similarly, regulation of the Hd1–Hd3a pathway is mediated by OsGI, a rice homolog of GI (Hayama et al., 2003). These findings reveal that a floral induction pathway from GI to FT in photoperiodic flowering is conserved between Arabidopsis (LD) and rice (SD), but that the photoperiod response differs between these plants.
A unique rice pathway with no obvious ortholog in Arabidopsis is also involved in photoperiodic flowering (Figure 1 and Table 1). Ehd1 is a flowering promoter that encodes a B-type response regulator. Ehd1 functions upstream of Hd3a and RFT1 (Doi et al., 2004). Ghd7, which encodes a CCT (CO, CO-LIKE, and TIMING OF CAB1)-domain protein, was isolated by analysis of natural variations in flowering time. Ghd7 affects the levels of Ehd1 and Hd3a transcripts, but does not affect Hd1 mRNA levels (Xue et al., 2008). Ghd7 represses Ehd1, Hd3a, and RFT1 under LD conditions, thereby delaying flowering.
Thus, two independent flowering pathways are present in rice, the conserved Hd1–Hd3a pathway and a unique Ghd7–Ehd1–Hd3a/RFT1 pathway, which may integrate environmental photoperiod signals in the control of flowering. In the following sections, we describe additional QTGs that were more recently cloned.
Hd17, a Rice Homolog of Arabidopsis ELF3
Subspecies japonica cultivars “Nipponbare” and “Koshihikari” differ in their flowering time and flowering responses to photoperiod. QTL analyses revealed that two QTLs on chromosomes 3 and 6 are involved in the difference in heading date between the two cultivars (Matsubara et al., 2008). The QTL mapped on chromosome 3 was designated as Hd16, and the QTL mapped on chromosome 6 was designated as Hd17. Both Hd16 and Hd17 are involved in photoperiod response, as revealed by observation of heading date in near-isogenic lines (NILs) under SD and LD conditions.
Hd17 explained a small proportion of the variance in flowering time between “Nipponbare” and “Koshihikari.” Map-based cloning demonstrated that this difference may result in part from a single-nucleotide polymorphism (SNP) within a putative gene encoding a rice homolog of the Arabidopsis EARLY FLOWERING 3 (ELF3) protein (Matsubara et al., 2012). The SNP was observed among Asian rice cultivars, mainly in japonica cultivars. It seems that the wild-type allele has the “Koshihikari” SNP (i.e., the “Nipponbare” allele is a natural variant), because almost all indica cultivars and wild accessions surveyed in the study carry the “Koshihikari” SNP.
The amino acid change (serine to leucine) caused by this SNP in Hd17 may reduce the mRNA level of the floral repressor Ghd7 under LD conditions, causing ‘Nipponbare’ to flower earlier than NIL-Hd17, which carries a chromosomal segment including the “Koshihikari” allele in the “Nipponbare” background. On the other hand, a loss-of-function mutation ef7 in the rice ELF3-like gene (= Hd17) seems to increase the Ghd7 transcription level, and the mutants flower later than wild-type plants under both SD and LD conditions (Saito et al., 2012). This suggests that the ELF3-like gene acts as a floral promoter by attenuating the Ghd7 transcription level (Figure 1).
Arabidopsis ELF3 regulates circadian rhythms by affecting the transcription of clock-associated genes such as LATE ELONGATED HYPOCOTYL (LHY), CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1), PSEUDO-RESPONSE REGULATORs (PRRs), and GI; the clock output gene CHLOROPHYLL A/B BINDING 2 (CAB2); and the floral promoter CO (reviewed by McClung, 2011) (Figure 1). The circadian clock function is conserved also in rice (Murakami et al., 2007). In the ef7 mutant, the expression of the luciferase gene driven by the CAB PROTEIN (a rice homolog of Arabidopsis CAB2) promoter under constant darkness was not affected, but the period of free-running rhythms under constant light was slightly shortened, suggesting that Ef7 mediates light input to the circadian clock but is not required for the clock function in the dark (Saito et al., 2012). By using an ELF3-like gene knockout and down-regulation of the expression of ELF3-like gene by RNAi, other groups revealed that the ELF3-like gene affects the mRNA expression of clock-associated genes and Hd1 (Yang et al., 2012; Zhao et al., 2012) (Figure 1). These observations suggest that the rice ELF3-like gene is also involved in the function of the circadian clock.
Hd16, a Gene Encoding Casein Kinase I
The “Koshihikari” allele of Hd16 decreased photoperiod response in comparison with the “Nipponbare” allele. Map-based cloning revealed an SNP in the gene encoding casein kinase I (CKI), which has no obvious ortholog in Arabidopsis (Hori et al., 2013). The SNP resulted in a non-synonymous substitution of an alanine (“Nipponbare”) with threonine (“Koshihikari”). CKI is a protein serine/threonine kinase that is highly conserved among plant and animal species (reviewed by Tuazon and Traugh, 1991; Gross and Anderson, 1998). CKI has various functions in both the cytoplasm and the nucleus, such as DNA repair, and regulation of the cell cycle and circadian rhythm (Liu et al., 2003; Rumpf et al., 2010). Phosphorylation of the clock components by CKI is the key step that initiates and regulates the circadian rhythm. The tau gene encodes CKIε in golden hamster (Mesocricetus auratus), and a missense mutation in this gene drastically reduces the period of the circadian rhythm (Ralph and Menaker, 1988). The non-synonymous substitution in the “Koshihikari” Hd16 allele is located at a site close to the tau mutation site, and is within the activation loop of the catalytic domain of CKI (Hori et al., 2013). However, the expression patterns of clock-associated genes are similar in the presence of the “Nipponbare” and “Koshihikari” Hd16 alleles. Therefore, Hd16 regulates flowering time mediated by the photoperiodic flowering pathway without affecting the regulation of the circadian rhythm.
To reveal the role of Hd16 in the photoperiodic flowering pathway, we investigated the genetic interactions between Hd16 and other flowering time QTLs, and the expression levels of the latter (Hori et al., 2013). In rice NILs with functional or deficient alleles of flowering-time genes, significant pairwise interactions were observed between Hd16 and four other QTLs: Ghd7, Hd1, DTH8, and Hd2 (= OsPRR37). The transcription levels of Ehd1, Hd3a, and RFT1 differed between NILs carrying the “Nipponbare” and “Koshihikari” Hd16 alleles. Biochemical characterization indicated that the Hd16 recombinant protein encoded by the “Nipponbare” allele specifically phosphorylated Ghd7 (but not Hd1) in vitro. The kinase activity of “Koshihikari” Hd16 was strongly decreased relative to that of “Nipponbare” (Hori et al., 2013). Thus, Hd16 acts as a Ghd7 inhibitor in the rice flowering-time pathway by enhancing the photoperiod response as a result of Ghd7 phosphorylation (Figure 1).
Another missense mutation was found in the kinase domain of Hd16 in the early-heading Korean cultivar “H143” (Kwon et al., 2014). In vitro kinase assays revealed that the “H143” Hd16 allele is also defective. Thus, there are two defective natural variants of Hd16. The “Koshihikari” and “H143” alleles were found only among japonica cultivars in temperate areas (Hori et al., 2013; Kwon et al., 2014).
Hd16 was previously identified as Early flowering 1 (EL1) (Dai and Xue, 2010), which controls rice flowering time by down-regulating the gibberellin (GA) signaling pathway mediated by phosphorylation of SLR1, encoded by Slender rice 1. SLR1 phosphorylation suppresses the GA response, whereas Ghd7 phosphorylation enhances the photoperiod response. In both cases, phosphorylation leads to delayed flowering under LD conditions. Thus, Hd16/EL1 appears to be associated with both photoperiod and GA responses in rice flowering.
Other QTGs Underlying Natural Flowering Time Variation in Rice
In addition to the QTGs described above, four rice homologs of the Arabidopsis flowering-related genes have been cloned by using genotypes that show natural variation in flowering time (Table 1). DTH8 encodes a rice homolog of the Arabidopsis HEME ACTIVATOR PROTEIN (YEAST) HOMOLOG 3 subunit of the CCAAT-box-binding transcription factor. Under LD conditions, DTH8 down-regulates Ehd1 and its downstream target Hd3a and therefore acts as a flowering suppressor (Wei et al., 2010) (Figure 1). A DTH8 variant also promotes flowering under SD conditions (Yan et al., 2011). Most recently, interaction between DTH8 and Hd1 was demonstrated by yeast-two-hybrid assay (Chen et al., 2014).
DTH3 encodes a rice homolog of Arabidopsis SUPPRESSOR OF OVEREXPRESSION OF CO1, a MIKC-type MADS-box protein. DTH3 up-regulates Ehd1 and RFT1 under both SD and LD conditions and thereby promotes flowering (Bian et al., 2011) (Figure 1). At this QTL, there is functional allelic variation between O. sativa and O. glaberrima, but probably not among O. sativa cultivars.
DTH2 encodes a CONSTANS-like protein. DTH2 up-regulates Hd3a and RFT1 under LD conditions and thus promotes flowering (Wu et al., 2013) (Figure 1). OsPRR37 encodes a rice homolog of Arabidopsis PRR7, and down-regulates Hd3a expression to suppress flowering under LD conditions (Murakami et al., 2007; Koo et al., 2013) (Figure 1).
Conservation and Diversification of Flowering Time Regulation between Rice and Arabidopsis
Much progress has been made in understanding the natural genetic variation in rice flowering. 12 QTGs have been isolated and assigned to specific photoperiod flowering pathways. Among them, 9 have homologs in the Arabidopsis genome, suggesting that the genetic basis of photoperiodic flowering has an ancient origin in flowering plants (Table 1).
On the other hand, it is evident that there are differences in the photoperiodic flowering pathways between the two species (Figure 1). The Ghd7–Ehd1–Hd3a/RFT1 pathway, which is modulated by Hd16, is not present in Arabidopsis. However, in many cases even orthologous genes have divergent functions, as exemplified by CO (flowering promotion in LD) and Hd1 (flowering promotion in SD), and by ELF3 (flowering repression in SD) and Hd17 (flowering promotion in both SD and LD). The differences between these orthologs may be associated with neofunctionalization, which is the evolution of new function of a duplicated gene, as suggested about the divergent function of orthologs of FLOWERING LOCUS VE and FT in sugar beet (Beta vulgaris, Pin et al., 2010; Abou-Elwafa et al., 2011). Actually, both Hd1 and ELF3 have putative paralog(s) in the rice genome.
Such differences may have resulted from evolution of these lineages in different geographic regions: rice in equatorial regions characterized by stable temperature and daylength all year round, Arabidopsis in temperate regions with fluctuating temperatures and changing daylength. As a result, rice has developed photoperiodic flowering, while Arabidopsis acquired an additional vernalization requirement as adaptation to the cold season (reviewed by Ballerini and Kramer, 2011).
Probably, this hypothesis should be examined among cereals, such as maize (Zea mays), wheat (Triticum aestivum) and barley (Hordeum vulgare) as well as rice, because they are more closely related to each other as rice and Arabidopsis (Magallón and Sanderson, 2005). Additionally, a similar difference in flowering phenology has been observed between tropical-origin (rice and maize) and temperate-origin (wheat and barley) cereals as between rice and Arabidopsis (e.g., Greenup et al., 2009; Jung and Müller, 2009).
We suggest that emphasizing divergence among plant species (particularly cereals) rather than conservation would help to better explain the genetic basis of flowering and adaptive divergence in rice.
Application of Natural Variation in Flowering Time Genes in Breeding Programs
Adjustment of photoperiod response changes flowering time and enhances adaptability to local environmental conditions in many plant species, including rice (e.g., Jung and Müller, 2009; Hori et al., 2012). To introduce rice at high latitudes, breeders have selected lines with a weaker response to photoperiod to produce early-flowering cultivars and to ensure maturation during the optimal period (e.g., Izawa, 2007; Ebana et al., 2011). The weak allele of Hd16 may have permitted the extension of the rice cultivation area into northern regions (Hori et al., 2013; Kwon et al., 2014). Deficient or weak alleles of Hd1, Ghd7, DTH8, DTH2, and OsPRR37 are also distributed in northern rice cultivation areas at high latitudes (Xue et al., 2008; Takahashi et al., 2009; Wei et al., 2010; Koo et al., 2013; Wu et al., 2013), strongly suggesting that such alleles are involved in the expansion of rice cultivation areas.
In addition to several major flowering time QTGs isolated in previous studies, we have identified QTLs with minor effects: Hd4 (likely allelic to Ghd7), Hd7, Hd9 (Lin et al., 2002, 2003), and Hd17 (Matsubara et al., 2012). These QTLs are necessary for breeders in fine tuning of the flowering time in rice cultivars. Breeders sometimes want to slightly change the flowering time of rice cultivars. Natural variations are observed at many loci (at least more than 20, according to a publicly available database, Q-TARO: http://qtaro.abr.affrc.go.jp/) involved in rice photoperiodic flowering pathways. These variations are available for producing a number of allelic combinations of flowering time QTLs and for developing rice cultivars adjusted to diverse cultivation areas.
Conclusions and Perspectives
Molecular cloning of QTGs underlying natural variation in flowering time of rice has improved our understanding of the genetic basis and provides insights into adaptive evolution and breeding in rice. However, we still know little about the distribution of QTG associated with flowering time in rice cultivars. We are still unable to predict exactly the relative effects of individual QTG on flowering time of rice cultivars in diverse natural field conditions, owing to limited knowledge of QTG × QTG and QTG x environmental interaction. This leads to the gap between genotype and phenotype (discussed by Benfey and Mitchell-Olds, 2008; Olsen and Wendel, 2013). To resolve this problem, we will need to assess the effect of each allele on flowering time in different allele combinations and in various environments by the combination of experimental populations, such as recombinant inbred lines (RILs), chromosome segment substitution lines (CSSLs), nested association mapping (NAM) population and multi-parent advanced generation inter-cross (MAGIC) population, and sequencing the alleles from each line. Furthermore, the transcriptome analysis of flowering-time genes under natural field conditions may also pave the way for the prediction of flowering time (Nagano et al., 2012).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Ministry of Agriculture, Forestry and Fisheries of Japan (Integrated Research Project for Plant, Insect, and Animal using Genome Technology, IP1001 to Masahiro Yano; Genomics for Agricultural Innovation, GPN0001 to Masahiro Yano), and by the Ministry of Education, Culture, Sports, Science and Technology of Japan through a Grant-in-Aid For Young Scientists (B) (20780008 to Kiyosumi Hori).
References
Abou-Elwafa, S. F., Büttner, B., Chia, T., Schulze-Buxloh, G., Hohmann, U., Mutasa-Göttgens, E., et al. (2011). Conservation and divergence of autonomous pathway genes in the flowering regulatory network of Beta vulgaris. J. Exp. Bot. 62, 3359–3374. doi: 10.1093/jxb/erq321
Andrés, F., and Coupland, G. (2012). The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13, 627–639. doi: 10.1038/nrg3291
Ballerini, E. S., and Kramer, E. M. (2011). In the light of evolution: a reevaluation of conservation in the CO–FT regulon and its role in photoperiodic regulation of flowering time. Front. Plant Sci. 2:81. doi: 10.3389/fpls.2011.00081
Benfey, P. N., and Mitchell-Olds, T. (2008). From genotype to phenotype: systems biology meets natural variation. Science 320, 495–497. doi: 10.1126/science.1153716
Bentley, A. R., Jensen, E. F., Mackay, I. J., Hönicka, H., Fladung, M., Hori, K., et al. (2013). “Flowering time,” in Genomics and Breeding for Climate-Resilient Crops: vol. 2 Target Traits, ed K. Chittaranjan (Berlin: Springer Berlin Heidelberg), 1–66.
Bian, X. F., Liu, X., Zhao, Z. G., Jiang, L., Gao, H., Zhang, Y. H., et al. (2011). Heading date gene, dth3 controlled late flowering in O. Glaberrima Steud. by down-regulating Ehd1. Plant Cell Rep. 30, 2243–2254. doi: 10.1007/s00299-011-1129-4
Brambilla, V., and Fornara, F. (2013). Molecular control of flowering in response to day length in rice. J. Integr. Plant Biol. 55, 410–418. doi: 10.1111/jipb.12033
Chen, J., Li, X., Cheng, C., Wang, Y., Qin, M., Zhu, H., et al. (2014). Characterization of epistatic interaction of QTLs LH8 and EH3 controlling heading date in rice. Sci. Rep. 4, 4263. doi: 10.1038/srep04263
Dai, C., and Xue, H. W. (2010). Rice early flowering1, a CKI, phosphorylates DELLA protein SLR1 to negatively regulate gibberellin signalling. EMBO J. 29, 1916–1927. doi: 10.1038/emboj.2010.75
Dai, X., Ding, Y., Tan, L., Fu, Y., Liu, F., Zhu, Z., et al. (2012). LHD1, an allele of DTH8/Ghd8, controls late heading date in common wild rice (Oryza rufipogon). J. Intgr. Plant Biol. 54, 790–799. doi: 10.1111/j.1744-7909.2012.01166.x
Doi, K., Izawa, T., Fuse, T., Yamanouchi, U., Kubo, T., Shimatani, Z., et al. (2004). Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 18, 926–936. doi: 10.1101/gad.1189604
Ebana, K., Shibaya, T., Wu, J., Matsubara, K., Kanamori, H., Yamane, H., et al. (2011). Uncovering of major genetic factors generating naturally occurring variation in heading date among Asian rice cultivars. Theor. Appl. Genet. 122, 1199–1210. doi: 10.1007/s00122-010-1524-1
Fowler, S., Lee, K., Onouchi, H., Samach, A., Richardson, K., Morris, B., et al. (1999). GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 18, 4679–4688. doi: 10.1093/emboj/18.17.4679
Fujino, K., Yamanouchi, U., and Yano, M. (2013). Roles of theHd5 gene controlling heading date for adaptation to the northern limits of rice cultivation. Theor. Appl. Genet. 126, 611–618. doi: 10.1007/s00122-012-2005-5
Greenup, A., Peacock, W. J., Dennis, E. S., and Trevaskis, B. (2009). The molecular biology of seasonal flowering-responses in Arabidopsis and the cereals. Ann. Bot. 103, 1165–1172. doi: 10.1093/aob/mcp063
Gross, S. D., and Anderson, R. A. (1998). Casein kinase I: spatial organization and positioning of a multifunctional protein kinase family. Cell. Signal. 10, 699–711. doi: 10.1016/S0898-6568(98)00042-4
Hayama, R., Yokoi, S., Tamaki, S., Masahiro Yano, M., and Shimamoto, K. (2003). Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422, 719–722. doi: 10.1038/nature01549
Hori, K., Kataoka, T., Miura, K., Yamaguchi, M., Saka, N., Nakahara, T., et al. (2012). Variation in heading date conceals quantitative trait loci for other traits of importance in breeding selection of rice. Breed. Sci. 62, 223–234. doi: 10.1270/jsbbs.62.223
Hori, K., Ogiso-Tanaka, E., Matsubara, K., Yamanouchi, U., Ebana, K., and Yano, M. (2013). Hd16, a gene for casein kinase I, is involved in the control of rice flowering time by modulating the day-length response. Plant J. 76, 36–46. doi: 10.1111/tpj.12268
Ishikawa, R., Aoki, M., Kurotani, K., Yokoi, S., Shinomura, T., Takano, M., et al. (2011). Phytochrome B regulates Heading date 1 (Hd1)-mediated expression of rice florigen Hd3a and critical day length in rice. Mol. Genet. Genomics 285, 461–470. doi: 10.1007/s00438-011-0621-4
Itoh, H., and Izawa, T. (2013). The coincidence of critical day length recognition for florigen gene expression and floral transition under long-day conditions in rice. Mol. Plant 6, 635–649. doi: 10.1093/mp/sst022
Izawa, T. (2007). Adaptation of flowering-time by natural and artificial selection in Arabidopsis and rice. J. Exp. Bot. 58, 3091–3097. doi: 10.1093/jxb/erm159
Jung, C., and Müller, A. E. (2009). Flowering time control and applications in plant breeding. Trends Plant Sci. 14, 563–573. doi: 10.1016/j.tplants.2009.07.005
Kojima, S., Takahashi, Y., Kobayashi, Y., Monna, L., Sasaki, T., Araki, T., et al. (2002). Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 43, 1096–1105. doi: 10.1093/pcp/pcf156
Komiya, R., Ikegami, A., Tamaki, S., Yokoi, S., and Shimamoto, K. (2008). Hd3a and RFT1 are essential for flowering in rice. Development 135, 767–774. doi: 10.1242/dev.008631
Komiya, R., Yokoi, S., and Shimamoto, K. (2009). A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development 136, 3443–3450. doi: 10.1242/dev.040170
Koo, B. H., Yoo, S. C., Park, J. W., Kwon, C. T., Lee, B. D., An, G., et al. (2013). Natural variation in OsPRR37 regulates heading date and contributes to rice cultivation at a wide range of latitudes. Mol. Plant 6, 1877–1888. doi: 10.1093/mp/sst088
Kwon, C. T., Yoo, S. C., Koo, B. H., Cho, S. H., Park, J. W., Zhang, Z., et al. (2014). Natural variation in Early flowering1 contributes to early flowering in japonica rice under long days. Plant Cell Environ. 37, 101–112. doi: 10.1111/pce.12134
Lee, S., Kim, J., Han, J. J., Han, M. J., and An, G. (2004). Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO 1/AGAMOUS-LIKE 20 (SOC1/AGL20) ortholog in rice. Plant J. 38, 754–764. doi: 10.1111/j.1365-313X.2004.02082.x
Lin, H., Ashikari, M., Yamanouchi, U., Sasaki, T., and Yano, M. (2002). Identification and characterization of a quantitative trait locus, Hd9, controlling heading date in rice. Breed. Sci. 52, 35–41. doi: 10.1270/jsbbs.52.35
Lin, H., Liang, Z. W., Sasaki, T., and Yano, M. (2003). Fine mapping and characterization of quantitative trait loci Hd4 and Hd5 controlling heading date in rice. Breed. Sci. 53, 51–59. doi: 10.1270/jsbbs.53.51
Liu, W., Xu, Z. H., Luo, D., and Xue, H. W. (2003). Roles of OsCKI1, a rice casein kinase I, in root development and plant hormone sensitivity. Plant J. 36, 189–202. doi: 10.1046/j.1365-313X.2003.01866.x
Magallón, S. A., and Sanderson, M. J. (2005). Angiosperm divergence times: the effects of genes, codon positions, and time constraints. Evolution 59, 1653–1670. doi: 10.1554/04-565.1
Matsubara, K., Kono, I., Hori, K., Nonoue, Y., Ono, N., A. Shomura, A., et al. (2008). Novel QTLs for photoperiodic flowering revealed by using reciprocal backcross inbred lines from crosses between japonica rice cultivars. Theor. Appl. Genet. 117, 935–945. doi: 10.1007/s00122-008-0833-0
Matsubara, K., Ogiso-Tanaka, E., Hori, K., Ebana, K., Ando, T., and Yano, M. (2012). Natural variation in Hd17, a homolog of Arabidopsis ELF3 that is involved in rice photoperiodic flowering. Plant Cell Physiol. 53, 709–716. doi: 10.1093/pcp/pcs028
McClung, C. R. (2011). The genetics of plant clock. Adv. Genet. 74, 105–139. doi: 10.1016/B978-0-12-387690-4.00004-0
Murakami, M., Tago, Y., Yamashino, T., and Mizuno, T. (2007). Comparative overviews of clock-associated genes of Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol. 48, 110–121. doi: 10.1093/pcp/pcl043
Nagano, A. J., Sato, Y., Mihara, M., Antonio, B. A., Motoyama, R., Itoh, H., et al. (2012). Deciphering and prediction of transcriptome dynamics under Fluctuating Field Conditions. Cell 151, 1358–1369. doi: 10.1016/j.cell.2012.10.048
Ogiso, E., Takahashi, Y., Sasaki, T., Yano, M., and Izawa, T. (2010). The role of casein kinase II in flowering time regulation has diversified during evolution. Plant Physiol. 152, 808–820. doi: 10.1104/pp.109.148908
Ogiso-Tanaka, E., Matsubara, K., Yamamoto, S., Nonoue, Y., Wu, J., Fujisawa, H., et al. (2013). Natural variation of the RICE FLOWERING LOCUS T 1 contributes to flowering time divergence in rice. PLoS ONE 8:e75959. doi: 10.1371/journal.pone.0075959
Olsen, K. M., and Wendel, J. F. (2013). Crop plants as models for understanding plant adaptation and diversification. Front. Plant Sci. 4:290. doi: 10.3389/fpls.2013.00290
Park, D. H., Somers, D. E., Kim, Y. S., Choy, Y. H., Lim, H. K., Soh, M. S., et al. (1999). Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285, 1579–1582. doi: 10.1126/science.285.5433.1579
Pin, P. A., Benlloch, R., Bonnet, D., Wremerth-Weich, E., Kraft, T., Gielen, J. J. L., et al. (2010). An antagonistic pair of FT homologs mediates the control of flowering time in sugar beet. Science 330, 1397–1400. doi: 10.1126/science.1197004
Putterill, J., Robson, F., Lee, K., Simon, R., and Coupland, G. (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80, 847–857. doi: 10.1016/0092-8674(95)90288-0
Ralph, M. R., and Menaker, M. (1988). A mutation of the circadian system in golden hamsters. Science 241, 1225–1227. doi: 10.1126/science.3413487
Rumpf, C., Cipak, L., Dudas, A., Benko, Z., Pozgajova, M., Riedel, C. G., et al. (2010). Casein kinase 1 is required for efficient removal of Rec8 during meiosis I. Cell Cycle 9, 2655–2660. doi: 10.4161/cc.9.13.12146
Saito, H., Ogiso-Tanaka, E., Okumoto, Y., Yoshitake, Y., Izumi, H., Yokoo, T., et al. (2012). Ef7 encodes an ELF3-like protein and promotes rice flowering by negatively regulating the floral repressor gene Ghd7 under both short- and long-day conditions. Plant Cell Physiol. 53, 717–728. doi: 10.1093/pcp/pcs029
Saito, H., Yuan, Q., Okumoto, Y., Doi, K., Yoshimura, A., Inoue, H., et al. (2009). Multiple alleles at Early flowering 1 locus making variation in the basic vegetative growth period in rice (Oryza sativa L.). Theor. Appl. Genet. 119, 315–323. doi: 10.1007/s00122-009-1040-3
Takahashi, Y., Shomura, A., Sasaki, T., and Yano, M. (2001). Hd6, a rice quantitative trait locus involved in photoperiod sensitivity, encodes the α subunit of protein kinase CK2. Proc. Natl. Acad. Sci. U.S.A. 98, 7922–7927. doi: 10.1073/pnas.111136798
Takahashi, Y., Teshima, K. M., Yokoi, S., Innan, H., and Shimamoto, K. (2009). Variations in Hd1 proteins, Hd3a promoters, and Ehd1 expression levels contribute to diversity of flowering time in cultivated rice. Proc. Natl. Acad. Sci. U.S.A. 106, 4555–4560. doi: 10.1073/pnas.0812092106
Tamaki, S., Matsuo, S., Wong, H. L., Yokoi, S., and Shimamoto, K. (2007). Hd3a protein is a mobile flowering signal in rice. Science 316, 1033–1036. doi: 10.1126/science.1141753
Thomas, B., and Vince-Prue, D. (1997). Photoperiodism in Plants. 2nd Edn. San Diego, CA: Academic Press.
Tsuji, H., Taoka, K. I., and Shimamoto, K. (2011). Regulation of flowering in rice: two florigen genes, a complex gene network, and natural variation. Curr. Opin. Plant Biol. 14, 45–52. doi: 10.1016/j.pbi.2010.08.016
Tuazon, P., and Traugh, J. (1991). Casein kinase I and II—multipotential serine protein kinases: structure, function, and regulation. Adv. Second Messenger Phosphoprotein Res. 23, 123–164.
Wei, X., Xu, J., Guo, H., Jiang, L., Chen, S., Yu, C., et al. (2010). DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol. 153, 1747–1758. doi: 10.1104/pp.110.156943
Wu, W., Zheng, X. M., Lu, G., Zhong, Z., Gao, H., Chen, L., et al. (2013). Association of functional nucleotide polymorphisms at DTH2 with the northward expansion of rice cultivation in Asia. Proc. Natl. Acad. Sci. U.S.A. 110, 2775–2780. doi: 10.1073/pnas.1213962110
Xue, W., Xing, Y., Weng, X., Zhao, Y., Tang, W., Wang, L., et al. (2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40, 761–767. doi: 10.1038/ng.143
Yan, W. H., Wang, P., Chen, H. X., Zhou, H. J., Li, Q. P., Wang, C. R., et al. (2011). A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol. Plant 4, 319–330. doi: 10.1093/mp/ssq070
Yang, Y., Peng, Q., Chen, G. X., Li, X. H., and Wu, C. Y. (2012). OsELF3 is involved in circadian clock regulation for promoting flowering under long-day conditions in rice. Mol. Plant 6, 202–215. doi: 10.1093/mp/sss062
Yano, M., Katayose, Y., Ashikari, M., Yamanouchi, U., Monna, L., Fuse, T., et al. (2000). Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12, 2473–2483. doi: 10.1105/tpc.12.12.2473
Zhao, D., Zhang, C., Feng, G., Yang, Q., Gu, M., and Liu, Q. (2013). Hd-q, a novel allele of Ef7 from a Chinese rice landrace, confers weak photoperiod sensitivity and improves local adaptability and yield potential. Mol. Breed. 32, 651–662. doi: 10.1007/s11032-013-9898-9
Keywords: breeding, natural variation, photoperiodic flowering, quantitative trait gene, rice
Citation: Matsubara K, Hori K, Ogiso-Tanaka E and Yano M (2014) Cloning of quantitative trait genes from rice reveals conservation and divergence of photoperiod flowering pathways in Arabidopsis and rice. Front. Plant Sci. 5:193. doi: 10.3389/fpls.2014.00193
Received: 28 December 2013; Accepted: 23 April 2014;
Published online: 13 May 2014.
Edited by:
Christian Jung, Christian Albrechts University of Kiel, GermanyReviewed by:
Peter Langridge, Australian Centre for Plant Functional Genomics, AustraliaChristian Jung, Christian Albrechts University of Kiel, Germany
Rita Sharma, Jawaharlal Nehru University, India
Copyright © 2014 Matsubara, Hori, Ogiso-Tanaka and Yano. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Masahiro Yano, Agrogenomics Research Center, National Institute of Agrobiological Sciences, 2-1-2 Kannondai, Tsukuba, Ibaraki 305-8602, Japan e-mail:bXlhbm9Abmlhcy5hZmZyYy5nby5qcA==
†Present address: Masahiro Yano, NARO Institute of Crop Science, Tsukuba, Japan
 Kazuki Matsubara
Kazuki Matsubara Kiyosumi Hori
Kiyosumi Hori Eri Ogiso-Tanaka
Eri Ogiso-Tanaka Masahiro Yano
Masahiro Yano